Abstract
Polymorphisms in genes that control immune function and regulation may influence susceptibility to pulmonary tuberculosis (TB). In this study, 14 polymorphisms in 12 key genes involved in the immune response (VDR, MR1, TLR1, TLR2, TLR10, SLC11A1, IL1B, IL10, IFNG, TNF, IRAK1, and FOXP3) were tested for their association with pulmonary TB in 271 patients with TB and 251 community-matched controls from the Republic of Moldova. In addition, gene–gene interactions involved in TB susceptibility were analyzed for a total of 43 genetic loci. Single nucleotide polymorphism (SNP) analysis revealed a nominal association between TNF rs1800629 and pulmonary TB (Fisher exact test P = 0.01843). In the pairwise interaction analysis, the combination of the genotypes TLR6 rs5743810 GA and TLR10 rs11096957 GT was significantly associated with an increased genetic risk of pulmonary TB (OR = 2.48, 95% CI = 1.62–3.85; Fisher exact test P value = 1.5 × 10−5, significant after Bonferroni correction). In conclusion, the TLR6 rs5743810 and TLR10 rs11096957 two-locus interaction confers a significantly higher risk for pulmonary TB; due to its high frequency in the population, this SNP combination may serve as a novel biomarker for predicting TB susceptibility.
Keywords: Gene polymorphisms, host genetics, immunity, pulmonary tuberculosis, susceptibility/resistance
Introduction
Tuberculosis (TB), caused by Mycobacterium tuberculosis, is a major cause of morbidity and mortality in many developing countries and is a significant threat to health in the developed world. One-third of the world’s population is infected with M. tuberculosis, but only a minor fraction (5–10%) develop an active form of the disease.1 Although several environmental and clinical conditions, such as diabetes mellitus, malnutrition, alcohol abuse, smoking, age, AIDS, and immunosuppressive therapies, are known to promote the development of the disease, many TB patients have no obvious risk factors. Evidence from animal and human studies indicates the importance of host genetic factors in the development of TB.2
To date, a considerable number of genetic candidates for TB susceptibility have been detected and characterized across populations by means of candidate gene approaches and genome-wide association studies (GWAS).3,4 Particular interest has focused on genetic factors of the immuno-inflammatory response that modulate infectivity and the progression of infection, including TLRs (TLR1–TLR10), natural resistance-associated macrophage protein (NRAMP1; also called solute carrier 11a1, SLC11A1), vitamin D receptor (VDR), cytokines TNF-α (TNF), IL-1 (IL1B), IFN-γ (IFNG), and IL-10 (IL10). Polymorphisms in these and other genes involved in the immune response have been reported to be associated with TB in different populations, although the results are inconsistent.3–5
We previously evaluated the effect of common genetic variations in the TLR pathway on the risk of pulmonary TB in a Moldavian population and identified variants in TLR2, TLR8, and TLR9 as being associated with TB.6 In the present work, we extended the analysis to 14 additional polymorphisms from 12 TB immune response candidate genes and investigated all pairwise genetic interactions for these and the previous genetic variants (43 polymorphisms altogether). The selected polymorphisms in this study have previously been shown to change the level or function of corresponding gene products and influence susceptibility/resistance to infections (Table 1).
Table 1.
Candidate genes and polymorphisms that are of interest in the present study.
| Gene | Locus | Protein | Gene class | Protein function | Polymorphism | Nucleotide substitution | Location | Frequency a | Reference |
|---|---|---|---|---|---|---|---|---|---|
| TLR1 | 4p14 | TLR 1 | PRR | Recognizes bacterial lipopeptides, activates macrophages and initiates immune response | rs5743618 | C>A | Missense (p.Ser602Ile) | 0.172–0.500 | Uciechowski et al., 2011,7 Naderi et al., 20168 |
| TLR2 | 4q31.3 | TLR 2 | PRR | Recognizes bacterial lipopeptides, activates macrophages and initiates immune response | rs111200466 | 23 bp del | 5′ UTR | 0.116-0.172 | Velez et al., 20109 |
| TLR10 | 4p14 | TLR 10 | PRR | Recognizes bacterial ligands, activates macrophages and initiates immune response | rs11096957 | T>G | Missense (p.Asn241His) | 0.324-0.486 | Oosting et al., 2014,10 Bulat-Kardum et al., 201511 |
| IRAK1 | Xq28 | IL-1 receptor-associated kinase 1 | Serine/threonine kinase | Regulates TLR and IL-1R mediated signaling | rs1059703 | G>A | Missense (p.Ser532Leu) | 0.088-0.211 | Sampath et al., 2013,12 Hu et al., 201513 |
| IFNG | 12q15 | IFN-γ | Cytokine | Activates macrophages to eliminate intracellular pathogens, including Mtb | rs2430561 | A>T | Intron | 0.354-0.528 | Pravica et al., 2000,14 Wei et al., 201715 |
| TNF | 6p21.3 | TNF-α | Cytokine | Activates macrophages to eliminate intracellular pathogens, attracts immunity cells to the site of infection | rs1800629 | G>A | Promoter | 0.094-0.187 | Correa et al., 2005,16 Elahi et al., 2009,17 de Arellano et al., 202018 |
| IL1B | 2q14.1 | IL-1β | Cytokine | Activates T-cells and promotes the production of IFN-γ | rs1143643 | C>T | Intron | 0.286-0.388 | Hall et al., 201519 |
| IL10 | 1q32.1 | IL-10 | Cytokine | Inhibits IFN-γ production and MHC class II expression on macrophages | rs1800896 | A>G | Promoter | 0.456-0.601 | Turner et al., 1997,20 Areeshi et al., 201721 |
| SLC11A1 | 2q35 | Natural Resistance-Associated Macrophage Protein 1 | Metal ion transporter | Influences Mtb survival by regulating cation levels in the macrophage | rs2276631 | C>T | Synonymous | 0.202-0.332 | Freĭdin et al. 2006,22 Yuan et al., 201723 |
| FOXP3 | Xp11.23 | Forkhead box protein P3 (FoxP3) | Transcriptional regulator | Inhibits cytokine production (IFN-γ) and T-cell effector function | rs2232365 | T>C | Intron | 0.338-0.450 | Beiranvand et al., 201724 |
| VDR | 12q13.11 | Vitamin D receptor | Ligand activated transcription factor | Regulates expression of a number of genes involved in killing of Mtb | rs7975232 | C>A | Intron | 0.404-0.539 | Uitterlinden et al., 2004,25 Xu and Shen, 201926 |
| rs1544410 | C>T | Intron | 0.338-0.470 | Uitterlinden et al., 2004,25 Xu and Shen, 2019,26 Chen et al., 201327 | |||||
| rs2228570 | A>G | Missense (p.Met1Thr) | 0.327-0.429 | Uitterlinden et al. 2004,25 Xu and Shen 2019,26 Chen et al. 201327 | |||||
| MR1 | 1q25.3 | MHC class I-related gene protein | Ag-presenting molecule | Presents metabolites of microbial vitamin B to MAITs | rs1052632 | G>A | Intron | 0.266-0.303 | Seshadri et al., 201728 |
a Allele frequency range in European populations (CEU, FIN, GBR, IBS, TSI) according to 1000 Genomes (http://www.1000genomes.org); minor allele in CEU was used as the reference. Minor allele in CEU population is underlined.
PRR: pattern recognition receptor. SNP rs2276631 in SLC11A1 is non-functional (synonymous) variant, which is in strong LD with the nearby TB associated SNP rs3731865 (INT4) (D′ = 0.9788 and r2 = 0.9479 in European populations).
The Republic of Moldova is a country with an unfavorable epidemiological situation regarding TB, ranking first for TB incidence (86/100,000 in 2018) in the European region.29 The identification of host genetic factors may provide a biological and theoretical basis for better understanding the high prevalence of TB within the Moldavian population and ultimately for the development of effective TB prevention and control strategies.
Materials and methods
Samples
The investigated case-control cohort included 272 unrelated patients (120 women and 152 men; mean age at recruitment = 40.7 ± 12.7 yr; mean age at diagnosis = 39.1 ± 12.2 yr) with infiltrative pulmonary TB. The diagnosis was based on clinical symptoms and chest radiographic findings and was confirmed by bacteriological assessment. None of the patients had a clinical history of diabetes mellitus, HIV infection, or receipt of immunosuppressive therapy. All patients were Caucasian and predominantly Moldavians (91.5%). The control group included 251 unrelated and ethnically matched (Moldavians, 87%) healthy individuals (150 women and 101 men; mean age at recruitment = 47.6 ± 14.1 yr) without history of TB and with normal radiographic examination findings. Control group subjects were recruited from the same high-burden TB-affected community as patients. Venous blood samples (EDTA anticoagulant added) were collected from TB patients and controls, and genomic DNA was extracted from peripheral blood leukocytes using a standard salting-out method.30 All cases and controls were vaccinated with the BCG vaccine, which was confirmed by the presence of a scar in the left shoulder. This study was approved by the Ethics Committee of the Institute of Phthisiopneumology (Republic of Moldova), and the Declaration of Helsinki protocols were followed. Each participant was thoroughly informed and provided written informed consent prior to enrollment in the study.
Polymorphism selection and genotyping
The following polymorphisms, VDR rs7975232, VDR rs1544410, VDR rs2228570, MR1 rs1052632, TLR1 rs5743618, TLR2 rs111200466, TLR10 rs11096957, SLC11A1 rs2276631, IL1B rs1143643, IL10 rs1800896, IFNG rs2430561, TNF rs1800629, IRAK1 rs1059703, and FOXP3 rs2232365 were investigated. Polymorphisms were selected based on previously published associations with TB, thus increasing the chance of selecting polymorphisms with phenotypic effects (Table 1). All polymorphisms except the insertion/deletion variant rs111200466 in TLR2 (–196 to –174 ins/del) were single nucleotide polymorphisms (SNPs).
The genotypes of the 12 SNPs (VDR rs7975232, VDR rs1544410, VDR rs2228570, MR1 rs1052632, TLR10 rs11096957, SLC11A1 rs2276631, IL1B rs1143643, IL10 rs1800896, IFNG rs2430561, TNF rs1800629, IRAK1 rs1059703, and FOXP3 rs2232365) were determined using Agena iPlex assays using a MALDI-TOF mass array spectrometer (Agena, San Diego, CA) following the manufacturer’s recommendations. Primers were designed using the Assay Design Suite v2.0 (https://agenacx.com). Two of the SNPs were not in Hardy–Weinberg equilibrium (HWE) in the control group: rs11096957 in TLR10 (P = 0.0071) and rs2430561 in IFNG (P = 0.013). Genotyping errors were excluded by random re-genotyping of the respective SNPs. Confirmatory genotyping of TLR10 rs11096957 was performed in 52 control samples (21%) using PCR–RFLP. PCR products were digested with the restriction endonuclease NlaIII (New England Biolabs; Arundel, Australia) and subjected to 2% agarose gel electrophoresis.
Confirmatory genotyping of IFNG rs2430561 was carried out by PCR with sequence-specific primers in 105 control samples (42%), and amplified SNP products were electrophoresed on 2% agarose gel. Additionally, SNPs TNF rs1800629 and TLR6 rs5743810 (the latter from our previous study)6, which exhibited a relationship with TB, were Sanger-sequenced (primers are in Supplemental Table S1) in 50 (∼10%) randomly selected samples, to confirm the quality of MALDI-TOF genotyping. All sequenced SNPs were 100% consistent with those obtained by MALDI-TOF.
Two polymorphisms, TLR1 rs5743618 and TLR2 rs111200466, could not be included in the Agena iPlex assay, and therefore, were genotyped using PCR and PCR–RFLP. Genotyping of rs5743618 was performed using PCR–RFLP. PCR products were digested with the restriction endonuclease AluI (New England Biolabs; Arundel, Australia) and subjected to 2% agarose gel electrophoresis. Genotyping of the rs111200466 polymorphism was conducted by standard PCR and agarose gel electrophoresis (2%).
The primer sequences, PCR conditions, and restriction enzymes for genotyping/sequencing of TLR2 rs111200466, IFNG rs2430561, TLR1 rs5743618, TLR10 rs11096957, TNF rs1800629, and TLR6 rs5743810 are outlined in Supplemental Table S1. The methodology used for the genotyping of TLR2 rs111200466, IFNG rs2430561, and TLR1 rs5743618 polymorphisms has been previously described.14,31,32
Statistical analysis
HWE tests were performed in controls and cases using Fisher’s exact test. Fisher’s exact test was also used to compare differences in allele frequencies between the groups. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated to measure the associations between SNPs and the risk of pulmonary TB. Minor alleles were assumed to be effect-associated alleles in all the tests. In addition, logistic regression was used to assess the association of each SNP with the risk of pulmonary TB using a log-additive genetic model. To account for possible bias due to statistically significant differences in age and sex distributions between the case and control groups (P < 0.0001 and P = 0.0004, respectively), the variables age and sex were included as covariates in the logistic regression analysis. All genetic tests described above were based on single SNP tests and were performed using the PLINK software package (version 1.9, http://pngu.mgh.harvard.edu/purcell/plink/).33 For IRAK1 rs1059703 and FOXP3 rs2232365 SNPs located on the X chromosome, HWE P values were calculated in females only and association tests were performed separately in men and women; therefore, 16 tests were conducted in total. To correct the significance level for multiple testing, Bonferroni correction was applied, and the significance level for all statistical tests was set at P < 0.003125 (0.05/16).
Haplotype analysis was performed for five polymorphisms in the TLR1-TLR6-TLR10 gene cluster, two polymorphisms in the TLR2 gene and three VDR gene polymorphisms using Haploview version 4.2 software.34 SNPs TLR1 rs4833095, TLR2 rs3804099, TLR6 rs5743810, and TLR10 rs11466657 were taken from the previous study conducted on the same cohort of TB patients and healthy controls. The solid spine of the linkage disequilibrium (LD) algorithm was applied to define the haplotype blocks. Differences in haplotype frequencies were tested using the χ2 test. Haplotypes with frequencies below 1% were excluded from the analyses. Multiple comparisons of haplotype frequencies were corrected by generating empirical P values via 10,000 permutations.
Pairwise epistatic interactions associated with susceptibility to TB were investigated via an exhaustive search of all pairwise combinations. There were 1065 combinations (741 common and 2 × 162 sex-specific) and an exhaustive search was the easiest way to find the best combination. Nonparametric methods such as multifactor dimensionality reduction (MDR) or k-nearest neighbors (KNN) check only a small part of the entire search space to be computationally effective and are applied mostly when a large number of markers are to be tested (e.g. in GWAS experiments).35,36 Another drawback of such methods is the interpretation of results, for example, when a strong marginal effect of one SNP makes the effects of several other polymorphisms in the interaction negligible.35,37 To address the latter, we restricted the search to the interactions of only two SNPs and evaluated the added value of the pairwise effects over individual effects.
Overall, pairwise epistatic analysis was performed as follows. First, we found SNP combinations associated with TB using logistic regression adjusted for sex and age under log-additive genetic model (implemented in software package SNPstats)38 for all pairs of the 43 genetic markers from this and our previous study.6 We used the log-additive model because it is a generalized model that is the best choice when the true inheritance pattern is unknown. Second, we found the exact genotype combination(s) that most contributed to the associations revealed in the previous step. Two methods were applied to all significant pairs (P < 0.05): logistic regression adjusted for sex and age, and Fisher’s exact test.39,40 Here, no genetic model was required for regression, since in both methods, one specific combination was tested against all other genotypes. The advantage of the Fisher test is its clear biological interpretation, but it is not trivial to adjust it to sex and age differences in the datasets. Therefore, logistic regression was used, which could easily incorporate such adjustment parameters.
To account for multiple testing, we used the Bonferroni correction and counted all tests of the first and second steps as independent, yielding a multiplication factor of 1/(1065 + 543) = 0.000622.
Power calculations were done using CaTS41 with a log-additive genetic model, assuming significance of 0.05 and the reported TB disease prevalence of 0.0020.29 The study had sufficient power (> 80%) to detect effect sizes (ORs) of 1.7 and 1.5 for high-risk allele frequencies of > 0.10 and > 0.20, respectively.
Results
Single polymorphism analysis
The overall genotyping performance in the study population was 83.2–100.0% depending on the SNP (Supplemental Table S2). Distributions of alleles of the investigated SNPs were in accordance with HWE in both groups except for TLR10 rs11096957 (Supplemental Table S2), which showed some deviation from HWE in controls (P = 0.0078). The minor allele frequencies of all polymorphisms in our controls were similar those in the populations of European descent from the 1000 Genomes Project database (Table 1). Association analysis between individual SNPs and TB revealed a moderate association between rs1800629 in TNF and pulmonary TB risk (Fisher exact test: OR = 0.63, P value = 0.01843; logistic regression: OR = 0.64, P = 0.03643) (Table 2), which did not remain significant after Bonferroni correction for multiple testing (P > 0.003125). None of the other loci showed an association with pulmonary TB risk (Table 2).
Table 2.
Association analysis of individual polymorphisms with pulmonary TB risk. For genotype data, see Supplementary Table 2.
| Polymorphism | Major/Minor allele | MAF in controls | MAF in patients | Fisher exact test |
Logistic regressiona |
||
|---|---|---|---|---|---|---|---|
| P Value | OR (95% CI) | P Value | OR (95% CI) | ||||
| Both sexes | |||||||
| VDR rs7975232 | C/A | 0.481 | 0.509 | 0.3797 | 1.12 (0.87–1.43) | 0.1819 | 1.20 (0.92–1.57) |
| VDR rs1544410 | C/T | 0.379 | 0.374 | 0.8977 | 0.98 (0.76–1.26) | 0.9695 | 0.99 (0.76–1.30) |
| VDR rs2228570 | G/A | 0.438 | 0.400 | 0.2262 | 0.85 (0.66–1.10) | 0.3169 | 0.88 (0.67–1.14) |
| MR1 rs1052632 | G/A | 0.271 | 0.259 | 0.7118 | 0.94 (0.70–1.26) | 0.7854 | 0.96 (0.71–1.30) |
| TLR1 rs5743618 | G/T | 0.402 | 0.407 | 0.8991 | 1.02 (0.80–1.31) | 0.9359 | 1.01 (0.78–1.32) |
| TLR2 rs111200466 | 23 bp ins/del | 0.175 | 0.165 | 0.6813 | 0.93 (0.68–1.29) | 0.5575 | 0.90 (0.65–1.27) |
| TLR10 rs11096957 | T/G | 0.474 | 0.472 | 0.9504 | 0.99 (0.78–1.27) | 0.7115 | 1.05 (0.82–1.35) |
| SLC11A1 rs2276631 | C/T | 0.242 | 0.249 | 0.8266 | 1.04 (0.78–1.39) | 0.9478 | 0.99 (0.73–1.35) |
| IL1B rs1143643 | C/T | 0.363 | 0.343 | 0.5127 | 0.92 (0.71–1.18) | 0.2367 | 0.85 (0.65–1.11) |
| IL10 rs1800896 | T/C | 0.447 | 0.447 | 1.0000 | 1.00 (0.77–1.31) | 0.9803 | 1.00 (0.75–1.32) |
| IFNG rs2430561 | T/A | 0.464 | 0.442 | 0.4925 | 0.92 (0.72–1.17) | 0.8409 | 1.02 (0.79–1.32) |
| TNF rs1800629 | G/A | 0.138 | 0.091 | 0.01843 | 0.63 (0.42–0.92) | 0.03643 | 0.64 (0.43–0.97) |
| Males | |||||||
| IRAK1 rs1059703 | A/G | 0.192 | 0.242 | 0.4356 | 1.34 (0.72–2.51) | 0.3563 | 1.34 (0.72–2.51) |
| FOXP3 rs2232365 | C/T | 0.404 | 0.456 | 0.4353 | 1.24 (0.74–2.07) | 0.4156 | 1.24 (0.74–2.07) |
| Females | |||||||
| IRAK1 rs1059703 | A/G | 0.250 | 0.233 | 0.686 | 0.91 (0.61–1.36) | 0.6544 | 0.91 (0.61–1.36) |
| FOXP3 rs2232365 | C/T | 0.463 | 0.413 | 0.2562 | 0.81 (0.58–1.15) | 0.2431 | 0.81 (0.58–1.15) |
MAF, minor allele frequency; OR, odds ratio; CI, confidence interval.
a Log-additive model adjusted for sex and age at recruitment.
Bold: significant associations (nominal P value < 0.05).
Haplotype analysis
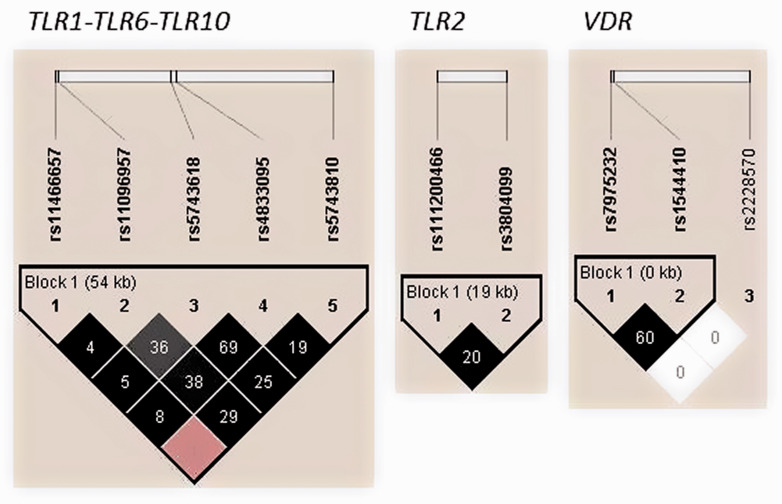
Nine common haplotypes in the gene cluster TLR1-TLR6-TLR10, four in TLR2, and three in VDR were identified at a frequency ≥1% by LD analysis (Figure 1). The rs11466657-rs11096957-rs5743618-rs4833095-rs5743810 haplotype A-T-G-T-A of the gene cluster TLR1-TLR6-TLR10 was significantly higher in the TB group compared with the controls (P = 0.0485; Table 3). However, after 10,000-fold permutation testing, this haplotype was not significant (P = 0.4463). In addition, marginal differences were observed between cases and controls for the haplotype A-T-G-T-G (rs11466657-rs11096957-rs5743618-rs4833095-rs5743810) in the gene cluster TLR1-TLR6-TLR10 (P = 0.0688) and haplotypes ins-T and ins-C (rs111200466-rs3804099) in TLR2 (P = 0.0732 and P = 0.0987, respectively). None of the remaining haplotypes were associated with the risk of pulmonary TB in this study (Table 3).
Figure 1.
Linkage disequilibrium (LD) plots of TLR2, VDR and TLR1-TLR6-TLR10 polymorphisms in cases and controls. The colours of the squares represent D’ values, with black being D’=1, and white D’=0. r2 values (%) are indicated on the squares.
Table 3.
Haplotype frequencies and associations with pulmonary TB.
| Haplotypesa | Frequency inPatientsb | Frequency inControls | P Valuec | PermutationP value |
|---|---|---|---|---|
| TLR2 | ||||
| insT | 0.591 | 0.536 | 0.0732 | 0.6576 |
| insC | 0.244 | 0.289 | 0.0987 | 0.7665 |
| delC | 0.154 | 0.160 | 0.7951 | 1 |
| delT | 0.012 | 0.016 | 0.5744 | 1 |
| TLR1-TLR6-TLR10 | ||||
| ATGTA | 0.298 | 0.243 | 0.0485 | 0.4463 |
| AGTCG | 0.255 | 0.263 | 0.7784 | 1 |
| ATGTG | 0.170 | 0.215 | 0.0688 | 0.6201 |
| AGGTG | 0.110 | 0.121 | 0.5933 | 1 |
| AGTTG | 0.045 | 0.041 | 0.7844 | 1 |
| GGTCG | 0.044 | 0.028 | 0.1705 | 0.9125 |
| ATTTG | 0.030 | 0.033 | 0.7762 | 1 |
| ATTCG | 0.023 | 0.034 | 0.2712 | 0.9818 |
| AGGTA | 0.010 | 0.018 | 0.27 | 0.9816 |
| VDR | ||||
| CC | 0.492 | 0.515 | 0.4556 | 1 |
| AT | 0.375 | 0.371 | 0.8939 | 1 |
| AC | 0.133 | 0.113 | 0.3484 | 0.9917 |
aHaplotypes: TLR2 [rs111200466, rs3804099]; TLR1-TLR6-TLR10 [rs11466657, rs11096957, rs5743618, rs4833095, rs5743810]; VDR [rs7975232, rs1544410]
bOnly haplotypes with a frequency > 0.01 were included in this table.
CP Values were calculated using χ2 test.
Bold: significant differences (nominal P value < 0.05) in haplotype distribution.
Gene–gene interaction analysis
Analysis of SNP–SNP combinations using a logistic regression model revealed 59 significant (P value < 0.05) genetic interactions (Supplemental Table S3), with the smallest P values observed for the marker pairs TOLLIP rs3793964–IRAK2 rs3844283 (P = 0.0025), TLR2 rs111200466–TOLLIP rs5743899 (P = 0.0044), and TLR10 rs11096957–TLR6 rs5743810 (P = 0.0052). Although none of these associations exceeded the conservative Bonferroni-corrected threshold of P value = 4.7 × 10−5 (0.05/1065), they suggest a potential role in conferring TB risk and therefore were pipelined into the in-depth genotype combination analysis using Fisher’s exact test and logistic regression adjusted for sex and age differences in the datasets. Genotype combinations reported by both methods were considered significant.
This resulted in 62 nominally significant associations (P value < 0.05 for either logistic regression or Fisher’s exact test) out of 543 possible genotype combinations (Supplemental Table S4). The strongest association was observed between TB and the combination of TLR6 rs5743810 GA and TLR10 rs11096957 GT genotypes (Fisher exact test P value = 1.5 × 10−5; logistic regression P value = 1.9 × 10−5; Table 4). This association remained significant after Bonferroni correction (P value = 0.05/(1065 + 543) = 3.5 × 10−5). Remarkably, the effect of this genotype combination on TB risk (OR = 2.48) was much greater than that of the individual genotypes (heterozygous model, OR = 1.72, P = 0.0042 and OR = 1.33, P = 0.13 for rs5743810 and rs11096957, respectively), implying a strong synergistic interaction between the two polymorphisms. None of the other genotype combinations were significant after the Bonferroni correction.
Table 4.
Association of combined genotypes TLR6 rs5743810 and TLR10 rs11096957 with pulmonary TB.
| Genotype |
Counts, n (%) |
Association (P value) |
OR (95% CI)a | |||
|---|---|---|---|---|---|---|
| TLR6 rs5743810 | TLR10 rs11096957 | Controls | Patients | Fisher test | Logistic regressiona | |
| GG | TT | 58 (23.8) | 56 (21.1) | 0.56 | 0.83 | 0.94 (0.60–1.47) |
| GG | GT | 57 (23.4) | 43 (16.2) | 0.059 | 0.033 | 0.61 (0.38–0.96) |
| GG | GG | 19 (7.8) | 16 (6.0) | 0.55 | 0.59 | 0.80 (0.37–1.72) |
| GA | TT | 7 (2.9) | 2 (0.8) | 0.14 | 0.094 | 0.25 (0.025–1.36) |
| GA | GT | 42 (17.2) | 91 (34.3) | 1.5 × 10−5 | 1.9 × 10−5 | 2.48 (1.62–3.85) |
| GA | GG | 42 (17.2) | 38 (14.3) | 0.45 | 0.34 | 0.79 (0.48–1.30) |
| AA | TT | 17 (7.0) | 18 (6.8) | 0.94 | 0.86 | 0.91 (0.43–1.91) |
| AA | GT | 2 (0.8) | 1 (0.4) | 0.94 | 0.60 | 0.45 (0.0077–8.86) |
a Adjusted for sex and age at recruitment.
Bold: significant association after Bonferroni correction (P value < 3.10945 × 10−5).
Discussion
In the present study, we applied a candidate gene approach and tested the association of 14 polymorphisms in genes VDR, MR1, TLR1, TLR2, TLR10, SLC11A1, IL1B, IL10, IFNG, TNF, IRAK1, and FOXP3 with the risk of pulmonary TB in Moldavian population. Although these genes are critical components of human immunity and their polymorphisms have been implicated in susceptibility to TB, we could not find convincing statistical evidence for their association with the risk of pulmonary TB. A borderline association was revealed for polymorphism rs1800629 in the TNF gene only, but this was not significant after corrections for multiple testing by the Bonferroni method. Overall, the results presented here do not support a major role of the analyzed common variants in conferring susceptibility or resistance to pulmonary TB in the Moldavian population.
The inability to reach significance level after Bonferroni correction may be explained by a fairly small sample size and inadequate statistical power to produce convincing associations for polymorphisms with low and moderate effects (OR < 1.5). For this reason, we cannot completely rule out the possibility of true functional effects for variants with nominal associations, particularly for the TNF rs1800629 polymorphism. The TNF gene encodes a multifunctional pro-inflammatory cytokine, TNF-α, which is mainly produced by activated monocytes, macrophages, and T-lymphocytes when stimulated by mycobacterial antigens. TNF-α acts synergistically with IFN-γ to induce macrophage killing of M. tuberculosis.42 In addition, TNF-α is involved in the recruitment of leukocytes to the site of infection and contributes to the formation of TB granuloma, preventing the spread of infection.42 It is reported that treatment with TNF-α inhibitors leads to reactivation of latent TB infection, indicating TNF-α is a key cytokine toward resistance to M. tuberculosis.43 SNP rs1800629 (−308G>A) is located within regulatory hotspot region and thus influences transcription critically.44,45 The minor allele A of rs1800629 has been associated with increased expression of TNF and higher plasma levels of TNF-α.17 In agreement with published functional studies, our results demonstrated a higher frequency of allele A in controls than in cases, suggesting its protective role against TB (OR = 0.63). Similarly, allele A was protectively associated with TB in the Colombian16 and Mexican18 populations. However, other genetic epidemiologic studies involving patient cohorts from various population groups, including Malawi,46 Iran,47 Indian,48 Cambodian,49 Chinese Han, and Tibetan,50 did not confirm these findings. The disparity in results across studies may be explained by certain factors, such as inadequate sample sizes and differences in environmental, demographic, cultural, host genetic, and bacterial characteristics of M. tuberculosis strains.
Additionally, a nominal yet suggestive association was demonstrated for haplotype A-T-G-T-A of the block rs11466657-rs11096957-rs5743618-rs4833095-rs5743810 in the cluster TLR1-TLR6-TLR10. TLR1, TLR6, and TLR10 are located in a 54-kb genomic region on chromosome 4p14 and encode proteins that share a high degree of homology in their amino acid sequences. All three genes belong to the TLR2 subfamily of TLRs, which plays a critical role in the early recognition of M. tuberculosis and subsequent activation of immune responses.51 Individual polymorphisms and haplotypes within the TLR10-TLR1-TLR6 locus have been associated with altered susceptibility to infectious diseases, including mycobacterial infections of leprosy and TB.11,52–55 Unfortunately, different sets of SNPs used in this and other studies complicate direct comparisons of the results. Even so, the identified haplotype A-T-G-T-A and, more generally, variations in genes TLR10, TLR1, and TLR6 could be promising replication targets for future studies in larger cohorts.
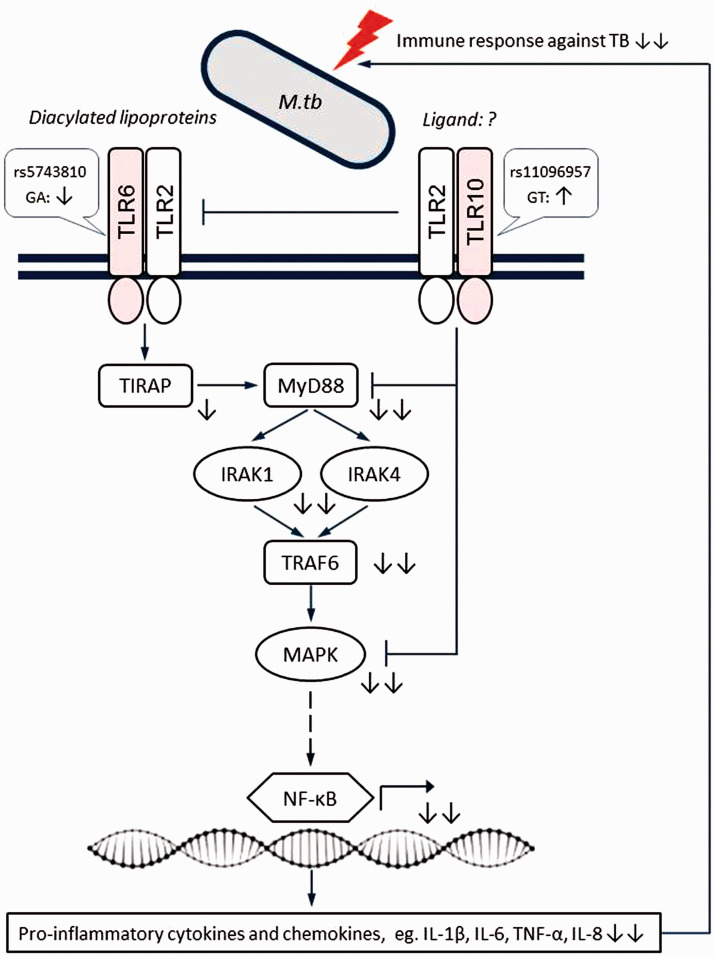
Genetic interactions are thought to underlie susceptibility or resistance to TB,5,56 so they could explain some of the missing heritability in this study. Therefore, we also analyzed the impact of allele combinations on the risk of TB. The strongest evidence for interaction in our data was between SNPs rs5743810 and rs11096957 located in TLR6 and TLR10, respectively. Interestingly, the two SNPs showed no or only a weak effect on TB susceptibility when evaluated alone, indicating a synergetic mechanism of TLR6 rs5743810 and TLR10 rs11096957 in conferring risk for pulmonary TB. The interactions between these SNPs are biologically plausible. First, TLRs are key players in host defense against infections. Specifically, TLR6 functionally interacts with TLR2 to mediate the cellular response to bacterial lipoproteins and activate the NF-κB pathway and inflammatory events through MyD88-dependent signaling.10,51,55 TLR10 also can form heterodimers with TLR2, but its specific ligands have not yet been identified and its downstream signaling is not fully understood. It is thought to act through both MyD88-dependent and -independent signaling pathways with inhibitory effects on inflammation.10 The genetic interaction between TLR6 and TLR10 observed in this study may reflect their mutual functional contribution to M. tuberculosis recognition and subsequent downstream signaling (Figure 2). Second, the SNPs in TLR6 and TLR10 genes were previously shown to be of functional significance. In fact, the two SNPs are non-synonymous variants located in the extracellular (leucine-rich repeat) domains of the encoded proteins. Both ex vivo and in vitro experiments showed that SNPs rs5743810 (Ser249Pro) and rs11096957 (Asn241His) may influence pro-inflammatory cytokine production in humans.10,57,58 In addition, rs5743810 was observed to affect NF-κB signaling activity, thereby modulating inflammatory responses.59 Furthermore, these two polymorphisms have been associated with several immune-related pathologic conditions and infectious diseases, including TB.11,54,55,60 These data support the relevance of additive interaction between SNPs rs5743810 and rs11096957 and suggest a molecular mechanism by which genetic variations in TLR6 and TLR10 genes might increase susceptibility to TB (Figure 2).
Figure 2.
Schematic representation of the proposed epistatic/synergistic interaction between SNPs TLR6 rs5743810 and TLR10 rs11096957 in conferring susceptibility to TB based on the present results.TLR6 and TLR10 have largely opposite effects on the immune response, with TLR10 mainly having a suppressive function. Simultaneous suppression of TLR6 signaling and activation of TLR10 signaling caused by genetic variations may result in decreased pro-inflammatory responses against M. tuberculosis, thus increasing the risk of TB. Symbols indicate the following: sharp arrow – positive interaction; blunt arrow – negative interaction; solid line – direct interaction; dashed line – indirect interaction; question mark – unknown ligand partner; up/down arrows – activation/suppression; double arrows – synergistic effects of two mutations. Figure adapted from Oosting et al. (2014).10
The present study is the first to identify an interaction between TLR6 rs5743810 and TLR10 rs11096957 gene variants in TB risk. Further larger case-control studies followed by functional tests are warranted to validate this initial finding and eventually translate it into clinical practice. Particularly, given the high spread of the combined TLR6 rs5743810 GA–TLR10 rs11096957 GT genotype in the European population (∼15–20%), it might be used as a novel predictive biomarker for identification of individuals at high risk for active TB disease.
Some limitations of our study should be considered. First, it was limited in power to detect weak association signals, so our negative results should be treated with caution. Second, healthy controls were not tested for latent M. tuberculosis infection, and therefore, it was not possible to discriminate between TB-infected and TB-uninfected individuals. However, as mentioned in the “Materials and methods,” the controls were recruited from TB communities where they were permanently exposed to TB and were therefore expected to be infected. Third, the number of polymorphisms in the immune system genes analyzed was limited. Given their key role in TB pathogenesis, additional TB risk variants, haplotypes, and allele combinations may exist.
A potential limitation of this study is the significant deviation from the HWE of the interacting SNP TLR10 rs11096957 (Asn241His) in the controls. Such deviations can result from genotyping errors, recruiting biases, natural selection, or simply chance. We excluded genotyping errors by random re-genotyping of TLR10 rs11096957 in 21% of the samples. In addition, our study design prevented the recruitment of relatives. Natural selection could be the reason for the observed deviation, which may be supported by the evidence of similar heterozygosity deficiency in Toscani in Italy, TSI (HWE P value = 0.02; 1000 Genomes Project data), and considerable intra-population variation of SNP TLR10 rs11096957 within Europe (G allele frequency range: 32.4% in British, GBR to 48.6% in Iberian population, IBS; 1000 Genomes Project data). Moreover, the recruitment of healthy controls from TB communities used in this project may have a similar impact on TLR10 to natural selection, contributing to the deficiency of rs11096957 heterozygotes. Taken together, these arguments justify the inclusion of TLR10 rs11096957 in association tests.
Conclusion
In the current study, we again found a significant association between rs1800629 in the TNF gene and pulmonary TB. In addition, haplotypes in the gene cluster TLR1-TLR6-TLR10 showed a weakly yet suggestive association. Furthermore, we provide convincing statistical evidence for a synergistic effect between polymorphic variants in the TLR6 and TLR10 genes on TB risk, which further supports the importance of TLR signaling in the genetic basis of TB and the concept of genetic interactions accounting for missing heritability. Further studies are warranted to validate the interaction between TLR6 and TLR10 and to elucidate its role in TB risk, which would be beneficial for human health.
Supplemental Material
Supplemental material, sj-pdf-1-ini-10.1177_17534259211029996 for Synergistic effect of genetic polymorphisms in TLR6 and TLR10 genes on the risk of pulmonary tuberculosis in a Moldavian population by Alexander Varzari, Igor V. Deyneko, Elena Tudor, Harald Grallert and Thomas Illig in Innate Immunity
Supplemental material, sj-pdf-2-ini-10.1177_17534259211029996 for Synergistic effect of genetic polymorphisms in TLR6 and TLR10 genes on the risk of pulmonary tuberculosis in a Moldavian population by Alexander Varzari, Igor V. Deyneko, Elena Tudor, Harald Grallert and Thomas Illig in Innate Immunity
Supplemental material, sj-xlsx-3-ini-10.1177_17534259211029996 for Synergistic effect of genetic polymorphisms in TLR6 and TLR10 genes on the risk of pulmonary tuberculosis in a Moldavian population by Alexander Varzari, Igor V. Deyneko, Elena Tudor, Harald Grallert and Thomas Illig in Innate Immunity
Supplemental material, sj-xlsx-4-ini-10.1177_17534259211029996 for Synergistic effect of genetic polymorphisms in TLR6 and TLR10 genes on the risk of pulmonary tuberculosis in a Moldavian population by Alexander Varzari, Igor V. Deyneko, Elena Tudor, Harald Grallert and Thomas Illig in Innate Immunity
Acknowledgements
We thank all the patients and control subjects for participating in this study and providing blood samples. We would like to thank Editage (www.editage.com) for English language editing.
Footnotes
Declaration of conflicting interests: The authors declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethics approval and consent to participate: The study was approved by the Research Ethic Committee of the Institute of Phthisiopneumology (Republic of Moldova) and was conducted in accordance with the Declaration of Helsinki. All participants gave their written informed consent.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The study was supported by the Academy of Sciences of Moldova and Hannover Unified Biobank. Alexander Varzari was sponsored by the Alexander von Humboldt Foundation.
ORCID iD: Alexander Varzari https://orcid.org/0000-0003-1855-3095
Supplemental material: Supplemental material for this article is available online.
References
- 1.Lawn SD, Zumla AI.Tuberculosis. Lancet 2011; 378: 57–72. [DOI] [PubMed] [Google Scholar]
- 2.Orlova M, Schurr E.Human genomics of Mycobacterium tuberculosis infection and disease. Curr Genet Med Rep 2017; 5: 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stein CM, Sausville L, Wejse C, et al. Genomics of human pulmonary tuberculosis: From genes to pathways. Curr Genet Med Rep 2017; 5: 149–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai L, Li Z, Guan X, et al. The research progress of host genes and tuberculosis susceptibility. Oxid Med Cell Longev 2019; 2019: 9273056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abel L, Fellay J, Haas DW, et al. Genetics of human susceptibility to active and latent tuberculosis: Present knowledge and future perspectives. Lancet Infect Dis 2018; 18: e64–e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varzari A, Deyneko IV, Vladei I, et al. Genetic variation in TLR pathway and the risk of pulmonary tuberculosis in a Moldavian population. Infect Genet Evol 2019; 68: 84–90. [DOI] [PubMed] [Google Scholar]
- 7.Uciechowski P, Imhoff H, Lange C, et al. Susceptibility to tuberculosis is associated with TLR1 polymorphisms resulting in a lack of TLR1 cell surface expression. J Leukoc Biol 2011; 90: 377–388. [DOI] [PubMed] [Google Scholar]
- 8.Naderi M, Hashemi M, Mirshekari H, et al. Toll-like receptor 1 polymorphisms increased the risk of pulmonary tuberculosis in an Iranian population sample. Biomed Environ Sci 2016; 29: 825–828. [DOI] [PubMed] [Google Scholar]
- 9.Velez DR, Wejse C, Stryjewski ME, et al. Variants in toll-like receptors 2 and 9 influence susceptibility to pulmonary tuberculosis in Caucasians, African-Americans, and West Africans. Hum Genet 2010; 127: 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oosting M, Cheng SC, Bolscher JM, et al. Human TLR10 is an anti-inflammatory pattern-recognition receptor. Proc Natl Acad Sci U S A 2014; 111: E4478–E4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bulat-Kardum LJ, Etokebe GE, Lederer P, et al. Genetic polymorphisms in the Toll-like Receptor 10, Interleukin (IL)17A and IL17F genes differently affect the risk for tuberculosis in Croatian population. Scand J Immunol 2015; 82: 63–69. [DOI] [PubMed] [Google Scholar]
- 12.Sampath V, Mulrooney NP, Garland JS, et al. Toll-like receptor genetic variants are associated with Gram-negative infections in VLBW infants. J Perinatol 2013; 33: 772–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu CY, Zhang XA, Meyer CG, et al. Polymorphism of X-linked CD40 ligand gene associated with pulmonary tuberculosis in the Han Chinese population. Genes Immun 2015; 16: 399–404. [DOI] [PubMed] [Google Scholar]
- 14.Pravica V, Perrey C, Stevens A, et al. A single nucleotide polymorphism in the first intron of the human IFN-g gene: Absolute correlation with a polymorphic CA microsatellite marker of high IFN-g production. Hum Immunol 2000; 61: 863–866. [DOI] [PubMed] [Google Scholar]
- 15.Wei Z, Wenhao S, Yuanyuan M, et al. A single nucleotide polymorphism in the interferon-γ gene (IFNG +874 T/A) is associated with susceptibility to tuberculosis. Oncotarget 2017; 8: 50415–50429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Correa PA, Gomez LM, Cadena J, et al. Autoimmunity and tuberculosis. Opposite association with TNF polymorphism. J Rheumatol 2005; 32: 219–224. [PubMed] [Google Scholar]
- 17.Elahi MM, Asotra K, Matata BM, et al. Tumor necrosis factor alpha -308 gene locus promoter polymorphism: an analysis of association with health and disease. Biochim Biophys Acta 2009; 1792: 163–172. [DOI] [PubMed] [Google Scholar]
- 18.de Arellano ITR, Lara CS, Espíndola LMT, et al. Exposure to biomass smoke, cigarettes, and alcohol modifies the association between tumour necrosis factor (-308G/A, -238G/A) polymorphisms and tuberculosis in Mexican carriers. Arch Med Sci 2020; 16: 672–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall NB, Igo RP, Jr, Malone LL, et al. Polymorphisms in TICAM2 and IL1B are associated with TB. Genes Immun 2015; 16: 127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turner DM, Williams DM, Sankaran D, et al. An investigation of polymorphism in the interleukin-10 genepromoter. Eur J Immunogenet 1997; 24: 1–8. [DOI] [PubMed] [Google Scholar]
- 21.Areeshi MY, Mandal RK, Dar SA, et al. IL-10 -1082 A>G (rs1800896) polymorphism confers susceptibility to pulmonary tuberculosis in Caucasians but not in Asians and Africans: a meta-analysis. Biosci Rep 2017; 37: BSR20170240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freĭdin MB, Rudko AA, Kolokolova OV, et al. A comparative analysis of tuberculosis susceptibility genetic make-up in Tuvinians and Russians. Mol Biol (Mosk) 2006; 40: 252–262. [PubMed] [Google Scholar]
- 23.Yuan L, Ke Z, Guo Y, et al. NRAMP1 D543N and INT4 polymorphisms in susceptibility to pulmonary tuberculosis: A meta-analysis. Infect Genet Evol 2017; 54: 91–97. [DOI] [PubMed] [Google Scholar]
- 24.Beiranvand E, Abediankenari S, Khani S, et al. G allele at -924 A > G position of FoxP3 gene promoter as a risk factor for tuberculosis. BMC Infect Dis 2017; 17: 673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uitterlinden AG, Fang Y, Van Meurs JB, et al. Genetics and biology of vitamin D receptor polymorphisms. Gene 2004; 338: 143–156. [DOI] [PubMed] [Google Scholar]
- 26.Xu X, Shen M.Associations between vitamin D receptor genetic variants and tuberculosis: a meta-analysis. Innate Immun 2019; 25: 305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C, Liu Q, Zhu L, et al. Vitamin D receptor gene polymorphisms on the risk of tuberculosis, a meta-analysis of 29 case-control studies. PLoS One 2013; 8: e83843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seshadri C, Thuong NT, Mai NT, et al. A polymorphism in human MR1 is associated with mRNA expression and susceptibility to tuberculosis. Genes Immun 2017; 18: 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.World Health Organization. Global tuberculosis report 2019. Geneva: WHO, 2019. [Google Scholar]
- 30.Miller SA, Dykes DD, Polesky HF.A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16: 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tahara T, Arisawa T, Wang F, et al. Toll-like receptor 2 -196 to 174del polymorphism influences the susceptibility of Japanese people to gastric cancer. Cancer Sci 2007; 98: 1790–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leoratti FM, Farias L, Alves FP, et al. Variants in the toll-like receptor signaling pathway and clinical outcomes of malaria. J Infect Dis 2008; 198: 772–780. [DOI] [PubMed] [Google Scholar]
- 33.Chang CC, Chow CC, Tellier LC, et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015; 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barrett JC, Fry B, Maller J, et al. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21: 263–265. [DOI] [PubMed] [Google Scholar]
- 35.Musani SK, Shriner D, Liu N, et al. Detection of gene x gene interactions in genome-wide association studies of human population data. Hum Hered 2007; 63: 67–84. [DOI] [PubMed] [Google Scholar]
- 36.Abo Alchamlat S, Farnir F.KNN-MDR: A learning approach for improving interactions mapping performances in genome wide association studies. BMC Bioinform 2017; 18: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edwards TL, Turner SD, Torstenson ES, et al. A general framework for formal tests of interaction after exhaustive search methods with applications to MDR and MDR-PDT. PLoS One 2010; 5: e9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Solé X, Guinó E, Valls J, et al. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006; 22: 1928–1929. [DOI] [PubMed] [Google Scholar]
- 39.Varzari A, Deyneko IV, Tudor E, et al. Polymorphisms of glutathione S-transferase and methylenetetrahydrofolate reductase genes in Moldavian patients with ulcerative colitis: Genotype-phenotype correlation. Meta Gene 2015; 7: 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varzari A, Tudor E, Bodrug N, et al. Age-specific association of CCL5 gene polymorphism with pulmonary tuberculosis: A case-control study. Genet Test Mol Biomarkers 2018; 22: 281–287. [DOI] [PubMed] [Google Scholar]
- 41.Skol AD, Scott LJ, Abecasis GR, et al. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet 2006; 38: 209–213. [DOI] [PubMed] [Google Scholar]
- 42.Cavalcanti YV, Brelaz MC, Neves JK, et al. Role of TNF-Alpha, IFN-Gamma, and IL-10 in the development of pulmonary tuberculosis. Pulm Med 2012; 2012: 745483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Long R, Gardam M.Tumour necrosis factor-alpha inhibitors and the reactivation of latent tuberculosis infection. CMAJ 2003; 168: 1153–1156. [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson AG, Symons JA, McDowell TL, et al. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci U S A 1997; 94: 3195–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ponomarenko M, Rasskazov D, Chadaeva I, et al. Candidate SNP markers of atherogenesis significantly shifting the affinity of TATA-binding protein for human gene promoters show stabilizing natural selection as a sum of neutral drift accelerating atherogenesis and directional natural selection slowing it. Int J Mol Sci 2020; 21: 1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fitness J, Floyd S, Warndorff DK, et al. Large-scale candidate gene study of tuberculosis susceptibility in the Karonga district of northern Malawi. Am J Trop Med Hyg 2004; 71: 341–349. [PubMed] [Google Scholar]
- 47.Jafari M, Nasiri MR, Sanaei R, et al. The NRAMP1, VDR, TNF-α, ICAM1, TLR2 and TLR4 gene polymorphisms in Iranian patients with pulmonary tuberculosis: A case-control study. Infect Genet Evol 2016; 39: 92–98. [DOI] [PubMed] [Google Scholar]
- 48.Selvaraj P, Sriram U, Mathan Kurian S, et al. Tumour necrosis factor alpha (-238 and -308) and beta gene polymorphisms in pulmonary tuberculosis: Haplotype analysis with HLA-A, B and DR genes. Tuberculosis (Edinb) 2001; 81: 335–341. [DOI] [PubMed] [Google Scholar]
- 49.Delgado JC, Baena A, Thim S, et al. Ethnic-specific genetic associations with pulmonary tuberculosis. J Infect Dis 2002; 186: 1463–1468. [DOI] [PubMed] [Google Scholar]
- 50.Wu S, Wang MG, Wang Y, et al. Polymorphisms of cytokine genes and tuberculosis in two independent studies. Sci Rep 2019; 9: 2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Casanova JL, Abel L, Quintana-Murci L.Human TLRs and IL-1Rs in host defense: Natural insights from evolutionary, epidemiological, and clinical genetics. Annu Rev Immunol 2011; 29: 447–491. [DOI] [PubMed] [Google Scholar]
- 52.Ma X, Liu Y, Gowen BB, et al. Full-exon resequencing reveals toll-like receptor variants contribute to human susceptibility to tuberculosis disease. PLoS One 2007; 2: e1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dittrich N, Berrocal-Almanza LC, Thada S, et al. Toll-like receptor 1 variations influence susceptibility and immune response to Mycobacterium tuberculosis. Tuberculosis (Edinb) 2015; 95: 328–335. [DOI] [PubMed] [Google Scholar]
- 54.Barletta-Naveca RH, Naveca FG, de Almeida VA, et al. Toll-Like Receptor-1 single-nucleotide polymorphism 1805T/G is associated with predisposition to multibacillary tuberculosis. Front Immunol 2018; 9: 1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mukherjee S, Huda S, Sinha Babu SP.Toll-like receptor polymorphism in host immune response to infectious diseases: A review. Scand J Immunol 2019; 90: e12771. [DOI] [PubMed] [Google Scholar]
- 56.de Wit E, van der Merwe L, van Helden PD, et al. Gene-gene interaction between tuberculosis candidate genes in a South African population. Mamm Genome 2011; 22: 100–110. [DOI] [PubMed] [Google Scholar]
- 57.Kormann MS, Depner M, Hartl D, et al. Toll-like receptor heterodimer variants protect from childhood asthma. J Allergy Clin Immunol 2008; 122: 86–92. [DOI] [PubMed] [Google Scholar]
- 58.Randhawa AK, Shey MS, Keyser A, et al. Association of human TLR1 and TLR6 deficiency with altered immune responses to BCG vaccination in South African infants. PLoS Pathog 2011; 7: e1002174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shey MS, Randhawa AK, Bowmaker M, et al. Single nucleotide polymorphisms in toll-like receptor 6 are associated with altered lipopeptide- and mycobacteria-induced interleukin-6 secretion. Genes Immun 2010; 11: 561–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schurz H, Daya M, Möller M, et al. TLR1, 2, 4, 6 and 9 variants associated with tuberculosis susceptibility: A systematic review and meta-analysis. PLoS One 2015; 10: e0139711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ini-10.1177_17534259211029996 for Synergistic effect of genetic polymorphisms in TLR6 and TLR10 genes on the risk of pulmonary tuberculosis in a Moldavian population by Alexander Varzari, Igor V. Deyneko, Elena Tudor, Harald Grallert and Thomas Illig in Innate Immunity
Supplemental material, sj-pdf-2-ini-10.1177_17534259211029996 for Synergistic effect of genetic polymorphisms in TLR6 and TLR10 genes on the risk of pulmonary tuberculosis in a Moldavian population by Alexander Varzari, Igor V. Deyneko, Elena Tudor, Harald Grallert and Thomas Illig in Innate Immunity
Supplemental material, sj-xlsx-3-ini-10.1177_17534259211029996 for Synergistic effect of genetic polymorphisms in TLR6 and TLR10 genes on the risk of pulmonary tuberculosis in a Moldavian population by Alexander Varzari, Igor V. Deyneko, Elena Tudor, Harald Grallert and Thomas Illig in Innate Immunity
Supplemental material, sj-xlsx-4-ini-10.1177_17534259211029996 for Synergistic effect of genetic polymorphisms in TLR6 and TLR10 genes on the risk of pulmonary tuberculosis in a Moldavian population by Alexander Varzari, Igor V. Deyneko, Elena Tudor, Harald Grallert and Thomas Illig in Innate Immunity