Abstract
Background
The emergence of COVID‐19 pandemic resulted in an urgent need for the development of therapeutic interventions. Of which, neutralizing antibodies play a crucial role in the prevention and resolution of viral infection.
Methods
We generated antibody libraries from 18 different COVID‐19 recovered patients and screened neutralizing antibodies to SARS‐CoV‐2 and its mutants. After 3 rounds of panning, 456 positive phage clones were obtained with high affinity to RBD (receptor binding domain). Clones were then reconstituted into whole human IgG for epitope binning assay and all 19 IgG were classified into 6 different epitope groups or Bins.
Results
Although all antibodies were found to bind RBD, the antibodies in Bin2 had superior inhibitory ability of the interaction between spike protein and angiotensin converting enzyme 2 receptor (ACE2). Most importantly, the antibodies from Bin2 showed stronger binding affinity or ability to mutant RBDs (N501Y, W463R, R408I, N354D, V367F, and N354D/D364Y) derived from different SARS‐CoV‐2 strains as well, suggesting the great potential of these antibodies in preventing infection of SARS‐CoV‐2 and its mutations. Furthermore, such neutralizing antibodies strongly restricted the binding of RBD to hACE2 overexpressed 293T cells. Consistently, these antibodies effectively neutralized wildtype and more transmissible mutant pseudovirus entry into hACE2 overexpressed 293T cells. In Vero‐E6 cells, one of these antibodies can even block the entry of live SARS‐CoV‐2 into cells at 12.5 nM.
Conclusions
These results indicate that the neutralizing human antibodies from the patient‐derived antibody libraries have the potential to fight SARS‐CoV‐2 and its mutants in this global pandemic.
Keywords: neutralizing antibodies, patient‐derived antibody libraries, SARS‐CoV‐2, Spike protein
The antibody libraries were generated from different COVID‐19 recovered patients for panning neutralizing antibodies to SARS‐CoV‐2 and its mutants. Then the positive clones were sequenced and reconstituted into whole human IgG for epitope binning assay. Finally, the antibodies were demonstrated to have great potential in blocking the infection of SARS‐CoV‐2 and its mutants.
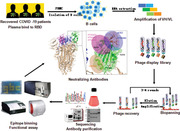
1. INTRODUCTION
A newly emerged pathogen spreading worldwide named severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is now causing a pandemic and more than 172.84 million cases of infection and 3.71 million deaths worldwide.[ 1 ] This global pandemic threatens public health in a way more devastating than ever before. SARS‐CoV‐2 belongs to Sarbecovirus subgenus and shares substantial genetic and functional similarity with other pathogenic human betacoronaviruses, including Severe Acute Respiratory Syndrome Coronavirus (SARS‐CoV) and Middle East Respiratory Syndrome Coronavirus (MERS‐CoV),[ 2 , 3 ] whereas the unique pathogenesis and rapid international transmission of SARS‐CoV‐2 make it the most infectious and destructive coronavirus compared to SARS‐CoV and MERS‐CoV.[ 4 , 5 , 6 ] At present, several vaccines have been reported to be effective in phase III trials and have received emergency approval in numerous countries.[ 7 ] However, due to the mutations and complexity of SARS‐CoV‐2, the current global epidemic has not been completely restrained. Thus, therapeutic drugs that are effective and specific to SARS‐CoV‐2 and its mutants are still in urgent demand to put an end to the pandemic.
Similar to other coronaviruses, SARS‐CoV‐2 utilizes its envelope spike (S) glycoprotein to interact with a cellular receptor for entry into target cells. Angiotensin converting enzyme 2 (ACE2) has been identified as the main receptor for viral entry through triggering a cascade of cell membrane fusion events.[ 8 ] The spike glycoprotein is composed of two functional subunits, which is responsible for binding to the host cell receptor (S1 subunit) and fusion of the viral and cellular membranes (S2 subunit). The host range and cellular tropism of SARS‐CoV‐2 are determinated by the receptor‐binding domain (RBD) within the S1 subunit.[ 9 ] Although the entry of SARS‐CoV‐2 into susceptible cells is a complex process and needs to be further explored, the receptor‐binding and proteolytic processing of the S protein are regarded as the key events in promoting virus‐cell fusion. While, an increasing amount of RBD mutants have been found under high positive selection pressure during the spread. Notably, new SARS‐CoV‐2 lineages carrying the mutation N501Y in the RBD of the spike protein which are more transmissible spread rapidly in the United Kingdom (UK).[ 10 ] The fact that such mutations increased the SARS‐CoV‐2 binding affinity to its host receptor ACE2 reveals a greater risk of more severe infections during a sustained pandemic of COVID‐19.[ 11 ] Thus, common neutralization of interaction between ACE2 and RBD with different mutations could serve as a promising candidate for prophylactic and therapeutic interventions against SARS‐CoV‐2 infection.[ 12 ]
During the last epidemic, SARS patients greatly benefited from convalescent serum collected from recovered subjects.[ 13 ] In a similar manner, the blood from COVID‐19 patients who have recently become virus‐free displayed serum neutralizing activities in a pseudotype entry assay.[ 14 ] However, the plasma neutralizing activity is low in most convalescent individuals. Nevertheless, rare but recurring RBD‐specific antibodies with potent antiviral activity were found in all individuals tested suggesting that humans are intrinsically have B cell clones that potently neutralize SARS‐CoV‐2.[ 15 ] Thus, it would be ideal to explore antibody cooperativity to develop a therapeutic antibody cocktail against SARS‐CoV‐2.[ 16 ] Different approaches can be developed to get full human antibodies against SARS‐CoV, such as single B cell antibody cloning,[ 17 ] XenoMouse[ 18 ] and phage display,[ 19 , 20 , 21 ] of which phage display library facilitates the identification and development of specific affinity antibodies in a rapid and cost‐effective manner. Several full human antibodies against the S1 domain have been generated and all of which can neutralize the virus in vitro and in vivo.[ 22 ] A panel of human neutralizing monoclonal antibodies targeting the SARS‐CoV‐2 RBD were selected from a phage display library constructed using peripheral circulatory lymphocytes collected from patients at the acute phase of the disease.[ 23 ] Furthermore, a naïve human semisynthetic phage library was also been used for identification of antibodies targeting the receptor‐binding domain (RBD) of SARS‐CoV‐2.[ 24 ] In order to develop better neutralizing human antibodies against SARS‐CoV‐2, single chain antibody fragment (scFv)‐phage libraries were constructed using B cells from COVID‐19 recovered patients which are thought to have more neutralizing potency. Several neutralizing human antibodies were then identified and characterized with the ability to bind with RBD and its mutants (N501Y, R408I, W463R, N354D, V367F and N354D/D364Y) and block the interaction with hACE2. Eventually, these antibodies dramatically inhibited SARS‐CoV‐2 RBD mediated entry into cells using both pseudovirus (wildtype or its mutants) and live SARS‐CoV‐2 virus, indicating that patient‐derived phage display antibody libraries could be an attractive source of neutralizing antibodies to SARS‐CoV‐2 and these antibodies are specific prophylactic and therapeutic agents that can be utilized against the ongoing SARS‐CoV‐2 pandemic.
2. RESULTS
2.1. Generation of patient‐derived antibody library
As the source of therapeutic antibodies to COVID‐19, the anti‐SARS‐CoV‐2 B cells were noticeably enriched in COVID‐19 convalescent patients. The PBMC from 18 different COVID‐19 recovered patients were isolated to generate phage displayed scFv libraries for panning the neutralizing antibodies (Figure 1a). Prior to constructing the phage display library, we examined the binding ability of plasma from 18 recovered COVID‐19 patients to SARS‐CoV‐2 RBD. It is observed that most convalescent patients were able to produce high titer of SARS‐CoV‐2 RBD‐specific antibodies compared with healthy donors using enzyme‐linked immunosorbent assay (ELISA) (Figure 1b). Then, the phage display libraries from 18 patients were constructed and quality assessment of each library was undertaken by using RT‐PCR clone counting and phage display ELISA. The correlation between the size of an antibody library and the likelihood of selecting for the desired antibodies is somewhat intuitive. As shown in Figure 1c, the size of primary libraries from patients ranged from 1.01 × 107 to 1.43 × 108, which is consistent with other human immunized scFv libraries.[ 25 ] The total size of all libraries is larger than 109. Ideally, the size of a library should be equal to its effective size, meaning that all the 109 antibody variants comprising the library are displayed as functional scFv molecules on the phage surface. While, the quality of gene amplification by RT‐PCR affects the effective size of the libraries. Thus, we appraised the positive rate and scFv frequency of each library by PCR, sequencing and phage ELISA. Sequence analysis revealed that the framework region (FR) and complementarity determining region (CDR) of selected clones showed the greatest difference in amino acid sequences and the mean positive rate is 95.83% and 88.38% (Figure 1d,e). Taken together, our data revealed that the scFv library from COVID‐19 recovered patients was constructed successfully.
FIGURE 1.
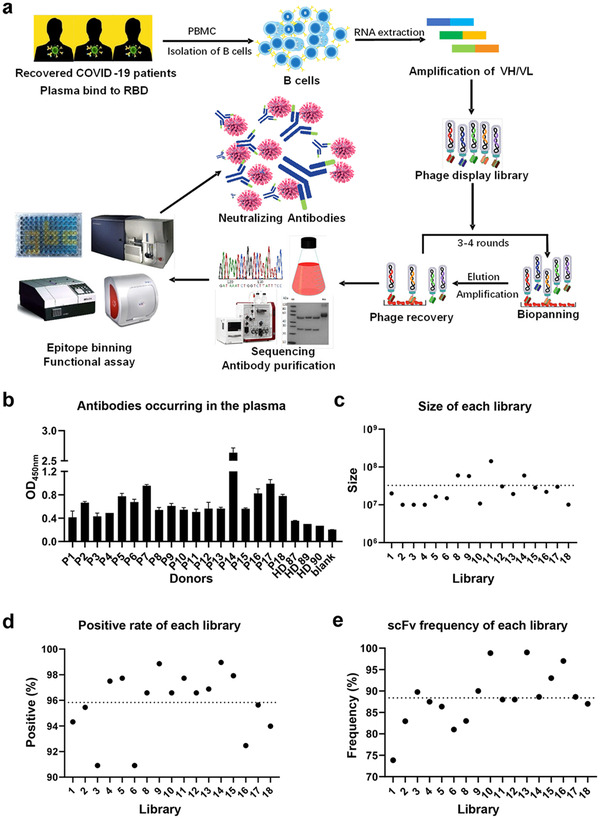
Construction of Patient‐derived scFv library. (a) The flow chart of construction and panning of patient‐derived scFv libraries. (b) The binding affinity of plasma from each patient to RBD by ELISA. (P1‐18, Recovered COVID 19 patients. HD87‐90, healthy donors). (c) The size of each library from 18 different recovered COVID‐19 patients (7# failed). (d) The clone quality assessment of each scFv library. (e) The scFv display rate of each library. Data are shown as the mean ± SEM of three independent experiments
2.2. Panning against SARS‐CoV‐2 RBD
Affinity selection of patient‐derived antibody library was performed using solid‐phase‐bound RBD. In order to exclude the potential bias, the 18 libraries were selected to solid‐phase‐bound RBD individually. After three rounds of panning, 1728 phage clones were randomly selected to induce the expression of soluble scFv for binding assay by ELISA. As can be seen from Figure 2a, 456 positive clones were identified to bind with RBD specifically. Subsequently, 19 scFv clones with highest affinity to RBD were reconstituted into whole human IgG for functional assay (Table S1) and S2 protein was used as a negative control to exclude non‐specific binding (Figure 2b). In order to characterize and group these IgG by the epitope binding regions generated against RBD, the “epitope binning” assay was performed on Octet systems. As shown in Figure 2c, 19 IgG can be divided into 6 different Bins. Of which, Bin1 and Bin2 are the biggest groups with 14 and 16 clones suggesting that these two epitopes on RBD are most important for recognizing by humoral immune system recognition. Therefore, engineering antibody that targets a specific functioning epitope on an RBD is more important than finding high‐affinity, tight‐binding mAb just to a non‐functional site.
FIGURE 2.
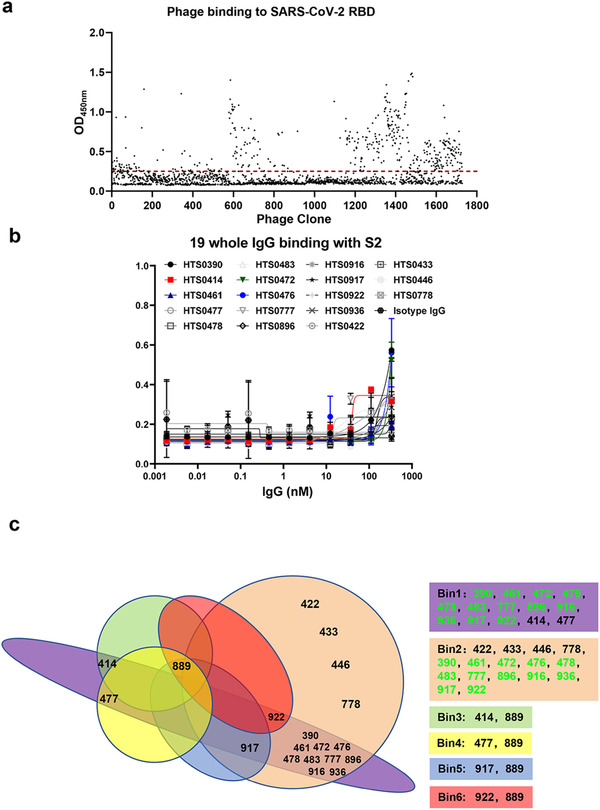
Screening for specific scFv to RBD. (a) The binding affinity of each clone after three rounds of enrichment. (b) The binding of 19 antibodies to S2 domain of SARS‐CoV‐2. (c) The 19 antibodies were divided into 6 bins by epitope binning assay
2.3. In vitro binding assay to RBD
To further confirm the binding affinity of each IgG in Bin1 and Bin2, the RBD was bound in 96‐well plates to complete an IgG binding assay. Consistent with scFv binding assay results (data not shown), the IgG in both Bin1 and Bin2 had similar binding affinity to RBD (Figure 3a,b). As the main step of SARS‐CoV‐2 entry to the host cells, the interaction between spike and ACE2 is quintessential for the viral infection. The blocking of this interaction by a neutralizing antibody has a vast potential in treating COVID‐19. Thus, we sought to investigate whether these IgG can block the binding of RBD to hACE2 overexpressed 293T cells by FACS inhibition assay. As shown in Figure 3c,d, all the IgG in Bin1 and Bin2 can block the binding of RBD to hACE2 overexpressed 293T cells. To further substantiate this hypothesis, the top four IgG in Bin2 were selected for further investigation. Although the SARS‐CoV‐2 is similar to SARS, the amino acid sequence of RBD of SARS‐CoV‐2 is only about 74% homologous to that of SARS‐CoV.[ 26 ] Therefore, we used the IgG specific to SARS (CR3022) as a control to do the functional assay. We demonstrated that all the IgG (including HTS0422, HTS0433, HTS0446, HTS0483 and CR3022) bind to RBD specifically (Figure 3e). By contrast, only the four IgG (HTS0422, HTS0433, HTS0446 and HTS0483) from Bin2 could inhibit the interaction between RBD and hACE2 (Figure 3f), suggesting that these antibodies may be bound to the key sites involved in the SARS‐CoV‐2 infection.
FIGURE 3.
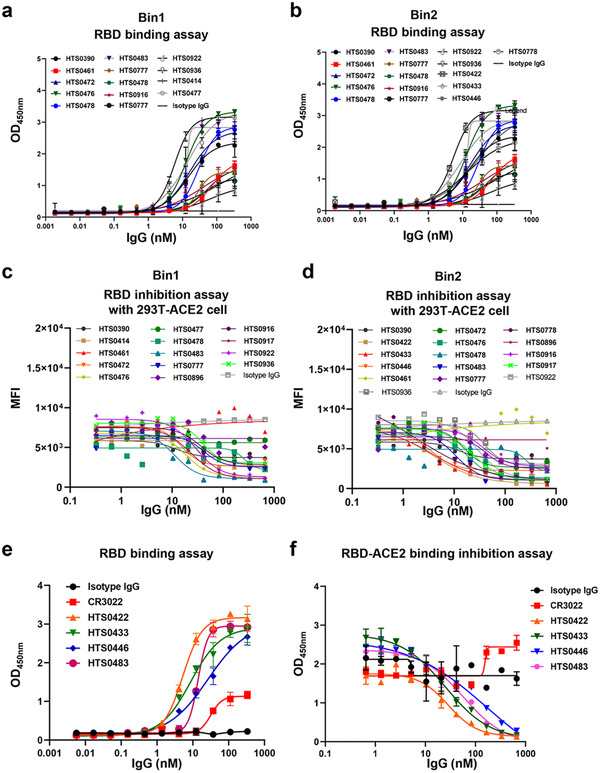
Binding and blocking assays for different scFv. (a, b) Binding of antibodies from Bin1 and Bin2 to RBD protein of SARS‐COV‐2 detected by ELISA. (c, d) Blocking the binding of RBD to ACE2 by antibodies from two groups detected by FACS. Abs were diluted as indicated in the figure. (e) The binding affinity of four positive antibodies from Bin2. (f) The blocking ability of four positive antibodies from Bin2. Data are shown as the mean ± SEM of three independent experiments
2.4. Cross‐neutralization against SARS‐CoV‐2 RBD mutations
As an RNA virus, mutations in SARS‐CoV‐2 are not unusual, and can be found in various countries globally. Previous studies demonstrated that some mutations in RBD enhanced the binding affinity to the host cells.[ 27 ] Therefore, cross‐neutralization against SARS‐CoV‐2 RBD mutations is essential for the development of an effective therapeutic antibody. In this study, six different SARS‐CoV‐2 RBD mutants (N501Y, R408I, W463R, N354D, V367F and N354D/D364Y) which were found in different regions all around the world were obtained from ACRO BIOSYSTEMS. As shown in Figure 4a, HTS0422, HTS0433 and HTS0446 bind with both wildtype and mutant (N501Y, W463R, N354D, V367F, both N354D and D364Y) RBD significantly. In addition, only HTS0483 binds with R408I, suggesting that R408I is essential for the binding of HTS0422, HTS0433 and HTS0446, and mutations at R408I abolished the binding of antibodies to the virus RBD, and HTS0483 may have broader cross‐neutralization effect against SARS‐CoV‐2 RBD mutations. Accordingly, only HTS0483 inhibits the binding between ACE2 and all RBD mutants including W436R and R408I. While the other antibodies (HTS0422, HTS0433 and HTS0446) can only restrict the interaction between ACE2 and wildtype, N501Y, N354D, V367F or N354D/D364Y (Figure 4b), implying that the mutations at W436R and R408I in RBD may cause the virus to escape from antibody blocking and, as a result, make the developing of broad vaccine become a considerable challenge. Meanwhile, it can also be found that even though all antibodies bind to N501Y significantly, the affinity of HTS0433 is much lower than the others as well as the inhibition of the binding between ACE2 and N501Y. What's more, although the CDR region is same between HTS0433 and HTS0446, there are three amino acids in the frame region of VL are different (DIV→AIR), suggesting that the frame region is also important in binding to N501Y mutation. This information will aid the affinity maturation of antibody to the more transmissible SARS‐CoV‐2 lineages with N501Y mutation.
FIGURE 4.

Cross‐neutralization against SARS‐CoV‐2 RBD mutations by different antibodies. (a) The binding affinity of four positive antibodies from Bin2 to different SARS‐CoV‐2 RBD mutations. (b) The blocking ability of four positive antibodies from Bin2 to different SARS‐CoV‐2 RBD mutations
2.5. Antiviral properties of human antibodies
To explore the potential of the top four IgG (HTS0422, HTS0433, HTS0446 and HTS0483) as therapeutic drugs, we tested whether these IgG can block the binding of RBD to hACE2 expressed cells by flow cytometry analysis. We found that these top four IgG from Bin2 could restrict the binding of RBD to hACE2‐293T cells but not CR3022, which bind to both SARS and SARS‐CoV‐2 RBD (Figure 5a). Furthermore, we determined the neutralization ability of these four IgG using a SARS‐CoV‐2 S pseudotyped lentiviral particle. Consistent with ELISA and FACS‐based blockade result, all 4 antibodies effectively neutralized pseudovirus entry to host cells ectopically expressing hACE2, with EC50 from 12.80 to 16.54 nM (Figure 5b,c). Moreover, SARS‐CoV‐2 strain hCoV‐19/Hangzhou/ZJU‐05/2020 was obtained to do the live virus neutralization assay. As shown in Figure 5d, these four IgG from Bin2 inhibited the infection of SARS‐CoV‐2 significantly at 12.5 nM. Finally, authentic infection of Vero‐E6 cells with live SARS‐CoV‐2 was neutralized with these IgG antibodies with IC50 from 12.5 to 50 nM (Figure 5e), suggesting that these neutralizing antibodies can effectively inhibit infection of the virus to host cells by targeting to the epitope on RBD, and these antibodies are potential therapeutic agents for the treatment of COVID‐19. To further substantiate that the antibody has cross‐neutralization activity, a neutralization assay against different mutant strains (N501Y, W436R, R408I, N354D, V367F and N354D/D364Y) by these antibodies was performed. As shown in Figure 6a, the infection of mutants N354D, V367F and N354D/D364Y could be inhibited by all 4 antibodies. While the mutants W436R and R408I could only be inhibited by HTS0483, suggesting great potential of HTS0483 in opposing mutant SARS‐CoV‐2 (Figure 6b). Consistent with the binding assay data, the neutralization of HTS0433 to N501Y mutant strain was also reduced obviously, suggesting that the point mutation in CDR3 of VL is important to block the infection of host cells. To further explore the cross‐neutralization of these antibodies to SARS‐CoV‐2, SARS‐CoV and MERS‐CoV, the four antibodies were used to do in vitro binding assay to RBD for different coronaviruses. As shown in Figure S1A, although HTS0422, HTS0433 and HTS0446 bind with RBD of SARS‐CoV‐2, SARS‐CoV and MERS‐CoV, HTS0483 only specifically bind to the RBD of SARS‐CoV‐2, suggesting that the binding site of HTS0483 is unique to SARS‐CoV‐2.
FIGURE 5.
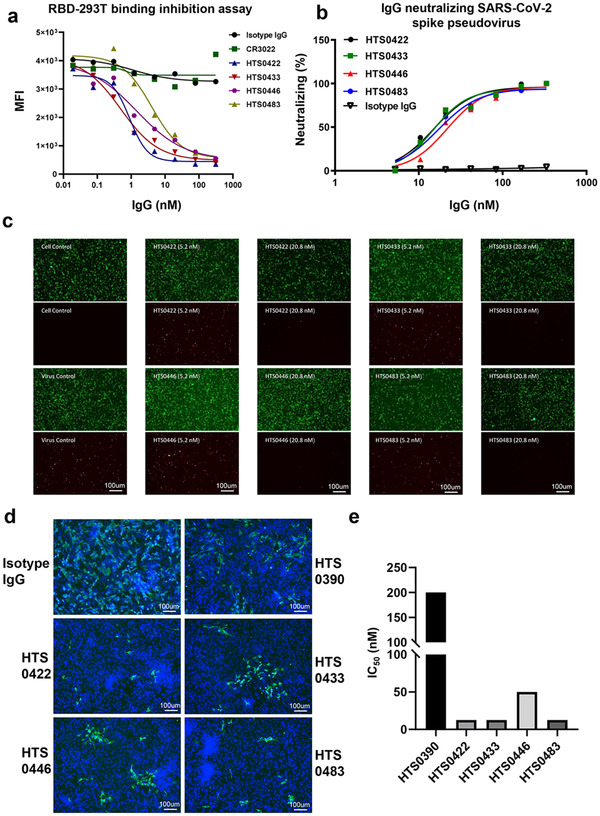
Antiviral properties of human IgG antibodies. (a) The inhibition of four positive antibodies from Bin2 to the binding of RBD to hACE2‐293T cells. (b and c) HIVs pseudotyped with the S glycoprotein (red) were incubated with anti‐RBD antibodies (HTS0422, HTS0433, HTS0446, HTS0483) diluted as indicated in the figure for 1 h before infection. The infected 293T‐ACE2 cells were observed under microscope and the neutralizing rate was calculated after FACS detection. (d) The authentic infection of SARS‐CoV‐2 (green) to Vero‐E6 cells was neutralized by Anti‐RBD antibodies (HTS0390, HTS0422, HTS0433 and HTS0483 are all at 1.875 μg ml‐1, HTS0446 is at 7.5 μg ml‐1). (e) The IC50 of each antibody from Bin2 was calculated by CPE assay
FIGURE 6.
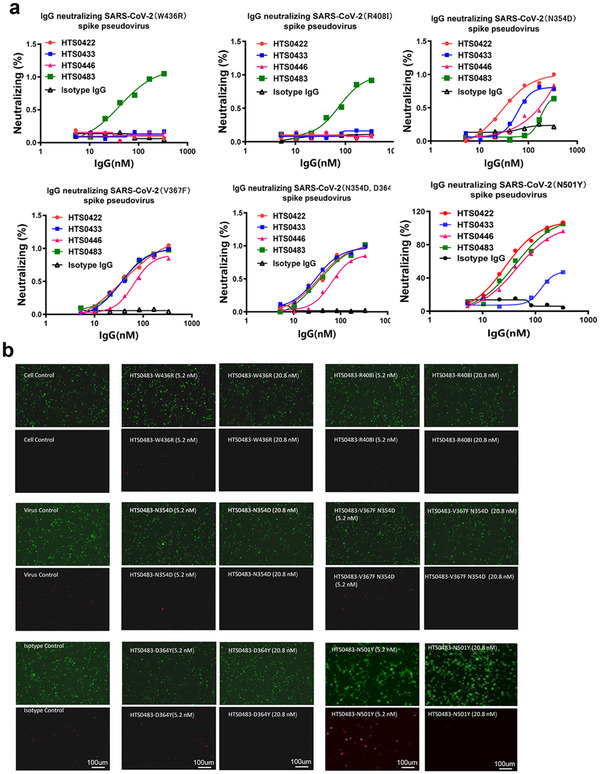
Antiviral properties of human IgG antibodies to pseudotyped SARS‐CoV‐2 RBD mutations. (a) Anti‐RBD antibodies (HTS0422, HTS0433, HTS0446 and HTS0483) neutralizes viruses pseudotyped with S glycoproteins from the SARS‐COV‐2 mutants. The infected 293T‐ACE2 cells were observed under microscope and the neutralizing rate was calculated after FACS detection. (b) HIVs pseudotyped with the S glycoprotein from the SARS‐COV‐2 mutants were incubated with HTS0483 at different concentration as indicated in the figure for 1 h before infection. Fluorescence intensity in target cells were measured, and the percent neutralization was calculated
3. DISCUSSION
Although different approaches have been taken to develop therapeutic antibodies for infectious diseases, phage display library persists as the common method for the generation of recombinant antibodies against various targets for biomedical applications and research.[ 28 ] Up till now, 9 fully human therapeutic antibodies discovered via phage display were approved by the Food and Drug Administration (FDA).[ 29 ] In this study, we constructed phage displayed human scFv libraries from 18 different COVID‐19 recovered patients to get neutralizing antibodies to SARS‐CoV‐2 infection. Four human antibodies with similar binding epitope were identified to have the potential as therapeutic antibodies for treating COVID‐19. Of which, the clone HTS0483 may have broader cross‐neutralization against not only wildtype SARS‐CoV‐2 RBD but also four other mutations.
Similar to other coronaviruses, SARS‐CoV‐2 uses the homotrimeric spike glycoprotein (comprising a S1 subunit and S2 subunit in each spike monomer) on the envelope to bind to their cellular receptors ACE2. The previous study demonstrated that the binding of SARS‐CoV and SARS‐CoV‐2 spike glycoproteins with ACE2 induces the dissociation of S1 with ACE2, prompting the S2 to transit from a metastable pre‐fusion to a more‐stable post‐fusion state that is essential for membrane fusion.[ 9 , 30 , 31 ] Furthermore, in vitro measurements indicate that the RBD is a key functional component within the S1 subunit and is responsible for binding of SARS‐CoV‐2 by ACE2.[ 32 ] Although computer modelling of the interaction between the SARS‐CoV‐2 RBD and ACE2 has identified some residues that are potentially involved in the interaction, the actual residues involved remained unclear. Thus, we cloned the receptor binding domain in S1 subunit of SARS‐CoV‐2 spike glycoproteins as a target for panning neutralizing antibodies from patient‐derived antibody libraries.
Although more than 400 positive clones were identified to bind with RBD by ELISA assay, only a part of them could block the interaction between RBD and ACE2 significantly, which is consistent with previous data that not all antibodies against RBD have the neutralizing ability to interact with ACE2.[ 33 ] Therefore, we sorted panel of selected antibodies according to epitope specificities by epitope binning assay. The more antibodies analyzed for cross‐blocking in a pairwise and combinatorial manner against their specific antigen, the higher the probability of discriminating their epitopes. Six different bins were identified, but most antibodies were divided into Bin1 and Bin2. Then the binding affinity and blocking ability of each antibody in Bin1 and Bin2 were measured. We demonstrated that the antibodies in Bin1 and Bin2 had similar binding affinity to RBD. However, the neutralizing or blocking ability of antibodies in Bin2 was much better than in Bin1, suggesting that the epitope of Bin2 is essential for ACE2 binding. Next, we checked the sequence of each antibody and found that there were 12 antibodies could be found with sequence pattern **GMDV in CDR3 of VH among all 19 antibodies including HTS0483 (Table S1). To our surprise, this sequence pattern is very similar to a newly published paper about stereotypic neutralizing VH antibodies against SARS‐CoV‐2 RBD in COVID‐19 patients and healthy individuals, suggesting the antibodies identified are expeditious and stereotypic expanded in SARS‐CoV‐2 infection.[ 20 ] Meanwhile, the sequence of HTS0422, HTS 0433 and HTS0446 are very similar with only a little difference in CDR3 of VL and frame region suggested that they may have the identical binding motif in the RBD domain. While the sequence of HTS0483 is totally different with the other 3 antibodies which may partially explain why only HTS0483 could blocking the entry of the mutants W436R and R408I. These data suggested that the W436R and R408I are essential for the bind of HTS0422, HTS0433 and HTS0446 (Figure S7b). Accordingly, the R408I mutation within RBD exhibiting low antigenicity which will eliminate the neutralization to SARS‐CoV‐2 by those antibodies.[ 34 ] Although the RBD domain is important for the viral infection, it seems that the lack of linear epitopes makes it hard to get neutralizing antibodies. Furthermore, the linear‐epitope‐specific antibodies which immunized mice failed to neutralize the authentic virus implied that conformational epitopes are crucial for developing neutralizing antibodies.[ 35 ] Therefore, if we can make some point mutation in RBD to explore the conformational epitopes in Bin2, the actual residues that mediate the interaction between RBD and ACE2 will be discovered.
So far, 36 nonsynonymous mutations were identified in RBD of the spike protein with a low prevalence (<1%) across all genomes.[ 36 ] And some of them such as N501Y in the United Kingdom has been proven to increase the infectivity and became the dominant lineage. In addition, the S protein is also important for antigen cognition. Thus, the variation of S protein may change the antigen of virus and influence the vaccine immune efficiency. It has been demonstrated that N439K mutation which has been found in more than 30 countries confer resistance against several neutralizing monoclonal antibodies, including one authorized for emergency use by the FDA.[ 37 ] Therefore, neutralization antibodies against more SARS‐CoV‐2 RBD mutations will have better potential in clinical application. In this study, although four antibodies are all from Bin2, the binding motif is identical to each other. For example, only HTS0483 bound with R408I and W436R mutant in RBD, suggesting that the binding of HTS0483 to RBD is independent of R481 and W436. Meanwhile, although the sequence of HTS0433 and HTS0466 is almost identical, with only three amino acids different in frame region, the affinity to mutant N501Y is totally different, which demonstrated that these amino acids in frame region of VL is critical to bind with N501Y. Furthermore, only HTS0483 blocked the binding of both W436R and R408I mutants to ACE2 implied that the mutation in RBD may eliminate some antibodies mediated blocking. Interestingly, the mutations W436R and R408I were isolated from Wuhan and India, respectively. While the recovered patients were all from Zhejiang province without the mutation in RBD. Therefore, the convalescent patients or vaccine immunized people without antibody to block the mutant virus may still have the possibility of SARS‐COV‐2 infection with W436R and R408I mutation. This could pose a huge challenge when developing effective vaccine against COVID‐19.
Some evidences have shown that the convalescent serum from a patient with a SARS‐CoV infection and horse anti‐SARS‐CoV serum could cross‐neutralize the infection of SARS‐CoV‐2, which implies that the structures of RBDs in SARS‐CoV‐2 and SARS‐CoV are similar.[ 3 , 8 ] While most antibodies targeting SARS‐CoV have little cross‐binding and neutralization activity against spike protein or RBD of SARS‐CoV‐2 (except SARS‐CoV antibody CR3022 which binds to the SARS‐CoV‐2 RBD with a KD of 6.2 nM).[ 38 , 39 ] In this study, we demonstrated that the binding affinity of CR3022 is lower than 4 antibodies from Bin2. Furthermore, the neutralization activity of CR3022 against RBD of SARS‐CoV‐2 to ACE2 is vanished when compared to our antibodies, implying that the binding motif of SARS‐CoV may be different from SARS‐CoV‐2. In addition, the pseudovirus entry and authentic virus infection of Vero‐E6 are both inhibited significantly by these antibodies. Then, the RBD sequence between SARS‐CoV‐2 and SARS‐CoV was compared and we found that the W436R and R408I are same in both, which suggested that the binding to these sites couldn't distinguish SARS‐CoV‐2 and SARS‐CoV.[ 40 ] Most importantly, the specific binding of HTS0483 to SARS‐CoV‐2 makes it possible to find a novel motif only displayed on SARS‐CoV‐2 but not SARS‐CoV or MERS‐CoV. Thus, the HTS0483 could be used as a tool to explore the unique properties of SARS‐CoV‐2.
In summary, patient‐derived antibody libraries were constructed for developing therapeutic antibodies against COVID‐19 pandemic and the neutralizing monoclonal antibodies not only have the potential for clinical application, but will also benefit the mechanism in exploring SARS‐CoV‐2 entry to host cells.
4. MATERIALS AND METHODS
4.1. Patients and blood samples
The study enrolled a total of 18 COVID‐19 patients (#202000004771, #202000004776, #202000004781, #202000004782, #202000004803, #202000004811, #202000004770, #202000004772, #202000004814, #202000004830, #202000004872, #202000005327, #202000005527, #202000004769, #202000004774, #202000004778, #202000004784 and #202000004786) and 3 healthy donors (HD87, HD89, HD90), and was approved by the Ethics Committee of the First Affiliated Hospital, Zhejiang University School of Medicine (2020‐llT‐42), the designated city hospital for treatment of COVID‐19 patients. All patients received antiviral and corticosteroid treatment, recovered and were discharged after SARS‐CoV‐2 infection status verified by RT‐PCR of routine nasal and pharyngeal swab specimens three times separately. Five milliliter peripheral blood samples from 18 recovered COVID‐19 patients were collected and separated into plasma and peripheral blood mononuclear cells (PBMCs) by Ficoll density gradient method. All plasma samples were heat‐inactivated at 56°C for 30 min before being stored at ‐80°C. PBMCs were maintained in RNAiso Plus (TAKARA) and stored at ‐80°C.
4.2. Anti‐RBD antibody measurement
18 plasma samples from COVID‐19 patients and 3 plasma samples from healthy donors were enrolled in this study. The SARS‐CoV‐2 antibody ELISA was performed according to the standard ELISA protocol. Briefly, 30 μl diluted plasma (1:200) were added to 384‐well plate coated with recombinant SARS‐CoV‐2 RBD and incubated for 1 h at 4°C. Wells were washed three times with PBST (0.1% Tween 20 in PBS) followed by the addition of HRP‐conjugated antibody against human IgG and subsequent incubation for 1 h at room temperature. Wells were washed three times and then TMB was added. Following 20 min of incubation at room temperature, the reaction was stopped and the color reaction was read at 450 nm on an ELISA reader.
4.3. scFv phage display library construction and quality assessment
Total RNA was prepared from PBMCs of 18 convalescent COVID‐19 patients followed by cDNA synthesis with a reverse transcription kit (RR036B, Takara) according to the manufacturer's instructions. The genes of variable regions of heavy chain and light chain (VH and VL) were amplified separately with the primers listed in Table S2 and then were further cloned into the phage‐display vector pCANTAB‐5E (GE Healthcare) with SfiI/XhoI or NheI/NotI digestion. The recombinant DNAs were transformed into Escherichia coli (E. coli) TG1 by electroporation. The transformants were plated on dishes containing 2‐YT agar (1.6% tryptone, 1% yeast extract, 0.5% NaCl, 1.5% bacteriological agar) with 100 μg ml‐1 ampicillin and 2% glucose. Following overnight incubation, all the colonies were scraped off the dishes, counted and finally stored at −80°C as a primary library. Ninety six single colonies from the primary library were randomly picked out, then the scFv gene were amplified with polymerase chain reaction (PCR) to detect the insert rate with electrophoresis and these colonies were also send out for DNA sequencing to calculate the correct rate of insert genes. Subsequently, the same phages TG1 transformants were superinfected with M13KO7 helper phage in a MOI (multiplicity of infection) ratio of 5:1, and scFv‐phages produced in overnight culture were purified with 20% PEG8000 and 2.5 M NaCl. ScFv molecules displayed by rescued phages were detected via the Myc tag in the scFv‐pIII fusion protein using ELISA.
4.4. Selection of phage scFv to SARS‐CoV‐2
The 96‐well plate were coated with RBD in PBS, blocked for 2 h with 2% BSA, and incubated for 3 h at room temperature with 1012 phage scFv (the libraries were panned individually) in 100 μl PBS containing 2% non‐fat milk. After intensive washes with PBST (0.1% Tween 20 in PBS), bound phage antibodies were eluted with 0.1 M Glycine‐HCl, pH 2.2, and immediately neutralized with 1.0 M Tris–Cl, pH 8.8. Eluted phage scFv were subjected to the next round of infection, rescue, and selection. After three rounds of panning, the higher binders to SARS‐CoV‐2 were selected by ELISA.
4.5. Phage ELISA to identify RBD binders
For ELISA assay, 96‐well ELISA plates (3923, Corning) were coated with anti‐his antibodies (100 μg 100 μl‐1 PBS) (A00186‐100, GenScript) overnight at 4°C. Blocking was conducted with 200 μl of 3% MPBS each well and Incubated at RT for 1 h. After washing three times with PBST, the wells were incubated with 50 μl of 3% MPBS and 50 μl phage supernatant each well at 37°C for 2 h. The wells were then washed four times with PBST and incubated with a 5000‐fold diluted HRP‐conjugated anti‐M13 monoclonal antibody (11973‐MM05T‐H, Sino) at 37°C for 1 h. The wells were washed four times with PBST again, and incubated with HRP chromogenic substrate tetramethylbenzidine (TMB, A610644‐0100, BBI) at room temperature over 10–20 min. The resulting optical density was then measured and absorbance was read at 450 nm.
4.6. Expression and purification of whole IgG antibodies
In order to obtain a large number of antibodies, the genes for VH of selected scFv were cloned into pTT5‐hIgG1 vector (containing the DNA coding sequence for IgG1‐CH) while VL genes were cloned into pTT5‐hKappa vector separately. The recombinant plasmids were then co‐transfected into 293F cells together with PEI. After 5 days’ culture, the antibodies in supernatant were collected and purified by protein A resin for whole IgG. At last, the antibodies were dialyzed to PBS for 16 h and filtration sterilized with 0.22 μm filter for long term preservation.
4.7. Antibody binding ELISA with RBD or RBD mutants
96‐well plates were coated with RBD or other RBD mutants in PBS, blocked for 2 h with 2% BSA, and incubated with a serial dilution of purified whole IgG antibodies. After three washes with PBST, the bound antibodies were detected by HRP‐conjugated anti‐human Fc antibody followed by incubation with TMD before the color reaction was measured.
To detect the binding of antibodies to the RBD of SARS‐CoV‐2, SARS‐CoV and MERS‐CoV, 96‐well plates were coated with SARS and MERS RBD in PBS, blocked 2 h with 2% BSA, and incubated with a serial dilution of purified whole IgG antibodies. After three washes with PBST, the bound antibodies were detected by HRP‐conjugated anti‐human Fc antibody followed by incubation with TMD and the color reaction was measured.
4.8. Competitive inhibition ELISA assay
RBD or RBD mutants were incubated for 1 h at room temperature with a serial dilution of purified whole IgG antibodies. The above mixture was subjected to 96‐well plate coated with ACE2 and incubated for another 1 h at 4°C. HRP‐conjugated anti‐mouse Fc was added as secondary antibody. The following procedures were the same as described in ELISA.
4.9. Competitive inhibition FACS assay
The human ACE2 gene (NM_021804.1) was synthesized and cloned into pCDH‐CMV‐MCS‐EF1‐CopGFP‐T2A‐Puromycin. Then the hACE2 containing plasmid together with psPAX2 and pMD2.G were packaged into lentivirus to infect 293T cells. And the positive clones were selected by puromycin to get hACE2 expressing cells (293T‐ACE2). 293T‐ACE2 cells were plated at 1.5 × 105 cells per well in 96‐well plates. The antibodies were diluted in PBS starting at a ratio of 1:4. Serial 4‐fold dilutions were mixed with 0.1 μg ml‐1 RBD equally, and incubated at 4°C for 1 h. The mixture was added to 293T‐ACE2 cells, incubated at 4°C for 1 h, washed 3 times with PBS before APC‐conjugated anti‐mouse Fc antibody was added. Following 1 h of incubation at 4°C, plates were washed with PBS then developed with FACS.
4.10. BLI assays
Octet systems (Octet RED96e, ForteBio) equipped with amine‐reactive, streptavidin, and anti‐species sensors were purchased from ForteBio Analytics (Shanghai) Co., Ltd. Epitope binning experiments of anti‐SARS‐CoV‐2 Antibodies were performed in 96‐channel mode with in tandem format. HIS1K Biosensors were loaded SARS‐CoV‐2 RBD protein, His Tag (MALS verified) (Cat. No. S1N‐C52H4, ACROBiosystems). Interactions with the first and second antibodies were completed in sequence; then, the detection of the binding signal of the second antibody determined whether the two antibodies recognize the same epitope. The in tandem style assay comprised a five‐step binding cycle; 1) a buffer baseline was established for 1 min, 2) 5 μg ml‐1 SARS‐CoV‐2 RBD protein was captured about 0.4 nm, 3) 7.5‐15 μg ml‐1 mAb array was loaded to saturate the immobilized antigen for 3 min, 4) 15 μg ml‐1 of the test mAb was bound for 1 min, and 5) the capture surfaces were regenerated for 30 s. HIS1K biosensors were regenerated with 10 mM Glycine‐HCl, pH1.5 for 5 s with three times and neutralized for 5 s with three times in neutralization buffer immediately after each regeneration. In tandem assays were conducted depending on the experiment.
4.11. Preparation of RFP/SARS‐CoV‐2 spike pseudovirus
Production of HIV pseudotyped with SARS‐CoV‐2 spike (wildtype and mutants N501Y, W436R, R408I, N354D, V367F and N354D/D364Y were all purchased from Acro biosystems) was performed using the standard lentivirus production method. Briefly, 293T cells were transfected with psPAX2 and vectors encoding SARS‐CoV‐2 spike protein as well as a core plasmid expressing RFP. After 48 h post‐infection, RFP/SARS‐CoV‐2 spike pseudoviruses were packaged and collected from the culture supernatant. All the sequence of RBD and S protein used in this study was listed in the Table S3.
4.12. Neutralization activity of mAbs against pseudovirus
RFP/SARS‐CoV‐2 spike pseudovirus system was used as an infection model to evaluate the neutralization activity of antibodies. First, the RFP/SARS‐CoV‐2 spike pseudoviruses were pre‐incubated with a serial dilution of purified whole IgG antibodies. Then, 293T‐ACE2 cells were infected by the RFP/SARS‐CoV‐2 spike pseudovirus with or without antibodies. After 2 h incubation at 37°C, the medium was refreshed with DMEM containing 10% FBS and continuously cultured for another 72 h. The infected 293T‐ACE2 cells were observed under microscope and the neutralizing IC50 was calculated after FACS detection.
4.13. Neutralization activity of mAbs against live SARS‐CoV‐2
SARS‐CoV‐2 obtained from a sputum sample was amplified in Vero‐E6 to make working stocks of the virus. To analyze the mAb neutralizing activities, two‐fold serial dilutions of mAbs were added to the same volume of 100 TCID50 of SARS‐CoV‐2 and incubated for 1 h at 37°C. The mixture was added to a monolayer of Vero‐E6 cells in a 96‐well plate and incubated at 37°C. Cytopathic effects (CPE) were observed and recorded from day 4 to day 6. For immunofluorescence (IF) analysis, Vero‐E6 cells were transfected and fixed on day 6 post‐transfection in 80% acetone for 10 min at room temperature. Cells were immunolabeled for 1.5 h at room temperature with the rabbit anti SARS‐CoV‐2 spike antibody, washed three times with PBS, and followed by the addition of Alexa Fluor 488‐conjugated goat anti‐rabbit IgG antibody. Wells were washed three times and then DAPI was added. Following 30 min of incubation at room temperature, the reaction was reading in fluorescent microscopy.
4.14. Statistics and data availability
Statistical analyses were analyzed by GraphPad Prism 8.0.2 (GraphPad Software). Data are shown as the mean ± SEM of three independent experiments. The authors declare that the main data supporting the findings of this study are available within the article and its Supporting Information files. Extra data are available from the corresponding author upon request.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Mingyao Liu, Yunqing Qiu, Tingbo Liang and Bing Du supervised the project. Bing Du, Ruihua Chao and Wenxiang Zhao conceived and designed the experiments. Yan Lou, Wenxiang Zhao, Haitao Wei, Min Chu, Ruihua Chao, Hangping Yao, Junwei Su, Yanan Li, Xiulan Li, Yu Cao, Yanyan Feng, Ping Wang, Yongyang Xia, Yushuan Shang, Fengping Li, Pingju Ge, Xinglin Zhang and Wenjing Gao performed the experiments. Bing Du, Wenxiang Zhao, Gaojie Song, Ruihua Chao, and Mingyao Liu analyzed the data. Bing Du and Ruihua Chao wrote the manuscript.
Supporting information
Supplementary figure 1. Cross reaction of human IgG antibodies to S protein from coronavirus. (A) Binding of antibodies to S protein of SARS‐CoV‐2, SARS‐CoV and MERS‐CoV is detected by ELISA. (B) The trimeric S protein is shown as yellow (PDB: 6VSB) with three RBDs colored red, blue and green, respectively. One of the RBD domain is zoomed‐in to show the positions of the introduced mutations. These mutations are shown as spheres in the original figure (left) and sticks in the zoom‐in figure (right). The pink circle indicates binding area of antibodies HTS0422, HTS0433, HTS0446, based on binding and inhibition assays.
ACKNOWLEDGMENTS
This work was supported by National Key R&D Program of China [2018YFA0507001, 2019YFA0802800]; Zhejiang University special scientific research fund for COVID‐19 prevention and control (2020XGZX086); Zhejiang Provincial Science and technology department key R&D plan emergency project [2020c03123‐8]; Innovation Program of Shanghai Municipal Education Commission [2017‐01‐07‐00‐05‐E00011]; Fundamental Research Funds for the Central Universities.
Lou, Y. , Zhao, W. , Wei, H. , Chu, M. , Chao, R. , Yao, H. , Su, J. , Li, Y. , Li, X. , Cao, Yu , Feng, Y. , Wang, P. , Xia, Y. , Shang, Y. , Li, F. , Ge, P. , Zhang, X. , Gao, W. , Song, G. , … Liu, M. . (2021). Cross‐neutralization of RBD mutant strains of SARS‐CoV‐2 by convalescent patient derived antibodies. Biotechnol J, 16, e202100207. 10.1002/biot.202100207
Yan Lou and Wenxiang Zhao contributed equally to this study.
Contributor Information
Bing Du, Email: bdu@bio.ecnu.edu.cn.
Tingbo Liang, Email: liangtingbo@zju.edu.cn.
Yunqing Qiu, Email: qiuyq@zju.edu.cn.
Mingyao Liu, Email: myliu@bio.ecnu.edu.cn.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Feng, Y. , Jiang, H. , Qiu, M. , Liu, L. , Zou, S. , Li, Y. , Guo, Q. , Han, N. , Sun, Y. , Wang, K. , Lu, L. , Zhuang, X. , Zhang, S. , Chen, S. , & Mo, F. (2021). Multi‐epitope vaccine design using an immunoinformatic approach for SARS‐CoV‐2. Pathogens, 10, 737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu, A. , Peng, Y. , Huang, B. , Ding, X. , Wang, X. , Niu, P. , Meng, J. , Zhu, Z. , Zhang, Z. , Wang, J. , Sheng, J. , Quan, L. , Xia, Z. , Tan, W. , Cheng, G. , & Jiang, T. (2020). Genome composition and divergence of the novel coronavirus (2019‐nCoV) originating in China. Cell Host Microbe, 27, 325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou, P. , Yang, X. L. , Wang, X. G. , Hu, B. , Zhang, L. , Zhang, W. , Si, H. R. , Zhu, Y. , Li, B. , Huang, C. L. , Chen, H. D. , Chen, J. , Luo, Y. , Guo, H. , Jiang, R. D. , Liu, M. Q. , Chen, Y. , Shen, X. R. , Wang, X. … Shi, Z. L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579, 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chinazzi, M. , Davis, J. T. , Ajelli, M. , Gioannini, C. , Litvinova, M. , Merler, S. , Pastore, Y. P. A. , Mu, K. , Rossi, L. , Sun, K. , Viboud, C. , Xiong, X. , Yu, H. , Halloran, M. E. , Longini, I. M., Jr. , & Vespignani, A. (2020). The effect of travel restrictions on the spread of the 2019 novel coronavirus (COVID‐19) outbreak. Science, 368, 395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xu, Z. , Shi, L. , Wang, Y. , Zhang, J. , Huang, L. , Zhang, C. , Liu, S. , Zhao, P. , Liu, H. , Zhu, L. , Tai, Y. , Bai, C. , Gao, T. , Song, J. , Xia, P. , Dong, J. , Zhao, J. , & Wang, F. S. (2020). Pathological findings of COVID‐19 associated with acute respiratory distress syndrome. The lancet. Respiratory Medicine, 8, 420–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chan, J. F. , Yuan, S. , Kok, K. H. , To, K. K. , Chu, H. , Yang, J. , Xing, F. , Liu, J. , Yip, C. C. , Poon, R. W. , Tsoi, H. W. , Lo, S. K. , Chan, K. H. , Poon, V. K. , Chan, W. M. , Ip, J. D. , Cai, J. P. , Cheng, V. C. , Chen, H. … Yuen, K. Y. (2020). A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person‐to‐person transmission: A study of a family cluster. Lancet, 395, 514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sadarangani, M. , Marchant, A. , & Kollmann, T. R. (2021). Immunological mechanisms of vaccine‐induced protection against COVID‐19 in humans. Nature Reviews Immunology, 21, 475–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hoffmann, M. , Kleine‐Weber, H. , Schroeder, S. , Krüger, N. , Herrler, T. , Erichsen, S. , Schiergens, T. S. , Herrler, G. , Wu, N. H. , Nitsche, A. , Müller, M. A. , Drosten, C. , & Pöhlmann, S. (2020). SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 181, 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Walls, A. C. , Park, Y. J. , Tortorici, M. A. , Wall, A. , McGuire, A. T. , & Veesler, D. (2020). Structure, function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell, 181, 281–292 e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Leung, K. , Shum, M. H. , Leung, G. M. , Lam, T. T. , & Wu, J. T. (2021). Early transmissibility assessment of the N501Y mutant strains of SARS‐CoV‐2 in the United Kingdom, October to November 2020. Euro Surveillance, 26, 2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen, J. , Wang, R. , Wang, M. , & Wei, G. W. (2020). Mutations strengthened SARS‐CoV‐2 infectivity. Journal of Molecular Biology, 432, 5212–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ju, B. , Zhang, Q. , Ge, J. , Wang, R. , Sun, J. , Ge, X. , Yu, J. , Shan, S. , Zhou, B. , Song, S. , Tang, X. , Yu, J. , Lan, J. , Yuan, J. , Wang, H. , Zhao, J. , Zhang, S. , Wang, Y. , Shi, X. … Zhang, L. (2020). Human neutralizing antibodies elicited by SARS‐CoV‐2 infection. Nature, 584, 115–119. [DOI] [PubMed] [Google Scholar]
- 13. Cheng, Y. , Wong, R. , Soo, Y. O. , Wong, W. S. , Lee, C. K. , Ng, M. H. , Chan, P. , Wong, K. C. , Leung, C. B. , & Cheng, G. (2005). Use of convalescent plasma therapy in SARS patients in Hong Kong. European Journal of Clinical Microbiology & Infectious Diseases, 24, 44–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ni, L. , Ye, F. , Cheng, M. L. , Feng, Y. , Deng, Y. Q. , Zhao, H. , Wei, P. , Ge, J. , Gou, M. , Li, X. , Sun, L. , Cao, T. , Wang, P. , Zhou, C. , Zhang, R. , Liang, P. , Guo, H. , Wang, X. , Qin, C. F. … Dong, C. (2020). Detection of SARS‐CoV‐2‐specific humoral and cellular immunity in COVID‐19 convalescent individuals. Immunity, 52, 971–977 e973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Robbiani, D. F. , Gaebler, C. , Muecksch, F. , Lorenzi, J. C. C. , Wang, Z. , Cho, A. , Agudelo, M. , Barnes, C. O. , Gazumyan, A. , Finkin, S. , Hagglof , T., Oliveira, T. Y. , Viant, C. , Hurley, A. , Hoffmann, H. H. , Millard, K. G. , Kost, R. G. , Cipolla, M. … Nussenzweig, M. C. (2020). Convergent antibody responses to SARS‐CoV‐2 in convalescent individuals. Nature, 584, 437–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yao, H. , Sun, Y. , Deng, Y. Q. , Wang, N. , Tan, Y. , Zhang, N. N. , Li, X. F. , Kong, C. , Xu, Y. P. , Chen, Q. , Cao, T. S. , Zhao, H. , Yan, X. , Cao, L. , Lv, Z. , Zhu, D. , Feng, R. , Wu, N. , Zhang, W. … Wang, X. (2021). Rational development of a human antibody cocktail that deploys multiple functions to confer Pan‐SARS‐CoVs protection. Cell Research, 31, 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shi, R. , Shan, C. , Duan, X. , Chen, Z. , Liu, P. , Song, J. , Song, T. , Bi, X. , Han, C. , Wu, L. , Gao, G. , Hu, X. , Zhang, Y. , Tong, Z. , Huang, W. , Liu, W. J. , Wu, G. , Zhang, B. , Wang, L. … Yan, J. (2020). A human neutralizing antibody targets the receptor‐binding site of SARS‐CoV‐2. Nature, 584, 120–124. [DOI] [PubMed] [Google Scholar]
- 18. Coughlin, M. , Lou, G. , Martinez, O. , Masterman, S. K. , Olsen, O. A. , Moksa, A. A. , Farzan, M. , Babcook, J. S. , & Prabhakar, B. S. (2007). Generation and characterization of human monoclonal neutralizing antibodies with distinct binding and sequence features against SARS coronavirus using XenoMouse. Virology, 361, 93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou, G. , & Zhao, Q. (2020). Perspectives on therapeutic neutralizing antibodies against the novel Coronavirus SARS‐CoV‐2. International Journal of Biological Sciences, 16, 1718–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim, S. I. , Noh, J. , Kim, S. , Choi, Y. , Yoo, D. K. , Lee, Y. , Lee, H. , Jung, J. , Kang, C. K. , Song, K. H. , Choe, P. G. , Kim, H. B. , Kim, E. S. , Kim, N. J. , Seong, M. W. , Park, W. B. , Oh, M. D. , Kwon, S. , & Chung, J. (2021). Stereotypic neutralizing VH antibodies against SARS‐CoV‐2 spike protein receptor binding domain in patients with COVID‐19 and healthy individuals. Science Translational Medicine, 13, eabd6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marks, J. D. , Hoogenboom, H. R. , Bonnert, T. P. , McCafferty, J. , Griffiths, A. D. , Winter, G. (1991). By‐passing immunization. Human antibodies from V‐gene libraries displayed on phage. Journal of Molecular Biology, 222, 581–597. [DOI] [PubMed] [Google Scholar]
- 22. Sui, J. , Li, W. , Murakami, A. , Tamin, A. , Matthews, L. J. , Wong, S. K. , Moore, M. J. , Tallarico, A. S. , Olurinde, M. , Choe, H. , Anderson, L. J. , Bellini, W. J. , Farzan, M. , & Marasco, W. A. (2004). Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proceedings of the National Academy of Sciences of the United States of America, 101, 2536–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Noy‐Porat, T. , Makdasi, E. , Alcalay, R. , Mechaly, A. , Levy, Y. , Bercovich‐Kinori, A. , Zauberman, A. , Tamir, H. , Yahalom‐Ronen, Y. , Israeli, M. , Epstein, E. , Achdout, H. , Melamed, S. , Chitlaru, T. , Weiss, S. , Peretz, E. , Rosen, O. , Paran, N. , Yitzhaki, S. … Rosenfeld, R. (2020). A panel of human neutralizing mAbs targeting SARS‐CoV‐2 spike at multiple epitopes. Nature Communications, 11, 4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Parray, H. A. , Chiranjivi, A. K. , Asthana, S. , Yadav, N. , Shrivastava, T. , Mani, S. , Sharma, C. , Vishwakarma, P. , Das, S. , Pindari, K. , Sinha, S. , Samal, S. , Ahmed, S. , & Kumar, R. (2020). Identification of an anti‐SARS‐CoV‐2 receptor‐binding domain‐directed human monoclonal antibody from a naive semisynthetic library. Journal of Biological Chemistry, 295, 12814–12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen, Y. , Huang, K. , Li, X. , Lin, X. , Zhu, Z. , & Wu, Y. (2010). Generation of a stable anti‐human CD44v6 scFv and analysis of its cancer‐targeting ability in vitro. Cancer Immunology, Immunotherapy, 59, 933–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang, Y. Y. , Li, B. R. , & Ning, B. T. (2020). The Comparative Immunological Characteristics of SARS‐CoV, MERS‐CoV, and SARS‐CoV‐2 Coronavirus Infections. Frontiers in Immunology, 11, 2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zeng, L. , Li, D. , Tong, W. , Shi, T. , & Ning, B. (2021). Biochemical features and mutations of key proteins in SARS‐CoV‐2 and their impacts on RNA therapeutics. Biochemical Pharmacology, 189, 114424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao, A. , Tohidkia, M. R. , Siegel, D. L. , Coukos, G. , & Omidi, Y. (2016). Phage antibody display libraries: a powerful antibody discovery platform for immunotherapy. Critical Reviews in Biotechnology, 36, 276–289. [DOI] [PubMed] [Google Scholar]
- 29. Frenzel, A. , Kugler, J. , Helmsing, S. , Meier, D. , Schirrmann, T. , Hust, M. , & Dubel, S. (2017). Designing human antibodies by phage display. Transfusion Medicine and Hemotherapy, 44, 312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gui, M. , Song, W. , Zhou, H. , Xu, J. , Chen, S. , Xiang, Y. , & Wang, X. (2017). Cryo‐electron microscopy structures of the SARS‐CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Research, 27, 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yuan, Y. , Cao, D. , Zhang, Y. , Ma, J. , Qi, J. , Wang, Q. , Lu, G. , Wu, Y. , Yan, J. , Shi, Y. , Zhang, X. , & Gao, G. F. (2017). Cryo‐EM structures of MERS‐CoV and SARS‐CoV spike glycoproteins reveal the dynamic receptor binding domains. Nature Communications, 8, 15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lan, J. , Ge, J. , Yu, J. , Shan, S. , Zhou, H. , Fan, S. , Zhang, Q. , Shi, X. , Wang, Q. , Zhang, L. , & Wang, X. (2020). Structure of the SARS‐CoV‐2 spike receptor‐binding domain bound to the ACE2 receptor. Nature, 581, 215–220. [DOI] [PubMed] [Google Scholar]
- 33. Barnes, C. O. , Jette, C. A. , Abernathy, M. E. , Dam, K. A. , Esswein, S. R. , Gristick, H. B. , Malyutin, A. G. , Sharaf, N. G. , Huey‐Tubman, K. E. , Lee, Y. E. , Robbiani, D. F. , Nussenzweig, M. C. , West, A. P., Jr. , & Bjorkman, P. J. (2020). SARS‐CoV‐2 neutralizing antibody structures inform therapeutic strategies. Nature, 588, 682–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Singh, P. K. , Kulsum, U. , Rufai, S. B. , Mudliar, S. R. , & Singh, S. (2020). Mutations in SARS‐CoV‐2 leading to antigenic variations in spike protein: A challenge in vaccine development. Journal of laboratory physicians, 12, 154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li, Y. , Ma, M. L. , Lei, Q. , Wang, F. , Hong, W. , Lai, D. Y. , Hou, H. , Xu, Z. W. , Zhang, B. , Chen, H. , Yu, C. , Xue, J. B. , Zheng, Y. X. , Wang, X. N. , Jiang, H. W. , Zhang, H. N. , Qi, H. , Guo, S. J. , Zhang, Y. … Tao, S. C. (2021). Linear epitope landscape of the SARS‐CoV‐2 spike protein constructed from 1,051 COVID‐19 patients. Cell Reports, 34, 108915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mastriani, E. , Rakov, A. V. , & Liu, S. L. (2021). Isolating SARS‐CoV‐2 strains from countries in the same meridian: Genome evolutionary analysis. JMIR Bioinform Biotech, 2, e25995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thomson, E. C. , Rosen, L. E. , Shepherd, J. G. , Spreafico, R. , da Silva Filipe, A. , Wojcechowskyj, J. A. , Davis, C. , Piccoli, L. , Pascall, D. J. , Dillen, J. , Lytras, S. , Czudnochowski, N. , Shah, R. , Meury, M. , Jesudason, N. , De Marco, A. , Li, K. , Bassi, J. , O'Toole, A. … Snell, G. (2021). Circulating SARS‐CoV‐2 spike N439K variants maintain fitness while evading antibody‐mediated immunity. Cell, 184, 1171–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tian, X. , Li, C. , Huang, A. , Xia, S. , Lu, S. , Shi, Z. , Lu, L. , Jiang, S. , Yang, Z. , Wu, Y. , & Ying, T. (2020). Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus‐specific human monoclonal antibody. Emerg Microbes Infect, 9, 382–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van den Brink, E. N. , Ter Meulen, J. , Cox, F. , Jongeneelen, M. A. , Thijsse, A. , Throsby, M. , Marissen, W. E. , Rood, P. M. , Bakker, A. B. , Gelderblom, H. R. , Martina, B. E. , Osterhaus, A. D. , Preiser, W. , Doerr, H. W. , de Kruif, J. , & Goudsmit, J. (2005). Molecular and biological characterization of human monoclonal antibodies binding to the spike and nucleocapsid proteins of severe acute respiratory syndrome coronavirus. Journal of Virology, 79, 1635–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yuan, M. , Wu, N. C. , Zhu, X. , Lee, C. D. , So, R. T. Y. , Lv, H. , Mok, C. K. P. , & Wilson, I. A. (2020). A highly conserved cryptic epitope in the receptor binding domains of SARS‐CoV‐2 and SARS‐CoV. Science, 368, 630–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1. Cross reaction of human IgG antibodies to S protein from coronavirus. (A) Binding of antibodies to S protein of SARS‐CoV‐2, SARS‐CoV and MERS‐CoV is detected by ELISA. (B) The trimeric S protein is shown as yellow (PDB: 6VSB) with three RBDs colored red, blue and green, respectively. One of the RBD domain is zoomed‐in to show the positions of the introduced mutations. These mutations are shown as spheres in the original figure (left) and sticks in the zoom‐in figure (right). The pink circle indicates binding area of antibodies HTS0422, HTS0433, HTS0446, based on binding and inhibition assays.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


