Abstract
The transcription of genes encoding proteins involved in the hepatic synthesis of lipids from glucose is strongly stimulated by carbohydrate feeding. It is now well established that in the liver, glucose is the main activator of the expression of this group of genes, with insulin having only a permissive role. While ADD1/SREBP-1 has been implicated in lipogenic gene expression through temporal association with food intake and ectopic gain-of-function experiments, no genetic evidence for a requirement for this factor in glucose-mediated gene expression has been established. We show here that the transcription of ADD1/SREBP-1c in primary cultures of hepatocytes is controlled positively by insulin and negatively by glucagon and cyclic AMP, establishing a link between this transcription factor and carbohydrate availability. Using adenovirus-mediated transfection of a powerful dominant negative form of ADD1/SREBP-1c in rat hepatocytes, we demonstrate that this factor is absolutely necessary for the stimulation by glucose of l-pyruvate kinase, fatty acid synthase, S14, and acetyl coenzyme A carboxylase gene expression. These results demonstrate that ADD1/SREBP-1c plays a crucial role in mediating the expression of lipogenic genes induced by glucose and insulin.
Feeding rodents with a high-carbohydrate diet induces in liver and adipose tissue the expression of a number of genes coding for proteins involved in energy storage. In the liver, the presence of elevated concentrations of both glucose and insulin is necessary to induce the expression of genes involved in fatty acid synthesis from glucose, namely, the l-pyruvate kinase (L-PK), fatty acid synthase (FAS), acetyl coenzyme A (acetyl-CoA) carboxylase (ACC), and S14 genes (10, 33). However, their roles are different. Glucose stimulates the transcriptional activity of the promoter of these genes. We have shown previously that glucose must be metabolized to have its transcriptional effect and that glucose-6-phosphate is a candidate as the signal metabolite (9, 23, 26). Insulin is not a direct mediator of the transcriptional response but is permissive to glucose action in hepatocytes through the induction of glucokinase gene expression, allowing glucose phosphorylation (5, 26).
Glucose response elements (GlRE) have been identified in the L-PK and S14 promoters. They consist of two copies of a motif related to the consensus binding site 5′-CACGTG-3′ (E box) separated by five nucleotides (33). E boxes found in the L-PK and S14 GlRE bind to a transcription factor of the basic–helix-loop-helix–leucine zipper (b-HLH-LZ) family. Among the transcription factors belonging to this class of proteins, upstream stimulatory factor/major late transcription factor (designated USF) has been implicated as the mediator of glucose action. USF was first found as a protein binding to the adenovirus (Ad) late promoter and subsequently found to bind to many other gene promoters. The tissue distribution of USF is ubiquitous and does not seem to be altered by the nutritional or hormonal status. For the two genes considered (L-PK and S14), binding of USF on the E box of the GlRE has been demonstrated in vitro, exclusive of other transcription factors (33). In hepatoma cells, overexpression of native USF proteins synthesized from expression vectors can act as transactivators of the L-PK promoter via the GlRE (20). However, a role for USF as the factor responsible for the glucose responsiveness of the L-PK and S14 genes is disputed since both supportive and contradictory data have been presented (15, 21). Moreover, to our knowledge, no link has been established between glucose activation of gene expression and a change in the transcriptional activity of USF. The reasons for these conflicting results are not entirely clear but could indicate that a factor other than USF is involved in the glucose responsiveness of glycolytic and lipogenic genes.
We reasoned that this factor might be ADD1/SREBP-1c (adipocyte determination and differentiation factor 1/sterol regulatory element binding protein). ADD1/SREBP-1c belongs to a family of transcription factors originally identified as involved in the regulation of genes by the cellular availability in cholesterol (2). Three members of the SREBP family have been described. SREBP-1a and SREBP-1c are encoded by a single gene through the use of alternative transcription start sites and differ in the first exon (31). In rat adipocytes, SREBP-1c was independently described as a factor involved in adipocyte determination and differentiation and called ADD1 (32). The third member of the family, SREBP-2, is derived from a different gene (2).
SREBPs are bound in a precursor form to the endoplasmic reticulum and nuclear membranes when the membrane concentration of cholesterol is high. When the concentration of cholesterol decreases, the precursor form is cleaved by a complex mechanism involving two proteolytic cleavages and a protein sensor for the cholesterol concentration (2). The mature form migrates inside the nucleus, where it binds to the sterol response element (SRE), 5′-ATCACCCAC-3′, on the promoters of genes involved in cholesterol uptake, such as the low-density lipoprotein receptor gene, or in cholesterol synthesis, such as the cytoplasmic hydroxymethylglutaryl-CoA synthase or hydroxymethylglutaryl-CoA reductase gene (34, 35). SREBPs are b-HLH-LZ transcription factors, but in contrast to other members of the same family, they are able to bind both to SRE and to E boxes due to the presence of a tyrosine in place of an arginine in their basic domain (18).
Various results suggest that ADD1/SREBP-1c, the most abundant form of SREBPs in liver and adipose tissue, also controls the genes involved in fatty acid synthesis. Overexpression of this factor in adipocyte cell lines stimulates the expression of FAS (16, 17). Similarly, its forced expression in transgenic mice led to a dramatically increased expression of hepatic FAS and ACC and to liver steatosis, whereas mRNAs for genes involved in cholesterol synthesis were barely affected (29) and precluded the usual decrease in the mRNAs of lipogenic enzymes observed during starvation (13). Finally, it was recently reported that the expression of ADD1/SREBP-1c itself is controlled positively by insulin in adipose tissue (16) and by the nutritional status in mouse liver (high expression and presence of the mature form of ADD1/SREBP-1c in the nucleus in refed compared to starved animals) (13), thus establishing a link between this transcription factor and the dietary carbohydrate status.
The aims of this study were thus (i) to identify in primary cultures of hepatocytes the factors which regulate the expression of ADD1/SREBP-1c and (ii) to test the hypothesis that ADD1/SREBP-1c is involved in the regulation of glucose-induced genes of the glycolytic/lipogenic pathway. For this purpose, an Ad vector was used, allowing us to express a dominant negative form of ADD1/SREBP-1c in cultured hepatocytes and subsequently monitor the endogenous expression of glucose-induced genes. We show that insulin and glucagon are, respectively, positive and negative transcriptional regulators of ADD1/SREBP-1c expression and that this transcription factor is indeed necessary for the glucose-induced expression of the FAS, S14, ACC, and L-PK genes.
MATERIALS AND METHODS
Animals.
Animal studies were conducted according to French guidelines for the care and use of experimental animals. Female Wistar rats (200 to 300 g [body weight]) from Iffa-Credo, L’Arbresle, France, were used for isolation of hepatocytes. They were housed in plastic cages at a constant temperature (22°C) with light from 0700 to 1900 h for at least 1 week before the experiments.
Preparation of recombinant Ad.
The dominant negative form of ADD1 (ADD1-DN) (18) was subcloned into the EcoRI-linearized pDK6 shuttle vector (7) under the control of the cytomegalovirus immediate-early promoter. Plasmid pDK6 ADD1-DN was cotransfected in 293 cells together with the ClaI-cut DNA of the E1a− Ad vector, Ad gal-nls (25). Recombinant Ad ADD1-DN plaques were detected by PCR amplification of viral DNA using ADD1 primers, and one clone was further amplified in 293 cells. The Ad vector Ad null, for which the expression cassette contains the major late promoter with no exogenous gene, was used as a control (19). Ad β-gal, which contains the Escherichia coli lacZ gene coding for β-galactosidase, driven by the Rous sarcoma virus promoter (19) was also used to determine the efficiency of Ad-mediated gene transfer in cultured hepatocytes. The Ad vectors were propagated in 293 cells, purified by cesium chloride density centrifugation, and stored as previously described (19).
Hepatocyte isolation, culture, and treatment with recombinant Ad.
Hepatocytes were isolated from fed rats by the collagenase method (1). Cell viability was assessed by the trypan blue exclusion test and was always higher than 85%. Hepatocytes were seeded at a density of 8 × 106 cells/dish in 100-mm-diameter petri dishes in medium 199 (M199) with Earle’s salts (Gibco/BRL, Paisley, United Kingdom) supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml), 0.1% (wt/vol) bovine serum albumin, 2% (vol/vol) Ultroser G (Gibco/BRL), 100 nM dexamethasone (Sigma, St. Louis, Mo.), 1 nM insulin (Actrapid; Novo Nordisk, Copenhagen, Denmark), and 100 nM triiodothyronine (T3; Sigma). After cell attachment (4 h), the medium was replaced by a medium similar to the plating medium but free of Ultroser G and albumin and the cells were cultured in various conditions as described in the figure legends. In the experiments in which gene expression was induced by a high glucose concentration, hepatocytes were first cultured for 16 h in the presence of 100 nM dexamethasone, 100 nM insulin, and 100 nM T3 and a nonstimulatory glucose concentration (5 mM) in order to induce the expression of glucokinase. Then a high (25 mM) glucose concentration was used.
For the experiments involving Ad, hepatocytes were cultured for 16 h after cell attachment in the presence of hormones and 5 mM glucose (glucokinase induction). Then, hepatocytes were incubated for 120 min at 37°C in M199 with hormones either without Ad (control incubation) or with various titers of Ad null, Ad β-gal, and Ad ADD1-DN ranging from 0.3 to 30 PFU/cell. The medium was then replaced by fresh medium containing hormones and either 5 or 25 mM glucose.
Staining techniques.
To detect the presence of lipid droplets, hepatocytes were fixed at the end of the culture period by 10% formaldehyde in phosphate-buffered saline and stained with oil red O as described elsewhere (11). To assess the efficiency of gene transfer when the Ad vector was used, hepatocytes were cultured for 48 h after a 2-h treatment with Ad β-gal (30 PFU/cell). At the end of the culture, expression of the β-galactosidase gene was demonstrated by incubating fixed hepatocytes at 37°C for 30 min in a chromogenic solution containing 35 mM K3Fe(CN)6-K4Fe(CN)6, 2 mM MgCl2, and 1 mg of 5-bromo-4-chloro-2-indolyl-β-d-galactopyranoside per ml. The efficiency of gene transfer was determined by counting clear and blue cells.
Glucose-6-phosphate assay.
The intracellular concentration of glucose-6-phosphate in cultured hepatocytes was measured by previously described spectrophotometric assay (23). Results are expressed as means ± standard errors of the means for two different cultures, each set up in triplicate.
Isolation of total RNA and Northern blot hybridization.
Total cellular RNAs were extracted from cultured hepatocytes by using guanidine thiocyanate (3) and prepared for Northern blot hybridization as previously described (4). Labeling of each DNA probe with [α-32P]dCTP was performed by random priming (Rediprime labeling kit; Amersham, Little Chalfont, United Kingdom). Autoradiograms of Northern blots were scanned and quantified with an image processor program. FAS and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNAs were as previously described (4, 6). Albumin and S14 cDNAs were kind gifts from J. L. Danan (Meudon, France) and R. Plannels (Marseille, France), respectively. ADD1/SREBP-1c cDNA is as previously described (16). Rat angiotensinogen (AGE) cDNA was purchased from the American Type Culture Collection (Rockville, Md.). cDNA probes for rat ACC, L-PK, and apolipoprotein AI (ApoAI) were prepared by reverse transcriptase-mediated PCR from rat liver total RNA. The PCR primers used were as follows: for ACC, upper primer 5′-TGAAGGCTGTGGTGATGGAT-3′ and lower primer 5′-CCGTAGTGGTTGAGGTTGGA-3′; for ApoAI, upper primer 5′-CCGGAATTCATGAAAGCTGCAGTGTTGGCTGTG-3′ and lower primer 5′-GCTCTAGACTCTAGGTTGCCCAGAACTCCTGAGT-3′; for L-PK, upper primer 5′-ATGGAAGGGCCAGCGGGATACCTTCGA-3′ and lower primer 5′-GGATACGCTCAGCACCCGCATGATGTT-3′.
Nuclear run-on transcription assay.
Nucleus isolation and nuclear run-on transcription experiments were performed as described by Dugail et al. (6).
Gel mobility shift assay.
The full-length cDNA encoding the 43-kDa human USF (12) was a kind gift from P. Pognonec (Nice, France). USF cDNA subcloned into pBluescript KS+ and ADD1-403 (18) and ADD1-403-DN cloned into pSVSport1 were translated by using the TNT SP6 (ADD1) or T7 (USF) coupled reticulocyte lysate system (Promega). The DNA probe used was double-stranded oligonucleotides derived from the wild-type sequence found between −95 and −53 relative to the transcription start site in the FAS promoter sequence 5′-GCCTGGGCGGCGCAGCCAAGCTGTCAGCCCATGTGGCGTGGC-3′, containing an E-box-like sequence (underlined). Only the upper strand is shown. Oligonucleotides were end labeled by using [γ-32P]ATP and T4 polynucleotide kinase and gel purified. For binding assays, reactions were performed for 30 min at 37°C in a 20-μl volume containing 20 mM Tris HCl (pH 7.5), 5% glycerol, 0.02% Nonidet P-40, 100 mM NaCl, 1 mM dithiothreitol, 5 mM MgCl2, 1 μg of poly(dI/dC), and 50 to 100 fmol (typically 30,000 cpm) of 32P-labeled probe. Similar amounts of ADD1-403 and USF proteins (estimated by [35S]methionine incorporation performed in separate reactions) and increasing amounts of ADD1-DN (1 to 6 μl of reticulocyte lysate) were added to the binding reaction.
RESULTS
The cDNA probe used recognizes both the SREBP-1c and SREBP-1a isoforms. In rodent liver, the expression of ADD1/SREBP-1c predominates over that of SREBP-1a by a 9:1 ratio (31), and SREBP-1a mRNA abundance is usually quantified by RNase protection. Moreover the expression of SREBP-1a is not affected by a fasting-refeeding experiment (13). We have thus considered that the signal obtained in Northern blots was representative of ADD1/SREBP-1c expression.
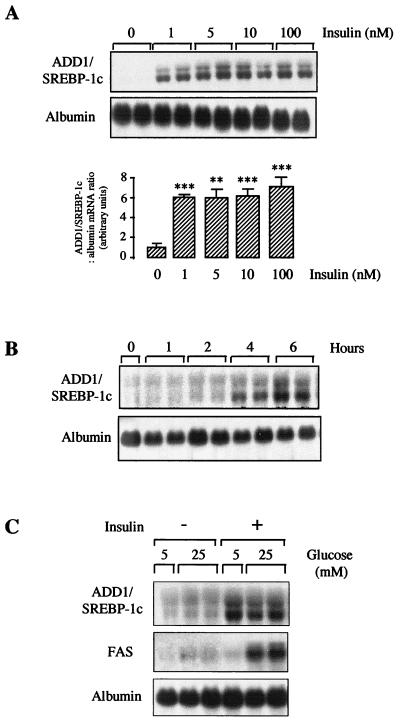
In rats, as previously described for mice (13), a 24-hour starvation period induces a nearly total disappearance of ADD1/SREBP-1c mRNA whereas refeeding with a high-carbohydrate diet induces its strong appearance (results not shown). To identify the factors (hormones and substrates) which control ADD1/SREBP-1c gene expression, we conducted a series of experiments with primary cultures of rat hepatocytes. Since in vivo a high-carbohydrate diet induces ADD1/SREBP-1c gene expression, obvious candidates were insulin, a hormone involved in metabolic adaptations in the fed state, and glucose itself. Hepatocytes cultured for 16 h in the absence of insulin did not express ADD1/SREBP-1c mRNA. Addition of insulin led in 6 h to a clear-cut increase in its expression, with a nearly maximal effect for an insulin concentration of 1 nM (Fig. 1A). This finding strongly suggests that insulin acts through its own receptor to stimulate SREBP-1c gene expression. A time course performed at the highest dose of insulin indicated that between 2 and 4 h after insulin addition, SREBP-1c gene expression was increased (Fig. 1B). To test a potential effect of glucose, hepatocytes were cultured for 16 h in the absence or presence of insulin and in the presence of a basal glucose concentration (5 mM). Switching the medium glucose concentration from a basal (5 mM) to a high (25 mM) level did not increase in 6 h SREBP-1c expression in the absence or in the presence of insulin (Fig. 1C), whereas a high glucose concentration had the usual stimulating effect on FAS gene expression in the presence of insulin. Thus, insulin but not glucose is likely responsible for the stimulating effect of a high-carbohydrate diet observed in vivo on ADD1/SREBP-1c expression.
FIG. 1.
Effects of insulin and glucose on ADD1/SREBP-1c gene expression in cultured hepatocytes. (A) Hepatocytes were cultured for 16 h in the presence of 5 mM glucose and then incubated for 6 h with or without insulin at a concentration of 1 to 100 nM in the presence of 5 mM glucose. A representative Northern blot as well as the quantification of blots obtained in three independent experiments is shown. (B) Hepatocytes were cultured for 16 h in the presence of 5 mM glucose and then incubated with 100 nM insulin for 1 to 6 h. (C) Hepatocytes were cultured for 16 h in the presence of 5 mM glucose and then incubated for 6 h with 5 or 25 mM glucose in the absence or presence of 100 nM insulin. Total RNAs were extracted and analyzed for the concentration of ADD1/SREBP-1c, FAS, and albumin mRNAs. The Northern blots are representative of three culture experiments. Statistical significance was analyzed by Student’s t test for unpaired data. When quantified, the concentrations of ADD1/SREBP-1c mRNA are expressed as a ratio to the corresponding albumin signal. ∗∗ and ∗∗∗, difference statistically significant for P < 0.01 and P < 0.001, respectively.
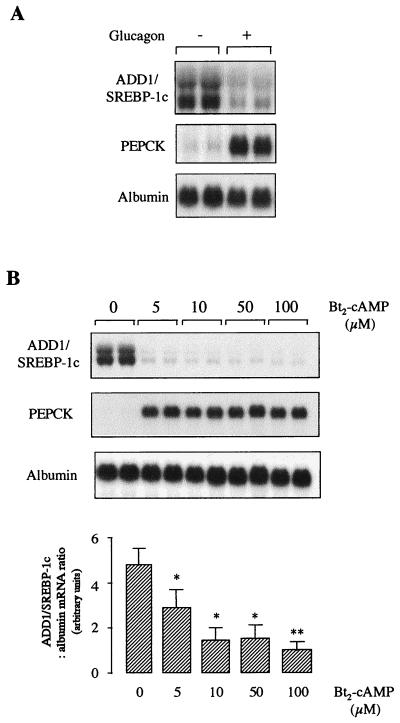
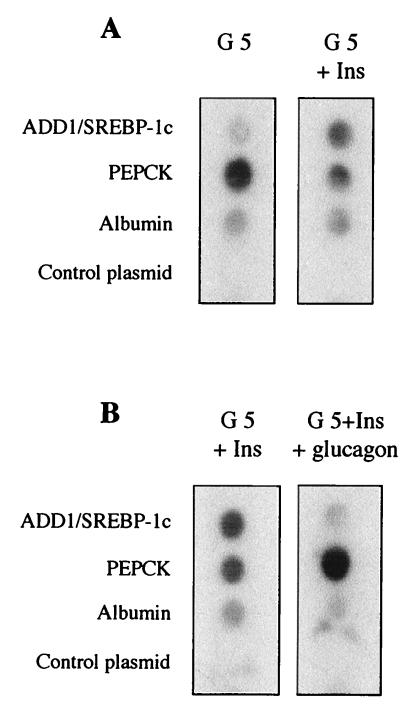
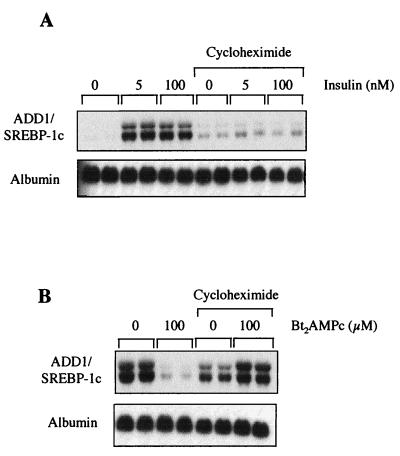
Glucagon inhibits the insulin effect on many hepatic processes, including expression of specific genes through the generation of cyclic AMP (cAMP) and the activation of protein kinase A. For instance, expression of the phosphoenolpyruvate carboxykinase (PEPCK) gene is inhibited by insulin and activated by glucagon (27). We therefore tested the effect of glucagon on insulin-induced ADD1/SREBP-1c expression. Hepatocytes were cultured for 16 h in the presence of 5 mM glucose and 100 nM insulin and then treated for a further 24 h in the absence or presence of 1 μM glucagon. As shown in Fig. 2A, ADD1/SREBP-1c expression is dramatically decreased by glucagon, whereas expression of the PEPCK gene, taken as a positive control, is strongly induced. The same effect is observed with dibutyryl-cAMP (Fig. 2B): in 6 h, 5 μM dibutyryl-cAMP is sufficient to profoundly inhibit ADD1/SREBP-1c gene expression and to markedly stimulate PEPCK gene expression. These results show that glucagon, through the production of cAMP, inhibits the insulin effect on ADD1/SREBP-1c expression. Run-on experiments were then performed with nuclei isolated from hepatocytes cultured for 1 h in the presence of insulin or glucagon (after previous insulin induction). The PEPCK gene was taken as a control. As shown previously, insulin decreases and glucagon increases the PEPCK gene transcription rate (Fig. 3) (27). On the same nuclei, insulin increases (Fig. 3A) and glucagon decreases (Fig. 3B) the ADD1/SREBP-1c transcription rate. This finding demonstrates that the hormonal effects described above involve a transcriptional mechanism, although we cannot exclude the possibility that the ADD1/SREBP-1c mRNA half-life is also affected by the hormones. To assess whether the effects of insulin and dibutyryl-cAMP on ADD1/SREBP-1c gene expression were dependent on ongoing protein synthesis, experiments were performed in the presence of cycloheximide. As shown in Fig. 4, inhibition of protein synthesis precluded the respective activatory and inhibitory effects of insulin and glucagon on ADD1/SREBP-1c gene expression, whereas the basal expression of albumin was not affected. This result suggests that ongoing protein synthesis is necessary for the hormone actions.
FIG. 2.
Effect of glucagon and dibutyryl-cAMP on ADD1/SREBP-1c gene expression in cultured hepatocytes. (A) Hepatocytes were cultured for 16 h in the presence of 5 mM glucose and 100 nM insulin. Cells were then incubated for 24 h with 5 mM glucose and 100 nM insulin in the absence or presence of 1 μM glucagon. (B) Hepatocytes were cultured for 16 h in the presence of 5 mM glucose and 100 nM insulin and then incubated for 6 h with 5 mM glucose and 100 nM insulin in the absence or presence of increasing concentrations of 5 to 100 μM dibutyryl-cAMP (Bt2-cAMP). Total RNAs were extracted and analyzed for the expression of ADD1/SREBP-1c, PEPCK, and albumin mRNAs. A representative Northern blot as well as the quantification of blots obtained in three independent experiments is shown. When quantified, the concentrations of ADD1/SREBP-1c mRNA are expressed as a ratio to the corresponding albumin signal. ∗ and ∗∗, difference statistically significant for P < 0.05 and P < 0.01, respectively.
FIG. 3.
Effects of insulin and glucagon on ADD1/SREBP-1c gene transcription in cultured hepatocytes. (A) Hepatocytes were cultured for 16 h in the presence of 5 mM glucose (G 5) and then incubated for 1 h with 5 mM glucose in the presence or absence of 100 nM insulin (Ins). (B) Hepatocytes were cultured for 16 h in the presence of 5 mM glucose and 100 nM insulin and then incubated for 1 h in the presence of 5 mM glucose and 100 nM insulin with or without 1 μM glucagon. Nuclei were isolated from cultured cells as described in Materials and Methods. The radiolabeled transcripts were hybridized to dot-blotted cDNAs for ADD1/SREBP-1c, PEPCK, and albumin and to pSV Sport1, the plasmid into which ADD1/SREBP-1c cDNA was cloned. The blot is representative of two experiments.
FIG. 4.
Effects of insulin and dibutyryl-cAMP on ADD1/SREBP-1c gene expression in the presence of cycloheximide. (A) Hepatocytes were cultured for 16 h in the presence of 5 mM glucose and then incubated 15 min in the presence of 5 μM cycloheximide before addition of 100 nM insulin for 6 h. (B) Hepatocytes were cultured for 16 h in the presence of 5 mM glucose and insulin and then incubated for 15 min in the presence of cycloheximide before addition of 100 μM dibutyryl-cAMP (Bt2AMPc) for 6 h. The Northern blot presented is representative of two experiments.
Changes in the insulin/glucagon ratio are thus powerful regulators of ADD1/SREBP-1c expression and hence of genes which depend on the presence of ADD1/SREBP-1c for transcriptional activation. However, it is also clear from previous results and those of the present experiment (Fig. 1C) that the presence of insulin and thus the expression of ADD1/SREBP-1c are not sufficient by themselves to induce the expression of lipogenic genes such as the FAS gene in hepatocytes.
To test the hypothesis that the mature form of ADD1/SREBP-1c is involved in the glucose activation of glycolytic and lipogenic genes, we used a strategy involving the expression in cultured hepatocytes of a dominant negative form of ADD1/SREBP-1c. The dominant negative form of ADD1/SREBP-1c consists of the amino-terminal fragment of ADD1/SREBP-1c (ADD1-403), for which the wild type has a maximal transcriptional activity, but containing an alanine mutation at amino acid 320 (17). This mutation abolishes the binding of ADD1-403 to both SRE and E boxes but still allows dimerization, leading to a decreased availability of endogenous ADD1/SREBP-1c. The efficiency of this dominant negative form to counteract the transcriptional activity of ADD1/SREBP-1c has previously been established in transient transfections in cell lines (17).
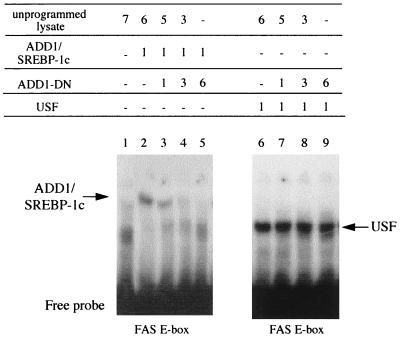
Since ADD1/SREBP-1c, like USF, is a b-HLH-LZ protein, it was important to demonstrate first that ADD1-DN was not able to dimerize with USF, thus impeding USF binding on an E box. Figure 5 shows results of a gel shift experiment using an oligonucleotide probe containing the E-box sequence of the proximal promoter of the FAS gene. Both ADD1/SREBP-1c and USF are able to bind to this oligonucleotide, as shown by the retarded complex observed in both cases compared to the nonprogrammed reticulocyte lysate. Increasing amounts of ADD1-DN in the presence of ADD1/SREBP-1c are able to decrease the retarded complex as previously demonstrated (18), whereas no effect was observed in the presence of USF. This shows that ADD1-DN does not interfere with USF binding.
FIG. 5.
Effect of the dominant negative form of ADD1/SREBP-1c on the binding of ADD1-403 and USF on the E box of the FAS gene in a gel mobility shift assay. Similar amounts of in vitro-translated ADD1/SREBP-1c (lanes 2 to 5) and human USF 1 (lanes 6 to 9) were added to labeled FAS oligonucleotide in the absence (lanes 2 and 6) or presence (lanes 3 to 5 and 7 to 9) of increasing amounts (microliters) of in vitro-translated ADD1-DN.
ADD1-DN was incorporated into an Ad vector in order to transduce primary cultures of hepatocytes. In preliminary experiments, in which the same Ad containing a β-galactosidase gene rather than ADD1-DN was used, more than 90% of the hepatocytes expressed β-galactosidase at an infection titer of 30 PFU/cell (results not shown).
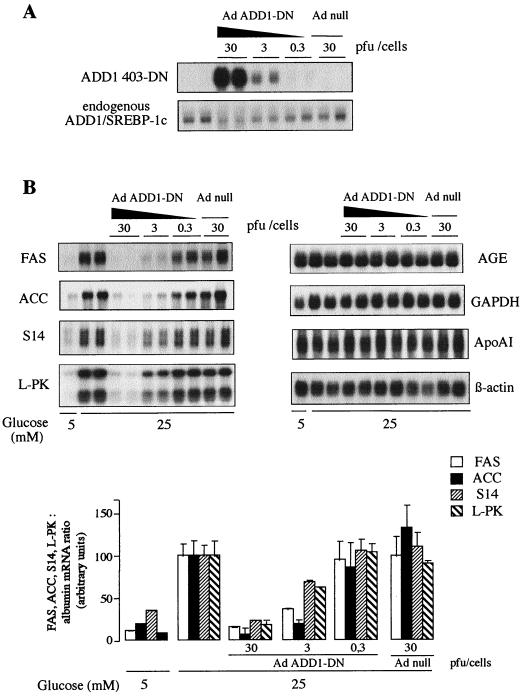
As shown in Fig. 6A, transduction of cultured hepatocytes with increasing titers of Ad led to a proportional expression of ADD1-DN. Expression of the endogenous wild-type ADD1/SREBP-1c was slightly decreased in the presence of the highest viral titer. Transduction of hepatocytes with an Ad which does not contain the ADD1-DN gene had no effect by itself (Fig. 6A). It must be pointed out that it is not possible to show in gel shift experiments that the binding of ADD1/SREBP-1c to its site is decreased when nuclear extracts from the infected cells are used, since the abundance of ADD1/SREBP-1 is too low to be detected.
FIG. 6.
Effect of a dominant negative form of ADD1/SREBP-1c on glycolytic and lipogenic gene expression in cultured rat hepatocytes. Hepatocytes were cultured for 16 h in the presence of 5 mM glucose, 100 nM dexamethasone, 100 nM T3, and 100 nM insulin. Hepatocytes were then incubated for 120 min with hormones either without Ad (control incubation) or with Ad null (30 PFU/cell) or Ad ADD1-DN (0.3, 3, and 30 PFU/cell). The medium was replaced by fresh medium containing hormones and 5 or 25 mM glucose. After 24 h, total RNAs were extracted and analyzed for the expression of ADD1-DN and endogenous ADD1/SREBP-1c expression (A) and for the expression of FAS, ACC, S14, L-PK, AGE, GAPDH, ApoAI, and β-actin (B). A representative Northern blot as well as the quantification of blots for FAS, ACC, S14, and L-PK expression, obtained in two independent experiments, is shown. When quantified, the concentrations of mRNAs are expressed as a ratio to the corresponding albumin signal and as a percentage of the value obtained in the presence of 25 mM glucose.
We then used the Ad-mediated ADD1-DN expression to test in cultured hepatocytes the specific involvement of the ADD1/SREBP-1c transcription factor in the control by glucose of glycolytic and lipogenic gene expression.
As shown in Fig. 6B, switching the medium glucose concentration from a basal (5 mM) to a high (25 mM) concentration induces in 24 h the expression of FAS, ACC, S14, and L-PK genes as previously described, whereas it has no effect on control genes such as the AGE, GAPDH, ApoAI, or β-actin gene. Expression of the dominant negative form of ADD1/SREBP-1c in a dose-dependent fashion prevents in a proportional manner the effect of glucose on FAS, ACC, S14, and L-PK gene expression but has no effect on control genes (Fig. 6B). Transduction of hepatocytes with an Ad which does not contain the ADD1-DN gene has no effect (Fig. 6B).
Since the effect of glucose on gene expression is dependent on its phosphorylation by glucokinase, we also checked that the expression of ADD1-DN was not affecting this step. Switching the glucose medium from 5 to 25 mM induced after 2 h an increase in glucose-6-phosphate concentration from 0.39 ± 0.01 to 1.89 ± 0.01 nmol/106 hepatocytes in control hepatocytes. In hepatocytes transduced previously with ADD1-DN and cultured for 24 h in the presence of 5 mM glucose, a switch for 2 h to 25 mM glucose led to a similar increase in glucose-6-phosphate concentration: 2.34 ± 0.16 nmol/106 hepatocytes (difference not significant).
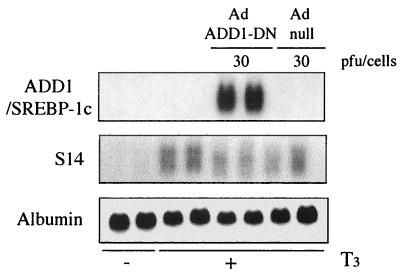
To confirm that ADD1/SREBP-1c is involved in the glucose-mediated activation of gene expression and does not impair a general transcription mechanism, we took advantage of the fact that S14 gene expression can be induced independently by glucose and by T3, the two effects being additive (22). In hepatocytes cultured in a low-glucose (5 mM) medium and in the absence of T3, S14 gene expression was barely detectable (Fig. 7). Addition of T3 led to a clear-cut induction of S14 gene expression. Interestingly, this effect of T3 was not affected by the expression of ADD1-DN (Fig. 7) at a dose which totally prevented the glucose effect (Fig. 6B).
FIG. 7.
Effect of a dominant negative form of ADD1/SREBP-1c on T3-induced S14 expression in cultured rat hepatocytes. Hepatocytes were cultured for 16 h in the presence of 5 mM glucose and then incubated for 120 min at 37°C in M199–5 mM glucose either without Ad (control incubation) or with Ad null (30 PFU/cell) or Ad ADD1-DN (30 PFU/cell). The medium was replaced by fresh medium containing 100 nM T3. After 24 h, total RNAs were extracted and analyzed for the expression of S14 and albumin. A representative Northern blot of two culture experiments is shown.
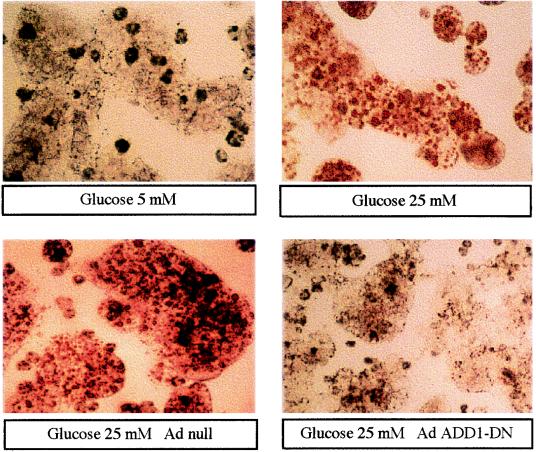
Finally, we investigated whether the inhibition of ADD1/SREBP-1c had functional effects in terms of lipid synthesis in hepatocytes. Culture for 48 h in the presence of 5 mM glucose does not lead to lipid accumulation in hepatocytes (Fig. 8). In contrast, in the presence of 25 mM glucose, red-colored lipid droplets are clearly visible in hepatocytes. Transduction of hepatocytes with an Ad which does not contain the ADD1-DN gene has no effect on lipid accumulation in the presence of 25 mM glucose whereas lipid accumulation is prevented by the expression of ADD1-DN. This finding demonstrates that the absence of a functional ADD1/SREBP-1c totally precludes lipid synthesis even in the presence of a high glucose concentration.
FIG. 8.
Effect of a dominant negative form of ADD1/SREBP-1c on lipid accumulation in cultured rat hepatocytes. Hepatocytes were cultured for 16 h in the presence of 5 mM glucose, 100 nM dexamethasone, 100 nM T3, and 100 nM insulin and then incubated for 120 min at 37°C in M199–5 mM glucose and the same hormones either without Ad (control incubation) or with Ad null (30 PFU/cell) or Ad ADD1-DN (30 PFU/cell). The medium was then replaced by fresh medium containing 5 or 25 mM glucose. After 48 h, hepatocytes were stained for the presence of lipid droplets. Microscopic views of cells at a magnification of ×60 are shown.
DISCUSSION
Adaptation of energy metabolism to the nutritional environment is essential for survival. The liver has a central role in these metabolic adaptations: it is the main organ which produces glucose when this substrate is not provided by the diet, and in a period of glucose availability, it will synthesize glycogen or lipids, the latter being ultimately stored as triglycerides in adipose tissue. Regulation of liver metabolism by a high-carbohydrate diet involves both posttranslational and transcriptional mechanisms. Genes coding for proteins involved in the synthesis of fatty acids from glucose such as L-PK, FAS, ACC, and S14 are among those which are particularly responsive in the liver to feeding large amounts of carbohydrates. Insulin has long been considered the major regulator of their expression. More recently, a specific transcriptional role for glucose has been recognized, with insulin having only a permissive role. However, nuclear factors allowing establishment of a link between glucose and the transcription processes were largely unknown (10, 33).
ADD1/SREBP-1c has been recently proposed as a factor essential to the expression of lipogenic enzymes; among the various isoforms of SREBPs, ADD1/SREBP-1c is expressed primarily in the liver and adipose tissue of rodents fed a normal chow diet (31), whereas the expression of SREBP-2, which controls the expression of genes involved in cholesterol synthesis (14), is increased mainly in conditions of strong cellular cholesterol depletion (28) and SREBP-1a is expressed primarily in cell lines (31). In the liver, the expression of ADD1/SREBP-1c and the expression of lipogenic enzymes in starve/refeed cycles follow the same pattern. The forced expression of the mature form of ADD1/SREBP-1c in both an adipocyte cell line and livers of transgenic mice stimulates the transcriptional activation of genes involved in lipid synthesis (16, 17, 29). However, loss-of-function experiments using knockouts of the SREBP-1 gene in mice were less informative: most mice died, and the expression of lipogenic enzymes in the survivors was normal or only slightly decreased (30).
We have now analyzed the role of ADD1/SREBP-1c, taking advantage of a very effective dominant negative form which could be expressed in primary cultured hepatocytes by using an Ad vector, thus allowing us to study endogenous rather than transfected promoter activity. We demonstrate that ADD1/SREBP-1c is essential for the induction by glucose of genes of fatty acid metabolism and that in its absence, lipid synthesis in the liver is dramatically blunted. This result establishes for the first time a link between a transcription factor and the well-known glucose effect on lipogenic enzyme gene expression. Insulin has a permissive role in the system by stimulating the transcription of ADD1/SREBP-1c in the liver. During carbohydrate feeding, this effect would be favored by the decreased glucagon concentration since the transcription of ADD1/SREBP-1c is down-regulated by this hormone. This hormonal regulation explains why ADD1/SREBP-1c expression is low in the livers of starved rodents but greatly increased after a carbohydrate refeeding (13).
The presence of insulin is not sufficient to induce the expression of these genes, which raises an intriguing question: what is the specific role of glucose in ADD1/SREBP-1c activation? As for other members of the SREBP family, ADD1/SREBP-1c is synthesized as a precursor form bound in the membranes of endoplasmic reticulum and nuclear membranes. The precursor forms of SREBP-2 and SREBP-1a are then cleaved by specific proteases in case of cholesterol cellular depletion, and the mature form is translocated into the nucleus (2). As noted previously, ADD1/SREBP-1c cleavage may be regulated by different signals linked to the nutritional environment (13, 16). The finding that refeeding starved animals with a carbohydrate-rich diet induces a large increase in the amount of mature ADD1/SREBP-1c in the liver nuclei (13) suggests that glucose can activate the proteolytic cleavage of SREBP-1c and increase its nuclear availability. Alternatively, ADD1/SREBP-1c might be constitutively cleaved or cleaved in the presence of insulin and transferred into the nucleus, but its full transcriptional activity would require a further activation linked in an unknown way to glucose metabolism. Along this line, we have recently suggested that the activation of glycolytic and lipogenic gene expression by glucose requires a dephosphorylation process (8).
The next question which arises is the identity of the DNA response element by which ADD1/SREBP-1c could control the expression of these genes. Since one of their common features is a responsiveness to glucose, one obvious possibility is the involvement of the GlRE itself. For the two genes in which it has been clearly identified, L-PK and S14, the GlRE consists of two motifs related to E boxes (33). ADD1/SREBP-1c is indeed able to bind to E boxes (18). Furthermore, in NIH 3T3 cells, cotransfection of an ADD1/SREBP-1c expression vector stimulated transcription of a chloramphenicol acetyltransferase reporter gene linked to four copies of the S14 GlRE (18). Although it has never been demonstrated that the L-PK gene promoter is transactivated by SREBP, its GlRE is very similar to that described for the S14 gene, and we have verified in gel retardation experiments that an oligonucleotide corresponding to this GlRE is able to bind the purified active form of ADD1/SREBP-1c (results not shown). For the FAS gene, a GlRE has not yet been identified. However, in its proximal promoter an insulin response element involving an E box at −64 to −59 has been identified in adipocyte cell lines (24). Since these cells were cultured in the presence of glucose, it cannot be ruled out that the insulin effect on the FAS gene promoter is in fact secondary to a stimulation of glucose metabolism as described for adipose tissue (9). ADD1/SREBP-1c was identified as the factor involved in insulin responsiveness through its binding to this E box (16), emphasizing its potential role in the control of FAS expression by nutritional conditions.
Although ADD1/SREBP-1c and SREBP-2 can bind to both SRE and the E box, ADD1/SREBP-1c would control the expression of genes involved in lipid synthesis through the presence of an E box whereas SREBP-2 controls the genes involved in cholesterol metabolism through the presence of an SRE. The reasons for this difference remain to be elucidated.
In conclusion, we propose the following scheme to explain the large increase in the expression of glycolytic and lipogenic gene expression in the liver after carbohydrate feeding. The high concentrations of insulin and glucose in the portal vein cooperate in a subtle interplay. Insulin stimulates the transcription of two factors, glucokinase and ADD1/SREBP-1c, necessary for the effect of glucose on gene expression. Glucokinase in the presence of a high glucose concentration then allows the generation of a signal metabolite for which the ultimate target would be the activation of ADD1/SREBP-1c, either through increased proteolytic cleavage or through activation of the mature form.
Finally, we point out that the use of Ad-mediated expression of the dominant negative form of ADD1/SREBP-1c in primary cultures of hepatocytes is a valuable tool for identifying other genes which might be regulated by this transcription factor and/or by glucose.
ACKNOWLEDGMENTS
We thank Jack Gauldie (McMaster University) for the generous gift of PDK6 and Marc Eloit (URA INRA, ENVA, Maisons-Alfort, France) for Ad βgal-nls.
Marc Foretz is a recipient of a doctoral fellowship from the Ministère de l’Enseignement Supérieur et de la Recherche. Pascal Ferré is financed by the Centre National de la Recherche Scientifique. This work was supported in part by the grant 97/3011 from the EEC Fair program, by the Institut Necker, and by the Association Francaise des Myopathies.
REFERENCES
- 1.Berry M N, Friend D S. High yield preparation of isolated rat liver parenchymal cells. J Cell Biol. 1969;43:506–520. doi: 10.1083/jcb.43.3.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown M S, Goldstein J L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 3.Chomczynski P, Sacchi N. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-choroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 4.Coupé C, Perdereau D, Ferré P, Hitier Y, Narkewicz M, Girard J. Lipogenic enzyme activities and mRNA in rat adipose tissue at weaning. Am J Physiol. 1990;258:E126–E133. doi: 10.1152/ajpendo.1990.258.1.E126. [DOI] [PubMed] [Google Scholar]
- 5.Doiron B, Cuif M H, Kahn A, Diaz-Guerra M J. Respective roles of glucose, fructose, and insulin in the regulation of the liver-specific pyruvate kinase gene promoter. J Biol Chem. 1994;269:10213–10216. [PubMed] [Google Scholar]
- 6.Dugail I, Quignard-Boulangé A, Le Liepvre X, Ardouin B, Lavau M. Gene expression of lipid storage-related enzymes in adipose tissue of the genetically obese Zucker rat. Biochem J. 1992;281:607–611. doi: 10.1042/bj2810607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emtage P C R, Wan Y, Bramson J L, Graham F L, Gauldie J. A double recombinant adenovirus expressing the costimulatory molecule B7-1 (murine) and human IL-2 induces complete tumor regression in a murine breast adenocarcinoma model. J Immunol. 1998;160:2531–2538. [PubMed] [Google Scholar]
- 8.Foretz M, Carling D, Guichard G, Ferré P, Foufelle F. AMP-activated protein kinase inhibits the glucose-activated expression of fatty acid synthase gene in rat hepatocytes. J Biol Chem. 1998;273:14767–14771. doi: 10.1074/jbc.273.24.14767. [DOI] [PubMed] [Google Scholar]
- 9.Foufelle F, Gouhot B, Pégorier J P, Perdereau D, Girard J, Ferré P. Glucose stimulation of lipogenic enzyme gene expression in cultured white adipose tissue. J Biol Chem. 1992;267:20543–20546. [PubMed] [Google Scholar]
- 10.Girard J, Ferré P, Foufelle F. Mechanisms by which carbohydrates regulate expression of genes for glycolytic and lipogenic enzymes. Annu Rev Nutr. 1997;17:325–352. doi: 10.1146/annurev.nutr.17.1.325. [DOI] [PubMed] [Google Scholar]
- 11.Green H, Kehinde O. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell. 1975;5:19–26. doi: 10.1016/0092-8674(75)90087-2. [DOI] [PubMed] [Google Scholar]
- 12.Gregor P D, Sawadogo M, Roeder R G. The adenovirus major late transcription factor USF is a member of the helix-loop-helix group of regulatory proteins and binds to DNA as a dimer. Genes Dev. 1990;4:1730–1740. doi: 10.1101/gad.4.10.1730. [DOI] [PubMed] [Google Scholar]
- 13.Horton J D, Bashmakov Y, Shimomura I, Shimano H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc Natl Acad Sci USA. 1998;95:5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horton J D, Shimomura I, Brown M, Hammer R, Goldstein J L, Shimano H. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element binding protein 2. J Clin Investig. 1998;101:2331–2339. doi: 10.1172/JCI2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaytor E N, Shih H M, Towle H C. Carbohydrate regulation of hepatic gene expression: evidence against a role for the upstream stimulatory factor. J Biol Chem. 1997;272:7525–7531. doi: 10.1074/jbc.272.11.7525. [DOI] [PubMed] [Google Scholar]
- 16.Kim J B, Sarraf P, Wright M, Yao K M, Mueller E, Solanes G, Lowell B B, Spiegelman B. Nutritional and insulin regulation of fatty acid synthetase and leptin gene expression through ADD1/SREBP1. J Clin Investig. 1998;101:1–9. doi: 10.1172/JCI1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J B, Spiegelman B M. ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev. 1996;10:1096–1107. doi: 10.1101/gad.10.9.1096. [DOI] [PubMed] [Google Scholar]
- 18.Kim J B, Spotts G D, Halvorsen Y D, Shih H M, Ellenberger T, Towle H C, Spiegelman B M. Dual DNA binding specificity of ADD1/SREBP1 controlled by a single amino acid in the basic helix-loop-helix domain. Mol Cell Biol. 1995;15:2582–2588. doi: 10.1128/mcb.15.5.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lafont A, Loirand G, Pacaud P, Vilde F, Lemarchand P, Escande D. Vasomotor dysfunction early after exposure of normal rabbit arteries to an adenoviral vector. Hum Gene Ther. 1997;8:1033–1040. doi: 10.1089/hum.1997.8.9-1033. [DOI] [PubMed] [Google Scholar]
- 20.Lefrançois-Martinez A M, Diaz-Guerra M J, Vallet V, Kahn A, Antoine B. Glucose-dependent regulation of the l-pyruvate kinase gene in a hepatoma cell line is independent of insulin and cyclic AMP. FASEB J. 1994;8:89–96. doi: 10.1096/fasebj.8.1.8299894. [DOI] [PubMed] [Google Scholar]
- 21.Lefrancois-Martinez A M, Martinez A, Antoine B, Raymondjean M, Kahn A. Upstream stimulatory factor proteins are major components of the glucose response complex of the L-type pyruvate kinase gene promoter. J Biol Chem. 1995;270:2640–2643. doi: 10.1074/jbc.270.6.2640. [DOI] [PubMed] [Google Scholar]
- 22.Mariash C N, Seelig S, Schwartz H L, Oppenheimer J H. Rapid synergistic interaction between thyroid hormone and carbohydrate on mRNAS14 induction. J Biol Chem. 1986;261:9583–9586. [PubMed] [Google Scholar]
- 23.Mourrieras F, Foufelle F, Foretz M, Morin J, Bouché S, Ferré P. Induction of fatty acid synthase and S14 gene expression by glucose, xylitol and dihydroxyacetone in cultured rat hepatocytes is closely correlated with glucose 6-phosphate concentrations. Biochem J. 1997;323:345–349. doi: 10.1042/bj3260345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moustaïd N, Beyer R S, Sul H S. Identification of an insulin response element in the fatty acid synthase promoter. J Biol Chem. 1994;269:5629–5634. [PubMed] [Google Scholar]
- 25.Oualikene O, Gonin P, Eloit M. Lack of evidence of phenotypic complementation of E1A/E1B-deleted adenovirus type 5 upon superinfection by wild-type virus in the cotton rat. J Virol. 1995;69:6518–6524. doi: 10.1128/jvi.69.10.6518-6524.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prip-Buus C, Perdereau D, Foufelle F, Maury J, Ferré P, Girard J. Induction of fatty acid synthase gene expression by glucose in primary culture of rat hepatocytes. Eur J Biochem. 1995;230:309–315. doi: 10.1111/j.1432-1033.1995.0309i.x. [DOI] [PubMed] [Google Scholar]
- 27.Sasaki K, Cripe T P, Koch S R, Andreone T L, Petersen D D, Beale E G, Granner D K. Multihormonal regulation of PEPCK gene transcription. The dominant role of insulin. J Biol Chem. 1984;259:15242–15251. [PubMed] [Google Scholar]
- 28.Sheng Z, Otani H, Brown M S, Goldstein J L. Independent regulation of sterol regulatory element binding proteins 1 and 2 in hamster liver. Proc Natl Acad Sci USA. 1995;92:935–938. doi: 10.1073/pnas.92.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimano H, Horton J D, Shimomura I, Hammer R E, Brown M S, Goldstein J L. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J Clin Investig. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimano H, Shimomura I, Hammer R E, Herz J, Goldstein J L, Brown M S, Horton J D. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Investig. 1997;100:2115–2124. doi: 10.1172/JCI119746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimomura I, Shimano H, Horton J D, Goldstein J L, Brown M. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J Clin Investig. 1997;99:838–845. doi: 10.1172/JCI119247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tontonoz P, Kim J B, Graves R A, Spiegelman B M. ADD1: a novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol Cell Biol. 1993;13:4753–4759. doi: 10.1128/mcb.13.8.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Towle H C, Kaytor E N, Shih H M. Regulation of the expression of lipogenic enzyme genes by carbohydrate. Annu Rev Nutr. 1997;17:405–433. doi: 10.1146/annurev.nutr.17.1.405. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Sato R, Brown M S, Hua X, Goldstein J L. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 35.Yokoyama C, Wang X, Briggs M R, Admon A, Wu J, Hua X, Goldstein J L, Brown M S. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993;75:187–197. [PubMed] [Google Scholar]