Abstract
Somatostatin receptor type II expression in the mammalian brain displays a spatially and temporally very restricted pattern. In an investigation of the molecular mechanisms controlling these patterns, we have recently shown that binding of the transcription factor SEF-2 to a novel initiator element in the SSTR-2 promoter is essential for SSTR-2 gene expression. Further characterization of the promoter identified a species-conserved TC-rich enhancer element. By screening a mouse brain cDNA expression library, we cloned a cDNA encoding the transcription factor MIBP1. MIBP1 interacts specifically with both the TC box in the SSTR-2 promoter and with the SEF-2 initiator-binding protein to enhance transcription from the basal SSTR-2 promoter. We then investigated SSTR-2, SEF-2, and MIBP1 mRNA expression patterns in the developing and adult murine brain by Northern blotting and in situ hybridization. While SEF-2 is widely expressed in many neuronal and nonneuronal tissues, MIBP1 expression overlapped precisely with expression of SSTR-2 in the frontal cortex and hippocampus. In summary, our data for the first time define a regulatory role for the transcription factor MIBP1 in mediating spatially and temporally regulated SSTR-2 expression in the brain.
The effects of somatostatin (SST) hormones are mediated by a family of five different, highly conserved receptors, SSTR-1 to SSTR-5 (2, 13, 18, 21, 32, 33), which belong to the class of seven helix transmembrane-spanning receptors. A series of detailed studies, employing Northern blotting, reverse transcriptase PCR and in situ hybridization, revealed complex gene type-specific expression patterns in many regions of the central nervous system. In particular, regions of intense SSTR-2 expression in the brain comprise the hypothalamic-hypophyseal system, which regulates the adenohypophyseal release of growth hormone, thyroid-stimulating hormone, and prolactin and the widespread somatostatinergic system in the frontal cortex and the hippocampus, modulating many cognitive and vegetative functions (5, 10). Furthermore, spatially and temporally regulated SSTR-2 expression patterns have been observed during embryonic brain and peripheral nerve development, suggesting that somatostatin signalling exerts functions during neurogenesis (9, 12).
In an investigation of molecular mechanisms controlling the expression pattern of SSTR-2, we recently provided an initial study of the human SSTR-2 promoter (20). We demonstrated that initiation of mRNA transcription is dependent on the presence of an E-box-binding site for the basic helix-loop-helix (bHLH) transcription factor SEF-2. SEF-2 function involves tethering of transcription factor IIB (TFIIB), a component of the basal transcription machinery, to the SSTR-2 initiator. Despite being an essential factor for basal promoter activity, SEF-2 alone is insufficient to mediate enhanced transcriptional activity of the SSTR-2 gene.
Therefore, we extended our SSTR-2 promoter analysis. In the present study, we demonstrate that a TC-rich species-conserved binding site, which is located 5′ adjacent to the E box, is required for enhanced promoter activity. Expression cloning revealed that a large zinc finger nuclear protein of previously unknown function, MIBP1, binds specifically to the TC box and activates transcription from the SSTR-2 promoter.
MATERIALS AND METHODS
Tissue culture and transient transfections.
N2A, NGP, Lan-1, GHFT-1, HeLa, and COS-1 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. Transient-transfection assays were performed by a standard calcium phosphate coprecipitation method (8). Luciferase activity was quantified as recommended by the manufacturer (Promega) in a luminometer (ML 3000; Dynatech). Relative light units were normalized to the protein concentration determined by the Bradford dye assay (Bio-Rad, Richmond, Calif.). All experiments were repeated at least five times, and standard deviations were <10%.
Reporter and expression plasmids.
The SSTR-2 promoter-LUC reporter series was generated by cloning single copies of the listed oligonucleotides into the multiple-cloning sites of plasmids pGL2LUC or TK-LUC (Promega, Madison, Wis.). Identical oligonucleotides were used for generation of the reporter plasmids and as probes for gel shifts. The reporter plasmids are as follows (the SSTR-2 promoter E box is highlighted in boldface, and point mutations are underlined): Inr, AATCTTCCTCTTTTCCTTCCAGATGTCACACTGGATCC; M1, AATCTTCCTCTTTTCCTTC; M2, CAGATGTCACACTGGATCC; M6, AATCTTCCTCTTTTCCTTCCAAATGTCACACTGGATCC; M13, TTTTCCTTCCAGATGTCACACTGGATCC; M14, CCAGATGTCACACTGGATCC; M15, AATCTTCCTCTTTTCCTTCCAG; M16, AATCTTGGTCTTTTGGTTCCAGATGTCACACTGGATCC; M17, AATCTTCCTCTTTTGGTTCCAGATGTCACACTGGATCC; M18, AATCTTGGTCTTTTCCTTCCAGATGTCACACTGGATCC; M19, TCTTCCTCTTTTCCTTCCAG; M20, TCTTCCTCTTT; M21, TTTTCCTTCCAGATG; and M50, CATATCTAGGTCATGACCTAGATATGAGCT. The oligomer M50, which was used as a nonspecific competitor, contains an unrelated RZRβ-binding site.
Cytomegalovirus promoter-based expression plasmids pCMX.PL1, pCMX.PL2 (29), pCMX-SEF-2, GST–SEF-2, and GST–bHLH–SEF-2 were described previously (20). To construct GST–Δ-bHLH–SEF-2, the entire SEF-2 cDNA deleted by the C-terminal bHLH domain was PCR amplified with a T7 primer and a specific EcoRI-modified SEF-2 primer (5′-TTGGAATTCATGCATCACC) and ligated into the EcoRI-SmaI sites of pGEX 4T1 (Pharmacia, Freiburg, Germany). To construct pCMX-MIBP1, a 7.5-kb DraI-NsiI-digested rat cDNA fragment (a generous gift from Kenshi Hayashi, Fukuoka, Japan) encoding the full-length MIBP1 protein was inserted at the EcoRV-PstI site of pCMX.PL2. To construct the GST-MZF expression plasmid, a cDNA fragment of rat MIBP1 coding for the C-terminal zinc finger (amino acids 1740 to 2110) was PCR amplified and inserted at the EcoRI-SmaI site of pGEX4T1 (Pharmacia).
DNA-binding studies.
Preparation of whole-cell extract and purification of glutathione S-transferase (GST) fusion proteins was described by Buettner et al. (3). In vitro translation reactions were performed with reticulocyte lysate as specified by the manufacturer (TNT; Promega). Purified GST fusion protein (0.5 μg), 4 μl of whole-cell extract, or 2 μl of in vitro translation product was mixed with 10 mM HEPES (pH 7.8)–80 mM KCl–10% glycerol–5 mM MgCl2–1 mM dithiothreitol–0.5 μg of poly(dG-dC) and incubated for 15 min on ice. Subsequently 1 ng of 32P-labelled probe was added. For supershift assays, the following antibodies were added to the reaction mix: anti-GST mouse monoclonal immunoglobulin G1 (Santa Cruz Biotechnology, Santa Cruz, Calif.) or anti-MIBP1 (α-P-M) rabbit polyclonal serum (26). After incubation for 30 min on ice, the samples were separated on a 4% polyacrylamide gel in 0.5× Tris-borate-EDTA (TBE) at 4°C. After electrophoresis, the gels were dried and subjected to autoradiography.
In vitro protein-binding assay.
Purified recombinant GST, GST–full-length SEF-2, GST–bHLH–SEF-2, or GST–Δ-bHLH–SEF-2 (2 μg) was coupled for 1 h at 4°C to 50 μl of glutathione-Sepharose 4B beads (Pharmacia) and then washed three times with 1 ml of GST binding buffer (20 mM HEPES [pH 7.5], 150 mM KCl, 25 mM MgCl2, 10 mM dithiothreitol, 0.1 mM EDTA, 0.15% Nonidet P-40). The matrix was resuspended in 500 μl of GST binding buffer, incubated for 1 h at 4°C with 5 μl of [35S]methionine-labelled MIBP1 protein, and again washed three times. Finally, the proteins were eluted, and denatured in Laemmli’s buffer at 95°C for 10 min, and separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (6% polyacrylamide).
Screening of genomic and λgt11 cDNA expression libraries.
The murine SSTR-2 promoter was isolated from a murine SVJ129 genomic library (Stratagene, La Jolla, Calif.) with the human SSTR-2 cDNA as a probe by using standard methods (23). A total of 1.8 × 105 plaques of a commercially available murine brain cDNA library in the expression vector λgt11 were screened by using a tetrameric SSTR-2 Inr-binding site as a probe. A detailed protocol for expression screening has been published recently (20). The C-terminal clone of MIBP1, isolated by expression screening, was used to rescreen and isolate the murine full-length cDNA.
Northern blots and in situ hybridizations.
Northern blots (Clontech) were hybridized in 50% formamide–5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–1% SDS–150 μg of tRNA per ml–250 μg of single-stranded DNA per ml at 42°C for 16 h. Final washes were done in 0.1× SSC–0.01% SDS at 65°C for 45 min. As probes, cDNA fragments of SEF-2 (nucleotides 1 to 1795) and MIBP1 (4469 to 5388) were used. A detailed protocol for in situ hybridization of paraffin-embedded mouse embryo slides has been published previously (15). As probes, the same cDNA fragments used for Northern blots were transcribed as sense and antisense 35S-UTP-labelled cRNAs.
Reverse transcriptase PCR.
Reverse transcription was performed with 3 μg of total cellular RNA as described previously (3), except that pd(N)6-primers (2 μg; Pharmacia) were used instead of sequence-specific primers. PCR amplification, as described previously (16), was performed with the following profile: SSTR-2 (35 cycles of 1 min at 94°C, 1 min at 56°C, and 1 min at 72°C), SEF-2 (30 cycles of 1 min at 94°C, 1 min at 63°C, and 1 min at 72°C), MIBP1 (30 cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C), and β-actin (30 cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C). The following specific PCR primers were used: human SSTR-2 sense, GGC TCC TCT AAG AGG AAG AAG; rodent SSTR-2 sense, GGG TCG TCC AAG AGG AAA AAG; SSTR-2 reverse, TCT CCA TTG AGG AGG GTC CTC; SEF-2 sense, CGC CAG GCT ATC CTT CCT CCA; SEF-2 reverse, CCT GTC CTC CAT TTC TAG ACC; MIBP1 sense, GCT TCA TGG TGC ATT AGT TCC; MIBP1 reverse, GGC TCG GTT TGT TTG GAT CCA; β-actin sense, TGA CGG GGT CAC CCA CAC TGT G; β-actin reverse, CTA GAA GCA TTT GCG GTG GAC. Then 10% of the PCR products were fractionated on 2% agarose gels and subjected to Southern blot analysis.
RESULTS
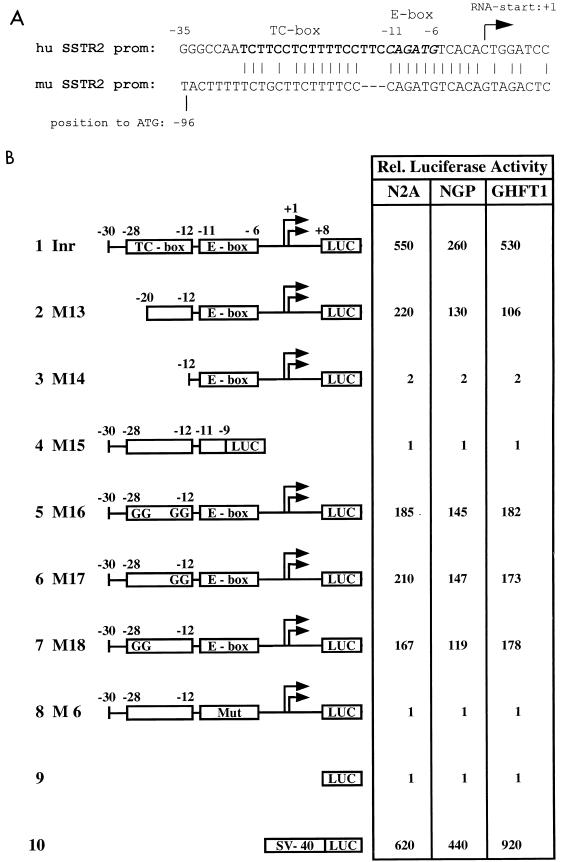
To identify species-conserved motifs in the SSTR-2 promoter, we aimed to determine the murine SSTR-2 promoter sequence for comparison with the previously characterized human promoter (20). Therefore, the murine SSTR-2 gene was isolated and sequenced in the 5′-flanking region. As displayed in Fig. 1A, two sequence motifs highly conserved between the human and murine promoters were detected: (i) an E box spanning residues −11 to −6, which was previously identified as the core initiator element of the human promoter indispensible for basal SSTR-2 promoter function (20); and (ii) a TC box 5′ adjacent to the E box, with the core sequence TCTTTTCC and a further TCT(T/G)C(C/T) 5′ extension. Further upstream, no other significant homology between the human and murine 5′-flanking regions was detected.
FIG. 1.
(A) Nucleotide sequence of the human (hu) SSTR-2 promoter (top) from positions +8 to −35 with respect to the mRNA initiation site and comparison to the murine (mu) SSTR-2 promoter (bottom). A conserved TC-rich sequence (TC box) is printed in boldface, and the E box from residues −11 to −6 is shown in italics. (B) Identification of cis-regulatory elements in the human SSTR-2 promoter. The indicated reporter plasmids (500 ng) were transfected into N2A, NGP, and GHFT-1 cells (rows 1 to 10). The mRNA initiation sites are indicated by two arrows. The E box and the TC box are shown as rectangles. The numbers indicate positions of the respective nucleotides within the human SSTR-2 promoter. The reporter M6-LUC, which contains a single point mutation in the E box (MUT), is shown in row 8, and reporters which contain the double point mutations in the TC box (CC to GG) are shown in rows 5 to 7.
To investigate which of these sequence motifs harbor critical cis-regulatory elements, we cloned a series of human SSTR-2 promoter deletion mutants into the promoterless luciferase reporter plasmid pGL2basic and tested these reporters in transient-transfection assays (Fig. 1B). Consistent with previous results, a luciferase reporter driven by the SSTR-2 promoter fragment spanning residues −30 to +8 revealed high levels of promoter activity in murine and human neuroblastoma cells (N2A and NGP) and also in rat pituitary cells (GHFT-1). This SSTR-2 promoter fragment (designated Inr in Fig. 1B) has been referred to as the SSTR-2 initiator (20). Deletion of the first half of the TC box (mutant M13) reduced SSTR-2 promoter activity significantly, whereas further deletion of the entire TC box (M14) caused reduction of activity to background levels. Introduction of CC-to-GG point mutations into the TC elements (M16 to M18) significantly impaired promoter function, indicating that an intact TC box is required for enhanced activity. In addition to the TC elements, SSTR-2 promoter function was critically dependent on the E-box core element. An intact TC box in the context of a partially deleted E box (M15) or an E box harboring a G-to-A-point mutation (M6) entirely failed to activate the expression of the luciferase reporters. In summary, we concluded from these results that the TC box harbors an important, species-conserved cis-regulatory element, which confers enhanced activity to the basal SSTR-2 promoter.
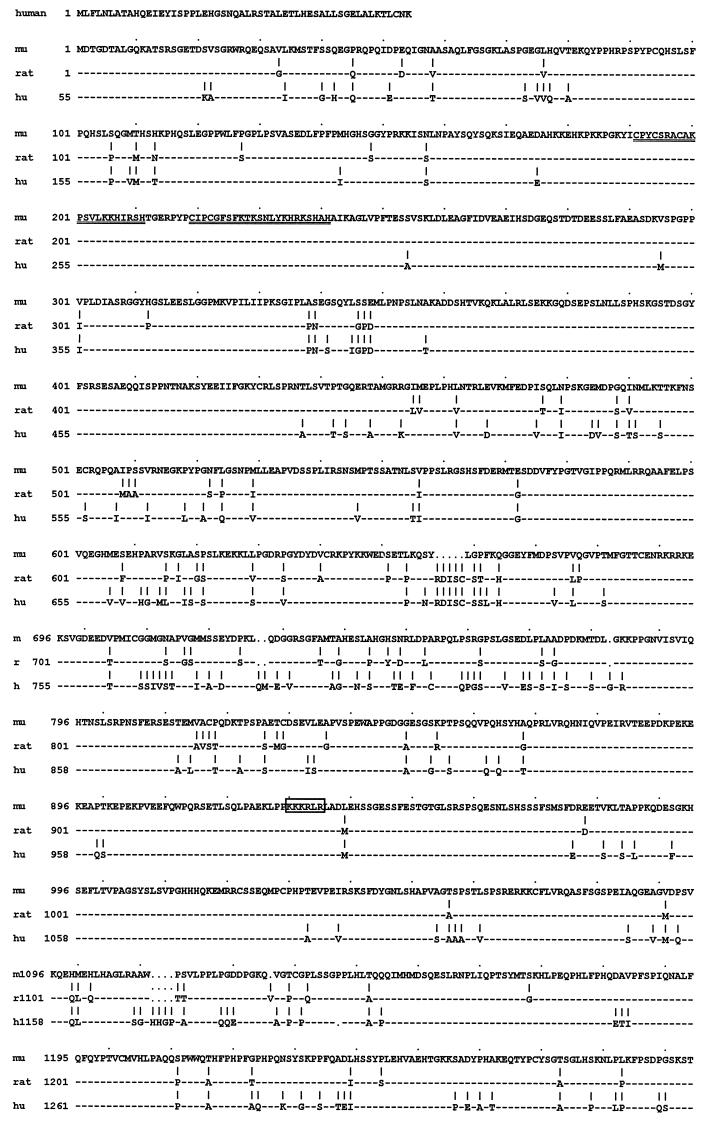
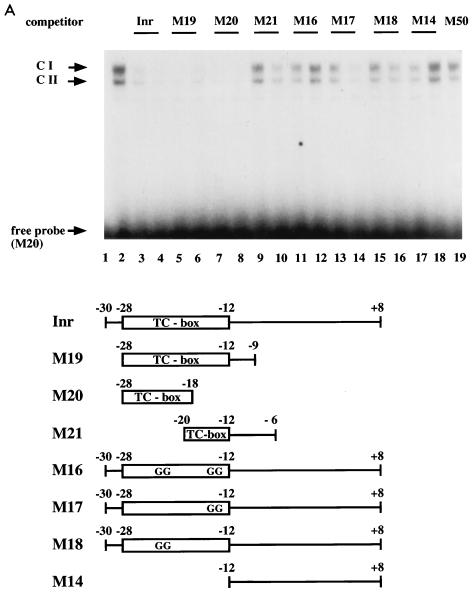
We therefore aimed to clone proteins interacting specifically with the TC-box element in the SSTR-2 promoter. A murine brain λgt11 expression library was screened by using the SSTR-2 promoter fragment shown in Fig. 1A. By screening 2 × 105 phage plaques, a cDNA clone was isolated, which encoded the C-terminal zinc finger domain of the murine MIBP1 homolog. MIBP1 codes for a large sequence-specific DNA-binding zinc finger protein of unknown physiological function. MIBP1 was previously isolated from rat (11) and human cDNA libraries and was also referred to as MBP-2/HIV-EP2 (17, 31). Furthermore, two partial MIBP1 cDNA fragments were isolated previously and designated AGIE-BP1 and AT-BP1 (14, 22). The cDNA insert from our expression screen was then used to rescreen a brain cDNA library. A total of 16 independent phage inserts, spanning the entire MIBP1 open reading frame, were isolated. The open reading frame codes for a protein of 2,431 amino acids with an expected molecular mass of approximately 270 kDa. We determined a very high degree of homology, with only 140 amino acid exchanges between the mouse and rat sequences, 86 of which are conservative. The entire murine, rat, and human MIBP1 (MBP-2/HIV-EP2) peptide sequences are displayed in Fig. 2. As noted previously (11), MIBP1 harbors two clusters of C2H2 zinc fingers in the N terminus (amino acids 191 to 241) and in the C terminus (amino acids 1785 to 1835), respectively. A putative nuclear translocation signal is located between amino acids 930 and 935, and an acidic region is located between amino acids 1883 and 1910. To determine whether MIBP1 binds to the SSTR-2 promoter in a sequence-specific manner, we expressed either the full-length MIBP1 protein by in vitro translation or a C-terminal fragment spanning amino acids 1740 to 2110 including the C-terminal zinc finger cluster as a GST fusion protein. These proteins were then used in a series of gel shift experiments. Two specific retarded complexes resulted from coincubation of in vitro-translated MIBP1 with the SSTR-2 TC box (Fig. 3A, lane 2). Both complexes were specifically competed by a 50- or 100-fold molar excess of the unlabelled SSTR-2 wild-type promoter sequence (lanes 3 and 4, Inr). We then performed a series of competition experiments with various mutated binding sites. Mutants M19 and M20 but not a 3′ fragment harboring the E box (mutant M14) or a central fragment (mutant M21) were able to compete, indicating that the TC box is the specific target of MIBP1 binding (Fig. 3A). Furthermore, the same mutations that significantly reduced (M16 to M18) or completely abolished (M14) SSTR-2 Inr function were also drastically impaired in their ability to compete MIBP1 DNA binding.
FIG. 2.
Comparison of the murine (mu), rat, and human (hu) MIBP1 peptide sequences. The rat and human amino acid residues differing from the respective residues of the murine protein are indicated below the murine sequence. Especially well conserved motifs include the N- and C-terminal zinc fingers (doubly underlined), the putative nuclear localization signal (boxed), and the acidic region (underlined).
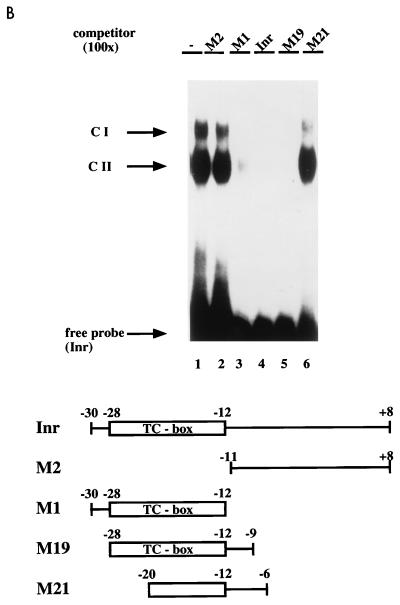
FIG. 3.
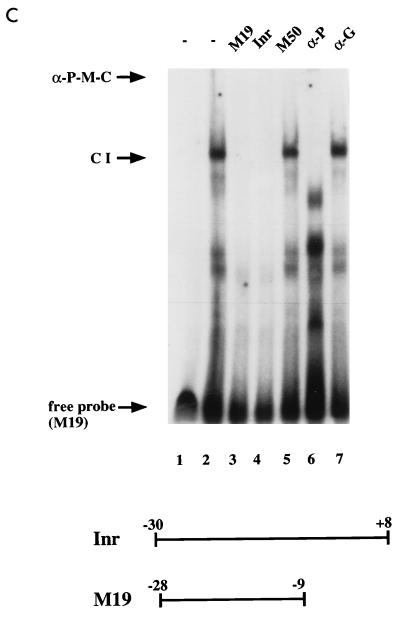
Sequence-specific binding of MIBP1 to the human SSTR-2 promoter. (A) Electrophoretic mobility shift assays were performed with a 32P-labelled oligonucleotide containing the promoter sequence from positions −28 to −18 (M20) and in vitro-translated MIBP1 protein (lane 2). Competition experiments were performed with 50- and 100-fold molar excesses of the indicated competitors. The MIBP1 TC-box complexes CI and CII are competed by the oligonucleotides Inr, M19, and M20. Oligonucleotides M21, containing a partially deleted TC box, and M14, representing the 3′ promoter region harboring the E box, fail to compete. Competition by oligonucleotides M16, M17, and M18, representing point-mutated TC boxes, is drastically impaired. As a control an unrelated competitor, M50, also failed to compete. (B) The C-terminal MIBP1 zinc finger binds to the SSTR-2 promoter. Electrophoretic mobility shift assays were performed with the Inr oligonucleotide from nucleotides −30 to +8 and purified recombinant GST fusion protein (lane 1). A 100-fold molar excess of unlabelled Inr oligonucleotide, oligonucleotide M1, or oligonucleotide M19 competed with formation of the MIBP1-Inr complexes CI and CII. The control oligonucleotides M2 and M21 failed to compete. (C) MIBP1 binds to the SSTR-2 promoter in vivo. Whole-cell extract prepared from GHFT-1 cells (4 μl) was incubated with 1 ng of 32P-labelled oligonucleotide M19 (lanes 2 to 7). Complex CI is specifically competed by a 100-fold molar excess of oligonucleotides M19 and Inr. In contrast, the unrelated oligonucleotide M50 does not compete. To establish the composition of complex CI, the electrophoretic mobility shift assay mixture was challenged with an antiserum directed against the C-terminal zinc finger domain of MIBP1 (lane 6) or an unrelated anti-GST control antibody (lane 7). The very faint supershifted MIBP1 complex is labelled α-P-M-C.
We further observed two specific DNA-protein complexes resulting from coincubation of the SSTR-2 Inr with the bacterially expressed C-terminal MIBP1-GST fusion protein (Fig. 3B). Consistently, 5′ SSTR-2 fragments spanning either the entire TC box (mutants M1 and M19) or the full Inr sequence competed for specific binding of the GST-MIBP1 protein. In contrast, both a 3′ fragment spanning the E box (mutant M2) and a central fragment (M21) failed to compete. Taken together, these data reveal that the TC box of the SSTR-2 promoter represents a sequence-specific binding site for MIBP1 and that the C-terminal MIBP1 zinc finger cluster is sufficient for binding to the TC box.
Next we addressed whether MIBP1 binding to the SSTR-2 promoter also occurs in vivo and performed gel mobility shift assays with protein extracts from cells that express SSTR-2 mRNA. We coincubated GHFT-1 extracts with the SSTR-2 promoter fragment M19 (harboring the intact TC box) and challenged the band shift with a polyclonal serum raised against the C-terminal zinc finger domain of MIBP1. The most prominent band shift complex, labelled CI in Fig. 3C, was specifically competed by a 100-fold molar excess of unlabelled oligonucleotides M19 and Inr, respectively (Fig. 3C, lanes 2 to 4). In contrast, an unrelated competitor (M50) failed to compete (lane 5). Addition of the polyclonal MIBP1 antiserum interfered with formation of the band shift complex CI. A small portion of complex CI supershifted as a very faint and large DNA-protein complex, which was difficult to visualize (labelled α-P-M-C in Fig. 3C, lane 6). Disruption of complex CI was a specific effect of the MIBP1 antiserum and was not observed by challenging the electrophoretic mobility shift assay mixture with an unrelated anti-GST control antiserum (lane 7). In summary, these data provide further evidence for sequence-specific binding of MIBP1 protein to the SSTR-2 promoter both in vitro and in vivo.
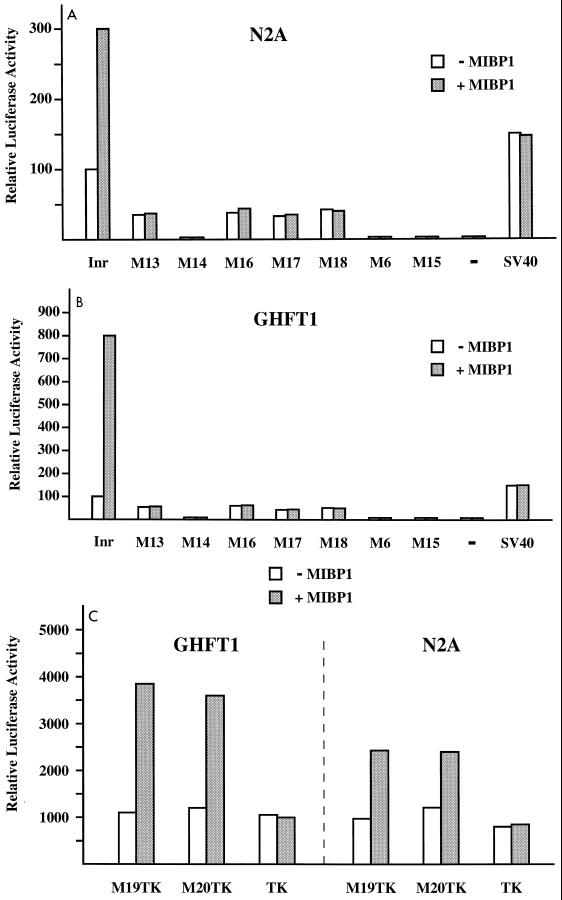
To investigate whether MIBP1 acts as a transcriptional activator of the SSTR-2 gene, we measured the effect of MIBP1 overexpression on the activity of the SSTR-2 promoter. The entire MIBP1 open reading frame was cloned into the expression plasmid pCMX.PL1 and transfected together with the SSTR-2 promoter luciferase reporter construct (nucleotides −30 to +8 as shown in Fig. 1A) into the neuroblastoma cell line N2A and the pituitary cell line GHFT-1. We chose these two cell lines because they represent tissues, i.e., neurons and pituitary gland cells, that physiologically express SSTR-2 mRNA (10, 24, 25, 28). Cotransfection of MIBP1 expression plasmid with wild-type SSTR-2 promoter reporter resulted in significant induction of luciferase expression in both N2A and GHFT-1 cells (Fig. 4A and B). In contrast, MIBP1 failed to activate luciferase expression from all SSTR-2 promoter constructs harboring either deletions (mutants M13 and M14) or point mutations (M16 to M18) in the MIBP1 TC-rich binding site. Consistent with previous results that mRNA initiation from the SSTR-2 promoter is critically dependent on the interaction of the bHLH protein SEF-2 with the E box, a single point mutation within the E-box sequence (mutant M6) or partial deletion of the 3′ half of the E box resulted in complete loss of promoter activity, irrespective of MIBP1 expression. Importantly, MIBP1 overexpression did not change luciferase expression from a simian virus 40 promoter control plasmid, indicating that the effect of MIBP1 as a transcriptional activator of the SSTR-2 Inr in N2A and GHFT-1 cells was promoter specific.
FIG. 4.
(A and B) MIBP1 mediates transcriptional activation of the human SSTR-2 promoter in N2A cells (A) and GHFT-1 cells (B). The luciferase activity of the reporter constructs Inr-LUC, the mutated Inr reporter constructs M13-LUC to M18-LUC, M6-LUC, and the promoterless control plasmids basic pGL (−) or SV40-LUC is displayed as open bars. Shaded bars represent promoter activity in cells cotransfected with 200 ng of pCMX-MIBP1 expression plasmid. (C) MIBP1 mediates transcriptional activation via the TC box of the human SSTR-2 promoter. Samples (500 ng) of the reporter construct M19-TK-LUC or M20-TK-LUC containing either the Inr sequence from nucleotides −28 to −9 (TC box [Fig. 1]) or −28 to 18 in front of the thymidine kinase (TK) promoter were cotransfected with 200 ng of pCMX-MIBP1 expression plasmid (shaded bars) or with empty pCMX plasmid (open bars). As a control, the pCMX-MIBP1 expression plasmid was also cotransfected with the promoterless TK-LUC reporter.
In addition, the SSTR-2 promoter TC box mediated transcriptional activation by MIBP1 in the context of an unrelated promoter. When we cloned single copies of the SSTR-2 promoter fragments M19 and M20, harboring a functional TC box, in front of a TK-LUC reporter, we observed three- to fourfold MIBP1-dependent reporter activation. Importantly, the activity of the parental TK-LUC reporter, lacking a TC box, was not changed by MIBP1 (Fig. 4C).
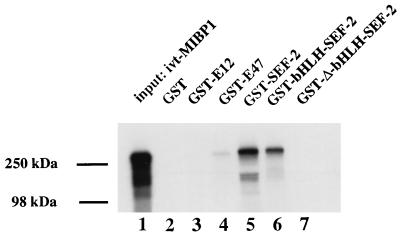
Since the MIBP1- and SEF-2-binding sites in the human and murine SSTR-2 promoter are located in very close proximity, we further examined a potential protein-protein interaction between MIBP1 and SEF-2. Therefore, GST fusion proteins containing full-length SEF-2 protein, the C-terminal bHLH domain of SEF-2, or SEF-2 with the bHLH domain deleted (Δ-bHLH–SEF-2) were immobilized to glutathione-coupled Sepharose and incubated with in vitro-translated [35S]methionine-labelled MIBP1 protein. As shown in Fig. 5, both the C-terminal bHLH domain and the full-length SEF-2 protein, but not Δ-bHLH–SEF-2, retained specifically in vitro-translated MIBP1 protein. The interaction was specific for the bHLH domain of SEF-2, since GST fusion proteins of E12 and E47 did not interact with MIBP1. These data strongly suggest that the evolutionarily conserved close proximity of the SSTR-2 TC-box and E-box elements in the human and murine promoters facilitate direct protein-protein interaction between MIBP1 and SEF-2.
FIG. 5.
MIBP1 and SEF-2 proteins interact in vitro. The GST pull-down experiment was performed with 5 μl of in vitro-translated [35S]methionine-labelled MIBP1 protein (input) and immobilized, recombinant GST–SEF-2 fusion protein, GST–bHLH–SEF-2, GST–Δ-bHLH–SEF-2, GST-E12, or GST-E47. The control protein GST failed to interact with ivt-MIBP1.
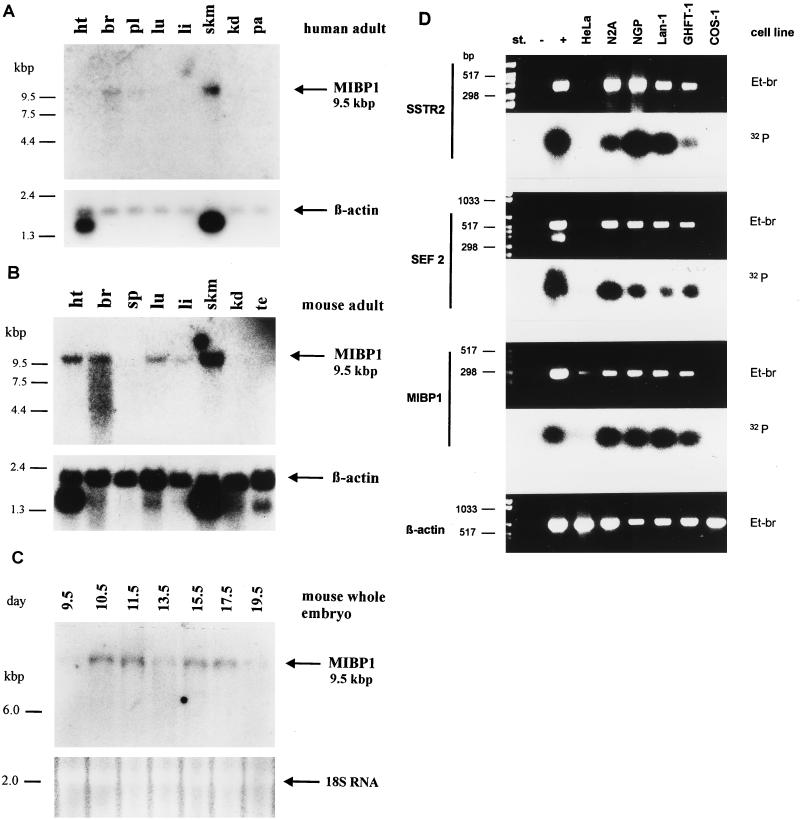
To extend our studies beyond the analysis of cell lines in vitro, we investigated the pattern of MIBP1 mRNA in comparison with expression of SEF-2 and SSTR-2 in vivo. Therefore, Northern blots of adult human and embryonic murine tissues were hybridized with a MIBP1 probe. A single MIBP1 mRNA approximately 9.5 kb long was detected in human brain and skeletal muscle and further in murine heart, brain, lung, and skeletal muscle and at very low levels in murine liver (Fig. 6A and B). Interestingly, mRNA expression of MIBP1 in brain, heart, skeletal muscle, and lung overlapped precisely with expression of SSTR-2 (21, 32) and SEF-2 (20) mRNA. During murine embryogenesis, MIBP1 expression started at stage E10.5 and persisted through all later stages (Fig. 6C). Finally, we examined by reverse transcriptase PCR analysis the expression patterns of SSTR-2, SEF-2, and MIBP1 mRNA in a panel of selected neural, pituitary gland, and epithelial cell lines (Fig. 6D). As expected, coexpression of SSTR-2, SEF-2, and MIBP1 was detected in neural cell lines (N2A, NGP, and Lan-1) and in pituitary gland cells (GHFT-1) but not in epithelial cell lines (HeLa and COS-1).
FIG. 6.
Multiple tissue Northern blots of adult human (A) and adult murine (B) tissues and developmental stages E9.5 to E19.5 of murine embryogenesis (C). A single MIBP1 transcript of approximately 9.5 kb is indicated by an arrow. Abbreviations: ht (heart), br (brain), pl (placenta), lu (lung), li (liver), skm (skeletal muscle), kd (kidney), pa (pancreas), sp (spleen), te (testis). β-Actin control hybridizations or ethidium bromide-stained 18S RNA are shown below the blots. (D) Reverse transcriptase PCR amplification of SSTR-2, SEF-2, and MIBP1 transcripts with 3 μg of total cellular RNA from the neuroblastoma cell lines (N2A, NGP, and Lan-1), pituitary gland cells (GHFT-1), and epithelial cells (HeLa and COS-1). Ethidium bromide (Et-br)-stained gels and Southern blots probed with the respective phospholabelled probes are shown in parallel.
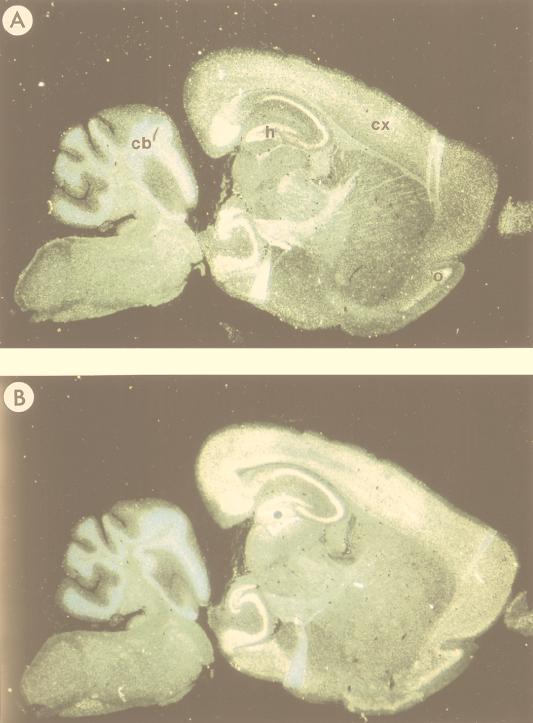
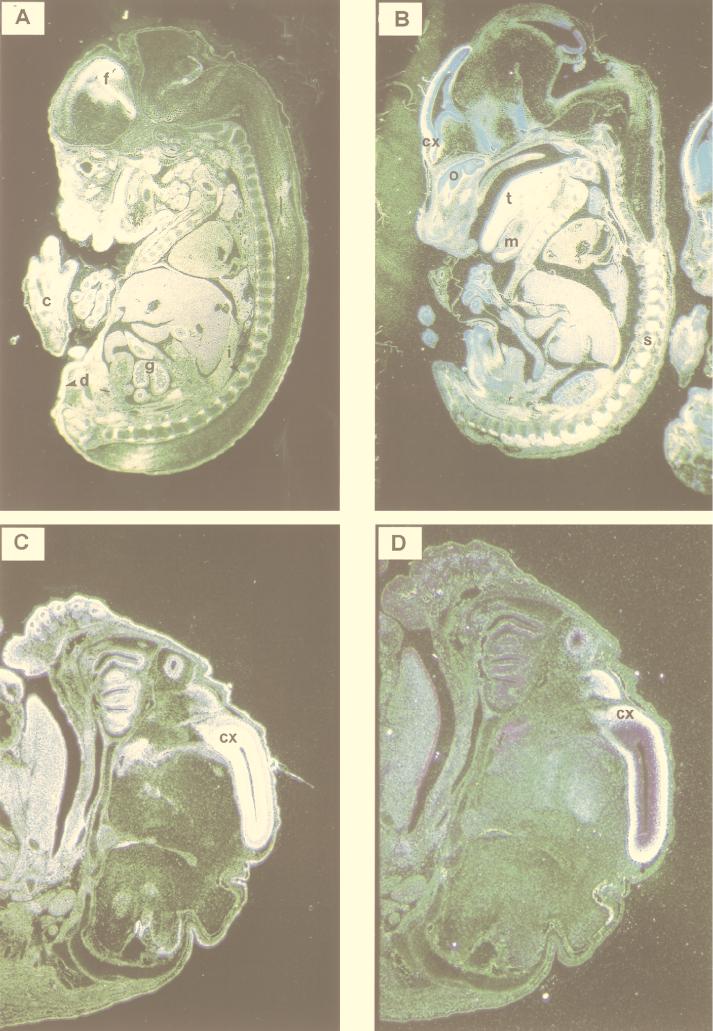
To explore in greater detail the expression patterns of MIBP1, SEF-2, and SSTR-2 mRNA in the adult brain and during embryonic development, we performed a series of in situ hybridizations to tissue sections of paraffin-embedded murine brain and embryo specimens. To avoid signals resulting from cross-hybridization, we prepared cRNA probes from the same MIBP1 partial clone that we used for the Northern blots shown in Fig. 6. In the adult brain (Fig. 7), specific signals resulting from MIBP1 and SEF-2 mRNA expression were colocalized to the cerebral cortex, the hippocampus, and corpora amygdala and, further, to a discrete layer of the cerebellar cortex overlapping precisely with regions of SSTR-2 mRNA distribution (10). In mouse embryos dissected at stages E13.5 (Fig. 8A and B) and E15.5 (Fig. 8C and D), specific MIBP1 signals were detected in restricted areas of the anterior neural tube over the primordial frontal cortex, in the spinal cord, in the dorsal root ganglia, and in developing skeletal muscle. As described previously (27) and further confirmed by our in situ hybridizations (Fig. 8A), SEF-2 mRNA expression patterns were much less restricted and highly abundant in many tissues, including the anterior neural tube, hindbrain, spinal cord, dorsal root ganglia, skeletal muscle, and subepidermal connective tissue. In summary, these studies revealed that SSTR-2-positive tissues in embryonic and adult mice overlap precisely with patterns of MIBP1 and SEF-2 coexpression.
FIG. 7.
Expression of SEF-2 and MIBP1 in adult mouse brain (sagittal sections) analyzed by in situ hybridization. The specificity of all the reactions was controlled by hybridizations with sense cRNA probes under identical conditions (data not shown). (A) Specific SEF-2 signals were obtained in cortex (cx), hippocampus (h), cerebellum (cb), and olfactory bulb (o). (B) Specific MIBP1 signals were obtained overlapping with all of the SEF-2-positive structures.
FIG. 8.
In situ hybridization of mouse embryos of gestational stage E13.5 (A and B) and E15.5 (C and D) with SEF-2 and MIBP1 cRNA probes. The specificity of all the reactions was controlled by performing hybridization with sense probes under identical conditions (data not shown). (A and C) SEF-2 expression in the dermis (d), forebrain (f), intervertebral discs (i), gut (g), cartilage (c), and cortex (cx). (B and D) MIBP1 expression in the forebrain (primordium of the cortex [cx]), tongue (t), spinal ganglia (s), lower-jaw mesenchyme (m), and olfactory epithelium (o).
DISCUSSION
During the last few years, a family of five G-protein-coupled, transmembrane-spanning receptors, which mediate the pleotrophic effects of SSTs, have been isolated (reviewed in reference 19). First, functional studies have implicated SST as a key regulatory molecule in the hypothalamic-hypophyseal axis through central control of growth hormone-releasing hormone-producing neurons in the arcuate nucleus (28). Second, SST modulates a large variety of cognitive and vegetative functions, including sleep behavior (1a), which involve the widespread somatostatinergic system in cortical and subcortical brain regions. Third, a variety of effects on peripheral target tissues, including release of hormones from the gastroenteropancreatic endocrine system and modulation of muscle contraction, especially in smooth muscle and vascular wall muscle cells, have been identified (4). All of these functions involve interactions between SST and the receptor subtype SSTR-2, which displays specific expression patterns on the surface of the respective target cells.
Furthermore, specific SSTR expression patterns have been detected during embryonal development of the rodent central and peripheral nervous systems. The precise role of somatostatinergic signalling during embryogenesis remains to be determined; however, transient changes in these expression patterns during development suggest that SSTRs modulate the differentiation and plasticity of neural cells (9). Although SSTR expression patterns in developing and adult nervous systems have been described in detail (7, 25) and reveal a high degree of conservation even among different amphibian and mammalian species (30), the transcriptional mechanisms which specify these patterns are entirely unknown.
The results of our study clearly show that a combinatorial set of two different transcription factors, the bHLH factor SEF-2 and the zinc finger protein MIBP1, bind in a sequence-specific manner to the SSTR-2 Inr and mediate enhanced transcriptional activity. Overlapping expression patterns of SEF-2 and MIBP1 in both embryonal and adult tissues coincide with spatially and temporally restricted expression patterns of SSTR-2, which have been studied in detail previously by using both in situ hybridization and receptor-specific radiolabelled ligands (1, 10, 12, 28a, 28b). These data define for the first time a transcriptional function of MIBP1 and imply that this transcription factor plays an important role in the development of nervous and neuroendocrine tissues.
The characterization of MIBP1 as a transcription factor that exhibits sequence-specific DNA-binding properties to the SSTR-2 TC box came as a surprise. A number of previous studies have shown that the rat MIBP1 homolog, the human MBP2 homolog, the partial MIBP1 clone AGIE-BP1, and the sequence-related transcription factor PRDII-BP1 bind specifically to NF-κB-like motifs with the consensus GGG N(4–5)CC (6, 11, 14, 17, 22). We therefore speculate that MIBP1 utilizes the multiple zinc finger clusters to bind to a number of promiscuous GC- or TC-rich binding sites. The huge MIBP1 peptide surface could thereby serve multiple functions, possibly as an adapter to other proteins or as an unwinding factor for TC- or GC-rich promoter regions. Clearly, the close proximity between the MIBP1- and the SEF-2-binding sites in the SSTR-2 has been evolutionarily conserved. These data suggest strongly that a direct interaction between the two proteins, which we observed in vitro, is functionally relevant.
We have previously shown that specific binding of SEF-2 to the E box is sufficient to initiate basal transcription from the SSTR-2 initiator. SEF-2 was able to recruit the basal transcription machinery via direct interaction with TFIIB and thereby to mediate gene transcription independently of a TATA element (20). We now show that MIBP1 binds on DNA in close proximity to SEF-2 and that the two proteins can physically interact in vitro. In addition, MIBP1 mediates enhanced SEF-2-dependent SSTR-2 promoter activity in vivo. Since SEF-2 expression occurs in many tissues, including neural, inflammatory, and muscle cells, MIBP1 seems to direct enhanced transcriptional activity to a specific promoter. The combinatorial activation of SSTR-2 gene expression by SEF-2 and MIBP1 may therefore specify SSTR-2 expression not only in neural cells but also under various physiological or pathophysiological conditions in smooth muscle and inflammatory cells.
ACKNOWLEDGMENTS
We are indebted to Kenshi Hayashi (Fukuoka, Japan) for providing the rat MIBP1 cDNA, to Richard B. Gaynor (University of Texas, Dallas) for providing the MIBP1/PRDII-BF1 zinc finger antiserum, to Pamela Mellon (San Diego, Calif.) for providing the cell line GHFT-1, and to Silvia Seegers for help with sequencing.
This work was supported by grants from the DFG to R.B. and R.S.
Equal contributions to this work were made by U.D. and A.P. Therefore, both should be considered first authors.
REFERENCES
- 1.Beaudet A, Greenspun D, Raelson J, Tannenbaum G S. Patterns of expression of SSTR1 and SSTR2 somatostatin receptor subtypes in the hypothalamus of the adult rat: relationship to neuroendocrine function. Neuroscience. 1995;65:551–561. doi: 10.1016/0306-4522(94)00486-o. [DOI] [PubMed] [Google Scholar]
- 1a.Beranek L, Obal F, Jr, Taishi P, Bodosi B, Laczi F, Krueger J M. Changes in rat sleep after single and repeated injections of the long-acting somatostatin analog octreotide. Am J Physiol. 1997;273:1484–1491. doi: 10.1152/ajpregu.1997.273.4.R1484. [DOI] [PubMed] [Google Scholar]
- 2.Bruno J F, Xu Y, Song J, Berelowitz M. Molecular cloning and functional expression of a brain-specific somatostatin receptor. Proc Natl Acad Sci USA. 1992;89:11151–11155. doi: 10.1073/pnas.89.23.11151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buettner R, Kannan P, Imhof A, Bauer R, Yim S O, Glockshuber R, Van Dyke M W, Tainsky M A. An alternatively spliced mRNA from the AP-2 gene encodes a negative regulator of transcriptional activation by AP-2. Mol Cell Biol. 1993;13:4174–4185. doi: 10.1128/mcb.13.7.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dimech J, Feniuk W, Latimer R D, Humphrey P P. Somatostatin-induced contraction of human isolated saphenous vein involves sst2 receptor-mediated activation of L-type calcium channels. J Cardiovasc Pharmacol. 1995;26:721–728. doi: 10.1097/00005344-199511000-00008. [DOI] [PubMed] [Google Scholar]
- 5.Epelbaum J, Dournaud P, Fodor M, Viollet C. The neurobiology of somatostatin. Crit Rev Neurobiol. 1994;8:25–44. [PubMed] [Google Scholar]
- 6.Fan C M, Maniatis T. A DNA-binding protein containing two widely separated zinc finger motifs that recognize the same DNA sequence. Genes Dev. 1990;4:29–42. doi: 10.1101/gad.4.1.29. [DOI] [PubMed] [Google Scholar]
- 7.Fodor M, Slama A, Guillaume V, Videau C, Csaba Z, Oliver C, Epelbaum J. Distribution and pharmacological characterization of somatostatin receptor binding sites in the sheep brain. J Chem Neuroanat. 1997;12:175–182. doi: 10.1016/s0891-0618(96)00199-8. [DOI] [PubMed] [Google Scholar]
- 8.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katz D M, He H, White M. Transient expression of somatostatin peptide is a widespread feature of developing sensory and sympathetic neurons in the embryonic rat. J Neurobiol. 1992;23:855–870. doi: 10.1002/neu.480230707. [DOI] [PubMed] [Google Scholar]
- 10.Kong H, DePaoli A M, Breder C D, Yasuda K, Bell G I, Reisin T. Differential expression of messenger RNAs for somatostatin receptor subtypes SSTR1, SSTR2 and SSTR3 in adult rat brain: analysis by RNA blotting and in situ hybridization histochemistry. Neuroscience. 1994;59:175–184. doi: 10.1016/0306-4522(94)90108-2. [DOI] [PubMed] [Google Scholar]
- 11.Makino R, Akiyama K, Yasuda J, Mashiyama S, Honda S, Sekiya T, Hayashi K. Cloning and characterization of a c-myc intron binding protein (MIBP1) Nucleic Acids Res. 1994;22:5679–5685. doi: 10.1093/nar/22.25.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maubert E, Slama A, Ciofi P, Viollet C, Tramu G, Dupouy J P, Epelbaum J. Developmental patterns of somatostatin-receptors and somatostatin-immunoreactivity during early neurogenesis in the rat. Neuroscience. 1994;62:317–325. doi: 10.1016/0306-4522(94)90335-2. [DOI] [PubMed] [Google Scholar]
- 13.Meyerhof W, Wulfsen I, Schönrock C, Fehr S, Richter D. Molecular cloning of a somatostatin-28 receptor and comparison of its expression pattern with that of a somatostatin-14 receptor in rat brain. Proc Natl Acad Sci USA. 1992;89:10267–10271. doi: 10.1073/pnas.89.21.10267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitchelmore C, Traboni C, Cortese R. Isolation of two cDNAs encoding zinc finger proteins which bind to the alpha 1-antitrypsin promoter and to the major histocompatibility complex class I enhancer. Nucleic Acids Res. 1991;19:141–147. doi: 10.1093/nar/19.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moser M, Imhof A, Pscherer A, Bauer R, Amselgruber W, Sinowatz F, Hofstadter F, Schule R, Buettner R. Cloning and characterization of a second AP-2 transcription factor: AP-2 beta. Development. 1995;121:2779–2788. doi: 10.1242/dev.121.9.2779. [DOI] [PubMed] [Google Scholar]
- 16.Moser M, Pscherer A, Roth C, Becker J, Mucher G, Zerres K, Dixkens C, Weis J, Guay-Woodford L, Buettner R, Fassler R. Enhanced apoptotic cell death of renal epithelial cells in mice lacking transcription factor AP-2beta. Genes Dev. 1997;11:1938–1948. doi: 10.1101/gad.11.15.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nomura N, Zhao M J, Nagase T, Maekawa T, Ishizaki R, Tabata S, Ishii S. HIV-EP2, a new member of the gene family encoding the human immunodeficiency virus type 1 enhancer-binding protein. Comparison with HIV-EP1/PRDII-BF1/MBP-1. J Biol Chem. 1991;266:8590–8594. [PubMed] [Google Scholar]
- 18.O’Carroll A M, Lolait S J, König M, Mahan L C. Molecular cloning and expression of a pituitary somatostatin receptor with preferential affinity for somatostatin-28. Mol Pharmacol. 1992;42:939–946. [PubMed] [Google Scholar]
- 19.Patel Y C, Greenwood M T, Panetta R, Demchyshyn L, Niznik H, Srikant C B. The somatostatin receptor family. Life Sci. 1995;57:1249–1265. doi: 10.1016/0024-3205(95)02082-t. [DOI] [PubMed] [Google Scholar]
- 20.Pscherer A, Dörflinger U, Kirfel J, Gawlas K, Ruschoff J, Buettner R, Schüle R. The helix-loop-helix transcription factor SEF-2 regulates the activity of a novel initiator element in the promoter of the human somatostatin receptor II gene. EMBO J. 1996;15:6680–6690. [PMC free article] [PubMed] [Google Scholar]
- 21.Rohrer L, Raulf F, Bruns C, Buettner R, Hofstaedter F, Schuele R. Cloning and characterization of a fourth human somatostatin receptor. Proc Natl Acad Sci USA. 1993;90:4196–4200. doi: 10.1073/pnas.90.9.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ron D, Brasier A R, Habener J F. Angiotensinogen gene-inducible enhancer-binding protein 1, a member of a new family of large nuclear proteins that recognize nuclear factor κB-binding sites through a zinc finger motif. Mol Cell Biol. 1991;11:2887–2895. doi: 10.1128/mcb.11.5.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Schindler M, Harrington K A, Humphrey P P, Emson P C. Cellular localisation and co-expression of somatostatin receptor messenger RNAs in the human brain. Brain Res Mol Brain Res. 1995;34:321–326. doi: 10.1016/0169-328x(95)00191-t. [DOI] [PubMed] [Google Scholar]
- 25.Schindler M, Holloway S, Humphrey P P, Waldvogel H, Faull R L, Berger W, Emson P C. Localization of the somatostatin sst2(a) receptor in human cerebral cortex, hippocampus and cerebellum. Neuroreport. 1998;9:521–525. doi: 10.1097/00001756-199802160-00027. [DOI] [PubMed] [Google Scholar]
- 26.Seeler J S, Muchardt C, Suessle A, Gaynor R B. Transcription factor PRDII-BF1 activates human immunodeficiency virus type 1 gene expression. J Virol. 1994;68:1002–1009. doi: 10.1128/jvi.68.2.1002-1009.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soosaar A, Chiaramello A, Zuber M X, Neuman T. Expression of basic-helix-loop-helix transcription factor ME2 during brain development and in the regions of neuronal plasticity in the adult brain. Brain Res Mol Brain Res. 1994;25:176–180. doi: 10.1016/0169-328x(94)90297-6. [DOI] [PubMed] [Google Scholar]
- 28.Tannenbaum G S, Zhang W H, Lapointe M, Zeitler P, Beaudet A. Growth hormone-releasing hormone neurons in the arcuate nucleus express both Sst1 and Sst2 somatostatin receptor genes. Endocrinology. 1998;139:1450–1453. doi: 10.1210/endo.139.3.5977. [DOI] [PubMed] [Google Scholar]
- 28a.Thoss V S, Perez J, Duc D, Hoyer D. Embryonic and postnatal mRNA distribution of five somatostatin receptor subtypes in the rat brain. Neuropharmacology. 1995;34:1673–1688. doi: 10.1016/0028-3908(95)00135-2. [DOI] [PubMed] [Google Scholar]
- 28b.Thoss V S, Duc D, Hoyer D. Somatostatin receptors in the developing rat brain. Eur J Pharmacol. 1996;297:145–155. doi: 10.1016/0014-2999(95)00736-9. [DOI] [PubMed] [Google Scholar]
- 29.Umesono K, Murakami K K, Thompson C C, Evans R M. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vallarino M, Mathieu M, D’Aniello B, Rastogi R K. Distribution of somatostatin-like immunoreactivity in the brain of the frog, Rana esculenta, during development. Brain Res Dev Brain Res. 1998;106:13–23. doi: 10.1016/s0165-3806(97)00162-4. [DOI] [PubMed] [Google Scholar]
- 31.van’t-Veer L J, Lutz P M, Isselbacher K J, Bernards R. Structure and expression of major histocompatibility complex-binding protein 2, a 275-kDa zinc finger protein that binds to an enhancer of major histocompatibility complex class I genes. Proc Natl Acad Sci USA. 1992;89:8971–8975. doi: 10.1073/pnas.89.19.8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamada Y, Post S R, Wang K, Tager H, Bell G, Seino S. Cloning and functional characterization of a family of human and mouse somatostatin receptors expressed in brain, gastrointestinal tract, and kidney. Proc Natl Acad Sci USA. 1992;89:251–255. doi: 10.1073/pnas.89.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yasuda K, Rens-Domiano S, Breder C D, Law S F, Saper C B, Reisine T, Bell G I. Cloning of a novel somatostatin receptor, SSTR3, coupled to adenylylclase. J Biol Chem. 1992;267:20422–20428. [PubMed] [Google Scholar]