Abstract
Here, we describe a new, simple, highly multiplexed serological test that generates a more complete picture of seroconversion than single antigen‐based assays. Flow cytometry is used to detect multiple Ig isotypes binding to four SARS‐CoV‐2 antigens: the Spike glycoprotein, its RBD fragment (the main target for neutralizing antibodies), the nucleocapsid protein, and the main cysteine‐like protease in a single reaction. Until now, most diagnostic serological tests measured antibodies to only one antigen and in some laboratory‐confirmed patients no SARS‐CoV‐2‐specific antibodies could be detected. Our data reveal that while most patients respond against all the viral antigens tested, others show a marked bias to make antibodies against either proteins exposed on the viral particle or those released after cellular infection. With this assay, it was possible to discriminate between patients and healthy controls with 100% confidence. Analysing the response of multiple Ig isotypes to the four antigens in combination may also help to establish a correlation with the severity degree of disease. A more detailed description of the immune responses of different patients to SARS‐CoV‐2 virus might provide insight into the wide array of clinical presentations of COVID‐19.
Keywords: coronavirus, diagnostics, serology, COVID‐19, FACS, microbeads
Magnetic fluorescent beads coated with different SARS‐CoV‐2 antigens can be mixed to test simultaneously for antibodies present in patient plasma in highly multiplexed flow cytometry‐based assays. This sensitive method allows identification, in a single reaction, of several Ig‐isotypes and specificities, revealing differential responses between COVID‐19 patients.
Introduction
Severe Acute Respiratory Syndrome‐Coronavirus‐2 (SARS‐CoV‐2) causes the respiratory disease referred to as COVID‐19 that was recognized by the WHO as a pandemic in 2020 [1, 2]. Antibodies generated against viral proteins can be used in diagnostics to complement assays for viral nucleic acids and to follow the evolution of the infection. Moreover, as the pandemic advances and new clinical manifestations are described, it is important to evaluate the quality, quantity, and duration of the immune response in patients with different severity and symptoms [3, 4, 5]. SARS‐CoV‐2 serology usually tests for antibodies to the envelope glycoprotein, Spike (S), which mediates attachment to host cells and virus cell entry via its Receptor Binding Domain (RBD) [6, 7], the major target for virus‐neutralizing antibodies [8] and nucleocapsid protein (NP). Several formats, such as ELISA, CLIA, and lateral flow devices are available. However, commercial tests generally detect antibodies only to one antigen [9] and antibodies against other viral antigens, like the 3CL main protease (Mpro), which is only synthesized once the virus has infected the cell, but leads to strong IgG concentration in saliva [10], are not usually evaluated. The use of combinations of antigens seems likely to more fully describe the magnitude and quality of the immune response in SARS‐CoV‐2‐infected patients. In this regard, various multiplex serological assays, based on Luminex or micro‐array technology, have been described [11, 12, 13, 14, 15, 16]. These assays achieve high sensitivity and specificity, however, in general, they only test for antigens present in the viral particle and require specialized equipment. Recently, however, a flow cytometric bead assay that detects IgG and IgM antibodies against the S1 gp subunit and NP with around 90% sensitivity and specificity has been described [16].
We describe here a multiplex bead‐based flow cytometry assay that assesses, in a single reaction, sero‐reactivity to four SARS‐CoV‐2 antigens (Spike, RBD, NP, and MPro) analyzing IgA, IgG, IgM, simultaneously on any standard flow cytometer. The technique yields results with low background signals and has specificity and sensitivity near 100%, therefore providing a powerful tool to achieve a more complete view of patient humoral immunity.
Results and discussion
A flow cytometry‐based, bead‐assisted multi‐antigen serological assay
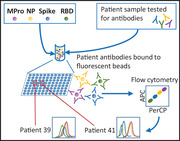
A high‐throughput, flow cytometry‐based multiplex assay to characterize COVID‐19 patient antibody responses was developed. To detect antibodies to multiple SARS‐CoV‐2 antigens in a single reaction, different viral proteins were immobilized on beads with different fluorescence intensities in the APC/PerCP channels (Figure 1A). After incubation with patient plasma, fluorescently labeled secondary antibodies were used to quantify IgG, IgA, and IgM bound to the viral antigens.
Figure 1.
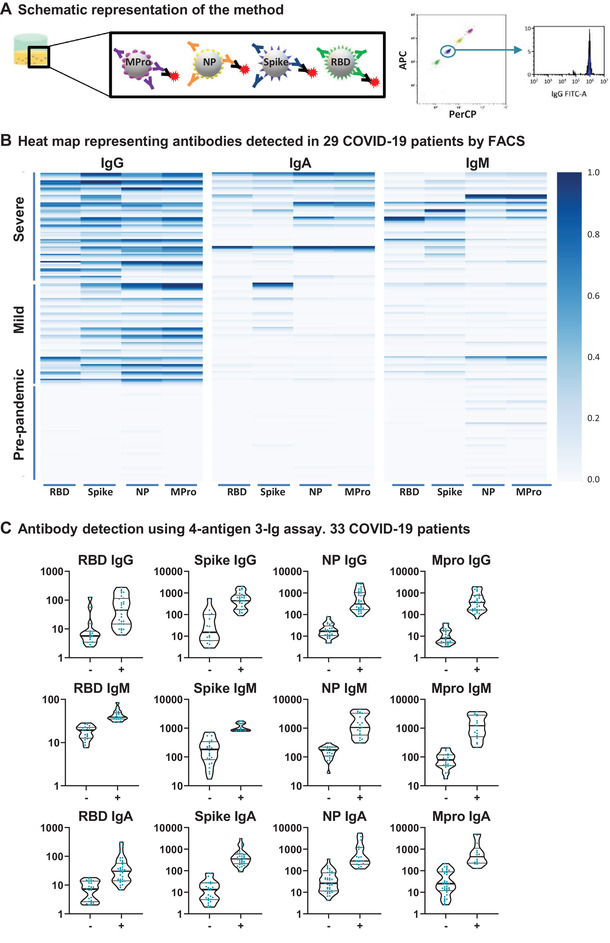
A multi‐antigen assay to detect SARS‐CoV‐2 specific antibodies. (A) Schematic representation of the method. Four different SARS‐CoV‐2 antigens (Mpro, NP, S, and RBD) were covalently coupled to magnetic beads with different fluorescence intensity in the APC and PerCP channels. Equal amounts of the different beads mixed in the same tube were incubated with plasma. SARS‐CoV‐2‐specific antibodies were visualized with fluorophore‐conjugated secondary antibodies followed by flow cytometry. (B) Heat map representing antibody titers from multi‐antigen COVID‐19 assays. Plasma from 15 healthy controls and 29 COVID‐19 patients were incubated with S‐, RBD‐, NP‐, and Mpro‐ coated beads. Specific IgG‐PE, IgA‐FITC+ IgM‐PE responses were analyzed by flow cytometry. The data are summarized in a heat map. Each column corresponds to one antigen while rows show four different plasma dilutions (1/100, 1/200, 1/600, 1/1800) for each individual. The intensity of the blue color depicts the amount of antibody. (C) Multi‐antigen assay for SARS‐CoV‐2 antibody detection in a single reaction. The assay was done, as in B, but only 1/100 dilution was tested and antibodies were developed with three combined anti‐human Ig antibodies (IgG‐FITC, IgM‐PE, and IgA‐PE‐Cyanine7). Statistic comparison was carried out using a Mann‐Whitney test.
Initial experiments were conducted to define detection limits and estimate the signal for known concentrations of antibody against the single His‐tag present in the antigen constructs and to test the signal obtained after plasma titration (Supporting Information Figures 1 and 2). These analyses showed good sensitivity and a clear discrimination between control and convalescent plasma samples over a wide range of dilutions. Combinations of either anti‐IgM‐PE and IgA‐FITC; IgG‐FITC, and IgA‐PE; or IgG‐FITC, IgA‐PE, and IgM‐PE/Cyanine7 were used with comparable results (Supporting Information Figure 3). Thus, antibodies of three isotypes, specific for four viral antigens could be determined in a single reaction.
The use of magnetic beads and flow cytometry therefore provides a method for serological analyses that is easily adaptable to hospital practice and clinical laboratories using any standard flow cytometer.
Multi‐antigen, bead‐assisted flow cytometry identifies COVID‐19 patients with 100% confidence
The presence of antibodies against SARS‐CoV‐2 S, RBD, NP, and Mpro antigens was tested in 44 plasma samples, including 29 COVID‐19 patients, 14 clinically classified as mild, that showed clinical symptoms but did not require hospitalization, and 15 with severe disease requiring intensive care (Supplementary Table 1). Testing each sample for IgA, IgG, IgM isotypes (Figure 1B) showed healthy controls and COVID‐19 patients clearly differed in the signal obtained for IgG antibodies against the four antigens. As expected, IgA and IgM SARS‐CoV‐2‐specific antibodies were not detected in all the plasma tested. IgA provided very clean and specific data. IgM seemed to have a higher background, however, due to relative bead fluorescence. These data were confirmed in analyses of an independent cohort of 33 patients, in a single dilution using a single reaction with 3‐Ig staining (Figure 1C).
The specificity and sensitivity of this methodology were determined using machine learning techniques. A random forest classifying algorithm was developed to evaluate the capacity of single‐antigen and multi‐antigen techniques to distinguish seronegative from COVID seropositive individuals (Supporting Information Table 2). When data for the four antigens and four dilutions of IgG were combined, 99.94% true positive and 100% true negative rates were defined. True positive rates near to 100% were also obtained when only three antigens (RBD, S, and Mpro) and one dilution were analyzed, highlighting the predictive power of the technique. When the sensitivity and specificity of each single‐antigen ELISA test were compared with the multi‐antigen FACS technique by ROC curve analysis (Figure 2A), the multi‐antigen assay was again superior. As expected, analysis of the data from the single‐antigen ELISAs in combination showed sensitivity and specificity comparable to the multiplex assay. Testing using a panel of plasma from patients with other respiratory viral/bacterial diseases confirmed the specificity of SARS‐CoV‐2‐specific antibody detection (Supporting Information Figure 4).
Figure 2.
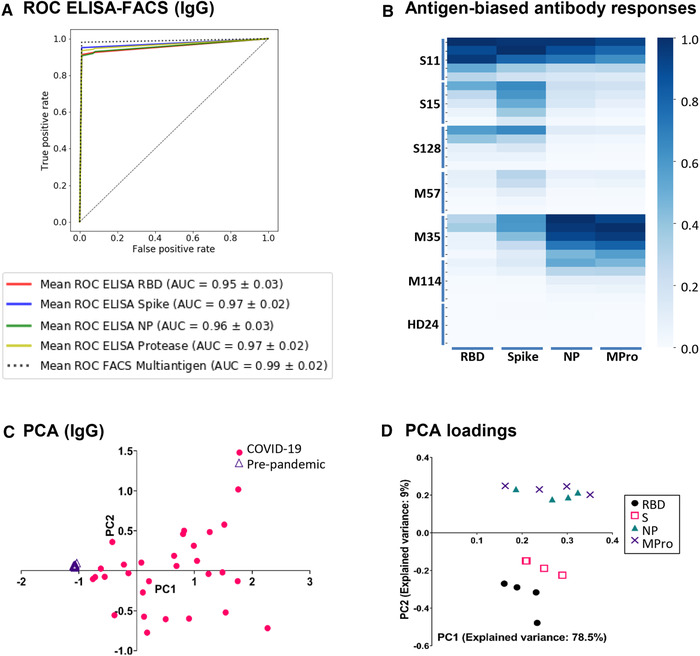
(A) ROC curves comparing single‐antigen ELISA and multi‐antigen FACS assays. A random forest classifier, trained with IgG values from one healthy and two COVID samples was used to predict the rest of the samples. The mean ROC curve after 15‐fold cross‐validation is shown for each condition. (B) Antigen‐biased antibody responses. Heat map of patients with biased IgG response against either S/RBD or NP/Mpro viral antigens. Data from 6 patients and 1 healthy donor are shown. (B) PCA of IgG. Triangles and circles represent pre‐pandemic controls and COVID‐19 patients, respectively. (C) PCA loadings. Visual representation of the loadings of the two first principal components. Each dilution of IgG titer against RBD, Spike, NP, and MPro is represented as a separate variable.
COVID‐19 patients respond differentially to the four viral antigens
The enhanced efficiency of the multi‐antigen test likely relates to the observation that some patients clearly responded preferentially to antigens exposed in the viral particle (S, RBD), while other patients responded mainly to antigens normally only exposed once cells have been infected (NP, Mpro) (Figure 2B) [10]. PCA independently confirmed the existence of this bias. Inspection of PCA loadings (Figure 2C‐D) showed that, for IgG, the second principal component corresponded to preferential production of antibodies against either NP+MPro or S+RBD. Analysis of IgA and IgM responses had similar patterns (not shown). This phenomenon, observed in 5 of 29 patients, suggests that biased antibody responses are likely common, emphasizing the value of multi‐antigen serological assays to minimize false‐negative results. Since not all patients respond similarly after infection by SARS‐CoV‐2, and different immune responses may contribute to the distinct disease manifestations, characterization of multi‐antigen antibody responses may yield insights into the different clinical presentations of SARS‐CoV‐2 infection.
Antibody response, disease severity, and vaccination
In general, patients with higher antibody titers are more likely to have suffered severe disease, indicating that infection severity could be linked to humoral immunity. However, analysis of only IgG responses did not clearly discriminate between patients who had suffered severe or mild disease (Figure 1B). Since statistically significant increased antibody responses in severe compared to mildly affected patients were observed for virus‐specific IgA antibodies, we used these variables to build a random forest that classified mild vs severe disease with 92% accuracy when IgG data (dilutions 1:600‐1800) and the IgA (dilution 1:100) responses were analyzed simultaneously (Figure 3A). Analysis of more patients is required, so that this approach could be analyzed on a prospective basis to evaluate its prognostic value.
Figure 3.
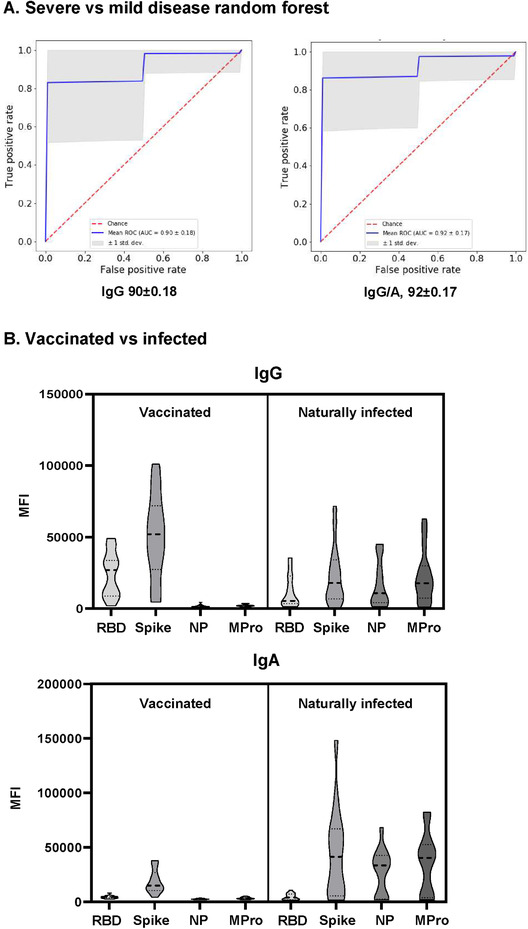
(A) Algorithm for classification of COVID‐19 patient severity. A random forest was trained to discriminate between COVID‐19 patients with severe or mild disease, using either IgG data alone or including data from other isotypes, and then used to predict unseen patients. The mean ROC curve after 300 random repetitions is shown for each condition. (B) Comparison of vaccinated individuals and COVID‐19 convalescent patient antibody responses. Plasma IgG and IgA from 15 vaccinated donors (Pfizer/BioNTech), were compared to the antibodies produced by 15 naturally infected individuals.
The multi‐antigen assay was also used to compare the antibody responses of 15 vaccinated individuals with those of naturally infected individuals (Supporting Information Table 3). As expected, vaccinated individuals only showed antibodies reactive with the S and RBD antigens, while plasma from naturally infected donors presented antibodies against all four viral antigens (Figure 3B). IgG was the predominant isotype in vaccinated donors who, in contrast to naturally infected individuals, made only minimal IgA responses.
Concluding remarks
Here, we report the development of a robust, quantitative, multiplex methodology that provides a detailed description of the humoral immune response to infection with SARS‐CoV‐2 with excellent sensitivity and specificity. The multi‐isotype, multi‐antigen assay permits screenings for SARS‐CoV‐2‐specific antibody responses for both research and diagnostics. This technique can easily be put into practice and can be automated. The simultaneous detection of antibodies to multiple viral proteins in a single tube greatly facilitates sample handling and comparison of the antibody response to different viral antigens. The method can be easily modified for the detection of antibodies in other fluids like saliva or breast milk and to include other antigens of potential interest. Importantly, detection of antibody responses to antigens absent from the vaccine allows discrimination between immunized and infected individuals. Moreover, since one dose of vaccine stimulates antibody responses in previously infected individuals compared to that observed in naïve individuals immunized twice [17], effective identification of previously infected individuals in the unvaccinated population would allow these individuals to be given the vaccine as a single booster, reducing the possibility of adverse reactions after a second dose and freeing up urgently needed vaccine doses to be given to non‐immune individuals.
Material and methods
Patient selection, samples, and Institutional Review Board permits
Experiments were carried out following the ethical principles established in the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. Patients (or their representatives) were informed about the study and gave a written informed consent. This study used samples from several hospitals. For optimization experiments, samples from the research project “Immune response dynamics as a predictor of COVID‐19 disease evolution. Implications for therapeutic decision‐making” approved by La Princesa Health Research Institute Research Ethics Committee (register # 4070) were used; samples and data from patients with severe versus mild disease were provided by the Biobank Hospital Universitario Puerta de Hierro Majadahonda (HUPHM)/Instituto de Investigación Sanitaria Puerta de Hierro‐Segovia de Arana (IDIPHISA) (PT17/0015/0020 in the Spanish National Biobanks Network), they were processed following standard operating procedures with the appropriate approval of the Ethics and Scientific Committees. For comparison of disease severity, 29 COVID‐19 patients, diagnosed by PCR, were recruited. Fourteen patients, classified as mild disease or asymptomatic, did not require treatment after diagnosis. Fifteen patients, classified as severe disease, required ICU hospitalization (Supporting Information Table 1). Plasma samples were obtained 33–40 days after diagnostic PCR and, separated by blood centrifugation after collection in EDTA tubes, 15 plasma samples were collected from healthy blood donors before June 2019 (PRE‐COVID‐19) in the Puerta de Hierro hospital biobank, were used as negative controls.
Fifteen vaccinated (Pfizer BioNTech) individuals were recruited at the Centro de Hemoterapia y Hemodonación de Castilla y León (ChemCyL) for comparison with SARS‐CoV‐2 infected patients (Supporting Information Table 2). These samples were obtained as part of the project “Development of serological assays for detection of viral antigens (SARS‐COV2)". The protocol was approved by the Bioethics Committees: CEIm Área de Salud Valladolid Este, Hospital Clínico Universitario de Valladolid, with the number/BIO 2020‐98‐COVID.
CPD respiratory panel human plasma samples were obtained from a commercial source (BioIVT ‐ West Sussex, United Kingdom).
Expression of the SARS‐CoV‐2 Cys‐like protease (Mpro), nucleocapsid (NP), Spike (S), and RBD proteins
Recombinant SARS‐CoV‐2 proteins were expressed with a histidine tag. Cys‐like protease (Mpro) and nucleocapsid (NP) proteins constructs were expressed in the E. coli strain BL21 Star (DE3) pLysS (ThermoFisher) and purified as described [10].
Recombinant cDNAs coding for soluble S (residues 1 to 1208) and RBD (332 to 534) proteins were cloned in the pcDNA3.1 vector for expression in HEK‐293F cells using standard transfection methods. The two constructs contained the S signal sequence at the N‐terminus, and a T4 fibritin trimerization sequence, a Flag epitope, and an 8xHis‐tag at the C‐terminus. In the S protein, the furin‐recognition motif (RRAR) was replaced by the GSAS sequence and it contained the A942P, K986P, and V987P substitutions in the S2 portion. Proteins were purified by Ni‐NTA affinity chromatography from transfected cell supernatants and they were transferred to 25 mM Hepes‐buffer and 150 mM NaCl, pH 7.5, during concentration.
Bead‐based flow cytometry assay for detection of antibodies to SARS‐CoV‐2
Magnetic fluorescent beads (106), with a mean diameter 5.5 μm and high‐density carboxyl functional groups on the surface (QuantumPlex™M COOH – Bangs Laboratories, Inc.), were covalently coupled with 30 μg of viral protein through their primary amines by two‐step EDC/NHS protocol. Beads were resuspended in a solution of PBS containing 1% casein and a stabilizer (Biorad 1x PBS blocker). To distinguish the beads coated with different antigens, different fluorescence intensity combinations in the APC and PerCP channels were used (Figure 1A).
Beads were incubated with either rabbit anti‐His‐tag antibody (Proteintech Group) or plasma from patients or healthy donors in a final volume of 50 μl in 96‐well‐plates (Nunc™ MicroWell™ 96‐Well, Thermo Fisher Scientific) using the dilutions indicated in each experiment. Patient plasma samples were diluted in PBS‐casein (Biorad,1× PBS blocker), and incubated with the beads for 40 min at room temperature under agitation. Beads were washed three times by addition of PBS, placing the tubes or plates on a magnet (MagneSphere® Mag. Sep. Stand 12‐ hole, 12 × 75 mm, Promega; Handheld Magnetic Separator Block for 96‐well plate, Merck, Millipore) and decantation of supernatant.
To visualize antibody bound to antigen‐coated beads, either PE‐conjugated anti‐rabbit antibody (0.25 μg/ml, Southern Biotech), PE‐conjugated anti‐human IgG and IgM, or FITC‐conjugated antihuman IgA antibody (Immunostep S.L.) were added (30 μL/well) and incubated for 20 min at room temperature under agitation. After three washes, data were acquired by flow cytometry using either CytoFLEX or Cytomics FC 500 (Beckman Coulter).
For large screenings performed on different days, data were normalized to the values of a positive control serum included in every assay.
ELISA for detection of antibodies to SARS‐CoV‐2
ELISA assays for the detection of antibodies directed against the four SARS‐CoV‐2 antigens were carried out as described [10].
Statistical analysis
To assess the prediction capacity of the new methodology, an algorithm was built using Scikit‐learn python package [18] (code available on request). Samples were stratified and randomly spliced into a training and a test set. The training samples were used to fit a random forest classifier which then predicted the healthy vs disease category of unseen test samples (1/7 of total samples). This was repeated n = 10,000 times. For each patient, accuracy was calculated as the proportion of correct predictions divided by the number of predictions made. As a complementary approach, a mean Receiver Operating Characteristic (ROC) curve was built for the random forest classifier by stratified 15‐fold cross‐validation, using the smaller set (2–3 samples) to train the model and then predicting the remaining ones.
For heatmap representation, each variable was scaled to a range (0,1) using the MixMaxScaler command from Scikit‐learn and visualized using heatmap command from seaborn python packages. For Principal Component Analysis, each variable was scaled as described, and the PCA command from Scikit‐learn was used to fit and transform the data. Principal components up to a 95% of accumulated explained variance were saved.
Comparison between severe and mild patients in each variable was performed by multiple t‐tests followed by False Discovery Rate (1%) correction by two‐stage step‐up method in Graph Pad Prism 8 Software (GraphPad Software, USA, www.graphpad.com).
Ethics approval statement for human and/or animal studies
La Princesa Health Research Institute Research Ethics Committee (register # 4070); Biobank Hospital Universitario Puerta de Hierro Majadahonda (HUPHM)/Instituto de Investigación Sanitaria Puerta de Hierro‐Segovia de Arana (IDIPHISA) (PT17/0015/0020 in the Spanish National Biobanks Network); CEIm Área de Salud Valladolid Este, Hospital Clínico Universitario de Valladolid, with the number/BIO 2020‐98‐COVID.
Patient consent statement
Experiments were carried out following the ethical principles established in the Declaration of Helsinki. Patients (or their representatives) were informed about the study and gave written informed consent.
Author's contributions
Y.C.M., D.F.S., C.C.S., E.M.G.C., D.N.H., A.B.M., P.M.F., A.A., J.M.R.F., M.V.G., J.M.C., and H.T.R. prepared reagents, performed experiments, and analyzed data; P.M.F., A.A., F.S.M., G.E.L., and C.V. selected and clinically characterized patients; D.F.S. carried out statistical analyses; R.J.A., J.M.R.F., M.V.G., F.S.M., and H.T.R. obtained financial support, conceived and designed the study; D.F.S., H.T.R., J.M.R.F., and M.V.G. wrote the manuscript with revisions from all authors.
Conflict of interest
J.M.R.F., J.M.C., H.T.R., and M.V.G. are inventors on the European patent “Assay for the detection of the Cys‐like protease (Mpro) of SARS‐CoV‐2” [EP20382495.8]. RJA is CEO of Immunostep, S.L. D.N.H. and A.B.M. are employed by Immunostep, S.L. The rest of the authors declare no potential conflict of interest.
Peer review
The peer review history for this article is available at https://publons.com/publon/10.1002/eji.202149319
Abbreviations
- CLIA
Chemiluminescence immunoassay
- ELISA
enzyme‐linked immunosorbent assay
- gp
glycoprotein
- PCA
principal component analysis
- ROC
receiver operating characteristic
Supporting information
Supporting Information
Acknowledgements
The authors thank the HUPHM/IDIPHISA Biobank and Centro de Hemoterapia y Hemodonación de Castilla y León (ChemCyL) for human specimens; the HUPHM Infectious Diseases and Intensive Care Units for organization of samples and clinical data; Sofía Garrido, Miriam García, (Immunology, HUPHM), for processing blood samples in difficult circumstances; and Mario Mellado (CNB director). This work was supported by: Spanish National Research Council (CSIC‐202020E079, CSIC‐COVID19‐028); Madrid Regional Government “IMMUNOTHERCAN” [S2017/BMD‐3733‐2 (MVG)]; Spanish Ministry of Science and Innovation [(MCIU/AEI/FEDER, EU, RTI2018‐093569‐B‐I00 (MVG), SAF2017‐82940‐R (JMRF), SAF2017‐83265‐R (HTR); SAF2017‐82886‐R (FSM)]; Health Institute Carlos III (ISCIII) [RETICS Program RD16/0012/0006; RIER (EMGC); PI19/00549 (AA)]; “La Caixa Bank Foundation” (HR17‐00016), Fondo Supera COVID (CRUE‐Banco de Santander), both to FSM.
Contributor Information
José M. Rodríguez‐Frade, Email: jmrfrade@cnb.csic.es.
Mar Valés‐Gómez, Email: mvales@cnb.csic.es.
Data availability statement
All data generated during this study are included in this publication.
References
- 1. Zhu, N. , Zhang, D. , Wang, W. , Li, X. , Yang, B. , Song, J. , Zhao, X. et al., China Novel Coronavirus, I. and Research, T., A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020. 382: 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Coronaviridae Study Group of the International Committee on Taxonomy of Viruses . The species Severe acute respiratory syndrome‐related coronavirus: classifying 2019‐nCoV and naming it SARS‐CoV‐2. Nature Microbiology. 2020. 5: 536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dudley, J. P. and Lee, N. T. , Disparities in Age‐specific Morbidity and Mortality From SARS‐CoV‐2 in China and the Republic of Korea. Clin. Infect. Dis. 2020. 71: 863–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gadi, N. , Wu, S. C. , Spihlman, A. P. and Moulton, V. R. , What's Sex Got to Do With COVID‐19? Gender‐Based Differences in the Host Immune Response to Coronaviruses. Front. Immunol. 2020. 11: 2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tay, M. Z. , Poh, C. M. , Renia, L. , MacAry, P. A. and Ng, L. F. P. , The trinity of COVID‐19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020. 20: 363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lan, J. , Ge, J. , Yu, J. , Shan, S. , Zhou, H. , Fan, S. , Zhang, Q. et al., Structure of the SARS‐CoV‐2 spike receptor‐binding domain bound to the ACE2 receptor. Nature 2020. 581: 215–220. [DOI] [PubMed] [Google Scholar]
- 7. Amanat, F. , Stadlbauer, D. , Strohmeier, S. , Nguyen, T. H. O. , Chromikova, V. , McMahon, M. , Jiang, K. et al., A serological assay to detect SARS‐CoV‐2 seroconversion in humans. Nat. Med. 2020. 26: 1033–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Azkur, A. K. , Akdis, M. , Azkur, D. , Sokolowska, M. , van de Veen, W. , Bruggen, M. C. , O'Mahony, L. et al., Immune response to SARS‐CoV‐2 and mechanisms of immunopathological changes in COVID‐19. Allergy 2020. 75: 1564–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harvey, R. , Mattiuzzo, G. , Hassall, M. , Sieberg, A. , Muller, M. A. , Drosten, C. , Rigsby, P. et al., Comparison of Serologic Assays for Middle East Respiratory Syndrome Coronavirus. Emerg. Infect. Dis. 2019. 25: 1878–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martinez‐Fleta, P. , Alfranca, A. , Gonzalez‐Alvaro, I. , Casasnovas, J. M. , Fernandez‐Soto, D. , Esteso, G. , Caceres‐Martell, Y. et al., SARS‐CoV‐2 Cysteine‐like Protease Antibodies Can Be Detected in Serum and Saliva of COVID‐19‐Seropositive Individuals. J. Immunol. 2020. 205: 3130–3140. [DOI] [PubMed] [Google Scholar]
- 11. den Hartog, G. , Schepp, R. M. , Kuijer, M. , GeurtsvanKessel, C. , van Beek, J. , Rots, N. , Koopmans, M. P. G. et al., SARS‐CoV‐2‐Specific Antibody Detection for Seroepidemiology: A Multiplex Analysis Approach Accounting for Accurate Seroprevalence. J. Infect. Dis. 2020. 222: 1452–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dawson, E. D. , Kuck, L. R. , Blair, R. H. , Taylor, A. W. , Toth, E. , Knight, V. and Rowlen, K. L. , Multiplexed, microscale, microarray‐based serological assay for antibodies against all human‐relevant coronaviruses. J. Virol. Methods 2021. 291: 114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Butt, J. , Murugan, R. , Hippchen, T. , Olberg, S. , van Straaten, M. , Wardemann, H. , Stebbins, E. et al., From Multiplex Serology to Serolomics‐A Novel Approach to the Antibody Response against the SARS‐CoV‐2 Proteome. Viruses 2021. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fotis, C. , Meimetis, N. , Tsolakos, N. , Politou, M. , Akinosoglou, K. , Pliaka, V. , Minia, A. et al., Accurate SARS‐CoV‐2 seroprevalence surveys require robust multi‐antigen assays. Sci. Rep. 2021. 11: 6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Klupfel, J. , Koros, R. C. , Dehne, K. , Ungerer, M. , Wurstle, S. , Mautner, J. , Feuerherd, M. et al., Automated, flow‐based chemiluminescence microarray immunoassay for the rapid multiplex detection of IgG antibodies to SARS‐CoV‐2 in human serum and plasma (CoVRapid CL‐MIA). Anal. Bioanal. Chem. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Egia‐Mendikute, L. , Bosch, A. , Prieto‐Fernandez, E. , Lee, S. Y. , Jimenez‐Lasheras, B. , Garcia Del Rio, A. , Antonana‐Vildosola, A. et al., Sensitive detection of SARS‐CoV‐2 seroconversion by flow cytometry reveals the presence of nucleoprotein‐reactive antibodies in unexposed individuals. Commun Biol 2021. 4: 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krammer, F. , Srivastava, K. , Alshammary, H. , Amoako, A. , Awawda, M. , Beach, K. , Bermúdez‐González, C. et al., Robust spike antibody responses and increased reactogenicity in seropositive individuals after a single dose of SARS‐CoV‐2 mRNA vaccine. medRxiv 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pedregosa, F. , Varoquaux, G. , Gramfort, A. , Michel, V. , Thirion, B. , Grisel, O. , Blondel, M. et al., Scikit‐learn: machine learning in python. J. Mach. Learn. Res. 2011. 12: 2825–2830. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
All data generated during this study are included in this publication.


