Abstract
The large intestine, with its array of crypts lining the epithelium and diverse luminal contents, regulates homeostasis throughout the body. In vitro crypts formed from primary human intestinal epithelial stem cells on a three-dimensional shaped hydrogel scaffold replicate the functional and architectural features of in vivo crypts. Collagen scaffolding assembly methods are provided, along with the microfabrication and soft lithography protocols necessary to shape these hydrogels to match the dimensions and density of in vivo crypts. Additionally, stem-cell scale-up protocols are provided so that even ultrasmall primary samples can be used as starting material. Initially, these cells are seeded as a proliferative monolayer over the shaped scaffold and cultured as stem/proliferative cells to expand the cell number and cover the scaffold surface with the crypt-shaped structures. To convert these immature crypts into fully polarized, functional units with a basal stem cell niche and luminal differentiated cell zone, stable, linear gradients of growth factors are formed across the crypts. These gradients imposed along the crypt long axis support a basal stem cell niche with transit amplifying cells migrating upward along this axis to form a luminal differentiated cell zone as in vivo. This platform supports the formation of a multitude of chemical gradients across the crypts including those of growth and differentiation factors, inflammatory compounds, bile and food metabolites, and bacterial products. All microfabrication and device assembly steps are expected to take 8 days if performing for the first time, with the primary cells cultured for 12 days to form mature in vitro crypts. Compatibility of the in vitro crypt arrays with standard microscopy methods, as well as the accessible luminal and basal reservoirs, facilitate mechanistic studies and screenings with sufficient replicates and statistical power to test hypotheses or survey large numbers of compounds (and bacteria) for their impact on epithelial homeostasis.
Introduction
The intestines house a multitude of growth factor, metabolite, gas, and extracellular matrix gradients across the epithelium that govern the composition and organization of various cell types, thus influencing organ-level self-renewal and function.1,2 The colonic epithelium is organized into a series of crypts, invaginations present throughout the length of the organ that are responsible for protection and maintenance of a stem cell niche at each crypt base.3,4 Intestinal stem cells are supported by Wnt (Wingless-related integration site) signaling agonists supplied by underlying stromal cells in vivo.5,6 The stem cells give rise to a population of proliferative transit amplifying cells that divide a finite number of times before terminally differentiating into absorptive or secretory (e.g., goblet and enteroendocrine) lineages at the luminal surface.7,8 These stem cell fate decisions are guided by food and bacterial metabolites that regulate Notch signaling in the epithelial cells.9 For example, butyrate, a short chain fatty acid produced through bacterial fermentation, has been shown to increase Notch signaling and absorptive colonocyte differentiation while decreasing proliferative stem cell activity.10,11 These studies connecting diet, gut microbiota, and crypt homeostasis have far-reaching implications in our understanding of the pathophysiology of inflammatory bowel diseases and colorectal cancer.12,13 As such, the development of physiologically relevant and analytically accessible intestinal models that enable increased throughput and high content information regarding crypt cellular behaviors is of high importance.14,15
The advent of protocols for long-term culture of primary intestinal stem cells and organoids has significantly advanced our understanding of stem cell-driven epithelial renewal.16,17 Compared to in vitro models based on immortalized cell lines, which remain gold standards in the pharmaceutical industry, primary cell-derived organoids recapitulate the complex interplay between different epithelial cell types and can be biobanked for population screens, as the cells retain the genotype and replicate physiologic properties displayed by the original host.18 However, even though organoids contain both stem and differentiated intestinal cells, they lack phenotypic compartments that can be experimentally modulated or controlled, rendering studies of the orderly spatial and temporal transitioning of cells between these regions challenging. Moreover, these culture systems do not possess accessible luminal regions (representing the lumen of the intestine) that are easily sampled or assayed. Future innovations and breakthroughs within the field will be strengthened by models in which these specialized culture techniques are engineered to accurately replicate intestinal architecture and physiology, and in a reliable and robust format that is usable in standard biomedical research laboratories. Primary roles of the large intestine include housing the microbiome, maintaining salt and water homeostasis, and serving as a waste storage compartment.2 These attributes are supported by low to zero flow rates of the luminal contents, a differentiated epithelial cell zone providing a secreted mucus barrier while supporting ion transport across the epithelium, and a protected stem cell niche in which rapidly dividing cells supply replacements for the short-lived differentiated cells. An in vitro culture system that supports these functions and enables their investigation as well as ready customization for a range of experimental questions is critical. Desired platform attributes include assay compatibility (such as optical transparency for microscopy and accessible basal/luminal tissue compartments for sampling and environmental manipulation), proper architecture to enable distinct cell zones and direct cell flow between the zones, and facile establishment of stable chemical and gas gradients to support tissue maturation while hosting luminal microbes.
Overview of the procedure
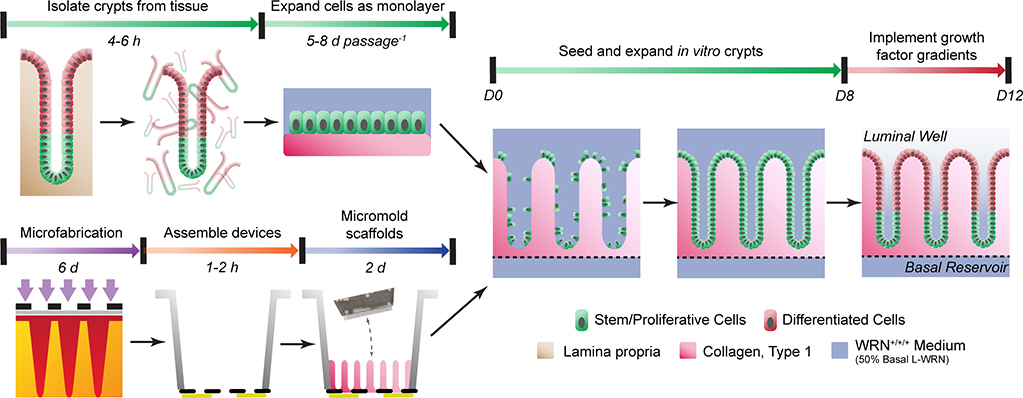
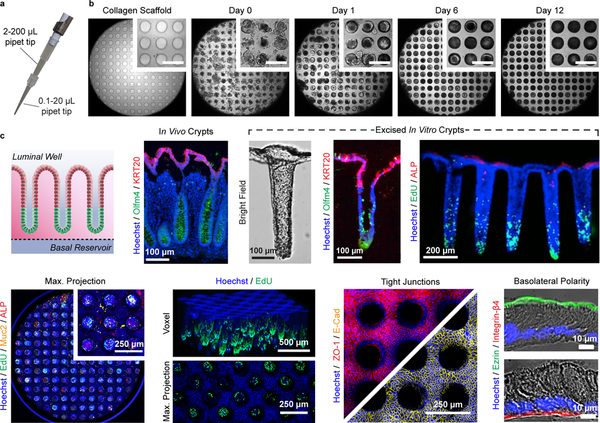
Due to longstanding obstacles in the culture and analysis of in vitro intestinal epithelia, we have developed a shaped scaffold supporting a primary intestinal epithelial cell monolayer to recapitulate both the functional and architectural features of in vivo intestinal crypts. The platform provides a microscale yet three-dimensional physiological mimic of the intestine that is suitable for established analytical assays and high-content information acquisition. This protocol will detail the microfabrication and soft lithography approaches utilized to fabricate the three-dimensional shaped hydrogel scaffolding and device cassettes, as well as the corresponding primary cell culture methods necessary to achieve a full simulacrum of the intestinal epithelium within each device (Fig. 1).
Fig. 1 |.
Workflow and timeline of the protocol.
Primary intestinal epithelial cells must initially be isolated and cultivated from in vivo tissue prior to the formation of in vitro crypts. While the organoid technique is the mainstream method for in vitro expansion of primary intestinal epithelial cells,19 we recommend the monolayer hydrogel culture technique for its consistency and ease of use (Steps 1–27).20 Compared to early monolayer propagation techniques, hydrogel monolayer culture enables serial passaging and in vitro expansion of primary intestinal epithelial cells without loss of proliferative capacity or genomic stability, and moreover does not require a feeder layer of cells to be co-cultured.21 Beyond intestinal epithelial cells, these expansion protocols may have applications to other primary cell and tissue types that are sensitive to the physicomechanical properties of their local environment and are supported by a collagen-rich basement membrane. Although not tested, these tissues types include lung epithelia, renal tissue (e.g., proximal tubule), hepatocytes, bile and pancreatic ducts, and endothelial tissue (e.g., blood and lymph). Collagen hydrogel scaffolds for primary intestinal cell expansion are formed by neutralizing and gelling acid-solubilized collagen within 6-well culture plates (Box 1), over which, living crypts isolated from primary intestinal tissue are plated (Box 2). These neutralized collagen scaffolds possess the appropriate biochemical cues and mechanical properties that, in conjunction with appropriate media supplementation (Table 1), enable long-term propagation and self-renewal of in vitro cultures. These cultures are ready for passage and sub-cultivation every 5–8 d when the cells reach ≥80% confluency over each scaffold. In brief, this process involves physical removal of the scaffold with cells from the culture plate, followed by its digestion in Type IV collagenase (Steps 10–21). The cells are broken up into smaller colonies by mechanical agitation in the presence of EDTA (Steps 22–24), and thereafter plated over fresh neutralized collagen hydrogels for further expansion (Steps 25–27). This process may be repeated indefinitely until the desired number of cells are obtained, though we recommend discarding cultures after 15 passages (ca. 75–120 days in culture), before the possible onset of genetic drift or chromosomal alterations.
Table 1 |.
Formulations of primary cell culture media
| Component | Stock Concentration | Volume Required for 500 mL (Final Concentration) | |||
|---|---|---|---|---|---|
| Maintenance Medium (MM) | Expansion Medium (EM) | Stem Medium (SM) | Differentiation Medium (DM) | ||
| L-WRN conditioned mediumb | N/A | 250 mL (50 % v/v) | 250 mL (50 % v/v) | 250 mL (50 % v/v) | --- |
| Advanced DMEM/F12 | 1× | 250 mL (50 % v/v) | 250 mL (50 % v/v) | 250 mL (50 % v/v) | 500 mL (100 % v/v) |
| Glutamax | 100× | 5 mL (1×) | 5 mL (1×) | 5 mL (1×) | 5 mL (1×) |
| HEPES | 1 M | 5 mL (10 mM) | 5 mL (10 mM) | 5 mL (10 mM) | 5 mL (10 mM) |
| Primocin | 50 mg mL−1 | 500 μL (50 μg mL−1) | 500 μL (50 μg mL−1) | 500 μL (50 μg mL−1) | 500 μL (50 μg mL−1) |
| N-acetylcysteine | 1 M | 500 μL (1 mM) | 500 μL (1 mM) | 500 μL (1 mM) | 500 μL (1 mM) |
| Epidermal growth factor, murine | 250 μg mL−1 | 100 μL (50 ng mL−1) | 100 μL (50 ng mL−1) | 100 μL (50 ng mL−1) | 100 μL (50 ng mL−1) |
| Nicotinamide | 1 M | --- | 5 mL (10 mM) | --- | --- |
| B27 | 50× | 10 mL (1×) | 10 mL (1×) | --- | --- |
| Gastrin | 1 mg mL−1 | 12.5 μL (10 nM) | 12.5 μL (10 nM) | --- | --- |
| Prostaglandin E2 | 1 mM | --- | 5 μL (10 nM) | --- | --- |
| A 83–01 | 5 mM | 50 μL (500 nM) | --- | 50 μL (500 nM) | 50 μL (500 nM) |
| SB202190 | 30 mM | 50 μL (3 μM) | 50 μL (3 μM) | --- | --- |
| Y-27632 | 10 mM | 500 μL (10 μM) | 500 μL (10 μM)c | --- | --- |
| Heat-inactivated FBS | N/A | 0 mL (10 % v/v) d | 0 mL (10 % v/v) d | 0 mL (10 % v/v) d | 50 mL (10 % v/v) |
Stock concentration in parentheses. Refer to Reagent Preparation section for aliquoting instructions.
Prepared by culture of the L-WRN cell line (CRL-3276, ATCC, RRID:CVCL_DA06, https://scicrunch.org/resolver/RRID:CVCL_DA06) and harvesting of media containing Wnt3a, R-spondin 3, and noggin. Refer to the protocols published by Stappenbeck and colleagues for details.19,70
Y-27632 is always present in MM, though is only included during the initial 48 h of culture in EM.
FBS is included in L-WRN conditioned medium at a concentration of 20 %, and no additional FBS is added to MM, EM, or SM. Heat inactivation is conducted in-house by incubating at 56 °C for 25 min.
Assembly of the in vitro crypt device cassettes requires several key manufacturing steps that can be grouped into the following categories: fabrication of the polydimethylsiloxane (PDMS) stamps that will be utilized for shaping the scaffolds (Steps 27–64), modification of cell culture inserts (Steps 65–72), and shaping of crosslinked collagen via PDMS micromolding (Steps 73–91). Fabrication of the PDMS stamps is based on conventional photolithography using a negative photoresist formulated in-house (Steps 27–45),22 and soft lithography for replica molding of the PDMS stamps (Steps 46–59). The PDMS stamps are surface modified with poly(ethylene glycol) to minimize adhesion of collagen during the micromolding process (Steps 60–64).23 For each cell culture insert, a hydrophilic polytetrafluoroethylene (PTFE) membrane is installed along with a superimposed impermeable film possessing a central 7 mm2 diffusion window (Steps 65–72). These modifications enable a self-sustaining, steep gradient of stem-cell supporting growth factors to be formed across the depth of the diffusion window, establishing a stem/proliferative niche at the base of each in vitro crypt and a differentiated cell zone at the crypt luminal end. Collagen hydrogel shaping is conducted within each modified insert by pressing the PDMS stamps into a liquid collagen solution in the presence of covalent crosslinking reagents at elevated pressure (Steps 73–91). These shaped scaffolds match the architecture of human in vivo crypts, and analogous to the neutralized collagen hydrogels, possess the requisite properties to support primary intestinal cell expansion and self-renewal along their surface.
Once primary intestinal epithelial cells are seeded within the in vitro crypt device cassettes (Steps 92–100), the cells are expanded for 8 d. Over this time, the cells should become 100% confluent over the surface of the scaffold and grow as a monolayer along the internal microwell surface (Steps 101–102). Once confluent, stem cell supporting growth factors are removed from the luminal compartment of each cell culture insert. The large volume of the basal and luminal reservoirs relative to that of the diffusion window and overlying shaped hydrogel enable these reservoirs to act as an infinite sink and source on time scales of 1–2 d. Thus a steep, linear, growth-factor gradient is established along the in vitro crypt long axis between the source and sink, or basal and luminal, reservoirs. After 4 d of growth under this gradient (with reservoir medium replenishments occurring every 24 h), the in vitro crypts have formed and are ready for assays (Steps 103–105). The final product is an array of living in vitro crypts that possess a stem/proliferative niche within the base of each microwell, and a differentiated cell zone at the luminal plane. If desired, the in vitro crypts can be cultured for greater lengths of time (at least 32 d, the longest time tested) in this manner, as they exhibit tissue-level self-renewal capabilities.24
Experimental design
Selection and sourcing of primary intestinal epithelial cells
Human adult intestinal epithelial stem cells are the recommended cell source for generating the model detailed in this protocol. While our primary focus is on colonic crypts, human small intestinal stem cells can also be used for in vitro crypt generation with slight modifications in medium formulation and scaffold design.25 The stem cells can be isolated from patient biopsy specimens obtained from the clinic, or cadaveric donor specimens obtained from a tissue or organ bank. To date, our research group has successfully expanded cells from 19 clinical biopsy specimens utilizing the methods described herein,20,24–26 including those from diseased patients,27,28 in addition to five cadaveric donor specimens (one published, remaining are unpublished).29 In general, cadaveric donor specimens can yield far more cells than the average clinical biopsy, where specimens are on the order of 7.5–50 mm2 and contain ca. 500–2000 crypts, as larger tissue samples or even whole organs can be obtained. However, variable post-mortem intervals (PMIs) between tissue banks and repositories will affect the yield of viable cells, which must be considered during cadaveric tissue acquisition. Although the stem cells can be expanded indefinitely in either organoid or monolayer formats,16,20 we recommend discarding the cells after a certain passage number (e.g., 15–20) due to the onset of genetic drift and chromosomal abnormalities observed at higher passage numbers. Primary cells derived from different donors are known to behave differently in culture depending on the age, gender, and health of individuals from whom the tissue was derived. Furthermore, region-specific (e.g., ascending, transverse, descending, sigmoid, and rectal) cellular behaviors are likely to be observed. Each of these factors must be considered during the interpretation of experimental results. In addition to primary human cells, intestinal stem cells derived from animal donors (e.g., murine, porcine, rat) can be utilized though they will behave differently in their self-renewing rate, life span after differentiation, and response to growth factors. For example, we have utilized murine colonic epithelial stem cells to generate in vitro crypt arrays, with the murine in vitro crypts exhibiting a much shorter life span (2 d) than the human in vitro crypts (≥32 d).24,30 Induced pluripotent stem cells (iPSCs), which can be differentiated into intestinal cells, are an enticing source as they can be obtained from primary subjects via minimally invasive procedures, thereby potentially expanding the diversity of experimental subjects. However, once differentiated into colonic epithelial cells, genetic profiles of the obtained tissue more closely resemble those of fetal human intestine rather than those of adult intestine.31 To date, iPSC-derived intestinal epithelial cells have not been tested as a cell source in formation of in vitro crypts, through merit exploration. Tumor cell lines should be avoided because they are genetically mutated, may not respect cell-cell boundaries, do not possess normal stem cells, and generally do not respond to physiological stimuli in the same manner as primary cells.
Utilization of collagen hydrogels
Native extracellular matrix (ECM) is composed of water, polysaccharides, and structural proteins, which predominantly include fibrous collagens, elastins, fibronectin, and laminin.32 Integrin proteins displayed at the basal surface of the cells are known to recognize these matrix proteins through common Arg-Gly-Asp (RGD) motifs, with their interactions dynamically modulated through mechanical stimuli.33 Here, a supportive ECM provides the necessary biophysical cues for primary intestinal epithelial stem cells to attach, self-renew, proliferate, and differentiate,1 which must possess the following essential parameters: presence of native ECM ligands, proper mechanical stiffness, and a high porosity that permits diffusion of growth factors to the basal side of the cells.2 A variety of isolated hydrogel matrices have been examined by our research group and others, with collagen ultimately being selected as meeting the above requirements and supporting the self-renewal of intestinal epithelial stem cells in a planar monolayer format.17,20,34 However, when these unmodified collagen scaffolds are shaped into three-dimensional microwell structures, the hydrogel does not possess sufficient mechanical strength to resist cell-induced deformation.24,25,30 Therefore, a carbodiimide coupling (EDC/NHS) strategy is utilized to strengthen the collagen hydrogel by crosslinking between adjacent carboxylic acid and amine groups of collagen fibers during the micromolding process.35 The unique advantage of EDC/NHS chemistry is its zero-length product (i.e., molecules are not incorporated into the collagen hydrogel), resulting in the scaffolds being virtually free of reactive or toxic byproducts after extensive washing in deionized water.36 Once this process is complete, the crosslinking reaction may modify the ECM ligands that mediate cell adhesion. To compensate for this loss, a monolayer of ECM molecules (e.g., human collagen) is coated over the surface of the crosslinked hydrogel to render it suitable for attachment and propagation of primary intestinal epithelial cells.24
Formation of stable chemical gradients
To establish stable and steep chemical gradients across the in vitro crypt array, the basal and luminal reservoirs must function as an infinite source and sink of stem cell supporting growth factors. Continuous perfusion through the basal and luminal reservoirs is frequently used to achieve these conditions, though the use of a fluidic flow/pump system while maintaining sterility represents a substantial obstacle for many research groups of varying technical backgrounds. To avoid constant perfusion, we used a simple strategy to reduce the available area for transport across the scaffold/membrane from 113 mm2 (standard 12-well cell culture insert diameter) to 7 mm2 through attachment of an impermeable film.24 This film, possessing a 3 mm dia. window in the center, decreases the area available for diffusion so that the basal and luminal chambers can be approximated as an infinite source and sink so long as media replenishments occur every 24 h. This gradient was confirmed by empirically measuring the diffusion of a model growth factor surrogate compound, fluorescein-dextran (40 kDa), across the scaffold and membrane. For reference, the molecular weights of Wnt3a, R-spondin, and noggin are: 39.7 kDa, 40.0 kDa, and 46 kDa, respectively. At 0 h, the concentration of fluorescein-dextran compound was 100 μg mL−1 in the basal compartment (source) and 0 μg mL−1 in the luminal compartment (sink), while at 24 h, the concentrations were 96.6 ± 3.0 μg mL−1 in the basal compartment (source) and 2.9 ± 0.7 μg mL−1 in the luminal compartment (sink).24 Thus, stable and steep chemical gradients can be established across the scaffold by simply limiting the diffusion area and replenishing the basal and luminal reservoirs every 24 h, an operation not requiring active fluid flow or mechanical pumps – only pipetting. Alterations of this optimized diffusion area are not recommended for 12-well insert dimensions, as the conditions for a semi-infinite boundary between compartments will not be satisfied and the formation of steep chemical gradients supporting a basal stem cell niche will not be possible. Adaptions to other insert dimensions (e.g., 6-well, 24-well, etc.) will require optimization, which can be modeled by calculating the transport of diluted species between compartments using Fick’s law.30,37
Comparison with alternative models
The human gastrointestinal tract has been modeled by animal systems, explanted tissue cultured ex vivo, and in vitro platforms generated from both immortalized and primary cells. Animal models are powerful tools for studying intestinal physiology, function, and disease, though suffer from ethical considerations, time consuming protocols, high operating costs, and in some cases, questionable relevance to human physiology.38,39 Ex vivo platforms can be readily generated from explanted human intestinal tissue, and are also capable of preserving complex, tissue-level interactions that are absent in simpler culture systems, though rapid cell death and deterioration limit their use.40 Due to these shortcomings, in vitro models have undergone aggressive development, facilitating the study of complex in vivo phenomena in a simplified and controlled manner. Early efforts were made to create experimental platforms with immortalized cell lines derived from intestinal tumors (e.g., Caco-2), which have been extensively utilized to predict human intestinal functions, such as permeability to drugs, toxins, and dietary metabolites.41 These immortalized cell lines are cost effective, easy to use, and consistent in their function, though they are not representative of all of the major cellular subtypes of the intestinal epithelium, nor can they replicate the important interactions between these subtypes, and thus do not accurately replicate physiology.42 Primary intestinal stem cells derived from human tissue specimens are capable of offering more physiologically relevant data for in vitro intestinal models, as they are capable of differentiating into all of the major cell types present in the in vivo tissue.2 The organoid model was the first in vitro platform to offer sustained culture of human intestinal stem cells, transforming intestinal research and opening exciting new possibilities for investigating intestinal physiology.16 Within this platform, primary intestinal stem cells are embedded within an extracellular matrix (ECM) enriched with growth factors (e.g., Wnt3a, R-spondin, noggin, epidermal growth factor) and nutrients. These cells proliferate and assemble into spherical structures possessing a hollow lumen, which fills with mucus and cellular debris as some of the cells differentiate into distinct compartments of absorptive and secretory lineages. However, accessing the interior of the organoid is severely restricted without specialized microinjection equipment, rendering the precise sampling and manipulation of luminal contents difficult.
The luminal compartment can be made accessible by culturing these cells in a planar format over a porous substrate. Such monolayer systems have been developed by culturing primary intestinal stem cells over hydrogel scaffolds formed within cell culture inserts (e.g., neutralized or crosslinked collagen hydrogels)25,43 and ECM-coated porous membranes,29,44,45 followed by spontaneous or forced differentiation. However, in most of these platforms the spatial organization and temporal transitioning of cells between phenotypic regions known to exist in vivo is absent. Intriguingly, monolayer culture of primary intestinal epithelial cells over porous membranes in the presence of an air-liquid interface (ALI) has been demonstrated by multiple research groups to produce differentiated lineages with a highly columnar morphology, similar to in vivo, with randomly dispersed proliferative cell regions.21,46 These cells become “squamous” or flattened in appearance once fluid is re-added to the culture system. Though proven useful, the necessity of maintaining an ALI for proper cell morphology and physiology results in significant experimental constraints.46
Recent organ-on-chip and microphysiological system (MPS) devices have been engineered to study more complex features of intestinal physiology, including the influences chemical gradients, mechanical forces, cellular migration, and three-dimensional anatomical structure on tissue-level function.24,25,37,47 The most established are microfluidic intestine-on-chip platforms, created by culture of tumor cells or organoid-derived stem cells within a polydimethylsiloxane (PDMS)-based microfluidic device.47 This platform separates an epithelial cell layer from an endothelial cell layer via a porous PDMS membrane, and applies mechanical forces through fluid and gas regulation systems.48 Microfluidic intestine platforms have been utilized for a number of applications, including the study of radiation injury,49 investigation of microbiome interactions,50,51 analysis of mucus accumulation,52 and interactions with MPS that mimic other tissues.53 On the other hand, the human in vitro crypt platform described herein does not focus on mechanical influences, but was designed to study how the microscale architecture of the intestine and its numerous and dynamic chemical gradients influence cellular behavior, stem cell fate/differentiation, and directional migration.24 While several culture systems possessing scaffolds that architecturally mimic crypts and/or villi have been reported, most utilize immortalized cell lines and are not compatible with primary cell culture.54–59 In particular the scaffolding does not provide appropriate environmental conditions, for example stiffness, to support growth of primary human cells. The in vitro crypt platform possesses a micro-engineered, three-dimensional scaffold that mimics the architecture of the epithelium and permits sustained culture of primary intestinal stem cells along with their differentiated lineages. The open and accessible luminal and basal compartments support the establishment and manipulation of chemical gradients across the tissue, a feature absent in most MPS, and without the need for fluid or gas control systems.
Each of the above intestinal models present their own advantages and limitations (Fig. 2). In general, high throughput proliferation, cytotoxicity, and viability assays can be readily performed using the organoid model as they are easily generated in standardized 96-well plate formats.10,60 However, organoids fall short for transport, metabolism, secretion and infection studies, which monolayer culture systems, in conjunction with porous membranes, are more suited for due to their accessible luminal and basal compartments.20,45,61,62 High-content image acquisition is also more easily performed on planar monolayers of cells, as they lack the need for multiplane imaging and z-stack image reconstruction. MPS platforms are generally more sophisticated than either organoid or monolayer culture systems, and as such, are more suited for specialized studies where the unique data available justify the increased effort in their setup and execution. For example, the human in vitro crypt system is the only one of the above examples that can be used to study how the perturbation of chemical gradients impact stem cell compartmentalization and migration.24 End users will be responsible for leveraging these considerations depending on the hypotheses being tested.
Fig. 2 |.
Comparison of in vitro culture platforms for primary intestinal epithelial cells.
Target audience and applications
This three-dimensional replica of the intestinal epithelium was developed to investigate questions relevant to intestinal biology in the areas of basic science and personalized medicine, and as such, researchers in the fields of stem cell biology, mucosal immunology, gastrointestinal research, and regenerative medicine utilize the system. These fields have long relied on immortalized cell lines (e.g., Caco-2, IEC-6, HT-29) and animal models (e.g., rat, murine, porcine) for molecular and physiological understandings of intestinal disease states, though each of these systems face significant challenges, most notably in their relevance to human physiology. The in vitro crypt model reports physiologically relevant outcomes from primary cells that retain native genetic information, cellular identity and physiologic functions of their original host tissue. This will be attractive not only to academic researchers, but also to nutrition, and pharmaceutical industry sectors that seek to improve the accuracy of pre-clinical trials.15
The influence of microbes and their metabolites, such as short chain fatty acids (SCFAs), on organismal health and tissue self-renewal is of tremendous interest among gastrointestinal researchers, and these features have been successfully modeled using the in vitro crypt platform. SCFAs are small, organic molecules produced through bacterial fermentation of dietary fiber, and include acetate, propionate, and butyrate, which are observed at millimolar levels within the colonic lumen in vivo.63 SCFAs are known to inhibit histone deacetylase activity, activate G-protein coupled receptors, and increase Notch signaling, ultimately acting as a primary energy source for differentiated colonocytes and inhibiting stem/progenitor cell activity.10 Within the in vitro crypt platform, butyrate increased the expression of absorptive colonocytes at the luminal surface to the greatest extent of the SCFAs tested, and diminished the activity of stem/progenitor cells, localizing their compartmentalization toward the base of the crypts.24,37 These results are in agreement with previous studies on the proliferation-inhibitory and neoplasia-preventing effects of butyrate.64 Beyond these selected SCFAs, a plethora of other metabolites and dietary supplements could be analyzed for their effects on crypt cell behavior, transport, and biotransformation.
Gut associated microbes including probiotic, commensal, or pathogenic microbes (e.g., bacteria, fungi, virus, archaea) can be co-cultured to directly on the intestinal platform to investigate their biological or physical interactions with host cells with respect to bacterial adhesion, motility, or viral life cycles. To maintain viability of both human colonic epithelial cells and the co-cultured microbes, a sharp oxygen gradient must be formed along the crypt length as is present in in vivo since the majority of gut associated bacteria are obligate anaerobes that cannot grow in the presence of oxygen. Using the planar and shaped collagen hydrogel culture systems, an obligate anaerobic commensal and probiotic bacteria Bifidobacterium adolescentis was successfully co-cultured with human colon cells on a modified platform with an anaerobic luminal reservoir but normoxic basal reservoir.65 Since the gut microbiome is altered in a wide range of human diseases, co-culture of autologous or therapeutic gut microbiota with the human colon cells has the potential to provide a platform for personalized medicine.
Pathogenesis of intestinal diseases such as colorectal cancer or inflammatory bowel disease can also be modeled with this platform, with comparisons to normal (i.e., healthy) conditions possible using patient-derived cells. Colonic carcinogenesis has previously been recapitulated using primary human colonic epithelial cells in organoid culture, in which colon cancer associated genes were selectively mutated using CRISPR/Cas9 gene editing.66,67 The in vitro crypt model could provide a more physiologically relevant platform in comparison by implementing the crypt microstructure and/or chemical gradients during healthy in vitro culture, carcinogenesis, and tumor progression. The long-term culture capabilities permit the observation of cancer-related mutations and DNA instabilities over time, and with respect to crypt location. Various factors that are known or suspected to be involved in tumorigenesis, including nutrients, metabolites and microbes, can be manipulated within the luminal or basal compartments for testing within this platform, owing to its open and accessible design. Using patient-derived healthy and diseased cells, the design and testing of personalized treatments can be envisioned.
Advantages
The in vitro crypt platform enables the study of a complex, self-renewing tissue that possesses the architectural and phenotypic cell distributions present within the large intestine in vivo while preserving the ability to sample and manipulate the basal and luminal tissue compartments for tailored experiments and analyses. The cell phenotypic compartmentalization that is observed and controlled within the model can be reliably reproduced through the use of easily formed growth factor and metabolite gradients, which is uncontrolled in other intestinal models where various cell types are randomly distributed, exist in inappropriate relationships, and in nonphysiologic dimensions. The self-renewing monolayer and in vitro crypt platforms are modular in nature, with tailorable cellular microenvironments for optimal hypothesis testing. For example, the platform has been used to form and assay barrier function of thick human mucus layers,68 to investigate epithelium-anaerobe interactions under an oxygen gradient across the in vitro crypts,65 and to examine peripheral blood mononuclear cell behavior in the presence of an immune-competent intestinal epithelium.68
In regards to its analytical merits, the fully accessible luminal and basal reservoirs ensure flexibility for sampling, transfer, and manipulation of culture media at any time point during culture. The platform uses industry-standard dimensions which permit high-throughput assays to be designed, as well as compatibility with those that employ automated fluid/liquid handling. We have intentionally integrated optically transparent materials throughout the device to render the platform compatible with fluorescence assays including various imaging modalities, as both the crosslinked hydrogel and basal PTFE membrane exhibit transparencies that support in situ imaging. The cassettes are demonstrated to be compatible with confocal fluorescence microscopy in conjunction with long-working distance objectives, z-stacking, and appropriate post-processing (i.e., deconvolution).37 Further scaling and the adoption of rapid, three-dimensional microscopy techniques (e.g., spinning disc confocal, light sheet) will undoubtedly increase data acquisition speeds. If these setups are not available, or greater resolution is desired, the in vitro crypts can be imaged individually by manual excision from the scaffold using a dissecting needle, followed by their supine placement over a glass coverslip.24,37,65 Beyond imaging, we have coupled this platform with several downstream analytical assays, including transport/permeability studies,4,8–10,12 drug metabolism,43,69 commercial plate reader assays (e.g., ELISA, CellTiter Glo),20,24–26,30 mass spectrometry,26 and RNAseq.43
Limitations and development opportunities
Throughout the maintenance of primary intestinal epithelial cells over neutralized collagen hydrogels and the establishment of in vitro crypt arrays, we utilize L-WRN conditioned medium as the source of Wnt3a, R-spondin 3, and noggin.19 These growth factors, along with recombinant EGF and other media supplements, function to maintain stem/proliferative activity of primary cells, enabling their long-term propagation in vitro. To date, this source of growth factors has been most consistent in regards to potency and reproducibility for primary cell expansion,70 and to be more cost-effective than purchasing recombinant Wnt3a, R-spondin, and noggin. However, we recognize that utilization of this source does limit control over the cell culture medium formulation, particularly if users wish to separately control these growth factor levels. Furthermore, these proteins are difficult to quantify directly in the presence of high serum levels and users therefore rely on activity-based assays for assessing inter-batch variability.70 Taken together with the consideration that conditioned medium likely contains an appreciable number of undefined components, compliance with industrial good manufacturing practices (GMP) is restricted with these formulations.71 Alternatively, these growth factors can be isolated from separate cell lines or obtained as a commercial formulation (e.g., IntestiCult Organoid Growth Medium), though these have not been optimized for human in vitro crypt culture.
While several advances in scaling fabrication and analysis of the in vitro crypt device cassettes have been made,23,40 there are still several challenges to their mass production and imaging related to the collagen scaffolding. Up to 25 micromolded scaffolds, possessing a total of 3875 crypts, are readily fabricated in parallel using optimized dimensions and environmental conditions.37 This limitation exists due to the fast gelation (i.e., <5 min) of collagen in the presence of covalent crosslinking reagents. Further scaffold manufacturing scalability could be enabled by the exploration of alternative crosslinking reagents or roll-to-roll molding methods. Three-dimensional bioprinting may also hold promise for this task, as many collagenous bioinks and gelation strategies have been proposed for extrusion-based printing of collagen scaffolds.72–76 While many of these strategies do not provide sufficient mechanical strength or resolution for formation of crypt architecture, one recently proposed method, freeform reversible embedding of suspended hydrogels (FRESH), achieved a collagen filament resolution of 20 μm and was capable of printing a wide range of tissue architectures, from capillaries to a full organ.76 With optimization, strategies such as this could eventually mature to create the reproducible crypt scaffolds accessible to users with bioprinting experience. However, print speeds and resolution will have to substantially improve for commercial or batch scalability to be feasible (i.e., fabrication of more than one in vitro crypt device at a time).
In regards to in situ analysis through the hydrogels, phenotypic compartmentalization can be readily assessed by fluorescence microscopy, though refractive index mismatches between collagen and the surrounding media result in sufficient light scattering such that high-resolution imaging (e.g., of subcellular structures) is challenging.37 Preliminary attempts to normalize the refractive indices throughout the sample using optical clearing solutions have been met with mixed results, as both formamide and organic solvent based solutions have resulted in hydrogel degradation. We have observed high compatibility of the hydrogels with glycerol-based mounting media, and therefore posit that several glycerol-based optical clearing solutions will yield improved imaging data.77,78 Ultimately, the move from biologically derived matrices to fully synthetic matrices may be the most advantageous route forward, as synthetic matrices are fully defined and mechanically tunable, thereby minimizing batch variability and roadblocks to GMP compliance. While noteworthy successes in primary intestinal organoid culture have been demonstrated with synthetic hydrogels,17,79 areas for further investigation include the ability to shape these materials, scale their fabrication, modulate their optical transmission for in situ imaging, and optimize their mechanical properties for long-term culture of primary cells as a monolayer over their surface.
Materials
Biological materials
Cell culture
L-WRN cells (ATCC, cat. no. CRL-3726, RRID:CVCL_DA06, https://scicrunch.org/resolver/RRID:CVCL_DA06) ▲CRITICAL This cell line is used for producing Wnt3a, R-spondin-3, and noggin conditioned medium under established methods.19 !CAUTION The cell lines used in your research should be regularly checked to ensure they are authentic and are not infected with mycoplasma.
Primary intestinal epithelial cells from patient biopsy, fresh cadaveric tissue, animal donor, or cell/tissue biobank. !CAUTION All relevant international and institutional regulations relating to the acquirement and usage of primary tissue must be followed. Consult your funding agency and institutional review board for any questions or clarifications regarding these policies. !CAUTION Appropriate validation of the cultured cells must be performed to ensure that genetic drift is minimized, so that the cells remain representative of their original host tissue. The culture methods below will result in cells that possess normal karyotypes between P0 and P15. !CAUTION The cell lines used in your research should be regularly checked to ensure they are authentic, possess a normal karyotype and are not infected with mycoplasma. ▲CRITICAL The human colonic biopsy specimen (male, 52 y) utilized to generate the data in this protocol was obtained during a routine screening colonoscopy performed at the University of North Carolina (UNC) Hospitals Meadowmount Endoscopy Center under UNC IRB #14–2013. All in vitro crypt experiments utilized colonic epithelial cells between P5 and P15 that were expanded over neutralized collagen hydrogels (RRID:CVCL_ZL23, https://scicrunch.org/resolver/RRID:CVCL_ZL23).
Reagents
Common Reagents
Deionized (DI) water (≥18.0 MΩ·cm)
Phosphate-buffered saline (PBS), 10× solution, pH 7.4 ± 0.1, without calcium and magnesium, RNase-/DNase- and protease-free (Corning, cat. no. 46–013-CM)
(4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) (HEPES) buffer, 1 M solution (Corning, cat. no. 25–060-CI)
Sodium bicarbonate, 7.5% solution (0.893 M, Corning, cat. no. 25–035-CI)
Sodium hydroxide, 1 M solution (Millipore-Sigma, cat. no. S2770)
Collagen Type I, rat tail (Corning, cat. no. 356236)
Ethylenediaminetetraacetic acid (EDTA), 0.5 M solution (ThermoFisher Scientific, cat. no. 15575–020)
Dimethyl sulfoxide (DMSO, Sigma Life Sciences, cat. no. D2650)
Hanks’ balanced salt solution (HBSS), no calcium, no magnesium (ThermoFisher Scientific, cat. no. 14170112)
Collagenase, Type 4 (Worthington Biochemical Corporation, cat. no. LS004189)
Ethanol, 200 proof (Fisher Scientific, cat. no. 04–355-223)
Crypt Isolation Reagents
Dithiothreitol (DTT, Millipore-Sigma, cat. no. D5545)
Sucrose (Fisher Scientific, cat. no. BP220–1)
D-sorbitol (Fisher Scientific, cat. no. BP439–500)
Culture Medium Components
Advanced DMEM/F-12 (ThermoFisher Scientific, cat. no. 12634010)
GlutaMAX Supplement (ThermoFisher Scientific, cat. no. 35050061)
Fetal bovine serum (FBS, R&D Systems, cat. no. S11150)
Primocin (InvivoGen, cat. no. ant-pm-2)
N-Acetyl-L-cysteine, cell culture reagent, ≥96% (MP Biomedicals, cat. no. 194603)
Recombinant Murine EGF (PeproTech, cat. no. 315–09)
Nicotinamide (Millipore-Sigma, cat. no. N0636)
B-27 Supplement (50×), serum free (ThermoFisher Scientific, cat. no. 17504044)
(Leu15)-Gastrin-1, human (AnaSpec, cat. no. AS-64149)
Prostaglandin E2 (PGE2, Cayman Chemical, cat. no. 14010)
A 83–01 (Millipore-Sigma, cat. no. SML0788)
SB 202190, Free Base, >99% (LC Laboratories, cat. no. S-1700)
Y-27632 dihydrochloride (MedChemExpress, cat. no. HY-10583)
Microfabrication Reagents
1002F epoxy resin (Miller-Stephenson, cat. no. EPON 1002F) !CAUTION Use a chemical fume hood and wear protective goggles, respirator mask, gloves, and any other appropriate personal protective equipment when handling this material.
Triarylsulfonium hexafluoroantimonate salts, mixed 50 % in propylene carbonate (Millipore-Sigma, cat. no. 654027) ▲CRITICAL The photoresist is not compatible with other photoacid generators.
Gamma-butyrolactone (Millipore-Sigma, cat. no. B103608) !CAUTION Use a chemical fume hood and wear protective goggles, respirator mask, gloves, and any other appropriate personal protective equipment when handling this material. !CAUTION Gamma-butyrolactone is a controlled substance and all corresponding government regulations must be followed for its acquisition and use.
Propylene glycol methyl ether acetate (Millipore-Sigma, cat. no. 484431) !CAUTION Use a chemical fume hood and wear protective goggles, respirator mask, gloves, and any other appropriate personal protective equipment when handling this material.
Isopropyl alcohol (Millipore-Sigma, cat. no. 109827)
Acetone (Millipore-Sigma, cat. no. 650501)
Trichloro(octyl)silane (Millipore-Sigma, cat. no. 235725) !CAUTION Use a chemical fume hood and wear protective goggles, respirator mask, gloves, and any other appropriate personal protective equipment when handling this material.
Poly(ethylene glycol) methyl ether acrylate (Millipore-Sigma, cat. no. 454990)
Benzyl alcohol (Millipore-Sigma, cat. no. 305197)
Sodium periodate (Millipore-Sigma, cat. no. 311448)
Dow Corning Sylgard 184 silicone elastomer base and curing agent (Ellsworth, cat. no. 184 SIL ELAST KIT 0.5KG)
Shaped Hydrogel Formation
2-(N-morpholino)ethanesulfonic acid (MES, Millipore-Sigma, cat. no. M8250)
N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC, Oakwood Chemical, cat. no. 25952–53-8)
N-hydroxysuccinimide (NHS, Millipore-Sigma, cat. no. 130672)
VitroCol, human collagen, type 1 (Advanced BioMatrix, cat. no. 5007–20ML)
Equipment
General laboratory equipment
Class II, type A2 biological safety cabinet
Humidified CO2 incubator (37 °C, 5 % CO2)
General purpose water bath (37 °C), filled with treated water (Millipore-Sigma, cat. no. S5525) or Lab Armor Beads (ThermoFisher Scientific, cat. no. A1254301)
Inverted phase contrast microscope for tissue culture (Nikon Eclipse TS100)
Benchtop centrifuge, capable of 600–2000 ×g
pH meter, calibrated between pH 4–7 and pH 7–10
Lyophilizer/Freeze Dry System (Labconco, cat. no. 7670021)
500 mL vacuum filter/storage bottle systems (Corning, cat. no. 430773)
50 mL conical tube vacuum filtration systems (VWR, cat. no. 89220–722)
15 mL conical tubes (Corning, cat. no. 352097)
50 mL conical tubes (Corning, cat. no. 352098)
0.6 mL microcentrifuge tubes (Fisher Scientific, cat. no. 02–681-311)
1.5 mL microcentrifuge tubes (Fisher Scientific, cat. no. 02–681-320)
Ice or chilled Lab Armor Beads (ThermoFisher Scientific, cat. no. A1254301)
Micropipettes (P20, P200, and P1000) and pipette tips
Razor blades (Ted Pella, cat. no. 121–32)
Scalpel and/or surgical scissors
Forceps
Parafilm
Tissue culture consumables
6-well polystyrene cell culture plate (Corning, cat. no. 353046)
12-well Corning Costar cell culture plate (Corning, cat. no. 3513)
Sterile sampling bags (MTC Bio, cat. no. B5911)
Serological pipettes (5 mL, 10 mL, and 25 mL)
Cryogenic storage vials (Corning, cat. no. 430487)
Microfabrication Equipment
Chrome/soda lime glass photomask (Front Range Photomask, high resolution direct write, critical dimension tolerance ±0.3 μm, Fig. 4)
Glass slides, 75×50 mm (Corning, cat. no. 2947–75X50)
Spin coater (Laurell Technologies, WS-650MZ-23NPP)
Plasma cleaner (Harrick Plasma, cat. no. PDC-001)
Collimated UV exposure system (Newport, cat. no. 92530)
350 nm long-pass filter (Omega Optical, cat. no. PL-360LP)
ZETA 7411-S 400-W Metal Halide UV Flood System (Loctite, cat. no. 630560)
Bottle roller (Fisher Scientific, cat. no. 11–676-250)
Oven, set to 95 °C (Fisher Scientific, cat. no. 11–475-154)
Hotplate (VWR, cat. no. 97042–698)
Orbital shaker (Bellco Glass Inc., cat. no. 7744–01010)
Oil-free diaphragm vacuum pump (BrandTech Scientific, ME2C NT 120V)
Vacuum desiccator (Fisher Scientific, cat. no. 08–642-7)
Round bottom glass tube, 50 mL (Millipore-Sigma, cat. no. CLS842250) with PTFE protected seal screw-cap (Millipore-Sigma, cat. no. Z232343)
Wide-mouth amber glass bottle, 1000 mL (Cole-Parmer, cat. no. UX-99540–57)
Nonwoven dry cleanroom wipers (TechniCloth, cat. no. TX604)
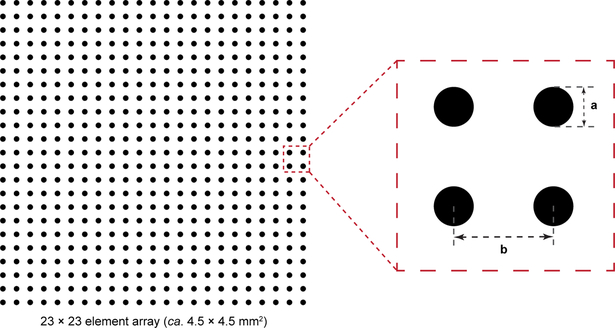
Fig. 4 |.
Design of in vitro crypt photomask. For human in vitro crypt fabrication, the diameter of each array element (a) is 80 μm with a pitch (b, center-to-center distance) of 200 μm. For mouse in vitro crypt fabrication, the diameter of each array element (a) is 75 μm with a pitch (b) of 125 μm. The mask can be designed in any standard CAD or vector graphics software, and read in “dark field” format (chrome background, features will be clear) during mask fabrication. High resolution (critical dimension tolerance ±0.3 μm) direct write in chrome over a soda lime glass support is sufficient for the features outlined in the protocol.
Device Assembly
12-well cell culture inserts (Corning, cat. no. 354236)
Biocompatible transfer adhesive (3M, cat. no. 1504XL)
Biopore membrane filter roll, hydrophilic PTFE (Millipore-Sigma, cat. no. BGCM00010)
Polycarbonate film, 0.127 mm thickness (McMaster-Carr, cat. no. 85585K102)
Biopsy punch, 3 mm dia. (Sklar Instruments, cat. no. 961105)
Scalpel, craft knife, and/or scissors
Self-healing cutting mat
Pneumatic curing vessel/pressure pot (Handler, model no. 448PP) equipped with nitrogen gas source capable of generating 30 psi (200 kPa) of pressure
Reagent setup
Phosphate-buffered saline, 1× solution (1× PBS)
Combine 100 mL of 10× PBS with 900 mL of DI water. Vacuum filter and store at room temperature (ca. 20–22 °C) for up to 6 months.
Neutralization buffer
This buffer is utilized during the preparation of neutralized collagen hydrogel scaffolds for monolayer propagation of intestinal epithelial cells. The concentrations of acid-solubilized collagen within manufacturer-provided solutions vary between lots, while the acetic acid concentrations remain constant (0.02 M); therefore, neutralization buffer components must be customized for each batch. For each batch of neutralization buffer, combine pre-determined volumes of 10× PBS, 1 M HEPES, 7.5% sodium bicarbonate, and 1 M NaOH so that the final concentrations within the neutralized collagen mixture equate to: 1 mg mL−1 collagen, 1× PBS, 20 mM HEPES, 53 mM sodium bicarbonate, and NaOH equimolar to acetic acid from the added collagen solution. Titrate the mixture to pH 7.4 prior to the addition of NaOH, vacuum filter, and store at 4 °C. As an example, consider that the collagen solution is supplied by the manufacturer at a concentration of 3.36 mg mL−1 in 0.02 M acetic acid. Dilution to 1 mg mL−1 would require a volumetric mixture of 29.7 % collagen stock and 70.3 % neutralization buffer. 50 mL of this batch of neutralization buffer would be comprised of 7.118 mL of 10× PBS (1.42× working concentration), 1.424 mL of 1 M HEPES (28.5 mM working concentration), 4.225 mL of 7.5% sodium bicarbonate (75.5 mM working concentration), 36.809 mL of DI water, and 0.424 mL of 1 M NaOH (8.5 mM working concentration). The Supplementary Calculations spreadsheet may be used for assistance. ?TROUBLESHOOTING
Preparation of stock reagents for tissue culture
All frozen stock reagents can be frozen and stored at −20 °C or −80 °C for up to 1 y (see individual items for recommended temperature). L-WRN conditioned medium: Aliquot from pooled collection at 250 mL, freeze at −20 °C for 24–48 h. Transfer to −80 °C for long-term storage. Primocin (50 mg mL−1): Aliquot from manufacturer as-is at 500 μL, store at −20 °C. N-acetyl-L-cysteine (NAC, 1 M): Combine 10 g of NAC and 61.28 g of 1× PBS. Stir at 37 °C until dissolved. Aliquot at 625 μL, store at −20 °C. Epidermal growth factor (EGF, 250 μg mL−1): Dissolve 1 mg of EGF into 4 mL of 0.1 wt % cell-culture grade BSA and 1× PBS. Aliquot at 100 μL, store at −20 °C. Nicotinamide (1 M): Dissolve 12.212 g of nicotinamide into 100 mL of 1× PBS. Stir at 37 °C until dissolved. Aliquot at 5 mL, store at −20 °C. Gastrin (1 mg mL−1): Dissolve 1 mg of gastrin into 1 mL of 0.1 wt % cell-culture grade BSA and 1× PBS. Aliquot at 12.5 μL, store at −20 °C. Prostaglandin E2 (PGE2, 1 mM): Dissolve 1 mg of PGE2 into 2.837 mL of DMSO. Aliquot at 5 μL, store at −20 °C. A 83–01 (5 mM): Dissolve 25 mg of A 83–01 in 11.86 mL DMSO. Aliquot at 50 μL, store at −20 °C. SB202190 (30 mM): Dissolve 200 mg of SB202190 into 20.12 mL of DMSO. Aliquot at 50 μL, store at −20 °C. Y-27632 dihydrochloride (10 mM): Dissolve 200 mg of Y-27632 into 62.449 mL of 1× PBS. Aliquot at 500 μL, store at −20 °C. Dithiothreitol (DTT, 50 mM): Combine 77.125 mg of dithiothreitol and 10 mL of 1× PBS. Vacuum filter and store at −20 °C.
Tissue rinse buffer
This buffer is utilized during the isolation of crypts from living tissue. Combine 100 mL of 10× PBS, 7.428 g of sucrose (43.4 mM working concentration), and 5.001 g of D-sorbitol (54.9 mM working concentration). Add DI water to a final volume of 500 mL using a volumetric flask. Vacuum filter and store at room temperature (ca. 20–22 °C) for up to 6 months.
Crypt isolation buffer
This buffer is utilized during the isolation of crypts from living tissue and must be prepared within 1 hour before intestinal crypt isolation. Transfer 10 mL of “Tissue Rinse Buffer” into a 15 mL conical tube within a biosafety cabinet. Combine with 100 μL of 50 mM DTT stock solution and 40 μL of 0.5 M EDTA.
Culture media formulations
Maintenance Medium (MM) is utilized for monolayer propagation of intestinal epithelial cells, while Expansion Medium (EM) is utilized for seeding cells over in vitro crypt scaffolds, and Stem Medium (SM) and Differentiation Medium (DM) are utilized for in vitro crypt phenotypic compartmentalization (Table 1). For MM, EM, and SM, thaw L-WRN conditioned medium overnight at 4 °C one day prior to medium preparation. On the day of medium preparation, thaw remaining frozen stock components to room temperature, then transfer all components into a sterile biosafety cabinet. Combine components according to the formulations in Table 1. Vacuum filter and store at 4 °C for up to 1 month.
Cryopreservation medium formulation
This medium is utilized for cryopreservation of isolated crypts or cultured intestinal epithelial cells for long-term storage. Combine 50 % (v/v) of cold (ca. 4 °C) Maintenance Medium, 40 % (v/v) fetal bovine serum, and 10 % (v/v) DMSO to obtain the total desired volume, and use immediately.
Collagenase stock solution
Dissolve lyophilized collagenase (Type 4) powder into HBSS (without Ca2+ or Mg2+) to a concentration of 5000 U mL−1. Vacuum filter, aliquot at 5 mL, and store at −20 °C for up to 1 y. Working solutions can be thawed and stored at 4 °C for up to 1 month.
Dissociation buffer
This buffer is utilized for gentle fragmentation of the cultured intestinal monolayers during passaging. Combine 49.9 mL of 1× PBS, 50 μL of 0.5 M EDTA, and 50 μL of 10 mM Y-27632. Vacuum filter, aliquot at 5 mL, and store at −20 °C for up to 1 y. Working solutions can be thawed and stored at 4 °C for up to 1 month.
Formulation of 1002F epoxy photoresist
1002F epoxy photoresist consists of 1002F resin, a photoacid generator (triarylsulfonium hexafluoroantimonate salts), and solvent (gamma-butyrolactone).22 Two different formulations (i.e., 1002F-10 and 1002F-100) are utilized to prepare master in vitro crypt molds. 1002F-10 is composed of resin, photoacid, and solvent at a weight ratio of 49:4.9:46.1, respectively. 1002F-100 is composed of resin, photoacid, and solvent at a weight ratio of 64:6.4:29.6, respectively. Sequentially combine 1002F resin, gamma-butyrolactone, and photoacid into a 1 L amber bottle. Tightly cap the bottle and seal with Parafilm. The bottle is then mechanically mixed on a bottle roller set to 30 rotations per minute (rpm) to facilitate complete dissolution of the resin for 1–4 weeks (required time depends on the viscosity of the formulation). This photoresist can then be stored in a dark location at room temperature (20–22 °C) for up to 3 y. !CAUTION Use a chemical fume hood and wear protective goggles, respirator mask, gloves, and any other appropriate personal protective equipment when combining these materials.
!CAUTION Gamma-butyrolactone is a controlled substance and all corresponding government regulations must be followed for its acquisition and use.
Poly(ethylene glycol) photografting solution
This solution, consisting of 10 wt % PEG monomer, 0.5 wt % benzyl alcohol, and 0.5 mM sodium periodate, is utilized to deposit a branched poly(ethylene glycol) coating on the surface of the PDMS stamps and minimize collagen adhesion during the micromolding process.23 Dissolve 53.473 mg of sodium periodate into ca. 300 mL of DI water. Combine with 50 g of PEG methyl ether acrylate monomer and 2.5 g of benzyl alcohol, then bring the mixture to a final volume of 500 mL with DI water using a 500 mL volumetric flask. Invert to mix the components. Store in an amber bottle at 4 °C for up to 6 months.
MES buffer, 0.1 M, pH 5
Dissolve 1.086 g of 2-(N-morpholino)ethanesulfonic acid (MES) into 50 mL of DI water. Titrate to pH 5. Vacuum filter and store at room temperature (20–22 °C) for up to 6 months.
EDC aliquots for micromolding, 0.6 M
Dissolve 232.5 mg of N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) into 2 mL of DI water. Use immediately, or aliquot at 100 μL and store at −20 °C for up to 1 y.
NHS aliquots for micromolding, 0.15 M
Dissolve 34.5 mg of N-hydroxysuccinimide (NHS) into 2 mL of DI water. Use immediately, or aliquot at 100 μL and store at −20 °C for up to 1 y.
MES-solubilized collagen, 5 mg mL−1
Dispense the contents of one bottle (100 mg) of Type 1 collagen, rat tail into a 50 mL conical tube. Incubate at −80 °C for ≥3 h. Apply a vented cap to the conical tube and lyophilize for ≥72 h. Re-solubilize the collagen in 20 mL of MES buffer (0.1 M, pH 5) to a final concentration of 5 mg mL−1. Incubate at 4 °C. Complete re-solubilization will take several days. Store at 4 °C for up to 6 months.
Procedure
[Box 1 |. Neutralized collagen hydrogel fabrication for monolayer expansion ●Timing 2 h].
Procedure
Place the “Neutralization Buffer” and collagen stock solution (in 0.02 M acetic acid) on ice within a sterile biosafety cabinet and incubate for 30 min, or until thoroughly chilled.
Within a 50 mL conical tube, dilute collagen stock solution in “Neutralization Buffer” to a final collagen concentration of 1 mg mL−1. First add the buffer, then the collagen stock solution, and invert five times to mix.
Centrifuge the neutralized collagen mixture at 2000 ×g for 30 s to remove any air bubbles that may have formed during mixing.
Dispense 1 mL of neutralized collagen (1 mg mL−1) solution into each well of a 6-well cell culture plate using a serological pipet.
Vigorously swirl and tap the edges of the plate against the work surface so that the collagen mixture spreads to the cover the entire horizontal base of each well. Visually inspect to ensure that no air bubbles are present, as these may cause the scaffolds to spontaneously lift during storage or culture.
Incubate within a humidified CO2 incubator at 37 °C for 1 h to set the hydrogel scaffolds.
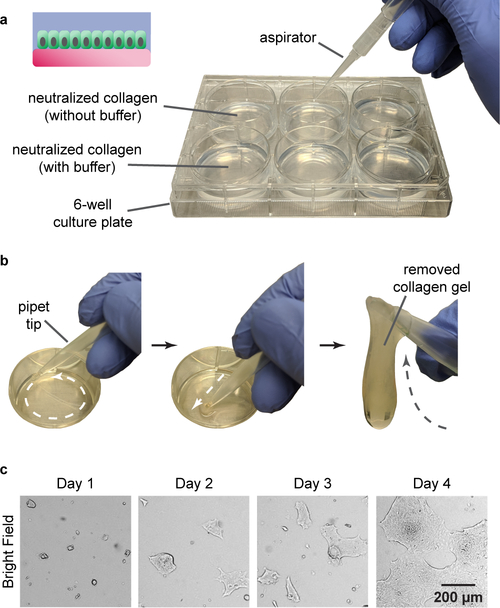
Overlay each hydrogel scaffold with 4 mL of 1× PBS. The hydrogel scaffolds should appear optically transparent, and maintain their shape as buffer is dispensed over their surface (Fig. 3a).
Fig. 3 |.
Monolayer hydrogel maintenance of primary intestinal epithelial cells. a, Neutralized collagen hydrogels are formed within 6-well culture plates and utilized for monolayer propagation of primary intestinal cells. b, During passaging, the collagen hydrogel scaffolds are manually removed from the culture plate using a pipet tip. c, Expansion of intestinal cells as monolayer patches over the hydrogel surface after passaging. Panel c is adapted from ref. 20 with permission from Elsevier. The human colonic biopsy specimen (male, 52 y) utilized to generate the data panel c was obtained during a routine screening colonoscopy performed at the University of North Carolina (UNC) Hospitals Meadowmount Endoscopy Center under UNC IRB #14–2013.
■PAUSE POINT The collagen hydrogel scaffolds can be used immediately for intestinal epithelial cell culture, though note that for some collagen batches, optimal attachment and spreading of these cells occurs best after room temperature (ca. 20–22 °C) incubation of the scaffold for 15 d. We speculate this may be related to changes in mechanical properties (e.g., stiffness) of the hydrogel during incubation, or the diffusion of unwanted contaminants (e.g., hormones or endotoxins) from the hydrogels that may be present in specific collagen batches. To prevent contamination, transfer the cell culture plates into sterile sampling bags, seal according to manufacturer-provided instructions within the biosafety cabinet, and store in a dark location. Scaffold fabrication may be scaled up into larger batches by this manner, with the pre-made hydrogels remaining viable for at least 12 months. !CAUTION Check the plates every 2–3 months to make sure the water has not evaporated. In case that substantial water evaporation is observed, add additional 4 mL PBS to each well.
Fig. 3 |.
Monolayer hydrogel maintenance of primary intestinal epithelial cells. a, Neutralized collagen hydrogels are formed within 6-well culture plates and utilized for monolayer propagation of primary intestinal cells. b, During passaging, the collagen hydrogel scaffolds are manually removed from the culture plate using a pipet tip. c, Expansion of intestinal cells as monolayer patches over the hydrogel surface after passaging. Panel c is adapted from ref. 20 with permission from Elsevier. The human colonic biopsy specimen (male, 52 y) utilized to generate the data panel c was obtained during a routine screening colonoscopy performed at the University of North Carolina (UNC) Hospitals Meadowmount Endoscopy Center under UNC IRB #14–2013.
[Box 2 |. Isolation of living crypts from primary intestinal tissue ●Timing 4–6 h].
Procedure
Transfer “Tissue Rinse Buffer,” DTT stock solution, and (4) 15 mL conical tubes into a sterile biosafety cabinet. For the first three conical tubes, aliquot 10 mL of “Tissue Rinse Buffer” into each and sequentially label them, “1,” “2,” and “3.” Within the final conical tube, prepare the “Crypt Isolation Buffer.” ▲CRITICAL Each subject/tissue sample will require its own set of rinse and isolation buffers, so scale up buffer preparations accordingly.
Transfer tissue sample into the biosafety cabinet, along with scalpel/surgical scissors, if needed for processing the tissue for crypt extraction.
Rinse away any debris, mucus or luminal contents from the tissue using a gentle stream of tissue rinse buffer. For smaller animal subjects (e.g., murine or rat) the tissue will most likely be supplied as a longitudinal section instead of a biopsied tissue punch, which must be cut open using surgical scissors or a scalpel to form a planar sheet of tissue. If a larger piece of human tissue is obtained, the tissue may need to be cut down to a size of ca. 500 mm2 for efficient processing. Human intestinal tissues from donors are normally associated with a thick mucus layer, and for cadaveric donors, bacteria and stool may also be present. To remove these luminal contents, the tissues are placed in a 50-mL conical tube, and 25 mL tissue rinse buffer is added. The tube was shaken to release the mucus layer. Repeat this procedure until the buffer becomes clear.
Transfer the tissue into the “Crypt Isolation Buffer,” and incubate at room temperature (20–22 °C) for 60 min.
Remove the tissue from the isolation buffer, and sequentially transfer to the first, second, and third “Tissue Rinse Buffers.” Incubation in the first and second “Tissue Rinse Buffers” is not necessary, with the tissue simply dropped in, removed, and transferred to the next solution.
Once the tissue has been placed in the third “Tissue Rinse Buffer,” seal tightly and apply 30 vigorous shakes to the sample to release crypts from the tissue.80
Withdraw 10 μL of this suspension, and deposit over a small petri dish or glass slide. Count all visible and intact crypts under a microscope, then multiply this number by 100 to determine density in units of [crypts mL−1]. ▲CRITICAL Aim for 3000–4000 crypts mL−1 at this stage. Additional or more vigorous shakes may need to be applied to release the desired number of crypts.
Repeat steps 4 through 7 for a second round of isolation. The “Crypt Isolation Buffer,” as well as the first and second “Tissue Rinse Buffers” can be reused from the first round. The third “Tissue Rinse Buffer” will need to be replaced with a fresh aliquot for each round of isolation. The incubation time in step 4 can be reduced to 30 min. ▲CRITICAL Two rounds of isolation are often sufficient to release all crypts, though additional isolation rounds may be performed until no crypts are observed within the 10 μL droplet under the microscope. Additional conical tube shakes (i.e., 50 shakes) may need to be applied with each successive round to release all crypts.
Calculate the total number of crypts obtained for each round of isolation and pool into a single 50 mL conical tube for each subject/tissue sample.
Centrifuge suspension at 600 ×g for 1 min to obtain a pellet of crypts. Discard the supernatant.
-
Resuspend crypts in “Maintenance Medium” (MM), warmed to 37 °C, at a concentration of 240 crypts mL−1 (human) or 2400 crypts mL−1 (murine or rat) for monolayer propagation over pre-made neutralized collagen hydrogels prepared in 6-well plates (see Box 1).
■PAUSE POINT The crypts may alternatively be cryogenically preserved for future expansion or analysis. If this is desired, resuspend in “Cryopreservation Medium,” at a concentration of 960 crypts mL−1 (human) or 9600 crypts mL−1 (murine or rat). Aliquot at 1 mL per cryogenic storage vial. Slowly freeze (−1 °C min−1) to −80 °C within an appropriate vessel for 24–48 h. Transfer to liquid nitrogen cryogenic system for long-term storage. Crypt densities are optimized so that a single vial may be seeded per neutralized collagen hydrogel (i.e., well) within a 6-well plate.
Rinse the pre-made neutralized collagen scaffolds with 4 mL 1× PBS for 3 times (5 min interval) and aspirate prior to plating the crypts. The rinses remove the accumulated salts caused by water evaporation during storage. !CAUTION The collagen hydrogel is very fragile, and touching the aspirator to the gel should be avoided. We recommend gently tipping the plate during this process and aspirating from the edge of the well. It is acceptable if a small amount of liquid remains.
Dispense 4 mL of crypts in MM solution per scaffold (i.e., well) within the 6-well plate.
Place scaffolds with crypts into a humidified CO2 incubator. We refer to this initial plating as passage 0 (P0).
Two days after seeding the crypts, verify the status of the cultures for signs of cell growth or infection. Small patches of cells should be forming at the luminal surface of the hydrogel and there will likely be debris present. Aspirate medium, and replenish with 4 mL of fresh MM. Repeat this process every two days until cells are ready to passage (≥80 % confluency, typically 5–8 d). ▲CRITICAL It may be necessary to increase the concentration of primocin within MM to 100 μg mL−1 and/or add 0.5 μg mL−1 of Amphotericin B during this initial passage to mitigate contamination.
Monolayer culture and passaging of primary intestinal epithelial cells ●Timing 5–8 d
▲CRITICAL Monolayer propagation of primary intestinal epithelial cells may be conducted once neutralized collagen hydrogel scaffolds are fabricated (Box 1) and primary cells are isolated (Box 2). The cells may be expanded over the scaffolds from fresh crypts (960 crypts well−1), cryopreserved crypt samples (1 vial well−1, ca. 960 crypts vial−1), or cryopreserved primary cell stocks (1 vial well−1, see below for cell density). !CAUTION Appropriate validation of the cultured cells must be performed to ensure that genetic drift is minimized, so that the cells remain representative of their original host tissue. The culture methods below will result in cells that possess grossly normal karyotypes between P0 and P15.
-
1
If starting from a cryopreserved stock of crypts or cultured cells, incubate Maintenance Medium (MM) in a water bath at 37 °C for 30 min, or until warm.
-
2
Transfer 1× PBS, (1) 15 mL conical tube, and (1) 6-well plate of neutralized collagen hydrogel scaffolds into a sterile biosafety cabinet.
-
3
Aspirate buffer storage solution from the collagen scaffolds that will be used for plating (1 vial well−1). Dispense 4 mL of 1× PBS and incubate for 5 min. Repeat this process two additional times (Fig. 3a).
?TROUBLESHOOTING
-
4
Retrieve cryopreserved cell or crypt vial(s) from liquid nitrogen storage. Thaw to room temperature within 2 min.
-
5
Dispense 4 mL of MM per thawed vial into the 15 mL conical tube, followed by the thawed cell stock solution (1 mL vial−1).
-
6
Centrifuge at 600 ×g for 1 min. Discard the supernatant, and resuspend the pellet in 4 mL of MM per thawed vial.
-
7
Aspirate 1× PBS from each collagen scaffold that will be used for plating, then transfer 4 mL of the cell suspension in MM to each well.
-
8
Place the collagen scaffold plate with cells into a humidified CO2 incubator. ▲CRITICAL In this and all following procedures, the day of cell seeding or passage is referred to as Day 0.
-
9
Replenish media every 48 h (e.g., Days 2, 4, 6, etc.). Aspirate spent medium from each well, and dispense 4 mL of fresh, 37 °C MM. ▲CRITICAL Verify, using a phase contrast microscope, that the cultures are expanding into larger patches over the hydrogel surface. The cells will be ready for passage once confluency is ≥80 % (typically Day 5–8). It is normal for the cells to be more confluent near the center of the scaffold than the perimeter due to the concave meniscus of the hydrogel.
?TROUBLESHOOTING
-
10
When the cells are ready for passage (≥80 % confluent), incubate MM and “Dissociation Buffer” in a water bath at 37 °C, or until warm. Retrieve collagenase stock solution (5000 U mL−1) from 4 °C storage and transfer into a sterile biosafety cabinet, along with 1× PBS, (1) 15 mL conical tube, and neutralized collagen scaffolds prepared in 6-well plates.
-
11
Aspirate buffer storage solution from the new collagen scaffolds that the cells will be passaged to. Dispense 4 mL of 1× PBS and incubate for 5 min. Repeat this process two additional times (Fig. 3a). ▲CRITICAL The cells will be passaged at a surface area ratio of 1:3 for monolayer expansion (i.e., (1) confluent well can be subcultured to (3) new wells of the same surface area during passaging). Plan according to the number of wells that are being passaged, and experimental requirements. The spreadsheet file in the Supplementary Information may be utilized for assistance.
-
12
Dispense 100 μL of collagenase stock solution for each well that will be passaged into the 15 mL conical tube.
-
13
Retrieve the cultured cells from the CO2 incubator, then transfer 1 mL of spent medium from each well that will be passaged into the 15 mL conical tube. Aspirate the remaining medium.
-
14
Using a 100–1000 μL pipet tip, scrape around the perimeter the collagen scaffold, against the walls of the well, to dislodge the hydrogel from the plate. Gently lift the scaffold, using the pipet tip, and transfer it into the conical tube (Fig. 3b).
!CAUTION The collagen hydrogel is very fragile, and may fragment during transfer. The conical tube should be held in close proximity to the 6-well plate to minimize any chance of cell loss. Gentle suction from a 1 mL micropipette may be utilized to secure the scaffold against the pipet tip during this transfer.
-
15
Using a 5 mL serological pipet, pipet the collagenase solution with scaffolds up/down 5–10 times. The scaffolds should be roughly broken up and transfer freely through the pipet tip by the end of this process.
-
16
Using a P1000 micropipette, pipet the collagenase solution with scaffolds up/down 10–20 times. The scaffolds should break up to a greater extent and transfer freely through the pipet tip by the end of this process.
-
17
Incubate at 37 °C for 10 min.
-
18
Centrifuge the cells at 600 ×g for 1 min and inspect the pellet. If a layer of collagen remains above the cells, resuspend the pellet and incubate for 1 additional min at 37 °C. Re-centrifuge using the same parameters above.
?TROUBLESHOOTING
-
19
Aspirate the supernatant from the cell pellet.
-
20
Re-suspend the pellet in 14 mL of 1× PBS to rinse the cells.
-
21
Centrifuge the cells at 600 ×g for 1 min, and aspirate the supernatant from the pellet.
-
22
Re-suspend the pellet in 150 μL of “Dissociation Buffer.” Using a P200 micropipette, break up the cell pellet by pipetting up/down 20 times.
-
23
Incubate at 37 °C for 5 min.
-
24
Using a P200 micropipette, break up the cell pellet into smaller fragments by pipetting up/down 40 times.
-
25
Add 4 mL of 37 °C MM to the conical tube for each new scaffold that will have cells plated.
■PAUSE POINT The cultured cells may alternatively be cryogenically preserved for future expansion or analysis. If this is desired, re-suspend in “Cryopreservation Medium,” noting that one confluent six-well scaffold can be expanded to three cryopreserved vials of cells. Aliquot at 1 mL per cryogenic storage vial. Slowly freeze (−1 °C min−1) to −80 °C within an appropriate vessel for 24–48 h. Transfer to liquid nitrogen cryogenic system for long-term storage. Cell densities are optimized so that a single vial may be seeded per neutralized collagen hydrogel (i.e., well) within a 6-well plate.
-
26
Aspirate the buffer from each new collagen scaffold, and dispense 4 mL of the cell suspension into each well.
-
27
Place the collagen scaffold plate with cells into a humidified CO2 incubator, and replenish media with fresh MM every 48 h until the next passage (Fig. 3c).
?TROUBLESHOOTING
1002F crypt master template fabrication (photolithography) ●Timing 2 d
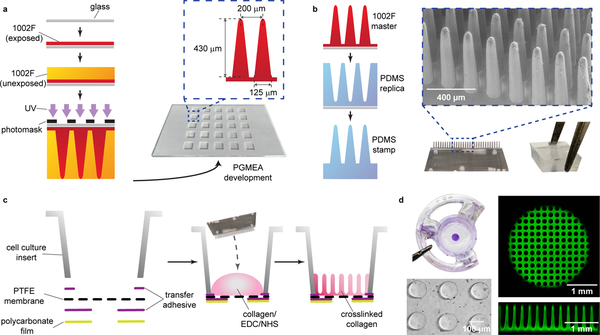
▲ CRITICAL 1002F formulation 10 (1002F-10), a custom-formulated epoxy photoresist, is utilized to generate a thin film of fully crosslinked 1002F epoxy over the glass support (Steps 28–34), which in turn serves to improve adhesion of the high aspect ratio micropillars fabricated in Steps 36–45. Skipping Steps 28–34 may result in the micropillars detaching from the glass support. 1002F formulation 100 (1002F-100) is utilized to generate the slanted, high aspect ratio, rounded apex micropillars that mimic the shape of human colon crypts (Steps 36–45). While it is possible to use other epoxy photoresists (e.g., SU-8), attempts to do so may not result in pillars with this exact shape. To achieve the best results for fabrication of the master mold, the user should understand the basic procedures and concepts of photolithography. Due to interlaboratory equipment and environmental variations, the user may need to conduct laboratory-specific process optimizations, including adjustment of spin coat speed, bake time, development time, and UV exposure dose.
-
28
Rinse the glass slides sequentially with acetone and isopropyl alcohol (IPA), then dry with compressed air (Fig. 4a).
-
29
Plasma treat the slides within a plasma cleaner set to 30 W for 10 min to eliminate any remaining surface contaminants and improve adhesion of the 1002F photoresist.
-
30
Transfer the cleaned substrates to a spin coater. Dispense ca. 2 mL of 1002F-10 photoresist over the center of the slide. Spread the dispensed 1002F-10 over the slide at 500 rpm for 10 s with an acceleration of 100 rpm s−1, then spin at 3000 rpm for 30 s with an acceleration of 300 rpm s−1 for a final film thickness of ca. 5 μm.
-
31
Soft bake: Incubate the slide for 30 min in an oven set to 95 °C.
-
32
Expose the entire film to a 1000 mJ cm−2 dose of UV light using a collimated UV exposure system. ▲CRITICAL No photomask is utilized in this step.
-
33
Post-exposure bake: Incubate the slide for 10 min in a 95 °C oven or hot plate to allow the photoacid to initiate crosslinking of the epoxy.
-
34
Hard bake: Incubate the slide for an additional 30 min on a hot plate set to 150 °C to harden the epoxy film (Fig. 4a).
-
35
Remove the slide from the hot plate and cool to room temperature.
-
36
Plasma treat the surface within a plasma cleaner set to 30 W for 10 min to cleanse the crosslinked 1002F of surface contaminants and improve the adhesion of additional 1002F layers.
-
37
Transfer the substrates to a spin coater. Dispense ca. 2 mL of 1002F-100 photoresist over the center of the slide. Spread the dispensed 1002F-100 over the slide at 500 rpm for 10 s with an acceleration of 100 rpm s−1, then spin at 1000 rpm for 30 s with an acceleration of 300 rpm s−1 to generate a film that is ca. 150 μm thick.
-
38
Soft bake: Incubate the slide for 2 h in an oven set to 95 °C.
-
39
Repeat Steps 37–38 two additional times to generate three total layers. The final thickness of the multilayer 1002F film is 420–450 μm (Fig. 4a).
■PAUSE POINT UV exposure and development steps can be performed on the next day. Remove the slide(s) from the oven at the end of the final baking step, and store them in the dark at room temperature (20–22 °C).
-
40
Transfer the substrate(s) to the UV exposure system. Remove any debris from the back (i.e., glass-exposed face) of the slide by wiping with an acetone-soaked cleanroom wiper. Invert the substrate and place the 1002F-coated side facing down. The photomask (Fig. 4) is then placed with the chrome side of the mask facing down, in conformal contact with the glass-exposed face of the photoresist-coated slide. Expose the assembly to a 1200 mJ cm−2 dose of UV light. ▲CRITICAL This backside exposure strategy (Fig. 5a) is used to generate slanted, rounded-apex micropillars, enabled by light scattering within the 1 mm gap (thickness of the glass slide) between the chrome mask and the photoresist film.
-
41
Post-exposure bake: Incubate the slide in a 95 °C oven for 30 min, followed by a 120 °C hot plate for 10 min.
-
42
Develop by incubating for 60 min in propylene glycol methyl ether acetate (PGMEA) developer under mild agitation (ca. 100 rpm) using an orbital shaker.
-
43
Sequentially rinse with PGMEA and IPA, then dry with compressed air (Fig. 5a). ▲CRITICAL The development time can be variable depending on degree of agitation. Utilize 60 min as a starting point and check the micropillars under a microscope. If they do not appear fully developed (e.g., remaining 1002F is connecting pillars, or other irregularities are present), place the slide back in PGMEA and continue to develop, rinse, dry, and inspect in 10 min increments.
-
44
Debris may become trapped in the micropillars during processing, which may be removed using PDMS. Within a disposable plastic weighing dish, combine Sylgard 184 silicone elastomer base and curing agent in a 10:1 weight ratio. Mix the solution using a spatula and degas in a vacuum desiccator until bubbles are completely removed. Dispense ca. 3 mL of the mixture over the slide and degas to remove air bubbles trapped in between the micropillars. Cure the mixture in an oven set to 95 °C for 10 min. Peel away PDMS to remove trapped debris, and discard. Repeat this PDMS application and delamination process one more time.
-
45
Hard bake: Incubate the master mold on a hot plate set to 150 °C for 30 min.
Fig. 5 |.
Microfabrication and cell culture insert modification for in vitro crypts. a, Photolithography workflow to produce micropillar master molds of 1002F resin. b, Soft lithography replica molding of PDMS to produce stamps that will be used for micromolding collagen. c, Cell culture insert modification and in situ micromolding of collagen. d, Completed cell culture inserts for in vitro crypts, and characterizations of the micromolded collagen scaffolds. Panels b and d are adapted from refs. 24 and 37 with permission from Elsevier and the American Chemical Society (Copyright 2019), respectively.
?TROUBLESHOOTING
■PAUSE POINT The 1002F master mold can be stored indefinitely at room temperature.
PDMS crypt stamp fabrication (soft lithography) ●Timing 2 d
-
46
Combine Sylgard 184 silicone elastomer base and the curing agent in a 10:1 weight ratio, and degas the mixture.
-
47
Dispense ca. 3 mL of the PDMS mixture over the 1002F master mold, and degas to remove air bubbles trapped between the micropillars.
-
48
Cure the mixture in an oven set to 95 °C for 30 min.
-
49
Peel the PDMS replica from the 1002F master mold. This demolded PDMS possesses an array of microwells (Fig. 5b).
-
50
Place the PDMS replica over a glass slide, functioning as a solid support, with the microwells facing upward, away from the glass slide. Verify that no air bubbles are trapped between the PDMS and the glass slide.
-
51
Incubate the substrate on a hot plate set to 150 °C for 2 h to harden the PDMS.
-
52
Plasma treat PDMS within a plasma cleaner set to 30 W for 2 min. This step generates an oxide layer over the PDMS surface to facilitate subsequent silane attachment.
-
53
Within a chemical fume hood, transfer the PDMS replica into a vacuum desiccator chamber with the microwells facing upward. Dispense 100 μL of trichloro(octyl)silane into a clean petri dish placed at the center of the chamber. Vacuum the chamber for 2 min using an oil-free vacuum pump. Close the main valve of the chamber and disconnect from the vacuum pump. Incubate for 16 h.
!CAUTION This silane is corrosive. Use a chemical fume hood and wear protective goggles, gloves and suitable personal protective equipment when handling.
-
54
Slowly vent the chamber within the chemical fume hood. Remove the petri dish with silane residue from the chamber and discard. Vacuum the chamber for 2 min using an oil-free pump, then vent. Repeat this vacuum/vent process two additional times.
-
55
Dispense ca. 2 mL of the Sylgard 184 mixture (Step 46) over the PDMS replica, and degas to remove air bubbles trapped within the microwells. Cure the mixture in an oven set to 95 °C for 10 min.
-
56
Repeat Step 55 until the desired height of the final stamp is reached (2–3 additional times). We have found that a height of 2–4 mm is relatively easy to hold and manipulate using forceps.
-
57
Demold the PDMS stamp from the PDMS replica. This demolded PDMS stamp possesses an array of micropillars that is identical to that on the 1002F master mold (Fig. 5b). The PDMS replica possessing microwells can be used for ca. 10 future stamp fabrications, after which, wrinkles and distortions begin to appear.
-
58
Harden the PDMS stamp by incubating on a 150 °C hot plate for 30 min.
-
59
Cool the PDMS stamp to room temperature, then trim with a razor blade to final dimensions of 4×4×4–5 mm (length×width×height, Fig. 5b).
■ PAUSE POINT PDMS stamps can be stored indefinitely at room temperature.
Surface treatment of PDMS stamps ●Timing 2 d
▲CRITICAL The surface of the PDMS stamps must be treated with poly(ethylene glycol) (PEG) to prevent adhesion of collagen and facilitate straightforward demolding during the subsequent hydrogel micromolding steps.
-
60
Prepare the “PEG Photografting Solution” fresh, or retrieve stock from 4 °C storage.
-
61
Place PDMS stamps in a round bottom glass tube, and fill with PEG grafting solution. Seal tightly with a PTFE screw cap.
-
62
Place the glass tube into a glass petri dish for support, and transfer onto the Loctite 400 W Metal Halide UV Flood System platform. Expose to UV radiation for 4 h. ▲CRITICAL Shake the tube every 30 min to dislodge any bubbles that become trapped within the micropillars, allowing for their uniform exposure to the photografting solution.
!CAUTION To prevent exposure of any part of your body to ultraviolet radiation, wear UV light protective goggles, gloves and suitable protective clothing.
!CAUTION The glass tube becomes very hot after UV irradiation, and heat-insulating gloves may be required for handling.
-
63
Thoroughly rinse the stamps with DI water, then incubate in DI water overnight to remove any non-covalently attached photografting solution components.
-
64
Rinse with 75% ethanol, and store in a conical tube filled with 75% ethanol until use.
?TROUBLESHOOTING
■PAUSE POINT The PEG-grafted PDMS stamps can be stored indefinitely in 75% ethanol at room temperature. The PEG coating is effective for at least 10 cycles of collagen hydrogel micromolding.
Assembly of device cassettes ●Timing 1–2 h
-
65
Remove the pre-installed membrane from a commercial 12-well cell culture insert (e.g., Transwell, Falcon, etc.) using a cutting tool. Cleanse with DI water and dry with a Kimwipe or compressed air (Fig. 5c). ▲CRITICAL Track-etched poly(ethylene terephthalate) (PET) and polycarbonate membranes, which are most commonly installed in commercial cell culture inserts, do not possess sufficient permeability to stem-cell supporting growth factors for in vitro crypts. These must be removed and replaced with fibrous polytetrafluoroethylene (PTFE) to permit in vitro crypt establishment.
-
66
Unfold the adhesive tape from the roll, place the inserts and cut the tape/backing around the inserts to separate from the adhesive roll or sheet. Run a craft knife along the outside and inside edges of the insert, taking care to only cut through the adhesive, but not the backing. Remove residual pieces of adhesive using a Kimwipe and/or forceps and discard.
!CAUTION Try not to touch the adhesive, as the adhesive possesses a very high tack and is exceptionally difficult to separate from nitrile gloves or skin.
-
67
Place adhesive coated basal rim in contact with a fresh, hydrophilic PTFE membrane (i.e., Biopore). Fold up any overhanging areas of the membrane toward the outside wall of insert to facilitate a tight seal between the membrane, the adhesive, and the insert base.
-
68
Attach transfer adhesive to the polycarbonate film, leaving adhesive backing on one face of the adhesive. Cut to squares of ca. 15 mm in length that can be easily manipulated around each cell culture insert. ▲CRITICAL The polycarbonate films often ship with transparent protective layers on one or both faces, which should be removed prior to assembly.
-
69
Using a biopsy punch, create a 3 mm dia. hole in the center of each polycarbonate/adhesive square. ▲CRITICAL Microscopy imaging may be a challenge if the window is placed sufficiently close to the wall of the cell culture insert.
-
70
Remove the adhesive backing and center the 3 mm hole with the base of the cell culture insert. Firmly press the surfaces together.
▲CRITICAL STEP Remove any air pockets and confirm that the adhesive is in conformal contact with all surfaces.
-
71
Any remaining material overhanging from the edge of the inserts can be trimmed with a craft knife or scissors.
-
72
Transfer the modified inserts into a 12-well plate.
■PAUSE POINT The modified inserts can be prepared in large batches and stored indefinitely at room temperature.
Micromolding of shaped hydrogel scaffolding ●Timing 2 d
-
73
Remove the PEG-grafted PDMS stamps from their storage solution and dry for 1 h before use.
-
74
Inspect the micropillars using a bright-field microscope. Verify that no defects are present in the micropillar features, and that they are free of dust, debris, and collagen (if the stamp is being reused from previous moldings). If any dust or debris is present, cleanse the stamps under a stream of 75% ethanol, dry, and inspect again.
?TROUBLESHOOTING
-
75
Prepare the collagen mixture that will be used for forming the shaped hydrogel scaffolds. Combine 50 μL of 0.6 M EDC and 50 μL of 0.15 M NHS within a microcentrifuge tube over ice and mix thoroughly by pipetting. Dispense 400 μL of MES-solubilized collagen (5 mg mL−1, pH 5) into the same vessel, and mix by gently pipetting up and down 40 times. Centrifuge at 15,000 ×g for 1 min if air bubbles form within the mixture.
!CAUTION This mixture will form a stiff gel within 5 min. Keep the solution cold (i.e., on ice) to limit the reaction rate and carry out the solution preparation and molding steps as quickly as possible.
-
76
Dispense 50 μL of the collagen/EDC/NHS mixture at the center of the luminal compartment within each modified cell culture insert (i.e., directly above the 3 mm dia. diffusion window).
-
77
Using forceps, apply and press a PDMS micropillar stamp into the gel within each insert, micropillar-side down. If there are any visible bubbles caught between the stamp and gel, the stamp can be lifted with forceps and repositioned (Fig. 5c).
▲CRITICAL STEP PDMS stamps should appear by naked eye to be in contact, or nearly in contact, with the PTFE membrane (i.e., not floating on top of the gel, Fig. S1, Supplementary Information).
-
78
Transfer the cell culture inserts with PDMS molds into a 12-well cell culture plate (if they are not already) and place the plate into a pneumatic curing vessel/pressure pot.
-
79
Apply 20 psi (200 kPa) of pressure from a nitrogen source. Start a timer for 5 min, and maintain an active supply of gas. The elevated pressure serves to eliminate trapped air pockets within the crosslinked hydrogel and force the viscous collagen mixture into the micropost void areas (Fig. S1, Supplementary Information).
!CAUTION The manufacturer of the pressure chamber utilized in this protocol recommends a maximum internal pressure of 22 psi. Use of higher pressures may compromise the integrity of the seals and/or gaskets.
-
80
After 5 min, terminate the gas flow and incubate the cassettes in the chamber for 2 h. The chamber will gradually de-pressurize during this time.
-
81
Open the quick-vent valve on the chamber to vent any residual gas before opening. Remove the 12-well plate with cell culture inserts.
-
82
Add 1 mL 1× PBS to the luminal compartment of each insert, and incubate for at least 10 min.
-
83
Carefully lift each PDMS stamp vertically from each cell culture insert with forceps. Using a bright-field microscope, verify that the microwells are present in each collagen scaffold without any distortions (Fig. 5c,d). ▲CRITICAL PDMS stamps must be lifted vertically (i.e., straight up) from the collagen to minimize tilting or bending of the microwells (Fig. S1, Supplementary Information).
?TROUBLESHOOTING
-
84
Transfer the cell culture inserts into a 2 L vessel of DI water to cleanse the scaffolds of residual crosslinking reagents. Incubate overnight at room temperature (20–22 °C).
-
85
Decant the DI water from the vessel and add enough 75% ethanol solution to cover the cell culture inserts.
-
86
Transfer the vessel with cell culture inserts into to a sterile biosafety cabinet, and incubate in the 75% ethanol solution for 5 min.
-
87
Transfer the cell culture inserts from the vessel into a 12-well cell culture plate. Decant any ethanol solution from the inserts back into the 2 L vessel.
-
88
Dispense 1 mL of 1× PBS into each luminal compartment, and 2 mL of 1× PBS into each basal compartment. Incubate for 30 min with the lid off.
-
89
Aspirate the solution from each compartment and dispense the same volumes as Step 88 of fresh buffer into each compartment.
■PAUSE POINT If not using immediately, the 12-well plate with cell culture inserts can be stored within a sterile sampling bag at 4 °C until use.
-
90
The day before passaging cells onto the shaped hydrogel scaffolds, dilute VitroCol human collagen to a concentration of 10 μg mL−1 in 1× PBS.
-
91
Aspirate the storage solution from the luminal and basal compartments of the cell culture inserts. Dispense 1 mL of the diluted human collagen solution into each luminal compartment, and 1 mL of the diluted human collagen into each basal compartment. Incubate overnight at room temperature (20–22 °C).
▲CRITICAL STEP Crosslinking of the hydrogel scaffolds is necessary for increasing their mechanical strength so that their three-dimensional shape is maintained throughout culture. However, this process also eliminates surface binding sites that are recognized by the cells as they attach to the matrix and establish focal adhesions. Incubation of diluted collagen overnight deposits a monolayer of collagen over the scaffold and restores these binding sites.
Culture of in vitro human colon crypts ●Timing 12 d
-
92
Aspirate human collagen solution from the luminal and basal compartments of the cell culture inserts with neutralized collagen scaffolds (i.e., not shaped with crypt structures). Dispense 1 mL of 1× PBS into each luminal compartment, and 2 mL of 1× PBS into each basal compartment. Incubate room temperature until cells are ready to be transferred.
-
93
Incubate Expansion Medium (EM), Dissociation Buffer, and 10 mM Y-27632 stock to 37 °C in a water or bead bath for 30 min, or until warm. Retrieve collagenase stock solution (5000 U mL−1) from 4 °C storage and transfer into a sterile biosafety cabinet, along with 1× PBS and (1) 15 mL conical tube.
-
94
Aliquot the volume of EM required for initial plating (both luminal and basal compartments, 3 mL total per cell culture insert). Dilute stock Y-27632 in EM to a concentration of 10 μM. ▲CRITICAL The cells will be passaged at a basal surface area ratio of 2:1 for seeding over the shaped hydrogel scaffolds (i.e., (1) confluent 6-well can be subcultured to (4) modified 12-well cell culture inserts possessing shaped hydrogel scaffolds). Plan according to the number of wells that are being passaged and experimental requirements.
-
95
Passage primary human colon epithelial cells from neutralized collagen hydrogel scaffolds that are ≥80 % confluent, following Steps 10–23 of this protocol.
-
96
Prepare a double pipet tip for fragmentation of the cell colonies: Place a 2–200 μL pipet tip over the end of a P200 micropipette, then place a 0.1–20 μL pipet tip over end of the 2–200 μL pipet tip (Fig. 6a). ▲CRITICAL The shear forces generated by the double pipet tip result in more efficient fragmentation of the cell colonies than the 2–200 μL pipet tip alone, with minimal effect on cell viability. The increased fragmentation allows for a higher yield of in vitro crypts, but is not necessary for routine maintenance over neutralized collagen hydrogels (Fig. S2, Supplementary Information).
-
97
Using a P200 micropipette equipped with the double pipet tip, break up the cell pellet into smaller fragments by pipetting up/down 40 times.
-
98
Dispense the required volume (1 mL luminal compartment−1) of EM+Y-27632 to the fragmented cell suspension.
-
99
Aspirate storage buffer from the cell culture inserts with shaped hydrogel scaffolds, beginning with the basal compartments, and ending with the luminal compartments.
!CAUTION The time during which collagen scaffolds are exposed to air should be minimized.
-
100
Plate cells within EM+Y-27632 into the luminal compartment of each cell culture insert. (1 mL luminal compartment−1). Dispense 2 mL of EM+Y-27632 (without cells, remaining from Step 94) into each basal compartment.
-
101
Transfer the plate into a humified CO2 incubator at 37 °C. Refrain from adjusting or disturbing the plate until Day 2 to allow the cells to settle into the bases of the microwells (Fig. 6b).
-
102Replenish the medium every 48 h with 3 mL of fresh, 37 °C EM (without Y-27632): Aspirate the spent medium, and dispense 1 mL of EM into each luminal compartment, and 2 mL of EM into each basal compartment at Days 2, 4, and 6 (Fig. 6b).
- (Optional) The cells can be fixed and stained for proliferative biomarkers (e.g., Ki67, EdU incorporation – see Table S1, Supporting Information) at any point during expansion (Days 0–8) to verify that cells are maintaining proliferative activity in all crypt regions.
-
103By Day 8, the intestinal epithelial cells should be 100 % confluent across the micromolded collagen scaffold array (Fig. 6b). Aspirate spent EM from each compartment and initiate polarization of the in vitro crypts into stem/proliferative and differentiated phenotypic compartments by dispensing 0.5 mL of Differentiation Medium (DM) into each luminal compartment and 1.5 mL of Stem Medium (SM) into each basal compartment. ▲CRITICAL These volumes are optimized for stable gradient formation and matching of hydrostatic pressures between compartments.
- (Optional) Gradient formation may be confirmed by sampling of the medium every 24 h and quantifying Wnt3a, R-spondin, and noggin in the basal and luminal compartments. Alternatively, a 40 kDa fluorescent dextran (100 μg mL−1) can be utilized as a surrogate for these growth factors and measured directly via fluorescence.
- (Optional) To eliminate compartmentalization, thus establishing a negative control for crypt polarization, DM or SM can be added to both basal and luminal compartments, resulting in crypt scaffolds that are covered with entirely differentiated or stem/proliferative cells, respectively.
- (Optional) The level of L-WRN conditioned medium within SM can be titrated up or down (from 50% v/v) to vary stem/proliferative compartment size along the crypt long axis.
- (Optional) Additional growth factors, metabolites, drugs, and/or pharmacological inhibitors can be added during this period to elicit desired effects or test experimental hypotheses.
-
104
Replenish the medium from each compartment every 24 h for 4 d: Aspirate the spent medium, and dispense 0.5 mL of DM into each luminal compartment and 1.5 mL of SM into each basal compartment at Days 9, 10, and 11.
-
105
Assay the in vitro crypts and/or subject to chemical fixation for staining on Day 12 or beyond (i.e., ≥4 d of chemical gradient conditions). The in vitro crypts exhibit tissue-level self-renewing capabilities, analogous to in vivo tissue, and can be cultured in this manner for at least 32 d (Fig. 6b,c).
?TROUBLESHOOTING
Fig. 6 |.
Formation and characterization of in vitro crypts. a, Assembly of the double pipet that is used for fragmenting cell colonies prior to in vitro crypt seeding. b, Characterization of in vitro crypt growth during expansion and chemical gradient phases of culture. Inset scale bars represent 250 μm. c, Assay and location of several key biomarkers for stem/proliferative and differentiated cell compartments. Panel b is adapted from ref. 37 with permission from the American Chemical Society (Copyright 2019). Panel c is adapted from refs. 24 and 37 with permission from Elsevier and the American Chemical Society (Copyright 2019), respectively. The human colonic biopsy specimen (male, 52 y) utilized to generate the data in this figure was obtained during a routine screening colonoscopy performed at the University of North Carolina (UNC) Hospitals Meadowmount Endoscopy Center under UNC IRB #14–2013.
Troubleshooting
Timing
Box 1, Neutralized collagen hydrogel fabrication for monolayer expansion: 2 h
Box 2, Isolation of living crypts from primary intestinal tissue: 4–6 h
Steps 1–27, Monolayer culture and passaging of primary intestinal epithelial cells: 5–8 d
Steps 28–45, 1002F crypt master template fabrication (photolithography): 2 d
Steps 46–59, PDMS crypt stamp fabrication (soft lithography): 2 d
Steps 60–64, Surface treatment of PDMS stamps: 2 d
Steps 65–72, Assembly of device cassettes: 1–2 h
Steps 73–91, Micromolding of shaped hydrogel scaffolding: 2 d
Steps 92–102, Expansion of primary intestinal epithelial cells over shaped scaffolds: 8 d
Steps 103–105, Generation of stem/proliferative and differentiated cell zones to form in vitro crypts: 4 d
Anticipated Results
Neutralized collagen hydrogel scaffolds possess the appropriate mechanical properties and biochemical cues for primary intestinal stem cell attachment and propagation (Fig. 3c). Throughout storage and culture, the hydrogels should remain securely attached to the polystyrene cell culture plate, without any evidence of lifting or degradation. At the density (1 mg mL−1) and depth (ca. 1–2 mm central thickness) fabricated, the neutralized collagen hydrogels should appear colorless and optically transparent (Fig. 3a). In conjunction with the formulated Maintenance Medium, containing Wnt3a, R-spondin 3, and noggin from the L-WRN cell line,19 in addition to recombinant EGF, the epithelial cells should expand as monolayer patches over the hydrogel surface with doubling times of ca. 24 h (Fig. 3c).20 These patches should reach ≥80 % confluency 5–8 d after plating over the hydrogels, after which, they are ready for further subcultivation. Within our laboratory, we prefer passaging on Day 5–6, as there is a higher population of stem/proliferative cells in culture that will rapidly expand upon plating over fresh hydrogels. These culture methods yield primary cell lines that exhibit minimal genetic drift for at least 15 passages (i.e., 75–120 days in culture) before exhibiting any chromosomal abnormalities.
During fabrication of the 1002F master template, the micropillars should firmly attach to the glass substrate (Fig. 5a, Steps 36–45), previously superimposed with a continuous thin film of crosslinked 1002F (Fig. 5a, Steps 28–34). Using the process parameters defined in the protocol, the micropillars should exhibit the following dimensions: 435 ± 15 μm height, 65 ± 6 μm basal diameter, and 128 ± 6 μm luminal diameter, along with angled side walls that taper to a convex apex (Fig. 5a).24,37 These dimensions should transfer precisely to the PDMS replica (microwells), PDMS stamps (micropillars), and micromolded collagen scaffolds (microwells) (Fig. 5b). Throughout ultraviolet surface treatment of the PDMS stamps, the PEG grafting solution will visibly change from cloudy to clear. The PDMS stamps will also exhibit higher hydrophilicity and become slippery after the 4 h treatment. No collagen should remain attached to the PDMS micropillars during shaping of the scaffolds or demolding. These shaped scaffolds, in conjunction with the crosslinking process and at the density (4 mg mL−1) and depth (450–500 μm) fabricated, should appear colorless and optically transparent (Fig. 5d).
Once plated over the shaped collagen hydrogel scaffolds, the intestinal epithelial cells will grow over the luminal surface and down to the base of the microwells when cultured in Expansion Medium (Fig. 6b). Forming a contiguous monolayer of cells that is 100% confluent should take no more than 8 d, but may be accomplished sooner. Over this time, the shape of the microwells should remain fully intact, without any sign of degradation or flattening, as a result of the crosslinking process. Due to the higher stiffness of the crosslinked scaffold, it is normal for some three-dimensional cellular macrostructures (e.g., quasi-organoids) to extend from the luminal surface during the first 2–3 d, though these will flatten into planar monolayers over the course of the expansion phase (8 d). During in vitro crypt polarization, in which opposing chemical gradients are introduced from the basal Stem Medium and luminal Differentiation Medium, liquid levels should remain at matching heights in the basal and luminal compartments, and no gross changes in cell morphology should occur. The polarized crypts are viable for at least 32 d, during which, epithelial cells will proliferate within the bases of the in vitro crypts, migrating up toward the luminal surface and differentiating into absorptive and secretory lineages, as in vivo. The differentiated cells will die off and slough into the lumen, being constantly replaced by upwardly migrating progeny of the stem cells, mimicking the self-renewal characteristics of the in vivo tissue. There should be no large-scale delamination of the cultured cells from the luminal plane as this indicates that the cells are dying in mass without continual replacement. Our laboratory typically characterizes this compartmental phenotypic polarization using fluorescence microscopy (Fig. 6c), with each of the markers below applicable to cells from the small or large intestine. The stem/proliferative compartment can be identified by immunofluorescent staining for stem/proliferative biomarkers, such as olfactomedin 4 (Olfm4), SRY-box transcription factor 9 (Sox9), and Antigen KI-67 (Ki67).24,37,81,82 However, we have found incubation (i.e., pulsing) of 5-ethynyl-2’-deoxyuridine (EdU) during the final 24 h of culture, followed by alkyne-azide click labeling with an appropriate fluorophore to be the most reproducible method for stem/proliferative compartment identification.83 The differentiated compartment can be generally identified by immunofluorescent staining for cytokeratin-20 (KRT20), along with more specific markers for absorptive and secretory lineages, including mucin-2 (Muc2), alkaline phosphatase (ALP), serotonin (5-HT), and chromogranin A (ChgA).24,37 These stem/proliferative and differentiated populations are easily modulated by the presence of known Notch signaling regulators in the luminal compartment.24,37 Antibodies and labeling kits routinely used within our laboratory that have been validated for this platform are provided in Supplementary Table 1, with respective labeling methods for the images in Fig. 6c available in our previous publications.24,37
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data Availability
The data that support the findings of this study are reported in previous publications.
Supplementary Material
Fig. S1. Effects of improper micromolding.
Fig. S2. Images of stunted and dead in vitro crypts.
Fig. S3. Unpolarized in vitro crypts.
Table S1. Routine solutions validated for labeling primary human intestinal epithelial cells cultured over collagen hydrogel scaffolds.
Sheet 1) Collagen Neutralization Volumes. Calculator for neutralization buffer formulations and mixing volumes during collagen hydrogel formation.
Sheet 2) Subcultivation Ratios. Calculator for subcultivating primary intestinal epithelial cells from 6-well collagen hydrogel scaffolds to various culture sizes/formats
Table 2 |.
Troubleshooting
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 3 | Neutralized collagen hydrogels have lifted from the culture plate base during storage or culture | Air bubbles were trapped within the hydrogel during formation | Centrifuge at 2000 ×g or higher for at least 30 s to remove any visible or microscopic air bubbles from the mixture during hydrogel formation |
| Neutralization buffer is incorrectly formulated | Verify calculations so that the amount of NaOH present in the collagen/buffer mixture is equimolar to the amount of acetic acid. Final collagen density should be 1 mg mL−1. | ||
| Neutralization buffer is too old | The presence of sodium bicarbonate within the neutralization buffer causes the pH to gradually rise during storage. This can be mitigated by adjusting the pH of the buffer prior to neutralization of collagen if an ideal target pH is known. Otherwise, make the neutralization buffer fresh each time. | ||
| Insufficient incubation time | Attempt longer incubation of the collagen/neutralization buffer mixture at 37 °C (up to 2 h is acceptable). | ||
| Culture medium has become too acidic, and the collagen is re-solubilizing | Ensure proper buffering of the Maintenance Medium: HEPES must be present at a concentration of 10 mM. | ||
| 9, 27 | Cells are not propagating and becoming confluent over the neutralized collagen hydrogels | Scaffolds were not incubated at room temperature for a sufficient amount of time prior to use | Incubate the scaffolds in 1× PBS at room temperature for at least 15 d prior to culture |
| Plating density of crypts or subcultivation ratio is too low | Increase crypt plating density to >1,000 crypts well−1 or modify subcultivation surface area ratio from 1:3 to 1:2. | ||
| Maintenance Medium is greater than 1 month old, and growth factors have degraded | Re-formulate Maintenance Medium from fresh stock components. | ||
| 27 | Cells have died after passaging | Dissociation Buffer was formulated incorrectly | Re-formulate dissociation buffer, ensuring that it contains no more than 0.5 mM EDTA and no less than 10 μM Y-27632. |
| 18 | Cloudy layer above the cell pellet after centrifugation | Collagenase is old/less active, leading to less breakdown of the hydrogel | Resuspend the pellet and the collagen layer by shaking and further incubate at 37 °C for an additional 5 min. Check if collagenase is >1 month old. If so, thaw new working aliquot. |
| 45 | Micropillars are uneven and different heights are observed | Spin coater is not level during each deposition of 1002F 100 | Relevel the spin coater and attempt again. |
| Micropillars are detaching from the surface | Steps 28–34 were skipped | A thin layer of 1002F 10 film must be applied and fully cured. | |
| Substrate was overdeveloped with PGMEA | Shorten the PGMEA development time. | ||
| Micropillars are bent | Overdevelop with PGMEA | Re-attempt photolithography process, and use a shorter development time (or dry/check development in shorter intervals). | |
| Underexposure with UV | If problem persists with shorter development times, attempt higher UV exposure dose. | ||
| Sidewall of micropillar is not slanted | Improper UV exposure dose (too high/low) | Optimize UV exposure dose. Use 1,000 mJ cm−2 as the starting point and try increasing/decreasing the UV exposure dose in 100 mJ cm−2 increments. | |
| UV exposure was performed on the front (i.e., 1002F) face of the glass support | Verify that the UV exposure is performed through the back (i.e., glass) face of the substrate coated with 1002F. | ||
| 64 | PEG is not grafted on PDMS stamps | UV irradiation is not powerful enough | The grafting of PEG requires a powerful UV irradiation, equivalent to Loctite ZETA 7411-S UV Flood System. |
| Photografting solution components have expired | Verify expiration dates of the solution components, and replace if necessary. | ||
| 74, 83 | Collagen is trapped within the PDMS pillars | PEG coating has degraded over time | The stamps can be rescued by re-performing the surface treatment (Steps 46–59) in which UV irradiation will completely degrade the entrapped collagen. |
| 83 | Collagen gel is trapped within the PDMS pillars | Insufficient rehydration time | Incubate in 1× PBS for a longer period of time (up to 2 h). |
| Collagen hydrogel did not set or appears unstable | EDC or NHS stocks reacted before mixing | Re-formulate the crosslinking reagents, and use immediately. | |
| Acetic acid was not completely removed during lyophilization, and the reaction environment is too acidic | Lyophilize a new batch of collagen for a greater length of time. The lyophilized product should appear completely desiccated with no visible traces of ice or moisture. | ||
| Sidewalls of the shaped hydrogel microwells appear bent or distorted (Fig. S1, Supplementary Information) | PDMS stamp was not lifted vertically from the gel, causing its deformation | Re-attempt the hydrogel micromolding process, with focus on a gentle and controlled vertical de-molding. | |
| PDMS stamp had defects prior to micromolding | Inspect the PDMS stamp to see if complementary distortions or bent micropillars are also present. If so, discard and re-make. | ||
| 101–103 | Cells not expanding over micromolded collagen scaffold | Human collagen coating not efficient | Incubation time and temperature may need to be optimized. |
| Expansion Medium is greater than 1 month old, and growth factors have degraded | Re-formulate Expansion Medium from fresh stock components. | ||
| 105 | In vitro crypts do not polarize into stem/proliferative and differentiated cell zones: proliferative cells (EdU+, Ki67+) are present in all regions (Fig. S3, Supplementary Information) | Growth factors have leaked from the basal compartment into the luminal compartment outside of the diffusion window | Ensure conformal contact and a tight seal between the adhesive/cell culture insert rim as well as the PTFE membrane/adhesive/polycarbonate film. |
| Growth factor (Wnt3a, R-spondin 3, noggin) concentrations in SM are too high | Test lower dilution ratios of L-WRN conditioned medium during preparation of SM (e.g., 50%, 20%, 10%) | ||
| Dimension of the crypt scaffold may be shorter than the design constraints allow | Modify fabrication of the 1002F master mold to achieve optimized crypt length (420–450 μm) | ||
| In vitro crypts do not polarize into stem/proliferative and differentiated cell zones: proliferative cells (EdU+, Ki67+) are absent in all regions | Growth factor (Wnt3a, R-spondin 3, noggin) concentrations in SM are too low | Test higher dilution ratios of L-WRN conditioned medium during preparation of SM (e.g., 50%, 60%, 70%) | |
| Stem Medium is greater than 1 month old, and growth factors have degraded | Re-formulate Stem Medium from fresh stock components. | ||
| >30% of in vitro crypts are filling with dead cells (Fig. S2, Supplementary Information) | Propagated cells contained a higher differentiated population than ideal for in vitro crypt formation | Passage cells from neutralized hydrogels 1–2 d earlier to minimize population of spontaneously differentiated cells | |
| In vitro crypts only grow halfway down the microwells (i.e., appear stunted) or are imperforate at the luminal plane (i.e., sheet of cells cover the crypt opening) (Fig. S2, Supplementary Information) | Cell colonies were subjected to insufficient fragmentation and clog the microwells instead of settling to their base | Using the double pipet tip method, pipet up/down an additional 10–20 times in Dissociation Buffer before plating the cells over the shaped scaffolds. 150 μL of Accutase (Corning, cat. no. 25–058-CI) containing 10 μM Y-27632 can alternatively be utilized in place of Dissociation Buffer for gentle, enzymatic breakdown of the colonies, though incubation time must be extended (e.g., 15–20 min) |
Acknowledgements
The authors gratefully acknowledge financial support from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) of the National Institutes of Health (R01 DK109559) and the University of Washington (start-up funding). We additionally wish to thank Prof. Scott T. Magness for procurement of the intestinal biopsy specimen, and Drs. Johanna S. Dutton, Jennifer C. Huling, and Angela Proctor for assistance in assembling the working versions of these protocols that are used within our research group.
Footnotes
Competing interests
N.L.A. and Y.W. have a financial interest in Altis Biosystems, Inc. The remaining authors declare no competing financial interests.
Key References
20 Wang, Y. L. et al. Self-renewing Monolayer of Primary Colonic or Rectal Epithelial Cells. Cell. Mol. Gastroenterol. Hepatol. 4, 165–182.e167, doi:10.1016/j.jcmgh.2017.02.011 (2017).
24 Wang, Y. et al. Formation of human colonic crypt array by application of chemical gradients across a shaped epithelial monolayer. Cell. Mol. Gastroenterol. Hepatol. 5, 113–130, doi:10.1016/j.jcmgh.2017.10.007 (2018).
25 Wang, Y. L. et al. A microengineered collagen scaffold for generating a polarized crypt-villus architecture of human small intestinal epithelium. Biomaterials 128, 44–55, doi:10.1016/j.biomaterials.2017.03.005 (2017).
References
- 1.Wang Y et al. Bioengineered Systems and Designer Matrices that Recapitulate the Intestinal Stem Cell Niche. Cell. Mol. Gastroenterol. Hepatol, doi: 10.1016/j.jcmgh.2018.01.008 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dutton JS, Hinman SS, Kim R, Wang YL & Allbritton NL Primary Cell-Derived Intestinal Models: Recapitulating Physiology. Trends Biotechnol. 37, 744–760, doi: 10.1016/j.tibtech.2018.12.001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker N et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–U1001, doi: 10.1038/nature06196 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Sangiorgi E & Capecchi MR Bmi1 is expressed in vivo in intestinal stem cells. Nat. Genet 40, 915–920, doi:DOI 10.1038/ng.165 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greicius G et al. PDGFRalpha(+) pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo. Proc. Natl. Acad. Sci. U. S. A 115, E3173–E3181, doi: 10.1073/pnas.1713510115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shoshkes-Carmel M et al. Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature 557, 242–246, doi: 10.1038/s41586-018-0084-4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu YC, Li LS & Fuchs E Transit-Amplifying Cells Orchestrate Stem Cell Activity and Tissue Regeneration. Cell 157, 935–949, doi: 10.1016/j.cell.2014.02.057 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker AM et al. Quantification of crypt and stem cell evolution in the normal and neoplastic human colon. Cell Rep. 8, 940–947, doi: 10.1016/j.celrep.2014.07.019 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.VanDussen KL et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development 139, 488–497, doi: 10.1242/dev.070763 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaiko GE et al. The Colonic Crypt Protects Stem Cells from Microbiota-Derived Metabolites. Cell 167, 1137−1137, doi: 10.1016/j.cell.2016.10.034 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Verma MS et al. A Common Mechanism Links Activities of Butyrate in the Colon. ACS Chem. Biol 13, 1291–1298, doi: 10.1021/acschembio.8b00073 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Bultman SJ Butyrate consumption of differentiated colonocytes in the upper crypt promotes homeostatic proliferation of stem and progenitor cells near the crypt base. Transl. Cancer Res 5, S526–S528, doi: 10.21037/tcr.2016.08.36 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parada Venegas D et al. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol 10, 277, doi: 10.3389/fimmu.2019.00277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gareau MG, Sherman PM & Walker WA Probiotics and the gut microbiota in intestinal health and disease. Nat. Rev. Gastroenterol. Hepatol 7, 503–514, doi: 10.1038/nrgastro.2010.117 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peters MF et al. Developing in vitro assays to transform gastrointestinal safety assessment: potential for microphysiological systems. Lab Chip 20, 1177–1190, doi: 10.1039/c9lc01107b (2020). [DOI] [PubMed] [Google Scholar]
- 16.Jung P et al. Isolation and in vitro expansion of human colonic stem cells. Nat. Med 17, 1225–1227, doi: 10.1038/nm.2470 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Glorevski N et al. Designer matrices for intestinal stem cell and organoid culture. Nature 539, 560–564, doi: 10.1038/nature20168 (2016). [DOI] [PubMed] [Google Scholar]
- 18.van de Wetering M et al. Prospective Derivation of a Living Organoid Biobank of Colorectal Cancer Patients. Cell 161, 933–945, doi: 10.1016/j.cell.2015.03.053 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyoshi H & Stappenbeck TS In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat. Protoc 8, 2471–2482, doi: 10.1038/nprot.2013.153 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang YL et al. Self-renewing Monolayer of Primary Colonic or Rectal Epithelial Cells. Cell. Mol. Gastroenterol. Hepatol 4, 165–182.e167, doi: 10.1016/j.jcmgh.2017.02.011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X et al. Cloning and variation of ground state intestinal stem cells. Nature 522, 173–178, doi: 10.1038/nature14484 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pai JH et al. Photoresist with low fluorescence for bioanalytical applications. Anal. Chem 79, 8774–8780, doi: 10.1021/ac071528q (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu S et al. Surface modification of poly(dimethylsiloxane) microfluidic devices by ultraviolet polymer grafting. Anal. Chem 74, 4117–4123 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Wang Y et al. Formation of human colonic crypt array by application of chemical gradients across a shaped epithelial monolayer. Cell. Mol. Gastroenterol. Hepatol 5, 113–130, doi: 10.1016/j.jcmgh.2017.10.007 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang YL et al. A microengineered collagen scaffold for generating a polarized crypt-villus architecture of human small intestinal epithelium. Biomaterials 128, 44–55, doi: 10.1016/j.biomaterials.2017.03.005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Speer JE et al. Molecular transport through primary human small intestinal monolayers by culture on a collagen scaffold with a gradient of chemical cross-linking. J. Biol. Eng 13, doi: 10.1186/s13036-019-0165-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keith BP et al. Colonic epithelial miR-31 associates with the development of Crohn’s phenotypes. JCI Insight 3, doi: 10.1172/jci.insight.122788 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toyonaga T et al. P149 Decreased Colonic Activin Receptor-like Kinase 1 Disrupts Epithelial Barrier Integrity and is Associated with a Poor Clinical Outcome in Crohn’s Disease. Gastroenterology 158, S46, doi: 10.1053/j.gastro.2019.11.133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y et al. Analysis of Interleukin 8 Secretion by a Stem-Cell-Derived Human-Intestinal-Epithelial-Monolayer Platform. Anal. Chem 90, 11523–11530, doi: 10.1021/acs.analchem.8b02835 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang YL et al. In Vitro Generation of Mouse Colon Crypts. ACS Biomater. Sci. Eng 3, 2502–2513, doi: 10.1021/acsbiomaterials.7b00368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finkbeiner SR et al. Transcriptome-wide Analysis Reveals Hallmarks of Human Intestine Development and Maturation In Vitro and In Vivo. Stem Cell Rep. 4, 1140–1155, doi: 10.1016/j.stemcr.2015.04.010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frantz C, Stewart KM & Weaver VM The extracellular matrix at a glance. J. Cell Sci 123, 4195–4200, doi: 10.1242/jcs.023820 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plow EF, Haas TA, Zhang L, Loftus J & Smith JW Ligand binding to integrins. J. Biol. Chem 275, 21785–21788, doi: 10.1074/jbc.R000003200 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Dosh RH, Jordan-Mahy N, Sammon C & Le Maitre CL Use of l-pNIPAM hydrogel as a 3D-scaffold for intestinal crypts and stem cell tissue engineering. Biomater Sci 7, 4310–4324, doi: 10.1039/c9bm00541b (2019). [DOI] [PubMed] [Google Scholar]
- 35.Liu Y et al. A simple, cross-linked collagen tissue substitute for corneal implantation. Invest. Ophthalmol. Vis. Sci 47, 1869–1875, doi: 10.1167/iovs.05-1339 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Song E, Yeon Kim S, Chun T, Byun HJ & Lee YM Collagen scaffolds derived from a marine source and their biocompatibility. Biomaterials 27, 2951–2961, doi: 10.1016/j.biomaterials.2006.01.015 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Hinman SS, Wang Y & Allbritton NL Photopatterned Membranes and Chemical Gradients Enable Scalable Phenotypic Organization of Primary Human Colon Epithelial Models. Anal. Chem 91, 15240–15247, doi: 10.1021/acs.analchem.9b04217 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martić-Kehl MI, Schibli R & Schubiger PA Can animal data predict human outcome? Problems and pitfalls of translational animal research. Eur. J. Nucl. Med. Mol. Imaging 39, 1492–1496, doi: 10.1007/s00259-012-2175-z (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.González-González M et al. Investigating Gut Permeability in Animal Models of Disease. Front. Physiol 9, 1962−1962, doi: 10.3389/fphys.2018.01962 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwerdtfeger LA, Nealon NJ, Ryan EP & Tobet SA Human colon function ex vivo: Dependence on oxygen and sensitivity to antibiotic. PLoS One 14, e0217170–e0217170, doi: 10.1371/journal.pone.0217170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Artursson P, Palm K & Luthman K Caco-2 monolayers in experimental and theoretical predictions of drug transport. Adv. Drug Del. Rev 46, 27–43, doi:Doi 10.1016/S0169-409x(00)00128-9 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Chow ECY, Liu S, Du Y & Pang KS The Caco-2 cell monolayer: usefulness and limitations. Expert Opin. Drug Metab. Toxicol 4, 395–411, doi: 10.1517/17425255.4.4.395 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Gunasekara DB et al. A Monolayer of Primary Colonic Epithelium Generated on a Scaffold with a Gradient of Stiffness for Drug Transport Studies. Anal. Chem 90, 13331–13340, doi: 10.1021/acs.analchem.8b02845 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moon C, VanDussen KL, Miyoshi H & Stappenbeck TS Development of a primary mouse intestinal epithelial cell monolayer culture system to evaluate factors that modulate IgA transcytosis. Mucosal Immunol. 7, 818–828, doi: 10.1038/mi.2013.98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.VanDussen KL et al. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut 64, 911–920, doi: 10.1136/gutjnl-2013-306651 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y et al. Long-Term Culture Captures Injury-Repair Cycles of Colonic Stem Cells. Cell 179, 1144–1159 e1115, doi: 10.1016/j.cell.2019.10.015 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kasendra M et al. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci. Rep 8, doi: 10.1038/s41598-018-21201-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim HJ & Ingber DE Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr. Biol. (Camb.) 5, 1130–1140, doi: 10.1039/c3ib40126j (2013). [DOI] [PubMed] [Google Scholar]
- 49.Jalili-Firoozinezhad S et al. Modeling radiation injury-induced cell death and countermeasure drug responses in a human Gut-on-a-Chip. Cell Death Dis. 9, 223, doi: 10.1038/s41419-018-0304-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jalili-Firoozinezhad S et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 3, 520–531, doi: 10.1038/s41551-019-0397-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tovaglieri A et al. Species-specific enhancement of enterohemorrhagic E. coli pathogenesis mediated by microbiome metabolites. Microbiome 7, 43, doi: 10.1186/s40168-019-0650-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sontheimer-Phelps A et al. Human Colon-on-a-Chip Enables Continuous In Vitro Analysis of Colon Mucus Layer Accumulation and Physiology. Cell Mol Gastroenterol Hepatol 9, 507–526, doi: 10.1016/j.jcmgh.2019.11.008 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herland A et al. Quantitative prediction of human pharmacokinetic responses to drugs via fluidically coupled vascularized organ chips. Nat Biomed Eng 4, 421–436, doi: 10.1038/s41551-019-0498-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang L, Murthy SK, Barabino GA & Carrier RL Synergic effects of crypt-like topography and ECM proteins on intestinal cell behavior in collagen based membranes. Biomaterials 31, 7586–7598, doi: 10.1016/j.biomaterials.2010.06.036 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Esch MB et al. On chip porous polymer membranes for integration of gastrointestinal tract epithelium with microfluidic ‘body-on-a-chip’ devices. Biomed. Microdevices 14, 895–906, doi: 10.1007/s10544-012-9669-0 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Yi B et al. Three-dimensional in vitro gut model on a villi-shaped collagen scaffold. BioChip Journal 11, 219–231, doi: 10.1007/s13206-017-1307-8 (2017). [DOI] [Google Scholar]
- 57.Costello CM et al. Microscale Bioreactors for in situ characterization of GI epithelial cell physiology. Sci. Rep 7, 12515, doi: 10.1038/s41598-017-12984-2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim W & Kim G An innovative cell-printed microscale collagen model for mimicking intestinal villus epithelium. Chem. Eng. J 224, 2308–2318, doi: 10.1016/j.cej.2017.12.001 (2018). [DOI] [Google Scholar]
- 59.Kim W & Kim G Intestinal Villi Model with Blood Capillaries Fabricated Using Collagen-Based Bioink and Dual-Cell-Printing Process. ACS Appl Mater Interfaces 10, 41185–41196, doi: 10.1021/acsami.8b17410 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Fujimichi Y, Otsuka K, Tomita M & Iwasaki T An Efficient Intestinal Organoid System of Direct Sorting to Evaluate Stem Cell Competition in Vitro. Sci. Rep 9, 20297, doi: 10.1038/s41598-019-55824-1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kozuka K et al. Development and Characterization of a Human and Mouse Intestinal Epithelial Cell Monolayer Platform. Stem Cell Rep. 9, 1976–1990, doi: 10.1016/j.stemcr.2017.10.013 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.In J et al. Enterohemorrhagic Escherichia coli Reduces Mucus and Intermicrovillar Bridges in Human Stem Cell-Derived Colonoids. Cell. Mol. Gastroenterol. Hepatol 2, 48–62.e43, doi: 10.1016/j.jcmgh.2015.10.001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.D’Argenio G & Mazzacca G Short-chain fatty acid in the human colon - Relation to inflammatory bowel diseases and colon cancer. Advances in Nutrition and Cancer 2 472, 149–158 (1999). [DOI] [PubMed] [Google Scholar]
- 64.Goncalves P & Martel F Butyrate and colorectal cancer: the role of butyrate transport. Curr. Drug Metab 14, 994–1008, doi: 10.2174/1389200211314090006 (2013). [DOI] [PubMed] [Google Scholar]
- 65.Kim R et al. An in vitro intestinal platform with a self-sustaining oxygen gradient to study the human gut/microbiome interface. Biofabrication, doi: 10.1088/1758-5090/ab446e (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Drost J et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 521, 43–47, doi: 10.1038/nature14415 (2015). [DOI] [PubMed] [Google Scholar]
- 67.Matano M et al. Modeling colorectal cancer using CRISPR-Cas9–mediated engineering of human intestinal organoids. Nat. Med 21, 256–262, doi: 10.1038/nm.3802 (2015). [DOI] [PubMed] [Google Scholar]
- 68.Wang Y, Kim R, Sims CE & Allbritton NL Building a Thick Mucus Hydrogel Layer to Improve the Physiological Relevance of In Vitro Primary Colonic Epithelial Models. Cell Mol Gastroenterol Hepatol, doi: 10.1016/j.jcmgh.2019.07.009 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Speer JE, Wang Y, Fallon JK, Smith PC & Allbritton NL Evaluation of human primary intestinal monolayers for drug metabolizing capabilities. J. Biol. Eng 13, 82, doi: 10.1186/s13036-019-0212-1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.VanDussen KL, Sonnek NM & Stappenbeck TS L-WRN conditioned medium for gastrointestinal epithelial stem cell culture shows replicable batch-to-batch activity levels across multiple research teams. Stem Cell Res. 37, doi: 10.1016/j.scr.2019.101430 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hinman SS et al. Microphysiological system design: simplicity is elegance. Curr Opin Biomed Eng 13, 94–102, doi: 10.1016/j.cobme.2019.12.010 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Noor N et al. 3D Printing of Personalized Thick and Perfusable Cardiac Patches and Hearts. Adv Sci (Weinh) 6, 1900344, doi: 10.1002/advs.201900344 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Drzewiecki KE et al. Methacrylation induces rapid, temperature-dependent, reversible self-assembly of type-I collagen. Langmuir 30, 11204–11211, doi: 10.1021/la502418s (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Diamantides N et al. Correlating rheological properties and printability of collagen bioinks: the effects of riboflavin photocrosslinking and pH. Biofabrication 9, 034102, doi: 10.1088/1758-5090/aa780f (2017). [DOI] [PubMed] [Google Scholar]
- 75.Rhee S, Puetzer JL, Mason BN, Reinhart-King CA & Bonassar LJ 3D Bioprinting of Spatially Heterogeneous Collagen Constructs for Cartilage Tissue Engineering. ACS Biomater. Sci. Eng 2, 1800–1805, doi: 10.1021/acsbiomaterials.6b00288 (2016). [DOI] [PubMed] [Google Scholar]
- 76.Lee A et al. 3D bioprinting of collagen to rebuild components of the human heart. Science 365, 482–487, doi: 10.1126/science.aav9051 (2019). [DOI] [PubMed] [Google Scholar]
- 77.Dekkers JF et al. High-resolution 3D imaging of fixed and cleared organoids. Nat. Protoc 14, 1756–1771, doi: 10.1038/s41596-019-0160-8 (2019). [DOI] [PubMed] [Google Scholar]
- 78.Zhu X et al. Ultrafast optical clearing method for three-dimensional imaging with cellular resolution. Proc. Natl. Acad. Sci. U. S. A 116, 11480–11489, doi: 10.1073/pnas.1819583116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hernandez HG et al. Fully synthetic matrices for in vitro culture of primary human intestinal enteroids and endometrial organoids. Biomaterials, 120125, doi: 10.1016/j.biomaterials.2020.120125 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ahmad AA et al. Optimization of 3-D organotypic primary colonic cultures for organ-on-chip applications. J. Biol. Eng 8, doi: 10.1186/1754-1611-8-9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Van der Flier LG, Haegebarth A, Stange DE, Van de Wetering M & Clevers H OLFM4 Is a Robust Marker for Stem Cells in Human Intestine and Marks a Subset of Colorectal Cancer Cells. Gastroenterology 137, 15–17, doi: 10.1053/j.gastro.2009.05.035 (2009). [DOI] [PubMed] [Google Scholar]
- 82.Ramalingam S, Daughtridge GW, Johnston MJ, Gracz AD & Magness ST Distinct levels of Sox9 expression mark colon epithelial stem cells that form colonoids in culture. Am. J. Physiol.: Gastrointest. Liver Physiol 302, G10–G20, doi: 10.1152/ajpgi.00277.2011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Salic A & Mitchison TJ A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. U. S. A 105, 2415–2420, doi: 10.1073/pnas.0712168105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Effects of improper micromolding.
Fig. S2. Images of stunted and dead in vitro crypts.
Fig. S3. Unpolarized in vitro crypts.
Table S1. Routine solutions validated for labeling primary human intestinal epithelial cells cultured over collagen hydrogel scaffolds.
Sheet 1) Collagen Neutralization Volumes. Calculator for neutralization buffer formulations and mixing volumes during collagen hydrogel formation.
Sheet 2) Subcultivation Ratios. Calculator for subcultivating primary intestinal epithelial cells from 6-well collagen hydrogel scaffolds to various culture sizes/formats
Data Availability Statement
The data that support the findings of this study are reported in previous publications.