Abstract
Poly(ADP-ribosyl)ation (PARylation) is a multifaceted post-translational modification, carried out by poly(ADP-ribosyl)transferases (poly-ARTs, PARPs), which play essential roles in (patho-) physiology, as well as cancer therapy. Using NAD+ as a substrate, acceptors, such as proteins and nucleic acids, can be modified with either single ADP-ribose units or polymers, varying considerably in length and branching. Recently, the importance of PAR structural heterogeneity with regards to chain length and branching came into focus. Here, we provide a concise overview on the current knowledge of the biochemical and physiological significance of such differently structured PAR. There is increasing evidence revealing that PAR’s structural diversity influences the binding characteristics of its readers, PAR catabolism, and the dynamics of biomolecular condensates. Thereby, it shapes various cellular processes, such as DNA damage response and cell cycle regulation. Contrary to the knowledge on the consequences of PAR’s structural diversity, insight into its determinants is just emerging, pointing to specific roles of different PARP members and accessory factors. In the future, it will be interesting to study the interplay with other post-translational modifications, the contribution of natural PARP variants, and the regulatory role of accessory molecules. This has the exciting potential for new therapeutic approaches, with the targeted modulation and tuning of PARPs’ enzymatic functions, rather than their complete inhibition, as a central premise.
Graphical Abstract
Graphical Abstract.
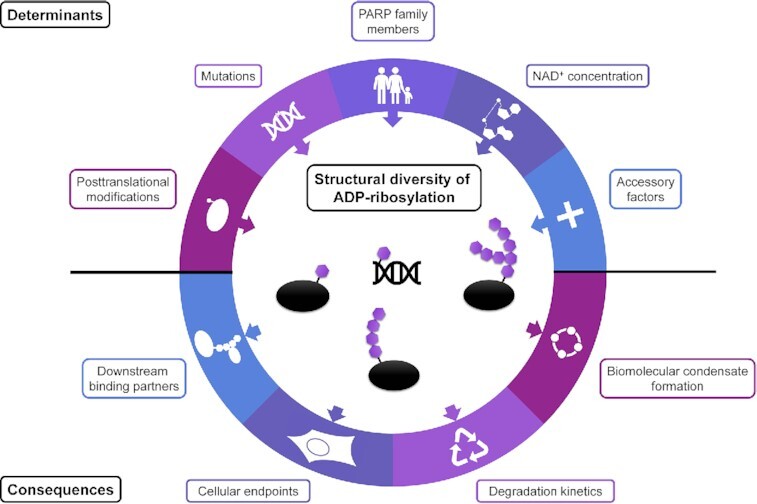
Determinants and consequences of the structural diversity of ADP-ribosylation and poly(ADP-ribosyl)ation.
INTRODUCTION
The genome provides a basic blueprint for the synthesis of proteins that are needed for cellular life. Yet, what adds tremendous further complexity to proteomes in terms of spatio-temporal regulation of dynamics, function, and crosstalk are post-translational modifications (1). Poly(ADP-ribosyl)ation (PARylation) is a complex and essential post-translational modification in most eukaryotes that is catalysed by certain enzymes of the family of ADP-ribosyltransferases diphtheria toxin-like (ARTDs), aka PARPs and tankyrases (TNKSs) [Lüscher et al., manuscript submitted]. So far, PARP1, PARP2, TNKS1 and TNKS2 have unambiguously been identified to synthesise poly(ADP-ribose) (PAR), i.e. representing poly-ARTs, while other members of the ARTD family act as mono-ADP-ribosyltransferases (mono-ARTs) or are catalytically inactive (2,3) [Lüscher et al., manuscript submitted]. PARylation plays crucial roles in diverse cellular processes, including DNA damage response and genome maintenance (4–7), chromatin regulation and telomere biology (8–12), replication and cell cycle control (13,14), transcription and RNA metabolism (15), inflammation and immunity (16–19) and cell death (20–23). Due to their versatile cellular functions, PARPs can be involved in several pathophysiological processes, the most prominent ones being carcinogenesis (24,25), ischemia reperfusion damage, inflammatory and neurodegenerative diseases (16,22,26), as well as metabolic diseases (16,27). Consequently, inhibitors of PARylation have been studied extensively with regards to their therapeutic benefit and by now, four different ones, i.e. olaparib, rucaparib, niraparib, and talazoparib, have been approved for the treatment of certain types of ovarian, breast, prostate and pancreatic cancer (28–33). In general, the rationale for the use of PARP inhibitors in cancer therapy follows the concept of synthetic lethality in cancers deficient in homologous recombination repair, as well as their use as chemosensitisers in combination with classical chemotherapeutics (34,35). Furthermore, PARP inhibition may open up new possibilities for the treatment of other types of cancer, like small-cell lung cancer (36), but also inflammatory diseases and ischemia reperfusion injury (16,37–39), as well as neurodegenerative diseases like Parkinson's disease (22,26,40). Recently, the importance and influence of PAR structural heterogeneity with regards to chain length and branching came into focus. Here, we review the biochemical nature of such differently structured PAR molecules, provide a concise overview on the current knowledge on its biochemical and physiological significance, and give an outlook on open questions to be addressed in the future.
Biochemistry of PARylation
PARPs use nicotinamide adenine dinucleotide (NAD+) as a substrate to covalently modify acceptor proteins, leading to the formation of linear or branched polymers varying in branching frequency (41) and extending to up to 200 ADP-ribose units in length (Figure 1) (42). The ADP-ribosyltransferase domain as a catalytic centre is highly conserved in all members of the ARTD family and consists of a donor site for NAD+ binding, as well as an acceptor site for binding of a target molecule or the distal moiety of an extending ADP-ribose chain (2,43). The nicotinamide binding pocket within the donor site is characterised by a conserved H–Y–E triad, consisting of a histidine, tyrosine and glutamate residue, respectively. While His862 and Tyr896 of PARP1 are involved in NAD+ binding, Glu988 is also essential for catalysing the elongation reaction, as natural replacement of this amino acid leads to the loss of PARylation activity, as seen in mono-ARTs (44–46). Interestingly, there is evidence suggesting a vivid interplay between mono-ADP-ribosylation (MARylation) and PARylation, as certain ARTs may MARylate and PARylate substrates, which then serve as an initiation and elongation point for further PARylation by other poly-ARTs, thereby potentially extending the substrate range and leading to PAR modifications synthesised by the collaborative action of different PARPs and other ARTs as well as certain sirtuins (47–49). Target proteins can be ADP-ribosylated at aspartate, arginine, glutamate, lysine, serine, or tyrosine residues (50–54), however, serine residues have recently emerged as essential acceptor sites for PARylation within the DNA damage response and this specific modification has shown to be mainly dependent on the presence of histone PARylation factor 1 (HPF1) as an accessory factor (55–57). By binding of HPF1 to PARP1 or PARP2 through a negatively charged region, a composite active site is formed by both proteins, remodelling the original active site, and thereby contributing HPF1 Glu284 as a key catalytic residue. Besides directing ADP-ribosylation towards serine modification, the interaction with HPF1 also substantially reduces PARP1 automodification, possibly through steric hindrance, thereby providing an additional level of regulation (58–60). After attachment of the first ADP-ribose moiety, subsequent PAR formation can be subdivided into linear elongation and the introduction of branching points. While elongation of the PAR chain is conveyed via (2′-1’’) ribose-ribose glycosidic bonds between ADP-ribose units, the branching reaction is characterised by the formation of (2’’-1’’’) ribose-ribose glycosidic bonds (61,62). It has been proposed that branching, rather than elongation, takes place, when the orientation of PAR bound by the PARP catalytic centre is rotated 180°, respectively (44). The tyrosine residue Y986 within PARP1 might play an essential role in this process, as an amino acid exchange to histidine (Y986H) enhances the affinity of the PAR binding site to the pyrophosphate residue, thereby weakening adenine binding, in turn enhancing the symmetry of the acceptor site and ultimately tipping the scales towards the branching reaction (44,63).
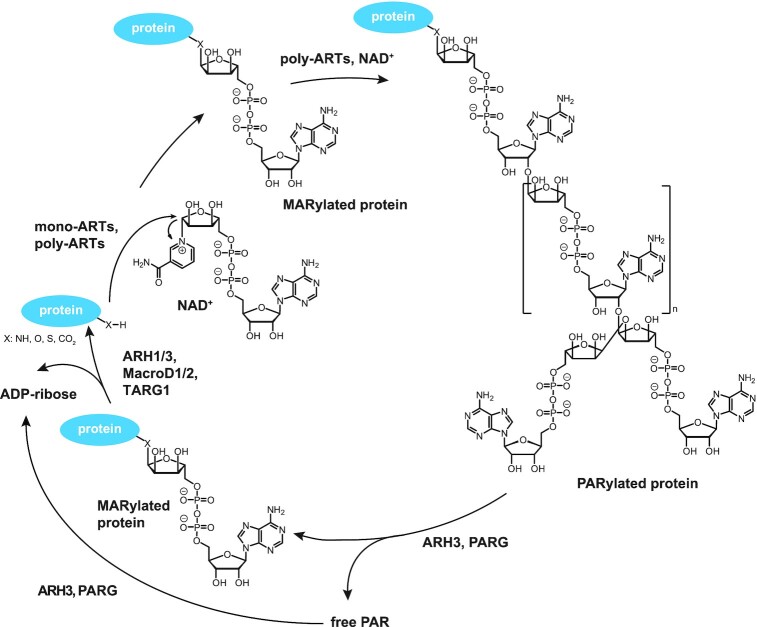
Figure 1.
The poly(ADP-ribosyl)ation cycle. Substrate proteins are ADP-ribosylated using NAD+ as substrate, resulting in MARylated or PARylated proteins. Degradation and removal of ADP-ribose units is carried out by hydrolases. ARH, ADP-ribose hydrolase; MARylated protein, mono(ADP-ribos)ylated protein; mono-ART, mono-ADP-ribosyltransferase; NAD+, nicotinamide adenine dinucleotide; PAR, poly(ADP-ribose); PARG, poly(ADP-ribose) glycohydrolase; poly-ART, poly-ADP-ribosyltransferase; PARylated protein, poly(ADP-ribosyl)ated protein. Figure modified with courtesy of Maike Lehner (University of Konstanz).
Overall, PARylation is a tightly regulated and highly dynamic transient modification, as PAR synthesis is counteracted by its hydrolysis by various catabolising enzymes like PAR glycohydrolase (PARG) and others (see below). While PARG is considered the main cellular PAR degrading enzyme and efficiently hydrolyses ribose-ribose bonds, it is unable to remove the terminal ADP-ribose unit from acceptor sites (64,65). The removal of mono-ADP-ribose from proteins has been identified as the rate-limiting step in PAR hydrolysis (66) and can therefore also occur as a persistent intermediate during PAR catabolism, in addition to being catalysed by mono-ARTs, e.g., PARP3 and PARP10 (47,67–69). Complete reversal of the modification is, in turn, mediated by several amino acid-specific hydrolases, like the macrodomain-containing proteins MacroD1/D2, terminal ADP-ribose glycohydrolase 1 (TARG1), and members of the ADP-ribose hydrolase (ARH) family (70–72). While MacroD1/D2 and TARG1 both have been shown to cleave the ester bond of modified aspartate and glutamate residues, MacroD1/D2 are only able to remove mono-ADP-ribose, while TARG1 can facilitate the removal of whole PAR chains (73). As for the ARH family members, ARH1 is only able to reverse mono-ADP-ribosylation and additionally displays an amino acid specificity for arginine (74), while ARH3 can hydrolyse PAR exoglycolytically and facilitate the removal of ADP-ribose, specifically from serine-modified targets (75–77). Furthermore, several phosphodiesterases like NUDT9, NUDT16 and ENPP1 have also been implicated in the degradation of PAR chains (78,79).
PARylation and non-covalent PAR binding
Besides covalent modification, proteins can also non-covalently interact with PAR chains through a diverse array of PAR reader modules, the most common one being the ‘classical’ PAR-binding motif (PBM), which can be found in over 800 human proteins (80). The motif consists of a loosely conserved sequence of ∼20 hydrophobic and basic amino acid residues (81). The positively charged amino acids within the PBM consensus most likely convey affinity to negatively charged PAR chains via electrostatic interactions, reaching KD values in the nanomolar range (82). Although it has been hypothesised that PAR chains interact with the PBM between the second phosphate of one ADP-ribosyl moiety and the first phosphate of the next (83), up to now the exact binding mechanism remains to be identified. Furthermore, it is possible that multiple PBMs within one protein potentially cooperate to bind especially long PAR chains (84). Interestingly, PAR binding regions often overlap with DNA and/or RNA binding domains. The resulting competitive binding between those molecules is likely to exert regulatory functions within the cell (85–91).
Another strongly-binding reader module are PAR-binding zinc fingers (PBZ), identified in the DNA damage response proteins APLF (aprataxin and PNK-like factor) and CHFR (checkpoint with forkhead and ring finger domains) (92), as well as an alleged variation identified in the checkpoint kinase Chk1 (93). They consist of two conserved cysteine and two histidine residues (Cys2-His2 type zinc fingers) and can bind two successive ADP-ribose units within a PAR chain (94). Contrary to CHFR, APLF possesses a tandem PBZ motif, where both zinc fingers cooperate in PAR binding reaching affinities with KD values in the low nanomolar range (48,93,95).
In addition, the considerably larger macrodomains (∼100–200 aa) act as PAR recognition modules, interacting with the terminal ADP-ribose moiety on acceptor residues reaching affinities with KD values in the micro- to nanomolar range (82,84). To date, macrodomains have been identified in certain PARP family members, histone variants, as well as the PAR degrading enzymes MacroD1/D2, TARG1, and PARG, thereby establishing a connection to PAR catabolism (43).
WWE domains, consisting of conserved tryptophan (W) and glutamate (E) residues, on the other hand, are found in several PARP family members and ubiquitin ligases (96). While their affinity for ADP-ribose is relatively low with KD values within the millimolar range, KD values for binding to iso-ADP-ribose lay within the micro- to nanomolar range (82)—however specific binding characteristics and affinities also appear to be dependent on the protein context (97).
Furthermore, forkhead-associated (FHA) and BRCA1 C-terminal (BRCT) domains have been implicated in PAR binding, most likely through electrostatic interactions of their phosphor-binding pockets with negatively charged PAR. While FHA domains have been shown to recognise iso-ADP-ribose, BRCT domains interact with ADP-ribosyl moieties (98,99).
Finally, several DNA- and RNA-binding motifs have also been identified as PAR-recognising modules, possibly due to the similarity between PAR and the nucleic acids. Amongst them are RNA recognition motifs (RRMs) as most abundant RNA-binding domains, serine/arginine (SR) repeats and lysine/arginine (KR)-rich motifs, oligonucleotide/oligosaccharide-binding (OB)-folds, PIN domains, as well as arginine/glycine-rich (RG/RGG) motifs (84,100,101).
The diversity of existing PAR binding modules, their abundance in a plethora of cellular proteins, and the wide spectrum of different binding mechanisms underscore the ubiquitous nature of PAR–protein interactions and point to highly regulated and specific modes of interaction with PAR of different structural qualities. Furthermore, it stands to reason that non-covalent PAR interaction and covalent PARylation do not function as independent mechanisms but are rather tightly connected on a functional level. First evidence for this assumption was obtained from recent studies, investigating the interplay between PARP1 and the tumour suppressor p53 or the RNA helicase DDX21, both of which are binding to PAR from activated, automodified PARP1, which in turn mediates their covalent modification (85,101,102). While the initial, non-covalent interaction of DDX21 with PAR is most likely conveyed by a C-terminal RNA-binding domain (101), PAR-binding of p53 is mediated by its intrinsically disordered C-terminal domain (CTD) containing a PBM-like motif. Interestingly, another candidate for an analogous PARylation mechanism is the multifunctional YB-1 protein, whose intrinsically disordered C-terminal domain has been demonstrated to stimulate PARP1 activity in a PAR-dependent manner (103). This is consistent with the finding that intrinsically disordered CTD-like regions are highly enriched among the PARylated proteome (85,104,105), suggesting that the observed interplay between non-covalent PAR interaction and covalent PARylation extends to several other proteins.
Viewed together, PARylation is a multifaceted post-translational modification that influences a plethora of cellular processes and is therefore inherently involved in a multitude of (patho-) physiological functions. However, as much as is known about the modification in general, as elusive the biological function and significance of its structural heterogeneity remain. In the following, we provide a collective overview of what is known about the determinants and the consequences of PAR chain length and branching (Figure 2).
Figure 2.
Determinants and consequences of ADP-ribose and poly(ADP-ribose) structural diversity. ADP-ribosylation is a multifaceted post-translational modification carried out by the family of ADP-ribosyltransferases diphtheria toxin-like (ARTDs, aka PARPs). Acceptor molecules, such as proteins, DNA, and RNA, are covalently modified with ADP-ribose units, and modifications can consist of either a single moiety (mono-ADP-ribosylation, MARylation)—mediated by mono-ARTs—or linear and/or branched chains (poly(ADP-ribosyl)ation, PARylation)—mediated by poly-ARTs. Determinants such as the activity or naturally occurring mutations of different PARP family members, the interplay with different accessory factors, other post-translational modifications, or the intracellular NAD+ concentration might determine the structure of the ADP-ribose modification. Consequently, the different structure of the modification can influence its degradation, downstream binding partners, the formation of biomolecular condensates, and thereby diverse cellular endpoints.
DETERMINANTS OF PAR STRUCTURAL HETEROGENEITY
Looking into a possible significance of PAR branching in vivo, we analysed PAR levels and quality in a comprehensive spectrum of 12 different mouse organs via MS, revealing a strong tissue-dependent effect, where high PAR levels did not correlate with high levels of branching (106). This finding raises the question about specific determinants of PAR structure, which influence PAR’s structural heterogeneity in physiological settings, e.g. in different cell types or under specific (patho-) physiological conditions (Figure 2).
It has long since been known that some members of the PARP family only catalyse mono- or oligo-ADP-ribosylation (46), contributing evidence that different PARP family members are able to direct PAR structure. This is supported by studies reporting that in case of PARP1 branching occurs about once every 20–50 ADP-ribose units (41,42), whereas no branching points have been detected in case of TNKS1 (62). Furthermore, in a recent study by Chen et al., the comparison of wildtype and Parp2–/– mouse embryonic fibroblasts and U2OS cells via UV spectroscopy and liquid chromatography tandem mass spectrometry (LC–MS/MS) revealed only a slight reduction in overall PAR formation in Parp2–/– cells, while R2-Ado levels, which are indicative of branched PAR, were significantly reduced. The authors concluded that PARP2 preferentially synthesises branched PAR upon being activated by binding via its N-terminal region to pre-existing PAR molecules, e.g. synthesised by PARP1. A detailed analysis of available expression data of PARPs in different tissues and cell types may shed some light onto the question, whether different PARP family members are responsible for the PAR branching variations we have observed across different mouse organs. In addition to the inherent activity of different PARP family members, naturally occurring mutations and polymorphisms might also be a determining factor with regards to PAR structure. In a recent study, we showed that differently structured PAR, as produced by different (non-natural) PARP1 variants, influenced cellular outcomes (106) (see below).
Furthermore, accessory factors can influence the quality of the formed PAR polymers. In this regard, HPF1 has been shown to not only switch the amino acid specificity for PARylation towards serine modification, but at the same time also influence PAR structure. The detection of PAR chain length via enzymatic labelling of terminal ADP-ribose (ELTA) revealed a reduction of both the amount and the length of PAR in the presence of HPF1 (107). In accordance, using novel HPLC assays quantifying ADP-ribose, NAD+ and nicotinamide, Rudolph et al. could recently show that the presence of HPF1 leads to the release of ADP-ribose, resulting in distinctly shorter PAR chains (60). Additionally, activation of PARP1 by the Y-box-binding protein 1 (YB-1) has been shown to increase total protein PARylation but at the same time decrease the length of PAR (108). It stands to reason that there are other, so far unidentified, accessory factors that influence PARylation in a similar manner.
Besides accessory factors, post-translational modifications of poly-ARTs could also influence PAR formation by allosteric mechanisms or by directly affecting catalytic domains of PARPs. Various modification sites, including phosphorylation sites within the catalytic domain, have already been identified for PARP1 and have shown to affect its activity (109–112). It is therefore reasonable to assume that some modifications might also influence PAR structure.
And finally, PARPs are known to affect the intracellular NAD+ metabolism through consumption of NAD+ (113), but an inverted relationship, where cellular NAD+ levels influence the formation of PAR with regards to chain length and branching, is certainly also feasible (114).
CONSEQUENCES OF PAR HETEROGENEITY: WHY STRUCTURE AND CHAIN LENGTH MATTER
PAR structure could affect biochemical, cellular, and organismic outcomes in several ways (Figure 2). For example: (i) PAR chain length and branching can determine the number of reader molecules that bind to a single PAR chain, thereby influencing local concentrations of PAR readers and/or bridging the formation of homo and hetero-protein complexes. Also, highly branched polymer may limit the available space for PAR binding domains of readers that recognise linear parts of the polymer. (ii) Structure-specific binding of PAR readers could lead to their selective subcellular recruitment and localisation, functional regulation, as well as degradation via PARylation-mediated ubiquitination. (iii) The synthesis of structurally heterogeneous PAR can consume variable amounts of intracellular NAD+; therefore, this can significantly influence local and ubiquitous metabolic processes. (iv) Since PAR is a highly charged molecule, PAR’s structural heterogeneity is likely to affect the biophysical properties of local subcellular environments, e.g., by regulating phase separating processes during DNA repair and stress granule formation. And (v), PAR molecules of different structural quality exhibit different stabilities/degradation kinetics and can give rise to a variable spectrum of catabolic metabolites with specific consequences in downstream cellular signalling processes. Overall, PAR’s structural diversity is likely to influence the spatio-temporal regulation of a multitude of cellular factors and mechanisms.
Evidence from cellular and animal studies
Recently, we analysed the consequences of PAR heterogeneity on cellular physiology, employing PARP1 variants, first identified in a random mutagenesis screen by Rolli et al. (63), as tools to mimic different PAR chain lengths and branching frequencies in cellular environments (106). For this, HeLa PARP1 knockout cells were reconstituted with different PARP1 variants, namely (i) PARP1\Y986H, producing hyperbranched PAR; (ii) PARP1\Y986S, producing especially short, and slightly hyperbranched PAR and (iii) PARP1\G972R, producing hypobranched PAR chains. Using those PARP1-reconstituted cells as a model system, we then comprehensively analysed the variants with regards to a spectrum of cellular endpoints, from clonogenic survival, cell viability, proliferation, and cell cycle progression, to the overall resistance to genotoxic stress after treatment with hydrogen peroxide (H2O2) or the topoisomerase I inhibitor camptothecin (CPT). While hyperbranched or short PAR only slightly—and even advantageously, in the case of PARP1\Y986H—affected the cellular phenotype, hypobranched PAR synthesised by the PARP1\G972R variant on the other hand adversely affected several cellular outcomes. Analysing gene expression profiles, we also observed a significant upregulation of genes coding for cell cycle inhibitors, apoptosis-related proteins, and DNA damage repair factors in PARP1\G972R-reconstituted cells, directly in line with the observed cellular outcomes of G2 arrest, increased cell death, and sensitivity to genotoxic treatment (106). These results suggest that PAR generated by wild-type PARP1, exhibits PAR chain lengths and branching already at quite the optimal length and branching distribution with regards to their functions in cellular stress responses. However, if the balance in chain lengths and branching frequencies is shifted towards shorter and hypobranched PAR, this considerably affects cellular physiology. This hypothesis is supported by results obtained from a knock-in mouse model carrying the D993A mutation within the catalytic domain of PARP1, functionally resembling the G972R mutation. Here, primary mouse embryonic fibroblasts (MEFs) isolated from the mouse model displayed a decreased, as well as delayed PARylation reaction after treatment with H2O2, the alkylating agents N-methyl-N’-nitro-N-nitrosoguanidine (MNNG) and N-methyl-N-nitrosourea (MNU), or CPT. Additionally, PAR branching was reduced in PARP1\D993A MEFs compared to wildtype (WT) MEFs after H2O2 treatment, as determined by mass spectrometry (MS). Those results could be confirmed by an in vitro auto-PARylation assay with recombinant protein, demonstrating a decelerated PAR formation and detecting shorter and less-branched PAR chains for PARP1\D993A compared to PARP1\WT. Consistent with data obtained for PARP1\G972R, the observed hypo-PARylation in MEFs had detrimental effects on cellular and organismic levels, leading to an increased sensitivity towards alkylating agents, as well as a compromised DNA damage response during DNA replication and subsequent cell death or senescence (115). Taken together, these results strongly suggest the importance of ‘optimal’ structured PAR with regards to chain length and branching in physiological settings. A limitation of these studies (106,115) is that PARP1 mutants analysed therein produce PAR chains with alterations in both - chain lengths and branching frequencies at the same time. It has yet to be shown, if it is possible to disentangle both parameters by engineering PARP1 mutants with more specific activities regarding the formation of PAR of defined chain length and branching.
When talking about structural heterogeneity, we also need to consider mono-ADP-ribosylation (MARylation), either as an intermediate in PAR catabolism or as a distinct modification catalysed by specific mono-ARTs. In a previous study, utilising the mono-ADP-ribosylating PARP1 mutant E988K, we have been able to demonstrate the influence of PARP1-mediated MARylation on cellular physiology. Already in unchallenged conditions, PARP1\E988K-reconstituted HeLa PARP1 knockout cells displayed altered cell morphology with enlarged nuclei, increased G2 arrest, and higher rates of cell death. Additionally, cells were more sensitive to genotoxic treatment with the topoisomerase I inhibitor camptothecin (CPT) (116). Furthermore, ARH3-defective cells have been shown to accumulate mono-ADP-ribose on histones in untreated conditions, as well as in response to DNA damage, which is potentially linked to pathological neurodegenerative outcomes in mice, as well as humans (117–120). While the persisting MARylation on chromatin has been shown to be non-toxic in this case, persistent PARylation on the other hand elicited distinct physiological effects, in line with synthetic lethality in ARH3- and PARG-deficient cells (47). To date, MARylation has been implicated in a wide variety of (patho-) physiological processes, ranging from cellular signalling, transcription and DNA repair to immunity, inflammation, anti-viral defence, neurodegeneration, and cancer biology. Here, we would like to refer to comprehensive reviews, further detailing and discussing the various functions of mono-ADP-ribose (121,122).
The influence of PAR chain length on non-covalent PAR-protein binding
To establish a mechanistic connection between PAR structural heterogeneity and the observed cellular effects, it is essential to investigate non-covalent interactions of PAR with specific binding proteins. On this basis, potential downstream signalling pathways could be identified that depend on distinctly structured PAR, providing additional insight into the biological relevance. So far, several studies have been published, analysing PAR chain length-specific binding behaviour (Table 1).
Table 1.
Poly(ADP-ribose) chain length- and branching-specific binding preferences of select proteins. LC–MS/MS (liquid chromatography tandem mass spectrometry), R-Ado (ribosyl adenosine), R2-Ado (diribosyl-adenosine), EMSA (electrophoretic mobility shift assay), SPR (surface plasmon resonance), DSF (differential scanning fluorimetry), MST (microscale thermophoresis), TR-FRET (time-resolved fluorescence resonance energy transfer), KD (dissociation constant), Tm (melting temperature), EC50 (half maximal effective concentration).
| Protein | Method | PAR tested | Strength of interaction | Preferred PAR structure | Ref. |
|---|---|---|---|---|---|
| APE1 | EMSA | 6–10-mer | Weak binding | Long, linear | (132) |
| 11–20-mer | Weak binding | ||||
| >20-mer (linear) | EC50 0.77 ± 0.08 μM | ||||
| >20-mer (branched) | EC50 1.2 ± 0.1 μM | ||||
| APLF | PAR digestion and LC–MS/MS analysis | Linear (R-Ado) | Binding of ∼4% of input | Branched | (48) |
| Branched (R2-Ado) | Binding of ∼24% of input | ||||
| Polβ | EMSA | 6- to 10-mer | Weak binding | Long, linear | (132) |
| 11- to 20-mer | Weak binding | ||||
| >20-mer (linear) | EC50 0.57 ± 0.06 μM | ||||
| >20-mer (branched) | EC50 1.3 ± 0.1 μM | ||||
| DEK | EMSA | 18-mer | No binding | Long | (90,131) |
| PAR overlay assay | 54-mer | KD 6.1×10–8 ± 5.2×10–9 M (formation of 1 complex) | |||
| ≤34-mer | Very weak binding | ||||
| Photoaffinity-based proteomics | 34 to 45-mer | Intermediate binding | |||
| ≥57-mer Unfractionated | Strong binding | ||||
| 8-mer, 40-mer | Log2 40-mer/8-mer: 7.56 | ||||
| H1 | PAR overlay assay | 5–10-mer | Weak binding | Long | (90) |
| 10–15-mer | Intermediate binding | ||||
| ≥15-mer /unfractionated | Strong binding | ||||
| Histones | PAR binding assay | Short, linear Long, linear Branched |
Binding hierarchy: branched polymers > long, linear polymers > short, linear polymers | Branched, long | (129) |
| NONO | SPR | ≤30-mer | No binding | Long | (127,131) |
| ≥60-mer | KD 2.01×10–8 M | ||||
| Unfractionated | KD 2.32×10–8 M | ||||
| Photoaffinity-based proteomics | 8-mer, 40-mer | Log2 40-mer/8-mer: 0.71 | |||
| p53 | EMSA | 16-mer | KD 2.5×10–7 ± 3.8×10–8 M (Formation of 1 complex) | Long | (123) |
| 55-mer | KD 1.3×10–7 ± 4.2×10–9 M (Formation of 3 complexes) | ||||
| SPR | 14-mer | KD 3.4×10–9 ± 1.0×10–11 M | |||
| 63-mer | N/A, due to complex binding behaviour | ||||
| DSF | 13–20-mer | Tm ∼ 42.5°C | (85) | ||
| 28–35-mer | Tm ∼ 43°C | ||||
| 36–60-mer | Tm ∼ 43.5°C | ||||
| PARG | Photoaffinity-based proteomics | 8-mer, 40-mer | Log2 40-mer/8-mer: –3.07 | Short | (131) |
| PARP1 | Photoaffinity-based proteomics | 8-mer, 40-mer | Log2 40-mer/8-mer: 3.76 | Long | (131) |
| EMSA | 4-mer | KD 4.4×10–7 M | |||
| 8-mer | KD 1.8×10–7 M | ||||
| 16-mer | KD 2.7×10–8 M | ||||
| 32-mer | KD 7.9×10–8 M | ||||
| RNF146 | PAR digestion and LC-MS/MS analysis | Linear (R-Ado) Branched (R2-Ado) | Binding of ∼29% of input Binding of ∼2% of input | Linear | (48) |
| Competition assay | 40-mer (average) | EC50 14.5 ± 0.13 nM KD 1.2×10–8 M | No chain length preference | (133) | |
| WWE-domain | Filter binding assay | 10-mer | KD 7.5×10–8 M | (107) | |
| 20-mer | KD 6.7×10–8 M | ||||
| MST | 20-mer | KD 1.0×10–7 M | |||
| WRN | PAR overlay assay | ≤10-mer | Strong binding | No preference | (89) |
| ≥10-mer | Very strong binding | ||||
| XPA | EMSA | 16-mer | No binding | Long | (123) |
| 55-mer | KD 3.2×10–7 ± 7.7×10–9 M (formation of 1 complex) | ||||
| SPR | 16-mer | No binding | |||
| 63-mer | KD 6.5×10–9 ± 1.3×10–10 M | ||||
| XRCC1 | TR-FRET | Mono-ADP-ribose | EC50 1.39 ± 0.06 mM | ? | (126) |
| 2–7-mer | EC50 110 ± 13 nM | ||||
| 8–15-mer | EC50 28 ± 2 nM | ||||
| 16–23-mer | EC50 17 ± 1 nM | ||||
| 24–41-mer | EC50 22 ± 2 nM | ||||
| Recruitment to sites of laser-induced DNA damage | Cells reconstituted with PARP1/WT and PARP1\Y986S (producing short PAR) | Impaired recruitment to sites of DNA damage in cells reconstituted with PARP1\Y986S compared to cells reconstituted with PARP1/WT | (106) | ||
| Photoaffinity-based proteomics | 8-mer, 40-mer | Log2 40-mer/8-mer: -2.06 | (131) |
One important interaction partner of PAR is p53 (80), which has been extensively studied with regards to PAR binding (85,86,123–125). Electrophoretic mobility shift assays (EMSAs) and surface plasmon resonance (SPR) analyses using high performance liquid chromatography (HPLC)-fractionated PAR chains of defined lengths have revealed that short (14–16-mers), as well as long PAR chains (55–63-mers) are bound by p53. While short polymer led to the formation of only one defined complex with p53, longer PAR chains promoted the formation of three distinct complexes. Generally, the affinity of the non-covalent interaction of PAR with p53 was found to be within the nanomolar range (KD values from 3.4 × 10–9 to 2.5 × 10–7 M) (123). Additionally, differential scanning fluorimetry (DSF) was employed to determine the melting temperature (Tm) of PAR-bound p53, as a measure of thermodynamic stability. Consistent with previous findings, Tm increased in a PAR chain length-dependent manner, again indicative of a higher affinity of p53 towards longer PAR chains (85). Furthermore, analysis of the PAR–p53 interaction via attenuated total reflection Fourier-transform infrared spectroscopy (ATR-FTIR) supported the previously obtained results and revealed preferential binding of p53 to PAR compared to DNA, which holds true for both sequence-dependent and -independent binding of p53 to DNA (86,124). In line with this finding, EMSA experiments showed that PAR was able to inhibit and even reverse binding of p53 to its consensus sequence (125). These results point to chain length-dependent regulatory functions of non-covalent p53-PAR binding with respect to p53′s complex DNA binding properties.
In contrast to p53, the DNA repair protein XPA (xeroderma pigmentosum complementation group A) was shown to promote the formation of a single complex with long PAR chains (55-mers), while no interaction could be determined with short polymer (16-mers) in EMSA and SPR experiments. The binding affinity of XPA to long polymers is comparable to the one of p53 (KD values from 6.5 × 10–9 to 3.2 × 10–7 M). In line with the lack of interaction between XPA and short PAR molecules, the DNA-binding ability of XPA was only significantly influenced by the addition of longer PAR chains (88,123).
For XRCC1 (X-ray repair cross-complementing 1), an important scaffold protein within base excision repair (BER), a preference for PAR chains longer than 7 ADP-ribose units could be shown in time-resolved fluorescence resonance energy transfer (TR-FRET) experiments, using the terbium-labelled BRCT1 domain of XRCC1 as a FRET donor and FITC-NAD+ within PAR chains as an acceptor. While BRCT1-binding to mono-ADP-ribose (EC50 of 1.39 ± 0.06 mM) and short ADP-ribose oligomers (2- to 7-mers; EC50 of 110 ± 13 nM) was relatively weak, longer PAR chains (8- to 41-mers) were bound with higher affinity (EC50 of 16–30 nM) (126). Furthermore, it could be shown that especially short PAR polymer, as produced by the PARP1 variant PARP1\Y986S, failed to efficiently recruit XRCC1 to sites of laser-induced DNA damage in HeLa cells, both in comparison to longer hyper-/hypobranched or wildtype PAR (106).
Another protein that has been shown to preferentially bind longer PAR chains is the oncoprotein DEK. While longer PAR chains (54-mers) promoted the formation of one defined complex with DEK in EMSA experiments with high binding affinities in the nanomolar range (KD value of 6 × 10–8 M), no complex formation could be observed in combination with short PAR molecules (18-mers). These results could be confirmed in PAR overlay assays with purified recombinant DEK and pooled HPLC-fractionated PAR chains. Here, binding affinities for long PAR chains (≥57-mers or unfractionated) were strong, while slightly weaker for medium sized PAR chains (34–54-mers) and very weak for shorter PAR chains (≤34-mers). In contrast to p53 and XPA, non-covalent interaction with either short, long or unfractionated PAR chains did not significantly influence the ability of DEK to bind to DNA, as shown by South-Western analyses with immobilised DEK (90).
The nuclear RNA-binding protein NONO (non-POU domain-containing octamer-binding protein) represents yet another non-covalent interaction partner of PAR that shows high binding affinities towards long PAR chains (≥ 60-mers). They are bound by NONO with similar affinity than unfractionated PAR (KD value of 2.32 × 10–8 for unfractionated PAR and 2.01 × 10-8 for long PAR chains 60-mers), while interactions with short PAR chains (≤ 30-mers) could not be detected in PAR-binding and SPR experiments (127).
Histones have long since been shown to bind PAR chains with high affinity. In different cell types, namely keratinocytes and rat hepatocytes, H1 has been found to be the predominant PAR binding protein, followed by the core histones H2A, H2B, H3 and H4 (128). For histone H1, PAR overlay assays showed a chain length-dependent increase in affinity, where short chains (5–10-mers) were only bound weakly and longer polymers (≥15-mers or unfractionated) were bound relatively tightly to the immobilised histone (90). Accordingly, binding assays in combination with DNA sequencing gels showed that all analysed histones preferentially bound to longer over shorter PAR polymer (129).
The RecQ helicase WRN (Werner syndrome protein) on the other hand seems to bind PAR chains independent of their length. While WRN showed a marginally higher affinity towards longer (and branched) PAR chains in PAR overlay assays, it still bound very short PAR chains (≤ 10-mers) strongly and with a similar affinity to unfractionated PAR. DNA binding of WRN, as well as its helicase and exonuclease activities, are abolished with increasing concentrations of PAR (89).
Based on those results, we can summarise that p53, XPA, XRCC1, DEK, NONO and histones show a preference for binding to longer PAR chains, but while p53, XPA, XRCC1, DEK and histones non-covalently interact with PAR via a conserved PAR binding motif (PBM) comprised of basic and hydrophobic amino acids (81,88,90,123,129), NONO has been shown to bind PAR via an RNA recognition motif (RRM) (127). Reversing the argument, p53, XPA, XRCC1, DEK, histones and WRN all share the common PBM consensus sequence (81,88–90,123,129), but show different binding behaviour, especially to short PAR chains. Taken together, this leads to the assumption that other regulatory mechanisms, or other so far unidentified PAR binding motifs or structural nuances, have to be at play. Apparently, the mode of PAR binding does not necessarily seem to be the sole determining factor for affinity to short or long PAR chains. Also, the functional significances of the specific PAR binding behaviours of the factors mentioned above are largely unknown and have to be elucidated in the future. In the following we give an example which provides some first hints of such functional significance of PAR of variable chain lengths.
One example of how PAR chain length can influence downstream cellular processes was provided by Min et al., analysing the serine/threonine-protein kinase Chk1, which is able to bind PAR chains via its PAR-binding regulatory (PbR) motif (93). While a possible chain length-specific binding of Chk1 to PAR was not investigated, the modulation of its kinase activity by chain length-dependent PAR binding could be demonstrated. Employing an in vitro kinase activity assay with GST-tagged Chk1 and subsequent analysis of Chk1 autophosphorylation as an indication for its activity, it could be shown that long PAR chains (>65-mers) stimulate Chk1 kinase activity, while shorter PAR chains (26–30-mers) do not (93). Additionally, it could be shown that parthanatos, as a PAR-induced and caspase-independent cell death mechanism, is more strongly induced by long polymer with an average length of 60 ADP-ribose units, than by shorter polymer (16- or 30-mers). At an equal concentration of 80 nM, long polymer induced >80% cell death in cortical neurons (an increase of 40–50% compared to shorter polymer), as determined by Hoechst33342 and PI staining and subsequent quantitative, computer-assisted cell counting (130).
In a recent study, Dasovich et al. significantly extended the spectrum of PAR chain length-specific interacting proteins using photoaffinity probes consisting of either short or long PAR (8-mer or 40-mer, respectively) and an incorporated photo-inducible crosslinker (131). While some of the insights obtained from the previously mentioned studies could be verified, others could not. In line with earlier results, DEK could be identified as a long-PAR binder (log2 ratio of long to short PAR of 7.56). Contrary to previous results, however, XRCC1 and NONO were identified as short or indifferent PAR binders (log2 ratios of –2.06 and 0.71, respectively). How these seemingly contradictory results can be reconciled awaits clarification. It may be possible that some of the proteins identified in cell lysates by Dasovich et al. have different protein modifications or accessory binding partners that modify their PAR-binding specificities which cannot be recapitulated in vitro with recombinant proteins alone. Interestingly, in the study by Dasovich et al. also PARP1 itself was shown to preferentially bind long PAR chains (log2 ratio of 3.76), which could also be verified in EMSA experiments using recombinant PARP1. There, the binding affinity increased approximately 16-fold from 4 to 16-mers, with nanomolar affinities for 16–32-mers (KD values of 1.1 × 10–8 M to 1.1 × 10–7 M). As PAR is rapidly formed by PARP1 after detection of DNA damage and subsequently degraded by catabolising enzymes, chain length may therefore present a regulatory mechanism to direct PARP1 dissociation. Additionally, also the PAR catabolising enzyme PARG has been shown to rather bind to short PAR chains (log2 ratio –3.07) (131), possibly indicating a preferential degradation of short polymer (cf. section on ‘The influence of PAR chain length and branching on PAR catabolism’).
The influence of PAR branching on non-covalent PAR-protein binding
While the influence of PAR chain length on protein binding and downstream processes has been studied for several different interaction partners, the understanding of PAR branching is still in its infancy.
As mentioned previously, histones have been shown to preferentially bind long PAR chains over shorter ones (90,129). Employing binding assays in combination with sequencing gels and subsequent analysis of degradation products of polymer fractions for branching frequency, all analysed histones (i.e. H1, H2A, H2B, H3 and H4) could be shown to bind with higher affinity to branched polymer over long or short linear polymer (129).
Another protein that has recently been connected to binding of branched PAR is the histone chaperone APLF. To analyse its binding properties, PAR chains bound by recombinant APLF were digested by PARG, pyrophosphatase and alkaline phosphatase and subsequently analysed via LC-MS/MS. This revealed an enrichment of diribosyl-adenosine (R2-Ado; indicative of branched parts of the polymer) over ribosyl-adenosine (R-Ado; indicative of linear parts of the polymer), suggesting preferential binding of the protein to branched PAR chains (48). Structural studies have revealed two tandem PAR-binding zinc finger (PBZ) motifs within APLF, of which the first presents with a high affinity for PAR (KD value of 5 × 10–8 M), whereas the second displays a considerably lower affinity (KD value of 8 × 10–6 M). This is in line with the theory that the first PBZ motif is able to bind two ADP-ribose molecules, while the second one only binds one ADP-ribose molecule (94,95,134) and permits the hypothesis that those tandem PBZ motifs might function as specific readers for branched PAR molecules (48). The study by Chen et al. also suggests a functional significance by reporting that the synthesis of branched PAR by PARP2 (see above) in combination with its binding to APLF leads to histone H3 removal at sites of DNA damage (48).
Two additional proteins, where binding to branched PAR chains has recently been investigated, are the apurinic/apyrimidinic endonuclease 1 (APE1) and DNA polymerase β (Polβ), both involved in the BER pathway. EMSA experiments, performed after fractionation of PAR chains by anion exchange chromatography, revealed effective binding of both proteins to fractions containing long and branched PAR chains (>20-mers) over fractions containing mostly small (6 to 10-mers) or medium sized (11 to 20-mers) PAR polymer. Separation of fractions into long and mostly linear or long and mostly branched polymers revealed a moderate binding preference of APE1 and Polβ to linear chains (EC50 of 0.77 ± 0.08 and 0.57 ± 0.06 μM, respectively) over branched PAR chains (EC50 of 1.2 ± 0.1 and 1.3 ± 0.1 μM, respectively). Using the N-terminally truncated APE1NΔ35 variant, the positively charged N-terminal amino acids 1–35 could be implicated in the binding of APE1 to linear PAR polymer, as affinity to PAR decreased in general, but to a higher extent for the fraction containing mostly linear PAR chains (EC50 of 3.4 ± 0.3 μM for branched chains and EC50 of 9.0 ± 1.2 μM for linear chains) (132).
Another PAR binding protein of interest, which might have a preference for linear PAR chains, is represented by the RING-domain E3 ligase RNF146 (also known as iduna). Similar to WRN, RNF146 has been shown to bind PAR chains of variable chain lengths, with nanomolar affinity for PAR polymers with an average length of 40 ADP-ribose units (KD value of 1.2 × 10–8 M) (133). A filter-binding assay revealed similar binding constants for the interaction of the RNF146 WWE domain with either 10-mer (7.5 × 10–8 M) or 20-mer (6.7 × 10-8 M) PAR chains (107). While there appears to be no major chain length specificity, interestingly, data from Chen et al. (48) suggest that RNF146 preferentially binds linear PAR chains, which are thought to be produced by tankyrases (62). In this regard, it is important to note that RNF146 acts as a positive regulator of Wnt signalling in the cytoplasm, where it interacts with PAR produced by TNKS1 and promotes the poly-ubiquitination and degradation of axin (135,136), thereby bridging PARylation and the ubiquitin system. Mechanistically, this PARylation-dependent ubiquitination is mediated by binding of PAR or iso-ADP-ribose to the WWE domain of RNF146, thereby inducing the allosteric activation of the RNF146 ubiquitin ligase. In addition to its functions in the cytoplasm, RNF146 is also activated in the nucleus in a PARP1-dependent manner to target proteins, such as PARP1 itself, XRCC1, DNA ligase III, and KU70 for proteasomal degradation (137). As a perspective, it will be interesting to test the hypothesis, whether such PARylation-dependent ubiquitination is exerted specifically by PAR molecules of defined structural characteristics. Furthermore, it will be interesting to analyse, if and how the interaction of RNF146 with structurally defined PAR chains will influence the degradation efficiencies and kinetics of the respective RNF146 target proteins.
Evidently, the influence of PAR branching on non-covalent protein binding is considerably less explored than the influence of PAR chain length, therefore presenting an interesting topic for further research, in particular in view of PAR regulating processes of liquid-liquid demixing and biomolecular condensate formation as discussed in the following section.
The potential influence of PAR chain length and branching on biomolecular condensate formation
Through binding to different interaction partners, PAR chain length and branching might also influence the dynamics of so-called biomolecular condensates by regulating liquid-liquid phase separation. Up to now, no study specifically explored how PAR chain length and branching affects the dynamics of biomolecular condensates. Yet, as phase separation occurs as a function of affinity, binding preferences of proteins towards PAR chains of different lengths and branching frequencies might serve as means to regulate the assembly of biomolecular condensates and at the same time determine protein interactions within. It has been suggested that PAR functions as a scaffold, where valency and specificity of protein binding are controlled by (i) the number of PARylated sites on a single acceptor molecule, (ii) the number of ADP-ribose units within one PAR chain, and (iii) the (linear or branched) structure of the modification (100). What has been revealed in this respect by now is that PAR can initiate liquid-liquid phase separation, resulting in a fast and fully reversible accumulation of different intrinsically disordered proteins at DNA strand breaks (105). For example, the fused in sarcoma (FUS) protein can bind PAR via its C-terminal RGG domains, which drives phase separation (105). FUS is involved in the regulation of RNA metabolism and has been implicated in the repair of DNA strand breaks. PAR-mediated FUS-containing assemblies might be vital in the formation of transient DNA repair compartments (138). Furthermore, the PAR-binding heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) undergoes phase separation into protein-rich droplets and this process is strongly promoted by binding of PAR to the PBM motif of hnRNP A1 (87,139). Besides seeding of biomolecular condensates, PAR has also been connected to their maintenance in context with apoptosis signal-regulating kinase 3 (ASK3) during osmotic stress. ASK3 bidirectionally responds to changes in osmolality, being phosphorylated and therefore activated under hypoosmotic conditions and in turn dephosphorylated and inactivated under hyperosmotic conditions, consequently regulating cell volume recovery. During hyperosmosis, liquid phase separation of ASK3 is necessary for its inactivation and PAR has been shown to maintain liquidity of those condensates, therefore presenting a supporting factor in osmotic stress regulation (140). For more detailed information on the involvement of PAR in biomolecular condensate formation, we would like to refer to a recent review article by Leung (100).
In conclusion, considering the biophysical nature of their regulation, it can be expected that PAR structure-specific properties play a decisive role in the spatio-temporal regulation of liquid-liquid demixing processes.
The influence of PAR chain length and branching on PAR catabolism
Besides its previously mentioned effects, PAR structure also influences its catabolism by different PAR degrading enzymes.
Monitoring the degradation of long PAR chains by purified PARG via the release of monomeric ADP-ribose, as well as the decrease of acid-insoluble ADP-ribose molecules, a biphasic course of PAR degradation could be demonstrated. While large polymers were degraded by PARG early and rapidly, smaller polymers and ADP-ribose monomers were accumulated and subsequently degraded in a later and approximately 20-fold slower second phase (141). By incubating free PAR with PARG, followed by protein digestion, phenol extraction, and polyacrylamide gel electrophoresis (PAGE), it could be shown that linear PAR chains were degraded slightly faster than branched ones (129). The same conclusion was achieved by in situ radiolabelling of cellular NAD+ and isolation of ADP-ribose polymers from MNNG-treated human HaCaT keratinocytes via boronate affinity chromatography and subsequent analysis via PAGE. While large, linear polymers (≥ 36-mers) were degraded rapidly, branched ones could still be detected for a prolonged period of time (142). Supporting previous results, analysis of reaction products of PARG degradation via high-resolution DNA sequencing gels and subsequent HPLC revealed the release and accumulation of oligomeric ADP-ribose (in accordance with endoglycosidic activity of PARG), which was later further degraded to monomers. Additionally, it could be shown that the percentage of branching residues increased with ongoing degradation, while simultaneously, the number of branches per polymer decreased, leading to the assumption that PARG digestion produces smaller, unbranched degradation products, while leaving the branching residues themselves intact (143). Of further note, linear PAR chains were degraded at a similar rate either as free polymer or bound to histone H2B in vitro, while protein-bound branched polymer seemed to be protected from degradation for longer periods of time, potentially due to mechanisms of steric hinderance (129).
Recently, we analysed PAR levels and branching frequencies in HeLa cells after treatment with H2O2 and observed an increase in branched PAR molecules between 5 and 7.5 min post treatment, followed by a gradual decline to basal levels up until the 20 min time point (106). This is in line with results suggesting the preferential degradation of linear PAR chains, accompanied by a relative increase in branching residues (142,143). To validate our findings, we performed an in vitro PAR degradation assay utilising human recombinant PARG and differently structured PAR as produced by our PARP1 variants and quantified generated ADP-ribose amounts. While short, as well as hypobranched PAR chains, were degraded relatively fast – in line with findings that PARG preferentially binds short polymer (131) – the degradation of long or hyperbranched PAR was considerably slower (106). As for the comparison of rather linear to strongly branched PAR, these findings are in line with previously obtained results, pointing towards an increased stability of branched PAR chains (142). Yet, for the comparison of longer to shorter PAR, these results seem to run counter to the model proposed by Hatakeyama et al., where large polymers are degraded more rapidly than smaller ones (141). One explanation for these at first glance contradictory results, could be that the in vitro assay of (106), for technical reasons, does not allow for the detection of endoglycosidic cleavage products. Thus, shorter linear oligomers might have been formed from branched polymers that are then further degraded later (106). In conclusion, by now there is strong biochemical and cellular evidence that PAR catabolism is highly structure-specific, yet potential functional consequences of these findings are so far largely unknown. One obvious, intriguing hypothesis to follow here, is the idea that structure-specific degradation intermediates exhibit specific downstream signalling functions via discriminative binding to specific factors.
HETEROGENEITY BEYOND PAR STRUCTURE
Heterogeneity with regards to PAR is not limited to the structure of the modification itself, but extends to the variety of modified substrates. Thus, in addition to the amino acid-specific modification of proteins, in recent years the addition of ADP-ribose units to DNA and RNA has been identified as a new type of post-replicative modification, catalysed by various members of the PARP family as well as certain toxin-related ARTs in non-mammalian organisms (144–146). For a comprehensive overview on this topic, the reader is referred to a recent review by Weixler et al. (146).
Mainly responsible for cellular PARylation of proteins, PARP1 has been shown to directly modify the ends of DNA oligonucleotides, preferentially at 3′-terminal phosphates on double-strand breaks (DSBs) and at 5′-terminal phosphates on single-stranded oligonucleotides (147,148). Similar to PARP1, PARP2 and PARP3 have also been shown to modify 3′- and 5′-terminal phosphates on SSBs and DSBs, with PARP3 preferentially modifying 5′-phosphates on blunt ends (149–151). Depending on the specific properties of the DNA substrate, DNA modifications by all three PARPs might even be preferred over the respective automodification (148,150). Of note, PARP3-ADP-ribosylated DNA can serve as a primed DNA substrate for PAR chain elongation by PARP1 and PARP2 (151). Moreover, PARP1, PARP2 or PARP3-dependent DNA modifications have been shown to be efficiently and fully reversed by PARG (147,150), while PARP3-dependent mono-ADP-ribosylation has additionally been shown to be efficiently removed by TARG1, MacroD2 and also ARH3 in case of 5′-terminal phosphate modification (149). Adding to the several studies investigating ADP-ribose modifications on DNA, a recent study detailed the modification of phosphorylated RNA ends by PARP10 (152), which has previously been shown to contain a specific RNA-recognition motif (153). Resembling the ADP-ribosylation of DNA, this process is reversible by the commonly known ADP-ribose catabolising enzymes PARG, TARG1, MacroD1/D2, as well as ARH3 (152).
In short, the reversible MARylation and PARylation on DNA and RNA, thereby forming hybrids of two nucleic acid-like molecules, could function as a physiologically relevant biological signal. The observation that DNA and RNA ends showed increased resistance towards phosphatase treatment (149,152) could point towards a protective mechanism, preventing the nucleic acid molecules from premature degradation. In addition, DNA ADP-ribosylation potentially plays an active role in DNA repair processes (146,151).
DRAWING PARALLELS TO THE UBIQUITIN SYSTEM
Besides PARylation, ubiquitination is another very important and well-studied post-translational modification. It is mediated through the interplay of ubiquitin (Ub)-activating enzymes (E1) with Ub-transferring enzymes (E2) and Ub ligases (E3). Most frequently, E3 ligases modify acceptor proteins at lysine (Lys) residues via an isopeptide bond to the C-terminal glycine of Ub (154). Substrates can either be (multi) monoubiquitinated (155) or polyubiquitinated via homotypic or heterotypic linkages through any of the seven lysine or a methionine residue within Ub. While homotypic linkages are characterised by conjugation via one specific linkage type, heterotypic polyubiquitination can include mixed linkages, branching sites, conjugation with Ub-like modifiers, and even post-translational modifications of Ub itself (156). The different Ub chains are recognised by Ub binding domains within specific Ub binding proteins (UBPs), thereby forming the connection between the modified substrates and distinct downstream processes (157,158). In contrast to PARylation, the biological function of different polymer lengths and linkages is somewhat better understood for the Ub system. While (multi) monoubiquitination is for instance involved in protein trafficking and endocytosis (159,160), it can also target substrates for degradation via the Ub-proteasome system (161). Lys48-linked Ub chains are the canonical signal to target substrates for proteasomal degradation (162), whereas Lys63 linkage is predominantly implicated in non-degrading pathways like DNA repair (163), protein transport (164), kinase activation and cellular signalling (165). Furthermore, a linkage via Lys11 is also involved in proteasomal degradation (166) and has additionally been implicated in the regulation of cell division (167). Amongst the more atypical linkages, Lys27 has been shown to be involved in the DNA damage response (168), while Lys29 and Lys33 contribute to the regulation of AMPK-related kinases (169). Further, it could recently be shown that chain length also plays an important role for recognition by UBPs for Lys29- and Lys33-linked chains. While longer chains (≥6-mers) were preferentially bound to enzymes involved in metabolic conversion (e.g. hydrolases, transferases, and oxidoreductases), shorter chains (i.e. dimers/tetramers) rather interacted with enzymes involved in protein modification (e.g. kinases, proteases, and also E3 ligases) (170). In line with this, previous studies already demonstrated that different deubiquitinating enzymes are influenced by different chain lengths. While isopeptidase T displays a distinctly lower affinity for linear dimeric Ub over the tri- or tetrameric forms (171), the cleavage efficiency of the hydrolase UCH-L3 has been shown to decrease with increasing chain lengths for Lys48- and Lys63-linked chains (172).
Transferring those insights obtained from the Ub system, it is reasonable to assume that PAR chain length and branching are of similar importance for downstream cellular processes and, taken together with the results we obtained from analysing differently structured PAR, this points towards a physiological significance of PAR structural heterogeneity.
CONCLUSION
As reviewed here, the biological significance of PAR’s structural heterogeneity is just emerging. This concerns both its determinants as well as its consequences (Figure 2). On the one hand, determinants of PAR structural diversity are certainly different PARP family members that catalyse the addition of different PAR chains or mono-ADP-ribose to target molecules, as well as accessory factors that influence PAR structure, but they might also be found in other post-translational modifications, naturally occurring PARP variants and polymorphisms, as well as the intracellular concentration of NAD+. Consequences of PAR’s structural diversity, on the other hand, comprise the selectivity towards downstream binding partners, the formation of biomolecular condensates, PAR catabolism, and a multitude of cellular endpoints. All this illustrates the intricate and multi-level system necessary for directed control of PARylation.
Future research needs to address the mechanistic details, potential (patho-) physiological consequences as well as therapeutic potential of PAR structure-specific biology. This includes open questions like: (i) What are the exact and specific natural molecular determinants that induce PAR formation of different chain lengths and branching frequencies in cells and organs? In this regard, it will also be interesting to see if different environmental stimuli and cellular stressors can lead to differences in the quality of the formed PAR molecules. (ii) What are further PAR chain length and, in particular, branching-specific downstream factors during cellular stress response? (iii) What are the molecular binding mechanisms of those PAR structure-specific interaction partners? (iv) Exactly how do PAR chain length and branching, as well as the formation of structure-specific degradation products, regulate cellular processes under physiological conditions, as well as under conditions of different forms of cellular stress? (v) How do different PAR structures and PAR heterogeneity affect biological functions on multicellular and organismic levels? And (vi), does dysregulation of PAR structure contribute to disease development, such as e.g., cancer and neurodegenerative diseases?
In order to address such questions, it will be necessary to develop more advanced molecular biological, bioanalytical, genetic, and pharmacological tools. For example, this comprises (i) proteomics approaches for the identification of branching- and chain length-specific interactomes, (ii) metabolomics approaches to monitor PAR structure-specific catabolic products, (iii) chemical synthesis of PAR chains of defined chain length and branching to conduct specific interaction studies and as baits for proteomics approaches, (iv) biophysical approaches to determine the structural characteristics of PAR-protein interactions, (v) genetic engineering of further PARP variants that produce PAR molecules of more defined branching and/or chain lengths and (vi) identification of pharmacological modulators of PARP activity, which go beyond the mere enzymatic inhibition of PARPs. Concerning the latter, by modulating the quality of PAR produced under certain conditions, it may be possible not only to inhibit PARP-related cellular processes more specifically, but potentially also boost and improve such processes, e.g. rendering genome maintenance and cellular stress response more efficient. This will potentially open up new possibilities, e.g. for cancer therapy, treatment of neurodegenerative diseases, and aging intervention strategies.
ACKNOWLEDGEMENTS
We thank and Prof. Dr Alexander Bürkle (University of Konstanz) for critically reading of the manuscript and Dr Maike Lehner (University of Konstanz) for providing Figure 1.
Contributor Information
Julia M Reber, Department of Biology, University of Konstanz, 78467 Konstanz, Germany.
Aswin Mangerich, Department of Biology, University of Konstanz, 78467 Konstanz, Germany.
FUNDING
German Research Foundation (DFG) [MA-4905/4-1]; Young Scholar Fund of the University of Konstanz funded by the DFG Excellence Initiative; J.M.R. was a fellow of the DFG-funded Konstanz Research School ‘Chemical Biology’ [GSC218]. Funding for open access charge: University of Konstanz/DFG.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lin H., Begley T.. Protein posttranslational modifications: chemistry, biology, and applications. Mol. Biosyst. 2011; 7:14–15. [DOI] [PubMed] [Google Scholar]
- 2.Hottiger M.O., Hassa P.O., Lüscher B., Schuler H., Koch-Nolte F.. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 2010; 35:208–219. [DOI] [PubMed] [Google Scholar]
- 3.Vyas S., Chesarone-Cataldo M., Todorova T., Huang Y.H., Chang P.. A systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nat. Commun. 2013; 4:2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hou W.H., Chen S.H., Yu X.. Poly-ADP ribosylation in DNA damage response and cancer therapy. Mutat. Res. 2019; 780:82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azarm K., Smith S.. Nuclear PARPs and genome integrity. Genes Dev. 2020; 34:285–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisemann T., Pascal J.M.. Poly(ADP-ribose) polymerase enzymes and the maintenance of genome integrity. Cell. Mol. Life Sci. 2020; 77:19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pandey N., Black B.E.. Rapid detection and signaling of DNA damage by PARP-1. Trends Biochem. Sci. 2021; 10.1016/j.tibs.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ray Chaudhuri A., Nussenzweig A.. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017; 18:610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slade D.Mitotic functions of poly(ADP-ribose) polymerases. Biochem. Pharmacol. 2019; 167:33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrision D., Gravells P., Thompson R., Bryant H.E.. Poly(ADP-Ribose) glycohydrolase (PARG) vs. poly(ADP-Ribose) polymerase (PARP) - function in genome maintenance and relevance of inhibitors for anti-cancer therapy. Front. Mol. Biosci. 2020; 7:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beneke S., Cohausz O., Malanga M., Boukamp P., Althaus F., Bürkle A.. Rapid regulation of telomere length is mediated by poly(ADP-ribose) polymerase-1. Nucleic Acids Res. 2008; 36:6309–6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harvey A., Mielke N., Grimstead J.W., Jones R.E., Nguyen T., Mueller M., Baird D.M., Hendrickson E.A.. PARP1 is required for preserving telomeric integrity but is dispensable for A-NHEJ. Oncotarget. 2018; 9:34821–34837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanzlikova H., Kalasova I., Demin A.A., Pennicott L.E., Cihlarova Z., Caldecott K.W.. The importance of poly(ADP-ribose) polymerase as a sensor of unligated okazaki fragments during DNA replication. Mol. Cell. 2018; 71:319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanzlikova H., Caldecott K.W.. Perspectives on PARPs in S phase. Trends Genet. 2019; 35:412–422. [DOI] [PubMed] [Google Scholar]
- 15.Kim D.S., Challa S., Jones A., Kraus W.L.. PARPs and ADP-ribosylation in RNA biology: from RNA expression and processing to protein translation and proteostasis. Genes Dev. 2020; 34:302–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ke Y., Wang C., Zhang J., Zhong X., Wang R., Zeng X., Ba X.. The role of PARPs in inflammation-and metabolic-related diseases: molecular mechanisms and beyond. Cells. 2019; 8:1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunze F.A., Hottiger M.O.. Regulating immunity via ADP-Ribosylation: Therapeutic implications and beyond. Trends Immunol. 2019; 40:159–173. [DOI] [PubMed] [Google Scholar]
- 18.Pazzaglia S., Pioli C.. Multifaceted role of PARP-1 in DNA repair and inflammation: pathological and therapeutic implications in cancer and non-cancer diseases. Cells. 2019; 9:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Morcillo F.J., Canton-Sandoval J., Martinez-Menchon T., Corbalan-Velez R., Mesa-Del-Castillo P., Perez-Oliva A.B., Garcia-Moreno D., Mulero V.. Non-canonical roles of NAMPT and PARP in inflammation. Dev. Comp. Immunol. 2021; 115:103881. [DOI] [PubMed] [Google Scholar]
- 20.Ying Y., Padanilam B.J.. Regulation of necrotic cell death: p53, PARP1 and cyclophilin D-overlapping pathways of regulated necrosis. Cell. Mol. Life Sci. 2016; 73:2309–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodriguez-Vargas J.M., Oliver-Pozo F.J., Dantzer F.. PARP1 and poly(ADP-ribosyl)ation signaling during autophagy in response to nutrient deprivation. Oxid. Med. Cell Longev. 2019; 2019:2641712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park H., Kam T.I., Dawson T.M., Dawson V.L.. Poly (ADP-ribose) (PAR)-dependent cell death in neurodegenerative diseases. Int. Rev. Cell Mol. Biol. 2020; 353:1–29. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Y., Liu L., Tao S., Yao Y., Wang Y., Wei Q., Shao A., Deng Y.. Parthanatos and its associated components: promising therapeutic targets for cancer. Pharmacol. Res. 2021; 163:105299. [DOI] [PubMed] [Google Scholar]
- 24.Dona F., Chiodi I., Belgiovine C., Raineri T., Ricotti R., Mondello C., Scovassi A.I.. Poly(ADP-ribosylation) and neoplastic transformation: effect of PARP inhibitors. Curr. Pharm. Biotechnol. 2013; 14:524–536. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez M.I., Majuelos-Melguizo J., Marti Martin-Consuegra J.M., Ruiz de Almodovar M., Lopez-Rivas A., Javier Oliver F.. Deciphering the insights of poly(ADP-ribosylation) in tumor progression. Med. Res. Rev. 2015; 35:678–697. [DOI] [PubMed] [Google Scholar]
- 26.Mao K., Zhang G.. The role of PARP1 in neurodegenerative diseases and aging. FEBS J. 2021; 10.1111/febs.15716. [DOI] [PubMed] [Google Scholar]
- 27.Hopp A.K., Gruter P., Hottiger M.O.. Regulation of glucose metabolism by NAD(+) and ADP-Ribosylation. Cells. 2019; 8:890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slade D.PARP and PARG inhibitors in cancer treatment. Genes Dev. 2020; 34:360–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Curtin N.J., Szabo C.. Poly(ADP-ribose) polymerase inhibition: past, present and future. Nat. Rev. Drug Discov. 2020; 19:711–736. [DOI] [PubMed] [Google Scholar]
- 30.Boussios S., Moschetta M., Karihtala P., Samartzis E.P., Sheriff M., Pappas-Gogos G., Ozturk M.A., Uccello M., Karathanasi A., Tringos M.et al.. Development of new poly(ADP-ribose) polymerase (PARP) inhibitors in ovarian cancer: Quo Vadis. Ann. Transl. Med. 2020; 8:1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cortesi L., Rugo H.S., Jackisch C.. An overview of PARP inhibitors for the treatment of breast cancer. Target Oncol. 2021; 16:255–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grewal K., Grewal K., Tabbara I.A.. PARP inhibitors in prostate cancer. Anticancer Res. 2021; 41:551–556. [DOI] [PubMed] [Google Scholar]
- 33.Singh H.M., Bailey P., Hubschmann D., Berger A.K., Neoptolemos J.P., Jager D., Siveke J., Springfeld C.. Poly(ADP-ribose) polymerase inhibition in pancreatic cancer. Genes Chromosomes Cancer. 2021; 60:373–384. [DOI] [PubMed] [Google Scholar]
- 34.Lord C.J., Ashworth A.. PARP inhibitors: synthetic lethality in the clinic. Science. 2017; 355:1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin K.Y., Kraus W.L.. PARP inhibitors for cancer therapy. Cell. 2017; 169:183. [DOI] [PubMed] [Google Scholar]
- 36.Knelson E.H., Patel S.A., Sands J.M.. PARP inhibitors in small-cell lung cancer: rational combinations to improve responses. Cancers (Basel). 2021; 13:727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koh S.H., Park Y., Song C.W., Kim J.G., Kim K., Kim J., Kim M.H., Lee S.R., Kim D.W., Yu H.J.et al.. The effect of PARP inhibitor on ischaemic cell death, its related inflammation and survival signals. Eur. J. Neurosci. 2004; 20:1461–1472. [DOI] [PubMed] [Google Scholar]
- 38.Teng F., Zhu L., Su J., Zhang X., Li N., Nie Z., Jin L.. Neuroprotective effects of poly(ADP-ribose)polymerase inhibitor olaparib in transient cerebral ischemia. Neurochem. Res. 2016; 41:1516–1526. [DOI] [PubMed] [Google Scholar]
- 39.Kim Y., Kim Y.S., Noh M.Y., Lee H., Joe B., Kim H.Y., Kim J., Kim S.H., Park J.. Neuroprotective effects of a novel poly (ADP-ribose) polymerase-1 inhibitor, JPI-289, in hypoxic rat cortical neurons. Clin. Exp. Pharmacol. Physiol. 2017; 44:671–679. [DOI] [PubMed] [Google Scholar]
- 40.Olsen A.L., Feany M.B.. PARP inhibitors and parkinson's disease. N. Engl. J. Med. 2019; 380:492–494. [DOI] [PubMed] [Google Scholar]
- 41.Miwa M., Saikawa N., Yamaizumi Z., Nishimura S., Sugimura T.. Structure of poly(adenosine diphosphate ribose): identification of 2′-[1′-ribosyl-2′-(or 3′-)(1′-ribosyl)]adenosine-5′,5′,5′-tris(phosphate) as a branch linkage. Proc. Natl. Acad. Sci. U.S.A. 1979; 76:595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alvarez-Gonzalez R., Jacobson M.K.. Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo. Biochemistry. 1987; 26:3218–3224. [DOI] [PubMed] [Google Scholar]
- 43.Barkauskaite E., Jankevicius G., Ahel I.. Structures and mechanisms of enzymes employed in the synthesis and degradation of PARP-dependent protein ADP-Ribosylation. Mol. Cell. 2015; 58:935–946. [DOI] [PubMed] [Google Scholar]
- 44.Ruf A., Rolli V., de Murcia G., Schulz G.E.. The mechanism of the elongation and branching reaction of poly(ADP-ribose) polymerase as derived from crystal structures and mutagenesis. J. Mol. Biol. 1998; 278:57–65. [DOI] [PubMed] [Google Scholar]
- 45.Steffen J.D., Brody J.R., Armen R.S., Pascal J.M.. Structural implications for selective targeting of PARPs. Front. Oncol. 2013; 3:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vyas S., Matic I., Uchima L., Rood J., Zaja R., Hay R.T., Ahel I., Chang P.. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat. Commun. 2014; 5:4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prokhorova E., Agnew T., Wondisford A.R., Tellier M., Kaminski N., Beijer D., Holder J., Groslambert J., Suskiewicz M.J., Zhu K.et al.. Unrestrained poly-ADP-ribosylation provides insights into chromatin regulation and human disease. Mol. Cell. 2021; 81:2640–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Q., Kassab M.A., Dantzer F., Yu X.. PARP2 mediates branched poly ADP-ribosylation in response to DNA damage. Nat. Commun. 2018; 9:3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mao Z., Hine C., Tian X., Van Meter M., Au M., Vaidya A., Seluanov A., Gorbunova V.. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011; 332:1443–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y., Wang J., Ding M., Yu Y.. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat. Methods. 2013; 10:981–984. [DOI] [PubMed] [Google Scholar]
- 51.Martello R., Leutert M., Jungmichel S., Bilan V., Larsen S.C., Young C., Hottiger M.O., Nielsen M.L.. Proteome-wide identification of the endogenous ADP-ribosylome of mammalian cells and tissue. Nat. Commun. 2016; 7:12917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leslie Pedrioli D.M., Leutert M., Bilan V., Nowak K., Gunasekera K., Ferrari E., Imhof R., Malmstrom L., Hottiger M.O.. Comprehensive ADP-ribosylome analysis identifies tyrosine as an ADP-ribose acceptor site. EMBO Rep. 2018; 19:e45310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palazzo L., Leidecker O., Prokhorova E., Dauben H., Matic I., Ahel I.. Serine is the major residue for ADP-ribosylation upon DNA damage. eLife. 2018; 7:e34334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laing S., Unger M., Koch-Nolte F., Haag F.. ADP-ribosylation of arginine. Amino Acids. 2011; 41:257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gibbs-Seymour I., Fontana P., Rack J.G.M., Ahel I.. HPF1/C4orf27 Is a PARP-1-Interacting protein that regulates PARP-1 ADP-Ribosylation activity. Mol. Cell. 2016; 62:432–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leidecker O., Bonfiglio J.J., Colby T., Zhang Q., Atanassov I., Zaja R., Palazzo L., Stockum A., Ahel I., Matic I.. Serine is a new target residue for endogenous ADP-ribosylation on histones. Nat. Chem. Biol. 2016; 12:998–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bonfiglio J.J., Fontana P., Zhang Q., Colby T., Gibbs-Seymour I., Atanassov I., Bartlett E., Zaja R., Ahel I., Matic I.. Serine ADP-ribosylation depends on HPF1. Mol. Cell. 2017; 65:932–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suskiewicz M.J., Zobel F., Ogden T.E.H., Fontana P., Ariza A., Yang J.C., Zhu K., Bracken L., Hawthorne W.J., Ahel D.et al.. HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation. Nature. 2020; 579:598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun F.H., Zhao P., Zhang N., Kong L.L., Wong C.C.L., Yun C.H.. HPF1 remodels the active site of PARP1 to enable the serine ADP-ribosylation of histones. Nat. Commun. 2021; 12:1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rudolph J., Roberts G., Muthurajan U.M., Luger K.. HPF1 and nucleosomes mediate a dramatic switch in activity of PARP1 from polymerase to hydrolase. eLife. 2021; 10:e65773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alemasova E.E., Lavrik O.I.. Poly(ADP-ribosyl)ation by PARP1: reaction mechanism and regulatory proteins. Nucleic Acids Res. 2019; 47:3811–3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rippmann J.F., Damm K., Schnapp A.. Functional characterization of the poly(ADP-ribose) polymerase activity of tankyrase 1, a potential regulator of telomere length. J. Mol. Biol. 2002; 323:217–224. [DOI] [PubMed] [Google Scholar]
- 63.Rolli V., O’Farrell M., Menissier-de Murcia J., de Murcia G.. Random mutagenesis of the poly(ADP-ribose) polymerase catalytic domain reveals amino acids involved in polymer branching. Biochemistry. 1997; 36:12147–12154. [DOI] [PubMed] [Google Scholar]
- 64.Slade D., Dunstan M.S., Barkauskaite E., Weston R., Lafite P., Dixon N., Ahel M., Leys D., Ahel I.. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature. 2011; 477:616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barkauskaite E., Brassington A., Tan E.S., Warwicker J., Dunstan M.S., Banos B., Lafite P., Ahel M., Mitchison T.J., Ahel I.et al.. Visualization of poly(ADP-ribose) bound to PARG reveals inherent balance between exo- and endo-glycohydrolase activities. Nat. Commun. 2013; 4:2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wielckens K., Schmidt A., George E., Bredehorst R., Hilz H.. DNA fragmentation and NAD depletion. Their relation to the turnover of endogenous mono(ADP-ribosyl) and poly(ADP-ribosyl) proteins. J. Biol. Chem. 1982; 257:12872–12877. [PubMed] [Google Scholar]
- 67.Kleine H., Poreba E., Lesniewicz K., Hassa P.O., Hottiger M.O., Litchfield D.W., Shilton B.H., Lüscher B.. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol. Cell. 2008; 32:57–69. [DOI] [PubMed] [Google Scholar]
- 68.Loseva O., Jemth A.S., Bryant H.E., Schuler H., Lehtio L., Karlberg T., Helleday T.. PARP-3 is a mono-ADP-ribosylase that activates PARP-1 in the absence of DNA. J. Biol. Chem. 2010; 285:8054–8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lüscher B., Butepage M., Eckei L., Krieg S., Verheugd P., Shilton B.H.. ADP-Ribosylation, a multifaceted posttranslational modification involved in the control of cell physiology in health and disease. Chem. Rev. 2018; 118:1092–1136. [DOI] [PubMed] [Google Scholar]
- 70.Jankevicius G., Hassler M., Golia B., Rybin V., Zacharias M., Timinszky G., Ladurner A.G.. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat. Struct. Mol. Biol. 2013; 20:508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rosenthal F., Feijs K.L., Frugier E., Bonalli M., Forst A.H., Imhof R., Winkler H.C., Fischer D., Caflisch A., Hassa P.O.et al.. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat. Struct. Mol. Biol. 2013; 20:502–507. [DOI] [PubMed] [Google Scholar]
- 72.O'Sullivan J., Tedim Ferreira M., Gagne J.P., Sharma A.K., Hendzel M.J., Masson J.Y., Poirier G.G.. Emerging roles of eraser enzymes in the dynamic control of protein ADP-ribosylation. Nat. Commun. 2019; 10:1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rack J.G.M., Palazzo L., Ahel I.. (ADP-ribosyl)hydrolases: structure, function, and biology. Genes Dev. 2020; 34:263–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ishiwata-Endo H., Kato J., Stevens L.A., Moss J.. ARH1 in health and disease. Cancers (Basel). 2020; 12:479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fontana P., Bonfiglio J.J., Palazzo L., Bartlett E., Matic I., Ahel I.. Serine ADP-ribosylation reversal by the hydrolase ARH3. eLife. 2017; 6:e28533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Abplanalp J., Leutert M., Frugier E., Nowak K., Feurer R., Kato J., Kistemaker H.V.A., Filippov D.V., Moss J., Caflisch A.et al.. Proteomic analyses identify ARH3 as a serine mono-ADP-ribosylhydrolase. Nat. Commun. 2017; 8:2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mueller-Dieckmann C., Kernstock S., Lisurek M., von Kries J.P., Haag F., Weiss M.S., Koch-Nolte F.. The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:15026–15031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Palazzo L., Thomas B., Jemth A.S., Colby T., Leidecker O., Feijs K.L., Zaja R., Loseva O., Puigvert J.C., Matic I.et al.. Processing of protein ADP-ribosylation by nudix hydrolases. Biochem. J. 2015; 468:293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Daniels C.M., Thirawatananond P., Ong S.E., Gabelli S.B., Leung A.K.. Nudix hydrolases degrade protein-conjugated ADP-ribose. Sci. Rep. 2015; 5:18271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gagne J.P., Isabelle M., Lo K.S., Bourassa S., Hendzel M.J., Dawson V.L., Dawson T.M., Poirier G.G.. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008; 36:6959–6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pleschke J.M., Kleczkowska H.E., Strohm M., Althaus F.R.. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 2000; 275:40974–40980. [DOI] [PubMed] [Google Scholar]
- 82.Krietsch J., Rouleau M., Pic E., Ethier C., Dawson T.M., Dawson V.L., Masson J.Y., Poirier G.G., Gagne J.P.. Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol. Aspects Med. 2013; 34:1066–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kamaletdinova T., Fanaei-Kahrani Z., Wang Z.Q.. The enigmatic function of PARP1: from PARylation activity to PAR readers. Cells. 2019; 8:1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Teloni F., Altmeyer M.. Readers of poly(ADP-ribose): designed to be fit for purpose. Nucleic Acids Res. 2016; 44:993–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fischbach A., Krüger A., Hampp S., Assmann G., Rank L., Hufnagel M., Stockl M.T., Fischer J.M.F., Veith S., Rossatti P.et al.. The C-terminal domain of p53 orchestrates the interplay between non-covalent and covalent poly(ADP-ribosyl)ation of p53 by PARP1. Nucleic Acids Res. 2018; 46:804–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krüger A., Stier A., Fischbach A., Bürkle A., Hauser K., Mangerich A.. Interactions of p53 with poly(ADP-ribose) and DNA induce distinct changes in protein structure as revealed by ATR-FTIR spectroscopy. Nucleic Acids Res. 2019; 47:4843–4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Duan Y., Du A., Gu J., Duan G., Wang C., Gui X., Ma Z., Qian B., Deng X., Zhang K.et al.. PARylation regulates stress granule dynamics, phase separation, and neurotoxicity of disease-related RNA-binding proteins. Cell Res. 2019; 29:233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fischer J.M., Popp O., Gebhard D., Veith S., Fischbach A., Beneke S., Leitenstorfer A., Bergemann J., Scheffner M., Ferrando-May E.et al.. Poly(ADP-ribose)-mediated interplay of XPA and PARP1 leads to reciprocal regulation of protein function. FEBS J. 2014; 281:3625–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Popp O., Veith S., Fahrer J., Bohr V.A., Bürkle A., Mangerich A.. Site-specific noncovalent interaction of the biopolymer poly(ADP-ribose) with the Werner syndrome protein regulates protein functions. ACS Chem. Biol. 2013; 8:179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fahrer J., Popp O., Malanga M., Beneke S., Markovitz D.M., Ferrando-May E., Bürkle A., Kappes F.. High-affinity interaction of poly(ADP-ribose) and the human DEK oncoprotein depends upon chain length. Biochemistry. 2010; 49:7119–7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ganz M., Vogel C., Czada C., Jorke V., Gwosch E.C., Kleiner R., Pierzynska-Mach A., Zanacchi F.C., Diaspro A., Kappes F.et al.. The oncoprotein DEK affects the outcome of PARP1/2 inhibition during mild replication stress. PLoS One. 2019; 14:e0213130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ahel I., Ahel D., Matsusaka T., Clark A.J., Pines J., Boulton S.J., West S.C.. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008; 451:81–85. [DOI] [PubMed] [Google Scholar]
- 93.Min W., Bruhn C., Grigaravicius P., Zhou Z.W., Li F., Krüger A., Siddeek B., Greulich K.O., Popp O., Meisezahl C.et al.. Poly(ADP-ribose) binding to Chk1 at stalled replication forks is required for S-phase checkpoint activation. Nat. Commun. 2013; 4:2993. [DOI] [PubMed] [Google Scholar]
- 94.Eustermann S., Brockmann C., Mehrotra P.V., Yang J.C., Loakes D., West S.C., Ahel I., Neuhaus D.. Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose). Nat. Struct. Mol. Biol. 2010; 17:241–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li G.Y., McCulloch R.D., Fenton A.L., Cheung M., Meng L., Ikura M., Koch C.A.. Structure and identification of ADP-ribose recognition motifs of APLF and role in the DNA damage response. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:9129–9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang Z., Michaud G.A., Cheng Z., Zhang Y., Hinds T.R., Fan E., Cong F., Xu W.. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 2012; 26:235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.He F., Tsuda K., Takahashi M., Kuwasako K., Terada T., Shirouzu M., Watanabe S., Kigawa T., Kobayashi N., Guntert P.et al.. Structural insight into the interaction of ADP-ribose with the PARP WWE domains. FEBS Lett. 2012; 586:3858–3864. [DOI] [PubMed] [Google Scholar]
- 98.Breslin C., Hornyak P., Ridley A., Rulten S.L., Hanzlikova H., Oliver A.W., Caldecott K.W.. The XRCC1 phosphate-binding pocket binds poly (ADP-ribose) and is required for XRCC1 function. Nucleic Acids Res. 2015; 43:6934–6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li M., Lu L.Y., Yang C.Y., Wang S., Yu X.. The FHA and BRCT domains recognize ADP-ribosylation during DNA damage response. Genes Dev. 2013; 27:1752–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Leung A.K.L.Poly(ADP-ribose): a dynamic trigger for biomolecular condensate formation. Trends Cell Biol. 2020; 30:370–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim D.S., Camacho C.V., Nagari A., Malladi V.S., Challa S., Kraus W.L.. Activation of PARP-1 by snoRNAs controls ribosome biogenesis and cell growth via the RNA helicase DDX21. Mol. Cell. 2019; 75:1270–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dasovich M., Leung A.K.L.. A nucleolar PARtnership expands PARP roles in RNA biology and the clinical potential of PARP inhibitors. Mol. Cell. 2019; 75:1089–1091. [DOI] [PubMed] [Google Scholar]
- 103.Alemasova E.E., Naumenko K.N., Kurgina T.A., Anarbaev R.O., Lavrik O.I.. The multifunctional protein YB-1 potentiates PARP1 activity and decreases the efficiency of PARP1 inhibitors. Oncotarget. 2018; 9:23349–23365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Leung A.K.Poly(ADP-ribose): an organizer of cellular architecture. J. Cell Biol. 2014; 205:613–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Altmeyer M., Neelsen K.J., Teloni F., Pozdnyakova I., Pellegrino S., Grofte M., Rask M.D., Streicher W., Jungmichel S., Nielsen M.L.et al.. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun. 2015; 6:8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Aberle L., Krüger A., Reber J.M., Lippmann M., Hufnagel M., Schmalz M., Trussina I., Schlesiger S., Zubel T., Schutz K.et al.. PARP1 catalytic variants reveal branching and chain length-specific functions of poly(ADP-ribose) in cellular physiology and stress response. Nucleic Acids Res. 2020; 48:10015–10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ando Y., Elkayam E., McPherson R.L., Dasovich M., Cheng S.J., Voorneveld J., Filippov D.V., Ong S.E., Joshua-Tor L., Leung A.K.L.. ELTA: Enzymatic labeling of terminal ADP-Ribose. Mol. Cell. 2019; 73:845–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Naumenko K.N., Sukhanova M.V., Hamon L., Kurgina T.A., Alemasova E.E., Kutuzov M.M., Pastre D., Lavrik O.I.. Regulation of poly(ADP-Ribose) polymerase 1 activity by Y-Box-Binding protein 1. Biomolecules. 2020; 10:1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Du Y., Yamaguchi H., Wei Y., Hsu J.L., Wang H.L., Hsu Y.H., Lin W.C., Yu W.H., Leonard P.G., Lee G.R.t.et al.. Blocking c-Met-mediated PARP1 phosphorylation enhances anti-tumor effects of PARP inhibitors. Nat. Med. 2016; 22:194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kauppinen T.M., Chan W.Y., Suh S.W., Wiggins A.K., Huang E.J., Swanson R.A.. Direct phosphorylation and regulation of poly(ADP-ribose) polymerase-1 by extracellular signal-regulated kinases 1/2. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:7136–7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wright R.H., Castellano G., Bonet J., Le Dily F., Font-Mateu J., Ballare C., Nacht A.S., Soronellas D., Oliva B., Beato M.. CDK2-dependent activation of PARP-1 is required for hormonal gene regulation in breast cancer cells. Genes Dev. 2012; 26:1972–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Piao L., Fujioka K., Nakakido M., Hamamoto R.. Regulation of poly(ADP-Ribose) polymerase 1 functions by post-translational modifications. Front. Biosci. 2018; 23:13–26. [DOI] [PubMed] [Google Scholar]
- 113.Hurtado-Bages S., Knobloch G., Ladurner A.G., Buschbeck M.. The taming of PARP1 and its impact on NAD(+) metabolism. Mol Metab. 2020; 38:100950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Alvarez-Gonzalez R., Mendoza-Alvarez H.. Dissection of ADP-ribose polymer synthesis into individual steps of initiation, elongation, and branching. Biochimie. 1995; 77:403–407. [DOI] [PubMed] [Google Scholar]
- 115.Schuhwerk H., Bruhn C., Siniuk K., Min W., Erener S., Grigaravicius P., Krüger A., Ferrari E., Zubel T., Lazaro D.et al.. Kinetics of poly(ADP-ribosyl)ation, but not PARP1 itself, determines the cell fate in response to DNA damage in vitro and in vivo. Nucleic Acids Res. 2017; 45:11174–11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rank L., Veith S., Gwosch E.C., Demgenski J., Ganz M., Jongmans M.C., Vogel C., Fischbach A., Buerger S., Fischer J.M.et al.. Analyzing structure-function relationships of artificial and cancer-associated PARP1 variants by reconstituting TALEN-generated HeLa PARP1 knock-out cells. Nucleic Acids Res. 2016; 44:10386–10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hanzlikova H., Prokhorova E., Krejcikova K., Cihlarova Z., Kalasova I., Kubovciak J., Sachova J., Hailstone R., Brazina J., Ghosh S.et al.. Pathogenic ARH3 mutations result in ADP-ribose chromatin scars during DNA strand break repair. Nat. Commun. 2020; 11:3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ghosh S.G., Becker K., Huang H., Dixon-Salazar T., Chai G., Salpietro V., Al-Gazali L., Waisfisz Q., Wang H., Vaux K.K.et al.. Biallelic mutations in ADPRHL2, encoding ADP-Ribosylhydrolase 3, lead to a degenerative pediatric stress-induced epileptic ataxia syndrome. Am. J. Hum. Genet. 2018; 103:431–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Danhauser K., Alhaddad B., Makowski C., Piekutowska-Abramczuk D., Syrbe S., Gomez-Ospina N., Manning M.A., Kostera-Pruszczyk A., Krahn-Peper C., Berutti R.et al.. Bi-allelic ADPRHL2 mutations cause neurodegeneration with developmental delay, ataxia, and axonal neuropathy. Am. J. Hum. Genet. 2018; 103:817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mashimo M., Bu X., Aoyama K., Kato J., Ishiwata-Endo H., Stevens L.A., Kasamatsu A., Wolfe L.A., Toro C., Adams D.et al.. PARP1 inhibition alleviates injury in ARH3-deficient mice and human cells. JCI insight. 2019; 4:e124519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lüscher B., Butepage M., Eckei L., Krieg S., Verheugd P., Shilton B.H.. ADP-Ribosylation, a multifaceted posttranslational modification involved in the control of cell physiology in health and disease. Chem. Rev. 2017; 118:1092–1136. [DOI] [PubMed] [Google Scholar]
- 122.Hopp A.K., Hottiger M.O.. Uncovering the invisible: mono-ADP-ribosylation moved into the spotlight. Cells. 2021; 10:680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fahrer J., Kranaster R., Altmeyer M., Marx A., Bürkle A.. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res. 2007; 35:e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Krüger A., Bürkle A., Hauser K., Mangerich A.. Real-time monitoring of PARP1-dependent PARylation by ATR-FTIR spectroscopy. Nat. Commun. 2020; 11:2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Malanga M., Pleschke J.M., Kleczkowska H.E., Althaus F.R.. Poly(ADP-ribose) binds to specific domains of p53 and alters its DNA binding functions. J. Biol. Chem. 1998; 273:11839–11843. [DOI] [PubMed] [Google Scholar]
- 126.Kim I.K., Stegeman R.A., Brosey C.A., Ellenberger T.. A quantitative assay reveals ligand specificity of the DNA scaffold repair protein XRCC1 and efficient disassembly of complexes of XRCC1 and the poly(ADP-ribose) polymerase 1 by poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 2015; 290:3775–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Krietsch J., Caron M.C., Gagne J.P., Ethier C., Vignard J., Vincent M., Rouleau M., Hendzel M.J., Poirier G.G., Masson J.Y.. PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic Acids Res. 2012; 40:10287–10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Panzeter P.L., Zweifel B., Malanga M., Waser S.H., Richard M., Althaus F.R.. Targeting of histone tails by poly(ADP-ribose). J. Biol. Chem. 1993; 268:17662–17664. [PubMed] [Google Scholar]
- 129.Panzeter P.L., Realini C.A., Althaus F.R.. Noncovalent interactions of poly(adenosine diphosphate ribose) with histones. Biochemistry. 1992; 31:1379–1385. [DOI] [PubMed] [Google Scholar]
- 130.Andrabi S.A., Kim N.S., Yu S.W., Wang H., Koh D.W., Sasaki M., Klaus J.A., Otsuka T., Zhang Z., Koehler R.C.et al.. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:18308–18313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dasovich M., Beckett M.Q., Bailey S., Ong S.E., Greenberg M.M., Leung A.K.L.. Identifying poly(ADP-ribose)-Binding proteins with photoaffinity-based proteomics. J. Am. Chem. Soc. 2021; 143:3037–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Moor N.A., Vasil’eva I.A., Kuznetsov N.A., Lavrik O.I.. Human apurinic/apyrimidinic endonuclease 1 is modified in vitro by poly(ADP-ribose) polymerase 1 under control of the structure of damaged DNA. Biochimie. 2020; 168:144–155. [DOI] [PubMed] [Google Scholar]
- 133.Andrabi S.A., Kang H.C., Haince J.F., Lee Y.I., Zhang J., Chi Z., West A.B., Koehler R.C., Poirier G.G., Dawson T.M.et al.. Iduna protects the brain from glutamate excitotoxicity and stroke by interfering with poly(ADP-ribose) polymer-induced cell death. Nat. Med. 2011; 17:692–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Oberoi J., Richards M.W., Crumpler S., Brown N., Blagg J., Bayliss R.. Structural basis of poly(ADP-ribose) recognition by the multizinc binding domain of checkpoint with forkhead-associated and RING Domains (CHFR). J. Biol. Chem. 2010; 285:39348–39358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang Y., Liu S., Mickanin C., Feng Y., Charlat O., Michaud G.A., Schirle M., Shi X., Hild M., Bauer A.et al.. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat. Cell Biol. 2011; 13:623–629. [DOI] [PubMed] [Google Scholar]
- 136.Huang S.M., Mishina Y.M., Liu S., Cheung A., Stegmeier F., Michaud G.A., Charlat O., Wiellette E., Zhang Y., Wiessner S.et al.. Tankyrase inhibition stabilizes axin and antagonizes wnt signalling. Nature. 2009; 461:614–620. [DOI] [PubMed] [Google Scholar]
- 137.Kang H.C., Lee Y.I., Shin J.H., Andrabi S.A., Chi Z., Gagne J.P., Lee Y., Ko H.S., Lee B.D., Poirier G.G.et al.. Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:14103–14108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sukhanova M.V., Singatulina A.S., Pastre D., Lavrik O.I.. Fused in sarcoma (FUS) in DNA Repair: Tango with poly(ADP-ribose) polymerase 1 and compartmentalisation of damaged DNA. Int. J. Mol. Sci. 2020; 21:7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A.P., Kim H.J., Mittag T., Taylor J.P.. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015; 163:123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Watanabe K., Morishita K., Zhou X., Shiizaki S., Uchiyama Y., Koike M., Naguro I., Ichijo H.. Cells recognize osmotic stress through liquid-liquid phase separation lubricated with poly(ADP-ribose). Nat. Commun. 2021; 12:1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hatakeyama K., Nemoto Y., Ueda K., Hayaishi O.. Purification and characterization of poly(ADP-ribose) glycohydrolase. Different modes of action on large and small poly(ADP-ribose). J. Biol. Chem. 1986; 261:14902–14911. [PubMed] [Google Scholar]
- 142.Malanga M., Althaus F.R.. Poly(ADP-ribose) molecules formed during DNA repair in vivo. J. Biol. Chem. 1994; 269:17691–17696. [PubMed] [Google Scholar]
- 143.Braun S.A., Panzeter P.L., Collinge M.A., Althaus F.R.. Endoglycosidic cleavage of branched polymers by poly(ADP-ribose) glycohydrolase. Eur. J. Biochem. 1994; 220:369–375. [DOI] [PubMed] [Google Scholar]
- 144.Nakano T., Takahashi-Nakaguchi A., Yamamoto M., Watanabe M.. Pierisins and CARP-1: ADP-ribosylation of DNA by ARTCs in butterflies and shellfish. Curr. Top. Microbiol. Immunol. 2015; 384:127–149. [DOI] [PubMed] [Google Scholar]
- 145.Jankevicius G., Ariza A., Ahel M., Ahel I.. The toxin-antitoxin system DarTG catalyzes reversible ADP-Ribosylation of DNA. Mol. Cell. 2016; 64:1109–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Weixler L., Scharinger K., Momoh J., Lüscher B., Feijs K.L.H., Zaja R.. ADP-ribosylation of RNA and DNA: from in vitro characterization to in vivo function. Nucleic Acids Res. 2021; 49:3634–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Talhaoui I., Lebedeva N.A., Zarkovic G., Saint-Pierre C., Kutuzov M.M., Sukhanova M.V., Matkarimov B.T., Gasparutto D., Saparbaev M.K., Lavrik O.I.et al.. Poly(ADP-ribose) polymerases covalently modify strand break termini in DNA fragments in vitro. Nucleic Acids Res. 2016; 44:9279–9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Matta E., Kiribayeva A., Khassenov B., Matkarimov B.T., Ishchenko A.A.. Insight into DNA substrate specificity of PARP1-catalysed DNA poly(ADP-ribosyl)ation. Sci. Rep. 2020; 10:3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Munnur D., Ahel I.. Reversible mono-ADP-ribosylation of DNA breaks. FEBS J. 2017; 284:4002–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zarkovic G., Belousova E.A., Talhaoui I., Saint-Pierre C., Kutuzov M.M., Matkarimov B.T., Biard D., Gasparutto D., Lavrik O.I., Ishchenko A.A.. Characterization of DNA ADP-ribosyltransferase activities of PARP2 and PARP3: new insights into DNA ADP-ribosylation. Nucleic Acids Res. 2018; 46:2417–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Belousova E.A., Ishchenko capital A.C., Lavrik O.I.. Dna is a new target of Parp3. Sci. Rep. 2018; 8:4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Munnur D., Bartlett E., Mikolcevic P., Kirby I.T., Rack J.G.M., Mikoc A., Cohen M.S., Ahel I.. Reversible ADP-ribosylation of RNA. Nucleic Acids Res. 2019; 47:5658–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yu M., Schreek S., Cerni C., Schamberger C., Lesniewicz K., Poreba E., Vervoorts J., Walsemann G., Grotzinger J., Kremmer E.et al.. PARP-10, a novel Myc-interacting protein with poly(ADP-ribose) polymerase activity, inhibits transformation. Oncogene. 2005; 24:1982–1993. [DOI] [PubMed] [Google Scholar]
- 154.Hershko A., Ciechanover A.. The ubiquitin system. Annu. Rev. Biochem. 1998; 67:425–479. [DOI] [PubMed] [Google Scholar]
- 155.Livneh I., Kravtsova-Ivantsiv Y., Braten O., Kwon Y.T., Ciechanover A.. Monoubiquitination joins polyubiquitination as an esteemed proteasomal targeting signal. Bioessays. 2017; 39: 10.1002/bies.201700027. [DOI] [PubMed] [Google Scholar]
- 156.Ohtake F., Tsuchiya H.. The emerging complexity of ubiquitin architecture. J. Biochem. 2017; 161:125–133. [DOI] [PubMed] [Google Scholar]
- 157.Dikic I., Wakatsuki S., Walters K.J.. Ubiquitin-binding domains - from structures to functions. Nat. Rev. Mol. Cell Biol. 2009; 10:659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhao X., Lutz J., Hollmuller E., Scheffner M., Marx A., Stengel F.. Identification of proteins interacting with ubiquitin chains. Angew. Chem. Int. Ed. Engl. 2017; 56:15764–15768. [DOI] [PubMed] [Google Scholar]
- 159.Su Y.T., Gao C., Liu Y., Guo S., Wang A., Wang B., Erdjument-Bromage H., Miyagi M., Tempst P., Kao H.Y.. Monoubiquitination of filamin B regulates vascular endothelial growth factor-mediated trafficking of histone deacetylase 7. Mol. Cell. Biol. 2013; 33:1546–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Haglund K., Di Fiore P.P., Dikic I.. Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem. Sci. 2003; 28:598–603. [DOI] [PubMed] [Google Scholar]
- 161.Braten O., Livneh I., Ziv T., Admon A., Kehat I., Caspi L.H., Gonen H., Bercovich B., Godzik A., Jahandideh S.et al.. Numerous proteins with unique characteristics are degraded by the 26S proteasome following monoubiquitination. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E4639–E4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Grice G.L., Nathan J.A.. The recognition of ubiquitinated proteins by the proteasome. Cell. Mol. Life Sci. 2016; 73:3497–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Messick T.E., Greenberg R.A.. The ubiquitin landscape at DNA double-strand breaks. J. Cell Biol. 2009; 187:319–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Erpapazoglou Z., Walker O., Haguenauer-Tsapis R.. Versatile roles of k63-linked ubiquitin chains in trafficking. Cells. 2014; 3:1027–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chen Z.J., Sun L.J.. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell. 2009; 33:275–286. [DOI] [PubMed] [Google Scholar]
- 166.Xu P., Duong D.M., Seyfried N.T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., Peng J.. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009; 137:133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wickliffe K.E., Williamson A., Meyer H.J., Kelly A., Rape M.. K11-linked ubiquitin chains as novel regulators of cell division. Trends Cell Biol. 2011; 21:656–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gatti M., Pinato S., Maiolica A., Rocchio F., Prato M.G., Aebersold R., Penengo L.. RNF168 promotes noncanonical K27 ubiquitination to signal DNA damage. Cell Rep. 2015; 10:226–238. [DOI] [PubMed] [Google Scholar]
- 169.Al-Hakim A.K., Zagorska A., Chapman L., Deak M., Peggie M., Alessi D.R.. Control of AMPK-related kinases by USP9X and atypical Lys(29)/Lys(33)-linked polyubiquitin chains. Biochem. J. 2008; 411:249–260. [DOI] [PubMed] [Google Scholar]
- 170.Lutz J., Hollmuller E., Scheffner M., Marx A., Stengel F.. The length of a ubiquitin chain: A general factor for selective recognition by ubiquitin-binding proteins. Angew. Chem. Int. Ed. Engl. 2020; 59:12371–12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Reyes-Turcu F.E., Shanks J.R., Komander D., Wilkinson K.D.. Recognition of polyubiquitin isoforms by the multiple ubiquitin binding modules of isopeptidase T. J. Biol. Chem. 2008; 283:19581–19592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bavikar S.N., Spasser L., Haj-Yahya M., Karthikeyan S.V., Moyal T., Kumar K.S., Brik A.. Chemical synthesis of ubiquitinated peptides with varying lengths and types of ubiquitin chains to explore the activity of deubiquitinases. Angew. Chem. Int. Ed. Engl. 2012; 51:758–763. [DOI] [PubMed] [Google Scholar]