Abstract
We measured SARS-CoV-2 RNA load in raw wastewater in Attica, Greece, by RT-qPCR for the environmental surveillance of COVID-19 for 6 months. The lag between RNA load and pandemic indicators (COVID-19 hospital and intensive care unit (ICU) admissions) was calculated using a grid search. Our results showed that RNA load in raw wastewater is a leading indicator of positive COVID-19 cases, new hospitalization and admission into ICUs by 5, 8 and 9 days, respectively. Modelling techniques based on distributed/fixed lag modelling, linear regression and artificial neural networks were utilized to build relationships between SARS-CoV-2 RNA load in wastewater and pandemic health indicators. SARS-CoV-2 mutation analysis in wastewater during the third pandemic wave revealed that the alpha-variant was dominant. Our results demonstrate that clinical and environmental surveillance data can be combined to create robust models to study the on-going COVID-19 infection dynamics and provide an early warning for increased hospital admissions.
Keywords: COVID-19, Wastewater-based epidemiology, RT-qPCR, Method validation, Hospital admission rates, ICU admission rates
Graphical abstract
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has grown rapidly worldwide, infecting more than 186 million people and claiming 4 million lives as of June 2021 (WHO, 2021). It can be transmitted via inhalation of airborne droplets and its main manifestation is infection of the respiratory system that ranges from innocuous to severe (Anand et al., 2021; Zhu et al., 2020). Limited diagnostic testing capacity, asymptomatic infections and pandemic fatigue to public health measures have hindered the ability to track the spread of SARS-CoV-2 and the Coronavirus disease 2019 (COVID-19) pandemic. In response, the surveillance of SARS-CoV-2 ribonucleic acid (RNA) measurements in wastewater has been used to study COVID-19 epidemiology because it is excreted into sewer system via feces, saliva, swabs and/or sputum of infected individuals (Anand et al., 2021; Peccia et al., 2020). Such excretion into wastewater can be described using the RNA load shedding profile from the total amount of virus RNA in wastewater at several time points after infection (Chen et al., 2020; Wölfel et al., 2020; Xu et al., 2020; Zhang et al., 2020).
Wastewater-based epidemiology (WBE) has been found to be a useful method to track COVID-19 and potentially other infectious diseases (Daughton, 2020). It has several advantages over individual patient testing. First, it is able to identify infections in asymptomatic, presymptomatic or mild cases (i.e., individuals unlikely to be diagnosed and tested clinically) (Kaplan et al., 2021; Peccia et al., 2020). Second, WBE is more efficient in that it reduces the number of tests required to evaluate a large population, costs considerably less, does not require patient consent, and test results are available earlier. Third, it is especially useful in locations where clinical testing is restricted, such as in poor countries in which the monitoring programs for COVID-19 are not developed or such developments are not a priority. Finally, the emergence of new variants within the population can be detected (Bar-Or et al., 2021). Reflective of these benefits, WBE has been utilized to monitor and track SARS-CoV-2 RNA within communities in many countries (Agrawal et al., 2021; Ahmed et al., 2020b; Bivins et al., 2020b; Gonzalez et al., 2020; Graham et al., 2021; Haramoto et al., 2020; Hokajarvi et al., 2021; Kumar et al., 2020; La Rosa et al., 2020; Randazzo et al., 2020; Wolfe et al., 2021).
While the number of studies reporting the detection of SARS-CoV-2 RNA in wastewater collection systems continues to grow, few reports have attempted to develop environmental surveillance tools or epidemiological models that relate SARS-CoV-2 RNA concentrations in wastewater with meaningful public health endpoints, such as hospital admission rates (Kaplan et al., 2021; Peccia et al., 2020). As such, there is a need to correlate SARS-CoV-2 RNA levels and/or SARS-CoV-2 variants in wastewater with reported COVID-19 cases to find leading indicators, e.g., time delay. Such information would allow inferences to be made about the progress of infection within the community and inform stakeholders regarding the implementation of regulations or policy measures (Bivins et al., 2020b). For example, the prediction of hospital admission rates from viral load can serve as an early-warning system for the health care infrastructure. Peccia et al. created epidemiological models after measuring the concentration of SARS-CoV-2 RNA in primary sludge over a 3 month period (Peccia et al., 2020). They reported a 2 to 8 day lag time between RNA load in the sludge and the manifestation of positive cases (as well as hospitalizations). However, due to the small sampling size, and positivity rates greater than 50% in the early pandemic case data, a direct correlation between absolute SARS-CoV-2 RNA concentrations in sludge and COVID-19 cases was not examined. Kaplan et al. used a differential equation-based epidemiological model and assumed SARS-CoV-2 shedding distributions to demonstrate that hospitalizations could be anticipated from the SARS-CoV-2 RNA load in primary sludge with a 3 to 5 day time lag (Kaplan et al., 2021). They used basic reproductive number (R0) RNA versus hospitalization rate and found the lagging indicator in a more epidemiologically meaningful way. The developed model provided a maximum error of 15 cases (from a total of 30 hospitalizations). Huisman et al. reported the lag indicators between SARS-CoV-2 RNA load in wastewater and pandemic indicators from data collected over a 4 month sampling period (Huisman et al., 2021). This study provided a computational framework to optimize fit between RNA load data and pandemic clinical indicators, specifically adjusting the testing-cases inconsistencies. These investigators reported a time delay between RNA load in wastewater/primary sludge and pandemic clinical indicators of 4 to 9 days. Using linear correlation, Medema et al. (2020) related cumulative COVID-19 cases to SARS-CoV-2 RNA load in wastewater (RNA copies/mL) with a time lag of 6 days.
Causes of the variation in the lag times between RNA detection in sewage and clinical manifestation of infection include societal responses to the pandemic, daily variations in population size, limitations and differences in the sampling in wastewater, as well as analytical approaches and inconsistencies with the clinical COVID-19 testing or the changes in the time required to report case data as the pandemic progresses (Medema et al., 2020; Peccia et al., 2020). This makes the correlation between the absolute SARS-CoV-2 RNA load and COVID-19 prevalence data less reliable (Medema et al., 2020). Normalization of wastewater RNA load to population size may partially resolve the variation in the cases due to increases/decreases in population of the city (Ahmed et al., 2020a); however more work is needed to improve the analytical methods (Ahmed et al., 2020d), population estimation, and epidemiological modelling methods for sewage surveillance of SARS-CoV-2. All of these factors affect conclusions about the lag time between RNA load in wastewater and pandemic clinical indicators. With the emergence of new SARS-Cov-2 variants that demonstrate different transmission and COVID-19 severity (Singh et al., 2021; Wang et al., 2021), their detection and quantification are important during sewage surveillance of SARS-CoV-2 to explain accurately the infection dynamics. Last but not least, the time lag variation between SARS-CoV-2 RNA load data and COVID-19 pandemic clinical indicators needs to be investigated over a longer period to validate the application of sewage surveillance of SARS-CoV-2 as an early warning system.
The aims of the present study are to: (1) use an optimized analytical method with a strict quality assurance (QA)/quality control (QC) system to determine SARS-CoV-2 RNA concentrations in 192 consecutive days of wastewater samples; (2) detect the SARS-CoV-2 variants in wastewater and the effects on pandemic clinical indicators; (3) create advanced computational workflows based on distributed lag modelling and artificial neural networks to estimate the new admission rates to hospitals or ICUs from wastewater viral loads.
2. Material and methods
2.1. Wastewater sampling
Daily composite flow proportional raw wastewater samples were collected from the wastewater treatment plant of Attica, the region of Greece that includes Athens metropolitan area and suburbs; this includes primary sedimentation, activated sludge processed with biological nitrogen and phosphorus removal and secondary sedimentation (Thomaidis et al., 2016). The wastewater treatment plant of Attica serves a large percentage of the population of Greece. All the information and the details about the studied wastewater treatment plant are provided in Table S1A of the Supporting Information (SI) document. The number of inhabitants was estimated daily based on the concentrations of total phosphorus (P), total nitrogen (N), biochemical oxygen demand (BOD), chemical oxygen demand (COD) and ammonium-nitrogen (NH4-N) (as described elsewhere) (Been et al., 2014; van Nuijs et al., 2011). The raw wastewater samples were collected daily (from August 31, 2020 through March 21, 2021) in pre-cleaned high-density polyethylene (HDPE) 2 L bottles, and transported at 4 °C to the laboratory. All samples were processed immediately upon arrival at the laboratory. Biosafety guidelines were followed during sampling, transportation and the analytical procedure (as described below). Additional details regarding sample preparation and analytical methods used for SARS-CoV-2 RNA extraction and analysis (using RT-qPCR) are provided in the Supplementary Material.
2.2. Stability study
There are only a few studies that have investigated the stability of SARS-CoV-2 concentration in wastewater under various conditions (Ahmed et al., 2020c; Bivins et al., 2020a; Hokajarvi et al., 2021). In the present study, the stability of qPCR targets for SARS-CoV-2 and Mengo Virus (MgV) in wastewater samples was investigated by measuring the levels of the N1 and N2 gene of SARS-CoV-2 and the exogenous control MgV at three different storage temperatures, i.e., 4 °C, −20 °C, and −80 °C. One wastewater sample positive for SARS-CoV-2 was mixed and divided into five aliquots of 50 mL, with 10 μL of MgV (Biomerieux, France) being spiked into each aliquot. The first aliquot was immediately analyzed. The second aliquot was stored at −20 °C for one day before being analyzed. The third aliquot was stored at −20 °C for 7 days and then analyzed. The fourth aliquot was stored for one day at 4 °C before being analyzed again. The fifth aliquot was stored at −80 °C and analyzed one day later. All experiments were performed in duplicate for the whole analytical procedure (Fig. S1).
2.3. Validation and quality control measures
To ensure the quality of the measurements and the overall analytical process, the following QA/QC measures were applied to every batch of analyses: analysis of a procedural blank sample (PCR Grade water) to evaluate cross-contamination, analysis of a positive quality control sample to insure run reliability, addition and determination of synthetic DNA as internal control (Magnetic Bead kit (IDEXX)) to assess inhibition and RNA purification, analysis of a PCR positive control and a PCR negative control and construction of a five-point calibration curve in each run. More details about QA/QC can be found in the Supplementary Material and Table S2.
2.4. Viral load
The population served by the wastewater treatment plant was calculated in real time based on the concentration levels of five physicochemical parameters (i.e., total phosphorus, total nitrogen, BOD, COD and NH4-N) for each sampling day from the beginning of the study period. In addition, flow rates from the wastewater treatment plant were provided daily (Table S1A, SIF). After the determination of virus genome copies per liter (copies/L, Section 3, SIF), the concentration was normalized to estimated population and the flow rates. In cases in which inhibition was observed (i.e., ΔCq < 2 between undiluted sample and 1:4 dilution or ΔCq < 3.3 between undiluted sample and 1:10), the viral load was corrected using the following equation:
where: RNAtotal is the total volume of RNA eluted from magnetic-bead extraction (0.1 mL); RNAPCR is the volume of purified RNA tested in PCR (0.005 mL); concentratetotal is the total volume of wastewater concentrate (0.5 mL); concentrateextracted is the volume of wastewater concentrate from which RNA was extracted (0.2 mL); wastewater is the volume of original wastewater sample processed with PEG procedure (50 mL); DF = 4 when the viral load was corrected based on Cq values for 1:4 dilution or 10 for 1:10 dilution.
2.5. SARS-CoV-2 variant analysis with high-throughput sequencing
Next-generation sequencing (NGS) was utilized for the investigation of existing variants of SARS-CoV-2 in wastewater samples in February 2021 (as described in our previous study) (Avgeris et al., 2021). Briefly, library preparation was carried out using the Ion Xpress Plus Fragment Library Kit (Ion Torrent, Thermo Fisher Scientific Inc.). Adapter ligation, nick-repair and clean-up of the ligated dsDNA were carried out according to the manufacturer's protocol. Quantification of the adapter-ligated library was performed using the Ion Library TaqMan Quantitation Kit (Ion Torrent) in an ABI 7500 Real-Time PCR system (Applied Biosystems). Emulsion PCR was employed for the template preparation process on an Ion OneTouch 2 System, while enrichment was carried out on the Ion OneTouch ES instrument, using the Ion PGM Hi-Q View OT2 kit (Ion Torrent). Finally, NGS based on the semi-conductor sequencing methodology was performed in the Ion Torrent PGM system.
Bioinformatic evaluation of the derived NGS datasets included alignment of the sequencing reads to the SARS-CoV-2 reference genome (NC_045512.2) with the Burrows-Wheeler Aligner (BWA-MEM) (Li and Durbin, 2009). The successfully aligned sequencing reads were visualized using the Integrative Genomics Viewer (IGV) (Thorvaldsdottir et al., 2013). Finally, variant calling of both SNVs and insertions/deletions was implemented using the iVar algorithm with the recommended parameters (Grubaugh et al., 2019).
2.6. Development of hospital admission rates model
The data including normalized RNA copies SARS-CoV-2/100K inhabitants, number of positive cases in Attica, and the number of new patients admitted to hospitals and ICUs were compiled for the period August 31, 2020 through March 21, 2021. The pandemic indicators (number of positive cases, new admissions to hospitals and new admissions to ICUs) were gathered from daily reports developed by the National Public Health Organization of Greece, i.e., EODY and the Ministry of Health. The coefficient of variation for log-normal distributed data (CVln) (Forootan et al., 2017) was plotted against log10(RNA copies/L) for more than 40 data points with at least 3 replicates to find a threshold at which the wastewater-based epidemiology data was statistically meaningful and they could be used for modelling purpose.
where E is efficiency of PCR method, the Cq is the quantification cycle. Each variable (RNA load in wastewater and pandemic clinical data) was treated as time-series data and was checked to detect turning points using the mean and linear slope difference test in the timeSeries R-package. The significance of turning points was evaluated by a probability value (P) (0.95) and quantity of information (I) according to Kendall's information theory (Kendall et al., 1994). If I and P gets large and small, there is a high possibility that the time-series data contain a longer sequence of decreasing or increasing trends around the detected turning point. This was used to evaluate a relationship between the variables by having similar changes in their turning points (Gocic and Trajkovic, 2013). The data was normalized between 0 and 100 for internal comparability in their turning point detection. The turning points were also used to investigate the lag between measurement of RNA copies of SARS-CoV-2 in wastewater and positive COVID-19 cases. To evaluate strength of Pearson's (linear) correlation coefficient between variables (positive cases and new admissions to hospitals or to ICUs versus RNA copies of SARS-CoV-2), several levels of averaging terms (n = 3, n = 4, n = 5 and n = 7) were applied. This was done to establish linear regression models between time-series data. The averaging was not based on a moving average, and averaging was performed independently for n consecutive days without being including in the next averaging batch. This was to decrease the effect of variation between daily activity and life/working style. The forecasting ability of the linear model was evaluated externally by the data collected between February 15 and 21, 2021. This approach was compared to the distributed lag measurement error time-series model (DLM). Although turning points allow the detection of several top (lowest p-value) lags, they may not represent the relationship between whole data and are useful only for detecting peaks, not valleys. The second method (which is used to estimate the lag value and averaging term) was based on root mean square error of leave-one-out cross validation (RMSECV). The RMSECV was calculated for averaging term and lag values between 0 and 10 (which is representative for long-term modelling of pandemic data). To check by-chance correlation, RNA load in wastewater data were randomly shuffled 10,000 times. The RMSECV, R2 and Q2 values of these shuffled models should be less than the main model to verify that the relationship between averaged SARS-CoV-2 RNA load in wastewater data and the pandemic clinical data is not random.
In addition to linear models, a multilayer artificial neural network (ANN) model was developed using a backpropagation algorithm, the leave-one-out cross validation technique and a test set to predict the pandemic clinical indicators. To construct the ANN, SARS-CoV-2 RNA copies in wastewater, fingerprint data (if RNA copies in wastewater were above limit of quantification (LOQ), SARS-CoV-2 RNA fingerprint gives a value of 1; when the inverse scenario occurs, it provides a value of 0) and positive cases of SARS-CoV-2 infections were treated as independent variables to model new admissions to hospitals. In the ANN model structure, a fixed optimized lag value was applied to the input data. ANN models were optimized and constructed in the R environment using the neuralnet R package. The third method used for evaluating the COVID-19 pandemic data was based on the association between the logarithm of the normalized SARS-CoV-2 RNA copies/100K inhabitants using a Bayesian Distributed-Lag Nonlinear Model of Poisson family with log-link (Zanobetti et al., 2000). To explore the association over a long period, a maximum lag of +50 days and minimum of −30 days were used, together with an imputation model with vague prior variance for the viral load in wastewater beyond the limits of the study period. A gamma-shaped lag-response association was assumed (Kaplan et al., 2021; Lewnard et al., 2020), i.e., the regression coefficients were constrained to be positive and to follow a gamma distribution with unknown shape and scale. This approximates the shape of both the incubation period of SARS-CoV-2 and the viral shedding in feces (Wölfel et al., 2020), making it a rational choice. Analyses were undertaken using the JAGS R package. All codes are available in http://trams.chem.uoa.gr/covid-19/.
3. Results and discussions
3.1. Optimization and validation of the analytical method
The effect of pre-analytical factors on the measurement of SARS-CoV-2 RNA load in wastewater samples was performed to allow the development of an optimized and validated method that would minimize day-to-day variations in the analytical measurements (Gerrity et al., 2021; Graham et al., 2021; Huisman et al., 2021; Peccia et al., 2020). Details about optimization and validation of the analytical methods can be found in the supplementary file which includes: (1) the effect of storage conditions on the stability of SARS-CoV-2 RNA concentrations (Fig. S1); (2) comparison of analytical methods (Table S3); and (3) method validation (Table S2). The optimized protocol was validated for limit of detection (LOD), method sensitivity, repeatability, trueness and precision, and proved to be fit for purpose. A strict QA/QC protocol was established and followed every day. In addition to these conventional method validation criteria (which can be found in the literature, e.g., Pérez-Cataluña et al., 2021; Philo et al., 2021), the storage conditions were evaluated. The results clearly revealed that the ideal storage temperature for SARS-CoV-2 detection and mutational/variant analyses in wastewater samples was 4 °C and the RNA load remained stable for up to seven days. Very low temperatures (i.e., −20° and −80 °C) were shown to rapidly destroy the genetic material, likely due to destabilization of the capsid of the virus and consequent exposure of the RNA to RNAses present in the wastewater.
3.2. Monitoring the SARS-CoV-2 load in wastewater treatment plants of Attica
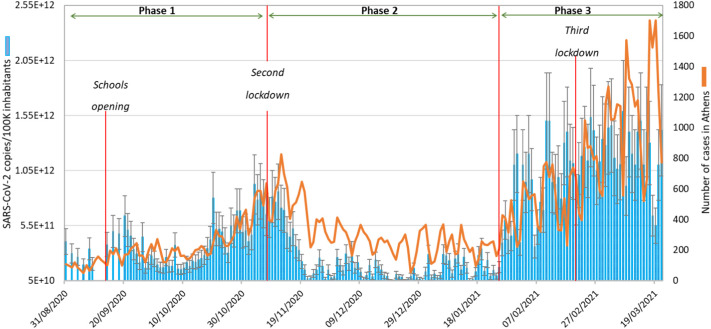
The first COVID-19 case in Greece was recorded on February 26, 2020 and the highest number of confirmed cases (during the sampling campaign) in Attica (1701 infections) occurred on March 17, 2021 (National Public Health Organization, 2021). Since the start of the pandemic, Greece has implemented three lockdowns (the last one being on February 20, 2021). Using the final validated method, the viral load was investigated and detected in all wastewater samples. The time-course of viral load and the measured COVID-19 positive cases data by National Public Health Organization (NPHO) of Greece are presented in Fig. 1 .
Fig. 1.
Viral load in wastewater and surveillance data; the SARS-CoV-2 load (RNA copies SARS-CoV-2/100K inhabitants) in the wastewater from wastewater treatment plants in Athens (blue bars) and NPHO-reported COVID-19 confirmed cases (orange line) are shown for the period August 31, 2020 through March 21, 2021. SARS-CoV-2 data are presented as mean ± SE from 191 wastewater samples. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The reported COVID-19 cases can be separated into 3 phases (Fig. 1). The first was August 31st through November 7th. At the beginning of this phase, the viral load was relatively steady, with no large fluctuations. There was an increased viral load after the first week of opening schools (September 14). This period also included the return from summer vacation, likely resulting in the number of inhabitants served by the wastewater treatment plants of Attica being lower than normal. There was a steep increase in the wastewater viral load after October 20. This phase ended with the announcement of the second lockdown on November 7. The second phase, November 8 through January 25, reflects the effectiveness of restrictions during the lockdown period (which included the Christmas holidays). During this phase, the viral load was the lowest for the whole study period, with values below the LOQ most of the time. However, a gradual increase was observed during the last days of January. The third phase, January 26 through March 21, almost coincides with the third wave of the pandemic. During this period, the largest increase in the number of cases occurred, and was likely related to the increasing prevalence of the alpha variant (B.1.1.7) of SARS-CoV-2 (Hellenic National Public Health Organization, 2021). Although the third lockdown started on February 20, there was no reduction in viral load or cases immediately after the lockdown announcement. During this period, the viral load was the highest for the entire study. It is notable that the viral load was usually lower on days of the weekend than weekdays. This pattern may have resulted from commuters in and out of the Attica peninsula. This trend was observed during all of the study period.
3.3. SARS-CoV-2 variants analysis in the wastewater treatment plant of Attica
The surge in viral load in wastewater and confirmed COVID-19 cases recorded in Attica (the region of Greece that includes Athens) from the beginning of February 2021 prompted us to analyze the mutational/variant profile of SARS-CoV-2 in wastewater samples collected in February 2021. S gene-related deletions (del69-70HV, del144Y), and missense mutations (L18F (C21614T), T20N (C21621T/A), P26S (C21638T), D80A (A21801C), K417N (G22813T), E484K (G23012A), N501Y (A23063T), D614G (A23403G), H655Y (C23523T), P681H (C23604A), A701V (C23664T), T716I (C23709T), S982A (T24506G)) were targeted for the detection and quantification in the following SARS-CoV-2 variants of concern (VOC): Β.1.1.7/UK lineage (Variant VOC_202012/01; alpha variant), B.1.351/South Africa lineage (Variant 20H/501.V2; beta variant) and.1/Japan-Brazil lineage (Variant 20J/501Y.V3; gamma variant). These analyses led to the conduct of 5.38 million sequencing reads, with a median read length of 339 bp. The findings of these analyses are summarized in Table 1, Table 2 .
Table 1.
SARS-CoV-2 variant analysis in the wastewater treatment plant of Athens; targeted DNA-seq analysis of genetic markers for detection and quantification of SARS-CoV-2 variants of concern.
| Position | Reference base | Alternative base | Alternative frequency (%) | Total depth | Reference amino acid | Alternative amino acid |
|---|---|---|---|---|---|---|
| del69-70HV | TACATG | −N(6) | 98.63 | 252,808 | HV | – |
| del144Y | TTA | −N(3) | 98.31 | 225,804 | Y | – |
| N501Y (A23063T) | A | T | 99.88 | 244,198 | N | Y |
| D614G (A23403G) | A | G | 84.26 | 261,583 | D | G |
| P681H (C23604A) | C | A | 93.29 | 344,390 | P | H |
| T716I (C23709T) | C | T | 99.85 | 338,016 | T | I |
| S982A (T24506G) | T | G | 99.90 | 257,801 | S | A |
Table 2.
SARS-CoV-2 variants present in the wastewater treatment plant of Athens; frequencies of analyzed SARS-CoV-2 variants of concern.
| Variant of concern | % frequency† | Genetic markers analyzed |
|---|---|---|
| Β.1.1.7/UK lineage (variant VOC_202012/01) | 96.3 ± 2.2 | N501Ya, D614Ga, P681H, T716I, S982A, del69-70 HV, del144Y |
| B.1.351/South Africa lineage (variant 20H/501.V2) | ND | D80A, K417Nb, E484Kb, N501Ya, D614Ga, A701V |
| P.1/Japan-Brazil lineage (variant 20J/501Y.V3) | ND | L18F, T20N, P26S, K417Nb, E484Kb, N501Ya, D614Ga, H655Y |
ND: not detected.
% proportion of SARS-CoV-2 variants of concern in wastewater samples. Data are presented as the mean ± SE.
Common genetic marker for Β.1.1.7/UK, B.1.351/South Africa & P.1/Japan-Brazil variants.
Common genetic marker for B.1.351/South Africa & P.1/Japan-Brazil variants.
Based on the frequency of the genetic markers analyzed, the Β.1.1.7/alpha lineage variant of concern was detected in 96.3% ± 2.2 (mean ± SE) of the total sequencing reads, while genetic markers specific for the B.1.351/beta (i.e., D80A, K417N, E484K and A701V) and P.1/gamma (i.e., L18F, T20N, P26S, D138Y, K417N and E484K) lineages were not detected. The Β.1.1.7/UK lineage (Variant VOC_202012/01), which emerged in southeast England in November 2020, has been associated with ≈50% increased transmissibility and mortality rates (Davies et al., 2021a; Davies et al., 2021b; European Centre for Disease Prevention and Control, 2021). The prevalence of the Β.1.1.7 variant in of February 2021 wastewater samples agrees with the significantly increased new COVID-19 cases and hospitalization cases in Attica for this period onwards.
3.4. Estimation of hospital admission rates: short-term modelling
An inherent uncertainty associated with environmental surveillance of SARS-CoV-2 RNA in wastewater is that the measurements might fall below the LOQ of the qPCR analytical method leading to the estimation of new hospitalizations not being accurate. For instance, Huisman et al. excluded few samples due to low quality and included the samples that had a threshold greater than 10% of the BCoV concentration in their study (Huisman et al., 2021). In our study, a threshold of 2.60e+11 SARS-CoV-2 RNA copies/100K inhabitants was applied to the normalized RNA load. This value was calculated from the CVln versus log10(RNA copies/L) curve of PCR results. As can be seen in Fig. S2, data with a CVln higher than 35% (which equates to SARS-CoV-2 RNA copies/100K inhabitants below 2.6e+11) would show great variation and therefore should not be used for modelling of COVID-19 indicators. Two modelling workflows were used: 1) models that use an averaging term in addition to lag and 2) models that do not include averaging terms and are based on probability modelling. For a first modelling approach based on linear regression, use of several levels of averaging terms (n = 3, n = 4, n = 5 and n = 7) was found to be necessary to improve the ability to predict pandemic clinical data. This could be due to day-to-day variations in life and working activities of Athens inhabitants, as well as weekly public restriction measures. Another factor affecting the correlation between WBE and pandemic clinical data was the time lag between wastewater RNA concentration and patient cases/hospitalizations. The aim of the present study was to improve the resolution of forecasting to several days ahead and determine the offset between RNA load in wastewater and pandemic clinical data.
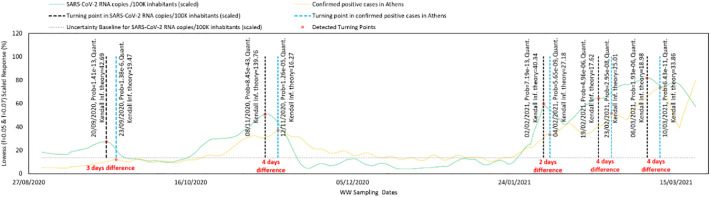
Temporal differences in the turning points of these data (i.e., between a large increase or decrease in the number of SARS-CoV-2 RNA copies/100K inhabitants and in the number of positive cases/new admissions to hospitals or ICUs) provide information about the delay (or lag time) between the RNA load in wastewater and pandemic clinical data. The most significant turning points are shown in Fig. 2 . Most of the detected turning points revealed low p-values ranging from 1.26e−05 to 8.45e−43. This implies that at the detected turning points and dates, the turning points followed a normal distribution. Five potential turning points were detected in the time series data of normalized SARS-CoV-2 RNA copies/100K inhabitants. These preceded the turning points in the number of pandemic clinical cases by an average of 2 to 4 days (Fig. 2). These results can be used qualitatively in that pandemic clinical cases would be expected to increase 3–4 days after a rise in the SARS-CoV-2 RNA copies/100K inhabitants in wastewater.
Fig. 2.
Identifying the time lag between RNA load in wastewater and SARS-CoV-2 pandemic clinical cases; turning points and difference between changes in the scaled SARS-Cov-2 RNA copies in wastewater/100k inhabitants and number of positive cases are presented. The grey dashed line is the CVln (%) threshold (i.e., 2.6e+11 normalized SARS-CoV-2 RNA copies/100K inhabitants (scaled value is 14%)). The black dashed lines are the top five detected turning points in the scaled SARS-Cov-2 RNA copies in wastewater/100k inhabitants. The blue dashed lines are the turning points detected in the trend profile of COVID-19 positive cases. The temporal separation between the blue dashed lines and the black dashed lines were used to derive lag time (delay) between turning points and peaks. Kendall information theory is calculated from −log2 (probability value |t) at given time (P is the probability to observe a turning point at time (t)). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The data were averaged (maximum for 4 days) after applying 3 or 4 days lags. The models of normalized SARS-CoV-2 RNA copies/100K inhabitants and pandemic clinical cases (i.e., positive clinical cases), new patients admitted to hospital or to ICU after 4 days lag followed by 3–4 days average are shown in Fig. S3. Using these averaged and lagged linear regression models and the data from 3 or 4 days of normalized SARS-CoV-2 RNA copies/100K inhabitants, the positive clinical cases, new patients admitted to hospital or to ICU can be estimated 3–4 days in advance. The predictive utility of these models was evaluated for the period February 19 to 22, 2021 (test set) (Table 3 ). This period of data collection was intentionally not included during the modelling procedure in the training set. Despite the increase in viral loads, the number of reported positive clinical cases was lower than the model predicted. This was mainly due to the number of tests being performed decreasing because of heavy snow and bad weather conditions which hindered clinical testing. However, the numbers of new admissions to hospitals and the ICU were predicted (confirmed n = 156, estimated n = 180 (165–195)). This supports the contention that reliable wastewater data may be used to predict the dynamics of SARS-CoV-2 infections, especially when clinical testing is restricted. However, from the R2 value results (R2 = 0.888 (lag = 4 days, averaging = 4 days) for new hospitalizations and R2 = 0.877 (lag = 4 days, averaging = 4 days) for new ICU admissions), it is apparent that admissions to hospitals or to ICUs don't have similar lag and averaging values as those derived for COVID-19 positive cases (R2 = 0.947 (lag = 4 days, averaging = 4 days)).
Table 3.
Predicting pandemic clinical cases in Athens using the averaged and lagged linear regression models; prediction of the number of SARS-CoV-2 positive clinical cases, new admissions to hospital or new admissions to ICU cases in Athens using normalized SARS-CoV-2 RNA copies/100K inhabitants identified in wastewater from February 19 to 22 (2021) by 4 days lag and 4 days average data.
| Variable | Experimental data |
Predicted datab |
|---|---|---|
| (Averaged by 4 days)a | (Averaged by 4 days and +4 day lag) | |
| Positive clinical cases | 564c | 930 (874–987) |
| New hospital admissions | 156 | 180 (165–194) |
| New ICU admissions | 13 | 16 (14–17) |
Averaged data between February 15 and 18, 2021.
Numbers in parentheses represent the lower and upper 95% confidence intervals.
The number of people being tested for SARS-CoV-2 decreased due to adverse weather conditions occurring between February 15 and 21, 2021.
While other important studies have established the correlation between viral RNA loads in wastewater and the rise in positive clinical cases and new admissions to hospital via distributed lag regression (Peccia et al., 2020; Zulli et al., 2021) or sewage surveillance of SARS-CoV-2 (Medema et al., 2020), only qualitative conclusions have been made thus far. Peccia et al. found that the rise in copies of RNA in primary sludge from wastewater treatment plants were reflected in reported COVID-19 cases within 6–8 days (Peccia et al., 2020). It is important to appreciate that the lead time after detection of RNA loads in wastewater as a pandemic indicator is under debate due to high variability (Bibby et al., 2021; Olesen et al., 2021). Previously, the lead time for WBE of COVID-19 in early detection had been estimated to be a maximum of four days (Bibby et al., 2021). In addition to pre-analytical factors, the behaviors of individuals (e.g., work attendance and lifestyle during the pandemic) and changes in the population size can influence the SARS-CoV-2 RNA loads in wastewater samples and lead to variability in the predictive models (Huisman et al., 2021; Olesen et al., 2021; Thomas et al., 2017). Therefore, the use of an averaging term (in addition to WBE lead time) to decrease the effect of such variations is vital. The present study serves as a proof of concept that the positive clinical cases and new admissions to hospitals and ICUs can be predicted using a linear regression analysis during the rise of a pandemic wave if the daily RNA copies/L measurement in wastewater has a CVln value below 35% and is higher than LOQ.
3.5. Estimation of hospital admission rates: long-term modelling
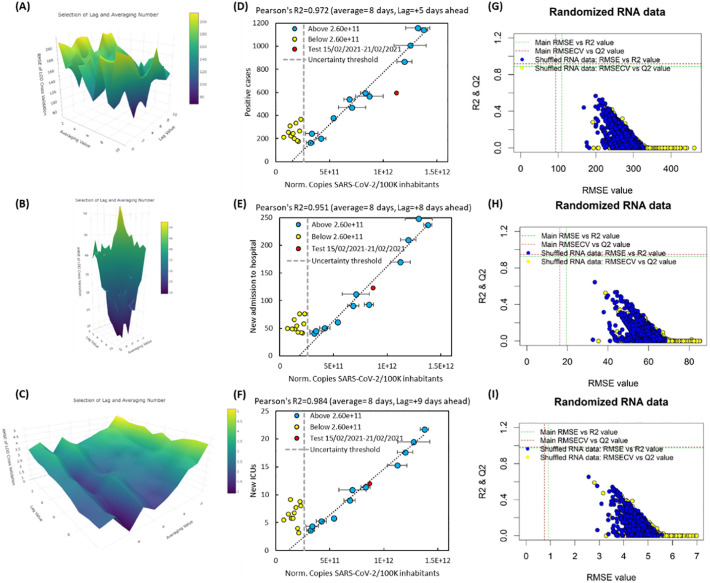
It is also possible to predict SARS-CoV-2 positive clinical cases, and new admissions to hospitals and ICUs 5 to 9 days ahead. The lag duration between copies of SARS-CoV-2 RNA in wastewater and pandemic clinical data and averaging values were optimized by root mean square error (RMSE) of leave-one-out cross validation technique (Fig. 3 ). The positive clinical cases can be estimated 5 days ahead if the number of RNA copies in wastewater are averaged by 8 days. The new admissions to hospitals and ICUs can be estimated 8 and 9 days (respectively) ahead if the number of RNA copies in wastewater are averaged by 8 days. As can be seen in Fig. 3, if the averaging factor is being neglected and only the lag value is considered, the RMSECV values become unacceptably high in the modelling of pandemic clinical data. Therefore, the present results support the inclusion of an averaging term in addition to the lag value during the modelling of SARS-CoV-2 pandemic clinical data when using normalized SARS-Cov-2 RNA copies/100K inhabitants. This is understandable given that with every new measure, there were varying lifestyle and working patterns throughout the recorded period. Such variations could alter the means by which individuals were exposed to SARS-CoV-2 within the week; averaging the data compensates for these variations. However, the number of data entries recorded over a longer period of time may be needed to enhance the predictive accuracy. It is noteworthy that using lag values between 2 and 8 provides lower RMSECV than those models without a lag value (Fig. 3). This supports the results in the previous “short-term modelling” section. However, as seen before, the models include several outliers (compare Fig. S3 with Fig. 3). The correlation and shuffling of RNA load in wastewater are presented in Fig. 3. All of the shuffled models provide RMSECV, R2 and Q2 values less than the main models developed for positive cases, and new admissions to hospitals and ICUs.
Fig. 3.
Grid search for optimal averaged lagged linear regression model; optimization of lag and averaging term using RMSE of leave-one-out cross validation in the estimation of positive cases (A), and new admissions to hospitals (B) or ICUs (C). The lower RMSECV results in better prediction performance. Each surface plot (A–C) shows the changes in RMSECV value around lag and averaging values. In plots D–F (positive cases, new admissions to hospitals, and new admissions to ICUs, respectively), data were subjected to linear regression analysis, with the line of best fit being shown as a dotted line. The test date with bad weather is shown with red marker. The grey dashed line is the CVln (%) threshold (i.e., 2.6e+11 normalized SARS-CoV-2 RNA copies/100K inhabitants). In the plots G–I (positive cases, new admissions to hospitals and new admissions to ICUs, respectively), the RNA load data were shuffled randomly and then subjected to linear regression analysis. R2 and Q2 values were calculated for the randomized data and compared with the main model (shown as red and green dotted lines, respectively). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
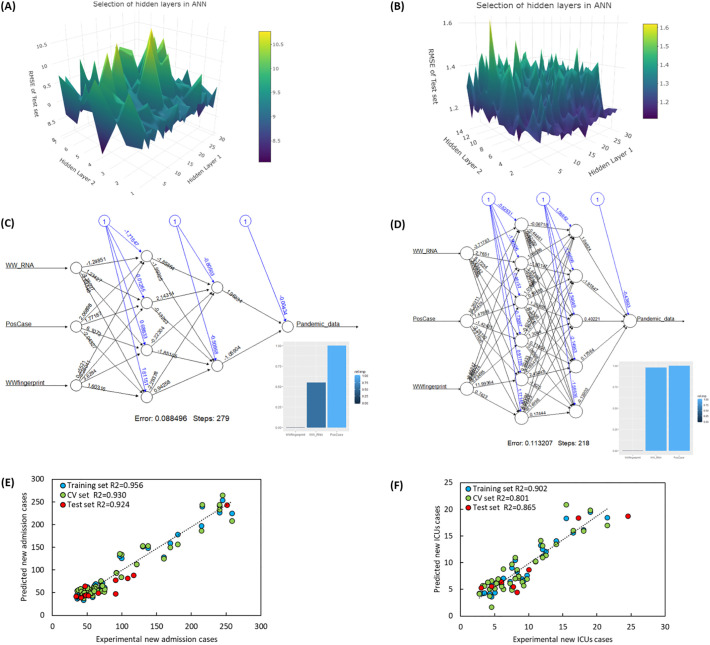
Estimation of RNA load in wastewater can be made independently of clinical testing indicators, and, in specific circumstances, such as adverse weather conditions and decreases in testing, provide a better representation of the status of SARS-CoV-2 infections in a population (Fernandez-Cassi et al., 2021). On the other hand, SARS-CoV-2 RNA loads in wastewater can be significantly influenced by sewer transportation, the analytical method used, environmental conditions (e.g., temperature) (Ahmed et al., 2020d) and insufficient sensitivity (e.g., levels fall below 35% CVln). To address these limitations, an artificial neural network model was used in the present study that combined the results from clinical testing (COVID-19-positive cases) and SARS-CoV-2 RNA load in wastewater to model new admissions to hospitals and to ICUs. The resultant analyses revealed the most optimal lags (taking into account their RMSECV) to be used to correlate SARS-CoV-2 RNA copies/100k inhabitants with positive clinical cases, new admissions to hospitals and ICUs to be 5, 8 and 9 days, respectively (Fig. 3). As such, the ANN models (Fig. 4 ) were also developed that included the averaging terms of 3 and 4 days for new admissions to hospitals and ICUs, respectively. These were the minimum averaging terms found to have lowest number of outliers and RMSEtest values.
Fig. 4.
Artificial Intelligence for modelling of SARS-CoV-2 pandemic data; the grid search results for selection of hidden layers in ANN in new admissions to hospitals (A) and new admissions to ICUs (B). The best combination of the hidden layer is the region where it showed the lowest RMSE value. Plots C and D show the ANN structure for new admissions to hospital and ICUs, respectively. The blue lines are bias in each node and the black line is the combination of layers and the weights used for each input data. The bar charts in the plots C and D show the importance of variables in ANN structure. Plots E and F show the predicted versus experimental data for new admissions to hospitals and ICUs, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
ANN models are also useful for estimating the new admissions to hospitals and ICUs from RNA load from 8 to 9 days ahead with lower averaging terms if combined with positive cases (3 and 4 days averaging for new admissions to hospitals and ICUs, respectively). The results indicate that inclusion of the 3 day lag for positive cases data and 8 day lag for SARS-CoV-2 RNA load in wastewater data results in very accurate prediction of the new admissions to hospitals, i.e., training set R2 = 0.956 and test set R2 = 0.924 (Fig. 4E). Acceptable results (training set R2 = 0.902 and test set R2 = 0.865) were also obtained in modelling new admissions to ICUs using a 4 day lag for positive cases data and 9 day lag for RNA load in wastewater data (Fig. 4F). However, the importance of variables in ANN structure (bar chart in Fig. 4C and D) indicates that the use of positive cases in modelling new admissions to hospitals is more significant than modelling new admissions to ICUs (equal variable importance for positive case and RNA load in wastewater data). This result may have been anticipated, given that not all of the positive cases would be expected to end up in the ICU. Our results relating to new admissions to hospitals are consistent with Peccia et al. (2020) who reported that sewage sludge results are not a leading indicator of progress of SARS-CoV-2 infection compared to positive test results. Finally, we have not reported the use of new hospital admission data in addition to positive cases and RNA load in wastewater data because the lag between new admissions to hospitals and new admissions to ICUs is 1 day. Such a model would have limited applications in planning actions to be taken to deal with the pandemic. Another issue associated with the use of positive cases in the ANN model is that any factors that disrupt the clinical testing process (such as inclement weather or pandemic-related restrictions on movement by the general public) can adversely impact the reliability of the model. This would reduce its forecasting ability and lead to inaccurate estimation of new admissions to hospitals or to ICUs. On the other hand, the advantage of these ANN models is that it can reduce the error of prediction of new admissions to hospitals or to ICUs for any SARS-CoV-2 RNA load in wastewater measurement that is below LOQ or CVln 35% by using data from positive clinical cases. The developed ANN and linear regression models were applied on the pandemic data recorded between February 15 and 22, 2021. Table S4 lists the estimated and actual observed data for each pandemic indicator (i.e., positive cases, new admissions to hospitals or ICUs). The new admissions to hospitals during this week were 102 (February 15–17, 2021) and 150 (February 18–20, 2021) which are very close to the estimated values of 86 (95% CI: 82–90) and 153 (95% CI: 148–159), respectively. Over these same periods, the number of new admission to ICUs, 12 and 13, are also close to the estimated values of 9 (95% CI: 8–10) and 14 (95% CI: 13–15) (Table S3).
3.6. Estimation of hospital admission rates; Bayesian distributed-lag nonlinear modelling
The aforementioned models were based on fixed lags and they included an averaging term. Although exclusion of the averaging terms results in somewhat less accurate prediction, it remains valid to track the changes in pandemic data in accordance with changes in RNA load in wastewater (Fig. S4). The distributed finite lag models provided mean errors of 141.08, 23.61 and 3.024 cases for positive cases, new admissions to hospitals, and new admissions to ICUs, respectively. In an attempt to decrease the error, the data was subjected to Bayesian Distributed-Lag Nonlinear Model (DLNM) analyses. The cumulative regression coefficients were 0.74 (95% CI: 0.63–0.89) for positive cases, 0.95 (95% CI: 0.80–1.29) for new admissions to hospitals, and 0.72 (95% CI: 0.59–0.91) for new admissions to ICUs. This indicates that a 10% increase in viral load results in 7.3% (95% CI: 6.2–8.9%), 9.5% (95% CI: 7.9–13.1%) and 7.1% (95% CI: 5.8–9.0%) increases in cases, new admissions to hospitals and to ICUs (respectively) spread out over a long time period. Despite the diffuse lag-response association, the model had a good fit to the data, as illustrated in Fig. S5. The DLNM analyses revealed viral load measurements to be associated with pandemic clinical indicators and, therefore, could be used to predict the burden on healthcare services. However, without adopting an averaging term, the substantial day-to-day variation in viral load in wastewater limits the practicality of predictions using DLNM, and the alternative aforementioned modelling approach would be anticipated to be a more reliable means of estimating COVID-19 pandemic indicators.
4. Conclusions
In the present study, we developed and showed that an optimized wastewater-based epidemiology measurements for SARS-CoV-2 RNA load in raw wastewater that accounted for the uncertainty derived from various sources (population estimation, viral load, feces quantity per person, quantity of SARS-CoV-2 shed for symptomatic and asymptomatic cases and measurement of various SARS-CoV-2 variants present in wastewater samples) can be used as a means to estimate the progression of the CovidID-19 pandemic within a community. We showed that daily measurements of wastewater samples correlate well with the clinical data. This enables real-time monitoring of COVID-19 pandemic indicators, improves prevalence prediction, and thereby facilitates the decisions by stakeholders, such as health departments and health care systems. A long period of monitoring SARS-CoV-2 load (and analyzed its variants) in raw wastewater samples was performed. This was needed to reveal important epidemiological information about the trends of infection and causes of rapid changes in both environmental and clinical data. Based on the frequency of the genetic markers analyzed, the Β.1.1.7/alpha lineage VOC was detected in 96.3% ± 2.2 (mean ± SE) of the total sequencing reads. The prevalence of the Β.1.1.7 variant in wastewater samples collected in February 2021 supports the rapid variation and increases in the new COVID-19 cases and hospitalization cases seen in Attica, at the time. The use of LOQ as a threshold was found to result in some outliers (measurements with insufficient sensitivity), whereas the use of the coefficient of variation for log-normally distributed data 35% (CVln) during environmental surveillance of SARS-CoV-2 seems to be a better way to measure threshold. Novel modelling approaches under epidemiological constraints were developed to estimate new admissions rates to hospitals and intensive care units from population-based, normalized SARS-CoV-2 RNA loads in wastewater samples. Two modelling workflows were developed: 1) models that use an averaging term in addition to lag time to remove variations; 2) models that do not include averaging terms and are based on probability modelling. Using an averaging term between 3 and 8 days, the new admissions to hospital and to ICUs can be accurately estimated from 2 to 8 days ahead with 95% confidence. Day-to-day variations in SARS-CoV-2 RNA load and clinical data (e.g., changes in testing frequency throughout the week) introduces variability into the modelling results. Although their estimation accuracy for pandemic indicators may not be comparable to the averaged-lag regression analysis presented here, the mean error derived from lag regression analysis remains low. For example, in the present study, mean errors (%) of 7.72%, 8.29% and 10.8% (mean error divided by maximum number of cases observed for each pandemic indicator) were observed for new admissions to hospital, confirmed positive cases and new admissions to ICUs, respectively. The grid search approach to find optimum lag times between SARS-CoV-2 RNA load in wastewater sample and pandemic clinical indicators provided even better results than the turning point-based method. The two ANN-based models revealed that both clinical and environmental surveillance data are complementary and can be used together with other epidemiological indices to better understand the status of COVID-19 in the general population. Especially in the case of new admissions to ICUs, environmental surveillance data appeared to be as important an indicator as community-based clinical surveillance. The findings of the present study provide valuable new approaches for predicting SARS-CoV-2 outbreaks and estimating the risk of SARS-CoV-2 transmission from symptomatic and presymptomatic cases. We anticipate the conditional uses of SARS-CoV-2 RNA load in wastewater (e.g., lag, averaging terms, and filtering out less meaningful analytical measurements by 35% CVln value) may advance the development of new approaches under epidemiological rational constraints.
Availability statement
All code and COVID-19 pandemic data for Athens are publicly available through http://trams.chem.uoa.gr/covid-19/.
CRediT authorship contribution statement
Study design: Nikolaos S. Thomaidis; Wastewater sampling and Sample concentration: Apostolos Karagiannidis and Aikaterini Kontou; RNA extraction and RT-qPCR detection: Aikaterini Galani and Marios Kostakis; NGS sequencing and mutational analysis: Andreas Scorilas, Margaritis Avgeris and Panagiotis G. Adamopoulos; Formal Analysis: Athina Markou, Nikiforos Alygizakis and Evi S. Lianidou; Development of hospital admission rates model and computational workflows: Reza Aalizadeh, Dimitrios Paraskevis and Theodore Lytras; Literature investigation-Data discussion: Reza Aalizadeh, Aikaterini Galani, Jordan Peccia and Nikolaos S. Thomaidis; Original draft writing: Reza Aalizadeh, Aikaterini Galani and Margaritis Avgeris; Review and editing: Jordan Peccia, David C. Thompson, Vasilis Vasiliou, Sotirios Tsiodras, Meletios-Athanasios Dimopoulos and Nikolaos S. Thomaidis. All authors have read and agreed to the published version of the manuscript.
Declaration of competing interest
Provisional patent applications entitled “Detection and mutational analysis of an RNA virus in an environmental sample” were filed by the National and Kapodistrian University of Athens, and the authors Andreas Scorilas, Margaritis Avgeris, Panagiotis G. Adamopoulos and Nikolaos S. Thomaidis on February 11, 2021 to the Hellenic Industrial Property Organisation (Case Number: 231-0004089064) and on March 22, 2021 to the Patent Cooperation Treaty (PCT application number: PCT/EP2021/057316). All other authors declare no competing interests.
Acknowledgements
Authors would like to acknowledge Athens Water Supply & Sewerage Company (EYDAP S.A.) and especially Mr. Konstantinos Vougiouklakis, Mr. Spyridon Dimoulas and Mr. Iraklis Karayiannis for granting permission for the collection of the wastewater samples and the Athens wastewater treatment plant operators for the collection of the samples. Dora Londra is acknowledged for her initial contribution to the qPCR analyses.
Editor: Damià Barceló
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2021.150151.
Appendix A. Supplementary data
Supplementary material
References
- Agrawal S., Orschler L., Lackner S. Long-term monitoring of SARS-CoV-2 RNA in wastewater of the Frankfurt metropolitan area in Southern Germany. Sci. Rep. 2021;11:5372. doi: 10.1038/s41598-021-84914-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., et al. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Choi P.M., Farkas K., et al. Surveillance of SARS-CoV-2 RNA in wastewater: methods optimization and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health. 2020;17:82–93. doi: 10.1016/j.coesh.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand U., Adelodun B., Pivato A., Suresh S., Indari O., Jakhmola S., et al. A review of the presence of SARS-CoV-2 RNA in wastewater and airborne particulates and its use for virus spreading surveillance. Environ. Res. 2021;196 doi: 10.1016/j.envres.2021.110929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avgeris M., Adamopoulos P.G., Galani A., Xagorari M., Gourgiotis D., Trougakos I.P., et al. Novel nested-seq approach for SARS-CoV-2 real-time epidemiology and in-depth mutational profiling in wastewater. Int. J. Mol. Sci. 2021;22:8498. doi: 10.3390/ijms22168498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Or I., Weil M., Indenbaum V., Bucris E., Bar-Ilan D., Elul M., et al. Detection of SARS-CoV-2 variants by genomic analysis of wastewater samples in Israel. Sci. Total Environ. 2021;789 doi: 10.1016/j.scitotenv.2021.148002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Been F., Rossi L., Ort C., Rudaz S., Delemont O., Esseiva P. Population normalization with ammonium in wastewater-based epidemiology: application to illicit drug monitoring. Environ. Sci. Technol. 2014;48:8162–8169. doi: 10.1021/es5008388. [DOI] [PubMed] [Google Scholar]
- Bibby K., Bivins A., Wu Z., North D. Making waves: plausible lead time for wastewater based epidemiology as an early warning system for covid-19. Water Res. 2021;202 doi: 10.1016/j.watres.2021.117438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., et al. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett. 2020;7:937–942. doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., North D., Ahmad A., Ahmed W., Alm E., Been F., et al. Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against covid-19. Environ. Sci. Technol. 2020;54:7754–7757. doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- Chen Y., Chen L., Deng Q., Zhang G., Wu K., Ni L., et al. The presence of SARS-CoV-2 RNA in the feces of covid-19 patients. J. Med. Virol. 2020;92:833–840. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- Daughton C.G. Wastewater surveillance for population-wide covid-19: the present and future. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372(6538) doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Jarvis C.I., Group CC-W C.I., Edmunds W.J., Jewell N.P., Diaz-Ordaz K., et al. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021;593:270–274. doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control . Stockholm; ECDC: 2021. SARS-CoV-2 - Increased Circulation of Variants of Concern and Vaccine Rollout in the EU/EEA, 14th Update – 15 February 2021. [Google Scholar]
- Fernandez-Cassi X., Scheidegger A., Bänziger C., Cariti F., Corzon A.T., Ganesanandamoorthy P., et al. Wastewater monitoring outperforms case numbers as a tool to track covid-19 incidence dynamics when test positivity rates are high. Water Res. 2021;200:117252. doi: 10.1016/j.watres.2021.117252. 2021.03.25.21254344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forootan A., Sjöback R., Björkman J., Sjögreen B., Linz L., Kubista M. Methods to determine limit of detection and limit of quantification in quantitative real-time PCR (qPCR) Biomol. Detect. Quantif. 2017;12:1–6. doi: 10.1016/j.bdq.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrity D., Papp K., Stoker M., Sims A., Frehner W. Early-pandemic wastewater surveillance of SARS-CoV-2 in southern Nevada: methodology, occurrence, and incidence/prevalence considerations. Water Res. 2021;10 doi: 10.1016/j.wroa.2020.100086. X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocic M., Trajkovic S. Analysis of changes in meteorological variables using Mann-Kendall and Sen's slope estimator statistical tests in Serbia. Glob Planet. Chang. 2013;100:172–182. [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., et al. Covid-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham K.E., Loeb S.K., Wolfe M.K., Catoe D., Sinnott-Armstrong N., Kim S., et al. SARS-CoV-2 RNA in wastewater settled solids is associated with covid-19 cases in a large urban sewershed. Environ. Sci. Technol. 2021;55:488–498. doi: 10.1021/acs.est.0c06191. [DOI] [PubMed] [Google Scholar]
- Grubaugh N.D., Gangavarapu K., Quick J., Matteson N.L., De Jesus J.G., Main B.J., et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019;20:8. doi: 10.1186/s13059-018-1618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellenic National Public Health Organization Information on the results of the Genomic Surveillance Network for SARS-CoV-2 mutations. 2021. https://eody.gov.gr/enimerosi-schetika-me-ta-apotelesmata-diktyoy-gonidiomatikis-epitirisis-gia-tis-metallaxeis-toy-sars-cov-2/ Available at.
- Hokajarvi A.M., Rytkonen A., Tiwari A., Kauppinen A., Oikarinen S., Lehto K.M., et al. The detection and stability of the SARS-CoV-2 RNA biomarkers in wastewater influent in Helsinki, Finland. Sci. Total Environ. 2021;770 doi: 10.1016/j.scitotenv.2021.145274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman J.S., Scire J., Caduff L., Fernandez-Cassi X., Ganesanandamoorthy P., Kull A., et al. Wastewater-based estimation of the effective reproductive number of SARS-CoV-2. medRxiv. 2021 doi: 10.1101/2021.04.29.21255961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan E.H., Wang D., Wang M., Malik A.A., Zulli A., Peccia J. Aligning SARS-CoV-2 indicators via an epidemic model: application to hospital admissions and RNA detection in sewage sludge. Health Care Manag. Sci. 2021;24:320–329. doi: 10.1007/s10729-020-09525-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall M.G., Stuart A., Ord K., Forster J., Arnold S., O'Hagan A. Wiley; 1994. Kendall's Advanced Theory of Statistics, Classical Inference and the Linear Model. [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., et al. First proof of the capability of wastewater surveillance for covid-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., et al. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewnard J.A., Liu V.X., Jackson M.L., Schmidt M.A., Jewell B.L., Flores J.P., et al. Incidence, clinical outcomes, and transmission dynamics of severe coronavirus disease 2019 in California and Washington: prospective cohort study. BMJ. 2020;369 doi: 10.1136/bmj.m1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of sars-coronavirus-2 rna in sewage and correlation with reported covid-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- National Public Health Organization 2021. https://eody.gov.gr/en/ Available at. Last accessed 26th February.
- van Nuijs A.L., Mougel J.F., Tarcomnicu I., Bervoets L., Blust R., Jorens P.G., et al. Sewage epidemiology-a real-time approach to estimate the consumption of illicit drugs in Brussels, Belgium. Environ. Int. 2011;37:612–621. doi: 10.1016/j.envint.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Olesen S.W., Imakaev M., Duvallet C. Making waves: defining the lead time of wastewater-based epidemiology for covid-19. Water Res. 2021;202 doi: 10.1016/j.watres.2021.117433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Cataluña A., Cuevas-Ferrando E., Randazzo W., Falcó I., Allende A., Sánchez G. Comparing analytical methods to detect SARS-CoV-2 in wastewater. Sci. Total Environ. 2021;758 doi: 10.1016/j.scitotenv.2020.143870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philo S.E., Keim E.K., Swanstrom R., Ong A.Q.W., Burnor E.A., Kossik A.L., et al. A comparison of SARS-CoV-2 wastewater concentration methods for environmental surveillance. Sci. Total Environ. 2021;760 doi: 10.1016/j.scitotenv.2020.144215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Cuevas-Ferrando E., Sanjuan R., Domingo-Calap P., Sanchez G. Metropolitan wastewater analysis for covid-19 epidemiological surveillance. Int. J. Hyg. Environ. Health. 2020;230 doi: 10.1016/j.ijheh.2020.113621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J., Rahman S.A., Ehtesham N.Z., Hira S., Hasnain S.E. SARS-CoV-2 variants of concern are emerging in India. Nat. Med. 2021;27:1131–1133. doi: 10.1038/s41591-021-01397-4. [DOI] [PubMed] [Google Scholar]
- Thomaidis N.S., Gago-Ferrero P., Ort C., Maragou N.C., Alygizakis N.A., Borova V.L., et al. Reflection of socioeconomic changes in wastewater: licit and illicit drug use patterns. Environ. Sci. Technol. 2016;50:10065–10072. doi: 10.1021/acs.est.6b02417. [DOI] [PubMed] [Google Scholar]
- Thomas K.V., Amador A., Baz-Lomba J.A., Reid M. Use of mobile device data to better estimate dynamic population size for wastewater-based epidemiology. Environ. Sci. Technol. 2017;51:11363–11370. doi: 10.1021/acs.est.7b02538. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- WHO Coronavirus (covid-19) Dashboard. 2021. https://covid19.who.int/?gclid=EAIaIQobChMIl76i0Mju8AIVmeR3Ch000Q6rEAAYASABEgI59vD_BwE
- Wolfe M.K., Archana A., Catoe D., Coffman M.M., Dorevich S., Graham K.E., et al. Scaling of SARS-CoV-2 RNA in settled solids from multiple wastewater treatment plants to compare incidence rates of laboratory-confirmed covid-19 in their sewersheds. Environ. Sci. Technol. Lett. 2021;8:398–404. doi: 10.1021/acs.estlett.1c00184. [DOI] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., et al. Virological assessment of hospitalized patients with covid-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020;26:502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanobetti A., Wand M.P., Schwartz J., Ryan L.M. Generalized additive distributed lag models: quantifying mortality displacement. Biostatistics. 2000;1:279–292. doi: 10.1093/biostatistics/1.3.279. [DOI] [PubMed] [Google Scholar]
- Zhang W., Du R.-H., Li B., Zheng X.-S., Yang X.-L., Hu B., et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulli A., Pan A., Bart S.M., Crawford F.W., Kaplan E.H., Cartter M., et al. Predicting daily covid-19 case rates from SARS-cov-2 RNA concentrations across a diversity of wastewater catchments. medRxiv. 2021 doi: 10.1101/2021.04.27.21256140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material