Abstract
A disease similar to ulcerative colitis in humans has been identified in cotton-top tamarins (CTTs) in captivity. The clinical signs include weight loss, diarrhea, and rectal bleeding with the pathological features and biochemical abnormalities of ulcerative colitis. Approximately 25 to 40% of these animals develop colon cancer after 2 to 5 years of captivity. An infectious etiology has been proposed; however, no microbial agent to date has been identified. Helicobacter spp. have been associated with enterocolitis and inflammatory bowel disease (IBD) in humans and animals. Infection with Helicobacter pylori or Helicobacter mustelae is associated with an increased risk of gastric adenocarcinoma and lymphoma of the mucosa-associated lymphoid tissue. Helicobacter hepaticus causes hepatitis, hepatic adenomas, and hepatocellular carcinomas in susceptible strains of mice. The aim of this study was to assess a colony of CTTs with a high incidence of IBD and colon cancer for the presence of colonic Helicobacter spp. A fusiform, gram-negative bacterium with bipolar flagella and periplasmic fibers was isolated from the feces of CTTs. The bacterium grew under microaerobic conditions at 37 and 42°C but not at 25°C, did not hydrolyze urea, was positive for catalase and oxidase, did not reduce nitrate to nitrite, did not hydrolyze indoxyl acetate or alkaline phosphatase, and was resistant to nalidixic acid, cephalothin, and trimethoprim-sulfamethoxazole. On the basis of 16S rRNA gene sequence analysis, the organism was classified as a novel Helicobacter species. This is the first Helicobacter isolated from CTTs. Further studies are needed to elucidate the role of this novel Helicobacter sp. in the pathogenesis of ulcerative colitis and colonic adenocarcinoma in CTTs.
Cotton-top tamarins (CTTs; Saguinus oedipus) are New World primates native to the rain forests of Colombia. In the 1960s they were imported for use in biomedical research to study Herpes samirii, Herpes ateles, and Epstein-Barr virus (26, 27). The CTT was placed on the endangered species list in 1977 due to destruction of its native habitat and capture for the pet trade and biomedical research. With the subsequent creation and stabilization of CTT breeding colonies, chronic ulcerative colitis (UC) and colonic adenocarcinoma were recognized as major health problems (4). Approximately 50% of colony-maintained animals develop active colitis, with the disease in 25 to 40% of those with active colitis progressing to colonic adenocarcinoma (21, 23, 41). Although extensive studies with animals in the wild have not been done, it appears that animals in their native habitat are free of the disease (41).
The clinical features and pathology of colitis in CTTs closely resemble those of ulcerative colitis in humans, and CTTs have been used as an animal model of the disease (4, 6). Clinical signs include chronic wasting, bloody diarrhea, and weight loss. In both humans and CTTs, UC is a spontaneous disease that affects both sexes equally and is responsive to sulfasalazine and steroid therapy. It also appears that UC in either species predisposes the individual to colonic adenocarcinoma (7). The disease waxes and wanes over time and fluctuates between normal, active, and chronic colitis before progressing to adenocarcinoma (5).
The histopathological lesions of acute colitis in CTTs include hyperplasia of the colonic epithelium with decreased numbers of goblet cells, crypt abscesses, and an inflammatory cell infiltrate in the lamina propria. In contrast to human UC, which involves the rectal area and progresses to proximal portions of the colon, the colonic mucosa of the CTT is usually diffusely involved. Chronic mucosal changes include loss of crypts, atrophy of the mucosa, and infiltration of the lamina propria by chronic inflammatory cells (5). Colonic adenocarcinoma in CTTs appears to arise spontaneously in association with chronic colitis. Unlike most human colonic cancers, the tumor is multicentric and is seldom preceded by dysplastic changes in the mucosa. Signet ring cells which contain mucin are often present.
The etiologies of UC and colonic adenocarcinoma are unknown, but they are probably multifactorial. In the CTT there is strong evidence to support both a species-related susceptibility and an infectious cause (1, 2). These infectious agents previously incriminated as a cause of but not proven to cause ulcerative colitis in CTTs are coronaviruses and Campylobacter spp. (1, 2). A recent study at the New England Regional Primate Research Center supports the suggestion that environmental factors, including an infectious agent, may be responsible for the disease. In this study CTTs were reared under identical conditions either in an isolation unit or in the conventional colony (21). Animals living in the conventional colony were statistically more likely to develop colitis (21). Disorders of the immune system and environmental stresses have also been proposed as possible etiologies (36, 39, 41).
The genus Helicobacter has expanded rapidly in recent years to include organisms that inhabit the gastric mucosa of humans and numerous species of animals (15). The type species, Helicobacter pylori, causes chronic gastritis and peptic ulcer disease in humans and has been linked to the development of gastric mucosa-associated lymphoma and gastric adenocarcinoma (20, 28, 42). Other species of Helicobacter, including Helicobacter felis and Helicobacter mustelae, also cause gastritis in their animal hosts (12, 25). In addition, H. mustelae has been associated with gastric adenocarcinoma and mucosa-associated lymphoid tissue lymphoma in ferrets (8, 13).
Numerous Helicobacter spp. have also been isolated from the intestinal tracts of humans, animals, and birds (14, 18, 32, 35, 37). Of particular interest are Helicobacter bilis and Helicobacter hepaticus because of their association with hepatitis and inflammatory bowel disease in several strains of mice (3, 17, 19, 33, 38). Male A/JCr and B6C3F1 mice infected with H. hepaticus also develop hepatic adenocarcinoma and are an important model for bacterially induced carcinogenesis (16, 17).
Because of the association between several Helicobacter spp. and inflammatory bowel disease and because infection with certain Helicobacter spp. predisposes the host to development of cancer, we examined CTTs for the presence of Helicobacter spp.
MATERIALS AND METHODS
Animals.
Thirty-four CTTs from an established colony of ∼200 animals with endemic colitis were surveyed. The animals ranged from 2 to 19 years of age. They were maintained in accordance with the guidelines of the Committee on Animals of the Harvard Medical School and the Guide for the Care and Use of Laboratory Animals (26a).
Bacterial culture.
Fecal samples from CTTs were homogenized with phosphate-buffered saline. A portion of the mixture was passed through 0.8-μm-pore-size filters onto CVA medium (Remel Laboratories, Lenexa, Kans.), which contains cefoperazone, vancomycin, and amphotericin B, and a Helicobacter-selective medium containing nalidixic acid, polymyxin B, amphotericin B, bacitracin, and vancomycin. The remaining slurry was streaked without filtration onto the CVA medium and the Helicobacter-selective medium. The cultures were incubated at 37 and 42°C under microaerobic conditions for 14 days in vented jars containing N2, H2, and CO2 (90:5:5). Pure cultures of Helicobacter spp. were subsequently passaged onto sheep blood agar plates for further characterization (Remel Laboratories).
Biopsy sample collection.
Colonic biopsy samples were obtained with a 3-mm biopsy forceps, and the samples were then placed in brucella broth (Difco Laboratories, Detroit, Mich.) with 10% glycerol and were frozen at −20°C before DNA extraction. The biopsy instrument was sanitized between animals by submersion and agitation in 10% glutaraldehyde (Wavicide; Wave Energy Systems, Wayne, N.J.) for 10 min followed by rinsing with sterile water (9).
Electron microscopy.
Isolate MIT 97-6194-5 was examined by electron microscopy. Cells grown on blood agar plates were centrifuged and gently suspended in 10 mM Tris-HCl buffer (pH 7.4) at a concentration of about 108 cells per ml. Samples were negatively stained with 1% (wt/vol) phosphotungstic acid (pH 6.5) for 20 to 30 s. The specimens were examined with a JEOL model JEM-1200EX transmission electron microscope operating at 100 kV.
Biochemical and phenotypical characterization.
Eight isolates were subjected to a detailed biochemical characterization as previously described by Shen et al. (32). The isolates were examined for catalase, oxidase, and urease activities. With the RapID NH System (Innovative Diagnostic Systems. Inc., Norcross, Ga.), the isolates were examined for the presence of alkaline phosphatase hydrolysis, indoxyl acetate hydrolysis, and gamma-glutamyl transpeptidase and for the hydrolysis of urea. The isolates were also tested for their ability to reduce nitrate by using nitrate broth (GIBCO Laboratories, Grand Island, N.Y.) and diagnostic reagents as described previously (14). Growth at 25, 37, and 42°C under aerobic, microaerobic, and anaerobic conditions was examined at 3- to 4-day intervals for up to 2 weeks. The organisms were also grown in the presence of 1% glycine. Susceptibility to cephalothin (30 μg/disc), nalidixic acid (30 μg/disc), and trimethoprim-sulfamethoxazole (23.75 μg/disc) was determined by culturing the organisms in the presence of discs impregnated with the antibiotic (Difco Laboratories). The bacteria were also Gram stained and examined for motility in sterile phosphate-buffered saline by phase-contrast microscopy.
DNA extraction for PCR analysis.
DNA was extracted from the biopsy samples and the cultured organisms with the High Pure PCR Template Preparation Kit (Boehringer Mannheim Biochemicals, Indianapolis, Ind.) according to the manufacturer’s directions. Briefly, the samples were lysed and incubated with 40 μl of proteinase K for 1 h at 55°C. A total of 200 μl of binding buffer was added to each sample, and the mixture was allowed to incubate for 10 min at 72°C before the addition of 100 μl of isopropanol. The samples were placed in a filter tube and centrifuged at 5,500 × g for 1 min. The flowthrough was discarded, 500 μl of wash buffer was added to the samples, and the mixture was centrifuged as described above. This washing step was repeated three times. Elution of the DNA was achieved by adding 200 μl of elution buffer to the filter tube and centrifuging the sample for 1 min at 8,000 rpm.
PCR amplification of bacterial DNA.
Two sets of primer sequences chosen for PCR amplification recognize a region of the 16S rRNA gene specific for members of the Helicobacter genus. One set of primers produces an amplified product of 422 bp, while the other produces an amplified product of 1.2 kb. PCR amplification was achieved by a previously described method (18). Briefly, 20 μl of the DNA preparation was added to 100 μl of a reaction mixture containing 1× Taq polymerase buffer (supplied by the manufacturer but supplemented with 1 M MgCl2 to a final concentration of 2.25 mM), 0.5 μM each primer, 200 μM each deoxynucleotide, and 200 μg of bovine serum albumin per ml. The samples were heated at 94°C for 4 min, briefly centrifuged, and cooled to 61°C. At this time, 2.5 U of Taq polymerase (Pharmacia, Uppsala, Sweden) and 1.0 U of polymerase enhancer (Perfect Match; Stratagene, La Jolla, Calif.) were added, and then 100 μl of mineral oil was laid over the samples. The following conditions were used for amplification of the 422-bp fragment: 35 cycles of denaturation at 94°C for 1 min, annealing at 61°C for 2 min, and elongation at 72°C for 2 min, followed by an elongation step of 7 min at 72°C. For amplification of the 1.2-kb fragment the following conditions were used: 35 cycles of denaturation at 94°C for 1 min, annealing at 58°C for 3 min, and elongation at 72°C for 3 min, followed by an elongation step of 8 min at 72°C. Fifteen microliters of the sample was then electrophoresed through a 1% agarose gel, followed by ethidium bromide staining and viewing by UV illumination.
Purification of PCR products for 16S sequencing.
A 1.2-kb piece of amplified DNA from each of two biopsy samples was purified by precipitation with polyethylene glycol 8000 (24). After removal of Ampliwax, 0.6 volume of 20% polyethylene glycol 8000 (Sigma Chemical Co., St. Louis, Mo.) in 2.5 M NaCl was added, and the mixture was incubated at 37°C for 10 min. The sample was centrifuged at 15,000 × g for 15 min, and the pellet was washed with 80% ethanol and pelleted as before described above. The pellet was air dried, dissolved in 30 μl of distilled water, and used for cycle sequencing as described below.
Genomic DNA extraction for 16S rRNA gene sequencing.
Bacteria isolated from the feces of three CTTs were cultured on blood agar plates, and the cells were harvested and washed twice with 1 ml of double-distilled H2O. The pellets were suspended in STET buffer (8% sucrose, 50 mM EDTA, 0.1% Triton X-100, 50 mM Tris-HCl [pH 8.0]), and lysozyme (hen egg white; Boehringer Mannheim Biochemicals) was added to a final concentration of 3 mg/ml. The suspension was incubated for 12 min at 37°C and was then lysed with 1% sodium dodecyl sulfate. RNase A (bovine pancreas; Boehringer Mannheim Biochemicals) was added to a final concentration of 0.05 mg/ml, and the solution was incubated for 1 h at 37°C. Then 0.1 volume of a 5% cetyltrimethylammonium bromide–0.5 M NaCl solution (Sigma Chemical Co.) was added, and the solution was gently mixed and incubated at 65°C for 10 min. The DNA was extracted with an equal volume of phenol-chloroform (1:1; vol/vol), precipitated overnight in 0.3 M sodium acetate with 2 volumes of absolute ethanol at −20°C, and pelleted by centrifugation at 13,000 × g for 1 h at 4°C. The ethanol was decanted, and the pellet was air dried and suspended in sterile distilled water.
16S rRNA gene sequencing.
The sequences of the 16S rRNA genes of three isolates from bacterial culture (MIT 97-6194-3, MIT 97-6194-4, and MIT 97-6194-5) and two PCR products from CTT colonic biopsy samples from the same animals from which isolates MIT 97-6194-3 and MIT 97-6194-4 were retrieved were determined. For amplification of 16S rRNA citrons, 16S rRNA gene sequencing, and 16S rRNA data analysis, we used the methods described by Fox et al. (18). Briefly, primers C70 and B37 (18) were used to amplify the 16S rRNA genes. The amplicons were purified and directly sequenced by using a TAQuence cycle sequencing kit (U.S. Biochemicals, Cleveland, Ohio). The 16S rRNA gene sequences were entered into RNA, a program for analysis of 16S rRNA data in Microsoft Quickbasic for use with International Business Machines personal computer-compatible computers, and were aligned as described previously (29). The database used contains approximately 100 Helicobacter, Wolinella, Arcobacter, and Campylobacter sequences and more than 900 sequences for other bacteria. Similarity matrices were constructed from the aligned sequences by using only those base positions for which data were available for 90% of the strains and were corrected for multiple base changes by the method of Jukes and Cantor (22). Phylogenetic trees were constructed by the neighbor-joining method (30).
RFLP analysis.
DNA fragments of 1.2 kb from five of the bacterial isolates were subjected to restriction fragment length polymorphism (RFLP) analysis. DNA digestion was accomplished by the addition of 10 U of the restriction endonuclease AluI (New England Biolabs, Beverly, Mass.) and 1 μl of restriction buffer (New England Biolabs) to 16 μl of DNA and incubation at 37°C for 2 h. The samples were then electrophoresed through a 3% agarose gel followed by ethidium bromide staining and viewing by UV illumination.
Histopathology.
Colonic biopsy samples were fixed in neutral buffered 10% formalin, processed by standard methods, and embedded in paraffin. Sections of 5 μm were stained with hematoxylin and eosin and Warthin-Starry silver stains. These sections were examined by light microscopy for evidence of lesions and for the presence of a bacterium with a morphology consistent with those of members of the genus Helicobacter.
Nucleotide sequence accession numbers.
The 16S rRNA sequence for strain MIT 97-6194-5 has been deposited in GenBank under accession no. AF107494.
RESULTS
Isolation, growth, and biochemical and physical characteristics.
After 3 to 5 days of incubation, a thin, spreading film developed on the agar surfaces. The bacteria were gram negative and, under a phase-contrast microscope, appeared fusiform and motile. The biochemical and physical characteristics of eight isolates from CTTs were compared to those of previously described Helicobacter spp. (Table 1). The bacteria grew under microaerobic conditions at 37 and 42°C but not at 25°C. All isolates were oxidase and catalase positive but urease negative. The isolates did not reduce nitrate or hydrolyze alkaline phosphatase or indoxyl acetate, and they did not have gamma-glutamyl transpeptidase activity. They were also resistant to nalidixic acid, cephalothin, and trimethoprim-sulfamethoxazole.
TABLE 1.
Characteristics which differentiate Helicobacter strains from CTTs from other Helicobacter speciesa
| Taxon | Catalase production | Nitrate reduction | Alkaline phosphatase hydrolysis | Urease activity | Indoxyl acetate hydrolysis | Gamma-glutamyl transpeptidase activity | Growth at 42°C | Growth with 1% glycine | Susceptibility
|
Periplasmic fibers | No. of flagella | Distribution of flagella | G+C content (mol%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nalidixic acid (30-μg disc) | Cephalothin (30-μg disc) | |||||||||||||
| Helicobacter from CTTs | + (8/8)b | − (0/8) | − (0/8) | − (0/8) | − (0/8) | − (0/8) | + (8/8) | + (8/8) | R (8/8) | R (8/8) | + | 6–12 | Bipolar | ND |
| Helicobacter rodentium | + | + | − | − | − | − | + | + | R | R | − | 2 | Bipolar | ND |
| Helicobacter pullorum | + | + | − | − | − | ND | + | ND | R | S | − | 1 | Monopolar | 34–35 |
| Helicobacter sp. strain CLO-3 | + | − | + | − | + | − | + | + | I | R | − | 45 | ||
| Helicobacter pylori | + | − | + | + | − | + | − | − | R | S | − | 4–8 | Bipolar | 35–37 |
| Helicobacter nemestrinae | + | − | + | + | − | ND | + | − | R | S | − | 4–8 | Bipolar | 24 |
| Helicobacter acinonyx | + | − | + | + | − | + | − | − | R | S | − | 2–5 | Bipolar | 30 |
| Helicobacter felis | + | + | + | + | − | + | + | − | R | S | + | 14–20 | Bipolar | 42 |
| Helicobacter fennelliae | + | − | + | − | + | − | − | + | S | S | − | 2 | Bipolar | 35 |
| Helicobacter trogontum | + | + | − | + | ND | + | + | ND | R | R | + | 5–7 | Bipolar | ND |
| Helicobacter muridarum | + | − | + | + | + | + | − | − | R | R | + | 10–14 | Bipolar | 34 |
| Helicobacter hepaticus | + | + | ND | + | + | ND | − | + | R | R | − | 2 | Bipolar | ND |
| Helicobacter canis | − | − | + | − | + | ND | + | ND | S | I | − | 2 | Bipolar | 48 |
| Helicobacter bilis | + | + | ND | + | − | ND | + | + | R | R | + | 3–14 | Bipolar | ND |
| “Flexispira rappini” | + | − | − | + | ND | + | + | − | R | R | + | 10–20 | Bipolar | 34 |
| Helicobacter cinaedi | + | + | − | − | − | − | − | + | S | I | − | 1–2 | Bipolar | 37–38 |
| Helicobacter pametensis | + | + | + | − | − | − | + | + | S | S | − | 2 | Bipolar | 38 |
| Helicobacter sp. strain Bird-C | + | + | + | + | + | − | + | + | S | R | − | 2 | Bipolar | 30 |
| Helicobacter sp. strain Bird-B | + | + | + | + | − | + | + | + | S | R | − | 2 | Bipolar | 31 |
| Helicobacter mustelae | + | + | + | + | + | + | + | − | S | R | − | 4–8 | Peritrichous | 36 |
Data were obtained from reference 32 and this study. Symbols and abbreviations: +, positive reaction; −, negative reaction; S, susceptible; R, resistant; I, intermediate; ND, not determined.
Numbers in parentheses are number of strains with the indicated result/number of strains tested.
Ultrastructure.
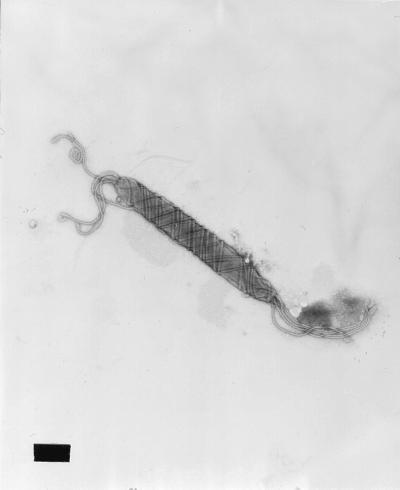
Cells had a fusiform appearance and measured approximately 0.5 by 4 to 5 μm (Fig. 1). They possessed periplasmic fibers and 6 to 12 bipolar, sheathed flagella.
FIG. 1.
Transmission electron micrograph of the novel Helicobacter sp. The typical bacterium is fusiform to slightly spiral and possesses periplasmic fibers and several sheathed flagella at each end. Bar, 0.5 μm.
PCR identification of strains.
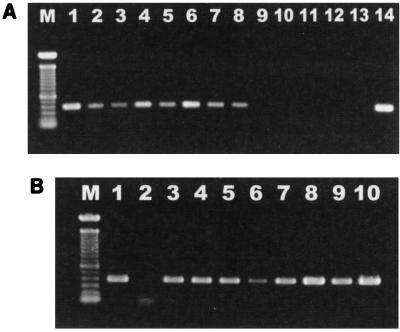
DNAs from colonic biopsy samples and pure fecal cultures were amplified with a Helicobacter genus-specific primer set. A 422-base fragment was amplified from 18 of 34 biopsy samples (Fig. 2A) and eight of eight of the fecal cultures analyzed (Fig. 2B).
FIG. 2.
(A) Electrophoresis of DNA isolated from colonic biopsy samples, amplified by PCR with Helicobacter genus-specific primers, and run on a 1% agarose gel. Lane M, 100-bp DNA ladder; lane 1, MIT 97-6194-6; lane 2, MIT R97-6194-7; lane 3, MIT 97-6194-5; lane 4, MIT 97-6194-4; lane 5, MIT 97-6194-3; lane 6, MIT R97-6837; lane 7, MIT R97-6834; lane 8, MIT 97-6196-8; lane 9, MIT R97-6841; lane 10, MIT R97-6835; lane 11, MIT R97-6832; lane 12, MIT R97-6836; lane 13, blank; lane 14, positive control. (B) Electrophoresis of DNA isolated from fecal cultures, amplified by PCR with Helicobacter genus-specific primers, and run on a 1% agarose gel. The faint band in lane 2 at approximately 100 bases represents the PCR primers. Lane M, 100-bp DNA ladder; lane 1, positive control; lane 2, blank; lane 3, MIT 97-6194-4; lane 4, MIT 97-6194-3; lane 5, MIT 97-6194-5; lane 6, MIT R97-6834; lane 7, MIT R97-6841; lane 8, MIT R97-6837; lane 9, MIT R97-6840; lane 10, MIT R97-6842.
Phylogenetic analysis.
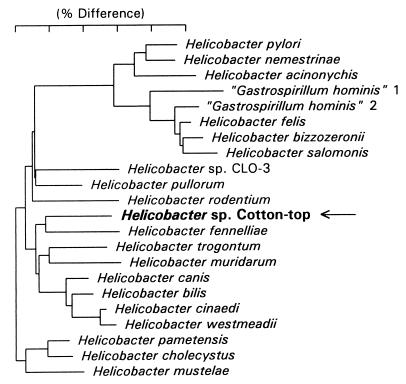
Full 16S rRNA sequences (approximately 1,500 bases) were determined for two isolates (MIT 97-6194-4 and MIT 97-6194-5), which were identical to one another. Comparison of this sequence with more than 100 Helicobacter sequences in our database indicated that the isolates represented a new Helicobacter species. A phylogenetic tree is shown in Fig. 3. The sequence is most similar to that of Helicobacter canis (a short-branch organism) but branches in this tree with Helicobacter fennelliae. The sequence of strains from CTTs contains a 350-base intervening sequence (IVS) at approximately position 210 (using Escherichia coli numbering). Unique IVSs are present in several Helicobacter species, including H. fennelliae, H. bilis, Helicobacter sp. strain CLO-3, some “Flexispira rappini” strains, and an H. canis-like strain, CCUG 29176. A partial sequence (850 bases), including the IVS, was obtained for a third isolate (MIT 97-6194-3), and this sequence was identical to the full sequences of strains from CTTs. Two 1,000-base PCR products obtained from colonic biopsy samples were also sequenced. One was identical to the other sequences of strains from CTTs and one was unique, indicating that the CTTs may harbor a second Helicobacter species.
FIG. 3.
Phylogenetic tree constructed on the basis of 16S rRNA sequence similarity values. The scale bar is equal to a 5% difference in nucleotide sequences as determined by measuring the lengths of the horizontal lines connecting two species.
RFLPs.
All of the bacterial isolates subjected to RFLP analysis gave identical banding patterns (Fig. 4). Included among the isolates analyzed were MIT 97-6194-3 and MIT 97-6194-5, which were shown by 16S rRNA gene sequence analysis to be novel Helicobacter species.
FIG. 4.
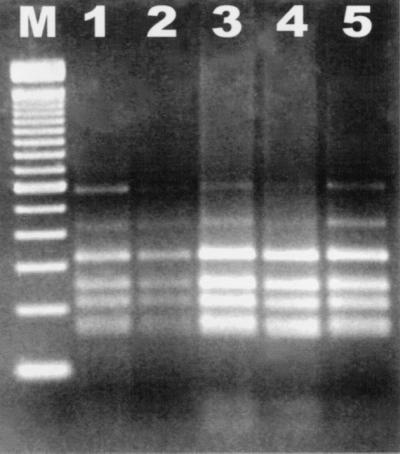
RFLP analysis of DNA isolated from pure fecal cultures, amplified by PCR with Helicobacter genus-specific primers, digested with AluI, and electrophoresed through a 3% agarose gel. Lane M, 100-bp DNA ladder; lane 1, MIT 97-6194-5; lane 2, MIT R97-6837; lane 3, MIT 97-6194-3; lane 4, MIT R97-6842; lane 5, MIT R97-6840.
Histopathology.
Chronic colitis, characterized by various degrees of inflammatory cell infiltration, fibrosis, and mucosal hyperplasia, was present in many of the colonic biopsy specimens. Hyperplastic colonic crypts had diminished goblet cell differentiation and closely packed, basophilic epithelial cells. The mucosa contained foci of interstitial fibrosis and histiocytes. Granulocytes were present in some mucosal sites, often infiltrating through mucosal epithelium and formed small crypt abscesses. Prominent submucosal lymphoid foci were also present in some specimens.
DISCUSSION
In this study we identified a novel urease-negative, fusiform organism in the intestines and feces of CTTs with chronic colitis. On the basis of its morphology, its biochemical traits, and 16S rRNA gene sequence analysis, it was characterized as a member of the genus Helicobacter. This is the first Helicobacter species to be identified in New World primates, although H. pylori and Helicobacter nemistrinae have been isolated from two Old World species: rhesus macaques (Macaca mulatta) and pigtail macaques (Macaca nemistrina), respectively (11). Several species of nonhuman primates are also commonly colonized with large gastric spiral organisms which have been given the provisional name Helicobacter heilmannii (31, 34), although to our knowledge they have never been reported in CTTs.
The novel Helicobacter species was compared biochemically, morphologically, and phylogenetically to other members of the genus Helicobacter. The Helicobacter sp. isolated from CTTs can be distinguished biochemically from other intestinal helicobacters by its lack of urease activity and its inability to hydrolyze alkaline phosphatase. Ultrastructurally, the novel bacterium possesses periplasmic fibers and bipolar sheathed flagella and is morphologically similar to the “F. rappini” group, although it can be distinguished from members of the latter by its lack of urease activity. Phylogenetically, it is most closely related to H. fennelliae, which has been isolated primarily from homosexual men with proctitis and colitis (37). RFLP analysis of the bacterial isolates showed that the CTTs examined were all infected with the same novel Helicobacter species.
Members of the genus Helicobacter can be difficult to culture, so it is not surprising that we were able to obtain pure cultures of the novel Helicobacter sp. from only 8 of 34 fecal samples analyzed. PCR, however, has been shown to be a more sensitive method for detection (17). Using primers specific to the 16S rRNA gene of Helicobacter, we were able to amplify a 422-bp fragment from 18 of 34 biopsy samples. The percentage of positive samples would probably increase with repeated sampling of animals and the analysis of more than one biopsy sample per animal.
Several species of helicobacters have been associated with gastritis, enteritis, and neoplasia in their hosts (11). Humans infected with H. pylori often develop a chronic gastritis, and some infected individuals are also at an increased risk for the development of gastric mucosa-associated lymphoma and gastric adenocarcinoma (20, 28, 42). Recent studies have also shown that immunodeficient mice infected with H. bilis or H. hepaticus develop an inflammatory bowel disease and that male A/JCr mice infected with H. hepaticus develop hepatitis, transmural typhlitis, and hepatic adenocarcinoma due to the chronicity of infection (3, 19, 33, 38, 40). Male B6C3F1 mice have also recently been shown to develop hepatic adenocarcinomas when infected with H. hepaticus (17). Two additional Helicobacter spp., Helicobacter cinaedi and H. fennelliae, are associated with proctitis, colitis, diarrhea, and bacteremia in humans (37); and experimental studies have shown that these species can colonize and cause diarrhea and bacteremia in macaques (10). Although the pathogenic potential of this newly identified Helicobacter species is unknown, given the evidence that other members of the genus Helicobacter may promote inflammation and hyperplasia of gut epithelium and predispose an individual to neoplasia, it is conceivable that the novel Helicobacter sp. isolated from CTTs may be contributing to the UC and subsequent progression to colonic adenocarcinoma that is common to these animals. Additional studies are needed to explore this possibility.
ACKNOWLEDGMENTS
This work was supported in part by NIH grants R01CA67529, PO1CA26731, and RR010146 (to J.G.F.), DE-10374 (to F.E.D.), and DE-11443 (to B.J.P.).
We thank Danielle Hatchadoorian, Melanie Ihrig, Lynn Jackson, Mary Patterson, and Nirah Shomer for help with sample collection and processing. We also thank Rebecca Ericson and Carol Lau for performing the 16S rRNA sequencing and David Lee-Parritz, Keith Mansfield, and Prabhat Seghal for allowing us access to the animals.
REFERENCES
- 1.Bertone E R, Giovannucci E L, King N W, Jr, Petto A J, Johnson L D. Family history as a risk factor for ulcerative colitis-associated colon cancer in cotton-top tamarin. Gastroenterology. 1998;114:669–674. doi: 10.1016/s0016-5085(98)70580-3. [DOI] [PubMed] [Google Scholar]
- 2.Brian D A, Shockley L J. Coronaviruses in tamarins and marmoset colitis. In: Clapp N K, editor. A primate model for the study of colitis and colonic carcinoma the cotton-top tamarin Saguinus oedipus. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 145–160. [Google Scholar]
- 3.Cahill R J, Foltz C J, Fox J G, Dangler C A, Powrie F, Schauer D B. Inflammatory bowel disease: an immune mediated condition triggered by bacterial infection with Helicobacter hepaticus. Infect Immun. 1997;65:3126–3131. doi: 10.1128/iai.65.8.3126-3131.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chalifoux L V, Brieland J K, King N W. Evolution and natural history of colonic disease in cotton-top tamarins (Saguinus oedipus) Dig Dis Sci. 1985;30:54S–58S. doi: 10.1007/BF01296976. [DOI] [PubMed] [Google Scholar]
- 5.Clapp N K, Henke M A, Hansard R M, Carson R L, Adams L J, Nardi R V. Natural history, time course, and pathogenesis of idiopathic colitis in cotton-top tamarins Saguinus oedipus. In: Clapp N K, editor. A primate model for the study of colitis and colonic carcinoma the cotton-top tamarin Saguinus oedipus. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 83–100. [Google Scholar]
- 6.Clapp N K, Nardi R V, Tobi M. Future directions for colon disease research using cotton-top tamarins. In: Clapp N K, editor. A primate model for the study of colitis and colonic carcinoma the cotton-top tamarin Saguinus oedipus. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 319–324. [Google Scholar]
- 7.Dobbins W O., III Dysplasia and malignancy in inflammatory bowel disease. Annu Rev Med. 1984;35:33–48. doi: 10.1146/annurev.me.35.020184.000341. [DOI] [PubMed] [Google Scholar]
- 8.Erdman S E, Correa P, Li X, Coleman L A, Fox J G. Helicobacter mustelae-associated mucosa associated lymphoid tissue (MALT) lymphoma in ferrets. Am J Pathol. 1997;151:273–280. [PMC free article] [PubMed] [Google Scholar]
- 9.Fantry G T, Zheng Q X, James S P. Conventional cleaning and disinfection techniques eliminate the risk of endoscopic transmission of Helicobacter pylori. Am J Gastroenterol. 1995;90:227–232. [PubMed] [Google Scholar]
- 10.Flores B M, Fennell C L, Kuller L. Experimental infection of pig-tailed macaques (Macaca nemestrina) with Campylobacter cinaedi and Campylobacter fennelliae. Infect Immun. 1990;58:3947–3953. doi: 10.1128/iai.58.12.3947-3953.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox J G. The expanding genus of Helicobacter: pathogenic and zoonotic potential. In: Sleisenger M H, Fordtram J S, editors. Seminars in gastrointestinal diseases. Vol. 8. Philadelphia, Pa: The W. B. Saunders Co.; 1997. pp. 124–141. [PubMed] [Google Scholar]
- 12.Fox J G, Correa P, Taylor N S, Lee A, Otto G, Murphy J C, Rose R. Helicobacter mustelae associated gastritis in ferrets: an animal model of Helicobacter pylori gastritis in humans. Gastroenterology. 1990;99:352–361. doi: 10.1016/0016-5085(90)91016-y. [DOI] [PubMed] [Google Scholar]
- 13.Fox J G, Dangler C A, Sager W, Borkowski R, Gliatto J M. Helicobacter mustelae associated gastric adenocarcinoma in ferrets (Mustela putorius furo) Vet Pathol. 1997;34:225–229. doi: 10.1177/030098589703400308. [DOI] [PubMed] [Google Scholar]
- 14.Fox J G, Dewhirst F E, Tully J G, Paster B J, Yan L, Taylor N S, Collins M J, Gorelick P L, Ward J M. Helicobacter hepaticus sp. nov, a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J Clin Microbiol. 1994;32:1238–1245. doi: 10.1128/jcm.32.5.1238-1245.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fox J G, Lee A. The role of Helicobacter species in newly recognized gastrointestinal tract diseases of animals. Lab Anim Sci. 1997;47:222–255. [PubMed] [Google Scholar]
- 16.Fox J G, Li X, Yan L, Cahill R J, Hurley R, Lewis R, Murphy J C. Chronic proliferative hepatitis in A/JCr mice associated with persistent Helicobacter hepaticus infection: a model of Helicobacter-induced carcinogenesis. Infect Immun. 1996;64:1548–1558. doi: 10.1128/iai.64.5.1548-1558.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fox J G, MacGregor J, Shen Z, Li X, Lewis R, Dangler C A. Comparison of methods to identify Helicobacter hepaticus in B6C3F1 used in a carcinogenesis bioassay. J Clin Microbiol. 1998;36:1382–1387. doi: 10.1128/jcm.36.5.1382-1387.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fox J G, Yan L, Dewhirst F E, Paster B J, Shames B, Murphy J C, Hayward A, Belcher J C, Mendes E N. Helicobacter bilis sp. nov., a novel Helicobacter isolated from bile, livers, and intestines of aged, inbred mouse strains. J Clin Microbiol. 1995;33:445–454. doi: 10.1128/jcm.33.2.445-454.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fox J G, Yan L, Shames B, Campbell J, Murphy J C, Li X. Persistent hepatitis and enterocolitis in germfree mice infected with Helicobacter hepaticus. Infect Immun. 1996;64:3673–3681. doi: 10.1128/iai.64.9.3673-3681.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham D Y. Campylobacter pylori and peptic ulcer disease. Gastroenterology. 1989;96:615–623. doi: 10.1016/s0016-5085(89)80057-5. [DOI] [PubMed] [Google Scholar]
- 21.Johnson L D, Ausman L M, Sehgal P K, King N W., Jr A prospective study of the epidemiology of colitis and colon cancer in coton-top tamarins (Saguinus eodipus) Gastroenterology. 1996;110:102–115. doi: 10.1053/gast.1996.v110.pm8536845. [DOI] [PubMed] [Google Scholar]
- 22.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press, Inc.; 1969. pp. 21–132. [Google Scholar]
- 23.Kirkwood J K, Pearson G R, Epstein M A. Adenocarcinoma of the large bowel and colitis in captive cotton-top tamarins Saguinus o. oedipus. J Comp Pathol. 1986;96:507–515. doi: 10.1016/0021-9975(86)90071-x. [DOI] [PubMed] [Google Scholar]
- 24.Kusukawa N, Uemori T, Asada K, Kato I. Rapid and reliable protocol for direct sequencing of material amplified by the polymerase chain reaction. BioTechniques. 1990;9:66–72. [PubMed] [Google Scholar]
- 25.Lee A, Hazell S L, O’Rourke J. Isolation of a spiral-shaped bacterium from the cat stomach. Infect Immun. 1988;56:2843–2850. doi: 10.1128/iai.56.11.2843-2850.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melendez L V, Hunt R D, Daniel M D, Fraser C E O, Barahona H H, Garcia F G, King N W. Lymphoma viruses of monkeys: herpesvirus and Herpesvirus ateles, the first oncogenic herpesviruses of primates—a review. In: Biggs P M, de-The G, Payne L N, editors. Oncogenesis and herpesviruses. Lyon, France: International Agency for Research on Cancer; 1972. pp. 451–461. [Google Scholar]
- 26a.National Research Council. Guide for the care and use of laboratory animals. Washington, D.C: National Academy Press; 1996. [Google Scholar]
- 27.Neubauer R H, Rabin H, Hopkins III R, Levy B M. Characteristics of cell lines established from Epstein-Barr virus induces marmoset tumors. Prim Med. 1978;10:156–162. [PubMed] [Google Scholar]
- 28.Parsonnet J. Bacterial infection as a cause of cancer. Environ Health Perspect. 1995;103:263–268. doi: 10.1289/ehp.95103s8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paster B J, Dewhirst F E. Phylogeny of Campylobacter, wolinellas, Bacteroides gracilis, and Bacteroides ureolyticus by 16S ribosomal ribonucleic acid sequencing. Int J Syst Bacteriol. 1988;38:56–62. [Google Scholar]
- 30.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 31.Sato, T., and T. A. Takeuchi. 1982. Infection by spirilla in the stomach of the rhesus monkey. Vet. Pathol. 19(Suppl. 7):17–25. [PubMed]
- 32.Shen Z, Fox J G, Dewhirst F E, Paster B J, Foltz C J, Yan L, Shames B, Perry L. Helicobacter rodentium sp. nov., a urease negative Helicobacter species isolated from laboratory mice. Int J Syst Bacteriol. 1997;47:627–634. doi: 10.1099/00207713-47-3-627. [DOI] [PubMed] [Google Scholar]
- 33.Shomer N H, Dangler C A, Fox J G. Helicobacter bilis induced inflammatory bowel disease (IBD) in defined flora scid mice. Infect Immun. 1997;65:4858–4864. doi: 10.1128/iai.65.11.4858-4864.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Solnick J V, O’Rourke J, Lee A, Paster B J, Dewhirst F E, Tompkins L S. An uncultured gastric spiral organism is a newly identified Helicobacter in humans. J Infect Dis. 1993;168:379–385. doi: 10.1093/infdis/168.2.379. [DOI] [PubMed] [Google Scholar]
- 35.Stanley J, Linton D, Burens A P, Dewhirst F E, On S L, Porter A, Owen R J, Costas M. Helicobacter pullorum sp. nov.—genotype and phenotype of a new species isolated from poultry and from human patients with gastroenteritis. Microbiology. 1994;140:3441–3449. doi: 10.1099/13500872-140-12-3441. [DOI] [PubMed] [Google Scholar]
- 36.Stonerook M J, Weiss H S, Rodriguez M A, Rodriguez J V, Hernandez J I, Peck O C, Wood J D. Temperature-metabolism relations in the cotton-top tamarin (Saguinus oedipus) model for ulcerative colitis. J Med Primatol. 1994;23:16–22. doi: 10.1111/j.1600-0684.1994.tb00090.x. [DOI] [PubMed] [Google Scholar]
- 37.Totten P A, Fennel C L, Tenover F C. Campylobacter cinaedi (sp. nov.) and Campylobacter fennelliae (sp. nov.): two new Campylobacter species associated with enteric disease in homosexual men. J Infect Dis. 1985;151:131–139. doi: 10.1093/infdis/151.1.131. [DOI] [PubMed] [Google Scholar]
- 38.Ward J M, Anver M R, Haines D C, Melhorn J M, Gorelick P, Yan L, Fox J G. Inflammatory large bowel disease in immunodeficient mice naturally infected with Helicobacter hepaticus. Lab Anim Sci. 1996;46:15–20. [PubMed] [Google Scholar]
- 39.Watkins P E, Warren B F, Stephens P, Ward P, Foulkes R. Treatment of ulcerative colitis in the cotton-top tamarin using antibody to tumour necrosis actor alpha. Gut. 1997;40:628–633. doi: 10.1136/gut.40.5.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whary M T, Morgan T J, Dangler C A, Gaudes K J, Taylor N S, Fox J G. Chronic active hepatitis induced by Helicobacter hepaticus in the A/JCr mouse is associated with a Th1 cell-mediated immune response. Infect Immun. 1998;66:3142–3148. doi: 10.1128/iai.66.7.3142-3148.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wood J D, Peck O C, Tefend K S, Rodriguez M A, Rodriguez J V, Hernandez J I, Stonebrook M J, Sharma H M. Colitis and colon cancer in cotton-top tamarins (Saguinus oedipus oedipus) living wild in their natural habitat. Dig Dis Sci. 1998;43:1443–1453. doi: 10.1023/a:1018842210330. [DOI] [PubMed] [Google Scholar]
- 42.Wotherspoon A C, Doglioni C, Diss T C, Pan L, Moschini A, de Boni M, Isaacson P G. Regression of primary low-grade B-cell gastric lymphoma of mucosa-associated lymphoid tissue type after eradication of Helicobacter pylori. Lancet. 1993;342:575–577. doi: 10.1016/0140-6736(93)91409-f. [DOI] [PubMed] [Google Scholar]