Abstract
Background
Infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) induces a complex antibody response that varies by orders of magnitude between individuals and over time.
Methods
We developed a multiplex serological test for measuring antibodies to 5 SARS-CoV-2 antigens and the spike proteins of seasonal coronaviruses. We measured antibody responses in cohorts of hospitalized patients and healthcare workers followed for up to 11 months after symptoms. A mathematical model of antibody kinetics was used to quantify the duration of antibody responses. Antibody response data were used to train algorithms for estimating time since infection.
Results
One year after symptoms, we estimate that 36% (95% range, 11%–94%) of anti-Spike immunoglobulin G (IgG) remains, 31% (95% range, 9%–89%) anti-RBD IgG remains, and 7% (1%–31%) of anti-nucleocapsid IgG remains. The multiplex assay classified previous infections into time intervals of 0–3 months, 3–6 months, and 6–12 months. This method was validated using data from a seroprevalence survey in France, demonstrating that historical SARS-CoV-2 transmission can be reconstructed using samples from a single survey.
Conclusions
In addition to diagnosing previous SARS-CoV-2 infection, multiplex serological assays can estimate the time since infection, which can be used to reconstruct past epidemics.
Keywords: antibody kinetics, SARS-SoV-2, seroprevalence, surveillance, time since infection
Longitudinal follow-up of individuals infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reveals differences in the duration of antibodies to multiple antigens. Mathematical models of antibody waning and statistical algorithms can estimate an individual’s time since SARS-CoV-2 infection, and previous transmission waves in a population.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), causing coronavirus disease 2019 (COVID-19), has led to widespread morbidity and mortality since its emergence. The response to the SARS-CoV-2 pandemic is critically dependent on surveillance data, most notably numbers of COVID-19–associated hospital admissions and deaths recorded through health systems surveillance, and cases confirmed SARS-CoV-2 positive by polymerase chain reaction (PCR)–based testing [1]. Serology, based on the detection of antibodies induced by infection with SARS-CoV-2, represents another category of surveillance information [2, 3]. Appropriately designed seroprevalence studies can provide estimates of the previously infected proportion of a population. Although no substitute for health system surveillance, seroprevalence studies have the advantage of accounting for asymptomatic cases and symptomatic individuals who do not present to health systems. Seroprevalence studies also provide information on the status of SARS-CoV-2 epidemics in situations where record-keeping by health systems is not possible, a common challenge in many low- and middle-income countries [4].
Infection with SARS-CoV-2 induces diverse humoral and cellular immune responses [5, 6]. Humoral immunity includes antibodies of several immunoglobulin isotypes targeting SARS-CoV-2 proteins, most notably spike (S) and nucleocapsid (N). The concentration of antibodies in blood varies substantially between individuals and with time since infection [6–11]. Studies of the duration of immunity to SARS-CoV-1 demonstrated that antibodies remain detectable 6 years after infection [12]. Longitudinal follow-up of individuals infected with SARS-CoV-2 indicates a pattern of waning antibody responses consistent with other coronaviruses [13, 14]. Within the first 3 months, antibody levels boost sharply and wane rapidly. Over a longer interval of 8 months, antibody levels wane more slowly [6]. These observations can be explained by the biphasic nature of antibody kinetics [15].
Serological diagnostics typically classify a sample as positive if a measured antibody level is greater than a defined cutoff. Analysis of quantitative rather than binary antibody levels provides additional information; for example, antibody levels are associated with time since infection, symptom severity, and sex [16]. However, large variation in antibody levels between individuals prevents this from having predictive value: Detected antibodies could be from a recent infection, or due to immunological memory of older infections. This limitation has recently been overcome for a range of pathogens through the combination of multiplex assays and classification algorithms. Using machine learning algorithms to analyze quantitative antibody responses to multiple antigens, the time since previous infection can be estimated for Plasmodium falciparum malaria [17, 18], Plasmodium vivax malaria [19], and cholera [20].
In this study, we use multiplex assays to measure antibodies to SARS-CoV-2 in healthcare workers and hospitalized patients followed for up to 11 months after infection, and apply mathematical models to characterize antibody kinetics in the first year following infection. Classification algorithms were developed that minimize the reduction in the sensitivity of serological tests over time, in addition to estimating time since previous infection from a single blood sample. Finally, we present a method for serological reconstruction of past SARS-CoV-2 transmission and validate its use with samples collected from a cross-sectional survey.
MATERIALS AND METHODS
Samples
A panel of 407 negative control serum or plasma samples was assembled from prepandemic cohorts (before December 2019) with ethical approval for broad antibody testing (Table 1). A panel of 407 positive control serum samples was assembled from individuals with recent SARS-CoV-2 infection. This included 72 samples from Paris hospitals that cared for patients with COVID-19 [21, 22]; 161 samples from healthcare workers in Strasbourg University Hospital who developed symptomatic PCR-confirmed COVID-19 [23]; and 174 samples from community members of Crépy-en-Valois, France, collected between February and March 2020, and confirmed seropositive by flow cytometry–based testing [24, 25].
Table 1.
Panels of Samples
| Cohort | Status | Participants, No. | Samples, No. | PCR Positive, No. | Age, y, Median (Range) | Sex (% Male) | Days Post–Symptom Onset, Median (Range) |
|---|---|---|---|---|---|---|---|
| Établissement Français du Sang 1 | Negative panel: prepandemic controls | 45 | 45 | … | >18 | … | … |
| Établissement Français du Sang 2 | Negative panel: prepandemic controls | 213 | 213 | … | 42 (18–81) | 40% | … |
| Thai Red Cross | Negative panel: prepandemic controls | 68 | 68 | … | >18 | … | … |
| Peruvian donors | Negative panel: prepandemic controls | 81 | 81 | … | >18 | … | … |
| Hôpital Bichat (Paris, France) |
Positive panel: hospitalized patients | 2 | 8 | 2 | 31 (30–32) | 100% | 14 (8–24) |
| Hôpital Cochin (Paris, France) |
Positive panel: hospitalized patients | 64 | 64 | 64 | 55 (25–79) | 76% | 17 (10–28) |
| Strasbourg hospitals 1 | Positive panel: infected HCWs | 161 | 161 | 161 | 32 (20–62) | 31% | 24 (13–39) |
| Crépy-en-Valois, Feb–Mar 2020 | Positive panel: infected community members (flow cytometry positive) | 154 | 174 | 0 | 17 (15–56) | 34% | … |
| Dublin hospitals | Hospitalized patients | 194 | 213 | 194 | 55 (21–92) | 47% | 13 (1–126) |
| Strasbourg hospitals 2 | Follow-up of infected HCWs | 347 | 724 | 347 | 41 (21–74) | 23% | 132 (11–284) |
| Institut Mutualiste Montsouris (Paris, France) |
Seroprevalence survey in HCWs (unknown status) | 769 | 769 | 20 | 41 (18–72) | 27% | … |
| Institut Mutualiste Montsouris (Paris, France) |
Follow-up of seropositive HCWs | 29 | 29 | 12 | 37 (24–63) | 41% | 304 (285–336) |
| Crépy-en-Valois, Nov–Dec 2020 | Seroprevalence survey in community members (unknown status) | 725 | 725 | NA | NA | NA | NA |
Abbreviations: HCW, healthcare worker; NA, not available; PCR, polymerase chain reaction.
The duration of antibody responses following SARS-CoV-2 infection was studied in longitudinal cohorts of hospitalized patients and healthcare workers. Two hundred thirteen serum samples from 194 patients in Dublin hospitals were collected, with date post–symptom onset extending up to 4 months. Seven hundred twenty-four serum samples from 347 healthcare workers in Strasbourg hospitals were collected, with date post–symptom onset extending up to 9 months [26].
In April 2020, our team implemented a study of the seroprevalence of SARS-CoV-2 in healthcare workers from Institut Mutualiste Montsouris, a hospital in Paris. Serum samples were collected from 769 healthcare workers, and tested with our multiplex assay (Supplementary Figure 1). Healthcare workers who tested seropositive in April 2020 were invited to present a second sample in January 2021. In total we obtained follow-up samples from 29 healthcare workers.
To demonstrate the serological reconstruction of past COVID-19 epidemics, 725 samples were collected from residents of the town of Crépy-en-Valois in France from 13 November to 17 December 2020. Past transmission levels estimated from serological data were validated using an independent source of data on intensive care unit (ICU) admissions for COVID-19 in Oise (the department containing the town of Crépy-en-Valois) extracted from the national public data on daily hospital admissions related to COVID-19 published by Sante Publique France, available on data.gouv.fr (file downloaded 18 April 2021 at https://www.data.gouv.fr/fr/datasets/donnees-hospitalieres-relatives-a-lepidemie-de-covid-19/#). Daily records cover the period from 18 March 2020 to 18 April 2021.
Serological Assays
We established a 9-plex bead-based assay allowing simultaneous detection of antibodies to 5 SARS-CoV-2 antigens and 4 seasonal coronaviruses (spike proteins of NL63, 229E, HKU1, OC43) in 1 µL serum or plasma samples that were heat-inactivated at 56°C for 30 minutes. SARS-CoV-2 antigens were from spike (whole trimeric spike [S], its receptor-binding domain [RBD], and spike subunit 2 [S2]), nucleocapsid protein (N), and a membrane-envelope fusion protein (ME). ME and S2 antigens were purchased from Native Antigen (Oxford, United Kingdom) and all other antigens were produced as recombinant proteins at Institut Pasteur. The mass of proteins coupled on beads was optimized to generate a log-linear standard curve with a pool of 27 positive sera prepared from patients with reverse-transcription quantitative PCR–confirmed SARS-CoV-2. We measured the levels of immunoglobulin G (IgG), immunoglobulin A (IgA), and immunoglobulin M (IgM) of each sample in 3 separate assays. In brief, serum was diluted 1:200 for IgG or 1:400 for IgA and IgM, and incubated with mixed antigen–coupled beads for 30 minutes. Secondary antibodies conjugated to R-phycoerythrin (Jackson Immunoresearch) were used at 1:120, 1:200, or 1:400 for detection of specific IgG, IgA, and IgM, respectively. All dilutions and cycles of washing steps were done in phosphate-buffered saline supplemented with 1% bovine serum albumin and 0.05% (v/v) Tween-20. On each assay plate, 2 blanks (only beads, no serum) were included to control for background signal as well as a standard curve prepared from 2-fold serial dilutions (1/50 to 1/102 400) of a pool of positive controls. Plates were read using a Luminex MAGPIX system and the median fluorescence intensity (MFI) was used for analysis. A 5-parameter logistic curve was used to convert MFI to relative antibody units, relative to the standard curve performed on the same plate to account for interassay variations. The data from our multiplex assay was compared against data from 2 different neutralization assays with live virus using a subset of serum samples [26] (Supplementary Methods).
Statistical Methods
SARS-CoV-2 antibody kinetics are described using a previously published mathematical model of the immunological processes underlying the generation and waning of antibody responses following infection (Supplementary Methods) [27].
For the estimation of time since previous infection, measurements of antibodies of 3 isotypes (IgG, IgM, IgA) to multiple SARS-CoV-2 antigens were used to create a training dataset. Samples where the time post–symptom onset was ≤14 days or unknown were excluded. In total we had 407 samples from prepandemic negative controls and 1402 positive samples. A random forests multiway classification algorithm was developed for categorizing samples into 4 classes: (1) negative; (2) infected <3 months ago; (3) infected 3–6 months ago; and (4) infected 6–12 months ago. The algorithm was calibrated to have 99% specificity for correctly classifying negative samples. Positive samples were classified according to the maximum number of votes. Uncertainty in classification performance was assessed via cross-validation with a training set comprising two-thirds of the data and a disjoint testing set comprising a third of the data, repeated 1000 times. Classification algorithms were implemented in R (version 3.4.3).
Ethical Considerations
Serum samples were biobanked at the Clinical Investigation and Access to BioResources platform at Institut Pasteur (Paris, France). Samples were obtained from consenting individuals through the CORSER study (Sero-epidemiological Study of the SARS-CoV-2 Virus Responsible for COVID-19 in France: ClinicalTrials.gov identifier NCT04325646), directed by Institut Pasteur and approved by the Comité de Protection des Personnes Ile de France III (on 19 February 2020) and the French COVID cohort (ClinicalTrials.gov identifier NCT04262921), sponsored by Inserm and approved by the Comité de Protection des Personnes Ile de France VI. Samples from French blood donors were approved for use by Etablissement Français du Sang (Lille, France) and approved through the CORSER study by the Comité de Protection des Personnes Ile de France VI. Sample collection in Hôpital Cochin was approved by the Research Ethics Commission of Necker-Cochin Hospital. Samples from healthcare workers in Strasbourg University Hospitals followed longitudinally were collected as part of an ongoing clinical trial (ClinicalTrials.gov identifier NCT04441684) that received ethical approval from the Comité de Protection des Personnes Ile de France III. Samples collected from patients in Dublin received ethical approval for study from the Tallaght University Hospital/St James’s Hospital Joint Research Ethics Committee (reference REC 2020-03). Use of the Peruvian negative controls was approved by the Institutional Ethics Committee from the Universidad Peruana Cayetano Heredia (SIDISI 100873). The Human Research Ethics Committee at the Walter and Eliza Hall Institute of Medical Research and the Ethics Committee of the Faculty of Tropical Medicine, Mahidol University, Thailand, approved the use of the Thai negative control samples. Informed written consent was obtained from all participants or their next of kin in accordance with the Declaration of Helsinki.
RESULTS
SARS-CoV-2 Antibodies Over Time
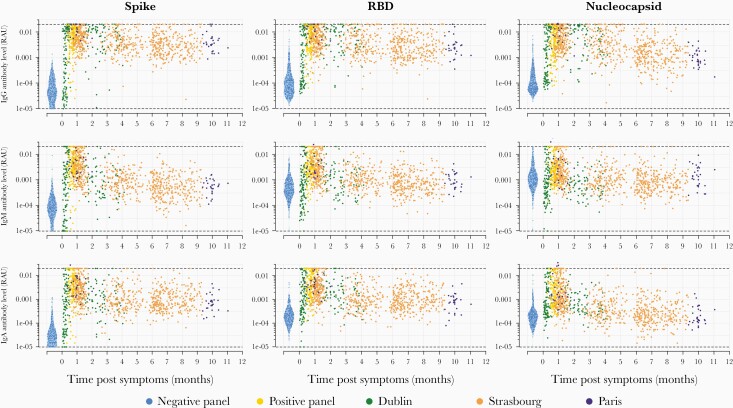
Antibodies of 3 isotypes (IgG, IgM, IgA) to 9 coronavirus antigens were measured in 407 prepandemic serum samples, and 1402 serum samples from individuals with previous SARS-CoV-2 infection. Nine hundred sixty-one of 1402 of the positive samples were from individuals with SARS-CoV-2 infection confirmed by PCR-based testing with available data on time post–symptom onset. Figure 1 presents the IgG, IgM, and IgA antibody responses to SARS-CoV-2 S, RBD, and N measured over time. Supplementary Figures 2 and 3 present the antibody responses to S2, ME, and the spike proteins of the 4 human seasonal coronaviruses. Notably, there is substantial interindividual variation in antibody responses, with antibody levels varying by orders of magnitude between individuals. As a measure of functional immunity, we applied live virus neutralizing assays to a subset of samples. There was substantial variation in neutralizing activity between individuals and over time (Supplementary Figure 4).
Figure 1.
Antibody kinetics in the first year following infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). A bead-based multiplex Luminex assay was used to measure antibodies of multiple isotypes (immunoglobulins G, M, and A) to multiple antigens in serum samples from individuals with polymerase chain reaction–confirmed SARS-CoV-2 infection and prepandemic negative controls. Abbreviations: IgA, immunoglobulin A; IgG, immunoglobulin G; IgM, immunoglobulin M; RAU, relative antibody unit; RBD, receptor-binding domain.
Modeled SARS-CoV-2 Antibody Kinetics
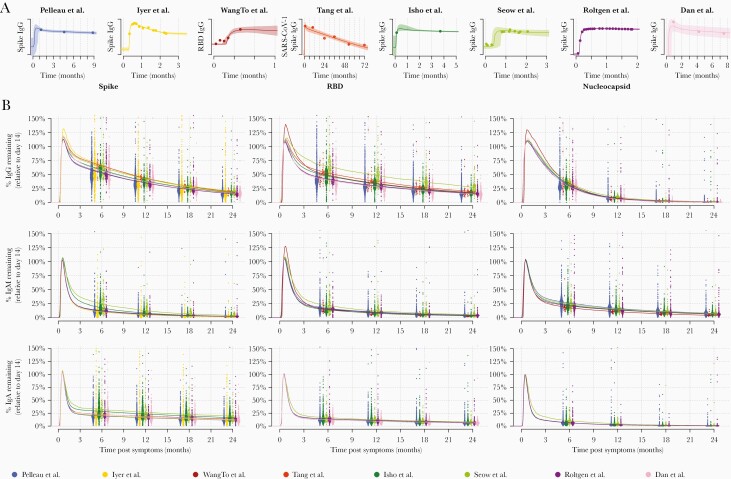
Our data on measured SARS-CoV-2 antibody responses were supplemented with data from 6 other longitudinal studies of the SARS-CoV-2 antibody response [6–11] and 1 longitudinal study of the SARS-CoV-1 antibody response [12]. Supplementary Figure 5 provides a comparison of the measured antibody responses between the 8 studies. The data were rescaled so that the mean antibody response for each study equals 1 at day 14 after symptom onset. In addition to the large interindividual variation, there is notable variation in antibody levels between studies. A mathematical model of antibody kinetics was simultaneously fitted to data from all 8 studies. Supplementary Figure 6 provides an overview of the model fit to the data. Figure 2A provides example fits to data from 1 representative individual from each of the 8 studies. Figure 2B provides model predictions of the percentage antibody level remaining over the first 2 years postinfection, where 100% is assumed to be the antibody response at day 14 following symptom onset. There are considerable differences in the pattern of waning between isotypes and between antigens. Table 2 summarizes the duration of the antibody response. One year following symptom onset, we estimate that 36% (95% range, 11%–94%) anti-S IgG remains, 31% (9%–89%) anti-RBD IgG remains, and 7% (1%–31%) anti-N IgG remains. The uncertainty represents the considerable degree of interindividual variation in the duration of the antibody responses. Antibodies of the IgM isotype waned more rapidly, with 6% (0%–27%) anti-S IgM remaining after 1 year, 9% (2%–32%) anti-RBD IgM remaining after 1 year, and 15% (4%–50%) anti-N IgM remaining after 1 year. Antibodies of the IgA isotype also waned rapidly, with 18% (4%–67%) anti-S IgA remaining after 1 year, 10% (3%–38%) anti-RBD IgA remaining after 1 year, and 3% (0%–13%) anti-N IgA remaining after 1 year.
Figure 2.
Modeled severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibody kinetics. A mathematical model of SARS-CoV-2 antibody kinetics was simultaneously fitted to data from 7 studies of SARS-CoV-2 [6-11] and 1 study of severe acute respiratory syndrome coronavirus (SARS-CoV-1) [12]. A, top row, Examples of the model fit to the data for 1 individual from each study. Data are represented as points, posterior median model prediction as lines, and 95% credible intervals as shaded areas. B, middle and bottom rows, Model-predicted duration of antibodies within the first 2 years following infection. Antibody levels are shown relative to the expected antibody level at day 14 post–symptom onset. Each point represents the prediction from an individual at 6, 12, 18, and 24 months post–symptom onset. The median predictions for each of the 8 studies are presented as lines. Abbreviations: IgA, immunoglobulin A; IgG, immunoglobulin G; IgM, immunoglobulin M; RBD, receptor-binding domain; SARS-CoV-1, severe acute respiratory syndrome coronavirus.
Table 2.
Estimated Duration of Antibody Responses Following Severe Acute Respiratory Syndrome Coronavirus 2 Infection
| Antibody | 6 Months | 12 Months | 24 Months |
|---|---|---|---|
| Spike IgG | 55% (16%–100%) | 36% (11%–94%) | 16% (5%–55%) |
| RBD IgG | 43% (13%–100%) | 31% (9%–89%) | 16% (5%–48%) |
| Nucleocapsid IgG | 30% (8%–92%) | 7% (1%–31%) | 0.8% (0%–7%) |
| Spike IgM | 12% (1%–52%) | 6% (0%–27%) | 2% (0%–9%) |
| RBD IgM | 16% (4%–51%) | 9% (2%–32%) | 4% (1%–16%) |
| Nucleocapsid IgM | 23% (6%–75%) | 15% (4%–50%) | 7% (2%–24%) |
| Spike IgA | 21% (4%–82%) | 18% (4%–67%) | 12% (3%–47%) |
| RBD IgA | 12% (4%–49%) | 10% (3%–38%) | 6% (2%–24%) |
| Nucleocapsid IgA | 6% (1%–30%) | 3% (0%–13%) | 0.6% (0%–4%) |
The percentage antibody level remaining over time is compared to the measured antibody level 14 days after symptom onset. Estimates are presented as the population median, with the 95% range due to interindividual variation.
Abbreviations: IgA, immunoglobulin A; IgG, immunoglobulin G; IgM, immunoglobulin M; RBD, receptor-binding domain.
We also observed comparable reductions in titers for viral neutralization over time (Supplementary Figure 4). The small number of samples prevented application of the antibody kinetic model to viral neutralization data, but measuring the reduction in paired samples we estimate a rate of decrease of –0.94 (interquartile range [IQR], –2.41 to 0.26) year-1. Assuming simple exponential decay in neutralization activity over time, this corresponds to 39.3% (IQR, 8.9%–125.2%) neutralizing activity remaining after 1 year, broadly consistent with the long-term kinetics of anti-RBD IgG.
Estimating Time Since Previous SARS-CoV-2 Infection
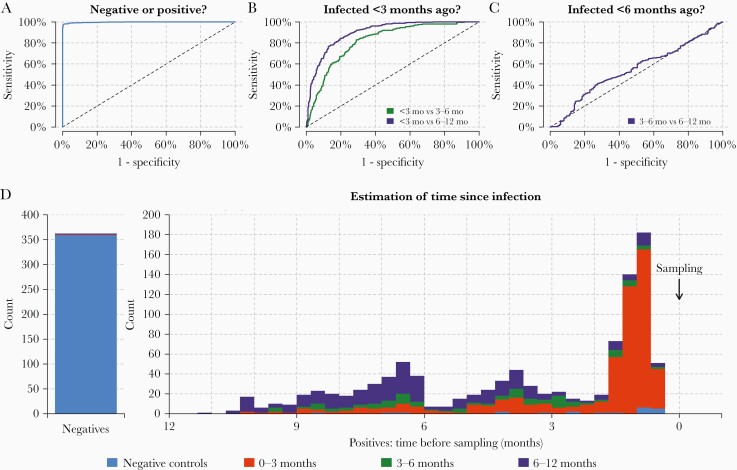
Using a dataset on measured IgG, IgM, and IgA antibody levels from prepandemic negative controls and samples from individuals with SARS-CoV-2 infection confirmed by PCR-based testing and followed for up to 11 months after symptom onset, random forest binary classification algorithms were trained to identify individuals with previous SARS-CoV-2 infection (Supplementary Figures 7 and 8). Next, a random forest multiway classification was trained to simultaneously identify previous infection and estimate the time since infection (Figure 3). The diagnostic algorithm identified samples from individuals with previous SARS-CoV-2 infection with 99% specificity and 98% (95% confidence interval [CI], 94%–99%) sensitivity (Figure 3A). The diagnostic test classified samples from individuals infected within the previous 3 months (Figure 3B). It was easier to distinguish recent infections (<3 months) from older infections (6–12 months), compared to infections that occurred 3–6 months ago. There was limited statistical signal to distinguish between infections that occurred 3–6 months ago and older infections occurring >6 months ago (Figure 3C). A breakdown of classification performance by time since infection is provided in Figure 3D. The diagnostic test had predictive power for samples of all categories, with the exception of samples from individuals infected 3–6 months ago. Many of these samples were incorrectly classified in the neighboring infection categories of 0–3 months or 6–12 months.
Figure 3.
Classification of time since previous severe acute respiratory syndrome coronavirus 2 infection. A cross-validated multiway classification algorithm was trained to estimate time since infection. A, The algorithm can differentiate between positive and negative samples. B, The algorithm can classify individuals infected within the previous 3 months. C, There is limited diagnostic power to distinguish between infections that occurred 3–6 months ago vs 6–12 months ago. D, Breakdown of classification performance according to time since previous infection. Colors represent model predicted classification. More than 99% of negative samples are correctly classified as negative (blue). For the positive samples, the distribution shows the time since previous infection. Samples with time since infection <3 months are mostly classified in the category 0–3 months (red). Samples with time since infection >6 months ago are mostly classified in the category 6–12 months (purple). There is a substantial degree of misclassification of samples with time since infection 3–6 months ago. This is due to the temporal imbalance in the training data.
Serological Reconstruction of Past COVID-19 Transmission in Oise Department, France
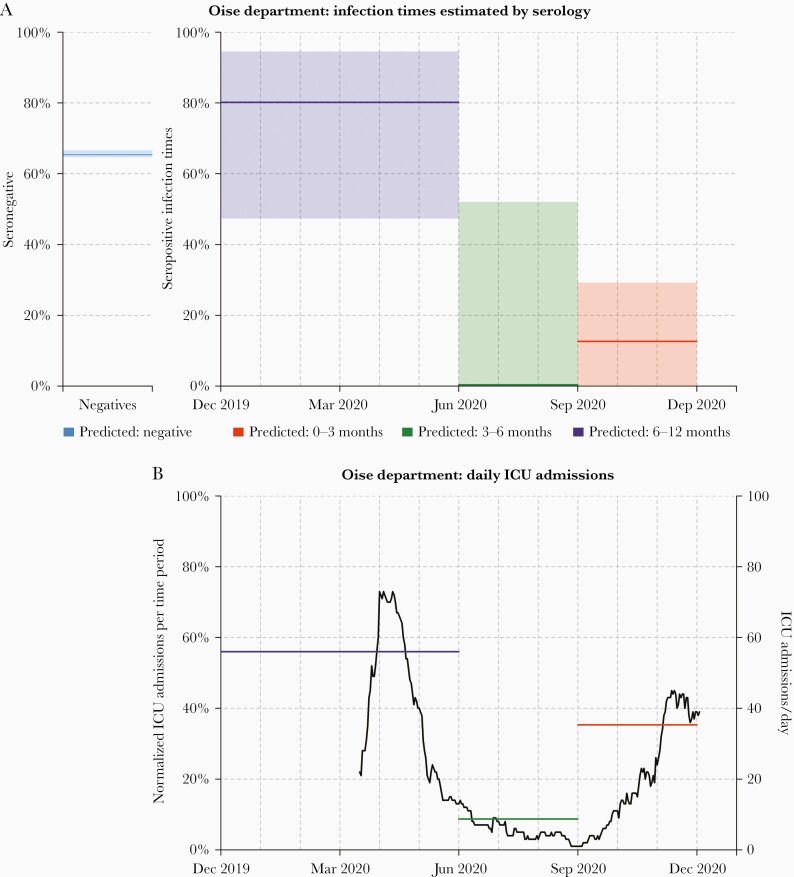
One of the first reported clusters of SARS-CoV-2 transmission in France occurred in Crépy-en-Valois in Oise Department from late February to early March 2020 [24]. A team of scientists led by Institut Pasteur are conducting repeat cross-sectional surveys in this community. Seven hundred twenty-five serum samples collected between 13 November and 17 December 2020 were analyzed with our serological assay. Two hundred fifty-one of 725 (34.6%) samples were seropositive, indicating previous infection with SARS-CoV-2.
Statistical algorithms for estimation of time since infection were applied to data from the 251 seropositive individuals. We estimated that 80.2% (95% CI, 47.3%–94.5%) were infected in the 6 months from December 2019 to May 2020, 0.0% (95% CI, 0.0%–52.1%) were infected in the 3 months from June to August, and 12.6% (95% CI, 0.0%–29.2%) were infected in the 3 months from September to November (Figure 4). The serological estimates are consistent with a large first wave before June 2020, and a smaller second wave (or the beginning of a larger one) between September and December 2020. To assess the validity of these estimates, we compared with data on reported ICU admissions in Oise Department between 18 March 2020 and 1 December 2020. Of the ICU admissions, 56.0% were reported in the 3 months from March to May, 8.7% were reported in the 3 months from June to August, and 35.3% were reported in the 3 months from September to November.
Figure 4.
Serological reconstruction of past coronavirus disease 2019 transmission in Oise Department, Franch. A, Of 725 samples collected from residents of Oise Department between 13 November and 17 December 2020, 65% (474/725) were severe acute respiratory syndrome coronavirus 2 seronegative. For the 251 seropositive individuals, we estimated that 80.2% (95% confidence interval [CI], 47.3%–94.5%) were infected in the 6 months from December 2019 to May 2020, 0.0% (95% CI, .0%–52.1%) were infected in the 3 months from June to August 2020, and 12.6% (95% CI, .0%–29.2%) were infected in the 3 months from September to November 2020. Proportions are presented as median estimates and do not necessarily sum to 100%. B, Reported intensive care unit (ICU) admissions in Oise Department between 18 March 2020 and 1 December 2020. Of the ICU admissions, 56.0% were reported in the 3 months from March to May, 8.7% were reported in the 3 months from June to August, and 35.3% were reported in the 3 months from September to November.
DISCUSSION
Based on data from a range of studies with up to 11 months of follow-up after symptom onset, we estimate that 31% (95% credible interval [CrI], 9%–89%) of anti-RBD IgG antibody levels remain 1 year after infection. Antibodies of other isotypes waned more rapidly, with 9% (95% CrI, 2%–32%) of anti-RBD IgM antibody levels remaining after 1 year, and 10% (95% CrI, 3%–38%) of anti-RBD IgA antibody levels remaining after 1 year. There was considerable variation in kinetic profiles between different SARS-CoV-2 antigens, with 7% (95% CrI, 1%–31%) of anti-N IgG antibody levels remaining after 1 year. Although the determinants of the duration of antigen-specific antibody responses remain poorly understood [16], the diversity in patterns of antibody kinetics can be quantified using epidemiological studies, yielding valuable information for serological diagnostics.
The majority of commercially available serological tests classify individuals as having previous SARS-CoV-2 infection if a measured antibody response is greater than a defined cutoff [28, 29]. Instead of reducing a complex antibody response to a binary data point, a more detailed serological signature based on quantitative measurements of multiple antibody responses provides 2 notable advantages [5]. First, the reduction in diagnostic sensitivity associated with waning antibodies is minimized—no reduction in sensitivity over the 11 months of follow-up was observed. Second, the time since previous infection can be estimated providing additional epidemiological information. For the current assay, time since previous infection was categorized into intervals of 0–3 months, 3–6 months, and 6–12 months. More precise classification is possible in theory, but this must be balanced against the statistical signal. For example, there was little statistical signal to discriminate between infections that occurred 6–9 months ago vs infections that occurred 9–12 months ago. It is anticipated that further assay improvements such as the incorporation of new antigens, more training samples with a range of time since infection, and better algorithms will lead to improvements in accuracy.
Existing seroprevalence studies estimate the proportion of a population previously infected with SARS-CoV-2. The addition of a diagnostic tool capable of estimating time since infection allows for the serological reconstruction of past incidence trends. Thus, using samples from a seroprevalence study collected at a single time point, we can discriminate between a scenario of constant SARS-CoV-2 transmission and a scenario where transmission occurs in distinct epidemic waves. This diagnostic technology has a range of possible applications. For countries that experienced a double wave of SARS-CoV-2 epidemics, it has been challenging to quantify the relative magnitude of these waves due to the challenges of implementing PCR-based diagnostic testing [30]. Furthermore, there are many epidemiological settings where it is unknown if SARS-CoV-2 transmission was constant over time, or occurred as distinct epidemic waves [4].
Estimates of time since previous SARS-CoV-2 infection in individual samples can be combined to provide a serological reconstruction of previous SARS-CoV-2 transmission in a population. Following validation of our statistical method on simulated data, we demonstrated that our test could accurately identify the 2 epidemic peaks in the town of Crépy-en-Valois using samples collected in a single cross-section. This finding was consistent with daily reported ICU admissions data collected continuously over 1 year in the Oise Department. The minor differences between the serologically reconstructed epidemic profile and that from public health surveillance data may be because the town of Crépy-en-Valois is not necessarily representative of the wider Oise Department, as the town was specifically targeted by the study team because of its early levels of SARS-CoV-2 transmission. Furthermore, we did not analyze data on COVID-19–specific ICU admissions before March 2020, and ICU admissions may not necessarily be representative of community-level infection events [31].
There are several limitations to this study. Estimates of the duration of antibody responses are based on data from multiple studies, each using a unique immunoassay [6–12]. Every immunoassay may differ in terms of background reactivity, cross-reactivity with other pathogens, protein formulation, dynamic range, and reproducibility. We believe that the benefit of drawing on multiple data sources outweighs the benefit of having a smaller, more homogeneous database, especially since the mathematical model of antibody kinetics is sufficiently flexible to incorporate data from multiple assays. Our selection of a mechanistic mathematical model of antibody kinetics is a potential limitation. The model is based on a mechanistic understanding of the immunological processes underlying the generation and persistence of antibodies, and imposes a flexible functional form on how antibody levels change over time. Although this approach has been validated in a range of applications [16–19], there will be instances where the model fails to capture the true pattern of antibody kinetics, for example in immunodeficient individuals. An advantage of a mechanistic model vs a nonparametric statistical model is the ability to make projections forward in time. We have provided predictions up to 2 years following infection, for example, by estimating that 16% (95% range, 5%–48%) of anti-RBD IgG antibodies remain after 2 years. There is a risk to providing predictions beyond the timescale of the data, but these predictions can be easily refuted via continued longitudinal studies.
Seroprevalence studies are playing a critical role in monitoring the progress of the SARS-CoV-2 pandemic. In the early stages of the pandemic, immunoassays had the advantage of measuring high antibody levels in the initial months after infection. As the pandemic progresses, seroprevalence will become more challenging to accurately measure due to waning antibody responses and increased vaccine-induced immunity. Multiplex assays and algorithms accounting for how antibody levels change over time may be an important tool for ensuring the ongoing utility of serosurveillance.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. The French COVID cohort is supported by the REACTing consortium and by the French Directorate General for Health. Jessica Vanhomwegen (Institut Pasteur) is thanked for provision of a MAGPIX machine. Jérôme Hadjadj and Laura Barnabei (Institut Imagine, Paris) are thanked for their work on the Hôpital Cochin study. Dionicia Gamboa (Cayetano Heredia University, Lima) is thanked for sharing negative control samples from Peru. Jetsumon Sattabongkot (Mahidol University, Bangkok) is thanked for sharing negative control samples from Thailand. Marie-Noelle Ungeheuer and Blanca Liliana Perlaza are thanked for processing samples at the Clinical Investigation and Access to BioResources platform in Institut Pasteur. We thank all patients and healthcare workers who kindly agreed for samples to be used for medical research purposes.
Data availability. All data and code used for reproducing the results are freely available on GitHub: https://github.com/MWhite-InstitutPasteur/SARSCoV2_AB_kinetics.
Financial support. This work was supported by the European Research Council (MultiSeroSurv ERC Starting Grant 852373 to M. W.); l’Agence Nationale de la Recherche and Fondation pour la Recherche Médicale (CorPopImm to M. W.); and the Institut Pasteur International Network (CoronaSeroSurv to M. W.). J. R. was supported by the Pasteur Paris University International PhD Program. C. C. was supported by the European Research Council 771813. M. B., A. L., and H. A. were supported by the URGENCE COVID-19 fundraising campaign of Institut Pasteur. H. A. is supported by the Centre for International Migration and Development. N. C. and C. N. C. are partly funded by a Science Foundation Ireland grant (grant code 20/SPP/3685). L. T. has been awarded the Irish Clinical Academic Training program, supported by the Wellcome Trust and the Health Research Board (grant number 203930/B/16/Z), the Health Service Executive, National Doctors Training and Planning and the Health and Social Care, Research and Development Division, Northern Ireland (https://icatprogramme.org/). M. B. and A. M. were supported by Institut Pasteur TaskForce funding (TooLab project). I. M. is supported by a National Health and Medical Research Council Principal Research Fellowship. S. F.-K.’s laboratory is funded by Strasbourg University Hospitals (SeroCoV-HUS; PRI 7782), the Agence Nationale de la Recherche (ANR-18-CE17-0028), Laboratoire d’Excellence TRANSPLANTEX (ANR-11-LABX-0070_TRANSPLANTEX), and Institut National de la Santé et de la Recherche Médicale (UMR_S 1109).
Potential conflicts of interest. M. T. W., I. M., J. R., S. Pel., M. B., and S. Pet. are inventors on provisional patent PCT/US 63/057.471 on a serological antibody-based diagnostics of SARS-CoV-2 infection. T. B. and O. S. are coinventors on provisional patent PCT/US 63/020 063 entitled “S-Flow: a FACS-based assay for serological analysis of SARS-CoV-2 infection” submitted by Institut Pasteur. S. K. reports personal fees from Accelerate Diagnostics, Astellas, Merck Sharp & Dohme, Pfizer, and Menarini, and grants and personal fees from bioMérieux, outside of the submitted work. All other authors report no potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Salje H, Tran Kiem C, Lefrancq N, et al. . Estimating the burden of SARS-CoV-2 in France. Science 2020; 369:208–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Le Vu S, Jones G, Anna F, et al. . Prevalence of SARS-CoV-2 antibodies in France: results from nationwide serological surveillance. Nat Commun 2021; 12:3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ward H, Cooke G, Atchison C, et al. . Declining prevalence of antibody positivity to SARS-CoV-2: a community study of 365,000 adults. medRxiv [Preprint]. Posted online 27 October 2020. doi: 10.1101/2020.10.26.20219725. [DOI] [Google Scholar]
- 4. Kagucia EW, Gitonga JN, Kalu C, et al. . Seroprevalence of anti-SARS-CoV-2 IgG antibodies among truck drivers and assistants in Kenya. medRxiv [Preprint]. Posted online 17 February 2021. doi: 10.1101/2021.02.12.21251294. [DOI] [Google Scholar]
- 5. Rosado J, Pelleau S, Cockram C, et al. . Multiplex assays for the identification of serological signatures of SARS-CoV-2 infection: an antibody-based diagnostic and machine learning study. Lancet Microbe 2021; 2:e60–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dan JM, Mateus J, Kato Y, et al. . Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 2021; 371:eabf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. To KK, Tsang OT, Leung WS, et al. . Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis 2020; 20:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iyer AS, Jones FK, Nodoushania A, et al. . Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci Immunol 2020;5:eabe0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Isho B, Abe KT, Zuo M, et al. . Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci Immunol 2020;5:eabe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Röltgen K, Powell AE, Wirz OF, et al. . Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci Immunol 2020;5:eabe0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Seow J, Graham C, Merrick B, et al. . Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol 2020; 5:1598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tang F, Quan Y, Xin ZT, et al. . Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J Immunol 2011; 186:7264–8. [DOI] [PubMed] [Google Scholar]
- 13. Callow KA, Parry HF, Sergeant M, Tyrrell DA. The time course of the immune response to experimental coronavirus infection of man. Epidemiol Infect 1990; 105:435–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Choe PG, Perera RAPM, Park WB, et al. . MERS-CoV antibody responses 1 year after symptom onset, South Korea, 2015. Emerg Infect Dis 2017; 23:1079–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Slifka MK, Antia R, Whitmire JK, Ahmed R. Humoral immunity due to long-lived plasma cells. Immunity 1998; 8:363–72. [DOI] [PubMed] [Google Scholar]
- 16. Scepanovic P, Alanio C, Hammer C, et al. ; Milieu Intérieur Consortium. Human genetic variants and age are the strongest predictors of humoral immune responses to common pathogens and vaccines. Genome Med 2018; 10:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Helb DA, Tetteh KK, Felgner PL, et al. . Novel serologic biomarkers provide accurate estimates of recent Plasmodium falciparum exposure for individuals and communities. Proc Natl Acad Sci U S A 2015; 112:E4438–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yman V, Tuju J, White MT, et al. . Distinct kinetics in antibody responses to 111 Plasmodium falciparum antigens identifies novel serological markers of recent malaria exposure. medRxiv [Preprint]. Posted online 16 December 2020. doi: 10.1101/2020.12.14.20242768. [DOI] [Google Scholar]
- 19. Longley RJ, White MT, Takashima E, et al. . Development and validation of serological markers for detecting recent exposure to Plasmodium vivax infection. Nature Med 2020; 26:741–9. [DOI] [PubMed] [Google Scholar]
- 20. Azman AS, Lessler J, Luquero FJ, et al. . Estimating cholera incidence with cross-sectional serology. Sci Transl Med 2019; 11:eaau6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lescure FX, Bouadma L, Nguyen D, et al. . Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect Dis 2020; 20:697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hadjadj J, Yatim N, Barnabei L, et al. . Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020; 369:718–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fafi-Kremer S, Bruel T, Madec Y, et al. . Serologic responses to SARS-CoV-2 infection among hospital staff with mild disease in eastern France. EBioMedicine 2020; 59:102915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grzelak L, Temmam S, Planchais C, et al. . A comparison of four serological assays for detecting anti-SARS-CoV-2 antibodies in human serum samples from different populations. Sci Transl Med 2020; 12:eabc3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dufloo J, Grzelak L, Staropoli I, et al. . Asymptomatic and symptomatic SARS-CoV-2 infections elicit polyfunctional antibodies. Cell Rep Med 2021;2:100275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Planas D, Bruel T, Grzelak L, et al. . Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med 2021; 27:917–24. [DOI] [PubMed] [Google Scholar]
- 27. White MT, Griffin JT, Akpogheneta O, et al. . Dynamics of the antibody response to Plasmodium falciparum infection in African children. J Infect Dis 2014; 210:1115–22. [DOI] [PubMed] [Google Scholar]
- 28. National SARS-CoV-2 Serology Assay Evaluation Group. Performance characteristics of five immunoassays for SARS-CoV-2: a head-to-head benchmark comparison. Lancet Infect Dis 2020; 20:1390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peluso MJ, Takahashi S, Hakim J, et al. . SARS-CoV-2 antibody magnitude and detectability are driven by disease severity, timing, and assay. medRxiv [Preprint]. Posted online 5 March 2021. doi: 10.1101/2021.03.03.21251639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pullano G, Di Domenico L, Sabbatini CE, et al. . Underdetection of cases of COVID-19 in France threatens epidemic control. Nature 2021; 590: 134–9. [DOI] [PubMed] [Google Scholar]
- 31. Hozé N, Paireau J, Lapidus N, et al. . Monitoring the proportion of the population infected by SARS-CoV-2 using age-stratified hospitalisation and serological data: a modelling study. Lancet Public Health 2021; 6:e408–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.