Abstract
Objective
To assess the performance of a combination of three quantitative MRI markers (iron deposition, basal neuronal metabolism, and regional atrophy) for differential diagnosis between amyotrophic lateral sclerosis (ALS) and primary lateral sclerosis (PLS).
Methods
In total, 33 ALS, 12 PLS, and 28 healthy control (HC) subjects underwent a 3T MRI study including single‐ and multi‐echo sequences for gray matter (GM) volumetry and quantitative susceptibility mapping (QSM) and a pseudo‐continuous arterial spin labeling (ASL) sequence for cerebral blood flow (CBF) measurement. Mean values of QSM, CBF, and GM volumes were extracted in the motor cortex, basal ganglia, thalamus, amygdala, and hippocampus. A generalized linear model was applied to the three measures to binary discriminate between groups. The diagnostic performances were evaluated via receiver operating characteristic analyses.
Results
A significant discrimination was obtained: between ALS and HCs in the left and right motor cortex, where QSM increases were respectively associated with disability scores and disease duration; between PLS and ALS in the left motor cortex, where PLS patients resulted significantly more atrophic; between ALS and HC in the right motor cortex, where GM volumes were associated with upper motor neuron scores. Significant discrimination between ALS and HC was achieved in subcortical structures only combining all three parameters.
Interpretation
While increased QSM values in the motor cortex of ALS patients is a consolidated finding, combining QSM, CBF, and GM volumetry shows higher diagnostic potential for differentiating ALS patients from HC subjects and, in the motor cortex, between ALS and PLS.
Introduction
Amyotrophic lateral sclerosis (ALS) is a progressive fatal disease of the motor system, affecting brain upper motor neurons (UMN) and spinal or brainstem lower motor neurons (LMN), with variable involvement of extra‐motor brain regions.1 Each patient differs in region of disease onset, progression, and phenotype, which is related to the combination of motor neurons involvement, meaning that ALS has a marked clinical heterogeneity.2 The “classic” ALS phenotype is characterized by both UMN and LMN impairment, while primary lateral sclerosis (PLS) and progressive muscular atrophy (PMA) represent the extremities of the spectrum of motor neuron disease (MND) phenotypes, respectively, characterized by pure UMN or LMN syndromes.2, 3 Other ALS phenotypes (bulbar, flail limbs, and pyramidal phenotypes) are (respectively) characterized by the decline of voluntary bulbar functions, progressive wasting in the upper or lower limbs, and degeneration of the pyramidal tract.2, 3
An early diagnosis and characterization of the ALS phenotypes would help clinicians to frame a more personalized therapy and possibly increase life quality and expectancy.2, 3, 4 Differentiation of PLS from UMN‐dominant phenotype of ALS remains a significant challenge in the early symptomatic phase of both disorders, with ongoing debate as to whether they form a clinical and histopathological continuum, the latter being recently analyzed by the review of Finegan et al. 2019.5 In this regard, conventional brain MRI fails to differentiate ALS from PLS due to a lack of a way to combine signs of the pathology into a combined clinical MRI diagnostic framework.6, 7 The development of advanced MRI techniques has allowed the proliferation of quantitative MRI (qMRI) markers, MRI‐derived measures that can be calibrated to standard physical units, whose diagnostic potential is yet to be fully investigated in the context of clinical MRI studies on the ALS pathology. For example, the involvement of gray matter (GM) structures in ALS neurodegeneration has been extensively studied using quantitative MRI markers, not only as morphometric indices of GM atrophy, but also as biophysical surrogates of structural and metabolic tissue properties, such as iron deposition8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18 and cerebral perfusion.19, 20, 21
Among these, abnormal iron accumulation in the motor cortex seems to be the most consistent finding in ALS patients, probably due to the activation of a proinflammatory mechanism that causes a microglial iron deposition in the middle and deep cortical layers, a phenomenon that was originally associated with ALS disease severity.22, 23 However, only one previous work has investigated the regional cerebral blood flow (CBF) concurrently with QSM, in ALS patients.21 As CBF provides a regional index of basal neuronal metabolism, it could in principle provide an independent quantitative marker of neuronal degeneration complementing the other ALS structural markers. In fact, similar to a positron emission tomography (PET) scan, a CBF measure would possibly address if pathological iron deposition, as resulting from cell death,24 is also accompanied by metabolic dysregulation (eventually before structural atrophy becomes manifest).
The aim of this work was to explore the diagnostic potential of combining QSM and CBF measures with GM volumetry (regional atrophy) in the discrimination of ALS/PLS patients from age‐ and sex‐matched healthy controls and between ALS and PLS phenotypes. Thus, a regional analysis was conducted for motor cortex, basal ganglia, thalamus, and hippocampus and amygdala complex.
The set of cortical and subcortical regions of interest was chosen with the following rationale: (i) we had strong prior expectations on the basis of previous pathological findings,8, 25, 26 (ii) we made use of a precise and validated GM parcellation to obtain a reliable and quantitative GM volumetry of the brain in each individual native imaging space. Particularly, the latter choice was operated to avoid any spatial transformation of the reconstructed QSM and CBF images prior to individual marker extractions, as well as to provide an additional quantitative marker for the regional brain atrophy that could possibly inconsistently covary with either QSM or CBF estimation, because of different neurodegeneration processes. We hypothesized that combining multiple qMRI features would possibly increase the discrimination power with respect to ALS/PLS pathology and phenotype, compared to the separate analysis of each feature.
Materials and Methods
Subjects
Thirty‐three right‐handed patients (19 males, 14 females; mean age 59 ± 10), with definite, clinical or laboratory‐supported probable ALS, according to El‐Escorial revised criteria,27 were consecutively recruited at the First Division of Neurology of the University of Campania “Luigi Vanvitelli” (Naples, Italy) from November 2018 to March 2020. ALS patients were required to meet the following criteria: classic, bulbar, LMN, or UMN dominant phenotypes3; disease onset not earlier than 36 months from enrollment; age of onset of 40 years or older. Twelve right‐handed patients (9 males, 3 females; mean age 59 ± 10) met the new consensus diagnostic criteria of probable (n = 2) and definite (n = 10) PLS.28
The clinical assessment included: disability status evaluation, measuring the ALSFRS‐R score (0–48, with lower total reflecting higher disability)29 and the UMN score, index of pyramidal dysfunction through the evaluation of the number of pathologic reflexes elicited from 15 body sites30; assessment of global cognitive functioning, administering the Italian version of Edinburgh Cognitive and Behavioural ALS Screen (ECAS).31, 32
Disease duration was calculated from symptom onset to scan date in months, and the rate of progression determined by: (48 − current ALSFRS‐R)/disease duration. Disease stage was assessed according to King’s clinical staging system.33 Genetic analysis was performed in all patients, exploring C9orf72 repeat expansion and mutations of SOD1, TARDBP, and FUS/TLS in all patients and, additionally, SPAST and SPG7, in PLS patients. No mutations of these genes were reported.
Twenty‐eight right‐handed healthy control subjects (HCs) (13 males, 15 females; mean age 56 ± 16) were enrolled by “word of mouth” and among caregivers’ friends. They were age‐, sex‐, and education‐matched with the enrolled ALS patients and unrelated to them. Moreover, they had no comorbid neurological, psychiatric, or medical conditions. They underwent MMSE and their scores were ≥27.
Exclusion criteria were: medical illnesses or substance abuse that could interfere with cognitive functioning; any (other) major systemic, psychiatric, or neurological diseases; other causes of brain damage, including lacunae and extensive cerebrovascular disorders at MRI; a vital capacity lower than 70% of the predicted value.
For more details, the demographic and clinical characterization is presented in Table 1. All participants provided written informed consent to participate in the study according to the Declaration of Helsinki. The study was approved by the Ethics Committee of the University of Campania “L. Vanvitelli” (Protocol nr. 591/2018).
Table 1.
Demographic, clinical, and neuropsychological measures of patients, divided into ALS and PLS, and healthy controls (HCs); data are shown as mean (standard deviation) or count (percentage)
| Variables | HC (n = 28) | ALS (n = 33) | PLS (n = 12) | HC versus ALS t‐test | HC versus ALS p‐value | HC versus PLS t‐test | HC versus PLS p‐value | ALS versus PLS t‐test | ALS versus PLS p‐value |
|---|---|---|---|---|---|---|---|---|---|
| Demographics | |||||||||
| Age, years | 56 (16) | 59 (10) | 59 (10) | −0.833 | 0.4 | −0.602 | 0.5 | −0.36 | 0.97 |
| Education, years | 12 (3) | 11 (3) | 12 (4) | 0.624 | 0.535 | −0.052 | 0.96 | −0.48 | 0.63 |
| Sex, male | 13 (46.4%) | 19 (57.6%) | 9 (75%) | 0.86 | 0.39 | 1.109 | 0.28 | 1.76 | 0.09 |
| Clinical features | |||||||||
| Disease duration, months | n.a. | 15 (11) | 30 (11) | −3.75 | 0.001 | ||||
| Disease onset (bulbar/upper limbs/lower limbs) | n.a. | 8/12/13 | 3/0/9 | ||||||
| Disease phenotype (classic/bulbar/LMN/pyramidal/PLS) | n.a. | 3/6/11/13 | 12 | ||||||
| ALSFRS−R total score | n.a. | 42 (5) | 41 (5) | 1.21 | 0.23 | ||||
| Disease progression rate1 | n.a. | 0.64 (0.57) | 0.28 (0.16) | 3.23 | 0.002 | ||||
| UMN score | n.a. | 8 (4) | 11 (3) | −1.92 | 0.062 | ||||
| King’s clinical staging:2 | n.a. | 9/19/5 | 0/8/4 | ||||||
| Neuropsychological measures | |||||||||
| MMSE | 29 (1) | n.a. | n.a. | ||||||
| ECAS total score | n.a. | 91 (25) | 96 (18) | −0.65 | 0.52 | ||||
Bold text formatting indicates statistical significant comparisons.
ALS, Amyotrophic Lateral Sclerosis; ALSFRS−R, ALS Functional Rating Scale Revised; ECAS, Edinburgh Cognitive and Behavioural ALS Screen; MMSE, Mini Mental State Examination; HCs, Healthy Controls; χ 2, Chi‐square test; LMN, Lower Motor Neuron; UMN, upper motor neuron.
Disease Progression Rate was computed as: 48 − ALSFRS‐R/Disease Duration.
According to the King’s clinical staging system, the number of regions involved gives the stage.
MRI data acquisition
MRI images were acquired on a 3 Tesla scanner equipped with a 32‐channel parallel head coil (General Electric Healthcare, Milwaukee, Wisconsin). The imaging protocol included:
-
‐
Three‐dimensional T1‐weighted images (gradient‐echo sequence Inversion Recovery prepared Fast Spoiled Gradient Recalled‐echo): repetition time (TR) = 6900 msec, echo time (TE) = 3.0 msec, resolution = 1 × 1 × 1 mm3, matrix size = 256 × 256, inversion time (TI) = 650 msec. Duration: 6.49 min.
-
‐
3D‐PCASL sequence: TR = 5306 msec, TE = 10.5 msec, field of view 128 × 128 mm2, slice thickness 4 mm, in plane resolution = 3.8 × 3.8 × 4 mm3, post labeling delay 2525 msec for a total of 30 subjects, 2025 msec for a total of 40 subjects, and 1525 msec for a total of 3 subjects, 34 slices. Duration: 5.09 min.
-
‐
Three‐dimensional multi‐echo gradient echo sequence: TR = 47.3 msec, first TE = 3.2 msec, echo spacing = 3.75 msec, total number of echoes = 12, resolution = 1 × 1 × 1 mm3, matrix size = 256 × 256, anterior–posterior phase‐encoding direction. Number of signal averages (NEX) = 0.7. Duration: 10.18 min.
-
‐
FLuid‐Attenuated Inversion Recovery (FLAIR) sequence: TR = 11000 msec, TE = 122.7 msec, Echo train length = 18, voxel dimension 0.5 × 0.5 × 3 mm3, anterior–posterior phase‐encoding direction. Fat Saturation. Duration: 3.30 min.
MRI data processing
Structural data were processed using FreeSurfer (FS) v 6.0 (https://surfer.nmr.mgh.harvard.edu/) using a Dell Inc. PowerEdge T430 workstation equipped with an Intel® Xenon(R) CPU ES‐2620 v4 @2.10 GHz processor and 8 GB of RAM and running Linux Ubuntu 18.04.5 LTS.
Anatomical data were imported in FS and submitted to the standard structural image preprocessing and reconstruction pipeline via the “recon‐all" FS command.34, 35, 36, 37, 38, 39, 40
After preprocessing, two atlases, namely the aparc.a2009s+aseg and the aseg, were considered to extract the cortical and subcortical regions of interests (ROIs) in the subject native space, respectively the left and right precentral gyri for the motor cortex, and the left and right caudate, putamen, pallidum, thalamus, hippocampus, and amygdala.
Volumes of these structures were derived from the FS commands asegstats2table and aparcstats2table.
Single‐subject whole‐brain CBF maps were calculated from the 3D‐PCASL raw images using the prescan proton density (PD) volume and the perfusion‐weighted series using the calibration formula in.41
QSM maps were reconstructed in native space in MATLAB using the toolbox “STI Suite”42 with the following preprocessing steps: three‐dimensional phase unwrapping using Laplacian based method,43 background phase removal using the V‐SHARP method44 and the dipole convolution for the final QSM map computation.45
For each subject, the two atlases were coregistered to the CBF and QSM maps in a two‐step procedure using FSL software (https://fsl.fmrib.ox.ac.uk/fsl/): for CBF and QSM analyses respectively, the prescan PD image and the magnitude image of the multi‐echo gradient echo acquisition were coregistered to the T1 images using FLIRT command (9 degree of freedom and using a trilinear interpolation). The obtained transformation matrices were then inverted and applied to the two atlases in native space using the FLIRT command (with nearest neighbor interpolation). In this way no transformations were applied to the CBF and QSM maps before extracting the regional mean values.
On axial FLAIR images, white matter hyperintensities (WMH) were rated according to the age‐related white matter changes scale (ARWMC)46 and Fazekas’ scale (4 point scale) for assessing periventricular and deep WMH.47
Statistical analysis
Mean CBF and QSM values within the ROIs were then computed for each subject. Using linear regression, the GM volumes were adjusted for age, sex and total intracranial volumes, the QSM values were adjusted for age and sex whereas the CBF values were adjusted for age, sex, post‐labelling delay (PLD), and global CBF, the latter computed as mean CBF value in the entire brain mask.
In all regions, a receiver operating characteristic (ROC) curve analysis was performed in MATLAB using the built‐in function “perfcurve” to assess the binary discrimination power of all three MRI measures combined together. First, a generalized linear model (GLM) with binomial distribution and logit link function was used to predict the membership to one out of two groups (ALS vs. HC, PLS vs. HC, ALS vs. PLS) from the regional QSM, CBF, and GM volumes, which were used as predictors. For each ROI, the GLM was fitted to the data and the ROC curves were calculated by comparing the fitted class values (predictions) with the true class memberships by applying a threshold varying between zero and one. This returned an area under the curve (AUC) as a diagnostic index and its confidence intervals and p‐value, the latter adjusted for multiple comparisons using false discovery rate (FDR) correction across all the considered ROIs.
The same discriminative analysis was also performed considering the MRI parameters separately.
Post hoc statistical analyses in the regions allowing significant between‐group discrimination were conducted by performing a one‐way between‐group ANOVA (three levels for HCs, ALS, and PLS) and Student’s t‐tests. In the same regions, linear regression analysis via robustfit MATLAB function was performed between the MR parameters and the clinical scores of disease duration, ALSFRS‐R (measured at time of image acquisition), and UMN.
Results
Demographics, clinical, and neuropsychological variables
Patients and controls characteristics are reported in the Table 1. ALS/PLS patients and HCs did not statistically differ on age, gender, and education. The t‐test revealed no significant difference in ALSFRS‐R total score and in global cognitive performances in ALS patients compared to PLS patients. On the basis of ECAS subscores,31, 32 according to the Strong criteria for frontotemporal spectrum disorder of ALS,48 8 ALS patients and 4 PLS patients had cognitive impairment (ci), with executive dysfunction in all cases, except for one ALS patient with both executive and language dysfunction. No differences between HC and PT were found in terms of WMH and none of patients reported severe periventricular and deep WMH according to the Fazekas scale.
ROC curve analysis
The ROC analyses (Fig. 1) performed considering all three qMRI measures as predictors significantly discriminated ALS patients from HC, in the motor cortex bilaterally (left precentral gyrus: AUC = 0.68, p < 0.05, q(FDR) < 0.05; right precentral gyrus: AUC = 0.68, p < 0.01, q(FDR) <0.05) and in several subcortical structures, preferentially from the right hemisphere (left caudate: AUC = 0.69, p < 0.01, q(FDR) < 0.05; right caudate: AUC = 0.66, p < 0.05, q(FDR) < 0.05; right putamen: AUC = 0.66, p < 0.05, q(FDR) < 0.05; right pallidum: AUC = 0.67, p < 0.05, q(FDR) < 0.05; right hippocampus: AUC = 0.69, p < 0.01, q(FDR) < 0.05; right amygdala: AUC = 0.70, p < 0.01, q(FDR) < 0.05).
Figure 1.
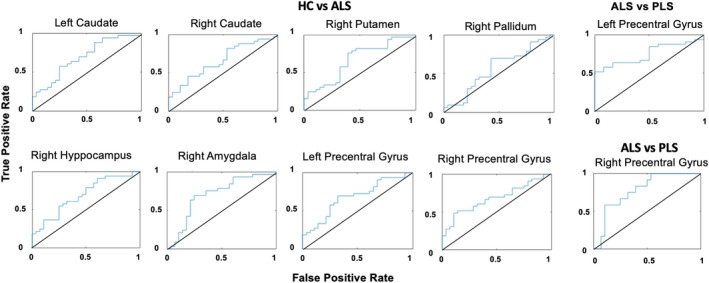
ROC curves for combined QSM, CBF, and GM volumes.
Moreover, PLS patients were significantly discriminated from HC in the right precentral gyrus (AUC = 0.78, p < 0.001, q(FDR) < 0.01) and from ALS patients in the left precentral gyrus (AUC = 0.75, p < 0.001, q(FDR) < 0.05).
No significant discriminations were found when considering the CBF and QSM separately in any groups’ comparisons and in any regions. When considering the volume measure alone, significant discrimination was found in the right precentral gyrus comparing HC and PLS (Table S1–S3).
QSM analysis
The ANOVA analysis of the QSM measures revealed a significant group effect only in the left (p < 0.05) and right (p < 0.05) precentral gyrus (Fig. 2). In the left precentral gyrus, post hoc t‐tests disclosed a significant QSM increase in the ALS versus HC (p < 0.05) and versus PLS (p < 0.05) groups, but not in the PLS versus HC (p > 0.05) groups (Fig. 2A). In this region, QSM values were significantly correlated to ALSFRS scores (t = 2.36, p < 0.05) in the ALS group (Fig. 2B).
Figure 2.
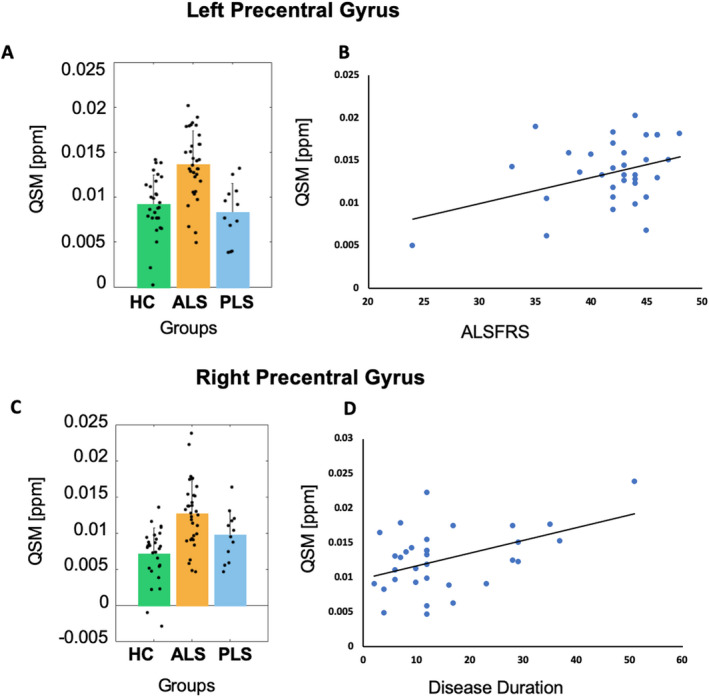
QSM analysis. Histogram left‐right precentral, scatter plots (with liner trend).
In the right precentral gyrus, post hoc t‐test disclosed a significant QSM increase in the ALS versus HC (p < 0.05), but not in the ASL versus PLS (p > 0.05) or PLS versus HC (p > 0.05), groups (Fig. 2C). In this region, QSM values were significantly correlated to disease durations (t = 2.74, p < 0.05) in the ALS group (Fig. 2D).
CBF analysis
The ANOVA analysis of the CBF value did not reveal any significant group effects in the considered ROIs.
GM volume analysis
In the motor cortex, the ANOVA analysis of the GM volumes revealed a significant group effect in the right precentral gyrus (p < 0.05) and in the left caudate (p < 0.05) (Fig. 3). Post hoc t‐test in the right precentral gyrus did not disclose GM volume differences between the ALS and HC (p > 0.05) groups but revealed a significant decrease of GM volume in the PLS versus HC (p < 0.01) and in the PLS versus ALS (p < 0.05) groups (Fig. 3A). A significant correlation was found between GM volume of the right precentral gyrus and UMN scores (t = −2.68, p < 0.05) for the ALS group (Fig. 3B).
Figure 3.
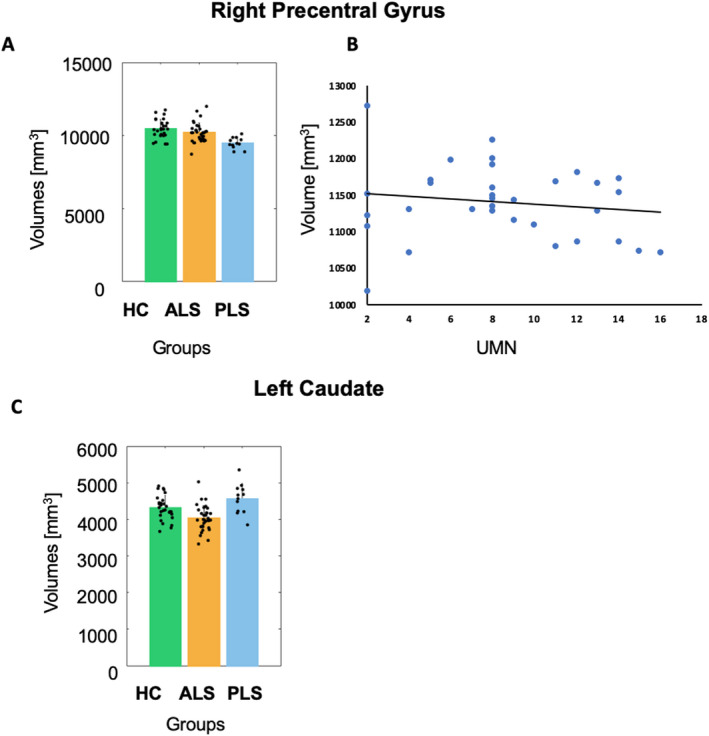
GM volume analysis. Histogram right precentral gyrus and left caudate, scatter plot (with linear trend).
Among subcortical structures, in the left caudate, post hoc t‐test revealed a significant decrease of GM volumes in the ALS versus HC (p < 0.05) and versus PLS (p < 0.05) groups, but no significant differences between the PLS and HC (p > 0.05) groups (Fig. 3C). No significant correlations were found between GM volumes and the clinical variables.
Discussion
The aim of this study was to address the differential diagnostic potential of combining three validated qMRI markers for the potential discrimination of ALS and PLS phenotypes in a clinical 3 T MRI study. This entailed obtaining regional quantities with established physical meaning from advanced MRI techniques that are widely available in clinical settings and can provide different views on some morphological and metabolic aspects of pathological brain alterations in ALS patients. Considering a number of GM regions previously reported as most implicated in the neurodegeneration processes of ALS/PLS pathology, we highlight that the combination of QSM and CBF, as independent cerebral markers of local iron deposition and blood flow metabolism, with GM volumetry, as marker of brain atrophy, has significant discrimination capabilities in differentiating ALS and PLS phenotypes in the motor cortex and features some interesting ALS‐specific alterations in the investigated subcortical GM structures.
From the ROC analysis of combined QSM, CBF, and GM volumes, a significant discrimination of ALS patients from HC subjects was obtained in the left and right motor cortices, and in subcortical structures, including left and right caudate, right hippocampus, right putamen, right pallidum, and right amygdala. Moreover, a discrimination between PLS and HC was statistically significant in the right motor cortex whereas a statistically significant discrimination between ALS and PLS was obtained in the left precentral gyrus.
While the ANOVA analyses of the individual parameters illustrated that the discrimination power of the three parameters in the bilateral motor cortex and left caudate is at least partly (if not mostly) explained by the concurrent QSM increases (iron accumulation) and GM volume decreases (atrophy), the results from the right subcortical structures clearly suggest that the three measures might have constructively interacted among them toward providing a more powerful discrimination of ALS patients from HC subjects. In fact, in those regions, none of the individual parameters significantly explained the between‐group variance, nor were these found significantly altered, in ALS patients.
Indeed, basal ganglia structures would be affected by ALS pathology not simply as relay stations of the motor pathway that supports the voluntary movements, but also in relation to the more complex functioning of the fronto‐striatal circuit that regulates higher‐order (cognitive) aspects of motor control.26, 49, 50, 51 In this regard, Finegan et al.26 revealed by volumetric and morphometric analyses, different patterns of subcortical GM degeneration in motor neuron diseases, showing preferential medial bi‐thalamic pathology in PLS compared to the predominant putaminal and amygdala degeneration detected in ALS. Hence, the fact that in our study these subcortical structures significantly discriminate between HCs and ALS when considering QSM, CBF, and GM volumes could suggest that different levels of atrophy and iron deposition may interact with the local metabolism within these structures.
The significant role of the hippocampus and amygdala in the discrimination between ALS and HC is also not surprising, given the multi‐systemic nature of this disorder that usually presents a variety of clinical frontotemporal manifestations as extra‐motor signatures.52, 53, 54 As for caudate, putamen and pallidum, in our analysis both hippocampus and amygdala did not show any significant atrophy, perfusion alteration, nor a significant variation of iron content, in the analyzed ALS patients, albeit, when all these parameters were concurrently included in a multiparametric discriminative analysis, this led to a significant discrimination from HCs. These findings corroborate previous neuropathological55 and neuroimaging20, 26, 56, 57, 58 evidence supporting the involvement of mesial temporal lobe pathology in ALS, with particular regard to hippocampal and amygdala pathology within the limbic system.
The lack of significant alterations in the subcortical structures, especially in PLS patients, could be ascribed to the reduced sample size or to the fact that the information from considered MR parameters was not sufficient to discriminate this clinical form from both the ALS and HC groups. A possible improvement would be the introduction, in the same model, of additional surrogate MR parameter, for example, describing the white matter damage frequently observed in the pathology.59, 60, 61
The right lateralization of the results could be linked to the manifestation of different types of ALS motor impairment that usually end up differentially affecting the two hemispheres,62 eventually with a different impact of atrophy across the two hemispheres.63, 64 In addition, right hemispheric microstructural and functional connectivity impairments, particularly in the frontotemporal‐limbic circuit, have been related to emotional dysfunctions in ALS,65, 66, 67 underlining the key role of the right limbic circuit in emotional processing in ALS.
Our analyses of the individual measures revealed that QSM values were significantly increased in ALS patients, compared to HC subjects, in both the left and right motor cortex. However, this was not the case for PLS patients, either due to a lack of statistical power or to a higher impact of the GM cortical atrophy in PLS patients which turned out to be significantly more atrophic than both ALS patients and HC subjects. The finding of a more marked atrophy of the precentral gyri in PLS patients in comparison to ALS patients is in agreement with previous neuropathological evidence reporting focal “knife edge” atrophy of the precentral gyrus in PLS which resulted strikingly absent in even advanced cases of ALS.68
Importantly, QSM values in the motor cortex of ALS patients were associated with disease‐specific functional disability in the left precentral gyrus and with disease duration in the right precentral gyrus. The different pattern of clinical correlates between left and right motor cortex is likely to be also ascribed to the different impact of GM atrophy on the two cortical hemispheres, which, in turn, could be related to the clinical type. In fact, only in the right motor cortex, we found that GM volumes were associated with the upper motor neuron disease burden and, only there, the increased QSM values in ALS patients were correlated with disease duration (and not with disease‐related disability). In contrast, only in the left motor cortex, the QSM increases in ALS patients were correlated with disease‐related disability and these measures also resulted to differentiate between ALS and PLS phenotypes. To the best of our knowledge, the finding that in the left precentral gyrus the combination of perfusion, volumetry, and QSM allows to significantly differentiate between ALS and PLS is novel and may thus represent a potential new marker for the diagnosis validation between the two pathologies, albeit future studies on larger cohorts are needed to confirm this finding. Thus, our results would confirm (i) that the primary motor cortex is a cortical place of pathological iron deposition in ALS patients,16 and (ii), that the QSM increase is a valid cortical marker for the disease.25
Interestingly, the negative mean values of QSM in the right precentral gyrus observed in some healthy subjects, suggested the possible concomitant contribution of different substances: paramagnetic substances like iron, which determine positive values of QSM, and diamagnetic substances, like myelin or calcium, which determine the negative values of the QSM, as already observed in previous report.69
Two previous QSM studies, using either a voxel‐based approach or a regional analysis along the posterior and precentral gyri, found an increase of iron deposition levels within the motor cortex of ALS patients, in comparison to either HCs8 or patients with ASL mimics and non‐motor neuron symptoms.13 In addition, the study of Donatelli et al.,14 which specifically targeted the orofacial portion of the primary motor cortex, also reported that the hypointensity of T2* signal and the corresponding higher susceptibility values, albeit non‐quantitatively, had significant discriminative power in diagnosing ALS with functional bulbar impairment. Similarly, the recent study of Conte et al.11 found higher QSM values in ALS patients with predominant UMN impairment, compared to those with predominant LMN impairment, no clinically defined motor neuron impairment, ALS mimics and HCs.
Thus, in contrast to some previous findings, we disclosed an inverse correlation between UMN scores and GM volumes, rather than QSM values, in both left and right precentral gyri, suggesting that GM atrophy also plays a role in the morphological neurodegeneration of the motor cortex in ALS patients. While the composition of our sample did not allow to compare between bulbar and UMN or LMN dominant phenotypes, it should be also considered that the two morphological aspects of iron deposition and neuronal loss likely interact with each other on different spatial scales. Indeed, it is believed that a QSM increase in the motor cortex may actually derive from an unbalanced distribution of iron deposition between middle and deep layers of the cortex, as demonstrated by an ultra‐high spatial resolution study performed at 7T.12 Thus, the lack of GM volume reduction in the ALS group would suggest that the stage of ALS disease of the studied patients was probably too early for disclosing a significant atrophy of the motor cortex but still not too early for disclosing a significantly abnormal iron deposition.
We did not find any significant CBF differences between groups in both cortical and subcortical structures. This was not surprising given the known impact of GM atrophy on CBF estimates which would expectedly be reduced in all groups, not only in response to local GM volume reduction, but also as a related consequence of partial volume effects on the reconstructed CBF maps.70 Indeed, the spatial resolution of CBF maps is a factor of three worse in each direction, resulting in partial volume effect operating at the single voxel level. Nonetheless, as we intended to keep all three measures as independent as possible, we did not correct the original voxel‐wise CBF estimates with surrogated and non‐quantitative voxel‐wise GM volume estimates, as is usually done in whole‐brain voxel‐wise analyses. On the other hand, similar to our study, Welton et al.21 also reported significant increases of QSM values with no concurrent alterations of regional CBF values, in the motor cortex of ALS patients, and even the study of Shen et al.,20 that was specifically designed to reveal localized changes in whole‐brain perfusion and GM volume, reported no significant CBF changes in the comparison between ALS patients (with or without cognitive impairment) and HCs, although in ALS with frontotemporal dementia (FTD) a distinctive pattern of GM loss and hypoperfusion (with more significant alterations in the left frontal and temporal lobe) was revealed, in comparison to both ALS patients and HCs. Differently, in our sample we did not include FTD patients, although 8 ALS patients and 4 PLS patients had cognitive impairment with a prevalent dysexecutive profile. Thus, the results of Shen et al.,20 together with our findings, would suggest that CBF is not strongly affected by ALS pathology but other clinical factors, like the cognitive status and the co‐occurrence of FTD, would prevail in determining the variance of CBF measures.71 Among potential confounding methodological features, the inter‐subject variability in the arterial input function, possibly resulting in a suboptimal setting of the post‐label delay, certainly represents an important limitation of used clinical version of the ASL sequence. Thus, apart from the partial volume issue, which calls for ultra‐high magnetic field implementations, this particular methodological aspect could be possibly addressed by future studies employing a multi‐delay version of the clinical ASL sequence.72
This study has some limitations. First, the lack of significant differences in the studied regions when volumetric, CBF, and QSM data have been considered separately, could well be due to the heterogeneity of the studied population (i.e., classic, UMN‐ and LMN‐dominant, bulbar) which could have produced less systematic alterations in one (or more) of the considered parameters. Thus, studies with larger sample sizes, not only for the underrepresented PLS group, but also for each subtype of ALS patients, are needed to confirm the present data. Second, another limitation of the present work resides in the lack of an additional qMRI marker accounting for myelin content.73 However, while we acknowledge that additional qMRI markers will likely increase the discrimination power on a regional level, we did not include this type of measurement in the presented protocol due to the need to keep the total duration of the exam within the typical time constraints of a clinical study (about 20 min). Another limitation of the current work is the exclusion of the corticostriatal pathway (CST) from our analysis, that instead has been reported to be altered in these pathologies.5, 8 This was essentially due to technical reasons: first, because we obtained the ROI masks in the native anatomical space of each subject, we could not reach the same volumetric accuracy for the CST, for which a tractography analysis would have been needed. On the other hand, to obtain meaningful values from diffusion MRI in the (cortical and subcortical) grey matter structures, more advanced diffusion MRI acquisitions (e.g., multi‐shell diffusion MRI) and therefore longer scanning would have been needed. Second, to perform the described multi‐parametric discriminative analysis, the combined use of all three MRI parameters was not possible in the white matter due to the low reliability of CBF values in these regions.
Last, we excluded from our analysis more disabled patients hindered to undergo an MRI exam because of diaphragmatic weakness and therefore we cannot exclude that this might have biased the cohort towards more severely disabled ALS patients.
Thereby, future studies are needed to possibly integrate additional views to the framework because of the expected gain in combining multiple markers within the same diagnostic framework.
In conclusion, we propose that combining QSM with additional qMRI measures like CBF and GM volumetry makes it possible to significantly discriminate between ALS and PLS phenotypes in the motor cortex and between ALS and HCs in both cortical and subcortical structures. Moreover, we confirmed that the QSM increases in the motor cortex in ALS compared to HCs can be significantly associated with either ALS disease severity or duration probably depending on the level of cortical atrophy. Future studies, with more advanced machine learning tools, on larger samples of ALS/PLS phenotypes (as previously discussed in the study of Grollemund et al. 201974) eventually integrating myelination‐related regional markers from more regions of interest, are needed to increase the regional diagnostic power of a qMRI protocol for clinical 3 Tesla MRI studies of patients with probable ALS/PLS and to help the classification of patients, even at single subject level.
Conflict of Interest
The authors have no conflict of interest to declare.
Table 2.
Confidence range and statistical p‐value of the ROC curve analyses considering the volumes, CBF and QSM values as input data
| ROI | ALS versus HC | ALS versus PLS | PLS versus HC |
|---|---|---|---|
| Left‐thalamus |
[0.42 0.71] >0.05 |
[0.40 0.75] >0.05 |
[0.50 0.84] >0.05 |
| Right‐thalamus |
[0.39 0.68] >0.05 |
[0.41 0.77] >0.05 |
[0.41 0.79] >0.05 |
| Left‐caudate |
[0.54 0.81] <0.05 |
[0.50 0.86] >0.05 |
[0.43 0.81] >0.05 |
| Right‐caudate |
[0.50 0.78] <0.05 |
[0.37 0.78] >0.05 |
[0.40 0.82] >0.05 |
| Left‐putamen |
[0.45 0.75] >0.05 |
[0.36 0.78] >0.05 |
[0.43 0.80] >0.05 |
| Right‐putamen |
[0.51 0.79] <0.05 |
[0.39 0.79] >0.05 |
[0.46 0.84] >0.05 |
| Left‐pallidum |
[0.41 0.71] >0.05 |
[0.47 0.83] >0.05 |
[0.39 0.79] >0.05 |
| Right‐pallidum |
[0.52 0.80] <0.05 |
[0.34 0.76] >0.05 |
[0.43 0.85] >0.05 |
| Left‐hippocampus |
[0.39 0.69] >0.05 |
[0.35 0.72] >0.05 |
[0.35 0.80] >0.05 |
| Right‐hippocampus |
[0.54 0.81] <0.05 |
[0.34 0.75] >0.05 |
[0.48 0.84] >0.05 |
| Left‐amygdala |
[0.46 0.75] >0.05 |
[0.49 0.85] >0.05 |
[0.49 0.88] >0.05 |
| Right‐amygdala |
[0.55 0.83] <0.05 |
[0.52 0.84] >0.05 |
[0.38 0.81] >0.05 |
| Left precentral |
[0.53 0.80] <0.05 |
[0.58 0.88] <0.05 |
[0.50 0.87] >0.05 |
| Right precentral |
[0.53 0.80] <0.05 |
[0.48 0.82] >0.05 |
[0.59 0.90] <0.05 |
Bold text formatting indicates statistical significant comparisons.
Supporting information
Table S1. Statistical p‐value of the ROC curve analyses considering the QSM as input data. Bold text formatting indicates statistical significant comparisons.
Table S2. Statistical p‐value of the ROC curve analyses considering the CBF as input data. Bold text formatting indicates statistical significant comparisons.
Table S3. Statistical p‐value of the ROC curve analyses considering the volume as input data. Bold text formatting indicates statistical significant comparisons.
Funding Information
No funding information provided.
References
- 1.Hardiman O, Al‐Chalabi A, Chio A, et al. Amyotrophic lateral sclerosis. Nat Rev Dis Primer 2017;3:17071. [DOI] [PubMed] [Google Scholar]
- 2.Swinnen B, Robberecht W. The phenotypic variability of amyotrophic lateral sclerosis. Nat Rev Neurol 2014;10:661–670. [DOI] [PubMed] [Google Scholar]
- 3.Chiò A, Calvo A, Moglia C, et al. Phenotypic heterogeneity of amyotrophic lateral sclerosis: a population based study. J Neurol Neurosurg Psychiatry 2011;82:740–746. [DOI] [PubMed] [Google Scholar]
- 4.Benatar M, Turner MR, Wuu J. Defining pre‐symptomatic amyotrophic lateral sclerosis. Amyotroph Lateral Scler Front Degener 2019;20:303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finegan E, Chipika RH, Shing SLH, et al. Primary lateral sclerosis: a distinct entity or part of the ALS spectrum? Amyotroph Lateral Scler Front Degener 2019;20:133–145. [DOI] [PubMed] [Google Scholar]
- 6.Fabes J, Matthews L, Filippini N, et al. Quantitative FLAIR MRI in amyotrophic lateral sclerosis. Acad Radiol 2017;24:1187–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singer MA, Statland JM, Wolfe GI, Barohn RJ. Primary lateral sclerosis. Muscle Nerve 2007;35:291–302. [DOI] [PubMed] [Google Scholar]
- 8.Acosta‐Cabronero J, Machts J, Schreiber S, et al. Quantitative susceptibility MRI to detect brain iron in amyotrophic lateral sclerosis. Radiology 2018;289:195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhattarai A, Chen Z, Ward PGD, et al. Serial assessment of iron in the motor cortex in limb‐onset amyotrophic lateral sclerosis using quantitative susceptibility mapping. Quant Imaging Med Surg 2020;10:1465–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhattarai A, Egan GF, Talman P, et al. Magnetic resonance iron imaging in amyotrophic lateral sclerosis. J Magn Reson 2021. [Online ahead of print]. [DOI] [PubMed] [Google Scholar]
- 11.Conte G, Contarino VE, Casale S, et al. Amyotrophic lateral sclerosis phenotypes significantly differ in terms of magnetic susceptibility properties of the precentral cortex. Eur Radiol 2021;31:5272–5280. [DOI] [PubMed] [Google Scholar]
- 12.Costagli M, Donatelli G, Biagi L, et al. Magnetic susceptibility in the deep layers of the primary motor cortex in amyotrophic lateral sclerosis. NeuroImage Clin 2016;12:965–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean KE, Shen B, Askin G, et al. A specific biomarker for amyotrophic lateral sclerosis: quantitative susceptibility mapping. Clin Imaging 2021;75:125–130. [DOI] [PubMed] [Google Scholar]
- 14.Donatelli G, Caldarazzo Ienco E, Costagli M, et al. MRI cortical feature of bulbar impairment in patients with amyotrophic lateral sclerosis. NeuroImage Clin 2019;24:101934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JY, Lee Y‐J, Park DW, et al. Quantitative susceptibility mapping of the motor cortex: a comparison of susceptibility among patients with amyotrophic lateral sclerosis, cerebrovascular disease, and healthy controls. Neuroradiology 2017;59:1213–1222. [DOI] [PubMed] [Google Scholar]
- 16.Schweitzer AD, Liu T, Gupta A, et al. Quantitative susceptibility mapping of the motor cortex in amyotrophic lateral sclerosis and primary lateral sclerosis. Am J Roentgenol 2015;204:1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheelakumari R, Madhusoodanan M, Radhakrishnan A, et al. A potential biomarker in amyotrophic lateral sclerosis: can assessment of brain iron deposition with SWI and corticospinal tract degeneration with DTI Help? Am J Neuroradiol 2016;37:252–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weidman EK, Schweitzer AD, Niogi SN, et al. Diffusion tensor imaging and quantitative susceptibility mapping as diagnostic tools for motor neuron disorders. Clin Imaging 2019;53:6–11. [DOI] [PubMed] [Google Scholar]
- 19.Rule RR, Schuff N, Miller RG, Weiner MW. Gray matter perfusion correlates with disease severity in ALS. Neurology 2010;74:821–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen D, Hou BO, Xu Y, et al. Brain structural and perfusion signature of amyotrophic lateral sclerosis with varying levels of cognitive deficit. Front Neurol 2018;9:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welton T, Maller JJ, Lebel RM, et al. Diffusion kurtosis and quantitative susceptibility mapping MRI are sensitive to structural abnormalities in amyotrophic lateral sclerosis. NeuroImage Clin 2019;24:101953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ignjatović A, Stević Z, Lavrnić S, et al. Brain iron MRI: a biomarker for amyotrophic lateral sclerosis. J Magn Reson Imaging 2013;38:1472–1479. [DOI] [PubMed] [Google Scholar]
- 23.Kwan JY, Jeong SY, Van Gelderen P, et al. Iron accumulation in deep cortical layers accounts for MRI signal abnormalities in ALS: correlating 7 tesla MRI and pathology. PLoS One 2012;7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersen HH, Johnsen KB, Moos T. Iron deposits in the chronically inflamed central nervous system and contributes to neurodegeneration. Cell Mol Life Sci 2014;71:1607–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Contarino VE, Conte G, Morelli C, et al. Toward a marker of upper motor neuron impairment in amyotrophic lateral sclerosis: a fully automatic investigation of the magnetic susceptibility in the precentral cortex. Eur J Radiol 2020;124:108815. [DOI] [PubMed] [Google Scholar]
- 26.Finegan E, Hi Shing L, Chipika RH, et al. Widespread subcortical grey matter degeneration in primary lateral sclerosis: a multimodal imaging study with genetic profiling. NeuroImage Clin 2019;24:102089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brooks BR, Miller RG, Swash M, et al. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Mot Neuron Disord 2000;1:293–299. [DOI] [PubMed] [Google Scholar]
- 28.Turner MR, Barohn RJ, Corcia P, et al. Primary lateral sclerosis: consensus diagnostic criteria. J Neurol Neurosurg Psychiatry 2020;91:373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cedarbaum JM, Stambler N, Malta E, et al. The ALSFRS‐R: a revised ALS functional rating scale that incorporates assessments of respiratory function. J Neurol Sci 1999;169:13–21. [DOI] [PubMed] [Google Scholar]
- 30.Turner MR, Cagnin A, Turkheimer FE, et al. Evidence of widespread cerebral microglial activation in amyotrophic lateral sclerosis: an [11C](R)‐PK11195 positron emission tomography study. Neurobiol Dis 2004;15:601–609. [DOI] [PubMed] [Google Scholar]
- 31.Poletti B, Solca F, Carelli L, et al. The validation of the Italian Edinburgh Cognitive and Behavioural ALS Screen (ECAS). Amyotroph Lateral Scler Front Degener 2016;17:489–498. [DOI] [PubMed] [Google Scholar]
- 32.Siciliano M, Trojano L, Trojsi F, et al. Edinburgh Cognitive and Behavioural ALS Screen (ECAS)‐Italian version: regression based norms and equivalent scores. Neurol Sci 2017;38:1059–1068. [DOI] [PubMed] [Google Scholar]
- 33.Roche JC, Rojas‐Garcia R, Scott KM, et al. A proposed staging system for amyotrophic lateral sclerosis. Brain 2012;135:847–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dale AM, Fischl B, Sereno MI. Cortical surface‐based analysis: I. segmentation and surface reconstruction. NeuroImage 1999;9:179–194. [DOI] [PubMed] [Google Scholar]
- 35.Dale AM, Sereno MI. Improved localizadon of cortical activity by combining EEG and MEG with MRI cortical surface reconstruction: a linear approach. J Cogn Neurosci 1993;5:162–176. [DOI] [PubMed] [Google Scholar]
- 36.Fischl B, Sereno MI, Dale AM. Cortical surface‐based analysis: II: inflation, flattening, and a surface‐based coordinate system. NeuroImage 1999;9:195–207. [DOI] [PubMed] [Google Scholar]
- 37.Fischl B, Liu A, Dale AM. Automated manifold surgery: constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE Trans Med Imaging 2001;20:70–80. [DOI] [PubMed] [Google Scholar]
- 38.Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci USA 2000;97:11050–11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salat DH, Buckner RL, Snyder AZ, et al. Thinning of the cerebral cortex in aging. Cereb Cortex 2004;14:721–730. [DOI] [PubMed] [Google Scholar]
- 40.Ségonne F, Dale AM, Busa E, et al. A hybrid approach to the skull stripping problem in MRI. NeuroImage 2004;22:1060–1075. [DOI] [PubMed] [Google Scholar]
- 41.Alsop DC, Detre JA, Golay X, et al. Recommended implementation of arterial spin‐labeled perfusion MRI for clinical applications: a consensus of the ISMRM perfusion study group and the European consortium for ASL in dementia. Magn Reson Med 2015;73:102–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li W, Avram AV, Wu B, et al. Integrated Laplacian‐based phase unwrapping and background phase removal for quantitative susceptibility mapping. NMR Biomed 2014;27:219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li W, Wu B, Liu C. Quantitative susceptibility mapping of human brain reflects spatial variation in tissue composition. NeuroImage 2011;55:1645–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu B, Li W, Guidon A, Liu C. Whole brain susceptibility mapping using compressed sensing. Magn Reson Med 2012;67:137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu C, Wei H, Gong N‐J, et al. Quantitative susceptibility mapping: contrast mechanisms and clinical applications. Tomography 2015;1:3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wahlund LO, Barkhof F, Fazekas F, et al. A new rating scale for age‐related white matter changes applicable to MRI and CT. Stroke 2001;32:1318–1322. [DOI] [PubMed] [Google Scholar]
- 47.Fazekas F, Chawluk JB, Alavi A, et al. MR signal abnormalities at 1.5 T in Alzheimer’s dementia and normal aging. Am J Roentgenol 1987;149:351–356. [DOI] [PubMed] [Google Scholar]
- 48.Strong MJ, Abrahams S, Goldstein LH, et al. Amyotrophic lateral sclerosis ‐ frontotemporal spectrum disorder (ALS‐FTSD): revised diagnostic criteria. Amyotroph Lateral Scler Front Degener 2017;18:153–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bede P, Elamin M, Byrne S, et al. Basal ganglia involvement in amyotrophic lateral sclerosis. Neurology 2013;81:2107–2115. [DOI] [PubMed] [Google Scholar]
- 50.Machts J, Loewe K, Kaufmann J, et al. Basal ganglia pathology in ALS is associated with neuropsychological deficits. Neurology 2015;85:1301–1309. [DOI] [PubMed] [Google Scholar]
- 51.Tremblay L, Worbe Y, Thobois S, et al. Selective dysfunction of basal ganglia subterritories: from movement to behavioral disorders. Mov Disord 2015;30:1155–1170. [DOI] [PubMed] [Google Scholar]
- 52.Christidi F, Karavasilis E, Rentzos M, et al. Hippocampal pathology in amyotrophic lateral sclerosis: selective vulnerability of subfields and their associated projections. Neurobiol Aging 2019;84:178–188. [DOI] [PubMed] [Google Scholar]
- 53.Trojsi F, Di Nardo F, Caiazzo G, et al. Hippocampal connectivity in amyotrophic lateral sclerosis (ALS): more than Papez circuit impairment. Brain Imaging Behav 2020. [Online ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Westeneng H‐J, Verstraete E, Walhout R, et al. Subcortical structures in amyotrophic lateral sclerosis. Neurobiol Aging 2015;36:1075–1082. [DOI] [PubMed] [Google Scholar]
- 55.Brettschneider J, Tredici KD, Toledo JB, et al. Stages of pTDP‐43 pathology in amyotrophic lateral sclerosis. Ann Neurol 2013;74:20–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chipika RH, Christidia F, Finegana E, et al. Amygdala pathology in amyotrophic lateral sclerosis and primary lateral sclerosis. J Neurol Sci 2020;417:117039. [DOI] [PubMed] [Google Scholar]
- 57.Ferraro PM, Jester C, Olm CA, et al. Perfusion alterations converge with patterns of pathological spread in transactive response DNA‐binding protein 43 proteinopathies. Neurobiol Aging 2018;68:85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Machts J, Keute M, Kaufmann J, et al. Longitudinal clinical and neuroanatomical correlates of memory impairment in motor neuron disease. NeuroImage Clin 2021;29:102545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Agosta F, Galantucci S, Riva N, et al. Intrahemispheric and interhemispheric structural network abnormalities in PLS and ALS. Hum Brain Mapp 2014;35:1710–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Canu E, Agosta F, Galantucci S, et al. Extramotor damage is associated with cognition in primary lateral sclerosis patients. PLoS One 2013;8:e82017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Finegan E, Li Hi Shing S, Siah WF, et al. Evolving diagnostic criteria in primary lateral sclerosis: the clinical and radiological basis of “probable PLS”. J Neurol Sci 2020;417:117052. [DOI] [PubMed] [Google Scholar]
- 62.Turner MR, Wicks P, Brownstein CA, et al. Concordance between site of onset and limb dominance in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry 2011;82:853–854. [DOI] [PubMed] [Google Scholar]
- 63.Devine MS, Pannek K, Coulthard A, et al. Exposing asymmetric gray matter vulnerability in amyotrophic lateral sclerosis. NeuroImage Clin 2015;7:782–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mezzapesa DM, Ceccarelli A, Dicuonzo F, et al. Whole‐brain and regional brain atrophy in amyotrophic lateral sclerosis. Am J Neuroradiol 2007;28:255–259. [PMC free article] [PubMed] [Google Scholar]
- 65.Crespi C, Cerami C, Dodich A, et al. Microstructural white matter correlates of emotion recognition impairment in amyotrophic lateral sclerosis. Cortex 2014;53:1–8. [DOI] [PubMed] [Google Scholar]
- 66.Palmieri A, Naccarato M, Abrahams S, et al. Right hemisphere dysfunction and emotional processing in ALS: an fMRI study. J Neurol 2010;257:1970–1978. [DOI] [PubMed] [Google Scholar]
- 67.Sedda A. Disorders of emotional processing in amyotrophic lateral sclerosis. Curr Opin Neurol 2014;27:659–665. [DOI] [PubMed] [Google Scholar]
- 68.Pringle CE, Hudson AJ, Munoz DG, et al. Primary lateral sclerosis: clinical features. Neurophatology and diagnostic criteria. Brain 1992;115:495–520. [DOI] [PubMed] [Google Scholar]
- 69.Betts MJ, Acosta‐Cabronero J, Cardenas‐Blanco A, et al. High‐resolution characterisation of the aging brain using simultaneous quantitative susceptibility mapping (QSM) and R2* measurements at 7T. NeuroImage 2016;138:43–63. [DOI] [PubMed] [Google Scholar]
- 70.Asllani I, Habeck C, Borogovac A, et al. Separating function from structure in perfusion imaging of the aging brain. Hum Brain Mapp 2009;30:2927–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Canosa A, Moglia C, Manera U, et al. Metabolic brain changes across different levels of cognitive impairment in ALS: a 18F‐FDG‐PET study. J Neurol Neurosurg Psychiatry 2021;92:357–363. [DOI] [PubMed] [Google Scholar]
- 72.Qin Q, Huang AJ, Hua J, et al. Three‐dimensional whole‐brain perfusion quantification using pseudo‐continuous arterial spin labeling MRI at multiple post‐labeling delays: accounting for both arterial transit time and impulse response function. NMR Biomed 2014;27:116–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee J, Hyun J‐W, Lee J, et al. So you want to image myelin using MRI: an overview and practical guide for myelin water imaging. J Magn Reson Imaging 2021;53:360–373. [DOI] [PubMed] [Google Scholar]
- 74.Grollemund V, Pradat P‐F, Querin G, et al. Machine learning in amyotrophic lateral sclerosis: achievements, pitfalls, and future directions. Front Neurosci 2019;13:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Statistical p‐value of the ROC curve analyses considering the QSM as input data. Bold text formatting indicates statistical significant comparisons.
Table S2. Statistical p‐value of the ROC curve analyses considering the CBF as input data. Bold text formatting indicates statistical significant comparisons.
Table S3. Statistical p‐value of the ROC curve analyses considering the volume as input data. Bold text formatting indicates statistical significant comparisons.


