Abstract
Epstein–Barr virus (EBV)-encoded latent membrane protein 1 (LMP1) plays an important oncogenic role in the viral latent infection. Recently, increasing evidence indicates that the high expression of LMP1 during EBV lytic cycle is related to the viral lytic replication. However, the mechanism by which LMP1 regulates EBV lytic replication remains unclear. (−)-Epigallocatechin-3-gallate (EGCG) prevents carcinogenesis by directly targeting numerous membrane proteins and effectively inhibits EBV lytic cascade. Here, we demonstrated that LMP1 promotes EBV lytic replication through the downstream signal molecules MAPKs, including ERKs, p38, and JNKs. LMP1 induces the phosphorylation of p53 through MAPKs to enhance the ability of wild-type p53 (wt-p53) to activate expression of BZLF1 gene, while the JNKs/c-Jun signal axis appears to be involved in EBV lytic replication induced by LMP1 in p53 mutant manner. We provided the first evidence that EGCG directly targets the viral membrane LMP1 (K d = 0.36 μM, n = 1) using fluorescence quenching, isothermal titration calorimetry (ITC) assay, and CNBR-activated Sepharose 4B pull-down affinity chromatography. Furthermore, we revealed that EGCG inhibits EBV lytic replication via suppressing LMP1 and thus blocking the downstream MAPKs/wt-p53 signal axis in AGS-EBV cells and JNKs/c-Jun signal axis in p53 mutant B95.8 cells. Our study, for the first time, reports the binding and inhibitory efficacy of EGCG to the LMP1, which is a key oncoprotein encoded by EBV. These findings suggest the novel function of LMP1 in the regulation of EBV lytic cycle and reveal the new role of EGCG in EBV-associated malignancies through suppressing viral reactivation.
Key words: Latent membrane protein 1 (LMP1), Epstein–Barr virus (EBV) lytic replication, MAPKs, p53, (−)-Epigallocatechin-3-gallate (EGCG)
INTRODUCTION
Epstein–Barr virus (EBV) is the first human tumor virus identified and known to be linked to several types of human malignancies, including nasopharyngeal carcinoma (NPC), Hodgkin’s lymphoma (HL), Burkitt’s lymphoma (BL), and EBV-associated gastric carcinoma (EBVaGC)1–3. EBVaGC, especially gastric adenocarcinomas, was first categorized by EBV positivity4. EBVaGC may represent the most common form of EBV-associated malignancy, which has distinct phenotypic and clinical characteristics, and accounts for an average of 10% in all gastric cancer cases, and its incidence depends on geographical distribution and environmental factors5,6.
In the host, EBV exhibits two alternative modes of infection: latent and lytic. In latency, EBV only expresses a limited number of gene products such as latent membrane protein 1 (LMP1), LMP2A/2B, EBV nuclear antigens (EBNAs), and EBV-encoded small RNAs (EBERs). When stimulated by certain chemicals or biological reagents, EBV undergoes three consecutive lytic stages, including immediate early (IE), early (E), and late (L) stages. Zta and Rta that are encoded by the lytic IE genes BZLF1 and BRLF1, respectively, activate transcription from the promoters of lytic E genes, which trigger EBV genomic DNA replication. Then the lytic L genes that encode structural proteins are expressed, followed by viral genome packaging into infectious virion particles. The switch from latency to the lytic cycle is known as EBV lytic reactivation, which can be triggered by the both IE transactivators Zta and Rta. In addition to activating the expression of lytic E genes7,8, Zta functions as a replication factor for EBV genomic DNA by binding the lytic origin of replication, oriLyt9. Furthermore, Zta can also directly activate transcription from its own promoter of BZLF1 gene (Zp)10 and the promoter of BRLF1 gene11, greatly amplifying the inductive effects on EBV lytic replication12. Earlier studies indicated that the latent infection is a major cause of EBV-associated malignancies13,14. Recently, EBV lytic infection has been shown to significantly contribute to carcinogenesis15–17. Evidence has supported that there is a close association of varying degrees of EBV lytic replication with EBV-associated cancers, including EBVaGC17. Also, clinical and epidemiological studies have revealed that individuals with elevated antibody titers against EBV lytic antigens and increased plasma EBV DNA load have a higher risk of developing EBV-associated tumors18.
EBV-encoded LMP1 is a classic oncoprotein, which activates multiple signaling pathways, including mitogen-activated protein kinases (MAPKs)19, nuclear factor κB (NF-κB)20, PKC21, Janus kinase (JAK)/signal transducer and activator of transcription (STAT)22, and phosphatidylinositol 3-kinase (PI3-K)/Akt23, that affect cell proliferation and survival. LMP1 is also abundantly expressed during the lytic cycle of viral replication. A previous study found that LMP1 plays a critical role in the production of EBV24. A recent research has shown that differentiation-dependent LMP1 expression is required for efficient EBV lytic replication in epithelial cells25. Additionally, LMP1 appears to facilitate EBV entry into the lytic cycle by leading to severe cellular stresses such as hypoxia, oxidative stress, and inflammation that induce EBV lytic cycle initiation16,26–29. LMP1 can also enhance transcriptional activity and stability of p53 by inducing phosphorylation of p53 through MAPKs in NPC cells19. Furthermore, some studies indicated that p53 might be a prerequisite for EBV lytic replication by facilitating the expression of Zta30,31. However, the mechanism by which LMP1 promotes EBV lytic replication is still not clear.
Blocking EBV lytic replication by natural compounds is valuable for the prevention and treatment of EBV-associated malignancies and helps improve clinical outcome. Epigallocatechin-3-gallate (EGCG), the most abundant anticancer polyphenol in green tea32–35, has antiviral effects against diverse viruses36 and is reported to inhibit EBV lytic cascade37–39. Accumulated researches have demonstrated that EGCG directly interacts with numerous membrane proteins such as Fas, ZAP-70, Fyn, insulin-like growth factor-I receptor (IGF-1R), and the 67-kDa laminin receptor (67LR)40–42. EGCG exerts the anticancer actions by modulating the activities or functions of these enzymes, receptors, and signaling molecules that affect cell growth and proliferation. Our group revealed that EGCG inhibits EBV lytic replication via downregulating LMP1 or inactivation of mitogen-activated protein kinase kinase (MEK)/extracellular signal-regulated kinases (ERKs) and PI3-K/Akt signaling pathways37,38. Based on these findings, we speculate that LMP1 might promote EBV lytic replication via its downstream oncogenic signaling axes, and EGCG might inhibit EBV lytic replication by targeting the LMP1-mediated signaling axes.
In this study, we first demonstrated that LMP1 promotes EBV lytic replication through differential regulation of the MAPK signal axes. In AGS-EBV cells, LMP1-induced phosphorylation of p53 through ERKs, p38, and c-Jun NH2-terminal kinases (JNKs) enhances the ability of wild-type p53 (wt-p53) to activate Zp. Phosphorylation of c-Jun by LMP1 through JNKs is involved in EBV lytic replication in p53 mutant B95.8 cells. Importantly, we provided the first evidence that EGCG could directly target EBV-encoded membrane protein LMP1 (K d = 0.36 μM, n = 1). Furthermore, we revealed that EGCG inhibits EBV lytic replication via downregulation of LMP1, thus blocking the MAPKs/wt-p53 signal axis in AGS-EBV cells and JNKs/c-Jun signal axis in p53 mutant B95.8 cells. Taken together, we identified that EGCG inhibits EBV lytic replication by directly targeting LMP1-mediated MAPK signal axes, which suggests EGCG as a potential therapeutic treatment for EBV-associated malignancies.
MATERIALS AND METHODS
Cell Lines and Cell Culture
The human gastric adenocarcinoma cell line AGS-EBV (wt-p53) containing recombinant EBV was cultured in HAM’s/F-12 medium (HyClone, Logan, UT, USA) supplemented with 10% fetal calf serum (Invitrogen, Carlsbad, CA, USA), penicillin (100 U), and streptomycin (100 mg/ml). B95.8 (mutant p53, VR-1492™), an EBV-positive B-lymphoma cell line, was cultured in RPMI-1640 medium (HyClone) supplemented with 10% newborn calf serum (Invitrogen), penicillin (100 U), and streptomycin (100 mg/ml). The human NPC cell lines CNE1, CNE1-LMP1, HNE2, and HNE2-LMP1, which have been described previously43–45, can reflect the genuine type II latency of NPC. The NPC cell lines and non-small cell lung carcinoma cell line H1299 (p53-null, CRL-5803™) were cultured in RPMI-1640 medium (HyClone) supplemented with 10% fetal calf serum, penicillin (100 U), and streptomycin (100 mg/mL). All cells were incubated in a humidified atmosphere of 5% CO2 at 37°C.
Chemicals and Cell Treatment
EGCG, PD98059, and SB202190 were purchased from Sigma-Aldrich (St. Louis, MO, USA). SP600125 was purchased from MedChemExpress (MCE, Princeton, NJ, USA), and protease inhibitor cocktail was purchased from Selleck Chemicals (Houston, TX, USA). All chemicals were dissolved in dimethyl sulfoxide (DMSO; Sigma-Aldrich) and used at the indicated concentrations. Detailed treatment procedures are described in the figure legends. The final concentration of DMSO in medium was maintained at <0.1%, which had no significant effect on cell growth. Vehicle controls were included for all treatments.
Plasmids and Small Interfering RNA (siRNA)/DNAzyme
The p53 siRNAs were designed and synthesized by Thermo Fisher Scientific (Waltham, MA, USA), and the specific sequence was GAGGGATGTTTGGGAGATGTA (3′-UTR). The pSG5-based expression vector for LMP1 and pEGFP-based expression vector for p53 were provided by Dr. Kenneth M. Izumi (Brigham and Women’s Hospital, Boston, MA, USA) and Prof. Qiao Wu (Xiamen University, Xiamen, China), respectively. DZ1, a designed DNAzyme that specifically targets transcripts of LMP1 was provided by Prof. Lunquan Sun (Xiangya Hospital, Central South University, Changsha, China), and the specific sequence was 5′-GCAAAGGAAGGCTAGCTACAACGAAGAGGACAA-3′. The transfections of plasmid/siRNA/DNAzyme were performed using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s protocol.
Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was isolated from cultured cells with TRIzol (Invitrogen) according to the manufacturer’s instructions, followed by RT, which was performed on 2 μg of total RNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo). qPCR was performed using Fast Start Universal SYBR Green Master (ROX) and 2 μl (10× dilution) of RT reaction products (Roche, Basel, Switzerland) with the primers of BZLF1 and BMRF1. The primers used here were as follows: BZLF1, 5′-CATGTTTCAACCGCTCCGACTGG-3′ (forward) and 5′-GCGCAGCCTGTCATTTTCAGATG-3′ (reverse); BMRF1, 5′-CTAGCCGTCCTGTCCAAGTGC-3′ (forward) and 5′-AGCCAAACGCTCCTTGCCCA-3′ (reverse). The following PCR program was used: 95°C for 10 min; 95°C for 15 s, 60°C for 1 min (plate read), 40 cycles; melt curve 60–95°C, increments of 0.5°C, for 15 s. Relative gene expression was calculated using the 2−ΔΔct method after normalization to the reference gene β-actin.
Western Blotting Analysis and Antibodies
Cell lysates (50 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred to polyvinylidene difluoride membranes. The membranes were incubated with blocking buffer and each primary antibody overnight at 4°C. After having been washed with phosphate buffer solution with Tween® 20 (PBST), the membrane was incubated with the conjugated secondary antibody at room temperature for 1 h. Finally, the protein bands were visualized using enhanced chemiluminescence (ECL; Beijing Solarbio Science & Technology Co. Ltd., Beijing, P.R. China) reagents. The primary antibodies used here were as follows: anti-β-actin antibody (sc-8432), anti-p53 antibody (sc-126), anti-p-p53 (ser15) antibody (sc-101762), anti-p-p53 (ser392) antibody (sc-7997), anti-p38 antibody (sc-81621), and anti-Sp1 antibody (sc-17824) (purchased from Santa Cruz Biotechnology, Santa Cruz, CA, USA); anti-LMP1 antibody (CS1-4) (purchased from DAKO, Copenhagen, Denmark); anti-Zta antibody (ab21134) and anti-Ea-D antibody (ab30541) (purchased from Abcam, Cambridge, UK); p-MAPK Family Antibody Sampler Kit (#9910), anti-Erks antibody (#4695), anti-p-p53 (ser20) antibody (#9287), anti-JNKs antibody (#9252), anti-p-c-Jun (ser73) antibody (#3270), anti-c-Jun antibody (#9165), anti-p-c-Jun (ser63) antibody (#91952), and anti-Smad2/3 antibody (#8685) (purchased from Cell Signaling Technology, Boston, MA, USA).
Fluorescence Quenching Assay
Fluorescence quenching assay was performed as previously described46 using a fluorescence spectrophotometer (model F-4600, HITACHI, Tokyo, Japan). A His-LMP1 fusion protein was expressed in animal cell, followed by purification using Ni-NTA (nickel–nitrilotriacetic acid; Qiagen, Chatsworth, CA, USA). The purified LMP1 was dialyzed against phosphate-buffered saline (PBS). The His-LMP1 fusion protein was incubated with different concentrations of EGCG. Fluorescence quenching was monitored at 25°C and 37°C by using 5 nm of excitation and emission slit width. The wavelength of excitation light was 280 nm, and the emission spectra were measured between 300 and 450 nm.
Isothermal Titration Calorimetry Assay
Isothermal titration calorimetry (ITC) assay was performed on a NANO ITC microcalorimeter (TA instruments, New Castle, PA, USA) to investigate the binding of EGCG to LMP1. Titration calorimetry was performed at 25°C in the assay buffer (PBS, pH 6.5). Briefly, the sample and syringe cell were filled with LMP1 (1.8 μM) and EGCG (20 μM), respectively, which were degassed prior to use. The titrations were conducted using 25 identical injections of 2 μl with duration of 11 s, and with a 175-s delay between each injection. The thermodynamics parameters of interaction between EGCG and purified LMP1 can be calculated by fitting the raw ITC data. The dissociation constant (K d) and chemical reaction quantity (n) were determined after analysis of the normalized ITC curve by the MicroCal PEAQ-ITC Analysis Software.
In Vitro Pull-Down Affinity Chromatography Assay
According to previous description47, a cellular supernatant fraction (500 μg) was incubated with EGCG-cyanogen bromide (CNBR)-activated Sepharose 4B or CNBR-activated Sepharose 4B as control beads (50 μl, 50% slurry) in reaction buffer [50 mM tris (pH 7.5), 5 mM EDTA, 150 mM NaCl, 1 mM DTT, 0.01% NP40, 0.02 mM phenylmethylsulfonyl fluoride (PMSF), 4 Ag/ml bovine serum albumin, 1× protease inhibitor cocktail]. After incubation with gentle rocking overnight at 4°, the beads were washed five times with washing buffer [50 mM tris (pH 7.5), 5 mM EDTA, 150 mM NaCl, 1 mM DTT, 0.01% NP40, 0.02 mM PMSF], and the obtained composite of beads and proteins was analyzed by Western blotting.
Coimmunoprecipitation (Co-IP) Assay
The whole-cell lysates (500 μg) prepared with IP lysis buffer (Pierce, Rockford, IL, USA) were incubated with 20 μl of protein A Sepharose beads (Sigma-Aldrich) for 2 h at 4°C on a rotating rocker and centrifuged for 10 min at 2000 rpm. The recovered supernatant fraction was incubated with p53 antibody (sc-126 X; Santa Cruz Biotechnology) in the presence of 1× protease inhibitors at 4°C overnight on a rotating rocker. Then 40 μl of protein A Sepharose beads was added, and the incubation was continued for 2 h at 4°C on a rotating rocker. The immunocomplexes bound to the Sepharose beads were recovered by a brief centrifugation followed by three washes with cold 1× PBS. The harvested beads were resuspended in 20 μl of 5× SDS-PAGE sample buffer and boiled for 5 min. A 50-μg cell lysate was used as an input control. The samples were analyzed by Western blotting.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assays were performed using the Chromatin Immunoprecipitation Kit (Merck Millipore, Billerica, MA, USA) according to the manufacturer’s instructions. The DNA isolated from the ChIP assay with p53 antibody or normal IgG (negative control), along with an input control, was amplified using PCR to determine p53-binding elements around the promoter region of the BZLF1 gene (Zp). ChIP primers used here were as follows: BZLF1 promoter (−221-+12), 5′-GCAAGGTGCAATGTTTAGTGAG-3′ (forward) and 5′-CCATGCATATTTCAACTGGGC-3′ (reverse); p21 promoter (p53 REs), 5′-CTGGACTGGGCACTCTTGTC-3′ (forward) and 5′-CTCCTACCATCCCCTTCCTC-3′ (reverse). p21 promoter was used as a positive control.
DNA Extraction and Quantification of EBV Copy Numbers
DNA was extracted from the EBV-positive cells using the QIAamp® DNA Mini Kit (Qiagen, Hilden, Germany) according to the kit handbook. Following the instructions of the EBV Polymerase PCR Fluorescence Quantitative Diagnostic Kit (DA, Guangzhou, China), a 10-fold dilution from 107 to 104 copies was first used to create the standard curve. Then 2 μl of each DNA sample was added to PCR tubes, and PCR was performed. The EBV copy numbers of the cells can be calculated by the corresponding threshold cycle with the aid of the standard curve.
Flow Cytometer Analysis
The cells were washed with PBS, followed by fixing with 4% paraformaldehyde for 30 min and permeabilization for 15 min with PBS containing 0.1% Triton X-100 at 4°C. Then the cells were washed with PBS and blocked with 2% BSA and 0.1% Triton X-100 in PBS for 30 min at room temperature. The cells were washed with PBS and incubated with 1:100 diluted monoclonal Zta antibody (Abcam) and monoclonal Ea-D antibody (Abcam) overnight at 4°C. Next, the cells were washed with PBS and incubated with 1:100 diluted Alexa Fluor® 633-conjugated goat anti-mouse IgG (Life Technologies, Carlsbad, CA, USA) for 1 h at room temperature. Incubation with PBS, without primary antibody followed by incubation with the secondary antibody, was used as a negative control. Finally, the cells were washed with PBS and resuspended in 1% paraformaldehyde for analysis using a FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Transfection and Dual-Luciferase Reporter Assay
The cells were cotransfected with pEGFP-C3-p53 (200 ng, 800 ng), pZp-luc (400 ng), and the pRL-TK internal control (10 ng). After transfection for 48 h, the cells were disrupted and firefly luciferase activity was measured using a Dual-Luciferase Reporter Gene Assay Kit (Promega, Madison, WI, USA) following the manufacturer’s instructions.
Statistical Analysis
Analysis of the experimental data was performed using the statistical software GraphPad Prism 8 (GraphPad Software Inc., La Jolla, CA, USA). Values are expressed as the mean ± standard deviation (SD). Groups were compared using the one-way analysis of variance (ANOVA) test. Differences between various groups were considered statistically significant (p < 0.05).
RESULTS
LMP1 Promotes EBV Lytic Replication
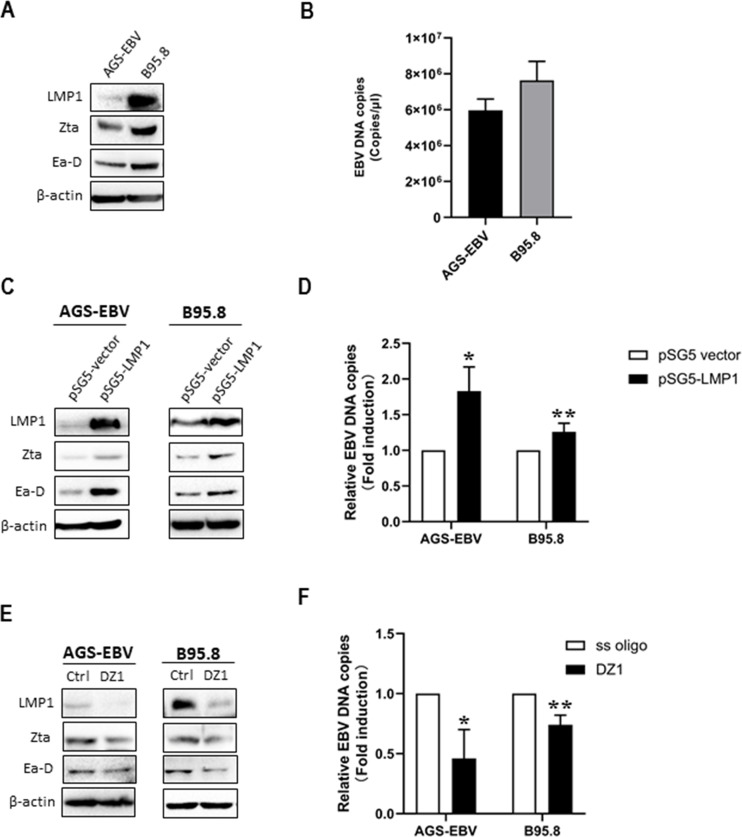
To investigate the role of LMP1 in the EBV lytic cycle, we firstly detected the related EBV-encoded products in the AGS-EBV cell line, which is permissive for EBV lytic replication and in well-established EBV lytically infected B95.8 cell line. Western blotting analysis showed that LMP1 and the lytic proteins Zta and Ea-D were coexpressed in both AGS-EBV and B95.8 cells (Fig. 1A). By using a qPCR, the EBV DNA copy numbers were detected at significant levels in both AGS-EBV cells (5.34 × 106 copies/μl) and B95.8 cells (6.27 × 106 copies/μl) (Fig. 1B). The results show that EBV spontaneous lytic replication exists in both cell lines, and LMP1 is also abundantly expressed.
Figure 1.
Latent membrane protein 1 (LMP1) promotes Epstein–Barr virus (EBV) lytic replication. (A) Western blotting analysis was performed to determine the expression of LMP1, Zta, and Ea-D in AGS-EBV and B95.8 cells. (B) EBV genomic copies were determined by using quantitative polymerase chain reaction (qPCR) assay with specific primers and a TaqMan probe targeting the BamHI W region of the EBV genome. (C) AGS-EBV and B95.8 cells were transfected with a plasmid containing pSG5-LMP1 or a control pSG5 vector. Expression of LMP1, Zta, and Ea-D was detected. (D) EBV genome copies were determined by qPCR. AGS-EBV and B95.8 cells were transfected with DZ1 or single-stranded oligonucleotides (ss oligo). (E) The levels of LMP1, Zta, and Ea-D and (F) numbers of EBV genome copies were detected. For each experiment, EBV DNA copy number levels from control group were set to 1. β-Actin was used as an internal control. *p < 0.05, **p < 0.01 versus control group.
Next, we performed LMP1 gain-of-function and loss-of-function studies in these two cell lines. When the cells were transfected with a plasmid containing pSG5-LMP1, Zta and Ea-D were markedly upregulated and EBV DNA copy numbers increased compared with the control group, the cells transfected with a pSG5 empty plasmid (Fig. 1C and D). As expected, LMP1 knockdown by DZ1, a designed DNAzyme that specifically targets transcripts of LMP1, significantly reduced expression of Zta and Ea-D and decreased EBV DNA copies compared with the control group, which is transfected with single stranded oligonucleotide (ss oligo) (Fig. 1E and F).
MAPKs Are Required for LMP1-Induced EBV Lytic Replication
MAPK signaling pathways, including ERKs, p38, and JNKs, are strongly activated in response to LMP1 induction19,48,49. We found that LMP1 overexpression increased the levels of phosphorylated ERKs, p38, and JNKs, followed by upregulation of EBV lytic proteins in AGS-EBV and B95.8 cells (Fig. 2A). To further determine whether MAPKs are involved in EBV lytic replication induced by LMP1, we treated the cells with the MAPK-specific inhibitors, including PD98059, which is a specific inhibitor of MEK1 that acts by suppressing activation of ERK1/2, SB202190, a specific inhibitor of p38 kinase, and SP600125, an inhibitor of JNKs that exhibits a 300-fold greater selectivity for JNKs as compared to ERKs or p38. The results showed that pretreatment with PD98059, SB202190, or SP600125 markedly decreased the levels of phosphorylated ERKs, P38, or JNKs and attenuated the expression of Zta and Ea-D (Fig. 2B–D). Subsequently, the cells were pretreated with PD98059, SB202190, and Sp600125 alone or in combination and simultaneously transfected with a plasmid containing pSG5-LMP1 or pSG5 control. We found that blocking MAPKs attenuated the inductive effects of LMP1 on expression of EBV lytic proteins (Fig. 2E). Similar changes were also observed in the numbers of EBV genomic copy in the cells pretreated with all three inhibitors in combination with pSG5-LMP1 transfection (Fig. 2F). The results reveal that blocking MAPKs can counteract LMP1-induced EBV lytic protein expression, suggesting that MAPKs are required for LMP1-induced EBV lytic replication.
Figure 2.
Mitogen-activated protein kinases (MAPKs) are required for EBV lytic replication induced by LMP1. (A) AGS-EBV and B95.8 cells were transfected with a plasmid containing pSG5-LMP1 or a control pSG5 vector. Western blotting analysis for LMP1, total and phosphorylated extracellular signal-regulated kinases (ERKs), p38, and c-Jun NH2-terminal kinases (JNKs), and EBV lytic proteins Zta and Ea-D were performed. AGS-EBV and B95.8 cells were pretreated with the MAPK-specific inhibitors, including PD98059 (B), SB202190 (C), and Sp600125 (D), at the concentrations as indicated for 24 h, respectively. Western blotting analysis was performed to detect total and phosphorylated ERKs, p38, and JNKs, and EBV lytic proteins Zta and Ea-D. AGS-EBV and B95.8 cells were transfected with the plasmid expressing LMP1 for 4 h prior to incubation with the MAPK-specific inhibitors alone or in combination, and then these cells were continuously cultured for 48 h. (E) The levels of total and phosphorylated MAPKs, lytic proteins, and (F) EBV DNA copy numbers were analyzed using Western blotting and qPCR, respectively. For each experiment, EBV DNA copy number levels from the control group were set to 1. β-Actin was used as an internal control. *p < 0.05, **p < 0.01 versus control group.
LMP1 Promotes EBV Lytic Replication Through MAPKs in a wt-p53-Dependent Manner
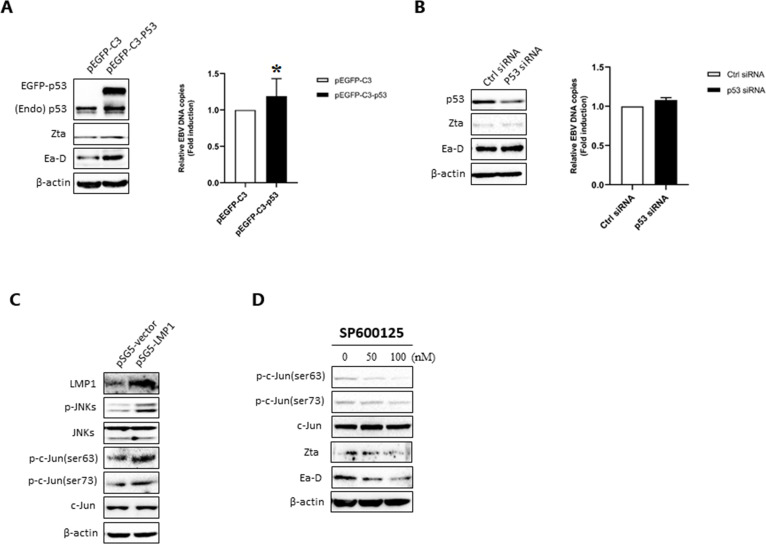
As known, one of the features of EBVaGC is the wt-p534. Furthermore, we identified the role of p53, an important downstream transcription factor of the MAPK signaling pathways, in EBV lytic replication. We found that transfection with the plasmid expressing wt-p53 significantly upregulated EBV lytic proteins Zta and Ea-D and increased EBV DNA copy numbers in AGS-EBV cells (Fig. 3A). When wt-p53 was knocked down by siRNA in AGS-EBV cells, the levels of EBV lytic proteins and EBV DNA copy numbers both decreased (Fig. 3B). To investigate whether p53 is involved in LMP1-induced EBV lytic replication, the cells were cotransfected with pSG5-LMP1/pSG5 and p53 siRNA/control siRNA. The results showed that p53 knockdown significantly inhibited LMP1-induced EBV lytic protein expression and EBV DNA replication (Fig. 3C). Next, we tested the effect of p53 on the activity of BZLF1 promoter (Zp). The p53-null H1299 cells were cotransfected with a Zp/dual-luciferase reporter system and the plasmid expressing wt-p53. The results showed that p53 activated Zp in a dose-dependent manner (Fig. 3D). A ChIP assay showed that p53 could directly bind to the nucleotide (nt) −221 to +12 region of Zp (Fig. 3E), which harbors the necessary cis-elements of Zp for maintaining low basal activity and activation by lytic cycle-inducing agents50,51 in AGS-EBV cells.
Figure 3.
Wild-type p53 (wt-p53) promotes EBV lytic replication by directly binding to and activating Zp in AGS-EBV cells. (A) AGS-EBV cells were transfected with a plasmid containing pEGFP-C3-p53 or a control pEGFP-C3 vector for 48 h. Western blotting analysis was performed to determine EGFP-p53, endogenous p53 (Endo p53), and EBV lytic proteins Zta and Ea-D, and EBV genome copies were analyzed using qPCR. (B) The cells were transfected with siRNA against p53 or control siRNA (ctrl siRNA) for 48 h. Western blotting analysis was performed to determine EGFP-p53, endo p53, and EBV lytic proteins, and EBV genome copies were analyzed using qPCR. (C) AGS-EBV cells were cotransfected with plasmid expressing LMP1 and p53 siRNA for 48 h. pSG5 empty vector and control siRNA were added as controls. Western blotting analysis for LMP1, p53, and lytic proteins and qPCR analysis for EBV genomic copy numbers were performed. β-Actin was used as an internal control. For each experiment, EBV DNA copy number levels from control group were set to 1. (D) The effect of p53 on BZLF1 promoter (Zp) activity in H1299 cells. After cotransfection with Zp-luc and pEGFP-C3-53 for 48 h, firefly luciferase activity reflecting Zp activity was measured and normalized to Renilla luciferase activity. (E) The interaction of p53 with Zp was analyzed using ChIP-PCR. P21 promoter was used as a positive control. *p < 0.05 and **p < 0.01 versus control group.
Our previous data have shown that LMP1 induces phosphorylation of p53 through MAPKs in NPC cells19. Here, the results showed that overexpression of LMP1 increased the levels of total p53 and phosphorylated p53 at Ser15, Ser392, and Ser20 in AGS-EBV cells (Fig. 4A). Treatment with PD98059 and SB202190 reduced the levels of phosphorylated p53 at Ser15 and Ser392, respectively, followed by downregulation of EBV lytic proteins, whereas no significant changes in the levels of total p53 were detected (Fig. 4B and C). The levels of total p53, phosphorylated p53 at Ser20, and EBV lytic proteins decreased after inactivation of JNKs by SP600125 (Fig. 4D). Moreover, we demonstrated that LMP1 overexpression increased the levels of Sp1 or SMAD2 precipitated by p53 (Fig. 4E). The results suggest that LMP1-induced phosphorylation of wt-p53 through MAPKs might enhance the ability of p53 to promote EBV lytic replication.
Figure 4.
LMP1 enhances the ability of wt-p53 to induce expression of EBV lytic proteins through ERKs, p38, and JNKs in AGSEBV cells. (A) AGS-EBV cells were transfected with a plasmid containing pSG5-LMP1 or a control pSG5 vector for 48 h. Western blotting analysis was performed to detect the levels of LMP1, total MAPKs and p53, and phosphorylated MAPKs and p53. (B–D) Western blotting analysis for EBV lytic proteins and phosphorylated p53 in AGS-EBV cells pretreated with PD98059, SB202190, or Sp600125 for 24 h at the concentrations indicated. (E) AGS-EBV cells were transfected with plasmid expressing LMP1 or empty vector for 48 h. Coimmunoprecipitation (Co-IP) assay using anti-p53 antibody was performed to analyze the interactions of p53 with Sp1 and SMAD2/3. The resulting immunocomplexes were recovered and analyzed by Western blotting analysis. β-Actin was used as an internal control.
Phosphorylation of c-Jun Mediated by JNKs Is Involved in EBV Lytic Replication Induced by LMP1 in p53 Mutant Cells
In B95.8 cells, transfection with plasmid expressing wt-p53 significantly upregulated EBV lytic proteins Zta and Ea-D and increased the numbers of EBV DNA copy (Fig. 5A). However, p53 knockdown did not change the levels of EBV lytic proteins and EBV DNA copy numbers (Fig. 5B). Notably, p53 gene mutations mainly in exons 5 through 8 have been previously reported in various kinds of malignant lymphomas such as BL52, non-Hodgkin’s lymphoma (NHL)53,54, diffuse large B-cell lymphoma (DLBCL)55, and B95.856–58. Therefore, there must be other LMP1-mediated MAPK signal axes involved in EBV lytic replication in p53 mutant cell lines. c-Jun is a component of the transcription factor activator protein-1 (AP-1) complex, which binds ZII motif of Zp for activation of the promoter59,60, and its transcriptional activity is regulated by phosphorylation at Ser63 and Ser73 through JNKs61. Our results further showed that LMP1 overexpression increased the levels of phosphorylated JNKs and phosphorylated c-Jun at these two sites (Fig. 5C). Blocking JNKs by SP600125 inhibited phosphorylation of c-Jun at Ser63 and Ser73, followed by downregulation of Zta and Ea-D (Fig. 5D). These results suggest that activation of c-Jun induced by LMP1 through JNKs is involved in EBV lytic replication in the cell line that has p53 mutation.
Figure 5.
Phosphorylation of c-Jun mediated by JNKs is involved in LMP1-induced EBV lytic replication in p53 mutant B95.8 cells. (A) AGS-EBV cells were transfected with a plasmid containing pEGFP-C3-p53 or a control pEGFP-C3 vector for 48 h. Western blotting analysis was performed to determine EGFP-p53, endogenous p53 (endo p53), and EBV lytic proteins Zta and Ea-D, and EBV genome copies were analyzed using qPCR. *p < 0.05 versus negative control. (B) The cells were transfected with siRNA against p53 or ctrl siRNA for 48 h. The levels of EGFP-p53, endo p53, and EBV lytic proteins were detected by Western blotting analysis, and EBV genome copies were analyzed using qPCR. For each experiment, EBV DNA copy number levels from control group were set to 1. (C) Western blotting analysis for LMP1, total c-Jun and JNKs, and phosphorylated c-Jun and JNKs in B95.8 cells transfected with pSG5-LMP1 or pSG5 vector. (D) B95.8 cells were pretreated with Sp600125 at the concentrations indicated for 24 h. Western blotting analysis for c-Jun, phosphorylated c-Jun, and EBV lytic proteins was performed. β-Actin was used as an internal control.
EGCG Directly Targeting LMP1 by Hydrophobic Interaction
Our data demonstrated that LMP1 promoted EBV lytic replication through differential regulation of the MAPK signal axes, and ECGC is commonly used to modulate MAPK signaling, so we tested the effect of EGCG on LMP1 during lytic replication.
Fluorescence Quenching of LMP1 by EGCG
LMP1 is an EBV-encoded membrane protein with molecular weight 66 kDa. We speculate that EGCG might inhibit EBV lytic replication by targeting the LMP1-mediated signaling axes. First, we identified the interaction between EGCG and LMP1 using fluorescence quenching. As shown in Figure 6A, LMP1 had a strong fluorescence emission peak at about 340 nm, and the fluorescence intensity of LMP1 decreased gradually with the addition of various concentrations of EGCG, which indicates that EGCG interacts with LMP1 and thus causes the quenching of LMP1 endogenous fluorescence.
Figure 6.
(−)-Epigallocatechin-3-gallate (EGCG) directly targets LMP1 by hydrophobic effect. (A) Effect of EGCG on fluorescence quenching of purified LMP1 at 37°C. The concentrations of EGCG range from 0 to 0.5 μM in approximately 0.05-μM steps. (B) Stern–Volmer plots for fluorescence quenching of purified LMP1 caused by EGCG at 25°C and 37°C. (C) The plot of lg(F 0 − F)/F as a function of lg[Q] for the determination of the number of bound EGCG molecules (n) per LMP1. (D) EGCG binding to LMP1 analyzed by isothermal titration calorimetry (ITC). (E) Western blotting analysis of whole-cell lysates following purification by a CNBR-activated Sepharose 4B and EGCG-CNBR-activated Sepharose 4B. An EGCG-CNBR-activated Sepharose 4B or CNBR-activated Sepharose 4B affinity column was each incubated with whole-cell lysates from AGS-EBV and B95.8 cells. After washing, the LMP1/EGCG complex was confirmed by Western blotting. (F) EGCG inhibits the proliferation of LMP1-positive cancer cells. CNE1/CNE1-LMP1 and HNE2/HNE2-LMP1 cells were treated with 1 μM EGCG for 4 days, respectively. The results are shown as the relative cell number to untreated control, and the data presented are means ± standard deviation (SD) (n = 3) (Student’s t-test, *p < 0.05, **p < 0.01).
In order to determine the quenching mechanism of EGCG on LMP1, the Stern–Volmer equation (F 0/F = 1 + K SV[Q] = 1 + k q τ 0[Q]) was used to analyze the fluorescence quenching values, where F 0 and F are the fluorescence intensities of LMP1 in the absence and presence of EGCG, K q is the quenching rate constant of the biomolecule, τ0 is the average lifetime of the biomolecule in the absence of quencher, which equals 10−8 s 62, K SV is the Stern–Volmer quenching constant, and [Q] is the concentration of EGCG. As can be seen in Figure 6B and Table 1, the plots of F 0/F versus [Q] at 25°C and 37°C both show good linear relationship, the values of K SV and K q decrease with the increasing of temperature, and the values of K q are much greater than 1010 L mol−1 s−1 63, suggesting that the possible quenching mechanism of EGCG interaction with LMP1 originated from static quenching rather than dynamic collision64.
Table 1.
Stern–Volmer Quenching Constants (K SV) and Bimolecular Quenching Rate Constant (K q) for the Binding of EGCG to LMP1 at Two Different Temperatures
| T | KSV/(L mol−1 S−1) | Kq/(L mol−1 S−1) | R 2 |
|---|---|---|---|
| 25°C | 2.570 × 106 | 2.570 × 1014 | 0.9208 |
| 37°C | 2.093 × 106 | 2.093 × 1014 | 0.91 |
Determination of the Number of Binding Sites and Thermodynamic Parameters
For static quenching, fluorescence quenching intensity and quenching agent concentration follow double logarithmic equation (lg[(F 0 − F)/F] = lgK A + nlg[Q])65, where K A value represents the binding constant, and n represents the number of bonding bits in each fluorescent substance. As can be seen in Figure 6C and Table 2, the values of K A decrease with the increase in temperature, indicating that the protein structure can accommodate better the ligand at 25°C than at 37°C. The number of binding sites (n) obtained is less than 2, suggesting that there is only a single high-affinity binding site available for the interaction between EGCG and LMP1. The thermodynamic parameters of binding, including Gibbs free energy change (ΔG), enthalpy change (ΔH), and entropy change (ΔS), are used as evidence for the nature of the acting forces66,67. When ΔH does not fluctuate considerably in the temperature range considered, both ΔH and ΔS can be obtained from the Van’t Hoff equation (lnK = -ΔH/RT + ΔS/R). ΔG could be calculated from the thermodynamic equation (ΔG = -RTlnK = ΔH − TΔS)68, where K is the binding constant at a certain temperature, R is the gas constant (8.314 J mol−1 K−1), and T is the temperature (K). As seen in Table 2, the negative value for ΔG is indicative of the binding process for the EGCG–LMP1 complex, which is spontaneous. According to the theoretical analysis of Ross and Subramanian69, positive values for ΔH and ΔS suggest that hydrophobic interaction is the main binding force in the EGCG–LMP1 complex.
Table 2.
Modified Stern–Volmer Association Constant (K A) for EGCG–LMP1 at Two Different Temperatures and Calculated ΔH, ΔS, and ΔG as Thermodynamic Parameters for the Binding of EGCG to LMP1
| T | KA/L mol−1 | n | R 2 | ΔH/kJ mol−1 | ΔG/kJ mol−1 | ΔS/J mol−1 K−1 |
|---|---|---|---|---|---|---|
| 298.15 K | 1.7989 × 1012 | 1.8249 | 0.9728 | 408.3 | −69.9478 | 1603.9 |
| 310.15 K | 3.0839 × 109 | 1.4472 | 0.9658 | −56.3407 |
EGCG Physically Binding to LMP1
To further investigate the interaction between EGCG and LMP1, we performed ITC, which confirmed the binding site number (N, 1.05) and revealed the dissociation constant (K d, 0.36 μM) in the low micromolar (Fig. 6D). In agreement with the fluorescence quenching binding results, the positive values achieved for ΔH (48.57 kJ/mol) and ΔS (286.3 J mol−1 K−1) show that the interaction between EGCG and LMP1 is predominantly due to hydrophobic interaction69. Next, we confirmed the interaction between EGCG and LMP1 using CNBR-activated Sepharose 4B pull-down affinity chromatography in combination with Western blotting. The results showed that LMP1 in whole-cell lysates from AGS-EBV and B95.8 cells could be specifically pulled down by EGCG-CNBR-activated Sepharose 4B (Fig. 6E). These results suggest that EGCG directly binds to LMP1 with a strong affinity (K d, 0.36 μM); the binding site number (n) is 1, and this binding occurs mostly through hydrophobic interaction. Interestingly, we found that there was a significant inhibition of proliferation without inducing cell death in the LMP1-positive NPC cells upon treatment with EGCG at the low concentration of 1 μM, a physiologically relevant concentration70 (Fig. 6F), indicating that this binding might confer EGCG responsiveness to the proliferation of cancer cells.
EGCG Inhibits EBV Lytic Replication by Targeting LMP1 and the Downstream MAPK Signaling Axes
After identifying that EGCG directly targets LMP1, our data further showed that EGCG could reduce the levels of LMP1 and p53 and inhibit the phosphorylation of ERKs, p38, JNKs, and p53 at ser392, ser20, and ser15 in a dose-dependent manner in AGS-EBV cells (Fig. 7A). Also, EGCG downregulated LMP1 and inhibited the phosphorylation of JNKs and c-Jun at ser63 and ser73 in B95.8 cells (Fig. 7B). We further demonstrated that EGCG significantly inhibited the transcription of lytic genes BZLF1 and BMRF1 and thus the expression of Zta and Ea-D (Fig. 7C and D). Flow cytometry analysis showed that EGCG treatment decreased Zta-positive rates from 16.24% to 10.93% and Ea-D-positive rates from 34.80% to 15.18% in AGS-EBV cells, and decreased Zta-positive rates from 17.51% to 5.79% and Ea-D-positive rates from 27.67% to 14.77% in B95.8 cells (Fig. 7E). Additionally, EBV genome copy numbers reduced markedly after EGCG treatment in both cells (Fig. 7F). These findings indicate that EGCG can directly target LMP1 and inhibit EBV lytic replication via blocking the LMP1/MAPKs/wt-p53 signal axis in AGS-EBV cells and LMP1/JNKs/c-Jun axis in p53 mutant B95.8 cells, respectively.
Figure 7.
EGCG inhibits EBV lytic replication by suppressing LMP1-mediated MAPK signal axes in AGS-EBV and B95.8 cells. AGS-EBV and B95.8 cells were starved in 0.1% FBS HAM’s/F-12 or RPMI-1640 medium for 24 h and then treated with different concentrations of EGCG as indicated or vehicle [dimethyl sulfoxide (DMSO)] for 24 h. (A, B) Western blotting analysis was performed to determine LMP1, total and phosphorylated ERKs, p38, and JNKs, total and phosphorylated p53, and total and phosphorylated c-Jun. (C) Effects of EGCG on the expression of Zta and Ea-D. β-Actin was used as an internal control. (D) Reverse transcription (RT)-qPCR quantification of mRNA expression levels for BZLF1 and BMRF1 genes. (E) Flow cytometry was performed to analyze the effects of EGCG on the numbers of Zta- or Ea-D-positive cells. Incubation with vehicle but no primary antibody followed by incubation with the secondary antibody was used as a negative control. (F) The changes in EBV genome copies were analyzed using qPCR. The levels of EBV DNA copy number from the control group were set to 1. *p < 0.05, **p < 0.01, ***p < 0.001 versus control group.
DISCUSSION
The association of varying degrees of EBV lytic replication with EBV-associated malignancies such as carcinomas and lymphomas is based on the enrichment of viral strains with enhanced lytic replication with the related tumors, detection of serum viral loads in affected patients, and decreased tumorigenesis of certain lymphomas upon abortive EBV lytic infection of preclinical in vivo models17. As one subtype of molecular classifications of gastric carcinoma4, several lytic EBV proteins expressed in EBVaGC have functions that contribute to tumorigenesis8,71. There is growing evidence that the biological significance of the viral oncoprotein LMP1 expression in the viral lytic stage is related to EBV lytic replication24,25,29. Here, we first demonstrated that LMP1 promotes EBV lytic replication via the MAPK signal axes. In AGS-EBV cells, LMP1 enhances the ability of wt-p53 to activate Zp through MAPKs, including ERKs, p38, and JNKs (Fig. 4). As known, p53 gene mutations have been reported in various kinds of malignant lymphomas. We further explored other LMP1-mediated MAPK signal axes involved in EBV lytic replication and found that phosphorylation of c-Jun by LMP1 through JNKs is involved in EBV lytic replication in p53 mutant B95.8 cells (Fig. 5).
The major polyphenol of green tea, EGCG, has been shown to prevent carcinogenesis by interacting with and binding numerous membrane proteins that affect cell growth and proliferation40,41, including fibronectin, Fas, 67-LR, vimentin, and IGF-IR. The EBV-encoded oncoprotein LMP1 is composed of a short cytoplasmic amino-terminal domain, six hydrophobic transmembrane domains, and a cytoplasmic carboxy-terminal domain72. Using fluorescence quenching, ITC assay, and CNBR-activated Sepharose 4B pull-down affinity chromatography, we first observed that EGCG directly binds to EBV-encoded membrane protein LMP1 (K d, 0.36 μM, n = 1), and the binding process is predominantly due to hydrophobic interaction (Fig. 6). It is interesting that expression of LMP1 increases the responsiveness of epithelial cancer cells to EGCG at physiological concentration (Fig. 6F), implying that this binding seems to regulate the biological functions of LMP1 that have obvious implications for cell proliferation and growth. Therefore, it is reasonable to assume that the hydrophobic benzene ring of EGCG might interact with the hydrophobic transmembrane domains of LMP1, thereby changing the excitation of the receptor signals. In addition, we speculated that EGCG inhibiting the signaling pathways through LMP1 might relate to other beneficial effects, including the inhibition of EBV lytic replication.
Recently, inhibiting lytic replication by EGCG has been developed as an effective strategy for the prevention and treatment of oncovirus-associated malignancies. Also, anticancer polyphenol EGCG has a broad spectrum of antiviral effects73,74 and can effectively inhibit EBV lytic replication38,39. Our previous studies have demonstrated that EGCG inhibits EBV spontaneous lytic replication by suppressing the activation of MEK/ERK1/2 signaling pathways or downregulation of LMP137,38. To this end, we further found that EGCG downregulates LMP1 and thus blocks the MAPKs/wt-p53 signal axis in AGS-EBV cells and JNKs/c-Jun signal axis in p53 mutant B95.8 cells, respectively, followed by decreased expression of EBV lytic proteins and reduced levels of EBV DNA copy numbers (Fig. 7).
Finally, we clarify molecular mechanisms of LMP1 promoting EBV lytic replication and reveal that EGCG inhibits EBV lytic replication by directly targeting LMP1-mediated MAPK signal axes (Fig. 8), raising the possibility that EGCG can be developed as an agent to prevent tumorigenesis of EBV-associated malignancies.
Figure 8.
Schematic illustrating EGCG inhibition of EBV lytic replication by directly targeting LMP1-mediated MAPK signal axes. LMP1 recruits TRAFs to constitutively activate MAPKs, including ERKs, p38, and JNKs, promoting EBV lytic replication. LMP1-induced phosphorylation of wt-p53 at Ser15, Ser20, and Ser392 through MAPKs can enhance the ability of p53 to activate Zp by directly binding to Zp as well as recruiting transactivators Sp1 and SMAD2. When p53 is in a mutated state, phosphorylation of c-Jun at Ser63 and Ser73 induced by LMP1 through JNKs is involved in regulating Zta expression. Polyphenol compound EGCG can directly target LMP1 to block the MAPKs/wt-p53 and JNKs/c-Jun signal axes, effectively inhibiting EBV lytic replication.
ACKNOWLEDGMENTS
We thank Prof. Qiao Wu for giving p53 expression plasmid as a gift, Prof. Wenhai Feng for giving the AGS-EBV cells, and Prof. Chung S. Yang for valuable suggestions. This study was supported by the National Natural Science Foundation of China (81430064, 81602402, 81874172, and 81874195) and the National Natural Science Foundation of China/National Institutes of Health (81110346).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1.Young LS, Yap LF, Murray PG. Epstein–Barr virus: More than 50 years old and still providing surprises. Nat Rev Cancer 2016;16(12):789–802. [DOI] [PubMed] [Google Scholar]
- 2.Lieberman PM. Epstein–Barr virus turns 50. Science 2014;343(6177):1323–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi SJ, Ryu E, Lee S, Huh S, Shin YS, Kang BW, Kim JG, Cho H, Kang H. Adenosine induces EBV lytic reactivation through ADORA1 in EBV-associated gastric carcinoma. Int J Mol Sci. 2019;20(6):1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513(7517):202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khan G, Hashim MJ. Global burden of deaths from Epstein–Barr virus attributable malignancies 1990–2010. Infect Agent Cancer 2014;9(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacome AA, Lima EM, Kazzi AI, Chaves GF, Mendonca DC, Maciel MM, Santos JS. Epstein–Barr virus-positive gastric cancer: A distinct molecular subtype of the disease? Rev Soc Bras Med Trop. 2016;49(2):150–7. [DOI] [PubMed] [Google Scholar]
- 7.Petosa C, Morand P, Baudin F, Moulin M, Artero JB, Muller CW. Structural basis of lytic cycle activation by the Epstein–Barr virus ZEBRA protein. Mol Cell 2006;21(4):565–72. [DOI] [PubMed] [Google Scholar]
- 8.Kalla M, Schmeinck A, Bergbauer M, Pich D, Hammerschmidt W. AP-1 homolog BZLF1 of Epstein–Barr virus has two essential functions dependent on the epigenetic state of the viral genome. Proc Natl Acad Sci USA 2010;107(2):850–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rennekamp AJ, Wang P, Lieberman PM. Evidence for DNA hairpin recognition by Zta at the Epstein–Barr virus origin of lytic replication. J Virol. 2010;84(14):7073–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flemington E, Speck SH. Autoregulation of Epstein–Barr virus putative lytic switch gene BZLF1. J Virol. 1990;64(3):1227–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Packham G, Economou A, Rooney CM, Rowe DT, Farrell PJ. Structure and function of the Epstein–Barr virus BZLF1 protein. J Virol. 1990;64(5):2110–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feederle R, Kost M, Baumann M, Janz A, Drouet E, Hammerschmidt W, Delecluse HJ. The Epstein–Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 2000;19(12):3080–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fiorina L, Ricotti M, Vanoli A, Luinetti O, Dallera E, Riboni R, Paolucci S, Brugnatelli S, Paulli M, Pedrazzoli P, Baldanti F, Perfetti V. Systematic analysis of human oncogenic viruses in colon cancer revealed EBV latency in lymphoid infiltrates. Infect Agent Cancer 2014;9:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ko YH. EBV and human cancer. Exp Mol Med. 2015;47:e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Hu J, Luo X, Bode AM, Dong Z, Cao Y. Therapies based on targeting Epstein–Barr virus lytic replication for EBV-associated malignancies. Cancer Sci. 2018;109(7):2101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Liu S, Hu J, Luo X, Li N, A MB, Cao Y. Epstein–Barr virus lytic reactivation regulation and its pathogenic role in carcinogenesis. Int J Biol Sci. 2016;12(11):1309–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munz C. Latency and lytic replication in Epstein–Barr virus-associated oncogenesis. Nat Rev Microbiol. 2019;17(11):691–700. [DOI] [PubMed] [Google Scholar]
- 18.Jia WH, Qin HD. Non-viral environmental risk factors for nasopharyngeal carcinoma: A systematic review. Semin Cancer Biol. 2012;22(2):117–26. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Guo L, Tao Y, Zhou S, Wang Z, Luo W, Hu D, Li Z, Xiao L, Tang M, Yi W, Tsao S, Cao Y. Latent membrane protein 1 of Epstein–Barr virus regulates p53 phosphorylation through MAP kinases. Cancer Lett. 2007;255(2):219–31. [DOI] [PubMed] [Google Scholar]
- 20.Ma X, Yang L, Xiao L, Tang M, Liu L, Li Z, Deng M, Sun L, Cao Y. Down-regulation of EBV-LMP1 radio-sensitizes nasal pharyngeal carcinoma cells via NF-kappaB regulated ATM expression. PLoS One 2011;6(11):e24647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan G, Luo W, Lu Z, Luo X, Li L, Liu S, Liu Y, Tang M, Dong Z, Cao Y. Epstein–Barr virus latent membrane protein 1 mediates phosphorylation and nuclear translocation of annexin A2 by activating PKC pathway. Cell Signal. 2007;19(2):341–8. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, Luo F, Li L, Yang L, Hu D, Ma X, Lu Z, Sun L, Cao Y. STAT3 activation induced by Epstein–Barr virus latent membrane protein1 causes vascular endothelial growth factor expression and cellular invasiveness via JAK3 And ERK signaling. Eur J Cancer 2010;46(16):2996–3006. [DOI] [PubMed] [Google Scholar]
- 23.Xiao L, Hu ZY, Dong X, Tan Z, Li W, Tang M, Chen L, Yang L, Tao Y, Jiang Y, Li J, Yi B, Li B, Fan S, You S, Deng X, Hu F, Feng L, Bode AM, Dong Z, Sun L, Cao Y. Targeting Epstein–Barr virus oncoprotein LMP1-mediated glycolysis sensitizes nasopharyngeal carcinoma to radiation therapy. Oncogene 2014;33(37):4568–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahsan N, Kanda T, Nagashima K, Takada K. Epstein–Barr virus transforming protein LMP1 plays a critical role in virus production. J Virol. 2005;79(7):4415–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nawandar DM, Ohashi M, Djavadian R, Barlow E, Makielski K, Ali A, Lee D, Lambert PF, Johannsen E, Kenney SC. Differentiation-dependent LMP1 expression is required for efficient lytic Epstein–Barr virus reactivation in epithelial cells. J Virol. 2017;91(8): e02438–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai TT, Ye SB, Liu YN, He J, Chen QY, Mai HQ, Zhang CX, Cui J, Zhang XS, Busson P, Zeng Y, Li J. LMP1-mediated glycolysis induces myeloid-derived suppressor cell expansion in nasopharyngeal carcinoma. PLoS Pathog. 2017;13(7):e1006503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun J, Hu C, Zhu Y, Sun R, Fang Y, Fan Y, Xu F. LMP1 increases expression of NADPH oxidase (NOX) and its regulatory subunit p22 in NP69 nasopharyngeal cells and makes them sensitive to a treatment by a NOX inhibitor. PLoS One 2015;10(8):e0134896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Termini JM, Gupta S, Raffa FN, Guirado E, Fischl MA, Niu L, Kanagavelu S, Stone GW. Epstein Barr virus Latent Membrane Protein-1 enhances dendritic cell therapy lymph node migration, activation, and IL-12 secretion. PLoS One 2017;12(9):e0184915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu J, Li Y, Li H, Shi F, Xie L, Zhao L, Tang M, Luo X, Jia W, Fan J, Zhou J, Gao Q, Qiu S, Wu W, Zhang X, Liao W, Bode AM, Cao Y. Targeting Epstein–Barr virus oncoprotein LMP1-mediated high oxidative stress suppresses EBV lytic reactivation and sensitizes tumors to radiation therapy. Theranostics 2020;10(26):11921–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang SS, Lo YC, Chua HH, Chiu HY, Tsai SC, Chen JY, Lo KW, Tsai CH. Critical role of p53 in histone deacetylase inhibitor-induced Epstein–Barr virus Zta expression. J Virol. 2008;82(15):7745–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chua HH, Chiu HY, Lin SJ, Weng PL, Lin JH, Wu SW, Tsai SC, Tsai CH. p53 and Sp1 cooperate to regulate the expression of Epstein–Barr viral Zta protein. J Med Virol. 2012;84(8):1279–88. [DOI] [PubMed] [Google Scholar]
- 32.Niedzwiecki A, Roomi MW, Kalinovsky T, Rath M. Anticancer efficacy of polyphenols and their combinations. Nutrients 2016;8(9):552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Negri A, Naponelli V, Rizzi F, Bettuzzi S. Molecular targets of epigallocatechin-gallate (EGCG): A special focus on signal transduction and cancer. Nutrients 2018;10(12):1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng C, Li Z, Zhao X, Liao C, Quan J, Bode AM, Cao Y, Luo X. Natural alkaloid and polyphenol compounds targeting lipid metabolism: Treatment implications in metabolic diseases. Eur J Pharmacol. 2020;870:172922. [DOI] [PubMed] [Google Scholar]
- 35.Cheng C, Zhuo S, Zhang B, Zhao X, Liu Y, Liao C, Quan J, Li Z, Bode AM, Cao Y, Luo X. Treatment implications of natural compounds targeting lipid metabolism in nonalcoholic fatty liver disease, obesity and cancer. Int J Biol Sci. 2019;15(8):1654–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu J, Xu Z, Zheng W. A review of the antiviral role of green tea catechins. Molecules 2017;22(8):1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu S, Li H, Tang M, Cao Y. (−)-Epigallocatechin-3-gallate inhibition of Epstein–Barr virus spontaneous lytic infection involves downregulation of latent membrane protein 1. Exp Ther Med. 2018;15(1):1105–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu S, Li H, Chen L, Yang L, Li L, Tao Y, Li W, Li Z, Liu H, Tang M, Boda AM, Dong Z, Cao Y. (−)-Epigallocatechin-3-gallate inhibition of Epstein–Barr virus spontaneous lytic infection involves ERK1/2 and PI3-K/Akt signaling in EBV-positive cells. Carcinogenesis 2013;34(3):627–37. [DOI] [PubMed] [Google Scholar]
- 39.Chang LK, Wei TT, Chiu YF, Tung CP, Chuang JY, Hung SK, Li C, Liu ST. Inhibition of Epstein–Barr virus lytic cycle by (−)-epigallocatechin gallate. Biochem Biophys Res Commun. 2003;301(4):1062–8. [DOI] [PubMed] [Google Scholar]
- 40.Bode AM, Dong Z. Epigallocatechin 3-gallate and green tea catechins: United they work, divided they fail. Cancer Prev Res. (Phila) 2009;2(6):514–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang CS, Wang H. Cancer therapy combination: Green tea and a phosphodiesterase 5 inhibitor? J Clin Invest. 2013;123(2):556–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimizu M, Shirakami Y, Moriwaki H. Targeting receptor tyrosine kinases for chemoprevention by green tea catechin, EGCG. Int J Mol Sci. 2008;9(6):1034–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hui KF, Chiang AK. Suberoylanilide hydroxamic acid induces viral lytic cycle in Epstein–Barr virus-positive epithelial malignancies and mediates enhanced cell death. Int J Cancer 2010;126(10):2479–89. [DOI] [PubMed] [Google Scholar]
- 44.Lo AK, Lo KW, Tsao SW, Wong HL, Hui JW, To KF, Hayward DS, Chui YL, Lau YL, Takada K, Huang DP. Epstein–Barr virus infection alters cellular signal cascades in human nasopharyngeal epithelial cells. Neoplasia 2006;8(3):173–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie L, Shi F, Li Y, Li W, Yu X, Zhao L, Zhou M, Hu J, Luo X, Tang M, Fan J, Zhou J, Gao Q, Wu W, Zhang X, Liao W, Bode AM, Cao Y. Drp1-dependent remodeling of mitochondrial morphology triggered by EBV-LMP1 increases cisplatin resistance. Signal Transduct Target Ther. 2020;5(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen ZP, Shemshedini L, Durand B, Noy N, Chambon P, Gronemeyer H. Pure and functionally homogeneous recombinant retinoid X receptor. J Biol Chem. 1994;269(41):25770–6. [PubMed] [Google Scholar]
- 47.Luo X, Yang L, Xiao L, Xia X, Dong X, Zhong J, Liu Y, Li N, Chen L, Li H, Li W, Liu W, Yu X, Chen H, Tang M, Weng X, Yi W, Bode AM, Dong Z, Liu J, Cao Y. Grifolin directly targets ERK1/2 to epigenetically suppress cancer cell metastasis. Oncotarget 2015;6(40):42704–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morris MA, Laverick L, Wei W, Davis AM, O’Neill S, Wood L, Wright J, Dawson CW, Young LS. The EBV-encoded oncoprotein, LMP1, induces an epithelial-to-mesenchymal transition (EMT) via its CTAR1 domain through integrin-mediated ERK-MAPK signalling. Cancers (Basel) 2018;10(5):130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kieser A, Sterz KR. The latent membrane protein 1 (LMP1). Curr Top Microbiol Immunol. 2015;391:119–49. [DOI] [PubMed] [Google Scholar]
- 50.Flemington E, Speck SH. Identification of phorbol ester response elements in the promoter of Epstein–Barr virus putative lytic switch gene BZLF1. J Virol. 1990;64(3):1217–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimizu N, Takada K. Analysis of the BZLF1 promoter of Epstein–Barr virus: Identification of an anti-immunoglobulin response sequence. J Virol. 1993;67(6):3240–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhatia KG, Gutierrez MI, Huppi K, Siwarski D, Magrath IT. The pattern of p53 mutations in Burkitt’s lymphoma differs from that of solid tumors. Cancer Res. 1992;52(15):4273–6. [PubMed] [Google Scholar]
- 53.Adamson DJ, Thompson WD, Dawson AA, Bennett B, Haites NE. p53 mutation and expression in lymphoma. Br J Cancer 1995;72(1):150–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen PM, Chiou TJ, Hsieh RK, Fan FS, Chu CJ, Lin CZ, Chiang H, Yen CC, Wang WS, Liu JH. p53 gene mutations and rearrangements in non-Hodgkin’s lymphoma. Cancer 1999;85(3):718–24. [DOI] [PubMed] [Google Scholar]
- 55.Mitani S, Kamata H, Fujiwara M, Aoki N, Okada S, Watanabe M, Tango T, Mori S. Missense mutation with/without nonsense mutation of the p53 gene is associated with large cell morphology in human malignant lymphoma. Pathol Int. 2007;57(7):430–6. [DOI] [PubMed] [Google Scholar]
- 56.Forte E, Luftig MA. MDM2-dependent inhibition of p53 is required for Epstein–Barr virus B-cell growth transformation and infected-cell survival. J Virol. 2009;83(6):2491–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li L, Zhou S, Chen X, Guo L, Li Z, Hu D, Luo X, Ma X, Tang M, Yi W, Tsao S, Cao Y. The activation of p53 mediated by Epstein–Barr virus latent membrane protein 1 in SV40 large T-antigen transformed cells. FEBS Lett. 2008;582(5):755–62. [DOI] [PubMed] [Google Scholar]
- 58.Farrell PJ, Allan GJ, Shanahan F, Vousden KH, Crook T. p53 is frequently mutated in Burkitt’s lymphoma cell lines. EMBO J. 1991;10(10):2879–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kenney SC, Mertz JE. Regulation of the latent-lytic switch in Epstein–Barr virus. Semin Cancer Biol. 2014;26:60–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murata T. Regulation of Epstein–Barr virus reactivation from latency. Microbiol Immunol. 2014;58(6):307–17. [DOI] [PubMed] [Google Scholar]
- 61.Morton S, Davis RJ, McLaren A, Cohen P. A reinvestigation of the multisite phosphorylation of the transcription factor c-Jun. EMBO J. 2003;22(15):3876–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dewey TG. Biophysical and biochemical aspects of fluorescence spectroscopy. New York: Plenum Press; 1991. p. xvii, 294. [Google Scholar]
- 63.Lakowicz JR, Weber G. Quenching of fluorescence by oxygen. A probe for structural fluctuations in macromolecules. Biochemistry 1973;12(21):4161–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shaikh SM, Seetharamappa J, Kandagal PB, Manjunatha DH. In vitro study on the binding of anti-coagulant vitamin to bovine serum albumin and the influence of toxic ions and common ions on binding. Int J Biol Macromol. 2007;41(1):81–6. [DOI] [PubMed] [Google Scholar]
- 65.Belatik A, Hotchandani S, Bariyanga J, Tajmir-Riahi HA. Binding sites of retinol and retinoic acid with serum albumins. Eur J Med Chem. 2012;48:114–23. [DOI] [PubMed] [Google Scholar]
- 66.Chaves OA, Jesus CS, Cruz PF, Sant’Anna CM, Brito RM, Serpa C. Evaluation by fluorescence, STD-NMR, docking and semi-empirical calculations of the o-NBA photo-acid interaction with BSA. Spectrochim Acta A Mol Biomol Spectrosc. 2016;169:175–81. [DOI] [PubMed] [Google Scholar]
- 67.Jattinagoudar LN, Nandibewoor ST, Chimatadar SA. Binding of fexofenadine hydrochloride to bovine serum albumin: Structural considerations by spectroscopic techniques and molecular docking. J Biomol Struct Dyn. 2017;35(6):1200–14. [DOI] [PubMed] [Google Scholar]
- 68.Jahanban-Esfahlan A, Panahi-Azar V. Interaction of glutathione with bovine serum albumin: Spectroscopy and molecular docking. Food Chem. 2016;202:426–31. [DOI] [PubMed] [Google Scholar]
- 69.Ross PD, Subramanian S. Thermodynamics of protein association reactions: Forces contributing to stability. Biochemistry 1981;20(11):3096–102. [DOI] [PubMed] [Google Scholar]
- 70.Yang CS, Wang X, Lu G, Picinich SC. Cancer prevention by tea: Animal studies, molecular mechanisms and human relevance. Nat Rev Cancer 2009;9(6):429–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Borozan I, Zapatka M, Frappier L, Ferretti V. Analysis of Epstein–Barr virus genomes and expression profiles in gastric adenocarcinoma. J Virol. 2018;92(2):e01239–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fennewald S, van Santen V, Kieff E. Nucleotide sequence of an mRNA transcribed in latent growth-transforming virus infection indicates that it may encode a membrane protein. J Virol. 1984;51(2):411–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Steinmann J, Buer J, Pietschmann T, Steinmann E. Anti-infective properties of epigallocatechin-3-gallate (EGCG), a component of green tea. Br J Pharmacol. 2013;168(5):1059–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang ZY, Li YQ, Guo ZW, Zhou XH, Lu MD, Xue TC, Gao B. ERK1/2-HNF4alpha axis is involved in epigallocatechin-3-gallate inhibition of HBV replication. Acta Pharmacol Sin. 2020;41(2):278–85. [DOI] [PMC free article] [PubMed] [Google Scholar]