Abstract
Corona Virus Disease 2019 (COVID-19) caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has become a global pandemic. Additionally, the SARS-CoV-2 infection in the patients of Gastric Cancer (GC; the third leading cause of death in the world) pose a great challenge for the health management of the patients. Since there have been uncertainties to develop a new drug against COVID-19, there is an urgent need for repurposing drugs that can target key proteins of both SARS-CoV-2 and GC. The SARS-CoV-2-RdRp protein contains the NiRAN domain, which is known to have kinase-like folds. A docking study of the FDA approved drugs against GC was performed using AutoDock 4.2 and Glide Schrodinger suite 2019 against SARS-CoV-2-RdRp protein. MMGBSA and MD simulation studies were performed to investigate the binding and stability of the inhibitors with the target protein. In this study, we have found 12 kinase inhibitors with high binding energies namely Baricitinib, Brepocitinib, Decernotinib, Fasudil, Filgotinib, GSK2606414, Peficitinib, Ruxolitinib, Tofacitinib, Upadacitinib, Pamapimod and Ibrutinib. These FDA approved drugs against GC can play a key role in the treatment of COVID-19 patients along with GC as comorbidity. We also hypothesize that JAK, ITK, Rho-associated kinases, FGFR2, FYN, PERK, TYK2, p38-MAPK and SYK kinases can be considered as key therapeutic targets in COVID-19 treatment. Taken altogether, we have proposed the SARS-CoV-2-RdRp as a potential therapeutic target through in-silico studies. However, further in-vitro and in-vivo studies are required for the validation of the proposed targets and drugs for the treatment of COVID-19 patients already suffering from GC.
Keywords: SARS-CoV-2-RdRp, Gastric cancer, Kinase inhibitors, Molecular docking, MD simulation
1. Introduction
Corona Virus Disease 2019 (COVID-19) caused by a novel coronavirus (2019-nCoV) emerged in December 2019 [1,2]. Later, it was formally named Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by the International Committee on Taxonomy of Viruses (ICTV). SARS-CoV-2 belongs to betacoronavirus genus, Nidovirales order and is closely related to Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV). This virus displays human to human transmission who are in close contact through air dpolets, hence WHO declared it as worldwide public health emergency [3]. The cumulative number of globally reported cases exceeds 180 million and the number of global deaths is almost 4 million [4]. To date, no precise treatment is available for COVID-19 associated diseases. With the current pace of infection and the high mortality rate, COVID-19 has caused the scarcity of adequate health care worldwide. Several COVID-19 patients are subjected to quarantine and require rapid diagnosis and treatment. To combat this disease, constant efforts have been underway to find the cure and to halt the virus's spread [5]. Because development of new drug from scratch takes very long time, several drugs used for Severe Acute Respiratory Syndrome Coronavirus (SARS), Middle East respiratory syndrome coronavirus (MERS) and malaria are under clinical trials for the treatment of COVID-19. Hence, the drug repurposing strategies is fastest way to find drug against COVID-19 [6].
To the best of our knowledge, there are very few studies on the impact of COVID-19 on GC patients. However, there have been reports of high incidences of oesophageal and non-cardiac GC in the regions with high COVID-19 infections, such as China, Japan, Central, and South America [7,8]. According to a recent study, out of 212 COVID-19 patients, 3% of patients have been diagnosed with GC. These patients who were tested positive for SARS-CoV-2 were either under treatment or under follow-up for any malignant tumour. This also indicates that GC patients are more vulnerable for the SARS-CoV-2. About 30% mortality rate was found in these patients. It is also assumed that patients with gastrointestinal cancer were more susceptible where immunotherapy and radiotherapy within 3 months of COVID-19 diagnosis were considered as risk factors for death [9]. Hence, GC patients were considered as a vulnerable group for COVID-19 infection.
Importantly, cancer patients are among the most susceptible group in this (COVID-19) pandemic and the GC patients pose a significant challenge for the treatment [10]. GC itself is a global health problem, each year over 1 million people are diagnosed with GC worldwide [11]. To date, the clinical characteristics of GC patients who are infected with COVID-19 are remotely known [12]. The presence of ACE2, a SARS-CoV-2 receptor, is also observed in gastric mucosa and GI tract cells, which may be considered as a vulnerable site for SARS-CoV-2 infection [13,14]. The lasting presence of the virus in gastric mucosa shows the possibility of gastric glandular epithelial cells as a site of the virus infection [13,15]. Hence, the treatment of GC patients with COVID-19 is of utmost concern. Therefore, there is an urgent need to find out GC medicines that can also target COVID-19.
Coronavirus uses RNA-dependent RNA polymerase (RdRp) for the transcription of their genes and replication of their genome [16]. Coronavirus replication occurs through non-structural proteins (nsps) which are encoded by open reading frame (orf) 1a and 1b which translates nsps into polyprotein [17,18]. Further, this polyprotein undergoes maturation through proteolysis to form polymerase complex. The SARS-CoV-2 polymerase complex comprises an nsp12 subunit which is connected with the nsp7-nsp8 heterodimer and nsp8 subunit at a distinct binding site [19]. This nsp12 is the catalytic subunit that has RdRp activity. Though its polymerase efficiency is quite low [20]. It has been reported that the active site of nsp12 binds to the first turn of RNA and arbitrates the RdRp activity [21]. It plays the central role in the replication of virus genome with the help of two co-factors namely nsp7 and nsp8. Furthermore, it possesses the nucleotidyltransferase activity due to presence of nidovirus RdRp-associated nucleotidyltransferase (NiRAN) domain, which has an atypical kinase-like fold with GTP and UTP binding efficiency [22].
The cryo-EM structure of SARS-CoV-2 RdRp also provides structural evidence for nucleotidyltransferase activity of the NiRAN domain [23,24]. Further, SARS-CoV-2 RdRp showed a bond with the NiRAN domain in the presence of ADP-BeF3 or GDP-BeF3 structures, which suggests that this domain binds to nucleotides [23,25]. Recent studies suggest that nsp9 and nsp13 also interact with the NiRAN domain of nsp12 protein. Nsp9 acts as an RNA-binding protein that is necessary for SARS-CoV-2 replication, however, its molecular function is still unknown [26]. Nsp9 binds as a monomer to the NiRAN domain, and its N-terminus binds to the supposedly active site of the NiRAN domain where GDP is bound. There are reports which suggest that the aforementioned domain could catalyse the transfer of GMP to the 50 end of RNA in nsp13 dependent manner and inhibited by nsp9 [25]. This led to the hypothesis that the NiRAN domain could act as GTPase which is necessary for transcript capping which is regulated by nsp9. Moreover, NiRAN is also considered to have a role in protein-mediated RNA priming [27]. Taken all together, it can be hypothesized that the nsp12 NiRAN domain acts as a nucleotidyltransferase, but if it is involved in transcript capping, protein-mediated priming or some other replication-associated process remains to be determined [28].
In another study, SARS-CoV-2 proteins are considered to upregulate or downregulate ∼100 human kinases which are involved in cellular physiology, metabolism, and immune activation [29]. Reports suggest when SARS-CoV-2 enters the host cell; cell division halts and inflammation pathways get activated through tentacles like structure (filopodia) which ultimately helps the virus to spread to neighbouring cells. Thus, the host cell becomes a virus factory [30]. Therefore, we hypothesized that many FDA approved kinases inhibitors may hinder the life cycle of SARS-CoV-2. About 30 kinase inhibitors have been characterized in terms of antiviral potential, affecting viral entry metabolism and replication [31]. Similarly, many kinases were found to be overexpressed in cancer cells, also leading to uncontrolled growth in the cells. Furthermore, various kinase inhibitors have been found effective against these kinases. Both cancer and viruses alter the cellular machinery, which is mostly regulated by kinases [32].
Arteriviruses are positive-stranded RNA viruses which can infect mammals. They can cause disease associated with respiratory distress syndrome, lethal haemorrhagic fever or abortion [33]. In arterivirus, it is well documented that self-GMPylation/UMPylation activities of an NiRAN-RdRp are considered to generate a transient state primed for transferring nucleoside monophosphate (NMP) to unknown viral or cellular biopolymers [27]. In SARS-CoV-2, nsp12 (NiRAN-RdRp) also has Mn2+dependent NMPylation activity [27]. This NMPylation activity catalyses the transfer of a single NMP to the cognate nsp9. Both NiRAN activity and nsp9 NMPylation have an pivotal role in coronavirus replication [27]. Characteristics like singular phyletic association with nidoviruses and its genetic segregation with the RdRp, makes NiRAN a plausible key regulator enzyme of nidoviruses [34]. Moreover, SARS-CoV-2 nsp12 displays structural features of kinase-like folds [35]. Hence, targeting NiRAN-RdRp becomes a favourable target for both SARS-CoV-2 and GC.
According to Martin et al., 2021, GC patients are vulnerable for COVID-19 infection [9]. Several kinases like ABL, SRC and FGFR (fibroblast growth factor receptor) serve as pivotal therapeutic targets to control both GC and COVID-19 [28,31,[36], [37], [38]]. Further, various kinase inhibitors are effective against these kinases [28,31,[36], [37], [38]]. Hence, we wanted to evaluate the possibility to target COVID-19 with GC kinase inhibitors to prevent drug over-usage. Through this approach, we wanted to observe the effect of already known GC kinase inhibitors against SARS-CoV-2-RdRp-NiRAN domain. This study would further aid in determining COVID-19 comorbidity specific therapeutics and reducing drug-induced complications in the patient. To the best of our knowledge, this is the first study regarding the repurposing of FDA approved drugs against GC comorbidity specific to COVID-19. Moreover, this is also the first study that has attempted to target NIRAN-RdRp against drugs in COVID-19 concerning GC.
In this study, with in-silico approaches, we proposed that N- terminal of RdRp of SARS-CoV-2 possess the kinase-like folds and is involved in the phosphotransferase catalysis. We report the docking of several FDA approved GC kinase inhibitors against nsp12 (NiRAN-RdRp). Further, we performed the MMGBSA and MD simulation studies of four selected compounds, which gave the best binding energies during our investigation. This study intends to find out the medicine/compounds that could treat COVID-19 patients suffering from GC as comorbidity. We have also provided the schematic representation of the workflow in Fig. 1 .
Fig. 1.
Schematic representation of the study.
SARS-CoV-2-RdRp (PDB-ID: 7BTF) protein was retrieved from RCSB protein data bank, which was later trimmed to 1–400 amino acids. These amino acids constitute the SARS-CoV-2-RdRp-NiRAN domain. This structure was further used for in-silico studies. First, it was docked with 12 compounds using AutoDock 4.2, and the best four compounds were re-docked by using Glide Schrodinger suite. Complex of these four compounds were further proceeded for MD simulations on Maestro Schrodinger Suite. Finally, the compounds brepocitinib, decernocitinib, filgotinib, and ibrutinib were found appropriate for drug repurposing.
2. Methods
2.1. Data source
The protein sequence of SARS-CoV-2-RNA dependent RNA polymerase (PDB ID:7BTF) was downloaded from the RCSB protein data bank. The first 400 amino acids from 934 amino acids were used for analysis. The criteria for selection of 1–400 amino acids from the full-length sequence of SARS-CoV-2-RdRp was according to existing literature, as this N-terminal part of the RdRp contains NiRAN domain [19,39]. The trimmed N-terminal 400 amino acids of protein SARS-CoV-2-RdRp was first checked for the stability on the ProSA web server [40]. Further, this SARS-CoV-2-RdRp (N-terminal 400 amino acids) was used for all our docking and simulation studies. In the study this N-terminal 400 amino acids sequence of the protein is termed as SARS-CoV-2-RdRp-NiRAN domain. A total of 16 compounds (Supp. Table. 1) were downloaded from the PubChem data base in 3D sdf format and later converted into pdb format. We did not use any large data set of the compounds.
2.2. Active site prediction using CASTp
Binding pockets and the probable ligand-binding residues in the SARS-CoV-2-RdRp-NiRAN domain were identified using CASTp 3.0 software with default constant probe radius of 1.4 Å. CASTp 3.0 uses computational geometry algorithms consisting of Delaunay triangulation, alpha shape and discrete flow [41]. The software measured the volume and surface areas (SA) of the computed cavities using a solvent accessible surface model (Richards' surface) and the molecular surface model (Connolly's surface) and provided the outcome in decreasing order of binding pocket volume and areas (Supp. Table. 2). Also, the output was visualized using discovery studio software.
2.3. Molecular docking using AutoDock 4.2
Blind docking of 16 compounds were performed using AutoDock 4.2 to check the binding of our compounds at the CASTp predicted active site. Protein structure of SARS-CoV-RdRp-NiRAN domain already prepared in PyMol was used for the docking studies. Further for grid generation and docking, we edited as well as prepared protein using AutoDock 4.2 suite. Removal of water, the addition of polar hydrogens, and addition of Kollman charges were done. Protein and ligands both were edited and prepared and saved in pdbqt format. Then grid box was generated using Graphical Interface program AutoDock tools (ADT). Grid was prepared using the Autogrid and the grid size was set to 50 × 50 × 50 xyz points with grid spacing of 1.000 Å and grid center was designated at dimensions (x, y, and z): 120.047, 116.829 and 99.308. After grid preparation it was saved as. gpf format, the default docking parameters (Genetic algorithm for search and Lamarkian 4.2 for output) were selected and saved as. dpf format. After that Autogrid and Autodock were run followed by result analysis. Further, the docking result was analysed for conformation with best dock score and the complex were further visualized using discovery studio.
2.4. Redocking using Glide Schrodinger 2019-2
To validate the docking studies done by AutoDock4.2, we have revalidated it through Glide v8.3, Schrodinger, LLC, New York, NY, 2019-2. First, ligands and protein were prepared and then docking were performed.
2.4.1. Ligand preparation
Energy minimization of all the 16 compounds were done using the OPLS-2005 force field (LigPrep, Glide-v8.3 Schrodinger, LLC, New York, NY, 2019-2). Further, ligands were processed using LigPrep module (a part of the Schrodinger suite) which can generate different types of structures from each input structure with various tautomer's, ionization states, ring conformations, and stereochemical characteristics [42].
2.4.2. Protein and grid preparation
SARS-CoV-2-RdRp-NiRAN domain was used for protein preparation using the protein preparation wizard of Maestro program v10.2, a part of the Schrodinger suite (Schrodinger, LLC, New York, NY, 2019-2). Default parameters (missing atoms update, optimization and minimization) were used to prepare the protein/receptor. The grid box was created using receptor grid generation program Glide v8.3, Schrodinger, LLC, New York, NY, 2019-2 [43]. Grid was generated after optimization and minimization of protein. The dimensions of the inner box were kept X = 20, Y = 20, Z = 20, and dimensions of the outer box were kept as X = 36, Y = 36, Z = 36 around 5A° of the residues predicted by CASTp in the cavity.
2.4.3. Molecular docking
Flexible docking was performed using Glide v8.3, Schrodinger 2019-2 to evaluate the docking score, Glide score and Glide e-model of all the prepared ligands against the SARS-CoV-2-RdRp-NiRAN domain.
Different conformations of the ligands were allowed to interact and generate docking score, Glide score, and Glide e-model with protein. Best conformation of ligand based on docking score was evaluated against the SARS-CoV-2-RdRp-NiRAN domain. Docking score is a collective scoring of Van der Waals energy, Coulomb energy, lipophilic term, hydrogen bond term, metal-binding term, rewards, and penalties.
Docking results were evaluated based on the scoring function given by Glide G-score, which can be represented as:
| GScore = 0.065 x Vander Waals energy +0.130 x Coulomb energy + Lipo + H bond + Metal + BuryP + RotB + Site |
Where in the formula: Lipo = hydrophobic interactions, Metal = metal binding, BuryP = buried polar group penalty, RotB = penalty for freezing rotatable bonds and Site = polar interactions existing in the active site.
2.5. Molecular mechanics-generalized born surface area (MMGBSA)
After docking the compounds which have the best docking scores and suitable ADMET properties were selected for Prime molecular mechanics-generalized born surface area (MMGBSA). The structure of receptor and ligand complex which were obtained from molecular docking were used for the MMGBSA study. MMGBSA is useful for the determination of the ligand binding energy calculation (Prime, version 2.1, Schrodinger, LLC, New York, 2011) (Hayes &Archontis 2012). Minimizations of the receptor and ligand complex poses were done using the local optimization feature in the Prime. OPLS-2005 is the force field used in the Generalized-Born/Surface Area continuum solvent model for the calculation of the energies of the complexes. Ligand strain energy was also calculated during the MMGBSA.
2.6. Molecular dynamics simulation study
In this study, MD simulations were done using Desmond a module of Schrodinger suite to evaluate the stability of the protein-ligand complex. Protein-ligand RMSD, root mean square fluctuation (RMSF), and protein-ligand contact. Bar graphs were analysed to assess the stability and conformational behaviour of the protein-ligand complex along the entire the 100 ns simulation.
2.6.1. System building
System building of the complex was done using System builder, desmond, Schrodinger, LLC, New York, NY, 2019-1. As the solvent model, the TIP3P water model was used. For the neutralization and stabilization of the solution system [44], we used the default parameters of recalculating the addition of ions and salt addition (0.15 M of NaCl) in an orthorhombic solvent box of dimensions (the distance of 10 × 10 × 10 Å and angle of 90 × 90 × 90°) to cover the complex completely and provide the environment for Molecular dynamics simulation.
2.6.2. Molecular dynamic simulation
MD simulation was performed using Desmond, Schrodinger, LLC, New York, NY, 2019-1. Molecular dynamics simulation of the complex was performed after the system-building of the SARS-CoV-2-RdRp-NiRAN domain-ligand complex was completed. The build system of the complex was loaded for MD simulation each for 100 ns. All the parameters like recording interval (ps), energy, trajectory, NPT (temperature- 300 K, pressure = 1.01325 bar) were followed as the default value, and the checkpoint interval of the simulation was kept 240.06 ps [45,46]. The post MD simulation analysis like RMSD, RMSF, Protein-ligand interaction, ligand torsions etc. was analysed using the ‘Simulation Interactions Diagram’ tool (Maestro-Desmond Interoperability Tools, version 4.1, Schrodinger, New York, 2019-1). Further structures were extracted using the same SID tool by following default steps that are open SID, load system; build.out cms file of a protein-ligand complex. Further, the options (RMSD, Protein RMSF/SSE, Ligand RMSF, Protein-Ligand interactions, Ligand torsions, and Ligand properties) were selected and results were generated and analysed. Finally, the results were obtained in the pdf format and converted in tiff format.
3. Results
A set of non-structural proteins control the replication machinery of coronavirus (nsps) which are encoded by open reading frame 1a (ORF1a) and 1 ab in its genome. Both the ORFs are translated as a single polypeptide chain which undergoes proteolytic cleavage to generate multiple proteins that assemble to form a multi-subunit polymerase complex [47]. Further, this complex mediates viral genome transcription and replication. RNA dependent RNA polymerase (RdRp) is one of the most multipurpose enzymes of retroviruses. This is an important enzyme for the replication of the genome and translation. The main cofactor of this complex is the catalytic subunit nsp12/RdRp [48]. The nsp12 subunit can conduct the polymerase reaction but with very low efficiency, whereas nsp7 and nsp8 cofactors remarkably stimulates its polymerase activity [49]. Furthermore, nsp12 is known to have N-terminal extension that possesses a kinase-like fold, possibly nidovirus RdRp-associated nucleotidyltransferase (NiRAN) [22]. The NiRAN and the interface domains approximately span over residues 51–398 amino acids of the SARS-Cov2-RdRp polypeptide sequence [19,39]. Moreover, to study the kinase activity of the RdRp along with NiRAN activity, this domain was trimmed from 1 to 400 amino acids (Fig. 2 ). Its conserved amino acids, active site and binding pockets are also stated in Supp. Table 2.
Fig. 2.
Structural representation of SARS-CoV2-RdRp-NiRAN domain. Figures generated on PyMol. (a) Green color cartoon representation. (b) Green color solid surface representation showing putative active site residues (Asp36, Lys73, Asp126, Asp218, Phe219, and Asp221) with red color.
The trimmed protein structure of the SARS-CoV-2-RdRp-NiRAN domain (1–400 amino acids) was run on the ProSA web server to check their stability and structure validation [40]. Recognition of errors and validation of the model quality of protein structure is a major part of the 3D model preparation. In our study, we used ProSA web server which is frequently used for the 3D model validation. This server calculates overall quality scores for a specific input structure. For a good protein model, this score should be within the range. If the score is out of range the structure may have error. In our case, we got the Z-Score −7.16 which is within range and shows the model is of good quality [ Supp. Fig. 1] [40]. A plot of residues scores of a native protein structure also generated during ProSA run, which represents local model quality. This plot shows local model quality by plotting energies as a function of amino acid sequence position. Generally, positive values represent to problematic or erroneous parts of the input structure. In our case most of the residues are correspond to negative values which showed that the quality of our model is quite good [ Supp. Fig. 2].
As per our analysis through CASTp on the SARS-CoV-2-RdRp-NiRAN domain, we got our putative active sites in pocket 3. Out of mentioned amino acids, 6 amino acids (Asp36, Lys73, Asp126, Asp218, Phe219, and Asp221) showed stable interactions during molecular docking through Glide v8.3 Schrodinger, LLC software code. While no stable protein-ligand interactions were found with the residues of pockets 1, 2, 4 and 5. Importantly, all the 16 compounds interacted with the residues of pocket 3. Therefore, pocket 3 was predicted as our real active site.
3.1. Molecular docking studies
Recent pathophysiological understanding of SARS-CoV-2 infection stipulates that those infected with the virus could experience cytokine release syndrome (CRS). Further, this CRS is distinguished by elevated interleukin IL-6, IL-2, IL-7, IL-10, etc [50]. Hence, the treatment of cytokine storm could play a crucial role in the treatment of severe COVID-19. Several cytokines involved in COVID-19 is mediated by Janus kinases (JAKs) [51]. ITK is highly expressed in T cells and regulates the activation and function of both CD4+ and CD8+ T cells, including cytokine production and cytotoxic function [52]. Hence JAKs and ITK inhibition present potent therapeutic strategies against COVID-19. As SARS-CoV-2 is an RNA virus and for RNA synthesis, RdRp is an important enzyme. It functions by catalysing the RNA template-dependent development of phosphodiester bonds within the RNA genome. Therefore, RdRp is an important therapeutic target in RNA virus-caused diseases [53].
3.1.1. Molecular docking studies using AutoDock 4.2
The molecular docking of FDA approved drugs (for JAKs and other kinases) was performed against the SARS-CoV-2-RdRp-NiRAN domain with the help of software AutoDock 4.2. Further, the binding affinity of the drugs was chosen based on the lowest RMSD value of 0.00. RMSD value 0 represents identical structures and when this value increases, the two structures are considered to be more different [54]. Out of 16, 12 compounds were selected, and the remaining four kinases' inhibitors were excluded from the list because they violated the Lipinski rule. These four compounds were erdafitinib (PubChem CID: 67462786), fedratinib (PubChem CID:16722836), saracatinib (PubChem CID: 10302451) and fostamatinib (PubChem CID: 11671467) (Supp. Table 3). As erdafitinib and fedratinib had molar refractivity over 130 and molecular weight over 500 g/mol, respectively. Further, saracatinib and fostamatinib had molecular weight of more than 500 g/mol and hydrogen bind acceptor more than 5. The selected 12 compounds were included in our study considering no cut-off value. The binding energy for these compounds were baricitinib (−7.3 kcal/mol), brepocitinib (−8.7 kcal/mol), decernotinib (−7.9 kcal/mol), fasudil (−7.2 kcal/mol), - filgotinib (−8.0 kcal/mol), GSK2606414 (- 8.6 kcal/mol), peficitinib (−7.6 kcal/mol), ruxolitinib (−7.4 kcal/mol) and ibrutinib (−8.8 kcal/mol), Tofacitinib (−6.9 kcal/mol), Upadacitinib (−6.6 kcal/mol) and Pamapimod (−6.9 kcal/mol) (Supp. Table 4). These binding energies indicate the presence of kinase like domain in the NiRAN domain of nsp12 or RdRp. The interactions and the residues involved in the compound's binding with the SARS-CoV-2-RdRp-NiRAN domain are also represented in Supp. Table 2. Moreover, the 2D and 3D interaction diagrams of the selected 12 kinase inhibitors with the SARS-CoV-2-RdRp-NiRAN domain are represented in Suppl. Table 5.
3.1.2. Molecular docking studies using Glide Schrodinger suite
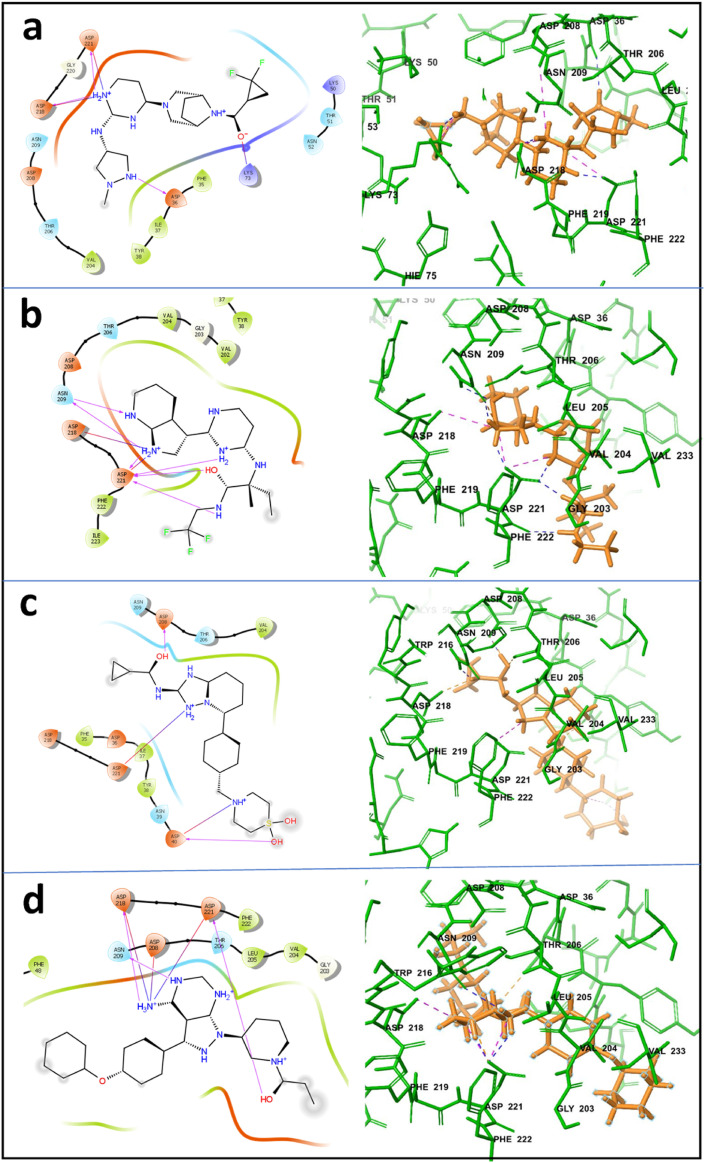
Out of twelve compounds, the four best compounds brepocitinib, decernocitinib, filgotinib, and ibrutinib have been selected for further re-docking and MD simulation studies. These compounds have been selected based on the highest docking score obtained after docking with AutoDock 4.2 and the cut off has been limited to −7.9. The information regarding docking and its score is mentioned in Supp. Table 4. Further, GSK2606414 showed AMES toxicity. Since it showed the ability to be carcinogenic, it was also excluded from the study. Re-docking of these compounds was performed using Glide module of Schrodinger Suite 2019-2. 2D and 3D interactions of these compounds with the SARS-CoV-2-RdRp-NiRAN domain were analysed and figures were generated on Glide platform of Schrodinger Suit 2019-2. 2D and 3D interaction diagrams are shown in Fig. 3 .
Fig. 3.
2D and 3D interaction diagram of inhibitory compounds with the SARS-CoV2-RdRp-NiRAN domain protein-based Dock; (a) with brepocitinib (b) with decernocitinib (c) with filgotinib, and (d) ibrutinib are shown. 2D diagrams were prepared using GLIDE and only H-bond interactions are presented here. In the 3D diagrams, SARS-CoV2-RdRp-NiRAN domain is presented with thin tubes; residues that directly contact with inhibitory compounds are depicted in thin stick models.
In these interactions, we mainly focused on H-bond interactions. The study of Ahmed et al., 2020 revealed active sites and the residues involved in the binding of the UTP and GTP within the SARS-CoV-2-RdRp-NiRAN domain [55]. Further, the residues aspartate, glutamate, lysine and arginine showed important interactions with the UTP and GTP in the NiRAN domain [34]. It was also revealed that these residues are at the active site and important conserved residues. Through CASTp, the important residues of the active site in pocket 3 are Arg33, Ala34, Phe35, Asp36, Ile37, Tyr38, Asn39, Lys50, Cys53, Arg55, Val71, Lys73, His75, Glu83, Arg116, Leu119, Thr120, Lys121, Tyr122, Thr123, Asp126, Val204, Thr206, Asp208, Asn209, Asp211, Tyr217, Asp218, Phe219 and Asp221.
In docking studies with Glide, brepocitinib shows strong interactions with the Asp36, Lys73, Asp218, and Asp22. The binding affinity for the brepocitinib was calculated to be −5.543 kcal/mol. In case of decernocitinib, residues Asn209, Asp218, and Asp221 showed strong interactions. The binding affinity for the decernocitinib was calculated as −6.694 kcal/mol. Filgotinib showed strong interactions with the residues Asp40, Asp208, and Asp221, and the binding affinity for the complex was calculated as −4.917 kcal/mol. Similarly, in the case of ibrutinib, the residues which showed strong interactions are Asp208, Asp218, and Asp221, and the binding affinity for this complex was calculated to be −6.137 kcal/mol. The docking scores, Glide G-score, and are Glide E-model are represented in Table 1 .
Table 1.
Molecular docking scores for Brepocitinib, Decernocitinib, Filgotinib and Ibrutinib against SARS-CoV-2-RdRp-NiRAN domain. Docking performed on Glide, Schrodinger suite.
| S.No. | Compound ID/name | Docking Score | Glide G-score | Glide E-model |
|---|---|---|---|---|
| 1 | Brepocitinib | - 5.543 | - 6.047 | - 68.281 |
| 2 | Decernocitinib | - 6.694 | - 6.991 | - 75.203 |
| 3 | Filgotinib | - 4.917 | - 4.931 | - 60.430 |
| 4 | Ibrutinib | - 6.137 | - 6.704 | - 73.405 |
On comparison of docking interactions obtained from AutoDock 4.2 and Glide module of Schrodinger Suite 2019-2. It was observed that brepocitinib have common interactions with Asp36 and Asp218 in the results obtained from both software's. Decernocitinib also showed interactions with Asp218 and Asp221 with both docking program AutoDock 4.2 and Glide. Similarly, ibrutinib also showed interactions with Asp36, Asp 218 and Asp221 with both AutoDock 4.2 and Glide. However, in case of filgotinib, both the software predicted different interaction sites. Glide showed interactions with Asp 40, Asp 208, Asp 221 whereas AutoDock 4.2 showed interactions with Arg33, Tyr 38, Phe48, Val 204, Asn 209, and Ser 236. All these results generated from Glide or AutoDock 4.2 show that all the compounds are binding in the same pocket or cavity of the SARS-CoV-2-RdRp-NiRAN domain. Data is shown in the Supplementary section as Supp. Table 6.
3.2. Molecular mechanics-generalized born surface area (MMGBSA)
MMGBSA study of the selected compounds brepocitinib, decernocitinib, filgotinib and ibrutinib were performed with the SARS-CoV-2-RdRp-NiRAN domain and it was observed that all the compounds have a good affinity towards SARS-CoV-2-RdRp-NiRAN domain (Table 2 ). MMGBSA scores for the brepocitinib, decernocitinib, filgotinib, and ibrutinib were calculated as −60.1315, −47.6964, −44.1200 and −77.1748 respectively. Among the four compounds, ibrutinib showed better affinity as compared to other compounds. The other individual energies of the MMGBSA wiz coulomb, covalent, H bond, solvation, Vander walls were also determined, which are given in Table 2.
Table 2.
The MM/GBSA binding energy scores and individual contributing energies.
|
Compounds |
MMGBSA dG Bind | MMGBSA dGBindContribution |
||||
|---|---|---|---|---|---|---|
| Coulomb | Covalent | H-bond | solvent GB | vdw | ||
| Brepocitinib | −60.1315 | −61.7106 | 3.9950 | −4.3891 | 55.5333 | −34.0907 |
| Decernocitinib | −47.6964 | −76.8374 | 14.4035 | −3.4601 | 65.5055 | −22.3610 |
| Filgotinib | −44.1200 | −59.6305 | 3.3934 | −2.7584 | 65.9576 | −26.1201 |
| Ibrutinib | −77.1748 | −167.7890 | 5.8154 | −1.3588 | 147.4467 | −28.3242 |
3.3. Molecular dynamics simulation analysis
Based on all the criteria of docking scores, ADMET and drug likeliness, we selected four compounds for further MD simulation studies. MD simulation studies were performed for the compounds brepocitinib, decernocitinib, filgotinib, and ibrutinib in complex with the SARS-CoV-2-RdRp-NiRAN domain. MD simulations were performed for 100 ns for each complex on the Desmond platform of Schrodinger suite 2019. For each protein-ligand complex, MD simulation was performed (Fig. 6 ). Post-MD simulation, 2D interaction diagram, protein-ligand contact analysis with different types of bonds, RMSD and RMSF analysis were also performed for understanding the stability of the ligand with the protein at the site of interaction during the simulation.
Fig. 6.
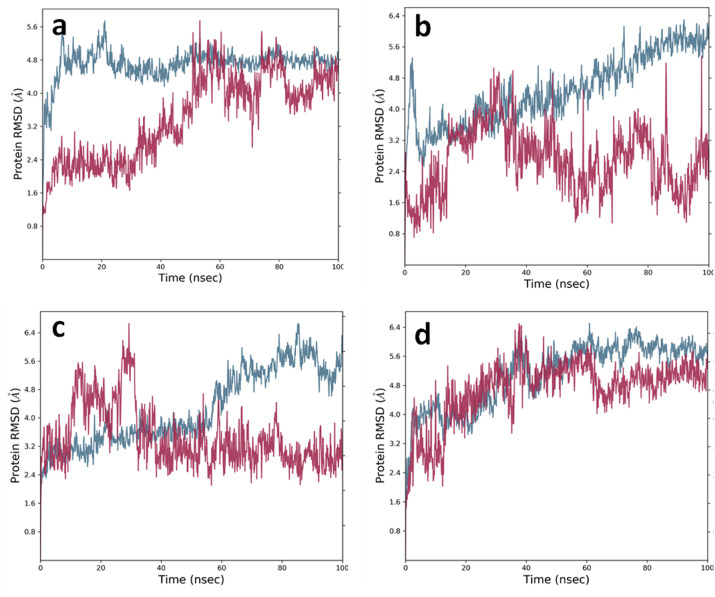
RMSD results of protein and protein-ligand complexes. Blue graph represents protein RMSD and maroon graph represents protein-ligand complex RMSD (a) SARS-CoV2-RdRp-NiRAN domain (residues 1–400) alone and with brepocitinib in a complex (b) SARS-CoV2-RdRp-NiRAN domain (residues 1–400) alone and with decernocitinib in a complex (c) SARS-CoV2-RdRp-NiRAN domain (residues 1–400) alone and with filgotinib in complex (d) SARS-CoV2-RdRp-NiRAN domain (residues 1–400) alone and with ibrutinib in complex.
3.3.1. SARS-CoV-2-RdRp-NiRAN domain-kinase inhibitors complex analysis
Post-MD simulation, 2D interaction diagrams show the interactions of the selected inhibitory compounds with the SARS-CoV-2-RdRp-NiRAN domain (Fig. 4). The analysis suggested during the simulation, all the selected inhibitory compounds are binding at the same active site. Protein-ligand contacts showing different interaction fractions are represented in Fig. 5. H bond occupancy, showing the H-bonds throughout the simulations are represented in Suppl. Fig. 3-6. Root mean square deviation (RMSD) plots of all these complexes were explained and shown in Fig. 5. RMSF of these complexes were also analysed and no considerable fluctuations were observed.
Fig. 4.
Post-MD simulations 2D representation of inhibitory compound interactions with the SARS-CoV-2-RdRp-NiRAN domain.
Fig. 5.
Protein-ligands contacts histogram representing important interacting residues of SARS-CoV2-RdRp-NiRAN domain with inhibitory compounds during MD simulation. X-axis showing residues and Y-Axis showing interactions fraction. (a) Interactions with brepocitinib (b) interactions with decernocitinib (c) interactions with filgotinib (d) interactions with ibrutinib. Green color represents H-bond, violet representing hydrophobic, pink representing ionic and blue representing water bridges, interactions fraction.
3.3.1.1. (SARS-CoV-2-RdRp-NiRAN domain/brepocitinib)
Post-MD simulation 2D interactions, in the case of brepocitinib show the interactions with Thr51, Lys73, Arg116, Asp208, Asp218, and Asp221. Lys73 and Arg116 donate H-bonds to brepocitinib while Thr51, Asp208, Asp218, and Asp221 receive H bond from brepocitinib. Lys73 and Asp221 make direct H-bond while Thr51, Arg116, Asp208, and Asp218 make H-bond through water molecules. Lys73 and Arg116 make strong H-bond as compared to other residues (Fig. 4a).
Protein-ligand contact (SARS-CoV-2-RdRp-NiRAN domain/Brepocitinib) analysis revealed different types of bond formation during the simulation (Fig. 5a). It was observed that Thr51, Lys73, Arg116, Asp208, Asn209, Asp218, and Asp221 make strong interaction with the brepocitinib. Lys73 and Arg116 make H-bond and water bridges while Asp218 and Asp221 make H-bond, water bridges, and ionic interactions. Thr51 and Asn209 make only water bridges while Asp208 makes water bridges as well as show ionic interaction.
Interactions that occur over 20.0% of the simulation time in the selected trajectory (0.00 through 100 ns) are shown. SARS-CoV-2-RdRp-NiRAN domain showing interaction with (a) brepocitinib (b) decernocitinib (c) filgotinib and (d) ibrutinib. Orange circle: charged (negative), Blue circle: charged (positive), White circle: water and red circle with bar represents Pi-cation.
Total contact analysis during the entire simulation time of 100 ns showed that maximum contact remained stable till 40 ns after that number of contacts slightly decreased and remained constant till the end of the simulation ( Supp. Fig. 3a). The timeline representation of contacts showed that Thr51, Lys73, Arg116, and Asp218 made strong interactions throughout the 100 ns of simulation time. While Asp208, Asn209, and Asp221 made interactions around up to 40 nanoseconds of simulation time ( Supp. Fig. 3b).
RMSD analysis of the SARS-CoV-2-RdRp-NiRAN domain-Brepocitinib complex showed some fluctuation up to 25 ns of simulation after that it became stable till the end of 100 ns of simulation. While ligand RMSD, after initial fluctuation of up to 6 ns became stable up to 30 ns. After that, no considerable changes in the ligand RMSD were observed (Fig. 6Aa). RMSD mean for backbone sidechain and ligands were calculated, and these were 4.743 ± 0.397 Å, 5.665 ± 0.415 Å and 1.588 ± 0.298 Å respectively.
3.3.1.2. (SARS-CoV-2-RdRp-NiRAN domain/decernocitinib)
2D interaction diagram after MD simulation showed the interactions of decernocitinib with the SARS-CoV-2-RdRp-NiRAN domain (Fig. 4b). The main interacting residues were Thr206, Asp208, Asp209, Asp218, and Asp221. These residues showed interaction more than 20% of the simulation time. Among these residues, only Asn209 donated an H-bond to the decernocitinib while the other residues Thr206, Asp208, Asp218, and Asp221 received H-bond from the decernocitinib.
Histogram of protein-ligand contacts (SARS-CoV-2-RdRp-NiRAN domain-decernocitinib) represents different types of bond interaction fractions (Fig. 5b). The main interacting residues.
Val204, Thr206, Asp208, Asn209, Asp218, Asp221, Phe222, and Ile223. Thr206, Asn209, Asp218, and Asp221 showed the direct H-bond interaction and the H-bond through water molecules, i.e., called water bridges. Val204 showed hydrophobic and water bridges interaction while Phe222 and Ile223 showed only hydrophobic interactions.
Total contact analysis suggests the total number of specific contacts formed during the simulation. SARS-CoV-2-RdRp-NiRAN domain formed bonds with the ligands. In the case of decernocitinib, total contacts remained high up to 50 ns after that it dropped slightly up to 75 ns and after that, it again increased and remained constant till the end of 100 ns of simulation (Supp. Fig. 4a). Supp. Fig. 4b represents specific residues that interact with the ligand in each trajectory frame. In the case of decernocitinib, Val204, Thr206, Asp208, Asn209, Asp218, Asp221 and Ile223 made contacts throughout the simulation. Out of these Val204, Asp208, Asn209 and Ile223 established good interactions whereas Asp218 and Asp221 showed very strong interactions (Supp. Fig. 4b).
RMSD graph of the SARS-CoV-2-RdRp-NiRAN domain-decernocitinib complex suggests that fluctuations in the protein RMSD remained stable from 5 to 20 ns after that the RMSD values increased with simulation time, whereas fluctuations in the ligand RMSD were observed throughout the simulation time (Fig. 6B.a). The RMSD mean for backbone sidechain and ligand were calculated to be 4.482 ± 0.888 Å, 5.490 ± 0.899 Å, 0.961 ± 0.475 Å respectively.
3.3.1.3. (SARS-CoV-2-RdRp-NiRAN domain/filgotinib)
Post-MD simulation 2D interaction diagram of SARS-CoV-2-RdRp-NiRAN domain with the filgotinib showed the main interactions with the Asp218 and Asp221. Asp218, and Asp21 both received the H-bonds from the filgotinib. And these were the only interactions that remained present for more than twenty percent of simulation time (Fig. 4c).
Histogram of protein-ligand contacts showed that the residues taking part in the interaction of the SARS-CoV-2-RdRp-NiRAN domain with the filgotinib (Fig. 5c). This also represents the bond fractions. The main interacting residues which show considerable effect were Tyr38, Val204, Thr206, Asp208, Asn209, Asp218 and Asp221. Asp218 and Asp221 showed strong H-bond interactions. Asp218 and Asp221 also showed water bridges while Asp221 showed ionic interaction. Tyr38 and Val204 showed hydrophobic and water bridge interactions. Asn209 showed H-bond, water bridge, and ionic interactions (Fig. 5c).
Total contact analysis during the simulation showed highest number of contacts during 12–22 ns and 53–56 ns and rest of the simulation time remained constant (Supp. Fig. 5a). Supp. Fig. 5b indicates that filgotinib interacts with the Tyr38, Val204, Asn209, Asp218 and Asp221. Out of these residues, Asp218 and Asp221 showed strong interactions while Tyr38, Val204 and Asn209 showed good interactions.
RMSD plot of SARS-CoV-2-RdRp-NiRAN domain-filgotinib complex showed stable protein RMSD graph till 50 ns of MD simulation and after that an elevation in the graph was observed. In the case of ligand RMSD up to 38 nanoseconds fluctuations were observed and after that stability in the RMSD was observed till the end of the simulation (Fig. 6C.a). The RMSD mean for backbone, sidechain, and ligands were calculated to be 4.292 ± 1.034 Å, 5.275 ± 1.008 Å and 1.308 ± 0.256 Å respectively.
3.3.1.4. (SARS-CoV-2-RdRp-NiRAN domain/ibrutinib)
2D interactions diagram after MD simulation showed stable residues for more than 20% of simulation time. Fig. 4d shows the interacting residues of the SARS-CoV-2-RdRp-NiRAN domain with the ibrutinib. The main interacting residues were Asp36, Thr296, Asp208, Asp218 and Asp221. All the residues received the H-bond from the ibrutinib and the residues Asp36, Thr296 and Asp208 displayed direct H-bond while Asp218 and Asp22 made H-bond through water molecules.
Histogram of protein-ligand (SARS-CoV-2-RdRp-NiRAN domain with ibrutinib) contacts showed the main interacting residues and bond fractions (Fig. 5d). The main residues of the SARS-CoV-2-RdRp-NiRAN domain showing interaction with the ibrutinib were Asp36, Tyr38, Val204, Thr206, Asp208, Asn209, Asp218 and Asp221. Asp36, Asp208, Asp218 and Asp221 showed strong H-bond interactions, as well as these residues also showed water bridges and ionic interactions. Thr206 and Asn209 also showed H-bond and water bridge interactions. Val204 also showed water bridge and hydrophobic interaction (Fig. 5d).
Total contact analysis of the SARS-CoV-2-RdRp-NiRAN domain showed maximum number of contacts between 50 and 60 ns and rest of the time contacts remained almost similar (Supp. Fig. 6a). Timeline representation of the contacts showed that residues Phe35, Asp36, Thr38, Val204, Thr206, Asn209, Asp218, and Asp221 interacted with ligands throughout the simulation. Out of these, Asp36, Asp208, Asp218 and Asp221 showed strong interactions as compared to other residues (Supp. Fig. 6b).
RMSD plot of the SARS-CoV-2-RdRp-NiRAN domain and ibrutinib is represented in Fig. 6D.a. Protein RMSD became stable after 50 ns of simulation. Similarly, ligand RMSD also became stable after 50 ns of simulation. The RMSD mean for backbone sidechain and ligand were calculated to be 5.118 ± 0.893 Å, 5.966 ± 0.869 Å and 1.497 ± 0.565 Å respectively.
4. Discussion
Kinases play a pivotal role in GC progression. Through structural similarity analyses using DALI, it was believed that the NiRAN domain of SARS-CoV-2-RdRp possesses structural features of kinase-like folds. However, its exact role is still unknown [35]. Hence, repurposing kinase inhibitors of GC for SARS-CoV-2 could aid in designing comorbidity specific therapeutics and reducing drug-induced complications. Furthermore, to the best of our knowledge, this is the first study using kinase inhibitors for GC comorbidity of COVID-19.
There are many available drugs are recommended for the treatment of COVID-19 under drug repurposing strategy. Drugs like lopinavir, darunavir, arabidol, nafamostat and chloroquine inhibit the viral entry by different mechanisms like inhibiting protease, hemagglutinin fusion machinery by preventing membrane fusion, changing endosomal pH etc. [[56], [57], [58], [59], [60], [61], [62], [63]]. Drugs azithromycin and doxycyclin inhibit the IL-6 production while tocilizumab inhibits the release of IL-6 [58,64,65]. Ruxolitinib and baricitinib target the Janus-Kinase pathway and inhibit cytokine release [66,67]. The other repurposing drugs have miscellaneous effects on the viral pathogenesis and inhibit the viral growth by different mechanisms, for instance, ivermectin inhibits the nuclear transport activity, and statins targets the angiotensin-converting enzyme 2 [68,69]. The remdesivir, the most accepted repurposed drug for the SARS-CoV-2, inhibits the RdRp protein, therefore, inhibits the viral replication [70].
In our case, we targeted the SARS-CoV-2- RdRp-NiRAN domain, as this is a very important protein for viral replication. We used gastric cancer kinase inhibitors to target the SARS-CoV-2-RdRp-NiRAN domain because these compounds showed a strong affinity with the active site of the SARS-CoV-2-RdRp-NiRAN domain. Moreover, these inhibitors may also inhibit the kinases involved in viral pathogenesis. The purpose of our study was to identify the inhibitors that can target the SARS-CoV-2-RdRp-NiRAN domain which has a kinase-like domain as well as they can also inhibit the kinases involved in the pathogenesis of SARS-CoV-2 and gastric cancer.
Since these kinases play an important role in GC cells, they become natural therapeutic target that could facilitate the treatment in patients with GC infected with COVID-19. Hence, inhibitors against these kinases were used for the present study. Further, the nsp12/SARS-CoV-2-RdRp contains the NiRAN domain. The NiRAN and the interface domains span over residues 51–398 of the SARS-CoV-2 RdRp polypeptide sequence which is proposed to have kinase-like folds. Moreover, corresponding residues which are important for the enzymatic activity of this domain mostly occur in pocket 3 are Phe35, Asp36, Ile37, Tyr38, Asn39, Phe48, Leu49, Lys50, Thr51, Asn52, Arg55, Val71, Lys73, His75, Glu83, Arg116, Leu119, Thr120, Lys121, Tyr122, Thr123, Asp 126, Val204, Thr206, Asp208, Asn209, Tyr217, Asp218, Phe219, Gly220, Asp221, and Ser236. The residues which are highlighted are considered to be strictly conserved across SARS-CoV-2-RdRp, whereas other were present in the active site of the domain [71].
For the MD simulation studies, four drugs were selected which showed the best binding energy and other docking parameters. These are brepocitinib, decernotinib, filgotinib, and ibrutinib. Brepocitinib is a potent JAK1/TYK 2 inhibitor. JAK pathway, as stated earlier, plays an important role in the intracellular signalling of cytokines for the various intracellular processes, which is deemed to be pivotal in both normal and pathological conditions [72]. Hence the loss of functions of TYK2 can lead to inhibition of the signalling of several cytokines such as IL-6, IL-10, IL-12, and IL-23. These cytokines are crucial for the pathogenesis of autoimmune disease [73]. Brepocitinib, which was formerly known as PF-06700841, is currently under investigation for several autoimmune indications [74]. It is known to directly inhibit TYK2-dependent IL-12, IL-23 signalling and JAK2 dependent signalling, which includes T-cells and keratinocytes [75].
Our docking studies using Glide Schrodinger Suites showed binding of brepocitinib at the active site of the SARS-CoV-2-RdRp-NiRAN domain. It showed strong H-bond interactions with the Asp36, Lys73, Asp218 and Asp221 (Fig. 3a). Post MD simulation 2D interaction diagram showed Thr51, Lys73, Arg116, Asp208, Asn209, Asp218, and Asp221 as main interacting residues that remained stable for more than 20% of simulation time. (Fig. 4a). When compared, residues Lys73, Asp218 and Asp221 were found common in both docking and post MD simulation interactions. This suggests that the common bonds were quite strong and remained stable during the MD simulation. Histogram diagram of protein-ligand (SARS-CoV-2-RdRp-NiRAN domain-brepocitinib) contacts also confirmed the above results along with other types of bond fractions and bonds formed during the simulation (Fig. 5a). RMSD results also suggested that during the simulation the protein and ligand remained stable because of the minimal fluctuations observed in the RMSD values. (Fig. 6A.a).
Decernocitinib, also known as VX‐509, is a selective inhibitor of JAK3. Decernocitinib showed about 25–120 fold higher with JAK3 in comparison to JAK1, JAK2, and TYK2, in cell-based assays [76]. The four JAK’S namely JAK1, JAK2, JAK3, and TYK2 share common subunits that are used by small cytokines like IL-2, IL-4, IL-7, IL-9, IL-15 and IL-21. Loss of function of JAK3 or the common gamma chain can have an immense effect on the immune system without affecting other organs. Decernocitinib is used against a variety of auto-immune diseases [77]. So far, this is the only JAK3 inhibitor known that has been evaluated in the clinical studies of rheumatoid arthritis. It is also reported that decernocitinib reduced T-cell mediated pro-inflammatory response in immune-mediated diseases such as collagen-induced arthritis and delayed-type hypersensitivity [78].
Docking studies showed decernocitinib interacted with the Asn209, Asp218 and Asp221 (Fig. 3b). Post MD simulation 2D interactions results showed strong H-bond interactions of residues Thr206, Asp208, Asn209, Asp218 and Asp221 with decernocitinib. Both the docking and the post MD simulation results showed common residues Asn209, Asp218 and Asp221 that made strong interactions during the simulation process (Fig. 4b). Histogram diagram of protein-ligand (SARS-CoV-2-RdRp-NiRAN domain-decernocitinib) contacts also showed similar results with different bond fractions, mainly H-bond and water bridges (Fig. 5b). Minimum fluctuations in the protein RMSD also suggested the stability of the protein-ligand complex during the simulation (Fig. 6B.a).
Filgotinib is an oral selective JAK1 inhibitor, which was proved effective and safe in the TORTUGA trial for patients suffering from ankylosing spondylitis, rheumatoid arthritis, Crohn's disease, and psoriatic arthritis. Several successful phase-III trials have been completed for Filgotinib against these diseases [79]. Filgotinib inhibits JAK1 for longer duration, this is evident from its pharmacodynamic and pharmacokinetic studies. These studies indicate that filgotinib and its active metabolite contribute to its pharmacodynamic properties [80]. Filgotinib showed approximately 30-fold higher efficacies towards JAK1 than JAK2 in human whole blood assay. Moreover, its metabolite also target JAK2 with lower potency [81].
Our docking results showed that the filgotinib interacts with the Asp40, Asp208 and Asp221 of the SARS-CoV-2-RdRp-NiRAN domain (Fig. 3c). Post MD simulation 2D interaction results showed its main interactions with residues Asp218 and Asp221. This suggested that the Asp221 residue of SARS-CoV-2-RdRp-NiRAN domain showed strongest interaction with nilotinib. (Fig. 4c). Histogram diagram of protein-ligand (SARS-CoV-2-RdRp-NiRAN domain-filgotinib) contact also showed the same results, and it represented the different bond fractions like H-bond, water bridges, and hydrophobic interactions (Fig. 5c). RMSD results suggested that the protein RMSD remained stable from 5 to 55 ns and the ligand RMSD were remained stable from 30 till the end of 100 ns of simulation (Fig. 6C.a). RMSD results also suggested that the fluctuations in the RMSD values were minimal, which suggested the stability of the protein-ligand complex during the simulation.
Ibrutinib is an oral bioavailable potent inhibitor for Bruton's tyrosine kinase (BTK) enzyme. It covalently binds to the 481 amino acid Cysteine inhibits the function of the BTK enzyme. Several preclinical studies have reported that it also inhibits many cellular processes like ERK signalling, NF-κB DNA binding, and tumour cell migration. However, it doesn't affect normal T-cell [82]. Various molecular and phenotypic analyses have confirmed that Ibrutinib also irreversibly inhibit interleukin-2-inducible T-cell kinase (ITK) by targeting T-cell. ITK is a potent therapeutic target that contributes to various pathogenesis, autoimmune and neoplastic diseases. Since there is a significant homology between BTK and ITK, through in-silico studies, Ibrutinib is considered as an immunomodulatory inhibitor of both the kinases [83]. It can also inhibit gastric carcinoma cell growth by targeting BTK as BTK is highly expressed in GC cells and ITK is highly expressed in GC tissues [84,85].
Although ibrutinib is a very effective drug against GC but with chronic lymphoid leukaemia (CLL) it showed an increased risk for second primary malignancies (SPM) [86]. However, in patients suffering from GC, ibrutinib is found to have additive inhibitory effects against Bruton's tyrosine kinase (Btk). Btk is a member of the Tec-family non-receptor tyrosine kinases family. It is over-expressed in gastric carcinoma tissues and gastric cancer cells. Its inhibition by ibrutinib impedes the growth of gastric cancer cells and is involved in effective cell killing mediated by inhibition of Btk. Further, it also induced the apoptosis of gastric carcinoma cells and is involved as a chemo-sensitizer for docetaxel (DTX) which is a standard care for gastric carcinoma patients [84]. Hence, to the best of our knowledge, the side effect is validated only for CLL patients and it has several beneficial effects on GC.
In our study, ibrutinib showed strong H-bond interactions with the SARS-CoV-2-RdRp-NiRAN domain and interacted with the residues Asp208, Asn209, Asp218 and Asp221through H-bonds from ibrutinib (Fig. 3d). Post MD simulation 2D interaction diagram showed the residues that remained stable for more than 20% time of simulation (Fig. 4d). Residues Asp36, Thr206, Asp208, Asp218 and Asp221 showed H-bond interaction during the 100 ns of MD simulation. The common residues obtained before and after simulation suggested that these residues have strong interactions. Histogram diagram of protein-ligand (SARS-CoV-2-RdRp-NiRAN domain-ibrutinib) contact also confirmed the result and the bond fractions were also represented. The most important interactions during the simulation were H-bond, water bridges, and ionic interactions (Fig. 5d). RMSD results of protein-ligand complex (SARS-CoV-2-RdRp-NiRAN domain-ibrutinib) showed that after 40 ns of simulation protein RMSD became stable and similar results were observed with the ligand RMSD (Fig. 6D.a). In both the cases, fluctuations in the RMSD values were minimal suggesting that during the simulation protein and ligand both remained stable.
The docking scores showed strong affinity of brepocitinib, decernocitinib, filgotinib, and ibrutinib with the SARS-CoV-2-RdRp-NiRAN domain (Table 1). The MMGBSA study also supported docking interactions. The docking scores for the brepocitinib, decernocitinib, filgotinib, and ibrutinib were calculated to be −5.543, - 6.694, - 4.917, and - 6.137 respectively (Table 1). In this study, it was observed that all four inhibitory compounds have almost the same affinity towards the SARS-CoV-2-RdRp-NiRAN domain. After docking studies, MMGBSA studies were performed and the binding energies were calculated for all the four compounds and these were found to be −60.1315, −47.6964, −44.1200, and −77.1748, for brepocitinib, decernocitinib, filgotinib, and ibrutinib respectively (Table 2). Besides this, the individual MMGBSA energies like coulomb, covalent, H bond, solvation, and Vander walls were also calculated which also demonstrated strong affinity of the compounds with the SARS-CoV-2-RdRp-NiRAN domain. Thus, MMGBSA study results showed strong affinity of all the compounds with the SARS-CoV-2-RdRp-NiRAN domain.
Nidovirus RdRp like SARS-CoV-2-RdRp contains an N-terminal NiRAN domain that is absent in other viral RdRPs. This domain was found essential for some viruses’ propagation like the equine arteritis virus and SARS-CoV (Lehmann et al., 2015a). Chen et al., 2020 showed the binding of the ADP-Mg2+ with the SARS-CoV-2-RdRp-NiRAN domain (Chen et al., 2020). It was observed that the ADP-Mg2+ binds to the NiRAN domain with the residues K73, E83, R116, D208, N209, G214, D218, F219, and F222.
In our docking and simulation studies, it was observed that brepocitinib binds to residues Asp36, Lys73, Asp218 and Asp221 at the active site before simulation and during simulation brepocitinib interacted with the residues Arg55, Arg116, Asp218 and Asp221. Further, decernocitinib docking studies showed that the main interacting residues were Asn209, Asp218, and Asp221, while during the simulation the main interacting residues were Asp208, Asn209, Asp218, and Asp221. In the case of filgotinib, the main docking interacting residues were Asp40, Asp208 and Asp221, while during simulation the main interacting residues were Asn209, Asp218, Asp221. Similarly, in case of ibrutinib, the main interacting residues in docking were Asp208, Asn209, Asp218, and Asp221, while during the simulation the main interacting residues were Asp36, Asp208, Asp218, and Asp221. These results suggest that the residues, which are involved in the binding of the ADP-Mg2+ with the SARS-CoV-2-RdRp-NiRAN domain, are also common with the residues that are binding with the studied inhibitor. Further, these results showed that all the inhibitory compounds in our study bound to the same binding pocket where ADP-Mg2+binds with the SARS-CoV-2-RdRp-NiRAN domain. These results strongly suggest that the inhibitors used in the study are binding at the active site. Docking scores and MMGBSA and MD simulation results also confirms that the binding is quite strong. Besides this, the surrounding residues were also common before and after simulation in all the four selected inhibitory compounds. These results also suggest that the compounds are binding at their active site, and not leaving the active site during the simulation.
5. Conclusion
Our study proposes the drugs against kinases that can be repurposed for the therapeutic treatment of GC patients infected with COVID-19. We propose nine drugs namely baricitinib, brepocitinib, decernotinib, fasudil, filgotinib, GSK2606414, peficitinib, ruxolitinib, and ibrutinib, which are kinase inhibitors in GC, can target the SARS-CoV-2 nsp-12/RdRp region. These drugs have shown strong binding affinity with the aforementioned SARS-CoV-2 region and eight, except GSK2606414 drugs, had no AMES toxicity. Out of these drugs, we selected four potent drugs: brepocitinib, decernocitinib, filgotinib, and ibrutinib based of their docking score and performed their redocking, MMGBSA, and MD simulation studies and discussed the results. In brief, these four drugs showed very good binding affinities with the SARS-CoV-2-RdRp-NiRAN domain. This also suggested that the SARS-CoV-2-RdRp-NiRAN domain might have similar structure and folds as the kinases. Taken all together, since there is no precise treatment available until now for the COVID-19 patients suffering from GC, drug repurposing is among the current achievable choice, thus these drugs can be validated in-vitro and in-vivo before clinical trials. These drugs can be utilised for GC patients infected with SARS-CoV-2. Although we do consider the fact that further in-vitro and in-vivo studies would be required for drawing more profound conclusions. These drugs have the potential to target potent active site of SARS-CoV-2.
Declaration of competing interest
No potential conflict of interest has been reported by authors.
Acknowledgments
We are grateful to Department of Science and Technology as Ramanujan fellowship grant no. SB/S2/RIN-132/20/5. DST-EMR: EMR/2017/001637. Ms. Charu Sonkar would like to express her sincere gratitude to CSIR, India, for her doctoral fellowship (09/1022(0035)/2017-EMR-I. Dr. Pawan Kumar Doharey is thankful to UGC-New Delhi for supporting him in the form of a Dr. D.S. Kothari Post-Doctoral Fellowship.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2021.104826.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jakhmola S., Indari O., Baral B., Kashyap D., Varshney N., Das A., Chatterjee S., Jha H.C. Comorbidity assessment is essential during COVID-19 treatment. Front. Physiol. 2020;11:984. doi: 10.3389/fphys.2020.00984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weekly Epidemiological Update on COVID-19 - 29 June 2021, (n.d.). https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---29-june-2021 (accessed June 30, 2021).
- 5.Ghazwani M.Y., Bakheit A.H., Hakami A.R., Alkahtani H.M., Almehizia A.A. Virtual screening and molecular docking studies for discovery of potential RNA-dependent RNA polymerase inhibitors. Crystals. 2021;11:471. doi: 10.3390/cryst11050471. [DOI] [Google Scholar]
- 6.Kifle Z.D., Ayele A.G., Enyew E.F. Drug repurposing approach, potential drugs, and novel drug targets for COVID-19 treatment. J. Environ. Public Health. 2021;2021 doi: 10.1155/2021/6631721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J.J., Kopetz S., Vilar E., Shen J.P., Chen K., Maitra A. Relative abundance of SARS-CoV-2 entry genes in the enterocytes of the lower gastrointestinal tract. Genes. 2020;11:645. doi: 10.3390/genes11060645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Apostolou K., Vogli S., Frountzas M., Syllaios A., Tolia M., Papanikolaou I.S., Schizas D. Upper gastrointestinal cancer management in the COVID-19 era: risk of infection, adapted role of endoscopy, and potential treatment algorithm alterations. J. Gastrointest. Canc. 2021;52:407–413. doi: 10.1007/s12029-020-00557-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.S. Martin, C. Kaeuffer, P. Leyendecker, N. Tuzin, Y. Tazi, F. Schaff-Wendling, T. Kleinheny, S. Husson-Wetzel, G. Pamart, J.-M. Limacher, O. Clerc, E. Dicop, J.-E. Kurtz, P. Barthélémy, J. Gantzer, COVID-19 in patients with cancer: a retrospective study of 212 cases from a French SARS-CoV-2 cluster during the first wave of the COVID-19 pandemic, Oncologyn/a (n.d.). 10.1002/onco.13831. [DOI] [PMC free article] [PubMed]
- 10.Moujaess E., Kourie H.R., Ghosn M. Cancer patients and research during COVID-19 pandemic: a systematic review of current evidence. Crit. Rev. Oncol. Hematol. 2020;150:102972. doi: 10.1016/j.critrevonc.2020.102972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thrift A.P., El-Serag H.B. Burden of gastric cancer. Clin. Gastroenterol. Hepatol. 2020;18:534–542. doi: 10.1016/j.cgh.2019.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L., Zhu F., Xie L., Wang C., Wang J., Chen R., Jia P., Guan H.Q., Peng L., Chen Y., Peng P., Zhang P., Chu Q., Shen Q., Wang Y., Xu S.Y., Zhao J.P., Zhou M. Clinical characteristics of COVID-19-infected cancer patients: a retrospective case study in three hospitals within Wuhan, China. Ann. Oncol. 2020;31:894–901. doi: 10.1016/j.annonc.2020.03.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao R., Qiu Y., He J.-S., Tan J.-Y., Li X.-H., Liang J., Shen J., Zhu L.-R., Chen Y., Iacucci M., Ng S.C., Ghosh S., Chen M.-H. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: a systematic review and meta-analysis. The Lancet Gastroenterol. Hepatol. 2020 doi: 10.1016/S2468-1253(20)30126-6. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sonkar C., Kashyap D., Varshney N., Baral B., Jha H.C. Impact of gastrointestinal symptoms in COVID-19: a molecular approach, SN compr. Clin. Med. 2020 doi: 10.1007/s42399-020-00619-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung K.S., Hung I.F., Chan P.P., Lung K.C., Tso E., Liu R., Ng Y.Y., Chu M.Y., Chung T.W., Tam A.R., Yip C.C., Leung K.-H., Fung A.Y.-F., Zhang R.R., Lin Y., Cheng H.M., Zhang A.J., To K.K., Chan K.-H., Yuen K.-Y., Leung W.K. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from the Hong Kong cohort and systematic review and meta-analysis. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.03.065. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snijder E.J., Decroly E., Ziebuhr J. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv. Virus Res. 2016:59–126. doi: 10.1016/bs.aivir.2016.08.008. Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beg M.A., Athar F. Anti-HIV and Anti-HCV drugs are the putative inhibitors of RNA-dependent-RNA polymerase activity of NSP12 of the SARS CoV-2 (COVID-19) Pharm. Pharmacol. Int. J. 2020;8:163–172. [Google Scholar]
- 18.Laha S., Chakraborty J., Das S., Manna S.K., Biswas S., Chatterjee R. Characterizations of SARS-CoV-2 mutational profile, spike protein stability and viral transmission, Infection. Genet. Evol. 2020;85:104445. doi: 10.1016/j.meegid.2020.104445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng Q., Peng R., Yuan B., Zhao J., Wang M., Wang X., Wang Q., Sun Y., Fan Z., Qi J., Gao G.F., Shi Y. Structural and biochemical characterization of the nsp12-nsp7-nsp8 core polymerase complex from SARS-CoV-2. Cell Rep. 2020;31:107774. doi: 10.1016/j.celrep.2020.107774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584:154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 22.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., Maruthi K., Eng E.T., Vatandaslar H., Chait B.T., Kapoor T.M., Darst S.A., Campbell E.A. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell. 2020;182:1560–1573. doi: 10.1016/j.cell.2020.07.033. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sreelatha A., Yee S.S., Lopez V.A., Park B.C., Kinch L., Pilch S., Servage K.A., Zhang J., Jiou J., Karasiewicz M., Łobocka M., Grishin N., Orth K., Kucharczyk R., Pawłowski K., Tomchick D.R., Tagliabracci V.S. Protein AMPylation by an evolutionarily conserved pseudokinase. Cell. 2018;175:809–821. doi: 10.1016/j.cell.2018.08.046. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan L., Ge J., Zheng L., Zhang Y., Gao Y., Wang T., Huang Y., Yang Y., Gao S., Li M., Liu Z., Wang H., Li Y., Chen Y., Guddat L.W., Wang Q., Rao Z., Lou Z. Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis. Cell. 2021;184:184–193. doi: 10.1016/j.cell.2020.11.016. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frieman M., Yount B., Agnihothram S., Page C., Donaldson E., Roberts A., Vogel L., Woodruff B., Scorpio D., Subbarao K., Baric R.S. Molecular determinants of severe acute respiratory syndrome coronavirus pathogenesis and virulence in young and aged mouse models of human disease. J. Virol. 2012;86:884–897. doi: 10.1128/JVI.05957-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slanina H., Madhugiri R., Bylapudi G., Schultheiß K., Karl N., Gulyaeva A., Gorbalenya A.E., Linne U., Ziebuhr J. Coronavirus replication–transcription complex: vital and selective NMPylation of a conserved site in nsp9 by the NiRAN-RdRp subunit. Proc. Natl. Acad. Sci. Unit. States Am. 2021;118 doi: 10.1073/pnas.2022310118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hillen H.S. Structure and function of SARS-CoV-2 polymerase. Curr. Opin. Virol. 2021;48:82–90. doi: 10.1016/j.coviro.2021.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suryawanshi R.K., Koganti R., Agelidis A., Patil C.D., Shukla D. Dysregulation of cell signaling by SARS-CoV-2. Trends Microbiol. 2021;29:224–237. doi: 10.1016/j.tim.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bouhaddou M., Memon D., Meyer B., White K.M., Rezelj V.V., Correa Marrero M., Polacco B.J., Melnyk J.E., Ulferts S., Kaake R.M., Batra J., Richards A.L., Stevenson E., Gordon D.E., Rojc A., Obernier K., Fabius J.M., Soucheray M., Miorin L., Moreno E., Koh C., Tran Q.D., Hardy A., Robinot R., Vallet T., Nilsson-Payant B.E., Hernandez-Armenta C., Dunham A., Weigang S., Knerr J., Modak M., Quintero D., Zhou Y., Dugourd A., Valdeolivas A., Patil T., Li Q., Hüttenhain R., Cakir M., Muralidharan M., Kim M., Jang G., Tutuncuoglu B., Hiatt J., Guo J.Z., Xu J., Bouhaddou S., Mathy C.J.P., Gaulton A., Manners E.J., Félix E., Shi Y., Goff M., Lim J.K., McBride T., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., De wit E., Leach A.R., Kortemme T., Shoichet B., Ott M., Saez-Rodriguez J., tenOever B.R., Mullins R.D., Fischer E.R., Kochs G., Grosse R., García-Sastre A., Vignuzzi M., Johnson J.R., Shokat K.M., Swaney D.L., Beltrao P., Krogan N.J. The global phosphorylation landscape of SARS-CoV-2 infection. Cell. 2020;182:685–712. doi: 10.1016/j.cell.2020.06.034. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weisberg E., Parent A., Yang P.L., Sattler M., Liu Q., Liu Q., Wang J., Meng C., Buhrlage S.J., Gray N., Griffin J.D. Repurposing of kinase inhibitors for treatment of COVID-19. Pharm. Res. (N. Y.) 2020;37 doi: 10.1007/s11095-020-02851-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.N. Krogan, Coronavirus and Cancer Hijack the Same Parts in Human Cells to Spread – and Our Team Identified Existing Cancer Drugs that Could Fight COVID-19, The Conversation. (n.d.). http://theconversation.com/coronavirus-and-cancer-hijack-the-same-parts-in-human-cells-to-spread-and-our-team-identified-existing-cancer-drugs-that-could-fight-covid-19-139955 (accessed September 19, 2020).
- 33.Snijder E.J., Kikkert M., Fang Y. Arterivirus molecular biology and pathogenesis. J. Gen. Virol. 2013;94:2141–2163. doi: 10.1099/vir.0.056341-0. [DOI] [PubMed] [Google Scholar]
- 34.Lehmann K.C., Gulyaeva A., Zevenhoven-Dobbe J.C., Janssen G.M.C., Ruben M., Overkleeft H.S., van Veelen P.A., Samborskiy D.V., Kravchenko A.A., Leontovich A.M., Sidorov I.A., Snijder E.J., Posthuma C.C., Gorbalenya A.E. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015;43:8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells. 2020;9 doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Becker J., Müller-Tidow C., Serve H., Domschke W., Pohle T. Role of receptor tyrosine kinases in gastric cancer: new targets for a selective therapy. World J. Gastroenterol. 2006;12:3297–3305. doi: 10.3748/wjg.v12.i21.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nam H.-J., Im S.-A., Oh D.-Y., Elvin P., Kim H.-P., Yoon Y.-K., Min A., Song S.-H., Han S.-W., Kim T.-Y., Bang Y.-J. Antitumor activity of saracatinib (AZD0530), a c-src/abl kinase inhibitor, alone or in combination with chemotherapeutic agents in gastric cancer. Mol. Canc. Therapeut. 2013;12:16–26. doi: 10.1158/1535-7163.MCT-12-0109. [DOI] [PubMed] [Google Scholar]
- 38.Huang T., Liu D., Wang Y., Li P., Sun L., Xiong H., Dai Y., Zou M., Yuan X., Qiu H. FGFR2 promotes gastric cancer progression by inhibiting the expression of Thrombospondin4 via PI3K-Akt-Mtor pathway. Chem. Pharm. Bull. 2018;50:1332–1345. doi: 10.1159/000494590. [DOI] [PubMed] [Google Scholar]
- 39.Neogi U., Hill K.J., Ambikan A.T., Heng X., Quinn T.P., Byrareddy S.N., Sönnerborg A., Sarafianos S.G., Singh K. Feasibility of known RNA polymerase inhibitors as anti-SARS-CoV-2 drugs. Pathogens. 2020;9:320. doi: 10.3390/pathogens9050320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian W., Chen C., Lei X., Zhao J., Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46:W363–W367. doi: 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shan Y., Klepeis J.L., Eastwood M.P., Dror R.O., Shaw D.E. Gaussian split Ewald: a fast Ewald mesh method for molecular simulation. J. Chem. Phys. 2005;122:54101. doi: 10.1063/1.1839571. [DOI] [PubMed] [Google Scholar]
- 43.Halgren T.A., Murphy R.B., Friesner R.A., Beard H.S., Frye L.L., Pollard W.T., Banks J.L. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 44.Gupta S., Singh V., Varadwaj P.K., Chakravartty N., Katta A.V.S.K.M., Lekkala S.P., Thomas G., Narasimhan S., Reddy A.R., Reddy Lachagari V.B. Secondary metabolites from spice and herbs as potential multitarget inhibitors of SARS-CoV-2 proteins. J. Biomol. Struct. Dyn. 2020:1–20. doi: 10.1080/07391102.2020.1837679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aduri N.G., Montefiori M., Khalil R., Gajhede M., Jørgensen F., Mirza O. Molecular dynamics simulations reveal the proton:peptide coupling mechanism in the bacterial proton-coupled oligopeptide transporter YbgH. ACS Omega. 2019;4:2040–2046. doi: 10.1021/acsomega.8b02131. [DOI] [Google Scholar]
- 46.Chaudhary A., Singh V., Varadwaj P.K., Mani A. Screening natural inhibitors against upregulated G-protein coupled receptors as potential therapeutics of Alzheimer's disease. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1817784. [DOI] [PubMed] [Google Scholar]
- 47.Ziebuhr J. In: Coronavirus Replication and Reverse Genetics. Enjuanes L., editor. Springer Berlin Heidelberg; Berlin, Heidelberg: 2005. The coronavirus replicase; pp. 57–94. [DOI] [Google Scholar]
- 48.Pandeya K.B., Ganeshpurkar A., Mishra M.K. Natural RNA dependent RNA polymerase inhibitors: molecular docking studies of some biologically active alkaloids of Argemone mexicana. Med. Hypotheses. 2020;144:109905. doi: 10.1016/j.mehy.2020.109905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subissi L., Imbert I., Ferron F., Collet A., Coutard B., Decroly E., Canard B. SARS-CoV ORF1b-encoded nonstructural proteins 12–16: replicative enzymes as antiviral targets. Antivir. Res. 2014;101:122–130. doi: 10.1016/j.antiviral.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Costela-Ruiz V.J., Illescas-Montes R., Puerta-Puerta J.M., Ruiz C., Melguizo-Rodríguez L. SARS-CoV-2 infection: the role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020 doi: 10.1016/j.cytogfr.2020.06.001. S135961012030109X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo W., Li Y.-X., Jiang L.-J., Chen Q., Wang T., Ye D.-W. Targeting JAK-STAT signaling to control cytokine release syndrome in COVID-19. Trends Pharmacol. Sci. 2020;41:531–543. doi: 10.1016/j.tips.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.M.C. McGee, A. August, W. Huang, BTK/ITK dual inhibitors: modulating immunopathology and lymphopenia for COVID-19 therapy, J. Leukoc. Biol.. n/a (n.d.). 10.1002/JLB.5COVR0620-306R. [DOI] [PMC free article] [PubMed]
- 53.Tian L., Qiang T., Liang C., Ren X., Jia M., Zhang J., Li J., Wan M., YuWen X., Li H., Cao W., Liu H. RNA-dependent RNA polymerase (RdRp) inhibitors: the current landscape and repurposing for the COVID-19 pandemic. Eur. J. Med. Chem. 2021;213:113201. doi: 10.1016/j.ejmech.2021.113201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carugo O., Pongor S. A normalized root-mean-spuare distance for comparing protein three-dimensional structures. Protein Sci. 2001;10:1470–1473. doi: 10.1110/ps.690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ahmad M., Dwivedy A., Mariadasse R., Tiwari S., Kar D., Jeyakanthan J., Biswal B.K. Prediction of small molecule inhibitors targeting the severe acute respiratory syndrome coronavirus-2 RNA-dependent RNA polymerase. ACS Omega. 2020 doi: 10.1021/acsomega.0c02096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choy K.-T., Wong A.Y.-L., Kaewpreedee P., Sia S.F., Chen D., Hui K.P.Y., Chu D.K.W., Chan M.C.W., Cheung P.P.-H., Huang X., Peiris M., Yen H.-L. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020;178:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu X., Wang X.-J. Potential inhibitors against 2019-nCoV coronavirus M protease from clinically approved medicines. J. Genet. Genom. 2020;47:119–121. doi: 10.1016/j.jgg.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Meyer S., Bojkova D., Cinatl J., Van Damme E., Buyck C., Van Loock M., Woodfall B., Ciesek S. Lack of antiviral activity of darunavir against SARS-CoV-2. Int. J. Infect. Dis. 2020;97:7–10. doi: 10.1016/j.ijid.2020.05.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vankadari N. Arbidol: a potential antiviral drug for the treatment of SARS-CoV-2 by blocking trimerization of the spike glycoprotein. Int. J. Antimicrob. Agents. 2020;56:105998. doi: 10.1016/j.ijantimicag.2020.105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu Z., Lu Z., Xu T., Chen C., Yang G., Zha T., Lu J., Xue Y. Arbidol monotherapy is superior to lopinavir/ritonavir in treating COVID-19. J. Infect. 2020;81:e21–e23. doi: 10.1016/j.jinf.2020.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoffmann M., Schroeder S., Kleine-Weber H., Müller M.A., Drosten C., Pöhlmann S. Nafamostat mesylate blocks activation of SARS-CoV-2: new treatment option for COVID-19. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00754-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jang S., Rhee J.-Y. Three cases of treatment with nafamostat in elderly patients with COVID-19 pneumonia who need oxygen therapy. Int. J. Infect. Dis. 2020;96:500–502. doi: 10.1016/j.ijid.2020.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P., Liu X., Zhao L., Dong E., Song C., Zhan S., Lu R., Li H., Tan W., Liu D. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2020;71:732–739. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sargiacomo C., Sotgia F., Lisanti M.P. COVID-19 and chronological aging: senolytics and other anti-aging drugs for the treatment or prevention of corona virus infection? Aging. 2020;12:6511–6517. doi: 10.18632/aging.103001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu X., Han M., Li T., Sun W., Wang D., Fu B., Zhou Y., Zheng X., Yang Y., Li X., Zhang X., Pan A., Wei H. Effective treatment of severe COVID-19 patients with tocilizumab. Proc. Natl. Acad. Sci. U. S. A. 2020;117:10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cao Y., Wei J., Zou L., Jiang T., Wang G., Chen L., Huang L., Meng F., Huang L., Wang N., Zhou X., Luo H., Mao Z., Chen X., Xie J., Liu J., Cheng H., Zhao J., Huang G., Wang W., Zhou J. Ruxolitinib in treatment of severe coronavirus disease 2019 (COVID-19): a multicenter, single-blind, randomized controlled trial. J. Allergy Clin. Immunol. 2020;146:137–146. doi: 10.1016/j.jaci.2020.05.019. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cantini F., Niccoli L., Nannini C., Matarrese D., Natale M.E.D., Lotti P., Aquilini D., Landini G., Cimolato B., Pietro M.A.D., Trezzi M., Stobbione P., Frausini G., Navarra A., Nicastri E., Sotgiu G., Goletti D. Beneficial impact of Baricitinib in COVID-19 moderate pneumonia; multicentre study. J. Infect. 2020;81:647–679. doi: 10.1016/j.jinf.2020.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wösten-van Asperen R.M., Lutter R., Specht P.A., Moll G.N., van Woensel J.B., van der Loos C.M., van Goor H., Kamilic J., Florquin S., Bos A.P. Acute respiratory distress syndrome leads to reduced ratio of ACE/ACE2 activities and is prevented by angiotensin-(1-7) or an angiotensin II receptor antagonist. J. Pathol. 2011;225:618–627. doi: 10.1002/path.2987. [DOI] [PubMed] [Google Scholar]
- 69.Wehbe Z., Wehbe M., Iratni R., Pintus G., Zaraket H., Yassine H.M., Eid A.H. Repurposing ivermectin for COVID-19: molecular aspects and therapeutic possibilities. Front. Immunol. 2021 doi: 10.3389/fimmu.2021.663586. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Elfiky A.A. Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253:117592. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang W.-F., Stephen P., Thériault J.-F., Wang R., Lin S.-X. Novel coronavirus polymerase and nucleotidyl-transferase structures: potential to target new outbreaks. J. Phys. Chem. Lett. 2020;11:4430–4435. doi: 10.1021/acs.jpclett.0c00571. [DOI] [PubMed] [Google Scholar]
- 72.Nogueira M., Puig L., Torres T. JAK inhibitors for treatment of psoriasis: focus on selective TYK2 inhibitors. Drugs. 2020;80:341–352. doi: 10.1007/s40265-020-01261-8. [DOI] [PubMed] [Google Scholar]
- 73.Minegishi Y., Saito M., Morio T., Watanabe K., Agematsu K., Tsuchiya S., Takada H., Hara T., Kawamura N., Ariga T., Kaneko H., Kondo N., Tsuge I., Yachie A., Sakiyama Y., Iwata T., Bessho F., Ohishi T., Joh K., Imai K., Kogawa K., Shinohara M., Fujieda M., Wakiguchi H., Pasic S., Abinun M., Ochs H.D., Renner E.D., Jansson A., Belohradsky B.H., Metin A., Shimizu N., Mizutani S., Miyawaki T., Nonoyama S., Karasuyama H. Human tyrosine kinase 2 deficiency reveals its requisite roles in multiple cytokine signals involved in innate and acquired immunity. Immunity. 2006;25:745–755. doi: 10.1016/j.immuni.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 74.Gerstenberger B.S., Ambler C., Arnold E.P., Banker M.-E., Brown M.F., Clark J.D., Dermenci A., Dowty M.E., Fensome A., Fish S., Hayward M.M., Hegen M., Hollingshead B.D., Knafels J.D., Lin D.W., Lin T.H., Owen D.R., Saiah E., Sharma R., Vajdos F.F., Xing L., Yang X., Yang X., Wright S.W. Discovery of tyrosine kinase 2 (TYK2) inhibitor (PF-06826647) for the treatment of autoimmune diseases. J. Med. Chem. 2020 doi: 10.1021/acs.jmedchem.0c00948. acs.jmedchem.0c00948. [DOI] [PubMed] [Google Scholar]
- 75.Page K.M., Suarez-Farinas M., Suprun M., Zhang W., Garcet S., Fuentes-Duculan J., Li X., Scaramozza M., Kieras E., Banfield C., Clark J.D., Fensome A., Krueger J.G., Peeva E. Molecular and cellular responses to the TYK2/JAK1 inhibitor PF-06700841 reveal reduction of skin inflammation in plaque psoriasis. J. Invest. Dermatol. 2020;140:1546–1555. doi: 10.1016/j.jid.2019.11.027. e4. [DOI] [PubMed] [Google Scholar]
- 76.Genovese M.C., van Vollenhoven R.F., Pacheco‐Tena C., Zhang Y., Kinnman N. VX-509 (decernotinib), an oral selective JAK-3 inhibitor, in combination with methotrexate in patients with rheumatoid arthritis. Arthritis Rheumatol. 2016;68:46–55. doi: 10.1002/art.39473. [DOI] [PubMed] [Google Scholar]
- 77.Gadina M., Schwartz D.M., O'Shea J.J. Decernotinib: a next-generation jakinib. Arthritis Rheum. 2016;68:31–34. doi: 10.1002/art.39463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mahajan S., Hogan J.K., Shlyakhter D., Oh L., Salituro F.G., Farmer L., Hoock T.C. VX-509 (decernotinib) is a potent and selective Janus kinase 3 inhibitor that attenuates inflammation in animal models of autoimmune disease. J. Pharmacol. Exp. Therapeut. 2015;353:405–414. doi: 10.1124/jpet.114.221176. [DOI] [PubMed] [Google Scholar]
- 79.van der Heijde D., Baraliakos X., Gensler L.S., Maksymowych W.P., Tseluyko V., Nadashkevich O., Abi-Saab W., Tasset C., Meuleners L., Besuyen R., Hendrikx T., Mozaffarian N., Liu K., Greer J.M., Deodhar A., Landewé R. Efficacy and safety of filgotinib, a selective Janus kinase 1 inhibitor, in patients with active ankylosing spondylitis (TORTUGA): results from a randomised, placebo-controlled, phase 2 trial. Lancet. 2018;392:2378–2387. doi: 10.1016/S0140-6736(18)32463-2. [DOI] [PubMed] [Google Scholar]
- 80.Westhovens R., Taylor P.C., Alten R., Pavlova D., Enríquez-Sosa F., Mazur M., Greenwald M., der Aa A.V., Vanhoutte F., Tasset C., Harrison P. Filgotinib (GLPG0634/GS-6034), an oral JAK1 selective inhibitor, is effective in combination with methotrexate (MTX) in patients with active rheumatoid arthritis and insufficient response to MTX: results from a randomised, dose-finding study (Darwin 1) Ann. Rheum. Dis. 2017;76:998–1008. doi: 10.1136/annrheumdis-2016-210104. [DOI] [PubMed] [Google Scholar]
- 81.Vanhoutte F., Mazur M., Voloshyn O., Stanislavchuk M., der Aa A.V., Namour F., Galien R., Meuleners L., van ’t Klooster G. Efficacy, safety, pharmacokinetics, and pharmacodynamics of filgotinib, a selective JAK-1 inhibitor, after short-term treatment of rheumatoid arthritis: results of two randomized phase IIa trials. Arthritis Rheumatol. 2017;69:1949–1959. doi: 10.1002/art.40186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Byrd J.C., Furman R.R., Coutre S.E., Flinn I.W., Burger J.A., Blum K.A., Grant B., Sharman J.P., Coleman M., Wierda W.G., Jones J.A., Zhao W., Heerema N.A., Johnson A.J., Sukbuntherng J., Chang B.Y., Clow F., Hedrick E., Buggy J.J., James D.F., O'Brien S. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N. Engl. J. Med. 2013;369:32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dubovsky J.A., Beckwith K.A., Natarajan G., Woyach J.A., Jaglowski S., Zhong Y., Hessler J.D., Liu T.-M., Chang B.Y., Larkin K.M., Stefanovski M.R., Chappell D.L., Frissora F.W., Smith L.L., Smucker K.A., Flynn J.M., Jones J.A., Andritsos L.A., Maddocks K., Lehman A.M., Furman R., Sharman J., Mishra A., Caligiuri M.A., Satoskar A.R., Buggy J.J., Muthusamy N., Johnson A.J., Byrd J.C. Ibrutinib is an irreversible molecular inhibitor of ITK driving a Th1-selective pressure in T lymphocytes. Blood. 2013;122:2539–2549. doi: 10.1182/blood-2013-06-507947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang J.D., Chen X.Y., Ji K.W., Tao F. Targeting Btk with ibrutinib inhibit gastric carcinoma cells growth. Am. J. Transl. Res. 2016;8:3003–3012. [PMC free article] [PubMed] [Google Scholar]
- 85.Lin W., Kao H.-W., Robinson D., Kung H.-J., Wu C.-W., Chen H.-C. Tyrosine kinases and gastric cancer. Oncogene. 2000;19:5680–5689. doi: 10.1038/sj.onc.1203924. [DOI] [PubMed] [Google Scholar]
- 86.Bond D.A., Huang Y., Fisher J.L., Ruppert A.S., Owen D.H., Bertino E.M., Rogers K.A., Bhat S.A., Grever M.R., Jaglowski S.M., Maddocks K.J., Byrd J.C., Woyach J.A. Second cancer incidence in CLL patients receiving BTK inhibitors. Leukemia. 2020;34:3197–3205. doi: 10.1038/s41375-020-0987-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.