Abstract
Distributed spatial infrastructures leveraging cloud computing technologies can tackle issues of disparate data sources and address the need for data‐driven knowledge discovery and more sophisticated spatial analysis central to the COVID‐19 pandemic. We implement a new, open source spatial middleware component (libgeoda) and system design to scale development quickly to effectively meet the need for surveilling county‐level metrics in a rapidly changing pandemic landscape. We incorporate, wrangle, and analyze multiple data streams from volunteered and crowdsourced environments to leverage multiple data perspectives. We integrate explorative spatial data analysis (ESDA) and statistical hotspot standards to detect infectious disease clusters in real time, building on decades of research in GIScience and spatial statistics. We scale the computational infrastructure to provide equitable access to data and insights across the entire USA, demanding a basic but high‐quality standard of ESDA techniques. Finally, we engage a research coalition and incorporate principles of user‐centered design to ground the direction and design of Atlas application development.
1. INTRODUCTION
In a quickly changing infectious disease landscape, access to current data and spatiotemporal insights are essential to the surveillance, monitoring, and detection of areas that are hard hit by the pandemic, so that planning efforts can be supported with solid understanding of needs. In the 1990s, infectious disease surveillance weakened in the United States, perhaps due to “false perceptions that such threats had dwindled or disappeared” (Berkelman, Bryan, Osterholm, LeDuc, & Hughes, 1994). State and local support for disease surveillance diminished as a result of budget restrictions, and the federal network relied on voluntary reporting statistics, varying diagnostic reporting standards, and a fragile computational ecosystem (Berkelman et al., 1994). On the one hand, along with the emergence of new types of data and computational power in the twenty‐first century, this perhaps facilitated tremendous growth in innovation across data surveillance techniques. City health departments brainstormed new ways of tracking disease outbreaks, from rapidly detecting new outbreaks of measles in New York City (Greene, Peterson, Kapell, Fine, & Kulldorff, 2016) to tracking food poisoning symptoms via social media to inform restaurant inspections in Chicago (Harris et al., 2014). Research exploded in using big data for public health, from integrating electronic health records and participatory surveillance systems to mining social media, web searches, and cellphone data (Bansal, Chowell, Simonsen, Vespignani, & Viboud, 2016; Lee et al., 2016; Mooney & Pejaver, 2018; Simonsen, Gog, Olson, & Viboud, 2016; Wong, Zhou, & Zhang, 2019). Yet this impactful body of work took time to shape and implement, often necessitating years of development, fine‐tuning, extensive funding, and a network of collaborators and connections. This also resulted in a geographic patchwork of technological capacity and expertise, often privileging large metropolitan areas with more advantageous access to research partners, developers, entrepreneurs, and clinical/healthcare institutions.
The novel coronavirus‐19 (COVID‐19) pandemic of 2020 tested all the stress points of infectious disease surveillance system work. By early March, much research had already emerged to provide insight into the virus, though often driven by localized interest and specialized expertise. The Johns Hopkins project had already been tracking total counts since December 2019 with international research partners; the Centers for Disease Control and Prevention (CDC) was reporting state totals daily; and other sites such as 1Point3Acres (1P3A) were reporting using their own data collection efforts. However, these approaches did not effectively capture the primary, critical need of infectious disease surveillance: universal, persistent access to data and statistical disease clusters at the finest temporal and spatial resolution available. The complex reporting structure, varying diagnostics, complex policies, and fragile computational ecosystem of the federal surveillance network made daily public data reporting below state geographic resolution implausible. Groups of volunteers, journalists, and academics emerged to scrape county data across local health departments with varying approaches and degrees of accessibility (New York Times, 2021; Centers for Disease Control and Prevention, 2020; Dong, Du, & Gardner, 2020; Johns Hopkins University, 2020; USAFacts, 2020; Yang et al., 2020). While some regions, predominantly metropolitan areas, had over a decade of practice, domain knowledge, and technological expertise to wrangle data and extract additional spatiotemporal insights, many more areas did not. Rural locales that are vulnerable to the pandemic because they have fewer hospital beds and ventilators, and mixed testing and policy environments, are also less likely to have access to the computational and analytic expertise often centered in more populated areas (Bettencourt, 2013; Bettencourt, Lobo, Helbing, Kuhnert, & West, 2007; Lobo, Bettencourt, Strumsky, & West, 2013).
A novel pandemic landscape provides crucial challenges for computational frameworks supporting surveillance structures: a rapidly changing environment, need for exploration across multiple spatial and temporal scales, variety and veracity of data sources, and need for more sophisticated spatial analysis techniques to detect emerging clusters and trends. As a basic standard, users need to quickly access updated data on a regular basis with temporal exploration enabled to track change. There is a further need for finer geographic resolution to find hotspots early using more robust spatial statistics that capture such nuances. Detecting locally high rates of COVID‐19 transmission at the county level, when compared to rates of neighboring counties, gives health workers across local health department jurisdictions the ability to detect, trace, and isolate before spreading and growing into state‐wide hotspots. Adjusting for population is also critical to detecting rural counties vulnerable to COVID‐19. At the heart of this matter is the complexity of space, and how to rapidly and effectively integrate spatial thinking in the conceptual model, methodology, and technical implementation of surveillance infrastructure.
As geographic information systems (GIS) move from siloed desktop environments to modern, heterogeneous networks, more flexible and dynamic infrastructures are required. Modern cloud mapping platforms like Google Maps, Mapbox, and Carto have become powerful alternatives to traditional GIS software but remain limited in their spatial analytical functionalities (Li, Anselin, & Koschinsky, 2015). Spatial cyberinfrastructures or cyberGIS combine distributed spatial data, distributed geoprocessing implementations, and integrated software systems (as spatial middleware) to perform spatial analysis, facilitate geovisualization, and coordinate knowledge discovery within web environments (Anselin, 2012; Wang, 2010; Wright & Wang, 2011; Yang, Kafatos, Wang, Wolf, & Yang, 2010). These cyberinfrastructures specialized for geospatial data and applications serve as technology‐agnostic extensions of service‐oriented architectures, grid, and cloud computing architectures essential for integrating data across distributed settings. Health applications and extensions of spatially explicit cyberGIS and distributed infrastructures are still in early stages, albeit with much promise and recent advancement. Clinical system application examples often attempt to integrate individual health data and environmental sensors with real‐time geoprocessing to assess exposure and activity trends (Goldberg et al., 2013; Jankowska, Schipperijin, Kerr, & Altintas, 2016). Several projects have additionally leveraged social‐spatial data from geotagged social media to track flu pandemics by incorporating spatial cyberinfrastructures to capture and process data streams, model spatiotemporal patterns, and explore flu trajectories with computational elegance (Bansal et al., 2016; Hay, George, Moyes, & Brownstein, 2013; Khoury & Ioannidis, 2014; Lee et al., 2016; Mooney & Pejaver, 2018; Simonsen et al., 2016; Wong et al., 2019). With COVID‐19, an explosion of geospatial perspectives facilitated new understandings of the pandemic throughout its multiple waves. Spatial diffusion of the virus traced in early stages highlighted the importance of spatiotemporal trends (Thakar, 2020). Building on geocomputational work done prior to the pandemic, social media were leveraged to monitor, measure, and understand different dimensions of the pandemic (Gray, Anyane‐Yeboa, Balzora, Issaka, & May, 2020; Huang et al., 2020; Xu, Zhang, Wang, 2020). At the same time, systemic inequities in healthcare and economic activity—often with distinct geospatial patterns—were linked with social, racial, and ethnic disparities in COVID‐19 cases, hospitalization, and death (Gray et al., 2020).
Distributed spatial infrastructures leveraging cloud computing technologies can tackle issues of disparate data sources and address the need for data‐driven knowledge discovery and more sophisticated spatial analysis central to the COVID‐19 pandemic. Recent efforts integrating spatial analysis software and libraries in a cyberinfrastructure to power cloud mapping or webGIS are extensive, with emerging promise in health applications. However, it would often take a significant amount of time and cost to build such a system and require many hardware resources to serve online users at a scale of hundreds of thousands. The US COVID Atlas serves as a new, unique application of cloud mapping with spatial analysis capabilities, extending cyberinfrastructure concepts. We implement a new, open source spatial middleware component (libgeoda) and system design to scale development quickly to effectively meet the needs of surveilling county‐level metrics in a rapidly changing pandemic landscape. We incorporate, wrangle, and analyze multiple data streams from volunteered and crowdsourced environments to leverage multiple data perspectives. We integrate explorative spatial data analysis (ESDA) and statistical hotspot standards to detect infectious disease clusters in real time, building on decades of research in GIScience and spatial statistics. We scale the computational infrastructure to provide equitable access to data and insights across the entire USA, demanding a basic but high‐quality standard of ESDA techniques. Finally, we engage a research coalition and incorporate principles of user‐centered design to ground the direction and design of Atlas application development.
The US COVID Atlas proves to be innovative in multiple ways, even among the crowded and competitive landscape of pandemic dashboards that emerged in 2020. From a design perspective, the Atlas integrates participatory design techniques to iteratively refine functionality using formal and informal discussions with a variety of users and a growing research coalition. From a data perspective, the Atlas is unique in integrating multiple disparate data sources, giving users the ability to visualize many aspects of data from different data sources, and also creating new visualizations using advanced spatial analytic features. From an infrastructure perspective, the Atlas is the first web application (to our knowledge) that integrates WebAssembly technology to manage computationally intensive spatial analysis functions (written in C++) directly in the web browser, opening wide new possibilities of browser‐based geoprocessing and GIScience.
In Section 2 we review the user‐oriented approach, data utilized (including dynamic and static components), and analytic strategies of the Atlas application. We also review the system infrastructure and dynamic data management strategy. In Section 3 we review the final application user interface and highlight examples of knowledge discovery facilitated through the application. In Section 4 we discuss and conclude.
2. INFRASTRUCTURE FRAMEWORK
The Atlas application was initiated in early March 2020 in response to the need for county‐level, dynamic visualizations of COVID‐19 infections across the United States. The objectives of the Atlas application were defined to meet the needs of the following goals: first, to provide access to real‐time data on the COVID‐19 pandemic at the county level; second, to incorporate ESDA and statistical hotspot standards to detect infectious disease; and third, to link the pandemic data to local community contexts to assist with rapid policy responses. The primary users of the Atlas were intended to be public health practitioners and planners at regional scales, clinical health professionals, and health researchers interested in tracking the pandemic in real time to facilitate knowledge discovery and refine hypotheses in a rapidly changing landscape. This later expanded to translate basic insights to public and business‐oriented audiences, though a primary audience of regional science and health planners in resource‐limited settings persists. To practice participatory design, we incorporated multiple research groups and health organizations in early conversations and focus groups and iteratively updated the Atlas application over time with ongoing feedback. Participatory design is characterized by user input and generally follows three stages: initial exploration of work, discovery process, and prototyping (Spinuzzi, 2005).
While led by the core group at the University of Chicago in spatial infrastructure, contributions to the Atlas persist across a network of collaborators at the University of Wisconsin‐Madison COVID Data Science Team, County Health Rankings & Roadmaps, Yu Research Group at the Department of Statistics of University of California Berkeley, CSI Solutions (a private sector health organization linking a national network of rural health practitioners), multiple volunteers and contributors, and multiple focus groups incorporating dozens more health and planning decision‐makers. A graphic design and communications team was integral to brainstorming scenarios and solutions, and a library/information science group was critical in developing working tutorials to make complex tasks accessible for users. Because the users of the Atlas also contributed in its development and thus shaped the goals of the final application (from data environment to visualization styles), we view the Atlas as a dynamic and shared systems infrastructure. The Atlas was developed as a free, open source application that leverages and links multiple open source tools and products. Open source software continues to grow in the spatial analysis community, facilitating methodological innovation and refining building blocks for more advanced analysis (Anselin, 2012; Rey, 2009). The Atlas code repository can be viewed at https://github.com/GeoDaCenter/covid, with core spatial analytics library built on GeoDa open software (Li et al., 2015).
2.1. Data and scale
We extract dynamic and static data across multiple domains for integration into the Atlas (see Table 1), including COVID‐19 health outcomes, local community healthcare capacity, and population, demographic, and socioeconomic status.
TABLE 1.
Data, scale, sources, and update process of the Atlas data project1
| Domain | Data (*Atlas calculation) | Source | Spatial resolution | Temporal resolution | Atlas update process | Atlas integration |
|---|---|---|---|---|---|---|
| COVID health outcomes | Cases | USAFacts (2020) and 1Point3Acres (2020) | County and state | Daily | Python scripts to update from source. Updated daily since March 2020 | Variable calculation, LISA statistical analysis, thematic maps, time slider, info. window, graph |
| *New cases | ||||||
| *7‐day ave. new | ||||||
| *7‐day ave. new per 10,000 ppl | ||||||
| Cumulative cases | ||||||
| *Per 10,000 ppl | ||||||
| *Per licensed bed | ||||||
| Deaths | ||||||
| *New deaths | ||||||
| cumulative | ||||||
| *Per 10,000 ppl | ||||||
| *Per licensed bed | ||||||
| Testing data | Li et al. (2020) | County and state | Daily | Updated daily since September 2020 | Thematic maps, time slider, data dashboard | |
| Total testing; | *Multiple sources (see Appendix A) | |||||
| *Positivity rate; | ||||||
| Testing criterion | ||||||
| COVID statistical insights | *Statistical hotspots (LISA analysis) | Li et al. (2020) | County | Daily |
Libgeoda on‐the‐fly calculation. Updated daily since March 2020 Python scripts to update from source. Updated daily since April 2020 |
Thematic maps, time slider |
| Hospital Severity Index (5‐day) | Altieri et al. (2021) | County | Daily | Data dashboard | ||
| Predicted death for 7 days | ||||||
| Healthcare capacity | Licensed hospital beds | CovidCareMap (Su et al., 2020), HRSA (pull all the related) (2018 facility report, U.S. Department of Health and Human Services, 2020) | County | Most recent available | Updated in March 2020 | Variable calculation, data dashboard |
| Community contextual factors | Population | U.S. Census Bureau (2019) | County and state | 5‐year average | Updated in March 2020 | Variable calculation, data dashboard |
| Community health factors: children in poverty; income inequality; median household income; food insecurity; unemployment; uninsured; primary care physicians; preventable hospital stays; residential segregation; severe housing problems | County Health Rankings and Roadmaps (2020) | County | Multiple | Updated in March 2020 | Data dashboard, thematic maps for (select variables) | |
| Community health context: 65 and older; adult obesity; diabetes prevalence: adult smoking; excessive drinking; drug overdose deaths | *Multiple sources (see Appendix B) | |||||
| Length and quality of life: life expectancy; self‐rated health | ||||||
| Geographic boundaries | US counties | US Census, TIGER (2018a) | County | Most recent available | Updated in March 2020 | Primary display (default), info. window |
| US Congressional districts | US Census, TIGER (2018b) | Congressional district | Updated in September 2020 | Overlay | ||
| US states | US Census, TIGER (2018a) | State | Updated in March 2020 | Primary display, info. window | ||
| Native American reservations | US Census, TIGER (2017) | Reservation | Updated in April 2020 | Overlay | ||
| Hypersegregated cities | Massey and Tannen (2015) | Metropolitan areas | Updated in May 2020 | Overlay | ||
| Black Belt counties | Southern US counties that were at least 30% Black or African American in the 2000 Census | County | Updated in May 2020 | Overlay |
2.1.1. Dynamic data
COVID‐19 health outcomes
To approximate daily COVID‐19 health outcomes, we extract daily confirmed COVID‐19 cases and deaths from USAFacts (2020) and 1P3A (Yang et al., 2020). We started with 1P3A in early March 2020 as it was the initial, crowdsourced data project aggregating county‐level data that served as a volunteer project. We access this data stream using a token provided by the group. We then added USAFacts data, considering that their county‐level data is confirmed by referencing state and local agencies directly (USAFacts, 2020).
While the CDC eventually reported county‐level data on COVID‐19 cases and death, it relies on the data collected by USAFacts, a non‐profit organization. That is, the CDC does not certify the quality of any individual data source reporting on COVID‐19 at the county level and, in fact, no federal governmental agency currently assumes this quality control role with near real‐time data estimates. Instead, the CDC refers readers to USAFacts to learn more about their data collection strategies and methodological decisions (Centers for Disease Control and Prevention, 2020). At the same time, the CDC also recognizes that discrepancies exist between national, state‐, county‐, and city‐level data pertaining to COVID‐19 and encourages readers to refer to local sources in cases of any discrepancy. In this context, multiple organizations, both formal and informal groups, have emerged to survey the local pandemic by reporting county‐level COVID‐19 cases and deaths on a daily basis. In addition to USAFacts, volunteers of 1P3A launched a crowdsourced data project in which they collect and disseminate county‐level COVID‐19 data. The New York Times has also assumed a major role in monitoring the spread of this pandemic, providing daily updates on COVID‐19 cases and deaths at the county level. Combining data from these sources and beyond, a group from Johns Hopkins University also releases a daily report on COVID‐19 cases and deaths. Importantly, each organization applies varied data collection and processing methodologies, resulting in inconsistencies in the daily case and death numbers associated with COVID‐19. Considering the discrepancies among different data sets, we provide more than one COVID‐19 data set in the Atlas to allow exploration and comparison.
Health indicator variable calculation
From confirmed case and death data, we calculate the following variables: daily new cases, 7‐day average new cases, cumulative cases, cases per 10,000 persons, 7‐day average of new cases per 10,000 persons, cases per licensed bed, daily new deaths, cumulative deaths, deaths per 10,000 persons, and deaths per licensed bed. We calculate the 7‐day average variable by taking the difference between the current day's confirmed count and the confirmed count 7 days ago, and then dividing this difference by 7. For example, we took the difference between the confirmed count as of June 30 and that as of June 23, divided the difference by 7, then used this as an estimate for the “7 day average daily confirmed count” variable for June 30. This measures the daily average growth for the week before June 30. This calculation is only available with USAFacts data because of data completeness, as 1P3A has missing data in January and February 2020.
Forecasting statistics
We integrate a COVID‐19 Pandemic Severity Index (cPSI) and 7‐day predicted death counts for each county from Altieri et al. (2021). These data were originally compiled, cleaned, and continue to update a large corpus of hospital‐ and county‐level data from a variety of public sources to aid data science efforts to combat the COVID‐19 pandemic. Specifically, the cPSI takes on three values (low, medium, high) to suggest the severity of the local COVID‐19 outbreak, based on cumulative deaths so far and predicted new deaths.
2.1.2. Static data
Populations at risk
Population data from the 2014–2018 5‐year American Community Survey were included to generate population‐adjusted COVID‐19 numbers (United States Census Bureau, 2020). We also extracted data reflecting healthcare system capacity from the 2018 facility reports of COVIDCareMap (Su et al., 2020), including the number of licensed hospital beds and number.
Community risk factors
We integrate county‐level social, economic, and health indicators to better contextualize the local COVID‐19 pandemic based on coalition discussions with County Health Rankings & Roadmaps (Remington, Catlin, & Gennuso, 2015), including percentage of children in poverty, income inequality (measured as the ratio of household income at the 80th percentile to income at the 20th percentile), median household income, food insecurity rate (measured as percentage of population who lack adequate access to food), unemployment rate, uninsured rate, primary care physicians (measured as ratio of population to primary care physicians), preventable hospital stays, residential black–white segregation, severe housing problems rate (measured as percentage of households with at least one of four housing problems: overcrowding, high housing costs, lack of kitchen facilities, or lack of plumbing facilities), percentage of population aged 65 or over, adult obesity (measured as percentage of the adult population (age 20 and older) who report a body mass index greater than or equal to 30 kg/m2), diabetes prevalence (measured as percentage of adults aged 20 or over with diagnosed diabetes), adult smoking (measured as percentage of adults who are current smokers), excessive drinking (measured as percentage of adults reporting binge or heavy drinking), drug overdose deaths (measured as drug poisoning deaths per 100,000 population), life expectancy (measured as average number of years a person can expect to live), and self‐rated health (measured as percentage of adults reporting fair or poor health).
Geographic risk factors
Hypersegregated cities, Native American reservations, and Black Belt areas were identified as vulnerable to the pandemic based on research coalition and focus group discussions. Hypersegregated cities include 52 metropolitan areas identified by Massey and Tannen (2015) that satisfied the criteria for black hypersegregation from 1970 to 2010. Recognizing the critical importance of spatial segregation in understanding racial stratification, Massey and Denton (1989) developed the concept of hypersegregation to describe metropolitan areas in which African Americans were highly segregated on at least four of the five dimensions of segregation they had identified (unevenness, isolation, clustering, concentration, and centralization). The Southern Black Belt areas have been variously defined (Webster & Bowman, 2008). We include southern US counties that were at least 30% black or African American in the 2000 Census. To highlight these vulnerable areas, the Atlas features each of these areas as one optional map overlay or highlight so that users can explore the local pandemic in these areas themselves.
2.2. Analytic methods
The Atlas incorporates multiple visual analytic methods to explore data across space and time, as well as spatial statistical products to measure spatial clusters and outliers.
2.2.1. Choropleth maps
The Atlas provides choropleth maps for geovisualization, a thematic map that represents data through various shading patterns on predetermined geographic areas (e.g., counties, states). We adopt Jenks natural breaks maps (Jenks, 1977) to classify the values of the pandemic data for each county/state. Specifically, a natural break map uses a nonlinear algorithm to maximize within‐group homogeneity so that we can group similar observations and highlight outliers. Importantly, the Jenks natural breaks are applied in two different ways to better visualize spatial temporal patterns of COVID‐19 cases. In the default setting (natural breaks—fixed bins), the algorithm is applied to the data for the most recent date and the resulting breaks are used to group observations for historical data. In other words, the legend stays the same as users explore the time slider, allowing better visualization of the pandemic spreading over time. The other option (natural breaks) applies the nonlinear algorithm to group observations for every day's data, generating maximized within‐group homogeneity for each day's data, resulting in different break values for every day. In the second case, the algorithm is applied to the most recent day's data and the resulting break values.
2.2.2. Spatiotemporal interactive exploration
The thematic maps on the Atlas are interactive and quickly provide access to multiple sources of COVID‐19 data over time for historical exploration using a time slider tool. For example, applying the natural breaks with fixed bins on accumulative confirmed COVID‐19 cases displays how the disease has spread over time since the very beginning: from just a few counties in March 2020 to almost the entire country by June. Alternatively, visualizing the temporal change based on the daily new confirmed cases (or 7‐day average) helps detect areas with emerging cases. Interactive, explorative temporal detection like this allows us to capture not only the base rate, but also the changes that are both critical to tracking areas of concern.
Individual counties or states are also interactive. An information window is enabled on hover, providing details on the location (i.e., state name or county name) and basic COVID‐19 case and death data. Upon clicking, a county‐specific data dashboard opens (on the right‐hand side) with detailed health indicators and community risk factors. Data are updated by clicking on a new county. A graph showing new cases and smoothed trends for the selected county is updated on the left‐hand side accordingly (see Figure 5b for a demonstration of clicking on Panola County, MS).
FIGURE 5.
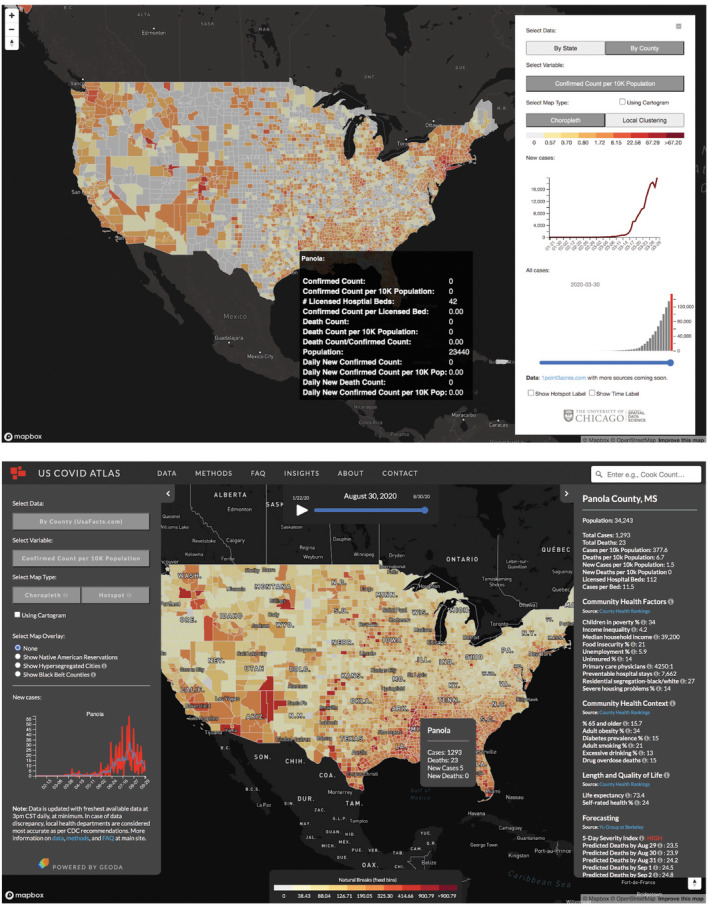
Screenshots of COVID Atlas web application on (top) March 30, 2020, and (bottom) August 30, 2020. In the matured application, a county‐level dashboard links dozens of contextual variables for each area selected
2.2.3. Spatial statistical analysis
Statistical spatial cluster identification based on local indicators of spatial association (LISA; Anselin, 2010, 2019; Anselin & Li, 2019) is implemented on case and death variables daily using an on‐the‐fly libgeoda service (see Section 2.3 for details). The clustering technique allows users to identify groups of contiguous counties that are statistically similar based on the COVID‐19 indicator (accumulative COVID‐19 cases, daily new cases, etc.). The “high‐high” cluster, for example, can be of particular interest as it allows identification of a group of areas (i.e., counties, or states, in Atlas) where not only their own COVID‐19 indicators are high but also they are surrounded by neighboring areas where the COVID‐19 indicators are high.
Note that we intentionally designed Atlas to be accessible to a wide audience. Therefore, we preset it with certain hyperparameters. Specifically, we use a first‐order queen contiguity weight and one CPU core12 to estimate the local Moran's I, with 999 Monte Carlo permutations and a 0.05 threshold for statistical significance. However, using GeoDa,3 others can replicate these results or employ different hyperparameters, such as different spatial weights to measure geographical similarity, different randomization options (e.g., number of Monte Carlo permutations and specific seeds), or different thresholds for statistical significance.
2.3. Systems infrastructure
2.3.1. System architecture
Because of the rapid development of the COVID‐19 pandemic, the US COVID Atlas was developed in a timely manner based on the libgeoda library, which is an existing library that provides the core features of the open source spatial analysis software GeoDa (Anselin, Syabri, & Kho, 2010). The core spatial analysis features, which include spatial weights, spatial clustering, spatial statistics, and spatial regression, were written in C++ and have been wrapped by SWIG (Beazley, 1996) for easy integration in Python and R (see Figure 1). In this research, WebAssembly (Haas et al., 2017) is used to compile the libgeoda code into a WebAssembly module (libgeoda.wasm) that can be executed in the web browser using JavaScript (see the red‐outlined boxes in Figure 1). A JavaScript module geoda.js is developed to bridge the communication between COVID Atlas user interface components and libgeoda.wasm. Combining the Mapbox platform, the COVID Atlas is able to do spatial exploration analysis of COVID‐19 data in a browser instead of a heavily equipped server with spatial database. This allows the COVID Atlas to be built and published as a cloud mapping application with spatial analytics in a short time period at a fraction of the cost of a traditional webGIS system.
FIGURE 1.
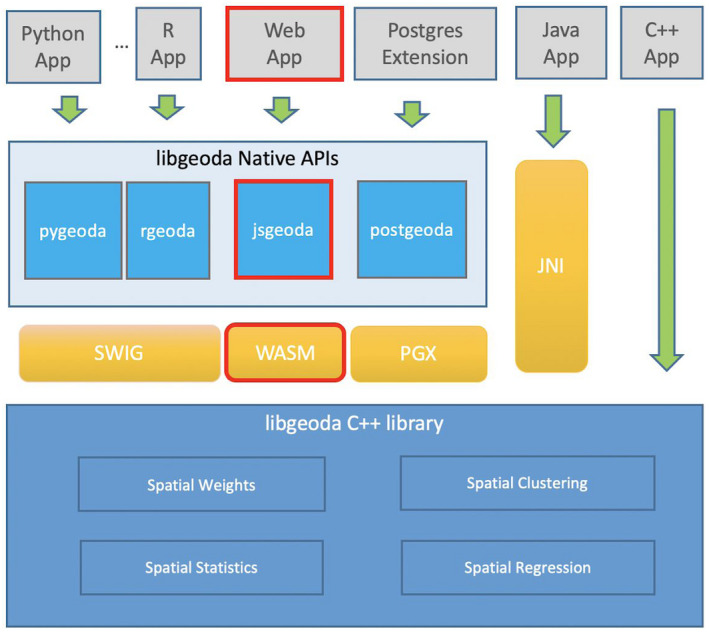
Software architecture of the libgeoda library
The COVID Atlas is designed as a web application that integrates a Mapbox platform, COVID‐19 data cleaning module, and libgeoda (see Figure 2). The COVID‐19 data clean module is an Extract, Transform, and Load module (see section below) developed to collect data automatically from several crowdsourced and volunteer‐contributed COVID‐19 data sources, clean the data automatically by removing noises that are not matched to any geographical units, validate the data manually if there are any issues that cannot be solved by the automatic scripts, and format the data to be consumed by the COVID Atlas web application. These data are aggregated separately at county and state level. Specifically, these data include longitudinal cumulative confirmed cases, longitudinal cumulative death counts, population, number of licensed beds, and other static data.
FIGURE 2.
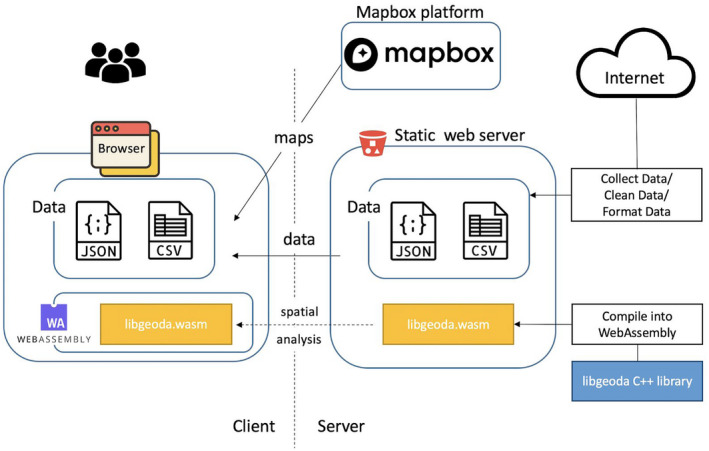
System design of Covid Atlas
The cleaned and formatted data are hosted on a static website in CSV and GeoJSON format, and are sent to users’ browsers on request. Then the libgeoda.wasm module is downloaded by and executed in the browser asynchronously to create the natural breaks map, box map or cartogram map for a selected variable, construct contiguous (queen) spatial weights and apply local spatial autocorrelation analysis (e.g., local Moran statistics) to locate hot‐ and cold‐spots. The results of spatial analysis are rendered as different choropleth maps using mapbox.gl and deck.gl. Because WebAssembly provides near‐native code execution speed in most modern browsers, the spatial analysis functions in the COVID Atlas run almost instantaneously. This enables us to implement a feature of spatiotemporal exploration of the COVID‐19 data, which allows users to examine how the patterns of COVID‐19 data change over time.
2.3.2. Shared data management system
We implement a combined managed and shared data handling system (as defined in Kolak et al., 2020), integrating dynamic data from multiple sources daily, linking with static sources, and then pushing cleaned data to the GitHub server to enable the automated ESDA processing and mapping (see Figure 3). Mapping and analytic services are made available for both upstream and downstream users from the Atlas. Current data files (flat and spatially enabled) are also available on the GitHub repository for download. Data are rerun daily for the entire temporal scale in the central processing module, as corrections to recent case or death data are not uncommon across third‐party data services. While the data processing pipeline was initially manual, it was gradually shifted to an automated system (see Figure 4) implementing an AWS Elastic Container Registry (ECR), AWS Elastic Container Service (ECS), and AWS Simple Storage Service (S3). First a docker image is deployed in the ECR and scheduled tasks are executed on the ECS. Data and processing scripts are retrieved from GitHub, and then recent COVID‐19 data sources are pulled, processed, and trigger email warnings if not completed successfully. A backup of cleaned data is stored on the S3 instance, and final data are pushed to GitHub, and will later be made available as an application programming interface (API) for the public. The data can also be piped through an alternate ESDA route, using pygeoda to calculate identical LISA clusters that could be sent to the core mapping service, and/or the API. As the scale of COVID‐19 data grows over time, this alternative may facilitate a more traditional approach of pre‐calculating results before pushing them to the front‐end.
FIGURE 3.
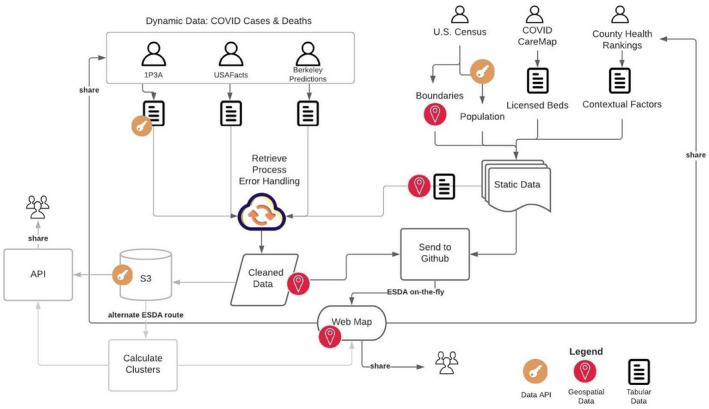
Data management system
FIGURE 4.
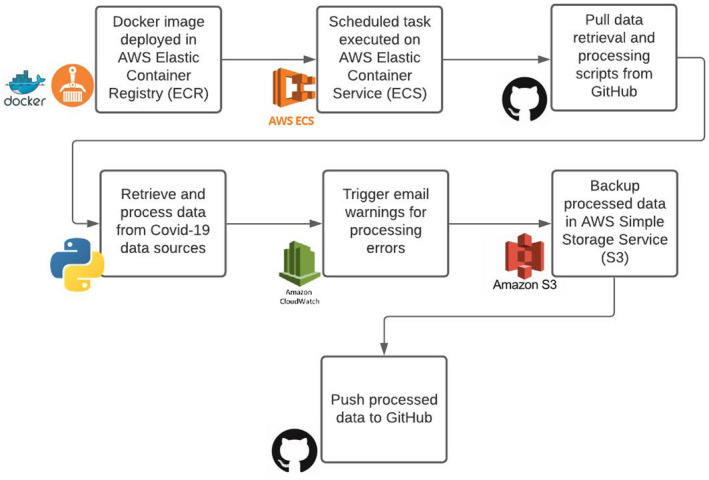
Data processing pipeline
3. RESULTS
The application was launched in late March 2020, and by September grew to more than 72,000 users and more than a dozen reviews in various news media outlets. The mapping interface went through multiple design iterations based on research coalition and user feedback, reflecting adherence to a participatory design framework. For example, addition of the popup information window with county clicks, including overlays reflecting geographic risk (like the Black Belt), and linking county‐level community variables dashboard features were all integrated following active discussions and informal user testing sessions with the research coalition (see Figure 5). Through these discussions, we identified and addressed key points from the coalition, including: translating spatial analysis terminology into more user‐friendly terms; adding icons to link to more information for complex topics; shifting the user workflow from left to right (rather than right to left); updating text and typography settings to elevate key terms and concepts; and much more. The coalition and user meetings also helped workshop which variables were considered priority (and thus needing to be styled and elevated accordingly), and which variables to integrate next. The mapping application further facilitated multiple insights into the pandemic, including highlighting the importance of spatial scale of disease patterns, interactive temporal exploration to detect emerging trends, and detection of geographic regions of risk reflecting underlying disparities.
Evaluating the spatial scale of the pandemic is essential for evaluating geographic risk over time. While healthcare policy is often designed at a higher level, say state or federal, the movement of people, sharing of resources and henceforth spread of infection are more local phenomena. For a cluster to be detectable at the state level, the prevalence rate needs to be very high, not only in that state but also in its neighboring states, and by that time it might be too late to contain further spread. County‐level data provide more granular information about the direction and magnitude of the activity. (Note that the true spatial scale of the COVID‐19 pandemic is arguably at an even finer resolution; however, case data below the county‐level resolution remain unavailable for the entire country for wider analysis.) In Figure 6 we illustrate disease patterns as a choropleth map of states and counties. At the state level, Illinois is shown as an area of concern. At the county level, it becomes clear that the Illinois flag was driven by a surge of cases in the Chicagoland area, constituting only a geographic fraction of the entire state. At the same time, we find hotspots in First Nation communities in the Southwest, and several rural poor counties in Alabama, that have some of the highest disease rates in the country; though because they were averaged out, they are missed at the state level. Finally, when LISA cluster analyses are implemented, county cluster regions become even clearer. The spread of COVID‐19 across the Southeastern counties pointed towards a persistent hot spot (using population‐adjusted total cases) in the region since the end of May. This hotspot initially spanned across counties in four states (Alabama, Georgia, Louisiana, and Mississippi). It did not emerge as a COVID‐19 hotspot at the state level till mid‐July. By that time the activity had spread out far enough to parts of eastern Arkansas and western Florida (see Figure 7).
FIGURE 6.
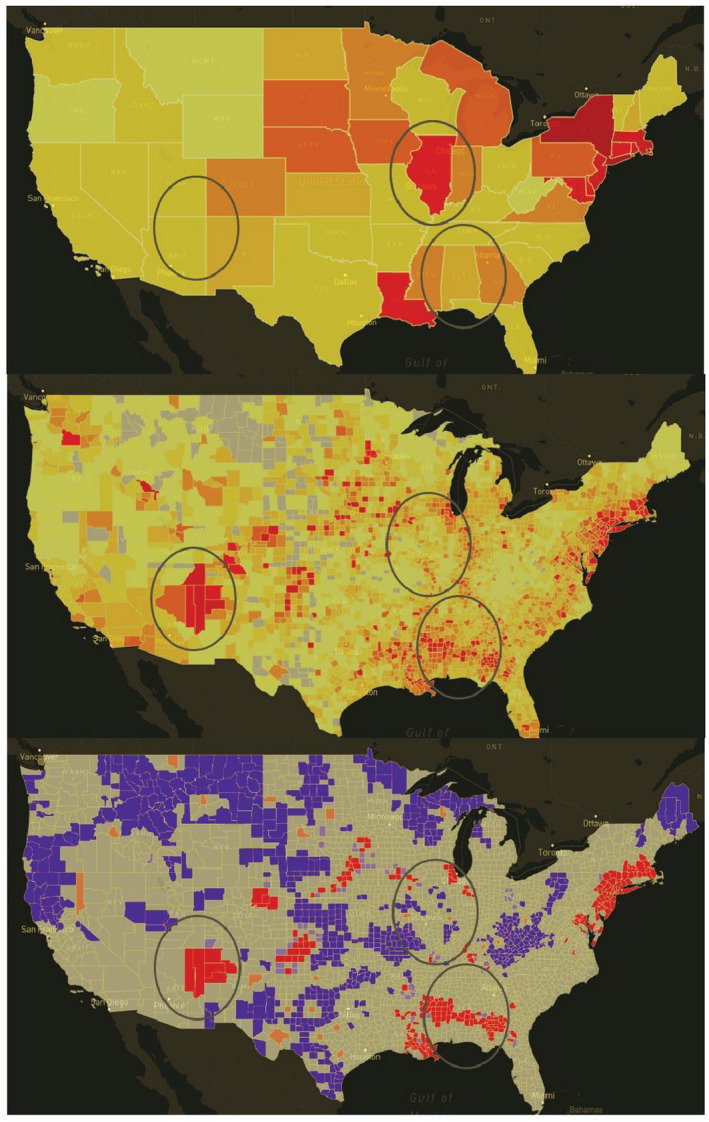
COVID case rates across different scales, using confirmed cases per 10,000 persons via USAFacts data on June 1, 2020
FIGURE 7.
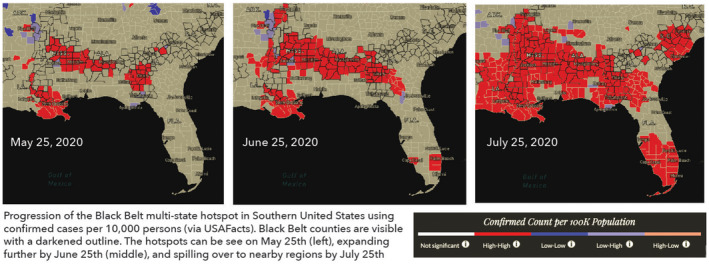
Black Belt multi‐state hotspot progression
Border effects or spillover across administrative lines can also create local clusters that might not be significant enough to detect at a state level. The interactive exploration enabled by the COVID Atlas also facilitates the exploration of trends over time and borders. Visualizing county‐level data at different times using measures like the 7‐day average daily new confirmed count per 10,000 population can help identify smaller emerging clusters that demonstrate increasing levels of disease activity but have not reached enough mass like other more mature and persistent hotspots generally identified using confirmed count per 10,000 population. For example, using the daily new variable adjusted by population, we found emerging hotspot areas days or weeks before they were communicated across national media outlets, such as New Orleans in March and the southern borders of Arizona and California in June. A steady rise in population‐adjusted cases was apparent after Memorial Day using the application, showing a lateral spread across the country in predominantly rural locales that prefaced the dramatic nation‐wide jump known as the second COVID‐19 wave. While not all emerging hotspot trends mature into larger and more mature hotspots, early detection remains essential to disease transmission mitigation. With urban areas attracting much focus early on in the pandemic, identifying these smaller clusters (often in less populated, rural areas) can support local officials with resource allocation and planning needs.
Together with temporal exploration, trends across multiple maps helped us uncover important trends of COVID‐19 spread and highlight regions of geographic risk. We identified consistent hotspots in Native American reservations and Black Belt counties by looking at how hotspots changed over time based on COVID‐19 case rates. These areas experience the direct legacies of colonialism within the United States with persistent community de‐investment and limited access to essential infrastructures (Power et al., 2020; Webster & Bowman, 2008). Within Native American communities, healthcare capacity is limited, with many required to travel hours to the nearest facility, and many communities do not have access to basic water or electricity (Brosemer et al., 2020; van Dorn, Conney, & Sabin, 2020). In the case of the Black Belt, unequal access to quality wastewater treatment has been connected to parasitic worm outbreaks in children—known as diseases of poverty (Bradbury et al., 2020; Meza, 2018). We found that hotspots have clearly emerged within this band and grown over time, especially across the Mississippi Delta (see Figure 7). A patchwork of policies between states made a unified public health approach even more difficult to achieve. While some of the highest rates of COVID‐19 have persisted here since spring, they have received less attention in the national discussion on COVID‐19. By adding the Black Belt as an overlay in the Atlas, we intend to shed more insight into the context of the story driving disease patterns over time.
4. DISCUSSION AND CONCLUSIONS
Exploring COVID‐19 data in near‐real time is critical to generating hypotheses and facilitating knowledge discovery in a rapidly evolving landscape characterized by inherent uncertainty of novel virus behaviors, heterogeneous policy and intervention approaches, and limited data settings. An ESDA approach enabled the exploration of data using multiple data sets and techniques to uncover real trends that remain consistent across differing analyses. To leverage ESDA capabilities of existing spatial statistical software, an inverted infrastructure and use of the libgeoda library were successfully developed in a fraction of the time and cost required of more standard and traditional approaches.
The US COVID Atlas application facilitated several insights as the pandemic matured across the country, including highlighting the importance of the spatial scale and necessity for finer‐grained analysis; need for interactive, temporal exploration of data to uncover spillover behaviors and hotspot changes over time; and illuminating regions of geographic risk that are driven from historical social, economic, and demographic disparities. ESDA techniques including LISA analyses have been successfully employed to better understand the COVID‐19 crises at finer spatial scales in Germany (Scarpone et al., 2020) and China (Kang, Choi, Kim, & Choi, 2020; Li et al., 2020; Yao et al., 2020). In the United States, daily surveillance using a space‐time scan statistic in the Covid19 Scan application similarly highlighted temporal trends towards smaller and more numerous clusters below state levels (Desjardins, Hohl, & Delmelle, 2020; Hohl, Delmelle, & Desjardins, 2020; Hohl, Delmelle, Desjardins, & Lan, 2020).
When uncovering areas of geographic risk, recent research is also in agreement with our finding that “geography matters.” Sun and colleagues also uncovered significant spatial clustering in the Black Belt, and went on to demonstrate that spatial models outperform aspatial approaches, especially when accounting for complex spatial heterogeneity patterns at the county scale (Sun, Matthews, Yang, & Hu, 2020). Identifying geographic boundaries that represent legacies of racist policies and community de‐investment like redlined neighborhoods have been linked to worse COVID‐19 health outcomes (Bertocchi & Dimico, 2020). Structural inequities and healthcare gaps have also been found to contribute to worse outcomes for communities of color in the 1918 influenza epidemic (Krishnan, Ogunwole, & Cooper, 2020). While access to data below county level and across multiple racial and ethnic groups is necessary to better understand the complexity of COVID‐19 health disparities, in its absence, integrating historical regions of persistent inequity may serve to approximate risk.
The COVID Atlas aims to create a unique data visualization highlighting local disease clusters and linking community context. There are many other dashboards that serve to capture some, if not all, aspects of the COVID‐19 pandemic. COVID‐19 dashboards could be categorized into three main types in terms of the functionalities that they serve: data source dashboards, data presentation dashboards and data analyzer dashboards. The first category, data source dashboards, includes dashboards produced by official/third‐party proprietary data sources, and they serve mostly to visualize raw data about the fundamentals of the COVID‐19 pandemic, usually with a particular geography in mind. These dashboards are hosted on state health department websites, and by the institutions (JHU, 1P3A, USAFacts, etc.) that aggregate the data. The second category, data presentation dashboards, serves mainly to reproduce existing data sources from data providers in the first category, usually publishing their own version of data as well. Examples include the COVID Tracking Project, which integrates and conforms existing state‐level data, especially on racial distribution and testing, and republishes through its own API (COVID Tracking Project, 2020), Worldometer (2020) and the University of Virginia's COVID‐19 Surveillance Dashboard (2020), which also serve similar functions. Dashboards of the third type go beyond data presentation and integration, in that they generally will produce some of their own metrics to measure one or more aspects of the pandemic. There are tools that try to reinterpret all aspects of existing data, by creating metrics for testing, infection rate, positive rate, and hospital headrooms, such as Covid Act Now, which systematically defines risk levels for the 50 US states (Covid Act Now, 2020), and Chris Glazner's COVID‐19 Dashboard and Tools, which provides more decision support and forecasting modeling based on existing data (Glazner, 2020). Other such dashboards try to capture one or two aspects of the pandemic, such as Covid19 Scan, which tracks significant space‐time clusters of outbreaks in the US (Desjardins et al., 2020; Hohl et al., 2020), and UW‐Madison's GeoDS Lab's Mapping Mobility Changes in Response to COVID‐19 (Gao, Rao, Kang, Liang, & Kruse, 2020). The COVID Atlas sits uniquely between the second and third category, where we integrate existing data sources, giving users the ability to visualize many aspects of existing data from different data sources (in our case 1P3A and USAFacts) at the state and county level, and also creating our own geographical visualization of local hotspots of many metrics (confirmed cases, confirmed deaths, etc.) using local Moran algorithms. The COVID Atlas is a data integration and repackaging tool, but, more importantly, it provides proprietary analysis based on existing data, confirming and extending findings shown in other geographic COVID‐19 dashboards. The Atlas is further distinguished by its research coalition integrating multiple disciplinary approaches, participatory design, and participatory goals resulting in new insight and linked contextual variables. Our open source approach also enabled rapid iterations, testing environments, and sharing of outcomes.
From another perspective, the computational infrastructure of the US COVID Atlas is unique, even among the thousands of COVID‐19 related online dashboards that emerged in the past year. (As of March 1, 2021, more than 20,000 results are returned from a Google Scholar search using keyword “COVID‐19 dashboards”.) Some dashboards are created to track the raw COVID‐19 cases and deaths in real time (Dong et al., 2020), the clinical trials (Thorlund et al., 2020), and the vaccination distribution (Berry, Soucy, Tuite, & Fisman, 2020; Bingham, 2021). Other dashboards provide expert knowledge based on COVID‐19 and related health, social and economic data, such as the Pandemic Vulnerability Index dashboard (Marvel et al., 2021) and the CoRisk‐Index to track the industry‐specific risk assessments (Stephany et al., 2020). Other applications help assess COVID‐19 risk with real‐time, geolocalized information (Chande et al., 2020), or map county‐level mobility patterns to detect new trends in the outbreak (Gao et al., 2020).
These online COVID‐19 dashboards use information management techniques to visually track, analyze, and display COVID‐19 related data, which are typically visualized as maps, tables, and line charts. To allow the user to interact with the dashboards, the data are normally pre‐processed and stored in a rational database or other structured data formats (e.g., JSON, XML) on a server that is efficient on data query and data filtering. A common approach during the pandemic was to use proprietary software such as ArcGIS (Esri, Redlands, CA) to manage both information integration and storage, as well as design the actual dashboard (Dong et al., 2020; Kamel Boulos & Geraghty, 2020; Sha et al., 2020). For open source options, developers could visualize the data on a map with web mapping libraries (e.g., Mapbox GL, OpenLayers) used in the front‐end to render the customized maps in a web browser and provide basic GIS functions like zoom, pan, and select, and geospatial analysis tasks like calculating area and distance. For time‐consuming data analysis tasks like pattern recognition using data mining or machine learning, the jobs are pre‐computed and results are stored on a server to feed the dashboards (Marvel et al., 2021; Stephany et al., 2020).
The spatial analysis, specifically local Moran statistics and cartogram functions, implemented in US COVID Atlas is computationally expensive (Hardisty & Klippel, 2010) and is commonly deployed on the server side to analyze data (Wang, 2010). To save the pre‐processing time and resources of applying spatial analysis methods on dozens of variables over hundreds of days, this system uses WebAssembly technology to run the spatial analysis function written in C++ in the web browser instead of pre‐computing on a server in the dashboard system mentioned above. With a novel design of a caching system, each run of local Moran statistics with 999 permutations only took around 400 ms in most modern browsers, which leads to a smooth user experience of space‐time exploratory spatial data analysis. In recent years, the WebAssembly technology has been gradually used in web‐based mapping applications (Florinsky, Garov, & Karachevtseva, 2018; Sit, Sermet, & Demir, 2019). To the best of our knowledge, this system is the first application that uses WebAssembly to run local Moran statistics and cartograms in web browsers.
There are multiple limitations in the analytic approach of the Atlas, though we attempt to minimize them. LISA cluster analyses rely on computational inference which can generate sensitive results. Depending on the mechanism of disease spread (e.g., outbreaks in a small concentrated population versus slow community spread) and how the testing resources are utilized, the new case count time series can be clumped at times or suffer from seasonality. In such instances, LISA clusters can vary from one day to another. To alleviate this, we added the 7‐day moving average of new cases, but these patterns could persist in the data and impact the calculation of LISA clusters. While our dashboard provides a nuanced view of how the spatial patterns change over time and across the USA, at the moment it does not supplement that information with what could be the potential drivers, such as state‐level policy and local mobility, and whether the pandemic is affecting certain groups of population disproportionately. An effective and rapid policy response is highly dependent on having a good understanding of the underlying transmission mechanisms and vulnerable populations. Thus, as the next step, we plan to include data sets regarding non‐pharmaceutical policies, social mobility, hospitalization, and infection rates by race and ethnicity, to improve the policy response and adaptation. Additionally, as we have noted before, the true spatial scale of the COVID‐19 pandemic is arguably at an even finer resolution than county level, but the data below the county‐level resolution currently remain unavailable for the entire country. We will look to incorporate the data into the Atlas if they become available in the future. Furthermore, adding a composite vulnerability index (beyond cPSI forecasting) to condense multiple dimensions of data for a more holistic perspective was considered, following discussions with coalition members and stakeholders. In the early stages of the pandemic, individual variables were thought to better capture the different phenomena that were considered important and interesting to the widest range of users that we worked with. As the pandemic shifts towards recovery and equilibrium, we have already been discussing how to implement a more multivariate approach next, including building a new index or typology that would be tailored to unique features of the pandemic directly.
In conclusion, the COVID Atlas project serves as a complex but efficient infrastructure to track, study, and better understand the current pandemic landscape. By integrating libgeoda and an inverted structure, it was able to be developed in days with minimal cost, rather than months or years. Use cases have included public health officials in rural counties reviewing regional trends, business owners seeking to safely reopen facilities that span multiple counties or states, and clinical health professors integrating the Atlas into coursework to understand the pandemic. Furthermore, the open source nature of the project opens possibilities for further developments and iterations outside the initial intended goals, allowing the technological infrastructure to bend and flex according to future needs still unknown. While we successfully integrated a dynamic infrastructure system that communicates multiple views of COVID‐19 change over time with a GIScience perspective, the Atlas also matured as we learned from the community and coalition it was designed with/for. We are reminded that future iterations of the Atlas and the impressive number of spatial decision support systems in the field must remain vigilant in careful consideration of the needs, context, and stories of the people behind the numbers, the millions of lives and ever‐growing number of deaths impacted by COVID‐19.
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
ACKNOWLEDGEMENTS
The authors would like to acknowledge and extend tremendous gratitude for the extensive support, guidance, and invaluable expertise gained from the Atlas research coalition, contributors, and many volunteers during all phases of the application development.
APPENDIX A. Testing data source
TABLE A1.
County‐level testing data sources
| Data source | States |
|---|---|
| Corona Data Scraper | AR, CA, FL, IL, ND, NE, NV, OR, TN, WI |
| Worldometer | NY, PA, TX, WA |
| CSDS Crawler | AK, AL, IA, IN, MD, ME, NM |
APPENDIX B. CHR data variables
TABLE B1.
Community level variables by county health rankings and roadmap
| Category label | Measure name | Var. unit | Measure definition | Sources | Year(s) |
|---|---|---|---|---|---|
| Community health factors | Health factors represent those things we can modify to improve community conditions and the length and quality of life for residents | ||||
| Children in poverty | % | Percentage of children under age 18 living in poverty | Small Area Income and Poverty Estimates | 2018 | |
| Income inequality | Rate | Ratio of household income at the 80th percentile to income at the 20th percentile | American Community Survey, 5‐year estimates | 2014–2018 | |
| Median household income | Dollars | The income where half of households in a county earn more and half of households earn less | Small Area Income and Poverty Estimates | 2018 | |
| Food insecurity | % | Percentage of population who lack adequate access to food | Map the Meal Gap | 2017 | |
| Unemployment | % | Percentage of population aged 16 and over unemployed but seeking work | Bureau of Labor Statistics | 2018 | |
| Uninsured | % | Percentage of population under age 65 without health insurance | Small Area Health Insurance Estimates | 2017 | |
| Primary care physicians | Ratio | Ratio of population to primary care physicians | Area Health Resource File/American Medical Association | 2017 | |
| Preventable hospital stays | Rate | Rate of hospital stays for ambulatory‐care sensitive conditions per 100,000 Medicare enrollees | Mapping Medicare Disparities Tool | 2017 | |
| Residential segregation‐ black/white | Rate | Index of dissimilarity where higher values indicate greater residential segregation between black and white county residents | American Community Survey, 5‐year estimates | 2014–2018 | |
| Severe housing problems | % | Percentage of households with at least 1 of 4 housing problems: overcrowding, high housing costs, lack of kitchen facilities, or lack of plumbing facilities | Comprehensive Housing Affordability Strategy (CHAS) data | 2012–2016 | |
| Community health context | Community health context reflects the existing health behaviors and demographics of individuals in the community that are influenced by the opportunities to live long and well | ||||
| % 65 and over | % | Percentage of population ages 65 and over | Census Population Estimates | 2018 | |
| Adult obesity | % | Percentage of the adult population (aged 20 and over) who report a body mass index (BMI) greater than or equal to 30 kg/m2 | United States Diabetes Surveillance System | 2016 | |
| Diabetes prevalence | % | Percentage of adults aged 20 and over with diagnosed diabetes | United States Diabetes Surveillance System | 2016 | |
| Adult smoking | % | Percentage of adults who are current smokers | Behavioral Risk Factor Surveillance System | 2017 | |
| Excessive drinking | % | Percentage of adults reporting binge or heavy drinking | Behavioral Risk Factor Surveillance System | 2017 | |
| Drug overdose deaths | Rate | Number of drug poisoning deaths per 100,000 population | National Center for Health Statistics—mortality files | 2016–2018 | |
| Length and quality of life | Length and quality of life reflects the physical and mental well‐being of residents within a community through measures representing how long and how well residents live | ||||
| Life expectancy | Rate | Average number of years a person can expect to live | National Center for Health Statistics—mortality files | 2016–2018 | |
| Self‐rated health | % | Percentage of adults reporting fair or poor health | Behavioral Risk Factor Surveillance System | 2017 | |
Kolak M, Li X, Lin Q, et al. The US COVID Atlas: A dynamic cyberinfrastructure surveillance system for interactive exploration of the pandemic. Transactions in GIS. 2021;25:1741–1765. 10.1111/tgis.12786
Marynia Kolak, Xun Li, and Qinyun Lin contributed equally to this manuscript as co‐lead authors.
ENDNOTES
As more COVID‐19 data become available, some variables have been updated and new variables are included in the Atlas. Check https://theuscovidatlas.org/ for the most updated version.
We set the number of CPU cores to be 1 so that Atlas can run smoothly across various browsers.
Users can go to “Preference” to change the number of CPU cores manually. The default in GeoDa is 6. Users can specify other hyperparameters mentioned here after choosing the “Cluster Maps” tool icon and selecting the univariate local Moran's I.
REFERENCES
- Anselin, L. (2010). Local indicators of spatial association—LISA. Geographical Analysis, 27(2), 93–115. 10.1111/j.1538-4632.1995.tb00338.x [DOI] [Google Scholar]
- Anselin, L. (2012). From SpaceStat to cyberGIS: Twenty years of spatial data analysis software. International Regional Science Review, 35(2), 131–157. 10.1177/0160017612438615 [DOI] [Google Scholar]
- Anselin, L. (2019). A local indicator of multivariate spatial association: Extending Geary's c . Geographical Analysis, 51(2), 133–150. 10.1111/gean.12164 [DOI] [Google Scholar]
- Anselin, L., & Li, X. (2019). Operational local join count statistics for cluster detection. Journal of Geographical Systems, 21(2), 189–210. 10.1007/s10109-019-00299-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anselin, L., Syabri, I., & Kho, Y. (2010). GeoDa: An introduction to spatial data analysis. In Fischer M. M. & Getis A. (Eds.), Handbook of applied spatial analysis (pp. 73–89). Berlin, Germany: Springer. 10.1007/978-3-642-03647-7_5 [DOI] [Google Scholar]
- Altieri, N., Barter, R. L., Duncan, J., Dwivedi, R., Kumbier, K., Li, X., … Yu, B. (2021). Curating a COVID‐19 data repository and forecasting county‐level death counts in the United States. Harvard Data Science Review. 10.1162/99608f92.1d4e0dae [DOI] [Google Scholar]
- Beazley, D. M. (1996). SWIG: An easy to use tool for integrating scripting languages with C and C++. In Proceedings of the USENIX Fourth Annual Tcl/Tk Workshop, Monterey, CA. [Google Scholar]
- Bansal, S., Chowell, G., Simonsen, L., Vespignani, A., & Viboud, C. (2016). Big data for infectious disease surveillance and modeling. Journal of Infectious Diseases, 214(Suppl. 4), S375–S379. 10.1093/infdis/jiw400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkelman, R., Bryan, R., Osterholm, M., LeDuc, J., & Hughes, J. (1994). Infectious disease surveillance: A crumbling foundation. Science, 264(5157), 368–370. 10.1126/science.8153621 [DOI] [PubMed] [Google Scholar]
- Berry, I., Soucy, J.‐P.‐R., Tuite, A., & Fisman, D. (2020). Open access epidemiologic data and an interactive dashboard to monitor the COVID‐19 outbreak in Canada. Canadian Medical Association Journal, 192(15), E420. 10.1503/cmaj.75262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertocchi, G., & Dimico, A. (2020). Covid‐19, race, and redlining (SSRN Scholarly Paper). Retrieved from https://papers.ssrn.com/abstract=3650128 [Google Scholar]
- Bettencourt, L. M. A. (2013). The origins of scaling in cities. Science, 340(6139), 1438–1441. 10.1126/science.1235823 [DOI] [PubMed] [Google Scholar]
- Bettencourt, L. M. A., Lobo, J., Helbing, D., Kuhnert, C., & West, G. B. (2007). Growth, innovation, scaling, and the pace of life in cities. Proceedings of the National Academy of Sciences of the United States of America, 104(17), 7301–7306. 10.1073/pnas.0610172104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham, K. (2021). The UK Government's Vaccine Taskforce: Strategy for protecting the UK and the world. The Lancet, 397(10268), 68–70. 10.1016/S0140-6736(20)32175-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury, R. S., Arguello, I., Lane, M., Cooley, G., Handali, S., Dimitrova, S. D., … Hobbs, C. V. (2020). Parasitic infection surveillance in Mississippi Delta children. American Journal of Tropical Medicine and Hygiene, 103(3), 1150–1153. 10.4269/ajtmh.20-0026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosemer, K., Schelly, C., Gagnon, V., Arola, K. L., Pearce, J. M., Bessette, D., & Schmitt Olabisi, L. (2020). The energy crises revealed by COVID: Intersections of indigeneity, inequity, and health. Energy Research & Social Science, 68, 101661. 10.1016/j.erss.2020.101661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention . (2020). CDC COVID Data Tracker. Retrieved from http://covid.cdc.gov/covid‐data‐tracker [Google Scholar]
- Chande, A., Lee, S., Harris, M., Nguyen, Q., Beckett, S. J., Hilley, T., … Weitz, J. S. (2020). Real‐time, interactive website for US‐county‐level COVID‐19 event risk assessment. Nature Human Behaviour, 4(12), 1313–1319. 10.1038/s41562-020-01000-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- County Health Rankings and Roadmaps . (2020). County health rankings. Retrieved from https://www.countyhealthrankings.org/ [Google Scholar]
- Covid Act Now . (2020). Covid act now. Retrieved from https://covidactnow.org/about [Google Scholar]
- COVID Tracking Project . (2020). The COVID tracking project. Retrieved from https://covidtracking.com/about [Google Scholar]
- Desjardins, M. R., Hohl, A., & Delmelle, E. M. (2020). Rapid surveillance of COVID‐19 in the United States using a prospective space‐time scan statistic: Detecting and evaluating emerging clusters. Applied Geography, 118, 102202. 10.1016/j.apgeog.2020.102202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, E., Du, H., & Gardner, L. (2020). An interactive web‐based dashboard to track COVID‐19 in real time. The Lancet Infectious Diseases, 20(5), 533–534. 10.1016/S1473-3099(20)30120-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florinsky, I. V., Garov, A. S., & Karachevtseva, I. P. (2018). A web system of virtual morphometric globes for Mars and the Moon. Planetary and Space Science, 159, 105–109. 10.1016/j.pss.2018.05.011 [DOI] [Google Scholar]
- Gao, S., Rao, J., Kang, Y., Liang, Y., & Kruse, J. (2020). Mapping county‐level mobility pattern changes in the United States in response to COVID‐19 (SSRN Scholarly Paper). Retrieved from https://papers.ssrn.com/abstract=3570145 [Google Scholar]
- Glazner, C. (2020). COVID‐19 dashboard and tools. Retrieved from https://dashboards.c19hcc.org/ [Google Scholar]
- Goldberg, D. W., Cockburn, M. G., Hammond, T. A., Jacquez, G. M., Janies, D., Knoblock, C., … Raubal, M. (2013). Envisioning a future for a spatial‐health CyberGIS marketplace. In Proceedings of the Second ACM SIGSPATIAL International Workshop on the Use of GIS in Public Health, Orlando, FL (pp. 27–30). New York, NY: ACM. 10.1145/2535708.2535716 [DOI] [Google Scholar]
- Gray, D. M., Anyane‐Yeboa, A., Balzora, S., Issaka, R. B., & May, F. P. (2020). COVID‐19 and the other pandemic: Populations made vulnerable by systemic inequity. Nature Reviews Gastroenterology & Hepatology, 17(9), 520–522. 10.1038/s41575-020-0330-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene, S. K., Peterson, E. R., Kapell, D., Fine, A. D., & Kulldorff, M. (2016). Daily reportable disease spatiotemporal cluster detection, New York City, New York, USA, 2014–2015. Emerging Infectious Diseases, 22(10), 1808–1812. 10.3201/eid2210.160097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, A., Rossberg, A., Schuff, D. L., Titzer, B. L., Holman, M., Gohman, D., … Bastien, J. (2017). Bringing the web up to speed with WebAssembly. In Proceedings of the 38th ACM SIGPLAN Conference on Programming Language Design and Implementation, Barcelona, Spain (pp. 185–200). New York, NY: ACM. 10.1145/3062341.3062363 [DOI] [Google Scholar]
- Hardisty, F., & Klippel, A. (2010). Analysing spatio‐temporal autocorrelation with LISTA‐Viz. International Journal of Geographical Information Science, 24(10), 1515–1526. 10.1080/13658816.2010.511717 [DOI] [Google Scholar]
- Harris, J. K., Mansour, R., Choucair, B., Olson, J., Nissen, C., & Bhatt, J. (2014). Health department use of social media to identify foodborne illness: Chicago, Illinois, 2013–2014. Morbidity and Mortality Weekly Report, 63(32), 681–685. [PMC free article] [PubMed] [Google Scholar]
- Hay, S. I., George, D. B., Moyes, C. L., & Brownstein, J. S. (2013). Big data opportunities for global infectious disease surveillance. PLoS Medicine, 10(4), e1001413. 10.1371/journal.pmed.1001413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl, A., Delmelle, E., & Desjardins, M. (2020). Rapid detection of COVID‐19 clusters in the United States using a prospective space‐time scan statistic: An update. SIGSPATIAL Special, 12(1), 27–33. 10.1145/3404820.3404825 [DOI] [Google Scholar]
- Hohl, A., Delmelle, E. M., Desjardins, M. R., & Lan, Y. (2020). Daily surveillance of COVID19 using the prospective space‐time scan statistic in the United States. Spatial and Spatiotemporal Epidemiology, 34, 100354. 10.1016/j.sste.2020.100354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., … Cao, B. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet, 395(10223), 497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowska, M., Schipperijn, J., Kerr, J., & Altintas, I. (2016). Applied CyberGIS in the age of complex spatial health data. In Proceedings of the Ninth International Conference on Geographic Information Science, Montreal, Quebec, Canada (pp. 147–152). [Google Scholar]
- Jenks, G. F. (1977). Optimal data classification for choropleth maps (Occasional paper no. 2). Lawrence, KS: Department of Geography, University of Kansas. [Google Scholar]
- Johns Hopkins University . (2020). Johns Hopkins Coronavirus Resource Center. Retrieved from http://coronavirus.jhu.edu [Google Scholar]
- Kamel Boulos, M. N., & Geraghty, E. M. (2020). Geographical tracking and mapping of coronavirus disease COVID‐19/severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) epidemic and associated events around the world: How 21st century GIS technologies are supporting the global fight against outbreaks and epidemics. International Journal of Health Geographics, 19(1), 8. 10.1186/s12942-020-00202-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, D., Choi, H., Kim, J.‐H., & Choi, J. (2020). Spatial epidemic dynamics of the COVID‐19 outbreak in China. International Journal of Infectious Diseases, 94, 96–102. 10.1016/j.ijid.2020.03.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury, M. J., & Ioannidis, J. P. A. (2014). Big data meets public health. Science, 346(6213), 1054–1055. 10.1126/science.aaa2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolak, M., Steptoe, M., Manprisio, H., Azu‐Popow, L., Hinchy, M., Malana, G., & Maciejewski, R. (2020). Extending Volunteered Geographic Information (VGI) with geospatial software as a service: Participatory asset mapping infrastructures for urban health. In Lu Y. & Delmelle E. (Eds.), Geospatial technologies for urban health (pp. 209–230). Cham, Switzerland: Springer. 10.1007/978-3-030-19573-1_11 [DOI] [Google Scholar]
- Krishnan, L., Ogunwole, S. M., & Cooper, L. A. (2020). Historical insights on coronavirus disease 2019 (COVID‐19), the 1918 influenza pandemic, and racial disparities: Illuminating a path forward. Annals of Internal Medicine, 173(6), 474–481. 10.7326/M20-2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, E. C., Asher, J. M., Goldlust, S., Kraemer, J. D., Lawson, A. B., & Bansal, S. (2016). Mind the scales: Harnessing spatial big data for infectious disease surveillance and inference. Journal of Infectious Diseases, 214(Suppl. 4), S409–S413. 10.1093/infdis/jiw344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Anselin, L., & Koschinsky, J. (2015). GeoDa web: Enhancing web‐based mapping with spatial analytics. In Proceedings of the 23rd SIGSPATIAL International Conference on Advances in Geographic Information Systems, Seattle, WA (pp. 1–4). New York, NY: ACM. 10.1145/2820783.2820792 [DOI] [Google Scholar]
- Li, H., Li, H., Ding, Z., Hu, Z., Chen, F., Wang, K., … Shen, H. (2020). Spatial statistical analysis of coronavirus disease 2019 (Covid‐19) in China. Geospatial Health, 15(1), 1. 10.4081/gh.2020.867 [DOI] [PubMed] [Google Scholar]
- Li, X., Lin, Q., Kolak, M., Martin, R., Yang, S., Menghaney, M., … Sihan‐Mao. (2020). GeoDaCenter/covid: Beta (Version beta). Zenodo. 10.5281/zenodo.4081869 [DOI] [Google Scholar]
- Lobo, J., Bettencourt, L. M. A., Strumsky, D., & West, G. B. (2013). Urban scaling and the production function for cities. PLoS ONE, 8(3), e58407. 10.1371/journal.pone.0058407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvel, S. W., House, J. S., Wheeler, M., Song, K., Zhou, Y.‐H., Wright, F. A., … Reif, D. M. (2021). The COVID‐19 Pandemic Vulnerability Index (PVI) dashboard: Monitoring county‐level vulnerability using visualization, statistical modeling, and machine learning. Environmental Health Perspectives, 129(1), 017701. 10.1289/EHP8690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey, D. S., & Denton, N. A. (1989). Hypersegregation in U.S. metropolitan areas: Black and Hispanic segregation along five dimensions. Demography, 26(3), 373–391. 10.2307/2061599 [DOI] [PubMed] [Google Scholar]
- Massey, D. S., & Tannen, J. (2015). A research note on trends in Black hypersegregation. Demography, 52(3), 1025–1034. 10.1007/s13524-015-0381-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meza, E. (2018). Examining wastewater treatment struggles in Lowndes County, AL (Unpublished MA thesis). Durham, NC: Duke University. [Google Scholar]
- Mooney, S. J., & Pejaver, V. (2018). Big data in public health: Terminology, machine learning, and privacy. Annual Review of Public Health, 39(1), 95–112. 10.1146/annurev-publhealth-040617-014208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- New York Times . (2021). Coronavirus (Covid‐19) data in the United States. Retrieved from https://github.com/nytimes/covid‐19‐data [Google Scholar]
- Power, T., Wilson, D., Best, O., Brockie, T., Bearskin, L. B., Millender, E., & Lowe, J. (2020). COVID‐19 and Indigenous Peoples: An imperative for action. Journal of Clinical Nursing, 29(15–16), 2737–2741. 10.1111/jocn.15320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington, P. L., Catlin, B. B., & Gennuso, K. P. (2015). The county health rankings: Rationale and methods. Population Health Metrics, 13(1), 11. 10.1186/s12963-015-0044-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey, S. J. (2009). Show me the code: Spatial analysis and open source. Journal of Geographical Systems, 11(2), 191–207. 10.1007/s10109-009-0086-8 [DOI] [Google Scholar]
- Scarpone, C., Brinkmann, S. T., Große, T., Sonnenwald, D., Fuchs, M., & Walker, B. B. (2020). A multimethod approach for county‐scale geospatial analysis of emerging infectious diseases: A cross‐sectional case study of COVID‐19 incidence in Germany. International Journal of Health Geographics, 19(1), 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha, D., Miao, X., Lan, H., Stewart, K., Ruan, S., Tian, Y., Tian, Y., … Yang, C. (2020). Spatiotemporal analysis of medical resource deficiencies in the U.S. under COVID‐19 pandemic. PLoS ONE, 15(10), e0240348. 10.1371/journal.pone.0240348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen, L., Gog, J. R., Olson, D., & Viboud, C. (2016). Infectious disease surveillance in the big data era: Towards faster and locally relevant systems. Journal of Infectious Diseases, 214(Suppl. 4), S380–S385. 10.1093/infdis/jiw376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sit, M., Sermet, Y., & Demir, I. (2019). Optimized watershed delineation library for server‐side and client‐side web applications. Open Geospatial Data, Software and Standards, 4(1), 8. 10.1186/s40965-019-0068-9 [DOI] [Google Scholar]
- Spinuzzi, C. (2005). The methodology of participatory design. Technical Communication, 52(2), 163–174. [Google Scholar]
- Stephany, F., Stoehr, N., Darius, P., Neuhäuser, L., Teutloff, O., & Braesemann, F. (2020). The CoRisk‐Index: A data‐mining approach to identify industry‐specific risk assessments related to COVID‐19 in real‐time. Preprint: arXiv:2003.12432. [Google Scholar]
- Su, A., Luo, D., Castro, H., Frankl, J., Moos, L., McFarland, M., … Yi, Z. N. (2020). CovidCareMap. Retrieved from https://www.covidcaremap.org [Google Scholar]
- Sun, F., Matthews, S. A., Yang, T.‐C., & Hu, M.‐H. (2020). A spatial analysis of the COVID‐19 period prevalence in U.S. counties through June 28, 2020: Where geography matters? Annals of Epidemiology, 52, 54–59. 10.1016/j.annepidem.2020.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakar, V. (2020). Unfolding events in space and time: Geospatial insights into COVID‐19 diffusion in Washington State during the initial stage of the outbreak. ISPRS International Journal of Geo‐Information, 9(6), 382. 10.3390/ijgi9060382 [DOI] [Google Scholar]
- Thorlund, K., Dron, L., Park, J., Hsu, G., Forrest, J. I., & Mills, E. J. (2020). A real‐time dashboard of clinical trials for COVID‐19. The Lancet Digital Health, 2(6), e286–e287. 10.1016/S2589-7500(20)30086-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- United States Census Bureau . (2020). American Community Survey population data. Retrieved from https://www.census.gov/programs‐surveys/acs [Google Scholar]
- University of Virginia, Biocomplexity Institute . (2020). COVID‐19 surveillance dashboard. Retrieved from https://nssac.bii.virginia.edu/covid‐19/dashboard [Google Scholar]
- USAFacts . (2020). US coronavirus cases and deaths. Retrieved from https://usafacts.org/visualizations/coronavirus‐covid‐19‐spread‐map [Google Scholar]
- U.S. Census Bureau . (2017). TIGER/Line Shapefile, 2017, nation, U.S., Current American Indian/Alaska Native/Native Hawaiian Areas National (AIANNH) National. Retrieved from https://catalog.data.gov/dataset/tiger‐line‐shapefile‐2017‐nation‐u‐s‐current‐american‐indian‐alaska‐native‐native‐hawaiian‐area [Google Scholar]
- U.S. Census Bureau . (2018a). Cartographic boundary files (county and states). Retrieved from https://www.census.gov/geographies/mapping‐files/time‐series/geo/carto‐boundary‐file.html [Google Scholar]
- U.S. Census Bureau . (2018b). TIGER/Line Shapefile, 2018, nation, U.S., 116th Congressional District National. Retrieved from https://catalog.data.gov/dataset/tiger‐line‐shapefile‐2018‐nation‐u‐s‐116th‐congressional‐district‐national [Google Scholar]
- U.S. Census Bureau . (2019). 2014–2018 American community survey 5‐year estimates. [Google Scholar]
- U.S. Department of Health and Human Services . (2020). HRSA Data Warehouse online location finder. Retrieved from https://findahealthcenter.hrsa.gov/ [Google Scholar]
- van Dorn, A., Cooney, R. E., & Sabin, M. L. (2020). COVID‐19 exacerbating inequalities in the US. The Lancet, 395(10232), 1243–1244. 10.1016/S0140-6736(20)30893-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. (2010). A cyberGIS framework for the synthesis of cyberinfrastructure, GIS, and spatial analysis. Annals of the Association of American Geographers, 100(3), 535–557. 10.1080/00045601003791243 [DOI] [Google Scholar]
- Webster, G. R., & Bowman, J. (2008). Quantitatively delineating the Black Belt geographic region. Southeastern Geographer, 48(1), 3–18. 10.1353/sgo.0.0007 [DOI] [Google Scholar]
- Wong, Z. S. Y., Zhou, J., & Zhang, Q. (2019). Artificial intelligence for infectious disease Big Data analytics. Infection, Disease & Health, 24(1), 44–48. 10.1016/j.idh.2018.10.002 [DOI] [PubMed] [Google Scholar]
- Worldometer . (2020). Worldometer coronavirus update. Retrieved from https://www.worldometers.info/coronavirus/ [Google Scholar]
- Wright, D. J., & Wang, S. (2011). The emergence of spatial cyberinfrastructure. Proceedings of the National Academy of Sciences of the United States of America, 108(14), 5488–5491. 10.1073/pnas.1103051108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, C., Zhang, X., & Wang, Y. (2020). Mapping of health literacy and social panic via web search data during the COVID‐19 public health emergency: Infodemiological study. Journal of Medical Internet Research, 22(7), e18831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C., Kafatos, M., Wang, D. W., Wolf, H. D., & Yang, R. (2010). US Patent No. 7,725,529 B2. Washington, DC: US Patent and Trademark Office. Retrieved from https://patents.google.com/patent/US7725529B2/en [Google Scholar]
- Yang, T., Shen, K., He, S., Li, E., Sun, P., Chen, P., … Guo, Y. (2020). CovidNet: To bring data transparency in the era of COVID‐19. Preprint, arXiv:2005.10948. [Google Scholar]
- Yao, Y. E., Pan, J., Wang, W., Liu, Z., Kan, H., Qiu, Y., Meng, X., … Wang, W. (2020). Association of particulate matter pollution and case fatality rate of COVID‐19 in 49 Chinese cities. Science of the Total Environment, 741, 140396. 10.1016/j.scitotenv.2020.140396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1Point3Acres . (2020). 1Point3Acres.com. Retrieved from https://coronavirus.1point3acres.com/en/data [Google Scholar]


