Abstract
Background Several platelet-derived microRNAs are associated with platelet reactivity (PR) and clinical outcome in cardiovascular patients. We previously showed an association between miR-204-5p and PR in stable cardiovascular patients, but data on functional mechanisms are lacking.
Aims To validate miR-204-5p as a regulator of PR in platelet-like structures (PLS) derived from human megakaryocytes and to address mechanistic issues.
Methods Human hematopoietic stem cells were differentiated into megakaryocytes, enabling the transfection of miR-204-5p and the recovery of subsequent PLS. The morphology of transfected megakaryocytes and PLS was characterized using flow cytometry and microscopy. The functional impact of miR-204-5p was assessed using a flow assay, the quantification of the activated form of the GPIIbIIIa receptor, and a fibrinogen-binding assay. Quantitative polymerase chain reaction and western blot were used to evaluate the impact of miR-204-5p on a validated target, CDC42. The impact of CDC42 modulation was investigated using a silencing strategy.
Results miR-204-5p transfection induced cytoskeletal changes in megakaryocytes associated with the retracted protrusion of proPLS, but it had no impact on the number of PLS released. Functional assays showed that the PLS produced by megakaryocytes transfected with miR-204-5p were more reactive than controls. This phenotype is mediated by the regulation of GPIIbIIIa expression, a key contributor in platelet–fibrinogen interaction. Similar results were obtained after CDC42 silencing, suggesting that miR-204-5p regulates PR, at least in part, via CDC42 downregulation.
Conclusion We functionally validated miR-204-5p as a regulator of the PR that occurs through CDC42 downregulation and regulation of fibrinogen receptor expression.
Keywords: miRNAs, CDC42, GPIIbIIIa, platelet reactivity, biomarkers
MicroRNAs as Biomarkers for Personalized Antiplatelet Therapy
Introduction
Platelets are involved in several human processes, including the regulation of inflammation, the immune system, and cancer. 1 2 They play a key role in regulating hemostasis and are involved in cardiovascular ischemic events. Platelet reactivity (PR) is a variable phenotype, either with 3 or without 4 antiplatelet drugs, and monitoring platelet function is now being investigated as a means to tailor antiplatelet drug strategies. 5 Understanding the mechanisms driving PR could lead to the development of new biomarkers or new therapeutic targets in diseases where platelets play a major role.
Human platelets express more than 450 megakaryocyte-derived microRNA (miRNA) molecules. 6 Over the past years, platelet-derived miRNAs have been identified as biologically and clinically relevant. 7 miRNAs are a class of short, noncoding RNAs of approximately 17 to 25 nucleotides. miRNAs bind by imperfect complementarity to a sequence of the three-prime untranslated region (3′UTR) of mRNAs, repressing the translation and/or degradation of endogenous RNA and thus modulating protein expression. Several miRNAs expressed in platelets are supposed to regulate platelet function, such as miR-223, 8 miR-200b, 9 miR-107, 9 miR-96, 10 or miR-126-3p, 11 12 and they are associated with PR and/or cardiovascular outcome. 7 9 However, their exact role in the modulation of human platelet function remains poorly understood.
In a previous study, we pointed out miR-204-5p as a putative biomarker of platelet function. 13 Recently, Ding et al 14 showed a positive correlation between miR-204-5p and patients with high PR. Databases such as TargetScan or miRanda indicate that miR-204-5p has between 700 to 3,000 potential target genes (TargetScan release 7.2 and miRanda August 2010 release, respectively). However, the combined analysis of proteomic and transcriptomic data from patients with high or low PR identified differentially expressed genes, targets of miR-204-5p, including THBS1 , CDC42 , CORO1C , SPTBN1 , TPM3 , GTPBP2 , and MAPRE2 . 13 Among these putative targets, CDC42 15 has been validated as a direct target of miR-204-5p using reporter gene assay.
CDC42 belongs to the RHO family of GTPases that contributes to cytoskeletal changes resulting in the regulation of cell morphogenesis and cell adhesion. Furthermore, CDC42 regulates actin polymerization, demarcation membrane system (DMS) formation, proplatelet biogenesis, 16 17 and platelet function. 18 We thus addressed the issue of the impact of miR-204-5p and CDC42 on platelet biogenesis and function using an in vitro model allowing to generate megakaryocytes and platelet-like structures (PLS) from human hematopoietic stem cells. 12 19
Methods
Human CD34+ Hematopoietic Progenitor Cells
CD34+ purification, differentiation, transfection, and measurement of transfection efficiency were performed as previously described. 12 19 Overexpression of miR-204-5p and the knock-down of CDC42 were performed using an hsa-miR-204-5p mimic (ThermoFisher, Waltham, United States) and small-interfering RNA (siRNA) (Qiagen, Hilden, Germany), respectively. Morphological and functional characterizations were performed on day (D) 15, 48 hours after transfection unless otherwise specified.
Megakaryocytes were recovered and centrifuged at 100 g for 10 minutes at 37°C in the presence of 25 μM prostaglandin I2 (PGI2; Cayman Chemical Company, Ann Arbor, United States) and 0.02 U/mL apyrase (Sigma, St Louis, United States) to avoid activation. The supernatant containing the PLS was centrifuged at 1,000 g for 10 minutes and resuspended in dedicated buffer. The concentration was monitored using Tali Image-Based Cytometer (ThermoFisher); megakaryocytes were defined as elements >6 μm and PLS were defined as elements <6 μm.
miRNA Quantification
miRNA content in PLS was quantified using TaqMan quantitative polymerase chain reaction (qPCR) and normalized using a panel of stably expressed miRNAs (miR-28, miR-29c, and miR-151) previously identified using the geNorm algorithm. 20 21
RNA Extraction and mRNA Analysis
mRNA analysis was performed as previously described. 12 The oligonucleotide sequences used for qPCR were:
HB2M-forward: 5′-TGCTCGCGCTACTCTCTCTTT-3′
reverse: 5′-TCTGCTGGATGACGTGAGTAAAC-3′
CDC42-forward: 5′-ACATCTGTTTGTGGATAACTCA-3′
reverse: 5′-GGGAGCCATATACTCTTGGA-3′
RHOA-forward: 5′- TGGAAAGACATGCTTGCTCAT -3′
reverse: 5′-GCCTCAGGCGATCATAATCTTC-3′
RAC1-forward: 5′- ATCCGCAAACAGATGTGTTCT -3′
reverse: 5′-CGCACCTCAGGATACCACT -3′.
Western Blot Analysis
Megakaryocytes and PLS were activated by stirring (1,100 rpm) for 6 minutes with thrombin receptor activator peptide (TRAP) 20 μM at 37°C and were then lysed in a dedicated buffer. The protein extracts were separated onto 12% polyacrylamide gel and electrophoretically transferred to a nitrocellulose membrane (Amersham Biosciences, New York, United States). Proteins of interest were labeled with primary anti-GAPDH and anti-CDC42 (Cell Signaling Technology, Danvers, United States). Band intensities were quantified using ImageJ software, and the CDC42 protein level was normalized to GAPDH.
Megakaryocyte Differentiation
In order to assess the possible role of miR-204-5p transfection on megakaryocyte differentiation, megakaryocyte pellet was resuspended in 200 μL phosphate-buffered saline (PBS) and incubated for 1 hour at 4°C in 70% ethanol. The cells were centrifuged at 1,000 g for 10 minutes, resuspended in 100 μL propidium iodide (PI) (BD Biosciences, Allschwil, Switzerland) at 50 μg/mL in PBS, and then incubated for 30 minutes at 37°C. PI labeling was measured using fluorescence-activated cell sorting (FACS) to evaluate the megakaryocytes' ploidy. Analyses were performed using an Accuri C6 flow cytometer (BD Biosciences).
Megakaryocyte differentiation was also analyzed using FACS with selected antibodies against specific markers of differentiation (CD41, CD42b, and CD42d) as previously described. 12
Megakaryocyte Polarization
Megakaryocytes were fixed and permeabilized using 4% paraformaldehyde (PAF) and 0.05% triton in PBS. They were subsequently centrifuged at 250 g for 10 minutes onto coverslips previously coated with 100 μg/mL poly-L-lysine (Sigma) for 5 minutes and then dried at room temperature. The DMS was visualized using colocalization of the filamentous (F)-actin and GP1bβ 22 23 in differentiated megakaryocytes, defined as megakaryocytes exhibiting at least two nuclear lobes. GP1bβ was labeled using a mouse antibody anti-GP1bβ (CD42c, Santa Cruz Biotechnology, Dallas, United States) and then stained with AF-488 anti-mouse antibody (Life Technologies, Carlsbad, United States). F-actin was visualized using TRITC-phalloidin (Sigma). Quantification was performed blindly on at least 30 mature megakaryocytes per condition.
The coverslips were mounted using the Vectashield Hardset mounting medium with DAPI (Reactolab SA, Servion, Switzerland). The cells were visualized using an LSM700 microscope (Zeiss, Oberkochen, Germany), and the images were analyzed blindly using ImageJ software ( http://rsb.info.nih.gov/ij/ ). The three-dimensional reconstitution of z-stacks was performed using a background subtraction and the application of a Gaussian filter using Imaris 9.5.1 software (Bitplane AG, Zurich, Switzerland).
ProPLS Formation
On D13, 5 hours after transfection, megakaryocytes were seeded at 5,000 cells/well in a 96-well plate. On D15, pictures were taken at magnification × 100 and analyzed using ImageJ software. Megakaryocytes bearing proPLS were defined as megakaryocytes with at least one protrusion (500 megakaryocytes per condition were analyzed). The surface areas of the protrusions were also evaluated for 50 megakaryocytes bearing proPLS.
PLS Production
The PLS suspension (300 μL) released from 500,000 megakaryocytes was labeled with Hoechst (ThermoFischer) and anti-CD41-APC antibody (BioLegend, San Diego, United States). PLS were washed and resuspended in 300 μL PBS and 50 μL Precision Count Beads (BioLegend). Samples were analyzed on an Attune NxT flow cytometer (ThermoFischer), and data processing was performed using FlowJo software. PLS were defined as Hoechst −/CD41+ events. The concentration of PLS was evaluated using the following equation:
CPLS = (PLS count × V Beads × C Beads )/Bead count × V PLS , where PLS count = number of PLS quantified, V Beads = volume of Precision Count Beads, C Beads = concentration of Precision Count Beads, bead count = number of beads quantified, and V PLS = volume of the PLS suspension.
PLS Size
To study the PLS size at rest, PLS were recovered and incubated for 1 hour at 37°C on poly-L-lysine-coated coverslips. Non-adherent PLS were removed by washing with PBS-0.1% BSA. Adherent PLS were fixed using 4% PAF and then permeabilized using 0.5% Triton X-100 in Hank's Balanced Salt Solution.
F-actin was stained with TRITC-phalloidin, and coverslips were mounted with Vectashield Hardset mounting medium with DAPI. Images were acquired using an LSM700 and analyzed blindly to the experimental condition using ImageJ software. The PLS area was measured in arbitrary units relative to the mock condition. At least 100 PLS were analyzed per condition.
PLS Receptor Expression
PLS were adjusted at 1.3 × 10 6 PLS/mL in PBS. PLS were labeled with anti-CD41-V450 (BD Biosciences), anti-CD61-APC (Miltenyi Biotec), CD42a (Abcam, Cambridge, England), anti-CD42b-BV650 (BioLegend), and monoclonal CD42d antibody (Santa Cruz, Dallas, United States), followed by secondary PE goat anti-mouse antibody (BioLegend), anti-CD49b-FITC (BD Biosciences) antibodies, and GPVI (BD Pharmingen, Allschwil, Switzerland) or with their corresponding isotypic antibodies. PLS were washed in PBS and fixed in 2% PAF. Flow cytometry was performed using an Attune NxT flow cytometer. The data were analyzed using FlowJo software.
PLS Activation
To investigate the potential effect of miR-204-5p on GPIIbIIIa activation, PLS were adjusted at 1 × 10 6 PLS/mL in Tyrode's albumin (TA) buffer and then activated with 1 U/mL thrombin (Sigma) for 6 minutes at 37°C without stirring. PLS were labeled with anti-CD31-APC (BioLegend), anti-CD62P-PE (BD Pharmingen) and PAC1-FITC (BD Biosciences), and antibodies or with their corresponding isotypic antibodies. PLS were then fixed with 2% PAF. Flow cytometry was performed using an Accuri C6 flow cytometer, and the data were analyzed using BD Accuri C6 Plus software (BD Biosciences).
PLS Adhesion to Fibrinogen
A spreading assay was performed by adhesion of PLS onto fibrinogen-coated coverslips (100 µg/mL) for 1 hour at 37°C. PLS were fixed, permeabilized, and labeled with TRITC and DAPI, as described above. Pictures were taken using an LSM700, and the PLS spreading areas were quantified using ImageJ software.
To investigate the binding of fibrinogen on activated PLS surfaces, PLS were adjusted to 1.3 × 10 6 cells/mL in TA buffer. PLS were then activated with 0.05 U/mL of human thrombin (Calbiochem, Los Angeles, United States) in the presence of 50 µg/mL of AF-488-labeled fibrinogen (ThermoFisher) for 10 minutes at 37°C without stirring. PLS were labeled with anti-CD31-APC antibody and fixed with 2% PAF. Flow cytometry was performed using an Accuri C6 flow cytometer with an FL-1 90% filter, and the data were analyzed using BD Accuri C6 Plus software. Double-labeled anti-CD31-APC and AF-488 events were considered to be PLS binding to fibrinogen.
Megakaryocyte Cytoskeleton
To study the impact of miR-204-5p on megakaryocyte cytoskeleton, these were put onto fibrinogen-coated coverslips and examined using confocal microscopy. Briefly, coverslips were coated with 100 μg/mL of fibrinogen (Sigma) at 4°C overnight. Coverslips were washed with PBS-0.1% BSA and blocked with 1% BSA. Megakaryocyte adhesion occurred at 37°C for 1 hour. Megakaryocytes were then permeabilized and labeled with TRITC and DAPI, as described above. Pictures were taken using an LSM700, and at least 20 mature megakaryocytes per condition were analyzed, blindly to the experimental condition, using ImageJ software.
Transmission Electron Microscopy
At D15, megakaryocytes and PLS were centrifuged in the presence of PGI2 at 1,000 g for 10 minutes without brake. After washing with PBS, the cells were prefixed in glutaraldehyde 0.2% for 10 minutes. The PLS were then centrifuged for 10 min at 2,000 g and fixed for 1 hour in glutaraldehyde 2%. The samples were postfixed in osmium, dehydrated with alcohol, and embedded in a plastic resin. Thin sections of the samples were stained with heavy metals and examined under a transmission electron microscope (CM10 Philips TM) at 80 kV.
F/G Actin Ratio Measurement
Megakaryocytes were activated by stirring (1,100 rpm) for 6 minutes at 37°C with 20 μM TRAP-6 (Bachem, Dubendorf, Switzerland). F and G (globular) actin fractions were recovered using the G-actin/F-actin in vivo assay kit (Cytoskeleton, Denver, United States) according to the manufacturer's instructions. Briefly, the megakaryocytes were harvested by centrifugation at 1,000 g for 10 minutes, resuspended in a lysis buffer, and homogenized by passing them through a 25G needle. After 10 minutes of incubation at 37°C, the lysates were ultra-centrifuged at 100,000 g for 2 hours at 37°C. The supernatants containing G-actin were retrieved, and the pellets containing F-actin were resuspended for 1 hour on ice in a volume of depolymerization buffer equal to the corresponding supernatant. Equal volumes of G- and F-fraction were loaded onto a 12% polyacrylamide gel and subjected to western blot analysis using rabbit anti-actin antibody (Cytoskeleton). G- and F-actin bands intensity was quantified separately using ImageJ software.
Flow Chamber Assay
A flow chamber assay was performed as previously described. 12 Briefly, megakaryocytes and PLS were perfused at 50 s −1 for 5 minutes onto fibrinogen-coated slides (Vena8 Fluoro + , Cellix, Dublin, Ireland). Pictures were taken at five different points along the channel (Si-3000 camera, Ceti, Medline Scientific, Chalgrove, England), and image analysis was performed using a custom-made framework written in MATLAB (The MathWorks, Inc. Natick, United States) and dedicated to quantifying PLS individually and megakaryocytes according to their size.
Statistical Analysis
To avoid batch effect from different donors of buffy coat, all the data are expressed relatively to mock control. All the data are presented as the mean ± standard error of the mean of at least three independent experiments. Statistical significance was evaluated using one-way ANOVA followed by a posthoc Tukey test or a Student t -test, when appropriate. Data were analyzed using GraphPad Prism 7 software (GraphPad Software Inc., San Diego, United States). p -Values < 0.05 were considered statistically significant.
Results
miR-204-5p Expression Profile during Differentiation
The relative level of miR-204-5p decreased during the differentiation process of the CD34+ cells in megakaryocytes and subsequently in PLS release ( Fig. 1A ).
Fig. 1.
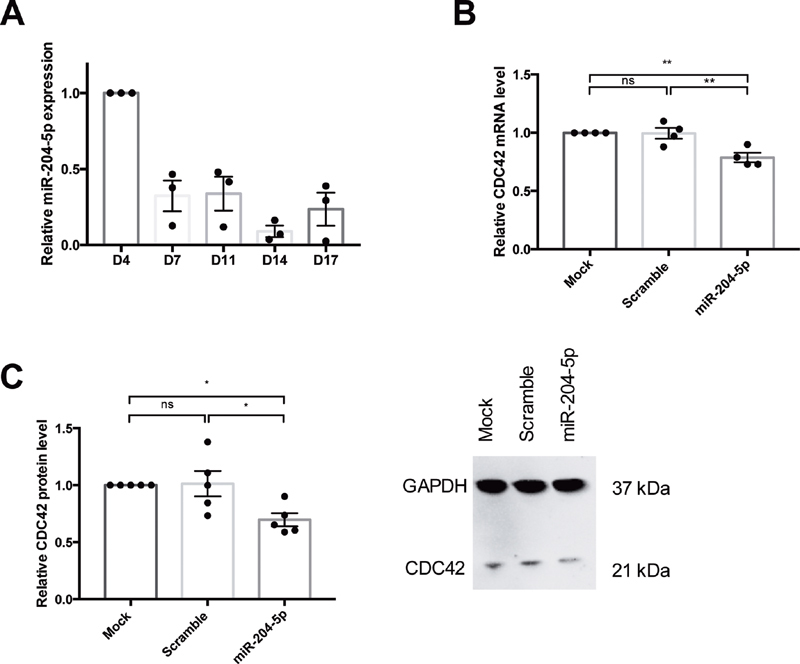
miR-204-5p level during the differentiation process and after overexpression. ( A ) miR-204-5p is decreased over the differentiation process from D4 to D14. miR-204-5p transfection induced a decrease of the relative expression of both ( B ) CDC42 mRNA and ( C ) protein level in megakaryocytes and PLS. n = 4 to 5 independent experiments; * p < 0.05, ** p < 0.01. PLS, platelet-like structure.
PLS Function
PLS activation with thrombin induced an increase in P-selectin expression (31.8 ± 5%), activation of GPIIbIIIa (47.8 ± 13%), and binding of AF-488 soluble fibrinogen (42.3 ± 10%) ( Supplementary Fig. S1 , available in the online version). These data are in line with the characteristics of PLS produced elsewhere with the same protocol. 12 19
The Impact of miR-204-5p Transfection on CDC42 Downregulation and on Other Rho GTPases
As expected, miR-204-5p transfection was associated with a 21.2 ± 4% downregulation of the mRNA level ( Fig. 1B ) and a 31 ± 6% downregulation of the protein level ( Fig. 1C ) of CDC42, which is a validated target of miR-204-5p. However, miR-204-5p transfection had no impact on RhoA and Rac 1 expression (0.89 ± 0.01, p = 0.97 and 0.94 ± 0.09, p = 1.00, respectively).
miR-204-5p Transfection Did Not Affect Megakaryocyte Differentiation but Modulated Megakaryocyte Polarization
FACS analysis showed that miR-204-5p transfection at D13 had no effect on the ploidy of megakaryocytes at D15 ( Fig. 2A ). In addition, the percentage of megakaryocytes expressing the differentiation markers CD41, CD42b, and CD42d was unchanged 48 hours after transfection showing that miR-204-5p transfection does not impact the differentiation process ( Fig. 2B ). Labeling of megakaryocytes with DAPI and GPIbβ/F-actin enables the study of the localization of both the nucleus and DMS, respectively. In both mock and scramble conditions, the nuclei were located at the periphery of the megakaryocytes, whereas their DMS was present at the opposite edge. Conversely, in megakaryocytes overexpressing miR-204-5p, nuclei were in the center of the cells and were surrounded by DMS, indicating an absence of polarization in these megakaryocytes ( Fig. 2C ). In mock condition, 85 ± 2.3% of megakaryocytes were polarized, whereas after transfection with miR-204-5p, polarization was observed in only 49 ± 6.4% of megakaryocyte cells ( Fig. 2D ). This indicates that transfection of miR-204-5p 48 hours before the end of the differentiation had no effect on the differentiation markers nor on the ploidy, but it did modulate megakaryocyte polarization.
Fig. 2.
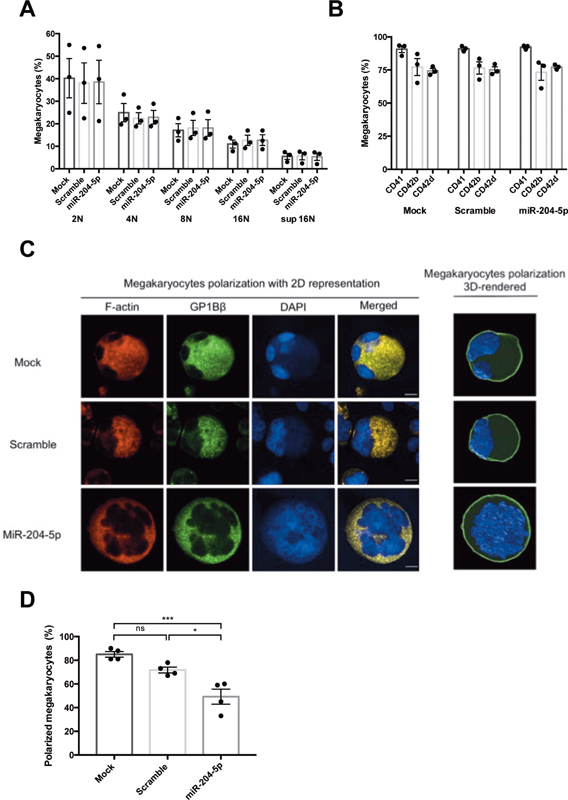
Effect of miR-204-5p on megakaryocyte differentiation. Fluorescence-activated cell sorting analysis at D15 showed that miR-204-5p ( A ) had no effect on megakaryocyte ploidy, and ( B ) no effect on the expression of the specific differentiation markers CD41, CD42b, and CD42d. miR-204-5p decreased megakaryocyte polarization defined as the colocalization of F-actin and GP1bβ ( C and D : images obtained using an LSM700 microscope, background subtraction, and application of a Gaussian filter using Imaris 9.5.1 software for 3D rendering; scale bars = 10 μm). n = 3 to 5 independent experiments; * p < 0.05, *** p < 0.001.
miRNA-204-5p Overexpression Modified the DMS Network
Thin section electron microscopy analysis showed that PLS obtained after miR-204-5p transfection of megakaryocytes displayed a similar ultrastructure as compared to PLS from control or scramble transfected megakaryocytes ( Fig 3A ), while in miR-204-5p-transfected megakaryocytes, the DMS structure showed major modifications with a vacuolated aspect and a reduced membrane invaginations ( Fig 3B ).
Fig. 3.
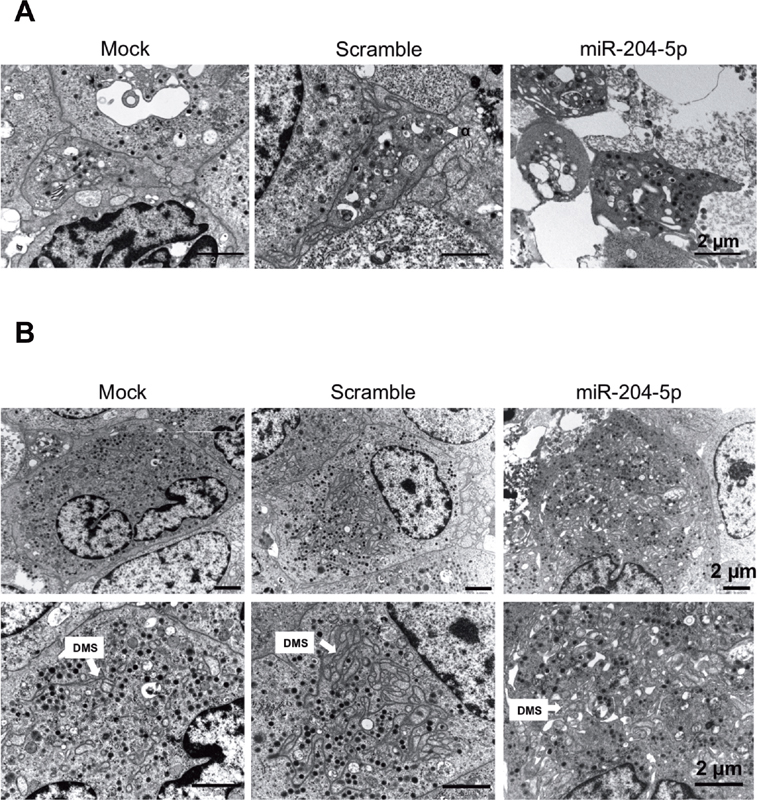
Representative TEM pictures of megakaryocytes and PLS. miR-204-5p transfection ( A ) does not impact PLS ultrastructure but ( B ) dramatically disturbed the DMS organization of megakaryocytes. Scale bars = 2 μm. DMS, demarcation membrane system; PLS, platelet-like structure; TEM, transmission electron microscopy.
miR-204-5p Overexpression Modulated the Production of PLS without Affecting Their Size
miR-204-5p transfection had no effect on the proportion of megakaryocytes releasing proPLS (data not shown). However, the megakaryocytes transfected with miR-204-5p produced abnormal protrusions with retracted networks of proPLS tips. Protrusions were more compact after miR-204-5p transfection, showing a 50% decrease in proPLS protrusion areas ( Fig. 4A and B ).
Fig. 4.
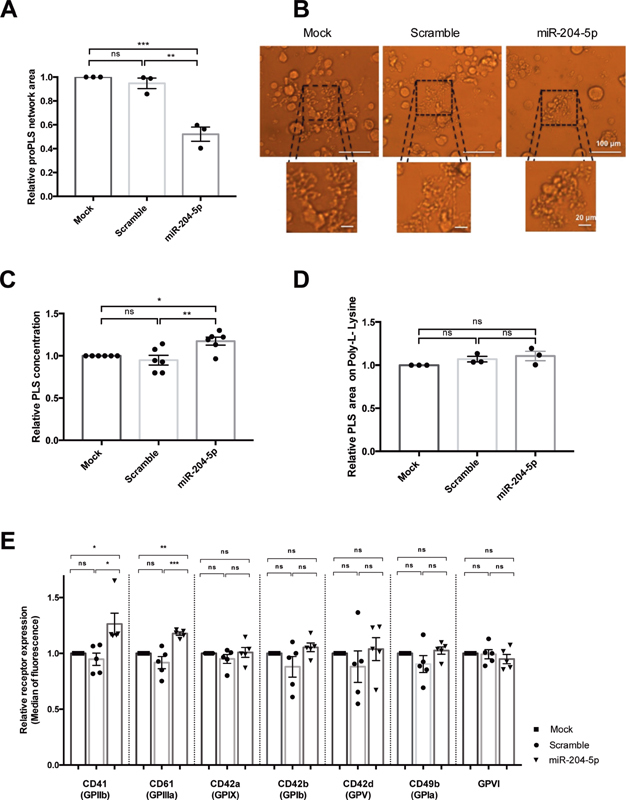
Characterization of PLS overexpressing miR-204-5p. miR-204-5p overexpression ( A ) significantly decreased the relative proPLS network area, and ( B ) affected the pattern of proPLS elongation (Si-3000 camera). ( C ) The number of PLS produced was slightly but significantly higher, whereas ( D ) no differences in PLS size were observed after adhesion on poly-L-lysine. ( E ) miR-204-5p increased expression of CD61 and CD41 receptors without affecting CD42b, CD42a, CD42d, CD49b, and GPVI expression. The results are expressed relative to the mock condition. n = 3 to 6 independent experiments; * p < 0.05, ** p < 0.01, *** p < 0.001. PLS, platelet-like structure.
In addition, miR-204-5p overexpression slightly increased the production of PLS (282 ± 120 PLS/μL in miR-204-5p condition) as compared to the control conditions (238 ± 95 and 201 ± 74 PLS/μL in mock and scramble, respectively, Fig. 4C ), without affecting their size ( Fig. 4D ). This was confirmed by an analysis of the median forward scatter of the PLS in flow cytometry ( Supplementary Fig. S2 , available in the online version).
miRNA-204-5p Overexpression Increased GPIIb and GPIIIa Expression in PLS
To further characterize the PLS produced by miR-204-5p-transfected megakaryocytes, we quantified the expression of selected platelet receptors. Fig. 4E shows that miR-204-5p transfection had no effect on CD42b (GPIb) expression, a late marker of PLS differentiation, neither on CD42a (GPIX), CD42d (GPV), CD49b (GPIa), and on GPVI expression. In contrast, miR-204-5p transfection enhanced the expression level of the two subunits of the fibrinogen receptor: CD41 (GPIIb) and CD61 (GPIIIa). Their median fluorescence increased by 26 ± 9.6 and 18 ± 1.5%, respectively.
miR-204-5p Overexpression Increased PLS Reactivity and Adhesion
Since we evidenced an increase in GPIIbIIIa expression in PLS derived from miR-204-5p-transfected megakaryocytes, we focused functional assays using fibrinogen as a matrix. Thrombin stimulation did not induce a significant modulation in P-selectin (CD62P) expression after miR-204-5p transfection ( Fig. 5A ). Among activated PLS (defined as CD31 +/CD62P+ PLS), the median fluorescence of PLS positive for the activated form of GPIIbIIIa was 30 ± 9% higher after miR-204-5p transfection than under the mock or scramble conditions ( Fig. 5B ). In static conditions, PLS adhesion on immobilized fibrinogen was associated with a 45 ± 13% greater PLS spreading area after miR-204-5p transfection than in the mock or scramble conditions ( Fig. 5C and D ). Finally, stimulation of PLS in the presence of soluble AF-488-labeled fibrinogen showed a 51 ± 15% greater capacity of miR-204-5p-transfected PLS to bind soluble fibrinogen than in the mock condition ( Fig. 5E ).
Fig. 5.
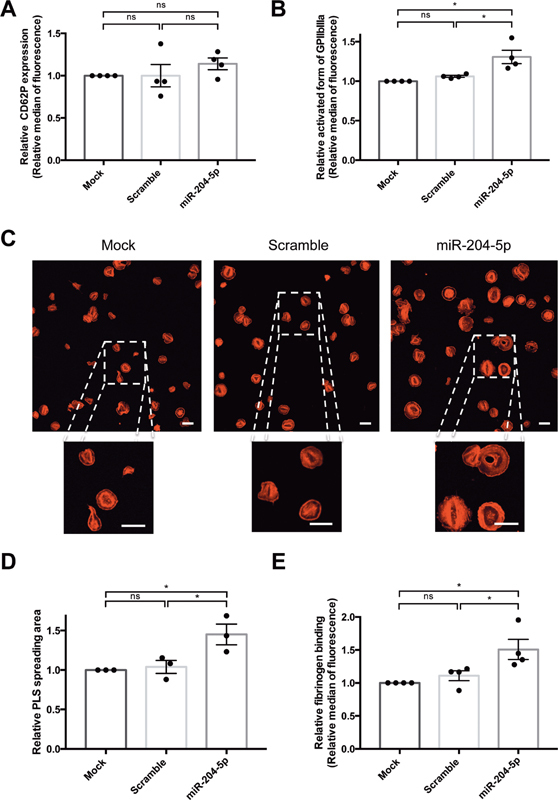
miR-204-5p overexpression affected PLS reactivity. miR-204-5p overexpression ( A ) did not impact CD62P expression after thrombin stimulation but ( B ) induced a higher PAC-1 relative median of fluorescence. miR-204-5p transfection ( C and D ) was associated with an increase in PLS spreading area on fibrinogen-coated coverslips (LSM700 microscope, scale bars = 20 μm) and ( E ) enhanced the binding of soluble fibrinogen to PLS. Results are expressed relative to mock. n = 3 to 4 independent experiments; * p < 0.05. PLS, platelet-like structure.
miR-204-5p Modulated Megakaryocyte Adhesion in Dynamic Conditions and Induced Cytoskeletal Reorganization
In flow conditions, we found 50 ± 16% more megakaryocytes adhering to the fibrinogen-coated channel after miR-204-5p transfection than in the mock condition ( Fig. 6A ). However, PLS adhesion did not differ significantly with this assay (data not shown). Since cytoskeletal rearrangement plays a critical role in cell adhesion and is regulated by CDC42, the actin cytoskeleton of megakaryocytes was analyzed using confocal microscopy. This revealed a markedly disturbed cytoskeletal organization. In control cells, actin filaments were located at cell periphery, whereas transfection with miR-204-5p increased the content of stress fibers ( Fig. 6B ). Analysis of actin dynamics showed that upon activation with TRAP, the F/G actin protein ratio increased more in megakaryocytes transfected with miR-204-5p than in mock or scrambled conditions ( Fig. 6C ). This indicates that miR-204-5p overexpression promotes the polymerization of G-actin in F-actin.
Fig. 6.
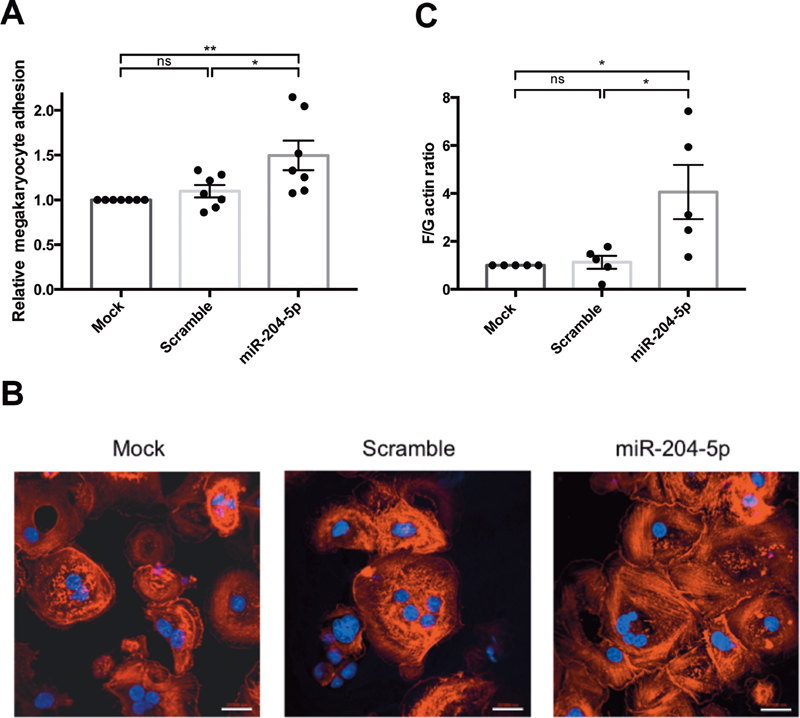
miR-204-5p modulated megakaryocyte adhesion and induced cytoskeletal reorganization. ( A ) Flow chamber experiments showed that miR-204-5p transfection increased the adhesion of megakaryocytes onto a fibrinogen-coated channel. ( B ) Confocal analysis of the megakaryocytes adhering to fibrinogen-coated coverslips in static conditions showed that miR-204-5p induced modification in actin cytoskeleton organization, with an increase in stress-fiber content. Polylobed nuclei were stained with DAPI ( blue ), and F-actin was stained with TRITC-phalloidin ( red , LSM700 microscope, scale bars = 20 μm). ( C ) miR-204-5p transfection enhances the F/G actin ratio in megakaryocytes. n = 3 to 6 independent experiments; * p < 0.05.
CDC42 Mediated the Effect of miR-204-5p on PLS
SiCDC42 transfection represses CDC42 protein expression by about 42 ± 7% ( Supplementary Fig. S3 , available in the online version). Downregulation of CDC42 produced very similar data to that obtained from the miR-204-5p transfection of megakaryocytes. Using our flow chamber assay, CDC42 silencing induced a 46 ± 14% increase in megakaryocyte adhesion ( Fig. 7A ) but had no effect on PLS adhesion (data not shown). Silencing of CDC42 and the overexpression of miR-204-5p showed similar F-actin organization, with greater stress fiber formation in megakaryocytes ( Fig. 7B ). The area of the proPLS network decreased by 38 ± 7% after CDC42 silencing ( Fig. 7C ). The pattern was similar to that observed after the overexpression of miR-204-5p, with a less expansive proPLS network and shorter protrusions than in control conditions ( Figs. 7D and 4B ). Finally, CDC42 silencing had no effect on P-selectin secretion ( Fig. 7E ) but increased the activated form of GPIIbIIIa by 22 ± 3% in PLS ( Fig. 7F ) and the PLS spreading area by 34 ± 6% ( Fig 7G and H ).
Fig. 7.
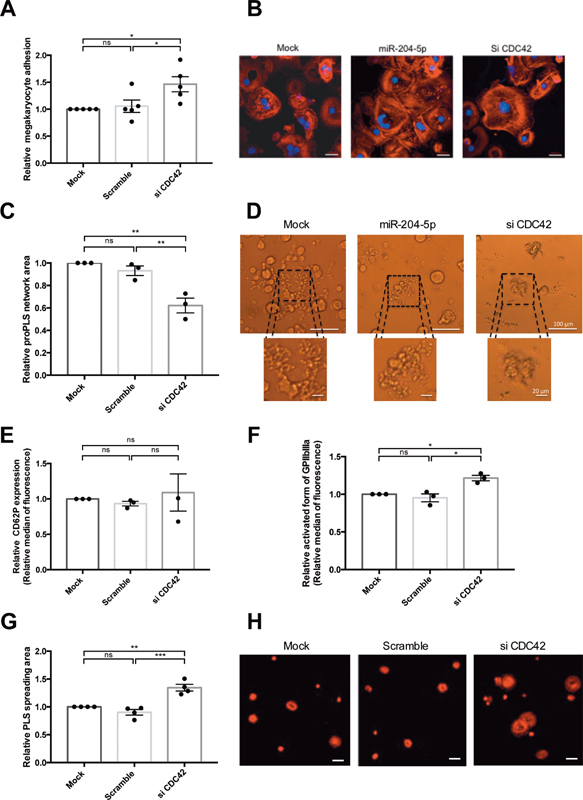
CDC42 mediated the effect of miR-204-5p on PLS. ( A ) CDC42 knock-down increased the adhesion of megakaryocytes in a fibrinogen-coated channel in dynamic conditions and ( B ) led to an actin cytoskeleton organization similar to that observed after miR-204-5p transfection with an increase in the stress-fiber content. Images were taken using confocal microscopy of megakaryocytes, with polylobed nucleus labeled with DAPI ( blue ) and F-actin labeled with TRITC-phalloidin ( red ) (scale bars = 20 μm). ( C ) Knock-down of CDC42 decreased the relative proPLS network area and ( D ) displayed a similar pattern to that observed after miR-204-5p transfection (Si-3000 camera, scale bars = 100 μm). ( E ) CDC42 silencing did not impact CD62P expression and ( F ) increased relative GPIIbIIIa expression in activated PLS and ( G and H ) enhanced the PLS spreading area on fibrinogen-coated coverslips (LSM700 microscope, scale bars = 20 μm). Results are expressed relative to the mock condition. n = 3 to 5 independent experiments; * p < 0.05, ** p < 0.01. PLS, platelet-like structure.
Discussion
This study investigated the functional impact of miR-204-5p in regulating PR using human cells. This step is of utmost importance to validate platelet-derived miRNA identified through association studies and proposed as biomarker of PR and/or recurrence of cardiovascular events. 24 PLS were derived from human hematopoietic stem cells according to a protocol which enables the production of elements ultra-structurally and functionally similar to circulating platelets. 19 25
During the differentiation process of hematopoietic stem cells, the level of miR-204-5p decreases over time, suggesting that its targets (including CDC42) are overexpressed in the last stages of the differentiation process. We first studied the effect of miR-204-5p on megakaryocyte differentiation and PLS formation. Our results showed that miR-204-5p had no effect on the polyploidization of megakaryocytes, nor on the proportion of megakaryocytes expressing the differentiation markers. However, transfection of megakaryocytes with miR-204-5p resulted in a radical modification of the nucleus polarization and DMS structure, an abnormal generation of the PLS network, and a retraction of the proPLS buds. The DMS is a membrane reservoir for proplatelet formation and extension, 23 26 and abnormal development of the DMS has been implicated in several hereditary diseases associated with macrothrombocytopenia. 27 28 Moreover, Mountford et al 29 showed that alteration of the DMS structure did not affect platelet production but dysregulated platelet adhesion, resulting in the formation of large, unstable thrombi. Taken together, those studies clearly indicated that DMS reorganization in megakaryocytes is critical for platelet production and function; therefore, modifications in the localization and organization of the DMS, induced by miR-204-5p overexpression, may lead to the production of functionally altered PLS.
Confocal microscopy analysis of megakaryocytes showed that miR-204-5p transfection was associated with a marked modification of cytoskeletal organization and an increase in stress-fiber content. Stress fibers are mainly composed by polymerized actin. G-actin polymerizes to form F-actin. In resting cells, about 40% of actin is in F form and this proportion rises to 80% after activation. 30 Dysregulation of the actin cytoskeleton was also shown to impair the generation of mature platelets, 31 32 indicating an effect of F-actin on both megakaryocyte architecture and platelet formation. The stimulated megakaryocytes in our study exhibited a greater F/G actin ratio in megakaryocytes transfected with miR-204-5p, indicating that miR-204-5p enhances F-actin polymerization. Actin polymerization is regulated by the Rho GTPases family, including RHOA, RAC1, and CDC42, which control the formation of stress fibers, lamellipodia, and filopodia, respectively. 33 Since CDC42 is a direct target of miR-204-5p, 15 we used a siRNA to downregulate its expression and we recapitulated the phenotype obtained with miR-204-5p overexpression. This suggests that the changes in megakaryocyte morphology associated with miR-204-5p transfection were mediated, at least in part, via CDC42 downregulation. Palazzo et al 34 showed that CDC42 is involved in cytoskeletal changes during differentiation via its effector N-WASP. The knock-down of N-WASP enhanced stress-fiber formation, further supporting the hypothesis that CDC42 regulates stress-fiber formation. Moreover, Antkowiak et al 17 showed that a deficiency in CDC42 dramatically impairs DMS biogenesis in megakaryocytes. They observed a similar pattern with the formation of abortive protrusions, bearing enlarged tips, which retracted into the cell after drug-induced inhibition of CDC42 in megakaryocytes. Lastly, they showed that formation of mature DMS is intimately linked to F-actin turnover through a microtubule-independent mechanism. Taken together, these results suggest that miR-204-5p modulates DMS formation through downregulation of CDC42 and an effect on F-actin polymerization into stress fibers.
The mechanism by which CDC42 increases stress fiber formation remains unclear. Due to several experimental limitations—notably the low number of megakaryocytes and PLS obtained in vitro and subsequently the small amount of protein recovered—it was not possible to use dedicated techniques allowing the study of protein phosphorylation, or the degree of activation of the Rho GTPases, the detection limits being reached. However, at least two hypotheses can be put forward. Stress-fiber formation passes through a RHOA-dependent process, and Rho GTPases interact with each other. 35 For example, in the initial steps of cell adhesion and spreading, the RHOA-GTP level is inhibited whereas RAC1 and CDC42 activation increases, resulting in suppressed actomyosin contractility and enhanced actin-mediated protrusion. At a later phase, RAC1 and CDC42 decrease and the activity of RHOA increases gradually inducing the formation of stress fibers and the maturation of focal adhesion. 36 In addition, RAC GTPase negatively regulates the RHOA-induced stress-fiber formation and focal adhesion assembly through its effector p21-activated kinases (PAK) at the leading edge. 37 Those studies indicate that Rho GTPases ensure a coordinated control of cytoskeleton modification. We observed a direct downregulation of CDC42 by miR-204-5p that does not impact RhoA and Rac1 mRNA expression level. However, this does not exclude a modification of the activity of these latter Rho GTPase family members, in particular RhoA, leading to the increased formation of stress fibers.
An alternative hypothesis is based on the actin depolymerization factor (ADF/cofilin) function. This family of actin-binding proteins acts as a negative regulator of actin stress fibers by depolymerizing and severing the existing actin filaments. Phosphorylation of cofilin by LIM kinases inhibits cofilin-induced actin depolymerization resulting in the formation of stress fibers. 38 LIM kinases are common downstream effectors of Rho GTPases and are activated by p65 PAK and p160 ROCK downstream of CDC42, RAC1, and RHOA. We can thus speculate that in miR-204-5p-transfected cells, downregulation of CDC42 induces an increased LIM kinases activity, leading to an enhanced amount of F-actin. Further studies are needed to understand the molecular pathways underlying the overexpression of stress fibers in miR-204-5p-transfected cells.
In our study, human megakaryocyte transfection with miR-204-5p had no impact on the number of megakaryocytes releasing a proPLS network, but it induced a slight but significant increase in the number of PLS produced. Conversely to our in vitro observations, proplatelet formation in Cdc42 −/− mice was moderately reduced. 39 40 These conflicting results might be explained by the PLS release in static conditions and the collection via flushing through a pipette tip leading to the mechanical release of all the PLS present in the megakaryocyte, whereas in the in vivo model, the elongation process allows the gradual release of proplatelets into the bloodstream. In addition, Pleines et al 39 showed that murine Cdc42 −/− platelets display a dramatic decreased life span in vivo, resulting in an increased turnover that can at least in part explain the moderate thrombocytopenia observed in these animals.
The functional impact of miR-204-5p was firstly investigated using a model of cell adhesion in dynamic conditions already described in our previous study. 12 Flow experiments showed an increase in megakaryocyte adhesion on fibrinogen after miR-204-5p transfection that was recapitulated after silencing. This indicates that miR-204-5p plays a role in megakaryocyte adhesion in flow conditions, at least partly due to its effect on CDC42. Surprisingly, PLS adhesion in dynamic conditions, after either miR-204-5p transfection or silencing CDC42, remained unaffected. These results may be due to the absence of red blood cells that contribute to PLS margination and enhance contact between the platelets and the channel, 41 while this margination effect is less important for larger cells such as megakaryocytes. It is of note that megakaryocyte function is often used as a proxy for platelet function, 42 suggesting that the functional data obtained with megakaryocytes might be translatable to PLS.
miR-204-5p transfection had no effect on the size of the PLS produced but induced an upregulation of CD41 and CD61, the two subunits of the fibrinogen receptor. Although the magnitude of this modulation may be considered as modest, it is noteworthy that it may have functional consequences. 43 The magnitude of this upregulation is in line with the 30% increase of the GPIIbIIIa active form after PLS activation. In addition, we demonstrated that miR-204-5p transfection of megakaryocytes produced PLS with an increase of fibrinogen binding and spreading area by approximately 50%. This suggests that miR-204-5p mediates its effect on PLS binding to fibrinogen at least in part via GPIIbIIIa upregulation, and via an increase of its affinity for fibrinogen. Taken together, these data demonstrated that miR-204-5p increases PLS binding to fibrinogen, and this may contribute to the higher PR phenotype.
Since silencing CDC42 recapitulates the phenotype associated with miR-204-5p transfection, this validated target of miR-204-5p plays a major role in the regulation of PR. The role of CDC42 on PR has been studied in mice, using different knockout approaches and leading to conflicting results. Using an Mx-cre; Cdc42 lox/lox mouse model, Akbar et al 44 showed that Cdc42 knockout was associated with an inhibition of filopodia formation, a decrease in P-selectin release, and an inhibition of agonist-induced platelet aggregation. Conversely, Pleines et al, 39 using Pf4-cre Cdc42 −/− knockout mice, found normal filopodia formation, increased aggregation upon low agonist stimulation, a reduced GPIIbIIIa activation, and an unaltered platelet spreading on human fibrinogen. The authors attribute these discrepancies to the use of different methodologies. 44 In our study we observed that miR-204-5p transfection resulted in an increase of GPIIbIIIa activation and PLS spreading on fibrinogen without affecting P-selectin expression. The discrepancies observed with the study of Pleines et al 39 may be due to the difference between the models used. Indeed, the platelets from Cdc42 KO mice seem less mature than platelets from wild-type mice with an increased in size and a decrease in GPIIbIIIa expression. This suggests that constitutive and total defect of CDC42 in mouse have an impact on megakaryocyte differentiation and platelet production and subsequently on platelet functionality. In contrast, miR-204-5p transfection of human-derived megakaryocyte induced a partial downregulation of CDC42 and has no impact on megakaryocyte differentiation, neither on surface receptor expression.
The exact mechanism allowing an increase in GPIIbIIIa expression after miR-204-5p transfection remains unclear. Since the Rho family GTPase proteins have been described as modulators of GPIIbIIIa activity, 16 45 we speculate that miR-204-5p may modulate GPIIbIIIa expression and activity, either directly or indirectly via the modulation of other members of the Rho family of GTPases. The small Rho GTPase RAP1b is known to play a key role in regulating outside-in signaling and could be a link between GPIIbIIIa activation and the cytoskeleton. 35 46 47 Indeed, inhibition of actin polymerization by cytochalasin D or latrunculin A impaired RAP1b-dependent fibrinogen binding in stimulated megakaryocytes 46 and RAP1b can enhance agonist-induced ligand binding to GPIIbIIIa possibly by modulating the interactions of GPIIbIIIa with the actin cytoskeleton. 46 We can thus speculate that in our study the increased actin polymerization induced by miR-204-5p transfection could result in an enhanced integrin affinity mediated at least in part by RAP1.
In conclusion, our findings demonstrated that the overexpression of miR-204-5p has an impact on the actin cytoskeleton and the localization of DMS in megakaryocytes which produce PLS. In addition, miR-204-5p overexpression upregulates PR through, at least in part, an increased expression of the GPIIbIIIa receptor. These structural and functional effects are mediated via the downregulation of CDC42. Since miR-204-5p is predicted to target thousands of mRNAs, our results do not exclude the involvement of other genes. To the best of our knowledge, this study is the first dedicated to the functional validation of miR-204-5p as a regulator of PR in human-derived cells, and it supports the idea that miR-204-5p has a potential role as a biomarker for PR, a phenotype of importance in a variety of clinical conditions.
Acknowledgements
The authors wish to thank Nicolas Liaudet (Bioimaging Core Facility, University of Geneva) for the image-processing of pictures from confocal microscopy, as well as Cecile Gamero, Jean-Pierre Aubry (Flow Cytometry Core Facility, University of Geneva), Solange Moll, and Phillippe Henchoz (Division of Clinical Pathology, University Hospitals of Geneva). We also wish to thank Julie Bodart for excellent technical assistance, Thomas Lecompte for helpful discussions, and the production and quality control staff at Geneva's blood transfusion center.
Funding Statement
Funding This work was supported by the Private Foundation of the University Hospitals of Geneva (grant RC04-05).
Conflict of Interest None declared.
Author Contribution
A.G., S.D-.G., C.S., J.-L.R., and P.F. designed the study and analyzed the data. A.G., S.D-G., and S. N. performed the experiments and analyzed the data. A.G., S.D-.G., and P.F. wrote the first draft of the manuscript, and all authors revised the intellectual content and approved the final version.
What is known about this topic?
miR-204-5p is a candidate biomarker of platelet reactivity.
CDC42 is a direct target of miR-204-5p.
What does this paper add?
miR-204-5p modulates megakaryocyte structure and platelet biogenesis via CDC42.
miR-204-5p transfection enhances platelet reactivity via the regulation of GPIIbIIIa expression.
CDC42 silencing induces a similar phenotype to miR-204-5p transfection suggesting that miR-204-5p increases platelet reactivity at least in part through regulation of CDC42.
Supplementary Material
References
- 1.Franco A T, Corken A, Ware J. Platelets at the interface of thrombosis, inflammation, and cancer. Blood. 2015;126(05):582–588. doi: 10.1182/blood-2014-08-531582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Semple J W, Italiano J E, Jr, Freedman J. Platelets and the immune continuum. Nat Rev Immunol. 2011;11(04):264–274. doi: 10.1038/nri2956. [DOI] [PubMed] [Google Scholar]
- 3.Mallouk N, Labruyère C, Reny J L. Prevalence of poor biological response to clopidogrel: a systematic review. Thromb Haemost. 2012;107(03):494–506. doi: 10.1160/TH11-03-0202. [DOI] [PubMed] [Google Scholar]
- 4.Framingham Heart Study . O'Donnell C J, Larson M G, Feng D. Genetic and environmental contributions to platelet aggregation: the Framingham heart study. Circulation. 2001;103(25):3051–3056. doi: 10.1161/01.cir.103.25.3051. [DOI] [PubMed] [Google Scholar]
- 5.Fontana P, Roffi M, Reny J L. Platelet function test use for patients with coronary artery disease in the early 2020s. J Clin Med. 2020;9(01):9. doi: 10.3390/jcm9010194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plé H, Landry P, Benham A, Coarfa C, Gunaratne P H, Provost P. The repertoire and features of human platelet microRNAs. PLoS One. 2012;7(12):e50746. doi: 10.1371/journal.pone.0050746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sunderland N, Skroblin P, Barwari T. MicroRNA biomarkers and platelet reactivity: the clot thickens. Circ Res. 2017;120(02):418–435. doi: 10.1161/CIRCRESAHA.116.309303. [DOI] [PubMed] [Google Scholar]
- 8.Landry P, Plante I, Ouellet D L, Perron M P, Rousseau G, Provost P. Existence of a microRNA pathway in anucleate platelets. Nat Struct Mol Biol. 2009;16(09):961–966. doi: 10.1038/nsmb.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagalla S, Shaw C, Kong X. Platelet microRNA-mRNA coexpression profiles correlate with platelet reactivity. Blood. 2011;117(19):5189–5197. doi: 10.1182/blood-2010-09-299719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kondkar A A, Bray M S, Leal S M. VAMP8/endobrevin is overexpressed in hyperreactive human platelets: suggested role for platelet microRNA. J Thromb Haemost. 2010;8(02):369–378. doi: 10.1111/j.1538-7836.2009.03700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaudewitz D, Skroblin P, Bender L H. Association of microRNAs and YRNAs with platelet function. Circ Res. 2016;118(03):420–432. doi: 10.1161/CIRCRESAHA.114.305663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia A, Dunoyer-Geindre S, Zapilko V, Nolli S, Reny J L, Fontana P. Functional validation of microRNA-126-3p as a platelet reactivity regulator using human haematopoietic stem cells. Thromb Haemost. 2019;119(02):254–263. doi: 10.1055/s-0038-1676802. [DOI] [PubMed] [Google Scholar]
- 13.Zufferey A, Ibberson M, Reny J L. New molecular insights into modulation of platelet reactivity in aspirin-treated patients using a network-based approach. Hum Genet. 2016;135(04):403–414. doi: 10.1007/s00439-016-1642-1. [DOI] [PubMed] [Google Scholar]
- 14.Ding T, Zeng X, Cheng B. Platelets in acute coronary syndrome patients with high platelet reactivity after dual antiplatelet therapy exhibit upregulation of miR-204-5p. Ann Clin Lab Sci. 2019;49(05):619–631. [PubMed] [Google Scholar]
- 15.Ma L, Deng X, Wu M, Zhang G, Huang J. Down-regulation of miRNA-204 by LMP-1 enhances CDC42 activity and facilitates invasion of EBV-associated nasopharyngeal carcinoma cells. FEBS Lett. 2014;588(09):1562–1570. doi: 10.1016/j.febslet.2014.02.039. [DOI] [PubMed] [Google Scholar]
- 16.Pleines I, Cherpokova D, Bender M. Rho GTPases and their downstream effectors in megakaryocyte biology. Platelets. 2019;30(01):9–16. doi: 10.1080/09537104.2018.1478071. [DOI] [PubMed] [Google Scholar]
- 17.Antkowiak A, Viaud J, Severin S. Cdc42-dependent F-actin dynamics drive structuration of the demarcation membrane system in megakaryocytes. J Thromb Haemost. 2016;14(06):1268–1284. doi: 10.1111/jth.13318. [DOI] [PubMed] [Google Scholar]
- 18.Aslan J E, McCarty O J. Rac and Cdc42 team up for platelets. Blood. 2013;122(18):3096–3097. doi: 10.1182/blood-2013-08-516906. [DOI] [PubMed] [Google Scholar]
- 19.Strassel C, Brouard N, Mallo L. Aryl hydrocarbon receptor-dependent enrichment of a megakaryocytic precursor with a high potential to produce proplatelets. Blood. 2016;127(18):2231–2240. doi: 10.1182/blood-2015-09-670208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kok M G, Halliani A, Moerland P D, Meijers J C, Creemers E E, Pinto-Sietsma S J. Normalization panels for the reliable quantification of circulating microRNAs by RT-qPCR. FASEB J. 2015;29(09):3853–3862. doi: 10.1096/fj.15-271312. [DOI] [PubMed] [Google Scholar]
- 21.Vandesompele J, De Preter K, Pattyn F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(07):H0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dütting S, Gaits-Iacovoni F, Stegner D. A Cdc42/RhoA regulatory circuit downstream of glycoprotein Ib guides transendothelial platelet biogenesis. Nat Commun. 2017;8:15838. doi: 10.1038/ncomms15838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eckly A, Heijnen H, Pertuy F. Biogenesis of the demarcation membrane system (DMS) in megakaryocytes. Blood. 2014;123(06):921–930. doi: 10.1182/blood-2013-03-492330. [DOI] [PubMed] [Google Scholar]
- 24.Garcia A, Dunoyer-Geindre S, Fish R J. Methods to Investigate miRNA function: focus on platelet reactivity. Thromb Haemost. 2021;121(04):409–421. doi: 10.1055/s-0040-1718730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Six K R, Sicot G, Devloo R, Feys H B, Baruch D, Compernolle V. A comparison of haematopoietic stem cells from umbilical cord blood and peripheral blood for platelet production in a microfluidic device. Vox Sang. 2019;114(04):330–339. doi: 10.1111/vox.12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schulze H, Korpal M, Hurov J. Characterization of the megakaryocyte demarcation membrane system and its role in thrombopoiesis. Blood. 2006;107(10):3868–3875. doi: 10.1182/blood-2005-07-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strassel C, Eckly A, Léon C. Intrinsic impaired proplatelet formation and microtubule coil assembly of megakaryocytes in a mouse model of Bernard-Soulier syndrome. Haematologica. 2009;94(06):800–810. doi: 10.3324/haematol.2008.001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eckly A, Strassel C, Freund M. Abnormal megakaryocyte morphology and proplatelet formation in mice with megakaryocyte-restricted MYH9 inactivation. Blood. 2009;113(14):3182–3189. doi: 10.1182/blood-2008-06-164061. [DOI] [PubMed] [Google Scholar]
- 29.Mountford J K, Petitjean C, Putra H W. The class II PI 3-kinase, PI3KC2α, links platelet internal membrane structure to shear-dependent adhesive function. Nat Commun. 2015;6:6535. doi: 10.1038/ncomms7535. [DOI] [PubMed] [Google Scholar]
- 30.Barkalow K L, Italiano J E, Jr, Chou D E, Matsuoka Y, Bennett V, Hartwig J H. Alpha-adducin dissociates from F-actin and spectrin during platelet activation. J Cell Biol. 2003;161(03):557–570. doi: 10.1083/jcb.200211122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartwig J H, Italiano J E., Jr Cytoskeletal mechanisms for platelet production. Blood Cells Mol Dis. 2006;36(02):99–103. doi: 10.1016/j.bcmd.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 32.Bender M, Eckly A, Hartwig J H. ADF/n-cofilin-dependent actin turnover determines platelet formation and sizing. Blood. 2010;116(10):1767–1775. doi: 10.1182/blood-2010-03-274340. [DOI] [PubMed] [Google Scholar]
- 33.Nayak R C, Chang K H, Vaitinadin N S, Cancelas J A. Rho GTPases control specific cytoskeleton-dependent functions of hematopoietic stem cells. Immunol Rev. 2013;256(01):255–268. doi: 10.1111/imr.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palazzo A, Bluteau O, Messaoudi K. The cell division control protein 42-Src family kinase-neural Wiskott-Aldrich syndrome protein pathway regulates human proplatelet formation. J Thromb Haemost. 2016;14(12):2524–2535. doi: 10.1111/jth.13519. [DOI] [PubMed] [Google Scholar]
- 35.Aslan J E, McCarty O J. Rho GTPases in platelet function. J Thromb Haemost. 2013;11(01):35–46. doi: 10.1111/jth.12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huveneers S, Danen E H.Adhesion signaling - crosstalk between integrins, Src and Rho J Cell Sci 2009122(Pt 8):1059–1069. [DOI] [PubMed] [Google Scholar]
- 37.Sanders L C, Matsumura F, Bokoch G M, de Lanerolle P.Inhibition of myosin light chain kinase by p21-activated kinase Science 1999283(5410):2083–2085. [DOI] [PubMed] [Google Scholar]
- 38.Sumi T, Matsumoto K, Takai Y, Nakamura T. Cofilin phosphorylation and actin cytoskeletal dynamics regulated by rho- and Cdc42-activated LIM-kinase 2. J Cell Biol. 1999;147(07):1519–1532. doi: 10.1083/jcb.147.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pleines I, Eckly A, Elvers M. Multiple alterations of platelet functions dominated by increased secretion in mice lacking Cdc42 in platelets. Blood. 2010;115(16):3364–3373. doi: 10.1182/blood-2009-09-242271. [DOI] [PubMed] [Google Scholar]
- 40.Pleines I, Dütting S, Cherpokova D. Defective tubulin organization and proplatelet formation in murine megakaryocytes lacking Rac1 and Cdc42. Blood. 2013;122(18):3178–3187. doi: 10.1182/blood-2013-03-487942. [DOI] [PubMed] [Google Scholar]
- 41.Weisel J W, Litvinov R I. Red blood cells: the forgotten player in hemostasis and thrombosis. J Thromb Haemost. 2019;17(02):271–282. doi: 10.1111/jth.14360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Basak I, Bhatlekar S, Manne B K. miR-15a-5p regulates expression of multiple proteins in the megakaryocyte GPVI signaling pathway. J Thromb Haemost. 2019;17(03):511–524. doi: 10.1111/jth.14382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dupont A, Fontana P, Bachelot-Loza C. An intronic polymorphism in the PAR-1 gene is associated with platelet receptor density and the response to SFLLRN. Blood. 2003;101(05):1833–1840. doi: 10.1182/blood-2002-07-2149. [DOI] [PubMed] [Google Scholar]
- 44.Akbar H, Shang X, Perveen R. Gene targeting implicates Cdc42 GTPase in GPVI and non-GPVI mediated platelet filopodia formation, secretion and aggregation. PLoS One. 2011;6(07):e22117. doi: 10.1371/journal.pone.0022117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goggs R, Williams C M, Mellor H, Poole A W. Platelet Rho GTPases-a focus on novel players, roles and relationships. Biochem J. 2015;466(03):431–442. doi: 10.1042/BJ20141404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bertoni A, Tadokoro S, Eto K. Relationships between Rap1b, affinity modulation of integrin alpha IIbbeta 3, and the actin cytoskeleton. J Biol Chem. 2002;277(28):25715–25721. doi: 10.1074/jbc.M202791200. [DOI] [PubMed] [Google Scholar]
- 47.Chrzanowska-Wodnicka M, Smyth S S, Schoenwaelder S M, Fischer T H, White G C., II Rap1b is required for normal platelet function and hemostasis in mice. J Clin Invest. 2005;115(03):680–687. doi: 10.1172/JCI22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


