Abstract
CRISPR–Cas9 generates double-stranded DNA breaks (DSBs) to activate cellular DNA repair pathways for genome editing. The repair of DSBs leads to small insertions or deletions (indels) and other complex byproducts, including large deletions and chromosomal translocations. Indels are well understood to disrupt target genes, while the other deleterious byproducts remain elusive. We developed a new in silico analysis pipeline for the previously described primer-extension-mediated sequencing assay to comprehensively characterize CRISPR–Cas9-induced DSB repair outcomes in human or mouse cells. We identified tremendous deleterious DSB repair byproducts of CRISPR–Cas9 editing, including large deletions, vector integrations, and chromosomal translocations. We further elucidated the important roles of microhomology, chromosomal interaction, recurrent DSBs, and DSB repair pathways in the generation of these byproducts. Our findings provide an extra dimension for genome editing safety besides off-targets. And caution should be exercised to avoid not only off-target damages but also deleterious DSB repair byproducts during genome editing.
INTRODUCTION
Genome editing technologies based on engineered nucleases not only greatly change the way we study life sciences but also cast light on the treatment of human genetic diseases (1,2). Among these powerful editing toolboxes, the clustered regularly interspaced short palindromic repeats (CRISPR) and associated protein (Cas) engineered from the bacterial defense system are the most widely used ones. The Streptococcus pyogenes Cas9 (referred to as Cas9 hereafter) from type II CRISPR–Cas systems is the earliest Cas protein to be engineered for performing genome editing in human cells (3–5). CRISPR–Cas9 is a two-component editing system, comprising of a Cas9 protein with cleavage activity and a single guide RNA (sgRNA) to bind both Cas9 and target DNA (6). CRISPR–Cas9 is in principle able to induce double-stranded DNA breaks (DSBs) at any locus 3 base pairs (bp) upstream of an NGG protospacer adjacent motif (PAM). Besides inducing mutations at target sites, the DNA repair process triggered by DSBs may also generate unintended damages at homologous off-target sites, raising great safety concerns. These off-target activities of CRISPR–Cas9 can be largely minimized by using high-fidelity Cas9 variants, choosing a better target sequence instead, or rapid activity shut-off by anti-CRISPR (7–9).
The first step for CRISPR–Cas9 editing is to initiate DSBs at DNA target sites that are complementary to the sgRNAs. The endogenous DNA repair pathways are subsequently activated to create a variety of DNA repair outcomes, including a large number of insertions and deletions (10). There are two main DSB repair pathways in mammalian cells, non-homologous end joining (NHEJ) and homologous recombination (HR). To repair Cas9-induced DSBs, NHEJ directly fuses two broken ends to seal DSBs, frequently accompanied by small insertions or deletions (indels) that are less than 20 bp; while HR requires external homologous donor DNA to introduce intended mutations (11). Besides, the microhomology-mediated end joining (MMEJ) is also involved in DSB repair after the exposure of microhomologies at juxtaposed broken ends following end processing (12,13). MMEJ requires microhomologies that range from 2–20 bp while NHEJ might also utilize microhomologies less than 4 bp (14,15). Both NHEJ and MMEJ may generate deleterious DSB repair byproducts, including large chromatin deletions and chromosomal translocations, resulting in chromosomal abnormality or tumorigenesis (12,15,16). In this context, large deletions, chromosomal translocations, or even chromosome loss have been detected by different research groups in mouse and human stem cells after CRISPR–Cas9 editing (10,17–20).
Approaches to manipulate DSB repair pathways have been developed to enhance genome editing (11,21). For instance, inhibitors for key NHEJ factors KU or LIG4 are used to increase the incorporated rate of donor fragments by enhancing HR (22–25); RAD51 has been fused with Cas9 nickase to facilitate the insertion of double-stranded oligonucleotide (26). In contrast, inhibitors for CtIP or RAD52 suppress HR and promote single-stranded oligonucleotide-mediated editing (22). However, blocking the DSB repair pathway would alter the spectrum of CRISPR–Cas9-induced repair outcomes and may threaten genome integrity. For this reason, the danger of using inhibitors for key DNA repair factors on genome integrity during genome editing remains to be elucidated.
Comprehensive assessment of global DSB repair outcomes would facilitate our understanding of the origins of the deleterious byproducts including large deletions and chromosomal translocations as well as help improve genome-editing safety (10,18,27,28). Here, we developed a new in silico analysis pipeline to identify genome-wide DSB repair outcomes based on the high-throughput sequencing data generated via previously described primer-extension-mediated sequencing (PEM-seq) (28). We find that large deletions heavily depend on microhomologies and large insertions contain substantial vector integrations. Chromosomal translocations distribute widely in the genome and are often dominated by off-target or other recurrent DSBs. Furthermore, we also detect an increased level of chromosomal abnormality in the absence of the NHEJ repair pathway.
MATERIALS AND METHODS
Plasmid construction
All sgRNAs used for HEK293T cells and mESCs targeting have been cloned into the double BbsI sites of pX330-vector (Addgene ID 42230). The plasmid used for CH12F3 cell targeting was an optimized vector in which we removed the AAV2 ITR sequence and introduced a mCherry gene with CMV promoter by Gibson assembly into pX330 vector.
Cell culture and plasmid transfection
The mESCs were cultured in ES-DMEM medium (Millipore) with 15% fetal bovine serum (FBS, ExCell Bio), Penicillin/Streptomycin (Corning), nucleotides (Millipore), l-glutamine (Corning), nonessential amino acids (Corning), PD0325901 (Selleck), CHIR99021 (Selleck) and LIF (Millipore) at 37°C with 5% CO2. mESCs in 6-cm dishes were transfected with 7.2 μg pX330-Cas9 plus 1.8 μg GFP expression vector by 4D-nucleofector X (Lonza, solution Cytomix, program GC104), then harvested for genomic DNA 3 days after transfection.
The wild-type, Ku80–/–, Lig4–/–, Parp1–/– and AID–/– CH12F3 cells were cultured in RPIM1640 medium (Corning) with 15% Fetal Bovine Serum (FBS, ExCell Bio), HEPES (Corning), penicillin–streptomycin (Corning), l-glutamine (Corning), nonessential amino acids (Corning), sodium pyruvate (Corning) and β-mercaptoethanol (Sigma-Aldrich) at 37°C with 5% CO2. Growing CH12F3 cells were transfected with 1.5 μg pX330-Cas9 or pX330-Cas9-mCherry expression vector per million by 4D-nucleofector X (Lonza, solution M1, procedure DN100) and seeding at 0.5 million cells/ml in fresh medium with 1 μg/ml anti-CD40, 5 ng/ml IL-4 and 0.5 ng/ml TGF-β. After 72 h stimulation, the cells were harvested and genomic DNA was extracted for PEM-seq library construction.
PEM-seq and 3C-HTGTS
The primers and sgRNAs used for library construction are listed in Supplementary Tables S1 and S2, respectively. The PEM-seq libraries were constructed according to the standard procedure described previously (28). About 20 μg genome DNA from edited cells was used for each library. Primer control libraries were done with Cas9-infected cells with no sgRNA.
The 3C-HTGTS libraries were constructed following the previously described procedures (29). Briefly for preparing the 3C-HTGTS libraries, 5–6 million cells were incubated with 1% formaldehyde for 10 min at room temperature and glycine was added to a final concentration of 125 mM to stop the cross-linking reaction. Then cell lysis buffer containing 10 mM Tris–HCl (pH 8.0), 10 mM NaCl, 0.2% NP-40, 10 mM EDTA was used to lysis cell and prepare nuclei. Then the nuclei restriction enzyme (RE) digestion was performed by incubating with 700 units of Dpn II restriction enzyme overnight at 37°C, and the digestion efficiency was checked by DNA gel electrophoresis. Re-ligate the DNA sequence at 16°C for 4 h to overnight under dilute conditions. De-crosslink the nuclei by incubating the DNA with Proteinase K at 56°C by rotating overnight. Finally, the purified DNA after RNase A treatment was the ‘3C templates’ and then subsequently prepared the 3C library as the same as PEM-seq library construction.
All the libraries were sequenced by Hiseq.
PEM-Q analysis
Before PEM-Q analysis, raw reads were pre-processed as we did in the previous method (28). We used cutadapt (http://cutadapt.readthedocs.io/en/stable/) to remove the universal adapters. Reads ending with QC <30 were trimmed; remaining reads larger than 25 bp were kept for library demultiplex by fastq-multx (https://github.com/brwnj/fastq-multx). Reads after demultiplex were analyzed by PEM-Q in five steps.
Reads alignment
To begin with, R1 and R2 of pair-end reads generated by Hiseq were stitched using flash 1.2.11 (https://ccb.jhu.edu/software/FLASH/) with default parameters. Then the stitched reads, along with unstitched R1 reads were aligned to reference genome (hg38 for human, mm10 for mouse) by bwa-mem. Reads were kept if their alignment start sites were around primer start with an error less than 4 bp. Meanwhile, R2 reads were aligned to the blue adapter, which was used to find random molecular barcode (RMB, equal to unique molecular index) in step 2. Mapped reads with the wrong primer location were discarded in this step.
RMB extract
We kept reads with the correct blue adapter allowing at most 2 bp truncation. Then, RMB within 2-bp loss in length were extracted according to blue adapter location. RMB was recorded in a separate file with sequence name (Qname). Reads with multiple tandem adapters were filtered in this step.
Find chimeric alignment
Chimeric reads were reported in SA tag in bwa-mem. Sequence aligned to primer was bait while the other side was prey. We then kept reads that only reported one chimeric junction and recorded their information as prey in a tab file. Reads with bait alignment not exceeding 10 bp after primer binding site were discarded. Extra bases between bait and prey were extracted and recorded as insertions. For those without insertions, we identified overlapped bases as microhomology between the end of bait and the start of prey. Reads that did not have chimeric alignment were further analyzed in step 4.
Find indels
Reads without chimeric alignment were linear alignment. Linear alignment length not exceeding 10 bp after cleavage site of CRISPR was discarded. The remaining were processed to find indels. Insertions and deletions were reported by ‘I’ and ‘D’ from CIGAR reported by bwa-mem. The same bases at the ends of deletions were identified as microhomology. Substitutions were also aware of PEM-Q and we identified substitutions according to MD tags reported by bwa-mem. The remaining reads without chimeric alignment or indels were recorded as germline.
Classify and deduplicate
Reads that have both bait and prey aligning to target chromosomes with inserted sequences were classified as insertions. Those without inserted sequences but with a distance between bait and prey no more than 500 kb were classified as deletions in this study. Reads with a distance between bait and prey exceeded 500 kb were classified as intra-chromosomal translocations, while those with prey from other chromosomes were classified as inter-chromosomal translocations. RMB extract in step 2 was relocated to reads according to their sequence name. Within each type of variants we classified, duplicates were removed according to prey's alignment information including chromosome, strand, junction, and bait end together with RMB. Specifically, for junctions close to genomic repetitive regions with mapping quality of the prey sequence lower than 30, we removed duplicates by RMB.
Additional program: vector (plasmid) analysis
There are two main types of vector integrations as described in the text. One is short vector insertions that the entire inserted fragments can be aligned to the vector backbone. The others with too long inserted fragments are discarded in PEM-Q. However, the second type still has potential large vector integrations. Therefore, we remapped these discarded reads to the genome and then the vector backbone to find missed vector integrations. We used bwa-mem to do the alignment with a default seed length of 20 bp.
Off-target and TSS analysis
Off-target identification was described previously (28), using MACS2 callpeak and a commonly used criteria. For TSS analysis, we used computeMatrix (deeptools 3.1.3) to calculate the signals in RNA Pol II ChIP-seq data, using parameters ‘-a 50000 -b 50000 -bs 1000’. As for PEM-seq data, we used the same algorithm described before (30) to assign junctions to the nearest TSSs.
RESULTS
Detecting global DNA repair outcomes of CRISPR–Cas9 by PEM-Q pipeline
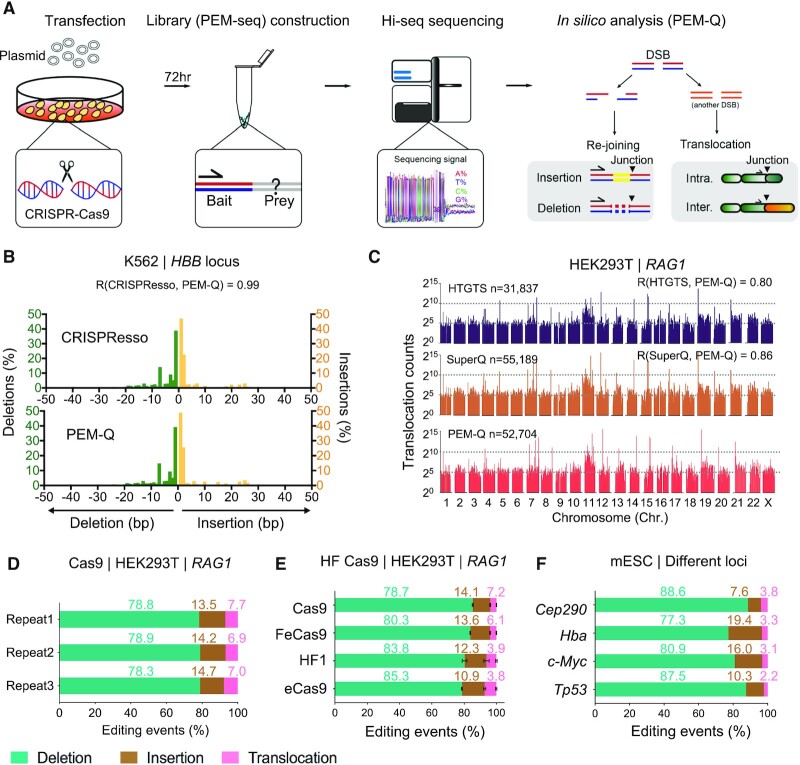
To gain insight into the full spectrum of DNA repair products resulted from genome editing exerted by CRISPR–Cas9, we have developed the PEM-seq to capture unknown broken ends (prey) fused with the target DSBs (bait) in cells 72 h post-transfection (Figure 1A). The identified repair outcomes are further categorized into re-joinings of the target broken ends, leading to insertions and deletions, and intra- or inter-chromosomal translocations (Figure 1A). In order to quantify indels that are invisible to the previous SuperQ pipeline (28), we employed bwa mem instead of bowtie2 for genome alignment and optimized the in silico analysis flow to develop a new pipeline, termed as PEM-Q (Supplementary Figure S1A; see Methods for details). Then we used PEM-Q to analyze the deep sequencing data from CRISPR–Cas9-edited K562 cells at HBB locus in parallel with CRISPResso (31). The distribution pattern of indels identified by PEM-Q was almost identical to that identified by CRISPResso (Figure 1B). The SuperQ and high-throughput genome-wide translocation sequencing (HTGTS) pipelines have been used to identify translocation junctions and off-targets of CRISPR–Cas9 in HEK293T cells at various loci (28,32,33). We re-analyzed the same sequencing data from RAG1 locus by PEM-Q and found highly similar genome-wide translocation patterns among these pipelines (Figure 1C). Moreover, PEM-Q identified 6 more off-targets than SuperQ and 10 more off-targets than HTGTS, showing a higher sensitivity of detecting off-targets (Supplementary Figure S1B and S1C). Therefore, PEM-Q is a unified in silico analysis pipeline for detecting global DNA repair outcomes of CRISPR–Cas9.
Figure 1.
Detecting global DNA repair outcomes of CRISPR–Cas9 by PEM-Q pipeline. (A) Experimental procedures. Sequencing libraries were generated by PEM-seq with Hi-seq sequencing. The identified re-joining events contain insertions and deletions, while translocation junctions beyond upstream and downstream 500 kb from the target site in the target chromosome are defined as intra-chromosomal translocations (Intra.) and junctions from different chromosomes are inter-chromosomal translocations (Inter.) Black triangles represent identified junctions. (B) Re-analysis the published data (31) by PEM-Q to compare with CRISPResso at the HBB locus in K562 cells. Green bars represent the frequency of deletions at indicated length normalized to total deletions; while yellow bars represent insertions. Pearson correlation (R) between CRISPResso and PEM-Q is 0.99. (C) Re-analysis of PEM-seq data (28) by PEM-Q versus HTGTS and SuperQ at the RAG1 locus in HEK293T cells. Total translocation junctions from three replicates are shown with 2-Mb bins on a log scale. Total numbers (n) of translocations and Pearson correlation (R) are indicated. (D–F) Bar charts showing percentages of deletions (cyan), insertions (brown) and translocations (pink) among total editing events. (D) Three repeat PEM-seq libraries at the RAG1 locus in HEK293T cells. (E) High-fidelity (HF) Cas9 enzymes at RAG1 locus in HEK293T cells. (F) Different target loci (Cep290, Hba, c-Myc and Tp53) in mESCs.
We next employed PEM-Q to systematically analyze public PEM-seq data from CRISPR–Cas9-edited HEK293T cells at the RAG1 gene (28).We divided the repair outcomes into re-joinings and translocations for further analysis. The re-joinings of the target DSB result in deletions and insertions, while chromosomal translocations are derived from the fusion of target DSB with another DSB either in the same chromosome or other chromosomes (Figure 1A). The PEM-Q-identified deletions, insertions, or translocations were highly reproducible from three repeat libraries (Supplementary Figure S1D and E). Furthermore, deletions were the main repair outcomes concentrated in the upstream and downstream 15 base pairs (bp) around the cleavage site, approximately 78.3–78.9% of total editing events; insertions were about 13.5–14.7%, enriched within a 5-bp region around the cleavage site; while translocations distributed widely in the genome at a rate of 6.9–7.7% (Figure 1D and E, Supplementary Figure S1D and E). A number of high-fidelity variants have been developed to reduce the off-target activity of Cas9 (8,28). We re-analyzed the PEM-seq data of three high-fidelity variants eCas9, HF1 and FeCas9 versus Cas9 from HEK293T cells at the RAG1 gene (28). High-fidelity variants showed similar levels of different repair outcomes, including high levels of translocations (Figure 1E), in line with a recent report (34).
We also used CRISPR–Cas9 to target the Cep290, Hba, c-Myc and Tp53 loci in mouse embryonic stem cells (mESCs) and then generated PEM-seq libraries for PEM-Q analysis. Inheritable repair outcomes in mESCs showed similar compositions as those in HEK293T cells despite the percentages of different repair outcomes varied at examined target sites (Figure 1F). Collectively, these data suggest that Cas9 repair outcomes are probably nonrandom, so we sought to further explore these repair outcomes of CRISPR–Cas9 with comprehensive analysis capability of PEM-Q.
Prevalent microhomologies at large deletions of CRISPR–Cas9
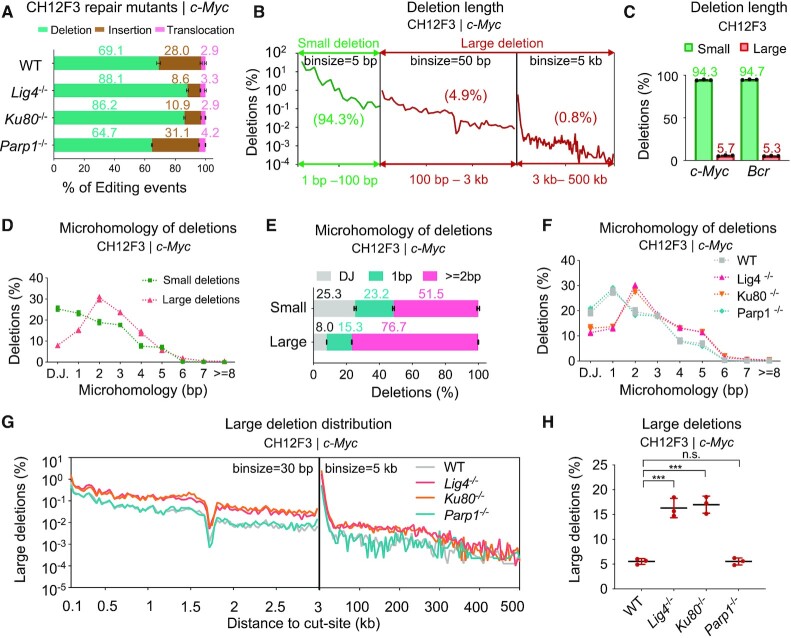
We also performed PEM-Q analysis on CRISPR–Cas9 editing at the c-Myc and Bcr locus in the mouse CH12F3 B cells. The two loci exhibited very distinct mutation patterns that CRISPR–Cas9 showed a higher editing efficiency with more various editing products at the c-Myc than at the Bcr locus (Supplementary Figure S2A-B). Exemplified by the top 10 editing products, a T-insertion target sequence induced by staggered cleavage of Cas9 (35,36) occupied 22.3% of all editing events from the c-Myc pool, while the similar C-insertion target sequence was only 4.2% (Supplementary Figure S2C), indicating that the editing locus affected the compositions of editing outcomes. Whereas, chromosomal deletions were the main editing events at both loci regardless of the genetic backgrounds (Figure 2A and Supplementary Figure S2B).
Figure 2.
Prevalent microhomologies at large deletions of CRISPR–Cas9. (A) Bar charts showing percentages of deletions (cyan), insertions (brown) and translocations (pink) among total editing events. The c-Myc locus in CH12F3 cells with indicated backgrounds. Error bars, mean ± SD. (B) The distribution pattern of deletions at the c-Myc locus in CH12F3 cells. Total junctions of three repeats are plotted on a log scale. Percentages of deletions within each region among total deletions are shown in the brackets. Please note that 5 bp, 50 bp, and 5 kb bin-sizes are used for the three regions, respectively. (C) Percentages of small and large deletions among total deletions at the c-Myc and Bcr loci in CH12F3 cells. Error bars, mean ± SD. (D and E) Line plot (D) and bar chart (E) of microhomologies with indicated length in small or large deletions among total deletions at the c-Myc locus in CH12F3 cells. Only deletions cross the cleavage sites are used for analysis. D.J., direct joining. Error bars, mean ± SD. (F) Line plot of microhomologies with indicated length in total deletions at the c-Myc locus in CH12F3 cells with indicated backgrounds. Only deletions cross the cleavage sites are used for analysis. D.J., direct joining. (G) The distribution patterns of large deletions at the c-Myc locus in CH12F3 cells with indicated backgrounds. Please note that 30 and 5 kb bin-sizes are used for two regions, respectively. (H) Percentages of large deletions among total deletions at the c-Myc locus in CH12F3 cells with indicated backgrounds. One-tailed t-test, ***P < 0.0005; n.s., not significant. Error bars, mean ± SD.
Chromosomal deletions were widely distributed downstream of the cloning primer binding site and expanded as long as hundreds of kb at the c-Myc locus while tens of kb at the Bcr locus in CH12F3 cells, depending on the cutting efficiency at two loci (Figure 2B and Supplementary Figure S3A). In this context, we divided Cas9-induced deletions into two parts: small deletions within 100 bp and large deletions larger than 100 bp. Small deletions were the main deletional events at a percentage of more than 94%, while large deletions were also unignorable, more than 5% at both c-Myc and Bcr loci (Figure 2B, C, and Supplementary Figure S3A). Specifically, 0.8% and 0.3% deletions were larger than 3 kb at the c-Myc or Bcr locus, respectively (Figure 2B and Supplementary Figure S3A). Similar findings were obtained in four different target sites in mESCs, and, notably, large deletions were increased to 11.7% and 14.5% at the c-Myc and Hba loci, respectively (Supplementary Figure S3B).
Given that the formation of deletion requires end processing that may promote the usage of microhomologies, we next examined the usage of microhomologies in these deletional events. Approximately 25% of small deletions preferred direct joining and around half of them used microhomologies longer than 2 bp with a decreasing trend over length (Figure 2D and E). Different from small deletions, large deletions heavily depended on microhomologies and over 76.7% of large deletions used microhomologies longer than 2 bp while direct joinings were only about 8.0% (Figure 2D and E), consistent with previous findings (35). Similar findings were obtained in CRISPR–Cas9-edited CH12F3 cells at the Bcr locus and mESCs at four different loci (Supplementary Figure S3C and S3D). For further verification, we examined the microhomology usage in CH12F3 cells deficient for Ku80 or Lig4. NHEJ predisposes to direct joining and is suppressed in Ku80- or Lig4-deficient cells (15). As anticipated, we detected an elevated level of microhomology usage in Cas9-induced deletions from Ku80- or Lig4-deficient CH12F3 cells (Figure 2F and Supplementary Figure S3E). Correspondingly, the level of large deletions from 100 bp to 300 kb was increased significantly in Ku80- or Lig4-deficient background (Figure 2G and Supplementary Figure S3F). However, the deficiency for Parp1 resulted in no significant effect on the deletion pattern at c-Myc locus despite a minor declined level of large deletions at the Bcr locus in CH12F3 cells (Figure 2G, H and Supplementary Figure S3F, G).
Recurrent small insertions and deleterious plasmid integrations
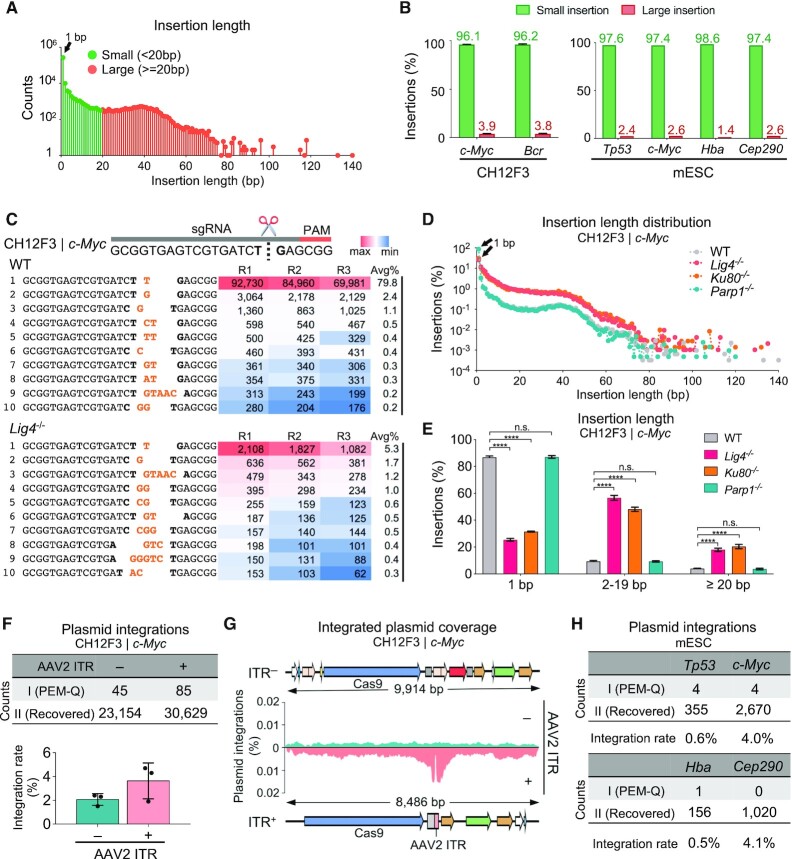
We also divided CRISPR–Cas9-induced insertions identified by PEM-Q into two parts: small insertions less than 20 bp and large insertions for 20 bp or more. Of note, repair outcomes with simultaneous insertions and deletions are categorized into insertions but not deletions in this study. Small insertions were 96.1% of all identified Cas9-induced insertions and 1-bp insertion dominated all insertions in CH12F3 cells at the c-Myc locus (Figure 3A and B). Dominant small insertions were also detected in CRISPR–Cas9-edited CH12F3 cells at Bcr locus and mESCs at four different loci (Figure 3B). In small insertions, the 1-bp insertions identical to the fourth nucleotide T upstream of NGG occurred most frequently, up to 79.8% of total insertions at the c-Myc locus (Figure 3C, #1 in top panel), resulted from the staggered cleavage of Cas9 (36,37). In this context, we found more examples of inserted nucleotides between the fiurth and fifth nucleotides upstream of NGG, ranking high in all small insertions (Figure 3C, #3, #6 and #10 in top panel). In the absence of Ku80 and Lig4, the 1-bp insertions declined significantly, especially for the top T insertions, from 79.8% in WT cells to 5.3% and 13.9% in Lig4- and Ku80-deficient cells, respectively (Figure 3C–E and Supplementary Figure S4A). Conversely, the level of insertions longer than 1 bp increased dramatically in both Lig4- and Ku80-deficient cells (Figure 3D and E). Whereas in the absence of Parp1, both frequency and order of top 10 insertions were highly similar to those in WT cells. The reproducible patterns suggest that the sequence and frequency of small insertions are recurrent with an unexplored mechanism (38).
Figure 3.
Recurrent small insertions and deleterious plasmid integrations. (A) The distribution of insertions with indicated length at the c-Myc locus in CH12F3 cells. Total junctions of three repeats are plotted on a log scale. 1-bp insertion is indicated by the black arrow. (B) Percentages of small (<20 bp) and large (≥20 bp) insertions among total insertions at the c-Myc and Bcr locus in CH12F3 cells (left) and four indicated loci in mESCs (right). Error bars, mean ± SD. (C) Top 10 most frequent insertions at the c-Myc locus in WT and Lig4–/- CH12F3 cells. The target site is shown on the top. Bases at cleavage site are in bold and inserted bases are in orange. The numbers from three repeats (R1, R2, R3) and average percentages (Avg%) of each type of insertions among total insertions are listed in the table, filled with gradient color from the maximum (red) to the minimum (blue) frequencies. (D) The distribution patterns of insertion length at the c-Myc locus in CH12F3 cells with indicated backgrounds. Total junctions of three repeats are plotted on a log scale. 1-bp insertions are pointed out by the black triangles. (E) Bar chart showing the percentages of insertions with indicated length among total insertions in CH12F3 cells with indicated backgrounds. Error bars, mean ± SD. One-tailed t-test, ****P < 0.0001; n.s., not significant. (F) The numbers of plasmid integrations with (+) or without (–) AAV2 ITR at the c-Myc locus in CH12F3 cells (top). Frequencies of integrated plasmid fragments among total editing events are shown at the bottom. Error bars, mean ± SD. (G) Coverage of plasmid integrations at the c-Myc locus in CH12F3 cells with (bottom) or without (top) AAV2 ITR. (H) The numbers and rates of plasmid integrations normalized to total editing events at indicated loci in mESCs.
We also noticed a pileup of large insertions around 40 bp in CRISPR–Cas9-edited CH12F3 cells at the c-Myc locus (Figure 3A). We extracted the inserted sequences from CH12F3 cells at the c-Myc locus to align to the Cas9-carrying plasmid and found 45 distinct inserted sequences originated from the transfected plasmids (Figure 3F, Supplementary Figure S4B, Structure I; Supplementary Table S3), indicating potential plasmid integrations into the genome during CRISPR–Cas9 editing. To gain deep insight into plasmid integrations, we performed PEM-Q analysis against the mouse genome and then the plasmid backbone sequence for sequence alignment. Three types of plasmid integrations with no overlap between the R1 and R2 sequencing reads were recovered at a frequency of ∼2% of total editing events at the c-Myc locus (Figure 3F and Supplementary Figure S4B, Structure II). The inserted sequences evenly covered the whole plasmid backbone (Figure 3G, top in cyan).
Interestingly, when using another Cas9-carrying plasmid with an adeno-associated virus 2 (AAV2) inverted terminal repeat (ITR) sequence for transfection, we observed an elevated level, although not statistically significant, of total plasmid integrations (Figure 3F and Supplementary Figure S4C). The ITR region forms a hairpin structure that affects the vector stability and, therefore, the ITR regions become a hotpot for plasmid integration (Figure 3G, bottom in salmon) (39). Moreover, the ITR-carrying plasmid is integrated into the genome at the CRISPR–Cas9-edited mESCs at different target sites (Figure 3H). This finding had striking similarities with viral integrations when using AAV to deliver Cas9 (39,40), which indicates that ITR or similar fragile sites are a significant cause for vector integrations.
Distribution profile of translocations induced by CRISPR–Cas9
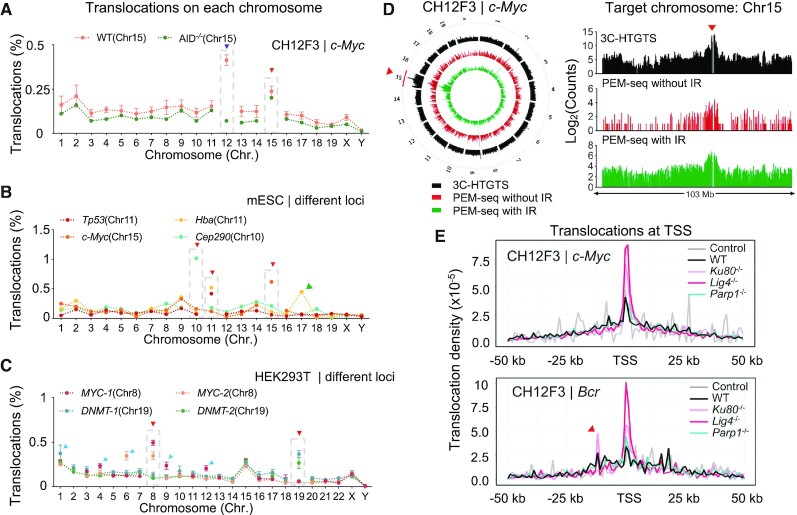
Translocation links the bait DSBs at the target site to genome-wide prey DSBs and thereby the translocation junctions represent the breakpoints of prey DSB in PEM-seq (28). Since large deletions can expand to the downstream region as long as 500 kb as revealed above (Figure 2B), we excluded identified junctions within upstream and downstream 500 kb of target sites for translocation analysis. Translocation junctions distributed widely in the genome with an obvious enrichment at the target chromosome Chr15 when editing the CH12F3 cells at c-Myc locus via CRISPR–Cas9 (Figure 4A). Similar enrichment in the target chromosome was also detected in PEM-seq libraries from CRISPR/Cas9-edited mESCs or human HEK293T cells at various loci (Figure 4B and C). Translocation requires the proximity of two DSBs and chromatin interaction plays an indispensable role in the formation of translocation as revealed by the I-SceI-induced translocations (30). We employed the 3C-HTGTS (29) to check the global interactions with the Cas9-target site. The distribution profile of translocation junctions was highly correlated with the interaction intensity revealed by 3C-HTGTS globally or within the target chromosome (Figure 4D and Supplementary Figure S5A). With this regard, the target chromosome Chr15 showed the most robust interaction intensity with the target site and thereby had the most translocations (Figure 4D). We also used 5-Gy ionizing radiation (IR) to generate genome-wide DSBs that are independent of Cas9. IR-induced translocations captured by Cas9-induced target DSBs at the c-Myc locus were also correlated to interaction intensity (Figure 4D and Supplementary Figure S5A).
Figure 4.
Distribution profile of translocations induced by CRISPR–Cas9. (A) The distribution patterns of translocation junctions on each chromosome at the c-Myc locus in WT and AID–/– CH12F3 cells. The target chromosome Chr15 is highlighted by dashed-line boxes and indicated by the red triangle. Chr12 is indicated by a purple triangle. (B) The distribution patterns of translocation junctions at indicated loci in mESCs. The target chromosome is highlighted by dashed-line boxes and indicated by the red triangle. The green triangle indicates Hba pseudogene on Chr17. (C) The distribution patterns of translocation junctions at indicated loci in HEK293T cells. The target chromosome is highlighted by dashed-line boxes and indicated by the red triangle. Chromosomes harboring robust off-target sites are pointed by blue triangles. (D) The distribution patterns of identified junctions in the entire genome (circos plot) or Chr15 (bar graph) by 3C-HTGTS (black), PEM-seq with or without (red) IR at the c-Myc locus in AID–/- CH12F3 cells. Signals were binned into 2 Mb intervals and plotted on a log scale. The upstream and downstream 500 kb region of the c-Myc locus (indicated by a red triangle) is removed. (E) The distribution patterns of translocation junctions around TSSs in CH12F3 cells with indicated backgrounds at the c-Myc locus (top) or Bcr locus (bottom). Translocations within the IgH region are excluded for analysis. Control represents primer control libraries without Cas9 cutting. The red triangle indicates a cluster of translocation junctions within the AT rich interactive domain 1B (Arid1b) gene on Chr17.
Chr12 also exhibited an enrichment of translocations in the CRISPR–Cas9-edited CH12F3 cells at the c-Myc locus (Figure 4A). CH12F3 cells can undergo class switch recombination stimulated by anti-CD40/Interleukin 4/TGF-β and activation-induced deaminase (AID) initiates substantial DSBs in the switch (S) regions (41). We examined the hotspot region in Chr12 and the vast majority of the DSBs enriched at S regions as anticipated (Supplementary Figure S5B). Moreover, the knock-out of AID resulted in a fallback of translocation level in Chr12 (Figure 4A, in green). S regions are only activated in B lymphocytes, however, recurrent DSBs often occur at off-target sites during genome editing in non-lymphocytes. We checked the translocation junctions in CRISPR–Cas9-edited HEK293T cells and detected an elevated level of translocations in chromosomes harboring robust off-target sites (Figure 4C). Besides off-target sites, the transcribed regions are also fragile for DSBs (42). In this context, translocations enriched at the transcription start sites (TSSs) of active genes were significantly higher than those of inactive genes (Figure 4E and Supplementary Figure S5C). In the PEM-seq libraries from Ku80- or Lig4-deficient CH12F3 cells, translocation levels at TSSs were significantly higher than those from WT cells, indicating elevated levels of DSBs in transcribed regions (Figure 4E and Supplementary Figure S5D-G). Of note, the percentages of total translocations were not significantly changed between Ku80- or Lig4-deficient cells and WT cells at both c-Myc and Bcr locus, while Parp1 deficiency led to a mild increase of total translocation levels, indicating a redundant role of NHEJ and MMEJ in translocation formation(Supplementary Figure S5H-I) (12).
DISCUSSION
The off-target activity has been considered to be the main obstacle to clinical applications of CRISPR–Cas9 and similar Cas-associated genome editing toolboxes (33,43–46). Recently, more and more abnormal chromosomal structures including large deletions and translocations induced by CRISPR–Cas9 have been observed by different laboratories (10,17–20,27,28,33). These inevitable deleterious repair byproducts are generated by endogenous DNA repair pathways and cannot be easily overcome by developed high-fidelity Cas9 variants (Figure 1E). It has become another dimension of threat to genome stability besides off-targets during genome editing. In order to comprehensively assess DNA repair outcomes during genome editing, we here develop a new in silico analysis pipeline PEM-Q for the linear amplification-based sequencing method PEM-seq. Compared to previous CRISPR evaluation assays, PEM-Q is equally suited for detecting insertions and deletions but more sensitive than HTGTS and SuperQ for identifying off-targets (Figure 1B and C). Several teams have developed different methods to identify DNA repair outcomes induced by CRISPR–Cas9. However, the previous assays could not cover all the editing outcomes, especially for the deleterious byproducts, induced by CRISPR–Cas9 (27,28,31,33,47–49). Here, our newly developed pipeline PEM-Q can capture and quantify all the editing outcomes in one experiment. Importantly, PEM-Q also integrates a convenient analysis tool for vector integration (Supplementary Table S4). Of note, though PEM-Q can sensitively identify valid off-targets indirectly by translocation capture, the robustness of off-target needs to be further determined by T7 endonuclease I assay (34), while the methods including DISCOVER-seq (46) and GUIDE-seq (43) can directly measure the robustness of off-target activity at identified sites. Moreover, given that the bait primers are located around 100 bp from the cleavage sites, some junctions followed the deletions of primer binding sites may escape from PEM-seq and PEM-Q. We placed bait primers at different distances from the c-Myc target site and found that only a limited number of junctions were missed by our assay (Supplementary Figure S6).
Small deletions facilitate the disruption of target genes and are the preferred products of CRISPR–Cas9. However, large deletions may disturb neighbor genes within even hundreds of kb from the target sites (10,18,38). We identified thousands of large deletions in each PEM-seq library from mouse or human cells, lymphocytes, or embryonic stem cells and found that microhomologies are prevalent at large deletions. Besides, we also found that NHEJ deficiency would increase large deletions (Figure 2H and Supplementary Figure S3G). Different from deletions, small insertions are recurrent as revealed in this study and also described previously (36,37). The source of large insertions is mainly some DNA fragments that co-exist in the cell during CRISPR–Cas9 editing, including damaged plasmids or virus backbones (34,39,50,51). Moreover, the level of large insertions can vary at different target sites and depends on the integrity of the used vector (Figure 3H); and the deficiency of NHEJ leads to significantly elevated levels of large insertions (Figure 3E), which is not reported previously. To suppress deleterious large insertions in clinical applications, DNA-based transfection methods for Cas9 delivery should be avoided. Besides, we want to emphasize that the impact of NHEJ deficiency on total editing outcomes may vary at different sites due to sequence-dependent microhomology usage, as exhibited at c-Myc versus Bcr sites. In this context, 1-nt insertions as the most frequent editing event occupy about 22.3% of total editing events at the c-Myc locus (Supplementary Figure S2C). In contrast, 1-nt insertions are not dominant at the Bcr locus (Supplementary Figure S2C). Therefore, NHEJ deficiency leads to a dramatic decrease of 1-nt insertions and shows different extents to the profile changes of editing outcomes at these two sites (Figure 2A, Supplementary Figure S2B, and Table S5).
Translocations occur in one out of hundreds of CRISPR–Cas9-edited cells extrapolated from our findings. Translocations required two simultaneous DSBs that can interact with each other before being fused. In this context, in parallel multiple-gene targeting would induce tremendous translocations between any two target sites. Moreover, recurrent DSBs induced by off-target activity or other physiologic or pathological situations also pose a great threat to genome integrity during genome editing. For instance, translocations induced during V(D)J recombination and class switch recombination usually cause lymphoid tumorigenesis (12,16,52,53). We also showed in this study that the deficiency for NHEJ factor Ku80 or Lig4 leads to a significant increase of large deletions, large insertions, and DSBs around TSSs (Figures 2H, 3E and 4E). Therefore, previously developed methods to employ NHEJ inhibitors for promoting HR are not applicable and may pose a great threat to genome integrity during genome editing. Furthermore, besides providing the guidelines for further improving the fidelity of genome editing, PEM-seq also shows great potential to distinguish various DNA repair products in studying DNA repair pathways.
DATA AVAILABILITY
Sequencing data were deposited into NODE (National Omics Data Encyclopedia, OEP001736) and the PEM-Q pipeline is available at the GitHub site: https://github.com/liumz93/PEM-Q. Other data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Kefei Yu and Dr Xiong Ji for their generous gifts of Lig4-deficient CH12F3 cells and mESCs. We thank the other lab members for their helpful comments. We thank the Core Facility at the National Center for Protein Sciences, Peking University, particularly the Flow Cytometry Core, for technical help.
Contributor Information
Mengzhu Liu, The MOE Key Laboratory of Cell Proliferation and Differentiation, School of Life Sciences, Center for Life Sciences, Genome Editing Research Center, Peking University, Beijing 100871, China.
Weiwei Zhang, The MOE Key Laboratory of Cell Proliferation and Differentiation, School of Life Sciences, Center for Life Sciences, Genome Editing Research Center, Peking University, Beijing 100871, China.
Changchang Xin, The MOE Key Laboratory of Cell Proliferation and Differentiation, School of Life Sciences, Center for Life Sciences, Genome Editing Research Center, Peking University, Beijing 100871, China.
Jianhang Yin, The MOE Key Laboratory of Cell Proliferation and Differentiation, School of Life Sciences, Center for Life Sciences, Genome Editing Research Center, Peking University, Beijing 100871, China.
Yafang Shang, State Key Laboratory of Molecular Biology, CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences; University of Chinese Academy of Sciences, 320 Yueyang Road, Shanghai 200031, China.
Chen Ai, The MOE Key Laboratory of Cell Proliferation and Differentiation, School of Life Sciences, Center for Life Sciences, Genome Editing Research Center, Peking University, Beijing 100871, China.
Jiaxin Li, The MOE Key Laboratory of Cell Proliferation and Differentiation, School of Life Sciences, Center for Life Sciences, Genome Editing Research Center, Peking University, Beijing 100871, China.
Fei-Long Meng, State Key Laboratory of Molecular Biology, CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences; University of Chinese Academy of Sciences, 320 Yueyang Road, Shanghai 200031, China.
Jiazhi Hu, The MOE Key Laboratory of Cell Proliferation and Differentiation, School of Life Sciences, Center for Life Sciences, Genome Editing Research Center, Peking University, Beijing 100871, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key R&D Program of China [2017YFA0506700 to J.H., F.-L.M.]; NSFC [31771485 to J.H.]. Funding for open access charge: National Key R&D Program of China [2017YFA0506700 to J.H., F.-L.M.]; NSFC [31771485 to J.H.].
Conflict of interest statement. None declared.
This paper is linked to: doi.org/10.1093/nar/gkab507.
REFERENCES
- 1.Wang D., Zhang F., Gao G.. CRISPR-based therapeutic genome editing: strategies and in vivo delivery by AAV vectors. Cell. 2020; 181:136–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doudna J.A.The promise and challenge of therapeutic genome editing. Nature. 2020; 578:229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J.. RNA-programmed genome editing in human cells. Elife. 2013; 2:e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via Cas9. Science. 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A.et al.. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anzalone A.V., Koblan L.W., Liu D.R.. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020; 38:824–844. [DOI] [PubMed] [Google Scholar]
- 8.Hendriks D., Clevers H., Artegiani B.. CRISPR–Cas tools and their application in genetic engineering of human stem cells and organoids. Cell Stem Cell. 2020; 27:705–731. [DOI] [PubMed] [Google Scholar]
- 9.Pawluk A., Davidson A.R., Maxwell K.L.. Anti-CRISPR: discovery, mechanism and function. Nat. Rev. Microbiol. 2018; 16:12–17. [DOI] [PubMed] [Google Scholar]
- 10.Kosicki M., Tomberg K., Bradley A.. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018; 36:765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeh C.D., Richardson C.D., Corn J.E.. Advances in genome editing through control of DNA repair pathways. Nat. Cell Biol. 2019; 21:1468–1478. [DOI] [PubMed] [Google Scholar]
- 12.Alt F.W., Zhang Y., Meng F.L., Guo C., Schwer B.. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013; 152:417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sfeir A., Symington L.S.. Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway?. Trends Biochem. Sci. 2015; 40:701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Truong L.N., Li Y., Shi L.Z., Hwang P.Y., He J., Wang H., Razavian N., Berns M.W., Wu X.. Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:7720–7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang H.H.Y., Pannunzio N.R., Adachi N., Lieber M.R.. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017; 18:495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao B., Rothenberg E., Ramsden D.A., Lieber M.R.. The molecular basis and disease relevance of non-homologous DNA end joining. Nat. Rev. Mol. Cell Biol. 2020; 21:765–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuccaro M.V., Xu J., Mitchell C., Marin D., Zimmerman R., Rana B., Weinstein E., King R.T., Palmerola K.L., Smith M.E.et al.. Allele-specific chromosome removal after Cas9 cleavage in human embryos. Cell. 2020; 183:1650–1664. [DOI] [PubMed] [Google Scholar]
- 18.Cullot G., Boutin J., Toutain J., Prat F., Pennamen P., Rooryck C., Teichmann M., Rousseau E., Lamrissi-Garcia I., Guyonnet-Duperat V.et al.. CRISPR–Cas9 genome editing induces megabase-scale chromosomal truncations. Nat. Commun. 2019; 10:1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adikusuma F., Piltz S., Corbett M.A., Turvey M., McColl S.R., Helbig K.J., Beard M.R., Hughes J., Pomerantz R.T., Thomas P.Q.. Large deletions induced by Cas9 cleavage. Nature. 2018; 560:E8–E9. [DOI] [PubMed] [Google Scholar]
- 20.Zuo E., Huo X., Yao X., Hu X., Sun Y., Yin J., He B., Wang X., Shi L., Ping J.et al.. CRISPR/Cas9-mediated targeted chromosome elimination. Genome Biol. 2017; 18:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ling X., Xie B., Gao X., Chang L., Zheng W., Chen H., Huang Y., Tan L., Li M., Liu T.. Improving the efficiency of precise genome editing with site-specific Cas9-oligonucleotide conjugates. Sci. Adv. 2020; 6:eaaz0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riesenberg S., Maricic T.. Targeting repair pathways with small molecules increases precise genome editing in pluripotent stem cells. Nat. Commun. 2018; 9:2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Z., Shi Z., Guo X., Jiang B., Wang G., Luo D., Chen Y., Zhu Y.S.. Ligase IV inhibitor SCR7 enhances gene editing directed by CRISPR–Cas9 and ssODN in human cancer cells. Cell Biosci. 2018; 8:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kuhn R.. Increasing the efficiency of homology-directed repair for CRISPR–Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015; 33:543–548. [DOI] [PubMed] [Google Scholar]
- 25.Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R., Ploegh H.L.. Increasing the efficiency of precise genome editing with CRISPR–Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015; 33:538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rees H.A., Yeh W.H., Liu D.R.. Development of hRad51-Cas9 nickase fusions that mediate HDR without double-stranded breaks. Nat. Commun. 2019; 10:2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turchiano G., Andrieux G., Klermund J., Blattner G., Pennucci V., El Gaz M., Monaco G., Poddar S., Mussolino C., Cornu T.I.et al.. Quantitative evaluation of chromosomal rearrangements in gene-edited human stem cells by CAST-Seq. Cell Stem Cell. 2021; 28:1136–1147. [DOI] [PubMed] [Google Scholar]
- 28.Yin J., Liu M., Liu Y., Wu J., Gan T., Zhang W., Li Y., Zhou Y., Hu J.. Optimizing genome editing strategy by primer-extension-mediated sequencing. Cell Discov. 2019; 5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jain S., Ba Z., Zhang Y., Dai H.Q., Alt F.W.. CTCF-Binding elements mediate accessibility of RAG substrates during chromatin scanning. Cell. 2018; 174:102–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y., McCord R.P., Ho Y.J., Lajoie B.R., Hildebrand D.G., Simon A.C., Becker M.S., Alt F.W., Dekker J.. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012; 148:908–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinello L., Canver M.C., Hoban M.D., Orkin S.H., Kohn D.B., Bauer D.E., Yuan G.C.. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol. 2016; 34:695–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu J., Meyers R.M., Dong J., Panchakshari R.A., Alt F.W., Frock R.L.. Detecting DNA double-stranded breaks in mammalian genomes by linear amplification-mediated high-throughput genome-wide translocation sequencing. Nat. Protoc. 2016; 11:853–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frock R.L., Hu J., Meyers R.M., Ho Y.J., Kii E., Alt F.W.. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 2015; 33:179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang W., Yin J., Zhang-Ding Z., Xin C., Liu M., Wang Y., Ai C., Hu J.. In-depth assessment of the PAM compatibility and editing activities of Cas9 variants. Nucleic Acids Res. 2021; 10.1093/nar/gkab507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Owens D.D.G., Caulder A., Frontera V., Harman J.R., Allan A.J., Bucakci A., Greder L., Codner G.F., Hublitz P., McHugh P.J.et al.. Microhomologies are prevalent at Cas9-induced larger deletions. Nucleic Acids Res. 2019; 47:7402–7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shou J., Li J., Liu Y., Wu Q.. Precise and predictable CRISPR chromosomal rearrangements reveal principles of Cas9-Mediated nucleotide insertion. Mol. Cell. 2018; 71:498–509. [DOI] [PubMed] [Google Scholar]
- 37.Chakrabarti A.M., Henser-Brownhill T., Monserrat J., Poetsch A.R., Luscombe N.M., Scaffidi P.. Target-specific precision of CRISPR-Mediated genome editing. Mol. Cell. 2019; 73:699–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen W., McKenna A., Schreiber J., Haeussler M., Yin Y., Agarwal V., Noble W.S., Shendure J.. Massively parallel profiling and predictive modeling of the outcomes of CRISPR/Cas9-mediated double-strand break repair. Nucleic Acids Res. 2019; 47:7989–8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanlon K.S., Kleinstiver B.P., Garcia S.P., Zaborowski M.P., Volak A., Spirig S.E., Muller A., Sousa A.A., Tsai S.Q., Bengtsson N.E.et al.. High levels of AAV vector integration into CRISPR-induced DNA breaks. Nat. Commun. 2019; 10:4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen G.N., Everett J.K., Kafle S., Roche A.M., Raymond H.E., Leiby J., Wood C., Assenmacher C.A., Merricks E.P., Long C.T.et al.. A long-term study of AAV gene therapy in dogs with hemophilia A identifies clonal expansions of transduced liver cells. Nat. Biotechnol. 2021; 39:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu X., Liu T., Shang Y., Dai P., Zhang W., Lee B.J., Huang M., Yang D., Wu Q., Liu L.D.et al.. ERCC6L2 promotes DNA orientation-specific recombination in mammalian cells. Cell Res. 2020; 30:732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiarle R., Zhang Y., Frock R.L., Lewis S.M., Molinie B., Ho Y.J., Myers D.R., Choi V.W., Compagno M., Malkin D.J.et al.. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011; 147:107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V., Wyvekens N., Khayter C., Iafrate A.J., Le L.P.et al.. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR–Cas nucleases. Nat. Biotechnol. 2015; 33:187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho S.W., Kim S., Kim Y., Kweon J., Kim H.S., Bae S., Kim J.S.. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014; 24:132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim D., Kim D.E., Lee G., Cho S.I., Kim J.S.. Genome-wide target specificity of CRISPR RNA-guided adenine base editors. Nat. Biotechnol. 2019; 37:430–435. [DOI] [PubMed] [Google Scholar]
- 46.Wienert B., Wyman S.K., Richardson C.D., Yeh C.D., Akcakaya P., Porritt M.J., Morlock M., Vu J.T., Kazane K.R., Watry H.L.et al.. Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science. 2019; 364:286–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brinkman E.K., Chen T., Amendola M., van Steensel B.. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014; 42:e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Connelly J.P., Pruett-Miller S.M.. CRIS.py: A versatile and high-throughput analysis program for CRISPR-based genome editing. Sci. Rep. 2019; 9:4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Labun K., Guo X., Chavez A., Church G., Gagnon J.A., Valen E.. Accurate analysis of genuine CRISPR editing events with ampliCan. Genome Res. 2019; 29:843–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nguyen Tran M.T., Mohd Khalid M.K.N., Wang Q., Walker J.K.R., Lidgerwood G.E., Dilworth K.L., Lisowski L., Pebay A., Hewitt A.W.. Engineering domain-inlaid SaCas9 adenine base editors with reduced RNA off-targets and increased on-target DNA editing. Nat. Commun. 2020; 11:4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Norris A.L., Lee S.S., Greenlees K.J., Tadesse D.A., Miller M.F., Lombardi H.A.. Template plasmid integration in germline genome-edited cattle. Nat. Biotechnol. 2020; 38:163–164. [DOI] [PubMed] [Google Scholar]
- 52.Hu J., Tepsuporn S., Meyers R.M., Gostissa M., Alt F.W.. Developmental propagation of V(D)J recombination-associated DNA breaks and translocations in mature B cells via dicentric chromosomes. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:10269–10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu J., Zhang Y., Zhao L., Frock R.L., Du Z., Meyers R.M., Meng F.L., Schatz D.G., Alt F.W.. Chromosomal loop domains direct the recombination of antigen receptor genes. Cell. 2015; 163:947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data were deposited into NODE (National Omics Data Encyclopedia, OEP001736) and the PEM-Q pipeline is available at the GitHub site: https://github.com/liumz93/PEM-Q. Other data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.