Abstract
Hepatitis C virus (HCV) cure after all-oral direct-acting antiviral (DAA) therapy greatly improves the liver and immune system. We aimed to assess the impact of this HCV clearance on immune system-related markers in plasma and the gene expression profile in human immunodeficiency virus (HIV)/HCV-coinfected patients with advanced cirrhosis. We performed a prospective study on 33 HIV/HCV-coinfected patients at baseline and 36 weeks after the sustained virological response. Gene expression was evaluated by RNA-seq analysis on peripheral blood mononuclear cells (PBMCs) and plasma biomarkers by multiplex immunoassays. We found a decrease in plasma biomarkers (PD1, PDL1, CXCL10, CXCL8, IL12p70, IL10, and TGFβ) and liver disease markers (stiffness measurement (LSM), hepatic venous pressure gradient (HVPG), and transaminases, among others). Furthermore, decreased plasma levels of CXCL8, CXCL10, IL10, and PD1 were associated with reduced LSM values. We also found two upregulated (HAS1 and IRG1) and 15 downregulated (CXCL11, CCL8, CCL7, CCL2, ADARB2, RRAD, MX1, SIGLEC1, IFI44L, IFI44, IFI27, IFI6, IFIT3, IFIT1B, and IFIT1) genes at the end of follow-up, all interferon-stimulated genes (ISGs) grouped into four pathways (“cytokine-cytokine receptor interaction”, “viral protein interaction with cytokine and cytokine receptor”, “chemokine signaling pathway”, and “hepatitis C”). Additionally, the decrease in most of these ISGs was significantly related to reduced LSM and HVPG values. In conclusion, HIV/HCV-coinfected patients with advanced-HCV-related cirrhosis who eradicated HCV following DAA therapy exhibited an improvement in liver disease markers and a significant decrease in plasma biomarkers and gene expression related to antiviral/inflammatory response, particularly in levels of several chemokines and ISGs.
Keywords: HIV/HCV coinfection, immune system, cirrhosis, DAA therapy, plasma biomarkers, PBMCs, gene expression
Introduction
Hepatitis C virus (HCV) infection causes chronic liver disease worldwide (1). Moreover, many people are coinfected with HCV and human immunodeficiency virus (HIV) (2). These individuals develop liver cirrhosis over decades that progress faster than in HCV-monoinfected patients (3). Likewise, HIV/HCV-coinfected individuals have higher rates of liver-related events (LRE) that included liver decompensation, chronic liver failure, hepatocellular carcinoma (HCC), liver transplantation, and liver-related deaths (4–7). Immunosuppression can explain this negative impact of HIV on chronic hepatitis C, but also because HIV can contribute to liver inflammation and fibrosis by the direct action of the virus itself on the hepatic stellate cells (8, 9), as well as factors related to lifestyle, such as alcohol consumption. Additionally, different extrahepatic manifestations associated with HCV, such as autoimmune, lymphoproliferative, metabolic, renal, cardiovascular, and central nervous system disorders, may also contribute to the morbimortality of HIV/HCV-coinfected patients (10).
HCV infection causes dysregulation of immune functions such as inflammation and immune activation and, together with other comorbidities, promotes cirrhosis progression and the development of LRE (7, 11, 12). Chronic HCV infection is characterized by an abundant hepatic and systemic production of inflammatory mediators and interferon-stimulated genes (ISGs) that cannot control the virus, indicating exhaustion of the interferon (IFN) system (13). Additionally, alterations in immune cells such as CD4+ and CD8+ T cells and natural killer (NK) cells follow HCV infection (14). For example, CD4+ and CD8+ lymphocytes show an exhausted phenotype with high levels of programmed death protein 1 (PD1), which stimulates their apoptosis (15–17). Also, a T-helper cell type 2 (Th2) predominance over a Th1 response has been proposed to promote chronic hepatitis C (18, 19). Moreover, HIV patients on suppressive antiretroviral therapy (ART) also show dysregulation of the immune system (7, 11), with very diverse alterations, including immune activation (20), inflammation (21), a deficit in T-cell functions (20, 22–26), and dysbiosis (27–29), which increase the risk of acquired immunodeficiency syndrome (AIDS), non-AIDS-related events, and death (30, 31).
Direct-acting antivirals (DAA) have transformed HCV therapy because almost all treated patients achieve a sustained virological response (SVR) (32–35). HCV eradication after DAA therapy decreases the risk of LRE in HCV-monoinfected patients with compensated cirrhosis, although this decrease has not been observed among those with decompensated cirrhosis (36–38). HIV/HCV-coinfected patients have a similar risk of LRE after successful DAA therapy as HCV-monoinfected individuals; however, the former group has a higher chance of non-liver-related death (39, 40). All this makes it necessary to study and monitor the cirrhotic patient after HCV eradication.
Peripheral blood transcriptome data provide crucial information on host immune response against pathogens, including HCV infection (41). For example, transcriptome analysis following successful DAA therapy offers a unique opportunity to analyze the possible normalization of the host response after years of chronic infection (42, 43). So far, only a few studies have explored the long-term impact of HCV clearance from DAA therapy on the peripheral blood transcriptome of HIV/HCV-coinfected patients (44, 45).
Objective
We aimed to evaluate the impact of HCV eradication following all-oral DAA therapy on immune system-related markers in plasma and the gene expression profile in peripheral blood mononuclear cells (PBMCs) in HIV/HCV-coinfected patients with advanced cirrhosis.
Patients and Methods
Study Subjects
We carried out a prospective study on 33 HIV/HCV-coinfected patients with advanced HCV-related cirrhosis who started anti-HCV therapy with all-oral DAA from four hospitals in Madrid (Spain) between January 2015 and June 2016 (ESCORIAL study; see Appendix). Our study was approved by the Research Ethics Committee of the Instituto de Salud Carlos III (CEI PI 41_2014) and was conducted according to Helsinki’s Declaration. All participants gave their signed written consent at the start of the study.
The inclusion criteria were: 1) chronic HCV and HIV infection; 2) prior history of liver decompensation (bleeding esophageal varices, ascites, hepatic encephalopathy) or existence of advanced cirrhosis (hepatic venous pressure gradient (HVPG) ≥10 mmHg, liver stiffness measurement (LSM) ≥25 kPa, or Child-Turcotte-Pugh (CTP) ≥7); 3) starting all-oral DAA therapy; 4) achieving SVR, defined as an undetectable HCV load at 12 weeks after completion of anti-HCV therapy; and 5) frozen PBMC samples available to perform RNA-seq at baseline and at the end of follow-up (36 weeks after SVR). Samples and clinical data are from baseline (HIV/HCV-b) and 36 weeks after SVR (HIV/HCV-f).
Additionally, we included two control groups to compare with HIV/HCV-f group: 1) 9 Chronic HCV-monoinfected patients (HCV-mono-f) with advanced HCV-related cirrhosis who achieved SVR and had the same follow-up (36 weeks after SVR). The HCV-monoinfected patients were selected as the control group for HIV/HCV-f because HCV-monoinfected patients become patients without chronic viral infection (both HIV and HCV negative) after reaching SVR but maintaining severe liver damage. 2) 26 HIV-monoinfected patients (HIV-mono) with undetectable HIV viral load and CD4+ >500 cells/µL (normal standard for HIV-infected patients). We selected the HIV-monoinfected patients as the control group for HIV/HCV-f because HIV/HCV-coinfected patients after reaching SVR become HIV-monoinfected patients.
Clinical Data And Samples
We collected clinical data prospectively using an online form. Later, the information was monitored to verify that the data collected matched the patient record. We calculated the CTP score from five factors (total bilirubin, international normalized ratio, albumin, ascites, and encephalopathy), ranging from 5 to 15 points. Trained operators assessed LSM using transient elastography (FibroScan®, Echosens, Paris, France), as previously described (46). LSM ranged from 2.5 to 75 kPa. After overnight fasting, the hemodynamic study to measure the HVPG was performed under light sedation with intravenous midazolam, as we previously described (47). The HVPG (expressed in mmHg) was the difference between wedged hepatic venous pressure and free hepatic venous pressure.
Peripheral venous blood samples were collected by venipuncture in ethylenediaminetetraacetic acid (EDTA) tubes. The same day, samples were sent to the HIV HGM BioBank (http://hivhgmbiobank.com/?lang=en) and were immediately processed by Ficoll-Paque density gradient, and PBMCs were stored in cryopreservation conditions (-180°C) in liquid nitrogen until analysis. Plasma was stored at -80°C.
Multiplex ELISA Assays
Plasma biomarkers (PD1, programmed cell death ligand 1 (PDL1), C-X-C motif chemokine ligand (CXCL)10, CXCL8, C-C motif chemokine ligand (CCL)2, IFNγ, interleukin (IL)12p70, IL2, IL10, and transforming growth factor-beta (TGFβ)) were evaluated with ProcartaPlex assays (Thermo Fisher Scientific Inc, Waltham, MA, USA) using a Bio-plex 200™ system (BioRad Laboratories Hercules, CA, USA) and according to the manufacturer’s specifications. As a high proportion of samples evaluated were below the lower limit of detection for absolute quantification, we used the raw fluorescence intensity values (a.u., arbitrary units) as a relative quantification of the analyte abundances, as previously described (48).
RNA Extraction, Library Preparation, and Sequencing
After blood extraction, PBMCs were isolated, and the RNeasy Minikit (Qiagen, Hilden, Germany) was used for total RNA purification. The RNA quantity and quality were assessed by Nanodrop 2000 and 2100 bioanalyzer RNA NANO assay (Agilent Technologies, CA, USA). The samples with an RNA integrity number over 7.5 were selected for sequencing. RNA samples were treated with RNase-free DNase set (Qiagen) following the manufacturer’s instructions. Library generation and sequencing of poly-A RNA were performed at the Centre for Genomic Regulation in Barcelona (Spain). Briefly, 500 nanograms of total RNA were used with Illumina’s TruSeq Stranded mRNA Sample Prep Kit v2 for library synthesis. This process allows capturing polyadenylated coding and non-coding RNAs. Ten libraries were multiplexed and pooled on each line of an Illumina HiSeq2500 sequencer, and a single read of 50 nts (1x50) was performed. We obtained an average of 25 million reads per sample.
Sequences were analyzed with a bioinformatic pipeline described in Supplementary Table 1. Briefly, for the read quality control, FastQC (v. 0.11.8) was used. Then, Trimmomatic (v. 0.38) was implemented for trimming the adapters. Reads were aligned to GRCH38 with STAR (v. 2.6.1d) software, and the Subread package of the FeatureCounts (v. 1.6.4) software was used for gene count estimation.
Genes Selected for Analysis
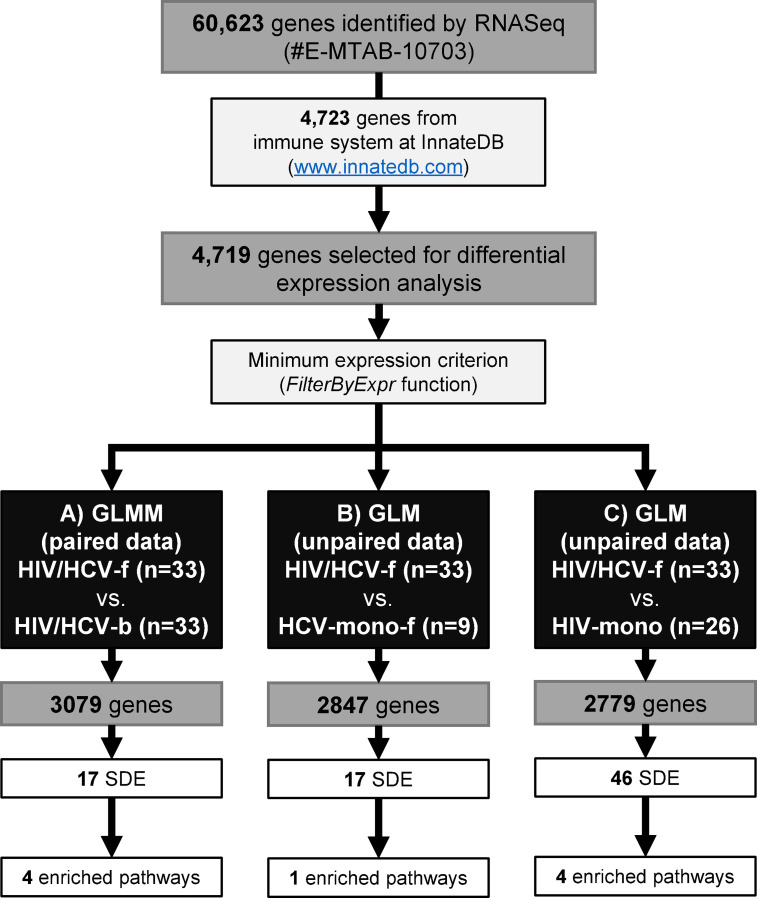
Figure 1 illustrates the workflow for the gene selection process. The bioinformatics pipeline identified a total of 60,623 different genes. Sequences are publicly available at ArrayExpress repository (EMBL-EBI; https://www.ebi.ac.uk/arrayexpress/) in raw format with the accession number E-MTAB-10703. Next, we selected 4,723 genes related to the immune system according to the public InnateDB database (www.innatedb.com), and 4,719 (99.9%) genes were identified in our sample database (Figure 1).
Figure 1.
Study design flowchart. RNA sequencing identified a total of 60,623 genes. Of these, we selected 4,723 genes of the immune system from the InnateDB. A total of 4,719 genes were selected for the differential expression analysis. Next, the FilterByExpr function was applied for each comparison: (A) HIV/HCV-f vs. HIV/HCV-b, 3,079 genes were selected, 17 were SDE genes involved in four biological pathways; (B) HIV/HCV-f vs. HCV-mono-f, 2,847 genes were selected, 17 were SDE genes involved in one biological pathway; (C) HIV/HCV-f vs. HIV-mono, 2,779 genes were selected, 46 were SDE genes involved in four biological pathways. HIV, human immunodeficiency virus; HCV, hepatitis C virus; HIV/HCV-f, HIV/HCV-coinfected patients 36 weeks after the sustained virological response (SVR); HCV-mono-f, HCV-monoinfected patients 36 weeks after SVR; HIV-mono, HIV-monoinfected patients; SDE, significantly differentially expressed; GLM, generalized linear model; GLMM, Generalized linear mixed model.
Statistical Analysis
The Statistical Package for the Social Sciences (SPSS) v22.0 software (IBM Corp., Chicago, IL, USA) was used to describe epidemiological and clinical variables among study groups at baseline and the end of follow-up.
We carried out the gene expression analysis using the R statistical package version v. 4.0.2 (R Foundation for Statistical Computing, Vienna, Austria). First, we normalized the counts by the Trimmed Mean of M (TMM) method for each of the comparisons and established an expression filter using the FilterByExpr function (R-package edgeR v. 3.32.1). Next, we analyzed the gene expression between groups by generalized linear models (GLM) for non-paired comparison and generalized linear mixed models (GLMM) for paired samples, using the R-package lme4 V. 1.1-23 and a negative binomial distribution. The differences between groups were evaluated by the fold-change (FC) and log2-FC for each gene. All p-values were two-tailed and corrected using the false discovery rate (FDR) with the Benjamini and Hochberg method (q-values) for multiple testing. We selected the significantly differentially expressed (SDE) genes according to FDR ≤0.05 and log2-FC ≤ -1 (2-fold downregulated) or log2-FC ≥1 (2-fold upregulated). Finally, we used TargetMine v2102 (https://targetmine.mizuguchilab.org/) to find enriched pathways and networks.
GLMM test was also used to evaluate the association between changes in biomarkers (plasma and PBMC gene expression) and liver disease scores during the follow-up in HIV/HCV-coinfected patients. The regression coefficient (β) gives the effect’s size and direction according to a positive or negative value.
Results
Patient Characteristics
Table 1 shows the epidemiological and clinical characteristics at baseline of 33 HIV/HCV-coinfected patients with advanced HCV-related cirrhosis, nine HCV-monoinfected patients, and 26 HIV-monoinfected patients. All patients infected with HIV were on ART and had an undetectable HIV viral load (<50 copies/mL).
Table 1.
Summary of patient characteristics at baseline.
| HIV/HCV-coinfected | HCV-mono | HIV-mono | |
|---|---|---|---|
| No. | 33 | 9 | 26 |
| Gender (male) | 25 (75.8%) | 7 (77.8%) | 16 (61.5%) |
| Age (years) | 51.5 (48.8; 53.1) | 63.5 (55.8; 71.2) | 51 (46; 53) |
| High alcohol intake | |||
| Previously (≥ 6 months) | 15 (45.5%) | 6 (66.7%) | 0 (0%) |
| Currently | 14 (42.4%) | 3 (33.3%) | 1 (3.8%) |
| IVDU | |||
| Previously (≥ 6 months) | 27 (81.8%) | 0 (0%) | 0 (0%) |
| Currently | 0 (0%) | 0 (0%) | 0 (0%) |
| Treatments | |||
| Previous IFNα therapy | 15 (45.5%) | 8 (88.9%) | – |
| DAAs regimens | |||
| Sofosbuvir + Ledipasvir | 14 (42.4%) | 3 (33%) | – |
| Sofosbuvir + Daclatasvir | 9 (27.3%) | 1 (11.1%) | – |
| Sofosbuvir + Daclatasvir + Simeprevir | 2 (6.1%) | 0 (0%) | – |
| Sofosbuvir + Simeprevir | 7 (21.2%) | 2 (22.2%) | – |
| Ombitasvir + Paritaprevir + Ritonavir + Dasabuvir | 1 (3%) | 3 (33%) | – |
| Antiretroviral therapy | |||
| NRTI+NNRTI-based | 5 (15.2%) | – | 16 (61.5%) |
| NRTI+II-based | 13 (39.4%) | – | 2 (7.7%) |
| NRTI+PI-based | 3 (9.1%) | – | 0 (0%) |
| PI+II+others-based | 4 (12.1%) | – | 0 (0%) |
| Others | 8 (24.2%) | – | 8 (30.8%) |
| HIV markers | |||
| Prior AIDS | 13 (39.4%) | – | 10 (38.5%) |
| Nadir CD4+ T-cells (cells/mm3) | 104.7 (70; 150) | – | 204 (99; 343) |
| Nadir CD4+ T-cells <200 cells/mm3 | 27 (90%) | – | 11 (44%) |
| CD4+ T-cells (cells/mm3) | 393 (234; 764) | – | 818 (685; 1036) |
| CD4+ T-cells <500 cells/mm3 | 19 (57.6%) | – | 0 (0%) |
| HIV-RNA >50 copies/mL | 0 (0%) | – | 0 (0%) |
| HCV markers | |||
| HCV genotype | |||
| 1 | 22 (68.8%) | 8 (88.9%) | – |
| 3 | 3 (9.4%) | 1 (11.1%) | – |
| 4 | 7 (21.9%) | 0 (0%) | – |
| Log10 HCV-RNA (IU/mL) | 6.3 (5.8; 6.7) | 6 (5.5; 6.2) | - |
| HCV-RNA ≥850.000 IU/mL | 24 (72.7%) | 5 (55.6%) | – |
| Liver disease | |||
| LSM (kPa) | 26.7 (19.2; 39.5) | 32.6 (22.3; 64) | – |
| HVPG (mmHg) | 14,5 (10; 17) | 17 (10; 20) | – |
Values are expressed as absolute number (percentage) and median (interquartile range).
HCV, hepatitis C virus; HIV, human immunodeficiency virus; IVDU, intravenous drug user; IFNα, interferon-alpha; DAAs, direct-acting antivirals; NRTI, nucleoside analogue HIV reverse transcriptase inhibitor; NNRTI, non-nucleoside analogue HIV reverse transcriptase inhibitor; PI, protease inhibitor; II, integrase inhibitor; AIDS, acquired immune deficiency syndrome; HCV-RNA, HCV plasma viral load; LSM, liver stiffness measures; kPa, kilopascals; HVPG, hepatic venous pressure gradient; mmHg, millimeter of mercury.
Changes in Disease Markers in HIV/HCV-Coinfected Patients After SVR
At the end of follow-up after all-oral DAA therapy, HIV/HCV-coinfected patients exhibited a significant decrease (q-value <0.05) in liver disease severity markers (LSM, HVPG, bilirubin, aspartate transaminase (AST), alanine aminotransferase (ALT), gamma-glutamyl transpeptidase (GGT), and alkaline phosphatase) and immune-related markers (PD1, PDL1, CXCL10, CXCL8, IL12p70, IL10, and TGFβ) (Table 2). By contrast, a substantial increase in hematologic biomarkers (leukocytes, neutrophils, hematocrit, hemoglobin, and platelets) was observed (Table 2). However, they did not show changes in CD4+ T-cell and CD8+ T-cell values and had an undetectable HIV viral load during the whole follow-up.
Table 2.
Summary of changes in disease markers in HIV-infected patients with advanced HCV-related cirrhosis during follow-up.
| HIV/HCV-b | HIV/HCV-f | p-values | q-values | |
|---|---|---|---|---|
| Liver profile | ||||
| LSM (kPa) | 26.7 (18.6; 39.6) | 21.1 (14.1; 31.6) | 0.010 | 0.021 |
| HVPG (mmHg) | 14.5 (10; 17) | 12 (9.5; 14) | 0.030 | 0.047 |
| Bilirubin (mg/dL) | 1.1 (0.8; 1.4) | 0.7 (0.4; 1.1) | <0.001 | 0.001 |
| AST (IU/L) | 70 (37.3; 101.3) | 30 (25.8; 43.5) | 0.006 | 0.016 |
| ALT (IU/L) | 54.5 (30.5; 69.5) | 23 (17.5; 32.5) | <0.001 | <0.001 |
| GGT (IU/L) | 111.5 (73.5; 153) | 45 (30; 62) | 0.002 | 0.007 |
| Alkaline phosphatase (IU/L) | 113 (87.3; 152.5) | 84.5 (57.5; 106) | <0.001 | <0.001 |
| Hematologic profile | ||||
| Leukocytes (106/mL) | 4.2 (3.4; 5.1) | 5.1 (3.5; 6.4) | 0.033 | 0.049 |
| Neutrophils (106/mL) | 2.2 (1.5; 2.7) | 2.6 (2; 3.6) | 0.007 | 0.016 |
| Hematocrit (%) | 41.6 (38.1; 46.8) | 43.1 (40.4; 46.6) | 0.201 | 0.264 |
| Hemoglobin (g/dL) | 14 (12.8; 15.7) | 14.4 (13.2; 15.5) | 0.308 | 0.335 |
| Platelets (109/L) | 80 (52.5; 104.5) | 91 (67.5; 118) | 0.012 | 0.023 |
| Immunologic profile | ||||
| CD4+ T cells (cells/mm3) | 393 (233.5; 806) | 347 (218; 698) | 0.235 | 0.294 |
| CD8+ T cells (cells/mm3) | 610 (376.5; 916) | 609 (357; 928.5) | 0.940 | 0.979 |
| HIV viral load >50 copies/mL (%) | 0/33 (0%) | 0/33 (0%) | 0.999 | 0.999 |
| PD1 (a.u.) | 118.5 (80.8; 147.5) | 61 (46.5; 81.5) | <0.001 | 0.002 |
| PDL1 (a.u.) | 29 (21; 43.3) | 22 (17.5; 38.8) | <0.001 | 0.002 |
| CXCL10 (a.u.) | 1193.5 (771; 1604) | 741.5 (307.8; 994) | <0.001 | 0.002 |
| CXCL8 (a.u.) | 127 (79.5; 188.8) | 73.5 (47; 99.3) | 0.003 | 0.008 |
| CCL2 (a.u.) | 470 (272.8; 677) | 455 (267.5; 644.8) | 0.292 | 0.332 |
| IFNγ (a.u.) | 15 (12; 26) | 16 (13; 23) | 0.285 | 0.332 |
| IL12p70 (a.u.) | 12 (10.3; 14.3) | 11 (9; 13) | 0.024 | 0.040 |
| IL2 (a.u.) | 11 (9.3; 12.5) | 10 (8.8; 12.5) | 0.053 | 0.074 |
| IL10 (a.u.) | 37 (28.8; 46.8) | 29 (25.3; 34.5) | 0.020 | 0.036 |
| TGFβ (a.u.) | 12.2 (8.9; 18.7) | 8.6 (6.7; 14.2) | 0.001 | 0.004 |
Values are expressed as median (P25th; P75th). Differences were calculated by Wilcoxon signed-rank test for paired samples. P-values, raw p-values; q-values, p-values corrected for multiple testing using the false discovery rate (FDR) with Benjamini and Hochberg procedure. The statistically significant differences are shown in bold.
HCV, hepatitis C virus; HIV, human immunodeficiency virus; HIV/HCV-b, HIV/HCV-coinfected at baseline; HIV/HCV-f, HIV/HCV-coinfected at week 36 after SVR; LSM, liver stiffness measure; kPa, kilopascal; HVPG, hepatic venous pressure gradient; mmHg, millimeter of mercury; AST, aspartate transaminase; ALT, alanine aminotransferase; GGT, gamma-glutamyl transpeptidase; g, gram; mg, milligram; L, liter, dL, deciliter; mL, milliliter; IU, international unit; a.u., arbitrary units of fluorescence; PDL1, programmed death-ligand 1; PD1, programmed death-1; CXCL10, C-X-C Motif Chemokine Ligand 10; CXCL8, C-X-C Motif Chemokine Ligand 8; CCL2, C-C Motif Chemokine Ligand 2; IFNγ, interferon-gamma; IL, interleukin; TGFβ, transforming growth factor-beta.
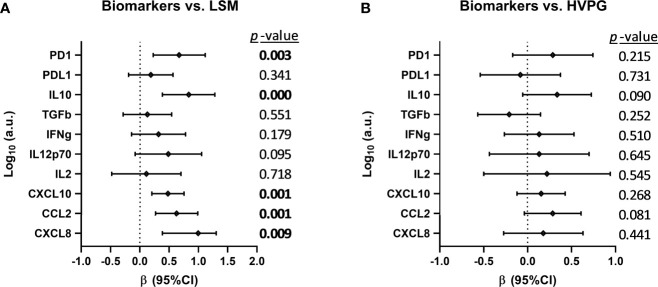
We analyzed the association between changes in plasma biomarkers and liver disease severity scores (LSM and HVPG) during the follow-up (Figure 2), but we only found a positive association (p-value <0.05) between plasma variations of PD1, IL10, CXCL10, CCL2, and CXCL8 and the change in LSM values (Figure 2A). We also performed this association analysis in HCV-monoinfected patients (see Supplementary Figure 1). In this case, we found a positive association (p-value <0.05) between plasma variations of IL-2, CXCL10, and CXCL8 and the change in LSM values, and between CXCL8 and HVGP values.
Figure 2.
Association between changes in plasma biomarkers and liver disease severity scores (A) [liver stiffness measurement (LSM) and (B) hepatic venous pressure gradient (HVPG)] after successful all-oral direct-acting antiviral treatment in HIV/HCV-coinfected patients with advanced cirrhosis. Data were calculated by GLMM models. Values are expressed as regression coefficient (β) and 95% of confidence interval (95%CI). The statistically significant differences are shown in bold.
Changes in Gene Expression in HIV/HCV-Coinfected Patients After SVR
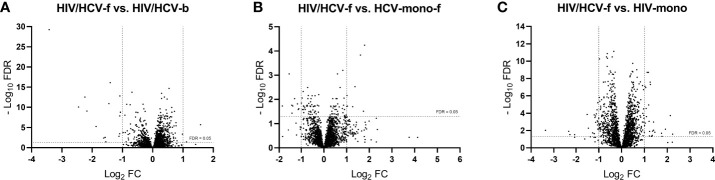
A total of 3,079 genes fulfilled the expression criteria for the statistical analysis, but only 17 SDE genes were found following all-oral DAA therapy (Figure 3A; full description in Table 3A). The HIV/HCV-f group showed two upregulated (HAS1 and IRG1) and 15 downregulated (CXCL11, CCL8, CCL7, CCL2, ADARB2, RRAD, MX1, SIGLEC1, IFI44L, IFI44, IFI27, IFI6, IFIT3, IFIT1B, and IFIT1) genes compared to the HIV/HCV-b group. Most of these downregulated genes were chemokines or ISGs that were classified into four significantly enriched Kyoto encyclopedia of genes and genomes (KEGG) pathways (q-value <0.05) (Table 4A): “cytokine-cytokine receptor interaction”, “viral protein interaction with cytokine and cytokine receptor”, “chemokine signaling pathway”, and “hepatitis C”.
Figure 3.
Volcano plots showing differentially expressed genes between groups: (A) HIV/HCV-f vs. HIV/HCV-b, (B) HIV/HCV-f vs. HCV-mono-f, and (C) HIV/HCV-f vs. HIV-mono. The vertical lines represent the cut-off of FC=2, and the horizontal line indicates the cut-off of FDR=0.05. FDR, false discovery rate for multiple comparisons using Benjamini and Hochberg procedure; FC, fold-change; HIV, human immunodeficiency virus; HCV, hepatitis C virus; HIV/HCV-f, HIV/HCV-coinfected patients 36 weeks after the sustained virological response (SVR); HCV-mono-f, HCV-monoinfected patients 36 weeks after SVR; HIV-mono, HIV-monoinfected patients.
Table 3.
Summary of significant differentially expressed genes (absolute fold-change ≥2; FDR ≤0.05) in peripheral blood mononuclear cells between: A) HIV/HCV-coinfected at week 36 after SVR (HIV/HCV-f) versus HIV/HCV-coinfected at baseline (HIV/HCV-b). B) HIV/HCV-coinfected at week 36 after SVR (HIV/HCV-f) versus HCV-monoinfected at week 36 after SVR (HCV-mono-f); C) HIV/HCV-coinfected at week 36 after SVR (HIV/HCV-f) and HIV-monoinfected (HIV-mono).
| A) HIV/HCV-f versus HIV/HCV-b | B) HIV/HCV-f versus HCV-mono-f | C) HIV/HCV-f versus HIV-mono | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene symbol | FC | Log2 (FC) | p-value * | q-value ** | Gene symbol | FC | Log2 (FC) | p-value * | q-value ** | Gene symbol | FC | Log2 (FC) | p-value * | q-value ** |
| HAS1 | 2.99 | 1.58 | ≤ 0.001 | ≤ 0.001 | CD8A | 3.46 | 1.79 | ≤ 0.001 | ≤ 0.001 | IL1A | 4.72 | 2.24 | 0.009 | 0.026 |
| IRG1 | 2.16 | 1.11 | 0.009 | 0.036 | TRGV2 | 3.35 | 1.75 | ≤ 0.001 | 0.031 | TNFAIP6 | 4.39 | 2.13 | ≤0.001 | ≤0.001 |
| IFIT3 | 0.48 | -1.07 | ≤0.001 | ≤0.001 | CD8B | 3.06 | 1.61 | ≤0.001 | ≤0.001 | SERPINB2 | 3.79 | 1.92 | 0.002 | 0.007 |
| MX1 | 0.47 | -1.08 | ≤0.001 | ≤0.001 | LAG3 | 2.59 | 1.37 | ≤0.001 | 0.003 | OLR1 | 3.41 | 1.77 | 0.019 | 0.048 |
| IFI6 | 0.47 | -1.09 | ≤0.001 | ≤0.001 | TNFRSF13C | 2.41 | 1.27 | ≤0.001 | 0.020 | CCL20 | 3.36 | 1.75 | 0.007 | 0.021 |
| ADARB2 | 0.47 | -1.09 | ≤0.001 | ≤0.001 | KLRK1 | 2.05 | 1.03 | ≤0.001 | 0.019 | SIK1 | 2.75 | 1.46 | 0.005 | 0.016 |
| RRAD | 0.46 | -1.11 | ≤0.001 | 0.001 | IFNLR1 | 2.02 | 1.02 | ≤0.001 | 0.007 | CXCL8 | 2.65 | 1.41 | 0.002 | 0.009 |
| IFI44 | 0.38 | -1.41 | ≤0.001 | ≤0.001 | CDKN2A | 2.01 | 1.01 | ≤0.001 | 0.019 | KCNMA1 | 2.64 | 1.40 | ≤0.001 | 0.001 |
| IFIT1 | 0.37 | -1.45 | ≤0.001 | ≤0.001 | CD180 | 0.47 | -1.10 | ≤0.001 | 0.024 | TNFSF15 | 2.57 | 1.36 | 0.003 | 0.012 |
| IFIT1B | 0.34 | -1.55 | 0.009 | 0.035 | CDKN1C | 0.46 | -1.13 | ≤0.001 | 0.026 | CXCL3 | 2.54 | 1.34 | 0.013 | 0.036 |
| CCL7 | 0.34 | -1.57 | ≤0.001 | 0.003 | FCGR3A | 0.45 | -1.17 | ≤0.001 | 0.015 | LAMB3 | 2.50 | 1.32 | 0.019 | 0.047 |
| CCL8 | 0.33 | -1.62 | 0.001 | 0.004 | SEMA3A | 0.41 | -1.29 | ≤0.001 | 0.009 | CFD | 2.40 | 1.27 | ≤0.001 | ≤0.001 |
| CCL2 | 0.27 | -1.87 | ≤0.001 | ≤0.001 | NRXN2 | 0.39 | -1.37 | ≤0.001 | 0.019 | CD68 | 2.39 | 1.26 | ≤0.001 | ≤0.001 |
| SIGLEC1 | 0.22 | -2.17 | ≤0.001 | ≤0.001 | CEACAM3 | 0.38 | -1.41 | 0.001 | 0.037 | EDN3 | 2.34 | 1.23 | 0.004 | 0.013 |
| IFI44L | 0.21 | -2.24 | ≤0.001 | ≤0.001 | CX3CR1 | 0.37 | -1.42 | ≤0.001 | 0.017 | DSP | 2.33 | 1.22 | 0.016 | 0.042 |
| CXCL11 | 0.18 | -2.45 | ≤0.001 | ≤0.001 | FAT4 | 0.35 | -1.53 | ≤0.001 | ≤0.001 | LRG1 | 2.31 | 1.21 | ≤0.001 | ≤0.001 |
| IFI27 | 0.09 | -3.42 | ≤0.001 | ≤0.001 | PPP1R17 | 0.28 | -1.86 | ≤0.001 | 0.019 | FOXD2 | 2.28 | 1.19 | ≤0.001 | ≤0.001 |
| CCDC85B | 2.26 | 1.18 | ≤0.001 | ≤0.001 | ||||||||||
| FANCL | 2.18 | 1.12 | ≤0.001 | ≤0.001 | ||||||||||
| RNF144B | 2.13 | 1.09 | ≤0.001 | ≤0.001 | ||||||||||
| FKBP1C | 2.11 | 1.08 | ≤0.001 | ≤0.001 | ||||||||||
| CDKN2A | 2.08 | 1.06 | ≤0.001 | ≤0.001 | ||||||||||
| CAVIN3 | 2.06 | 1.04 | ≤0.001 | 0.001 | ||||||||||
| ADAT3 | 2.03 | 1.02 | ≤0.001 | ≤0.001 | ||||||||||
| TNFSF9 | 2.01 | 1.01 | 0.001 | 0.003 | ||||||||||
| TRAV25 | 0.50 | -1.00 | ≤0.001 | ≤0.001 | ||||||||||
| EDAR | 0.49 | -1.04 | 0.002 | 0.007 | ||||||||||
| PARD3 | 0.49 | -1.04 | ≤0.001 | 0.001 | ||||||||||
| CNTNAP1 | 0.49 | -1.04 | ≤0.001 | ≤0.001 | ||||||||||
| PF4V1 | 0.48 | -1.05 | ≤0.001 | 0.002 | ||||||||||
| CMTM5 | 0.48 | -1.06 | ≤0.001 | 0.001 | ||||||||||
| HAVCR1 | 0.48 | -1.07 | ≤0.001 | ≤0.001 | ||||||||||
| ALOX12 | 0.47 | -1.10 | ≤0.001 | ≤0.001 | ||||||||||
| FSTL1 | 0.46 | -1.13 | ≤0.001 | ≤0.001 | ||||||||||
| PDZD2 | 0.45 | -1.14 | ≤0.001 | ≤0.001 | ||||||||||
| PF4 | 0.45 | -1.17 | ≤0.001 | ≤0.001 | ||||||||||
| IGKV1-5 | 0.44 | -1.19 | 0.007 | 0.023 | ||||||||||
| GP1BA | 0.44 | -1.20 | ≤0.001 | ≤0.001 | ||||||||||
| CLEC4F | 0.43 | -1.20 | 0.007 | 0.022 | ||||||||||
| COL5A3 | 0.41 | -1.27 | 0.006 | 0.019 | ||||||||||
| ADARB2 | 0.36 | -1.46 | 0.001 | 0.005 | ||||||||||
| ROBO1 | 0.35 | -1.51 | ≤0.001 | ≤0.001 | ||||||||||
| IGLV8-61 | 0.24 | -2.06 | 0.010 | 0.030 | ||||||||||
| IGLV2-23 | 0.20 | -2.29 | 0.010 | 0.029 | ||||||||||
| IGLV2-18 | 0.20 | -2.34 | 0.004 | 0.012 | ||||||||||
| IGHG2 | 0.10 | -3.37 | 0.003 | 0.010 | ||||||||||
Values are expressed as fold-change (FC) and its log2. (*), raw p-values; (**), p-values corrected for multiple testing using the false discovery rate (FDR) with Benjamini and Hochberg procedure.
HCV, hepatitis C virus; HIV, human immunodeficiency virus; HIV/HCV-f, HIV/HCV-coinfected at week 36 after SVR; HIV/HCV-b, HIV/HCV-coinfected at baseline; HCV-mono-f, HCV-monoinfected at week 36 after SVR; HIV-mono, HIV-monoinfected patients.
Table 4.
Summary of significant pathways (FDR ≤0.05) in peripheral blood mononuclear cells between: A) HIV/HCV-coinfected at week 36 after SVR (HIV/HCV-f) versus HIV/HCV-coinfected at baseline (HIV/HCV-b). B) HIV/HCV-coinfected at week 36 after SVR (HIV/HCV-f) versus HCV-monoinfected at week 36 after SVR (HCV-mono-f); C) HIV/HCV-coinfected at week 36 after SVR (HIV/HCV-f) and HIV-monoinfected (HIV-mono).
| A) HIV/HCV-f versus HIV/HCV-b | B) HIV/HCV-f versus HCV-mono-f | C) HIV/HCV-f versus HIV-mono | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| KEGG | Hits | Genes | q-values | KEGG | Hits | Genes | q-values | KEGG | Hits | Genes | q-values |
| Cytokine-cytokine receptor interaction | 4 | CCL2, CCL7, CCL8, CXCL11 | 0.021 | Primary immunodeficiency | 3 | CD8A, CD8B, TNFRSF13C | 0.009 | Cytokine-cytokine receptor interaction | 9 | CCL20, CXCL3, CXCL8, EDAR, IL1A, PF4, PF4V1, TNFSF15, TNFSF9 | <0.001 |
| Viral protein interaction with cytokine and cytokine receptor | 4 | CCL2, CCL7, CCL8, CXCL11 | 0.001 | Viral protein interaction with cytokine and cytokine receptor | 5 | CCL20, CXCL3, CXCL8, PF4, PF4V1 | 0.002 | ||||
| Chemokine signaling pathway | 4 | CCL2, CCL7, CCL8, CXCL11 | 0.006 | Chemokine signaling pathway | 6 | CCL20, CXCL3, CXCL8, PARD3, PF4, PF4V1 | 0.003 | ||||
| Hepatitis C | 3 | IFIT1, IFIT1B, MX1 | 0.047 | Rheumatoid arthritis | 4 | CCL20, CXCL3, CXCL8, IL1A | 0.017 | ||||
q-values, p values corrected for multiple comparisons using Benjamini and Hochberg procedure. In red, upregulated genes in HIV/HCV-f group; in green, downregulated genes in HIV/HCV-f group.
KEGG, Kyoto Encyclopedia of Genes and Genomes; HIV, human immunodeficiency virus; HCV, hepatitis C virus; HIV/HCV-b, HIV/HCV-coinfected patients at baseline; HIV/HCV-f, HIV/HCV-coinfected patients 36 weeks after SVR; HCV-mono-f, HCV-monoinfected at week 36 after SVR; HIV-mono, HIV-monoinfected patients.
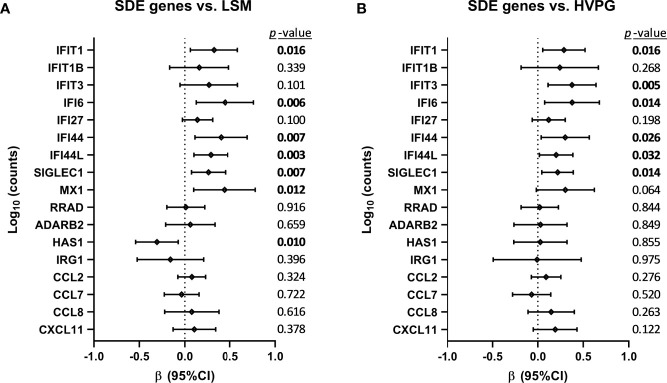
The association between changes in SDE genes and liver disease severity scores (LSM and HVPG) is shown in Figure 4. We found positive associations (p-value <0.05) of IFIT1, IFI6, IFI44, IFI44L, SIGLEC1, and MX1 with LSM values, and HAS1 showed a negative association (Figure 4A). Additionally, we found positive associations (p-value <0.05) of IFIT1, IFIT3, IFI6, IFI44, IFI44L, and SIGLEC1 with HVPG values (Figure 4B).
Figure 4.
Association between changes in significant differentially expressed (SDE) genes and liver disease severity scores (A) [liver stiffness measurement (LSM) and (B) hepatic venous pressure gradient (HVPG)] after successful all-oral direct-acting antiviral treatment in HIV/HCV-coinfected patients with advanced cirrhosis. Data were calculated by GLMM models. Values are expressed as regression coefficient (β) and 95% of confidence interval (95%CI). The statistically significant differences are shown in bold.
We also evaluated SDE genes in HCV-monoinfected patients (see Supplementary Table 2). In this case, we found 4 SDE (HAS1, IFI44L, SIGLEC1, and IFI27), which were also differentially expressed in HIV/HCV-coinfected patients after SVR. However, we also found seven other SDE genes (SNAI1, MAPK8IP1, SHB, IFNG, NRCAM, ZBTB32, and CYP1A1) that are ISGs.
Gene Expression in HIV/HCV-Coinfected Patients After SVR Compared to Control Groups
By comparing the HIV/HCV-f with the HCV-mono-f, 2,847 genes met the expression criteria, and 17 SDE genes were found (Figure 3B, full description in Table 3B). The HIV/HCV-f group had eight upregulated and nine downregulated genes compared to the HCV-mono-f group. The analysis of the altered pathways related to these genes (Table 4B) showed the only significant KEGG pathway identified was “primary immunodeficiency pathway” (q-value <0.05), with three SDE genes upregulated (CD8A, CD8B, and TNFRSF13C) in HIV/HCV-f.
In the HIV/HCV-f versus HIV-mono comparison, 2,779 genes met the expression criteria, and 46 SDE genes were found (Figure 3C, full description in Table 3C). The HIV/HCV-f group had 25 upregulated and 21 downregulated genes compared with the HIV-mono group. The analysis of the altered pathways (Table 4C) gave four significant results (q-value <0.05) in the KEGG enrichment analysis (“cytokine-cytokine receptor interaction”, “viral protein interaction with cytokine and cytokine receptor”, “chemokine signaling pathway”, and “rheumatoid arthritis”), with six SDE genes upregulated (CCL20, CXCL3, CXCL8, IL1A, TNFSF15, and TNFSF9) and four downregulated (EDAR, PF4, PARD3, and PF4V1) in HIV/HCV-f. The HIV/HCV-f vs. HCV-mono-f and the HIV/HCV-f vs. HIV-mono comparisons showed only one upregulated gene in common (CDKN2A).
Discussion
In this study, our significant findings for HIV/HCV-coinfected patients after SVR following all-oral DAA therapy were: i) an improvement in immunological and liver disease markers, with an association between decreased plasma levels of PD1, IL10, CXCL10, CXCL8, and CCL2 and a reduction in LSM values; ii) a decrease in the expression of ISGs, mainly encoding for chemokines and antiviral proteins, which were grouped into four altered pathways (“cytokine-cytokine receptor interaction”, “viral protein interaction with cytokine and cytokine receptor”, “chemokine signaling pathway”, and “hepatitis C”); additionally, the decrease in most of these genes was also related to a reduction in LSM or HVPG values; and iii) only the “primary immunodeficiency pathway” was altered compared to HCV-mono-f, while four pathways (“cytokine-cytokine receptor interaction”, “viral protein interaction with cytokine and cytokine receptor”, “chemokine signaling pathway”, and “rheumatoid arthritis”) were altered compared to HIV-monoinfected patients.
Little is known about how HCV elimination after all-oral DAA therapy affects the immune system of HIV/HCV-coinfected patients. This population is of particular interest since they suffer from higher inflammation and accelerated fibrogenesis than HCV-monoinfected patients (49, 50), and despite ART, residual inflammation remains in HIV-monoinfected patients (51, 52). Chronic hepatitis C promotes hepatic and systemic inflammation, with high production of CXCL8, CCL2, CXCL10, IFNγ, and IL12p70, among others (13), and activation and exhaustion of the immune system, with increased levels of PD1 and PDL1 (53). Our data show an improvement in these immune and liver disease biomarkers in plasma (immune exhaustion (PD1), chemokines (CXCL10, CCL2, and CXCL8), and cytokines (IL10)) after achieving SVR with DAA therapy in HIV/HCV-coinfected individuals. These results are similar to those found in HCV-monoinfected persons, in whom immune and liver function improved after HCV clearance by DAA, although normalization of the immune system after SVR is not always achieved (54, 55). Regarding HIV/HCV-coinfected patients, there are limited data on their status following DAA therapy. However, some of our findings are similar to those described in previous studies showing improved liver function (56–61) and a decrease in plasma chemokine levels (CXCL10, CCL2, and CXCL8) (45, 62–66). Chemokines attract leucocytes to the liver, promoting cirrhosis and the appearance of LRE (67), and they are surrogate markers of liver inflammation (68). Thus, the decline of CXCL10, CCL2, and CXCL8 would indicate a reduction of liver and systemic inflammation after HCV clearance. Chronic hepatitis C upregulates exhaustion markers, such as PD1 (53, 66), which is an immune checkpoint receptor that inhibits the adaptive immune response. Thus, a decrease in plasma levels of PD1 would indicate recovery of the exhausted adaptive immune response, as seen in HCV-monoinfected patients (53, 66). IL10 has a central role in chronic hepatitis C, limiting the inflammatory response (69) and reducing liver fibrosis (70). However, we found a decrease in plasma IL10 after SVR, which may be a consequence of HCV clearance and a general reduction in inflammation.
Gene expression in PBMCs was also analyzed in HIV/HCV-coinfected patients, where we mainly found a decrease in ISGs after achieving SVR with all-oral DAA therapy. ISGs are related to the degree of host immune dysregulation in chronic hepatitis C, affecting the control of virus replication and spontaneous HCV clearance (13). Chronic hepatitis C is also associated with increased levels of several relevant IFN-sensitive chemokines that promote the development of liver fibrosis and cirrhosis (67). In HCV-monoinfected patients, several studies have reported the downregulation of many chemokines, cytokines, and other ISG genes in PBMCs after SVR following DAA therapy (43, 71–75), but normal levels of these genes are not always restored, indicating incomplete normalization of the immune system. Information on the impact of HCV clearance after DAA therapy on PBMC gene expression in HIV/HCV-coinfected patients is scarce, but changes in ISGs and pathways related to antiviral defense, innate immune response, cytokine signals, and inflammatory response/chemotaxis have been previously observed (44, 45). The prolonged and sustained down-regulation of ISGs suggests a reversal of the exhausted and inflammatory immune phenotype after hepatitis C cure with all-oral DAA therapy. Furthermore, in our study, reduced ISG expression was associated with an improvement in liver disease severity scores (LSM and HVPG) in HIV/HCV-coinfected patients who achieved SVR following all-oral DAA therapy.
We also performed a statistical analysis in HCV-monoinfected patients during follow-up (baseline vs. after-SVR) to evaluate whether there is a pattern similar to HIV/HCV-coinfected patients. On the one hand, we found that plasma CXCL10 and CXCL8 remained positively associated with LSM values in HCV-monoinfected and HIV/HCV-coinfected patients, but IL-2 was also associated with LSM and CXCL8 with HVPG in HCV-monoinfected patients. On the other hand, 4 SDE (HAS1, IFI44L, SIGLEC1, and IFI27) were common in HCV-monoinfected and HIV/HCV-coinfected patients, but seven other SDE genes (SNAI1, MAPK8IP1, SHB, IFNG, NRCAM, ZBTB32, and CYP1A1) were only found in HCV-monoinfected patients; which are all ISGs (http://www.interferome.org/). These findings show that the changes in HCV monoinfected patients after SVR were not precisely the same as in HIV/HCV-coinfected patients, although almost equivalent.
Moreover, we found that the PBMC gene expression of CD8A, CD8B, and TNFRSF13C was higher in HIV/HCV-coinfected patients after achieving SVR (HIV/HCV-f) than HCV-monoinfected patients after SVR (HCV-mono-f). These three genes are grouped into the “primary immunodeficiency pathway”, the only KEGG pathway with a q-value lower than 0.25. The overexpression of CD8A and CD8B may be linked to increased CD8 T-cell count, an intrinsic feature of HIV infection. HIV-infected patients have the CD4/CD8 ratio usually diminished in most patients, and it is related to higher inflammation, activation, immune deregulation, and increased risk of non-AIDS morbidity and mortality (76). A similar finding was found in HIV-infected patients with sepsis, who showed overexpression of genes involved in cytotoxic T-cell signaling (CD8A, CD8B) (77). These findings reveal an effect of HIV infection per se, which persists after SVR. However, this does not answer why HIV/HCV-coinfected patients have higher rates of liver-related events than HCV-monoinfected patients. Besides, TNFRSF13C is a regulator of the peripheral B-cell population that enhances B-cell survival, which is decreased by HCV infection, and it has been related to HCV-induced B cells clonal disorders, such as mixed cryoglobulinemia and non-Hodgkin’s lymphoma (78). Also, an increase in TNFRSF13C expression has been described after HCV eradication with antiviral treatment, together with complete clinical remission of HCV-induced B-cell clonal disorders (78). Overexpression of TNFRS13C in HIV/HCV-coinfected patients may be related to continuous stimulation of B cells by HIV chronic infection. The other two genes in the top 5 of overexpression were TRGV2 and LAG3. TRGV2 is a T-cell receptor gamma chain that participates in HIV control, indicating a recovery of the immune response against the virus (79). However, LAG3 is upregulated in T-cells of HIV-monoinfected patients and point to immune exhaustion, characterized by functional unresponsiveness of T-cells (80).
HIV/HCV-f and HCV-mono-f showed no differences in the PBMC gene expression levels of chemokines and ISGs and plasma levels of biomarkers related to immune exhaustion, chemokines, and Th1 and Th2 cytokines (data not shown) after HCV elimination by DAA. Overall, these data indicate that HCV treatment had a similar impact on peripheral blood markers in both groups.
When compared to HIV-monoinfected individuals, some cytokine genes (CCL20, CXCL3, CXCL8, IL1A, TNFSF15, and TNFSF9) were upregulated in HIV/HCV-coinfected patients after HCV elimination. This, again, indicates that although DAA treatment leads to a broad reduction in cytokine and ISG expression, the restoration of the immune response is not complete. Particularly worrying are the elevated levels of CCL20, which is a pro-angiogenic factor that promotes HCC hypervascularization (81) and might be behind the increased probability of developing HCC following SVR (82–84). High gene expression levels of CXCL3, CXCL8, and IL1A also correlate with HCV-associated liver inflammation, cirrhosis, and HCC (85–89). Overexpression of TNFSF15 increases inflammation and both liver and intestinal fibrosis, promoting chronic immunological diseases (90, 91). Moreover, we observed lower expression of PF4 and PF4V1 in HIV/HCV-coinfected patients after SVR than in HIV-monoinfected patients. PF4 and PF4V1 genes encode two chemokines (CXCL4 and CXCL4L1, respectively) related to platelet activation, and their expression promotes inflammation and wound repair. In the liver, CXCL4 and CXCL4L1 stimulate proliferation, chemotaxis, and chemokine expression of hepatic stellate cells, promoting liver fibrosis (92, 93). The lowest values found in HIV/HCV-coinfected patients after SVR may reflect HCV elimination.
Study Limitations
Our conclusions should be interpreted with caution. i) The limited sample size can reduce the statistical power to detect small differences between groups. In addition, this can also increase the risk of false-positive results. ii) The design of our study was prospective, and biases could have been introduced. Nevertheless, the principal analysis of the study was carried out with a GLMM model for paired data, which reduces the impact of study biases, limits the false positive rate, and increases statistical power. iii) Our work was an association study and lacked mechanistic exploration or functional validation.
Final Remarks
HCV elimination by DAA treatment in HIV/HCV-coinfected individuals improves liver inflammation and HCV-related extrahepatic diseases in most cases (94). Nevertheless, while some aspects of the immune response are restored, others remain altered. Therefore, long-term chronic HCV infection might lead to an irreversible impairment of the immune system, perhaps by inducing epigenetic changes that persist after virus eradication (95). However, since virus-host interactions are very complex, it is difficult to know which gene expression changes are clinically relevant.
Due to an unbalanced immunity after SVR, HCV elimination poses some concerns regarding reactivation of latent coinfections, such as HBV (96) or herpesvirus (97, 98), or the development of HCC (82–84). For instance, despite reducing inflammation, an increase in angiogenesis-related gene expression, which may favor HCC development, has been observed in cured patients (81, 99). Interestingly, long-term epigenetic modifications induced by HCV are associated with an increased risk for HCC development even after HCV cure with DAA (95). However, other studies have not identified any effect of DAA on the recurrence of HCC (100).
Knowing the extent and causes of the partial recovery of the immune system in HIV patients on ART after HCV chronic infection might be relevant to understanding the susceptibility of cured patients to the development of immune-mediated diseases, cancer, and infections with different pathogens (including re-infections), as well as contributing to drug design and vaccine.
Conclusions
HIV/HCV-coinfected patients with advanced cirrhosis who eradicated HCV infection with all-oral DAA therapy exhibited significant decreases in plasma biomarkers and gene expression related to antiviral/inflammatory response, particularly several chemokines and ISGs, along with an improvement in liver disease markers. However, complete normalization of the immune system was not achieved, as observed by comparison with HCV and HIV-monoinfected patients.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding authors upon reasonable request. Patients’ raw sequence data are publicly available at the ArrayExpress repository (EMBL-EBI; https://www.ebi.ac.uk/) under the accession number E-MTAB-10703.
Ethics Statement
The studies involving human participants were reviewed and approved by The Institutional Review Board and the Research Ethics Committee of the Instituto de Salud Carlos III (ISCIII) approved the study (CEI PI 41_2014). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
Conceptualization: SR and MA, and AF-R. Data curation: JB, JG-G, CD, VH, LI-S, and LP-L. Formal analysis: OB-K, SR, IM, and AF-R. Funding acquisition: JB, JG-G, MA, and SR. Investigation and methodology: SS, OB-K, MA, and AF-R. Project administration: JB and SR. Supervision and visualization: MA and SR. Writing – original draft preparation: IM, AF-R, MA, and SR. Writing – Review and editing: DS-C. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by grants from Instituto de Salud Carlos III (ISCII; grant numbers PI20/00474 and PI17/00657 to JB, PI20/00507 and PI17/00903 to JGG, PI18CIII/00020 to AF-R, PI18CIII/00028 to MA, and PI20CIII/00004 and PI17CIII/00003 to SR). AF-R and MA are Miguel Servet researchers supported and funded by ISCIII (grant numbers: CP14CIII/00010 to AFR and CP17CIII/00007 to MAJS). The study was also funded by the RD16/0025/0017, RD16/0025/0018 and RD16CIII/0002/0002 projects as part of the Plan Nacional R + D + I and co-funded by ISCIII- Subdirección General de Evaluación and the Fondo Europeo de Desarrollo Regional (FEDER). JB is an investigator from the Programa de Intensificación de la Actividad Investigadora en el Sistema Nacional de Salud (I3SNS), Ref. INT16/00100. DS-C is a ‘Sara Borrell’ researcher from ISCIII (grant number CD20CIII/00001) and has also been supported through Fundación SEIMC-GESIDA by a fellowship award from Fundación ONCE ‘Oportunidad al Talento, 2019/20 and 2020/21’ co-financed by Fondo Social Europeo.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We want to acknowledge the patients in this study for their participation and the Spanish HIV HGM BioBank integrated into the Spanish AIDS Research Network (RIS) and collaborating Centers for the generous gifts of clinical samples used in this work. This study would not have been possible without the collaboration of all medical and nursing staff and data managers who have taken part in the project (see (Appendix, which shows all collaborators). We also want to thank the Bioinformatics Unit at the Institute of Health Carlos III for their valuable support for the bioinformatics analysis.
Appendix
The ESCORIAL study group.
Hospital General Universitario Gregorio Marañón (Madrid, Spain): Cristina Díez, Luis Ibáñez, Leire Pérez-Latorre, Diego Rincón, Teresa Aldámiz-Echevarría, Vega Catalina, Pilar Miralles, Teresa Aldámiz-Echevarría, Francisco Tejerina, María C Gómez-Rico, Esther Alonso, José M Bellón, Rafael Bañares, and Juan Berenguer.
Hospital Universitario La Paz/IdiPAZ (Madrid, Spain): José Arribas, José I Bernardino, Carmen Busca, Ana Delgado, Javier García-Samaniego, Víctor Hontañón, Luz Martín-Carbonero, Rafael Micán, María L Montes-Ramírez, Victoria Moreno, Antonio Olveira, Ignacio Pérez-Valero, Eulalia valencia, and Juan González-García.
Hospital Universitario Puerta de Hierro (Madrid, Spain): Elba Llop and José Luis Calleja.
Hospital Universitario Ramón y Cajal (Madrid, Spain): Javier Martínez and Agustín Albillos.
Fundación SEIMC/GeSIDA (Madrid, Spain): Marta de Miguel, María Yllescas, and Herminia Esteban.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.723196/full#supplementary-material
References
- 1.The Polaris Observatory HCV Collaborators . Global Prevalence and Genotype Distribution of Hepatitis C Virus Infection in 2015: A Modelling Study. Lancet Gastroenterol Hepatol (2017) 2(3):161–76. 10.1016/s2468-1253(16)30181-9 [DOI] [PubMed] [Google Scholar]
- 2.Platt L, Easterbrook P, Gower E, McDonald B, Sabin K, McGowan C, et al. Prevalence and Burden of HCV Co-Infection in People Living With HIV: A Global Systematic Review and Meta-Analysis. Lancet Infect Dis (2016) 16(7):797–808. 10.1016/S1473-3099(15)00485-5 [DOI] [PubMed] [Google Scholar]
- 3.Vallet-Pichard A, Pol S. Natural History and Predictors of Severity of Chronic Hepatitis C Virus (HCV) and Human Immunodeficiency Virus (HIV) Co-Infection. J Hepatol (2006) 44(1 Suppl):S28–34. 10.1016/j.jhep.2005.11.008 [DOI] [PubMed] [Google Scholar]
- 4.Lo Re V, 3rd, Kallan MJ, Tate JP, Localio AR, Lim JK, Goetz MB, et al. Hepatic Decompensation in Antiretroviral-Treated Patients Co-Infected With HIV and Hepatitis C Virus Compared With Hepatitis C Virus-Monoinfected Patients: A Cohort Study. Ann Intern Med (2014) 160(6):369–79. 10.7326/M13-1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.López-Diéguez M, Montes ML, Pascual-Pareja JF, Quereda C, Von Wichmann MA, Berenguer J, et al. The Natural History of Liver Cirrhosis in HIV-Hepatitis C Virus-Coinfected Patients. AIDS (2011) 25(7):899–904. 10.1097/QAD.0b013e3283454174 [DOI] [PubMed] [Google Scholar]
- 6.Macias J, Berenguer J, Japon MA, Giron JA, Rivero A, Lopez-Cortes LF, et al. Fast Fibrosis Progression Between Repeated Liver Biopsies in Patients Coinfected With Human Immunodeficiency Virus/Hepatitis C Virus. Hepatology (2009) 50(4):1056–63. 10.1002/hep.23136 [DOI] [PubMed] [Google Scholar]
- 7.Ingiliz P, Rockstroh JK. Natural History of Liver Disease and Effect of Hepatitis C Virus on HIV Disease Progression. Curr Opin HIV AIDS (2015) 10(5):303–8. 10.1097/COH.0000000000000187 [DOI] [PubMed] [Google Scholar]
- 8.Sherman KE, Peters MG, Thomas D. Human Immunodeficiency Virus and Liver Disease: A Comprehensive Update. Hepatol Commun (2017) 1(10):987–1001. 10.1002/hep4.1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuyama AC, Hong F, Saiman Y, Wang C, Ozkok D, Mosoian A, et al. Human Immunodeficiency Virus (HIV)-1 Infects Human Hepatic Stellate Cells and Promotes Collagen I and Monocyte Chemoattractant Protein-1 Expression: Implications for the Pathogenesis of HIV/hepatitis C Virus-Induced Liver Fibrosis. Hepatology (2010) 52(2):612–22. 10.1002/hep.23679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soriano V, Berenguer J. Extrahepatic Comorbidities Associated With Hepatitis C Virus in HIV-Infected Patients. Curr Opin HIV AIDS (2015) 10(5):309–15. 10.1097/COH.0000000000000175 [DOI] [PubMed] [Google Scholar]
- 11.Naggie S. Hepatitis C Virus, Inflammation, and Cellular Aging: Turning Back Time. Top Antivir Med (2017) 25(1):3–6. [PMC free article] [PubMed] [Google Scholar]
- 12.Shin EC, Sung PS, Park SH. Immune Responses and Immunopathology in Acute and Chronic Viral Hepatitis. Nat Rev Immunol (2016) 16(8):509–23. 10.1038/nri.2016.69 [DOI] [PubMed] [Google Scholar]
- 13.Heim MH, Thimme R. Innate and Adaptive Immune Responses in HCV Infections. J Hepatol (2014) 61(1 Suppl):S14–25. 10.1016/j.jhep.2014.06.035 [DOI] [PubMed] [Google Scholar]
- 14.Wedemeyer H, Khera T, Strunz B, Bjorkstrom NK. Reversal of Immunity After Clearance of Chronic HCV Infection-All Reset? Front Immunol (2020) 11:571166. 10.3389/fimmu.2020.571166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radziewicz H, Ibegbu CC, Fernandez ML, Workowski KA, Obideen K, Wehbi M, et al. Liver-Infiltrating Lymphocytes in Chronic Human Hepatitis C Virus Infection Display an Exhausted Phenotype With High Levels of PD-1 and Low Levels of CD127 Expression. J Virol (2007) 81(6):2545–53. 10.1128/JVI.02021-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kasprowicz V, Schulze Zur Wiesch J, Kuntzen T, Nolan BE, Longworth S, Berical A, et al. High Level of PD-1 Expression on Hepatitis C Virus (HCV)-Specific CD8+ and CD4+ T Cells During Acute HCV Infection, Irrespective of Clinical Outcome. J Virol (2008) 82(6):3154–60. 10.1128/JVI.02474-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Radziewicz H, Ibegbu CC, Hon H, Osborn MK, Obideen K, Wehbi M, et al. Impaired Hepatitis C Virus (HCV)-Specific Effector CD8+ T Cells Undergo Massive Apoptosis in the Peripheral Blood During Acute HCV Infection and in the Liver During the Chronic Phase of Infection. J Virol (2008) 82(20):9808–22. 10.1128/JVI.01075-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prezzi C, Casciaro MA, Francavilla V, Schiaffella E, Finocchi L, Chircu LV, et al. Virus-Specific CD8(+) T Cells With Type 1 or Type 2 Cytokine Profile are Related to Different Disease Activity in Chronic Hepatitis C Virus Infection. Eur J Immunol (2001) 31(3):894–906. [DOI] [PubMed] [Google Scholar]
- 19.Eisen-Vandervelde AL, Waggoner SN, Yao ZQ, Cale EM, Hahn CS, Hahn YS. Hepatitis C Virus Core Selectively Suppresses Interleukin-12 Synthesis in Human Macrophages by Interfering With AP-1 Activation. J Biol Chem (2004) 279(42):43479–86. 10.1074/jbc.M407640200 [DOI] [PubMed] [Google Scholar]
- 20.Fernandes JR, Berthoud TK, Kumar A, Angel JB. IL-23 Signaling in Th17 Cells is Inhibited by HIV Infection and is Not Restored by HAART: Implications for Persistent Immune Activation. PloS One (2017) 12(11):e0186823. 10.1371/journal.pone.0186823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Paula HHS, Ferreira ACG, Caetano DG, Delatorre E, Teixeira SLM, Coelho LE, et al. Reduction of Inflammation and T Cell Activation After 6 Months of cART Initiation During Acute, But Not in Early Chronic HIV-1 Infection. Retrovirology (2018) 15(1):76. 10.1186/s12977-018-0458-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pandiyan P, Younes SA, Ribeiro SP, Talla A, McDonald D, Bhaskaran N, et al. Mucosal Regulatory T Cells and T Helper 17 Cells in HIV-Associated Immune Activation. Front Immunol (2016) 7:228. 10.3389/fimmu.2016.00228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miles B, Miller SM, Connick E. CD4 T Follicular Helper and Regulatory Cell Dynamics and Function in HIV Infection. Front Immunol (2016) 7:659. 10.3389/fimmu.2016.00659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Biasi S, Bianchini E, Nasi M, Digaetano M, Gibellini L, Carnevale G, et al. Th1 and Th17 Proinflammatory Profile Characterizes Invariant Natural Killer T Cells in Virologically Suppressed HIV+ Patients With Low CD4+/CD8+ Ratio. AIDS (2016) 30(17):2599–610. 10.1097/QAD.0000000000001247 [DOI] [PubMed] [Google Scholar]
- 25.DaFonseca S, Niessl J, Pouvreau S, Wacleche VS, Gosselin A, Cleret-Buhot A, et al. Impaired Th17 Polarization of Phenotypically Naive CD4(+) T-Cells During Chronic HIV-1 Infection and Potential Restoration With Early ART. Retrovirology (2015) 12:38. 10.1186/s12977-015-0164-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahnke YD, Fletez-Brant K, Sereti I, Roederer M. Reconstitution of Peripheral T Cells by Tissue-Derived CCR4+ Central Memory Cells Following HIV-1 Antiretroviral Therapy. Pathog Immun (2016) 1(2):260–90. 10.20411/pai.v1i2.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merlini E, Tincati C, Biasin M, Saulle I, Cazzaniga FA, d’Arminio Monforte A, et al. Marchetti: Stimulation of PBMC and Monocyte-Derived Macrophages via Toll-Like Receptor Activates Innate Immune Pathways in HIV-Infected Patients on Virally Suppressive Combination Antiretroviral Therapy. Front Immunol (2016) 7:614. 10.3389/fimmu.2016.00614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mudd JC, Brenchley JM. Gut Mucosal Barrier Dysfunction, Microbial Dysbiosis, and Their Role in HIV-1 Disease Progression. J Infect Dis (2016) 214 Suppl:2, S58–66. 10.1093/infdis/jiw258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tincati C, Merlini E, Braidotti P, Ancona G, Savi F, Tosi D, et al. Impaired Gut Junctional Complexes Feature Late-Treated Individuals With Suboptimal CD4+ T-Cell Recovery Upon Virologically Suppressive Combination Antiretroviral Therapy. AIDS (2016) 30(7):991–1003. 10.1097/QAD.0000000000001015 [DOI] [PubMed] [Google Scholar]
- 30.Hunt PW, Lee SA, Siedner MJ. Immunologic Biomarkers, Morbidity, and Mortality in Treated HIV Infection. J Infect Dis (2016) 214(Suppl_2):S44–50. 10.1093/infdis/jiw275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hart BB, Nordell AD, Okulicz JF, Palfreeman A, Horban A, Kedem E, et al. Inflammation-Related Morbidity and Mortality Among HIV-Positive Adults: How Extensive Is it? J Acquir Immune Defic Syndr (2018) 77(1):1–7. 10.1097/QAI.0000000000001554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang TJ, Ghany MG. Therapy of Hepatitis C–back to the Future. N Engl J Med (2014) 370(21):2043–7. 10.1056/NEJMe1403619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.A.-I. H. G. Panel . Hepatitis C Guidance 2018 Update: AASLD-IDSA Recommendations for Testing, Managing, and Treating Hepatitis C Virus Infection. Clin Infect Dis (2018) 67(10):1477–92. 10.1093/cid/ciy585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.e. e. e. European Association for the Study of the Liver . Electronic Address and L. European Association for the Study of the: EASL Recommendations on Treatment of Hepatitis C 2018. J Hepatol (2018) 69(2):461–511. 10.1016/j.jhep.2018.03.026 [DOI] [PubMed] [Google Scholar]
- 35.Berenguer J, Gil-Martin A, Jarrin I, Moreno A, Dominguez L, Montes M, et al. All-Oral Direct-Acting Antiviral Therapy Against Hepatitis C Virus (HCV) in Human Immunodeficiency Virus/HCV-Coinfected Subjects in Real-World Practice: Madrid Coinfection Registry Findings. Hepatology (2018) 68(1):32–47. 10.1002/hep.29814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berkan-Kawinska A, Piekarska A, Janczewska E, Lorenc B, Tudrujek-Zdunek M, Tomasiewicz K, et al. Real-World Effectiveness and Safety of Direct-Acting Antivirals in Patients With Cirrhosis and History of Hepatic Decompensation: Epi-Ter2 Study. Liver Int (2021) 41(8):1789–801. 10.1111/liv.14858 [DOI] [PubMed] [Google Scholar]
- 37.Krassenburg LAP, Maan R, Ramji A, Manns MP, Cornberg M, Wedemeyer H, et al. Clinical Outcomes Following DAA Therapy in Patients With HCV-Related Cirrhosis Depend on Disease Severity. J Hepatol (2020) 74(5):1053–63. 10.1016/j.jhep.2020.11.021 [DOI] [PubMed] [Google Scholar]
- 38.Mendizabal M, Pinero F, Ridruejo E, Herz Wolff F, Anders M, Reggiardo V, et al. Awareness: Disease Progression in Patients With Hepatitis C Virus Infection Treated With Direct-Acting Antiviral Agents. Clin Gastroenterol Hepatol (2020) 18(11):2554–63.e3. 10.1016/j.cgh.2020.02.044 [DOI] [PubMed] [Google Scholar]
- 39.Corma-Gomez A, Morano L, Tellez F, Rivero-Juarez A, Real LM, Alados JC, et al. HIV Infection Does Not Increase the Risk of Liver Complications in Hepatitis C Virus-Infected Patient With Advanced Fibrosis, After Sustained Virological Response With Direct-Acting Antivirals. AIDS (2019) 33(7):1167–74. 10.1097/QAD.0000000000002186 [DOI] [PubMed] [Google Scholar]
- 40.Chalouni M, Pol S, Sogni P, Fontaine H, Lacombe K, Marc-Lacombe J, et al. Increased Mortality in HIV/HCV-Coinfected Compared to HCV-Monoinfected Patients in the DAA Era Due to non-Liver-Related Death. J Hepatol (2021) 74(1):37–47. 10.1016/j.jhep.2020.08.008 [DOI] [PubMed] [Google Scholar]
- 41.Rosenberg BR, Freije CA, Imanaka N, Chen ST, Eitson JL, Caron R, et al. Genetic Variation at IFNL4 Influences Extrahepatic Interferon-Stimulated Gene Expression in Chronic HCV Patients. J Infect Dis (2018) 217(4):650–5. 10.1093/infdis/jix593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramamurthy N, Marchi E, Ansari MA, Pedergnana V, McLean A, Hudson E, et al. Impact of Interferon Lambda 4 Genotype on Interferon-Stimulated Gene Expression During Direct-Acting Antiviral Therapy for Hepatitis C. Hepatology (2018) 68(3):859–71. 10.1002/hep.29877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alao H, Cam M, Keembiyehetty C, Zhang F, Serti E, Suarez D, et al. Baseline Intrahepatic and Peripheral Innate Immunity are Associated With Hepatitis C Virus Clearance During Direct-Acting Antiviral Therapy. Hepatology (2018) 68(6):2078–88. 10.1002/hep.29921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun B, Abadjian L, Monto A, Freasier H, Pulliam L. Hepatitis C Virus Cure in Human Immunodeficiency Virus Coinfection Dampens Inflammation and Improves Cognition Through Multiple Mechanisms. J Infect Dis (2020) 222(3):396–406. 10.1093/infdis/jiaa109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carlton-Smith C, Holmes JA, Naggie S, Lidofsky A, Lauer GM, Kim AY, et al. IFN-Free Therapy is Associated With Restoration of Type I IFN Response in HIV-1 Patients With Acute HCV Infection Who Achieve SVR. J Viral Hepat (2018) 25(5):465–72. 10.1111/jvh.12836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Medrano LM, Garcia-Broncano P, Berenguer J, Gonzalez-Garcia J, Jimenez-Sousa MA, Guardiola JM, et al. Elevated Liver Stiffness is Linked to Increased Biomarkers of Inflammation and Immune Activation in HIV/hepatitis C Virus-Coinfected Patients. AIDS (2018) 32(9):1095–105. 10.1097/QAD.0000000000001787 [DOI] [PubMed] [Google Scholar]
- 47.Rincon D, Lo Iacono O, Tejedor M, Hernando A, Ripoll C, Catalina MV, et al. Prognostic Value of Hepatic Venous Pressure Gradient in Patients With Compensated Chronic Hepatitis C-Related Cirrhosis. Scand J Gastroenterol (2013) 48(4):487–95. 10.3109/00365521.2012.711848 [DOI] [PubMed] [Google Scholar]
- 48.Breen EJ, Tan W, Khan A. The Statistical Value of Raw Fluorescence Signal in Luminex xMAP Based Multiplex Immunoassays. Sci Rep (2016) 6:26996. 10.1038/srep26996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benhamou Y, Bochet M, Di Martino V, Charlotte F, Azria F, Coutellier A, et al. Liver Fibrosis Progression in Human Immunodeficiency Virus and Hepatitis C Virus Coinfected Patients. Multivirc Group Hepatol (1999) 30(4):1054–8. 10.1002/hep.510300409 [DOI] [PubMed] [Google Scholar]
- 50.Poynard T, Mathurin P, Lai CL, Guyader D, Poupon R, Tainturier MH, et al. A Comparison of Fibrosis Progression in Chronic Liver Diseases. J Hepatol (2003) 38(3):257–65. 10.1016/s0168-8278(02)00413-0 [DOI] [PubMed] [Google Scholar]
- 51.Klatt NR, Chomont N, Douek DC, Deeks SG. Immune Activation and HIV Persistence: Implications for Curative Approaches to HIV Infection. Immunol Rev (2013) 254(1):326–42. 10.1111/imr.12065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deeks SG, Lewin SR, Havlir DV. The End of AIDS: HIV Infection as a Chronic Disease. Lancet (2013) 382(9903):1525–33. 10.1016/S0140-6736(13)61809-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szereday L, Meggyes M, Berki T, Miseta A, Farkas N, Gervain J, et al. Direct-Acting Antiviral Treatment Downregulates Immune Checkpoint Inhibitor Expression in Patients With Chronic Hepatitis C. Clin Exp Med (2020) 20(2):219–30. 10.1007/s10238-020-00618-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Polyak SJ, Crispe IN, Baumert TF. Liver Abnormalities After Elimination of HCV Infection: Persistent Epigenetic and Immunological Perturbations Post-Cure. Pathogens (2021) 10(1):44. 10.3390/pathogens10010044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rockey DC. Fibrosis Reversal After Hepatitis C Virus Elimination. Curr Opin Gastroenterol (2019) 35(3):137–44. 10.1097/MOG.0000000000000524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schwabl P, Mandorfer M, Steiner S, Scheiner B, Chromy D, Herac M, et al. Interferon-Free Regimens Improve Portal Hypertension and Histological Necroinflammation in HIV/HCV Patients With Advanced Liver Disease. Aliment Pharmacol Ther (2017) 45(1):139–49. 10.1111/apt.13844 [DOI] [PubMed] [Google Scholar]
- 57.Fernandes FF, Piedade J, Guimaraes L, Nunes EP, Chaves U, Goldenzon RV, et al. Effectiveness of Direct-Acting Agents for Hepatitis C and Liver Stiffness Changing After Sustained Virological Response. J Gastroenterol Hepatol (2019) 34(12):2187–95. 10.1111/jgh.14707 [DOI] [PubMed] [Google Scholar]
- 58.Lledo GM, Carrasco I, Benitez-Gutierrez LM, Arias A, Royuela A, Requena S, et al. Regression of Liver Fibrosis After Curing Chronic Hepatitis C With Oral Antivirals in Patients With and Without HIV Coinfection. AIDS (2018) 32(16):2347–52. 10.1097/QAD.0000000000001966 [DOI] [PubMed] [Google Scholar]
- 59.Navarro J, Laguno M, Vilchez HH, Guardiola JM, Carrion JA, Force L, et al. Efficacy and Safety of Direct Antiviral Agents in a Cohort of Cirrhotic HCV/HIV-Coinfected Patients. J Antimicrob Chemother (2017) 72(10):2850–6. 10.1093/jac/dkx223 [DOI] [PubMed] [Google Scholar]
- 60.Olveira A, Dominguez L, Troya J, Arias A, Pulido F, Ryan P, et al. Persistently Altered Liver Test Results in Hepatitis C Patients After Sustained Virological Response With Direct-Acting Antivirals. J Viral Hepat (2018) 25(7):818–24. 10.1111/jvh.12883 [DOI] [PubMed] [Google Scholar]
- 61.Diez C, Berenguer J, Ibanez-Samaniego L, Llop E, Perez-Latorre L, Catalina MV, et al. Persistence of Clinically Significant Portal Hypertension After Eradication of Hepatitis C Virus in Patients With Advanced Cirrhosis. Clin Infect Dis (2020) 71(10):2726–9. 10.1093/cid/ciaa502 [DOI] [PubMed] [Google Scholar]
- 62.Medrano LM, Berenguer J, Salguero S, Gonzalez-Garcia J, Diez C, Hontanon V, et al. Successful HCV Therapy Reduces Liver Disease Severity and Inflammation Biomarkers in HIV/HCV-Coinfected Patients With Advanced Cirrhosis: A Cohort Study. Front Med (Lausanne) (2021) 8:615342. 10.3389/fmed.2021.615342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mascia C, Vita S, Zuccala P, Marocco R, Tieghi T, Savinelli S, et al. Changes in Inflammatory Biomarkers in HCV-Infected Patients Undergoing Direct Acting Antiviral-Containing Regimens With or Without Interferon. PloS One (2017) 12(6):e0179400. 10.1371/journal.pone.0179400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anthony DD, Sulkowski MS, Smeaton LM, Damjanovska S, Shive CL, Kowal CM, et al. Hepatitis C Virus (HCV) Direct-Acting Antiviral Therapy in Persons With Human Immunodeficiency Virus-HCV Genotype 1 Coinfection Resulting in High Rate of Sustained Virologic Response and Variable in Normalization of Soluble Markers of Immune Activation. J Infect Dis (2020) 222(8):1334–44. 10.1093/infdis/jiaa254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kostadinova L, Shive CL, Judge C, Zebrowski E, Compan A, Rife K, et al. During Hepatitis C Virus (HCV) Infection and HCV-HIV Coinfection, an Elevated Plasma Level of Autotaxin Is Associated With Lysophosphatidic Acid and Markers of Immune Activation That Normalize During Interferon-Free HCV Therapy. J Infect Dis (2016) 214(9):1438–48. 10.1093/infdis/jiw372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ghiglione Y, Polo ML, Urioste A, Rhodes A, Czernikier A, Trifone C, et al. Hepatitis C Virus (HCV) Clearance After Treatment With Direct-Acting Antivirals in Human Immunodeficiency Virus (HIV)-HCV Coinfection Modulates Systemic Immune Activation and HIV Transcription on Antiretroviral Therapy. Open Forum Infect Dis (2020) 7(5):ofaa115. 10.1093/ofid/ofaa115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marra F, Tacke F. Roles for Chemokines in Liver Disease. Gastroenterology (2014) 147(3):577–94.e1. 10.1053/j.gastro.2014.06.043 [DOI] [PubMed] [Google Scholar]
- 68.Askarieh G, Alsio A, Pugnale P, Negro F, Ferrari C, Neumann AU, et al. Systemic and Intrahepatic Interferon-Gamma-Inducible Protein 10 kDa Predicts the First-Phase Decline in Hepatitis C Virus RNA and Overall Viral Response to Therapy in Chronic Hepatitis C. Hepatology (2010) 51(5):1523–30. 10.1002/hep.23509 [DOI] [PubMed] [Google Scholar]
- 69.Sato T, Terai M, Tamura Y, Alexeev V, Mastrangelo MJ, Selvan SR. Interleukin 10 in the Tumor Microenvironment: A Target for Anticancer Immunotherapy. Immunol Res (2011) 51(2-3):170–82. 10.1007/s12026-011-8262-6 [DOI] [PubMed] [Google Scholar]
- 70.Aroucha DC, do Carmo RF, Moura P, Silva JL, Vasconcelos LR, Cavalcanti MS, et al. High Tumor Necrosis Factor-Alpha/Interleukin-10 Ratio is Associated With Hepatocellular Carcinoma in Patients With Chronic Hepatitis C. Cytokine (2013) 62(3):421–5. 10.1016/j.cyto.2013.03.024 [DOI] [PubMed] [Google Scholar]
- 71.Carlin AF, Aristizabal P, Song Q, Wang H, Paulson MS, Stamm LM, et al. Temporal Dynamics of Inflammatory Cytokines/Chemokines During Sofosbuvir and Ribavirin Therapy for Genotype 2 and 3 Hepatitis C Infection. Hepatology (2015) 62(4):1047–58. 10.1002/hep.27971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hengst J, Falk CS, Schlaphoff V, Deterding K, Manns MP, Cornberg M, et al. Direct-Acting Antiviral-Induced Hepatitis C Virus Clearance Does Not Completely Restore the Altered Cytokine and Chemokine Milieu in Patients With Chronic Hepatitis C. J Infect Dis (2016) 214(12):1965–74. 10.1093/infdis/jiw457 [DOI] [PubMed] [Google Scholar]
- 73.Saraiva GN, do Rosario NF, Medeiros T, Leite PEC, Lacerda GS, de Andrade TG, et al. Restoring Inflammatory Mediator Balance After Sofosbuvir-Induced Viral Clearance in Patients With Chronic Hepatitis C. Mediators Inflamm (2018) 2018:8578051. 10.1155/2018/8578051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Holmes JA, Carlton-Smith C, Kim AY, Dumas EO, Brown J, Gustafson JL, et al. Dynamic Changes in Innate Immune Responses During Direct-Acting Antiviral Therapy for HCV Infection. J Viral Hepat (2019) 26(3):362–72. 10.1111/jvh.13041 [DOI] [PubMed] [Google Scholar]
- 75.Sung PS, Lee EB, Park DJ, Lozada A, Jang JW, Bae SH, et al. Interferon-Free Treatment for Hepatitis C Virus Infection Induces Normalization of Extrahepatic Type I Interferon Signaling. Clin Mol Hepatol (2018) 24(3):302–10. 10.3350/cmh.2017.0074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bruno G, Saracino A, Monno L, Angarano G. The Revival of an "Old" Marker: CD4/CD8 Ratio. AIDS Rev (2017) 19(2):81–8. [PubMed] [Google Scholar]
- 77.Huson MA, Scicluna BP, van Vught LA, Wiewel MA, Hoogendijk AJ, Cremer OL, et al. The Impact of HIV Co-Infection on the Genomic Response to Sepsis. PloS One (2016) 11(2):e0148955. 10.1371/journal.pone.0148955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Landau DA, Rosenzwajg M, Saadoun D, Klatzmann D, Cacoub P. The B Lymphocyte Stimulator Receptor-Ligand System in Hepatitis C Virus-Induced B Cell Clonal Disorders. Ann Rheum Dis (2009) 68(3):337–44. 10.1136/ard.2007.085910 [DOI] [PubMed] [Google Scholar]
- 79.Juno JA, Kent SJ. What Can Gamma Delta T Cells Contribute to an HIV Cure? Front Cell Infect Microbiol (2020) 10:233. 10.3389/fcimb.2020.00233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Khaitan A, Unutmaz D. Revisiting Immune Exhaustion During HIV Infection. Curr HIV/AIDS Rep (2011) 8(1):4–11. 10.1007/s11904-010-0066-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Benkheil M, Van Haele M, Roskams T, Laporte M, Noppen S, Abbasi K, et al. CCL20, a Direct-Acting Pro-Angiogenic Chemokine Induced by Hepatitis C Virus (HCV): Potential Role in HCV-Related Liver Cancer. Exp Cell Res (2018) 372(2):168–77. 10.1016/j.yexcr.2018.09.023 [DOI] [PubMed] [Google Scholar]
- 82.Conti F, Buonfiglioli F, Scuteri A, Crespi C, Bolondi L, Caraceni P, et al. Early Occurrence and Recurrence of Hepatocellular Carcinoma in HCV-Related Cirrhosis Treated With Direct-Acting Antivirals. J Hepatol (2016) 65(4):727–33. 10.1016/j.jhep.2016.06.015 [DOI] [PubMed] [Google Scholar]
- 83.Ravi S, Axley P, Jones D, Kodali S, Simpson H, McGuire BM, et al. Unusually High Rates of Hepatocellular Carcinoma After Treatment With Direct-Acting Antiviral Therapy for Hepatitis C Related Cirrhosis. Gastroenterology (2017) 152(4):911–2. 10.1053/j.gastro.2016.12.021 [DOI] [PubMed] [Google Scholar]
- 84.Reig M, Marino Z, Perello C, Inarrairaegui M, Ribeiro A, Lens S, et al. Unexpected High Rate of Early Tumor Recurrence in Patients With HCV-Related HCC Undergoing Interferon-Free Therapy. J Hepatol (2016) 65(4):719–26. 10.1016/j.jhep.2016.04.008 [DOI] [PubMed] [Google Scholar]
- 85.Tawfik AK, Amin AM, Yousef M, El-Sayd NM, Elashry H, Elkadeem M, et al. IL-1alpha Correlates With Severity of Hepatitis C Virus-Related Liver Diseases. J Inflammation Res (2018) 11:289–95. 10.2147/JIR.S166564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shimoda K, Begum NA, Shibuta K, Mori M, Bonkovsky HL, Banner BF, et al. Interleukin-8 and hIRH (SDF1-Alpha/PBSF) mRNA Expression and Histological Activity Index in Patients With Chronic Hepatitis C. Hepatology (1998) 28(1):108–15. 10.1002/hep.510280116 [DOI] [PubMed] [Google Scholar]
- 87.Tachibana Y, Nakamoto Y, Mukaida N, Kaneko S. Intrahepatic Interleukin-8 Production During Disease Progression of Chronic Hepatitis C. Cancer Lett (2007) 251(1):36–42. 10.1016/j.canlet.2006.10.028 [DOI] [PubMed] [Google Scholar]
- 88.Han KQ, He XQ, Ma MY, Guo XD, Zhang XM, Chen J, et al. Inflammatory Microenvironment and Expression of Chemokines in Hepatocellular Carcinoma. World J Gastroenterol (2015) 21(16):4864–74. 10.3748/wjg.v21.i16.4864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang S, Liu Z, Wu D, Chen L, Xie L. Single-Cell RNA-Seq Analysis Reveals Microenvironmental Infiltration of Plasma Cells and Hepatocytic Prognostic Markers in HCC With Cirrhosis. Front Oncol (2020) 10:596318. 10.3389/fonc.2020.596318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Valatas V, Kolios G, Bamias G. TL1A (TNFSF15) and DR3 (TNFRSF25): A Co-Stimulatory System of Cytokines With Diverse Functions in Gut Mucosal Immunity. Front Immunol (2019) 10:583. 10.3389/fimmu.2019.00583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guo J, Luo Y, Yin F, Huo X, Niu G, Song M, et al. Overexpression of Tumor Necrosis Factor-Like Ligand 1 A in Myeloid Cells Aggravates Liver Fibrosis in Mice. J Immunol Res (2019) 2019:7657294. 10.1155/2019/7657294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zaldivar MM, Pauels K, von Hundelshausen P, Berres ML, Schmitz P, Bornemann J, et al. CXC Chemokine Ligand 4 (Cxcl4) is a Platelet-Derived Mediator of Experimental Liver Fibrosis. Hepatology (2010) 51(4):1345–53. 10.1002/hep.23435 [DOI] [PubMed] [Google Scholar]
- 93.Li J, Liu B, Yan LN, Lau WY. The Roles and Potential Therapeutic Implications of CXCL4 and its Variant CXCL4L1 in the Pathogenesis of Chronic Liver Allograft Dysfunction. Cytokine Growth Factor Rev (2015) 26(1):67–74. 10.1016/j.cytogfr.2014.11.004 [DOI] [PubMed] [Google Scholar]
- 94.van der Meer AJ, Berenguer M. Reversion of Disease Manifestations After HCV Eradication. J Hepatol (2016) 65(1 Suppl):S95–108. 10.1016/j.jhep.2016.07.039 [DOI] [PubMed] [Google Scholar]
- 95.Perez S, Kaspi A, Domovitz T, Davidovich A, Lavi-Itzkovitz A, Meirson T, et al. Hepatitis C Virus Leaves an Epigenetic Signature Post Cure of Infection by Direct-Acting Antivirals. PloS Genet (2019) 15(6):e1008181. 10.1371/journal.pgen.1008181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Holmes JA, Yu ML, Chung RT. Hepatitis B Reactivation During or After Direct Acting Antiviral Therapy - Implication for Susceptible Individuals. Expert Opin Drug Saf (2017) 16(6):651–72. 10.1080/14740338.2017.1325869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Perello MC, Fernandez-Carrillo C, Londono MC, Arias-Loste T, Hernandez-Conde M, Llerena S, et al. Reactivation of Herpesvirus in Patients With Hepatitis C Treated With Direct-Acting Antiviral Agents. Clin Gastroenterol Hepatol (2016) 14(11):1662–1666 e1. 10.1016/j.cgh.2016.05.016 [DOI] [PubMed] [Google Scholar]
- 98.El Kassas M, Wifi MN, Mahdy R, Afify S, Hafez E, El Latif YA, et al. Herpes Zoster Reactivation in Patients With Chronic Hepatitis C Under Treatment With Directly Acting Antiviral Agents: A Case Series. Arab J Gastroenterol (2017) 18(1):39–41. 10.1016/j.ajg.2017.02.003 [DOI] [PubMed] [Google Scholar]
- 99.Villani R, Facciorusso A, Bellanti F, Tamborra R, Piscazzi A, Landriscina M, et al. DAAs Rapidly Reduce Inflammation But Increase Serum VEGF Level: A Rationale for Tumor Risk During Anti-HCV Treatment. PloS One (2016) 11(12):e0167934. 10.1371/journal.pone.0167934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.ANRS Collaborative Study Group on Hepatocellular Carcinoma . Lack of Evidence of an Effect of Direct-Acting Antivirals on the Recurrence of Hepatocellular Carcinoma: Data From Three ANRS Cohorts. J Hepatol (2016) 65(4):734–40. 10.1016/j.jhep.2016.05.045 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding authors upon reasonable request. Patients’ raw sequence data are publicly available at the ArrayExpress repository (EMBL-EBI; https://www.ebi.ac.uk/) under the accession number E-MTAB-10703.