Abstract
Histone recognition constitutes a key epigenetic mechanism in gene regulation and cell fate decision. PHF14 is a conserved multi-PHD finger protein that has been implicated in organ development, tissue homeostasis, and tumorigenesis. Here we show that PHF14 reads unmodified histone H3(1–34) through an integrated PHD1-ZnK-PHD2 cassette (PHF14PZP). Our binding, structural and HDX-MS analyses revealed a feature of bipartite recognition, in which PHF14PZP utilizes two distinct surfaces for concurrent yet separable engagement of segments H3-Nter (e.g. 1–15) and H3-middle (e.g. 14–34) of H3(1–34). Structural studies revealed a novel histone H3 binding mode by PHD1 of PHF14PZP, in which a PHF14-unique insertion loop but not the core β-strands of a PHD finger dominates H3K4 readout. Binding studies showed that H3-PHF14PZP engagement is sensitive to modifications occurring to H3 R2, T3, K4, R8 and K23 but not K9 and K27, suggesting multiple layers of modification switch. Collectively, our work calls attention to PHF14 as a ‘ground’ state (unmodified) H3(1–34) reader that can be negatively regulated by active marks, thus providing molecular insights into a repressive function of PHF14 and its derepression.
INTRODUCTION
Histone recognition is a key epigenetic mechanism for gene expression, chromatin organization, cell division and development. Plant homeodomain finger protein 14 (PHF14) is a multi-PHD finger protein that is conserved from worm to human. PHF14 is about 500–900 residues and contains 3–4 PHD finger modules with implicated functions in histone readout. Previous studies revealed roles of PHF14 in development, tissue homeostasis, and tumorigenesis. For example, PHF14-null mice died shortly after birth with severe developmental defects of multiple organs (1,2). PHF14 has been proposed as a negative regulator of platelet-derived growth factor receptor-α (PDGFRα), p14ARF, p16INK4a, p15INK4b and E-cadherin, and thus promotes cell cycle progression and cancer cell invasion (1–8). Despite these findings, the molecular functions of PHF14, especially its PHD finger modules, remain obscure.
PHF14 features a conserved PZP domain that is integrated by two flanking PHD fingers and a zinc finger knuckle (ZnK). The PZP domain has been identified in JADE1/2/3 (gene for apoptosis and differentiation in epithelia 1/2/3), BRPF1/2/3 (bromodomain and plant homeodomain finger containing proteins1/2/3), AF10/17 (ALL1-fused gene from chromosome 10/17), KDM4A/B/C (Lysine demethylase 4 A/B/C) and PHF14 (9). Among them, JADEPZP and BRPFPZP act as readers of unmodified histone H3K4 as well as nucleosomal DNA to regulate the acetyltransferase activity of human HBO1 (histone acetyltransferase binding to hORC1), MOZ (monocytic leukemia zinc-finger protein), and MORF (MOZ-related factor) (10–13); AF10PZP and AF17PZP recognize unmodified H3K27 rather than H3K27me3, and further recruit the DOT1L (histone H3K79 methyltransferase) complex to promote H3K79 methylation (14). Thus far, the histone binding activities of PHF14 and KDM4 subfamily PZP domains have not been characterized.
Here, we characterized PHF14 as a reader of unmodified histone H3(1–34) through its PZP domain. Our binding, structural, and HDX-MS analyses revealed a bipartite engagement feature between PHF14PZP and H3(1–34), in which two distinct reader surfaces are exploited for concurrent yet separable recognition of different H3(1–34) segments. Remarkably, our structural studies revealed a previously uncharacterized H3K4 recognition mode by PHF14PZP. Histone H3 engagement and modification crosstalk were also examined through binding and mutagenesis studies, which revealed that PHF14PZP is sensitive to active marks such as H3K4me3, H3R8me2a, and H3K23ac but not repressive marks such as H3K27me3. This distinguishes PHF14PZP from AF10/AF17 PZP domains that are sensitive to H3K27me3 and BRPF1/2/3 PZP domains that are insensitive to H3R8me2a. Thus, our work revealed novel histone binding activity of the PZP family domains. Collectively, our work establishes the molecular function of PHF14PZP, which paves the way for future functional studies of PHF14 in development and disease.
MATERIALS AND METHODS
Plasmids and peptides
Zebrafish PHF14 cDNA was amplified from genomic cDNA and constructed into pSUMOH10 (a modified PET28b vector with an N-terminal 10×His-SUMO tag) vectors. H3(1–13)-PHF14PZP was generated by fusing histone H3(1–13) to the N-terminus of PHF14PZP with a ‘SGSGS’ linker in the frame of pSUMOH10. BRPF1 and AF10 cDNAs were amplified from genomic cDNA and constructed into pGood6P (a modified pGEX-6P-1 vector with altered multiple cloning sites) and pSUMOH10 vectors, respectively. Point mutants were generated according to the ‘QuikChange’ site-directed mutagenesis strategy. Histone peptides were synthesized at Beijing SciLight Biotechnology Ltd. Co. and the detailed sequence information is listed in Supplementary Table S1.
Protein expression and purification
Zebrafish PHF14PZP (278–487), H3(1–13)-PHF14PZP (278–487) and human PHF14PHD3 (723–793), PHF14PHD4 (863–920) were overexpressed with an N-terminal 10×His-SUMO tag in Escherichia coli strain BL21 (DE3). The expression was induced by 0.2 mM isopropyl β-d-thiogalactoside (IPTG) in the presence of 0.2 mM ZnCl2 at 18°C in LB medium. After overnight induction, cells were harvested by centrifugation and re-suspended in lysis buffer: 500 mM NaCl, 20 mM Tris–HCl, pH 7.5 and 2 mM β-mercaptoethanol. The cells were lysed by a UH-12 high-pressure homogenizer (Union-Biotech), then the lysates were cleared by centrifugation at 16,770 × g. The protein was affinity purified by a HisTrap column with the peak eluted at an imidazole concentration of 150 mM. After SUMO tag cleavage by ULP1, the His-SUMO tag was removed by the HisTrap column. Target proteins in the flow-through were pooled and purified by HiTrap Q columns, then further polished by the Superdex 75 gel filtration columns under an elution buffer containing 100 mM NaCl and 20 mM HEPES-Na, pH 7.5. The peak fractions were pooled and concentrated to 5–14 mg/ml, and stored at −80°C for future use. All PHF14PZP mutants were purified using essentially the same procedures described above. BRPF1PZP (266–454) and AF10PZP (1–208) were prepared according to previous reports (11,14).
Isothermal titration calorimetry (ITC) assays
For ITC measurements, synthetic histone peptides (>95% purity) and recombinant proteins were all prepared under the same titration buffer containing 100 mM NaCl, 20 mM HEPES-Na and pH 7.5. Protein concentration was measured based on UV 280 nm absorption. Peptide concentrations were determined by weighing in large quantities. After weighing and quantification, each peptide was dissolved in Milli-Q water, aliquoted and freeze-dried for future use. ITC experiments were performed at 25°C using a MicroCal PEAQ-ITC instrument (Malvern Panalytical). Usually, histone H3 peptides at 1.0 mM were titrated into proteins at 0.1 mM, and each ITC assay consisted of 17 successive injections, with 0.4 μl used for the first and 2.4 μl used for the rest. The acquired calorimetric titration data were analysed with Origin version 7.0 software (OriginLab) using the ‘One Set of Binding Sites’ fitting model. ITC experiments were repeated at least twice, and all the thermodynamic data are summarized in Supplementary Table S2.
Crystallization, data collection and structural determination
Crystallization was performed by the sitting-drop vapor diffusion method at 18°C by mixing 0.2 μl protein solution with 0.2 μl reservoir solution for the primary screening using the Mosquito® crystal. The volume of protein and reservoir solution were adjusted to 2 μl during optimization. Free state PHF14PZP (12 mg/ml) was crystallized in reservoir solution containing 25% PEG3350, 0.2 M MgCl2, 0.1 M Bis–Tris, and pH 6.5. The crystals of PHF14PZP with H3(1–25) complex (1:1 ratio, 12 mg/ml) were grown in the condition containing 0.1 M carboxylic acids, 0.1 M buffer system 2, pH 7.5 (Morpheus® 1–46, Molecular Dimensions). The H3(1–13)-PHF14PZP fusion protein (12 mg/ml) was crystalized in reservoir solution: 18% PEG4000, 0.1 M HEPES-Na, pH 7.5 and 12% isopropanol buffer.
For data collection, crystals were flash frozen in liquid nitrogen under cryoprotectant solutions. For the H3(1–25)-PHF14PZP complex crystal, the reservoir solution can directly serve as cryoprotectant; for free and fusion crystals, the cryoprotectant is composed of reservoir solution supplemented with 10% (w/v) glycerol. Diffraction data were collected at wavelength 1.2818 or 0.9793 Å on beam lines BL17U or BL19U at the National Facility for Protein Science in Shanghai. All data were indexed, integrated, and merged using the HKL2000 software package (HKL Research Inc.). The structure of free PHF14PZP was solved using the zinc single-wavelength anomalous dispersion (Zn-SAD) method. The complex structures of PHF14PZP and H3(1–13)-PHF14PZP were solved by molecular replacement with the Molrep program (15) using the free PHF14PZP structure as the search model. The structures were refined using PHENIX (16) with iterative manual model building using COOT. Detailed structural collection and refinement statistics are shown in Supplementary Table S3.
Hydrogen-deuterium exchange mass spectrometry (HDX-MS) analysis
Free and H3-bound PHF14PZP proteins were prepared at 100 μM concentration in buffer containing 100 mM NaCl and 20 mM HEPES-Na, at pH 7.5. To initiate deuterium labelling, 5 μl of each 100 μM protein solution was diluted with 45 μl of labelling buffer (100 mM NaCl, 20 mM HEPES-Na, 99% D2O, pH 7.5) at room temperature. The labelling reaction was quenched by adding 50 μl of ice-cold quench buffer (formic acid in water solution at pH 1.3, 100% H2O) after 5 min. Then 5 μl pepsin (5 μM) was added into the sample for protein digestion for 5 min. Then the samples were injected into a Waters NanoACQUITY UPLC system for mass spectrometry analyses. The Q Exactive mass spectrometer was operated in the data-dependent acquisition mode using Xcalibur 2.0.0.0 software (Thermo Fisher Scientific), acquiring over an m/z of 350–2000. Peptides were identified using Proteome Discoverer (version PD1.4, Thermo Fisher Scientific), and HDX-MS data were processed by HD Examiner from Thermo Fisher Scientific.
Thermal shift assay (TSA)
For TSA assays, 2 mg/ml PHF14PZP and H3(1–13)-PHF14PZP were mixed with 5× Sypro Orange (Invitrogen) in 25 μl buffer (100 mM NaCl, 20 mM HEPES-Na, pH 7.5). Samples were heated from 25°C to 90°C at a rate of 0.5°C per minute. Assays were performed with a CFX96 real-time PCR instrument (Bio-Rad). Fluorescence signal changes were recorded and analysed by the software CFX-Manager (Bio-Rad).
Co-immunoprecipitation (Co-IP) assay
About 1×107 HEK293T cells were transiently and separately transfected with 3×Flag-PHF14(1–914)-WT (wild type), 3×Flag-PHF14-ΔPZP (PZP domain was deleted), 3×Flag-PHF14-Δloop (insertion loop was replaced by ‘SS’ linker), and 3×Flag-PHF14-I386M (I386 was mutated to M) plasmids. At 72 h post-transfection, cells were washed with PBS and then lysed in 1.5 ml cold lysis buffer (Beyotime P0013 buffer with EDTA-free Selleck protease inhibitor). The cell lysates were then pre-cleared with pre-washed protein G agarose beads (Invitrogen,15920010) for 30 min. A total of 350 μl pre-clear lysate was incubated with 5 μl anti-H3 antibody (CST, 4620S) or anti-Flag antibody (Sigma, F1084) and 30 μl protein G agarose beads, or 5 μl corresponding IgG (CST, 3900S) and 30 μl protein G agarose beads overnight. Beads were then washed three times with lysis buffer, followed by boiling in sample buffer then loaded for gel electrophoresis and immunoblot. Anti-H3 and anti-Flag antibody used in immunoblot were obtained from ABclonal (A2348) and Sigma (F1804), respectively. All uncropped images can be found in Supplementary Figure S6D.
RESULTS
The PZP domain of PHF14 binds unmodified histone H3(1–34)
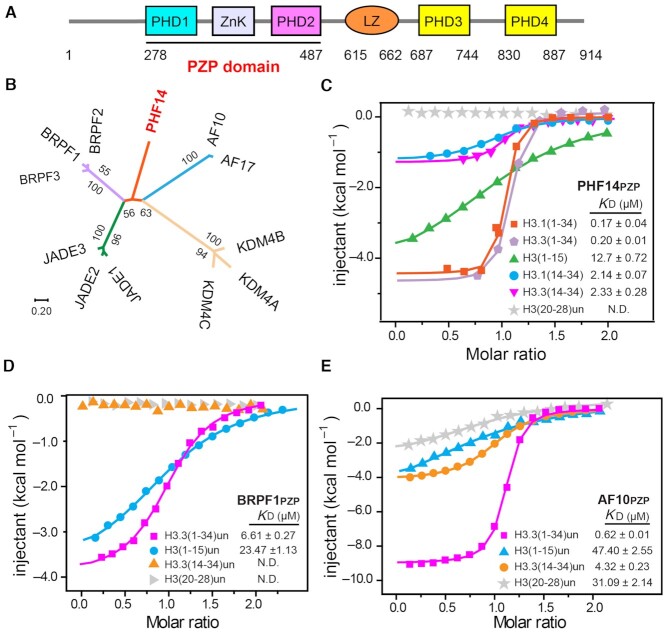
Full length PHF14 is composed of an N-terminal PZP domain (PHD1-ZnK-PHD2), a leucine zipper, and two other PHD fingers (PHD3 and PHD4) from its N- to C-termini (Figure 1A). The PZP domain is a signature module among the PHF14 orthologues from worm to human (Supplementary Figure S1A). Phylogenetic analysis showed that PHF14PZP stands out as an evolutionarily unique subfamily member among its paralogs (Figure 1B). The PZP domain of BRPF1 was reported to recognize unmodified histone H3(1–15) (10,11), while the AF10/AF17 subfamily PZPs were shown to recognize unmodified histone H3(15–34) (14). The conservation and divergence of PZP family members raise an interesting question regarding the histone binding activity of PHF14PZP.
Figure 1.
PHF14PZP is an unmodified histone H3(1–34) reader. (A) Schematic diagram showing the domain architecture of human PHF14. (B) PZP motif based-evolutionary analyses of PHF14 homologs using the Neighbor-Joining method. The evolutionary distances were computed using the Poisson correction method and are drawn to scale in the diagram. The numbers next to each node were given as percentages and represent bootstrap support for the node (500 replicates). Evolutionary analysis was conducted in MEGA7 (37–40). (C–E) ITC fitting curves of zebrafish PHF14PZP (C), BRPF1PZP (D) and AF10PZP (E) titrated with histone H3 peptides in different frames. N.D. = not detected.
To address this question, we first performed expression screening of PHF14PZP orthologs including species such as human, zebrafish, and fly, and were able to purify the zebrafish PHF14PZP at high quality and yield. Next, we performed isothermal titration calorimetry (ITC) assays using different histone H3 peptides within the range of residues 1–34 (sequences derived from H3.1 and H3.3). We observed that PHF14PZP binds to unmodified histone H3.1(1–34) and H3.3(1–34) peptide at affinities of 0.17 and 0.20 μM, respectively (Figure 1C). The similar binding profiles suggest that H3-PHF14PZP engagement is insensitive to type of histone H3 variants despite their residue differences at position 31 (Ser31 for H3.3 and Ala31 for H3.1). PHF14PZP displayed micromolar binding to H3(1–15), H3.1(14–34) and H3.3(14–34) at KD values of 12.7, 2.14 and 2.33 μM, respectively (Figure 1C). Therefore, PHF14PZP displays a histone H3 binding feature that combines H3-Nter (e.g. 1–15) and H3-middle (e.g. 14–34) recognition activities into an event of long H3 N-terminal tail engagement. Moreover, the binding affinity of PHF14PZP toward H3(1–34) is ∼14–75-fold higher than those toward H3-Nter or H3-middle, suggesting a synergistic effect for H3(1–34) readout by PHF14PZP. The PHF14 contains one or two more PHD fingers at the C-terminal region (PHD3 and PHD4) depending on the mode of alternative splicing (Supplementary Figure S1B). Sequence alignment suggested that both PHD3 and PHD4 lack key residues responsible for histone binding (Supplementary Figure S1C). This is confirmed by ITC assays using unmodified H3(1–15) and H3(1–15) K4me3 peptides (Supplementary Figure S1D). Collectively, these results establish that PHF14 is an unmodified long histone H3 N-terminal tail reader through its PZP domain.
Unmodified histone H3(1-34) readout by other PZP domains
BRPF1PZP recognizes histone H3(1–15) but not H3(15–34) according to heteronuclear single-quantum coherence (HSQC) nuclear magnetic resonance (NMR) experiments (10,11), while AF10PZP was shown to recognize histone H3(15–34) (ITC assay) but not H3(1–15) (pull down assay) (14). To uniformly compare H3(1–34) binding abilities among PHF14PZP, AF10PZP and BRPF1PZP, we purified AF10PZP and BRPF1PZP and further performed ITC assays with H3 peptides of interest. Consistent with previous reports, BRPF1PZP did not bind to H3(14–34). However, the binding affinity of BRPF1PZP toward H3(1–34) (KD = 6.61 μM) was ∼3.5-fold higher than towards H3(1–15) (KD = 23.47 μM) (Figure 1D), suggesting an assistant role of H3(14–34) during BRPF1PZP-H3(1–15) binding. As for AF10PZP-H3 binding, the KD value of AF10PZP toward H3(14–34) was 4.32 μM, which accorded closely with previous work (14). AF10PZP could also recognize H3(1–15) with KD values of 47.40 μM by our ITC assays. AF10PZP recognized H3(1–34) with much stronger binding affinities (KD = 0.62 μM) compared to its ability to recognize H3-Ner or H3-middle, which behaved similarly to PHF14PZP in general (Figure 1E). Unlike AF10PZP, further analysis showed that PHF14PZP did not bind to a shorter region of H3-middle, H3(20–28), suggesting divergent H3-middle binding properties between AF10PZP and PHF14PZP. It appeared that the PZP domains of BPPF1, AF10 and PHF14 family members are bona fide H3(1–34) readers that share a common unmodified H3-Nter binding activity but diverge on H3-middle recognition, highlighting concordant but twisted histone readout features among the PZP family proteins for elaborate molecular regulation.
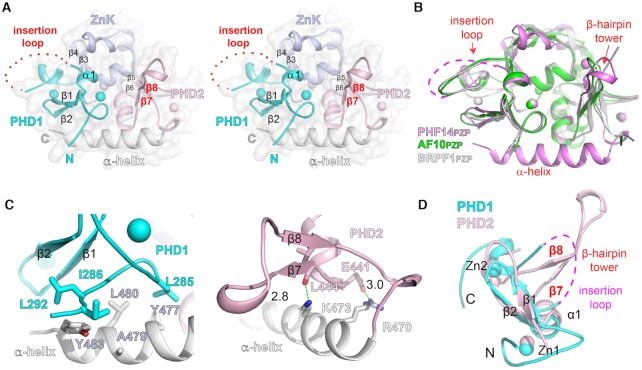
Crystal structure of the PZP domain of PHF14 in free state
To explore the molecular function of PHF14PZP, we first crystallized the PZP domain (aa. 278–487) of zebrafish PHF14 and solved its structure in a free state at 1.85 Å (Supplementary Table S3; PDB, 7D86). Most residues were modelled based on the electron density map except for 278–283, 486–487, and an insertion loop (aa 320–336) within PHD1. PHF14PZP is a highly compact module that integrates the ‘PHD1-ZnK-PHD2’ cassette and a C-terminal α-helix through extensive hydrophobic or polar interactions among the subdomains (Figure 2A and Supplementary Figure S2A).
Figure 2.
Crystal structure of PHF14PZP in a peptide-free state. (A) Wall-eye stereo view of the overall structure of free PHF14PZP. PHD1 (cyan), ZnK (light blue), PHD2 (light pink), and α-helix (white) are shown as ribbons. Zinc ions are depicted as spheres. Red dotted lines indicate the invisible insertion loop. Key secondary structural elements are labeled. (B) Structural alignment of PHF14PZP (magenta), AF10PZP (green) and BRPF1PZP (white). (C) Interactions between the C-terminal α-helix with PHD1 (left) and PHD2 (right). Key residues are shown as sticks. Hydrogen bonds are shown as green dashes with distances labeled in Å. (D) Structural alignment of PHF14PHD1 (cyan) and PHF14PHD2 (light pink). Note that the insertion loop is topologically equivalent to the β-hairpin tower.
The core structure of PHF14PZP clearly superimposes with AF10 (14) and BRPF1 (10) PZP domains with calculated Cα RMS deviations of 0.81 and 0.55 Å, respectively (Figure 2B). Despite this, structure-based sequence alignment showed that PHF14PZP is characteristic of an insertion loop within PHD1, a β-hairpin tower within PHD2, and an extended α-helix at the C-terminus (Figure 2B and Supplementary Figure S2B). In particular, the long C-terminal α-helix of PHF14PZP extends from PHD2 to PHD1 and stabilizes a compact PZP module through hydrophobic interactions with PHD1 (Figure 2C, left) and hydrogen bonding interactions with PHD2 (Figure 2C, right). The long C-terminal α-helix is unique to PHF14PZP and key residues involved in the PHD1/2 interaction are not conserved in other PZP domains (Supplementary Figure S2B).
It is worth noting that the insertion loop of PHD1 and the β-hairpin tower of PHD2 are topologically equivalent over a PHD finger scaffold. Both elements are variable insertions between the second core β-strand (β2) and the last zinc-coordinating ‘CXXH’ motif, which is spatially next to the first core β-strand (β1) of a canonical PHD finger (Figure 2D). Sequences of these insertions are quite divergent among PHD fingers, thus laying the molecular foundation for functional diversity of a PHD finger or its integrated form (Supplementary Figure S2B).
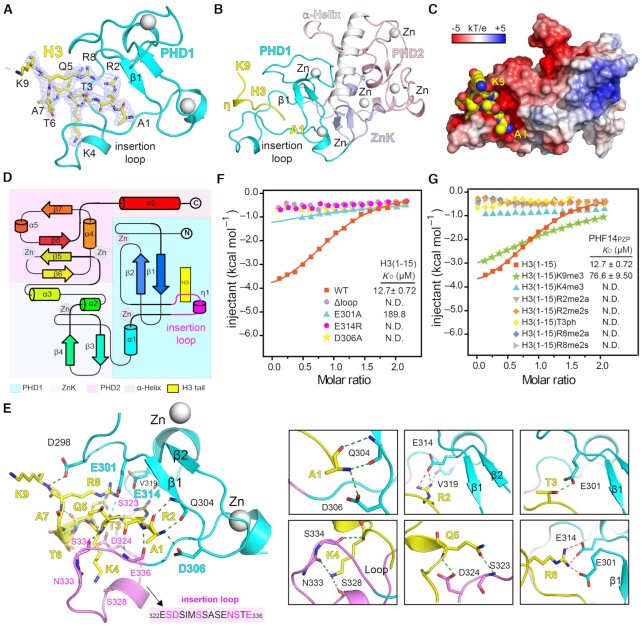
Structural basis for H3(1–9) recognition by PHF14PZP
To explore the molecular basis for histone H3 binding by PHF14PZP, we performed extensive crystallization trials of the H3(1–34)-PHF14PZP complex but they failed to work. Then, we turned to a shorter peptide H3(1–25), which displayed decent binding with PHF14PZP at an affinity of 2.20 μM (Supplementary Figure S3A). The H3(1–25)-PHF14PZP complex was successfully crystallized, and the corresponding crystal structure was solved at 2.1 Å resolution (Supplementary Table S3; PDB,7D87).
Despite using H3(1–25) for crystallization, we were only able to model H3(1–9) according to the electron densities (Figure 3A). The basic histone H3(1–9) peptide adopts a ‘η-loop’ (η, 310 helix) conformation (Figure 3B), and binds intimately to an acidic cleft formed between β1 and the insertion loop of PHD1 (Figure 3C and D). Upon complex formation, the invisible insertion loop becomes ordered with traceable electron densities (Supplementary Figure S3B, left). The reordered insertion loop contains a short 310 helix (η1) in the middle, and its conformation is stabilized by multiple intra-chain hydrogen bonding interactions (Supplementary Figure S3B, right).
Figure 3.
Molecular details for H3(1–9) readout by PHF14PZP. (A) 2Fo−Fc omit map (blue meshes) around the H3 peptide (yellow sticks) is contoured at the 0.7σ level. PHD1 finger is shown as cyan ribbon. (B) Overall structure of the H3(1–25)-PHF14PZP complex. PHD1 finger (cyan), ZnK (light blue), PHD2 finger (light pink), and H3 peptide (yellow) are shown as ribbons. Zinc ions are depicted as white spheres. (C) Electrostatic surface view of PHF14PZP bound to H3 peptides. Electrostatic potential is shown as a spectrum ranging from − 5 kT/e (red) to + 5 kT/e (blue). (D) Topology diagram of the H3(1–9)-PHF14PZP complex. Helices and strands are labelled and rainbow-colored from the N- (blue) to C- (red) termini. H3 is depicted in yellow and the insertion loop is highlighted in magenta. (E) Binding details of PHF14PZP with H3 peptide. The PHD1 finger of PHF14PZP is shown as a cyan and magenta (loop) ribbon. The residues of H3 peptide are depicted as yellow sticks. Key residues of PHF14PZP are depicted as purple or cyan sticks. Green dashes and red dashes represent the hydrogen bonds and ionic bonds, respectively. (F) ITC fitting curves of PHF14PZP mutants titrated with the H3(1–15) peptide. N.D. = not detected. (G) ITC fitting curves of PHF14PZP titrated with the H3(1–15) peptide bearing different modifications. N.D. = not detected.
Extensive hydrogen bonding and electrostatic interactions ensure a stable association of PHF14PZP with H3(1–9) (Figure 3E; Supplementary Figure S3C). In detail, the backbone of H3(1–9) is anchored by eight hydrogen bonding interactions, involving residue pairs H3A1:Q304, H3A1:D306, H3R2:E336, H3K4:S334, H3Q5:D324, and H3K9:D298 (Figure 3E, left). As for histone sidechain registration, H3R2 interacts with residues E314 and V319 between the β2 strand and the insertion loop; H3T3 interacts with residue E301 before the β1 strand; H3K4 interacts with residues S328 and N333 of the insertion loop; H3Q5 interacts with residue S323 of the insertion loop; and H3R8 interacts with residues E301 and E314 (Figure 3E, right). Three acidic residues, D306, E314, and E301 respectively contribute to H3 N-terminal α-amine, and H3R2 and H3R8 recognition through electrostatic interactions. The positively charged H3K4 side chain is largely recognized by main chain–mediated hydrogen bonding interactions. By contrast, the H3K9 side chain is largely solvent exposed (Figure 3E, left).
Mutagenesis and modification crosstalk studies of H3(1–15)-PHF14PZP interaction
To validate the above observations, we first generated point mutants of PHF14PZP around the H3 binding surface: E301A, D306A, and E314R. ITC experiments showed that E314R and D306A abolished binding and E301A led to ∼28-fold decreased affinity. The dramatic affinity drop by single point mutation suggests a highly coordinated binding feature between PHF14PZP and H3(1–15). We also generated an insertion loop deletion mutant (Δloop) by replacing a long loop (322ESDSIMSSASENST336) with an ‘SS’ linker. As expected, this mutant displayed no binding toward H3(1–15) peptide (Figure 3F), highlighting the importance of this insertion loop that is unique to PHF14PZP among its paralogues (Supplementary Figure S2B). This insertion loop is conserved from zebrafish to human but not in C. elegans and fly, suggesting an evolutionarily acquired histone reader activity of PHF14PZP in vertebrates (Supplementary Figure S1A).
The very N-terminal tail of H3 is subject to various covalent modifications, including methylations of histone R2, K4, R8 and K9 as well as phosphorylation of T3, laying the molecular foundation for histone modification crosstalk (17). Next, we examined binding of PHF14PZP with modified H3(1–15) peptides (Figure 3G). ITC assays revealed that methylations of H3 R2, K4, R8 and phosphorylation of H3T3 abolished binding; the binding KD values dropped from 12.7 to 76.6 μM for H3K9me3. Unmodified H3 R2, K4 and R8 are buried in the complex, which explains why their modifications disrupted binding (Supplementary Figure S3D). By contrast, H3K9 is rather solvent exposed, which may account for a mild tolerance of H3K9me3 by PHF14PZP. H3T3 is next to acidic residue E301, which may repel T3ph mark from binding due to charge repulsion. Collectively, these results are consistent with the fact that unmodified H3 R2, T3, K4 and R8 are directly involved in binding, and suggest a mechanism of modification switch in downregulating H3 readout of PHF14.
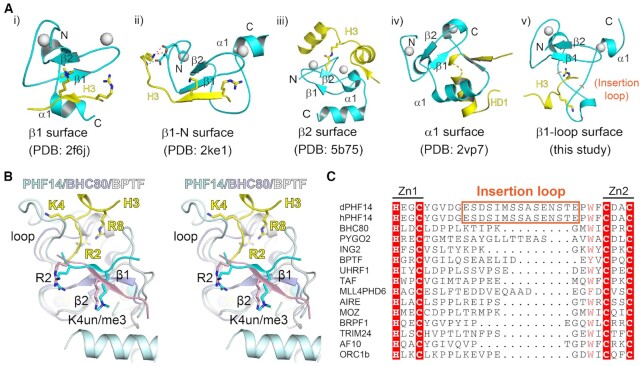
Comparison of PHF14 PHD1 finger with other histone binding PHD fingers
The PHD finger is a functionally diverse module and may achieve different ligand-binding activities by exploiting distinct functional surfaces over a common scaffold (18). These surfaces have been divided into four groups: (i) the β1 surface (19); (ii) β1-N surface (20); (iii) β2-surface (21) and (iv) α1-surface (22), and they can recognize ligands such as H3(1–6) K4me3, an extended H3(1–10), H3K14cr (crotonylation) and BCL9, respectively (Figure 4Ai–iv).
Figure 4.
A novel recognition model for H3 peptide readout by PHF14PZP. (A) A summary of different recognition modes of PHD fingers. PHD fingers and their ligands are depicted by cyan and yellow ribbons, respectively. Zinc ions are shown as white spheres and the corresponding secondary structure are labelled. (B) Structural alignment of H3 (yellow)-PHF14 (pale cyan), H3 (light pink)-BHC80 (blue white) and H3 (cyan)-BPTF (white) complexes in wall-eye stereo view. H3 R2, K4, and R8 are depicted as sticks. (C) Sequence alignment of different PHD finger proteins: BHC80 (2PUY), PYGO (2VP7), ING2 (2G6Q), BPTF (2F6J), UHRF1 (3SOX), TAF (5WXG), MLL4PHD6 (6O7G), AIRE (2KFT), MOZ (5B78), BRPF1 (6U04), TRIM24 (3O37), AF10 (5DAH), ORC1b (5HH7).
The H3 recognition mode by PHF14PZP does not belong to any of the above four groups, which we named the ‘β1-loop surface’ mode (Figure 4Av). In the classical ‘β1-surface’ mode, the H3 tail forms an anti-parallel β-sheet with the β1 stand and the R2 and K4 residues extend towards the β2 strand in cis (Figure 4B). In the case of PHF14PZP, the insertion loop next to the β1 strand dominates H3 recognition, and H3K4 flips away from its classical binding site and points in an opposite direction (Figure 4B). To adapt to this, the histone H3(1–9) tail takes on an ‘η-loop’ structure to ensure a concurrent readout of R2, K4 and R8 of histone H3.
Sequence analysis revealed that despite being an unmodified H3(1–9) reader, PHF14PZP is quite divergent from classical H3K4me3 or unmodified H3K4 readers. Key aromatic or acidic residues that are responsible for H3K4me3 or unmodified H3K4 recognition are not conserved in PHF14PZP (Supplementary Figure S4A and B). Moreover, the insertion loop of PHF14PZP is evolutionarily conserved among its vertebrate orthologues but not its paralogues, suggesting a unique function of PHF14 family members (Figure 4C; Supplementary Figures S1A and S2B). Despite being a close paralog of PHF14PZP, the BRPF family PZPs adopt a classical ‘β1 surface’ mode for unmodified H3-Nter recognition, in which unmodified H3 R2 and K4 but not R8 dominate H3-Nter recognition (Supplementary Figure S4C) (10,11,23). Accordingly, although both PHF14PZP and BRPF1PZP are sensitive to H3K4me3, BRPF1PZP can tolerate another active mark, H3R8me2a, but PHF14PZP is strictly sensitive to all types of H3R8 methylations.
Crystal structure of H3(1–13)-PHF14PZP fusion protein
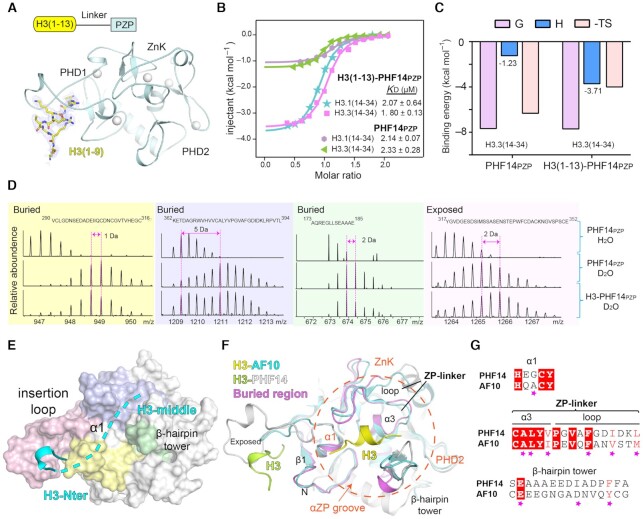
PHF14PZP recognizes histone H3(14–34) (Figure 1C), however, our current crystallographic efforts failed to elucidate the underlying molecular basis. We thus designed a chimeric construct, H3(1–13)-PHF14PZP, by fusing H3(1–13) to PHF14PZP (aa 278–487) from the N-terminus through an ‘SGSGS’ linker and tried to crystallize the complex of H3(1–13)-PHF14PZP and H3(14–34). The resultant chimeric protein, H3(1–13)-PHF14PZP, was expressed with high yield. As reflected by thermal shift assay, the H3(1–13)-PHF14PZP displayed better thermal stability than the free state PHF14PZP with a positive Tm shift of 3.2°C (Supplementary Figure S4D). Next, we successfully crystallized H3(1–13)-PHF14PZP and solved its structure at 1.95 Å resolution (Supplementary Table S3; PDB,7D8A). In the crystal structure, residues H3(1–9) were modelled according to the electron density map; meanwhile, residues H3(10–13), the ‘SGSGS’ linker, and N-terminal residues 278–284 of PHF14PZP were disordered (Figure 5A). The overall structure of H3(1–13)-PHF14PZP and its H3(1–9) engagement mode are essentially the same as those of the H3(1–25)-PHF14PZP complex. We calculated a Cα RMSD of 0.117 Å between the two structures (Supplementary Figure S4E), which validates the molecular design of the chimeric protein.
Figure 5.
Molecular basis for H3(14–34) recognition by PHF14PZP and HDX-MS analyses of H3(1–34) recognition by PHF14PZP. (A) Overall structure of H3(1–13)-PHF14PZP. PHF14PZP is shown as a light blue ribbon. Histone H3 peptide is shown as yellow sticks. Zinc ions are depicted as white spheres. Blue meshes, 2Fo−Fc omit map of histone H3 peptide contoured at 0.7 σ level. (B) ITC fitting curves of H3(1–13)-PHF14PZP and PHF14PZP titrated with H3.1/3(14–34) peptides. (C) Thermodynamic parameters of PHF14PZP and H3(1–13)-PHF14PZP titrated with H3.3(14–34) peptides. (D) Mapping H3(1–34) binding regions on PHF14PZP based on the HDX-MS data. (E) Structure of PHF14PZP is shown in surface view with the corresponding H3(1–34) binding regions color coded as light pink, blue, yellow, and green corresponding to panel (D). Histone H3-Nter and H3-middle are displayed in cyan ribbon and dashes, respectively. (F) The structural alignment of H3-AF10PZP and H3-PHF14PZP complexes. PHF14PZP and AF10PZP are shown as grey and cyan ribbons, respectively. H3 peptides are show as lime (H3-PHF14) and yellow (H3-AF10) ribbons, respectively. The corresponding key elements forming the ‘αZP’ groove are marked. (G) Sequence alignment of PHF14PZP and AF10PZP. Magenta stars represent the key residues for H3(20–28) recognition in AF10PZP.
Unfortunately, we did not get the complex crystal of H3(1–13)-PHF14PZP with H3(14–34) despite many trials. Nevertheless, we performed binding studies using the chimeric protein to explore impact of H3-Nter engagement on H3-middle binding. As shown in Figure 5, while the binding affinities of H3(1–13)-PHF14PZP toward H3.1/3.3-middle were nearly the same as the binding affinities of PHF14PZP toward H3.1/3.3-middle (Figure 5B), the binding enthalpy (ΔH) increased from −1.2 kcal/mol for PHF14PZP to −3.7 kcal/mol for H3(1–13)-PHF14PZP (Figure 5B and C). Enthalpic contributions to the free energy usually reflects the specificity and strength of the interactions between two partners (24). Therefore, a gain of ΔH in the case of H3(1–13)-PHF14PZP suggested that H3-Nter engagement by the PHD1 finger of PHF14PZP promoted H3-middle recognition by PHF14PZP likely through an allosteric effect. Moreover, the entropic contributions are less pronounced (−TΔS: −6.3 kcal/mol versus −4.0 kcal/mol), suggesting existence of entropy-enthalpy compensation that may result from effects such as solvent reorganization (25).
Structure-based HDX-MS analyses of H3(1–34) recognition by PHF14PZP
To figure out the location of the functional pocket for H3-middle recognition of PHF14PZP, we performed hydrogen-deuterium exchange mass spectrometry (HDX-MS) assays (26,27). By comparing the HDX-based mass shift of PHF14PZP with and without H3(1–34) peptide, four peptide fragments of PHF14PZP were identified as responsible for histone H3 binding (Figure 5D and Supplementary Figure S5A–C). Three fragments spanning ‘173–185’, ‘290–316’ and ‘362–394’ of PHF14PZP displayed reduced exchange rates (the m/z recovered as H3-PHF14PZP engagement), indicating that they were relatively buried in the presence of H3(1–34). Moreover, the fragment ‘317–352’ displayed an enhanced exchange rate (the m/z increased as a result of H3-PHF14PZP engagement), suggesting that it was exposed to solvent upon histone binding.
We then mapped the four fragments onto the complex structure of PHF14PZP with H3(1–9) (Figure 5E). The more exposed fragment ‘317–352’ belongs to PHD1 and contains the insertion loop (322–336) that dominates H3(1–9) readout (Figure 3E). Consistent with the resultant HDX-MS data, the insertion loop achieved an ordered conformation upon H3(1–9) (H3-Nter) binding and became more solvent exposed in the complex structure (Figure 5E, F). The other three fragments were mapped to an ‘α1’ element of PHD1 (α1), an ‘α3-loop’ linker element between ZnK and PHD2 (ZP-linker), and the β-hairpin tower of PHD2, collectively creating a surface groove, designated as the ‘αZP’ surface, being positioned for H314-34 (H3-middle) engagement (Figure 5F). In support of this finding, structural alignment of PHF14PZP with AF10PZP-H3(20–28) revealed that AF10PZP utilizes a similar ‘αZP’ surface for H3-middle engagement (Figure 5F). Sequence alignment between PHF14 and AF10 revealed that key residues involved in H3-middle segment binding are largely conserved in the ‘αZP’ groove of PHF14 (Figure 5G). In sum, our structure-based HDX-MS analyses established that PHF14PZP adopts two distinct functional surfaces, namely the ‘β1-loop’ surface and the ‘αZP’ groove, for concurrent recognition of the H3-Nter and H3-middle segments encompassing residues 1 to 34 of histone H3.
Allosteric regulation of H3-middle binding by H3-Nter engagement
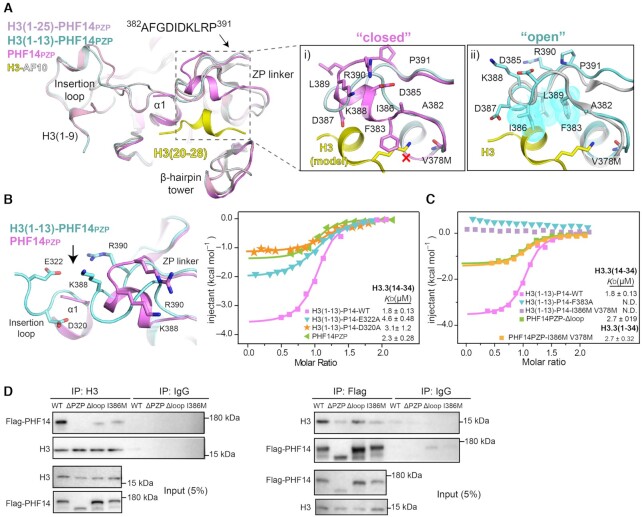
Despite similar binding KD values, our ITC assays revealed changes in binding features during PHF14PZP and H3(1–13)-PHF14PZP recognizing H3-middle, which goes from largely entropy-driven to enthalpy-driven (Figure 5B and C). This suggests the existence of an allosteric effect in which H3-Nter binding by the ‘β1-loop’ surface may affect H3-middle binding by the ‘αZP’ groove. Consistent with this speculation, comparison of H3(1–13)-PHF14PZP with PHF14PZP in both free and H3(1–25)-bound states revealed a conformational reorganization of the ‘αZP’ groove upon H3(1–9) engagement (Figure 6A). In the free state, the linker element ‘382–391’ of ‘αZP’ adopts an α-helical structure; the hydrophobic residue F383 points out and sterically clashes with histone H3K27 that is modelled based on the H3-AF10PZP complex structure (Figure 6Ai and Supplementary Figure S6A). Whereas, in both H3(1–13)-PHF14PZP and the H3(1–25)-PHF14PZP complex, the ‘382–391’ linker takes on a random coil conformation and the F383 side chain is flipped into a buried hydrophobic core with residues I386 and L389 (Figure 6Aii and Supplementary Figure S6B, left). Structural alignment with H3-AF10PZP suggests that the ‘αZP’ conformation of H3(1–13)-PHF14PZP represents an ‘open’ state that is compatible with histone H3-middle binding, while the ‘αZP’ conformation of free PHF14PZP represents a ‘closed’ one.
Figure 6.
Molecular basis for H3(14–34) recognition by PHF14PZP. (A) Structural alignment of PHF14PZP (magenta), H3(1–13)-PHF14PZP (pale cyan), H3(1–25)-PHF14PZP (light pink) and H3-AF10PZP (yellow-grey). Close-up views are a comparison of a ‘closed’ (i) and an ‘open’ (ii) conformation of the ZP linker. Key residues of ZP linker and H3 are shown as sticks and labelled as indicated. The red cross denotes a clash between F383 and a lysine residue of H3; hydrophobic residues are also shown as dotted spheres in cyan. (B) The interaction of insertion loop between ZP-linker region. Key residues are shown as sticks (left). ITC fitting curves of indicated H3(1–13)-PHF14PZP wild type, mutants and PHF14PZP, titrated with H3.3(14–34) peptides (right). P14 is short for PHF14PZP. (C) ITC fitting curves of indicated H3(1–13)-PHF14PZP mutants, PHF14PZP-Δloop and PHF14-I386M V378M titrated with H3.3(14–34) or H3.3(1–34) peptides. P14 is short for PHF14PZP. (D) Co-immunoprecipitation results from HEK293T cells with 3×Flag-PHF14-WT, 3×Flag-PHF14-ΔPZP, 3×Flag-PHF14-Δloop, and 3× Flag-PHF14-I386M overexpression showing bipartite recognition of the H3 tail by PHF14PZP.
Crystal packing analysis showed that the ZP-linkers in both free and complex PHF14PZP are in essentially the same packing environment (Supplementary Figure S6C), therefore the observed conformational difference of the ZP-linkers is less likely a consequence of different crystal packing interactions. The insertion loop of the PHD1 finger is spatially next to the ZP-linker (Figure 6B, left). Mechanistically, the conformational rearrangement of the ZP-linker is likely trigged by H3-Nter engagement by PHD1. We were able to detect charge-driven interactions between two acidic residues (D320 and E322) within the reordered insertion loop and two basic residues (K388 and R390) that flip towards the insertion loop in the rearranged ZP linker (Figure 6B, left and Supplementary Figure S6B, right). These favourable interactions likely account for the observed allosteric effect between H3-Nter binding and H3-middle binding by PHF14PZP. Supporting this finding, we generated D320A and E322A mutants of H3(1–13)-PHF14PZP and revealed obviously compromised H3(14–34) binding by ITC assays, notably in term of enthalpy change (Figure 6B, right).
Bipartite engagement of histone H3(1-34) by PHF14PZP
Previous ITC and structural studies revealed cooperative engagement of H3(1–34) by two distinct surfaces of the highly integrated PZP domain of PHF14. On the other hand, to explore the separability of H3-Nter and H3-middle engagement by PHF14PZP, we generated PHF14PZP mutants by disrupting the H3-Nter or the H3-middle binding surfaces. Firstly, we generated F383A and I386M/V378M mutants from the ‘αZP’ groove of both H3(1–13)-PHF14PZP and PHF14PZP. As shown in Figure 6C, ITC assays revealed that both F383A and I386M/V378M completely disrupted H3(1–13)-PHF14PZP binding with H3(14–34), which supports critical roles of F383/I386/V378 and conceivably an intact ‘αZP’ groove for H3-middle engagement. Remarkably, when H3(1–34) peptide was used for titration, PHF14PZP I386M/V378M returned a binding KD of 2.70 μM owing to the remaining H3-Nter binding by PHF14PZP. Similarly, the Δloop (insertion loop replaced by ‘SS’ linker) mutant of PHF14PZP retained 2.70 μM binding affinities with H3(14–34), respectively, though its binding with H3(1–15) was totally disrupted (Figure 6C and 3F). Taken together, these results suggest a bipartite feature for H3-PHF14PZP binding, in which PHF14PZP has evolved with two functional surfaces for concurrent yet separable engagement with either H3-Nter or H3-middle segments of H3(1–34).
To explore bipartite engagement of H3-PHF14PZP in cellular context, we performed co-immunoprecipitation (Co-IP) assays by ectopically expressing WT (wild type), ΔPZP (PZP deleted), Δloop, and I386M of Flag-tagged full-length PHF14 in HEK293T cells. Co-IP assays showed that PHF14-WT interacted with H3 efficiently while PHF14-ΔPZP abolished its interaction with H3. Consistent with the in vitro binding data, both PHF14-Δloop and PHF14-I386M partly disturbed their interactions with H3 (Figure 6D). Collectively, these results support an important role of the PZP domain in determining the chromatin binding activity of PHF14.
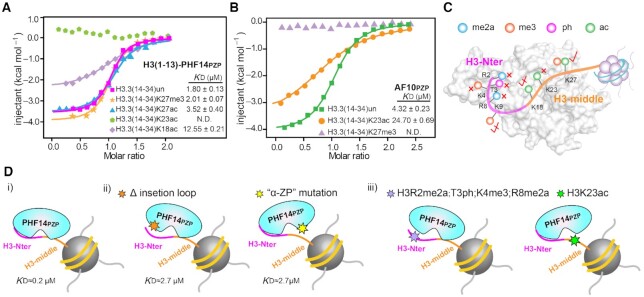
PHF14PZP is sensitive to H3K23 but not H3K27 modifications
It has been reported that AF10PZP is an unmodified H3K27 reader, and H3K27 modifications (H3K27ac or H3K27me3) could disrupt AF10PZP-H3(14–34) interaction (14). Surprisingly, our binding studies showed that PHF14PZP is strictly sensitive to H3K23ac, mildly sensitive to H3K18ac, but tolerates H3K27ac and H3K27me3 (Figure 7A). By contrast, AF10PZP is mildly sensitive to H3K23ac and strictly sensitive to H3K27me3 (Figure 7B). These results suggest that, despite the similarity of their ‘αZP’ surfaces (Figure 5E, F), PHF14PZP is set apart from AF10PZP by serving as an unmodified H3K23 but not H3K27 reader (Figure 7C). Mechanistically, this may reflect sequence motif similarity between ‘K18····K23’ and ‘K23···K27’ of H3, and a twisted reader activity of PHF14PZP towards the former motif. In support, we were not able to detect binding of H3(20–28) with PHF14PZP likely due to missing of K18 (Figure 1C), but the binding between H3(20–28) and AF10PZP is retained (Figure 1D). Distinct set of enzymes are often involved in type- and site-specific installation of histone modifications for epigenetic regulation (17). For example, global histone H3K23ac is dependent on the MOZ/MORF (KAT6) family histone acetyltransferases (HAT) that play key roles in cell cycle progression, stem cell maintenance, and human disease (28). By contrast, H3K27ac and H3K27me3 are mainly installed by p300/CBP (KAT3) family members and Polycomb repressive complex 2 (PRC2 complex), respectively. Therefore, the observed histone H3K23ac sensitivity distinguishes PHF14PZP from other reported PZP domains such as AF10/AF17PZP and BRPF1/2/3PZP and highlights distinct regulatory pathways of PHF14 function.
Figure 7.
Modification crosstalk and hierarchical switch of H3-PHF14PZP engagement. (A,B) ITC fitting curves of H3(1–13)-PHF14PZP (A) and AF10PZP (B) titrated with H3(14–34) peptides with indicated modifications. N.D. = not detected. (C) A model illustrating modification switch features of H3(1–34)-PHF14PZP engagement. H3-Nter and H3-middle are colored magenta and orange, respectively. Indicated modifications are shown as color-coded spheres. Strictly sensitive, mildly sensitive, and tolerated modifications are denoted by red cross, half-tick, and tick signs, respectively. (D) A model highlighting hierarchical modification switch features for downregulation of the H3(1–34)-PHF14PZP complex formation. H3-Nter and H3-middle are colored magenta and orange, respectively. Hexagonal stars of different colors indicate protein mutations at insertion loop region, mutation at ‘α-ZP’ surface, histone modifications in H3-Nter, and histone modification in H3-middle region, respectively.
A hierarchical modification switch model for PHF14 regulation
Our current work revealed unique histone H3 binding features of PHF14PZP, in which a long N-terminal tail of unmodified histone H3 is recognized by two reader surfaces of an integrated PZP module in a bipartite and synergistic manner (Figure 7Di, ii). Adding to the hierarchy, a leucine zipper (LZ) next to PHF14PZP may further promote chromatin association of PHF14 by increasing multivalent engagement through LZ dimerization (Figure 1A). Meanwhile, our modification crosstalk studies revealed that H3-PHF14PZP engagement is sensitive to methylations of histone H3 R2, K4, R8, as well as H3T3ph or H3K23ac, while it tolerates H3K9me3 and H3K27me3 (Figure 7C and Diii), suggesting multiple lines of modification switch or tolerance. Moreover, considering the bipartite H3 recognition feature of PHF14PZP, the observed modification switch may take place in a hierarchical manner, in which switching off H3-Nter or H3-middle engagement by one reader surface may retain decent binding by another (Figure 7Diii). Such a molecular setup may ensure better regulatory potentials through sequential downregulation of chromatin engagement of PHF14 under physiological conditions.
DISCUSSION
In the past two decades, a wealth of ‘reader’ proteins have been characterized to recognize histones and regulate diverse nuclear processes (29–31). Yet, these advances pose more challenges for characterizing novel readers and to define their functional importance. PHF14 is a conserved metazoan protein that contains multiple PHD fingers with uncharacterized molecular activities. Here, our biochemical and structural studies established that PHF14 is a bipartite histone H3(1–34) reader through its N-terminal PZP domain (the ‘PHD1-ZnK-PHD2’ cassette) but not its C-terminal PHD3 and PHD4 fingers. Notably, an insertion loop unique to PHF14 contributes to H3-binding by the PZP domain through a novel ‘β1-loop surface’ mode. Moreover, H3-Nter-induced reorganization of the insertion loop could allosterically produce a conformational switch of the H3-middle binding surface to promote H3(1–34) engagement synergistically. The recognition of histone H3(1–34) by PHF14 is sensitive to modifications occurred to R2, T3, K4, R8 and K23, suggesting a mechanism of hierarchical modification switch. Given roles of PHF14 in development, tissue homeostasis and tumorigenesis, our work calls attention to a molecular link between histone readout by PHF14 and these biological processes.
The PHD finger is a versatile epigenetic reader module (18); interestingly, it often pairs or integrates with other modules to achieve elevated reader activities. For example, the ADD domain is an integration of a PHD finger and a GATA-like finger, and such an integration endows ATRX, a heterochromatin remodelling factor, with an H3K9me3 reader activity in addition to unmodified H3K4 readout (32). The PZP domain represents another highly integrated reader module with elevated reader activities. Here we first showed that PHF14PZP is a long H3 N-terminal tail (1-34) reader, an activity shared by the AF10 and likely the BRPF1 family members. Despite this, these PZP domains displayed twisted histone binding activities in term of key recognition site and modification sensitivity for PZP subtype-specific function.
Compared with its paralogues, PHF14PZP acquired its reader activity with evolved functional elements, including a unique insertion loop of its PHD1 finger and an H3K23-specific ‘αZP’ surface. Interestingly, these functional elements are not absolutely conserved among PHF14 orthologues. It appears that the histone reader activity of PHF14PZP is acquired in species of vertebrates but not fly and C. elegans (Supplementary Figure S1A). Such a fact is reminiscent of the CHD1 protein, in which only the double chromodomain of human but not yeast CHD1 displays H3K4me3 reader activity, in accordance with a need for H3K4me3-mediated epigenetic regulation (33). Conceivably, a gain of histone reader function in vertebrate PHF14PZP reflects a need for ‘H3-PHF14’ engagement in gene regulation to ensure the phenotypic complexity of some eukaryotic organisms. On the other hand, the existence of PHF14 across a ‘broader’ spectrum of species from C. elegans to human suggests a conserved function of PHF14 beyond histone readout by PHF14PZP, which await further investigation.
PHF14 has been reported to negatively regulate expression of PDGFRα among other genes as a transcriptional co-repressor (1). Our biochemical and structural studies established that PHF14 is an unmodified histone H3 reader that is sensitive to active histone marks such as H3K4me3, H3R8me2a, and H3K23ac, but tolerates repressive marks such as H3K9me3 and H3K27me3. Other groups also found that PHF14 is repelled by H3K4me3 (34–36). These results together support a primary function of PHF14 as an epigenetic repressor. Conceivably, as a transcriptional co-repressor, PHF14 may associate with chromatin through submicromolar binding to an unmodified ‘ground state’ histone H3 so as to maintain a repressive or ‘poised’ state of its target genes. To turn on these genes, the cell will establish a particular set of histone modifications as part of the epigenetic remodelling events. Thus, the observed modification switch may serve as a mechanism to derepress PHF14 in response to upstream signals that may lead to PHF14-sensitive modifications. In the interim, the bipartite recognition feature of PHF14 may permit a ‘hierarchical modification switch’ layer of regulation so as to coordinate an ‘OFF-ON’ gene fate transition in development and disease.
DATA AVAILABILITY
The atomic coordinates and structure factors for zebrafish PHF14PZP, H3(1–25)-PHF14PZP, and H3(1–13)-PHF14PZP have been deposited into Protein Data Bank under accession codes 7D86, 7D87 and 7D8A, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff members at beamline BL17U/19U of the National Facility for Protein Science in Shanghai and Dr Shilong.Fan. at the Tsinghua Center for Structural Biology for their data collection assistance and the China National Center for Protein Sciences in Beijing for facility support. We thank Xiaolin Tian, Songbiao Zhu and Dr Haiteng Deng in Center of Protein Analysis Technology of Tsinghua University for MS analysis.
Author contributions: H.L. conceived and supervised the project; S.Z. designed and performed the experiments with assistance from Y.B., and H.C. B.G. and S.J. offered expert comments; H.L. and S.Z. wrote the paper.
Contributor Information
Shuangping Zheng, MOE Key Laboratory of Protein Sciences, Beijing Frontier Research Center for Biological Structure, Beijing Advanced Innovation Center for Structural Biology, Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing 100084, China.
Yucong Bi, MOE Key Laboratory of Protein Sciences, Beijing Frontier Research Center for Biological Structure, Beijing Advanced Innovation Center for Structural Biology, Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing 100084, China.
Haining Chen, MOE Key Laboratory of Protein Sciences, Beijing Frontier Research Center for Biological Structure, Beijing Advanced Innovation Center for Structural Biology, Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing 100084, China.
Bo Gong, Key Laboratory of Biomembrane and Membrane Engineering, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Shunji Jia, Key Laboratory of Biomembrane and Membrane Engineering, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Haitao Li, MOE Key Laboratory of Protein Sciences, Beijing Frontier Research Center for Biological Structure, Beijing Advanced Innovation Center for Structural Biology, Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing 100084, China; Tsinghua-Peking Center for Life Sciences, Beijing 100084, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31725014, 91753203 to H.L.]; National Key R&D Program of China [2020YFA0803300 to H.L.]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kitagawa M., Takebe A., Ono Y., Imai T., Nakao K., Nishikawa S., Era T.. Phf14, a novel regulator of mesenchyme growth via platelet-derived growth factor (PDGF) receptor-alpha. J. Biol. Chem. 2012; 287:27983–27996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang Q., Zhang L., Wang Y., Zhang C., Zhou S., Yang G., Li Z., Gao X., Chen Z., Zhang Z.. Depletion of PHF14, a novel histone-binding protein gene, causes neonatal lethality in mice due to respiratory failure. Acta Biochim. Biophys. Sin. (Shanghai). 2013; 45:622–633. [DOI] [PubMed] [Google Scholar]
- 3.Zhao Y., He J., Li Y., Xu M., Peng X., Mao J., Xu B., Cui H.. PHF14 promotes cell proliferation and migration through the AKT and ERK1/2 pathways in gastric cancer cells. Biomed. Res. Int. 2020; 2020:6507510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu S., Luo C., Li F., Hameed N.U.F., Jin Q., Zhang J.. Silencing expression of PHF14 in glioblastoma promotes apoptosis, mitigates proliferation and invasiveness via Wnt signal pathway. Cancer Cell Int. 2019; 19:314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miao L., Liu H.Y., Zhou C., He X.. LINC00612 enhances the proliferation and invasion ability of bladder cancer cells as ceRNA by sponging miR-590 to elevate expression of PHF14. J. Exp. Clin. Cancer Res. 2019; 38:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eun Park J., Wilford Tse S., Xue G., Assisi Maqueda C., Ramon G.P.X., Low J.K., Kon OiL, Tay C.Y., Tam JP., Sze S.K.. Pulsed SILAC-based proteomic analysis unveils hypoxia- and serum starvation-induced de novo protein synthesis with PHD finger protein 14 (PHF14) as a hypoxia sensitive epigenetic regulator in cell cycle progression. Oncotarget. 2019; 10:2136–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L., Huang Q., Lou J., Zou L., Wang Y., Zhang P., Yang G., Zhang J., Yu L., Yan D.et al.. A novel PHD-finger protein 14/KIF4A complex overexpressed in lung cancer is involved in cell mitosis regulation and tumorigenesis. Oncotarget. 2017; 8:19684–19698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akazawa T., Yasui K., Gen Y., Yamada N., Tomie A., Dohi O., Mitsuyoshi H., Yagi N., Itoh Y., Naito Y.et al.. Aberrant expression of the PHF14 gene in biliary tract cancer cells. Oncol. Lett. 2013; 5:1849–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perry J.The Epc-N domain: a predicted protein-protein interaction domain found in select chromatin associated proteins. BMC Genomics. 2006; 7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein B.J., Cox K.L., Jang S.M., Cote J., Poirier M.G., Kutateladze T.G.. Molecular basis for the PZP domain of BRPF1 association with chromatin. Structure. 2020; 28:105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein B.J., Muthurajan U.M., Lalonde M.E., Gibson M.D., Andrews F.H., Hepler M., Machida S., Yan K., Kurumizaka H., Poirier M.G.et al.. Bivalent interaction of the PZP domain of BRPF1 with the nucleosome impacts chromatin dynamics and acetylation. Nucleic. Acids. Res. 2016; 44:472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lalonde M.E., Avvakumov N., Glass K.C., Joncas F.H., Saksouk N., Holliday M., Paquet E., Yan K., Tong Q., Klein B.J.et al.. Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Genes Dev. 2013; 27:2009–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saksouk N., Avvakumov N., Champagne K.S., Hung T., Doyon Y., Cayrou C., Paquet E., Ullah M., Landry A.J., Cote V.et al.. HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol. Cell. 2009; 33:257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen S., Yang Z., Wilkinson A.W., Deshpande A.J., Sidoli S., Krajewski K., Strahl B.D., Garcia B.A., Armstrong S.A., Patel D.J.et al.. The PZP domain of AF10 senses unmodified H3K27 to regulate DOT1L-mediated methylation of H3K79. Mol. Cell. 2015; 60:319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vagin A., Teplyakov A.. MOLREP: an automated program for molecular replacement. J. Appl. Crystalphy. 1997; 30:1022–1025. [Google Scholar]
- 16.Liebschner D., Afonine P.V., Baker M.L., Bunkoczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J.et al.. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019; 75:861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bannister A.J., Kouzarides T.. Regulation of chromatin by histone modifications. Cell Res. 2011; 21:381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y., Li H.. Many keys to push: diversifying the ‘readership’ of plant homeodomain fingers. Acta Biochim. Biophys. Sin. (Shanghai). 2012; 44:28–39. [DOI] [PubMed] [Google Scholar]
- 19.Li H., Ilin S., Wang W., Duncan E.M., Wysocka J., Allis C.D., Patel D.J.. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006; 442:91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chignola F., Gaetani M., Rebane A., Org T., Mollica L., Zucchelli C., Spitaleri A., Mannella V., Peterson P., Musco G.. The solution structure of the first PHD finger of autoimmune regulator in complex with non-modified histone H3 tail reveals the antagonistic role of H3R2 methylation. Nucleic. Acids. Res. 2009; 37:2951–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiong X., Panchenko T., Yang S., Yan P., Zhang W., Xie W., Li Y., Zhao Y., David Allis C., Li H.et al.. Selective recognition of histone crotonylation by double PHD fingers of MOZ and DPF2. Nat. Chem. Biol. 2016; 12:1111–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiedler M., Sanchez-Barrena M.J., Nekrasov M., Mieszczanek J., Rybin V., Muller J., Evans P., Bienz M.. Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signaling complex. Mol. Cell. 2008; 30:507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin S., Jin L., Zhang J., Liu L., Ji P., Wu M., Wu J., Shi Y.. Recognition of unmodified histone H3 by the first PHD finger of bromodomain-PHD finger protein 2 provides insights into the regulation of histone acetyltransferases monocytic leukemic zinc-finger protein (MOZ) and MOZ-related factor (MORF). J. Biol. Chem. 2011; 286:36944–36955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bronowska A.K.Thermodynamics of ligand-protein interactions: Implications for molecular design. IntechOpen. 2011; [Google Scholar]
- 25.Chodera J.D., Mobley D.L.. Entropy-enthalpy compensation: role and ramifications in biomolecular ligand recognition and design. Annu. Rev. Biophys. 2013; 42:121–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konermann L., Pan J., Liu Y.H.. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem. Soc. Rev. 2011; 40:1224–1234. [DOI] [PubMed] [Google Scholar]
- 27.Marcsisin S.R., Engen J.R.. Hydrogen exchange mass spectrometry: what is it and what can it tell us?. Anal. Bioanal. Chem. 2010; 397:967–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang F., Abmayr S.M., Workman J.L.. Regulation of KAT6 acetyltransferases and their roles in cell cycle progression, stem cell maintenance, and human disease. Mol. Cell. Biol. 2016; 36:1900–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taverna S.D., Li H., Ruthenburg A.J., Allis C.D., Patel D.J.. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 2007; 14:1025–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Musselman C.A., Lalonde M.E., Cote J., Kutateladze T.G.. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 2012; 19:1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrews F.H., Strahl B.D., Kutateladze T.G.. Insights into newly discovered marks and readers of epigenetic information. Nat. Chem. Biol. 2016; 12:662–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwase S., Xiang B., Ghosh S., Ren T., Lewis P.W., Cochrane J.C., Allis C.D., Picketts D.J., Patel D.J., Li H.et al.. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat. Struct. Mol. Biol. 2011; 18:769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sims R.J. III, Chen C.-F., Santos-Rosa H., Kouzarides T., Patel SS., Reinberg D.. Human but not yeast CHD1 binds directly and selectivly to histone H3 methylation at lyscine 4 via its tandem Chromo Domains. J. Biol. Chem. 2005; 280:41789–41792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burton A.J., Haugbro M., Gates L.A., Bagert J.D., Allis C.D., Muir T.W.. In situ chromatin interactomics using a chemical bait and trap approach. Nat. Chem. 2020; 12:520–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eberl H.C., Spruijt C.G., Kelstrup C.D., Vermeulen M., Mann M.. A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol. Cell. 2013; 49:368–378. [DOI] [PubMed] [Google Scholar]
- 36.Bartke T., Vermeulen M., Xhemalce B., Robson S.C., Mann M., Kouzarides T.. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010; 143:470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Felsenstein J.Confidence limits on phylogenies:An approach using the bootsrap. Evolution. 1985; 39:783–791. [DOI] [PubMed] [Google Scholar]
- 38.Saitou N., Nei M.. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987; 4:406–425. [DOI] [PubMed] [Google Scholar]
- 39.Tamura K., Nei M., Kumar S.. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:11030–11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar S., Stecher G., Tamura K.. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016; 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors for zebrafish PHF14PZP, H3(1–25)-PHF14PZP, and H3(1–13)-PHF14PZP have been deposited into Protein Data Bank under accession codes 7D86, 7D87 and 7D8A, respectively.