Abstract
TRIANNI mice carry an entire set of human immunoglobulin V region gene segments and are a powerful tool to rapidly isolate human monoclonal antibodies. After immunizing these mice with DNA encoding the spike protein of SARS‐CoV‐2 and boosting with spike protein, we identified 29 hybridoma antibodies that reacted with the SARS‐CoV‐2 spike protein. Nine antibodies neutralize SARS‐CoV‐2 infection at IC50 values in the subnanomolar range. ELISA‐binding studies and DNA sequence analyses revealed one cluster of three clonally related neutralizing antibodies that target the receptor‐binding domain and compete with the cellular receptor hACE2. A second cluster of six clonally related neutralizing antibodies bind to the N‐terminal domain of the spike protein without competing with the binding of hACE2 or cluster 1 antibodies. SARS‐CoV‐2 mutants selected for resistance to an antibody from one cluster are still neutralized by an antibody from the other cluster. Antibodies from both clusters markedly reduced viral spread in mice transgenic for human ACE2 and protected the animals from SARS‐CoV‐2‐induced weight loss. The two clusters of potent noncompeting SARS‐CoV‐2 neutralizing antibodies provide potential candidates for therapy and prophylaxis of COVID‐19. The study further supports transgenic animals with a human immunoglobulin gene repertoire as a powerful platform in pandemic preparedness initiatives.
Keywords: COVID‐19, neutralizing antibodies, spike protein, SARS‐CoV‐2, variants of concern
Transgenic mice with a complete human antibody repertoire (Trianni platform) were immunized with the spike protein (S protein) of SARS‐CoV‐2 and hybridoma clones were established. Two clusters of clonally related SARS‐CoV‐2‐neutralizing monoclonal antibodies were isolated. Cluster 1 binds to the N‐terminal domain (NTD) and Cluster 2 to the receptor‐binding domain (RBD) of the CoV‐2 spike protein. The antibodies also significantly reduce the viral spread in hACE2‐transgenic mice.
Introduction
Since December 2019 [1], SARS‐CoV‐2 has rapidly spread throughout the world, leading to more than 133,971,287 confirmed cases of COVID‐19 and 2.902.493 million deaths [2]. Several vaccines have been developed and licensed worldwide at an unprecedented speed [3, 4]. In countries or age groups with high vaccine coverage, case counts have decreased substantially [5, 6]. For all licensed vaccines, the vaccine antigen is the ectodomain of the spike protein. Its receptor‐binding domains (RBDs) interacting with the cellular receptor human Angiotensin‐converting enzyme 2 (hACE2) and its N‐terminal domain (NTD) were identified as primary targets of the neutralizing activity of convalescent sera and monoclonal antibodies [7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19]. Clinical development of some of these monoclonal antibodies is ongoing, already providing evidence of their utility in therapeutic trials [20, 21, 22]. Some have since been licensed for the treatment of COVID‐19 (reviewed in [23]).
After crossing species barriers, zoonotic viruses adapt to new species. Early after the new entry, adaptation to efficient transmission is most likely the most substantial selective pressure, explaining the rapid emergence of SARS‐CoV‐2 variants such as those carrying the D614G mutation [24] or more complex mutational signatures in the spike protein [25, 26]. Once a larger percentage of a population has overcome the first infection with an emerging virus, immune escape variants of this virus will have a selective advantage. Evidence that immune escape may already contribute to the emergence of new SARS‐CoV‐2 variants has been reported for the Beta (B.1.351), the Gamma (P.1), and the Delta variant [27, 28, 29, 30]. Since the antibody response to current mRNA vaccines mimics the antibody response to natural infection [31], the vaccines' efficacy against these variants may be impaired [27, 31‐33]. Similarly, the neutralizing potency of monoclonal antibodies may be reduced or completely lost [15, 34, 35].
Concerning the loss of vaccine efficacy against emerging variants, it has been argued that mRNA and viral vector vaccines can be rapidly adapted to include the spike proteins of the variants of concern (VOC). While this should work for individuals with no prior exposure to SARS‐CoV‐2 or COVID‐19 vaccines, the outcome is less predictable in the presence of pre‐existing memory B cells specific for the founder spike protein. Therefore, more targeted vaccine approaches employing structure‐based designs of spike vaccines against emerging VOCs and reliable animal models for validation and optimization are required. Transgenic mice with a complete human antibody repertoire could be such a model.
Here, we report the rapid isolation of fully human SARS‐CoV‐2 neutralizing monoclonal antibodies from immunized TRIANNI mice harboring the complete human antibody repertoire (patent US 2013/0219535 A1; Fig. 1A) [36, 37]. The neutralizing antibodies target the RBD or the NTD of the spike protein, providing a pair of noncompeting human monoclonal antibodies for clinical development. Further characterization of the antibodies´ human VH gene usage and divergence from germline sequences support the concept of mouse models with a human antibody repertoire in pandemic preparedness efforts and suggest the use of these models in structure‐based optimization of vaccine antigens against emerging SARS‐CoV‐2 variants.
Figure 1.
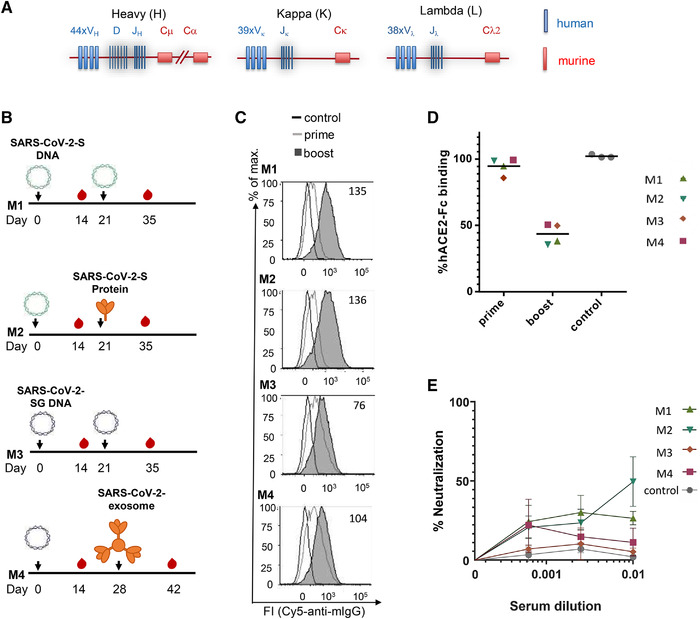
Immunization of TRIANNI mice for induction of SARS‐CoV‐2 neutralizing antibodies. TRIANNI mice harboring the entire human Ig variable region repertoire (A) were primed by intramuscular electroporation with expression plasmids for WT SARS‐CoV‐2‐S (M1, M2) or a hybrid SARS‐CoV‐2‐S containing the intracytoplasmic domain of VSV‐G (M3, M4). (B) Mice were boosted with the expression plasmids used for priming (M1, M3), soluble trimeric S protein (M2), or exosomes carrying the hybrid SARS‐CoV‐2‐S protein (M4). (C) A flow cytometric assay assessed the binding of sera at a 1:200 dilution to the SARS‐CoV‐2‐S protein with HEK‐293T cells transiently expressing the S protein. Numbers indicate the relative mean fluorescence intensities of sera drawn 2 weeks after the booster immunizations. (D) Competitive inhibition of hACE2‐Fc binding to trimeric S protein by sera (1:200) from control mice and mice at the indicated time points after the first immunization. The mean percentage of binding compared to control binding is shown (two experiments each performed in triplicates). (E) For the neutralization assay, Vero‐E6 cells were infected with the SARS‐CoV‐2 isolate MUC‐IMB‐1 in the presence or absence of week 5, sera, and three control sera from TRIANNI mice immunized with an irrelevant immunogen. SARS‐CoV‐2 infection was quantitated after 20 to 24 h by staining with purified IgG from a convalescent COVID‐19 patient and a fluorescence‐labeled anti‐human IgG using an ELISPOT reader. The mean and SEM of triplicates of one experiment are shown. The control sera represent the mean and SEM of the mean of three control sera each tested in triplicates.
Results
Immunization
Based on the published nucleotide sequence of SARS‐CoV‐2 [38], the first immunogens available early during the current pandemic were expression plasmids encoding the spike protein, the primary target of neutralizing antibodies to coronaviruses [39, 40]. In addition to a DNA vaccine encoding the WT S protein (SARS‐CoV‐2‐S DNA), we generated a DNA vaccine in which the coding region of the cytoplasmic domain of the SARS‐CoV‐2 spike protein was replaced by the cytoplasmic domain of the G protein of VSV (SARS‐CoV‐2‐SG DNA). For SARS‐CoV, it was previously shown that such a modification increases S protein expression at the cell surface and induces higher neutralizing antibody responses after immunization with exosomal vaccines [41]. The time interval between the DNA priming immunization and the booster immunization allowed for the production and purification of a recombinant S protein stabilized in the prefusion state (SARS‐CoV‐2‐S protein) and an exosomal vaccine presenting a membrane‐anchored form of the SARS‐CoV‐2‐S protein (SARS‐CoV‐2‐Exosome) (Fig. 1B).
TRIANNI mice were selected for these immunization experiments (Fig. 1A) because they produce antibodies with human Ig variable (V) region and murine constant (C) regions. One unique feature of the TRIANNI mice is that the intronic regions between the exons of the variable segments of Ig genes are of murine origin. This design aims to maintain proper regulation of expression and splicing of the V regions of the immunoglobulin exons as a prerequisite for proper affinity maturation. After selecting antibodies of interest, the human V region gene segments can be recombinantly fused to human C region gene segments allowing the expression of fully human antibodies for clinical applications.
Two groups of TRIANNI mice were either immunized by intramuscular electroporation with the SARS‐CoV‐2‐S DNA and/or the SARS‐CoV‐2‐SG DNA (Fig. 1B). Three weeks later, the SARS‐CoV‐2‐S DNA primed mice were either boosted with the SARS‐CoV‐2‐S DNA or the SARS‐CoV‐2‐S protein adjuvanted with Monophosphoryl Lipid A (MPLA) liposomes. The SARS‐CoV‐2‐SG DNA primed mice were boosted with SARS‐CoV‐2‐SG or SARS‐CoV‐2 exosomes at weeks 3 and 4, respectively (Fig. 1B). Using a flow cytometric assay with cells transiently transfected with SARS‐CoV‐2‐S DNA [41], antibody responses were analyzed 2 weeks after the priming and the boosting immunization (Fig. 1C). Antibodies to SARS‐CoV‐2‐S were detectable in all four animals after the DNA priming immunization and further increased after the booster immunizations.
Sera of the immunized mice were also analyzed for competition with the binding of the cellular receptor hACE2 to the SARS‐CoV‐2‐S protein (Fig. 1D). Two weeks after the booster immunization, all four sera reduced hACE2 binding. Sera from the mice primed with SARS‐CoV‐2‐S DNA and boosted with SARS‐CoV‐2‐S protein or SARS‐CoV‐2‐S DNA also neutralized WT SARS‐CoV‐2 at a 1:100 dilution by 50 and 27%, respectively (Fig 1E).
Hybridoma screening for spike binding, ACE2 competing, and neutralizing antibodies
The TRIANNI mouse (M2) with the best neutralizing activity was boosted with the adjuvant SARS‐CoV‐2‐S protein on week 6 and sacrificed 5 days later to generate hybridoma lines. A 1:100 dilution of the serum from this time point neutralized WT SARS‐CoV‐2 by 86%. Cells from the spleen and draining LNs were fused with Sp2/0‐Ag14 cells, and hybridoma cells were grown in 106 96‐well plates seeded at a density of 1000 cells per well. Between 10 to 20 days after the fusion, antibodies secreted by the hybridoma cells were screened by flow cytometry for binding to the S protein. A total of 29 hybridomas could be identified that secreted antibodies binding to the S protein. These antibodies were designated TRES antibodies (TRIANNI‐Erlangen anti‐SARS‐CoV‐2‐ Spike). The screening results of 10 representative hybridoma supernatants containing TRES antibodies are displayed in Fig. 2A (for a list of all binding antibodies and their characteristics, see Supporting information Table S1). Intracellular staining revealed that most hybridomas expressed the IgG2c subtype.
Figure 2.
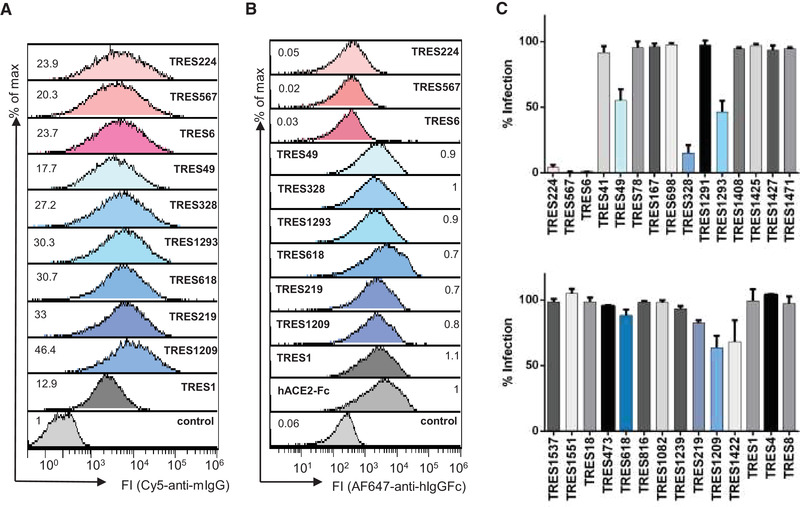
Screening of hybridoma supernatants. (A) The binding of antibodies from undiluted TRES hybridoma supernatants to HEK‐293T cells expressing the SARS‐CoV‐2 spike protein was detected with a fluorescence‐conjugated murine pan IgG antibody. Numbers indicate the relative mean fluorescence intensity. (B) Detection of hACE2‐competing TRES antibodies. Numbers depict the relative mean fluorescence intensity. (C) Vero‐E6 cells were infected with the SARS‐CoV‐2 MUC‐IMB‐1 isolate in the presence or absence of undiluted TRES hybridoma supernatants. SARS‐CoV‐2 infection was quantified after 20 to 24 h by staining as described in Fig. 1F. The mean and standard deviation of triplicates of one experiment are shown.
The binding antibodies were further characterized in an hACE2 competition assay. Three (TRES6, 224, 567) of the 29 binding antibodies were able to outcompete hACE2 binding very efficiently (Fig. 2B), indicating that these antibodies might bind the RBD and possibly neutralize SARS‐CoV‐2.
Subsequently, all supernatants were tested in a virus neutralization assay with SARS‐CoV‐2 MUC‐IMB‐1, an early isolate from the Webasto cluster in Bavaria carrying the D614G mutation in the spike protein [42, 43]. Supernatants from the hybridomas competing with ACE2‐binding (TRES antibodies 224, 567, and 6) strongly neutralized SARS‐CoV‐2 (Fig. 2C) and, indeed, an RBD‐specific ELISA confirmed the RBD binding of these three antibodies (Supporting information Fig. S1A). Three additional hybridoma supernatants could be identified to inhibit virus infection by at least 40% (TRES49, 328, 1293). They do not bind to the RBD, recombinant S1 proteins, or the S2 protein (Supporting information Fig. S1A‐C). TRES49, 328, 1293 bind to a stabilized S protein trimer and recombinant NTD (Supporting information Fig. S1D and E). A similar binding profile was also observed for the TRES antibodies 618, 219, and 1209. Thus, these six antibodies differ in their binding profile from TRES6, TRES224, and TRES567. We also identified one hybridoma supernatant (TRES1) that strongly binds to S2 (Supporting information Fig. S1B).
Characterization of monoclonal TRES antibodies
Based on the screening results, hybridomas were subcloned, and antibodies from the supernatants of hybridoma subclones of the TRES antibodies 6, 224, 567, 49, 219, 328, 618, 1209, 1293, and 1 were purified and further characterized. Because the TRES antibodies 6, 224, and 567 compete with hACE2 for binding to RBD (Fig. 2A), EC50 (50% effective concentration) was determined in an hACE2 competition ELISA (Fig. 3A). Surface plasmon resonance (SPR) analyses further revealed KD values of purified RBD‐ and NTD‐binding antibodies against the S protein stabilized in the prefusion state ranging from 9.5 to 1.6 nM (Supporting information Table S1).
Figure 3.
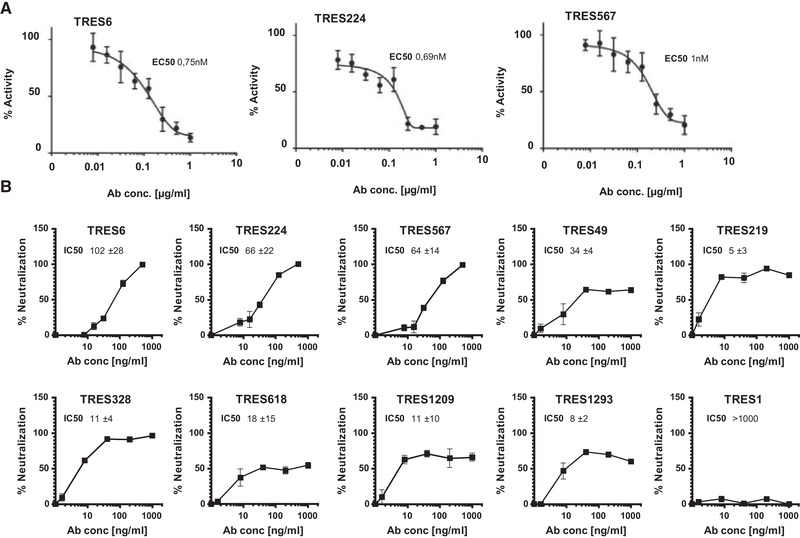
Characterization of monoclonal TRES antibodies. (A) ELISA‐based hACE2‐competition assay with TRES antibodies. Plates were coated with RBD and incubated with serial dilutions of TRES antibodies and soluble hACE2 (400 ng/mL). Bound hACE2 was quantitated with HRP‐coupled antibodies against the hFcγ1‐Tag of hACE2. The mean and standard deviations of quadruplicates of one representative experiment out of two are shown in the graphs. The mean EC50s of all experiments are also shown. (B) Vero‐E6 cells were incubated with the CoV‐2 ER‐1 isolate with increasing concentrations of the respective TRES antibodies. SARS‐CoV‐2 infection after 20 to 24 h was quantitated as described in Fig. 1 F. Graphs show the mean and SEM of triplicates of one representative experiment of at least three experiments. The mean IC50 and standard deviations, in ng/mL, of all experiments are also given. IC50s were calculated with inhibitor versus variable slope fitting curve with GraphPad Prism 7.02.
The neutralizing activity of purified antibodies was determined against a second WT SARS‐CoV‐2 isolate, CoV‐2‐ER1 (Fig. 3B) which also carries the D614G mutation dominating the SARS‐CoV‐2 populations in Europe [44, 45]. Antibodies that compete with hACE2 (TRES6, 224, and 567) had IC50s ranging from 64 to 102 ng/mL. The difference in the IC50s between TRES6 and 567 reflects the experimental variation of the assay and the quantification of antibody concentrations as both antibodies share the same amino acid sequence (Sequence analysis of variable regions of TRES antibodies). The IC50s of antibodies not interfering with hACE2 binding (TRES49, 328, 618, and 1293) ranged between 5 and 34 ng/mL. Although the initial screening of hybridoma supernatants had not detected neutralization by TRES618, 219, 1209, the antibodies had an IC50 of 18, 5, and 11 ng/mL, respectively. The S2‐binding antibody TRES1 did not display neutralizing activity (Fig. 3B). When the antibodies were assessed in a cell‐cell fusion assay using Vero‐E6 cells and HEK‐293T cells expressing the S protein, the RBD‐binding antibodies seemed to inhibit cell‐cell fusion more efficiently than the neutralizing antibodies that do not bind to RBD (Supporting information Fig. S2). In general, cell‐cell fusion inhibition required higher antibody concentrations, and total inhibition of cell‐cell fusion was, therefore, not achieved.
Sequence analysis of variable regions of TRES antibodies
Sequencing of productive immunoglobulin variable regions expressed by hybridomas encoding TRES antibodies revealed a broad usage of V regions of H and L chains with a preference for κL chains (Supporting information Table S1). Compared to the corresponding germline sequences, TRES VH and VL nucleotide sequences showed similarities as low as 94.44 and 97.85%, respectively, indicating the presence of somatic mutations and suggesting affinity maturation. Consistent with their similar binding profiles (Supporting information Fig. S1), TRES6, 224, and 567 H and L chain exons contain the same VHDJH and VLJL recombination joining sequences and are, therefore, designated cluster 1 antibodies. All three antibodies express the VH3‐33*01 or VH3‐33*06 and KV1D‐17*01 gene segments (Supporting information Table S1). An alignment of amino acid sequences of VH and VL of cluster 1 antibodies with the inferred germline‐encoded antibody revealed an identical L chain with five nonsilent somatic mutations in the antigen bindings loop (complementary determining regions, CDR) and one in the framework (FR) (Supporting information Fig. S3) TRES6 and 567 utilize the same VDJ H chain exon with four somatic mutations in the FR. TRES224 contains two additional mutations in the CDR1 and CDR2 and an unusual change of a tyrosine conserved in 98% of all human VH segments. A second cluster comprises six TRES antibodies (Supporting information Table S1; TRES49, 219, 328, 618, 1209, and 1293) that share the VHDJH and VLJL recombination joining sequences. Various somatic mutations are also present in the CDRs and the FR of their VH and VL segments (Supporting information Fig. S3). The non‐neutralizing antibodies of a third cluster bind to S1, but not to RBD, and are composed of an H chain with a human VH and an L chain with an unusual mouse VλX region (Supporting information Table S1) [46], which could be explained by the fact that the mouse VλX region is still present in TRIANNI mice.
Generation of fully humanized TRES antibodies
Gene segments encoding the human V exons of the H and L chains of all neutralizing TRES antibodies were fused with the human γ1 H and the κL chain constant regions, respectively. Recombinant antibodies entirely composed of human Ig regions were produced in HEK‐293F cells and purified by affinity chromatography. As expected, all recombinant TRES antibodies bound to the S protein in the flow cytometric assay (Fig. 4A).
Figure 4.
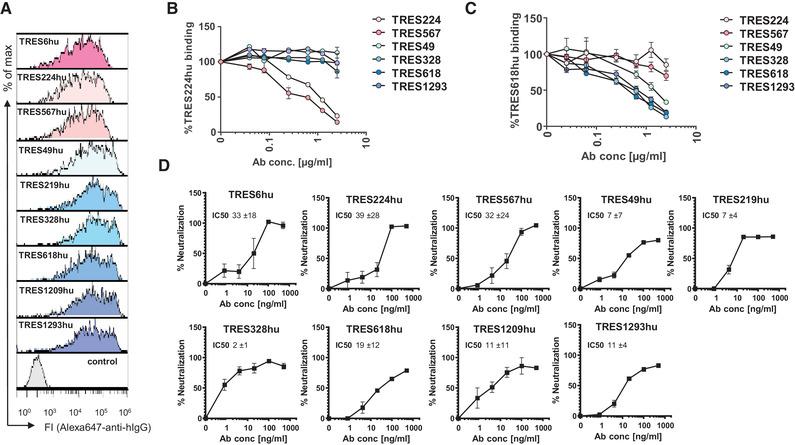
Characterization of recombinant human TRES antibodies. (A) Flow cytometric analysis of HEK‐293T cells expressing the SARS‐CoV‐2‐S protein and stained with recombinant humanized IgG1 TRES (TREShu) antibodies and a fluorochrome‐labeled secondary antibody against human IgG‐Fc. A non‐S binding human antibody served as a negative control. (B, C) HEK‐293T cells expressing the SARS‐CoV‐2 spike protein were incubated with recombinant TRES antibodies with a human Fcγ1 region and serially diluted TRES hybridoma antibodies with a murine Fcγ. Bound recombinant human TRES224hu (B) or TRES618hu (C) were detected with a mouse Alexa647‐labeled antibody directed against the human Fcγ region. The mean percentages of binding and SEM of one experiment performed in triplicates are shown. (D) The SARS‐CoV‐2 neutralizing activity of the human recombinant TRES antibodies was analyzed as described in Fig. 3B. Shown are means and SEM of triplicates of one representative experiment out of three. Also given are the mean and standard deviation of IC50s, given in ng/ml, of the three independent experiments, calculated as described in Fig. 3B
To explore a potential overlap of epitopes between TRES antibodies of cluster 1 and cluster 2, cells expressing the SARS‐CoV‐2‐S protein were incubated with TRES224hu or TRES618hu in the presence of increasing concentrations of TRES224, 567, 49, 328, 618, and 1293 hybridoma antibodies containing mouse C regions (Fig. 4B and C).
No competition between the cluster 1 and cluster 2 antibodies could be observed. In contrast and as expected, binding of the recombinant TRES224 with human C regions (TRES224hu) was efficiently blocked by TRES567 and 224 (Fig. 4B), indicating that these two neutralizing antibodies from cluster 1 recognize the same epitope. Accordingly, the binding of TRES618hu was blocked by TRES49, 328, 618, and 1293, verifying that cluster 2 antibodies bind the same epitope. Most importantly, however, the humanized TRES antibodies also neutralized SARS‐CoV‐2 with similar IC50s as the parental hybridoma TRES antibodies (Fig. 4D), confirming that the identified antibody sequences confer neutralization.
The breadth of neutralization, escape mutants, and structural modeling
Neutralization assays and spike protein binding assays (Supporting information Fig. S4A to C) were also performed for two emerging SARS‐CoV‐2 VOC and three antibodies from each cluster. The IC50s of the cluster 1 antibodies against the Alpha (B.1.1.7) variant were in the range of 6 –49 ng/mL (Supporting information Fig. S4D, left panel) and were similar to those for the WT virus. IC50s consistently below 10 ng/mL (Supporting information Fig. S4E, right panel) indicate that the Beta (B.1.351) isolate might be more susceptible to neutralization by cluster 1 antibodies than the WT virus. In contrast, cluster 2 antibodies lost their neutralizing activity against both VOC (Supporting information Fig. S4).
To further explore the possibility for de novo emergence of resistant variants to cluster 1 and cluster 2 antibodies, the CoV‐2 ER‐1 isolate carrying the D614G mutation was passaged in the presence of increasing concentrations of TRES6 and TRES328 antibodies. After five passages, neutralization assays were performed. While the passaged virus became resistant to antibody neutralization when the antibody was present during passaging, the virus remained sensitive to the TRES antibody when the antibodies were not included during passaging (Supporting information Fig. S4E). Whole‐genome sequencing of the passaged SARS‐CoV‐2 viruses revealed the emergence of a TRES6 antibody escape variant with an I68R mutation in the NTD and a T478K mutation in the RBD (Supporting information Fig. S4F, displayed in pink). In contrast, the TRES328 escape variant harbored an L241‐Y248 deletion with a phenylalanine insertion in the NTD (Supporting information Fig. S4F, displayed in blue). Additional point mutations and deletions were observed in proximity to the S1‐S2 cleavage sites. These mutations probably reflect adaptation to the replication in the Vero‐E6 host cells [47], since these sites were also deleted in the virus variant (P5) passaged in the absence of a neutralizing antibody (Supporting information Fig. S4F, displayed in white).
The experimental characterization has shown that TRES328 binds to the NTD. Modeling of the respective complex suggests that TRES328 contacts the sequence stretch 241–248 that is deleted in the escape variant (Supporting information Fig. S5A). In the WT virus, NTD residues R246 and Y248 play an essential role for antibody binding by forming specific polar interactions with Y27 and E31 of the CDRH1 (Supporting information Fig. S5B). Replacement of residues 241–248 by a single phenylalanine in the escape variant significantly reduces the length of the N5‐loop and causes a loss of the stabilizing interactions mediated by R246/Y248 (Supporting information Fig. S5C). In addition, the deletion places D246 (corresponding to D253 of the WT) in the vicinity of E31 (Supporting information Fig. S5C), thereby further destabilizing the interaction due to electrostatic repulsion between the two negatively charged residues. Interestingly, as described above, TRES328 did not neutralize the Alpha (B.1.1.7) variant, although this variant carries no mutation in the putative TRES328‐binding site. This loss of binding might, however, be due to a deletion of the amino acids 69–70 in the Alpha (B.1.1.7) NTD, which might lead to a change in the allosteric conformation of the S1 epitope by pulling the loop of amino acids 69–76 inward [30, 33, 48]. This conformational change in the NTD might result in a concealed TRES328 binding site and a subsequent loss of neutralization.
TRES6 exhibits only limited sequence similarity to other spike‐binding antibodies of known 3D‐structure, thus, impeding molecular modeling of the RBD‐TRES6 complex. However, the observation that a T478K mutation results in a loss of TRES6 binding indicates that T478 is part of the binding epitope. Since this epitope is also mutated in the recently isolated Delta (B.1.617.2) variant [49], this variant of concern is very likely able to escape TRES6‐ mediated neutralization. Inspection of other known antibody structures reveals that a subset of RBD‐binding antibodies also recognizes T478. One example for this group of antibodies is COVOX‐253 (Supporting information Fig. S5D and E), which recognizes T478 by Y33 of CDRL1 and D108 of CDRH3. These two amino acids are also present in TRES6 at the respective sequence positions suggesting that TRES6 might use similar structural principles for RBD recognition. However, a more detailed analysis will require an experimental structure determination of the TRES6‐RBD complex.
Efficacy in a murine challenge model
To assess whether the antibodies generated in this study can confer protection from disease, we evaluated the efficacy of one antibody from each cluster in a stringent hACE2 transgenic mouse [50] viral challenge model under postexposure prophylactic settings. Three groups of mice (n = 12/group) were infected intranasally with 300 FFUs of SARS‐CoV‐2 (MUC‐IMB‐1). One day later, mice were injected intravenously with 5.25 mg/kg body weight of the cluster 1 antibody TRES6, the cluster 2 antibody TRES328, or the isotype‐matched control antibody TRES480.
Six animals from each group were sacrificed on day 4 or 10 or according to humane endpoints, and viral loads were determined in the lungs, BAL (bronchoalveolar lavages), brain, liver, and spleen. Both antibodies reduced the amount of viral RNA in the lungs and BAL samples approximately 30‐ to 200‐fold after 4 days (Fig. 5A) or approximately 100‐ o 450‐fold after 10 days (Fig. 5B). Viral load in the other organs was reduced close to the level of detection. Interestingly, the titer of infected virus in the BAL samples from TRES6‐ and TRES328‐treated animals was below the detection level and, therefore, at least 1000‐fold lower than in mice receiving the control antibody (Fig. 5C). Compared to the isotype control, TRES480, treatment of mice with TRES6 and TRES328 prevented the loss of body weight induced by the viral infection (Fig. 5) and reduced clinical symptoms assessed by a clinical score (Fig. 5E). Importantly, none of the TRES antibody‐treated mice reached clinical endpoints requiring euthanasia, while four of six SARS‐CoV‐2‐infected mice receiving the isotype control antibody TRES480 had to be euthanized (Fig. 5F).
Figure 5.
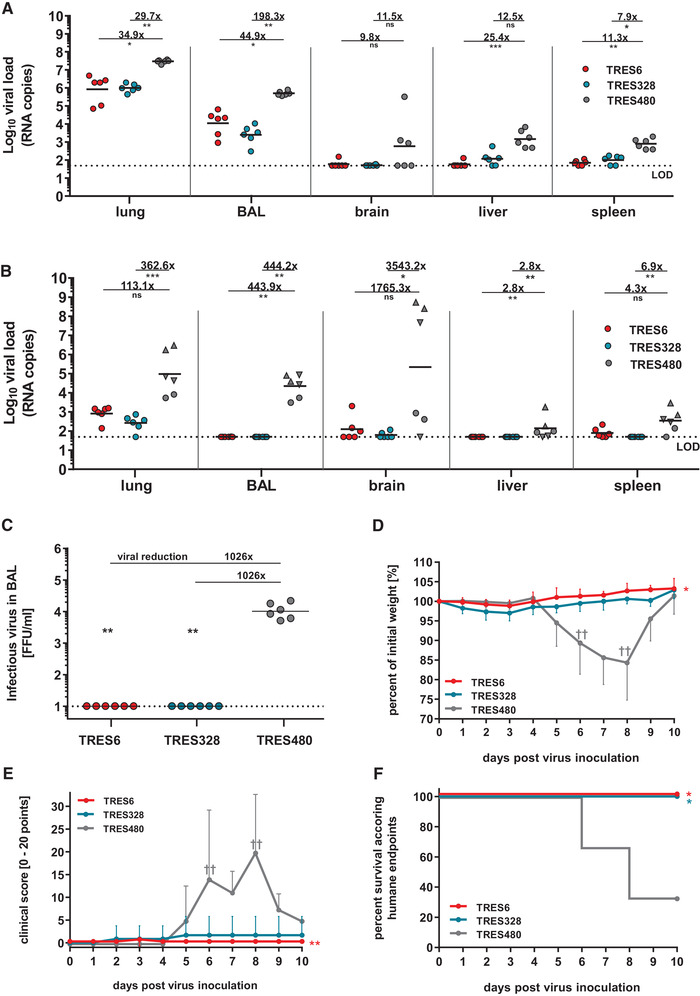
Efficacy of TRES6 and TRES328 in a postexposure prophylactic model. Reduction of viral load in hACE2‐transgenic mice treated with TRES6, TRES328, or TRES480 isotype control antibody. Mice were inoculated intranasally with 300 FFU of SARS‐CoV‐2 on day 0. One day later, mice were treated intravenously with 5.25 mg/kg TRES6, TRES328, or TRES480 control antibody. Viral loads were determined on day 4 (A) or day 10 (B) after virus inoculation by RT‐qPCR in the indicated organ samples. Data points represent the viral copy number of individual animals with the geometric means of each group depicted as lines, circles (●) indicate the survival of 4‐ or 10‐day post‐infection, and triangles indicate euthanized mice according to humane endpoints at day 6 (▲) or day 8 (▼). Calculated reduction of viral RNA is shown in comparison to the TRES480 control group. (C) Infectious virus load in BAL samples from antibody and isotype‐treated mice. Infectious virus was measured by focus‐forming assay. (D) Bodyweight and (E) clinical score of antibody‐ and isotype‐treated mice. Animals reaching humane endpoints were euthanized and are marked by a cross (†). (F) Survival curve of antibody‐treated and isotype control antibody‐treated animals. Percent survival as the fraction of animals surviving humane endpoints (Kaplan–Meier analysis). The experiment was performed once. Statistical analysis of the presented data was performed by Kruskal–Wallis test (one‐way ANOVA) and Dunn´s Pairwise Multiple Comparison Procedures as post‐hoc test in comparison to the TRES‐480 control (ns: nonsignificant, *p < 0.05, **p < 0.01, ***p < 0.001).
Discussion
By immunizing TRIANNI mice with a DNA prime protein boost regimen, 29 monoclonal antibodies were obtained that bind to the S protein of SARS‐CoV‐2 expressed on the cell surface of transfected HEK‐293T cells. The use of immunization and screening approaches based on the expression of the WT S protein avoids the need for prior knowledge of domains targeted by neutralizing antibodies and allows obtaining a panel of antibodies binding to different epitopes. Indeed, different binding patterns were observed, including antibodies that (i) bind to the RBD and compete with ACE2 binding, (ii) bind to the S1 subunit without binding to RBD, (iii) only bind to the ectodomain of the S protein stabilized in a trimeric form and the NTD, and (iv) bind to the S2 subunit. In those mice that we first challenged with SARS‐CoV‐2 and subsequently treated with the monoclonal antibodies, we could also assess any potential disease‐enhancing properties. This issue has been suggested as a potential outcome of infections occurring in the presence of vaccine‐induced antiviral antibodies not providing sterilizing immunity [51]. However, no evidence for such an enhancing effect could be observed. A combination therapy with an antibody from cluster 1 and cluster 2 seems especially attractive because this will extend the neutralization breadth and enhance the genetic barrier for the emergence of antibody‐resistant SARS‐CoV‐2 variants. This is further supported by the observation that the Alpha (B.1.1.7) and the Beta (B.1.351) variants maintain neutralization sensitivity to at least one of the two clusters of antibodies. In addition, virus escape mutants from one antibody of each cluster are still sensitive to a representative TRES antibody from the other cluster.
Most of the neutralizing antibodies in convalescent COVID‐19 patients are close to the germline sequence [52]. However, it was recently shown that antibodies derived from convalescent COVID‐19 patients, 3–6‐month post symptom onset, show significant somatic hypermutation [53, 54]. It was also shown, that this affinity maturation further enhances the neutralization capacity of RBD‐binding antibodies [31]. Accordingly, the TRES antibodies had mutations in the VH and VL regions, including the CDRs (Fig. S3), suggesting that affinity maturation can also occur in TRIANNI mice after a DNA prime protein boost immunization regimen. Although mice encoding an entire [11] or partial [55] human immunoglobulin repertoire have been used previously to generate monoclonal antibodies targeting the spike protein of SARS‐CoV‐2, direct evidence for affinity maturation, in the context of SARS‐CoV‐2, has not yet been provided. Using a 3‐week immunization regimen consisting of a single intradermal priming immunization with DNA encoding the S protein and booster immunizations with an RBD‐Fc fusion protein in VelocImmuneTM mice, Hansen et al. showed induction of RBD‐specific antibody responses and also reported the isolation of neutralizing RBD‐binding antibodies from B cells that were sorted based on RBD binding [11]. Wang et al. performed sequential immunizations of H2L2TM (Harbour Antibodies) mice with spike ectodomains of different human coronaviruses and derived a monoclonal antibody that cross‐neutralized WT SARS‐CoV‐2 moderately with an IC50 of 570 ng/mL [55].
Although extensive affinity maturation does not seem to be necessary for the formation of potent neutralizing antibodies against SARS‐CoV‐2 [52], this may be relevant for elicitation of neutralizing antibodies against other pathogens as exemplified by HIV‐1 [56, 57, 58]. As observed for four of seven other potent neutralizing antibodies targeting the NTD and recovered from convalescent patients [7], our TRIANNI mouse‐derived NTD antibodies also use the VH1‐24 gene segment. Therefore, transgenic animal models supporting affinity maturation of antibodies with human variable regions should be an essential part of future pandemic preparedness efforts and be explored in structure‐guided vaccine antigen design against SARS‐CoV‐2.
Besides generating potent neutralizing antibodies that also confer protection in the hACE2 mouse model, a key criterion for applying TRIANNI mice in future pandemic responses is the time needed to obtain such antibodies. The immunization schedule we used took 7 weeks until hybridomas were generated. Growing the hybridomas, their screening and subcloning took approximately 6 weeks. Sequence analyses from these hybridomas and gene synthesis for the generation of fully human antibodies took another 6 weeks. Accelerating the development time would certainly be desirable. Whether shortening the immunization schedule still results in potent neutralizing antibodies, it is difficult to predict and may depend on the degree of affinity maturation needed to generate such antibodies. Since, as described above, an extensive antibody affinity maturation is needed for the elicitation of neutralizing antibodies against some pathogens, for example, HIV, a shortened immunization schedule, in this case, might not be advantageous [56, 57, 58]. The general time needed for the isolation of antibodies against such pathogens, however, could be accelerated by staining antigen‐specific memory B cells or GC‐selected surface IgG‐positive plasma blasts with fluorescence‐labeled antigen and sorting them by flow cytometry instead of the generation of hybridomas (Supporting information Fig. S6). Paired Ig VH and VL sequences from single cells could then be amplified by PCR and directly cloned into expression plasmids described for memory B cells from convalescent COVID‐19 patients [10, 59]. Transiently expressed paired recombinant antibodies could then be screened for binding and neutralization, shortening the development time by several weeks.
Material and methods
Mice
The TRIANNI C57/Bl6 mouse line HHKKLL was established in cooperation with TRIANNI (Patent US 2013/0219535 A1). Mice were maintained under specific pathogen‐free conditions in the Franz–Penzoldt Center animal facility of the University of Erlangen–Nürnberg following institutional and national regulations and the Federation of European Laboratory Animal Science Associations. All immunization animal experiments were approved by the Regierung von Unterfranken (TVA 55.2.2‐2532‐2‐961).
Immunization of TRIANNI mice and the establishment of SARS‐CoV‐2‐specific hybridoma clones
Two 9–12‐week‐old TRIANNI mice were primed with pCG1‐CoV‐2019‐S ([60] designated SARS‐CoV2‐S DNA) encoding WT SARS‐COV‐2 spike protein (position 21580 –25400 from GenBank NC_045512). Two additional TRIANNI mice were primed by SARS‐CoV‐2‐SG DNA encoding a chimeric protein in which the intracytoplasmic domain of SARS‐CoV‐2‐S was replaced by the intracytoplasmic domain of VSV‐G, as previously described for SARS‐CoV [41]. The DNA vaccines were delivered by intramuscular electroporation as previously described [61]. Briefly, a total of 30 μg DNA diluted in 60 μL PBS was injected in both hind legs under constant isoflurane (CP‐Pharma, Burgdorf, Germany) anesthesia. Immediately after the intramuscular injection, electrical impulses were applied to the injection site with a TriGrid electrode array (provided by Drew Hannaman, Ichor Medical Systems, Inc., San Diego, USA). Mice were boosted intramuscularly either with (i) 5 μg of the S protein of SARS‐CoV‐2 stabilized in a prefusion conformation (designated SARS‐CoV‐2‐S protein) and adjuvanted with 25 μg MPLA liposomes (Polymun Scientific GmbH, Klosterneuburg, Austria) into the hind leg, (ii) by electroporation of the DNA vaccines used for priming, or (iii) with exosomes purified from HEK‐293T cells transiently transfected with SARS‐CoV‐2 DNA as described previously [41]. Details for the immunization schedule are summarized in Figure 1B. Mice were bled by punctuation of the retro‐orbital sinus using nonheparinized capillaries (Hirschmann Laborgeräte, Eberstadt, Germany). Serum was collected, inactivated for 30 min at 60°C, and kept at −20°C for long‐term storage.
One of the immunized mice received a second booster immunization with SARS‐CoV‐2‐S protein at week 5. Five days later, spleen and inguinal LNs were harvested and fused with the azaguanine‐resistant SP2/0 Ag14 hybridoma line (ATCC#CRL‐1581) using the PEG (Sigma Aldrich, Taufkirchen, Germany) method [62]. Briefly, 2 × 108 spleen cells and inguinal LN cells (about 1 × 108 B cells) were mixed with 108 Sp2/0 cells and washed three times with RPMI1640 medium (Gibco, Thermo Fisher Scientific, Waltham, USA) without FCS. A total of 2 mL PEG was added dropwise within 1 min to the suspended cell pellet. The mix of fused cells was resuspended in R10+ medium (RPMI 1640 with 10% FCS, 0.05 mM mercaptoethanol, 2 mM l‐glutamine, 1 mM sodium pyruvate, and 1% penicillin/streptomycin (all from Gibco, Thermo Fisher Scientific); enriched with HAT (Gibco, Thermo Fisher Scientific) and OPI Media supplement (Sigma Aldrich). Cells were seeded into 96‐well plates with approximately 1 × 104 cells per well. A total of 10–20 days postfusion, hybridoma clones were tested for spike‐binding antibodies. Positive clones were expanded in R10+ supplemented with HT (Sigma Aldrich) and subcloned by the limiting dilution method.
Flow cytometric screening of hybridoma clones for SARS‐CoV‐2‐specific antibodies and Ig isotypes
To detect antibody binding to SARS‐CoV‐2‐S protein, HEK‐293T cells were cotransfected with SARS‐CoV‐2‐S DNA and a GFP reporter plasmid (e.g., pEGFP‐C1) using the PEI method as described previously [63]. A total of 105 thawed or freshly transfected cells were incubated first in 96‐well plate with 100 μL undiluted hybridoma supernatant or 100 μL mouse serum (1:200 dilutions in R10+ medium), bound antibodies were detected with a Cy5‐conjugated goat anti‐pan‐mouse IgG antibody (Southern Biotechnology, Birmingham, USA, #SBA‐1030‐15). Cells were washed in FACS buffer and analyzed with an Attune Nxt, CytoFlex, or a Gallios flow cytometer (Thermo Fisher Scientific or Beckman Coulter, Brea, USA, respectively) and evaluated with Flow LogicTM (IInivai Technologies Mentone, Australia). Flow cytometric analysis adhered to “Guidelines for the use of flow cytometry and cell sorting in immunological studies” [64].
Hybridoma cells were stained intracellularly for the assessment of the monoclonality of the colonies and the determination of the IgG subtype. Briefly, 105 cells were fixed for 20 min in 2% paraformaldehyde (Morphisto, Frankfurt am Main, Germany) diluted in PBS, washed twice with FACS buffer, resuspended in permeabilization buffer (0.5% Saponin Sigma Aldrich, in FACS buffer) containing fluorochrome‐conjugated murine H and L isotype‐specific antibodies (anti‐mouse IgG1‐APC # 550874 and anti‐mouse IgG3‐bio # 020620 from BD, Franklin Lakes, USA, anti‐mouse IgG2b‐PE SBA‐1090‐09 and anti‐mouse IgG2c‐bio SBA‐1079‐08 from Southern Biotech, anti‐mouse Ig light chain lambda APC # 407306 and anti‐mouse IgG light chain kappa PE both from Biolegend, San Diego, USA) and incubated for 1 h at 4°C. Cells were again washed in FACS buffer and analyzed with an Attune Nxt, CytoFlex, or a Gallios flow and evaluated with Flow LogicTM.
Virus neutralization assay
Neutralizing activities of hybridoma supernatants, antibodies, and sera were assessed in a microneutralization assay based on detecting the number of virus‐producing cells via immunofluorescence. For this, 1 × 104 Vero‐E6 cells were seeded per well of a 96‐flat bottom plate one day before the infection. Purified antibodies, sera, or hybridoma supernatants were first diluted and preincubated for 1 h with 1.88 × 10E4 infectious units of SARS‐CoV‐2 stocks per well in a final volume of 100 μL. All dilutions of hybridoma supernatants or purified antibodies were prepared in a cell OptiPROTM culture medium. After removing the cell culture medium, seeded Vero‐E6 cells were incubated for 1 h with 100 μL per well of the preincubated virus‐antibody mix. Afterward, the supernatant was discarded, the cells washed once with 100 μL PBS, and 100 μL fresh cell culture medium added. The cells were incubated for 20–24 h, washed with PBS, fixed with 4% paraformaldehyde for 20 min, and subsequently stained. They were permeabilized for 15 min with 0.5% TritonX in PBS and blocked with 5% skimmed milk diluted in PBS for 1 h. Subsequently, the plates treated with murine antibodies were stained with protein G‐purified sera from a convalescent patient diluted 1:100 in PBS containing 2% skimmed milk. Plates that were treated with human antibodies were stained with an antibody mix of murine origin directed against SARS‐CoV‐2 at a concentration of 250 ng/mL. After 1 h, the cells were washed, and a goat anti‐human IgG FITC (Jackson ImmunoResearch, West Grove, USA #109‐096‐088) antibody or a FITC‐conjugated goat anti‐mouse IgG serum (Jackson ImmunoResearch # 115‐095‐003) was applied. After 1 h and a washing step, positive wells were identified by a CTL‐ELISPOT reader (Immunospot; CTL Europe GmbH, Bonn, Germany). The signal was analyzed with the ImmunoSpot® fluoro‐X™ suite (Cellular Technology Limited, Cleveland, USA). IC50 values were calculated by plotting the virus activity in percent against the antibody concentrations and using the normalized response versus inhibitor equation (variable slope) or inhibitor versus response—Variable slope (four parameters) of GraphPad Prism 7.02.
Quantitative antibody competition assay
For the assessment of the quantitative antibody competition, SARS‐CoV‐2‐S DNA‐transfected HEK‐293T cells were incubated with 100 μL of a humanized protein G purified TRES antibody at a concentration of 250 ng/mL and serial dilutions of TRES antibodies with a murine Fc region at final concentrations ranging from 2.5 μg/mL to 0.01 ng/mL. The cells were incubated for 30 min on ice, washed, and bound antibodies were detected with a mouse IgG2a Alexa647‐conjugated antibody directed against human IgG‐Fc (BioLegend #409320). After washing, the mean fluorescence intensities of transfected cells were determined with an Attune Nxt and the Flow Logic softwareTM.
Small animal challenge experiments
Animal experiments were performed following the EU Directive 2010/63/EU for animal experiments and were approved by local authorities after review by an ethical commission (TVV 21/20). Thirty‐six female K18‐hACE2 mice (Jackson Laboratory, Bar Harbor, USA) were infected intranasally under isoflurane anesthesia with 300 FFU of SARS‐CoV‐2 strain MUC‐IMB‐1 p.1 in a total volume of 50 μL. Twenty‐four hours after virus inoculation, 12 mice per group were injected intravenously either with 5.25 mg/kg body weight of TRES6 or TRES328 antibody or an isotype control antibody (TRES480) in a total volume of 100 μL. The animals were scored daily, and the survival and disease incidence were measured over a maximal 10‐day period. One cohort (n = 6) was euthanized at day 4, while the second cohort (n = 6) was scored for humane endpoints and euthanized at day 10 post virus inoculation. After euthanasia, BALs were collected, and lungs, brain, liver, and spleen were taken. The organs were homogenized in 2 mL PBS using gentleMACS M tubes and gentleMACS Octo Dissociator (Miltenyi Biotec, Bergisch Gladbach, Germany). Afterward, the tubes were centrifuged at 2000 × g for 5 min at 4°C to discard cell debris. Viral RNA was isolated from 140 μL of homogenated supernatants or BAL using QIAamp Viral RNA Mini Kit (Qiagen, Aarhus, Denmark). The viral load in indicated organs and BAL was analyzed by RT‐qPCR [65]. Reactions were performed using TaqMan® Fast Virus 1‐Step Master Mix (Thermo Fisher Scientific) and 5 μL of isolated RNA as a template. Synthetic SARS‐CoV‐2 RNA (Twist Bioscience, San Francisco, USA) was used as a quantitative standard to obtain viral copy numbers. The viral load reduction was calculated in comparison to the isotype control. The clinical scoring system included the following items: weight loss and body posture (0–20 points), general conditions including the appearance of fur and eye closure (0–20 points), reduced activity, and general behavior changes (0–20 points), and limb paralysis (0–20 points). Mice were euthanized when reaching a cumulative score of 20.
For the detection of infectious virus in BAL Vero‐E6 cells were seeded at 2 × 104 cells per well in a 96‐well plate in 200 μL DMEM, 10 % FCS, 1 × penicillin/streptomycin (Thermo Fisher Scientific) 24 h before infection with 20‐100 μL of BAL diluted in DMEM with 1× penicillin/streptomycin for 3 h. After replacing the supernatant with an overlay medium (DMEM with 1 % methylcellulose, 2 % FCS, and 1× penicillin/streptomycin), cells were incubated for 27 h. SARS‐CoV‐2‐infected cells were visualized using SARS‐CoV‐2 S protein‐specific immunochemistry staining with anti‐SARS‐CoV‐2 spike glycoprotein S1 antibody (Abcam, Cambridge, Great Britain) as described previously [65]. Statistical evaluation of the data was performed by Kruskal–Wallis test (one‐way ANOVA) and Dunn's Pairwise Multiple Comparison Procedures as post‐hoc test.
Detection of antibody binding, cloning, and sequencing of Ig V exon sequences, generation of human IgG expression vectors, purification of the murine and human antibodies, and the virus propagation were performed as described in the Supporting information.
Conflict of Interest
T.W., K.Ü., and H.‐M.J. have filed a patent application on TRES antibodies and hold shares of COVER Antibodies GmbH, a company that has licensed IP rights on TRES antibodies. The other authors declare no commercial or financial conflict of interest.
Peer review
The peer review history for this article is available at https://publons.com/publon/10.1002/eji.202149374.
Abbreviations
- BAL
bronchoalveolar lavages
- CDR
complementary determining regions
- EC50
50% effective concentration
- FR
framework
- hACE2
human Angiotensin‐converting enzyme 2
- MPLA
monophosphoryl Lipid A
- NTD
N‐terminal domain
- RBDs
receptor‐binding domains
- SPR
surface plasmon resonance
- VOC
variants of concern
Supporting information
Supporting information
Acknowledgments
We would like to thank Isabell Schulz and Doris Jungnickl for their excellent technical assistance. We kindly thank Jasmin Fertey and Rosina Ehmann for providing high titer virus stocks of SARS‐CoV‐2 and Alexandra Rockstroh for the optimized SARS‐CoV‐2 detection protocol. The expression plasmid for the spike ectodomain was kindly provided by Jason McLellan, Austin, USA, the TriGrid electrode array for DNA electroporation by Drew Hannaman, Ichor Medical Systems, Inc., and S2 and S1 protein by Thomas Schumacher, Virion/Serion GmbH, Würzburg. The ELISPOT Analyzer was obtained with financial support from Foundation Dormeur, Vaduz.
This work was supported by a Grant (01KI2043) from the Bundesministerium für Bildung und Forschung (BMBF) and funds from the Bavarian State Ministry for Science and the Arts to K.Ü., H‐M J., and T.H.W. Further support was provided by B‐FAST and COVIM, two BMBF‐funded projects of the Netzwerk Universitätsmedizin (NaFoUniMedCovid19; FKZ: 01KX2021) and funds from the Deutsche Forschungsgemeinschaft (DFG) through the research training groups RTG 2504 and RTG1660 and the DFG‐funded TRR130 (to HMJ and TW).
The authors declare that they complied with all relevant ethical regulations. All relevant data supporting the findings of this study are available within the article and its Supplementary information files. Open Access funding enabled and organized by Projekt DEAL.
Contributor Information
Klaus Überla, Email: klaus.ueberla@fau.de.
Hans‐Martin Jäck, Email: hans-martin.jaeck@fau.de.
Data availability statement
The Sequences of the Viruses used in this study are available on GISAID under the following accession numbers: GISAID EPI ISL 406862, GISAID EPI_ISL_755639, EPI_ISL_610249, EPI_ISL_2718549, EPI_ISL_2718548, EPI_ISL_2718547. The Sequence of the Beta isolate can be found under the GenBank accession number: MW822592.1. The sequences of neutralizing CoV‐2 antibodies have been submitted to GenBank (accession no. in legend of Supporting information Fig. S3).
References
- 1. Zhu, N. , Zhang, D. , Wang, W. , Li, X. , Yang, B. , Song, J. , Zhao, X. et al., A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020. 382: 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.[Internet], Center for Systems Science and Engineering (CSSE) at Johns Hopkins University. COVID‐19 Dashboard. 2021. [cited 2021 April 09], Available from https://www.arcgis.com/apps/opsdashboard/index.html#/bda7594740fd40299423467b48e9ecf6.
- 3. Forni, G. , Mantovani, A. , Forni, G. , Mantovani, A. , Moretta, L. , Rappuoli, R. , Rezza, G. et al., COVID‐19 vaccines: where we stand and challenges ahead. Cell Death & Differentiation 2021. 28: 626–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kyriakidis, N. C. , López‐Cortés, A. , González, E. V. , Grimaldos, A. B. and Prado, E. O. , SARS‐CoV‐2 vaccines strategies: a comprehensive review of phase 3 candidates. NPJ Vaccines 2021. 6: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aran, D. Estimating real‐world COVID‐19 vaccine effectiveness in Israel using aggregated counts. medRxiv 2021. 10.1101/2021.02.05.21251139. [DOI] [Google Scholar]
- 6. Bernal, J. L. , Andrews, N. , Gower, C. , Stowe, J. , Robertson, C. , Tessier, E. , Simmons, R. et al., Early effectiveness of COVID‐19 vaccination with BNT162b2 mRNA vaccine and ChAdOx1 adenovirus vector vaccine on symptomatic disease, hospitalisations and mortality in older adults in England. medRxiv 2021. 10.1101/2021.03.01.21252652 2021.03.01.21252652. [DOI] [Google Scholar]
- 7. Cerutti, G. , Guo, Y. , Zhou, T. , Gorman, J. , Lee, M. , Rapp, M. , Reddem, E R. et al., Potent SARS‐CoV‐2 neutralizing antibodies directed against spike N‐terminal domain target a single supersite. Cell Host & Microbe 2021. 29: 819–833.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chi, X. , Yan, R. , Zhang, J. , Zhang, G. , Zhang, Y. , Hao, M. , Zhang, Z. et al., A neutralizing human antibody binds to the N‐terminal domain of the Spike protein of SARS‐CoV‐2. Science 2020. 369: 650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang, N. , Sun, Y. , Feng, R. , Wang, Y. , Guo, Y. , Zhang, L. , Deng, Y. Q. et al., Structure‐based development of human antibody cocktails against SARS‐CoV‐2. Cell Res. 2021. 31: 101–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brouwer, P. J. M. , Caniels, T. G. , van der Straten, K. , Snitselaar, J. L. , Aldon, Y. , Bangaru, S. , Torres, J. L. et al., Potent neutralizing antibodies from COVID‐19 patients define multiple targets of vulnerability. Science 2020. 369: 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hansen, J. , Baum, A. , Pascal, K. E. , Russo, V. , Giordano, S. , Wloga, E. , Fulton, B. O. et al., Studies in humanized mice and convalescent humans yield a SARS‐CoV‐2 antibody cocktail. Science 2020. 369: 1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Voss, W. N. , Hou, Y. J. , Johnson, N. V. , Kim, J. E. , Delidakis, G. , Horton, A. P. , Bartzoka, F. et al., Prevalent, protective, and convergent IgG recognition of SARS‐CoV‐2 non‐RBD spike epitopes in COVID‐19 convalescent plasma. bioRxiv 2020. 10.1101/2020.12.20.423708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu, L. , Wang, P. , Nair, M. S. , Yu, J. , Rapp, M. , Wang, Q. , Luo, Y. et al., Potent neutralizing antibodies against multiple epitopes on SARS‐CoV‐2 spike. Nature 2020. 584: 450–456. [DOI] [PubMed] [Google Scholar]
- 14. Cao, Y. , Su, B. , Guo, X. , Sun, W. , Deng, Y. , Bao, L. , Zhu, Q. et al., Potent neutralizing antibodies against SARS‐CoV‐2 identified by high‐throughput single‐cell sequencing of convalescent patients’ B cells. Cell 2020. 182: 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wibmer, C. K. , Ayres, F. , Hermanus, T. , Madzivhandila, M. , Kgagudi, P. , Oosthuysen, B. , Lambson, B E. et al., SARS‐CoV‐2 501Y.V2 escapes neutralization by South African COVID‐19 donor plasma. Nat. Med. 2021. 27: 622–625. [DOI] [PubMed] [Google Scholar]
- 16. Chen, X. , Li, R. , Pan, Z. , Qian, C. , Yang, Y. , You, R. , Zhao, J. et al., Human monoclonal antibodies block the binding of SARS‐CoV‐2 spike protein to angiotensin converting enzyme 2 receptor. Cellular & Molecular Immunology 2020. 17: 647–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ju, B. , Zhang, Q. , Ge, J. , Wang, R. , Sun, J. , Ge, X. , Yu, J. et al., Human neutralizing antibodies elicited by SARS‐CoV‐2 infection. Nature 2020. 584: 115–119. [DOI] [PubMed] [Google Scholar]
- 18. Robbiani, D. F. , Gaebler, C. , Muecksch, F. , Lorenzi, J. C. C. , Wang, Z. , Cho, A. , Agudelo, M. et al., Convergent antibody responses to SARS‐CoV‐2 in convalescent individuals. Nature 2020. 584: 437–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zost, S. J. , Gilchuk, P. , Case, J. B. , Binshtein, E. , Chen, R. E. , Reidy, J. X. , Trivette, A. et al., Potently neutralizing human antibodies that block SARS‐CoV‐2 receptor binding and protect animals. bioRxiv: The Preprint Server For Biology 2020. 10.1101/2020.05.22.111005 2020.05.22.111005. [DOI] [Google Scholar]
- 20.[Internet], ClinicalTrials.gov. Study Assessing the Efficacy and Safety of Anti‐Spike SARS CoV‐2 Monoclonal Antibodies for Prevention of SARS CoV‐2 Infection Asymptomatic in Healthy Adults and Adolescents Who Are Household Contacts to an Individual With a Positive SARS‐CoV‐2 RT‐PCR Assay, Identifier: NCT04452318. 2020. [cited 2020 Nov 19]. Available from https://clinicaltrials.gov/ct2/show/NCT04452318.
- 21.[Internet], ClinicalTrials.gov. A Study of LY3819253 (LY‐CoV555) in Preventing SARS‐CoV‐2 Infection and COVID‐19 in Nursing Home Residents and Staff (BLAZE‐2) Identifier: NCT04497987. 2020. [cited 2020 Nov 19]. Available from https://clinicaltrials.gov/ct2/show/NCT04497987?term=NCT04497987&draw=2.
- 22.[Internet], ClinicalTrials.gov. Safety, Tolerability, and Efficacy of Anti‐Spike (S) SARS‐CoV‐2 Monoclonal Antibodies for the Treatment of Ambulatory Adult Patients With COVID‐19, NCT04425629. 2020. [cited 2020 Nov 19]. Available from https://clinicaltrials.gov/ct2/show/NCT04425629?term=Regeneron&cond=COVID&draw=2&rank=1.
- 23. Deb, P. , Molla, Md M. A. and Saif‐Ur‐Rahman, K. M. , An update to monoclonal antibody as therapeutic option against COVID‐19. Biosaf. Health. 2021. 3: 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hou, Y. J. , Chiba, S. , Halfmann, P. , Ehre, C. , Kuroda, M. , Dinnon, K. H. , Leist, S. R. et al., SARS‐CoV‐2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 2020. 370: 1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang, Q. , Qiu, Y. , Li, J. Y. , Liao, C.‐H. , Zhou, Z. J. and Ge, X.‐Y. , Receptor utilization of angiotensin‐converting enzyme 2 (ACE2) indicates a narrower host range of SARS‐CoV‐2 than that of SARS‐CoV. Transboundary and Emerging Diseases 2020. 68: 1046–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ramanathan, M. , Ferguson, I. D. , Miao, W. and Khavari, P. A. SARS‐CoV‐2 B.1.1.7 and B.1.351 Spike variants bind human ACE2 with increased affinity. bioRxiv: The Preprint Server For Biology 2021. 10.1101/2020.05.22.111005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sabino, E. C. , Buss, L. F. , Carvalho, M. P. S. , Prete, C. A. Jr. , Crispim, M. A. E. , Fraiji, N. A. , Pereira, R. H. M. et al., Resurgence of COVID‐19 in Manaus, Brazil, despite high seroprevalence. Lancet (London, England) 2021. 397: 452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abdelnabi, R. , Boudewijns, R. , Foo, C. S. , Seldeslachts, L. , Sanchez‐Felipe, L. , Zhang, X. , Delang, L. et al., Comparative infectivity and pathogenesis of emerging SARS‐CoV‐2 variants in Syrian hamsters. bioRxiv 2021. 10.1101/2021.02.26.433062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ko, S. H. , Mokhtari, E. B. , Mudvari, P. , Stein, S. , Stringham, C D. , Wagner, D. , Ramelli, S. et al., High‐throughput, single‐copy sequencing reveals SARS‐Cov‐2 spike variants coincident with mounting humoral immunity during acute COVID‐19. bioRxiv: The Preprint Server For Biology 2021. 10.1371/journal.ppat.1009431, https://www.ncbi.nlm.nih.gov/pubmed/33831133/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meng, B. , Kemp, S. A. , Papa, G. , Datir, R. , Ferreira, I. A. T. M. , Marelli, S. , Harvey, W. T. et al., Recurrent emergence of SARS‐CoV‐2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep., 2021. 35: 109292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang, Z. , Schmidt, F. , Weisblum, Y. , Muecksch, F. , Barnes, C. O. , Finkin, S. , Schaefer‐Babajew, D. et al., mRNA vaccine‐elicited antibodies to SARS‐CoV‐2 and circulating variants. Nature 2021: 592: 616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu, K. , Werner, A. P. , Koch, M. , Choi, A. , Narayanan, E. , Stewart‐Jones, G. B. E. , Colpitts, T. et al., Serum neutralizing activity elicited by mRNA‐1273 vaccine. N. Engl. J. Med. 2021. 384: 1468–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xie, X. , Liu, Y. , Liu, J. , Zhang, X. , Zou, J. , Fontes‐Garfias, C. R. , Xia, H. et al., Neutralization of SARS‐CoV‐2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine‐elicited sera. Nat. Med. 2021. 27: 620–621. [DOI] [PubMed] [Google Scholar]
- 34. Chen, R. E. , Zhang, X. , Case, J. B. , Winkler, E. S. , Liu, Y. , VanBlargan, L. A. , Liu, J. et al., Resistance of SARS‐CoV‐2 variants to neutralization by monoclonal and serum‐derived polyclonal antibodies. Nat. Med. 2021. 27: 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tada, T. , Dcosta, B. M. , Zhou, H. , Vaill, A. , Kazmierski, W. and Landau, N. R. , Decreased neutralization of SARS‐CoV‐2 global variants by therapeutic anti‐spike protein monoclonal antibodies. bioRxiv 2021. 10.1101/2021.02.18.431897. [DOI] [Google Scholar]
- 36. Asensio, M. A. , Lim, Y. W. , Wayham, N. , Stadtmiller, K. , Edgar, R. C. , Leong, J. , Leong, R. et al., Antibody repertoire analysis of mouse immunization protocols using microfluidics and molecular genomics. mAbs 2019. 11: 870–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Medina‐Cucurella, A. V. , Mizrahi, R. A. , Asensio, M. A. , Edgar, R. C. , Leong, J. , Leong, R. , Lim, Y. W. et al., Preferential identification of agonistic OX40 antibodies by using cell lysate to pan natively paired, humanized mouse‐derived yeast surface display libraries. Antibodies 2019. 8: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu, F. , Zhao, S. , Yu, B. , Chen, Y. M. , Wang, W. , Song, Z. G. , Hu, Y. et al., A new coronavirus associated with human respiratory disease in China. Nature 2020. 579: 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang, Y. , Yang, C. , Xu, X. , Xu, W. and Liu, S. Structural and functional properties of SARS‐CoV‐2 spike protein: potential antivirus drug development for COVID‐19. Acta Pharmacol. Sin. 2020. 41: 1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li, F. Structure, function, and evolution of coronavirus spike proteins. Annual Review of Virology 2016. 3: 237–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kuate, S. , Cinatl, J. , Doerr, H. W. and Uberla, K. Exosomal vaccines containing the S protein of the SARS coronavirus induce high levels of neutralizing antibodies.Virology 2007. 362: 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Böhmer, M. M. , Buchholz, U. , Corman, V. M. , Hoch, M. , Katz, K. , Marosevic, D. V. , Böhm, S. et al., Investigation of a COVID‐19 outbreak in Germany resulting from a single travel‐associated primary case: a case series. Lancet Infectious diseases 2020. 20: 920–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wölfel, R. , Corman, V. M. , Guggemos, W. , Seilmaier, M. , Zange, S. , Müller, M. A. , Niemeyer, D. et al., Virological assessment of hospitalized patients with COVID‐2019. Nature 2020. 581: 465–469. [DOI] [PubMed] [Google Scholar]
- 44. Korber, B. , Fischer, W. M. , Gnanakaran, S. , Yoon, H. , Theiler, J. , Abfalterer, W. , Hengartner, N. et al., Tracking changes in SARS‐CoV‐2 spike: evidence that D614G increases infectivity of the COVID‐19 virus. Cell 2020. 182: 812–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Long, Q. X. , Tang, X. J. , Shi, Q. L. , Li, Q. , Deng, H. J. , Yuan, J. , Hu, J. L. et al., Clinical and immunological assessment of asymptomatic SARS‐CoV‐2 infections. Nat. Med. 2020. 26: 1200–1204. [DOI] [PubMed] [Google Scholar]
- 46. Gerdes, T. and Wabl, M. , Physical map of the mouse λ light chain and related loci. Immunogenetics 2002. 54: 62–65. [DOI] [PubMed] [Google Scholar]
- 47. Liu, Z. , Zheng, H. , Lin, H. , Li, M. , Yuan, R. , Peng, J. , Xiong, Q. et al., Identification of common deletions in the spike protein of severe acute respiratory syndrome coronavirus 2. J. Virol. 2020. 94: e00790–e00820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wrapp, D. , Wang, N. , Corbett, K. S. , Goldsmith, J A. , Hsieh, C. L. , Abiona, O. , Graham, B. S. et al., Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science (New York, N.Y.) 2020. 367: 1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dhar, M. S. , Marwal, R. , Vs, R. , Ponnusamy, K. , Jolly, B. , Bhoyar, R. C. , Fatihi, S. et al., Genomic characterization and epidemiology of an emerging SARS‐CoV‐2 variant in Delhi, India. medRxiv 2021. 10.1101/2021.06.02.21258076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hassan, A. O. , Kafai, N. M. , Dmitriev, I. P. , Fox, J. M. , Smith, B. K. , Harvey, I. B. , Chen, R. E. , Winkler, E. S. et al., A single‐dose intranasal ChAd vaccine protects upper and lower respiratory tracts against SARS‐CoV‐2. Cell 2020. 183: 169–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lee, W. S. , Wheatley, A. K. , Kent, S. J. and DeKosky, B. J. , Antibody‐dependent enhancement and SARS‐CoV‐2 vaccines and therapies. Nature Microbiology 2020. 5: 1185–1191. [DOI] [PubMed] [Google Scholar]
- 52. Kreer, C. , Zehner, M. , Weber, T. , Ercanoglu, M. S. , Gieselmann, L. , Rohde, C. , Halwe, S. et al., Longitudinal isolation of potent near‐germline SARS‐CoV‐2‐neutralizing antibodies from COVID‐19 patients. Cell 2020. 182: 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gaebler, C. , Wang, Z. , Lorenzi, J. C. C. , Muecksch, F. , Finkin, S. , Tokuyama, M. , Cho, A. et al., Evolution of antibody immunity to SARS‐CoV‐2. Nature 2021. 591: 639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sokal, A. , Chappert, P. , Barba‐Spaeth, G. , Roeser, A. , Fourati, S. , Azzaoui, I. , Vandenberghe, A. et al., Maturation and persistence of the anti‐SARS‐CoV‐2 memory B cell response. Cell 2021. 184: 1201–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang, C. , Li, W. , Drabek, D. , Okba, N. M. A. , van Haperen, R. , Osterhaus, A. D. M. E. , van Kuppeveld, F. J. M. et al., A human monoclonal antibody blocking SARS‐CoV‐2 infection. Nat. Commun. 2020. 11: 2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wu, X. , Zhou, T. , Zhu, J. , Zhang, B. , Georgiev, I. , Wang, C. , Chen, X. et al., Focused evolution of HIV‐1 neutralizing antibodies revealed by structures and deep sequencing. Science 2011. 333: 1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Walker, L. M. , Huber, M. , Doores, K. J. , Falkowska, E. , Pejchal, R. , Julien, J. ‐. P. , Wang, S. ‐. K. et al., Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 2011. 477: 466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Klein, F. , Diskin, R. , Scheid, J. F. , Gaebler, C. , Mouquet, H. , Georgiev, I. S. , Pancera, M. et al., Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV‐1 neutralization. Cell 2013. 153: 126–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tiller, T. , Meffre, E. , Yurasov, S. , Tsuiji, M. , Nussenzweig, M. C. and Wardemann, H. , Efficient generation of monoclonal antibodies from single human B cells by single cell RT‐PCR and expression vector cloning. J. Immunol. Methods 2008. 329: 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hoffmann, M. , Kleine‐Weber, H. , Schroeder, S. , Krüger, N. , Herrler, T. , Erichsen, S. , Schiergens, T. S. et al., SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020. 181: 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Klessing, S. , Temchura, V. , Tannig, P. , Peter, A. S. , Christensen, D. , Lang, R. and Überla, K. , CD4(+) T cells induced by tuberculosis subunit vaccine H1 can improve the HIV‐1 Env humoral response by intrastructural help. Vaccines 2020. 8: 604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Brachs, S. , Lang, C. , Buslei, R. , Purohit, P. , Fürnrohr, B. , Kalbacher, H. , Jäck, H. M. et al., Monoclonal antibodies to discriminate the EF hand containing calcium binding adaptor proteins EFhd1 and EFhd2. Monoclonal Antibodies in Immunodiagnosis and Immunotherapy 2013. 32: 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lapuente, D. , Maier, C. , Irrgang, P. , Hübner, J. , Peter, A. S. , Hoffmann, M. , Ensser, A. et al., Rapid response flow cytometric assay for the detection of antibody responses to SARS‐CoV‐2. Eur. J. Clin. Microbiol. Infect. Dis. 2020. 40: 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cossarizza, A. , Chang, H. D. , Radbruch, A. , Acs, A. , Adam, D. , Adam‐Klages, S. , Agace, W. W. et al., Guidelines for the use of flow cytometry and cell sorting in immunological studies (second edition). Eur. J. Immunol. 2019. 49: 1457–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Case, J. B. , Bailey, A. L. , Kim, A. S. , Chen, R. E. and Diamond, M. S. , Growth, detection, quantification, and inactivation of SARS‐CoV‐2. Virology 2020. 548: 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Data Availability Statement
The Sequences of the Viruses used in this study are available on GISAID under the following accession numbers: GISAID EPI ISL 406862, GISAID EPI_ISL_755639, EPI_ISL_610249, EPI_ISL_2718549, EPI_ISL_2718548, EPI_ISL_2718547. The Sequence of the Beta isolate can be found under the GenBank accession number: MW822592.1. The sequences of neutralizing CoV‐2 antibodies have been submitted to GenBank (accession no. in legend of Supporting information Fig. S3).


