Abstract
Adhesion G protein-coupled receptors (aGPCRs) form a sub-group within the GPCR superfamily. Their distinctive structure contains an abnormally large N-terminal, extracellular region with a GPCR autoproteolysis-inducing (GAIN) domain. In most aGPCRs, the GAIN domain constitutively cleaves the receptor into two fragments. This process is often required for aGPCR signalling. Over the last two decades, much research has focussed on aGPCR-ligand interactions, in an attempt to deorphanize the family. Most ligands have been found to bind to regions N-terminal to the GAIN domain. These receptors may bind a variety of ligands, ranging across membrane-bound proteins and extracellular matrix components. Recent advancements have revealed a conserved method of aGPCR activation involving a tethered ligand within the GAIN domain. Evidence for this comes from increased activity in receptor mutants exposing the tethered ligand. As a result, G protein-coupling partners of aGPCRs have been more extensively characterised, making use of their tethered ligand to create constitutively active mutants. This has led to demonstrations of aGPCR function in, for example, neurodevelopment and tumour growth. However, questions remain around the ligands that may bind many aGPCRs, how this binding is translated into changes in the GAIN domain, and the exact mechanism of aGPCR activation following GAIN domain conformational changes. This review aims to examine the current knowledge around aGPCR activation, including ligand binding sites, the mechanism of GAIN domain-mediated receptor activation and how aGPCR transmembrane domains may relate to activation. Other aspects of aGPCR signalling will be touched upon, such as downstream effectors and physiological roles.
Keywords: adhesion receptors, agonists, G-protein-coupled receptors, G-proteins, signal transduction
Introduction
G protein-coupled receptors (GPCRs) are currently the most successfully targeted superfamily of receptors in modern medicine [1]. GPCRs are classified into five main families; Glutamate, Rhodopsin, Frizzled/Taste, Secretin and importantly for this review, Adhesion [2]. They are responsible for a large variety of cellular responses with a diverse selection of stimuli, resulting in a complex network of interactions between the ligands, the receptors and the signalling cascade. Whilst GPCRs in general are the most targeted receptor superfamily, historically very little pharmaceutical research has been conducted on adhesion GPCRs (aGPCRs). Despite their importance in adhesion, cell migration, paracrine signalling and numerous disease implications [3], aGPCR research has been hampered by the orphan status of many receptors. Nonetheless, aGPCRs provide an intriguing potential alternative drug target compared with many other families, in particular, within oncology and fertility. Several recent reviews have highlighted the emerging role of these receptors in therapeutics therefore it is not the aim of this review to reiterate these points [4,5]. Instead, here we aim to discuss the current understanding of known aGPCR ligands, their activation, structure and function.
aGPCR nomenclature has drastically changed since their discovery
What we know today as aGPCRs were first characterised in leukocytes in the 1980s. They were identified as the glycoproteins targeted by the mouse monoclonal antibody for F4/80, the mouse equivalent of the human GPCR EMR1 [6]. F4/80, EMR1 and CD97 were the first members of the GPCR subfamily originally known as EGF-TM7; named for the appearance of F4/80 as a chimera of 7-transmembrane receptors and epidermal growth factor (EGF) [7,8].
A misleading alternative name for this family was LNB-TM7, denoting its long N-terminal region, but also a close association with Class B1 GPCRs [9]. Early reviews often listed these GPCRs as a subfamily of Class B GPCRs, due to their sequence similarity in the 7 transmembrane helix domain (7TM) [10,11]. However, analysis of the entire GPCR superfamily revealed distinctions between this family and Class B1, in particular in the extracellular domain (ECD). This new family, with 24 members at the time, was named ‘adhesion’, for their apparent role in cell adhesion due to the mucin-like stalks in their N-terminal region [12,13]. Subsequently, all 33 human family members were divided further into nine clusters (Table 1), with each having a relatively high sequence similarity that the family lacks as a whole [14]. The International Union of Basic and Clinical Pharmacology defined the family fully in 2015 [15].
Table 1. A summary of known endogenous ligands, receptor activation mechanisms, G protein couplings and domains contained in the NTF, N-terminal to the GAIN domain, of every human aGPCR.
| Cluster | aGPCR | Determined ligand(s) | Activation mechanism1 | Established G proteins couplings | N-terminal domain(s) | Source |
|---|---|---|---|---|---|---|
| I | ADGRL1 (Latrophilin-1) | Teneurin-2, FLRT1, FLRT3, neurexin-1α, -1β, -2β | Tethered agonist (A)/constitutively active mutants (C) | Gs, Gi | Lectin, olfactomedin, STP, HomR | [77–86] |
| ADGRL2 (Latrophilin-2) | Teneurin-2, FLRT3 | Unknown | Unknown | |||
| ADGRL3 (Latrophilin-3) | Teneurin-3, FLRT1, FLRT3, Unc5D | Tethered agonist (A) | G12, G13 | |||
| ADGRL4 (ELTD1) | - | Unknown but not tethered agonist or constitutive activity | Unknown | Lectin, EGF-Like, 2× Ca2+-binding EGF | ||
| II | ADGRE1 (EMR1) | - | Unknown | Unknown | EGF-Like, 5× Ca2+-binding EGF | [16,40,49,53,76,87–93] |
| ADGRE2 (EMR2) | Chondroitin sulfate B, FHR1 | Unknown/constitutive activity (C) | G16 | EGF-Like, 4× Ca2+-binding EGF | ||
| ADGRE3 (EMR3) | - | Unknown | Unknown | EGF-Like, 1× Ca2+-binding EGF | ||
| ADGRE4 (EMR4) | - | Unknown/not expressed at cell surface | Unknown | |||
| ADGRE5 (CD97) | CD55, chondroitin sulfate B, integrins α5β1 and αvβ3, CD90 | Tethered agonist (A)/constitutive activity (C) | G12, G13, G14, Gz | EGF-Like, 4× Ca2+-binding EGF, RGD motif | ||
| III | ADGRA1 (GPR123) | - | No GAIN domain present therefore not tethered agonist | Unknown | - | [14,22,94–97] |
| ADGRA2 (GPR124) | Integrin αvβ3, glycosaminoglycans, syndecan-1,2 | Unknown | Unknown | LRR, IG, RGD motif, HomR | ||
| ADGRA3 (GPR125) | - | Constitutive activity (C) | Unknown | LRR, IG, HomR | ||
| IV | ADGRC1 (CELSR1) | - | Unknown but not tethered agonist | Unknown | EC, 5× Ca2+-binding EGF, 2× LamG, EGF-Lam, HomR | [39,98–101] |
| ADGRC2 (CELSR2) | - | Tethered agonist (A)/constitutive activity (C) | Potentially Gq | |||
| ADGRC3 (CELSR3) | Dystroglycan | Tethered agonist (A) | Potentially Gq | EC, 5× Ca2+-binding EGF, 2× LamG, 2× EGF-Lam, HomR | ||
| V | ADGRD1 (GPR133) | Plxdc2 | Tethered agonist (A)/constitutive activity (C) | Gs | - | [14,102–104] |
| ADGRD2 (GPR144) | - | Unknown | Unknown | PTX | ||
| VI | ADGRF1 (GPR110) | Synaptamide | Soluble ligand allosteric binding (B) | Gq, Gs | SEA | [14,23,96,105–112] |
| ADGRF2 (GPR111) | - | Unknown but not tethered agonist | Unknown | - | ||
| ADGRF3 (GPR113) | - | Unknown | Unknown | HomR, EGF | ||
| ADGRF4 (GPR115) | - | Unknown but not tethered agonist | Unknown | - | ||
| ADGRF5 (GPR116) | Surfactant protein D | Tethered agonist (A) | Gq, G11 | SEA, 2× IG | ||
| VII | ADGRB1 (BAI1) | Phosphatidylserine, integrin αvβ5, lipopolysaccharide, RTN4R, CD36 | Tethered agonist (A) | G12, G13 | RGD motif, 5× TSR, HomR | [14,31,95,113–123] |
| ADGRB2 (BAI2) | Glutaminase interacting protein | Tethered agonist (A) | Gz, Gi | 4× TSR, HomR | ||
| ADGRB3 (BAI3) | C1ql-1,4 | Unknown | Unknown | CUB, 4× TSR, HomR | ||
| VIII | ADGRG1 (GPR56) | Collagen III, tissue transglutaminase 2, laminin | Tethered agonist (A) | Gi, Gq | - | [46,54,62,67,96,106,124–134,134–138] |
| Progastrin | ||||||
| ADGRG2 (GPR64, HE6) | - | Constitutive activity (C) | Gq, G11 | - | ||
| ADGRG3 (GPR97) | Cortisol* | Soluble ligand (TM binding) (B)/constitutive activity (C) | Gs, Gi, Go | - | ||
| ADGRG4 (GPR112) | - | Unknown | Unknown | PTX, RGD motif | ||
| ADGRG5 (GPR114) | - | Tethered agonist (A)/constitutive activity (C) | Gs | - | ||
| ADGRG6 (GPR126, VIGR, DREG) | Collagen IV, laminin-211 | Tethered agonist (A) | Gs, Gi, Go | CUB, PTX, SEA, HomR | ||
| Cellular prion protein | ||||||
| ADGRG7 (GPR128) | - | Unknown | Unknown | - | ||
| IX | ADGRV1 (GPR98, VLGR1) | - | Tethered agonist (A) | Unknown | 35× CB, PTX, EAR | [42,109] |
Letters in brackets denote the panel from Figure 3 that illustrates the activation mechanism used by each aGPCR;
CB: Calx-beta motif; CUB: Complement C1r/C1s, Uegf, Bmp1; EAR: Epilepsy-associated repeat; EC: Extracellular cadherin domains (9 cadherin repeats); EGF: Epidermal growth factor; FHR: Factor H-related protein; FLRT: fibronectin leucine-rich transmembrane protein; HomR: Hormone receptor; IG: Immunoglobulin; LamG: laminin-G like domain; LRR: Leucine-rich repeat domain; PTX: Pentraxin; SEA: Sperm protein, enterokinase and agrin; STP: Ser/Thr/Pro-rich domain; TSR: Thrombospondin type 1 repeat. Ligands shown in red are soluble and act while not anchored to a cell or extracellular matrix. List of ligands adapted from Vizurraga et al. [22].
Endogenous ligands of aGPCR
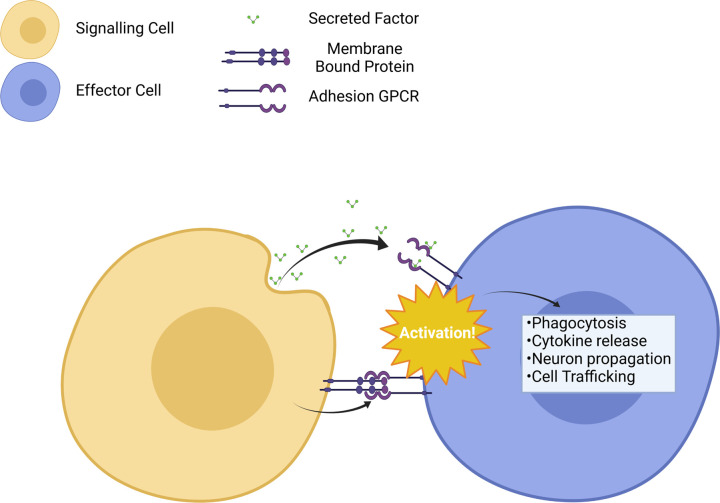
Due to their role in cell-to-cell adhesion, it is unsurprising that several ‘anchor points’ such as receptors and proteins typically found in the plasma membrane can potentially activate aGPCRs [16]. A schematic representing this endogenous paracrine activation is portrayed in Figure 1. Phospholipids, such as phosphatidylserine (PS), are an integral part of the plasma membrane, involved in numerous cell signalling events, whilst exofacial PS is a key marker of apoptotic events [17]. PS can activate the brain specific angiogenesis inhibitor 1 (BAI1, ADGRB1) found on microglial cell surfaces and cause engulfment of the presenting cell. Further membrane-bound proteins associated with aGPCRs include the lysophosphatidic acid receptor (LPA1) present on the vast majority of mammalian tissues which can bind to CD97 (ADGRE5) found on lymphoid and myelinoid cells increasing the signalling of LPA1 [18]. Activation promotes adhesion and migration to sites of inflammation.
Figure 1. Types of signalling between cells using aGPCRs.
aGPCRs are mainly utilised in paracrine or autocrine signalling via either secreted factors (top) or membrane-bound proteins and proteoglycans on adjacent cells (bottom). Activation through either of these two methods can lead to a cellular response. Created with Biorender.
Aside from membrane bound proteins, secreted factors are also known to activate aGPCRs (Figure 1) [19]. These include secreted peptides and proteoglycans typically found in the extracellular fluid or the tissue stroma around the body. This is where the original classification of the aGPCRs, and their most closely related family, the Class B1 GPCRs showed their similarity with both classes activated through hydrophilic peptides. Whilst this is the case for some aGPCRs, many other non-peptide ligands have already been documented for aGPCRs (Table 1) [20]. Soluble aGPCR ligands are typically glycosaminoglycans, such as chondroitin sulfate found in lung and pancreatic tissue [21]. Other soluble ligands include proteins such as glutaminase interacting protein (GIP) as well as small molecules such as synaptamide, an endocannabinoid-like derivative [22,23]. This varied subset of ligands suggests a multifaceted role for these receptors outside of simply cell-to-cell adhesion and paracrine signalling. Despite their broad distribution and novel screening techniques, 17 of the 33 known aGPCRs are still without known endogenous ligands (Table 1), membrane-bound or unbound [24], with significant efforts focused on deorphanisation.
aGPCR structure is separated into two fragments, each with conserved and variable regions
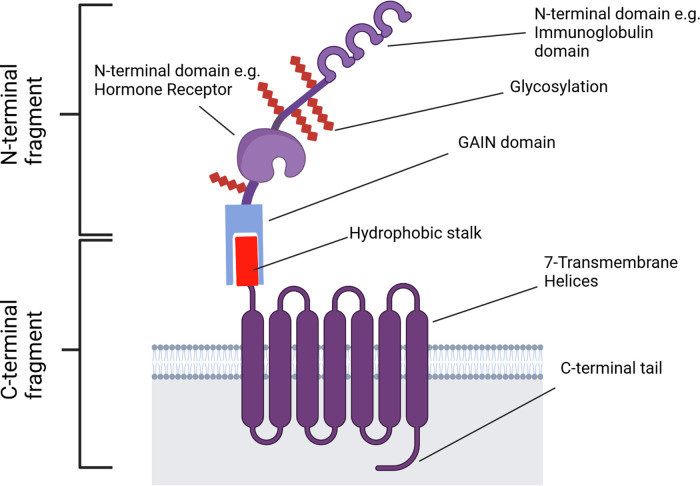
aGPCRs are made up of two major components: N- and C-terminal fragments (NTF and CTF, respectively). The NTF encompasses most of the protein's ECD, comprising the GPCR autoproteolysis-inducing (GAIN) domain and a large, heavily glycosylated N-terminal region that varies in structure between each individual aGPCR and aGPCR sub-group. The CTF is C-terminal to the GAIN domain's GPCR proteolysis site (GPS), comprising the 7TM domain and an intracellular C-terminal tail (Figure 2).
Figure 2. Example aGPCR structure.
The GPS, dividing the N- and C-terminal fragments, lies between the hydrophobic stalk and GAIN domain. Created with Biorender.
aGPCR activation mechanisms suggest stalk and lever function
Due to the initial lack of endogenous ligands and modern techniques, initial exploration into the signalling of aGPCRs was slow. Despite having initially been placed into the Class B1 GPCR family, the large ECD lent itself to the ligand-binding site theory, like Class C GPCRs [25]. However, removal of part of the ECD increased receptor activity, contrary to the initial prediction of it decreasing due to the loss of the orthosteric site [26]. This led to the proposition of the disinhibition model of signalling where the N-terminal domain inhibits constitutive activity through locking to the 7-TM domain, which upon activation, moves away from the receptor to increase signalling. This theory was challenged by the discovery of protease-activated receptor (PAR) activation mechanisms through the tethered agonist model [27]. PARs are cleaved by a number of endogenous proteases as well as proteases found in other species, resulting in a shorter N-terminal peptide [28] that can fold into the activation domain on the receptor. In 2014, two independent teams observed activation of aGPCRs by polypeptide fragments exposed post-cleavage, indicating that the tethered agonist model also applies to aGPCRs [29,30]. This further pointed to the conserved GAIN domain being responsible for autoproteolysis, contrary to PARs which require external proteolytic action. The GAIN domain helped explain the initial results of an increase in activity following cleavage as the cut sites were coincidentally located within the GAIN domain itself, mimicking the typical response of aGPCR activation [31].
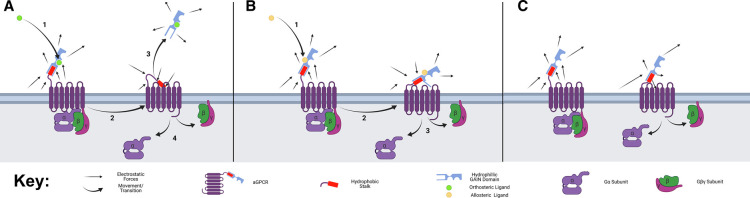
Although initial experimentation was difficult due to the hydrophobic nature of the fragments, cleavage and subsequent treatment using stalk fragments was successful. Single amino acid changes in the post cleavage stalk-peptides were discovered to have a variety of responses in aGPCRs, including inverse agonism [32]. This was partially explained with the stalk behaving like a lever and activity depending on its placement into the activation domain [33] (Figure 3A). This was given more credit thanks to the discovery of predicted β-turn elements within the stalk regions resulting in the stalk fragment bending into the receptor following cleavage [31]. The most recent theory suggests that the hydrophobic nature of the stalk contributes to the activity of the receptor, pushing it away from the aqueous ECD and into the relatively hydrophobic activation domain [22]. This does not explain all receptor activity however, with many noncleaved receptors still being able to signal in some capacity. Class B1 secretin-like GPCRs are also activated by soluble peptides such as glucagon and parathyroid hormone, exhibiting several activation states depending on the agonist present [34]. Therefore, it is likely that aGPCRs are also activated by allosteric agonists binding to the typically cleaved NTD. Currently, the exact mechanism is not known, but it may be postulated that agonist binding to an allosteric site could result in a conformational change of the NTD to push the stalk domain far enough into the activation domain of the receptor, eliciting activity through TMD to stabilse the active state (Figure 3B). Alternatively, there could be binding directly to the TMD stabilising it such as with cortisol and ADGRG3 (Table 1). This is further supported by the autoinhibitory nature of the GAIN domain with ligand binding to allosteric sites on the NTD relieving this action. Whilst orthosteric agonist activation is considered to result in full activation of the aGPCR, allosteric ligand binding can produce a graded response depending on the extent that the active site is stabilised. Many of these receptors have some degree of constitutive activity, without the need of an agonist present to elicit activity (Table 1 and Figure 3C). Finally, ligands may also bind to allosteric or orthosteric sites resulting in conformational changes that cause the opposite effect of typical agonism, also known as inverse agonism. These could move the stalk fragment away from the activation domain within the receptor, or destabilise the active site [35].
Figure 3. Proposed activation states of aGPCRs and the corresponding electrostatic forces.
Inactive aGPCRs have their G proteins bound and stalks away from the activation domain in the centre of the GPCR. This is due to the hydrophilic GAIN domain still being attached and the hydrophobic stalk being hidden within it. (A) Full activation of the aGPCR is achieved by autoproteolysis of the GAIN domain, to expose the hydrophobic stalk to the ECM, pushing it toward the hydrophobic centre of the activation domain. This activates the GPCR releasing the G protein causing further downstream effects. (B) Partial allosteric activation can result in a conformational change of the GAIN domain resulting in the exposure of part of the hydrophobic stalk. This pushes the stalk toward the activation domain resulting in a higher chance of the G protein subunit dissociating. (C) Some receptors have constitutive activity, and this is likely due to the exposure of some of the hydrophobic residues on the stalk, resulting in more forces pushing the stalk away from the water rich ECM and toward the hydrophobic centre of the aGPCR. This can partially activate the aGPCR resulting in a higher chance of G protein subunit dissociation and downstream effects. Created using Biorender.
N-terminal motifs of aGPCRs vary heavily and may determine ligand binding
The N-terminal regions of aGPCRs are consistently longer than those of Class A GPCRs, hence their initial grouping with Class B1. Their high Ser/Thr content, with many being glycosylated, gives the region a rigid, extended structure with high solubility, similar to mucin. Hence, one of the first identified aGPCRs was termed the EGF module-containing mucin-like hormone receptor (EMR1/ADGRE1) [36]. However, this feature is unlikely to directly influence ligand binding, as shown for GPR56 (ADGRG1) and one of its ligands, collagen III [37].
Many domains found in the aGPCR NTF are conserved features found in other proteins and across the aGPCR subfamilies (Table 1 — readers are directed to Hamann et al. [15] for a pictural representation of these NTFs). For instance, the most common feature is the hormone receptor domain (HomR), most commonly proximal to the GAIN domain [14]. This bears a striking sequence similarity to HomRs found in Class B GPCRs, to the extent that the latter may have descended from aGPCRs [38]. However, the GAIN domain has been shown to block the hormone-binding site of the ADGRL1 HomR [39], making their use for binding hormone ligands unlikely.
All cluster II aGPCRs contain EGF-like domains that vary only subtly between individuals. For example, the 3 amino acids that differ between ADGRE2 and 5 cause a huge bias of the ligand CD55 towards ADGRE5 [40]. Some EGF-like domains bind Ca2+, which is important for maintaining their structure and mediating protein ligand binding [41]. The Calx-beta motif, found in ADGRV1, also binds Ca2+ in the NTF, as demonstrated by their presence in Na+/Ca2+ exchangers [42]. From this, some have inferred that Calx-beta motifs could use Ca2+ to bind ligands, similarly to Ca2+-binding EGF domains [43]. Moreover, Complement C1r/C1s, Uegf, Bmp1 (CUB) domains have been demonstrated to use Ca2+ to bind ligands in various proteins [44,45], and have recently been shown to mediate intramolecular interactions in the ADGRG6 ECD. This gives a closed conformation by giving an interface between the CUB domain's tip and the more distal HomR that may contribute to the signalling state of the receptor [46].
aGPCR NTFs contain other domains and motifs found in a variety of proteins that are known to bind specific protein partners. Arginine-glycine-aspartate (RGD) motifs are known to bind integrins and are notably found in ADGRE5 [47–49]. Pentraxin (PTX) domains are found in a variety of aGPCRs, with variations between individuals that allow recruitment of specific ligands to specific receptors. For instance, the PTX and CUB domains of ADGRG6 have been shown to bind collagen IV, but not other collagen subtypes [50]. While not every identified NTF domain has been matched to a binding partner, the expansive repertoire of motifs and structures present demonstrate the heterogeneity of the ligands with which this family may interact.
Alternative splicing also expands this repertoire. The most variable part of aGPCR transcripts is the region N-terminal to the GAIN domain. Here, the position of individual domains can be altered, by addition of Ser/Thr stretches that vary NTF structure, or excluded entirely [51]. This is demonstrated in ADGRG6, where inclusion of 23 amino acids, many of which are glycosylated, disrupts this receptor's closed conformation, instead giving the receptor a more extended conformation that disrupts its ability to facilitate myelination in vivo [46].
The GAIN domain separates aGPCRs into two fragments and may bind ligands
The GAIN domain is found almost ubiquitously in aGPCRs, between the variable N-terminal domains and the 7TM region, with only ADGRA1 lacking this region [14]. Its primary function is to allow receptor autoproteolysis at the GPS site, located proximally to the final β-strand (β13) of the GAINB subdomain (consensus: HLT, cleaving between L and T). These residues form a sharp turn, created by a disulfide bridge located proximally to the GPS and the Leu R-group being trapped in a hydrophobic pocket. Proteolysis is achieved by nucleophilic attack on the L-T peptide bond by the Thr R-group, with the resulting ester hydrolysing to give two separate fragments of the original protein [22,39,52].
There is also evidence for ligands binding to the GAIN domain to trigger aGPCR activation, such as CD90 binding ADGRE5 [49,53]. More recently, a small molecule agonist of ADGRF1, synaptamide, has been shown to interact with its GAIN domain [23]. These observations could explain the finding that cancer-causing mutations are found on the surface of the GAIN domain [39].
The aGPCR 7TM domain retains recognised GPCR functions with novel motifs
Cryo-electron microscopy (cryo-EM) was recently used to elucidate the first full-length structure of an active aGPCR (ADGRG3) in complex with small molecule agonists (glucocorticoids cortisol and beclomethasone) and Gαo [54]. The resultant structure had a 7TM region overall resembling that of a Class A GPCR, other than a greater separation between TM6 and TM7, giving a larger ligand-binding site. Extracellular Loop 2 (ECL2) forms a hydrophilic β-sheet that has a weak constitutive interaction with ECL3. This could act as a mechanism for relaying conformational changes from the NTF to the CTF upon ligand binding, allowing removal of the ECL2 ‘flexible lid’. This would expose the ligand binding pocket between TM6 and TM7, with the hydrophobic cores of glucocorticoid ligands packing against TM7. Alternatively, this feature may act to prevent dissociation of small molecule ligands by blocking their exit from the orthosteric site, slowing their dissociation rate and increasing the length of time over which the receptor signals, as seen in Class A GPCRs such as the endothelin-1 receptor B [55,56].
Unlike Class A GPCRs [57,58], ADGRG3 did not contain a core triad (IPF) motif or an NPxxY motif in TM7 that are involved in signal transduction across the GPCR. Instead of the core triad, ADGRG3 contained upper quaternary and lower triad cores (UQC and LTC) of hydrophobic residues that performed equivalent functions. A vital ‘toggle switch’ residue (W4906.53), contained within the UQC, recognises ligand binding and causes a conformational change that leads to its coupling to Gαo. Moreover, ADGRG3 lacks an ionic lock motif, normally found at the base of TM3 in Class A GPCRs (consensus: E/DRY), replacing it with a hydrophobic lock (HLY motif) that may perform similar roles in stabilising the receptor conformation on its cytoplasmic surface [54]. The differences in these key functional motifs between aGPCRs and other GPCR families further demonstrate the evolutionary distance between them, justifying the classification of aGPCRs as their own subfamily. The separation of the core triad into UQC and LTC between Family A and aGPCRs suggests a relatively distant common ancestor exists between the two. This structural difference also suggests the existence of novel methods for the design of small-molecule drugs targeting aGPCRs.
aGPCRs activate a variety of effectors
GPCRs typically propagate their activation signal through two main classes of effectors: heterotrimeric G proteins, and β-arrestins resulting in an incredibly varied intracellular response profile [59]. ADGRF1 (GPR110), for example, is activated by synaptamide and can increase intracellular cAMP in a Gs dependent-manner as well as mobilise intracellular Ca2+ in a Gq/11 dependent-manner, resulting in neurite growth and neurogenesis [60,61]. Interestingly, many of the downstream mediators and effectors of aGPCRs were discovered before their ligands, due to the self-cleavage aspect of their function. For example, ADGRG2 (GPR64) is currently an orphan receptor but due to manual cleavage of its NTD it has been observed to activate Gq mobilising intracellular Ca2+ [62] and Gs stimulating intracellular cAMP production [63]. ADGRG2 was also found to undergo β-arrestin-mediated endocytosis, further increasing its signalling repertoire by acting as a scaffold for downstream effectors.
β-arrestins are well known to mediate GPCR internalisation and activate numerous intracellular effectors for downstream signalling pathways, dependent on both the receptor itself as well as the ligand bound. Interestingly, β-arrestins can function in aGPCR signalling without full activation, which is atypical for many GPCRs [31]. The presence of the activating stalk in ADGRG1 was found to not be required for β-arrestin association, and therefore signalling. This could mean that allosteric agonism or even constitutive activity could be explained by arrestin recruitment and signal propagation. A further class of accessory protein recently discovered to interact with aGPCRs are the receptor activity-modifying proteins (RAMPs). RAMPs are a family of three single-pass transmembrane spanning proteins which were initially discovered to allow functional membrane expression and alter ligand specificity of the Class B1 GPCR calcitonin-like receptor (CALCRL) [64]. Since then, they have been discovered to interact with more GPCRs affecting receptor trafficking, downstream signalling and recycling [65,66]. Whilst the repertoire of RAMP-interacting GPCRs has expanded across Class A, B1 and C, in 2019, it was discovered that ADGRF5 (GPR116) interacts with RAMP2 and 3 [67]. Whilst the role of RAMPs in aGPCR function is currently unknown, this opens another avenue of research into aGPCR activity that may aide in the discovery of endogenous ligands which can only activate aGPCRs in the presence of RAMPs.
aGPCRs have a multitude of physiological effects
As mentioned previously and reviewed extensively by Monk et al. [68], aGPCRs have significant function in paracrine signalling. They have a major role in the immune system, demonstrated by the large variety of aGPCRs found on immune cells [69]. These include ADGRB1 (BAI1) described above, which is required for the phagocytosis of apoptotic cells and pathogens in the brain. In addition to this, other aGPCRs such as ADGRG1 (GPR56) have been shown to be present in inflammatory natural killer cells along with cytotoxic lymphocytes [70]. Paracrine signalling is not limited to the immune system however, with several aGPCRs including ADGRL1 (Latrophilin-1) being suggested to increase synapse formation and function [71]. ADGRC1 (CELSR1) is another aGPCR responsible for dendritogenesis and axon guidance where KO studies have shown impaired migration of branchiomotor neurons during development [72]. One other area in which aGPCR function is also seemingly is required in the trafficking of stem cells to the bone marrow and their retention therein to produce haematopoietic cells, likely using soluble ligands due to the systemic trafficking of these cells [73].
Clinical significance of aGPCR malfunction
aGPCRs have been implicated in numerous diseases, in particular, various types of cancers where a lack of function results in increased cell growth and metastasis [5]. One of the deadliest forms of cancer, lung cancer, can be severely affected by aGPCR mutations. ADGRB3 is an angiogenesis inhibitor, which has been found to be the most significantly mutated gene in 13% of lung squamous tumours, with mutations resulting in decreased activity of the receptor, increasing blood flow to the tumour [74]. Furthermore, it was discovered that in many lung cancers, the translation of ADGRB3 is decreased, resulting in reduced tumour suppressive effects provided by the receptor. Breast cancer was the second most diagnosed form of cancer in 2018 [75] and similarly to lung cancer, showed altered expression or mutation in aGPCRs. While typically not expressed in breast epithelial cells, ADGRE2 was shown to be up-regulated in invasive breast carcinomas and negatively correlated with survival and patient prognosis [76]. Previous research suggested ADGRE2 to have functions in the immune system therefore indicating further exploration is required in determining the secondary function in carcinoma progression. Recently, it was discovered that a further aGPCR, ADGRL4, promoted angiogenesis during both the development of the endothelium, as well as in several cancers where it is overexpressed. Of note is the lack of canonical GPCR signalling by this aGPCR, although several genes were found to have altered expression following activation suggesting an unusual method of signal transduction [24]. aGPCRs are quickly becoming a target of interest for other diseases outside of cancer, hopefully allowing for further harnessing of aGPCRs as therapeutic targets [73].
Perspectives
aGPCR research is a rapidly changing field with many orphan aGPCRs and an emerging picture of how agonists cause receptor activation. Further insights into these may help in rational drug design for aGPCRs. This would aid in treatments of diseases which aGPCRs are involved in, such as cancer, due to their control over angiogenesis and up-regulation in breast cancer.
The current consensus on aGPCR activation involves the GAIN domain acting in an autoinhibitory way to occlude a tethered ligand, as demonstrated by constitutively active aGPCR mutants. Ligands may bind to the NTF to cause the tethered ligand to be exposed, allowing its binding to the aGPCR orthosteric site and receptor activation. Recent cryo-EM studies have shown that, from here, aGPCR conformational changes reflect those in Class A GPCRs, but make use of different motifs.
Future directions may include the cryo-EM analysis of more aGPCRs to allow comparative structural studies demonstrating the importance of sites such as the UQC; further elucidation of how the binding of ligands to the NTF can cause receptor activation; and using this newfound knowledge of aGPCR activation to design drugs to alter their activity.
Acknowledgements
This work was supported an AstraZeneca Scholarship awarded to MR, a Cambridge Trust and Christ's College Scholarship awarded to T.N. and a Medical Research Council Confidence in Concept award to G.L. and M.H. (MC_PC_17156).
Abbreviations
- CUB
C1r/C1s, Uegf, Bmp1
- ECD
extracellular domain
- ECL2
extracellular loop 2
- EGF
epidermal growth factor
- GAIN
GPCR autoproteolysis-inducing
- GPS
GPCR proteolysis site
- LPA1
lysophosphatidic acid receptor
- PS
phosphatidylserine
- PTX
Pentraxin
- RAMPs
receptor activity-modifying proteins
- TM
transmembrane
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Open Access Statement
Open access for this article was enabled by the participation of University of Cambridge in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society under a transformative agreement with JISC.
Author Contributions
M.R. and T.N. wrote the manuscript. M.H. and G.L. revised and edited the manuscript.
References
- 1.Sriram, K. and Insel, P.A. (2018) G protein-coupled receptors as targets for approved drugs: how many targets and how many drugs? Mol. Pharmacol. 93, 251–258 10.1124/mol.117.111062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lagerström, M.C. and Schiöth, H.B. (2008) Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 7, 339–357 10.1038/nrd2518 [DOI] [PubMed] [Google Scholar]
- 3.Bondarev, A.D., Attwood, M.M., Jonsson, J., Chubarev, V.N., Tarasov, V.V. and Schiöth, H.B. (2020) Opportunities and challenges for drug discovery in modulating adhesion G protein-coupled receptor (GPCR) functions. Expert Opin. Drug Discov. 15, 1291–1307 10.1080/17460441.2020.1791075 [DOI] [PubMed] [Google Scholar]
- 4.Yona, S., Lin, H.-H., Siu, W.O., Gordon, S. and Stacey, M. (2008) Adhesion-GPCRs: emerging roles for novel receptors. Trends Biochem. Sci. 33, 491–500 10.1016/j.tibs.2008.07.005 [DOI] [PubMed] [Google Scholar]
- 5.Gad, A.A. and Balenga, N. (2020) The emerging role of adhesion GPCRs in cancer. ACS Pharmacol. Transl. Sci. 3, 29–42 10.1021/acsptsci.9b00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Starkey, P.M., Turley, L. and Gordon, S. (1987) The mouse macrophage-specific glycoprotein defined by monoclonal antibody F4/80: characterization, biosynthesis and demonstration of a rat analogue. Immunology 60, 117–122 PMID: [PMC free article] [PubMed] [Google Scholar]
- 7.McKnight, A.J. and Gordon, S. (1998) The EGF-TM7 family: unusual structures at the leukocyte surface. J. Leukoc. Biol. 63, 271–280 10.1002/jlb.63.3.271 [DOI] [PubMed] [Google Scholar]
- 8.McKnight, A.J. and Gordon, S. (1996) EGF-TM7: a novel subfamily of seven-transmembrane-region leukocyte cell-surface molecules. Immunol. Today 17, 283–287 10.1016/0167-5699(96)80546-9 [DOI] [PubMed] [Google Scholar]
- 9.Stacey, M., Lin, H.-H., Gordon, S. and McKnight, A.J. (2000) LNB-TM7, a group of seven-transmembrane proteins related to family-B G-protein-coupled receptors. Trends Biochem. Sci. 25, 284–289 10.1016/S0968-0004(00)01583-8 [DOI] [PubMed] [Google Scholar]
- 10.Flower, D.R. (1999) Modelling G-protein-coupled receptors for drug design. Biochim. Biophys. Acta Rev. Biomembr. 1422, 207–234 10.1016/S0304-4157(99)00006-4 [DOI] [PubMed] [Google Scholar]
- 11.Harmar, A.J. (2001) Family-B G-protein-coupled receptors. Genome Biol. 2, reviews3013.1–reviews3013.10 10.1186/gb-2001-2-12-reviews3013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crocker, P.R., Morris, L. and Gordon, S. (1988) Novel cell surface adhesion receptors involved in interactions between stromal macrophages and haematopoietic cells. J. Cell Sci. Suppl. 9, 185–206 10.1242/jcs.1988.Supplement_9.10 [DOI] [PubMed] [Google Scholar]
- 13.Fredriksson, R., Lagerström, M.C., Lundin, L.-G. and Schiöth, H.B. (2003) The G-protein-coupled receptors in the human genome form five main families. phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 63, 1256–1272 10.1124/mol.63.6.1256 [DOI] [PubMed] [Google Scholar]
- 14.Bjarnadóttir, T.K., Fredriksson, R., Höglund, P.J., Gloriam, D.E., Lagerström, M.C. and Schiöth, H.B. (2004) The human and mouse repertoire of the adhesion family of G-protein-coupled receptors. Genomics 84, 23–33 10.1016/j.ygeno.2003.12.004 [DOI] [PubMed] [Google Scholar]
- 15.Hamann, J., Aust, G., Araç, D., Engel, F.B., Formstone, C., Fredriksson, R.et al. (2015) International union of basic and clinical pharmacology. XCIV. adhesion G protein–coupled receptors. Pharmacol. Rev. 67, 338–367 10.1124/pr.114.009647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stacey, M., Chang, G.-W., Davies, J.Q., Kwakkenbos, M.J., Sanderson, R.D., Hamann, J.et al. (2003) The epidermal growth factor–like domains of the human EMR2 receptor mediate cell attachment through chondroitin sulfate glycosaminoglycans. Blood 102, 2916–2924 10.1182/blood-2002-11-3540 [DOI] [PubMed] [Google Scholar]
- 17.Naeini, M.B., Bianconi, V., Pirro, M. and Sahebkar, A. (2020) The role of phosphatidylserine recognition receptors in multiple biological functions. Cell. Mol. Biol. Lett. 25, 23 10.1186/s11658-020-00214-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riaz, A., Huang, Y. and Johansson, S. (2016) G-Protein-Coupled lysophosphatidic acid receptors and their regulation of AKT signaling. Int. J. Mol. Sci. 17, 215 10.3390/ijms17020215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paavola, K.J. and Hall, R.A. (2012) Adhesion G protein-coupled receptors: signaling, pharmacology, and mechanisms of activation. Mol. Pharmacol. 82, 777–783 10.1124/mol.112.080309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arimont, M., van der Woude, M., Leurs, R., Vischer, H.F., de Graaf, C. and Nijmeijer, S. (2019) Identification of key structural motifs involved in 7 transmembrane signaling of adhesion GPCRs. ACS Pharmacol. Transl. Sci. 2, 101–113 10.1021/acsptsci.8b00051 [DOI] [Google Scholar]
- 21.Chiang, N.-Y., Chang, G.-W., Huang, Y.-S., Peng, Y.-M., Hsiao, C.-C., Kuo, M.-L.et al. (2016) Heparin interacts with the adhesion GPCR GPR56, reduces receptor shedding, and promotes cell adhesion and motility. J. Cell Sci. 129, 2156–2169 10.1242/jcs.174458 [DOI] [PubMed] [Google Scholar]
- 22.Vizurraga, A., Adhikari, R., Yeung, J., Yu, M. and Tall, G.G. (2020) Mechanisms of adhesion G protein–coupled receptor activation. J. Biol. Chem. 295, 14065–14083 10.1074/jbc.REV120.007423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, B.X., Hu, X., Kwon, H.-S., Fu, C., Lee, J.-W., Southall, N.et al. (2020) Synaptamide activates the adhesion GPCR GPR110 (ADGRF1) through GAIN domain binding. Commun. Biol. 3, 109 10.1038/s42003-020-0831-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Favara, D.M., Liebscher, I., Jazayeri, A., Nambiar, M., Sheldon, H., Banham, A.H.et al. (2021) Elevated expression of the adhesion GPCR ADGRL4/ELTD1 promotes endothelial sprouting angiogenesis without activating canonical GPCR signalling. Sci. Rep. 11, 8870 10.1038/s41598-021-85408-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pin, J.-P., Galvez, T. and Prézeau, L. (2003) Evolution, structure, and activation mechanism of family 3/C G-protein-coupled receptors. Pharmacol. Ther. 98, 325–354 10.1016/S0163-7258(03)00038-X [DOI] [PubMed] [Google Scholar]
- 26.Paavola, K.J., Stephenson, J.R., Ritter, S.L., Alter, S.P. and Hall, R.A. (2011) The N terminus of the adhesion G protein-coupled receptor GPR56 controls receptor signaling activity. J. Biol. Chem. 286, 28914–28921 10.1074/jbc.M111.247973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nieberler, M., Kittel, R.J., Petrenko, A.G., Lin, H.H. and Langenhan, T. (2016) Control of Adhesion GPCR Function Through Proteolytic Processing. In Adhesion G Protein-Coupled Receptors: Molecular, Physiological and Pharmacological Principles in Health and Disease (Langenhan, T. and Schöneberg, T., eds), pp. 83–109, Springer International Publishing, Cham: [DOI] [PubMed] [Google Scholar]
- 28.Heuberger, D.M. and Schuepbach, R.A. (2019) Protease-activated receptors (PARs): mechanisms of action and potential therapeutic modulators in PAR-driven inflammatory diseases. Thromb. J. 17, 4 10.1186/s12959-019-0194-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liebscher, I., Schön, J., Petersen, S.C., Fischer, L., Auerbach, N., Demberg, L.M.et al. (2014) A tethered agonist within the ectodomain activates the adhesion G protein-coupled receptors GPR126 and GPR133. Cell Rep. 9, 2018–2026 10.1016/j.celrep.2014.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo, R., Jeong, S.-J., Yang, A., Wen, M., Saslowsky, D.E., Lencer, W.I.et al. (2014) Mechanism for adhesion G protein-coupled receptor GPR56-Mediated rhoA activation induced By collagen III stimulation. PLoS ONE 9, e100043 10.1371/journal.pone.0100043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoveken, H.M., Hajduczok, A.G., Xu, L. and Tall, G.G. (2015) Adhesion G protein-coupled receptors are activated by exposure of a cryptic tethered agonist. Proc. Natl Acad. Sci. U.S.A. 112, 6194–6199 10.1073/pnas.1421785112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kishore, A., Purcell, R.H., Nassiri-Toosi, Z. and Hall, R.A. (2016) Stalk-dependent and stalk-independent signaling by the adhesion G protein-coupled receptors GPR56 (ADGRG1) and BAI1 (ADGRB1). J. Biol. Chem. 291, 3385–3394 10.1074/jbc.M115.689349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell, R.H. and Hall, R.A. (2018) Adhesion G protein–coupled receptors as drug targets. Annu. Rev. Pharmacol. Toxicol. 58, 429–449 10.1146/annurev-pharmtox-010617-052933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krumm, B. and Roth, B.L. (2020) A structural understanding of class B GPCR selectivity and activation revealed. Structure 28, 277–279 10.1016/j.str.2020.02.004 [DOI] [PubMed] [Google Scholar]
- 35.Salzman, G.S., Zhang, S., Gupta, A., Koide, A., Koide, S. and Araç, D. (2017) Stachel -independent modulation of GPR56/ADGRG1 signaling by synthetic ligands directed to its extracellular region. Proc. Natl Acad. Sci. U.S.A. 114, 10095–10100 10.1073/pnas.1708810114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baud, V., Chissoe, S.L., Viegas-Péquignot, E., Diriong, S., N'guyen, V.C., Roe, B.A.et al. (1995) EMR1, an unusual member in the family of hormone receptors with seven transmembrane segments. Genomics 26, 334–344 10.1016/0888-7543(95)80218-B [DOI] [PubMed] [Google Scholar]
- 37.Luo, R., Jin, Z., Deng, Y., Strokes, N. and Piao, X. (2012) Disease-Associated mutations prevent GPR56-collagen III interaction. PLoS ONE 7, e29818 10.1371/journal.pone.0029818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nordstrom, K.J.V., Lagerstrom, M.C., Waller, L.M.J., Fredriksson, R. and Schioth, H.B. (2008) The secretin GPCRs descended from the family of adhesion GPCRs. Mol. Biol. Evol. 26, 71–84 10.1093/molbev/msn228 [DOI] [PubMed] [Google Scholar]
- 39.Araç, D., Boucard, A.A., Bolliger, M.F., Nguyen, J., Soltis, S.M., Südhof, T.C.et al. (2012) A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis: Cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 31, 1364–1378 10.1038/emboj.2012.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin, H.-H., Stacey, M., Hamann, J., Gordon, S. and McKnight, A.J. (2000) Human EMR2, a novel EGF-TM7 molecule on chromosome 19p13.1, Is closely related to CD97. Genomics 67, 188–200 10.1006/geno.2000.6238 [DOI] [PubMed] [Google Scholar]
- 41.Rao, Z., Handford, P., Mayhew, M., Knott, V., Brownlee, G.G. and StuartZ, D. (1995) The structure of a Ca2+-binding epidermal growth factor-like domain: Its role in protein-protein interactions. Cell 82, 131–141 10.1016/0092-8674(95)90059-4 [DOI] [PubMed] [Google Scholar]
- 42.Nikkila, H., McMillan, D.R., Nunez, B.S., Pascoe, L., Curnow, K.M. and White, P.C. (2000) Sequence similarities between a novel putative G protein-coupled receptor and Na+/Ca2+ exchangers define a cation binding domain. Mol. Endocrinol. 14, 1351–1364 10.1210/mend.14.9.0511 [DOI] [PubMed] [Google Scholar]
- 43.McMillan, D.R. and White, P.C. (2010) Studies on the very large g protein-coupled receptor: from initial discovery to determining its role in sensorineural deafness in higher animals. In Adhesion-GPCRs: Structure to Function (Yona, S. and Stacey, M., eds), pp. 76–86, Springer, Boston, MA: [DOI] [PubMed] [Google Scholar]
- 44.Andersen, C.B.F., Madsen, M., Storm, T., Moestrup, S.K. and Andersen, G.R. (2010) Structural basis for receptor recognition of vitamin-B12–intrinsic factor complexes. Nature 464, 445–448 10.1038/nature08874 [DOI] [PubMed] [Google Scholar]
- 45.Venkatraman Girija, U., Gingras, A.R., Marshall, J.E., Panchal, R., MdA, S., Gál, P.et al. (2013) Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation. Proc. Natl Acad. Sci. U.S.A. 110, 13916–13920 10.1073/pnas.1311113110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leon, K., Cunningham, R.L., Riback, J.A., Feldman, E., Li, J., Sosnick, T.R.et al. (2020) Structural basis for adhesion G protein-coupled receptor Gpr126 function. Nat. Commun. 11, 194 10.1038/s41467-019-14040-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plow, E.F., Haas, T.A., Zhang, L., Loftus, J. and Smith, J.W. (2000) Ligand binding to integrins. J. Biol. Chem. 275, 21785–8 10.1074/jbc.R000003200 [DOI] [PubMed] [Google Scholar]
- 48.Tjong, W.-Y. and Lin, H.-H. (2019) The RGD motif is involved in CD97/ADGRE5-promoted cell adhesion and viability of HT1080 cells. Sci. Rep. 9, 1517 10.1038/s41598-018-38045-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, T., Ward, Y., Tian, L., Lake, R., Guedez, L., Stetler-Stevenson, W.G.et al. (2005) CD97, an adhesion receptor on inflammatory cells, stimulates angiogenesis through binding integrin counterreceptors on endothelial cells. Blood 105, 2836–2844 10.1182/blood-2004-07-2878 [DOI] [PubMed] [Google Scholar]
- 50.Paavola, K.J., Sidik, H., Zuchero, J.B., Eckart, M. and Talbot, W.S. (2014) Type IV collagen is an activating ligand for the adhesion G protein-coupled receptor GPR126. Sci. Signal. 7, ra76 10.1126/scisignal.2005347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knierim, A.B., Röthe, J., Çakir, M.V., Lede, V., Wilde, C., Liebscher, I.et al. (2019) Genetic basis of functional variability in adhesion G protein-coupled receptors. Sci. Rep. 9, 11036 10.1038/s41598-019-46265-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin, H.-H., Chang, G.-W., Davies, J.Q., Stacey, M., Harris, J. and Gordon, S. (2004) Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J. Biol. Chem. 279, 31823–31832 10.1074/jbc.M402974200 [DOI] [PubMed] [Google Scholar]
- 53.Wandel, E., Saalbach, A., Sittig, D., Gebhardt, C. and Aust, G. (2012) Thy-1 (CD90) Is an interacting partner for CD97 on activated endothelial cells. J. Immunol. 188, 1442–1450 10.4049/jimmunol.1003944 [DOI] [PubMed] [Google Scholar]
- 54.Ping, Y.-Q., Mao, C., Xiao, P., Zhao, R.-J., Jiang, Y., Yang, Z.et al. (2021) Structures of the glucocorticoid-bound adhesion receptor GPR97–Go complex. Nature 589, 620–626 10.1038/s41586-020-03083-w [DOI] [PubMed] [Google Scholar]
- 55.Clozel, M., Fischli, W. and Guilly, C. (1989) Specific binding of endothelin on human vascular smooth muscle cells in culture. J. Clin. Invest. 83, 1758–1761 10.1172/JCI114078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shihoya, W., Nishizawa, T., Okuta, A., Tani, K., Dohmae, N., Fujiyoshi, Y.et al. (2016) Activation mechanism of endothelin ETB receptor by endothelin-1. Nature 537, 363–368 10.1038/nature19319 [DOI] [PubMed] [Google Scholar]
- 57.Maeda, S., Qu, Q., Robertson, M.J., Skiniotis, G. and Kobilka, B.K. (2019) Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes. Science 364, 552–557 10.1126/science.aaw5188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rasmussen, S.G.F., DeVree, B.T., Zou, Y., Kruse, A.C., Chung, K.Y., Kobilka, T.S.et al. (2011) Crystal structure of the β2 adrenergic receptor–Gs protein complex. Nature 477, 549–555 10.1038/nature10361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simundza, J. and Cowin, P. (2013) Adhesion G-Protein-Coupled receptors: elusive hybrids come of Age. Cell Commun. Adhes. 20, 213–225 10.3109/15419061.2013.855727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee, J.-W., Huang, B.X., Kwon, H., Rashid, M.A., Kharebava, G., Desai, A.et al. (2016) Orphan GPR110 (ADGRF1) targeted by N-docosahexaenoylethanolamine in development of neurons and cognitive function. Nat. Commun. 7, 13123 10.1038/ncomms13123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Demberg, L.M., Winkler, J., Wilde, C., Simon, K.-U., Schön, J., Rothemund, S.et al. (2017) Activation of adhesion G protein-coupled receptors. J. Biol. Chem. 292, 4383–4394 10.1074/jbc.M116.763656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, D.-L., Sun, Y.-J., Ma, M.-L., Wang, Y., Lin, H., Li, R.-R.et al. (2018) Gq activity- and β-arrestin-1 scaffolding-mediated ADGRG2/CFTR coupling are required for male fertility. eLife 7, e33432 10.7554/eLife.33432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ahn, J.I., Yoo, J.-Y., Kim, T.H., Kim, Y.I., Broaddus, R.R., Ahn, J.Y.et al. (2019) G-protein coupled receptor 64 (GPR64) acts as a tumor suppressor in endometrial cancer. BMC Cancer 19, 810 10.1186/s12885-019-5998-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McLatchie, L.M., Fraser, N.J., Main, M.J., Wise, A., Brown, J., Thompson, N.et al. (1998) RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature 393, 333–339 10.1038/30666 [DOI] [PubMed] [Google Scholar]
- 65.Mackie, D.I., Nielsen, N.R., Harris, M., Singh, S., Davis, R.B., Dy, D.et al. (2019) RAMP3 determines rapid recycling of atypical chemokine receptor-3 for guided angiogenesis. Proc. Natl Acad. Sci. U.S.A. 116, 24093–24099 10.1073/pnas.1905561116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Routledge, S.J., Ladds, G. and Poyner, D.R. (2017) The effects of RAMPs upon cell signalling. Mol. Cell Endocrinol. 449, 12–20 10.1016/j.mce.2017.03.033 [DOI] [PubMed] [Google Scholar]
- 67.Huang, K.-Y. and Lin, H.-H. (2018) The activation and signaling mechanisms of GPR56/ADGRG1 in melanoma cell. Front. Oncol. 8, 304 10.3389/fonc.2018.00304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Monk, K.R., Hamann, J., Langenhan, T., Nijmeijer, S., Schöneberg, T. and Liebscher, I. (2015) Adhesion G protein–Coupled receptors: From In vitro pharmacology to In vivo mechanisms. Mol. Pharmacol. 88, 617–623 10.1124/mol.115.098749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin, H.-H., Hsiao, C.-C., Pabst, C., Hébert, J., Schöneberg, T. and Hamann, J. (2017) Adhesion GPCRs in regulating immune responses and inflammation. Adv. Immunol. 136, 163–201 10.1016/bs.ai.2017.05.005 [DOI] [PubMed] [Google Scholar]
- 70.Chang, G.-W., Hsiao, C.-C., Peng, Y.-M., Vieira Braga, F.A., Kragten, N.A.M., Remmerswaal, E.B.M.et al. (2016) The adhesion G protein-coupled receptor GPR56/ADGRG1 is an inhibitory receptor on human NK cells. Cell Rep. 15, 1757–1770 10.1016/j.celrep.2016.04.053 [DOI] [PubMed] [Google Scholar]
- 71.Sando, R. and Südhof, T.C. (2021) Latrophilin GPCR signaling mediates synapse formation. eLife 10, e65717 10.7554/eLife.65717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Langenhan, T., Piao, X. and Monk, K.R. (2016) Adhesion G protein-coupled receptors in nervous system development and disease. Nat. Rev. Neurosci. 17, 550–561 10.1038/nrn.2016.86 [DOI] [PubMed] [Google Scholar]
- 73.Bassilana, F., Nash, M. and Ludwig, M.-G. (2019) Adhesion G protein-coupled receptors: opportunities for drug discovery. Nat. Rev. Drug Discov. 18, 869–884 10.1038/s41573-019-0039-y [DOI] [PubMed] [Google Scholar]
- 74.Aust, G., Zhu, D., Van Meir, E.G. and Xu, L. (2016) Adhesion GPCRs in Tumorigenesis. In Adhesion G Protein-Coupled Receptors: Molecular, Physiological and Pharmacological Principles in Health and Disease (Langenhan, T. and Schöneberg, T., eds), pp. 369–396, Springer International Publishing, Cham [Google Scholar]
- 75.Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R.L., Torre, L.A. and Jemal, A. (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 76.Bhudia, N., Desai, S., King, N., Ancellin, N., Grillot, D., Barnes, A.A.et al. (2020) G protein-coupling of adhesion GPCRs ADGRE2/EMR2 and ADGRE5/CD97, and activation of G protein signalling by an anti-EMR2 antibody. Sci. Rep. 10, 1004 10.1038/s41598-020-57989-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lelianova, V.G., Davletov, B.A., Sterling, A., Rahman, M.A., Grishin, E.V., Totty, N.F.et al. (1997) α-Latrotoxin receptor, latrophilin, Is a novel member of the secretin family of G protein-coupled receptors. J. Biol. Chem. 272, 21504–8 10.1074/jbc.272.34.21504 [DOI] [PubMed] [Google Scholar]
- 78.Matsushita, H., Lelianova, V.G. and Ushkaryov, Y.A. (1999) The latrophilin family: multiply spliced G protein-coupled receptors with differential tissue distribution. FEBS Lett. 443, 348–352 10.1016/S0014-5793(99)00005-8 [DOI] [PubMed] [Google Scholar]
- 79.Silva, J.-P., Lelianova, V.G., Ermolyuk, Y.S., Vysokov, N., Hitchen, P.G., Berninghausen, O.et al. (2011) Latrophilin 1 and its endogenous ligand lasso/teneurin-2 form a high-affinity transsynaptic receptor pair with signaling capabilities. Proc. Natl Acad. Sci. U.S.A. 108, 12113–8 10.1073/pnas.1019434108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.O'Sullivan, M.L., de Wit, J., Savas, J.N., Comoletti, D., Otto-Hitt, S., Yates, J.R.et al. (2012) FLRT proteins are endogenous latrophilin ligands and regulate excitatory synapse development. Neuron 73, 903–910 10.1016/j.neuron.2012.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boucard, A.A., Ko, J. and Südhof, T.C. (2012) High affinity neurexin binding to cell adhesion G-protein-coupled receptor CIRL1/Latrophilin-1 produces an intercellular adhesion complex. J. Biol. Chem. 287, 9399–9413 10.1074/jbc.M111.318659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jackson, V.A., Mehmood, S., Chavent, M., Roversi, P., Carrasquero, M., del Toro, D.et al. (2016) Super-complexes of adhesion GPCRs and neural guidance receptors. Nat. Commun. 7, 11184 10.1038/ncomms11184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nechiporuk, T., Urness, L.D. and Keating, M.T. (2001) ETL, a novel seven-transmembrane receptor that Is developmentally regulated in the heart. J. Biol. Chem. 276, 4150–4157 10.1074/jbc.M004814200 [DOI] [PubMed] [Google Scholar]
- 84.Nazarko, O., Kibrom, A., Winkler, J., Leon, K., Stoveken, H., Salzman, G.et al. (2018) A comprehensive mutagenesis screen of the adhesion GPCR latrophilin-1/ADGRL1. iScience 3, 264–278 10.1016/j.isci.2018.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mathiasen, S., Palmisano, T., Perry, N.A., Stoveken, H.M., Vizurraga, A., McEwen, D.P.et al. (2020) G12/13 is activated by acute tethered agonist exposure in the adhesion GPCR ADGRL3. Nat. Chem. Biol. 16, 1343–1350 10.1038/s41589-020-0617-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Müller, A., Winkler, J., Fiedler, F., Sastradihardja, T., Binder, C., Schnabel, R.et al. (2015) Oriented cell division in the C. elegans embryo Is coordinated by G-Protein signaling dependent on the adhesion GPCR LAT-1. PLoS Genet. 11, e1005624 10.1371/journal.pgen.1005624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gordon, S., Lin, H.-H., Hamann, J., Kwakkenbos, M.J., Kop, E.N., Stacey, M.et al. (2004) The EGF-TM7 family: a postgenomic view. Immunogenetics. 55, 655–666 10.1007/s00251-003-0625-2 [DOI] [PubMed] [Google Scholar]
- 88.Hamann, J., Vogel, B., van Schijndel, G.M. and van Lier, R.A. (1996) The seven-span transmembrane receptor CD97 has a cellular ligand (CD55. DAF). J. Exp. Med. 184, 1185–1189 10.1084/jem.184.3.1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Irmscher, S., Brix, S.R., Zipfel, S.L.H., Halder, L.D., Mutlutürk, S., Wulf, S.et al. (2019) Serum FHR1 binding to necrotic-type cells activates monocytic inflammasome and marks necrotic sites in vasculopathies. Nat. Commun. 10, 2961 10.1038/s41467-019-10766-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boyden, S.E., Desai, A., Cruse, G., Young, M.L., Bolan, H.C., Scott, L.M.et al. (2016) Vibratory urticaria associated with a missense variant in ADGRE2. N. Engl. J. Med. 374, 656–663 10.1056/NEJMoa1500611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hamann, J., Hsiao, C.C., Lee, C.S., Ravichandran, K.S. and Lin, H.H. (2016) Adhesion GPCRs as Modulators of Immune Cell Function. In Adhesion G Protein-Coupled Receptors: Molecular, Physiological and Pharmacological Principles in Health and Disease (Langenhan, T. and Schöneberg, T., eds), pp. 329–150, Springer International Publishing, Cham [Google Scholar]
- 92.Hilbig, D., Dietrich, N., Wandel, E., Gonsior, S., Sittig, D., Hamann, J.et al. (2018) The interaction of CD97/ADGRE5 With β-Catenin in adherens junctions Is lost during colorectal carcinogenesis. Front. Oncol. 8, 182 10.3389/fonc.2018.00182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.I, K.-Y., Huang, Y.S, Hu, C.-H., Tseng, W.-Y., Cheng, C.-H., Stacey, M.et al. (2017) Activation of adhesion GPCR EMR2/ADGRE2 induces macrophage differentiation and inflammatory responses via Gα16/Akt/MAPK/NF-κB signaling pathways. Front. Immunol. 8, 373 10.3389/fimmu.2017.00373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vallon, M. and Essler, M. (2006) Proteolytically processed soluble tumor endothelial marker (TEM) 5 mediates endothelial cell survival during angiogenesis by linking integrin αvβ3 to glycosaminoglycans. J. Biol. Chem. 281, 34179–34188 10.1074/jbc.M605291200 [DOI] [PubMed] [Google Scholar]
- 95.Chong, Z.-S., Ohnishi, S., Yusa, K. and Wright, G.J. (2018) Pooled extracellular receptor-ligand interaction screening using CRISPR activation. Genome Biol. 19, 205 10.1186/s13059-018-1581-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Krishnan, A., Nijmeijer, S., de Graaf, C. and Schiöth, H.B. (2016) Classification, nomenclature, and structural aspects of adhesion GPCRs. In Adhesion G Protein-Coupled Receptors (Langenhan, T. and Schöneberg, T., eds), pp. 15–41, Springer International Publishing, Cham: [DOI] [PubMed] [Google Scholar]
- 97.Spiess, K., Bagger, S.O., Torz, L.J., Jensen, K.H.R., Walser, A.L., Kvam, J.M.et al. (2019) Arrestin-independent constitutive endocytosis of GPR125/ADGRA3. Ann. N. Y. Acad. Sci. 1456, 186–199 10.1111/nyas.14263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang, X.-J., Zhang, D.-L., Xu, Z.-G., Ma, M.-L., Wang, W.-B., Li, L.-L.et al. (2014) Understanding cadherin EGF LAG seven-pass G-type receptors. J. Neurochem. 131, 699–711 10.1111/jnc.12955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lindenmaier, L.B., Parmentier, N., Guo, C., Tissir, F. and Wright, K.M. (2019) Dystroglycan is a scaffold for extracellular axon guidance decisions. eLife 8, e42143 10.7554/eLife.42143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Formstone, C.J., Moxon, C., Murdoch, J., Little, P. and Mason, I. (2010) Basal enrichment within neuroepithelia suggests novel function(s) for Celsr1 protein. Mol. Cell Neurosci. 44, 210–222 10.1016/j.mcn.2010.03.008 [DOI] [PubMed] [Google Scholar]
- 101.Shima, Y., Kengaku, M., Hirano, T., Takeichi, M. and Uemura, T. (2004) Regulation of dendritic maintenance and growth by a mammalian 7-pass transmembrane cadherin. Dev. Cell 7, 205–216 10.1016/j.devcel.2004.07.007 [DOI] [PubMed] [Google Scholar]
- 102.Bianchi, E., Sun, Y., Almansa-Ordonez, A., Woods, M., Goulding, D., Martinez-Martin, N.et al. (2021) Control of oviductal fluid flow by the G-protein coupled receptor Adgrd1 is essential for murine embryo transit. Nat. Commun. 12, 1251 10.1038/s41467-021-21512-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fischer, L., Wilde, C., Schöneberg, T. and Liebscher, I. (2016) Functional relevance of naturally occurring mutations in adhesion G protein-coupled receptor ADGRD1 (GPR133). BMC Genomics 17, 609 10.1186/s12864-016-2937-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bohnekamp, J. and Schöneberg, T. (2011) Cell adhesion receptor GPR133 couples to Gs protein. J. Biol. Chem. 286, 41912–6 10.1074/jbc.C111.265934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lum, A.M., Wang, B.B., Beck-Engeser, G.B., Li, L., Channa, N. and Wabl, M. (2010) Orphan receptor GPR110, an oncogene overexpressed in lung and prostate cancer. BMC Cancer 10, 40 10.1186/1471-2407-10-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fredriksson, R., Lagerström, M.C., Höglund, P.J. and Schiöth, H.B. (2002) Novel human G protein-coupled receptors with long N-terminals containing GPS domains and Ser/Thr-rich regions. FEBS Lett. 531, 407–414 10.1016/S0014-5793(02)03574-3 [DOI] [PubMed] [Google Scholar]
- 107.LopezJimenez, N.D, Sainz, E., Cavenagh, M.M, Cruz-Ithier, M.A, Blackwood, C.A, Battey, J.Fet al. . Two novel genes, Gpr113, which encodes a family 2 G-protein-coupled receptor, and Trcg1, are selectively expressed in taste receptor cells. Genomics 2005;85:472–482. 10.1016/j.ygeno.2004.12.005 [DOI] [PubMed] [Google Scholar]
- 108.Abe, J., Suzuki, H., Notoya, M., Yamamoto, T. and Hirose, S. (1999) Ig-Hepta, a novel member of the G protein-coupled hepta-helical receptor (GPCR) family that Has immunoglobulin-like repeats in a long N-terminal extracellular domain and defines a New subfamily of GPCRs. J. Biol. Chem. 274, 19957–19964 10.1074/jbc.274.28.19957 [DOI] [PubMed] [Google Scholar]
- 109.Fukuzawa, T., Ishida, J., Kato, A., Ichinose, T., Ariestanti, D.M., Takahashi, T.et al. (2013) Lung surfactant levels are regulated by Ig-Hepta/GPR116 by monitoring surfactant protein D. PLoS ONE 8, e69451 10.1371/journal.pone.0069451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Prömel, S., Waller-Evans, H., Dixon, J., Zahn, D., Colledge, W.H., Doran, J.et al. (2012) Characterization and functional study of a cluster of four highly conserved orphan adhesion-GPCR in mouse. Dev. Dyn. 241, 1591–1602 10.1002/dvdy.23841 [DOI] [PubMed] [Google Scholar]
- 111.Bridges, J.P., Safina, C., Pirard, B., Brown, K., Filuta, A., Bouhelal, R.et al. (2021) Activation of GPR116/ADGRF5 by its tethered agonist requires key amino acids in extracellular loop 2 of the transmembrane region. Cell Biol. 10.1101/2021.04.01.438115 [DOI] [Google Scholar]
- 112.Tang, X., Jin, R., Qu, G., Wang, X., Li, Z., Yuan, Z.et al. (2013) GPR116, an adhesion G-Protein–Coupled receptor, promotes breast cancer metastasis via the Gαq-p63RhoGEF-Rho GTPase pathway. Cancer Res. 73, 6206–6218 10.1158/0008-5472.CAN-13-1049 [DOI] [PubMed] [Google Scholar]
- 113.Duman, J.G., Tu, Y.-K. and Tolias, K.F. (2016) Emerging roles of BAI adhesion-GPCRs in synapse development and plasticity. Neural Plast. 2016, 8301737 10.1155/2016/8301737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Koh, J.T., Lee, Z.H., Ahn, K.Y., Kim, J.K., Bae, C.S., Kim, H.-H.et al. (2001) Characterization of mouse brain-specific angiogenesis inhibitor 1 (BAI1) and phytanoyl-CoA alpha-hydroxylase-associated protein 1, a novel BAI1-binding protein. Mol. Brain Res. 87, 223–237 10.1016/S0169-328X(01)00004-3 [DOI] [PubMed] [Google Scholar]
- 115.Park, D., Tosello-Trampont, A.-C., Elliott, M.R., Lu, M., Haney, L.B., Ma, Z.et al. (2007) BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 450, 430–434 10.1038/nature06329 [DOI] [PubMed] [Google Scholar]
- 116.Das, S., Sarkar, A., Ryan, K.A., Fox, S., Berger, A.H., Juncadella, I.J.et al. (2014) Brain angiogenesis inhibitor 1 is expressed by gastric phagocytes during infection with helicobacter pylori and mediates the recognition and engulfment of human apoptotic gastric epithelial cells. FASEB J. 28, 2214–2224 10.1096/fj.13-243238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Koh, J.T., Kook, H., Kee, H.J., Seo, Y.-W., Jeong, B.C., Lee, J.H.et al. (2004) Extracellular fragment of brain-specific angiogenesis inhibitor 1 suppresses endothelial cell proliferation by blocking αvβ5 integrin. Exp. Cell Res. 294, 172–184 10.1016/j.yexcr.2003.11.008 [DOI] [PubMed] [Google Scholar]
- 118.Kaur, B., Cork, S.M., Sandberg, E.M., Devi, N.S., Zhang, Z., Klenotic, P.A.et al. (2009) Vasculostatin inhibits intracranial glioma growth and negatively regulates in vivo angiogenesis through a CD36-Dependent mechanism. Cancer Res. 69, 1212–1220 10.1158/0008-5472.CAN-08-1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zencir, S., Ovee, M., Dobson, M.J., Banerjee, M., Topcu, Z. and Mohanty, S. (2011) Identification of brain-specific angiogenesis inhibitor 2 as an interaction partner of glutaminase interacting protein. Biochem. Biophys. Res. Commun. 411, 792–797 10.1016/j.bbrc.2011.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kakegawa, W., Mitakidis, N., Miura, E., Abe, M., Matsuda, K., Takeo, Y.H.et al. (2015) Anterograde C1ql1 signaling Is required in order to determine and maintain a single-Winner climbing fiber in the mouse cerebellum. Neuron 85, 316–329 10.1016/j.neuron.2014.12.020 [DOI] [PubMed] [Google Scholar]
- 121.Bolliger, M.F., Martinelli, D.C. and Sudhof, T.C. (2011) The cell-adhesion G protein-coupled receptor BAI3 is a high-affinity receptor for C1q-like proteins. Proc Natl Acad Sci. U S A. 108, 2534–2539 10.1073/pnas.1019577108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sigoillot, S.M., Iyer, K., Binda, F., González-Calvo, I., Talleur, M., Vodjdani, G.et al. (2015) The secreted protein C1QL1 and Its receptor BAI3 control the synaptic connectivity of excitatory inputs converging on cerebellar purkinje cells. Cell Rep. 10, 820–832 10.1016/j.celrep.2015.01.034 [DOI] [PubMed] [Google Scholar]
- 123.Purcell, R.H., Toro, C., Gahl, W.A. and Hall, R.A. (2017) A disease-associated mutation in the adhesion GPCR BAI2 (ADGRB2) increases receptor signaling activity. Hum. Mutat. 38, 1751–1760 10.1002/humu.23336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fredriksson, R., Gloriam, D.E.I., Höglund, P.J., Lagerström, M.C. and Schiöth, H.B. (2003) There exist at least 30 human G-protein-coupled receptors with long Ser/Thr-rich N-termini. Biochem. Biophys. Res. Commun. 301, 725–734 10.1016/S0006-291X(03)00026-3 [DOI] [PubMed] [Google Scholar]
- 125.Luo, R., Jeong, S.-J., Jin, Z., Strokes, N., Li, S. and Piao, X. (2011) G protein-coupled receptor 56 and collagen III, a receptor-ligand pair, regulates cortical development and lamination. Proc Natl Acad Sci. U S A. 108, 12925–12930 10.1073/pnas.1104821108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xu, L., Begum, S., Hearn, J.D. and Hynes, R.O. (2006) GPR56, an atypical G protein-coupled receptor, binds tissue transglutaminase, TG2, and inhibits melanoma tumor growth and metastasis. Proc. Natl Acad. Sci. U.S.A. 103, 9023–9028 10.1073/pnas.0602681103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhu, B., Luo, R., Jin, P., Li, T., Oak, H.C., Giera, S.et al. (2019) GAIN domain-mediated cleavage is required for activation of G protein-coupled receptor 56 (GPR56) by its natural ligands and a small-molecule agonist. J. Biol. Chem. 294, 19246–19254 10.1074/jbc.RA119.008234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jin, G., Sakitani, K., Wang, H., Jin, Y., Dubeykovskiy, A., Worthley, D.L.et al. (2017) The G-protein coupled receptor 56, expressed in colonic stem and cancer cells, binds progastrin to promote proliferation and carcinogenesis. Oncotarget 8, 40606–40619 10.18632/oncotarget.16506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Moriguchi, T., Haraguchi, K., Ueda, N., Okada, M., Furuya, T. and Akiyama, T. (2004) DREG, a developmentally regulated G protein-coupled receptor containing two conserved proteolytic cleavage sites. Genes Cells 9, 549–560 10.1111/j.1356-9597.2004.00743.x [DOI] [PubMed] [Google Scholar]
- 130.Stehlik, C., Kroismayr, R., Dorfleutner, A., Binder, B.R. and Lipp, J. (2004) VIGR - a novel inducible adhesion family G-protein coupled receptor in endothelial cells. FEBS Lett. 569, 149–155 10.1016/j.febslet.2004.05.038 [DOI] [PubMed] [Google Scholar]
- 131.Petersen, S.C., Luo, R., Liebscher, I., Giera, S., Jeong, S.-J., Mogha, A.et al. (2015) The adhesion GPCR GPR126 Has distinct, domain-Dependent functions in schwann cell development mediated by interaction with laminin-211. Neuron 85, 755–769 10.1016/j.neuron.2014.12.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Küffer, A., Lakkaraju, A.K.K., Mogha, A., Petersen, S.C., Airich, K., Doucerain, C.et al. (2016) The prion protein is an agonistic ligand of the G protein-coupled receptor Adgrg6. Nature 536, 464–468 10.1038/nature19312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Peeters, M.C., Fokkelman, M., Boogaard, B., Egerod, K.L., van de Water, B., IJzerman, A.P.et al. (2015) The adhesion G protein-coupled receptor G2 (ADGRG2/GPR64) constitutively activates SRE and NFκB and is involved in cell adhesion and migration. Cell Signal. 27, 2579–2588 10.1016/j.cellsig.2015.08.015 [DOI] [PubMed] [Google Scholar]
- 134.Wilde, C., Fischer, L., Lede, V., Kirchberger, J., Rothemund, S., Schöneberg, T.et al. (2016) The constitutive activity of the adhesion GPCR GPR114/ADGRG5 is mediated by its tethered agonist. FASEB J. 30, 666–673 10.1096/fj.15-276220 [DOI] [PubMed] [Google Scholar]
- 135.Yeung, J., Adili, R., Stringham, E.N., Luo, R., Vizurraga, A., Rosselli-Murai, L.K.et al. (2020) GPR56/ADGRG1 is a platelet collagen-responsive GPCR and hemostatic sensor of shear force. Proc. Natl Acad. Sci. U.S.A. 117, 28275–28286 10.1073/pnas.2008921117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hsiao, C.-C., Chu, T.-Y., Wu, C.-J., van den Biggelaar, M., Pabst, C., Hébert, J.et al. (2018) The adhesion G protein-Coupled receptor GPR97/ADGRG3 Is expressed in human granulocytes and triggers antimicrobial effector functions. Front. Immunol. 9, 2830 10.3389/fimmu.2018.02830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gupte, J., Swaminath, G., Danao, J., Tian, H., Li, Y. and Wu, X. (2012) Signaling property study of adhesion G-protein-coupled receptors. FEBS Lett. 586, 1214–1219 10.1016/j.febslet.2012.03.014 [DOI] [PubMed] [Google Scholar]
- 138.Mogha, A., Benesh, A.E., Patra, C., Engel, F.B., Schoneberg, T., Liebscher, I.et al. (2013) Gpr126 functions in schwann cells to control differentiation and myelination via G-Protein activation. J. Neurosci. 33, 17976–17985 10.1523/JNEUROSCI.1809-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]