Abstract
Background
Antibiotic resistance (ABR) is recognized as an increasing threat to global health. Haiti declared ABR an emerging public health threat in 2018, however, the current surveillance system is limited. We described the microbiological data from a Médecins Sans Frontières trauma hospital, to increase knowledge on ABR in Haiti for similar facilities.
Methods
A retrospective cross-sectional analysis of routine microbiological data of samples taken from patients admitted to the inpatient ward or followed up in the outpatient clinic of the trauma hospital from March 2012 to December 2018. Resistance trends were analysed per isolate and compared over the 7 year period.
Results
Among 1742 isolates, the most common samples were pus (53.4%), wound swabs (30.5%) and blood (6.9%). The most frequently detected bacteria from these sample types were Staphylococcus aureus (21.9%), Pseudomonas aeruginosa (20.9%) and Klebsiella pneumoniae (16.7%). MDR bacteria (32.0%), ESBL-producing bacteria (39.1%), MRSA (24.1%) and carbapenem-resistant Enterobacteriaceae (CRE) species (2.6%) were all detected. Between 2012 and 2018 the number of ESBL isolates significantly increased from 3.2% to 42.9% (P = 0.0001), and resistance to clindamycin in MSSA isolates rose from 3.7% to 29.6% (P = 0.003). Two critical WHO priority pathogens (ESBL-producing CRE and carbapenem-resistant P. aeruginosa) were also detected.
Conclusions
Over a 7 year period, a high prevalence of MDR bacteria was observed, while ESBL-producing bacteria showed a significantly increasing trend. ABR surveillance is important to inform clinical decisions, treatment guidelines and infection prevention and control practices.
Introduction
Antibiotic resistance (ABR) is an ever-increasing threat to human health. MDR organisms (MDROs) are difficult to treat with conventional antibiotics, resulting in increased morbidity and mortality.1,2 Low and middle-income countries (LMICs) often have the additional burdens of poor sanitation, substandard facilities and insufficient infection and control practices contributing to the high rate of MDRO infections.3,4
Globally, MDROs have spread rapidly, including extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae and methicillin-resistant Staphylococcus aureus (MRSA). Healthcare-associated infections (HAI) are on the rise, with resource-limited settings reporting twice the average prevalence compared with Europe (15.5 versus 7.1 per 100 patients).5 There are several factors behind this difference, including lack of infection prevention and control (IPC) programmes, trained staff and basic sanitary materials as a consequence of weak health systems with inadequate infrastructure and limited resources.3,6
Haiti, a country of 11 million people on the island of Hispaniola, has suffered in recent years from multiple crises: an earthquake in 2010, followed by a massive cholera outbreak that continued through to early 2019, a hurricane in 2016, ongoing political instability and another earthquake in 2021. As a result, the public health infrastructure has collapsed. There are limited data on ABR prevalence from this context, especially studies that follow the trend over time.
In February 2012, Médecins Sans Frontières (MSF) opened an advanced trauma care centre to cater for trauma cases in the capital of Haiti, Port-au-Prince. This study aimed to determine the prevalence and pattern of MDR bacteria amongst patients admitted to the MSF Nap Kembe Acute Trauma Hospital, Tabarre, Port-au-Prince, Republic of Haiti from March 2012 to December 2018.
Methods
Study design and setting
This was a retrospective, cross-sectional analysis of routine microbiological and clinical data.
The Nap Kembe Trauma Hospital opened in 2012 and admitted patients until the end of 2018. At the peak of operations, the hospital had four operating theatres. In 2018 there were 44 hospital beds, including five solely dedicated to infected wounds, and a six bed ICU with mechanical ventilation capacity. The hospital provided care free of charge as well as rehabilitation services, mental health care, health education, health promotion and psychosocial support. Antibiotic susceptibility results influenced clinical decisions regarding case management for the specific patients. An antibiotic stewardship programme was introduced in 2017. The antibiotic stewardship and IPC committees worked together to monitor HAIs through a case-based surveillance system and to set strategies to improve antibiotic use.
Study population and period
All samples taken from patients admitted from 12 March 2012 to 31 December 2018 were eligible for inclusion into this study.
Inclusion and exclusion criteria
Isolates with unknown sample type or patient identifiers were excluded. For multiple samples from the same patient on the same date from the same site, only one sample was included in the data analysis.
Data collection and entry
Routine data from microbiological hard copy results were double entered into an EpiData database (EpiData Association, Odense, Denmark) and validated. Programme data from the laboratory and hospital registers were used for this study and matched using an identifying key.
Microbiological analysis
Samples were sent to a private external microbiology laboratory, validated by MSF and accredited by the College of American Pathologists. Conventional microbiological methods were used for bacteriological identification. Antibiotic susceptibility testing was performed by VITEK 2 (bioMérieux, Marcy-l’Étoile, France) or the Kirby–Bauer disc diffusion method and standardized as per CLSI. Quality control testing was performed by using appropriate ATCC control strains.
Data analysis
All isolates with organism growth were included in the overall cumulative antibiogram analyses. However, only isolates that could be matched to a patient ID in the hospital registers were included in the analysis of clinical characteristics. Analysis was done using Microsoft Excel and R 3.4.0 Software with the AMR package.7 An MDRO was defined as non-susceptible to at least one agent in three or more antibiotic categories8 and identified with the ‘mdro’ function of the package. Descriptive statistics were used to describe the patient population by proportions and means and presented with 95% CI. Overall, cumulative antibiograms were analysed for bacteria with at least 30 isolates, or a minimum of one isolate if a rare species. Prevalence of resistance to various antibacterial agents was presented as the proportion of intermediate (I) and resistant (R) combined. Comparisons of categorical variables were performed by χ2 and Fisher’s exact tests with a two-tailed P value <0.05 chosen for statistical significance. Trend analysis was performed using the Cochran–Armitage test.
Ethics
This research fulfilled the exemption criteria set by the MSF Ethics Review Board (ERB) for a posteriori analyses of routinely collected clinical data and thus did not require MSF ERB review. It was conducted with permission from the Medical Director, Operational Centre Brussels, MSF. Local ethics approval was given by the Ministere de la Santé Publique et la Population Comité National de Bioethique ref number: 1920-31.
Results
Patient and isolate characteristics
Over the 7 year period, a total of 1742 isolates were detected from 1275 samples. Samples were taken from at least 694 patients, while 530 isolates from 420 samples did not have a patient ID recorded. Most samples were taken from men (74.7%) and the median age was 30.0 (IQR: 22–41) years (Table 1).
Table 1.
Characteristics of patients sampled with identification between March 2012 and December 2018 at the MSF Nap Kembe Trauma Hospital, Tabarre, Haiti
| 2012, n (%) | 2013, n (%) | 2014, n (%) | 2015, n (%) | 2016, n (%) | 2017, n (%) | 2018, n (%) | Total, n (%) | |
|---|---|---|---|---|---|---|---|---|
| Total patients with identification matched to medical data | 40 | 35 | 65 | 88 | 184 | 147 | 135 | 694 |
| sex | ||||||||
| female | 7 (17.5) | 4 (11.4) | 16 (24.6) | 25 (28.4) | 54 (29.3) | 37 (25.2) | 26 (23.9) | 169 (25.3) |
| male | 33 (82.5) | 31 (88.6) | 49 (75.4) | 63 (71.6) | 130 (70.7) | 110 (74.8) | 83 (76.1) | 499 (74.7) |
| age | ||||||||
| median age, years [IQR]) | 34 [26–47] | 31 [26–41] | 30 [24–39] | 30 [20–40] | 30 [18–45] | 29 [20–38] | 32 [25–44] | 30 [22–41] |
| reason for admission from ER | ||||||||
| trauma from accident | 28 (71.8) | 11 (31.4) | 46 (70.8) | 64 (72.7) | 123 (68.3) | 99 (69.2) | 72 (63.2) | 443 (66.7) |
| trauma from violence | 8 (20.5) | 2 (5.7) | 5 (7.7) | 15 (17.0) | 29 (16.1) | 26 (18.2) | 37 (32.5) | 122 (18.4) |
| non-traumaa | 3 (7.7) | 2 (5.7) | 4 (6.2) | 4 (4.5) | 13 (7.2) | 1 (0.7) | 2 (1.7) | 29 (4.4) |
| others | 0 (0.0) | 20 (57.1) | 10 (15.4) | 5 (5.7) | 15 (8.3) | 17 (11.9) | 3 (2.6) | 70 (10.5) |
| severity of patient at admission | ||||||||
| mild | 0 (0.0) | 0 (0.0) | 0 (0.0) | 6 (6.8) | 2 (1.1) | 3 (2.0) | 1 (0.7) | 12 (1.7) |
| moderate | 13 (32.5) | 16 (45.7) | 25 (38.5) | 17 (19.3) | 30 (16.3) | 22 (15.0) | 8 (5.9) | 131 (18.9) |
| severe | 21 (52.5) | 17 (48.6) | 31 (47.7) | 52 (59.1) | 111 (60.3) | 73 (49.7) | 63 (46.7) | 368 (53.0) |
| critical | 5 (12.5) | 2 (5.7) | 8 (12.3) | 12 (13.6) | 37 (20.1) | 45 (30.6) | 63 (46.7) | 172 (24.8) |
| not recorded | 1 (2.5) | 0 (0.0) | 1 (1.5) | 1 (1.1) | 4 (2.2) | 4 (2.7) | 0 (0.0) | 11 (1.6) |
| Total isolates | 79 | 137 | 226 | 322 | 398 | 369 | 262 | 1742 |
| bacteria | ||||||||
| S. aureus | 35 (44.3) | 38 (27.7) | 49 (21.7) | 66 (20.5) | 70 (17.6) | 42 (11.4) | 40 (19) | 340 (19.5) |
| P. aeruginosa | 15 (19.0) | 23 (16.8) | 44 (19.5) | 39 (12.1) | 65 (16.3) | 53 (14.4) | 20 (9.5) | 259 (14.9) |
| E. coli | 6 (7.6) | 16 (11.7) | 20 (8.8) | 35 (10.9) | 44 (11.1) | 52 (14.1) | 29 (13.7) | 202 (11.6) |
| E. cloacae | 5 (6.3) | 12 (8.8) | 24 (10.6) | 32 (9.9) | 35 (8.8) | 26 (7.0) | 22 (10.4) | 156 (9.0) |
| K. pneumoniae | 2 (2.5) | 14 (10.2) | 12 (5.3) | 25 (7.8) | 25 (6.3) | 33 (8.9) | 18 (8.5) | 129 (7.4) |
| E. faecalis | 1 (1.3) | 6 (4.4) | 14 (6.2) | 17 (5.3) | 31 (7.8) | 21 (5.7) | 23 (10.9) | 113 (6.5) |
| Acinetobacter baumannii | 3 (3.8) | 3 (2.2) | 11 (4.9) | 11 (3.4) | 12 (3.0) | 18 (4.9) | 4 (1.9) | 62 (3.6) |
| P. mirabilis | 2 (2.5) | 4 (2.9) | 3 (1.3) | 9 (2.8) | 15 (3.8) | 13 (3.5) | 3 (1.4) | 49 (2.8) |
| Serratia marcescens | 1 (1.3) | 2 (1.5) | 10 (4.4) | 12 (3.7) | 10 (2.5) | 8 (2.2) | 5 (2.4) | 48 (2.8) |
| Aeromonas hydrophilia | 2 (2.5) | 2 (1.5) | 4 (1.8) | 4 (1.2) | 12 (3.0) | 12 (3.3) | 7 (3.3) | 43 (2.5) |
| other | 7 (8.9) | 17 (12.4) | 35 (15.5) | 72 (22.4) | 79 (19.8) | 91 (24.7) | 40 (19) | 341 (19.6) |
| specimen type | ||||||||
| pus | 21 (26.6) | 41 (29.9) | 125 (55.3) | 208 (64.6) | 222 (55.8) | 196 (53.1) | 117 (55.5) | 930 (53.4) |
| wound swabs | 50 (63.3) | 81 (59.1) | 74 (32.7) | 62 (19.3) | 103 (25.9) | 108 (29.3) | 54 (25.6) | 532 (30.5) |
| blood cultures | 2 (2.5) | 6 (4.4) | 7 (3.1) | 21 (6.5) | 35 (8.8) | 27 (7.3) | 23 (10.9) | 121 (6.9) |
| fluidsb | 1 (1.3) | 7 (5.1) | 7 (3.1) | 22 (6.8) | 21 (5.3) | 22 (6.0) | 8 (3.8) | 88 (5.1) |
| bone | 0 (0.0) | 0 (0.0) | 6 (2.7) | 4 (1.2) | 7 (1.8) | 4 (1.1) | 5 (2.4) | 26 (1.5) |
| tissue | 2 (2.5) | 0 (0.0) | 0 (0.0) | 2 (0.6) | 4 (1.0) | 6 (1.6) | 0 (0.0) | 14 (0.8) |
| othersc | 3 (3.8) | 2 (1.5) | 7 (3.1) | 3 (0.9) | 6 (1.6) | 6 (1.6) | 4 (1.9) | 31 (1.8) |
| location where isolates were taken | ||||||||
| IPD | 52 (65.8) | 69 (50.4) | 117 (51.8) | 177 (55.0) | 269 (67.6) | 144 (39.0) | 100 (47.4) | 928 (53.3) |
| ICU | 2 (2.5) | 35 (25.5) | 31 (13.7) | 45 (14) | 70 (17.6) | 60 (16.3) | 43 (20.4) | 286 (16.4) |
| OPD | 15 (19) | 9 (6.6) | 8 (3.5) | 11 (3.4) | 7 (1.8) | 22 (6) | 17 (8.1) | 89 (5.1) |
| othersd | 9 (11.4) | 15 (10.9) | 32 (14.2) | 17 (5.3) | 13.3 (3.3) | 5 (1.4) | 0 (0.0) | 91 (5.2) |
| not recorded | 1 (1.3) | 9 (6.6) | 38 (16.8) | 72 (22.4) | 39 (9.8) | 138 (37.4) | 51 (24.2) | 358 (20.0) |
ER, emergency room; IPD, inpatient department; OPD, outpatient department.
Non-trauma reasons include non-trauma surgical pathologies, cardiovascular emergencies and other non-trauma reasons not defined.
Including secretions.
Urine, faeces and other types of swab.
Other is procedure room in ER.
The largest proportion of isolates was detected from patients admitted to the inpatient department (53.3%) and were mostly pus samples (53.4%) followed by wound swabs (30.5%) and blood samples (6.9%) (Table 1). The most frequently isolated species was S. aureus, not only in total over the 7 year period, but for each year of the study, except in 2017 when Pseudomonas aeruginosa was more common (Table 1).
The most frequently isolated bacteria from pus samples were S. aureus (21.9%), of which 27.0% were MDR. P. aeruginosa (20.9%) was the most frequently isolated species from wound swabs (28.8% MDR), and Klebsiella pneumoniae (16.7%) was most the isolated from blood cultures (90.0% MDR) (Table 2).
Table 2.
Number of bacteria isolated by specimen type and number of MDR isolates among them between 2012 and 2018 at the MSF Nap Kembe Trauma Hospital, Tabarre, Haiti
| Specimen type/Bacteria | Isolates, n (%) | MDR isolates, n (%) | MRSA isolates, n (%) | ESBL isolates, n (%) | CRE isolates, n (%) |
|---|---|---|---|---|---|
| Pus | |||||
| S. aureus | 204 (21.9) | 55 (27.0) | 50 (24.5) | ||
| E. coli | 118 (12.7) | 62 (52.5) | 51 (43.2) | 4 (3.4) | |
| P. aeruginosa | 118 (12.7) | 26 (22.0) | |||
| E. cloacae | 86 (9.2) | 41 (47.7) | 28 (32.6) | 4 (4.7) | |
| E. faecalis | 64 (6.9) | 0 (0.0) | |||
| K. pneumoniae | 58 (6.2) | 40 (69.0) | 41 (70.7) | ||
| P. mirabilis | 33 (3.5) | 10 (30.3) | 6 (18.2) | 1 (3.0) | |
| other | 249 (26.2) | ||||
| Wound swab | |||||
| P. aeruginosa | 111 (20.9) | 32 (28.8) | |||
| S. aureus | 101 (19.0) | 28 (27.7) | 25 (24.8) | ||
| E. cloacae | 57 (10.7) | 28 (49.1) | 16 (28.1) | 2 (3.5) | |
| E. coli | 44 (8.3) | 26 (59.1) | 20 (45.5) | ||
| K. pneumoniae | 34 (6.4) | 23 (67.6) | 26 (76.5) | ||
| E. faecalis | 31 (5.8) | 0 (0.0) | |||
| other | 154 (29.6) | ||||
| Blood | |||||
| K. pneumoniae | 20 (16.7) | 18 (90.0) | 17 (85.0) | ||
| Staphylococcus haemolyticus | 15 (12.5) | 0 (0.0) | |||
| E. coli | 14 (11.7) | 10 (71.4) | 10 (71.4) | ||
| Staphylococcus hominis | 11 (9.2) | 0 (0.0) | |||
| S. aureus | 10 (8.3) | 5 (50.0) | 5 (50.0) | ||
| other | 50 (41.4) |
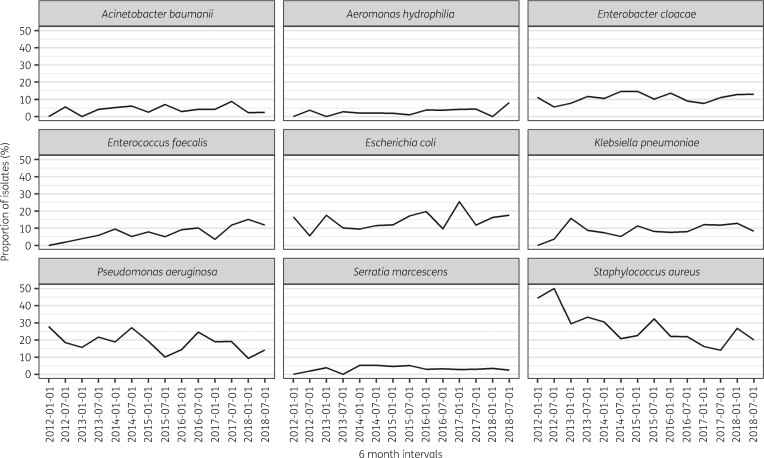
The proportion of S. aureus detected in every type of specimen decreased substantially over the study period, while other bacteria including Escherichia coli and Enterococcus faecalis were isolated more frequently (Figure 1).
Figure 1.
Evolution of detection of the most frequent nine bacteria over 6 monthly intervals between 2012 and 2018 at the MSF Nap Kembe Trauma Hospital, Tabarre, Haiti.
Patterns of MDR
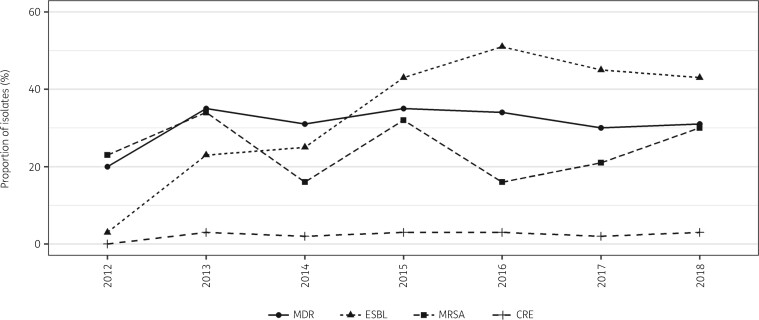
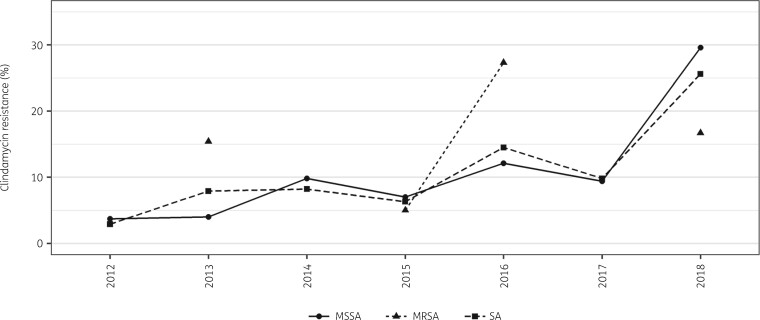
Overall, 32.0% of all isolates were MDR and this did not significantly change over the 7 year period, from 22.9% in 2012 to 37.8% in 2018 (P = 0.424), peaking in 2015 at 40.2% (Figure 2). Of the 340 S. aureus isolates, 82 (24.1%) were MRSA with no significant change in proportion over the 7 year period (P = 0.578) (Figure 2). Resistance to clindamycin increased significantly for all S. aureus isolates between 2012 and 2018 from 2.9% to 25.6% (P = 0.001), including MSSA isolates from 3.7% to 29.6% for the same period (P = 0.003) (Figure 3).
Figure 2.
Evolution of the proportion of MDR bacteria, ESBL-producing bacteria, MRSA and carbapenem-resistant Enterobacteriaceae (CRE) between 2012 and 2018 at the MSF Nap Kembe Trauma Hospital, Tabarre, Haiti.
Figure 3.
Resistance trends of S. aureus (SA), MSSA and MRSA to clindamycin between 2012 and 2018 at the MSF Nap Kembe Trauma Hospital, Tabarre, Haiti. Minimum number of isolates per year for SA >30, for MSSA >25 and for MRSA >10. MRSA did not have enough isolates in 2012, 2013 and 2014.
ESBL-producing Enterobacteriaceae increased significantly from 3.2% in 2012 to 42.9% in 2018 (P ≤ 0.001). The prevalence of ESBL-producing strains was 66.7% for K. pneumoniae, 35.6% for E. coli, 32.7% for E. cloacae and 43.8% for S. marcescens. The proportion of ESBL-producing strains of K. pneumoniae and E. coli was found to be higher in blood samples than in pus and wound swabs, but the difference was not statistically significant (Table 2). Resistance to carbapenems was detected in 2.6% of Enterobacteriaceae and did not significantly change over the study period (P = 0.493) (Figure 2).
Three carbapenem-resistant ESBL-producing Enterobacteriaceae were detected over the 7 year period (two E. coli and one Proteus vulgaris isolate)—these have been defined as critical priority pathogens by WHO. Enterobacteriaceae with more than 30 isolates (Enterobacter cloacae, E. coli, K. pneumoniae, Proteus mirabilis and Serratia marcescens) were highly resistant to third-generation cephalosporins (cefotaxime and/or ceftazidime): almost 80% were resistant to cefotaxime (79.8%); 85.5% of K. pneumoniae were resistant to ceftazidime.
Approximately three-quarters of P. aeruginosa isolates (74.5%) were tested against a carbapenem antibiotic (imipenem), with 26.9% overall resistance. The trend of resistance over the years could not be tested because of the low number of isolates for some years.
The resistance patterns of the 10 most commonly isolated bacteria to clinically relevant antibiotics over the 7 year study period are presented in Table S1 (available as Supplementary data at JAC-AMR Online).
Discussion
Studies exploring antibiotic resistance in Haiti are scarce and are usually focused on cholera and TB. To our knowledge, this is the largest study exploring antibiotic resistance trends in trauma cases in Haiti during the post-crisis phase after the 2010 earthquake.
From the samples taken in our trauma hospital over 7 years, the prevalence 32.0% of all isolates being MDROs is a source of concern; whilst the trend of MDRO prevalence did not increase significantly, it is still a worrying trend. The reported prevalence of MDROs in the literature varies widely depending on setting, hospital size and patient profile; the level of care influences the prevalence of MDROs, with ICUs often having a higher prevalence of MDROs than non-ICU settings. Nevertheless, MDROs cause increased morbidity and mortality, especially in LMICs.1–4
The prevalence of MRSA across the Latin American region is generally high; the ReLAVRA surveillance data indicate 44% of S. aureus isolates from across 15 countries in Latin America were MRSA.9 There is a range from 57.3% MRSA from all S. aureus isolates across three hospitals in the Dominican Republic,10 to our finding of 24.1% MRSA in our trauma hospital in Haiti, to a lower prevalence reported in Nicaragua (20%) and Cuba (6%).11,12
It is concerning that there is increasing clindamycin resistance over time, not only in all S. aureus isolates, but also in MSSA (Figure 3). This is in concordance with the findings of Carrel et al.,13 which showed an increasing resistance to clindamycin from 2003 to 2010 in the USA (from 5.7% to 16.1%) and Stein et al.,14 reporting an increase in MSSA in Israel between 2009 and 2012 (from 5% to 29%). Furtado et al.15 reported 64% resistance to clindamycin in MRSA isolates in Brazil. This increasing resistance to a widely used antibiotic with several advantages—including high bioavailability, good skin and soft tissue permeability, availability in both parenteral and oral formulations and being relatively cheap—is an alarming situation. It gives emphasis to the need for the implementation of IPC, antibiotic stewardship and ABR surveillance programmes in low-resource settings.13
Latin America reports the highest rates of ESBL producers10,16 and we documented an overall increase of ESBL-producing Enterobacteriaceae (from 3.2% in 2012 to 42.9% in 2018) in Haiti. We found a higher ESBL rate in K. pneumoniae (66.7%) than current Latin America region estimates of 58%16 and 36.3%;17 however, this may be due to the differences in sample types.
The resistance of E. coli to the third generation cephalosporins was 60% in the neighbouring country, Dominican Republic, in 2016,11 which is consistent with our findings of 51% resistance to cefotaxime and 70% resistance to ceftazidime in E. coli isolates.
In 2017, WHO published a list18 of priority pathogens of antibiotic-resistant bacteria that were classified as critical, high and medium priority according to the urgency of need for new antibiotics. We identified two critical-priority pathogens (carbapenem-resistant ESBL-producing Enterobacteriales and carbapenem-resistant P. aeruginosa) in our clinical setting between 2012 and 2018. Two E. coli and one P. vulgaris isolate were found to be carbapenem-resistant (imipenem) ESBL-producing Enterobacterales species over the 7 year period. Chaintarli et al.19 reported less than 10% resistance to imipenem in their ESBL isolates in a smaller study in Haiti; we detected 1.4% imipenem resistance in our ESBL isolates, which is significantly lower. However, in a hospital with such high rates of ESBL positivity, if associated with resistance to other antibiotic classes (e.g. fluoroquinolones, tetracyclines) and depending on the patient profile and severity of the infection, this may translate into a need for more carbapenem use. In this type of scenario, a stricter ABR stewardship system is required.
Sader et al.20 found 5.8% carbapenem-resistant Enterobacteriaceae (CRE) in six Latin American countries in 2019, which is slightly higher than our finding of 2.6% over 7 years. Carbapenem resistance in P. aeruginosa isolates was reported at 20% in the Dominican Republic in 2016,9 which is consistent with our findings (26.9%). While we found no resistance to carbapenems in K. pneumoniae isolates, Correa et al.21 reported 8.6% resistance in Brazil and de Luna et al.10 reported 51% resistance to meropenem. There are several factors that limit healthcare facilities in low-resource settings from tackling their ABR problem, such as resource limitations, lack of trained personnel and weak infection control measures.6 While the Haitian Government announced ABR as an emerging public health threat at the end of 2018,22 there is still no specific national plan or microbiological surveillance system in place and the country was still not a part of the PAHO Latin American Network for Antimicrobial Resistance Surveillance—the ReLAVRA network—at the time of writing. Although the laboratory capacity in Haiti has increased dramatically since the 2010 earthquake and cholera outbreak, the main areas strengthened have been molecular testing for TB, HIV, malaria and diarrhoeal and respiratory diseases.23 Access to quality microbiological diagnostics is still limited, and the guided use of antibiotics through microbiological results and the surveillance of ABR in the country is minimal. Boulos et al.24 reported that not having access to microbiological culture was associated with increased fatality among neonatal sepsis cases in Haiti. Moreover, self-treatment with easily accessible, substandard antibiotics in street markets is a common practice in LMICs contributing to antibiotic resistance.25,26 The current overall prevalence of ABR in Haiti is not well known, since only a few scientific publications exist, and they focus on neonatal sepsis,19,24,27 surgical burns28 and disaster-related wound infections with very small sample sizes.29,30
There were some strengths to this study. It comprised a large sample size over a long period that allowed for the evolution of resistance trends. Data were doubled-entered and validated. However, there are some limitations related to the nature of retrospective observational studies and data quality. The data were taken from patients admitted to a trauma hospital and therefore the results represent a narrow patient profile. The exact total number of samples is not known because of missing data on sterile samples. Furthermore, the majority of samples were either pus or wound swabs that can be prone to cutaneous colonization. Also, approximately 30% of microbiological data were unable to be matched with patient data. Nevertheless, this study provides evidence drawn from largest sample from Haiti thus far. Further research is needed to inform treatment guidelines in the shadow of increasing resistance patterns and to show the impact of antibiotic stewardship and IPC programmes in resource-limited settings.
Conclusions
This study provides baseline information for bacterial resistance patterns for trauma infections in Haiti, highlighting the importance of ABR surveillance in hospitals and contributing to the knowledge of ABR in Haiti.
Overall, a high prevalence of MDR bacteria was observed among patients in a trauma hospital during a 7 year period in Port-au-Prince, Haiti. Additionally, we documented a high prevalence of ESBL-producing bacteria and resistance to clindamycin in S. aureus that increased significantly during that time; of note, there were several isolates of WHO critically resistant bacteria. These findings were from a trauma hospital with a structured IPC programme and access to microbiological culture. Without a strong push to implement good IPC practices, develop antibiotic stewardship and improve surveillance, antibiotic resistance will continue to rise, leading to narrowing therapeutic choices for HAIs in LMICs where the resources are already scarce.
Supplementary Material
Acknowledgements
We thank Dr Mohamad Khalife and Dr Tony Reid for editorial support and Dr Dorothee Orbach and hospital staff who contributed to the data collection throughout the years, as well as the patients who were part of the study.
Funding
This work was supported by routine programme funding of Médecins Sans Frontières.
Transparency declarations
None to declare.
Supplementary data
Tables S1 is available as Supplementary data at JAC-AMR Online.
References
- 1.WHO. Global Antimicrobial Resistance Surveillance System (GLASS) Report, Early Implementation 2020. https://www.who.int/publications/i/item/9789240005587.
- 2.Friedman ND, Temkin E, Carmeli Y.. The negative impact of antibiotic resistance. Clin Microbiol Infect 2016; 22: 416–22. [DOI] [PubMed] [Google Scholar]
- 3.Ogunsola FT, Mehtar S.. Challenges regarding the control of environmental sources of contamination in healthcare settings in low-and middle-income countries - a narrative review. Antimicrob Resist Infect Control 2020; 9: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Founou RC, Founou LL, Essack SY.. Clinical and economic impact of antibiotic resistance in developing countries: a systematic review and meta-analysis. PLoS One 2017; 12: e0189621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allegranzi B, Nejad SB, Combescure C. et al. Burden of endemic health-care-associated infection in developing countries: systematic review and meta-analysis. Lancet 2011; 377: 228–41. [DOI] [PubMed] [Google Scholar]
- 6.Bardossy AC, Zervos J, Zervos M.. Preventing hospital-acquired infections in low-income and middle-income countries: impact, gaps, and opportunities. Infect Dis Clin North Am 2016; 30: 805–18. [DOI] [PubMed] [Google Scholar]
- 7.Berends MS, Luz CF, Friedrich AW. et al. AMR - an R package for working with antimicrobial resistance data. bioRxiv 2019; 10.1101/810622. [DOI] [Google Scholar]
- 8.Magiorakos A-P, Srinivasan A, Carey RB. et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18: 268–81. [DOI] [PubMed] [Google Scholar]
- 9.Pan American Health Organization Antimicrobial Resistance Special Program. Magnitude and Trends of Antimicrobial Resistance in Latin America. ReLAVRA 2014, 2015, 2016. Summary Report. 2020. https://www.paho.org/en/documents/magnitude-and-trends-antimicrobial-resistance-latin-america-relavra-2014-2015-2016.
- 10.de Luna D, Sánchez JJ, Peguero M. et al. Antimicrobial resistance profiles of microorganisms isolated from hospitalized patients in Dominican Republic. Rev Panam Salud Publica 2020; 44: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guzmán-Blanco M, Mejía C, Isturiz R. et al. Epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) in Latin America. Int J Antimicrob Agents 2009; 34: 304–8. [DOI] [PubMed] [Google Scholar]
- 12.Pan American Health Organization. Annual Report of the Monitoring/Surveillance Network for Resistance to Antibiotics, 2004. 2005. https://www3.paho.org/English/AD/DPC/CD/amr-2004.htm.
- 13.Carrel M, Goto M, Schweizer ML. et al. Diffusion of clindamycin-resistant and erythromycin-resistant methicillin-susceptible Staphylococcus aureus (MSSA), potential ST398, in United States Veterans Health Administration Hospitals, 2003-2014. Antimicrob Resist Infect Control 2017; 6: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stein M, Komerska J, Prizade M. et al. Clindamycin resistance among Staphylococcus aureus strains in Israel: implications for empirical treatment of skin and soft tissue infections. Int J Infect Dis 2016; 46: 18–21. [DOI] [PubMed] [Google Scholar]
- 15.Furtado GH, Rocha J, Hayden R. et al. Early switch/early discharge opportunities for hospitalized patients with methicillin-resistant Staphylococcus aureus complicated skin and soft tissue infections in Brazil. Braz J Infect Dis 2019; 23: 86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guzmán-Blanco M, Labarca JA, Villegas MV. et al. Extended spectrum β-lactamase producers among nosocomial Enterobacteriaceae in Latin America. Braz J Infect Dis 2014; 18: 421–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vega S, Dowzicky MJ.. Antimicrobial susceptibility among Gram-positive and Gram-negative organisms collected from the Latin American region between 2004 and 2015 as part of the Tigecycline Evaluation and Surveillance Trial. Ann Clin Microbiol Antimicrob 2017; 16:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.WHO. Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis. 2017. https://apps.who.int/iris/handle/10665/311820.
- 19.Chaintarli K, Lenglet A, Beauzile BD. et al. High prevalence of ESBL-positive bacteria in an obstetrics emergency hospital and neonatal care unit—Haiti, 2016. Infect Control Hosp Epidemiol 2018; 39: 1381–3. [DOI] [PubMed] [Google Scholar]
- 20.Sader HS, Carvalhaes CG, Arends SJR. et al. Aztreonam/avibactam activity against clinical isolates of Enterobacterales collected in Europe, Asia and Latin America in 2019. J Antimicrob Chemother 2021; 76: 659–66. [DOI] [PubMed] [Google Scholar]
- 21.Correa L, Martino MDV, Siqueira I. et al. A hospital-based matched case–control study to identify clinical outcome and risk factors associated with carbapenem-resistant Klebsiella pneumoniae infection. BMC Infect Dis 2013; 13: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laboratoire National de Santé Publique - Haïti. Le Bulletin du Laboratoire National de Santé Publique - Haïti. Laboratoire National de Santé Publique, 2018.
- 23.Jean Louis F, Buteau J, Boncy J. et al. Building and rebuilding: the national public health laboratory systems and services before and after the earthquake and cholera epidemic, Haiti, 2009–2015. Am J Trop Med Hyg 2017; 97: 21–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boulos A, Rand K, Johnson JA. et al. Neonatal sepsis in Haiti. J Trop Pediatr 2017; 63: 70–3. [DOI] [PubMed] [Google Scholar]
- 25.Moise K, Bernard JJ, Henrys JH.. Evaluation of antibiotic self-medication among outpatients of the state university hospital of Port-Au-Prince, Haiti: a cross-sectional study. Pan Afr Med J 2017; 28: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jean-Baptiste T, Carpenter JF, Dahl K. et al. Substandard quality of the antimicrobials sold in the street markets in Haiti. Antibiotics (Basel) 2020; 9: 407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lenglet A, Faniyan O, Hopman J.. A nosocomial outbreak of clinical sepsis in a neonatal care unit (NCU) in Port-Au-Prince Haiti, July 2014 – September 2015. PLoS Curr 2018; 10: ecurrents.outbreaks.58723332ec0de952adefd9a9b6905932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy RA, Nisenbaum L, Labar AS. et al. Invasive infection and outcomes in a humanitarian surgical burn program in Haiti. World J Surg 2016; 40: 1550–7. [DOI] [PubMed] [Google Scholar]
- 29.Miskin IN, Nir-Paz R, Block C. et al. Antimicrobial therapy for wound infections after catastrophic earthquakes. N Engl J Med 2010; 363: 2571–3. [DOI] [PubMed] [Google Scholar]
- 30.Marra AR, Valle Martino MD, Ribas MR. et al. Microbiological findings from the Haiti disaster. Travel Med Infect Dis 2012; 10: 157–61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.