Abstract
Background:
The presence of invasive cribriform adenocarcinoma (ICC), an expanse of cells containing punched-out lumina uninterrupted by stroma, in radical prostatectomy (RP) specimens has been associated with biochemical recurrence (BCR). However, ICC identification has only moderate inter-reviewer agreement.
Objective:
To investigate quantitative machine-based assessment of the extent and prognostic utility of ICC, especially within individual Gleason grade groups.
Design, setting, and participants:
A machine learning approach was developed for ICC segmentation using 70 RP patients and validated in a cohort of 749 patients from four sites whose median year of surgery was 2007 and with median follow-up of 28 mo. ICC was segmented on one representative hematoxylin and eosin RP slide per patient and the fraction of tumor area composed of ICC, the cribriform area index (CAI), was measured.
Outcome measurements and statistical analysis:
The association between CAI and BCR was measured in terms of the concordance index (c index) and hazard ratio (HR).
Results and limitations:
CAI was correlated with BCR (c index 0.62) in the validation set of 411 patients with ICC morphology, especially those with Gleason grade group 2 cancer (n = 192; c index 0.66), and was less prognostic when patients without ICC were included (c index 0.54). A doubling of CAI in the group with ICC morphology was prognostic after controlling for Gleason grade, surgical margin positivity, preoperative prostate-specific antigen level, pathological T stage, and age (hazard ratio 1.19, 95% confidence interval 1.03–1.38; p = 0.018).
Conclusions:
Automated image analysis and machine learning could provide an objective, quantitative, reproducible, and high-throughput method of quantifying ICC area. The CAI performance for grade group 2 cancer suggests that for patients with little Gleason 4 pattern, the ICC fraction has a strong prognostic role.
Keywords: Prostate cancer, Cribriform, Machine learning, Digital pathology, Gleason grading, Biochemical recurrence
Patient summary:
Machine-based measurement of a specific cell pattern (cribriform; sieve-like, with lots of spaces) in images of prostate specimens could improve risk stratification for patients with prostate cancer. In the future, this could help in expanding the criteria for active surveillance.
1. Introduction
Estimates of prostate cancer aggressiveness are based on several clinical factors, including tumor stage, prostate specific antigen (PSA) levels, and tissue morphology evaluated via Gleason grading [1]. Using the Gleason grading system, a pathologist categorizes all morphological patterns seen in tumor tissue into one of five patterns, with the Gleason score for a radical prostatectomy (RP) specimen being the sum of the two most common patterns [2]. One pattern, cribriform, is graded as Gleason pattern 4, and appears as an expanse of carcinoma cells containing multiple gland lumina and no intervening stroma [3–5]. The presence of any amount of cribriform morphology has been correlated with worse outcomes compared to other types of pattern 4 [4,6–10], although some groups have reported that cribriform morphology tends to be prognostic only for patients with low Gleason scores [11,12].
Despite its importance, studies of cribriform morphology have been hampered by the high degree of interobserver disagreement for cribriform identification [13]. A variety of patterns in both benign and malignant tissue have been described as cribriform in the literature, and not all such patterns carry the same risk [14]. The close resemblance of ill-formed and fused glands to cribriform morphology and the distinction made by some groups between small and large cribriform patterns further complicate diagnostic agreement [15]. The extent of these challenges raises the possibility that a computationally derived index could provide a more reproducible assessment of cribriform extent on pathology slides. As a result of the increase in the consistency of cribriform area quantification, this index could be more robustly associated with biochemical recurrence (BCR) than area estimations by a pathologist. In particular, cribriform area measurement has the potential to add prognostic value within Gleason grade groups.
Machine learning has been applied to several problems in prostate pathology, including diagnosis [16,17], Gleason grading [18–20], and outcome prognostication [21]. These approaches rely on extraction of computerized morphology features for elements such as glands, nuclei, and image texture. By contrast, our computational approach leverages previous work on the prognostic power of invasive cribriform adenocarcinoma (ICC) morphology to use automated delineation of that pattern for risk assessment. In addition, while previous work has used machine learning for Gleason grading, relatively few studies have attempted to correlate automated pattern assessment with disease outcome. In particular, computerized grading approaches have not explicitly accounted for the potential role of ICC morphology in determining outcome or how ICC content may have a differential prognostic value for different grade groups.
Here we present an automated method for quantification of ICC area, called the cribriform area index (CAI), which is a measure of the proportion of specimen tumor tissue that is composed of ICC. Our approach uses deep learning (DL), a type of machine learning in which an artificial neural network is given images and annotations and then learns to replicate the annotations on new images [22], to identify ICC patterns on digitized pathology slides. Via CAI, we evaluated the association between ICC area and BCR risk in the study population overall, in each Gleason grade group, and as an additional marker after patients were stratified by a machine learning model based on lumen morphology. A large, multi-institutional, retrospectively collected cohort was used to validate CAI across variations in specimen preparation and digitization.
2. Patients and methods
2.1. Data set description
Data for a total of 819 patients were retrospectively collected from four institutions: the University of Pennsylvania (UPenn), New York-Presbyterian/Weill Cornell Medical Center (WCMC), The Cancer Genome Atlas (TCGA), and University Hospitals Cleveland Medical Center (UHCMC), in accordance with institutional review board–approved protocols at each site. The data set is described in Table 1. A single slide from each patient was used in accordance with the design of companion diagnostic validation studies in prostate cancer, in which a small tissue sample is selected for molecular analysis [23–25]. To select this slide, all slides from each case were obtained from the archives at the source institution and reviewed by a pathologist there. A diagnostic slide representing all salient features of a particular cancer case was then selected. For each case, the selected slide therefore reflected the grade group, morphology, and stage to the best possible degree. This slide generally also contained the dominant focus of the cancer. A pathologist then annotated a single representative tumor region on each digital image. Inclusion criteria for the study were a successfully digitized hematoxylin and eosin slide, at least 30 d of post-RP PSA follow-up, PSA <0.2 ng/ml after surgery, and no history of neoadjuvant or adjuvant therapy. BCR-free survival was measured from the date of surgery to the date of the second consecutive PSA test result >0.2 ng/ml for patients with BCR, and censored at the date of last PSA test for those without BCR.
Table 1–
Clinical parameters for the 819 patients in the study cohort
| Variable | Training set | Validation set | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| UPenn | UHCMC | UPenn | TCGA | WCMC | Total | |
| Patients (n) | 70 | 146 | 350 | 174 | 79 | 749 |
| Race, n (%) | ||||||
| Caucasian | 63 (90.0) | 92 (63.0) | 231 (66.0) | 69 (39.7) | 0 (0.0) | 392 (52.3) |
| African American | 7 (10.0) | 35 (24.0) | 111 (31.7) | 3 (1.7) | 79 (100.0) | 228 (30.4) |
| Other | 0 (0.0) | 6 (4.1) | 8 (2.3) | 1 (0.6) | 0 (0.0) | 15 (2.0) |
| Unknown | 0 (0.0) | 13 (8.9) | 0 (0.0) | 101 (58.0) | 0 (0.0) | 114 (15.2) |
| Median age, yr (IQR) | 59 (55–65) | 61 (57–64) | 61 (56–66) | 61 (55–65) | 61 (56–66) | 61 (55–66) |
| Unknown (n) | 0 | 131 | 0 | 0 | 0 | 131 |
| pT stage, n (%) | ||||||
| pT2 | 36 (51.4) | 9 (6.2) | 163 (46.6) | 90 (51.7) | 63 (79.7) | 325 (43.4) |
| pT3 a | 0 (0.0) | 2 (1.4) | 1 (0.3) | 0 (0.0) | 0 (0.0) | 3 (0.4) |
| pT3a | 23 (32.9) | 5 (3.4) | 139 (39.7) | 52 (29.9) | 11 (13.9) | 207 (27.6) |
| pT3b | 11 (15.7) | 2 (1.4) | 47 (13.4) | 27 (15.5) | 5 (6.3) | 81 (10.8) |
| pT4 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (1.1) | 0 (0.0) | 2 (0.3) |
| Unknown | 0 (0.0) | 128 (87.7) | 0 (0.0) | 3 (1.7) | 0 (0.0) | 131 (17.5) |
| N stage, n (%) | ||||||
| N0 | 70 (100.0) | 11 (7.5) | 345 (98.6) | 125 (71.8) | 78 (98.7) | 559 (74.6) |
| N1 | 0 (0.0) | 0 (0.0) | 2 (0.6) | 24 (13.8) | 1 (1.3) | 27 (3.6) |
| Unknown | 0 (0.0) | 135 (92.5) | 3 (0.9) | 25 (14.4) | 0 ( 0.0) | 163 (21.8) |
| Median PSA, ng/ml (IQR) | 8 (5–11) | 6 (5–8) | 6 (5–9) | 6 (5–9) | 6 (4–8) | 6 (5–9) |
| Unknown | 0 | 66 | 4 | 5 | 3 | 78 |
| RP grade group, n (%) | ||||||
| 1 | 8 (11.4) | 35 (24.0) | 74 (21.1) | 20 (11.5) | 17 (21.5) | 146 (19.5) |
| 2 | 38 (54.3) | 81 (55.5) | 163 (46.6) | 72 (41.4) | 40 (50.6) | 356 (47.5) |
| 3 | 16 (22.9) | 13 (8.9) | 68 (19.4) | 43 (24.7) | 15 (19.0) | 139 (18.6) |
| 4 | 4 (5.7) | 2 (1.4) | 22 (6.3) | 23 (13.2) | 1 (1.3) | 48 (6.4) |
| 5 | 4 (5.7) | 4 (2.7) | 21 (6.0) | 16 (9.2) | 6 (7.6) | 47 (6.3) |
| Unknown | 0 (0.0) | 11 (7.5) | 2 (0.6) | 0 (0.0) | 0 (0.0) | 13 (1.7) |
| PSM, n (%) | 30 (42.9) | 9 (6.2) | 207 (59.1) | 29 (16.7) | 8 (10.1) | 253 (33.8) |
| Unknown | 0 (0.0) | 131 (89.7) | 1 (0.3) | 16 (9.2) | 0 (0.0) | 148 (19.8) |
| Median FU for CPS, mo (IQR) | 22 (10–54) | 79 (59–104) | 27 (18–55) | 17 (8–30) | 22 (4–45) | 28 (15–62) |
| Patients with BCR, n (%) | 35 (50.0) | 46 (31.5) | 114 (32.6) | 7 (4.0) | 10 (12.7) | 177 (23.6) |
BCR = biochemical recurrence; CPS = censored patients; FU = follow-up; IQR = interquartile range; PSA = prostate-specific antigen (preoperative); PSM = positive surgical margin; RP = radical prostatectomy; TCGA = The Cancer Genome Atlas; UHCMC = University Hospitals Cleveland Medical Center; UPenn = University of Pennsylvania; WCMC = New York-Presbyterian/Weill Cornell Medical Center.
Substaging information for pT3 unavailable for these cases.
The training set (ST) was chosen to contain enough patients to train the ICC segmentation model while maximizing the size of the validation set, and consisted of 70 patients from UPenn whose slides were scanned on a different scanner than for the other UPenn patients. ST was used to train both the ICC DL segmentation model and the machine learning model that CAI augmented. The prognostic value of CAI was then evaluated using the validation set (SV) consisting of the remaining 749 patients.
2.2. DL-based detection and segmentation of ICC
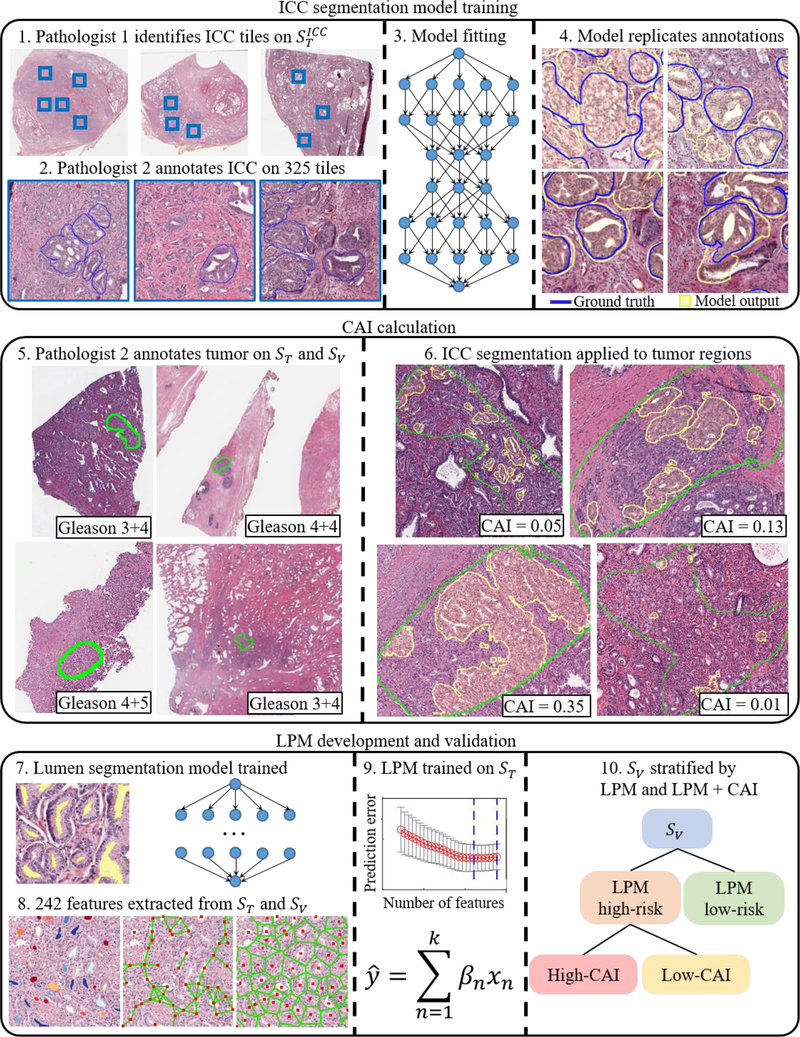
ST was used to train and test the UNet-inspired [26] DL segmentation model. Images were searched for fields of view containing ICC, which were then annotated and used for model training. In total, 325 tiles from 36 patients, comprising the ICC set, were used to train the ICC segmentation model, and for every tile with ICC annotations, both pathologists agreed that ICC was present. This process is shown in Figure 1. This model produced a pixel-wise true positive rate of 0.94 and true negative rate of 0.79.
Fig. 1 –
Overall study workflow with development of the ICC segmentation model, CAI calculation, and LPM development and validation.
CAI = cribriform area index; ICC = invasive cribriform adenocarcinoma; LPM = lumen-based prognosis model; ST = training set; SV = validation set
The ICC segmentation model was applied to the annotated tumor region of each image in SV and the results were reviewed to qualitatively assess the model performance. CAI was calculated for each patient as the proportion of the annotated tumor area that was composed of ICC. Further details on the model training and validation are provided in the Supplementary material.
2.3. Statistical analysis
The association between BCR and CAI was analyzed in SV with CAI as both a categorical and a continuous variable in. First, outcomes of a high-CAI subset, defined as CAI >0.10, were compared with the CAI ≤0.10 set using log-rank p value and hazard ratios (HRs). Second, the added risk correlated with an increase in CAI was evaluated with CAI as a continuous variable using by Harell’s concordance index (c index) in SV overall and in each Gleason grade group. The third analysis compared the risk of increasing CAI on a categorical basis using HRs between four groups: no ICC (CAI 0), and a small (CAI 0–0.05), moderate (CAI 0.05–0.15), or large (CAI >0.15) amount of ICC. CAI was then assessed for prognostic independence in a Cox multivariable proportional-hazards model with Gleason grade, surgical margin positivity, preoperative PSA, pathological T stage, and age at surgery. For the multivariable analysis, only patients with CAI >0 were considered, and the hazard associated with CAI was assessed for doubling of CAI to model how small absolute differences can represent large relative differences at small CAI values.
CAI was also combined with a machine learning model based on gland lumen morphology, the lumen-based prognosis model (LPM), to investigate the prognostic value of CAI added to the machine learning model. The development of this model was described by Leo et al [27], although that study used a larger training set, while the LPM here was trained only on the 70 patients in ST. Patients identified as LPM high-risk were further stratified by the presence of substantial ICC, defined as CAI >0.10. The HR was then computed between the LPM low-risk, LPM high-risk low-CAI, and LPM high-risk high-CAI groups. Analyses were performed using Python v3.6.6 and MATLAB v2019b.
3. Results
3.1. Association between CAI and BCR in SV
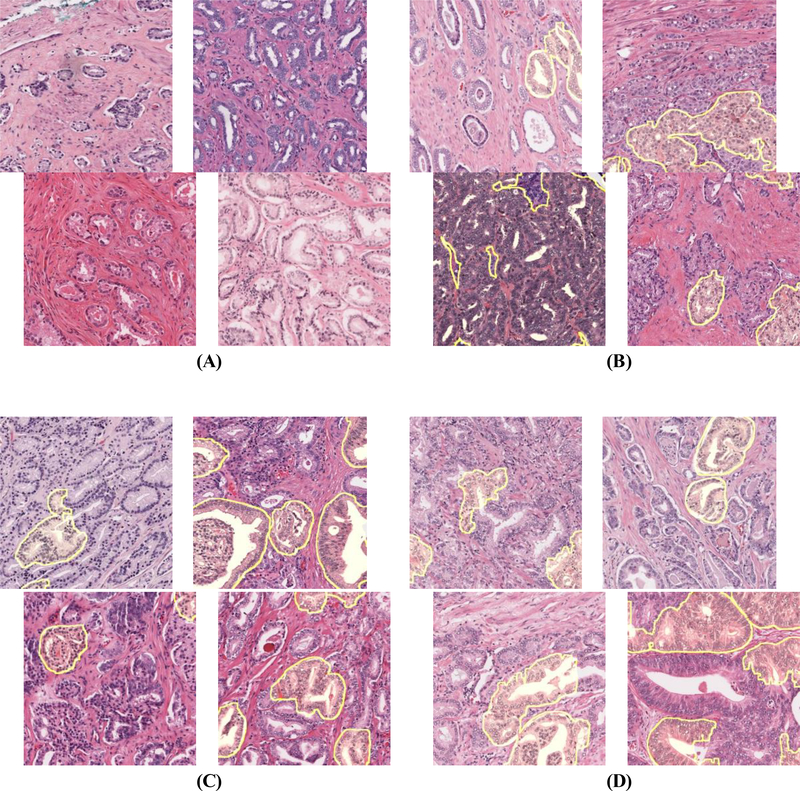
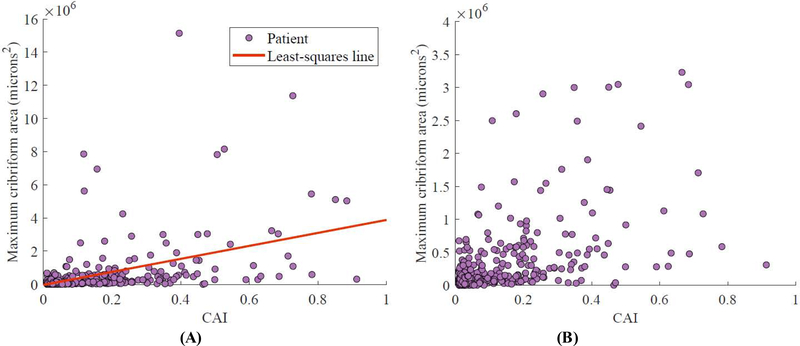
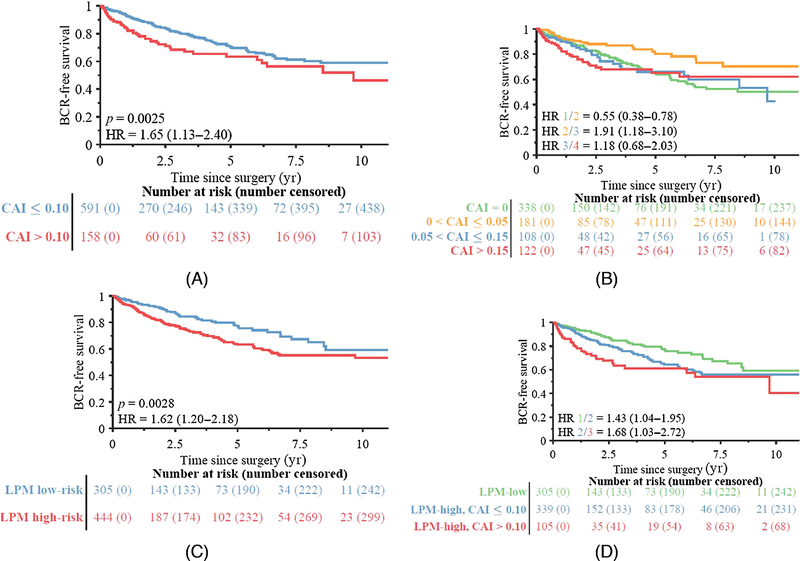
Figure 2 shows ICC segmentation results for patients in SV. CAI was moderately correlated with the size of the largest ICC area in an image (Spearman correlation coefficient 0.65; p < 0.001). As shown in Figure 3, owing to variability in tumor area, there were some patients with high CAI with only small ICC regions, and vice versa. CAI was prognostic in the 411 SV patients who had ICC morphology (defined as CAI >0), with a c index of 0.62, but was weakly correlated with BCR in SV overall (c index 0.54). Doubling of CAI was prognostic independent of Gleason grade, surgical margin positivity, preoperative PSA, pathological T stage, and age at surgery among the 298 patients with ICC (HR 1.19, 95% confidence interval [CI] 1.03–1.38; p = 0.018; Table 2). Patients with a substantial amount of ICC (CAI >0.10) were at much higher risk of BCR than patients with CAI below this threshold (HR 1.65, 95% CI 1.13–2.40; p = 0.003), as seen in Figure 4.
ig. 2 –
Automated ICC segmentation results, shown as yellow shading, for patients in the validation set categorized by cribriform area index and outcome. (A) Patients without ICC and long BCR-free survival. (B) Patients with ICC and early BCR. (C) Patients with ICC and early BCR in grade group 2. (D) Patients with ICC and long survival. False-positives in ICC detection are most apparent in D, suggesting that improved ICC segmentation could further stratify patients by risk.
BCR = biochemical recurrence; ICC = invasive cribriform adenocarcinoma.
Fig. 3 –
Comparison of CAI and size of largest ICC region for the 411 patients in the validation set with CAI >0, with each patient represented as a dot. (A) All 411 patients and (B) 401 patients with ten outliers excluded to aid in visualization. CAI was moderately correlated with maximum ICC area (Spearman correlation coefficient 0.65; p < 0.001).
CAI = cribriform area index; ICC = invasive cribriform adenocarcinoma.
Table 2 –
Cox proportional-hazards univariable and multivariable analysis of risk factors for biochemical recurrence for the 298 patients in the validation set who had detectable invasive cribriform carcinoma (CAI >0) and data available for all covariates
| Variable | Univariable | Multivariable | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| HR (95% CI) | p value | CAI continuous | CAI categorical | |||
|
| ||||||
| HR (95% CI) | p value | HR (95% CI) | p value | |||
| log2 CAI | 1.31 (1.14–1.51) | <0.001 | 1.19 (1.03–1.38) | 0.018 | ||
| CAI >0.10 vs ≤0.10 | 2.30 (1.40–3.76) | <0.001 | – | – | 1.66 (0.97–2.85) | 0.063 |
| Gleason grade group | ||||||
| 1 | Reference | Reference | Reference | |||
| 2 | 1.40 (0.51–3.79) | 0.512 | 0.60 (0.21–1.69) | 0.332 | 0.62 (0.22–1.74) | 0.362 |
| 3 | 2.55 (0.95–6.84) | 0.063 | 0.88 (0.30–2.56) | 0.816 | 0.96 (0.33–2.80) | 0.942 |
| ≥4 | 3.46 (2.06–5.81) | <0.001 | 1.77 (0.60–5.17) | 0.299 | 1.85 (0.63–5.46) | 0.265 |
| PSM | 2.46 (1.48–4.10) | <0.001 | 1.59 (0.92–2.77) | 0.098 | 1.54 (0.88–2.71) | 0.129 |
| log2 PSA in ng/ml | 1.92 (1.50–2.45) | <0.001 | 1.63 (1.25–2.12) | <0.001 | 1.62 (1.24–2.10) | <0.001 |
| Stage ≥pT3 vs <pT3 | 4.02 (2.22–7.28) | <0.001 | 2.23 (1.13–4.37) | 0.020 | 2.18 (1.11–4.30) | 0.024 |
| Age at surgery in years | 1.05 (1.01–1.09) | 0.009 | 1.03 (0.99–1.06) | 0.125 | 1.03 (0.99–1.06) | 0.108 |
CAI = cribriform area index; CI = confidence interval; HR = hazard ratio; PSA = prostate-specific antigen (preoperative); PSM = positive surgical margin.
Fig. 4 –
BCR-free survival in groups stratified by CAI among the 749 patients in the validation set. (A) Using a single threshold, the group with CAI ≤0.10 had significantly better survival than the group with CAI >0.10. (B) Using multiple thresholds, the group with CAI between 0 and 0.05 had significantly better survival than the other groups; the other groups had no significant differences in survival. (C) Validation set patients stratified by LPM. (D) LPM high-risk patients further stratified by CAI >0.10.
BCR = biochemical recurrence; CAI = cribriform area index; HR = hazard ratio; LPM = lumen-based prognosis model.
Figure 4 also shows survival profiles for patients with no ICC (CAI 0) and small (CAI 0–0.05), moderate (CAI 0.05–0.15), and large (CAI >0.15) amounts of ICC. While there was no clear difference in risk between patients with CAI 0 and CAI >0, the low-CAI group had significantly better survival than the moderate-CAI group, and the latter had a nonsignificant difference in survival from the high-CAI group.
While the proportion of patients for whom ICC was detected ranged between 39% and 72% across sites (Table 3), the median CAI and c index for CAI among patients with CAI >0 were similar across sites, with the exception of the TCGA data, which had only seven BCR events, complicating site-specific analysis of those patients.
Table 3 –
Results for the validation set by site and by Gleason grade group for all patients in each cohort and for the subgroup with CAI >0
| All patients | Patients with CAI >0 | ||||
|---|---|---|---|---|---|
|
|
|||||
| n | c index | n (%) | c index | Median CAI (IQR) | |
| Site | |||||
| UHCMC | 146 | 0.48 | 92 (63) | 0.59 | 0.08 (0.14) |
| WCMC | 79 | 0.56 | 31 (39) | 0.63 | 0.05 (0.05) |
| TCGA | 174 | 0.78 | 125 (72) | 0.90 | 0.07 (0.15) |
| UPenn | 350 | 0.56 | 163 (47) | 0.60 | 0.06 (0.18) |
| Gleason grade group | |||||
| 1 | 146 | 0.47 | 57 (39) | 0.52 | 0.04 (0.06) |
| 2 | 356 | 0.52 | 193 (54) | 0.66 | 0.04 (0.09) |
| 3 | 139 | 0.43 | 93 (67) | 0.51 | 0.11 (0.22) |
| 4 | 48 | 0.51 | 31 (65) | 0.62 | 0.14 (0.23) |
| 5 | 47 | 0.60 | 31 (66) | 0.57 | 0.13 (0.21) |
CAI = cribriform area index; IQR = interquartile range; TCGA = The Cancer Genome Atlas; UHCMC = University Hospitals Cleveland Medical Center; UPenn = University of Pennsylvania; WCMC = New York-Presbyterian/Weill Cornell Medical Center.
3.2. Prognostic value of CAI in specific Gleason grade groups
Table 3 shows that the prognostic value of CAI varied between Gleason grade groups, with the highest c index (0.66) observed for patients with Gleason grade group 2 cancer with ICC. Notably, ICC was detected in 57 patients (39%) with Gleason grade group 1 disease; however, all of these patients had surgery before 2016 and therefore may have had some cribriform patterns graded as pattern 3 instead of pattern 4 [28].
3.3. CAI further stratifies the LPM high-risk group
For the purpose of developing the LPM, a DL model for gland lumen segmentation was trained on 41 1 mm × 1 mm tiles containing 4927 annotated gland lumens from 37 slides from ST. This model yielded a per-pixel true positive rate of 0.94, true negative rate of 0.97, and F1 score of 0.90 on the four holdout regions used for model testing.
Gland lumens were then segmented in the tumor regions of all 819 images. On the basis of previous work in prostate cancer [16], 216 descriptors of morphology and architecture were extracted from lumen segmentations and a further 26 Haralick texture features from the entire tumor region. As ST was composed of two 35-patient cohorts collected at different times, features that were unstable between these two cohorts [29] were removed.
The 115 stable features were used to train a Cox regression model and perform feature selection via tenfold elastic-net regularization (α = 0.5). The final LPM, containing five features, was then applied to each slide to calculate a risk score for each patient. A risk score threshold was learned on ST to maximize the difference in survival time between predicted low-risk and high-risk patients.
The LPM was prognostic in SV (HR 1.62, 95% CI 1.20–2.18; p = 0.003) by itself, and its prognostic power was improved by addition of CAI. The group of LPM high-risk patients with CAI >0.10 had a much higher BCR rate than the group with LPM low-risk or the group of LPM high-risk patients with CAI ≤0.10, especially within 2.5 yr of surgery. Of the five features selected for the LPM, three described the range in lumen shape across the tumor, one was a measure of uniformity in lumen orientation, and the last was the average distance between lumens. All of these features were positively correlated with BCR-free survival.
4. Discussion
According to some estimates, 40% of RP patients experience BCR [30] and the associated higher risk of metastasis and disease-specific mortality [31]. The current gold standard for BCR prognosis—nomograms—relies heavily on Gleason scoring, which has known inter-reviewer variability [32,33]. Therefore, there have been efforts to go beyond Gleason scoring and directly correlate specific architectural patterns with outcome. Cribriform morphology has been correlated with poor outcomes, and was found in 16% of non-BCR cases but 61% of BCR cases [34] and in 81% of all metastatic cases [12], with an odds ratio for BCR of 1.173 per additional 1 mm2 in cribriform area [6]. However, these studies relied on manual identification of cribriform morphology and usually did not examine the relationship between the amount of cribriform pattern and outcome [4,6]. In part because of the time-intensive nature and limited reproducibility of manual identification of cribriform morphology [13], large, multi-institutional studies relating the amount of cribriform pattern to outcome have not been conducted.
To mitigate these challenges, we used a combination of quantitative image analysis and machine learning for ICC segmentation in this study. The DL-based method allowed us to study the prognostic value of cribriform morphology in a validation set of 749 patients from four institutions. The association of CAI with elevated BCR risk is consistent with literature on the prognostic value of cribriform morphology [7,11,12], including studies that found that the presence of large cribriform foci was more prognostic than the presence of small foci [4,6,35]. This suggests that the automated method was sufficiently robust to be useful for BCR prognosis. The prognostic value of CAI was consistent between sites, which may imply that CAI is resilient and robust to site-specific preanalytic variations and batch effects. With the recent adoption of guidelines that include the reporting of cribriform morphology in pathology reports [36,37], these findings may assist in the development of digital pathology platforms to aid in identifying high-risk morphological patterns.
An increase in CAI was most strongly correlated with BCR for patients with Gleason grade group 2 disease, with a c index of 0.66 among patients with ICC morphology. It may be the case that for patients with a small amount of Gleason pattern 4, the fraction of the pattern 4 morphology that is ICC is especially prognostic of BCR. Kir et al [11] found that the presence of cribriform pattern was significantly associated with BCR in Gleason score 3 + 3 cases, but not in other cases. That study used data predating the adoption of standards to grade all cribriform morphology as Gleason pattern 4, but supports the findings here that ICC is most prognostic in cases with very little overall pattern 4 morphology. Similarly, Kweldam et al [12] found that the presence of cribriform morphology was significantly associated with disease-specific death for patients with Gleason score 3 + 4, but not for those with Gleason score 4 + 3.
The prognostic value of ICC in Gleason grade group 2 is especially relevant in the context of identifying patients who would be candidates for active surveillance. While active surveillance has traditionally been restricted to patients with grade group 1 cancer [38], there is evidence that patients with grade group 2 disease may also benefit from more conservative management [39]. However, patients in this group have diverse outcomes, and identification of which patients are truly at low risk remains a challenge [40]. Automated ICC analysis could potentially serve as an additional determinant of active surveillance eligibility, as previous studies using human readers have recommended [41].
There has recently been a surge in interest in machine learning applications, both handcrafted and DL-based, for digital pathology. These include automated prostate cancer detection and Gleason grading [17–20] and outcome prognosis [21,22,42–44]. The black-box nature of DL approaches poses a challenge to their validation and certification, especially since site-specific differences in specimen preparation may affect the model in unexpected ways that are difficult to detect. Although DL was used in this study, it was not applied for directly prognosticating outcome, but for defining CAI. In this way, the prognostic power of CAI is rooted in previous work establishing the utility of ICC, with the segmentation results being straightforward to scrutinize and evaluate.
CAI also added value to a BCR prognosis model, the LPM based on features of gland lumen morphology, and revealed an ultra-high-risk group among patients who were both identified as at high risk by the LPM and had CAI >0.10. In this case, the ability to simultaneously quantify lumen morphology and recognize a specific tissue pattern—ICC—improved the prognostic performance.
This study does have some limitations. While the training data set was annotated for ICC morphology, immunohistochemistry was not available for differentiating ICC from other cribriform patterns or intraductal carcinoma. As ICC identification has imperfect concordance, it is possible that a model trained on a different pathologist’s annotations would perform differently. However, this may not pose a problem, since CAI was validated for prognosis rather than agreement with pathologists. Although a head-to-head comparison was not performed, CAI is more efficient and reproducible in quantifying ICC extent than pathologist assessments. In addition, since the original Gleason grade was used for each slide, not all slides were graded according to the latest guidelines [28]. This potentially affected the results for CAI within grade groups, but appears unlikely to have had an impact on the overall conclusion that CAI was prognostic of BCR. Finally, cribriform content was not a criterion for slide selection, and it is possible that some patients would have had very different CAI values if all slides had been considered. The results of this study may encourage undertaking of the large effort needed to digitize all the slides for many cases for a more comprehensive assessment and characterization of cribriform patterns.
5. Conclusions
In this study, tumor ICC content, as quantified via a deep learning model, was prognostic of BCR, with more ICC associated with higher risk of BCR. In addition, ICC content was most prognostic in Gleason grade group 2, an intermediate-risk group that may benefit from additional prognostic markers, especially in the active surveillance setting. This suggests that analysis of cribriform morphology may be included in future prognostic tools for prostate cancer, particularly ones that rely solely on automated visual analysis. CAI may also add prognostic value to existing postoperative nomograms [1,45] through reproducible quantification of ICC area.
Supplementary Material
Acknowledgments
Funding/Support and role of the sponsor: Research reported in this publication was supported by the National Cancer Institute (award numbers 1U24CA199374- 01, R01CA202752-01A1, R01CA208236-01A1, R01 CA216579-01A1, R01 CA220581-01A1, 1U01 CA239055-01, 1U01CA248226-01, and 1U54CA254566-01), the National Heart, Lung and Blood Institute (1R01HL15127701A1), the National Institute for Biomedical Imaging and Bioengineering (1R43EB028736-01), the National Center for Research Resources (1 C06 RR12463-01), VA Merit Review award IBX004121A from the United States Department of Veterans Affairs Biomedical Laboratory Research and Development Service, the Office of the Assistant Secretary of Defense for Health Affairs, through the Breast Cancer Research Program (W81XWH-19-1-0668), the Prostate Cancer Research Program (W81XWH-15-1-0558, W81XWH-20-1-0851), the Lung Cancer Research Program (W81XWH-18-1-0440, W81XWH-20-1-0595), the Peer Reviewed Cancer Research Program (W81XWH-18-1-0404), the Kidney Precision Medicine Project (KPMP) Glue Grant, the Ohio Third Frontier Technology Validation Fund, the Clinical and Translational Science Collaborative of Cleveland (UL1TR0002548) from the National Center for Advancing Translational Sciences (NCATS) component of the National Institutes of Health and NIH roadmap for Medical Research, The Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering at Case Western Reserve University, and the National Science Foundation Graduate Research Fellowship Program (CON501692). The sponsors played no direct role in the study. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the U.S. Department of Veterans Affairs, the Department of Defense, the National Science Foundation, or the United States Government.
Financial disclosures: Anant Madabhushi certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: Andrew Janowczyk reports a consulting or advisory role with Merck. Michael Feldman reports consulting fees from Philips Digital Pathology. Anant Madabhushi reports holding equity in Elucid Bioimaging and in Inspirata Inc., has served as a scientific advisory board member for Inspirata Inc, AstraZeneca, Bristol Meyers-Squibb, and Merck, currently serves on the advisory board of Aiforia Inc., has sponsored research agreements with Philips, AstraZeneca, Boehringer-Ingelheim, and Bristol Meyers-Squibb, has technology licensed to Elucid Bioimaging, and is involved in an NIH U24 grant with PathCore Inc. and three different R01 grants with Inspirata Inc. Sacheth Chandramouli, Patrick Leo, Andrew Janowczyk, Kaustav Bera, Michael Feldman, Sanjay Gupta, and Anant Madabhushi report patents pending or awarded for automated cancer diagnosis and prognosis from digital pathology images. The remaining authors have nothing to disclose.
Footnotes
Data sharing statement: With the exception of TCGA cases, images used in this study are covered by material transfer agreements precluding sharing of this material. Model results for TCGA cases can be requested from the corresponding author.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stephenson AJ, Scardino PT, Eastham JA, et al. Postoperative nomogram predicting the 10-year probability of prostate cancer recurrence after radical prostatectomy. J Clin Oncol 2005;23:7005–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gleason DF, Mellinger GT. Prediction of prognosis for prostatic adenocarcinoma by combined histological grading and clinical staging. J Urol 1974;111:58–64. [DOI] [PubMed] [Google Scholar]
- 3.Epstein JI, Allsbrook WC, Amin MB, Egevad LL. The 2005 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason grading of prostatic carcinoma. Am J Surg Pathol 2005;29:1228–42. [DOI] [PubMed] [Google Scholar]
- 4.Trudel D, Downes MR, Sykes J, Kron KJ, Trachtenberg J, van der Kwast TH. Prognostic impact of intraductal carcinoma and large cribriform carcinoma architecture after prostatectomy in a contemporary cohort. Eur J Cancer 2014;50:1610–6. [DOI] [PubMed] [Google Scholar]
- 5.Kweldam CF, Wildhagen MF, Steyerberg EW, Bangma CH, van der Kwast TH, van Leenders GJ. Cribriform growth is highly predictive for postoperative metastasis and disease-specific death in Gleason score 7 prostate cancer. Mod Pathol 201428:457–64. [DOI] [PubMed] [Google Scholar]
- 6.Iczkowski KA, Torkko KC, Kotnis GR, et al. Digital quantification of five high-grade prostate cancer patterns, including the cribriform pattern, and their association with adverse outcome. Am J Clin Pathol 2011;136:98–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong F, Yang P, Wang C, et al. Architectural heterogeneity and cribriform pattern predict adverse clinical outcome for Gleason grade 4 prostatic adenocarcinoma. Am J Surg Pathol 2013;37:1855–61. [DOI] [PubMed] [Google Scholar]
- 8.Choy B, Pearce SM, Anderson BB, et al. Prognostic significance of percentage and architectural types of contemporary Gleason pattern 4 prostate cancer in radical prostatectomy. Am J Surg Pathol 2016;40:1400–6. [DOI] [PubMed] [Google Scholar]
- 9.McKenney JK, Wei W, Hawley S, et al. Histologic grading of prostatic adenocarcinoma can be further optimized. Am J Surg Pathol 2016;40:1439–56. [DOI] [PubMed] [Google Scholar]
- 10.Siadat F, Sykes J, Zlotta AR, et al. Not all Gleason pattern 4 prostate cancers are created equal: a study of latent prostatic carcinomas in a cystoprostatectomy and autopsy series. Prostate 2015;75:1277–84. [DOI] [PubMed] [Google Scholar]
- 11.Kir G, Sarbay B, Gümüş E, Topal C. The association of the cribriform pattern with outcome for prostatic adenocarcinomas. Pathol Res Pract 2014;210:640–4. [DOI] [PubMed] [Google Scholar]
- 12.Kweldam CF, Kmmerlin IP, Nieboer D, et al. Disease-specific survival of patients with invasive cribriform and intraductal prostate cancer at diagnostic biopsy. Mod Pathol 2016;29:630–6. [DOI] [PubMed] [Google Scholar]
- 13.Kweldam CF, Nieboer D, Algaba F, et al. Gleason grade 4 prostate adenocarcinoma patterns: an interobserver agreement study among genitourinary pathologists. Histopathology 2016;69:441–9. [DOI] [PubMed] [Google Scholar]
- 14.Lee TK, Ro JY. Spectrum of cribriform proliferations of the prostate: from benign to malignant. Arch Pathol Lab Med 2018;142:938–46. [DOI] [PubMed] [Google Scholar]
- 15.Kweldam CF, van Leender GJ, van der Kwast T. Grading of prostate cancer: a work in progress. Histopathology 2018;74:146–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leo P, Elliott R, Shih NNC, Gupta S, Feldman M, Madabhushi A. Stable and discriminating features are predictive of cancer presence and Gleason grade in radical prostatectomy specimens: a multi-site study. Sci Rep 2018;8:14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pantanowitz L, Quiroga-Garza GM, Bien L, et al. An artificial intelligence algorithm for prostate cancer diagnosis in whole slide images of core needle biopsies: a blinded clinical validation and deployment study. Lancet Digital Health 2020;2:e407–16. [DOI] [PubMed] [Google Scholar]
- 18.Nagpal K, Foote D, Liu Y, et al. Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. NPJ Digit Med 2018;2:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ström P, Kartasalo K, Olsson H, et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: a population-based, diagnostic study. Lancet Oncol 2020;21:222–32. [DOI] [PubMed] [Google Scholar]
- 20.Bulten W, Pinckaers H, van Boven H, et al. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: a diagnostic study. Lancet Oncol 2020;21:233–41. [DOI] [PubMed] [Google Scholar]
- 21.Lee G, Sparks R, Ali S, et al. Co-occurring gland angularity in localized subgraphs: predicting biochemical recurrence in intermediate-risk prostate cancer patients., PLoS One 2014;9:e97954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Artificial intelligence in digital pathology — new tools for diagnosis and precision oncology. Nat Rev Clin Oncol 2019;16:703–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knezevic D, Goddard AD, Natraj N, et al. Analytical validation of the Oncotype DX prostate cancer assay - a clinical RT-PCR assay optimized for prostate needle biopsies, BMC Genomics 2013;14:690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein EA, Cooperberg MR, Magi-Galluzzi C, et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of Gleason grade heterogeneity, tumor multifocality, and biopsy undersampling, Eur Urol 2014;66:550–60. [DOI] [PubMed] [Google Scholar]
- 25.Marrone M, Potosky AL, Penson D, Freedman AN. A 22 gene-expression assay, Decipher® (GenomeDx Biosciences) to predict five-year risk of metastatic prostate cancer in men treated with radical prostatectomy. PLoS Curr 2015;7:ecurrents.eogt.761b81608129ed61b0b48d42c04f92a4. 10.1371/currents.eogt.761b81608129ed61b0b48d42c04f92a4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. Lect Notes Comput Sci 2015;9351:234–41. [Google Scholar]
- 27.Leo P, Janowczyk A, Elliott R, et al. Computer extracted gland features from H&E predicts prostate cancer recurrence comparably to a genomic companion diagnostic test: a large multi-site study. NPJ Precis Oncol. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Humphrey PA, Moch H, Cubilla AL, Ulbright TM, Reuter VE. The 2016 WHO classification of tumours of the urinary system and male genital organs—part B: prostate and bladder tumours. Eur Urol 2016;70:106–19. [DOI] [PubMed] [Google Scholar]
- 29.Leo P, Lee G, Shih NNC, Elliott R, Feldman MD, Madabhushi A. Evaluating stability of histomorphometric features across scanner and staining variations: prostate cancer diagnosis from whole slide images. J Med Imaging 2016;3:047502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolla M, van Poppel H, Tombal B, et al. Postoperative radiotherapy after radical prostatectomy for high-risk prostate cancer: long-term results of a randomised controlled trial (EORTC trial 22911). Lancet 2012;380:2018–27. [DOI] [PubMed] [Google Scholar]
- 31.Dignam JJ, Hamstra DA, Lepor H, et al. Time interval to biochemical failure as a surrogate end point in locally advanced prostate cancer: analysis of randomized trial NRG/RTOG 9202. J Clin Oncol 2019:37:213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKenney JK, Simko J, Bonham M, et al. The potential impact of reproducibility of Gleason grading in men with early stage prostate cancer managed by active surveillance: a multi-institutional study. J Urol 2011;186:465–9. [DOI] [PubMed] [Google Scholar]
- 33.Ozkan TA, Eruyar AT, Cebeci OO, Memik O, Ozcan L, Kuskonmaz I. Interobserver variability in Gleason histological grading of prostate cancer, Scand J Urol 2016;50:420–4. [DOI] [PubMed] [Google Scholar]
- 34.Iczkowski KA, Paner GP, van der Kwast T. The new realization about cribriform prostate cancer. Adv Anat Pathol 2018;25:31–7. [DOI] [PubMed] [Google Scholar]
- 35.Hollemans E, Verhoef EI, Bangma CH, et al. Large cribriform growth pattern identifies ISUP grade 2 prostate cancer at high risk or recurrence and metastasis, Mod Pathol 2018;32:139–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Leenders GJ, van der Kwast TH, Grignon DJ, et al. The 2019 International Society of Urological Pathology (ISUP) Consensus Conference on grading of prostatic carcinoma. Am J Surg Pathol 2020;44:e87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Epstein JI, Amin MB, Fine SW, et al. The 2019 Genitourinary Pathology Society (GUPS) white paper on contemporary grading of prostate cancer, Arch Pathol Lab Med 2021;145:461–93. [DOI] [PubMed] [Google Scholar]
- 38.Newcomb LF, Brooks JD, Carroll PR, et al. Canary Prostate Active Surveillance Study: design of a multi-institutional active surveillance cohort and biorepository. Urology 2010;75:407–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ploussard G, Isbarn H, Briganti A, et al. Can we expand active surveillance criteria to include biopsy Gleason 3+4 prostate cancer? A multi-institutional study of 2,323 patients. Urol Oncol 2015;33:71.e1–9. [DOI] [PubMed] [Google Scholar]
- 40.Morlacco A, Cheville JC, Rangel LJ, Gearman DJ, Karnes RJ. Adverse disease features in Gleason score 3 + 4 “favorable intermediate-risk” prostate cancer: implications for active surveillance. Eur Urol 2017;72:442–7. [DOI] [PubMed] [Google Scholar]
- 41.Keefe DT, Schieda N, Hallani SE, et al. Cribriform morphology predicts upstaging after radical prostatectomy in patients with Gleason score 3 + 4 = 7 prostate cancer at transrectal ultrasound (TRUS)-guided needle biopsy. Virchows Arch 2015;467:437–42. [DOI] [PubMed] [Google Scholar]
- 42.Cruz-Roa AA, Arevalo Ovalle JE, Madabhushi A, González Osorio FA. A deep learning architecture for image representation, visual interpretability and automated basal-cell carcinoma cancer detection. Lect Notes Comput Sci 2013;8150:403–10. [DOI] [PubMed] [Google Scholar]
- 43.Basavanhally A, Ganesan S, Feldman MD, et al. Multi-field-of-view framework for distinguishing tumor grade in ER+ breast cancer from entire histopathology slides. IEEE Trans Biomed Eng 2013;60:2089–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis JS, Ali S, Luo J, Thorstad WL, Madabhushi A. A quantitative histomorphometric classifier (QuHbIC) identifies aggressive versus indolent p16-positive oropharyngeal squamous cell carcinoma. Am J Surg Pathol 2014;38:128–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cooperberg MR, Hilton JF, Carroll PR. The CAPRA-S score. Cancer 2011;117:5039–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.