Abstract
Macrophages react to microbial and endogenous danger signals by activating a broad panel of effector and homeostatic responses. Such responses entail rapid and stimulus‐specific changes in gene expression programs accompanied by extensive rewiring of metabolism, with alterations in chromatin modifications providing one layer of integration of transcriptional and metabolic regulation. A systematic and mechanistic understanding of the mutual influences between signal‐induced metabolic changes and gene expression is still lacking. Here, we discuss current evidence, controversies, knowledge gaps, and future areas of investigation on how metabolic and transcriptional changes are dynamically integrated during macrophage activation. The cross‐talk between metabolism and inflammatory gene expression is in part accounted for by alterations in the production, usage, and availability of metabolic intermediates that impact the macrophage epigenome. In addition, stimulus‐inducible gene expression changes alter the production of inflammatory mediators, such as nitric oxide, that in turn modulate the activity of metabolic enzymes thus determining complex regulatory loops. Critical issues remain to be understood, notably whether and how metabolic rewiring can bring about gene‐specific (as opposed to global) expression changes.
Keywords: epigenetics, inflammation, macrophages, metabolism, transcription
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; Immunology; Metabolism
Macrophages react to danger signals by triggering a broad panel of effector and homeostatic responses. This review highlights the dynamic crosstalk between metabolism and inflammatory gene expression during macrophage activation.

Glossary
- ACLY
ATP citrate lyase
- ACO2
Aconitase
- αKT
alpha‐ketoglutarate
- AKT
Protein Kinase B
- ALDOA
Fructose‐bisphosphate aldolase A
- AP‐1
Activator protein 1
- ARG1
Arginase 1
- ATM
Adipose tissue macrophage
- CBP
CREB‐binding protein
- CD36
Cluster of differentiation 36
- CD206
Cluster of differentiation 206
- COBRA
Constraint‐based reconstruction and analysis
- CXCL1
Chemokine (C‐X‐C motif) ligand 1
- DHAP
Dihydroxyacetone phosphate
- DIC
Mitochondrial dicarboxylate carrier
- elF5A
Eukaryotic translation initiation factor 5A
- ETC
Electron transport chain
- FAO
Fatty acid oxidation
- G3P
Glycerol 3‐phosphate
- GAPDH
Glyceraldehyde 3‐phosphate dehydrogenase
- GCN5
General control non‐repressed 5
- GPD1
Glycerol‐3‐phosphate dehydrogenase 1
- GPD2
Glycerol‐3‐phosphate dehydrogenase 2
- GPS
Glycerol phosphate shuttle
- H3K27ac
Histone H3 acetylation at lysine 27
- H3K18la
Histone H3 lactylation at lysine 18
- H3K27me3
Histone 3 tri‐methylation at Lysine 27
- HAT
Histone acetyl transferase
- HDAC
Histone deacetylase
- HIF1α
Hypoxia‐inducible factor 1 alpha
- IDH
Isocitrate dehydrogenase
- IFN
Interferon
- IL‐4
Interleukin 4
- IL12β
Interleukin 12 beta
- IL1β
Interleukin 1 beta
- IL6
Interleukin 6
- iNOS
inducible nitric oxide synthase
- IRF
Interferon regulatory factor (1‐9)
- IRG1
Immune responsive gene 1
- JMJD3
Jumonji domain‐containing protein D3
- Km
Michaelis–Menten constant
- LAL
Lysosomal acid lipase
- LDH
Lactate dehydrogenase
- LPS
Lipopolysaccharide
- MGL
Macrophage galactose C‐type Lectin
- MPC
Mitochondrial pyruvate carrier
- mTORC
Mammalian target of rapamycin
- NF‐κB
Nuclear factor kappa B
- NO
Nitric oxide
- OGDC
Oxoglutarate/α‐ketoglutarate dehydrogenase complex
- oxLDL
Oxidized low‐density lipoproteins
- oxPAPC
Oxidized phospholipids
- OXPHOS
Oxidative phosphorylation
- P/CAF
P300/CBP‐associated factor
- PDC
Pyruvate dehydrogenase complex
- PDK
Pyruvate dehydrogenase kinase
- PFK1
Phosphofructokinase 1
- PFK2
6‐phosphofructo‐2‐kinase
- PGC1β
Peroxisome proliferator‐activated receptor gamma (PPAR‐gamma) coactivator‐1 beta
- PGE2
Prostaglandin E2
- PPARγ
Peroxisome proliferator‐activated receptor gamma
- ROS
Reactive oxygen species
- SAM
S‐adenosyl‐methionine
- SLC
Solute carrier family protein
- SSP
Serine‐synthesis pathway
- STAT
Signal transducer and activator of transcription
- SUCNR1
Succinate receptor 1
- TCA
Tricarboxylic acid cycle
- TET
Ten‐eleven translocation
- TNFα
Tumor necrosis factor alpha
Introduction
The different functions exerted by macrophages are regulated and tuned by the signals they are exposed to in different tissues, during homeostasis or in response to acute or chronic damage. In the past few years, the availability of a large panel of unbiased, high‐throughput data, including transcriptomic and epigenomic datasets, has greatly increased our understanding of the mechanisms by which tissue‐microenvironmental signals determine distinct transcriptional programs in macrophages already early in organ development (Gautier et al, 2012; Yona et al, 2013; Gosselin et al, 2014; Lavin et al, 2014; Mass et al, 2016; Bonnardel et al, 2019; Sakai et al, 2019). Different macrophage gene expression programs are enabled by signal‐regulated transcription factors (TFs) binding to thousands of cis‐regulatory regions maintained accessible and functional by macrophage lineage‐determining TFs such as PU.1 and IRF8 (Barish et al, 2010; Ghisletti et al, 2010; Heinz et al, 2010; Mancino et al, 2015). The complex interplay among TFs, coregulators, and chromatin modifications implicated in the control of specific macrophage functions and the overall principles regulating inducible gene expression have been extensively reviewed elsewhere (Heinz et al, 2015; Amit et al, 2016; Glass & Natoli, 2016; Monticelli & Natoli, 2017; Barrat et al, 2019; Stadhouders et al, 2019).
Concomitantly to the induction of gene expression changes, the detection of danger signals or other regulatory stimuli rapidly activates an extensive reprogramming of macrophage metabolism (Artyomov et al, 2016; Jung et al, 2019; O'Neill et al, 2016; O'Neill & Pearce, 2016; Russell et al, 2019; van Teijlingen & Pearce, 2020). How such metabolic reprogramming impacts macrophage phenotypes has only recently begun to emerge. As discussed below, it is now clear that metabolic rewiring not only aims at fulfilling energy requirements, but also supports the distinctive changes in macrophage functions determined by different stimuli.
Macrophage activation and polarization
Classical in vitro models for the elucidation of signal‐dependent gene expression and metabolic reprogramming include treatment of mouse bone marrow‐ or human monocyte‐derived macrophages with lipopolysaccharide (LPS) and/or interferon γ (IFNγ) to induce a “classically” activated (M1) phenotype or treatment with interleukin‐4 (IL‐4) to induce an “alternatively” activated (M2) phenotype (Murray et al, 2014). M1 macrophages express proinflammatory cytokines and chemokines and produce reactive oxygen species (ROS) and molecules with microbicidal activity such as nitric oxide (NO), while M2 macrophages activate the transcription of genes involved in wound healing and defense from parasitic and fungal infections. M1‐ and M2‐polarized states are an oversimplification of more complex biological responses (Mantovani, 2016) whereby combinations of stimuli transiently and usually reversibly instruct macrophages to acquire a variety of functional states (Murray et al, 2014). However, for the sake of simplicity, the M1/M2 nomenclature is still used to describe functionally opposite macrophage states.
Mechanistically, similar but also clearly distinct M1 phenotypes can be triggered by IFNγ or LPS. In the first case, the response is mainly driven by STAT1 activation and a few partner TFs such as IRF1/2 (Interferon Regulatory Factors1/2), while in the case of LPS it is driven by multiple signal‐induced TFs, such as NF‐kB, AP‐1, and IRFs that activate the IFNβ1 gene, followed by IFNβ release and a secondary response driven by IFNβ itself and by other inflammatory cytokines released upon activation, acting in a paracrine and/or autocrine manner. Although most changes in epigenomic features and TF binding induced by LPS or IFNγ occur at pre‐accessible regulatory regions, initially inactive genomic loci can acquire competence to activate transcription upon stimulation, so‐called “latent” or “de novo” enhancers (Kaikkonen et al, 2013; Ostuni et al, 2013). Therefore, the usage of the genomic regulatory elements responsible for macrophage activation is flexibly adjusted in response to stimulation.
Macrophage activation and metabolic reprogramming
Upregulation of glucose utilization and decrease in oxygen consumption are hallmarks of classically activated macrophages (O'Neill & Pearce, 2016). The metabolic state of an activated macrophage after ca. 24 h LPS treatment is characterized by impaired mitochondrial oxidative phosphorylation (OXPHOS) coupled with increased aerobic glycolysis (Fig 1) (Rodríguez‐Prados et al, 2010; Freemerman et al, 2014). In this setting, pyruvate produced as an end product of glycolysis is reduced to lactate instead of being oxidized to acetyl‐CoA by the pyruvate dehydrogenase complex (PDC) to be eventually fed into the tricarboxylic acid (TCA) cycle in mitochondria (Fig 1). An important player in this metabolic rewiring is nitric oxide (NO), whose increased production is ascribed to the transcriptional activation of the inducible nitric oxide synthase (encoded by the Nos2 gene). NO and NO‐derived reactive nitrogen species can inactivate the iron‐sulfur‐containing complexes of the mitochondrial electron transport chain, thus eventually promoting mitochondrial dysfunction (Fig 1) (Chouchani et al, 2013; Clementi et al, 1998; Van den Bossche et al, 2016). The increase in glycolytic metabolism is thus first of all an adaptive response aimed at maintaining a sufficient level of ATP production in cells with impaired mitochondrial respiration (Everts et al, 2012). In parallel, the response to LPS or IFNγ results in three main alterations in the TCA cycle that can be summarized as follows.
Figure 1. An overview of metabolic and transcriptional reprogramming in LPS‐activated macrophages.

Macrophages respond to proinflammatory stimuli by extensively reprogramming their transcriptional and metabolic activity. The glycolytic flux is enhanced, and oxygen consumption is reduced, while several pathways, such as the TCA cycle, are altered and specific metabolites accumulate over time. Both glucose uptake and metabolism are rapidly upregulated, and the pyruvate produced is converted into lactate. The induction of the Nos2 gene leads to a massive burst of NO production. NO acts as an antimicrobial molecule and impairs the activity of the electron transport chain (ETC), eventually reducing cellular oxygen consumption. The TCA cycle is remodeled because of the impairment of IDH activity, leading to the accumulation of citrate which is exported to the cytoplasm via the mitochondrial citrate carrier (SLC25A1). Cleavage of cytoplasmic citrate by ACLY equilibrates the mitochondrial and the cytosolic acetyl‐CoA pool. ACOD1 induction leads to aconitate being diverted to itaconate, which functions as an immunomodulatory molecule and also affects succinate dehydrogenase (SDH), blocking its activity and thus promoting succinate accumulation. Among key transcription factors produced, HIF1α is stabilized by the accumulation of succinate and it is able to induce several key metabolic genes. Together with key transcription factors, the second wave of inflammatory mediators results from the activity of histone acetyltransferases, which rely on the acetyl‐CoA produced by the cell to support activation of gene expression.
First, downregulation of the activity of isocitrate dehydrogenases, which convert isocitrate to α‐ketoglutarate, leads to the accumulation of citrate (Fig 1) (Tannahill et al, 2013; Jha et al, 2015). Accumulation of citrate inside mitochondria favors its export to the cytoplasm by the mitochondrial citrate transporter (encoded by SLC25A1) (Infantino et al, 2013). In the cytoplasm, citrate undergoes cleavage by the ATP citrate lyase (ACLY) leading to the production of oxaloacetate and acetyl‐coenzyme A (acetyl‐CoA), a central metabolic intermediate for the synthesis of fatty acids and prostaglandins (Fig 1). In addition, a most prominent signaling function of acetyl‐CoA is to provide the acetyl groups required for acetylation of histones (Lauterbach et al, 2019), which undergo hyperacetylation at thousands of cis‐regulatory elements activated in response to the recruitment of TFs induced by macrophage stimulation (Ghisletti et al, 2010; Heinz et al, 2010; Kaikkonen et al, 2013; Ostuni et al, 2013).
Second, the strong LPS‐mediated induction of the ACOD1 gene (previously known as IRG1, Immune Responsive Gene 1) encoding aconitate decarboxylase 1 (Michelucci et al, 2013) allows the synthesis of the metabolite itaconate (Strelko et al, 2011), from the TCA cycle intermediate cis‐aconitate (Fig 1). Itaconate, a cysteine‐reactive electrophile (Bambouskova et al, 2018; Mills et al, 2018), starts accumulating within 4 h after in vitro LPS stimulation of macrophages, reaches concentrations in the low millimolar range at approx. 24 h (Lauterbach et al, 2019; Seim et al, 2019), and exerts complex anti‐bacterial (McFadden & Purohit, 1977; Michelucci et al, 2013) as well as various immunoregulatory effects (Hooftman et al, 2020; Swain et al, 2020) that are outside of our main focus and have been extensively covered in recent review articles (Hooftman & O'Neill, 2019; O'Neill & Artyomov, 2019; Zaslona & O'Neill, 2020; Cordes & Metallo, 2021).
Accumulation of itaconate also causes extensive metabolic reprogramming at least at two distinct levels: It impairs glycolysis mainly by modifying and inhibiting ALDOA (fructose‐bisphosphate aldolase A) (Qin et al, 2019) and attenuates fluxes in the TCA cycle by inhibiting the succinate dehydrogenase (SDH), eventually resulting in succinate accumulation (Fig 1) (Jha et al, 2015; Cordes et al, 2016; Lampropoulou et al, 2016). Accumulation of succinate has two consequences. Extracellularly, released succinate triggers succinate receptor 1 (SUCNR1), eventually causing context‐dependent anti‐inflammatory effects (Peruzzotti‐Jametti et al, 2018; Keiran et al, 2019; Harber et al, 2020; Wu et al, 2020). Intracellularly, the increased concentrations of succinate inhibit prolyl hydroxylases that generate succinate as reaction product and are thus subjected to product inhibition (Tannahill et al, 2013). These oxygen‐regulated dioxygenases induce prolyl hydroxylation of hypoxia‐inducible factor 1 alpha (HIF1α), targeting it to degradation in normoxic conditions; therefore, their inhibition by succinate increases HIF1α protein levels. In turn, the accumulation of HIF1α leads to transcriptional activation of the interleukin 1β (IL‐1β) gene (Tannahill et al, 2013) and other HIF1α‐dependent genes, including those encoding several glycolytic enzymes (Cheng et al, 2014). HIF1α accumulation and downstream changes in gene expression thus represent a paradigm of how a metabolic alteration can bring about gene‐specific transcriptional effects. Notably, itaconate has been recently shown to covalently modify (“itaconation”) cysteine residues of proteins (Qin et al, 2019; Qin et al, 2020). Given the abundance of this metabolite in M1‐polarized macrophages, it will be crucial to understand whether itaconate can also directly modify histones and regulate the transcription of inflammatory genes.
Compared with inflammatory macrophages, alternatively activated (M2) macrophages show a milder metabolic reprogramming, with an intact TCA cycle and a prevalent use of fatty acid oxidation (FAO, also known as β‐oxidation) to fuel mitochondrial oxidative phosphorylation (OXPHOS) (Fig 2A) (Jha et al, 2015). Mechanistically, IL‐4 potently enhances the uptake of fatty acids and induces the transcription of oxidative metabolism genes by the direct activation of STAT6 and its partner PPARγ with its coactivator PGC1β (Vats et al, 2006; Odegaard et al, 2007; Nomura et al, 2016; Piccolo et al, 2017; Czimmerer et al, 2018; Nelson et al, 2018). The substrates for FAO are exogenous lipids taken up by CD36, a scavenger receptor transcriptionally regulated by PPARγ and with high affinity for long‐chain fatty acids and triacylglycerols (Fig 2A) (Feng et al, 2000; Chawla et al, 2001). In turn, triacylglycerols must be metabolized by the lysosomal acid lipase (LAL), and the impairment of this lipolysis step decreases macrophage oxidative respiration and impairs M2 polarization (Huang et al, 2014). Essential for OXPHOS and FAO metabolism in M2 macrophages is the generation of α‐ketoglutarate from glutamine, the most abundant extracellular amino acid (Liu et al, 2017). α‐ketoglutarate is a co‐substrate required for the activity of the histone demethylase Jmjd3, which removes the repressive histone mark H3K27me3 (De Santa et al, 2007), thus promoting the activation of M2 genes, as discussed below (Fig 2B) (Liu et al, 2017). Glutamine has also been reported to support M2 macrophage polarization through the glutamine‐UDP‐GlcNac pathway, which is fundamental for glycosylation of proteins relevant for M2 macrophage function, such as the macrophage mannose receptor and the macrophage galactose‐binding lectin (Fig 2C; Jha et al, 2015).
Figure 2. IL‐4‐induced changes in metabolism and their effects on gene expression.
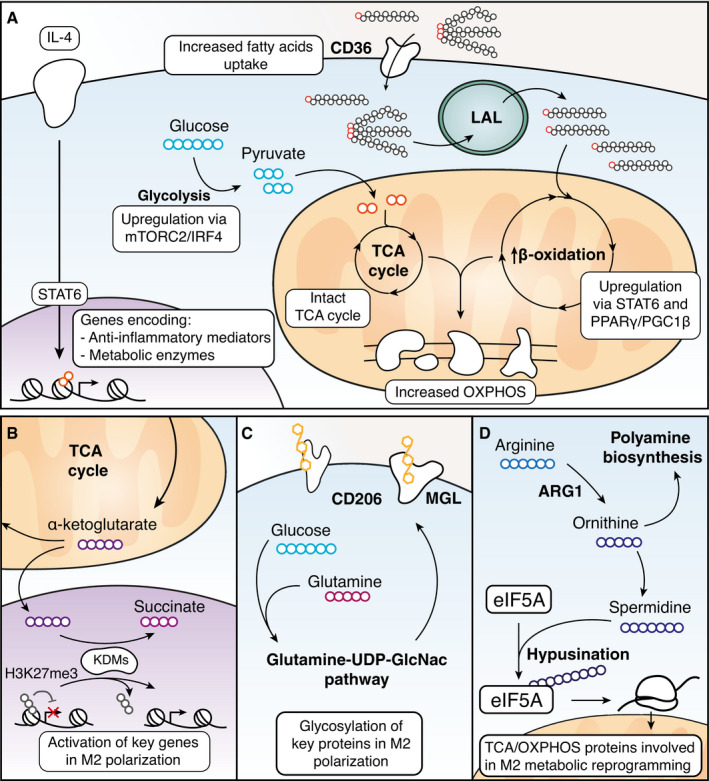
(A) In response to IL‐4 stimulation, glycolysis is increased; the TCA cycle remains intact and both fatty acid uptake and metabolism are enhanced via regulation of metabolic genes by STAT6 and PPARγ. (B) Alpha‐ketoglutarate, an intermediate of the TCA cycle, acts as a co‐substrate of lysine demethylases such as JMJD3 to remove the repressive H3K27me3 mark, eventually activating gene transcription. (C) Glutamine and glucose are employed via the Glutamine‐UDP‐GlcNac pathway to glycosylate proteins involved in M2 macrophage function such as CD206 and MGL. (D) Arginase 1, upregulated by IL‐4, converts arginine to ornithine which is used in polyamine biosynthesis. Ornithine can be also further metabolized to spermidine which was found to be involved in the hypusination of translation factor eIF5A, which controls the production of key mitochondrial proteins involved in the TCA cycle and OXPHOS.
The role of glycolysis in M2 macrophage functions is not completely clear yet. Macrophage stimulation with IL‐4 leads to the activation of the mTORC2 pathway and the upregulation of glycolysis by IRF4, and the decrease in this pathway may inhibit M2 polarization and function (Fig 2A) (Huang et al, 2016). On the contrary, more recent data suggest that glycolysis is not required for M2 functions, as long as OXPHOS remains intact (Wang et al, 2018).
A hallmark of mouse M2 macrophages is the expression of the arginase 1 (Arg1) gene (Munder et al, 1998), which converts arginine to ornithine, resulting in several effects: (i) It provides the substrate for the synthesis of polyamines, which negatively impact pathogen fitness (Esser‐von Bieren et al, 2013); (ii) it depletes arginine from the extracellular milieu, preventing deleterious consequences of sustained activation of Th2 lymphocytes (Pesce et al, 2009); and (iii) it promotes efferocytosis (engulfment of apoptotic cells) in a feed‐forward loop in which arginine and ornithine taken up from engulfed apoptotic cells are converted into putrescine, which in turn enhances subsequent rounds of efferocytosis (Yurdagul et al, 2020). A complete understanding of how arginine metabolism impacts M2 macrophage activation is still lacking. However, arginine‐derived spermidine was recently shown to be involved in the hypusination of the translation factor elF5A, which controls the production of a subset of mitochondrial proteins involved in the TCA cycle and OXPHOS: Inactivation of this pathway selectively impairs OXPHOS‐dependent alternative macrophage activation, hinting at a close link between IL‐4‐mediated Arg1 gene induction and metabolic reprogramming in of M2 macrophages (Fig 2D) (Puleston et al, 2019).
Of note, most of the findings on M1 and M2 macrophage activation and metabolic reprogramming reported so far come from studies using mouse macrophages. However, it is important to consider that substantial differences have been reported in the response of human and mouse macrophages to inflammatory stimuli in vitro. Particularly, M1 stimulation of mouse macrophages leads to Nos2 gene expression and NO release, while human macrophages do not seem to release NO in vitro (Schneemann et al, 1993). Moreover, human peripheral blood monocyte‐derived macrophages (hMDMs) do not exhibit the switch to glycolysis and mitochondrial dysfunction that has been reported for mouse macrophages upon LPS treatment (Vijayan et al, 2019). Thus, activated hMDMs rely on OXPHOS rather than glycolysis for ATP production unlike mouse macrophages (Vijayan et al, 2019). Finally, human monocyte‐derived macrophages do not upregulate ARG1 expression upon IL‐4 treatment (Raes et al, 2005). However, there may exist inflammatory settings in vivo characterized by elevated NOS2 and ARG1 expression in human macrophages (Anstey et al, 1996; Nicholson et al, 1996; St Clair et al, 1996; Thomas & Mattila, 2014). In conclusion, it is crucial to be aware of substantial differences between mouse and human macrophages and to verify to what extent in vitro experimental models recapitulate in vivo phenomena.
Coordination of transcriptional and metabolic changes
As discussed above, on the one hand stimulus‐dependent changes in transcriptional programs can result in differential expression of genes encoding components of metabolic pathways; on the other hand, metabolic reprogramming can bring about gene‐specific transcriptional changes (such as those due to HIF1α accumulation) as well as changes in the abundance of intermediates such as acetyl‐CoA that are required for chromatin modifications and may thus exert a pervasive impact on transcription. Availability of metabolites required for chromatin modifications appears to play a central role in the integration of metabolic information and transcriptional control. Specifically, the enzymatic activity of many chromatin modifiers is regulated, at least in part, by the concentration of substrates and cofactors, whose availability may change depending on the stimulus macrophages are exposed to. For example, metabolic intermediates serve as cofactors (e.g., NAD+) or substrates (e.g., acetyl‐CoA) for numerous histone deacetylases (HDACs) and histone acetyl transferases (HATs), respectively (Houtkooper et al, 2012; Kinnaird et al, 2016). Moreover, the activity of lysine demethylases and cytosine‐hydroxylases is influenced by levels of α‐ketoglutarate (αKG) and succinate. αKG is a required cofactor for Jumonji‐C (JmJC) domain‐containing histone lysine demethylases (JMJDs) and ten‐eleven translocation (TET) families of 5mC hydroxylases (Tsukada et al, 2006; Tahiliani et al, 2009) and is converted into succinate during catalysis. Generally, these enzymes are subjected to product‐mediated inhibition by succinate, and therefore, their activity is dependent on the cofactor‐to‐product (αKG/succinate) ratio (Carey et al, 2015; Sciacovelli et al, 2016). For example, IL‐4 stimulation promotes a high αKG/succinate ratio, substrate, and inhibitor, respectively, of histone H3K27me3 demethylase JMJD3 (De Santa et al, 2007) thus potentiating gene induction in response to this cytokine (Liu et al, 2017). More recently, type I IFN has been linked to epigenetic regulation in macrophages via control of amino acid metabolism. Type I IFN was shown to suppress the serine‐synthesis pathway (SSP), thus negatively impacting on the production of S‐adenosyl‐methionine (SAM), the only substrate for histone methylation, and eventually deposition of H3K27me3 on gene promoters (Shen et al, 2021).
The link between metabolites and chromatin modifiers, and more in general the effects of metabolic reprogramming on macrophage function, have been extensively reviewed (O'Neill et al, 2016; Phan et al, 2017; Williams & O'Neill, 2018; Jung et al, 2019). In this review, we will instead focus on recent studies providing novel aspects of the interplay between metabolism and transcription, and we will highlight the most relevant knowledge gaps in the field. We will first discuss novel data on glucose metabolism changes over the course of the inflammatory response and their impact on inflammatory gene transcription. We will then explore new roles of the inflammatory mediator nitric oxide (NO) in the regulation of the inflammatory response through the modulation of metabolite production, and we will speculate on how this may influence epigenomic modifications and transcriptional programs. In the last part, we will examine selected examples of the influence of tissue environment on macrophage metabolic and transcriptional reprogramming, showing how its dysregulation can lead to disease states characterized by chronic inflammation. Finally, we will provide a long‐term perspective on this field of investigation.
Glucose metabolism as driver of inflammatory gene expression
Glucose utilization is crucial for classical macrophage activation, and its inhibition affects many functions typical of their inflammatory phenotype, including phagocytosis, ROS production, and secretion of proinflammatory cytokines. Upgrade of glycolysis in LPS‐activated macrophages occurs in sequential phases involving multiple mechanisms. First, a rapid increase in glucose uptake that reaches a plateau already 20 min after LPS stimulation (Fukuzumi et al, 1996) was reported to be mediated by the early induction of SLC2A1 (GLUT1), the main glucose transporter in myeloid cells (Fig 1) (Freemerman et al, 2014). Although the transcriptional regulation of SLC2A1 has historically been linked to HIF1α (Ebert et al, 1995), the rapid kinetics of SLC2A1 induction by LPS is not compatible with a role for HIF1α and imposes that alternative mechanisms are considered.
Second, transcriptional induction of PFK2, encoding the 6‐phosphofructo‐2‐kinase, determines the accumulation of fructose 2,6‐bisphosphate, an allosteric activator of the rate‐limiting glycolytic enzyme PFK1 (phosphofructokinase 1), eventually leading to increased glycolytic fluxes (Rodríguez‐Prados et al, 2010). Third, HIF1α was shown to play a critical role in inflammatory macrophage activation in vivo (Cramer et al, 2003), an effect that also involves regulation of glycolytic metabolism.
HIF1α accumulation in LPS‐activated macrophages is a two‐step process involving first its transcriptional induction by NF‐kB (Rius et al, 2008), and then the stabilization of the HIF1α protein via succinate‐mediated inactivation of prolyl hydroxylases (Fig 1) (Tannahill et al, 2013). The essential role of HIF1α in inflammatory macrophage activation may in part be linked to the control of the expression of multiple glycolytic enzymes (Cheng et al, 2014) and in part to the control of IL‐1β expression (Tannahill et al, 2013) (Fig 1). However, the slow kinetics of HIF1α accumulation implies that early reprogramming of energy metabolism in LPS‐activated macrophages must be HIF1α‐independent, as already reported in the specific case of PFK2 induction (Rodríguez‐Prados et al, 2010). HIF1α also induces the expression of the lactate dehydrogenase (LDH), which produces lactate from pyruvate, and of the pyruvate dehydrogenase kinase (PDK), which inactivates the pyruvate dehydrogenase thus limiting pyruvate conversion to acetyl‐CoA (Fig 1) (Semenza et al, 1996; Kim et al, 2006; Palsson‐McDermott et al, 2015). Enhanced LDH activity appears to have a critical role in this context: Since in classically activated macrophages OXPHOS is progressively impaired, maintenance of the flux through the glycolytic pathway is critical and this involves the regeneration of NAD+ from NADH in the LDH‐mediated conversion of pyruvate into lactate (Fig 1).
Glucose oxidation controls transcriptional regulation of inflammatory mediators
It has been recently reported that oxidative metabolism of macrophages is increased within the first hour and until eight hours post‐LPS stimulation, in contrast to the reduced oxygen consumption occurring at later time points (Langston et al, 2019; Lauterbach et al, 2019). This boost in oxidative metabolism in the early phases of LPS stimulation depends on increased glycolysis fueling the TCA cycle and is regulated by the mitochondrial glycerol 3‐phosphate dehydrogenase (GPD2) (Fig 3A). GPD2 is a rate‐limiting enzyme in the glycerol phosphate shuttle (GPS), which includes the glycolytic enzyme GAPDH and GPD1, located on the cytoplasmic side of the mitochondrial membrane. In the GPS, the NADH produced by GAPDH during glycolysis is used by GPD1 to reduce dihydroxyacetone phosphate (DHAP) to glycerol 3‐phosphate (G3P), thus regenerating NAD+ required for glycolysis. Inside mitochondria, electrons from G3P are directly transferred to the electron transport chain (ETC) through the FAD cofactor of GPD2 (Fig 3A). Upon LPS‐induced activation, the expression of GDP2 and the activity of the GPS are increased, resulting in both an increased NAD+ regeneration, which maximizes glycolysis, and an enhanced flux of electrons in the ETC (Langston et al, 2019). The relevance of this enzyme for macrophage activation is underscored by the observation that loss of GPD2 impairs the secretion of critical inflammatory cytokines upon LPS activation (Langston et al, 2019).
Figure 3. Novel aspects of metabolic remodeling and its transcriptional consequences in macrophages.

(A) The glycerol phosphate shuttle plays a key role in linking increased glycolytic flux to increased oxygen consumption in the early phases of the LPS response. (B) ACLY, a key gene able to regulate global acetyl‐CoA levels in the cell, is activated via AKT in the early phase (< 3 h) after LPS exposure, and its activity is crucial for the upregulation of several proinflammatory mediators. (C) Lactate produced from pyruvate is used to generate a novel post transcriptional modification on histones, H3K18la (lactylation of K18 on histone H3), which is deposited on genes activated at late time points in the LPS response. (D) The burst of NO production inactivates PDC, ACO2, and OGDC at late time points in the LPS response by nitrosylating one of their subunits (blue circles), leading to the normalization of citrate, itaconate, and succinate levels over time.
Interestingly, a GPD2‐dependent metabolic shift from increased (within the first 3 h of LPS‐induced macrophage activation) to decreased oxidative metabolism occurs over the course of 24 h of LPS exposure (Langston et al, 2019). This process appears to be relevant for LPS tolerance, the process whereby macrophages exposed to LPS for an extended time are subsequently unable to mount a full transcriptional response upon restimulation (Foster et al, 2007). A variety of mechanisms has been shown to contribute to the LPS‐tolerant state of macrophages, including receptor desensitization, secondary activation of negative signaling molecules, and induction of anti‐inflammatory microRNAs (Medvedev et al, 2000; Mages et al, 2008; Nahid et al, 2009; Doxaki et al, 2015; Seeley et al, 2018). In this context, the lack of re‐induction of tolerized genes correlates with defective deposition of histone marks associated with active transcription, chromatin accessibility, and nucleosome remodeling at promoters and enhancers of tolerized genes (Foster et al, 2007; Chen & Ivashkiv, 2010; Saeed et al, 2014). Emerging evidence indicates that an additional mechanism involves the metabolic regulation of acetyl‐CoA production by GPD2 (Langston et al, 2019). Upon prolonged exposure to LPS, GPD2‐dependent shutdown of oxidative metabolism limits the ability of glucose oxidation to support acetyl‐CoA production, thus contributing to the inability to induce specific proinflammatory mediators. However, whether and how GPD2‐mediated glucose oxidation and acetyl‐CoA production can modulate histone acetylation and the expression of a specific subset of LPS‐inducible genes as opposed to a global reduction in transcriptional activity remains unclear.
Links between glucose metabolism and histone acetylation
Histone acetyl transferases (HATs) catalyze the addition of acetyl groups to the histone N‐terminal lysines using acetyl‐CoA as donor (Choudhary et al, 2009). There are two separate pools of cellular acetyl‐CoA: the mitochondrial pool, generated by pyruvate oxidization to acetyl‐CoA by the mitochondrial pyruvate dehydrogenase complex (PDC), and the cytosolic pool. Since the inner mitochondrial membrane does not contain an acetyl‐CoA transporter, acetyl units are transported into the cytosol as part of a larger molecule, citrate. Briefly, immediately after LPS stimulation, increased glycolysis leads to augmented production of pyruvate that is fed into the Krebs’ cycle by the PDC, leading to increased citrate synthesis (Lauterbach et al, 2019). Citrate is then exported from the mitochondria to the cytosol by the citrate‐malate transporter, SLC25A1. Subsequently, the ATP citrate lyase (ACLY) cleaves citrate into oxaloacetate and acetyl‐CoA which then reaches an equilibrium in the cytoplasm and the nucleus. Cellular acetyl‐CoA fluctuates in response to a number of factors and changes in cytoplasmic acetyl‐CoA levels produced by ACLY‐mediated cleavage of citrate are sufficient to regulate global HAT activity (Wellen et al, 2009).
In various systems, high levels of glucose have been shown to increase the cellular amount of acetyl‐CoA, eventually affecting global histone acetylation (Lee et al, 2014; Moussaieff et al, 2015; Sivanand et al, 2017). ACLY appears to be an important bridge linking glucose metabolism to macrophage inflammatory gene expression programs (Langston et al, 2019; Lauterbach et al, 2019). These recent studies show that treatment of primary macrophages with LPS for 0.5‐2h results in AKT‐dependent activation of ACLY (Fig 3B) (Langston et al, 2019; Lauterbach et al, 2019). Blocking ACLY activity with structurally distinct inhibitors alters the transcription of specific subsets of inflammatory genes in vitro as well as in an in vivo model of peritonitis (Lauterbach et al, 2019). Along the same line, upregulation of ACLY by LPS and the consequent increase in cytosolic acetyl‐CoA levels were previously shown to be required for the transcriptional upregulation of proinflammatory mediators, such as nitric oxide (NO), ROS, and prostaglandin E2 (PGE2) (Infantino et al, 2013). Mitochondrial export of citrate and ACLY activity are also induced in an AKT‐dependent manner in response to IL‐4 treatment of macrophages, and they contribute to the activation of signature M2 genes (Covarrubias et al, 2016).
A key question is how fluctuations in acetyl‐CoA levels mediated by ACLY trigger a gene‐specific transcriptional response rather than global effects on chromatin acetylation and accessibility. ACLY inhibition seems to preferentially impact “late” genes, whose promoter undergoes extensive de novo acetylation in response to stimulation (Hargreaves et al, 2009; Ramirez‐Carrozzi et al, 2009). This observation has been interpreted as evidence that constitutively acetylated early genes may be less dependent on this pathway. Indeed, the genes regulated by ACLY, such as IL6 and IL12b, are secondary response genes, namely genes induced in a protein synthesis‐dependent manner, while primary response genes, such as CXCL1, remain unaffected (Lauterbach et al, 2019). However, IL1b, an early response gene, in one case was found to be ACLY dependent (Langston et al, 2019), while in another report it did not change in response to ACLY inhibition (Lauterbach et al, 2019). Moreover, TNF, an early gene, was also found to be regulated by ACLY inhibition (Lauterbach et al, 2019). Clearly, whereas activation of early LPS‐inducible genes is comparatively less demanding than that of late genes, as they are constitutively acetylated and bound by RNA Polymerase II, the mere induction kinetics and the basal chromatin status at a gene locus do not suffice to explain the gene‐selective effects of ACLY inhibition. Indeed, histone acetylation is an extremely dynamic modification and the high levels observed at the promoters of constitutively acetylated, early inducible genes reflect a dynamic equilibrium of acetylation and deacetylation rather than a static acetylated state. Overall, it seems unlikely that higher amounts of acetyl‐CoA are selectively required to acetylate the promoters of late but not early inducible genes (Escoubet‐Lozach et al, 2011). Another mechanism proposed to explain the different ACLY dependence of specific inflammatory genes is the selective recruitment of distinct HATs with different Km to distinct regulatory regions (Fig 4, upper panel) (Lauterbach et al, 2019). In this framework, selectivity may result from the gene‐specific recruitment of HATs that have a differential response to fluctuating acetyl‐CoA concentrations. In mammalian cell lines, the concentration of acetyl‐CoA has been reported to range between 2 and 20 μM (Lee et al, 2014) which is well above the Km of several HATs including Gcn5 and P/CAF (Tanner et al, 2000; Langer et al, 2002), whose activity would thus be unaffected by acetyl‐CoA fluctuations within such range. On the contrary, the concentration of acetyl‐CoA is near the Km of p300/CBP, which is 6.7 μM (Lau et al, 2000; Liu et al, 2008), which makes this specific HAT a candidate transducer of acetyl‐CoA fluctuations into gene expression changes (Fan et al, 2015). Indeed, the autoacetylation of p300 (which reflects its acetyltransferase activity), as well as its capacity to acetylate other substrates, is influenced by cytoplasmic acetyl‐CoA concentrations (Marino et al, 2014). p300 was previously found to associate in an LPS‐regulated manner to early and late inducible gene promoters (Ghisletti et al, 2010; Escoubet‐Lozach et al, 2011). Moreover, p300 may similarly be involved in alternative macrophage activation as it has been shown to preferentially regulate a subset of Akt‐dependent M2 genes (Covarrubias et al, 2016). Overall, it appears that specific gene subsets may be more sensitive than others to acetyl‐CoA fluctuation and this differential sensitivity may in turn be a major determinant of differential ACLY dependence.
Figure 4. Putative mechanisms linking global or local changes in metabolites concentration to changes in gene expression.
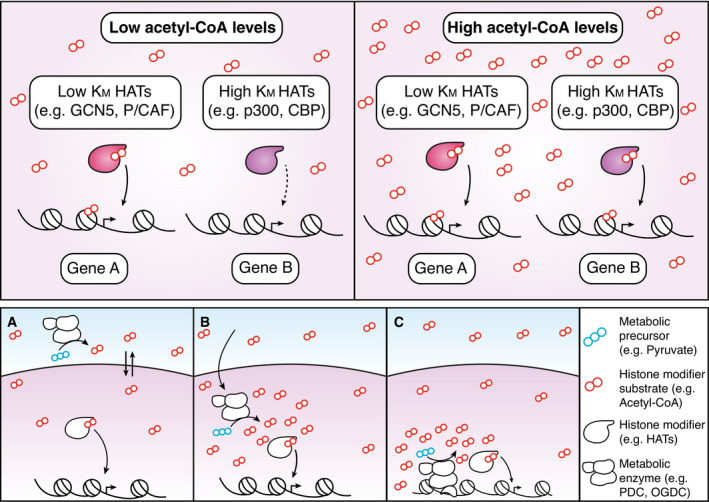
Upper panel. Changes in global availability of the substrate (e.g., acetyl‐CoA) of a histone modifier (e.g., a histone acetyl transferase, HAT) differentially regulate enzymes with distinct properties. For instance, HATs such as GCN5 or P/CAF have a low Km and are thus largely insensitive to broad changes in cellular substrate concentration. Conversely, HATs such as p300/CBP have a Km which is within the range of cellular concentration of the enzyme, making them able to transduce an increase in acetyl‐CoA concentration into increased histone acetylation levels. Lower panel. Three possible mechanisms explaining the impact of metabolite concentration on gene activity. (A) A metabolite produced outside of the nucleus diffuses through the nuclear membrane and equilibrates between different compartments. Changes in its cellular concentration thus affect histone modification activity inside the nucleus. (B) Translocation of a metabolic enzyme into the nucleus increases local production of a metabolite (e.g., acetyl‐CoA), modulating the activity of histone‐modifying enzymes. (C) Direct recruitment of a metabolic enzyme onto chromatin increases the local production of substrate, enhancing the deposition of histone modifications at specific loci.
Importantly, while acetyl‐CoA synthesis in the nucleus is not required as it can diffuse freely through the nuclear pores, multiple acetyl‐CoA producing enzymes, such as ACLY and PDC, have also been found in the nucleus (Fig 4, lower panel) (Takahashi et al, 2006; Wellen et al, 2009; Ariyannur et al, 2010; Comerford et al, 2014). These observations suggest that histone acetylation may also depend on acetyl‐CoA produced locally and possibly in proximity of genomic cis‐regulatory elements.
A key question that needs to be addressed is whether and how these enzymes are recruited onto specific genomic regulatory elements by sequence‐specific TFs, which could provide a simple mechanism for selective dependencies. In this model, genes not requiring a cytoplasmic source of acetyl‐CoA would be those able to recruit enzymes to their cis‐regulatory elements to locally generate acetyl‐CoA (Fig 4, lower panel). In conclusion, ACLY preferentially regulates the expression of specific inflammatory gene subsets but the mechanisms accounting for this selectivity remain unclear.
Lactate regulates macrophage phenotypic states
As discussed above, at late stages (24 h) of LPS activation, macrophages exhibit high rates of aerobic glycolysis resulting in high production of lactate (Zaslona & O'Neill, 2020). While the shutdown of oxidative metabolism is a central component of the metabolic reprogramming during prolonged exposure to LPS, the induction of aerobic glycolysis was previously considered just a compensatory mechanism to support ATP production. However, lactate production has important biological consequences linked to its activity as a signaling mediator (Brooks, 2009), an energy source (Bergman et al, 1999; Hirschhaeuser et al, 2011), and an immunosuppressive molecule capable of inhibiting proliferation, activation, and cytokine production in T cells, dendritic cells, and macrophages (Gottfried et al, 2006; Fischer et al, 2007; Dietl et al, 2010; Goetze et al, 2011; Colegio et al, 2014; Peter et al, 2015; Pucino et al, 2017; Nolt et al, 2018).
High lactate amounts produced by enhanced glycolysis in M1‐polarized cells also enable a recently described modification of histones, lactylation (Zhang et al, 2019) (Fig 3C). Histone H3 lactylation at lysine 18 (H3K18la) is deposited at the promoters of genes involved in tissue repair and wound healing at late time points (16‐24h) after stimulation, promoting their activation (Fig 3C). By contrast, histone lactylation does not occur at promoters of early inducible genes whose induction peaks when lactate levels are still low (Fig 3C) (Zhang et al, 2019). Thus, metabolic rewiring leads to increased production and accumulation of lactate, histone lactylation, and eventually a switch to a homeostatic gene expression program favoring the resolution of inflammation. Along the same line, increased intracellular lactate in macrophages was shown to induce an M2‐like tumor growth‐promoting phenotype (Bohn et al, 2018; Morioka et al, 2018; Liu et al, 2019). Mechanistically, the p300 HAT seems to be responsible for histone lactylation (Fig 3C) (Zhang et al, 2019) although the contribution of additional enzymes cannot be ruled out at this stage. Moreover, a most interesting question relates to the accumulation of lactate in the nucleus. Lactate can freely diffuse from the cytoplasm, where it is produced, into the nucleus (Zheng et al, 2003) but there is also evidence of LDH in the nucleus (Ronai, 1993), suggesting the possibility of a local production. It is still unclear what are the nuclear lactate concentrations required for this epigenetic modification and what mechanisms account for its selective deposition at a subset of genomic regions. Finally, it is worth mentioning that also non‐histone proteins are lactylated on lysine residues; specifically, glycolytic enzymes have been found to be the major targets of this modification, suggesting that the effects of increased lactate production in classically activated macrophages may be pervasive (Gaffney et al, 2020).
Nitric oxide‐dependent metabolic remodeling and chromatin regulation
Central to the metabolic switch of mouse M1 macrophages is the expression of the inducible nitric oxide synthase (iNOS) encoded by Nos2, a secondary response gene. iNOS produces large amounts of nitric oxide (NO), whose antimicrobial activity is linked to the nitrosation or nitrosylation and the consequent inactivation of essential enzymatic systems of intracellular bacterial pathogens (Fig 1) (Richardson et al, 2011). A metabolic side effect of increased NO production is the inhibition of mitochondrial respiration by nitrosation of the NADH dehydrogenase (Complex I) of the ETC and the competitive inhibition of O2‐consuming cytochrome c oxidase (Complex IV) (Clementi et al, 1998; Chouchani et al, 2013). In addition, NO nitrosylates and inactivates all iron‐sulfur proteins of the ETC, thereby inhibiting electron transport and mitochondrial ATP production (Fig 1) (Van den Bossche et al, 2016).
Recently, NO has also been shown to inhibit the pyruvate dehydrogenases complex (PDC), the oxoglutarate/α‐ketoglutarate dehydrogenase complex (OGDC), which converts α‐ketoglutarate to succinyl‐CoA in the Krebs’ cycle, and aconitase (ACO2) (Seim et al, 2019; Palmieri et al, 2020) (Fig 3D). Moreover, isocitrate dehydrogenase (IDH) activity has also been shown to be inhibited by NO (Yang et al, 2002; Lee et al, 2003; Bailey et al, 2019). At late stages (48–72 h) of LPS and IFNγ stimulation, an impaired flux through PDC and OGDC was observed and this reduced flux through the TCA cycle leads to the normalization of citrate, itaconate, and succinate levels and eventually decreased levels of acetyl‐CoA and succinyl‐CoA (Seim et al, 2019). Mechanistically, NO‐dependent suppression of PDC occurs via direct S‐nitrosation of one of its subunits during macrophage polarization (Palmieri et al, 2020). These data suggest that NO‐mediated inhibition of PDC may contribute to the late decline in acetyl‐CoA production (Fig 3D; Palmieri et al, 2020). Thus, NO takes center stage not only as an orchestrator of changes in macrophage mitochondrial metabolism during prolonged stimulation, but also as a potential regulator of downstream epigenomic changes.
As discussed above, metabolic enzymes, including glycolytic enzymes, have been observed in the nucleus (Boukouris et al, 2016). Among them, PDC and OGDC have been shown to generate acetyl‐CoA and succinyl‐CoA in the nucleus and local synthesis appears to be important for histone acetylation and succinylation (Sutendra et al, 2014). A functional PDC, in spite of its considerable size, has been found to be able to translocate from the mitochondria to the nucleus in response to diverse stimuli, and the nuclear localization seems to be required for basal histone acetylation (Fig 3D) (Sutendra et al, 2014). Pyruvate can diffuse into the nucleus from the cytosol and then serves as a substrate for nuclear PDC. The detailed mechanisms involved in the regulation of nuclear processes by these enzymes and their possible involvement in dynamic gene expression changes associated with macrophage activation remain to be defined.
Endogenous inflammatory signals reprogram macrophage metabolism
Each tissue‐resident macrophage population contributes to tissue‐specific homeostatic functions (Ginhoux & Guilliams, 2016). Consistent with this functional specialization, different tissue‐resident macrophages express distinct and to some extent tissue‐specific gene expression programs (Gautier et al, 2012). These differences in gene expression are associated with the selection and usage of partially non‐overlapping sets of tissue‐specific enhancers (Gosselin et al, 2014; Lavin et al, 2014; Sakai et al, 2019).
Notably, tissue‐resident macrophages can also be distinguished by differential expression of genes regulating metabolism, as different metabolic genes are associated with distinct populations. For example, genes regulating oxidative metabolism are enriched in microglia, where they play fundamental roles for microglia homeostatic functions such as trophic factor release and debris clearance by phagocytosis. Lipid metabolism signatures are enriched in alveolar macrophages, in which the high lipid content of surfactant imposes dedicated control mechanisms. Finally, eicosanoid and lipid signaling are enriched in peritoneal macrophages (Gautier et al, 2012; Gautier et al, 2014; Amit et al, 2016). Recently, the analysis of transcriptomes of six tissue‐resident macrophage populations also showed distinct responses to short‐ and long‐term metabolic challenge (Brykczynska et al, 2020). In this study, distinct or similar responses of macrophage subtypes to acute vs. chronic nutritional stress were identified, such as in the case of adipose tissue macrophages (ATMs), where feeding upregulated a local, low‐grade inflammatory response selectively in ATMs (Brykczynska et al, 2020).
Overall, the tissue environment is a critical determinant of tissue‐specific macrophage gene expression (Gautier et al, 2012; Gosselin et al, 2014; Lavin et al, 2014; Sakai et al, 2019) and its dysregulation contributes to a diverse range of inflammatory associated diseases. A paradigmatic example is represented by the atherosclerotic plaque microenvironment, where macrophages are exposed to oxidized lipids, cytokines, and cholesterol crystals that can greatly affect their activation state. In this peculiar environment, macrophages ingest and accumulate oxidized lipid particles and become overloaded foam cells (Ross, 1993). Foam cells display an activated state as (oxidized) lipids act as TLR agonists, leading to the activation of a proinflammatory response. In a recent report, the oxidized phospholipids (collectively known as oxPAPC) have been shown to induce a unique hypermetabolic state in macrophages exposed to LPS (Di Gioia et al, 2020). oxPAPC are produced at injury sites and are also part of the oxidized low‐density lipoproteins (oxLDL) that promote atherosclerosis (Bochkov et al, 2002; Berliner et al, 2009; Que et al, 2018). When primary macrophages are primed with LPS and exposed to oxPAPC, glycolysis is increased but without the classical decrease in OXPHOS that occurs in response to LPS alone (Di Gioia et al, 2020). From a mechanistic point of view, macrophages exposed to oxPAPC and LPS are able to feed the TCA cycle with glutamine and favor the accumulation of oxaloacetate (OAA) in the cytoplasm, as a result of increased cytoplasmic metabolism of citrate. In turn, OAA accumulation stabilizes HIF1α and increases IL‐1β production, by inactivating HIF prolyl hydroxylases which target HIF1α for rapid proteasomal degradation (Koivunen et al, 2007; Di Gioia et al, 2020). In addition, oxPAPC exposure promotes the transcriptional activation of Acly, driving a hyperinflammatory phenotype (Di Gioia et al, 2020). This mechanism is relevant in vivo, as similar metabolic adaptations occur in atherosclerotic mice and in hypercholesterolemic human subjects (Baardman et al, 2020; Di Gioia et al, 2020).
Overall, the main concept discussed above is that the exposure to oxidized phospholipids, altering metabolism and consequently inflammatory gene transcription, can drive a hypermetabolic, hyperinflammatory macrophage phenotype, which is critical in inflammation‐associated diseases (Di Gioia et al, 2020).
A long‐term perspective: modeling macrophage activation
While major technical advances led to the identification of mechanisms controlling signal‐induced transcriptional and metabolic changes in activated macrophages, a rigorous, quantitative, and system‐wide integration of these two regulatory layers is yet to be established.
While extremely valuable for the systematic description of cellular behavior, genome‐wide, and high‐throughput data are usually combined and interpreted using associative statistical approaches (such as correlations among variables, clustering, and grouping), that are useful to provide an unbiased description of biological systems but is also intrinsically not suitable to predict changes in cell states. Notably, data‐driven associations do not necessarily imply causality among the variables under investigation and therefore they are not necessarily informative of the molecular mechanism underlying a given phenotype. The application of these methodologies is in fact most appropriate to describe and interpret genome‐wide transcriptional changes. Thus, TF‐mediated gene expression, within a specific timeframe, can be considered as not constrained by most other changes observed in the cell. To clarify this concept, the activation of one gene (e.g., TNF) does not generate any intrinsic limitation to the activation of a second gene (e.g., IL1B). However, such approximation is not suitable to interpret changes in metabolites detected in high‐throughput metabolomics as those result from the adjustment of thousands of interlinked reactions all participating in the consumption and production of common metabolite pools. Indeed, while high‐throughput transcriptomics assesses the absolute and relative levels of expression gene by gene, scalable high‐throughput techniques reporting flux rates for all individual biochemical reactions within a cell at once are yet to be established.
Nevertheless, recent advances have highlighted the possibility to integrate a variety of omics datasets in the context of interpretable and predictive human models (Bordbar et al, 2014; Brunk et al, 2018; Robinson et al, 2020). A central tenet in these approaches is that all simultaneous molecular circuits occurring in a cell function coherently as they are all operating under countless constraints. Constraints can be divided into specific categories, namely physical–chemical (e.g., conservation of mass, elements, energy), spatial (e.g., cellular compartmentalization of molecules), environmental (e.g., nutrients, temperature, pH), and regulatory constraints, and they are all inviolable. Constraint‐based modeling and reconstruction analysis (COBRA) relates to approaches aiming to collect, integrate, and analyze in a bottom‐up (i.e., operator‐supervised) fashion all known biochemical transformations belonging to the network under investigation (Palsson, 2015). COBRA approaches have been used mostly for metabolic networks, including those of macrophages, as the thousands of known biochemical reactions occurring in cells can be explicitly formulated and combined in mathematical terms. These strategies have already been adopted in key works to reconstruct macrophage metabolic networks and coherently integrate condition‐specific changes (Bordbar et al, 2010; Bordbar et al, 2012; Jha et al, 2015).
Far from being a mere technological and modeling tour de force, this approach would instead provide researchers with accurate predictions of system‐level changes in macrophage biology that may also have translational impact, in principle enabling controlled and highly predictable reprogramming of macrophage properties. However, stoichiometric COBRA models face challenges when modeling dynamic cellular states. In this context, it is critical to complement COBRA models with kinetic models assembled to simulate changes in metabolite concentrations over time combining stoichiometric, reaction rate laws, kinetic parameters, and enzyme concentrations (Volkova et al, 2020). Nevertheless, given the amount of information required to construct a kinetic model, this strategy has so far been successfully adopted only to model cells with low complexity such as prokaryotes. Recent computational efforts provide promising solutions to model dynamic cellular states for complex genome‐wide networks exploiting the integration of time‐resolved multi‐omics datasets with constrained models, in order to predict reaction flux rates (for an overview see Basler et al (2018)). Time‐series metabolomics have been integrated to model flux distribution in red blood cells (Bordbar et al, 2017) and similar approaches have been applied to model changes in the transition from naïve to primed murine pluripotent stem cells (Chandrasekaran et al, 2017).
The development and application of similar strategies to time‐resolved multi‐omics datasets describing the process of macrophage activation are yet to be tested. These approaches will shed light on cause–effect relationships between changes in signaling, transcriptional, and metabolic changes from the early activation phase to the long‐term phenotype of polarized macrophages, providing a critical contribution to unraveling outstanding questions in this field (Box 1).
Conflict of interest
The authors declare that they have no conflict of interest.
Box 1. In need of answers.
What are the mechanisms by which dynamic changes in concentration of metabolites during macrophage activation trigger gene‐specific transcriptional responses rather than global effects?
What is the subcellular localization of key metabolites impacting epigenomic and transcriptional responses of macrophages exposed to different stimuli? Since the abundance and/or the distribution of metabolites might considerably change, it is crucial to measure metabolite fluctuations within distinct subcellular compartments.
Given the heterogeneity of macrophage functional states, to what extent do metabolic and transcriptional programs show heterogeneity in dynamic in vivo settings?
What is the full range and functional impact of chromatin modifications in the nuclear compartment that are impacted by dynamic changes in metabolite availability? A wide variety of novel modifications is increasingly being discovered, but their functional impact on transcriptional regulation requires extensive additional work.
How will integrated system‐level analyses change our understanding of macrophage activation? While a plethora of transcriptomics studies has been performed in many different macrophage types, also at single‐cell levels, metabolomic approaches reporting flux rates for all individual biochemical reactions at once are yet to be generated. In this way, system‐level changes can be accurately predicted, explaining the different behavior of polarized macrophages in the early activation phase compared with their long‐term phenotype.
Acknowledgments
The authors regret for being unable, due to length restriction, to cite all relevant studies and thank the reviewers for their helpful comments. The work related to the topic of this review in our laboratory is supported by the European Research Council (ERC grant 692789 MEDICI to G.N.) and Marie Sklodowska‐Curie Actions (MSCA‐IF “MetChromTx” ID 789792 to F.G.). F.P. is a PhD student within the European School of Molecular Medicine (SEMM).
EMBO reports (2021) 22: e53251.
See the Glossary for abbreviations used in this article.
Contributor Information
Gioacchino Natoli, Email: gioacchino.natoli@ieo.it.
Serena Ghisletti, Email: serenamarialuisa.ghisletti@ieo.it.
References
- Amit I, Winter DR, Jung S (2016) The role of the local environment and epigenetics in shaping macrophage identity and their effect on tissue homeostasis. Nat Immunol 17: 18–25 [DOI] [PubMed] [Google Scholar]
- Anstey NM, Weinberg JB, Hassanali MY, Mwaikambo ED, Manyenga D, Misukonis MA, Arnelle DR, Hollis D, McDonald M, Granger DL (1996) Nitric oxide in Tanzanian children with malaria: inverse relationship between malaria severity and nitric oxide production/nitric oxide synthase type 2 expression. J Exp Med 184: 557–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariyannur PS, Moffett JR, Madhavarao CN, Arun P, Vishnu N, Jacobowitz DM, Hallows WC, Denu JM, Namboodiri AM (2010) Nuclear‐cytoplasmic localization of acetyl coenzyme a synthetase‐1 in the rat brain. J Comp Neurol 518: 2952–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artyomov MN, Sergushichev A, Schilling JD (2016) Integrating immunometabolism and macrophage diversity. Semin Immunol 28: 417–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baardman J, Verberk SGS, van der Velden S, Gijbels MJJ, van Roomen CPPA, Sluimer JC, Broos JY, Griffith GR, Prange KHM, van Weeghel Met al (2020) Macrophage ATP citrate lyase deficiency stabilizes atherosclerotic plaques. Nat Commun 11: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JD, Diotallevi M, Nicol T, McNeill E, Shaw A, Chuaiphichai S, Hale A, Starr A, Nandi M, Stylianou Eet al (2019) Nitric Oxide modulates metabolic remodeling in inflammatory macrophages through TCA cycle regulation and itaconate accumulation. Cell Rep 28: 218–230.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bambouskova M, Gorvel L, Lampropoulou V, Sergushichev A, Loginicheva E, Johnson K, Korenfeld D, Mathyer ME, Kim H, Huang LHet al (2018) Electrophilic properties of itaconate and derivatives regulate the IkappaBzeta‐ATF3 inflammatory axis. Nature 556: 501–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barish GD, Yu RT, Karunasiri M, Ocampo CB, Dixon J, Benner C, Dent AL, Tangirala RK, Evans RM (2010) Bcl‐6 and NF‐kappaB cistromes mediate opposing regulation of the innate immune response. Genes Dev 24: 2760–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrat FJ, Crow MK, Ivashkiv LB (2019) Interferon target‐gene expression and epigenomic signatures in health and disease. Nat Immunol 20: 1574–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler G, Fernie AR, Nikoloski Z (2018) Advances in metabolic flux analysis toward genome‐scale profiling of higher organisms. Biosci Rep 38: BSR20170224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman BC, Butterfield GE, Wolfel EE, Casazza GA, Lopaschuk GD, Brooks GA (1999) Evaluation of exercise and training on muscle lipid metabolism. Am J Physiol 276: E106–E117 [DOI] [PubMed] [Google Scholar]
- Berliner JA, Leitinger N, Tsimikas S (2009) The role of oxidized phospholipids in atherosclerosis. J Lipid Res 50: S207–S212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkov VN, Kadl A, Huber J, Gruber F, Binder BR, Leitinger N (2002) Protective role of phospholipid oxidation products in endotoxin‐induced tissue damage. Nature 419: 77–81 [DOI] [PubMed] [Google Scholar]
- Bohn T, Rapp S, Luther N, Klein M, Bruehl T‐J, Kojima N, Aranda Lopez P, Hahlbrock J, Muth S, Endo Set al (2018) Tumor immunoevasion via acidosis‐dependent induction of regulatory tumor‐associated macrophages. Nat Immunol 19: 1319–1329 [DOI] [PubMed] [Google Scholar]
- Bonnardel J, T’Jonck W, Gaublomme D, Browaeys R, Scott CL, Martens L, Vanneste B, De Prijck S, Nedospasov SA, Kremer Aet al (2019) Stellate cells, hepatocytes, and endothelial cells imprint the kupffer cell identity on monocytes colonizing the liver macrophage niche. Immunity 51: 638–654.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordbar A, Lewis NE, Schellenberger J, Palsson BO, Jamshidi N (2010) Insight into human alveolar macrophage and M. tuberculosis interactions via metabolic reconstructions. Mol Syst Biol 6: 422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordbar A, Mo ML, Nakayasu ES, Schrimpe‐Rutledge AC, Kim Y‐M, Metz TO, Jones MB, Frank BC, Smith RD, Peterson SNet al (2012) Model‐driven multi‐omic data analysis elucidates metabolic immunomodulators of macrophage activation. Mol Syst Biol 8: 558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordbar A, Monk JM, King ZA, Palsson BO (2014) Constraint‐based models predict metabolic and associated cellular functions. Nat Rev Genet 15: 107–120 [DOI] [PubMed] [Google Scholar]
- Bordbar A, Yurkovich JT, Paglia G, Rolfsson O, Sigurjónsson ÓE, Palsson BO (2017) Elucidating dynamic metabolic physiology through network integration of quantitative time‐course metabolomics. Sci Rep 7: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukouris AE, Zervopoulos SD, Michelakis ED (2016) Metabolic enzymes moonlighting in the nucleus: metabolic regulation of gene transcription. Trends Biochem Sci 41: 712–730 [DOI] [PubMed] [Google Scholar]
- Brooks GA (2009) Cell‐cell and intracellular lactate shuttles. J Physiol 587: 5591–5600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunk E, Sahoo S, Zielinski DC, Altunkaya A, Dräger A, Mih N, Gatto F, Nilsson A, Preciat Gonzalez GA, Aurich MKet al (2018) Recon3D enables a three‐dimensional view of gene variation in human metabolism. Nat Biotechnol 36: 272–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska U, Geigges M, Wiedemann SJ, Dror E, Boni‐Schnetzler M, Hess C, Donath MY, Paro R (2020) Distinct transcriptional responses across tissue‐resident macrophages to short‐term and long‐term metabolic challenge. Cell Rep 30: 1627–1643 [DOI] [PubMed] [Google Scholar]
- Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB (2015) Intracellular alpha‐ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518: 413–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran S, Zhang J, Sun Z, Zhang L, Ross CA, Huang YC, Asara JM, Li H, Daley GQ, Collins JJ (2017) Comprehensive mapping of pluripotent stem cell metabolism using dynamic genome‐scale network modeling. Cell Rep 21: 2965–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A, Barak Y, Nagy L, Liao D, Tontonoz P, Evans RM (2001) PPAR‐gamma dependent and independent effects on macrophage‐gene expression in lipid metabolism and inflammation. Nat Med 7: 48–52 [DOI] [PubMed] [Google Scholar]
- Chen J, Ivashkiv LB (2010) IFN‐gamma abrogates endotoxin tolerance by facilitating Toll‐like receptor‐induced chromatin remodeling. Proc Natl Acad Sci USA 107: 19438–19443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S‐C, Quintin J, Cramer RA, Shepardson KM, Saeed S, Kumar V, Giamarellos‐Bourboulis EJ, Martens JH, Rao NA, Aghajanirefah A (2014) mTOR‐and HIF‐1α–mediated aerobic glycolysis as metabolic basis for trained immunity. Science 345: 1250684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouchani ET, Methner C, Nadtochiy SM, Logan A, Pell VR, Ding S, James AM, Cochemé HM, Reinhold J, Lilley KSet al (2013) Cardioprotection by S‐nitrosation of a cysteine switch on mitochondrial complex I. Nat Med 19: 753–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M (2009) Lysine acetylation targets protein complexes and co‐regulates major cellular functions. Science 325: 834–840 [DOI] [PubMed] [Google Scholar]
- Clementi E, Brown GC, Feelisch M, Moncada S (1998) Persistent inhibition of cell respiration by nitric oxide: crucial role of S‐nitrosylation of mitochondrial complex I and protective action of glutathione. Proc Natl Acad Sci USA 95: 7631–7636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colegio OR, Chu N‐Q, Szabo AL, Chu T, Rhebergen AM, Jairam V, Cyrus N, Brokowski CE, Eisenbarth SC, Phillips GMet al (2014) Functional polarization of tumour‐associated macrophages by tumour‐derived lactic acid. Nature 513: 559–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comerford S, Huang Z, Du X, Wang Y, Cai L, Witkiewicz A, Walters H, Tantawy M, Fu A, Manning Het al (2014) Acetate dependence of tumors. Cell 159: 1591–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes T, Metallo CM (2021) Exploring the evolutionary roots and physiological function of itaconate. Curr Opin Biotechnol 68: 144–150 [DOI] [PubMed] [Google Scholar]
- Cordes T, Wallace M, Michelucci A, Divakaruni AS, Sapcariu SC, Sousa C, Koseki H, Cabrales P, Murphy AN, Hiller Ket al (2016) Immunoresponsive gene 1 and itaconate inhibit succinate dehydrogenase to modulate intracellular succinate levels. J Biol Chem 291: 14274–14284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covarrubias AJ, Aksoylar HI, Yu J, Snyder NW, Worth AJ, Iyer SS, Wang J, Ben‐Sahra I, Byles V, Polynne‐Stapornkul Tet al (2016) Akt‐mTORC1 signaling regulates Acly to integrate metabolic input to control of macrophage activation. Elife 5: e11612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer T, Yamanishi Y, Clausen BE, Förster I, Pawlinski R, Mackman N, Haase VH, Jaenisch R, Corr M, Nizet Vet al (2003) HIF‐1α is essential for myeloid cell‐mediated inflammation. Cell 112: 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czimmerer Z, Daniel B, Horvath A, Rückerl D, Nagy G, Kiss M, Peloquin M, Budai MM, Cuaranta‐Monroy I, Simandi Zet al (2018) The transcription factor STAT6 mediates direct repression of inflammatory enhancers and limits activation of alternatively polarized macrophages. Immunity 48: 75–90.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietl K, Renner K, Dettmer K, Timischl B, Eberhart K, Dorn C, Hellerbrand C, Kastenberger M, Kunz‐Schughart LA, Oefner PJet al (2010) Lactic acid and acidification inhibit TNF secretion and glycolysis of human monocytes. J Immunol 184: 1200–1209 [DOI] [PubMed] [Google Scholar]
- Doxaki C, Kampranis SC, Eliopoulos AG, Spilianakis C, Tsatsanis C (2015) Coordinated regulation of miR‐155 and miR‐146a genes during induction of endotoxin tolerance in macrophages. J Immunol 195: 5750–5761 [DOI] [PubMed] [Google Scholar]
- Ebert BL, Firth JD, Ratcliffe PJ (1995) Hypoxia and mitochondrial inhibitors regulate expression of glucose transporter‐1 via distinct Cis‐acting sequences. J Biol Chem 270: 29083–29089 [DOI] [PubMed] [Google Scholar]
- Escoubet‐Lozach L, Benner C, Kaikkonen MU, Lozach J, Heinz S, Spann NJ, Crotti A, Stender J, Ghisletti S, Reichart Det al (2011) Mechanisms establishing TLR4‐responsive activation states of inflammatory response genes. PLoS Genet 7: e1002401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser‐von Bieren J, Mosconi I, Guiet R, Piersgilli A, Volpe B, Chen F, Gause WC, Seitz A, Verbeek JS, Harris NL (2013) Antibodies trap tissue migrating helminth larvae and prevent tissue damage by driving IL‐4Ralpha‐independent alternative differentiation of macrophages. PLoS Pathog 9: e1003771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts B, Amiel E, van der Windt GJ, Freitas TC, Chott R, Yarasheski KE, Pearce EL, Pearce EJ (2012) Commitment to glycolysis sustains survival of NO‐producing inflammatory dendritic cells. Blood 120: 1422–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Krautkramer KA, Feldman JL, Denu JM (2015) Metabolic regulation of histone post‐translational modifications. ACS Chem Biol 10: 95–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Han J, Pearce SF, Silverstein RL, Gotto AM Jr, Hajjar DP, Nicholson AC (2000) Induction of CD36 expression by oxidized LDL and IL‐4 by a common signaling pathway dependent on protein kinase C and PPAR‐gamma. J Lipid Res 41: 688–696 [PubMed] [Google Scholar]
- Fischer K, Hoffmann P, Voelkl S, Meidenbauer N, Ammer J, Edinger M, Gottfried E, Schwarz S, Rothe G, Hoves Set al (2007) Inhibitory effect of tumor cell‐derived lactic acid on human T cells. Blood 109: 3812–3819 [DOI] [PubMed] [Google Scholar]
- Foster SL, Hargreaves DC, Medzhitov R (2007) Gene‐specific control of inflammation by TLR‐induced chromatin modifications. Nature 447: 972–978 [DOI] [PubMed] [Google Scholar]
- Freemerman AJ, Johnson AR, Sacks GN, Milner JJ, Kirk EL, Troester MA, Macintyre AN, Goraksha‐Hicks P, Rathmell JC, Makowski L (2014) Metabolic reprogramming of macrophages: glucose transporter 1 (GLUT1)‐mediated glucose metabolism drives a proinflammatory phenotype. J Biol Chem 289: 7884–7896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuzumi M, Shinomiya H, Shimizu Y, Ohishi F, Utsumi S (1996) Endotoxin‐induced enhancement of glucose influx into murine peritoneal macrophages via GLUT1. Infect Immun 64: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney DO, Jennings EQ, Anderson CC, Marentette JO, Shi T, Schou Oxvig A‐M, Streeter MD, Johannsen M, Spiegel DA, Chapman Eet al (2020) Non‐enzymatic lysine lactoylation of glycolytic enzymes. Cell Chem Biol 27: 206–213.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier EL, Ivanov S, Williams JW, Huang S‐C, Marcelin G, Fairfax K, Wang PL, Francis JS, Leone P, Wilson DBet al (2014) Gata6 regulates aspartoacylase expression in resident peritoneal macrophages and controls their survival. J Exp Med 211: 1525–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov Set al (2012) Gene‐expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol 13: 1118–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei C‐Let al (2010) Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Ginhoux F, Guilliams M (2016) Tissue‐resident macrophage ontogeny and homeostasis. Immunity 44: 439–449 [DOI] [PubMed] [Google Scholar]
- Di Gioia M, Spreafico R, Springstead JR, Mendelson MM, Joehanes R, Levy D, Zanoni I (2020) Endogenous oxidized phospholipids reprogram cellular metabolism and boost hyperinflammation. Nat Immunol 21: 42–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Natoli G (2016) Molecular control of activation and priming in macrophages. Nat Immunol 17: 26–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetze K, Walenta S, Ksiazkiewicz M, Kunz‐Schughart LA, Mueller‐Klieser W (2011) Lactate enhances motility of tumor cells and inhibits monocyte migration and cytokine release. Int J Oncol 39: 453–463 [DOI] [PubMed] [Google Scholar]
- Gosselin D, Link VM, Romanoski C, Fonseca G, Eichenfield D, Spann N, Stender J, Chun H, Garner H, Geissmann Fet al (2014) Environment drives selection and function of enhancers controlling tissue‐specific macrophage identities. Cell 159: 1327–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottfried E, Kunz‐Schughart LA, Ebner S, Mueller‐Klieser W, Hoves S, Andreesen R, Mackensen A, Kreutz M (2006) Tumor‐derived lactic acid modulates dendritic cell activation and antigen expression. Blood 107: 2013–2021 [DOI] [PubMed] [Google Scholar]
- Harber KJ, de Goede KE, Verberk SGS, Meinster E, de Vries HE, van Weeghel M, de Winther MPJ, Van den Bossche J (2020) Succinate Is an Inflammation‐Induced Immunoregulatory Metabolite in Macrophages. Metabolites 10: 372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R (2009) Control of inducible gene expression by signal‐dependent transcriptional elongation. Cell 138: 129–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK (2010) Simple combinations of lineage‐determining transcription factors prime cis‐regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, Glass CK (2015) The selection and function of cell type‐specific enhancers. Nat Rev Mol Cell Biol 16: 144–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhaeuser F, Sattler UG, Mueller‐Klieser W (2011) Lactate: a metabolic key player in cancer. Cancer Res 71: 6921–6925 [DOI] [PubMed] [Google Scholar]
- Hooftman A, Angiari S, Hester S, Corcoran SE, Runtsch MC, Ling C, Ruzek MC, Slivka PF, McGettrick AF, Banahan Ket al (2020) The Immunomodulatory metabolite itaconate modifies NLRP3 and inhibits inflammasome activation. Cell Metab 32: 468–478.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooftman A, O'Neill LAJ (2019) The Immunomodulatory Potential of the Metabolite Itaconate. Trends Immunol 40: 687–698 [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Pirinen E, Auwerx J (2012) Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol 13: 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S‐C, Everts B, Ivanova Y, O'Sullivan D, Nascimento M, Smith AM, Beatty W, Love‐Gregory L, Lam WY, O'Neill CMet al (2014) Cell‐intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat Immunol 15: 846–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SCC, Smith AM, Everts B, Colonna M, Pearce EL, Schilling JD, Pearce EJ (2016) Metabolic reprogramming mediated by the mTORC2‐IRF4 signaling axis is essential for macrophage alternative activation. Immunity 45: 817–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infantino V, Iacobazzi V, Palmieri F, Menga A (2013) ATP‐citrate lyase is essential for macrophage inflammatory response. Biochem Biophys Res Comm 440: 105–111 [DOI] [PubMed] [Google Scholar]
- Jha A, Huang S‐C, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, Chmielewski K, Stewart K, Ashall J, Everts Bet al (2015) Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42: 419–430 [DOI] [PubMed] [Google Scholar]
- Jung J, Zeng H, Horng T (2019) Metabolism as a guiding force for immunity. Nat Cell Biol 21: 85–93 [DOI] [PubMed] [Google Scholar]
- Kaikkonen M, Spann N, Heinz S, Romanoski C, Allison K, Stender J, Chun H, Tough D, Prinjha R, Benner Cet al (2013) Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell 51: 310–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiran N, Ceperuelo‐Mallafré V, Calvo E, Hernández‐Alvarez MI, Ejarque M, Núñez‐Roa C, Horrillo D, Maymó‐Masip E, Rodríguez MM, Fradera Ret al (2019) SUCNR1 controls an anti‐inflammatory program in macrophages to regulate the metabolic response to obesity. Nat Immunol 20: 581–592 [DOI] [PubMed] [Google Scholar]
- Kim JW, Tchernyshyov I, Semenza GL, Dang CV (2006) HIF‐1‐mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab 3: 177–185 [DOI] [PubMed] [Google Scholar]
- Kinnaird A, Zhao S, Wellen KE, Michelakis ED (2016) Metabolic control of epigenetics in cancer. Nat Rev Cancer 16: 694–707 [DOI] [PubMed] [Google Scholar]
- Koivunen P, Tiainen P, Hyvärinen J, Williams KE, Sormunen R, Klaus SJ, Kivirikko KI, Myllyharju J (2007) An endoplasmic reticulum transmembrane prolyl 4‐hydroxylase is induced by hypoxia and acts on hypoxia‐inducible factor α. J Biol Chem 282: 30544–30552 [DOI] [PubMed] [Google Scholar]
- Lampropoulou V, Sergushichev A, Bambouskova M, Nair S, Vincent E, Loginicheva E, Cervantes‐Barragan L, Ma X, Huang S‐C, Griss Tet al (2016) Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab 24: 158–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer MR, Fry CJ, Peterson CL, Denu JM (2002) Modulating acetyl‐CoA binding in the GCN5 family of histone acetyltransferases. J Biol Chem 277: 27337–27344 [DOI] [PubMed] [Google Scholar]
- Langston PK, Nambu A, Jung J, Shibata M, Aksoylar HI, Lei J, Xu P, Doan MT, Jiang H, MacArthur MRet al (2019) Glycerol phosphate shuttle enzyme GPD2 regulates macrophage inflammatory responses. Nat Immunol 20: 1186–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OD, Courtney AD, Vassilev A, Marzilli LA, Cotter RJ, Nakatani Y, Cole PA (2000) p300/CBP‐associated factor histone acetyltransferase processing of a peptide substrate. Kinetic analysis of the catalytic mechanism. J Biol Chem 275: 21953–21959 [DOI] [PubMed] [Google Scholar]
- Lauterbach MA, Hanke JE, Serefidou M, Mangan MSJ, Kolbe C‐C, Hess T, Rothe M, Kaiser R, Hoss F, Gehlen Jet al (2019) Toll‐like receptor signaling rewires macrophage metabolism and promotes histone acetylation via ATP‐citrate lyase. Immunity 51: 997–1011.e7 [DOI] [PubMed] [Google Scholar]
- Lavin Y, Winter D, Blecher‐Gonen R, David E, Keren‐Shaul H, Merad M, Jung S, Amit I (2014) Tissue‐resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159: 1312–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Carrer A, Shah S, Snyder N, Wei S, Venneti S, Worth A, Yuan Z‐F, Lim H‐W, Liu Set al (2014) Akt‐dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab 20: 306–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Yang ES, Park JW (2003) Inactivation of NADP+‐dependent isocitrate dehydrogenase by peroxynitrite. Implications for cytotoxicity and alcohol‐induced liver injury. J Biol Chem 278: 51360–51371 [DOI] [PubMed] [Google Scholar]
- Liu N, Luo J, Kuang D, Xu S, Duan Y, Xia Y, Wei Z, Xie X, Yin B, Chen Fet al (2019) Lactate inhibits ATP6V0d2 expression in tumor‐associated macrophages to promote HIF‐2alpha‐mediated tumor progression. J Clin Invest 129: 631–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P‐S, Wang H, Li X, Chao T, Teav T, Christen S, Di Conza G, Cheng W‐C, Chou C‐H, Vavakova Met al (2017) α‐ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat Immunol 18: 985–994 [DOI] [PubMed] [Google Scholar]
- Liu X, Wang L, Zhao K, Thompson PR, Hwang Y, Marmorstein R, Cole PA (2008) The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature 451: 846–850 [DOI] [PubMed] [Google Scholar]
- Mages J, Dietrich H, Lang R (2008) A genome‐wide analysis of LPS tolerance in macrophages. Immunobiology 212: 723–737 [DOI] [PubMed] [Google Scholar]
- Mancino A, Termanini A, Barozzi I, Ghisletti S, Ostuni R, Prosperini E, Ozato K, Natoli G (2015) A dual cis‐regulatory code links IRF8 to constitutive and inducible gene expression in macrophages. Genes Dev 29: 394–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A (2016) Reflections on immunological nomenclature: in praise of imperfection. Nat Immunol 17: 215–216 [DOI] [PubMed] [Google Scholar]
- Mariño G, Pietrocola F, Eisenberg T, Kong Y, Malik S, Andryushkova A, Schroeder S, Pendl T, Harger A, Niso‐Santano Met al (2014) Regulation of autophagy by cytosolic acetyl‐coenzyme A. Mol Cell 53: 710–725 [DOI] [PubMed] [Google Scholar]
- Mass E, Ballesteros I, Farlik M, Halbritter F, Gunther P, Crozet L, Jacome‐Galarza Ce, Handler K, Klughammer J, Kobayashi Yet al (2016) Specification of tissue‐resident macrophages during organogenesis. Science 353: aaf4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden BA, Purohit S (1977) Itaconate, an isocitrate lyase‐directed inhibitor in Pseudomonas indigofera. J Bacteriol 131: 136–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedev AE, Kopydlowski KM, Vogel SN (2000) Inhibition of lipopolysaccharide‐induced signal transduction in endotoxin‐tolerized mouse macrophages: dysregulation of cytokine, chemokine, and toll‐like receptor 2 and 4 gene expression. J Immunol 164: 5564–5574 [DOI] [PubMed] [Google Scholar]
- Michelucci A, Cordes T, Ghelfi J, Pailot A, Reiling N, Goldmann O, Binz T, Wegner A, Tallam A, Rausell Aet al (2013) Immune‐responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc Natl Acad Sci USA 110: 7820–7825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EL, Ryan DG, Prag HA, Dikovskaya D, Menon D, Zaslona Z, Jedrychowski MP, Costa ASH, Higgins M, Hams Eet al (2018) Itaconate is an anti‐inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 556: 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli S, Natoli G (2017) Transcriptional determination and functional specificity of myeloid cells: making sense of diversity. Nat Rev Immunol 17: 595–607 [DOI] [PubMed] [Google Scholar]
- Morioka S, Perry JSA, Raymond MH, Medina CB, Zhu Y, Zhao L, Serbulea V, Onengut‐Gumuscu S, Leitinger N, Kucenas Set al (2018) Efferocytosis induces a novel SLC program to promote glucose uptake and lactate release. Nature 563: 714–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussaieff A, Rouleau M, Kitsberg D, Cohen M, Levy G, Barasch D, Nemirovski A, Shen‐Orr S, Laevsky I, Amit Met al (2015) Glycolysis‐mediated changes in acetyl‐CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab 21: 392–402 [DOI] [PubMed] [Google Scholar]
- Munder M, Eichmann K, Modolell M (1998) Alternative metabolic states in murine macrophages reflected by the nitric oxide synthase/arginase balance: competitive regulation by CD4+ T cells correlates with Th1/Th2 phenotype. J Immunol 160: 5347–5354 [PubMed] [Google Scholar]
- Murray P, Allen J, Biswas S, Fisher E, Gilroy D, Goerdt S, Gordon S, Hamilton J, Ivashkiv L, Lawrence Tet al (2014) Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41: 14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahid MA, Pauley KM, Satoh M, Chan EKL (2009) miR‐146a Is critical for endotoxin‐induced tolerance implication in innate immunity. J Biol Chem 284: 34590–34599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson VL, Nguyen HC, Garcìa‐Cañaveras JC, Briggs ER, Ho WY, DiSpirito JR, Marinis JM, Hill DA, Lazar MA (2018) PPARγ is a nexus controlling alternative activation of macrophages via glutamine metabolism. Genes Dev 32: 1035–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson S, Bonecini‐Almeida MdG, Lapa e Silva Jr, Nathan C, Xie Qw, Mumford R, Weidner Jr, Calaycay J, Geng J, Boechat Net al (1996) Inducible nitric oxide synthase in pulmonary alveolar macrophages from patients with tuberculosis. J Exp Med 183: 2293–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolt B, Tu F, Wang X, Ha T, Winter R, Williams DL, Li C (2018) Lactate and immunosuppression in sepsis. Shock 49: 120–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M, Liu J, Rovira II, Gonzalez‐Hurtado E, Lee J, Wolfgang MJ, Finkel T (2016) Fatty acid oxidation in macrophage polarization. Nat Immunol 17: 216–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegaard JI, Ricardo‐Gonzalez RR, Goforth MH, Morel CR, Subramanian V, Mukundan L, Red Eagle A, Vats D, Brombacher F, Ferrante AWet al (2007) Macrophage‐specific PPARgamma controls alternative activation and improves insulin resistance. Nature 447: 1116–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill LAJ, Artyomov MN (2019) Itaconate: the poster child of metabolic reprogramming in macrophage function. Nat Rev Immunol 19: 273–281 [DOI] [PubMed] [Google Scholar]
- O'Neill LA, Kishton RJ, Rathmell J (2016) A guide to immunometabolism for immunologists. Nat Rev Immunol 16: 553–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill LA, Pearce EJ (2016) Immunometabolism governs dendritic cell and macrophage function. J Exp Med 213: 15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G (2013) Latent enhancers activated by stimulation in differentiated cells. Cell 152: 157–171 [DOI] [PubMed] [Google Scholar]
- Palmieri EM, Gonzalez‐Cotto M, Baseler WA, Davies LC, Ghesquière B, Maio N, Rice CM, Rouault TA, Cassel T, Higashi RMet al (2020) Nitric oxide orchestrates metabolic rewiring in M1 macrophages by targeting aconitase 2 and pyruvate dehydrogenase. Nat Commun 11: 698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson BO (2015) Systems biology: constraint‐based reconstruction and analysis. Cambridge: Cambridge University Press; [Google Scholar]
- Palsson‐McDermott E, Curtis A, Goel G, Lauterbach M, Sheedy F, Gleeson L, van den Bosch M, Quinn S, Domingo‐Fernandez R, Johnston Det al (2015) Pyruvate kinase M2 regulates Hif‐1α activity and IL‐1β induction and is a critical determinant of the warburg effect in LPS‐activated macrophages. Cell Metab 21: 65–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peruzzotti‐Jametti L, Bernstock JD, Vicario N, Costa ASH, Kwok CK, Leonardi T, Booty LM, Bicci I, Balzarotti B, Volpe Get al (2018) Macrophage‐derived extracellular succinate licenses neural stem cells to suppress chronic neuroinflammation. Cell Stem Cell 22: 355–368.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesce JT, Ramalingam TR, Mentink‐Kane MM, Wilson MS, El Kasmi KC, Smith AM, Thompson RW, Cheever AW, Murray PJ, Wynn TA (2009) Arginase‐1‐expressing macrophages suppress Th2 cytokine‐driven inflammation and fibrosis. PLoS Pathog 5: e1000371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter K, Rehli M, Singer K, Renner‐Sattler K, Kreutz M (2015) Lactic acid delays the inflammatory response of human monocytes. Biochem Biophys Res Commun 457: 412–418 [DOI] [PubMed] [Google Scholar]
- Phan AT, Goldrath AW, Glass CK (2017) Metabolic and epigenetic coordination of T cell and macrophage immunity. Immunity 46: 714–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo V, Curina A, Genua M, Ghisletti S, Simonatto M, Sabo A, Amati B, Ostuni R, Natoli G (2017) Opposing macrophage polarization programs show extensive epigenomic and transcriptional cross‐talk. Nat Immunol 18: 530–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucino V, Bombardieri M, Pitzalis C, Mauro C (2017) Lactate at the crossroads of metabolism, inflammation, and autoimmunity. Eur J Immunol 47: 14–21 [DOI] [PubMed] [Google Scholar]
- Puleston DJ, Buck MD, Klein Geltink RI, Kyle RL, Caputa G, O’Sullivan D, Cameron AM, Castoldi A, Musa Y, Kabat AMet al (2019) Polyamines and eIF5A hypusination modulate mitochondrial respiration and macrophage activation. Cell Metab 30: 352–363.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Qin KE, Zhang Y, Jia W, Chen Y, Cheng BO, Peng L, Chen N, Liu Y, Zhou Wet al (2019) S‐glycosylation‐based cysteine profiling reveals regulation of glycolysis by itaconate. Nat Chem Biol 15: 983–991 [DOI] [PubMed] [Google Scholar]
- Qin W, Zhang Y, Tang H, Liu D, Chen Y, Liu Y, Wang C (2020) Chemoproteomic profiling of itaconation by bioorthogonal probes in inflammatory macrophages. J Am Chem Soc 142: 10894–10898 [DOI] [PubMed] [Google Scholar]
- Que X, Hung M‐Y, Yeang C, Gonen A, Prohaska TA, Sun X, Diehl C, Määttä A, Gaddis DE, Bowden Ket al (2018) Oxidized phospholipids are proinflammatory and proatherogenic in hypercholesterolaemic mice. Nature 558: 301–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes G, Van den Bergh R, De Baetselier P, Ghassabeh GH (2005) Arginase‐1 and Ym1 are markers for murine, but not human, alternatively activated myeloid cells. J Immunol 174: 6561–6562 [DOI] [PubMed] [Google Scholar]
- Ramirez‐Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST (2009) A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 138: 114–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AR, Payne EC, Younger N, Karlinsey JE, Thomas VC, Becker LA, Navarre WW, Castor ME, Libby SJ, Fang FC (2011) Multiple targets of nitric oxide in the tricarboxylic acid cycle of Salmonella enterica serovar typhimurium. Cell Host Microbe 10: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rius J, Guma M, Schachtrup C, Akassoglou K, Zinkernagel AS, Nizet V, Johnson RS, Haddad GG, Karin M (2008) NF‐κB links innate immunity to the hypoxic response through transcriptional regulation of HIF‐1α. Nature 453: 807–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JL, Kocabaş P, Wang H, Cholley P‐E, Cook D, Nilsson A, Anton M, Ferreira R, Domenzain I, Billa Vet al (2020) An atlas of human metabolism. Sci Signal 13: eaaz1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Prados J‐C, Través PG, Cuenca J, Rico D, Aragonés J, Martín‐Sanz P, Cascante M, Boscá L (2010) Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J Immunol 185: 605–614 [DOI] [PubMed] [Google Scholar]
- Ronai Z (1993) Glycolytic‐enzymes as DNA‐binding proteins. Int J Biochem 25: 1073–1076 [DOI] [PubMed] [Google Scholar]
- Ross R (1993) The pathogenesis of atherosclerosis: a perspective for the 1990s. Nature 362: 801–809 [DOI] [PubMed] [Google Scholar]
- Russell DG, Huang L, VanderVen BC (2019) Immunometabolism at the interface between macrophages and pathogens. Nat Rev Immunol 19: 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed S, Quintin J, Kerstens HHD, Rao NA, Aghajanirefah A, Matarese F, Cheng S‐C, Ratter J, Berentsen K, van der Ent MAet al (2014) Epigenetic programming of monocyte‐to‐macrophage differentiation and trained innate immunity. Science 345: 1251086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai M, Troutman TD, Seidman JS, Ouyang Z, Spann NJ, Abe Y, Ego KM, Bruni CM, Deng Z, Schlachetzki JCMet al (2019) Liver‐derived signals sequentially reprogram myeloid enhancers to initiate and maintain kupffer cell identity. Immunity 51: 655–670.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G (2007) The histone H3 lysine‐27 demethylase Jmjd3 links inflammation to inhibition of polycomb‐mediated gene silencing. Cell 130: 1083–1094 [DOI] [PubMed] [Google Scholar]
- Schneemann M, Schoedon G, Hofer S, Blau N, Guerrero L, Schaffner A (1993) Nitric oxide synthase is not a constituent of the antimicrobial armature of human mononuclear phagocytes. J Infect Dis 167: 1358–1363 [DOI] [PubMed] [Google Scholar]
- Sciacovelli M, Gonçalves E, Johnson TI, Zecchini VR, da Costa ASH, Gaude E, Drubbel AV, Theobald SJ, Abbo SR, Tran MGBet al (2016) Fumarate is an epigenetic modifier that elicits epithelial‐to‐mesenchymal transition. Nature 537: 544–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley JJ, Baker RG, Mohamed G, Bruns T, Hayden MS, Deshmukh SD, Freedberg DE, Ghosh S (2018) Induction of innate immune memory via microRNA targeting of chromatin remodelling factors. Nature 559: 114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seim GL, Britt EC, John SV, Yeo FJ, Johnson AR, Eisenstein RS, Pagliarini DJ, Fan J (2019) Two‐stage metabolic remodelling in macrophages in response to lipopolysaccharide and interferon‐γ stimulation. Nat Metab 1: 731–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL, Jiang BH, Leung SW, Passantino R, Concordet JP, Maire P, Giallongo A (1996) Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia‐inducible factor 1. J Biol Chem 271: 32529–32537 [DOI] [PubMed] [Google Scholar]
- Shen L, Hu P, Zhang Y, Ji Z, Shan X, Ni L, Ning Na, Wang J, Tian He, Shui Get al (2021) Serine metabolism antagonizes antiviral innate immunity by preventing ATP6V0d2‐mediated YAP lysosomal degradation. Cell Metab 33: 971–987.e6 [DOI] [PubMed] [Google Scholar]
- Sivanand S, Rhoades S, Jiang Q, Lee JV, Benci J, Zhang J, Yuan S, Viney I, Zhao S, Carrer Aet al (2017) Nuclear Acetyl‐CoA production by ACLY promotes homologous recombination. Mol Cell 67: 252–265.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Clair E, Wilkinson WE, Lang T, Sanders L, Misukonis MA, Gilkeson GS, Pisetsky DS, Granger D, Weinberg JB (1996) Increased expression of blood mononuclear cell nitric oxide synthase type 2 in rheumatoid arthritis patients. J Exp Med 184: 1173–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadhouders R, Filion GJ, Graf T (2019) Transcription factors and 3D genome conformation in cell‐fate decisions. Nature 569: 345–354 [DOI] [PubMed] [Google Scholar]
- Strelko CL, Lu WY, Dufort FJ, Seyfried TN, Chiles TC, Rabinowitz JD, Roberts MF (2011) Itaconic acid is a mammalian metabolite induced during macrophage activation. J Am Chem Soc 133: 16386–16389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutendra G, Kinnaird A, Dromparis P, Paulin R, Stenson TH, Haromy A, Hashimoto K, Zhang N, Flaim E, Michelakis ED (2014) A nuclear pyruvate dehydrogenase complex is important for the generation of Acetyl‐CoA and histone acetylation. Cell 158: 84–97 [DOI] [PubMed] [Google Scholar]
- Swain A, Bambouskova M, Kim H, Andhey PS, Duncan D, Auclair K, Chubukov V, Simons DM, Roddy TP, Stewart KMet al (2020) Comparative evaluation of itaconate and its derivatives reveals divergent inflammasome and type I interferon regulation in macrophages. Nat Metab 2: 594–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind Let al (2009) Conversion of 5‐methylcytosine to 5‐hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, McCaffery JM, Irizarry RA, Boeke JD (2006) Nucleocytosolic acetyl‐coenzyme a synthetase is required for histone acetylation and global transcription. Mol Cell 23: 207–217 [DOI] [PubMed] [Google Scholar]
- Tannahill GM, Curtis AM, Adamik J, Palsson‐McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NHet al (2013) Succinate is an inflammatory signal that induces IL‐1β through HIF‐1α. Nature 496: 238–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner KG, Langer MR, Denu JM (2000) Kinetic mechanism of human histone acetyltransferase P/CAF. Biochemistry 39: 11961–11969 [DOI] [PubMed] [Google Scholar]
- van Teijlingen BN, Pearce EJ (2020) Cell‐intrinsic metabolic regulation of mononuclear phagocyte activation: findings from the tip of the iceberg. Immunol Rev 295: 54–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas AC, Mattila JT (2014) “Of mice and men”: arginine metabolism in macrophages. Front Immunol 5: 479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument‐Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y (2006) Histone demethylation by a family of JmjC domain‐containing proteins. Nature 439: 811–816 [DOI] [PubMed] [Google Scholar]
- Van den Bossche J, Baardman J, Otto N, van der Velden S, Neele A, van den Berg S, Luque‐Martin R, Chen H‐J, Boshuizen M, Ahmed Met al (2016) Mitochondrial dysfunction prevents repolarization of inflammatory macrophages. Cell Rep 17: 684–696 [DOI] [PubMed] [Google Scholar]
- Vats D, Mukundan L, Odegaard JI, Zhang L, Smith KL, Morel CR, Greaves DR, Murray PJ, Chawla A (2006) Oxidative metabolism and PGC‐1β attenuate macrophage‐mediated inflammation. Cell Metab 4: 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan V, Pradhan P, Braud L, Fuchs HR, Gueler F, Motterlini R, Foresti R, Immenschuh S (2019) Human and murine macrophages exhibit differential metabolic responses to lipopolysaccharide‐a divergent role for glycolysis. Redox Biol 22: 101147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkova S, Matos MRA, Mattanovich M, de Mas IM (2020) Metabolic modelling as a framework for metabolomics data integration and analysis. Metabolites 10: 303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Zhang S, Vuckovic I, Jeon R, Lerman A, Folmes CD, Dzeja PP, Herrmann J (2018) Glycolytic stimulation is not a requirement for M2 macrophage differentiation. Cell Metab 28: 463–475.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB (2009) ATP‐citrate lyase links cellular metabolism to histone acetylation. Science 324: 1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams NC, O'Neill LAJ (2018) A role for the krebs cycle intermediate citrate in metabolic reprogramming in innate immunity and inflammation. Front Immunol 9: 141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JY, Huang TW, Hsieh YT, Wang YF, Yen CC, Lee GL, Yeh CC, Peng YJ, Kuo YY, Wen HTet al (2020) Cancer‐derived succinate promotes macrophage polarization and cancer metastasis via succinate receptor. Mol Cell 77: 213–227.e215 [DOI] [PubMed] [Google Scholar]
- Yang ES, Richter C, Chun JS, Huh TL, Kang SS, Park JW (2002) Inactivation of NADP(+)‐dependent isocitrate dehydrogenase by nitric oxide. Free Radic Biol Med 33: 927–937 [DOI] [PubMed] [Google Scholar]
- Yona S, Kim K‐W, Wolf Y, Mildner A, Varol D, Breker M, Strauss‐Ayali D, Viukov S, Guilliams M, Misharin Aet al (2013) Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38: 79–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurdagul A Jr, Subramanian M, Wang X, Crown SB, Ilkayeva OR, Darville L, Kolluru GK, Rymond CC, Gerlach BD, Zheng Zeet al (2020) Macrophage metabolism of apoptotic cell‐derived arginine promotes continual efferocytosis and resolution of injury. Cell Metab 31: 518–533.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaslona Z, O'Neill LAJ (2020) Cytokine‐like roles for metabolites in immunity. Mol Cell 78: 814–823 [DOI] [PubMed] [Google Scholar]
- Zhang DI, Tang Z, Huang HE, Zhou G, Cui C, Weng Y, Liu W, Kim S, Lee S, Perez‐Neut Met al (2019) Metabolic regulation of gene expression by histone lactylation. Nature 574: 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Roeder RG, Luo Y (2003) S phase activation of the histone H2B promoter by OCA‐S, a coactivator complex that contains GAPDH as a key component. Cell 114: 255–266 [DOI] [PubMed] [Google Scholar]


