Abstract
COVID‐19 is the disease evoked by a new breed of coronavirus called the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). Recently, COVID‐19 has become a pandemic by infecting more than 152 million people in over 216 countries and territories. The exponential increase in the number of infections has rendered traditional diagnosis techniques inefficient. Therefore, many researchers have developed several intelligent techniques, such as deep learning (DL) and machine learning (ML), which can assist the healthcare sector in providing quick and precise COVID‐19 diagnosis. Therefore, this paper provides a comprehensive review of the most recent DL and ML techniques for COVID‐19 diagnosis. The studies are published from December 2019 until April 2021. In general, this paper includes more than 200 studies that have been carefully selected from several publishers, such as IEEE, Springer and Elsevier. We classify the research tracks into two categories: DL and ML and present COVID‐19 public datasets established and extracted from different countries. The measures used to evaluate diagnosis methods are comparatively analysed and proper discussion is provided. In conclusion, for COVID‐19 diagnosing and outbreak prediction, SVM is the most widely used machine learning mechanism, and CNN is the most widely used deep learning mechanism. Accuracy, sensitivity, and specificity are the most widely used measurements in previous studies. Finally, this review paper will guide the research community on the upcoming development of machine learning for COVID‐19 and inspire their works for future development. This review paper will guide the research community on the upcoming development of ML and DL for COVID‐19 and inspire their works for future development.
Keywords: 2019‐nCoV, deep learning, COVID‐19, COVID‐19 dataset, machine learning
1. INTRODUCTION
The coronavirus disease (COVID‐19) is defined as a disease or infection evoked by a new breed of coronavirus called the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), which was previously named 2019‐nCoV (Chung et al., 2020; Gralinski & Menachery, 2020; Rothe et al., 2020). It was first discovered in Wuhan Hubei City in China. The World Health Organization (WHO) reported the first COVID‐19 case on 31 December 2019. The COVID‐19 outbreak was declared as a global outbreak on 30 January 2020 (Sohrabi et al., 2020). Since 2009, the WHO considered H1N1 as an internationally pandemic disease; COVID‐19 was the second pandemic disease declared (Cucinotta & Vanelli, 2020; Zarocostas, 2009). The disease spread mainly through close contact with infected individuals, although researchers are still investigating potential infection routes of the disease (Corman et al., 2020; G. Li & De Clercq, 2020).
The major signs of COVID‐19 infection are fever, dry cough and breathing difficulty. Some patients may have muscle aches and experience fatigue and loss of taste or smell (anosmia), and up to 10% have GI‐related symptoms, such as diarrhoea (Tsatsakis et al., 2020). As initially thought, one of the potential ways for the virus to spread between people is direct contact. Thus, social distancing can reduce the possibility of being infected. Disease transmission can occur at a distance of as far as 6 feet. As a result of that, the breathing drops generated from the infected person while talking or sneezing can be taken as one of the primary reasons to cause the disease to spread. Furthermore, the symptoms of COVID‐19 in some cases are not noticeable (Rothe et al., 2020). Figure 1 shows how COVID‐19 is spread from a person to another person when there is no social distancing.
FIGURE 1.
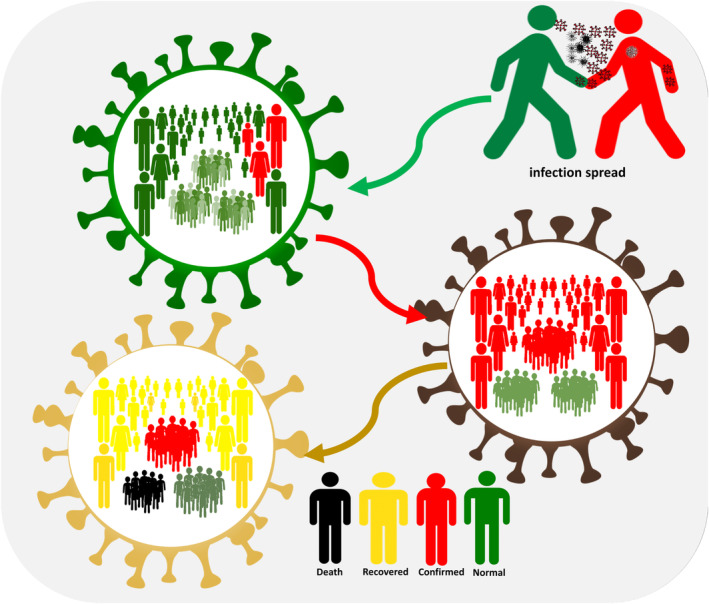
COVID‐19 Person to person spread (Al‐Betar et al., 2021)
In general, several approaches for diagnosing COVID‐19 are available, such as nucleic acid‐based methods using polymerase chain reaction (PCR) (Abdulkareem, Mohammed, et al., 2021; Esbin et al., 2020), next‐generation sequencing (Harris et al., 2013), computed tomography (CT) scan (Akram et al., 2021), chest X‐ray (CXR) (Mohammed et al., 2020; Pan, Guan, et al., 2020; Sahlol et al., 2020; Shi et al., 2020) and paper‐based detection (K. Mao et al., 2020). These methods are used in monitoring changes in organs, and patients may need to undergo these pathological tests. The most popular of these pathological tests are CT scan and CXR (Dansana et al., 2020; Kumar, Nagpal, et al., 2020; Kumari et al., 2020; Mohammed et al., 2021; Waheed et al., 2020). Regarding CT and X‐ray chest techniques used for COVID‐19 diagnosis, many advantages distinguished these technologies. For instance, the CT provides more details about the patient's status and relatively quick compared with the other technologies. Using the X‐ray chest can get the results at a lower price and lower radiation. However, these technologies have some drawbacks that may affect their performance and usage, such as the CT scans of the brain can be affected by bone nearby, and the X‐ray chest does not provide 3D information.
Unfortunately, a precise and quick method for COVID‐19 diagnosis is still unavailable. Usually, medical images (CT scan and X‐ray) are used in COVID‐19 diagnosis (Pan, Guan, et al., 2020; Pan, Ye, et al., 2020; Shi et al., 2020). These images are observed by an expert, who analyses content according to his or her experience in making diagnosis. In general, doctors can experience fatigue because of long working hours and make wrong diagnosis. Patients with COVID‐19 can present irregularities in their CT or CXR results, and many lung problems are identical to COVID‐19. Moreover, normality on either a CT scan or CXR does not necessarily indicate a negative COVID‐19 case. For this reason, assistant tools in the health care sector are required to ensure correct diagnosis. For faster and more accurate results, new approaches for detecting COVID‐19 on the basis of the principles of artificial intelligence (AI) have been proposed, specifically deep learning (DL) and machine learning (ML) (Alimadadi et al., 2020; Goodfellow et al., 2016; LeCun et al., 2015; Michie et al., 1994).
Traditional ML and DL techniques have been developed by various scholars to assist doctors in making correct diagnosis. Such techniques can help in classifying X‐ray or CT scans of the chest into two classes: infected and normal. A decision is made after several steps, such as reading of an X‐ray image, prepossessing and extraction of unique features from input images, then features are input into the ML or DL model for a final prediction decision. The other purposes of using such techniques are predicting an outbreak separation by analysing COVID‐19 data and predicting red zones and number of infected cases with AI. Several AI techniques (DL and ML) are used in examining patients with COVID‐19 according to X‐ray and CT scans:
Supervised learning (Ong, Wong, et al., 2020; Siddiqui et al., 2020)
Unsupervised learning (Abdulkareem, Mohammed, et al., 2021; Carrillo‐Larco & Castillo‐Cara, 2020; Cui et al., 2020)
The DL techniques can be used to diagnose coronavirus, predicted the virus infection, and diagnose the virus. The most prominent DL techniques are:
Convolutional Neural Network (CNN) (Alazab et al., 2020; Apostolopoulos et al., 2020)
Deep Neural Network (DNN) (Das et al., 2020; Ni et al., 2020)
Recurrent Neural Network (RNN) (Liang et al., 2020; Zeroual et al., 2020)
Generative Adversarial Networks (GANs) (Jamshidi et al., 2020; Loey, Smarandache, et al., 2020; Waheed et al., 2020)
The literature is carefully selected based on impact and quality. Figure 2 shows the main source of COVID‐19 scientific publications using ML and DL. In the figure, the selected works are classified according to the digital database used to retrieve the papers, such as Elsevier, IEEE Xplore, MDPI, Springer, IOP Press and Wiley. Figure 2 shows the distribution of COVID‐19 work based on the type of publication, such as open access and close access.
FIGURE 2.
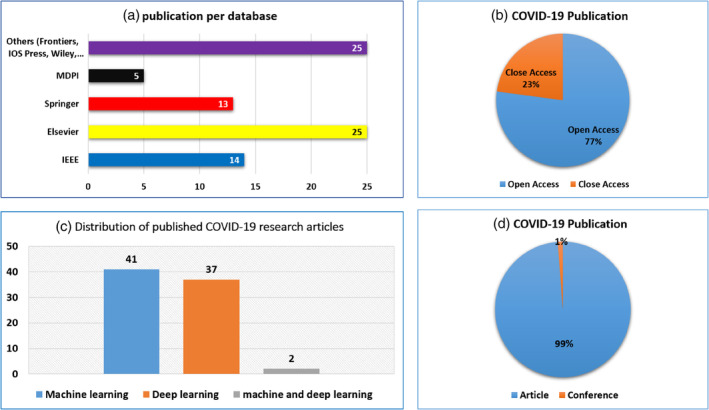
publications statistics on ML and DL works in COVID‐19. (a) Publication per database. (b) COVID‐19 publication. (c) Distribution of published COVID‐19 research articles. (d) COVID‐19 publication
The main contributions of this work are to contribute a comprehensive review of the most recent ML and DL approaches used in assisting doctors and scientists in COVID‐19 diagnosis. More than 200 papers from high‐impact‐factor journals published in IEEE, Elsevier, Springer and Wiley were reviewed. The present paper can provide information on the most used public COVID‐19 datasets which can be utilized by other researchers. Also, this review paper will guide the research community on the upcoming development of machine learning for COVID‐19 and inspire their works for future development. This review paper will guide the research community on the upcoming development of ML and DL for COVID‐19 and inspire their works for future development.
This paper answers the following questions:
Q1‐ What are ML and DL mechanisms applied for COVID‐19 diagnosis?
Q2‐ Are there any public standard datasets that can be applied to examine ML and DL techniques?
Q3‐ What are the most important measures that can be used to evaluate ML and DL approaches?
Q4‐ Are there any end‐to‐end application available for COVID‐19 diagnosis?
Q5‐ Which countries have the most number of researchers who publish work about ML and DL techniques to diagnose COVID‐19?
Q6‐ What is the suitable ML or DL mechanism for determining different performance indicators for COVID‐19 diagnosis?
The remaining part of this review consists of the following sections: Section 2 presents the proposed protocol for selecting studies about COVID‐19. Section 3.1 presents the research related to ML algorithms for COVID‐19, and Section 3.2 presents works related to DL algorithms for COVID‐19. Hybrid approaches are presented in Section 3.3. Public COVID‐19 datasets provided in Section 4. Section 5 provides the most commonly used measures for ML and DL approaches for COVID‐19. The review is concluded and future work is recommended in Section 7.
2. PROTOCOL FOR SELECTING COVID‐19‐RELATED STUDIES
Studies were selected using the most relevant keywords, namely, ‘COVID‐19’, ‘machine learning’ and ‘deep learning’. Only studies written in English were collected, and digital databases were used, such as Elsevier, IEEE Xplore, MDPI, Springer, IOP Press and Wiley. The procedure of our protocol included five steps. The first step was searching for studies on the COVID‐19 pandemic. A total of 255 papers were retrieved. The search was carried out in April 2021, and studies regarding the analysis of ML and DL techniques for the COVID‐19 pandemic were included. Three keywords were used in this step, namely, ‘COVID‐19’, ‘machine learning’ and ‘deep learning’ in various combinations and merged with the ‘AND’ operator.
In the second step, duplicate papers were excluded. A total of 55 papers were excluded. In the third step, selected papers were filtered based on titles and abstracts, and papers with topics outside the scope of our domain were excluded. More than 100 articles were excluded, and only 100 papers were selected and passed to the next step. In step four, we skimmed each paper to ensure that its topic is within the defined scope. As a result, 10 articles were excluded. Finally, 90 papers related to ML and DL techniques for the COVID‐19 pandemic were included. A flowchart of the paper selection procedure is shown in Figure 3. The same eligibility criteria were used in the three steps.
FIGURE 3.
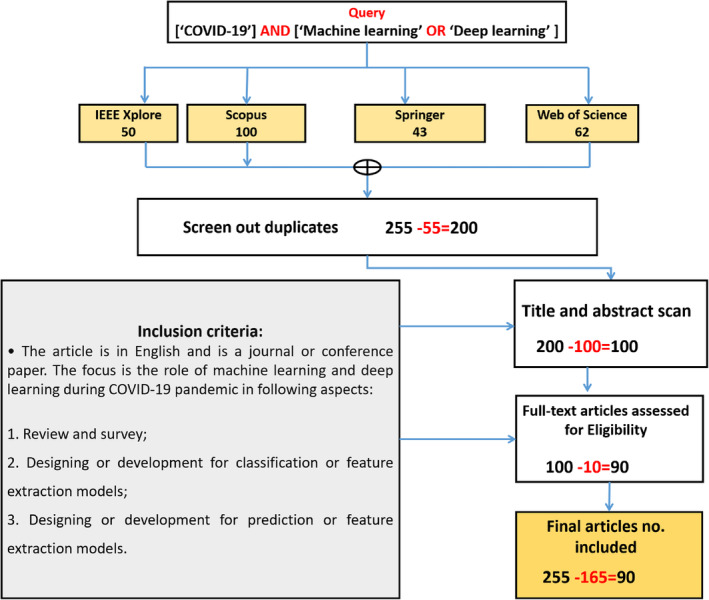
Flowchart of selected studies involving the query and inclusion criteria
3. MACHINE LEARNING AND DEEP LEARNING FOR COVID‐19
Given the widespread infection of the coronavirus, ML and DL have been used in improving the performance of traditional techniques for COVID‐19 detection or prediction. ML approaches for COVID‐19 detection has been used, such as supervised and unsupervised learning, which will be discussed in Section 3.1. Furthermore, several DL approaches have been used for COVID‐19 detection, which will be discussed in Section 3.2. Section 3.3 describes the use of hybrid approaches that combine ML and DL for detection of COVID‐19 infection. Figure 4 presents the categorized AI techniques (ML and DL) selected in this survey paper.
FIGURE 4.
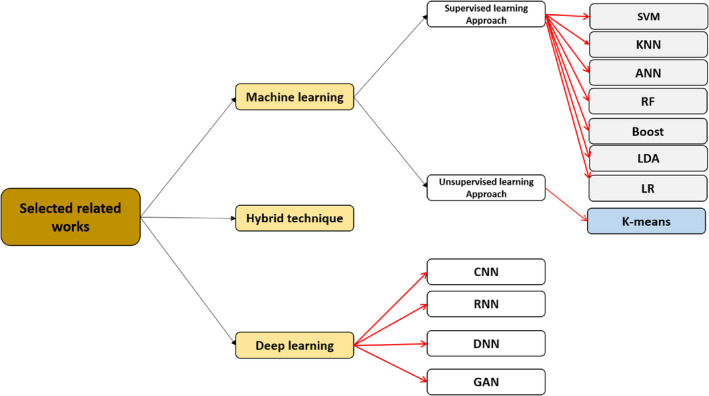
Categorizing of related works
3.1. Machine learning approaches
ML instructs computers on what to do and train them to perform actions independently. It is a data analysis method that involves the development and fitting of models and allows machines to ‘learn’ by practice and make predictions.
ML is used for COVID‐19 detection by analysing the content of the input image of an X‐ray or CT scan and extract unique features from them. According to these features, the prediction of the input image will be provided as a normal case or as an infected case.
In general, ML algorithms that are used for COVID‐19 diagnosis can be categorized into two approaches which are supervised learning approaches describes in Section 3.1.1 and unsupervised learning approaches describes in Section 3.1.2. Figure 5 shows the ML research for COVID‐19.
FIGURE 5.
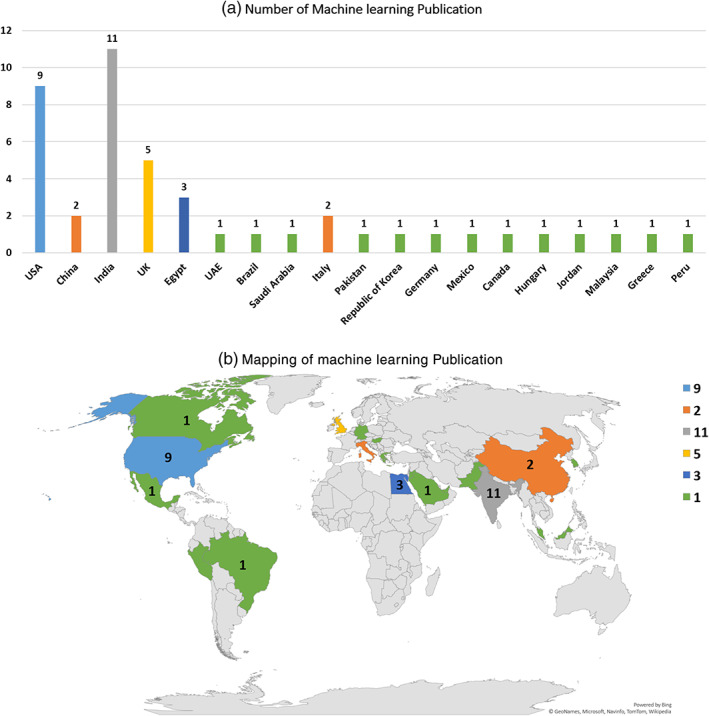
Machine learning research for COVID‐19. (a) Number of machine learning publication. (b) Mapping of machine learning publication
3.1.1. Supervised learning approaches
Many regression, classification and feature extraction methods have been used in the detection of COVID‐19 (Peng & Nagata, 2020). These methods are used to accomplish the following: (i) determination of how the epidemic will end, (ii) prediction of the coronavirus transmission over regions, (iii) analysis of the expansion rate and forms of cure over different countries, (iv) correlation of the effect of weather condition and coronavirus and (v) analysis of the transmission rate of the virus (M. Yadav et al., 2020). Recent studies focused on the use of a logistic forecasting model (Wang, Zheng, et al., 2020), neural network‐based prediction model (Wieczorek et al., 2020), a hybrid ensemble nonlinear autoregressive neural network ensemble model combining neural networks with type‐2 fuzzy and the firefly algorithm (Melin et al., 2020a), prediction of COVID 19 (Kavadi et al., 2020), prediction of respiratory decompensation in Covid‐19 patients with ML (Burdick et al., 2020), development of association rules between weather data and COVID‐19 pandemic for the prediction of death rate (Malki et al., 2020), classification of images with target outputs, such as pneumonia, COVID‐19 and healthy lungs in using Q‐deformed entropy and DL features (Hasan et al., 2020), supervised ML models for predicting COVID‐19 cases (Rustam et al., 2020), analysis of the spatial relationship in the spread of COVID‐19 (Melin et al., 2020b), evaluation of health opinions and online contents relevant to COVID‐19 with ML (Sear et al., 2020) and ML techniques for investigating pandemic COVID 19 effects on young students activities, mental health and learning styles (Khattar et al., 2020).
Siddiqui et al. (2020) investigated the correlation between patient temperature and COVID‐19 case status (i.e., suspected, confirmed and death), using the k‐means algorithm. The methodology encompasses three phases: database design, clustering and data collection. In the first phase, the dataset used is ‘coronavirus disease (COVID‐2019) situation reports’ obtained from the WHO. This dataset involves the infection rate in various regions in China. The second phase provides a description of the dataset, which consists of seven features (i.e., region, population [10,000 s], suspected cases, confirmed cases and death). Two features were added to the dataset, namely, lowest temperature and highest temperature. The reason is that patient temperature is considered one of the main factors for identifying COVID‐19 case status. In the last phase, clustering algorithms based on k‐means is used in finding new trends. The trends demonstrated the impact of temperature on each region in the three COVID‐19 states (i.e., suspected, confirmed and death).
Ong, Wong et al. (2020) analysed the existing COVID‐19 vaccine candidates and then proposed a computational model based on ML and reverse vaccinology (RV) for the prediction of candidate proteins for vaccine design. The main role of RV is to study the bioinformatics of pathogen genomes. Therefore, potential vaccine candidates are identified. The dataset used in this study was collected from the UniProt (Bairoch et al., 2005) and NCBI databases. It consists of SARS‐CoV‐2 sequences and all proteins extracted from known human coronavirus strains. In the present study, the authors relied on Vaxign and Vaxign‐ML (Y. He et al., 2010; Ong, Wang, et al., 2020) to predict proteome biological characteristics and enhanced the Vaxign‐ML model based on RV and ML, using XGBoost, RF, support vector machine (SVM), KNN and logistic regression (LR) methods in predicting the protein levels of all SARS‐CoV‐2 proteins.
Randhawa et al. (2020) used intrinsic COVID‐19 genomic signatures as patterns to establish a decision tree ML alignment‐free approach for predicting gene sequence of the COVID‐19 virus. The alignment‐free approach process only raw DNA sequence data and rapidly deliver the taxonomic classifications of novel pathogens. The proposed methodology was tested on a large dataset comprising more than 5000 unique viral genomic sequences. These data were collected from a well‐known repository called Virus‐Host DB. The results showed that the proposed method is an alternative way for analysing pathogen genome sequences and provide precise taxonomic classifications for unseen sequences in real time.
Pinter et al. (2020) explored the potential of using a hybridisation model of network‐based fuzzy inference system and multi‐layered perceptron‐imperialist competitive algorithm to predict the outbreak of COVID‐19 according to time series data compiled from the Hungary statistical reports of infected cases and death rates. Three metrics were used in evaluating the performance of the proposed prediction model, namely, were mean absolute percentage error, root mean square error (RMSE) and determination coefficient. The proposed prediction model showed promising performance in estimating the total mortality and prediction of the outbreak of COVID‐19.
Elaziz et al. (2020) used CXR images to build a visual diagnostic tool for differentiating between COVID‐19 and normal cases. Initially, on the basis of fractional multichannel exponent moments, features were extracted from CXR images. This process is time consuming and costly; a parallel computational multicore framework was used to boost the computation of images and data processing. In the present study, a new feature selection method was proposed, and a modified manta‐ray foraging optimization algorithm based on differential evolution (MRFODE) was used in identifying relevant subset features from extracted features. Iteratively, MRFODE generates a candidate subset of features, which are evaluated with a KNN approach. Two COVID‐19 X‐ray datasets were used in the study. The first dataset was collected from two resources, which were images published by Joseph Paul Cohenand, Paul Morrison and Lan Dao and images extracted from 43 different publications. These images were classified into two types: normal and viral pneumonia. This dataset encompassed 216 COVID‐19 positive images and 1675 negative COVID‐19 images. The second dataset was collected by a research team composed of members from different countries, including Qatar, Bangladesh, Pakistan and Malaysia (Chowdhury, Rahman, et al., 2020). The research team added new images from the COVID‐19 database. These images were collected by the Interventional Radiology and Italian Society of Medical Radiology. This dataset comprised 219 COVID‐19 positive images and 1341 negative COVID‐19 images. The proposed method (i.e., MRFODE) was evaluated using classification accuracy, recall and precision. The results showed that MRFODE was managed for the purpose of achieving promising classification accuracy in classifying COVID‐19 patients samples.
Fayyoumi et al. (2020) designed an online questionnaire for normal and COVID‐19 cases in Jordan. Data from the questionnaire was used in determining the presence of signs and symptoms in both groups. The researchers created a COVID‐19 dataset of signs and symptoms of different patients. Thereafter, the researchers used this dataset as input to a set of ML models (SVM, multi‐layer perceptron [MLP]) and statistical approaches (i.e., LR) to predict potential patients with COVID‐19. The classification accuracy showed that MLP had the best performance (91.62%). SVM had the best performance (91.67%) in terms of precision.
Kavadi et al. (2020) proposed an alternative approach for preventing the outbreak of COVID‐19 in India. The researchers used the COVID‐19 Indian database. The proposed approach is combining two well‐regarded approaches, which were nonlinear machine learning (NML) and partial derivative regression. PDL was used in normalizing the dataset, and then NML was used for prediction. Experimental results demonstrated that this technique is superior with regard to classification accuracy and prediction time compared with previous works.
Wang, Zheng et al. (2020) exerted considerable efforts in predicting the COVID‐19 trend using logistic modelling method and FbProphet ML algorithm. The dataset used in this study is the most recent COVID‐19 epidemical dataset related to time series data at a country level. Many countries were involved in this study, including Brazil, Russia, India, Peru and Indonesia. In practice, the cap value of the epidemic trend was estimated through logistic modelling. Then, the result was used in creating the learning model of the FbProphet model for the prediction of the epidemic trend of COVID‐19. According to results of the experiment, the proposed model will enable decision‐makers in single country to act rapidly during a COVID‐19 outbreak.
Burdick et al. (2020) proposed a prediction model for ventilation needs among COVID‐19 patients based on XGBoost classifier. XGBoost classifier utilizes a couple of decision trees and aggregates their results into one score. In this study, the dataset is collected from five united states health care systems for the patients who enrolled or were admitted to these five hospitals between 24 March 2020 and 4 May 2020. For each patient, 12 variables are measured, such as diastolic blood pressure (DBP), temperature and blood urea nitrogen (BUN). The proposed model produces a diagnostic ratio for predicting ventilation higher than the Modified Early Warning Score (MEWS). Along with this achievement, the proposed model produced a good compromise between sensitivity and specificity, where it has higher sensitivity (0.90) and higher specificity (p < 0.05) than MEWS.
De Felice and Polimeni (2020) used a MLbibliometric methodology to analyse and evaluate research trends in COVID‐19. Research papers were obtained from Scopus and contained information regarding countries, outputs, journals, institutions, funding, keywords and citation counts. The results showed a significant increment in the number of published documents for the studied period, determined the clinical features of patients with COVID‐19 and defined COVID‐19 as the most common topic.
Samuel et al. (2020) determine public opinion about the pandemic using tweets that mention coronavirus. In addition, the authors presented insights about the relationship between the progress of worry‐sentiment and the time as COVID‐19 reached maximum levels in the United States. These insights were demonstrated by utilizing analytics that explains the text with the aid of data visualization of the text. In the context of textual analytics, two essential ML methods are provided and their effectiveness is compared in categorization of coronavirus tweets with of variable lengths. The results showed high performance of the two ML methods, as they obtained a high classification accuracy for short Tweets. However, the accuracy obtained by the two methods are not promising for longer Tweets.
The ML models' ability to anticipate patients affected by COVID‐19 was demonstrated in Rustam et al. (2020). Four standard models were studied, including least absolute shrinkage and selection operator (LASSO), exponential smoothing (ES), linear regression (LR) and SVM to anticipate the threatening factors of COVID‐19. Each model made three prediction types, such as number of newly infected cases, number of deaths and the number of recoveries in the next 10 days. The current scenario of the COVID‐19 is used to evaluate these methods. The results show the ES model achieves high performance as it achieves the best results in predicting the new confirmed cases when it is compared against other comparable models.
Sedik et al. (2020) presented two data augmentation tactics in order to enhance the learning process for each of the neural network (CNN) and LSTM. Consequently, increasing the accuracy of COVID‐19 detection. The results prove that the proposed strategies improve logarithmic loss, the accuracy of detection and testing time.
Cobb and Seale (2020) studied trends in COVID‐19 growth trends in US counties, considering the existence of shelter‐in‐place (SIP) in these counties. They calculated the rates of compound growth, using cumulative confirmed COVID‐19 cases for a duration of 2 months and 10 days from 21 January 2020 to 31 March 2020. The calculation provided a single number, which was used by the forest ML model that showed the speed of the virus during the studied period. The results showed that SIP orders effectively reduced COVID‐19 cases rate, particularly in large population counties. Figure 6 shows an example of Supervised learning approaches for COVID‐19.
FIGURE 6.
Supervised learning approaches for COVID‐19 (Asnaoui et al., 2020)
3.1.2. Unsupervised learning approaches
In contrast to supervised learning, we have unlabelled data in unsupervised learning (i.e., clustering). The aim is to gather the data into similar clusters without any additional knowledge. In general, the major clustering techniques can be split into two categories: hierarchical clustering and partition clustering. The hierarchical methods can be classified as divisive (top‐down approach) or agglomerative (bottom‐up approach). The divisive clustering starts with all objects in one cluster and then attempt to divide them into smaller clusters until a stopping criterion is satisfied. In contrast, The agglomerative clustering starts by considering each object as an separate cluster and then consolidate them into larger clusters until a stopping criterion is satisfied (Abasi, Khader, Al‐Betar, Naim, Makhadmeh, & Alyasseri, 2020a).
Partition algorithms are intended to classify text documents into separate clusters. Typically, these methods use each cluster‐centroid to attract similar data (Abasi, Khader, Al‐Betar, Naim, & Alyasseri, et al., 2020). The ultimate aim of these methods is to efficiently disperse a massive amount of data into a collection of heterogeneous clusters, each with homogeneous data (Abasi et al., 2021). Figure 7a shows example of agglomerative and divisive hierarchical clustering to a dataset of five clusters, {a, b, c, d, e} and Figure 7b shows structure of partition clustering to a dataset of fourteen clusters.
FIGURE 7.
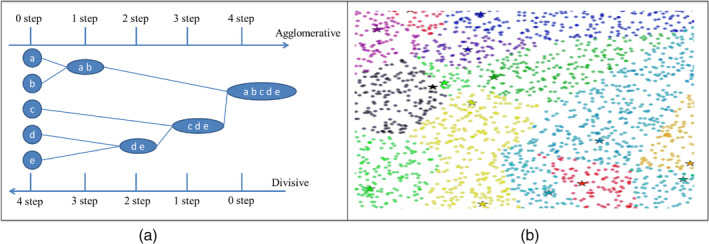
(a) Application of agglomerative and divisive to a dataset of five objects, {a, b, c, d, e} (b) Application of partition clustering to a dataset of fourteen clusters
Partition clustering technique is the most applied in data clustering by researchers because it requires low execution time when compared with hierarchical clustering techniques (Abasi et al., 2019b). In addition, the evaluation results of when apply clustering emphasizes that partition clustering produce the best performance results with different multi‐scale text collections (Abasi, Khader, Al‐Betar, Naim, Makhadmeh, & Alyasseri, 2020b).
A clustering model is proposed in Cui et al. (2020) to identify the latent clusters in COVID‐19 patients. The study was conducted on a dataset of more than six thousand adult patients screened positive with SARS‐CoV‐2 infection at the Mount Sinai Health System in New York, USA. Chronicity and one of the 18 body structures are mapped to patient diagnoses, and the optimum count of clusters was estimated by applying K‐means algorithm and elbow approach. They identified four clusters. Any patients who had positive COVID‐19 scans had respiratory problems. COVID‐19 patients are the most commonly affected with high comorbidity and chronic diseases. Age is also a major factor, but comorbidity and chronicity are strongly associated. Moreover, patients with a history of immune system disorders and metabolic or genitourinary diseases are more vulnerable to severe problems or medical conditions in the circulatory system when they are infected. The discovery of these four clusters is a significant step in identifying the path of the disease and patient treatment and subsequently improving disease prevention.
In order to identify data‐driven countries clusters for predicting COVID‐19 effect, the k‐means are used in Carrillo‐Larco and Castillo‐Cara (2020). It was informed by health system coverage, socio‐economic status, metrics of air pollution and estimated disease prevalence. The researchers compare the clusters in terms of case fatality rate, number of deaths, number of confirmed COVID‐19 cases and the order in which the first case was identified by the country. With 155 countries, the model was developed and used in defining clusters. Three principal component analysis (PCA) parameters are used in the model and five or six clusters achieve the optimal result by combining countries to the related sets. The findings indicate that a model of five or six clusters can stratify countries according to reported numbers of COVID‐19 cases. However, in terms of number of fatalities the model could not stratify countries. Figure 8 shows an example of Deep learning steps for COVID‐19.
FIGURE 8.

Deep learning steps for COVID‐19 (Ozturk et al., 2020)
3.2. Deep learning approaches for COVID‐19
The main DL algorithms used for COVID‐19 are CNN, which is described in Section 3.2.1, DNN, which is described in Section 3.2.2, RNN, which described in Section 3.2.3, GANs which is described in Section 3.2.4, and hybrid approach between ML and DL, which is described in Section 3.3. Figure 9 shows the DL publications for COVID‐19 in different countries.
FIGURE 9.
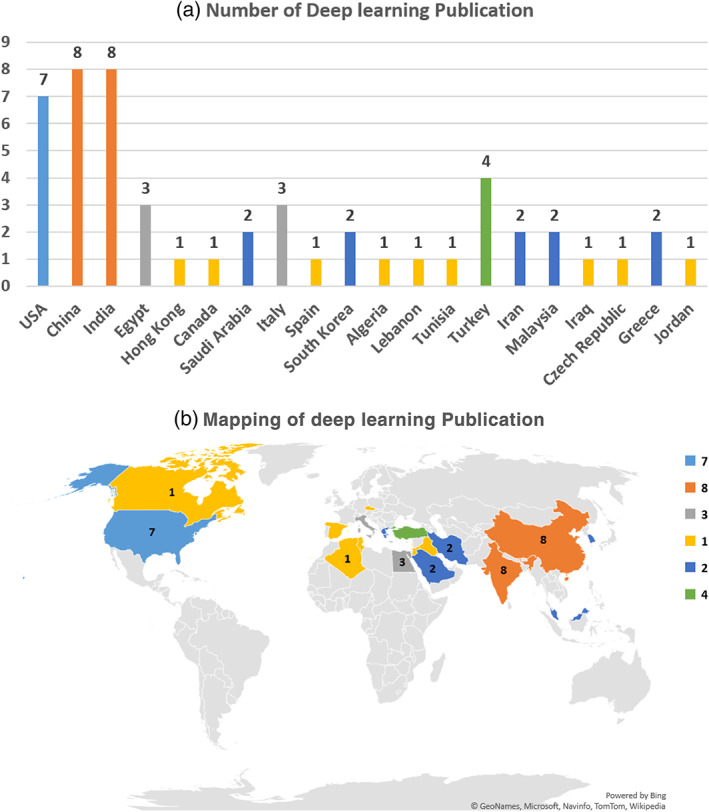
Deep learning publication in different countries for COVID‐19. (a) Number of deep learning publication. (b) Mapping of deep learning publication
The DL algorithms are widely utilized to handle COVID‐19 disease with different perspectives. Rahman et al. (2020) argue that the 5G RAN is incorporated with edge computing. The local DL mechanism was distributed for COVID‐19 edges, and the method used the global DL framework incorporated with three‐ phase reconciliation for the management of the cloud environment. Their proposed DL model supported experts in the COVID‐19 domain in adding semantic to key decision‐making.
Patients with COVID‐19 are also classified with a DL mechanism called DenseNet201 (Jaiswal et al., 2020). This classification method is based on the chest CT scans, and a DL system is pretrained to diagnose coronavirus infection. The ImageNet database was used in assessing the proposed method. The classification model showed competitive results. In another study, the Covid‐19 cases were classified with entropy and DL mechanism, and CT scans were used (Hasan et al., 2020). Initially, CT images were sliced for the reduction of intensity variants. Thereafter, the backgrounds of the CT images were isolated with a histogram thresholding mechanism. Next, features were extracted from each CT lung scan with a Q‐deformed entropy algorithm combined with a DL mechanism. The extracted features were then utilized for classifying CT images with a neural network with LSTM.
COVID‐19 infection was predicted using DL approaches (Alakus & Turkoglu, 2020). Laboratory data and DL were used in determining which patient has COVID‐19 infection. The proposed method was able to recognize patients with COVID‐19, with an accuracy rate of 86.66%.
Three classes can be used for COVID‐19 detection from CXR images, namely, coronavirus, pneumonia and normal (Toğaçar, Ergen, & Cömert, 2020). The fuzzy colour method is used in restructuring data, and structured data are stacked. Then, DL methods (i.e., MobileNetV2 and SqueezeNet) are used with a feature selection method called Social Mimic optimisation. Finally, an SVM is used in classifying efficient features. Their proposed approach was able to reach an accuracy rate of 99.27%. CXR images are used in diagnosing COVID‐19 with patch‐based CNN architectures. This approach provides interpretable saliency maps for COVID‐19 diagnosis and triage of patients. In another study (Yoo et al., 2020), the decision tree classifier with DL was used in diagnosing COVID‐19 infection with CXR images. The proposed model contained three binary decision tree (BDT) classifiers trained based on a CNN model. The first BDT was used to classify a CXR image as either normal or abnormal. Signs of tuberculosis were discovered using a second BDT with abnormal images. COVID‐19 was discovered using the third BDT of abnormal images. The rate of accuracy reached 95%. Similarly, the model in Wang, Liu, et al. (2020) used CXR images and three types of data: normal, viral pneumonia and COVID‐19. The accuracy rate achieved by the model was 96.1%.
CNNs are used in determining the severity of lung disease during COVID‐19 infection (Abdulkareem, Sani, et al., 2021; Zhu et al., 2020). This is achieved through a radiologist score of illness asperity taken from portable CXR images. Their CNN can determine the severity of stage lung disease, prognosticate, and anticipate response to medication. Spatial Transformer Networks which is a kind of neural network is adapted for analysing the lung ultrasonography (LUS) images of different COVID‐19 cases in different stages (Roy et al., 2020). This is done to predict the disease severity score and thus diagnoses the COVID‐19 infection based on LUS images.
The sequence prediction of the DL method is used for monitoring COVID‐19 infection and recovering process (Heni, 2020). This is to measure the impact of Bacille Calmette‐Guerin and infection rates of tuberculosis in such populations. The DL methods used for the COVID‐19 outbreak is provided in Hurt et al. (2020). Chest radiographs obtained from five patients with COVID‐19 treated in the US and China were used.
Deep transfer learning utilizes learned model knowledge to resolve a new similar task with slightest retraining along with Edge Devices (ED) for instance, Intelligent Medical Equipment, Webcam, Drone, IoT, Robot, etc. A survey of such techniques is presented by Sufian et al. (2020) to point out how such infrastructure can help in the automation when there is an outbreak. Such techniques categorise COVID‐19 by utilizing X‐ray images, CT Images and viral genome sequences.
Predicting COVID‐19 is tackled by some researchers using transfer learning in which a model trained on one task is reused into another task after some tuning in to the new task. Minaee et al. (2020) apply transfer learning to figure out COVID‐19 by examining CXR images. Authors trained four CNNs namely, DenseNet‐121, ResNet50, ResNet18 and SqueezeNet. The evaluation results show that most models achieve a sensitivity rate of around 98% and a specificity rate of about 90%. Similarly, Vaid, Kalantar, and Bhandari (2020) use CNNs with the deployment of transfer learning method to enhance results accuracy when recognizing COVID‐19 from CXR scans. The evaluation results are (96.3%) accuracy and loss (0.151 binary cross‐entropy).
In a recent survey, Farhat et al. (2020) summarizes the state‐of‐the‐art methods based on DL and pulmonary medical images. They mentioned that for a better prediction CT scans were preferred more than CXRs.
Lung ultrasound (LUS) imaging is considered a cheaper and safe real‐time imaging technique that can be used in COVID‐19 diagnosis. Roy et al. (2020) develop a fully‐annotated dataset of LUS images. The proposed dataset is used with Spatial Transformer Networks to anticipate the asperity score of the illness. In addition, they introduce a new uninorms‐based approach to provide video‐level score aggregation. The proposed dataset is benchmarked on different DL models for predicting pixel‐level segmentation of COVID‐19 imaging biomarkers.
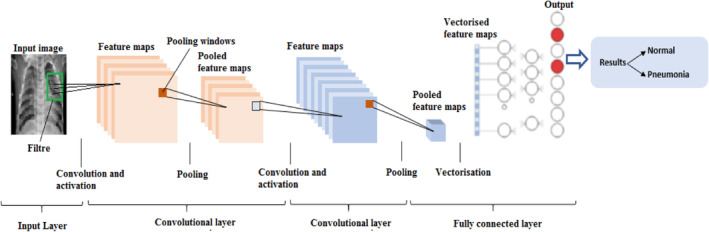
3.2.1. Convolutional neural network
As stated in Albawi et al. (2017), the word ‘deep learning’ applies to multi‐layered AI neural networks (ANN). It has become one of the most effective instruments in the last few decades and is very prominent in literature as it manages an immense quantity of data. The interest in deeper secret layers in various fields, particularly in pattern recognition, recently overcame classical methods' performances. CNNs are the most common DNNs. It is called convolution by that name, a linear mathematical action between matrices . A CNN has many levels, including convolutionary, nonlinear, pooling and completely linked levels. Convolutionary and completely linked layers have parameters that need to be set. In ML problems, a CNN has outstanding results, particularly in applications dealing with image data, such as largest image recognition data collection, computer vision and natural language processing. In this article, all the elements and the main CNN problems were clarified and described, as well as how these elements work. Moreover, the parameters which affect CNN efficiency were defined. This paper assumed that readers recognize computer and the artificial neural network in a fairly strong way.
Alazab et al. (2020) presented an AI technique which utilized a deep CNN to recognize patients with COVID‐19 patients with two real‐world datasets collected from Australia and Jordan. Their algorithm was tested using 1000 X‐ray images of real patients. Experimentally, their technique detected COVID‐19 cases with an accuracy of 95%˘99%. Their technique was also used in predicting the number of patients with COVID‐19 and recovered and deaths cases over the next 7 days with two forecasting methods, namely, autoregressive integrated moving average model and LSTM. Datasets from Australia and Jordan were used for training and testing purposes. The number of confirmed COVID‐19, recovered and death cases in Australia and Jordan were detected, and the average accuracy was 94.80% (Australia) and 88.43% (Jordan).
Image biomarkers from X‐ray images for COVID‐19 can be extracted automatically using deep CNN. Apostolopoulos et al. (2020) used a CNN called Mobile Net to analyse how extracted features are successful in classifying COVID‐19 biomarkers from X‐ray images. The outcome showed a classification accuracy of 87.66% between seven classes of COVID‐19, with 99.18% accuracy in identifying COVID‐19 from non‐COVID‐19 and sensitivity and specificity of 97.36% and 99.42%, respectively. In addition, Civit‐Masot et al. (2020) utilized the VGG16 architecture (Simonyan & Zisserman, 2014) to create a DL model to identify COVID‐19 from X‐ray pulmonary images. The result showed a high sensitivity of around 100% and high specificity. Brunese et al. (2020) presented a DL approach for COVID‐19 recognition from X‐rays. The approach had three steps. First, sign of pneumonia was checked using CXR. Second, COVID‐19 was differentiated from pneumonia. Finally, the location of COVID‐19 in the X‐ray was identified. The obtained results were promising as the time needed for detection was approximately 2.5 s and the average accuracy was 97.
To address the problem of the availability of only a small collection of CXR images for COVID‐19, Oh et al. (2020) introduced a patch‐based CNN that can work by tuning a few parameters. In this method, the classification outcome was obtained by majority voting multiple patch locations with inference results. The method overall accuracy was 91.9% which is close to COVID‐Net (Wang & Wong, 2020) with 92.4%. Sedik et al. (2020) addressed this issue by proposing two data‐augmentation models to improve learning process for each of CNN and the convolutional LSTM in the classification of medical images (X‐ray and CT). The aim was to enhance the accuracy of COVID‐19 detection. The obtained outcome revealed enhancement in terms of logarithmic loss, testing time and detection accuracy, compared with the outcomes of DL models that do not utilize data augmentation.
Islam et al. (2020) developed an automated system for disease identification. The system helped physicians in diagnosing COVID 19 and offered reliable and clear swift outcomes, which showed decrease in mortality risk. This work attempted to present a profound learning strategy focused on LSTM and CNN models to automatically diagnose COVID‐19 according to CXR images. CNN was used in extracting deep functions, and LSTM was used in identifying extracted elements. The machine was fitted with a series of 4575 X‐ray pictures, including 1525 COVID‐19 pictures. The findings of the tests indicated that the proposed method hit 99.4% precision, 99.9% specificity, 99.2% sensitivity, 99.3% and 98.9 F1 score.
An invasive and insightful online research model based on CNN and real‐time data with adaptive algorithms was presented in Farooq and Bazaz (2020). Another difficulty in constantly improving training data is modelling and simulating certain issues by tuning model parameters over time. The key function of the algorithm is the removal of the necessity of redesigning the model when a new collection is obtained, as compared to traditional DL approaches in a constantly evolving training scenario . According to the concept of validation, the authors used it to research the effects of numerous disease response techniques. Finally, the authors suggest and model a strategy for managed natural immunization by means of risk dependent population division (PC), which divides the population into low‐risk (LR) and high‐risk (HR) compartments depending on risk factors (e.g., comorbidities and age).
Panwar et al. (2020) employed CT scans and X‐ray for the lung for COVID‐19 detection. The authors proposed a DL nCOVnet neural network system which can be used in examining the X‐rays of patients as an alternate fast testing tool for detecting COVID‐19. The proposed model achieved a true positive rate of 97% after the use of successive layers of CNN and unbiased sets of data.
Ardakani et al. (2020) stated that rapid diagnostic approaches can be helpful in treating patients in situations that are labour intensive and facilitate the monitoring and prevention of the spread of pandemic illnesses, such as COVID‐19. In this work, 1020 CT images were obtained from 108 COVID‐19 patients and 86 non‐COVID‐19 patients with typical and viral pneumonia. GoogleNet, AlexNet VGG‐16, SqueezeNet, ResNet‐18, MobileNet‐V2, ResNet‐101, Xception and ResNet‐50 were used in differentiating between COVID‐19 infection and non‐COVID‐19 classes in 10 recognized convolutional neural networks. ResNet‐101 and Xception provided the highest results among the networks. ResNet‐101 differentiated COVID‐19 cases with an AUC of 0.994 from non‐COVID‐19 cases with specificity of 99.02% and sensitivity of 100%. A sensitivity of 98.04% and specificity of 100% were attained by the Xception model. However, the radiologist's validation result was consistent with each AUC in terms of sensitivity, specificity and accuracy . They concluded that ResNet101 can be used as an alternative instrument in radiology department and can be used as a high sensitivity model for COVID‐19 infections prediction.
Al‐Waisy et al. (2021) proposed a parallel architecture (COVID‐DeepNet) based on the incorporation of a deep belief network and a convolutional deep belief network trained from scratch with a large‐scale dataset was then integrated. The system accurately diagnosed patients with COVID‐19, with a detection accuracy rate of 99.93%, sensitivity of 99.90%, specificity of 100%, precision of 100%, F1‐score of 99.93%, MSE of 0.021% and RMSE of 0.016%.
Paluru et al. (2021) proposed anamorphic depth embedding‐based lightweight CNN, called Anam‐Net, to segment anomalies in COVID‐19 chest CT images. The number of parameters of the state‐of‐the‐art UNet (or its variants) was 7.8 times that of the proposed Anam‐Net. Thus, Anam‐Net is lightweight and infers mobile or resource constraint (point‐of‐care) platforms. The suitability of Anam‐Net for point‐of‐care platforms was demonstrated by using it in embedded systems, such as Raspberry Pi 4, NVIDIA Jetson Xavier and mobile‐based Android application (CovSeg) embedded with Anam‐Net.
Monshi et al. (2021) proposed a CovidXrayNet model based on EfficientNet‐B0 to optimize the data augmentation and the CNN hyperparameters for detecting COVID‐19 from CXRs in terms of validation accuracy. CovidXrayNet achieves 95.82% on the COVIDx dataset, with only 30 epochs of training. Also, this optimization increases the accuracy of the popular CNN architectures, such as the Visual Geometry Group network (VGG‐19) and the Residual Neural Network (ResNet‐50), by 11.93% and 4.97%, respectively.
Ismael and Şengür (2021) presented deep‐learning‐based approaches, namely deep feature extraction, fine‐tuning of pretrained CNNs and end‐to‐end training of a developed CNN model, classify COVID‐19 and normal (healthy) CXR images. For deep feature extraction, pretrained deep CNN models (ResNet18, ResNet50, ResNet101, VGG16 and VGG19) were used. A dataset containing 180 COVID‐19 and 200 normal (healthy) CXR images was used. The deep features extracted from the ResNet50 model and SVM classifier with a linear kernel function produced a 94.7% accuracy score, which was the highest among the obtained results. The achievement of the fine‐tuned ResNet50 model was 92.6%, whereas end‐to‐end training of the developed CNN model produced a 91.6% result.
3.2.2. Deep neural network
DL strategies have been proposed since 2006 (Liu et al., 2017) for a quick learning algorithm for deep beliefs. Owing to their inherent ability to overcome the downside of traditional neural networks. DL strategies were also identified as suitable for comprehensive computer vision application testing, pattern recognition, speech recognition, reading and recommendation systems in natural language. Some DL architectures and practical configurations were discussed in Liu et al. (2017). Four DL architectures were presented, namely, auto encoders, neural networks, Boltzmann machine and deep belief networks. A clear rationale was provided for the selection of possible DNN models for solving problems in certain cases, such as speech processing, pattern detection and computer vision.
According to Das et al. (2020), 6–9 h are required in checking contamination with RT‐PCR kits. Owing to the lower sensitivity of RT‐PCR, it shows high incorrect results. To address this dilemma, COVID‐19's marker and diagnosis focus on X‐ray and computed tomography (CT) radiology. An automated recognition technique that utilizes deep ML is developed. CXR are analysed directly using DL techniques to improve the algorithm operation. The methods train network weights using large datasets and maximize weight with pretrained networks in small datasets.
In a work by Wang, Zheng et al. (2020) retrospectively 5372 patients from seven cities or provinces with CT images were investigated. Firstly, the DL method was pretrained using CT images from 4106 patients. The images was useful in exploring lung properties. On the basis of the duration of qualified and externally evaluated success of DL framework, 1266 patients (924 with COVID‐19; 471 had follow‐up of more than 5 days) and 342 with pneumonia from six municipalities or provinces were enrolled. The DL method was effective in evaluating COVID‐19 from other pneumonia (AUC 0.87 and 0.88) and viral pneumonia (AUC 0.86) cases in the four previous sets of validation. The patients were stratified with the DL method, and a substantial difference in period spent in the hospital was observed. The DL device positioned itself automatically on the centres of the suspicious areas without human intervention, exhibiting characteristics that are similar to documented findings in radiology. DL offers a simple way of rapid COVID‐19 screening and recognizes possible high‐risk patients, thereby maximizing care services and preventing serious symptoms.
According to Hu et al. (2020), dying from severe alveolar damage and progressive respiratory failure can be attributed to the onset of a serious illness. RT‐PCR is the universal standard for clinical evaluation but can trigger false adverse effects. Furthermore, the lack of RT‐PCR services for testing may postpone the resulting clinical decision and care in a pandemic situation. Chest visualization in CT is a powerful testing and forecasting method for patients with COVID‐19. This work proposed the recognition and characterization of COVID‐19 infections with CT images through a weakly controlled profound learning technique. The technique may help in decreasing manual markings in CT images and distinguishing COVID‐19 instances from non‐COVID‐19 instances. Based on the positive findings, the authors claimed that their work can predict the large‐scale application of proven technologies in clinical trials. To comply with shifts in size and place of lesions, the authors suggested a multi‐scale learning system. They fed intermediate CNN representations (i.e., Conv3, Conv4 and Conv5) with function charts and classification layers which are used with 1X1 convolution. The results obtained with the proposed technique was compared with the results obtained by radiologists (0.994 vs. 0.873).
The optimisation algorithm salp swarm algorithm has been used with DL for the gene selection of COVID‐19 (Altan & Karasu, 2020), and methods that reduces death rate has attracted considerable interest (Farooq & Bazaz, 2020). Altan and Karasu (2020) proposed a hybrid method between a chaotic salp swarm algorithm (CSSA) and CNN approach to recognize COVID‐19 pneumonia infection using X‐ray datasets. The main purpose of using CSSA is to optimize the diagnosed coronavirus pneumonia. The efficiency of the proposed hybrid technique has shown a high accuracy rate to spot COVID‐19 disease from X‐ray images.
3.2.3. Recurrent neural network
Predicting newly contaminated and recovered COVID‐19 cases are important in planning resource distribution and updating curfew rules for downturning disease progression. Zeroual et al. (2020) compared five DL methods to predict the number of new COVID‐19 cases and recovered COVID‐19 cases within 17 days. The compared methods were LSTM, simple RNN, gated recurrent units, Variational AutoEncoder (VAE) and bidirectional LSTM (BiL‐STM) algorithms. The study utilized collected data from Spain, Italy, China, France, Australia and USA. The obtained results showed that VAE was superior to the other algorithms in terms of Loss, MAE, RMSE, MAPE, EV and RMSLE.
Clinical survival analysis can be used in forecasting the probability of a clinical outcome. Liang et al. (2020) utilized a DL‐based survival model to forecast the risk level of patients with COVID‐19 in progressing to severe illness by using clinical characteristics obtained at entrance. The proposed model was evaluated using three separate cohorts data from Guangdong, Hubei and Wuhan, China. The developed model was used by an online calculation platform https://aihealthcare.tencent.com/COVID19-Triage_en.html, which was used in determining patient prioritization at admission. The patients with a high risk of developing a serious illness was able to receive needed care as soon as possible.
Arora et al. (2020) applied DL models to anticipate the positive cases of COVID‐19 in India. In their method, the RNN used LSTM variants. The following variants of LSTM were utilized: deep LSTM, convolutional LSTM and bi‐directional LSTM models in predicting positive COVID‐19 cases. Experimentally, the bi‐directional LSTM obtained the best results, whereas and convolutional LSTM had the worst results in terms of prediction errors. In terms of daily and weekly predictions, the bi‐directional LSTM achieved astonishing results, with 3% or less for short‐term prediction.
3.2.4. Generative adversarial networks
In Loey, Smarandache et al. (2020), the authors introduced deep transfer learning and GAN for COVID‐19 cases recognition using CXR images. This algorithm was tested using a dataset with 307 images. These images were classified into four classes as follows: pneumonia virus, COVID‐19, bacterial pneumonia and normal. Three models of deep transfer were used, namely, Alexnet, Googlenet and Restnet18. Three experimental scenarios were proposed for the recognition of COVID‐19 cases. The first scenario included four classes of images, whereas the second scenario included three classes of images. Two classes of images are included in the third scenario. The COVID‐19 images were included in the dataset used in each experimental scenario. Experimentally, the Googlenet as a deep transfer model achieved the best accuracy results in the first experimental scenario. Alexnet as a deep transfer model showed the highest accuracy in the second experimental scenario, and Googlenet obtained the best accuracy in the third experimental scenario.
Jamshidi et al. (2020) utilized DL techniques, such as GANs, intense learning machine and LSTM, to diagnose patients with COVID‐19. They proposed an applied bioinformatics strategy integrating numerous knowledge facets from a range of organized and unstructured data sources into user‐friendly interfaces for doctors and scientists. The key benefit of these AI systems is the increased speed of evaluation and care phase of COVID‐19.
3.3. Hybrid approach combining machine and deep learning for COVID‐19
An iteratively pruned DL model was introduced in Rajaraman, Siegelman, et al. (2020). X‐ray images with pulmonary manifestations of COVID‐19 were used. In this method, a custom CNN and a trained model using ImageNet was used in learning the specific feature representations of COVID‐19. Then, the learned knowledge was used in categorizing patients as COVID‐19‐viral abnormality, normal and bacterial pneumonia cases. Experimentally, the proposed model achieves excellent results, with an accuracy of 99.01% and AUC value of 0.9972.
Rajaraman and Antani (2020) increased the data used for training to detect COVID‐19 with weakly labelled data. The reason was that COVID‐19 has similar characteristics to pulmonary viral pathogens. The training data was enlarged to include the weakly labelled X‐ray pathogens for bacterial or viral pneumonia. The selected images were used for training a CNN algorithm and contrast results against non‐augmented data‐trained model. Six datasets were used for evaluation. Experimentally, the weakly labelled data augmentation was better than the baseline non‐augmentation on training in identifying COVID‐19 manifestations as viral pneumonia.
Similarly, the authors in Zhu et al. (2020) applied a deep‐learning CNN to determine the severity of lung illness of patients with COVID‐19. Their approach was tested using a real‐world dataset with 131 CXRs obtained from 84 patients with COVID‐19 in US hospitals. The dataset was divided. Approximately 80% of the data were used for training, and 20% were used for testing. The correlation analysis and mean square error analysis are utilized to assess the proposed approach. The results were satisfactory in a small dataset, but the authors mentioned that their approach should be tested with a larger dataset. Furthermore, the authors mentioned that their approach may be used in identifying the severity of lung diseases in patients with COVID‐19 and examining illness development and response to treatment.
Z. Li et al. (2020) developed a DL‐based AI system to recognize COVID‐19‐infected lung regions and evaluate illness severity and development with thick‐section chest CT images. Their system was tested using a dataset with 531 CT scans collected from 204 patients with COVID‐19. The simulation results of their system were compared with the patient diagnosis reports and key features obtained from radiology result obtained using the receiver operating characteristic curve and Cohen's kappa. The results showed the ability of their system to segment lung infection regions, diagnosing, and follow‐up treatment for COVID‐19 patients using CT scans . The authors mentioned several limitations of their system: 1) organ movement due to breathing and heart motion may cause wrong diagnosis, 2) their system is tested based on the changes in imaging biomarkers at the whole lung level and 3) their system is tested using CT scans of COVID‐19 patients only.
Another DNN algorithm for recognizing and categorizing COVID‐19 infection from CT images was developed in Hu et al. (2020). Patients without pneumonia and those with community‐acquired pneumonia were categorized as COVID‐19 by the proposed algorithm. This algorithm identified the precise locations of lesions or inflammation regions caused by COVID‐19 and thus facilitated in determining the severity of illness and was useful in triage and treatment. The algorithm is evaluated using 60 instances published in the TCIA dataset. The number of samples used was varied. For instance, 40 were used in the training, 10 for validation and 10 in the testing stage. In the simulation, the algorithm showed promising results in terms of accuracy, precision and area under the receiver operating characteristic curve.
Zhang et al. (2020) employed DL‐based software to detect, localize and quantify COVID‐19 pneumonia. The proposed AI program utilized a 3D CNN combined with V‐Net bottleneck structures. Their method was tested using 2460 images collected from Huoshenshan Hospital in Wuhan, China. Experimentally, their method achieved excellent results, which were useful in disease assessment and formulation of treatment plans.
Rahman, Hossain et al. (2021) studied nine different versions of a DL algorithm used in detecting COVID‐19 phenomena with medical IoT devices to collect raw data. The DL algorithms were tested for the design of adversarial examples for each type and identification of the accountability of these algorithms. The authors pointed out that DL algorithms consider defensive models against adversarial perturbations, which remain vulnerable to adversarial cases.
4. PUBLIC COVID‐19 DATASETS
One of the most challenging tasks in implementing ML and DL approaches is obtaining suitable datasets for the training, testing and evaluation of proposed techniques. On this basis, this section presents the most used recent COVID‐19 datasets, which have different characteristics, sizes and data types, such as CT scan or X‐ray images. Table 1 provides a full demonstration of these datasets and how they can be accessed.
TABLE 1.
Existing public COVID‐19 datasets
| Dataset | Data type | Classification Output | Dataset type | Characteristics | Primary dataset | Secondary dataset | Techniques | Achievement | Available online |
|---|---|---|---|---|---|---|---|---|---|
| World Health Organization (Siddiqui et al., 2020) | Outbreak | Statistical report | ‘Coronavirus disease (COVID‐19) situation reports’ | Rate of infection for temperature in various provinces of China. | ✓ | (Kumar, Arora, et al., 2020; Bandyopadhyay & Dutta, 2020; Alzubaidi et al., 2021; Chaudhary & Singh, 2021; Loey, Smarandache, et al., 2020) | Accuracy Scenario 1: 80.6%, Scenario 2: 85.2%, Scenario 3: 99.9% | WHO. Coronavirus disease (COVID‐2019) situation reports. ‘https://www.who.int/publications/m/item/weekly‐epidemiological‐update—28‐september‐2020, 2020’ | |
| Protegen database (Randhawa et al., 2020) | Laboratory findings | Diagnosis | annotated proteins | Positive samples of 397 bacterial and 178 viral protective antigens (PAgs) and 4979 negative samples. | ✓ | (Ong, Wong, et al., 2020; Ong, Wang, et al., 2020) | ‘https://www.ncbi.nlm.nih.gov/’ | ||
| (NCBI) database (DNA sequence data) (Randhawa et al., 2020) | Laboratory findings | Diagnosis | COVID‐19 virus sequence | 5000 unique viral genomic sequences ( 61.8 million bp) | ✓ | (Kannan et al., 2020; Touati et al., 2021; Derecichei & Atikukke, 2020; Khairkhah et al., 2020; Y.‐F. Mao et al., 2021) | Method 1: 72.7%, Method 2: 68.7%, Method 3: 91.45% | ‘https://sourceforge.net/projects/mldsp‐gui/files/COVID19Dataset/’ | |
| Hungary‐COVID‐Data (Pinter et al., 2020) | Outbreak | Statistical report | Time series data | Statistical analyses of the cases of COVID‐19 and the fatality rate in Hungary | ✓ | (Chandra et al., 2021; Guasti, 2020; Pinter et al., 2020) | ‘https://www.worldometers.info/coronavirus/country/hungary/’ | ||
| First dataset: (pneumonia) database, Second dataset: COVID‐19 images (Elaziz et al., 2020) | Medical image | Diagnosis | chest x‐ray image | Dataset part 1: 216 COVID‐19 and 1,675 normal cases. Dataset part 2: 219 COVID‐19 and 1,341 normal cases. | ✓ | (Kassani et al., 2020; Qi et al., 2020; Barstugan et al., 2020; Afshar et al., 2020; Elaziz et al., 2020; El‐Kenawy et al., 2020; Luján‐García, Yáñez‐Márquez, et al., 2020; Rasheed et al., 2021; Toğaçar, Ergen, Cömert, & Özyurt, 2020) | Accuracy Dataset1: 96.09% Dataset2: 98.09% | Dataset 1: ‘https://github.com/ieee8023/covid‐chestxray‐dataset’. Dataset 2: ‘https://www.sirm.org/category/senza‐categoria/covid‐19/’ | |
| real novel COVID‐19 data (Fayyoumi et al., 2020) | Laboratory findings | Diagnosis | Signs and symptoms of the patients | 64 negative PCR test and 41 Positive PCR test | ✓ | (Bandyopadhyay & Dutta, 2020; Albahri et al., 2020; Fayyoumi et al., 2020; Chakraborty & Ghosh, 2020; Loey, Manogaran, et al., 2020) | Accuracy: 91.67% | unavailable | |
| Indian COVID‐19 dataset (Kavadi et al., 2020) | Outbreak | Statistical report | Time series data | Statistical reports of COVID‐19 cases in India | ✓ | (Prakash et al., 2020; Sujatha et al., 2020; Shastri et al., 2020; Sujath et al., 2020; R. S. Yadav, 2020) | Case 1: 97.82%, Case 2: 98%, Case 3: 96.66% Case 4: 97.50% | ‘https://www.kaggle.com/sudalairajkumar/covid19‐in‐india’ | |
| COVID‐19 epidemiological data (Wang, Zhengm et al., 2020) | Outbreak | Statistical report | Time series data | COVID‐19 infected cases in numerous countries ( Brazil, Russia, India, Peru and Indonesia) | ✓ | (Punn et al., 2020; Mohamadou et al., 2020; Muhammad et al., 2020; Muhammad et al., 2021; Tuli et al., 2020) | Accuracy: 94.99%, Sensitivity 93.34%, Specificity 94.30% | ‘https://www.who.int/emergencies/diseases/novel‐coronavirus‐2019/situation‐reports’ | |
| U.S. hospitals COVID‐19 admission data (Burdick et al., 2020) | Laboratory findings | Diagnosis | Patient Vital Sign and Lab Measurement | 197 confirmed cases based on 12 factors, for example systolic blood pressure, Blood pressure, and heart rate. | ✓ | (Cheng et al., 2020; Qian et al., 2021; Sáez et al., 2021) | Accuracy: 76.2%, Sensitivity 95%, Specificity 76.3% | ‘https://doi.org/10.1016/j.compbiomed.2020.103949’ | |
| Facebook‐Covid‐19 data (Sear et al., 2020) | Outbreak | Statistical report | COVID‐19 related comments | Data on common Facebook posts from the date 1/17/2020 until 2/28/2020. | ✓ | (Ahmed, Shahbaz, et al., 2020; Raamkumar et al., 2020; Azizan et al., 2020; Cannata et al., 2021) | Sensitivity Case 1: 94.3% and 90.9%, Case 2: 79.6% and 81.5% | ‘https://www.cdc.gov/media/releases/2020/s0229‐COVID‐19‐first‐death.html’ | |
| COVID‐XRay‐5K DATASET (Minaee et al., 2020) | Medical image | Diagnosis | 5000 images | COVID‐19 X‐ray samples and for Non‐COVID samples | ✓ | (Ahammed et al., 2020; Chowdhury, Kabir, et al., 2020; Khan, Sohail, et al., 2020; Nugroho, 2021; Imad et al., 2020; Rezaee et al., 2020; Satu et al., 2021) | Sensitivity rate 98% and specificity rate 90% | ‘https://github.com/shervinmin/DeepCovid.git’ | |
| ChestX‐ray8 database (Wang et al., 2017a) | Medical image | Diagnosis | x‐ray images | totally of 108,948 images which 24,636 include one or more patient images. The rest are 84,312 normal cases images. last Updated 20 July 2020 | ✓ | (Sohan, 2020; Zhao et al., 2020; Crosby et al., 2020; Sathitratanacheewin et al., 2020; Silva et al., 2020; Verma & Tayeb, 2021) | Sensitivity 72%, Specificity 82% | ‘https://nihcc.app.box.com/v/ChestXray‐NIHCC’ | |
| Deep‐Learning‐COVID‐19‐on‐CXR‐using‐Limited‐Training‐Data‐Sets (Oh et al., 2020) | Medical image | Diagnosis | x‐ray images | Public CXR datasets are available JSRT, SCR, NLM(MC), Pneumonia, and COVID‐19 | ✓ | (Cohen et al., 2020) | Accuracy is 91.9 % | ‘https://github.com/jongcye/Deep‐Learning‐COVID‐19‐on‐CXR‐using‐Limited‐Training‐Data‐Sets’ | |
| COVID‐19 and Pneumonia Scans Dataset (Brunese et al., 2020) | Medical image | Diagnosis | x‐ray images | 5887 images | ✓ | (Afshar et al., 2020; Amyar et al., 2020; Harmon et al., 2020; Maghdid et al., 2021; Sharma, 2020) | Area under the ROC curve higher than 97% | ‘https://public.roboflow.com/classification/covid‐19‐and‐pneumonia‐scans’ | |
| Random Sample of NIH Chest X‐ray Dataset (Wang, Peng, et al., 2017b) | Medical image | Diagnosis | x‐ray images | total of 5,606 images and labels are extracted from the dataset of NIH Chest X‐ray | ✓ | (Antin et al., 2017; Basu et al., 2020; Bharati et al., 2020; Filice et al., 2020; Tang et al., 2021) | Accuracy 90.13% | ‘https://www.kaggle.com/nih‐chest‐xrays/sample’ | |
| ICLUS ‐ Italian Covid‐19 Lung Ultra‐sound project (Wang, Peng, et al., 2017b) | Medical video | Diagnosis | Lung ultrasound video | a database of ultrasounds images that can possibly used for identifying patient status in different stages | ✓ | (Roy et al., 2020; Che et al., 2021; Carrer et al., 2020; Dastider et al., 2021) | Accuracy convex: 84%, linear: 94% | ‘https://www.disi.unitn.it/iclus’ | |
| COVID‐CT (Zhao et al., 2020) | Medical image | Diagnosis | CT images | Tongji Hospital, Wuhan, China COVID‐19 data patients between January and April. Total of 349 CT images that included clinical features extracted from 216 patient of COVID‐19 disease. | ✓ | (Yang et al., 2020; Afshar et al., 2020; Al‐Karawi et al., 2020; Roberts et al., 2020; X. He et al., 2020) | Accuracy 95.37%, Sensitivity 95.99%, Specificity 94.76% | ‘https://github.com/UCSD‐AI4H/COVID‐CT’ | |
| PEDIATRIC CXR DATASET (Kermany et al., 2018) | Medical image | Diagnosis | chest X‐ray images | Dataset are collected from Guangzhou Women and Children's Medical Center in Guangzhou which include different classes of medical images such non‐COVID‐19 viral pneumonia, bacterial pneumonia, and normal lungs. | ✓ | (Haghanifar et al., 2020; Longjiang et al., 2019; Rajaraman, Siegelman, et al., 2020; Rajaraman, Sornapudi¸ et al., 2020; Rajaraman and Antani (2020); Siddiqi, 2020) | Accuracy rate 92.8%, sensitivity rate 93.2% and specificity rate 90.1% | ‘https://data.mendeley.com/datasets/rscbjbr9sj/2’ | |
| RSNA CXR DATASET (Shih et al., 2019) | Medical image | Diagnosis | chest X‐ray images | Multi‐expert curated and chest X‐ray images dataset contains samples from the National Institutes of Health (NIH) CXR‐14 | ✓ | (Rahman, Khandakar, et al., 2021; Rajaraman, Siegelman, et al., 2020; Rajaraman, Sornapudi¸ et al., 2020; Rajaraman & Antani, 2020; Zamzmi et al., 2020) | Accuracy rate 93.08%, sensitivity rate 97.53, precision rate 93.15%, and F‐score of 94.57% | ‘https://www.rsna.org/education/ai‐resources‐and‐training/ai‐image‐challenge’ | |
| TWITTER COVID‐19 CXR DATASET | Medical image | Diagnosis | chest X‐ray images | A collection of 134 CXRs with 2K2K pixel resolution in JFIF format that provided by a cardiothoracic radiologist from Spain via Twitter of SARS‐CoV‐2 confirmed cases | ✓ | (Haghanifar et al., 2020; Rajaraman, Siegelman, et al., 2020; Rajaraman, Sornapudi¸ et al., 2020; Rajaraman & Antani, 2020) | Accuracy rate 99.01%, and area under the curve 99.72% | ‘https://twitter.com/ChestImaging’ | |
| MONTREAL COVID‐19 CXR DATASET (Cohen et al., 2020) | Medical image | Diagnosis | chest X‐ray images | A collection of 179 CXRs | ✓ | (Rajaraman, Siegelman, et al., 2020; Rajaraman, Sornapudi¸ et al., 2020; Rajaraman & Antani, 2020) | Accuracy rate 99.01%, and area under curve 99.72% | ‘https://arxiv.org/abs/2003.11597’ | |
| COVID‐19 Chest X‐Rays for Lung Severity Scoring (Cohen et al., 2020) | Medical image | Diagnosis | chest X‐ray images | 131 CXR images that extracted from 84 COVID‐19 cases | ✓ | (Mangal et al., 2020; Hall et al., 2020; Minaee et al., 2020; Mukherjee et al., 2021; Zhu et al., 2020; Rahimzadeh & Attar, 2020), (Ahmed et al., 2021; Apostolopoulos & Mpesiana, 2020; Khan, Shah, et al., 2020; Luján‐García, Moreno‐Ibarra, et al., 2020; Pham, 2021; Tartaglione et al., 2020; Tizhoosh & Fratesi, 2021) | Accuracy rate 96.6%, precision rate 93.17%, recall rate 98.25% and F‐measure rate 95.6% | ‘https://github.com/ieee8023/covid‐chestxray‐dataset’ | |
| TCIA dataset (Hu et al., 2020) | Medical image | Diagnosis | 3D CT lung scans | 60 3D CT images that retrieved based on manual delineations of the lung anatomy | ✓ | (Choi et al., 2020; Dai et al., 2020; Le et al., 2021; Moitra & Mandal, 2019; Suter et al., 2020) | CI:0.501–0.756, iAUC: 0.620; 95% | ‘http://doi.org/10.7937/K9/TCIA.2017.3r3fvz08’ | |
| large‐scale COVID‐19 CX‐R image dataset (Al‐Waisy et al., 2020) | Medical image | Diagnosis | chest X‐ray image | 800 images that include four main classes specifically normal, COVID‐19, pneumonia virus, and pneumonia bacterial. | ✓ | (Al‐Waisy et al., 2020) | RMSE: 0.012%, MSE: 0.011%, specificity: 100%, sensitivity: 99.98%, precision: 100%, F1‐score: 99.99%, and accuracy rate: 99.99% | ‘https://github.com/AlaaSulaiman/COVID19‐vs‐Normal‐dataset’ | |
| The World Mortality Dataset (Karlinsky & Kobak, 2021) | Outbreak | Statistical report | Time series data | data have collected weekly, monthly, or quarterly all‐cause mortality data from 77 countries, openly available as the regularly‐updated World Mortality Dataset | ✓ | (Karlinsky & Kobak, 2021; Ahmad, Ali, et al., 2020; Chowdhury et al., 2021; Elfiky et al., 2018; Malki et al., 2020) | AUC score: 81.38% and accuracy score: 81.30% | ‘https://github.com/akarlinsky/world_mortality’ |
These datasets have been carefully selected from the literature by a well‐designed query in collecting the related works. A combination of keywords involved ML and DL and COVID‐19 diagnosis used in searching for the most relevant papers. The collecting of these papers selected from well‐regarded digital publisher databases (i.e., Elsevier, IEEE Xplore, MDPI, Springer, IOP Press, Wiley, and others). The papers retrieved from the query are subjected to filtering and screening processes to select only the COVID‐19 diagnoses dataset and excluded other datasets such as mask detection.
According to Table 1, authors have made significant achievements by providing different volumes and types of datasets that can significantly support COVID‐19 diagnosis. However, the majority of available datasets are dedicated for diagnosis purposes, especially in terms of medical images. This situation is understandable, given that data acquisition based on medical images can be less complex and time consuming than the process using COVID‐19 RT‐PCR samples. Sample size varies among available datasets as it relies on the number of involved patients. However, most datasets are primarily generated through several examination experiments, and few datasets are collected from other available datasets regarded as secondary datasets (i.e., after processing the original dataset). However, secondary datasets have greatly contributed to the diagnosis of COVID‐19. For instance, large‐scale COVID‐19 CX‐R image datasets have been applied to the CLAHE algorithm (Al‐Waisy et al., 2020) as an enhancement technique for the resolution of medical images in the three primary datasets. The classification rate increased by 8% in the proceed dataset relative to that in raw images.
5. COVID‐19 EVALUATION MEASURES USING MACHINE LEARNING AND DEEP LEARNING
The performance of DL and ML techniques used in detecting COVID‐19 is normally evaluated against several measurements. The most popular measure is classification accuracy. In this section, other measures are used and fully described with mathematical formulation. Table 2 presents the evaluation measures used in evaluating the results of DL and ML methods for COVID‐19 classification. As shown in Table 1, each dataset uses one or more evaluation measures.
TABLE 2.
Evaluation measures for COVID‐19 diagnosis models
| Measures | Equation | References | |
|---|---|---|---|
| Accuracy |
|
(Apostolopoulos et al., 2020; Minaee et al., 2020; Randhawa et al., 2020; Vaid, Kalantar, & Bhandari, 2020; Prakash et al., 2020; Loey, Manogaran, et al., 2020; Fayyoumi et al., 2020; Kavadi et al., 2020; Pinter et al., 2020; Alazab et al., 2020; Paul et al., 2020; Rajaraman, Siegelman, et al., 2020; Altan & Karasu, 2020; Zhu et al., 2020; Rahman, Hossain, et al., 2021; Hu et al., 2020; Rajaraman & Antani, 2020; Loey, Smarandache, et al., 2020; Rahman et al., 2020; Lalmuanawma et al., 2020; Hasan et al., 2020; Jaiswal et al., 2020; Roy et al., 2020; Oh et al., 2020; Pathak et al., 2020; Das et al., 2020; Alakus & Turkoglu, 2020; Hurt et al., 2020; Ni et al., 2020; Toğaçar, Ergen, & Cömert, 2020; Wang, Liu, et al., 2020; Yoo et al., 2020) | |
| Sensitivity |
|
(Fayyoumi et al., 2020; Roy et al., 2020; Pathak et al., 2020; Oh et al., 2020; Das et al., 2020; Alakus & Turkoglu, 2020; Altan & Karasu, 2020; Apostolopoulos et al., 2020; Burdick et al., 2020; Civit‐Masot et al., 2020; Hu et al., 2020; Jaiswal et al., 2020; Loey, Smarandache, et al., 2020; Loey, Manogaran, et al., 2020; Mantas et al., 2020b; Minaee et al., 2020; Ni et al., 2020; Rahman et al., 2020; Rajaraman, Siegelman, et al., 2020; Rajaraman & Antani, 2020; Toğaçar, Ergen, & Cömert, 2020; Yoo et al., 2020) | |
| Specificity |
|
(Fayyoumi et al., 2020; Apostolopoulos et al., 2020; Burdick et al., 2020; Civit‐Masot et al., 2020; Loey, Manogaran, et al., 2020; Minaee et al., 2020; Rajaraman, Siegelman, et al., 2020; Altan & Karasu, 2020; Hu et al., 2020; Rajaraman & Antani, 2020; (Loey, Smarandache, et al., 2020; Rahman et al., 2020; Pathak et al., 2020; Oh et al., 2020; Das et al., 2020; Jaiswal et al., 2020; Ni et al., 2020; Toğaçar, Ergen, & Cömert, 2020; Yoo et al., 2020) | |
| G_Mean |
|
(Fayyoumi et al., 2020) | |
| Precision |
|
(Fayyoumi et al., 2020; Loey, Manogaran, et al., 2020; Mantas et al., 2020b; Rajaraman, Siegelman, et al., 2020; Altan & Karasu, 2020; Mantas et al., 2020b; Oh et al., 2020; Roy et al., 2020; Alakus & Turkoglu, 2020; Hu et al., 2020; Jaiswal et al., 2020; Loey, Smarandache, et al., 2020; Rajaraman & Antani, 2020; Toğaçar, Ergen, & Cömert, 2020; Pathak et al., 2020; Yoo et al., 2020) | |
| F1‐score |
|
(Alazab et al., 2020; Loey, Manogaran, et al., 2020; Roy et al., 2020; Das et al., 2020; Oh et al., 2020; Alakus & Turkoglu, 2020; Altan & Karasu, 2020; Jaiswal et al., 2020; Loey, Smarandache, et al., 2020; Ni et al., 2020; Rajaraman, Siegelman, et al., 2020; Rajaraman & Antani, 2020; Toğaçar, Ergen, & Cömert, 2020) | |
| Root mean square error |
|
(Alazab et al., 2020; Pinter et al., 2020; Zeroual et al., 2020) | |
| Mean absolute percentage error |
|
(Arora et al., 2020; Pinter et al., 2020; Zeroual et al., 2020) | |
| Mean absolute error |
|
(Peng & Nagata, 2020; Zeroual et al., 2020; Heni, 2020; Zhu et al., 2020) | |
| Explained variance |
|
(Zeroual et al., 2020), | |
| Root Mean squared log error |
|
(Zeroual et al., 2020), | |
| Receiver operating characteristic |
|
(Z. Li et al., 2020; Das et al., 2020; Altan & Karasu, 2020; Hu et al., 2020; Ni et al., 2020; Rajaraman, Siegelman, et al., 2020) | |
| Logarithmic loss |
|
(Vaid, Kalantar, & Bhandari, 2020) | |
| Testing the execution time | TET | (Brunese et al., 2020; Kavadi et al., 2020) | |
| Area under curve |
|
(Z. Li et al., 2020; Alakus & Turkoglu, 2020; Burdick et al., 2020; Heni, 2020; Hu et al., 2020; Jaiswal et al., 2020; Rajaraman, Siegelman, et al., 2020; Rajaraman & Antani, 2020; Wang, Zgeng, et al., 2020; Yoo et al., 2020) | |
| Average time for COVID‐19 detection |
|
(Brunese et al., 2020) | |
| Matthews correlation coefficient |
|
(Cole et al., 2020; Rajaraman & Antani, 2020) | |
| Others Evaluation (cluster visualization, p‐value, correlation coefficient) | ‐ | (Vaid, Cakan¸ et al., 2020; Massie et al., 2020; Sear et al., 2020; Siddiqui et al., 2020; Kannan et al., 2020; Ahmad, Garhwal, et al., 2020; Carrillo‐Larco & Castillo‐Cara, 2020; Cui et al., 2020; Jamshidi et al., 2020; Ni et al., 2020; Rajaraman, Siegelman, et al., 2020; Sujath et al., 2020; M. Yadav et al., 2020; Zhang et al., 2020; Zhu et al., 2020) |
Figure 10 shows the measures used in evaluating diagnosis models for COVID‐19. More than 18 evaluation measures are used by state‐of‐the‐art. Clearly, accuracy is the main evaluation criterion used by most researchers, followed by sensitivity.
FIGURE 10.
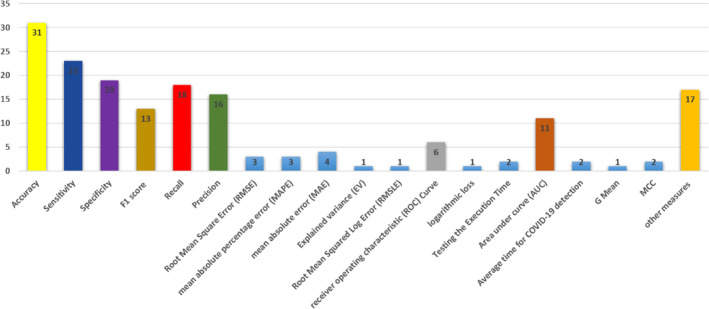
Evaluation measures
Figure 11 shows the classifiers used in evaluating ML and DL methods for COVID‐19. As shown in Figure 11, the SVM classifier is the main technique used by most researchers in the COVID‐19 domain. Boost, K‐means and logistic regression are ranked second according to the percentages of using these classifiers in research papers.
FIGURE 11.

The percentage of using machine learning approaches for COVID‐19
6. CHALLENGES AND FUTURE DIRECTIONS
As aforementioned, the ML and DL methods have been intensively utilized for COVID‐19. Although the ML and DL methods show successful outcomes for their tested COVID19 cases, there are countable challenges that can be considered to enhance the research quality of such direction. Figure 12 shows the percentage of using deep learning approaches for COVID‐19. These challenges can be summarized as follows:
Under usage of ultrasound dataset: Most of the datasets used in the literature are for CT scan and x‐ray, where the ultrasound datasets are rarely developed and used. It may also directly assist in the triage of patients; the first‐look estimation of the disease's severity and the urgency at which a patient needs further care. The utilization of ultrasound for COVID‐19 detection can be beneficial for low and middle‐income countries as the diagnosis through RT‐PCR or CT may not always be available. Interestingly, the usage of ultrasound imaging in real‐time, when combined with DL methods, demonstrates valid results instantly (Roy et al., 2020).
Heterogeneity and size of the datasets: The literature has datasets with different sizes, but the limitation is that the images for COVID‐19 (i.e., not images for a general chest infection) are scarce (Wang, Peng, et al., 2017a; Wang, Peng, et al., 2017b). Such a small size of COVID‐19 images can be an obstacle to obtain accurate results. Another issue related to dataset collection is that they are obtained from limited regions such as Argentina, Brazil, Canada, China, India, Italy, and Japan (Elaziz et al., 2020; Fayyoumi et al., 2020; Minaee et al., 2020; Pinter et al., 2020; Randhawa et al., 2020; Wang, Zhengm et al., 2020). This can reduce the possibility of having results related to other features related to COVID‐19 such as race, region, and food, etc.
Severity level: Most of the datasets used to have two labels for the possible output classes (i.e., infected or not infected) (Jaiswal et al., 2020). However, there is a need to have a dataset that is annotated to show the severity level of the illness to monitor patient healing progress.
End‐to‐end system: There is no complete real‐time system that utilizes the research outcomes in machine learning methods (Jaiswal et al., 2020). The researchers can build a real‐time system using deep learning and machine learning mechanisms to diagnose COVID‐19 virus infection. Such a system can help in checking hospitals, airports, markets, schools, and any other area that is considered a host spot those areas which are the primary hotshots for extending the pandemics.
Effect on different body organs: Most of the studies rely on investigating lung or chest images to predict the infection with COVID‐19. Other organs affected by the virus need to be investigated further as this may ease the virus detection and reduce the risk of being infected and not knowing that. Especially because when the virus reaches the patient's lung is considered a late severe infection stage, and at an earlier stage of infection there are no symptoms on the virus carrier (Bernheim et al., 2020).
FIGURE 12.
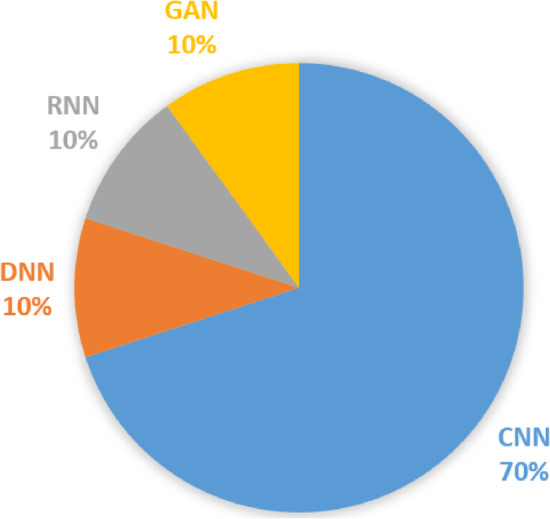
The percentage of using deep learning approaches for COVID‐19
With respect to the previous deep learning and machine learning studies related to COVID‐19, this review paper can be considered as a rich resource that inspires future studies to turn their attentions to study the COVID‐19 concerns in different perspectives such as:
Historical record of the patient: The deep learning and machine learning mechanisms can be tweaked to predict COVID‐19 patients with constraints related to their historical records (e.g., chronic diseases).
Monitoring the COVID‐19 vaccine: Deep learning and machine learning approaches can be used to monitor and observe the effect of the vaccine or other medicine recipes on the patient healing or body chemistry.
The COVID‐19 disease lifespan: Deep learning and machine learning can be used to monitor the development of the COVID‐19 virus in the infected cases using x‐ray, CT scan, or ultrasound images to determine the critical infected cases.
Other deep learning and machine learning mechanisms for COVID‐19: For example, the reinforcement learning approach can be used to improve the efficiency of the deep learning and machine learning approaches.
Utilizing the internet of things (IOT): There is no much work related to employing IOT for COVID‐19 monitoring. The use of a cloud server along with optimized sensor networking can help in achieving such a goal. The system can be used for remote health monitoring or elderly care. Such a system requires more attention to be given to security and privacy issues of the data collected.
Gene expression: The connection between the genes expression of the confirmed COVID‐19 case can be studied to determine the most informative genes related to the infection. The ML and DL techniques can be used to extract these genes and describe their effect on the possibility of being infected by the virus and the severity degree of the infected cases.
Effect of infection on body chemistry: There is a need to have a dataset that provides information related to being infected with COVID‐19 and the changes that happened to the chemistry of the body (e.g., blood group, RNA sequence, oxygen level, etc.). Such data can help in using other means of diagnosing patients which could help in finding other ways to detect the infected cases.
7. CONCLUSION
In this paper, a comprehensive review of deep learning and machine learning techniques used to diagnose and to recognize the COVID‐19 outbreak is conducted. The main purpose of this work is to summarize the previous studies and their applications applied for COVID‐19. The reviewed studies have been selected from several public and highly‐reputed research databases such as IEEE, Springer, Elsevier, MDPI, etc. The selection of these papers passes through several filters to remove the duplicate ideas with relevant content related to COVID‐19 using Machine learning and Deep learning strategies. In summary, India has the most COVID‐19 based machine learning studies, while China has the most COVID‐19 based deep learning studies.
In general, the summarized studies related to COVID‐19 are analyzed and discussed based on three categories, namely, machine learning, deep learning, and hybrid approaches. The studies related to Machine learning are classified into two classes: supervised learning and unsupervised learning. Furthermore, the studies related to the deep learning are categorized into CNN, DNN, RNN, GAN and Optimization with deep learning approaches. All of summarized studies either used for diagnosing the COVID‐19 by means of analyzing the X‐ray images and CT scan images. Furthermore, few studies used either machine learning or deep learning to anticipate the COVID‐19 outbreak in the countries. In total, SVM, LDA, KNN, ANN, Boost, RF, K‐means, and LR as a machine learning algorithm and CNN, DNN, RNN, and GANs as a deep learning algorithm are used for COVID‐19 diagnosis and outbreak prediction. In conclusion, for COVID‐19 diagnosing and outbreak prediction, SVM is the most widely used machine learning mechanism, and CNN is the most widely used deep learning mechanism.
In this review paper, the most recent and public COVID‐19 datasets used by other researchers to evaluate and compare their methods are reported and their accessibility are provided in Table 1.
The evaluation measurements used to diagnose the COVID‐19 based machine learning and deep learning strategies are summarized. Accuracy, sensitivity and specificity are the most widely used measurements in the previous studies. This is inline with the previous studies using the deep learning and machine learning strategies for the prediction process.
Biographies
Zaid Abdi Alkareem Alyasseri is currently working as a Senior Lecturer, in the Center for Artificial Intelligence, Faculty of Information Science and Technology, Universiti Kebangsaan Malaysia (UKM). He received the B.Sc. degree in computer science from Babylon University, Babylon‐Iraq, in 2007, and the M.Sc. degree in computer science from University Science Malaysia (USM), Penang‐Malaysia, in 2013, and the Ph.D. from USM in the field of Artificial Intelligence (Brain‐Inspired Computing) in 2020. He is serving as an associate editor and reviewer in more than 35 high reputation journals with different publishers such as IEEE, Elsevier, Springer, Wiley, Hindawi, etc. His main research interests include Optimization, Pattern recognition, EEG, Brain‐Computer Interface, Signal and Image Processing, Machine and Deep Learning.
Mohammed Azmi Al‐Betar is an associate professor at the Master of Artificial Intelligence (MSAI) program in the College of Engineering and IT at Ajman University and previously was at the Department of Computer Science in Al‐Balqa Applied University, Irbid, Jordan. He is currently the director of the Artificial Intelligence Research Center (AIRC) and a Head of the Evolutionary computation research group which publish more than 175 scientific publications in high quality and well‐reputed journals and conferences. Dr. Al‐Betar ranked in Stanford study of the world's top 2% of scientists. His research interests are mainly directed to metaheuristic optimization methods and hard combinatorial optimisation problems including Scheduling, engineering and Timetabling.
Iyad Abu Doush is an Associate Professor in the department of Computer Science and Information Systems at American University of Kuwait. He obtained his PhD from the Computer Science Department at New Mexico State University / USA in 2009. Dr. Abu Doush completed his B.Sc. in computer science from Yarmouk University, Jordan, and his M.Sc. in Computer Science and Information Systems from Yarmouk University, Jordan. Dr. Abu Doush has supervised, advised and referred senior projects, master theses and number of journals. Dr. Abu Doush served as coach and committee member in the ACM Jordanian Collegiate Programming Contest for three years. Dr. Abu Doush has been funded several times to conduct research in his areas of expertise from different agencies including: USAID, Microsoft, King Abdullah II Design & Development Bureau, Deanship of Research & Graduate Studies at Yarmouk University and Jordanian Scientific Research Support Fund. Dr. Abu Doush has published more than 60 articles in international journals and conferences. Dr. Abu Doush was selected to serve as a visiting researcher in universities of Malaysia and Lithuania. His research interests include: Evolutionary Algorithms, Optimization, Accessibility, and Human Computer Interaction.
Mohammed A. Awadallah is an Associate Professor of artificial intelligence, in the department of computer & information sciences, Al‐aqsa University, Gaza, Palestine. He was appointed as adjunct research associate (ARA) at Artificial Intelligence Research Center (AIRC) ‐ Ajman University, United Arab Emirates since February 2021. The main research interest of Dr. Awadallah is in the area of applied computing where many real‐world complex problems such as nurse rostering, timetabling, economic load dispatch, feature selection, gene selection etc. are being solve using metaheuristic‐based optimization methods like harmony search, artificial bee colony, grey wolf optimizer, bat algorithm, krill herd algorithm and so on. He has published more than 60 research papers in international journals and conferences such as IEEE, Elsevier, Springer, Taylor & Francis's publishers. Furthermore, he serves as a reviewer and editor to numerous conference and journals of high repute such as applied soft computing, information sciences, soft computing, memetic computing, swarm and evolutionary computation etc.
Ammar Kamal Abasi received B. Sc.in computer information system from Jordan University of Science and Technology, and M.Sc. in the international business from the University of Jordan. He obtained his Ph.D. in artificial intelligence & software engineering from Universiti Sains Malaysia, in 2021. His research interests include evolutionary algorithms, nature‐inspired computation, and their applications to optimization problems. His current position is a software engineer and artificial intelligence researcher at BrainTech Malaysia.
Sharif Naser Makhadmeh adjunct research associate (ARA) at Artificial Intelligence Research Center (AIRC) ‐ Ajman University, United Arab Emirates. He received his B.Sc. in Computer Science from Yarmouk University, Jordan, in 2013 and MSc in Information Technology from the Universiti Utara Malaysia (UUM), Malaysia, in 2015. He obtained a PhD degree in Artificial Intelligence from Universiti Sains Malaysia (USM), Malaysia, in 2020. His research interests include Optimization Algorithms, Artificial Intelligence, Power Scheduling Problem, Smart Home.
Osama Ahmad Alomari is an assistant professor in the Dept of Computer Engineering at Istanbul Gelisim University, Turkey. He received his B.Sc. from Al‐al Bayt University, Computer Science, Jordan, in 2005 and MSc in computer science from the National University of Malaysia (UKM) in 2012. He obtained a Ph.D. degree from Universiti Sains Malaysia, Computer Science (Artificial Intelligence), in 2018. In general, his research interests are Optimization, Pattern recognition, Feature selection, Microarray data analysis, Machine, and Deep Learning, and Network security.
Karrar Hameed Abdulkareem received the B.S. degree in computer science (artificial intelligence) from the University of Technology, Iraq, in 2007, and the M.S. degree in computer science (internetworking technology) from the Universiti Teknikal Malaysia Melaka (UTeM), Malaysia, in 2016. The Ph.D. degree in computer science and information technology with the Universiti Tun Hussein Onn Malaysia (UTHM), Malaysia in 2021. His research interests multi‐criteria decision making, image dehazing, and computer security.
Afzan Adam is a senior lecturer in the Center for Artificial Intelligence and Technology, Universiti Kebangsaan Malaysia. Awarded PhD from Leeds University, her research interest mainly in digital pathology, but also applying her machine learning and image processing skills on other domain. She is also a member of Digital Pathology Association and Malaysian Board of Technologist.
Robertas Damaševičius (Member, IEEE) received the Ph.D. degree in informatics engineering from the Kaunas University of Technology, Lithuania, in 2005. He is currently a Professor with the Department of Applied Informatics, Vytautas Magnus University, Lithuania, and an Adjunct Professor with the Faculty of Applied Mathematics, Silesian University of Technology, Poland. He also lectures software maintenance, human–computer interface, and robot programming courses. His research interests include sustainable software engineering, human–computer interfaces, assisted living, data mining, and machine learning. He is the author of more than 300 articles as well as a monograph published by Springer. He is also the Editor‐in‐Chief of the Information Technology and Control Journal. He has been the Guest Editor of several invited issues of international journals, such as BioMed Research International, Computational Intelligence and Neuroscience, Journal of Healthcare Engineering, IEEE Access, and Electronics.
Mazin Abed Mohammed completed his B.Sc. Degree in Computer Science from College of Computer, University of Anbar, Iraq, Master of IT from Universiti Tenaga Nasional Malaysia and Ph.D., from Universiti Teknikal Malaysia Melaka, Malaysia. He is currently working as an Associated professor in department of Information Systems in College of Computer Science and Information Technology, University of Anbar, Ramadi, Iraq. He teaches a variety of university courses in Computer Science, such as, Operating Systems, Database design, Mobile systems Programing, Software Project Management, Web technologies and Software Requirements and design. His areas of research interest include Artificial Intelligence, Medical image processing, machine learning, Computer Vision, computational intelligence, IoT, Biomedical Computing, bio‐informatics, and fog Computing. He has published more than 90 papers International Journals and conferences. Dr. Mohammed outstanding scientific production spans over 90+ contributions published in high standard ISI journals, such as IEEE Access, Future Generation Computer Systems, Medical Informatics, Computers & Electrical Engineering, Computational Science, and Medical Systems.
Raed Abu Zitar has joined SCAI‐SUAD on Feb‐2‐2021. Prior to that date he was working with Ajman University‐UAE from 2018 until 2021, as a Professor, founding Coordinator for the Master of Artificial Intelligence Program (one of the first in UAE), Head of the Center of Artificial Intelligence, and Manager for the Teaching and Learning Center. He has more than 28 years of experience in teaching, research, and administration, starting as an Instructor at the United Arab Emirates University back in 1993. He also was in the position of chairperson and Dean of several colleges and Scientific Research Deanships at several universities. He has taught wide spectrum of computer science, computer engineering, and electrical engineering courses both undergraduate and graduate. He has supervised many Ph.D. and Master Students theses.
Alyasseri, Z. A. A. , Al‐Betar, M. A. , Doush, I. A. , Awadallah, M. A. , Abasi, A. K. , Makhadmeh, S. N. , Alomari, O. A. , Abdulkareem, K. H. , Adam, A. , Damasevicius, R. , Mohammed, M. A. , & Zitar, R. A. (2022). Review on COVID‐19 diagnosis models based on machine learning and deep learning approaches. Expert Systems, 39(3), e12759. 10.1111/exsy.12759
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- Abasi, A. K. , Khader, A. T. , Al‐Betar, M. A. , Naim, S. , Alyasseri, Z. A. A. , & Makhadmeh, S. N. (2020). A novel hybrid multi‐verse optimizer with K‐means for text documents clustering. Neural Computing & Applications, 32, 17703–17729. [Google Scholar]
- Abasi, A. K. , Khader, A. T. , Al‐Betar, M. A. , Naim, S. , Makhadmeh, S. N. , & Alyasseri, Z. A. A. (2021). An improved text feature selection for clustering using binary grey wolf optimizer. In Proceedings of the 11th National Technical Seminar on Unmanned System Technology 2019 (pp. 503–516). Singapore: Springer. [Google Scholar]
- Abasi, A. K. , Khader, A. T. , Al‐Betar, M. A. , Naim, S. , Makhadmeh, S. N. , & Alyasseri, Z. A. A. (2019b). A text feature selection technique based on binary multi‐verse optimizer for text clustering. In 2019 IEEE Jordan International Joint Conference on Electrical Engineering and Information Technology (JEEIT) (pp. 1–6). IEEE. [Google Scholar]
- Abasi, A. K. , Khader, A. T. , Al‐Betar, M. A. , Naim, S. , Makhadmeh, S. N. , & Alyasseri, Z. A. A. (2020a). Link‐based multi‐verse optimizer for text documents clustering. Applied Soft Computing, 87, 106002. 10.1016/j.asoc.2019.106002 [DOI] [Google Scholar]
- Abasi, A. K. , Khader, A. T. , Al‐Betar, M. A. , Naim, S. , Makhadmeh, S. N. , & Alyasseri, Z. A. A. (2020b). A novel ensemble statistical topic extraction method for scientific publications based on optimization clustering. Multimedia Tools and Applications, 80(1), 1–46. [Google Scholar]
- Abdulkareem, A. B. , Sani, N. S. , Shahnorbanun, S. , Alyasseri, Z. A. A. , Afzan, A. , Abdul Hadi, A. R. , & Abdulkarem, B. A. (2021). Predicting COVID‐19 based on environmental factors with machine learning. Intelligent Automation & Soft Computing, 28(2), 305–320. 10.32604/iasc.2021.015413 [DOI] [Google Scholar]
- Abdulkareem, K. H. , Mohammed, M. A. , Salim, A. , Arif, M. , Geman, O. , Gupta, D. , & Khanna, A. (2021). Realizing an effective COVID‐19 diagnosis system based on machine learning and IOT in smart hospital environment. IEEE Internet of Things Journal, 1, 1–1. 10.1109/JIOT.2021.3050775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afshar, P. , Heidarian, S. , Enshaei, N. , Naderkhani, F. , Rafiee, M. J. , Oikonomou, A. , Fard, F. B. , Samimi, K. , Plataniotis, K. N. , & Mohammadi, A. (2020). COVID‐CT‐MD: COVID‐19 computed tomography (CT) scan dataset applicable in machine learning and deep learning. arXiv preprint arXiv:2009.14623 . [DOI] [PMC free article] [PubMed]
- Ahammed, K. , Satu, M. S. , Abedin, M. Z. , Rahaman, M. A. , & Islam, S. M. S . (2020). Early detection of coronavirus cases using chest X‐ray images employing machine learning and deep learning approaches. medRxiv .
- Ahmad, A. , Garhwal, S. , Ray, S. K. , Kumar, G. , Malebary, S. J. , & Barukab, O. M. (2020). The number of confirmed cases of covid‐19 by using machine learning: Methods and challenges. Archives of Computational Methods in Engineering, 28(4), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad, F. S. , Ali, L. , Raza‐Ul‐Mustafa, Khattak, H. A. , Hameed, T. , Wajahat, I. , Kadry, S. , & Bukhari, S. A. C. (2020). A hybrid machine learning framework to predict mortality in paralytic ileus patients using electronic health records (EHRs). Journal of Ambient Intelligence and Humanized Computing, 1(1), 1–11. [Google Scholar]
- Ahmed, N. , Shahbaz, T. , Shamim, A. , Khan, K. S. , Hussain, S. , & Usman, A. (2020). The COVID‐19 infodemic: A quantitative analysis through Facebook. Cureus, 12(11), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed, S. , Hossain, M. F. , & Noor, M. B. T. (2021). Convid‐Net: An enhanced convolutional neural network framework for COVID‐19 detection from X‐ray images. In Proceedings of International Conference on Trends in Computational and Cognitive Engineering (pp. 671–681). Springer. [Google Scholar]
- Akram, T. , Attique, M. , Gul, S. , Shahzad, A. , Altaf, M. , Naqvi, S. S. R. , Damaševičius, R. , & Maskeliūnas, R. (2021). A novel framework for rapid diagnosis of COVID‐19 on computed tomography scans. Pattern Analysis and Applications, 1, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alakus, T. B. , & Turkoglu, I. (2020). Comparison of deep learning approaches to predict COVID‐19 infection. Chaos, Solitons & Fractals, 140, 110120. 10.1016/j.chaos.2020.110120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alazab, M. , Awajan, A. , Mesleh, A. , Abraham, A. , Jatana, V. , & Alhyari, S. (2020). COVID‐19 prediction and detection using deep learning. International Journal of Computer Information Systems and Industrial Management Applications, 12, 168–181. [Google Scholar]
- Albahri, A. , Hamid, R. A. , Alwan, J. K. , Al‐qays, Z. T. , Zaidan, A. A. , Zaidan, B. B. , Albahri, A. O. S. , AlAmoodi, A. H. , Khlaf, J. M. , Almahdi, E. M. , Thabet, E. , Hadi, S. M. , Mohammed, K. I. , Alsalem, M. A. , Al‐Obaidi, J. R. , & Madhloom, H. T. (2020). Role of biological data mining and machine learning techniques in detecting and diagnosing the novel coronavirus (COVID‐19): A systematic review. Journal of Medical Systems, 44(7), 122. 10.1007/s10916-020-01582-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albawi, S. , Mohammed, T. A. , & Al‐Zawi, S. (2017). Understanding of a convolutional neural network. In 2017 International Conference on Engineering and Technology (ICET) (pp. 1–6). IEEE. [Google Scholar]
- Al‐Betar, M. A. , Alyasseri, Z. A. A. , Awadallah, M. A. , & Doush, I. A. (2021). Coronavirus herd immunity optimizer (CHIO). Neural Computing and Applications, 33(10), 5011–5042. 10.1007/s00521-020-05296-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alimadadi, A. , Aryal, S. , Manandhar, I. , Munroe, P. B. , Joe, B. , & Cheng, X. (2020). Artificial intelligence and machine learning to fight COVID‐19. Physiological Genomics, 52(4), 200–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Karawi, D. , Al‐Zaidi, S. , Polus, N. , & Jassim, S. (2020). Machine learning analysis of chest CT scan images as a complementary digital test of coronavirus (COVID‐19) patients. medRxiv .
- Altan, A. , & Karasu, S. (2020). Recognition of COVID‐19 disease from X‐ray images by hybrid model consisting of 2D curvelet transform, chaotic salp swarm algorithm and deep learning technique. Chaos, Solitons & Fractals, 140, 110071. 10.1016/j.chaos.2020.110071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Waisy, A. , Mohammed, M. A. , Al‐Fahdawi, S. , Maashi, M. , Garcia‐Zapirain, B. , Abdulkareem, K. H. , Mostafa, S. , Kumar, N. M. , & Le, D. N. (2021). COVID‐DEEPNet: Hybrid multimodal deep learning system for improving COVID‐19 pneumonia detection in chest X‐ray images. Computers, Materials and Continua, 67(2). 1–10. 10.32604/cmc.2021.012955 [DOI] [Google Scholar]
- Al‐Waisy, A. S. , Al‐Fahdawi, S. , Mohammed, M. A. , Abdulkareem, K. H. , Mostafa, S. A. , Maashi, M. S. , Arif, M. , & Garcia‐Zapirain, B. (2020). COVID‐CheXNet: Hybrid deep learning framework for identifying COVID‐19 virus in chest X‐rays images. Soft Computing, 21(28), 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Alzubaidi, M. A. , Otoom, M. , Otoum, N. , Etoom, Y. , & Banihani, R. (2021). A novel computational method for assigning weights of importance to symptoms of COVID‐19 patients. Artificial Intelligence in Medicine, 112, 102018. 10.1016/j.artmed.2021.102018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amyar, A. , Modzelewski, R. , Li, H. , & Ruan, S. (2020). Multi‐task deep learning based CT imaging analysis for COVID‐19 pneumonia: Classification and segmentation. Computers in Biology and Medicine, 126, 104037. 10.1016/j.compbiomed.2020.104037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antin, B. , Kravitz, J. , & Martayan, E. (2017). Detecting pneumonia in chest X‐rays with supervised learning. Semantic scholar.org. [Google Scholar]
- Apostolopoulos, I. D. , Aznaouridis, S. I. , & Tzani, M. A. (2020). Extracting possibly representative COVID‐19 biomarkers from X‐ray images with Deep Learning approach and image data related to pulmonary diseases. Journal of Medical and Biological Engineering, 40, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolopoulos, I. D. , & Mpesiana, T. A. (2020). Covid‐19: Automatic detection from X‐ray images utilizing transfer learning with convolutional neural networks. Physical and Engineering Sciences in Medicine, 43(2), 635–640. 10.1007/s13246-020-00865-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardakani, A. A. , Kanafi, A. R. , Acharya, U. R. , Khadem, N. , & Mohammadi, A. (2020). Application of deep learning technique to manage COVID‐19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Computers in Biology and Medicine, 121, 103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora, P. , Kumar, H. , & Panigrahi, B. K. (2020). Prediction and analysis of COVID‐19 positive cases using deep learning models: A descriptive case study of India. Chaos, Solitons & Fractals, 139, 110017. 10.1016/j.chaos.2020.110017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asnaoui, K. E. , & Chawki, Y. , & Idri, A. (2020). Automated methods for detection and classification pneumonia based on x‐ray images using deep learning, arXiv preprint arXiv:2003.14363 .
- Azizan, M. , Ismail, H. H. , & Qaiwer, S. N. (2020). Power and solidarity in positive facebook postings amidst COVID‐19 in Malaysia. Journal of Nusantara Studies (JONUS), 5(2), 329–364. 10.24200/jonus.vol5iss2pp329-364 [DOI] [Google Scholar]
- Bairoch, A. , Apweiler, R. , Wu, C. H. , Barker, W. C. , Boeckmann, B. , Ferro, S. , Gasteiger, E. , Huang, H. , Lopez, R. , Magrane, M. , Martin, M. J. , Natale, D. A. , O'Donovan, C. , Redaschi, N. , & Yeh, L.‐S. L. (2005). The universal protein resource (UniProt). Nucleic Acids Research, 33(suppl_1), D154–D159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay, S. K. , & Dutta, S. (2020). Machine learning approach for confirmation of covid‐19 cases: Positive, negative, death and release. medRxiv .
- Barstugan, M. , Ozkaya, U. , & Ozturk, S. (2020). Coronavirus (COVID‐19) classification using CT images by machine learning methods. arXiv preprint arXiv:2003.09424 .
- Basu, S. , Mitra, S. , & Saha, N. (2020). Deep learning for screening COVID‐19 using chest X‐ray images. In 2020 IEEE Symposium Series on Computational Intelligence (SSCI) (pp. 2521–2527). IEEE. [Google Scholar]
- Bernheim, A. , Mei, X. , Huang, M. , Yang, Y. , Fayad, Z. A. , Zhang, N. , Diao, K. , Lin, B. , Zhu, X. , Li, K. , Li, S. , Shan, H. , Jacobi, A. , & Chung, M. (2020). Chest CT findings in coronavirus disease‐19 (COVID‐19): Relationship to duration of infection. Radiology, 200463. 1, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharati, S. , Podder, P. , & Mondal, M. R. H. (2020). Hybrid deep learning for detecting lung diseases from X‐ray images. Informatics in Medicine Unlocked, 20, 100391. 10.1016/j.imu.2020.100391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunese, L. , Mercaldo, F. , Reginelli, A. , & Santone, A. (2020). Explainable deep learning for pulmonary disease and coronavirus COVID‐19 detection from X‐rays. Computer Methods and Programs in Biomedicine, 196, 105608. 10.1016/j.cmpb.2020.105608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick, H. , Lam, C. , Mataraso, S. , Siefkas, A. , Braden, G. , Dellinger, R. P. , McCoy, A. , Vincent, J.‐L. , Green‐Saxena, A. , Barnes, G. , Hoffman, J. , Calvert, J. , Pellegrini, E. , & Das, R. (2020). Prediction of respiratory decompensation in Covid‐19 patients using machine learning: The READY trial. Computers in Biology and Medicine, 124, 103949. 10.1016/j.compbiomed.2020.103949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannata, A. , Cannavò, F. , Di Grazia, G. , Aliotta, M. , Cassisi, C. , De Plaen, R. S. , Gresta, S. , Lecocq, T. , Montalto, P. , & Sciotto, M. (2021). Seismic evidence of the COVID‐19 lockdown measures: A case study from eastern Sicily (Italy). Solid Earth, 12(2), 299–317. 10.5194/se-12-299-2021 [DOI] [Google Scholar]
- Carrer, L. , Donini, E. , Marinelli, D. , Zanetti, M. , Mento, F. , Torri, E. , Smargiassi, A. , Inchingolo, R. , Soldati, G. , Demi, L. , Bovolo, F. , & Bruzzone, L. (2020). Automatic pleural line extraction and COVID‐19 scoring from lung ultrasound data. IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control, 67(11), 2207–2217. 10.1109/TUFFC.2020.3005512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo‐Larco, R. M. , & Castillo‐Cara, M. (2020). Using country‐level variables to classify countries according to the number of confirmed COVID‐19 cases: An unsupervised machine learning approach. Wellcome Open Research, 5, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty, T. , & Ghosh, I. (2020). Real‐time forecasts and risk assessment of novel coronavirus (COVID‐19) cases: A data‐driven analysis. Chaos, Solitons & Fractals, 135, 109850. 10.1016/j.chaos.2020.109850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra, R. , Jain, A. , & Chauhan, D. S. (2021). Deep learning via LSTM models for COVID‐19 infection forecasting in India. arXiv preprint arXiv:2101.11881 . [DOI] [PMC free article] [PubMed]
- Chaudhary, L. , & Singh, B. (2021). Community detection using unsupervised machine learning techniques on COVID‐19 dataset. Social Network Analysis and Mining, 11(1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che, H. , Radbel, J. , Sunderram, J. , Nosher, J. L. , Patel, V. M. , & Hacihaliloglu, I. (2021). Multi‐feature multi‐scale CNN‐derived COVID‐19 classification from lung ultrasound data. arXiv preprint arXiv:2102.11942 . [DOI] [PubMed]
- Cheng, F.‐Y. , Joshi, H. , Tandon, P. , Freeman, R. , Reich, D. L. , Mazumdar, M. , Kohli‐Seth, R. , Levin, M. , Timsina, P. , & Kia, A. (2020). Using machine learning to predict ICU transfer in hospitalized COVID‐19 patients. Journal of Clinical Medicine, 9(6), 1668. 10.3390/jcm9061668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Y. S. , Ahn, S. S. , Chang, J. H. , Kang, S.‐G. , Kim, E. H. , Kim, S. H. , Jain, R. , & Lee, S.‐K. (2020). Machine learning and radiomic phenotyping of lower grade gliomas: Improving survival prediction. European Radiology, 1, 1–9. [DOI] [PubMed] [Google Scholar]
- Chowdhury, M. E. , Rahman, T. , Khandakar, A. , Al‐Madeed, S. , Zughaier, S. M. , Doi, S. A. , Hassen, H. , & Islam, M. T. (2021). An early warning tool for predicting mortality risk of COVID‐19 patients using machine learning. Cognitive Computation, 1, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury, M. E. , Rahman, T. , Khandakar, A. , Mazhar, R. , Kadir, M. A. , Mahbub, Z. B. , Islam, K. R. , Khan, M. S. , Iqbal, A. , Al‐Emadi, N. , Reaz, M. B. I. , & Islam, M. T . (2020). Can AI help in screening viral and covid‐19 pneumonia? arXiv preprint arXiv:2003.13145 .
- Chowdhury, N. K. , Kabir, M. A. , Rahman, M. , & Rezoana, N . (2020). ECOVNet: An ensemble of deep convolutional neural networks based on efficientnet to detect COVID‐19 from chest X‐rays. arXiv preprint arXiv:2009.11850 .
- Chung, M. , Bernheim, A. , Mei, X. , Zhang, N. , Huang, M. , Zeng, X. , Cui, J. , Xu, W. , Yang, Y. , Fayad, Z. A. , Jacobi, A. , Li, K. , Li, S. , & Shan, H. (2020). CT imaging features of 2019 novel coronavirus (2019‐nCoV). Radiology, 295(1), 202–207. 10.1148/radiol.2020200230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civit‐Masot, J. , Luna‐Perejón, F. , Domínguez Morales, M. , & Civit, A. (2020). Deep learning system for COVID‐19 diagnosis aid using X‐ray pulmonary images. Applied Sciences, 10(13), 4640. 10.3390/app10134640 [DOI] [Google Scholar]
- Cobb, J. , & Seale, M. (2020). Examining the effect of social distancing on the compound growth rate of COVID‐19 at the county level (United States) using statistical analyses and a random forest machine learning model. Public Health, 185, 27–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, J. P. , & Morrison, P. , & Dao, L. (2020). COVID‐19 image data collection. arXiv preprint arXiv:2003.11597 .
- Cole, M. A. , Elliott, R. J. , & Liu, B. (2020). The impact of the Wuhan Covid‐19 lockdown on air pollution and health: A machine learning and augmented synthetic control approach. Environmental and Resource Economics, 76(4), 553–580. 10.1007/s10640-020-00483-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman, V. M. , Landt, O. , Kaiser, M. , Molenkamp, R. , Meijer, A. , Chu, D. K. , Bleicker, T. , Brünink, S. , Schneider, J. , Schmidt, M. L. , Mulders, D. G. J. C. , Haagmans, B. L. , van der Veer, B. , van den Brink, S. , Wijsman, L. , Goderski, G. , Romette, J.‐L. , Ellis, J. , Zambon, M. , … Drosten, C. (2020). Detection of 2019 novel coronavirus (2019‐nCoV) by real‐time RT‐PCR. Eurosurveillance, 25(3), 2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby, J. , Chen, S. , Li, F. , MacMahon, H. , & Giger, M. (2020). Network output visualization to uncover limitations of deep learning detection of pneumothorax. In Medical Imaging 2020: Image Perception, Observer Performance, and Technology Assessment (Vol. 11316, 113160O). International Society for Optics and Photonics. [Google Scholar]
- Cucinotta, D. , & Vanelli, M. (2020). Who declares COVID‐19 a pandemic. Acta Biomedica: Atenei Parmensis, 91(1), 157–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, W. , Robins, D. , & Finkelstein, J. (2020). Unsupervised machine learning for the discovery of latent clusters in covid‐19 patients using electronic health records. Studies in Health Technology and Informatics, 272, 1–4. [DOI] [PubMed] [Google Scholar]
- Dai, C. , Wang, S. , Mo, Y. , Zhou, K. , Angelini, E. , Guo, Y. , & Bai, W. (2020). Suggestive annotation of brain tumour images with gradient‐guided sampling. In International Conference on Medical Image Computing and Computer‐Assisted Intervention (pp. 156–165). Springer. [Google Scholar]
- Dansana, D. , Kumar, R. , Bhattacharjee, A. , Hemanth, D. J. , Gupta, D. , Khanna, A. , & Castillo, O. (2020). Early diagnosis of COVID‐19‐affected patients based on X‐ray and computed tomography images using deep learning algorithm. Soft Computing, 1, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Das, N. N. , Kumar, N. , Kaur, M. , Kumar, V. , & Singh, D. (2020). Automated deep transfer learning‐based approach for detection of COVID‐19 infection in chest X‐rays. IRBM, 1, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastider, A. G. , Sadik, F. , & Fattah, S. A. (2021). An integrated autoencoder‐based hybrid CNN‐LSTM model for COVID‐19 severity prediction from lung ultrasound. Computers in Biology and Medicine, 132, 104296. 10.1016/j.compbiomed.2021.104296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Felice, F. , & Polimeni, A. (2020). Coronavirus disease (COVID‐19): A machine learning bibliometric analysis. In Vivo, 34(3 suppl), 1613–1617. 10.21873/invivo.11951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derecichei, I. , & Atikukke, G. (2020). Machine learning model to track SARS‐CoV‐2 viral mutation evolution and speciation using next‐generation sequencing data. In Proceedings of the 11th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics (pp. 1–1). ACM. [Google Scholar]
- Elaziz, M. A. , Hosny, K. M. , Salah, A. , Darwish, M. M. , Lu, S. , & Sahlol, A. T. (2020). New machine learning method for image‐based diagnosis of COVID‐19. PLoS One, 15(6), e0235187. 10.1371/journal.pone.0235187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky, A. A. , Pany, M. J. , Parikh, R. B. , & Obermeyer, Z. (2018). Development and application of a machine learning approach to assess short‐term mortality risk among patients with cancer starting chemotherapy. JAMA Network Open, 1(3), e180926. 10.1001/jamanetworkopen.2018.0926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Kenawy, E.‐S. M. , Ibrahim, A. , Mirjalili, S. , Eid, M. M. , & Hussein, S. E. (2020). Novel feature selection and voting classifier algorithms for COVID‐19 classification in CT images. IEEE Access, 8, 179317. 10.1109/ACCESS.2020.3028012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esbin, M. N. , Whitney, O. N. , Chong, S. , Maurer, A. , Darzacq, X. , & Tjian, R. (2020). Overcoming the bottleneck to widespread testing: A rapid review of nucleic acid testing approaches for COVID‐19 detection. RNA, 26(7), 771–783. 10.1261/rna.076232.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhat, H. , Sakr, G. E. , & Kilany, R. (2020). Deep learning applications in pulmonary medical imaging: Recent updates and insights on COVID‐19. Machine Vision and Applications, 31(6), 1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq, J. , & Bazaz, M. A. (2020). A novel adaptive deep learning model of Covid‐19 with focus on mortality reduction strategies. Chaos, Solitons & Fractals, 138, 110148. 10.1016/j.chaos.2020.110148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayyoumi, E. , Idwan, S. , & AboShindi, H. (2020). Machine learning and statistical modelling for prediction of novel COVID‐19 patients case study: Jordan. International Journal of Advanced Computer Science and Applications, 11(5), 1–6. [Google Scholar]
- Filice, R. W. , Stein, A. , Wu, C. C. , Arteaga, V. A. , Borstelmann, S. , Gaddikeri, R. , Galperin‐Aizenberg, M. , Gill, R. R. , Godoy, M. C. , Hobbs, S. B. , Jeudy, J. , Lakhani, P. C. , Laroia, A. , Nayak, S. M. , Parekh, M. R. , Prasanna, P. , Shah, P. , Vummidi, D. , Yaddanapudi, K. , & Shih, G. (2020). Crowdsourcing pneumothorax annotations using machine learning annotations on the NIH chest X‐ray dataset. Journal of Digital Imaging, 33(2), 490–496. 10.1007/s10278-019-00299-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow, I. , Bengio, Y. , Courville, A. , & Bengio, Y. (2016). Deep learning (Vol. 1). MIT Press. [Google Scholar]
- Gralinski, L. E. , & Menachery, V. D. (2020). Return of the coronavirus: 2019‐nCoV. Viruses, 12(2), 135. 10.3390/v12020135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasti, P. (2020). The impact of the COVID‐19 pandemic in Central and Eastern Europe: The rise of autocracy and democratic resilience. Democratic Theory, 7(2), 47–60. 10.3167/dt.2020.070207 [DOI] [Google Scholar]
- Haghanifar, A. , Majdabadi, M. M. , Choi, Y. , Deivalakshmi, S. , & Ko, S. (2020). COVID‐CXNET: Detecting COVID‐19 in frontal chest x‐ray images using deep learning, arXiv preprint arXiv:2006.13807 . [DOI] [PMC free article] [PubMed]
- Hall, L. O. , Paul, R. , Goldgof, D. B. , & Goldgof, G. M. (2020) Finding covid‐19 from chest X‐rays using deep learning on a small dataset, arXiv preprint arXiv:2004.02060 .
- Harmon, S. A. , Sanford, T. H. , Xu, S. , Turkbey, E. B. , Roth, H. , Xu, Z. , Yang, D. , Myronenko, A. , Anderson, V. , Amalou, A. , Blain, M. , Kassin, M. , Long, D. , Varble, N. , Walker, S. M. , Bagci, U. , Ierardi, A. M. , Stellato, E. , Plensich, G. G. , … Turkbey, B. (2020). Artificial intelligence for the detection of COVID‐19 pneumonia on chest ct using multinational datasets. Nature Communications, 11(1), 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, S. R. , Cartwright, E. J. , Török, M. E. , Holden, M. T. , Brown, N. M. , Ogilvy‐Stuart, A. L. , Ellington, M. J. , Quail, M. A. , Bentley, S. D. , Parkhill, J. , & Peacock, S. J. (2013). Whole‐genome sequencing for analysis of an outbreak of meticillin‐resistant Staphylococcus aureus: A descriptive study. The Lancet Infectious Diseases, 13(2), 130–136. 10.1016/S1473-3099(12)70268-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan, A. M. , AL‐Jawad, M. M. , Jalab, H. A. , Shaiba, H. , Ibrahim, R. W. , & AL‐Shamasneh, A. R. (2020). Classification of Covid‐19 coronavirus, pneumonia and healthy lungs in CT scans using Q‐deformed entropy and deep learning features. Entropy, 22(5), 517. 10.3390/e22050517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X. , Yang, X. , Zhang, S. , Zhao, J. , Zhang, Y. , Xing, E. , & Xie, P . (2020). Sample‐efficient deep learning for COVID‐19 diagnosis based on CT scans. MedRxiv .
- He, Y. , Xiang, Z. , & Mobley, H. L. (2010). Vaxign: The first web‐based vaccine design program for reverse vaccinology and applications for vaccine development. Journal of Biomedicine and Biotechnology, 2010, 1–16. 10.1155/2010/297505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heni, B. (2020). COVID‐19, Bacille Calmette‐Guérin (bcg) and tuberculosis: Cases and recovery previsions with deep learning sequence prediction, 25(2), 165–172. [Google Scholar]
- Hu, S. , Gao, Y. , Niu, Z. , Jiang, Y. , Li, L. , Xiao, X. , Wang, M. , Fang, E. F. , Menpes‐Smith, W. , Xia, J. , Ye, H. , & Yang, G. (2020). Weakly supervised deep learning for COVID‐19 infection detection and classification from CT images. IEEE Access, 8, 118869. 10.1109/ACCESS.2020.3005510 [DOI] [Google Scholar]
- Hurt, B. , Kligerman, S. , & Hsiao, A. (2020). Deep learning localization of pneumonia: 2019 Coronavirus (COVID‐19) outbreak. Journal of Thoracic Imaging, 35(3), W87–W89. 10.1097/RTI.0000000000000512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imad, M. , Khan, N. , Ullah, F. , Hassan, M. A. , Hussain, A. , & Faiza, Faiza . (2020). COVID‐19 classification based on chest X‐ray images using machine learning techniques. Journal of Computer Science and Technology Studies, 1, 1–11. [Google Scholar]
- Islam, M. Z. , Islam, M. M. , & Asraf, A. (2020). A combined deep CNN‐LSTM network for the detection of novel coronavirus (COVID‐19) using X‐ray images. Informatics in Medicine Unlocked, 20, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismael, A. M. , & Şengür, A. (2021). Deep learning approaches for COVID‐19 detection based on chest X‐ray images. Expert Systems with Applications, 164, 114054. 10.1016/j.eswa.2020.114054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal, A. , Gianchandani, N. , Singh, D. , Kumar, V. , & Kaur, M. (2020). Classification of the COVID‐19 infected patients using DenseNet201 based deep transfer learning. Journal of Biomolecular Structure and Dynamics, 1, 1–8. 10.1080/07391102.2020.1788642 [DOI] [PubMed] [Google Scholar]
- Jamshidi, M. , Lalbakhsh, A. , Talla, J. , Peroutka, Z. , Hadjilooei, F. , Lalbakhsh, P. , Jamshidi, M. , La Spada, L. , Mirmozafari, M. , Dehghani, M. , Sabet, A. , Sa, R. , So, R. , Bayat‐Makou, N. , Mohamadzade, B. , Malek, Z. , Jamshidi, A. , Kiani, S. , Hashemi‐Dezaki, H. , & Mohyuddin, W. (2020). Artificial intelligence and COVID‐19: Deep learning approaches for diagnosis and treatment. IEEE Access, 8, 109581. 10.1109/ACCESS.2020.3001973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan, S. , Subbaram, K. , Ali, S. , & Kannan, H. (2020). The role of artificial intelligence and machine learning techniques: Race for COVID‐19 vaccine. Archives of Clinical Infectious Diseases, 15(2), 1–8. 10.5812/archcid.103232 [DOI] [Google Scholar]
- Karlinsky, A. , & Kobak, D. (2021). The world mortality dataset: Tracking excess mortality across countries during the COVID‐19 pandemic, medRxiv . [DOI] [PMC free article] [PubMed]
- Kassani, S. H. , Kassasni, P. H. , Wesolowski, M. J. , Schneider, K. A. , & Deters, R. (2020) Automatic detection of coronavirus disease (COVID‐19) in X‐ray and CT images: A machine learning‐based approach. arXiv preprint arXiv:2004.10641 . [DOI] [PMC free article] [PubMed]
- Kavadi, D. P. , Patan, R. , Ramachandran, M. , & Gandomi, A. H. (2020). Partial derivative Nonlinear Global Pandemic Machine Learning Prediction of COVID 19. Chaos, Solitons & Fractals, 139, 110056. 10.1016/j.chaos.2020.110056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermany, D. S. , Goldbaum, M. , Cai, W. , Valentim, C. C. , Liang, H. , Baxter, S. L. , McKeown, A. , Yang, G. , Wu, X. , Yan, F. , Dong, J. , Prasadha, M. K. , Pei, J. , Ting, M. Y. L. , Zhu, J. , Li, C. , Hewett, S. , Dong, J. , & Zhang, K. (2018). Identifying medical diagnoses and treatable diseases by image‐based deep learning. Cell, 172(5), 1122–1131. 10.1016/j.cell.2018.02.010 [DOI] [PubMed] [Google Scholar]
- Khairkhah, N. , Aghasadeghi, M. R. , Namvar, A. , & Bolhassani, A. (2020). Design of novel multiepitope constructs‐based peptide vaccine against the structural S, N and M proteins of human COVID‐19 using immunoinformatics analysis. PLoS ONE, 15(10), e0240577. 10.1371/journal.pone.0240577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, A. I. , Shah, J. L. , & Bhat, M. M. (2020). CoroNet: A deep neural network for detection and diagnosis of COVID‐19 from chest x‐ray images. Computer Methods and Programs in Biomedicine, 196, 105581. 10.1016/j.cmpb.2020.105581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, S. H. , Sohail, A. , & Khan, A. (2020). COVID‐19 detection in chest X‐ray images using a new channel boosted CNN. arXiv preprint arXiv:2012.05073 . [DOI] [PMC free article] [PubMed]
- Khattar, A. , Jain, P. R. , & Quadri, S. (2020). Effects of the disastrous pandemic COVID 19 on learning styles, activities and mental health of young Indian students ‐ A machine learning approach. In 2020 4th International Conference on Intelligent Computing and Control Systems (ICICCS) (pp. 1190–1195). IEEE. [Google Scholar]
- Kumar, R. , Arora, R. , Bansal, V. , Sahayasheela, V. J. , Buckchash, H. , Imran, J. , Narayanan, N. , Pandian, G. N. , & Raman, B. (2020). Accurate prediction of COVID‐19 using chest X‐ray images through deep feature learning model with smote and machine learning classifiers. MedRxiv .
- Kumar, R. , Nagpal, S. , Kaushik, S. , & Mendiratta, S. (2020). COVID‐19 diagnostic approaches: Different roads to the same destination. VirusDisease, 31(2), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari, P. , Singh, A. , Ngasainao, M. R. , Shakeel, I. , Kumar, S. , Lal, S. , Singhal, A. , Sohal, S. S. , Singh, I. K. , & Hassan, M. I. (2020). Potential diagnostics and therapeutic approaches in COVID‐19. Clinica Chimica Acta, 510, 488–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalmuanawma, S. , Hussain, J. , & Chhakchhuak, L. (2020). Applications of machine learning and artificial intelligence for covid‐19 (SARS‐CoV‐2) pandemic: A review. Chaos, Solitons & Fractals, 139, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, N. Q. K. , Hung, T. N. K. , Do, D. T. , Lam, L. H. T. , Dang, L. H. , & Huynh, T.‐T. (2021). Radiomics‐based machine learning model for efficiently classifying transcriptome subtypes in glioblastoma patients from MRI. Computers in Biology and Medicine, 132, 104320. 10.1016/j.compbiomed.2021.104320 [DOI] [PubMed] [Google Scholar]
- LeCun, Y. , Bengio, Y. , & Hinton, G. (2015). Deep learning. Nature, 521(7553), 436–444. [DOI] [PubMed] [Google Scholar]
- Li G., De Clercq E., Therapeutic options for the 2019 novel coronavirus (2019‐nCoV) (2020). [DOI] [PubMed]
- Li, Z. , Zhong, Z. , Li, Y. , Zhang, T. , Gao, L. , Jin, D. , Sun, Y , Ye, X. , Yu, L. , Hu, Z. , Xiao, J. , Huang, L. , & Tang, Y . (2020). From community acquired pneumonia to COVID‐19: A deep learning based method for quantitative analysis of COVID‐19 on thick‐section CT scans. medRxiv . [DOI] [PMC free article] [PubMed]
- Liang, W. , Yao, J. , Chen, A. , Lv, Q. , Zanin, M. , Liu, J. , Wong, S. , Li, Y. , Lu, J. , Liang, H. , Chen, G. , Guo, H. , Guo, J. , Zhou, R. , Ou, L. , Zhou, N. , Chen, H. , Yang, F. , Han, X. , … He, J. (2020). Early triage of critically ill COVID‐19 patients using deep learning. Nature Communications, 11, 3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. , Wang, Z. , Liu, X. , Zeng, N. , Liu, Y. , & Alsaadi, F. E. (2017). A survey of deep neural network architectures and their applications. Neurocomputing, 234, 11–26. 10.1016/j.neucom.2016.12.038 [DOI] [Google Scholar]
- Loey, M. , Manogaran, G. , Taha, M. H. N. , & Khalifa, N. E. M. (2020). A hybrid deep transfer learning model with machine learning methods for face mask detection in the era of the COVID‐19 pandemic. Measurement, 167, 108288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loey, M. , Smarandache, F. , & Khalifa, N. E. M. (2020). Within the lack of chest COVID‐19 X‐ray dataset: A novel detection model based on gan and deep transfer learning. Symmetry, 12(4), 651. 10.3390/sym12040651 [DOI] [Google Scholar]
- Longjiang, E. , Zhao, B. , Guo, Y. , Zheng, C. , Zhang, M. , Lin, J. , Luo, Y. , Cai, Y. , Song, X. , & Liang, H. (2019). Using deep‐learning techniques for pulmonary‐thoracic segmentations and improvement of pneumonia diagnosis in pediatric chest radiographs. Pediatric Pulmonology, 54(10), 1617–1626. [DOI] [PubMed] [Google Scholar]
- Luján‐García, J. E. , Moreno‐Ibarra, M. A. , Villuendas‐Rey, Y. , & Yáñez‐Márquez, C. (2020). Fast COVID‐19 and pneumonia classification using chest X‐ray images. Mathematics, 8(9), 1423. 10.3390/math8091423 [DOI] [Google Scholar]
- Luján‐García, J. E. , Yáñez‐Márquez, C. , Villuendas‐Rey, Y. , & Camacho‐Nieto, O. (2020). A transfer learning method for pneumonia classification and visualization. Applied Sciences, 10(8), 2908. 10.3390/app10082908 [DOI] [Google Scholar]
- Maghdid, H. S. , Asaad, A. T. , Ghafoor, K. Z. , Sadiq, A. S. , Mirjalili, S. , & Khan, M. K. (2021). Diagnosing COVID‐19 pneumonia from X‐ray and CT images using deep learning and transfer learning algorithms. In Multimodal Image Exploitation and Learning 2021 (Vol. 11734, 117340E). International Society for Optics and Photonics. [Google Scholar]
- Malki, Z. , Atlam, E.‐S. , Hassanien, A. E. , Dagnew, G. , Elhosseini, M. A. , & Gad, I. (2020). Association between weather data and COVID‐19 pandemic predicting mortality rate: Machine learning approaches. Chaos, Solitons & Fractals, 138, 110137. 10.1016/j.chaos.2020.110137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangal, A. , Kalia, S. , Rajgopal, H. , Rangarajan, K. , Namboodiri, V. , Banerjee, S. , & Arora, C. (2020). CovidAID: Covid‐19 detection using chest X‐ray, arXiv preprint arXiv:2004.09803 .
- Mao, K. , Zhang, H. , & Yang, Z. (2020). Can a paper‐based device trace COVID‐19 sources with wastewater‐based epidemiology?. [DOI] [PubMed]
- Mao, Y.‐F. , Yuan, X.‐G. , & Cun, Y.‐P. (2021). A novel machine learning approach (svmSomatic) to distinguish somatic and germline mutations using next‐generation sequencing data. Zoological research, 42(2), 246. 10.24272/j.issn.2095-8137.2021.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massie, A. B. , Boyarsky, B. J. , Werbel, W. A. , Bae, S. , Chow, E. K. , Avery, R. K. , Durand, C. M. , Desai, N. , Brennan, D. , Garonzik‐Wang, J. M. , & Segev, D. L. (2020). Identifying scenarios of benefit or harm from kidney transplantation during the COVID‐19 pandemic: A stochastic simulation and machine learning study. American Journal of Transplantation, 20(11), 2997–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melin, P. , Monica, J. C. , Sanchez, D. , & Castillo, O. (2020a). A new prediction approach of the COVID‐19 virus pandemic behavior with a hybrid ensemble modular nonlinear autoregressive neural network. Soft Computing, 19, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melin, P. , Monica, J. C. , Sanchez, D. , & Castillo, O. (2020b). Analysis of spatial spread relationships of coronavirus (COVID‐19) pandemic in the world using self organizing maps. Chaos, Solitons and Fractals, 138, 109917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michie, D. , Spiegelhalter, D. J. , Taylor, C. , & Campbell, J. (1994). Machine learning, neural and statistical classification (Vol. 13, pp. 1–298).Citeseer. [Google Scholar]
- Minaee, S. , Kafieh, R. , Sonka, M. , Yazdani, S. , & Jamalipour Soufi, G. (2020). Deep‐COVID: Predicting COVID‐19 from chest X‐ray images using deep transfer learning. Medical Image Analysis, 65, 101794. 10.1016/j.media.2020.101794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamadou, Y. , Halidou, A. , & Kapen, P. T. (2020). A review of mathematical modeling, artificial intelligence and datasets used in the study, prediction and management of COVID‐19. Applied Intelligence, 50(11), 3913–3925. 10.1007/s10489-020-01770-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed, M. A. , Abdulkareem, K. H. , Al‐Waisy, A. S. , Mostafa, S. A. , Al‐Fahdawi, S. , Dinar, A. M. , Alhakami, W. , Baz, A. , Al‐Mhiqani, M. N. , Alhakami, H. , Arbaiy, N. , Maashi, M. S. , Mutlag, A. A. , & Díez, B. G.‐Z. I. D. L. T. D. L. T. (2020). Benchmarking methodology for selection of optimal covid‐19 diagnostic model based on entropy and topsis methods. IEEE Access, 8, 99115–99131. [Google Scholar]
- Mohammed, M. A. , Abdulkareem, K. H. , Garcia‐Zapirain, B. , Mostafa, S. A. , Maashi, M. S. , Al‐Waisy, A. S. , Subhi, M. A. , Mutlag, A. A. , & Le, D.‐N. (2021). A comprehensive investigation of machine learning feature extraction and classification methods for automated diagnosis of COVID‐19 based on X‐ray images. Computers, Materials and Continua, 66, 3289–3310. 10.32604/cmc.2021.012874 [DOI] [Google Scholar]
- Moitra, D. , & Mandal, R. K. (2019). Automated AJCC (7th edition) staging of non‐small cell lung cancer (NSCLC) using deep convolutional neural network (CNN) and recurrent neural network (RNN). Health Information Science and Systems, 7, 1, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshi, M. M. A. , Poon, J. , Chung, V. , & Monshi, F. M. (2021). CovidXrayNet: Optimizing data augmentation and CNN hyperparameters for improved COVID‐19 detection from CXR. Computers in Biology and Medicine, 133, 104375. 10.1016/j.compbiomed.2021.104375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammad, L. , Algehyne, E. A. , Usman, S. S. , Ahmad, A. , Chakraborty, C. , & Mohammed, I. (2021). Supervised machine learning models for prediction of COVID‐19 infection using epidemiology dataset. SN computer science, 2(1), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammad, L. , Islam, M. M. , Usman, S. S. , & Ayon, S. I. (2020). Predictive data mining models for novel coronavirus (COVID‐19) infected patients' recovery. SN Computer Science, 1(4), 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, H. , Ghosh, S. , Dhar, A. , Obaidullah, S. M. , Santosh, K. , & Roy, K. (2021). Shallow convolutional neural network for COVID‐19 outbreak screening using chest X‐rays. Cognitive Computation, 1, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, Q. , Sun, Z. Y. , Qi, L. , Chen, W. , Yang, Y. , Wang, L. , Zhang, X. , Yang, L. , Fang, Y. , Xing, Z. , Zhou, Z. , Yu, Y. , Lu, G. M. , & Zhang, L. J. (2020). A deep learning approach to characterize 2019 coronavirus disease (COVID‐19) pneumonia in chest CT images. European Radiology, 30(12), 6517–6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugroho, C. A. (2021). Klasifikasi k‐nearest neighbor chest X‐ray pasien Covid‐19 dengan haralick features dan histogram of oriented gradient
- Oh, Y. , Park, S. , & Ye, J. C. (2020). Deep learning COVID‐19 features on CXR using limited training data sets. IEEE Transactions on Medical Imaging, 39(8), 2688–2700. [DOI] [PubMed] [Google Scholar]
- Ong, E. , Wang, H. , Wong, M. U. , Seetharaman, M. , Valdez, N. , & He, Y. (2020). Vaxign‐ml: Supervised machine learning reverse vaccinology model for improved prediction of bacterial protective antigens. Bioinformatics, 36(10), 3185–3191. 10.1093/bioinformatics/btaa119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong, E. , Wong, M. U. , Huffman, A. , & He, Y. (2020). Covid‐19 coronavirus vaccine design using reverse vaccinology and machine learning. BioRxiv . [DOI] [PMC free article] [PubMed]
- Ozturk, T. , Talo, M. , Yildirim, E. A. , Baloglu, U. B. , Yildirim, O. , & Acharya, U. R. (2020). Automated detection of COVID‐19 cases using deep neural networks with X‐ray images. Computers in Biology and Medicine, 121, 103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluru, N. , Dayal, A. , Jenssen, H. B. , Sakinis, T. , Cenkeramaddi, L. R. , Prakash, J. , & Yalavarthy, P. K. (2021). Anam‐net: Anamorphic depth embedding‐based lightweight CNN for segmentation of anomalies in COVID‐19 chest CT images. IEEE Transactions on Neural Networks and Learning Systems, 32(3), 932–946. 10.1109/TNNLS.2021.3054746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, F. , Ye, T. , Sun, P. , Gui, S. , Liang, B. , Li, L. , Zheng, D. , Wang, J. , Hesketh, R. L. , Yang, L. , & Zheng, C. (2020). Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID‐19) pneumonia. Radiology, 295, 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Y. , Guan, H. , Zhou, S. , Wang, Y. , Li, Q. , Zhu, T. , Hu, Q. , & Xia, L. (2020). Initial CT findings and temporal changes in patients with the novel coronavirus pneumonia (2019‐nCoV): A study of 63 patients in Wuhan, China. European Radiology, 30(6), 3306–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panwar, H. , Gupta, P. , Siddiqui, M. K. , Morales‐Menendez, R. , & Singh, V. (2020). Application of deep learning for fast detection of COVID‐19 in X‐Rays using nCOVnet. Chaos, Solitons & Fractals, 138, 109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak, Y. , Shukla, P. K. , Tiwari, A. , Stalin, S. , Singh, S. , & Shukla, P. K. (2020). Deep transfer learning based classification model for COVID‐19 disease. IRBM, 1, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, H. Y. , Kim, T. K. , & Lin, C. T. (2020). Generalizability of deep learning tuberculosis classifier to COVID‐19 chest radiographs: New tricks for an old algorithm? Journal of Thoracic Imaging, 35(4), W102–W104. [DOI] [PubMed] [Google Scholar]
- Peng, Y. , & Nagata, M. H. (2020). An empirical overview of nonlinearity and overfitting in machine learning using COVID‐19 data. Chaos, Solitons & Fractals, 139, 110055. 10.1016/j.chaos.2020.110055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham, T. D. (2021). Classification of COVID‐19 chest X‐rays with deep learning: New models or fine tuning? Health Information Science and Systems, 9(1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter, G. , Felde, I. , Mosavi, A. , Ghamisi, P. , & Gloaguen, R. (2020). COVID‐19 pandemic prediction for hungary; a hybrid machine learning approach. Mathematics, 8(6), 890. 10.3390/math8060890 [DOI] [Google Scholar]
- Prakash, K. B. , Imambi, S. S. , Ismail, M. , Kumar, T. P. , & Pawan, Y. N. (2020). Analysis, prediction and evaluation of COVID‐19 datasets using machine learning algorithms. International Journal of Emerging Trends in Engineering Research, 8(5), 2199–2204. [Google Scholar]
- Punn, N. S. , Sonbhadra, S. K. , & Agarwal, S. (2020). COVID‐19 epidemic analysis using machine learning and deep learning algorithms. MedRxiv .
- Qi, X. , Jiang, Z. , Yu, Q. , Shao, C. , Zhang, H. , Yue, H. , Ma, B. , Wang, Y. , Liu, C. , Meng, X. , Huang, S. , Wang, J. , Xu, D. , Lei, J. , Xie, G. , Huang, H. , Yang, J. , Ji, J. , Pan, H. , … & Ju, S . (2020). Machine learning‐based CT radiomics model for predicting hospital stay in patients with pneumonia associated with SARS‐CoV‐2 infection: A multicenter study. Medrxiv . [DOI] [PMC free article] [PubMed]
- Qian, Z. , Alaa, A. M. , & van der Schaar, M. (2021). CPAS: The UK's national machine learning‐based hospital capacity planning system for COVID‐19. Machine Learning, 110(1), 15–35. 10.1007/s10994-020-05921-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raamkumar, A. S. , Tan, S. G. , & Wee, H. L. (2020). Use of health belief model‐based deep learning classifiers for classifying COVID‐19 social media content to examine public behaviors towards physical distancing. JMIR Public Health and Surveillance, 6(3), e20493. 10.2196/20493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimzadeh, M. , & Attar, A. (2020). A modified deep convolutional neural network for detecting COVID‐19 and pneumonia from chest X‐ray images based on the concatenation of Xception and ResNet50V2. Informatics in Medicine Unlocked, 19, 100360. 10.1016/j.imu.2020.100360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman, A. , Hossain, M. S. , Alrajeh, N. A. , & Alsolami, F. (2021). Adversarial examples–security threats to COVID‐19 deep learning systems in medical IoT devices. IEEE Internet of Things Journal, 8(12), 9603–9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman, M. A. , Hossain, M. S. , Alrajeh, N. A. , & Guizani, N. (2020). B5G and explainable deep learning assisted healthcare vertical at the edge: COVID‐I9 perspective. IEEE Network, 34(4), 98–105. 10.1109/MNET.011.2000353 [DOI] [Google Scholar]
- Rahman, T. , Khandakar, A. , Qiblawey, Y. , Tahir, A. , Kiranyaz, S. , Kashem, S. B. A. , Islam, M. T. , Al Maadeed, S. , Zughaier, S. M. , Khan, M. S. , & Chowdhury, M. E. H. (2021). Exploring the effect of image enhancement techniques on COVID‐19 detection using chest X‐ray images. Computers in Biology and Medicine, 132, 104319. 10.1016/j.compbiomed.2021.104319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaraman, S. , & Antani, S. (2020). Weakly labeled data augmentation for deep learning: A study on COVID‐19 detection in chest X‐rays. Diagnostics, 10(6), 358. 10.3390/diagnostics10060358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaraman, S. , Siegelman, J. , Alderson, P. O. , Folio, L. S. , Folio, L. R. , & Antani, S. K. (2020). Iteratively pruned deep learning ensembles for COVID‐19 detection in chest X‐rays. IEEE Access, 8, 115041. 10.1109/ACCESS.2020.3003810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaraman, S. , Sornapudi, S. , Alderson, P. O. , Folio, L. R. , & Antani, S. K. (2020). Analyzing inter‐reader variability affecting deep ensemble learning for COVID‐19 detection in chest radiographs. PLoS One, 15(11), e0242301. 10.1371/journal.pone.0242301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randhawa, G. S. , Soltysiak, M. P. , El Roz, H. , de Souza, C. P. , Hill, K. A. , & Kari, L. (2020). Machine learning using intrinsic genomic signatures for rapid classification of novel pathogens: COVID‐19 case study. PLoS One, 15(4), e0232391. 10.1371/journal.pone.0232391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed, J. , Hameed, A. A. , Djeddi, C. , Jamil, A. , & Al‐Turjman, F. (2021). A machine learning‐based framework for diagnosis of COVID‐19 from chest X‐ray images. Interdisciplinary Sciences: Computational Life Sciences, 13(1), 103–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaee, K. , Badiei, A. , & Meshgini, S. (2020). A hybrid deep transfer learning based approach for COVID‐19 classification in chest X‐ray images. In 2020 27th National and 5th International Iranian Conference on Biomedical Engineering (ICBME) (pp. 234–241). IEEE. [Google Scholar]
- Roberts, M. , Driggs, D. , Thorpe, M. , Gilbey, J. , Yeung, M. , Ursprung, S. , Aviles‐Rivero, A. I. , Etmann, C. , McCague, C. , Beer, L. , Weir‐McCall, J. , Teng, Z. , Rudd, J. H. , Sala, E. , & Schönlieb, C.‐B . (2020). Machine learning for COVID‐19 detection and prognostication using chest radiographs and CT scans: A systematic methodological review, arXiv preprint arXiv:2008.06388 .
- Rothe, C. , Schunk, M. , Sothmann, P. , Bretzel, G. , Froeschl, G. , Wallrauch, C. , Zimmer, T. , Thiel, V. , Janke, C. , Guggemos, W. , Seilmaier, M. , Drosten, C. , Vollmar, P. , Zwirglmaier, K. , Zange, S. , Wölfel, R. , & Hoelscher, M. (2020). Transmission of 2019‐nCoV infection from an asymptomatic contact in Germany. New England Journal of Medicine, 382(10), 970–971. 10.1056/NEJMc2001468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, S. , Menapace, W. , Oei, S. , Luijten, B. , Fini, E. , Saltori, C. , Huijben, I. , Chennakeshava, N. , Mento, F. , Sentelli, A. , Peschiera, E. , Trevisan, R. , Maschietto, G. , Torri, E. , Inchingolo, R. , Smargiassi, A. , Soldati, G. , Rota, P. , Passerini, A. , … Demi, L. (2020). Deep learning for classification and localization of COVID‐19 markers in point‐of‐care lung ultrasound. IEEE Transactions on Medical Imaging, 39(8), 2676–2687. [DOI] [PubMed] [Google Scholar]
- Rustam, F. , Reshi, A. A. , Mehmood, A. , Ullah, S. , On, B. , Aslam, W. , & Choi, G. S. (2020). COVID‐19 future forecasting using supervised machine learning models. IEEE Access, 8, 101489–101499. [Google Scholar]
- Sáez, C. , Romero, N. , Conejero, J. A. , & García‐Gómez, J. M. (2021). Potential limitations in COVID‐19 machine learning due to data source variability: A case study in the nCov2019 dataset. Journal of the American Medical Informatics Association, 28(2), 360–364. 10.1093/jamia/ocaa258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlol, A. T. , Yousri, D. , Ewees, A. A. , Al‐qaness, M. A. A. , Damasevicius, R. , & Elaziz, M. A. (2020). COVID‐19 image classification using deep features and fractional‐order marine predators algorithm. Scientific Reports, 10(1), 15364. 10.1038/s41598-020-71294-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakagianni, A. , Feretzakis, G. , Kalles, D. , Koufopoulou, C. , & Kaldis, V. (2020). Setting up an easy‐to‐use machine learning pipeline for medical decision support: A case study for COVID‐19 diagnosis based on deep learning with CT scans. Studies in Health Technology and Informatics, 272, 13–16. [DOI] [PubMed] [Google Scholar]
- Samuel, J. , Ali, G. , Rahman, M. , Esawi, E. , & Samuel, Y. (2020). COVID‐19 public sentiment insights and machine learning for tweets classification. Information, 11(6), 314. 10.3390/info11060314 [DOI] [Google Scholar]
- Sathitratanacheewin, S. , Sunanta, P. , & Pongpirul, K. (2020). Deep learning for automated classification of tuberculosis‐related chest X‐ray: Dataset distribution shift limits diagnostic performance generalizability. Heliyon, 6(8), e04614. 10.1016/j.heliyon.2020.e04614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satu, M. S. , Ahammed, K. , Abedin, M. Z. , Rahman, M. A. , Islam, S. M. S. , Azad, A. , Alyami, S. A. , & Moni, M. A. (2021). Convolutional neural network model to detect COVID‐19 patients utilizing chest X‐ray images. medRxiv .
- Sear, R. F. , Velasquez, N. , Leahy, R. , Restrepo, N. J. , El Oud, S. , Gabriel, N. , Lupu, Y. , & Johnson, N. F. (2020). Quantifying COVID‐19 content in the online health opinion war using machine learning. IEEE Access, 8, 91886–91893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedik, A. , Iliyasu, A. M. , El‐Rahiem, A. , Abdel Samea, M. E. , Abdel‐Raheem, A. , Hammad, M. , Peng, J. , El‐Samie, E. F. A. , & El‐Latif, A. A. A. (2020). Deploying machine and deep learning models for efficient data‐augmented detection of COVID‐19 infections. Viruses, 12(7), 769. 10.3390/v12070769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, S. (2020). Drawing insights from COVID‐19‐infected patients using CT scan images and machine learning techniques: A study on 200 patients. Environmental Science and Pollution Research, 27(29), 37155–37163. 10.1007/s11356-020-10133-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shastri, S. , Singh, K. , Kumar, S. , Kour, P. , & Mansotra, V. (2020). Time series forecasting of Covid‐19 using deep learning models: India‐USA comparative case study. Chaos, Solitons & Fractals, 140, 110227. 10.1016/j.chaos.2020.110227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H. , Han, X. , & Zheng, C. (2020). Evolution of CT manifestations in a patient recovered from 2019 novel coronavirus (2019‐nCoV) pneumonia in Wuhan, China. Radiology, 295(1), 20–20. 10.1148/radiol.2020200269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih, G. , Wu, C. C. , Halabi, S. S. , Kohli, M. D. , Prevedello, L. M. , Cook, T. S. , Sharma, A. , Amorosa, J. K. , Arteaga, V. , Galperin‐Aizenberg, M. , Gill, R. R. , Godoy, M. C. B. , Hobbs, S. , Jeudy, J. , Laroia, A. , Shah, P. N. , Vummidi, D. , Yaddanapudi, K. , & Stein, A. (2019). Augmenting the National Institutes of Health Chest Radiograph Dataset with expert annotations of possible pneumonia. Radiology: Artificial Intelligence, 1(1), e180041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi, R. (2020). Efficient pediatric pneumonia diagnosis using depthwise separable convolutions. SN Computer Science, 1(6), 1–15. [Google Scholar]
- Siddiqui, M. K. , Morales‐Menendez, R. , Gupta, P. K. , Iqbal, H. , Hussain, F. , Khatoon, K. , & Ahmad, S. (2020). Correlation between temperature and COVID‐19 (suspected, confirmed and death) cases based on machine learning analysis. Journal of Pure and Applied Microbiology, 14, 1017–1024. [Google Scholar]
- Silva, J. , Zilberman, J. , Patiño, Y. P. , Varela, N. , & Lezama, O. B. P. (2020). Classification of chest diseases using deep learning. In International Symposium on Distributed Computing and Artificial Intelligence (pp. 152–158). Springer. [Google Scholar]
- Simonyan, K. , & Zisserman, A. (2014). Very deep convolutional networks for large‐scale image recognition. arXiv preprint arXiv:1409.1556 .
- Sohan, M. F. (2020). So you need datasets for your COVID‐19 detection research using machine learning? arXiv preprint arXiv:2008.05906 .
- Sohrabi, C. , Alsafi, Z. , O'Neill, N. , Khan, M. , Kerwan, A. , Al‐Jabir, A. , Iosifidis, C. , & Agha, R. (2020). World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID‐19). International Journal of Surgery, 76, 71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sufian, A. , Ghosh, A. , Sadiq, A. S. , & Smarandache, F. (2020). A survey on deep transfer learning to edge computing for mitigating the COVID‐19 pandemic. Journal of Systems Architecture, 108, 101830. 10.1016/j.sysarc.2020.101830 [DOI] [Google Scholar]
- Sujath, R. , Chatterjee, J. M. , & Hassanien, A. E. (2020). A machine learning forecasting model for COVID‐19 pandemic in India. Stochastic Environmental Research and Risk Assessment, 34, 959–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujatha, R. , Chatterjee, J. , & Hassanien, A. E . (2020). A machine learning methodology for forecasting of the COVID‐19 cases in India.
- Suter, Y. , Knecht, U. , Alão, M. , Valenzuela, W. , Hewer, E. , Schucht, P. , Wiest, R. , & Reyes, M. (2020). Radiomics for glioblastoma survival analysis in pre‐operative MRI: Exploring feature robustness, class boundaries, and machine learning techniques. Cancer Imaging, 20(1), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, S. , Ghorbani, A. , Yamashita, R. , Rehman, S. , Dunnmon, J. A. , Zou, J. , & Rubin, D. L. (2021). Data valuation for medical imaging using Shapley value and application to a large‐scale chest X‐ray dataset. Scientific Reports, 11(1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglione, E. , Barbano, C. A. , Berzovini, C. , Calandri, M. , & Grangetto, M. (2020). Unveiling COVID‐19 from CHEST X‐ray with deep learning: A hurdles race with small data. International Journal of Environmental Research and Public Health, 17(18), 6933. 10.3390/ijerph17186933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tizhoosh, H. R. , & Fratesi, J. (2021). COVID‐19, AI enthusiasts, and toy datasets: Radiology without radiologists. European radiology, 31(5), 3553–3554. 10.1007/s00330-020-07453-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toğaçar, M. , Ergen, B. , & Cömert, Z. (2020). COVID‐19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X‐ray images using fuzzy color and stacking approaches. Computers in Biology and Medicine, 121, 103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toğaçar, M. , Ergen, B. , Cömert, Z. , & Özyurt, F. (2020). A deep feature learning model for pneumonia detection applying a combination of mRMR feature selection and machine learning models. IRBM, 41(4), 212–222. 10.1016/j.irbm.2019.10.006 [DOI] [Google Scholar]
- Touati, R. , Messaoudi, I. , Oueslati, A. , Lachiri, Z. , & Kharrat, M. (2021). New intraclass helitrons classification using DNA‐image sequences and machine learning approaches. IRBM, 42(3), 154–164. [Google Scholar]
- Tsatsakis, A. , Calina, D. , Falzone, L. , Petrakis, D. , Mitrut, R. , Siokas, V. , Pennisi, M. , Lanza, G. , Libra, M. , Doukas, S. G. , Doukas, P. G. , Kavali, L. , Bukhari, A. , Gadiparthi, C. , Vageli, D. P. , Kofteridis, D. P. , Spandidos, D. A. , Paoliello, M. M. B. , Aschner, M. , & Docea, A. O. (2020). SARS‐CoV‐2 pathophysiology and its clinical implications: An integrative overview of the pharmacotherapeutic management of COVID‐19. Food and Chemical Toxicology, 146, 111769. 10.1016/j.fct.2020.111769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuli, S. , Tuli, S. , Tuli, R. , & Gill, S. S. (2020). Predicting the growth and trend of COVID‐19 pandemic using machine learning and cloud computing. Internet of Things, 11, 100222. 10.1016/j.iot.2020.100222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaid, S. , Cakan, C. , & Bhandari, M. (2020). Using machine learning to estimate unobserved COVID‐19 infections in North America. The Journal of Bone and Joint Surgery, 8, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaid, S. , Kalantar, R. , & Bhandari, M. (2020). Deep learning COVID‐19 detection bias: Accuracy through artificial intelligence. International Orthopaedics, 44, 1539–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma, Y. , & Tayeb, S. (2021). Evaluation of machine learning architectures in healthcare. In 2021 IEEE 11th Annual Computing and Communication Workshop and Conference (CCWC) (pp. 1377–1382). IEEE. [Google Scholar]
- Waheed, A. , Goyal, M. , Gupta, D. , Khanna, A. , Al‐Turjman, F. , & Pinheiro, P. R. (2020). CovidGAN: Data augmentation using auxiliary classifier GAN for improved Covid‐19 detection. IEEE Access, 8, 91916–91923. 10.1109/ACCESS.2020.2994762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , & Wong, A. (2020). COVID‐net: A tailored deep convolutional neural network design for detection of COVID‐19 cases from chest X‐ray images. arXiv preprint arXiv:2003.09871 . [DOI] [PMC free article] [PubMed]
- Wang, N. , Liu, H. , & Xu, C. (2020). Deep learning for the detection of COVID‐19 using transfer learning and model integration. In 2020 IEEE 10th International Conference on Electronics Information and Emergency Communication (ICEIEC) (pp. 281–284). IEEE. [Google Scholar]
- Wang, P. , Zheng, X. , Li, J. , & Zhu, B. (2020). Prediction of epidemic trends in COVID‐19 with logistic model and machine learning technics. Chaos, Solitons & Fractals, 139, 110058. 10.1016/j.chaos.2020.110058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Peng, Y. , Lu, L. , Lu, Z. , Bagheri, M. , & Summers, R. M. (2017a). Chestx‐ray8: Hospital‐scale chest X‐ray database and benchmarks on weakly‐supervised classification and localization of common thorax diseases. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (pp. 2097–2106). IEEE. [Google Scholar]
- Wang, X. , Peng, Y. , Lu, L. , Lu, Z. , Bagheri, M. , & Summers, R. M. (2017b). Chestx‐ray8: Hospital‐scale chest X‐ray database and benchmarks on weakly‐supervised classification and localization of common thorax diseases. CoRR, 1, 2097–2106. [Google Scholar]
- Wieczorek, M. , Siłka, J. , Połap, D. , Woźniak, M. , & Damaševičius, R. (2020). Real‐time neural network based predictor for cov19 virus spread. PLoS ONE, 15(12), e0243189. 10.1371/journal.pone.0243189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav, M. , Perumal, M. , & Srinivas, M. (2020). Analysis on novel coronavirus (COVID‐19) using machine learning methods. Chaos, Solitons & Fractals, 139, 110050. 10.1016/j.chaos.2020.110050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav, R. S. (2020). Data analysis of COVID‐2019 epidemic using machine learning methods: A case study of India. International Journal of Information Technology, 12, 1321–1330. 10.1007/s41870-020-00484-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , He, X. , Zhao, J. , Zhang, Y. , Zhang, S. , & Xie, P. (2020). COVID‐CT‐dataset: A CT image dataset about COVID‐19. arXiv preprint arXiv:2003.13865 .
- Yoo, S. H. , Geng, H. , Chiu, T. L. , Yu, S. K. , Cho, D. C. , Heo, J. , Choi, M. S. , Choi, I. H. , Cung, C. , Nhung, N. V. , Min, B. J. , & Lee, H. (2020). Deep learning‐based decision‐tree classifier for COVID‐19 diagnosis from chest X‐ray imaging. Frontiers in Medicine, 7, 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamzmi, G. , Rajaraman, S. , & Antani, S. (2020). Accelerating super‐resolution and visual task analysis in medical images. Applied Sciences, 10(12), 4282. 10.3390/app10124282 [DOI] [Google Scholar]
- Zarocostas, J. (2009). World Health Organization declares A (H1N1) influenza pandemic. BMJ: British Medical Journal (Online), 338, 1–10. [DOI] [PubMed] [Google Scholar]
- Zeroual, A. , Harrou, F. , Dairi, A. , & Sun, Y. (2020). Deep learning methods for forecasting COVID‐19 time‐series data: A comparative study. Chaos, Solitons & Fractals, 140, 110121. 10.1016/j.chaos.2020.110121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H.‐t. , Zhang, J.‐s. , Zhang, H.‐h. , Nan, Y.‐d. , Zhao, Y. , Fu, E.‐q. , Xie, Y.‐h. , Liu, W. , Li, W.‐p. , Zhang, H.‐j. , Jiang, H. , Li, C.‐m. , Li, Y.‐y. , Ma, R.‐n. , Dang, S.‐k. , Gao, B.‐b. , Zhang, X.‐j. , & Zhang, T. (2020). Automated detection and quantification of COVID‐19 pneumonia: CT imaging analysis by a deep learning‐based software. European Journal of Nuclear Medicine and Molecular Imaging, 47(11), 2525–2532. 10.1007/s00259-020-04953-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. , Zhang, Y. , He, X. , & Xie, P. (2020). COVID‐CT‐dataset: A CT scan dataset about COVID‐19. arXiv preprint arXiv:2003.13865 .
- Zhu, J. , Shen, B. , Abbasi, A. , Hoshmand‐Kochi, M. , Li, H. , & Duong, T. Q. (2020). Deep transfer learning artificial intelligence accurately stages COVID‐19 lung disease severity on portable chest radiographs. PLoS ONE, 15(7), e0236621. 10.1371/journal.pone.0236621 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


