Abstract
The Caenorhabditis elegans genome encodes nineteen functional Argonaute proteins that use 22G-RNAs, 26G-RNAs, miRNAs or piRNAs to regulate target transcripts. Only one Argonaute is essential under normal laboratory conditions: CSR-1. While CSR-1 has been studied widely, nearly all studies have overlooked the fact that the csr-1 locus encodes two isoforms. These isoforms differ by an additional 163 amino acids present in the N-terminus of CSR-1a. Using CRISPR-Cas9 genome editing to introduce GFP::3xFLAG into the long (CSR-1a) and short (CSR-1b) isoforms, we found that CSR-1a is expressed during spermatogenesis and in several somatic tissues, including the intestine. CSR-1b is expressed constitutively in the germline. small RNA sequencing of CSR-1 complexes shows that they interact with partly overlapping sets of 22G-RNAs. Phenotypic analyses reveal that the essential functions of csr-1 described in the literature coincide with CSR-1b, while CSR-1a plays tissue specific functions. During spermatogenesis, CSR-1a integrates into an sRNA regulatory network including ALG-3, ALG-4 and WAGO-10 that is necessary for fertility at 25°C. In the intestine, CSR-1a silences immunity and pathogen-responsive genes, and its loss results in improved survival from the pathogen Pseudomonas aeruginosa. Our findings functionally distinguish the CSR-1 isoforms and highlight the importance of studying each AGO isoform independently.
INTRODUCTION
From the discovery of miRNAs to understanding the molecular mechanisms of RNA interference (RNAi), the nematode Caenorhabditis elegans has been a champion of sRNA biology (1). At the core of small RNA (sRNA) mediated gene regulatory pathways are the conserved Argonaute (AGO) proteins, which are guided in a sequence specific manner by sRNAs to impart a variety of gene regulatory outcomes on target transcripts (2). C. elegans possesses a remarkable expanded family of nineteen AGO proteins which function in association with four types of sRNAs (including miRNAs, piRNAs, 22G-RNAs and 26G-RNAs). These sRNA pathways regulate gene expression to orchestrate normal developmental and differentiation programs, as well as to navigate stressful environmental conditions, such as elevated temperature (3) (Seroussi, in preparation). The scope of sRNA mediated gene regulatory potential in the worm grows more complex when considering that several AGOs, including ALG-1, ALG-2, ERGO-1, PPW-1 and CSR-1 are predicted to be expressed as multiple isoforms, which have not been characterized.
The AGO CSR-1 (Chromosome Segregation and RNAi Deficient) is one of the most intensively studied AGOs in C. elegans, owing in part to the fact that it is the only solely essential ago, and to its potentially conflicting and varied roles in gene regulation throughout development (3,4). The csr-1 locus encodes two isoforms, CSR-1a and CSR-1b, that vary only in their N-terminus by 163 amino acids (4). Within this extended N-terminal region of CSR-1a are fifteen arginine/glycine RGG/RG motifs, many of which are arranged into di- or tri-RGG/RG motifs (5). In AGOs of other species, including Drosophila Piwi, RGG/RG motifs are the sites of arginine di-methylation, which is required for the recruitment of additional factors, such as Tudor proteins, to ensure functionality of the AGO/sRNA complex (called the RISC; RNA Induced Silencing Complex) (6–8). These motifs are also typical of RNA binding proteins, and have been identified in association with phase separated RNA rich cytoplasmic granules, such as germ granules (9). The role and importance of the RGG/RG motifs in CSR-1a have not been determined, and CSR-1 has been widely studied as a single protein, thus any differential functions of CSR-1 isoforms during development are currently unknown.
Previous studies indicated that CSR-1 is expressed in both the germline and soma in hermaphrodites (4,10). Immunostaining with antibodies that recognize both CSR-1 isoforms showed that CSR-1 is present in the hermaphrodite germline throughout all stages of germline development, from the two-cell embryo to spermatogenesis and oogenesis (4). Within the germline, CSR-1 is found in the cytoplasm, and is enriched in phase separated germ granules known as P granules (4). In oocytes and during early embryonic cell divisions, CSR-1 is present within the nucleus and associates with condensed chromosomes upon nuclear envelope breakdown (4). In the soma, CSR-1 was observed within intestinal cells, where it was detected in both the cytoplasm and nucleus (4,10). The germline expression pattern of CSR-1 highlights its role in fertility, as homozygous null mutants that eliminate both csr-1 isoforms are nearly sterile, and the few embryos produced display chromosome segregation defects during early embryonic divisions that result in lethality (3,4,11). However, which CSR-1 isoform is expressed in which tissues, and which isoform is necessary for fertility are unexplored.
While most AGOs are thought of as gene silencers, CSR-1 has been shown to play both positive (licensing) and negative (silencing) gene regulatory roles (11–13). CSR-1 associates with a type of endogenous sRNA known as 22G-RNAs that target nearly 5000 germline-expressed protein coding genes (about 25% of the genome) (4,14). CSR-1-associated 22G-RNAs are produced by the RNA dependent RNA polymerase (RdRP), EGO-1, one of four RdRPs present in the worm (4,15). Microarray and GRO-seq studies demonstrated that loss of csr-1 does not result in the global up-regulation of the majority of target transcripts (4,11,14,16). Instead, most CSR-1 target transcripts decrease in expression levels in csr-1 mutants, pointing to its role in the protection or licensing of target transcripts, rather than their destruction or silencing.
There are several ways in which CSR-1 has been implicated in promoting the expression of its target transcripts. First, CSR-1 has been shown to promote the expression of histones. In this capacity, CSR-1 is thought to act as an endonuclease or ‘Slicer,’ where it cleaves the 3′ stem loop of histone RNA transcripts enabling them to be translated. Loss of csr-1 leads to decreased levels of histone transcripts and proteins (10). Second, CSR-1 licenses the expression of germline GFP transgenes in opposition to the silencing effects of the piRNA pathway, in a process related to paramutation, termed RNA activation (RNAa) (17,18). Third, CSR-1 promotes the formation of euchromatin at its numerous target gene loci throughout the genome, and loss of csr-1 leads to aberrant accumulation of heterochromatic histone modifications and decreased transcription at target gene loci (16,19).
CSR-1 is one of only a few AGOs which possess a predicted Slicer motif, DEDH, within its PIWI domain, rendering it capable of endonucleolytic cleavage of target transcripts. CSR-1 exhibits Slicer activity in vitro (20), and mutation of the Slicer residues leads to embryonic lethality, although at a later developmental stage than null mutants (11,13). Slicer activity is required for several context-specific functions, including fine tuning kinetochore and cell cycle gene expression during embryogenesis and clearing maternal mRNAs at the oocyte to embryo transition, RNAi, processing histone mRNA transcripts, and stimulating the production of 22G-RNAs (4,10,11,13,20,21).
While most studies of CSR-1 have focused on the oogenic gonad and embryos, CSR-1 has also been implicated in spermatogenesis. Spermatogenesis occurs in the L4 larval stage of development, prior to the onset of oogenesis (22). In spermatogenesis, the AGOs ALG-3 and -4 regulate a set of spermatogenesis transcripts both positively and negatively via endogenous sRNAs called 26G-RNAs (14,23). 26G-RNAs are produced by the RdRP RRF-3, which synthesizes dsRNA that is processed by the endonuclease Dicer and the phosphatase PIR-1 (24). The targeting of transcripts by 26G-RNAs leads to the amplification of 22G-RNAs by the RdRPs EGO-1 and RRF-1, and the subsequent targeting of the same transcripts by different 22G-RNA/AGO RISCs (23,25–27). alg-3; alg-4 double mutants display a progressive loss of male and hermaphrodite fertility due to spermatogenesis defects (14,23). Curiously, while loss of csr-1 in hermaphrodites leads to sterility in one generation (3,4), loss of csr-1 in males leads progressive loss of fertility similar to that of alg-3, alg-4 double mutants (14), suggesting a distinct role for CSR-1 in spermatogenesis.
The characterization of CSR-1 associated 22G-RNAs in males showed that CSR-1 targets a set of protein coding genes in common with CSR-1 targets in hermaphrodites undergoing oogenesis, plus an additional set of genes. Furthermore, a subset of the genes targeted by CSR-1 during spermatogenesis overlap with genes targeted by ALG-3 and ALG-4. Because 26G-RNAs associated with ALG-3 and -4 are thought to lead to the production of 22G-RNAs, this places CSR-1 downstream of ALG-3 and -4 during spermatogenesis (14). Consistent with its role in oogenesis, CSR-1 positively regulates its spermatogenic targets at the level of transcription during spermatogenesis (14), and depletion of csr-1 by RNAi led to the upregulation of spermatogenesis genes in oogenesis (28). Why CSR-1 regulates the same subsets of genes in the spermatogenic and oogenic gonad, and which CSR-1 isoform(s) affect male fertility remain to be determined. Finally, although CSR-1 has been detected in the germline throughout development, transcriptomic analyses of isolated male and female C. elegans gonads showed that the csr-1a mRNA is abundant in the spermatogenic gonad, but is not highly expressed during oogenesis. These data point to a potential role for CSR-1a specifically during spermatogenesis that has not been explored (29).
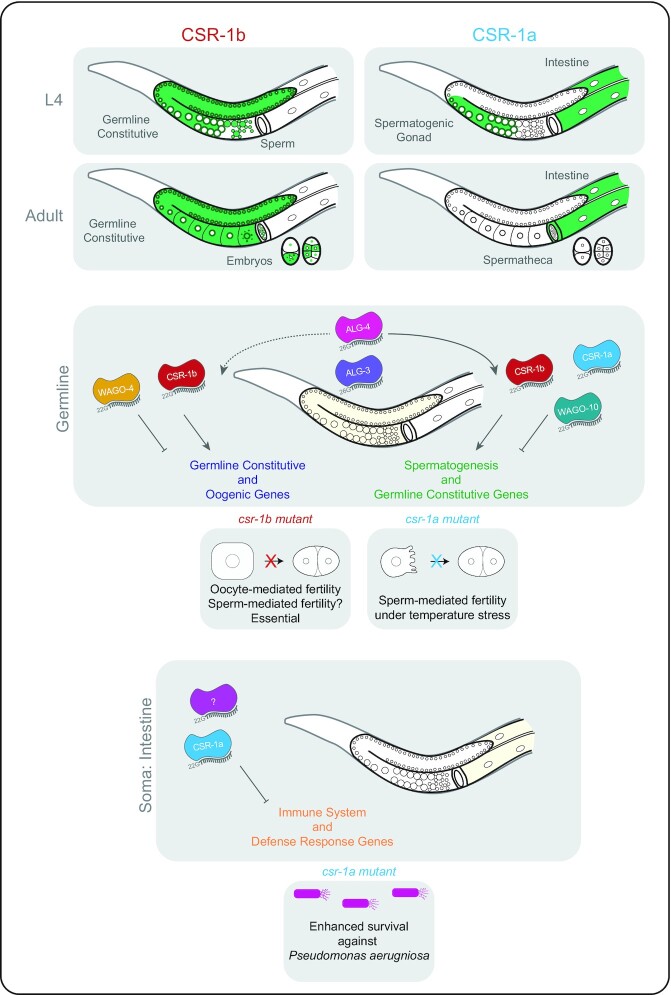
Here, we show that the two isoforms of CSR-1 have distinct expression profiles throughout development. The long isoform, CSR-1a, is expressed in the germline only during spermatogenesis, while the short isoform, CSR-1b, is expressed constitutively in the germline throughout all stages of development. During spermatogenesis, CSR-1a and CSR-1b integrate into a larger sRNA network to regulate many facets of spermatogenesis. While CSR-1b, in conjunction with WAGO-4, associates with 22G-RNAs to regulate germline constitutive (i.e. expressed both during spermatogenesis and oogenesis) and oogenic transcripts (29), CSR-1a associates with 22G-RNAs to target a set of spermatogenesis-enriched transcripts (29). CSR-1a and another uncharacterized AGO, WAGO-10, integrate into the ALG-3 and -4 26G-RNA pathway to enable male fertility during temperature stress. CSR-1b also plays a role in regulating spermatogenesis genes, but is essential for hermaphrodite fertility and embryonic viability. CSR-1a is also expressed in several somatic tissues, including the intestine, where it regulates immunity and pathogen response genes. Remarkably, loss of csr-1a renders worms more resistant to death by the bacterial pathogen Pseudomonas aeruginosa. Collectively, our data demonstrate that different AGO isoforms can display distinct expression profiles, and are capable of different functions, highlighting the importance of studying each AGO isoform as a separate entity.
MATERIALS AND METHODS
C. elegans genetics and culture
C. elegans strains were derived from the wild type Bristol N2 strain and cultured according to standard growth conditions (30) on Nematode Growth Medium (NGM) plates inoculated with E. coli OP50 at 20°C, unless otherwise stated. Synchronised worms were reared at 20°C and harvested as follows: L1s at 8 h after plating, L2s at 26 h, L3s at 34.5 h, L4s at 48 h, young adults at 60 h and gravid adults at 72 h. Embryos were collected from gravid adult worms by standard bleaching 72 hours after plating.
Strain construction and validation
Strains were constructed using the following methods. Genotypes are presented in the Strains Table 1.
Table 1.
Strains used in this study
| Experimental models: organisms/strains | Source | Strain name |
|---|---|---|
| Wild-type C. elegans, Bristol Strain | CGC | N2 |
| csr-1(tor67[gfp::3xflag::csr-1]) IV | (37) | JMC101 |
| csr-1(tor160[csr-1 exon1::gfp::3xflag IV:7957568]) IV | This study | JMC151 |
| csr-1(tor67[csr-1 exon2::gfp::3xflag IV:7958598], csr-1(mg660[G120*])) IV | This study | JMC164 |
| csr-1(tm892) IV/nT1 [unc-?(n754) let-?] (IV;V) | (3); (4) | WM182 |
| unc-119(ed3) III; mgTi13[rpl-28::gfp::csr-1b]; csr-1(tm892) | This study | JMC160 |
| unc-119(ed3) III; mgTi11[rpl-28::csr-1a exon1::gfp]; csr-1(tm892) | This study | JMC161 |
| mgIs75[rpn-2p::gfp] IV; mgTi14[pbs-5::mcherry] | This study | GR3035 |
| unc-119(ed3) III; mgTi11[rpl-28::csr-1a exon1::gfp::csr-1] | This study | GR3037 |
| csr-1(mg657[G91R]) IV | This study | GR3039 |
| unc-119(ed3) III; mgTi13[rpl-28::gfp::csr-1b] | This study | GR3041 |
| csr-1(mg660[G120*]) IV | This study | GR3042 |
| csr-1(mg661[G120*]) mgIs75[rpn-2p::gfp] IV; mgTi14[pbs-5::mcherry] | This study | GR3043 |
| wago-10(tor133) V | This study | JMC192 |
| alg-4(tm1184) III | (3) | |
| alg-3(tm1155) IV | (3) | |
| alg-4(tm1184) III; csr-1(tor67[csr-1 exon2::gfp::3xflag IV:7958598], csr-1(mg660[G120*])) IV, alg-3(tm1155) IV; wago-10(tor133) V | This study | JMC245 |
| alg-3(tor141[gfp::3xflag::alg-3]) IV | This study | JMC205 |
| alg-4(tor143[gfp::3xflag::alg-4]) III | This study | JMC207 |
| wago-10(tor127[gfp::3xflag::wago-10]) V | This study | JMC233 |
| wago-10(tor162[3xflag::wago-10]) V | This study | JMC248 |
| wago-4(tor117[gfp::3xflag::wago-4]) II | (38) | JMC223 |
| pha-1(e2123) | (31) | GE24 |
| znfx-1(gg634[HA::tagRFP::znfx-1]) II. | (65) | YY1446 |
| pgl-1(gg640[pgl-1::flag::mCardinal]) IV | (65) | YY1492 |
| pgl-1(zuIs252[Pnmy-21::PGL-1::mRFP; unc119(+)] unc119(ed3)) III; | (95) | JJ2213 |
| wago-4(tor117[gfp::3xflag::wago-4]) II, znfx-1(gg634[HA::tagRFP::znfx-1]) II | This study | JMC255 |
| znfx-1(gg634[HA::tagRFP::znfx-1]) II; csr-1(tor160[csr-1 exon1::gfp::3xflag IV:7957568]) IV | This study | JMC259 |
| znfx-1(gg634[HA::tagRFP::znfx-1]) II; csr-1(tor67[csr-1 exon2::gfp::3xflag IV:7958598], csr-1(mg660[G120*])) IV | This study | JMC260 |
| pgl-1(zuIs252[Pnmy-21::PGL-1::mRFP; unc119(+)] unc119(ed3)) III; csr-1(tor160[csr-1 exon1::gfp::3xflag IV:7957568]) IV | This study | JMC277 |
| pgl-1(zuIs252[Pnmy-21::PGL-1::mRFP; unc119(+)] unc119(ed3)) III; csr-1(tor67[csr-1 exon2::gfp::3xflag IV:7958598], csr-1(mg660[G120*])) IV | This study | JMC278 |
| znfx-1(gg634[HA::tagRFP::znfx-1]) II, wago-4(tm1019) II; csr-1(tor160[csr-1 exon1::gfp::3xflag IV:7957568]) IV | This study | JMC279 |
| znfx-1(gg634[HA::tagRFP::znfx-1]) II, wago-4(tm1019) II; csr-1(tor67[csr-1 exon2::gfp::3xflag IV:7958598], csr-1(mg660[G120*])) IV | This study | JMC276 |
| znfx-1(gg634[HA::tagRFP::znfx-1]) II, wago-4(tor117[gfp::3xflag::wago-4]) II; csr-1(mg660[G120*]) IV | This study | JMC275 |
| wago-4(tor117[gfp::3xflag::wago-4]) II; pgl-1(zuIs252[Pnmy-21::PGL-1::mRFP; unc119(+)] unc119(ed3)) III; csr-1(mg660[G120*]) IV | This study | JMC283 |
| wago-4(tm1019) II; pgl-1(zuIs252[Pnmy-21::PGL-1::mRFP; unc119(+)] unc119(ed3)) III; csr-1(tor160[csr-1 exon1::gfp::3xflag IV:7957568]) IV | This study | JMC284 |
| wago-4(tm1019) II; pgl-1(zuIs252[Pnmy-21::PGL-1::mRFP; unc119(+)] unc119(ed3)) III; csr-1(tor67[csr-1 exon2::gfp::3xflag IV:7958598], csr-1(mg660[G120*])) IV | This study | JMC285 |
| znfx-1(gg634[HA::tagRFP::znfx-1]) II; alg-4(tm1184) III; csr-1(tor67[csr-1 exon2::GFP::Flag IV:7958598], csr-1(mg660[G120*])) IV; alg-3(tm1155) IV; wago-10(tor133) V | This study | JMC286 |
Co-CRISPR
Strains JMC164, GR3042, GR3043 were constructed from JMC101, GE24 [pha-1(e2123) III] (31), and GR3042 respectively, using the Co-CRISPR technique. All sgRNA sequences were inserted into pDD162 by New England Biolabs Q5 site-directed mutagenesis as described (32). Repair template single-stranded DNA oligonucleotides were designed as described (33,34). Injections were performed using editing of dpy-10 (to generate cn64 rollers) as a phenotypic co-CRISPR marker (34,35). Injection mixes contained 60ng/μl each of the co-CRISPR and gene of interest sgRNA/Cas9 construct, and 50ng/μl each of the co-CRISPR and gene of interest repair template oligonucleotides. To generate the wago-10(tor133) allele, which deletes the region between nucleotides 695–2394 (1699nt deletion), multiple sgRNAs spanning the wago-10 genomic region were designed and injected along with the dpy-10 co-CRISPR plasmid as described above. Insertion/deletion events were screened by PCR. sgRNA and homologous repair template sequences are listed in Supplemental Table S3. All CRISPR edits were confirmed by sequencing.
CRISPR/Cas9 homologous recombination with self-excising cassette
Strains JMC101 and JMC151 were generated using this method. CRISPR was performed as described (36–38). The repair templates were amplified using primers listed in the Supplemental Table S3 and introduced into the vector pDD282 (Addgene #66823) by isothermal assembly (39). Plasmids expressing guide RNAs were generated with the NEBuilder kit (New England BioLabs) using pDD162 (Addgene #47549) and the primers listed in the Supplemental Table S3.
miniMos transgenesis
Strains GR3037 and GR3041 were made using this method. Cloning was performed by isothermal/Gibson assembly (39). All plasmids used for transgenesis are listed in Supplemental Table S3, and were assembled using pNL43 (40). MiniMos injections were performed as described in (41). To make the strain GR3037, exon 1 of csr-1a (a 474 bp fragment beginning at the start codon of csr-1a) was placed downstream of the rpl-28 promoter (605 bp immediately upstream of the rpl-28 start codon) and fused in-frame at the 5′ end of GFP. The 3′ end of the GFP coding sequence was fused in-frame with a DNA fragment containing the remaining exons of csr-1a and the csr-1 3′UTR (3829 bp beginning at the first codon of exon 2 of csr-1a). This construct (assembled in pNL201 and used to generate mgTi11 and mgTi12) drives the ubiquitous expression of CSR-1a with an internal GFP tag inserted after K158. To make strain GR3041, the DNA fragment containing GFP fused in-frame to csr-1a exons 2 through the csr-1 3′UTR (4697 bp, from pNL201) was placed downstream of the rpl-28 promoter (605 bp immediately upstream of the rpl-28 start codon). This construct (assembled in pNL213 and used to generate mgTi13) drives the ubiquitous expression of CSR-1b fused to GFP at its N-terminus via a short linker with the sequence ‘GLNSD’.
Generation of the rpn-2p::gfp transgene
The integrated mgIs75[rpn-2p::gfp::unc-54] transgene was generated from sEx10255 (42). EMS mutagenesis was used to induce integration of the extrachromosomal array onto chromosome IV.
Mutagenesis, screening and genome sequencing
Mutagenesis was performed by treatment of L4 animals in 47 mM EMS for 4 h at 20°C. Genomic DNA was prepared using the Gentra Puregene Tissue kit (Qiagen) according to the manufacturer's instructions. Genomic DNA libraries were prepared using the NEBNext genomic DNA library construction kit (New England Biololabs), and sequenced on an Illumina Hiseq. Deep sequencing reads were analyzed using Cloudmap (43).
Microscopy
Live fluorescence microscopy of CSR-1::GFP
Staged animals were picked into a 1× M9 (22 mM KH2PO4, 42 mM Na2HPO4, 86 mM NaCl) droplet with 2 μl of 2 mM levamisole on standard microscopy slides with 2% agarose pads. Imaging was performed on a Nikon TiE inverted microscope and Nikon C2 confocal system with a PL Apochromat Lambda 60× Oil Immersion objective. Images were acquired using NIS Elements AR software. For time course images, larvae and embryo images were taken every 60 seconds. All images were acquired with the same settings and exported via NIS-Elements AR Software. Any post-acquisition adjustments made to images were applied equally between control and experimental sets using Fiji (44) and Nikon NIS viewer.
Imaging released sperm
Males were moved to plates without hermaphrodites 1–2 days prior to imaging to maximize sperm retention. Males were soaked in 2 ml of 1× Sperm Buffer (50 mM HEPES, 25 mM KCl, 35 mM NaCl, 1 mM MgSO4, 5 mM CaCl2, 10 mM dextrose/D-glucose, pH 7.8, filter sterilized) with 0.5 μl of 5 μg/ml DAPI for 5 min in a crystallization dish, then transferred to poly-L-lysine coated slides. 2 mM levamisole was added, and worm tails were cut with a 17-gauge needle to release sperm/germline. Samples were imaged immediately using Sequential Scan Mode on a Leica DMi8 TCS SP8 confocal microscope using a 63×/1.40 HC PL APO CS2 oil objective.
CSR-1 zygotic expression assay
Male worms homozygous for GFP::3XFLAG::CSR-1 were crossed to N2 hermaphrodites. After 24 h, hermaphrodites were collected and dissected to obtain early embryos. Later staged embryos and L1 larvae were obtained by washing plates with 1× M9. Embryos and larvae were transferred to slides with 2% agarose pads. 2 μl of 2 mM levamisole was added to immobilize larvae. Samples were imaged at 60× on a Nikon TiE inverted microscope and Nikon C2 confocal system utilizing the NIS-Elements AR Software.
Imaging and quantification of germ granules
Worms were washed from plates in M9 and immobilized on positively charged glass slides with M9 and 10 μM levamisole. Germlines were dissected with a 17-gauge needle and then imaged immediately on a Leica DMi8 TCS SP8 confocal microscope using a 63×/1.40 HC PL APO CS2 oil objective. All images were collected using the same acquisition settings. Colocalization of proteins was calculated with the ImageJ plugin JaCoP (45). One germline from each of six different animals was imaged per strain and developmental time point. Regions of interest (ROI) were generated using the 3D objects counter plugin in ImageJ (46) by adjusting the threshold until only germ granule pixels are detected. Mander's co-localization coefficients are calculated using JaCoP (45). To count the number of germ granules per nucleus, images were split into separate channels using ImageJ, and the ‘Find Maxima’ function was used to identify granules around each nucleus in each channel. Z-slices corresponding to the center of the nucleus were counted from 20 nuclei per germline in five germlines per genotype (100 nuclei total). To analyze any differences in the number of granules per nucleus among strains, mixed-effects ANOVA was used, as calculated by ‘lmer()’ in R (version 3.6.3). Hypothesis testing was performed with the ‘car’ package (47) using Type II sums of squares. Residual data points were normally distributed.
Imaging and analysis of rpn-2p::gfp transgenic worms
Bright field and GFP fluorescence images showing rpn-2p::GFP expression were collected using a Zeiss AxioZoom V16, equipped with a Hammamatsu Orca flash 4.0 digital camera, using Axiovision software. Images for all strains were collected using the same acquisition settings and then processed identically using ImageJ (46). To quantify rpn-2p::GFP expression, the maximum pixel intensity within the intestine of each animal was measured using ImageJ. Values were then normalized such that wild type expression was set to one.
Immunofluorescence microscopy
Gonads and embryos were excised in 1× PBS with 2 mM levamisole on poly-L-lysine coated slides from adult hermaphrodites grown at 20°C. Slides were frozen on dry-ice for at least 10 min or stored at –80°C for up to 2 weeks. Fixation and incubation with primary and secondary antibodies were performed as described by (4). DNA was stained with 1 μg/ml DAPI in 1× PBS. Imaging was performed on a Nikon TiE inverted microscope and Nikon C2 confocal system with a PL Apochromat Lambda 60× Oil Immersion objective and a Leica DMi8 TCS SP8 using a 63×/1.40 HC PL APO CS2 oil objective. Images were processed using ImageJ software (46).
DAPI staining and imaging L4 germlines
Wild-type and mutant worms were shifted from 20°C to 25°C 24 h before imaging. L4-stage worms were methanol fixed and DNA was stained with 1 μg/ml DAPI in 1× PBS. Imaging was performed on a Nikon TiE inverted microscope and Nikon C2 confocal system with a PL Apochromat Lambda 60x Oil Immersion objective. Images were acquired using NIS Elements AR software and maximum intensity projections were generated with ImageJ software (46).
Brood size assays
All brood size assays were conducted on N2, tagged strains and mutants reared at 20°C and 25°C. Strains were propagated at 20°C prior to the assay. 10–20 L4 individuals (1 worm/plate) were placed at the temperature the assay was performed. Every 24 h the adult was transferred to a fresh plate and embryos were counted. The number of larvae was scored the following day. This continued until the adult hermaphrodite was no longer laying eggs. The total number of embryos laid by a single hermaphrodite over the course of the egg-laying period was determined (total brood size), as well as the total number of larvae that hatched (viable brood size). Data were plotted using GraphPad Prism or the ggplot2 package in R (48). Statistical analyses in GraphPad Prism used unpaired t-tests with an alpha of 0.05 for pair-wise comparisons, and ordinary one-way ANOVA for comparisons of more than two samples, followed by Tukey's multiple comparisons tests. ANOVA, calculated using ‘lmer()’ in R (version 3.6.3), was used to analyze any differences in the number of viable progeny among strains. Hypothesis testing was performed with the ‘car’ package (47) using Type II sums of squares. Residual data points were normally distributed.
Pseudomonas aeruginosa survival assay
Psuedomonas aeruginosa strain PA14 was streaked on standard Luria-Bertani (LB) plates supplemented with 100 μg/ml carbenicillin and grown overnight at 37°C. Single colonies were picked and grown in 3 ml of LB overnight. 20 μl of PA14 was seeded on 3.5 cm slow killing (SK) NGM plates as previously described (49). These SK plates were subsequently incubated overnight at 37°C and then equilibrated for 2 days at 25°C. All strains used for the PA14 survival assay were grown to the gravid adult stage on 3.5 cm NGM plates at 20°C and bleached. Synchronized progeny were then grown to the L4-stage on NGM plates at 20°C. Approximately 50 L4s were picked to SK plates and moved to 25°C. Worms were transferred to new SK plates every 24 h. Dead worm carcasses were counted at 48 h, and both carcasses and live worms were counted at 72 h.
Male mating rescue assay
N2 and mutant hermaphrodites were reared at 20°C. Ten L4 hermaphrodites of each genotype were transferred to plates with either three or zero N2 males, and maintained at 25°C. After 24 h, and at every 24 h thereafter, the adult hermaphrodite was transferred to a fresh plate (males were removed), and the number of larvae was scored the following day until no more progeny were produced. Embryos/oocytes were imaged on an Olympus SZX16 Fluorescent Stereomicroscope with a Nikon DS-Fi1 camera. At 60 hours post temperature shift plates were assessed for the presence of male progeny to indicate successful mating.
Antibodies used in this study
See Table 2.
Table 2.
Antibodies used in this study
| Antibody | Reference | Name |
|---|---|---|
| Rabbit anti-CSR-1 (total) (peptide antigen: VDYNAPKDPEFRQKYPNLKFP) | (4) | 49-C1 |
| Rabbit anti-CSR-1 (total) (peptide antigen: QRCKDKGMHIGSYSMDQHNGERGSENFL) | (4) | 49-C2 |
| Rabbit anti-CSR-1a (peptide antigen: YQGKVKYRGGDRHDFNYEKTPTGSY) | This study | 56 |
| Mouse IgM anti-PGL-1 | (96) | K76 |
| Rabbit anti-GFP | Torrey Pines | RRRID: AB_2313770 |
| Mouse anti-alpha Tubulin | Sigma-Aldrish | RRID: AB_477593 |
| Monoclonal Anti-FLAG M2 antibody | Sigma-Aldrich | RRID: AB_262044 |
Protein lysate preparation
Synchronous populations of animals were grown at a density of approximately 100,000 animals per 15 cm plate on NGM with 5× concentrated OP50 Escherichia coli, harvested at staged time points in 1× M9 buffer, and washed with ddH2O. The worm pellets were flash frozen in a dry ice/ethanol bath and stored at –80°C. The frozen pellets were resuspended 1:1 (v/v) in ice-cold DROSO complete buffer (30 mM HEPES, 100 mM potassium acetate, 2 mM magnesium acetate, 0.1% Igepal CA 630 (Sigma-Aldrich), 2 mM DTT, 4× concentration complete protease inhibitor (Roche) and 1% (v/v) phosphatase inhibitors 2 and 3 (Sigma-Aldrich)) or EDTA Complete buffer (30 mM HEPES, 100 mM potassium acetate, 10 mM EDTA, 10% glycerol, 0.1% NP-40 or Igepal CA630 (Sigma-Aldrich), 2 mM DTT, 1 tablet/5 ml protease inhibitor (Roche), 1:100 phosphatase inhibitor 2 (Sigma-Aldrich), 1:100 phosphatase inhibitor 3 (Sigma-Aldrich)) and homogenized either with a stainless steel dounce homogenizer (Wheaton Incorporated) until worms and embryos were no longer visible under the microscope or by sonication (Bioruptor Pico) for 10 cycles of 30 s ON/30 s OFF. In cases where RNA was to be isolated from immunoprecipitations using the protein lysates, 1% (v/v) SUPERaseIn RNase Inhibitor (ThermoFisher) was added to the buffer prior to homogenization. Lysates were centrifuged at 13 000 × g for 10min at 4°C, and the supernatant transferred to a fresh tube. A Lowry Assay (Bio-Rad) was used to determine the total protein concentration using a Nanodrop 1800C spectrophotometer.
Western blotting
Samples (typically 10–40 μg of protein lysate) were resolved by SDS-PAGE on precast gradient gels (4–12% Bis–Tris Bolt gels, ThermoFisher) and transferred to Hybond-C membrane (Amersham Biosciences) using a Bio-Rad semi-dry transfer apparatus set at 25 V for 45 min. Membranes were washed 3 × 5 min with PBST (0.1% Tween-20 in PBS) and blocked for 1 h at room temperature with 5% milk-PBST (PBST with 5% w/v dried nonfat milk). Membranes were incubated overnight at 4°C in primary antibodies (Antibodies Table). Membranes were washed 3 × 10 min in PBST, blocked with 5% milk-PBST for 1 h at room temperature, then incubated for 1 h with secondary antibodies conjugated to HRP (Cell Signaling Technology). Membranes were washed 3 × 5 min in PBST and visualized using Luminata Forte Western HRP substrate (Sigma-Aldrich). Primary antibodies for FLAG [1 mg/ml], CSR-1 (antibody 49C1) [0.25 mg/ml] were used at 1:3000 and CSR-1a (antibody 56) [0.25–0.57 mg/ml] was used at 1:1000. Secondary antibodies listed in the Antibodies Table [1 mg/ml] were used at a concentration of 1:1000.
Immunoprecipitation
For each IP and co-IP experiment, 5 mg total protein lysate was used per reaction. For small RNA cloning 3 to 5 × 5 mg IPs were performed and beads were pooled at the final wash step. GFP-Trap Magnetic Agarose beads (ChromoTek) equilibrated in EDTA Complete buffer were used for anti-GFP IPs and RFP-Trap beads or unconjugated beads (ChromoTek) were used for the Mock IP control. Dynabead Protein G beads (Invitrogen) were used for all non GFP/RFP IPs. CSR-1 rabbit peptide antibodies used for each IP were 49C2 [1.25 mg/ml] for total CSR-1 and 56 [0.25–0.57 mg/ml] for CSR-1a (4). Dynabeads were resuspended in PBS with 0.1% Tween and incubated while rotating with 5 μg of CSR-1 antibody (or rabbit IgG for the matched Mock IP control) for 10 min at RT, then washed 3 × 5 min with EDTA complete buffer. Protein lysates were incubated with 25 μl of bead slurry for 1 h while rotating at 4°C. Beads were then washed with EDTA Complete buffer at least 5 × 10 min each. Protein was eluted from the beads and denatured by incubation with sample buffer (4× Bolt Buffer with reducing agent, ThermoFisher) for 10 min at 70°C. Input samples were prepared from the same lysate and diluted to a concentration of 2 mg/ml in sample buffer. For small RNA library samples, 10% of the total IP was taken for western blot to determine efficacy. The remaining sample was mixed with 4× Tri-Reagent (Molecular Research Center, Inc.) and RNA was extracted as described (4).
Small RNA library construction
RNA for sRNA libraries, was isolated from either total worm lysate (input) or IP samples. Up to 4 μg of RNA from the IP samples were treated with 5′ polyphosphatase (Lucigen) for 30 min at 30°C followed by a standard phenol:chloroform:isoamyl alcohol extraction and ethanol precipitation. 1 μg of total RNA from each sample was used for sRNA library preparation using the NEBNext Small RNA Library Kit for Illumina (New England Biolabs). The resulting PCR product was visualized using 10% PAGE stained with SYBRGold (Life Technologies) and a region of 140–160 bp was excised. DNA was eluted overnight at room temperature in elution buffer (3.3 M NaCl, 100 mM Tris pH 7.5, 1 mM EDTA), and precipitated with 20 μg glycogen and 1× volume isopropanol overnight at -20°C. The resulting DNA was resuspended in 20 μl Ultrapure™ water, and quantified using a Qubit HS DNA kit (ThermoFisher). All samples were pooled in equal amounts into a 20 nM solution and sequenced as 50 bp single end reads on an Illumina HiSeq 2500 or Miniseq. At least two biological replicates were prepared for each sample.
mRNA library construction
Synchronized staged animals were collected in 1× M9 and washed in 3× in M9 and 1× in ddH2O. 4× pellet volumes of Tri-Reagent (Molecular Research Center, Inc.) were added to samples, and samples were flash frozen in a dry ice/ethanol bath, then stored at –80°C for later RNA extraction. Total RNA extraction and ethanol precipitation were performed as described (4), followed by quantification using a Nanodrop 1800C spectrophotometer. 2 μg of each sample was treated with DNase I (Ambion), phenol:chloroform extracted, ethanol precipitated, resuspended in Ultrapure™ Distilled Water (ThermoFisher) and quantified using a Qubit HS RNA kit (ThermoFisher). 1 μg of total DNase treated RNA was used for library preparation using the NEBNext Ultra II Directional RNA Kit for Illumina with Poly-A selection, according to the manufacturer's manual. Samples were quantified with the Qubit HSDNA kit (ThermoFisher). Libraries were pooled in equal amounts to 20 nM final concentration and sequenced 50 bp single end reads on an Illumina HiSeq 2500 Sequencing System. For each genotype and temperature, three biological replicates were prepared.
qRT-PCR
RNA extraction was performed as described for mRNA-seq experiments above. cDNA synthesis was performed using SuperScript VILO Master Mix (Thermo Fisher), as per the manufacturers’ requirements. qRT-PCR was performed as described (4,18) on a QuantStudio 3 Real-Time PCR System. Primer sets are listed in Supplemental Table S3. All data are expressed as a ratio of the experimental transcript to the control transcript gpd-2. Error is standard deviation.
Data analysis
Small RNA enrichment analysis
Raw read quality was assessed via FASTQC (version 0.11.5; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and adapters were trimmed via Cutadapt (version 1.15) (50) using the adapter sequence ‘AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC’. Any reads that were not trimmed and were outside of the 16–30 nucleotide range were discarded ($ cutadapt -a adapter sequence -f fastq -m 16 -M 30 --discard-untrimmed). Reads were then mapped to the C. elegans genome (WormBase release WS262) using STAR-aligner (version 2.6.0c) (51) with the following command:
$ --runThreadN 12 --outSAMtype BAM SortedByCoordinate --outFilterMultimapNmax 50 --outFilterMultimapScoreRange 0 --outFilterMismatchNoverLmax 0.05 --outFilterMatchNmin 16 --outFilterScoreMinOverLread 0 --outFilterMatchNminOverLread 0 --alignIntronMax 1.
A custom R script (version 3.6.3) was used to count the number of reads for each feature (e.g. protein coding genes, pseudogenes, etc.). WormBase version WS262 PRJNA13758 ce11 canonical geneset annotations were used (excluding miRNAs). C. elegans miRNA annotations were obtained from miRBase (release 22.1). To determine targets enriched for sRNAs in Argonaute IPs, reads were first normalized to library size (reads per million - RPM). Enrichment was defined as genes having a minimum of 5 RPM in both IP samples, and greater than or equal to 2-fold enrichment in both IPs relative to Input samples. Similarly, sRNA-seq data were determined to be depleted (15) by the presence of > 5RPM in Input, and greater than or equal to 2-fold decrease in the number of reads in the mutant versus Input. All sRNA samples, including those published by other groups were analyzed using our own computational pipeline for consistency.
The size and first nucleotide distribution plots and enrichment scatter plots were made using the ggplot2 package (48). Heatmaps were generated using the pheatmap package (https://CRAN.R-project.org/package=pheatmap). Venn plots were made using VennDiagram package (https://CRAN.R-project.org/package=VennDiagram) and VennDetail Package 1.2.0 (https://github.com/guokai8/VennDetail). Statistical analysis for all Venn-pie Gene set enrichment comparisons was conducted using Fisher's Exact Test. The overlap between enriched sRNAs and various published gene sets was determined and plotted using the VennDetail Package (https://github.com/guokai8/VennDetail) using percentage parameters. Additional genes and gene groups of interest were examined using WormMine (52,53), WormExp (54) and WormBase Enrichment Suite (55). The significance in overlap between gene sets was calculated using Fisher's Exact Test. Non-templated nucleotide additions to sRNA 3′ ends were identified and quantified using a custom script. Metagene plots of sRNA distributions along target genes were made using a custom script. Replicates were aggregated and normalized by RPM. For clustering diagrams, sRNA targets were grouped via hierarchical clustering by binary distance using the pheatmap package (https://CRAN.R-project.org/package=pheatmap).
mRNA differential gene expression analysis
mRNA analysis was conducted in the same way as the small RNA analysis described above, except that reads were trimmed with the following command (retaining only reads without adapters): $ cutadapt -a adapter sequence -f fastq -m 51 -M 51. Differential expression and statistical analysis were performed using DESeq2 (56) and significantly-changed genes were designated by the P-adjusted value of <0.1. Gene overlaps and enrichment set analyses were conducted as described above.
Gene ontology analysis
Gene lists generated via our custom pipelines were subjected to Gene Ontology Analysis using the WormBase Enrichment Suite (55).
Sequence alignment and cladogram construction
csr-1 sequences for other Caenorhabditis species were obtained from the NCBI database (57), then aligned with MUSCLE (58) and custom coloured to highlight particular amino acids relevant to this study in SeaView (59). The cladogram was constructed based on the alignment.
RESULTS
A screen for proteasomal regulators uncovers csr-1a alleles
The csr-1 locus encodes two isoforms that vary by the addition of 163 amino acids to the N-terminal region of CSR-1a. In contrast to the rest of the CSR-1 protein, the N-terminal region is poorly conserved among related nematode species, with the notable exception of the RG motifs (Figure 1A-B). We became interested in the two isoforms of CSR-1 for two major reasons. First, several reports demonstrated that the two isoforms of CSR-1 were differentially expressed throughout development, suggesting they could play different roles in gene regulation in different tissues (4,29). Second, a forward genetic screen for factors that regulate the proteasome pathway serendipitously uncovered several alleles that only affect csr-1a, suggesting a somatic function specifically for this isoform (Supplementary Figure S1). In this screen, we generated a genome-integrated repetitive transgene array that drives GFP expression under control of the promoter of the rpn-2 gene (rpn-2p::gfp). rpn-2 encodes the C. elegans orthologue of Rpn2/PSMD1, a non-ATPase subunit of the 19S proteasome that is essential for viability of eukaryotic cells (Supplementary Figure S1A) (60). For reasons that are not entirely clear, the rpn-2p::gfp fusion gene was generally silenced throughout the worm, but could be activated in the intestine by proteasome inhibitors or mutations in proteasome subunits, like another proteasomal fusion gene rpt-3p::gfp (data not shown and (40)). EMS mutagenesis of this strain yielded eleven mutants that showed elevated expression of GFP in intestinal cells, including mutations in: rrf-1, rsd-2 and rde-1, which encode RNAi pathway factors (61–63); four independent alleles of csr-1a that lie within the first exon; and four alleles which remain uncharacterized (Supplementary Figure S1B-E). We verified the effects of these mutations with independent alleles (data not shown).
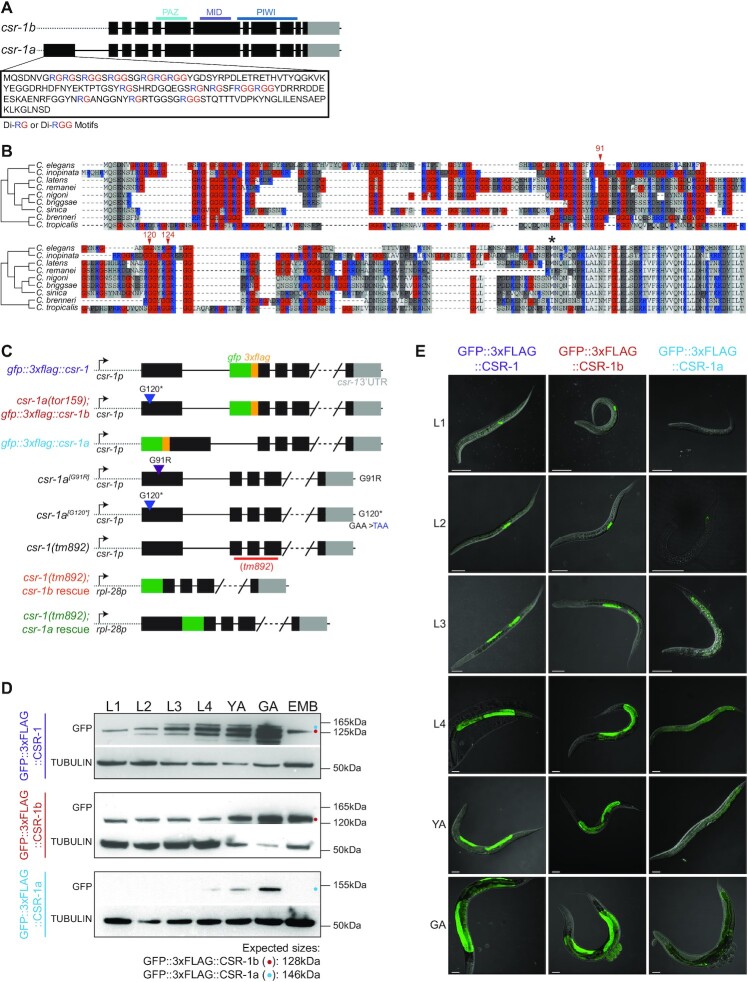
Figure 1.
CSR-1a and CSR-1b isoforms are differentially expressed throughout development. (A) Schematic representation of CSR-1 isoforms. Black boxes are exons, gray boxes are 3′UTRs. (B) Multiple sequence alignment of the N-terminal region of csr-1 for nine Caenorhabditis species. Arrows indicate C. elegans amino acid positions mutated as a result of the forward genetic screen (as shown in Supplementary Figure S1B and C). The asterisk indicates the first amino acid of CSR-1b in C. elegans. (C) Schematic representations of all csr-1 mutant strains, endogenously tagged strains and single copy transgene insertion strains used in this study. (D) Western blot using GFP antibodies to detect GFP::3xFLAG::CSR-1, GFP::3xFLAG::CSR-1b and GFP::3xFLAG::CSR-1a expression in each life stage (larval stages 1 to 4 (L1–L4), young adult (YA), gravid adult (GA) and embryo (EMB) stages). (E) Fluorescence micrographs of whole animals expressing endogenously tagged CSR-1 isoforms during each life stage (L1-L4, YA and GA). Scale bar, 50 μm.
All of the csr-1a alleles recovered cause amino acid substitutions at conserved glycine residues (G91R, two alleles of G120E and G124E) within or nearby conserved RGG/RG repeats of the N-terminus (Figure 1B, Supplementary Figure S1C). We hypothesized that CSR-1a and the other RNAi factors recovered may play a role in sRNA-mediated silencing of the rpn-2p::gfp transgene, therefore we asked whether CSR-1a associates with transgene-derived sRNAs via CSR-1a Immunoprecipitation (IP) with a CSR-1-specific antibody, followed by sRNA sequencing. In parallel, we assessed whether transgene-derived sRNAs were depleted in csr-1a mutants, as loss of AGOs generally destabilizes sRNA pools. We observed no enrichment of transgene sRNAs in CSR-1a complexes, nor did we detect a depletion of transgene sRNAs in the csr-1a mutant strain, suggesting that the role for CSR-1a in regulating rpn-2p::gfp expression was not sRNA-mediated, and could be indirect (Supplementary Figure S1F and G). Despite these results, this screen left us with a rich set of alleles to assess the isoform-specific functions of CSR-1a throughout development.
CSR-1 isoforms have distinct expression patterns throughout development
In addition to the csr-1a alleles recovered in our screen, we generated an extensive set of csr-1 mutants and epitope-tagged strains using CRISPR-Cas9 genome editing (36,64) and MiniMos single copy transgene insertions (41) (Figure 1C). We introduced GFP::3xFLAG into the N-terminus of the long isoform, CSR-1a, via CRISPR-Cas9 genome editing at the endogenous csr-1 locus (36). Because the csr-1b locus is entirely shared with the csr-1a locus, epitope tagging CSR-1b invariably results in the internal epitope tagging of CSR-1a, thus we used this to our advantage to epitope tag both isoforms in a single strain (37). However, having both isoforms tagged precludes our ability to study CSR-1b separately. Therefore, we also introduced a stop codon into the first exon of csr-1a, to generate a third strain that is a null mutant for csr-1a, and expresses only GFP::3xFLAG::CSR-1b (referred to as csr-1a(tor159) in figures where csr-1a loss of function is assessed).
We evaluated the tissues and developmental time points in which each isoform is expressed using western blotting (Figure 1D) and confocal microscopy (Figures 1E, 2, 3A, Supplementary Figure S2-S3). In these experiments, CSR-1a and CSR-1b displayed clear differences in the timing and tissue-specificity of expression. Western blotting on the GFP::3xFLAG::CSR-1b strain demonstrated that, consistent with published data, CSR-1b is expressed during all developmental stages in hermaphrodites, with the highest levels of protein present in adults and in embryos (Figure 1D) (4). In contrast, CSR-1a displays a more restricted expression profile, and is detected only in L4, young adult, and gravid adult developmental stages (Figure 1D).
Figure 2.
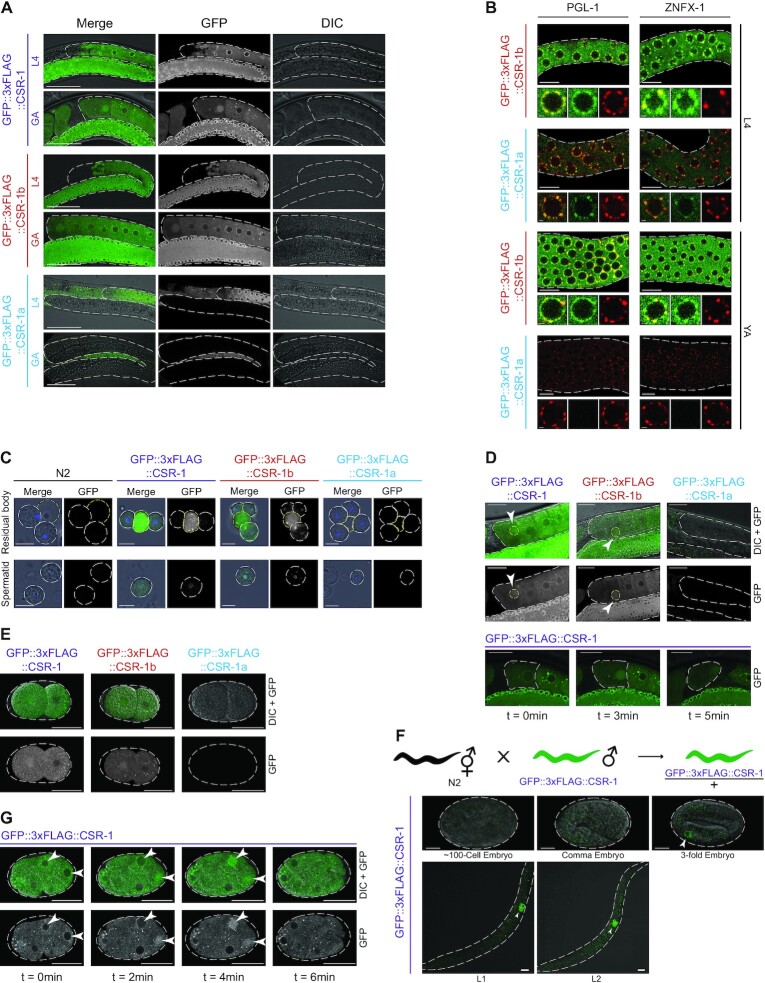
CSR-1a and CSR-1b isoforms are differentially expressed in the germline. (A) Fluorescence micrographs of animals in L4 and gravid adult (GA) stages expressing GFP::3xFLAG::CSR-1, GFP::3xFLAG::CSR-1b and GFP::3xFLAG::CSR-1a in the germlines. Scale bar, 50 μm. (B) Fluorescence micrographs of L4 and young adult (YA) germlines expressing GFP::3xFLAG::CSR-1b and GFP::3xFLAG::CSR-1a (green), along with either PGL-1::mRFP or HA::TagRFP::ZNFX-1 (red). Top panels show the pachytene region of each germline, and bottom panels show a single nucleus. Note that CSR-1a is not expressed in the YA stage. Scale bar, 10 μm in top images, 1 μm in bottom images. (C) Fluorescence micrographs of residual bodies (outlined in yellow) and spermatocytes (top) spermatids(bottom) (both outlined in white) from animals expressing GFP::3xFLAG::CSR-1, GFP::3xFLAG::CSR-1b and GFP::3xFLAG::CSR-1a and stained with DAPI (blue in Merge), compared to wild-type (N2) animals. Scale bar, 5 μm. (D) Fluorescence micrographs showing localization of endogenously tagged CSR-1 isoforms in oocytes (top) and time lapse of endogenously tagged CSR-1 localization in the -1 oocyte over the course of five minutes (bottom). Scale bar, 25 μm. (E) Fluorescence micrographs showing expression of endogenously tagged CSR-1 isoforms and total CSR-1 (reflecting CSR-1b expression) in 2-cell embryos prior to the onset of zygotic expression. Scale bar, 25 μm. (F) Fluorescence micrographs showing zygotic expression of GFP::3xFLAG::CSR-1 in embryos, and L1 and L2 animals. Embryos were generated from crosses of wild-type hermaphrodites to gfp::3xflag::csr-1 males. Scale bar, 10 μm. (G) Fluorescence micrographs showing a time lapse of nuclear localization of endogenously tagged CSR-1 (reflecting CSR-1b expression) in a developing 8-cell embryo over the course of 6 min. Scale bar, 25 μm.
Figure 3.
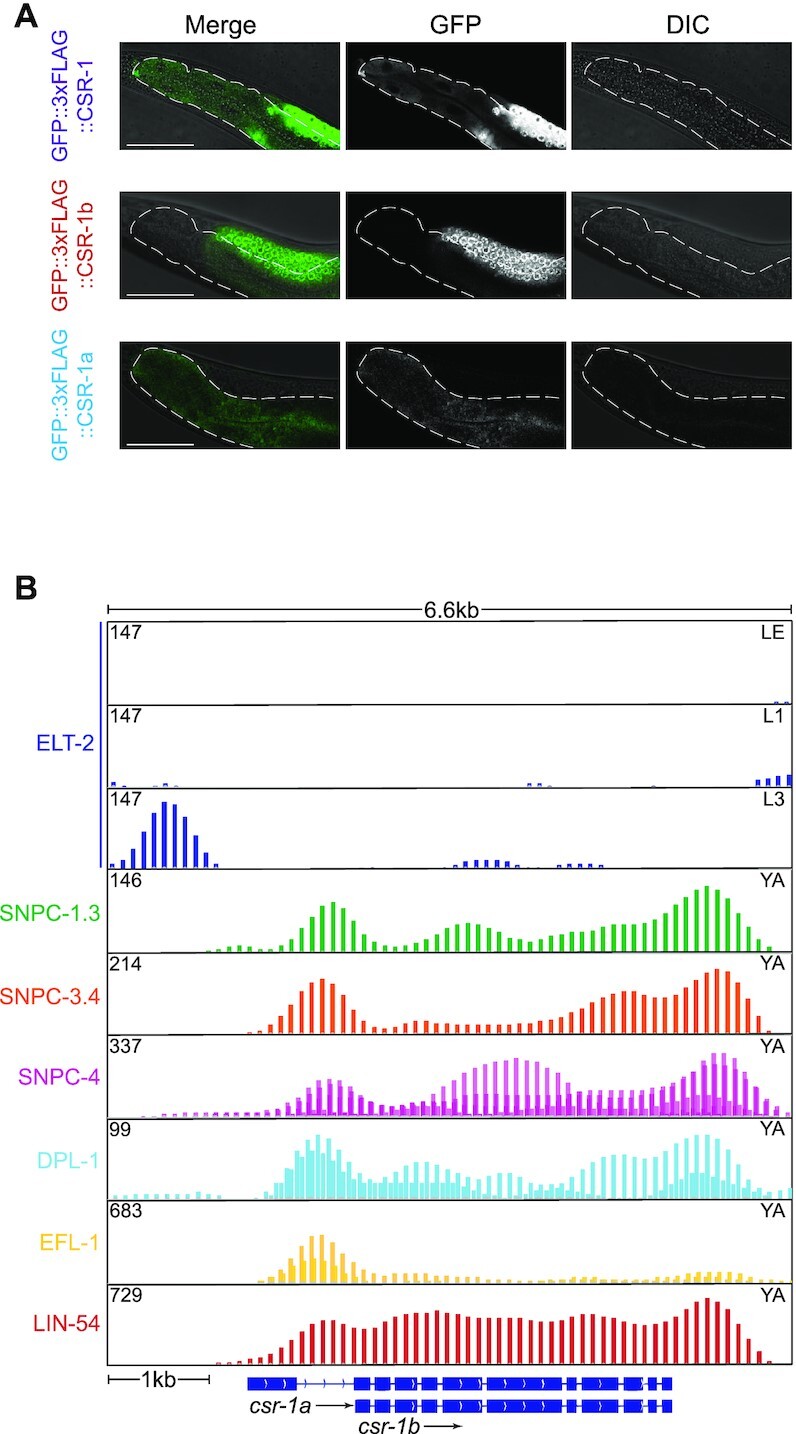
CSR-1a is expressed in the intestine. (A) Fluorescence micrographs of animals expressing GFP::3xFLAG::CSR-1, GFP::3xFLAG::CSR-1b and GFP::3xFLAG::CSR-1a in the intestine. Scale bar, 50 μm. (B) Notable transcription factors enriched in binding within the promoters of each csr-1 isoform, from the ModERN project (68).
In examining each strain by confocal microscopy throughout development, we observed that CSR-1b is constitutively present in the germline at all developmental stages in hermaphrodites, including the primordial germ cells in embryos, the spermatogenic gonad, and the oogenic gonad (Figures 1E, 2A, Supplementary Figure S2). Consistent with expression during spermatogenesis in hermaphrodites (Figure 2A), CSR-1b is also expressed in the male gonad (Supplementary Figure S2). In contrast, CSR-1a is only detected in the proximal germline at the L4 larval stage in hermaphrodites, coincident with sperm production (Figures 1E, 2A, Supplementary Figure S3). CSR-1a is also expressed in the male gonad, similar to its expression pattern in L4 hermaphrodites (Supplementary Figure S3).
Within all germline tissues, CSR-1b is present primarily in the cytoplasm and robustly localizes to germ granules (Figures 1E, 2A, B, Supplementary Figure S2). Similarly, in the L4 germline, CSR-1a is also present in the cytoplasm and enriched in germ granules (Figure 2A and B, Supplementary Figure S3, Figure 9A). CSR-1 had previously been shown to localize to P granules (4), but in the meanwhile, several other types of germ granules, including Z granules have been identified (65). To determine which type of germ granules each CSR-1 isoform associates with, we crossed RFP-tagged versions of PGL-1 (a P granule marker) and ZNFX-1 (a Z granule marker) into the GFP::3xFLAG::CSR-1a/b strains and determined the extent of pixel overlap in L4 and young adult hermaphrodite germlines. We found that both isoforms of CSR-1 overlap with PGL-1 and ZNFX-1 to a similar extent in the L4 germline and that CSR-1b overlaps with both PGL-1 and ZNFX-1 in the oogenic germline (Figure 2B, see Figure 8E for quantification).
Figure 9.
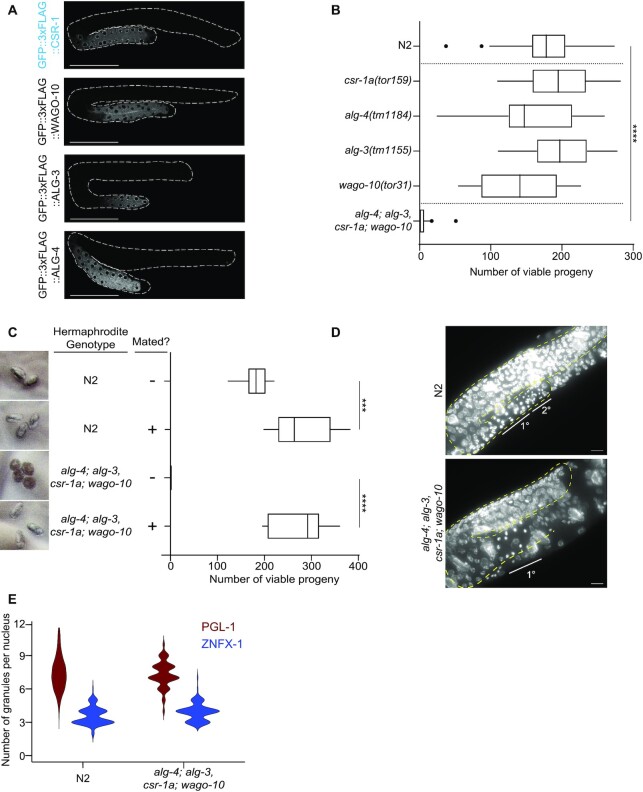
Four spermatogenesis AGOs contribute to sperm-mediated fertility. (A) Fluorescence micrographs of L4 hermaphrodite germlines showing GFP::3xFLAG tagged CSR-1a, ALG-3, ALG-4, and WAGO-10 expression. Scale bar, 50 μm. (B) Box plot of median viable brood size for spermatogenesis ago mutants (with Tukey whiskers) at 25°C. Single mutants were outcrossed before brood analysis and the quadruple ago mutant was outcrossed in the course of generating the strain. Differences are not significant unless marked. Significance was determined with a one-way ANOVA (alpha = 0.05) with Tukey's multiple comparison test. **** indicates significance of P< 0.0001, and refers to all pairwise contrasts with the quadruple ago mutant. Number of individual P0 hermaphrodites counted for each genotype: N2 = 67; csr-1(tor159); gfp::3xflag::csr-1b = 12; alg-4(tm1184) = 17; alg-3(tm1155) = 16; wago-10(tor31) = 17; alg-4; alg-3, csr-1a; wago-10 = 18 (C) Box plot of median viable brood size for the quadruple spermatogenesis ago mutant vs. N2 (with Tukey whiskers) with or without mating to N2 males at 25°C. Number of individual P0 hermaphrodites counted for each genotype: N2 = 10; N2 mated = 10; alg-4; alg-3, csr-1a; wago-10 = 10; alg-4; alg-3, csr-1a; wago-10 mated = 9. Significance was determined with a one-way ANOVA (alpha = 0.05) with Tukey's multiple comparison test. ***indicates significance of P< 0.001 and **** indicates significance of P< 0.0001. Representative images of progeny produced by each genotype are shown at the left (unfertilized oocytes or embryos). (D) DAPI staining of whole L4 N2 or the quadruple spermatogenesis ago mutants. The spermatogenic gonad is outlined, and primary and secondary spermatocytes (defined by the appearance of the DNA and position), are marked. Scale bar is 25 μm. (E) Quantification of PGL-1::mRFP (burgundy) or HA::TagRFP::ZNFX-1 (blue) in N2 and quadruple spermatogenesis ago mutant strains. For each data set, 20 nuclei from the germlines of five different animals were counted (N = 100 nuclei). Comparisons between strains for each type of granule are not significant, as evaluated as a mixed-effects ANOVA.
Figure 8.
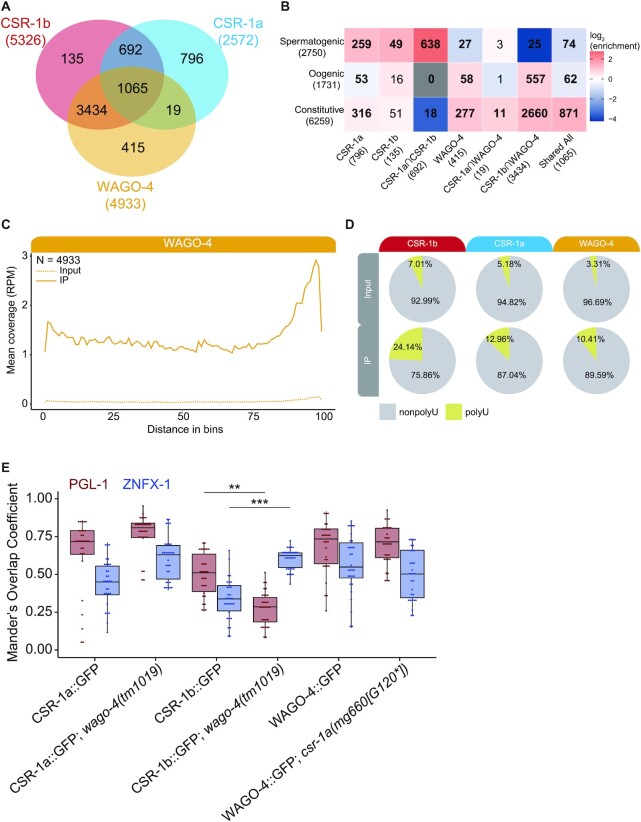
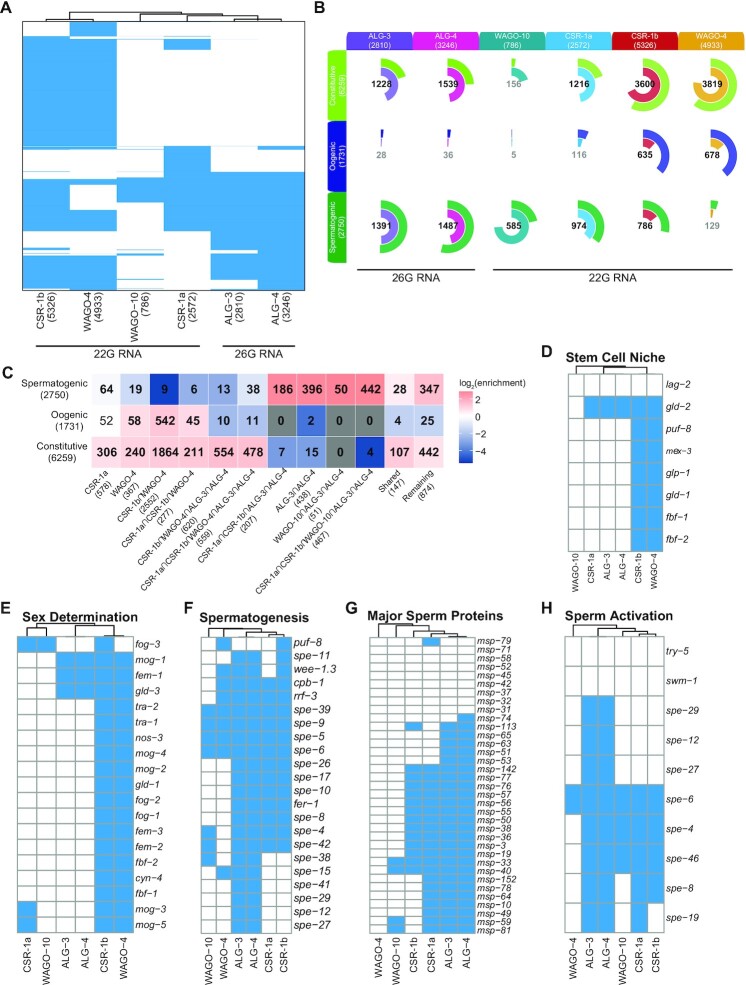
CSR-1 isoforms intersect differently with the WAGO-4 22G-RNA pathway. (A) Venn diagram showing the overlap between CSR-1a/b 22G-RNA targets and WAGO-4 22G-RNA targets from L4 AGO IP samples. (B) Heatmap depicting the overlap between 22G-RNA protein coding targets of CSR-1b only, CSR-1a only, WAGO-4 only, and various shared target groups with mutually exclusive germline constitutive, oogenic, and spermatogenic enriched transcript sets (29). Bold numbers indicate significant enrichment in overlapping genes (Fisher's Exact Test, P< 0.05). Enrichment = number of overlapping genes divided by the expected number of overlapping genes drawn from two independent groups. (C) Metagene plot of the distribution of WAGO-4 enriched 22G-RNAs antisense to protein coding genes along the gene body. N = Number of genes. (D) Pie charts of percentage of total reads with U-tailing (chartreuse; addition of one or more non-templated uridines) for Input (top) and IP (bottom) samples of CSR-1b, CSR-1a, and WAGO-4 sRNAs (all genome matching sRNAs are included) (E) Quantification of the number of GFP::3xFLAG::CSR-1a, GFP::3xFLAG::CSR-1b, and WAGO-4::GFP pixels that overlap with PGL-1::mRFP (burgundy) or HA::TagRFP::ZNFX-1 (blue) pixels in wild-type or mutant worms, using Mander's Correlation. For each data set, five Z stacks of proximal germline regions from six different animals per strain were counted (N = 30 slices, approximately 80–100 nuclei per worm). Significance was analyzed as a mixed-effects ANOVA. ** indicates significance of P< 0.01, *** indicates significance of P< 0.001.
Because of their germline expression patterns, we next explored whether either CSR-1 isoform could be transmitted to progeny via sperm (CSR-1a, b) or oocytes (CSR-1b only). First, we performed confocal microscopy on sperm released from males. We observed that much of the CSR-1b pool was present in the residual body, a structure where sperm cytoplasmic contents are discarded during spermatogenesis (Figure 2C) (22). We also detected CSR-1b in mature spermatids, where it was present both in the nucleus and cytoplasmic foci (Figure 2C) (14). In contrast, CSR-1a is not segregated into the residual body, nor is it packaged into mature spermatids. These observations indicate that CSR-1b alone has the potential to be passed from males to progeny via sperm.
In oocytes, CSR-1b localizes to the cytoplasm, germ granules, and oocyte chromatin (Figure 2A, B, D), and therefore could be maternally provisioned (4,66). In self-fertilizing GFP::3xFLAG::CSR-1b hermaphrodites, we observed GFP expression in early embryos, prior to the onset of zygotic transcription (Figure 2E). In contrast, we did not observe GFP expression in early embryos produced from crosses between GFP::3xFLAG::CSR-1 males and wild-type hermaphrodites (Figure 2F). Instead, we first detected zygotic GFP::3xFLAG::CSR-1 in the primordial germ cells, Z2 and Z3. Finally, during early embryogenesis until approximately the 50 cell stage, maternally provisioned CSR-1b is detected in both somatic and germ cells (Figure 2E, G). In both cell types, CSR-1b is present in the cytoplasm and localizes to the nucleus in a cell-cycle dependent manner, coincident with mitosis (Figure 2G). Collectively, these data indicate that CSR-1b is maternally, and potentially paternally inherited, and that zygotic expression of CSR-1b initiates when the primordial germ cells become transcriptionally active during late embryonic development. In contrast, CSR-1a is not maternally or paternally inherited, and is not expressed in embryos.
In adult hermaphrodites, we were unable to detect CSR-1a in the oogenic germline (Figure 2A-B D, Supplementary Figure S3), nevertheless, western blotting data demonstrated the presence of CSR-1a in adults (Figure 1D). This suggested that CSR-1a may be expressed in the soma. Indeed, we observed CSR-1a in several adult somatic tissues of both hermaphrodites and males (Supplementary Figure S3), including the intestine (Figure 3A), spermatheca (Supplementary Figure S3), and several cells within the tail (Supplementary Figure S3). In adult males, CSR-1a is also present in the vas deferens and seminal vesicle (Supplementary Figure S3). In fact, the earliest expression of CSR-1a was in the intestine from the L3 stage onward (Figure 1C). In contrast, CSR-1b was not observed in any post-embryonic somatic tissues.
Consistent with this intestinal expression pattern, we observed enrichment of the intestinal master transcription factor ELT-2 (67) upstream of the csr-1a transcription start site at the L3 stage of development by examining published transcription factor binding data (Figure 3B) (68). The first intron of the csr-1 locus possesses potential regulatory elements for csr-1b. Within this intron, we did not observe ELT-2 binding, but instead observed enrichment of several germline enriched transcription factors, including members of the SNAP-C (69) and DRM complexes (70), which we predict regulate the expression of csr-1b in the germline. Taken together, these results highlight the potential for CSR-1a to play novel roles during spermatogenesis and within specific somatic tissues, including the intestine.
CSR-1b is the essential CSR-1 isoform but CSR-1a can compensate
csr-1 is the only singly essential ago in C. elegans. Homozygous zygotic null mutants of csr-1 (which affect both isoforms) that are generated from heterozygous hermaphrodites develop to adulthood, but display defects in oogenesis (3,4). Consequently, they produce few embryos, and those few embryos that are generated display chromosome segregation defects and arrest early in embryogenesis, prior to the 50 cell stage (4,11). In contrast, csr-1 null mutant males are fertile under normal lab culture conditions, but display a progressive loss of fertility over several generations at a stressful temperature (25°C) (14). The different expression profiles of CSR-1 isoforms in the germline suggest that each isoform could be responsible for different facets of fertility. For instance, because CSR-1a is not expressed in the hermaphrodite germline or in early embryos, we expected that loss of csr-1a would likely have little impact on oocyte development or embryo viability. Therefore, we asked whether either isoform was sufficient for fertility, or if both were redundantly required for proper germline development and gamete function.
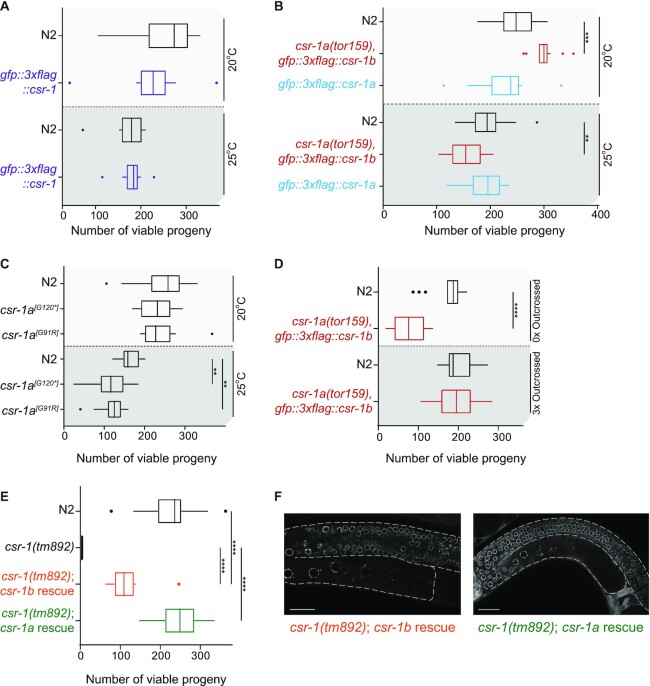
We assessed the fertility of our set of csr-1a mutants and epitope tagged strains, along with two strains in which csr-1 null mutants were rescued with ubiquitous expression of either CSR-1a or CSR-1b (Figure 4). First, we verified that each of our GFP::CSR-1a, GFP::CSR-1b, and GFP::CSR-1 strains displayed wild type fertility under the normal lab culture temperature of 20°C (Figure 4A-B). Next, we tested whether these strains displayed any fertility defects under the stressful temperature of 25°C. At 25°C, GFP::CSR-1b, which also possesses a null mutation in csr-1a (csr-1a(tor159)), displayed a slight but reproducible reduction in viable brood size (Figure 4A-B). We also tested a strain possessing only the csr-1a premature stop allele, but no gfp insertion in csr-1b (csr-1a[G120*]), as well as a missense allele affecting a conserved RG repeat of the CSR-1a recovered from the proteasomal regulators screen (csr-1a[G91R]) (Figure 1B). While csr-1a[G120*] was a null allele, CSR-1a[G91R] was expressed at slightly lower levels than wild-type CSR-1a, but localized to the intestine and P granules normally (Supplementary Figure S4A-D). Consistent with our results using csr-1a(tor159), both of these alleles displayed a normal brood size at 20°C, and reduced brood size at 25°C (Figure 4C).
Figure 4.
CSR-1b is the essential CSR-1 isoform, but CSR-1a is required for full fertility under stress. (A-D) Box plot of viable brood size counts for the strains expressing endogenously tagged total CSR-1 or csr-1a mutants compared to wild type N2, when reared at 20°C and 25°C. Median is represented, with Tukey whiskers. Differences are not significant unless marked. Significance was determined with a one-way ANOVA (alpha = 0.05) with Tukey's multiple comparison test. ** indicates significance of P< 0.01, *** indicates significance of P< 0.001, **** indicates significance of P< 0.0001 (A) Viable brood size for the strain endogenously tagged total CSR-1 vs. N2, when reared at 20°C and 25°C. Number of individual P0 hermaphrodites counted for each genotype: N2 20°C = 14; gfp::3xflag::csr-1 20°C = 15; N2 25°C = 14; gfp::3xflag::csr-1 25°C = 13. (B) Viable brood size counts for the strains expressing endogenously tagged CSR-1b (red) and CSR-1a (blue) vs. N2, when reared at 20°C and 25°C. Note that the endogenously tagged CSR-1b strain also carries a stop mutation in csr-1a. Number of individual P0 hermaphrodites counted for each genotype: N2 20°C = 16; csr-1a(tor159), gfp::3xflag::csr-1b 20°C = 16; gfp::3xflag::csr-1a 20°C = 16; N2 25°C = 19; csr-1a(tor159), gfp::3xflag::csr-1b 25°C = 17; gfp::3xflag::csr-1a 25°C = 18. (C) Viable brood size for csr-1a mutants vs. N2 reared at 20°C and 25°C. Number of individual P0 hermaphrodites counted for each genotype: N2 20°C = 30; csr-1a[G120*] 20°C = 16; csr-1a[G91R] = 15; N2 25°C = 14; csr-1a[G120*] 25°C = 16; csr-1a[G91R] 25°C = 16. (D) Viable brood size of csr-1a(tor159), gfp::3xflag::csr-1b animals vs. N2 reared at 25°C. 0x Outcrossed worms were reared for > 25 generations at 20°C before shifting to 25°C for brood assessment. 3x Outcrossed worms were crossed to wild-type animals before shifting to 25°C for brood assessment. Number of individual P0 hermaphrodites counted for each genotype: 0x outcrossed N2 = 18; 0x outcrossed csr-1a(tor159); gfp::3xflag::csr-1b = 12; 3x outcrossed N2 = 17; csr-1a(tor159), gfp::3xflag::csr-1b = 17. (E) Viable brood size for the csr-1(tm892) null mutant and N2 vs. the csr-1(tm892) null mutant rescued with csr-1b and csr-1a transgenes expressed under control of the rpl-28 promoter and reared at 20°C. Number of individual P0 hermaphrodites counted for each genotype: N2 = 33; csr-1(tm892) = 20; csr-1(tm892); csr-1b rescue = 13; csr-1(tm892); csr-1a rescue=18. (F) Fluorescence micrographs showing germline expression of GFP::CSR-1 in animals rescued with each csr-1 isoform-specific transgene. Scale bar, 25 μm.
The subtle and temperature-dependent fertility defect led us to pursue the role of csr-1a at 25°C further. Because CSR-1a germline expression occurs only during spermatogenesis, and loss of both CSR-1 isoforms in males causes a progressive loss of fertility over many generations (14), we tested whether loss of CSR-1a causes a progressive decline in fertility. We cultured the csr-1a(tor159) null mutant at 20°C for > 25 generations, then shifted it to 25°C to perform brood analysis. In parallel, we freshly outcrossed these ‘late generation’ csr-1a(tor159) null mutants, shifted them to 25°C and assayed their brood size in parallel. We found that csr-1a(tor159) null mutants in culture for many generations displayed a smaller brood size than wild-type worms at 25°C, but that this phenotype was reversed by outcrossing to wild-type (N2) before selfing (Figure 4D). These data suggest that loss of CSR-1a from the spermatogenic germline results in progressive loss of fertility through a transgenerational epigenetic mechanism. This is reminiscent of transgenerational epigenetic inheritance defects displayed by other germline ago mutants, including csr-1(tm892) null mutant males (14).
Next, we asked if ubiquitous expression of either CSR-1 isoform is sufficient to rescue the csr-1(tm892) null mutant. To do this, we constructed single copy genomically-integrated transgenes, in which the coding sequence of each csr-1 isoform was tagged with gfp and was expressed using a constitutive rpl-28 promoter and csr-1 3′UTR in a csr-1(tm892) null mutant background (Figure 1B). Remarkably, both strains were viable, indicating complementation (Figure 4E). We measured the extent to which expression of each isoform rescued fertility by performing total brood counts versus viable brood counts. In contrast to csr-1(tm892), which produces very few embryos, the csr-1(tm892) strain possessing the rpl-28::gfp::csr-1b transgene produced 73.1% the total brood of wild-type worms (data not shown). However, despite this robust brood size, we found that many of the embryos were not viable (Figure 4E; csr-1b rescue viability was 47.4% of wild-type worms), and died late in embryogenesis as malformed larvae (Supplementary Figure S4F). This embryonic arrest occurs later in development than embryos produced by csr-1(tm892) homozygous hermaphrodites, suggesting that the phenotype could be due to different causes, including the developmental timing and cell types in which CSR-1b was expressed via this ubiquitous promoter, or the level to which it was expressed. The brood size may also be reduced due to a lack of CSR-1a in this strain.
To ensure that only csr-1a was expressed in the second transgenic rescue strain, we eliminated the first intron. Western blotting on protein lysates from this strain showed that only GFP::CSR-1a was expressed from this transgene and there was no CSR-1b protein present (Supplementary Figure S4E). By using this constitutive rpl-28 promoter, CSR-1a is expressed in many tissues and at developmental time points where and when it would not normally be expressed, including in the oogenic germline and in early embryos (Figure 4F). Remarkably, this strain displayed a viable brood size comparable to wild type (Figure 4E), indicating that GFP::CSR-1a can compensate for loss of csr-1b when ectopically expressed in the oogenic germline and in embryos. Overall, this result was unexpected, and suggests that CSR-1a can execute the molecular functions of CSR-1b, but has differentiated itself from CSR-1b by its expression profile, and potentially other features of the protein. Conversely, the reduced brood size in the CSR-1b-expressing rescue strain may indicate that CSR-1b cannot perform the functions of CSR-1a, or that the timing, cell types, and levels of CSR-1b are not optimized in this strain.
By integrating these observations with our knowledge of CSR-1 expression profiles, we conclude that under normal culture conditions, CSR-1a is not required for embryonic viability. Instead csr-1b plays an essential role during oocyte development and in the early embryo. However, under stressful conditions, such as elevated temperature, csr-1a may play a role in fertility that is likely linked to a role in sperm development and differentiation (see below).
CSR-1a and CSR-1b associate with 22G-RNAs to target partly overlapping sets of protein coding genes
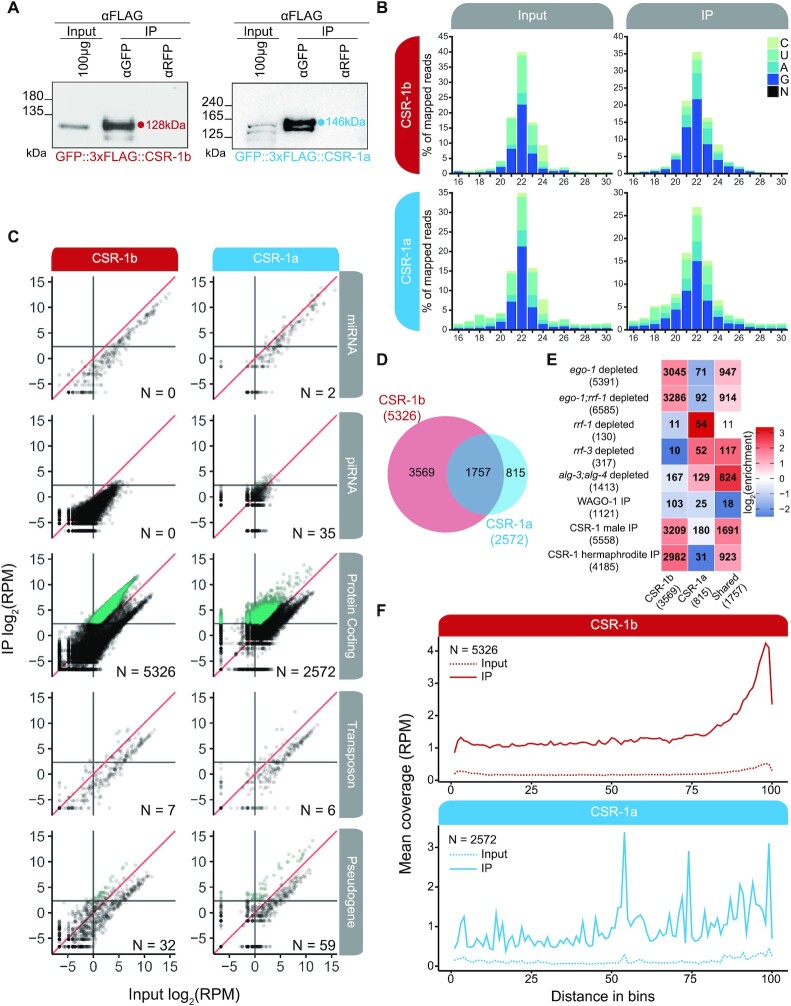
To understand whether each CSR-1 isoform has distinct or overlapping target transcripts and roles in gene regulation, we set out to determine the pools of sRNAs bound by each isoform. Past studies have shown that CSR-1 associates with 22G-RNAs, and 22G-RNAs display perfect complementarity to target transcripts (4,14,15). Therefore, by determining the sRNA complements associated with each CSR-1 isoform, we can computationally identify gene targets. In these experiments, we used GFP antibodies to perform immunoprecipitation (IP) of GFP::3xFLAG::CSR-1 (for CSR-1a, CSR-1b and CSR-1) in synchronized L4 worm populations maintained at 20°C (Figure 5, Supplementary Figure S5). We chose L4 worms because both isoforms are robustly expressed at this stage. For CSR-1b we expected that sRNA populations would reflect germline activities during spermatogenesis. For CSR-1a, we anticipated that we would recover both somatic and germline (spermatogenesis) sRNA populations, due to its expression profile. In parallel, we cloned and sequenced total sRNA samples from the same samples (Input). Two replicates were performed for each condition (Supplementary Tables S1-2).
Figure 5.
CSR-1b and CSR-1a associate with different pools of sRNAs. (A) Anti-FLAG western blots of GFP IPs used in sequencing GFP::3xFLAG::CSR-1a and GFP::3xFLAG::CSR-1b associated sRNAs. IPs were performed on recently outcrossed populations at 20°C. (B) Bar graph of first nucleotide and size distribution of normalized sRNA reads from Input and IP samples sequenced from GFP::3xFLAG::CSR-1b and GFP::3xFLAG::CSR-1a. (C) Scatter plots of sRNAs in Input and IP samples of CSR-1b (left) and CSR-1a (right) broken down by genomic features. Each dot represents a gene. Teal dots represent transcripts/genes which have a minimum of 5RPM in IP samples and are 2-fold or greater enriched relative to Input in both replicates. Sense reads are counted for miRNAs and piRNAs, and antisense reads are counted for protein coding genes, transposons, and pseudogenes. The number of features enriched in the IP is shown (N = #). For protein coding, transposons, and pseudogenes, only 22G-RNAs (defined as 21–23nt, with no first nucleotide bias) were considered for enrichment. RPM = reads per million. (D) Venn Diagram showing the overlap between the enriched 22G-RNAs antisense to protein coding genes from CSR-1b (red, 5326 total genes) and CSR-1a (blue, 2572 genes) IPs. (E) Heatmap depicting the overlap between 22G-RNA protein coding targets of CSR-1b only, CSR-1a only, and shared targets with the indicated data sets (4,14,15,23,71). Bold numbers indicate significant enrichment in overlapping genes (Fisher's Exact Test, P< 0.05). Enrichment = number of overlapping genes divided by the expected number of overlapping genes drawn from two independent groups. (F) Metagene plot of the distribution of enriched CSR-1b or CSR-1a 22G-RNAs antisense to protein coding genes along the gene body. N = Number of genes.
We focused our analysis on the CSR-1a and CSR-1b IPs (Figure 5A). (Note that total CSR-1 results are presented in Supplementary Figure S5, but not discussed further.) We first examined the size and first nucleotide distribution of sRNAs from the Input and IP samples, and observed a large proportion of 22G-RNA reads present in each CSR-1 IP (Figure 5B, Supplementary Tables S1-2). Next, we determined which reads were enriched in each GFP::3xFLAG::CSR-1 IP relative to Input (with a minimum of 5 reads per million (RPM) and a 2-fold or greater increase in IP versus Input) to define a set of target transcripts (genes for which antisense sRNAs are enriched in the IP) for each isoform (Figure 5C, Supplementary Table S2). Protein coding genes are the major targets of both isoforms (CSR-1a: 2572 genes; CSR-b: 5326 genes out of 20116 total genes in the genome) (Figure 5C). Both isoforms also target a small number of transposable elements (CSR-1a: 6 TE; CSR-b: 7 TE out of 285 total TE) and pseudogenes (CSR-1a: 59 pseudogenes; CSR1-b: 32 pseudogenes out of 1885 total pseudogenes) (Figure 5C). While there is extensive overlap between the CSR-1a and CSR-1b protein coding gene targets (1757 genes), there are also distinct sets of genes that are specifically targeted by one isoform (815 for CSR-1a, 3569 for CSR-1b) (Figure 5D).
We examined how the CSR-1a/b shared and isoform-specific 22G-RNA targets overlap with those of other sRNA pathways by comparing them to previously published RdRP mutant, ago mutant, and AGO IP/sRNA-seq datasets (Figure 5E) (4,14,15,71). Such comparisons could provide insights into any differences in 22G-RNA biogenesis pathways between the CSR-1 isoforms and their distinct and overlapping target gene sets. Striking patterns emerged from these comparisons. First, we found that CSR-1b-specific targets and CSR-1a/b shared targets are over-represented in genes that are depleted of 22G-RNAs in ego-1 mutants. In contrast, CSR-1a-specific targets are over-represented in genes that are depleted of sRNAs in rrf-1 and rrf-3 mutants, while CSR-1b-specific targets are not. CSR-1a/b shared targets are also over-represented in genes depleted of sRNAs in rrf-3 mutants (Figure 5E). The overlap with different RdRPs reflects CSR-1 isoform expression profiles: RRF-1 is mostly expressed in the soma (like CSR-1a), EGO-1 is expressed constitutively in the germline (like CSR-1b), and RRF-3 is important during spermatogenesis (like CSR-1a and CSR-1b) (4,14,15,23,27,71). Consistent with a link to spermatogenic sRNA pathways (below), CSR-1a-specific and CSR-1a/b shared targets are over-represented in genes depleted of sRNAs in alg-3; alg-4 spermatogenesis-specific ago mutants (Figure 5E). Interestingly, CSR-1b-specific and CSR-1a/b shared targets are over-represented in existing CSR-1(total) targets from males and hermaphrodites, while CSR-1a-specific targets are under-represented in these sets. This likely reflects the relative abundance of each isoform, as CSR-1b is expressed at higher levels than CSR-1a and can be more easily immunoprecipitated by CSR-1 antibodies. Of note, we observed a similar phenomenon with our GFP::3xFLAG::CSR-1 (both isoform) IPs (Supplementary Figure S5). All sets of CSR-1 target genes are under-represented in WAGO-1 targets, consistent with a functional distinction between the CSR-1 and WAGO-1 pathways (Figure 5E) (4,15). Collectively, these data suggest that different biogenesis mechanisms may contribute to the production of each subset of CSR-1 associated 22G-RNAs.
We asked if there are any differences in the distribution of sRNAs along the groups of target genes for each CSR-1 isoform. To assess this, we performed metagene analysis, examining the density of sRNAs along the set of CSR-1a vs. CSR-1b protein coding gene targets, after normalizing for gene length (Figure 5F). CSR-1b -associated sRNAs displayed a pattern consistent with previous reports, in which sRNAs were enriched at the 3′ end of the target transcripts (72). However, we observed a distinct pattern of sRNAs associated with CSR-1a, in which sRNAs were distributed along the entire length of the target transcripts. This result is reminiscent of the pattern of WAGO-1 sRNAs, which are generated by the RdRP RRF-1 (15). Collectively, these observations highlight similarities and differences in CSR-1a vs. CSR-1b 22G-RNAs, and hint that, at least in some tissues, there may be differences in the CSR-1 22G-RNA biogenesis pathways.
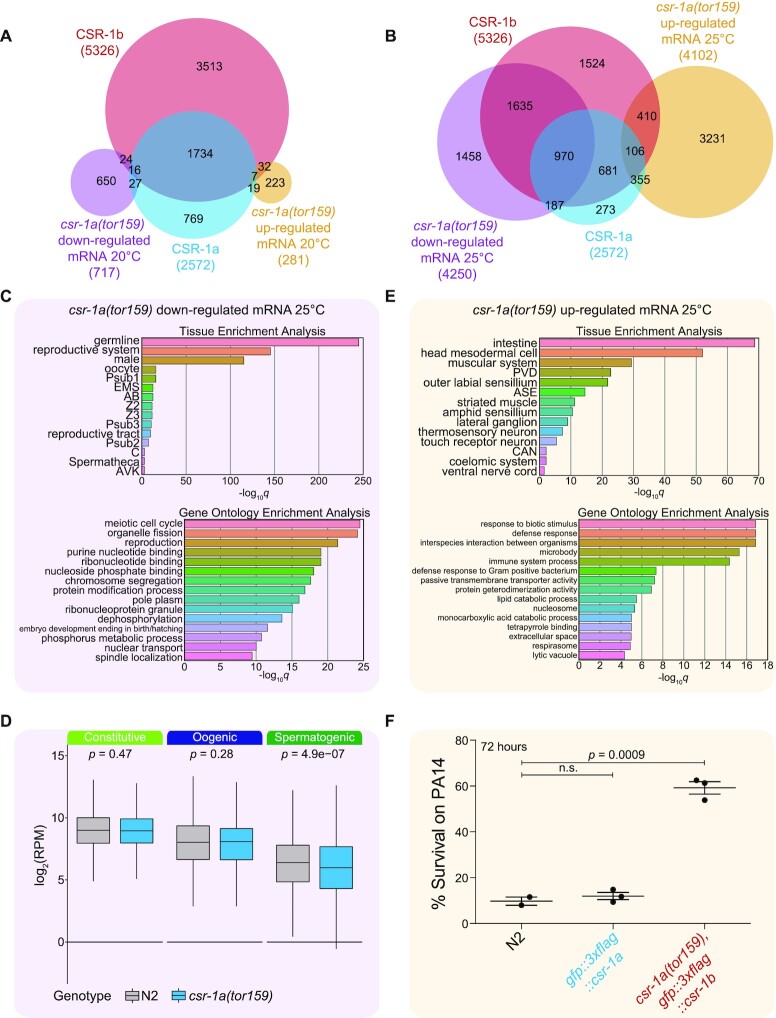
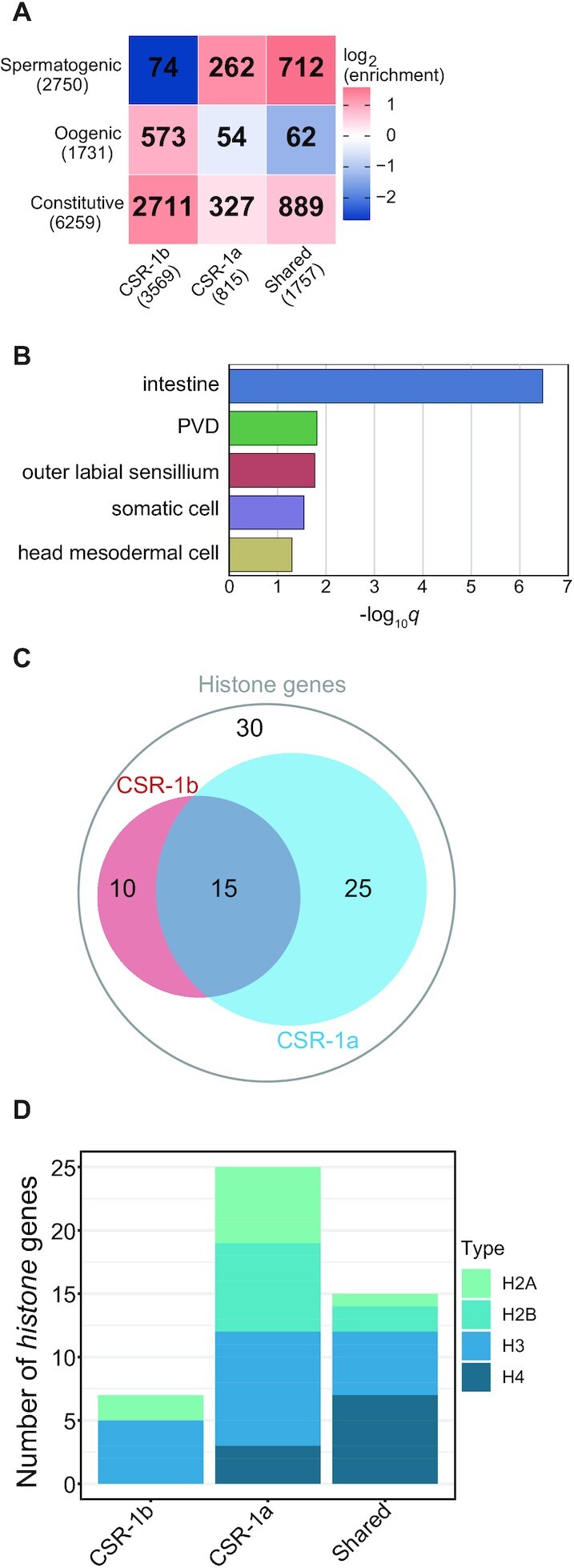
CSR-1a and CSR-1b target different subsets of germline genes
To further characterize the overlapping and distinct 22G-RNA gene targets of each CSR-1 isoform, we categorized targets based on existing data that designate mutually exclusive sets of sperm, germline constitutive, and oocyte enriched transcripts (29) (Figure 6A, Supplementary Tables S1-2). Because CSR-1a and CSR-1b only overlap in their expression in the spermatogenic gonad, we anticipated that the set of shared targets would be enriched in germline genes. Indeed, we observed that germline constitutive and spermatogenic gene sets were over-represented among the shared targets, and accounted for nearly all 1757 of these shared targets, while oogenic genes were under-represented (Figure 6A). The CSR-1b specific targets were over-represented in the germline constitutive and oogenic gene sets. This reflects the constitutive germline expression pattern of CSR-1b and highlights its broad role in germline gene regulation. It is important to note that the CSR-1b specific targets are under-represented in spermatogenesis genes, highlighting that any spermatogenesis targets of CSR-1b are generally shared with CSR-1a. Finally, CSR-1a specific targets were over-represented in germline constitutive and spermatogenic gene sets, and under-represented in oogenic genes, again, reflecting its expression during spermatogenesis. Notably, approximately one-third of the CSR-1a specific targets (268/815 genes) did not fall into any of these germline categories, and were enriched for targets expressed in somatic tissues. Of these 268 genes, 171 were enriched in the intestine (Figure 6B).
Figure 6.
CSR-1 isoforms target shared and distinct sets of genes. (A) Heatmap depicting the overlap between 22G-RNA protein coding targets of CSR-1b only, CSR-1a only, and shared targets with mutually exclusive germline constitutive, oogenic, and spermatogenic enriched transcript sets (29). Bold numbers indicate significant enrichment in overlapping genes (Fisher's Exact Test, P< 0.05). Enrichment = number of overlapping genes divided by the expected number of overlapping genes drawn from two independent groups. (B) Tissue enrichment analysis of the 268 CSR-1a targets that do not overlap with the sets of germline transcripts in (A). (C) Venn diagram of core histone genes enriched in the IPs for CSR-1b (red) and CSR-1a (blue), relative to all core histone genes in C. elegans. (D) Bar graph showing the distribution of types of core histone genes enriched in each CSR-1 isoform IP.
Based on a previously described role for CSR-1 in regulating histones, we examined histone targeting by the CSR-1 isoforms in more detail. Replication dependent histone mRNAs are generally not poly-adenylated, but possess a 3′ stem loop that must be cleaved for mRNA maturation and translation (73). CSR-1 has been implicated in histone mRNA processing in worms. In this mechanism, sRNAs guide CSR-1 to the stem loop cleavage site, where CSR-1 is proposed to act as the endonuclease (10). There are approximately 80 histone genes detectable in the C. elegans genome and we found that 50 of the histone genes are targeted by CSR-1a, CSR-1b, or both isoforms (Figure 6C-D, Supplementary Table S2). CSR-1a targets Histones H2A, H2B, and H3, while CSR-1b targets predominantly H3. Histone H4 is a prominent target of both isoforms. Because both CSR-1 isoforms possess the same catalytic tetrad within the PIWI domain, it is plausible that both isoforms could process the 3′ stem loop of different classes of histone genes in different tissues during development.
Gene expression is perturbed in csr-1a mutants
Our data collectively attribute the previously described expression pattern and essential functions of CSR-1 in the literature to CSR-1b, whereas CSR-1a plays an uncharacterized role that promotes fertility. We therefore focused our efforts on understanding how loss of CSR-1a affects the expression of its sRNA target genes to maintain fertility. Because csr-1a mutants exhibit temperature-dependent fertility defects after long-term culture, we performed mRNA-seq on late generation L4 stage csr-1a(tor159) mutants vs. wild-type worms at 20°C and 25°C.
We first compared gene expression changes in csr-1a(tor159) mutants to wild-type worms at 20°C. We found that very few genes that changed at this temperature were the targets of CSR-1a or CSR-1b 22G-RNAs (Figure 7A, Supplementary Table S2). In contrast, when we compared gene expression changes of csr-1a(tor159) mutants to wild-type worms at 25°C, we found that a greater number of genes display altered expression (Figure 7B, Supplementary Table S2). A large fraction of these mis-expressed genes are CSR-1a and/or CSR-1b targets. Interestingly, the majority (61%) of genes that display reduced expression are CSR-1b targets (including both shared CSR-1a/b targets or CSR-1b specific targets). Further, 66% of genes with reduced expression are targeted by either CSR-1 isoform, while only 21% of genes with increased expression are targeted by either CSR-1 isoform. Genes that are uniquely targeted by CSR-1a tended to display increased expression (355 up-regulated genes vs. 187 down-regulated genes). Overall, 63% of CSR-1a 22G-RNA targets are mis-regulated. We also verified expression changes for several genes using qRT-PCR (Supplementary Figure S6).
Figure 7.
Loss of csr-1a affects different sets of genes in different tissues. (A-B) Venn diagrams showing the overlap between genes that are differentially expressed in csr-1a(tor159), gfp::3xflag::csr-1b (simply referred to here as csr-1a(tor159)) vs. wild-type worms at 20°C (A) (up-regulated genes: orange, down-regulated genes: purple) and 25°C (B) with the 22G-RNA targets defined by CSR-1a and CSR-1b IPs. (C) Tissue enrichment analysis (top) and Gene Ontology analysis (bottom) for genes that are down-regulated in csr-1a(tor159) vs. wild-type worms at 25°C. (D) Analysis of RPM in csr-1a(tor159) (blue) vs. wild-type worms (pink) at 25°C for genes in the germline constitutive, oogenic, and spermatogenic groups (29). (E) Tissue enrichment analysis (top) and Gene Ontology analysis for genes that are over-expressed in csr-1a(tor159) vs. wild-type worms at 25°C (F) Survival of worms at 72 hours treatment with Pseudomonas aeruginosa (PA14) in slow killing assays. Each data point represents one experiment, starting from at least 50 worms. Comparisons between samples were made using two-tailed t-tests.
Tissue Enrichment and Gene Ontology (GO) analysis indicated that the genes that display decreased expression in csr-1a(tor159) mutants are expressed in the germline, reproductive system, and in males. GO terms including meiotic cell cycle, reproduction, chromosome segregation, and nucleotide binding were also enriched in this set, suggesting that these genes are involved in spermatogenesis (Figure 7C, Supplementary Table S2). Indeed, when we assessed the relative expression of germline constitutive, oogenic, and spermatogenic transcripts in wild-type and csr-1a(tor159) mutant worms, we observed that only spermatogenic genes were significantly decreased in the mutant (Figure 7D, Supplementary Table S2) (29). These results point to a positive gene regulatory role for CSR-1a in the spermatogenic gonad, and are consistent with the previously described positive, or licensing role for CSR-1 overall in regulating spermatogenic transcripts (14).
In sharp contrast, the genes that displayed increased expression in csr-1a(tor159) mutants at 25°C are largely expressed in the soma, including the intestine, where CSR-1a is expressed (Figure 7E, Supplementary Table S2). GO terms including response to biotic stimulus, stress response, interspecies interaction between organisms, and immune system process were enriched in this set, suggesting a role in pathogen responses. The intestine serves as an important interface with the environment, including pathogens. This prompted us to ask whether csr-1a(tor159) mutants respond better or worse than wild-type animals when faced with a pathogen, such as the gram-negative bacterium Pseudomonas aeruginosa (strain: PA14) (49,74). To our surprise, we observed that csr-1a(tor159) mutants were able to survive better on PA14 than wild-type animals (Figure 7F). We reason that the upregulation of immune response genes in csr-1a(tor159) mutants ‘primes’ worms to better defend against PA14, and possibly other pathogens. Collectively, our gene expression data indicate that CSR-1a promotes the expression of spermatogenesis genes in the L4 spermatogenic gonad. In contrast, somatic CSR-1a, likely acting in the intestine (soma), is required to dampen expression of immune and pathogen response genes. These roles are likely mediated, at least in part, by sRNA directed mechanisms. However, many of the mis-expressed genes are not direct targets of CSR-1a or CSR-1b, so it is important to note that indirect effects on gene expression are also possible.
CSR-1b and CSR-1a differentially intersect with the WAGO-4 22G-RNA pathway
Having established a positive gene regulatory role for CSR-1a alongside CSR-1b during spermatogenesis, we aimed to determine how CSR-1-dependent gene regulation intersects with other sRNA pathways during spermatogenesis. First, we assessed the germline AGO, WAGO-4.
Recent studies have demonstrated that WAGO-4 binds to a pool of 22G-RNAs that target nearly all of the previously defined CSR-1 protein coding gene targets (75). WAGO-4 is expressed constitutively in the germline, where it localizes to germ granules, including Z granules in adults and P granules in early embryos (65). WAGO-4 has been implicated in transgenerational epigenetic inheritance of exogenous RNAi, and its loss leads to a transgenerational loss of fertility at the stressful temperature of 25°C (Mortal germline, Mrt phenotype) (65,75,76). Because of this link between the CSR-1 and WAGO-4 targets, we aimed to delineate the relationship between the targets each CSR-1 isoform and WAGO-4.
We cloned and sequenced sRNAs associated with GFP::3xFLAG::WAGO-4 from L4 worms, and compared these data to the targets of CSR-1a and CSR-1b (37). Our WAGO-4 IPs identified 22G-RNAs that target 4933 protein coding genes, which overlap extensively with existing data sets (data not shown) (75). We asked if the targets of WAGO-4 overlap with CSR-1a or CSR-1b targets, and found that the greatest overlap was between CSR-1b and WAGO-4 targets (4499/5326 CSR-1b genes; 84.5%) (Figure 8A, Supplementary Tables S1-2). Among these were 1065 CSR-1a/b shared targets, which encompassed almost all of the CSR-1a and WAGO-4 overlap in targets. In fact, only 19 CSR-1a-specific targets overlapped with WAGO-4 targets. These data demonstrate that CSR-1b and WAGO-4 pathways intersect, while any relationship between the CSR-1a and WAGO-4 pathways is centered around targets shared with CSR-1b, rather than CSR-1a-specific targets.
We next asked what types of germline target genes fall into each AGO group (Figure 8B, Supplementary Table S2). Here, the distinction is clear. CSR-1a and CSR-1b specific targets are both enriched in spermatogenic and germline constitutive genes, while the CSR-1a/b shared targets are enriched in spermatogenic genes only. In contrast, CSR-1b/WAGO-4 shared targets are enriched in oogenic and germline constitutive genes. Genes targeted by all three AGOs were enriched in germline constitutive genes. These data distinguish CSR-1a, CSR-1b and WAGO-4 into two major germline regulatory modules: CSR-1a and CSR-1b target a set of spermatogenesis genes, while WAGO-4 operates with CSR-1b to target germline constitutive and, to a lesser extent oogenic transcripts.
To better understand the relationship between the CSR-1 isoforms and WAGO-4, we examined several other features of WAGO-4 22G-RNAs. First, we analyzed the pattern of 22G-RNAs along WAGO-4 targets using a metagene plot (Figure 8C). Here, we observed that the WAGO-4 22G-RNA pattern appears very similar to the CSR-1b 22G-RNA pattern and, as we had observed for CSR-1b, is distinct from the CSR-1a 22G-RNA pattern (compare Figure 8C to Figure 5F). This may reflect different 22G-RNA biogenesis mechanisms.
Next, we examined the extent to which WAGO-4 22G-RNAs display non-templated addition of uridine (Figure 8D). CSR-1 22G-RNAs are known to be uridylated at their 3′ ends by terminal uridyl-transferase, CDE-1, and this modification was originally thought to facilitate turnover of CSR-1-associated 22G-RNAs (4,77). Conversely, WAGO-4 is thought to favor an association with the uridylated 22G-RNAs. Hence, the uridylation state of the 22G-RNAs may control the balance in association between CSR-1 and WAGO-4 (75). First, we assessed to what extent CSR-1a and CSR-1b 22G-RNAs were uridylated. We found that CSR-1b 22G-RNAs were consistently U-tailed (24.14% of 22G-RNA reads in IP samples had one or more U’s added vs. 7.01% in Input samples). In contrast, CSR-1a 22G-RNAs displayed a more modest degree of U-tailing (12.96% of 22G-RNA reads in IP samples vs. 5.18% in Input samples). We next assessed the uridylation of WAGO-4 22G-RNAs and found them to be uridylated to a similar extent as CSR-1a (10.41% of 22G-RNA reads in IP samples vs. 3.31% in Input samples). These uridylation differences of CSR-1b vs. CSR-1a and WAGO-4 22G-RNAs uncover another important distinction between the CSR-1 isoforms, and suggest that CSR-1a and WAGO-4 22G-RNAs are regulated differently than those in complex with CSR-1b.
When examining the localization patterns of CSR-1 isoforms, we found that both CSR-1a and CSR-1b are present in at least two types of germ granules that are closely situated on the nuclear periphery: P granules and Z granules (Figure 2B). Z granules have been implicated in transgenerational epigenetic inheritance, while P granules have been implicated in sRNA biogenesis and exoRNAi (78). WAGO-4 and the helicase ZNFX-1 were identified as the founding members of Z granules, and only a few other proteins have been identified in association with Z granules so far (65,76,78,79). Loss of either wago-4 or znfx-1 did not influence the localization of each other to Z granules (65,76). However, loss of both isoforms of csr-1 disrupted ZNFX-1 foci in adult hermaphrodites, and was associated with a dissociation of PGL-1 from the nuclear periphery during oogenesis (4,28,76).
Given the relationships we established between WAGO-4 and CSR-1a/b targets above, we wanted to determine if loss of wago-4 affected the association of either CSR-1 isoform with germ granules. To do this, we first quantified the association of GFP::CSR-1a, GFP::CSR-1b and GFP::WAGO-4 with the P and Z granule markers PGL-1::RFP and RFP::ZNFX-1, respectively, in L4 wild-type worms (Figure 8E). We found that all three AGOs overlap with PGL-1 and ZNFX-1 pixels to a comparable extent. This implies that CSR-1a/b and WAGO-4 reside in both P and Z granules. We next performed the same quantification for each CSR-1 isoform in wago-4(tm1019) null mutants. While CSR-1a maintained an equivalent overlap with PGL-1 and ZNFX-1, CSR-1b displayed decreased overlap with PGL-1 and increased overlap with ZNFX-1. Loss of csr-1a led to no change in WAGO-4 overlap with either marker. These observations imply that loss of wago-4 shifts the balance of CSR-1b from P granules to Z granules, and reinforces the notion that CSR-1b and WAGO-4 act in concert to maintain germ granule homeostasis. In contrast, CSR-1a appears to function independently of WAGO-4, and does not influence germ granule morphology or the balance between CSR-1b and WAGO-4. In sum, our data point to a broad role in germline constitutive gene regulation and germline homeostasis mediated via CSR-1b and WAGO-4, while CSR-1a and CSR-1b promote the expression of spermatogenesis transcripts.
CSR-1a and a group of spermatogenesis-specific AGOs are required for sperm-mediated fertility
The AGOs ALG-3 and ALG-4 play an established role in spermatogenesis, and their functions have been linked to CSR-1 during sperm development (14,23). Therefore, we aimed to assess how the CSR-1 isoforms intersect with the ALG-3, ALG-4 pathway. In addition, we have evaluated the expression patterns and sRNA binding partners of every AGO in a comprehensive study of all C. elegans AGOs (Seroussi, in preparation). This study uncovered a fourth WAGO with a spermatogenesis-specific expression profile: WAGO-10 (Figure 9A). Consistent with its expression, WAGO-10 also associates with 22G-RNAs that target spermatogenesis transcripts (Seroussi, in preparation, and Figure 10, Supplementary Tables S1-2). ALG-3, ALG-4, CSR-1a, and WAGO-10 therefore define a set of AGOs with spermatogenesis-specific germline expression.
Figure 10.
Spermatogenesis AGOs target different subsets of germline genes. (A) Clustering diagram of genes targeted by 22G-RNA binding AGOs and 26G-RNA binding AGOs during the L4 stage. Each blue line represents a single gene targeted by an AGO/sRNA. There are 7137 genes in total represented. (B) Venn-pie diagrams comparing groups of genes targeted by each AGO (inner circle) with gene sets that define spermatogenesis-enriched transcripts, oogenesis-enriched transcripts, and germline constitutive (outer circle) (29). The size of each color-coded circle represents the proportion of that sample that overlaps with the other sample. Numbers in the center of the circle indicate the number of genes that overlap between the two data sets. Numbers in bold are statistically significant as calculated by Fisher's Exact Test (one sided), P< 0.05. (C) Heatmap depicting the overlap between protein coding targets of various combinations of AGOs with mutually exclusive germline constitutive, oogenic, and spermatogenic enriched transcript sets (29). Bold numbers indicate significant enrichment in overlapping genes (Fisher's Exact Test, P< 0.05). Enrichment = number of overlapping genes divided by the expected number of overlapping genes drawn from two independent groups. (D-H) Clustering diagrams for various germline and spermatogenesis pathways depicting which AGOs target which genes in each pathway: (D) Stem Cell Niche (81); (E) Sex Determination (82); (F) Spermatogenesis (22); (G) Major Sperm Proteins (83); (H) Sperm Activation (84).
The overlap in expression profiles and target gene sets (Figures 9A, 10A, Supplementary Table S2) suggested that these four AGOs could function redundantly to regulate spermatogenesis. Therefore, we built a quadruple null mutant for the spermatogenesis agos, and assessed male fertility. Double mutants of alg-3; alg-4 and csr-1 null mutants have previously been shown to exhibit a progressive loss of male fertility over several generations when cultured at the stressful temperature of 25°C (14,23). We also noticed that our csr-1a(tor159) allele showed a decrease in fertility at 25°C or after long-term culture at 20°C (Figure 2D). Thus, we outcrossed each single spermatogenesis ago mutant and compared the brood size of each freshly outcrossed single mutant to the quadruple spermatogenesis ago mutant in the first generation experiencing the temperature shift. For each of the outcrossed single mutants, the brood size was comparable to wild-type worms (Figure 9B). In sharp contrast, the quadruple ago mutant was nearly sterile in the first generation experiencing the temperature shift. Further, we noticed that the quadruple spermatogenesis ago mutant produced only unfertilized oocytes, which is consistent with a defect in spermatogenesis. To determine whether the fertility defect was due to non-functional sperm, we crossed the quadruple spermatogenesis ago mutant hermaphrodites to wild-type males at 25°C, and found that this rescued fertility to wild-type levels (Figure 9C).
We were curious as to the cause of the spermatogenesis defect, so we took two approaches to broadly examine the phenotype. First, we DAPI stained L4 hermaphrodite wild-type vs. quadruple spermatogenesis ago mutants reared at 25°C to examine whether there were any developmental arrests or delays. Remarkably, we observed a block in spermatogenesis at the primary spermatocyte developmental stage in the quadruple spermatogenesis ago mutant (Figure 9D). In fact, we did not detect mature spermatids in the majority of individuals examined from this strain. In parallel, we examined the number of germ granules (as measured by counting the number of ZNFX-1 and PGL-1 foci) present in quadruple ago mutants vs. wild-type worms at 25°C, because germ granule dysfunction has been associated with a transgenerational loss of fertility in hermaphrodites (80) (Figure 9E). However, we did not detect any differences in the number of ZNFX-1 or PGL-1 foci, suggesting that defects in germ granule homeostasis were not at the root of the loss of fertility in the quadruple spermatogenesis ago mutant. Collectively, these data indicate that the spermatogenesis-specific AGOs, ALG-3, ALG-4, CSR-1a, and WAGO-10 cooperate to produce functional sperm and are essential for fertility.
Interconnected AGO/sRNA modules target germline transcripts during spermatogenesis
The ALG-3, ALG-4 26G-RNA pathway is thought to act upstream of CSR-1 (14). However, this model does not take into account the roles of other spermatogenesis-expressed AGOs, including the individual isoforms of CSR-1, WAGO-10, or WAGO-4. As a part of our comprehensive study of all C. elegans AGOs (Seroussi, in preparation), we performed IP/sRNA-seq experiments on ALG-3 and ALG-4 to define their sets of 26G-RNA target transcripts, and WAGO-10 to define its 22G-RNA targets. Each of these AGOs targets protein coding genes (ALG-3: 2810; ALG-4 3246; WAGO-10: 786 genes) (Figure 10A-B, Supplementary Tables S1-2). Given this abundance of sRNA targeting data, we now have an important opportunity to define the network of spermatogenic gene regulation in greater detail.
We first performed an unsupervised clustering analysis to visualize subsets of genes that were coordinately targeted by multiple AGOs (Figure 10A). From this analysis, the overlap in WAGO-4 and CSR-1b targets discussed above is evident. Similarly, ALG-3 and ALG-4 show a high degree of overlap in their target transcripts, as was previously described (14,23). WAGO-10 targets almost entirely overlap with CSR-1a, and with ALG-3 and ALG-4 targets, providing a possible explanation for the quadruple ago mutant fertility phenotype.
We next compared the targets of each AGO to oogenic, spermatogenic and germline constitutive gene sets (29) (Figure 10B, Supplementary Table S2). We found that like CSR-1a, ALG-3 and ALG-4 targets are enriched, roughly equally, in germline constitutive and spermatogenic transcripts, while WAGO-10 targets are enriched in spermatogenic transcripts. Consistent with our analysis above, CSR-1b targets are most enriched for germline constitutive genes. However, a comparable number of CSR-1b targets are enriched in oogenesis and spermatogenesis genes, reflecting its distinct gene regulatory modules (WAGO-4 vs. CSR-1a). WAGO-4 targets are enriched in germline constitutive and oogenic gene sets, and are conspicuously under-represented in spermatogenesis genes. From the clustering visualization (Figure 10A), we can detect more specific subsets of sRNA target genes based on the combination of AGOs that target them (Figure 10C, Supplementary Table S2). This analysis reinforces our findings above. Gene sets targeted by WAGO-4 and any other AGO are over-represented in germline constitutive and oogenic genes. Genes targeted by various combinations of ALG-3, ALG-4, WAGO-10, CSR-1a, and to a lesser extent CSR-1b (but excluding WAGO-4) are over-represented in spermatogenic genes. Collectively, our findings implicate CSR-1b and WAGO-4 in a germline gene regulatory module that is likely to be important in both spermatogenesis and oogenesis (based on the nature of the genes targeted and the expression profiles of WAGO-4 and CSR-1b), while the CSR-1a, WAGO-10, ALG-3 and ALG-4 form a spermatogenesis gene regulatory module that is also likely to involve CSR-1b.
We homed in on genes involved in several important facets of spermatogenesis and spermiogenesis to understand how these different processes might be regulated. As anticipated, CSR-1b and WAGO-4 appear to play broad roles in the germline that are important for both spermatogenesis and oogenesis. All Delta/Notch signaling factors involved in maintaining the germline stem cell niche (81) are targeted almost exclusively by CSR-1b and WAGO-4 22G-RNAs (gld-2 is also a target of CSR-1a, ALG-3, and ALG-4) (Figure 10D, Supplementary Table S2). The exception in this pathway is lag-2, the Delta ligand expressed in the distal tip cell, which is not targeted by any sRNA pathways described here. This is likely due to the fact that the distal tip cell is somatic. Similarly, the sex determination pathway (82) is extensively targeted by CSR-1b and WAGO-4 22G-RNAs (Figure 10E, Supplementary Table S2), as are many chromatin associated DNA and RNA binding proteins expressed in mature spermatozoa (Supplementary Table S2) (66).
Sperm-specific functions are subject to regulation by the spermatogenesis AGOs. Genes involved in spermatogenesis—the differentiation of germ cells into sperm (22)—are predominantly targeted by ALG-3, ALG-4, CSR-1a and CSR-1b, and to a lesser extent WAGO-10 and WAGO-4 (Figure 10F, Supplementary Table S2). The Major Sperm Proteins (MSPs) are encoded by a group of about 30 highly homologous genes (along with about 20 pseudogenes) (83). These small (∼15kDa), basic proteins are expressed late in spermatogenesis, and play roles in sperm structure, motility, and signaling (84). The msp genes are targeted by ALG-3, ALG-4, CSR-1a, and to a lesser extent, CSR-1b and WAGO-10 (Figure 10G, Supplementary Table S2). Notably WAGO-4 does not target any msp genes. Finally, the spermiogenesis, or sperm activation pathway, yields motile, fertilization competent spermatozoa from immature spermatids (84), and most components of this pathway are targeted by ALG-3, ALG-4, CSR-1a, WAGO-10 and CSR-1b (Figure 10H, Supplementary Table S2). While these few examples only scratch the surface, they highlight the gene regulatory modules at play during spermatogenesis, and place CSR-1b at a critical intersection between the two major AGO networks.
DISCUSSION
Because it is the only essential AGO in C. elegans, exhibits several noteworthy developmental and molecular phenotypes, and regulates a large portion of the transcriptome, CSR-1 has been the focus of intense study since its initial characterization. Remarkably, CSR-1 is only conserved in clade III and V nematodes, yet in the only other nematode species in which its function has been studied, C. briggsae, it is also essential (85). The essential nature of CSR-1 points to important gene regulatory roles in gametogenesis and embryonic development, but it was unclear which isoforms of CSR-1 were responsible for these varied gene regulatory functions. Using CRISPR-Cas9 genome editing to insert GFP::3xFLAG tags each CSR-1 isoform, and mutant studies to understand the outcome of loss of each isoform on development, we have uncovered distinct expression patterns, sRNA binding partners, and gene regulatory functions for the two CSR-1 isoforms (Figure 11).
Figure 11.
Model for differential functions of CSR-1a and CSR-1b. A model summarizing the expression patterns, sRNA partners, pathways, and functions of CSR-1 isoforms.
CSR-1 isoforms regulate different facets of fertility
Our previous studies using antibodies that recognize both CSR-1 isoforms failed to distinguish the expression patterns of CSR-1a and CSR-1b (4). By GFP::3xFLAG tagging the individual CSR-1 isoforms, we found that CSR-1b is expressed constitutively in the germline throughout all stages, and is present in somatic cells of the early embryo, due solely to maternal deposition. In contrast, CSR-1a expression in the germline is restricted to spermatogenesis. Moreover, CSR-1a is the only isoform expressed in the adult soma, where it is predominantly present in the intestine and spermatheca. These expression profiles alone shine a light on which CSR-1 isoform may be responsible for which phenotypes. Our analysis shows that CSR-1b alone carries out essential gene regulatory functions in the hermaphrodite germline and within embryos, where the sterile/lethal phenotype manifests. Therefore, we conclude that csr-1b is the essential isoform under ‘luxurious’ lab culture conditions.
We were also surprised to find that the csr-1b rescue strain displayed lower fertility than the endogenously tagged GFP::3xFLAG::CSR-1b strain at 20°C (compare Figure 4B to E), as both strains express only CSR-1b. This fertility defect appears to be due in large part to a late embryonic developmental arrest that occurs later than that of csr-1(tm892) null mutant embryos (Supplementary Figure S4F). This indicates that ectopic expression of CSR-1b from the rpl-28 promoter does not fully recapitulate wild-type CSR-1b expression, and suggests that the levels, developmental timing, and/or cells in which CSR-1b is expressed are subject to developmentally-important regulation. In particular, we speculate that based on the expression pattern of CSR-1b during embryogenesis, ectopic expression in somatic cells in late embryonic development could be deleterious for development.
Conversely, expression of csr-1a from the same rpl-28 promoter rescues the fertility defect of a csr-1(tm892) null mutant, suggesting that CSR-1a can perform at least some of the functions of CSR-1b. Interestingly, when we sequenced sRNAs associated with CSR-1a in L4 hermaphrodites of the csr-1a rescue strain, we found that CSR-1a binds to 22G-RNAs that target almost all of the CSR-1b targets (data not shown). This result is consistent with the fertility data, and reinforces the idea that CSR-1a can compensate at the molecular level for CSR-1b. Although we did not sequence sRNAs associated with ectopically expressed CSR-1a during oogenesis in the csr-1a rescue, we anticipate that CSR-1a binds to CSR-1b 22G-RNAs, resulting in fertility.
On its own, CSR-1a plays an important role in sperm-based fertility within hermaphrodites. Loss of csr-1a alone leads to a transgenerational loss of sperm-based fertility in hermaphrodites. CSR-1a also plays an overlapping role with ALG-3, ALG-4 and WAGO-10 in spermatogenesis; collectively these AGOs are required for generating mature sperm, and hence sperm-mediated fertility, at stressful temperatures. Because CSR-1b is also expressed throughout the germline during spermatogenesis, it is conceivable that it too is required for sperm-based fertility. Unfortunately, our data do not specifically address this issue. Comparing the transgenerational loss of fertility between csr-1a and csr-1 null mutants is fraught by the additional defects in oogenesis and embryogenesis caused by loss of csr-1b. Instead, to assess a role for CSR-1b in sperm-based fertility, we would need to examine male fertility in a transgenerational mating assay similar to Conine et al. (14) or rescue the csr-1(tm892) null mutant with a csr-1a transgene expressed from the csr-1 promoter. Selective degradation of CSR-1b during spermatogenesis via a degron approach (86) may also enable such an assessment. Related to this, any male-specific roles for either CSR-1 isoform should also be explored in the future. While many aspects of spermatogenesis are consistent between male and hermaphrodite C. elegans, there are several features that are sex-specific (87).
CSR-1 isoforms associate with P and Z granules
Our previous work showed that CSR-1 co-localizes with PGL-1 and is present within germ granules known as P granules (4). We initially speculated that CSR-1a might be the only isoform capable of localizing to P granules due to its unique RGG/RG rich N-terminus. RGG/RG motifs have been implicated in RNA binding, recruiting RNA binding proteins, and inducing phase separation in vitro—all of which are functions relevant to germ granules (9). Loss of csr-1 has been shown to result in the mis-localization of P granules to the core cytoplasm of the germline, pointing to a role for CSR-1 in the maintenance and proper positioning of P granules (4,28). Remarkably, we observed that both CSR-1 isoforms overlap with P granule and Z granule markers to a similar extent in the L4 germline. Examination of CSR-1b overlap with PGL-1 and ZNFX-1 during oogenesis yielded the same pattern (data not shown). There are multiple mechanisms by which proteins can associate with phase separated granules, including association with RNA and other proteins that induce phase separation (9). The fact that CSR-1b, without an RGG/RG rich domain, and CSR-1a[G91R] localize to germ granules supports the notion that RNA and/or other proteins enable the association of CSR-1 with these granules.
Prior to our findings, there were six known Z granule components (65,79,88), thus the identification of CSR-1 isoforms in association with ZNFX-1 is important. Although there are some limitations of our analysis to consider, including the facts that we were not able to visualize PGL-1, ZNFX-1, and CSR-1 in the same strains, and that our imaging represents a snapshot (and not live tracking) of development, there are several important implications. First, there may be an uncharacterized region of overlap between Z and P granules in the L4 germline where the CSR-1 isoforms reside, as opposed to CSR-1 isoforms existing as discrete pools within each granule. Second, it is possible that CSR-1 isoforms could move between the granules. Third, if the patterns we observed represent localization to distinct granules, CSR-1 isoforms could play different roles in the different granules, as Z granules have been linked to transgenerational epigenetic inheritance and P granules are associated with maternal inheritance and germ cell fate (65,78).
CSR-1b and WAGO-4, a founding member of Z granules, possess extensive overlap in their 22G-RNA targets. Thus, it may not be surprising that CSR-1b is present in Z granules, or that loss of wago-4 perturbs the distribution of CSR-1b between P and Z granules. The loss of wago-4 does not perturb the CSR-1a distribution, and notably, the overlap in CSR-1a and WAGO-4 targets is almost exclusively via targets shared with CSR-1b. Why and how germline genes are subject to regulation by different combinations of CSR-1a, CSR-1b, and WAGO-4 is currently unknown, but there is clearly a deeper molecular link that demands further study.
How do CSR-1a's RG motifs contribute to its functions?
Regarding the function of the RGG/RG motifs within the first exon of CSR-1a, we recovered several alleles that affected these motifs from the screen for proteasomal regulators. This could point to a role for arginine methylation in regulating CSR-1a. Arginine methylation is important for the recruitment of tudor domain proteins to AGO, for example, TudorSN and Piwi in Drosophila (6–8). In the worm, we hypothesize that tudor domain proteins, such as RSD-6 or EKL-1 (a known CSR-1 pathway component) (4,15,63,89,90) may interact with CSR-1a in a methyl-arginine dependent manner.
We studied one of these alleles, csr-1a[G91R] in more detail. While mutation of a single RG motif in csr-1a[G91R] led to lower levels of CSR-1a expression, it did not perturb the formation of germ granules, nor did it prevent the association of CSR-1a with PGL-1 (Supplementary Figure S4). In addition, sequencing sRNAs associated with CSR-1a[G91R] showed a depletion for 22G-RNAs targeting approximately one-half (1279/2572) of the protein coding genes that are normally the targets of CSR-1a (data not shown). These genes are both germline and intestinal genes, suggesting that this particular RG motif could be important for gene regulation in both tissues. It is important to note that mutation of multiple RG residues may be necessary to perturb germ granules or cause substantial changes in sRNA association, thus a detailed mutagenesis study of these residues will be necessary in the future.
CSR-1b associates with chromatin
We also observed that CSR-1b associates with spermatid and oocyte nuclei and chromatin. These observations are consistent with a previous proteomic study that found CSR-1 peptides enriched on both oocyte and sperm chromatin (66), as well as microscopy and ChIP studies that point to an association of CSR-1 with chromatin (4,10,14,18). These collective findings point to a low but consistent level of CSR-1b present in the nucleus and/or associated with germ cell chromatin, despite the fact that the bulk of CSR-1b protein is present within the cytoplasm and germ granules (4). This pool of CSR-1b can be passed from parent to progeny via sperm or oocyte chromatin and presents the possibility for epigenetically marking chromosomes, influencing chromatin landscapes, and reinforcing patterns of transcription in progeny (14,19,91). CSR-1 has been implicated in transgenerational phenotypes, including Mrt and RNAa (14,17,18), and this chromatin association could be but one of several molecular mechanisms by which these phenomena are executed.
An extensive network of spermatogenesis sRNA pathways
In addition to CSR-1a, we identified a second novel spermatogenesis-expressed AGO, WAGO-10, and found that CSR-1a, WAGO-10, ALG-3 and ALG-4 that target spermatogenesis genes. This overlapping profile of gene targets for the spermatogenesis AGOs pointed to the potential for redundancy. While csr-1a, and alg-3; alg-4 mutants display a transgenerational loss of fertility at high temperatures (14), the quadruple spermatogenesis ago mutant displayed male-dependent infertility in a single generation at the stressful temperature, supporting that these AGOs function redundantly during spermatogenesis. Although we did not deeply examine the phenotype of quadruple spermatogenesis ago mutant germlines, we observed that germ granules appeared comparable in quantity to those of wild-type animals. However, germ cells were arrested in meiosis prior to differentiation into secondary spermatocytes, based on nuclear morphology. Further cell biological examination of this phenotype and characterization of any changes in gene regulation that result from loss of these four agos will be important next steps to understanding how these AGOs influence spermatogenesis.
We have stratified germline genes into different gene regulatory modules likely to be regulated by different AGOs. CSR-1b and WAGO-4 are constitutively expressed in the germline, and coordinately target germline constitutive and oogenic genes. CSR-1b is likely to play gene licensing rather than silencing roles (4,14,18), while WAGO-4 appears to play a silencing role (75). How these AGOs coordinate gene expression to maintain germline homeostasis and fertility remains unclear, and will undoubtedly be the focus of future studies.
CSR-1b also plays a role in regulating spermatogenic transcripts, this time alongside CSR-1a and the other spermatogenic AGOs. In this way, CSR-1b fills an important sRNA regulatory niche across all classes of germline genes. Perhaps this broad role for CSR-1b contributes to its essential nature. Exactly how CSR-1b regulates all of these different types of transcripts—at the co-transcriptional or post-transcriptional level—remains to be determined. There is some precedent for CSR-1’s capacity to differentially regulate subsets of targets in the embryo (10,11), but detailed molecular studies will need to be carried out in the germline to fully understand the scope of CSR-1b mechanisms.
The four spermatogenesis AGOs, ALG-3, ALG-4, CSR-1a, and WAGO-10, are expressed predominantly in the proximal germline, where they target mostly spermatogenic and some germline constitutive genes, but not oogenic genes. Perhaps not surprisingly, these AGOs target genes in all major pathways/components: spermatogenesis, spermiogenesis, and MSPs. Based on our current understanding of these pathways, it seems plausible that ALG-3 and ALG-4 26G-RNAs act in a primary capacity, while CSR-1a, WAGO-10, and to a lesser extent, CSR-1b 22G-RNAs act in a secondary capacity (14). ALG-3 and ALG-4 were previously implicated as licensing AGOs in conjunction with CSR-1 (14). Our mRNA-seq analysis in csr-1a(tor159) mutants supports this relationship: spermatogenic gene expression is decreased. It will be critical to examine gene expression changes in each single and combination spermatogenic ago mutant to determine the contribution of each AGO to fertility and gene regulation as well as to delineate functional distinctions and redundancies. Importantly, while these AGOs play an essential role in buffering the processes of spermatogenesis and spermiogenesis from stress to maintain fertility, their deposition into the residual body and depletion from maturing sperm indicates that they are unlikely to be direct couriers of transgenerational epigenetic inheritance.
Somatic gene silencing roles for CSR-1a
CSR-1a is the only CSR-1 isoform expressed in the adult soma, where it is required for repression of endogenous intestinal transcripts, including innate immunity and pathogen responsive genes. From our initial screen for proteasomal regulators, we isolated mutations affecting csr-1a and several known RNAi factors, including rde-1, rrf-1, and rsd-2. The screen was carried out using a repetitive array transgene (rpn-2p::gfp), and rde-1, rrf-1, and rsd-2 have all been implicated in exogenous and endogenous sRNA-mediated gene silencing (61–63). We therefore anticipated that CSR-1a may participate in somatic sRNA-mediated silencing of rpn-2p::gfp. However, sRNA sequencing from csr-1a[G120*]and from CSR-1a IPs in the rpn-2p::gfp strain showed no link between CSR-1a and sRNAs derived from the transgene, pointing to an indirect effect for CSR-1a in regulating the transgene. It is plausible that the other RNAi factors are involved in somatic transgene silencing, consistent with a previous report (92). However, at this point, it appears that CSR-1a affects the rpn-2p::gfp transgene indirectly, possibly by affecting other regulators such as transcription factors, histone modifiers, or proteasome pathway components.
Interestingly, in an attempt to identify other somatic functions for CSR-1a, we also tested whether loss of csr-1a resulted in RNAi deficiency (Rde) (62). Null mutants of csr-1 have been shown to have limited capacity to silence germline transcripts, including pos-1 and cdk-1 (4). However, when we performed RNAi on dpy-11, an intestinal-expressed gene, we observed no difference between csr-1a mutants and wild-type worms (data not shown). While we were surprised that loss of csr-1a did not result in RNAi deficiency, our systematic study of C. elegans AGOs points to several other AGOs that are expressed in the intestine and have been previously implicated in RNAi, including the secondary sRNA binding AGOs, SAGO-1 and SAGO-2 (3) (Seroussi, in preparation). One particular interesting candidate is SAGO-2, which has recently been shown to interact with CSR-1a by mass spectrometry (93). Thus, it is possible that these AGOs could function redundantly with CSR-1a to execute exogenous RNAi in this tissue.
Loss of csr-1a led to the increased expression of many somatic genes. This suggests that CSR-1a silences the expression of its targets in the intestine, and possibly other somatic tissues. Our gene expression analysis hinted that CSR-1a could play a role in pathogen and/or stress responses within the intestine, as genes related to defense responses, interspecies interaction and the immune system were upregulated in csr-1a(tor159) mutants. Despite this observation, we were still somewhat surprised to find that loss of csr-1a was beneficial to worm survival from the bacterial pathogen Pseudomonas aeruginosa. We reason that upregulation of such genes could enable the intestine to be ‘primed’ for encounters with pathogens or other environmental stressors. It will be important to test the response of csr-1a(tor159) mutants to a full array of pathogens and environmental stresses in the future, to determine if this is a generalized or specialized response.
Slicer activity of CSR-1 isoforms
Our studies uncover many new dimensions of CSR-1 biology, however they do not address whether Slicer function is important for both CSR-1 isoforms and in what tissues or gene regulatory contexts. Both isoforms possess the catalytic tetrad of residues within the PIWI domain that are required for Slicer activity, and CSR-1 is one of few AGOs in the worm for which Slicer activity has been studied in vitro and in vivo (11,20,21). Slicer activity has been implicated in fine tuning maternally deposited transcripts with high 22G-RNA density in the embryo (11). This function can be attributed to CSR-1b based on expression of the isoforms alone. Histone mRNA maturation is another activity for which Slicer activity is anticipated to be required. As both CSR-1 isoforms associate with 22G-RNAs that target half of all histone transcripts, Slicer activity may be important for both isoforms in this context. Clearly, detailed tissue-specific studies will be important to delineate the roles of Slicer activity for each isoform throughout development.
Summary: A tale of two Argonautes
Our studies disentangle a significant amount of complexity surrounding the essential C. elegans AGO, CSR-1, and generate a myriad of additional questions surrounding the tissue-specific functions of CSR-1 and other AGOs. Newly developed means of expressing and degrading or inactivating proteins in a tissue specific manner will enable us to understand the roles of each CSR-1 isoform at a finer resolution, in specific cells and tissues (86).
In other Caenorhabditis nematodes, particularly male/female mating species, csr-1 not only has multiple isoforms, but possesses duplicated gene copies (85,94). This expansion has been proposed to serve as a defense mechanism against viruses and other invasive nucleic acid transmitted between the members of obligatory mating species (94). We have shown that CSR-1a buffers the effects of temperature stress during spermatogenesis to maintain fertility and is involved in gene silencing in the intestine, a tissue that directly interfaces with the environment. Given its roles in C. elegans, it is possible that these additional copies of csr-1 have further specialized to enable other Caenorhabditis species to navigate an array of stresses encountered in their natural environment.
Importantly, CSR-1 is not the only Argonaute expressed as multiple isoforms. Several other AGOs, including ALG-1, ALG-2, ERGO-1, and PPW-1, also possess this capacity in C. elegans, but isoform-specific functions of these proteins have not been examined. Our work highlights the importance of dissecting each of these AGO isoforms’ roles to unravel how complex tissue- and developmental stage-specific sRNA networks safeguard the genome, germline, and soma.
DATA AVAILABILITY
Small RNA-seq and poly(A) selected mRNA-seq data generated in this study are available under the accession number GEO: GSE154678. Germline constitutive, oogenic and spermatogenic gene lists were defined as in (29) available from GEO under accession number GSE57109. RdRP mutant small RNA data are from (71) [GSE66344]. alg-3, alg-4 mutant sRNA data are from (23) [GSE49672]. WAGO-1 IP/sRNA-seq data was obtained from (15) available under accession number [GSE18231]. The CSR-1 IP/sRNA-seq data from males was obtained from (14), accession number [GSE49672], while the hermaphrodite data was obtained from (4), accession number [GSE18167]. The custom scripts used to generate the processed data are available at www.github.com/ClaycombLab/Charlesworth_2020.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs John Calarco and Arneet Saltzman, and members of the Claycomb lab for critical reading and constructive comments on this manuscript. We thank Dr Carolyn Phillips and her lab for thoughtful discussions about the CSR-1 isoforms. Thanks to the lab of Dr. Thomas Hurd for use of microscopes, and to WormBase and The Caenorhabditis Genetics Center for data and strains. We are also grateful to the Toronto Area Worm Community for reagents, protocols, and discussions.
Authors contributions: A.G.C., N.J.L., U.S., M.S.R., M.J.A., J.R.W., A.E.S., R.X.L., R.I.M. and A.J.D. performed the experiments. A.G.C., M.S.R., U.S. and A.J.D. performed computational analyses. J.M.C. wrote the manuscript, with the assistance of A.G.C., N.J.L. and R.I.M. J.M.C., A.G.C., U.S., N.J.L., M.S.R., A.E.S., R.I.M. and A.J.D. devised the figures.
Contributor Information
Amanda G Charlesworth, Department of Molecular Genetics, University of Toronto Toronto, ON M5S 1A8 Canada.
Uri Seroussi, Department of Molecular Genetics, University of Toronto Toronto, ON M5S 1A8 Canada.
Nicolas J Lehrbach, Department of Molecular Biology, Massachusetts General Hospital, Boston, MA 02114, USA; Department of Genetics, Harvard Medical School, Boston, MA 02115, USA.
Mathias S Renaud, Department of Molecular Genetics, University of Toronto Toronto, ON M5S 1A8 Canada.
Adam E Sundby, Department of Molecular Genetics, University of Toronto Toronto, ON M5S 1A8 Canada.
Ruxandra I Molnar, Department of Molecular Genetics, University of Toronto Toronto, ON M5S 1A8 Canada.
Robert X Lao, Department of Molecular Genetics, University of Toronto Toronto, ON M5S 1A8 Canada.
Alexandra R Willis, Department of Molecular Genetics, University of Toronto Toronto, ON M5S 1A8 Canada.
Jenna R Woock, Department of Molecular Genetics, University of Toronto Toronto, ON M5S 1A8 Canada.
Matthew J Aber, Department of Molecular Biology, Massachusetts General Hospital, Boston, MA 02114, USA; Department of Genetics, Harvard Medical School, Boston, MA 02115, USA.
Annette J Diao, Department of Molecular Genetics, University of Toronto Toronto, ON M5S 1A8 Canada.
Aaron W Reinke, Department of Molecular Genetics, University of Toronto Toronto, ON M5S 1A8 Canada.
Gary Ruvkun, Department of Molecular Biology, Massachusetts General Hospital, Boston, MA 02114, USA; Department of Genetics, Harvard Medical School, Boston, MA 02115, USA.
Julie M Claycomb, Department of Molecular Genetics, University of Toronto Toronto, ON M5S 1A8 Canada.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institutes of Health Research [PJT-156083 and PJG-175378 to J.M.C.]; CRC Tier II in Small RNA Biology (to J.M.C.); CIHR-CGSM fellowship (to M.S.R.); some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs [P40 OD010440]. Funding for open access charge: CIHR Project Grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Youngman E.M., Claycomb J.M.. From early lessons to new frontiers: the worm as a treasure trove of small RNA biology. Front. Genet. 2014; 5:416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meister G.Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 2013; 14:447–459. [DOI] [PubMed] [Google Scholar]
- 3.Yigit E., Batista P.J., Bei Y., Pang K.M., Chen C.C., Tolia N.H., Joshua-Tor L., Mitani S., Simard M.J., Mello C.C.. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006; 127:747–757. [DOI] [PubMed] [Google Scholar]
- 4.Claycomb J.M., Batista P.J., Pang K.M., Gu W., Vasale J.J., van Wolfswinkel J.C., Chaves D.A., Shirayama M., Mitani S., Ketting R.F.et al.. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009; 139:123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thandapani P., O’Connor T.R., Bailey T.L., Richard S.. Defining the RGG/RG motif. Mol. Cell. 2013; 50:613–623. [DOI] [PubMed] [Google Scholar]
- 6.Kirino Y., Kim N., de Planell-Saguer M., Khandros E., Chiorean S., Klein P.S., Rigoutsos I., Jongens T.A., Mourelatos Z.. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat. Cell Biol. 2009; 11:652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu K., Chen C., Guo Y., Lam R., Bian C., Xu C., Zhao D.Y., Jin J., MacKenzie F., Pawson T.et al.. Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain. Proc. Natl. Acad. Sci. USA. 2010; 107:18398–18403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vagin V.V., Wohlschlegel J., Qu J., Jonsson Z., Huang X., Chuma S., Girard A., Sachidanandam R., Hannon G.J., Aravin A.A.. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009; 23:1749–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chong P.A., Vernon R.M., Forman-Kay J.D.. RGG/RG Motif Regions in RNA Binding and Phase Separation. J. Mol. Biol. 2018; 430:4650–4665. [DOI] [PubMed] [Google Scholar]
- 10.Avgousti D.C., Palani S., Sherman Y., Grishok A.. CSR-1 RNAi pathway positively regulates histone expression in C. elegans. EMBO J. 2012; 31:3821–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerson-Gurwitz A., Wang S., Sathe S., Green R., Yeo G.W., Oegema K., Desai A.. A Small RNA-Catalytic argonaute pathway tunes germline transcript levels to ensure embryonic divisions. Cell. 2016; 165:396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wedeles C.J., Wu M.Z., Claycomb J.M.. A multitasking Argonaute: exploring the many facets of C. elegans CSR-1. Chromosome Res. 2013; 21:573–586. [DOI] [PubMed] [Google Scholar]
- 13.Quarato P., Singh M., Cornes E., Li B., Bourdon L., Mueller F., Didier C., Cecere G.. Germline inherited small RNAs facilitate the clearance of untranslated maternal mRNAs in C. elegans embryos. Nat. Commun. 2021; 12:1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conine C.C., Moresco J.J., Gu W., Shirayama M., Conte D.J., Yates J.R., Mello C.C.. Argonautes Promote Male Fertility and Provide a Paternal Memory of Germline Gene Expression in C. elegans. Cell. 2013; 155:1532–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu W., Shirayama M., Conte D.J., Vasale J., Batista P.J., Claycomb J.M., Moresco J.J., Youngman E.M., Keys J., Stoltz M.J.et al.. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell. 2009; 36:231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cecere G., Hoersch S., O’Keeffe S., Sachidanandam R., Grishok A.. Global effects of the CSR-1 RNA interference pathway on the transcriptional landscape. Nat. Struct. Mol. Biol. 2014; 21:358–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seth M., Shirayama M., Gu W., Ishidate T., Conte D., Mello C.C.. The C. elegans CSR-1 Argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Dev. Cell. 2013; 27:656–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wedeles C.J., Wu M.Z., Claycomb J.M.. Protection of germline gene expression by the C. elegans Argonaute CSR-1. Dev. Cell. 2013; 27:664–671. [DOI] [PubMed] [Google Scholar]
- 19.Gushchanskaia E.S., Esse R., Ma Q., Lau N.C., Grishok A.. Interplay between small RNA pathways shapes chromatin landscapes in C. elegans. Nucleic Acids Res. 2019; 47:5603–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aoki K., Moriguchi H., Yoshioka T., Okawa K., Tabara H.. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007; 26:5007–5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fassnacht C., Tocchini C., Kumari P., Gaidatzis D., Stadler M.B., Ciosk R.. The CSR-1 endogenous RNAi pathway ensures accurate transcriptional reprogramming during the oocyte-to-embryo transition in Caenorhabditis elegans. PLoS Genet. 2018; 14:e1007252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.L’Hernault S.W.Spermatogenesis. WormBook. 2006; 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conine C.C., Batista P.J., Gu W., Claycomb J.M., Chaves D.A., Shirayama M., Mello C.C.. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2010; 107:3588–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaves D.A., Dai H., Li L., Moresco J.J., Oh M.E., Conte D., Yates J.R., Mello C.C., Gu W.. The RNA phosphatase PIR-1 regulates endogenous small RNA pathways in C. elegans. Mol. Cell. 2021; 81:546–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han T., Manoharan A.P., Harkins T.T., Bouffard P., Fitzpatrick C., Chu D.S., Thierry-Mieg D., Thierry-Mieg J., Kim J.K.. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2009; 106:18674–18679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gent J.I., Schvarzstein M., Villeneuve A.M., Gu S.G., Jantsch V., Fire A.Z., Baudrimont A.. A Caenorhabditis elegans RNA-directed RNA polymerase in sperm development and endogenous RNA interference. Genetics. 2009; 183:1297–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vasale J.J., Gu W., Thivierge C., Batista P.J., Claycomb J.M., Youngman E.M., Duchaine T.F., Mello C.C., Conte D.J.. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc. Natl. Acad. Sci. USA. 2010; 107:3582–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell A.C., Updike D.L.. CSR-1 and P granules suppress sperm-specific transcription in the C. elegans germline. Development. 2015; 142:1745–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ortiz M.A., Noble D., Sorokin E.P., Kimble J.. A new dataset of spermatogenic vs. oogenic transcriptomes in the nematode Caenorhabditis elegans. G3 (Bethesda). 2014; 4:1765–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brenner S.The genetics of Caenorhabditis elegans. Genetics. 1974; 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schnabel H., Schnabel R.. An Organ-Specific Differentiation Gene, pha-1, from Caenorhabditis elegans. Science. 1990; 250:686–688. [DOI] [PubMed] [Google Scholar]
- 32.Dickinson D.J., Ward J.D., Reiner D.J., Goldstein B.. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods. 2013; 10:1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paix A., Wang Y., Smith H.E., Lee C.Y., Calidas D., Lu T., Smith J., Schmidt H., Krause M.W., Seydoux G.. Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 Sites in Caenorhabditis elegans. Genetics. 2014; 198:1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward J.D.Rapid and precise engineering of the Caenorhabditis elegans genome with lethal mutation co-conversion and inactivation of NHEJ repair. Genetics. 2015; 199:363–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arribere J.A., Bell R.T., Fu B.X., Artiles K.L., Hartman P.S., Fire A.Z.. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics. 2014; 198:837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dickinson D.J., Pani A.M., Heppert J.K., Higgins C.D., Goldstein B.. Streamlined Genome Engineering with a Self-Excising Drug Selection Cassette. Genetics. 2015; 200:1035–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ouyang J.P.T., Folkmann A., Bernard L., Lee C.Y., Seroussi U., Charlesworth A.G., Claycomb J.M., Seydoux G.. P Granules Protect RNA Interference Genes from Silencing by piRNAs. Dev. Cell. 2019; 50:716–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lev I., Toker I.A., Mor Y., Nitzan A., Weintraub G., Antonova O., Bhonkar O., Ben Shushan I., Seroussi U., Claycomb J.M.et al.. Germ Granules Govern Small RNA Inheritance. Curr. Biol. 2019; 29:2880–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gibson D.G.Synthesis of DNA fragments in yeast by one-step assembly of overlapping oligonucleotides. Nucleic Acids Res. 2009; 37:6984–6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lehrbach N.J., Ruvkun G.. Proteasome dysfunction triggers activation of SKN-1A/Nrf1 by the aspartic protease DDI-1. eLife. 2016; 5:e17721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frøkjær-Jensen C., Davis M.W., Sarov M., Taylor J., Flibotte S., LaBella M., Pozniakovsky A., Moerman D.G., Jorgensen E.M.. Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat. Methods. 2014; 11:529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunt-Newbury R., Viveiros R., Johnsen R., Mah A., Anastas D., Fang L., Halfnight E., Lee D., Lin J., Lorch A.et al.. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 2007; 5:e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minevich G., Park D.S., Blankenberg D., Poole R.J., Hobert O.. CloudMap: a cloud-based pipeline for analysis of mutant genome sequences. Genetics. 2012; 192:1249–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B.et al.. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bolte S., Cordelières F.P.. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006; 224:213–232. [DOI] [PubMed] [Google Scholar]
- 46.Schneider C.A., Rasband W.S., Eliceiri K.W.. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012; 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fox J., Weisberg S.. An R Companion to Applied Regression. Third edition. Sage. 2019; Thousand Oaks CA. [Google Scholar]
- 48.Wickham H.ggplot2: Elegant Graphics for Data Analysis. 2016; NY: Springer-Verlag. [Google Scholar]
- 49.Tan M.W., Rahme L.G., Sternberg J.A., Tompkins R.G., Ausubel F.M.. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA. 1999; 96:2408–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin M.Cutadapt removes adapter sequences from high-throughput sequencing reads. Bioinformatics in Action. 2011; 17:10–12. [Google Scholar]
- 51.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith R.N., Aleksic J., Butano D., Carr A., Contrino S., Hu F., Lyne M., Lyne R., Kalderimis A., Rutherford K.et al.. InterMine: a flexible data warehouse system for the integration and analysis of heterogeneous biological data. Bioinformatics. 2012; 28:3163–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kalderimis A., Lyne R., Butano D., Contrino S., Lyne M., Heimbach J., Hu F., Smith R., Stěpán R., Sullivan J.et al.. InterMine: extensive web services for modern biology. Nucleic Acids Res. 2014; 42:W468–W472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang W., Dierking K., Schulenburg H.. WormExp: a web-based application for a Caenorhabditis elegans-specific gene expression enrichment analysis. Bioinformatics. 2016; 32:943–945. [DOI] [PubMed] [Google Scholar]
- 55.Angeles-Albores D., Lee R., Chan J., Sternberg P.. Two new functions in the WormBase Enrichment Suite. MicroPubl. Biol. 2018; 2018: 10.17912/W25Q2N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W.. GenBank. Nucleic Acids Res. 2017; 45:D37–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edgar R.C.MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gouy M., Guindon S., Gascuel O.. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010; 27:221–224. [DOI] [PubMed] [Google Scholar]
- 60.Bard J.A.M., Goodall E.A., Greene E.R., Jonsson E., Dong K.C., Martin A.. Structure and Function of the 26S Proteasome. Annu. Rev. Biochem. 2018; 87:697–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sijen T., Fleenor J., Simmer F., Thijssen K.L., Parrish S., Timmons L., Plasterk R.H., Fire A.. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001; 107:465–476. [DOI] [PubMed] [Google Scholar]
- 62.Tabara H., Sarkissian M., Kelly W.G., Fleenor J., Grishok A., Timmons L., Fire A., Mello C.C.. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999; 99:123–132. [DOI] [PubMed] [Google Scholar]
- 63.Tijsterman M., May R.C., Simmer F., Okihara K.L., Plasterk R.H.. Genes required for systemic RNA interference in Caenorhabditis elegans. Curr. Biol. 2004; 14:111–116. [DOI] [PubMed] [Google Scholar]
- 64.Kim H., Ishidate T., Ghanta K.S., Seth M., Conte D., Shirayama M., Mello C.C.. A co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics. 2014; 197:1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wan G., Fields B.D., Spracklin G., Shukla A., Phillips C.M., Kennedy S.. Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance. Nature. 2018; 557:679–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chu D.S., Liu H., Nix P., Wu T.F., Ralston E.J., Yates J.R., Meyer B.J.. Sperm chromatin proteomics identifies evolutionarily conserved fertility factors. Nature. 2006; 443:101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fukushige T., Hawkins M.G., McGhee J.D.. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev. Biol. 1998; 198:286–302. [PubMed] [Google Scholar]
- 68.Kudron M.M., Victorsen A., Gevirtzman L., Hillier L.W., Fisher W.W., Vafeados D., Kirkey M., Hammonds A.S., Gersch J., Ammouri H.et al.. The ModERN Resource: Genome-Wide Binding Profiles for Hundreds of. Genetics. 2018; 208:937–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kasper D.M., Wang G., Gardner K.E., Johnstone T.G., Reinke V.. The C. elegans SNAPc component SNPC-4 coats piRNA domains and is globally required for piRNA abundance. Dev. Cell. 2014; 31:145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harrison M.M., Ceol C.J., Lu X., Horvitz H.R.. Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex. Proc. Natl. Acad. Sci. USA. 2006; 103:16782–16787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sapetschnig A., Sarkies P., Lehrbach N.J., Miska E.A.. Tertiary siRNAs mediate paramutation in C. elegans. PLoS Genet. 2015; 11:e1005078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Davis G.M., Tu S., Anderson J.W., Colson R.N., Gunzburg M.J., Francisco M.A., Ray D., Shrubsole S.P., Sobotka J.A., Seroussi U.et al.. The TRIM-NHL protein NHL-2 is a co-factor in the nuclear and somatic RNAi pathways in. eLife. 2018; 7:e35478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Keall R., Whitelaw S., Pettitt J., Müller B.. Histone gene expression and histone mRNA 3′ end structure in Caenorhabditis elegans. BMC Mol. Biol. 2007; 8:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mahajan-Miklos S., Tan M.W., Rahme L.G., Ausubel F.M.. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell. 1999; 96:47–56. [DOI] [PubMed] [Google Scholar]
- 75.Xu F., Feng X., Chen X., Weng C., Yan Q., Xu T., Hong M., Guang S.. A Cytoplasmic Argonaute Protein Promotes the Inheritance of RNAi. Cell Rep. 2018; 23:2482–2494. [DOI] [PubMed] [Google Scholar]
- 76.Ishidate T., Ozturk A.R., Durning D.J., Sharma R., Shen E.Z., Chen H., Seth M., Shirayama M., Mello C.C.. ZNFX-1 Functions within Perinuclear Nuage to Balance Epigenetic Signals. Mol. Cell. 2018; 70:639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Wolfswinkel J.C., Claycomb J.M., Batista P.J., Mello C.C., Berezikov E., Ketting R.F.. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell. 2009; 139:135–148. [DOI] [PubMed] [Google Scholar]
- 78.Sundby A.E., Molnar R.I., Claycomb J.M.. Connecting the Dots: Linking Caenorhabditis elegans Small RNA Pathways and Germ Granules. Trends Cell Biol. 2021; 31:387–401. [DOI] [PubMed] [Google Scholar]
- 79.Wan G., Bajaj L., Fields B., Dodson A.E., Pagano D., Fei Y., Kennedy S.. ZSP-1 is a Z granule surface protein required for Z granule fluidity and germline immortality in Caenorhabditis elegans. EMBO J. 2021; 40:e105612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Spichal M., Heestand B., Kretovich Billmyre K., Frenk S., Mello C.C., Ahmed S.. Germ granule dysfunction is a hallmark and mirror of Piwi mutant sterility. Nat. Commun. 2021; 12:1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.White-Cooper H., Bausek N.. Evolution and spermatogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010; 365:1465–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ellis R., Schedl T.. Sex determination in the germ line. WormBook. 2007; 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Scott A.L., Dinman J., Sussman D.J., Ward S.. Major sperm protein and actin genes in free-living and parasitic nematodes. Parasitology. 1989; 98:471–478. [DOI] [PubMed] [Google Scholar]
- 84.Smith H.E.Nematode sperm motility. WormBook. 2014; 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tu S., Wu M.Z., Wang J., Cutter A.D., Weng Z., Claycomb J.M.. Comparative functional characterization of the CSR-1 22G-RNA pathway in Caenorhabditis nematodes. Nucleic Acids Res. 2015; 43:208–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martinez M.A.Q., Kinney B.A., Medwig-Kinney T.N., Ashley G., Ragle J.M., Johnson L., Aguilera J., Hammell C.M., Ward J.D., Matus D.Q.. Rapid degradation of Caenorhabditis elegans proteins at single-cell resolution with a synthetic auxin. G3 (Bethesda). 2020; 10:267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.L’Hernault S.W.The genetics and cell biology of spermatogenesis in the nematode C. elegans. Mol. Cell. Endocrinol. 2009; 306:59–65. [DOI] [PubMed] [Google Scholar]
- 88.Placentino M., de Jesus Domingues A.M., Schreier J., Dietz S., Hellmann S., de Albuquerque B.F., Butter F., Ketting R.F.. Intrinsically disordered protein PID-2 modulates Z granules and is required for heritable piRNA-induced silencing in the Caenorhabditis elegans embryo. EMBO J. 2021; 40:e105280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang C., Montgomery T.A., Fischer S.E., Garcia S.M., Riedel C.G., Fahlgren N., Sullivan C.M., Carrington J.C., Ruvkun G.. The Caenorhabditis elegans RDE-10/RDE-11 complex regulates RNAi by promoting secondary siRNA amplification. Curr. Biol. 2012; 22:881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sakaguchi A., Sarkies P., Simon M., Doebley A.L., Goldstein L.D., Hedges A., Ikegami K., Alvares S.M., Yang L., LaRocque J.R.et al.. Caenorhabditis elegans RSD-2 and RSD-6 promote germ cell immortality by maintaining small interfering RNA populations. Proc. Natl. Acad. Sci. USA. 2014; 111:E4323–E4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.She X., Xu X., Fedotov A., Kelly W.G., Maine E.M.. Regulation of heterochromatin assembly on unpaired chromosomes during Caenorhabditis elegans meiosis by components of a small RNA-mediated pathway. PLoS Genet. 2009; 5:e1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Grishok A., Sinskey J.L., Sharp P.A.. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev. 2005; 19:683–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Barucci G., Cornes E., Singh M., Li B., Ugolini M., Samolygo A., Didier C., Dingli F., Loew D., Quarato P.et al.. Small-RNA-mediated transgenerational silencing of histone genes impairs fertility in piRNA mutants. Nat. Cell Biol. 2020; 22:235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shi Z., Montgomery T.A., Qi Y., Ruvkun G.. High-throughput sequencing reveals extraordinary fluidity of miRNA, piRNA, and siRNA pathways in nematodes. Genome Res. 2013; 23:497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wolke U., Jezuit E.A., Priess J.R.. Actin-dependent cytoplasmic streaming in C. elegans oogenesis. Development. 2007; 134:2227–2236. [DOI] [PubMed] [Google Scholar]
- 96.Strome S., Wood W.B.. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell. 1983; 35:15–25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Small RNA-seq and poly(A) selected mRNA-seq data generated in this study are available under the accession number GEO: GSE154678. Germline constitutive, oogenic and spermatogenic gene lists were defined as in (29) available from GEO under accession number GSE57109. RdRP mutant small RNA data are from (71) [GSE66344]. alg-3, alg-4 mutant sRNA data are from (23) [GSE49672]. WAGO-1 IP/sRNA-seq data was obtained from (15) available under accession number [GSE18231]. The CSR-1 IP/sRNA-seq data from males was obtained from (14), accession number [GSE49672], while the hermaphrodite data was obtained from (4), accession number [GSE18167]. The custom scripts used to generate the processed data are available at www.github.com/ClaycombLab/Charlesworth_2020.