SUMMARY
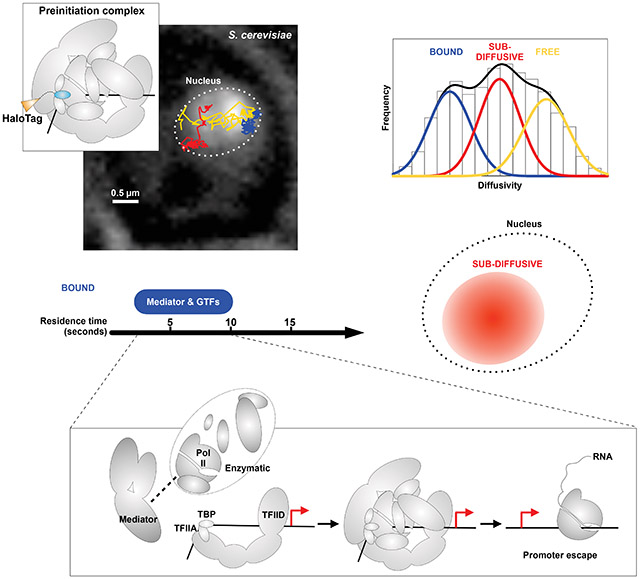
Transcription initiation by RNA polymerase II (Pol II) requires preinitiation complex (PIC) assembly at gene promoters. In the dynamic nucleus where thousands of promoters are broadly distributed in chromatin, it is unclear how multiple individual components converge on any target to establish the PIC. Here, we use live-cell, single-molecule tracking in S. cerevisiae to visualize constrained exploration of the nucleoplasm by PIC components and Mediator’s key functions in guiding this process. On chromatin, TFIID/TBP, Mediator, and Pol II instruct assembly of a short-lived PIC, which occurs infrequently but efficiently within a few seconds on average. Moreover, PIC exclusion by nucleosome encroachment underscores regulated promoter accessibility by chromatin remodeling. Thus, coordinated nuclear exploration and recruitment to accessible targets underlies dynamic PIC establishment in yeast. Collectively, our study provides a global spatio-temporal model for transcription initiation in live cells.
Graphical Abstract
eTOC BLURB
To characterize how transcription initiates in physiological space and time, Nguyen et. al. perform single-molecule tracking of all preinitiation complex (PIC) components in live yeast, illuminating spatio-temporal coordination underlying their convergence on chromatin to assemble the PIC and initiate transcription on a timescale of several seconds.
INTRODUCTION
RNA polymerase II (Pol II) transcribes the vast majority of eukaryotic genes. To initiate this process, Pol II interfaces with general transcription factors (GTFs) TFIID, TATA-binding protein TBP, TFIIA, TFIIB, TFIIE, TFIIF, TFIIH and associated kinase module TFIIK, as well as the coactivator Mediator to establish a pre-initiation complex (PIC) at gene promoters (Cramer, 2019; Robinson et al., 2016; Schilbach et al., 2017). Conserved DNA elements are directly recognized by TFIID and TBP in the presence of TFIIA (Patel et al., 2018; Zhang et al., 2016), while Pol II and other GTFs are generally tasked with enzymatic functions such as promoter melting and transcription start site (TSS) scanning (Cramer, 2019). The Mediator may transit between upstream activating sequences (UASs) and promoters in yeast (Jeronimo et al., 2016; Knoll et al., 2018; Petrenko et al., 2016; Wong et al., 2014) and form phase-separated condensates with sequence-specific transcription factors (TFs) at super-enhancers in embryonic stem cells (ESCs) (Boija et al., 2018; Shrinivas et al., 2019), but its specific role in transcription initiation is not well understood (Khattabi et al., 2019; Knoll et al., 2018; Petrenko et al., 2017). While PIC assembly has been well studied in vitro, the mechanism and kinetics underlying this process in vivo remain fundamental questions in gene regulation.
PIC components searching for promoters must contend with chromatin organization in the nucleus. These components tend to bind nucleosome-depleted regions (NDRs) adjoining the downstream “+1” nucleosome (Lauberth et al., 2013; Rhee and Pugh, 2012), and this association is sensitive to characteristic histone modifications (Joo et al., 2017; Kubik et al., 2018; Lauberth et al., 2013; Vermeulen et al., 2007). Thus, proximal nucleosomes contribute to the recognition of native promoters, many of which lack core motifs (Haberle and Stark, 2018). Beyond promoters, non-specific interactions with excess bulk chromatin can affect target-search efficiency of PIC components (Normanno et al., 2012). Furthermore, the abundances and distributions of individual components also influence PIC formation. Despite stoichiometric composition within the complex, PIC components are present at different concentrations (Borggrefe et al., 2001; Kimura et al., 1999) and at least two—Mediator and Pol II—may traffic in transient and stable clusters in mammalian cells (Cho et al., 2018; Cisse et al., 2013; Li et al., 2019). Understanding PIC establishment in vivo demands quantitative knowledge of how its components explore the nuclear environment to find promoter targets.
Single-molecule tracking (SMT) is a powerful approach to characterize the dynamic behaviors of factors in live cells at high spatiotemporal resolution (Lionnet and Wu, 2021; Mazza et al., 2012). SMT enables visualization of nuclear factors diffusing in space, transiently sampling chromatin and stably associating with specific targets, informing various modes of target search (Hansen et al., 2020; Izeddin et al., 2014). This dynamic view complements static snapshots acquired by genomics and structural approaches. However, despite increasing SMT implementation, systematic studies of factors that assemble a complex on chromatin are limited, precluding our understanding of this essential process in genome metabolism.
Here, we apply live-cell SMT in budding yeast to capture the nuclear dynamics of all ten PIC components and reveal different modes of spatial exploration in the nucleoplasm and hierarchical recruitment to chromatin. We also determine residence times on chromatin and extrapolate other parameters relevant to transcription initiation kinetics in live cells. Our findings suggest a model in which spatio-temporal coordination among key factors enables convergence of PIC components for transcription initiation in seconds.
RESULTS
Global dynamics of PIC components in live cells
To investigate PIC assembly in vivo, we first characterized the diffusive behaviors and chromatin association of all ten PIC components. We created individual strains expressing as the sole source a terminal fusion of HaloTag (Los et al., 2008) on Rpb1 (Pol II), Med14 (Mediator), Taf1 (TFIID), TBP, Toa1 (TFIIA), Sua7 (TFIIB), Tfg1 (TFIIF), Tfa1 (TFIIE), Ssl2 (TFIIH), or Kin28 (TFIIK) (Figure 1A). Wildtype-level growth of these strains confirmed functionality of the tagged essential proteins (Figures S1A-C).
Figure 1. Global dynamics of individual PIC components.
(A) To-scale schematics of N- or C-terminal HaloTag fusions. JF552, Janelia Fluor 552.
(B) Overlays of single TFIID and TFIIA trajectories, colored according to calculated diffusion coefficients. Nuclei are demarcated (ovals). N, number of trajectories.
(C) Diffusion coefficient (D, log10) histograms (bars) and multi-Gaussian fits (thick curves) of H2B and nuclear HaloTag (top, only fits shown) and PIC components, where resolved populations are shown (thin curves). Histograms contain data from three biological replicates.
(D) Fractions of populations by Spot-On kinetic modeling (Figure S1E and Table S1), with % chromatin-bound (FB) indicated. Results are means ± SD from three biological replicates.
(E) Top: fitted MED and TFIIE logD histograms. Trajectories with logD values within mean ± SD of respective population (shaded bars) were selected. B, chromatin-bound. UN, unbound. INT, intermediate. Middle: representative trajectories. Bottom: average mean-squared displacement (MSD) of each population with corresponding power-law fit.
(F) Violin plots of radii of confinement (Rc, μm) calculated for trajectories of each indicated population. Rc = 0.75 μm (horizontal dashed line) represents the yeast nuclear radius as indicated by the median Rc of HaloTag.
See also Figures S1 and S2.
We labeled each HaloTag fusion with Janelia Fluor 552 (JF552) (Zheng et al., 2019), imaged single molecules at 10 ms framerate and tracked their positions with ~30 nm precision to establish 2D projections of trajectories through nuclear space (Figure 1B, see Methods). This “fast-tracking” regime captured 10,000-50,000 trajectories for each PIC component as well as histone H2B and the nuclear HaloTag (HaloTag fused to a nuclear-localization signal), which served as standards for chromatin-bound and free behaviors, respectively. We extracted the apparent diffusion coefficient (D) based on the mean-squared displacement (MSD) of each trajectory (see Methods) then plotted the distribution of logD values for each factor (Figures 1C and S1D). Gaussian fitting resolved two dynamically distinct populations for Pol II, Mediator, TFIID, IIF, IIH and IIK, and unexpectedly, three for TBP, TFIIA, IIB and IIE (Figure 1C). Accordingly, we performed two- or three-state kinetic modeling of single displacements (Hansen et al., 2018) to achieve more robust quantitation of the average D (av. D) and fraction of molecules (F) comprising each population (Figures 1D, S1E and Table S1, see Methods).
As anticipated, the majority of H2B (F~70%) exhibited low mobility (av. D=0.01 μm2/s), taken as the average diffusivity of yeast chromatin (Figures 1C and 1D). The lowest-mobility populations of PIC components, assigned as chromatin-bound and subsequently validated using binding mutants (below), displayed several-fold faster diffusion (av. D=0.03-0.06 μm2/s). This diffusivity range was not observed for nuclear HaloTag (Figure S1D), informing that the HaloTag moiety was not responsible for chromatin binding by tagged PIC components. The minor fractions of bound Mediator and all GTFs (FB=21%-44%) indicate that most molecules underwent diffusive processes in the nucleoplasm. Note that FB values represent all binding at promoter targets and non-specific sites throughout chromatin. FB for TBP (34%) accounts for binding at Pol I and Pol III-transcribed genes, and relative to other PIC components, the notably large FB for Pol II (48%) reflects elongating and terminating activities occurring downstream of promoters.
Diffusivities of the most mobile populations (av. D=0.6-3.3 μm2/s), assigned as unbound, correlated negatively with the molecular weights (MW) of individual PIC components (Figure S1F), as expected of molecules diffusing in the same medium. Notably, unbound H2B diffused more slowly (av. D=1.6 μm2/s) than expected (>2 μm2/s) for a species of similar MW to H2A-Halo-H2B (~55 kDa), consistent with its biochemical association with various histone chaperones in the nucleoplasm (Hammond et al., 2017). The low av. D of Med14 and Taf1 (0.6 μm2/s) suggests that they diffuse in large MDa-sized Mediator and TFIID complexes. Intriguingly, the small GTFs TBP, TFIIA, IIB and IIE, whose unbound populations were the most mobile among PIC components (av. D > 2 μm2/s) (Figure S1F), also displayed a well-resolved, less mobile third population with av. D (0.4-0.6 μm2/s) resembling that of unbound Mediator and TFIID (Figures 1C, 1D and Table S1). These results indicate that PIC components infrequently interact with chromatin and more often undergo multiple diffusive processes to explore the nucleoplasm.
Subdiffusive nuclear exploration by PIC components
To acquire a spatial perspective on the three dynamic behaviors of PIC components, we subclassified trajectories into bound, intermediate and unbound populations according to the logD histograms (Figures 1E and S2A). The ensemble MSDs computed for bound populations of PIC components and H2B were well fit by a power law model (MSD ∝ tα) with anomalous coefficient α=0.5-0.6 (Figures S2B and S2C) characteristic of subdiffusion (α<1) (Woringer and Darzacq, 2018; Woringer et al., 2020). These results are quantitatively consistent with the reported mobility of a chromosomal tetO array in yeast (Miné-Hattab et al., 2017). To estimate the spatial constraint imposed by chromatin, we fit the MSDs of individual trajectories with a circular-confined diffusion model to extract the radius of confinement (Rc) (Lerner et al., 2020; Wieser and Schütz, 2008). As expected, bound PIC components and H2B exhibited similar median Rc~0.13 μm representing the spatial confinement of yeast chromatin (Figure 1F).
Remarkably, intermediate populations of TBP, TFIIA, IIB and IIE exhibited a subdiffusive behavior distinct from that of chromatin (α=0.8) (Figures S2B and S2C), with Rc values indicating a tendency to explore sub-nuclear areas with apparent median Rc~0.4 μm (the radius of the yeast nucleus is ~0.75 μm) (Figure 1F). By contrast, ensemble MSDs from the unbound populations of these four GTFs are characteristic of free diffusion, displaying plateaus consistent with confinement to the average dimensions of the yeast nucleus (Figure S2B) (Dion and Gasser, 2013). Thus, the three dynamic populations resolved for TBP, TFIIA, IIB and IIE correspond to chromatin-bound, subdiffusive and free diffusive states.
Unbound Mediator and TFIID also displayed subdiffusive dynamics with α=0.9 (Figures S2B and S2C) and median Rc~0.4 μm, notably similar to the intermediate populations of TBP, TFIIA, IIB and IIE (Figure 1F). To further investigate the manner by which these six PIC components explore apparently limited space, we analyzed the angles formed by consecutive displacements (Figure S2D) (Hansen et al., 2020). Trajectories of unbound Mediator and TFIID as well as intermediate populations of TBP, TFIIA, IIB and IIE exhibited almost two-fold enrichment (1.3-fold for TBP) of large angles (180°±30°) relative to small angles (0°±30°) (Figure S2E). This directional bias suggests a back-and-forth motion consistent with confined diffusion whereby molecules oversample a local environment, such as a transient “trapping zone” proposed for mammalian CTCF (Hansen et al., 2020; Izeddin et al., 2014). Unbound TBP, TFIIA, IIB and IIE displayed no such bias, indicating unconstrained mobility within the yeast nucleus.
Unbound Pol II, TFIIF, IIH and IIK also exhibited subdiffusion with α=0.8 (Figures S2B and S2C) and median Rc~0.5 μm (Figure 1F). The indistinguishable diffusive behaviors of unbound Pol II and TFIIF (Figures 1F, S1F and S2C) suggest that they associate in the nucleoplasm, consistent with biochemical properties of the two factors in yeast nuclear extracts (Rani et al., 2004). Similarly, the diffusive behaviors of TFIIH and IIK are also consistent with their biochemical interactions (Keogh et al., 2002). Our analysis reveals that all PIC components undergo subdiffusive processes to explore the yeast nucleus, with a tendency to sample 0.4-0.5 μm-radial areas.
TFIID/TBP, Mediator and Pol II instruct hierarchical PIC assembly
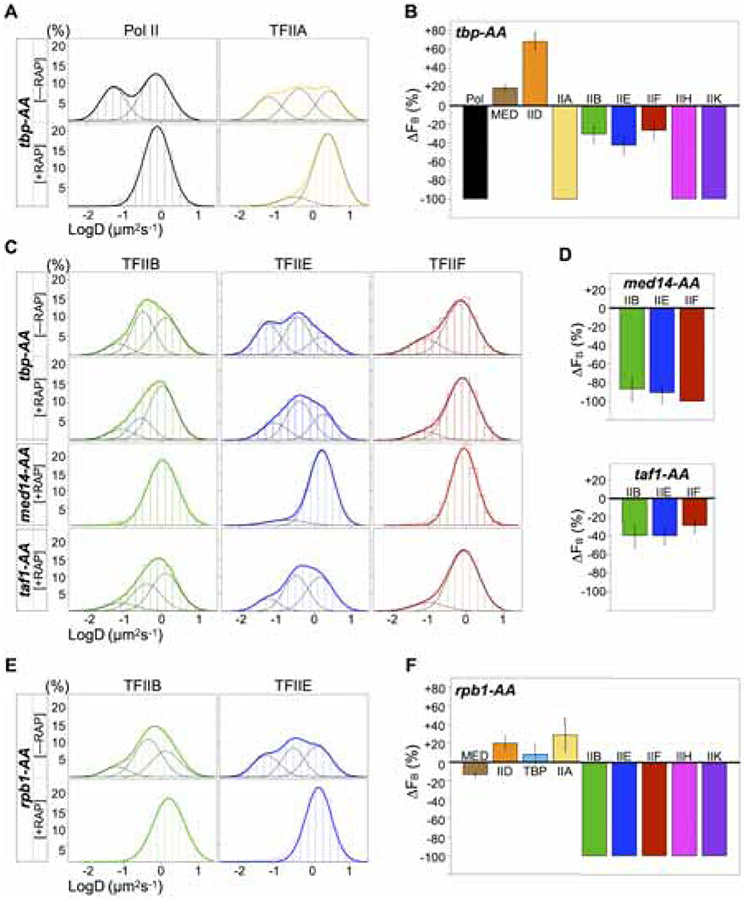
We next sought to determine how PIC components are recruited to chromatin in live cells by observing the effect of depleting key components on chromatin binding by the other components. To explore the role of TBP, we used the Anchor-Away (AA) technique to deplete it from the nuclei of growing cells by addition of rapamycin, which coupled ribosome processing with eviction of TBP tethered to the ribosomal protein Rpl13A via FRB and FKBP12 tags, respectively (Haruki et al., 2008). Efficient TBP depletion was achieved after one-hour rapamycin treatment, with little to no observable co-depletion of other PIC components (Figure S3A), which we then fast-tracked. Chromatin binding by Pol II, TFIIA, IIH, and IIK was reduced to minute, unquantifiable levels (Figures 2A, 2B, S3B, S3C and S3D, Table S2). By contrast, increased binding by Mediator (+20%) and especially TFIID (+70%) was consistent with genome-wide association at UASs and promoters, respectively, under similar TBP depletion conditions (Knoll et al., 2018, 2020). The dramatic response by TFIID also implicates an inhibitory effect of TBP on TFIID binding at promoters, as recently shown in vitro (Le et al., 2019).
Figure 2. TFIID/TBP, Mediator and Pol II instruct hierarchical PIC assembly.
(A) LogD histograms of Pol II and TFIIA before (−RAP, top) and after (+RAP, bottom) TBP depletion by Anchor Away (tbp-AA) (also see Figure S3C). Histograms contain data from one (−RAP) and two or three (+RAP) biological replicates. RAP, rapamycin.
(B) Changes in chromatin-bound fraction (ΔFB) after TBP depletion relative to wildtype, calculated using mean FB ± SD from two or three biological replicates of each condition (see Methods), and errors were propagated. [tbp-AA cells were imaged once without RAP to confirm wildtype-level FB]. See Figures S3D and Table S2 for Spot-On fits and results.
(C) LogD histograms of TFIIB, IIE and IIF before (−RAP, top row) and after depletion (+RAP) of TBP (tbp-AA, middle row), Med14 (med14-AA, third row), or Taf1 (taf1-AA, bottom row).
(D) ΔFB after Med14 or Taf1 depletion, computed and shown as in (B).
(E) LogD histograms of TFIIB and IIE in rpb1-AA cells before (−RAP, top) and after (+RAP, bottom) Pol II depletion.
(F) ΔFB after Rpb1 depletion, computed and shown as in (B). See Figures S3I and Table S2 for Spot-On fits and results
See also Figures S3 and S4.
Unexpectedly, after TBP depletion, we observed binding by TFIIB, IIE and IIF with more modest decreases (−27% to −43%) (Figures 2B and 2C). We next asked whether bound Mediator or TFIID was responsible for recruiting these three GTFs (Baek et al., 2006) in the absence of TBP. Depletion of Med14, a critical structural subunit of Mediator, resulted in full or near-complete abolishment of chromatin binding by TFIIB (−84%), TFIIE (−92%) and TFIIF (−100%) (Figures 2C and 2D). Double depletion of TBP and Med14 elicited identical responses (Figures S3E and S3F), further attributing TBP-independent binding by TFIIB, IIE and IIF (Figure 2C) to Mediator. In contrast, depletion of the TFIID subunit Taf1 caused less severe defects comparable TBP depletion (−29% to −40%), indicating Mediator’s dominant role in recruiting TFIIB, IIE and IIF. Notably, Med14 depletion, while causing dramatic defects in recruitment of the enzymatic components, showed no negative effect on chromatin binding by TFIID, TBP and TFIIA (Figures S4A, S4B and Table S2). Thus, promoter recognition by these three components appears to be a distinct step in PIC assembly that assists in recruitment of the enzymatic components, as evidenced by partial defect in this process upon depletion of TBP (Figure 2B) or Taf1 (Figure S4B).
TFIIB, IIE and IIF physically interface with the PIC via Pol II (Robinson et al., 2016; Schilbach et al., 2017). We explored Pol II’s involvement in PIC assembly by depleting its catalytic subunit Rpb1. Rpb1 co-depleted with another Pol II subunit Rpb9 and no other PIC component (Figure S3G). Under Rpb1 AA, we observed a modest effect on chromatin binding by Mediator, while that by the enzymatic components TFIIB, IIE, IIF, IIH and IIK was essentially abolished (Figures 2E, 2F, S3H, S3I and Table S2). Thus, both Mediator and Pol II are required for the recruitment of TFIIB, IIE and IIF. Our finding also indicates that in vivo, TFIIB does not stably associate with the “upstream complex” comprising DNA, TFIIA and TBP in the absence of Pol II, as demonstrated in vitro (Lee and Hahn, 1995). Similar observations for human TFIIB (Zhang et al., 2016) suggest that this aspect of PIC assembly may be conserved. Importantly, Rpb1 depletion elicited only modest changes in binding by TFIID, TBP and TFIIA, similar to Mediator and consistent with genome-wide ChIP studies (Joo et al., 2017; Knoll et al., 2018). These results support a model in which Mediator and TFIID-TBP-TFIIA together organize a platform at promoters for stepwise recruitment of Pol II and the remaining GTFs, as shown in vitro (Johnson and Carey, 2003).
Our AA experiments reveal a previously uncharacterized collaboration between Mediator and Pol II to coordinate recruitment of the enzymatic components of the PIC (Figure S4C). Furthermore, disruption of this hierarchy abolished chromatin binding for seven PIC components (Pol II, TFIIA, TFIIB, IIE, IIF, IIH and UK), suggesting a remarkable absence of detectable ‘non-specific’ chromatin interactions beyond promoter regions. Based on the strict dependencies demonstrated here, chromatin binding detected for the enzymatic GTFs also indicates presence of TBP, Mediator and Pol II in the bound entity taken as the PIC. This interpretation circumvents technical limitations on multi-color SMT imaging for direct PIC observation at any one location in live cells.
Notably, as with chromatin binding, subdiffusion by TFIIB and IIE was nearly ablated upon depletion of either Med14 (Figure 2C) or Rpb1 (Figure 2E), but not Taf1 (Figure 2C), suggesting that (1) Mediator, not TFIID, is specifically responsible for subdiffusion by TFIIB and IIE in the presence of Pol II and (2) this process is conducive to recruitment of TFIIB and IIE to chromatin. We also observed that chromatin binding and subdiffusion by TFIIA, while generally insensitive to Med14 depletion (Figure S4A), was nearly abolished or dramatically reduced upon TBP depletion (Figure 2A) or Taf1 depletion (Figure S4A), respectively. Thus, TFIIA subdiffusion is coordinated by TFIID in the presence of TBP during the process of promoter recognition (Kraemer et al., 2001; Patel et al., 2018). These findings indicate that nuclear exploration and chromatin binding by the four smallest and most dynamic PIC components—TFIIB, TFIIE, TFIIA and TBP—are guided by Mediator and TFIID with considerable molecular specificity.
The PIC is short-lived in live cells
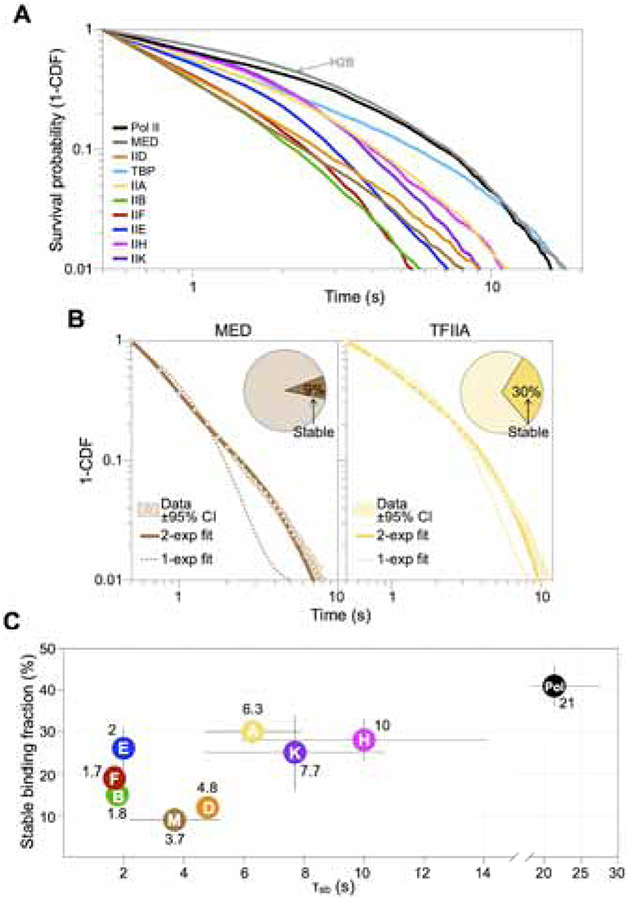
We next sought to determine the chromatin residence time of PIC components in live cells, which inform the lifetime of the PIC and provide key metrics for transcription initiation kinetics. We implemented a “slow-tracking” regime to selectively visualize and determine the dwell times of binding events. This imaging regime features long exposure time (250 ms) to blur out diffusing molecules and low laser power to limit fluorophore photobleaching (Chen et al., 2014). Individual survival probability curves (1-CDF), computed from the observed dwell times of several thousand binding events, showed that on average, all PIC components, except Pol II and TBP, were substantially shorter-lived on chromatin than H2B (Figure 3A), which indicated the temporal limit imposed by photobleaching and chromatin motions (Hansen et al., 2017). To accommodate longer dwell times of Pol II and TBP, we sought to further reduce the photobleaching limit by time-lapse imaging, which incorporates alternating excitation laser “on” (250 ms) and “off” (250-500 ms) periods (Gebhardt et al., 2013). This approach allowed better separation of the survival curve of Pol II from the bulk histone H2B standard for long-term chromatin residence (Figure S5E). However, time-lapse did not allow distinction between TBP and H2B, indicating that the factor’s dwell time is beyond resolution by SMT.
Figure 3. The PIC is short-lived in live cells.
(A) Log-log survival-probability curves (1-CDF) from dwell times of single binding events. Curves contain data from three biological replicates.
(B) 1-CDF data (white dots) of Mediator and TFIIA, with ±95% confidence interval (CI) obtained by resampling (shaded area). Double-exponential fit (solid line) resolves fractions of molecules engaged in stable (fsb) and transient binding (pie chart, fsb values shown) and respective dissociation rates (ksb and ktb, reported in Table S1). Single-exponential fit (dotted line) is shown for each dataset.
(C) fsb (%) and corrected average residence times of stable binding τsb (indicated in seconds) by individual PIC components. Results are means ± SD from three biological replicates. Values for Pol II were obtained by time-lapsed imaging (Figure S5E).
See also Figure S5.
Within the SMT resolvable range, each survival curve for PIC components minimally required a double-exponential fit corresponding to stable and transient binding characterized by dissociation rates (ksb and ktb, respectively) and fractions of all binding events (fsb and ftb) (Figures 3B and S5D, Table S1). We used H2B decay kinetics to correct k values (Figure S5A-C, see Methods) (Hansen et al., 2017) and computed the average residence times (τ=1/k) for stable (τsb) and transient (τtb) binding (Figure 3C and Table S1). Both time-lapse regimes (250 ms and 500 ms “off”) produced similar τsb of 20-23 s for Pol II (fsb~40%), consistent with a value of 26 s derived by fluorescence recovery after photobleaching (FRAP) (Figures S5F and S5G). Among GTFs, TFIIA, IIH and IIK exhibited the longest τsb, ranging between 6 s and 10 s (fsb=25-30%) (Figure 3C). It is worth noting that while TFIIH has a well-established role in DNA repair (Rimel and Taatjes, 2018) and has been implicated to participate in Pol I-mediated transcription (Iben et al., 2002), its strict dependence on Pol II for recruitment to chromatin under our experimental conditions (Figures 2F and S3H) indicates that these additional functions had little if any contribution to our SMT measurements. Mediator and TFIID displayed τsb of 4-5 s (fsb~10%, lowest among all components). The shortest-lived components TFIIB, IIE and IIF showed τsb ~ 2 s (fsb=15%-25%), indicating that on average, a full PIC assemblage lasts only 2 seconds on chromatin in living cells.
Importantly, TBP depletion resulted in miniscule fractions of detectable binding events (1-2%) and even shorter residence times for Pol II (2 s), TFIIA (4s), IIH and IIK (3s) (Figures S5H and S5I), confirming the link between their normal τsb to PIC formation at promoters and Pol II transcription. For TFIIB, IIE, and IIF, TBP depletion had little effect on τsb but reduced fsb by ~50% (Figures S5H and S5I). Because recruitment of these three GTFs requires Pol II (Figure 2F), the miniscule Pol II fraction with a similar residence time (~2 s) under TBP depletion may be an indication of that interaction with TFIIB, IIE, and IIF. Finally, consistent with fast-tracking results, TBP depletion had little effect on Mediator but substantially increased the τsb and fsb for TFIID (Figures S5H and S5I).
Kinetic coupling between promotor escape and PIC lifetime
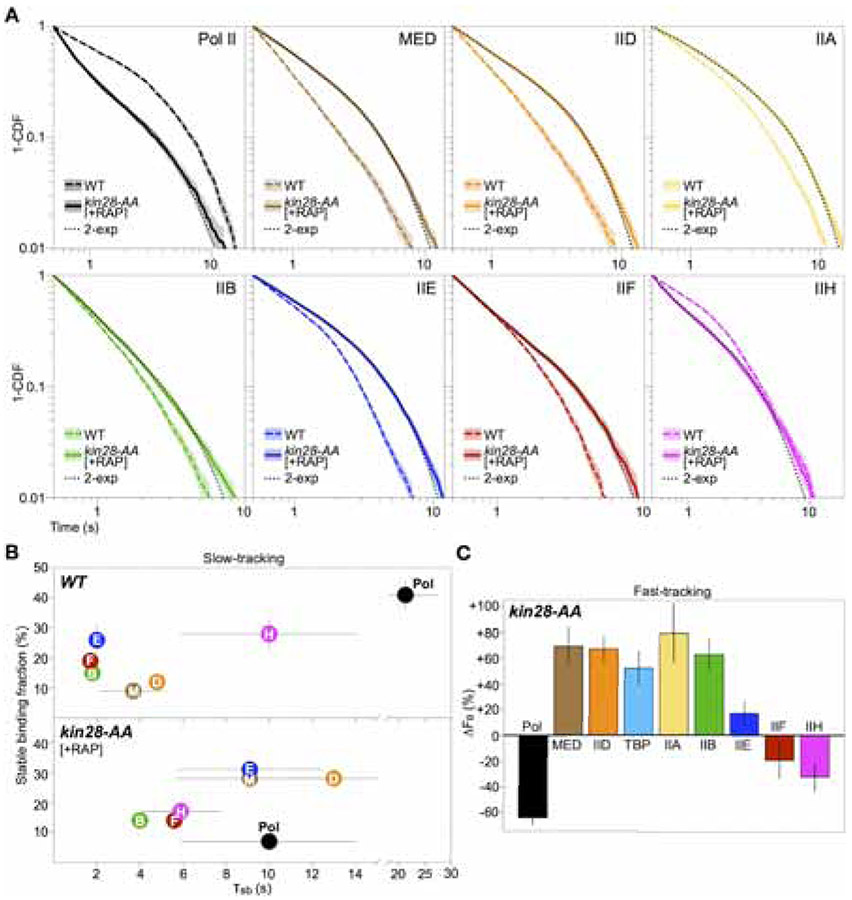
To explore a connection between the short lifetime of the PIC and the transcription initiation process in live cells, we disrupted a key initiation event involving serine-5 phosphorylation of the Rpb1 C-terminal domain (CTD) by the Kin28 kinase (TFIIK). This disruption is associated with delayed Pol II promoter escape and global reduction of nascent RNA synthesis (Jeronimo and Robert, 2014; Rodríguez-Molina et al., 2016; Wong et al., 2014). If initiation were driving PIC turnover in live cells, its delay should result in longer dwell times for PIC components. Indeed, we found that Kin28 depletion (Figure S6A) resulted in at least 2-fold longer τsb for Mediator and all GTFs except TFIIH (Figures 4A, 4B, S6B and S6C). The shorter TFIIH τsb suggests that its stable PIC engagement involves interfacing with TFIIK, as shown by cryo-EM (Schilbach et al., 2017). In addition, upon Kin28 depletion, Pol II exhibited substantially shorter residence time of 10 s (compared to 23 s in wildtype cells), which represents an average of heterogeneous molecules stalled near promoters and elongating or prematurely terminated along gene bodies (Jeronimo and Robert, 2014; Rodríguez-Molina et al., 2016; Wong et al., 2014). Fast-tracking experiments further validated these results, showing larger chromatin-bound fractions for Mediator, TFIID, TBP, TFIIA, IIB and IIE (20% to 80% increase), in contrast to Pol II (65% decrease) and TFIIH (30% decrease) (Figures 4C, S6D and S6E). Of interest, despite displaying longer τsb, global binding by TFIIF was modestly decreased (by 20%) under Kin28 depletion, which may reflect a global reduction in elongation complexes where TFIIF, unlike other PIC components, has been shown to be present and functional (Schweikhard et al., 2014; Zawel et al., 1995).
Figure 4. Delayed chromatin dissociation upon Kin28 depletion.
(A) 1-CDF data and double-exponential fit obtained for PIC components after Kin28 depletion, shown as in Figure 3B. Wildtype data (WT, without fit) is shown for comparison. [kin28-AA cells were imaged once without RAP to confirm wildtype-level dynamics (Figure S6C).] Curves contain data from three biological replicates.
(B) fsb (%) and τsb (s) of PIC components in wildtype cells (WT, top) and after Kin28 depletion (+RAP, bottom) [not reported for TFIIA because its extended survival approached the H2B limit (Figure S6B)]. Values are means ± SD from three biological replicates. See Table S2 for fit results.
(C) ΔFB after Kin28 depletion, computed and shown as in Figure 2B.
See also Figure S6.
These findings indicate a temporally stalled PIC in the absence of Kin28, thus linking transcription initiation and Pol II promoter escape to rapid PIC turnover in live cells.
Highly dynamic target search and low promoter occupancy by PIC components
We next sought to reconstruct the average trajectory of a molecule undergoing target search (Figure 5A). The duration of this process, or the τsearch, is the temporal sum of alternating transient chromatin binding (average dwell time τtb) and diffusion in the nucleoplasm (with average duration τfree) as the molecule samples Ntrials chromatin sites before reaching any one of the specific targets (Chen et al., 2014; Lionnet and Wu, 2021; Loffreda et al., 2017; Tatavosian et al., 2018). We found that τsearch of Mediator and the GTFs ranged between 8 s and 48 s (Figure 5B and see Methods), which is on the same order of magnitude as that of the yeast TF Ace1 (8 s) (Mehta et al., 2018) and generally ~10-fold shorter than reported values for mammalian nuclear factors (Chen et al., 2014; Hansen et al., 2017; Tatavosian et al., 2018), likely reflecting the smaller yeast nucleus. Notably, target search predominantly took place in the nucleoplasm and involved few transient binding events for most GTFs (Ntrials=5-8), while Mediator and TFIID appeared to sample chromatin more frequently (Ntrials>20) during PIC establishment (Figure 5C). Considering potential accessible binding sites at several thousand accessible NDRs genome-wide, the low Ntrials values suggest highly targeted recruitment with little non-specific interactions by PIC components.
Figure 5. Target search kinetics and occupancy by PIC components.
(A) Schematic of a single molecule’s trajectory between two promoter targets conceptualizing τsb, τsb, τfree. The exemplary molecule diffuses in the nucleoplasm and samples Ntrials=2 nonspecific sites before encountering a target. The τsearch equation is indicated.
(B) Average total τfree (white bars) and total τtb (colored bars) computed for each PIC component according to the equation in (A) (see Methods for τfree and Ntrials derivations). Errors were derived from three biological replicates in both fast and slow tracking.
(C) Average Ntrials of transient interactions between two specific binding events by each PIC component. Errors were derived from three biological replicates in both fast and slow tracking.
(D) Schematic of unbound and bound states of an average promoter target conceptualizing the sampling interval (SI), τsb and temporal occupancy (O). The SI and O equations are indicated.
(E) Average O (%) of each PIC component at a promoter, calculated according to equation in (D) (see Methods). SI values are reported in Table S1. Errors were obtained from fast- and slow-tracking results (means ± SD) and documented # molecules/cell (median ± SD).
(F) Association of PIC components at an average promoter assuming independent, uncoordinated binding, simulated over a 100 s time window based on SI and τsb values to represent occupancy levels close to those shown in (E). Bottom: cumulative count of co-localized components, where ‘8’ is required for PIC assembly (Pol II and TBP occupancies are assumed based on dependencies established in Figures 2B and 2F).
(G) Top: equation for calculating the observed association rate kobs (s−1) of a pseudo-first order binding reaction between PIC components and chromatin targets, where the latter are considered mostly free at equilibrium due to low occupancy levels shown in (E). The concentrations of the PIC component ([Factor]) and chromatin targets ([Targets]) were derived from published data. Bottom: kobs values obtained using the above equation. Errors were derived from τsearch results and SDs documented for cellular abundances of PIC components.
The % occupancy of the PIC at a promoter within a given time window is a critical metric of the gene’s potential for RNA synthesis. Occupancy by a factor is a function of its τsearch and τsb as well as its abundance and the number of available targets (Figure 5D and see Methods) (Chen et al., 2014). Levels of Mediator and the GTFs range between 2,000 and 5,000 molecules per cell (Ho et al., 2018) and ~6,000 PIC targets exist in budding yeast (Rhee and Pugh, 2012). These values indicate that on average, PIC components each occupy a promoter only 4-12% of the time (Figure 5E and Table S1). For example, the promoter would host Mediator for 4 s (τsb from Figure 3C) once every 100 s, resulting in 4% occupancy. These low individual occupancy levels indicate that without mechanisms to coordinate their recruitment, co-occupancy by components for PIC assembly would be highly improbable (p=10−9) (Figure 5F). Indeed, the strict recruitment hierarchy demonstrated in live cells (Figure 2) indicates that occupancy by individual PIC components must temporally overlap for full PIC assembly and function. This inference is further supported by the observed association rates kobs, which take into account the τsearch as well as the factor and target abundances, which were similar for Mediator, TFIID, IIH and IIK (kobs= 0.01 s−1, or one event every 100 s; Figure 5G). Intriguingly, TFIIB, IIE and IIF displayed ~7-fold more frequent association, which may reflect a tendency to dynamically sample a target site, similar to the in vitro behavior of human TFIIB (Zhang et al., 2016), or perhaps re-association with the early elongation complex, as shown for TFIIF (Schweikhard et al., 2014; Zawel et al., 1995).
Together, these results reveal dynamic target search by PIC components resulting in infrequent PIC formation at yeast promoters. Moreover, the extrapolated kobs of individual components suggest that once nucleated, this process may occur with remarkable efficiency.
Nucleosomes outcompete PIC at gene promoters
To examine how promoter chromatin architecture influences PIC establishment in live cells, we inactivated the essential remodeling enzyme RSC by AA depletion of its catalytic subunit Sth1, which leads to an upstream shift of the “+1” nucleosomes genome-wide and consequently occlusion of DNA elements at a large subset of promoters (Ganguli et al., 2014; Hartley and Madhani, 2009; Kubik et al., 2018) (Figures 6A and S7A). Fast-tracking results showed >50% reduction in chromatin binding by Pol II (Figures 6B, S7B and S7C), indicating a substantial transcriptional defect in line with ChIP findings (Kubik et al., 2018). We also found ~20-40% decreases in FB for Mediator, TFIIA, IIB, IIE, IIF, IIH and IIK (but not for TFIID), consistent with reduced TBP levels detected by ChIP at ~30% of Pol II-transcribed genes most affected by Sth1 depletion (Kubik et al., 2018). (High levels of TBP binding to Pol I and Pol III-transcribed genes generally unaffected by RSC inactivation (S. Kubik and D. Shore, personal communication) may account for modest effect on TBP binding in live cells.) Thus, the PIC is precluded from about a third of promoters after RSC inactivation, likely due to steric hindrance from the shifted +1 nucleosomes. The unexpected 50% increase in TFIID binding, shown for both Taf1 (Figures 6B, S7B and S7C) and Taf2 (Figure S7D), suggests that RSC normally inhibits TFIID recruitment, perhaps by directly competing for binding to the NDR and the +1 nucleosome (Brahma and Henikoff, 2018; Ramachandran et al., 2015).
Figure 6. Nucleosome encroachment precludes PIC from many chromatin targets in live cells.
(A) Top: Schematic of a nucleosome-depleted region (red) with flanking nucleosomes (ovals) positioned relative to TBP-binding site (TATA) and the transcription start site (TSS, red arrow). The PIC’s footprint (dashed oval) is informed by genome-wide mapping of components (Grünberg et al., 2016; Rhee and Pugh, 2012). Bottom: shifted flanking nucleosomes ensuing RSC inactivation may occlude the PIC binding site.
(B) ΔFB after Sth1 depletion, computed using mean FB ± SD from three biological replicates of (−RAP) and (+RAP) conditions (Figure S7C and Table S2), and errors were propagated.
(C) 1-CDF data of TFIID, IIE and IIH after Sth1 depletion, with corresponding wildtype data for comparison, shown as in Figure 4A (see Figures S7E for other components). [sth1-AA cells were imaged once without RAP to confirm wildtype-level dynamics (Figure S7E).]
(D) fsb (%) and τsb (s) of PIC components in wildtype cells (top, WT) and after (bottom, +RAP) Sth1 depletion, shown as in Figure 4B. See Table S2 for fit results.
See also Figure S7.
To investigate whether RSC inactivation affects PIC dynamics at targets that remained accessible, we performed slow tracking and found relatively unchanged τsb for Mediator, TFIID, IIA, IIB, IIE and IIF (Figures 6C, 6D, S7E and Table S2). Intriguingly, shorter τsb observed for TFIIH and IIK suggests that RSC activity is also important for PIC functions post-assembly. Shortened residence of TFIIH, which is tasked with promoter melting and TSS scanning (Murakami et al., 2015), is consistent with the correlation between TSS usage and changes in nucleosome positions upon RSC inactivation (Klein-Brill et al., 2019). Taken together, our results indicate that RSC activity is important for PIC recruitment to a subset of promoter targets, and post-recruitment PIC functions at other targets. Furthermore, these findings demonstrate that our SMT approach is amenable to dynamic changes following epigenetic perturbations in yeast.
DISCUSSION
Nuclear exploration by PIC components
All PIC components undergo subdiffusive processes in the yeast nucleus, with a tendency to explore 0.4 μm-0.5 μm radial regions. This behavior by TBP, TFIIA, IIB and IIE occurs in a fraction of the molecules, while unbound molecules may search the entire nucleus (0.75 μm radius) (Figure 1F). Importantly, subdiffusion by these small GTFs mirrors the diffusivity (Figure S2C), apparent confinement Rc (Figure 1F) and directional bias (Figure S2D) of Mediator and TFIID. Furthermore, demonstrated requirement for Mediator (Figures 2C) and TFIID/TBP (Figures 2A and S4A) suggests that the large complexes constrain spatial exploration by the most diffusive GTFs. This process may span the entire nucleus as individual molecules explore different subnuclear regions; alternately, it may be localized as molecules cluster. Ensemble fluorescence images indicate that the bulk of Mediator and TFIID co-occupy a subnuclear region consistent with the Rc of single molecules (~0.4 μm) (Figure S8). We thus favor a scenario wherein Mediator and TFIID guide a fraction of TBP, TFIIA, IIB and IIE to explore a common subnuclear region while not excluding a more dispersive process occurring globally. Intriguingly, the observed Mediator and TFIID distributions in yeast invite speculation of foci or condensates, perhaps nucleated by sequence-specific TFs (Boija et al., 2018; Shrinivas et al., 2019), as shown for Mediator in mouse ESCs (Cho et al., 2018), although we cannot discount volume exclusion by the substantial nucleolus and other chromatin substructures from giving rise to their compact organization (Woringer and Darzacq, 2018; Woringer et al., 2014). Such steric effects would explain limited diffusion of large components—Mediator, TFIID, Pol II-TFIIF and TFIIH-IIK (>700 kDa)—relative to the small GTFs TBP, TFIIA, IIB and IIE (<150 kDa) (Figure 1F).
TFIIB and IIE subdiffusion and chromatin binding also requires the presence of Pol II in addition to Mediator (Figures 2E and 2F). We speculate that TFIIB and IIE could co-localize with a Pol II-Mediator complex biochemically identified as a major form of Pol II in yeast nuclear extracts (Rani et al., 2004). Furthermore, Mediator and Pol II may enable TFIIB and IIE to dynamically interact with nearby chromatin, thereby constraining their diffusion to the local environment (Izeddin et al., 2014; McSwiggen et al., 2019). Such activity would be consistent with the relatively high chromatin association rates obtained for TFIIB and IIE (Figure 5G). Importantly, in addition to TFIIB and IIE, Pol II is also required for recruitment of TFIIF, IIH and IIK. Thus, Mediator and Pol II constitute a critical bridge between chromatin targets and all enzymatic components of the PIC.
Taken together, we propose that Mediator and TFIID sequester enzymatic and promoter-recognizing components, respectively, to a shared subnuclear territory (Figure 7A) sustained by weak, rather than strong, interactions as components did not appreciably co-deplete in AA experiments (Figures S3A and S3G). This organization may facilitate PIC assembly by locally concentrating individual components, as Mediator and TFIID are required for basal transcription using physiological levels of factors but dispensable in a reaction with purified components (Baek et al., 2002). It may also enhance target-search efficiency by merging individual components in a reduced search space (Kent et al., 2020). Furthermore, the remarkably similar search and association kinetics of Mediator and TFIID in live cells (Figures 5B, 5C and 5G) complement prior evidence for cooperative promoter engagement (Grünberg et al., 2016; Johnson and Carey, 2003; Johnson et al., 2002; Knoll et al., 2018). Mediator and TFIID may thus serve to unite PIC components spatially in the nucleoplasm and temporally on chromatin targets, underscoring their essential functions in vivo (Petrenko et al., 2017; Warfield et al., 2017). Our model for PIC components implicates a similar subnuclear localization of highly transcribed genes, and the relative distributions of these nuclear constituents may be illuminated by future multi-color imaging studies.
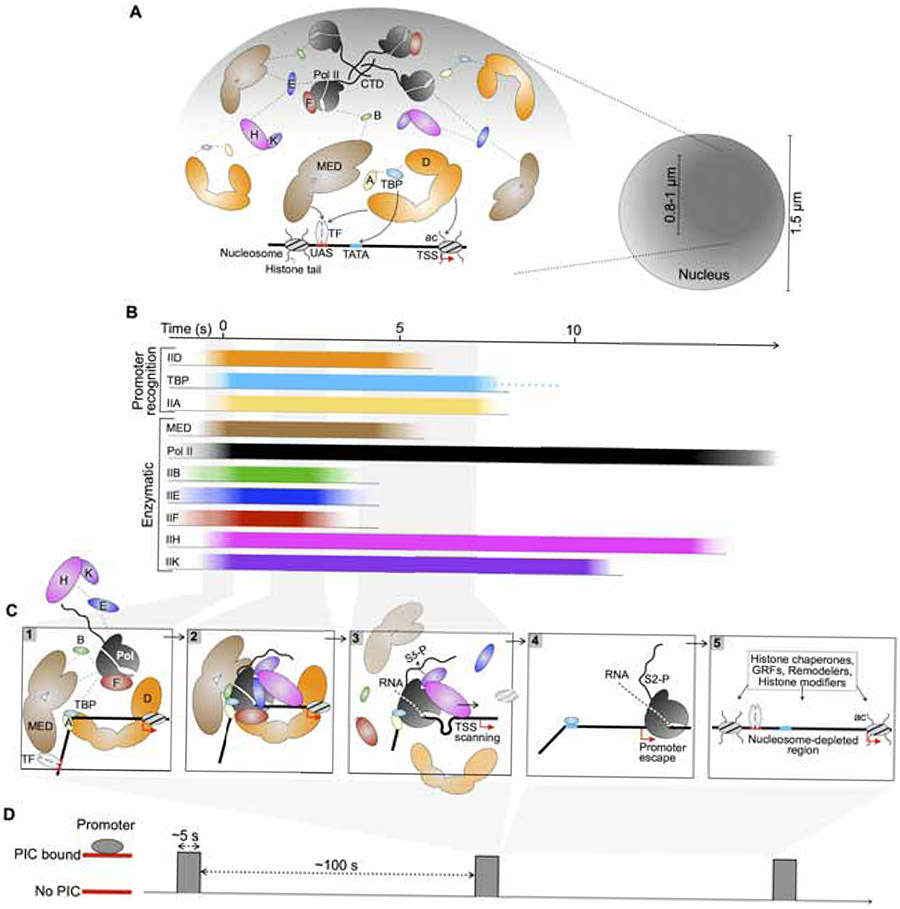
Figure 7. Spatio-temporal model of PIC establishment in vivo.
(A) Left: A model for spatial clustering supported by multi-valent interactions (dashed lines) among promoter-recognizing PIC components TFIID, TBP and TFIIA as well as Mediator and enzymatic components. The presence of active genes in this region is implicated, with one stereotypical promoter is shown. Mediator and TFIID may interact with DNA elements, bound sequence-specific transcription factor (TF), and the +1 nucleosome via acetylated histone tails (ac). Right: Schematic of a subnuclear environment (dark shade) explored by PIC components in a subdiffusive manner. The diameter of this region was estimated based on median Rc values (0.4-0.5 μm) (Figure 1F).
(B) An average timescale of association and dissociation of PIC components at a yeast promoter. Recruited components are shown to associate in a quasi-synchronous manner based on calculated kobs (Figure 5G). Binding events are temporally scaled according to average τsb (Figure 3C) and estimated for TBP and Pol II.
(C) Sequential stages of PIC assembly and disassembly according to binding and dissociation of individual components in (B).
(D) An average timescale depicting sparse PIC establishment at a promoter, based on the ~5 s assembly window outlined in (B) and the association rate kobs = 0.01 s−1 estimated for Mediator and TFIID (Figure 5G).
See also Figure S8.
A temporal model for PIC assembly and transcription initiation
We synthesize our data with prior knowledge to propose a temporal sequence for de-novo PIC establishment at a yeast promoter (Figure 7B). Mediator and TFIID survey the chromatin for accessible NDRs, bound sequence-specific TFs (Petrenko et al., 2016; Tuttle et al., 2018), GRFs (Ansari et al., 2009; Layer and Weil, 2013; Papai et al., 2010) and local histone marks (Joo et al., 2017) (Figure 7A). TFIID and TFIIA orchestrate TBP engagement with TATA or TATA-like element (Patel et al., 2018; Zhang et al., 2016), effectively reconfiguring promoter architecture (Figure 7C, Step 1). Mediator recruits Pol II/TFIIF, TFIIB and TFIIE, which enables TFIIH/IIK engagement (Compe et al., 2019; Maxon et al., 1994), to this reconfigured target to form a complete PIC (Figure 7C, Step 2). Extrapolated association rates kobs (Figure 5G) implicate the efficiency of this process, with individual components converging on a promoter in a near-synchronous manner. Intriguingly, although they have similar abundances (Ho et al., 2018) and exhibit similar association (Figure 5G) and dissociation (Figure 3C) kinetics, the global bound fraction is higher for TFIIK than IIH (Figure 1D), which may correspond to the former’s additional roles such as Mediator phosphorylation (Guidi et al., 2004) and transcription termination (Medler and Ansari, 2015). The coactivator SAGA may coordinate PIC assembly at a minor subset of promoters (Donczew et al., 2020), although its physical and functional overlap with Mediator is unclear.
TFIIK activity is associated with Pol II promoter escape as well as disengagement of Mediator (Jeronimo and Robert, 2014; Wong et al., 2014) and TFIID (Knoll et al., 2020). In live cells, Kin28 depletion resulted in similar dissociation kinetics for Pol II, Mediator and TFIID (Figure 4B), further indicating that promoter clearance by these components is kinetically coupled. Therefore, the normal ~5 s average residence time of Mediator and TFIID should encompass complete PIC assembly and initiation events up to Pol II escape (Figures 7B and 7C). Within this time window, rapid dissociation of TFIIB and IIF may reflect TSS-scanning activity (Qiu et al., 2019) and nascent RNA synthesis (Fujiwara et al., 2019) (Figure 7C, Step 3). The short TFIIE residence time (2 s) suggests that it is not required for retention of TFIIH/IIK, consistent with an in vitro study showing that human TFIIE releases while TFIIH is actively opening DNA (Compe et al., 2019). The residence time of TFIIH indicates that it may take ~10 s for complete PIC disassembly and Pol II escape, which may involve mobilization of the +1 nucleosome (Figure 7C, Step 4). As Pol II enters elongation, chromatin remodelers and regulators, including RSC and GRFs, collaboratively reestablish the NDRs before the next initiation event (Kubik et al., 2018) (Figure 7C, Step 5). Our model conveys the remarkable kinetic efficiency of PIC assembly and transcription initiation in yeast, which may be a conserved feature of eukaryotic transcription as studies in mouse and human cell lines have identified dynamic Pol II clusters whose 5-8 s lifetime may encompass transcription initiation (Cho et al., 2016; Cisse et al., 2013).
Based on the kobs of PIC components (Figure 5G), we estimate that on average, the PIC assembles once every 100 seconds at a yeast promoter (Figure 7D), which may thus receive ~36 PICs/hour. As mRNA synthesis rates during exponential growth in glucose range from 4 to 60 mRNAs/hour (Pelechano et al., 2010), questions remain as to whether such productivity range reflects varying PIC assembly and initiation kinetics across different promoters. These questions may be addressed by tracking and comparing PIC dynamics in the vicinity of specific genes, as demonstrated for the TFs Ace1 (Mehta et al., 2018) and Gal4 (Donovan et al., 2019) at their native targets CUP1 array and GAL10, respectively. From another perspective, considering the median synthesis rate of 7 mRNAs/hour genome-wide (Pelechano et al., 2010), our estimate of 36 PICs/hour may reflect prevalent non-productive, spurious initiation giving rise to pervasive cryptic transcripts (Neil et al., 2009; Xu et al., 2009). Furthermore, it will be important to determine how regulatory mechanisms, such as those involving TFs, chromatin regulators, fragile nucleosomes (Kubik et al., 2015), sub-nucleosome species (Brahma and Henikoff, 2018) or the Mot1-Ino80-NC2 complex (Xue et al., 2017), influence PIC dynamics globally as well as locally.
In summary, our study deconstructs global yeast transcription initiation in physiological space and time, revealing specific coordination on and off chromatin that enables efficient PIC assembly. In this context, our findings also offer insights relevant to other processes involving establishment of multi-component machineries at sparse chromatin targets.
LIMITATIONS OF THE STUDY
This work provides an average temporal guide to future studies of variations in the kinetics of PIC establishment and transcription initiation across several thousand Pol II-transcribed genes in yeast. Furthermore, by imaging yeast during logarithmic growth in rich medium, our measurements are likely predominated by transcription of highly expressed housekeeping genes. We anticipate application of the SMT approach outlined here to investigate PIC dynamics under various growth conditions to elucidate the kinetics underlying activation of inducible, stress-response genes.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Carl Wu (wuc@jhu.edu).
Materials Availability
Strains and plasmids listed in the Key Resources Table are available from the Lead Contact without restriction.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Rapamycin (Sirolimus) | LC Laboratories | Cat#R-5000 |
| HaloTag ligand - Janelia Fluor 552 (JF552) | Zheng et al., 2019 | N/A |
| HaloTag ligand - Janelia Fluor 646 (JF646) | Grimm et al., 2015 | N/A |
| Deposited data | ||
| Raw localization/tracking data (DiaTrack output) and main processing scripts | This paper | Mendeley Data DOI: 10.17632/3xktk72wbd.1 |
| Experimental models: Organisms/strains | ||
| S. cerevisiae YAR295 (Halo-H2B) | Ranjan et al., 2020 | N/A |
| S. cerevisiae YAR302 (Free Halo) | Ranjan et al., 2020 | N/A |
| S. cerevisiae YAR270 (rpb1-AA) | Ranjan et al., 2020 | N/A |
| See Table S3 for S. cerevisiae strains generated for this study. | This paper | N/A |
| Oligonucleotides | ||
| See Table S4 for oligonucleotides used for tagging S. cerevisiae PIC components for SMT imaging and Anchor Away. | This paper (from IDT) | N/A |
| Recombinant DNA | ||
| pFA6a-TRP1-pGAL1-HBH | Booher and Kaiser, 2008 | RRID:Addgene_26886 |
| N-terminal HaloTag: pFA6a-TRP1-pGAL1-HALO | This paper | N/A |
| C-terminal HaloTag: pBS-SK-Halo-NatMX | This paper | N/A |
| N-terminal FRB-GFP tag for AA: pFA6a-TRP1-pGAL1-FRB-GFP | This paper | N/A |
| C-terminal FRB-GFP tag for AA: pFA6a-FRB-GFP-kanMX | This paper | N/A |
| Software and algorithms | ||
| Sojourner | Wu Lab | https://github.com/sheng-liu/sojourner |
| Fiji | Schindelin et al., 2012 | http://fiji.sc |
| Prism version 8 for Mac | GraphPad Software | www.graphpad.com |
| DiaTrack version 3.05 | Vallotton and Olivier, 2013 | N/A |
Data and Code Availability
Trajectory coordinates have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the Key Resources Table. Original microscopy data (.cxd or .tif movies) will be shared by the Lead Contact upon request.
Custom scripts used to analyze SMT data in the Sojourner computational package have been deposited at Mendeley and are publicly available as of the date of publication. The DOI and Github addresses are listed in the Key Resources Table.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All strains used in this study are derived from haploid S. cerevisiae W303. Cells were grown in YAPD (1% yeast extract, 2% peptone, 2% glucose, 60 mg/L adenine hemisulfate) or CSM (Complete Supplement Mixture, 60 mg/L adenine hemisulfate) medium as described below.
METHOD DETAILS
Yeast strain construction
Proteins were chromosomally tagged in haploid W303 Saccharomyces cerevisiae by standard high-efficiency transformation and homologous recombination of PCR-amplified DNA (Gietz and Schiestl, 2007). All yeast strains harbored pdr5Δ to enhance retention of fluorescent ligands (Ball et al., 2016). To tag the C terminus, the HALOTAG sequence (Promega) was cloned into the pBluescript SK (−) vector upstream of a NatMX (nourseothricin) cassette, generating plasmid pBS-SK-Halo-NatMX. PCR-amplified DNAs for yeast transformation contained HALO-NATMX with a 5’ sequence coding for a SG4 amino-acid linker. To tag the N termini of TBP and Toal, the HALOTAG sequence was cloned between the PacI and AscI restriction sites on pFA6a-TRP1-pGAL1-HBH (Booher and Kaiser, 2008), generating plasmid pFA6a-TRP1-pGAL1-HALO. PCR-amplified DNAs for yeast transformation contained sequences coding for a GSG4 (TBP) and (G4S)2 (Toa1) linker. Tagging was carried out according to the strategy outlined by Booher & Kaiser for essential proteins (Booher and Kaiser, 2008).
For Anchor-Away (AA) experiments, pdr5Δ was implemented in strain HHY221 (Haruki et al., 2008). Then, the FRB-GFP-KANMX sequence was synthesized from plasmid pFA6a-FRB-GFP-kanMX to target RPB1, KIN28, and STH1 at the 3’ ends. Strains expressing Rpb1-FRB-GFP, Kin28-FRB-GFP, and Sth1-FRB-GFP were confirmed for rapamycin-dependent nuclear depletion my imaging GFP fluorescence. To create a strain expressing FRB-GFP-TBP for TBP AA, N-terminal tagging was carried out as described with PCR-amplified DNA containing FRB-GFP and a sequence coding for G4SG3SG4 linker (from plasmid pFA6a-TRP1-pGAL1-FRB-GFP). Subsequently, PIC components were HaloTagged in AA strains as described.
Yeast strains expressing Halo-H2B (YAR295) and nuclear HaloTag (YAR302) were generated as described by Ranjan et. al. (Ranjan et al., 2020).
Yeast cell growth and spot test
Cells were grown at 30°C in YAPD. Serial 6-fold dilutions in YAPD were prepared from OD600 = 1.0 cultures. Dilution series were spotted on YAPD plates and incubated in the dark at room temperature, 30°C, and 38°C for two days to assess the heat-shock response. We also spotted W303 strain harboring no Halo fusions and htz1Δ strain as negative and positive controls for growth defect, respectively.
Live-cell fluorescence imaging
Sample preparation
Cells were grown at 30°C in CSM in the presence of 3-30 nM (for fast tracking) or 1-10 nM (for slow tracking) JF552 to label Halo fusions. Mid-log cultures (OD600~0.6-0.8) were harvested and washed with ~1 mL CSM five times. For AA experiments, rapamycin (LC Laboratories), dissolved in DMSO, was added to the growing culture (OD=0.6) to a final 1 μg/mL concentration 1 hour before harvesting. All washes were carried out with CSM + rapamycin (1 μg/mL) medium. For control AA experiments, DMSO was added to the culture and wash medium.
A coverslip (#1.5, ø 25 mm, Electron Microscopy Services) was heat-treated, coated with Concanvalin A (0.5 mg/mL), and assembled in a metal Attofluor chamber (ø 35 mm, Invitrogen). Washed cells (1 mL) were added and allowed to attach to the coverslip for 2 minutes, followed by gentle rinses with fresh wash medium to achieve a monolayer of cells. Final sample contains 1 mL of medium in the chamber.
Single-molecule imaging
Imaging was carried out at room temperature on an Axio Observer Z1 microscope (Zeiss, Germany) equipped with an α-Plan-Apochromat 150x/1.35 glycerin-immersion objective (ZEISS, Germany). To excite JF552, we used CL555-100 555 nm laser (CrystaLaser, Reno, NV) and a filter cube containing a 561 nm BrightLine single-edge beamsplitter and a 612/69 nm BrightLine single-band bandpass emission filter (Semrock, Rochester, NY). Images were acquired by a C9100-13 EM-CCD camera (Hamamatsu Photonics, Japan) featuring 512x512 pixels with 16 μm pixel size, operating at ~−80°C (forced-air cooling) and EM gain 1200x. The pixel size of recorded images is 107 nm. A 750 nm blocking edge BrightLine multiphoton short-pass emission filter and a 405/488/561/635 nm StopLine quad-notch filter (Semrock, Rochester, NY) were placed in front of the camera. We used the ZEN imaging software (ZEISS, Germany) and HCImage (Hamamatsu Photonics, Japan) to operate the microscope and camera, respectively. Excitation laser was triggered by TTL exposure output signal from the camera.
For fast tracking, we excited the sample with ~1 kW/cm2 continuous laser and imaged a 128x128 pixel field of view (containing ~5 single cells) for ~1.5 minutes at 10 millisecond camera integration time. Irradiated cells, monitored every 30 minutes for up to 3 hours, exhibited no observable cellular damage and underwent bud growth and cell division similar to unexposed cells (data not shown).
For slow tracking, we used lower excitation power (~50 W/ cm2) and imaged a 256x256 pixel field for ~3 minutes at 250 ms/frame. For time-lapse imaging (Gebhardt et al., 2013) of H2B, Rpb1 and TBP, we modified the slow-tracking regime to alternate 250 ms excitation and 250 ms or 500 ms dark time and imaged each field of view for ~3 minutes at 500 ms or 750 ms/frame, respectively.
QUANTIFICATION AND STATISTICAL ANALYSIS
Localizing and tracking single molecules
Raw movies were pre-processed in Fiji (Schindelin et al., 2012) to bypass bleaching of initial nuclear fluorescence (first ~1,000 and 150 frames for fast and slow tracking, respectively) and generate a substack (5,000-6,000 and 600-1,000 frames for fast and slow-tracking files, respectively) in which each image contained sparse single molecules (<2 molecules per nucleus). We used a maximum-intensity Z projection of each file to locate and manually window nuclei to create a binary mask. Substacks were saved as 16-bit TIFFs for subsequent analysis.
To localize single molecules, we used DiaTrack v3.05 (Vallotton and Olivier, 2013) at the following settings: high precision (HWHM=1 pixel), remove blur 0.1, remove dim 50-100. Tracking was performed with 6 pixel (~0.65 μm) maximum allowance between consecutive localizations. This cut-off was informed by the smooth tail of the frequency histogram of displacements (Figure S1E). In some cases, such as free HaloTag, 8 pixels were allowed to achieve this feature. Localization and tracking results were saved as MATLAB files for subsequent analyses.
Slow-tracking data were similarly processed, with 2-pixel maximum allowance between consecutive localizations based on H2B displacement observed under the same imaging regime (Figures S4A and S4B).
Analyzing fast-tracking data
We used the Sojourner package (https://github.com/sheng-liu/sojourner) to analyze tracking results. Trajectories containing < 3 localizations (2 displacements) were discarded to reduce false detections. Binary masks generated during pre-processing were applied to select nuclear trajectories. Average length of selected data was 10-12 displacements (median = 5-7). To determine the diffusion coefficient (D), we performed linear fitting between Δt 20 – 50 ms of the MSD computed for trajectories containing ≥ 5 displacements. The slopes of the lines, subject to R2≥ 0.8 criterion, were divided by 4 to obtain D (Mazza et al., 2012). We then imported log10D values to Prism (version 8.00 for Mac, GraphPad Software, La Jolla, California, USA, www.graphpad.com) and plotted the frequency histogram with 0.2 μm2/s binning of logD values. We used the default “Sum of two Gaussians” and a custom “Sum of three Gaussians” function to fit the logD distributions without hard constraints. We did not quantify the resolved subpopulations due to limits of the MSD-based analysis, including the trajectory-length selection and unreliable linear fits of MSDs from very stably-bound molecules (data not shown), which omitted up to 50% of the data. We used this approach to inform the number of resolvable dynamic states for each factor. To perform quantitation of these states, we carried out two- or three-state kinetic modeling of displacements obtained from all trajectories (Hansen et al., 2018). We used the Spot-On web interface (https://spoton.berkeley.edu/) with the following settings: bin width = 0.01 μm, number of timepoints = 5, jumps to consider = 6, max jump = 1-1.2 μm and Z correction with dZ = 0.6 μm. Model fit was performed on the CDF of displacements for 3 iterations. The localization error, obtained from fitting the data, was ~25-40 nm. Data for HaloTag and chromatin-binding mutants where the average trajectories were shorter (4-6 displacements) were fitted to 3-4 timepoints and with 0.35 nm localization error constraint.
Subclassifying trajectories and MSD analysis
We separated trajectories from distinct subpopulations according to logD values within one standard deviation of the mean. After this process, two or three subsets were generated for each PIC component and H2B, without significant overlap (Figure S2A). We then computed the average MSD for each subset and performed power-law fit MSD=B(Δt)α between Δt = 10 ms and 100 ms (Figure S2B). A power-law behavior where α<1 indicates sub-diffusion (higher confinement ~ smaller α) and B is directly related to diffusion coefficient (Izeddin et al., 2014). The average MSD profiles of chromatin-free subpopulations were consistent with free diffusion in a confined space, where the MSD plateau is related to the diameter of the confinement (Dion and Gasser, 2013). The theoretical MSD plateau for free diffusion in the haploid yeast nucleus (diameter~1.5 μm) is 0.45 μm (Dion and Gasser, 2013) (Figure S2B, dotted line).
To calculate the apparent radius of confinement (Rc) for individual trajectories, we first fit a straight line for each MSD curve in a log-log plot, i.e. log(MSD(t)) vs. log(dt) to determine the α. Subdiffusive molecules are selected with α<1 and R2≥ 0.8. MSD curves from these molecules were then fit to the following confined diffusion model (Lerner et al., 2020; Wieser and Schütz, 2008),
Displacement angle analysis
We calculated the angles formed to consecutive displacements of single molecules according to Hansen et al. (Hansen et al., 2020). The analysis was performed on the subclassified intermediate and unbound populations of Mediator, TFIID, TBP, TFIIA, IIB and IIE. Backward bias of molecule movement, i.e. 180°±30° / 0°±30° > 1, indicates potential confinement.
Calculating change in chromatin-bound fraction after AA
For AA experiments, we calculated the change in chromatin-bound fraction FB (%) as follows,
FB,[+RAP] was determined for three biological replicates. For rpb1-AA, tbp-AA, and kin28-AA, we obtained FB,[−RAP] for one biological replicate to confirm wildtype-level association frequencies (Table S2). Then, we used FB,WT to calculate ΔFB. For sth1-AA, FB,[−RAP] was determined for three biological replicates. All variables applied were means ± s.d. from biological replicates and the error for ΔFB was obtained through propagation.
Analyzing slow-tracking data
Tracking results from DiaTrack were processed in Sojourner to determine the apparent dwell times (temporal lengths of trajectories). The cumulative frequency distribution (1-CDF) of dwell times was fitted with the double-exponential decay function, where fsb and ksb represent the frequency and apparent dissociation rate constant of stable binding, respectively, and ktb represents the apparent dissociation rate constant of transient binding (Mazza et al., 2012),
We used the bootstrap method (Efron and Tibshirani, 1986) to generate 100 resampled datasets and calculate 95% confidence interval (CI). We then computed 1-CDF and performed fitting for each dataset. The mean values for fsb, ksb, and ktb were taken, with their SDs providing an assessment of fit quality.
We adapted the approach described by Hansen et. al. (Hansen et al., 2017) to correct the apparent dissociation rates. First, we imaged chromosomal H2B under the same regime, with the assumption that its decay kinetics measures limits imposed by photobleaching and chromatin motions. The 1-CDF for H2B was fitted as described (Figure S4C), and its ksb was used for correction as follows,
Calculating search kinetics
Our derivations represent a hybrid of approaches described by Tatavosian et. al. (Tatavosian et al., 2018) and Loffreda et. al. (Loffreda et al., 2017). There are Nns nonspecific and Ns specific binding sites in the genome, associated with transient (average lifetime τtb) and stable (average lifetime τsb) binding events, respectively. A molecule’s typical trajectory cycle is composed of Ntrials nonspecific interactions interspersed with free diffusion (lasting on average τfree), followed by a specific binding event. Assuming equal on-rates at all genomic sites, the molecule samples all sites at random and the average search time between two consecutive specific binding events is,
Assuming equal accessibility and introducing the nonspecific-to-specific site ratio as rs = Nns/Ns,
Thus,
To obtain rs, we considered two scenarios underlying detection of binding events during slow tracking.
- Blinking-limited: molecules fluoresce in a bound state with probability Ps of occurring at a specific site, which is equal to the fraction of stable binding events obtained from slow tracking. Ps is proportional to the fraction of time the molecule spends at specific sites relative to the overall time it spends bound to chromatin.
- Diffusion-limited: molecules fluoresce in a diffusive state and are motion-blurred until they engage a binding site in focus. Here, Ps is proportional to the number of specific sites relative to all sites,
These physical processes can happen in the cell coincidentally and likely represent two extremes of a spectrum of behaviors single molecules can exhibit. We reasoned that the relative likelihood of detecting a blinking-limited binding event by slow tracking is proportional to the global fraction of bound molecules (FB). Therefore, we computed a weighted average value for rs as follows,
To obtain τfree we considered the chromatin-bound frequency FB, obtained by fast tracking, as proportional to the fraction of time the molecule is bound at specific and non-specific chromatin sites,
Therefore,
Calculating target occupancy
We used the approach described by Chen et. al. (Chen et al., 2014) to calculate the average temporal occupancy of each PIC component at an average promoter target. First, we estimated the sampling interval SI, which represents the average time between two consecutive binding events at a specific site,
We considered Ns=6,000 PIC targets based on published ChIP-exo results (Rhee and Pugh, 2012). Values (median±s.d) for cellular abundance (Ns, molecules/cell) were obtained from the Saccharomyces Genome Database (SGD) (Cherry et al., 1998)—Med14: 1,989±849; Taf1: 1,633±434; Toa1: 2,599±1,754; Sua7: 4,262±1,425; Tfg1: 4,780±2,343; Tfa1: 3,514±437; Ssl2: 2,137±600; Kin28: 2,151±704.
The average target occupancy (O) is,
Simulating target occupancy
We used experimentally determined τsb and calculated SI to simulate occupancy of each PIC component at an average promoter according to the null model of all PIC factors binding independently. For each factor, we simulated a sequence of on/off intervals during which the promoter was respectively occupied and unoccupied. During the simulation, the duration of individual on/off intervals were drawn at random from exponential distributions with respective average τsb and SI - τsb. We chose the exponential distribution form because it corresponds to the simplest possible biochemical scenario of a single rate-limiting step driving the lifetime of each state. Once we independently simulated on and off states for each of the PIC components at the promoter, we scanned through the time traces to calculate the number of factors bound at each given time. Simulations were performed in MATLAB.
Fluorescence Recovery after Photobleaching (FRAP)
Haploid strains expressing Halo-H2B, Rpb1-Halo, and Halo-TBP were grown and imaging samples prepared as described. We labeled Halo fusions with 20 nM JF646 (Grimm et al., 2015). FRAP experiments were performed at room temperature on an LSM 800 confocal microscope with Airyscan and a Plan-Apochromat 63x/1.40 oil-immersion objective (ZEISS, Germany). Microscope control and data acquisition were performed in Zen (ZEISS, Germany). Two frames of each cell were acquired before photobleaching. Then, a 640 nm laser was used at 100% power for ~10-30 to bleach ~50% of the nuclear region, and the cell was excited with 0.2% laser power and imaged at 1 s interval for about 2 minutes. For each sample, FRAP was performed on 20-25 cells.
Data were analyzed in Fiji. We used the StackReg plugin (Thevenaz et al., 1998) to correct for drift when applicable. For each frame, mean intensities were determined and background-subtracted for a reference area of unbleached fluorescence (REF) and the bleached area (BL). BL intensity relative to the REF (BL/REF) was normalized against pre-bleach BL/REF to account for the maximum intensity achievable by fluorescence recovery. Data from 15-20 cells were averaged to generate a single recovery curve for “Exponential Recovery” fit up to 30 s due to significant movement of cells past this time point. The fit function was y = a × (1 – e−bx) + c, where a represents the fraction of slow recovery and b the corresponding rate, and 1/b the estimated τ.
Deconvolution microscopy
Yeast cells were grown to mid-log (JF646 was added to label Taf1-Halo), harvested and fixed using 4% formaldehyde (EMS) in PBS for 15 minutes at room temperature. Fixed cells were washed with PBS and spread between a coverslip and a standard glass slide. The edges of the imaging sample were sealed with clear nail polish.
Imaging was performed at room temperature on a DeltaVision Elite system (GE Healthcare) equipped with an Olympus 100 × 1.4 NA oil-immersion objective and a scientific complementary metal-oxide semiconductor camera (PCO). For each field of view, GFP and JF646 fluorescence image stacks were acquired sequentially at 0.2-μm intervals. Individual stacks were subjected to signal ratio enhancement and 20 cycles of iterative deconvolution. Projection images were generated by the Volume Viewer tool using SoftWoRx suite (GE Healthcare). The resulting images were colored and merged in ImageJ.
Supplementary Material
Taf1-Halo motion by fast tracking in a live cell, Related toFigure 1. Raw movie of Taf1-Halo-JF552 in the nucleus of a live cell. Data was acquired at 10 ms framerate and rendered at 50 fps (frames per second). The nuclear outline (oval) was estimated from total nuclear fluorescence (not shown).
Kin28-Halo motion by slow tracking in a live cell, Related toFigure 3. Raw movie of Kin28-Halo-JF552 in the nucleus of a live cell. Two single binding events intervened by a motion-blurred diffusive episode, likely by the same molecule, are captured. Data was acquired at 250 ms framerate and rendered at 25 fps (frames per second). The nuclear outline (oval) was estimated from total nuclear fluorescence (not shown).
HIGHLIGHTS.
Dynamic PIC components explore a reduced nuclear space in a sub-diffusive manner
Mediator and Pol II coordinate target search and recruitment of enzymatic components
Full PIC assembly and initiation-coupled disassembly may occur within a few seconds
Nucleosomes abrogate PIC establishment on chromatin in live cells
ACKNOWLEDGMENTS
We dedicate this work to the memory of Maxime Dahan, former leader of the Transcription Imaging Consortium (TIC) at the HHMI-Janelia Research Campus. We thank Anders Hansen, Maxime Woringer and Xavier Darzacq for assistance with Spot-On and angular analyses; James Zhe Liu, Brian English and Brian Mehl for general advice on SMT; Slawomir Kubik and David Shore for sharing unpublished results; Diana Stavreva, David Garcia, Arpita Upadhyaya and Gordon Hager for input on statistical assessment of residence time data; Lu Bai for technical advice on FRAP in yeast; Xiaojun Ren for SMT discussions; Erin Pryce and the Integrated Imaging Center for training on the LSM 800 for FRAP experiments; Matt Hurlock and Yumi Kim for assistance with deconvolution microscopy, and Gisela Storz, Jessica Liu, Rogelio Hernandez-Lopez and Wu Lab members for comments. This study was supported by HHMI funding to the TIC (C.W., T.L., Q.Z., and L.L.), the Damon Runyon Cancer Research Foundation (V.Q.N.), a Johns Hopkins Bloomberg Distinguished Professorship (C.W.) and the National Institute of Health grant GM132290-01 (C.W.).
Footnotes
DECLARATION OF INTERESTS
T.L. holds intellectual property rights related to Janelia Fluor dyes used in this publication. L.D.L. and Q.Z. are listed as inventors on patents and patent applications whose values might be affected by publication. The remaining authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ansari SA, He Q, and Morse RH (2009). Mediator complex association with constitutively transcribed genes in yeast. Proc National Acad Sci 106, 16734–16739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek HJ, Malik S, Qin J, and Roeder RG (2002). Requirement of TRAP/Mediator for Both Activator-Independent and Activator-Dependent Transcription in Conjunction with TFIID-Associated TAFIIs. Mol Cell Biol 22, 2842–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek HJ, Kang YK, and Roeder RG (2006). Human Mediator Enhances Basal Transcription by Facilitating Recruitment of Transcription Factor IIB during Preinitiation Complex Assembly. Journal of Biological Chemistry 281, 15172–15181. [DOI] [PubMed] [Google Scholar]
- Ball DA, Mehta GD, Salomon-Kent R, Mazza D, Morisaki T, Mueller F, McNally JG, and Karpova TS (2016). Single molecule tracking of Ace1p in Saccharomyces cerevisiae defines a characteristic residence time for non-specific interactions of transcription factors with chromatin. Nucleic Acids Research 44, gkw744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista T, Grünberg S, Minoungou N, Koster MJE, Timmers HTM, Hahn S, Devys D, and Tora L (2017). SAGA Is a General Cofactor for RNA Polymerase II Transcription. Molecular Cell 68, 130–143.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, Zamudio AV, Li CH, Shrinivas K, Manteiga JC, Hannett NM, et al. (2018). Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 175, 1842–1855.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher KR, and Kaiser P (2008). A PCR-based strategy to generate yeast strains expressing endogenous levels of amino-terminal epitope-tagged proteins. [DOI] [PubMed] [Google Scholar]
- Borggrefe T, Davis R, Bareket-Samish A, and Kornberg RD (2001). Quantitation of the RNA polymerase II transcription machinery in yeast. The Journal of Biological Chemistry 276, 47150–47153. [DOI] [PubMed] [Google Scholar]
- Brahma S, and Henikoff S (2018). RSC-Associated Subnucleosomes Define MNase-Sensitive Promoters in Yeast. Molecular Cell 73, 238–249.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang Z, Li L, Chen B-C, Revyakin A, Hajj B, Legant W, Dahan M, Lionnet T, Betzig E, et al. (2014). Single-Molecule Dynamics of Enhanceosome Assembly in Embryonic Stem Cells. Cell 156, 1274–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Adler C, Ball C, Chervitz SA, Dwight SS, Hester ET, Jia Y, Juvik G, Roe T, Schroeder M, et al. (1998). SGD: Saccharomyces Genome Database. Nucleic Acids Res 26, 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W-K, Jayanth N, English BP, Inoue T, Andrews JO, Conway W, Grimm JB, Spille J-H, Lavis LD, Lionnet T, et al. (2016). RNA Polymerase II cluster dynamics predict mRNA output in living cells. ELife 5, 1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W-K, Spille J-H, Hecht M, Lee C, Li C, Grube V, and Cisse II (2018). Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361,412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse II, Izeddin I, Causse SZ, Boudarene L, Senecal A, Muresan L, Dugast-Darzacq C, Hajj B, Dahan M, and Darzacq X (2013). Real-Time Dynamics of RNA Polymerase II Clustering in Live Human Cells. Science 341, 664–667. [DOI] [PubMed] [Google Scholar]
- Compe E, Genes CM, Braun C, Coin F, and Egly J-M (2019). TFIIE orchestrates the recruitment of the TFIIH kinase module at promoter before release during transcription. Nature Communications 10, 2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P (2019). Organization and regulation of gene transcription. Nature 573, 45–54. [DOI] [PubMed] [Google Scholar]
- Dion V, and Gasser SM (2013). Chromatin movement in the maintenance of genome stability. Cell 152, 1355–1364. [DOI] [PubMed] [Google Scholar]
- Donczew R, Warfield L, Pacheco D, Erijman A, and Hahn S (2020). Two roles for the yeast transcription coactivator SAGA and a set of genes redundantly regulated by TFIID and SAGA. Elife 9, e50109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan BT, Huynh A, Ball DA, Patel HP, Poirier MG, Larson DR, Ferguson ML, and Lenstra TL (2019). Live- cell imaging reveals the interplay between transcription factors, nucleosomes, and bursting. The EMBO Journal 38, e51794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B, and Tibshirani R (1986). Bootstrap Methods for Standard Errors, Confidence Intervals, and Other Measures of Statistical Accuracy. Stat Sci 1, 54–75. [Google Scholar]
- Fujiwara R, Damodaren N, Wilusz JE, and Murakami K (2019). The capping enzyme facilitates promoter escape and assembly of a follow-on preinitiation complex for reinitiation. Proceedings of the National Academy of Sciences 116, 22573–22582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguli D, Chereji RV, Iben JR, Cole HA, and Clark DJ (2014). RSC-dependent constructive and destructive interference between opposing arrays of phased nucleosomes in yeast. Genome Research 24, 1637–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt JCM, Suter DM, Roy R, Zhao ZW, Chapman AR, Basu S, Maniatis T, and Xie XS (2013). Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nature Methods 10, 421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, and Schiestl RH (2007). High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nature Protocols 2, 31–34. [DOI] [PubMed] [Google Scholar]
- Grimaldi Y, Ferrari P, and Strubin M (2014). Independent RNA polymerase II preinitiation complex dynamics and nucleosome turnover at promoter sites in vivo. Genome Research 24, 117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm JB, English BP, Chen J, Slaughter JP, Zhang Z, Revyakin A, Patel R, Macklin JJ, Normanno D, Singer RH, et al. (2015). A general method to improve fluorophores for live-cell and single-molecule microscopy. Nature Methods 12, 244–50-3 p following 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünberg S, Henikoff S, Hahn S, and Zentner GE (2016). Mediator binding to UASs is broadly uncoupled from transcription and cooperative with TFIID recruitment to promoters. The EMBO Journal 35, 2435–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidi BW, Bjornsdottir G, Hopkins DC, Lacomis L, Erdjument-Bromage H, Tempst P, and Myers LC (2004). Mutual Targeting of Mediator and the TFIIH Kinase Kin28*. J Biol Chem 279, 29114–29120. [DOI] [PubMed] [Google Scholar]
- Haberle V, and Stark A (2018). Eukaryotic core promoters and the functional basis of transcription initiation. Nat Rev Mol Cell Bio 19, 621–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond CM, Strømme CB, Huang H, Patel DJ, and Groth A (2017). Histone chaperone networks shaping chromatin function. Nature Reviews Molecular Cell Biology 18, 141–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AS, Pustova I, Cattoglio C, Tjian R, and Darzacq X (2017). CTCF and cohesin regulate chromatin loop stability with distinct dynamics. ELife 6, 2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AS, Woringer M, Grimm JB, Lavis LD, Tjian R, and Darzacq X (2018). Robust model-based analysis of single-particle tracking experiments with Spot-On. ELife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AS, Amitai A, Cattoglio C, Tjian R, and Darzacq X (2020). Guided nuclear exploration increases CTCF target search efficiency. Nat Chem Biol 16, 257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley PD, and Madhani HD (2009). Mechanisms that Specify Promoter Nucleosome Location and Identity. Cell 137, 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, and Laemmli UK (2008). The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Molecular Cell 31, 925–932. [DOI] [PubMed] [Google Scholar]
- Ho B, Baryshnikova A, and Brown GW (2018). Unification of Protein Abundance Datasets Yields a Quantitative Saccharomyces cerevisiae Proteome. Cell Systems 6, 192–205.e3. [DOI] [PubMed] [Google Scholar]
- Iben S, Tschochner H, Bier M, Hoogstraten D, Hozák P, Egly J-M, and Grummt I (2002). TFIIH Plays an Essential Role in RNA Polymerase I Transcription. Cell 109, 297–306. [DOI] [PubMed] [Google Scholar]
- Izeddin I, Récamier V, Bosanac L, Cissé II, Boudarene L, Dugast-Darzacq C, Proux F, Bénichou O, Voituriez R, Bensaude O, et al. (2014). Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. ELife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C, and Robert F (2014). Kin28 regulates the transient association of Mediator with core promoters. Nature Structural & Molecular Biology 21, 449–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C, Langelier M-F, Bataille AR, Pascal JM, Pugh BF, and Robert F(2016). Tail and Kinase Modules Differently Regulate Core Mediator Recruitment and Function In Vivo. Molecular Cell 64, 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KM, and Carey M (2003). Assembly of a Mediator/TFIID/TFIIA Complex Bypasses the Need for an Activator. Curr Biol 13, 772–777. [DOI] [PubMed] [Google Scholar]
- Johnson KM, Wang J, Smallwood A, Arayata C, and Carey M (2002). TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA. Genes & Development 16, 1852–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo YJ, Ficarro SB, Soares LM, Chun Y, Marto JA, and Buratowski S (2017). Downstream promoter interactions of TFIID TAFs facilitate transcription reinitiation. Genes & Development 31, 2162–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent S, Brown K, Yang C, Alsaihati N, Tian C, Wang H, and Ren X (2020). Phase-Separated Transcriptional Condensates Accelerate Target-Search Process Revealed by Live-Cell Single-Molecule Imaging. Cell Reports 33, 108248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh M-C, Cho E-J, Podolny V, and Buratowski S (2002). Kin28 Is Found within TFIIH and a Kin28-Ccl1-Tfb3 Trimer Complex with Differential Sensitivities to T-Loop Phosphorylation. Mol Cell Biol 22, 1288–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattabi LE, Zhao H, Kalchschmidt J, Young N, Jung S, Blerkom PV, Kieffer-Kwon P, Kieffer-Kwon K-R, Park S, Wang X, et al. (2019). A Pliable Mediator Acts as a Functional Rather Than an Architectural Bridge between Promoters and Enhancers. Cell 178, 1145–1158.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H, Tao Y, Roeder RG, and Cook PR (1999). Quantitation of RNA Polymerase II and Its Transcription Factors in an HeLa Cell: Little Soluble Holoenzyme but Significant Amounts of Polymerases Attached to the Nuclear Substructure. Molecular and Cellular Biology 19, 5383–5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein-Brill A, Joseph-Strauss D, Appleboim A, and Friedman N (2019). Dynamics of Chromatin and Transcription during Transient Depletion of the RSC Chromatin Remodeling Complex. Cell Reports 26, 279–292.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll ER, Zhu ZI, Sarkar D, Landsman D, and Morse RH (2018). Role of the pre-initiation complex in Mediator recruitment and dynamics. ELife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll ER, Zhu ZI, Sarkar D, Landsman D, and Morse RH (2020). Kin28 depletionincreases association of TFIID subunits Taf1 and Taf4 with promoters in Saccharomyces cerevisiae. Nucleic Acids Res 48, 4244–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer SM, Ranallo RT, Ogg RC, and Stargell LA (2001). TFIIA Interacts with TFIID via Association with TATA-Binding Protein and TAF40. Mol Cell Biol 21, 1737–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubik S, Bruzzone MJ, Jacquet P, Falcone J-L, Rougemont J, and Shore D (2015). Nucleosome Stability Distinguishes Two Different Promoter Types at All Protein-Coding Genes in Yeast. Molecular Cell 60, 422–434. [DOI] [PubMed] [Google Scholar]
- Kubik S, O’Duibhir E, Jonge W.J. de, Mattarocci S, Albert B, Falcone J-L, Bruzzone MJ, Holstege FCP, and Shore D (2018). Sequence-Directed Action of RSC Remodeler and General Regulatory Factors Modulates +1 Nucleosome Position to Facilitate Transcription. Molecular Cell 71, 89–102.e5. [DOI] [PubMed] [Google Scholar]
- Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH, and Roeder RG (2013). H3K4me3 Interactions with TAF3 Regulate Preinitiation Complex Assembly and Selective Gene Activation. Cell 152, 1021–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer JH, and Weil PA (2013). Direct TFIIA-TFIID protein contacts drive budding yeast ribosomal protein gene transcription. The Journal of Biological Chemistry 288, 23273–23294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SN, Brown CR, Harvey S, Boeger H, Elmlund H, and Elmlund D (2019). The TAFs of TFIID Bind and Rearrange the Topology of the TATA-Less RPS5 Promoter. International Journal of Molecular Sciences 20, 3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, and Hahn S (1995). Model for binding of transcription factor TFIIB to the TBP-DNA complex. Nature 376, 609–612. [DOI] [PubMed] [Google Scholar]
- Lerner J, Gomez-Garcia PA, McCarthy RL, Liu Z, Lakadamyali M, and Zaret KS (2020). Two-Parameter Mobility Assessments Discriminate Diverse Regulatory Factor Behaviors in Chromatin. Mol Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Dong A, Saydaminova K, Chang H, Wang G, Ochiai H, Yamamoto T, and Pertsinidis A (2019). Single-Molecule Nanoscopy Elucidates RNA Polymerase II Transcription at Single Genes in Live Cells. Cell 178, 491–506.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim MK, Tang V, Saux AL, Schüller J, Bongards C, and Lehming N (2007). Gal11p Dosage-compensates Transcriptional Activator Deletions via Taf14p. J Mol Biol 374, 9–23. [DOI] [PubMed] [Google Scholar]
- Lionnet T, and Wu C (2021). Single-molecule tracking of transcription protein dynamics in living cells: seeing is believing, but what are we seeing? Curr Opin Genet Dev 67, 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffreda A, Jacchetti E, Antunes S, Rainone P, Daniele T, Morisaki T, Bianchi ME, Tacchetti C, and Mazza D (2017). Live-cell p53 single-molecule binding is modulated by C-terminal acetylation and correlates with transcriptional activity. Nature Communications 8, 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, et al. (2008). HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chemical Biology 3, 373–382. [DOI] [PubMed] [Google Scholar]
- Maxon ME, Goodrich JA, and Tjian R (1994). Transcription factor IIE binds preferentially to RNA polymerase IIa and recruits TFIIH: a model for promoter clearance. Gene Dev 8, 515–524. [DOI] [PubMed] [Google Scholar]
- Mazza D, Abernathy A, Golob N, Morisaki T, and McNally JG (2012). A benchmark for chromatin binding measurements in live cells. Nucleic Acids Research 40, e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSwiggen DT, Hansen AS, Teves SS, Marie-Nelly H, Hao Y, Heckert AB, Umemoto KK, Dugast-Darzacq C, Tjian R, and Darzacq X (2019). Evidence for DNA-mediated nuclear compartmentalization distinct from phase separation. Elife 8, e47098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medler S, and Ansari A (2015). Gene looping facilitates TFIIH kinase-mediated termination of transcription. Sci Rep-Uk 5, 12586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta GD, Ball DA, Eriksson PR, Chereji RV, Clark DJ, McNally JG, and Karpova TS (2018). Single-Molecule Analysis Reveals Linked Cycles of RSC Chromatin Remodeling and Ace1p Transcription Factor Binding in Yeast. Molecular Cell 72, 875–887.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miné-Hattab J, Recamier V, Izeddin I, Rothstein R, and Darzacq X (2017). Multi-scale tracking reveals scale-dependent chromatin dynamics after DNA damage. Mol Biol Cell 28, 3323–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K, Mattei P-J, Davis RE, Jin H, Kaplan CD, and Kornberg RD (2015). Uncoupling Promoter Opening from Start-Site Scanning. Molecular Cell 59, 133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil H, Malabat C, d’Aubenton-Carafa Y, Xu Z, Steinmetz LM, and Jacquier A (2009). Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457, 1038–1042. [DOI] [PubMed] [Google Scholar]
- Normanno D, Dahan M, and Darzacq X (2012). Intra-nuclear mobility and target search mechanisms of transcription factors: A single-molecule perspective on gene expression. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms 1819, 482–493. [DOI] [PubMed] [Google Scholar]
- Papai G, Tripathi MK, Ruhlmann C, Layer JH, Weil PA, and Schultz P (2010). TFIIA and the transactivator Rap1 cooperate to commit TFIID for transcription initiation. Nature 465, 956–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AB, Louder RK, Greber BJ, Grünberg S, Luo J, Fang J, Liu Y, Ranish J, Hahn S, and Nogales E (2018). Structure of human TFIID and mechanism of TBP loading onto promoter DNA. Science 362, eaau8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Chávez S, and Pérez-Ortín JE (2010). A Complete Set of Nascent Transcription Rates for Yeast Genes. PLoS ONE 5, e15442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrenko N, Jin Y, Wong KH, and Struhl K (2016). Mediator Undergoes a Compositional Change during Transcriptional Activation. Molecular Cell 64, 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrenko N, Jin Y, Wong KH, and Struhl K (2017). Evidence that Mediator is essential for Pol II transcription, but is not a required component of the preinitiation complex in vivo. ELife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu C, Jin H, Vvedenskaya I, Llenas JA, Zhao T, Malik I, Schwartz SL, Cui P, Čabart P, Han KH, et al. (2019). Promoter scanning during transcription initiation in Saccharomyces cerevisiae: Pol II in the “shooting gallery.” Biorxiv 810127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Zentner GE, and Henikoff S (2015). Asymmetric nucleosomes flank promoters in the budding yeast genome. Genome Research 25, 381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani PG, Ranish JA, and Hahn S (2004). RNA Polymerase II (Pol II)-TFIIF and Pol II-Mediator Complexes: the Major Stable Pol II Complexes and Their Activity in Transcription Initiation and Reinitiation. Mol Cell Biol 24, 1709–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan A, Nguyen VQ, Liu S, Wisniewski J, Kim JM, Tang X, Mizuguchi G, Elalaoui E, Nickels TJ, Jou V, et al. (2020). Live-cell single particle imaging reveals the role of RNA polymerase II in histone H2A.Z eviction. Elife 9, e55667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, and Pugh BF (2012). Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483, 295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimel JK, and Taatjes DJ (2018). The essential and multifunctional TFIIH complex. Protein Science 27, 1018–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PJ, Trnka MJ, Bushnell DA, Davis RE, Mattei P-J, Burlingame AL, and Kornberg RD (2016). Structure of a Complete Mediator-RNA Polymerase II Pre-Initiation Complex. Cell 166, 1411–1422.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Molina JB, Tseng SC, Simonett SP, Taunton J, and Ansari AZ (2016). Engineered Covalent Inactivation of TFIIH-Kinase Reveals an Elongation Checkpoint and Results in Widespread mRNA Stabilization. Molecular Cell 63, 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilbach S, Hantsche M, Tegunov D, Dienemann C, Wigge C, Urlaub H, and Cramer P (2017). Structures of transcription pre-initiation complex with TFIIH and Mediator. Nature 551, 204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweikhard V, Meng C, Murakami K, Kaplan CD, Kornberg RD, and Block SM (2014). Transcription factors TFIIF and TFIIS promote transcript elongation by RNA polymerase II by synergistic and independent mechanisms. Proc National Acad Sci 111, 6642–6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrinivas K, Sabari BR, Coffey EL, Klein IA, Boija A, Zamudio AV, Schuijers J, Hannett NM, Sharp PA, Young RA, et al. (2019). Enhancer Features that Drive Formation of Transcriptional Condensates. Molecular Cell 75, 549–561.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatavosian R, Duc HN, Huynh TN, Fang D, Schmitt B, Shi X, Deng Y, Phiel C, Yao T, Zhang Z, et al. (2018). Live-cell single-molecule dynamics of PcG proteins imposed by the DIPG H3.3K27M mutation. Nature Communications 9, 2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenaz P, Ruttimann UE, and Unser M (1998). A pyramid approach to subpixel registration based on intensity. Ieee T Image Process 7, 27–41. [DOI] [PubMed] [Google Scholar]
- Tuttle LM, Pacheco D, Warfield L, Luo J, Ranish J, Hahn S, and Klevit RE (2018). Gcn4-Mediator Specificity Is Mediated by a Large and Dynamic Fuzzy Protein-Protein Complex. Cell Reports 22, 3251–3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallotton P, and Olivier S (2013). Tri-track: Free Software for Large-Scale Particle Tracking. Microscopy and Microanalysis 19, 451–460. [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WWMP, van Schaik FMA, Varier RA, Baltissen MPA, Stunnenberg HG, Mann M, and Timmers H.Th.M. (2007). Selective Anchoring of TFIID to Nucleosomes by Trimethylation of Histone H3 Lysine 4. Cell 131, 58–69. [DOI] [PubMed] [Google Scholar]
- Warfield L, Ramachandran S, Baptista T, Devys D, Tora L, and Hahn S (2017). Transcription of Nearly All Yeast RNA Polymerase II-Transcribed Genes Is Dependent on Transcription Factor TFIID. Molecular Cell 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Werven FJ, van Teeffelen HAAM, Holstege FCP, and Timmers HTM (2009). Distinct promoter dynamics of the basal transcription factor TBP across the yeast genome. Nature Structural & Molecular Biology 16, 1043–1048. [DOI] [PubMed] [Google Scholar]
- Wieser S, and Schütz GJ (2008). Tracking single molecules in the live cell plasma membrane—Do’s and Don’t’s. Methods 46, 131–140. [DOI] [PubMed] [Google Scholar]
- Wong KH, Jin Y, and Struhl K (2014). TFIIH phosphorylation of the Pol II CTD stimulates mediator dissociation from the preinitiation complex and promoter escape. Molecular Cell 54, 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woringer M, and Darzacq X (2018). Protein motion in the nucleus: from anomalous diffusion to weak interactions. Biochem Soc T 46, BST20170310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woringer M, Darzacq X, and Izeddin I (2014). Geometry of the nucleus: a perspective on gene expression regulation. Current Opinion in Chemical Biology 20, 112–119. [DOI] [PubMed] [Google Scholar]
- Woringer M, Izeddin I, Favard C, and Berry H (2020). Anomalous Subdiffusion in Living Cells: Bridging the Gap Between Experiments and Realistic Models Through Collaborative Challenges. Aip Conf Proc 8, 134. [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Münster S, Camblong J, Guffanti E, Stutz F, Huber W, and Steinmetz LM (2009). Bidirectional promoters generate pervasive transcription in yeast. Nature 457, 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Pradhan SK, Sun F, Chronis C, Tran N, Su T, Van C, Vashisht A, Wohlschlegel J, Peterson CL, et al. (2017). Mot1, Ino80C, and NC2 Function Coordinately to Regulate Pervasive Transcription in Yeast and Mammals. Molecular Cell 67, 594–607.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawel L, Kumar KP, and Reinberg D (1995). Recycling of the general transcription factors during RNA polymerase II transcription. Gene Dev 9, 1479–1490. [DOI] [PubMed] [Google Scholar]
- Zhang Z, English BP, Grimm JB, Kazane SA, Hu W, Tsai A, Inouye C, You C, Piehler J, Schultz PG, et al. (2016). Rapid dynamics of general transcription factor TFIIB binding during preinitiation complex assembly revealed by single-molecule analysis. Genes & Development 30, 2106–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Ayala AX, Chung I, Weigel AV, Ranjan A, Falco N, Grimm JB, Tkachuk AN, Wu C, Lippincott-Schwartz J, et al. (2019). Rational Design of Fluorogenic and Spontaneously Blinking Labels for Super-Resolution Imaging. Acs Central Sci 5, 1602–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Taf1-Halo motion by fast tracking in a live cell, Related toFigure 1. Raw movie of Taf1-Halo-JF552 in the nucleus of a live cell. Data was acquired at 10 ms framerate and rendered at 50 fps (frames per second). The nuclear outline (oval) was estimated from total nuclear fluorescence (not shown).
Kin28-Halo motion by slow tracking in a live cell, Related toFigure 3. Raw movie of Kin28-Halo-JF552 in the nucleus of a live cell. Two single binding events intervened by a motion-blurred diffusive episode, likely by the same molecule, are captured. Data was acquired at 250 ms framerate and rendered at 25 fps (frames per second). The nuclear outline (oval) was estimated from total nuclear fluorescence (not shown).
Data Availability Statement
Trajectory coordinates have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the Key Resources Table. Original microscopy data (.cxd or .tif movies) will be shared by the Lead Contact upon request.
Custom scripts used to analyze SMT data in the Sojourner computational package have been deposited at Mendeley and are publicly available as of the date of publication. The DOI and Github addresses are listed in the Key Resources Table.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.