Abstract
Previous studies have shown that microRNA‐206 (miR‐206) exhibits anti‐tumour properties in various tumours. Nevertheless, diagnostic significance of miR‐206 in oral cancer is still poorly known. Our research was carried out to explore the performance of miR‐206 in the diagnosis of oral cancer. Quantitative real‐time polymerase chain reaction (qRT‐PCR) method was adopted to measure the level of miR‐206 in serum specimens from oral cancer cases and control individuals. Chi‐square test was performed to analyse the correlation between miR‐206 level and clinicopathological parameters of the cases. Receiver operating characteristic (ROC) curve was constituted to assess diagnostic accuracy of miR‐206 in oral cancer. Serum miR‐206 level in oral cancer patients was significantly lower than that in control individuals (P < .001). miR‐206 expression was obviously related to T classification (P = .033), TNM stage (P = .008) and lymph node metastasis (P = .028). The area under the curve (AUC) of the ROC curve was 0.846 (95% CI = 0.797‐0.896, P < .001) with a specificity of 72.7% and a sensitivity of 81.2%. It revealed that miR‐206 might be a non‐invasive indicator in differentiating oral cancer cases from control individuals. Down‐regulation of miR‐206 is related to the development of oral cancer. Serum miR‐206 might be an effective indicator for early detection of oral cancer.
Keywords: diagnosis, MiR‐206, oral cancer
1. INTRODUCTION
Oral cancer, a frequent tumour in the world, belongs to head and neck tumours.1 In recent years, the morbidity rate of oral cancer is increasing, which seriously affects the quality of the patients' life.2 The aetiology of oral cancer is complicated, and relevant aetiological factors of this tumour include smoking, alcohol consumption, the infection of human papillomavirus and genetic and environmental elements.3 However, since most cases are diagnosed at advanced stages, overall survival rate of the malignancy is still unsatisfactory.4 Surgery, chemotherapy and radiotherapy are main treatments for oral cancer, but there are some limitations in these methods.5 Thus, searching for novel diagnostic factors and therapeutic targets is an urgent need for oral cancer.
MicroRNAs (MiRNAs) are a group of short and conservative non‐coding RNAs, which play important roles in multiple biological process.6 Accumulated evidence supported that the dysregulation of miRNAs was related to the pathogenesis and progression of human cancers.7 MicroRNA‐206 (miR‐206), seating on human chromosome 6p12.2, was considered to be a tissue‐specific miRNA associated with the differentiation of skeletal muscle.8 More and more proofs unveiled that miR‐206 acted as an anti‐oncogene and its decreased level was detected in many human tumours.9, 10 Previous studies suggested that high levels of miR‐206 could suppress tumour cell growth, migration and invasion, and induced their apoptosis.11, 12 Besides, miR‐206 was proven to hold certain significance in the diagnosis, treatment and prognosis of tumours.13, 14 Nevertheless, diagnostic ability of miR‐206 in oral cancer has not been studied.
In our research, we detected the level of miR‐206 in serum among oral cancer cases and analysed the correlation between miR‐206 level and clinicopathological parameters of the cases. The potential of serum miR‐206 as a diagnostic indicator for oral cancer has also been studied.
2. MATERIALS AND METHODS
2.1. Participants and specimens
Our research was authorized by the Ethical Committee of Chinese PLA General Hospital (No. JSH2016OSCC002). All patients for our study signed written informed consent. We enrolled 132 oral cancer patients who were histopathologically diagnosed by experienced pathologists at Chinese PLA General Hospital. Patients were more than 18 years older and had no history of other cancers or oral diseases. Before our research, cases had not received any radio‐ or chemotherapy treatments. Besides, 85 healthy people served as controls.
5 mL peripheral blood was obtained from each participant after overnight fasting. Then, serum was separated from blood sample through centrifugation at 1500 g for 10 min. Supernate was at once kept at −80℃ until use. Clinical data of the cases are listed in Table 1, including age, gender, T classification, histological grade, TNM stage and lymph node metastasis.
TABLE 1.
Relationship between miR‐206 expression and clinical features of oral cancer patients
| Clinical features | Cases (n = 132) | miR‐206 expression | χ 2 | P | |
|---|---|---|---|---|---|
| High (n = 59) | Low (n = 73) | ||||
| Age (years) | |||||
| ≤57 | 70 | 32 | 38 | 0.062 | .803 |
| >57 | 62 | 27 | 35 | ||
| Gender | |||||
| Male | 87 | 36 | 51 | 1.136 | .286 |
| Female | 45 | 23 | 22 | ||
| Smoking | |||||
| No | 51 | 26 | 25 | 1.327 | .249 |
| Yes | 81 | 33 | 48 | ||
| Alcohol consumption | |||||
| No | 67 | 34 | 33 | 2.014 | .156 |
| Yes | 65 | 25 | 40 | ||
| T classification | |||||
| T1‐T2 | 76 | 40 | 36 | 4.563 | .033 |
| T3‐T4 | 56 | 19 | 37 | ||
| Histological grade | |||||
| Well/moderate | 79 | 37 | 42 | 0.364 | .546 |
| Poor | 53 | 22 | 31 | ||
| TNM stage | |||||
| Ⅰ‐Ⅱ | 68 | 38 | 30 | 7.099 | .008 |
| Ⅲ‐Ⅳ | 64 | 21 | 43 | ||
| Lymph node metastasis | |||||
| Negative | 85 | 44 | 41 | 4.824 | .028 |
| Positive | 47 | 15 | 32 | ||
2.2. QRT‐PCR
According to the protocol of the manufacturer, miRNeasy mini kit (Qiagen) was employed to isolate miRNAs from serum samples. The quality and quantity of RNA were detected via NanoDrop 2000 Spectrophotometer (NanoDrop Technologies). TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) was adopted to perform reverse transcription. Primers for reverse transcription contained 5′‐CTCAGCGGCTGTCGTGGACTGCGCGCTGCCGCTGAGCCACACAC‐3′ for miR‐206 and 5′‐CTCGCTTCGGCAGCACA‐3′ for U6. qRT‐PCR was conducted with 40 cycles of denaturing at 95°C for 15 s, annealing at 60°C for 20 s and extending at 72°C for 20 s, utilizing SYBR Real‐Time PCR kit (GenePharma) with 7500 Real‐Time PCR system (Applied Biosystems) based on instruction book. U6 acted as endogenous reference. Primer sequences were as follows: miR‐206, sense‐5′‐CGTCAGAAGGAATGATGCACAG‐3′ and anti‐sense‐5′‐ACCTGCGTAGGTAGTTTCATGT‐3′; and U6, sense‐5′‐CTCGCTTCGGCAGCACA‐3′ and anti‐sense‐5′‐AACGCTTCACGAATTTGCGT‐3′. Relative quantification of miR‐206 expression was analysed via 2−ΔΔCt method. All data analysed were from three independent experiments repeated in triplicate.
2.3. Statistical analysis
We used SPSS 18.0 software and GraphPad Prism 5.0 software to analyse all data. Data were presented as mean ± standard deviation (SD). Different levels of miR‐206 between oral cancer specimens and matched control specimens were compared with Student's t test. The correlation between miR‐206 level and clinicopathological parameters of the cases was assessed by chi‐square test. Receiver operating characteristic (ROC) curve was constructed to assess diagnostic ability of miR‐206 in oral cancer. P < .05 was seen as significant threshold.
3. RESULTS
3.1. Basic characteristics of enrolled participants
Clinical characteristics of oral cancer patients are shown in Table 1. 45 women and 87 men with a mean age of 57.39 ± 19.28 years (range, 41‐78 years) were enrolled in our research. There were 81 smokers and 65 drinkers. 76 patients had T classification of T1‐T2 and 56 at T3‐T4. Histological grade was well/moderate in 79 patients and poor in the remaining 53 ones. According to TNM staging system, 68 patients were at stage Ⅰ‐Ⅱ and 64 at stage Ⅲ‐Ⅳ. Moreover, among these patients, 47 had positive lymph node metastasis.
3.2. Serum miR‐206 level was reduced in oral cancer cases
Quantitative real‐time polymerase chain reaction was adopted to measure the level of serum miR‐206 in oral cancer cases and control volunteers. The results revealed that the expression of serum miR‐206 in oral cancer cases was significantly lower than that in control volunteers (P < .001; Figure 1).
FIGURE 1.
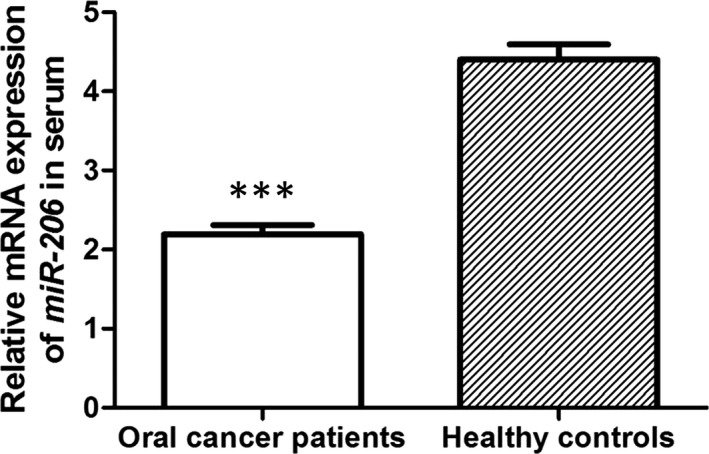
Relative expression levels of miR‐206 mRNA in oral cancer patients and healthy individuals. Compared with healthy controls, serum miR‐206 level was significantly down‐regulated in oral cancer patients at mRNA level. *** indicated P < .001
3.3. Association between miR‐206 level and clinicopathological parameters of oral cancer cases
Using the mean serum miR‐206 level as a cut‐off value, 132 oral cancer cases were classified into high miR‐206 level group (n = 59) and low miR‐206 level group (n = 73). The correlation between miR‐206 level and clinicopathological parameters of cancer cases was analysed via chi‐square test. The results indicated that miR‐206 level was obviously related to T classification (P = .033), TNM stage (P = .008) and lymph node metastasis (P = .028) (Table 1). Nevertheless, no significant correlation was found between miR‐206 level and age, gender, smoking, alcohol consumption or histological grade (all, P > .05; Table 1).
3.4. Diagnostic ability of miR‐206 in oral cancer cases
Receiver operating characteristic curve analysis was conducted to investigate whether miR‐206 could be adopted in the diagnosis of oral cancer. As shown in Figure 2, the area under the curve (AUC) was 0.846 (95% CI = 0.797‐0.896, P < .001) with a sensitivity of 81.2% and a specificity of 72.7%. The optimal cut‐off value for serum miR‐206 level to diagnose the disease was 2.88. Serum miR‐206 might be a valuable indicator for the diagnosis of oral cancer cases.
FIGURE 2.
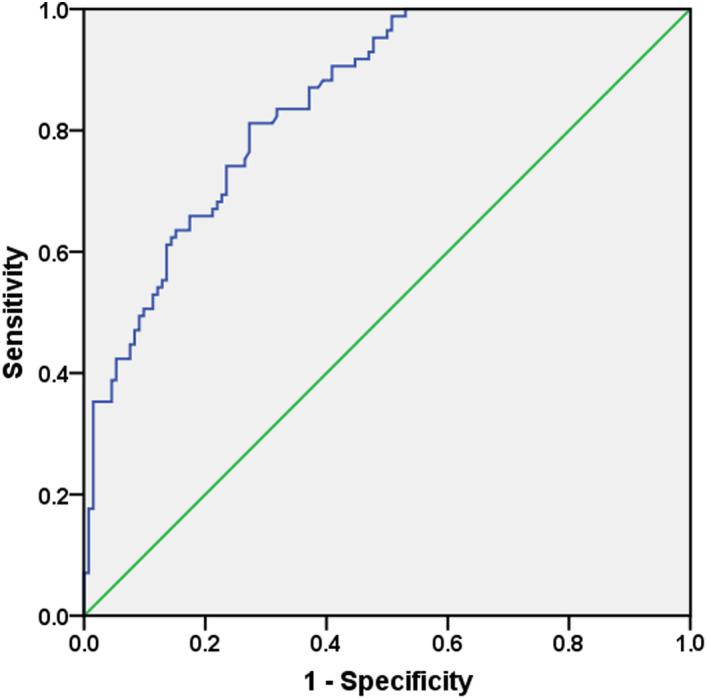
ROC curve analysis of serum miR‐206 in the diagnosis of oral cancer patients. The AUC was 0.846 with a sensitivity of 81.2% and a specificity of 72.7%
4. DISCUSSION
Accumulated documents reported that miRNAs were related to carcinogenesis and the progression of several tumours via acting as cancer genes or anti‐oncogenes.15 Nevertheless, their functions in oral cancer are not fully understood. In our research, we proved that serum miR‐206 level was significantly reduced in oral cancer patients compared to healthy controls. We also assessed the correlation between miR‐206 level and clinicopathological parameters of the cases. As a result, patients with low level of miR‐206 more frequently faced advanced T classification, advanced TNM stage and positive lymph node metastasis, all of which were aggressive clinicopathological parameters representing advanced development and metastasis of tumours. In ROC curve analysis, serum miR‐206 functioned as a forceful indicator to distinguish oral cancer cases from control people with satisfactory AUC, sensitivity and specificity.
miR‐206 belongs to miR‐1 family which includes muscle‐specific miRNAs and is involved in the development of muscle.16 More and more proofs indicated that miR‐206 was abnormally expressed in many cancers. For example, Chen et al reported that the level of miR‐206 was reduced in lung adenocarcinoma tissue samples and cells and that its up‐regulation could suppress cell viability, migration and angiogenesis.17 Liang et al suggested that miR‐206 was down‐regulated in triple‐negative breast cancer tissue samples and cells. Heightened level of miR‐206 might be involved in the invasion and angiogenesis of breast cancer.18 Moreover, Bao et al proved that decreased level of miR‐206 was closely related to advanced clinical stage and T classification and positive metastasis in osteosarcoma patients.19 According to Yunqiao et al, low miR‐206 level occurred more often in hepatocellular carcinoma patients with positive lymph node metastasis and advanced TNM stage.12 Similar to above‐mentioned achievements, our findings revealed that miR‐206 could serve as an anti‐oncogene in oral cancer and participate in the disease pathogenesis and development. However, serum miR‐206 level was not related to histological grade of oral cancer patients in our research, which was inconsistent with previous findings.20, 21 The role of miR‐206 in different cancers might be varied. Besides, sample size and source might cause differences as well. Considering these limitations, further studies are required.
Accumulated data suggested that miR‐206 was related to overall survival, disease‐free survival and prognosis of many cancers, such as oral squamous cell carcinoma.22, 23, 24 Nevertheless, diagnostic ability of miR‐206 in oral cancer has not been studied. MiRNAs are stable and easily detected in some body fluids, such as blood, serum and plasma, suggesting that they could become non‐invasive indicators for the diagnosis and prognosis of tumours.25 A research scheduled by Tan et al mentioned that circulating miR‐206 could be a potential diagnostic factor for hepatocellular carcinoma.26 Such result was consistent with ours that diagnostic ability of serum miR‐206 was strong in oral cancer.
In oestrogen receptor–positive breast cancer, TGF‐β signalling pathway was a target of miR‐206 in inhibiting epithelial mesenchymal transition.27 Cai et al proved that miR‐206 inhibited the growth, migration and invasion of renal cell carcinoma cells via targeting VEGF‐A directly.28 Based to earlier researches, we hypothesized that miR‐206 participated in tumorigenesis and disease progression via regulating diverse genes in different processes of human tumours. Nevertheless, exact molecular mechanism of miR‐206 functioning in oral cancer remains unclear.
In conclusion, serum miR‐206 level is reduced in oral cancer patients and related to the development of this tumour. Moreover, serum miR‐206 could be an effective bio‐marker for early diagnosis of oral cancer.
CONFLICT OF INTEREST
None.
AUTHOR CONTRIBUTIONS
L.W. conceived and designed the experiments; L.W., H.S. conceived and performed the experiments; S.Y. prepared figures. H.S., S.Y. wrote the main manuscript text. All authors reviewed the manuscript.
Wang L, Song H, Yang S. MicroRNA‐206 has a bright application prospect in the diagnosis of cases with oral cancer. J Cell Mol Med. 2021;25:8169–8173. 10.1111/jcmm.16598
Funding information
The 63rd batch of grants from China Post‐doctoral Science Fund (2018M633673). The 11th batch of special grants from China Post‐doctoral Science Fund (2018T111137)
Contributor Information
Lili Wang, Email: tejftnd@163.com.
Shiming Yang, Email: ppuirvgjfh@163.com.
DATA AVAILABILITY STATEMENT
All data generated or analysed in our research are included in this article.
REFERENCES
- 1.Chang CF, Chen SL, Sung WW, et al. PBK/TOPK expression predicts prognosis in oral cancer. Int J Mol Sci. 2016;17:1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faden AA. The potential role of microbes in oncogenesis with particular emphasis on oral cancer. Saudi Med J. 2016;37:607‐612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou Y, Zhang H, Zhuo X, Liu Y, Zhang G, Tan Y. Over‐expression of TWIST, an epithelial‐mesenchymal transition inducer, predicts poor survival in patients with oral carcinoma. Int J Clin Exp Med. 2015;8:9239‐9247. [PMC free article] [PubMed] [Google Scholar]
- 4.Lokesh K, Kannabiran J, Rao MD. Salivary lactate dehydrogenase (LDH)‐ A novel technique in oral cancer detection and diagnosis. J Clin Diagn Res. 2016;10:ZC34‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tachibana H, Sho R, Takeda Y, et al. Circulating miR‐223 in oral cancer: its potential as a novel diagnostic biomarker and therapeutic target. PLoS ONE. 2016;11:e0159693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu WF, Wang HM, Lu BC, Zhang GZ, Ma HM, Wu ZY. miR‐206 inhibits human laryngeal squamous cell carcinoma cell growth by regulation of cyclinD2. Eur Rev Med Pharmacol Sci. 2015;19:2697‐2702. [PubMed] [Google Scholar]
- 7.Sun C, Liu Z, Li S, et al. Down‐regulation of c‐Met and Bcl2 by microRNA‐206, activates apoptosis, and inhibits tumor cell proliferation, migration and colony formation. Oncotarget. 2015;6:25533‐25574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma G, Wang Y, Li Y, et al. MiR‐206, a key modulator of skeletal muscle development and disease. Int J Biol Sci. 2015;11:345‐352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao H, Xiao W, Cao J, et al. miR‐206 functions as a novel cell cycle regulator and tumor suppressor in clear‐cell renal cell carcinoma. Cancer Lett. 2016;374:107‐116. [DOI] [PubMed] [Google Scholar]
- 10.Chen QY, Jiao DM, Wu YQ, et al. MiR‐206 inhibits HGF‐induced epithelial‐mesenchymal transition and angiogenesis in non‐small cell lung cancer via c‐Met /PI3k/Akt/mTOR pathway. Oncotarget. 2016;7:18247‐18261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang YJ, Xu F, Li HB, Han JC, Li L. miR‐206 inhibits non small cell lung cancer cell proliferation and invasion by targeting SOX9. Int J Clin Exp Med. 2015;8:9107‐9113. [PMC free article] [PubMed] [Google Scholar]
- 12.Yunqiao L, Vanke H, Jun X, Tangmeng G. MicroRNA‐206, down‐regulated in hepatocellular carcinoma, suppresses cell proliferation and promotes apoptosis. Hepatogastroenterology. 2014;61:1302‐1307. [PubMed] [Google Scholar]
- 13.Mirzaei HR, Sahebkar A, Mohammadi M, et al. Circulating microRNAs in hepatocellular carcinoma: potential diagnostic and prognostic biomarkers. Curr Pharm Des. 2016;22:5257‐5269. [DOI] [PubMed] [Google Scholar]
- 14.Yang Q, Zhang C, Huang B, et al. Downregulation of microRNA‐206 is a potent prognostic marker for patients with gastric cancer. Eur J Gastro Hepatol. 2013;25:953‐957. [DOI] [PubMed] [Google Scholar]
- 15.Zhang C, Yao C, Li H, Wang G, He X. Serum levels of microRNA‐133b and microRNA‐206 expression predict prognosis in patients with osteosarcoma. Int J Clin Exp Pathol. 2014;7:4194‐4203. [PMC free article] [PubMed] [Google Scholar]
- 16.Tian R, Liu T, Qiao L, Gao M, Li J. Decreased serum microRNA‐206 level predicts unfavorable prognosis in patients with melanoma. Int J Clin Exp Pathol. 2015;8:3097‐3103. [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X, Tong ZK, Zhou JY, Yao YK, Zhang SM. MicroRNA‐206 inhibits the viability and migration of human lung adenocarcinoma cells partly by targeting MET. Oncol Lett. 2016;12:1171‐1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang Z, Bian X, Shim H. Downregulation of microRNA‐206 promotes invasion and angiogenesis of triple negative breast cancer. Biochem Biophys Res Comm. 2016;477:461‐466. [DOI] [PubMed] [Google Scholar]
- 19.Bao YP, Yi Y, Peng LL, et al. Roles of microRNA‐206 in osteosarcoma pathogenesis and progression. Asian Pac J Cancer Prev. 2013;14:3751‐3755. [DOI] [PubMed] [Google Scholar]
- 20.Ren XL, He GY, Li XM, et al. MicroRNA‐206 functions as a tumor suppressor in colorectal cancer by targeting FMNL2. J Cancer Res Clin Oncol. 2016;142:581‐592. [DOI] [PubMed] [Google Scholar]
- 21.Ling S, Ruiqin M, Guohong Z, Bing S, Yanshan C. Decreased microRNA‐206 and its function in cervical cancer. Eur J Gynaecol Oncol. 2015;36:716‐721. [PubMed] [Google Scholar]
- 22.Wang S, Lu S, Geng S, Ma S, Liang Z, Jiao B. Decreased expression of microRNA‐206 correlates with poor clinical outcome in patients with malignant astrocytomas. Pathol Oncol Res. 2014;20:343‐348. [DOI] [PubMed] [Google Scholar]
- 23.Sun P, Sun D, Wang X, Liu T, Ma Z, Duan L. miR‐206 is an independent prognostic factor and inhibits tumor invasion and migration in colorectal cancer. Cancer Biomark. 2015;15:391‐396. [DOI] [PubMed] [Google Scholar]
- 24.Lin F, Yao L, Xiao J, Liu D, Ni Z. MiR‐206 functions as a tumor suppressor and directly targets K‐Ras in human oral squamous cell carcinoma. Onco Targets Ther. 2014;7:1583‐1591. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Lu YC, Chang JT, Huang YC, et al. Combined determination of circulating miR‐196a and miR‐196b levels produces high sensitivity and specificity for early detection of oral cancer. Clin Biochem. 2015;48:115‐121. [DOI] [PubMed] [Google Scholar]
- 26.Tan Y, Ge G, Pan T, et al. A serum microRNA panel as potential biomarkers for hepatocellular carcinoma related with hepatitis B virus. PLoS ONE. 2014;9:e107986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin K, Yin W, Wang Y, et al. MiR‐206 suppresses epithelial mesenchymal transition by targeting TGF‐beta signaling in estrogen receptor positive breast cancer cells. Oncotarget. 2016;7:24537‐24548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai Y, Li H, Zhang Y. Downregulation of microRNA‐206 suppresses clear cell renal carcinoma proliferation and invasion by targeting vascular endothelial growth factor A. Oncol Rep. 2016;35:1778‐1786. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed in our research are included in this article.


