Abstract
Candida glabrata is a yeast frequently isolated from human specimens. Based upon its well-known ability to rapidly hydrolyze trehalose, we have developed a novel and cost-effective test incubating one yeast colony emulsified in 50 μl of citrate buffer (0.1 M [pH 5.0]) containing 4% (wt/vol) trehalose for 3 h at 37°C. Trehalase-generated glucose is detected with a commercially available dipstick (range, 1.0 to 50 g/liter). For evaluation, consecutive clinical isolates and several reference strains of C. glabrata (n = 160), C. albicans (n = 120), and other yeast species with potential ability for utilization of trehalose (C. dubliniensis, n = 11; C. famata, n = 15; C. guilliermondii, n = 5; C. lusitaniae, n = 16; C. parapsilosis, n = 20; C. tropicalis, n = 34; C. viswanathii, n = 5; Pichia angusta, n = 2; C. zeylanoides, n = 2; Saccharomyces cerevisiae, n = 16; C. neoformans, n = 7) were tested. Identification of C. glabrata is achieved within 3 h, with a specificity of 99.1% and a sensitivity of 98.8% when grown on Sabouraud dextrose agar supplemented with 4% glucose.
The incidence of fungal infections has been constantly increasing over the past decades, due in part to the increase of non-Candida albicans candidal species (8). In some institutions more than one-third of candidal bloodstream infections are caused by species other than C. albicans. C. glabrata is one of these species and has been reported to be the yeast species isolated second most frequently from clinical infections (2, 8). This species occurs as a saprophyte on the human body. It is emerging as an opportunistic pathogen also in neonates (3). Generally, colonization at multiple sites precedes infection. Portals of entry include the respiratory tract, the genitourinary tract, and wounds (12). Since this species can even be isolated from environmental surfaces, the potential nosocomial acquisition must be also taken into account. Two risk factors for nosocomial acquisition are long duration of hospitalization and prior antimicrobial therapy (12).
Since fungemia is associated with high mortality rates, as the majority of fungemia patients have a rapidly progressive or ultimately fatal disease (4), immediate implementation of an appropriate antimycotic therapy is mandatory. Due to the commonly occurring innate or acquired resistance of C. glabrata to fluconazole, the widespread use of this azole might result in selection for this yeast species (13). In one study, C. glabrata has even been reported to be responsible for 75% of fungemias in patients receiving fluconazole (13). Therefore, rapid identification is essential, but up to now identification of yeast species other than C. albicans by commercially available identification kits or conventional methods has been both time-consuming and expensive. Typical morphological characteristics such as glossy, smooth, and dome-shaped colonies and typical small spherical yeast cells seen in the germ tube test may be an aid for rapid identification of C. glabrata, but when applied as the sole basis for identification, misidentifications may easily result. However, thus far there has been no practicable rapid test available. Therefore, we have developed a rapid test allowing identification of C. glabrata within 3 h.
This test takes advantage of the well-known ability of C. glabrata to rapidly hydrolyze trehalose (1-α-d-glucopyranosyl-1,1-α-glucopyranoside). This enzyme activity is frequently encountered in other yeast species isolated from human specimens, but by no other yeast species is hydrolysis of trehalose performed as rapidly as it is by C. glabrata (1).
C. glabrata isolates tested (n = 160) comprised isolates from patients with clinically relevant infections, including two reference strains (ATCC 90030 and DSM 11950). The percentages of isolates from the indicated clinical sources were as follows: 45.5%, urine; 28.4%, tracheal secretion and sputum; 11.4%, vagina; 7.9%, wound; 3.4%, blood; 3.4%, other. For testing purposes, the majority of C. glabrata isolates (72.5%) were taken from primary culture plates (Sabouraud dextrose agar containing 4% glucose; Oxoid, Wesel, Germany) incubated in ambient air for 18 to 24 h at 37°C. The rest of the isolates comprised subcultures incubated under the same conditions. Confirmation of species identification of all isolates tested was done by applying API ID 32 C (bioMérieux, Nürtingen, Germany) and checking micromorphology (Dalmau technique) and colony morphology for each isolate tested. Glucose generated by cleavage due to cell-bound trehalase was detected with a commercially available dipstick (Diabur-Test 5000; Boehringer, Mannheim, Germany) commonly used for measuring glucose in urine (range, 1 to 50 g/liter), particularly in patients with diabetes mellitus. For a description of the dipstick test principle, see http://134.225.167.114/NCBE/PROTOCOLS/-PRACBK/glucdet.html on the Internet. One colony of each C. glabrata isolate was emulsified in 50 μl of citrate buffer (0.1 M [pH 5.0]) containing 4% (wt/vol) trehalose (Merck, Darmstadt, Germany) for 3 h at 37°C. In 158 of the tested strains (n = 160; sensitivity, 98.8%) glucose could be detected by spotting 10 μl of this suspension onto a dipstick. After an additional incubation period of 3 h, retesting of the two isolates that were C. glabrata negative resulted in an overall positivity rate of 100%. When dipsticks were read after 2 min, colorimetrically estimated glucose concentrations (Fig. 1) ranged from 2.5 to 50 g/liter (2.5 to 5.0, 10, and 20 to 50 g/liter for 50, 25, and 25% of isolates tested, respectively).
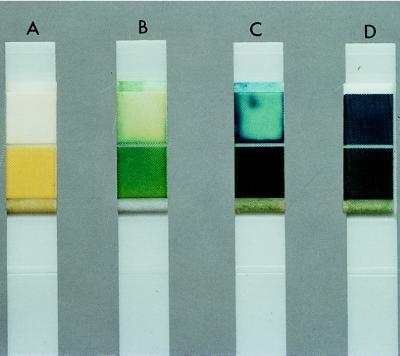
FIG. 1.
Typical results for dipsticks after 3 h of incubation. (A) Negative for non-C. glabrata yeasts (99.0%) tested (Table 1); (B and C) positive for C. glabrata, glucose concentration ranging from 2.5 to 50 g/liter; (D) positive control, i.e., glucose solution (50 g/liter).
To evaluate the specificity of our test system, commonly isolated yeast species potentially capable of utilizing trehalose as a sole carbohydrate source (5) were tested (Table 1). This ability was tested by determining the species ability to grow on yeast nitrogen base agar (Difco Laboratories, Detroit, Mich.) supplemented with 1% (wt/vol) trehalose after prior starvation of 3 days on a nonnutrient agar. The strains tested were isolated from patients with clinical infection or represented reference strains, and cultivation was performed as stated above. When testing these isolates by using the same procedure described above, only C. tropicalis isolates (8.8%), comprising 1.0% of all non-C. glabrata isolates, gave a positive result (range, 2.5 to 5 g/liter), thus resulting in an overall specificity of 99.1%. Elevation of the incubation temperature was without obvious influence (data not shown). Apart from C. tropicalis no other yeast species tested produced false-positive results under the described conditions, even when incubation was extended for up to 24 h.
TABLE 1.
Results of different yeast species tested for rapid hydrolysis of trehalose
| Yeast with potential to utilize trehalose (5) | Total no. of isolates tested | Included reference strain(s) | No. (%) of isolates showing:
|
|
|---|---|---|---|---|
| Growth on YNBa agar + 1% trehalose | Glucose production in our trehalase test (35°C, 3 h) | |||
| C. albicans | 120 | ATCC 10261, ATCC 48867, DSM 70014, DSM 5817 | 114 (94.9) | 0 (0) |
| C. dubliniensis | 11 | CBS 7987, CBS 7988 | 11 (100) | 0 (0) |
| C. famata | 15 | ATCC 26418 | 15 (100) | 0 (0) |
| C. glabrata | 160 | ATCC 90030, DSM 11950 | 160 (100) | 158b (98.8) |
| C. guilliermondii | 5 | ATCC 90877, DSM 70051, DSM 70052, DSM 6381 | 5 (100) | 0 (0) |
| C. lusitaniae | 16 | ATCC 34449 | 16 (100) | 0 (0) |
| C. parapsilosis | 20 | ATCC 90018, ATCC 22019, DSM 4237, DSM 70126, DSM 70125 | 20 (100) | 0 (0) |
| C. tropicalis | 34 | ATCC 90874, ATCC 90018, ATCC 750 | 34 (100) | 3 (8.8) |
| C. viswanathii | 5 | ATCC 28269 | 5 (100) | 0 (0) |
| C. zeylanoides | 2 | DSM 70185 | 2 (100) | 0 (0) |
| Pichia angusta | 2 | CBS 4732, CBS 1976 | 2 (100) | 0 (0) |
| Saccharomyces cerevisiae | 16 | DSM 1333, DSM 3797, DSM 4266 | 12 (75) | 0 (0) |
| Cryptococcus neoformans | 7 | CBS 950 | 7 (100) | 0 (0) |
| Total | 413 | 403 | 161 | |
YNB, yeast nitrogen base.
Retesting of the two negative isolates after an additional 3 h of incubation yielded positive test results.
To determine the influence of the culture medium on the test performance, we compared test results obtained with the following media (Table 1): Columbia blood agar base EH supplemented with 5% sheep blood, pH 7.5 (Difco Laboratories); Columbia blood agar base EH supplemented with 5% sheep blood, colistin, and nalidixic acid, pH 7.5 (Difco Laboratories); chocolate agar, pH 7.5 (Becton Dickinson, Cockeysville, Md.); Trypticase soy bouillon agar supplemented with 5% sheep blood, pH 7.5 (Becton Dickinson); CHROMagar, pH 6.6 (Mast Diagnostica, Rheinfeld, Germany); Sabouraud chloramphenicol agar, pH 6.3 (Sanofi Diagnostics Pasteur, Marnes-La-Coquette, France); Sabouraud dextrose agar, pH 5.6 (GIBCO BRL, Paisley, Scotland); Sabouraud dextrose agar containing 2% glucose, pH 5.6 (Merck, Darmstadt, Germany), with and without supplementation with 0.05% chloramphenicol; and Sabouraud dextrose agar containing 4% glucose, pH 5.6 (Oxoid), with and without supplementation with 0.05% chloramphenicol. Apart from Sabouraud dextrose agar from GIBCO BRL (lot 10E9440B), which gave negative results in 45 C. glabrata strains (28%), cultivation on these different isolation media was without any obvious influence on the test performance of C. glabrata isolates. From no other Sabouraud dextrose agar could false-negative results be observed when applying the rapid trehalase test. Therefore, we conclude that this effect is due to an unknown constituent present in this brand only.
Growth on different isolation media further showed influence on the test performance of C. tropicalis. Testing different Sabouraud dextrose media revealed that only strains of C. tropicalis are capable of producing glucose within 3 h. The positivity rate from Sabouraud dextrose media supplemented with 2% glucose increased slightly over time (23.6, 29.4, and 50.0% after 3, 6, and 24 h of incubation, respectively). The medium leading to the lowest positivity rate (8.8%) for C. tropicalis was, as mentioned above, Sabouraud dextrose agar supplemented with 4% glucose (pH 5.6). The addition of chloramphenicol showed no effect on the test’s outcome.
Usage of media containing blood resulted in an unacceptable rate of positive reactions for C. tropicalis (up to 68.5%) and C. albicans (2%), so yeasts cultivated on media supplemented with blood should not be used for performing the rapid trehalase test.
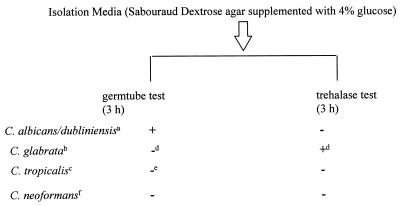
Therefore, evaluation on the isolation media to be used is strongly recommended when the rapid trehalase test is implemented. To avoid false-positive results, we recommend using the rapid trehalase test in combination with evaluation of the distinct morphological characteristics (small round cells without pseudohyphae production seen in the germ tube test) and typical colony appearance (smooth, glossy, and dome shaped) of C. glabrata. Thus, identification of C. glabrata can be achieved within 3 h. Only in cases of doubtful or conflicting results should further tests be necessary to achieve a definitive identification.
The most rapid means of presumptive identification of Candida species is to use commercially available chromogenic differential media for primary culture of human specimens, thereby particularly enhancing recognition of the presence of mixed yeast cultures. In a recently performed evaluation of CHROMagar, sensitivity and specificity for presumptive identification of C. albicans, C. krusei, and C. tropicalis in mixed yeast cultures exceeded 99% (7). However, since the color of C. glabrata colonies on CHROMagar varies from white to pink to purple, this medium is not suited for identification of this frequently encountered yeast species (7).
The two tests described so far for identification of C. glabrata are also based on its rapid hydrolysis of trehalose. The one developed by Stockman and Roberts has a yeast nitrogen base broth supplemented with trehalose, cycloheximide, and bromcresol green as an indicator of acidification due to utilization of trehalose (10). After incubation at 35°C for 1 h, an overall sensitivity of 89.3% and specificity of 96.8% for identification of C. glabrata was determined (10). This method has not been described in detail (size of inoculum, concentration of trehalose), and no further studies have been published so far. Our limited experience with this method revealed extreme difficulties in the adjustment of the buffer capacity of this system that was required to avoid false-positive results due to residual acid production induced by utilization of endogenically stored carbohydrates. Furthermore, preparation of the medium used is time-consuming and hazardous.
In the second assay, Durham yeast fermentation tubes (7.0 ml) supplemented with 1% (wt/vol) trehalose and bromcresol purple were inoculated with yeasts (McFarland 3 to 4) (5). The tubes, overlaid with melted paraffin, were incubated at 42°C. C. tropicalis and C. glabrata were the only two yeast species that fermented trehalose at this temperature within an incubation period of 24 h. Differentiation of C. glabrata from C. tropicalis should be achieved by demonstrating typical pseudohyphae on cornmeal medium in the case of the latter species. The overall sensitivity and specificity of this screening protocol were determined to be 97.8 and 95.8%, respectively. Major drawbacks of this test are (i) its complex, time-consuming performance, (ii) the large inoculum required (at least 50 colonies), and (iii) the relatively long time needed to perform it (at least 24 h).
In contrast to the disadvantages of the above-described assays, the striking advantages of our novel test for identification of C. glabrata are (i) results that are obtained within 3 h for the majority of strains, (ii) an easy-to-prepare and easy-to-perform format, (iii) small inoculum (permitting testing of as few as one colony), (iv) usage of commercially available products, and (v) cost-effectiveness (approximately $0.30 per isolate tested). For convenience the test can be performed in microtiter plates with removable strips. This represents an easy-to-perform and time-saving format, allowing simultaneous testing of a large number of isolates. Furthermore, respective plates can be prepared in advance and stored deep-frozen (≤−18°C) without loss of performance quality for at least 6 months.
Costs for identification of one clinical isolate of C. glabrata with commercially available identification kits vary (e.g., Vitek YBC card and API ID 32, approximately $5). The handling time needed for inoculation (transfer, inoculation, resealing the well: ∼10 s) and evaluation (removal of the dipstick, pipetting, reading: ∼30 s) of the rapid test is extremely short in comparison with conventional methods (3 min for inoculation, 3 min for evaluation [manual reading and searching of the code book] for API ID 32 C [bioMérieux]). Thus, implementation of the proposed screening scheme (Fig. 2) will greatly reduce (by up to 90%) both workload and costs for identification of yeasts.
FIG. 2.
Rapid identification scheme for C. glabrata. Footnote letters: a to c, color on CHROMagar (a, green [11]; b, varies from white to pink to purple; c, blue with pink halo); d, small (diameter, 3 to 5 μm) spherical yeast cells and typical colony morphology (6) have to be present; e, typical pseudohyphae; f, further testing, such as rapid urea broth test (9) (4 h), required. +, positive test; −, negative test.
Specificity is achieved by performing the reaction in a buffer of pH 5.0, thereby avoiding positive test results in the case of other trehalose-utilizing yeasts, and by using a detection method not influenced by traces of glucose present in each biological specimen. In the case of false-negative test results for C. glabrata (1.2%)—i.e., no production of glucose within 3 h but characteristics (colony morphology, germ tube test) indicative of C. glabrata (6)—prolongation of incubation for an additional 3 h and subsequent retesting are possible.
The proposed screening scheme (Fig. 2) offers an ideal supplement to rapid identification of C. albicans using the germ tube test and enables rapid identification of more than 95% of all yeast isolates from human specimens when the CHROMagar and/or germ tube test is used in combination with our trehalase test, thus facilitating the diagnosis of fungal infections.
Acknowledgments
We thank K. Tintelnot and M. L. Kerkmann for providing yeast strains.
H.P.-L. and N.S. contributed equally to this work.
REFERENCES
- 1.Barnett J A, Payne R W, Yarrow D. Yeasts: characteristics and identification. 2nd ed. Cambridge, Mass: Cambridge University Press; 1990. p. 167. [Google Scholar]
- 2.Borg-von Zepelin, M., H. Eiffert, M. Kann, and R. Rüchel. 1992. Changes in the pathogen spectrum of fungi: isolation results of clinical specimens in the period from October 1987 to March 1992 in the University Clinics of Göttingen. Mycoses 35(Suppl.):21–25.
- 3.Glick C, Graves G R, Feldman S. Torulopsis glabrata in the neonate: an emerging fungal pathogen. South Med J. 1993;86:969–970. doi: 10.1097/00007611-199308000-00026. [DOI] [PubMed] [Google Scholar]
- 4.Komshian S V, Uwaydah A K, Sobel J D, Crane L R. Fungemia caused by Candida species and Torulopsis glabrata in the hospitalized patient: frequency, characteristics, and evaluation of factors influencing outcome. Rev Infect Dis. 1989;2:379–390. doi: 10.1093/clinids/11.3.379. [DOI] [PubMed] [Google Scholar]
- 5.Land G, Burke J, Shelby C, Rhodes J, Collett J, Bennett I, Johnson J. Screening protocol for Torulopsis (Candida) glabrata. J Clin Microbiol. 1996;34:2300–2303. doi: 10.1128/jcm.34.9.2300-2303.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marks M I, O’Toole E. Laboratory identification of Torulopsis glabrata: Typical appearance on routine bacteriological media. Appl Microbiol. 1970;19:184–185. doi: 10.1128/am.19.1.184-185.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Odds F C, Bernaerts R. CHROMagar Candida, a new differential isolation medium for presumptive identification of clinically important Candida species. J Clin Microbiol. 1994;32:1923–1929. doi: 10.1128/jcm.32.8.1923-1929.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfaller, M. A. 1995. Epidemiology of candidiasis. J. Hosp. Infect. 30(Suppl.):329–338. [DOI] [PubMed]
- 9.Pincus D H, Salkin I F, McGinnis M R. Rapid methods in medical mycology. Lab Med. 1988;19:315–320. [Google Scholar]
- 10.Stockman L, Roberts G. Abstracts of the 85th Annual Meeting of the American Society for Microbiology, 1985. Washington, D.C: American Society for Microbiology; 1985. Rapid screening method for the identification of C. glabrata, abstr. F-80; p. 377. [Google Scholar]
- 11.Sullivan D, Coleman D. Candida dubliniensis: characteristics and identification. J Clin Microbiol. 1998;36:329–334. doi: 10.1128/jcm.36.2.329-334.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vazquez J A, Dembry L M, Sanchez V, Vazquez M A, Sobel J D, Dmuchowski C, Zervos M J. Nosocomial Candida glabrata colonization: an epidemiologic study. J Clin Microbiol. 1998;36:421–426. doi: 10.1128/jcm.36.2.421-426.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wingard J R, Merz W G, Rinaldi M G, Miller C B, Karp J E, Saral R. Association of Torulopsis glabrata infections with fluconazole prophylaxis in neutropenic bone marrow transplant patients. Antimicrob Agents Chemother. 1993;37:1847–1849. doi: 10.1128/aac.37.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]