Abstract
Lipid enveloped viruses contain a lipid bilayer coat that protects their genome to help facilitate entry into the new host cell. This lipid bilayer comes from the host cell which they infect. After viral replication, the mature virion hijacks the host cell plasma membrane where it is then released to infect new cells. This process is facilitated by the interaction between phospholipids that make up the plasma membrane and specialized viral matrix proteins. This step in the viral lifecycle may represent a viable therapeutic strategy for small molecules that aim to block enveloped virus spread. In this review, we summarize the current knowledge on the role of plasma membrane lipid–protein interactions on viral assembly and budding.
Keywords: Ebola virus, HIV, lipid–protein interaction, Marburg virus, virus assembly, virus budding
Introduction
Lipid-enveloped viruses possess a bilayer membrane that is acquired at the late stage of virus assembly and budding. During this process the nucleocapsid and accessory viral proteins become engulfed within the cellular membrane that is studded with viral transmembrane glycoproteins. The host lipid bilayer, which becomes the viral lipid envelope, surrounds the viral nucleocapsid and genome and harbors the transmembrane viral glycoproteins. Viral budding (i.e. the process of pinching the virion off from the host cell) occurs when the virus is released into the extracellular space [1]. The assembly and budding of new virions involves selective lipid–protein interactions between the host cellular membrane and viral proteins [2]. The main driver of lipid-enveloped virus budding is known as the matrix protein (or M) for a number of viruses harboring an RNA genome [3].
Several lipid-enveloped viruses, such as filoviruses (Ebola virus (EBOV) and Marburg virus (MARV), retroviruses (e.g. HIV-1) and influenza viruses, assemble and bud directly from the host cell plasma inner membrane. The plasma membrane bilayer is asymmetric in healthy cells and consists of an outer leaflet composed mainly of phosphatidylcholine (PC), sphingomyelin (SM) and glycosphingolipids and an inner leaflet enriched with phosphatidylethanolamine (PE), phosphatidylserine (PS), phosphatidylinositol (PI) and phosphoinositides (PIPs) [4]. The lipids enriched in the inner leaflet of the plasma membrane (PM) are predominantly anionic lipids, which attract proteins with cationic binding domains [5]. PS is the most abundant anionic lipid in human cells and plays a major role in the recruitment of proteins that contain PS binding domains or PS selective motifs as well as interactions with clusters of cationic residues on proteins [6]. PS and other anionic lipids are known to interact with several viral matrix proteins. These viral matrix proteins play an important role in the budding and assembly of enveloped viruses. Despite PS being predominantly localized to the PM inner leaflet in healthy cells, PS has been shown to be exposed at sites of virus budding and reside in the viral envelope outer leaflet, where it can play an important role in virus attachment and entry [7].
This review focuses on recent studies of the lipid–protein interactions that are involved in enveloped virus assembly and budding with an emphasis on viruses that bud from the plasma membrane. Here we highlight the role of viral matrix proteins, matrix–host protein interactions, and post-translational modifications of matrix proteins in facilitating the assembly and budding of filoviruses (EBOV and MARV), HIV-1 and influenza.
Filovirus assembly and budding
VP40 lipid binding and selectivity
EBOV and MARV are filoviruses and are some of the most dangerous pathogens on earth with high fatality rates, a negative sense RNA genome, and a lipid envelope derived from the host cell PM (Figure 1). Filoviruses encode the matrix protein VP40, which regulates viral assembly and budding at the PM through the formation of a large matrix of VP40 oligomers [7]. EBOV VP40 (eVP40) and MARV VP40 (mVP40) are dimeric proteins that harbor a N-terminal domain (NTD) and C-terminal domain (CTD) [8–10]. The NTD has a critical alpha-helical dimerization interface while the CTD is heavily involved in lipid binding and host protein interactions [8]. Mutation of residues in the NTD alpha-helical dimer interface not only inhibits eVP40 and mVP40 dimerization [8, 10] but also results in loss of VP40 plasma membrane localization [9,11].
Figure 1. Schematic depicting the Ebola virus (EBOV) matrix protein VP40 oligomeric forms and assembly, budding and scission of the EBOV.
(1) VP40 is synthesized and exists primarily as a dimer at very low protein concentrations that is mediated by alpha-helical NTD interactions. Mutation of residues in the dimer interface will significantly impair VP40 dimerization as well as its localization to the plasma membrane. (2) VP40 dimers are able to rearrange into an octameric form that is triggered by RNA binding [12]. The ring octamer is formed through an oligomerization interface in the NTD that is distinct from the N-terminal alpha-helical dimer interface. (3) VP40 dimers are trafficked to the plasma membrane and hijack the COPII vesicle protein, Sec24C for this process. VP40 also undergoes a number of post-translational modifications that are important for protein stability and viral budding including SUMOylation, ubiquitylation and phosphorylation. (4) Additionally, VP40 moves at the plasma membrane in a ballistic motion that is dependent on actin polymerization. Actin also plays an important role in increasing virus egress. (5) At the plasma membrane inner leaflet, VP40 binds PS and PI(4,5)P2 and can assemble into (6) oligomers via CTD–CTD interactions. VP40 oligomerization and formation of the virus matrix layer is an essential step in proper virion or VLP formation. (7) As VP40 oligomerization occurs, PS can be detected on the outer plasma membrane leaflet and exposed PS plays an important role in viral attachment and entry through host receptor interactions. (8) As formation of VP40 assembly and virus budding sites ensues, the virus nucleocapsid is recruited to sites of assembly through an actin-dependent mechanism. (9) A number of different host factors interact with VP40 at or near the plasma membrane including Tsg101, NEDD4, IQGAP1 and Amot. These interactions help facilitate the late stages of virus budding and membrane scission. (10) Mature virions or VLPs are released from host cell membrane giving rise to filamentous particles. Created with BioRender.com.
eVP40 binds membranes that contain PS and this binding regulates eVP40 localization and oligomerization at the plasma membrane. Lowering the levels of PS in mammalian cells inhibits assembly and egress of eVP40 into virus-like particles (VLPs) [7]. eVP40 binds PS in the PM inner leaflet through two cationic patches in the C-terminal domain (CTD1 contains Lys224 and Lys225 and CTD2 contains Lys274 and Lys275). When these lysine residues were substituted with alanine, PS binding affinity was significantly reduced and eVP40 localization to the plasma membrane was diminished [13]. The PS selectivity of eVP40 was evident when comparing PS binding of eVP40 to other glycerophospholipids such as phosphatidylglycerol or phosphatidic acid as well as when arginine residues replaced lysine residues in CTD1 and CTD2. While arginine residues in CTD1 and 2 could support binding of eVP40 to PS containing vesicles, the affinity was reduced ∼17-fold compared with WT VP40 [13].
PS binding is a critical and selective factor in budding of EBOV but is not the only lipid factor required for eVP40 oligomerization and viral budding [14]. Phosphatidylinositol-4,5-bisphosphate (PI(4,5)P2), which is found enriched in the PM inner leaflet, has been shown to promote and stabilize eVP40 oligomers for proper assembly and budding (Figure 1). Depletion of PI(4,5)P2 leads to a substantial reduction in large eVP40 oligomers and reduction in VLP formation [15]. Monitoring eVP40 protein dynamics in vitro indicated that PI(4,5)P2 stabilizes key contact residues that may be important for eVP40 oligomer stability. PI(4,5)P2 may also play this important role in eVP40 oligomer stability as eVP40 clusters PI(4,5)P2 [16] and PS becomes exposed at the outer PM during viral budding [14].
Since the discovery of MARV in 1967, little information is available on mVP40 interactions with host proteins and lipids. mVP40 underlies the inner leaflet of the virus and regulates budding from the host cell membrane [17]. In contrast with eVP40, mVP40 is promiscuous in its interaction with anionic phospholipids. mVP40 associated similarly with vesicles containing different types of phosphatidylinositols (PIPs) regardless of the position of the phosphate groups [17]. mVP40 acts as a charge sensor, its binding to the plasma membrane is dependent on the anionic charge density of the plasma membrane and when this charge density is neutralized there is a dramatic reduction in mVP40 localization [17]. Despite the promiscuity in mVP40 lipid binding, proper oligomerization of mVP40 dimers is mediated by PS and PI(4,5)P2 where PS is required for NTD–NTD interactions and PI(4,5)P2 for CTD–CTD interactions [18]. As with eVP40, mVP40 has two loop regions in the CTD that contain cationic residues and form a lipid binding basic patch. As the dimer approaches the membrane, the number of contacts it makes with anionic lipids increases while contacts with the neutral hydrophobic acyl chains do not [19]. This is consistent with the lack of membrane penetration observed for mVP40 dimers in vitro [20] and in Cryo ET studies [9].
VP40 trafficking
eVP40 rearranges into at least three different oligomeric structures: dimer, ring octamer, and oligomer [10]. Each has a distinct function required by the viral life cycle. The dimeric eVP40 is trafficked to the inner leaflet of the plasma membrane [21]. Once the dimers associate with the PM, they undergo structural rearrangements into an array of dimer–dimer interactions [9, 18, 22]. The oligomers form extended VP40 matrix layers via CTD–CTD interactions. eVP40 structures have different affinities for PS. Dimeric eVP40 has a higher affinity for PS than the monomer and although not fully elucidated, the dimer–dimer oligomers likely have a high avidity for the anionic membrane. In contrast, the RNA binding octameric ring has 10-fold lower affinity for PS containing membranes, which may explain the lack of eVP40 ring octamers at the PM [13].
eVP40 trafficking to the plasma membrane and subsequent budding is facilitated by interaction with different host proteins. eVP40 contains two late (L) domains (PTAP and PPxY) that hijack proteins that facilitate viral budding (i.e. the ESCRT machinery). The PPxY domain helps recruit ubiquitin E3 ligases such as NEDD4, ITCH and SMURF through interaction with their WW domains. VP40-E3 ligase interactions together with the host ESCRT pathway facilitate virus egress [23]. Notably, NEDD4 acetylation by the acetyltransferase P300 has been reported as critical for VP40 ubiquitylation and EBOV egress [24]. eVP40 also interacts with Tsg101 (Figure 1), a component of the ESCRT machinery at sites of viral assembly to facilitate the pinching off of budding vesicles allowing for the release of viral particles [25].
In addition to VP40 interactions with ubiquitin E3 ligases via the VP40 PPxY domain there is a functional relationship between eVP40 and host proteins that contain the PPxY domain. For instance, angiomotin (Amot) is a multifunctional protein that contains PPxY domains and regulates actin dynamics and cellular migration. The egress of eVP40 VLPs and authentic EBOV was significantly reduced in knockdown Amot cells. In addition, the co-expression of eVP40 and Amot rescued the inhibition of eVP40 VLP egress [26]. Amot is proposed to help facilitate the interaction of eVP40 with actin which is important for EBOV egress and oligomerization. Time lapse imaging showed that eVP40 colocalized with and moved on actin fibers in cells, this movement caused smaller eVP40 particles to form larger oligomers [27]. A truncated version of Amot which lacks all PPxY motifs and the F-actin binding domain failed to rescue the inhibition of eVP40 VLP egress [26]. TIRF microscopy images also showed that in the absence of Amot the actin cytoskeleton appears less organized and does not colocalize with eVP40 VLPs [26].
In addition to WW-domain and PPxY domain interactions, several other proteins have been identified to facilitate VP40 intracellular transport. eVP40 relies on the COPII transport system through interaction with Sec24C for its intracellular transport [28]. COPII is a coat protein (vesicle coat protein) that transports proteins from the rough endoplasmic reticulum to the Golgi apparatus. COPII is composed of five proteins: Sar1, Sec23, Sec24, Sec13 and Sec31. Yamayoshi et al. showed that Sec24C interacts with eVP40 via the region of amino acids 303–307, amino acid substitutions in eVP40 that abolish this interaction had a negative effect on eVP40 accumulation at the plasma membrane [28]. Rab14 has recently been identified to also play an important role in intracellular trafficking of eVP40. Rab14 is a small GTPase that belongs to the Ras superfamily and is involved in various aspects of membrane trafficking. Fan et, al used proximity-dependent biotin-identification (BioID) to identify Rab14 GTPase as one of the interacting partners for eVP40. eVP40 colocalized with Rab14 when expressed in HeLa cells and in the absence of or with decreased Rab14, VP40 plasma membrane localization was diminished [21]. While less information on the mVP40 trafficking mechanism is available, mutation of a hydrophobic patch in the mVP40 CTD greatly diminished mVP40 plasma membrane localization [11].
VP40 post-translational modifications
VP40 has been shown to undergo several post-translational modifications including phosphorylation, ubiquitylation, and SUMOylation (Small Ubiquitin-like Modifier (SUMO)), which are important for VP40 stability and/or virus replication. Baz-Martínez et al. [29] demonstrated that VP40 is modified by SUMO and that SUMO is included in VLPs. They further determined that the lysine residue 326 is involved in SUMOylation and that mutation of this residue reduced the stability of VP40 and incorporation of SUMO into VLPs [29]. This study suggests that EBOV hijacks the cellular SUMOylation system to modify and stabilize VP40. Ubiquitination of VP40 is important for the budding of eVP40 VLPs. The L-domain of VP40 interacts with the NEDD4 ubiquitin ligase leading to VP40 ubiquitination. The expression of Interferon-stimulated gene 15 (ISG15), another ubiquitin-like protein, inhibits the budding of eVP40 VLPs. ISG15 competes with eVP40 for interaction with Nedd4 ubiquitin ligase inhibiting ubiquitination of VP40 and consequently eVP40 VLP budding. When the L-domain of VP40 is mutated and VP40 cannot interact with NEDD4, ISG15 does not inhibit eVP40 VLP release [30].
Phosphorylation of VP40 facilitates efficient virus assembly and egress. Studies have shown that mVP40 is phosphorylated at tyrosine residues 7,10,13 and 29 and this phosphorylation is important for efficient assembly and production of infectious virions [31]. When these tyrosine residues are mutated diminishing mVP40 phosphorylation, mVP40 is still able to bind cellular membranes and produce filamentous VLPs. However, mutants have impaired ability to recruit nucleocapsid structures and released VLPs have low infectivity [31]. mVP40 Tyr phosphorylation is hypothesized to be an important factor for the efficient recruitment of nucleocapsid and proper assembly into infectious virions.
eVP40 has also been shown to be phosphorylated on tyrosine residues (Tyr13 and Tyr292) and while the mechanism is not very well understood, Tyr13 phosphorylation is thought to be important for VLP egress [32]. Mutation of Tyr13 to alanine decreased the release of Ebola VLPs. The phosphorylation of eVP40 was found to be modulated by the host c-Abl1 tyrosine kinase [32]. The release of EBOV VLPs was inhibited by c-Abl1 knockdown and inhibition with a small molecule [32]. Thus, c-Abl1 is proposed to regulate the proper budding and/or release of EBOV through a mechanism involving Tyr phosphorylation of eVP40. While a number of VP40 post-translational modifications have been described the structural consequences on the different VP40 structural forms are still unknown.
HIV assembly and budding
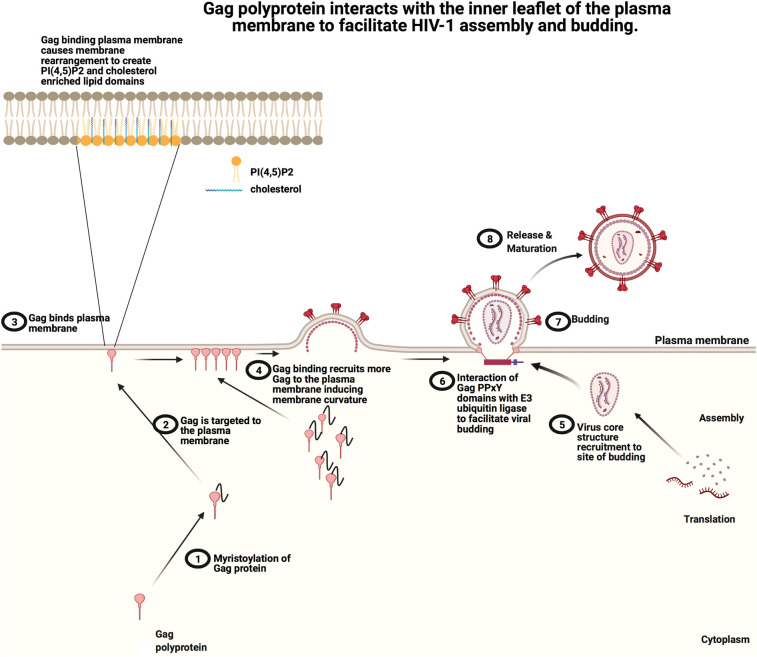
The human immunodeficiency virus (HIV) is the causative agent of acquired immune deficiency syndrome (AIDS). Currently, there are more than 40 million people in the world that are infected with HIV-1. HIV-1 egresses from an infected cell via assembly and budding at the plasma membrane. The virus encodes for the Gag polyprotein that is necessary for binding to the plasma membrane inner leaflet and making the protein–protein and lipid–protein interactions that are necessary for HIV-1 budding [33]. Gag consists of four domains dubbed p17 matrix (MA), p24 capsid (CA), p7 nucleocapsid (NC) and p6. HIV-1 creates its own specific lipid environment for virus assembly and budding through interactions of the Gag matrix domain and specific lipids in the plasma membrane.
Gag self assembles at the inner leaflet of the host cell plasma membrane where it binds the plasma membrane through an interaction of the Gag matrix domain and PI(4,5)P2 (Figure 2). Gag assembly leads to the formation of cholesterol enriched PIP2 liquid-disordered domains at an early stage of virus assembly [34]. Analysis of HIV-1 late stage assembly showed that once the virion has assembled, Gag selectively traps PI(4,5)P2 and cholesterol but does not trap PE or SM [34]. Studies have shown that cholesterol plays a role in Gag matrix protein binding to membranes as well as in the stabilization of PI(4,5)P2 domains. Molecular dynamics simulations have shown that Gag matrix protein also interacts better with the charged headgroups of PS in the membrane in the presence of cholesterol [35]. The trapping of cholesterol by Gag enhances PI(4,5)P2 clustering and makes the membrane surface more anionic due to the PI(4,5)P2 enrichment. This data suggests that Gag assembly promotes lipid rearrangement in the plasma membrane creating a PI(4,5)P2/cholesterol-enriched membrane environment to facilitate the recruitment of additional Gag proteins at the sites of budding.
Figure 2. Schematic of the HIV-1 Gag assembly and budding from the host cell plasma membrane.
(1) The Gag polyprotein is myristoylated on a N-terminal glycine, which (2) facilitates membrane insertion into the plasma membrane inner leaflet. (3) At the plasma membrane, Gag is able to interact with PI(4,5)P2 and PS, which helps to facilitate Gag oligomerization and cluster PI(4,5)P2 and cholesterol. (4) Gag oligomerization and clustering of PI(4,5)P2 and cholesterol into enriched regions of the plasma membrane, further induces recruitment of Gag proteins and likely facilitates changes in membrane curvature. (5) As the Gag mediated bud site matures, the virus core is recruited and (6) interactions with host proteins such as an E3 ligase (7) facilitate viral budding. (8) Finally, a mature HIV Gag virion is released from the plasma membrane. Adapted from ‘HIV Replication Cycle’ by BioRender.com (2021). Retrieved from https://app.biorender.com/biorender-templates.
Gag assembly at the plasma membrane is facilitated by myristoylation (a lipidation modification where a myristoyl group is covalently attached at the N-terminal glycine residue) and binding to NEDD4 E3 ubiquitin ligase family members through the PPxY domain. Gag is myristoylated at its N-terminus, this facilitates its trafficking and association to the plasma membrane allowing for protein–protein and protein–lipid interactions necessary for budding [36]. Studies in which the Gag N-terminal glycine was mutated demonstrated inhibition of myristoylation and production of viral particles [36]. Myristoylation is thought to help Gag bind the plasma membrane inner leaflet and is essential for efficient assembly of HIV into active virions. Retroviral Gag proteins also contain the PPxY domains that recruit proteins of the NEDD4 E3 ubiquitin ligase family to facilitate virus release. Mercenne et al. [37] showed that Amot can bind both NEDD4L and Gag and is required for the efficient budding of HIV-1. Using electron microscopy they observed that when Amot is present, Gag proteins can assemble into spheres and the virus can successfully bud, however, when Amot was depleted, incomplete spheres of Gag proteins accumulated on the inner membrane surface and the virus particles were not released [37]. Large amounts of actin have been detected in HIV-1 virions and this is proposed to be incorporated by interactions with the nucleocapsid domain of the Gag polyprotein [38]. Just as in EBOV, Amot may facilitate Gag polyprotein interactions with filamentous actin.
Influenza assembly and budding
The influenza virus causes seasonal epidemics where millions of people are infected globally each year. Pandemic strains of influenza have sometimes emerged as the segmented viral genome is prone to reassortment when co-infection occurs in cells. Influenza A is a single stranded negative-sense RNA virus with its viral envelope derived from the host cell plasma membrane. Influenza A encodes for the glycoproteins hemagglutinin (HA) and neuraminidase (NA) as well as the ion channel protein M2, all of which are important for viral entry as well as steps of assembly and egress [39]. Influenza A also encodes for the matrix protein (M1) which is important for virion budding and morphogenesis [40].
Influenza virus uses cholesterol enriched lipid domains in the plasma membrane as sites of virus assembly and budding. HA and NA are intrinsically associated with lipid domains, whereas the M2 and M1 proteins are excluded from these domains [41]. The exact mechanism of influenza virus assembly and budding is not currently known. In the VLP system, HA protein buds from cells in vesicles that resemble virions without requiring the expression of any other viral proteins. Similarly, the single expression of NA and M2 leads to the release of VLPs. Combining HA, NA, M2 and M1 significantly increased the amount of VLPs released [42]. One of the problems in studying influenza budding and assembly is that the VLP system does not accurately depict viral infection. During virus infection single HA expression is not efficient for virus budding, budding requires additional viral proteins, such as M2 [43]. In fact, deletion of the M2 protein or mutation of the M2 cytoplasmic tail inhibits virus budding [44]. This data suggests that HA may initiate influenza virus budding but it alone does not have the ability to complete budding and needs recruitment of other viral components like M2. It is hypothesized that HA and NA are targeted to lipid domains in the plasma membrane, this recruitment causes joining and enlargement of lipid domains and may cause membrane curvature and initiate the budding event. M1 binds to HA and NA providing a site for the recruitment of viral ribonucleoprotein particles and M2 to the site of virus budding. M2 further alters membrane curvature causing membrane scission and release of progeny virions.
Conclusions
Although enveloped virus replication pathways are now understood in considerable detail there are still gaps in understanding the role of protein–protein and lipid–protein interactions at the plasma membrane. IQGAP1 is a widely expressed scaffolding protein with the WW-domain that interacts with viral PPxY L-domains [45]. IQGAP1 interaction with eVP40 is required for EBOV budding. The L-domain regions of VP40 mediate this interaction and when IQGAP1 is suppressed there is reduced release of eVP40 VLPs [45]. IQGAP1 has also been detected in purified HIV-1 virions [46] where it interacts with the nucleocapsid and p6 domains of the HIV-1 Gag protein, negatively regulating Gag trafficking and virion release. Depletion of IQGAP1 increases HIV-1 viral particle release while its overexpression reduces viral particle release. In addition, the expression of IQGAP1 restricts Gag targeting to the plasma membrane [47]. IQGAP1 is involved in actin cytoskeletal remodeling during cell migration and formation of filopodia which are all important for viral budding. However, there is still no clear understanding on what IQGAP1, or actin specifically do, and it is still not clear why IQGAP1 expression has different effects on different viruses. In addition to this, not all enveloped viruses assemble at the plasma membrane. Coronaviruses such as SARS-CoV-2 are lipid enveloped viruses, however, their assembly occurs at the endoplasmic reticulum-Golgi intermediate compartment (ERGIC) [48] and their exocytosis through lysosomes [49].
Some progress has already been made in the development of therapeutics that target assembly and budding and these efforts will be enhanced by a greater understanding of the viral processes reviewed herein. Despite considerable progress, several key aspects of virus assembly and budding are not yet well understood and represent important research opportunities. The most pressing issues that need to be addressed include (1) characterization of the mechanism of membrane curvature generation from the plasma membrane, (2) defining the precise role of actin and accessory proteins such as IQGAP1 in viral assembly and budding, (3) determining the effects of post-translational modifications (e.g. phosphorylation and ubiquitylation) of matrix protein on structural changes, protein trafficking, lipid binding, and oligomerization and (4) defining the mechanism of membrane scission during virus budding.
Perspectives
Lipid-enveloped viruses that assemble and egress from the host cell plasma membrane can cause significant disease and mortality. There are still unmet clinical needs with respect to vaccines and therapeutics that can slow the spread or treat those who become infected for a number of these viral infections.
The last two decades have provided a basic understanding of some mechanisms by which viral matrix proteins interact with host lipids and oligomerize on both model and cellular membranes.
A better understanding of the molecular details (e.g. host-virus interactions) of virus assembly and budding from the plasma membrane may lead to pan-viral strategies to inhibit spread and dissemination of lipid-enveloped viruses.
Abbreviations
- CTD
C-terminal domain
- EBOV
Ebola virus
- HA
hemagglutinin
- ISG15
Interferon-stimulated gene 15
- MARV
Marburg virus
- NA
neuraminidase
- NTD
N-terminal domain
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
- PIPs
phosphoinositides
- PM
plasma membrane
- PS
phosphatidylserine
- SM
sphingomyelin
- SUMO
Small Ubiquitin-like Modifier
- VLPs
virus-like particles
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
These studies were supported by the NIH AI081077 to R.V.S.
Author Contributions
The review was conceived by B.B.M. and R.V.S. B.B.M. wrote the first draft and B.B.M. and R.V.S. edited and rewrote the final version.
References
- 1.Henrik, G., Roger, H. and Dirk-Jan, E.O. (1998) Virus maturation by budding. Microbiol. Mol. Biol. Rev. 62, 1171–1190 10.1128/MMBR.62.4.1171-1190.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villanueva, R.A., Rouillé, Y. and Dubuisson, J. (2005) Interactions between virus proteins and host cell membranes during the viral life cycle. Int. Rev. Cytol. 245, 171–244 10.1016/S0074-7696(05)45006-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lorizate, M. and Kräusslich, H.G. (2011) Role of lipids in virus replication. Cold Spring Harb. Perspect. Biol. 3, a004820 10.1101/cshperspect.a004820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skotland, T. and Sandvig, K. (2019) The role of PS 18:0/18:1 in membrane function. Nat. Commun. 10, 2752 10.1038/s41467-019-10711-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujimoto, T. and Parmryd, I. (2017) Interleaflet coupling, pinning, and leaflet asymmetry—major players in plasma membrane nanodomain formation. Front. Cell Dev. Biol. 4, 155 10.3389/fcell.2016.00155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soni, S.P. and Stahelin, R.V. (2014) The ebola virus matrix protein VP40 selectively induces vesiculation from phosphatidylserine-enriched membranes. J. Biol. Chem. 289, 33590–33597 10.1074/jbc.M114.586396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adu-Gyamfi, E., Johnson, K.A., Fraser, M.E., Scott, J.L., Soni, S.P., Jones, K.R.et al. (2015) Host cell plasma membrane phosphatidylserine regulates the assembly and budding of ebola virus. J. Virol. 89, 9440–9453 10.1128/JVI.01087-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oda, S.I., Noda, T., Wijesinghe, K.J., Halfmann, P., Bornholdt, Z.A., Abelson, D.M.et al. (2016) Crystal structure of Marburg virus VP40 reveals a broad, basic patch for matrix assembly and a requirement of the N-terminal domain for immunosuppression. J. Virol. 90, 1839–1848 10.1128/JVI.01597-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan, W., Clarke, M., Norris, M.J., Kolesnikova, L., Koehler, A., Bornholdt, Z.A.et al. (2020) Ebola and Marburg virus matrix layers are locally ordered assemblies of VP40 dimers. eLife 9, e59225 10.7554/eLife.59225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bornholdt, Z.A., Noda, T., Abelson, D.M., Halfmann, P., Wood, M.R., Kawaoka, Y.et al. (2013) Structural rearrangement of ebola virus VP40 begets multiple functions in the virus life cycle. Cell 154, 763–774 10.1016/j.cell.2013.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wijesinghe, K.J., McVeigh, L., Husby, M.L., Bhattarai, N., Ma, J., Gerstman, B.S.et al. (2020) Mutation of hydrophobic residues in the C-Terminal domain of the marburg virus matrix protein VP40 disrupts trafficking to the plasma membrane. Viruses 12, 482 10.3390/v12040482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landeras-Bueno, S., Wasserman, H., Oliveira, G., VanAernum, Z.L., Busch, F., Salie, Z.L.et al. (2021) Cellular mRNA triggers structural transformation of ebola virus matrix protein VP40 to its essential regulatory form. Cell Rep. 35, 108986 10.1016/j.celrep.2021.108986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Vecchio, K., Frick, C.T., Gc, J.B., Oda, S.I., Gerstman, B.S., Saphire, E.O.et al. (2018) A cationic, C-terminal patch and structural rearrangements in ebola virus matrix VP40 protein control its interactions with phosphatidylserine. J. Biol. Chem. 293, 3335–3349 10.1074/jbc.M117.816280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adu-Gyamfi, E., Soni, S.P., Xue, Y., Digman, M.A., Gratton, E. and Stahelin, R.V. (2013) The ebola virus matrix protein penetrates into the plasma membrane: a key step in viral protein 40 (VP40) oligomerization and viral egress. J. Biol. Chem. 288, 5779–5789 10.1074/jbc.M112.443960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, K.A., Taghon, G.J., Scott, J.L. and Stahelin, R.V. (2016) The Ebola Virus matrix protein, VP40, requires phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) for extensive oligomerization at the plasma membrane and viral egress. Sci. Rep. 6, 19125 10.1038/srep19125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gc, J.B., Gerstman, B.S., Stahelin, R.V. and Chapagain, P.P. (2016) The ebola virus protein VP40 hexamer enhances the clustering of PI(4,5)P2 lipids in the plasma membrane. Phys. Chem. Chem. Phys. 18, 28409–28417 10.1039/C6CP03776C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wijesinghe, K.J. and Stahelin, R.V. (2016) Investigation of the lipid binding properties of the Marburg virus matrix protein VP40. J. Virol. 906, 3074–3085 10.1128/JVI.02607-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amiar, S., Husby, M.L., Wijesinghe, K.J., Angel, S., Bhattarai, N., Gerstman, B.S.et al. (2021) Lipid-specific oligomerization of the Marburg virus matrix protein VP40 is regulated by two distinct interfaces for virion assembly. J. Biol. Chem. 296, 100796 10.1016/j.jbc.2021.100796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhattarai, N., Gerstman, B.S., Stahelin, R.V. and Chapagain, P.P. (2017) Plasma membrane association facilitates conformational changes in the marburg virus protein VP40 dimer. RSC Adv. 7, 22741–22748 10.1039/C7RA02940C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wijesinghe, K.J., Urata, S., Bhattarai, N., Kooijman, E.E., Gerstaman, B.S., Chapagain, P.P.et al. (2017) Dectection of lipid-induced structural changes of the Maburg virus matrix protein VP40 using hydrogen/deuterium exchange mass-spectrometry. J. Biol. Chem. 292, 6108–6122 10.1074/jbc.M116.758300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan, J., Liu, X., Mao, F., Yue, X., Lee, I. and Xu, Y. (2020) Proximity proteomics identifies novel function of Rab14 in trafficking of ebola virus matrix protein VP40. Biochem. Biophys. Res. Commun. 527, 387–392 10.1016/j.bbrc.2020.04.041 [DOI] [PubMed] [Google Scholar]
- 22.Gerstman, B.S. and Chapagain, P.P. (2017) Membrane association and localization dynamics of the ebola virus matrix protein VP40. Biochim. Biophys. Acta Biomembr. 1859, 2012–2020 10.1016/j.bbamem.2017.07.007 [DOI] [PubMed] [Google Scholar]
- 23.Han, Z., Sagum, C.A., Bedford, M.T., Sidhu, S.S., Sudol, M. and Harty, R.N. (2016) ITCH E3 ubiquitin ligase interacts with Ebola Virus VP40 to regulate budding. J. Virol. 90, 9163–9171 10.1128/JVI.01078-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang, L., Zhou, S., Chen, M., Yan, J., Yang, Y., Wu, L.et al. (2016) P300-mediated NEDD4 acetylation drives ebolavirus VP40 egress by enhancing NEDD4 ligase activity. PLoS Pathog. 17, e1009616 10.1371/journal.ppat.1009616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Votteler, J. and Sundquist, W.I. (2013) Virus budding and the ESCRT pathway. Cell Host Microbe 14, 232–241 10.1016/j.chom.2013.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han, Z., Ruthel, G., Dash, S., Berry, C.T., Freedman, B.D., Harty, R.N.et al. (2020) Angiomotin regulates budding and spread of ebola virus. J. Biol. Chem. 295, 8596–8601 10.1074/jbc.AC120.013171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adu-Gyamfi, E., Digman, M.A., Gratton, E. and Stahelin, R.V. (2012) Single-particle tracking demonstrates that actin coordinates the movement of the ebola virus matrix protein. Biophys. J. 103, L41–L43 10.1016/j.bpj.2012.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamayoshi, S., Noda, T., Ebihara, H., Goto, H., Morikawa, Y., Lukashevich, I.S.et al. (2008) Ebola virus matrix protein VP40 uses the COPII transport system for its intracellular transport. Cell Host Microbe 3, 168–177 10.1016/j.chom.2008.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baz-Martínez, M., El Motiam, A., Ruibal, P., Condezo, G.N., de la Cruz-Herrera, C.F., Lang, V.et al. (2016) Regulation of ebola virus VP40 matrix protein by SUMO. Sci. Rep. 6, 37258 10.1038/srep37258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okumura, A., Pitha, P.M. and Harty, R.N. (2008) ISG15 inhibits ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc. Natl Acad. Sci. U.S.A. 105, 3974–3979 10.1073/pnas.0710629105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolesnikova, L.M.E., Schudt, G., Shams-Eldin, H. and Becker, S. (2012) Phosphorylation of Marburg virus matrix protein VP40 triggers assembly of nucleocapsids with the viral envelope at the plasma membrane. Cell Microbiol. 14, 182–197 10.1111/j.1462-5822.2011.01709.x [DOI] [PubMed] [Google Scholar]
- 32.García, M., Cooper, A., Shi, W., Bornmann, W., Carrion, R., Kalman, D.et al. (2012) Productive replication of ebola virus is regulated by the c-Abl1 tyrosine kinase. Sci. Transl. Med. 4, 123ra24 10.1126/scitranslmed.3003500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sundquist, W.I. and Kräusslich, H.G. (2012) HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2, a006924 10.1101/cshperspect.a006924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Favard, C., Chojnacki, J., Merida, P., Yandrapalli, N., Mak, J., Eggeling, C.et al. (2019) HIV-1 Gag specifically restricts PI(4,5)P2 and cholesterol mobility in living cells creating a nanodomain platform for virus assembly. Sci. Adv. 5, eaaw8651 10.1126/sciadv.aaw8651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charlier, L., Louet, M., Chaloin, L., Fuchs, P., Martinez, J., Muriaux, D.et al. (2014) Coarse-Grained simulations of the HIV-1 matrix protein anchoring: Revisiting Its assembly on membrane domains. Biophys. J. 106, 577–585 10.1016/j.bpj.2013.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Resh, M.D. (2004) A myristoyl switch regulates membrane binding of HIV-1 Gag. Proc. Natl Acad. Sci. U.S.A. 101, 417–418 10.1073/pnas.0308043101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mercenne, G., Alam, S.L., Arii, J., Lalonde, M.S. and Sundquist, W.I. (2015) Angiomotin functions in HIV-1 assembly and budding. Elife 4, e03778 10.7554/eLife.03778.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rahman, S.A., Koch, P., Weichsel, J., Godinez, W.J., Schwarz, U., Rohr, K.et al. (2014) Investigating the role of F-actin in human immunodeficiency virus assembly by live-cell microscopy. J. Virol. 88, 7904–7914 10.1128/JVI.00431-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bouvier, N.M. and Palese, P. (2008) The biology of influenza viruses. Vaccine 26, D49–D53 10.1016/j.vaccine.2008.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shtykova, E.V., Baratova, L.A., Fedorova, N.V., Radyukhin, V.A., Ksenofontov, A.L., Volkov, V.V.et al. (2013) Structural analysis of influenza A virus matrix protein M1 and its self-assemblies at low pH. PLoS One 8, e82431 10.1371/journal.pone.0082431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rossman, J.S. and Lamb, R.A. (2011) Influenza virus assembly and budding. Virology 411, 229–236 10.1016/j.virol.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen, B.J., Leser, G.P., Morita, E. and Lamb, R.A. (2007) Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J.Virol. 81, 7111–7123 10.1128/JVI.00361-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rossman, J.S., Jing, X., Leser, G.P., Balannik, V., Pinto, L.H. and Lamb, R.A. (2010) Influenza virus m2 ion channel protein is necessary for filamentous virion formation. J. Virol. 84, 5078–5088 10.1128/JVI.00119-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCown, M.F. and Pekosz, A. (2005) The influenza A virus M2 cytoplasmic tail is required for infectious virus production and efficient genome packaging. J. Virol. 79, 3595–3605 10.1128/JVI.79.6.3595-3605.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu, J., Qu, J., Liu, Y., Jambusaria, Y., Han, R., Ruthel, Z.et al. (2013) Host IQGAP1 and ebola virus VP40 interactions facilitate virus-like particle egress. J. Virol. 87, 7777–7780 10.1128/JVI.00470-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chertova, E., Chertov, O., Coren, L.V., Roser, J.D., Trubey, C.M., Bess, J.W.et al. (2006) Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol. 80, 9039–9052 10.1128/JVI.01013-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sabo, Y., de Los Santos, K. and Goff, S.P. (2020) Iqgap1 negatively regulates HIV-1 gag trafficking and virion production. Cell Rep. 30, 4065–4081 10.1016/j.celrep.2020.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stertz, S., Reichelt, M., Spiegel, M., Kuri, T., Martínez-Sobrido, L., García-Sastre, A.et al. (2006) The intracellular sites of early replication and budding of SARS-coronavirus. Virology 361, 304–315 10.1016/j.virol.2006.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghosh, S., Dellibovi-Ragheb, T.A., Kerviel, A., Pak, E., Qiu, Q., Fisher, M.et al. (2020) β-coronaviruses use lysosomes for egress instead of the biosynthetic secretory pathway. Cell 183, 1520–1535.e14 10.1016/j.cell.2020.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han, Z., Madara, J.J., Herbert, A., Prugar, L.I., Ruthel, G., Lu, J.et al. (2015) Calcium regulation of hemorrhagic fever virus budding: mechanistic implications for host-oriented therapeutic intervention. PLoS Pathog. 11, e1005220 10.1371/journal.ppat.1005220 [DOI] [PMC free article] [PubMed] [Google Scholar]