Abstract
The mechanism of signal transduction by the estrogen receptor (ER) is complex and not fully understood. In addition to the ER, a number of accessory proteins are apparently required to efficiently transduce the steroid hormone signal. In the absence of estradiol, the ER, like other steroid receptors, is complexed with Hsp90 and other molecular chaperone components, including an immunophilin, and p23. This Hsp90-based chaperone complex is thought to repress the ER’s transcriptional regulatory activities while maintaining the receptor in a conformation that is competent for high-affinity steroid binding. However, a role for p23 in ER signal transduction has not been demonstrated. Using a mutant ER (G400V) with decreased hormone binding capacity as a substrate in a dosage suppression screen in yeast cells (Saccharomyces cerevisiae), we identified the yeast homologue of the human p23 protein (yhp23) as a positive regulator of ER function. Overexpression of yhp23 in yeast cells increases ER transcriptional activation by increasing estradiol binding in vivo. Importantly, the magnitude of the effect of yhp23 on ER transcriptional activation is inversely proportional to the concentration of both ER and estradiol in the cell. Under conditions of high ER expression, ER transcriptional activity is largely independent of yhp23, whereas at low levels of ER expression, ER transcriptional activation is primarily dependent on yhp23. The same relationship holds for estradiol levels. We further demonstrate that yhp23 colocalizes with the ER in vivo. Using a yhp23-green fluorescent protein fusion protein, we observed a redistribution of yhp23 from the cytoplasm to the nucleus upon coexpression with ER. This nuclear localization of yhp23 was reversed by the addition of estradiol, a finding consistent with yhp23’s proposed role as part of the aporeceptor complex. Expression of human p23 in yeast partially complements the loss of yhp23 function with respect to ER signaling. Finally, ectopic expression of human p23 in MCF-7 breast cancer cells increases both hormone-dependent and hormone-independent transcriptional activation by the ER. Together, these results strongly suggest that p23 plays an important role in ER signal transduction.
Estrogen is a steroid hormone responsible for the proper function of a variety of mammalian physiological processes. In addition to its central role in reproduction (6, 33), estrogen also affects the cardiovascular (19), skeletal (47), immune (9), and nervous (57) systems and plays a role in the initiation and progression of breast cancer (58).
The estrogen signal is mediated by the estrogen receptor (ER), a ligand-inducible transcription factor. In the absence of estradiol, the ER is found predominantly in the nucleus (32, 56), as part of a multiprotein complex consisting of a dimer of Hsp90 (6), a p23 monomer (42), and one of several immunophilins, including Cyp-40 (49) and FKBP52 (48). It has been proposed that this Hsp90-based chaperone complex inactivates the ER’s transcriptional regulatory capabilities and maintains the ER in a conformation competent for steroid binding (46). Upon binding estradiol, ER dissociates from this complex, dimerizes, and recognizes specific DNA sequences (35), termed estrogen-response elements (EREs), within the promoters of estrogen-responsive genes. Once bound to an ERE, ER is believed to modulate transcription of the linked gene through direct or indirect interactions with general transcriptional factors (15, 25).
Although many aspects of ER signaling are not yet understood, it appears that the proteins essential to ER function are conserved among eukaryotes to such an extent that introduction of ER into Saccharomyces cerevisiae, which lacks endogenous ER, is sufficient for the faithful reconstitution of estrogen signaling within these cells (18, 41). When expressed in yeast cells, the human ER will activate transcription from EREs located in reporter gene promoters in response to estradiol. This ability of ER to function within yeast cells allows a wide variety of genetic approaches to be taken toward defining the mechanisms of signal transduction and transcriptional regulation by the receptor.
To identify proteins that affect ER function, we have carried out a dosage suppression screen in yeast cells. In this technique, a mutant ER protein, with a reduced ability to activate transcription, is used as a substrate to isolate a yeast gene product(s) that is capable of overcoming this mutant phenotype by favoring the interaction between ER and these factors, thus reconstituting receptor transcriptional activity. The mutant ER used in our dosage suppression screen contains a glycine-to-valine substitution at position 400 (G400V ER). This mutation is believed to alter the conformation of the ligand-binding domain, resulting in decreased hormone binding by the receptor, with a corresponding reduction in G400V ER’s ability to activate transcription in response to estradiol (54). This mutant was selected as the substrate for the screen because it affects an early step in the ER signaling pathway, namely steroid binding, and therefore has the potential to result in the isolation of factors important for either steroid binding or transcriptional activation. We anticipate that characterization of these proteins will ultimately lead to a more complete understanding of the ER signal transduction pathway.
Using G400V ER as our dosage suppression screen substrate, we have isolated a yeast gene that, when overexpressed, is capable of increasing both G400V and wild-type (wt) ER’s ability to activate transcription in response to estradiol. This gene product is the yeast homologue of the vertebrate p23 protein (yhp23) (28), a component of the Hsp90-based molecular chaperone complex associated with unliganded steroid receptors as part of the aporeceptor complex. We have examined the functional relationship between p23 and ER in vivo under a range of receptor, estradiol, and p23 concentrations. Our findings suggest that p23 plays a role in ER signal transduction.
MATERIALS AND METHODS
Yeast strains and growth conditions.
The W303a (a ade2 leu2 his3 trp1 ura3) yeast strain was used in experiments where indicated. The yhp23/SBA1 knockout (KO) and parental (PA) strains are described elsewhere (2). Standard genetic methods were used for growth and manipulation. Cultures were propagated at 30°C in rich medium (yeast extract-peptone-dextrose) or selective minimal medium with 2% glucose (or raffinose or galactose) supplemented with amino acids. To induce gene expression from vectors containing the Gal1-10 inducible promoter, yeast cells were grown in selective medium containing either 2% galactose–1% raffinose or 2% raffinose.
Plasmid constructs.
The YKL117w gene was cloned from the 4.3 library plasmid by PCR with the following primers: 5′-GAAGATCTCCACCATGTACCCATACGATGTTCCTGACTATGCGTCCGATAAAGTTATTAACCCTCAAGTTGC-3′ (encoding a hemagglutin [HA] epitope and containing a BglII site) and 5′-CCCCATGGTTACTCATTCTAGCACTCCAGGTTGATTT-3′. The PCR product was gel purified and subcloned into the pGEM-T Easy vector (Promega). The entire fragment was sequenced to ensure the fidelity of the PCR product. HA-yhp23 was released from this vector by digesting it with BglII (within primer) and PstI (within the polylinker of pGEM-T Easy) and then subcloned into BglII/PstI-digested pCMV5. HA-yhp23 was released from pCMV5 by the BglII/BamHI digest and subcloned into the BamHI site of the yeast expression vectors containing the glycerol-phosphate dehydrogenase promoter (GPD) pRS314GPD(Trp1) and PRS315GPD(Leu2). The yhp23-green fluorescent protein (GFP) fusion protein was created by subcloning the HindIII/XbaI fragment of GFP from pEGFP-N3 (Clontech) into a BglII/HindIII fragment of HA-yhp23 in the pGEM-T Easy vector and placing the HA-yhp23-GFP fusion protein into pCMV5 at the BglII/XbaI sites. The HA-yhp23-GFP fusion protein was excised from pCMV5 by using BglII/BamHI and then subcloned into the BamHI site of the yeast expression vector pRS314GPD. The ER1–115–Lex DNA binding domain fusion protein was constructed by excising the EcoRI/PvuII fragment of pGEX-ER1–185 (a kind gift from P. Kushner) and inserting it into the EcoRI- and XhoI-digested pEG 202 two-hybrid vector (20). The XhoI sticky end was first blunted to make ligation with the PvuII blunt end possible.
An HA epitope was also placed at the amino terminus of the human p23 cDNA by PCR by using the following primers: 5′-GCGGATCCACCATGTACCCATACGATGTTCCTGACTATGCGCAGCCTGCTTCTGCAAAGTGGTACGATCG-3′ (containing a BglII site and encoding an HA epitope) and 5′-CACCACCCATGTTGTTCATCATCTCAGAG-3′. The same PCR and cloning strategy was used to create the HA-p23 yeast and mammalian expression vectors.
wt ER, G400V ER, and wt glucocorticoid receptor (GR) were expressed from vectors containing the Gal1-10 promoter, the 2μm plasmid replication origin from yeast cells, and either the TRP1 (p2T-GAL) or HIS3 (p2H-GAL) gene. Reporter plasmids ERE-CYC1-LacZ or GRE-CYC1-LacZ contain a single ERE or three glucocorticoid response elements (GREs) upstream of a truncated CYC1 promoter linked to the β-galactosidase gene; these plasmids also contain the URA3 gene as a selectable marker and the yeast 2μ plasmid replication origin. Full-length GRIP1 was expressed constitutively in yeast from the alcohol dehydrogenase promoter as described previously (21).
Transient transfections.
MCF-7 cells were seeded onto 60-mm dishes at 2.5 × 105 cells/dish in phenol red-free Dulbecco modified Eagle medium supplemented with 10% charcoal-stripped fetal bovine serum. The following day the cells were transfected by the liposome-mediated method as described by the manufacturer (Trans IT-100; Pan Vera, Madison, Wis.) with 5 μg of ERE-thymidine kinase-luciferase reporter plasmid (XETL) and 2 μg of pCMV-HA-p23. At 12 h posttransfection, cells were refed with the same medium containing 0.1 nM 17-β-estradiol or ethanol vehicle. A luciferase assay was performed 24 h later as previously described (50).
β-Galactosidase assays.
With yeast liquid cultures, quantitative β-galactosidase measurements were carried out as previously described (18). Cultures were grown overnight in selective media containing 2% glucose. Equal numbers of cells (as determined by measuring the optical density at 600 nm [OD600]) were pelleted and washed in sterile water to remove glucose medium and subcultured into 2 ml of selective medium containing either 2% galactose–1% raffinose or 2% raffinose. Steroid hormones were added to the medium as a 1,000-fold dilution in ethanol. Cultures were incubated for 12 h at 30°C. One-half of the cells were pelleted, washed in 0.5 ml of LacZ buffer (5 mM KCl, 0.5 mM MgSO4 · 7H2O, 60 mM Na2HPO4 · 7H2O, 60 mM NaH2PO4 · H2O [pH 7.0], with 0.025% β-mercaptoethanol added freshly), and pelleted again. Cells were resuspended in 50 μl of LacZ buffer and permeabilized by adding 50 μl of CHCl3–20 μl of 0.1% sodium dodecyl sulfate (SDS) and vortexing for 10 s. After 5 min, 0.5 ml of a 2-mg/ml concentration of o-nitrophenyl-β-d-galactopyranoside (ONPG) was added, and the reaction was allowed to continue for 1 to 10 min, after which the reaction was stopped by adding 0.5 ml of 1 M Na2CO3. Reactions were centrifuged for 1 min to pellet the cell debris, and levels of β-galactosidase activity were determined by measuring the OD420. The cell number was determined by measuring the OD600 of the remaining cells. Receptor activity is expressed as β-galactosidase units, determined by using the following equation: β-galactosidase units = (1,000 × OD420)/(reaction volume [milliliters] × reaction time [minutes] × OD600). All experiments were performed in triplicate, and the data shown are representative of multiple experiments. The experiments were performed in the linear range of the assay.
Plate assays were performed by replica plating colonies from glucose plates onto galactose X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) indicator plates containing 1 nM 17-β-estradiol.
Preparation of yeast extracts and immunoblotting.
Yeast extracts were prepared from 5-ml cultures. Equal numbers of cells (as determined by measuring the OD600) were pelleted and washed in 0.5 ml of 1× phosphate-buffered saline (PBS) supplemented with 3 mM dithiothreitol and protease inhibitors, including 1 mM phenylmethylsulfonyl fluoride and 1 μg each of aprotinin, pepstatin A, and leupeptin per ml. Subsequent steps were carried out at 4°C. Cells were again pelleted and resuspended in 200 μl of receptor buffer (10 mM Tris, pH 7.5; 1 mM EDTA; 50 mM NaCl; 20% glycerol) containing protease inhibitors. An equal volume of glass beads was added to each tube, and cells were lysed by shaking for 20 min in an Eppendorf horizontal shaker. Extracts were separated from glass beads by centrifuging the extracts through a small hole (made with a 20-gauge needle) at the bottom of the microcentrifuge tube. The lysates were subsequently cleared by centrifugation at 10,000 rpm for 15 min, after which the supernatants were transferred to a new tube. The protein concentration of each extract was measured by using the Bio-Rad Protein Assay and then standardized accordingly. For Western blotting, protein extracts (50 to 100 μg) were fractionated on SDS–10% polyacrylamide gels and transferred to Immobilon paper (Millipore). Blots were probed either with a combination of the ER monoclonal antibodies C311 and C314 (Santa Cruz) or with the anti-HA monoclonal antibody 12CA5 (Boehringer Mannheim). Rabbit polyclonal antiserum against endogenous yhp23 was the generous gift of B. Freeman (University of California at San Francisco). The blots were developed with horseradish peroxidase-coupled sheep anti-mouse or donkey anti-rabbit antibodies and enhanced chemiluminescence reagent (Amersham).
In vivo estradiol-binding assay.
Ligand-binding assays were carried out as previously described (34). Duplicate cultures of yeast cells containing either G400V ER with or without HA-yhp23 or wt ER with or without HA-yhp23 were inoculated into selective liquid medium containing 2% galactose–1% raffinose and grown overnight at 30°C to induce receptor expression. Equal numbers of cells were pelleted and inoculated into 3 ml of 2% galactose–1% raffinose medium containing either 1 nM (G400V ER) or 0.1 nM (wt ER) of 3H-labeled 17-β-estradiol (72 Ci/mmol) (NET317; NEN) such that the final OD600 was 2. Cells were incubated with the labeled estradiol for 1 h at 30°C, after which 1 ml of the cultures was pelleted and washed three times with 2% glucose in PBS, resuspended in 150 μl of the same solution, and then transferred to a scintillation vial. After the addition of scintillation fluid, the total 3H-labeled 17-β-estradiol was measured by scintillation counting. To account for any nonspecific binding, 3H-labeled 17-β-estradiol binding to a GR-containing strain was subtracted from the counts obtained with the ER-expressing strain. β-Galactosidase assays were carried out as described above with 1 ml of the remaining culture.
Subcellular localization of yhp23.
W303a was transformed with the pRS314GPD-yhp23-GFP expression vector; with the ERE-dependent β-galactosidase reporter plasmid; or with the p2H GAL expression vector containing G400V ER, wt ER, GR, or p2H GAL plasmid alone. Yeast strains were grown in 2 ml of 2% galactose–1% raffinose for 12 h. Cells were fixed by adding 240 μl of 37% formaldehyde to each 2-ml culture and incubated at 30°C for 90 min. Cultures were then pelleted by centrifugation and washed three times with 200 μl of solution A (1.2 M sorbitol, 50 mM KPO4). Cell walls were digested by resuspending cells in 100 μl of solution A plus 0.1% β-mercaptoethanol, 0.02% Glusulase (Dupont), and 5 μg of Zymolase (U.S. Biological) per ml for 1 h at 37°C. Cells were then washed twice in 100 μl of PBS and once in 100 μl of PBS plus 0.1% Nonidet P-40 (NP-40) and then blocked for 2 h in 5% bovine serum albumin in Tris-buffered saline (pH 7.4) at room temperature. Cells were then incubated with 100 μl of the appropriate anti-steroid receptor primary antibody (for ER, a mixture of monoclonal antibodies C311 and C314 [Santa Cruz], and for GR, monoclonal antibody BuGr2 diluted in blocking solution) for 2 h at room temperature. Cells were washed once in 100 μl of PBS, once in 100 μl of PBS plus 0.1% NP-40, and once again in 100 μl of PBS, followed by incubation with goat anti-mouse rhodamine-conjugated secondary antibody (Vector Labs), diluted in blocking solution, for 4 h at room temperature. Secondary antibody was removed by washing the cells twice in 100 μl of PBS and then twice in 100 μl of PBS plus 0.1% NP-40. In order to visualize the nuclei, cells were then incubated in 1 μg of Hoechst dye H334211 per ml for 10 min, followed by one wash with 100 μl of PBS. Cells were then resuspended in 30 μl of PBS, 5 μl of which was then plated onto a poly-d-lysine-treated microscope slide and allowed to settle for 10 min before excess fluid was removed by aspiration. Unattached cells were then removed by washing the slide with 20 μl of PBS. The cells were mounted by using 3 μl of Citifluor (Ted Pella, Reading, Calif.), and a coverslip was secured to the slide with rubber cement. GFP, rhodamine, and Hoechst signals were imaged and photographed by using a Zeiss Axioplan 2 microscope.
RESULTS
Yeast dosage suppression screen.
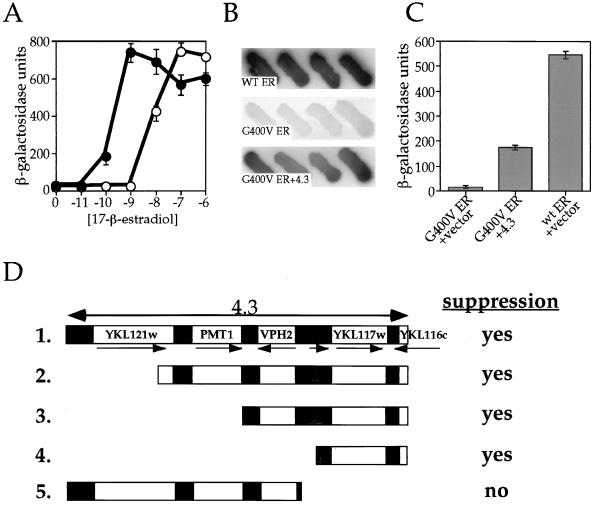
To compare wt ER and G400V ER function in yeast cells, we constructed two strains containing a galactose-inducible expression vector, encoding either wt ER or G400V ER, and a reporter plasmid containing an ERE located upstream of the β-galactosidase gene. The transcriptional activities of wt ER and G400V ER, as a function of hormone concentration, were measured. Compared to wt ER, G400V ER requires a 100-fold increase in 17-β-estradiol concentration before receptor transcriptional activation is observed in our yeast assay (Fig. 1A). At saturating ligand concentrations, however, G400V ER is able to reach the same maximal activity as wt ER, suggesting that once the block to steroid binding is overcome, the receptor is able to act as efficiently as wt ER in entering the functional interactions downstream of estradiol binding, including both protein-protein and protein-DNA interactions, that are necessary for transcriptional activation.
FIG. 1.
Isolation of a yeast genomic fragment that suppresses the G400V ER phenotype. (A) Transcriptional activity of wt ER and G400V ER as a function of 17-β-estradiol concentration. The W303a yeast strain was transformed with a galactose-inducible expression vector containing either wt or G400V ER, along with an ERE-containing β-galactosidase reporter plasmid. Transcriptional activation by wt ER (solid circles) and G400V ER (open circles) in response to increasing 17-β-estradiol concentration was determined by liquid β-galactosidase assay as described in Materials and Methods. Note that G400V ER requires a 100-fold-higher estradiol concentration to induce transcriptional activation than wt ER. The dosage suppression screen was carried out in the presence of 1 nM 17-β-estradiol, the conditions under which the G400V ER phenotype is most pronounced. (B) The relative activity of wt ER, G400V ER, and G400V ER plus suppressor 4.3. Four independent colonies on X-Gal indicator plates containing 1 nM 17-β-estradiol are shown and represent wt ER plus empty library plasmid (wt ER), G400V ER plus empty library plasmid (G400V ER), or G400V ER plus suppressor plasmid 4.3 (G400V ER+4.3). (C) Transcriptional activity of wt ER, G400V ER, and G400V ER in the presence of suppressor 4.3. Liquid β-galactosidase assays were performed on yeast strains containing wt ER plus empty library plasmid, G400V ER plus empty library plasmid, or G400V ER plus 4.3 at 1 nM 17-β-estradiol. Suppressor 4.3 increases G400V ER activity 10-fold, bringing its activity to nearly one-third that of wt ER. (D) Identification of YKL117w as the G400V ER suppressor. The sequence of the yeast genomic fragment contained within the suppressor 4.3 was determined by aligning the 5′ and 3′ ends of the insert to the yeast genomic sequence database. Suppressor 4.3 contains an 8,147-bp fragment comprising four complete ORFs (YKL121w, PMT1, VPH2, and YKL117w), a partial ORF (YKL116w), and a tRNA-Ala gene (shaded box). The relative positions and orientations of the genes within the 4.3 fragment are shown schematically (fragment 1). Identification of the gene responsible for suppressing the G400V ER phenotype was accomplished by constructing 5′ and 3′ deletion derivatives of the 4.3 suppressor (fragments 2 to 5) and assaying their ability to increase G400V ER transcriptional activity at 1 nM 17 β-estradiol. “yes” indicates that the fragment was capable of suppressing the G400V ER phenotype; “no” indicates that the fragment failed to increase G400V ER transcriptional activity. YKL117w was present within the suppressing fragments (fragments 1 to 4) but was absent within the fragment that did not suppress (fragment 5), suggesting that its gene product is responsible for increasing G400V ER transcriptional activation. YKL117w encodes the yeast homologue of the human p23 protein (yhp23).
The G400V ER phenotype is most apparent at a concentration of 1 nM 17-β-estradiol, where wt ER shows maximal transcriptional response, but G400V ER displays only minimal transcriptional activity (Fig. 1A). Exploiting this phenotypic difference, we carried out a dosage suppression screen to identify yeast proteins that, when overexpressed, would increase the transcriptional activity of G400V ER, thereby suppressing the mutant phenotype. The yeast strain containing G400V ER and an estrogen-responsive β-galactosidase reporter gene was transformed with a high-copy-number yeast genomic library and assayed for G400V ER transcriptional activity on X-Gal indicator plates containing 1 nM 17-β-estradiol. Under these conditions, yeast colonies expressing the wt ER are blue, while the G400V ER-expressing yeast colonies appear white (Fig. 1B). Blue colonies were considered to be potential suppressor candidates. Five candidate suppressors of the G400V ER phenotype were isolated after screening ∼6,000 colonies, which we estimate to represent about one-half of the yeast genome.
Identification of the yeast ORF YKL117w as a suppressor of the G400V ER phenotype.
One high-copy-number suppressor plasmid, designated 4.3, was found to increase G400V ER transcriptional activity 10-fold, bringing G400V ER transcriptional activation to one-third the wt ER level at 1 nM 17-β-estradiol (Fig. 1B and C). The 5′ and 3′ ends of the insert of the library plasmid were sequenced and aligned with the yeast genome. In this manner, we were able to identify the suppressor DNA as an 8,147-bp genomic fragment of chromosome XI (26) containing multiple open reading frames (ORFs) (Fig. 1D). To identify the suppressing ORF, a series of deletions were constructed and analyzed for their ability to increase G400V ER transcriptional activity. As seen in Fig. 1D, the suppression of the G400V ER phenotype correlates with the presence of ORF YKL117w.
Possible role of YKL117w in ER signaling.
A search of the Swissprot database revealed YKL117w to be the yeast homologue of the human p23 protein (yhp23) (28), a component of the Hsp90-based molecular chaperone complex. During completion of this study, two separate reports (2, 16) characterizing yeast strains with YKL117w deleted have called this gene SBA1, reflecting an increased susceptibility of steroid signaling to benzoquinone ansamycin antibiotics. Since the YKL117w gene product’s homology to the vertebrate p23 protein was of considerable importance in investigating its role in ER function, we have chosen to refer to it as yhp23 to maintain this emphasis.
Although p23’s specific function is not known, in vitro studies suggest that it is crucial to the stability of the aporeceptor complex of unliganded steroid receptors. Removal of p23 greatly reduces the formation of stable aporeceptor complexes of the GR (11, 12, 24) and the progesterone receptor (PR) (29, 30). In addition, in vitro studies have suggested that p23 possesses abilities typical of molecular chaperones, since it is capable of interacting with denatured β-galactosidase, suppressing its aggregation and maintaining it in an intermediate, folding-competent conformation (17). The relative importance of these two aspects of p23 function in ER signaling has not been determined.
Characterization of yhp23’s role in ER signaling.
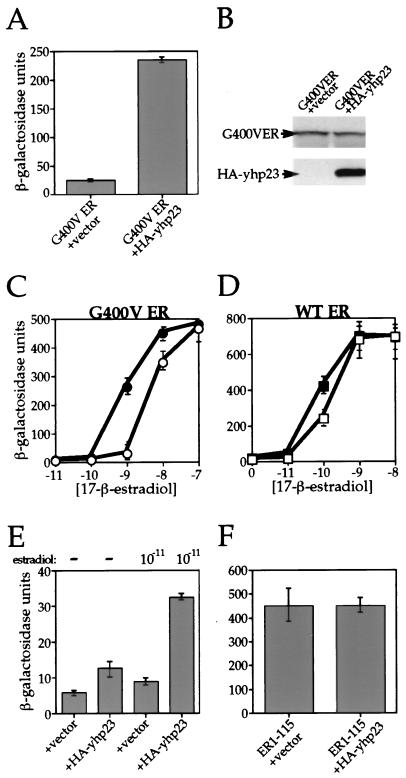
Having identified yhp23 as the suppressing ORF, we cloned the YKL117w gene with an amino-terminal HA epitope tag (HA-yhp23) into a yeast expression vector containing a constitutively active GPD promoter. We then established a yeast strain that overexpresses HA-yhp23 in the presence of G400V ER, along with a reporter plasmid containing the β-galactosidase gene under control of an ERE. As seen in Fig. 2A, overexpression of HA-yhp23 increases G400V ER transcriptional activity by 10-fold. Extracts from these yeast strains were prepared and analyzed by immunoblotting with antibodies specific for ER and HA. Figure 2B demonstrates that the overexpression of yhp23 does not affect the level of G400V ER protein and, therefore, the increase in transcriptional activity is not a result of increased receptor expression. The HA-tagged yhp23 protein migrates on an SDS-polyacrylamide gel at approximately 34 kDa. The higher molecular mass of the yeast protein (34 kDa versus the 23-kDa human protein) is expected, as the yhp23 is larger (216 residues) than its human counterpart (160 residues). Further comparison of G400V ER activity in the presence or absence of yhp23 overexpression over a range of hormone concentrations revealed that G400V ER function was also enhanced at 10 nM 17-β-estradiol (Fig. 2C). The 30% increase in G400V ER activity seen at this level of hormone, however, is relatively small compared to the 10-fold increase observed at the 1 nM 17-β-estradiol concentration used in the screen. This pattern continues at even higher hormone concentrations, so that no increase of G400V ER activity by yhp23 is observed at 100 nM concentrations of 17-β-estradiol (Fig. 2C) and 1 mM 17-β-estradiol (data not shown). These findings suggest that yhp23’s importance to G400V ER signaling decreases at high estradiol concentrations.
FIG. 2.
Effect of yhp23 overexpression on G400V ER and wt ER transcriptional activity. An HA-tagged yhp23 gene was cloned into the yeast expression vector pRS314 downstream of the GPD promoter. Yeast strains were then constructed which coexpress G400V ER or wt ER with the HA-tagged yhp23 in the presence of a β-galactosidase reporter gene under the control of the appropriate hormone response element (ERE). (A) Overexpression of HA-yhp23 increases G400V ER transcriptional activation. The activity of G400V ER in the presence or absence of overexpressed yhp23 was determined by liquid β-galactosidase assay at a 1 nM concentration of 17-β-estradiol. Overexpression of yhp23 increased G400V ER activity approximately 10-fold. (B) Increased transcriptional activation of G400V ER by yhp23 is not a function of increased ER levels. Whole-cell lysates were prepared from the yeast strains described in panel A. Equal amounts of proteins were separated on an SDS–4 to 20% gradient polyacrylamide gel, transferred to Immobilon paper, and probed with an ER-specific monoclonal antiserum (top panel) or a monoclonal antibody directed against the HA epitope on yhp23 (bottom panel) and visualized by enhanced chemiluminescence. (C) Dose-response curves for G400V ER. The activity of G400V ER in the absence (open circles) or presence (solid circles) of overexpressed yhp23 at the indicated 17-β-estradiol concentrations is shown. The increased transcriptional activity displayed by G400V ER in the presence of HA-yhp23 overexpression is seen to be greatest at low hormone concentrations and is lost completely at the highest hormone concentrations assayed. (D) Dose-response curves for wt ER. The activity of wt ER in the absence (open squares) or presence (solid squares) of overexpressed HA-yhp23 at the indicated 17-β estradiol concentrations is shown. The effect of HA-yhp23 overexpression is seen to be greatest at low hormone concentrations and is lost completely at the highest hormone concentrations assayed. (E) Overexpression of HA-yhp23 increases the ligand-independent activity of the ER. The activity of the ER in the absence or presence of 0.01 nM 17-β-estradiol is shown. Ligand-independent transcriptional activity by ER is approximately threefold higher in the presence of HA-yhp23 overexpression. The addition of 0.01 nM 17-β-estradiol resulted in a greater level of ER ligand-dependent activity in the HA-yhp23 overexpressing strain, suggesting that yhp23 overexpression increases the sensitivity of ER to low concentrations of 17-β-estradiol. (F) HA-yhp23 overexpression does not affect the AF-1 activity of ER. The amino-terminal 115 residues of ER, containing the AF-1 activity of the ER in yeast cells, were fused to a Lex DNA binding domain (see Materials and Methods). Constitutive AF-1 activity resulted in transcription of a reporter β-galactosidase gene downstream of a LexA binding site, which was unaffected by overexpression of HA-yhp23.
Given p23’s proposed chaperone-like activities in vitro, it could be argued that yhp23’s interaction with the mutant G400V ER might arise as a function of the receptor’s misfolded steroid-binding domain rather than reflect a true role in the ER signaling pathway. To determine whether yhp23 overexpression affects wt ER activity, we constructed an additional yeast strain that overexpresses yhp23 in the presence of wt ER, along with an ERE-controlled β-galactosidase gene reporter plasmid. The effect of yhp23 overexpression upon ER activity was then assayed over a range of hormone concentrations (Fig. 2D). wt ER activity was affected in a manner similar to that of G400V ER, albeit less dramatically. At 0.1 nM 17-β-estradiol, wt ER activity is increased by approximately 70% in the presence of overexpressed yhp23, with no increase observed at the higher concentrations of 1 and 10 nM 17-β-estradiol. In contrast to G400V ER, however, an increase in wt ER activity was observed at 0.01 nM 17-β-estradiol. To more closely examine the effect of yhp23 on wt ER activity at this low hormone concentration, we compared wt ER ligand-independent activity to wt ER activity at 0.01 nM 17-β-estradiol. As seen in Fig. 2E, overexpression of yhp23 increases wt ER ligand-independent activity by approximately twofold and ligand-dependent activation by threefold in the presence of 0.01 nM 17-β-estradiol. These findings suggest that overexpression of yhp23 greatly increases wt ER’s ability to respond to low levels of ligand. Without yhp23 overexpression, the fold induction of wt ER activity was only 50% upon administration of 0.01 nM 17-β-estradiol compared to a 250% induction of wt ER activity in the presence of overexpressed yhp23. These results suggest that increased levels of yhp23 can facilitate wt ER ligand-independent activity and potentiate ER signaling at low levels of hormone. Thus, the ability of yhp23 to functionally interact with wt ER and not just the G400V ER mutant strongly implicates yhp23 as a member of the ER signaling pathway in yeast cells.
Importantly, yhp23 does not appear to have any effect upon the activity of the ER AF-1 domain. When we fused the amino-terminal 115 residues of ER to a Lex DNA binding domain and expressed this construct in yeast cells, we observed constitutive activation of a Lex-responsive β-galactosidase reporter plasmid, a result consistent with earlier findings demonstrating that the AF-1 activity of ER is contained within this region of the receptor (39). As shown in Fig. 2F, this constitutive activation was not affected by yhp23 overexpression, suggesting that the increased function of both the mutant and the wt ER observed in Fig. 2C and D, respectively, does not appear to be mediated by an increase in AF-1 activity. This finding, taken together with the observed hormone concentration-dependent effect of yhp23 overexpression (Fig. 2C and D), suggests that yhp23 affects the ER signal transduction pathway at the step of ligand binding.
yhp23 overexpression increases ligand binding by both G400V ER and wt ER.
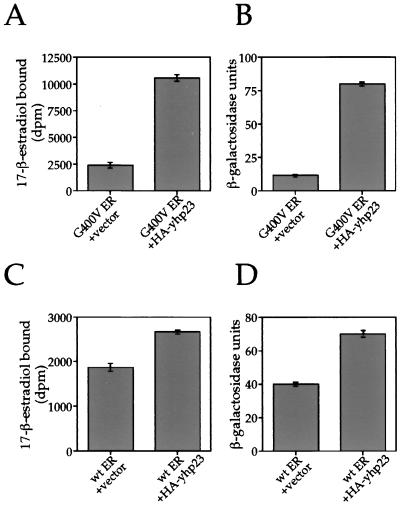
Considering p23’s proposed role in aporeceptor complex formation and given the nature of the G400V ER mutation, we examined whether the increase in G400V ER transcription in the presence of overexpressed yhp23 results from increased estradiol binding by the receptor. Estradiol binding by G400V ER and wt ER in the presence or absence of yhp23 overexpression was measured in vivo. The yeast strains were incubated for 1 h in medium containing 3H-labeled 17-β-estradiol and washed three times to remove unbound steroid, and the amount of estradiol bound to G400V ER and wt ER was measured by quantifying the 3H-labeled 17-β-estradiol content by liquid scintillation counting. β-Galactosidase assays were carried out in parallel on the same cells to correlate effects of ligand binding to transcriptional activation.
We first examined ligand binding by G400V ER at a 1 nM concentration of 3H-labeled 17-β-estradiol. As seen in Fig. 3A, ligand binding by G400V ER was increased by approximately fivefold in the presence of overexpressed yhp23. This increase in ligand binding was found to correlate with a sevenfold increase in G400V ER transcriptional activity (Fig. 3B). Immunoblot analysis showed that the levels of G400V ER were unchanged by yhp23 overexpression (data not shown).
FIG. 3.
Overexpression of yhp23 increases estradiol binding by G400V ER and wt ER in vivo. The total amount of estradiol bound by G400V ER and wt ER in the presence or absence of yhp23 overexpression was determined by an in vivo ligand-binding assay with the same yeast strains as described in Fig. 2. After a 1-h incubation in the presence of 3H-labeled 17-β-estradiol, cells were washed to remove unbound ligand, and the amount of bound estradiol was determined by liquid scintillation counting. Liquid β-galactosidase assays were carried out, in parallel, on an aliquot of the same 3H-labeled 17-β-estradiol-incubated cells in order to correlate the levels of ligand binding to the resulting levels of transcriptional activation. Whole-cell extracts from the assayed yeast strains were fractionated on SDS-polyacrylamide gel electrophoresis, and ER expression was examined by use of immunoblotting with an ER-specific rabbit polyclonal antiserum as described in Materials and Methods. No alteration in G400V ER or wt ER levels in response to increased HA-yhp23 expression were noted (data not shown). (A) HA-yhp23 overexpression increases ligand binding by G400V ER. Cells expressing G400V ER in the presence or absence of HA-yhp23 overexpression were assayed for ligand binding in the presence of 1 nM 3H-labeled 17-β-estradiol. Ligand binding by G400V ER is seen to increase approximately fivefold in the presence of overexpressed HA-yhp23. (B) Increased ligand binding by G400V ER in the presence of HA-yhp23 overexpression correlates with an increase in transcriptional activation by the receptor. Liquid β-galactosidase assays, carried out on an aliquot of the cells described in panel A, demonstrate a corresponding sevenfold increase in transcriptional activity by G400V ER. (C) HA-yhp23 overexpression increases ligand binding by wt ER. Cells expressing wt ER in the presence or absence of HA-yhp23 overexpression were assayed for ligand binding in the presence of 0.1 nM 3H-labeled 17-β-estradiol. Ligand binding by wt ER is seen to increase by approximately 50% in the presence of overexpressed HA-yhp23. (D) Increased ligand binding by wt ER in the presence of HA-yhp23 overexpression correlates with an increase in transcriptional activation by the receptor. Liquid β-galactosidase assays, carried out on an aliquot of the cells described in panel C, demonstrate a corresponding 70% increase in transcriptional activity by wt ER.
We then examined ligand binding by wt ER, which was also found to increase in the presence of yhp23 overexpression, although to a lesser extent. At a 0.1 nM concentration of 3H-labeled 17-β-estradiol, wt ER ligand binding was increased by approximately 50% (Fig. 3C). β-Galactosidase assay of these cells demonstrated a corresponding 70% increase in transcriptional activity (Fig. 3D). Again, immunoblot analysis showed no difference in the level of ER expression exhibited by the two strains (data not shown). These results indicate that yhp23 overexpression increases ER transcriptional activity by increasing the total number of ligand-bound receptors.
The magnitude of yhp23’s effect on wt ER signaling is a function of wt ER and estradiol concentrations.
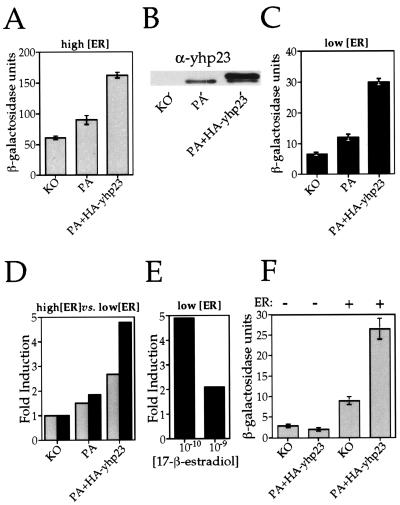
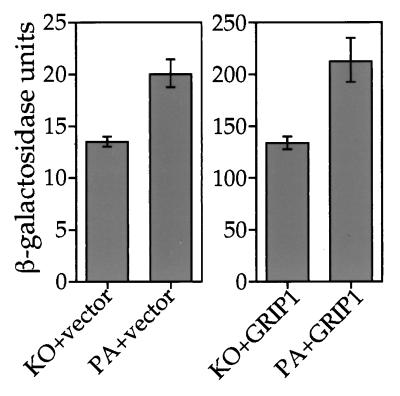
Having demonstrated that overexpression of yhp23 increases wt ER transcriptional activity, we next asked whether a decrease in yhp23 concentration would reduce wt ER activity. To this end, we compared wt ER transcriptional activity in the yhp23 knockout strain (KO) to that in the parental strain (PA) (2), as well as the parental strain overexpressing HA-yhp23 (PA+HA-yhp23 strain). In this way, we were able to assay wt ER function (i) in the absence of yhp23 (KO), (ii) in the presence of endogenous levels of yhp23 (PA), and (iii) in the presence of both endogenous yhp23 and overexpressed HA-yhp23 (PA+HA-yhp23) (Fig. 4B). A wt ER expression vector and an ERE–β-galactosidase reporter plasmid were introduced into these three yeast strains, and wt ER transcriptional activity was assayed at 0.1 nM 17-β-estradiol. The activity of wt ER within the three different strains at this hormone concentration is shown in Fig. 4A. In the absence of yhp23 (KO), wt ER signaling still occurs, demonstrating that wt ER is capable of functioning in a yhp23-independent manner. Endogenous levels of yhp23 (PA) result in a 50% increase in wt ER activity relative to the KO strain. The PA+HA-yhp23 strain, which contains the highest yhp23 levels (Fig. 4B), exhibits an even higher level of wt ER induction (2.7-fold) than that seen in the KO strain. This increase in wt ER activity is not a function of increased receptor levels, since wt ER levels were unchanged in the absence or presence of yhp23 (data not shown). Thus, wt ER transcriptional activity increases in direct proportion to the concentration of yhp23.
FIG. 4.
Yhp23’s effect on wt ER activity is a function of the yhp23, ER, and estradiol concentrations. wt ER transcriptional activation was compared in three yeast strains expressing yhp23 at different concentrations. The knockout strain (KO) is yhp23 deficient, the parental strain (PA) expresses yhp23 at endogenous levels, and the HA-yhp23-transformed parental strain (PA+HA-yhp23) expresses endogenous yhp23 and exogenous HA-yhp23. All three strains express wt ER under the control of a galactose-inducible promoter, along with an ERE–β-galactosidase reporter plasmid. wt ER activity in the KO and PA+HA-yhp23 strains was compared over a range of hormone concentrations as shown in Fig. 2D. The greatest effect of yhp23 levels was observed at 0.1 nM 17-β-estradiol. Data shown represent the results of β-galactosidase assays repeated with all three yeast strains (KO, PA, and PA+HA-yhp23) at this hormone concentration. (A) The effect of yhp23 on wt ER transcriptional activation at high ER concentrations. ER transcriptional activity as a function of yhp23 concentration was determined by liquid β-galactosidase assay in cells incubated in galactose-raffinose-supplemented medium containing 0.1 nM 17-β-estradiol. (B) KO, PA, and PA+HA-yhp23 strains express different levels of yhp23. Equal amounts of whole-cell lysates from the KO, PA, and PA+HA-yhp23 strains were analyzed by immunoblotting by using anti-yhp23 polyclonal antibody as described in Materials and Methods. The upper band seen in the PA+HA-yhp23 lane corresponds to the HA-tagged yhp23. (C) The effect of yhp23 on wt ER transcriptional activity at low wt ER concentration. ER transcriptional activity as a function of yhp23 concentration was determined by liquid β-galactosidase assay in cells incubated in raffinose-supplemented medium containing 0.1 nM 17-β-estradiol. Under these conditions ER expression is 10-fold lower than with cells grown in the presence of galactose (not shown). (D) yhp23 induction of wt ER transcriptional activity is inversely proportional to wt ER concentration. The fold induction of yhp23 on ER transcriptional activity at 0.1 nM 17-β-estradiol under conditions of high ER expression (galactose; shaded columns) or low ER expression (raffinose; solid columns) is standardized to the ER activity in the KO strain. (E) The effect of yhp23 on wt ER function is inversely proportional to the ligand concentration. The fold induction of ER transcriptional activation in the PA+HA-yhp23 versus KO strains at 0.1 and 1 nM 17-β-estradiol was determined under conditions of low ER expression (raffinose medium). The results indicate that the fold induction of ER transcriptional activation in the presence of yhp23 overexpression is significantly greater at 0.1 nM 17-β-estradiol than at 1 nM 17-β-estradiol. (F) yhp23 overexpression increases estradiol-independent wt ER transcriptional activation. The effect of yhp23 overexpression on ERE-dependent transcriptional activation in the presence or absence of ER is shown. KO and PA+HA-yhp23 strains containing the ERE reporter construct were transformed with either the empty expression vector or with the ER-containing expression vector and assayed for β-galactosidase activity in galactose-raffinose medium without estradiol.
Having shown that wt ER activity increases as a function of yhp23 concentration, we next asked if the effect of yhp23 upon ER signaling is also a function of wt ER concentration. This was accomplished by repeating the above experiment in medium containing only raffinose as a carbon source, resulting in low levels of wt ER expression. When wt ER transcriptional activity was assayed under these conditions, a similar pattern of yhp23-dependent ER activation was observed: wt ER functions in the KO strain, and this activity increases as a function of yhp23 concentration (Fig. 4C). Importantly, the magnitude of the effect of yhp23 on wt ER transcriptional activation is greater at the lower wt ER concentration, such that overexpression of the yhp23 in the PA+HA-yhp23 strain results in an almost fivefold increase in wt ER transcriptional activity relative to the KO strain, thereby doubling the induction seen at high wt ER concentrations (Fig. 4D). Thus, the magnitude of yhp23’s effect on wt ER transcriptional activity is greater at low, rather than high, wt ER concentrations.
Our earlier studies of yhp23 induction of G400V ER and wt ER activity (Fig. 2C and D) suggested that the importance of yhp23 to wt ER function correlates inversely to hormone concentration. This idea was further confirmed with the KO and PA+HA-yhp23 strains, when the activity of wt ER within these strains was assayed at both 0.1 and 1 nM 17-β-estradiol under conditions of low wt ER expression. As seen in Fig. 4E, the fivefold induction of wt ER transcriptional activity observed within the PA+HA-yhp23 strain at 0.1 nM 17-β-estradiol decreases to only a twofold induction at the higher concentration of 1 nM 17-β-estradiol. Again this trend continues such that, when activities are compared at yet higher hormone concentrations, no difference in activity is observed between the KO and PA+HA-yhp23 strains at 10 nM 17-β-estradiol (data not shown). Thus, the effect of yhp23 overexpression on wt ER transcriptional activation is inversely proportional to the hormone concentration.
Comparison of wt ER activity in the KO and PA+HA-yhp23 strains in the absence of estradiol again confirmed a role for yhp23 in wt ER ligand-independent transcriptional activation. As shown in Fig. 4F, yhp23 is not essential to wt ER-dependent activation in the absence of ligand. Interestingly, as was observed with ligand-dependent activity, wt ER function in the absence of ligand increases with yhp23 overexpression. This effect on transcription is not observed in the absence of wt ER expression, suggesting that yhp23 is operating via wt ER to induce estradiol-independent transcriptional activation.
Thus, the relationship of yhp23 to wt ER signal transduction is dependent not only on concentrations of yhp23 but also upon the levels of wt ER and estradiols. Our data demonstrate that the effect of yhp23 upon wt ER function is most pronounced when wt ER activity is examined at low estradiol and wt ER concentrations.
yhp23 is not essential for the formation of a functional ER-GRIP1 complex.
The ER activates transcription in mammalian cells through two transcriptional activation domains, termed AF-1 and AF-2 (36). The AF-1 domain, located within the amino terminus of the receptor, does not require steroid binding to achieve an active conformation (39). In contrast, the AF-2 domain lies within the steroid-binding domain and is dependent upon estradiol binding for its activity (55). The ER AF-2 region has been shown to activate transcription in yeast cells and in cultured mammalian cells through interaction with coactivator proteins, including GRIP1/TIF2 (21) and SRC1 (43). Although AF-2 activity is observed in some promoter contexts in yeast cells (40), the AF-2 coactivators have no yeast homologues, and thus ER transcriptional activity in yeast cells is greatly potentiated in the presence of ectopically expressed mammalian coactivators (22). We therefore sought to more specifically determine whether yhp23 might play a role in AF-2–coactivator interactions.
The recent determination of the structure of the ER ligand-binding domain bound to estradiol has suggested that proper folding of this region around the steroid hormone is crucial to the formation of an AF-2 domain competent for interaction with coactivators (3). Since p23 has been proposed to have chaperone-like activities (17), we hypothesized that it may play a role in the proper folding of the AF-2 domain around estradiol during the process of steroid binding. To determine whether yhp23 facilitates wt ER AF-2–coactivator interactions, we introduced the mammalian coactivator GRIP1 into yhp23 KO and PA yeast strains expressing wt ER (KO+GRIP1 and PA+GRIP1, respectively). Coexpression of GRIP1 should activate AF-2, thereby increasing wt ER transcriptional activation relative to the control strains lacking GRIP1 (22) and allowing us to compare GRIP1 induction of wt ER activity in the presence or absence of yhp23. If yhp23 were important for AF-2 interaction with GRIP1, then GRIP1-dependent ER transcriptional activation would be reduced in the KO versus the PA strain. However, as shown in Fig. 5, this is not the case. wt ER transcriptional activation in the PA and KO strains is enhanced to similar extents (approximately 10-fold) compared to the corresponding control strains lacking GRIP1 (Fig. 5). Thus, GRIP1 induction of wt ER transcriptional activation is not altered by yhp23 expression, suggesting that yhp23 is not required for the formation of a functional ER-GRIP1 complex.
FIG. 5.
yhp23 is not essential for the functional interaction between ER and the coactivator GRIP1. yhp23 KO and PA strains were constructed that express wt ER both in the presence or in the absence of the coactivator GRIP1. ER activity was assayed in the four strains after incubation in raffinose medium containing 0.1 nM 17-β-estradiol. Data were collected in the same experiment but are displayed with separate y axes to more clearly demonstrate GRIP1 induction of ER activity within each strain. Coexpression of GRIP1 increases ER transcriptional activation in both KO and PA strains approximately 10-fold, an effect that is independent of yhp23 expression. Note that yhp23 increased ER transcriptional activation to the same extent in the absence or presence of GRIP1.
ER and yhp23 colocalize within the nucleus of yeast cells.
Prior genetic and biochemical studies have demonstrated that aporeceptor complex formation is conserved in yeast cells (5). Hsp82, the yeast homologue of Hsp90, has been shown to associate with hormone-free ER and GR in yeast cells. In addition, genetic studies indicate that ER and GR signaling is reduced in yeast strains expressing only 5% of the wt level of Hsp82 (44). Compelling genetic evidence also exists for the role of the yeast Hsp70 (31), p60 (8), Hsp40 (31), and immunophilin (14) homologues in steroid signaling in yeast cells, with the majority of these proteins having been shown to associate with GR in the absence of hormone (2, 7).
Given human p23’s presence in the aporeceptor complexes of PR (28) and GR (11) and having shown that yhp23 affects ER function, we proceeded to determine whether yhp23 and ER colocalize in vivo. To determine the cellular distribution of yhp23 in yeast cells, we created a yhp23-GFP fusion protein by subcloning GFP at the carboxy terminus of the yhp23 protein. Expression of the fusion protein was confirmed by Western blotting. Importantly, the yhp23-GFP fusion protein is also able to suppress the G400V ER phenotype (not shown), proving that addition of the GFP moiety does not eliminate yhp23’s ability to functionally interact with G400V ER.
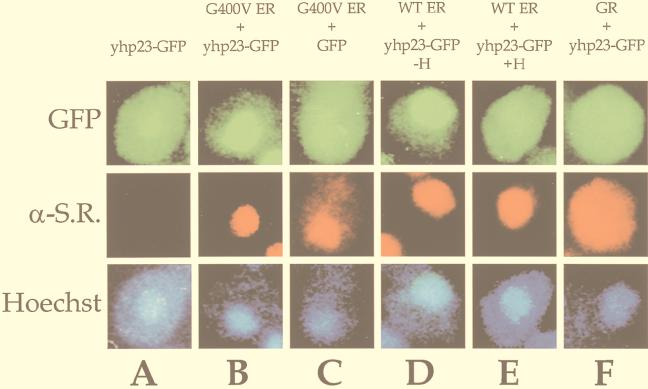
We constructed several yeast strains that express yhp23-GFP either alone or in combination with G400V ER, wt ER, or wt GR. Figure 6A demonstrates that the distribution of yhp23-GFP in the absence of steroid receptor expression is largely cytoplasmic, though a small proportion of signal corresponding to the nucleus is also evident. This pattern is consistent with the expression pattern described for the human p23 protein within mammalian cells (28). Strikingly, upon coexpression of G400V ER, yhp23-GFP becomes predominantly limited to the nucleus, thus colocalizing with G400V ER (Fig. 6B). Recall that ER is a steroid receptor that resides in the nucleus in the absence of hormone. Importantly, this pattern of nuclear localization was not seen when G400V ER was coexpressed with just the GFP protein, indicating that yhp23 is responsible for the localization of the fusion protein to the nucleus (Fig. 6C). Nuclear localization of yhp23-GFP was also observed when it was coexpressed with wt ER (Fig. 6D). Thus, the ability of G400V ER to colocalize yhp23-GFP in the same manner as wt ER suggests that the G400V ER phenotype is not a result of deficient aporeceptor complex formation. Additionally, when cells coexpressing wt ER and yhp23-GFP were incubated in 17-β-estradiol, yhp23-GFP redistributed to the cytoplasm, reestablishing the pattern seen in yeast cells lacking ER expression (Fig. 6A). As a final control, yhp23-GFP coexpressed with GR, a steroid receptor that exists outside the nucleus in the steroid-free state, did not localize to the nucleus (Fig. 6F) but instead showed a cytoplasmic distribution similar to that of GR.
FIG. 6.
ER and yhp23-GFP colocalize within the nucleus of yeast cells. To examine yhp23 subcellular localization in yeast cells, a yhp23-GFP fusion protein was constructed. Yeast strains were created that express yhp23-GFP (A) or GFP (C) either alone or in combination with G400V ER (B and C), wt ER (D and E), or GR (F). Cells were grown in galactose-raffinose-containing medium in the absence or the presence of 17-β-estradiol. Cells were fixed, permeabilized, and incubated with the appropriate receptor primary antibody, a corresponding Texas-red-conjugated secondary antibody, and the DNA in the nucleus was stained with Hoechst dye H334211. The GFP, Texas red, and Hoechst fluorescent signals were visualized by using a Zeiss Axioplan 2 fluorescence microscope. Note that yhp23-GFP is expressed throughout the cytoplasm in the absence of ER expression. In the presence of coexpressed G400V ER as well as wt ER, yhp23-GFP becomes localized to the nucleus. Incubation of the wt ER strain in 1 μM 17-β-estradiol results in the redistribution of yhp23-GFP from the nucleus to the cytoplasm, thereby reversing the ER-yhp23-GFP colocalization observed in the absence of estradiol. α-S.R., anti-steroid receptor primary antibody.
The colocalization of yhp23 and ER is consistent with the proposed role of yhp23 as a member of the ER aporeceptor complex. Upon coexpression with ER, aporeceptor complex formation causes yhp23, presumably through an interaction with Hsp82 (the yeast homologue of Hsp90), to become localized to the nucleus. The addition of hormone appears to result in the dissociation of the aporeceptor complex, allowing yhp23 to redistribute throughout the cell.
Complementation of yhp23 by human p23 in yeast cells.
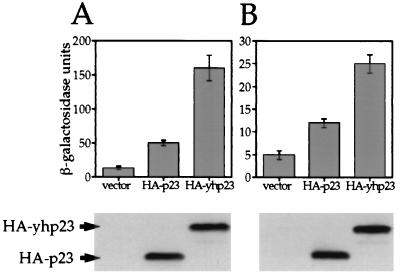
We next examined whether human p23, when ectopically expressed in yeast cells, functions like yhp23 to suppress the G400V ER phenotype. We established yeast strains that express G400V ER and an ERE-responsive reporter plasmid in the presence of HA-tagged human p23 (HA-p23) or HA-yhp23. A third strain containing the expression vector without an insert (vector) was used as a negative control. As shown in Fig. 7A, human p23 is capable of increasing hormone-dependent G400V ER transcriptional activation in yeast cells, although to a lesser degree than yhp23 (4-fold and 13-fold, respectively). The reduced G400V ER transcriptional activity is not a function of reduced human p23 expression relative to yhp23, since immunoblot analysis with an antibody directed against the HA epitope present on both proteins shows equal expression levels in yeast cells (Fig. 7A, bottom panel). In addition, G400V ER expression is unaffected by yeast or human p23 coexpression (data not shown). These findings suggest that human p23 can function like yhp23 in yeast cells, albeit less potently, to increase G400V ER transcriptional activity.
FIG. 7.
Complementation of yhp23 by human p23 in yeast cells. (A) Overexpression of HA-p23 suppresses the G400V ER phenotype. An HA epitope-tagged human p23 (HA-p23) was subcloned into the yeast expression vector pRS316GPD. The W303a yeast strain expressing G400V ER and an ERE–β-galactosidase reporter gene was transformed with expression vectors containing no insert (vector), HA-p23, or HA-yhp23. G400V ER transcriptional activity was measured by liquid β-galactosidase assay in galactose-containing medium with 1 nM 17-β-estradiol. Equal amounts of whole-cell lysates from the strain containing vector, HA-p23, and HA-yhp23 were analyzed by immunoblotting by using anti-HA antibody as described in Materials and Methods (bottom panel). (B) Human p23 partially complements the loss of yhp23 with respect to ER signaling. yhp23 KO yeast strain expressing wt ER and an ERE-responsive β-galactosidase reporter gene were transformed with expression vectors containing no insert (vector), HA-p23, or HA-yhp23. ER transcriptional activity was determined by liquid β-galactosidase assay in raffinose medium containing 0.1 nM 17-β-estradiol. Immunoblot analysis for HA-p23 and HA-yhp23 was performed as in panel A and demonstrates that HA-p23 and HA-yhp23 are expressed at similar levels (bottom panel).
We next evaluated whether human p23 could function in yeast cells to increase ER transcriptional activation in the absence of endogenous yhp23. The KO strain expressing wt ER and an ERE-responsive promoter were transformed with the empty expression vector, HA-p23, or HA-yhp23. When assayed at a 0.1 nM concentration of 17-β-estradiol, expression of human p23 increases ER transcriptional activity compared to the use of the vector only (Fig. 7B), but to a lesser extent than with yhp23 (twofold and fivefold, respectively). Again, the reduced activity of ER in the human p23-expressing strain is not a function of reduced p23 levels relative to yhp23 (Fig. 7B, bottom panel). These findings indicate that human p23 can partially complement the loss of yhp23 function in yeast cells with respect to ER signaling, thus suggesting that yhp23 and p23 are functional homologues.
Increased ER transcriptional activation by human p23 overexpression in MCF-7 cells.
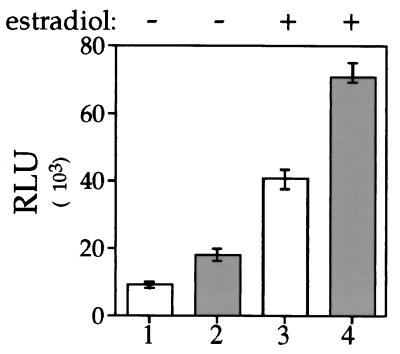
To establish whether p23 affects ER signal transduction in mammalian cells, we examined the ability of human p23 to increase ER-mediated transcriptional enhancement when overexpressed in cultured mammalian cells. ER-containing MCF-7 cells were transfected with the reporter plasmid ERE-thymidine kinase-luciferase and a plasmid encoding human p23. Transfected cells were treated with 0.1 nM 17-β-estradiol or ethanol vehicle for 24 h, and the transcriptional activity was quantified by measuring the luciferase activity. As shown in Fig. 8, both hormone-dependent and hormone-independent ER transcriptional activity are increased roughly twofold when p23 is overexpressed. This likely represents an underestimate of p23’s importance to ER function, since these results are obtained in a cell line that contains endogenous p23 (not shown). These findings suggest that p23 is a limiting factor for ER signal transduction and therefore subsequent ER-mediated transcriptional enhancement.
FIG. 8.
Activation of ER transcriptional enhancement by p23 overexpression. ER-containing MCF-7 cells (2.5 × 105 cells/6-cm dish) were transiently transfected by using the lipid Trans-IT 100 with 5 μg of the ERE-containing luciferase reporter plasmid and 2 μg of a pCMV expression vector (open columns) or pCMV-HA-p23 expression vector (shaded columns). Cells were incubated for 24 h in the presence of 0.01 nM 17-β-estradiol or ethanol vehicle and then harvested. ER transcriptional activation was measured by using a luciferase assay, normalized to total protein concentration in each sample, and expressed as relative luminescence units (RLU). The data represent the mean of an experiment done in triplicate, which was repeated four times.
DISCUSSION
Using dosage suppression analysis in yeast cells to isolate factors involved in ER signal transduction, we have identified the yeast homologue of the human p23 (yhp23) as a protein that, when overexpressed, results in a 10-fold increase in G400V ER transcriptional activation. In vivo estradiol-binding assays suggest that yhp23 overexpression increases G400V ER transcriptional activity by increasing the number of estradiol-bound receptors. The effect of yhp23 overexpression was not limited to G400V ER, as it also increases both ligand binding and transcriptional activation by wt ER. No effect of yhp23 overexpression was observed on the constitutive activity of ER1–115, thereby demonstrating that yhp23 does not affect AF-1 activity per se. We therefore conclude that yhp23 is a member of the reconstituted steroid receptor signaling pathway in yeast cells, acting at the step of ligand binding by the receptor. This role is consistent with the currently proposed function of human p23 in steroid receptor-aporeceptor complex formation.
By using a yeast strain deficient in yhp23 expression (KO), analyses of wt ER signaling as a function of yhp23, wt ER, and estradiol concentrations were carried out. Our studies demonstrate that the magnitude of the effect of yhp23 on wt ER transcriptional activation is inversely proportional to the concentration of both wt ER and estradiol. Thus, at low, subsaturating concentrations of estradiol, yhp23 overexpression markedly increases wt ER transcriptional activation. In contrast, at saturating concentrations of estradiol, the effect of yhp23 overexpression on wt ER transcriptional activation is comparatively small. Furthermore, the magnitude of the effect of yhp23 on wt ER transcriptional activation is greater at low, rather than high, wt ER expression levels. Taken together, our findings indicate that the effect of yhp23 on wt ER signaling varies depending on yhp23, wt ER, and estradiol concentrations.
Subcellular localization studies with a yhp23-GFP fusion protein indicate that yhp23 is largely cytoplasmic in the absence of ER expression. When coexpressed with either the mutant or wt ER, yhp23 colocalizes with the receptor to the nucleus in the absence of estradiol. This colocalization is reversed upon estradiol treatment, such that yhp23 is released into the cytoplasm. From these observations, we conclude that yhp23 is part of the ER aporeceptor complex in yeast cells and that the distribution of yhp23 within the cell is dynamic and affected both by ER expression and estradiol binding.
It has been proposed that the function of p23 in steroid receptor signaling is to promote, through its interaction with Hsp90, the maturation or stabilization of the aporeceptor complex (24, 30). This model, derived largely from in vitro experiments (45, 46), proposes that the heat shock proteins Hsp90, p60, Hsp70, and possibly Hsp40 (10) form a complex termed a “foldosome” (23), within which Hsp90 exists in a conformation incompatible with p23 binding (53) (Fig. 9). The foldosome binds to the free receptor, which exists in a conformation with low affinity for the ligand. In a process that requires ATP and monovalent cations, the Hsp90 component of the foldosome and the receptor undergo conformational changes (13), such that Hsp90 is now capable of binding p23 (53), and the receptor exhibits high-affinity steroid binding. p23 binding to Hsp90 appears to stabilize this immature aporeceptor complex in vitro. In the absence of p23, the Hsp90-receptor complex is inherently unstable and rapidly dissociates (13).
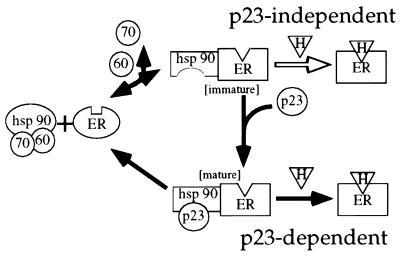
FIG. 9.
A model for p23-dependent and -independent ER signal transduction. A simplified version of the current model of aporeceptor complex formation as deduced from in vitro studies is indicated by the black arrows. Hsp90 refers to the Hsp90 dimer, 70 refers to Hsp70, 60 refers to p60, p23 refers to the mammalian p23 protein, ER refers to the estrogen receptor, and H refers to hormone. The immunophilins, which do not appear to be essential to aporeceptor complex formation but are isolated with the complex in vivo have been excluded from the model for simplicity. According to the model, p60, Hsp70 (and possibly Hsp40), and a dimer of Hsp90 preassociate to form the “foldosome” complex. The foldosome binds to the steroid-binding domain of the native receptor. Hsp70 and p60 are released from the complex in a process that requires ATP and a monovalent cation, while Hsp90 and the receptor undergo conformational changes, such that the receptor assumes a conformation with high affinity for steroid. This new complex (labeled “immature” in this figure) is inherently unstable and quickly disassociates unless p23 binds to Hsp90, thereby stabilizing the aporeceptor complex (labeled “mature” in this figure). Our data, however, suggests that a second, p23-independent pathway to ER ligand binding exists in vivo, occurring when ligand binds directly to the immature aporeceptor complex (open arrow), which is favored at high ER and/or estradiol concentrations. We suggest that the G400V ER mutation renders the receptor less capable of participating in the p23-independent pathway, thereby functionally uncoupling the p23-independent and -dependent pathways.
The ability of yhp23 to increase ER transcriptional activation might be expected if yhp23 is limiting for the formation of mature aporeceptor complexes. Although we cannot exclude the possibility that yhp23 overexpression increases the ratio of yhp23 binding to each molecule of ER, this idea runs contrary to the current model of p23 function. Increasing the concentration of yhp23, therefore, would be expected to result in a greater number of mature ER aporeceptor complexes in the cell. As a result, the total estradiol binding will increase, which is consistent with our in vivo estradiol binding assays (Fig. 3). Thus, overexpression of yhp23, by increasing the number of mature aporeceptor complexes, will manifest itself as an increase in transcriptional activation by ER at a given hormone concentration (Fig. 2 and 4). Thus, our in vivo findings are consistent with the current model of p23 function as derived from in vitro experiments.
Our results indicating that wt ER activity is detectable in the absence of yhp23 (Fig. 4), however, suggest that in addition to the p23-dependent pathway, there must also exist a p23-independent pathway leading to estradiol binding and signaling by the ER. We propose that p23-independent activation of the ER in vivo occurs through estradiol binding directly to the immature (p23-deficient) aporeceptor complex (Fig. 9). This hypothesis is consistent with in vitro observations that in the absence of p23, the foldosome proteins Hsp90, Hsp70, and p60 are sufficient to induce the hormone-binding conformation of steroid receptors (13). We therefore propose that estradiol binding by the ER is a composite of both the p23-independent and p23-dependent pathways. The relative contribution of each pathway to ER activation is dependent upon the concentration of p23, ER, and estradiol (see below).
One prediction of our model is that yhp23 becomes less relevant to ER activation as the ratio of immature to mature aporeceptor complexes increases. The ratio of the two types of aporeceptor complexes is, in turn, a reflection of both ER and yhp23 concentrations. Increasing ER expression when p23 levels are constant results in a greater number of the immature aporeceptor complexes, which favors hormone binding through the p23-independent pathway. Conversely, increasing yhp23 levels facilitates the formation of mature aporeceptor complexes and therefore the p23-dependent pathway. This model of ER signal transduction is consistent with our in vivo findings that indicate that the magnitude of the effect of yhp23 on ER transcriptional activation is inversely proportional to the concentration of ER (Fig. 4D).
The proposed model further envisions that the concentration of estradiol also affects the relative contribution of the p23-dependent and p23-independent steroid binding pathways. As suggested by Fig. 9, free steroid can be considered to be competing with yhp23 for binding to the immature aporeceptor complex. As a result, the p23-independent pathway becomes more prominent as estradiol concentrations rise or, conversely, as yhp23 levels fall. Consistent with this notion, our data demonstrate that the magnitude of yhp23’s effect on ER signaling is greatest at low subsaturating, rather than high saturating, estradiol concentrations (Fig. 4E). Thus, our model predicts that the balance among yhp23, ER, and estradiol, ultimately determines the relative contributions of the p23-dependent and p23-independent pathways to ER signal transduction.
Our model also provides insight into the observation that yhp23 overexpression induces G400V ER activity to a greater extent than that observed for wt ER (Fig. 2). We suggest that by altering the steroid-binding domain conformation, the G400V mutation largely eliminates the p23-independent pathway. G400V ER is temperature sensitive, relative to the wt ER, for estradiol binding in vitro, displaying reduced estradiol binding at 25°C but not at 4°C (54). This suggests that the G400V ER mutation destabilizes the conformation of the steroid-binding domain, such that the receptor is unable to bind steroid with high affinity at 25°C. This mutation does not inhibit G400V ER’s interaction with the aporeceptor complex since G400V ER has been reported to be complexed with Hsp90 (1) and since our subcellular localization studies suggest that G400V ER associates with yhp23 as efficiently as does wt ER. We therefore propose that the G400V mutation, by altering the structure of the steroid-binding domain, largely inhibits estradiol binding to the transient, immature aporeceptor complex, thereby diminishing estradiol binding through the p23-independent pathway. As a result, yhp23 competes more effectively with estradiol for binding to the complex, favoring the p23-dependent estradiol-binding pathway. The stability gained through yhp23 binding to the Hsp90-chaperone machinery, in turn, facilitates steroid binding by G400V ER.
It has been suggested by others that wt ER does not stably interact with Hsp90. This conclusion is based, in part, on studies with a VP16-Gal-ERLBD fusion protein (1, 37). Although these constructs continue to exhibit ligand-dependent activation, association of this construct with Hsp90 could not be demonstrated. While there are a number of possible reasons for this discrepancy, we feel that the most probable explanation is a difference in the inherent characteristics of the wt and fusion proteins (1). In addition, there are several reports that support a role for Hsp90 in the process of ligand binding by ER. (i) ER has been isolated with members of the aporeceptor complex from MCF-7 cells (52) and bovine uterus (49). (ii) Hsp90 has been demonstrated to colocalize with ER in the nucleus (38). (iii) ER function has been found to be affected by mutant Hsp90 molecules (44). (iv) ER function is also affected by the Hsp90-specific inhibitor geldanamycin (51). Although these studies do not exclude the possibility of additional transcriptional repressors of wt ER, these findings, along with our results, strongly support a role for the components of the Hsp90-based chaperone complex in ER signal transduction.
Elegant genetic studies of yhp23 function in yeast cells by Bohen (2) and Fang et al. (16) demonstrate that yhp23 associates with Hsp90 and is a part of the GR aporeceptor complex in yeast cells. In contrast to our findings with ER, analysis of androgen receptor (AR) signaling in yeast cells suggests that it is largely p23 independent. This may reflect inherent differences in the mechanism of signal transduction employed by the receptors. Alternatively, we would suggest that, although the analysis of AR signaling was performed under a range of steroid concentrations, the levels of AR expression used may have favored the p23-independent pathway. It would be interesting to reevaluate AR signaling as a function of yhp23 at both low AR and testosterone concentrations, conditions that would favor the p23-dependent pathway.
The partial complementation of human p23 in yeast cells lacking yhp23 strongly suggests that yhp23 functions as the p23 homologue with respect to ER signaling (Fig. 7). Although the yeast and human p23 proteins have regions of identity, significant sequence differences between the proteins also exist (2). We speculate that this reflects species-specific differences in p23-Hsp90 association and might therefore explain the inability of human p23 to fully complement the loss of yhp23 function. It would be interesting to examine whether yeast cells expressing human Hsp90 (44) and p23 increase ER activity to the same extent as their yeast counterparts.
Finally, our studies also provide insight into the possible mechanisms by which the ER communicates with other signaling pathways. Unexpectedly, a significant increase in estradiol-independent activation of ER was observed as a result of yhp23 overexpression (Fig. 4F). This estradiol-independent activation of wt ER was also observed upon p23 overexpression in MCF-7 cells (Fig. 8). Previous studies have proposed that estradiol-independent transcriptional activation by wt ER results, in part, from ER phosphorylation through an epidermal growth factor-dependent pathway (4, 27). Thus, the most direct interpretation of our data suggests that maintenance of ER within the aporeceptor complex facilitates (but is not essential to) estradiol-independent activity, perhaps by maintaining ER in a conformation amenable to phosphorylation.
Besides ER, several other signaling molecules (including c-Src and c-Raf) are dependent upon chaperone complexes for their function (45). Yeast cells deficient in the DnaJ homologue YDJ1, for instance, display both altered steroid receptor and Src kinase activity (31). Thus, molecular chaperones link diverse signaling pathways. Additional insight into the mechanism of this cross talk comes from our subcellular localization studies that reveal a striking colocalization of ER and yhp23 within the nucleus. Estradiol treatment was shown to liberate yhp23 (and presumably other chaperone proteins) from the nucleus, allowing it to redistribute throughout the cytoplasm, where it can potentially interact with other signaling proteins. Although our colocalization studies were carried out under conditions of overexpression, we speculate that estradiol activation of ER may, through the release of chaperone components, modulate the activity of a variety of chaperone-dependent pathways. In light of p23’s role in stabilizing chaperone complexes, it is likely to play a key regulatory role in any such “chaperone signaling.”
In conclusion, we have provided evidence that yhp23 is a member of the ER signaling pathway and a positive regulator of ER function. We also suggest that at high ER and/or estradiol concentrations, conditions often present in the yeast system or during transient overexpression of ER in cultured mammalian cells, ER signaling occurs largely through a p23-independent pathway. Under low physiological concentrations of ER and estradiol, p23 is likely to be an important contributor to ER signaling. Our results also indicate that alterations in the level or subcellular distribution of p23 are potential mechanisms for modulating estradiol-dependent and -independent ER transcriptional activation. We are currently examining whether the p23 levels, the subcellular distribution, or the modification state fluctuates between normal and tumor cells, during cellular proliferation, during differentiation, or upon growth factor treatment. As aporeceptor complex formation is also believed to be important for ligand binding by GR, PR, and AR, p23 likely plays an important role in these pathways as well.
ACKNOWLEDGMENTS
We thank S. Bohen and D. Toft for the yeast p23 deletion strain and human p23 cDNA, respectively; B. Freeman and K. Yamamoto for generously providing the antiserum against yeast p23 protein; and M. Stallcup and P. Kushner for the GRIP1 and ER expression vectors, respectively. We also thank I. Rogatsky, J. Trowbridge, A. Hittelman, and A. Caplan for critically reading the manuscript.
This work was supported by the Army Breast Cancer Research Fund grants DAMD17-94-4454, DAMD17-96-1-6032 (to M.J.G.), and DAMD-17-98-1-8134 and NIH Training Grant 2T32GM07308 from the National Institute of General Medical Sciences (to R.K.) and the Irma T. Hirschl Charitable Trust.
REFERENCES
- 1.Aumais J P, Lee H S, Lin R, White J H. Selective interaction of hsp90 with an estrogen receptor ligand-binding domain containing a point mutation. J Biol Chem. 1997;272:12229–12235. doi: 10.1074/jbc.272.18.12229. [DOI] [PubMed] [Google Scholar]
- 2.Bohen S P. Genetic and biochemical analysis of p23 and ansamycin antibiotics in the function of Hsp90-dependent signaling proteins. Mol Cell Biol. 1998;18:3330–3339. doi: 10.1128/mcb.18.6.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brzozowski A M, Pike A C, Dauter Z, Hubbard R E, Bonn T, Engstrom O, Ohman L, Greene G L, Gustafsson J A, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 4.Bunone G, Briand P A, Miksicek R J, Picard D. Activation of the unliganded estrogen receptor by EGF involves the MAP kinase pathway and direct phosphorylation. EMBO J. 1996;15:2174–2183. [PMC free article] [PubMed] [Google Scholar]
- 5.Caplan A J. Yeast molecular chaperones and the mechanism of steroid hormone action. Trends Endocrinol Metab. 1997;8:271–276. doi: 10.1016/s1043-2760(97)00079-9. [DOI] [PubMed] [Google Scholar]
- 6.Catelli M G, Binart N, Jung-Testas I, Renoir J M, Baulieu E E, Feramisco J R, Welch W J. The common 90-kd protein component of non-transformed ’8S’ steroid receptors is a heat-shock protein. EMBO J. 1985;4:3131–3135. doi: 10.1002/j.1460-2075.1985.tb04055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang H C, Lindquist S. Conservation of Hsp90 macromolecular complexes in Saccharomyces cerevisiae. J Biol Chem. 1994;269:24983–24988. [PubMed] [Google Scholar]
- 8.Chang H C, Nathan D F, Lindquist S. In vivo analysis of the Hsp90 cochaperone Sti1 (p60) Mol Cell Biol. 1997;17:318–325. doi: 10.1128/mcb.17.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cutolo M, Sulli A, Seriolo B, Accardo S, Masi A T. Estrogens, the immune response and autoimmunity. Clin Exp Rheumatol. 1995;13:217–226. [PubMed] [Google Scholar]
- 10.Dittmar K D, Banach M, Galigniana M D, Pratt W B. The role of DnaJ-like proteins in glucocorticoid receptor · hsp90 heterocomplex assembly by the reconstituted hsp90 · p60 · hsp70 foldosome complex. J Biol Chem. 1998;273:7358–7366. doi: 10.1074/jbc.273.13.7358. [DOI] [PubMed] [Google Scholar]
- 11.Dittmar K D, Demady D R, Stancato L F, Krishna P, Pratt W B. Folding of the glucocorticoid receptor by the heat shock protein (hsp) 90-based chaperone machinery. The role of p23 is to stabilize receptor · hsp90 heterocomplexes formed by hsp90 · p60 · hsp70. J Biol Chem. 1997;272:21213–21220. doi: 10.1074/jbc.272.34.21213. [DOI] [PubMed] [Google Scholar]
- 12.Dittmar K D, Hutchison K A, Owens-Grillo J K, Pratt W B. Reconstitution of the steroid receptor · hsp90 heterocomplex assembly system of rabbit reticulocyte lysate. J Biol Chem. 1996;271:12833–12839. doi: 10.1074/jbc.271.22.12833. [DOI] [PubMed] [Google Scholar]
- 13.Dittmar K D, Pratt W B. Folding of the glucocorticoid receptor by the reconstituted Hsp90-based chaperone machinery. The initial hsp90 · p60 · hsp70-dependent step is sufficient for creating the steroid binding conformation. J Biol Chem. 1997;272:13047–13054. doi: 10.1074/jbc.272.20.13047. [DOI] [PubMed] [Google Scholar]
- 14.Duina A A, Chang H C, Marsh J A, Lindquist S, Gaber R F. A cyclophilin function in Hsp90-dependent signal transduction. Science. 1996;274:1713–1715. doi: 10.1126/science.274.5293.1713. [DOI] [PubMed] [Google Scholar]
- 15.Elliston J F, Fawell S E, Klein-Hitpass L, Tsai S Y, Tsai M-J, Parker M G, O’Malley B W. Mechanism of estrogen receptor-dependent transcription in a cell-free system. Mol Cell Biol. 1990;10:6607–6612. doi: 10.1128/mcb.10.12.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang Y, Fliss A E, Rao J, Caplan A J. SBA1 encodes a yeast Hsp90 cochaperone that is homologous to vertebrate p23 proteins. Mol Cell Biol. 1998;18:3727–3734. doi: 10.1128/mcb.18.7.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freeman B C, Toft D O, Morimoto R I. Molecular chaperone machines: chaperone activities of the cyclophilin Cyp-40 and the steroid aporeceptor-associated protein p23. Science. 1996;274:1718–1720. doi: 10.1126/science.274.5293.1718. [DOI] [PubMed] [Google Scholar]
- 18.Garabedian M J, Yamamoto K R. Genetic dissection of the signaling domain of a mammalian steroid receptor in yeast. Mol Biol Cell. 1992;3:1245–1257. doi: 10.1091/mbc.3.11.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ginsburg G S, Douglas P S. Why cardiologists should be interested in estrogen. Am J Cardiol. 1996;78:559–561. doi: 10.1016/s0002-9149(96)00365-7. [DOI] [PubMed] [Google Scholar]
- 20.Golemis E A, Brent R. Searching for interacting proteins with the two-hybrid system III. In: Bartel P L, Fields S, editors. The yeast two-hybrid system. New York, N.Y: Oxford University Press; 1997. pp. 43–72. [Google Scholar]
- 21.Hong H, Kohli K, Garabedian M J, Stallcup M R. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong H, Kohli K, Trivedi A, Johnson D L, Stallcup M R. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutchison K A, Dittmar K D, Pratt W B. All of the factors required for assembly of the glucocorticoid receptor into a functional heterocomplex with heat shock protein 90 are preassociated in a self-sufficient protein folding structure, a “foldosome.”. J Biol Chem. 1994;269:27894–27899. [PubMed] [Google Scholar]
- 24.Hutchison K A, Stancato L F, Owens-Grillo J K, Johnson J L, Kirshna P, Toft D O, Pratt W B. The 23-kDa acidic protein in reticulocyte lysate is the weakly bound component of the hsp foldosome that is required for assembly of the glucocorticoid receptor into a functional heterocomplex with hsp90. J Biol Chem. 1995;270:18841–18847. doi: 10.1074/jbc.270.32.18841. [DOI] [PubMed] [Google Scholar]
- 25.Ing N H, Beekman J M, Tsai S Y, Tsai M J, O’Malley B W. Members of the steroid hormone receptor superfamily interact with TFIIB (S300-II) J Biol Chem. 1992;267:17617–17623. [PubMed] [Google Scholar]
- 26.Jacquier A, Legrain P, Dujon B. Sequence of a 10.7-kb segment of yeast chromosome XI identifies the APN1 and the BAF1 loci and reveals one tRNA gene and several new open reading frames including homologs to RAD2 and kinases. Yeast. 1992;8:121–132. doi: 10.1002/yea.320080207. [DOI] [PubMed] [Google Scholar]
- 27.Joel P B, Traish A M, Lannigan D A. Estradiol-induced phosphorylation of serine 118 in the estrogen receptor is independent of p42/p44 mitogen-activated protein kinase. J Biol Chem. 1998;273:13317–13323. doi: 10.1074/jbc.273.21.13317. [DOI] [PubMed] [Google Scholar]
- 28.Johnson J L, Beito T G, Krco C J, Toft D O. Characterization of a novel 23-kilodalton protein of unactive progesterone receptor complexes. Mol Cell Biol. 1994;14:1956–1963. doi: 10.1128/mcb.14.3.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson J L, Toft D O. Binding of p23 and hsp90 during assembly with the progesterone receptor. Mol Endocrinol. 1995;9:670–678. doi: 10.1210/mend.9.6.8592513. [DOI] [PubMed] [Google Scholar]
- 30.Johnson J L, Toft D O. A novel chaperone complex for steroid receptors involving heat shock proteins, immunophilins, and p23. J Biol Chem. 1994;269:24989–24993. [PubMed] [Google Scholar]
- 31.Kimura Y, Yahara I, Lindquist S. Role of the protein chaperone YDJ1 in establishing Hsp90-mediated signal transduction pathways. Science. 1995;268:1362–1365. doi: 10.1126/science.7761857. [DOI] [PubMed] [Google Scholar]
- 32.King W J, Greene G L. Monoclonal antibodies localize oestrogen receptor in the nuclei of target cells. Nature. 1984;307:745–747. doi: 10.1038/307745a0. [DOI] [PubMed] [Google Scholar]
- 33.Korach K S, Couse J F, Curtis S W, Washburn T F, Lindzey J, Kimbro K S, Eddy E M, Migliaccio S, Snedeker S M, Lubahn D B, Schomberg D W, Smith E P. Estrogen receptor gene disruption: molecular characterization and experimental and clinical phenotypes. Recent Prog Horm Res. 1996;51:159–186. [PubMed] [Google Scholar]
- 34.Kralli A, Bohen S P, Yamamoto K R. LEM1, an ATP-binding-cassette transporter, selectively modulates the biological potency of steroid hormones. Proc Natl Acad Sci USA. 1995;92:4701–4705. doi: 10.1073/pnas.92.10.4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar V, Chambon P. The estrogen receptor binds tightly to its responsive element as a ligand-induced homodimer. Cell. 1988;55:145–156. doi: 10.1016/0092-8674(88)90017-7. [DOI] [PubMed] [Google Scholar]
- 36.Kumar V, Green S, Stack G, Berry M, Jin J R, Chambon P. Functional domains of the human estrogen receptor. Cell. 1987;51:941–951. doi: 10.1016/0092-8674(87)90581-2. [DOI] [PubMed] [Google Scholar]
- 37.Lee H S, Aumais J, White J H. Hormone-dependent transactivation by estrogen receptor chimeras that do not interact with hsp90. Evidence for transcriptional repressors. J Biol Chem. 1996;271:25727–25730. [PubMed] [Google Scholar]
- 38.Meng X, Devin J, Sullivan W P, Toft D, Baulieu E E, Catelli M G. Mutational analysis of Hsp90 alpha dimerization and subcellular localization: dimer disruption does not impede “in vivo” interaction with estrogen receptor. J Cell Sci. 1996;109:1677–1687. doi: 10.1242/jcs.109.7.1677. [DOI] [PubMed] [Google Scholar]
- 39.Metzger D, Ali S, Bornert J M, Chambon P. Characterization of the amino-terminal transcriptional activation function of the human estrogen receptor in animal and yeast cells. J Biol Chem. 1995;270:9535–9542. doi: 10.1074/jbc.270.16.9535. [DOI] [PubMed] [Google Scholar]
- 40.Metzger D, Losson R, Bornert J M, Lemoine Y, Chambon P. Promoter specificity of the two transcriptional activation functions of the human oestrogen receptor in yeast. Nucleic Acids Res. 1992;20:2813–2817. doi: 10.1093/nar/20.11.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Metzger D, White J H, Chambon P. The human oestrogen receptor functions in yeast. Nature. 1988;334:31–36. doi: 10.1038/334031a0. [DOI] [PubMed] [Google Scholar]
- 42.Nair S C, Toran E J, Rimerman R A, Hjermstad S, Smithgall T E, Smith D F. A pathway of multi-chaperone interactions common to diverse regulatory proteins: estrogen receptor, Fes tyrosine kinase, heat shock transcription factor Hsf1, and the aryl hydrocarbon receptor. Cell Stress Chaperones. 1996;1:237–250. doi: 10.1379/1466-1268(1996)001<0237:apomci>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Onate S A, Tsai S Y, Tsai M J, O’Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 44.Picard D, Khursheed B, Garabedian M J, Fortin M G, Lindquist S, Yamamoto K R. Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature. 1990;348:166–168. doi: 10.1038/348166a0. [DOI] [PubMed] [Google Scholar]
- 45.Pratt W B. The hsp90-based chaperone system: involvement in signal transduction from a variety of hormone and growth factor receptors. Proc Soc Exp Biol Med. 1998;217:420–434. doi: 10.3181/00379727-217-44252. [DOI] [PubMed] [Google Scholar]
- 46.Pratt W B, Toft D O. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 47.Raisz L G. Estrogen and bone: new pieces to the puzzle. Nat Med. 1996;2:1077–1078. doi: 10.1038/nm1096-1077. [DOI] [PubMed] [Google Scholar]
- 48.Ratajczak T, Carrello A. Cyclophilin 40 (CyP-40), mapping of its hsp90 binding domain and evidence that FKBP52 competes with CyP-40 for hsp90 binding. J Biol Chem. 1996;271:2961–2965. doi: 10.1074/jbc.271.6.2961. [DOI] [PubMed] [Google Scholar]
- 49.Ratajczak T, Carrello A, Mark P J, Warner B J, Simpson R J, Moritz R L, House A K. The cyclophilin component of the unactivated estrogen receptor contains a tetratricopeptide repeat domain and shares identity with p59 (FKBP59) J Biol Chem. 1993;268:13187–13192. [PubMed] [Google Scholar]
- 50.Rogatsky I, Logan S K, Garabedian M J. Antagonism of glucocorticoid receptor transcriptional activation by the c-Jun N-terminal kinase. Proc Natl Acad Sci USA. 1998;95:2050–2055. doi: 10.1073/pnas.95.5.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Segnitz B, Gehring U. The function of steroid hormone receptors is inhibited by the hsp90-specific compound geldanamycin. J Biol Chem. 1997;272:18694–186701. doi: 10.1074/jbc.272.30.18694. [DOI] [PubMed] [Google Scholar]
- 52.Segnitz B, Gehring U. Subunit structure of the nonactivated human estrogen receptor. Proc Natl Acad Sci USA. 1995;92:2179–2183. doi: 10.1073/pnas.92.6.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sullivan W, Stensgard B, Caucutt G, Bartha B, McMahon N, Alnemri E S, Litwack G, Toft D. Nucleotides and two functional states of hsp90. J Biol Chem. 1997;272:8007–8012. doi: 10.1074/jbc.272.12.8007. [DOI] [PubMed] [Google Scholar]
- 54.Tora L, Mullick A, Metzger D, Ponglikitmongkol M, Park I, Chambon P. The cloned human oestrogen receptor contains a mutation which alters its hormone binding properties. EMBO J. 1989;8:1981–1986. doi: 10.1002/j.1460-2075.1989.tb03604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Webster N J, Green S, Jin J R, Chambon P. The hormone-binding domains of the estrogen and glucocorticoid receptors contain an inducible transcription activation function. Cell. 1988;54:199–207. doi: 10.1016/0092-8674(88)90552-1. [DOI] [PubMed] [Google Scholar]
- 56.Welshons W V, Lieberman M E, Gorski J. Nuclear localization of unoccupied oestrogen receptors. Nature. 1984;307:747–749. doi: 10.1038/307747a0. [DOI] [PubMed] [Google Scholar]
- 57.Wickelgren I. Estrogen stakes claim to cognition. Science. 1997;276:675–678. doi: 10.1126/science.276.5313.675. [DOI] [PubMed] [Google Scholar]
- 58.Yager J D, Liehr J G. Molecular mechanisms of estrogen carcinogenesis. Annu Rev Pharmacol Toxicol. 1996;36:203–232. doi: 10.1146/annurev.pa.36.040196.001223. [DOI] [PubMed] [Google Scholar]