Abstract
Severe acute respiratory syndrome coronaviruses (SARS-CoVs) caused worldwide epidemics over the past few decades. Extensive studies on various strains of coronaviruses provided a basic understanding of the pathogenesis of the disease. Presently, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is leading a global pandemic with unprecedented challenges. This is the third coronavirus outbreak of this century. A signaling pathway map of signaling events induced by SARS-CoV infection is not yet available. In this study, we present a literature-annotated signaling pathway map of reactions induced by SARS-CoV infected cells. Multiple signaling modules were found to be orchestrated including PI3K-AKT, Ras-MAPK, JAK-STAT, Type 1 IFN and NFκB. The signaling pathway map of SARS-CoV consists of 110 molecules and 101 reactions mediated by SARS-CoV proteins. The pathway reaction data are available in various community standard data exchange formats including Systems Biology Graphical Notation (SBGN). The pathway map is publicly available through the GitHub repository and data in various formats can be freely downloadable.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12079-021-00642-2.
Keywords: COVID-19, Post-translational modifications, Protein–protein interaction, Translocation, Differential expression
Introduction
Coronaviruses (CoVs) are enveloped, single stranded, positive sense RNA viruses that are phenotypically and genotypically diverse. CoVs can infect bats, birds, cats, dogs, pigs, mice, horses, whales, and humans (Zaki et al. 2012). The first detected human coronavirus can be traced back to studies carried out in 1965 (Kahn and McIntosh 2005). However, the field of coronavirology advanced significantly in recent years (Kahn and McIntosh 2005). Though the exact mechanism of species-to-species transmission of the virus has not been clear. From the turn of the twenty-first century three deadly CoVs: the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV; 2002), the Middle East Respiratory Syndrome coronavirus (MERS-CoV; 2012), and the ongoing 2019 novel coronavirus, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) made a mark in history by causing havoc in different parts of the world (Guo et al. 2020). The diversity in these viruses can be attributed to RNA dependent RNA polymerases, as they possess high frequency of RNA recombination and large genome size as compared to other RNA viruses (Zaki et al. 2012). The shared proteins among the different CoVs may vary in structure and function. However, the proteins seem to be multifunctional, indicating a common theme interconnecting CoVs (Wong and Saier 2021). The ability of SARS-CoVs to transfect hosts have been a major concern and eventually led from an epidemic to pandemic situation (Perlman and Netland 2009). SARS-CoV infection resulted in severe and potentially fatal lung disease. Majority of the patients infected with SARS-CoV showed febrile illness accompanied by weakness. Nevertheless, a considerable population of individuals developed severe inflammation of the lung, necessitating ventilator support and intensive care. Many patients from this group had acute respiratory distress syndrome (ARDS) with high mortality (Hui and Sung 2004). Individuals of this group also showed manifestations of infection in other organ systems. Lymphopenia (Wong et al. 2003a), gastrointestinal symptoms (Leung et al. 2003), impaired renal functions (Chu et al. 2005), and impaired liver function (Chan et al. 2004; Wong et al. 2003b) were other common conditions.
SARS-CoVs have a genome size of ~ 30 Kb with 14 open reading frames (ORFs) encoding the viral proteins during its infection cycle (Bruford et al. 2008; Gordon et al. 2020). These proteins mainly comprise of four structural proteins, spike (S), membrane (M), envelope (E), and nucleocapsid (N). Table 1 summarizes the functional receptors of various strains of CoVs. Broadly, CoVs infection cycle involves attachment/binding to the host cells, replicase protein expression, replication and transcription, assembly and release (Fehr and Perlman 2015). Of these, binding of CoVs to the host cell membrane receptors is a major determinant for the detection of infection cycle and host range of CoVs. To infect a new host species CoV must adapt to the receptor of the new host (Belouzard et al. 2012). Alternatively, host-factors such as, interferon-inducible transmembrane proteins can play essential role to restrict or facilitate the attachment and entry of CoVs (Bailey et al. 2014; Huang et al. 2011; Zhao et al. 2014; Zhao et al. 2018).
Table 1.
List of extensively studied coronavirus receptors and functions of non-structural proteins
| Coronavirus | Receptor | References |
|---|---|---|
| HCoV-229E | APN | Yeager et al. (1992) |
| HCoV-NL63 | ACE2 | Hofmann et al. (2005) |
| HCoV-OC43 | 9-O-acetylated sialic acid | Lu et al. (2020) |
| HCoV-HKU1 | 9-O-acetylated sialic acid | Lu et al. (2020) |
| CCoV | APN | Benbacer et al. (1997) |
| BCoV | N-acetyl-9-O-acetylneuraminic acid | Schultze and Herrler (1992) |
| SARS-CoV | ACE2 | Li et al. (2003) |
| MERS-CoV | DPP4 | Raj et al. (2013) |
Various studies determined CoV proteins modulating host signaling pathways through the interactions with host proteins (Khan and Islam 2021; Munjal et al. 2021). Particularly, studies have shown that activation of intracellular signaling cascades induced upon SARS-CoV infection leads to the post-translational modifications (PTMs) and activation of downstream molecules (Garrington and Johnson 1999; Kyriakis and Avruch 2001; Whitmarsh and Davis 2000). Predominantly, CoVs infection caused altered expression of host kinases, chemokines and transcription factors to evade host-immune responses and to aid replication and assembly of viral particle (Garrington and Johnson 1999; Kyriakis and Avruch 2001; Whitmarsh and Davis 2000). Upon SARS-CoV infection, cell death has been observed in cultured Vero E6 cells (Mizutani et al. 2004c). Supplementary Table 1 summarizes the list of cell lines used to investigate the biological functions induced by SARS-CoV (Kaye 2006). The signaling pathways regulating cell death and survival in SARS-infected cells were highly complex. The in-depth understanding of these regulatory mechanisms will help in underlying the pathophysiology of SARS-CoVs infection cycle and host-range.
Despite the immense need to understand the pathology of SARS-CoVs, the signaling pathway map of reactions induced by SARS-CoV proteins are not available. This study presents the signal transduction pathways of SARS-CoV-infected cells in Graphical Pathway Markup Language (GPML) and SBGN format. GPML format is a custom XML format compatible with pathway visualisation and analysis tools such as, Cytoscape (Shannon et al. 2003) and PathVisio (Kutmon et al. 2015). SBGN is a set of three complementary visual languages that helps in representing networks of biological interactions in a standard, unambiguous manner. This would further result in efficient representation, visualization, storage, exchange, and reuse of various types of biological knowledge (Bergmann et al. 2020). Nodes describe entity pools (genes, proteins, etc.) and process (associations, influences, etc.). Edges describe the relationship between the nodes.
The generation of SARS-CoV-host signaling pathway map
An extensive search of published literature was performed using PubMed to annotate the reactions induced by SARS-CoV proteins. Several query terms were used including, “SARS-CoV” AND “ACE2”, “SARS-CoV” AND “pathway” OR “signaling”. The articles were screened for information pertaining to protein–protein interactions (molecular association), PTMs (catalysis), transport (translocation of proteins between sub-cellular compartments), activation/inhibition events and gene regulation events (Sharma et al. 2015; Zhong et al. 2014) which were induced by SARS-CoV infection in host cells. The pathway map was generated using PathVisio (version 3.3.0), an open-source pathway drawing software and CellDesigner (version 4.4.2), a process diagram editor for drawing gene regulatory and biochemical networks.
Results and discussion
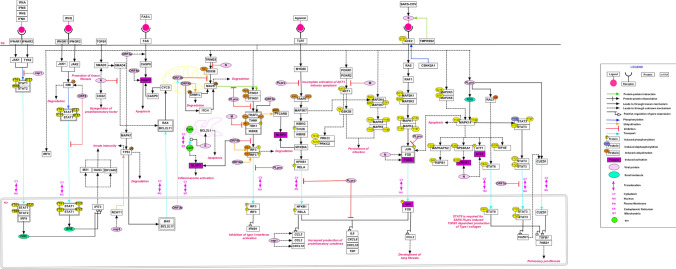
The signaling events orchestrated by SARS-CoV are shown in Fig. 1. A number of signaling cascades are triggered during the entire process of viral infection, from S protein-ACE2 binding for internalization into the host cells to apoptotic cell death (Li et al. 2003). S protein of SARS-CoV was cleaved at multiple motifs by host type II transmembrane serine protease (TMPRSS2) that in turn activated the process of cell–cell or virus-cell fusion (Glowacka et al. 2011). It was observed that overexpression of ACE2 allowed efficient SARS-CoV’s S driven cell–cell fusion (Glowacka et al. 2011). ACE2 and TMPRSS2 were co-expressed in type II pneumocytes (Glowacka et al. 2011). SARS-CoV was also found to be dependent on the activity of pH-dependent endosomal cysteine protease cathepsin L (CTSL) for virus-cell fusion (Glowacka et al. 2011). The binding of S with ACE2 activated CSNK2A1 mediated ACE2 phosphorylation. This led to severe immune response involving cytokines and chemokines in pneumocytes of infected patients. (Chen et al. 2010). On infection with SARS-CoV various proinflammatory factors such as CXCL8, CXCL10, CCL2, CCL3, CCL5 and COX2 were found to be triggered through RAS-MAPK-AP1 and NFκB pathways (Cinatl et al. 2005; Chen et al. 2010; Law et al. 2007; Hu et al. 2017). E protein, ORF3a and ORF8b were involved in the activation of NLRP3 inflammasomes (Nieto-Torres et al. 2015; Shi et al. 2019; Siu et al. 2019). Also, the activation of c-Fos, FosB, CREB1 and ATF2 by N protein showed its involvement in the AP1 pathway (He et al. 2003). The role of SARS-CoV proteins in innate immunity have been reported by various studies. M protein of SARS-CoV inhibited the activation of interferon regulatory factor 3 (IRF3) and interferon (IFN) synthesis by hampering the formation of a functional TRAF3-TANK-TBK1/IKKϵ complex (Siu et al. 2009). The N protein interfered in the association of TRIM25 and DDX58 that led to the suppression of Type I IFN production (Hu et al. 2017). ORF9b which was localized in the mitochondria (MT) induced ubiquitination and proteasomal degradation of DNM1L (DRP1), leading to the elongation of MT. ORF9b possibly appropriates ITCH (AIP4), a ubiquitin E3 ligase to trigger the degradation of MAVS accompanied by loss of TRAF3 and TRAF6 thus significantly suppressing the IFN responses (Shi et al. 2014). Co-expression of nsp1 in Type I IFN pathway decreased the phosphorylation levels at Y701 and S727 of STAT1 and Y690 of STAT2 (Law et al. 2007). PLpro of SARS-CoV inhibited stimulator of interferon genes (STING)-mediated activation and translocation of IRF3 thereby preventing the induction of Type I IFN (Sun et al. 2012). PLpro also inhibited IFN production mediated through agonist induced TLR7 signaling pathway by removing the K63 linked polyubiquitination of TRAF3 and TRAF6 (Li et al. 2016a). It disrupted the STING mediated signaling and then negatively regulated type I IFN induction (Chen et al. 2014; Sun et al. 2012). It also physically interacted with the STING-TRAF3-TBK1 complex, reducing the ubiquitinated forms of STING, DDX58, TRAF-3, TBK1 and IRF3.
Fig. 1.
A schematic depiction of reactions induced by severe acute respiratory syndrome coronavirus (SARS-CoV). The pathway reaction map depicts each type of reactions such as molecular associations, catalysis, and translocation events induced upon by SARS-CoV infection. Site and residue information of post-translational modifications are also provided whenever available in literature. Important pathways including, PI3K-AKT, Ras-MAPK, and NFκB were found to be activated. The edges representing the relationships between nodes are provided in the legend
ORF6 (P6) of SARS-CoV could prevent IFN-β production upon infection and may be involved in the suppression of the IFN immune signaling. It targeted N-Myc (and STAT) interactor (NMI) to induce its degradation, which led to NMI-enhanced IFN signaling suppression and was also involved in blocking STAT1 nuclear translocation(Cheng et al. 2015). ORF3b was accumulated in the nucleus during the initial phase. During the later phases it got translocated to the MT where it was involved in the inhibition of signaling for the production of Type I IFN possibly by inhibiting the MAVS response to DDX58 (Freundt et al. 2009). SARS-unique domain (SUD) of Nsp3 could enhance cellular E3 ubiquitin ligase known as ring-finger and CHY zinc-finger domain containing 1 (RCHY1) that led to proteasomal degradation of TP53 resulting in downregulation of TP53 (Ma-Lauer et al. 2016). ORF8b directly interacted with IRF3 and affected its dimerization and phosphorylation status thereby suppressing IFN-β. In addition, the expression of ORF8b and ORF8ab also regulated the stability and function of IRF3 (Wong et al. 2018).
SARS-CoV infection has been shown to induce apoptosis through DNA fragmentation and activation of caspase (Mizutani et al. 2004c; Yan et al. 2004). ORF8a induced apoptosis by activating caspase-3 (Chen et al. 2007). The cytopathic effects on entry of SARS-CoV into the host were partially inhibited by p38-MAPK14 specific inhibitor, which indicated that p38-MAPK14 pathway may be involved in cell death (Mizutani et al. 2004c). Also, the activation and phosphorylation of p38-MAPK14 and its downstream targets including MAPKAPK2, HSPB1, RPS6KA1, CREB1, ATF1 and EIF4E were observed (Mizutani et al. 2004a, 2004c, 2006). ORF3a inhibited the nuclear translocation of STAT3 leading to dysregulation in transcription of anti-apoptotic genes (Padhan et al. 2008). Infection of cells with SARS-CoV led to dephosphorylation of STAT3 at Y705 and increased the phosphorylation of STAT3 at S727 (Mizutani et al. 2004a). p38-MAPK14 pathway was shown to be associated with STAT3 dephosphorylation at Y705 (Mizutani et al. 2004a). SARS-CoV PLpro stimulated Egr-1 (CUZD1) dependent activation of pro-fibrotic genes, TGFB1 and THBS1 through ROS-MAPK-STAT3 pathway (Li et al. 2016b). TGFB1 dependent expression of Type I collagen was stimulated by SARS-CoV PLpro via STAT6 pathway (Wang et al. 2017). Additionally, JNK and PI3K-AKT pathways were essential for the persistence of SARS-CoV infection (Mizutani et al. 2005). AKT1 and its downstream targets, GSK3B1, PRKCI and PRKCZ were phosphorylated. AKT1 was phosphorylated at serine residue (S473) (Mizutani et al. 2004b). Also, the overexpression of M protein led to downregulation of AKT1 phosphorylation thus inducing apoptosis (Chan et al. 2007). E protein of SARS-CoV activated apoptosis by sequestering anti-apoptotic BCL2L1 (Bcl-xL) to endoplasmic reticulum (Yang et al. 2005). Upon infection with SARS-CoV, N protein enhanced TGF-β/Smad3-induced expression of plasminogen activator inhibitor-1 (PAI-1) (Zhao et al. 2008). Thus, N protein modulated TGF-β signaling to block apoptosis of SARS-CoV-infected host cells while promoting tissue fibrosis (Zhao et al. 2008). ORF3a was involved in apoptotic pathway through indirect activation of TP53 by p38-MAPK (Padhan et al. 2008).
Conclusions
SARS-CoV-host signaling modules will accelerate the understanding of various molecules and their roles in the pathogenesis of infection caused by SARS-CoV-2. Most likely, SARS-CoV-host signal transduction pathway may be useful for the development of potential molecular targets for antiviral treatment against different strains of CoVs. The pathway map available through this study will thus provide the scientific community with a platform that will further help to understand the pathology of various strains of SARS-CoVs.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Alexander Mazein for providing the SBGN training and Marek Ostaszewski for his valuable suggestions to enrich the manuscript. JS is a recipient of Bio-CARe Women Scientists award by Department of Biotechnology (DBT), Government of India (Grant number-BT/PR19924/BIC/101/568/2016). LB is a recipient of National-Post doctoral fellowship scheme by Science and Engineering Research Board (SERB), Department of Science and Technology, (Project No.PDF/2017/002992).
Authors’ contributions
JS and LB conceptualized and designed the study. KTSP, NSM, AP, LB and JS analyzed and interpreted the data. KTSP, NSM, GD, AK, AP, LB and JS contributed to the writing of the manuscript.
Funding
This work was funded by the intramural funds of Institute of Bioinformatics, India and Bio-CARe by Department of Biotechnology (DBT), Government of India, grant number BT/PR19924/BIC/101/568/2016.
Data availability
Data generated during this study are available at GitHub via following URLs:
https://github.com/js-iob/SARS-CoV-Pathway/blob/master/SARS_CoV_Signaling_pathway_map.gpml
https://github.com/js-iob/SARS-CoV-Pathway/blob/master/SARS-CoV_Signaling_pathway_map_GPML.pdf
https://github.com/js-iob/SARS-CoV-Pathway/blob/master/SARS-CoV_Signaling_pathway_map.xml
https://github.com/js-iob/SARS-CoV-Pathway/blob/master/SARS-CoV_Signaling_pathway_map_SBGN.pdf
Declarations
Conflict of interest
The authors declare that no competing financial interests exist.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
K. T. Shreya Parthasarathi and Nupur S. Munjal have contributed equally to this work
Contributor Information
Lavanya Balakrishnan, Email: lavanya.ibioinformatics@gmail.com.
Jyoti Sharma, Email: jyoti@ibioinformatics.org.
References
- Bailey CC, Zhong G, Huang IC, Farzan M. IFITM-family proteins: the cell's first line of antiviral defense. Annu Rev Virol. 2014;1:261–283. doi: 10.1146/annurev-virology-031413-085537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benbacer L, Kut E, Besnardeau L, Laude H, Delmas B. Interspecies aminopeptidase-N chimeras reveal species-specific receptor recognition by canine coronavirus, feline infectious peritonitis virus, and transmissible gastroenteritis virus. J Virol. 1997;71:734–737. doi: 10.1128/jvi.71.1.734-737.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann FT, et al. Systems biology graphical notation markup language (SBGNML) version 0.3. J Integr Bioinform. 2020 doi: 10.1515/jib-2020-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruford EA, Lush MJ, Wright MW, Sneddon TP, Povey S, Birney E. The HGNC database in 2008: a resource for the human genome. Nucleic Acids Res. 2008;36:D445–448. doi: 10.1093/nar/gkm881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HL, et al. Retrospective analysis of liver function derangement in severe acute respiratory syndrome. Am J Med. 2004;116:566–567. doi: 10.1016/j.amjmed.2003.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CM, Ma CW, Chan WY, Chan HY. The SARS-Coronavirus Membrane protein induces apoptosis through modulating the Akt survival pathway. Arch Biochem Biophys. 2007;459:197–207. doi: 10.1016/j.abb.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, et al. Open reading frame 8a of the human severe acute respiratory syndrome coronavirus not only promotes viral replication but also induces apoptosis. J Infect Dis. 2007;196:405–415. doi: 10.1086/519166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IY, et al. Upregulation of the chemokine (C–C motif) ligand 2 via a severe acute respiratory syndrome coronavirus spike-ACE2 signaling pathway. J Virol. 2010;84:7703–7712. doi: 10.1128/JVI.02560-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Yang X, Zheng Y, Yang Y, Xing Y, Chen Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell. 2014;5:369–381. doi: 10.1007/s13238-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Chen S, Li R, Chen Y, Wang M, Guo D. Severe acute respiratory syndrome coronavirus protein 6 mediates ubiquitin-dependent proteosomal degradation of N-Myc (and STAT) interactor. Virol Sin. 2015;30:153–161. doi: 10.1007/s12250-015-3581-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu KH, et al. Acute renal impairment in coronavirus-associated severe acute respiratory syndrome. Kidney Int. 2005;67:698–705. doi: 10.1111/j.1523-1755.2005.67130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinatl J, Jr, Michaelis M, Morgenstern B, Doerr HW. High-dose hydrocortisone reduces expression of the pro-inflammatory chemokines CXCL8 and CXCL10 in SARS coronavirus-infected intestinal cells. Int J Mol Med. 2005;15:323–327. [PubMed] [Google Scholar]
- Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freundt EC, Yu L, Park E, Lenardo MJ, Xu XN. Molecular determinants for subcellular localization of the severe acute respiratory syndrome coronavirus open reading frame 3b protein. J Virol. 2009;83:6631–6640. doi: 10.1128/JVI.00367-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrington TP, Johnson GL. Organization and regulation of mitogen-activated protein kinase signaling pathways. Curr Opin Cell Biol. 1999;11:211–218. doi: 10.1016/S0955-0674(99)80028-3. [DOI] [PubMed] [Google Scholar]
- Glowacka I, et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DE, et al. A SARS-CoV-2-human protein-protein interaction map reveals drug targets and potential drug-repurposing. Nature. 2020 doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Hu BJ, Yang XL, Zeng LP, Li B, Ouyang S, Shi ZL. Evolutionary arms race between virus and host drives genetic diversity in bat severe acute respiratory syndrome-related coronavirus spike genes. J Virol. 2020;94:e00902-20. doi: 10.1128/JVI.00902-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R, et al. Activation of AP-1 signal transduction pathway by SARS coronavirus nucleocapsid protein. Biochem Biophys Res Commun. 2003;311:870–876. doi: 10.1016/j.bbrc.2003.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H, Pyrc K, van der Hoek L, Geier M, Berkhout B, Pohlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci U S A. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, et al. The Severe Acute Respiratory Syndrome Coronavirus Nucleocapsid Inhibits Type I Interferon Production by Interfering with TRIM25-Mediated RIG-I Ubiquitination. J Virol. 2017;91:e02143-16. doi: 10.1128/JVI.02143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang IC, et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS Coronavirus, and Influenza A Virus. PLoS Pathog. 2011;7:e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui DS, Sung JJ. Treatment of severe acute respiratory syndrome. Chest. 2004;126:670–674. doi: 10.1378/chest.126.3.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn JS, McIntosh K. History and recent advances in coronavirus discovery. Pediatr Infect Dis J. 2005;24:S223–227. doi: 10.1097/01.inf.0000188166.17324.60. [DOI] [PubMed] [Google Scholar]
- Kaye M. SARS-associated coronavirus replication in cell lines. Emerg Infect Dis. 2006;12:128–133. doi: 10.3201/eid1201.050496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Islam A. SARS-CoV-2 proteins exploit host's genetic and epigenetic mediators for the annexation of key host signaling pathways. Front Mol Biosci. 2021;7:598583. doi: 10.3389/fmolb.2020.598583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutmon M, van Iersel MP, Bohler A, Kelder T, Nunes N, Pico AR, Evelo CT. PathVisio 3: an extendable pathway analysis toolbox. PLoS Comput Biol. 2015;11:e1004085. doi: 10.1371/journal.pcbi.1004085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol Rev. 2001;81:807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- Law AH, Lee DC, Cheung BK, Yim HC, Lau AS. Role for nonstructural protein 1 of severe acute respiratory syndrome coronavirus in chemokine dysregulation. J Virol. 2007;81:416–422. doi: 10.1128/JVI.02336-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung WK, et al. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003;125:1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SW, et al. SARS coronavirus papain-like protease inhibits the TLR7 signaling pathway through removing Lys63-linked polyubiquitination of TRAF3 and TRAF6. Int J Mol Sci. 2016;17:678. doi: 10.3390/ijms17050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SW, et al. SARS coronavirus papain-like protease induces Egr-1-dependent up-regulation of TGF-beta1 via ROS/p38 MAPK/STAT3 pathway. Sci Rep. 2016;6:25754. doi: 10.1038/srep25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma-Lauer Y, et al. p53 down-regulates SARS coronavirus replication and is targeted by the SARS-unique domain and PLpro via E3 ubiquitin ligase RCHY1. Proc Natl Acad Sci U S A. 2016;113:E5192–E5201. doi: 10.1073/pnas.1603435113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani T, Fukushi S, Saijo M, Kurane I, Morikawa S. Phosphorylation of p38 MAPK and its downstream targets in SARS coronavirus-infected cells. Biochem Biophys Res Commun. 2004;319:1228–1234. doi: 10.1016/j.bbrc.2004.05.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani T, Fukushi S, Saijo M, Kurane I, Morikawa S. Importance of Akt signaling pathway for apoptosis in SARS-CoV-infected Vero E6 cells. Virology. 2004;327:169–174. doi: 10.1016/j.virol.2004.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani T, Fukushi S, Murakami M, Hirano T, Saijo M, Kurane I, Morikawa S. Tyrosine dephosphorylation of STAT3 in SARS coronavirus-infected Vero E6 cells. FEBS Lett. 2004;577:187–192. doi: 10.1016/j.febslet.2004.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani T, Fukushi S, Saijo M, Kurane I, Morikawa S. JNK and PI3k/Akt signaling pathways are required for establishing persistent SARS-CoV infection in Vero E6 cells. Biochim Biophys Acta. 2005;1741:4–10. doi: 10.1016/j.bbadis.2005.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani T, Fukushi S, Saijo M, Kurane I, Morikawa S. Regulation of p90RSK phosphorylation by SARS-CoV infection in Vero E6 cells. FEBS Lett. 2006;580:1417–1424. doi: 10.1016/j.febslet.2006.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munjal NS, Sapra D, Goyal A, Parathasarthi KTS, Pandey A, Banerjee M, Sharma J. Deciphering the interactions of SARS-CoV-2 proteins with human ion channels using machine learning-based method. Res Square. 2021 doi: 10.21203/rs.3.rs-622770/v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres JL, et al. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology. 2015;485:330–339. doi: 10.1016/j.virol.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhan K, Minakshi R, Towheed MAB, Jameel S. Severe acute respiratory syndrome coronavirus 3a protein activates the mitochondrial death pathway through p38 MAP kinase activation. J Gen Virol. 2008;89:1960–1969. doi: 10.1099/vir.0.83665-0. [DOI] [PubMed] [Google Scholar]
- Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj VS, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze B, Herrler G. Bovine coronavirus uses N-acetyl-9-O-acetylneuraminic acid as a receptor determinant to initiate the infection of cultured cells. J Gen Virol. 1992;73(4):901–906. doi: 10.1099/0022-1317-73-4-901. [DOI] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma J, et al. A knowledgebase resource for interleukin-17 family mediated signalling. J Cell Commun Signal. 2015;9:291–296. doi: 10.1007/s12079-015-0297-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Qi HY, Boularan C, Huang NN, Abu-Asab M, Shelhamer JH, Kehrl JH. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J Immunol. 2014;193:3080–3089. doi: 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Nabar NR, Huang NN, Kehrl JH. SARS-coronavirus open reading frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov. 2019;5:101. doi: 10.1038/s41420-019-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu KL, Kok KH, Ng MH, Poon VK, Yuen KY, Zheng BJ, Jin DY. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J Biol Chem. 2009;284:16202–16209. doi: 10.1074/jbc.M109.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu KL, et al. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019;33:8865–8877. doi: 10.1096/fj.201802418R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, et al. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS ONE. 2012;7:e30802. doi: 10.1371/journal.pone.0030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, et al. SARS coronavirus papain-like protease up-regulates the collagen expression through non-Samd TGF-beta1 signaling. Virus Res. 2017;235:58–66. doi: 10.1016/j.virusres.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmarsh AJ, Davis RJ. A central control for cell growth. Nature. 2000;403:255–256. doi: 10.1038/35002220. [DOI] [PubMed] [Google Scholar]
- Wong NA, Saier MH., Jr The SARS-coronavirus infection cycle: a survey of viral membrane proteins, their functional interactions and pathogenesis. Int J Mol Sci. 2021;22:1308. doi: 10.3390/ijms22031308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong RS, et al. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. BMJ. 2003;326:1358–1362. doi: 10.1136/bmj.326.7403.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WM, et al. Temporal patterns of hepatic dysfunction and disease severity in patients with SARS. JAMA. 2003;290:2663–2665. doi: 10.1001/jama.290.20.2663. [DOI] [PubMed] [Google Scholar]
- Wong HH, Fung TS, Fang S, Huang M, Le MT, Liu DX. Accessory proteins 8b and 8ab of severe acute respiratory syndrome coronavirus suppress the interferon signaling pathway by mediating ubiquitin-dependent rapid degradation of interferon regulatory factor 3. Virology. 2018;515:165–175. doi: 10.1016/j.virol.2017.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, et al. SARS coronavirus induces apoptosis in Vero E6 cells. J Med Virol. 2004;73:323–331. doi: 10.1002/jmv.20094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, et al. Bcl-xL inhibits T-cell apoptosis induced by expression of SARS coronavirus E protein in the absence of growth factors. Biochem J. 2005;392:135–143. doi: 10.1042/BJ20050698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager CL, Ashmun RA, Williams RK, Cardellichio CB, Shapiro LH, Look AT, Holmes KV. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhao X, Nicholls JM, Chen YG. Severe acute respiratory syndrome-associated coronavirus nucleocapsid protein interacts with Smad3 and modulates transforming growth factor-beta signalling. J Biol Chem. 2008;283:3272–3280. doi: 10.1074/jbc.M708033200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Guo F, Liu F, Cuconati A, Chang J, Block TM, Guo JT. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc Natl Acad Sci U S A. 2014;111:6756–6761. doi: 10.1073/pnas.1320856111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, et al. Identification of residues controlling restriction versus enhancing activities of IFITM proteins on entry of human coronaviruses. J Virol. 2018;92:e01535-17. doi: 10.1128/JVI.01535-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, et al. TSLP signaling pathway map: a platform for analysis of TSLP-mediated signalling. Database Oxford. 2014;2014:bau007. doi: 10.1093/database/bau007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated during this study are available at GitHub via following URLs:
https://github.com/js-iob/SARS-CoV-Pathway/blob/master/SARS_CoV_Signaling_pathway_map.gpml
https://github.com/js-iob/SARS-CoV-Pathway/blob/master/SARS-CoV_Signaling_pathway_map_GPML.pdf
https://github.com/js-iob/SARS-CoV-Pathway/blob/master/SARS-CoV_Signaling_pathway_map.xml
https://github.com/js-iob/SARS-CoV-Pathway/blob/master/SARS-CoV_Signaling_pathway_map_SBGN.pdf