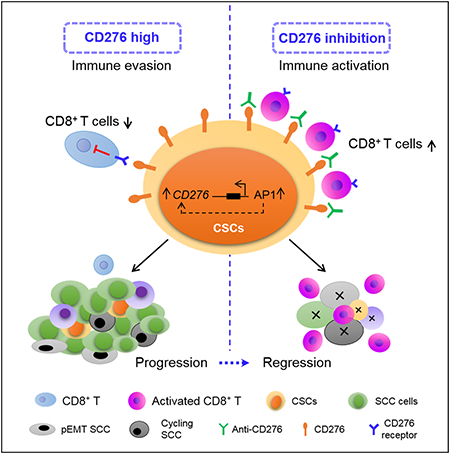
Summary
Immunosurveillance is a critical mechanism guarding against tumor development and progression. Checkpoint inhibitors have shown significant success in cancer treatment but expression of key factors such as PD-L1 in putative cancer stem cell (CSC) populations in squamous cell carcinoma has been inconclusive, suggesting CSCs may have developed alternate mechanisms to escape immune surveillance. Here we show CSCs upregulate the immune checkpoint molecule CD276 (B7-H3) to evade host immune responses. CD276 is highly expressed by CSCs in mouse and human head and neck squamous cell carcinoma (HNSCC) and can be used to prospectively isolate tumorigenic CSCs. Anti-CD276 antibodies eliminate CSCs in a CD8+ T cell-dependent manner, inhibiting tumor growth and lymph node metastases in a mouse HNSCC model. Single-cell RNAseq showed that CD276 blockade remodels SCC heterogeneity and reduces epithelial-mesenchymal transition. Together, these results show that CSCs utilize CD276 for immune escape and suggest targeting CD276 may reduce CSCs in NHSCC.
Keywords: Cancer stem cell, head and neck squamous cell carcinoma, lineage tracing, CD276, immune surveillance, BMI1
Graphical Abstract
eTOC BLURB
Wang et al. show that cancer stem cells (CSCs) utilize the immune checkpoint molecule CD276 (B7-H3) to evade immune surveillance during HNSCC initiation, development and metastasis. CD276 blockade eliminates CSCs and inhibits tumor growth and metastasis by increasing anti-tumor immunity.
Introduction
Head and neck squamous cell carcinoma (HNSCC) is highly invasive and frequently metastasizes to cervical lymph nodes (Chen and Wang, 2019; Hedberg et al., 2016). Current treatment modalities include surgical resection of the tumor combined with or without radiotherapy and/or chemotherapy (Sacco and Cohen, 2015). However, the prognosis for patients with HNSCC remains dismal due to rapid lymph node metastasis, local recurrence and a high incidence of therapeutic resistance (Hedberg et al., 2016; Wang et al., 2012). Therefore, there is an urgent need to identify novel therapeutic targets and develop effective treatment modalities for HNSCC. Accumulating evidence suggests that CSCs play a crucial role in tumor initiation, growth, therapy resistance and metastasis (Prager et al., 2019; Saygin et al., 2019). Recently, we provided important evidence for a hierarchical organization in HNSCC and demonstrated, by means of in vivo lineage tracing, that BMI1+ CSCs have exclusive tumorigenic and metastatic potential and are inherently resistant to chemotherapy (Chen et al., 2017). These findings indicate that targeting CSCs is a promising strategy to eradicate HNSCC.
A number of studies have focused on the inhibition of regulatory pathways that are critical for the self-renewal and tumorigenic potentials of CSCs. It is well-known that tumor cells are potentially immunogenic and the immune system plays a critical role in the surveillance against nascent cancerous cells from developing into tumors. Immune surveillance is also important for restraining tumor progression and metastasis (Sanmamed and Chen, 2018; Iglesias-Bartolome and Gutkind, 2020). Since CSCs are the origin of cancerous tissues, CSCs have to develop intrinsic mechanisms to evade immune surveillance during tumor development. Interestingly, HNSCC frequently metastasizes to cervical lymph nodes that are enriched with immune cells (Hedberg et al., 2016). Because disseminated CSCs mediate metastasis, CSCs should be able to escape immune cell killing first and then proliferate and differentiate in lymph nodes to form metastatic tumors. However, little is known about how CSCs can survive under immune surveillance during HNSCC initiation, progression and metastasis. Tumor cells expressing PD-L1 (B7-H1) have been identified as an important mechanism for tumor cells to evade immune challenge (Sanmamed and Chen, 2018). Recently, immune checkpoint inhibitors targeting PD1/PD-L1 and CTLA4 have achieved good success in suppressing several solid tumors including melanoma, non-small cell lung cancer, and HNSCC (Ferris et al., 2016; Harrington et al., 2017; Sanmamed and Chen, 2018; Sharma et al., 2017; Yang, 2015). However, little is known about whether immunotherapy could target CSCs. Studies on PD-L1 expression in CSCs have yielded contradictory results. Some studies showed that PD-L1 expression was increased in CSCs of HNSCC (Lee et al., 2016), and in breast and colon cancers (Hsu et al., 2018; Wu et al., 2017), whereas other studies indicated decreased levels of PD-L1 on CSCs (Prager et al., 2019; Tamai et al., 2014). In our previous study, we noted that there was no significant difference in PD-L1 (also known as CD274) mRNA levels between BMI1+ CSCs and BMI1− non-CSCs in murine HNSCC (Chen et al., 2017). Surprisingly, analysis of TCGA datasets that use of a machine-learning algorithm demonstrated that stemness signatures correlated with lower PD-L1 expression in HNSCC and lung SCC (Malta et al., 2018), suggesting that CSCs might utilize other immune checkpoints to mediate immune evasion during the tumor development. These findings further indicate that PD1/PD-L1 blockade might not effectively target CSCs, indicating that CSCs might use other immune checkpoints to fight against immunotherapy. Therefore, to improve the efficacy of HNSCC immunotherapy, it is critical to understand the immune mechanisms by which CSCs evade immune surveillance during HNSCC initiation, progression and metastasis.
Herein, we identified that CD276, an immune checkpoint molecule belonging to the B7 family (Leitner et al., 2009; Prasad et al., 2004; Suh et al., 2003), was highly expressed in CSCs of HNSCC and transcriptionally dependent on AP-1-associated super-enhancers (SEs). The high expression of CD276 helped CSCs escape immune surveillance in HNSCC initiation, progression and metastasis. The blockage of CD276 with monoclonal antibodies eliminated CSCs and inhibited tumor growth and metastasis by enhancing CD8+ T lymphocytes-mediated anti-tumor immunity.
Results
CD276 highly expressed in CSCs of HNSCC
Recently, we established a 4-nitroquinoline 1-oxide (4NQO)-induced Bmi1CreER;RosatdTomato mouse model of HNSCC that fully simulates human HNSCC initiation, development and metastasis and allows the performance of in vivo lineage tracing and isolation of BMI1+ CSCs in an intact tumor immune microenvironment (Chen et al., 2017). To explore whether anti-PD1 antibodies could kill CSCs of HNSCC, we took advantage of this syngeneic mouse model and treated Bmi1CreER;RosatdTomato mice with 4NQO in their drinking water for 16 weeks and then provided them with normal drinking water. At 22 weeks, the tumor-bearing mice were administered either anti-PD1 antibodies or vehicle with isotype IgG three times a week for four weeks. A single dose of tamoxifen was also administered 1 day prior to sacrificing the mice to label BMI1+ CSCs. Anti-PD1 antibodies could not significantly inhibit HNSCC growth and lymph node metastasis. In vivo lineage tracing demonstrated that anti-PD1 antibodies could not eliminate BMI1+ CSCs in HNSCC, indicating that BMI1+ CSCs might be intrinsically resistant to anti-PD1 immunotherapy (Jia et al., 2020). To explore whether other immune checkpoints might mediate CSC immune evasion, we analyzed the RNA-seq data generated from BMI1+ CSCs and BMI1− non-CSCs isolated from murine HNSCC and found that the mRNA levels of Cd276 and Cd80 were significantly higher in BMI1+ CSCs than in BMI1− non-CSCs (Figure S1A). Analysis of the cancer genome atlas (TCGA) database showed that CD276 mRNA expression was inversely correlated with mRNA expression of CD8A, CD8B, GZMB and IFNG, while CD80 expression was positively correlated with the expression of CD8A, CD8B, GZMB and IFNG in human HNSCC (Figure S1B), implicating that CD276 might function as an immune checkpoint to exclude the infiltration of immune cell in HNSCC. To confirm that CD276 was highly expressed in CSCs, we immunostained CD276 in HNSCC of Bmi1CreER;RosatdTomato mice with anti-CD276 antibodies. Although CD276 had a broader staining pattern, tomato-positive BMI1+ CSCs were strongly CD276-positive in HNSCC tissues. BMI1+ CSCs were mainly located near the interface of tumor and stroma, and most of them were found in the invasive tumor front (ITF) (Figures 1A, 1B and Figure S1C). Similarly, more CD276+ cancer cells resided in the ITF compared to the cells localized in the tumor bulk (TB) (Figure 1C). Importantly, the majority of BMI1+ CSCs (beyond 90%) strongly expressed CD276 compared to BMI1− non-CSCs (Figure 1D), suggesting that CSCs of HNSCC are mostly confined to CD276+ cancer cells, although not all CD276+ cells were BMI1+ CSCs.
Figure 1. The expression of CD276 is increased in CSCs and associated with HNSCC development and metastasis.
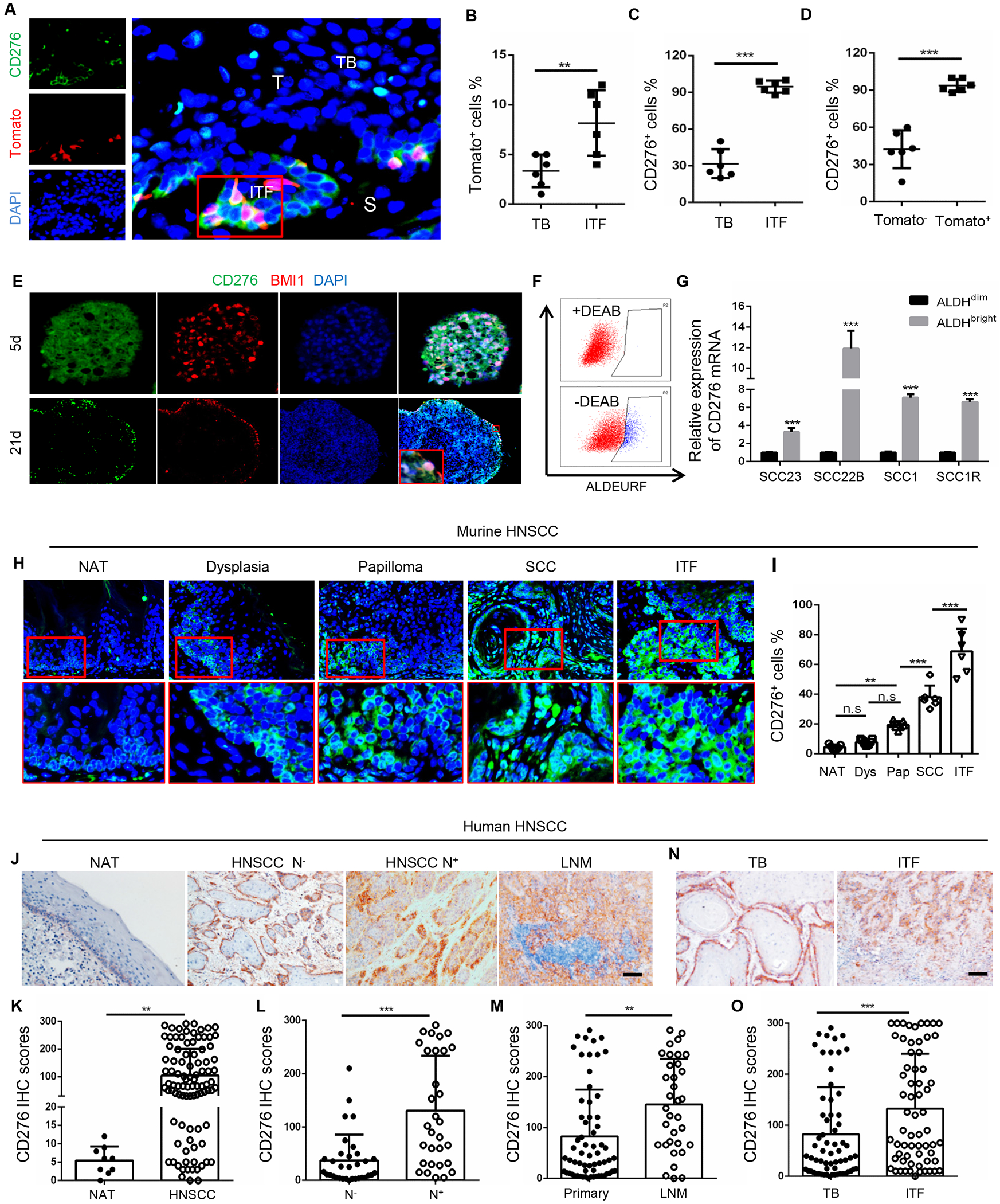
(A) Representative images of CD276 and Tomato+ CSCs in primary HNSCC from 4NQO-treated BmiCreER;Rosa26tdTomato mice. ITF, invasive tumor front; TB, tumor bulk; S, stromal; T, tumor. Scale bar, 10μm.
(B) Quantification of percentage of Tomato+ CSCs in TB and ITF. Values are mean ± SD., **p<0.01 by Student’s t test, n = 6.
(C) Quantification of percentage of CD276+ tumor cells in TB and ITF. Values are mean ± SD., ***p<0.001 by Student’s t test; n = 6.
(D) Quantification of percentage of CD276+ cells in Tomato+ CSCs and Tomato− non-CSCs. Values are mean ± SD., ***p<0.001 by Student’s t test, n=6.
(E) Representative images for CD276 and BMI1 immunostaining in tumorspheres. Scale bar, 50μm (upper panels), 100μm (lower panels).
(F) Representative fluorescence-activated cell sorting (FACS) plot of ALDHbright cancer cells isolated from HNSCC cell lines.
(G) RT-qPCR showed that the expression of CD276 was significantly upregulated in ALDHbright cancer cells than in ALDHdim cancer cells. Data represent mean ± SD., ***p<0.001 by Student’s t test, n = 3.
(H) Representative images of CD276 immunostaining in non-cancerous adjacent normal tongue epithelium (NAT), dysplasia, papilloma, SCC and invasive tumor front (ITF) of HNSCC from 4NQO-induced mouse HNSCC. Scale bar, 20μm (upper panels), 10μm (lower panels).
(I) Quantification of CD276 expression in NAT, dysplasia (Dys), papilloma (Pap), SCC and ITF of HNSCC from 4NQO-induced mouse HNSCC. **p<0.01, ***p<0.001 by one-way ANOVA; n = 6-7.
(J) Representative images of CD276 expression in human NAT, HNSCC without cervical lymph node metastasis (HNSCC N−), HNSCC with cervical lymph node metastasis (HNSCC N+) and lymph node metastatic HNSCC (LNM). Scale bar, 100μm.
(K) Quantification of CD276 expression in NAT and HNSCC from patients. **p<0.01 by Student’s t test.
(L) Quantification of CD276 expression in HNSCC without cervical lymph node metastasis (N−) and HNSCC with lymph node metastasis (N+). ***p<0.001 by Student’s t test.
(M) Quantification of CD276 expression in primary HNSCC (Primary) and lymph node metastatic HNSCC (LNM). **p<0.01 by Student’s t test.
(N) Representative image of CD276 expression in tumor bulk (TB) and ITF of HNSCC. Scale bar, 100μm
(O) Quantification of CD276 expression in tumor bulk (TB) and ITF of HNSCC.***p<0.001 by Student’s t test.
See also Figure S1
We found that CD276 was co-expressed with BMI1 in sphere-derived cells of HNSCC (Figure 1E). Interestingly, strong expression of CD276 was detected at the peripheral of the tumor spheres versus the expression found in the core of the spheres. Previously we and others have used aldehyde dehydrogenase (ALDH) activity to isolate CSCs from human HNSCC cell lines and primary tumor tissues (Chen et al., 2017). Using an ALDEFLUOR™ kit, ALDHbright CSC-like cells and ALDHdim non-CSC-like cells were sorted from HNSCC cells lines and found that the CD276 mRNA expression levels were also significantly higher in ALDHbright cells than in ALDHdim cells (Figures 1F and 1G).
4NQO-induced mouse HNSCC mimics human HNSCC and gradually progresses from dysplasia, papilloma (carcinoma in situ) to SCC in which BMI1+ CSCs were identified at the initial stage of HNSCC. Next, we further evaluated the expression of CD276 and investigated its correlation with HNSCC initiation, progression and metastasis. Immunostaining revealed that CD276 expression was readily detected in papilloma and significantly increased during mouse HNSCC development and progression (Figures 1H and 1I). In human HNSCC, CD276 expression was dramatically increased in HNSCC compared with adjacent non-cancerous epithelium (Figures 1J and 1K). When compared to human primary HNSCC without lymph node metastasis, CD276 expression was significantly increased in those with lymph node metastasis (Figure 1L). Moreover, cancer cells disseminated to the cervical lymph node had higher expression levels of CD276 than those localized in primary sites (Figure 1M). Within the same tumor, the expression of CD276 was also significantly increased in ITF as compared to tumor bulk (Figures 1N and 1O). These findings pointed towards a correlation of CD276 expression and the aggressiveness of the tumor. Very interestingly, we noted that CD276, in addition to ITF, was uniquely expressed at the periphery of the tumor nest in both mouse and human HNSCCs, indicating that CD276 might serve as a physical shield against immune cell infiltrations.
We sorted CD276high cancer cells from HNSCC patient-derived xenografts (PDXs) using EpCAM+CD276high markers (Figure 2A) and determined their self-renewal capacity via tumorsphere formation assays. EpCAM+CD276high cancer cells freshly isolated from two human HNSCC PDXs formed significantly more abundant and larger spheres compared to EpCAM+CD276low cancer cells, suggesting that CD276 is an enrichment marker for CSCs of HNSCC (Figures 2B–2D). To reinforce the correlation between CD276 expression and CSCs of HNSCC, we performed in vivo limiting dilution tumorigenicity assays. The EpCAM+CD276high fraction contained more tumorigenic cells than the EpCAM+CD276low fraction (Figure 2E). To validate these findings in murine HNSCC, murine HNSCC tissues were transplanted into NOG mice and cancer cells expressing high and low CD276 were isolated from murine HNSCC xenografts. EpCAM+CD276high cancer cells were enriched for CSCs and demonstrated higher tumorigenicity than EpCAM+CD276low cancer cells in murine HNSCC (Figures 2F–2H). The organoid formation assay also demonstrated that EpCAM+CD276high tumor cells isolated from the primary HNSCC formed significantly larger organoids than EpCAM+CD276low tumor cells (Figure S1D). To further characterize EpCAM+CD276high CSCs, we compared the gene expression profiles of EpCAM+CD276high and EpCAM+CD276low cancer cells by RNA sequencing (RNA-seq). We found that 842 genes were upregulated and 1081 genes were downregulated in EpCAM+CD276high cells by >2-fold compared to EpCAM+CD276low cells. Genes upregulated in EpCAM+CD276high were associated with known CSC signatures (BMI1, ABCG2, GATA4, SOX21, LHX5, FGFR1, RASD1 and ITGA6) and epithelial-mesenchymal transition (EMT) (CDH2, FAP, MMP9, MMP10, MMP1, SNAI2 and LAMA3). In contrast, genes associated with keratinocyte differentiation were downregulated in EpCAM+CD276high cells (Figure 2I). Importantly, the gene set enrichment analysis (GSEA) also revealed that the stemness gene signatures and EMT signatures were significantly enriched in EpCAM+CD276high cells (Figures 2J and 2K). GSEA of HNSCC in TCGA datasets also showed similar findings (Figures 2L and 2M).
Figure 2. CD276 is an enrichment surface marker for CSCs of HNSCC.
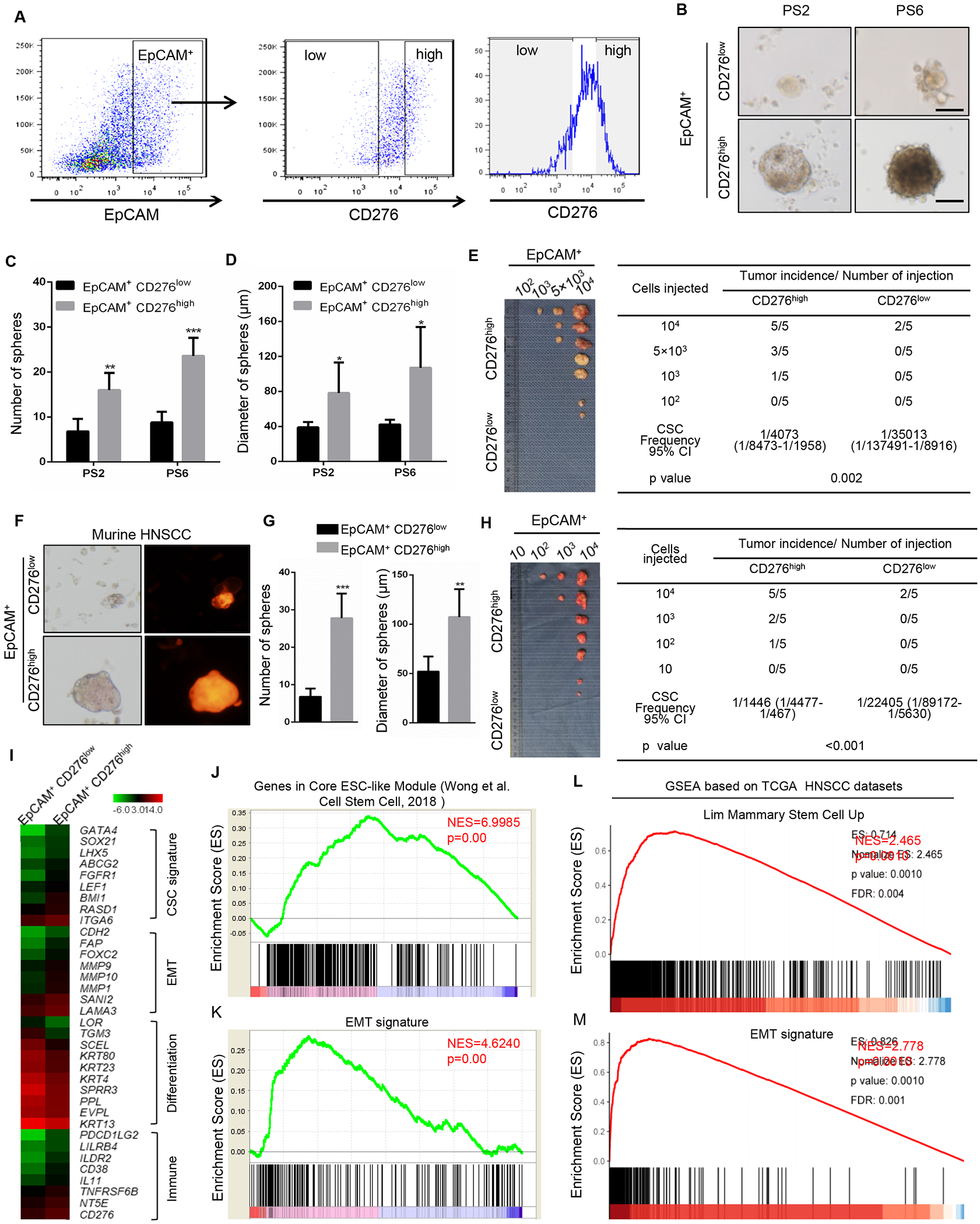
(A) Sorting strategy for CD276high and CD276low tumor cells.
(B) Representative image of tumorspheres derived from CD276high and CD276low tumor cells from HNSCC PDXs. PS2, patient sample 2; PS6, patient sample 6. Scale bar, 50 μM (upper panels), 100μm (lower panels).
(C) Quantification of number of tumorspheres from CD276high and CD276low tumor cells from HNSCC PDXs. **p<0.01, ***p<0.001 by Student’s t test.
(D) Quantification of diameter of tumorspheres from CD276high and CD276low tumor cells from HNSCC PDXs. **p<0.05 by Student’s t test.
(E) In vivo limiting dilution analysis of CD276high and CD276low tumor cells from HNSCC PDXs. The frequency of allograft formation at each cell dose injected is shown. The data were analyzed using ELDA software.
(F) Representative image of tumorspheres from CD276high and CD276low tumor cells isolated from murine HNSCC. Scale bar, 50μm
(G) Quantification of number and diameter of tumor spheres from mouse CD276high and CD276low tumor cells. ***p<0.001 and **p <0.01 by Student’s t test.
(H) In vivo limiting dilution analysis of murine CD276high and CD276low tumor cells. The frequency of allograft formation at each cell dose injected is shown. The data were analyzed using ELDA software.
(I) Heatmap showing the gene expression profiles of EpCAM+CD276high and EpCAM+CD276low tumor cells by RNA-seq.
(J) GSEA showed that the stemness gene signature was significantly enriched in EpCAM+CD276high cells.
(K) GSEA showed that the EMT signature was significantly enriched in EpCAM+CD276high cells.
(L) GSEA showed that the stemness gene signature was significantly enriched in human HNSCC with high expression of CD276 from TCGA datasets.
(M) GSEA showed that the EMT signature was significantly enriched in human HNSCC with high expression of CD276 from TCGA datasets.
Control of CD276 transcription by FOSL1-associated super-enhancers (SEs)
We further investigated the molecular mechanisms governing CD276 expression in HNSCC. Based on our MED1 and BRD4 ChIP-seq results in human SCC cells for a different study, we unexpectedly discovered that SEs were associated with CD276. MED1 and BRD4 occupancies on SEs in CD276 were disrupted by the well-known BET inhibitor JQ1 (Hnisz et al., 2013; Sengupta and George, 2017; Vaharautio and Taipale, 2014) (Figure 3A). Consistently, JQ1 treatment significantly inhibited the expression of CD276 at both mRNA and protein levels. I-BET-151, another BET inhibitor used in clinical trials (Sengupta and George, 2017; Vaharautio and Taipale, 2014), also potently inhibited the expression of CD276 (Figures 3B and 3C). Moreover, JQ1 treatment also inhibited HNSCC tumor growth and CD276 expression in vivo (Figures S1E–SI). To locate the upstream factors that are responsible for the upregulation of CD276 through the binding and activation of the distal cis-regulatory regions, we focused on potential transcription factors that might bind to the SE region. Interestingly, the AP1 binding site was observed within the SE region, upstream of the CD276 gene (Figure 3A). Co-incidentally, we identified that FOSL1, a key component of AP-1, was increased in CSCs of HNSCC and controlled CSC self-renewal and invasive growth (Chen et al., 2017). Therefore, we examined whether AP1/FOSL1 promoted CD276 expression mediated by SEs. The knockdown of FOSL1 significantly inhibited the expression of CD276 mRNA and protein in HNSCC cells (Figures 3D–3F). Moreover, ChIP-qPCR results demonstrated that the knockdown of FOSL1 significantly reduced the enrichment of FOSL1, MED1 and BRD4 on the SEs (Figures 3G–3I). While CD276 functions as an immune checkpoint, CD276 has also been found to activate intracellular signaling pathways to promote tumorigenesis (Lemke et al., 2012). We screened several oncogenic signaling pathways and found that the over-expression of CD276 potently induced the expression of FOSL1 and c-Jun in SCC1 and SCC23 cells (Figure 3J), indicating that CD276 could also activate AP-1. In contrast, knockdown of CD276 in HN6 cells by two different shRNAs for CD276, which have constitutive CD276 expression, inhibited the expression of FOSL1 and c-Jun (Figure 3K). Consistently, Matrigel invasion assays revealed that the knockdown of CD276 significantly inhibited SCC invasion in vitro (Figures 3L and 3M). Moreover, we also injected CD276 knockdown HN6 cells and control cells into mouse tongues and found that the knockdown of CD276 also attenuated HNSCC tumor growth (Figure 3N). Immunostaining with anti-human pan-keratin (PCK) revealed that the knockdown of CD276 also partially reduced lymph node metastasis (Figure 3O).
Figure. 3. FOSL1-assoicated SEs control CD276 transcription in HNSCC.
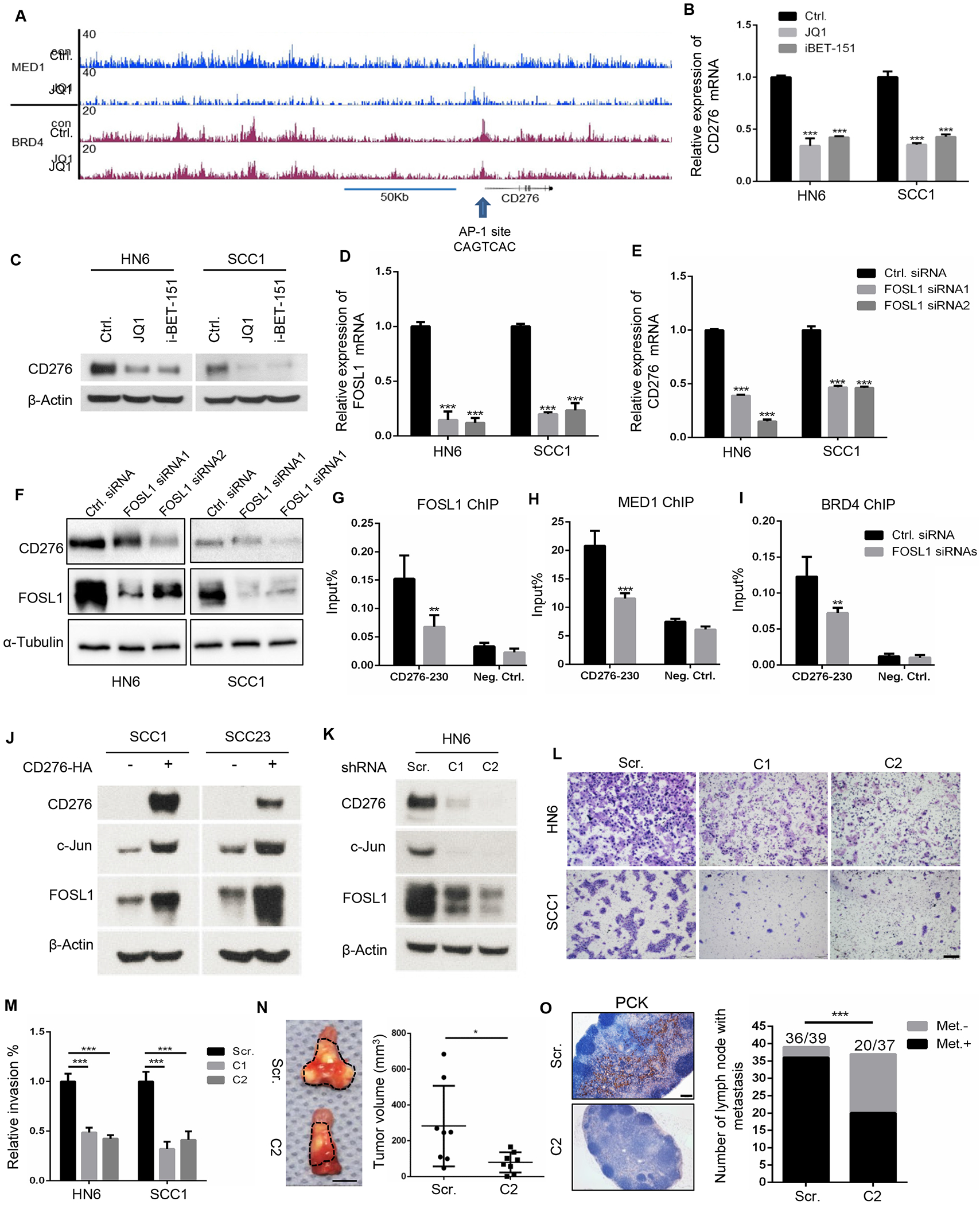
(A) MED1 and BRD4 enrichments show that SEs is associated with CD276 in HNSCC. Arrow indicates the AP1 binding site within SEs.
(B) Disrupting SEs by JQ1 and iBET-151 reduces CD276 mRNA expression in HNSCC cells. ***p<0.001 by one-way ANOVA.
(C) Disrupting SEs by JQ1 and iBET-151 reduces CD276 expression in HNSCC cells.
(D) RT-qPCR showing knockdown of FOSL1 by siRNA in HNSCC cells. ***p<0.001 by one-way ANOVA.
(E) Knockdown of FOSL1 inhibited CD276 mRNA expression in HNSCC cells. ***p<0.001 by one-way ANOVA.
(F) Knockdown of FOSL1 inhibited CD276 expression in HNSCC cells by Western blot.
(G) Knockdown of FOSL1 reduced FOSL1 occupancies on SEs in CD276. **p<0.01 by Student’s t test.
(H) Knockdown of FOSL1 reduced MED1 occupancies on SEs in CD276. ***p<0.001 by Student’s t test.
(I) Knockdown of FOSL1 reduced BRD4 occupancies on SEs in CD276. **p<0.01 by Student’s t test.
(J) Over-expression of CD276 induced the expression of c-Jun and FOSL1 in SCC1 and SCC23 cells.
(K) Knockdown of CD276 inhibited the expression of c-Jun and FOSL1 in HN6 cells. Scr, scramble shRNA; C1, CD276 shRNA vector 1, C2, CD276 shRNA vector 2.
(L,M) Representative image and quantification of invasive HN6 and SCC1 cells transduced with scramble shRNA (Scr.); CD276 shRNA1(C1); CD276 shRNA2 (C2). ***p<0.001 by one-way ANOVA. Scale bar, 200μm
(N) The knockdown of CD276 in HN6 cells inhibited tumor growth in nude mice. C2, HN6 cells transduced with CD276 shRNA2; Scr, HN6 cells transduced with scramble shRNA. *p < 0.05 by Student’s t test, n=8. Scale bar, 5mm.
(O) The knockdown of CD276 in HN6 cells inhibited lymph node metastasis. The percentages of lymph node with metastatic tumor cells were analyzed by Fisher’s exact test. ***p < 0.001. Met−, without lymph node metastasis; Met+, with lymph node metastasis. Scale bar, 200μm
Elimination of CSCs and inhibition of cancer progression by CD276 blockade
Because CD276 is an immune checkpoint enriched in CSCs, we tested whether the blockade of CD276 with anti-CD276 antibodies could inhibit HNSCC growth and metastasis using our syngeneic Bmi1CreER;RosatdTomato mouse model of HNSCC. HNSCC was induced as described before (Chen et al., 2017). At 22 weeks, the tumor-bearing mice were administered either anti-CD276 antibodies or vehicle control with isotype IgG three times a week for four weeks (Figure 4A). A single dose of tamoxifen was administered 1 day prior to sacrificing the mice to label CSCs. Mice developed visible lesions on the tongue upon 4NQO induction (Figure 4B). A significant decrease was detected in overall lesion number and lesion area in mice treated with anti-CD276 antibodies compared to the isotype IgG (Figures 4C and 4D). Another advantage of our HNSCC mouse model is that tumor initiation can be distinguished from dysplasia at new areas although HNSCC is already established in some locations, therefore allowing us to monitor immune surveillance during tumor initiation. Histologically, the dysplasia and SCC numbers were also significantly decreased after the administration of anti-CD276 antibodies in mice bearing HNSCC (Figures 4E–4G). Moreover, mice rarely developed highly invasive carcinomas when treated with CD276 antibody, and the invasive grade (Figure 4H) and invasive depth (Figure 4I) were significantly decreased. Importantly, a significant reduction of BMI1+ CSCs was detected in the primary HNSCC of the Bmi1CreER;RosatdTomato mice that were treated with anti-CD276 antibodies (Figure 4J). BMI1+ CSCs were active-caspase3-positive after CD276 blockade, indicating that the apoptosis was induced in BMI1+ CSCs of HNSCC (Figure 4K).
Figure 4. CD276 blockade suppresses HNSCC growth and eliminates cancer stem cell in HNSCC.
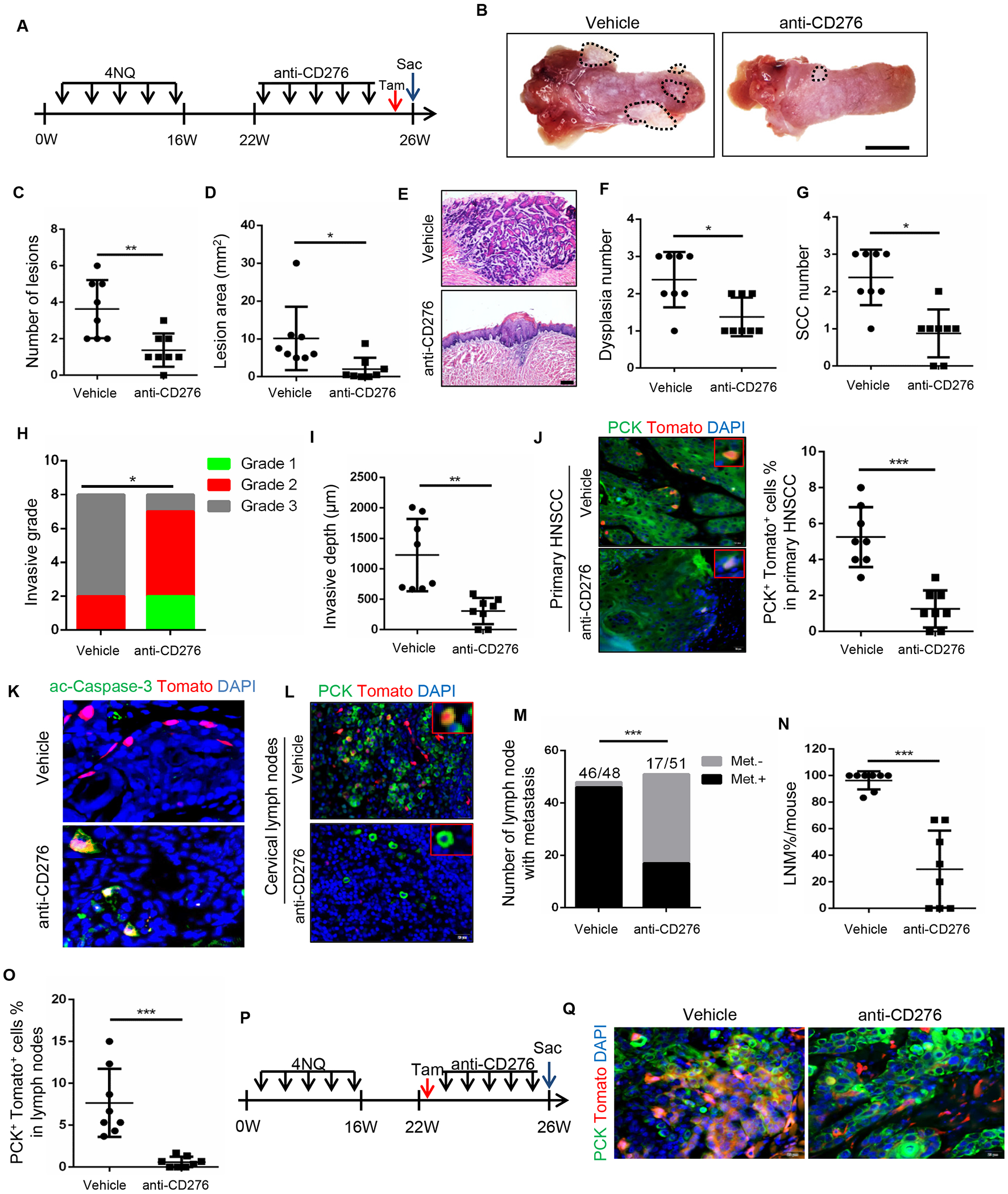
(A) Experimental design for the anti-CD276 treatment and lineage tracing of CSCs of HNSCC in Bmi1CreER;RosatdTomato mice.
(B) Representative image of tongue lesion 26 week in 4NQO-induced BmiCreER;RosatdTomato mice treated with anti-CD276 antibodies or vehicle with isotype IgG (vehicle). Scale bar, 3mm
(C) Quantification of lesion numbers visible in the mouse tongues. Values are mean ± SD from the pool of two independent experiments, n = 8, **p<0.01 by Student’s t test.
(D) Quantification of lesion areas visible in the mouse tongues. Values are mean ± SD from the pool of two independent experiments, n=8, *p<0.05 by Student’s t test.
(E) H&E staining of HNSCC in 4NQO-induced Bmi1CreER;RosatdTomato mice treated with anti-CD276 and vehicle with isotype IgG. Scale bar, 200μm
(F) Quantification of microscopic dysplasia numbers in mouse tongues. Values are mean ± SD from the pool of two independent experiments (n=8). *p<0.05 by Student’s t test.
(G) Quantification of microscopic SCC numbers in mouse tongues. Values are mean ± SD from the pool of two independent experiments (n=8). *p<0.05 by Student’s t test.
(H) Quantification of HNSCC invasive grades, n = 8, *p<0.05 by Cochran-Armitage test.
(I) Quantification of HNSCC invasive depths, n = 8, **p<0.01 by Student’s t test.
(J) Representative image and quantification of PCK+Tomato+ CSCs in primary HNSCC from mice treated with anti-CD276 or vehicle control with isotype IgG (n=8). Values are mean ± SD from the pool of two independent experiments (n=8). ***p<0.001 by Student’s t test. Scale bar, 20μm.
(K) Immunostaining of active caspase-3 (ac-Casp-3) in Tomato+ CSCs. Scale bar, 20μm.
(L) Representative image of PCK+ cancer cells and PCK+Tomato+ CSCs in cervical lymph nodes from mice treated with anti-CD276 antibodies. Scale bar, 20μm.
(M) Number of cervical lymph nodes with metastasis upon anti-CD276 treatment. ***p<0.001 by Fisher Chi square test. Met.−, without lymph node metastasis; Met.+, with lymph node metastasis.
(N) Percentage of lymph node metastasis in mice upon anti-CD276 treatment. ***p<0.001 by Student’s t test.
(O) Quantification of PCK+Tomato+ CSCs in cervical lymph node from mice treated with anti-CD276. ***p<0.001 by Student’s t test.
(P) Experimental design used to trace Bmi1+-derived tumor tissues in Bmi1CreER;RosatdTomato mice.
(Q) PCK immunostaining and Bmi1+-derived tumor tissues in BmiCreER;RosatdTomato mice upon anti-CD276 treatment. Scale bar, 20μm
To further investigate the role of CD276 blockade on metastasis, cervical lymph nodes were carefully dissected under a microcopy. To precisely compare lymph node metastasis, cervical lymph nodes were immunostained with anti-pan-cytokeratin (PCK) which specifically detected metastatic tumor cells in lymph nodes (Chen et al., 2017). We found that the total number of lymph nodes with metastasis and metastatic rate per mouse were all significantly reduced following anti-CD276 treatment (Figures 4L–4N). Importantly, BMI1+ CSCs residing in cervical lymph nodes were eliminated by anti-CD276 antibodies (Figure 4O). As expected, in vivo lineage tracing for one month further revealed that CD276 blockade potently inhibited BMI1+ CSCs-derived tumor cell growth (Figures 4P and 4Q).
Landscape alterations of HNSCC upon anti-CD276 treatment
To provide a more comprehensive and unbiased assessment of tumor responses upon anti-CD276 treatment, we performed scRNA-seq analysis on the HNSCC cells from the 4NQO-induced mice treated with anti-CD276 for 4 weeks. Because tumors were significantly reduced after anti-CD276 treatment, we pooled tumors from 3 mice for the anti-CD276-treated group. We obtained single-cell transcriptomes for 11658 cells, then identified and visualized transcriptional homogeneous clusters of cells using graph-based clustering and dimensionality reduction with Uniform Manifold Approximation and Projection (UMAP) (Figure S2A). Clusters were further annotated by comparing their transcriptional state using the Single R package and evaluation of known cell-type specific markers (Figures S2B–S2I). We identified cancer cells expressing the epithelial cell adhesion molecule Epcam); lymphoid population expressing Cd3d, Cd8a, Foxp3, and Cd4; stromal cells expressing Col1a1; endothelial cells expressing Vwf; and myeloid population expressing Itgam.
To clarify an overall landscape of cancer cell populations associated with anti-CD276 treatment, we characterized the alterations of the subpopulations of cancer cells by separating cancer cells (total 6274 cells, 4665 cells from vehicle group and 1609 cells from anti-CD276 group) and yielded 6 distinct cancer cell subpopulations defined by the transcriptional state of cells and known marker genes, representing high plasticity and complexity (Figures 5A–5I and Figures S2J–S2P). Notably, cluster 0 (c0) was enriched with genes associated with partial epithelial-mesenchymal transition (pEMT) or tumor-specific keratinocyte (Lamc2, Lama3, Lamb3, Itga6, Itgb1), representing the most invasive subpopulation of HNSCC cells with high metastatic potential located at the leading edge/invasive tumor front (Figures 5C, 5D and Figure S2K) (Ji et al., 2020; Puram et al., 2017). Cells in the cluster (c1) mainly consisted of immediate early genes implicated in stress response (Atf3, Erg1, and Dusp6) (Figures 5C, 5E and Figure S2L), and Cluster 2 (c2) consisted primary of epithelial genes, including Krt6b, Krt13, Krt16, Sprr1b, Sprr2a3, Sprr1a, representing the cancer cells with typical epithelial differentiation at the core of tumors (Figures 5C, 5F and Figure S2M) (Puram et al., 2017). Cluster 3 (c3) was characterized with high proliferative activity and reflected high cell-cycling cells, which expressed Mki67, Cdk1, Hmgb2, Birc5 (Figures 5C, 5G and Figure S2N) (Puram et al., 2017). Cluster 4 (c4) consisted of genes associated with detoxification and drug metabolism (Figures 5C, 5H and Figure S2O) (Adh7, Aldh3a1, Gstm1, Gsta1) (Puram et al., 2017). All these cluster of cancer cells were also identified in human primary HNSCC at single cell resolution (Puram et al., 2017). Interestingly, we also identified cluster 5 (c5) consisting of a small population cancer cells expressing MHCII molecules (H2-Aa, H2-Ab1, H2-Eb1, Cd74) (Figures 5C, 5I and Figure S2P), which was also identified in nasopharyngeal carcinoma as an immune suppressive cancer cells with high tumorigenic capability (Jin et al., 2020).
Figure 5. ScRNA-seq analysis identifies landscape alterations of murine HNSCC upon anti-CD276 treatment.
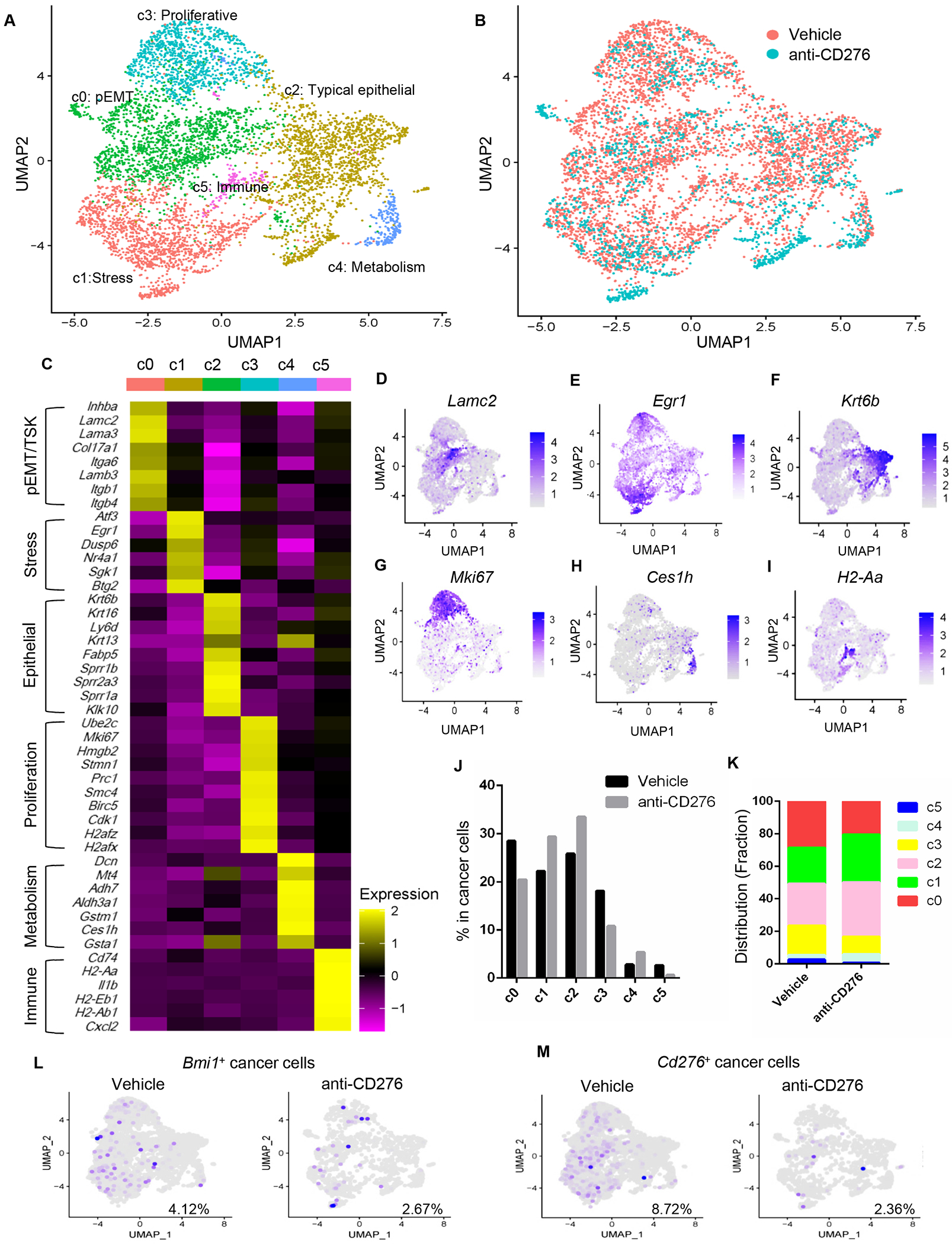
(A) UMAP plot showing identified cancer cell populations from two groups merged which were treated with control IgG and anti-CD276 for 4 weeks.
(B) UMAP plot showing distribution of annotated clusters within cancer cells upon control IgG or anti-CD276 treatment for 4 weeks.
(C) Heatmap displaying expression of selected marker genes in each cluster.
(D-I) UMAP plot of tumor cells displaying selected marker gene expression.
(J) Percentage of different cancer cell populations annotated in response of anti-CD276 treatment.
(K) Distribution fraction of different cancer cell populations annotated in response of anti-CD276 treatment.
(L) UMAP plot of Bmi1+ cancer cells upon anti-CD276 treatment.
(M) UMAP plot of Cd276+ cancer cells upon anti-CD276 treatment.
See also Figure S2
The percentage of pETM cancer cells (c0) decreased significantly upon anti-CD276 treatment in comparison with HNSCC treated with vehicles (Figures 5B, 5J and 5K). These findings support the notion that targeting CD276 inhibits lymph node metastasis of HNSCC in our in vivo studies. In response to anti-CD276 treatment, the proportion of highly proliferative (c3) and MHCII+ HNSCC cells (c5) were also significantly reduced as compared to control mice, implying a major impact on proliferation and activation of the immune environment. Anti-CD276 treatment led to an increase in frequency of c1 and c4, which might indicate a cellular response to stress from anti-tumor microenvironment and a feedback to detoxification. In comparison with vehicle, the proportion of cells in c2 (typical epithelial) was increased upon anti-CD276 treatment, suggesting the EMT program was blocked by anti-CD276 treatment. These findings revealed that the residual tumors are less aggressive and are characterized with low pEMT and cycling cells after anti-CD276 treatment. In line with our lineage tracing data, Bmi1+ and Cd276+ cancer cells were significantly decreased in mice treated with anti-CD276 (Figures 5L and 5M).
Modulating HNSCC immune microenvironment by CD276 blockade
To explore the role of immune cells in CD276 blockade-mediated tumor inhibition, NOG mice, without lymphocytes and NK cells, were inoculated with freshly isolated murine HNSCC cells and treated with anti-CD276 antibodies and vehicle with isotype IgG. CD276 blockade did not show any tumor-suppressive effects in NOG mice bearing murine HNSCC tumors (Figures 6A and 6B), indicating that the anti-tumor effect of CD276 blockade is dependent on immune cells. To reinforce these observations in vitro, HNSCC cells were treated with anti-CD276 antibodies, and we found that anti-CD276 did not affect sphere formation and cell invasion (Figures 6C and 6D). Consistently, Western blot analysis found that anti-CD276 neutralizing antibodies were unable to inhibit intracellular AP-1 activation in SCC cells (Figure S3A). Similarly, anti-CD276 did not affect primary human HNSCC organoid formation (Figure 6E). Immunostaining showed that anti-CD276 neutralizing antibodies did not appear to inhibit the expression of c-Jun and FOSL1 in HNSCC organoids (Figure S3B). These results suggest that, while the knockdown of CD276 might affect tumor growth in nude mice because AP-1 activation mediated by intracellular signaling of CD276 was inhibited, our anti-CD276 antibodies were unable to inhibit intracellular AP-1 activation. Furthermore, the co-culture assays demonstrated that anti-CD276 treatment significantly enhanced HNSCC cell apoptosis mediated by activated primary T lymphocytes as determined by immunostaining of the active caspase-3+ cancer cells (Figure 6F).
Figure 6. CD276 blockade modulates tumor immune microenvironment of HNSCC.
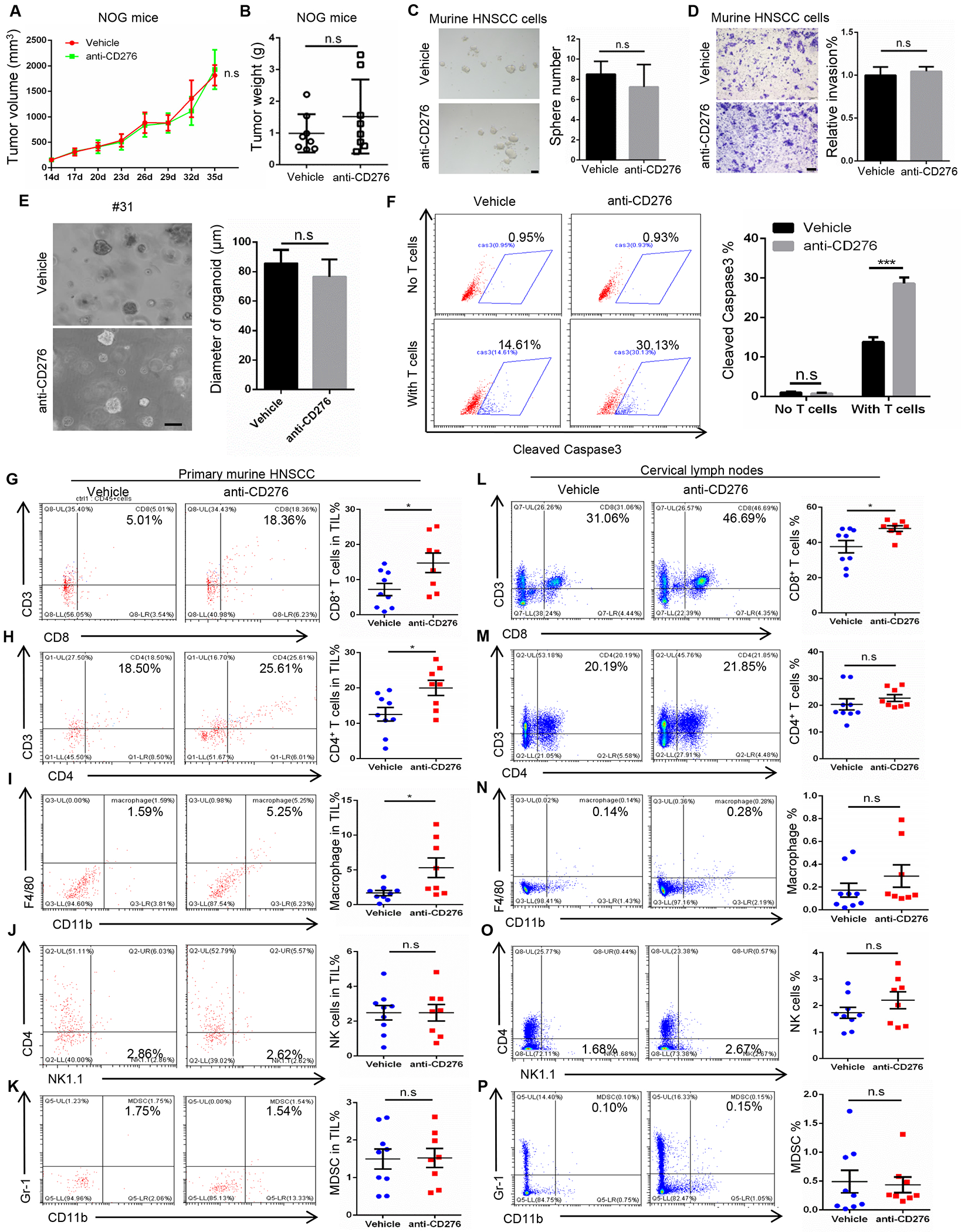
(A) Tumor volume in NOG mice bearing murine HNSCC treated with anti-CD276. Values are mean ± SEM., n = 8, two-way ANOVA, n.s., non-significant.
(B) Tumor weight in NOG mice bearing murine HNSCC treated with anti-CD276. Values are mean ± SD., n = 8, Student’s t-test, n.s., non-significant.
(C) Representative image and quantification of tumorspheres derived from murine HNSCC cells treated with control IgG and anti-CD276 antibodies. Student’s t-test, n.s., non-significant. Scale bar, 50μm
(D) Representative image and quantification of migrated murine HNSCC cells treated with control IgG and anti-CD276 antibodies. Student’s t-test, n.s., non-significant. Scale bar, 200μm
(E) Representative image and quantification of organoids derived from freshly isolated human HNSCC cells treated with control IgG and anti-CD276 antibodies. Student’s t-test, n.s., non-significant. Scale bar, 100μm
(F) The percentage of cleaved caspase 3 positive cells in HN6 cells treated with control IgG and anti-CD276 antibodies. HN6 cells were co-cultured with activated human peripheral blood T cells for 8h. ***p<0.001 by Student’s t test, n.s., non-significant.
(G-K) Frequencies of tumor-infiltrating CD8+ and CD4+ T lymphocytes, macrophages, NK cells and MDSCs in HNSCC upon anti-CD276 treatment for 10 days. Values are mean ± SD from the pool of three independent experiments. vehicle group, n=9; anti-CD276 group, n=8. *p<0.05 by Student’s t test, n.s., non-significant.
(L-P) Frequencies of CD8+ and CD4+ T lymphocytes, macrophages, NK cells and MDSCs in cervical lymph nodes upon anti-CD276 treatment for 10 days. Values are mean ± SD from the pool of three independent experiments. vehicle control, n=9; anti-276 group, n=8. *p<0.05 by Student’s t test, n.s., non-significant.
See also Figure S3
We further examined how anti-CD276 eliminated CSCs and inhibited HNSCC growth and metastasis in the syngeneic mouse model of HNSCC. Immunohistological examination of HNSCC revealed that anti-CD276 significantly increased the number of apoptotic cells (Figure S3C) and Granzyme B+ (GZMB+) cells (Figure S3D) compared with isotype IgG. Because GZMB, largely produced by CD8+ T and NK cells, induces tumor cell apoptosis (Lee et al., 2017; Yonesaka et al., 2018), we next investigated whether CD8+ T and NK cells were involved in CD276 blockade-mediated tumor inhibition. Minimal CD8+ T cells and NKp46+ NK cells were present in tumors treated with isotype IgG. In contrast, CD276 blockade strikingly increased the infiltration of CD8+ T cells (Figure S3E). While NKp46+ NK cells in tumor tissues were also found to be modestly increased (Figure S3F), myeloid-derived suppressor cells (MDSCs) in tumor tissues were not changed after anti-CD276 treatment (Figure S3G).
Recently, it has been shown that intratumoral immune cells undertake remodeling with high complexity, diversity and heterogeneity during immune checkpoint therapy (Gubin et al., 2018; Wei et al., 2017; Zhang et al., 2020). To better understand the overall immune landscape remodeling upon anti-CD276 treatment, we further characterized the early alterations of the subpopulation of the tumor-infiltrated immune cells in HNSCC treated with anti-CD276 antibodies by flow cytometry. Because mouse tumors were small and drastically reduced after anti-CD276 treatment, it was difficult to isolate immune cells from tumor tissues at the 4-week endpoint of our experiments. Therefore, immune cells were isolated from mouse tumors 10 days after anti-CD276 treatment. Flow cytometry analysis revealed that the frequency of intratumoral CD8+ and CD4+ T lymphocytes and macrophages were significantly increased upon anti-CD276 treatment, indicating anti-CD276 treatment provokes an antitumor immunity (Figures 6G–6I). Compared to immunostaining at the endpoint, the percentage of intratumoral NK cells were not significantly increased most likely due to the short treatment regimens (Figure 6J). MDSCs were not significantly changed upon anti-CD276 treatment (Figure 6K). We also examined whether anti-CD276 blockade enhanced antitumor immunity in cervical lymph nodes by using flow cytometer. Cells were harvested from cervical lymph nodes of HNSCC mice treated with anti-CD276. Flow cytometry analysis showed that the frequency of CD8+ T cells was significantly increased after anti-CD276 treatment in lymph nodes (Figure 6L). The percentage of CD4+ T cells, macrophages and NK1.1+ cells were increased although their changes were not statistically significant (Figures 6M–6O). There was no change in MDSCs (Figure 6P).
Impeding CD276 blockade-mediated CSC elimination by depleting CD8+ T cells
To determine the extent to which CD276 blockade promoted an antitumor reaction dependent on CD8+ T cells, CD8+ T cells were deleted using anti-CD8α antibodies (Lee et al., 2017) (Figure 7A). Depletion of CD8+ T cells significantly diminished the inhibition of tumor growth mediated by CD276 blockade based on analysis of overall lesions number and area (Figures 7B–7D). Histological analysis revealed that depletion of CD8+ T cells significantly reversed anti-CD276-mediated inhibited invasive growth (Figures 7E and 7F). Importantly, in vivo lineage tracing revealed that the elimination of BMI1+ CSCs induced by CD276 blockade was impaired after depletion of CD8+ T in primary HNSCC (Figures 7G and 7H). Similarly, the inhibition of cervical lymph node metastasis was also significantly impeded in mice by depletion of CD8+ T cells (Figures 7I and 7J). The elimination of BMI1+ CSCs in cervical lymph nodes was also significantly prevented by depletion of CD8+ T cells (Figures 7I and 7K). As expected, the infiltration of CD8+ T cells was increased in response of anti-CD276 treatment, but deleted upon anti-CD8α treatment (Figure 7L). To explore the functional role of NK cells in CD276-mediated anti-tumor immunity in HNSCC, depletion of NK single population were also performed. Single depletion of NK cells slightly affected tumor size and invasive depth but did not significantly impair anti-CD276-meidated tumor inhibition (Figures S4A–4D). The double depletion of CD8+ T and NK cells had similar effects as compared to single deletion of CD8+ T cells.
Figure 7. Depletion of CD8+ T lymphocytes impairs CD276 blockade-mediated inhibition of tumor growth and CSC elimination.
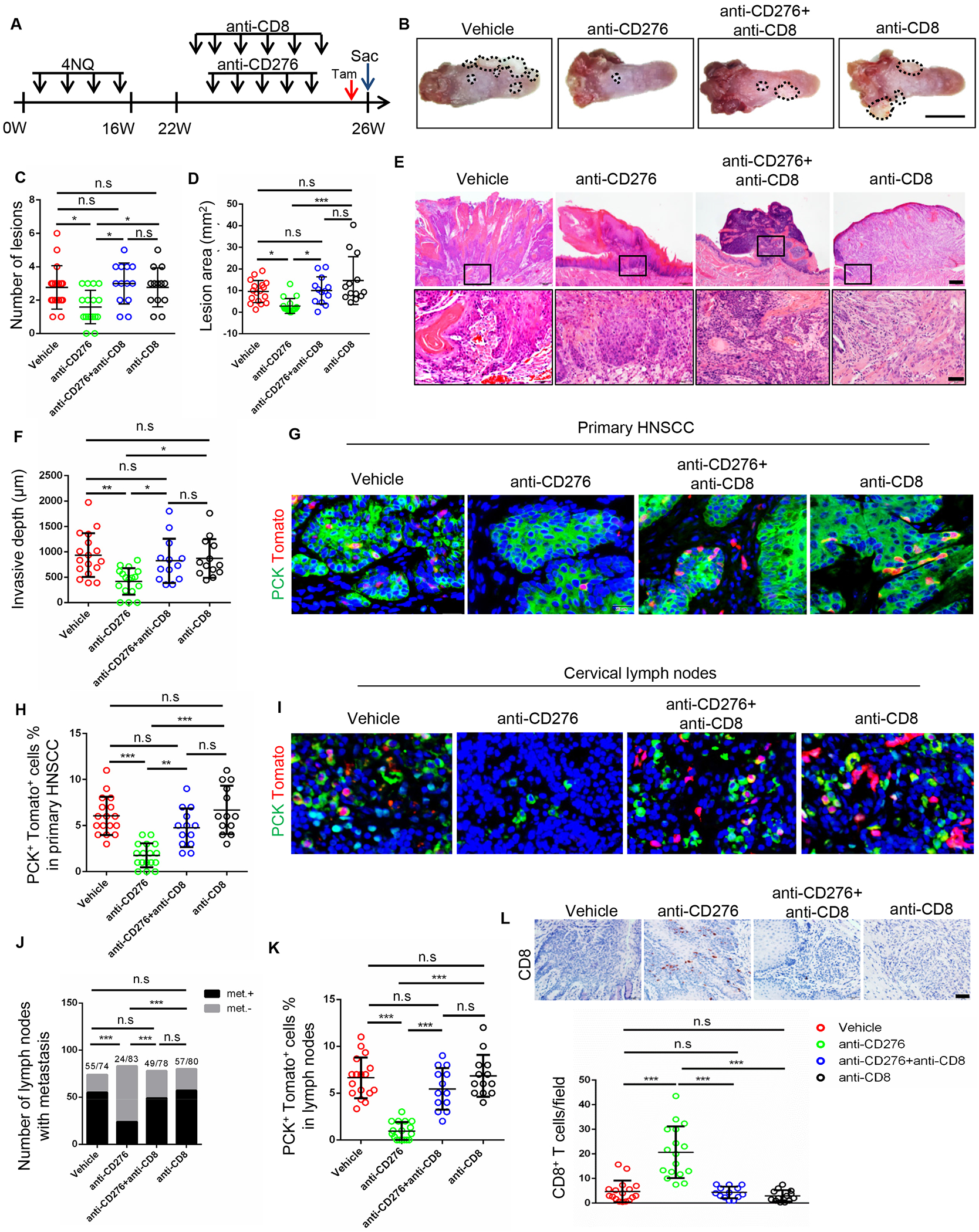
(A) Experimental design for depletion of CD8+ T lymphocytes and lineage tracing of CSCs of HNSCC in Bmi1CreER; RosatdTomato mice.
(B) Representative image of tongue lesions 26 week in Bmi1CreER;RosatdTomato mice treated with different antibodies. Vehicle, Isotype IgG; anti-CD276, anti-CD276 + Isotype IgG; anti-CD276+anti-CD8, anti-CD276 + anti-CD8α + Isotype IgG; anti-CD8, anti-CD8α + Isotype IgG. Scale bar, 5mm
(C) Quantification of lesion number visible in the mouse tongues. Values are mean ± SD from the pool of four independent experiments (n=13-17 as indicated); *p<0.05 by one-way ANOVA; n.s, non-significant.
(D) Quantification of lesion areas in the mouse tongues. Values are mean ± SD from the pool of four independent experiments. Vehicle, n = 17; anti-CD276, n = 17; anti-CD276 + anti-CD8, n = 13; anti-CD8, n = 13. *p<0.05 and ***p<0.001 by one-way ANOVA; n.s, non-significant.
(E) Representative image of H&E staining of HNSCC in Bmi1CreER;RosatdTomato mice. Scale bar, 200μm (upper panels), 50μm (lower panels)
(F) Quantification of HNSCC invasive depth. Vehicle, n = 17; anti-CD276, n = 17; anti-CD276 + anti-CD8, n = 13; anti-CD8, n = 13. *p<0.05 and **p<0.01 by one-way ANOVA. n.s indicated non-significant.
(G and H) Representative image and quantification of PCK+Tomato+ CSCs in anti-CD276-treated Bmi1CreER;RosatdTomato mice with or without the depletion of CD8+ T cells. Values are mean ± SD. Vehicle, n=17; anti-CD276, n=16; anti-CD276 + anti-CD8, n=13; anti-CD8, n=13. **p<0.01 and ***p<0.001 by one-way ANOVA. n.s, non-significant. Scale bar, 20μm
(I) Representative image of PCK+ cancer cells and PCK+Tomato+ CSCs in cervical lymph nodes in different groups. Scale bar, 20μm
(J) Number of cervical lymph nodes with metastasis in different groups. ***p<0.001 by Chi square test. Number of cervical lymph nodes indicated in the figure.
(K) Quantification of PCK+Tomato+ CSCs in cervical lymph nodes in different groups. Vehicle, n = 17; anti-CD276, n = 17; anti-CD276 + anti-CD8, n = 13; anti-CD8, n = 13. ***p<0.001 by one-way ANOVA.
(L) Representative image and quantification of CD8+ lymphocytes in HNSCC from anti-CD276-treated Bmi1CreER;RosatdTomato mice with or without the depletion of CD8+ T cells. Vehicle, n=17; anti-CD276, n = 17; anti-CD276 + anti-CD8, n = 13; anti-CD8, n = 13. ***p<0.001 by one-way ANOVA. n.s., non-significant. Scale bar, 50μm.
CD276 expression inversely correlated with infiltrated CD8+ T lymphocytes
We examined CD276 expression in a TCGA HNSCC patient cohort and found that CD276 expression was increased in HNSCC compared with normal tissues (Figure S5A). Importantly, HNSCC patients with high expression of CD276 had a poor prognosis compared to HNSCC patients with low expression of CD276 (Figure S5B). However, in contrast, there was no association between CD274 (also known as PD-L1) expression and HNSCC patient survival in the TCGA database (Figure S5C).
Since FOSL1 and CD276 formed a positive regulatory loop, we analyzed their expression in TCGA HNSCC patient cohort and found a positive correlation between FOSL1 and CD276 (Figure S5D). Furthermore, immunostaining confirmed that CD276 protein expression levels were positively correlated with FOSL1 expression levels in the human HNSCC samples (Figure S5E). Finally, we examined the relationship between CD276 expression and CD8+ T cells in HNSCC. Immunostaining confirmed that CD276 expression was inversely correlated with infiltration of CD8+ T cells in human HNSCC samples from 2 independent patient cohorts (Figures S6A–6C). Analysis of the TCGA HNSCC patient cohort revealed that the expression of CD276 was inversely correlated with the infiltration level of CD8+ T cells by 6 different algorithms (Figure S6D).
DISCUSSION
Immunotherapy has emerged as a new type of clinical treatment for cancer. However, the relevance of immune checkpoints and its potential therapeutic role in targeting CSCs is not well studied (Miao et al., 2019). Moreover, how CSCs escape immune surveillance is not well characterized during HNSCC initiation, development and metastasis. In this study, we demonstrated that CD276 as an immune checkpoint was highly expressed in CSCs using a syngeneic mouse model of HNSCC combined with in vivo lineage tracing of CSCs. Our results indicate that CSCs utilized CD276 to overcome anti-tumor immunity during HNSCC initiation, development and metastasis. CD276 blockade potently eliminated CSCs and inhibited HNSCC metastasis by enhancing CD8+ T cell-mediated antitumor immunity, thereby highlighting CD276 as a unique therapeutic target for HNSCC immunotherapy.
Immune surveillance is critical for preventing tumor development and progression. Since tumors are derived from CSCs, CSCs must develop important strategies to evade immune surveillance to effectively initiate tumor growth (Miao et al., 2019). Moreover, HNSCC tumor cells, most likely CSCs, frequently metastasize to and survive in cervical lymph nodes which are enriched with immune cells. It was reported that transforming growth factory-β-responding CSCs selectively acquired the expression of CD80, an immune cell surface ligand to interact with T lymphocytes (Miao et al., 2019). CD80-expressing CSCs directly inhibited cytotoxic T cell activity and mediated tumor resistance to adoptive cytotoxic T cell transfer (ACT)-based immunotherapy (Miao et al., 2019). Interestingly, we also identified that CD80 was highly expressed in BMI1+ CSCs based on our RNA-seq results. In the future, it will be important to determine whether BMI1+ CSCs are resistant to ACT-based immunotherapy. Since CD276 blockade reduced the number of both dysplasia and tumors, our results suggest that CSCs utilized CD276 to evade immune surveillance during HNSCC initiation. Notably, CD276+ cancer cells were mainly located at the ITF and the periphery of tumor nests, which was also observed previously (Mao et al., 2017), indicating that CD276 might function as a shield to protect CD276+ cells and inner tumor cells (CD276− cells) against killing by CD8+ T cells during HNSCC development. Moreover, because CD276 blockade potently inhibited lymph node metastasis in mice with established tumors, these findings suggest that CD276 also promotes CSC immune evasion during metastasis.
Emerging evidence indicates that HNSCC is characterized by high heterogeneity and plasticity (Cillo et al., 2020; Puram et al., 2017). Our scRNA-seq analysis confirms that murine HNSCC mimic human HNSCC with similar subpopulation of cancer cells, including subsets characterized with pEMT, stress, typical epithelial, proliferation, metabolism and immune features (Jin et al., 2020; Puram et al., 2017).
Of note, EMT/pEMT is associated with tumor metastasis and generation of CSCs (Mani et al., 2008; Puram et al., 2017). Importantly, the landscapes of HNSCC were remodeled in response of CD276 blockade, in which the subsets characterized with pEMT, proliferation and immunosuppresive features were significantly decreased. In line with our in vivo lineage tracing data, lymph node metastasis and BMI1+ CSCs were significantly reduced following anti-CD276 treatment. These findings strongly support that targeting CD276 eliminates CSCs and inhibits metastasis in HNSCC, implicating its promising preventive and therapeutic potentials.
CD276 expression was inversely correlated with CD8+ T cell infiltration in human HNSCC samples. CD276 blockade significantly increased infiltration of CD8+ T cells, and depletion of CD8+ T cells impaired anti-tumor immunity by CD276 blockade. Our immunostaining found that NKp46+ NK cells in residual tumor tissues were increased. The deletion of NK cells slightly affected CD276 blockade-mediated inhibition of HNSCC growth, but this finding was not statistically significant. These results support that CD8+ T cells are the major functional targets for CD276 in tumor immunity (Lee et al., 2017; Yonesaka et al., 2018). Importantly, CD276 blockade significantly inhibited lymph node metastasis of HNSCC which is one of the most challenging issues in HNSCC treatment, further demonstrating that targeting CD276 enhances anti-tumor immunity. Despite the success of anti-PD-1/PD-L1 immunotherapy, a significant proportion of patients failed to respond to these therapies (Ferris et al., 2016; Harrington et al., 2017). One of the major reasons for unresponsiveness to immunotherapy is insufficient infiltration of activated CD8+ T cells into the tumor microenvironment. Interestingly, CD276 expression was found to correlate with non-responsiveness to anti-PD-1 immunotherapy in non-small cell lung cancer and ovarian cancer due to the possible exclusion of CD8+ tumor-infiltrating lymphocytes by CD276-expressing tumors (Cai et al., 2020; Yonesaka et al., 2018). Our results suggest that abnormal expression of CD276 in HNSCC might be responsible for the exclusion and dysfunction of CD8+ T cell infiltration. CD276 is widely expressed in a variety of tumors, including osteosarcoma, brain tumors and others as well as tumor vasculature (Du et al., 2019; Majzner et al., 2019; Seaman et al., 2017). CD276-drug conjugates, but not CD276 alone, have been shown to strongly inhibit tumor growth and metastasis by using human lung, colon and breast cancer models (Seaman et al., 2017). Importantly, our in vivo studies demonstrated that CD276 blockade alone potently inhibited HNSCC growth, indicating that CD276 might play a more unique and critical role in HNSCC compared with other solid tumors. Very recently, chimeric antigen receptor (CAR) T cells targeting CD276 have been generated and have been shown to control the growth of pancreatic ductal adenocarcinoma, ovarian cancer and neuroblastoma (Du et al., 2019; Majzner et al., 2019). It will be interesting to test whether CAT T cell targeting CD276 could help to eliminate CSCs in HNSCC and inhibit metastasis.
Although CD276 was found to be upregulated in a variety of tumors, the molecular mechanism controlling CD276 expression remains unclear. Elucidation of its upstream molecular signaling may help to understand tumor immune evasion and aide the development of innovative strategies for targeting CD276. Previous studies have indicated that a miRNA-regulatory mechanism is responsible for the CD276 expression pattern in neuroblastoma and ovary cancer (Wang et al., 2018; Xu et al., 2009). CD276 was highly and homogenously expressed in multiple tumors, including pediatric solid tumors, brain tumors, and pancreatic ductal adenocarcinoma (Du et al., 2019; Majzner et al., 2019; Seaman et al., 2017). Unlike these solid tumors, we found that CD276 was preferentially expressed at the ITF and periphery of HNSCC. Both BMI1+ and CD276high CSCs were located at the ITF of HNSCC, suggesting that similar invasive niches and molecular mechanisms might govern the phenotype of BMI1+ and CD276high cancer cells. Interestingly, we found that overexpression of CD276 induced the expression of c-Jun and FOSL1, suggesting that CD276 might promote CSC invasion and metastasis by activating AP-1 (Ding et al., 2013). Notably, we found that the CD276 transcription was controlled by SEs, which has been shown to control cell identity and play a crucial role in tumorigenesis (Hnisz et al., 2013; Sengupta and George, 2017; Vaharautio and Taipale, 2014). Strikingly, FOSL1, which drives the invasive growth and metastasis of BMI1+ CSCs (Chen et al., 2017), was involved in the establishment of SEs in CD276. Our findings reveal that CD276 and AP-1 formed a positive feedback loop to promote CSC immune evasion in addition to CSC self-renewal and metastasis. While BET inhibitors have been shown to inhibit tumor growth by disrupting SEs, recently it was found that BET inhibitors cooperated with PD-1 blockade to facilitate antitumor response in non-small cell lung cancer by reducing suppressive regulatory T cells (Adeegbe et al., 2018). In the future, it will be important to test whether BET inhibitors could eliminate CSCs and inhibit HNSCC initiation, development and metastasis by increasing anti-tumor immune response through inhibition of CD276 expression.
Limitation of the Study
Although the mouse model allows in vivo lineage tracing of CSCs following immunotherapy, we are unable to observe the inhibition of tumor growth dynamically. HNSCC cells grow invasively underneath the mucosa so that these tumors cannot be measured directly without sacrificing mice. MDSCs play an important role in tumor growth and development (Sanmamed and Chen, 2018; Gubin et al., 2018). Although anti-CD276 did not affect the number of MDSCs, it remains possible that anti-CD276 might affect the function of MDSCs. The 4NQO-induced HNSCC model might also have some limitations which is unable to accurately examine changes in MDSCs. Furthers studies are necessary to dissect the heterogeneity and phenotype changes of MDSCs in HNSCC upon CD276 blockade using advanced technologies such as single cell RNA sequencing (scRNAseq) and spatial RNA sequencing (spRNAseq). Based on the status of human papillomavirus (HPV), HNSCC are mainly categorized into two subtypes: HPV− and HPV+. HPV+ and HPV− HNSCC might have distinct immune features. While our model simulates the molecular pathogenesis of HPV-negative HNSCC, we do not know whether CSCs in HPV+ HNSCC also utilize CD276 to evade Immune surveillance.
STAR★ METHODS
Detailed methods are provided in the online version of this paper and include the following:
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for reagents may be directed to, and will be fulfilled by the Lead Contact, Cun-Yu Wang (cwang@dentistry.ucla.edu).
Materials Availability
Cell lines and plasmids used in this study are described in the Key Resource Table and available upon request. Bmi1CreER;R26tdTomato mice are available upon request with the approved animal protocol. All requests need to execute a suitable Materials Transfer Agreement.
Data and Code Availability
RNA-seq and scRNA-seq data were deposited at the Gene Expression Omnibus (GEO) under the accession number GEO: GSE132627 and GSE164817.
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Mice
Bmi1CreER;R26tdTomato mice were generated by crossmating Bmi1CreER (Jackson Laboratory, JAX:010531) with R26tdTomato (Jackson Laboratory, JAX:007908) as described before (Chen et al., 2017). NOG mice were purchased from Taconic. Mice were housed under standard conditions in the animal facility of UCLA. All procedures were performed based on the UCLA Animal Research Committee-approved protocols. The induction of mouse HNSCC with 4-NQO was performed as described before (Jia et al., 2020).
Cell lines
Human HNSCC cell lines SCC1, SCC22B, and SCC23 cells were obtained from the University of Michigan, and HN6 cells were from Wayne State University. SCC1R was cisplatin-resistant and maintained in 5 μg/ml of cisplatin. These cells were culture in normal DMEM supplemented with 10% FBS at 37°C in 5% CO2 atmosphere.
Human HNSCC samples
The use of human HNSCC samples in this study was approved by the UCLA Institutional Review Board. Fresh human HNSCC primary tissue were obtained from Ronald Reagan UCLA Medical center and subcutaneously inoculated into flank of 6-week-old NOG mice (Taconic). The paraffin-embedded blocks of the human HNSCC were obtained from the UCLA Translational Pathological Core Laboratory and sectioned for immunohistochemistry.
METHOD DETAILS
4NQO model, lineage tracing and antibody treatment
The induction of HNSCC and lineage tracing were performed as previously described (Chen et al., 2017). 4-Nitroquinoline N-oxide (4-NQO; sc-256815, Santa Cruz,) was suspended in propylene glycol (Sigma-Aldrich, W294025) at 5 mg/ml and stored for at 4°C. Six-week-old Bmi1CreER;R26tdTomato mice were consecutively treated by 40 μg/ml 4-NQO (Cat#sc-256815, Santa Cruz)-containing drinking water for 16 weeks. The drinking water was changed every two weeks. Afterwards, mice were feed with normal drinking water for another 10-14 weeks.
For CD276 blockade treatment, mice at 22 weeks were randomly divided into two groups and intraperitoneally given the vehicle IgG control (InVivoMAb rat IgG1 isotype control, BioXcell Cat#BE0088, 10mg/kg body weight) and anti-CD276 antibodies (InVivoMAb anti-mouse CD276, BioXcell Cat#BE0124, 10mg/kg body weight) three times per week for 4 weeks. To label BMI1+ CSCs, Bmi1CreER;R26tdTomato mice were injected with tamoxifen intraperitoneally at a dose of 225 mg/kg body weight 24 h before sacrificing the mice. For depletion of CD8+ T lymphocytes, mice were intraperitoneally given anti-CD8 (InVivoPlus anti-mouse CD8α, BioXcell Cat#BP0061, 100μg/mouse) twice per week for 4 weeks. InVivoPlus rat IgG2b isotype (BioXcell Cat#BP0090, 100μg/mouse) as control antibodies were given twice per week for 4 weeks. For the depletion of CD8+ T and NK cells, mice were given anti-CD8 (100μg/mouse) and anti-NK1.1 antibodies (InVivoPlus anti-mouse NK1.1, BioXcell Cat#BP0036, 100μg/mouse), anti-CD8 (100μg/mouse), and anti-NK1.1 antibodies (100μg/mouse) twice per week for 4 weeks. Both InVivoPlus rat IgG2b isotype (BioXcell Cat#BP0090, 100μg/mouse) and InVivoPlus mouse IgG2a isotype (BioXcell CatBP0085, 100μg/mouse) as control antibodies were given twice per week for 4 weeks. For JQ1 treatment, the animals were randomly assigned into either control or JQ1 treatment group. JQ1 (APExBIO, cat#4499) stock solution was prepared in dimethylsulfoxide (Sigma-Aldrich, cat#D2650) at 200 mg/ml and administered at 50 mg/kg body weight.
HNSCC formation and invasive grades were examined by H&E staining and the invasive depth was measured in the H&E stained sections. The cervical lymph node metastasis was determined with anti-PCK antibodies and the percentage of lymph node with metastasis was evaluated (Chen et al., 2017).
Immunostaining
After treatment, mice were euthanized, and the neck skin was cut and detached. The cervical lymph nodes were harvested with sharp scissors under the Leica EZ4 Stereo Microscope and fixed with 4% paraformaldehyde (PFA; Cat#P6148, Sigma-Aldrich). After collecting all the lymph nodes, mouse jaws were opened with a bone scissor and the tongue tissues were harvested with a sharp scissor and fixed with 4% PFA. Paraffin-embedded HNSCC sections or frozen sections were processed and cut by the UCLA Translational Pathological Core (Chen et al., 2017; Ding et al., 2013). For immunofluorescent staining, sections were incubated with primary antibodies, including anti-PCK (Abcam, Cat#ab9377; 1:200), anti-active caspase3 (Abcam, Cat#ab13847; 1:100), anti-CD276 (Cell Signaling Technology, Cat#14058; 1:100), anti-Bmi1 (Cell Signaling Technology, Cat#6864; 1:100) and anti-CD276 conjugated with FITC (LS Bio, Cat#LS-C419560; 1:100). The antigens were visualized using secondary antibodies conjugated with Cy2 or Cy3 (Jackson ImmunoResearch Laboratories). Sections were then counterstained and mounted with ProLong™ Diamond Antifade Mountant with DAPI (Thermo Fisher Scientific, Cat# P36962). Images were acquired with a microscopy running Cellsens software. .
For immunohistochemistry of human or murine HNSCC samples, sections were incubated with the following primary antibodies at 4°C overnight, anti-CD276 (Cell Signaling Technology, Cat#14058; 1:100), anti-CD8α (Cell Signaling Technology, Cat#85336S; 1:100), anti-FOSL1 (Abcam, Cat#ab232745; 1:100), anti-CD8 (Abcam Cat#ab22378; 1:100), anti-NKp46 (Abcam, Cat#ab214468; 1:100), and anti-active caspase3 (Abcam, Cat#ab13847; 1:100). The slides then were then incubated with horseradish perioxidase-labeled polymers for 2h at room temperature and detected with AEC+ chromogen (Dako EnVision System, Cat# MP-6401-15). The immunohistochemistry scores were determined as previously described (Pirker et al., 2012).
HNSCC PDX model, cell isolation and flow cytometry
The use of human HNSCC tissues for this study was approved by the UCLA Institutional Review Board. The human HNSCC primary tissues were obtained from the UCLA Translational Pathological Core and subcutaneously inoculated into flank of 6-week-old NOG mice (Taconic) to generate the PDXs. The murine HNSCC primary tissues from 4NQO-induced Bmi1CreER;RosatdTomato mouse were transplanted to the flank of 6-week-old NOG mice to generate murine HNSCC xenografts. Human and murine HNSCC xenografted tumors were chopped and then digested into single cell suspensions by using a human tumor cell dissociation kit (Miltenyi Biotec, Cat#130095929) or a mouse tumor dissociation kit (Miltenyi Biotec, Cat# 130096730). To isolate CD276high and CD276low cancer cells from tumor tissues, the single cell suspension was incubated with anti-human EpCAM-PE (Miltenyi Biotec, Cat#130110999; 1:50), anti-human CD276-APC (Miltenyi Biotec, Cat#130095522; 1:100) or anti-mouse EpCAM-FITC (Miltenyi Biotec, Cat# 130102214; 1:50) and anti-mouse CD276-APC (BioLegend, Cat#135608; 1:50) for 30 min on ice and then sorted by a FACSVantage SE (Beckton Dickson). The results were analyzed with FlowJo software (https://www.flowjo.com). For ALDHbright and ALDHdim cell sorting, cancer cells were stained with an ALDHEFLUOR assay kit (STEMCELL Technologies, Cat#01700).
Immune cell profiling by flow cytometry
Primary murine HNSCC tissues and cervical lymph nodes were harvested from 4NQO-induced mice treated with vehicles or anti-CD276 antibodies for 10 days. Single cell suspensions were achieved as described above. For staining of cell surface markers, cells were incubated with indicated antibodies on ice for 30min and washed with staining buffer. Then, the stained cells were examined using a flow cytometer (Navios, Beckman Coulter) and the data was analyzed with CytExpert software (https://www.beckman.com/flow-cytometry/instruments/cytoflex/software). Antibodies used for flow cytometry analysis are listed below: anti-mouse FVS700 APC-700 (BD Biosciences, Cat#564997), anti-mouse CD45 Percp-cy5.5 (BioLegend, Cat#103132), anti-mouse CD4 PE-cy7 (BioLegend, Cat#100422), anti-mouse CD3 Alexa Flour488 (BioLegend, Cat#100210), anti-mouse CD8 BV786 (BD Biosciences, Cat#563332), anti-mouse NK1.1 BV510 (BioLegend, Cat#108738), anti-mouse Gr-1 PE/Dazzle 594 (BioLegend, Cat#108452), anti-mouse CD11b BV605 (BioLegend, Cat#101257), and anti-mouse F4/80 BV650 (BioLegend, Cat#123149).
Tumorsphere formation assays
For tumorsphere formation assay, FACS-sorted cells were seeded in ultra-low attachment plates and cultured in serum-free DMEM/F12 (Thermo Fisher Scientific, Cat#11330-032) supplemented with 1% B27 supplement (Thermo Fisher Scientific, Cat#17504044), 1% N2 supplement (Thermo Fisher Scientific, Cat#17502048), penicillin-streptomycin (100 μg/ml; Thermo Fisher Scientific, Cat#15140122), human recombinant epidermal growth factor (EGF; 20 ng/ml; R&D Systems, Cat#236-EG-01M), and human recombinant basic fibroblast growth factor (bFGF; 10 ng/ml; R&D Systems, Cat#233-FB-025/CF), in a humidified 5% CO2 incubator at 37°C.
Organoid culture
HNSCC organoids were cultured as described previously (Driehuis et al., 2019). In brief, SCC cells isolated from primary HNSCC were suspended with BioCoat MATRIGEL MATRIX (BioCoat, Cat#354253) mixed with organoid medium (1:1). The miscible liquid was plated on the 24-well culture plates and concreted in incubator at 37°C for 30 minutes, and then organoids were cultured in the self-configured medium as described before (Driehuis et al., 2019). The medium contained DMEM/F12, 1 × B27 supplement (Thermo Fisher, Cat#12587010), 1.25 mmol/L N-acetyl-l-cysteine (Sigma-Aldrich, Cat#A7250), 10 mmol/L Nicotinamide (Sigma-Aldrich, Cat#N0636), 50 ng/mL human EGF (PeproTech, Cat#AF-100-15), 500 nmol/L A83-01(PeproTech, Cat#9094360), 10 ng/mL human FGF10 (PeproTech, Cat#100-26), 5 ng/mL human FGF2 (Sino Biological, Cat#10014-HNAE), 1 μmol/L Prostaglandin E2 (MCE, Cat#HY-101952), 0.3 μmol/L CHIR 99021 (Sigma-Aldrich, Cat#SML1046), 1 μmol/L Forskolin (Abcam, Cat#ab120058), 50ng/ml R-spondin (R&D Systems, Cat#3266-RS), 25ng/ml Noggin (PeproTech, Cat#120-10C), and 10 μmol/L Rho-associated kinase (ROCK) inhibitor Y-27632 (TargetMol, Cat#T1725).
Limiting-dilution assays in vivo
For limiting-dilution assays in mice, different numbers of FACS-sorted tumor cells isolated from human and murine HNSCC xenografts were mixed with Matrigel and subcutaneously injected into the flank of NOG mice. The tumor growth in mice was monitored every day.
Cell invasion assays
The in vitro cell invasion was measured using the Corning™ BioCoat™ Matrigel™ Invasion Chamber (Corning; Cat#354480) in accordance with the manufacturer’s instructions. In brief, cells were seeded in the upper chambers, and culture medium with 5% FBS was added to the lower chambers. After 24 hours incubation, cells that invaded to the reverse side of inserts were stained with a Hema3 Staining kit (Fisher, #123-869) and quantified with a microscope.
Cytotoxicity assays in vitro
Human peripheral blood mononuclear cells (PBMC) were isolated from leukopheresis by Ficoll–Hypaque density gradient. T cells were isolated from PMBC by using Pan T Cell Isolation Kit (Miltenyi Biotech, Cat#130096535) in accordance with the manufacturer’s guidelines. The 24-well plates were coated with Ultra-LEAF™ Purified anti-human CD3 (10ug/ml, BioLegend, Cat#300331) and Ultra-LEAF™ Purified anti-human CD28 (2ug/ml Bioligand, Cat#302933) in PBS overnight at 4°C. The isolated T cells were plated into 24-well plates to be activated for 72h in RPMI medium supplemented with 10% FBS,1X MEM Non-Essential Amino Acids (ThermoFisher, Cat#11140050), 1mM sodium pyruvate (ThermoFisher, Cat#11360070), 100 U/ml penicillin, 100μg/ml streptomycin and 100 IU/mL human IL2 (Peprotech, Cat#20002). HNSCC cells and activated T cells were co-cultured in the presence of anti-CD276 (10ug/ml, MIH35 clone, Thermo Fisher Scientific, Cat#16593781) or the isotype control antibodies at a 1:10 ratio for 10h at 37°C. The cells were stained with APC-conjugated anti-human-CD3 antibodies (BioLegend, Cat#300412) for 30min on ice and washed with PBS plus 2% FBS. Cells were then fixed, permeabilized and stained with FITC-conjugated anti-human cleaved Caspase-3 antibodies (BD Biosciences, Cat#559341) for 30min on ice and washed with PBS plus 2% FBS. The percentages of cleaved Caspase-3 positive tumor cells were analyzed by flow cytometry.
Cell culture, siRNA or shRNA knockdown, transfection, and western blot
Human HNSCC cell lines were maintained in DMEM containing 10% FBS. For transient transfection of FOSL1 siRNAs, 10nM siRNAs were transfected with Lipofectamine RNAiMAX reagents (Thermo Fisher Scientific, Cat#13778150) following the manufacturer’s instructions. Two FOSL1 siRNA sequences are: 5’-GCUCAUCGCAAGAGUAGCA-3’ and 5’-GAGCUGCAGUGGAUGGUAC-3’. The control siRNA was purchased from Dharmacon (ON-TARGETplus Non-targeting siRNA, D-001810-01). For stable knockdown of CD276, cells were infected with lentiviruses expressing two different pGIPZ-shCD276 lentiviral vectors (Dharmacon, Cat# RHS4531-EG80381) and selected with puromycin (0.5 μg/mL) for 2 weeks. For transfection of CD276 plasmids, 3 μg control (GeneCopoeia, Cat#EX-NEG-Lv105) or CD276 (GeneCopoeia, Cat#EX-Z2350-Lv120, HA-tagged) plasmids were transfected with FuGENE 6 transfection reagent (Promega, Cat#E2691) following the manufacturer’s instructions. The knockdown or overexpression of CD276 was confirmed by Western blot.
The CelLytic buffer (Sigma-Aldrich, Cat#C3228) was utilized to extract proteins from HNSCC cells. Aliquots of protein extracts were separated on SDS-PAGE before being transferred to a PVDF membrane. Membranes were blocked with 5% milk for 1 hr and incubated with primary antibodies overnight, followed by incubation with the peroxidase-coupled secondary antibodies for 1 h at room temperature. Primary antibodies used in this study were: anti-CD276 (Cell Signaling Technology, Cat#14058, 1: 1000), anti-FOSL1 (Cell Signaling Technology, Cat#5281; 1:2000), anti-c-Jun (Cell Signaling Technology, Cat#9165; 1:2000), anti-HA-Tag (Cell Signaling Technology, Cat#3724; 1:1000), antβ-actin (Cell Signaling Technology, Cat#4970; 1: 5000), and anti-α-Tubulin (Sigma, Cat#T5168; 1:5000).
RT-qPCR and ChIP-qPCR
For RT-qPCR, total RNA was extracted using TRIzol reagents (Thermo Fisher Scientific, Cat#15596026) or RNeasy Micro Kit (QIAGEN, Cat# 74004), and 1-2 μg of RNA was used for the reverse transcription reaction with random primer (Thermo Fisher Scientific Cat#48190011), dNTP mix (Thermo Fisher Scientific, Cat#18427013), and M-MuLV Reverse Transcriptase (New England Biolabs, Cat#M0253L). The levels of mRNA were quantified using a SYBRGreen supermix (Bio-Rad, Cat#1708880). The primer sequences for qPCR are listed in Table S1. Relative expression levels of the indicated genes were compared with GAPDH expression using the 2ΔΔct method.
ChIP-qPCR assays were performed as described previously (Li and Wang, 2008; Li et al., 2017a). Cells were crosslinked with 1% formaldehyde in PBS at 37 °C for 10 min, rinsed with PBS and collected with a cell scraper. The cell pellet was resuspended and lysed with ChIP lysis buffer and sonicated to generate 200-500 bp DNA fragments with an ultrasonic sonicator. The chromatin complexes were immunoprecipitated with anti-FOSL1 (Cell Signaling Technology Cat#5281), anti-MED1 (Bethyl, Cat#A300-793A) and anti-BRD4 (Abacm, Cat#ab128874). The precipitated DNA-chromatin products were purified and the DNA levels were quantified by qPCR. The DNA levels are expressed as the percentage of input DNA. The primer sequences used for ChIP-qPCR are listed in Table S1.
RNA-seq
Quality of the RNA for sequencing was assessed using an Agilent 2100 Bioanalyzer. RNA library was prepared at UCLA sequencing core facilities using the KAPA RNA-Seq Library Preparation Kits (KAPA Biosystems, cat#07960140001), and RNAs were single-end sequenced on Illumina HiSeq 3000 machines. Cufflinks with Refseq mRNAs were utilized to determine transcript assembly and differential expression. RNA-seq data was analyzed using the cummeRbund package in R (http://cole-trapnell-lab.github.io/cufflinks/). The heatmap was generated with MeV 4.9 software (http://www.tm4.org/mev.html).
scRNA-seq
To identify the landscape alterations of HNSCC cells following anti-CD276 treatment, primary murine HNSCC tissues were carefully resected and harvested from the tongue of 4NQO-induced mice treated with vehicles or anti-CD276 antibodies for 4 weeks. To avoid inter-individual variability, we harvested pooled primary murine HNSCC samples from 3 mice in two independent cohorts of 4NQO-induced C57BL/6 mice treated with vehicle and anti-CD276 antibodies. Single cell suspensions were achieved as described above and lysed with Red Blood Cell Lysis Buffer (BioLegend, Cat#420301). Cells were then resuspended into single cells for 10x genomics processing. scRNA-seq libraries were prepared using Chromium Next GEM Single Cell 3’ GEM, Library & Gel Bead Kit v3.1(10X Genomics, Cat#PN-1000121) as per manufacturer’s protocol. Sequencing was performed on NovaSeq 6000 System (Illumina).
scRNA-seq raw data was generated from the 10x Genomics platform. Cell Ranger Single Cell Software Suite (version 3.1.0) was utilized to carry out sample de-multiplexing, barcode and UM1 processing, and single-cell 3’ gene counting. A gene expression matrix was constructed in sequential processing as provided in the Seurat toolkit (version 3.1.5) pipeline. Cells with >10% mitochondrial genes mapped and <200 genes were eliminated from downstream analysis. All samples were merged into one Seurat object that was normalized by regressing out total cellular UMI counts and percentage of mitochondrial genes. Principal component analysis (PCA) was used for dimensionality reduction, and UMAP plots were generated via the RunUMAP function.
Initial cells clustering was performed using the Louvain algorithm based on a shared nearest-neighbor network. The final resolution of all subsequent clustering analyses was determined along with the biological questions and the need for details as investigated in the subsequent annotation. To delegate cell types, we compared clusters with those of the Immgen database using SingleR (V1.0.6) package. Differential expression of marker genes was performed using the FindAllMarkers function in Seurat with default parameters. Clusters were further annotated by directly comparing their transcriptional state with the known clusters using Single R Package and top differentially expressed genes with cell type specific expression reported in the literature. We identified cancer cells expressing Epcam, fibroblasts expressing Col1a1, endothelial cells expressing Vwf, lymphoid cells expressing Cd3d, Cd4, Cd8a, Foxp3, and myeloid cells expressing Itgam. To further dissect the cancer subsets, annotations were settled by separating their respective clusters, repeating dimensionality reduction and unsupervised reclustering. Annotations were also performed based on the results of Single R and top differentially expressed genes. During all above-mentioned re-clustering, we observed small clusters of cells co-expressing keratinocyte and endothelial markers which were doublets and thereby removed from further analysis as described previously (Ji et al., 2020).
QUANTIFICIATION AND STATISTICAL ANALYSIS
We performed statistical analyses using Graphpad Prism 6.0 for windows (GraphPad software, Inc.). Statistical parameters of the analyses are reported in the Figure Legends. Limiting-dilutions were calculated using the Extreme Limiting Dilution Analysis software (http://bioinf.wehi.edu.au/software/elda/). GSEA and the statistical analyses were performed with GSEA software (http://www.broad.mit.edu/GSEA) and a two-tailed t-test, respectively. A customized gene set for core ESC-like module was from Wong’s study (Wong et al., 2008). TCGA data analyses are from TIMER (Li et al., 2017b) (https://cistrome.shinyapps.io/timer/) and TIMER2.0 (Li et al., 2020) (http://timer.cistrome.org).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| InVivoMAb rat IgG1 isotype control | BioXcell | Cat#BE0088; RRID: AB_1107775 |
| InVivoMAb anti-mouse CD276 (B7-H3) | BioXcell | Cat#BE0124; RRID: AB_10950149 |
| InVivoPlus anti-mouse CD8α | BioXcell | Cat#BP0061;RRID: AB_1125541 |
| InVivoPlus anti-mouse NK1.1 | BioXcell | Cat#BP0036; RRID: AB_1107737 |
| InVivoPlus rat IgG2b isotype | BioXcell | Cat#BP0090; RRID: AB_1107780 |
| InVivoPlus mouse IgG2a isotype | BioXcell | Cat#BP0085; RRID: AB_1107771 |
| Rabbit polyclonal anti-wide spectrum Cytokeratin | Abcam | Cat#ab9377; RRID: AB_307222 |
| Rabbit polyclonal anti-Caspase-3 | Abcam | Cat#ab13847; RRID: AB_443014 |
| Rabbit monoclonal anti-CD276 | Cell Signaling Technology | Cat#14058; RRID: AB_2750877 |
| Mouse monoclonal anti-BMI1 | Abcam | Cat#ab14389;RRID:AB_2065390 |
| Rabbit polyclonal anti-CD276 FITC | LS Bio | Cat#LS-C419560; RRID:AB_2888639 |
| Rabbit monoclonal anti-CD8α (D8A8Y) | Cell Signaling Technology | Cat#85336; RRID: AB_2800052 |
| Mouse monoclonal anti-CD57 | Abcam | Cat#ab187274; RRID:AB_2888640 |
| Rabbit polyclonal anti-FOSL1 | Abcam | Cat#ab232745, RRID:AB_2888631 |
| Rabbit polyclonal anti-GZMB | Abcam | Cat#ab4059; RRID:AB_304251 |
| Rat monoclonal anti-CD8 | Abcam | Cat#ab22378, RRID:AB_447033 |
| Rabbit polyclonal anti-NKp46 | Abcam | Cat#ab214468; RRID:AB_2814876 |
| Rabbit monoclonal anti-FOSL1 | Cell Signaling Technology | Cat#5281; RRID:AB_10557418: |
| Rabbit monoclonal anti-c-Jun | Cell Signaling Technology | Cat#9165; RRID:AB_2130165 |
| Rabbit monoclonal anti-HA-Tag | Cell Signaling Technology | Cat#3724; RRID:AB_1549585 |
| Rabbit monoclonal anti-β-actin | Cell Signaling Technology | Cat#4970; RRID:AB_2223172 |
| Mouse monoclonal anti-α-Tubulin | Sigma-Aldrich | Cat#T5168; RRID:AB_477579 |
| Rabbit polyclonal anti-MED1 | Bethyl | Cat#A300–793A; RRID:AB_577241 |
| Rabbit monoclonal anti-BRD4 | Abacm | Cat#ab128874; RRID:AB_11145462 |
| Ultra-LEAF™ Purified anti-human CD3 antibody | BioLegend | Cat#300331; RRID:AB_11147368 |
| Ultra-LEAF™ Purified anti-human CD28 antibody | BioLegend | Cat#302933; RRID:AB_11150591 |
| Rat IgG2a kappa Isotype Control (eBR2a), Functional Grade, eBioscience™ | Thermo Fisher Scientific | Cat#16432182, RRID:AB_470156 |
| CD276 (B7-H3) Monoclonal Antibody (MIH35), Functional Grade, eBioscience™ | Thermo Fisher Scientific | Cat#16593781; RRID: AB_1548799 |
| anti-human CD3-APC | BioLegend | Cat#300412; RRID:AB_314066 |
| anti-human cleaved Caspased-3-FITC | BD Biosciences | Cat#559341; RRID:AB_397234 |
| anti-human EpCAM-PE | Miltenyi Biotec | Cat#130110999; RRID:AB_2657495 |
| anti-human CD276-APC | Miltenyi Biotec | Cat#130095522; RRID:AB_10827690 |
| anti-mouse EpCAM-FITC | Miltenyi Biotec | Cat#130102214; RRID:AB_2657515 |
| anti-mouse CD276-APC | BioLegend, | Cat#135608; RRID:AB_2566063 |
| anti-mouse FVS700 APC-700 | BD Biosciences | Cat#564997; RRID:AB_2869637 |
| anti-mouse CD45 Percp-cy5.5 | BioLegend | Cat#103132; RRID:AB_893340 |
| anti-mouse CD4 PE-cy7 | BioLegend | Cat#100422; RRID:AB_312707 |
| anti-mouse CD3 Alexa Flour488 | BioLegend | Cat#100210; RRID:AB_389301 |
| anti-mouse CD8 BV786 | BD Biosciences | Cat#563332; RRID:AB_2721167 |
| anti-mouse NK1.1 BV510 | BioLegend | Cat#108738; RRID:AB_2562217 |
| anti-mouse Gr-1 PE/Dazzle 594 | BioLegend | Cat#108452; RRID:AB_2564249 |
| anti-mouse CD11b BV605 | BioLegend | Cat#101257; RRID:AB_2565431 |
| anti-mouse F4/80 BV650 | BioLegend | Cat#123149; RRID:AB_2564589 |
| Biological Samples | ||
| Human HNSCC embedded tumor samples | UCLA translational pathological core laboratory | http://pathology.ucla.edu/tpcl |
| Human HNSCC primary samples | UCLA translational pathological core laboratory | http://pathology.ucla.edu/tpcl |
| Human HNSCC Paraffin Embedded Tissue Array (OR601c) | US Biomax | https://www.biomax.us/ |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DMEM | Thermo Fisher Scientific | Cat#11995065 |
| DMEM/F-12 | Thermo Fisher Scientific | Cat#11330-032 |
| Fetal Bovine Serum | Thermo Fisher Scientific | Cat#10437028 |
| M-MuLV Reverse Transcriptase | New England Biolabs | Cat#M0253L |
| Random Primers | Thermo Fisher Scientific | Cat#48190011 |
| dNTP | Thermo Fisher Scientific | Cat#18427013 |
| Penicillin-Streptomycin (10,000 U/mL) | Thermo Fisher Scientific | Cat#15140122 |
| Trypsin-EDTA | Thermo Fisher Scientific | Cat#R001100 |
| Lipofectamine RNAiMAX Transfection Reagent | Thermo Fisher Scientific | Cat#13778150 |
| TRIzol Reagent | Thermo Fisher Scientific | Cat#15596026 |
| FuGENE 6 transfection reagent | Promega | Cat#E2691 |
| B-27 Supplement (50X), serum free | Thermo Fisher Scientific | Cat#17504044 |
| B-27 Supplement (50X), minus vitamin A | Thermo Fisher Scientific | Cat#12587010 |
| N-2 Supplement (100X) | Thermo Fisher Scientific | Cat#17502048 |
| Sodium Pyruvate | Thermo Fisher Scientific | Cat#11360070 |
| N-acetyl-l-cysteine | Sigma-Aldrich | Cat#A7250 |
| Nicotinamide | Sigma-Aldrich | Cat#N0636 |
| Paraformaldehyde | Sigma-Aldrich | Cat#P6148 |
| JQ1 | APExBIO | Cat#A1910 |
| iBET-151 | MCE | Cat#HY-13235 |
| Recombinant Human EGF protein | R&D Systems | Cat#236-EG-01M |
| Animal-Free Recombinant Human EGF | PeproTech, | Cat#AF-100-15 |
| A83-01 | PeproTech, | Cat#9094360 |
| FGF10 | PeproTech, | Cat#100-26 |
| Recombinant Human IL-2 | PeproTech | Cat#20002 |
| Recombinant Human FGF2 | Sino Biological | Cat#10014-HNAE |
| Prostaglandin E2 | MCE | Cat#HY-101952 |
| CHIR 99021 | Sigma-Aldrich, | Cat#SML1046 |
| Forskolin | Abcam | Cat#ab120058 |
| Recombinant Human R-spondin Protein | R&D Systems | Cat#3266-RS |
| Recombinant Human Noggin | PeproTech | Cat#120-10C |
| Y-27632 | TargetMol | Cat#T1725 |
| Recombinant Human FGF basic (146 aa) Protein | R&D Systems | Cat#233-FB-025/CF |
| CelLytic buffer | Sigma-Aldrich | Cat#C3228 |
| 4-Nitroquinoline N-oxide (4NQO) | Sigma-Aldrich | Cat#N8141 |
| ProLong™ Diamond Antifade Mountant with DAPI | Thermo Fisher Scientific | Cat#P36962 |
| Critical Commercial Assays | ||
| ALDEFLUOR Kit | STEMCELL Technologies | Cat#01700 |
| AEC+ chromogen | Dako EnVision System | Cat#MP-6401-15 |
| RNeasy Micro Kit | QIAGEN | Cat# 74004 |
| SYBRGreen supermix | Bio-Rad | Cat#1708880 |
| Clarity Western ECL Substrate | Bio-Rad | Cat#1705060 |
| KAPA RNA-Seq Library Preparation Kits | KAPA Biosystems | Cat#07960140001 |
| Human Tumor Cell Dissociation Kit | Miltenyi Biotech | Cat#130095929 |
| Mouse Tumor Cell Dissociation Kit | Miltenyi Biotech | Cat#130096730 |
| Human Pan T Cell Isolation Kit | Miltenyi Biotech | Cat#130096535 |
| Chromium Next GEM Single Cell 3’ GEM, Library & Gel Bead Kit v3.1 | 10X Genomics | Cat#PN-1000121) |
| Deposited Data | ||
| Raw data files for RNA sequencing | NCBI GEO | GEO:GSE132627 |
| Raw data file for scRNA sequencing | NCBI GEO | GEO:GSE164817 |
| Experimental Models: Cell Lines | ||
| SCC1 | University of Michigan | N/A |
| SCC22B | University of Michigan | N/A |
| SCC23 | University of Michigan | N/A |
| SCC1R | This laboratory | N/A |
| HN6 | Wayne State University | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: Bmi1Cre-ER | Jackson Laboratory | JAX:010531 |
| Mouse: R26tdTomato | Jackson Laboratory | JAX:007908 |
| Mouse: NOG | Taconic | NOG-F |
| Oligonucleotides | ||
| See Table S1 | This paper | N/A |
| Recombinant DNA | ||
| GIPZ Non-silencing Lentiviral shRNA Control | Dharmacon | Cat#RHS4346 |
| GIPZ Lentiviral Human CD276 shRNA | Dharmacon | Cat#RHS4531-EG80381 |
| OmicsLink™ ORF lentiviral expression plasmid Control | GeneCopoeia | Cat#EX-NEG-Lv105 |
| OmicsLink™ ORF lentiviral expression plasmid CD276 | GeneCopoeia | Cat#EX-Z2350-Lv120 |
| Software and Algorithms | ||
| Cellsens | Olympus | http://www.olympus-lifescience.com/en/software/cellsens/ |
| CytExpert software | Beckman Coulter | https://www.beckman.com/flow-cytometry/instruments/cytoflex/software |
| Cufflinks | Trapnell Lab | http://cole-trapnell-lab.github.io/cufflinks/ |
| ELDA: Extreme Limiting Dilution Analysis | The Walter and Eliza Hall | http://bioinf.wehi.edu.au/software/elda/ |
| MeV | N/A | http://www.tm4.org/mev.html |
| GSEA | Broad Institute | http://www.broad.mit.edu/GSEA |
| TIMER | Li et al., 2017b | https://cistrome.shinyapps.io/timer/ |
| CellRanger (version 3.1.0) | Cell Ranger Single Cell Software Suite |
https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger |
| Seurat (version 3.1.5) | Seurat R package | https://satijalab.org/seurat/ |
| SingleR (V1.0.6) | SingleR R package | https://github.com/dviraran/SingleR |
| GraphPad Prism 6 software | GraphPad Software, Inc. | https://www.graphpad.com/scientific-software/prism/ |
HIGHLIGHTS:
CSCs use CD276 to evade immune surveillance in SCC initiation and metastasis
CD276 is a functional cell surface marker to isolate CSCs from SCC
CD276 blockade remodels SCC heterogeneity with decrease of pEMT.
CD276 blockade kills CSCs in a CD8+ T cell-dependent manner.
ACKNOWLEDGEMENTS
We thank Mari Ekimyan Salvo for reading the manuscript. This work was supported by NIH/NIDCR grant R01DE029173.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adeegbe DO, Liu S, Hattersley MM, Bowden M, Zhou CW, Li S, Vlahos R, Grondine M, Dolgalev I, Ivanova EV, et al. (2018). BET bromodomain inhibition cooperates with PD-1 blockade to facilitate antitumor response in Kras-Mutant non-small cell lung cancer. Cancer Immunol. Res. 6, 1234–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, Li J, Liu D, Hong S, Qiao Q, Sun Q, Li P, Lyu N, Sun T, Xie S, et al. (2020). Tumor-expressed B7-H3 mediates the inhibition of antitumor T-cell functions in ovarian cancer insensitive to PD-1 blockade therapy. Cell Mol. Immunol. 17, 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, and Wang CY (2019). Targeting cancer stem cells in squamous cell carcinoma. Precis. Clin. Med 2, 152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Wu M, Li Y, Chang I, Yuan Q, Ekimyan-Salvo M, Deng P, Yu B, Yu Y, Dong J, et al. (2017). Targeting BMI1(+) cancer stem cells overcomes chemoresistance and inhibits metastases in squamous cell carcinoma. Cell Stem Cell 20, 621–634.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cillo AR, Kurten CHL, Tabib T, Qi Z, Onkar S, Wang T, Liu A, Duvvuri U, Kim S, Soose RJ, et al. (2020). Immune landscape of viral- and carcinogen-driven head and neck cancer. Immunity 52, 183–199. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Pan H, Li J, Zhong Q, Chen X, Dry SM, and Wang CY (2013). Epigenetic activation of AP1 promotes squamous cell carcinoma metastasis. Sci. Signal. 6, ra28.1–ra28.13, S0–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driehuis E, Kolders S, Spelier S, Lohmussaar K, Willems SM, Devriese LA, de Bree R, de Ruiter EJ, Korving J, Begthel H, et al. (2019). Oral mucosal organoids as a potential platform for personalized cancer therapy. Cancer Discov. 9, 852–871. [DOI] [PubMed] [Google Scholar]
- Du H, Hirabayashi K, Ahn S, Kren NP, Montgomery SA, Wang X, Tiruthani K, Mirlekar B, Michaud D, Greene K, et al. (2019). Antitumor responses in the absence of toxicity in solid tumors by targeting B7-H3 via chimeric antigen receptor T cells. Cancer Cell 35, 221–237.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris RL, Blumenschein G Jr., Fayette J, Guigay J, Colevas AD, Licitra L, Harrington K, Kasper S, Vokes EE, Even C, et al. (2016). Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N. Engl. J. Med 375, 1856–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubin MM, Esaulova E, Ward JP, Malkova ON, Runci D, Wong P, Noguchi T, Arthur CD, Meng W, Alspach E, et al. (2018). High-dimensional analysis delineates myeloid and lymphoid compartment remodeling during successful immune-checkpoint cancer therapy. Cell 175, 1014–1030.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington KJ, Ferris RL, Blumenschein G Jr., Colevas AD, Fayette J, Licitra L, Kasper S, Even C, Vokes EE, Worden F, et al. (2017). Nivolumab versus standard, single-agent therapy of investigator’s choice in recurrent or metastatic squamous cell carcinoma of the head and neck (CheckMate 141): health-related quality-of-life results from a randomised, phase 3 trial. Lancet Oncol. 18, 1104–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedberg ML, Goh G, Chiosea SI, Bauman JE, Freilino ML, Zeng Y, Wang L, Diergaarde BB, Gooding WE, Lui VW, et al. (2016). Genetic landscape of metastatic and recurrent head and neck squamous cell carcinoma. J. Clin. Invest 126, 169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, and Young RA (2013). Super-enhancers in the control of cell identity and disease. Cell 155, 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JM, Xia W, Hsu YH, Chan LC, Yu WH, Cha JH, Chen CT, Liao HW, Kuo CW, Khoo KH, et al. (2018). STT3-dependent PD-L1 accumulation on cancer stem cells promotes immune evasion. Nat. commun 9, 1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias-Bartolome R and Gutkind JS (2020). Unleashing immunotherapy by targeting cancer stem cells. Cell Stem Cell 27, 187–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji AL, Rubin AJ, Thrane K, Jiang S, Reynolds DL, Meyers RM, Guo MG, George BM, Mollbrink A, Bergenstrahle J, et al. (2020). Multimodal analysis of composition and spatial architecture in Human Squamous Cell Carcinoma. Cell 182, 497–514.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Zhang W, and Wang CY (2020). BMI1 inhibition eliminates residual cancer stem cells after PD1 blockade and activates antitumor immunity to prevent metastasis and relapse. Cell Stem Cell 27, 238–253.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Li R, Chen MY, Yu C, Tang LQ, Liu YM, Li JP, Liu YN, Luo YL, Zhao Y, et al. (2020). Single-cell transcriptomic analysis defines the interplay between tumor cells, viral infection, and the microenvironment in nasopharyngeal carcinoma. Cell Res. 30, 950–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Shin JH, Longmire M, Wang H, Kohrt HE, Chang HY, and Sunwoo JB (2016). CD44+ cells in head and neck squamous cell carcinoma suppress T-cell-mediated immunity by selective constitutive and inducible expression of PD-L1. Clin. Cancer Res. 22, 3571–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Martin-Orozco N, Zheng P, Li J, Zhang P, Tan H, Park HJ, Jeong M, Chang SH, Kim BS, et al. (2017). Inhibition of the B7-H3 immune checkpoint limits tumor growth by enhancing cytotoxic lymphocyte function. Cell Res. 27, 1034–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner J, Klauser C, Pickl WF, Stockl J, Majdic O, Bardet AF, Kreil DP, Dong C, Yamazaki T, Zlabinger G, et al. (2009). B7-H3 is a potent inhibitor of human T-cell activation: No evidence for B7-H3 and TREML2 interaction. Eur. J. Immunol 39, 1754–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke D, Pfenning PN, Sahm F, Klein AC, Kempf T, Warnken U, Schnolzer M, Tudoran R, Weller M, Platten M, et al. (2012). Costimulatory protein 4IgB7H3 drives the malignant phenotype of glioblastoma by mediating immune escape and invasiveness. Clin. Cancer Res 18, 105–117. [DOI] [PubMed] [Google Scholar]
- Li J, and Wang CY (2008). TBL1-TBLR1 and beta-catenin recruit each other to Wnt target-gene promoter for transcription activation and oncogenesis. Nat. Cell Biol 10, 160–169. [DOI] [PubMed] [Google Scholar]
- Li J, Yu B, Deng P, Cheng Y, Yu Y, Kevork K, Ramadoss S, Ding X, Li X, and Wang CY (2017a). KDM3 epigenetically controls tumorigenic potentials of human colorectal cancer stem cells through Wnt/beta-catenin signalling. Nat. Commun 8, 15146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, and Liu XS (2017b). TIMER: A web server for Ccomprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 77, e108–e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, and Liu XS (2020). TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 48, W509–W514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majzner RG, Theruvath JL, Nellan A, Heitzeneder S, Cui Y, Mount CW, Rietberg SP, Linde MH, Xu P, Rota C, et al. (2019). CAR T Cells Targeting B7-H3, a pan-cancer antigen, demonstrate potent preclinical activity against pediatric solid tumors and brain tumors. Clin. Cancer Res 25, 2560–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malta TM, Sokolov A, Gentles AJ, Burzykowski T, Poisson L, Weinstein JN, Kaminska B, Huelsken J, Omberg L, Gevaert O, et al. (2018). Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell 173, 338–354.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. (2008). The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L, Fan TF, Wu L, Yu GT, Deng WW, Chen L, Bu LL, Ma SR, Liu B, Bian Y, et al. (2017). Selective blockade of B7-H3 enhances antitumour immune activity by reducing immature myeloid cells in head and neck squamous cell carcinoma. J. Cell Mol. Med. 21, 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Yang H, Levorse J, Yuan S, Polak L, Sribour M, Singh B, Rosenblum MD, and Fuchs E (2019). Adaptive immune resistance emerges from tumor-initiating stem cells. Cell 177, 1172–1186.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirker R, Pereira JR, von Pawel J, Krzakowski M, Ramlau R, Park K, de Marinis F, Eberhardt WE, Paz-Ares L, Storkel S, et al. (2012). EGFR expression as a predictor of survival for first-line chemotherapy plus cetuximab in patients with advanced non-small-cell lung cancer: analysis of data from the phase 3 FLEX study. Lancet Oncol. 13, 33–42. [DOI] [PubMed] [Google Scholar]
- Prager BC, Xie Q, Bao S, and Rich JN (2019). Cancer stem cells: the architects of the tumor ecosystem. Cell Stem Cell 24, 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad DV, Nguyen T, Li Z, Yang Y, Duong J, Wang Y, and Dong C (2004). Murine B7-H3 is a negative regulator of T cells. J. Immunol. 173, 2500–2506. [DOI] [PubMed] [Google Scholar]
- Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, Rodman C, Luo CL, Mroz EA, Emerick KS, et al. (2017). Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 171, 1611–1624.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco AG, and Cohen EE (2015). Current treatment options for recurrent or metastatic head and neck squamous cell carcinoma. J. Clin. Oncol. 33, 3305–3313. [DOI] [PubMed] [Google Scholar]
- Sanmamed MF, and Chen L (2018). A paradigm shift in cancer immunotherapy: from enhancement to normalization. Cell 175, 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saygin C, Matei D, Majeti R, Reizes O, and Lathia JD (2019). Targeting cancer stemness in the clinic: from hype to hope. Cell Stem Cell 24, 25–40. [DOI] [PubMed] [Google Scholar]
- Seaman S, Zhu Z, Saha S, Zhang XM, Yang MY, Hilton MB, Morris K, Szot C, Morris H, Swing DA, et al. (2017). Eradication of tumors through simultaneous ablation of CD276/B7-H3-positive tumor cells and tumor vasculature. Cancer Cell 31, 501–515.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, and George RE (2017). Super-enhancer-driven transcriptional dependencies in cancer. Trends Cancer 3, 269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Hu-Lieskovan S, Wargo JA, and Ribas A (2017). Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168, 707–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh WK, Gajewska BU, Okada H, Gronski MA, Bertram EM, Dawicki W, Duncan GS, Bukczynski J, Plyte S, Elia A, et al. (2003). The B7 family member B7-H3 preferentially down-regulates T helper type 1-mediated immune responses. Nat. Immunol 4, 899–906. [DOI] [PubMed] [Google Scholar]
- Tamai K, Nakamura M, Mizuma M, Mochizuki M, Yokoyama M, Endo H, Yamaguchi K, Nakagawa T, Shiina M, Unno M, et al. (2014). Suppressive expression of CD274 increases tumorigenesis and cancer stem cell phenotypes in cholangiocarcinoma. Cancer Sci. 105, 667–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaharautio A, and Taipale J (2014). Cancer. Cancer by super-enhancer. Science 346, 1291–1292. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ow TJ, and Myers JN (2012). Pathways for cervical metastasis in malignant neoplasms of the head and neck region. Clin. Anat 25, 54–71. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang Z, Zhang C, Liu X, Li G, Liu S, Sun L, Liang J, Hu H, Liu Y, et al. (2018). Genetic and clinical characterization of B7-H3 (CD276) expression and epigenetic regulation in diffuse brain glioma. Cancer Sci. 109, 2697–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei SC, Levine JH, Cogdill AP, Zhao Y, Anang NAS, Andrews MC, Sharma P, Wang J, Wargo JA, Pe’er D, et al. (2017). Distinct cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 checkpoint blockade. Cell 170, 1120–1133.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, and Chang HY (2008). Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell 2, 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Chen M, Wu P, Chen C, Xu ZP, and Gu W (2017). Increased PD-L1 expression in breast and colon cancer stem cells. Clin. Exp. Pharmacol. Physiol. 44, 602–604. [DOI] [PubMed] [Google Scholar]
- Xu H, Cheung IY, Guo HF, and Cheung NK (2009). MicroRNA miR-29 modulates expression of immunoinhibitory molecule B7-H3: potential implications for immune based therapy of human solid tumors. Cancer Res. 69, 6275–6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y (2015). Cancer immunotherapy: harnessing the immune system to battle cancer. J. Clin. Invest 125, 3335–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonesaka K, Haratani K, Takamura S, Sakai H, Kato R, Takegawa N, Takahama T, Tanaka K, Hayashi H, Takeda M, et al. (2018). B7-H3 negatively modulates CTL-mediated cancer immunity. Clin. Cancer Res 24, 2653–2664. [DOI] [PubMed] [Google Scholar]
- Zhang H, Christensen CL, Dries R, Oser MG, Deng J, Diskin B, Li F, Pan Y, Zhang X, Yin Y, et al. (2020). CDK7 inhibition potentiates genome instability triggering anti-tumor immunity in small cell lung cancer. Cancer Cell 37, 37–54.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and scRNA-seq data were deposited at the Gene Expression Omnibus (GEO) under the accession number GEO: GSE132627 and GSE164817.


