Abstract
The current genetic strategies used to identify Tropheryma whippelii, the putative agent of Whipple’s disease, are based on PCR-mediated amplification of a part of its 16S rRNA gene (16S rDNA). Because there is very little intraspecies variation in these molecules, they are not suitable as targets for epidemiologic investigations. However, the intergenic spacer region between the 16S and 23S rDNAs is usually much more variable and has repeatedly been used for epidemiologic purposes. We have therefore amplified the spacer region of T. whippelii directly from clinical specimens from nine independent Swiss patients with Whipple’s disease by PCR with primers complementary to the 3′ and 5′ ends of the 16S and 23S rDNAs, respectively. The amplicons were directly sequenced and the sequences were compared to the T. whippelii reference sequence in GenBank/EMBL (accession no. X99636). Complete sequence homogeneity was found between the samples from our nine patients; the spacer sequence was also identical to the reference sequence. However, the sequences corresponding to the 3′ and 5′ ends of the 16S and the 23S rDNAs of T. whippelii, respectively, differed from the respective sequences in GenBank/EMBL. The same sequence found in our patients was then found in a sample from the German patient from which the published sequence had been derived. We conclude that the 16S-23S rDNA spacer region seems to be very conserved in T. whippelii and that the respective reference entry in public databases should be revised.
Whipple’s disease is a chronic systemic infection associated with gram-positive “Whipple bacilli” which are usually present in the lamina propria of the small intestine (7). Most patients suffer from persistent diarrhea, malabsorption, and weight loss. However, common extraintestinal manifestations such as arthritis or arthralgias may be present years before symptoms appear in the digestive tract (7). Since every attempt to reproducibly culture the Whipple bacillus failed until recently (19), the traditional laboratory diagnosis of Whipple’s disease is based on the detection of diastase-resistant periodic acid-Schiff (PAS)-positive, non-acid-fast rods in infected tissue by light microscopy (8). Nowadays, molecular identification methods are increasingly being applied when Whipple’s disease is suspected (2, 4, 17, 18, 21). Current strategies are based on PCR-mediated amplification of parts of the 16S rRNA gene(s) of the putative etiologic agent Tropheryma whippelii (17, 23).
In view of the widespread interest in this novel bacterium within a still expanding frame of obscure diseases possibly attributable to variant strains of T. whippelii (18), it would be useful to have a molecular approach for epidemiologic investigations. The intergenic region between the genes coding for the 16S and the 23S rRNAs (16S-23S rDNA spacer region) is known to be more variable than the flanking structural genes and was proposed as a promising tool for analysis of strains in various taxonomic groups (10). This spacer region has recently been amplified and sequenced directly from an organism in a duodenal biopsy specimen from a patient with Whipple’s disease (13) with a T. whippelii-specific and a broad-spectrum primer derived from the 3′ end of the 16S rRNA gene and the 5′ end of the 23S rRNA gene, respectively.
In this study, we have analyzed the 16S-23S rDNA spacer region of T. whippelii in nine independent Swiss patients with Whipple’s disease by nested PCR and direct sequencing. The initial failure to obtain amplicons with T. whippelii-specific primers is shown to be due to errors contained in the published reference sequence from which these primers had been derived.
(Parts of this study were presented at the 98th General Meeting of the American Society for Microbiology, Atlanta, Georgia, 17 to 21 May 1998 [10a].)
MATERIALS AND METHODS
Patients.
Specimens from nine patients with Whipple’s disease (Table 1) shown to contain T. whippelii DNA by PCR with primers TW-1 and TW-2 (2) were investigated. For seven of the nine patients the identity of the amplicons was confirmed by sequence analysis. No sequence information was available for organisms from the remaining two patients. Patients 1 to 3 have been described previously (2, 19).
TABLE 1.
Specimens from patients with Whipple’s disease used to analyze the 16S-23S rDNA spacer region of T. whippelii
| Patient no. | Sexa | Age (yr) | Clinical findings | Specimen | Date (day/mo/yr) | PASb |
T. whippelii PCR result
|
|
|---|---|---|---|---|---|---|---|---|
| 16S rDNAc | Spacer regiond | |||||||
| 1 | F | 30 | Spondylodiscitis | Biopsy of lumbar spine | 9/10/1995 | −e | + | + |
| 2 | M | 55 | Endocarditis | Heart valve | 9/01/1996 | + | + | + |
| 3 | M | 63 | Endocarditis | Heart valve | 1/12/1995 | + | + | + |
| 4 | M | 48 | Malabsorption, weight loss | Biopsy of duodenum | 23/09/1996 | NDf | + | + |
| 5 | M | 72 | Arthritis, chronic diarrhea | Biopsy of ileum | 2/4/1996 | ND | + | + |
| 6 | M | 50 | Relapsing oligoarthritis | Biopsy of duodenum | 24/4/1997 | − | + | + |
| 7 | F | 32 | Arthralgias, chronic colitis | Biopsy of duodenum | 3/3/1997 | −e | + | + |
| 8 | F | 30 | Chronic abdominal pain | Biopsy of duodenum | 20/2/1996 | ND | + | + |
| 9 | M | 59 | Arthritis | Joint fluid | 14/10/1996 | − | + | + |
F, female; M, male.
The presence (+) or absence (−) of PAS-positive inclusions in macrophages is indicated.
T. whippelii-specific amplification with primers TW-1 and TW-2 (2).
Detection of 16S-23S rDNA spacer region with T. whippelii-specific amplification system 6 followed by amplification system 8 (Fig. 1).
PAS positive with an ileum biopsy specimen.
ND, no data available.
Controls.
A DNA extract known to contain T. whippelii DNA (kindly provided by Matthias Maiwald, Heidelberg, Germany) was used as a positive control. This extract had previously been used to determine the nucleotide sequence of the 16S-23S spacer region of this organism (13).
Negative controls included DNA extracts from 10 duodenal biopsy specimens and 2 gastric aspirates negative for T. whippelii DNA by PCR with primers TW-1 and TW-2. These samples were taken from patients with PAS-negative biopsy specimens from the duodenum and without symptoms commonly found in Whipple’s disease such as diarrhea, weight loss, arthralgias, arthritis, or chronic fever.
Bacterial strains and cultivation.
One clinical isolate each of an Arthrobacter sp., an Aureobacterium sp., Brevibacterium casei, a Cellulomonas sp., Corynebacterium striatum, “Corynebacterium aquaticum,” Escherichia coli, a Microbacterium sp., Mycobacterium fortuitum, a Propionibacterium sp., and a Tsukamurella sp. was tested. Cultures were grown aerobically on Columbia agar (Becton Dickinson [BBL], Basel, Switzerland) with 5% sheep blood at 37°C for 1 to 3 days.
Extraction of DNA.
Biopsy specimens from patients 1 to 3 were processed as described previously (9). Briefly, the samples were suspended in digestion buffer (50 mM Tris-HCl [pH 8.5], 1 mM EDTA, 0.5% sodium dodecyl sulfate, 200 μg of proteinase K per ml), and the mixture was incubated at 55°C for 3 h with agitation. Proteinase K was heat inactivated at 95°C for 12 min, and 1 to 5 μl of the supernatant was directly used for PCR amplification as described below except for the addition of Tween 20 to a final concentration of 2% (9).
Synovial fluid and gastric aspirates were centrifuged at 14,000 × g for 10 min, and the resulting pellets were treated in the same way as the remaining biopsy samples. They were suspended in 200 μl of digestion buffer and incubated at 55°C for 1 h and 30 min with agitation. To isolate the genomic DNA, QIAamp DNA binding columns (QIAGEN, Hilden, Germany) were used according to the manufacturer’s protocol, and 5 μl of the eluted DNA was used for PCR amplification.
One loop of bacterial biomass was suspended in 200 μl of 4% CHELEX 100 (Bio-Rad, Richmond, Calif.). After incubation at 95°C for 15 min and centrifugation at 14,000 × g for 10 min, 1 μl of the supernatant of the lysate was used for PCR.
Amplification and direct sequencing of the 16S-23S rDNA spacer region.
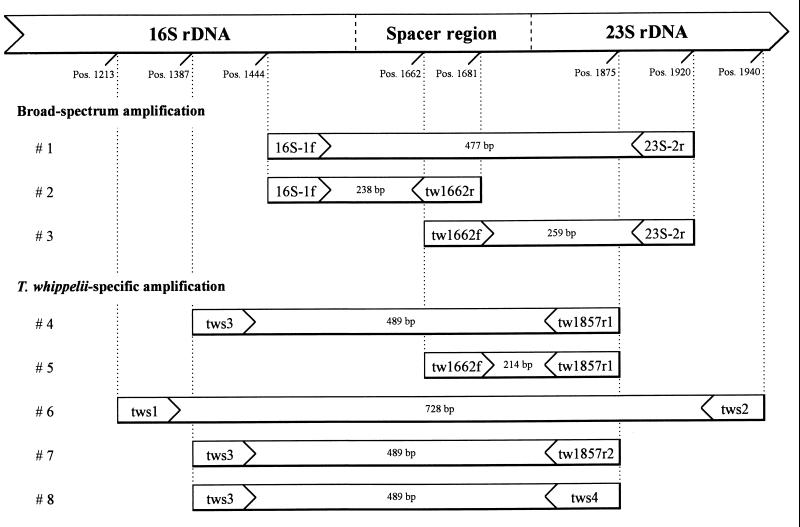
All oligonucleotides used for amplification and/or sequencing are listed in Table 2. Various primer combinations (Fig. 1) were chosen to allow broad-spectrum and T. whippelii-specific amplifications.
TABLE 2.
Oligonucleotides used to analyze the 16S-23S rDNA spacer region of T. whippelii
| Primer, sensea | Sequence (5′→3′) | Positionb | Target [rDNA/organism(s)] |
|---|---|---|---|
| 16S-1f, f | AAGTCGTAACAAGGT | 1444→1458 | 16S/bacteriac |
| 23S-2r, r | GGTTBCCCCATTCGGd | 1920→1906 | 23S/bacteriac |
| tw1662f, f | ACTATTGGGTTTTGAGAGGC | 1662→1681 | Spacer region/T. whippeliie |
| tw1662r, r | GCCTCTCAAAACCCAATAGT | 1681→1662 | Spacer region/T. whippeliie |
| tw1857r1, r | TCCCGAGCGTTATCCGAGA | 1875→1857 | 23S/T. whippeliie |
| tw1857r2, r | TCCCGAGGCTTATCGCAGA | 1875→1857 | 23S/T. whippeliie |
| tws1, f | ATCGCAAGGTGGAGCGAATCT | 1213→1233 | 16S/T. whippeliie |
| tws2, r | CGCATTCTGGCGCCCCAC | 1940→1923 | 23S/T. whippeliie |
| tws3, f | CCGGTGACTTAACCTTTTTGGAGA | 1387→1410 | 16S/T. whippeliie |
| tws4, r | TCCCGAGGCTTATCGCAGATTG | 1875→1854 | 23S/T. whippeliie |
f, forward; r, reverse.
16S-1f and 23S-2r correspond to E. coli 16S rRNA positions 1492 to 1506 and 23S rRNA positions 115 to 130, respectively (12).
B corresponds to C:G:T at a ratio of 1:1:1.
This study.
FIG. 1.
Primers and amplification systems used to analyze the 16S-23S rDNA spacer region of T. whippelii. The position (Pos.) numbering and expected product sizes are given according to those of Maiwald et al. (13).
Amplifications and reamplifications were both done in a final volume of 50 μl containing each deoxyribonucleoside triphosphate at a concentration of 200 μM, 25 pmol of the corresponding primers, 2.5 U of AmpliTaq Gold polymerase including the appropriate amount of its optimized buffer (Perkin-Elmer, Norwalk, Conn.), and 5 μl of DNA template. For reamplification 1 μl from the first amplification was used as a template. In each batch of PCR, H2O and E. coli DNA were included as controls.
PCR was performed on a Gene Amp PCR System 9600 (Perkin-Elmer) with an initial activation of the AmpliTaq Gold polymerase and denaturation of the templates at 95°C for 12 min, followed by 40 cycles of denaturation at 95°C for 1 min, annealing at either 50°C (systems 1 and 2) or 55°C (all other systems) for 1 min, and extension at 72°C for 1 min. Final extension was at 72°C for 10 min.
PCR products (10 μl) were separated by agarose gel electrophoresis, stained with ethidium bromide, and visualized under UV light.
Sequencing of the amplicons was done in both directions with a solid-phase and/or a cycle sequencing kit followed by analysis on an ALFexpress DNA Sequencer (Pharmacia Biotech, Uppsala, Sweden). Solid-phase sequencing was performed with products derived from broad-spectrum amplifications 2 and 3 (Fig. 1) with the appropriate 5′-biotinylated and 5′-fluorescence-labeled primers and the Autoload SPS Kit (Pharmacia Biotech) as described by the manufacturer. For cycle sequencing, the products of amplification systems 3, 7, and 8 (Fig. 1) were purified with the QIAquick PCR Purification Kit (QIAGEN) according to the provider’s protocol. The sequencing reaction was performed with the appropriate 5′-fluorescence-labeled primer and the Thermo Sequenase Kit with 7-deaza-dGTP (Amersham Life Science, Little Chalfont, England) under the same PCR conditions described above for the amplification except that an initial denaturation at 95°C for only 5 min was used. Raw data were automatically processed with ALFwin version 1.10 (Pharmacia Biotech) and were analyzed with the Genetics Computer Group (GCG) software package (University of Wisconsin, Madison) and DNASIS version 7.00 (Hitachi Software Engineering America, Brisbane, Calif.) for comparison with entries at GenBank/European Molecular Biology Laboratory (EMBL) databases.
Interpretation of sequencing results.
Sequences determined in one direction only were considered correct if they were in agreement with the reference sequence of T. whippelii (accession no. X99636) (13). In case of discrepancies from the reference sequence, concordant results in both sequencing directions were required for the sequence to be accepted as correct.
Nucleotide sequence accession number.
The T. whippelii sequence spanning the 3′ end of the 16S gene, the 16S-23S ribosomal intergenic spacer, and the 5′ end of the 23S gene determined in this study has been deposited in GenBank under accession no. AF074933.
RESULTS
Initial investigations.
Based on the reference sequence (13), two primers complementary to the 3′ and the 5′ ends of the 16S and 23S rDNAs of T. whippelii, respectively, that promised to be specific for this organism were designed (amplification 4; Fig. 1). When tested with DNA extracted from the heart valve from patient 2, no band of the expected size (489 bp) was visible. Even after reamplification with amplification system 5 (expected amplicon size, 214 bp), no products appeared, although the DNA used had previously given rise to a strong signal when it was amplified with other primer combinations targeting fragments of up to ±530 bp, as shown by Schoedon and coworkers (19). Considering the fact that some of our specimens were believed not to contain microorganisms other than T. whippelii, we decided to switch to a nonspecific approach known to amplify the spacer region(s) of many bacteria (10).
Broad-spectrum amplification and sequencing of the 16S-23S rDNA spacer region.
DNA extracts from heart valves from patients 2 and 3 were amplified by broad-spectrum amplification 1 (Fig. 1). Again, no products (477 bp) were visible on the agarose gel. However, after reamplification with either amplification system 2 or 3 (Fig. 1), bands of the expected length (238 and 259 bp, respectively) were visible for both patients. These fragments were then sequenced, and no differences were found between the sequences derived from the two patients. However, some discrepancies between our sequences (sequenced in both directions) and the T. whippelii reference sequence were observed in the 3′ and 5′ terminal regions of the 16S and 23S rRNA genes, respectively, but not within the spacer region. We therefore used our broad-spectrum approach with exactly the same DNA extract from which the reference sequence of the T. whippelii ribosomal intergenic spacer region had been derived originally (13). Astonishingly, the sequence determined was completely identical to the sequences derived from our two patients, with the following three regions differing from the published reference sequence (Fig. 2) (the capital letters and blank spaces in parentheses indicate the bases that were different between the two sequences) (i) gcGgCtGga instead of gcCgGt( )ga (positions 1475 to 1482 of reference sequence; 3′ terminal region of 16S rRNA gene), (ii) ctGCgataaGCct instead of ctCGgataaCGct (positions 1858 to 1870; 5′ terminal region of 23S rRNA gene), and (iii) aaGCGAgc instead of aaC( )AGgc (positions 1883 to 1889; 5′ terminal region of 23S rRNA gene).
FIG. 2.
Location of errors contained in the published T. whippelii reference sequence (GenBank/EMBL accession no. X99636). The base numbering is that used by Maiwald et al. (13); dots and hyphens indicate identity and alignment gaps, respectively. The 16S-23S rDNA spacer region is in boldface type. corr., corrected sequence (this study).
T. whippelii-specific amplification of the 16S-23S rDNA spacer region.
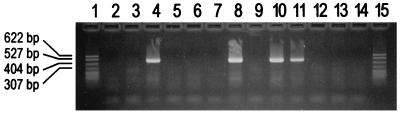
On the basis of the results presented above, primer tw1857r1 was revised to tw1857r2 (Table 2). DNA extracts from usually sterile (heart, spine, joint) and nonsterile (intestine, stomach) body sites were then amplified with the T. whippelii-specific amplification system 6 followed by nested reamplification with amplification systems 4 and 7. While no bands of the expected size (728 bp) were visible with amplification system 6 for patients and controls, reamplification with amplification system 7 (with the modified reverse primer tw1857r2) resulted in amplicons of 489 bp for all patients but none of the controls. In contrast, reamplification with amplification system 4 (with the original primer tw1857r1) was negative for both patients and controls (data not shown). Primer tw1857r2 was then further modified to tws4. Again, nested PCR (amplification system 6 followed by amplification system 8) was positive for all patient samples but negative for the controls (Fig. 3). In addition, all of the 11 bacterial species analyzed were negative for amplicons by nested PCR (as described above; data not shown).
FIG. 3.
Detection of the T. whippelii 16S-23S rDNA spacer region directly with human clinical specimens. Products of specific nested amplification (amplification system 6 followed by amplification system 8; Fig. 1) were analyzed on an ethidium bromide-stained agarose gel. Lanes 1 and 15, pBR322 DNA digested with MspI; lanes 4, 8, 10, and 11, patients positive for T. whippelii spacer amplicons; lanes 2, 3, 5 to 7, 9, and 12 to 14, patients negative for T. whippelii spacer amplicons (controls).
Comparative sequence analysis of the T. whippelii 16S-23S rDNA spacer region.
For each patient at least the spacer region generated either by amplification system 1 followed by amplification systems 2 and 3 or by amplification system 6 followed by amplification system 8 was sequenced and aligned with the T. whippelii reference sequence (Fig. 2). All the sequences (spacer region and flanking coding regions) were found to be identical to the modified reference sequence.
DISCUSSION
Epidemiologic investigations of Whipple’s disease are still very limited mainly due to difficulties in both the clinical and the (so far) non-culture-based laboratory diagnosis of this enigmatic disorder (7, 8, 22). Typing of possibly dissimilar Whipple bacilli may, however, be performed by molecular methods based on PCR-amplified DNA, provided that a suitable target sequence can be identified. In the present study we have examined the 16S-23S rDNA spacer region of T. whippelii directly from clinical specimens from independent Swiss patients by PCR and comparative sequence analysis. Assays with both broad-spectrum (universal eubacterial) and T. whippelii-specific amplification systems (Fig. 1) followed by direct sequencing have been used.
Initial attempts with specific primers derived from the published reference sequence (13) failed for a patient suffering from T. whippelii-associated endocarditis even when a seminested approach (amplification system 4 followed by amplification system 5) was used. However, the same heart valve specimen was positive by seminested broad-spectrum PCRs (amplification system 1 followed by amplification system 2 or 3). These results suggested that the lack of spacer products with T. whippelii-specific amplification systems 4 and 5 was not due to PCR inhibition or to the use of insufficient quantities of templates or poor-quality templates.
The sequences of the broad-spectrum amplicons were found to be identical to the published sequence of the spacer region of T. whippelii (13). However, they differed slightly from the reference sequence at the 3′ and the 5′ ends of the 16S and 23S rDNAs, respectively. Four of these differences were found at positions within the region from which primer tw1857r1 had been derived, thus explaining the failure to get any amplicons with amplification systems 4 and 5. Interestingly, in a 23S rDNA alignment the T. whippelii reference sequence differed at exactly the same four positions from the sequences of most other bacteria (gram-positive organisms with high or low GC contents, alpha to epsilon proteobacteria), all of which shared at these sites the nucleotides now found in our specimen (data not shown). Resequencing of the DNA from the German reference sample showed it to be identical to the DNA derived from our endocarditis patient. Thus, errors contained in the reference entry were assumed (Fig. 2).
On the basis of the revised reference sequence, novel T. whippelii-specific nested PCR systems were designed. These systems should also be applicable to specimens originating from usually nonsterile tissues with possibly low bacterial DNA contents. As expected, an assay with amplification system 6 followed by amplification system 7 (with the revised primer tw1857r2; Table 2) was positive for all nine patients with Whipple’s disease (Table 1), which again supported our revision of the reference sequence. On the basis of the most recent updates of GenBank and EMBL data (March 1998), primer tw1857r2 was then extended at its 3′ end with the intention to further improve the specificity of reamplification. The optimized nested PCR (amplification systems 6 and 8) was evaluated with specimens from patients with or without Whipple’s disease as well as with clinical isolates of actinomycetes, i.e., bacteria phylogenetically related to T. whippelii (13, 17, 23). Reamplification was exclusively positive for DNA in the specimens from the nine Whipple’s disease patients (Fig. 3), whereas all control specimens as well as all bacterial strains tested remained negative. To our surprise, sequencing of all these spacer fragments revealed no polymorphisms at all; i.e., all sequences determined for Swiss Whipple bacilli were found to be identical to the revised reference sequence.
The apparent homogeneity of the T. whippelii 16S-23S rDNA spacer sequence is remarkable because this intergenic region was useful for strain differentiation for many other bacteria (10, 15). However, most of such intraspecies variations seem to be due to recombination between individual rRNA operons present in a given organism (11, 16). According to this very recent hypothesis, one would expect almost negligible spacer polymorphism for bacteria with only a single rRNA operon. One such representative seems to be Mycobacterium leprae (20), another uncultured actinomycete pathogen. As a matter of fact, there was good molecular evidence for near identity among unrelated strains of leprosy bacilli (3, 6). However, whether the apparent conservation of the T. whippelii spacer sequence is indeed due to the presence of a single rRNA operon remains pure speculation. Further investigations ideally should include specimens from geographically more diverse patients with Whipple’s disease as well as from environmental habitats (14).
In four of the six specimens for which histopathological analysis was performed, no typical PAS-positive inclusions in macrophages were seen (Table 1). This is astonishing since the diagnosis of Whipple’s disease usually relies on the demonstration of such structures mainly in duodenal biopsy specimens. However, this has also been observed by others (4) and suggests that PCR is more sensitive than histopathological analysis. Probably the best-documented case in this regard is our patient 1 (Table 1), who was not only PCR positive and PAS negative according to the results of tests performed with a lumbar spine biopsy specimen but also PCR positive according to the results of tests with four of four intestinal biopsy specimens, only one of which was also PAS positive (2).
Although our study was restricted to a small number of clinical samples almost exclusively derived from Swiss patients, we conclude that the T. whippelii 16S-23S rDNA spacer region seems to be very conserved and that the respective reference sequence in public databases should be revised. Further investigations are clearly needed to establish a molecular method for differentiation of Whipple bacilli. Such a method could possibly answer the question of whether the different clinical manifestations of Whipple’s disease are a consequence of differences among the infecting strains. In addition, it remains to be shown whether host factors may also play an important role, as shown previously for mycobacterial infections (1, 5).
ACKNOWLEDGMENTS
This work was supported by a grant from the Swiss National Science Foundation (grant 32-50790.97) to M. Altwegg.
We thank M. Maiwald (Heidelberg, Germany) for providing the DNA extract from which the spacer sequence of T. whippelii had originally been determined and A. von Graevenitz for careful reading of the manuscript.
ADDENDUM IN PROOF
Since the submission of the manuscript we have analyzed the spacer region from 19 additional patients who were T. whippelii positive by PCR. By sequence analysis we have found two amplicons differing in only a few nucleotides from the published sequence. This phenomenon is now being further analyzed.
REFERENCES
- 1.Altare F, Durandy A, Lammas D, Emile J-F, Lamhamedi S, Le Deist F, Drysdale P, Jouanguy E, Döffinger R, Bernaudin F, Jeppsson O, Gollob J A, Meinl E, Segal A W, Fischer A, Kumararatne D, Casanova J-L. Impairment of mycobacterial immunity in human interleukin-12 receptor deficiency. Science. 1998;280:1432–1435. doi: 10.1126/science.280.5368.1432. [DOI] [PubMed] [Google Scholar]
- 2.Altwegg M, Fleisch-Marx A, Goldenberger D, Hailemariam S, Schaffner A, Kissling R. Spondylodiscitis caused by Tropheryma whippelii. Schweiz Med Wochenschr. 1996;126:1495–1499. [PubMed] [Google Scholar]
- 3.Clark-Curtiss J E, Walsh G P. Conservation of genomic sequences among isolates of Mycobacterium leprae. J Bacteriol. 1989;171:4844–4851. doi: 10.1128/jb.171.9.4844-4851.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dauga C, Miras I, Grimont P A D. Strategy for detection and identification of bacteria based on 16S rRNA genes in suspected cases of Whipple’s disease. J Med Microbiol. 1997;46:340–347. doi: 10.1099/00222615-46-4-340. [DOI] [PubMed] [Google Scholar]
- 5.de Jong R, Altare F, Haagen I-A, Elferink D G, de Boer T, van Breda Vriesman P J C, Kabel P J, Draaisma J M T, van Dissel J T, Kroon F P, Casanova J-L, Ottenhoff T H M. Severe mycobacterial and Salmonella infections in interleukin-12 receptor-deficient patients. Science. 1998;280:1435–1438. doi: 10.1126/science.280.5368.1435. [DOI] [PubMed] [Google Scholar]
- 6.de Wit M Y L, Klatser P R. Mycobacterium leprae isolates from different sources have identical sequences of the spacer region between the 16S and 23S ribosomal RNA genes. Microbiology. 1994;140:1983–1987. doi: 10.1099/13500872-140-8-1983. [DOI] [PubMed] [Google Scholar]
- 7.Dobbins W O., III . Whipple’s disease. Springfield, Ill: Charles C Thomas Publisher; 1987. [Google Scholar]
- 8.Dobbins W O., III The diagnosis of Whipple’s disease. N Engl J Med. 1995;332:390–392. doi: 10.1056/NEJM199502093320611. [DOI] [PubMed] [Google Scholar]
- 9.Goldenberger D, Perschil I, Ritzler M, Altwegg M. A simple “universal” DNA extraction procedure using SDS and proteinase K is compatible with direct PCR amplification. PCR Methods Appl. 1995;4:368–370. doi: 10.1101/gr.4.6.368. [DOI] [PubMed] [Google Scholar]
- 10.Gürtler V, Stanisich V A. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology. 1996;142:3–16. doi: 10.1099/13500872-142-1-3. [DOI] [PubMed] [Google Scholar]
- 10a.Hinrikson H P, Dutly F, Altwegg M. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. Homogeneity of the 16S-23S rDNA spacer region of Tropheryma whippelii, abstr. D-5; p. 213. [Google Scholar]
- 11.Lan R, Reeves P R. Recombination between rRNA operons created most of the ribotype variation observed in the seventh pandemic clone of Vibrio cholerae. Microbiology. 1998;144:1213–1221. doi: 10.1099/00221287-144-5-1213. [DOI] [PubMed] [Google Scholar]
- 12.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: John Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- 13.Maiwald M, Ditton H-J, von Herbay A, Rainey F A, Stackebrandt E. Reassessment of the phylogenetic position of the bacterium associated with Whipple’s disease and determination of the 16S-23S ribosomal intergenic spacer sequence. Int J Syst Bacteriol. 1996;46:1078–1082. doi: 10.1099/00207713-46-4-1078. [DOI] [PubMed] [Google Scholar]
- 14.Maiwald M, Schuhmacher F, Ditton H-J, von Herbay A. Environmental occurrence of the Whipple’s disease bacterium (Tropheryma whippelii) Appl Environ Microbiol. 1998;64:760–762. doi: 10.1128/aem.64.2.760-762.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Normand P, Ponsonnet C, Nesme X, Neyra M, Simonet P. ITS analysis of prokaryotes. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Norwell, Mass: Kluwer Academic Publishers; 1996. pp. 1–12. [Google Scholar]
- 16.Pérez Luz S, Rodriguez-Valera F, Lan R, Reeves P R. Variation of the ribosomal operon 16S-23S gene spacer region in representatives of Salmonella enterica subspecies. J Bacteriol. 1998;180:2144–2151. doi: 10.1128/jb.180.8.2144-2151.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Relman D A, Schmidt T M, MacDermott R P, Falkow S. Identification of the uncultured bacillus of Whipple’s disease. N Engl J Med. 1992;327:293–301. doi: 10.1056/NEJM199207303270501. [DOI] [PubMed] [Google Scholar]
- 18.Rickman L S, Freeman W S, Green W S, Feldman S T, Sullivan J, Russack V, Relman D A. Uveitis caused by Tropheryma whippelii (Whipple’s bacillus) N Engl J Med. 1995;322:363–366. doi: 10.1056/NEJM199502093320604. [DOI] [PubMed] [Google Scholar]
- 19.Schoedon G, Goldenberger D, Forrer R, Gunz A, Dutly F, Höchli M, Altwegg M, Schaffner A. Deactivation of macrophages with interleukin-4 is the key to the isolation of Tropheryma whippelii. J Infect Dis. 1997;176:672–677. doi: 10.1086/514089. [DOI] [PubMed] [Google Scholar]
- 20.Sela S, Clark-Curtiss J E, Bercovier H. Characterization and taxonomic implications of the rRNA genes of Mycobacterium leprae. J Bacteriol. 1989;171:70–73. doi: 10.1128/jb.171.1.70-73.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Herbay A, Ditton H-J, Maiwald M. Diagnostic application of a polymerase chain reaction assay for the Whipple’s disease bacterium to intestinal biopsies. Gastroenterology. 1996;110:1735–1743. doi: 10.1053/gast.1996.v110.pm8964398. [DOI] [PubMed] [Google Scholar]
- 22.von Herbay A, Otto H F, Stolte M, Borchard F, Kirchner T, Ditton H-J, Maiwald M. Epidemiology of Whipple’s disease in Germany. Scand J Gastroenterol. 1997;32:52–57. doi: 10.3109/00365529709025063. [DOI] [PubMed] [Google Scholar]
- 23.Wilson K H, Blitchington R, Frothingham R, Wilson J A P. Phylogeny of the Whipple’s-disease-associated bacterium. Lancet. 1991;338:474–475. doi: 10.1016/0140-6736(91)90545-z. [DOI] [PubMed] [Google Scholar]