Abstract
Architectural DNA-binding proteins (ADBPs) are abundant constituents of eukaryotic or bacterial chromosomes that bind DNA promiscuously and function in diverse DNA reactions. They generate large conformational changes in DNA upon binding yet can slide along DNA when searching for functional binding sites. Here we investigate the mechanism by which ADBPs diffuse on DNA by single-molecule analyses of mutant proteins rationally chosen to distinguish between rotation-coupled diffusion and DNA surface sliding after transient unbinding from the groove(s). The properties of yeast Nhp6A mutant proteins, combined with molecular dynamics simulations, suggest Nhp6A switches between two binding modes: a static state, in which the HMGB domain is bound within the minor groove with the DNA highly bent, and a mobile state, where the protein is traveling along the DNA surface by means of its flexible N-terminal basic arm. The behaviors of Fis mutants, a bacterial nucleoid-associated helix-turn-helix dimer, are best explained by mobile proteins unbinding from the major groove and diffusing along the DNA surface. Nhp6A, Fis, and bacterial HU are all near exclusively associated with the chromosome, as packaged within the bacterial nucleoid, and can be modeled by three diffusion modes where HU exhibits the fastest and Fis the slowest diffusion.
INTRODUCTION
A number of single-molecule studies have shown that DNA-binding proteins can travel along the DNA duplex by a thermal energy-driven (passive) process. One-dimensional diffusion occurs bidirectionally and allows the DNA-binding protein to interrogate DNA sequence or structural information over a DNA region. A combination of 3-D diffusion, involving complete dissociation and rebinding to a separate DNA segment, intersegmental transfer processes and 1-D diffusion enables DNA-binding proteins to rapidly search for their targets (1–7). Some DNA-binding proteins have been shown to undergo extensive 1-D diffusion or sliding (8–17), whereas others are believed to rely more on 3-D search processes to find their specific targets (18,19). In Escherichia coli, the well-studied Lac repressor spends most of its time searching for its operator by 1-D diffusion along the chromosome with frequent 3-D excursions (20–22).
1-D diffusion of proteins along DNA could occur by structurally distinct mechanisms (2–7). The protein may remain intimately associated with one or both DNA grooves, whereby it rotates around the DNA helix as it follows the grooves (Figure 1A). Support for rotation-coupled sliding by some proteins comes from the relative insensitivity of 1-D diffusion rates with monovalent ion concentration, implying low frequency dissociation, and by the relationship of sliding rates with protein size and frictional drag (10,11,13,23,24). Alternatively, a protein may unbind from the DNA groove(s) and move along the DNA surface with its positively charged DNA-binding surface continuously connected to the electronegative field of the DNA backbone (referred to here as DNA surface sliding) (Figure 1B). DNA surface sliding can include rotational movement along the DNA backbone rails and rotation-uncoupled translational motion across the rails. 1-D diffusion rates can be augmented by short-range hopping to separate segments along the DNA molecule (Figure 1C) (14,25–29). Hopping, whereby the protein transiently dissociates from DNA, is expected to be sensitive to monovalent ion concentration (2,30) and to hydrodynamic forces of buffer flow when present in vitro. Intersegmental transfer reactions, also referred to as DNA-facilitated exchange, have been shown to occur by the proteins studied in this work (31–34). Direct exchange to a nearby segment on the same DNA molecule, however, will require looping that will be inhibited on DNAs elongated in vitro, except for special systems employing juxtaposed DNAs (14,35).
Figure 1.
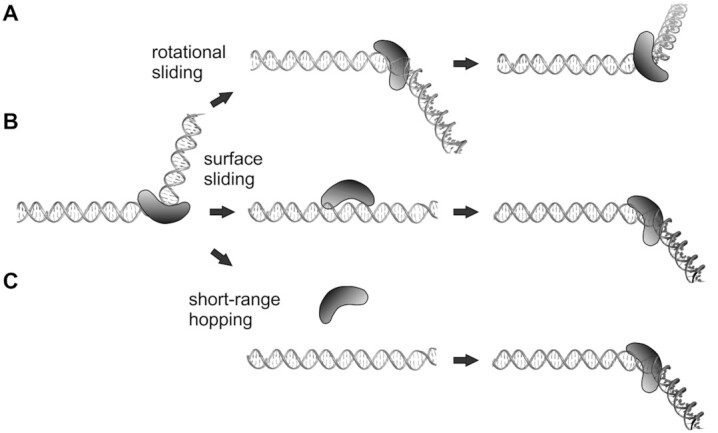
Mechanisms of 1-D diffusion by proteins along DNA. For schematic purposes the protein (ADBP) is depicted as the crescent-shaped HMGB domain, which binds within an expanded DNA minor groove and results in bending of the DNA helical axis. (A) Rotation-coupled sliding. The protein remains within the minor groove as it rotationally slides around the DNA helix. The protein in the middle and right models have traveled 4 and 9 bp, respectively, in a manner requiring continuous conformational changes to the DNA structure. (B) Sliding along the surface of DNA. The protein unbinds from the minor groove but remains electrostatically associated with the DNA surface as it travels to a new DNA segment where it rebinds within the minor groove. (C) Short-range hopping. The protein dissociates from the DNA electrostatic field, diffuses in solution (middle panel) and rebinds at a new location (right panel). Short-range hopping is considered unlikely to be observed under the hydrodynamic force of buffer flow used in the in vitro imaging studies reported here but can occur in vivo. An additional mode of diffusion that is not illustrated occurs by intersegmental transfer reactions, which, within the same DNA molecule, will require DNA looping. Although the ADBPs studied here engage in intersegmental transfer, these reactions will also be disfavored on DNA molecules extended by buffer flow.
Proteins that surround the DNA helix can diffuse on DNA by additional mechanisms. For example, TALE proteins, which form a superhelix that wraps around DNA, are able to rapidly slide over the DNA surface by a rotation-uncoupled mechanism and without intimate DNA contact, as highlighted by the ability of hydrodynamic flow to effectively push the protein unidirectionally along DNA under physiologic or higher salt concentrations (36,37). Proliferating Cell Nuclear Antigen (PCNA), which forms a donut structure around DNA, is believed to travel along DNA using both rotation-coupled and -uncoupled mechanisms (38,39). Other specialized proteins such as the MutS mismatch repair proteins (14,40) and a Type III restriction enzyme (41) have been shown to adopt rotation-uncoupled translational diffusion modes in an ATP-dependent manner.
We showed in a previous report that several abundant eukaryotic chromatin-associated or bacterial nucleoid-associated architectural DNA-binding proteins (ADBPs) support 1-D diffusion along DNA (17). These proteins have broad roles in DNA biology, including transcription, replication, recombination and DNA damage repair reactions, nucleosome dynamics (Nhp6A), and chromosome organization and packaging (42–48). The defining property of ADBPs is the distortion of DNA structure within stably bound complexes. The conformational changes include local bending of the DNA axis, changes in helical twist, and changes in major and/or minor groove widths. We noted that even though these proteins could slide over tens of kilobases with continuous DNA contact, they exhibited a high free-energy barrier of 1-D diffusion as compared to other DNA-binding proteins (17). The high free-energy barrier was attributed to the DNA conformational changes that are coupled with binding. Numerous studies have documented the impact of base sequence on DNA conformation (49–52). If ADBPs remain bound within the DNA grooves when diffusing along DNA, continual changes in DNA conformation would be required, leading to a rough and uneven energy landscape as the protein travels along mixed sequence DNA (Figure 1A). On the other hand, sliding along the electronegative surface of B-DNA, followed by re-insertion into the groove(s), would not have such constraints (Figure 1B). To account for the high free energy barrier, Levin and Levy (53) proposed that APDBs diffuse on DNA using a similar binding interface as in the stably bound state. Thus, protein residues involved in stable complex formation will be continually engaging with DNA during diffusion, thereby slowing diffusion rates. This model does not consider the energetic costs of the DNA conformational changes required for translocation over each base-pair step.
In this report, we investigate mechanisms by which Nhp6A, an abundant yeast chromatin-associated HMGB protein, and Fis, an abundant bacterial nucleoid-associated protein, support 1-D diffusion on DNA by examining the consequences of particular amino acid changes in vitro. The mutations were chosen to test protein-specific features of DNA interactions, particularly as they relate to DNA structure in the bound complexes, and were guided by atomic structures of the protein-DNA complexes and previously identified biochemical properties of the mutant proteins. The experimental work with Nhp6A was complemented with molecular dynamics simulations to provide a structural model of the 1-D diffusion process. We also examine the consequences of these mutations on diffusion in E. coli. The in vivo dynamics of Nhp6A and Fis are compared with the abundant bacterial nucleoid-associated protein HU, whose in vitro mobility properties have been determined previously (17). HU and Nhp6A can be considered functional homologs in some respects, even though they are structurally unrelated.
MATERIALS AND METHODS
Preparation of fluorescently labeled proteins for in vitro studies
Proteins without tags but having single or double cysteines for labeling at locations outside the DNA-binding interface were expressed from pET11a-derived plasmids in E. coli (Supplementary Table S1). Pseudo-WT Nhp6A and the double mutant M29A + F48A contain a cysteine substituted at residue 2 and a second cysteine added to the C-terminal end. The Nhp6A N-terminal deletion mutant Δ(2–16) contains a single cysteine added to the C-terminus. The Nhp6A proteins were purified as described in Yen et al. (54). The pseudo wild-type Fis and all Fis mutant derivatives contained a single cysteine substituted for Gln21 within the N-terminal β-hairpin arms that do not participate in DNA binding. Fis proteins were purified as described in Stella et al. (55). Proteins were labeled by Atto488 (ATTO-TEC) using maleimido chemistry as described in Kamagata et al. (17). Protein and dye concentrations were determined by the Bradford method and absorbance of dyes.
Ensemble dissociation and single-molecule sliding measurements in vitro
Extended DNA arrays with phage λ DNA (New England Biolabs) were prepared in a flow cell as described in Igarashi et al. (56). The flow cell was set on an objective-type total internal reflection fluorescence (TIRF) microscope (DMI6000, Leica). A 488-nm laser was introduced into the objective lens with N.A. = 1.49 in TIRF geometry, and the fluorescence was detected by an EM-CCD camera (DU-897, Andor). For the single molecule measurements of Fis mutants in 150 mM potassium glutamate (Kglu), we used another TIRF microscope with highly inclined thin illumination (HILO) setup of the 488-nm laser (IX-73, Olympus) and an EM-CCD camera (iXon Ultra 888) (17).
For ensemble lifetime measurements, 0.5–2 nM Nhp6A proteins or 0.3–0.5 nM Fis proteins in a solution containing 20 mM HEPES, 1 mM EDTA, 1 mM DTT, 2 mM trolox, 0.2 mg/mL BSA and 50 mM Kglu at pH 7.2 were introduced into the flow cell using a syringe pump (Chemyx). After binding of the fluorescent proteins to DNA, buffer containing the same components except for 100 mM Kglu was introduced, and the fluorescent intensity change of DNAs was measured under 0.6 ml/min of the flow at 0.1 s exposures with 1 s intervals at 22°C. The intensity of 488-nm laser in the TIRF setup was reduced to minimize photobleaching of Atto488. Average fluorescent intensity of at least four DNA molecules was analyzed using ImageJ (57).
For single-molecule measurements, 0.005–0.7 nM Fis mutants or 0.02–0.5 nM Nhp6A mutants in a solution containing 20 mM HEPES, 1 mM EDTA, 1 mM DTT, 2 mM trolox, 0.1–0.2 mg/ml BSA and 50 mM or 150 mM Kglu at pH 7.2 were introduced into the flow cell. The movements of the molecules along DNA were measured under 0.6 ml/min of the flow at 50 ms exposures with 104 ms intervals in 50 mM Kglu or at 20 ms exposures with 44 ms intervals in 150 mM Kglu at 22°C. Fluorescent spots of single proteins labeled by Atto488 were tracked using ImageJ with the plugin ‘Particle track and analysis’. In 50 mM Kglu experiments, molecules absorbed on the surface or stacked on the DNA end were removed by cropping the image area for tracking. Trajectories with at least six points were used in the analysis. To assess the effect of DNA fluctuation (58), we calculated a periodogram over the frequency for the transverse motion of Nhp6A-wt (Supplementary Figure S1). In the periodogram, there was little deviation from the flat spectrum, indicating that DNA fluctuation was averaged out in our experimental time range. In 150 mM Kglu, we tracked the molecules in the observation area and selected the molecules bound to DNAs as described in Kamagata et al. (59). Trajectories with at least 10 points were used in the analysis. MSD and distribution analyses were performed as described in Kamagata et al. (17,59). Average D values were calculated by fitting the ensemble-averaged MSD plots (5 data points in 50 mM Kglu or 9 data points in 150 mM Kglu) with 2Dt + a, where a is the offset corresponding to localization error and DNA fluctuation. The standard error of each data point was used as the fitting weight. Displacements were calculated from all pairs of positions of a molecule at time intervals of 312 ms in 50 mM Kglu or 176 ms in 150 mM Kglu for all trajectories. For the fitting of displacement distribution with two diffusion components, we used the equation:
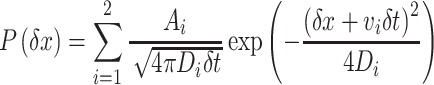 |
(1) |
where δt, δx, P(δx), Ai, vi and Di represent time interval, displacement in the time interval, occurrence of δx, amplitude of the ith mode, drift velocity of the ith mode and diffusion coefficient of the ith mode, respectively (17,60). The standard error for each displacement bin was estimated using a bootstrap method with 1000 replicates and was used as the fitting weight. The bin sizes were empirically selected in a manner consistent with the bin sizes estimated by N0.5 or 2N1/3, where N denotes the sample number. Note that Di values include the measurement error. In some cases, we used a one diffusion component equation. These analyses were conducted using an Excel macro and Igor. Levenberg-Marquardt least-squares fitting was used, and the errors were calculated as parameter uncertainties using Igor. The drift velocity of the mobile state of Nhp6A-wt was calculated to be 0.30 μm/s, which was ∼1000-fold smaller than the flow speed estimated in a similar condition (60). Welch’s t test was used to evaluate significant differences between the mutants and WT. It was assumed that the estimated parameters have a Gaussian distribution. The parameter errors estimated from the fitting were used as the parameter variance, and the sample number of each parameter was the total sample number multiplied by the mode fraction.
Single-molecule measurements in vivo
Low amounts of ADBPs fused with eGFP were generated by basal expression from pBR322 (lac promoter) or pET11a (T7 promoter) plasmids in E. coli MG1655-derived strains RJ3909 or RJ3455 (Δfis) containing lacIQ and lacking the T7 RNA polymerase (Supplementary Table S1). Nhp6A C-terminal GFP fusions (61) and Fis N-terminal fusions (62,63) have no demonstrable effect on DNA dynamics. C-terminal FP fusions to HupB (employed here) and HupA have been used in other studies (64–68). Cells grown in M9 media with 0.2% glucose, 0.1% casamino acids and 10 μM FeSO4 to OD600 ∼0.3 were concentrated about 5-fold by brief centrifugation, and 1–2 μl was applied to 1.5% agarose pads in the same media. In some experiments, cell growth and agarose pads employed MOPS-rich media (Teknova). After photobleaching excess fluorescent molecules to enable single-molecule imaging, a series of fluorescence images at 10 ms exposures with 30.8 ms intervals at 22°C were obtained using a TIRF microscope with a 488 nm laser in HILO illumination mode. Fluorescent spots were tracked using ImageJ with the plugin ‘Particle track and analysis’. Tracks with at least six points were used in the analysis. D was calculated by fitting the ensemble-averaged MSD plots (five data points) with 4D + a. The standard error of each data point was used as the fitting weight. The diffusion coefficients for primarily cytoplasmically-associated proteins that undergo rapid diffusion are likely underestimates because only a subset of molecules could be tracked over ≥6 consecutive images. For diffusion analysis of each molecule, we used initial five displacements of trajectories (trajectories longer than six points were truncated) and calculated apparent diffusion coefficient (D*) values of individual molecules as described by Stracy et al. (67,69). The distribution of D* values was fit with the following equation:
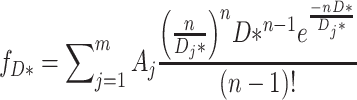 |
(2) |
where m, Aj, n and Dj denote the total number of the diffusion mode, the amplitude of the jth mode, the number of steps in the trajectory and the diffusion coefficient of the jth mode, respectively. We used n = 5. The standard error for each D* bin was estimated using a bootstrap method with 1000 replicates and was used as the fitting weight. Levenberg-Marquardt least-squares fitting was used, and the errors were calculated as parameter uncertainties using Igor. For determining m values for Equation (2), we initially used residuals between the experimental data and best-fitted curves. If several continuous residuals deviated from 0 in the positive or negative region, a larger m value was then tested. We also calculated AIC values for each m value and confirmed the lower AIC values with higher m values to support the use of a particular m value in the fittings. Finally, we used a chi-square test for evaluating the hypothesis that the observed frequency of each D value matches the expected one of theoretical fitted curves with alpha (significance level) = 0.01. The Welch’s t test, as performed for the in vitro data, was used to evaluate significant differences between the mutants and WT.
Molecular dynamics simulations
Coarse-grained (CG) molecular dynamics (MD) simulations were performed on Nhp6A-wt, Nhp6A-Δ(2–16) and Nhp6A-MF mutant proteins. For each system, we placed Nhp6A, modeled using PDB structure 1J5N (70), near a 50-bp random-sequence double-stranded DNA. The 1-bead-per-amino-acid AICG2 + model (71) was used for the interactions among protein particles. Specifically, a native-structure-dependent nonlocal potential was used to keep the HMGB domain folded, while flexible local potentials were used for the N-terminal arm (71). DNA was modeled with the 3SPN.2C model (72), where each nucleotide is represented by three CG particles, namely, base, sugar and phosphate. We used a variation of the recently-developed PWMcos method (73), which considers electrostatics, excluded volume interactions and semi-structure-based protein–base interactions, to model the binding between Nhp6A and DNA. Details regarding CG potentials can be found in the Supplementary Methods.
All production simulations were carried out with the MD package Cafemol (74). We conducted Langevin dynamics at the temperature T = 300 K and the ionic concentration IC = 150 mM. No periodic boundary condition was used; instead, an upper limit of 100 Å was set for the distances between the center-of-mass of Nhp6A and DNA. Twenty independent simulations of 108 MD steps were performed for each protein (wild-type or mutants).
To quantitatively analyze the binding pattern, we used a vector to describe the DNA-binding interface of Nhp6A for each simulated structure (Λ):
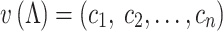 |
(3) |
where 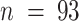 is the number of amino-acids in Nhp6A-wt, and
is the number of amino-acids in Nhp6A-wt, and 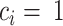 if residue
if residue 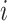 is within 10 Å of DNA, otherwise 0. Using this definition, a Euclidean distance matrix was constructed for 2000 structures randomly chosen from the MD trajectories. By applying the DBSCAN method (75), the 2000 selected structures were classified into two clusters (Supplementary Figure S2). Using the support vector classification method (76), we were able to assign a binding mode for any simulated structure with respect to the features of the two clusters. For more details of the clustering and classification methods, see the Supplementary Methods and Supplementary Figures S2–S5.
is within 10 Å of DNA, otherwise 0. Using this definition, a Euclidean distance matrix was constructed for 2000 structures randomly chosen from the MD trajectories. By applying the DBSCAN method (75), the 2000 selected structures were classified into two clusters (Supplementary Figure S2). Using the support vector classification method (76), we were able to assign a binding mode for any simulated structure with respect to the features of the two clusters. For more details of the clustering and classification methods, see the Supplementary Methods and Supplementary Figures S2–S5.
RESULTS
The yeast chromatin protein Nhp6A
Background
Saccharomyces cerevisiae Nhp6A is an abundant monomeric chromatin-associated protein whose nuclear levels approach those of nucleosomes (77–81). Like its mammalian counterpart HMGB1 and 2, Nhp6A collaborates with other specialized DNA-binding proteins to facilitate the assembly and function of multi-component nucleoprotein structures such as Pol II and Pol III transcription preinitiation complexes (79,82–86). Nhp6A has been shown to function in DNA repair by targeting MSH2 and MSH6 to DNA mismatches (87) and can substitute for HMGB1/2 in formation of RAG1/2 recombination complexes (88). It also functions with chromatin modifying complexes like yeast FACT and SWI/SNF to facilitate transcription and replication (80,89–94). In these diverse reactions, Nhp6A is believed to function largely in a DNA architectural manner where bending of DNA is involved.
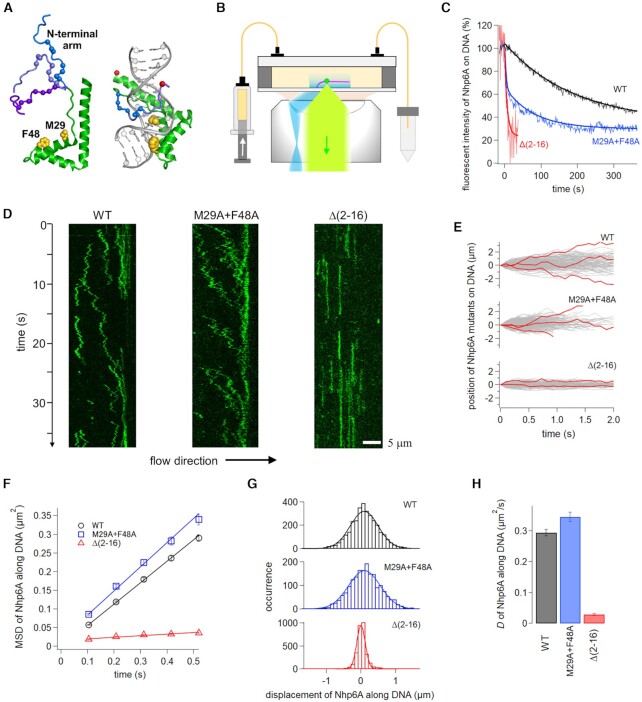
Nhp6A contains a 77 amino acid residue HMGB domain linked to a 16 residue unstructured N-terminal arm containing two clusters of basic residues (Figure 2A). Previous electrophoretic mobility shift assays (EMSA) have shown that Nhp6A binds DNA in a near sequence-neutral manner in vitro (78). However, NMR-based analyses of Nhp6A binding to different short DNA duplexes revealed short-range sequence preferences (95). In vivo, Nhp6A binds preferentially within nucleosome-depleted regions of yeast chromosomes, corresponding to intergenic promoter-regulatory regions of Pol II genes as well as in Pol III genes and localized regions in telomeres (96). The HMGB domain binds within an expanded minor groove, introducing considerable curvature into the DNA duplex, in part due to insertion of the Phe48 and Met29 side chains into the base stack (70,95) (Figure 2A). Mutants lacking either one or both of these side chains remain able to form DNA complexes stable to electrophoresis under low salt conditions with moderately lower affinities and with reduced DNA curvature as compared to Nhp6A-wt (70). Moreover, mutants lacking Phe48 are no longer targeted to cisplatin-induced DNA kinks (97). Binding of Nhp6A is strongly stabilized by the N-terminal arm that contains two patches of tandem arginines or lysines (residues 8–10 and 13–16) (54). NMR data indicate that the N-terminal arm dynamically wraps around the DNA complex primarily through the major groove (70). Without the N-terminal arm, Nhp6A cannot form DNA complexes stable to electrophoresis, and ligation of DNA microcircles, which require robust DNA bending, occurs only at very high protein concentrations (54).
Figure 2.
Single-molecule fluorescence measurements in vitro demonstrate that Nhp6A slides along DNA using its N-terminal basic arm. (A) Unbound and DNA-bound structures of Nhp6A (PDB codes: 1LWM and 1J5N, respectively). The HMGB domain is green with DNA inserting residues Met29 and Phe48 yellow. Basic residues within the flexible N-terminal arm (three examples from the NMR ensemble shown) are denoted with blue spheres, and positions of the fluorescent dyes in the DNA complex are shown as red spheres. (B) Fluorescence imaging set-up of proteins (green dot) in a flow cell with DNA array (pink) using TIRF illumination (blue) and detection (light green). (C) Time courses of dissociation of Nhp6A mutants from DNA in 100 mM Kglu. Solid curves are fitting curves by single or double exponentials. (D) Typical kymographs of Nhp6A molecules (green) along DNA in 50 mM Kglu. (E) Single-molecule traces of Nhp6A mutants along DNA. Red traces highlight individual trajectories. (F) Time courses of mean square displacements (MSDs) along DNA for Nhp6A mutants. The error denotes the standard error of all data pairs within a given interval. The solid lines represent the best fitted line with the slope of 2D. (G) Displacement distributions of Nhp6A mutants along DNA in 312 ms intervals. Solid curves were fitting curves by a single Gaussian function (Equation 1). (H) Average diffusion coefficients of Nhp6A mutants for 1-D sliding along DNA. Errors denotes the fitting error of the data in panel (F).
Residence times of Nhp6A mutants on DNA
DNA binding lifetimes by wild-type (wt) and mutant Nhp6A proteins were measured at the ensemble level by briefly incubating Atto488-labeled protein in buffer containing 50 mM K glutamate (Kglu) with an array of λ DNA molecules extended in a flow cell by hydrodynamic force (Figure 2B). The flow was then shifted to protein-free buffer with 100 mM Kglu, and bulk dissociation rates were followed by TIRF microscopy. Dissociation of Nhp6A-wt proteins fit a single exponential decay curve with a time constant of 191 ± 3 s (Figure 2C). The relatively long lifetime by the wild-type protein in protein-free buffer is in agreement with earlier studies employing single-DNA molecules (32,34,98). By contrast, lifetimes of DNA complexes by Nhp6A-Δ(2–16) that is missing the basic N-terminal arm were extremely short (7 ± 1 s). DNA complexes by the double alanine substitution mutant Nhp6A-M29A + F48A (Nhp6A-MF), which lacks both DNA intercalating side chains, decayed in a biphasic manner with about half the proteins dissociating rapidly (3.4 ± 0.2 s) and the remainder associated with DNA in a relatively stable manner. The stable population may reflect the mutant proteins bound to DNA segments whose sequence can readily adopt the curvature and groove width changes required for binding of the HMGB domain.
Single-molecule tracking of Nhp6A mutants on DNA
The effect of mutations on the movement of single Nhp6A proteins on DNA arrays was measured using TIRF imaging (Figure 2B). Previous studies on DNA mobility by Nhp6A-wt molecules were performed in buffer containing 150 mM Kglu (17). These experiments showed wild-type molecules bound DNA in a mobile mode, supporting bidirectional diffusion with a coefficient, D, of 0.336 ± 0.005 μm2/s or an average moving distance of 2.4 kb in 1 s. In 150 mM Kglu buffer, both Nhp6A mutants Δ(2–16) and MF did not form sufficiently stable complexes to enable tracking. However, tracking was possible by lowering the Kglu concentration to 50 mM. As shown in the kymographs (Figure 2D) and the single-molecule traces of travel times up to ∼2 s (Figure 2E), Nhp6A-wt and -MF mutant proteins travel bidirectionally on DNA. However, the Nhp6A-MF kymograph reveals a directional bias over long time frames that correlates with the direction of buffer flow. Remarkably, Nhp6A-Δ(2–16) molecules appear immobile on DNA (Figure 2D and E).
We quantified 272–308 single-molecule trajectories with a time resolution of 104 ms and a length of at least 0.52 s for Nhp6A-wt and the mutants. Mean square displacement (MSD) plots for Nhp6A-wt and -MF molecules are linearly increased against time, confirming diffusion along DNA (Figure 2F). For these proteins, displacement distributions of individual molecules fit well to a single Gaussian function, consistent with a single sliding mode (Figure 2G). Diffusion coefficients, D, were calculated from 2D of the slope of the MSD plots (Figure 2H). D of Nhp6A-MF is 20% greater than Nhp6A-wt, reflecting moderately enhanced mobility, whereas the very low D of Nhp6A-Δ(2–16), which is comparable with the spatial resolution of imaging (96 nm, calculated from the offset of the MSD plots), corresponds to near immobile molecules.
To summarize, Nhp6A mutants Δ(2–16) and MF bound sufficiently stably to DNA in vitro under reduced salt conditions (50 mM Kglu) to enable single-molecule tracking. Nhp6A-Δ(2–16) associated with DNA in a near stationary mode, leading us to conclude that the N-terminal arm is critical for DNA mobility, in addition to its established role in stabilizing binding. Nhp6A-MF, which no longer has the DNA intercalating side chains at the HMGB domain-minor groove interface, bound DNA in a mobile mode with diffusion rates greater than those of WT. We address below whether the increased mobility of Nhp6A-MF is because of destabilization of the HMGB domain in the minor groove or reduced friction of rotational sliding within the minor groove.
Dynamics of Nhp6A-wt and mutants in E. coli cells
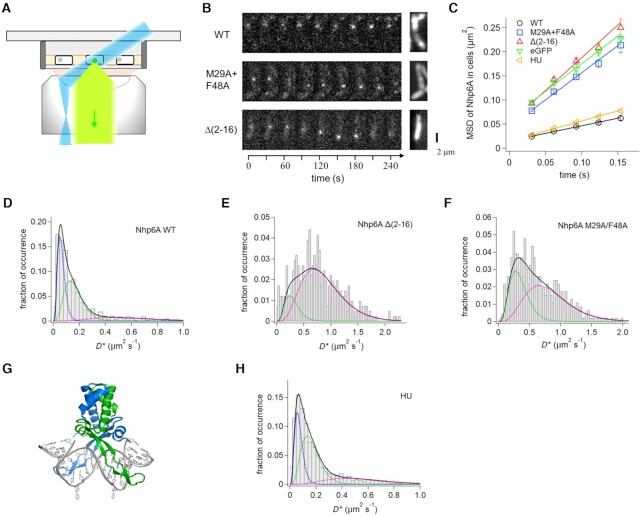
Nhp6A-wt and mutant proteins were fused to eGFP at their C-termini, expressed at low levels in E. coli, and tracked at the single-molecule level using fluorescence microscopy with HILO illumination (Figure 3A). Images from cells growing on agarose pads were acquired with 10 ms exposures at 30.8 ms intervals (Figure 3B). One-step disappearance of the fluorescence spots by photobleaching confirmed that they originated from single molecules. MSD plots obtained from 2D-tracked traces of Nhp6A-wt increase linearly with time confirming diffusional motion, albeit slow, in cells (Figure 3C). The distribution of D values, calculated from 4D of the slope of individual single-molecule trajectories, was fit by the sum of theoretical distributions with three average diffusion coefficients, suggesting three distinguishable mobility modes (Figure 3D). About 39% of the Nhp6A-wt molecules partitioned into a very slow mobility mode (D1 = 0.064 ± 0.004 μm2/s), 47% into a slow mobility mode (D2 = 0.164 ± 0.009 μm2/s), and 14% were dispersed into a faster mobility mode (D3 = 0.58 ± 0.05 μm2/s) (Table 1). The D1 mode corresponds to molecules associated with the chromosome in a near stationary manner, whereas molecules in the D2 mode demonstrate some localized movement. Although the molecules grouped into the D3 mode exhibit considerable mobility, their average rate of movement was less than cytoplasmically-associated eGFP (Figure 3C and Supplementary Figure S6) (99). The D3 molecules are probably transiently associated with the bacterial nucleoid. Overall, most of the Nhp6A-wt molecules (∼86%) appear intimately associated with the nucleoid and exhibit low mobility rates.
Figure 3.
Single-molecule dynamics of Nhp6A and HU in growing cells. (A) Fluorescence imaging set-up of ADBPs fused with eGFP (green dot) in E. coli cells (white rectangle) on an agarose pad (gray shading) using HILO illumination (blue) and detection (light green). (B) Time courses of typical fluorescent images of single molecules of Nhp6A wild-type and mutants (white dots) (left) and images averaged over the time series (right). (C) Time course MSDs of Nhp6A mutants, HU, and eGFP. The error denotes the standard error of all pairs within a given interval. The solid lines represent the best fitted line with the slope of 4D. (D–F) Distributions of average diffusion coefficients of individual molecules of (D) Nhp6A-wt, (E) Δ(2–16) and (F) M29A + F48A. Black solid curves represent the best fitted curves by Equation (2) with two or three components. Colored dashed curves denote each of the two or three components. (G) Structure of the Anabaena HU dimer bound to DNA (PDB code: 1P78). The flipped-out bases stabilized the complex for crystallography (47). (H) Distributions of average diffusion coefficients of individual HU molecules.
Table 1.
Diffusion coefficients and amplitudes for 3-D diffusion modes of proteins in vivo1
| Proteins | D1 (μm2 s–1) | D2 (μm2 s–1) | D3 (μm2 s–1) | A1: A2: A3 (%) |
|---|---|---|---|---|
| Nhp6A WT | 0.064 ± 0.004 | 0.164 ± 0.009 | 0.58 ± 0.05 | 39 ± 4: 47 ± 4: 14 ± 2 |
| Nhp6A M29A + F48A | 0.35 ± 0.03* | 0.82 ± 0.04* | 0: 37 ± 7: 63 ± 7* | |
| Nhp6A Δ(2–16) | 0.30 ± 0.05* | 0.86 ± 0.04* | 0: 15 ± 5*: 85 ± 9* | |
| HU3 | 0.071 ± 0.003 | 0.167 ± 0.007 | 0.54 ± 0.02 | 31 ± 3: 49 ± 3: 20 ± 1 |
| Fis WT (Δfis)2 | 0.036 ± 0.0009 | 0.096 ± 0.004 | 0.47 ± 0.03 | 49 ± 3: 42 ± 3: 9 ± 1 |
| Fis R71A (Δfis) | 0.032 ± 0.001* | 0.088 ± 0.005 | 0.26 ± 0.02* | 43 ± 3: 47 ± 3: 10 ± 2 |
| Fis N84A (Δfis) | 0.039 ± 0.001* | 0.127 ± 0.006* | 0.61 ± 0.05* | 43 ± 3: 41 ± 3: 17 ± 2* |
| Fis R85A (Δfis) | 0.20 ± 0.01* | 0.74 ± 0.02* | 0: 26 ± 3*: 74 ± 5* | |
| Fis K90A (Δfis) | 0.041 ± 0.001* | 0.140 ± 0.005* | 0.75 ± 0.07* | 43 ± 3: 46 ± 3: 10 ± 1 |
| Fis WT (fiswt) 3 | 0.042 ± 0.001 | 0.135 ± 0.004 | 0.66 ± 0.05 | 50 ± 2: 41 ± 2: 8 ± 1 |
| Fis R85A (fiswt) 3 | 0.065 ± 0.002* | 0.172 ± 0.007* | 0.57 ± 0.02 | 43 ± 2*: 41 ± 2: 16 ± 1* |
| eGFP 4 | 0.90 ± 0.04 |
1The parameters were obtained by fitting the distribution of diffusion coefficients for individual molecules in cells by Equation (2). The error denotes the fitting error. Bar graphs of the data are provided in Supplementary Figures S9 and S10A.
2Data obtained from a fis-null (Δfis) or fis-wt (fiswt) strain.
3For Fis WT and R85A (fiswt), the chi-square test suggested a small deviation between the model with m = 3 in Equation (2) and the data, implying the presence of additional modes or the conversion between modes in the observation time range (not included in our model). Due to the problematic fitting with m = 4, the result with m = 3 is presented.
4For eGFP, we used m = 1 in Equation (2) to calculate an average.
*Significant difference between mutants and WT was determined using Welch’s t test with α = 0.05; no asterisk indicates no significant difference. We note that statistically significant variations in the values of the mutants versus wild-type proteins obtained by the analyses of the imaging data may not necessarily reflect biologically meaningful differences.
MSD time course plots of eGFP fusions to Nhp6A mutants Δ(2–16) and MF show fast diffusion of both mutants in E. coli cells in a manner resembling eGFP (Figure 3C). Most (∼85%) of the Nhp6A-Δ(2–26) molecules distribute into a very fast mode implying little association with the nucleoid, whereas ∼15% partition into a moderately mobile mode (0.30 ± 0.05 μm2/s). Nhp6A-MF molecules distribute into two similar D modes except that 37% partition into the moderately mobile mode (0.35 ± 0.03 μm2/s) indicative of dynamic association with chromosomal DNA. The in vivo mobilities of the Nhp6A mutants are consistent with the absence of long-lived DNA complexes at 150 mM Kglu in vitro. The very poor binding of Nhp6A-Δ(2–16) in bacterial cells is also consistent with its severe growth phenotype in yeast where it almost mimics the null mutant (54,61). Individually, the Nhp6A-M29A and Nhp6A-F48A mutants exhibit broad transcriptional defects in yeast as well as compromised growth rates (54,96). The two single mutants still retain partial binding to nucleosome-free promoter-regulatory regions in yeast (96), which may relate to the moderately mobile (D2) population of Nhp6A-MF molecules in E. coli cells.
The bacterial nucleoid protein HU
Background
The HU protein is generally the most abundant and conserved non-specific DNA-binding protein in bacteria under diverse growth conditions (100–102). It participates in transcription (103–107), replication (108,109), transposition and site-specific recombination (110,111), recombination/repair (112,113) reactions, and is believed to play a prominent role in the organization and compaction of the chromosome into the nucleoid (48,114,115). HU is distributed throughout the chromosome in vivo (107) and exhibits preferential binding in vitro to bent and distorted DNA structures (116–119), consistent with the sensitivity of HU mutants to DNA damage. Like the non-specific binding class of HMGB proteins, HU binds and compacts DNA through DNA bending in a sequence-neutral manner (32,120–122), but the structures of the DNA complexes are unrelated (Figure 3G) (123). Nevertheless, Nhp6A and mammalian HMGB1/2 can substitute for HU as DNA architectural factors in bacterial transcription and recombination reactions and in chromosome compaction (78,124–126). Unlike Nhp6A, HU does not form complexes with B-DNA segments that are stable to gel electrophoresis, even in moderate to low salt concentrations (119,127). However, HU does bind random sequence DNA sufficiently strongly to enable single-molecule tracking at 150 mM Kglu (17). These in vitro studies revealed that HU associates with DNA in a mobile mode, similar to Nhp6A but with about a 50% greater 1-D diffusion coefficient (0.492 ± 0.007 μm2/s).
HU dynamics in E. coli
HU (HupAB) was imaged in E. coli cells at the single-molecule level by fusing eGFP to the C-terminal end of HupB. The MSD time course obtained from 2011 molecules in exponentially growing cells is shown in Figure 3C, which reveals slightly greater average mobility than Nhp6A-wt. Individual HU molecules were fit into three diffusion modes whose coefficients and distribution are remarkable similar to those of Nhp6A-wt, except that HU has 8% fewer very slow (D1 = 0.071 ± 0.003 μm2/s) molecules and 6% more fast mobility (D3 = 0.54 ± 0.02 μm2/s) molecules (Figure 3H and Table 1). The diffusion coefficients are consistent with most of HU being associated with the chromosome, consistent with earlier bulk microscopy data (64–67).
During the preparation of this manuscript, similar single-molecule tracking experiments on HU in E. coli were reported by Bettridge et al. (68). In this study, PAmCherry was fused to HupA, which can heterodimerize with HupB or homodimerize with itself. These authors modeled their data into two diffusion modes. Under growth conditions where the dominant species is predicted to be heterodimeric (101), as is the case in our experiments where only the heterodimer is expected to be fluorescently labeled, Bettridge et al. report very similar diffusion coefficients to our data when it is fit into two diffusion modes but with a more equal distribution of the populations (D1:D2 = 45:55%, as compared to 70:30% for our data).
The bacterial nucleoid protein Fis
Background
Fis, along with HU, is the most abundant DNA-binding protein in rapidly growing E. coli with steady-state levels of each at about 25 000–30 000 dimers/cell (102,128). Chromatin immunoprecipitation experiments have revealed Fis bound to >1000 sites that are distributed throughout the E. coli chromosome (129,130). Many, but not all, of these stable binding sites are intergenic and associated with regions involved in transcription, replication or site-specific recombination. Fis functions as a classical bacterial transcriptional activator or repressor, depending on the location of its binding site within a promoter region (46,131–133). In other contexts, targeted binding by Fis regulates site-specific DNA recombination reactions (134,135) and the timing of initiation of DNA replication at E. coli oriC (136). Fis also participates in chromosome organization and compaction (33,48,114,115). Stable Fis-binding sites share a 15 bp highly degenerate consensus motif marked by G/C bps at each end and an A/T-rich center (55,137). Fis also nonspecifically binds DNA at sub-nanomolar affinities, but these nonspecific complexes are readily dissociated by excess ‘competitor’ DNA through a facilitated dissociation mechanism (33,34).
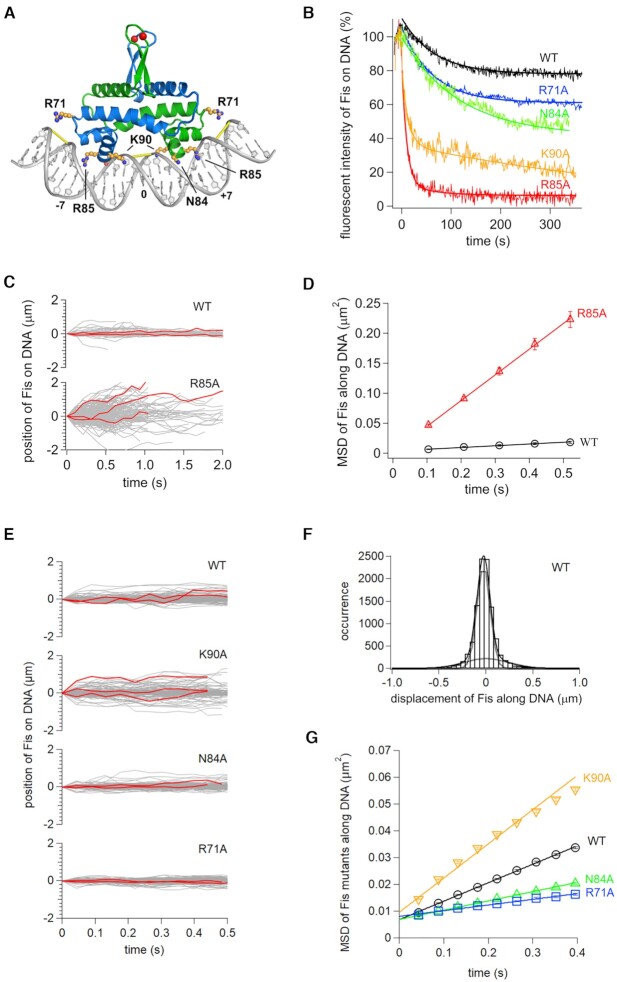
X-ray structures of Fis bound to DNA targets of varying affinities have shown that the helix-turn-helix (HTH) motifs on each subunit insert into adjacent major grooves on one side of the helix and induce considerable bending (Figure 4A) (55,137,138). Moreover, minor groove widths vary considerably as the center and flanking regions of the binding interface are compressed to about half of the canonical width, and the minor groove on the opposite side of the major groove interface is expanded by about 50% of the canonical width. Minor groove compression at the center of the binding site is required for Fis binding because of the unusually short spacing between the recognition α-helices between the HTH motifs in the dimer (139,140). Most Fis-DNA contacts are to the phosphate backbone and only one direct base contact (by Arg85) is critical for binding.
Figure 4.
Role of Fis residues on DNA diffusion in vitro. (A) Structure of the Fis-DNA complex (based on PDB code 3IV5) highlighting side chains studied in this work. Red spheres denote labeling positions (Q21C), and yellow bars denote where the minor groove is narrowed to about half its canonical width. High affinity Fis binding motifs can be written as G(-7) - - - - wwwww - - - - C(+7) where w is A or T (bold w is numbered 0), and - indicates small or no base preferences except that A/T at -4 and T/A at + 4 are disfavored (see (55,137) for details). (B) Time courses of dissociation of Fis mutants from DNA in 100 mM Kglu. Solid curves are fitting curves by single or double exponentials. (C) Single-molecule traces of Fis-wt and Fis-R85A along DNA in 50 mM Kglu with example trajectories in red. (D) Time courses of MSDs along DNA for Fis-wt and Fis-R85A in 50 mM Kglu. The errors denote the standard error of all pairs within a given interval. The solid lines represent the best fitted line with the slope of 2D. (E) Single-molecule traces of Fis-wt, K90A, N84A and R71A along DNA in 150 mM Kglu. (F) Displacement distribution of Fis-wt along DNA in 176 ms intervals in 150 mM Kglu. Solid curves are the best fitted curves by double Gaussian functions (Equation 1). Dashed curves were the best fitted curves by each of the double Gaussian functions. (G) Time course plot of MSDs for Fis-wt, K90A, N84A and R71A molecules in 150 mM Kglu.
Previous single-molecule tracking of Fis-wt revealed that most of the molecules bound DNA in a near stationary mode, but a minority were mobile for at least some of the imaging time (17). The low mobility complexes that were distributed along the λ genome under the non-equilibrium binding conditions probably correspond to complexes bound at DNA segments whose sequence readily conforms to the Fis binding surface, but these were not necessarily correlated with previously identified stable Fis-binding sites on λ DNA. Some of the mobile tracts were punctuated with pauses or complete stops. In the experiments performed in this work, the minority mobile population (24% of the total) had a calculated diffusion coefficient, D2, of 0.156 ± 0.007 μm2/s in 150 mM Kglu buffer (Table 2), which is significantly lower than the D2 population measured for Nhp6A or HU under the same conditions. In the present work we examine the mobility of four Fis mutants with changes at residues selected to test whether mobile Fis molecules remain bound within the major groove, or alternatively, travel along the DNA surface. Taken together, the mutant data are most consistent with mobile Fis molecules unbinding from the major groove and traveling along the DNA surface.
Table 2.
Diffusion coefficients and amplitudes for 1-D diffusion modes of Fis proteins along DNA in vitro1
| Proteins | [Kglu] (mM) | D1 (μm2 s–1) | D2 (μm2 s–1) | A1: A2 (%) |
|---|---|---|---|---|
| Fis WT | 50 | 0.0092 ± 0.0004 | ||
| Fis R85A | 50 | 0.040 ± 0.009* | 0.27 ± 0.02 | 32 ± 6: 68 ± 8 |
| Fis K90A | 50 | 0.0134 ± 0.0004* | ||
| Fis WT | 150 | 0.0138 ± 0.0004 | 0.156 ± 0.007 | 76 ± 2: 24 ± 1 |
| Fis R71A | 150 | 0.0259 ± 0.0006* | 0.24 ± 0.02* | 88 ± 2*: 12.0 ± 0.9* |
| Fis N84A | 150 | 0.0132 ± 0.0003 | 0.122 ± 0.007* | 85 ± 2*: 15.3 ± 0.9* |
| Fis K90A | 150 | 0.0098 ± 0.0002* | 0.242 ± 0.006* | 68.8 ± 0.9*: 31.2 ± 0.6* |
1The parameters were obtained by fitting the displacement distribution of single molecules along DNA in vitro by Equation (1). Bar graphs of the data are provided in Supplementary Figure S10B.
*Significant difference between mutants and WT was determined using Welch’s t test with α = 0.05; no asterisk indicates no significant difference. We note that statistically significant variations in the values of the mutants versus wild-type proteins obtained by the analyses of the imaging data may not necessarily reflect biologically meaningful differences.
The Fis-R85A mutant
The most important base contact for Fis-DNA binding is by Arg85, whose guanidino group hydrogen bonds with a purine corresponding to the conserved G at the ends of high affinity binding sites (Figure 4A) (55). Loss of Arg85 eliminates detectable binding to specific or nonspecific DNA substrates by EMSA and results in a fis-null phenotype in vivo (33,141–143). As expected, Fis-R85A forms extremely unstable complexes on DNA arrays as demonstrated by the rapid decay of bound molecules after the switch from binding buffer with 50 mM Kglu to protein-free buffer with 100 mM Kglu (Figure 4B). Fis-wt molecules exhibited a biphasic decay whereby ∼20% of the molecules dissociated within a time constant of 66 ± 5 s, and the remainder were stably associated over the 5 min time period measured. Stable association by Fis-wt to non-specific DNA targets in the absence of competing DNA has been well documented by earlier single-DNA molecule studies (32,34,62).
As predicted, the residence times of Fis-R85A complexes at 150 mM Kglu were too short to enable tracking. However, Fis-R85A bound sufficiently stably at 50 mM Kglu to obtain trajectories of at least 0.52 s. Fis-R85A was much more mobile than Fis-wt at 50 mM Kglu, exhibiting bidirectional movement on DNA with a small bias towards the buffer flow direction (Figure 4C). The MSD plots of Fis-R85A molecules measured over 104 ms intervals are linearly increased with time and highlight the high diffusion coefficients of the mutant relative to WT (Figure 4D). Displacement plots show a broad distribution of travel lengths by individual Fis-R85A molecules relative to the narrow distribution of wild-type molecules (Supplementary Figure S7A). The distribution of Fis-R85A molecules is best fit by a double Gaussian function where 68% are in a higher mobility mode (D2 = 0.27 ± 0.02 μm2/s) and 32% are in a lower mobility mode (D1 = 0.040 ± 0.009 μm2/s) (Table 2). The drift velocity of the higher mobility mode by Fis-R85A was 0.37 ± 0.05 μm/s in the direction of buffer flow. By contrast, Fis-wt molecules are best modeled by a single low mobility distribution (D = 0.0092 ± 0.0004 μm2/s) reflecting little, if any, mobility in 50 mM Kglu by most molecules (Table 2).
In summary, whereas Fis-R85A molecules bound DNA too unstably at physiological salt concentrations to enable tracking, at low salt concentrations (50 mM Kglu) most Fis-R85A molecules associated with DNA in a mobile mode. Under the low salt conditions, almost all Fis-wt complexes are immobile. As Arg85 on each Fis subunit makes 1–3 H-bonds per subunit to base(s) on the floor of the major groove (55), loss of these contacts unlocks the major connection within the major groove to destabilize binding and increase diffusion rates on DNA.
The Fis-K90A mutant
The Lys90 side chain Nϵ atom is positioned to form a salt link and thereby neutralize the backbone DNA phosphates across from each other where the minor groove width at the center of the binding site is at its narrowest (Figure 4A) (55). This fits with the Fis-K90A mutant having little effect on binding when the central 5 bp region is A/T-rich and thus an intrinsically narrow minor groove, but loss of Lys90 has a severe effect when multiple G/C bps are present within the center or when binding to random sequence DNA (33,138).
Fis-K90A exhibited a complex dissociation profile, with about half of the molecules dissociating very rapidly and about 35% in a much more stable complex with DNA (Figure 4B). The range of residence times can be explained by a large fraction of the mutant proteins initially associating with non-optimal binding sequences, specifically those without A/T-rich centers, whereas about 35% are relatively stably bound to more optimal binding sequences. If mobile Fis molecules rotationally travel along DNA while bound within the major groove, continual compression of the central minor groove would be required, and loss of Lys90 would be expected to reduce mobility. On the other hand, the destabilizing effect of the K90A substitution will promote unbinding from the major groove, which could lead to greater mobility by sliding on the DNA surface.
Although Fis-K90A binding lifetimes during imaging in buffer with 150 mM Kglu tend to be shorter than Fis-wt, we obtained 1976 trajectories of Fis-K90A molecules on DNA over time intervals of at least 0.52 s (Figure 4E). The MSD time course show that Fis-K90A molecules on average exhibit significantly greater travel speeds than Fis-wt (Figure 4G). Fis-K90A molecules were grouped into two classes by fitting the displacements into a double Gaussian function: 69% were in a low mobility mode that corresponds to near stationary molecules, and 31% were in a higher mobility mode (Table 2 and Supplementary Figure S7B). Fis-wt molecules analyzed in parallel had 24% in the higher mobility mode (Figure 4F and Table 2). The average diffusion coefficient, D2, for mobile K90A molecules, determined over 0.176 s time intervals, is 0.242 ± 0.006 μm2/s as compared to 0.156 ± 0.007 μm2/s for the mobile Fis-wt fraction (Table 2).
In summary, Fis-K90A complexes on λ DNA were less stable, contained slightly greater numbers of molecules in the mobile fraction, and the mobile molecules exhibited moderately faster travel rates on DNA than Fis-wt. As elaborated further in the Discussion section, we propose that these properties are consistent with mobile Fis-K90A molecules unbinding from the major groove and diffusing along the DNA surface without the requirement for continual minor groove narrowing.
The Fis-N84A mutant
The Asn84 side chain hydrogen bonds to a DNA backbone phosphate and to a purine N7 atom if present at the bottom strand of the ± 4 position of the binding site (Figure 4A) (55). Nevertheless, an alanine substitution has only a minor effect on binding to high affinity sites (31,55,143). The most dramatic phenotype of Fis-N84A is that it strongly enhances binding to otherwise poor binding sites that contain a T at position ± 4 (55,144). An X-ray structure has shown that if a thymine is present at this position, its 5-methyl group sterically clashes with the Asn84 side chain, thereby inhibiting binding (55). If Fis was rotationally sliding with its recognition helix in the major groove, N84A might be predicted to reduce sliding friction and thereby increase mobility because the mutant protein would no longer clash with T methyl groups protruding from the major groove floor.
Residence time assays on λ DNA show a small increase in the dissociation rate by Fis-N84A when compared to Fis-wt, but almost half the molecules are still bound after 5 min in 100 mM Kglu buffer (Figure 4B). The MSD time plot for Fis-N84A molecules in 150 mM Kglu buffer reveals slightly slower average diffusion rates relative to Fis-wt (Figure 4G). Most (∼85%) of the molecules are associated with DNA in a near stationary mode and the remainder are in a slowly diffusing mode (D2) that has a 22% lower diffusion coefficient than the Fis-wt D2 fraction (Table 2 and Supplementary Figure S7B). Thus, while the difference is small, the N84A mutation appears to lower sliding rates, perhaps because complexes containing a T at ± 4 in the interface are more stable over the short tracking intervals (0.176 s) used to calculate D. The behavior of Fis-N84A, therefore, does not support sliding by Fis within the major groove because the small non-polar mutant side chain would have been expected to reduce sliding friction and thereby increase diffusion rates.
The Fis-R71A mutant
The Arg71 side chain dynamically contacts DNA phosphates 5 bp outside the core interface and promotes the DNA segments flanking the core to wrap around the sides of the Fis dimer (Figure 4A) (31,137). Although stable DNA complexes formed by Fis-R71 mutants exhibit markedly reduced DNA curvatures, Fis-R71A binds to both high affinity and non-specific sites with only small decreases in equilibrium binding affinity. Thus, this mutant enables us to address the effects of Fis-induced DNA bending on 1-D diffusion.
Measurements of the stability of Fis-R71A binding to λ DNA after the shift to protein-free buffer showed a small decrease in residence times for about 30% of complexes in comparison with Fis-wt (Figure 4B). The remainder exhibit similar stability as Fis-wt and may reflect complexes where the flanking DNA sequences can more readily curve around the basic sides of Fis in comparison to those in the less stable complexes (137). Trajectories of 923 Fis-R71A molecules on DNA at 150 mM Kglu were analyzed (Figure 4E). The MSD time course reveals a slower average rate of mobility by Fis-R71A than Fis-wt molecules (Figure 4G). Fitting the individual displacements into a double Gaussian function gave a D2 value of about 1.5-fold and the D1 value of about double that of Fis-wt, but the D1 fraction was slightly increased along with a corresponding decrease in the D2 fraction (Table 2 and Supplementary Figure S7B). We conclude that the average mobility of the reduced bending mutant Fis-R71A is not increased over that of Fis-wt, as may have been expected if bending impeded diffusion by rotation when bound within the major groove.
Dynamics of Fis-wt and mutants in E. coli cells
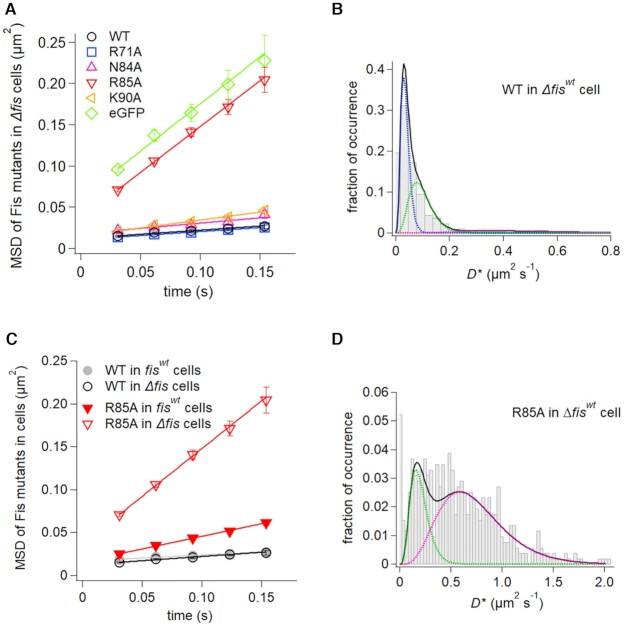
The mobility of Fis-wt and mutants in vivo were evaluated in E. coli Δfis cells using eGFP fusions to the Fis N-terminal end that is not close to the DNA binding surface. We emphasize that unlike the in vitro tracking experiments, the in vivo experiments primarily reflect steady-state association of Fis with the chromosome. The MSD time course of 2D-tracked traces of Fis-wt reveals an overall low rate of diffusion (Figure 5A). The distribution of diffusion coefficients from individual single-molecule traces (Figure 5B) was fit into three groups: 49% of the molecules partitioned into a very low mobility mode (D1 =0.036 ± 0.001 μm2/s), 42% partitioned into a slow mobility mode (D2 = 0.096 ± 0.004 μm2/s) and a small number (9%) were distributed into a faster mobility mode (D3 = 0.47 ± 0.03 μm2/s) (Table 1). Many of the molecules in the D1 group that appear near immobile within the nucleoid are probably associated with the numerous high affinity Fis-binding sites dispersed throughout the E. coli chromosome. Molecules in the D2 group may reflect proteins associated with weak or non-specific DNA-binding sites during the tracking time period and undergo localized sliding or intersegmental transfer reactions (see Discussion section). Diffusion coefficients of the faster mobility molecules (D3 fraction) are in the range of those for the D3 class of Nhp6A and HU molecules and represent molecules transiently associated with the nucleoid.
Figure 5.
In vivo dynamics of Fis mutants. (A) Time courses of MSDs of Fis mutants in Δfis cells. The errors denote the standard error of all pairs within a certain interval. The solid lines represent the best fitted lines with the slope of 4D. (B and D) Distributions of average diffusion coefficients of individual molecules of Fis-wt (B) and Fis-R85A (D) in Δfis cells. Black solid curves represent the best fitted curves by Equation (2) with two or three components. Colored dashed curves denote each of the two or three components. (C) Time courses of MSDs of Fis-wt and Fis-R85A in Δfis and fiswt cells. The solid lines represent the best fitted line with the slope of 4D.
We also imaged gfpFis-wt molecules in fiswt cells, where, under the growth conditions employed, about 20 000 dimers of native Fis proteins/cell are expressed (128). Under these conditions, a large fraction of the high-affinity Fis-binding sites are expected to be bound by native Fis. The low number of gfpFis subunits is expected to be heterodimerized with native Fis subunits, given the rapid rate of subunit switching by Fis ((145), and see below). Indeed, single step photobleaching of the molecules confirmed a single fluorescent molecule was present per unit (dimer). The MSD plot of gfpFis-wt molecules in fiswt cells is nearly identical to that in Δfis cells (Figure 5C). Although the D2 and D3 diffusion coefficients are slightly greater in fiswt cells, the numbers of molecules in the different populations are indistinguishable (Table 1 and Supplementary Figure S8A). The results suggest that the high affinity Fis-binding sites are not well saturated under the growth conditions employed where >2 chromosomes per cell is expected. We conclude that Fis is nearly exclusively associated with the bacterial nucleoid in fiswt cells where it exhibits a low rate of diffusion. Previous live imaging studies have also shown that Fis is essentially entirely associated with the nucleoid (63). Fluorescence recovery after photobleaching (FRAP) experiments have also demonstrated slow diffusion of Fis molecules within the nucleoid (63).
With the exception of gfpFis-R85A, all of the Fis mutants also exhibited low mobility in Δfis cells (Figure 5A and Table 1; Supplementary Figure S8B). The mobility of gfpFis-R85A molecules resembles cytoplasmic eGFP (Figure 5A,D and Supplementary Figure S6 and Table 1), indicating that most are not, or only weakly, associated with the nucleoid, as predicted from the highly unstable binding properties in vitro under physiological ionic strength and the near null phenotype of the mutant in vivo (33,141–143). The D2 and D3 populations of gfpFis-K90A molecules exhibit greater mobilities than the corresponding Fis-wt populations (Table 1), which account for the slightly greater overall mobility in the MSD time course (Figure 5A). Although small, the greater proportion of Fis-N84A molecules in the fast D3 mode explains the slightly greater average mobility of this mutant relative to Fis-wt (Table 1, Figure 5A). The average mobility of Fis-R71A molecules from the MSD time plot is nearly indistinguishable from Fis-wt (Figure 5A), even though the minority D3 population exhibits slower diffusion rates (Table 1). The Fis-R71A result indicates that bending of the DNA flanking the core interface does not demonstrably affect in vivo diffusion rates.
We investigated the in vivo dynamics of Fis heterodimers with only one subunit containing Arg85. These were generated by expressing low amounts of the Fis-R85A eGFP fusion in fiswt cells. All the gfpFis-R85A molecules appeared to be heterodimerized with a wild-type subunit as evidenced by the much slower mobility of labeled molecules as compared to gfpFis-R85A homodimers in Δfis cells (Figure 5C). The distribution and diffusion coefficients are remarkably similar to Fis-wt homodimers, albeit there are ∼2-fold more D3 molecules (Table 1 and Supplementary Figure S8A). The slow diffusion rate by the heterodimers in comparison to the Fis-R85A homodimers indicates that a single Arg85 connection to the floor of the major groove is sufficient to retard the mobility of the protein.
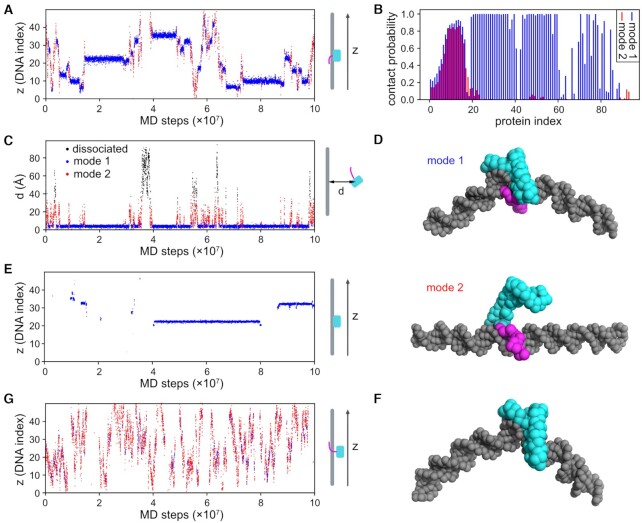
MD simulations of 1D diffusion along DNA by Nhp6A: switching between two DNA-bound conformations with different mobilities
Single-molecule tracking of Nhp6A-wt in vitro demonstrated that the HMGB protein bidirectionally travels along extended DNA molecules. Removal of the flexible N-terminal basic arm, however, severely reduced or eliminated its mobility. This result was unexpected because the NMR structure of the Nhp6A-DNA complex showed that the N-terminal arm functioned like a clamp as it wrapped around the DNA duplex, consistent with its large effect on stabilizing the complex. Deletion of the N-terminal arm was therefore predicted to increase mobility. Removal of the Met29 and Phe48 side chains, which protrude from the HMGB domain into the minor groove floor, moderately increased mobility of Nhp6A on DNA. This result could be explained by reduced sliding friction if the HMGB domain traveled within the minor groove, or the increased mobility could relate to the moderate decrease in complex stability by the two alanine substitutions. We performed molecular dynamics (MD) simulations to address the mechanism by which Nhp6A molecules travel on DNA and to understand the dynamics of the mutants.
The MD system contained a 50 bp random sequence DNA and Nhp6A, using PDB code 1J5N as the template structure. Coarse-grained MD simulations were performed as described in the Materials and Methods section. The simulations employed the PWMcos method (73) to model interactions between the protein and DNA bases within the minor groove that play important roles in Nhp6A-DNA binding. We performed 20 independent 108-step simulations for Nhp6A-wt. A representative time series of the binding positions of Nhp6A-wt on DNA is shown in Figure 6A. Nhp6A-wt molecules traveled bidirectionally along DNA, but their mobilities were transiently interrupted by pauses.
Figure 6.
MD simulations suggest that Nhp6A binds DNA in two distinct modes. (A) A representative time series of Nhp6A-wt binding positions on DNA (z). Blue and red dots denote the binding modes coupled with DNA bending (mode 1) and sliding (mode 2), respectively. (B) DNA contact probabilities of Nhp6A-wt residues to DNA for modes 1 (blue) and 2 (red). (C) A representative time series of the distance (d) between the center-of-mass of the HMGB domain and DNA. The blue and red dots represent the two binding modes, respectively, while the black dots represent dissociated states (no contact between Nhp6A and DNA). The plot was based on the same MD trajectory as (A). (D) Representative conformations of Nhp6A-wt binding to DNA. Residues 1–16 and 17–93 of Nhp6A are shown as magenta and cyan spheres, respectively; DNA is projected as gray spheres. In mode 1, the HMGB domain is inserted in the minor groove, and the N-terminal basic arm binds to the DNA backbone, in this example from within the major groove. In mode 2, only the N-terminal basic arm interacts with the DNA backbone, in this example largely from within the minor groove. (E) A representative time series of Nhp6A-Δ(2–16) in contact with DNA. Only mode 2 was observed (blue dots). The sections without data points represent dissociation of Nhp6A from DNA. (F) A representative conformation of Nhp6A-Δ(2–16) (cyan HMGB domain) bound within the minor groove of bent DNA. (G) A representative time series of the movement along DNA for Nhp6A-MF (M29A + F48A). Compared to Nhp6A-wt (panel A), the fraction of mode 2 increased, while that of mode 1 decreased.
To quantitatively analyze the binding configurations and diffusion patterns of Nhp6A-wt on DNA, clustering analysis was performed on the DNA-binding interface of the Nhp6A-wt structures obtained from the simulations. We found that the simulated structures could be classified into two distinct groups (Supplementary Figure S2), hereafter termed mode 1 (84%) and mode 2 (16%). For each binding mode, the DNA-contact probability of each Nhp6A-wt residue was computed and plotted in Figure 6B. In mode 1 (colored blue), Nhp6A-wt had most of its residues closely associated with DNA, whereas in mode 2 (colored red), Nhp6A contacted DNA only with its N-terminal segment. When each dot in Figure 6A was colored according to the binding mode, it becomes apparent that mode 1 correlates with molecules localized at a constant position whereas mode 2 correlates with molecules diffusing on the DNA. For the same trajectory, we also plotted in Figure 6C the minimum distance from the center-of-mass of the HMGB domain to DNA particles. In combination, these plots show that Nhp6A-wt proteins in mode 1 are immobile and closely associated with DNA, whereas those in mode 2 are mobile and less intimately associated with DNA. Figure 6C also shows the occasional complete dissociation of Nhp6A-wt from the DNA molecule, typically from the mode 2 phase.
Inspection of representative examples of mode 1 molecules revealed structures similar to the NMR-derived complex in which the folded HMGB domain was inserted into a widened minor groove with accompanying bending of the DNA helix (Figure 6D, mode 1). In contrast, mode 2 structures had the HMGB domain outside of the DNA grooves, but the proteins remained dynamically associated with the DNA phosphodiester backbone via basic residues within the flexible N-terminal arm (Figure 6D, mode 2). A video of a representative time trajectory illustrating switching between the two binding modes and the dynamic interaction of the N-terminal arm with DNA is provided in Supplementary Movie S1.
Simulations of Nhp6A-Δ(2–16) binding to DNA showed that the mutant without the N-terminal basic arm bound DNA with much lower affinity than Nhp6A-wt, as predicted (Figure 6E and Supplementary Table S2). When bound, Nhp6A-Δ(2–16) was exclusively in mode 1 (a representative structure is shown in Figure 6F) where the protein is immobile on DNA (Figure 6E). The DNA was still bent within the Nhp6A-Δ(2–16) complex, consistent with the ligation of 98 bp DNA circles in the presence of high Nhp6A-Δ(2–16) concentrations (54). The absence of mobility on DNA fits the single-molecule imaging studies performed in 50 mM Kglu buffer (Figure 2D–H).
Simulations of the Nhp6A-MF mutant showed weaker binding to DNA and a shift in the relative distribution of binding modes to only 18% in mode 1 and 82% in mode 2 (Figure 6G and Supplementary Table S2). Although the diffusion rate of molecules in mode 2 was the same as those for Nhp6A-wt, the greater proportion of Nhp6A-MF molecules in the mode 2 conformation resulted in greater overall mobility on DNA (Supplementary Table S2), consistent with the increased diffusion coefficient measured experimentally (Figure 2D–H).
DISCUSSION
ADBPs support 1-D diffusion along DNA molecules even though they introduce large conformational changes into DNA in stably bound complexes (17). The mobilities of yeast Nhp6A and bacterial HU and Fis proteins on otherwise naked DNA are slower than measured for many other DNA-binding proteins and are accompanied by a large free energy barrier. Here we examine the mechanism of 1-D diffusion by these ADBPs by analyzing the dynamics of mutant proteins both in vitro and in vivo. These proteins contain strategically selected mutations that affect different molecular aspects of binding based on the structures of the DNA complexes and the biochemical properties of the mutant proteins.
We consider two models for how ADBPs travel along elongated DNA. One mechanism is rotation-coupled sliding, whereby the protein remains intimately associated with one or both DNA grooves as it diffuses bidirectionally along the DNA helix. Continual DNA distortions by a rotational sliding mechanism by ADBPs would imply a rough energy landscape that is dependent upon the DNA sequence. A second mechanism involves transient unbinding of the protein from the DNA groove(s) and diffusion along the sugar-phosphate backbone on the DNA surface. A hallmark of many ADBPs is their highly basic DNA binding surface, often with flexible basic peptide segments, that can enable continuous and largely isoenergetic connections with the electronegative DNA surface. As summarized below, both mutant experimental data and MD simulations are most consistent with Nhp6A diffusion along the DNA surface via its unstructured basic arm. This mechanism also fits available data regarding diffusion by the HU protein where both flexible basic arms and the DNA-binding surface of the body are likely to be involved. Based on the behaviors of mutants, we propose that Fis also travels along the DNA surface, even though flexible basic peptide segments are probably not involved.
Additional mechanisms for protein mobility along DNA include direct transfer between DNA segments and hopping, which are particularly relevant to target searching by DNA-binding proteins in vivo where 1-D diffusion will be hindered by other bound proteins. Short-range hopping events, involving complete dissociation from DNA followed by rebinding to a nearby segment, is considered unlikely under our in vitro experimental conditions where there is a constant unidirectional hydrodynamic force due to buffer flow (but note exception below). Buffer flow, which extends individual DNA molecules to their near full contour length, will also inhibit intramolecular transfer events that require DNA looping. On the other hand, the high local concentration of folded DNA within the bacterial nucleoid offers ample opportunities for intersegmental transfer and localized hopping to proximal DNA segments. Nevertheless, the diffusion of ADBPs in bacterial cells is generally slow with many molecules appearing nearly immobile within the nucleoid. We discuss the in vivo dynamics of wild-type and mutant proteins in light of their in vitro properties and conclude that the highly compacted and protein-bound chromosome in vivo reduces the dynamics of even heterologous sequence-nonspecific ADBPs like yeast Nhp6A.
Mechanism of 1-D diffusion by the yeast HMGB protein Nhp6A
As is true for all HMGB-family proteins, the DNA minor groove must dramatically widen and the DNA duplex must curve away from the L-shaped structure in order to accommodate the HMGB domain of Nhp6A (Figures 2A and 7A) (45,70,95,146). Thus, 1-D diffusion by rotationally sliding within the minor groove would require continual expansion of the groove together with bending of the DNA axis (Figure 1A). A feature of Nhp6A that is critical for stable DNA binding is the N-terminal flexible arm. This 16 amino acid residue segment with its two patches of 3–4 tandem basic residues dynamically wraps around the DNA duplex to stabilize the complex, primarily through electrostatic interactions with the phosphate backbone from within the compressed major groove on the DNA side opposite to the inserted HMGB domain (Figures 2A and 7A). Whereas the N-terminal arm clearly stabilizes the bound complex (Figure 2C, (54)), we show here that the arm is also required for DNA sliding. Indeed, complexes formed by a mutant missing the N-terminal arm appear nearly immobile on DNA (Figure 2E–H). The very low amount of diffusion observed in vitro by this mutant could be due to unbinding by the HMGB domain and brief sliding of the basic domain along the DNA surface before it completely dissociates.
Figure 7.
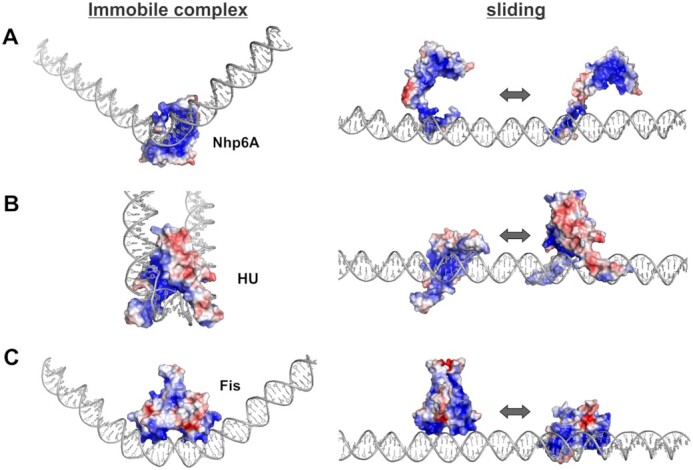
Diffusion of ADBPs along the DNA surface. Left panels are structures determined by X-ray or NMR with additional DNA appended to the ends. DNA curvatures in all of these bound DNA complexes are believed to be dynamic in solution. Right panels are models of the proteins with their basic DNA-binding surface sliding on the acidic surface of standard B-DNA. The protein re-inserts into the DNA groove(s) preferentially at segments whose sequence readily adopts the bound conformation. (A) Nhp6A (±4 kT/e) based on PDB codes: 1J5N (DNA complex, left) and 1LWM (protein-only ensemble, right). The DNA complex on the left is oriented to visualize the interface of the HMGB domain bound to DNA and the N-terminal arm (beginning at residue 6), which is threaded through the compressed major groove. When sliding, the basic HMGB domain, in addition to the N-terminal arm, may sometimes associate with the DNA surface, as occasionally observed in MD structures. (B) Anabaena HU-DNA complex (±4 kT/e, left) based on PDB code: 1P78 with the extended DNA following that in PDB code: 2NP2 (HU-family protein Hbb (163)). Models of sliding HU dimers are consistent with E. coli HU structures obtained by MD simulations (161). The sliding protein on the left is looking into the basic saddle of HU; the β-hairpin arms (see backbone cartoon in Figure 3G) are flexible. An alternative sliding mode for E. coli HU has been proposed where the protein diffuses along DNA on its side (68,167). (C) Fis (±3 kT/e) based on PDB code: 3IV5. The sliding protein on the right is looking into the helix-turn-helix DNA binding region. Surface electrostatic potentials were rendered in PyMol using APBS: blue is electropositive; red is electronegative.
A second prominent feature of Nhp6A, which is conserved among many HMGB-family proteins, are aliphatic side chains (Met29 and Phe48 in the case of Nhp6A) that protrude from the concave face of the HMGB domain into minor groove. Because these side chains insert into base pair steps within the DNA complex, rotational sliding within the groove would be expected to involve considerable frictional drag. Substitutions of these two residues with alanine both destabilize binding and increase 1-D diffusion rates. Like Nhp6A-wt, diffusion by Nhp6A-MF along DNA is bidirectional over short time frames (Figure 2E and G), but the Nhp6A-MF mutant uniquely exhibits a clear mobility bias in the direction of buffer flow over long time periods (Figure 2D). The behavior of Nhp6A-MF is consistent with sliding along DNA with the unbound HMGB domain exposed to solvent. For this mutant, short-range hopping, where buffer flow pushes the protein towards one end of the DNA molecule, may also contribute to its enhanced diffusion. Similar results were obtained for the DNA glycosylase Fpg, which inserts a phenylalanine into base pair steps as it scans for damaged DNA. An F111A substitution in E. coli Fpg enhanced diffusion rates along DNA (23).
MD simulations provide a structural mechanism that accommodates these experimental observations. The simulations reveal that Nhp6A oscillates between two binding modes: a stationary mode, whose structure fits the NMR-derived structure of the DNA complex, and a mobile mode in which the HMGB domain is not bound within the minor groove, but the protein is dynamically associated with DNA via ionic interactions between basic resides in the flexible N-terminal arm and the acidic phosphate backbone (Figures 6 and 7A and Supplementary Movie S1). During migration along DNA, the basic segment dynamically inserts within the highly electronegative minor groove (e.g. Figure 6D mode 2 (147)) or within the major groove (e.g. Supplementary Figure S3, top left structure) and may traverse the grooves.
The simulations further show that the Nhp6A mutant without the N-terminal arm forms only DNA complexes in the stationary mode where the HMGB domain is bound within the minor groove, albeit, in an unstable complex that frequently dissociates into solution. MD simulations also show that alanine substitutions in place of the two DNA intercalating residues destabilize the binding of the HMGB domain, thereby shifting the binding mode distribution to favor the mobile mode and overall faster diffusion rates. An alternative model, in which the loss of the intercalating side chains leads to less frictional drag during rotational sliding within the minor groove, is not supported by the MD simulations.
Given these data, we propose that Nhp6A slides on the surface of DNA (Figures 6D and 7A) rather than undergoing rotation-coupled diffusion where the HMGB domain remains bound within the minor groove. The HMGB domain of sliding proteins then re-inserts into the minor groove at DNA segments that are energetically favorable to adopt the bound structure. Certain damaged DNA, supercoiled DNA, or higher order nucleoprotein complexes where the DNA is bent would be especially favorable. The mobile mode, whereby the protein is dynamically connected to the DNA backbone through its flexible N-terminal basic arm, provides a mechanism by which it can bypass other DNA-bound proteins as it travels along chromatin and embed itself within, and thereby stabilize, large nucleoprotein complexes (59). This mechanism may also localize chromatin-modifying complexes like yeast FACT and SWI/SNF that utilize Nhp6A (80,91,93,94).
In support of the two-state model for Nhp6A diffusion, Sarangi et al. (148) have reported evidence for unbent as well as bent complexes of Nhp6A by time-resolved FRET, and McCauley et al. (149) have presented evidence for a loosely bound state by HMGB proteins, including Nhp6A, by force microscopy studies that may correspond to the mobile mode discussed here. The two-state model is also supported by studies of other DNA-binding proteins including TALE proteins (37), PCNA (38), hOGG1 (58), p53 (60) and cohesin SA2 (150).
Unstructured basic peptide segments are commonly associated with DNA-binding proteins and are a hallmark of the major class of nuclear localization signals (151). A well-studied example of a transcription factor that diffuses along DNA utilizing unstructured basic peptide segments is p53 (13,29,60,152–156). Removal of the C-terminal unstructured basic segment from p53 slows sliding (13,154) and prevents jumping along DNA (156). Also, the basic linker segment of p53 participates in the switching between the fast and slow sliding modes (157). Isolated basic peptides from a variety of proteins have been shown to slide on DNA, albeit under low salt conditions, and when fused, promote sliding of non-DNA binding proteins like streptavidin (158,159). Flexible basic segments on DNA binding proteins have also been proposed to promote intersegmental transfer reactions by the monkey-bar mechanism (160).
The bacterial nucleoid protein HU
A DNA surface diffusion mechanism is also applicable to the bacterial HU-family of proteins. E. coli HU travels along DNA in vitro at speeds greater than Nhp6A, yet dramatically distorts DNA structure when bound in a static complex. Previous course-grained MD simulations provided evidence that HU also oscillates between static and rotation-uncoupled mobile modes on DNA (161). The static mode reflects X-ray structures of bent DNA complexes of HU-family proteins (123,162,163) that are supported by solution studies (120,164). In these structures, the DNA is associated with a basic saddle on the dimer surface with flexible basic β-ribbon arms from each subunit threaded through an expanded minor groove (Figures 3G and 7B). Prolines near the tip of the β-arms insert into the base stack, and the flanking DNA is dynamically wrapped around the sides of the dimer to give overall curvatures that can exceed 140°. MD studies showed that the DNA is unbent and that the flexible β-arms are typically not threaded through the minor groove when HU is mobile on DNA (161). Nevertheless, the DNA backbone remains connected to the strongly electropositive surface of the saddle and arms as the protein travels along the DNA surface (Figure 7B). Multiple HU-DNA binding modes are supported by ITC, FRET and thermodynamics studies, which have concluded that E. coli HU interacts with a bent (140°) 34 bp DNA segment in a higher affinity binding state and to unbent interfaces of 10 and 6 bp, which involve the same region of the saddle and arms, in lower affinity states (165,166).
An alternative structural model for the sliding mode has been proposed by Bettridge et al. (68) based on X-ray structures by Hammel et al. that show HU dimers sandwiched between unbent DNA segments within the crystal lattice (167,168). In this model, HU slides on the DNA surface on its ‘side’ through interactions with main chain amides and a triad of conserved lysines. In support of the importance of one or more of these lysines, an HU mutant missing all three lysine side chains exhibited nearly exclusively fast diffusion rates in vivo, reflecting largely cytoplasmic localization (68). However, because interactions between multiple HU proteins stabilize the lattice in the co-crystals with unbent DNA, it is unclear whether sufficient binding energy exists in this mode to enable single HU molecules to remain connected to individual DNA molecules when traveling over thousands of base pairs.
1-D diffusion by the bacterial protein Fis
Taken together, we believe the wild-type and mutant data are also most compatible with the Fis protein diffusing along a conformationally-normal DNA surface by means of its electropositive DNA-binding surface (Figure 7C). Rotational tracking by the Fis dimer with its DNA recognition helix of each subunit bound within the major groove would require the minor groove at the center of the binding interface to be continually compressed to about half its canonical width along with bending of the DNA axis within the major groove interfaces (55,137,138). Although not directly contacted by the protein, compression of the central minor groove is essential because of the unusually short spacing between DNA recognition helices in the dimer, and bending within the major groove interface is required for main chain and side chain contacts to DNA phosphates that stabilize the complex.
In vitro tracking experiments show that Fis-wt exhibits two DNA-binding modes, static and mobile, and that the distribution of these modes is sensitive to the monovalent ion concentration. At 50 mM K+, nearly all proteins are bound in a static mode, but at 150 mM K+, 10–25% of the proteins bidirectionally travel along DNA, with periods of mobility occasionally interrupted by pausing (Figure 4C,E, Table 2 and (17)). A Fis mutant lacking the primary connection to the major groove floor, Fis-R85A, only transiently associates with DNA at 150 mM K+ but forms DNA complexes at 50 mM K+ with sufficient residence times for their dynamics to be measured. In the lower salt conditions, 72% of the Fis-R85A DNA complexes are mobile. Although this result could be compatible with the loss of hydrogen bonding to purines enhancing rotational sliding within the major groove, it seems more likely, given the properties of the other Fis mutants, that the instability of binding by loss of Arg85 results in its mobility because of unbinding from the major groove.
DNA binding by Fis is highly sensitive to the intrinsic width of the minor groove at the center of the binding interface (55,138). Lys90 from each Fis subunit forms a salt-link with DNA backbone phosphate groups that most closely approach each other across the central compressed minor groove (Figure 4A). DNA binding by Fis-K90A is extremely sensitive to minor groove widths as evidence by the 1000-fold poorer binding relative to Fis-wt to an otherwise high affinity binding site but lacking an intrinsically narrow minor groove at its center (138). Thus, Fis-K90A would be expected to be compromised for rotational sliding over random sequence DNA. However, Fis-K90A exhibits enhanced 1-D diffusion on DNA relative to Fis-wt at 150 mM K+ (Figure 4E and G, and Table 2). Like Fis-wt, Fis-K90A complexes at 50 mM K+ are almost entirely immobile (Table 2). The salt dependence for mobility by Fis-wt and Fis-K90A is compatible with unbinding of the proteins from the major groove in order to diffuse on DNA.
The Fis Asn84 side chain sterically clashes with the 5-methyl group of thymine when present at the ± 4 position in the binding site, as shown by the Fis-F23 DNA X-ray complex (55). The consequence of this clash is that Fis-N84A binds to a DNA site containing a T at position 4 on both half sites with about 25-fold greater affinity than Fis-wt (55). This predicts that the T 5-methyl group should interfere with rotational sliding by Fis-wt but that Fis-N84A may exhibit increased mobility due to reduced sliding friction. However, Fis-N84A does not exhibit greater 1-D diffusion rates as compared to Fis-wt (Figure 4E and G, and Table 2). In addition, Fis-R71A, which exhibits reduced bending of DNA flanking the core interface (31,137), was not found to increase the overall diffusion rate. The results of these two mutants are also most consistent with Fis traveling on the surface of DNA molecules without involving DNA conformational changes.
Diffusion of ADBPs in vivo
Each of the ADBPs studied here, including ectopically expressed yeast Nhp6A, nearly exclusively associates with the bacterial nucleoid. The distributions of individual molecules for each protein can be modeled by three diffusion modes that include a near statically bound state, a low mobility state, and a faster mobility state representing a small number (9–20%) of molecules transiently associated with the nucleoid. Nhp6A and HU dynamics are remarkably similar in E. coli cells, even though Nhp6A is not native to bacteria. The similarities include both the values of the three diffusion mode coefficients and the relative number of molecules in the three modes. Overall, the average mobility of HU molecules is slightly greater than Nhp6A in vivo, but the difference is much less than the 50% greater sliding rates by HU measured on elongated DNA molecules in vitro at 150 mM Kglu.
Perhaps surprisingly, slightly more (39%) of the Nhp6A molecules distribute into the D1 mode than HU (31%). These low mobility proteins probably reflect binding to folded or curved DNA, such as at the base or apex of plectonemically supercoiled DNA loops. It has also been proposed that HU proteins may collaborate to stabilize DNA bundles in a manner reflective of HU-DNA crystal lattices (68,167,168). Both HU and Nhp6A often associate within multi-component nucleoprotein complexes where curved DNA stabilizes their binding. Specific interactions by the yeast HMGB protein with E. coli chromosome-associated proteins are obviously unlikely, but Nhp6A, as well as other eukaryotic HMGB proteins, can facilitate assembly of bacterial nucleoprotein complexes by DNA architectural mechanisms (78,126). In native yeast chromosomes, Nhp6A is primarily bound to nucleosome-free regions, especially those within promoter regions of highly transcribed genes (96).
The number and mobility of the Nhp6A and HU molecules in the D2 mode, where they are experiencing localized mobility within the folded chromosome in the nucleoid, are nearly identical. In addition to 1-D sliding, proteins in this mode may undergo intersegmental transfer reactions, which have been shown to efficiently occur in vitro (33,34) and localized hopping. Fourteen (Nhp6A)-20% (HU) of the proteins partitioned into a broad class (D3) of faster moving molecules that are undergoing 3-D diffusion within the cell together with transient interactions with chromosomal DNA. In vitro, HU binds DNA with the lowest stability of the three wild-type ADBPs studied here, consistent with it showing the greatest proportion of D3 molecules of the wild-type proteins.
Without the N-terminal basic arm, Nhp6A mutants exhibit high diffusion rates that approach those measured for cytoplasmic eGFP and consistent with the poor binding observed in vitro and the yeast growth phenotype that approaches that of the null mutant (54). Moreover, the N-terminal truncated mutant is no longer preferentially located in the yeast nucleus, which is attributed to its poor DNA binding since the nuclear localization signal is within the HMGB domain (61,169). Most Nhp6A-MF molecules that are missing both residues that insert into the base stack in the minor groove also show fast diffusion in vivo, but 37% of the molecules exhibit intermediate mobility (D2 = 0.35 ± 0.03 μm2/s), consistent with partial association with the bacterial chromosome. Likewise, individual Nhp6A-M29A and Nhp6A-F48A mutants retain partial binding to nucleosome-free promoter regions in yeast by chromatin-immunoprecipitation analyses (96).
As expected from in vitro measurements, Fis molecules exhibit the lowest mobility of the ADBPs analyzed with over 90% of the molecules associated with the chromosome in a static or very low mobility mode. The immobile fraction is consistent with proteins bound to the many stable binding sites distributed around the chromosome, many of which function as regulatory sites in transcription and other DNA reactions (129,130). 1-D sliding and intersegmental transfer reactions (31,33,34) probably account for the very low mobility exhibited by the D2 population; hopping seems less likely because of the long binding lifetimes exhibited by Fis. Less than 10% of the Fis-wt molecules exhibit a faster mobility mode where substantial 3-D diffusion must be occurring.
With exception of R85A, each of the Fis mutants tested exhibited average MSDs in vivo that were similar or very slightly increased over the wild-type. Fis-R71A, N84A and K90A exhibit minor differences in diffusion coefficients and amplitudes that are mostly within the range of experimental and fitting accuracies and may reflect the small differences in binding affinities by the respective mutations. Most Fis-R85A molecules exhibited fast in vivo diffusion rates that approach those of cytoplasmic GFP, as expected given the unique importance of Arg85-mediated hydrogen bonding to the floor of the major groove on Fis binding. However, most (84%) heterodimers with only one subunit containing the Arg85 side chain associate with the nucleoid in a low mobility (D1 or D2) state. This result is consistent with some functional Fis-binding sites (e.g. one of the Hin recombinational enhancer sites) that do not have a purine on one side of the Fis-binding motif where Arg85 normally hydrogen bonds.
CONCLUDING REMARKS
Unbinding from the grooves and sliding on the DNA surface presents a smoother energy landscape than sliding within the groove(s) and thus provides a partial solution to the search-speed/binding stability paradox (170). One proposal for obtaining biologically relevant search speeds relies on conformational changes in the protein between the sliding and bound modes (170–173). For ADBPs, the DNA must undergo large conformational changes for stable binding. Protein conformational changes coupled to stable binding in the case of the Fis protein are unlikely as the Fis dimer (excluding the N-terminal arms that are not involved in DNA binding) can be thought of as a rigid body (55,145). The Fis dimer, with its narrow spacing between DNA recognition helices, can only sample the target sequence if the central minor groove of the interface is narrowed to about half its canonical width. Once the two recognition helixes are inserted into the major grooves, bending of the DNA is needed to establish the full constellation of the mostly backbone contacts to generate a stable complex.
For Nhp6A and HU, the DNA must also conform by large changes in groove widths and overall curvature to form a stable complex. In addition, Nhp6A and HU contain flexible peptide arms that adopt many conformations in solution and when translocating along the surface of conformationally normal DNA, but these arms acquire restrictive conformation(s) when in a stable complex. We suggest that sliding on the DNA surface after the primary DNA-binding domain has unbound from the groove(s) and is no longer contacting DNA bases may also be applicable to 1-D searches by other DNA binding proteins, especially when conformational changes in DNA that would cause free energy barriers for diffusion are coupled with binding.
Supplementary Material
ACKNOWLEDGEMENTS
K.K., E.M. and R.C.J. thank Prof. Margot E. Quinlan (UCLA) for use of her fluorescence microscope.
Contributor Information
Kiyoto Kamagata, Institute of Multidisciplinary Research for Advanced Materials, Tohoku University, Katahira 2-1-1, Aoba-ku, Sendai 980-8577, Japan.
Yuji Itoh, Institute of Multidisciplinary Research for Advanced Materials, Tohoku University, Katahira 2-1-1, Aoba-ku, Sendai 980-8577, Japan.
Cheng Tan, Department of Biophysics, Graduate School of Science, Kyoto University, Kyoto 606-8502, Japan.
Eriko Mano, Institute of Multidisciplinary Research for Advanced Materials, Tohoku University, Katahira 2-1-1, Aoba-ku, Sendai 980-8577, Japan.
Yining Wu, Institute of Multidisciplinary Research for Advanced Materials, Tohoku University, Katahira 2-1-1, Aoba-ku, Sendai 980-8577, Japan.
Sridhar Mandali, Department of Biological Chemistry, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, CA 90095-1737, USA.
Shoji Takada, Department of Biophysics, Graduate School of Science, Kyoto University, Kyoto 606-8502, Japan.
Reid C Johnson, Department of Biological Chemistry, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, CA 90095-1737, USA; Molecular Biology Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
MEXT/JSPS KAKENHI [16KK0157, 20K06571 to K.K.]; National Institutes of Health [GM038509 to R.C.J.]. Funding for open access charge: MEXT/JSPS KAKENHI [16KK0157, 20K06571 to K.K.]; National Institutes of Health [GM038509 to R.C.J.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Von Hippel P.H., Berg O.G.. Facilitated target location in biological systems. J. Biol. Chem. 1989; 264:675–678. [PubMed] [Google Scholar]
- 2.Halford S.E., Marko J.F.. How do site-specific DNA-binding proteins find their targets?. Nucleic Acids Res. 2004; 32:3040–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tafvizi A., Mirny L.A., van Oijen A.M.. Dancing on DNA: kinetic aspects of search processes on DNA. Chem. Phys. Chem. 2011; 12:1481–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monico C., Capitanio M., Belcastro G., Vanzi F., Pavone F.S.. Optical methods to study protein-DNA interactions in vitro and in living cells at the single-molecule level. Int. J. Mol. Sci. 2013; 14:3961–3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Redding S., Greene E.C.. How do proteins locate specific targets in DNA?. Chem. Phys. Lett. 2013; 570: 10.1016/j.cplett.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamagata K., Murata A., Itoh Y., Takahashi S.. Characterization of facilitated diffusion of tumor suppressor p53 along DNA using single-molecule fluorescence imaging. J. Photochem. Photobiol. C Photochem. Rev. 2017; 30:36–50. [Google Scholar]
- 7.Kamagata K., Itoh Y., Subekti D.R.G.. How p53 molecules solve the target DNA search problem: a review. Int. J. Mol. Sci. 2020; 21:1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kabata H., Kurosawa O., Arai I., Washizu M., Margarson S.A., Glass R.E., Shimamoto N.. Visualization of single molecules of RNA-polymerase sliding along DNA. Science. 1993; 262:1561–1563. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y.M., Austin R.H., Cox E.C.. Single molecule measurements of repressor protein 1D diffusion on DNA. Phys. Rev. Lett. 2006; 97:048302. [DOI] [PubMed] [Google Scholar]
- 10.Blainey P.C., van Oijent A.M., Banerjee A., Verdine G.L., Xie X.S.. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:5752–5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blainey P.C., Luo G., Kou S.C., Mangel W.F., Verdine G.L., Bagchi B., Xie X.S.. Nonspecifically bound proteins spin while diffusing along DNA. Nat. Struct. Mol. Biol. 2009; 16:1224–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorman J., Plys A.J., Visnapuu M.L., Alani E., Greene E.C.. Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat. Struct. Mol. Biol. 2010; 17:932–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tafvizi A., Huang F., Fersht A.R., Mirny L.A., van Oijen A.M.. A single-molecule characterization of p53 search on DNA. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:563–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorman J., Wang F., Redding S., Plys A.J., Fazio T., Wind S., Alani E.E., Greene E.C.. Single-molecule imaging reveals target-search mechanisms during DNA mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E3074–E3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson S.R., Dunn A.R., Kathe S.D., Warshaw D.M., Wallace S.S.. Two glycosylase families diffusively scan DNA using a wedge residue to probe for and identify oxidatively damaged bases. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E2091–E2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itoh Y., Murata A., Sakamoto S., Nanatani K., Wada T., Takahashi S., Kamagata K.. Activation of p53 facilitates the target search in DNA by enhancing the target recognition probability. J. Mol. Biol. 2016; 428:2916–2930. [DOI] [PubMed] [Google Scholar]
- 17.Kamagata K., Mano E., Ouchi K., Kanbayashi S., Johnson R.C.. High free-energy barrier of 1D diffusion along DNA by architectural DNA-binding proteins. J. Mol. Biol. 2018; 430:655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F., Redding S., Finkelstein I.J., Gorman J., Reichman D.R., Greene E.C.. The promoter-search mechanism of Escherichia coli RNA polymerase is dominated by three-dimensional diffusion. Nat. Struct. Mol. Biol. 2013; 20:174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sternberg S.H., Redding S., Jinek M., Greene E.C., Doudna J.A.. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014; 507:62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elf J., Li G.W., Xie X.S.. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007; 316:1191–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammar P., Leroy P., Mahmutovic A., Marklund E.G., Berg O.G., Elf J.. The Lac repressor displays facilitated diffusion in living cells. Science. 2012; 336:1595–1598. [DOI] [PubMed] [Google Scholar]
- 22.Garza de Leon F., Sellars L., Stracy M., Busby S.J.W., Kapanidis A.N.. Tracking low-copy transcription factors in living bacteria: the case of the lac repressor. Biophys. J. 2017; 112:1316–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunn A.R., Kad N.M., Nelson S.R., Warshaw D.M., Wallace S.S.. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic Acids Res. 2011; 39:7487–7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dikic J., Menges C., Clarke S., Kokkinidis M., Pingoud A., Wende W., Desbiolles P.. The rotation-coupled sliding of EcoRV. Nucleic Acids Res. 2012; 40:4064–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gowers D.M., Wilson G.G., Halford S.E.. Measurement of the contributions of 1D and 3D pathways to the translocation of a protein along DNA. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15883–15888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonnet I., Biebricher A., Porte P.-L., Loverdo C., Benichou O., Voituriez R., Escude C., Wende W., Pingoud A., Desbiolles P.. Sliding and jumping of single EcoRV restriction enzymes on non-cognate DNA. Nucleic Acids Res. 2008; 36:4118–4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sidorova N.Y., Scott T., Rau D.C.. DNA concentration-dependent dissociation of EcoRI: direct transfer or reaction during hopping. Biophys. J. 2013; 104:1296–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esadze A., Kemme C.A., Kolomeisky A.B., Iwahara J.. Positive and negative impacts of nonspecific sites during target location by a sequence-specific DNA-binding protein: origin of the optimal search at physiological ionic strength. Nucleic Acids Res. 2014; 42:7039–7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subekti D.R.G., Murata A., Itoh Y., Takahashi S., Kamagata K.. Transient binding and jumping dynamics of p53 along DNA revealed by sub-millisecond resolved single-molecule fluorescence tracking. Sci. Rep. 2020; 10:13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berg O.G., Winter R.B., Von Hippel P.H.. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981; 20:6929–6948. [DOI] [PubMed] [Google Scholar]
- 31.Pan C.Q., Finkel S.E., Cramton S.E., Feng J.A., Sigman D.S., Johnson R.C.. Variable structures of Fis-DNA complexes determined by flanking DNA-protein contacts. J. Mol. Biol. 1996; 264:675–695. [DOI] [PubMed] [Google Scholar]
- 32.Skoko D., Wong B., Johnson R.C., Marko J.F.. Micromechanical analysis of the binding of DNA-bending proteins HMGB1, NHP6A, and HU reveals their ability to form highly stable DNA-protein complexes. Biochemistry. 2004; 43:13867–13874. [DOI] [PubMed] [Google Scholar]
- 33.Skoko D., Yoo D., Bai H., Schnurr B., Yan J., McLeod S.M., Marko J.F., Johnson R.C.. Mechanism of chromosome compaction and looping by the Escherichia coli nucleoid protein Fis. J. Mol. Biol. 2006; 364:777–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giuntoli R.D., Linzer N.B., Banigan E.J., Sing C.E., de la Cruz M.O., Graham J.S., Johnson R.C., Marko J.F.. DNA-segment-facilitated dissociation of Fis and NHP6A from DNA detected via single-molecule mechanical response. J. Mol. Biol. 2015; 427:3123–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Itoh Y., Murata A., Takahashi S., Kamagata K.. Intrinsically disordered domain of tumor suppressor p53 facilitates target search by ultrafast transfer between different DNA strands. Nucleic Acids Res. 2018; 46:7261–7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cuculis L., Abil Z., Zhao H., Schroeder C.M.. Direct observation of TALE protein dynamics reveals a two-state search mechanism. Nat. Commun. 2015; 6:7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cuculis L., Abil Z., Zhao H., Schroeder C.M.. TALE proteins search DNA using a rotationally decoupled mechanism. Nat. Chem. Biol. 2016; 12:831–837. [DOI] [PubMed] [Google Scholar]
- 38.Kochaniak A.B., Habuchi S., Loparo J.J., Chang D.J., Cimprich K.A., Walter J.C., van Oijen A.M.. Proliferating cell nuclear antigen uses two distinct modes to move along DNA. J. Biol. Chem. 2009; 284:17700–17710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daitchman D., Greenblatt H.M., Levy Y.. Diffusion of ring-shaped proteins along DNA: case study of sliding clamps. Nucleic Acids Res. 2018; 46:5935–5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho W.K., Jeong C., Kim D., Chang M., Song K.M., Hanne J., Ban C., Fishel R., Lee J.B.. ATP alters the diffusion mechanics of MutS on mismatched DNA. Structure. 2012; 20:1264–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwarz F.W., Tóth J., van Aelst K., Cui G., Clausing S., Szczelkun M.D., Seidel R.. The helicase-like domains of type III restriction enzymes trigger long-range diffusion along DNA. Science. 2013; 340:353–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson R.C., Johnson L.M., Schmidt J.W., Gardner J.F., Higgins N.P.The bacterial chromosome. 2005; Washington, D.C.ASM Press; 65–132. [Google Scholar]
- 43.Bianchi M.E., Agresti A.. HMG proteins: dynamic players in gene regulation and differentiation. Curr. Opin. Genet. Dev. 2005; 15:496–506. [DOI] [PubMed] [Google Scholar]
- 44.Dillon S.C., Dorman C.J.. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010; 8:185–195. [DOI] [PubMed] [Google Scholar]
- 45.Stros M.HMGB proteins: interactions with DNA and chromatin. Biochim. Biophys. Acta. 2010; 1799:101–113. [DOI] [PubMed] [Google Scholar]
- 46.Browning D.F., Grainger D.C., Busby S.J.. Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr. Opin. Microbiol. 2010; 13:773–780. [DOI] [PubMed] [Google Scholar]
- 47.Reeves R.Nuclear functions of the HMG proteins. Biochim. Biophys. Acta. 2010; 1799:3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dame R.T., Rashid F.M., Grainger D.C.. Chromosome organization in bacteria: mechanistic insights into genome structure and function. Nat. Rev. Genet. 2020; 21:227–242. [DOI] [PubMed] [Google Scholar]
- 49.Berman H.M.Crystal studies of B-DNA: the answers and the questions. Biopolymers. 1997; 44:23–44. [DOI] [PubMed] [Google Scholar]
- 50.Olson W.K., Gorin A.A., Lu X.J., Hock L.M., Zhurkin V.B.. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:11163–11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dickerson R.E.DNA bending: the prevalence of kinkiness and the virtues of normality. Nucleic Acids Res. 1998; 26:1906–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rohs R., Jin X., West S.M., Joshi R., Honig B., Mann R.S.. Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 2010; 79:233–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leven I., Levy Y.. Quantifying the two-state facilitated diffusion model of protein-DNA interactions. Nucleic Acids Res. 2019; 47:5530–5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yen Y.M., Wong B., Johnson R.C.. Determinants of DNA binding and bending by the Saccharomyces cerevisiae high mobility group protein NHP6A that are important for its biological activities. Role of the unique N terminus and putative intercalating methionine. J. Biol. Chem. 1998; 273:4424–4435. [DOI] [PubMed] [Google Scholar]
- 55.Stella S., Cascio D., Johnson R.C.. The shape of the DNA minor groove directs binding by the DNA-bending protein Fis. Genes Dev. 2010; 24:814–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Igarashi C., Murata A., Itoh Y., Subekti D.R.G., Takahashi S., Kamagata K.. DNA garden: a simple method for producing arrays of stretchable DNA for single-molecule fluorescence imaging of DNA-binding proteins. Bull. Chem. Soc. Jpn. 2017; 90:34–43. [Google Scholar]
- 57.Schneider C.A., Rasband W.S., Eliceiri K.W.. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012; 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vestergaard C.L., Blainey P.C., Flyvbjerg H.. Single-particle trajectories reveal two-state diffusion-kinetics of hOGG1 proteins on DNA. Nucleic Acids Res. 2018; 46:2446–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kamagata K., Ouchi K., Tan C., Mano E., Mandali S., Wu Y., Takada S., Takahashi S., Johnson R.C.. The HMGB chromatin protein Nhp6A can bypass obstacles when traveling on DNA. Nucleic Acids Res. 2020; 48:10820–10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murata A., Ito Y., Kashima R., Kanbayashi S., Nanatani K., Igarashi C., Okumura M., Inaba K., Tokino T., Takahashi S.et al.. One-dimensional sliding of p53 along DNA is accelerated in the presence of Ca(2+) or Mg(2+) at millimolar concentrations. J. Mol. Biol. 2015; 427:2663–2678. [DOI] [PubMed] [Google Scholar]
- 61.Yen Y.M., Roberts P.M., Johnson R.C.. Nuclear localization of the Saccharomyces cerevisiae HMG protein NHP6A occurs by a Ran-independent nonclassical pathway. Traffic. 2001; 2:449–464. [DOI] [PubMed] [Google Scholar]
- 62.Graham J.S., Johnson R.C., Marko J.F.. Concentration-dependent exchange accelerates turnover of proteins bound to double-stranded DNA. Nucleic Acids Res. 2011; 39:2249–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hadizadeh Yazdi N., Guet C.C., Johnson R.C., Marko J.F.. Variation of the folding and dynamics of the Escherichia coli chromosome with growth conditions. Mol. Microbiol. 2012; 86:1318–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wery M., Woldringh C.L., Rouviere-Yaniv J.. HU-GFP and DAPI co-localize on the Escherichia coli nucleoid. Biochimie. 2001; 83:193–200. [DOI] [PubMed] [Google Scholar]
- 65.Wang W., Li G.W., Chen C., Xie X.S., Zhuang X.. Chromosome organization by a nucleoid-associated protein in live bacteria. Science. 2011; 333:1445–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fisher J.K., Bourniquel A., Witz G., Weiner B., Prentiss M., Kleckner N.. Four-dimensional imaging of E. coli nucleoid organization and dynamics in living cells. Cell. 2013; 153:882–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stracy M., Lesterlin C., Garza de Leon F., Uphoff S., Zawadzki P., Kapanidis A.N.. Live-cell superresolution microscopy reveals the organization of RNA polymerase in the bacterial nucleoid. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E4390–E4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bettridge K., Verma S., Weng X., Adhya S., Xiao J.. Single-molecule tracking reveals that the nucleoid-associated protein HU plays a dual role in maintaining proper nucleoid volume through differential interactions with chromosomal DNA. Mol. Microbiol. 2021; 115:12–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stracy M., Kapanidis A.N.. Single-molecule and super-resolution imaging of transcription in living bacteria. Methods. 2017; 120:103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Masse J.E., Wong B., Yen Y.M., Allain F.H., Johnson R.C., Feigon J.. The S. cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding. J. Mol. Biol. 2002; 323:263–284. [DOI] [PubMed] [Google Scholar]
- 71.Li W., Wang W., Takada S.. Energy landscape views for interplays among folding, binding, and allostery of calmodulin domains. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:10550–10555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Freeman G.S., Hinckley D.M., Lequieu J.P., Whitmer J.K., de Pablo J.J.. Coarse-grained modeling of DNA curvature. J. Chem. Phys. 2014; 141:165103. [DOI] [PubMed] [Google Scholar]
- 73.Tan C., Takada S.. Dynamic and structural modeling of the specificity in protein-DNA interactions guided by binding assay and structure data. J. Chem. Theory Comput. 2018; 14:3877–3889. [DOI] [PubMed] [Google Scholar]
- 74.Kenzaki H., Koga N., Hori N., Kanada R., Li W., Okazaki K., Yao X.Q., Takada S.. CafeMol: a coarse-grained biomolecular simulator for simulating proteins at work. J. Chem. Theory Comput. 2011; 7:1979–1989. [DOI] [PubMed] [Google Scholar]
- 75.Ester M., Kriegel H.-P., Sander J., Xu X.. A density-based algorithm for discovering clusters in large spatial databases with noise. KDD. 1996; 96:226–231. [Google Scholar]
- 76.Smola A.J., Schölkopf B.. A tutorial on support vector regression. Stat. Comput. 2004; 14:199–222. [Google Scholar]
- 77.Kolodrubetz D., Burgum A.. Duplicated NHP6 genes of Saccharomyces cerevisiae encode proteins homologous to bovine high mobility group protein 1. J. Biol. Chem. 1990; 265:3234–3239. [PubMed] [Google Scholar]
- 78.Paull T.T., Johnson R.C.. DNA looping by Saccharomyces cerevisiae high mobility group proteins NHP6A/B. Consequences for nucleoprotein complex assembly and chromatin condensation. J. Biol. Chem. 1995; 270:8744–8754. [DOI] [PubMed] [Google Scholar]
- 79.Paull T.T., Carey M., Johnson R.C.. Yeast HMG proteins NHP6A/B potentiate promoter-specific transcriptional activation in vivo and assembly of preinitiation complexes in vitro. Genes Dev. 1996; 10:2769–2781. [DOI] [PubMed] [Google Scholar]
- 80.Stillman D.J.Nhp6: a small but powerful effector of chromatin structure in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2010; 1799:175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McCullough L.L., Connell Z., Xin H., Studitsky V.M., Feofanov A.V., Valieva M.E., Formosa T.. Functional roles of the DNA-binding HMGB domain in the histone chaperone FACT in nucleosome reorganization. J. Biol. Chem. 2018; 293:6121–6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moreira J.M., Holmberg S.. Chromatin-mediated transcriptional regulation by the yeast architectural factors NHP6A and NHP6B. EMBO J. 2000; 19:6804–6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kruppa M., Moir R.D., Kolodrubetz D., Willis I.M.. Nhp6, an HMG1 protein, functions in SNR6 transcription by RNA polymerase III in S. cerevisiae. Mol. Cell. 2001; 7:309–318. [DOI] [PubMed] [Google Scholar]
- 84.Lopez S., Livingstone-Zatchej M., Jourdain S., Thoma F., Sentenac A., Marsolier M.C.. High-mobility-group proteins NHP6A and NHP6B participate in activation of the RNA polymerase III SNR6 gene. Mol. Cell. Biol. 2001; 21:3096–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Martin M.P., Gerlach V.L., Brow D.A.. A novel upstream RNA polymerase III promoter element becomes essential when the chromatin structure of the yeast U6 RNA gene is altered. Mol. Cell. Biol. 2001; 21:6429–6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Biswas D., Imbalzano A.N., Eriksson P., Yu Y., Stillman D.J.. Role for Nhp6, Gcn5, and the Swi/Snf complex in stimulating formation of the TATA-binding protein-TFIIA-DNA complex. Mol. Cell. Biol. 2004; 24:8312–8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Labazi M., Jaafar L., Flores-Rozas H.. Modulation of the DNA-binding activity of Saccharomyces cerevisiae MSH2-MSH6 complex by the high-mobility group protein NHP6A, in vitro. Nucleic Acids Res. 2009; 37:7581–7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dai Y., Wong B., Yen Y.M., Oettinger M.A., Kwon J., Johnson R.C.. Determinants of HMGB proteins required to promote RAG1/2-recombination signal sequence complex assembly and catalysis during V(D)J recombination. Mol. Cell. Biol. 2005; 25:4413–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Formosa T., Eriksson P., Wittmeyer J., Ginn J., Yu Y., Stillman D.J.. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 2001; 20:3506–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brewster N.K., Johnston G.C., Singer R.A.. A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol. Cell. Biol. 2001; 21:3491–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hepp M.I., Alarcon V., Dutta A., Workman J.L., Gutiérrez J.L.. Nucleosome remodeling by the SWI/SNF complex is enhanced by yeast high mobility group box (HMGB) proteins. Biochim. Biophys. Acta. 2014; 1839:764–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kurat C.F., Yeeles J.T.P., Patel H., Early A., Diffley J.F.X.. Chromatin controls DNA replication origin selection, lagging-strand synthesis, and replication fork rates. Mol. Cell. 2017; 65:117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hepp M.I., Smolle M., Gidi C., Amigo R., Valenzuela N., Arriagada A., Maureira A., Gogol M.M., Torrejón M., Workman J.L.et al.. Role of Nhp6 and Hmo1 in SWI/SNF occupancy and nucleosome landscape at gene regulatory regions. Biochim. Biophys. Acta Gene Regul. Mech. 2017; 1860:316–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Formosa T., Winston F.. The role of FACT in managing chromatin: disruption, assembly, or repair?. Nucleic Acids Res. 2020; 48:11929–11941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Allain F.H., Yen Y.M., Masse J.E., Schultze P., Dieckmann T., Johnson R.C., Feigon J.. Solution structure of the HMG protein NHP6A and its interaction with DNA reveals the structural determinants for non-sequence-specific binding. EMBO J. 1999; 18:2563–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dowell N.L., Sperling A.S., Mason M.J., Johnson R.C.. Chromatin-dependent binding of the S. cerevisiae HMGB protein Nhp6A affects nucleosome dynamics and transcription. Genes Dev. 2010; 24:2031–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wong B., Masse J.E., Yen Y.M., Giannikopoulos P., Feigon J., Johnson R.C.. Binding to cisplatin-modified DNA by the Saccharomyces cerevisiae HMGB protein Nhp6A. Biochemistry. 2002; 41:5404–5414. [DOI] [PubMed] [Google Scholar]
- 98.Kamar R.I., Banigan E.J., Erbas A., Giuntoli R.D., Olvera de la Cruz M., Johnson R.C., Marko J.F.. Facilitated dissociation of transcription factors from single DNA binding sites. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E3251–E3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Elowitz M.B., Surette M.G., Wolf P.E., Stock J.B., Leibler S.. Protein mobility in the cytoplasm of Escherichia coli. J. Bacteriol. 1999; 181:197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rouvière-Yaniv J., Gros F.. Characterization of a novel, low-molecular-weight DNA-binding protein from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1975; 72:3428–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Claret L., Rouviere-Yaniv J.. Variation in HU composition during growth of Escherichia coli: the heterodimer is required for long term survival. J. Mol. Biol. 1997; 273:93–104. [DOI] [PubMed] [Google Scholar]
- 102.Ishihama A., Kori A., Koshio E., Yamada K., Maeda H., Shimada T., Makinoshima H., Iwata A., Fujita N.. Intracellular concentrations of 65 species of transcription factors with known regulatory functions in Escherichia coli. J. Bacteriol. 2014; 196:2718–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Aki T., Choy H.E., Adhya S.. Histone-like protein HU as a specific transcriptional regulator: co-factor role in repression of gal transcription by GAL repressor. Genes Cells. 1996; 1:179–188. [DOI] [PubMed] [Google Scholar]
- 104.Becker N.A., Kahn J.D., Maher L.J. 3rd. Effects of nucleoid proteins on DNA repression loop formation in Escherichia coli. Nucleic Acids Res. 2007; 35:3988–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Oberto J., Nabti S., Jooste V., Mignot H., Rouviere-Yaniv J.. The HU regulon is composed of genes responding to anaerobiosis, acid stress, high osmolarity and SOS induction. PLoS One. 2009; 4:e4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Berger M., Farcas A., Geertz M., Zhelyazkova P., Brix K., Travers A., Muskhelishvili G.. Coordination of genomic structure and transcription by the main bacterial nucleoid-associated protein HU. EMBO Rep. 2010; 11:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Prieto A.I., Kahramanoglou C., Ali R.M., Fraser G.M., Seshasayee A.S., Luscombe N.M.. Genomic analysis of DNA binding and gene regulation by homologous nucleoid-associated proteins IHF and HU in Escherichia coli K12. Nucleic Acids Res. 2012; 40:3524–3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dixon N.E., Kornberg A.. Protein HU in the enzymatic replication of the chromosomal origin of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1984; 81:424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Leonard A.C., Grimwade J.E.. Initiation of DNA replication. EcoSal Plus. 2010; 4: 10.1128/ecosalplus.4.4.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Craigie R., Arndt-Jovin D.J., Mizuuchi K.. A defined system for the DNA strand-transfer reaction at the initiation of bacteriophage Mu transposition: protein and DNA substrate requirements. Proc. Natl. Acad. Sci. U.S.A. 1985; 82:7570–7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Johnson R.C., Bruist M.F., Simon M.I.. Host protein requirements for in vitro site-specific DNA inversion. Cell. 1986; 46:531–539. [DOI] [PubMed] [Google Scholar]
- 112.Boubrik F., Rouviere-Yaniv J.. Increased sensitivity to gamma irradiation in bacteria lacking protein HU. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:3958–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li S., Waters R.. Escherichia coli strains lacking protein HU are UV sensitive due to a role for HU in homologous recombination. J. Bacteriol. 1998; 180:3750–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lioy V.S., Cournac A., Marbouty M., Duigou S., Mozziconacci J., Espéli O., Boccard F., Koszul R.. Multiscale structuring of the E. coli chromosome by nucleoid-associated and condensin proteins. 2018; 172:771–783. [DOI] [PubMed] [Google Scholar]
- 115.Verma S.C., Qian Z., Adhya S.L.. Architecture of the Escherichia coli nucleoid. PLoS Genet. 2019; 15:e1008456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pontiggia A., Negri A., Beltrame M., Bianchi M.E.. Protein HU binds specifically to kinked DNA. Mol. Microbiol. 1993; 7:343–350. [DOI] [PubMed] [Google Scholar]
- 117.Castaing B., Zelwer C., Laval J., Boiteux S.. HU protein of Escherichia coli binds specifically to DNA that contains single-strand breaks or gaps. J. Biol. Chem. 1995; 270:10291–10296. [DOI] [PubMed] [Google Scholar]
- 118.Kamashev D., Rouviere-Yaniv J.. The histone-like protein HU binds specifically to DNA recombination and repair intermediates. EMBO J. 2000; 19:6527–6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Swinger K.K., Rice P.A.. Structure-based analysis of HU-DNA binding. J. Mol. Biol. 2007; 365:1005–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hodges-Garcia Y., Hagerman P.J., Pettijohn D.E.. DNA ring closure mediated by protein HU. J. Biol. Chem. 1989; 264:14621–14623. [PubMed] [Google Scholar]
- 121.van Noort J., Verbrugge S., Goosen N., Dekker C., Dame R.T.. Dual architectural roles of HU: formation of flexible hinges and rigid filaments. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:6969–6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xiao B., Johnson R.C., Marko J.F.. Modulation of HU-DNA interactions by salt concentration and applied force. Nucleic Acids Res. 2010; 38:6176–6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Swinger K.K., Lemberg K.M., Zhang Y., Rice P.A.. Flexible DNA bending in HU-DNA cocrystal structures. EMBO J. 2003; 22:3749–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Paull T.T., Haykinson M.J., Johnson R.C.. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993; 7:1521–1534. [DOI] [PubMed] [Google Scholar]
- 125.Segall A.M., Goodman S.D., Nash H.A.. Architectural elements in nucleoprotein complexes: interchangeability of specific and non-specific DNA binding proteins. EMBO J. 1994; 13:4536–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Becker N.A., Kahn J.D., Maher L.J. III. Eukaryotic HMGB proteins as replacements for HU in E. coli repression loop formation. Nucleic Acids Res. 2008; 36:4009–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pinson V., Takahashi M., Rouviere-Yaniv J.. Differential binding of the Escherichia coli HU, homodimeric forms and heterodimeric form to linear, gapped and cruciform DNA. J. Mol. Biol. 1999; 287:485–497. [DOI] [PubMed] [Google Scholar]
- 128.Ball C.A., Osuna R., Ferguson K.C., Johnson R.C.. Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J. Bacteriol. 1992; 174:8043–8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cho B.K., Knight E.M., Barrett C.L., Palsson B.O.. Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts. Genome Res. 2008; 18:900–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kahramanoglou C., Seshasayee A.S., Prieto A.I., Ibberson D., Schmidt S., Zimmermann J., Benes V., Fraser G.M., Luscombe N.M.. Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucleic Acids Res. 2011; 39:2073–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.McLeod S.M., Aiyar S.E., Gourse R.L., Johnson R.C.. The C-terminal domains of the RNA polymerase alpha subunits: contact site with Fis and localization during co-activation with CRP at the Escherichia coli proP P2 promoter. J. Mol. Biol. 2002; 316:517–529. [DOI] [PubMed] [Google Scholar]
- 132.Aiyar S.E., McLeod S.M., Ross W., Hirvonen C.A., Thomas M.S., Johnson R.C., Gourse R.L.. Architecture of Fis-activated transcription complexes at the Escherichia coli rrnB P1 and rrnE P1 promoters. J. Mol. Biol. 2002; 316:501–516. [DOI] [PubMed] [Google Scholar]
- 133.Typas A., Stella S., Johnson R.C., Hengge R.. The -35 sequence location and the Fis-sigma factor interface determine sigmas selectivity of the proP (P2) promoter in Escherichia coli. Mol. Microbiol. 2007; 63:780–796. [DOI] [PubMed] [Google Scholar]
- 134.Johnson R.C.Site-specific DNA inversion by serine recombinases. Microbiol. Spect. 2015; 3: 10.1128/microbiolspec.MDNA3-0047-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Landy A.The λ Integrase site-specific recombination pathway. Microbiol. Spect. 2015; 3: 10.1128/microbiolspec.MDNA3-0051-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kaur G., Vora M.P., Czerwonka C.A., Rozgaja T.A., Grimwade J.E., Leonard A.C.. Building the bacterial orisome: high-affinity DnaA recognition plays a role in setting the conformation of oriC DNA. Mol. Microbiol. 2014; 91:1148–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hancock S.P., Stella S., Cascio D., Johnson R.C.. DNA sequence determinants controlling affinity, stability and shape of DNA complexes bound by the nucleoid protein Fis. PLoS One. 2016; 11:e0150189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hancock S.P., Ghane T., Cascio D., Rohs R., Di Felice R., Johnson R.C.. Control of DNA minor groove width and Fis protein binding by the purine 2-amino group. Nucleic Acids Res. 2013; 41:6750–6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kostrewa D., Granzin J., Koch C., Choe H.W., Raghunathan S., Wolf W., Labahn J., Kahmann R., Saenger W.. Three-dimensional structure of the E. coli DNA-binding protein FIS. Nature. 1991; 349:178–180. [DOI] [PubMed] [Google Scholar]
- 140.Yuan H.S., Finkel S.E., Feng J.A., Kaczor-Grzeskowiak M., Johnson R.C., Dickerson R.E.. The molecular structure of wild-type and a mutant Fis protein: relationship between mutational changes and recombinational enhancer function or DNA binding. Proc. Natl. Acad. Sci. U.S.A. 1991; 88:9558–9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Koch C., Ninnemann O., Fuss H., Kahmann R.. The N-terminal part of the E.coli DNA binding protein FIS is essential for stimulating site-specific DNA inversion but is not required for specific DNA binding. Nucleic Acids Res. 1991; 19:5915–5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Osuna R., Finkel S.E., Johnson R.C.. Identification of two functional regions in Fis: the N-terminus is required to promote Hin-mediated DNA inversion but not lambda excision. EMBO J. 1991; 10:1593–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Feldman-Cohen L.S., Shao Y., Meinhold D., Miller C., Colón W., Osuna R.. Common and variable contributions of Fis residues to high-affinity binding at different DNA sequences. J. Bacteriol. 2006; 188:2081–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shao Y., Feldman-Cohen L.S., Osuna R.. Functional characterization of the Escherichia coli Fis-DNA binding sequence. J. Mol. Biol. 2008; 376:771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Merickel S.K., Sanders E.R., Vazquez-Ibar J.L., Johnson R.C.. Subunit exchange and the role of dimer flexibility in DNA binding by the Fis protein. Biochemistry. 2002; 41:5788–5798. [DOI] [PubMed] [Google Scholar]
- 146.Thomas J.O., Travers A.A.. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 2001; 26:167–174. [DOI] [PubMed] [Google Scholar]
- 147.Rohs R., West S.M., Sosinsky A., Liu P., Mann R.S., Honig B.. The role of DNA shape in protein-DNA recognition. Nature. 2009; 461:1248–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sarangi M.K., Zvoda V., Holte M.N., Becker N.A., Peters J.P., Maher L.J., Ansari A.. Evidence for a bind-then-bend mechanism for architectural DNA binding protein yNhp6A. Nucleic Acids Res. 2019; 47:2871–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.McCauley M.J., Rueter E.M., Rouzina I., Maher L.J. 3rd, Williams M.C.. Single-molecule kinetics reveal microscopic mechanism by which High-Mobility Group B proteins alter DNA flexibility. Nucleic Acids Res. 2013; 41:167–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Countryman P., Fan Y., Gorthi A., Pan H., Strickland J., Kaur P., Wang X., Lin J., Lei X., White C.et al.. Cohesin SA2 is a sequence-independent DNA-binding protein that recognizes DNA replication and repair intermediates. J. Biol. Chem. 2018; 293:1054–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Vuzman D., Levy Y.. Intrinsically disordered regions as affinity tuners in protein-DNA interactions. Mol. Biosyst. 2012; 8:47–57. [DOI] [PubMed] [Google Scholar]
- 152.Khazanov N., Levy Y.. Sliding of p53 along DNA can be modulated by its oligomeric state and by cross-talks between its constituent domains. J. Mol. Biol. 2011; 408:335–355. [DOI] [PubMed] [Google Scholar]
- 153.Terakawa T., Kenzaki H., Takada S.. p53 searches on DNA by rotation-uncoupled sliding at C-terminal tails and restricted hopping of core domains. J. Am. Chem. Soc. 2012; 134:14555–14562. [DOI] [PubMed] [Google Scholar]
- 154.Murata A., Itoh Y., Mano E., Kanbayashi S., Igarashi C., Takahashi H., Takahashi S., Kamagata K.. One-dimensional search dynamics of tumor suppressor p53 regulated by a disordered C-terminal domain. Biophys. J. 2017; 112:2301–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kamagata K., Mano E., Itoh Y., Wakamoto T., Kitahara R., Kanbayashi S., Takahashi H., Murata A., Kameda T.. Rational design using sequence information only produces a peptide that binds to the intrinsically disordered region of p53. Sci. Rep. 2019; 9:8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Subekti D.R.G., Kamagata K.. The disordered DNA-binding domain of p53 is indispensable for forming an encounter complex to and jumping along DNA. Biochem. Biophys. Res. Commun. 2021; 534:21–26. [DOI] [PubMed] [Google Scholar]
- 157.Subekti D.R.G., Murata A., Itoh Y., Fukuchi S., Takahashi H., Kanbayashi S., Takahashi S., Kamagata K.. The disordered linker in p53 participates in nonspecific binding to and one-dimensional sliding along DNA revealed by single-molecule fluorescence measurements. Biochemistry. 2017; 56:4134–4144. [DOI] [PubMed] [Google Scholar]
- 158.Mangel W.F., McGrath W.J., Xiong K., Graziano V., Blainey P.C.. Molecular sled is an eleven-amino acid vehicle facilitating biochemical interactions via sliding components along DNA. Nat. Commun. 2016; 7:10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Xiong K., Blainey P.C.. Molecular sled sequences are common in mammalian proteins. Nucleic Acids Res. 2016; 44:2266–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Vuzman D., Azia A., Levy Y.. Searching DNA via a “Monkey Bar” mechanism: the significance of disordered tails. J. Mol. Biol. 2010; 396:674–684. [DOI] [PubMed] [Google Scholar]
- 161.Tan C., Terakawa T., Takada S.. Dynamic coupling among protein binding, sliding, and DNA bending revealed by molecular dynamics. J. Am. Chem. Soc. 2016; 138:1520–5126. [DOI] [PubMed] [Google Scholar]
- 162.Rice P.A., Yang S., Mizuuchi K., Nash H.A.. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996; 87:1295–1306. [DOI] [PubMed] [Google Scholar]
- 163.Mouw K.W., Rice P.A.. Shaping the Borrelia burgdorferi genome: crystal structure and binding properties of the DNA-bending protein Hbb. Mol. Microbiol. 2007; 63:1319–1330. [DOI] [PubMed] [Google Scholar]
- 164.Lavoie B.D., Shaw G.S., Millner A., Chaconas G.. Anatomy of a flexer-DNA complex inside a higher-order transposition intermediate. Cell. 1996; 85:761–771. [DOI] [PubMed] [Google Scholar]
- 165.Koh J., Saecker R.M., Record M.T. Jr. DNA binding mode transitions of Escherichia coli HU alpha beta: evidence for formation of a bent DNA - protein complex on intact, linear duplex DNA. J. Mol. Biol. 2008; 383:324–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Koh J., Shkel I., Saecker R.M., Record M.T. Jr. Nonspecific DNA binding and bending by HU alpha beta: interfaces of the three binding modes characterized by salt-dependent thermodynamics. J. Mol. Biol. 2011; 410:241–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hammel M., Amlanjyoti D., Reyes F.E., Chen J.H., Parpana R., Tang H.Y., Larabell C.A., Tainer J.A., Adhya S.. HU multimerization shift controls nucleoid compaction. Sci. Adv. 2016; 2:e1600650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Remesh S.G., Verma S.C., Chen J.H., Ekman A.A., Larabell C.A., Adhya S., Hammel M.. Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling. Nat. Commun. 2020; 11:2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hanover J.A., Love D.C., DeAngelis N., O’Kane M.E., Lima-Miranda R., Schulz T., Yen Y.M., Johnson R.C., Prinz W.A.. The High Mobility Group Box transcription factor Nhp6Ap enters the nucleus by a calmodulin-dependent, Ran-independent pathway. J. Biol. Chem. 2007; 282:33743–33751. [DOI] [PubMed] [Google Scholar]
- 170.Slutsky M., Mirny L.A.. Kinetics of protein-DNA interaction: facilitated target location in sequence-dependent potential. Biophys. J. 2004; 87:4021–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Winter R.B., Berg O.G., Von Hippel P.H.. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli Lac repressor–operator interaction: kinetic measurements and conclusions. Biochemistry. 1981; 20:6961–6977. [DOI] [PubMed] [Google Scholar]
- 172.Spolar R.S., Record M.T. Jr. Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994; 263:777–784. [DOI] [PubMed] [Google Scholar]
- 173.Bruinsma R.F.Physics of protein-DNA interaction. Phys. a Stat. Mech. Its Appl. 2002; 313:211–237. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.