Abstract
Nucleocapsid protein (N protein) is the most abundant protein in SARS-CoV2 and is highly conserved, and there are no homologous proteins in the human body, making it an ideal biomarker for the early diagnosis of SARS-CoV2. However, early detection of clinical specimens for SARS-CoV2 remains a challenge due to false-negative results with viral RNA and host antibodies based testing. In this manuscript, a microfluidic chip with femtoliter-sized wells was fabricated for the sensitive digital detection of N protein. Briefly, β-galactosidase (β-Gal)-linked antibody/N protein/aptamer immunocomplexes were formed on magnetic beads (MBs). Afterwards, the MBs and β-Gal substrate fluorescein-di-β-d-galactopyranoside (FDG) were injected into the chip together. Each well of the chip would only hold one MB as confined by the diameter of the wells. The MBs in the wells were sealed by fluorocarbon oil, which confines the fluorescent (FL) product generated from the reaction between β-Gal and FDG in the individual femtoliter-sized well and creates a locally high concentration of the FL product. The FL images of the wells were acquired using a conventional inverted FL microscope. The number of FL wells with MBs (FL wells number) and the number of wells with MBs (MBs wells number) were counted, respectively. The percentage of FL wells was calculated by dividing (FL wells number) by (MBs wells number). The higher the percentage of FL wells, the higher the N protein concentration. The detection limit of this digital method for N protein was 33.28 pg/mL, which was 300 times lower than traditional double-antibody sandwich based enzyme-linked immunosorbent assay (ELISA).
Keywords: Digital detection, Microfluidic chip, COVID-19, SARS-CoV2, Nucleocapsid protein, Aptamer/antibody sandwich
Graphical abstract
An aptamer/antibody sandwich method for digital detection of SARS-CoV2 nucleocapsid protein based on microfluidic chip with femtoliter-sized wells was proposed for the first time.
1. Introduction
The outbreak of coronavirus disease 2019 (COVID-19) caused a global health crisis. COVID-19 is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV2), and it has high infectivity and high lethality [1]. More than 190 million people have been diagnosed with COVID-19, and millions died from COVID-19 [2]. Although COVID-19 vaccines have been administered worldwide, early diagnosis remains an effective way for early isolation of patients with COVID-19 and control the spread of SARS-CoV-2 in the context of the limited effectiveness of the vaccine and the fast mutations of SARS-CoV-2 [3].
At present, the reverse transcription-polymerase chain reaction (RT-PCR) test is the gold standard for the diagnosis of SARS-CoV-2 [4]. Nevertheless, there is growing evidence demonstrating that this technique may generate false-negative results [5]. Direct detection of host antibodies (IgM) from a small volume of serum or plasma is generally less sensitive than RT-PCR, and SARS-COV-2 may be detected within the first week after symptoms onset, while the viral load is typically higher. When detected 10–14 days after the onset of symptoms, the viral load is low or undetectable, the performance of these tests decreases significantly [6,7].
In addition to viral RNA and host antibodies, viral structural proteins are also alternative targets for SARS-CoV2 detection, such as nucleocapsid protein (N protein), spike protein (S protein), membrane protein, and envelope protein [8]. N protein is the most abundant protein in SARS-CoV2 and is highly conserved [9], and there are no homologous proteins in the human body. Therefore, N protein is an ideal biomarker for the early diagnosis of SARS-CoV2.
Aptamers are high-affinity single-stranded DNA or RNA molecules to specific targets and screened using an in vitro selection procedure called Systematic Evolution of Ligands by EXponential enrichment (SELEX) discovered by Gold and Szostak in the 1990s [10]. Up till now, many nucleic acid aptamers have been identified using SELEX, including aptamers that are specific to pathogenic organisms [11], proteins [12], cells [13], heavy metal ions [14], and small molecules [15]. Compared with antibodies widely used in biomedicine, the aptamer is a novel molecular recognition element. The specificity and affinity of aptamer binding to target are equal to or even stronger than that of antibody. Furthermore, aptamer has the advantages of a short screening cycle, low immunogenicity, simple and rapid synthesis, low cost, easy modification, good stability and can be transported at room temperature [16]. After the outbreak of COVID-19, Zhaofeng Luo's group quickly screened out the aptamers of N protein [17] and demonstrated that the antibody/aptamer sandwich method has a higher sensitivity for N protein detection than double-antibody or double-aptamer sandwich methods. The usage of aptamers also reduces the complexity and cost of the detection system. However, the detection sensitivity still needs to be improved if applied in early diagnosis.
A digital assay is one in which the sample is partitioned into many containers so that each partition contains a discrete number of biological molecules. The digital assays, usually based on microfluidic compartmentalized techniques, such as digital enzyme-linked immunosorbent assay (dELISA) or digital PCR, bring new levels of precision for the sensitive quantitation of nucleic acids, proteins, and the exploration of single-cell genotype and phenotype. Absolute quantitative information can be obtained with qualitative measurements [18,19]. Digital ELISA using magnetic beads (MBs) labelled specific antibodies. At a low concentration of biomarkers, each MB can capture one or zero target protein molecules. After an immune reaction, an immune complex is formed, and an enzyme that produces an FL product is used to report the signal. A final quantification was performed using FL imaging to detect the FL signal from the individual microwells by enclosing the single MB in a 50 fL reaction well [20]. Digital ELISA is about 100–1000 times more sensitive than traditional ELISA [21] and plays an irreplaceable role in the detection of protein markers in tumours [22], neurological [23], cardiovascular [24], infectious diseases [25] and immune inflammation [26].
To improve the sensitivity for N protein detection, a microfluidic chip with femtoliter-sized wells was fabricated for the digital detection of N protein (Fig. 1 ) in this study. With this method combining aptamer/antibody, we were able to detect N protein with the limit of 33.28 pg/mL, 300 times lower than traditional double-antibody sandwich ELISA.
Fig. 1.
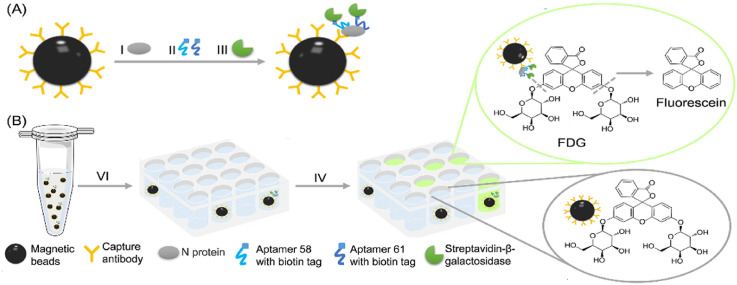
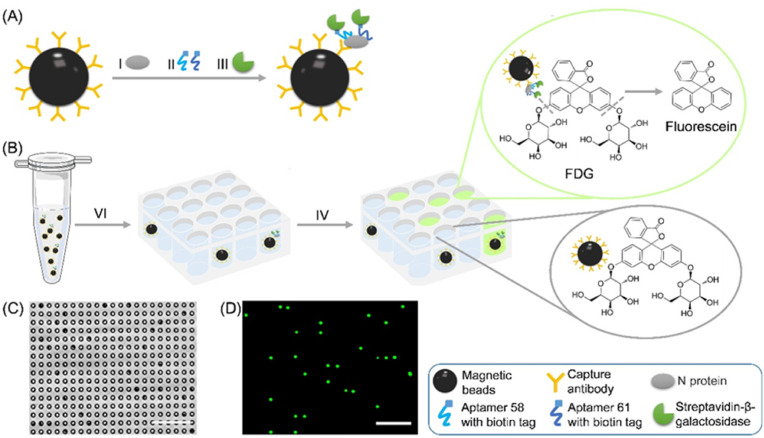
Digital detection of N protein by a microfluidic chip with femtoliter-sized wells. (A) Formation of SA-β-Gal-linked antibody/N protein/aptamer immunocomplex on MBs. (B) (Ⅵ) MBs with β-Gal substrate (FDG) was loaded together into the wells of the microfluidic chip. (Ⅳ) After being flushed with FC-40, the FL product generated from the reaction between β-Gal and FDG into individual femtoliter-sized wells. (C) The representative bright-field image and (D) FL image of the array. Scale bar: 50 μm.
2. Materials and methods
2.1. Reagents
PDMS base and curing agent were purchased from Dow Chemical Company. N-(3-Dimethylaminopropyl)-N′-ethyl carbodiimide Hydrochloride (EDC) was purchased from Sigma-Aldrich. SARS-CoV-2 nucleocapsid monoclonal antibody and biotinylated polyclonal antibody, N protein and S protein were purchased from Yitaijian (Suzhou) Biotechnology Co., Ltd. 96 well plate was purchased from Corning Inc. Carboxylated MBs, and SA coated MBs were purchased from SuZhou Nanomicro Technologies Inc. Streptavidin-β-galactosidase (SA-β-Gal) and biotin-β-galactosidase (bio-β-Gal) were purchased from Millipore. Fluorescein di-beta-d-galactopyranoside (FDG) was purchased from AAT Bioquest. Horseradish peroxidase-conjugated streptavidin (SA-HRP), 3,3 ′,5,5 ′-tetramethylbenzidine (TMB), and stop solution were purchased from Dakewe Biotech Co., Ltd. 4%–12% PAGE precast protein gel was purchase from Genscript Corporation. HRP conjugated goat anti-mouse IgG (H + L) was purchased from Proteintech Group, Inc. Ultra-sensitive ECL substrate was purchased from Thermo Fisher Scientific. Sodium carbonate and sodium hydrogen carbonate were purchased from Shanghai Macklin Biochemical Co., Ltd. Bovine serum albumin (BSA) was purchased from Beyotime Biotechnology. Tween-20 and MgCl2 were purchased from Aladdin Chemical Co., Ltd. Absolute ethyl alcohol (EtAc) was purchased from Aladdin Company. Phosphate-buffered saline (PBS) was purchased from Beijing Solar Science & Technology Co., Ltd. Fluorinert FC-40 was purchased from 3 M. SU-8 2005 was purchased from Microchem. Biotinylated aptamer 58 (Apt 58) and 61(Apt 61) were synthesized by Sangon Biotech (Shanghai, China). The sequences of Apt 58 and 61 were listed in Table 1 .
Table 1.
The sequences of apt 58 and 61.
| Name | DNA Sequence (5′-3′) |
|---|---|
| Apt 58 | biotin-GCT GGA TGT CAC CGG ATT GTC GGA CAT CGG ATT GTC TGA GTC ATA TGA CAC ATC CAG C |
| Apt 61 | biotin-GCT GGA TGT TGA CCT TTA CAG ATC GGA TTC TGT GGG GCG TTA AAC TGA CAC ATC CAG C |
2.2. Fabrication of the moulds of the channel layer and the array layer
The fabrication of the moulds of the channel layer and the array layer were referred to the previously reported article [27]. The mould of the channel layer was manufactured by computer numerical control technology. The mould of the array layer was manufactured by photolithography. Briefly, the photoresist SU-8 2005 was spin-coated to a 4-inch silicon wafer using a glue homogenizer. The first stage rotation speed is 500 rpm/min and acceleration is 100 (rpm/min)/s, spin coating for 10 s. In the second stage, the rotation speed is 4000 rpm/min and the acceleration is 500 (rpm/min)/s, spin coating for 40 s. The silicon wafer was baked at 95 °C for 3 min and return to room temperature. Load the chromium mask and silicon wafer using a SUSS MA6 lithography machine and expose them to UV for 19 s. The silicon wafer was baked at 95 °C for 3 min and return to room temperature. The silicon wafer was immersed in a SU-8 developer solution for 30 s, and rinsed with isopropyl alcohol and blow-dried with an air gun. The silicon wafer was baked at 150 °C for 20 min and return to room temperature. The moulds of the channel layer and the array layer were kept in dry and clean conditions before use.
2.3. Fabrication of the microfluidic chip
PDMS pre-polymer was prepared by mixing the base and curing agent in a ratio of 10:1 (m/m). The PDMS pre-polymer was poured over the channel layer mould and the array layer mould respectively and cured for 40 min at 70 °C. The PDMS was then peeled off from the moulds and cut into appropriate pieces. Before surface modification, inlet and outlet ports were created in the channel layer using a 0.75 mm micro punch. Surface modification was performed at 80% power for 30 s using a vacuum plasma generator (Diener Plasma, ZEPTO). The channel layer and the array layer were manually aligned immediately and pressed together to promote bonding. The assembly of the two layers was further bonded together by baking at 70 °C for 5 min.
2.4. The performance of the microfluidic chip
SA-β-Gal was added to PBST with 1 mM MgCl2 and 250 mM FDG to a final concentration of 147 nM and incubated at room temperature (RT) for 30 min. The solution was injected into the chip with a flow rate of 8 mL/cm through the inlet port using a peristaltic pump (WH-SP-02, Suzhou Wenhao). The array's bright-field images and FL images were acquired using an inverted FL microscope (ECLIPSE Ti2, Nikon).
2.5. Digital detection of the activity of β-gal
One μL streptavidin-coated MBs (3 μm) was washed twice with PBS and suspended in 100 μL PBS. Bio-β-Gal was added to a final concentration of 1 nM. React for 30 min at 25 °C, and wash MBs 5 times with PBST. The MB-β-Gal complex was resuspended in 50 μL PBST with 1 mM MgCl2 and 250 mM FDG and injected into the microfluidic chip with a flow rate of 8 mL/cm using the peristaltic pump. Place the chip at RT for 5 min. FC-40 was injected with a flow rate of 12 mL/cm to flush away MBs on the surface of the array. The chip was incubated at RT for 30 min, the bright-field and FL images of the array were acquired using the inverted FL microscope.
2.6. Aptamer-N protein interaction studies using biolayer interferometry technology
The aptamer-N protein interaction studies were carried out on the biolayer interferometry technology-based instrument (FortéBio Octet K2), using a 96-well plate in kinetics mode. Before loading apt 58 and apt 61 onto SA sensors, the sensors were hydrated for 10 min in a biosensor tray inside the Octet instrument set at 30 °C. The kinetics parameters used in the study were listed in Table 2 . The steps were carried out in different wells in a microplate, baseline: PBS, loading: apt 58 or 61 (100 nM) in PBS, baseline 2: PBST, association: N protein (0.65 nM) in PBST, dissociation: PBST. The wells were filled with 200 μL of the respective solutions for each step.
Table 2.
The kinetics parameters were used in the experiment.
| Step | Type | Time (s) | Shake speed (rpm) |
|---|---|---|---|
| 1 | Baseline | 300 | 1000 |
| 2 | Loading | 300 | 1000 |
| 3 | Baseline 2 | 300 | 1000 |
| 4 | Association | 300 | 1000 |
| 5 | Dissociation | 600 | 1000 |
2.7. Western blot
Different concentrations of N protein (100 ng and 50 ng) were separated in 4%–12% SDS-PAGE, electric-transferred to a membrane and probed with the antibody against N protein (dilution: 1:1000). The membrane was then incubated with horseradish peroxidase modified goat anti-mouse IgG antibody (dilution: 1:2000) and the bands on the PVDF membrane were developed using ECL detection reagents according to the manufacturer's instructions.
2.8. Conjugation of antibody to MBs
Carboxylated MBs (2.7 μm) were functionalized with an anti-N protein monoclonal antibody using the EDC coupling method. Briefly, 400 μg carboxylated MBs was washed twice with PBS and suspended in 100 μL PBS. 560 μg EDC was added to activated carboxyl groups at 25 °C for 20 min. Wash MBs twice quickly with pre-cooled PBS. 20 μg monoclonal antibody against N protein was added to react with activated carboxyl groups at 25 °C for 60 min. Wash MBs three times with PBS, add Tris/BSA buffer (50 mM Tris, 0.01% Tween-20, 0.5% BSA, pH 7.4) to block the unreacted activated carboxyl groups at 25 °C for 30 min. Wash antibody functionalized MBs (MBs-Ab) three times with PBS, and resuspend 400 μL Tris/BSA buffer at 4 °C before use.
2.9. Detection of N protein using ELISA method
SARS-CoV-2 nucleocapsid monoclonal antibody was added into 50 mM carbonate buffer (pH 9.6) to a final concentration of 8 mg/mL. Add 50 μL of the solution to each well and incubate at 4 °C overnight. Wash each well three times with PBST, and block with 10% BSA at 37 °C for 2 h. Wash each well three times with PBST, add 50 μL of different concentrations of N protein (3.16 mg/mL, 1 mg/mL, 316 ng/mL, 100 ng/mL, 31.6 ng/mL, 10 ng/mL, 0) and incubate at 37 °C for 1.5 h. Wash each well three times with PBST, add biotinylated polyclonal antibody (ELISA dilution: 1: 5000) respectively and incubate at 37 °C for 1 h. Wash each well three times with PBST, add SA-HRP (ELISA dilution: 1: 500) and incubate at 37 °C for 45min. Wash each well five times with PBST, add 50 μL of TMB into each well and incubate at 37 °C for 5 min. Add 50 μL of stop solution into each well, the OD values of each well at 450 nm was acquired using a microplate reader (Synergy H1, BioTek).
2.10. Digital detection of N protein
Ten μL MBs-Ab was washed three times with PBS. The MBs were resuspended and incubated with different concentrations of N protein (1000 ng/mL, 100 ng/mL, 10 ng/mL, 1 ng/mL, 100 pg/mL, 0) in PBS at 25 °C for 1 h. The MBs were washed three times with PBS, resuspended and incubated with biotinylated apt 58 and 61 (each 10 nM) in PBS at 25 °C for 1 h. The MBs were washed three times with PBS, resuspended and incubated with SA-β-Gal (14.7 nM) in PBS at 25 °C for 1 h. The MBs were washed six times in PBS, the MBs pellet was resuspended in 50 μL PBST with 1 mM MgCl2 and 250 mM FDG and injected into the microfluidic chip using the peristaltic pump. Place the chip at RT for 5 min. FC-40 was then injected to flush away MBs on the surface of the array. The chip was incubated at RT for 30 min, the bright-field and FL images of the array were acquired using the inverted FL microscope.
2.11. Image analysis
The bright-field and FL images were analyzed using self-programmed software. The number of FL wells with MBs (FL wells number) and the number of wells with MBs (MBs wells number) were counted. The percentage of FL wells was calculated by dividing (FL wells number) by (MBs wells number). The concentration of N protein can be calculated by substituting the percentage of FL wells into the standard curve equation.
3. Results and discussion
3.1. The principle of this assay for digital detection of N protein
Briefly, SA-β-Gal-linked antibody/N protein/aptamer immunocomplexes were formed on MBs (Fig. 1A). The MBs and fluorescein-di-β-d-galactopyranoside (FDG) were injected into the chip together. FDG is one of the most sensitive fluorogenic substrates of β-Gal that has no fluorescence unless hydrolyzed by β-Gal. The hydrolysis processes of the two glycosidic bonds of FDG by β-Gal were showed in Fig. S1. FDG is sequentially hydrolyzed by β-gal, first to fluorescein-mono-β-d-galactopyranoside (FMG) and then to highly fluorescent fluorescein for detection [28,29].
The chip was fabricated with polydimethylsiloxane (PDMS) using well-developed moulding techniques. There are a million femtoliter-sized wells on the chip, which form an array, and each well would only hold one MB as confined by the diameter of wells. The working process of the MBs separated into the wells of the developed microfluidic chip is as follows. Briefly, the MBs and FDG mixture was aspirated into a 1 mL syringe, the syringe needle was inserted into a narrow pipe that connected to the inlet port of the chip (Fig. S2A). The top and side views of the chip are shown in Fig. S2B. The piston of the syringe was driven by the pump to push the MBs into the chip chamber. The MBs slowly sank to the bottom of the well due to gravity. After 5 min, FC-40 was injected, and the excess MBs on the surface of the array were flushed away (Fig. S2C) through the outlet port of the chip. Finally, the MBs was separated into the well of the developed microfluidic chip, and each well of the chip only hold one MB by confining the diameter of the wells. The MBs in the wells were sealed by FC-40, which confines the FL product generated from the reaction between β-Gal and FDG in the individual femtoliter-sized well and creates a locally high concentration of the FL product (Fig. 1B). The bright-field and FL images of the wells were acquired using a conventional inverted FL microscope (Fig. 1C and D). The number of FL wells with MBs (FL wells number) and the number of wells with MBs (MBs wells number) were counted, respectively. The percentage of FL wells was calculated by dividing (FL wells number) by (MBs wells number). MBs wells with fluorescence were defined as “1”, and MBs wells with no fluorescence were defined as “0”. That is distinct from but analogous to the use of the word “digital” in computing, where it refers to the binary code of 0s and 1s on which computers operate. That is why it was called digital detection.
3.2. The performance of the manufactured microfluidic chip
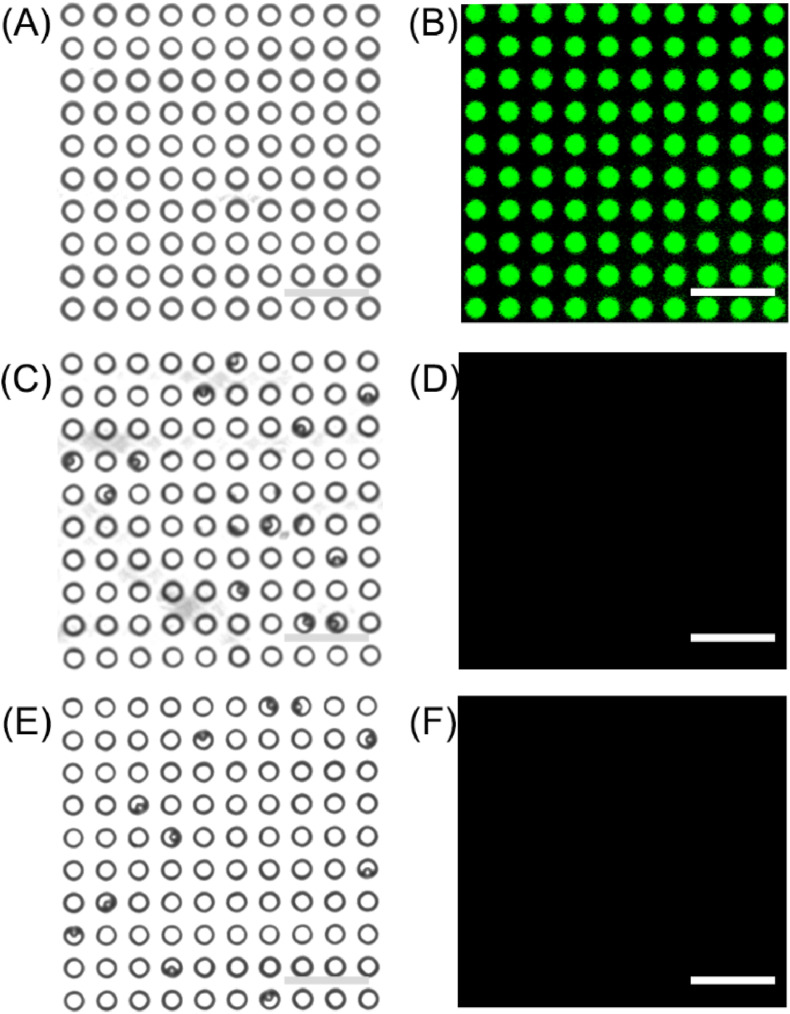
The PDMS polymer microfluidic chip consists of the channel layer and the array layer. The array layer contains 200 arrays, and each array contains 5000 wells, the width and height of each well was 4.5 μm and 3.5 μm, respectively. The central distance between the two wells was 10 μm (Fig. 2 A). The FL solution of the SA-β-Gal and FDG mixture was injected into the chip to verify its performance. The wells were filled with the FL solution under 488 nm laser radiation using the inverted FL microscope (Fig. 2B), which confirmed that the wells have good hydrophilicity. Afterwards, carboxylated MBs with a diameter of 2.7 μm and SA-coated MBs with a diameter of 3 μm were injected into the chip, respectively. The beads were seeded into the wells under the action of gravity, and one single well only holds one MB (Fig. 2C and E). Moreover, the two kinds of MBs did not have any background fluorescence under 488 nm laser radiation (Fig. 2D and F), which avoids the difficulty of distinguishing background fluorescence from weak positive fluorescence.
Fig. 2.
Manufactured microfluidic chips with an array of wells. (A) The representative light-field image of the array. (B) The representative FL image of the array after injection of an FL solution of SA-β-Gal and FDG mixture. (C) The representative light-field image of the array after injection of carboxylated MBs and (E) SA coated MBs. (D) The background fluorescence of carboxylated MBs and (F) SA coated MBs. Scale bar: 25 μm.
3.3. The feasibility of this assay
The activity of β-Gal was detected as a proof-of-concept study. Bio-β-Gal was labelled on the surface of SA coated MBs and formed the MB-β-Gal complex. MB-β-Gal and β-Gal's substrate FDG were injected into the microfluidic chip together. Fig. 3 A and B shows the representative FL images of the array in the presence of 10 nM bio-β-Gal and without bio-β-Gal. The percentage of FL wells is 97.18% in the presence of 10 nM bio-β-Gal, and the percentage of FL wells is 0.011% without-bio-β-Gal (Fig. 3C). The results proved that the principle for digital detection is feasible.
Fig. 3.
Performance verification of the manufactured microfluidic chip for digital detection. (A) The representative FL images of the array in the presence of 10 nM bio-β-Gal and (B) without-bio-β-Gal. (C) The percentage of FL wells in the presence of 10 nM bio-β-Gal and without bio-β-Gal. The error bars represent the standard deviation of three independent measurements. Scale bar: 50 μm.
3.4. The sensitivity and specificity of this assay for N protein detection
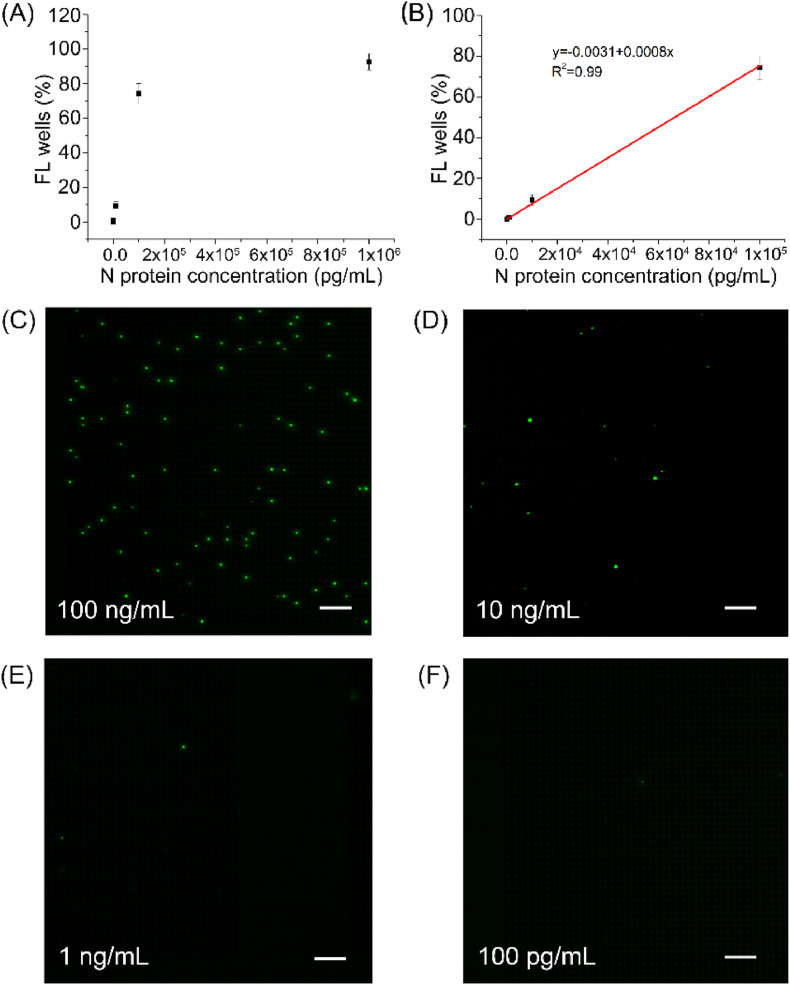
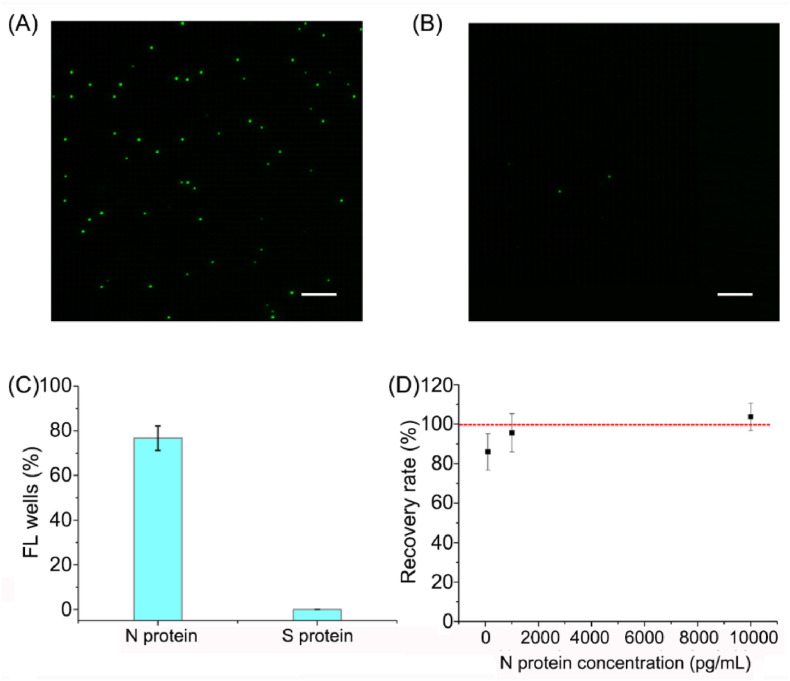
As reported, the order of the interaction force between molecules is covalent bond > electrostatic attraction > hydrogen bond > hydrophobic action > van der Waals force. The interaction forces between antigen and antibody are mainly van der Waals force and hydrogen bond. The interaction forces between antigen and DNA are mainly electrostatic attraction, hydrogen bond and van der Waals force. When DNA molecules with negative polarity bind to proteins, they can provide more oxygen atoms, enabling more electron pairs to generate more hydrogen bonds, increasing the bond energy of interaction, and maintaining a more stable structure. That is also the reason why aptamers have great potential in the field of scientific research. Therefore, an antibody/aptamer sandwich assay was used for the digital detection of N protein. Firstly, the interaction between N protein and apt 58 or 61 was verified by biolayer interferometry technology, and the interaction between N protein and anti-N protein antibody was verified by Western blot (Fig. S3). The percentage of FL wells (y-axis) presented a dose-dependent increase with the concentration of N protein (Fig. 4 A). Calibration curve (Fig. 4B) shows that y-axis value is proportional to the concentration of N protein in the range of 100 pg/mL to 100 ng/mL with a linear equation of y = −0.0031 + 0.0088x (R2 = 0.995). The limit of detection (LOD) of 33.28 pg/mL was calculated using this equation, based on triple standard deviation plus the mean of blanks. While the detection limit of conventional antibody sandwich ELISA for N protein is 10 ng/mL (Fig. S4), which is about 300 times higher than the digital detection assay used in this manuscript. The usage of aptamer reduces the complexity and cost of the digital detection system. The representative original light-field and fluorescent images with different concentrations of N protein ranging from 100 pg/mL, 1 ng/mL, 10 ng/mL, to 100 ng/mL were displayed in Fig. S5A-5H. The FL images in the dashed box of Fig. S5 was selected and exhibited in Fig. 4C–F. To verify the specificity of the assay, SARS-CoV-2 S protein was used as a control. The percentage of FL wells was only 0.62% with 100 ng/mL S protein under this assay setting, while the percentage was 76.71% with 100 ng/mL N protein (Fig. 5 A–C), which confirmed the excellent specificity of this assay.
Fig. 4.
Application of microfluidic chip with aptamer/antibody sandwich for N protein detection. (A) Plots of the percentage of FL wells vs different concentrations of N protein (1000 ng/mL, 100 ng/mL, 10 ng/mL, 1 ng/mL, 100 pg/mL, 0). (B) Calibration curve of the percentage of FL wells vs N protein concentration. The error bars represent the standard deviation of three independent measurements. (C–F) Representative FL images with different concentrations of N protein. Scale bar: 50 μm.
Fig. 5.
The specificity and recovery experiments for digital detection of N protein. (A) The representative FL images for digital detection of N protein and (B) S protein. (C) The difference in the percentage of FL wells for digital detection of N protein and S protein. (D) Distribution of the recovery with different concentrations of spiked N protein (1 ng/mL, 10 ng/mL, and 100 ng/mL) in 20% human serum. The error bars represent the standard deviation of three independent measurements. Scale bar: 50 μm.
As far as we know, there are several pre-existing technologies for N protein detection, such as lateral flow biosensor [17], gold nanoparticles (AuNPs)-based ELISA assay [30], and chemiluminescence enzyme immunoassay method (CL-ELISA) [31]. Lateral flow biosensor is an easy-to-use platform and is often used as a preliminary screening method for viruses, the LOD for N protein is 1 ng/mL. The AuNPs-based ELISA used AuNPs coated monoclonal antibody to enhance HRP concentration, which can improve the detection limit of N protein compared with conventional ELISA, and the LOD for N protein is 0.9 pg/mL. CL-ELISA was capable of detecting N protein at 1.56 pg/mL. Although the LOD of the AuNPs-based ELISA method and CL-ELISA methods is lower than that of our method, these optimized ELISA methods are the relative quantitative method rather than absolute quantitative methods. The biggest advantage of absolute quantification over relative quantification is that absolute abundance of the analytes can be obtained. The parallel reaction monitoring (PRM) quantitation of mass spectrometry assay is an absolute quantitative method with a lower detection limit for N protein [32]. Nonetheless, it requires complex instrumentation and laboratory operations which are more suitable to be used in scientific research. In this manuscript, the aptamer/antibody sandwich method for digital detection of N protein is also an absolute quantitative method. The absolute quantitative information can be obtained with qualitative measurements. The operation process is much simpler than that of mass spectrometry, and only a fluorescence detection device is needed to observe the detection results which is more suitable for practical applications.
3.5. The recovery experiments for digital detection of N protein in real sample
To verify this assay for practical application without the risk of patient samples, we performed recovery experiments by spiking different concentrations of N protein (the final concentrations are 1 ng/mL, 10 ng/mL, and 100 ng/mL) into diluted serum from a healthy volunteer (The SARS-CoV2 nucleic acid test was negative.). Meanwhile, the native N protein cannot be detected in the serum using this method. Acceptable recovery rates (85.9%–103.7%) for N protein were obtained (Fig. 5D), which confirmed that this assay detected N protein in human serum samples with little interference.
3.6. The reusability of the microfluidic chip
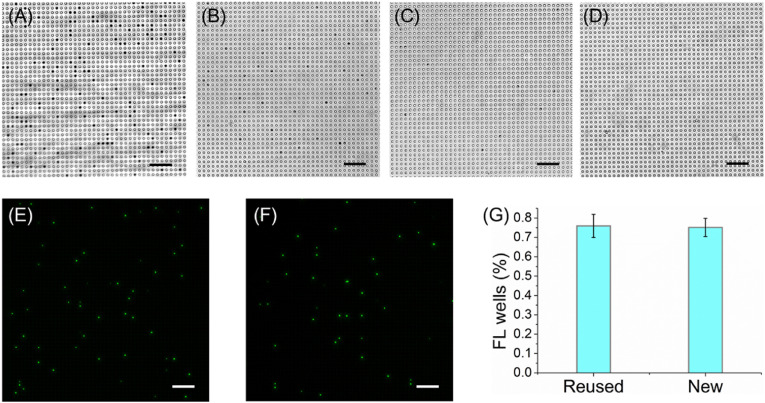
We also investigated the reusability of the microfluidic chip. Absolute EtAc was injected into the used chip, and the chip was vibrated in an ultrasonic bath for 120 s to make the MBs out of the wells. The MBs enter the chip chamber from the wells were immediately flushed away by absolute EtAc. Repeat the process twice, except to replace the absolute EtAc with double distilled water (ddH2O). Finally, the chips were baked in an oven at 37 °C for 2 h to dry any excess ddH2O in the wells. After three times of ultrasound and washing, almost all the MBs in the wells of the chip were all washed away (Fig. 6 A–D). And there is almost no difference between the percentage of FL wells when detection of N protein using the new and the cleaned chip (Fig. 6E–G). These results demonstrated the reusability of the PDMS-based chip, and would greatly reduce the experimental time.
Fig. 6.
The reusability of the microfluidic chip. (A) The bright-field image of the used chip without washing. (B) The bright-field images of the used chip were washed once with absolute EtAc, (C) twice and (D) three times with ddH2O. (E) The FL images for detection of 100 ng/mL N protein using the reused chip and (F) the new chip. (G) The difference in the percentage of FL wells (%) for detection of 100 ng/mL N protein using the reused and the new chip. The error bars represent the standard deviation of three independent measurements. Scale bar: 50 μm.
4. Conclusion
In this assay, the wells of the microfluidic chip were used to isolate a single MB that carrying target molecules. To ensure MBs can capture all the molecules, the number of beads was largely excessive over that of the target molecules. However, only about 3%–5% of the total number of MBs could be entered into the wells, and the majority of targets cannot be detected and analyzed. The major limitation of this assay for detecting rare molecules is the low efficiency of beads loading. Meanwhile, the sensitivity of this assay for N protein detection was directly related to the affinity of the antibody and aptamer. The higher the affinity, the lower the sensitivity. Future work will focus on screening better antibodies and aptamers.
Credit author statement
Chenchen Ge and Juan Feng: Conceptualization, Methodology, Investigation, Writing-original draft. Dou Wang: Methodology, Investigation. Jiaming Zhang and Kai Hu: Software, Investigation. Ling Zha: Validation. Rongsong Li and Xuejuan Hu: Supervision, Funding acquisition, Project administration, Writing-Review & Editing.
Fundings
This work was supported by the Special anti-COVID-19 Project Fund of Guangdong Provincial Education Department [grant number 2020KZDZX1214], Natural Science Foundation of Top Talent of SZTU [grant number 2020102], Shenzhen Natural Science Foundation [grant number [JCYJ20190813141001745], Startup Research Fund of Shenzhen Technology University (RL), and the Shenzhen Pengcheng Scholar Program (JH). Special Fund for Science and Technology Innovation of Pingshan District, Shenzhen [grant number PSKG202006].
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.talanta.2021.122847.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Meselson M. Droplets and aerosols in the transmission of SARS-CoV-2. N. Engl. J. Med. 2020;382(21):2063. doi: 10.1056/NEJMc2009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang F., Kream R.M., Stefano G.B. Long-term respiratory and neurological sequelae of COVID-19. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. 2020;26 doi: 10.12659/MSM.928996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taleghani N., Taghipour F. Diagnosis of COVID-19 for controlling the pandemic: a review of the state-of-the-art. Biosens. Bioelectron. 2021;174:112830. doi: 10.1016/j.bios.2020.112830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kruttgen A., Cornelissen C.G., Dreher M., Hornef M.W., Imohl M., Kleines M. Comparison of the SARS-CoV-2 Rapid antigen test to the real star Sars-CoV-2 RT PCR kit. J. Virol. Methods. 2021;288:114024. doi: 10.1016/j.jviromet.2020.114024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boger B., Fachi M.M., Vilhena R.O., Cobre A.F., Tonin F.S., Pontarolo R. Systematic review with meta-analysis of the accuracy of diagnostic tests for COVID-19. Am. J. Infect. Contr. 2021;49(1):21–29. doi: 10.1016/j.ajic.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suhandynata R.T., Hoffman M.A., Kelner M.J., McLawhon R.W., Reed S.L., Fitzgerald R.L. Longitudinal monitoring of SARS-CoV-2 IgM and IgG seropositivity to detect COVID-19. J. Appl. Lab. Med. 2020;5(5):908–920. doi: 10.1093/jalm/jfaa079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qu J., Wu C., Li X., Zhang G., Jiang Z., Li X., Zhu Q., Liu L. Profile of immunoglobulin G and IgM antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2020;71(16):2255–2258. doi: 10.1093/cid/ciaa489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srinivasan S., Cui H., Gao Z., Liu M., Lu S., Mkandawire W., Narykov O., Sun M., Korkin D. Structural genomics of SARS-CoV-2 indicates evolutionary conserved functional regions of viral proteins. Viruses. 2020;12(4) doi: 10.3390/v12040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savastano A., Ibanez de Opakua A., Rankovic M., Zweckstetter M. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat. Commun. 2020;11(1):6041. doi: 10.1038/s41467-020-19843-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 11.Davydova A., Vorobjeva M., Pyshnyi D., Altman S., Vlassov V., Venyaminova A. Aptamers against pathogenic microorganisms. Crit. Rev. Microbiol. 2016;42(6):847–865. doi: 10.3109/1040841X.2015.1070115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiong H., Yan J., Cai S., He Q., Peng D., Liu Z., Liu Y. Cancer protein biomarker discovery based on nucleic acid aptamers. Int. J. Biol. Macromol. 2019;132:190–202. doi: 10.1016/j.ijbiomac.2019.03.165. [DOI] [PubMed] [Google Scholar]
- 13.Gao T., Ding P., Li W., Wang Z., Lin Q., Pei R. Isolation of DNA aptamers targeting N-cadherin and high-efficiency capture of circulating tumor cells by using dual aptamers. Nanoscale. 2020;12(44):22574–22585. doi: 10.1039/d0nr06180h. [DOI] [PubMed] [Google Scholar]
- 14.Wang D., Ge C., Lv K., Zou Q., Liu Q., Liu L., Yang Q., Bao S. A simple lateral flow biosensor for rapid detection of lead(ii) ions based on G-quadruplex structure-switching. Chem. Commun. (Camb) 2018;54(97):13718–13721. doi: 10.1039/c8cc06810k. [DOI] [PubMed] [Google Scholar]
- 15.Biniuri Y., Luo G.F., Fadeev M., Wulf V., Willner I. Redox-switchable binding properties of the ATP-aptamer. J. Am. Chem. Soc. 2019;141(39):15567–15576. doi: 10.1021/jacs.9b06256. [DOI] [PubMed] [Google Scholar]
- 16.Ding F., Gao Y., He X. Recent progresses in biomedical applications of aptamer-functionalized systems. Bioorg. Med. Chem. Lett. 2017;27(18):4256–4269. doi: 10.1016/j.bmcl.2017.03.032. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L., Fang X., Liu X., Ou H., Zhang H., Wang J., Li Q., Cheng H., Zhang W., Luo Z. Discovery of sandwich type COVID-19 nucleocapsid protein DNA aptamers. Chem. Commun. (Camb) 2020;56(70):10235–10238. doi: 10.1039/d0cc03993d. [DOI] [PubMed] [Google Scholar]
- 18.Witters D., Sun B., Begolo S., Rodriguez-Manzano J., Robles W., Ismagilov R.F. Digital biology and chemistry. Lab Chip. 2014;14(17):3225–3232. doi: 10.1039/c4lc00248b. [DOI] [PubMed] [Google Scholar]
- 19.Cohen L., Hartman M.R., Amardey-Wellington A., Walt D.R. Digital direct detection of microRNAs using single molecule arrays. Nucleic Acids Res. 2017;45(14):e137. doi: 10.1093/nar/gkx542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rissin D., Kan C., Campbell T., Howes S., Fournier D., Song L., Piech T., Patel P., Chang L., Rivnak A., Ferrell E., Randall J., Provuncher G., Walt D., Duffy D. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 2010;28(6):595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basu A. Digital assays Part II: digital protein and cell assays. SLAS Technol. 2017;22(4):387–405. doi: 10.1177/2472630317705681. [DOI] [PubMed] [Google Scholar]
- 22.Wilson D.H., Rissin D.M., Kan C.W., Fournier D.R., Piech T., Campbell T.G., Meyer R.E., Fishburn M.W., Cabrera C., Patel P.P., Frew E., Chen Y., Chang L., Ferrell E.P., von Einem V., McGuigan W., Reinhardt M., Sayer H., Vielsack C., Duffy D.C. The simoa HD-1 analyzer: a novel fully automated digital immunoassay analyzer with single-molecule sensitivity and multiplexing. J. Lab. Autom. 2016;21(4):533–547. doi: 10.1177/2211068215589580. [DOI] [PubMed] [Google Scholar]
- 23.O'Connell G., Alder M., Smothers C., Still C., Webel A., Moore S. Use of high-sensitivity digital ELISA improves the diagnostic performance of circulating brain-specific proteins for detection of traumatic brain injury during triage. Neurol. Res. 2020;42(4):346–353. doi: 10.1080/01616412.2020.1726588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jing W., Wang Y., Chen C., Zhang F., Yang Y., Ma G., Yang E.H., Snozek C.L.N., Tao N., Wang S. Gradient-based rapid digital immunoassay for high-sensitivity cardiac troponin T (hs-cTnT) detection in 1 muL plasma. ACS Sens. 2021;6(2):399–407. doi: 10.1021/acssensors.0c01681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stuelke E., James K., Kirchherr J., Allard B., Baker C., Kuruc J., Gay C., Margolis D., Archin N. Measuring the inducible, replication-competent HIV reservoir using an ultra-sensitive p24 readout, the digital ELISA viral outgrowth assay. Front. Immunol. 2020;11:1971. doi: 10.3389/fimmu.2020.01971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song Y., Ye Y., Su S.H., Stephens A., Cai T., Chung M.T., Han M.K., Newstead M.W., Yessayan L., Frame D., Humes H.D., Singer B.H., Kurabayashi K. A digital protein microarray for COVID-19 cytokine storm monitoring. Lab Chip. 2021;21(2):331–343. doi: 10.1039/d0lc00678e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun J., Hu J., Gou T., Ding X., Song Q., Wu W., Wang G., Yin J., Mu Y. Power-free polydimethylsiloxane femtoliter-sized arrays for bead-based digital immunoassays. Biosens. Bioelectron. 2019;139:111339. doi: 10.1016/j.bios.2019.111339. [DOI] [PubMed] [Google Scholar]
- 28.Fieldler F., Hinz H. No intermediate channelling in stepwise hydrolysis of fluorescein di-β-D-galactoside by β-galactosidase. Eur. J. Biochem. 1994;222(1):75–81. doi: 10.1111/j.1432-1033.1994.tb18843.x. [DOI] [PubMed] [Google Scholar]
- 29.Huang Z. Kinetic fluorescence measurement of fluorescein di-beta-D-galactoside hydrolysis by beta-galactosidase: intermediate channeling in stepwise catalysis by a free single enzyme. Biochemistry. 1991;30(35):8535–8540. doi: 10.1021/bi00099a006. [DOI] [PubMed] [Google Scholar]
- 30.Duan Y., Wu W., Zhao Q., Liu S., Liu H., Huang M., Wang T., Liang M., Wang Z. Enzyme-antibody-modified gold nanoparticle probes for the ultrasensitive detection of nucleocapsid protein in SFTSV. Int. J. Environ. Res. Publ. Health. 2020;17(12):4427. doi: 10.3390/ijerph17124427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujimoto K., Chan K., Takeda K., Lo K., Leung R., Okamoto T. Sensitive and specific enzyme-linked immunosorbent assay using chemiluminescence for detection of severe acute respiratory syndrome viral infection. J. Clin. Microbiol. 2008;46(1):302–310. doi: 10.1128/JCM.01006-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cazares L., Chaerkady R., Samuel Weng S., Boo C., Cimbro R., Hsu H., Rajan S., Dall'Acqua W., Clarke L., Ren K., McTamney P., Kallewaard-LeLay N., Ghaedi M., Ikeda Y., Hess S. Development of a parallel reaction monitoring mass spectrometry assay for the detection of SARS-CoV-2 spike glycoprotein and nucleoprotein. Anal. Chem. 2020;92(20):13813–13821. doi: 10.1021/acs.analchem.0c02288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.