Abstract
The kidney tropism of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) has been well‐validated clinically and often leads to various forms of renal damage in coronavirus disease‐2019 (COVID‐19) patients. However, the underlying mechanisms and diagnostic approaches remain to be determined. We interrogated the expression of virus‐related host factors in single‐cell RNA sequencing (scRNA‐seq) datasets of normal human kidneys and kidneys with pre‐existing diseases and validated the results with urinary proteomics of COVID‐19 patients and healthy individuals. We also assessed the effects of genetic variants on kidney susceptibility using expression quantitative trait loci (eQTLs) databases. We identified a subtype of tubular cells, which we named PT‐3 cells, as being vulnerable to SARS‐CoV‐2 infections in the kidneys. PT‐3 cells were enriched in viral entry factors and replication and assembly machinery but lacked antiviral restriction factors. Immunohistochemistry confirmed positive staining of PT‐3 cell marker SCL36A2 on kidney sections from COVID‐19 patients. Urinary proteomic analyses of COVID‐19 patients revealed that markers of PT‐3 cells were significantly increased, along with elevated viral receptor angiotensin‐converting enzyme 2. We further found that the proportion of PT‐3 cells increased in diabetic nephropathy but decreased in kidney allografts and lupus nephropathy, suggesting that kidney susceptibility varied among these diseases. We finally identified several eQTLs that regulate the expression of host factors in kidney cells. PT‐3 cells may represent a key determinant for the kidney tropism of SARS‐CoV‐2, and detection of PT‐3 cells may be used to assess the risk of renal infection during COVID‐19.
Keywords: chronic kidney disease, COVID‐19, diabetic kidney disease, SARS‐CoV‐2, scRNA‐seq
Through single‐cell analysis, we identified PT‐3 cells, a proximal tubule epithelial cell subtype in human kidney, that are highly susceptible to severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection. We validated these results using immunostaining and urinary proteomics in coronavirus disease‐2019 (COVID‐19) patients. Our findings provide not only new insights into the mechanism involving SARS‐CoV‐2 infection of the kidney cells, but also a potential strategy for risk assessment of kidney infection among COVID‐19 patients.
Abbreviations
- ACE2
angiotensin‐converting enzyme 2
- ANPEP
alanyl aminopeptidase
- BSG
basign
- CD
collecting duct
- CFH+
complement factor H
- CKD
chronic kidney disease
- CLEC4M
C‐type lectin domain family 4 member M
- COVID‐19
coronavirus disease‐2019
- CTSB/L
cathepsins B and L
- DCT
distal convoluted tubule
- DN
diabetic nephropathy
- DPP4
dipeptidyl peptidase 4
- ENDO
endothelium
- eQTLs
expression quantitative trait loci
- IC
intercalated cell
- LEUK
leukocyte
- LN
lupus nephropathy
- LOH
loop of Henle
- LY6E
lymphocyte antigen 6 family member E
- MES
mesangial cell
- PC
principal cell
- PODO
podocyte
- PTECs
proximal tubular epithelial cells
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- scRNA-seq/snRNA-seq
single-cell/single-nucleus RNA sequencing
- SNP
single nucleotide polymorphism
- TMPRSS2
transmembrane serine protease 2
- TOP3B
DNA topoisomerase III beta
- ZCRB1
zinc finger CCHC-type and RNA-binding motif-containing 1
Introduction
Coronavirus disease‐2019 (COVID‐19) is an infectious disease caused by a novel discovered coronavirus, the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). It has been reported that the kidney is one of the most commonly affected extrapulmonary organs of SARS‐CoV‐2 infection and varying degrees of renal damage is often observed in COVID‐19 patients [1, 2]. In addition to indirect factors (e.g., cytokine storm, hypoxia), the direct kidney infection of SARS‐CoV‐2 could also lead to kidney injury [3, 4, 5, 6]. Autopsy reports have provided evidence that the virus could directly infect kidney cells of COVID‐19 patients and patient‐derived SARS‐CoV‐2 were able to replicate in nonhuman primate kidney tubular epithelial cells [7]. However, why kidney cells are permissive to SARS‐CoV‐2 infection and which renal cell types are more susceptible remain poorly understood.
The successful infection of SARS‐CoV‐2 depends critically on its interaction with target cells, which involves various host factors such as cell surface receptors, cellular proteases, antiviral restriction factors, factors participate viral replication, and assembly [8, 9, 10]. Angiotensin‐converting enzyme 2 (ACE2) has been identified as the SARS‐CoV‐2 receptor, and the transmembrane serine protease 2 (TMPRSS2) has been confirmed to cleave the viral spike (S) protein, which facilitates viral entry [11]. However, several candidate receptors and proteases may serve similar functions. For example, basign (BSG, also known as CD147), dipeptidyl peptidase 4 (DPP4), and alanyl aminopeptidase (ANPEP) reportedly interact with the S protein to promote SARS‐CoV‐2 or other human coronavirus entry into cells [12, 13, 14]. Other members of the TMPRSS family, such as TMPRSS4, may function similarly to TMPRSS2 [15]. In addition, cellular proteases, including cathepsins B and L (CTSB/L) and furin, can mediate S protein cleavage [11, 16]. In contrast, lymphocyte antigen 6 family member E (LY6E) and interferon‐induced transmembrane proteins had been identified as potent coronavirus restriction factors [9, 17]. Thus, comprehensive characterizations of these host factors in kidney cells will greatly improve our understanding on the renal tropism and pathogenicity of SARS‐CoV‐2.
Single‐cell RNA sequencing (scRNA‐seq) analysis is a powerful approach to characterize complex tissues at single‐cell resolution and reveal diverse cellular responses in different cell types. Many studies have applied scRNA‐seq to predict the tissue tropism of SARS‐CoV‐2, including the kidney. The transcripts of ACE2 were primarily detected in proximal tubular epithelial cells (PTECs), while the mRNA of TMPRSS2 was robustly expressed in the distal convoluted tubule (DCT) and intercalated cells rather than the PTECs [18, 19]. Few studies have comprehensively evaluated the expression patterns of other host factors (such as restriction factors and replication/assembly factors) in different kidney cells.
The expression of host factors determines SARS‐CoV‐2 susceptibility, which is partially regulated by genetic variations, especially expression quantitative trait loci (eQTLs) [20]. eQTLs refer to genetic variants that regulate transcript expression levels. Although several studies have described eQTLs associated with the expression of ACE2 in multiple tissues [20, 21], little is known regarding the association of eQTLs with other host factors that underlie the SARS‐CoV‐2 kidney tropism.
In this study, we surveyed the expression patterns of host factors in published scRNA‐seq datasets of healthy human kidneys and clarified the distribution of ACE2 and TMPRSS2 in PTECs. We also identified and characterized a PTEC subtype that was highly susceptible to SARS‐CoV‐2 infection, which we named PT‐3 cells. Due to the limitation of scRNA‐seq data, such as dropout events, and possible differences between the transcriptome and the proteome, urinary proteomics was applied to validate the results obtained via single‐cell analysis. We confirmed that the marker proteins of PT‐3 cells were significantly elevated in the urine of COVID‐19 patients. Moreover, we assessed the expression pattern of host factors in scRNA‐seq datasets of kidneys with kidney diseases, to predict the tropism of SARS‐CoV‐2. Finally, we explored the eQTLs regulating the expression of host factors and identified the eQTLs associated with host proteases and restriction factors, providing a potential interpretation for the differences in kidney susceptibility among individuals.
Results
Identification of a specific SARS‐CoV‐2‐susceptible cell type in the kidney
We first evaluated the autopsy kidney specimens from 26 COVID‐19 patients and found that spherical particles surrounded by crown‐like projections, which were highly suspected as SARS‐CoV‐2 virus, scattered within the apical side of renal PTECs by electron microscopy (Fig. 1A). This finding led us to the question that there could be differences in susceptibility between distinct kidney cell types to SARS‐CoV‐2 infection. To answer this question, we systematically surveyed the expression patterns of host factors required for the SARS‐CoV‐2 replication cycle in publicly available scRNA‐seq datasets of normal human kidneys (GSE131882) [22], which included 15 728 cells from three normal human kidney samples. Our analysis identified 11 distinct kidney cell types (Fig. 1B), in concordance with the original report. Although ACE2 mRNA was predominantly expressed in PTECs, it was undetectable in other cell types, such as podocytes (PODO) and endothelial cells. In contrast, the expression of BSG, a potential SARS‐CoV‐2 receptor, was broadly detected in kidney cells, with the highest expression levels observed in collecting duct (CD) cells (Fig. 1B). We also surveyed the receptors of other known coronaviruses, such as C‐type lectin domain family 4 member M (CLEC4M; SARS‐CoV) [23], DPP4 (Middle East respiratory syndrome coronavirus), and ANPEP (human coronavirus 229E) and found that the expression of these proteins was predominantly observed on PTECs, except for CLEC4M (Fig. 1B). When examining cellular proteases, we found that CTSB was broadly expressed in different cell types, including PTECs; in contrast, TMPRSS2 was strongly expressed in CD and DCT cells, rather than PTECs. Other proteases, such as CTSL and furin, could also be detected in various kidney cells but were almost undetectable in PTECs (Fig. 1B). These results indicated that PTECs were the only kidney cell type that showed appreciable ACE2 mRNA levels co‐expressed with varying levels of proteases. We next investigated the gene expression patterns of restriction factors. As shown in Fig. 1B, LY6E and IFITM2/3 were highly expressed in glomerular cells, while only minimal transcripts for these genes were detected in renal tubular cells, especially in PTECs. Finally, we surveyed the expression patterns of factors involved in viral replication and assembly. The factors required for viral replication (ZCRB1, zinc finger CCHC‐type and RNA‐binding motif‐containing 1; TOP3B, DNA topoisomerase III beta) could be detected in most kidney cell types. In addition, the factors engaged in viral assembly and trafficking were detected across the whole dataset, with moderate to high levels of expression (Fig. 1b).
Fig. 1.
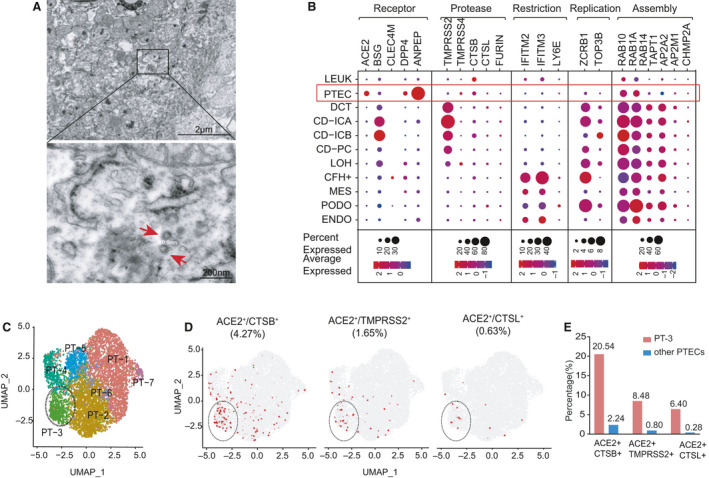
Gene expression profile of host factors related to SARS‐CoV‐2 infection in normal human kidneys (GSE131882). (A) Electron microscopy from autopsy kidney specimens of COVID‐19 patients showing cytoplasm of proximal tubules containing spherical particles measuring around 80 nm (arrow) and surrounded by spikes. (B) Dot plot showing expression patterns of host factors within 11 cell types in the kidneys (sample size = 3). Dot size indicates the proportion of cells expressing a gene within particular cell types, and color intensity indicates the average gene expression level (blue to red denotes low to high expression). CFH+, complement factor H; ENDO, endothelium; IC, intercalated cell; LEUK, leukocyte; LOH, loop of Henle; PC, principal cell. Restriction, restriction factors; Replication, factors engaged in viral replication; Assembly, factors engaged in viral assembly. (C) UMAP plot displaying seven subclusters of PTECs identified by unsupervised clustering analysis, which have been distinctively colored and labeled (PT‐1 to PT‐7). (D) Feature plots displaying distribution of ACE2+/CTSB+ cells, ACE2+/TMPRSS2+ cells, and ACE2+/CTSL+ cells over the UMAP of PTEC subclusters. ACE2 and proteases co‐expressing PTECs are colored as red. (E) Histograms showing the percentage of ACE2+/CTSB+ (left panel), ACE2+/TMPRSS2+ (middle panel), and ACE2+/CTSL+ (right panel) cells in PT‐3 cells and other PTEC subtypes.
Since only a proportion of PTECs are ACE2‐positive, we speculated that there might be some subtypes of PTECs that were more vulnerable to SARS‐CoV‐2 infection. Hence, we performed an unsupervised clustering analysis of 6071 PTECs from the same dataset and obtained seven subclusters of PTECs (PT‐1 to PT‐7; Fig. 1C). We then delineated the distribution of double‐positive cells for various combinations of ACE2 with TMPRSS2, CTSB, and CTSL in PTECs (Fig. 1D). Intriguingly, we indeed identified that only the PT‐3 subcluster demonstrated a higher proportion of ACE2+/CTSB+ double‐positive cells compared with other PTECs (20.54% vs 2.24%; Fig. 1E). In addition, the ACE2+/TMPRSS2+ and ACE2+/CTSL+ double‐positive cells also primarily appear among PT‐3 cells, with lower proportions than the ACE2+/CTSB+ cells (Fig. 1D,E). Then, we characterized the expression of host factors in these PTEC subclusters (Fig. 2A). Overall, PT‐3 cells showed the highest expression of AEC2 and co‐expressed with two other human coronavirus receptors, DPP4 and ANPEP. Compared with other PTEC subtypes, PT‐3 cells exhibited higher levels of proteases (e.g., CTSB) and cellular machinery required for viral replication (ZCRB1, TOP3B) and assembly, but lacked viral restriction factors (LY6E, IFITM2, and IFITM3; Fig. 2A). Meanwhile, gene set enrichment analysis (GSEA) revealed that PT‐3 cells were not only significantly enriched in various pathways related to virus infection, but also enriched in endocytosis and autophagy pathways (Fig. 2B). The cell identities and distribution of proximal tubule segment‐specific markers [24] in these PTECs were described in Fig. 2C,D. PT‐3 cells primarily expressed specific markers for Segment 3, and SLC36A2, PEPD were identified as the signature genes of PT‐3 cells.
Fig. 2.
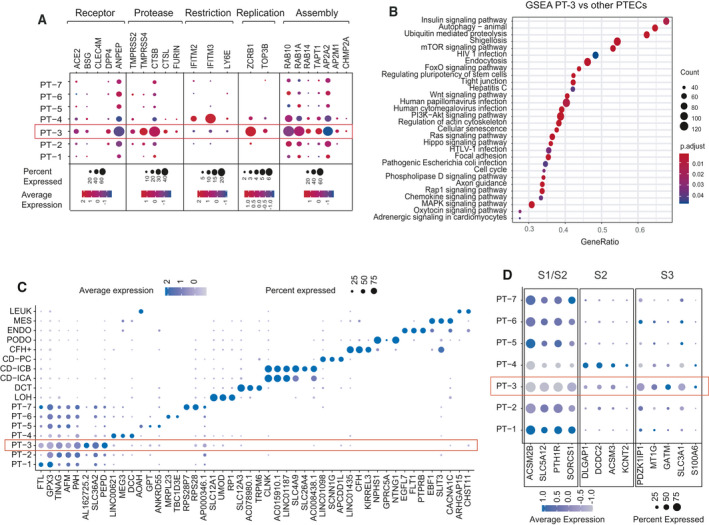
Characterization of PT‐3 cells that may be highly susceptible to SARS‐CoV‐2 infection. (A) Dot plot showing expression intensity and density of host factors within seven PTEC cell subtypes. (B) Dot plot showing enriched KEGG pathways of differentially expressed genes between PT‐3 cells and other PTECs by GSEA analysis. (C) Dot plot showing signature genes of different kidney cell types and PTEC subtypes. Dot size represents proportion of cells expressing a gene within respective cell subtypes. Darker indicates higher expression levels. (D) Expression pattern of marker genes for three segments (S1–S3) of proximal tubule in seven PTEC subclusters. Dot size represents proportion of cells expressing a gene within respective cell subtypes. Darker indicates higher expression levels.
Considering the optimal environment for coronavirus replication provided by PT‐3 cells, it strongly indicated that these cells were the target cells of SARS‐CoV‐2 infection in the kidney.
Proteomics verification of PT‐3 cells in urine of COVID‐19 patients
To verify our findings derived from scRNA‐seq data analysis, we first performed immunohistochemistry (IHC) staining for PT‐3 cell marker SCL36A2 on kidney sections from COVID‐19 patients and controls. In control kidney, SLC36A2 was detected in some of the proximal tubules. This focal or scattered staining pattern indicated that PT‐3 cells were dispersed among the proximal tubules. In kidney from COVID‐19 patients, light microscopy showed decrease in SLC36A2‐positive cells in the proximal tubules which had typical features of acute kidney injury (AKI), including loss of brush border, flattening, and focal loss of tubular epithelial cells (Fig. 3A).
Fig. 3.
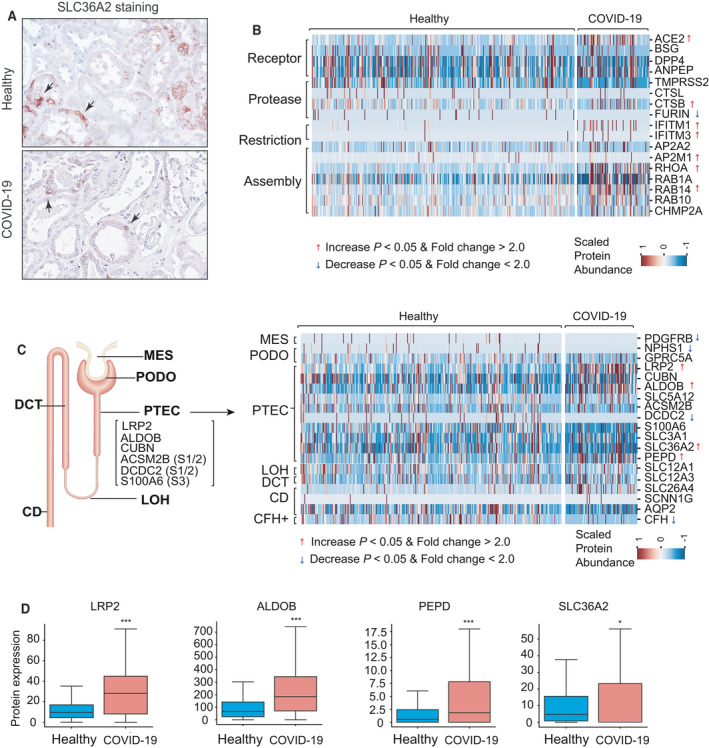
Urinary proteomic profiles of healthy individuals and COVID‐19 patients. (A) Immunohistochemistry staining showing the distribution of PT‐3 marker protein SLC36A2 in proximal tubules from control (upper panel) and patient with COVID‐19 (lower panel). Arrow indicates SLC36A2‐positive PTECs. (B) Heatmap showing protein abundance changes in host factors in urine samples of healthy individuals (n = 350) and COVID‐19 patients (n = 97). The color scale represents the protein abundance. Red arrow indicates increased proteins in urine of COVID‐19 patients (Mann–Whitney U‐test P < 0.05, fold change > 2.0). Blue arrow indicates decreased proteins in urine of COVID‐19 patients (Mann–Whitney U‐test P < 0.05, fold change > 2.0). (C) Left panel: A schematic representation of nephron showing the signature genes for PTECs and markers of three segments (S1–S3) of proximal tubule. Right panel: Heatmap showing differences in urinary protein abundance of cell type‐specific markers between healthy individuals and COVID‐19 patients. (D) Boxplots displaying medians (midline), interquartile range (IQR) (box), 1.5× IQR (whisker) of marker proteins levels of PTECs and PT‐3 cells in healthy individuals and COVID‐19 patients. Mann–Whitney U‐test, *P < 0.05, ***P < 0.001.
We next performed urinary proteomic analysis and focused on protein abundance changes in various host factors, as well as that of specific markers for kidney cells. Urine samples were collected from 97 COVID‐19 patients and 350 age‐ and gender‐matched healthy individuals (Table 1). The median of proteins identified were 1370 and 1024 in healthy individuals and COVID‐19 patients, respectively [false discovery rate (FDR) < 1%]. We observed a significantly increased level of ACE2 in the urine of COVID‐19 patients, compared with healthy individuals (Fig. 3B). Since ACE2 is predominantly expressed in PTECs in kidney, we speculated that the increased ACE2 protein abundance may be due to the shedding of PTECs. The level of CTSB was also significantly increased. In contrast, TMPRSS2 and CTSL appeared unchanged. Moreover, the protein abundance of the restriction factors IFITM1 and IFITM3 was increased in COVID‐19 group compared with the healthy group. And the protein levels of cellular factors involved in virus assembly, such as AP2M1, RHOA, and RAB14, were also increased in COVID‐19 patients (Fig. 3B).
Table 1.
Clinical features of healthy individuals and COVID‐19 patients underwent urinary proteomics. IQR, interquartile range; ‐, not available.
| Characteristics | Healthy controls (n = 350) | COVID‐19 (n = 97) |
|---|---|---|
| Baseline and demographic | ||
| Age, median (IQR), years | 49 (37–60) | 53 (37–62) |
| Gender | ||
| Female (%) | 226 (64.6%) | 66 (68.0%) |
| Male (%) | 124 (35.4%) | 31 (32.0%) |
| Comorbidities | ||
| Hypertension (%) | – | 27(27.8%) |
| Diabetes (%) | – | 15(15.5%) |
| Cardiovascular diseases (%) | – | 9(9.3%) |
| CKDs (%) | – | 10(10.3%) |
As for cell type‐specific markers (Fig. 3C), the levels of PTEC marker LRP2 and ALDOB in the urine of COVID‐19 patients were significantly higher than that of healthy individuals. Whereas markers associated with complement factor H cells, PODOs, and mesangial cells (MESs) decreased, markers for loop of Henle cells, DCT cells, and CD cells remained unchanged. No markers for endothelial cells were detected in either group. Importantly, a significantly elevated level of PT‐3 cell‐specific markers (PEPD, SLC36A2) was also detected (Fig. 3D). Thus, our urinary proteomic results provided supporting evidence that PT‐3 cells represent the preferred targeting cell type for SARS‐CoV‐2 in the kidney.
Diabetic nephropathy increases the PT‐3 cell proportions in the kidneys
Although COVID‐19 patients with comorbidities, especially those with chronic kidney disease (CKD) are at higher risk of kidney injury, it is still unclear whether underlying kidney disease, per se, increases the vulnerability of kidney cells to SARS‐CoV‐2 infection. To predict kidney susceptibility to SARS‐CoV‐2 infection, we assessed changes in the expression patterns of host factors and proportions of PT‐3 cells in kidneys with pre‐existing kidney diseases, including diabetic nephropathy (DN), kidney allografts, and lupus nephropathy (LN; Table 2). We first analyzed the scRNA‐seq dataset (GSE131882), which included 9883 cells from 3 DN biopsy samples and 15 728 cells from three normal human kidney samples [22]. The expression of ACE2 was predominantly detected in PTECs, and its level was significantly increased in diabetic kidneys (Fig. 4A,B). This result was in line with previous studies at the mRNA and protein levels [25, 26]. Compared with controls, the expression levels of protease CTSB and factors related to virus assembly were also significantly increased in diabetic PTECs (Fig. 4A,B). In diabetic kidneys, although the overall number of PTECs decreased, the proportion of PT‐3 cells was significantly higher compared with controls (28.5% vs 11.1%; Fig. 4C). Concomitantly, the proportions of ACE2+/CTSB+, ACE2+/TMPRSS2+, and ACE2+/CTSL+ cells among PTECs were higher than those in controls (Fig. 4D). We also surveyed changes in the expression levels of host factors in PT‐3 cells (Fig. 4E). There was no upregulation of ACE2 mRNA in diabetic PT‐3 cells, although a significant increase in DPP4 expression was observed. Except furin, the expression of major cellular proteases was also upregulated in diabetic PT‐3 cells. Compared with control kidneys, the expression levels of IFITM2 and IFITM3 in PT‐3 cells from diabetic kidneys were increased, while LY6E remained unchanged (Fig. 4E).
Table 2.
Information of the included datasets in our single‐cell analysis. c, cell counts; CTL, control; n, sample sizes.
| First author (year) | Clinical feature | Sequencing method | Accession number | Sample sizes (CTL) | Sample sizes (CKD) |
|---|---|---|---|---|---|
| Wilson (2019) | Normal kidney | snRNA‐seq | GSE131882 | n = 3, c = 15 728 | / |
| Wilson (2019) | DN | snRNA‐seq | GSE131882 | n = 3, c = 15 728 | n = 3, c = 9883 |
| Rajasree Menon (2020) | Kidney allograft | scRNA‐seq | GSE140989 | n = 19, c = 17 578 | n = 5, c = 4690 |
| Evan Der (2019) | Lupus nephritis | scRNA‐seq | SDY997 | n = 3, c = 160 | n = 21, c = 2629 |
Fig. 4.
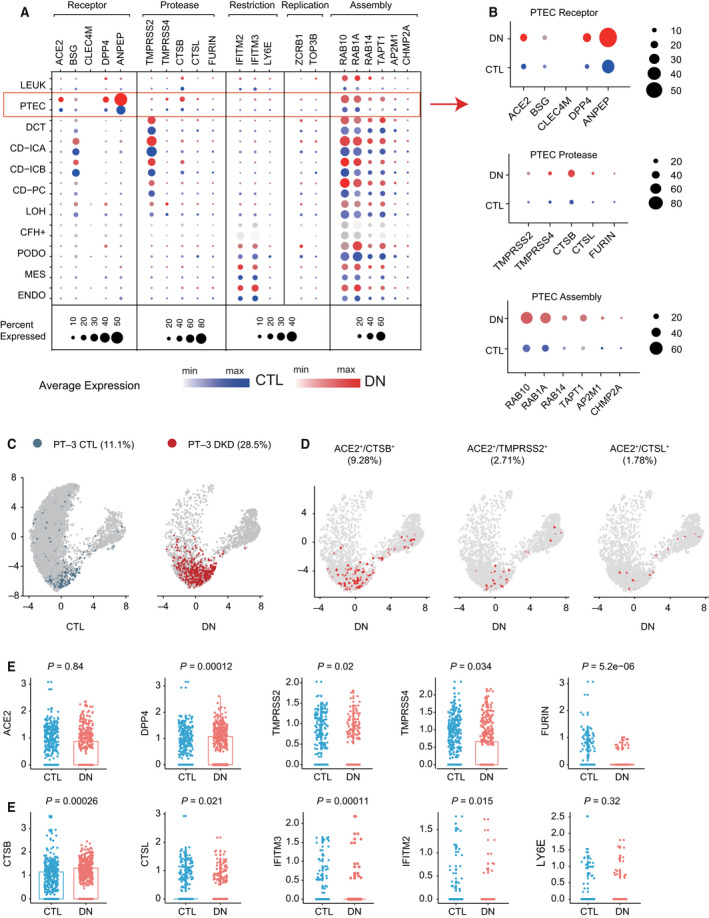
Single cell expression profiling of host factors related to SARS‐CoV‐2 infection in diabetic kidneys. (A) Dot plot showing the expression patterns of host factors in all cell types from normal kidney controls (in blue, sample size = 3) and diabetic kidneys (in red, sample size = 3). Dot size represents the percentage of cells expressing a gene within the cell cluster. CTL, control. (B) Dot plots showing the expression levels of ACE2, cellular proteases and factors related to virus assembly in PTECs of controls (in blue) and DN patients (in red). (C) Feature plot showing distribution and number differences in PT‐3 cells over the UMAP of PTEC subclusters from controls (in blue) and DN patients (in red). (D) Feature plot showing distribution of ACE2+/CTSB+ cells, ACE2+/TMPRSS2+ cells, and ACE2+/CTSL+ cells over the UMAP of PTECs from DN patients. ACE2 and proteases co‐expressing PTECs are colored as red. (E) Scatter‐box plots illustrating the average expression levels of host factors in PT‐3 cells. Comparisons between DN and controls were performed using Mann–Whitney U‐test. The significance level was set as 0.05.
In summary, the proportion of PT‐3 cells in diabetic kidney was increased, which was accompanied by upregulation of protease expression. These findings indicated a higher risk of SARS‐CoV‐2 entry into the kidneys of DN patients, which consequently increases the incidence of kidney infection.
Characteristic of PT‐3 cells in renal allograft and lupus nephritis
We next interrogated a public scRNA‐seq dataset GSE140989, which included 4690 cells from five kidney allograft biopsy samples and 17 578 cells from 19 normal kidney tissue samples [27]. In PTECs from kidney allografts, the proportions of cells expressing receptors (ACE2, DPP4, and ANPEP) and cells expressing CTSB were lower than that in healthy kidneys. Compared with healthy kidneys, restriction factors, especially LY6E, in kidney allografts were increased, while there was no difference in the expression of factors involved in virus assembly (Fig. 5A). Then, we found the fraction of PT‐3 cells in kidney allografts was significantly decreased (Fig. 5B), and the expression of ACE2 was downregulated in these PT‐3 cells, while proteases CTSB/CTSL and restriction factor LY6E were upregulated (Fig. 5C).
Fig. 5.
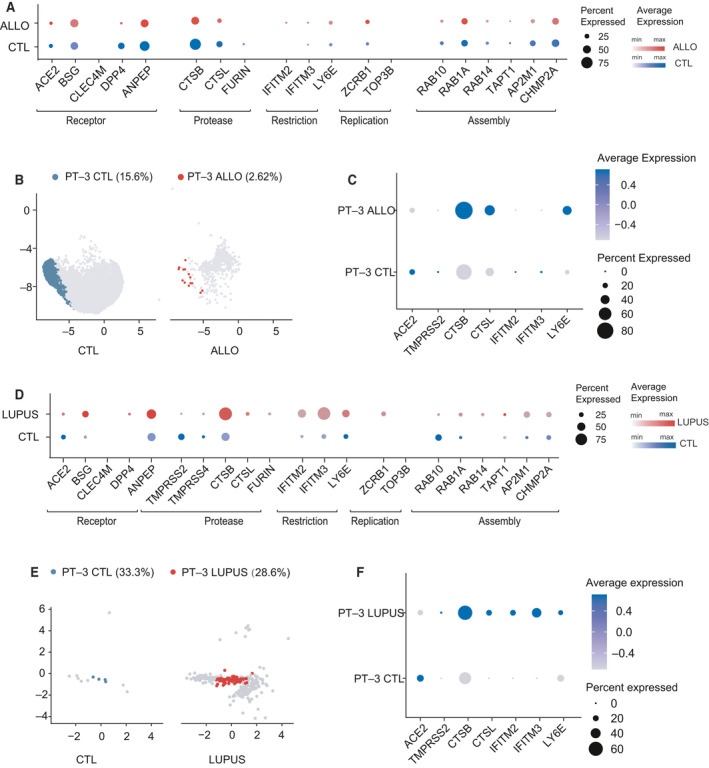
Single cell expression profiling of host factors in kidney allograft and lupus nephritis kidneys. (A) Dot plot showing the expression patterns of host factors in PTECs from normal kidneys (in blue, sample size = 19) and kidney allografts (in red, sample size = 5). Dot size represents the proportion of cells expressing a gene within the cell cluster. CTL, control; ALLO, kidney allograft. (B) Feature plot showing changes in distribution and proportion of PT‐3 cells over the UMAP of PTEC subclusters from controls (in blue) and kidney allograft (in red). (C) Dot plot showing the expression pattern of ACE2, proteases and restriction factors in PT‐3 cells from kidney allografts and normal kidneys. Dot size represents the proportion of cells expressing a gene within the cell type. Darker indicates higher expression level. (D) Dot plot showing the expression patterns of host factors in PTECs from control kidneys (in blue, sample size = 3) and LN kidneys (in red, sample size = 21). Dot size represents the proportion of cells expressing a gene within the cell cluster. CTL, control; LUPUS, lupus nephritis. (E) Feature plot showing difference in distribution and proportion of PT‐3 cells over the UMAP of PTEC subclusters from controls (in blue) and LN patients (in red). (F) Dot plot showing the expression pattern of ACE2, proteases and restriction factors in PT‐3 cells and other PTECs from LN kidneys. Dot size represents the proportion of cells expressing a gene within the cell type. Darker indicates higher expression level.
We further re‐analyzed a scRNA‐seq dataset for LN (Immport, SDY997), which included 2629 cells from 21 LN kidney biopsy samples and 160 cells from three healthy donor kidney samples [28]. We observed that both ACE2 expression and the proportion of ACE2+ cells decreased in LN PTECs. Compared with controls, we found a higher proportion of cells expressing CTSB, restriction factors, and factors for viral replication (ZCRB1) and assembly in LN PTECs (Fig. 5D). We also found that PT‐3 cells decreased slightly in LN kidneys compared with healthy kidneys (28.6% vs 33.33%; Fig. 5E). And in these lupus PT‐3 cells, ACE2 mRNA level decreased while the levels of proteases and restriction factors increased (Fig. 5F).
Overall, the decreased proportion of PT‐3 cells in kidney allografts and LN suggests that kidney susceptibility varied among different kidney diseases.
Evidence of genetic variations determining kidney susceptibility to SARS‐CoV‐2 infection
Since a considerable proportion of COVID‐19 patients present no evidence of kidney infection, even among those with multiple organ infections, suggesting the effects of genetic variants on kidney susceptibility. To delineate this genetic basis, we examined eQTLs of genes encoding host factors, using the Human Kidney eQTL Atlas [29] and GTEx database. We did not identify any significant eQTL for ACE2 gene in the kidney (Table S1), consistent with previous study [21]. However, we did find some eQTLs associated with the kidney expression of proteases and restriction factors, such as IFITM2 and LY6E. Among these, five eQTL‐single nucleotide polymorphisms (SNPs; rs3809111, rs111797074, rs75117940, rs909097, and rs909098) were significantly associated with the low expression level of IFITM2. And these associations were only detected in the tubules, suggesting individual differences in IFITM2 expression in renal tubules. In kidney cortex samples from GTEx database, another significant eQTL rs111817998 was found to contribute to high IFITM2 expression.
Among the six eQTLs identified, three of them (rs3809111, rs75117940, and rs111817998) located in the promoter region, and two located in intronic regions (rs909097 and rs909098) (Fig. 6A). A single nucleotide change in transcription factor‐binding sites (TFBSs) can affect interactions with transcription factors, altering downstream gene expression. To explore the regulatory mechanisms associated with these eQTL‐SNPs, we examined the potential TFBSs in the promoter region of IFITM2, using PROMO [30]. The T allele of SNP rs3809111 might disrupt the binding motifs of PXR‐1: RXR‐alpha [T05671]. The C allele of SNP rs111817998 disrupted the binding motifs of 4 transcription factors, including signal transducer and activator of transcription 4 (STAT4 [T01577]), TFII‐I [T00824], c‐Ets‐1 [T00112], and glucocorticoid receptor (GR)‐alpha [T00337] (Fig. 6B). STAT4 and TFII‐I have been reported to participate in the host innate immune response against viral infection [31, 32]. Although the association and regulatory mechanism need to be further confirmed, special attention should be paid to these genetic variants in future studies of COVID‐19 patients.
Fig. 6.
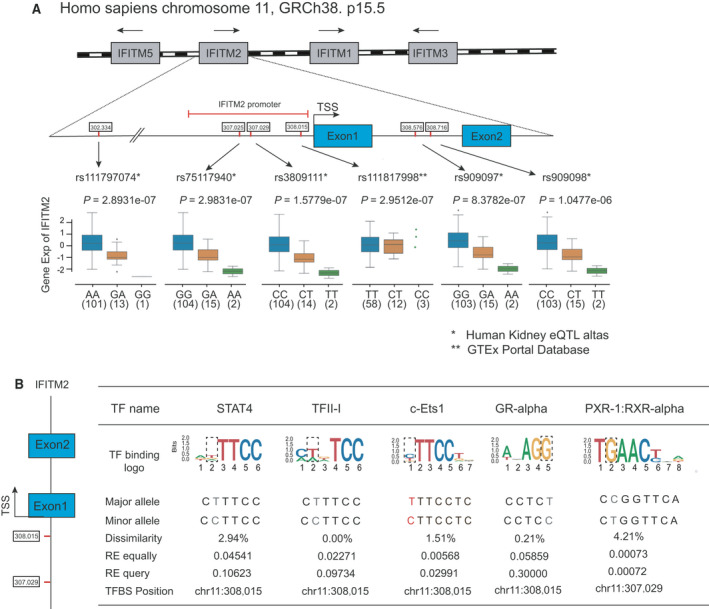
eQTLs associated with IFITM2 transcript expression level in human kidney. (A) Schematic representation of four genes (IFITM1, IFITM2, IFITM3, and IFITM5, in gray) located in one IFITM locus in chromosome 11p15.5. The blue boxes represent the exons of IFITM2 gene, and red marks represent the position of six eQTL‐SNPs for IFITM2 detected in the two databases. Box plots below show the normalized IFITM2 gene expression levels related to SNP genotypes, and the P value for each eQTL association. TSS, transcription start sites. (B) Sequence logo plots visualizing TFBS of transcription factors could be disrupted by SNP rs3809111 and SNP rs111817998 in the promoter of the IFITM2 gene.
Discussion
Although the high prevalence of kidney involvement is well‐documented in patients with COVID‐19, limited information exists regarding host factors that determine this kidney tropism of SARS‐CoV‐2. In the current study, we comprehensively characterized the expression patterns and expression levels of host factors in kidney cells, by mining human kidney scRNA‐seq datasets, and identified PT‐3 cells, a special subtype of PTECs that may function as a “gateway” cell type for the SARS‐CoV‐2 invasion of the kidney. Our analysis revealed that PT‐3 cells not only enriched for ACE2 expression, but also expressed two other human coronavirus receptors, DPP4 and ANPEP, at high levels than other kidney cells. Additionally, the expression levels of proteases and host factors involved in virus replication and assembly in PT‐3 cells were higher than other PTECs. PT‐3 cells were also significantly enriched in various virus infection‐related pathways, as well as endocytosis and autophagy pathways. These pathways play important roles in viral infection [8, 33]. Although the expression of restriction factors was also observed in PT‐3 cells, the basal expression level was lower than that in other PTECs. Based on the transcription signature of PT‐3 cells, we predict that these cells are highly permissive to infection by SARS‐CoV‐2 and other human coronaviruses. Urinary proteomics confirmed that the protein levels of PT‐3 markers, in addition to ACE2, CTSB, and restriction factors, were significantly increased in the urine of COVID‐19 patients. This result further supported the findings derived from our scRNA‐seq analyses, which suggested that PT‐3 cells were the target cells of SARS‐CoV‐2 and provided new insights to understand the pathogenesis of kidney infections. Moreover, the detection of PT‐3 cell‐specific markers in the urine could be utilized as a noninvasive and dynamic method for kidney infection risk assessment.
TMPRSS2 transcripts were most abundantly expressed in CD cells and DCT cells and rarely expressed in the PTECs that highly expressed ACE2. This expression pattern is distinct from the pattern observed in nasal epithelial cells, where the protease TMPRSS2 is highly expressed on the cell membranes of goblet cells and ciliated cells and is co‐expressed with ACE2 [19]. In contrast, the proportion of ACE2 and TMPRSS2 co‐expressing cells was much lower than that of ACE2 and CTSB co‐expressing cells among PT‐3 cells. For both SARS‐CoV and SARS‐CoV‐2, S protein priming by TMPRSS2 has been shown to be crucial for entry into target cells [11]. Compared with SARS‐CoV, the S protein of SARS‐CoV‐2 contains a four‐amino acid insertion at the S1/S2 site, which allows it to be proteolytically cleaved by a variety of cellular proteases, including CTSB and CTSL [34]. Previous studies also verified that CTSB had high proteolytic activity against SARS‐CoV‐2, whereas CTSL showed lower proteolytic activity against SARS‐CoV‐2 than SARS‐CoV [34]. These findings indicated that SARS‐CoV‐2 might use both TMPRSS2 and CTSB/L for S protein priming. Thus, the prominent co‐expression of ACE2 and CTSB in PT‐3 cells suggests that the invasion of SARS‐CoV‐2 into these cells is likely mediated by CTSB and that the routes of viral entry may be cell type‐specific. However, the specific mechanism requires further study.
Our survey identified an increased proportion of PT‐3 cells in diabetic kidneys, which could lead to enhanced kidney susceptibility to SARS‐CoV‐2 infection. This conclusion is consistent with a previous study and clinical observations [2, 3, 25]. We also found lower proportion of PT‐3 cells in kidney allografts than healthy controls, accompanied by the upregulation of restriction factors. However, it has been reported that kidney transplant recipients with COVID‐19 are at high risk of acute kidney injury [35]. The kidney abnormalities in kidney transplant patients could be due to other mechanisms such as immune‐mediated effects. We also predicted the decreased viral susceptibility of kidneys with LN, based on the decreased proportion of PT‐3 cells observed in LN kidneys compared with healthy kidneys. Whether patients with systemic lupus erythematosus have a higher risk of SARS‐CoV‐2 infection than the general population remains unclear [36]. These findings provided useful information regarding the SARS‐CoV‐2 infection of kidneys in LN patients. In addition, evidence on the kidney infectivity of SARS‐CoV‐2 virus has been largely derived from autopsy studies [4, 7, 37, 38]. Some studies also reported the detection of SARS‐CoV‐2 RNA or protein in the urine of COVID‐19 patients [39, 40]. Despite these positive findings, many other studies did not find the presence of SARS‐CoV‐2 virus in the kidneys of COVID‐19 patients [41]. Therefore, caution is needed to draw conclusions about the direct kidney infection by SARS‐CoV‐2 [38].
Indeed, there are discrepancies between transcriptome and proteome, and this issue could be more significant in single‐cell transcriptomics due to shallow sequencing depth and low mRNA capture efficiency of this approach. For instance, at single‐cell transcriptomics level, the mRNA levels of ACE2 in major glomeruli cell types such as PODOs and MESs were hardly detected. While at protein level, low level of ACE2 was confirmed in the glomeruli [26]. Regarding TMPRSS2, it was hardly detected in the ACE2‐positive PTECs based on the results of single‐cell analysis, whereas immunostaining confirmed that it also colocalized with ACE2 in the proximal tubules of kidney organoids [42]. Meanwhile, posttranscriptional modification is an important factor contributing to this discrepancy [43]. In this context, accurate prediction based on results from scRNA‐seq analysis alone could be challenging, and a multi‐omics data integrative strategy can provide a more definitive conclusion.
Overall, our analysis identified PT‐3 cells, a PTEC subtype, as key determinant for SARS‐CoV‐2 infection in the kidney. These findings provide new insights into the mechanism underlying the SARS‐CoV‐2 infection of kidney cells and present a potential strategy for risk assessment of renal infection during COVID‐19.
Materials and methods
Study population
We first collected autopsy kidney specimens from 26 cases of patients died from COVID‐19. We also collected the urine samples of 97 COVID‐19 patients. Laboratory confirmed cases of COVID‐19 were defined as a positive result on viral gene sequencing highly similar with known SARS‐CoV‐2 or real‐time polymerase chain reaction assay of respiratory tract specimens or blood specimens. The control group consisted of 350 healthy individuals who were matched on age, gender with the 97 COVID‐19 patients.
Study approval
Autopsies were performed in the Union Hospital of Tongji Medical College of Huazhong University of Science and Technology. The study was approved by the local ethics committee. Urinary proteomics study of healthy individuals and COVID‐19 patients was approved by institutional review boards of Beijing Proteome Research Center and ethics committee of Guangdong Provincial Hospital of Chinese Medicine (BF2020‐052‐01). All clinical investigations were conducted according to the Declaration of Helsinki. Written informed consent was obtained from enrolled subjects or next of kin (for deceased subjects).
Tissue sampling and processing
Kidney tissue specimens were fixed in 2.5% glutaraldehyde or 10% formalin for 48–72 h before transmission electron microscope (TEM) examination or histological staining. For evidence of SARS‐CoV‐2 infection in kidney samples, we performed electron microscope examination. After the osmium tetroxide postfixation and gradient dehydration, the samples were embedded in Epon for TEM (HT‐7800; Hitachi, Tokyo, Japan) observation, as previously described [5].
To determine the distribution and localization of SLC36A2 in kidney, immunohistochemical staining was performed in autopsied kidney specimens obtained from four COVID‐19 patients and four healthy controls, using SLC36A2 antibody from NOVUS (Catalog Number: NBP1‐59418; Centennial, CO, USA).
Urinary proteomic sample preparation and NanoHPLC‐MS analysis
The midstream of the first morning urine was collected and put in 56 °C water bath for 30 min. The urine samples were then prepared, sterilized, and stored at −80 °C. Briefly, one milliliter urine sample was centrifuged at 176 000 g for 70 min to save the pellets. The samples were then processed according to a published streamlined workflow with minor modification, in which the centrifuged pellets were digested in solution with 1 μg of trypsin at 37 °C for 4 h [44].
Tryptic peptides were separated using a self‐packed capillary column packed with C18 reverse‐phase particles, and the treated samples were analyzed with Thermo Fisher Orbitrap mass spectrometer (MS) coupled with online Easy‐nLC 1000 nano (Nano HPLC) system (Thermo Fisher Scientific, Waltham, MA, USA). Raw LC‐MS/MS files were uploaded onto FIRMIANA platform for further processing and analysis, and the mascot v.2.5.1 (Matrix Science, Boston, MA, USA) was used for peptide identification with peptide FDR < 1%.
We used the intensity‐based absolute quantification (iBAQ) algorithm for protein quantification. For batch‐to‐batch comparisons, iBAQ was further transformed into iFOT (intensity‐based Fraction of Total), which represented the normalized intensity of the protein identified in the LC‐MS/MS analysis. For visualization purposes, the value of iFOTs is multiplied by 105. Trypsin digests of human HEK293T cells were used as control quality samples and routinely assessed by the LC‐MS/MS to ensure instrument reproducibility.
Mann–Whitney U‐test was performed to compare protein abundance between two groups.
Data source
We obtained publicly available scRNA‐seq/single‐nucleus RNA sequencing (snRNA‐seq) datasets of human kidney with CKDs from Gene Expression Omnibus database or ImmPort database (Table 2). Additional data related to this paper may be requested from the authors.
Data processing, quality control, and normalization
The original gene‐barcode matrix was downloaded and processed by Seurat3 r package (https://satijalab.org/seurat/). Cells/nuclei with < 500 or more than 5000 expressed genes were filtered out, and genes expressed in < 3 cells or nuclei were removed [22]. Then, a log‐normalize step was performed using the method “LogNormalize.”
Cell unsupervised clustering and cell type identification
We used the top 2000 most variable genes to generate clusters at multiple resolutions and selected the best matching clusters according to the instruction in Seurat [45]. Then, we performed principal component analysis for dimensionality reduction and the Louvain method for detection of cell subgroups. The first 10 principal components were selected and used for the Uniform Manifold Approximation and Projection (UMAP). Specific marker genes of different cell types were identified by likelihood ratio test in Seurat package. PTECs which enriched in the expression of ACE2 were extracted for further clustering analysis, and PTEC subclusters were identified using the FindCluster function in Seurat with the resolution parameter of 0.3. DEGs within clusters were identified using FindMarkers function with logfc.threshold = 0.25 and min.pct = 0.25.
Gene set enrichment analysis
Gene set enrichment analysis was conducted using clusterProfiler package (v3.5) with default parameters [46]. Differential expressed genes in PT‐3 subtype (compared to other PTEC subtypes) were identified with the limma r package using a log‐fold‐change threshold of 1.25 and P value threshold of 0.05. The preranked gene list according to fold change values was input for GSEA on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. P value < 0.05 and adjusted P value < 0.05 were set up as the significant cutoff for GSEA.
Expression quantitative trait loci inquiry
We inquired the eQTLs which might regulate virus‐related host factor gene expression in the Human Kidney eQTL Atlas (http://susztaklab.com/eqtl) and Genotype‐Tissue Expression (GTEx) database.
Statistics
All statistical analyses were conducted using r or python (https://www.python.org/). For comparison between two independent groups, we performed two‐tailed, unpaired Student's t‐test for data that were normally distributed. Otherwise, we performed Mann–Whitney U‐test. P < 0.05 was considered statistically significant. The level of significance was assigned as *P < 0.05, **P < 0.01, ***P < 0.001, or ****P < 0.0001.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
HP conceived the project and designed the study. HL, FX, and XM conducted all single‐cell analysis and prepared the figures. HS carried out the histological examination and immunohistochemical staining. LS carried out experiments and analyzed the urinary proteomics data. CZ and ZZ supervised these works. XM, HL, FX, YS, and YL participated in results interpretation. XM, HL, and HP wrote the manuscript, and all the authors approved for the manuscript submission.
Peer Review
The peer review history for this article is available at https://publons.com/publon/10.1111/febs.16114.
Supporting information
Table S1. eQTLs associated with gene expression of host factors in kidney.
Acknowledgements
HP was supported by grants from the Science and Technology Program of Guangzhou, China (International Science & Technology Cooperation Program; 201807010037), National Natural Science Foundation of China (NSFC 81873613) and Special Support Plan for High‐Level Talents of Guangdong Province. ZZ was supported by grants from Guangdong Provincial Key Laboratory of Research on Emergency in TCM (2017B030314176) and National Key Research and Development Project (2020YFA0708001). HL was supported by grants from Foundation for Talent Scientific Research People in colleges and universities of Guangdong, China.
Hongchun Lin, Xinxin Ma and Fang Xiao contributed equally to this work.
Contributor Information
Zhongde Zhang, Email: doctorzzd99@gzucm.edu.cn.
Chun Zhang, Email: drzhangchun@hust.edu.cn.
Hui Peng, Email: pengh@mail.sysu.edu.cn.
References
- 1. Bradley BT, Maioli H, Johnston R, Chaudhry I, Fink SL, Xu H, Najafian B, Deutsch G, Lacy JM, Williams T et al. (2020) Histopathology and ultrastructural findings of fatal COVID‐19 infections in Washington State: a case series. Lancet 396, 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Richardson S, Hirsch JS, Narasimhan M, Crawford JM, McGinn T, Davidson KW, the Northwell COVID‐19 Research Consortium , Barnaby DP, Becker LB, Chelico JD et al. (2020) Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID‐19 in the New York City Area. JAMA 323, 2052–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pei G, Zhang Z, Peng J, Liu L, Zhang C, Yu C, Ma Z, Huang Y, Liu W, Yao Y et al. (2020) Renal involvement and early prognosis in patients with COVID‐19 pneumonia. J Am Soc Nephrol 31, 1157–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Puelles VG, Lutgehetmann M, Lindenmeyer MT, Sperhake JP, Wong MN, Allweiss L, Chilla S, Heinemann A, Wanner N, Liu S et al. (2020) Multiorgan and renal tropism of SARS‐CoV‐2. N Engl J Med 383, 590–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Su H, Yang M, Wan C, Yi LX, Tang F, Zhu HY, Yi F, Yang HC, Fogo AB, Nie X et al. (2020) Renal histopathological analysis of 26 postmortem findings of patients with COVID‐19 in China. Kidney Int 98, 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Farkash EA, Wilson AM & Jentzen JM (2020) Ultrastructural evidence for direct renal infection with SARS‐CoV‐2. J Am Soc Nephrol 31, 1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Braun F, Lutgehetmann M, Pfefferle S, Wong MN, Carsten A, Lindenmeyer MT, Norz D, Heinrich F, Meissner K, Wichmann D et al. (2020) SARS‐CoV‐2 renal tropism associates with acute kidney injury. Lancet 396, 597–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fung TS & Liu DX (2019) Human coronavirus: host‐pathogen interaction. Annu Rev Microbiol 73, 529–557. [DOI] [PubMed] [Google Scholar]
- 9. Huang IC, Bailey CC, Weyer JL, Radoshitzky SR, Becker MM, Chiang JJ, Brass AL, Ahmed AA, Chi X, Dong L et al. (2011) Distinct patterns of IFITM‐mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog 7, e1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Millet JK & Whittaker GR (2015) Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res 202, 120–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hoffmann M, Kleine‐Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A et al. (2020) SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Raj VS, Mou H, Smits SL, Dekkers DHW, Müller MA, Dijkman R, Muth D, Demmers JAA, Zaki A, Fouchier RAM et al. (2013) Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus‐EMC. Nature 495, 251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang K, Chen W, Zhang Z, Deng Y, Lian JQ, Du P, Wei D, Zhang Y, Sun XX, Gong L et al. (2020) CD147‐spike protein is a novel route for SARS‐CoV‐2 infection to host cells. Signal Transduct Target Ther 5, 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yeager CL, Ashmun RA, Williams RK, Cardellichio CB, Shapiro LH, Look AT & Holmes KV (1992) Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357, 420–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zang R, Gomez Castro MF, McCune BT, Zeng Q, Rothlauf PW, Sonnek NM, Liu Z, Brulois KF, Wang X, Greenberg HB et al. (2020) TMPRSS2 and TMPRSS4 promote SARS‐CoV‐2 infection of human small intestinal enterocytes. Sci Immunol 5, eabc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT & Veesler D (2020) Structure, function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell 181, 281–292.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pfaender S, Mar KB, Michailidis E, Kratzel A, Boys IN, V'Kovski P, Fan W, Kelly JN, Hirt D, Ebert N et al. (2020) LY6E impairs coronavirus fusion and confers immune control of viral disease. Nat Microbiol. 5, 1330–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Batlle D, Soler MJ, Sparks MA, Hiremath S, South AM, Welling PA & Swaminathan S; COVID‐19 and ACE2 in Cardiovascular, Lung, and Kidney Working Group (2020) Acute kidney injury in COVID‐19: emerging evidence of a distinct pathophysiology. J Am Soc Nephrol 31, 1380–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sungnak W, Huang N, Becavin C, Berg M, Queen R, Litvinukova M, Talavera‐Lopez C, Maatz H, Reichart D, Sampaziotis F et al. (2020) SARS‐CoV‐2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med 26, 681–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cao Y, Li L, Feng Z, Wan S, Huang P, Sun X, Wen F, Huang X, Ning G & Wang W (2020) Comparative genetic analysis of the novel coronavirus (2019‐nCoV/SARS‐CoV‐2) receptor ACE2 in different populations. Cell Discov 6, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang YM & Zhang H (2020) Genetic roadmap for kidney involvement of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) Infection. Clin J Am Soc Nephrol 15, 1044–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wilson PC, Wu H, Kirita Y, Uchimura K, Ledru N, Rennke HG, Welling PA, Waikar SS & Humphreys BD (2019) The single‐cell transcriptomic landscape of early human diabetic nephropathy. Proc Natl Acad Sci USA 116, 19619–19625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chan VS, Chan KY, Chen Y, Poon LL, Cheung AN, Zheng B, Chan KH, Mak W, Ngan HY, Xu X et al. (2006) Homozygous L‐SIGN (CLEC4M) plays a protective role in SARS coronavirus infection. Nat Genet 38, 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lake BB, Chen S, Hoshi M, Plongthongkum N, Salamon D, Knoten A, Vijayan A, Venkatesh R, Kim EH, Gao D et al. (2019) A single‐nucleus RNA‐sequencing pipeline to decipher the molecular anatomy and pathophysiology of human kidneys. Nat Commun 10, 2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Menon R, Otto EA, Sealfon R, Nair V, Wong AK, Theesfeld CL, Chen X, Wang Y, Boppana AS, Luo J et al. (2020) SARS‐CoV‐2 receptor networks in diabetic and COVID‐19‐associated kidney disease. Kidney Int 98, 1502–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ye M, Wysocki J, William J, Soler MJ, Cokic I & Batlle D (2006) Glomerular localization and expression of Angiotensin‐converting enzyme 2 and Angiotensin‐converting enzyme: implications for albuminuria in diabetes. J Am Soc Nephrol 17, 3067–3075. [DOI] [PubMed] [Google Scholar]
- 27. Menon R, Otto EA, Hoover P, Eddy S, Mariani L, Godfrey B, Berthier CC, Eichinger F, Subramanian L, Harder J et al. (2020) Single cell transcriptomics identifies focal segmental glomerulosclerosis remission endothelial biomarker. JCI Insight 5, e133267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Der E, Suryawanshi H, Morozov P, Kustagi M, Goilav B, Ranabothu S, Izmirly P, Clancy R, Belmont HM, Koenigsberg M et al. (2019) Tubular cell and keratinocyte single‐cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat Immunol 20, 915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qiu C, Huang S, Park J, Park Y, Ko YA, Seasock MJ, Bryer JS, Xu XX, Song WC, Palmer M et al. (2018) Renal compartment‐specific genetic variation analyses identify new pathways in chronic kidney disease. Nat Med 24, 1721–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Messeguer X, Escudero R, Farré D, Núñez O, Martinez J & Alba MM (2002) PROMO: detection of known transcription regulatory elements using species‐tailored searches. Bioinformatics 18, 333–334. [DOI] [PubMed] [Google Scholar]
- 31. Nguyen KB, Watford WT, Salomon R, Hofmann SR, Pien GC, Morinobu A, Gadina M, O'Shea JJ & Biron CA (2002) Critical role for STAT4 activation by type 1 interferons in the interferon‐gamma response to viral infection. Science 297, 2063–2066. [DOI] [PubMed] [Google Scholar]
- 32. Roy AL (2012) Biochemistry and biology of the inducible multifunctional transcription factor TFII‐I: 10 years later. Gene 492, 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang N & Shen H‐M (2020) Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID‐19. Int J Biol Sci 16, 1724–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jaimes JA, Millet JK & Whittaker GR (2020) Proteolytic cleavage of the SARS‐CoV‐2 spike protein and the role of the novel S1/S2 site. iScience 23, 101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fava A, Cucchiari D, Montero N, Toapanta N, Centellas J, Vila‐Santandreu A, Coloma A, Meneghini M, Manonelles A, Sellares J et al. (2020) Clinical characteristics and risk factors for severe COVID‐19 in hospitalized kidney transplant recipients: a multicentric cohort study. Am J Transplant, 20, 3030–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gartshteyn Y, Askanase AD, Schmidt NM, Bernstein EJ, Khalili L, Drolet R, Broderick RJ, Geraldino‐Pardilla L & Kapoor T (2020) COVID‐19 and systemic lupus erythematosus: a case series. Lancet Rheumatol 2, e452–e454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hanley B, Naresh KN, Roufosse C, Nicholson AG, Weir J, Cooke GS, Thursz M, Manousou P, Corbett R, Goldin R et al. (2020) Histopathological findings and viral tropism in UK patients with severe fatal COVID‐19: a post‐mortem study. Lancet Microbe 1, e245–e253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hassler L, Reyes F, Sparks M, Welling P & Batlle D (2021) Evidence for and against direct kidney infection by SARS‐CoV‐2 in patients with COVID‐19. Clin J Am Soc Nephrol. Online ahead of print. CJN.04560421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peng L, Liu J, Xu W, Luo Q, Chen D, Lei Z, Huang Z, Li X, Deng K, Lin B et al. (2020) SARS‐CoV‐2 can be detected in urine, blood, anal swabs, and oropharyngeal swabs specimens. J Med Virol 92, 1676–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. George S, Pal AC, Gagnon J, Timalsina S, Singh P, Vydyam P, Munshi M, Chiu JE, Renard I, Harden CA et al. (2021) Evidence for SARS‐CoV‐2 spike protein in the urine of COVID‐19 patients. Kidney360 2, 922–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kudose S, Batal I, Santoriello D, Xu K, Barasch J, Peleg Y, Canetta P, Ratner LE, Marasa M, Gharavi AG et al. (2020) Kidney biopsy findings in patients with COVID‐19. J Am Soc Nephrol 31, 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wysocki J, Ye M, Hassler L, Gupta AK, Wang Y, Nicoleascu V, Randall G, Wertheim JA & Batlle D (2021) A novel soluble ACE2 variant with prolonged duration of action neutralizes SARS‐CoV‐2 infection in human kidney organoids. J Am Soc Nephrol 32, 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lores E, Wysocki J & Batlle D (2020) ACE2, the kidney and the emergence of COVID‐19 two decades after ACE2 discovery. Clin Sci (Lond) 134, 2791–2805. [DOI] [PubMed] [Google Scholar]
- 44. Leng W, Ni X, Sun C, Lu T, Malovannaya A, Jung SY, Huang Y, Qiu Y, Sun G, Holt MV et al. (2017) Proof‐of‐concept workflow for establishing reference intervals of human urine proteome for monitoring physiological and pathological changes. EBioMedicine 18, 300–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Butler A, Hoffman P, Smibert P, Papalexi E & Satija R (2018) Integrating single‐cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu G, Wang L‐G, Han Y & He Q‐Y (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. eQTLs associated with gene expression of host factors in kidney.


