Abstract
R-loops, which consist of a DNA/RNA hybrid and a displaced single-stranded DNA (ssDNA), are increasingly recognized as critical regulators of chromatin biology. R-loops are particularly enriched at gene promoters, where they play important roles in regulating gene expression. However, the molecular mechanisms that control promoter-associated R-loops remain unclear. The epigenetic ‘reader’ Tudor domain-containing protein 3 (TDRD3), which recognizes methylarginine marks on histones and on the C-terminal domain of RNA polymerase II, was previously shown to recruit DNA topoisomerase 3B (TOP3B) to relax negatively supercoiled DNA and prevent R-loop formation. Here, we further characterize the function of TDRD3 in R-loop metabolism and introduce the DExH-box helicase 9 (DHX9) as a novel interaction partner of the TDRD3/TOP3B complex. TDRD3 directly interacts with DHX9 via its Tudor domain. This interaction is important for recruiting DHX9 to target gene promoters, where it resolves R-loops in a helicase activity-dependent manner to facilitate gene expression. Additionally, TDRD3 also stimulates the helicase activity of DHX9. This stimulation relies on the OB-fold of TDRD3, which likely binds the ssDNA in the R-loop structure. Thus, DHX9 functions together with TOP3B to suppress promoter-associated R-loops. Collectively, these findings reveal new functions of TDRD3 and provide important mechanistic insights into the regulation of R-loop metabolism.
INTRODUCTION
Eukaryotic gene expression is regulated through the recruitment of transcription factors, chromatin-modifying enzymes, and RNA polymerases, at enhancers and promoters (1,2). Upon transcription initiation, the separated DNA strands and nascent RNA transcript can adopt numerous non-B DNA structures that, if left unresolved, may interfere with the movement of the transcription machinery and impede gene expression (3–6). One of the most common non-B DNA structures that arise during transcription is the three-stranded R-loop structure, which consists of a DNA/RNA hybrid and a displaced non-template strand (7–10). Recent studies using DNA/RNA-specific antibodies (S9.6) and next-generation sequencing approaches have revealed the widespread presence of R-loops along human genomes (11–13). Specifically, more than half of all R-loops are formed at mammalian gene promoter regions (11,14,15), where they can positively or negatively influence gene expression. For example, in mouse embryonic stem cells, promoter-associated R-loops differentially modulate the binding of two key chromatin-regulatory complexes, Tip60-p400 and polycomb repressive complex 2 (PRC2), to promote the expression of genes important for differentiation (16). R-loops can also repel the binding of DNA methyltransferases to gene promoters, thus protecting the underlying DNA from methylation (13,17). However, excessive and prolonged R-loop formation can block RNA polymerase II (RNAPII) elongation and interfere with productive transcription (18). Persistent R-loops can also promote heterochromatin formation and lead to gene silencing (19). Importantly, because the exposed single-stranded DNA (ssDNA) is vulnerable to DNA damage, unprogrammed R-loops have been increasingly recognized as a source of genomic instability, a hallmark of human cancers (7,10,20–22).
Cells employ various strategies to prevent or limit unprogrammed R-loop formation. These mechanisms include: (i) DNA topoisomerases that act to relax negatively supercoiled DNA and thereby prevent R-loop formation (23–26); (ii) DNA/RNA helicases that unwind the DNA/RNA hybrid and resolve R-loops (27–32); (iii) ribonuclease (RNase) H enzymes that degrade the RNA portion of R-loops (33–35) and (iv) pre-mRNA processing factors that, through interactions with mRNA transcripts, prevent the re-hybridization of the nascent RNA with template DNA (36,37). However, as the functions of R-loops are dependent on the genomic context, how these seemingly redundant R-loop-managing pathways are discriminately targeted to specific genomic regions is unclear.
Posttranslational modifications of histones play a pivotal role in regulating transcription, primarily by acting as docking sites for effector proteins that recognize or ‘read’ these marks (38–40). The Tudor domain-containing protein 3 (TDRD3) is one such effector molecule that ‘reads’ methylarginine marks on histones and on the C-terminal domain of RNAPII (41–43). Importantly, the genome-wide distribution of TDRD3 is strongly associated with gene promoters and coincides with the formation of promoter-proximal R-loops (11,13–15). Mechanistically, the C-terminal Tudor domain of TDRD3 mediates its interactions with arginine-methylated substrates and its N-terminal oligonucleotide/oligosaccharide-binding (OB)-fold recruits the DNA topoisomerase 3B (TOP3B) through direct protein-protein interactions (25,42). Thus, TOP3B is directed by TDRD3 to the promoters of actively transcribed genes, including c-MYC and NRAS, to resolve negatively supercoiled DNA in the wake of RNAPII and prevent R-loop formation (25). Although several studies have reported TOP3B as a major interaction partner of TDRD3 (25,44–46), it is likely that TDRD3 recruits additional proteins for R-loop resolution. Furthermore, TDRD3 can also bind single-stranded nucleic acids (47). Whether and how this activity contributes to its function in R-loop regulation is not known.
In this study, to further investigate the molecular pathways involved in the regulation of promoter-associated R-loops, we identified the DExH-box helicase 9 (DHX9) as a new interaction partner of TDRD3. DHX9 unwinds a variety of double-stranded DNA and RNA structures, including R-loops, and is involved in various cellular processes, including transcription, replication, and RNA processing (27,48,49). Here, we show that DHX9 forms a protein complex with TDRD3 and TOP3B, within which TDRD3 functions as a scaffold to bridge their interactions. The interaction of TDRD3 with DHX9 relies on its functional Tudor domain, which is critical for the recruitment of DHX9 to its target gene promoters. In addition to directing DHX9 to promoters, TDRD3 also stimulates DHX9 helicase activity to resolve R-loops, likely through its N-terminal OB-fold-mediated interaction with ssDNA in the R-loop structure. Furthermore, as demonstrated using both cellular and in vitro transcription assays, DHX9 and TOP3B function cooperatively to resolve co-transcriptional R-loops at TDRD3 target genes. These results reveal new functions of the methylarginine effector molecule TDRD3 and provide novel mechanistic insights into the regulation of promoter-associated R-loops.
MATERIALS AND METHODS
Cell lines and reagents
HEK293 and MCF7 cells were obtained from ATCC. Both cell lines were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and maintained at 37°C. Two TDRD3 knockout (KO) MCF7 cell lines (KO1 and KO2) were generated using CRISPR/Cas9 technology, using two sgRNAs against TDRD3 (sgTDRD3-1: CTGCGATTACAGATGACTGA and sgTDRD3-2: GACTCTAACACCACAGTTCT). TOP3B KO MCF7 cells were generated using sgTOP3B: CCACTGAGAGCGCCTCGTTG. The sgRNAs were cloned into the PX330 vector. Transfection was carried out using Lipofectamine 2000 (11668-019; Thermo Fisher Scientific). Individual clones were screened for deletion of TDRD3 and TOP3B by Western blot analysis. Anti-FLAG M2 Magnetic Beads (M8823) was purchased from Sigma. The type I PRMT inhibitor MS023 and the 3xFLAG peptide were purchased from APExBIO. The site-directed mutagenesis kit (#200521) was purchased from Agilent Technologies. The siRNA targeting the 3′-UTR of DHX9 (A-009950-16-0005) and the siRNA targeting TOP3B (L-005282-00-0005) were purchased from Dharmacon. The restriction enzymes and RNase H (M0297L) were purchased from New England BioLabs. RNase A (EN0531) was purchased from Thermo Fisher Scientific.
Antibodies and plasmids
Anti-TDRD3 polyclonal and monoclonal antibodies were prepared as described previously (42). Anti-TDRD3 rabbit monoclonal antibody (D3O2G #5492) was purchased from Cell Signaling Technology. Anti-TOP3B mouse monoclonal (ab56445) and anti-DHX9 rabbit polyclonal (ab26271) antibodies were purchased from Abcam. Anti-DHX9 rabbit polyclonal antibody (A300-854A) was purchased from Bethyl Laboratories. Anti-GFP mouse monoclonal antibody (B-2) was purchased from Santa Cruz Biotechnology. Mouse monoclonal anti-Flag M2 (F1804) and anti-ACTIN (A5316) antibodies were purchased from Sigma. Mouse monoclonal anti-DNA/RNA hybrid S9.6 antibody used for the DNA/RNA immunoprecipitation (DRIP) assay has been described before (25,50). The Asymmetric 26 (ASYM26) antibody was kindly provided by Dr. Stéphane Richard (McGill University).
GST-TDRD3, GST-OB, GST-UBA, GST-Tudor and GST-TOP3B were prepared by sub-cloning the open reading frames from full-length TDRD3 and TOP3B cDNA into a pGEX-6P-1 vector, as described previously (25,42). GFP-DHX9, GFP-DHX9 RBD (amino acids 1–262), GFP-DHX9 RGG (amino acids 1064–1270), and GFP-DHX9 HD (amino acids 255–1077) were generated by sub-cloning open reading frames from full-length DHX9 cDNA into a pEGFP-C1 vector. Flag-TDRD3, Flag-TOP3B, and Flag-DHX9 were generated by sub-cloning TDRD3, TOP3B, and DHX9 cDNA into p3XFLAG-CMV-7.1 vectors. All point mutations were generated using site-directed mutagenesis. The pFC53 plasmid used in the in vitro DNA/RNA immunoprecipitation (DRIP) assay have been described before (25,50).
Tandem affinity purification
HEK293 cells were transfected with an NTAP-TOP3B plasmid. A total of 3 × 108 cells were used for tandem affinity purification. In brief, cells were first lysed in lysis buffer containing 50 mM Tris–HCl (pH 7.5), 125 mM NaCl, 5% glycerol, 0.2% NP-40, 1.5 mM MgCl2, 25mM NaF, 1 mM Na3VO4 and protease inhibitors. After centrifugation, the supernatant was incubated with rabbit-IgG Sepharose (GE17-0969-01; GE Healthcare Life Sciences) at 4°C for 4 h. The bound beads were first washed with lysis buffer and then TEV protease cleavage buffer (10 mM Tris–HCl [pH 7.5], 100 mM NaCl and 0.2% NP-40). The bound protein complexes were eluted by the addition of 50 mg TEV protease (4°C, overnight). TEV protease cleavage products were then incubated with Streptavidin agarose (69203–3; Millipore) at 4°C for 2 h. The bound proteins were eluted by boiling in SDS sample buffer. Samples were loaded on SDS-PAGE gels followed by either silver staining or SYPRO Ruby staining. After comparison with control samples, differential bands were cut from the gel, and proteins were identified by LC–MS/MS.
Recombinant protein expression and purification
GST-tagged constructs were transformed into Escherichia coli BL21 (DE3) cells and grown to an OD600 of 0.6. Expression was induced by adding 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), and cells were cultured for 16 h at 16°C. Cells were lysed by sonication in binding buffer (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH 7.4) and centrifuged at 21 000 × g for 10 min. The supernatant was incubated with Glutathione Sepharose (17075601; GE Healthcare Life Sciences), and GST-tagged proteins were purified according to the manufacturer's instructions. Flag-tagged constructs were transfected into HEK293 cells with Lipofectamine 2000. Forty-eight hours post-transfection, cells were lysed in co-immunoprecipitation (co-IP) buffer containing 0.5 M NaCl. Flag-tagged proteins were immunoprecipitated and purified with anti-FLAG M2 magnetic beads and eluted with 3xFLAG peptide.
Reverse transcription quantitative PCR (RT-qPCR)
Total RNA was extracted using TRIzol reagent (15596-018; Thermo Fisher Scientific). Reverse transcription was performed using a High-Capacity cDNA Reverse Transcription Kit (4368814; Thermo Fisher Scientific). 2 μl of 10-fold diluted cDNA was used for qPCR analysis using Power SYBR Green PCR Master Mix (43–687-06; Thermo Fisher Scientific). Real-time qPCR was performed on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad), according to the manufacturer's instructions. The comparative cycle threshold (CT) method (ΔΔCT) was used to quantify relative changes in gene expression. CT values were normalized by subtracting β-actin CT values from target gene CT values for each sample. All amplifications were done in triplicate.
Co-immunoprecipitation (co-IP)
Cells from a 10-cm plate were washed with 1× PBS and lysed with 1 ml of co-IP buffer (50 mM Tris–Cl [pH 7.4], 150 mM NaCl, 15 mM MgCl2, 5 mM EDTA, and 0.1% Nonidet P-40), containing a cocktail of protease inhibitors (A32965; Thermo Fisher Scientific). Cell extracts were briefly sonicated and centrifuged at 21 000 × g for 10 min to remove insoluble debris. Cell lysates were incubated with antibodies overnight at 4°C, followed by incubation with Protein A/G polyacrylamide beads (53133; Thermo Fisher Scientific) for 2 h. Beads were then washed three times with co-IP buffer, and bound proteins were eluted by SDS-PAGE loading buffer. Proteins were resolved on an SDS-PAGE gel and analyzed by western blot analysis.
GST pull-down
For GST pull-down using recombinant proteins, GST-tagged proteins purified from E. coli were incubated with Flag-tagged proteins purified from HEK293 cells in the co-IP lysis buffer. For GST pull-down using total cell lysates, cells were lysed in lysis buffer containing 20 mM Tris–HCl (pH 7.4), 150 mM NaCl, 0.1% NP-40 and protease inhibitors. After removing insoluble debris, the cell lysates were incubated with purified GST-tagged recombinant proteins with gentle rocking overnight at 4°C. Glutathione Sepharose beads were added to the protein and lysate mixture and incubated with gentle rocking at 4°C for 2 h. The mixture was centrifuged, the supernatant was discarded, and the beads were washed three times with the cell lysis buffer. After centrifuging again, the pellet was eluted in 30 μl 2× SDS sample buffer. The samples were loaded on SDS-PAGE gels and analyzed by western blot using the indicated antibodies.
Chromatin immunoprecipitation (ChIP)
Cells from two 10-cm confluent plates were cross-linked with 1% formaldehyde for 10 min at 37°C. Cross-linking was terminated by adding glycine to a final concentration of 0.125 M, followed by incubation at RT for 5 min. Cells were washed with 1× PBS and lysed with 2 ml of ChIP lysis buffer (5 mM PIPES [pH 8.0], 85 mM KCl, 0.5% Nonidet P-40 and protease inhibitor cocktail) for 15 min. The lysate was centrifuged at 2400 × g for 5 min, and the nuclear pellet was resuspended in 900 μl of nuclei lysis buffer (50 mM Tris–Cl [pH 8.0], 10 mM EDTA, 1% SDS, and protease inhibitor cocktail) for 5 min. Nuclear extracts were transferred to 1.5 ml Bioruptor Microtubes (C30010016; Diagenode), and chromatin was fragmented by sonication using the Bioruptor Pico Sonication System (B01060003; Diagenode) for 7–8 cycles (30 s on, 30 s off/cycle). Fragmented chromatin was centrifuged at 21 000 × g for 10 min to remove insoluble debris, and supernatant was diluted 5-fold in ChIP dilution buffer (20 mM Tris–Cl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.01% SDS and protease inhibitor cocktail). Four percent of the diluted fragmented chromatin was used as input. Antibodies were added to the diluted chromatin and incubated overnight at 4°C, followed by incubation with Protein A magnetic beads (10002D; Thermo Fisher Scientific) for 2 h. Proteins and non-specific chromatin were washed off beads for 3–5 min as follows: 1× wash with 1 ml of low salt wash buffer (20 mM Tris–Cl [pH 8.0], 150 mM NaCl, 2 mM EDTA, 1% Triton X-100 and 0.1% SDS), 1× wash with high salt wash buffer (20 mM Tris–Cl [pH 8.0], 500 mM NaCl, 2 mM EDTA, 1% Triton X-100 and 0.1% SDS), 1× wash with LiCl buffer (20 mM Tris [pH 8.0], 250 mM LiCl, 1 mM EDTA, 1% NP40, and 1% Na-deoxycholate), and 2× wash with TE buffer (10 mM Tris–Cl [pH 8.0] and 1 mM EDTA). Chromatin was eluted, and DNA/protein cross-links were reversed by resuspending beads in 200 ul of elution buffer (1% SDS and 0.1M NaHCO3), followed by incubation in a 65°C water bath overnight. Reverse cross-linked DNA-protein samples were digested with proteinase K at 45°C for 45 min, and the immunoprecipitated DNA was purified by phenol:chloroform:isoamyl alcohol (25:24:1) extraction and ethanol precipitation. Precipitated DNA was resuspended in 100 μl of distilled water, and 2 μl was used for qPCR analysis.
For ChIP-reChIP experiments, chromatin isolated from MCF7 cells (three 10-cm plates) was subjected to two ChIPs. In the first ChIP, chromatin was equally divided for immunoprecipitation with 4 μg of control IgG or anti-TDRD3 antibodies. Immunoprecipitated chromatin was eluted with 75 μl TE/10 mM DTT solution and diluted 20 times with ChIP dilution buffer, and a second ChIP was carried out with 4 μg of control IgG and anti-DHX9 or anti-TOP3B antibodies. Purified DNA was analyzed by qPCR using primers for the indicated regions. The comparative CT method (ΔΔCT) was used to quantify relative changes in DNA levels. CT values were normalized by subtracting input CT values from target gene CT values for each sample. All amplifications were performed in triplicate.
DNA/RNA immunoprecipitation (DRIP)-qPCR
The DRIP assay was adapted from multiple studies (13,25,29) with the following modifications: After restriction enzyme cocktail digestion, DNA was incubated in RNase A buffer (10 μg/ml RNase A, 0.5 M NaCl, and 10 mM Tris–Cl [pH 7.4]) at 37°C for 1 h, followed by phenol:chloroform:isoamyl alcohol (25:24:1) extraction and ethanol precipitation. DNA was resuspended in distilled water and treated with or without RNase H (10 U/6 μg DNA) at 37°C overnight, followed by phenol:chloroform extraction and ethanol precipitation. For each DRIP, 4 μg of genomic DNA was incubated with 10 μg of S9.6 antibody in the binding buffer (10 mM NaPO4 [pH 7.0], 140 mM NaCl, and 0.05% Triton X-100). Precipitated DNA was resuspended in 100 μl of distilled water, and 2 μl was used for qPCR analysis. All amplifications were performed in triplicate.
Quantification of nascent RNA transcription
Chromatin RNA immunoprecipitation assay was performed to quantify the expression level of nascent RNA (51). Briefly, cells from two 10-cm plates were cross-linked with 1% formaldehyde for 10 min at RT, and terminated by the addition of Glycine to a final concentration of 0.2 M for 5 min at RT. Cells were washed twice with PBS and collected by centrifugation at 800 × g for 10 min. The pellets were re-suspended in 4 ml of lysis buffer (1× PBS, pH 7.5, 1 mM PMSF, 0.2% NP40, supplemented with protease inhibitors), and incubated on ice for 20 min. The nuclei were pelleted by centrifugation at 500 × g for 10 min at 4°C. The collected nuclei were re-suspended in PBS and sonicated using the Bioruptor Pico Sonication System (B01060003; Diagenode) for 12 cycles (30 s on, 30 s off/cycle). Supernatant was harvested by centrifugation at 18 000 × g for 35 min at 4°C, followed by overnight antibody incubation at 4°C. The immunocomplex was bound by the Protein A magnetic beads, followed by washing for three times with 1 ml RIPA-1000 buffer (1 M NaCl, 1 mM PMSF, 1% NP40, 0.1% sodium deoxycholate, 0.05% SDS, in 1× PBS), one time with 1 ml LiCl–wash buffer (250 mM LiCl, 1% NP40, 0.1% sodium deoxycholate, 0.05% SDS, in 1× PBS), and one time with TE buffer (10 mM Tris–HCl, pH 8.0, 0.1 mM EDTA). The beads were then re-suspended in 100 ml elution buffer (10 mM dithiothreitol (DTT), 1% SDS, 1mM PMSF and 1× PBS) and incubated for 5 h at 65°C. After elution and reverse crosslinking, 500 μl of TRIzol reagent (15596-018; Thermo Fisher Scientific) was added for RNA extraction, followed by the DNase I (M0303L, New England Biolabs) treatment to remove contaminating DNA fraction in the final RNA samples. After DNase I inactivation, the RNA samples were reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (4368814; Thermo Fisher Scientific). Before using qPCR to quantify the target gene nascent transcription, all cDNA samples (Input, IgG control, RNAPII IP, No-RT control) were tested with ribosomal protein S19 and tRNA primers. RNase inhibitor (M0314L, New England Biolabs) was added to all the buffers described above.
In vitro R-loop detection using DRIP-qPCR
In vitro transcription for R-loop formation has been described before (25). In brief, R-loop-prone pFC53 plasmids were transcribed in vitro with T3 RNA polymerase in the presence of individual proteins at 37°C for 30 min. The reaction was then inactivated at 65°C for 10 min. Samples were equally divided and treated with either RNase A plus RNase H or only RNase A at 37°C for 30 min, followed by proteinase K treatment at 37°C for 30 min. Samples were then purified using phenol:chloroform extraction. Precipitated DNA was dissolved in water. In vitro transcription products were incubated with S9.6 antibody in binding buffer (10 mM NaPO4 pH 7.0, 140 mM NaCl, and 0.05% Triton X-100) at 4°C overnight. The binding mixture was then incubated with protein A/G agarose for 1 h. The bound DNA was eluted with buffer containing 50 mM Tris pH 8.0, 10 mM EDTA, 0.5% SDS and 300 μg of proteinase K at 50°C for 30 min. Samples were then purified by phenol:chloroform extraction and ethanol precipitation. Immunoprecipitated DNA was analyzed by qPCR using pFC53 R-loop primers: Forward, TTTAGAGCTTGACGGGGAAA; Reverse, CAACAGTTGCGTAGCCTGAA.
In vitro R-loop detection using Dot-Blot
Total nucleic acid from the in vitro pFC53 transcription reaction was extracted by a standard SDS/Proteinase K digestion, followed by phenol/chloroform extraction and ethanol precipitation. Indicated amount of purified nucleic acid was applied to the Hybond N+/Positive nylon membrane (RPN303B, GE Healthcare) assembled in the Bio-Dot Apparatus (1706545, Bio-Rad). The membrane was subsequently UV cross-linked (0.12 J/m2) and blocked with 5% milk/TBST (0.1% Tween 20) for 1 h at RT. The level of R-loops was detected using the mouse monoclonal S9.6 antibody and the loading of the nucleic acid was visualized by the Methylene blue staining.
Helicase assay
The R-loop substrates were generated by annealing 5′ 6-carboxyfluorescein (FAM)-labeled RNA oligonucleotides (UCGAAUCUCAUCAUCGCGCGAAAUUUCACA), R-loop forward DNA oligonucleotides (CCTCGCAAACTACTTAGATGTCATCCGCGAGC), and R-loop reverse DNA oligonucleotides (GCTCGCAACGCGCGATGATGAGATTCGAAAGCGAGG) at a molar ratios of 1:5:2.5 (52). The annealing was carried out by heating the oligonucleotides mixture to 95°C for 5 min followed by slow cooling to RT. The annealing product was then purified with MicroSpin™ S-200 HR columns. The helicase assay was performed by incubating the R-loop substrates (5 nM) with various recombinant proteins, as indicated, at 37°C for 10 min in the helicase buffer (20 mM Tris [pH 7.5], 3.5 mM MgCl2, 3.5 mM ATP, 0.1 mg/ml BSA, 5 mM DTT, and 1% [v/v] glycerol). The reaction was initiated by adding ATP and terminated by chilling on ice for 5 min (27). The products were separated by electrophoresis through 12% (w/v) nondenaturing polyacrylamide gels. After electrophoresis, the FAM signal in the gels was detected using a Chemi Doc™ Imaging System (Bio-Rad).
Electrophoretic mobility shift assay (EMSA)
The ssDNA probe used in the assay was a 5′ 6-FAM-labeled DNA oligonucleotides (CCTCGCAAACTACTTAGATGTCATCCGCGAGC). EMSA was performed by incubating the ssDNA probe (5 nM) with various recombinant TDRD3 proteins at 25°C for 20 min in binding buffer (20 mM Tris [pH 7.6], 10 mM MgCl2 and 1 mM DTT). The resulting protein–DNA complexes were resolved on 6% nondenaturing polyacrylamide gels at 80 V for 65 min using 1× TBE buffer (89 mM Tris, 89 mM boric acid, and 2 mM disodium EDTA) (53). After electrophoresis, the FAM signal in the gels was detected using a Chemi Doc™ Imaging System (Bio-Rad).
RNA-seq and data analysis
RNA was isolated from wild type (WT) and TDRD3 KO (KO1 and KO2) MCF7 cells (each with two biological replicates) using TRIzol reagent according to the manufacturer's instructions (Life Technologies). Poly(A) RNA-seq libraries were constructed using a KAPA mRNA HyperPrep Kit (Illumia), and purified libraries were validated using a Bioanalyzer 2100 system with DNA High Sensitivity Chip (Agilent) and quantified using a Qubit fluorometer (Life Technologies). RNA sequencing was performed in the City of Hope Integrative Genomics Core facility on an Illumina HiSeq 2500 in 51-bp single-end read mode, following the manufacturer's recommendations.
The 51-bp-long single-ended sequence reads were mapped to the human genome (hg19) using TopHat (v2.0.8). The raw counts of each gene were generated by HTSeq (v0.6.1p1) in ‘-m union’ mode, normalized using the trimmed mean of M values (TMM) method, and compared using the Bioconductor package ‘edgeR (v3.20.9)’. Genes with RPKM (reads per kilobase of transcript, per million mapped reads) ≥1 in at least one sample were retained, and differentially expressed genes were identified by log2-based fold change ≥1 or ≤–1 for up- and down-regulated genes (FDR < 0.05), respectively.
To generate the heatmap plot, RPKM values were generated to normalize for sequencing depth and gene length per each gene in a sample during edgeR running. Genes with standard deviations of observed values ≤1 and without at least three observations with absolute values greater than four were removed. After filtration, 7950 out of 20 452 genes remained. Log2-based RPKM values were used to generated clustering. Pearson correlations were used as the distance measure for average linkage clustering in Cluster3. TreeView was used to generate the heatmap plot using the clustering outputs.
For gene set enrichment analysis (GSEA), genes were ranked by the signed P-value score, which is − log10 (P) with the sign of the log2-based fold change. The pre-ranked data were uploaded to GSEA v3.0 and analyzed against the cancer hallmark pathway gene set.
Comparing TDRD3-regulated and DHX9-regulated gene expression
2336 commonly up- and 1903 down-regulated genes identified in both TDRD3 KO1 and KO2 MCF7 cells were analyzed in comparison with differentially expressed genes in control and DHX9 siRNA knockdown H1299 cells (54). Enrichr with hallmark database (55–57) was used to perform enrichment analyses on TDRD3-regulated and DHX9-regulated genes. The bubble plot was generated based on combined scores and common hallmark pathways between both datasets from enrichment analyses.
Comparing TDRD3 target genes with R-loop forming promoters
The 121 target genes that show TDRD3 occupancy at promoters were searched against the R-loop mapping DRIP-seq dataset (GSE70189) (15) uploaded onto the R-loop database: http://rloop.bii.a-star.edu.sg
Statistical analysis
All experiments were performed at least three times. Statistical analyses were performed using Student's t-tests. P < 0.05 was considered statistically significant. Quantification of agarose gel images and immunoblotting images was performed using ImageJ software.
RESULTS
TDRD3 interacts with DHX9
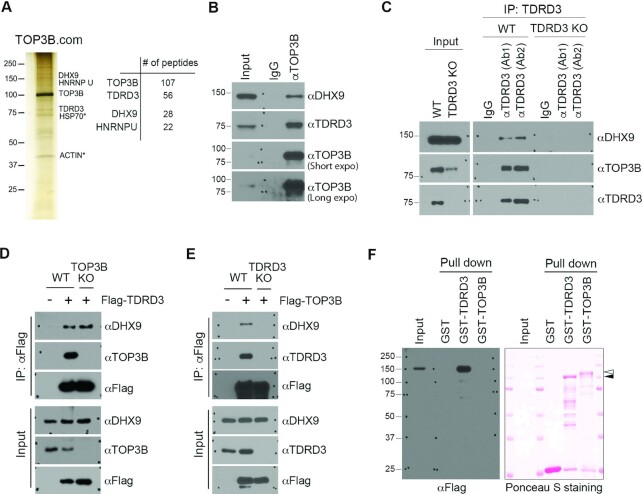
To investigate the transcriptional regulatory mechanism(s) mediated by the TDRD3–TOP3B protein complex, we performed tandem affinity purification and mass spectrometry (TAP-MS) analysis of TOP3B to identify additional protein components of this complex. We identified DHX9 and heterogeneous nuclear ribonucleoprotein U (hnRNP U) as novel interaction partners of TOP3B (Figure 1A and Supplementary Table 1). DHX9 and hnRNP U have been reported to interact with each other, and both proteins are components of several messenger ribonucleoprotein (mRNP) complexes, including the coding region determinant (CRD)-mediated complex (58). To validate the protein interactions identified by TAP-MS, we immunoprecipitated endogenous TOP3B from MCF7 breast cancer cell lysates and detected interacting proteins using western blot analysis. Both TDRD3 and DHX9 were detected in TOP3B-enriched protein samples (Figure 1B), suggesting that these three proteins likely exist in the same protein complex. To further confirm their interaction, we performed co-immunoprecipitation (co-IP) of TDRD3 from both wild type (WT) and TDRD3 knockout (KO) MCF7 cells using two anti-TDRD3 antibodies from different sources (Ab1 and Ab2) and detected its interaction with DHX9 and TOP3B. Both anti-TDRD3 antibodies were able to co-IP DHX9 and TOP3B in WT, but not TDRD3 KO, MCF7 cells, demonstrating that the interaction detected is not caused by non-specific recognition by the TDRD3 antibody (Figure 1C). Consistent with our previous reports (25), loss of TDRD3 destabilizes TOP3B (Figure 1C), whereas it does not affect the level of DHX9.
Figure 1.
TDRD3 interacts with DHX9. (A) Tandem affinity purification of the TOP3B protein complex (TOP3B.com) from HEK293 cells. The eluted protein complex was separated by SDS-PAGE and silver-stained (left panel). The number of unique peptides from the top four purified proteins is shown (right panel). (B) The interaction of DHX9 with TDRD3 and TOP3B was confirmed by co-immunoprecipitation (co-IP). MCF7 cells were immunoprecipitated with control IgG and αTOP3B antibodies. The eluted protein samples were detected by western blot analysis using αDHX9, αTDRD3 and αTOP3B antibodies. (C) The interaction of DHX9 with TDRD3 and TOP3B was detected by co-IP in wild type (WT) and TDRD3 knockout (KO) MCF7 cells using two different αTDRD3 antibodies (Ab1 and Ab2). TDRD3, TOP3B and DHX9 were detected in the input samples by western blot analysis. (D) TOP3B is dispensable for the TDRD3-DHX9 interaction. Both WT and TOP3B KO MCF7 cells were transfected with Flag-TDRD3. The interaction of TDRD3 with DHX9 and TOP3B was detected by co-IP. (E) TDRD3 is essential for the TOP3B-DHX9 interaction. Both WT and TDRD3 KO MCF7 cells were transfected with Flag-TOP3B. The interaction of TOP3B with DHX9 and TDRD3 was detected by co-IP. (F) TDRD3 directly interacts with DHX9. Flag-DHX9 recombinant proteins were purified from HEK293 cells. GST, GST-TDRD3, and GST-TOP3B recombinant proteins were purified from E. coli. The interactions of GST-tagged recombinant proteins with Flag-DHX9 were assessed by GST pull-down and western blot analysis using an αFlag antibody. The amount of proteins used in the binding was visualized by the Ponceau S staining of the PVDF membrane.
To characterize the interactions among these three proteins, we tested the hypothesis that TDRD3 functions as a scaffold for the TOP3B–DHX9 interaction. First, we performed co-IP assays and compared the interaction of TDRD3 with DHX9 in WT and TOP3B KO MCF7 cells. The interaction of TDRD3 with DHX9 remains unaffected in the absence of TOP3B (Figure 1D), suggesting that TOP3B is not essential for the TDRD3–DHX9 interaction. Next, we examined the interaction of TOP3B with DHX9 in WT and TDRD3 KO MCF7 cells. The interaction of TOP3B with DHX9 was abolished in the absence of TDRD3 (Figure 1E), suggesting that TDRD3 is required for the TOP3B-DHX9 interaction. Lastly, to determine if DHX9 directly interacts with TDRD3, we incubated recombinant GST-tagged TDRD3 and TOP3B, purified from E. coli, with Flag-tagged DHX9, purified from HEK293 cells (Supplementary Figure S1A), and conducted a GST pull-down experiment. GST-TDRD3, but not GST-TOP3B, directly interacted with DHX9 (Figure 1F). Thus, these approaches have provided independent lines of evidence supporting a direct and physiologically relevant interaction between TDRD3 and DHX9.
The C-terminal Tudor domain of TDRD3 interacts with DHX9
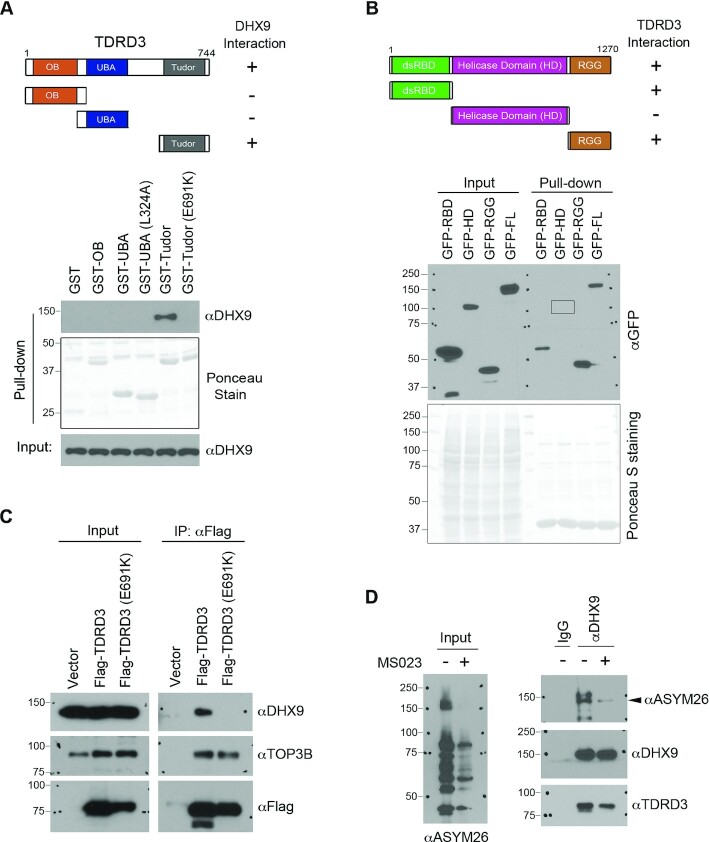
Next, we mapped the interaction between TDRD3 and DHX9. TDRD3 contains three functional domains: the OB-fold, the ubiquitin-associated (UBA) domain, and the Tudor domain, which are critical for mediating its interaction with protein partners (Figure. 2A, upper panel). The OB-fold interacts with TOP3B, and the Tudor domain interacts with arginine-methylated protein substrates, including histones and the C-terminal domain of RNAPII (42,43). The UBA domain of TDRD3 was reported to interact with ubiquitin chains (59). Through sequence analysis and by performing a GST pull-down assay, we demonstrated that a conserved UBA domain amino acid, leucine 324 (L324), is essential for the interaction between the UBA domain and tetra-ubiquitin (Supplementary Figure S1B and S1C). To determine which domain of TDRD3 interacts with DHX9, we performed GST pull-down assays by incubating MCF7 cell lysates with recombinant GST-tagged TDRD3 truncations containing only the N-terminal OB-fold, UBA domain, or C-terminal Tudor domain. The Tudor domain of TDRD3, but not the OB-fold or UBA domain, interacted with DHX9 (Figure 2A, lower panel). Importantly, mutation of glutamic acid to lysine at amino acid 691 (E691K) of TDRD3, which was reported to abolish Tudor domain interactions with arginine-methylated protein substrates (42), completely disrupted the interaction between the Tudor domain and DHX9 (Figure 2A, lower panel).
Figure 2.
Characterization of the interaction between DHX9 and TDRD3. (A) Mapping the region of TDRD3 that interacts with DHX9. GST-tagged TDRD3 truncation constructs were generated, containing the oligonucleotide/oligosaccharide-binding (OB)-fold, the ubiquitin-associated domain (UBA; wild type or with L324A mutation), and the Tudor domain (Tudor; wild type or with E691K mutation), respectively. A graphic summary of their interactions with DHX9 is shown. A GST pull-down assay was performed by incubating the recombinant GST-fusion proteins with MCF7 cell lysates. The pull-down samples were detected by western blot analysis using an αDHX9 antibody. The GST-fusion proteins were visualized by Ponceau staining. (B) Mapping the region of DHX9 that interacts with TDRD3. GFP-tagged full-length or truncations of DHX9 constructs were generated. The locations of the N-terminal double-stranded RNA binding domain (dsRBD), the helicase domain (HD), and the C-terminal RGG-containing domain (RGG) are indicated (upper panel). A graphic summary of their interactions with TDRD3 is shown. A GST pull-down assay was performed by incubating the recombinant GST-Tudor with MCF7 lysates that were transfected with different GFP-DHX9 fusion vectors. Both the input and pull-down samples were detected by western blot analysis using an αGFP antibody. The GST-Tudor recombinant protein used in the binding was visualized by Ponceau staining. (C) The interaction of TDRD3 with DHX9 requires a functional Tudor domain. A co-IP assay was performed to assess the interaction of Flag-tagged WT and methylarginine binding-deficient (E691K) TDRD3 with endogenous DHX9 in MCF7 cells. The input and αFlag antibody-immunoprecipitated samples were analyzed by western blot using indicated antibodies. (D) DHX9 interacts with TDRD3 in an arginine methylation-dependent manner. The interaction of DHX9 with TDRD3 was detected by co-IP in MCF7 cells treated with vehicle (−) or with the type I PRMT inhibitor MS023 (+). The level of cellular ADMA was detected by western blot using a pan-ADMA antibody (ASYM26). The DHX9-immunoprecipitated samples were probed for methylation and interactions with TDRD3 using the ASYM26 and αTDRD3 antibodies.
To determine which segments of DHX9 interact with TDRD3, we expressed three truncations of DHX9: the N-terminal RNA-binding domain (RBD; amino acids 1–262) that harbors two double-stranded RNA-binding domains (dsRBD), the helicase domain (HD; amino acids 255–1077), and the C-terminal fragment that contains nuclear localization/export signals (NLS/NES) and an RGG-box (49) (RGG; amino acids 1064–1270) (Figure 2B, upper panel). The GFP-tagged truncations of DHX9, as well as full-length (FL) DHX9, were expressed in MCF7 cells. Individual cell lysates were incubated with the recombinant GST-Tudor domain of TDRD3, and their interactions were detected by GST pull-down and western blot analysis. Both the N-terminal RBD and C-terminal RGG fragments of DHX9 interacted with the Tudor domain of TDRD3 (Figure 2B, lower panel). To further demonstrate that the functional Tudor domain is important for the TDRD3-DHX9 interaction, we performed a co-IP experiment to compare the interactions of WT and Tudor domain-mutant (E691K) TDRD3 with DHX9 in MCF7 cells. Consistent with the results from the GST pull-down assays (Figure 2A), mutation of the TDRD3 Tudor domain abolished its interaction with DHX9, whereas its interaction with TOP3B, which is mainly mediated through the OB-fold, was largely unaffected (Figure 2C). These results suggested that the TDRD3-DHX9 interaction might be regulated by arginine methylation.
It was reported that DHX9 is arginine methylated by the protein arginine methyltransferase 1 (PRMT1) (60). To determine the extent to which arginine methylation contributes to the DHX9-TDRD3 interaction, we performed a co-IP experiment in MCF7 cells treated with DMSO or the type I arginine methyltransferase inhibitor MS023 to inhibit PRMT1 activity (61). As shown in Figure 2D, MS023 treatment dramatically reduced the cellular asymmetrical dimethylation (ADMA) levels, including the arginine methylation of DHX9, as detected by a pan-ADMA antibody ASYM26. Concomitantly, the interaction between DHX9 and TDRD3 was also reduced. Altogether, these results demonstrate that the Tudor domain of TDRD3 mediates its interaction with DHX9, likely by recognizing the methylarginine modification on DHX9.
TDRD3 recruits DHX9 to the promoters of its target genes
TDRD3 functions as an methylarginine effector molecule and plays important roles in transcription regulation and breast cancer tumorigenesis (25,42,62,63). To further investigate the significance of its interaction with DHX9, we tested the hypothesis that DHX9 can be recruited by TDRD3 to regulate target gene expression. First, we performed RNA-seq analysis to identify differentially expressed genes in WT MCF7 cells and two independent TDRD3 KO MCF7 cell lines generated using CRISPR/Cas9 technology (Supplementary Figure S2A). Heatmap clustering analysis showed that the KO clones (KO1 and KO2) exhibited strongly correlated up- and down- regulation of gene expression (Supplementary Figure S2B), suggesting that the observed changes in gene expression are due to TDRD3 KO and not likely caused by off-target effects of the guide RNAs. We identified 2336 commonly up-regulated and 1903 commonly down-regulated genes in both KO clones compared to the WT MCF7 cells (Supplementary Figure S2C and Supplementary Table 2). Gene set enrichment analysis (GSEA) of these differentially expressed genes revealed that loss of TDRD3 resulted in a significant down-regulation of estrogen-responsive gene expression (Supplementary Figure S2D), consistent with its previously reported function as a co-activator of estrogen receptor-mediated transcription (42). Next, to determine the extent to which DHX9 contributes to TDRD3-regulated gene expression, we analyzed a set of recently published RNA-seq data from DHX9 knockdown human non-small cell lung carcinoma H1299 cells (54). Despite cell type differences, the majority of down-regulated cancer hallmark pathways in TDRD3 KO MCF7 cells, including estrogen response and mTORC1 signaling, showed similar down-regulation in DHX9 knockdown H1299 cells (Supplementary Figure S2E). Together with the results demonstrating their direct protein-protein interaction, this result further suggests that DHX9 functions together with TDRD3 in regulating gene expression. To further test this, we attempted to determine the genome-wide co-localization of DHX9 with TDRD3 using chromatin immunoprecipitation followed by sequencing (ChIP-seq). However, we were not able to obtain reliable DHX9 ChIP-seq signals using DHX9 antibodies from two different sources, likely because of its weak and transient association with chromatin. Unfortunately, there was also no publicly available DHX9 ChIP-seq datasets from any mammalian cells. Thus, we decided to take a candidate approach to examine DHX9 function in the regulation of a few selected TDRD3 target genes.
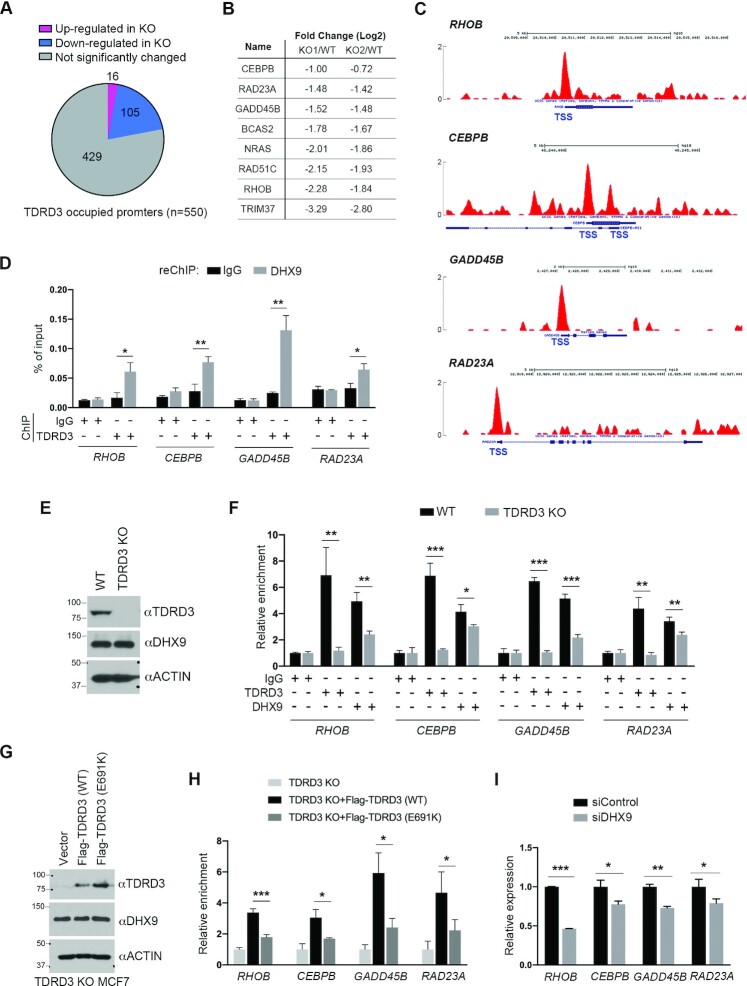
To identify direct TDRD3 target genes, we analyzed genes that showed TDRD3 enrichment at their promoters, identified by TDRD3 ChIP-seq analysis (42), and genes that exhibited differential levels of expression in WT vs. TDRD3 KO MCF7 cells (Supplementary Figure S2C). Among these bona fide TDRD3 target genes (Supplementary Table 3), the majority (105 out of 121) showed significantly reduced gene expression in TDRD3 KO MCF7 cells (Figure 3A), supporting its function as a transcription co-activator (25,42). To investigate how DHX9 and TDRD3 function together in transcription regulation, we focused on eight genes that showed similarly reduced expression in both KO clones (Figure 3B) and exhibited strong TDRD3 enrichment at their promoters (Figure 3C and Supplementary Figure S3A).
Figure 3.
TDRD3 recruits DHX9 to its target gene promoters. (A) A pie-chart demonstration of genes with TDRD3 bound to their promoters and genes up- or down-regulated after TDRD3 knockout (KO) in MCF7 cells. (B) Genes showing more than a 2-fold reduction in TDRD3 KO versus wild type (WT) MCF7 cells were selected for further analysis. The relative expression of each gene in both KO clones is shown. (C) UCSC Genome Browser plots of TDRD3 ChIP-seq reads along the indicated genes in MCF7 cells. The y-axis represents the normalized number of reads; the thick blue boxes represent the open reading frames; and the transcription start site (TSS) is labeled. (D) A ChIP-reChIP assay was performed to detect the co-occupancy of TDRD3 and DHX9 at target gene promoters. The first round of ChIP was performed using control IgG and αTDRD3 antibodies, and the second round of ChIP (reChIP) was performed using IgG and αDHX9 antibodies. ChIP DNA was analyzed by qPCR using primers for the indicated gene promoters. (E) TDRD3 KO does not affect DHX9 protein expression. TDRD3 and DHX9 were detected in WT and TDRD3 KO MCF7 cells by western blot analysis. ACTIN was used as a loading control. (F) Loss of TDRD3 reduces DHX9 recruitment to target gene promoters. ChIP assays were performed in WT and TDRD3 KO MCF7 cells using control IgG, αTDRD3, and αDHX9 antibodies. (G) Rescued expression of Flag-tagged WT and methylarginine binding-deficient (E691K) TDRD3 in TDRD3 KO MCF7 cells, detected by western blot analysis. (H) The methylarginine binding function of TDRD3 is essential for promoting DHX9 recruitment to the target gene promoters. DHX9 ChIP assays were performed in TDRD3 KO MCF7 cells and TDRD3 KO MCF7 cells re-expressing Flag-TDRD3 (WT) and Flag-TDRD3 (E691K). The relative enrichment of DHX9 in TDRD3 KO MCF7 cells was used for normalization. (I) DHX9 knockdown reduces TDRD3 target gene expression. MCF7 cells were transfected with either control siRNA (siControl) or DHX9-specific siRNA (siDHX9). The expression of TDRD3 target genes was detected by RT-qPCR assays. Experiments were performed independently for three times. Data are represented as mean ± standard deviation of three technical qPCR replicates. Statistical analysis was performed using Student's t-tests. * P < 0.05; ** P < 0.01; *** P < 0.001.
To determine if DHX9 is recruited to the promoters of TDRD3 target genes, we performed ChIP experiments using control IgG and anti-DHX9 antibodies. Compared to the IgG control, DHX9 showed ∼2- to 6-fold greater enrichment on all of the promoters tested (Supplementary Figure S3B), indicating its role in promoter regulation. To determine if TDRD3 and DHX9 co-localize at these target gene promoters, we performed ChIP-reChIP experiments by using control IgG and anti-TDRD3 antibodies for the first round, and control IgG and anti-DHX9 antibodies for the second round (reChIP). Among the eight TDRD3 target genes tested, we detected significant co-occupancy of TDRD3 and DHX9 on the promoters of five, including RHOB, CEBPB, GADD45B, RAD23A and RAD51C (Figure 3D and Supplementary Figure S3C).
To test if TDRD3 is important for the recruitment of DHX9 to target gene promoters, we performed ChIP experiments and compared DHX9 enrichment levels in WT and TDRD3 KO MCF7 cells. Because of the high similarity between the two independent TDRD3 KO clones in terms of gene expression (Supplementary Figure S2B and S2C), we used only clone KO1, which we refer to as KO in the following study. The loss of TDRD3 in MCF7 cells does not affect DHX9 protein levels (Figure 3E). However, the enrichment of DHX9 was significantly reduced on the promoters of all target genes they co-occupied (Figure 3F), suggesting that TDRD3 facilitates DHX9 recruitment to these gene promoters. Because the functional Tudor domain is essential for the interaction of TDRD3 and DHX9 (Figure 2A and C), we next determined if the recruitment of DHX9 relies on the TDRD3 Tudor domain. To do that, we re-expressed either WT or Tudor domain-mutant (E691K) Flag-TDRD3 in TDRD3 KO MCF7 cells and performed ChIP experiments using aniti-DHX9 antibody. We found that DHX9 expression levels were similar in MCF7 cells expressing WT and E691K mutant TDRD3 (Figure 3G); however, the enrichment of DHX9 at TDRD3 target gene promoters was restored only by the expression of WT, but not E691K mutant TDRD3 (Figure 3H), demonstrating that the recruitment of DHX9 to TDRD3 target gene promoters depends on the functional Tudor domain. Consistent with the role of arginine methylation in facilitating their recruitment and interaction, treating cells with type I PRMT inhibitor MS023 significantly reduced the promoter recruitment of both TDRD3 and DHX9 (Supplementary Figure S3D). Furthermore, when we knocked down DHX9 expression using siRNA, we found that the enrichment of TDRD3 on two of the four target genes was decreased (Supplementary Figure S3E and S3F), indicating that the physical interaction between TDRD3 and DHX9 is important for their stable association with target gene promoters. As expected, DHX9 knockdown reduced the expression of TDRD3 target genes (Figure 3I), further supporting that these two proteins function together in regulating gene expression. Notably, when we examined TDRD3 recruitment at a few previously reported DHX9 target sites, including the promoter and transcription termination site of human β-actin gene (28,64), we only found a marginal TDRD3 enrichment at the termination region (Supplementary Figure S3G), suggesting that TDRD3 and DHX9 might only co-regulate a subset of target genes.
DHX9 resolves R-loops at TDRD3 target gene promoters
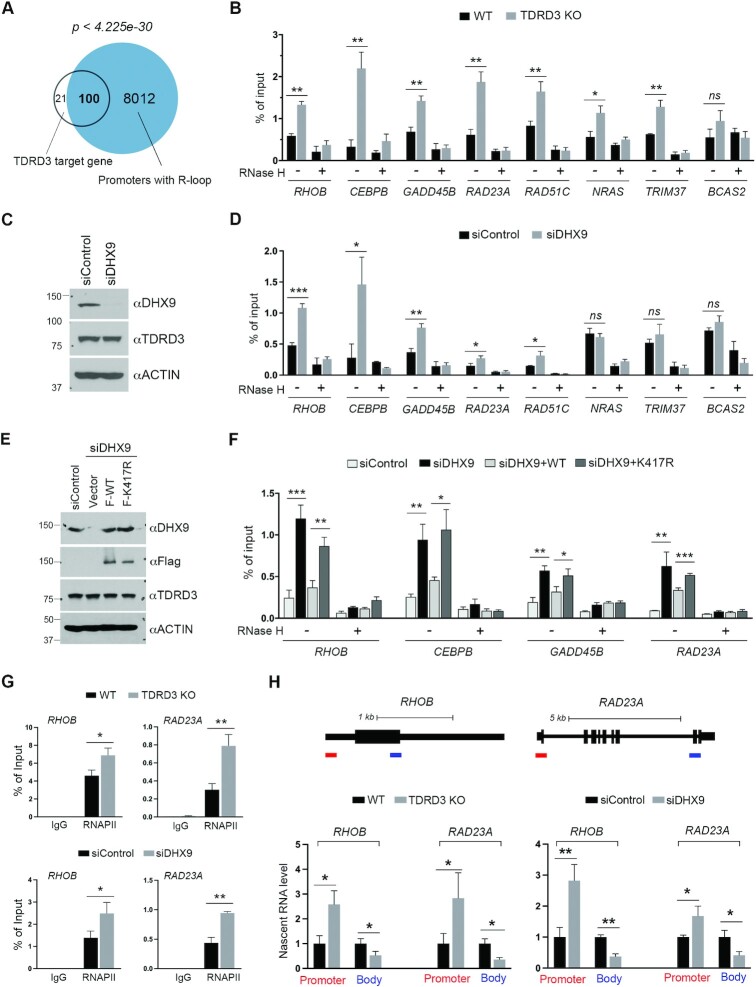
Our previous work demonstrated that about 50% of TDRD3-bound promoters are enriched for R-loop formation (25). We analyzed the bona fide TDRD3 target genes (Figure 3A), and found that 83% (100 out of 121) form promoter R-loops (Figure 4A), as revealed by the DNA/RNA immunoprecipitation followed by cDNA conversion coupled to high-throughput sequencing (DRIPc-seq) (15). The strong correlation between TDRD3 enrichment and R-loop formation (p-value < 4.225e-30) led us to test if TDRD3 regulates R-loop levels at these target gene promoters. We performed the DRIP-qPCR analysis on select TDRD3 target genes (Figure 3B) in WT and TDRD3 KO MCF7 cells. The loss of TDRD3 caused significant elevation of R-loops at almost all of the loci tested (Figure 4B), supporting the function of TDRD3 in suppressing promoter R-loops (25).
Figure 4.
The TDRD3-DHX9 protein complex resolves promoter-associated R-loops. (A) Venn diagram of TDRD3 target genes and genes that form R-loops at their promoters, as identified by DRIPc-Seq (15). (B) TDRD3 knockout (KO) increases the R-loop levels at the promoters of its target genes. DRIP-qPCR analysis was performed to compare R-loop levels at the promoters of the TDRD3 target genes in wild type (WT) and TDRD3 KO MCF7 cells. Samples treated with RNase H, which disrupts R-loops, served as negative controls. (C) DHX9 knockdown does not affect TDRD3 protein expression. DHX9 and TDRD3 were detected by western blot analysis in MCF7 cells transfected with control siRNA (siControl) or DHX9-specific siRNA (siDHX9). ACTIN was used as a loading control. (D) DHX9 knockdown increases R-loop levels at the promoters of TDRD3 target genes. DRIP-qPCR analysis was performed to compare R-loop levels at the promoters of the TDRD3 target genes in control and DHX9 knockdown MCF7 cells. Samples treated with RNase H served as negative controls. (E) MCF7 cells were transfected with control siRNA (siControl) or DHX9-specific siRNA (siDHX9). After 24 h, the siDHX9-transfected cells were transfected with either an empty vector, WT DHX9 or helicase activity-deficient (K417R) DHX9 for an additional 48 h. DHX9 and TDRD3 were detected in these cells by western blot analysis. The anti-ACTIN was used as a loading control. (F) The helicase activity of DHX9 is essential for resolving R-loops at TDRD3 target gene promoters. DRIP-qPCR analysis was performed to compare R-loop levels at the promoters of the TDRD3 target genes in control (siControl) or DHX9 knockdown (siDHX9) MCF7 cells or DHX9 knockdown MCF7 cells with re-expression of either WT DHX9 (siDHX9 + WT) or helicase activity-deficient DHX9 (siDHX9 + K417R), as described in (E). Samples treated with RNase H served as negative controls. (G) TDRD3 KO and DHX9 knockdown cause RNAPII accumulation at gene promoters. RNAPII ChIP assays were performed in WT and TDRD3 KO, as well as control and DHX9 knockdown MCF7 cells. Normal rabbit IgG was used as a negative control. (H) Impact of TDRD3 KO and DHX9 knockdown on the nascent RNA transcription. Diagram demonstration of RHOB and RAD23A gene locus. The bars at the bottom of the diagrams indicate the amplicon locations for nascent RNA transcripts (red: promoter; blue: gene body and 3′end). Chromatin RNA immunoprecipitation was performed in WT and TDRD3 KO, as well as control and DHX9 knockdown MCF7 cells. The levels of nascent RNA were quantified by RT-qPCR. Experiments were performed independently three times. Data are represented as mean ± standard deviation of three technical qPCR replicates. Statistical analysis was performed using Student's t-tests. * P < 0.05; ** P < 0.01; *** P < 0.001.
The DNA/RNA helicase activity of DHX9 has been reported to be involved in the resolution of various DNA and RNA structures, including R-loops, formed during transcription and DNA replication (27,48,49). To test the hypothesis that TDRD3 recruits DHX9 to resolve promoter R-loops, we compared the levels of R-loops formed at the promoters of TDRD3 target genes in control and siRNA-mediated DHX9 knockdown MCF7 cells. DHX9 knockdown was effective and did not affect TDRD3 expression levels (Figure 4C). Among the eight TDRD3 target genes that we examined, five (RHOB, CEBPB, GADD45B, RAD23A and RAD51C) showed increased R-loops at their promoter regions upon DHX9 knockdown (Figure 4D). To determine if the DNA/RNA helicase activity of DHX9 is essential for the resolution of promoter R-loops, we re-expressed Flag-tagged WT or helicase activity-deficient (K417R) DHX9 (65,66) in the DHX9 knockdown MCF7 cells. The DHX9-siRNA was designed to targets its 3′UTR region, thus allowing the successful expression of exogenous DHX9 (Figure 4E). The re-expression of WT DHX9, but not K417R mutant DHX9, suppressed the induction of R-loops caused by DHX9 knockdown at the promoters of all target genes tested (Figure 4F). Altogether, these results demonstrate that DHX9 is recruited by TDRD3 to resolve promoter R-loops in a helicase activity-dependent manner.
Formation of R-loops is often associated with RNAPII pausing (67,68). To further investigate the impact of increased R-loops caused by TDRD3 KO and DHX9 knockdown, we performed ChIP experiments and compared the levels of RNAPII at the promoters of RHOB and RAD23A. As expected, TDRD3 KO and DHX9 knockdown both led to increased RNAPII levels at target gene promoters (Figure 4G), supporting RNAPII pausing. Next, we performed chromatin RNA immunoprecipitation using the RNAPII antibody (Supplementary Figure S4) and compared the levels of nascent RNA transcripts (51). Consistent with the increased R-loop and RNAPII pausing, the promoter-associated nascent RNA was significantly increased upon TDRD3 KO and DHX9 knockdown, whereas the transcripts that extend to downstream gene body and 3′ end were significantly reduced (Figure 4H), supporting the reduced gene expression. Taken together, these results reveal the function of TDRD3 and DHX9 in suppressing promoter-associated R-loops to facilitate RNAPII elongation and gene expression.
TDRD3 promotes the helicase activity of DHX9 in R-loop resolution
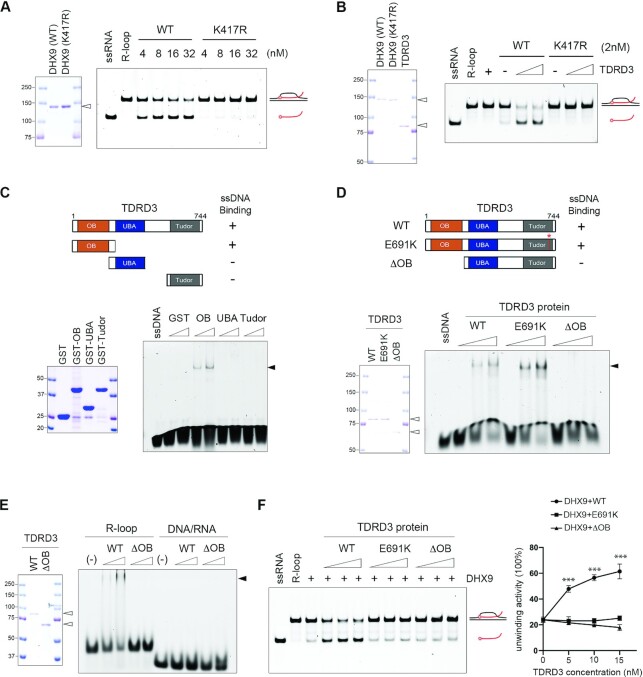
It was reported that, although the N-terminal dsRBD and C-terminal RGG-box of DHX9 are dispensable for its basic helicase activity, they can modulate such activity by regulating DHX9 interactions with DNA and RNA substrates (69,70). The interaction of TDRD3 with both domains of DHX9 (Figure 2B) led us to hypothesize that TDRD3 might regulate DHX9 helicase activity. To test this hypothesis, we first established an in vitro helicase assay in which recombinant DHX9 proteins purified from HEK293 cells were incubated with an R-loop substrate. Because DHX9 prefers substrates with a short single-stranded non-complementary 3′ tail (52,71), we assembled a unique R-loop structure, which contains a free 10-bp 3′ end that does not anneal to the complementary DNA sequences (Materials and Methods). As shown in Figure 5A, addition of increasing amounts of WT DHX9, but not the helicase activity-deficient (K417R) DHX9, led to the appearance of a faster-migrating species on native gels, representing the release of FAM (fluorescein)-labeled RNA oligonucleotides and thus demonstrating a helicase activity-dependent R-loop resolution. This activity requires ATP (Supplementary Figure S5A). Next, to test if TDRD3 promotes DHX9 helicase activity in R-loop resolution, we added increasing amounts of recombinant TDRD3 proteins to the helicase assay reactions containing low concentrations of either WT or K417R mutant DHX9. As shown in Figure 5B, the addition of TDRD3 strongly stimulated the R-loop resolution activity of WT DHX9, but not the K417R mutant. This stimulating effect can be observed in as early as 30 s from the start of the reaction (Supplementary Figure S5B). Notably, TDRD3 alone does not exhibit R-loop resolution activity.
Figure 5.
TDRD3 promotes DHX9 helicase activity in R-loop resolution. (A) DHX9 resolves R-loops in a helicase activity-dependent manner. Coomassie Blue staining of recombinant wild type (WT) and helicase activity-deficient (K417R) DHX9 purified from HEK293 cells (left). The helicase assay on R-loops was performed by incubating increasing amounts of recombinant WT or K417R DHX9 with 5′ 6-FAM-labeled R-loop substrates (5 nM) at 37°C for 10 min. The reaction products were analyzed by gel electrophoresis and fluorescence imaging (right). The open triangle indicates the recombinant proteins. (B) TDRD3 stimulates the helicase activity of WT DHX9, but not K417R mutant DHX9, in R-loop resolution. Coomassie Blue staining of recombinant WT, K417R mutant DHX9, and TDRD3 purified from HEK293 cells (left). The helicase assay on R-loops was performed by incubating 5′ 6-FAM-labeled R-loop substrates with constant amounts of either WT or K417R mutant DHX9 (2 nM) and increasing amounts (40 and 60 nM) of TDRD3 (right). The open triangles indicate the recombinant proteins. (C) The OB-fold of TDRD3 binds single-stranded DNA (ssDNA). A graphic summary of the interactions of truncated TDRD3 fragments with ssDNA is shown (upper panel). Coomassie Blue staining of recombinant GST-fusion proteins of TDRD3, including its OB-fold, UBA domain, and Tudor domain, purified from E. coli (lower left panel). The electrophoretic mobility shift assay (EMSA) was performed by incubating a 5′ 6-FAM-labeled ssDNA oligonucleotide (5 nM) with increasing amounts (25 and 50 nM) of the recombinant TDRD3 proteins (lower right panel). The solid triangle indicates the protein–nucleotide complex. (D) The OB-fold is essential for the interaction of TDRD3 with ssDNA. A graphic summary of the WT, methylarginine binding-deficient (E691K), and OB-fold-truncated (ΔOB) TDRD3 interaction with ssDNA is shown (upper panel). Coomassie Blue staining of all three recombinant TDRD3 proteins purified from HEK293 cells (lower left panel). EMSA was performed to detect the binding of increasing amounts (20, 40 and 60 nM) of recombinant proteins with a 5′ 6-FAM-labeled ssDNA oligonucleotide (5 nM) (lower right panel). The open triangle indicates the recombinant proteins. The solid triangle indicates the protein-nucleotide complex. (E) TDRD3 interacts with R-loops, but not DNA/RNA hybrids. EMSA was performed by incubating increasing amounts (40 and 60 nM) of recombinant WT and OB-fold-truncated (ΔOB) TDRD3 with 5′ 6-FAM-labeled R-loop or DNA/RNA hybrid oligonucleotide (5 nM). The open triangle indicates the recombinant proteins. The solid triangle indicates the protein-nucleotide complex. (F) Both the OB-fold and the functional Tudor domain are required for TDRD3 to stimulate the helicase activity of DHX9 in R-loop resolution. The helicase assay on R-loops was performed by incubating 5′ 6-FAM-labeled R-loop substrates (5 nM) with a constant amount of DHX9 (2 nM) and increasing amounts (20, 40 and 60 nM) of WT, methylarginine binding-deficient (E691K), and ssDNA binding-deficient (ΔOB) TDRD3 (left). Helicase activity was quantified by measuring the percentage of unwound substrates under the indicated assay conditions (right). Statistical analysis was performed using Student's t-tests of data from three independent experiments. *** P < 0.001.
TDRD3 was shown to enhance the processivity of TOP3B enzymatic activity through its interaction with ssDNA (47). Because R-loops contain a displaced, non-template ssDNA, we tested if the ssDNA-binding capability of TDRD3 is essential for its stimulation of DHX9 helicase activity. To do that, we first mapped the ssDNA-interacting region of TDRD3, which was not defined in the previous study (47). Briefly, three fragments of TDRD3, corresponding to its OB-fold, UBA domain, and Tudor domain, respectively, were purified as GST-tagged recombinant proteins (Figure 5C, left panel). Their interactions with FAM-labeled ssDNA probes were examined using the electrophoretic mobility shift assay (EMSA). The OB-fold of TDRD3, but not the UBA or Tudor domain, was sufficient to bind ssDNA, which caused slower migration of the ssDNA probes on native gels, representing the formation of protein-DNA complexes (Figure 5C, right panel). Second, to determine if the OB-fold is essential for TDRD3 to interact with ssDNA, we performed the EMSA assay and compared the ssDNA-binding ability of WT, methylarginine binding-deficient Tudor mutant (E691K), and OB-fold truncation mutant (ΔOB) TDRD3 purified from HEK293 cells (Figure 5D, left panel). The deletion of the OB-fold completely abolished the interaction of TDRD3 with ssDNA, which was not affected by the Tudor domain mutation (Figure 5D, right panel), suggesting that TDRD3 binds ssDNA through its OB-fold. Because the R-loop structure contains a non-templated ssDNA, we next tested if the ssDNA-binding capability enables TDRD3 to bind R-loops. We performed the EMSA assays to compare the interactions of TDRD3 with either R-loops or DNA/RNA hybrids that do not contain single-stranded oligonucleotides. As shown in Figure 5E, addition of TDRD3 caused slower migration of the R-loop probes on native gels but had no effects on the migration of the DNA/RNA hybrid probes, suggesting that TDRD3 binds R-loops, but not DNA/RNA hybrids. Furthermore, the OB-fold deletion truncation failed to bind R-loops, suggesting that the OB-fold-mediated ssDNA interaction is essential for TDRD3 to bind R-loops. Lastly, we performed the helicase assay and compared the efficiency of WT, DHX9 interaction-deficient (E691K) (Figure 2C), and the ssDNA binding-deficient (ΔOB) TDRD3 in stimulating DHX9-catalyzed R-loop resolution. Only the WT TDRD3 was able to promote DHX9 helicase activity (Figure 5F), suggesting that both protein-protein interactions and the OB-fold-mediated ssDNA interaction are essential for TDRD3 to promote DHX9 helicase activity in R-loop resolution. These results demonstrate that, in addition to recruiting DHX9 to target gene promoters, TDRD3 can also stimulate the biochemical activities of DHX9 on R-loop substrates.
DHX9 and TOP3B function together to resolve co-transcriptional R-loops
We previously reported that TDRD3 recruits TOP3B to gene promoters to resolve underwound and negatively supercoiled DNA and prevent R-loop formation (25,50). Consistent with this finding, we detected co-occupancy of TOP3B and TDRD3 at the promoters of TDRD3 target genes (Figure 6A and Supplementary Figure S6A), and TDRD3 knockout dramatically reduced the enrichment of TOP3B at these promoters (Figure 6B and Supplementary Figure S6B). Furthermore, siRNA-mediated knockdown of TOP3B expression reduced the expression of TDRD3 target genes (Supplementary Figure S6C and S6D) and increased the levels of R-loops at target gene promoters (Supplementary Figure S6E), which are associated with RNAPII promoter pausing (Supplementary Figure S6F) and the increase of promoter-associated nascent transcripts (Supplementary Figure S6G), suggesting that TOP3B is another critical component, in addition to DHX9, that is recruited by TDRD3 to regulate gene expression.
Figure 6.
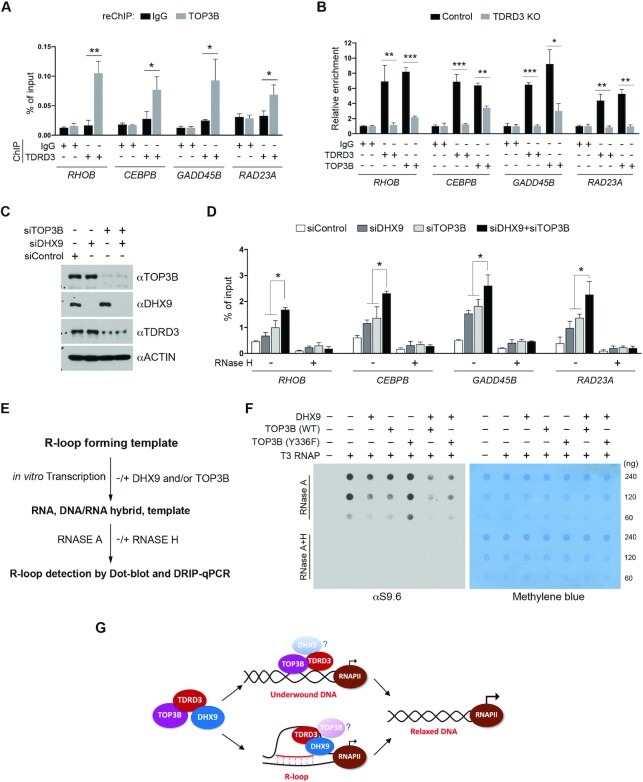
DHX9 and TOP3B function together to resolve co-transcriptional R-loops. (A) A ChIP-reChIP assay was performed to detect the co-occupancy of TDRD3 and TOP3B at target gene promoters. The first round of ChIP was performed using control IgG and αTDRD3 antibodies, and the second round of ChIP (reChIP) was performed using IgG and αTOP3B antibodies. ChIP DNA was analyzed by qPCR using primers for the indicated gene promoters. (B) Loss of TDRD3 reduces TOP3B recruitment to target gene promoters. ChIP assays were performed in WT and TDRD3 KO MCF7 cells using control IgG, αTDRD3, and αTOP3B antibodies. (C) MCF7 cells were transfected with control siRNA (siControl), DHX9-specific siRNA (siDHX9), or TOP3B-specific siRNA (siTOP3B), individually or in combination. TOP3B, DHX9 and TDRD3 were detected by western blot analysis using indicated antibodies. (D) TOP3B and DHX9 function together in resolving R-loops at TDRD3 target gene promoters. DRIP-qPCR analysis was performed to compare R-loop levels at the promoters of TDRD3 target genes in control (siControl), DHX9 knockdown (siDHX9), TOP3B knockdown (siTOP3B), and DHX9/TOP3B double knockdown (siDHX9 + siTOP3B) MCF7 cells. Samples treated with RNase H served as negative controls. (E) Schematic of the in vitro plasmid-based R-loop formation assay followed by quantification of R-loop levels using Dot-blot and DRIP-qPCR. pFC53 plasmids, which contain the R-loop-forming sequence of Airn (13), were transcribed in vitro using T3 RNA polymerase under standard conditions. The transcription products, including the DNA template, the free RNA product, and R-loop containing template, were then subjected to RNase A digestion to remove RNA. The remaining samples were equally divided and either left untreated (−) or treated with RNase H (+). The final products were purified and subjected to either Dot-blot analysis or DRIP-qPCR with DNA/RNA hybrid-specific antibody (S9.6) for R-loop quantification. (F) TOP3B and DHX9 function together to resolve co-transcriptional R-loops in vitro. The pFC53 plasmid was subjected to in vitro transcription in the presence of DHX9, WT TOP3B, and topoisomerase activity-deficient (Y336F) TOP3B, either individually or in combination, as indicated. The level of R-loops in each reaction was detected by Dot-Blot using the S9.6 antibody. The loading of the nucleic acid was visualized by methylene blue staining of the membrane. (G) TDRD3 functions as a scaffold that assembles a protein complex, containing TOP3B and DHX9, to regulate co-transcriptional R-loops at gene promoters. TDRD3 not only recruits TOP3B and DHX9 to specific genomic regions, but also stimulates their respective enzymatic activities to either resolve underwound DNA to prevent R-loop formation (with TOP3B) or resolve existing R-loops to avoid R-loop accumulation (with DHX9). This activity of TDRD3 is likely conferred by its interaction with ssDNA.
To investigate how DHX9 and TOP3B function together to facilitate TDRD3 target gene expression, we tested the hypothesis that both enzymes are involved in resolving co-transcriptional R-loops at TDRD3 target gene promoters. MCF7 cells were transfected with siRNAs to knockdown the expression of DHX9 and TOP3B, either individually or in combination (Figure 6C). We then performed DRIP-qPCR analysis in these cells and compared the levels of R-loops at the promoters of TDRD3 target genes. Knockdown of either DHX9 or TOP3B caused an increase in R-loops; however, knockdown of both DHX9 and TOP3B led to a much greater increase in R-loops than knockdown of either individually (Figure 6D), suggesting that DHX9 and TOP3B function non-redundantly in suppressing co-transcriptional R-loops.
To further define the cooperative function of DHX9 and TOP3B in R-loop resolution, we examined the effects of these two enzymes on R-loop formation during in vitro transcription of a GC-rich R-loop-prone plasmid (pFC53) (13). Briefly, the plasmid was subjected to in vitro transcription using T3 RNA polymerase, followed by RNase A digestion to remove the transcribed RNA product. RNase H treatment, which specifically degrades RNA in double-stranded DNA/RNA hybrids, was used as a negative control. Following nucleic acid purification, the amount of co-transcriptional R-loops can be quantified by Dot-Blot and DRIP-qPCR analysis (Figure 6E). Unlike the helicase assay using short R-loop substrates, this assay system introduces DNA topology and active transcription as additional variables that impact co-transcriptional R-loop dynamics, thus, more closely recapitulating in vivo situations. Using this assay, we previously showed that TOP3B suppresses the formation of co-transcriptional R-loops by resolving underwound and negatively supercoiled DNA (25,50). In this study, we observed that the addition of WT DHX9, but not the helicase activity-deficient (K417R) DHX9, reduced the formation of R-loops during in vitro transcription (Supplementary Figure S6H), consistent with results from the helicase assays (Figure 5A) and further demonstrating that the helicase activity of DHX9 is required to resolve co-transcriptional R-loops. To determine if DHX9 and TOP3B function together to suppress co-transcriptional R-loop formation, we added DHX9 and TOP3B to the reaction either individually or in combination and compared R-loop formation using S9.6 Dot-blot and DRIP-qPCR analyses. Notably, the addition of DHX9 with WT TOP3B, but not the topoisomerase activity-deficient (Y336F) TOP3B, cooperatively reduced the R-loop levels compared to the addition of DHX9 or TOP3B individually (Figure 6F and Supplementary Figure S6I). We noticed that the changes in R-loop levels detected by DRIP-qPCR are more prominent than those detected by total nucleic acid Dot-Blot analysis. This discrepancy likely arises because the DRIP assay involves several steps of stringent washing, which may disrupt/remove some short base-paired R-loops that contribute to total R-loop levels detected by S9.6 Dot-Blot assay. Altogether, these cellular and in vitro assays demonstrate that DHX9 and TOP3B can function together to suppress co-transcriptional R-loops at TDRD3 target gene promoters.
DISCUSSION
R-loops have emerged as critical regulators of chromatin biology, influencing both gene expression and genome stability. Because the formation of R-loops occurs at various functional elements in the genome (i.e. enhancers, promoters, termination sites, and intergenic regions), the molecular pathways involved in R-loop regulation are likely context-dependent. An ever-growing number of factors have been reported to regulate cellular R-loop levels, including topoisomerases, helicases, and nucleases; However, how these seemingly redundant molecular pathways are targeted to R-loop-prone genomic regions remains unclear. Our study reveals that recruitment of the DNA/RNA helicase DHX9 and the DNA topoisomerase TOP3B by the epigenetic reader protein TDRD3 could be a critical mechanism underlying the regulation of promoter-associated R-loops in transcription activation (Figure 6G).
TDRD3 directs TOP3B and DHX9 to suppress promoter-associated R-loops
Genome-wide R-loop mapping studies have revealed that promoter-proximal regions are hotspots for R-loop formation (11,13,14); however, the molecular mechanisms that control the formation of promoter-associated R-loops are still elusive. Mutations in serine/arginine-rich splicing factor 2 (SRSF2), which are found in a substantial proportion of patients with myelodysplastic syndrome, lead to the accumulation of R-loops at the promoters of genes involved in cell proliferation (37). It was proposed that SRSF2 mutations induce RNAPII pausing at transcription start sites, which potentially increases promoter-associated R-loops (37). However, how the mutation of a general splicing factor causes R-loop levels to increase at specific target genes remains unclear. Furthermore, similar to SRSF2 mutations, loss of function of the tumor suppressor gene BRCA2 also causes RNAPII pausing and R-loop accumulation at promoter-proximal pausing sites (67), suggesting that the formation of R-loops is associated with RNAPII pausing. However, whether paused RNAPII is the cause or consequence of R-loop accumulation remains to be further investigated. Results from this and our previous studies suggest that the protein complex assembled by the methylarginine effector molecular TDRD3, which contains the DNA topoisomerase TOP3B and the DNA/RNA helicase DHX9, could play a major role in regulating the promoter-associated R-loops. Importantly, knockout TDRD3 or knockdown either TOP3B or DHX9 leads to RNAPII promoter pausing and increases the levels of promoter-associated nascent transcripts (Figure 4G, H, and Supplementary Figure S6F, S6G)—similar effects caused by the knocking down of SRSF2 or BRCA2 (37,67). Thus, our study provides another strong evidence linking promoter R-loop formation with RNAPII pausing.
TDRD3 is a major methylarginine effector molecule (41,42) that is ubiquitously expressed in all tissues (59). More than 60% of chromatin-localized TDRD3 is at promoter regions (42), where it functions as a scaffold to assemble protein complexes to facilitate transcription activation. TDRD3 has the potential to form three different protein complexes: TDRD3/TOP3B, TDRD3/DHX9, and TDRD3/TOP3B/DHX9, involving in R-loop regulation. We envision that different combination of the three protein components are likely recruited at different gene promoters or at the same promoter but different stages of transcription activation. One such example is the recruitment of TDRD3-TOP3B complex to the NRAS gene promoter (Supplementary Figure S6A), where the TDRD3-DHX9 co-occupancy was not detected (Supplementary Figure S3C). We propose that TDRD3 recruits TOP3B to resolve underwound and negatively supercoiled DNA created by transcribing RNAPII to prevent R-loop formation, whereas recruits DHX9 to act directly on DNA/RNA hybrids to resolve existing R-loops. However, we cannot exclude the possibility that DHX9 exists in TDRD3/TOP3B complex to help altering DNA topology, and that TOP3B participates in TDRD3/DHX9 complex for R-loop resolution (Figure 6G). Nevertheless, the topoisomerase activity of TOP3B and the helicase activity of DHX9 assembled by TDRD3 provides a unique mechanism that enables the spatial-temporal modulation of R-loop dynamics at gene promoters. This is important because, despite the function of short-lived transient R-loops in promoting an open chromatin state for active transcription (13,16), persistent R-loops can block RNAPII progression (18) and lead to the epigenetic silencing of gene expression (19).
The stoichiometry of TDRD3 with its interaction partners remains to be determined to better understand how these different protein complex functions on chromatin. TDRD3 recruits TOP3B and DHX9 through direct protein-protein interactions: the OB-fold binds TOP3B and the Tudor domain binds DHX9. Because the Tudor domain is the ‘reader’ module that is primarily responsible for the chromatin localization of TDRD3, we speculate that the Tudor domain may be critical for the initial promoter recruitment of TDRD3, whereas other domains, such as the N-terminal OB-fold and the UBA domain (Figure 2A), may provide multivalent interactions that either synergistically or sequentially contribute to the chromatin engagement of TDRD3. In that case, the Tudor domain remains free to recruit other arginine-methylated proteins, such as DHX9, to the promoters.
Context-dependent function of DHX9 in R-loop metabolism
DHX9 is a multifunctional helicase that plays a central role in many cellular processes, including the regulation of transcription, DNA replication, and maintenance of genome stability (27,48,49). Although its activity in resolving R-loops has been characterized in vitro using biochemical assays, its functions in R-loop metabolism in vivo are likely context-dependent, considering the distinct genetic and epigenetic composition of promoters, gene bodies, and termination sites. At gene promoters, DHX9 has been characterized as a transcriptional co-activator that bridges the acetyltransferase CBP to RNAPII through direct protein-protein interactions (65). Importantly, the helicase activity of DHX9 has been reported to be critical for its function in transcription activation (66), but the natural DNA substrate for its helicase activity at promoter regions has not been determined. Results from our study suggest that promoter-associated R-loops are likely targets of DHX9 helicase activity. Although the interaction between DHX9 and TDRD3 is specific and direct (Figures 1 and 2), it is possible that only a portion of total cellular DHX9 is associated with TDRD3 and TOP3B at gene promoters. Due to potential technical difficulties, the extent to which TDRD3 contributes to the genome-wide distribution of DHX9 has yet to be determined by ChIP-seq analysis. However, the gene expression comparison supports an overall functional correlation between TDRD3 and DHX9 in transcription regulation (Supplementary Figure S2E). A recent study reported that DHX9 is involved in R-loop resolution at the transcription termination sites of β-actin and γ-actin genes (28). The formation of R-loops over G-rich pause sites downstream of poly(A) signals is an important feature of RNAPII pause-dependent transcriptional termination for a group of mammalian genes (8,22). The human helicase Senataxin has been found to resolve these R-loops to promote RNAPII release and transcription termination (29). It is of interest to determine whether Senataxin and DHX9 function redundantly or cooperatively in R-loop resolution at the transcription termination sites. Additionally, Chakraborty et al. reported that DHX9 depletion suppressed R-loop accumulation induced by the loss of the splicing factors SF3B3 and SFPQ (72), indicating that DHX9 may promote R-loop formation under certain circumstances. However, it should be noted that the level of R-loops is closely related to the activity of transcription (73,74). Thus, by altering overall transcriptional output, DHX9 can, in effect, promote R-loop formation indirectly. This may explain the global attenuation of R-loops in the absence of DHX9, despite its biochemical properties towards R-loop resolution (28,72).
Whether the TDRD3-regulated DHX9 function could contribute to R-loop metabolism at the transcription termination sites of the gene or under the splicing deficient conditions are intriguing questions that deserve further investigation. TDRD3 harbors a functional UBA domain (Supplementary Figure S1C), but its endogenous protein target has not been identified. It is possible that the UBA domain could contribute to the context-dependent R-loop regulation either by modulating DHX9 activity or by targeting the TDRD3–DHX9 protein complex to genomic regions enriched for ubiquitin signals, such as histone H2B ubiquitination, which has been linked to transcription elongation (75,76), RNA processing (77) and R-loop suppression (78).
TDRD3 binds ssDNA and R-loops
In addition to directing DHX9 to specific gene promoters, TDRD3 can also promote DHX9 helicase activity to resolve R-loops (Figure 5), further demonstrating the functional interaction between these two proteins. The N-terminal OB-fold of TDRD3, which is necessary and sufficient to bind ssDNA (Figure 5C and D), is required for stimulating the helicase activity of DHX9 (Figure 5F). Although the biochemical mechanisms underlying this stimulation are still under investigation, we propose to test a working model in which the OB-fold binds the displaced ssDNA within the R-loop structure and tethers it to DHX9, thereby increasing the affinity and processivity of DHX9 on the DNA/RNA hybrid substrate (Figure 6G). This model is supported by at least two pieces of evidence: First, TDRD3 binds ssDNA and R-loops, but not DNA/RNA hybrids (Figure 5E). The ssDNA-binding activity of TDRD3 was reported to stabilize the TOP3B-DNA complex and shift the DNA relaxation reaction from a distributive to processive mode, thus stimulating TOP3B topoisomerase activity (47). Second, the ssDNA-binding protein RPA can enhance the association of RNase H1 with DNA/RNA hybrids and stimulates its activity on R-loops (53), suggesting that enhancing R-loop engagement by the ssDNA-binding proteins could be a potential mechanism to stimulate R-loop processing. However, the ability of TDRD3 to regulate DHX9 helicase activity cannot simply be attributed to the binding of TDRD3 to ssDNA, because this regulatory activity is lacking in methylarginine binding-deficient (E691K) TDRD3, in which the DHX9 interaction is abolished (Figure 5F), demonstrating that both the physical protein-protein interaction and ssDNA-binding are required to stimulate DHX9 activity.
Identification of TDRD3 as an ssDNA and R-loop binding protein has another layer of significance in terms of understanding the molecular mechanisms by which TDRD3 associates with chromatin. Although the methylarginine signals, either from histones or non-histone proteins, seem essential for TDRD3 chromatin association (Supplementary Figure S3D), the mutation that disrupts the interactions of the Tudor domain with methylarginine substrates only partially abolishes its chromatin localization (25), indicating that other anchoring mechanisms, such as the OB-ssDNA interaction, could contribute, at least in part, to the stable association. Furthermore, this ssDNA-binding ability expands the functions of TDRD3 as a multivalent scaffold for both proteins and nucleic acids and suggests its potential functions in other DNA-templated processes.
DATA AVAILABILITY
RNA-seq data generated in this study are available at NCBI GEO database with the accession number GSE178905.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all members of the Yang laboratory for their help and constructive discussion. We also thank Kerin K. Higa for providing helpful comments on scientific writing.
Author contributions: Y.Y., W.Y., and Q.H. conceived the project and designed the experiments. W.Y. and Q.H. performed most of the experiments with help from Z.W., L.S. and Y.Y. H.C., and X.W. performed RNA-seq analysis. Y.Y., W.Y. and Q.H. wrote the manuscript with inputs from other co-authors.
Contributor Information
Wei Yuan, Department of Cancer Genetics and Epigenetics, Beckman Research Institute, City of Hope National Cancer Center, Duarte, CA 91010, USA.
Qais Al-Hadid, Department of Cancer Genetics and Epigenetics, Beckman Research Institute, City of Hope National Cancer Center, Duarte, CA 91010, USA.
Zhihao Wang, Department of Cancer Genetics and Epigenetics, Beckman Research Institute, City of Hope National Cancer Center, Duarte, CA 91010, USA.
Lei Shen, Department of Cancer Genetics and Epigenetics, Beckman Research Institute, City of Hope National Cancer Center, Duarte, CA 91010, USA.
Hyejin Cho, Department of Molecular and Cellular Biology, Beckman Research Institute, City of Hope National Cancer Center, Duarte, CA 91010, USA.
Xiwei Wu, Department of Molecular and Cellular Biology, Beckman Research Institute, City of Hope National Cancer Center, Duarte, CA 91010, USA.
Yanzhong Yang, Department of Cancer Genetics and Epigenetics, Beckman Research Institute, City of Hope National Cancer Center, Duarte, CA 91010, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
CONquer canCER Now Award from the Concern Foundation (to Y.Y.);V Scholar Award from the V Foundation for Cancer Research (to Y.Y.); Y.Y. is supported by an R01 grant from the National Institutes of Health [GM133850]; Q.H. is a postdoctoral fellow supported by the DNA Damage Response and Oncogenic Signaling (DNADRS) T32 training program funded by the National Cancer Institute [T32CA186895]; Integrative Genomics Core at City of Hope is supported by the National Cancer Institute [P30CA33572]. Funding for open access charge: City of Hope Start-up Fund.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lee T.I., Young R.A.. Transcriptional regulation and its misregulation in disease. Cell. 2013; 152:1237–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soutourina J.Transcription regulation by the Mediator complex. Nat. Rev. Mol. Cell Biol. 2018; 19:262–274. [DOI] [PubMed] [Google Scholar]
- 3.Duquette M.L., Handa P., Vincent J.A., Taylor A.F., Maizels N.. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004; 18:1618–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belotserkovskii B.P., De Silva E., Tornaletti S., Wang G., Vasquez K.M., Hanawalt P.C.. A triplex-forming sequence from the human c-MYC promoter interferes with DNA transcription. J. Biol. Chem. 2007; 282:32433–32441. [DOI] [PubMed] [Google Scholar]
- 5.Tous C., Aguilera A.. Impairment of transcription elongation by R-loops in vitro. Biochem. Biophys. Res. Commun. 2007; 360:428–432. [DOI] [PubMed] [Google Scholar]
- 6.Huertas P., Aguilera A.. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell. 2003; 12:711–721. [DOI] [PubMed] [Google Scholar]
- 7.Sollier J., Cimprich K.A.. Breaking bad: R-loops and genome integrity. Trends Cell Biol. 2015; 25:514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aguilera A., Garcia-Muse T.. R loops: from transcription byproducts to threats to genome stability. Mol. Cell. 2012; 46:115–124. [DOI] [PubMed] [Google Scholar]
- 9.Santos-Pereira J.M., Aguilera A.. R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 2015; 16:583–597. [DOI] [PubMed] [Google Scholar]
- 10.Groh M., Gromak N.. Out of balance: R-loops in human disease. PLos Genet. 2014; 10:e1004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L., Chen J.Y., Zhang X., Gu Y., Xiao R., Shao C., Tang P., Qian H., Luo D., Li H.et al.. R-ChIP using inactive RNase H reveals dynamic coupling of R-loops with transcriptional pausing at gene promoters. Mol. Cell. 2017; 68:745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El Hage A., Webb S., Kerr A., Tollervey D. Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLos Genet. 2014; 10:e1004716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ginno P.A., Lott P.L., Christensen H.C., Korf I., Chedin F.. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell. 2012; 45:814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dumelie J.G., Jaffrey S.R.. Defining the location of promoter-associated R-loops at near-nucleotide resolution using bisDRIP-seq. eLife. 2017; 6:e28306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanz L.A., Hartono S.R., Lim Y.W., Steyaert S., Rajpurkar A., Ginno P.A., Xu X., Chedin F.. Prevalent, dynamic, and conserved R-loop structures associate with specific epigenomic signatures in mammals. Mol. Cell. 2016; 63:167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen P.B., Chen H.V., Acharya D., Rando O.J., Fazzio T.G.. R loops regulate promoter-proximal chromatin architecture and cellular differentiation. Nat. Struct. Mol. Biol. 2015; 22:999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grunseich C., Wang I.X., Watts J.A., Burdick J.T., Guber R.D., Zhu Z., Bruzel A., Lanman T., Chen K., Schindler A.B.et al.. Senataxin mutation reveals how R-loops promote transcription by blocking DNA methylation at gene promoters. Mol. Cell. 2018; 69:426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loomis E.W., Sanz L.A., Chedin F., Hagerman P.J.. Transcription-associated R-loop formation across the human FMR1 CGG-repeat region. PLoS Genet. 2014; 10:e1004294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakama M., Kawakami K., Kajitani T., Urano T., Murakami Y.. DNA-RNA hybrid formation mediates RNAi-directed heterochromatin formation. Genes to cells : devoted to molecular & cellular mechanisms. 2012; 17:218–233. [DOI] [PubMed] [Google Scholar]
- 20.Beratan D.N., Liu C., Migliore A., Polizzi N.F., Skourtis S.S., Zhang P., Zhang Y.. Charge transfer in dynamical biosystems, or the treachery of (static) images. Acc. Chem. Res. 2015; 48:474–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aguilera A.The connection between transcription and genomic instability. EMBO J. 2002; 21:195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skourti-Stathaki K., Proudfoot N.J.. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014; 28:1384–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El Hage A., French S.L., Beyer A.L, Tollervey D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010; 24:1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuduri S., Crabbe L., Conti C., Tourriere H., Holtgreve-Grez H., Jauch A., Pantesco V., De Vos J., Thomas A., Theillet C.et al.. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 2009; 11:1315–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y., McBride K.M., Hensley S., Lu Y., Chedin F., Bedford M.T.. Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol. Cell. 2014; 53:484–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang T., Wallis M., Petrovic V., Challis J., Kalitsis P., Hudson D.F.. Loss of TOP3B leads to increased R-loop formation and genome instability. Open Biol. 2019; 9:190222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakraborty P., Grosse F.. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair (Amst.). 2011; 10:654–665. [DOI] [PubMed] [Google Scholar]
- 28.Cristini A., Groh M., Kristiansen M.S., Gromak N.. RNA/DNA hybrid interactome identifies DXH9 as a molecular player in transcriptional termination and R-loop associated DNA damage. Cell Rep. 2018; 23:1891–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skourti-Stathaki K., Proudfoot N.J., Gromak N.. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell. 2011; 42:794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mersaoui S.Y., Yu Z., Coulombe Y., Karam M., Busatto F.F., Masson J.Y., Richard S.. Arginine methylation of the DDX5 helicase RGG/RG motif by PRMT5 regulates resolution of RNA:DNA hybrids. EMBO J. 2019; 38:e100986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song C., Hotz-Wagenblatt A., Voit R., Grummt I.. SIRT7 and the DEAD-box helicase DDX21 cooperate to resolve genomic R loops and safeguard genome stability. Genes Dev. 2017; 31:1370–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hodroj D., Recolin B., Serhal K., Martinez S., Tsanov N., Abou Merhi R., Maiorano D. An ATR-dependent function for the Ddx19 RNA helicase in nuclear R-loop metabolism. EMBO J. 2017; 36:1182–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lockhart A., Pires V.B., Bento F., Kellner V., Luke-Glaser S., Yakoub G., Ulrich H.D., Luke B.. RNase H1 and H2 are differentially regulated to process RNA-DNA hybrids. Cell Rep. 2019; 29:2890–2900. [DOI] [PubMed] [Google Scholar]
- 34.Amon J.D., Koshland D. RNase H enables efficient repair of R-loop induced DNA damage. eLife. 2016; 5:e20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao H., Zhu M., Limbo O., Russell P.. RNase H eliminates R-loops that disrupt DNA replication but is nonessential for efficient DSB repair. EMBO Rep. 2018; 19:e45335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen H.D., Leong W.Y., Li W., Reddy P.N.G., Sullivan J.D., Walter M.J., Zou L., Graubert T.A.. Spliceosome mutations induce R loop-associated sensitivity to ATR inhibition in myelodysplastic syndromes. Cancer Res. 2018; 78:5363–5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen L., Chen J.Y., Huang Y.J., Gu Y., Qiu J., Qian H., Shao C., Zhang X., Hu J., Li H.et al.. The augmented R-loop is a unifying mechanism for myelodysplastic syndromes induced by high-risk splicing factor mutations. Mol. Cell. 2018; 69:412–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jenuwein T., Allis C.D.. Translating the histone code. Science. 2001; 293:1074–1080. [DOI] [PubMed] [Google Scholar]
- 39.Taverna S.D., Li H., Ruthenburg A.J., Allis C.D., Patel D.J.. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 2007; 14:1025–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yun M., Wu J., Workman J.L., Li B.. Readers of histone modifications. Cell Res. 2011; 21:564–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sikorsky T., Hobor F., Krizanova E., Pasulka J., Kubicek K., Stefl R.. Recognition of asymmetrically dimethylated arginine by TDRD3. Nucleic Acids Res. 2012; 40:11748–11755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y., Lu Y., Espejo A., Wu J., Xu W., Liang S., Bedford M.T.. TDRD3 is an effector molecule for arginine-methylated histone marks. Mol. Cell. 2010; 40:1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sims R.J. 3rd, Rojas L.A., Beck D., Bonasio R., Schuller R., Drury W.J. 3rd, Eick D., Reinberg D. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science. 2011; 332:99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goto-Ito S., Yamagata A., Takahashi T.S., Sato Y., Fukai S.. Structural basis of the interaction between Topoisomerase IIIbeta and the TDRD3 auxiliary factor. Sci. Rep. 2017; 7:42123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu D., Shen W., Guo R., Xue Y., Peng W., Sima J., Yang J., Sharov A., Srikantan S., Yang J.et al.. Top3beta is an RNA topoisomerase that works with fragile X syndrome protein to promote synapse formation. Nat. Neurosci. 2013; 16:1238–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stoll G., Pietilainen O.P.H., Linder B., Suvisaari J., Brosi C., Hennah W., Leppa V., Torniainen M., Ripatti S., Ala-Mello S.et al.. Deletion of TOP3beta, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat. Neurosci. 2013; 16:1228–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siaw G.E., Liu I.F., Lin P.Y., Been M.D., Hsieh T.S.. DNA and RNA topoisomerase activities of Top3beta are promoted by mediator protein Tudor domain-containing protein 3. PNAS. 2016; 113:E5544–E5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jain A., Bacolla A., Del Mundo I.M., Zhao J., Wang G., Vasquez K.M.. DHX9 helicase is involved in preventing genomic instability induced by alternatively structured DNA in human cells. Nucleic Acids Res. 2013; 41:10345–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee T., Pelletier J.. The biology of DHX9 and its potential as a therapeutic target. Oncotarget. 2016; 7:42716–42739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang L., Wang Z., Narayanan N., Yang Y.. Arginine methylation of the C-terminus RGG motif promotes TOP3B topoisomerase activity and stress granule localization. Nucleic Acids Res. 2018; 46:3061–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Percipalle P., Obrdlik A.. Analysis of nascent RNA transcripts by chromatin RNA immunoprecipitation. Methods Mol. Biol. 2009; 567:215–235. [DOI] [PubMed] [Google Scholar]
- 52.Lee C.G., Hurwitz J.. A new RNA helicase isolated from HeLa cells that catalytically translocates in the 3′ to 5′ direction. J. Biol. Chem. 1992; 267:4398–4407. [PubMed] [Google Scholar]
- 53.Nguyen H.D., Yadav T., Giri S., Saez B., Graubert T.A., Zou L.. Functions of replication protein A as a sensor of R loops and a regulator of RNaseH1. Mol. Cell. 2017; 65:832–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi L., Magee P., Fassan M., Sahoo S., Leong H.S., Lee D., Sellers R., Brulle-Soumare L., Cairo S., Monteverde T.et al.. A KRAS-responsive long non-coding RNA controls microRNA processing. Nat. Commun. 2021; 12:2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A.. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013; 14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A.et al.. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016; 44:W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie Z., Bailey A., Kuleshov M.V., Clarke D.J.B., Evangelista J.E., Jenkins S.L., Lachmann A., Wojciechowicz M.L., Kropiwnicki E., Jagodnik K.M.et al.. Gene set knowledge discovery with enrichr. Curr Protoc. 2021; 1:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weidensdorfer D., Stohr N., Baude A., Lederer M., Kohn M., Schierhorn A., Buchmeier S., Wahle E., Huttelmaier S.. Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. RNA. 2009; 15:104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Linder B., Plottner O., Kroiss M., Hartmann E., Laggerbauer B., Meister G., Keidel E., Fischer U.. Tdrd3 is a novel stress granule-associated protein interacting with the Fragile-X syndrome protein FMRP. Hum. Mol. Genet. 2008; 17:3236–3246. [DOI] [PubMed] [Google Scholar]
- 60.Smith W.A., Schurter B.T., Wong-Staal F., David M.. Arginine methylation of RNA helicase a determines its subcellular localization. J. Biol. Chem. 2004; 279:22795–22798. [DOI] [PubMed] [Google Scholar]
- 61.Eram M.S., Shen Y., Szewczyk M., Wu H., Senisterra G., Li F., Butler K.V., Kaniskan H.U., Speed B.A., Dela Sena C.et al.. A potent, selective, and cell-active inhibitor of human type I protein arginine methyltransferases. ACS Chem. Biol. 2016; 11:772–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Narayanan N., Wang Z., Li L., Yang Y.. Arginine methylation of USP9X promotes its interaction with TDRD3 and its anti-apoptotic activities in breast cancer cells. Cell Discov. 2017; 3:16048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morettin A., Paris G., Bouzid Y., Baldwin R.M., Falls T.J., Bell J.C., Cote J.. Tudor domain containing protein 3 promotes tumorigenesis and invasive capacity of breast cancer cells. Sci. Rep. 2017; 7:5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee T., Di Paola D., Malina A., Mills J.R., Kreps A., Grosse F., Tang H., Zannis-Hadjopoulos M., Larsson O., Pelletier J.. Suppression of the DHX9 helicase induces premature senescence in human diploid fibroblasts in a p53-dependent manner. J. Biol. Chem. 2014; 289:22798–22814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakajima T., Uchida C., Anderson S.F., Lee C.G., Hurwitz J., Parvin J.D., Montminy M.. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997; 90:1107–1112. [DOI] [PubMed] [Google Scholar]
- 66.Aratani S., Fujii R., Oishi T., Fujita H., Amano T., Ohshima T., Hagiwara M., Fukamizu A., Nakajima T.. Dual roles of RNA helicase A in CREB-dependent transcription. Mol. Cell. Biol. 2001; 21:4460–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shivji M.K.K., Renaudin X., Williams C.H., Venkitaraman A.R.. BRCA2 regulates transcription elongation by RNA polymerase II to prevent R-loop accumulation. Cell Rep. 2018; 22:1031–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zatreanu D., Han Z., Mitter R., Tumini E., Williams H., Gregersen L., Dirac-Svejstrup A.B., Roma S., Stewart A., Aguilera A.et al.. Elongation factor TFIIS prevents transcription stress and R-loop accumulation to maintain genome stability. Mol. Cell. 2019; 76:57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang S., Grosse F.. Domain structure of human nuclear DNA helicase II (RNA helicase A). J. Biol. Chem. 1997; 272:11487–11494. [DOI] [PubMed] [Google Scholar]
- 70.Xing L., Zhao X., Niu M., Kleiman L.. Helicase associated 2 domain is essential for helicase activity of RNA helicase A. Biochim. Biophys. Acta. 2014; 1844:1757–1764. [DOI] [PubMed] [Google Scholar]
- 71.Zhang S., Grosse F.. Nuclear DNA helicase II unwinds both DNA and RNA. Biochemistry. 1994; 33:3906–3912. [DOI] [PubMed] [Google Scholar]
- 72.Chakraborty P., Huang J.T.J., Hiom K.. DHX9 helicase promotes R-loop formation in cells with impaired RNA splicing. Nat. Commun. 2018; 9:4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stork C.T., Bocek M., Crossley M.P., Sollier J., Sanz L.A., Chedin F., Swigut T., Cimprich K.A.. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. eLife. 2016; 5:e17548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Crossley M.P., Bocek M., Cimprich K.A.. R-loops as cellular regulators and genomic threats. Mol. Cell. 2019; 73:398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tanny J.C., Erdjument-Bromage H., Tempst P., Allis C.D.. Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes Dev. 2007; 21:835–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu L., Li L., Zhou B., Qin Z., Dou Y.. H2B ubiquitylation promotes RNA Pol II processivity via PAF1 and pTEFb. Mol. Cell. 2014; 54:920–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Woloszynska M., Le Gall S., Neyt P., Boccardi T.M., Grasser M., Langst G., Aesaert S., Coussens G., Dhondt S., Van De Slijke E.et al.. Histone 2B monoubiquitination complex integrates transcript elongation with RNA processing at circadian clock and flowering regulators. PNAS. 2019; 116:8060–8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chernikova S.B., Razorenova O.V., Higgins J.P., Sishc B.J., Nicolau M., Dorth J.A., Chernikova D.A., Kwok S., Brooks J.D., Bailey S.M.et al.. Deficiency in mammalian histone H2B ubiquitin ligase Bre1 (Rnf20/Rnf40) leads to replication stress and chromosomal instability. Cancer Res. 2012; 72:2111–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data generated in this study are available at NCBI GEO database with the accession number GSE178905.