Abstract
Translation reinitiation is a gene-specific translational control mechanism. It is characterized by the ability of short upstream ORFs to prevent full ribosomal recycling and allow the post-termination 40S subunit to resume traversing downstream for the next initiation event. It is well known that variable transcript-specific features of various uORFs and their prospective interactions with initiation factors lend them an unequivocal regulatory potential. Here, we investigated the proposed role of the major initiation scaffold protein eIF4G in reinitiation and its prospective interactions with uORF’s cis-acting features in yeast. In analogy to the eIF3 complex, we found that eIF4G and eIF4A but not eIF4E (all constituting the eIF4F complex) are preferentially retained on ribosomes elongating and terminating on reinitiation-permissive uORFs. The loss of the eIF4G contact with eIF4A specifically increased this retention and, as a result, increased the efficiency of reinitiation on downstream initiation codons. Combining the eIF4A-binding mutation with that affecting the integrity of the eIF4G1–RNA2-binding domain eliminated this specificity and produced epistatic interaction with a mutation in one specific cis-acting feature. We conclude that similar to humans, eIF4G is retained on ribosomes elongating uORFs to control reinitiation also in yeast.
INTRODUCTION
Translation is a cyclical process, essentially divided into initiation, elongation, termination, and ribosome recycling. Translation of most mRNAs is cap dependent and follows the scanning mode of operation (reviewed in (1)). In eukaryotes, translation initiation commences with the assembly of the 43S preinitiation complex (43S PIC) comprising the small 40S ribosomal subunit, the eIF2-GTP-Met-tRNAiMet ternary complex (eIF2-TC), and eIF3, eIF5, eIF1 and eIF1A (reviewed in (2)). The eIF4F complex, which comprises eIF4E, eIF4A and eIF4G, then mediates the attachment of the 43S PIC to the 5′ cap structure of mRNA forming the 48S PIC. eIF4E is a cap-binding protein, eIF4A is a DEAD-box RNA helicase, and eIF4G represents a scaffold protein harboring binding sites for RNA (named RNA1, RNA2, RNA3 (3)), eIF4E, eIF4A, and the poly(A) binding protein (PABP) (1). eIF4G can also interact with eIF3 (in mammals) or eIF5 (in yeast) to facilitate 43S PIC recruitment to the mRNA (reviewed in (1)). Yeast eIF4G has two paralogues, eIF4G1 and G2, which can make similar contacts with RNA and initiation factors and thereby promote initiation (4). The higher-order 48S complex scans along the messenger for the initiation codon. Upon the start codon recognition, eIF5B promotes joining of a 60S subunit, which results in the ejection of most eIFs, forming the 80S ribosome poised for elongation.
In contrast to this generalized view, an earlier study suggested that some eIFs may remain transiently associated with ribosomes also throughout the elongation and termination phases (5). It was proposed that ribosomes terminating at short ORF preceding a second ORF that have still retained certain eIFs could then be able to recycle only the large 60S subunit, resume traversing downstream, and upon acquisition of a new eIF2-TC, they would continue scanning for the next AUG to eventually start translating the second ORF (6). Indeed, this incomplete recycling of post-termination complexes at the stop codon of an upstream short ORF (uORF) represents a basic principle of the gene-specific regulatory mechanism called reinitiation (REI). REI has been drawing a lot of attention in recent years mainly due to the underexplored role of trans-acting factors promoting its efficiency (reviewed in (7–10)).
Over the years it has been demonstrated that the efficiency of REI depends on: (i) cis-acting mRNA features surrounding a given uORF, (ii) duration of the uORF elongation, (iii) certain eIFs and (iv) the intercistronic distance between uORF and the main ORF needed for the acquisition of the new eIF2-TC (reviewed in (2)). We recently developed a novel in vivo RNA-protein Ni2+-pull down (RaP-NiP) assay and revealed that one of the eIFs that is transiently retained on the 80S ribosome terminating on short uORFs is yeast eIF3 (11). This result was very recently corroborated by selective translational complex profiling (Sel-TCP-seq) (12). We further showed that its post-termination retention at certain uORFs is required for efficient REI downstream, as suggested before based on yeast genetics (13,14). These findings thus solved a long-standing puzzle, which is why REI efficiency decreases with the increasing time to translate a short uORF: the more time it takes, the higher probability that eIF3 will drop off the ribosome. Importantly, recent reports from yeast and mammals provided the ultimate support for the post-initiation retention of eIF3 on elongating ribosomes, its role in promoting REI after short uORFs, proposing even a specific function for human eIF3 during early elongation on regular ORFs (12,15–17).
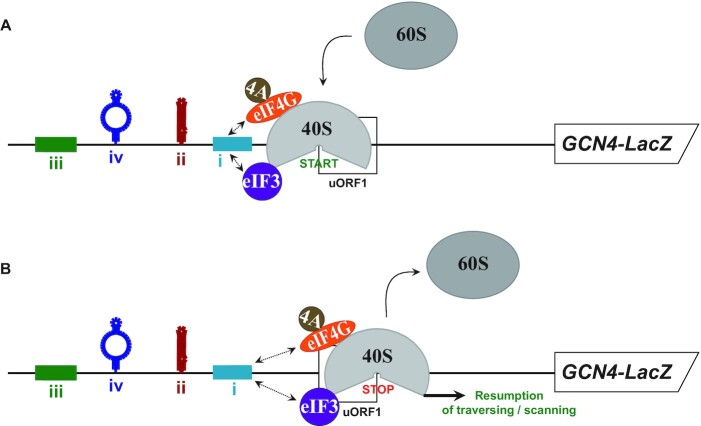
The classical example of the REI mechanism is the translational control of the yeast mRNA encoding the transcriptional activator GCN4 (reviewed in (7,18)), which is governed by four uORFs in a rather intricate fail-safe mechanism (19) (Supplementary Figure S1). This mechanism is very sensitive to the eIF2-TC levels that are changing in response to different nutrient conditions (20). The first of the four uORFs is efficiently translated under both nutrient replete and deplete conditions, and after its translation, it allows efficient resumption of traversing of the post-termination 40S subunit downstream. The second REI-permissive uORF, uORF2, serves as a backup of uORF1 to capture all ribosomes that eventually scanned through the uORF1 AUG (19). In non-stressed cells, where the eIF2-TC levels are high, nearly all of the traversing ribosomes can rebind the eIF2-TC before reaching one of the last two distant uORFs (uORFs 3 and 4), neither of which supports efficient REI; i.e. terminating ribosomes are efficiently recycled and the main GCN4 ORF is not expressed. Under starvation conditions, the GCN2 kinase phosphorylates eIF2, which suspends formation of new eIF2-TCs in the cytoplasm. Consequently, post-termination 40S ribosomes traversing from the uORF1 or uORF2 stop codon downstream will require more time to rebind the eIF2-TC. This will allow a large proportion of them to bypass REI-non-permissive uORF3 and uORF4 and reacquire the eIF2-TC only past uORF4 but still before the GCN4 start codon. Thus, whereas the global protein synthesis is significantly down-regulated, protein expression of GCN4 is concurrently induced.
We and others demonstrated that the high REI competence of uORF1 and uORF2 depends on several cis-acting mRNA features (reviewed in (7,18)). For the purpose of this work it suffices to mention only the GCN4 5′ enhancer (for the complete overview, please see (21)). The 5′ enhancer is formed by several REI-promoting elements (RPEs) with a specific structural arrangement occurring in the upstream regions of uORF1 and uORF2 (Supplementary Figure S1); for example uORF1 utilizes four RPEs (i–iv) (14,19). The RPE ii. functions autonomously, whereas RPEs i and iv cooperate with a/TIF32 subunit of eIF3 to stabilize the 40S-mRNA-eIF3 post-termination complex and allow it to resume traversing downstream (11,14,19).
Interestingly, yeast genetic analyses, experiments in mammalian reconstituted systems and selective 80S ribosomal profiling analyses in human cells suggested that also some eIF4F components might follow the eIF3 fate on the elongating ribosome and boost the efficiency of REI after translation of certain short uORFs, like eIF3 does (17,22–24). In particular, experiments with reporters driven by different internal ribosome entry sites (IRES) investigating factor requirements for REI after translation of short uORFs in rabbit reticulocyte lysates revealed that efficient REI occurred only if the original initiation event involved eIF4F, or at least eIF4A and the eIF4G’s middle domain (22). In mammals, this well conserved domain consists of binding regions for RNA, eIF4A, and eIF3 (25). Using the mammalian in vitro reconstituted translation system it was also demonstrated that the 3′ directionality of resumption of traversing for REI downstream is promoted by the eIF4F complex (24). Furthermore, yeast genetic experiments suggested that eIF4G2 mutations altering eIF4E- and eIF4A-binding sites increase the efficiency of REI after translation of some uORFs from the GCN4 mRNA leader, possibly by decreasing the migration rate of the traversing 40S subunits (23). And last but not least, a recent study employing selective 80S ribosomal profiling (17) clearly demonstrated that human eIF4G1 and eIF4E (together with eIF3 (12)) persist on elongating 80S ribosomes with a decay half-length of ∼12 codons. It could very well be that the post-initiation retention of eIF4G is mediated by eIF3 with which it directly interacts in mammals (26,27). Despite all this progress, in vivo evidence for the direct eIF4G involvement in the establishment of the REI competence is lacking.
To examine the prospective role of the eIF4F components in REI in the budding yeast S. cerevisiae, we employed our recently developed in vivo RaP-NiP assay (11). We show here that eIF4G and eIF4A but not eIF4E are specifically enriched on ribosomes engaged in translation of short REI-permissive uORFs and that the loss of contact between eIF4G and eIF4A impacts efficiency of REI, at least in the budding yeast.
MATERIALS AND METHODS
Construction of plasmids and yeast strains
Constructions of plasmids and yeast strains are described in the Supplementary Material and listed in Supplementary Tables S1-S3.
Yeast in vivo RNA-protein Ni2+-pull down (RaP-NiP) assay
To examine whether eIF4G in complex with eIF4A remain persistently associated with the elongating and post-termination ribosomes on uORF1 and uORF2 of GCN4, as well as on uORF1 of YAP1 and YAP2 to promote REI in vivo, we employed our well-established yeast in vivoRNA-Protein Ni2+-Pull Down (RaP-NiP) assay. This assay was developed, trouble-shot and described in great detail here (11). Just briefly, different GCN4 leader constructs, described below and in (11), were individually introduced into yeast cells that were: (i) deleted for both, chromosomal GCN4 along with its 5′ flanking sequences as described in (11), and a gene encoding one of the eIF4F components at a time and (ii) expressing the His-tagged version of the deleted eIF4F component from a plasmid as the sole allele of this gene. Exponentially growing transformants were first cross-linked with formaldehyde (1%) and the whole cell extracts (WCE) were then prepared and pre-incubated with a set of two sequence-specific oligonucleotides for the subsequent RNase H cleavage as described in (11) (please see Supplementary Figure S2A and B for a flowchart). Thus digested samples were incubated with Ni2+-sepharose beads to pull down the His-tagged eIF4F component and any proteins and RNA fragments co-purifying with it. Purified complexes were subsequently incubated with Proteinase K to digest all proteins. Co-purifying RNA fragments were then isolated by hot phenol extraction and subsequently treated with DNase I to remove any traces of DNA contamination. cDNAs were synthesized by RT-PCR and the relative co-purification yields of different uORF fragments were determined by qPCR. This way we ensured that most, if not all, of the material that we retrieved originated only from the eIF4F-bound ribosomes engaged with our segment of interest (please see Supplementary Figure S2C for an explanatory schematic). The relative quantities of individual uORF fragments were normalized to the amounts of corresponding input (WCE) mRNA levels as well as to the mRNA levels of ACT1 house-keeping gene, the mRNA of which was also recovered in trace amounts by this procedure. In particular, for the relative quantification of our qPCR data we applied the 2ΔΔCT or Delta-delta method (Pfaffl MW 2001) where the recovered uORF or ACT1 mRNA levels were normalized to the corresponding input mRNA levels. To obtain statistically significant data, we took into account the experimentally calculated primer efficiencies in our analysis. All presented experiments were repeated several times with at least 3 biological replicates.
Please note that in order to minimize the RNase H cutting and qPCR amplification errors among mRNAs carrying different uORFs, in our original paper (11) we took the uORF1-only construct shown in Figure 1A as a template, designed the specific 5′ and 3′ RNase H cutting and qPCR amplification primers, and subsequently replaced the uORF1 segment (encompassing the REI-promoting elements) bordered by both sets of primers (Y1 in Figure 1A) with the corresponding segment of uORF4 (Y4 in Figure 1A; please note that the uORF4-only construct has the AUG of uORF3 mutated out) or of uORF3 (shown in Figure 2A). This way the RNase H cutting, as well as qPCR primers, were the same for all three constructs and, intuitively, so was the length of the qPCR amplicon. The identical length of the RNase H digested fragment for all these uORFs is critical also because segments with varying lengths could introduce a ‘ribosome occupancy error’. In other words, we expected that besides the elongating/terminating eIF4F-bound 80S ribosomes, the eIF4F-bound 40S ribosomes scanning through the 5′ UTR will be also pulled down in our procedure, with the relatively same efficiency among all RaP-NiP constructs. Therefore, generating RNA segments of uneven size could result in varying ribosome occupancy that would artificially influence the amounts of the RNase H-cleaved RNAs co-purifying with the His-tagged eIF4F components, which would not directly reflect the true differences among the uORF constructs. Functionality (permissiveness for REI) of these artificial constructs was previously validated using the β-galactosidase assay (11).
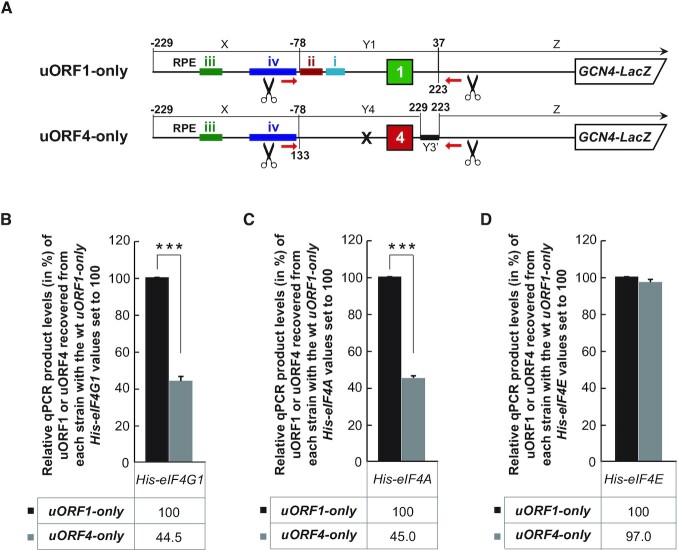
Figure 1.
eIF4G and eIF4A but not eIF4E from the eIF4F complex are preferentially retained on ribosomes elongating and terminating on REI-permissive uORF1 from the GCN4 mRNA leader. (A) Schematics showing uORF1- and uORF4-only RaP-NiP constructs. Colored bars indicate positions of individual RPEs of uORF1 (color-coding of both uORFs reflects their REI-permissiveness (green) or REI-non-permissiveness (red); the RNase H cutting sites are indicated by scissors; the RT-PCR primer binding sites are indicated by red arrows. For explanation of segments X, Yn, Z, and Y3’, and the construction details, please see (11). (B) The YMP53 (gcn4Δ tif4631Δ tif4632Δ His-eIF4G1) strain was introduced either with the uORF1 or uORF4-only RaP-NiP constructs shown in panel A and the resulting transformants were pre-cultured in minimal media overnight, diluted to OD600 ∼0.1 and further cultivated to OD600 ∼1. The exponentially growing cells were then subjected to RaP-NiP as described in Materials and Methods. Relative qPCR product levels (in %) of the uORF1 or uORF4 segments recovered from His-eIF4G1 expressing strains with standard deviations obtained from at least three independent experiments from three independent transformants (i.e. biological replicates) normalized to the amounts of corresponding input (WCE) mRNA levels, as well as to the mRNA levels of recovered reference ACT1 gene, are given with the values of uORF1-only set to 100%. Statistical significance was assessed using unpaired, two-sided, t-test (***P< 0.0005). (C) The YMP63 (gcn4Δ tif1Δ tif2Δ His-eIF4A) strain was introduced with the uORF1 or uORF4-only RaP-NiP constructs shown in panel A and treated as described in panel B. Relative qPCR product levels (in %) of the uORF1 or uORF4 segments recovered from the His-eIF4A expressing strains were processed as described in panel B with the values of His-eIF4A uORF1-only set to 100% (***P< 0.0005). (D) The YMP67 (gcn4Δ cdc33/tif45Δ His-eIF4E) strain was introduced with the uORF1 or uORF4-only RaP-NiP constructs shown in panel A and treated as described in panel B. Relative qPCR product levels (in %) of the uORF1 or uORF4 segments recovered from His-eIF4E expressing strains were processed as described in panel B with the values of His-eIF4E uORF1-only set to 100%.
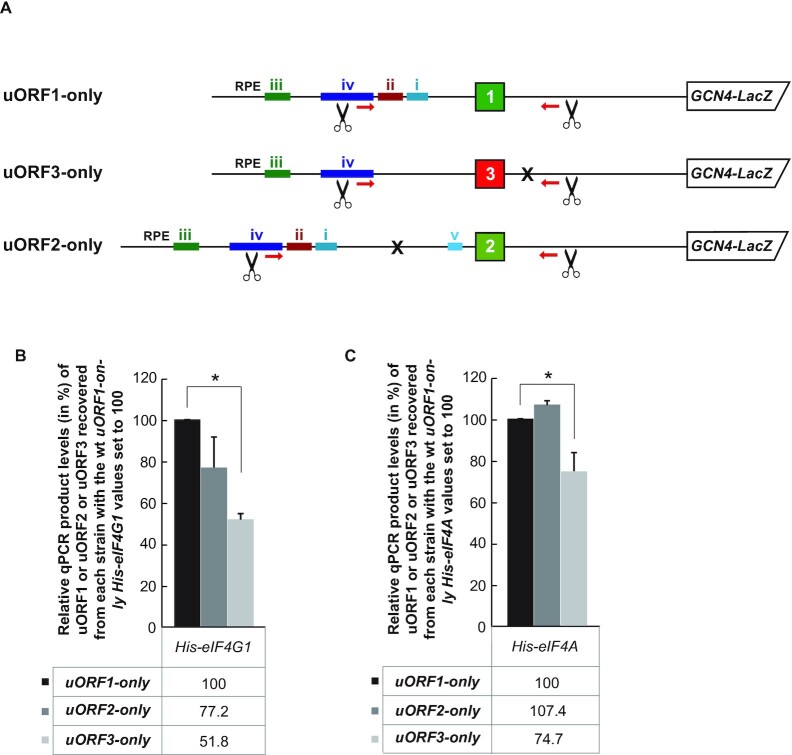
Figure 2.
eIF4G and eIF4A are preferentially retained on ribosomes elongating and terminating on both REI-permissive uORFs from the GCN4 mRNA leader. (A) Schematics showing individual uORF1-, uORF2- and uORF3-only RaP-NiP constructs. (B) The YMP53 strain described in Figure 1B and (C) the YMP63 strain described in Figure 1C were introduced with the uORF1-only or uORF2-only or uORF3-only RaP-NiP constructs shown in panel A and treated as described in Figure 1B. Relative qPCR product levels (in %) of the corresponding uORF1 or uORF2 or uORF3 segments recovered from His-eIF4G1 expressing strains were processed as described in Figure 1B with the values of His-eIF4G1 or His-eIF4A uORF1-only set to 100%. Statistical significance was assessed using ANOVA: Single Factor with Post Hoc testing (*P< 0.05).
Other techniques
The β-galactosidase assay was carried out as described before (28). Western blot analyses of the WCEs of Saccharomyces cerevisiae cells expressing eIF4G1 or its mutant forms were carried out as in (29).
RESULTS
eIF4G and eIF4A but not eIF4E from the eIF4F complex are preferentially retained on ribosomes elongating and terminating on REI-permissive uORFs in yeast
Previous studies suggested that at least some components of the eIF4F complex control REI after the translation of short uORFs in yeast and mammals, however, the mechanistic details remain poorly understood (17,22,23). Here, we employed our recently established RaP-NiP assay capable of capturing selected initiation factor-bound ribosomes translating short uORFs in yeast (11) to investigate whether or not the constituents of the eIF4F complex remain bound to ribosomes during the first few elongation cycles to promote REI in vivo, like eIF3 does (11,12,17). To do that, we first compared the occupancy of the eIF4A, 4G and 4E-bound post-termination 40S ribosomes individually at the REI-permissive uORF1 versus REI-non-permissive uORF4 of the GCN4 mRNA leader fragments (Figure 1A). As shown before for eIF3 (11), we hypothesized that in case of the REI involvement of any of the eIF4F components, co-purification of RNA segments containing uORF1—allowing efficient REI—should result in a higher yield compared to co-purification of RNA segments bearing REI-non-permissive uORF4. Consistent with our proposition, we detected >2-fold higher amounts of the uORF1-specific RNA segment co-purifying with His-tagged eIF4G1 and eIF4A compared to the uORF4-specific segment in S. cerevisiae strains deleted for both eIF4G-encoding genes (TIF4631 and TIF4632) or both eIF4A-encoding genes (TIF1 and TIF2), respectively (Figure 1B and C). Please note that control cells expressing an empty vector produced no signal, and a background value of ∼3% of the uORF1 segment was recovered from cells expressing untagged eIF4G1 compared to its His-tagged allele (Supplementary Figure S3A and B), as observed before in case of eIF3 (11). It should also be stressed that based on our thorough analysis conducted in the past (11), this assay is set up in such a way that the observed differences are created predominantly by the captured elongating/terminating ribosomes and not by 40S ribosomes that are still scanning towards the uORFs from the 5′ cap. This was evidenced by the fact that performing the RaP-NiP assay with the His-tagged eIF2; i.e. with the factor recently unambiguously demonstrated to dissociate from the PICs prior to subunit joining (17), no difference between uORF1- and uORF4-specific constructs was observed (11).
In contrast to eIF4G and eIF4A, no difference in the recovery of uORF1- versus uORF4-specific segments was observed with the His-tagged eIF4E protein in a strain deleted for an eIF4E-encoding gene (CDC33/TIF45) (Figure 1D). Since the overall amounts of uORF1-specific segments that we recovered with all three His-tagged strains were relatively the same (Supplementary Figure S3C), we conclude that the entire eIF4F complex is retained on ribosomes past the initiation phase, as recently shown for human cells (17). At the same time, however, it seems that the ribosomes elongating and terminating on REI-permissive uORFs are capable to preferentially increase retention of eIF4G and 4A factors over eIF4E.
To further document the specificity of our findings, we next examined the occupancy of eIF4G1-bound ribosomes at the second REI-non-permissive uORF of the GCN4 mRNA leader, uORF3, as well as at the second REI-permissive uORF, uORF2 (Figure 2A and B), and obtained similar results. A mild drop (by ∼20%) in the recovered RNA amounts of uORF2 compared to uORF1 (Figure 2B) could indicate that the REI mechanism of uORF2, relying on just two RPEs (14,19,21), might be somewhat less dependent on eIF4G as opposed to that operating on uORF1 utilizing four RPEs, as observed before for eIF3 (11). Analogous analysis with the His-tagged eIF4A allele revealed no differences between uORF1 and uORF2 and a significant, ∼25% drop at uORF3 (Figure 2C). In spite of these specific differences, perhaps attributable to specific contacts that these two factors make individually with the ribosome (30), overall our results nicely correlate with the well-described roles of uORFs 1 and 2 versus uORFs 3 and 4 in the GCN4 translational control mechanism (19), and further validate the specificity and accuracy of our RaP-NiP assay. They also suggest that both eIF4G and eIF4A - like eIF3 (11) but unlike eIF4E (Figure 1D) and eIF2 (11,12,17) – are preferentially retained on ribosomes elongating and terminating on REI-permissive uORFs, and that this retention might somehow stimulate REI downstream.
As a proof of principle, we also analyzed two short uORFs individually preceding yeast genes YAP1 and YAP2, both encoding stress related transcription factors, which were experimentally shown to allow efficient REI (31) (Supplementary Figure S4). To be able to exploit our RaP-NiP set-up with all its controls, we simply took the uORF1-only construct and replaced the uORF1 segment with the corresponding 5′ UTR sequences of both YAP genes (Supplementary Figure S5A and B), as we did with other uORFs of GCN4 here (Figures 1A and 2A) and in the past (11). Please only note that since the inserts of both YAP genes must have been taken longer, and their uORFs are also twice as long as uORF1, we could compare them only between each other and not with GCN4 uORFs to avoid the ‘ribosome occupancy error’ mentioned above. Nonetheless, as shown in Supplementary Figure S5C and D, whereas both His-tagged eIF4G and eIF4A pulled down varying amounts of RNA segments of both genes, perhaps reflecting the degree of their permissiveness for REI like in case of uORF1 versus uORF2 of GCN4 (Figure 2B), no differences were observed with His-tagged eIF4E (Supplementary Figure S5E).
The eIF4G1-1 mutant specifically increases the efficiency of REI past the uORF1 translation in vivo
To investigate the molecular mechanics of a potential involvement of eIF4G and eIF4A in REI, we took advantage of an existing battery of eIF4G2 mutants affecting eIF4G interactions with other eIF4F constituents. In particular, based on the protein sequence homology with its yeast paralog we created three eIF4G1 mutants: (i) three-point mutations in its HEAT domain (V648G, E796G, L823S) impairing eIF4G binding to eIF4A—designated eIF4G1-1 (32,33); (ii) four-point mutations (three in its RNA2 RNA-binding domain [R509I, K527R, R537S] and one in the HEAT domain [V648D]) also shown to impair eIF4A binding to eIF4G with no apparent effect on a model mRNA binding in vitro—designated eIF4G1-8 (23,32,33) and (iii) two-point mutations in its eIF4E-binding domain (L457A, L458A) eliminating eIF4G binding to eIF4E—designated eIF4G1-459 (32,34) (Figure 3A). We introduced these mutant eIFG1 alleles individually into a S. cerevisiae strain deleted for genes encoding both isoforms of eIF4G and tested them for growth phenotypes by a conventional spot assay. Please note that neither of the mutations significantly affected the eIF4G protein levels (Figure 3B), as observed before for eIF4G2 (23). As also reported before for eIF4G2 mutants (32–34), all three eIF4G1 mutants produced temperature sensitive (Ts–) phenotypes with the eIF4G1-459 allele displaying also a relatively mild slow growth (Slg–) (Figure 3C). As also noted before for eIF4G2 mutants (23,34), all observed phenotypes of the eIF4G1-1 and -8, and -459 mutants were specifically suppressible by high copy expression of eIF4A or eIF4E, respectively (Figure 3D and Supplementary Figure S3D). These results confirm the specificity of the binding domains for either eIF4A or eIF4E and thus clearly demonstrate mutual interchangeability of both eIF4G-encoding alleles, at least in the tested conditions.
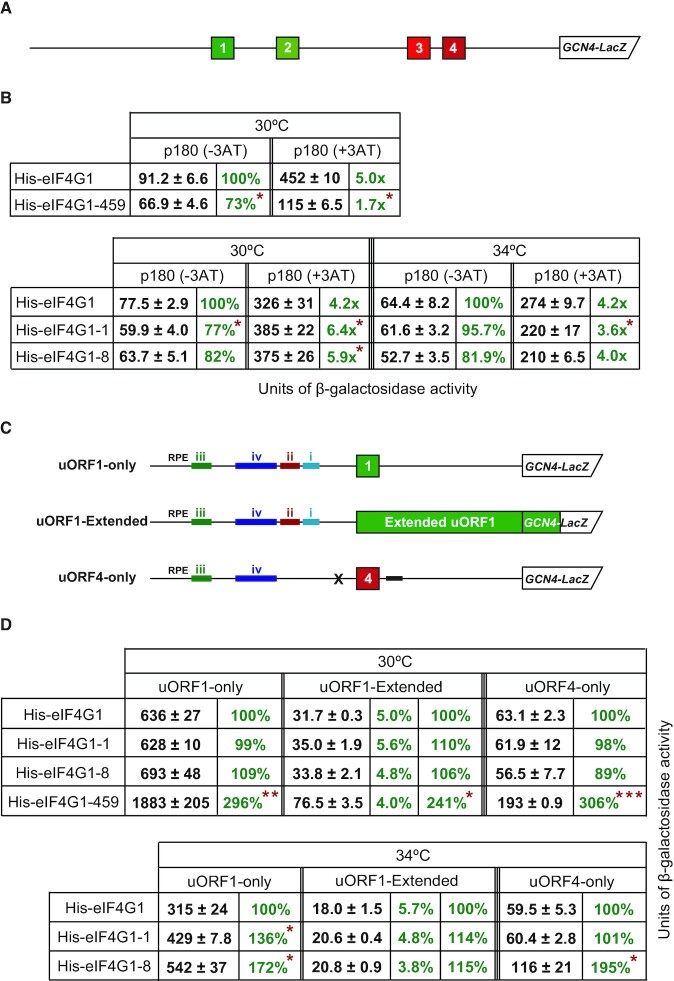
Figure 3.
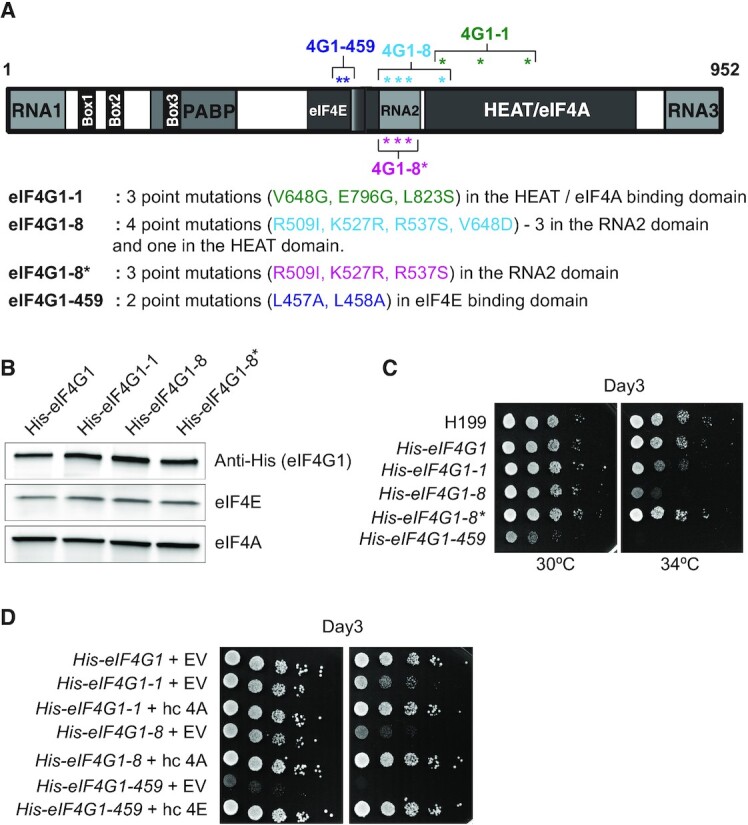
Growth analysis of the selected eIF4G mutants with or without overexpression of eIF4A or eIF4E. (A) Schematic representation of the eIF4G1 (adopted from (34)) showing the positions of introduced point mutations and changes in amino acid residues analyzed in this study. (B) Western blot analysis of WCEs from the strains described in panel C grown at 30°C, using anti-His, anti-eIF4A, and anti-eIF4E antibodies. (C) The eIF4G1-1, eIF4G1-8 and eIF4G1-459 mutant strains impart the Ts– and the latter also Slg– phenotypes. Strain YMP47 (GCN2 tif4631Δ, tif4632Δ) was transformed with individual YEplac181-based plasmids carrying the eIF4G1 alleles and the resident YEp-eIF4G1-URA plasmid was evicted on 5-FOA. The resulting strains, eIF4G1 (YMP53), eIF4G1-1 (YMP55), -8 (YMP56), -8* (YMP91) and -459 (YMP57), together with the isogenic H199 strain (GCN2 TIF4631, TIF4631) were then spotted in five serial 10-fold dilutions on SD plates and incubated at 30°C or 34°C for 3 days. (D) Growth defects of individual eIF4G mutants are suppressible by either high copy expression of eIF4E or eIF4A. The strains described in panel C were transformed individually with YEplac112-based plasmids carrying either eIF4A or eIF4E alleles, or an empty plasmid, and the resulting transformants were spotted in five serial 10-fold dilutions on SD plates and incubated at 30 or 34°C for 3 days.
To investigate a potential involvement of the eIF4A and eIF4E-binding domains of eIF4G1 in REI on GCN4, we next tested the aforementioned eIF4G mutants for the so-called Gcn– (general control non-derepressible) phenotype using our well-established GCN4-lacZ reporter assay with the intact GCN4 5′ mRNA leader (Figure 4A). The cells are grown in SD media overnight and then one half of them is supplemented with the drug 3-amino triazol (3-AT) (10 mM) that mimics amino acid starvation. Only cells that can derepress GCN4 expression by the delayed REI mechanism described in the Introduction can induce the β-galactosidase activity in the presence of 3-AT. As can be seen in Figure 4B, whereas eIF4G1-459 dramatically failed to derepress GCN4 expression at 30°C (1.7-fold versus 5-fold for wild type (wt) cells)—thus exhibiting the Gcn– phenotype, both eIF4G1-1 and -8 showed even more robust derepression (6.4- and 5.9-fold, respectively, versus 4.2-fold for wt). However, increasing the temperature by 4°C revealed a mild but statistically significant Gcn– phenotype also with the eIF4G1-1 allele (3.6.-fold versus 4.2-fold for wt) but not with eIF4G1-8 (Figure 4B).
Figure 4.
The eIF4G1-1 mutant imparts a modest Gcn– phenotype and specifically increases the efficiency of REI past the uORF1 translation in vivo. (A) Schematic showing the GCN4-LacZ construct with all four upstream uORFs. (B) The eIF4G1-1 and -459 mutants impart the Gcn– phenotype. Along with pMP121 (containing the wt GCN4 leader), the GCN4-lacZ construct from panel A was introduced individually into the strains carrying various eIF4G-encoding alleles described in Figure 3. The resulting transformants were pre-cultured in minimal media overnight, diluted to OD600 ∼0.3, grown for 2 h., and the same aliquots of cultures were either treated with 10 mM 3-AT (to induce the GCN4-lacZ expression) or without 3-AT for 6 h. The β-galactosidase activities were measured in the WCEs and expressed in units of nmol of o-nitrophenyl-b-d-galactopyranoside hydrolyzed per min per mg of protein. The mean values and standard deviations obtained from at least three independent measurements with three independent transformants, and activity in the mutant constructs relative to wt (in %), as well as the induced vs. induced fold-increases are given where applicable. Statistical significance of the differences between wt and mutant eIF4G1 alleles was assessed using Anova: Single Factor test with Post Hoc testing (*P< 0.05). (C) Schematics showing various modified GCN4-LacZ constructs described in the main text. (D) The eIF4G1-1 mutant specifically increases the efficiency of REI past the uORF1 translation in vivo. The GCN4-lacZ constructs from panel C were introduced individually into the strains carrying various eIF4G-encoding alleles described in Figure 3. The resulting transformants were pre-cultured in minimal media overnight, diluted to OD600 ∼0.35, grown for an additional 6 hrs. and the β-galactosidase activities were measure and analyzed as described in panel B (*P< 0.05).
To provide a deeper mechanistic insight into the defects caused by the eIF4G1 mutations in REI, we measured (i) the efficiency of REI after translation of solitary uORF1 or uORF4, as well as (ii) the extent of so-called leaky scanning past the AUG of uORF1 at 30 and 34°C, respectively, using a specifically designed GCN4-lacZ construct (Figure 4C). Leaky scanning was determined by using a construct where uORF1 extends out of frame into the GCN4-LacZ coding sequence and thus the measured β-galactosidase activity reflects only those ribosomes that bypass the AUG of uORF1 and initiate at the GCN4 AUG instead.
Our measurements revealed that eIF4G1-459 increased the efficiency of REI past both tested uORFs independently of their permissiveness for REI at 30°C, as well as the level of leaky scanning, pointing at some general, REI-non-specific defect in translation (Figure 4D, side columns). In agreement, this particular mutant was previously shown to robustly enhance translation of model mRNAs in cell-free extracts in a cap- and poly(A) tail-independent manner (32). This result is also consistent with no difference in the eIF4E retention on ribosomes elongating and terminating on REI-permissive vs. non-permissive uORFs (Figure 1D and Supplementary Figure S5). The eIF4G1-1 and -8 mutants showed no differences at this temperature (Figure 4D; side columns). At the elevated temperature, however, eIF4G1-1 displayed statistically significant increase in the REI efficiency specifically only with REI-permissive uORF1, whereas eIF4G1-8 mimicked the effect of eIF4G1-459 at 30°C (Figure 4D; side columns). Since no significant defects in leaky scanning were observed with any of the mutants (Figure 4D; middle column), the effect of eIF4G1-1 at 34°C could be directly attributed to some altered activity of post-termination ribosomes upon completing the uORF1 translation. The specific loss of the eIF4G-4A contact, documented before (32,33), could for example slow down the resumption of traversing of the post-termination ribosomes after uORF1 translation, and/or the traversing and subsequent scanning per se, to provide the 40S ribosome with more time to reacquire the ternary complex before arriving at the next (u)ORF, as proposed by others (23). This would elevate the REI activity on the downstream AUG (in this case the AUG of our GCN4-lacZ reporter), resulting in the Gcn– phenotype as we observed with the wt GCN4 leader (Figure 4B). Finally, the non-specific effect of eIF4G1-8 at the elevated temperature could suggest the importance of the intact RNA2-binding domain for the general REI competence (see discussion).
The eIF4G1-1 mutant specifically increases retention of ribosomes elongating and terminating on REI-permissive uORFs versus REI-non-permissive uORF4
Based on our β-galactosidase activity measurements described above we wished to explore the observed uORF1-specific effect of the eIF4G1-1 mutant in more detail. To do that, we subjected eIF4G1-1 (the eIF4A-binding mutant) and -8 (the eIF4A- and RNA-binding mutant) to our RaP-NiP assay. Please note that we did not include eIF4G1-459—the eIF4E-binding mutant—due to a very slow-growing character of these cells preventing us from gaining enough material to carry out the assay. In addition, due to the technical specifics of our RaP-NiP assay we neither could subject the eIF4G1-1 and -8 Ts– mutants to heat shock at 34°C. Nonetheless, both mutants grown at 30°C significantly increased the recovery of the uORF1-containing RNA segment (by 1.8–2-fold) (Figure 5B) and uORF2-containing RNA segment (by 1.5-fold) (Figure 5C), however, whereas eIF4G1-8 also partially increased the recovery of the uORF4-containing RNA segment (by ∼1.6-fold), the eIF4G1-1 mutant did not at all (Figure 5B). These findings nicely correspond to our β-galactosidase measurements at the elevated temperature (Figure 4B and D) further supporting our conclusion that the effect of eIF4G1-1 is uORF1-specific. It is important to emphasize that this effect is not attributable to loss of eIF4A-binding, but to the intactness of the eIF4G1-RNA2-binding domain, which distinguishes eIF4G1-1 from eIF4G1-8. In strong support, creating the stably expressed eIF4G1-8* mutant with no detectable growth phenotype (Figure 3C) where all three RNA2 domain mutations were preserved but V648G in the HEAT domain was fixed (Figure 3A and B) resulted in virtually no recovery difference between uORFs 1 and 4 (Figure 5B).
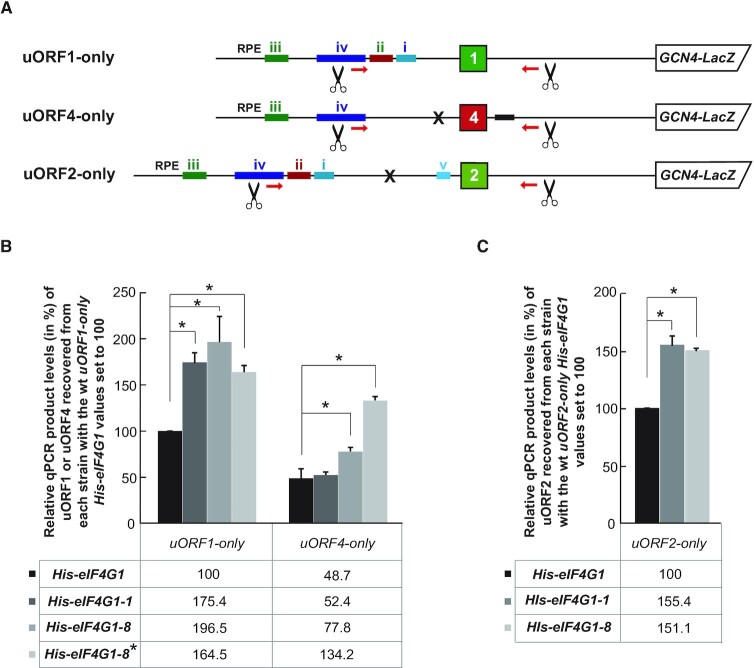
Figure 5.
The eIF4G1-1 mutant specifically increases retention of ribosomes elongating and terminating on REI-permissive uORFs versus REI-non-permissive uORF4. (A) Schematics showing simplified uORF1-, uORF2- and uORF4-only RaP-NiP constructs. (B) The YMP53, YMP55, YMP56 and YMP91 strains described in Figure 3 were introduced with either uORF1- or uORF4-only RaP-NiP constructs shown in panel A and treated as described in Figure 1B. Relative qPCR product levels (in %) of the uORF1 or uORF4 segments recovered from each strain with standard deviations were processed as described in Figure 2B with the values of the wt uORF1-only strain expressing His-eIF4G1 set to 100% (*P< 0.05). (C) The YMP53, YMP55 and YMP56 strains described in Figure 3 were introduced with uORF2-only RaP-NiP construct shown in panel A and treated as described in panel B (*P< 0.05).
All these genetic interactions can be explained as follows. Since the eIF4A helicase unwinds secondary structures in 5′ leaders of mRNAs, we speculate that the previously documented impairment of the eIF4G–4A interaction may increase the residency time of the post-termination 40S subunit at the stop codon of a REI-permissive uORF before its departure downstream, resulting in the observed higher RNA recovery in case of uORF1. This will not be important for REI-non-permissive uORFs, such as uORF4, because they lack RPEs and, therefore, the 40S is expected to be recycled rapidly irrespective of the presence or absence of eIF4A; i.e. the recovery does not differ from wt eIF4G.
Taking into account that no specific phenotype was observed with either of the mutants at 30°C in our reporter assays (Figure 4D), we also conclude that the RaP-NiP assay is more sensitive than β-galactosidase measurements, as noted before (11). It is noteworthy that this higher sensitivity may stem from a fact that with the RaP-NiP assay we directly capture all cross-linked post-termination species irrespective of their ability to eventually resume traversing downstream (which depends on eIF4A), whereas the β-galactosidase activity measurements naturally do rely on resumption of traversing. Therefore, the differences in the efficiency of the RNA segment recovery in the former case must be by definition more prominent when compared to the differences in activity measurements with the impaired eIF4A function in the latter case.
The eIF3-binding RPE i. of uORF1 genetically interacts with eIF4G in vivo
Having demonstrated that the mutated HEAT domain of eIF4G1 specifically increased co-purification of the uORF1-containing RNA segment with eIF4G1-1, we next examined the effect of eliminating the uORF1-specific cis-acting elements called RPEs (14) to find out if they closely cooperate with eIF4G1 or not. In particular, we focused our attention on previously identified mutations SUB40 and CAAII of RPE i and RPE ii, respectively; i.e. the RPEs that are utilized by uORF1 (14) (Figure 6A). As shown before in the His-eIF3a wt cells (11), both RPE mutations, when introduced into our uORF1-only Rap-NiP construct, significantly (by ∼35–50% and ∼70%) reduced the efficiency of the uORF1-containing RNA segment co-purification in His-eIF4G1 (Figure 6B), as well as His-eIF4A wt cells (Figure 6C). Interestingly, our analysis of eIF4G mutants revealed two things. First, the robust negative effect of the CAAII mutation on the co-purification efficiency is superior to the positive effect displayed by both eIF4G mutants, being perhaps even exacerbated (by additional ∼30%) when combined with His-eIF4G1-1 (Figure 6B) that we cannot explain. Nonetheless, it further supports our earlier conclusion that the structural integrity of RPE ii. is crucial for efficient REI and clearly demonstrates that there is neither direct nor functional interaction between RPE ii and eIF4G. And the second is that whereas there is also no genetic interaction between eIF4G1-1 and the SUB40 mutation of RPE i, altering the integrity of the RNA2 domain alone (in eIF4G1-8*) or combining it with the loss of the eIF4A binding (in eIF4G1-8) eliminated the negative effect of SUB40 on the recovered uORF1 mRNA fragment levels (Figure 6B). As RPE i. was shown to promote REI by binding to the a/Tif32 subunit of eIF3, this dominant genetic epistasis might indicate some functional interplay between eIF4G and eIF3 involving this specific RPE.
Figure 6.
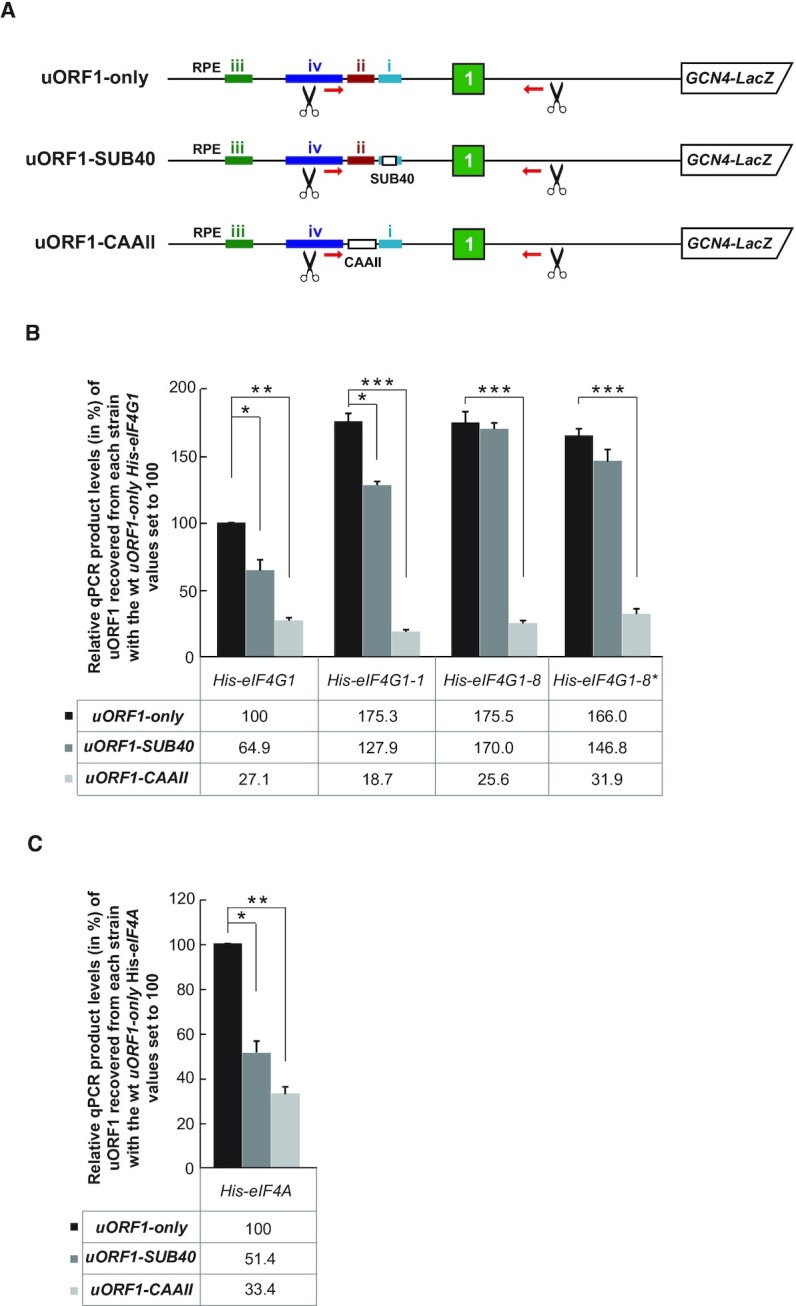
The eIF3-binding RPE i. of uORF1 genetically interacts with eIF4G in vivo. (A) Schematics showing uORF1-only RaP-NiP construct and its derivatives with substitutions in RPE i (uORF1-SUB40; substitution of nt -40 through -49 of RPE i. with the complementary sequence) or in RPE ii. (uORF1-CAAII; substitution of whole RPE ii. sequence [nt –55 through –76] with seven CAA repeats). (B) The YMP53, YMP55, YMP56 and YMP91 strains described in Figure 3 were introduced with the uORF1-only RaP-NiP construct or its derivatives shown in panel A and treated as described in Figure 1B. Relative qPCR product levels (in %) of the uORF1 segment recovered from each strain were processed as described in Figure 2B with the values of the wt uORF1-only strain expressing His-eIF4G1 set to 100% (*P< 0.05, **P< 0.005, ***P< 0.0005). (C) The strain YMP63 described in Figure 1C was introduced with the uORF1-only RaP-NiP construct or its derivatives shown in panel A and treated as described in panel B (*P< 0.05, **P< 0.005).
DISCUSSION
Several recent studies provided several hints indicating that eIF4F may play a specific role in REI, however, a direct evidence was lacking. Therefore, with the RaP-NiP technique that enabled us to unambiguously implicate eIF3 in the REI process in yeast by showing that it stays bound to ribosomes elongating and terminating on short upstream ORFs to render them REI-permissive (11), we wished to examine whether or not the components of the eIF4F complex promote REI by similar means in yeast too.
While this manuscript was in preparation, a seminal selective 40S/80S ribosome footprinting work from the Teleman's lab revealed that scanning in majority of tested human cell lines occurs in a cap-tethered way and that eIF4G1 and eIF4E components of human eIF4F remain bound to 80S ribosomes during an early elongation phase (with a decay half-length of ∼12 codons) (17). Since these authors together with the Wolf's lab and us expectedly demonstrated that, besides the eIF4F factors, early human elongation complexes also carry eIF3 (12,16), and earlier evidence suggested an active involvement of mammalian eIF3, eIF4G and eIF4A in promoting REI (15,22), they proposed a simple model explaining the molecular basis of REI in mammals. It postulates that since majority of mammalian uORFs are believed to be permissive for REI (6,35,36), the fact that the cap-tethered eIF4F complex and eIF3 are temporarily retained on 80S ribosomes as they translate short uORFs suffices to prevent full ribosomal recycling and allow prompt assembly of the post-termination 48S complex. This can then resume traversing downstream to reacquire eIF2-TC along the way to REI on the next AUG.
In yeast, however, the situation is different. (i) uORFs in yeast were shown to be primarily non-permissive for REI (35,37) and thus the ribosomes are generally not competent to reinitiate (35,37), unless they are surrounded by specific cis-acting elements, some of which act in concert with eIF3 (reviewed in (7,18)). In addition, (ii) scanning is believed to occur in the cap-severed and not tethered manner (12,38). Finally, (iii) yeast eIF4F binds the ribosome not via eIF3 but via eIF5 and eIF1 (39,40)}, which leave the ribosome upon 60S subunit joining (1). This raises the question of whether the yeast 4F complex can persist on elongating 80S ribosomes to promote REI like its mammalian counterpart, whose eIF4A component was recently shown to make direct contacts with the mammalian-specific e, k and l subunits of eIF3 near the mRNA exit pore on the solvent-exposed side of the 48S PIC (30). Based on the aforementioned mammalian model, the prospective lack of these factors and the contacts that they make on the yeast 80S ribosome could markedly weaken the affinity of eIF3 towards the early elongating 80S couple and thus dramatically compromise the efficiency of REI associated with ordinary uORFs (i.e. those lacking specific cis-acting elements).
However, findings presented in this study do not support this beautifully simple idea. First of all, we detected >2-fold higher amounts of the REI-permissive uORF1 and uORF2 RNA segments co-purifying with His-tagged eIF4G1 and eIF4A compared to the REI-non-permissive uORF3 and uORF4 segments in the corresponding deletion strains, but observed no difference with the His-tagged eIF4E cap-binding protein; analogous data were also obtained with uORFs from both YAP genes (Figures 1, 2 and Supplementary Figure S5). These findings plus the fact that the overall amounts of uORF1-specific segments that we recovered with all three His-tagged strains were relatively the same (Supplementary Figure S3C) have the following implications. (i) Similar to mammals (17), it seems that also in yeast the entire eIF4F complex is retained on ribosomes past the initiation phase. The scanning may occur in the cap-severed mode as proposed (38), our data do not imply anything in this respect, nonetheless, based on our results only the cap is most probably severed and not the eIF4E cap-binding protein. (ii) At the same time, however, it seems that the ribosomes elongating and terminating on REI-permissive uORFs are capable to preferentially increase retention of eIF4G and 4A factors over eIF4E. This strongly suggests that the former factors might actively promote REI after translation of short uORFs.
While there is no structural information on the yeast 48S PIC containing eIF4F available at present, taking into account the substantial conservation of the initiation pathway, it is conceivable to assume that the placement of eIF4F within the 48S PIC in yeast is similar to humans (30), only mediated by other actors (for example via eIF4G binding to eIFs 1 and 5 (39,40) and, perhaps, eIF4A binding to other eIF3 subunits). Therefore, we propose that the main players that could boost the efficiency of REI remain roughly the same in yeast and humans, perhaps only with a slightly modified mode of action. What makes the reported difference then? Could it be the opposing role of the post-termination 40S recycling factors eIF2D/DENR•MCTS1 in yeast versus mammals? Whereas they are believed to promote both ribosome recycling and REI in mammals (41,42), in yeast they drive terminating ribosomes into completion of a full ribosomal cycle (43). Therefore, one could postulate that in contrast to mammals, the post-termination 40S-bound eIFs in yeast must negotiate with eIF2D/DENR•MCTS1 to increase the odds of the post-termination decision making process towards REI.
Now let's return to the yeast case. Clearly, pure presence of a factor on early elongating ribosomes does not necessarily mean an active role in REI. The direct evidence implicating eIF4G in REI in yeast came with the eIF4G1-1 mutant that: (A) imparted a modest but statistically significant Gcn– phenotype (Figure 4), (B) specifically increased the efficiency of REI past the uORF1 over uORF4 translation in vivo (Figure 4) and (C) specifically increased retention of ribosomes elongating and terminating on REI-permissive uORFs 1 and 2 versus REI-non-permissive uORF4 (Figure 5). Similar results, however, lacking the specificity for REI-permissive versus non-permissive uORFs, were also obtained with the eIF4G1-8 mutant and its simpler variant eIF4G1-8*. Whereas 1–1 is deficient in recruiting eIF4A into the eIF4F complex and 1–8* has an altered integrity of the RNA2 RNA-binding domain, eIF4G1-8 is functionally the combination of both (23,32,33). Interestingly, RNA2 was shown to promote critical step(s) of initiation downstream of the eIF4F formation, most likely during scanning (34,44). Taken altogether, we propose that the fully intact RNA-binding ability of the eIF4G-RNA2 determines its specificity for REI-permissive versus non-permissive uORFs, whereas the preserved eIF4G-4A contact is required for (i) timely progression of partial ribosomal recycling and, subsequently, (ii) for resumption and processivity of traversing (later scanning). In other words, consistent, specific increases specified in B) and C) may suggest that the RNA2 binding domain of eIF4G makes direct contacts with some of the uORF1 or uORF2 RPEs, as demonstrated for eIF3 (11,14,19), that somehow modulate the progression of partial ribosomal recycling and/or resumption of traversing (Figure 7). Indeed, combining eIF4G1-8 and mainly 1–8* with the SUB40 mutation of RPE i. masked the negative effect of SUB40 on the REI efficiency (Figure 6). This example of genetic epistasis points to some molecular interplay between eIF4G and eIF3, because eIF3 was shown to specifically interact with RPE i. (11,14,19). What lies behind this genetic interaction from the mechanistic point of view is, however, unclear to us and remains to be elucidated.
Figure 7.
Model of the eIF4G contribution to the REI-permissiveness of uORF1 of yeast GCN4. The fully intact RNA-binding ability of the eIF4G-RNA2 domain allows for a specific interaction with the RPE i during uORF1 translation, perhaps in co-operation with eIF3 (A), whereas the preserved post-termination eIF4G-4A contact enables timely resumption and processivity of traversing (B).
An increase in the residency time of the post-termination 40S subunit at the stop codon of short uORFs in yeast and/or compromised processivity of traversing/scanning downstream, which would provide the 40S ribosome with more time to reacquire the eIF2-TC before arriving at the next (u)ORF, was—in connection with 1–1 and 1–8 mutations in the eIF4G2 isoform—already proposed by the Asano's lab (23). Besides the apparent increase in the REI efficiency, as observed by us, which was suppressible by increased levels of eIF4A, these authors also observed a defect in AUG recognition (increased leaky scanning) that we did not. This discrepancy may originate from the use of different eIF4G isoforms that, despite being functionally interchangeable (4), are only ∼50% identical in sequence to one another (45).
Taking altogether, here we provide, to our knowledge, the first direct in vivo evidence from yeast cells that, upon subunit joining, eIF4G along with eIF3 continue interacting with the 80S ribosome for a few elongation cycles to promote REI downstream. In addition, we conclude that since the basic REI players in yeast and humans seem to be relatively the same, more work is required to elucidate what else lies behind the difference in REI permissiveness of short uORFs between yeast and higher eukaryotes.
DATA AVAILABILITY
All data and reagents are freely available. Further information and requests for reagents should be direct to L.S.V. (valasekl@biomed.cas.cz).
Supplementary Material
ACKNOWLEDGEMENTS
We are thankful to Olga Krýdová and Kristína Koudelová for technical and administrative assistance, and to the members of the Valášek laboratory for helpful suggestions.
Contributor Information
Mahabub Pasha Mohammad, Laboratory of Regulation of Gene Expression, Institute of Microbiology AS CR, Prague, Videnska 1083, 142 20, Czech Republic.
Anna Smirnova, Laboratory of Regulation of Gene Expression, Institute of Microbiology AS CR, Prague, Videnska 1083, 142 20, Czech Republic.
Stanislava Gunišová, Laboratory of Regulation of Gene Expression, Institute of Microbiology AS CR, Prague, Videnska 1083, 142 20, Czech Republic.
Leoš Shivaya Valášek, Laboratory of Regulation of Gene Expression, Institute of Microbiology AS CR, Prague, Videnska 1083, 142 20, Czech Republic.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant of Excellence in Basic Research (EXPRO 2019) provided by the Czech Science Foundation [19-25821X]. Funding for open access charge: Grant of Excellence in Basic Research (EXPRO 2019) provided by the Czech Science Foundation [19-25821X].
Conflict of interest statement. None declared.
REFERENCES
- 1.Hinnebusch A.G.The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 2014; 83:779–812. [DOI] [PubMed] [Google Scholar]
- 2.Valášek L.S.Ribozoomin’ – translation initiation from the perspective of the ribosome-bound eukaryotic initiation factors (eIFs). Curr. Protein Pept. Sci. 2012; 13:305–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berset C., Zurbriggen A., Djafarzadeh S., Altmann M., Trachsel H.. RNA-binding activity of translation initiation factor eIF4G1 from Saccharomyces cerevisiae. RNA. 2003; 9:871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarkson B.K., Gilbert W.V., Doudna J.A.. Functional overlap between eIF4G isoforms in Saccharomyces cerevisiae. PLoS One. 2010; 5:e9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozak M.Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol. Cell. Biol. 1987; 7:3438–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kozak M.Constraints on reinitiation of translation in mammals. Nucleic Acids Res. 2001; 29:5226–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gunisova S., Hronova V., Mohammad M.P., Hinnebusch A.G., Valasek L.S.. Please do not recycle! translation reinitiation in microbes and higher eukaryotes. FEMS Microbiol. Rev. 2018; 42:165–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wethmar K., Barbosa-Silva A., Andrade-Navarro M.A., Leutz A.. uORFdb–a comprehensive literature database on eukaryotic uORF biology. Nucleic Acids Res. 2014; 42:D60–D67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Somers J., Poyry T., Willis A.E.. A perspective on mammalian upstream open reading frame function. Int. J. Biochem. Cell Biol. 2013; 45:1690–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinnebusch A.G., Ivanov I.P., Sonenberg N.. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science. 2016; 352:1413–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohammad M.P., Munzarova Pondelickova V., Zeman J., Gunisova S., Valasek L.S.. In vivo evidence that eIF3 stays bound to ribosomes elongating and terminating on short upstream ORFs to promote reinitiation. Nucleic Acids Res. 2017; 45:2658–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner S., Herrmannova A., Hronova V., Gunisova S., Sen N.D., Hannan R.D., Hinnebusch A.G., Shirokikh N.E., Preiss T., Valasek L.S.. Selective translation complex profiling reveals staged initiation and co-translational assembly of initiation factor complexes. Mol. Cell. 2020; 79:546–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szamecz B., Rutkai E., Cuchalova L., Munzarova V., Herrmannova A., Nielsen K.H., Burela L., Hinnebusch A.G., Valášek L.. eIF3a cooperates with sequences 5′ of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRNA. Genes Dev. 2008; 22:2414–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munzarová V., Pánek J., Gunišová S., Dányi I., Szamecz B., Valášek L.S.. Translation reinitiation relies on the interaction between eIF3a/TIF32 and progressively folded cis-Acting mRNA elements preceding short uORFs. PLos Genet. 2011; 7:e1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hronova V., Mohammad M.P., Wagner S., Panek J., Gunisova S., Zeman J., Poncova K., Valasek L.S.. Does eIF3 promote reinitiation after translation of short upstream ORFs also in mammalian cells. RNA biol. 2017; 14:1660–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin Y., Li F., Huang L., Polte C., Duan H., Fang J., Sun L., Xing X., Tian G., Cheng Y.et al.. eIF3 associates with 80S Ribosomes to promote translation elongation, mitochondrial homeostasis, and muscle health. Mol. Cell. 2020; 79:575–587. [DOI] [PubMed] [Google Scholar]
- 17.Bohlen J., Fenzl K., Kramer G., Bukau B., Teleman A.A.. Selective 40S footprinting reveals Cap-Tethered Ribosome scanning in human cells. Mol. Cell. 2020; 79:561–574. [DOI] [PubMed] [Google Scholar]
- 18.Hinnebusch A.G.Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005; 59:407–450. [DOI] [PubMed] [Google Scholar]
- 19.Gunisova S., Valasek L.S.. Fail-safe mechanism of GCN4 translational control-uORF2 promotes reinitiation by analogous mechanism to uORF1 and thus secures its key role in GCN4 expression. Nucleic Acids Res. 2014; 42:5880–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dever T.E., Feng L., Wek R.C., Cigan A.M., Donahue T.D., Hinnebusch A.G.. Phosphorylation of initiation factor 2a by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992; 68:585–596. [DOI] [PubMed] [Google Scholar]
- 21.Gunisova S., Beznoskova P., Mohammad M.P., Vlckova V., Valasek L.S.. In-depth analysis of cis-determinants that either promote or inhibit reinitiation on GCN4 mRNA after translation of its four short uORFs. RNA. 2016; 22:542–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pöyry T.A., Kaminski A., Jackson R.J.. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame. Genes Dev. 2004; 18:62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watanabe R., Murai M.J., Singh C.R., Fox S., Ii M., Asano K.. The eukaryotic initiation factor (eIF) 4G HEAT domain promotes translation re-initiation in yeast both dependent on and independent of eIF4A mRNA helicase. J. Biol. Chem. 2010; 285:21922–21933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skabkin M.A., Skabkina O.V., Hellen C.U., Pestova T.V.. Reinitiation and other unconventional posttermination events during eukaryotic translation. Mol. Cell. 2013; 51:249–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imataka H., Sonenberg N.. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cel Biol. 1997; 17:6940–6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LeFebvre A.K., Korneeva N.L., Trutschl M., Cvek U., Duzan R.D., Bradley C.A., Hershey J.W., Rhoads R.. Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J. Biol. Chem. 2006; 281:22917–22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Villa N., Do A., Hershey J.W., Fraser C.S.. Human eukaryotic initiation factor 4G (eIF4G) binds to eIF3c, -d, and -e to promote mRNA recruitment to the ribosome. J. Biol. Chem. 2013; 288:32932–32940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grant C.M., Hinnebusch A.G.. Effect of sequence context at stop codons on efficiency of reinitiation in GCN4 translational control. Mol. Cell. Biol. 1994; 14:606–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen K.H., Valášek L.. In vivo deletion analysis of the architecture of a multi-protein complex of translation initiation factors. Methods Enzymol. 2007; 431:15–32. [DOI] [PubMed] [Google Scholar]
- 30.Brito Querido J., Sokabe M., Kraatz S., Gordiyenko Y., Skehel J.M., Fraser C.S., Ramakrishnan V.. Structure of a human 48S translational initiation complex. Science. 2020; 369:1220–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vilela C., Linz B., Rodrigues-Pousada C., McCarthy J.E.. The yeast transcription factor genes YAP1 and YAP2 are subject to differential control at the levels of both translation and mRNA stability. Nucleic Acids Res. 1998; 26:1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tarun S.Z. Jr, Sachs A.B.. Binding of eukaryotic translation initiation factor 4E (eIF4E) to eIF4G represses translation of uncapped mRNA. Mol. Cell. Biol. 1997; 17:6876–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neff C.L., Sachs A.B.. Eukaryotic translation initiation factors eIF4G and eIF4A from Saccharomyces cerevisiae physically and functionally interact. Mol. Cell. Biol. 1999; 19:5557–5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park E.H., Walker S.E., Lee J.M., Rothenburg S., Lorsch J.R., Hinnebusch A.G.. Multiple elements in the eIF4G1 N-terminus promote assembly of eIF4G1*PABP mRNPs in vivo. EMBO J. 2011; 30:302–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson R.J., Hellen C.U., Pestova T.V.. Termination and post-termination events in eukaryotic translation. Adv. Protein Chem. Struct. Biol. 2012; 86:45–93. [DOI] [PubMed] [Google Scholar]
- 36.Bohlen J., Harbrecht L., Blanco S., Clemm von Hohenberg K., Fenzl K., Kramer G., Bukau B., Teleman A.A.. DENR promotes translation reinitiation via ribosome recycling to drive expression of oncogenes including ATF4. Nat. Commun. 2020; 11:4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yun D.F., Laz T.M., Clements J.M., Sherman F.. mRNA sequences influencing translation and the selection of AUG initiator codons in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 1996; 19:1225–1239. [DOI] [PubMed] [Google Scholar]
- 38.Archer S.K., Shirokikh N.E., Beilharz T.H., Preiss T.. Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature. 2016; 535:570–574. [DOI] [PubMed] [Google Scholar]
- 39.Asano K., Shalev A., Phan L., Nielsen K., Clayton J., Valášek L., Donahue T.F., Hinnebusch A.G.. Multiple roles for the carboxyl terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO J. 2001; 20:2326–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He H., von der Haar T., Singh C.R., Ii M., Li B., Hinnebusch A.G., McCarthy J.E., Asano K.. The yeast eukaryotic initiation factor 4G (eIF4G) HEAT domain interacts with eIF1 and eIF5 and is involved in stringent AUG selection. Mol. Cell. Biol. 2003; 23:5431–5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schleich S., Strassburger K., Janiesch P.C., Koledachkina T., Miller K.K., Haneke K., Cheng Y.S., Kuchler K., Stoecklin G., Duncan K.E.et al.. DENR-MCT-1 promotes translation re-initiation downstream of uORFs to control tissue growth. Nature. 2014; 512:208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skabkin M.A., Skabkina O.V., Dhote V., Komar A.A., Hellen C.U., Pestova T.V.. Activities of ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010; 24:1787–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Young D.J., Makeeva D.S., Zhang F., Anisimova A.S., Stolboushkina E.A., Ghobakhlou F., Shatsky I.N., Dmitriev S.E., Hinnebusch A.G., Guydosh N.R.. Tma64/eIF2D, Tma20/MCT-1, and Tma22/DENR recycle post-termination 40S subunits in vivo. Mol. Cell. 2018; 71:761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pestova T.V., Kolupaeva V.G.. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002; 16:2906–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goyer C., Altmann M., Lee H.S., Blanc A., Deshmukh M., Woolford J.L., Trachsel H., Sonenberg N.. TIF4631 and TIF4632: Two yeast genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor 4F) contain an RNA recognition motif-like sequence and carry out an essential function. Mol. Cell. Biol. 1993; 13:4860–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and reagents are freely available. Further information and requests for reagents should be direct to L.S.V. (valasekl@biomed.cas.cz).