Abstract
Differentiated cells across multiple species and organs can re‐enter the cell cycle to aid in injury‐induced tissue regeneration by a cellular program called paligenosis. Here, we show that activating transcription factor 3 (ATF3) is induced early during paligenosis in multiple cellular contexts, transcriptionally activating the lysosomal trafficking gene Rab7b. ATF3 and RAB7B are upregulated in gastric and pancreatic digestive‐enzyme‐secreting cells at the onset of paligenosis Stage 1, when cells massively induce autophagic and lysosomal machinery to dismantle differentiated cell morphological features. Their expression later ebbs before cells enter mitosis during Stage 3. Atf3 –/– mice fail to induce RAB7‐positive autophagic and lysosomal vesicles, eventually causing increased death of cells en route to Stage 3. Finally, we observe that ATF3 is expressed in human gastric metaplasia and during paligenotic injury across multiple other organs and species. Thus, our findings indicate ATF3 is an evolutionarily conserved gene orchestrating the early paligenotic autodegradative events that must occur before cells are poised to proliferate and contribute to tissue repair.
Keywords: acinar‐ductal metaplasia, BHLHA15, plasticity, RAB5, spasmolytic polypeptide‐expressing metaplasia
Subject Categories: Autophagy & Cell Death, Regenerative Medicine
Pancreatic and gastric exocrine cells regenerate after injury through a process called paligenosis. Injury‐induced ATF3 promotes autophagy and cell autodegradation via transcriptional activation of Rab7.
Introduction
Injury in adult organisms can trigger differentiated cells to reprogram to an embryonic‐like, progenitor state. The change from a differentiated cell to a proliferating progenitor‐like cell has been shown in nearly every organ, for example, kidney (Kusaba et al, 2014), lung (Hong et al, 2001; Tata et al, 2013), stomach (Nam et al, 2010; Leushacke et al, 2017; Radyk et al, 2018; Willet et al, 2018; Meyer et al, 2019), pancreas (Mills & Sansom, 2015; Willet et al, 2018), intestine (van Es et al, 2012; Jones et al, 2019), and liver (Raven et al, 2017; Chen et al, 2020). As early as 1900, it was proposed that such cellular retooling was a fundamental feature shared by cells, suggesting an evolutionarily conserved program that mature cells could invoke to reroute their energy from performing differentiated cell functions (like secretion of digestive enzymes) to instead undergo mitosis (Adami, 1900).
In the last few years, we have begun to delineate the shared features of injury‐induced conversion of mature cells to regenerating proliferative cells (a process termed paligenosis) (Willet et al, 2018). Paligenosis progresses via three distinct, sequential stages: (i) autodegradation of mature cell structures via lysosomal and autophagy pathways, (ii) shift to expression of a progenitor or embryonic gene network, and (iii) re‐entry into the cell cycle. Much evidence indicates metaplasias largely arise via cell plasticity (often paligenosis), in particular spasmolytic polypeptide‐expressing metaplasia (SPEM) in the stomach and acinar‐to‐ductal metaplasia (ADM) in the pancreas (Storz, 2017; Burclaff & Mills, 2018; Meyer & Goldenring, 2018). Metaplasia is a lesion characterized by expression of progenitor/embryonic genes, often with increased proliferation, ultimately leading to increased risk for tumorigenesis. Recently, it has become clear that metaplasia in healthy organisms is an evolutionarily conserved injury response that is usually simply a transient, regenerative state that allows tissue to recruit mature cells to help repair injury after large‐scale damage (Goldenring, 2018; Saenz & Mills, 2018; Saenz et al, 2018; Burclaff et al, 2020). It is likely that increased cancer risk occurs when metaplasia is chronic and/or with repeated injury (Amieva & Peek, 2016; Giroux & Rustgi, 2017; Storz, 2017; Jin & Mills, 2019; Wroblewski et al, 2019). Accordingly, loss of a key paligenosis gene, Ddit4, increases tumorigenesis by allowing cells to proliferate even with DNA damage present (Miao et al, 2021). This further demonstrates the need to better understand paligenosis as a process that guards against cancer initiation.
Given the evolutionarily conserved role of paligenosis as a mechanism underlying both regeneration and tumorigenesis, we attempted to identify the core set of genes that might be largely dedicated to executing paligenosis. Such paligenosis genes are expected to be evolutionarily conserved, shared across all tissue types, and required only for injury‐induced proliferation of differentiated cells but not in dedicated stem cells or during embryonic development. We recently showed that two genes meeting those criteria are Ddit4 (mentioned above) and Ifrd1. DDIT4 is invoked early in paligenosis to quench mTORC1, which must be shut off for the massive autophagy and lysosomal activity of Stage 1, while IFRD1 is essential to restore mTORC1 activity for entry into S phase (Miao et al, 2020b).
Here, we dissect the conserved genetic program that drives the activation of the autodegradation activity in Stage 1. We focus on mechanisms conserved across two exocrine cell populations in two different organs: gastric zymogenic chief cells (ZCs) and pancreatic acinar cells (ACs). These, almost entirely postmitotic, cells live months to years in the absence of injury. ZCs and ACs have large, polarized morphologies with abundant rough endoplasmic reticulum (rER) and numerous, large secretory granules. Their secretory architecture is maintained almost entirely by a two‐step transcriptional regulation program with X‐box‐binding protein 1 (Huh et al, 2010; Hess et al, 2011) transcriptionally activating the secretory granule trafficking transcription factor MIST1 (encoded by Bhlha15) (Pin et al, 2001; Ramsey et al, 2007; Lo et al, 2017; Dekaney et al, 2019). MIST1 induces and maintains expression of numerous transcriptional targets whose gene products in turn coordinate cellular organization, vesicular trafficking, and stress responses (Kowalik et al, 2007). For example, MIST1 binds to and directly upregulates the pro‐exocrine secretory apparatus Rab proteins RAB26 and RAB3D by direct transcriptional upregulation of their genes (Tian et al, 2010; Mills & Taghert, 2012; Jin & Mills, 2014). MIST1 is an unique transcription factor; in that, it acts as a scaling factor for cell architecture and maturity by regulating abundance of cell trafficking proteins (Mills & Taghert, 2012).
Rab proteins, like RAB26, RAB3D, and RAB27, are important in gastric zymogenic chief cells and pancreatic acinar cells because they help maintain secretory cell function and cell state (Ohnishi et al, 1996; Tian et al, 2010; Jin & Mills, 2014; Hou et al, 2015; Lo et al, 2017). Other Rab GTPases, like RAB5, RAB7, and RAB9, organize the endolysosomal system. RAB5 traffics the early endosome, RAB7 traffics the late endosome, lysosome, and autophagosome, and RAB9 traffics late endosomes as they transition to the trans‐Golgi network (Wandinger‐Ness & Zerial, 2014; Langemeyer et al, 2018). There are over 70 human Rab GTPases, which exhibit some overlap in function but can also exhibit distinct functions based on intrinsic characteristics or through protein cofactors. For example, RAB7A and RAB7B are both known to control late endosome maturation and lysosome biogenesis (Guerra & Bucci, 2016); however, they cannot be defined as isoforms because they do not share conserved motifs (Pereira‐Leal & Seabra, 2000). The function of RAB7A is generally restricted to late endosomes and facilitates the transition to lysosomes (Bucci et al, 2000), while RAB7B can control endosomal transport to the trans‐Golgi network (Guerra & Bucci, 2016) and can interact with cystine protease ATG4B to modulate autophagy (Kjos et al, 2017).
We will show that activating transcription factor 3 (ATF3) is induced during paligenotic injury in diverse organs, working to reverse the MIST1 cell maturation program early in Stage 1. ATF3 is a highly conserved transcription factor with clear orthologues even in primitive multicellular organisms like sea anemones (Nematostella vectensis; https://omabrowser.org/oma/home/) (Altenhoff et al, 2018). ATF3 is rapidly induced following injury, and Atf3 loss in multiple organisms and tissues generally exhibits phenotypes after injury (Hartman et al, 2004; Li et al, 2010; Wang et al, 2012; Brooks et al, 2015; Gey et al, 2016; Fazio et al, 2017; Zhao et al, 2017; Zhu et al, 2018). ATF3 has also been shown to transcriptionally repress MIST1 (Fazio et al, 2017).
Here, we show that ATF3 has a surprising role in vesicle trafficking: it binds to and transcriptionally activates Rab7b (also previously identified as Rab42; Itoh et al, 2006; Marubashi & Fukuda, 2020), which helps govern the maturation of late endosomes to lysosomes (Yang et al, 2004; Progida et al, 2010; Stroupe, 2018). We show RAB7 expression increases dramatically in early paligenotic exocrine cells, forming large accumulations of vesicles and co‐labeling, as expected, with autophagic and lysosomal vesicles. Using ATF3 overexpression and knockdown constructs in human epithelial cells and Atf3 −/− mice, we show that autodegradative machinery depends on ATF3, partly through transcriptional regulation of Rab7b. Owing to failed degradation in Stage 1, Atf3 −/− mice also have markedly aberrant later stages of paligenosis with increased cell death and decreased proliferation and repair in paligenosis Stage 3. We also find ATF3 expression focally in human SPEM, suggesting ATF3 is a staple in early transitions to metaplasia. These data indicate that ATF3 is a key evolutionarily conserved paligenosis gene that acts as one of the first proteins activated in paligenotic injury. It works principally to induce autodegradation, in part, by increasing the lysosome and autophagy coordinator Rab7b.
Results
Degradation of specialized cell features follows exocrine cell injury
To better characterize the early, autodegradation phase of paligenosis at ultrastructural resolution, we identified the organelles and trafficking features that characterize the earliest morphological changes after injury. To induce stomach injury, we used high‐dose tamoxifen (HDT), which has been described by our laboratory and others to quickly and synchronously elicit a stomach injury and induce SPEM independent of estrogen receptor activity or sex of mice (Huh et al, 2012; Saenz et al, 2016; Leushacke et al, 2017; Keeley et al, 2019). To induce pancreas injury, we repeatedly treated with the secretagogue cerulein, which has long been described to cause damage to the exocrine pancreas (Adler et al, 1985; Niederau et al, 1985; Saluja et al, 1985; Willet et al, 2018). Injury timelines and how the time points correspond to paligenosis stages are indicated in Fig EV1A and B.
Figure EV1. Schemes for gastric and pancreatic injury treatments, and hydroxychloroquine treatment in stomach tissue to inhibit paligenosis.
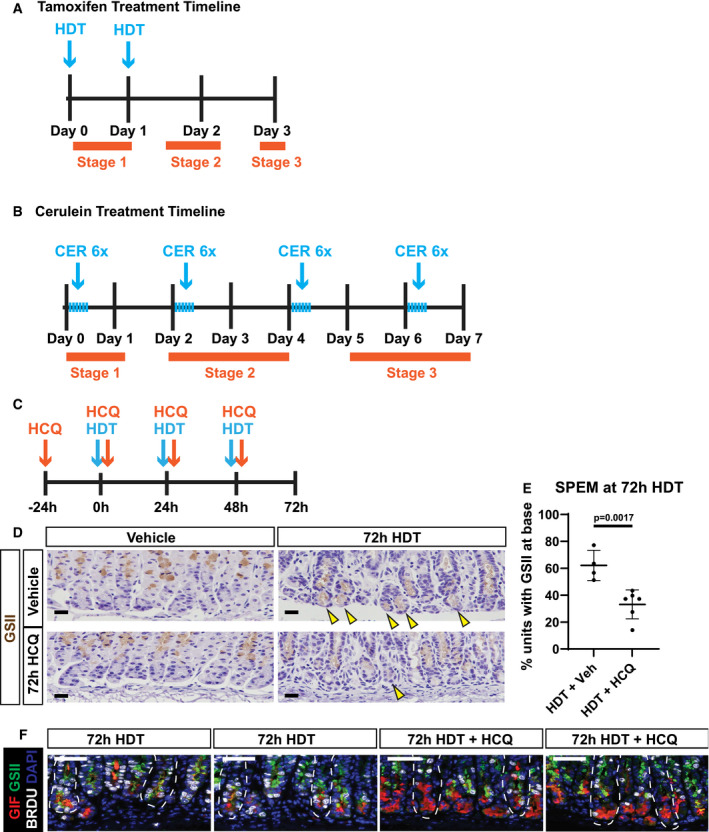
- Scheme of tamoxifen treatment timeline. Mice were administered high‐dose tamoxifen (HDT; 5 mg/20 g body weight) by intraperitoneal injection up to two times, and stomachs were harvested at the respective/indicated timepoint as per the experimental details. Day 3 of timeline is when peak SPEM is observed. Orange bars indicate each stage of paligenosis and where they correspond in the treatment timeline.
- Scheme of cerulein (CER) treatment timeline to induce pancreatitis. Mice were administered repeated cerulein injections by intraperitoneal injection. Six hourly injections of cerulein (50 μg/kg) were given every other day for up to 7 days (peak ADM). Orange bars indicate each stage of paligenosis and where they correspond in the treatment timeline.
- Scheme of hydroxychloroquine (HCQ) treatment timeline. Mice were pretreated with 60 mg/kg HCQ (or PBS) one day prior to high‐dose tamoxifen (HDT) administration alongside HCQ (60 mg/kg/day). Stomachs were harvested at day 3 (72 h) when peak SPEM is normally observed.
- Immunohistochemistry for GSII in gastric units to mark normal mucous neck cells (vehicle) and SPEM cells (72‐h HDT) in mice treated with vehicle, HDT, HCQ, or HDT + HCQ. Scale bars 20 µm. Counterstained with hematoxylin. Arrows mark GSII at the base of the stomach unit indicating SPEM.
- Quantification of the fraction of gastric units exhibiting SPEM (from EV1D). Each data point = mean of counts from 40 to 50 gastric glands from an individual mouse, n = 4–6 mice per treatment group; black line = mean of means from each mouse ± SEM. Significance by nonparametric two‐tailed t‐test.
- Immunofluorescence of gastric units; nuclei (DAPI, blue), chief cells (GIF, red), progenitor/metaplastic cells (GSII green + red GIF), and proliferative cells (BrdU, white) after treatment with HDT or HDT + HCQ for 72 h. 72‐h HDT units exhibit SPEM (GSII and GIF overlap), while 72‐h HDT+HCQ units show scant SPEM and full chief cell cytoplasm packed with GIF secretory granules. Scale bars 50 µm. Stomach unit base outlined by dashed white line.
We analyzed the initial downscaling stage of paligenosis via focused ion beam‐scanning electron microscopy (FIB‐SEM) three‐dimensional (3D) nanotomography (Fig 1A and Movies [Link], [Link], [Link], [Link]). FIB‐SEM of vehicle‐treated ZCs compared to 24‐h HDT‐treated ZCs revealed a dense accumulation of lysosomes and autolysosomes in the basal region of the cell, where the cell’s rER network and many mitochondria are concentrated. In addition to loss of rER, we saw exocrine granule degradation as well as swelling and fragmenting of mitochondria. The ultrastructural changes were corroborated by immunofluorescence at 6 and 24‐h HDT showing gradual loss of GIF+ exocrine granules coincident with increased LAMP1+ lysosomes (Fig 1B). Transmission electron microscopy (tEM) showed that ZCs following 24‐h HDT injury harbored increased lysosomes and double‐membrane bound vesicles (consistent with autophagosomes) filled with cell components, such as granules, ER, and cytosol (Fig 1C). Granular census was also reduced (Fig 1D), consistent with downscaling of mature cell architecture and an increase in single‐ and double‐membraned vesicles, which we summarize and quantify as autodegradation events (Fig 1E). Similarly, we saw decreased granule number and increased autodegradation events in pancreatic ACs following repeated cerulein (CER) injury for one day (Fig 1F–H). These results support our initial description (Willet et al, 2018) that Stage 1 of paligenosis consists of an increase in both lysosomal and autophagic degradation pathways and further clarifies which organelles are targeted for degradation.
Figure 1. Secretory cell structure is dismantled rapidly and in a choreographed pattern in paligenosis.
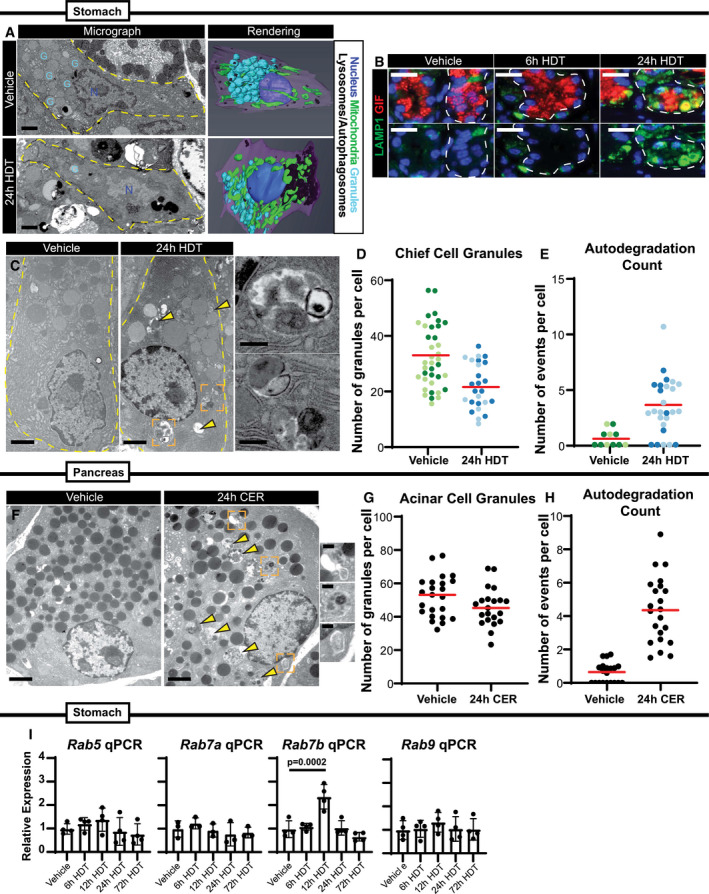
- Left—Single‐level electron micrograph images of zymogenic chief cells (ZCs) after 24‐h vehicle or high‐dose tamoxifen (HDT) treatment from FIB‐SEM 3D tomography. G = representative secretory granules; N = nuclei. Scale bar, 2 μm. Right—Stacked EM images rendered into 3D projections using Amira software. Cells outlined by dashed yellow line.
- Representative immunofluorescence images of stomach unit bases at homeostasis, 6‐hour, and 24‐h HDT stained with GIF (red, granules) and LAMP1 (green, lysosomes) Scale bar, 25 μm. Stomach unit base outlined by dashed white line.
- Representative TEM images of normal and injured ZCs (scale bar, 2 μm). Right (insets)‐higher magnification images of boxed regions from left show emergence of double‐membraned autophagic vesicles engulfing mature granules, mitochondria, and rough ER 24-h after HDT injury. Scale bar, 500 nm. Arrowheads mark vesicular degradation events in the cell lacking double membranes. Cells outlined by dashed yellow line.
- Graph of the number of zymogenic chief cell granules per cell in vehicle vs. 24‐h HDT treatment. Each data point = total granules in a cell in a single TEM section with cells from 2 separate mice plotted together (each cell is a data point; cells from the same mouse are grouped by color shade). Red line indicates mean across all the mice.
- Graph of the number of degradation events scored as in D. A degradation event was counted if electron dense‐cargo or organelles were surrounded by a single membrane (lysosome) or double membrane (autophagosome). Red line indicates mean across all the mice.
- Representative TEM images of normal and injured acinar cells. Scale bar, 2 μm. Right (insets)‐higher magnification images of boxed regions from left show emergence of double‐membraned autophagic vesicles engulfing mature granules, mitochondria, and rough ER 24-h after cerulein (CER) injury. Scale bar, 500 nm. Arrowheads mark degradation events without double membranes.
- Graph of the number of acinar cell granules per cell in vehicle vs. 24‐h CER treatment. Each data point = total granules in a cell in a single cell counted from one mouse. Red line indicates mean.
- Graph of the number of degradation events scored in vehicle vs. 24‐h CER treatment. Each data point = total events in a cell in a single cell counted from one mouse. Red line indicates mean.
- mRNA expression by qRT–PCR of Rab5, Rab7a, Rab7b, and Rab9. Significance determined by one‐way ANOVA, Tukey post‐hoc. Each data point represents the mean of triplicate technical replicates from a single mouse, n = 3–4 mice in independent experiments. Error bars denote standard deviation.
We wanted to determine whether lysosomes and autophagosomes are required for paligenosis progression in the stomach, so we treated mice with hydroxychloroquine (HCQ; 60 mg/kg/day). Intraperitoneal injection of HCQ dampens autophagic activity by blocking autophagosome–lysosome fusion (Rosenfeldt et al, 2013; Cao et al, 2017). A 3‐day course of HCQ significantly decreased the fraction of stomach units that exhibit SPEM (Fig EV1, EV2, EV3, EV4, EV5C–F). We also saw many ZCs that appeared to never downscale their granules (meaning they did not undergo the autodegradative processes Stage 1), as marked by sustained GIF expression and residually large apical cytoplasms (Fig EV1, EV2, EV3, EV4, EV5D–F). Thus, lysosomes and autophagosomes are necessary for Stage 1 of paligenosis and, consequently, for subsequent progression of cells to Stage 2, which is characterized by induction of SPEM markers.
Figure EV2. Protein expression of RABs in stomach and pancreas.
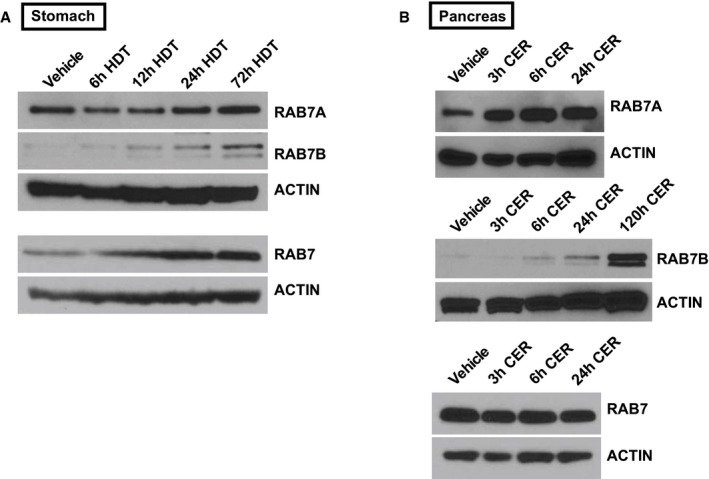
- RAB7A, RAB7B, RAB7 expression by Western blot in stomachs treated with high‐dose tamoxifen (HDT) compared to ACTIN loading control.
- RAB7A, RAB7B, RAB7 expression by Western blot in pancreata treated with cerulein (CER) compared to ACTIN loading control.
Figure EV3. Analysis of ATF3‐binding sites in the mouse Rab7b first intron.
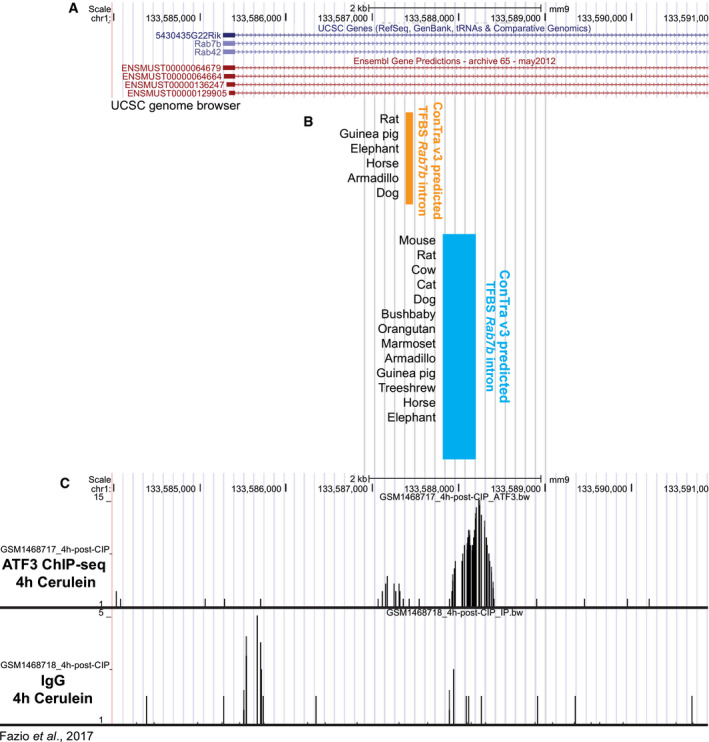
- Mouse Rab7b gene alignment using UCSC genome browser (NCBI37/mm9).
- Regions of predicted conserved ATF3 binding derived from ConTra v3. Orange indicates conserved ATF3 sites in one region of Rab7b and blue indicates a second region of conserved ATF3 sites downstream, but still in the first intron. Highlighted regions are aligned to the gene in (A). TFBS = transcription factor binding sites.
Figure EV4. Markers of Spasmolytic Polypeptide‐Expressing Metaplasia, cell proliferation, and cell death in the stomach following injury.
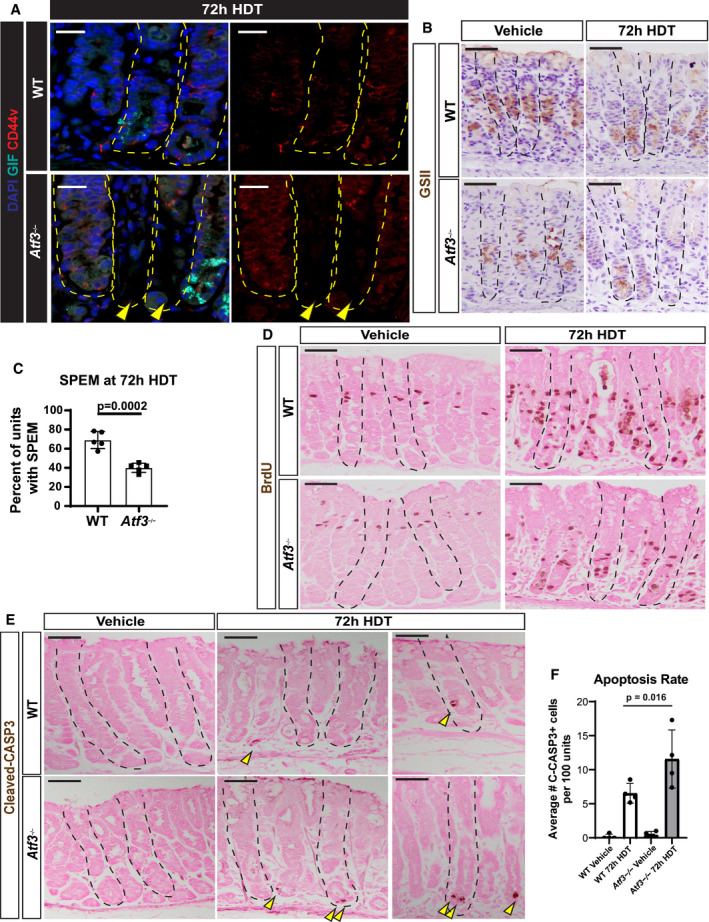
- Representative immunofluorescence of gastric units stained for DAPI (blue), gastric intrinsic factor (GIF, cyan), and CD44v (marker of spasmolytic polypeptide‐expressing metaplasia [SPEM] when expressed in cells at the unit base, red). WT (top) and Atf3 −/− (bottom) mice were treated with 72‐h high‐dose tamoxifen (HDT). Arrowhead indicates base dropout in Atf3 −/−stomach. Scale bars 20 µm. Stomach units outlined by dashed yellow line.
- Representative immunohistochemistry of GSII in gastric units to mark normal mucous neck cells (vehicle) and SPEM cells (72‐h HDT) in WT (top) and Atf3 −/− (bottom) mice. Scale bars 50 µm. Counterstained with hematoxylin. Stomach units outlined by dashed black line.
- Quantification of the percent of gastric units exhibiting SPEM as counted by the number of units with GSII at the unit base over the number of unit total. Each data point = mean of counts from 40 to 50 gastric glands from an individual mouse, n = 5 mice per treatment group. Significance by nonparametric two‐tailed t‐test. Error bars denote standard deviation.
- Representative immunohistochemistry of BrdU in WT (top) and Atf3KO (bottom) gastric units with vehicle or 72‐h high‐dose tamoxifen (HDT) treatment. BrdU administered 90 minutes prior to sacrifice to capture cells in S phase. Eosin counterstain. Scale bars 50 µm. Stomach units outlined by dashed black line.
- Representative immunohistochemistry of cleaved‐caspase 3 in gastric units of WT (top) and Atf3KO (bottom) mice when treated with vehicle or 72‐h high‐dose tamoxifen (HDT). Counterstained with eosin. Arrowheads point to ZCs with high expression of cleaved‐CASP3. Scale bars 50 µm. Stomach units outlined by dashed black line.
- Graph of the apoptosis rate following 72‐h HDT injury as counted by the number of cleaved‐caspase 3+ cells from a 40× field. Each data point = mean of counts from 40 to 50 gastric glands from an individual mouse, n = 4 mice per treatment group. Significance by nonparametric two‐tailed t‐test. Error bars denote standard deviation.
Figure EV5. Conservation of Atf3 expression across tissues after injury.
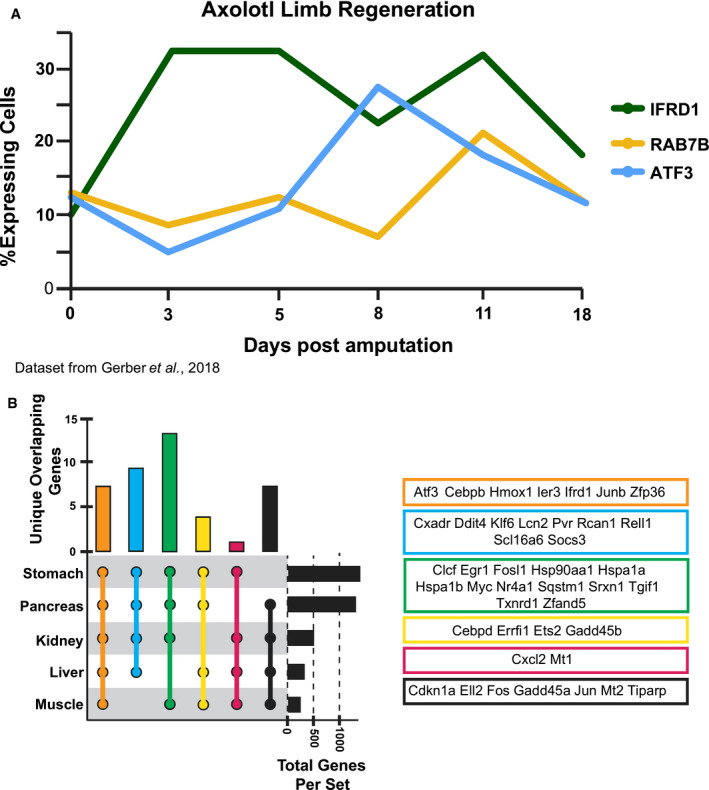
- Fraction of cells expressing ATF3, RAB7B, and IFRD1 in previously published single‐cell RNA‐sequencing of regenerating cells after axolotl limb amputation. IFRD1 is a previously published paligenosis gene known to be activated during Stage 1.
- Intersection of transcripts upregulated by paligenotic injury in 5 tissues. Only intersections of gene sets ≥ 4 and transcripts unique for that intersection are depicted. Unique genes from each intersection are listed above the UpSet plot depicting the intersection combination, the number of overlapping genes, and the total number of genes in the dataset.
The massive degradation events occurring early in paligenosis indicated that cellular membrane trafficking was dramatically altered and pointed to alterations in the Rab family of small GTPases. RABs facilitate vesicle and organelle movement and are especially important for cells with secretory functions, like exocrine cells in the stomach and pancreas (Johnson et al, 2004; Faust et al, 2008; Fukuda, 2008; Tian et al, 2010; Jin & Mills, 2014; Takahashi et al, 2017). RAB function can be regulated at the transcriptional level in exocrine cells (Johnson et al, 2004; Tian et al, 2010). Accordingly, we found a significant increase in Rab7b (though not Rab5, Rab7a, Rab9) mRNA at 12‐h HDT treatment: the time when the majority of ZCs are in Stage 1 of paligenosis (Fig 1I). Our analysis of a previously published dataset (Kowalik et al, 2007) also suggests that Rab7b is expressed in pancreatic acinar cells following cerulein‐induced pancreatitis. Together the findings suggest that, like Rab26 and Rab3d, Rab7b is transcriptionally controlled in exocrine cells.
ATF3 binds to and directly induces the Rab7b gene during paligenotic downscaling
While there was little RAB7 expression in vehicle‐treated stomach and pancreas, we observed large balloons and crescents of RAB7+ vesicles during stage 1 of paligenosis in stomach and pancreas (Fig 2A and D). Note that we have not found an antibody that can distinguish RAB7B from RAB7A in tissue sections, so we used total RAB7 antibody for these immunostaining studies. However, we favor the interpretation that the majority of the RAB7+ vesicles are likely RAB7B vesicles, given qRT–PCR and Western blot showed only Rab7b mRNA and RAB7B protein are induced consistently after paligenotic injury in pancreas and stomach (Fig EV2A and B). In contrast, there was no consistent pattern of increased expression in any of the following paligenotic injury scenarios in pancreas and stomach: total RAB7 protein by Western blot of whole tissue (Fig EV2A and B), Rab7a mRNA by qRT–PCR from whole tissue (Fig 1I), and RAB7A protein by Western blot of whole tissue (Fig EV2A and B).
Figure 2. ATF3 transcriptionally upregulates Rab7b early in paligenosis, and RAB7 protein is associated with large lysosomal aggregates.
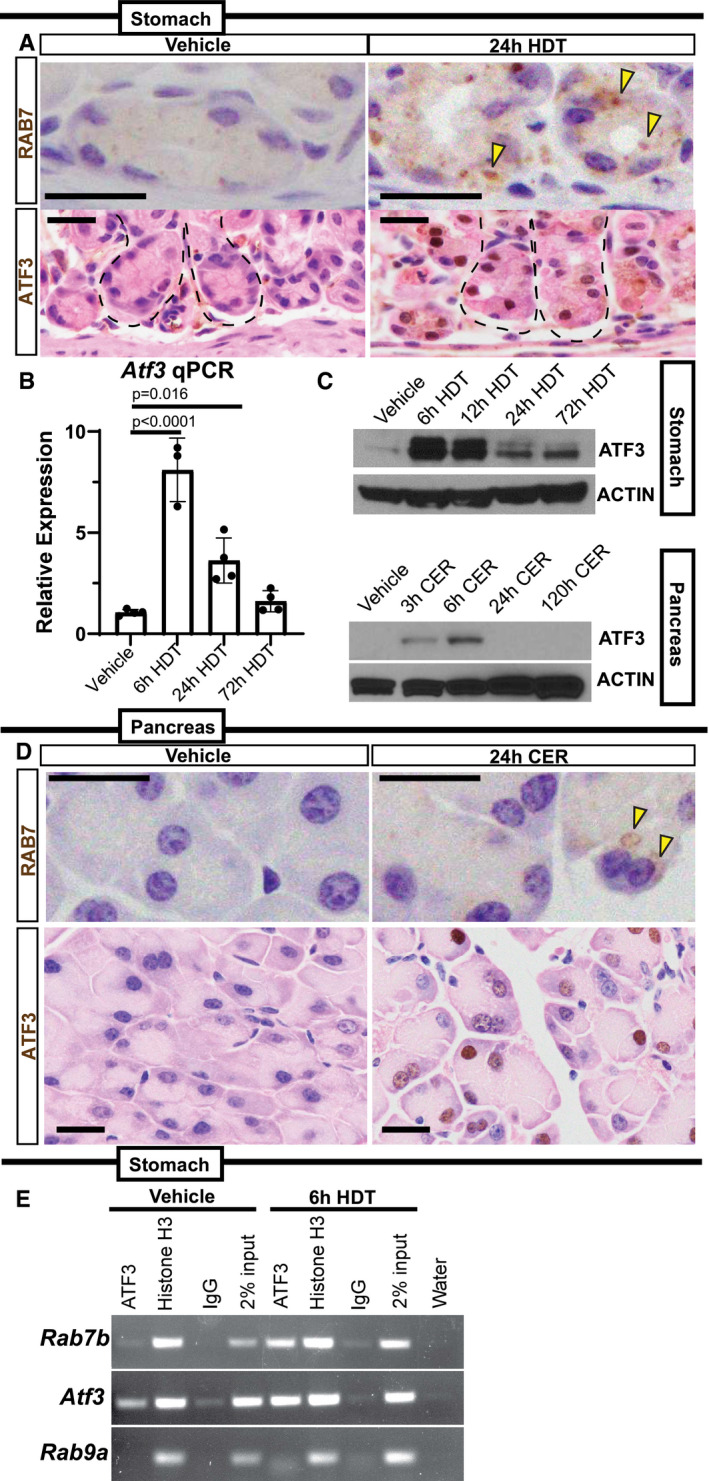
- Top—Representative RAB7 expression in base of stomach units (ZCs) in vehicle or 24‐h HDT by immunohistochemistry. Arrowheads point to RAB7 vesicle aggregates. Scale bar 20 µm. Bottom—Representative ATF3 expression in base of stomach units (ZCs) by immunohistochemistry. ATF3 appears in nuclei following 24‐h high‐dose tamoxifen (HDT). Scale bar 20 µm. Stomach unit base outlined by dashed black line.
- Atf3 mRNA expression by qRT–PCR in vehicle, 6, 24, and 72‐h HDT treatment. Each data point represents the mean of triplicate technical replicates from a single mouse, n = 3–4 mice in independent experiments. Significance determined by one‐way ANOVA, Tukey. Error bars denote standard deviation.
- Western blot of ATF3 expression over HDT and CER time courses with β‐Actin loading control.
- Top—Representative RAB7 staining in pancreatic acinar cells. Arrowheads indicate RAB7 vesicles. Scale bar 20 µm. Bottom—Representative ATF3 expression in pancreatic acinar cells. Nuclei express ATF3 after 24‐h cerulein (CER) injury. Scale bar 20 µm.
- ChIP‐PCR on mouse stomach tissue at vehicle and 6‐hour HDT. Chromatin probed with ATF3, Histone H3 antibodies, or a Rabbit IgG control. Also included is the 2% chromatin input and water to control for DNA contamination. Rab7b amplicon is from a conserved, putative ATF3‐binding site in the first intron. Amplicon from Atf3 is a positive control from a previously characterized ATF3‐binding site; negative control is from a site in Rab9a lacking putative ATF3‐binding motifs.
We reasoned that the transcriptional upregulation of Rab7b specifically within early paligenosis might give us a route to identify, for the first time, early transcriptional regulators of the paligenosis process. Our previous work showed that MIST1 activates Rab transcription (e.g., Rab3d and Rab26) by binding within well‐conserved cis‐regulatory regions within the first intron. Thus, we used the MEME Suite web server (Bailey et al, 2009) and FIMO (Find Individual Motif Occurrences) software (Grant et al, 2011) to identify transcription factor binding sites within the first intron of Rab7b. A putative ATF3‐binding motif was identified within this region. More detailed analysis using Contra v3 (Kreft et al, 2017) (http://bioit2.irc.ugent.be/contra/v3) (Fig EV3A–C) further corroborated that a potential ATF3 site was highly evolutionarily conserved across mammals. ATF3 was previously characterized in the early stages of cerulein‐induced pancreatitis in mice (Fazio et al, 2017; Data ref: Fazio et al, 2017) and our in silico‐predicted ATF3‐binding site matched a site from chromatin immunoprecipitation sequencing (ChIP‐Seq) analysis of genomic ATF3 binding early after cerulein injury from the published dataset (Fig EV3A–C). ATF3 was also an appealing target because it was previously described as one of the most highly upregulated genes in a mouse model of SPEM (Nozaki et al, 2008; Miao et al, 2020b). Thus, the data pointed to ATF3 as a key transcription factor early in paligenosis, potentially activating Rab7 to facilitate the massive autophagic changes in Stage 1.
Accordingly, ATF3 was rapidly induced specifically within ZCs and ACs following injury (Fig 2A and D), and Atf3 transcript and ATF3 protein were also both increased in whole tissue analysis of the stomach and pancreas (Fig 2B and C). ATF3 was dramatically increased early in paligenosis Stage 1, but its expression returned to almost homeostatic levels in paligenosis Stage 3. Moreover, ATF3 binding to the Rab7b gene in stomach tissue was confirmed using ChIP (Fig 2E) with primers designed to amplify genomic DNA including the consensus ATF3 intronic binding site described above (Fazio et al, 2017). We used an equivalent intronic region in the Rab9a gene (which has similar function and gene structure; Mackiewicz & Wyroba, 2009) as a negative control and a previously identified ATF3‐binding site within the Atf3 gene as a positive control (as ATF3 can transcriptionally regulate itself (Inoue et al, 2004)). As predicted, ATF3 specifically bound both to Atf3 and Rab7b (vs. non‐specific IgG and Rab9 negative controls) with binding greatly increased early (6 h) after injury. Thus, induction of ATF3 activity and the consequent activation of Rab7 occurs very early following injury; indeed, it is the earliest conserved molecular event we have so far identified in paligenosis.
ATF3 can act as a transcriptional activator or repressor depending on the gene and the context (Hai et al, 1999). To confirm that ATF3 activated Rab7b and to test if their molecular interaction was conserved across species, we used AGS cells (a human gastric epithelial line) to generate stable cell clones that overexpressed ATF3 or stably repressed ATF3 using shRNAs. AGS cells did not express substantial endogenous ATF3, though cells expressing anti‐ATF3 shRNA (ATF3‐KD) still expressed less (Fig 3A). ATF3‐KD caused statistically significant decrease in RAB7B mRNA and fewer cells with detectable RAB7 protein, and conversely, when ATF3 protein was overexpressed (ATF3‐OE), we saw an induction of RAB7 protein and mRNA (Fig 3A and B). Of note, many of the ATF3‐OE cells showed large RAB7+ globular and crescentic vesicular formations phenocopying those seen in vivo in paligenotic murine cells (Fig 3C). Our in vivo data suggested that ATF3 could induce autophagy in part via directly increasing RAB7 abundance. Supporting our hypothesis, we noted increased cathepsin D (CTSD)‐positive lysosomes in cells with higher expression of ATF3 correlating consequently with increased RAB7B (Fig 3C). To determine whether RAB7B was sufficient, even in the absence of ATF3, to induce these large vesicles observed in vivo and in vitro, we forced expression of FLAG‐tagged, WT RAB7B in ATF3‐KD cells. Figure 3D shows the large vesicular phenotype is even more dramatic with RAB7B overexpression. On the other hand, transfection with RAB7BT22N, a GTPase‐dead RAB7B mutant (GTPase cycling is critical for RABs to traffic and maintain vesicular compartments), abrogated the phenotype (Fig 3D). These results are consistent with previous reports that RAB7BT22N impairs endolysosomal trafficking necessary for maturation of cathepsin D, which is activated in its pro‐form in the lysosome (Progida et al, 2010). Taken together, ATF3 is sufficient to transcriptionally activate RAB7B, and RAB7B itself (whether ATF3‐induced or transfected) is sufficient to activate the endolysosomal pathway to facilitate cell autodegradation. While ATF3, a transcription factor, may also induce other genes that activate autodegradation pathways, our results indicate RAB7B appears to be a principal target in paligenosis.
Figure 3. ATF3 transcriptionally upregulates RAB7B in vitro to govern formation of autodegradative vesicles.
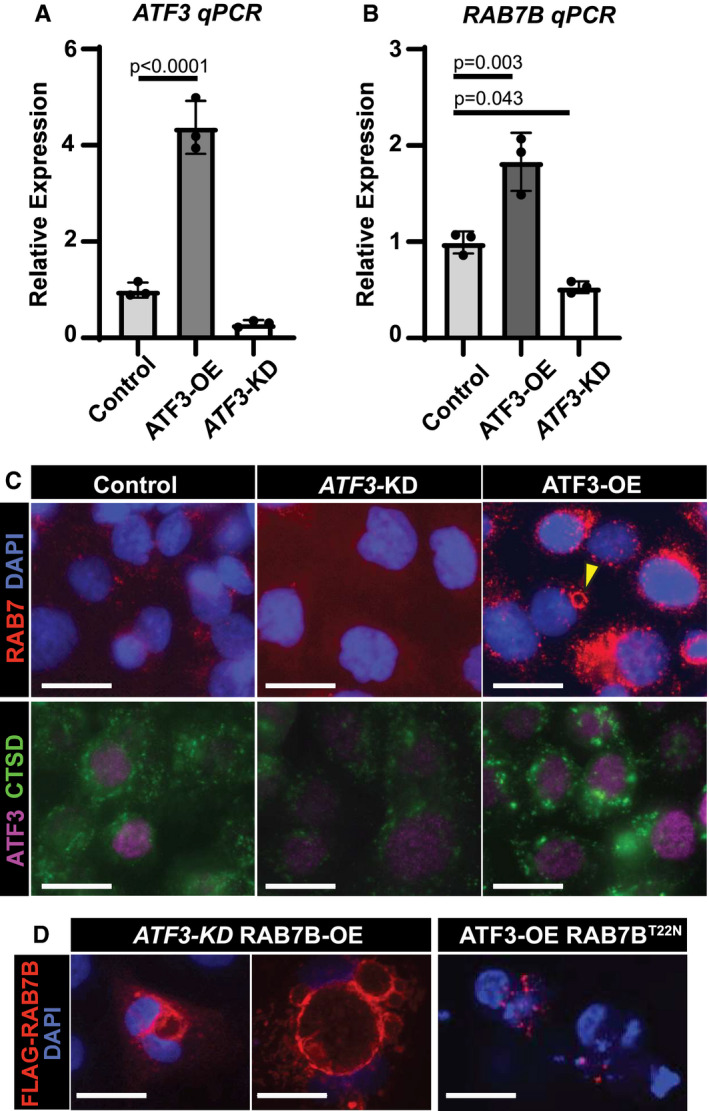
- mRNA expression of ATF3 by qRT–PCR in control, ATF3 knockdown (KD), and ATF3 overexpression (OE) AGS cell lines generated using a PiggybacTM transposon system. Significance by one‐way ANOVA, Dunnett post‐hoc. Data point = mean of technical triplicates in a single mouse. n = 3 mice. Error bars denote standard deviation.
- mRNA expression of RAB7B by qRT–PCR in AGS cell lines from 3A. Statistics and plotting as for panel A.
- Top—Immunofluorescence for RAB7 (red) and DAPI (blue, nuclei) in control, ATF3 knockdown (KD), and ATF3 overexpression (OE) AGS cells. Arrowhead indicates aggregated RAB7 vesicular formation similar to those observed in tissue. Bottom—Immunofluorescence for ATF3 (purple) and cathepsin D (CTSD, green, lysosomes) in AGS cells. Scale bar, 20 µm.
- Left—Immunofluorescence for anti‐FLAG (RAB7B) (red) and DAPI (blue, nuclei) in ATF3 knockdown; RAB7B overexpression (ATF3‐KD RAB7B‐OE) AGS cells show large vesicular structures. Right—Immunofluorescence for anti‐FLAG (RAB7B) (red) and DAPI (blue, nuclei) in ATF3 overexpression; RAB7B GTPase mutant (ATF3‐OE RAB7BT22N) AGS cells fail to make vesicles. Scale bar, 20 µm.
Atf3 is required for massive lysosome and autophagy induction in Stage 1 of paligenosis
To determine whether loss of Atf3 results in decreased Rab7 in vivo, we studied paligenosis in stomach and pancreas of Atf3 −/− mice (Hartman et al, 2004). It is important to note that these mice have a global loss of Atf3, meaning Atf3 is neither in differentiated nor in progenitor cells. However, since we only see ATF3 induction in differentiated cells after injury, Atf3’s main role is primarily in differentiated cells. In addition, we observed ATF3 induction in ACs in the pancreas, where there is no adult stem cell population, again suggesting that ATF3 mainly functions in differentiated cells after injury. In WT mice, we observed scant RAB7 in vehicle tissue of the stomach or pancreas, but following injury in WT animals, we saw the previously observed RAB7+ vesicles and large circular aggregates. Many of the large RAB7‐vesicles were lysosomes or autolysosomes as indicated by enclosed CTSD (Fig 4A and B). Although low‐abundance RAB7 could still be detected in ZCs and ACs of Atf3 −/− mice, the large RAB7‐positive vesicles were predominantly lacking. Injury induced RAB7+ vesicles in WT mice, but in Atf3 −/− mice, we did not observe upregulation of large RAB7+ vesicles (Fig 4A and B).
Figure 4. Atf3 coordinates cellular autodegradation after injury in paligenosis stage I.
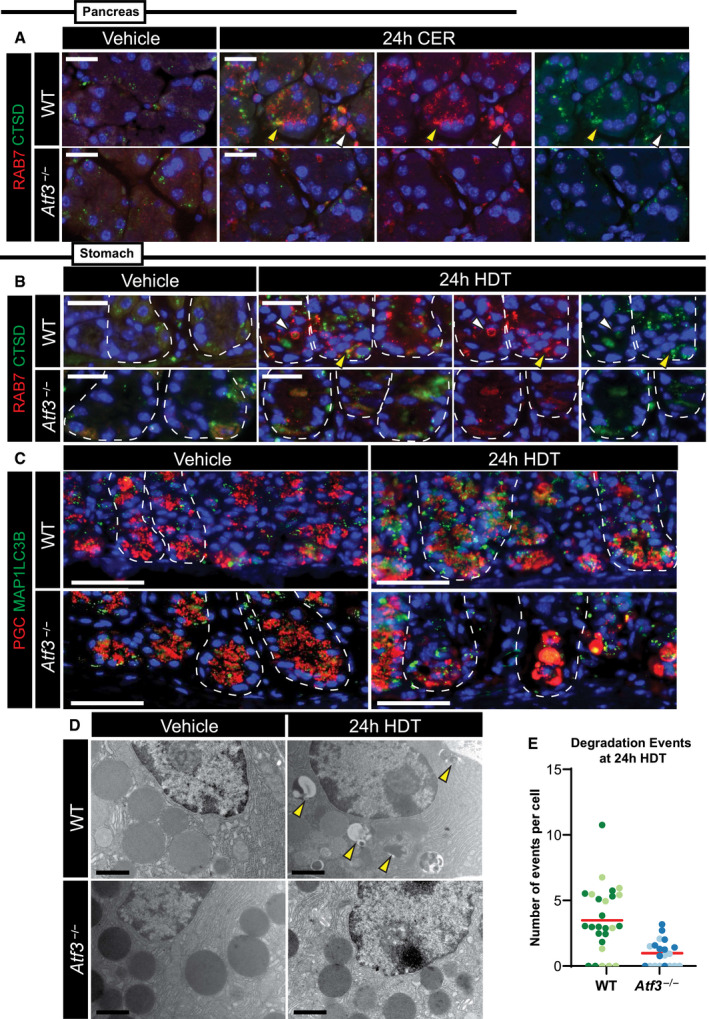
- Representative immunofluorescence images of pancreatic acinar cells after vehicle or 24‐h cerulein (CER) treatment stained for RAB7 (red) and cathepsin D (CTSD, green, lysosomes). Top row is wild‐type, bottom row is Atf3 −/−. Yellow arrowhead highlights RAB7 and CTSD co‐expression. White arrowhead points to an area of RAB7 expression only. Scale bar, 20 μm.
- Representative immunofluorescence images of gastric chief cells after vehicle or 24‐h high‐dose tamoxifen (HDT) treatment stained for RAB7 (red) and cathepsin D (CTSD, green, lysosomes). Top row is wild‐type, bottom row is Atf3 −/−. Yellow arrowhead highlights RAB7 and CTSD co‐expression. White arrowhead points to an area of RAB7 expression only. Scale bars 20 μm. Stomach unit base outlined by dashed white line.
- Representative immunofluorescence images of gastric chief cells after vehicle or 24‐h HDT treatment stained for pepsinogen C (PGC, red, granules) and MAP1LC3B (green, autophagy marker). Top row is wild‐type, bottom row is Atf3 −/−. Scale bar, 50 μm. Stomach unit base outlined by dashed white line.
- Transmission electron micrograph images of zymogenic chief cells (ZCs) after 24‐h vehicle or HDT treatment in WT (top) and Atf3 −/− mice (bottom). Scale bar, 1 μm. Arrowheads indicate cell components being degraded.
- Quantification of degradation events per cell counted from EM images of ZCs after 24‐h HDT treatment in WT and Atf3 −/−mice as shown in panel 3D. Each data point = total granules in a cell in a single TEM section with cells from 2 separate mice plotted together (data point colors indicate cells from a single mouse). Red line indicates mean across all the mice.
Cathepsin D is a marker of both lysosomes and autolysosomes, so we next examined whether loss of ATF3 and RAB7 affected autophagic trafficking. We observed that the autophagic vesicle marker MAP1LC3B (LC3) was induced in Stage 1 of paligenosis in the stomach, consistent with previous work and our EM analysis indicating increased autophagic trafficking (Willet et al, 2018; Meyer et al, 2019; Fig 4C). Such LC3+ autophagic puncta were not induced in the absence of ATF3. We confirmed ATF3‐dependent induction of lysosomes and autophagosomes using TEM (Fig 4D and E).
The results indicate ATF3 is induced rapidly in paligenosis and is required for RAB7‐associated autophagy/lysosomal trafficking. Thus, ATF3 is both necessary and sufficient to induce the endolysosome‐trafficking protein RAB7 and is critical for the massive induction of autodegradative machinery that cells use in Stage 1 of paligenosis to downscale much of their mature cell architecture.
Atf3 loss results in less repair and more cell death after secretory cell injury
Paligenosis is a stepwise process, that can be inhibited between each stage. We have previously shown that blocking autodegradative function during paligenosis impairs regeneration (Willet et al, 2018). We reasoned the aberrant induction of autodegradation in the absence of ATF3 would affect the fraction of cells able to progress to Stage 2 (expressing metaplastic/progenitor genes) and Stage 3 (entering the cell cycle). Stage 2 is easily measured by induction of the epitope for the lectin GSII in ZCs, with the resultant cells (known as SPEM cells) co‐labeling with GSII and ZC markers (like PGC or GIF). Atf3 −/− mice showed profound loss of progression to Stage 2, with only rare SPEM cells (Figs 5A and EV4A–C). Stage 2 failure was corroborated by a second SPEM marker CD44v (Fig EV4A; Khurana et al, 2013; Engevik et al, 2016; Meyer et al, 2019). Moreover, the failure to produce Stage 2 SPEM cells correlated with reduced proliferation (Figs 5B and C, and EV4D). Note that progenitors higher up in the gastric unit (in the isthmus and neck) are induced to proliferate via a non‐paligenotic mechanism (Willet et al, 2018; Choi et al, 2019; Miao et al, 2020a; Miao et al, 2020b). As expected, those non‐paligenotic cells were still induced to proliferate after injury, so overall proliferation in the injured stomach was less affected by loss of ATF3 than paligenotic proliferation specifically (Fig EV4D; cf: Fig 5B vs. Fig 5C showing basal, paligenotic proliferation).
Figure 5. Atf3 loss increases cell death and reduces proliferation after paligenotic injury.
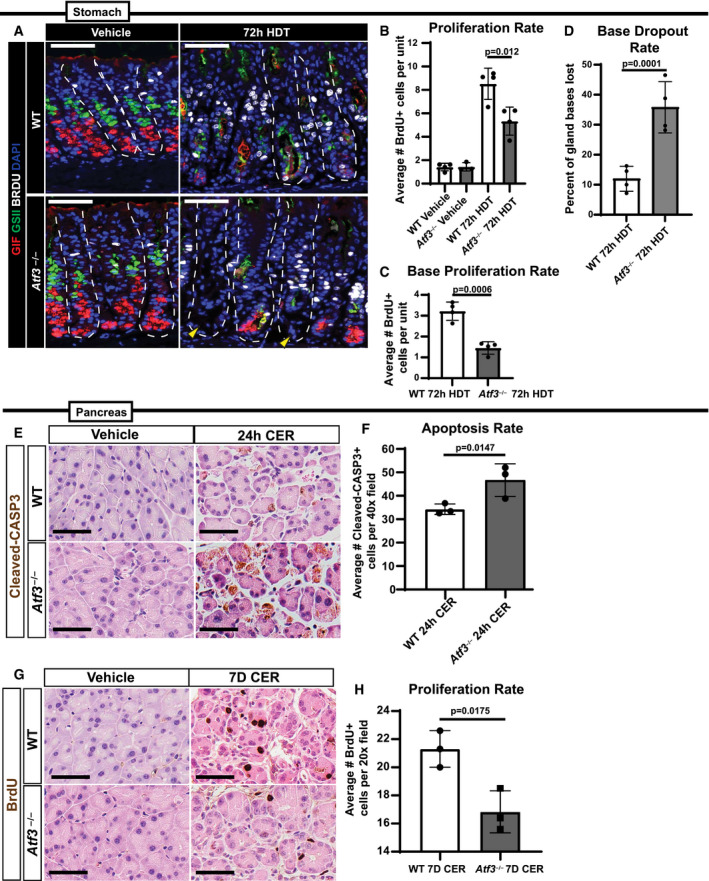
- Representative immunofluorescence images of gastric units after vehicle or 72‐h high‐dose tamoxifen (HDT) treatment stained for GIF (red, granules), GSII (green, neck cells and SPEM marker), BRDU (white, proliferation), and DAPI (blue, nuclei). Top row is wild‐type, bottom row is Atf3 −/−. Yellow arrowheads point to areas of base dropout where ZCs have been lost. Scale bar, 50 μm. Stomach units outlined by dashed white line.
- Proliferation as quantified by number of BrdU+ cells per unit in WT and Atf3 −/−vehicle ± HDT. Each data point = mean of counts from 40 to 50 gastric glands from an individual mouse, n = 3–4 mice per treatment group. Significance by unpaired two‐tailed t‐test. Error bars denote standard deviation.
- Plot and statistical analysis as for Panel B but quantifying BrdU+ cells only in the bottom 1/3 of the unit, which are derived mostly from paligenosis and not from isthmal and neck stem and progenitor cells.
- Death of ZCs after injury plotted by quantifying the base dropout rate, which is the fraction of glands that do not reach the muscularis mucosa from the lumen. Each data point is the mean of 80–100 glands from an individual mouse, n = 4 mice per treatment group. Significance by unpaired two‐tailed t‐test. Error bars denote standard deviation.
- Representative immunohistochemistry of cleaved‐caspase 3 in pancreatic acinar cells marking apoptosis in vehicle or 24‐h cerulein (CER) treatment. Top row is WT, and bottom row is Atf3 −/−. Scale bar, 50 μm.
- Apoptosis quantified following 24‐h CER injury by the number of cleaved‐caspase 3+ cells from a 40× field. Each data point is mean of positive cells in 4–5 40× fields in an individual mouse, n = 3 mice per treatment group. Significance by unpaired two‐tailed t‐test. Error bars denote standard deviation.
- Representative immunohistochemistry of BrdU in pancreatic acinar cells marking proliferation in vehicle or 7 days of repeated cerulein injury. Top row is WT, and bottom row is Atf3 −/−. Scale bar, 50 μm.
- Quantification of proliferation at peak ADM as counted by the number of BrdU+ cells from a 20× field following 7 days of cerulein treatment. Plotting and statistical analysis as per panel F.
We noticed that the lack of paligenotic progression to stages 2 and 3 did not indicate mature cells were delayed in the paligenotic process, as many of the bases of gastric units, where ZCs or paligenotic cells should be, were simply missing. This pattern was consistent with dropout of whole gastric unit bases, a phenomenon we have previously described when there is a failure to progress through paligenosis (Willet et al, 2018; Miao et al, 2020b). Inability to complete paligenosis resulted in marked base dropout with approximately 1/3 of gastric unit bases missing (Fig 5D). As might be expected, the increased base dropout in Atf3 −/− correlated with significantly increased cleaved‐CASP3 positivity, indicating cell death by apoptosis (Fig EV4E and F). In the pancreas, ATF3 was also required for paligenosis progression, as Atf3 −/−ACs had significantly higher cleaved‐CASP3 expression (Fig 5E and F) as well as decreased Stage 3 proliferation induction (Fig 5G and H).
Overall, the results indicate that the failure to upregulate autodegradation in Stage 1, in the absence of Atf3, led to the failure to progress through Stage 2 of paligenosis and finally resulted in increased cell death in Stage 3, in both stomach and pancreas.
ATF3 is critical for injury responses across organs
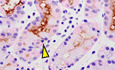
Our studies thus far showed that ATF3 could regulate autodegradative activity in a conserved way across tissue and species by regulating RAB7. In addition, loss of ATF3 caused the same Stage 1 failure with associated overall paligenosis failure in two different organs, suggesting that early ATF3 induction might be a conserved feature of paligenosis. HDT injury‐producing SPEM in mice is a model of the SPEM that occurs in chronic atrophic gastritis in humans (Petersen et al, 2017). Using human stomach samples of tissue adjacent to gastric tumor tissue, we have previously extensively characterized (Lennerz et al, 2010; Radyk et al, 2018; Willet et al, 2018), we observed expression of ATF3 in SPEM. However, as might be expected, given ATF3 expression was early and transient following injury in mice, and most SPEM is chronic and mitotically quiescent in humans (Willet et al, 2018; Burclaff et al, 2020) scattered, potentially transitional SPEM cells were ATF3‐positive (Fig 6). Morphologically normal human gastric epithelial cells lacked ATF3 expression (Fig 6).
Figure 6. ATF3 is expressed in human SPEM.
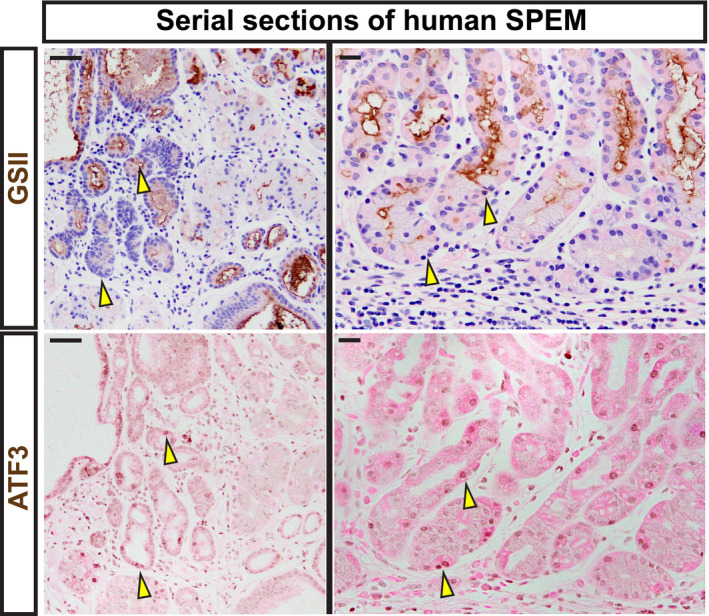
Serial sections of human tissue. Representative immunohistochemistry of GSII (top; spasmolytic polypeptide‐expressing metaplasia [SPEM] marker and neck cells) counterstained with hematoxylin (blue) and eosin (pink). Representative staining of ATF3 (bottom) counterstained with eosin (pink). Tissue was taken from regions adjacent to gastric adenocarcinoma. Scale bars left—50 μm and right—20 μm. Arrowheads indicate strongly expressing ATF3 cells and the corresponding region in the same gland labeled with GSII in a serial tissue section. GSII at the base of gastric glands in the body of the stomach indicate transition to SPEM.
As ATF3 is highly conserved, we took advantage of a previously published Axolotl single‐cell RNA‐sequencing analysis of limb regeneration (Gerber et al, 2018; Data ref: Gerber et al, 2018) that shows high correlation with gene expression in mouse paligenosis (Miao et al, 2020b). We found that, like the previously identified paligenosis gene IFRD1, ATF3 is induced during limb regeneration. RAB7B is induced at the next time point after ATF3 (Fig EV5A), as represented by the fraction of cells that express these genes throughout the course of repair after limb amputation. We previously showed that regenerative proliferation in kidney and liver occurs by paligenosis (Willet et al, 2018), and activation of muscle satellite cells following injury (Yue et al, 2020) also appears to involve a paligenosis‐like process involving emergence from quiescence. We assayed genes whose expression was increased after paligenotic injury in all 5 organs: stomach, pancreas, liver, kidney, and muscle to determine potential core paligenosis genes (Fig EV5B). We found only 7 genes were upregulated in all 5 datasets: Atf3, Cebpb, Hmox1, Ier3, Ifrd1 (Fahling et al, 2013; Data ref: Fahling et al, 2013; Kowalik et al, 2007; Data ref: Kowalik et al, 2007; Moore et al, 2015; Data ref: Moore et al, 2015; Otu et al, 2007; Data ref: Otu et al, 2007; Yue et al, 2020; Data ref: Yue et al, 2020), Junb, and Zfp36. Thus, the results indicate Atf3 may play a conserved role in mediating paligenosis across multiple tissues and species.
Discussion
Our work here shows that ATF3, induced early in Stage 1 of paligenosis, is required for initiation of the autodegradative machinery. Though there are likely additional targets critical for ATF3 actions in paligenosis, we show both in mouse paligenosis models and in human epithelial cells that ATF3 binds and activates Rab7b, which encodes a protein critical in turn for trafficking autophagic vesicles and lysosomes. ATF3 is necessary for RAB7 induction and for induction of a large complement of autophagolysosomes. ATF3 induction after paligenotic injury is conserved across tissues and species with ATF3 increased even in human metaplasia.
The concept of an evolutionarily conserved program, paligenosis, which mechanizes cell plasticity for mature cells, like pancreatic or gastric exocrine cells, is recent. Paligenosis, as originally defined, is a stepwise program that retools cell architecture to convert a mature cell to a regenerative, progenitor‐like phenotype. The three paligenosis stages (1. autodegradation, 2. increased progenitor/metaplastic expression, and 3. cell cycle re‐entry) can be genetically and morphologically defined, and progression from stage to stage can be blocked by inhibitors or deletion of key genes (Fig 7). The stepwise, conserved nature of the program allows us to determine whether there are dedicated, paligenosis genes and genetic machinery, independent of tissue and conserved across evolutionary phyla, as is the case with other cellular programs like apoptosis. We recently identified Ifrd1 and Ddit4 as paligenosis‐governing genes and proposed that paligenosis genes in general would be: (i) largely dispensable for normal development or proliferative activity; (ii) activated primarily during regeneration‐inducing injury; and (iii) highly evolutionarily conserved. We argue here that ATF3 is a similar such gene required for paligenosis. It is upregulated early in paligenosis in multiple tissues, is known to be induced by injury, and is conserved from sea anemones to humans, and, in general, organisms null for ATF3 have phenotypes only after injury. Furthermore, we show evidence here that ATF3 is expressed during transition to metaplasia in humans, which is representative of a much larger cohort (> 500) that we are currently analyzing for expression of paligenosis genes. We observe 11% of human metaplastic lesions have focal ATF3 in non‐proliferative foci, consistent with ATF3 being upstream of the decision for cells to proliferation.
Figure 7. Model of ATF3 function in paligenosis.
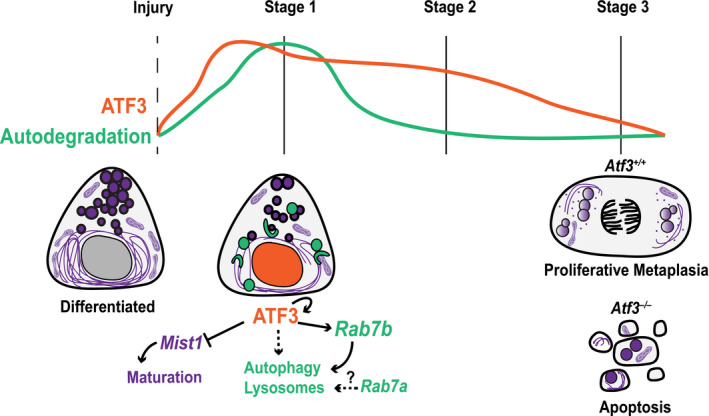
ATF3 peaks early after injury and helps to coordinate the induction of autodegradation by transcriptionally activating Rab7b and other targets to increase the number of lysosomes and induce autophagy. ATF3 also functions at this same time (Stage 1) to repress scaling and maturation factor Mist1.
We describe a model (Fig 7) in which ATF3 is induced early after injury to help transcriptionally upregulate Rab7b and other genes important for autophagy and lysosomal accumulation during paligenosis Stage 1. During this time ATF3 also likely transcriptionally represses genes involved in cell differentiation/maturation like MIST1, a relationship that has been shown before in pancreas (Fazio et al, 2017). Shortly after Stage 1, ATF3 decreases, and the cell progresses through the remainder of paligenosis. Without Atf3, the defects in Stage 1 (autodegradation) obstruct the cell’s ability to become regenerative, metaplastic cells. A greater proportion of cells die and tissue repair is compromised. In addition, we describe a new signaling axis for injury‐induced cell plasticity by ATF3 transcriptional regulation of Rab7b during paligenosis.
Atf3 joins aforementioned Ddit4 and Ifrd1 as paligenosis‐governing genes (Miao et al, 2020b). At the tissue level, loss of Atf3 appears most similar to loss of Ifrd1 as lack of either gene leads to increased cell death in paligenosis Stage 3. While IFRD1 functions to suppress p53‐mediated repression of mTORC1, allowing proliferation in Stage 3, ATF3 instead activates autodegradation pathways for proper clearance of differentiated/specialized cell features. Though both ATF3 and IFRD1 have different mechanisms to govern paligenosis, it is clear that failure to clear paligenosis checkpoints, either by inability to upregulate autodegradation (Atf3 −/−) or by inability to re‐activate mTORC1 (Ifrd1 −/−), leads to decreased tissue recovery after injury. ATF3 and DDIT4 both function in Stage 1 to control autodegradation. Unlike Atf3, however, Ddit4 −/− mice have increased proliferation. DDIT4 functions during initial stages to deactivate mTORC1; hence, Ddit4 −/− mice express constitutively high mTORC1 throughout paligenosis, which overrides a checkpoint (Miao et al, 2020b). Additionally, loss of Ddit4 and induction of metaplasia increases tumorigenesis in an N‐methyl‐N‐nitrosourea spontaneous gastric cancer model (Miao et al, 2021).
Activating transcription factor 3 is a member of the ATF/CREB family of transcription factors and shares similar binding sites with AP‐1 (also from the ATF/CREB and c‐Fos/c‐Jun families), which regulates immediate early genes following stress/injury. Atf3 has been implicated in injury settings in many organs including cerebral ischemia (Wang et al, 2012), axon regeneration (Hunt et al, 2012; Gey et al, 2016), acute lung injury (Zhao et al, 2017), heart failure (Brooks et al, 2015), alcohol liver damage (Tsai et al, 2015), liver ischemia‐reperfusion injury (Zhu et al, 2018), stress‐induced β‐cell apoptosis (Hartman et al, 2004), ADM (Fazio et al, 2017; Azizi et al, 2021), and acute kidney injury (Li et al, 2010). ATF3 is also important for injury responses in Drosophila (Zhou et al, 2017) and Axolotl (Gerber et al, 2018). In all those cases, Atf3 loss exacerbates the injury phenotype but is dispensable for homeostasis and embryonic development, just as we observed here in stomach and pancreas injury. A recent publication similarly showed that Atf3 loss leads to decreased ADM and that absence of Atf3 in a spontaneous cancer mouse model actually prevented KRASG12D‐driven pancreatic cancer (Azizi et al, 2021). That may be consistent with our results here, as mutant KRAS‐driven cancer depends on ADM, which depends on ATF3 and paligenosis. Cells lacking ATF3 may die rather than transform. Thus, the role of ATF3 in cancer is complex and under active study.
It has not been previously reported that injury‐responsive ATF3 accumulation induces autophagy and lysosomal activity, but ATF3 has been reported to interact with individual autophagy pathway members. For example, ATF3 regulates Beclin‐1 (Lin et al, 2014) and Atg5 (Sood et al, 2017). There may also exist an autophagy‐ATF3 feedback loop because defective autophagy (Atg4b loss) impairs ATF3 activity in a mouse lung injury model (Aguirre et al, 2014).
To our knowledge, there are no previous reports of ATF transcription factors transcriptionally regulating any Rabs, though potential ATF binding sites have been described to be present in the 5’‐flanking region of Rab7 (Bhattacharya et al, 2006). In fact, in general, transcriptional regulation of RABs or regulation of RAB function via titrating protein abundance is usually not studied, with some few exceptions such as MIST1 scaling RABs, and a report of c‐Myc helping drive melanoma by activating Rab7 (Alonso‐Curbelo et al, 2014). However, an ATF‐related protein, AP‐1, regulates Rab11a in B cells in mice (Lizundia et al, 2007), suggesting that ATF/CREB transcription factors may have an underappreciated and understudied role in transcriptionally regulating RABs.
RAB7A and RAB7B are regulators of endosomal trafficking and maturation. RAB7 proteins are largely localized to late endosomes and lysosomes to aid in the autophagosome–lysosome fusion and mitochondrion–lysosome contacts (Stroupe, 2018), events undoubtedly important during Stage 1 of paligenosis. As such, complete Rab7 loss in pancreatic acinar cells impairs autophagic and endocytic pathways in mouse models of pancreatitis (Takahashi et al, 2017). Here, we show that Atf3 loss impairs autophagic and lysosomal pathways, further supporting the notion of ATF3 function through Rab7b during paligenosis and injury. There are few studies specifically focused on RAB7B, identified because of its size and sequence similarity to RAB7A. RAB7B has similar functions as RAB7A and was, accordingly, first observed to be associated with lysosomes in monocytes (Yang et al, 2004). More recently, RAB7B has been shown to interact with ATG4B on vesicles and negatively regulate autophagy (Kjos et al, 2017), indicating it also, like RAB7A, regulates autophagic trafficking. There may not be complete concordance between the two RAB7 proteins, however, as RAB7B may have an added function in some circumstances of aiding retrograde transport from late endosomes or lysosomes to the Golgi (Progida et al, 2010).
Using AGS cells, we demonstrated that expression of RAB7B was sufficient to form the large vesicular compartments observed following injury in murine tissue. This effect was independent of ATF3 expression, as vesicles were readily observed with ATF3 knockdown and RAB7B overexpression. In addition, vesicle formation was dependent on RAB7B’s GTPase activity, as no vesicles were observed with the RAB7BT22N mutant, even with overexpression of ATF3.
Recent studies have shown the importance of degradation and autophagy in regulating cell plasticity in several organs (Saera‐Vila et al, 2016; Call & Nichenko, 2020; Li et al, 2020; Romermann et al, 2020). We speculate that Stage 1 of paligenosis is not only required to change the cell structure to one that is more physiologically progenitor‐like but also to generate enough energy for DNA replication and cell division. Ribosomes are an enormous pool for nucleotides, which may be one reason we see so much degradation machinery accumulate in the rER portion of the cell, where there may be release of nucleotides from ribosomes. As mentioned above, functional degradation machinery may also act as a checkpoint indicating a cell is healthy enough to commit to cell cycle re‐entry. This was the case when we administered hydroxychloroquine to inhibit lysosome–autophagosome fusion (Fig EV1, EV2, EV3, EV4, EV5C–F) and when we blocked lysosome activity during paligenosis (Willet et al, 2018) which, like Atf3 and Ifrd1 loss, also ultimately led to increased cell death.
Activating transcription factor 3 may transcriptionally regulate other targets during Stage 1 that do not control degradation. ATF3 can repress Mist1, the secretory cell scaling factor expressed only in mature ZCs and ACs; Mist1 downregulation is a key early step in ZC and AC paligenosis (Kowalik et al, 2007; Nozaki et al, 2008; Lennerz et al, 2010; Karki et al, 2015; Meyer et al, 2019). It also represses Sox9 (Fazio et al, 2017), a metaplastic gene whose induction defines transition from Stage 1 to Stage 2, supporting the hypothesis that ATF3 plays several roles in Stage 1 of paligenosis. It is not clear how ATF3 is upregulated for paligenosis and metaplasia initiation, but once accumulated, ATF3 acts in a positive feedback loop to regulate itself. Reactive oxygen species are also induced en route to SPEM and may contribute to autodegradation and antioxidant pathway activation, as shown by the induction of antioxidant gene xCT, a cysteine/glutamate transporter in SPEM (Meyer et al, 2019). Likewise, reactive oxygen species are important for ADM induction and progression to pancreatic adenocarcinoma (Liou et al, 2016; Cheung et al, 2020).
The current study stimulates several important questions. First, how does metabolism affect the cell fate transitions in paligenosis? We saw dramatic structural changes in mitochondria during paligenosis, which likely reflect changes in metabolism. The metabolic profile of cells as they transition from their normal state to a metaplastic state is not well characterized, though a recent study demonstrated disrupted acinar cell cholesterol metabolism underlies pancreatitis in mouse models (Mareninova et al, 2021). We showed a dependency on dynamic mTORC1 activity during metaplasia (Willet et al, 2018) and recent studies show acetyl‐CoA metabolism contributes to cell plasticity in the pancreas and potentially tumorigenesis via epigenetic regulation (Carrer et al, 2019). Epigenetics also plays a role in SPEM initiation, most recently shown by miR148a repression of DNA‐methylating gene DNMT1, a pro‐metaplasia gene (Shimizu et al, 2020). Second, which cells are poised to undergo paligenosis and become metaplastic? Not all mature cells in a tissue may have the ability to re‐enter the cell cycle after injury, but organs experiencing chronic damage (like GI organs) harbor cells that act as facultative progenitors if the injury is severe enough, though the age of cells in the GI system is a factor for their ability to contribute to plasticity (Engevik et al, 2016; Weis et al, 2017) and some cell types like acid‐secreting parietal cells seem unable to return to the cell cycle. Further study of general, conserved genes that govern paligenosis in diverse contexts should help us have a better understanding of regeneration, tumorigenesis, and also potentially how plasticity arose as a way for multicellular organisms to use cells for both specialized, differentiated function and as progenitors after damage.
Materials and Methods
Reagents and Tools table
| Reagent/Resource | Reference or Source | Identifier or Catalog number |
|---|---|---|
| Experimental Models | ||
| C57BL/6J (M. musculus) | Jackson Lab | Stock No: 000664 |
| Atf3– /– (M. musculus) | Hartman et al (2004) | |
| AGS cells (H. sapiens) | ATCC | CLR‐1739 |
| XL‐10 Gold Ultracompetent Cells | Agilent | Cat #200317 |
| Human patient samples | Lennerz et al (2010), Capoccia et al (2013), Khurana et al, (2013), Radyk et al (2018) | |
| Recombinant DNA | ||
| ATF3 (human) | Sequence cloned out of Addgene #26115 and inserted into a homemade PiggyBac Transposon vector for overexpression (vector also used in ref. Miao et al (2020) | Cat #26115 |
| RAB7B (human) |
Origene This sequence template was also used to generate the RAB7BT22N mutant by amplifying a truncated C‐terminal domain and a three‐part ligation using oligos encoding the mutant N‐terminal domain. |
Cat #RC202283 |
Methods and Protocols
Animal studies and reagents
All experiments involving animals were performed according to protocols approved by the Washington University School of Medicine Animal Studies Committee. Mice were maintained in a specified pathogen‐free barrier facility under a 12‐h light cycle. All mice used in experiments were 6−8 weeks old. Male and female mice were used for each experiment, and we did not note any differences due to sex. Statistical justification for animal numbers is based on our previous experience, consultation with biostatisticians, and using the sampling power analysis approach. Animals were randomized when allocating into each treatment groups. Experimental cohorts were randomly designated at birth for specific animals to minimize bias. Wild‐type C57BL/6 mice were purchased from Jackson Laboratories (Bar Harbor, ME). Atf3 –/– mice were bred onto a C57BL/6 background and received from Dr. Tsonwin Hai at the Ohio State University Medical Center. Atf3 –/– mice were first described in Ref. Hartman et al (2004).
Tamoxifen (250 mg/kg body weight; Toronto Research Chemicals, Inc, Toronto, Canada) was administered by intraperitoneal (IP) injection to induce SPEM in the stomach (see Fig EV1 for timeline; Saenz et al, 2016). Tamoxifen was prepared in a 10% ethanol and 90% sunflower oil solution by sonication (Sigma, St Louis, MO).
Pancreatitis was induced by 6 hourly IP injections of 50 μg/kg (in 0.9% saline) cerulein (Sigma‐Aldrich) given every other day for up to 7 days (see Fig EV1 for timeline). Mice were sacrificed 24‐h after the final cerulein injection.
Hydroxychloroquine sulfate (Acros Organics) was dissolved in PBS and administered by intraperitoneal injection at a dose of 60 mg/kg/day (Rosenfeldt et al, 2013). See Fig EV1C for injection timeline.
All mice were given an intraperitoneal injection containing BrdU (120 mg/kg) and 5‐fluoro‐2′‐deoxyuridine (12 mg/kg) 90 min before sacrifice to capture cells in S phase at the experimental endpoint.
Immunofluorescence and immunohistochemistry
Mouse tissues were excised, flushed with phosphate‐buffered saline (PBS), and then inflated with freshly prepared 4% paraformaldehyde. The stomach was clamped and suspended in fixative in a 50‐ml conical overnight, followed by rinses in PBS and 70% ethanol, arrangement in 3% agar in a tissue cassette, and paraffin processing. Sections (5 μm) were cut, deparaffinized, and rehydrated with graded Histo‐Clear (National Diagnostics), ethanol, and water, then antigen‐retrieved in sodium citrate buffer (2.94 g sodium citrate, 500 μl Tween 20, pH 6.0) using a pressure cooker. Slides were blocked in 5% normal serum, 0.2% Triton X‐100, in PBS. Slides were incubated overnight at 4°C in primary antibodies (see Reagent Table), then rinsed in PBS, incubated 1 h at room temperature in secondary antibodies, and/or fluorescently labeled lectin rinsed in PBS, mounted in ProLong Gold Antifade Mountant with 4′6‐diamidino‐2‐phenylindole (DAPI) (Molecular Probes).
For immunohistochemistry, steps were identical except the following. An extra quenching step was performed for 15 min in a methanol solution containing 1.5% H2O2 before antigen retrieval. Substrate reaction and detection was performed using ImmPACT VIP Peroxidase (horseradish peroxidase) Substrate Kit (Vector Laboratories, Burlingame, CA) as detailed per the manufacturer’s protocol, and sections were developed with DAB for desired time and counterstained with hematoxylin and/or eosin. Slides were mounted in Permount Mounting Medium.
Western blot
Approximately 75 mg mouse corpus stomach or pancreas tissue were sonicated in RIPA buffer with 1× protease inhibitor cocktail (Thermo). Protein was separated using NuPAGE 10% Bis‐Tris gels and transferred to an Invitrogen nitrocellulose membrane. Membranes were then blocked with 3% bovine serum albumin (BSA) and incubated overnight at 4°C with primary antibodies (see Reagent Table for antibodies). The membranes were rinsed in Tris‐buffered saline (TBS) pH 8.0, incubated 1 hour at room temperature in secondary antibody in 5% low‐fat milk. Actin antibody was used as a control to ensure equal loading of protein in each gel lane. Secondary antibody signals were imaged and detected by SuperSignal West Pico PLUS Chemiluminescent Substrate Kit.
Imaging and quantifications
Fluorescence microscopy was performed using a Zeiss Axiovert 200 microscope with an Axiocam MRM camera and Apotome II instrument for grid‐based optical sectioning. Post‐imaging adjustments, including contrast, fluorescent channel overlay, and pseudocoloring, were performed with AxioVision and Adobe Photoshop CS6. Histology of stomach and immunohistochemistry were imaged using an Olympus BX51 light microscope and Olympus SZX12 dissecting microscope w/12 MPixel Olympus DP70 camera. Images were analyzed, and post‐imaging adjustments were performed with Adobe Photoshop CS6.
Quantification of BrdU proliferation rates was done by counting 5 or more 20× fields from a whole‐slide scanned images from the Hamamatsu NanoZoomer 2.0‐HT system.
FIB‐SEM 3D Nanotomography
FIB‐SEM 3D nanotomography was performed as described in ref. Lo et al (2017). Mouse gastric tissues were bisected into fixative containing 2.5% glutaraldehyde and 2% paraformaldehyde in 0.15 M cacodylate buffer containing 2 mM CaCl2 (pH 7.4), fixed for 15 min at 37°C and then overnight at 4°C, sliced into ∼1.5‐mm‐thick pieces, rinsed in cacodylate buffer for 10 min at room temperature, and subjected to secondary fixation in 1% osmium tetroxide/0.3% potassium ferrocyanide in cacodylate buffer for 1 h on ice. The samples were then washed in ultrapure water and stained en bloc in 2% aqueous uranyl acetate for 1 h. After staining, samples were briefly washed in ultrapure water, dehydrated in a graded acetone series, and infiltrated with microwave assistance (Pelco BioWave Pro) into LX112 resin, which was cured in an oven for 48 h at 60°C. Cured blocks were trimmed and mounted onto SEM pins with silver epoxy and faced with a diamond knife. Toluidine blue‐stained sections (300 nm thick) were used to locate a region of interest. Blocks were sputter‐coated with 10 nm of iridium (Leica, ACE 600) with rotation on a planetary stage to ensure saturation. Regions of interest on a FIB‐SEM (Zeiss, Crossbeam 540) were located by secondary electron imaging at 10 KeV. Once a region was found, the sample was prepared using the ATLAS (Fibics) 3D nanotomography engine. In short, a platinum pad was deposited on a 40‐µm × 40‐µm region of interest with the FIB set to 30 KeV and 1.5 nA. Three vertical lines for focus and sigmation and two angled lines for z‐tracking were milled into the platinum pad at 50 pA, and a protective pad of carbon was deposited on top of the milled platinum at 1.5 nA. Following this, a rough trench 50 µm wide and 55 µm deep was milled at 30 nA and polished at 7 nA. Once polishing of the block face was complete, face detection, focusing, and z‐tracking were all performed on the fiducial marks that were milled into the platinum pad. Imaging was performed at 2 KeV and 1.1 nA using the EsB (energy selective backscatter) detector with a grid voltage of 1100 V. The block was milled at a current of 700 pA with 20 nm slices, and 2,000 × 1,750 pixel images were acquired at a resolution of 20 nm/pixel with a dwell of 8 or 10 µs and a line average of 3 for a total z‐depth of 35 µm.
3D reconstruction
3D reconstruction was performed as described in ref. Lo et al (2017). All EM serial images were imported into the Amira 6 3D software package for 3D reconstruction. Areas of interest were manually segmented into data objects with intervening unsegmented slices (“X”) based on object volume. For organelles with suboptimal contrast, the thresholding function was used for manual segmentation. The interpolation function was used to highlight areas of interest between manually segmented slices. 3D models were generated from labeled objects. The “Smooth” and “Simplify” functions were used for model generation, and pseudocoloring was used to enhance visualization for each organelle. Animations were created with the Animation Director in the software.
Electron microscopy
For TEM, stomachs were fixed, sectioned, stained, and imaged as previously described (Willet et al, 2018). Briefly, tissue was collected as described above, fixed overnight at 4°C in modified Karnovsky’s fixative, and sectioned. Tissue was processed by the Washington University in St. Louis Department of Pathology and Immunology Electron Microscopy Facility.
Human cancer cell culture
AGS cells (ATCC) were grown under standard conditions (Advanced DMEM/F12, 10% FBS, Nonessential amino acids, and Pen/Strep at 37°C and 5% CO2) and were cotransfected with a transposon expressing shRNA against ATF3 (TRCN0000329691) under a U6 promoter as well as a homemade plasmid expressing hyperactive PiggyBac transposase to generate stable incorporation of the vector into the genome. After selection with puromycin, passaged cells were obtained for experiments within the first 10 passages. To generate ATF3 overexpression lines, the human ATF3 sequence was cloned from Addgene #26115 and inserted into a homemade PiggyBac transposon vector for overexpression.
Following selection of stable ATF3 overexpression or ATF3 knockdown constructs, the cells were transfected with transposons containing either wild‐type or T22N point mutant of RAB7B containing an N‐terminal 3x‐FlagTag. RAB7B template was purchased from OriGene (Catalog #RC202283), and the mutant was generated by amplifying a truncated C‐terminal domain and a three‐part ligation using oligos encoding the mutant N‐terminal domain. All sequences were confirmed by Sanger sequencing services provided by Genewiz. Two days after transfection with RAB7B constructs, the cells were fixed and stained with an anti‐FLAG antibody, which permitted specific identification of those doubly transfected cells and to determine the cellular localization of mutated RAB7BT22N.
For immunofluorescence experiments, cells were grown on coverslips and blocking and staining was completed in 12‐well culture plates. For imaging, the coverslips were mounted onto slides with ProLong Gold Antifade Mountant with 4′6‐diamidino‐2‐phenylindole (DAPI) (Molecular Probes).
qRT–PCR
RNA was isolated using RNeasy (Qiagen) per the manufacturer’s protocol. The integrity of the mRNA was verified with a BioTek Take3 spectrophotometer and electrophoresis on a 2% agarose gel. RNA was treated with DNase I (Invitrogen), and 1 µg of RNA was reverse‐transcribed with SuperScript III (Invitrogen) following the manufacture’s protocol. Measurements of cDNA abundance were performed by qRT–PCR using Applied Biosystems QuantStudio 3 Real‐time PCR system. Power SYBR Green master mix (Thermo Scientific) fluorescence was used to quantify the relative amplicon amounts of each gene. Primer sequences are located in Table EV1.
ChIP‐PCR
Approximately 25 mg of stomach corpus tissue was flash‐frozen in liquid nitrogen until IP was performed. The SimpleChIP Plus Enzymatic Chromatic IP Kit (Cell Signaling Technology) was used for tissue prep and IP per the manufacturer’s protocol (see Reagent table for antibody concentrations). Tissue was fixed with PFA for 20 min and disaggregated with a Medimachine (BD Biosciences). Standard PCR was used to determine ATF3 binding at the Rab7b first intron (see Table EV2 for primer sequences).
Human patient samples
Human gastric pathological tissue specimens were obtained with approval by the Institutional Review Board of Washington University School of Medicine. Samples used are deidentified tissues archived from standard‐of‐care procedures. Informed consent was received from all patients for the use of their tissue and experiments conformed to the principles set out in the WMA Declaration of Helsinki and the Department of Health and Human Services Belmont Report. Our research team does not have contact with patients and patient information is deidentified. All specimens are stored behind double‐locked doors, and only approved members of the research team have access. Gastric clinical samples have been previously described in Ref. Lennerz et al (2010), Capoccia et al (2013), Radyk et al (2018), Willet et al (2018).
Graphing and statistical analysis
Graphs and statistics were done with GraphPad Prism (La Jolla, CA). In cases involving more than two samples to compare, significance was determined using ANOVA with post‐hoc correction: Tukey was used to assess statistical significance if multiple conditions were compared; Dunnett post‐hoc when comparing multiple samples to a single control. Otherwise, unpaired, two‐tailed Student’s t‐test was used to determine significance. Data that would violate normalcy by study design were analyzed by nonparametric Mann–Whitney U‐test. Most data (e.g., cell counts) were first analyzed as mean values of measurements from multiple fields across multiple sections of an organ for each animal, and then, the mean of those means was plotted. A P‐value of ≤ 0.05 was considered significant. Samples were randomized, and measurements were blinded to prevent the introduction of experimental bias. Formal tests for normalcy were not performed. At least the minimum numbers of animals are used to achieve statistical significance in any determinations.
Author contributions
MDR designed and performed experiments, analyzed and interpreted data, performed statistical analyses, and drafted and revised the manuscript; LBS and BLP designed and performed experiments, analyzed and interpreted data, performed statistical analyses, and drafted the manuscript; JWB, JB, CJC, and YK designed and performed experiments. C‐CS and JAJF designed the experiments. JCM designed the experiments, provided funding for the study, generated data, performed bioinformatics analyses, and drafted and revised the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Review Process File
Expanded View Figures PDF
Table EV1
Table EV2
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Acknowledgements
We thank Dr. Tsonwin Hai at the Ohio State University for the Atf3 −/− mice. We thank Karen Green, Lisa Snipes, and Kevin Boyer of the Department of Pathology Electron Microscopy Center. We acknowledge the AITAC of the Washington University Digestive Disease Center (DDRCC: P30 DK052574) and the NIH Shared Instrumentation Grant S10RR0227552 (for NanoZoomer slide scanning). FIB‐SEM nanotomography was performed in the Washington University Center for Cellular Imaging (WUCCI), which is funded, in part, by the Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital (CDI‐CORE‐2015‐505 and CDI‐CORE‐2019‐813), the Foundation for Barnes‐Jewish Hospital (3770), and Siteman Cancer Center (P30CA091842). MDR was funded by F31CA236506‐01 (NIH NCI), the Philip and Sima Needleman Student Fellowship in Regenerative Medicine, and 5T32GM007067‐43 (NIH); LBS was supported by Siteman Cancer Center Cancer Pathway Program (P30 CA091842 and T32CA113275). BLP by Washington University Biology Summer Undergraduate Research Fellowship Program (BioSURF) and the Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital; JWB was supported by the Department of Defense, through the PRCRP program under Award No. W81XWH2010630 (USAMRAA), T32‐DK007130 (NIH NIDDK), the Digestive Disease Research Core Centers Pilot and Feasibility Grant as part of P30 DK05257, NIH R21 AI156236, and the American Gastroenterological Association AGA2021‐5101; J.B. was supported by National Institutes of Health (NIH) training grant (GM007067) and the Philip and Sima Needleman Student Fellowship in Regenerative Medicine; CJC by T32‐CA009547 (NIH NCI); JCM by the National Institute of Diabetes and Digestive and Kidney Diseases (R21 DK111369, R01DK094989, R01DK105129, R01DK110406 R01120680), The National Institutes of Health National Cancer Institute (P30 CA09182, R01 CA239645, R01 CA246208), and the BETRNet (U54 CA163060).
EMBO reports (2021) 22: e51806.
Data availability
This study includes no data deposited in external repositories.
References
- Adami JG (1900) On Growth and Overgrowth. In: "Festschrift" in Honor of Abraham Jacobi, MD, LLD: To Commemorate the Seventieth Anniversary of His Birth, May Sixth, 1900, Huber F, Sondern FE (eds), pp 422–432. New Rochelle, NY: Knickerbocker Press; [Google Scholar]
- Adler G, Hahn C, Kern HF, Rao KN (1985) Cerulein‐induced pancreatitis in rats: increased lysosomal enzyme activity and autophagocytosis. Digestion 32: 10–18 [DOI] [PubMed] [Google Scholar]
- Aguirre A, López‐Alonso I, González‐López A, Amado‐Rodríguez L, Batalla‐Solís E, Astudillo A, Blázquez‐Prieto J, Fernández AF, Galván JA, dos Santos CCet al (2014) Defective autophagy impairs ATF3 activity and worsens lung injury during endotoxemia. J Mol Med (Berl) 92: 665–676 [DOI] [PubMed] [Google Scholar]
- Alonso‐Curbelo D, Riveiro‐Falkenbach E, Pérez‐Guijarro E, Cifdaloz M, Karras P, Osterloh L, Megías D, Cañón E, Calvo T, Olmeda Det al (2014) RAB7 controls melanoma progression by exploiting a lineage‐specific wiring of the endolysosomal pathway. Cancer Cell 26: 61–76 [DOI] [PubMed] [Google Scholar]
- Altenhoff AM, Glover NM, Train C‐M, Kaleb K, Warwick Vesztrocy A, Dylus D, de Farias TM, Zile K, Stevenson C, Long Jet al (2018) The OMA orthology database in 2018: retrieving evolutionary relationships among all domains of life through richer web and programmatic interfaces. Nucleic Acids Res 46: D477–D485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amieva M, Peek RM Jr (2016) Pathobiology of Helicobacter pylori‐Induced Gastric Cancer. Gastroenterology 150: 64–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizi N, Toma J, Martin M, Khalid MF, Mousavi F, Win PW, Borrello MT, Steele N, Shi J, Pasca di Maglinao Met al (2021) Loss of activating transcription factor 3 prevents KRAS‐mediated pancreatic cancer. Oncogene 40: 3118–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya M, Ojha N, Solanki S, Mukhopadhyay CK, Madan R, Patel N, Krishnamurthy G, Kumar S, Basu SK, Mukhopadhyay A (2006) IL‐6 and IL‐12 specifically regulate the expression of Rab5 and Rab7 via distinct signaling pathways. EMBO J 25: 2878–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks AC, DeMartino AM, Brainard RE, Brittian KR, Bhatnagar A, Jones SP (2015) Induction of activating transcription factor 3 limits survival following infarct‐induced heart failure in mice. Am J Physiol Heart Circ Physiol 309: H1326–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci C, Thomsen P, Nicoziani P, McCarthy J, van Deurs B (2000) Rab7: a key to lysosome biogenesis. Mol Biol Cell 11: 467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burclaff J, Mills JC (2018) Plasticity of differentiated cells in wound repair and tumorigenesis, part II: skin and intestine. Dis Model Mech 11: dmm035071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burclaff J, Willet SG, Saenz JB, Mills JC (2020) Proliferation and Differentiation of Gastric Mucous Neck and Chief Cells During Homeostasis and Injury‐induced Metaplasia. Gastroenterology 158: 598–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Call JA, Nichenko AS (2020) Autophagy: an essential but limited cellular process for timely skeletal muscle recovery from injury. Autophagy 16: 1344–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B, Parnell LA, Diamond MS, Mysorekar IU (2017) Inhibition of autophagy limits vertical transmission of Zika virus in pregnant mice. J Exp Med 214: 2303–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capoccia BJ, Jin RU, Kong YY, Peek RM Jr, Fassan M, Rugge M, Mills JC (2013) The ubiquitin ligase Mindbomb 1 coordinates gastrointestinal secretory cell maturation. J Clin Invest 123: 1475–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrer A, Trefely S, Zhao S, Campbell SL, Norgard RJ, Schultz KC, Sidoli S, Parris JLD, Affronti HC, Sivanand Set al (2019) Acetyl‐CoA metabolism supports multistep pancreatic tumorigenesis. Cancer Discov 9: 416–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Oh S, Gregory S, Shen X, Diehl AM (2020) Single‐cell omics analysis reveals functional diversification of hepatocytes during liver regeneration. JCI Insight 5: e141024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung EC, DeNicola GM, Nixon C, Blyth K, Labuschagne CF, Tuveson DA, Vousden KH (2020) Dynamic ROS Control by TIGAR Regulates the Initiation and Progression of Pancreatic Cancer. Cancer Cell 37: 168–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E, Means AL, Coffey RJ, Goldenring JR (2019) Active Kras expression in gastric isthmal progenitor cells induces foveolar hyperplasia but not metaplasia. Cell Mol Gastroenterol Hepatol 7: 251–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekaney CM, King S, Sheahan B, Cortes JE (2019) Mist1 expression is required for paneth cell maturation. Cell Mol Gastroenterol Hepatol 8: 549–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engevik AC, Feng R, Choi E, White S, Bertaux‐Skeirik N, Li J, Mahe MM, Aihara E, Yang Li, DiPasquale Bet al (2016) The development of spasmolytic polypeptide/TFF2‐expressing metaplasia (SPEM) during gastric repair is absent in the aged stomach. Cell Mol Gastroenterol Hepatol 2: 605–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahling M, Mathia S, Paliege A, Koesters R, Mrowka R, Peters H, Persson PB, Neumayer HH, Bachmann S, Rosenberger C (2013) Tubular von Hippel‐Lindau knockout protects against rhabdomyolysis‐induced AKI. J Am Soc Nephrol 24: 1806–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahling M, Mathia S, Paliege A, Koesters R, Mrowka R, Peters H, Persson PB, Neumayer HH, Bachmann S, Rosenberger C (2013) Gene Expression Omnibus GSE44925 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE44925). [DATASET]
- Faust F, Gomez‐Lazaro M, Borta H, Agricola B, Schrader M (2008) Rab8 is involved in zymogen granule formation in pancreatic acinar AR42J cells. Traffic 9: 964–979 [DOI] [PubMed] [Google Scholar]
- Fazio EN, Young CC, Toma J, Levy M, Berger KR, Johnson CL, Mehmood R, Swan P, Chu A, Cregan SPet al (2017) Activating transcription factor 3 promotes loss of the acinar cell phenotype in response to cerulein‐induced pancreatitis in mice. Mol Biol Cell 28: 2347–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazio EN, Young CC, Toma J, Levy M, Berger KR, Johnson CL, Mehmood R, Swan P, Chu A, Cregan SPet al (2017). Gene Expression Omnibus GSE60250 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE60250) [DATASET]
- Fukuda M (2008) Regulation of secretory vesicle traffic by Rab small GTPases. Cell Mol Life Sci 65: 2801–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber T, Murawala P, Knapp D, Masselink W, Schuez M, Hermann S, Gac‐Santel M, Nowoshilow S, Kageyama J, Khattak Set al (2018) Single‐cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science 362: eaaq0681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber T, Murawala P, Knapp D, Masselink W, Schuez M, Hermann S, Gac‐Santel M, Nowoshilow S, Kageyama J, Khattak Set al (2018). Gene Expression Omnibus GSE106269 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE106269). [DATASET] [DOI] [PMC free article] [PubMed]
- Gey M, Wanner R, Schilling C, Pedro MT, Sinske D, Knoll B (2016) Atf3 mutant mice show reduced axon regeneration and impaired regeneration‐associated gene induction after peripheral nerve injury. Open Biol 6: 160091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux V, Rustgi AK (2017) Metaplasia: tissue injury adaptation and a precursor to the dysplasia‐cancer sequence. Nat Rev Cancer 17: 594–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenring JR (2018) Pyloric metaplasia, pseudopyloric metaplasia, ulcer‐associated cell lineage and spasmolytic polypeptide‐expressing metaplasia: reparative lineages in the gastrointestinal mucosa. J Pathol 245: 132–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS (2011) FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra F, Bucci C (2016) Multiple roles of the small GTPase Rab7. Cells 5: 34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai T, Wolfgang CD, Marsee DK, Allen AE, Sivaprasad U (1999) ATF3 and stress responses. Gene Expr 7: 321–335 [PMC free article] [PubMed] [Google Scholar]
- Hartman MG, Lu D, Kim ML, Kociba GJ, Shukri T, Buteau J, Wang X, Frankel WL, Guttridge D, Prentki Met al (2004) Role for activating transcription factor 3 in stress‐induced beta‐cell apoptosis. Mol Cell Biol 24: 5721–5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess DA, Humphrey SE, Ishibashi J, Damsz B, Lee AH, Glimcher LH, Konieczny SF (2011) Extensive pancreas regeneration following acinar‐specific disruption of Xbp1 in mice. Gastroenterology 141: 1463–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong KU, Reynolds SD, Giangreco A, Hurley CM, Stripp BR (2001) Clara cell secretory protein‐expressing cells of the airway neuroepithelial body microenvironment include a label‐retaining subset and are critical for epithelial renewal after progenitor cell depletion. Am J Respir Cell Mol Biol 24: 671–681 [DOI] [PubMed] [Google Scholar]
- Hou Y, Ernst SA, Stuenkel EL, Lentz SI, Williams JA (2015) Rab27A is present in mouse pancreatic acinar cells and is required for digestive enzyme secretion. PLoS One 10: e0125596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WJ, Esen E, Geahlen JH, Bredemeyer AJ, Lee AH, Shi G, Konieczny SF, Glimcher LH, Mills JC (2010) XBP1 controls maturation of gastric zymogenic cells by induction of MIST1 and expansion of the rough endoplasmic reticulum. Gastroenterology 139: 2038–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WJ, Khurana SS, Geahlen JH, Kohli K, Waller RA, Mills JC (2012) Tamoxifen induces rapid, reversible atrophy, and metaplasia in mouse stomach. Gastroenterology 142: 21–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt D, Raivich G, Anderson PN (2012) Activating transcription factor 3 and the nervous system. Front Mol Neurosci 5: 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Zama T, Kamimoto T, Aoki R, Ikeda Y, Kimura H, Hagiwara M (2004) TNFalpha‐induced ATF3 expression is bidirectionally regulated by the JNK and ERK pathways in vascular endothelial cells. Genes Cells 9: 59–70 [DOI] [PubMed] [Google Scholar]
- Itoh T, Satoh M, Kanno E, Fukuda M (2006) Screening for target Rabs of TBC (Tre‐2/Bub2/Cdc16) domain‐containing proteins based on their Rab‐binding activity. Genes Cells 11: 1023–1037 [DOI] [PubMed] [Google Scholar]
- Jin RU, Mills JC (2014) RAB26 coordinates lysosome traffic and mitochondrial localization. J Cell Sci 127: 1018–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin RU, Mills JC (2019) The cyclical hit model: how paligenosis might establish the mutational landscape in Barrett's esophagus and esophageal adenocarcinoma. Curr Opin Gastroenterol 35: 363–370 [DOI] [PubMed] [Google Scholar]
- Johnson CL, Kowalik AS, Rajakumar N, Pin CL (2004) Mist1 is necessary for the establishment of granule organization in serous exocrine cells of the gastrointestinal tract. Mech Dev 121: 261–272 [DOI] [PubMed] [Google Scholar]
- Jones JC, Brindley CD, Elder NH, Myers MG Jr, Rajala MW, Dekaney CM, McNamee EN, Frey MR, Shroyer NF, Dempsey PJ (2019) Cellular plasticity of Defa4(Cre)‐expressing paneth cells in response to notch activation and intestinal injury. Cell Mol Gastroenterol Hepatol 7: 533–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki A, Humphrey SE, Steele RE, Hess DA, Taparowsky EJ, Konieczny SF (2015) Silencing Mist1 gene expression is essential for recovery from acute pancreatitis. PLoS One 10: e0145724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeley TM, Horita N, Samuelson LC (2019) Tamoxifen‐induced gastric injury: effects of dose and method of administration. Cell Mol Gastroenterol Hepatol 8: 365–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana SS, Riehl TE, Moore BD, Fassan M, Rugge M, Romero‐Gallo J, Noto J, Peek RM Jr, Stenson WF, Mills JC (2013) The hyaluronic acid receptor CD44 coordinates normal and metaplastic gastric epithelial progenitor cell proliferation. J Biol Chem 288: 16085–16097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjos I, Borg Distefano M, Saetre F, Repnik U, Holland P, Jones AT, Engedal N, Simonsen A, Bakke O, Progida C (2017) Rab7b modulates autophagic flux by interacting with Atg4B. EMBO Rep 18: 1727–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalik AS, Johnson CL, Chadi SA, Weston JY, Fazio EN, Pin CL (2007) Mice lacking the transcription factor Mist1 exhibit an altered stress response and increased sensitivity to caerulein‐induced pancreatitis. Am J Physiol Gastrointest Liver Physiol 292: G1123–G1132 [DOI] [PubMed] [Google Scholar]
- Kowalik AS, Johnson CL, Chadi SA, Weston JY, Fazio EN, Pin CL (2007) Gene Expression Omnibus GSE3644 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE3644) [DATASET]
- Kreft L, Soete A, Hulpiau P, Botzki A, Saeys Y, De Bleser P (2017) ConTra v3: a tool to identify transcription factor binding sites across species, update 2017. Nucleic Acids Res 45: W490–W494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba T, Lalli M, Kramann R, Kobayashi A, Humphreys BD (2014) Differentiated kidney epithelial cells repair injured proximal tubule. Proc Natl Acad Sci USA 111: 1527–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langemeyer L, Frohlich F, Ungermann C (2018) Rab GTPase function in endosome and lysosome biogenesis. Trends Cell Biol 28: 957–970 [DOI] [PubMed] [Google Scholar]
- Lennerz JKM, Kim S‐H, Oates EL, Huh WJ, Doherty JM, Tian X, Bredemeyer AJ, Goldenring JR, Lauwers GY, Shin Y‐Ket al (2010) The transcription factor MIST1 is a novel human gastric chief cell marker whose expression is lost in metaplasia, dysplasia, and carcinoma. Am J Pathol 177: 1514–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leushacke M, Tan SH, Wong A, Swathi Y, Hajamohideen A, Tan LT, Goh J, Wong E, Denil SLIJ, Murakami Ket al (2017) Lgr5‐expressing chief cells drive epithelial regeneration and cancer in the oxyntic stomach. Nat Cell Biol 19: 774–786 [DOI] [PubMed] [Google Scholar]
- Li HF, Cheng CF, Liao WJ, Lin H, Yang RB (2010) ATF3‐mediated epigenetic regulation protects against acute kidney injury. J Am Soc Nephrol 21: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wu J, Sun X, Wu Q, Li Y, Li K, Zhang Q, Li Y, Abel ED, Chen H (2020) Autophagy reprograms alveolar progenitor cell metabolism in response to lung injury. Stem Cell Reports 14: 420–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Li HF, Chen HH, Lai PF, Juan SH, Chen JJ, Cheng CF (2014) Activating transcription factor 3 protects against pressure‐overload heart failure via the autophagy molecule Beclin‐1 pathway. Mol Pharmacol 85: 682–691 [DOI] [PubMed] [Google Scholar]
- Liou GY, Doppler H, DelGiorno KE, Zhang L, Leitges M, Crawford HC, Murphy MP, Storz P (2016) Mutant KRas‐induced mitochondrial oxidative stress in acinar cells upregulates EGFR signaling to drive formation of pancreatic precancerous lesions. Cell Rep 14: 2325–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizundia R, Chaussepied M, Naissant B, Masse GX, Quevillon E, Michel F, Monier S, Weitzman JB, Langsley G (2007) The JNK/AP‐1 pathway upregulates expression of the recycling endosome rab11a gene in B cells transformed by Theileria. Cell Microbiol 9: 1936–1945 [DOI] [PubMed] [Google Scholar]
- Lo H‐Y, Jin RU, Sibbel G, Liu D, Karki A, Joens MS, Madison BB, Zhang Bo, Blanc V, Fitzpatrick JAJet al (2017) A single transcription factor is sufficient to induce and maintain secretory cell architecture. Genes Dev 31: 154–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackiewicz P, Wyroba E (2009) Phylogeny and evolution of Rab7 and Rab9 proteins. BMC Evol Biol 9: 101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mareninova OA, Vegh ET, Shalbueva N, Wightman CJ, Dillon DL, Malla S, Xie Y, Takahashi T, Rakonczay Z, French SWet al (2021) Dysregulation of mannose‐6‐phosphate dependent cholesterol homeostasis in acinar cells mediates pancreatitis. J Clin Invest 10.1172/JCI146870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marubashi S, Fukuda M (2020) Rab7B/42 is functionally involved in protein degradation on melanosomes in keratinocytes. Cell Struct Funct 45: 45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AR, Engevik AC, Willet SG, Williams JA, Zou Y, Massion PP, Mills JC, Choi E, Goldenring JR (2019) Cystine/Glutamate antiporter (xCT) is required for chief cell plasticity after gastric injury. Cell Mol Gastroenterol Hepatol 8: 379–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AR, Goldenring JR (2018) Injury, repair, inflammation and metaplasia in the stomach. J Physiol 596: 3861–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao ZF, Adkins‐Threats M, Burclaff JR, Osaki LH, Sun JX, Kefalov Y, He Z, Wang ZN, Mills JC (2020a) A metformin‐responsive metabolic pathway controls distinct steps in gastric progenitor fate decisions and maturation. Cell Stem Cell 26: 910–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z‐F, Lewis MA, Cho CJ, Adkins‐Threats M, Park D, Brown JW, Sun J‐X, Burclaff JR, Kennedy S, Lu Jet al (2020b) A dedicated evolutionarily conserved molecular network licenses differentiated cells to return to the cell cycle. Dev Cell 55: 178–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao ZF, Sun JX, Adkins‐Threats M, Pang MJ, Zhao JH, Wang X, Tang KW, Wang ZN, Mills JC (2021) DDIT4 licenses only healthy cells to proliferate during injury‐induced metaplasia. Gastroenterology 160: 260–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JC, Sansom OJ (2015) Reserve stem cells: Differentiated cells reprogram to fuel repair, metaplasia, and neoplasia in the adult gastrointestinal tract. Sci Signal 8: re8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JC, Taghert PH (2012) Scaling factors: transcription factors regulating subcellular domains. BioEssays 34: 10–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore BD, Jin RU, Osaki L, Romero‐Gallo J, Noto J, Peek RK, Mills JC (2015) Gene Expression Omnibus GSE71580 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE71580). [DATASET]
- Moore BD, Jin RU, Osaki L, Romero‐Gallo J, Noto J, Peek RK, Mills JC (2015) Identification of alanyl aminopeptidase (CD13) as a surface marker for isolation of mature gastric zymogenic chief cells. Am J Physiol Gastrointest Liver Physiol 309: G955–G964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KT, Lee H, Sousa JF, Weis VG, O'Neal RL, Finke PE, Romero–Gallo J, Shi G, Mills JC, Peek RMet al (2010) Mature chief cells are cryptic progenitors for metaplasia in the stomach. Gastroenterology 139: 2028–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederau C, Ferrell LD, Grendell JH (1985) Caerulein‐induced acute necrotizing pancreatitis in mice: protective effects of proglumide, benzotript, and secretin. Gastroenterology 88: 1192–1204 [DOI] [PubMed] [Google Scholar]
- Nozaki K, Ogawa M, Williams JA, Lafleur BJ, Ng V, Drapkin RI, Mills JC, Konieczny SF, Nomura S, Goldenring JR (2008) A molecular signature of gastric metaplasia arising in response to acute parietal cell loss. Gastroenterology 134: 511–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi H, Ernst SA, Wys N, McNiven M, Williams JA (1996) Rab3D localizes to zymogen granules in rat pancreatic acini and other exocrine glands. Am J Physiol 271: G531–G538 [DOI] [PubMed] [Google Scholar]
- Otu HH, Naxerova K, Ho K, Can H, Nesbitt N, Libermann TA, Karp SJ (2007) Gene Expression Omnibus GSE6998 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE6998). [DATASET] [DOI] [PubMed]
- Otu HH, Naxerova K, Ho K, Can H, Nesbitt N, Libermann TA, Karp SJ (2007) Restoration of liver mass after injury requires proliferative and not embryonic transcriptional patterns. J Biol Chem 282: 11197–11204 [DOI] [PubMed] [Google Scholar]
- Pereira‐Leal JB, Seabra MC (2000) The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J Mol Biol 301: 1077–1087 [DOI] [PubMed] [Google Scholar]
- Petersen CP, Mills JC, Goldenring JR (2017) Murine models of gastric corpus preneoplasia. Cell Mol Gastroenterol Hepatol 3: 11–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin CL, Rukstalis JM, Johnson C, Konieczny SF (2001) The bHLH transcription factor Mist1 is required to maintain exocrine pancreas cell organization and acinar cell identity. J Cell Biol 155: 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Progida C, Cogli L, Piro F, De Luca A, Bakke O, Bucci C (2010) Rab7b controls trafficking from endosomes to the TGN. J Cell Sci 123: 1480–1491 [DOI] [PubMed] [Google Scholar]
- Radyk MD, Burclaff J, Willet SG, Mills JC (2018) Metaplastic cells in the stomach arise, independently of stem cells, via dedifferentiation or Transdifferentiation of chief cells. Gastroenterology 154: 839–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey VG, Doherty JM, Chen CC, Stappenbeck TS, Konieczny SF, Mills JC (2007) The maturation of mucus‐secreting gastric epithelial progenitors into digestive‐enzyme secreting zymogenic cells requires Mist1. Development 134: 211–222 [DOI] [PubMed] [Google Scholar]
- Raven A, Lu W‐Y, Man TY, Ferreira‐Gonzalez S, O’Duibhir E, Dwyer BJ, Thomson JP, Meehan RR, Bogorad R, Koteliansky Vet al (2017) Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature 547: 350–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römermann D, Ansari N, Schultz‐Moreira AR, Michael A, Marhenke S, Hardtke‐Wolenski M, Longerich T, Manns MP, Wedemeyer H, Vogel Aet al (2020) Absence of Atg7 in the liver disturbed hepatic regeneration after liver injury. Liver Int 40: 1225–1238 [DOI] [PubMed] [Google Scholar]
- Rosenfeldt MT, O’Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, Au A, Rai TS, Zheng L, Ridgway Ret al (2013) p53 status determines the role of autophagy in pancreatic tumour development. Nature 504: 296–300 [DOI] [PubMed] [Google Scholar]
- Saenz JB, Burclaff J, Mills JC (2016) Modeling murine gastric metaplasia through tamoxifen‐induced acute parietal cell loss. Methods Mol Biol 1422: 329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz JB, Mills JC (2018) Acid and the basis for cellular plasticity and reprogramming in gastric repair and cancer. Nat Rev Gastroenterol Hepatol 15: 257–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz JB, Vargas N, Mills JC (2018) Tropism for spasmolytic polypeptide‐expressing metaplasia allows helicobacter pylori to expand its intragastric niche. Gastroenterology 156: 160–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saera‐Vila A, Kish PE, Louie KW, Grzegorski SJ, Klionsky DJ, Kahana A (2016) Autophagy regulates cytoplasmic remodeling during cell reprogramming in a zebrafish model of muscle regeneration. Autophagy 12: 1864–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saluja A, Saito I, Saluja M, Houlihan MJ, Powers RE, Meldolesi J, Steer M (1985) In vivo rat pancreatic acinar cell function during supramaximal stimulation with caerulein. Am J Physiol 249: G702–G710 [DOI] [PubMed] [Google Scholar]
- Shimizu T, Sohn Y, Choi E, Petersen CP, Prasad N, Goldenring JR (2020) Decrease in MiR‐148a expression during initiation of chief cell transdifferentiation. Cell Mol Gastroenterol Hepatol 9: 61–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood V, Sharma KB, Gupta V, Saha D, Dhapola P, Sharma M, Sen U, Kitajima S, Chowdhury S, Kalia Met al (2017) ATF3 negatively regulates cellular antiviral signaling and autophagy in the absence of type I interferons. Sci Rep 7: 8789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz P (2017) Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nat Rev Gastroenterol Hepatol 14: 296–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroupe C (2018) This is the end: regulation of Rab7 nucleotide binding in endolysosomal trafficking and autophagy. Fron Cell Dev Biol 6: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Mashima H, Miura K, Maeda D, Goto A, Goto T, Sun‐Wada GH, Wada Y, Ohnishi H (2017) Disruption of small GTPase Rab7 exacerbates the severity of acute pancreatitis in experimental mouse models. Sci Rep 7: 2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tata PR, Mou H, Pardo‐Saganta A, Zhao R, Prabhu M, Law BM, Vinarsky V, Cho JL, Breton S, Sahay Aet al (2013) Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature 503: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Jin RU, Bredemeyer AJ, Oates EJ, Blazewska KM, McKenna CE, Mills JC (2010) RAB26 and RAB3D are direct transcriptional targets of MIST1 that regulate exocrine granule maturation. Mol Cell Biol 30: 1269–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WW, Matsumura S, Liu W, Phillips NG, Sonntag T, Hao E, Lee S, Hai T, Montminy M (2015) ATF3 mediates inhibitory effects of ethanol on hepatic gluconeogenesis. Proc Natl Acad Sci USA 112: 2699–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es JH, Sato T, van de Wetering M, Lyubimova A, Yee Nee AN, Gregorieff A, Sasaki N, Zeinstra L, van den Born M, Korving Jet al (2012) Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol 14: 1099–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandinger‐Ness A, Zerial M (2014) Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harb Perspect Biol 6: a022616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Deng S, Lu Y, Zhang Y, Yang L, Guan Y, Jiang H, Li H (2012) Increased inflammation and brain injury after transient focal cerebral ischemia in activating transcription factor 3 knockout mice. Neuroscience 220: 100–108 [DOI] [PubMed] [Google Scholar]
- Weis VG, Petersen CP, Weis JA, Meyer AR, Choi E, Mills JC, Goldenring JR (2017) Maturity and age influence chief cell ability to transdifferentiate into metaplasia. Am J Physiol Gastrointest Liver Physiol 312: G67–G76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willet SG, Lewis MA, Miao ZF, Liu D, Radyk MD, Cunningham RL, Burclaff J, Sibbel G, Lo HG, Blanc Vet al (2018) Regenerative proliferation of differentiated cells by mTORC1‐dependent paligenosis. EMBO J 37: e98311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewski LE, Choi E, Petersen C, Delgado AG, Piazuelo MB, Romero‐Gallo J, Lantz TL, Zavros Y, Coffey RJ, Goldenring JRet al (2019) Targeted mobilization of Lrig1(+) gastric epithelial stem cell populations by a carcinogenic Helicobacter pylori type IV secretion system. Proc Natl Acad Sci USA 116: 19652–19658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Chen T, Han C, Li N, Wan T, Cao X (2004) Rab7b, a novel lysosome‐associated small GTPase, is involved in monocytic differentiation of human acute promyelocytic leukemia cells. Biochem Biophys Res Commun 318: 792–799 [DOI] [PubMed] [Google Scholar]
- Yue L, Wan R, Luan S, Zeng W, Cheung TH (2020) Dek modulates global intron retention during muscle stem cells quiescence exit. Dev Cell 53: 661–676 [DOI] [PubMed] [Google Scholar]
- Yue L, Wan R, Luan S, Zeng W, Cheung TH (2020) Gene Expression Omnibus GSE113631 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE113631). [DATASET]
- Zhao Y, Wu X, Qian L, Guo L, Liao J, Wu X (2017) Activating transcription factor 3 protects mice against pseudomonas aeruginosa‐induced acute lung injury by interacting with lipopolysaccharide binding protein. Mol Immunol 90: 27–32 [DOI] [PubMed] [Google Scholar]
- Zhou J, Edgar BA, Boutros M (2017) ATF3 acts as a rheostat to control JNK signalling during intestinal regeneration. Nat Commun 8: 14289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Wang H, Jiang B, Ni X, Jiang L, Li C, Wang X, Zhang F, Ke B, Lu L (2018) Loss of ATF3 exacerbates liver damage through the activation of mTOR/p70S6K/ HIF‐1alpha signaling pathway in liver inflammatory injury. Cell Death Dis 9: 910 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Review Process File
Expanded View Figures PDF
Table EV1
Table EV2
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Data Availability Statement
This study includes no data deposited in external repositories.


