Abstract
We describe severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)–specific immune responses in a patient with lymphoma and recent programmed death 1 (PD-1) inhibitor therapy with late onset of severe coronavirus disease 2019 disease and prolonged SARS-CoV-2 replication, in comparison to age-matched and immunocompromised controls. High levels of HLA-DR+/CD38+ activation, interleukin 6, and interleukin 18 in the absence of B cells and PD-1 expression was observed. SARS-CoV-2–specific antibody responses were absent and SARS-CoV-2–specific T cells were minimally detected. This case highlights challenges in managing immunocompromised hosts who may fail to mount effective virus-specific immune responses.
Keywords: cellular immunity, COVID-19, humoral immunity, immunocompromise, lymphoma, SARS-CoV-2
We describe defects in severe acute respiratory syndrome coronavirus 2–specific immune responses and persistent viral replication in a patient with lymphoma. This case highlights challenges in managing immunocompromised hosts who may fail to mount effective virus-specific immune responses.
Global research efforts are focused on understanding the optimal immune responses to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), to facilitate the development and evaluation of SARS-CoV-2 vaccines and treatments. SARS-CoV-2–specific functional neutralizing antibodies are considered important for viral neutralization and clearance [1], and most individuals develop CD4+ and CD8+ T-cell responses to SARS-CoV-2 [2]. However, studies determining the degree and duration of protective immune responses are still needed.
Observation of SARS-CoV-2–specific immune response in individuals with defined immunodeficiencies help reveal critical aspects of viral immune control. Defects in humoral immunity have been associated with incomplete immune control of SARS-CoV-2, prolonged viral culture, and shedding [3–6]. Whether the morbidity and mortality of SARS-CoV-2 infection is impacted by immune checkpoint inhibitor (ICI) therapy remains unclear. ICIs inhibit negative regulatory cell surface molecules, leading to sustained T-cell activation, which can be beneficial in cancer treatment. However, preclinical studies also demonstrate that the programmed death 1 (PD-1)/programmed death ligand 1 (PD-L1) axis regulates critical antiviral immune responses [7]. To investigate SARS-CoV-2 host immunological control further, including those associated with ICIs, we describe the clinical course and SARS-CoV-2–specific immune responses of a patient with functional B-cell deficiency and recent PD-1 inhibitor treatment, and persistent SARS-CoV-2 replication.
CASE REPORT
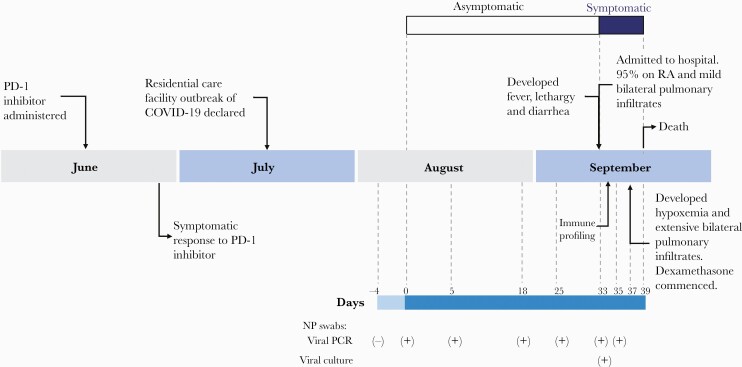
In mid-2020, an elderly male, with a 12-month history of multiple relapse and transformed indolent CD5–CD10– non-Hodgkin lymphoma receiving third-line PD-1 ICI therapy and a 10-year history of castrate-sensitive metastatic prostate cancer, transitioned to living in a residential facility. Prior lines of therapy included 6 cycles of rituximab and chlorambucil in late 2019 and a single cycle of vinblastine and prednisolone in early 2020. Clinically appreciable nodal response occurred within 14 days of cycle 1 of PD-1 ICI therapy. Unfortunately, his residential facility became the site of a coronavirus disease 2019 (COVID-19) outbreak and the facility was quarantined, precluding him from receiving further immune therapy. Seven days later he was found to be SARS-CoV-2 positive, despite reporting no symptoms (Figure 1, Supplementary Table 1). He remained asymptomatic but continued to test positive to SARS-CoV-2. On day 33 after the first positive test he was admitted to hospital after developing diarrhea, fever, and lethargy. On arrival, he had features consistent with mild COVID-19 [8] (Supplementary Table 2, Figure 1). Peripheral blood was sampled on day 1 of admission and SARS-CoV-2 immune responses were analyzed (Supplementary Methods). On day 4 of hospitalization, he developed severe COVID-19 [8] with sudden-onset of shortness of breath and hypoxia with escalating oxygen requirements (Supplementary Figure 1). Biochemical features consistent with COVID-19 cytokine release syndrome [8] were present (Supplementary Table 2), and a computed tomography pulmonary angiogram demonstrated extensive diffuse ground-glass opacity. Although he commenced dexamethasone (6 mg) intravenously daily per local guidelines, he progressed to critical COVID-19 [8] with type 1 respiratory failure and died (on day 6 of symptomatic COVID-19 illness and day 39 after first positive SARS-CoV-2 result).
Figure 1.
Timeline of clinical features, treatments, and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) tests of an immunocompromised individual with prolonged SARS-CoV-2 replication. Dates of relevant events such
as symptom onset, clinical progression, treatments, and SARS-CoV-2 tests (detailed in Supplementary Tables 1 and 2) are shown. Abbreviations: (-), not detected; (+), detected; COVID-19, coronavirus disease 2019; NP, nasopharyngeal; PCR, polymerase chain reaction; PD-1, programmed death 1; RA, room air.
RESULTS
Increased Interleukin 6 and Interleukin 18 in an Immunocompromised Individual During Acute COVID-19
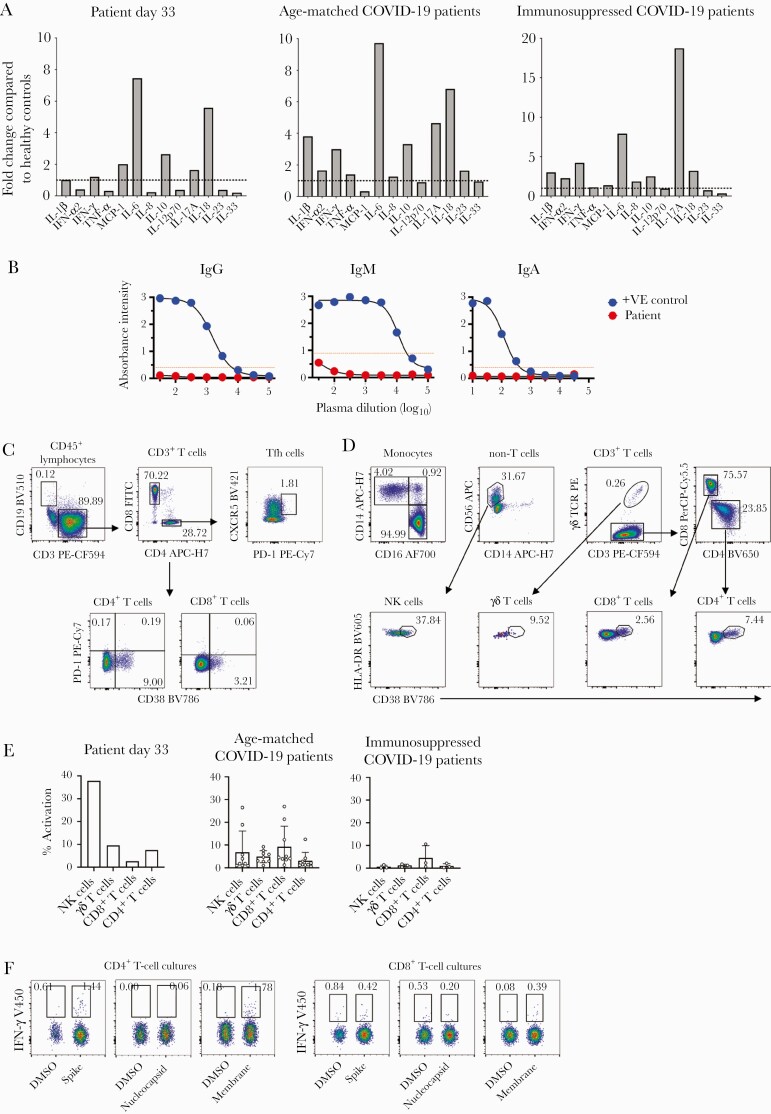
As high levels of proinflammatory cytokines and chemokines are predictive of severe COVID-19 [9], 13 proinflammatory cytokines and chemokines were quantified in the patient’s plasma. To define any potential dysregulation in cytokine/chemokine milieu in the COVID-19 immunocompromised patient, we have compared the inflammatory cytokine/chemokine levels to healthy age-matched controls. We used healthy age-matched non–COVID-19 controls to express the cytokine and chemokine levels as a fold change to allow for normalization of the data and ease of comparison across groups. At symptom onset (day 33), high levels of interleukin (IL) 6 and IL-18 were observed at 7.5- and 5.6-fold, respectively (Figure 2A), compared to healthy age-matched controls. Notably, several proinflammatory cytokines (interferon [IFN]–α2, tumor necrosis factor–α, IL-8, IL-12p70, IL-23, and IL-33) were detected at much lower levels compared to those observed in healthy individuals. In comparison, age-matched immunocompetent COVID-19 patients [10] (n = 9, median 7 days post–symptom onset [range, 3–19 days]) had similarly higher levels of IL-6 (9.7-fold) and IL-18 (6.8-fold), but also higher levels of anti-inflammatory cytokines of IL-17A, IL-10, IL-1β, and IFN-γ, compared with healthy age-matched controls. Interestingly, other immunosuppressed COVID-19 patients had higher IL-6 levels, but also showed very high levels of IL-17A (18.7-fold) (n = 4; 3 patients on >10 mg prednisolone for >1 month for liver transplant [n = 2] or psoriatic arthritis [n = 1], 1 diffuse large B-cell lymphoma patient; median, 9 days after symptom onset [range, 5–19 days]) [10]. Overall, our case patient showed typical IL-6 and IL-18 cytokine responses, which was similar to other COVID-19 patients of similar age. Immunomodulatory therapies targeting dysregulated cytokine responses may be indicated in some immunocompetent patients [8].
Figure 2.
Immune responses in an immunocompromised patient with coronavirus disease 2019 (COVID-19). A, Cytokine and chemokine responses in an immunocompromised patient with COVID-19, age-matched COVID-19 patients (n = 9), and other immunosuppressed COVID-19 patients (n = 4; 3 patients on >10 mg prednisolone for >1 month for liver transplant [n = 2] or psoriatic arthritis [n = 1], 1 diffuse large B-cell lymphoma patient). Cytokine and chemokine levels are expressed as fold-change compared to the levels observed in 8 healthy age-matched donors (2 males, 6 females; median age, 72 years). B, Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) receptor binding domain–specific IgM, IgG, and IgA antibodies as determined by enzyme-linked immunosorbent assay, compared to a patient with mild COVID-19. C and D, Flow cytometry plots and percentage frequencies for each immune population are shown using the T follicular helper and antibody secreting cell panel (C) and the immune cell activation panel (monocytes and T/B/natural killer/γδ T cells) (D). E, Bar graph of HLA-DR+CD38+–activated immune cell subsets from (D), as well as age-matched COVID-19 patients (n = 9) and other immunosuppressed COVID-19 patients (n = 3) where mean and standard deviation are shown. F, CD4+ and CD8+ T-cell responses to SARS-CoV-2 spike, nucleocapsid, and membrane peptide pools compared to dimethyl sulfoxide (DMSO) control after 10 days of expansion with each peptide pool. Interferon-γ frequencies are based on the CD4+ or CD8+ T-cell population before concatenating the DMSO control and peptide flow cytometry files. Abbreviations: COVID-19, coronavirus disease 2019; DMSO, dimethyl sulfoxide; FITC, fluorescein isothiocyanate; IFN, interferon; IgA, immunoglobulin A; IgG, immunoglobulin G; IgM, immunoglobulin M; IL, interleukin; MCP-1, monocyte chemoattractant protein-1; NK, natural killer; PE, R-phycoerythrin; TCR, T cell receptor; Tfh, T follicular helper; TNF, tumor necrosis factor; +VE, positive.
Lack of SARS-CoV-2–Specific Antibodies, B Cells, Activated T Follicular Helper Cells, and PD-1 Expression
We next determined whether our immunocompromised patient with COVID-19 could mount robust and broad immune responses toward SARS-CoV-2 virus, as we have observed in a nonsevere case of COVID-19 [11]. Unsurprisingly, at day 33, there were no detectable antibody responses toward receptor-binding domain (RBD) (Figure 2B), as this patient had no B cells detected in the blood (Figure 2C). Instead, the lymphocyte population was dominated by CD8+ T cells, which was 2.5 times higher than CD4+ T cells. Strikingly, PD-1 expression was absent in both CD8+, CD4+, and CD4+CXCR5+ T follicular helper subsets. On the other hand, we observed relatively high levels of HLA-DR+/CD38+–activated natural killer (NK) cells and CD4+ T cells, followed by reasonable activation of γδ T cells and CD8+ T cells (Figure 2D and 2E). These levels were comparable to other age-matched COVID-19 patients, although when compared to other immunosuppressed COVID-19 patients who were not on ICI, NK cell activation was markedly higher in the case patient, as well as γδ T-cell and CD4+ T-cell activation levels (Figure 2E). We also found a higher ratio of patrolling CD16+ monocytes (24:1) compared to conventional CD14+ monocytes (Figure 2D), in contrast to a healthy state, with the predominant monocyte population being classical CD14+ monocytes. Therefore, the immune profiles, although not prototypical, showed medium to high levels of HLA-DR+/CD38+ activation in the absence of B cells and PD-1 expression.
Minimal Detection of SARS-CoV-2–Specific T Cells
We assessed whether the immunocompromised individual could elicit functional SARS-CoV-2–specific CD4+ and CD8+ T-cell responses using overlapping SARS-CoV-2 peptide pools spanning the immunogenic regions of spike (S), nucleocapsid (N), and membrane (M) proteins, as previously [12]. Small SARS-CoV-2–reactive CD4+ T-cell responses toward S and M were detected (1.44% and 1.78%, respectively), and minimal CD8+ T-cell responses were observed for M (Figure 2F). Although lower in magnitude, these findings are similar to our previous study [12], where CD4+ T-cell responses mainly targeted S and M proteins, and to a lesser extent toward N protein, in COVID-19 convalescent individuals. Here, we were unable to detect any responses toward the N protein, and very low responses to S and M proteins.
Persistent SARS-CoV-2 Replication
A nasopharyngeal swab taken on the day of admission remained positive for SARS-CoV-2 by polymerase chain reaction (PCR), and SARS-CoV-2 was subsequently isolated and cultured in Vero cells, confirming persistent viral replication 33 days after the first SARS-CoV-2 PCR-positive result (Figure 1, Supplementary Methods). Genomic analysis of 5 samples collected from days 0 to 36 indicated the sequences from these samples were all from the same cluster within the D.2 lineage (formerly B1.1.25.2), consistent with ongoing viral shedding rather than reinfection. Minimal intrahost evolution was identified, with 1 mutation in orf1ab (P5371S) identified at day 26, and a deletion in orf1ab (del:11288:9) identified at day 33 after the first positive PCR result; several synonymous single-nucleotide polymorphisms were also identified without associated amino acid changes.
DISCUSSION
Our results add to a growing number of reports that demonstrate that patients with B-cell and immunoglobulin deficiencies are at risk of an inadequate antiviral immunity, likely due to lack of functional neutralizing antibodies [3, 4]. Not only are these patients at risk of an atypical COVID-19 phenotype, but they may also act as a prolonged source of transmission.
We hypothesize that our patient’s underlying lymphoma, with multiple lines of treatment including drugs targeting the humoral immune system, in combination with recent PD-1 therapy, resulted in profound immunosuppression and incomplete immune clearance of SARS-CoV-2. B cells, activated follicular T helper cells, and antibody responses toward RBD were absent in peripheral blood, consistent with a history of B-cell malignancy and associated B-cell–depleting therapy. SARS-CoV-2–specific T cells were only minimally detected, likely reflecting immunosuppression secondary to chemotherapy. High levels of IL-6, IL-18, and HLA-DR+/CD38+ activation were present, yet PD-1 expression was absent on CD8+, CD4+ and CD4+CXCR5+ T follicular helper subsets, in line with a persistent suppressive effect of PD-1 inhibitor treatment administered nearly 3 months prior. In chronic infection and cancer, prolonged antigen exposure leads to permanent PD-1 expression, which can impair immune-mediated clearance of infected and cancerous cells [13]. However, during acute infection, activation of the PD-1 pathway has a beneficial effect in dampening acute phase immune responses and preventing immunopathology [14]. We hypothesize that our patient’s rapid decline and immunopathology was contributed to by a lack of activation of the PD-1 pathway due to recent PD-1 inhibitor therapy leading to an unchecked acute inflammatory response, in addition to an overall state of heightened net immunosuppression.
Our case adds to the growing body of evidence that individuals with humoral immunodeficiency may shed infectious virus for prolonged periods of time [3, 4]. In immunocompetent hosts, SARS-CoV-2 RNA can be detected long after a patient becomes culture negative, yet culture positivity has not been identified past 20 days [15]. However, culture positivity can occur in immunocompromised hosts for up to 8 months [3, 4, 6], and immunocompromised hosts may need to meet additional criteria for clearance from isolation.
Our case illustrates defects in SARS-CoV-2–specific immune responses and persistent viral replication in a patient with lymphoma, B-cell depletion, and recent PD-1 inhibitor therapy. A limitation of our study is that it comprises a single case and immune analysis at a single time point, making it difficult to draw general conclusions regarding SARS-CoV-2 pathogenesis, viral kinetics, and immunity. Nevertheless, our analysis of SARS-CoV-2–specific immune parameters may shed new light on which immune pathways are important in controlling SARS-CoV-2 replication and preventing severe disease.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We would like to thank the staff of Austin Pathology, Melbourne Pathology and Victorian Infectious Diseases Reference Laboratory for performing the diagnostic SARS-CoV-2 nucleic acid testing assays, and the Microbiological Diagnostic Unit Public Health Laboratory for performing whole genome sequencing and analysis.
Patient consent statement. The patient’s written consent was obtained. This project was approved by Austin Health Human Ethics Committee (HREC/63201/Austin-2020).
Financial support. C. L. G. (GNT 1160963), J. A. T. (GNT 1160963), and J. C. K. (GNT 1142613) are supported by the National Health and Medical Research Council (NHMRC) Early Career Fellowships. This work was supported by the NHMRC Leadership Investigator Grant (number 1173871 to K. K.) and GNT1196103 to B. P. H.). T. H. O. N. is supported by NHMRC EL1 Fellowship (number 1194036) and W. Z. is supported by the Melbourne International Research Scholarship and the Melbourne International Fee Remission Scholarship from the University of Melbourne.
Potential conflicts of interest. All authors: No reported conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Ou X, Liu Y, Lei X, et al. . Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 2020; 11:1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grifoni A, Weiskopf D, Ramirez SI, et al. . Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 2020; 181:1489–1501.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avanzato VA, Matson MJ, Seifert SN, et al. . Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell 2020; 183:1901–12.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baang JH, Smith C, Mirabelli C, et al. . Prolonged severe acute respiratory syndrome coronavirus 2 replication in an immunocompromised patient. J Infect Dis 2021; 223:23–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckland MS, Galloway JB, Fhogartaigh CN, et al. ; CITIID-NIHR COVID-19 BioResource Collaboration; MRC-Toxicology Unit COVID-19 Consortium. Treatment of COVID-19 with remdesivir in the absence of humoral immunity: a case report. Nat Commun 2020; 11:6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sepulcri C, Dentone C, Mikulska M, et al. . The longest persistence of viable SARS-CoV-2 with recurrence of viremia and relapsing symptomatic COVID-19 in an immunocompromised patient—a case study [manuscript published online ahead of print 28 April 2021]. Open Forum Infect Dis 2021. doi:10.1093/ofid/ofab217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barber DL, Wherry EJ, Masopust D, et al. . Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 2006; 439:682–7. [DOI] [PubMed] [Google Scholar]
- 8.National Institutes of Health. COVID-19 treatment guidelines: clinical spectrum of SARS-CoV-2 infection.2020. https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum/. Accessed 22 December 2020.
- 9.Zhang X, Tan Y, Ling Y, et al. . Viral and host factors related to the clinical outcome of COVID-19. Nature 2020; 583:437–40. [DOI] [PubMed] [Google Scholar]
- 10.Copaescu A, James F, Mouhtouris E, et al. . The role of immunological and clinical biomarkers to predict clinical COVID-19 severity and response to therapy—a prospective longitudinal study. Front Immunol 2021; 12:646095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thevarajan I, Nguyen THO, Koutsakos M, et al. . Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat Med 2020; 26:453–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Habel JR, Nguyen THO, van de Sandt CE, et al. . Suboptimal SARS-CoV-2-specific CD8+ T cell response associated with the prominent HLA-A*02:01 phenotype. Proc Natl Acad Sci U S A 2020; 117:24384–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pauken KE, Wherry EJ. Overcoming T cell exhaustion in infection and cancer. Trends Immunol 2015; 36:265–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schönrich G, Raftery MJ. The PD-1/PD-L1 axis and virus infections: a delicate balance. Front Cell Infect Microbiol 2019; 9:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rhee C, Kanjilal S, Baker M, Klompas M. Duration of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infectivity: when is it safe to discontinue isolation?. Clin Infect Dis 2021; 72:1467–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.