Li et al. show with a new assay that NFKB1 variants disrupting the transcriptional activity of RELA-p50 dimers underlie an autosomal dominant form of common variable immunodeficiency by haploinsufficiency, thereby providing a functional approach to test new NFKB1 variants.
Abstract
Autosomal dominant (AD) NFKB1 deficiency is thought to be the most common genetic etiology of common variable immunodeficiency (CVID). However, the causal link between NFKB1 variants and CVID has not been demonstrated experimentally and genetically, as there has been insufficient biochemical characterization and enrichment analysis. We show that the cotransfection of NFKB1-deficient HEK293T cells (lacking both p105 and its cleaved form p50) with a κB reporter, NFKB1/p105, and a homodimerization-defective RELA/p65 mutant results in p50:p65 heterodimer–dependent and p65:p65 homodimer–independent transcriptional activation. We found that 59 of the 90 variants in patients with CVID or related conditions were loss of function or hypomorphic. By contrast, 258 of 260 variants in the general population or patients with unrelated conditions were neutral. None of the deleterious variants displayed negative dominance. The enrichment in deleterious NFKB1 variants of patients with CVID was selective and highly significant (P = 2.78 × 10−15). NFKB1 variants disrupting NFKB1/p50 transcriptional activity thus underlie AD CVID by haploinsufficiency, whereas neutral variants in this assay should not be considered causal.
Introduction
Common variable immunodeficiency (CVID) is a clinically and genetically heterogeneous disorder characterized by hypogammaglobulinemia, impaired production of specific antibodies, and susceptibility to various infections (Bonilla et al., 2016). Patients with CVID commonly have abnormal B and T cell phenotypes, such as small numbers of class-switched memory B cells and regulatory T cells (Cunningham-Rundles, 2019). Other features, such as autoimmunity, are seen in a subset of patients (Gathmann et al., 2014; Resnick et al., 2012). As one of the most frequently diagnosed inborn errors of immunity (IEI; Bousfiha et al., 2020; Tangye et al., 2020; Tangye et al., 2021), CVID is estimated to affect between 1 in 10,000 and 1 in 50,000 individuals, typically beginning in early adulthood and affecting a wide range of organs, including the respiratory and gastrointestinal tracts in particular (Bogaert et al., 2016; Resnick et al., 2012). Overall, 22 monogenic disorders have been reported to account, collectively, for at least 10% of CVID cases (Abolhassani et al., 2020; Cunningham-Rundles, 2019; Maffucci et al., 2016; van Schouwenburg et al., 2015). According to the International Union of Immunological Societies Expert Committee, the loci involved are ARHGEF1, ATP6AP1, CD19, CD20, CD21, CD81, IKZF1, IRF2BP2, MOGS, NFKB1, NFKB2, PIK3CD, PIK3R1, PTEN, RAC2, SEC61A1, SH3KBP1, TNFSF13, TNFRSF13B, TNFRSF13C, TNFSF12, and TRNT1 (Bousfiha et al., 2020; Tangye et al., 2020; Tangye et al., 2021). It has been suggested that monoallelic NFKB1 variants, including predicted loss-of-expression or predicted loss-of-function (pLOF) variants, first reported in 2015 (Fliegauf et al., 2015), are the most common cause of CVID in the European Union (4%; Tuijnenburg et al., 2018) and the second most prevalent cause of CVID in the United States of America (5%; Abolhassani et al., 2020). More recently, an association with monoallelic NFKB1 variants was identified in a cohort of 157 individuals displaying a broader clinical spectrum than CVID, including opportunistic infections, autoimmunity, noninfectious enteropathy, autoinflammation, lymphoproliferation, and cancer (Lorenzini et al., 2020).
The NFKB1 gene encodes nuclear factor of κ light polypeptide gene enhancer in B cells 1 (NF-κB1), one of the five REL homology domain (RHD)–containing proteins. The full-length precursor, p105 (amino acids 1–969), gives rise to p50 (amino acids 1–433), which, like the other four REL proteins (RELA/p65, RELB, c-REL, and NF-κB2/p100/p52), contains the RHD (Zhang et al., 2017). However, NF-κB1 and NF-κB2 differ from the other three REL proteins in not having a transactivation domain (Zhang et al., 2017). As a result, p50 and p52 have transcriptional activity only when dimerized with other REL proteins (Hoffmann et al., 2006). Following phosphorylation with the upstream trimeric IκB kinase (IKK) complex (composed of IKKα, IKKβ, and IKKγ/NF-κB essential modulator), p105 undergoes polyubiquitination, and its C-terminal portion is subjected to proteasomal degradation, with cleavage at approximately amino acid 433 to release the p50 subunit (Hayden and Ghosh, 2008). Various stimuli, such as cytokine and antigen receptors, activate the IKK complex (Taniguchi and Karin, 2018). This activation leads to translocation of the p50-containing dimers to the nucleus, where they bind to response elements in the promoter regions of specific target genes (Taniguchi and Karin, 2018). The most abundant and best-studied NF-κB dimers, p65:p50 heterodimers, usually promote gene expression (Siggers et al., 2011; Smale, 2012; Zhao et al., 2014). By contrast, p50:p50 homodimers have no transactivation domain and act as transcriptional repressors, unless bound to other proteins (Smale, 2012). The C-terminal part of the p105 precursor contains ankyrin repeats that can also repress expression by sequestering NF-κB dimers in the cytosol (Rice et al., 1992).
Heterozygous NFKB1 variants in patients with CVID or other clinical phenotypes have been reported in 15 studies (Boztug et al., 2016; Christiansen et al., 2020; Dieli-Crimi et al., 2018; Duan and Feanny, 2019; Fliegauf et al., 2015; Kaustio et al., 2017; Lorenzini et al., 2020; Lougaris et al., 2017a; Lougaris et al., 2017b; Maffucci et al., 2016; Maréchal et al., 2020; Rae et al., 2017; Schipp et al., 2016; Schröder et al., 2019; Tuijnenburg et al., 2018), but none of these studies experimentally characterized the function of these variants without interference from other REL proteins. Indeed, exogenously or endogenously expressed p65 can form p65:p65 homodimers with transcriptional activity indistinguishable from that of p65:p50 heterodimers (Ganchi et al., 1993; Phelps et al., 2000). In previous studies, the impact of NFKB1 variants on the transcriptional activity of NF-κB dimers was not measured in the absence of p65:p65 homodimers (Kaustio et al., 2017; Lorenzini et al., 2020). Moreover, the NFKB1 variants were tested in cells carrying WT NFKB1 alleles, resulting in a mixture of dimers, including p50mut:p65, p50wt:p65, and p65:65, in the same cells (Kaustio et al., 2017; Lorenzini et al., 2020). We therefore aimed to develop a new, robust, and accurate functional assay for classifying NFKB1 variants according to their ability to activate NF-κB–mediated transcription in a strictly p50-dependent manner. We used this assay to evaluate the biochemical impact of 365 reported nonsynonymous NFKB1 variants found in our database and other databases of healthy and sick individuals. Finally, we tested the hypothesis that patients with CVID may display enrichment in NFKB1 variants experimentally demonstrated to be deleterious. Indeed, a second limitation of previous studies is that no genetic evidence has been provided for an enrichment in pLOF, let alone experimentally proven loss-of-function (LOF) variants, exclusively or even selectively, in patients with CVID. Overall, there is suggestive, but not conclusive, evidence that deleterious variants of NFKB1 can cause CVID. We tested the validity of this hypothesis biochemically and genetically.
Results
NFKB1 variants present in our and other databases of healthy and sick individuals
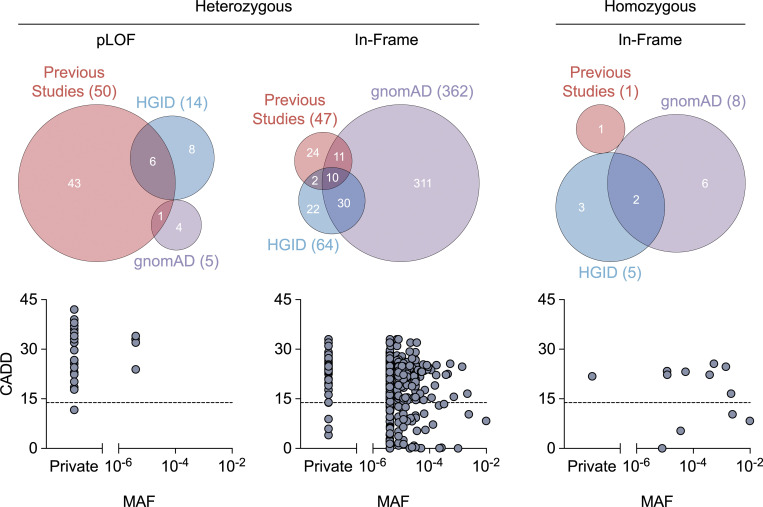
To date, 97 heterozygous variants of NFKB1 have been reported in patients with CVID, autoinflammation, autoimmunity, or unspecified phenotypes (Fig. 1; and Table S1 and Table S2; Boztug et al., 2016; Christiansen et al., 2020; Dieli-Crimi et al., 2018; Duan and Feanny, 2019; Fliegauf et al., 2015; Kaustio et al., 2017; Lorenzini et al., 2020; Lougaris et al., 2017a; Lougaris et al., 2017b; Maffucci et al., 2016; Maréchal et al., 2020; Rae et al., 2017; Schipp et al., 2016; Schröder et al., 2019; Tuijnenburg et al., 2018). One homozygous variant of NFKB1 was recently reported in a patient with combined immunodeficiency (Fig. 1 and Table S1; Mandola et al., 2021). All these reported variants were private (i.e., not found in public databases) or rare (i.e., with a minor allele frequency [MAF] < 0.01; Fig. 1 and Table S1). In our Human Genetics of Infectious Diseases (HGID) CVID (HGID-CVID) cohort of 305 patients with CVID, we identified six new private or rare heterozygous variants (Fig. 1 and Table S1). In addition, by searching our HGID infectious disease (HGID-ID) cohort of 7,966 individuals with various infectious diseases, we found 54 new heterozygous and five new homozygous variants of this gene, all of which were private or rare (Fig. 1 and Table S1). In addition to the published variants and those found in the HGID-CVID and HGID-ID cohorts, an additional 315 heterozygous and six homozygous rare variants are reported in the public Genome Aggregation Database (gnomAD; v2.1.1; Fig. 1 and Table S1). Thus, in total, 474 private or rare, heterozygous or homozygous variants have been found in private or public databases for patients or healthy individuals (Fig. 1 and Table S1). The large number of variants present in public databases demonstrates that NFKB1 variants are not uncommon in individuals and cannot therefore necessarily be considered causal for any disease, including CVID. The 474 variants identified comprised two large deletions, 60 pLOF variants (17 nonsense, 29 frameshift, and 14 splice site), and 412 in-frame variants (1 start loss, 4 in-frame indels, and 407 missense; Table S1). Only 5 of the 98 variants reported in patients (with CVID, combined immunodeficiency, or other phenotypes) and none of the 368 variants reported in the gnomAD were functionally tested. We therefore evaluated the expression and function of all 157 variants reported in previous studies or present in the HGID cohort, with the exception of the two large deletions. We also tested 208 variants reported in gnomAD that had a MAF ≥ 10−5, were reported in at least two individuals, or were reported in the gnomAD control cohort. In total, we selected 365 variants for functional testing.
Figure 1.
NFKB1 variants present in our and other databases of healthy and sick individuals. Heterozygous and homozygous NFKB1 variants reported in previous studies or present in our HGID cohorts (HGID-CVID and HGID-ID) or gnomAD v2.1.1. The numbers in parentheses indicate the total number of variants identified in each database. pLOF variants include nonsense, frameshift, and splice-site variants, whereas in-frame variants comprise start-loss, in-frame indel, and missense variants. 10 in-frame variants are present in both the heterozygous and homozygous states (CADD score v1.6; Kircher et al., 2014; Rentzsch et al., 2019). The black dashed line represents the mutation significance cutoff threshold for NFKB1 (13.85; Itan et al., 2016).
Expression of mutant NFKB1 alleles
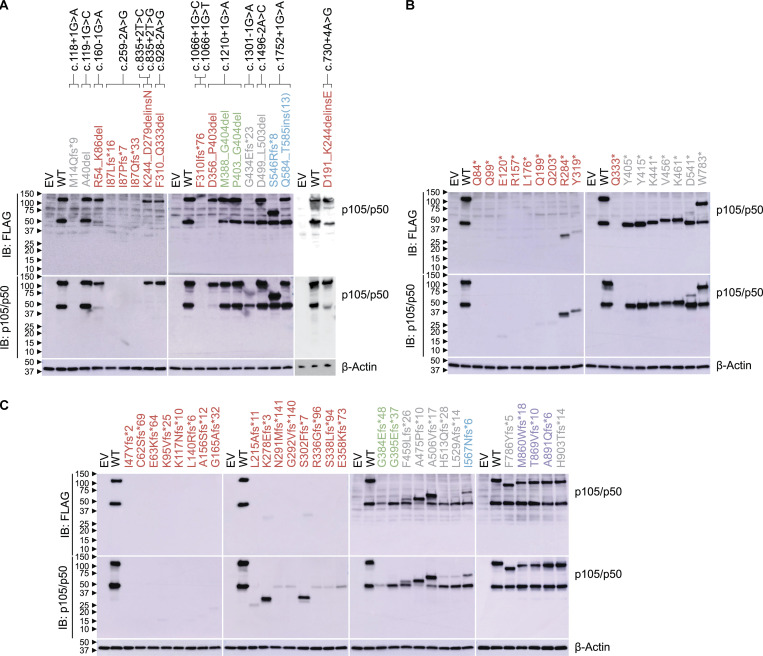
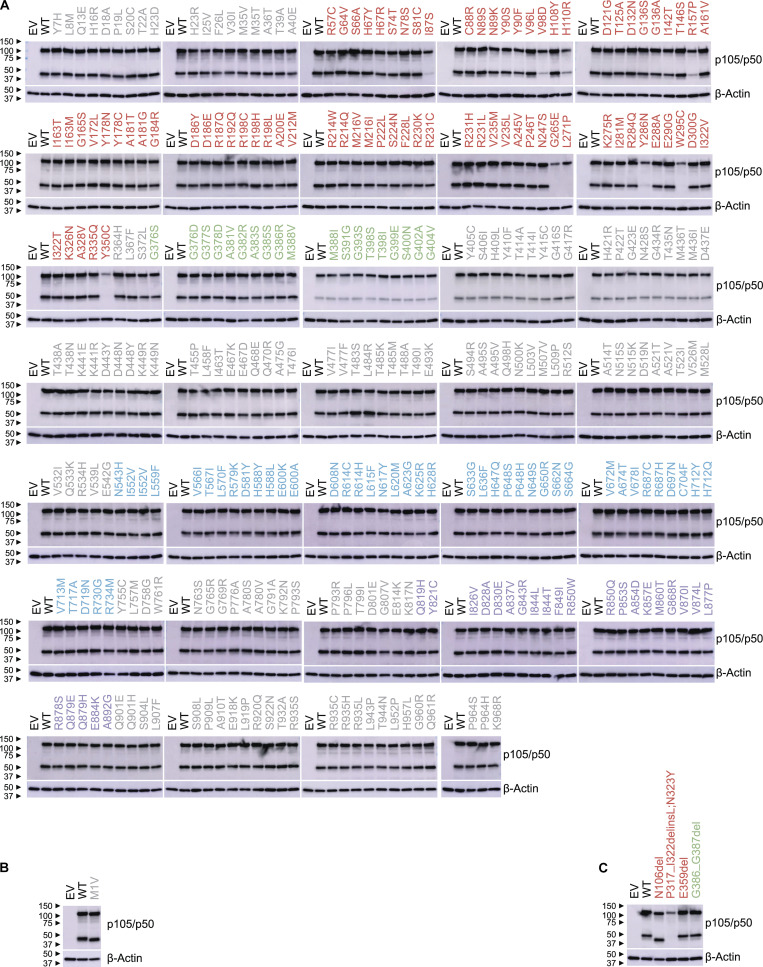
We first used an exon-trapping assay to determine whether the 14 splice-site variants generated abnormal transcripts. We showed that all 14 variants led to abnormally spliced products, including 10 in-frame and 7 frameshift transcripts (Table S3 and Data S1). We then cloned these abnormal transcripts, along with all the other 351 variants, to test their expression. We designed an assay for assessing the expression of mutant NFKB1 alleles without interference due to endogenous NFKB1 expression. For this purpose, we generated an NFKB1-deficient (NFKB1−/−) human embryonic kidney (HEK) 293T cell line with CRISPR-Cas9 technology (Fig. 2, A and B). We transfected NFKB1−/− cells with cDNAs encoding the WT p105 and all 368 mutant alleles. We used an intron-less overexpression plasmid for this purpose and were not, therefore, able to detect nonsense-mediated mRNA decay triggered by premature stop codons (Maquat, 2004). Our system would therefore be expected to underestimate defects due to variants introducing premature stop codons, particularly those occurring downstream from the proteolytic cleavage site (amino acid 433). We detected both p50 and p105 when the WT sequence was overexpressed in NFKB1−/− cells, demonstrating that proteolytic cleavage was functional (Fig. S1, A–C; and Fig. S2, A–C). As expected, 28 of the 63 mutants generated by the 60 pLOF variants produced neither p50 nor p105 (Fig. S1, A-C; and Table 1). 5 of the remaining 35 mutants produced normal amounts of p50 and p105 with molecular weights indistinguishable from those of the WT forms; 2 produced smaller amounts of p50 and p105; 25 produced a normal (15) or smaller (10) amount of p50 with no detectable p105; 2 produced a smaller amount of p105 with no detectable p50; and only 1 produced a smaller amount of p50, with a lower molecular weight than WT, and a normal amount of p105 (Fig. S1, A–C; and Table 1). For the 305 in-frame variants, 294 produced normal amounts of p50 and p105, 5 produced smaller amounts of p50 and p105, and 6 produced a normal (1) or smaller (5) amount of p105 with no detectable p50 (Fig. S2, A–C; and Table 1). Thus, 69 of the 368 mutants displayed abnormal expression of p50, p105, or both.
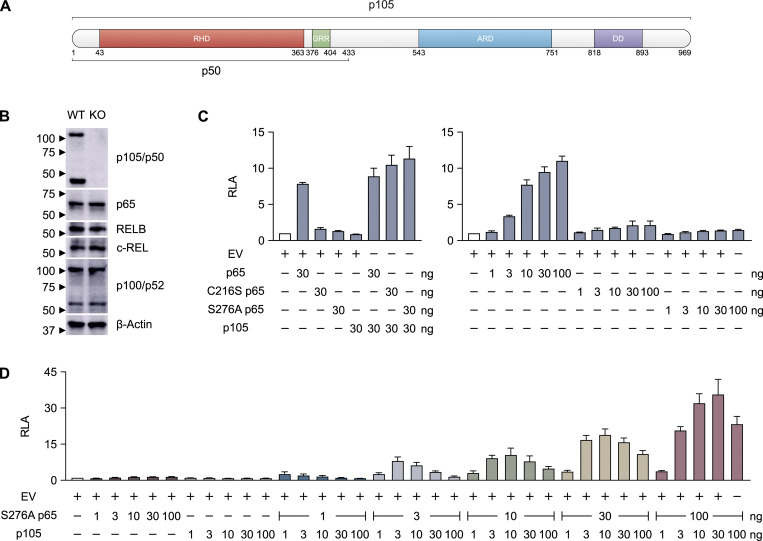
Figure 2.
A luciferase reporter assay for detecting p50-specific transcriptional activity. (A) Schematic illustration of the protein (p105) encoded by the NFKB1 gene. The p50 subunit spans amino acids 1–433. (B) Immunoblot of p105/p50, p65, RELB, c-REL, p100/p52, and β-actin in WT and NFKB1-deficient (KO) HEK293T cells. p105/p50 was detected with an anti-p105/p50 antibody recognizing the N terminus of p105. (C) Relative luciferase activity (RLA) of NFKB1-deficient HEK293T cells transfected with a κB reporter construct containing the HIV-1 κB enhancer motif (κB-luc) in the presence or absence of plasmids encoding WT, C216S, or S276A p65 or WT p105. The total amount of plasmid in each transfection was kept constant by adding EV. (D) RLA of NFKB1-deficient HEK293T cells transfected with κB-luc in the presence or absence of various amounts (1, 3, 10, 30, or 100 ng) of plasmids encoding S276A p65 or various amounts (1, 3, 10, 30, or 100 ng) of plasmids encoding WT p105. The total amount of plasmid in each transfection was kept constant by adding EV. The data shown are representative of two independent experiments (B). The values shown are the means ± SEM of three independent experiments (C and D).
Figure S1.
Expression of pLOF mutant NFKB1 alleles. (A–C) Immunoblot of p105/p50 and β-actin in NFKB1-deficient HEK293T cells transfected with plasmids encoding WT or mutant p105. WT and mutant p105 cDNAs were inserted into the pLenti-III-EF1α vector with an N-terminal FLAG tag (A–C). p105/p50 was detected with an anti-FLAG antibody (A–C) or an anti-p105/p50 antibody recognizing the N terminus of p105 (A–C). Variants in the RHD, GRR, ARD, DD, and other regions of p105 are colored red, green, blue, purple, and gray, respectively. The data shown are representative of two independent experiments. IB, immunoblot.
Figure S2.
Expression of in-frame mutant NFKB1 alleles. (A–C) Immunoblot of p105/p50 and β-actin in NFKB1-deficient HEK293T cells transfected with plasmids encoding WT or mutant p105. WT and mutant p105 cDNAs were inserted into the pLenti-III-EF1α vector without (B) or with an N-terminal FLAG tag (A and C). p105/p50 was detected with an anti-p105/p50 antibody recognizing the N terminus of p105 (A) or with a p105/p50 antibody recognizing residues surrounding amino acid 415 of p105 (B and C). Variants in the RHD, GRR, ARD, DD, and other regions of p105 are colored red, green, blue, purple, and gray, respectively. The data shown are representative of two independent experiments.
Table 1. Expression pattern of mutant NFKB1 alleles.
| p105 | ||||
|---|---|---|---|---|
| Normal | Reduced | Absent | ||
| p50 | Normal | 299 | 0 | 15 |
| Reduced | 1 | 7 | 10 | |
| Absent | 1 | 7 | 28 | |
A new luciferase reporter assay for detecting p50-specific transcriptional activity
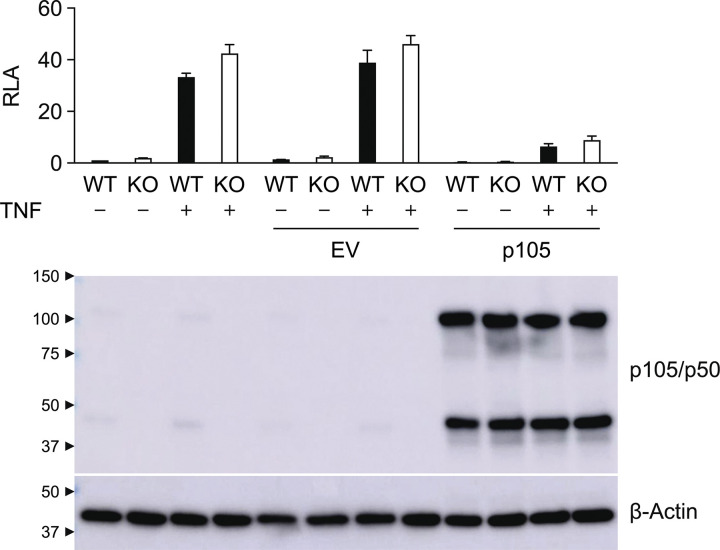
We then developed a luciferase reporter assay capable of determining the functional impact of NFKB1 variants. A lack of functional p65:p50 heterodimers can be compensated by other REL proteins, including p65:p65 homodimers in particular (Ganchi et al., 1993; Hoffmann et al., 2003). We first confirmed this result by showing that TNF stimulation results in similar levels of luciferase activity in NFKB1−/− cells and their parental NFKB1+/+ cells (Fig. S3). We then transfected NFKB1−/− cells with WT p105 to determine whether overexpression can up-regulate the transcriptional level of the luciferase. The overexpression of p105 did not induce detectable NF-κB activation, even though a large amount of p105 was cleaved to yield p50, as shown by Western blotting (Fig. S3). These results are consistent with the previous observation that p65 contains a transactivation domain whereas p50 does not, and that p50:p50 homodimers do not have transcriptional activity (Pereira and Oakley, 2008). We then transfected NFKB1−/− cells with p105 in the presence of p65 in an attempt to increase the dimerization of p65:p50 and drown out the effects of p50:p50 homodimers by sheer numbers. However, the addition of p65 alone resulted in high levels of luciferase activity (Fig. 2 C), consistent with the previous observation that p65:p65 homodimers have potent transcriptional activity (Ganchi et al., 1993). We therefore designed a new luciferase assay for assessing the function of NFKB1 alleles with p65:p50 heterodimers while minimizing interference from p65:p65 homodimers. We transfected NFKB1−/− cells with two p65 mutants, C216S and S276A, which have been reported to form p50:p65 heterodimers, but not p65:p65 homodimers (Ganchi et al., 1993). To test NF-κB activity, we used a reporter construct that contains an NF-κB consensus site (5′-GGGACTTTCC-3′) that not only is capable of binding p65:p50 heterodimers (Siggers et al., 2011) but also is found in several NF-κB–regulated genes with important immunological functions, such as NFKB2, CSF-1, TAP1, LMP2, and PLCD1 (https://bioinfo.lifl.fr/NF-KB/). Consistent with previous findings (Ganchi et al., 1993), both these p65 mutants displayed minimal NF-κB activity following transfection on their own (Fig. 2 C) and significant NF-κB activity following cotransfection with p105 (Fig. 2 C). S276A p65 elicited slightly stronger activity than C216S p65 (Fig. 2 C). We therefore chose to use S276A p65 in subsequent experiments. By transfecting NFKB1−/− cells with a combination of various amounts of WT p105 and S276A p65, we determined the optimal conditions for the assay (30 ng WT p105 with 100 ng S276A p65) as those yielding the highest fold-change difference in luciferase activity (30-fold) relative to empty vector (EV; Fig. 2 D). We therefore established this new luciferase assay, using a homodimer-defective p65 mutant and a NFKB1-deficient cell line, as a robust assay for measuring the specific transcription activity of p50 proteins in a strictly p50/p65 heterodimer–dependent manner.
Figure S3.
Effect of p105 on TNF-induced NF-κB activation. RLA of WT and NFKB1-deficient (KO) HEK293T cells transfected with pGL4.32[luc2P/NF-κB-RE/Hygro] in the presence or absence of plasmids encoding p105 and left unstimulated or stimulated with TNF (20 ng/ml) for 6 h. p105/p50 was detected with an anti-p105/p50 antibody recognizing the N terminus of p105. The values shown are the means ± SEM of two independent experiments.
Functional characterization of NFKB1 variants
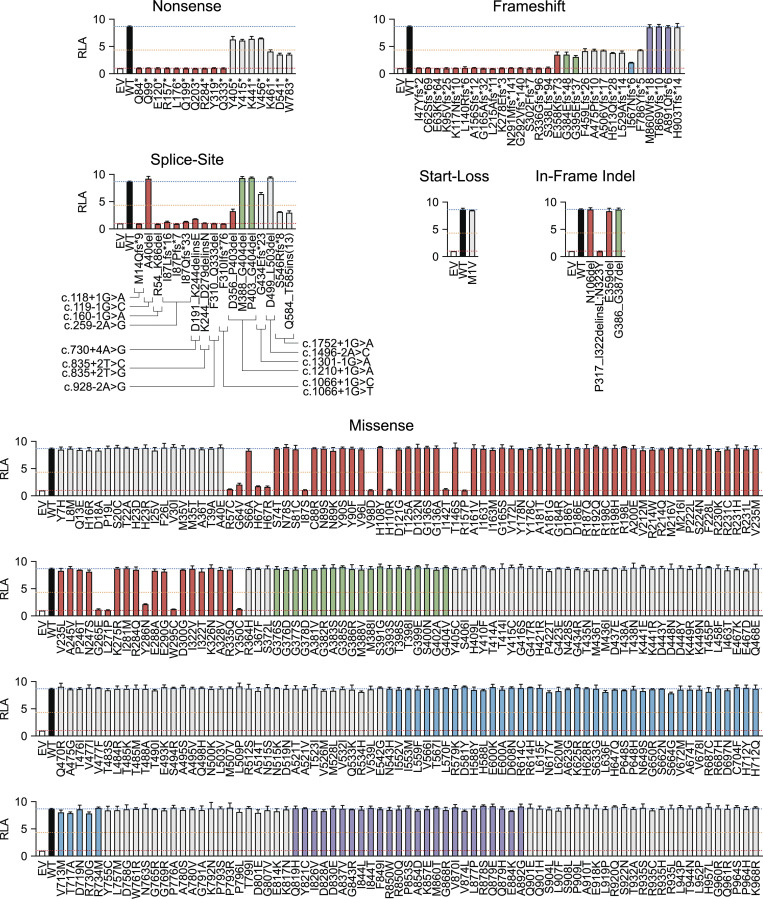
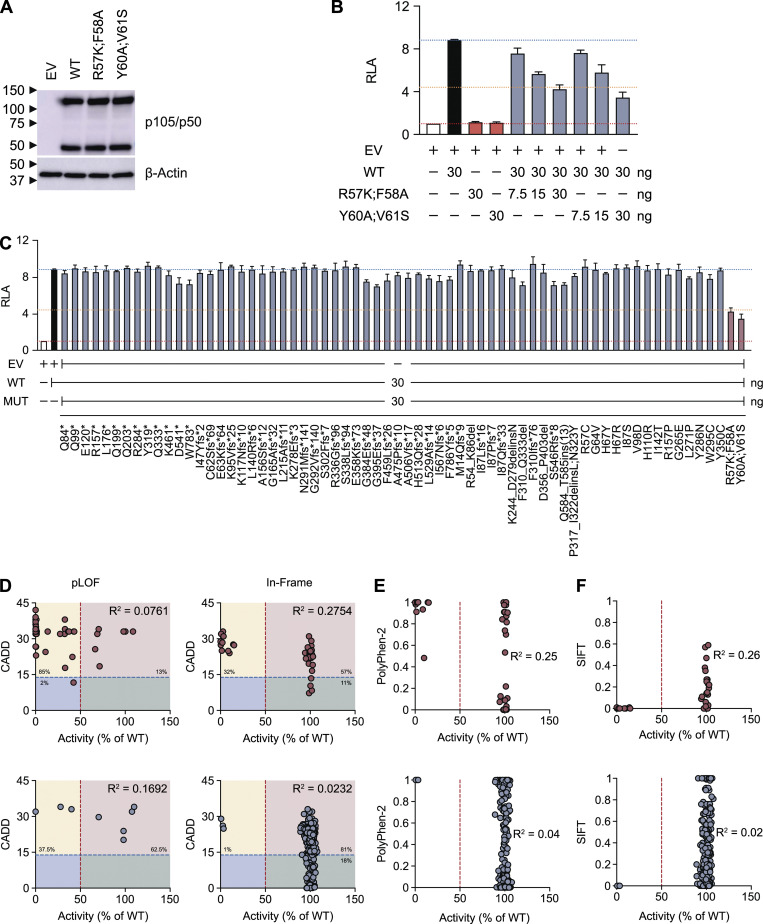
Using the new luciferase reporter assay described above, we investigated 365 nonsynonymous variants reported in previous studies or present in our HGID cohort or gnomAD (Table S1). We defined the deleterious variants as LOF if they had a luciferase activity equivalent to that of EV, and as hypomorphic if their luciferase activity was less than half that of the WT allele. We identified 44 LOF variants: 10 nonsense, 15 frameshift, 8 splice site, 10 missense, and 1 in-frame indel variants (Fig. 3 and Table 2). 3 nonsense, 10 frameshift, 2 splice site, and 4 missense variants were hypomorphic, and 5 of these variants were severely hypomorphic (<25% WT activity; Fig. 3 and Table 2). Therefore, 63 variants in total were found to be deleterious, 5 of which had normal levels of p105 and p50 expression and were located in the RHD of p50 (Fig. S1, A–C; and Fig. S2, A–C). The remaining 302 variants were neutral (Fig. 3 and Table 2). However, five pLOF variants with a premature stop codon downstream from the proteolytic cleavage site expressed a p50-like subunit (Fig. S1, A–C) and were found to be neutral in our luciferase assay (Fig. 3), possibly due to overexpression from intron-less plasmids. We then investigated whether LOF or hypomorphic mutant alleles could have dominant-negative (DN) effects on the WT allele. We assessed the impact of increasing the amount of cDNA for 66 mutant forms of p105 while keeping the amount of cDNA encoding WT p105 constant. Two p105 mutants (R57K;F58A and Y60A;V61S) were generated to serve as positive controls, as p50 harboring the same mutations had already been reported to have a DN effect on NF-κB activity (Bressler et al., 1993). As expected, both R57K;F58A and Y60A;V61S were LOF and DN (Fig. S4, A and B), and none of the mutant alleles tested had a DN effect on the WT allele at a mutant/WT ratio of 1:1 (Fig. S4 C). Finally, we found no significant correlation between the activity of mutant alleles and their combined annotation-dependent depletion (CADD), Polymorphism Phenotyping v2 (PolyPhen-2), and sorting tolerant from intolerant (SIFT) scores (Fig. S4, D–F). The discordance between the predicted and experimental deleteriousness of variants is consistent with previous reports (Miosge et al., 2015). These scores cannot, therefore, be used to predict whether a rare NFKB1 allele will be deleterious. The functional assay thus identified 63 NFKB1 variants as deleterious and suggested that CADD, PolyPhen-2, and SIFT do not accurately predict the functional activity of NFKB1 variants. We therefore established a new functional test indispensable for determining the deleteriousness and, by inference, pathogenicity of human NFKB1 variants.
Figure 3.
Functional characterization of NFKB1 variants. RLA of NFKB1-deficient HEK293T cells transfected with κB-luc in the presence of plasmids encoding S276A p65 (100 ng) plus WT (30 ng) or mutant p105 (30 ng). The red dashed line represents the activity of EV. The orange dashed line represents half the activity of WT p105. The blue dashed line represents the activity of WT p105. Columns corresponding to variants in the RHD, glycine-rich region (GRR), ankyrin repeat domain (ARD), death domain (DD), and other regions of p105 are colored red, green, blue, purple, and gray, respectively. The values shown are the means ± SEM of three independent experiments.
Table 2. Functional activity of NFKB1 variants.
| Variant | Tested | LOF | Hypomorphic | Neutral |
|---|---|---|---|---|
| Nonsense | 17 | 10 | 3 | 4 |
| Frameshift | 29 | 15 | 10 | 4 |
| Splice site | 14 | 8 | 2 | 4 |
| Start loss | 1 | 0 | 0 | 1 |
| In-frame indel | 4 | 1 | 0 | 3 |
| Missense | 300 | 10 | 4 | 286 |
| Total | 365 | 44 | 19 | 302 |
Figure S4.
Negative dominance and correlation between predicted score and the activity of the NFKB1 variants. (A) Immunoblot of p105/p50 and β-actin in NFKB1-deficient HEK293T cells transfected with WT, R57K;F58A, or Y60A;V61S p105. WT and mutant p105 cDNA was inserted into the pLenti-III-EF1α vector with an N-terminal FLAG tag. p105/p50 was detected with an anti-p105/p50 antibody recognizing the N terminus of p105. (B) RLA of NFKB1-deficient HEK293T cells transfected with κB-luc in the presence of plasmids encoding S276A p65 (100 ng) plus WT and/or mutant p105, as indicated. The total amount of plasmid in each transfection was kept constant by adding EV. (C) RLA of NFKB1-deficient HEK293T cells transfected with κB-luc in the presence of plasmids encoding S276A p65 (100 ng) plus WT and mutant p105, as indicated. The total amount of plasmid in each transfection was kept constant by adding EV. (D–F) The transcriptional activity of all mutant alleles was normalized against that of the WT allele. Red and blue dots denote the variants associated with CVID and other phenotypes, respectively. The red dashed line indicates half the activity of the WT allele. (D) CADD score. The blue dashed line represents the mutation significance cutoff threshold for NFKB1 (13.85; Itan et al., 2016). The yellow, pink, green, and blue rectangles represent the true-positive, false-positive, true-negative, and false-negative areas, respectively. The data shown are representative of two independent experiments (A). The values shown are the means ± SEM of three independent experiments (B and C). The R2 values were obtained in a correlation analysis performed with Prism software (D–F). No significant difference between the mutant and WT p105 by one-way ANOVA performed with Prism software (C).
Population genetics of human NFKB1
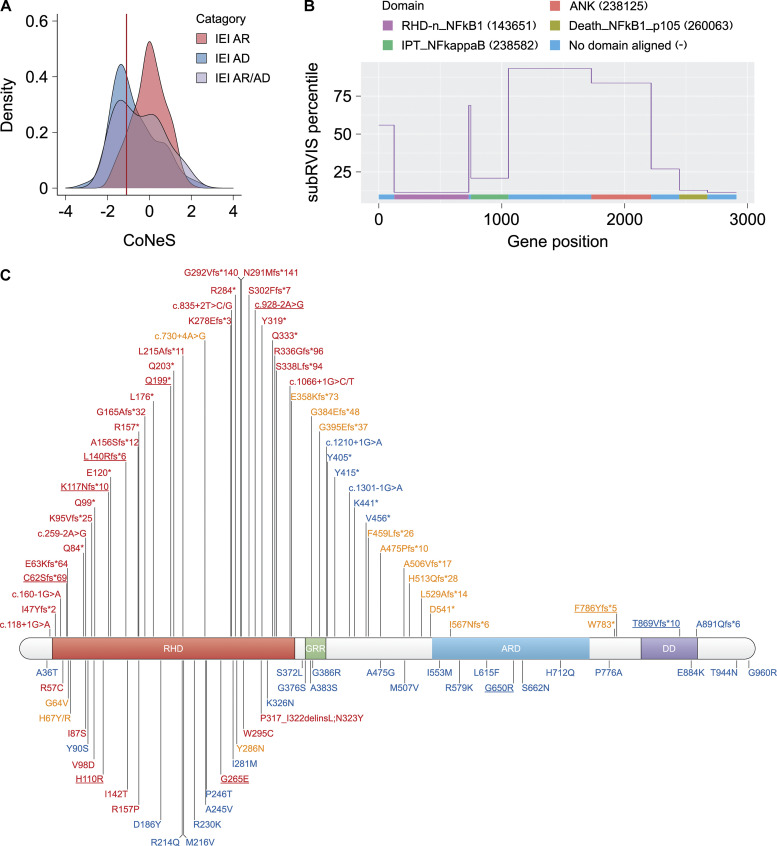
Consistent with the above findings, NFKB1 has a gene damage index of 1.96 (Itan et al., 2015), a neutrality index of 0.2 (McDonald and Kreitman, 1991), and a SnIPRE f parameter of 0.41 (within the top 33% of genes within the genome subject to the greatest constraints; Eilertson et al., 2012), indicating that it is highly conserved in human populations and has evolved under purifying selection. In addition, NFKB1 has a probability of LOF intolerance score of 1 (Lek et al., 2016) and an LOF observed/expected upper bound fraction of 0.044 (Karczewski et al., 2020), suggesting that it is extremely intolerant to LOF variants, with a probability of LOF intolerance > 0.9 considered to be associated with genes operating by haploinsufficiency. Moreover, NFKB1 has a consensus negative selection score of −1.11 (Rapaport et al., 2021), placing it within the group of IEI genes operating by haploinsufficiency (Fig. S5 A). Finally, subregion residual variation intolerance score (subRVIS) varies considerably between NFKB1 domains (SD percentile = 56.9; Gussow et al., 2016), but the RHD and death domain were found to be under stronger constraint (subRVIS percentiles = 11.48 and 12.86 relative to all protein domains; Fig. S5 B). This finding is consistent with our data suggesting that all the LOF variants are located in the RHD. Our results therefore suggest that heterozygous deleterious NFKB1 variants can cause autosomal dominant (AD) disease by haploinsufficiency.
Figure S5.
Population genetics of human NFKB1 and NFKB1 variants in patients with CVID or related conditions. (A) The distribution of consensus negative selection (CoNeS) for genes underlying autosomal recessive (AR) IEI, AD IEI, and AR/AD IEI. NFKB1 (red line) falls within the category of AD IEI. (B) Intolerance to functional variation along the NFKB1 genic region, as measured by the subRVIS percentiles. The protein domains were annotated with the Conserved Domain Database (CDD). The subregions were defined as the regions within the gene aligning with the CDD and the unaligned regions were the regions lying between CDD alignments. The numbers in parentheses indicate the ID of the sequence in the CDD. (C) Schematic illustration of NFKB1 variants in the NFKB1 gene. Variants tested LOF, hypomorphic, and neutral are colored red, orange, and blue, respectively. New variants identified in this study are underlined.
Enrichment in deleterious NFKB1 variants in patients with CVID
Following the functional characterization of all 365 variants, we tested the hypothesis of an enrichment in heterozygous deleterious NFKB1 variants in patients with CVID. For the enrichment analysis, we counted the number of experimentally proven deleterious NFKB1 variants in our HGID-CVID cohort of 305 patients with CVID and in the gnomAD control cohort of 60,146 individuals. We found seven deleterious variants in the HGID-CVID cohort and only two in the gnomAD control cohort (Table 3). There was a strong enrichment in deleterious NFKB1 variants in the HGID-CVID cohort relative to the gnomAD control cohort (P = 2.78 × 10−15; odds ratio [OR] = 697.9; 95% confidence interval [CI] = 133.2 to 8,192; Table 4). This enrichment was also observed when we restricted our analysis to Europeans (P = 5.73 × 10−11; OR = ∞; 95% CI = 102.2 to ∞), which account for 66% of the HGID-CVID cohort and 40% of the gnomAD control cohort, respectively (Table 4). Strikingly, we identified six deleterious variants in the HGID-ID cohort of patients with various infectious diseases, five of whom were later found to satisfy the diagnostic criteria for CVID (Table 3). We found that 55 of the 97 published variants (57%) were deleterious (including the 2 large deletions not tested with our assay) and 42 (43%) were neutral (Table 3). The 55 deleterious variants included 51 identified in patients with CVID, and 2 were identified in patients without CVID at the time of study, but with the related presentation of cytopenia, splenomegaly, lymphadenopathy, and autoinflammatory disorder (Table 3; Duan and Feanny, 2019; Lorenzini et al., 2020). The remaining 2 variants were identified in patients without specified phenotypes (Table 3; Lorenzini et al., 2020). Only 3 of the 63 deleterious variants identified were reported in the whole gnomAD, each with a MAF < 4 × 10−6 (Table 3). One of these three variants has also been reported in patients with CVID (Table 3; Rae et al., 2017; Schröder et al., 2019; Tuijnenburg et al., 2018). Finally, none of the variants with a MAF ≥ 10−5 or reported in at least two individuals in the whole gnomAD (n = 141,456 individuals) were deleterious (Table S1), consistent with our hypothesis that deleterious NFKB1 variants are extremely rare in the general population. Overall, these results firmly demonstrate that deleterious NFKB1 variants can cause AD CVID.
Table 3. Heterozygous NFKB1 variants experimentally demonstrated to be deleterious or neutral.
| Source | Phenotype | Total | Tested | LOF | Hypomorphic | Neutral | |
|---|---|---|---|---|---|---|---|
| Previous studies | CVID | 78 | 76 | 33 | 16 | 27 | |
| IDD | 4 | 4 | 2 | 0 | 2 | ||
| Unspecified | 15 | 15 | 1 | 1 | 13 | ||
| 97 | 95 | 36 | 17 | 42 | |||
| HGID | HGID-CVID | CVID | 15 | 15 | 6 | 1 | 8 |
| HGID-ID | CVID | 5 | 5 | 4 | 1 | 0 | |
| ID | 64 | 64 | 1 | 0 | 63 | ||
| gnomAD | Control | 209 | 209 | 2a | 0 | 207 | |
| Case | 158 | 51 | 0 | 1 | 50 | ||
| 367 | 260 | 2a | 1 | 257 | |||
IDD, immune dysregulation disorder.
One variant has been reported in patients with CVID.
Table 4. Analysis of enrichment in NFKB1 variants experimentally demonstrated to be deleterious.
| Cohort | Total exomes | Exomes with deleterious variant | Fisher’s test | OR (95% CI) |
|---|---|---|---|---|
| HGID-CVID | 305 | 7 | 2.78 × 10−15 | 697.9 (133.2–8,192) |
| gnomAD control | 60,146 | 2a | ||
| HGID-CVID EUR | 220 | 5 | 5.73 × 10−11 | ∞ (102.2–∞) |
| gnomAD control NFE | 24,146 | 0 |
EUR, European; NFE, non-Finnish European.
One variant has been reported in patients with CVID.
Discussion
We systematically studied the impact of 365 known NFKB1 variants with a newly developed NF-κB reporter assay specifically evaluating p65:p50-dependent transcription activity. This assay demonstrated that only 80% (48/60) of the pLOF variants were LOF or hypomorphic but also revealed that 5% (15/305) of the in-frame variants were LOF or hypomorphic (Fig. S5 C). We also showed that none of the LOF or hypomorphic variants tested exerted any detectable negative dominance. All LOF variants were found to be confined to the RHD of p105/p50, which is responsible for dimerization, nuclear localization, DNA binding, and interaction with IκB proteins (Pereira and Oakley, 2008). These findings clearly support the hypothesis that the disruption of p65:p50 heterodimers is the key molecular mechanism underlying the deleteriousness of NFKB1 variants. Together with population genetic data, they further show that AD NFKB1 deficiency operates by haploinsufficiency, as opposed to negative dominance.
Any new NFKB1 variant can be tested with this reporter assay to determine its functional significance. Nevertheless, this assay has three limitations: (1) It cannot evaluate variants that affect the function of p105 in a p50-independent manner, through the inhibition of TPL-2 kinase, for example (Beinke et al., 2003; Waterfield et al., 2003). I553M, the only mutation for which it has been suggested that disease is caused by alterations to the phosphorylation and stability of p105 rather than p50 function (Kaustio et al., 2017), was neutral in our assay. (2) It cannot evaluate the repressive transcriptional effect of p50:p50 homodimers. (3) It may underestimate the deleteriousness of variants generating premature stop codons, because the intron-less cDNAs used in our assay fail to elicit nonsense-mediated mRNA decay (Maquat, 2004). The five pLOF variants that were neutral in our in vitro assay may thus, nevertheless, be deleterious in vivo. Notwithstanding, this new in vitro functional assay can accurately identify deleterious NFKB1 variants. Additional ex vivo tests on leukocytes from patients can further delineate the biological impact of these and other variants in the heterozygous state.
Our study provides compelling evidence that heterozygosity for a deleterious NFKB1/p50 variant underlies AD CVID and that it does so by haploinsufficiency. Our study establishes a reference database of 365 known NFKB1 variants based on a new NF-κB reporter assay. Any new NFKB1 variant found in CVID patients can now be tested for deleteriousness and, by inference, for causality. Very few of the patients with NFKB1/p50 variants identified as deleterious in our assay had immunological conditions related to, but not designated as, CVID (Duan and Feanny, 2019; Lorenzini et al., 2020). It is tempting to speculate that these patients were misdiagnosed or diagnosed before the onset of bona fide CVID. Patients with any condition found to carry a deleterious NFKB1 variant should thus also be repeatedly tested for CVID phenotypes (e.g., hypogammaglobulinemia). Conversely, with the caveats mentioned above, patients with CVID carrying NFKB1 variants that are functionally neutral in our assay should not be diagnosed as NFKB1 deficient, even if the variant is rare and predicted to be deleterious in silico, unless further experimental investigation reveals molecular and biochemical defects.
Materials and methods
Patient cohorts
Patients enrolled in the HGID-CVID cohort were diagnosed with CVID according to established criteria, including serum IgG and IgA and/or IgM deficiency with a proven loss of antibody production (Bonilla et al., 2016; Tangye et al., 2020). Written informed consent was obtained from each patient, in accordance with local regulations and protocols approved by the institutional review board of Mount Sinai, Saint-Louis Hospital, and the National Institute of Allergy and Infectious Diseases of the National Institutes of Health. The individuals studied in the HGID-ID cohort had a wide range of different infectious diseases and recruited by clinicians for analysis in our laboratories. All study participants provided written informed consent for the use of their DNA in studies aiming to identify genetic risk variants for disease. Institutional review board approval was obtained from The Rockefeller University and the Institut National de la Santé et de la Recherche Médicale (INSERM) and various collaborating institutions.
Whole-exome sequencing
Genomic DNA was extracted from whole blood and sheared with an S2 focused ultrasonicator (Covaris). An adapter-ligated library was prepared with the TruSeq DNA Sample Prep Kit (Illumina). Exome capture was performed with the SureSelect Human All Exon V5 kit (Agilent Technologies). Paired-end sequencing was performed on a HiSeq 2500 System (Illumina) generating 100-base reads. The sequences were aligned with the GRCh37 build of the human genome reference sequence, with the Burrows-Wheeler Aligner (Li and Durbin, 2009). Downstream processing and variant calling were performed with the Genome Analysis Toolkit (McKenna et al., 2010), SAMtools (Li et al., 2009), and Picard Tools (Broad Institute). All variants were annotated with in-house annotation software.
Plasmids
RELA/p65 and NFKB1/p105 were amplified from cDNA derived from HEK293T cells (American Type Culture Collection). The full-length WT and mutant p65 sequences were inserted into the pLenti-III-EF1α vector (Applied Biological Materials) with a C-terminal HA tag. The full-length WT p105 and M1V mutant sequences were inserted into the pLenti-III-EF1α vector (Applied Biological Materials). The full-length WT and all other mutant p105 sequences were inserted into the pLenti-III-EF1α vector (Applied Biological Materials) with an N-terminal FLAG tag. The κB reporter construct (κB-luc) was generated by inserting the sequence (5′-GATCCGGAGGGACTTTCCGCTGGGGACTTTCCAGCCG-3′) containing HIV-1 κB enhancer motif (Stein et al., 1989) into the pGL4.23[luc2/minP] vector (E8411; Promega) between KpnI and BglII sites. The pGL4.32[luc2P/NF-κB-RE/Hygro] and pRL-SV40 vectors were obtained from Promega (E8491 and E2231, respectively). Site-directed mutagenesis was performed as previously described (Zheng et al., 2004).
Cell culture and transfection
HEK293T and COS-7 cells (American Type Culture Collection) were maintained in DMEM (Gibco) supplemented with 10% FBS (Gibco). Transient transfection was performed with Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific), in accordance with the manufacturer’s instructions.
Generation of NFKB1-deficient (NFKB1−/−) HEK293T cells
NFKB1−/− HEK293T cells were generated with the CRISPR-Cas9 system. Guide RNAs designed to target exon V (sense: 5′-CACCGGGCACCAGGTAGTCCACCAT-3′; antisense: 5′-AAACATGGTGGACTACCTGGTGCCC-3′) were inserted into the lentiCRISPR v2 vector (52961; Addgene). HEK293T cells were transfected with the resulting plasmid with the aid of Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific) and cultured at low density for clonal selection. Individual clones were picked, expanded, and analyzed by genomic PCR and subsequently tested for an absence of p105/p50 expression.
Exon-trapping assay
A genomic fragment containing the WT sequence corresponding to each variant and its surrounding exons and introns was amplified from HEK293T cells by PCR with specific primers (Table S4) and inserted into the pSPL3 vector (Life Technologies). Each variant was reintroduced by site-directed mutagenesis. COS-7 cells were transfected with the resulting plasmids in the presence of Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific). Cells were harvested 24 h after transfection, and total RNA was extracted with the RNeasy Mini Kit (QIAGEN). Reverse transcription was performed with the iScript Reverse Transcription Supermix (Bio-Rad), according to the manufacturer’s instructions. After cDNA synthesis, PCR was performed with the SD6 (5′-TCTGAGTCACCTGGACAACC-3′) and SA2 (5′-ATCTCAGTGGTATTTGTGAGC-3′) primers. The amplified cDNAs were inserted into the pCR4-TOPO vector (Thermo Fisher Scientific) and subsequently sequenced with the M13 forward (5′-GTAAAACGACGGCCAG-3′) and M13 reverse (5′-CAGGAAACAGCTATGAC-3′) primers. For each variant, we analyzed 100 colonies, to determine the ratio of spliced products from WT and mutant minigenes.
Luciferase reporter assay
The luciferase reporter assay was performed as previously described (Béziat et al., 2018). WT and NFKB1−/− HEK293T cells were transfected with a reporter plasmid (100 ng/well for a 96-well plate), the pRL-SV40 vector (10 ng/well), and a WT or mutant p105 and/or WT or mutant p65 in the presence of Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific). After 24 h, cells were harvested, and luciferase activity was measured with the Dual-Glo Luciferase Assay System (Promega).
Western blotting
Western blotting was performed as previously described (Li et al., 2019). Whole-cell lysates were prepared in RIPA buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P40, 0.5% sodium deoxycholate, and 0.1% SDS) supplemented with cOmplete Protease Inhibitor Cocktail (Roche). Proteins were separated by electrophoresis in 4–20% Mini-PROTEAN TGX Precast Protein Gels (Bio-Rad), and the resulting bands were transferred onto Immobilon-P polyvinylidene fluoride membrane (Millipore). All blots were incubated overnight with primary antibodies and developed with the Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific). The antibodies used in this study included antibodies against p105/p50 (N terminus; 3035; Cell Signaling Technology), p105/p50 (residues surrounding amino acid 415; 12540; Cell Signaling Technology), p65 (sc-372; Santa Cruz Biotechnology), RELB (sc-48366; Santa Cruz Biotechnology), c-REL (sc-6955; Santa Cruz Biotechnology), p100/p52 (4882; Cell Signaling Technology), FLAG (A8592; Millipore Sigma), and β-actin (5125; Cell Signaling Technology) and the following secondary antibodies: Amersham ECL mouse IgG, HRP-linked whole antibody (from sheep; NA931; GE Healthcare Life Sciences) and Amersham ECL rabbit IgG, HRP-linked whole antibody (from donkey; NA934; GE Healthcare Life Sciences).
Online supplemental material
Fig. S1 shows the protein expression of pLOF mutant NFKB1 alleles. Fig. S2 shows the protein expression of in-frame mutant NFKB1 alleles. Fig. S3 shows the effect of p105 on TNF-induced NF-κB activation. Fig. S4 shows that none of the deleterious NFKB1 variants exert negative dominance and the absence of correlation between the activity and CADD, PolyPhen-2, and SIFT scores of NFKB1 alleles. Fig. S5 shows population genetics of human NFKB1 and a summary of NFKB1 variants in patients with CVID or related conditions. Table S1 lists NFKB1 variants in our and other databases of healthy and sick individuals. Table S2 lists the genotype and the phenotype in individuals with NFKB1 variants. Table S3 shows the alternative transcripts generated by NFKB1 splice-site variants. Table S4 lists the primers used for the amplification of NFKB1 genomic DNA. Data S1 illustrates NFKB1 splice-site variants and their corresponding alternative transcripts (described in Table S3), based on the results of exon trapping assays.
Supplementary Material
lists all NFKB1 variants in our and other databases of healthy and sick individuals.
lists NFKB1 genotype–phenotype correlation in individuals with NFKB1 variants.
lists alternative transcripts generated by NFKB1 splice-site variants.
lists primers used for the amplification of NFKB1 genomic DNA.
illustrates NFKB1 splice-site variants and their corresponding alternative transcripts (described in Table S3), based on the results of exon trapping assays.
Acknowledgments
We warmly thank the patients and their families for participating in the study. We thank the members of the Laboratory of Human Genetics of Infectious Diseases for fruitful discussions. We thank Yelena Nemirovskaya, Dana Liu, Dominick Papandrea, Mark Woollett, Lazaro Lorenzo, Cécile Patissier, and Tatiana Kochetkov for administrative and technical assistance.
This work was funded in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (P01AI061093), the National Center for Research Resources of the National Institutes of Health, the National Center for Advancing Sciences of the National Institutes of Health (UL1TR001866), The Rockefeller University, the St. Giles Foundation, INSERM, University of Paris, the Qatar National Research Fund (NPRP10-0206-170359), the French Foundation for Medical Research (EQU201903007798), the SCOR Corporate Foundation for Science, the French National Research Agency under the “Investments for the future” program (ANR-10-IAHU-01), the “PNEUMOPID” project (ANR-14-CE15-0009-01), and the Integrative Biology of Emerging Infectious Diseases Laboratory of Excellence (ANR-10-LABX-62-IBEID). J. Rosain was supported by the MD-PhD program of the Imagine Institute, with the support of the Bettencourt-Schueller Foundation and the INSERM PhD program (poste d’accueil INSERM). A.N. Spaan was supported by the European Commission (Horizon 2020 Marie Skłodowska-Curie Individual Fellowship 789645), the Dutch Research Council (Rubicon Grant 019.171LW.015), and the European Molecular Biology Organization (long-term fellowship ALTF 84-2017, nonstipendiary). V.K. Rao, G. Uzel, A.F. Freeman, S.M. Holland, and H.C. Su were supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health Intramural Research Program.
Author contributions: J. Li, W.-T. Lei, B. Lyu, J. Rosain, Q. Zhang, and B. Boisson performed the experiments and analyzed the data. P. Zhang, F. Rapaport, Y. Seeleuthner, Y. Zhang, B. Bigio, L. Abel, and A. Cobat conducted whole-exome sequencing and computational and statistical analyses. B. Hammadi, M. Migaud, M. Chrabieh, V. Béziat, J. Bustamante, S.-Y. Zhang, E. Jouanguy, S. Boisson-Dupuis, J. El Baghdadi, V. Aimanianda, K. Thoma, M. Fliegauf, B. Grimbacher, A.-S. Korganow, C. Saunders, V.K. Rao, G. Uzel, A.F. Freeman, S.M. Holland, H.C. Su, C. Cunningham-Rundles, C. Fieschi, and A. Puel provided clinical samples and analyzed clinical data. T. Asano, S.J. Pelham, A.N. Spaan, and D. Hum provided expertise and feedback. J. Li, J.-L. Casanova, Q. Zhang, and B. Boisson designed the study and wrote the manuscript with the assistance of all coauthors.
References
- Abolhassani, H., Hammarström L., and Cunningham-Rundles C.. 2020. Current genetic landscape in common variable immune deficiency. Blood. 135:656–667. 10.1182/blood.2019000929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinke, S., Deka J., Lang V., Belich M.P., Walker P.A., Howell S., Smerdon S.J., Gamblin S.J., and Ley S.C.. 2003. NF-kappaB1 p105 negatively regulates TPL-2 MEK kinase activity. Mol. Cell. Biol. 23:4739–4752. 10.1128/MCB.23.14.4739-4752.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béziat, V., Li J., Lin J.X., Ma C.S., Li P., Bousfiha A., Pellier I., Zoghi S., Baris S., Keles S., et al. 2018. A recessive form of hyper-IgE syndrome by disruption of ZNF341-dependent STAT3 transcription and activity. Sci. Immunol. 3:eaat4956. 10.1126/sciimmunol.aat4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogaert, D.J., Dullaers M., Lambrecht B.N., Vermaelen K.Y., De Baere E., and Haerynck F.. 2016. Genes associated with common variable immunodeficiency: one diagnosis to rule them all? J. Med. Genet. 53:575–590. 10.1136/jmedgenet-2015-103690 [DOI] [PubMed] [Google Scholar]
- Bonilla, F.A., Barlan I., Chapel H., Costa-Carvalho B.T., Cunningham-Rundles C., de la Morena M.T., Espinosa-Rosales F.J., Hammarström L., Nonoyama S., Quinti I., et al. 2016. International Consensus Document (ICON): Common Variable Immunodeficiency Disorders. J. Allergy Clin. Immunol. Pract. 4:38–59. 10.1016/j.jaip.2015.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousfiha, A., Jeddane L., Picard C., Al-Herz W., Ailal F., Chatila T., Cunningham-Rundles C., Etzioni A., Franco J.L., Holland S.M., et al. 2020. Human Inborn Errors of Immunity: 2019 Update of the IUIS Phenotypical Classification. J. Clin. Immunol. 40:66–81. 10.1007/s10875-020-00758-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boztug, H., Hirschmugl T., Holter W., Lakatos K., Kager L., Trapin D., Pickl W., Förster-Waldl E., and Boztug K.. 2016. NF-κB1 Haploinsufficiency Causing Immunodeficiency and EBV-Driven Lymphoproliferation. J. Clin. Immunol. 36:533–540. 10.1007/s10875-016-0306-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressler, P., Brown K., Timmer W., Bours V., Siebenlist U., and Fauci A.S.. 1993. Mutational analysis of the p50 subunit of NF-kappa B and inhibition of NF-kappa B activity by trans-dominant p50 mutants. J. Virol. 67:288–293. 10.1128/jvi.67.1.288-293.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen, M., Offersen R., Jensen J.M.B., Petersen M.S., Larsen C.S., and Mogensen T.H.. 2020. Identification of Novel Genetic Variants in CVID Patients With Autoimmunity, Autoinflammation, or Malignancy. Front. Immunol. 10:3022. 10.3389/fimmu.2019.03022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham-Rundles, C. 2019. Common variable immune deficiency: Dissection of the variable. Immunol. Rev. 287:145–161. 10.1111/imr.12728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieli-Crimi, R., Martínez-Gallo M., Franco-Jarava C., Antolin M., Blasco L., Paramonov I., Semidey M.E., Álvarez Fernández A., Molero X., Velásquez J., et al. 2018. Th1-skewed profile and excessive production of proinflammatory cytokines in a NFKB1-deficient patient with CVID and severe gastrointestinal manifestations. Clin. Immunol. 195:49–58. 10.1016/j.clim.2018.07.015 [DOI] [PubMed] [Google Scholar]
- Duan, L., and Feanny S.. 2019. Novel heterozygous NFKB1 mutation in a pediatric patient with cytopenias, splenomegaly, and lymphadenopathy. LymphoSign J. 6:61–67. 10.14785/lymphosign-2019-0006 [DOI] [Google Scholar]
- Eilertson, K.E., Booth J.G., and Bustamante C.D.. 2012. SnIPRE: selection inference using a Poisson random effects model. PLOS Comput. Biol. 8:e1002806. 10.1371/journal.pcbi.1002806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf, M., Bryant V.L., Frede N., Slade C., Woon S.T., Lehnert K., Winzer S., Bulashevska A., Scerri T., Leung E., et al. 2015. Haploinsufficiency of the NF-κB1 Subunit p50 in Common Variable Immunodeficiency. Am. J. Hum. Genet. 97:389–403. 10.1016/j.ajhg.2015.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganchi, P.A., Sun S.C., Greene W.C., and Ballard D.W.. 1993. A novel NF-kappa B complex containing p65 homodimers: implications for transcriptional control at the level of subunit dimerization. Mol. Cell. Biol. 13:7826–7835. 10.1128/MCB.13.12.7826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gathmann, B., Mahlaoui N., Gérard L., Oksenhendler E., Warnatz K., Schulze I., Kindle G., Kuijpers T.W., van Beem R.T., Guzman D., et al. European Society for Immunodeficiencies Registry Working Party . 2014. Clinical picture and treatment of 2212 patients with common variable immunodeficiency. J. Allergy Clin. Immunol. 134:116–126. 10.1016/j.jaci.2013.12.1077 [DOI] [PubMed] [Google Scholar]
- Gussow, A.B., Petrovski S., Wang Q., Allen A.S., and Goldstein D.B.. 2016. The intolerance to functional genetic variation of protein domains predicts the localization of pathogenic mutations within genes. Genome Biol. 17:9. 10.1186/s13059-016-0869-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden, M.S., and Ghosh S.. 2008. Shared principles in NF-kappaB signaling. Cell. 132:344–362. 10.1016/j.cell.2008.01.020 [DOI] [PubMed] [Google Scholar]
- Hoffmann, A., Leung T.H., and Baltimore D.. 2003. Genetic analysis of NF-kappaB/Rel transcription factors defines functional specificities. EMBO J. 22:5530–5539. 10.1093/emboj/cdg534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, A., Natoli G., and Ghosh G.. 2006. Transcriptional regulation via the NF-kappaB signaling module. Oncogene. 25:6706–6716. 10.1038/sj.onc.1209933 [DOI] [PubMed] [Google Scholar]
- Itan, Y., Shang L., Boisson B., Patin E., Bolze A., Moncada-Vélez M., Scott E., Ciancanelli M.J., Lafaille F.G., Markle J.G., et al. 2015. The human gene damage index as a gene-level approach to prioritizing exome variants. Proc. Natl. Acad. Sci. USA. 112:13615–13620. 10.1073/pnas.1518646112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itan, Y., Shang L., Boisson B., Ciancanelli M.J., Markle J.G., Martinez-Barricarte R., Scott E., Shah I., Stenson P.D., Gleeson J., et al. 2016. The mutation significance cutoff: gene-level thresholds for variant predictions. Nat. Methods. 13:109–110. 10.1038/nmeth.3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski, K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. Genome Aggregation Database Consortium . 2020. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 581:434–443. 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaustio, M., Haapaniemi E., Göös H., Hautala T., Park G., Syrjänen J., Einarsdottir E., Sahu B., Kilpinen S., Rounioja S., et al. 2017. Damaging heterozygous mutations in NFKB1 lead to diverse immunologic phenotypes. J. Allergy Clin. Immunol. 140:782–796. 10.1016/j.jaci.2016.10.054 [DOI] [PubMed] [Google Scholar]
- Kircher, M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., and Shendure J.. 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46:310–315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek, M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., et al. Exome Aggregation Consortium . 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 536:285–291. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., and Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.. 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Ritelli M., Ma C.S., Rao G., Habib T., Corvilain E., Bougarn S., Cypowyj S., Grodecká L., Lévy R., et al. 2019. Chronic mucocutaneous candidiasis and connective tissue disorder in humans with impaired JNK1-dependent responses to IL-17A/F and TGF-β. Sci. Immunol. 4:eaax7965. 10.1126/sciimmunol.aax7965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzini, T., Fliegauf M., Klammer N., Frede N., Proietti M., Bulashevska A., Camacho-Ordonez N., Varjosalo M., Kinnunen M., de Vries E., et al. NIHR BioResource . 2020. Characterization of the clinical and immunologic phenotype and management of 157 individuals with 56 distinct heterozygous NFKB1 mutations. J. Allergy Clin. Immunol. 146:901–911. 10.1016/j.jaci.2019.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lougaris, V., Moratto D., Baronio M., Tampella G., van der Meer J.W.M., Badolato R., Fliegauf M., and Plebani A.. 2017a. Early and late B-cell developmental impairment in nuclear factor kappa B, subunit 1-mutated common variable immunodeficiency disease. J. Allergy Clin. Immunol. 139:349–352.e1. 10.1016/j.jaci.2016.05.045 [DOI] [PubMed] [Google Scholar]
- Lougaris, V., Patrizi O., Baronio M., Tabellini G., Tampella G., Damiati E., Frede N., van der Meer J.W.M., Fliegauf M., Grimbacher B., et al. 2017b. NFKB1 regulates human NK cell maturation and effector functions. Clin. Immunol. 175:99–108. 10.1016/j.clim.2016.11.012 [DOI] [PubMed] [Google Scholar]
- Maffucci, P., Filion C.A., Boisson B., Itan Y., Shang L., Casanova J.L., and Cunningham-Rundles C.. 2016. Genetic Diagnosis Using Whole Exome Sequencing in Common Variable Immunodeficiency. Front. Immunol. 7:220. 10.3389/fimmu.2016.00220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandola, A.B., Sharfe N., Nagdi Z., Dadi H., Vong L., Merico D., Ngan B., Reid B., and Roifman C.M.. 2021. Combined immunodeficiency caused by a novel homozygous NFKB1 mutation. J. Allergy Clin. Immunol. 147:727–733.e2. 10.1016/j.jaci.2020.08.040 [DOI] [PubMed] [Google Scholar]
- Maquat, L.E. 2004. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 5:89–99. 10.1038/nrm1310 [DOI] [PubMed] [Google Scholar]
- Maréchal, E., Beel K., Crols R., Hernalsteen D., and Willekens B.. 2020. Long-Term Survival after Progressive Multifocal Leukoencephalopathy in a Patient with Primary Immune Deficiency and NFKB1 Mutation. J. Clin. Immunol. 40:1138–1143. 10.1007/s10875-020-00862-y [DOI] [PubMed] [Google Scholar]
- McDonald, J.H., and Kreitman M.. 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 351:652–654. 10.1038/351652a0 [DOI] [PubMed] [Google Scholar]
- McKenna, A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., and DePristo M.A.. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miosge, L.A., Field M.A., Sontani Y., Cho V., Johnson S., Palkova A., Balakishnan B., Liang R., Zhang Y., Lyon S., et al. 2015. Comparison of predicted and actual consequences of missense mutations. Proc. Natl. Acad. Sci. USA. 112:E5189–E5198. 10.1073/pnas.1511585112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, S.G., and Oakley F.. 2008. Nuclear factor-kappaB1: regulation and function. Int. J. Biochem. Cell Biol. 40:1425–1430. 10.1016/j.biocel.2007.05.004 [DOI] [PubMed] [Google Scholar]
- Phelps, C.B., Sengchanthalangsy L.L., Malek S., and Ghosh G.. 2000. Mechanism of kappa B DNA binding by Rel/NF-kappa B dimers. J. Biol. Chem. 275:24392–24399. 10.1074/jbc.M003784200 [DOI] [PubMed] [Google Scholar]
- Rae, W., Ward D., Mattocks C.J., Gao Y., Pengelly R.J., Patel S.V., Ennis S., Faust S.N., and Williams A.P.. 2017. Autoimmunity/inflammation in a monogenic primary immunodeficiency cohort. Clin. Transl. Immunology. 6:e155. 10.1038/cti.2017.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport, F., Boisson B., Gregor A., Béziat V., Boisson-Dupuis S., Bustamante J., Jouanguy E., Puel A., Rosain J., Zhang Q., et al. 2021. Negative selection on human genes underlying inborn errors depends on disease outcome and both the mode and mechanism of inheritance. Proc. Natl. Acad. Sci. USA. 118:e2001248118. 10.1073/pnas.2001248118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentzsch, P., Witten D., Cooper G.M., Shendure J., and Kircher M.. 2019. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 47(D1):D886–D894. 10.1093/nar/gky1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick, E.S., Moshier E.L., Godbold J.H., and Cunningham-Rundles C.. 2012. Morbidity and mortality in common variable immune deficiency over 4 decades. Blood. 119:1650–1657. 10.1182/blood-2011-09-377945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, N.R., MacKichan M.L., and Israël A.. 1992. The precursor of NF-kappa B p50 has I kappa B-like functions. Cell. 71:243–253. 10.1016/0092-8674(92)90353-E [DOI] [PubMed] [Google Scholar]
- Schipp, C., Nabhani S., Bienemann K., Simanovsky N., Kfir-Erenfeld S., Assayag-Asherie N., Oommen P.T., Revel-Vilk S., Hönscheid A., Gombert M., et al. 2016. Specific antibody deficiency and autoinflammatory disease extend the clinical and immunological spectrum of heterozygous NFKB1 loss-of-function mutations in humans. Haematologica. 101:e392–e396. 10.3324/haematol.2016.145136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder, C., Sogkas G., Fliegauf M., Dörk T., Liu D., Hanitsch L.G., Steiner S., Scheibenbogen C., Jacobs R., Grimbacher B., et al. 2019. Late-Onset Antibody Deficiency Due to Monoallelic Alterations in NFKB1. Front. Immunol. 10:2618. 10.3389/fimmu.2019.02618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggers, T., Chang A.B., Teixeira A., Wong D., Williams K.J., Ahmed B., Ragoussis J., Udalova I.A., Smale S.T., and Bulyk M.L.. 2011. Principles of dimer-specific gene regulation revealed by a comprehensive characterization of NF-κB family DNA binding. Nat. Immunol. 13:95–102. 10.1038/ni.2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale, S.T. 2012. Dimer-specific regulatory mechanisms within the NF-κB family of transcription factors. Immunol. Rev. 246:193–204. 10.1111/j.1600-065X.2011.01091.x [DOI] [PubMed] [Google Scholar]
- Stein, B., Rahmsdorf H.J., Steffen A., Litfin M., and Herrlich P.. 1989. UV-induced DNA damage is an intermediate step in UV-induced expression of human immunodeficiency virus type 1, collagenase, c-fos, and metallothionein. Mol. Cell. Biol. 9:5169–5181. 10.1128/MCB.9.11.5169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangye, S.G., Al-Herz W., Bousfiha A., Chatila T., Cunningham-Rundles C., Etzioni A., Franco J.L., Holland S.M., Klein C., Morio T., et al. 2020. Human Inborn Errors of Immunity: 2019 Update on the Classification from the International Union of Immunological Societies Expert Committee. J. Clin. Immunol. 40:24–64. 10.1007/s10875-019-00737-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangye, S.G., Al-Herz W., Bousfiha A., Cunningham-Rundles C., Franco J.L., Holland S.M., Klein C., Morio T., Oksenhendler E., Picard C., et al. 2021. The Ever-Increasing Array of Novel Inborn Errors of Immunity: an Interim Update by the IUIS Committee. J. Clin. Immunol. 41:666–679. 10.1007/s10875-021-00980-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi, K., and Karin M.. 2018. NF-κB, inflammation, immunity and cancer: coming of age. Nat. Rev. Immunol. 18:309–324. 10.1038/nri.2017.142 [DOI] [PubMed] [Google Scholar]
- Tuijnenburg, P., Lango Allen H., Burns S.O., Greene D., Jansen M.H., Staples E., Stephens J., Carss K.J., Biasci D., Baxendale H., et al. NIHR BioResource–Rare Diseases Consortium . 2018. Loss-of-function nuclear factor κB subunit 1 (NFKB1) variants are the most common monogenic cause of common variable immunodeficiency in Europeans. J. Allergy Clin. Immunol. 142:1285–1296. 10.1016/j.jaci.2018.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schouwenburg, P.A., Davenport E.E., Kienzler A.K., Marwah I., Wright B., Lucas M., Malinauskas T., Martin H.C., Lockstone H.E., Cazier J.B., et al. WGS500 Consortium . 2015. Application of whole genome and RNA sequencing to investigate the genomic landscape of common variable immunodeficiency disorders. Clin. Immunol. 160:301–314. 10.1016/j.clim.2015.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterfield, M.R., Zhang M., Norman L.P., and Sun S.C.. 2003. NF-kappaB1/p105 regulates lipopolysaccharide-stimulated MAP kinase signaling by governing the stability and function of the Tpl2 kinase. Mol. Cell. 11:685–694. 10.1016/S1097-2765(03)00070-4 [DOI] [PubMed] [Google Scholar]
- Zhang, Q., Lenardo M.J., and Baltimore D.. 2017. 30 Years of NF-κB: A Blossoming of Relevance to Human Pathobiology. Cell. 168:37–57. 10.1016/j.cell.2016.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, B., Barrera L.A., Ersing I., Willox B., Schmidt S.C., Greenfeld H., Zhou H., Mollo S.B., Shi T.T., Takasaki K., et al. 2014. The NF-κB genomic landscape in lymphoblastoid B cells. Cell Rep. 8:1595–1606. 10.1016/j.celrep.2014.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, L., Baumann U., and Reymond J.L.. 2004. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32:e115. 10.1093/nar/gnh110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists all NFKB1 variants in our and other databases of healthy and sick individuals.
lists NFKB1 genotype–phenotype correlation in individuals with NFKB1 variants.
lists alternative transcripts generated by NFKB1 splice-site variants.
lists primers used for the amplification of NFKB1 genomic DNA.
illustrates NFKB1 splice-site variants and their corresponding alternative transcripts (described in Table S3), based on the results of exon trapping assays.