Abstract
Cytosine base editor (CBE) enables targeted C-to-T conversions at single base-pair resolution and thus has potential therapeutic applications in humans. However, the low efficiency of the system limits practical use of this approach. We reported a high-throughput human cells-based reporter system that can be harnessed for quickly measuring editing activity of CBE. Screening of 1813 small-molecule compounds resulted in the identification of Ricolinostat (an HDAC6 inhibitor) that can enhance the efficiency of BE3 in human cells (2.45- to 9.21-fold improvement). Nexturastat A, another HDAC6 inhibitor, could also increase BE3-mediated gene editing by 2.18- to 9.95-fold. Ricolinostat and Nexturastat A also boost base editing activity of the other CBE variants (BE4max, YE1-BE4max, evoAPOBEC1-BE4max and SpRY-CBE4max, up to 8.32-fold). Meanwhile, combined application of BE3 and Ricolinostat led to >3-fold higher efficiency of correcting a pathogenic mutation in ABCA4 gene related to Stargardt disease in human cells. Moreover, we demonstrated that our strategy could be applied for efficient generation of mouse models through direct zygote injection and base editing in primary human T cells. Our study provides a new strategy to improve the activity and specificity of CBE in human cells. Ricolinostat and Nexturastat A augment the effectiveness and applicability of CBE.
INTRODUCTION
The CRISPR/Cas system of bacterial adaptive immunity has been adapted to be a powerful and versatile tool for manipulating the genome, and has been widely applied in biological research and preclinical treatments with animal or cellular disease models (1–3). Base editing is a newly developed genome engineering tool that enables gene editing through irreversible base conversion without induction of double-stranded DNA breaks (DSBs) (4,5), which may be further explored as a novel gene therapy approach. Cytosine base editor (CBE), containing engineered cytosine deaminase with CRISPR/Cas9, can be used to modify genomes from different species and genetic screening by inducing C•G to T•A conversion (4,6,7).
Remarkable progress has been made to increase the targeting scope, editing specificity, and fidelity of CBE. Specifically, engineered Streptococcus pyogenes SpCas9 and Staphylococcus aureus SaCas9 variants with altered protospacer adjacent motif (PAM) compatibility have been adopted for CBE-mediated genome editing (8). Recently, intensive efforts have been made to address the target scope and/or specificity, including evolved or additional deaminase, Cas9 variants (or additional Cas enzymes (i.e. Cas12a)) and dual-deaminase CRISPR base editor (9–16). As half of reported pathogenic variants are point mutations and the majority (∼60%) of these are transition mutations (4), base editors are being widely utilized to study and treat human genetic diseases in animal models (5,17).
However, correction of a point mutation directed by CBE has remained inefficient, which greatly limits applications of this system. Therefore, identification of compounds that could improve the efficiency of CBE-mediated genome editing is highly desired. Small molecules can regulate DNA repair, and thus may be valuable for modulation of genome editing. SCR7, an inhibitor of LIG4 that could suppress nonhomologous end-joining (NHEJ), has been shown to improve the homology-directed repair efficiency of CRISPR/Cas9 (18). Similarly, RS-1, a small molecule stimulating RAD51, which is a key homologous recombination (HR) factor, increases CRISPR/SpCas9 editing efficiency (19,20), and BRD0539 can abolish CRISPR/SpCas9-mediated gene editing (21). Taken together, these lines of evidence show that there are likely to be small molecules that can increase the efficiency of CBE-mediated genome editing, and we sought to identify one or more of them in this study.
MATERIALS AND METHODS
Construction of BE3 and sgRNA expression vectors
Plasmids for BE3 were originally from Professor Caixia Gao (Chinese Academy of Sciences; Addgene #98164). We cloned the APOBEC1-nCas9-UGI fragment and U6-BFP-sgRNA to generate plasmids pBE3-sgRNA. Plasmids PX330-NG for the expression of SpCas9-NG was a gift from Osamu Nureki (Addgene #117919). pCMV-BE4max (Addgene #112093), YE1-BE4max (Addgene #138155), evoAPOBEC1-BE4max (Addgene #122611) and engineered SpRY-CBE4max vectors (original Addgene #139999) without EGFP fragment were used for the study.
The plasmids expressing BE3-NG were generated based on the replacement of partial coding sequence of nCas9 with the corresponding sequence of SpCas9-NG via PshAI. The vector plasmids (pSin-BFP-IRES-PURO) were generated based on the parental vector pSin-EFIα-EGFP-IRES-PURO with a point mutation. The pDonor-mT/mG plasmids were originally from Dr. Murry Charles (University of Washington) and contain the following: mTomato, mEGFP, and two homologous arms. We generated pDonor-BFP-IRES-PURO by replacing mTomato and mEGFP cassettes with BFP and PURO. pDonor-mT-ABCA4 was generated by replacing EGFP cassette with a partial ABCA4 coding region. Plasmid DNA was isolated by standard techniques. DNA sequencing confirmed the desired specific sequences in the constructs.
Cell culture
HEK-293 cells were obtained from ATCC (CAT#CRL-1573) and were cultured as previously described (22). To generate AAVS1 knock-in cell lines containing BFP expression cassettes, HEK-293 cells were seeded on day 0 at 2.5 × 105 cells in six-well plates, and on day 1, pDonor-BFP-IRES-PURO and pX330-AAVS1 plasmids were transfected by TurboFect Transfection Reagent (Thermo Fisher Scientific, MA). Cells were selected with puromycin (0.9 μg/ml) and individual colonies were picked under the microscope at day 15. PCR was used for the confirmation of knock-in at the AAVS1 locus with specific primers, which is shown in Supplementary Figure S2A and Table S2. The cell line with the BFP expression cassette knock-in at the AAVS1 locus was named HEK-293-AAVS1-BFP. The HEK-293-AAVS1-ABCA4 cell line was generated with a similar approach. Individual red fluorescent colonies were picked under the fluorescent microscope. HeLa and HT1080 cells were obtained from ATCC (CAT#CCL-2 & CCL-121) and cultured as HEK-293. The initial concentration for small molecule screening was 10 μM. After seeding cells for 24 h, cells were treated with small molecules for 48 or 72 h.
Chemical reagents
The compound library (Catalog No. L1000, Supplementary Table S3) was commercially available at Targetmol company. The HDAC inhibitors Ricolinostat, Nexturastat A, Citarinostat, Droxinostat, Entinostat, Panobinostat, Quisinostat 2HCl, TMP269, Tubacin, Tucidinostat were purchased from TargetMol (Boston, MA, USA).
Flow cytometry analysis
The flow cytometry (FCM) protocol was described previously (22). On day 0, 0.9 × 105 HEK-AB cells were seeded in 24-well plates. On day 1, the cells were transfected with 250 ng BE3 (harboring sgRNA and BE3 enzyme coding cassette) plasmids or 300 ng of plasmids (100 ng of sgRNA and 200 ng of CBE variants) with TurboFect Transfection Reagent (1.5 μl), and then treated with individual compounds. On day 3, cells were harvested for flow cytometry and genomic DNA isolation. The EGFP positive percentage was obtained via flow cytometry (BD Biosciences, NY) and CellQuest software was used to analyze data. To determine the optimized time point for cell collection, cells were harvested for flow cytometry on day 2, 3, 4, 5, and 6.
Sanger sequencing for pJET colonies
Sequence flanking the CRISPR target sites for BFP and ABCA4 was PCR amplified, and products were inserted into the vector pJET1.2 (CloneJET PCR Cloning Kit, Thermo Fisher Scientific). The ligated products were transformed into Escherichia coli. The corresponding plasmids were isolated and then sequenced on an ABI PRISM 3730 DNA Sequencer.
Animals
All animal procedures were approved by the Institutional Animal Care and Use Committee of the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, China. Mice were housed in individual ventilated cages (IVC) in an accredited specific pathogen free facility under a 12 h dark-light cycle condition. Zygotes were collected from female mice (B6D2F1 (C57BL/6♀× DBA2♂)) that were mated to male Oct4-eGFP (C57BL/6 background). ICR females were used as pseudo-pregnant foster mothers.
In vitro transcription
BE3 mRNA transcriptional templates were amplified by PCR using KOD-Plus-Neo (TOYOBO) from plasmids pCMV-BE3 (Addgene #73021), purified by the Universal DNA Purification Kit (TIANGEN) and then transcribed using the mMACHINE T7 ULTRA transcription Kit (Invitrogen) following the manufacturer's instructions. The transcriptional templates of sgRNAs were amplified from Px330-mCherry (Addgene #98750) and transcribed in vitro using the MEGAshortscript T7 kit (Invitrogen) following the manufacturer's instructions. mRNAs and sgRNAs were subsequently purified with the MEGAclear Transcription Clean-Up Kit (Invitrogen), resuspended in hot (95°C) RNase-free water and then stored at −80°C.
Zygote microinjection and culture
Eight-week-old B6D2F1 female mice were superovulated with 6 international units of pregnant mare's serum gonadotropin (PMSG) for 48 h and then injected human chorionic gonadotropin (hCG), subsequently mated to homozygous Oct4-eGFP males for 12 h. Zygotes were harvested from oviducts of B6D2F1 females with plug 24 h post hCG injection using hyaluronidase (Sigma). For microinjection, the mixture of BE3 mRNA (100 ng/μl) and sgRNA (10, 20, 50 and 100 ng/μl) was diluted in RNAase-free water, centrifuged at 4°C, 13 400g for 10 min, and then injected into the cytoplasm of zygotes in a droplet of HCZB medium containing 5 μg/ml cytochalasin B (CB, Sigma) using a micromanipulator (Olympus) and a FemtoJet microinjector (Eppendorf). The injected zygotes were cultured for 16 or 24 h to two-cell embryos in AA-KSOM (Millipore) medium with different concentration of Ricolinostat or Nexturastat A, then cultured in normal AA-KSOM medium for 72 h to blastocyst stage at 37 °C under 5% CO2 in air. Two-cell embryos were transferred into oviduct of pseudopregnant ICR females at 0.5 day post copulation for generating mouse models as previously described (23).
Blastocyst genotyping
Mouse blastocysts were directly lysed by 5 μl buffer from the Mouse Direct PCR Kit (Bimake), incubated at 55°C for 50 min and 95°C for 5 min. The targeted region was amplified by PCR using Phanta® Max Super-Fidelity DNA Polymerase (Vazyme) from blastocyst lysis (about 2 μl), purified by gel electrophoresis using Universal DNA Purification Kit and then Sanger sequencing and Next-generation sequencing (NGS).
NGS and data analysis
Amplicons for next-generation sequencing were generated using two rounds of PCR. Equal amounts of the PCR amplicons were subjected to paired-end read sequencing using the Hiseq 1500 platform (Illumina, San Diego, CA, USA) at Novogene (Tianjin, China). Adaptors and low-quality reads were removed from the resulting 150-bp paired-end reads using Trimmomatic (version 0.36) (24). Reads were then mapped to the template using Bowtie2 (version 2.3.3). All NGS-data obtained with this method in this paper.
Cell viability assay
Cell viability was evaluated by cell counting kit‐8 (CCK‐8; DojindoCo, Kumamoto, Japan). Approximately 1 × 104 HEK-293, HT1080 or HeLa cells were seeded in 96‐well plates and incubated for 24 h. Then, cells were treated with 1.25, 2.5, 5, 10 or 20 μM of Ricolinostat or Nexturastat A for 24 h. After washing the cells with PBS, 100 μl of DMEM containing 10 μl of CCK-8 solution was added to the cells of each well. The plates were then incubated for 1–2 h, and the absorbance per well was measured at 450 nm using a microplate reader (Thermo, Rockford). As to cell viability assay (data of Supplementary Figures S1, S3, S4 and S6), cells were treated as those collected for FCM.
BE3-mediated gene editing in human primary T cells
CD3+ T cells were isolated from umbilical cord blood (Beijing Cord Blood Bank), activated with anti-CD3/anti-CD28 Dynabeads (Thermo Fisher Scientific), and expanded in X-VIVO15 medium (Lonza) supplemented with 5% (v/v) heat-inactivated FBS (Gibco) and 300 IU/mL recombinant human IL-2 (Sino Biological Inc.) as previously described (25). After 3 days of activation, the activated T cells were collected for electroporation. After removing beads, 1 × 106 T cells were resuspended in 20 μl transfection buffer containing 1 μg endotoxin-free plasmid coding BE3 and sgRNA targeting EMX1 site and transferred into the electroporation cuvette. P3 Primary Cell 4D-Nucleofector X Kit (V4XP-3024, Lonza), and program EO-115 were used for T cell electroporation. After electroporation, cells were resuspended in pre-warmed medium containing 2.5 μM Ricolinostat or Nexturastat A or solvent (DMSO) only as control, transferred into culture plates, and incubated at 37°C in an atmosphere of 5% CO2. Three days later, the electroporated cells were collected for genomic DNA extraction (QIAGEN). Then, targeted gene fragments were amplified and purified for NGS.
Knock-out and detection of HDAC6
SpCas9 and two sgRNAs were used to knockout HDAC6 as previously described (26). The primers for constructing the vectors are listed in Supplementary Table S2. For Western blotting, a total of 40 μg proteins per lane were separated by SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF) membrane. The membrane was then incubated overnight at 4°C with anti-HDAC6 antibody (ABclonal, Wuhan, China, #A11259) or anti-β-Actin antibody (ABclonal, Wuhan, China, #AC026) at a dilution of 1:2000 or 1:50 000, respectively. Protein bands were visualized using a Typhoon FLA 7000 (GE, Pittsburgh, PA, USA).
Measurement of protein and sgRNA expression levels
To determine the expression levels of BE3 protein, 400 ng plasmids encoding BE3 and 200 ng plasmids encoding sgRNA (targeting HEK-Site 3 or RNF2) were transfected in 1.8 × 105 HEK-293 cells and 2.5 μM Ricolinostat or Nexturastat A or solvent (DMSO) was applied after transfection. Cells were collected 72 h after transfection to measure BE3 and sgRNA expression levels. For Western blotting, anti-Cas9 antibody (ABclonal, Wuhan, China, #A14997) or anti-β-Actin antibody (ABclonal, Wuhan, China, #AC026) were used at a dilution of 1:10 000 or 1:50 000, respectively. Western blotting results were analyzed with ImageJ. To determine the sgRNA expression levels, total RNA was isolated using MiniBEST Universal RNA Extraction Kit (TaKaRa) and 500 ng of total RNA was reverse transcribed using PrimeScript RT Master Mix (TaKaRa) according to the manufacturer's protocol. Quantitative PCR was preformed using Power SYBR Green PCR Master Mix (ABI) on a QuantStudio 3 Real-Time PCR System (ABI). The expression levels of sgRNAs were normalized with that of GAPDH. All primers used for qRT-PCR are listed in Supplementary Table S2.
ATAC-seq assay and data analysis
To detect the chromatin accessibility, 1.8 × 105 HEK-293 cells in 12-well plate were transfected with 600 ng of plasmids (200 ng of HEK-Site4-sgRNA and 400 ng of YE1-BE4max encoding plasmids) and treated with 2.5 μM Ricolinostat. Cells were collected 72 h after transfection for ATAC-seq assay using the NovaSeq 6000 platform (Illumina, San Diego, CA, USA) and analyzed with standard protocol.
Quantification and statistical analysis
All data were expressed as mean ± s.e.m. Differences were determined by 2-tailed Student's t-test between two groups, or one-way ANOVA followed by post-hoc Bonferroni test for multiple groups. The criterion for statistical significance was *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
RESULTS
Performance of the episomal BFP to GFP conversion reporter system
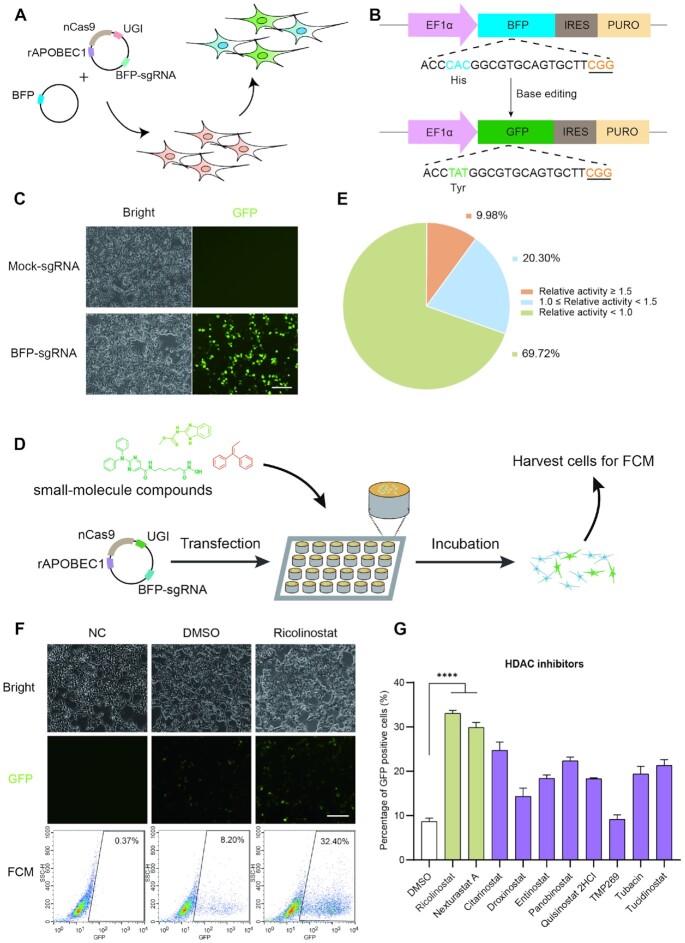
To identify small-molecule compounds that can boost CBE-mediated genome editing in human cells, we cannot directly employ our available reporter systems (22,27,28). Recently, we developed a simple and quick method to assess base editing in human cells (29), while, it still cannot be assigned as a high-throughput screening assay due to multiple steps in the whole process. Thus, a convenient reporter system should be established. To address it, we generated a CBE-mediated reporter system based on the BFP to GFP conversion (30), which could be easily tracked with flow cytometry (FCM). Two plasmids were transfected into the cells, including an all-in-one plasmid (pBE3-sgRNA) targeting a sequence harboring the CAC encoding residue (histidine) of BFP, and a plasmid encoding BFP (Figure 1A). Theoretically, base editing of codon 66 from CAC (histidine) to TAC or TAT (tyrosine) would be performed via CBE mediated editing at position 4 (C4) of the protospacer (counting the PAM as positions 21–23) with/without editing of C6 (Figure 1B). As we expected, the results showed that, after the editing, the percentage of GFP positive cells gradually increased over the first three days without obvious toxicity, especially in the first two days (Figure 1C and Supplementary Figure S1A–C). At 48 h post transfection, 33.04% cells are GFP-positive, compared with no GFP-positive cells in the control group. Collectively, the above results showed that CBE-mediated reporter system was amenable to high-throughput analysis via FCM.
Figure 1.
Establishment of a reporter system to identify small molecules increasing gene-editing efficiency of CBE in human cells. (A) Schematic diagram of BFP to GFP conversion reporter system for evaluation of BE3-mediated gene-editing efficiency. (B) Schematic illustration of BFP to GFP sequence editing by CBE. The target sequence is shown. The special residues of His and Tyr are highlighted in blue and green, respectively. The underlined nucleotide represents the PAM. (C) Fluorescence images of BFP to GFP conversion. Scale bar, 10 μm. (D) Illustration of drug screening platform in the present study. (E) The summary of activity of the tested drugs. (F) Representative images and raw flow cytometry data of Ricolinostat treatment group. Scale bar, 10 μm. (G) Additional HDAC inhibitors modulating BE3 efficiency. All of the compounds were used in 5 μM, except TMP269 (2.5 μM), Panobinostat (100 nM) and Quisinostat 2HCl (100 nM). Error bars, S.E.M.; n = 3; NC, negative control; Control, DMSO treated group; ****P < 0.0001.
Generation of the BFP to GFP conversion reporter system at the human AAVS1 locus
The study described above used a target sequence on an episomal plasmid. Next, to test whether the system would still be functional if the target sequence was located at a human chromosomal locus, we inserted a BFP to GFP reporter cassette at the human AAVS1 locus on chromosome 19 (31) (Supplementary Figure S2A). AAVS1 (also known as the PPP1R12C locus) is a well-validated ‘safe harbor’ for hosting DNA transgenes. It has an open chromatin structure and contains native insulators that prevent silencing of the integrated genes. Also, there are no known adverse effects on cells as a result of DNA fragment insertion (31). For these reasons, we selected AAVS1 as a targeting site for harboring the BFP to GFP reporter cassette.
We designed one sgRNA targeting the AAVS1 locus to introduce DSBs and the pDonor-BFP-IRES-Puro plasmid, which carries two arms homologous to the AAVS1 locus as well as BFP and the puromycin resistance gene. To screen for positive colonies, we picked the colonies after puromycin selection for one week and confirmed them with gene-specific PCR. Among 14 colonies, all except one are PCR-positive (Supplementary Figure S2B).
BE3-mediated BFP to GFP editing in HEK-293-AAVS1-BFP cell line
We selected four positive colonies (HEK-293-AAVS1-BFP-7, 8, 10 and 11) for testing. The plasmids (pBE3-sgRNA) were transfected into the cells, and the editing efficiency was measured by identifying GFP-positive cells with FCM on day 3. Cells from different colonies possess different editing efficiency for C4 (HEK-293-AAVS1-BFP-7, 10.82%; HEK-293-AAVS1-BFP-8, 6.41%; HEK-293-AAVS1-BFP-10, 6.64% and HEK-293-AAVS1-BFP-11, 14.06%; respectively). Thus, HEK-293-AAVS1-BFP-11 (also called HEK-AB) was chosen to perform further studies due to the highest level of base editing. Sanger sequencing results confirmed the base editing events (from BFP to GFP, Supplementary Figure S3A).
We observed a dose-dependent effect of plasmid transfection (Supplementary Figure S3B). Specifically, the higher the dose, the higher the editing efficiency we obtained (4.28% with 200 ng and 43.16% with 400 ng). Not surprisingly, excessive dosage leads to the cytotoxicity (Supplementary Figure S3C). Considering this toxicity and the potential for further improvement, we chose a 250 ng plasmid dose for further study.
Drug screening for improving efficiency of BE3-mediated gene editing
To screen for small-molecule compounds that increased BE3-mediated gene editing, we took advantage of the above method as a high-throughput screening platform via FCM. At day 2, HEK-AB cells were transfected with one plasmid for expressing BE3 and sgRNA, and then treated with individual compounds. At day 4, we harvested cells to assess the editing efficiency via FCM (Figure 1D). After initial testing, we found 181 compounds that increased the editing efficiency by at least 1.5-fold compared with the DMSO control (Figure 1E).
To confirm the results, we performed second-round screening for these 181 compounds, which led to the identification of 24 compounds with higher editing activity but without notable cytotoxicity. We tested two different concentrations (4 and 20 μM) of each compound to test dose-dependent effects. Only one compound (Ricolinostat) substantially improved the editing efficiency (3.18-fold at 4 μM) with slight toxicity (Supplementary Figure S4A–D). Compared with Ricolinostat, due to the relative low improvement effects of the remaining compounds, we did not perform additional studies on the rest. In summary, here we identified Ricolinostat as a candidate small-molecule compound to boost BE3-mediated gene editing (Figure 1F and Supplementary Figure S5A).
Ricolinostat enhances BE3-mediated gene editing in a dose-and time-dependent manner
To optimize the conditions for Ricolinostat action, we tested different concentrations and found it achieved maximal editing effects (editing efficiency of 32.41%) at 5 μM. Even at the concentration of 2.5 μM, it still increased the efficiency of BFP to GFP by 2.89-fold without notable cytotoxicity (Supplementary Figure S6A and C). The augmented effects were further confirmed with next-generation sequencing (NGS, Supplementary Figure S5B). We also observed the indels triggered with BE3, while there is no indels increase with the treatment of Ricolinostat (Supplementary Figure S5C).
To investigate the most effective timing of Ricolinostat treatment, we set the transfection at 0 h and treated the cells at different relative times (pre24, pre12, post0, 2, 4, 6, 8, 10, 12 and 24 h). The results revealed that Ricolinostat increases the editing efficiency significantly when added 0–12 h after transfection. The 24 h treatment group also showed 1.59-fold increase without notable toxicity (Supplementary Figure S6B and D). An understanding of toxicity is critical for clinical application of this system. With a CCK-8 assay, we found that Ricolinostat exhibited only slight toxicity in three different cell lines (HEK-293, HT1080 and HeLa) at the 2.5 μM concentration (Supplementary Figure S7A-C).
Because it is reported that Ricolinostat is an inhibitor of histone deacetylase (HDAC) (32), which is 10-fold more selective towards HDAC6 and also affects other class I HDACs, we then asked whether additional inhibitors of HDAC family members could have similar effects. We selected nine small-molecule inhibitors that target HDAC1-11 for testing. Not surprisingly, Nexturastat A and Citarinostat, both HDAC6 inhibitors, could also increase BE3-mediated gene editing by 3.44-fold and 2.84-fold, respectively (Figure 1G). Other HDAC inhibitors also increased the efficiency of BE3, although less effectively than Ricolinostat or Nexturastat A (Figure 1G). Cytotoxicity of Nexturastat A was very similar to that of Ricolinostat, only showing slight toxicity at 2.5 μM concentration (Supplementary Figure S7A–C). Collectively, here we demonstrated that two HDAC6 inhibitors (Ricolinostat and Nexturastat A) may be harnessed to increase BE3’s efficiency.
Ricolinostat and Nexturastat A enhance BE3-mediated gene editing at endogenous gene loci in human cells
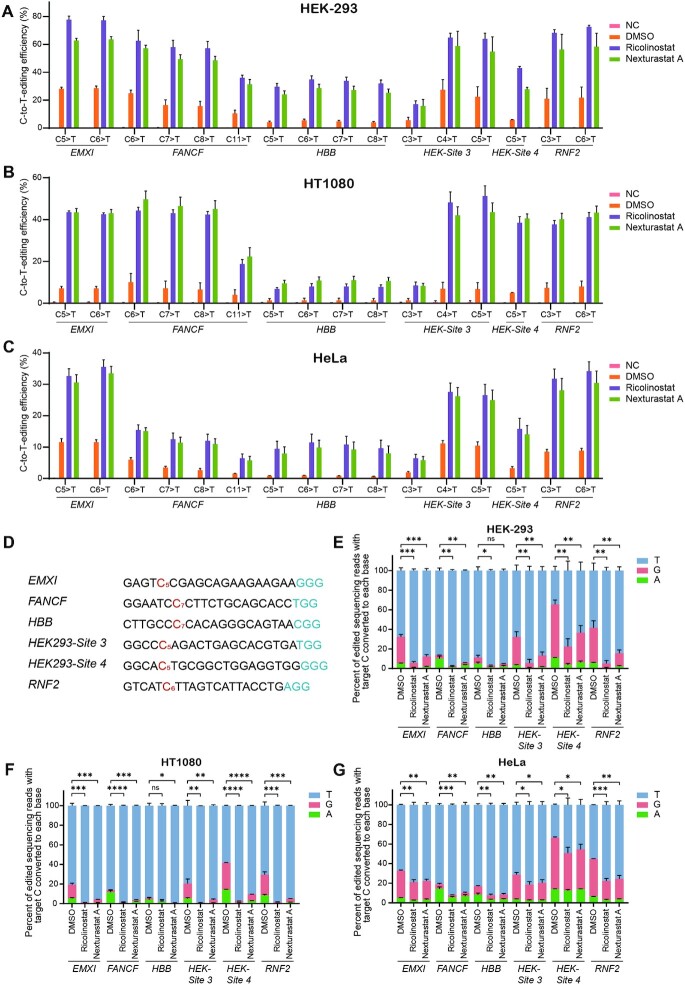
To further confirm the enhancement effects, 6 sites (EMX1, FANCF, HBB, HEK-Site3, HEK-Site4, RNF2, Supplementary Table S1) from human endogenous genes were selected and tested. Compared with DMSO, both Ricolinostat and Nexturastat A showed enhancing effects (an average of 4.24-fold and 3.46-fold increase, respectively) at these sites in HEK-293 cells (Figure 2A and Supplementary Figure S8A). Specifically, Ricolinostat achieved 5.50 and 5.90-fold higher editing in C7 and C8 (Supplementary Figure S8A). To exclude direct interactions between drugs and transfection reagents, we changed growth medium 10 h post-transfection and then added treatment compounds. Under these conditions, the enhancement effects were similar (2.61-fold for Ricolinostat and 2.28-fold for Nexturastat A; Supplementary Figure S8B). To test whether the system also works well in other cell types, we tested additional cell lines (HT1080 and HeLa cells) at these loci. Our results from HT1080 cells showed that, compared to control, Ricolinostat and Nexturastat A significantly improved the efficiency of gene editing averaging 7.28-fold and 7.96-fold, respectively (Figure 2B and Supplementary Figure S8C). Similar results were observed with HeLa cells (Ricolinostat and Nexturastat A increased editing 5.55-fold and 4.90-fold, respectively; Figure 2C and Supplementary Figure S8D).
Figure 2.
Ricolinostat and Nexturastat A increased BE3 efficiency and specificity at endogenous loci. (A–C) The C-to-T base editing efficiencies with treatment of Ricolinostat or Nexturastat A in HEK-293, HT1080 and HeLa cells, respectively. (D) Protospacers and PAM (blue) sequences of genomic loci studied (EMX1, FANCF, HBB, HEK-Site 3, HEK-Site 4 and RNF2), with the target C’s analyzed in (A–C) shown in red. After editing, one C with relative lower C to T conversion purity was chosen as target C of each site. (E–G) HEK-293, HT1080 and HeLa cells were treated with Ricolinostat, Nexturastat A and DMSO (control). The composition of edited DNA sequencing reads (reads in which the target C is mutated) is shown. Error bars, S.E.M.; n = 3; NC, negative control; Control, DMSO treated group; ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
With further analysis, surprisingly, we did observe that C-to-non-T (including C-to-A/G) editing was remarkably decreased (2.91- to 8.01-fold for HEK-293 cells, 2.43- to 21.78-fold for HT1080 cells, and 1.31- to 2.36-fold for HeLa cells; Figure 2D–G), which indicated the specificity of the editing has been considerably improved with Ricolinostat treatment. We also analyzed indel frequencies in three different cell lines, and found there is no notable difference between the treatment and control (Supplementary Figure S9A–C).
To test whether Ricolinostat and Nexturastat A may increase off-target effects, we amplified fragments harboring the target site by PCR with specific primers (Supplementary Table S2) and performed NGS to detect eight potential off-target sites (Supplementary Table S1) for one on-target site. We observed a slight increase in editing at four of the 48 off-target sites when comparing treatment to control (Supplementary Figure S10A), but the ratio of on-target to off-target editing was comparable at all four sites (Supplementary Figure S10B).
Ricolinostat and Nexturastat A increase base editing activity of the additional CBE variants and ABE
Engineered CBE variants were recently developed (9,11,33,34). We asked whether Ricolinostat and Nexturastat A could also improve the base editing efficiencies of additional CBE variants (BE4max, YE1-BE4max, evoAPOBEC1-BE4max and SpRY-CBE4max). We tested four CBE variants with BFP reporter system and three endogenous loci (FANCF, HBB, HEK-Site 4). The results showed that Ricolinostat can enhance the efficiency of CBE variants in BFP reporter system (1.57- to 6.57-fold improvement, Supplementary Figure S11A) and at endogenous loci (1.50- to 8.32-fold improvement, Supplementary Figure S12A–C). Nexturastat A has the silimiar results (Supplementary Figures S11A and S12A-C). Five mismatched sgRNAs for BFP plus BE4max or YE1-BE4max were transfected into the HEK-AB cells, respectively. Off-target effects were measured via FCM. We found that, with the treatment of Ricolinostat or Nexturastat A, off-targets have been slightly increased (Supplementary Figure S11B). We also investigated four potential DNA off-target sites as previously reported for FANCF and HEK-Site 4, respectively (9,10), four potential RNA off-target sites for RNF2 and HEK-Site 3 as well (35) (Supplementary Table S1). With the treatment of Ricolinostat or Nexturastat A, the DNA and RNA off-target effect triggered with YE1-BE4max was not detectable (Supplementary Figures S12D and S13A, B).
Of importance, consistent with our previous results, the editing product purity was also improved, especially with YE1-BE4max mediated editing at HEK-Site 4 (Supplementary Figure S14A-C). We sought to know whether Ricolinostat or Nexturastat A could increase the editing efficiency of ABE. To address it, we analyzed the base editing efficiencies of ABEmax-NG at six endogenous target sites (Supplementary Table S1) and found that Ricolinostat and Nexturastat A did enhance the base editing efficiencies of ABEmax-NG by up to 5.59-fold (46.75% versus 8.44% without treatment) at the ABE-Site 12 (Supplementary Figure S15). Collectively, these results indicated that Ricolinostat and Nexturastat A improved the base editing efficiencies of CBE and ABE.
Boosting activity with HDAC6 inhibitors for generation of mouse disease model
To evaluate the effect of these small molecules for the generation of mouse disease model, we designed an sgRNA to target an integrated eGFP in the mouse genome, introducing an early stop codon at amino acid 183 (Supplementary Figure S16A). Then we injected BE3 mRNA and GFP-sgRNA into eGFP expressing zygotes, and treated mouse embryos with two concentrations (1 μM and 2.5 μM) of Ricolinostat and Nexturastat A for 24 h (Supplementary Figure S16B and C). The eGFP signal of blastocysts indicates that Ricolinostat and Nexturastat A improved editing efficiency (Supplementary Figure S16D). Next, we subjected those embryos to Sanger sequencing and NGS, and found that Ricolinostat and Nexturastat A notably enhanced editing efficiency (Supplementary Figure S16E and F). Not surprisingly, we observed high concentrations of these compounds, especially Ricolinostat, to be toxic to pre-implantation embryo development (Supplementary Figure S16C and D). To minimize the toxicity to embryos and improve the efficiency of gene editing, we investigated different lengths of treatment time (16 h and 24 h) and concentrations (0.25, 0.5, 1 and 2.5 μM) of Nexturastat A (Supplementary Figure S17A). The fluorescence intensity and sequencing results illustrated that higher concentration does lead to the higher editing efficiency (Supplementary Figure S17B–D). Notably, we observed no embryo toxicity of treatment with 0.5 μM Nexturastat A for 24 h, while maintaining the high gene editing efficiency (1.57-fold improvement, Supplementary Figure S17A and D).
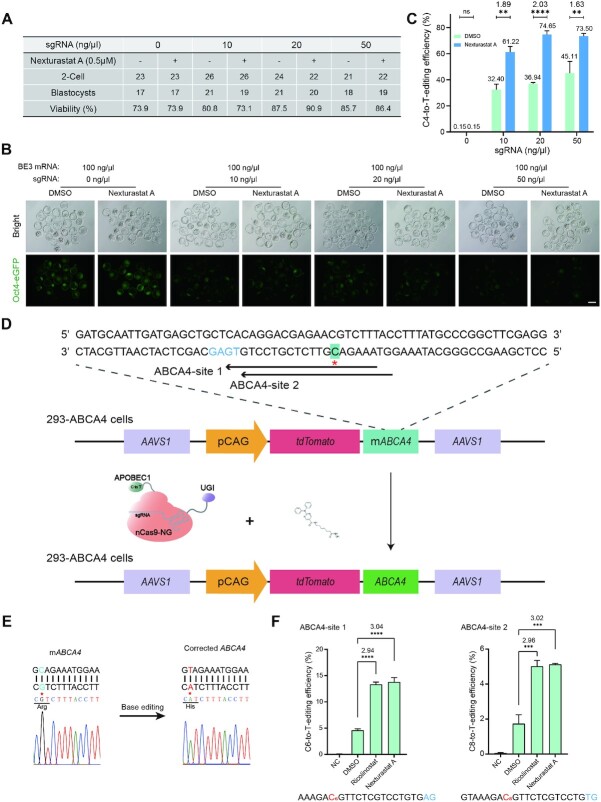
To further verify Nexturastat A’s augmenting effectiveness, we decreased the injection amounts of sgRNA (Figure 3A). The results showed that Nexturastat A had a better boosting effect (1.63- to 2.02-fold improvement, Figure 3B and C). To assess the feasibility of Nexturastat A for promoting generation of mouse disease model, we designed an sgRNA targeting the c.202 of Tyr, which would introduce a premature stop codon at the 68 amino acid and generate an albinism mouse model (Supplementary Figure S18A). Nexturastat A did not affect pre-implantation embryo development or birth rate in mice (Supplementary Figure S18B and C). Sanger sequencing and NGS results revealed that Nexturastat A significantly improved C-to-T conversion at the target site (Supplementary Figure S18D-G). On-target editing frequency with the treatment of Nexturastat A was 1.20- and 1.23-fold higher in blastocysts and mice, respectively, compared to control (Supplementary Figure S18E and G). Morphological analysis showed that albino mice were observed from 30 out of 40 (75.0%) or 34 out of 39 (87.2%) treated zygotes in the DMSO or Nexturastat A group, respectively (Supplementary Figure S18F). Importantly, we also observed that the number of mice containing C-to-non-T editing was remarkably decreased (from 10 to 6, Supplementary Figure S18H), which is consistent with the results of endogenous gene editing in three cell lines with Ricolinostat treatment (Figure 2D–G). We also investigated whether indel percentage would be affected with the treatment of Nexturastat A. We analyzed each mouse generated via BE3 with or without treatment of Nexturastat A. We found that the total number of mice harboring indels was not affected by Nexturastat A as well as the indel percentage of the mice with albinism phenotype (nine of each group, Supplementary Figure S18H). Thus, these results demonstrate that Nexturastat A can improve BE3-mediated base editing in the resultant mice through zygotic cytoplasm injection. We suspected that the relative lower augmenting effect of Nexturastat A may be due to mRNA and microinjections in mice experiments rather than plasmids and transfection in cultured cells studies.
Figure 3.
Nexturastat A significantly boosted BE3-mediated gene editing in mouse embryos and correction of ABCA4 mutation. (A) Summary of the viability of blastocysts from zygotes injected with BE3 mRNA and different concentration of eGFP-sgRNA, and treated with Nexturastat A (0.5 μM). (B) Fluorescence images of blastocysts with the editing. Scale bar, 100 μm. (C) NGS results showed C to T conversion rate in obtained embryos at eGFP locus (p.Q183X). Total embryos from each group were randomly divided into three groups. (D) A schematic of BE3-NG-mediated gene correction of ABCA4 (CAT to CGT missense mutation). The target base is shown in green, and the PAM is shown in blue. (E) Sanger sequencing results of the colonies showed that the mutant ABCA4 (mABCA4) gene was corrected by BE3-NG. The mutated and corrected nucleotide is highlighted with red star. (F) The C-to-T base editing frequencies at ABCA-site 1 and ABCA-site 2. mABCA4, pCAG represents partial human ABCA4 fragments harboring mutation and CAG promoter, respectively. Error bars, S.E.M.; n = 3; Control, DMSO treated group; ns, not significant; **P < 0.01, ***P < 0.001, ****P < 0.0001.
Correction of a pathogenic mutation and genome manipulation of primary human T cells
Next, we asked whether it could be utilized to correct disease-causative mutations. Fragment harboring one mutation (p.H2032R) in ABCA4 from Stargardt disease (STGD) patients has been inserted into the AAVS1 locus in HEK-293 cells (36), one of the most common inherited disorder that usually causes vision loss in childhood or adolescence (Figure 3D). Thus, the HEK-293-AAVS1-ABCA4 (also called 293-ABCA4) cell line harbors a partial ABCA4 coding region carrying a causative mutation at the AAVS1 locus. As there is no 5′-NGG-3′ PAM around the mutation, we designed two sgRNAs (C6 for ABCA4-site 1 and C8 for ABCA4-site 2) that contain a 5′-NG-3′ PAM to test whether Ricolinostat and Nexturastat A could improve the activity of BE3-NG mediated gene correction. Sanger sequencing results of pJET colony confirmed the base editing events (Figure 3E). The NGS results showed that Ricolinostat and Nexturastat A substantially enhanced BE3-NG efficiency at both sites. Specifically, we observed 3.09- and 3.20-fold improved activity at the ABCA4-site 1 of Ricolinostat and Nexturastat A treated groups compared with the DMSO group, respectively (Figure 3F). At the ABCA4-site 2, 3.44- and 3.52-fold improvement was found (Figure 3F). We also studied six potential DNA off-target sites (Supplementary Table S1) for ABCA4-site 1 and ABCA4-site 2, respectively. While, the off-target effects were not detectable with or without the treatment (Supplementary Figure S19). Taken together, here we illustrated a case of taking advantage of small-molecule compounds to augment BE3-NG efficiency for gene correction in human cells.
T-cell genome engineering holds great promise for human disease therapy, while, genetic manipulation of these cells is inefficient (37). We asked whether Ricolinostat and Nexturastat A could enhance editing efficiency in primary human T cells, and selected one endogenous gene (EMX1) to test its performance. The results showed that Ricolinostat improved the editing efficiency by 2.58- and 2.42-fold at C5 and C6, respectively (Supplementary Figure S20). With Nexturastat A treatment, the efficiency of gene editing was improved by 1.76- and 1.68-fold at C5 and C6, respectively (Supplementary Figure S20).
Knock-out of HDAC6 increases BE3-mediated gene editing efficiency
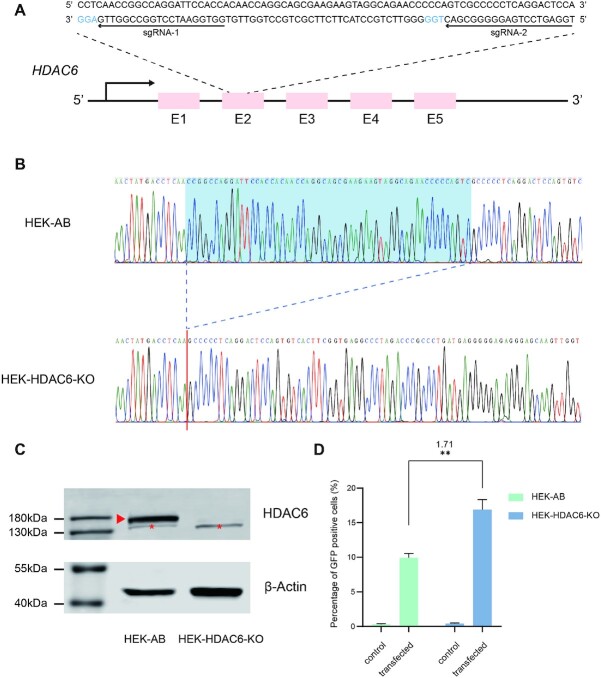
Because both HDAC6 inhibitors (Ricolinostat and Nexturastat A) enhanced CBE-mediated gene editing activity, we speculated that HDAC6 may play an important role in the gene editing process. To precisely dissect the role of HDAC6, we generated an HDAC6 knockout cell line (HEK-HDAC6-KO) by CRISPR/Cas9-based gene editing in HEK-AB cells (Figure 4A). We screened the candidate colonies via Sanger sequencing and found one colony containing a homozygous deletion mutation in the coding sequence of HDAC6 (Figure 4B). Western blot analysis revealed that HDAC6 protein is undetectable in the HEK-HDAC6-KO cell lines (Figure 4C). In agreement with the inhibitor results above, we observed that knockout of HDAC6 results in a significant increase (1.71-fold; P < 0.01) of BFP gene editing with BE3 (Figure 4D). We acknowledge that the boosting effects of this single gene knock-out are not as high as that of HDAC6 inhibitors (Ricolinostat or Nexturastat A), which may be due to the HDAC inhibitors targeting additional gene(s). Further studies are required to dissect the molecular mechanisms for this pathway. Collectively, these data reveal HDAC6 may be one of the target genes for Ricolinostat and Nexturastat A modulated CBE-mediated gene editing activity.
Figure 4.
Knock-out of HDAC6 increases BE3-mediated gene editing efficiency. (A) A schematic of SpCas9 mediated knock-out of HDAC6. The two sgRNAs are marked with arrows, and the PAMs are shown in blue. (B) Sanger sequencing results of HDAC6 in parental (HEK-AB) and HDAC6 knock-out cells (HEK-HDAC6-KO). Homozygous 55-bp deletion has been identified in HEK-HDAC6-KO cells. (C) Western blotting confirms KO of HDAC6. Band for HDAC6 has been highlighted with a red arrow. Non-specific band were marked with red-star. (D) Increased editing efficiency with knock-out of HDAC6 in HEK-AB cells. Error bars, S.E.M.; n = 3; control, non-transfected group; **P < 0.01.
Mechanism of HDAC6 increases CBE-mediated gene editing efficiency
We further determined the effects of Ricolinostat and Nexturastat A on BE3 and sgRNA expression by Western blotting and qRT-PCR. The expression of BE3 was increased with the treatment of Ricolinostat or Nexturastat A by 4.45- and 2.01-fold, respectively (Supplementary Figure S21A). Surprisingly, we observe the slightly low expression of sgRNA after treatment with either Ricolinostat or Nexturastat A (Supplementary Figure S21B). We further compared BE3 expression level in HEK- HDAC6-KO cells with that in HEK-AB via Western blotting and found that the expression of BE3 in HEK-HDAC6-KO cells was substantially increased (4.66-fold higher than HEK-AB) (Supplementary Figure S21C). These results revealed HDAC6 does play an important role for the enhanced editing efficiency.
It was reported that chromosomal structures and/or epigenetic modifications prevent Cas9 from accessing target DNA and reduce cutting frequencies (38). We speculated that HDAC6 inhibitors may change chromosomal structures and/or epigenetic modifications, which may improve chromatin accessibility and modulate the formation of the DNA-RNA-Cas9 editing complex. ATAC-seq assay was used to detect the change of chromatin accessibility with or without Ricolinostat treatment. While, there was no notable difference after the treatment (Supplementary Figure S22A–D). These results suggest that the enhancement of base editing by Ricolinostat and Nexturastat A may be involved in the increased BE3 expression, rather than sgRNA expression and chromatin accessibility.
DISCUSSION
As single-base mutations for human diseases are common, gene editing at single base-pair resolution is important for human gene therapies. Recently, C-to-T conversion via CBE or A-to-G conversion via ABE has been reported (4,39). However, there is no report of using small-molecule compounds to improve the activity of CBE, which may be a convenient, reversible, and effective approach to enhance CBE efficiency. Here we generated a reporter system for the rapid identification of small-molecule enhancers for CBE. Our screening system is based on the conversion of C (1st C of CAC, histidine codon) to T (TAT or TAC, tyrosine), which may be harnessed for screening high-activity or high-fidelity variants. Also, this system could be adopted for the identification of editing-window narrowed or shifted CBE variants. Our results reveal that HDAC6 inhibitors (Ricolinostat and Nexturastat A) enhanced CBE-mediated editing activity, which was further confirmed by targeted deep sequencing (Figures 2, 3 and Supplementary Figure S12). Also, we showed that HDAC6 inhibitors (Ricolinostat and Nexturastat A) also enhanced the base editing efficiencies of ABEmax-NG by up to 5.59-fold (Supplementary Figure S15), which is consistent with recent report from Kim's group (40). These studies revealed those HDAC inhibitors (HDAC6 inhibitors and HDAC1, 2 inhibitors) could both improve the editing efficiency of ABE and CBE, highlighting the role of HDAC modulating base editing.
As to the enhanced editing of HDAC6 inhibitors for CBE, here we showed, the BE3 expression level is increased (Supplementary Figure S21A), while, the detailed molecular mechanism is not clear. Studies demonstrated that HDAC6 is localized exclusively in the cytoplasm, where it is involved in the control of microtubule dynamics (41). With the inhibition of its activity, more HDAC6 can bind to microtubules or HDAC6 may remain binding to microtubules for a longer duration than usual, which results in the suppression of microtubule dynamics together with an increase in the stability of microtubules (42). Knockdown of HDAC6 thus increases the rate of cytoplasmic trafficking and nuclear localization of plasmid DNA (43). Also, HDAC6 deacetylates MLH1, a key DNA mismatch repair protein; with the inhibition of HDAC6, deacetylation of MLH1 is decreased, leading to more efficient DNA damage repair (44). Multiple factors including increased cytoplasmic trafficking and more efficient DNA repair may contribute to the enhanced efficiency of CBE by Ricolinostat and Nexturastat A.
Off-target effects remain a major concern that precludes safe and reliable application in genome editing, especially for clinical treatment. In the present study, we sought to enhance CBE mediated genome editing without increasing off-target activity by using small-molecule compounds. As our data suggested, HDAC6 inhibitors slightly increase the off-target editing at the target sequence, which can potentially introduce additional mutation(s) in the target sequence. At present Ricolinostat or Nexturastat A may not be directly treated as practical enhancer for CBE mediated gene therapy, additional studies, i.e, identification of high-fidelity CBE variant with single-base editing, would be required to address the off-target issue. Thus, it will be of great interest to further narrow the editing window to increase fidelity of CBE and identify high-fidelity CBE. Surprisingly, we found the editing products of CBE with Ricolinostat and Nexturastat A having less C-to-non-T (C-to-A/G) conversion at the target-nucleotide (Figure 2), which was further confirmed with mouse embryo data (Supplementary Figure S18H). A strategy was reported to improve product purity, i.e., CBE fused with Gam from bacteriophage Mu reduces indels and C-to-non-T conversions (45). So far, we don’t know which method (the present one or CBE fused with Gam) would achieve higher products purity. Further parallel comparison study would be required to address it. These results highlight the notable merits of HDAC6 inhibitors for CBE mediated genome editing application.
Collectively, using a novel BFP based directional screening system, we identified small-molecule compounds, Ricolinostat and Nexturastat A, would substantially advances the capabilities of CBE by increased efficiency and specificity for human gene therapy.
DATA AVAILABILITY
The raw data of NGS (next-generation sequencing) results have been submitted to the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/) under BioProject PRJNA675767 (SRA: SRR13021367-SRR13021369; sample accession numbers, SAMN16721847), BioProject PRJNA675913 (SRA: SRR13038042-SRR13038053, SRR13038456-SRR13038467, SRR13039567-SRR13039578; sample accession numbers, SAMN16730926), BioProject PRJNA678030 (SRA: SRR13051586-SRR13051588, SRR13052566-SRR13052571, SRR13052590-SRR13052593; sample accession numbers, SAMN16776053), BioProject PRJNA678183 (SRA: SRR13060619-SRR13060626, SRR13060761; sample accession numbers, SAMN16789435) and BioProject PRJNA727693 (SRA: SRR14460737- SRR14460748, SRR14469736- SRR14469751, SRR14470661- SRR14470675; sample accession numbers, SAMN19030120).
The data of the drug screening are summarized in Supplementary Table S4. Organized NGS data in figures and Supplementary figures are showed in Supplementary Table S5.
Supplementary Material
ACKNOWLEDGEMENTS
Author Contributions: F.G and J.L designed research, T.Z, Q.L, C.Z, XJ.L, H.L, T.T, N.T, Y.C, XY.L, performed research and T.Z, Q.L, C.L, J.Z, Z.S, H.W, J.L and F.G. performed data analyses, and T.Z, Q.L, J.L and F.G. wrote the manuscript. All authors have read and approved the final manuscript.
Contributor Information
Tianyuan Zhao, School of Ophthalmology and Optometry, Eye Hospital, Wenzhou Medical University, State Key Laboratory and Key Laboratory of Vision Science, Ministry of Health and Zhejiang Provincial Key Laboratory of Ophthalmology and Optometry, Wenzhou, Zhejiang, China.
Qing Li, State Key Laboratory of Cell Biology, Shanghai Key Laboratory of Molecular Andrology, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, Shanghai, China.
Chenchen Zhou, School of Ophthalmology and Optometry, Eye Hospital, Wenzhou Medical University, State Key Laboratory and Key Laboratory of Vision Science, Ministry of Health and Zhejiang Provincial Key Laboratory of Ophthalmology and Optometry, Wenzhou, Zhejiang, China.
Xiujuan Lv, School of Ophthalmology and Optometry, Eye Hospital, Wenzhou Medical University, State Key Laboratory and Key Laboratory of Vision Science, Ministry of Health and Zhejiang Provincial Key Laboratory of Ophthalmology and Optometry, Wenzhou, Zhejiang, China.
Hongyan Liu, School of Ophthalmology and Optometry, Eye Hospital, Wenzhou Medical University, State Key Laboratory and Key Laboratory of Vision Science, Ministry of Health and Zhejiang Provincial Key Laboratory of Ophthalmology and Optometry, Wenzhou, Zhejiang, China.
Tianxiang Tu, School of Ophthalmology and Optometry, Eye Hospital, Wenzhou Medical University, State Key Laboratory and Key Laboratory of Vision Science, Ministry of Health and Zhejiang Provincial Key Laboratory of Ophthalmology and Optometry, Wenzhou, Zhejiang, China.
Na Tang, State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, University of Chinese Academy of Sciences, Beijing, China.
Yanbo Cheng, State Key Laboratory of Cell Biology, Shanghai Key Laboratory of Molecular Andrology, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, Shanghai, China.
Xiaoyu Liu, School of Ophthalmology and Optometry, Eye Hospital, Wenzhou Medical University, State Key Laboratory and Key Laboratory of Vision Science, Ministry of Health and Zhejiang Provincial Key Laboratory of Ophthalmology and Optometry, Wenzhou, Zhejiang, China.
Changbao Liu, The Second Affiliated Hospital and Yuying Children's Hospital of Wenzhou Medical University, Wenzhou, Zhejiang, China.
Junzhao Zhao, The Second Affiliated Hospital and Yuying Children's Hospital of Wenzhou Medical University, Wenzhou, Zhejiang, China.
Zongming Song, Henan Eye Hospital, Henan Provincial People's Hospital and People's Hospital of Zhengzhou University, People's Hospital of Henan University, Zhengzhou, Henan, China.
Haoyi Wang, State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, University of Chinese Academy of Sciences, Beijing, China.
Jinsong Li, State Key Laboratory of Cell Biology, Shanghai Key Laboratory of Molecular Andrology, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, Shanghai, China.
Feng Gu, School of Ophthalmology and Optometry, Eye Hospital, Wenzhou Medical University, State Key Laboratory and Key Laboratory of Vision Science, Ministry of Health and Zhejiang Provincial Key Laboratory of Ophthalmology and Optometry, Wenzhou, Zhejiang, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key R&D Program of China [2018YFA0107304]; National Natural Science Foundation of China [81201181]; Natural Science Foundation of Zhejiang Province [2017C37176]; Project of State Key Laboratory of Ophthalmology, Optometry and Visual Science, Wenzhou Medical University [J02-20190201]; Lin He's Academician Workstation of New Medicine and Clinical Translation [17331209, 18331105]; China National Postdoctoral Program for Innovative Talents [BX20200348]; Chinese Postdoctoral Science Foundation [2020M681407]; Wenzhou City Key Innovation Team of Reproductive Genetics Grant, Zhejiang, China [C20170007] and Wenzhou city Grant [2021Y1893]. Funding for open access charge: National Key R&D Program of China.
Conflict of interest statement. All authors have stated explicitly that there are no conflicts of interest in connection with this article. The authors declare that they have a patent (pending) for the identified compounds for genome editing in this study.
REFERENCES
- 1.Fellmann C., Gowen B.G., Lin P.-C., Doudna J.A., Corn J.E.. Cornerstones of CRISPR–Cas in drug discovery and therapy. Nat. Rev. Drug Discov. 2017; 16:89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu H., Li C., Gao C.. Applications of CRISPR-Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 2020; 21:661–677. [DOI] [PubMed] [Google Scholar]
- 3.Beyret E., Liao H.-K., Yamamoto M., Hernandez-Benitez R., Fu Y., Erikson G., Reddy P., Izpisua Belmonte J.C.. Single-dose CRISPR–Cas9 therapy extends lifespan of mice with Hutchinson–Gilford progeria syndrome. Nat. Med. 2019; 25:419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R.. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016; 533:420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeh W.-H., Shubina-Oleinik O., Levy J.M., Pan B., Newby G.A., Wornow M., Burt R., Chen J.C., Holt J.R., Liu D.R.. In vivo base editing restores sensory transduction and transiently improves auditory function in a mouse model of recessive deafness. Sci. Transl. Med. 2020; 12:eaay9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang B., Yang L., Chen J.. Development and application of base editors. CRISPR J. 2019; 2:91–104. [DOI] [PubMed] [Google Scholar]
- 7.Li Q., Li Y., Yang S., Huang S., Yan M., Ding Y., Tang W., Lou X., Yin Q., Sun Z.et al.. CRISPR-Cas9-mediated base-editing screening in mice identifies DND1 amino acids that are critical for primordial germ cell development. Nat. Cell Biol. 2018; 20:1315–1325. [DOI] [PubMed] [Google Scholar]
- 8.Kim Y.B., Komor A.C., Levy J.M., Packer M.S., Zhao K.T., Liu D.R.. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 2017; 35:371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thuronyi B.W., Koblan L.W., Levy J.M., Yeh W.H., Zheng C., Newby G.A., Wilson C., Bhaumik M., Shubina-Oleinik O., Holt J.R.et al.. Continuous evolution of base editors with expanded target compatibility and improved activity. Nat. Biotechnol. 2019; 37:1070–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng T.L., Li S., Yuan B., Wang X., Zhou W., Qiu Z.. Expanding C-T base editing toolkit with diversified cytidine deaminases. Nat. Commun. 2019; 10:3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doman J.L., Raguram A., Newby G.A., Liu D.R.. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol. 2020; 38:620–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuo E., Sun Y., Yuan T., He B., Zhou C., Ying W., Liu J., Wei W., Zeng R., Li Y.et al.. A rationally engineered cytosine base editor retains high on-target activity while reducing both DNA and RNA off-target effects. Nat. Methods. 2020; 17:600–604. [DOI] [PubMed] [Google Scholar]
- 13.Huang T.P., Zhao K.T., Miller S.M., Gaudelli N.M., Oakes B.L., Fellmann C., Savage D.F., Liu D.R.. Circularly permuted and PAM-modified Cas9 variants broaden the targeting scope of base editors. Nat. Biotechnol. 2019; 37:626–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleinstiver B.P., Sousa A.A., Walton R.T., Tak Y.E., Hsu J.Y., Clement K., Welch M.M., Horng J.E., Malagon-Lopez J., Scarfò I.et al.. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 2019; 37:276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X., Ding C., Yu W., Wang Y., He S., Yang B., Xiong Y.C., Wei J., Li J., Liang J.et al.. Cas12a base editors induce efficient and specific editing with low DNA damage response. Cell Rep. 2020; 31:107723. [DOI] [PubMed] [Google Scholar]
- 16.Grünewald J., Zhou R., Lareau C.A., Garcia S.P., Iyer S., Miller B.R., Langner L.M., Hsu J.Y., Aryee M.J., Joung J.K.. A dual-deaminase CRISPR base editor enables concurrent adenine and cytosine editing. Nat. Biotechnol. 2020; 38:861–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z., Lu Z., Yang G., Huang S., Li G., Feng S., Liu Y., Li J., Yu W., Zhang Y.et al.. Efficient generation of mouse models of human diseases via ABE- and BE-mediated base editing. Nat. Commun. 2018; 9:2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kühn R.. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015; 33:543–548. [DOI] [PubMed] [Google Scholar]
- 19.Song J., Yang D., Xu J., Zhu T., Chen Y.E., Zhang J.. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat. Commun. 2016; 7:10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jayathilaka K., Sheridan S.D., Bold T.D., Bochenska K., Logan H.L., Weichselbaum R.R., Bishop D.K., Connell P.P.. A chemical compound that stimulates the human homologous recombination protein RAD51. PNAS. 2008; 105:15848–15853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maji B., Gangopadhyay S.A., Lee M., Shi M., Wu P., Heler R., Mok B., Lim D., Siriwardena S.U., Paul B.et al.. A high-throughput platform to identify small-molecule inhibitors of CRISPR-Cas9. Cell. 2019; 177:1067–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y., Ge X., Yang F., Zhang L., Zheng J., Tan X., Jin Z.B., Qu J., Gu F.. Comparison of non-canonical PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells. Sci. Rep. 2014; 4:5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Y., Liang D., Wang Y., Bai M., Tang W., Bao S., Yan Z., Li D., Li J. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell stem cell. 2013; 13:659–662. [DOI] [PubMed] [Google Scholar]
- 24.Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang N., Cheng C., Zhang X., Qiao M., Li N., Mu W., Wei X.F., Han W., Wang H.. TGF-β inhibition via CRISPR promotes the long-term efficacy of CAR T cells against solid tumors. JCI Insight. 2020; 5:e133977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie L., Huang J., Li X., Dai L., Lin X., Zhang J., Luo J., Zhang W.. Generation of a homozygous HDAC6 knockout human embryonic stem cell line by CRISPR/Cas9 editing. Stem Cell Res. 2019; 41:101610. [DOI] [PubMed] [Google Scholar]
- 27.Xie H., Ge X., Yang F., Wang B., Li S., Duan J., Lv X., Cheng C., Song Z., Liu C.et al.. High-fidelity SaCas9 identified by directional screening in human cells. PLoS Biol. 2020; 18:e3000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tu M., Lin L., Cheng Y., He X., Sun H., Xie H., Fu J., Liu C., Li J., Chen D.et al.. A ‘new lease of life’: FnCpf1 possesses DNA cleavage activity for genome editing in human cells. Nucleic Acids Res. 2017; 45:11295–11304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lv X., Qiu K., Tu T., He X., Peng Y., Ye J., Fu J., Deng R., Wang Y., Wu J.et al.. Development of a Simple and Quick Method to Assess Base Editing in Human Cells. Mol. Ther. Nucleic Acids. 2020; 20:580–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zong Y., Wang Y., Li C., Zhang R., Chen K., Ran Y., Qiu J.L., Wang D., Gao C.. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 2017; 35:438–440. [DOI] [PubMed] [Google Scholar]
- 31.Yang F., Liu C., Chen D., Tu M., Xie H., Sun H., Ge X., Tang L., Li J., Zheng J.et al.. CRISPR/Cas9-loxP-mediated gene editing as a novel site-specific genetic manipulation tool. Mol. Ther. Nucleic Acids. 2017; 7:378–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santo L., Hideshima T., Kung A.L., Tseng J.C., Tamang D., Yang M., Jarpe M., van Duzer J.H., Mazitschek R., Ogier W.C.et al.. Preclinical activity, pharmacodynamic, and pharmacokinetic properties of a selective HDAC6 inhibitor, ACY-1215, in combination with bortezomib in multiple myeloma. Blood. 2012; 119:2579–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koblan L.W., Doman J.L., Wilson C., Levy J.M., Tay T., Newby G.A., Maianti J.P., Raguram A., Liu D.R.. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 2018; 36:843–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walton R.T., Christie K.A., Whittaker M.N., Kleinstiver B.P.. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020; 368:290–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou C., Sun Y., Yan R., Liu Y., Zuo E., Gu C., Han L., Wei Y., Hu X., Zeng R.et al.. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature. 2019; 571:275–278. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X., Ge X., Shi W., Huang P., Min Q., Li M., Yu X., Wu Y., Zhao G., Tong Y.et al.. Molecular diagnosis of putative Stargardt disease by capture next generation sequencing. PLoS One. 2014; 9:e95528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schumann K., Lin S., Boyer E., Simeonov D.R., Subramaniam M., Gate R.E., Haliburton G.E., Ye C.J., Bluestone J.A., Doudna J.A.et al.. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lazzarotto C.R., Malinin N.L., Li Y., Zhang R., Yang Y., Lee G., Cowley E., He Y., Lan X., Jividen K.et al.. CHANGE-seq reveals genetic and epigenetic effects on CRISPR–Cas9 genome-wide activity. Nat. Biotechnol. 2020; 38:1317–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R.. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017; 551:464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin H.R., See J.E., Kweon J., Kim H.S., Sung G.J., Park S., Jang A.H., Jang G., Choi K.C., Kim I.et al.. Small-molecule inhibitors of histone deacetylase improve CRISPR-based adenine base editing. Nucleic Acids Res. 2021; 49:2390–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyake Y., Keusch J.J., Wang L., Saito M., Hess D., Wang X., Melancon B.J., Helquist P., Gut H., Matthias P.. Structural insights into HDAC6 tubulin deacetylation and its selective inhibition. Nat. Chem. Biol. 2016; 12:748–754. [DOI] [PubMed] [Google Scholar]
- 42.Asthana J., Kapoor S., Mohan R., Panda D. Inhibition of HDAC6 deacetylase activity increases its binding with microtubules and suppresses microtubule dynamic instability in MCF-7 cells. J. Biol. Chem. 2013; 288:22516–22526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vaughan E.E., Geiger R.C., Miller A.M., Loh-Marley P.L., Suzuki T., Miyata N., Dean D.A.. Microtubule acetylation through HDAC6 inhibition results in increased transfection efficiency. Mol. Ther. 2008; 16:1841–1847. [DOI] [PubMed] [Google Scholar]
- 44.Zhang M., Hu C., Moses N., Haakenson J., Xiang S., Quan D., Fang B., Yang Z., Bai W., Bepler G.et al.. HDAC6 regulates DNA damage response via deacetylating MLH1. J. Biol. Chem. 2019; 294:5813–5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Komor A.C., Zhao K.T., Packer M.S., Gaudelli N.M., Waterbury A.L., Koblan L.W., Kim Y.B., Badran A.H., Liu D.R.. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci. Adv. 2017; 3:eaao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of NGS (next-generation sequencing) results have been submitted to the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/) under BioProject PRJNA675767 (SRA: SRR13021367-SRR13021369; sample accession numbers, SAMN16721847), BioProject PRJNA675913 (SRA: SRR13038042-SRR13038053, SRR13038456-SRR13038467, SRR13039567-SRR13039578; sample accession numbers, SAMN16730926), BioProject PRJNA678030 (SRA: SRR13051586-SRR13051588, SRR13052566-SRR13052571, SRR13052590-SRR13052593; sample accession numbers, SAMN16776053), BioProject PRJNA678183 (SRA: SRR13060619-SRR13060626, SRR13060761; sample accession numbers, SAMN16789435) and BioProject PRJNA727693 (SRA: SRR14460737- SRR14460748, SRR14469736- SRR14469751, SRR14470661- SRR14470675; sample accession numbers, SAMN19030120).
The data of the drug screening are summarized in Supplementary Table S4. Organized NGS data in figures and Supplementary figures are showed in Supplementary Table S5.