Abstract
A flexible method to image unmodified transcripts and transcription in vivo would be a valuable tool to understand the regulation and dynamics of transcription. Here, we present a novel approach to follow native transcription, with fluorescence microscopy, in live C. elegans. By using the fluorescently tagged Argonaute protein NRDE-3, programmed by exposure to defined dsRNA to bind to nascent transcripts of the gene of interest, we demonstrate transcript labelling of multiple genes, at the transcription site and in the cytoplasm. This flexible approach does not require genetic manipulation, and can be easily scaled up by relying on whole-genome dsRNA libraries. We apply this method to image the transcriptional dynamics of the heat-shock inducible gene hsp-4 (a member of the hsp70 family), as well as two transcription factors: ttx-3 (a LHX2/9 orthologue) in embryos, and hlh-1 (a MyoD orthologue) in larvae, respectively involved in neuronal and muscle development.
INTRODUCTION
Understanding the dynamics of transcription is essential to deciphering cell fate specification and maintenance. While the use of fluorescent proteins as transcriptional reporters has been invaluable in this regard, it provides little information on the amount of endogenous transcripts and their dynamics. Methods such as single molecule fluorescent in situ hybridization (smFISH) or single-cell RNA sequencing, on fixed or dissociated tissues, allow for precise measurements of transcript abundance at the transcription site and in the nucleus, but only as a snapshot, missing information on the dynamics of transcription.
Over the last two decades, several methods were developed to label and quantify mRNA in living cells or animals (1). To this day, the most popular method for in vivo mRNA labelling, either of active transcription sites or individual mRNAs, is the phage-derived MS2-MCP system (2), which requires modifying the target transcript to add multiple stem-loop MS2 repeats, on which the fluorescently tagged MS2 coat protein (MCP) binds with high affinity. Thanks to this powerful method, studies have observed dynamics and bursting transcription in cultured cells; in recent years it has been applied to live animal studies in the Drosophila embryo (3), in the mouse (4), and Caenorhabditis elegans (5). It requires, however, editing the gene of interest to insert multiple MS2 repeats or using a reporter transgene, and is difficult to scale up to assay the expression of many different genes; as a result, it has only seen limited uses in whole organisms so far. Additionally, the numerous repeats have been reported to interfere with transcript processing and cytoplasmic export (6).
More recently, alternative methods that label unmodified transcripts have been developed, first using a fluorescently labelled Pumilio homology domain (7), which can be programmed at the protein level to bind to specific 8-nucleotide RNA sequences (8), then with the use of catalytically inactive Cas9 programmed with chimeric oligonucleotides (9), or Cas13 programmed with a long guide RNA (10,11), to bind to a transcript of interest. These newer methods have so far only been used in cultured cells, to image accumulation of transcripts in some cellular compartments (stress granules and paraspeckles). However, to image transcription sites or single transcripts, binding of multiple fluorescent proteins on one transcript becomes necessary. Since this would require multiple guide RNAs tiling over the transcript, it renders these new methods less powerful. While efforts have expanded significantly in recent years (1), there is still a need to develop more flexible systems to visualize native mRNAs in live cells and organisms. Particularly useful qualities would be easy programmability to bind to any RNA sequence; a good signal-to-noise ratio, to allow imaging down to single molecules; scalability up to the whole genome; and applicability to cells in culture as well as whole organisms.
Here, we report the development in the model organism C. elegans of a novel method to image transcription in vivo, relying on the sequence-specific binding of the Argonaute NRDE-3 to transcripts of interest, that improves on existing approaches. We demonstrate its usefulness to study the transcriptional dynamics of transcription factors and a heat shock gene, in the first reported live imaging of transcription of unmodified genes in an animal. This method allows imaging both at transcription sites and in the cytoplasm, and is inexpensive to deploy.
MATERIALS AND METHODS
Mutants
Alleles used in this study were eri-1(mg366), nrde-2(gg95), and nrde-1(gg88).
Transgenes
vbaIs52: peef-1A.1::YFP::nrde-3 II
vbaIs53: prps-27::mNeonGreen::flag::nrde-3 II
vbaIs54: peef-1A.1::YFP::nrde-3::SL2::sid-1 II
vbaIs55: peef-1A.1::VenusC::nrde-3 II
vbaIs56: peef-1A.1::VenusN::nrde-3 I
All transgenes were integrated as single copy by CRISPR, at loci ttTi5605 (chrII) or ttTi4348 (chrI).
Strains
VBS662 eri-1(mg366); nrde-2(gg95); vbaIs52
VBS663 eri-1(mg366); nrde-2(gg95); vbaIs53
VBS664 eri-1(mg366); nrde-2(gg95); vbaIs55; vbaIs56
VBS668 eri-1(mg366); nrde-2(gg95); vbaIs54
Plate preparation and dsRNA exposure
dsRNA targetting the gene of interest is expressed by bacteria, from the Ahringer library (12); bacterial clones were sequenced to verify identity, and we tested the specificity to the gene of interest with Clonemapper (13). The clone targetting the gene his-13 (ZK131.7) also matches other histone genes at several other loci across the genome.
dsRNA-expression plates were prepared fresh, at most one week before experiment. Bacteria culture expressing dsRNA against the gene of interest were plated, and the following day L3–L4 worms added on the plate. Nuclear localization and transcription spots were then scored on the next generation. If exposure to dsRNA persists for multiple generations, we found that transcription spots became more difficult to observe, possibly due to residual epigenetic silencing in the germline.
For certain genes, like elt-2 or ant-1.1, primary RNAi was sufficiently efficient to affect the fitness and prevent growth. To circumvent this knockdown, we diluted the bacteria expressing dsRNA targeting the gene of interest to 50% or 75% with bacteria carrying the negative control plasmid L4440.
Mounting and imaging
Worms were mounted on agar pads, and immobilized using levamisole or polystyrene beads. In our experience, immobilizing the worms with levamisole or azide negatively affects cellular processes and hinders the measurements of transcriptional dynamics. All live imaging was performed on a spinning disk confocal microscope, with laser power at 515 nm of 0.4 mW. For timelapse imaging, we adjusted the frequency of image acquisition to limit bleaching of weak signal while still achieving sufficient image resolution. For the timelapses presented here, we used frequencies of 4 min (for ttx-3 and hsp-4) or 2 min (for hlh-1).
α-amanitin treatment
Worms were exposed to dsRNA against hlh-1 for one generation, then L4 individuals were put in a liquid solution of α-amanitin at 20 μg/ml in M9, while the negative control individuals were exposed to M9 only, for 3 h before imaging.
smFISH
Because smFISH probes and NRDE-3 will bind to the same exonic sequences, competition could reduce efficiency of smFISH labelling. We therefore used a combination of smFISH probes targetting intronic as well as exonic sequences, since the secondary siRNA loaded by NRDE-3 will be restricted to exonic sequences. Worms were grown at 20°C on dsRNA-containing NGM plates, then fixed in 4% formaldehyde/PBS for 45 min at room temperature, washed twice with PBS then incubated in 70% ethanol overnight at 4°C to allow permeabilization. Embryos were equilibrated for 5 min with wash buffer (WB) containing 10% formamide and 2x saline-sodium citrate (SSC), then hybridized in hybridization buffer (10% formamide, 2× SSC, 100 mg/ml dextran sulfate, 1 mg/ml Escherichia coli tRNA, 2 mM vanadyl ribonucleoside complex, 0.2 mg/ml BSA) containing 0.125 μM of probes labelled with Quasar 670, overnight at 37°C. Following hybridization, samples were washed twice with WB including 30 min incubation at 37°C during the second wash then washed once in 2× SSC buffer before being mounted in Vectashield Mounting Medium with DAPI (centrifugation steps were done at 5000g). Imaging was performed on a spinning disk confocal microscope.
smiFISH
The protocol was adapted for C. elegans from the original publication (14).
As for smFISH, we used intronic and exonic probes; smiFISH secondary probes were labelled with cy5. Mixed stages worms were fixed in 4% formaldehyde in 1× PBS for 45 min; rinsed twice in PBS, then permeabilized in 70% ethanol at 4°C overnight. They were then rinsed in 15% formamide in 1× SSC, then incubated at room temperature for 30 min, before addition of the hybridization mix (2.5 μl 20× SSC, 0.8 μl 20 μg/μl S. cerevisiae tRNA, 7.5 μl formamide, 1μl probe duplexes, 24 μl H2O, 0.5 μl 200 mM vanadyl ribonucleoside complex, 13.2 μl 40% dextran sulfate) then incubated at 37°C overnight. Worms were then rinsed twice in 15% formamide in 1× SSC for 30 min, washed in PBS, and finally mounted in Vectashield. Images were acquired at laser power at 0.4 mW at 515 nm; 0.6 mW at 642 nm; 10.2 mW at 405 nm.
Image analysis
Image analysis was performed in Fiji; transcription spot measurements were performed in trackmate (15), on full stacks for ttx-3 and hsp-4 timelapses and hlh-1 images, and maximum intensity Z projection for hlh-1 timelapses. Fluorescent NRDE-3 spots were automatically segmented and tracked with a diameter of 1μm, with subsequent manual editing of tracks. For background subtraction, a similar sized spot was segmented in immediate proximity in the nucleus, and its mean intensity substracted from that of the segmented spot. To differentiate between tracking errors and transcription site disappearance, a spot was counted as absent if we were unable to observe it for more than two timeframes. Subsequent analysis was performed in R.
qPCR
Gravid adults were bleached, and synchronized larvae were plated on dsRNA-expression plates with bacteria expressing dsRNA against hlh-1 or GFP. Worms were harvested for RNA purification at the L4 stage using lysis reagent (Qiazol, Qiagen) according to manufacturers instructions. mRNA was reverse transcribed using SuperScript III Reverse Transcriptase (invitrogen). Quantitative PCR was carried out in CFX96 from Bio-Rad detection system with SYBR Green Master Mix (invitrogen) in 20 μl total volume reaction. Primers used to quantify hlh-1 span exons and intron boundaries, to be specific to pre mRNA, and do not match the dsRNA used.
tba-1: TCAACACTGCCATCGCCGCC/TCCAAGCGAGACCAGGCTTCAG; hlh-1: ACACTGACAAGTTTCGCTGC/GAGAGCTTGAGCTTCTCCCC
Three independent biological samples were carried, each of which was measured in 3 technical replicates.
RESULTS
Principle of the method
In the C. elegans exogenous RNAi pathway, exogenous double stranded RNA from the environment are recognized and processed into small interfering RNAs that are then loaded onto the primary Argonaute RDE-1 and bind to the target mRNA (Figure 1A). RDE-1 does not cleave the transcript (16), but recruits two protein complexes: RDE-8 (17) and RDE-12 (18,19), that degrade the target transcript, and induce recruitment of the RNA-dependent RNA polymerase (RdRP) RRF-1 (20). This polymerase synthesizes secondary single-stranded triphosphorylated siRNAs starting with a guanine, known as 22G siRNAs, complementary to the target mRNA, that tile over the transcript length, starting 5′ of the primary siRNA trigger (21,22). These 22G siRNAs are loaded into a nematode-specific family of secondary Argonautes (23,24) known as WAGO, mostly expressed in the germline, with the exception of the nuclear Argonaute NRDE-3 (25), expressed in the soma. Once NRDE-3 loads a 22G siRNA, it translocates from the cytoplasm to the nucleus, where it binds to the nascent transcript and recruits NRDE-2 (26), leading to blocking of transcript elongation, as well as chromatin modification through recruitment of NRDE-1 and NRDE-4 (27).
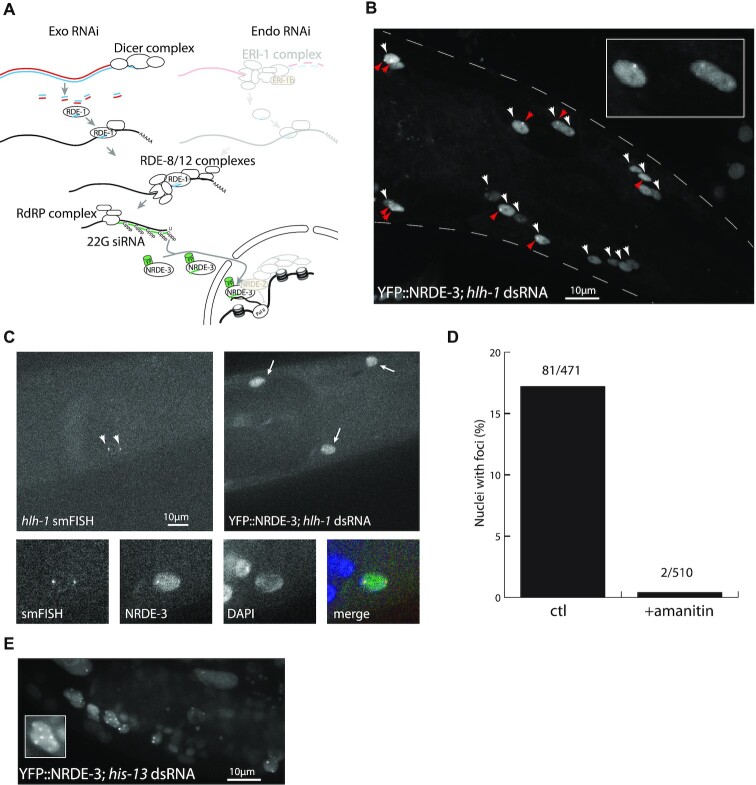
Figure 1.
Fluorescent NRDE-3 labels active transcription sites. (A) Simplified C. elegans RNAi pathway, as modified in our method. The exo-RNAi pathway is unaffected; the endo-RNAi pathway (upper right; greyed out) is abrogated by a mutation in ERI-1b (orange); and transcriptional silencing by the nuclear RNAi pathway (bottom right, greyed out) is blocked by a mutation in NRDE-2 (orange). NRDE-3 is labelled with a fluorescent protein (FP). (B) Labelling of hlh-1 transcription sites in larval head muscles (L4 stage). Muscle nuclei (white arrowheads) show nuclear localization of YFP::NRDE-3, while other tissues show cytoplasmic localization. In some of these nuclei, one or two bright foci (red arrowheads) are visible. Insert: enlargement of three nuclei. (C) Colocalization of smFISH signal and YFP::NRDE-3. SmFISH probes matching hlh-1 exonic and intronic sequences localize at the transcription sites. YFP::NRDE-3 localizes to the nucleus of muscle cells, and accumulates at foci coinciding with smFISH spots. (D) YFP::NRDE-3 foci are sensitive to amanitin. After exposure to α-amanitin, YFP::NRDE-3 foci were no longer observable. (E) dsRNA matching multicopy genes induce localization of YFP::NRDE-3 at multiple foci. dsRNA against the sequence of his-13 also matches the sequence of other histone genes, at multiple clusters across the genome. Worms fed with dsRNA matching multiple genes displayed more than two and up to eight YFP::NRDE-3 foci, in multiple tissues (somatic gonad, hypodermis). Insert: one developing somatic gonad nucleus with seven foci.
To label active transcription sites, we took advantage of this specific binding of NRDE-3 to nascent transcripts. We introduced two mutations (Figure 1A, orange): nrde-2(gg95), a putative null deletion allele to prevent the blocking of transcript elongation (26); and eri-1(mg366), to block the endogenous RNAi pathway that competes with the exogenous RNAi pathway for 22G siRNA synthesis (25). We do not expect these mutations to extensively interfere with biological processes: the eri-1(mg366) mutation, which blocks endogenous 26G RNAi pathway, is widely used to enhance RNAi efficiency and is therefore well characterized. 26G siRNAs can be divided in two classes: class I restricted to spermatogenesis, and class II present in embryos and larvae, mostly targetting recently duplicated genes and pseudogenes (28,29). The mutation in nrde-2, blocking the nrde pathway, has been described as reducing brood size (26) and inducing germline mortality at 25°C (30), but otherwise not associated with gross defects. Targets of the nrde pathway, downstream of 26G siRNAs, are limited and mostly represent repetitive genomic loci (31). NRDE-3 was tagged at the N-terminal end with a fluorescent protein (YFP or NeonGreen). After exposing worms to double-stranded RNA matching the sequence of a target transcript, and in the absence of transcriptional silencing due to the nrde-2(–) mutation, we expect to observe nuclear localization of fluorescently-labelled NRDE-3, and accumulation of NRDE-3 at transcription sites.
This method takes advantage of several specificities of C. elegans small RNA pathways: the dsRNA uptake pathway (through the SID-2 (32) and SID-1 (33) dsRNA transporters), which allows easy delivery of dsRNA against any gene of interest in the entire animal by feeding; the RdRP complex, which synthesizes a set of 22G siRNAs antisense to the transcript of interest, 5′ from the siRNA site; existence of the cleavage-deficient nuclear Argonaute NRDE-3, that can accumulate on the nascent transcript; and the availability of genome-wide dsRNA-synthesizing bacteria libraries (12).
Validation of the method
In the absence of endogenous RNAi due to eri-1(–), we observed cytoplasmic localization of NRDE-3 in the majority of tissues, as previously described (25). In the germline, early embryo and intestine, however, we observed nuclear localization in the absence of exogenous dsRNA, revealing the presence of an alternate source of 22G siRNAs in these tissues (Supplementary Figure S1). When worms were exposed to dsRNA matching the sequence of the muscle-specific transcription factor hlh-1, the C. elegans orthologue of MyoD, by feeding, we observed nuclear localization of NRDE-3 specifically in muscle cells (Figure 1B; white arrowheads), as previously described (25). While in an eri-1(–) background, nuclear localization of NRDE-3 spread to all cells after a few days (25), we did not observe such generalized nuclear localization in the eri-1(–); nrde-2(–) background. In a subset of muscle cells, we observed one or two bright spots in the nucleus, which we hypothesized to correspond to active hlh-1 transcription sites (Figure 1B, red arrowheads). To demonstrate that these nuclear spots are indeed active transcription sites, we performed fluorescent in situ hybridization targeting hlh-1 intronic and exonic sequences, and observed colocalization with YFP::NRDE-3 (Figure 1C; arrowheads), thus validating the principle of our method. To show that these foci were dependent on transcription, we tested the disappearance of foci after exposure to an RNA polymerase II inhibitor. In worms exposed to the Pol II inhibitor α-amanitin (34), 2/510 nuclei showed foci, while in untreated worms, we observed foci in 81/471 nuclei (Figure 1D). Finally, when worms were exposed to dsRNA matching the sequence of highly conserved histone genes at multiple locations across the genome (ZK131.7, his-13), we observed more than two, and up to eight spots per nucleus (Figure 1E), as expected from the transcription site labelling of multiple histone genes. The nuclear foci therefore reflect active transcription sites.
This labelling method, to be useful, should minimally interfere with the processes it aims to image. One possible concern is interference from the tagged NRDE-3 on transcription of the gene of interest. To verify the absence of transcriptional silencing in the nrde-2(–) genetic background, we performed a qRT PCR on the hlh-1 pre-mRNA, and found no evidence of downregulation in presence of dsRNA targeting hlh-1 (Supplementary Figure S2). Our results agree with previous observations showing little or no transcriptional silencing in an nrde-2(–) background (26). Additionally, we do not expect NRDE-3-based labelling to block splicing, since siRNAs are synthesized in the cytoplasm from mature transcripts and should not match the intron-exon boundaries present in a pre-mRNA. Finally, while Argonaute proteins do bind with high affinity to their targets, this should not be sufficient to block translation, as the similarly strong binding of MCP to the MS2 stem-loop is insufficient to block the progression of ribosomes (35). A mutant for NRDE-1, which is required for transcriptional silencing downstream of NRDE-2 (27), instead of a nrde-2 mutation, also allowed labelling of active transcription sites (Supplementary Figure S3).
We tested the applicability of this method on multiple genes, expressed at various levels in multiple tissues, and saw nuclear localization patterns and foci corroborating independently reported expression patterns (Figure 2): wnt ligand mom-2 in posterior cells in the mid embryo, matching expression pattern documented by smFISH (36); transcription factor hlh-1 in muscle cells in the late embryo, matching documented expression patterns (37); the highly expressed adenosine nuclear transporter ant-1.1 in all tissues; EGF ligand lin-3 in the anchor cell, matching previous data (38); GATA transcription factor elt-2 in the intestine (39); LIM homeobox transcription factor ttx-3 in AIY neurons in the embryo, matching previous data (40), (F. Soulavie and V. Bertrand, unpublished data). The method worked in most tissues; we found that we were generally able to observe nuclear localization, and in some of these nuclei we observed foci corresponding to active transcription sites (Supplementary Table S1). This method did not work, however, with every dsRNA tested, and we failed to observe specific nuclear localization or transcription foci for several genes, such as the notch-regulated gene sygl-1 in the germline (5). This likely reflects the variability of RNAi efficiency, depending on tissues and dsRNA sequence.
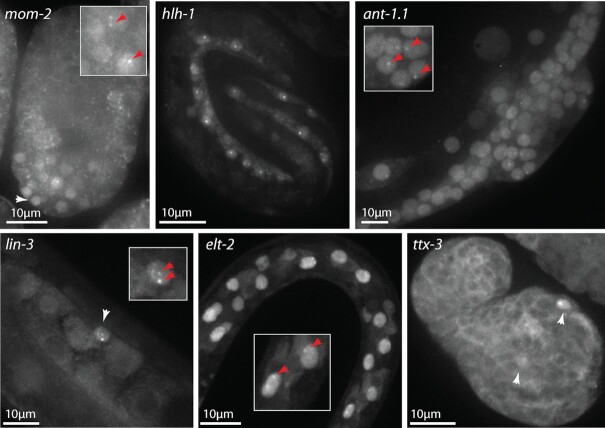
Figure 2.
NRDE-3 labelling of transcription sites works on multiple genes and tissues. After maternal exposure to mom-2 dsRNA, YFP::NRDE-3 is nuclear localized in the posterior part (white arrowhead) of the mid embryo, and accumulates at nuclear foci in some of these cells (red arrows). After maternal exposure to hlh-1 dsRNA, YFP::NRDE-3 is nuclear localized in muscle cells in the three-fold embryo, and accumulates at one or two transcription sites per nucleus. After exposure to ant-1.1 dsRNA, all cells in this L4 larva show nuclear localization of YFP::NRDE-3. In some of them (hypodermis, developing vulva, somatic gonad) transcription foci are visible (red arrows). After exposure to lin-3 dsRNA, the anchor cell at the third larval stage shows nuclear localization (white arrowhead) and two transcription foci (red arrows). After exposure to elt-2 dsRNA, nuclear localization is stronger in the intestine cells at the second larval stage, and some of them show transcription foci (red arrows). After maternal exposure to ttx-3 dsRNA, YFP::NRDE-3 is nuclear localized in AIY neurons in the comma stage embryo (white arrowheads) and, in one of them, a transcription site is visible.
Signal intensity will naturally be a function of the number of fluorescently tagged NRDE-3 bound to the target transcripts. It derives that a longer transcript, with more secondary siRNAs tiling over its length, will have a brighter signal than a shorter transcript. Similarily, since RRF-1 synthesizes siRNAs 5′ from the trigger site (Figure 1A), a short dsRNA, or one targetting the 5′ end of the transcript, imply a lesser accumulation of fluorescently tagged NRDE-3 on the transcript, and therefore a weaker signal than with a full-length dsRNA. Indeed, while using a full-length dsRNA against hlh-1 we observed clear nuclear foci (Figure 1B), when using a dsRNA trigger targetting only the first exon of hlh-1, we observed nuclear localization of NRDE-3, but failed to observe any nuclear foci.
Optimization of the method
Not all tissues were found to be equally amenable to transcriptional labelling. When a strain expressing YFP::NRDE-3 was exposed to dsRNA matching the sequence of the fluorescent protein, we would expect every fluorescent cell to have a strong nuclear signal; however, we observed that in neurons and pharynx, YFP::NRDE-3 still localized to the cytoplasm (Figure 3A). These tissues are known to have low RNAi efficiency, as they do not express the dsRNA transporter sid-1 (33). Since expressing sid-1 in these tissues could be sufficient to restore RNAi silencing by feeding (41), we co-expressed YFP::nrde-3 and sid-1 under the control of the same eef-1A.1 promoter and observed improved nuclear localization and labelling of transcription sites in neurons and the pharynx (Figure 3B). In addition, labelling of ttx-3 transcription, which already worked in neurons without sid-1 expression (Figure 2), was significantly improved with a stronger nuclear localization and clearer transcription spots (Figure 3C). Nonetheless, the general efficiency of labelling in neurons and pharynx still remained lower than in other somatic tissues.
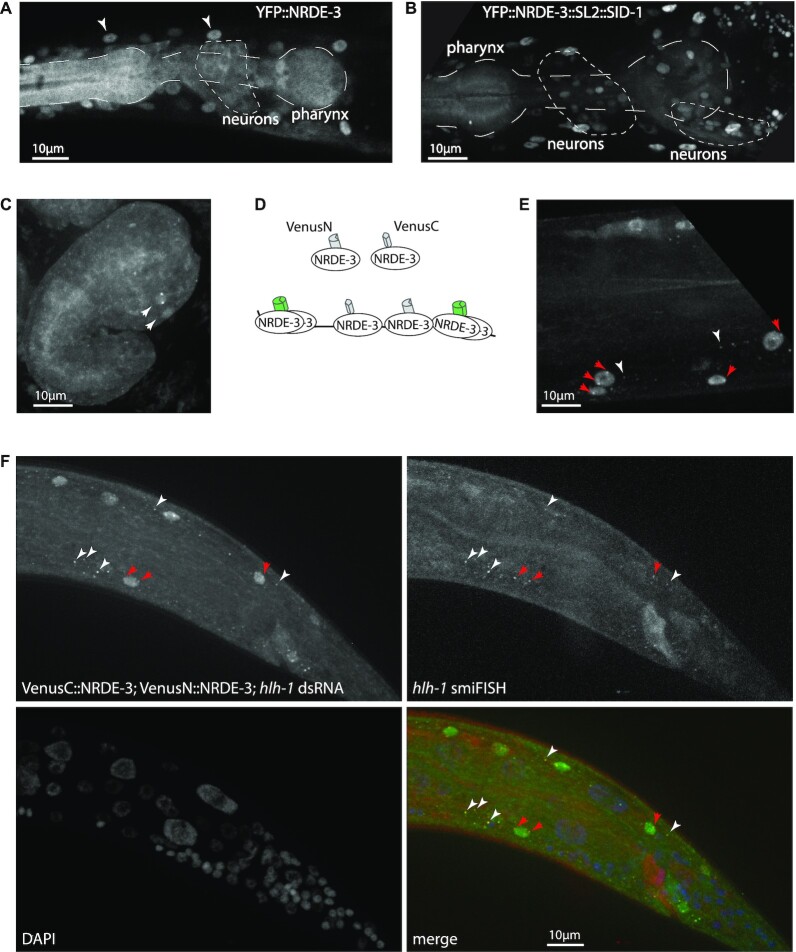
Figure 3.
Improvements on transcriptional labelling. (A) Neurons and pharynx have less effective RNAi. After exposure to dsRNA against GFP, YFP::NRDE-3 is nuclear localized in most tissues (white arrowheads: muscle nuclei), but localizes to the cytoplasm in pharynx and neurons. (B) Restoring expression of SID-1 in all tissues improves nuclear localization. After exposure to dsRNA against GFP, YFP::NRDE-3 is nuclear localized in neurons and, partially, in the posterior pharynx. (C) Improved transcriptional labelling in neurons. An embryo expressing SID-1 in all tissues, after maternal exposure to dsRNA against ttx-3, shows nuclear localization and transcriptional foci in AIY neurons (compare with Figure 2). (D) Principle of trimolecular fluorescence complementation. N-terminal and C-terminal fragments of the fluorescent protein Venus are fused to NRDE-3. In the cytoplasm or nucleus, local concentration of NRDE-3 molecules does not allow fluorescence complementation, thus reducing background fluorescence; once bound on the target transcript, VenusN::NRDE-3 and VenusC::NRDE-3 are in sufficient proximity to allow for fluorescence complementation. (E) Labelling of transcription with trimolecular fluorescence complementation. After exposure to dsRNA against hlh-1, bright transcription foci are visible in multiple nuclei (red arrows); additionally, some cytoplasmic foci are visible in the cytoplasm of muscle cells (white arrowheads), but not in other tissues. (F) Trimolecular fluorescence complementation labels cytoplasmic transcripts. Cytoplasmic spots (white arrowheads), due to fluorescence complementation between VenusN::NRDE-3 and VenusC::NRDE-3, as well as nuclear active transcription sites (red arrows), colocalize with smiFISH probes labelling hlh-1 transcripts.
For further optimization of the method, we focused mostly on the transcription factor hlh-1 for several reasons: first, it is transcribed throughout the life of the animal, for initiation and maintenance of the myocyte fate (42). Second, its expression starts relatively early in development, at least as early as 210 min (37,43), allowing for labelling in the embryo as well as larval stages. Third, as a transcription factor, its expression level is relatively low (44), providing a good test case to evaluate the applicability of our method to moderately expressed C. elegans genes.
While accumulation of fluorescently labelled NRDE-3 in the nucleus is useful to identify cells expressing the gene of interest, proteins not bound to a transcript have the drawback of increasing the background fluorescence above which the fluorescent signal at transcription sites needs to rise to be detectable. It is an important variable to control in transcriptional imaging: too much of the tagged protein, the transcription site or individual mRNAs cannot be seen over background fluorescence; too little, the availability of the protein may be a limiting factor (45). While sufficient for snapshot imaging, the YFP::nrde-3 transgene we used had the drawback of high expression under control of the eef-1A.1 promoter, and low photostability, restricting laser power and exposure time, thus limiting its use for time-lapse imaging. To improve on these points, we fused NRDE-3 with the more photostable and brighter fluorescent protein mNeonGreen (46), under control of the ubiquitous, medium-level expression rps-27 promoter. With the improved signal-to-noise ratio, we were able to observe more frequently transcriptional foci in several tissues in vivo.
To compare the transcriptional labelling efficiency at the transcription site between YFP and mNeonGreen tagged NRDE-3, we performed, in parallel, double labelling with smiFISH (single molecule inexpensive FISH) (14) targetting hlh-1 transcripts, and NRDE-3 fused to YFP or mNeonGreen with exposure to hlh-1 dsRNA (Supplementary Figures S4, S5). To limit degradation of fluorescent protein signal-to-noise ratio, we limited formaldehyde fixation to a maximum of 45 min. While different resistance of fluorophores to the fixation procedure may affect the results, it still provides useful information regarding the relative efficiency of labelling.
When scoring the brightest nuclear smiFISH spot in each nucleus, most likely representing active transcription sites, we observed 62% colabelling with YFP::NRDE-3, and 79% with mNeonGreen::NRDE-3 (Supplementary Figure S5A). When scoring the two brightest spots per nucleus, we observed 46% colabelling with YFP::NRDE-3 and 58% with mNeonGreen::NRDE-3 (Supplementary Figure S5B). mNeonGreen therefore appears to be superior to YFP in term of sensitivity. We observed positive and significant correlation of intensity between smiFISH and fluorescent NRDE-3 signal intensities at transcription sites (Supplementary Figure S5C; mean correlation: 0.42 for YFP; 0.46 for mNeonGreen). Although the fixation procedure and competition between NRDE-3 and smiFISH probes for transcript binding may reduce the amount of correlation, these data indicate that mNeonGreen is better than YFP to measure activity at transcription sites.
Trimolecular fluorescence complementation allows cytoplasmic mRNA labelling
Another approach to reduce background fluorescence is trimolecular fluorescence complementation, where a fluorescent protein is reconstituted when two proteins bind in close proximity on a common target, which has successfully been used to label transcription sites or transcripts with other RNA binding proteins (8,47–50). We therefore decided to implement it for the NRDE-3 system. Briefly, the fluorescent protein Venus was split in two halves: VenusC and VenusN, which were fused to the N-terminal end of NRDE-3. Only when bound in close proximity to the same transcript would the two halves of Venus be close enough to reconstitute a fluorescent protein (Figure 3D). Without exposure to dsRNA, we observed a low level of nonspecific fluorescence complementation in the nerve ring, intestinal nuclei, and associated with the cytoskeleton in seam cells, but not in other larval or adult tissues. After exposure to hlh-1 dsRNA, we observed fluorescence complementation in muscle nuclei, and bright foci in many of these nuclei (Figure 3E), with better contrast in vivo than observed with the simpler YFP::nrde-3 transgene.
Occasionally, we also observed additional fainter nuclear spots, possibly representing mRNAs being processed for export, as well as cytoplasmic spots, which we hypothesized to be cytoplasmic transcripts (Figure 3E). We therefore performed co-labelling with smiFISH targeting hlh-1, and observed colocalization in both nuclei and cytoplasm (Figure 3F), confirming their nature as cytoplasmic transcripts. We observed co-labelling by NRDE-3 and smiFISH probes for 82% of cytoplasmic transcripts; a ratio similar to those observed with double labelling with two smFISH probe sets targetting the same transcript (14,39,51), indicating efficient and specific labelling of single mRNAs with NRDE-3. Reconstituted Venus is however less photostable than the other fluorescent proteins; it is nonetheless possible to observe movement of cytoplasmic transcripts (Supplementary movie 1).
We also compared the efficiency of transcription site labelling between splitVenus, YFP and mNeonGreen. When scoring the brightest nuclear smiFISH spots in each nucleus, we observed colabelling with splitVenus::NRDE-3 of 56% and 52% of spots (Supplementary Figures S4, S5A, B), similar to the one observed with YFP::NRDE-3. However, we observed lower correlation of intensity between smiFISH and fluorescent NRDE-3 signal intensity at transcription sites than with YFP or mNeonGreen (Supplementary Figure S5C). The lower correlation could be due to the noise added by the reconstitution process. Reconstituted Venus is therefore not more efficient than mNeonGreen for labelling of active transcription sites, but has the advantage of allowing labelling of cytoplasmic transcripts.
Monitoring transcriptional dynamics
We next applied this method to follow the dynamics of transcription over time in living animals. To determine whether our method allows the quantification of transcriptional activity over time, we tested whether we could observe the induction of chaperone hsp-4 (a HSPA5 orthologue) expression by heat shock. While, at 20°C, some expression of hsp-4 is sufficient for nuclear localization of mNeonGreen::NRDE-3 after exposure to hsp-4 dsRNA, no transcription spot is visible (Figure 4A, top panel). After heat shock at 30°C, transcription sites become visible around 24 min after beginning of heat shock (Figure 4A, middle panel), and are bright and present in every cell by 68 min (Figure 4A, bottom panel; supplementary movie 2). Some nuclei had more than two spots, due to the polyploid nature of epidermal nuclei and existence of paralogues of hsp-4. The intensity of active hsp-4 transcription sites increases over the course of heat shock (Figure 4B). This demonstrate that our method can be used to monitor changes in gene expression over time.
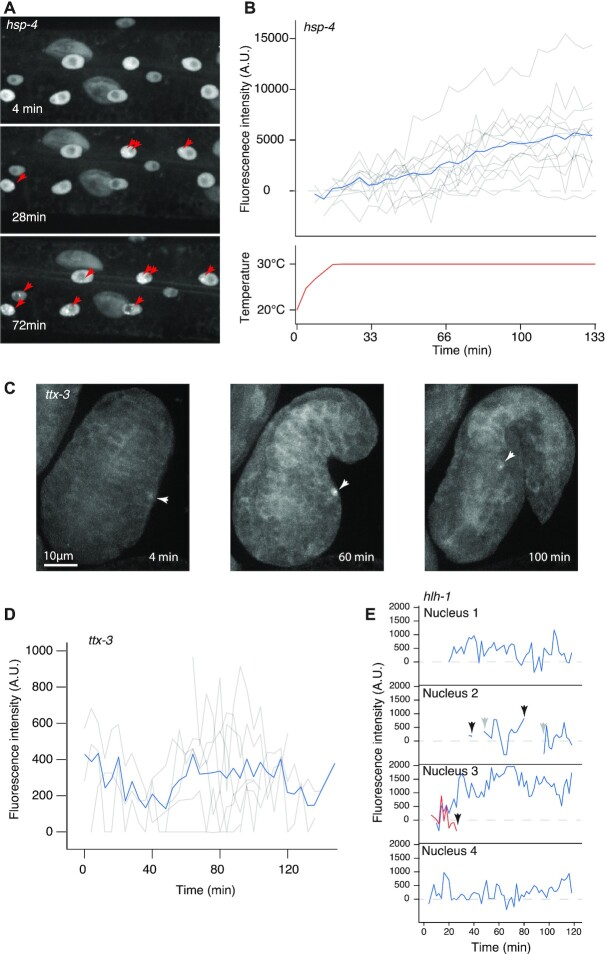
Figure 4.
NRDE-3 labelling reveals dynamics of transcription. (A) Induction of hsp-4 transcription in response to heat shock. Three frames showing apparition of hsp-4 transcription sites (red arrows) in an L4 larva over the course of heat shock at 30°C. (B) Dynamics of hsp-4 transcription sites in response to heat shock at 30°C. Fluorescence intensity of transcription foci in multiple nuclei of the same larva (top panel, grey lines) and their average (blue line) in response to temperature (bottom panel). (C) Dynamics of ttx-3 transcription in the embryo. Three frames showing the AIY nucleus (white arrowheads) before a transcription spot is visible, at 4 min; a bright transcription spot at 60 min, and a dimming spot, before it dropped below detection limit, at 100 min. (D) Fluorescence intensity of ttx-3 transcription spots of 10 embryos, temporally aligned based on embryo elongation. Average fluorescence intensity is represented as a blue line. (E) Fluorescence intensity of hlh-1 transcription spots in muscle nuclei in an L4 larva. In nucleus 2, the transcription spot dropped below detection limits for more than two timeframes (black arrows) before reappearing (grey arrows). In nucleus 3, two transcription spots were present, one disappeared.
We next tested whether our method was able to track moderately expressed, and temporally and spatially regulated genes, such as transcription factors. We first exposed nematodes expressing YFP::NRDE-3 and SID-1 in all tissues to dsRNA against the transcription factor ttx-3, an LHX-2/9 orthologue necessary for acquisition of the AIY neuronal fate, and imaged their progeny at intervals of 4 min, to minimize bleaching; we used this specific line to increase efficiency in the nervous system. We observed, in embryos, nuclear localization of the fluorescent signal in AIY neurons and followed transcriptional spots during embryonic development (Figure 4C; supplementary movie 3). By tracking the intensity over time, we were able to measure transcriptional activity in multiple embryos (Figure 4D) and observed a relative stability when considering the average over multiple embryos, while individual spots display important fluctuations. This suggests the existence of intrinsic noise at individual ttx-3 loci. We next analysed dynamics of the transcription factor hlh-1, to evaluate the possibility of tracking transcription in multiple nuclei simultaneously. To improve the signal-to-noise ratio, reduce phototoxicity and bleaching, we used a strain expressing mNeonGreen::NRDE-3. We imaged worms at the L4 stage for 2 h and recorded the intensity of nuclear foci over time. We were able to follow the appearance and disappearance of transcriptional spots, and quantify changes in intensity (Figure 4E; supplementary movie 4). There were also brief troughs in transcription or complete disappearance of transcription spots, which could mean an absence of transcription or a transcription level below detection limit (Figure 4E, nuclei 2 and 3). In nuclei where we observed two transcription spots (Figure 4E, nucleus 3), we saw no correlation in intensity fluctuation between the two loci. As for ttx-3, this suggests that transcription at individual hlh-1 loci undergoes random fluctuation.
To quantify expression noise, the coefficient of variation has been shown to be a reliable indicator (52). While the average coefficient of variation was similar in all three cases (hsp-4: 0.97; ttx-3: 0.85; hlh-1: 0.65), the dynamics were different for the three genes. For hsp-4, we observed that the coefficient of variation consistently decreased over time, as the average expression level increased (Supplementary Figure S6A), consistent with the general observation that expression noise tends to decrease when expression level increases (53,54). This is not observed for transcription factors ttx-3 and hlh-1, where the mean expression stays constant (Supplementary Figure S6B, C). Taken together, these data illustrate that this method can be used to monitor transcriptional dynamics.
DISCUSSION
Here, we developed and applied a novel method to image transcription in C. elegans. This is, to our knowledge, the first dynamic imaging of transcription, without modification of the gene of interest, in a live animal. We took advantage of the C. elegans RNAi pathways and tools to program NRDE-3 to bind to the transcript of interest. This approach makes it a fast and inexpensive method, bypassing the need for in vitro synthesis of antisense RNAs and their delivery into cells of interest. Our approach offers several advantages over current in vivo transcription imaging methods. It does not require modification of the transcript of interest, which could perturb the system and cannot be easily scaled up. In addition, our method induces binding of multiple fluorescent proteins to each target transcript from a single dsRNA trigger, allowing detection of low quantities, down to single mRNA molecules. This result would be difficult to obtain with methods relying on programmable proteins such as Pumilio, Cas9 or Cas13, which would require the introduction of either multiple programmed Pumilio transgenes or long RNAs.
Although powerful, this method has several identified limitations. Because synthesis of 22G siRNAs is downstream of primary RNAi, which requires transcript cleavage by RDE-8/12 complexes to recruit the RdRP complex (Figure 1A), some level of post-transcriptional silencing can still be present. To mitigate this silencing of target genes when necessary, such as with ant-1.1, we reduced the amount of dsRNA ingested by diluting the dsRNA-expressing bacteria of interest with bacteria expressing a dsRNA not matching any C. elegans sequence. Another limitation, as previously stated, is that this method requires mutations in eri-1 and nrde-2; while we do not expect it to severely affect biological processes, there are nonetheless events that would need to be studied in a wild-type background. Additionally, because the target gene needs to be already expressed for synthesis of 22G siRNAs to proceed, there exists a delay between the first initiation of transcription of the gene of interest and translocation of fluorescently labelled NRDE-3 to the nucleus, allowing for labelling of transcription sites. While this approach greatly facilitates the labelling of transcripts, it still requires image acquisition and analysis, and is therefore not a high throughput method. Finally, we cannot affirm that NRDE-3 does not affect export and stability of the target mRNA; however, transcripts are not blocked from export out of the nucleus (Figure 3F).
The expression patterns we observed with NRDE-3 labelling are consistent with data independently obtained by other teams. Comparisons with smFISH or smiFISH labelling of hlh-1 transcripts, the most accurate mRNA visualization methods, indicate that NRDE-3 labelling accurately represents endogenous expression and transcript localization (Figures 1C, 3F; Supplementary Figure S4), with good efficiency. In the absence of a preexisting method to follow transcriptional dynamics of unmodified genes in live animals, it is more difficult to establish directly validating comparisons for the transcriptional dynamics we observed. Nonetheless, transcriptional dynamics of other genes observed in whole animals with the MS2-MCP reporter transgene system revealed stochastic fluctuations of the notch target gene sygl-1 in the C. elegans germline (5); or the eve stripe 2 enhancer in the early D. melanogaster embryo (3). The dynamics here reported for transcription factors ttx-3 and hlh-1, with random fluctuation at individual loci, are consistent with those observed for other genes involved in fate determination. Our method will allow us, among other things, to study the dynamics of terminal transcription factors responsible for cell fate acquisition, and how variability in their transcription affects fate specification and maintenance.
While this method was developed in C. elegans, it could be transposed to other organisms, with adjustments to account for divergence of RNAi pathways. Nuclear RNAi pathways have been described in other organisms (55,56), and it may be possible to introduce different components of the pathway, such as synthetic triphosphorylated small RNAs. However, it remains to be seen whether it could be efficiently adapted to other organisms. Beyond its use as an imaging platform, we demonstrated the usefulness of the Argonaute NRDE-3 as a general purpose, highly specific, RNA programmable RNA binding protein, which opens multiple possible applications for transcriptome engineering (57). Argonautes bind with high affinity and specificity to their target mRNAs (58), making it a tool of choice to be developed further. While previous attempts at protein complementation with Argonaute proteins in vitro did not give specific results (59), the success of trimolecular fluorescence complementation on transcripts shows that NRDE-3 could also be used for split protein complementation in vivo, for example to activate proteins only in target cells or at specific genomic loci (60,61).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the CGC for strains and reagents; J. Ewbank, E. Bertrand and X. Pichon for extensive discussions and suggestions; members of our lab for comments on the manuscript; F. Soulavie for communicating unpublished results; and three anonymous reviewers for suggestions and criticism. Imaging was performed on PiCSL-FBI core facility (IBDM, AMU-Marseille), supported by the French National Research Agency through the ‘Investments for the Future’ program [France-BioImaging, ANR-10-INBS-04]; some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs [P40 OD010440].
Contributor Information
Amel Toudji-Zouaz, Aix Marseille University, CNRS, IBDM, Turing Centre for Living Systems, Marseille, France.
Vincent Bertrand, Aix Marseille University, CNRS, IBDM, Turing Centre for Living Systems, Marseille, France.
Antoine Barrière, Aix Marseille University, CNRS, IBDM, Turing Centre for Living Systems, Marseille, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agence Nationale de la Recherche [ANR-11-LABX-0054 and ANR-17-ERC2-0018]; Fondation pour la Recherche Médicale [DEQ20180339160]. Funding for open access charge: ANR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pichon X., Lagha M., Mueller F., Bertrand E.. A growing toolbox to image gene expression in single cells: sensitive approaches for demanding challenges. Mol. Cell. 2018; 71:468–480. [DOI] [PubMed] [Google Scholar]
- 2.Bertrand E., Chartrand P., Schaefer M., Shenoy S.M., Singer R.H., Long R.M.. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 1998; 2:437–445. [DOI] [PubMed] [Google Scholar]
- 3.Bothma J.P., Garcia H.G., Esposito E., Schlissel G., Gregor T., Levine M.. Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:10598–10603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park H.Y., Lim H., Yoon Y.J., Follenzi A., Nwokafor C., Lopez-Jones M., Meng X., Singer R.H.. Visualization of dynamics of single endogenous mRNA labeled in live mouse. Science. 2014; 343:422–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee C., Shin H., Kimble J.. Dynamics of Notch-Dependent Transcriptional Bursting in Its Native Context. Dev. Cell. 2019; 50:426–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lenstra T.L., Larson D.R.. Single-molecule mRNA detection in live yeast. Curr. Protoc. Mol. Biol. 2016; 113:14.24.1–14.24.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abil Z., Denard C., Zhao H.. Modular assembly of designer PUF proteins for specific post-transcriptional regulation of endogenous RNA. J. Biol. Eng. 2014; 8:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozawa T., Natori Y., Sato M., Umezawa Y.. Imaging dynamics of endogenous mitochondrial RNA in single living cells. Nat. Methods. 2007; 4:413–419. [DOI] [PubMed] [Google Scholar]
- 9.Nelles D.A., Fang M.Y., O’Connell M.R., Xu J.L., Markmiller S.J., Doudna J.A., Yeo G.W.. Programmable RNA tracking in live cells with CRISPR/Cas9. Cell. 2016; 165:488–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abudayyeh O.O., Gootenberg J.S., Essletzbichler P., Han S., Joung J., Belanto J.J., Verdine V., Cox D.B.T., Kellner M.J., Regev A.et al.. RNA targeting with CRISPR–Cas13. Nature. 2017; 550:280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang L., Wang Y., Li S., Yao R., Luan P., Wu H., Carmichael G.G., Chen L.. Dynamic imaging of RNA in living cells by CRISPR-Cas13 systems. Mol. Cell. 2019; 76:981–997. [DOI] [PubMed] [Google Scholar]
- 12.Kamath R., Fraser A., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M.et al.. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003; 421:231–237. [DOI] [PubMed] [Google Scholar]
- 13.Thakur N., Pujol N., Tichit L., Ewbank J.J.. Clone mapper: an online suite of tools for RNAi experiments in Caenorhabditis elegans. G3 Genes, Genomes, Genet. 2014; 4:2137–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsanov N., Samacoits A., Chouaib R., Traboulsi A., Gostan T., Weber C., Zimmer C., Zibara K., Walter T., Peter M.et al.. smiFISH and FISH-quant – a flexible single RNA detection approach with super-resolution capability. Nucleic Acid Res. 2016; 44:e165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tinevez J.Y., Perry N., Schindelin J., Hoopes G.M., Reynolds G.D., Laplantine E., Bednarek S.Y., Shorte S.L., Eliceiri K.W.. TrackMate: an open and extensible platform for single-particle tracking. Methods. 2017; 115:80–90. [DOI] [PubMed] [Google Scholar]
- 16.Steiner F.A., Okihara K.L., Hoogstrate S.W., Sijen T., Ketting R.F.. RDE-1 slicer activity is required only for passenger-strand cleavage during RNAi in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2009; 16:207–211. [DOI] [PubMed] [Google Scholar]
- 17.Tsai H.-Y., Chen C.-C.G., Conte D. Jr, Moresco J.J., Chaves D.A., Mitani S., Yates J.R. III, Tsai M.-D., Mello C.C.. A ribonuclease coordinates siRNA amplification and mRNA cleavage during RNAi. Cell. 2015; 160:407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang H., Vallandingham J., Shiu P., Li H., Hunter C.P., Mak H.Y.. The DEAD box helicase RDE-12 promotes amplification of RNAi in cytoplasmic foci in C. elegans. Curr. Biol. 2014; 24:832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shirayama M., Stanney W., Gu W., Seth M., Mello C.C.. The Vasa homolog RDE-12 engages target mRNA and multiple argonaute proteins to promote RNAi in C. elegans. Curr. Biol. 2014; 24:845–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aoki K., Moriguchi H., Yoshioka T., Okawa K., Tabara H.. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007; 26:5007–5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pak J., Fire A.. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007; 315:241–244. [DOI] [PubMed] [Google Scholar]
- 22.Sijen T., Steiner F., Thijssen K.L., Plasterk R.H.A.. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007; 315:244–247. [DOI] [PubMed] [Google Scholar]
- 23.Yigit E., Batista P.J., Bei Y., Pang K.M., Chen C.C.G., Tolia N.H., Joshua-Tor L., Mitani S., Simard M.J., Mello C.C.. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006; 127:747–757. [DOI] [PubMed] [Google Scholar]
- 24.Gu W., Shirayama M., Conte D., Vasale J., Batista P.J., Claycomb J.M., Moresco J.J., Youngman E.M., Keys J., Stoltz M.J.et al.. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell. 2009; 36:231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guang S., Bochner A.F., Pavelec D.M., Burkhart K.B., Harding S., Lachowiec J., Kennedy S.. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science. 2008; 321:537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guang S., Bochner A.F., Burkhart K.B., Burton N., Pavelec D.M., Kennedy S.. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010; 465:1097–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burkhart K.B., Guang S., Buckley B. a., Wong L., Bochner A.F., Kennedy S.. A Pre-mRNA–associating factor links endogenous siRNAs to chromatin regulation. PLos Genet. 2011; 7:e1002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischer S.E.J., Montgomery T.A., Zhang C., Fahlgren N., Breen P.C., Hwang A., Sullivan C.M., Carrington J.C., Ruvkun G.. The ERI-6/7 helicase acts at the first stage of an siRNA amplification pathway that targets recent gene duplications. PLoS Genet. 2011; 7:e1002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han T., Manoharan A.P., Harkins T.T., Bouffard P., Fitzpatrick C., Chu D.S., Thierry-Mieg D., Thierry-Miege J., Kim J.K.. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:18674–18679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buckley B.A., Burkhart K.B., Gu S.G., Spracklin G., Kershner A., Fritz H., Kimble J., Fire A., Kennedy S.. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012; 489:447–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou X., Xu F., Hui M., Ji J., Yin M., Feng X., Guang S.. Nuclear RNAi contributes to the silencing of off-target genes and repetitive sequences in Caenorhabditis elegans. Genetics. 2014; 197:121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mcewan D.L., Weisman A.S., Hunter C.P.. Uptake of Extracellular double-stranded RNA by SID-2. Mol. Cell. 2012; 47:746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winston W.M., Moldowitch C., Hunter C.P.. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002; 295:2456–2459. [DOI] [PubMed] [Google Scholar]
- 34.Lindell T.J., Weinberg F., Morris P.W., Roeder R.G., Rutter W.J.. Specific inhibition of nuclear RNA polymerase II by α-amanitin. Science. 1970; 170:447–449. [DOI] [PubMed] [Google Scholar]
- 35.Halstead J.M., Wilbertz J.H., Wippich F., Ephrussi A.. An RNA biosensor for imaging the first round of translation from single cells to living animals. Science. 2015; 347:1367–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harterink M., Kim D.H., Middelkoop T.C., Doan T.D., van Oudenaarden A., Korswagen H.C.. Neuroblast migration along the anteroposterior axis of C. elegans is controlled by opposing gradients of Wnts and a secreted Frizzled-related protein. Development. 2011; 138:2915–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nair G., Walton T., Murray J.I., Raj A.. Gene transcription is coordinated with, but not dependent on, cell divisions during C. elegans embryonic fate specification. Development. 2013; 140:3385–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barkoulas M., van Zon J.S., Milloz J., van Oudenaarden A., Félix M.-A.. Robustness and epistasis in the C. elegans vulval signaling network revealed by pathway dosage modulation. Dev. Cell. 2013; 24:64–75. [DOI] [PubMed] [Google Scholar]
- 39.Raj A., van den Bogaard P., Rifkin S.A., van Oudenaarden A., Tyagi S.. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods. 2008; 5:877–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bertrand V., Hobert O.. Linking asymmetric cell division to the terminal differentiation program of postmitotic neurons in C. elegans. Dev. Cell. 2009; 16:563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calixto A., Chelur D., Topalidou I., Chen X., Chalfie M.. Enhanced neuronal RNAi in C. elegans using SID-1. Nat. Methods. 2010; 7:554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harfe B.D., Branda C.S., Krause M., Stern M.J., Fire A.. MyoD and the specification of muscle and non-muscle fates during postembryonic development of the C. elegans mesoderm. Development. 1998; 125:2479–2488. [DOI] [PubMed] [Google Scholar]
- 43.Packer J.S., Zhu Q., Huynh C., Sivaramakrishnan P., Preston E., Dueck H., Stefanik D., Tan K., Trapnell C., Kim J.et al.. A lineage-resolved molecular atlas of C. elegans embryogenesis at single-cell resolution. Science (80-.). 2019; 365:eaax1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao J., Packer J.S., Ramani V., Cusanovich D.A., Huynh C., Daza R., Qiu X., Lee C., Furlan S.N., Steemers F.J.et al.. Comprehensive single cell transcriptional profiling of a multicellular organism by combinatorial indexing. Science. 2017; 357:661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferguson M.L., Larson D.R., Shav-Tal Y.Measuring transcription dynamics in living cells using fluctuation analysis. Imaging Gene Expression - Methods and Protocols. 2013; Springer; 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heppert J.K., Dickinson D.J., Pani A.M., Higgins C.D., Steward A., Ahringer J., Kuhn J.R., Goldstein B.. Comparative assessment of fluorescent proteins for in vivo imaging in an animal model system. Mol. Biol. Cell. 2016; 27:3385–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamada T., Yoshimura H., Inaguma A., Ozawa T.. Visualization of nonengineered single mRNAs in living cells using genetically encoded fluorescent probes. Anal. Chem. 2011; 83:5708–5714. [DOI] [PubMed] [Google Scholar]
- 48.Wu B., Chen J., Singer R.H.. Background free imaging of single mRNAs in live cells using split fluorescent proteins. Sci. Rep. 2014; 4:3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park S.Y., Moon H.C., Park H.Y.. Live-cell imaging of single mRNA dynamics using split superfolder green fluorescent proteins with minimal background. RNA. 2020; 26:101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valencia-Burton M., McCullough R.M., Cantor C.R., Broude N.E.. RNA visualization in live bacterial cells using fluorescent protein complementation. Nat. Methods. 2007; 4:421–427. [DOI] [PubMed] [Google Scholar]
- 51.Stapel L., Lombardot B., Broaddus C., Kainmueller D., Jug F., Myers E.W., Vastenhouw N.L.. Automated detection and quantification of single RNAs at cellular resolution in zebrafish embryos. Dev. 2016; 143:530–539. [DOI] [PubMed] [Google Scholar]
- 52.Chalancon G., Ravarani C.N.J., Balaji S., Martinez-Arias A., Aravind L., Jothi R., Babu M.M.. Interplay between gene expression noise and regulatory network architecture. Trends Genet. 2012; 28:221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bar-Even A., Paulsson J., Maheshri N., Carmi M., O’Shea E., Pilpel Y., Barkai N.. Noise in protein expression scales with natural protein abundance. Nat. Genet. 2006; 38:636–643. [DOI] [PubMed] [Google Scholar]
- 54.Faure A.J., Schmiedel J.M., Lehner B.. Systematic analysis of the determinants of gene expression noise in embryonic stem cells. Cell Syst. 2017; 5:471–484. [DOI] [PubMed] [Google Scholar]
- 55.Bajczyk M., Bhat S.S., Szewc L., Szweykowska-Kulinska Z., Jarmolowski A., Dolata J.. Novel nuclear functions of arabidopsis ARGONAUTE1: Beyond RNA interference. Plant Physiol. 2019; 179:1030–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schraivogel D., Meister G.. Import routes and nuclear functions of Argonaute and other small RNA-silencing proteins. Trends Biochem. Sci. 2014; 39:420–431. [DOI] [PubMed] [Google Scholar]
- 57.Mackay J.P., Font J., Segal D.J.. The prospects for designer single-stranded RNA-binding proteins. Nat. Struct. Mol. Biol. 2011; 18:256–261. [DOI] [PubMed] [Google Scholar]
- 58.Wee L.M., Flores-Jasso C.F., Salomon W.E., Zamore P.D.. Argonaute divides Its RNA guide into domains with distinct functions and RNA-binding properties. Cell. 2012; 151:1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Furman J.L., Badran A.H., Ajulo O., Porter J.R., Stains C.I., Segal D.J., Ghosh I.. Toward a general approach for RNA-templated hierarchical assembly of split-proteins. J. Am. Chem. Soc. 2010; 132:11692–11701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei H., Wang Z.. Engineering RNA-binding proteins with diverse activities. Wiley Interdiscip. Rev. RNA. 2015; 6:587–613. [DOI] [PubMed] [Google Scholar]
- 61.Shekhawat S.S., Ghosh I.. Split-protein systems: beyond binary protein-protein interactions. Curr. Opin. Chem. Biol. 2011; 15:790–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.