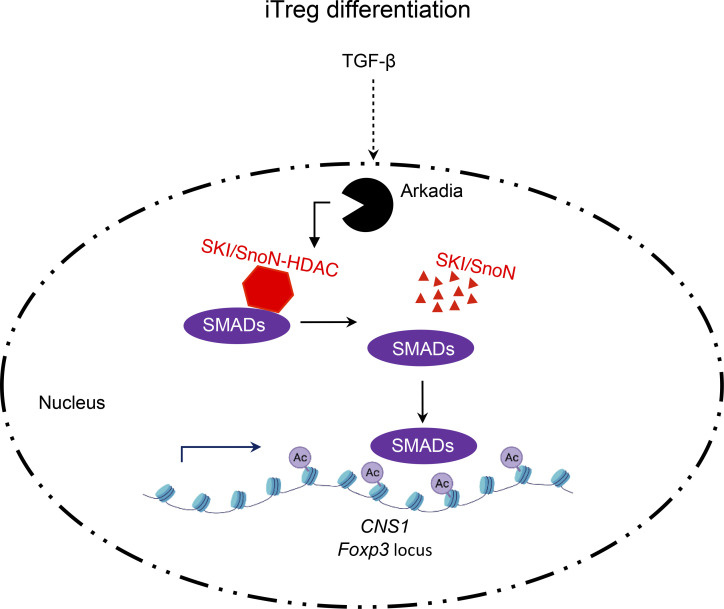
TGF-β signaling is indispensable for both Th17 and Treg cell differentiation. We show that Arkadia, an E3 ubiquitin ligase acting in TGF-β signaling, is selectively required for the differentiation of iTreg, but not Th17, cells both in vitro and in vivo.
Abstract
TGF-β signaling is fundamental for both Th17 and regulatory T (Treg) cell differentiation. However, these cells differ in requirements for downstream signaling components, such as SMAD effectors. To further characterize mechanisms that distinguish TGF-β signaling requirements for Th17 and Treg cell differentiation, we investigated the role of Arkadia (RNF111), an E3 ubiquitin ligase that mediates TGF-β signaling during development. Inactivation of Arkadia in CD4+ T cells resulted in impaired Treg cell differentiation in vitro and loss of RORγt+FOXP3+ iTreg cells in the intestinal lamina propria, which increased susceptibility to microbiota-induced mucosal inflammation. In contrast, Arkadia was dispensable for Th17 cell responses. Furthermore, genetic ablation of two Arkadia substrates, the transcriptional corepressors SKI and SnoN, rescued Arkadia-deficient iTreg cell differentiation both in vitro and in vivo. These results reveal distinct TGF-β signaling modules governing Th17 and iTreg cell differentiation programs that could be targeted to selectively modulate T cell functions.
Graphical Abstract
Introduction
T helper type 17 (Th17) and regulatory T (Treg) cells often act in opposition to each other to maintain immune homeostasis. Th17 cells are critical effectors in mucosal barrier homeostasis but can also contribute to autoimmune disease (Korn et al., 2009). Treg cells mediate tolerance to self- and nonself-antigens, preventing autoimmune disease and limiting tissue inflammation. Conversely, excessive Treg cell activity can suppress immunosurveillance and tumor cell elimination (Josefowicz et al., 2012a; Sakaguchi et al., 2008).
Cytokine-activated STAT3 and TGF-β signaling pathways are vital for Th17 cell differentiation (Bettelli et al., 2006; Harris et al., 2007; Ivanov et al., 2006; Mangan et al., 2006; Veldhoen et al., 2006a; Yang et al., 2007), while signaling through the IL-23 and IL-1 receptors is additionally required for Th17 cell–mediated pathogenicity (Ghoreschi et al., 2010; Langrish et al., 2005; McGeachy et al., 2009; Sutton et al., 2006). Under some conditions, however, TGF-β is arguably dispensable for Th17 cell differentiation. IL-6 alone can induce the Th17-regulating transcription factor retinoic acid–related orphan receptor gamma t (RORγt) and IL-17A/F expression in STAT6/T-box transcription factor TBX21 (T-bet) double KO (DKO) cells (Das et al., 2009; Yang et al., 2008b). Together with reciprocal suppression by IL-2/STAT5 of Th17 cells and IL-6/STAT3 of Treg cells (Bettelli et al., 2006; Laurence et al., 2007), these data highlight key roles of inhibitory pathways in restraining alternative differentiation programs. Nevertheless, TGF-β signaling is physiologically required for Th17 cell functions in vivo (Ghoreschi et al., 2010; Mangan et al., 2006; Veldhoen et al., 2006b).
TGF-β signaling is important for differentiation of both thymus-derived natural Treg cells (nTreg cells) and peripherally induced Treg cells (iTreg cells). In neonatal mice lacking Tgfbr2 in T cells, there was transient loss of nTreg cells (Liu et al., 2008). Differentiation of iTreg cells relies on both TGF-β (Chen et al., 2003; Zheng et al., 2004) and IL-2/STAT5 signaling pathways (Burchill et al., 2007; Yao et al., 2007) and the Foxp3 CNS1 enhancer (Josefowicz et al., 2012b; Zheng et al., 2010), which contains binding sites for SMAD2/3 transcriptional effectors of TGF-β signaling (Tone et al., 2008).
While both Th17 and iTreg cell differentiation relies on signaling through TGF-β receptors in vivo, deficiencies of downstream signaling components have divergent effects. SMAD4- or SMAD2/3-deficient T cells lose Foxp3 induction and fail to differentiate into iTreg cells (Takimoto et al., 2010; Yang et al., 2008a). However, Th17 cell differentiation in the SMAD4 KO background was similar to that of WT mice (Yang et al., 2008a), and comparable Th17 responses were induced in the absence of both Smad4 and Tgfbr2 (Zhang et al., 2017). SMAD2/3 DKO cell had normal RORγt levels but reduced IL-17A (Takimoto et al., 2010). Together, these data support the hypothesis that the SMAD (the homologies to Caenorhabditis elegans Sma and Drosophila Mad genes) complexes act as Foxp3 activators in iTreg cells and as potential inhibitors of Rorc(t) in Th17 cells (Zhang et al., 2017), but the differences in mechanisms between the cell types remain unclear.
To further explore differences between Th17 and iTreg cells in the TGF-β signaling pathway, we examined the role of Arkadia (RNF111), a RING-type E3 ligase involved in multiple signaling pathways mediated by TGF-β family members. We found that Arkadia was selectively required for iTreg, but not Th17, cell differentiation, and its absence in T cells resulted in increased susceptibility to inflammatory bowel disease. Absence of Arkadia resulted in failure of TGF-β–induced v-ski avian sarcoma viral oncogene homolog (SKI) and Ski-related novel protein N (SnoN) degradation, and targeted inactivation of these corepressors rescued Foxp3 induction in Arkadia-deficient T cells. Our results thus identify another key distinction in TGF-β signaling pathways in Th17 and iTreg cells, which could potentially be exploited for selective therapeutic targeting of these T cell subsets.
Results and discussion
Arkadia regulates in vitro differentiation of Treg, but not Th17, cells in response to TGF-β
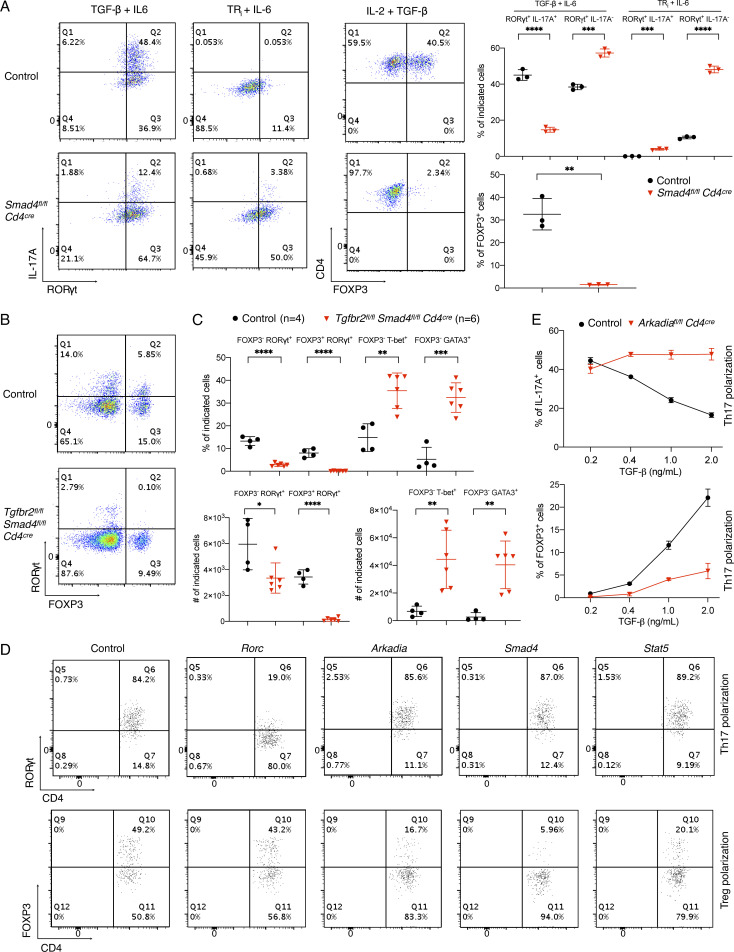
We examined Th17 cell differentiation with SMAD4-deficient T cells. Consistent with published data, loss of Smad4 had little effect on the induction of RORγt by TGF-β plus IL-6 or IL-6 alone (Fig. S1 A). However, in contrast to the earlier study, there was little expression of IL-17A in SMAD4-deficient cells after exposure to IL-6. Reflecting the importance of TGF-β signaling in Th17 cell differentiation, there was marked reduction in RORγt+FOXP3− Th17 cells in the large intestine lamina propria (LILP) of mice with T cell–specific inactivation of both SMAD4 and TGF-βR2 (Fig. S1, B and C). In light of intact Th17-dependent experimental allergic encephalomyelitis (EAE) in this genetic background (Zhang et al., 2017), these data suggest environment-dependent differences in Th17 cell regulation. We next used CRISPR-Cas9 gene targeting to interrogate the roles of multiple TGF-β pathway genes in Th17 and iTreg cell differentiation in vitro. As expected, inactivation of Smad4 or Stat5 inhibited the differentiation of iTreg, but not Th17, cells (Fig. S1 D). Among additional targeted genes, we were particularly interested in examining the role of Arkadia/RNF111, which enhances the TGF-β–SMAD2/3 signaling pathway (Mavrakis et al., 2007).
Figure S1.
Differential requirement of components of TGF-β signaling pathway in Treg and Th17 cell differentiation, related to Fig. 1. (A) Role of SMAD4 in Th17 and Treg cell differentiation. Left: Representative flow cytometry panels of T cells from control littermates and conditional Smad4 mutant mice, differentiated in vitro under Th17 and Treg cell conditions, or with blockade of TGF-β signaling with TRi (SB525334), an inhibitor of TGF-βR1. Right: Representative data of two independent experiments of T cells differentiated under the indicated conditions. Black circle, control T cells; red triangle, SMAD4-deficient T cells. P values were calculated by unpaired t test. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (B) Representative RORγt and FOXP3 staining of CD4+ TCRβ+ T lymphocytes from LILP of control littermates (with at least one allele expressing intact SMAD4 and TGF-βR2) and Tgfbr2fl/fl Smad4fl/fl Cd4cre conditional DKO mice (4–6 wk old) kept under SPF conditions. (C) Proportion and total cell number of indicated T cell populations in the LILP from control (black circle) and Tgfbr2fl/fl Smad4fl/fl Cd4cre mice (red triangle), as shown in B. Data in B and C were combined from two independent experiments. Differences were determined by unpaired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (D) Flow cytometry analysis of CD4+ T cells differentiated in vitro under Th17 and Treg conditions following transduction of retroviruses expressing the indicated gRNAs. Naive T cells were from Cas9 transgenic mice, and analysis was performed following gating on the Thy1.1 transduction marker. (E) Flow cytometry to assess percentage of IL-17A (top graph) and FOXP3 (bottom graph) expressing T cells from control littermates (black circle) and Arkadiaf/f Cd4Cre mice (red triangle) polarized in the presence of different concentration of TGF-β plus 20 ng/ml IL-6. Data in D and E are representative of three independent experiments. All error bars represent SD (A, C, and E).
Arkadia is an essential component of a subset of NODAL signaling responses and functions in embryonic development (Episkopou et al., 2001; Niederländer et al., 2001). TGF-β signaling was shown to activates Arkadia-mediated ubiquitination and degradation of SKI, SnoN, and SMADs (Koinuma et al., 2003; Levy et al., 2007; Liu et al., 2006; Nagano et al., 2007; Tsubakihara et al., 2015). SKI and SnoN bind to SMAD4 and ligand-activated SMAD2/3 and recruit transcriptional corepressor complexes containing histone deacetylases to suppress TGF-β–induced transcription (Akiyoshi et al., 1999; Luo et al., 1999; Sun et al., 1999). Because TGF-β signaling was proposed to relieve SMAD4- and SKI-dependent inhibition of Rorc(t) expression and Th17 cell differentiation (Zhang et al., 2017), we hypothesized that Arkadia may mediate the TGF-β–induced degradation of the SMAD4-associated repressive complex. In contrast to our expectation, inactivation of Arkadia had no effect on the induction of RORγt under Th17 cell differentiation conditions (Fig. S1 D). Instead, there was dramatic loss of FOXP3 under Treg differentiation conditions. Similar results were observed with multiple guide RNAs (gRNAs) targeting Arkadia (described in Materials and methods and data not shown).
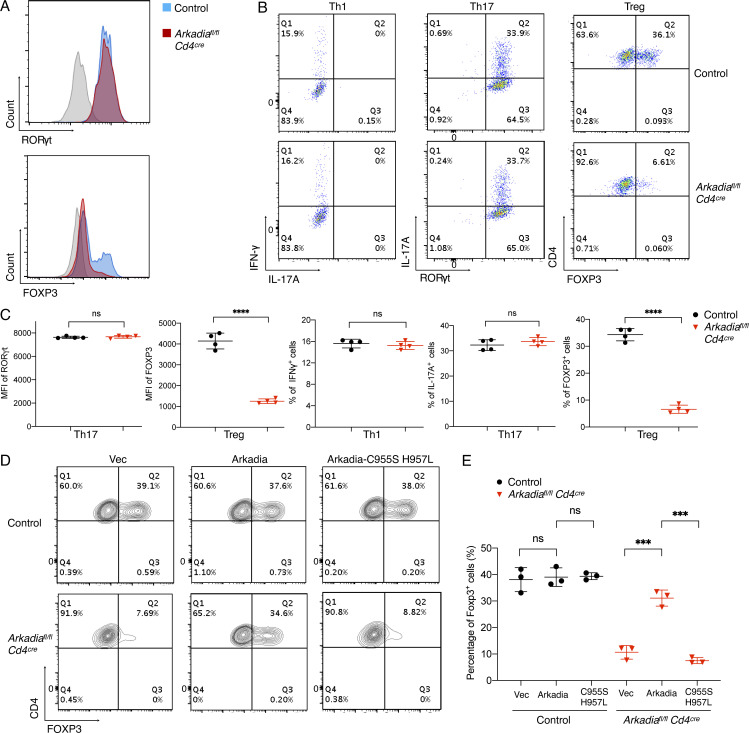
Since global KO of Arkadia in mice results in embryonic lethality, we generated Arkadiafl/fl mice and bred them to Cd4Cre transgenic mice to investigate the role of Arkadia in T cells. Arkadiafl/fl Cd4Cre mice were viable, fertile, and normal in size and did not display obvious signs of inflammation. Mutant mice had no discernible differences in cell number or composition in spleen and LNs. Arkadia-deficient T cells expressed levels of IFN-γ, IL-17A, and RORγt similar to control cells upon in vitro differentiation. In contrast, there was substantial reduction of Foxp3 expression in Arkadia-deficient T cells under Treg differentiation conditions (Fig. 1, A–C). To confirm that Arkadia function is required for Treg cell differentiation in vitro, exogenous WT or E3 ligase activity–dead Arkadia proteins were expressed in Arkadia-deficient T cells. WT, but not the mutant Akadia, protein rescued Foxp3 expression in T cells differentiated under Treg conditions (Fig. 1, D and E). Under suboptimal conditions for Th17 cell differentiation, achieved at higher concentrations of TGF-β, Arkadia-deficient T cells retained ability to produce IL-17A. By contrast, WT IL-17A+ cells were reduced with increasing concentration of TGF-β due to up-regulation of Foxp3 (Fig. S1 E; Zhou et al., 2008). Collectively, these results indicate that Arkadia is required for TGF-β induction of Foxp3.
Figure 1.
Arkadia is selectively required for in vitro Treg cell differentiation. (A) Expression of RORγt and FOXP3 in control and Cd4Cre Arkadiafl/fl T cells differentiated in vitro under Th17 and Treg conditions, respectively. (B) Representative flow cytometry profiles of IFN-γ, IL-17A, RORγt, and FOXP3 in T cells polarized under indicated conditions. (C) Statistical analysis of mean fluorescence intensity (MFI) of RORγt and FOXP3 and percentage of cells expressing IFN-γ, IL-17A, and FOXP3 following in vitro differentiation. (D and E) Rescue of Treg cell differentiation in Arkadia-deficient CD4+ T cells. Representative flow cytometry panels (D) and composite data (E) for FOXP3 expression in control and Arkadia KO Treg cells transduced with lentivirus expressing empty vector (Vec), WT Arkadia (Arkadia), and E3 ligase activity–dead mutant (Arkadia-C955S H957L). Data in A–E are representative of three independent experiments. Error bars represent SD; black circle, control T cells; red triangle, Arkadia-deficient T cells (C and E). Statistics were calculated using unpaired t test. ***, P < 0.001; ****, P < 0.0001.
Requirement for Arkadia in iTreg cell differentiation in vivo
We next analyzed the development of thymus-derived nTreg cells and the differentiation of peripheral iTreg cells in Arkadia conditional mutant mice and control littermates. Arkadiafl/fl Cd4Cre mice had normal numbers of thymocyte subsets but had a notable increase in proportion and number of Helios+ nTreg cells in lymphoid organs (Fig. S2, A and B).
Figure S2.
Loss of Arkadia in T cells results in elevated thymic Treg cells but no perturbations in the intestinal T cell compartment of SPF mice, related to Fig. 2. (A) Analysis of thymocyte subsets from control littermates and Arkadia conditional KO mice. Representative flow cytometry panels (left), and proportions and total numbers of double-positive thymocytes from multiple mice (right). (B) Analysis of nTreg cell proportions and numbers among CD4 single-positive T cells from different lymphoid organs of control littermates and Arkadia conditional KO mice. (C) Analysis of GATA3 and FOXP3 expression in CD4+ TCRβ+ T lymphocytes from LILP of control and Arkadia conditional KO mice under SPF conditions. (D) INF-γ and IL-17A production by CD4+ TCRβ+ T lymphocytes from LILP of control and Arkadia conditional KO mice in SPF conditions. Data for C and D are from one of three independent experiments, with n = 13 in the three experiments. Unpaired t test was used for statistical analysis. Black circle, control littermates; red triangle, Arkadia conditional KO mice (A–D). (E) Mice were the same as those in Fig. 2 C. CD4+ CD8− T lymphocytes from different tissues were stained for Helios and FOXP3 to analyze the development of nTreg cells. Relative ratios of frequency and cell number of nTreg cells (Helios+ FOXP3+) are shown, respectively. The relative ratios of cell number were normalized to the ratios of donor B cells in peripheral blood. C1 LN, cecal-colonic draining LN; control/WT, control cell frequency or cell number divided by that of WT cell (black circle); Ark KO/WT, Arkadia mutant cell frequency or cell number divided by that of WT cell (red triangle). Data were obtained from a single experiment with a total of 13 mice for two experimental groups. Ratios of cotransferred cells in each animal were calculated individually and combined for analysis with unpaired t test. All error bars represent SD (A–E). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
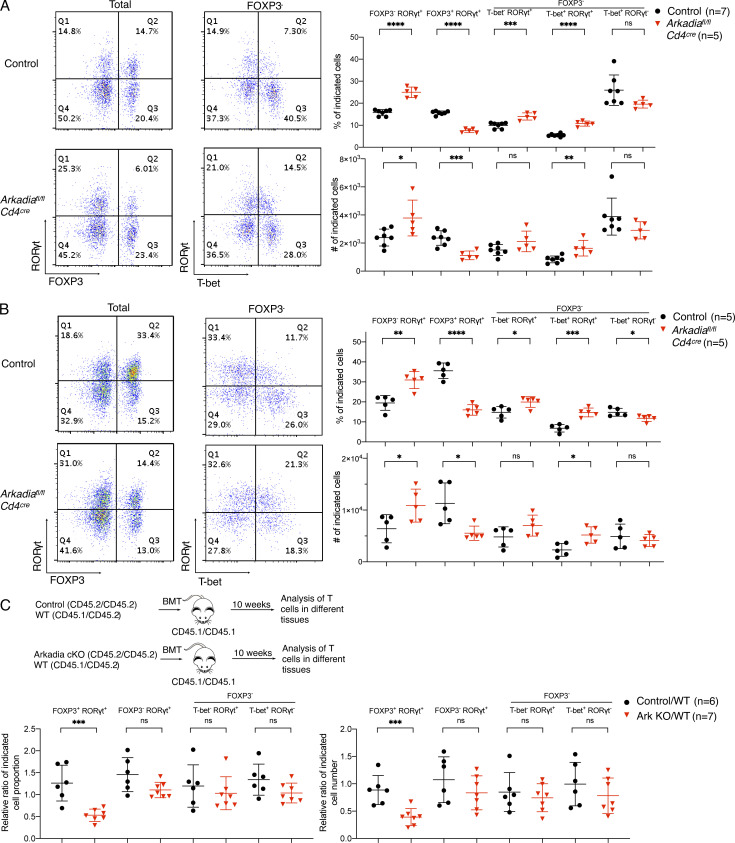
Consistent with in vitro results, RORγt+FOXP3+ iTreg cells were markedly reduced in the LILP of Arkadia conditional mutant mice. Notably, RORγt+FOXP3− Th17 cells were increased in both proportion and absolute cell number when Arkadia was absent in T cells (Fig. 2 A). There were no differences in GATA3+ cells among the FOXP3+ and FOXP3− populations or in IFNγ and IL-17A production among LILP CD4+ T cells from WT and Arkadiafl/fl Cd4Cre mice (Fig. S2, C and D). Together, these results suggest that Arkadia specifically regulates iTreg cell differentiation in vivo.
Figure 2.
Arkadia is required for iTreg, but not Th17, cell differentiation in vivo. (A) Analysis of CD4+ TCRβ+ T lymphocytes from LILP of control littermates and Arkadia conditional KO mice under SPF conditions. Left: Representative flow cytometry plots. Right: cell proportions (percentages) and numbers of indicated subpopulations. Data are from one of three independent experiments. n = 13 in the three experiments. (B) Analysis of CD4+ TCRβ+ T lymphocytes from LILP of control littermates and Arkadia mutant mice colonized with H. helicobacter. Data are representative of two independent experiments (n = 10). Statistical analyses in A and B were performed with unpaired t test. Black circle, control littermates; red triangle, Arkadia conditional KO mice (A and B). (C) Top: Experimental scheme for mixed bone marrow chimera experiment (see Materials and methods). Bottom: Relative ratios of iTreg, Th17, and Th1 lymphocytes in reconstituted mice (designated control/WT and Ark KO/WT). Relative ratios of cell number were normalized to the ratios of donor B cells in peripheral blood. Control/WT (black circle) and Ark KO/WT (red triangle): control or Arkadia mutant cell frequency or number divided by that of WT cells. Data are from one experiment with total 13 mice for two experimental groups. Ratios of cotransferred cells in each animal were calculated individually and combined for analysis with unpaired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Error bars represent SD.
Helicobacter hepaticus is a commensal bacterium that induces RORγt+ iTreg cells to maintain intestinal homeostasis in WT mice (Xu et al., 2018). Following colonization with H. hepaticus, the proportion and number of RORγt+FOXP3+ regulatory cells were reduced in Arkadiafl/fl Cd4Cre mice compared with control littermates, but, despite elevation in Th17 cells, spontaneous colitis was not detected (Fig. 2 B). To confirm that Arkadia-deficient T cells had a cell-intrinsic defect in iTreg cell differentiation, we performed mixed bone marrow chimera experiments, with CD45.2/CD45.2 control littermate or Arkadia mutant bone marrow mixed with that of CD45.1/CD45.2 mice (WT), followed by transfer into lethally irradiated CD45.1/CD45.1 recipient mice. 10 wk after transfer, there was reduction in proportion and number of iTreg cells from Arkadia-deficient bone marrow, confirming a cell-intrinsic Arkadia requirement for in vivo iTreg differentiation (Fig. 2 C). In contrast, there was no significant difference in Th17 and Th1 cells, further supporting a specific requirement for Arkadia in iTreg cell differentiation.
In the mixed bone marrow chimeras, thymic Arkadia-deficient nTreg cells were present in significantly higher proportions and numbers compared with control cells (Fig. S2 E). Disruption of thymic TGF-β signaling was reported to result in enhanced IL-2 expression in CD8− CD4+ CD25− thymocytes (Liu et al., 2008), but we detected no difference in Il2 from control and mutant thymocytes or in CD25 expression in control and Arkadia-deficient thymic nTreg or CD8− CD4+ FOXP3− cells from mixed bone marrow chimeras (data not shown). IL-2 signaling is thus unlikely to contribute to the nTreg increase in mutant mice. While the mechanism remains unclear, it is possible that loss of Arkadia promotes thymic nTreg proliferation, which has been suggested to be restrained by TGF-β signaling (Li et al., 2006).
Arkadia is required to maintain intestinal homeostasis
Mice lacking RORγt+ iTreg cells develop minor to pronounced microbiota-dependent intestinal inflammation. Furthermore, mice with partially compromised iTreg cells (e.g., with mutations in CNS1 of Foxp3, cMaf, or Tgfbr1) have no early pathology but develop intestinal inflammation with age or immunodeficiencies (Feng et al., 2014; Josefowicz et al., 2012b; Xu et al., 2018; Zheng et al., 2010). Therefore, we reasoned that mice with T cell–specific Arkadia deficiency may be more susceptible under conditions that sensitize for inflammation.
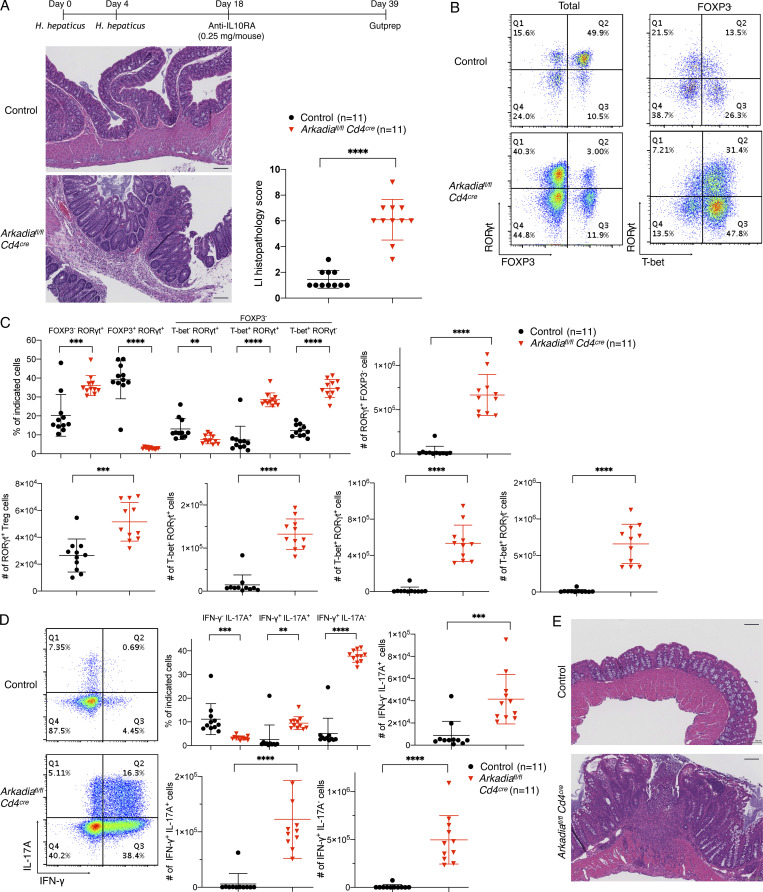
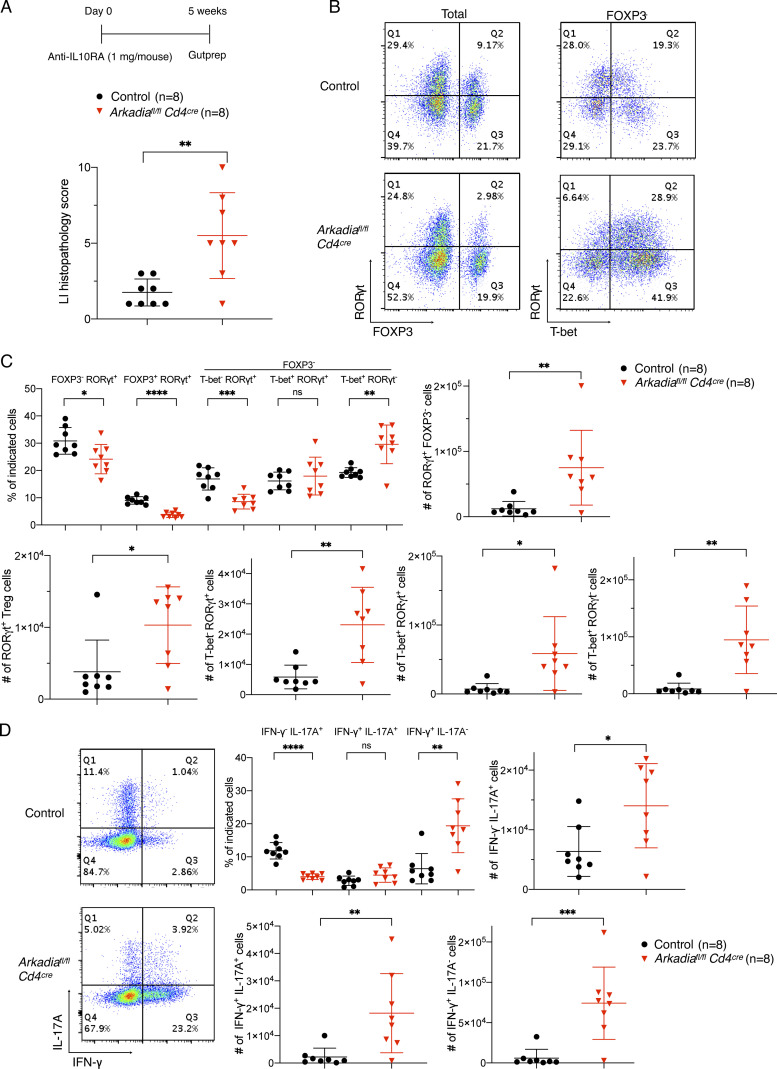
To test this hypothesis, we administered anti–IL-10RA antibody to mutant and control mice kept under specific pathogen–free (SPF) conditions with or without H. hepaticus colonization (Figs. 3 A and S3 A). In mice with H. hepaticus, injection of 0.25 mg antibody was well tolerated by control mice, but Arkadia mutant mice developed severe colitis (Fig. 3 A). Strikingly, among colonic T cells, there was much greater expansion of proinflammatory T cells, including Th1, Th17, and RORγt+ T-bet+ cells with Th1-like features, in Arkadiafl/fl Cd4Cre mice compared with control littermates. This was accompanied by marked reduction in proportion of RORγt+ FOXP3+ cells in the LILP in mutant mice (Fig. 3, B and C), even though there was some iTreg cell expansion under the inflammatory conditions (compare with Fig. 2 B). As expected, proinflammatory Th1 or Th1-like Th17 cells expressed high IFN-γ or both IL-17A and IFN-γ upon ex vivo restimulation (Fig. 3 D). Under SPF conditions, in the absence of H. hepaticus, there are fewer microbiota-induced iTreg cells and, upon blockade, fewer Th17/Th1 cells (Xu et al., 2018). Accordingly, there was little intestinal inflammation and pathology in WT mice even with a higher dose (1 mg) of anti–IL-10RA antibody. However, Arkadia mutant mice developed colitis under these conditions (Fig. 3 E). Differences in LILP T cells between control and mutant mice were similar to those observed with the H. hepaticus colonization conditions (Fig. S3, B–D). The overwhelming expansion of Th1 and Th1-like Th17 cells rather than iTreg cells in mice that developed inflammation in these colitis models further supports a specific requirement for Arkadia for iTreg cell differentiation.
Figure 3.
Arkadia is required in T cells to maintain mucosal homeostasis. (A) Top: Schematic of IL-10RA blockade in H. hepaticus–colonized control littermates and Arkadia mutant mice. Bottom: Representative large intestine sections (left) and colitis scores (right). Scale bars on the lower right in intestine sections represent 100 µm. (B) Representative RORγt, FOXP3, and T-bet staining in CD4+ TCRβ+ T lymphocytes isolated from LILP of indicated mice with IL-10RA blockade as shown in A. (C and D) Flow cytometry and statistical analysis of indicated T lymphocyte subpopulations with IL-10RA blockade shown in B. (C) Statistical analysis of indicated subpopulations. (D) Representative flow cytometry staining (left) and statistical analysis (right) of LILP lymphocytes expressing INF-γ and IL-17A. Data in A–D are from two independent experiments (n = 11) combined. (E) Representative H&E staining of large intestine sections from control littermates and Arkadia mutant mice with IL-10RA blockade maintained under SPF conditions. Data are from one experiment with eight mice for each group (n = 8). Scale bars represent 100 µm. Statistical analysis was performed with unpaired t test. (A, C, and D) Black circle, control littermates; red triangle, Arkadia mutant mice. Error bars represent SD. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Figure S3.
Arkadia-deficient mice are susceptible to immune system perturbation, related to Fig. 3. (A) Scheme of IL-10RA blockade under SPF conditions and analysis of colitis scores in colon of control and Arkadia conditional KO littermates. (B–D) Representative flow cytometry and indicated T cell subpopulations (proportions and numbers) following IL-10RA blockade as shown in A. (B) Representative RORγt, FOXP3, and T-bet expression in CD4+ TCRβ+ T lymphocytes isolated from LILP of indicated mice. (C) Proportions and numbers of indicated T lymphocyte subpopulations. (D) Representative flow cytometry analysis of INF-γ and IL-17A expression (left) and frequencies of cells in each biological replicate (right). Data in A–D represent a single experiment with eight mice for each group (n = 8). Unpaired t test was used for statistical analysis. (A, C, and D) Black circle, control littermates; red triangle, Arkadia conditional KO mice. Error bars represent SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Arkadia is dispensable for Th17 responses in vivo
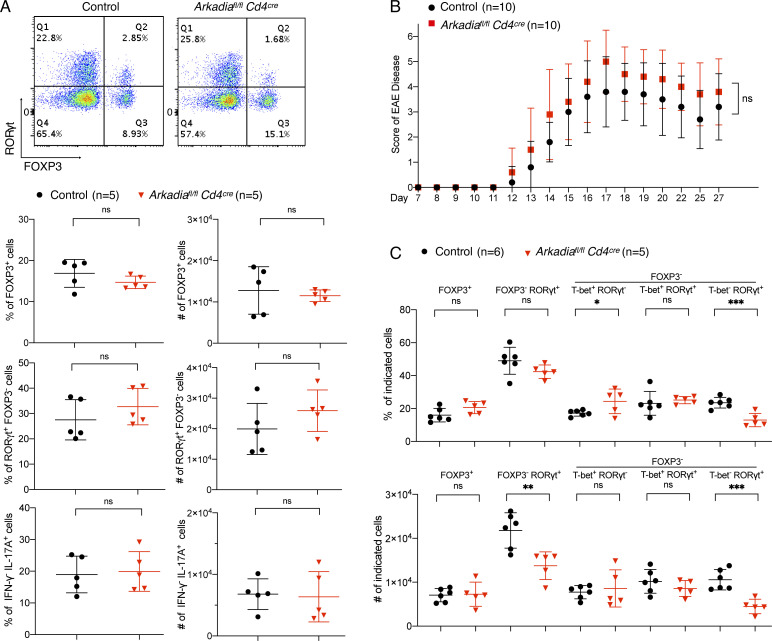
We further explored the role of Arkadia in Th17 responses induced by commensal bacteria and in EAE, a Th17 cell–mediated autoimmune disease. Th17 cell differentiation induced by segmented filamentous bacteria (SFB; Ivanov et al., 2009) was similar in mutant and control littermates, as were proportions and numbers of Treg and Th17 cells in small intestine (Fig. 4 A). Furthermore, there was no difference in Th17-mediated EAE severity between mice with Arkadia-deficient or sufficient T cells (Fig. 4 B). There was no significant difference in spinal cord Treg cell frequency and number, although there was a higher proportion of Th1 relative to Th17 cells in the mutant mice (Fig. 4 C), probably due to the reduced TGF-β signaling (Oh and Li, 2013). Taken together, these results are consistent with Arkadia being dispensable for in vivo Th17 cell differentiation.
Figure 4.
Arkadia does not influence SFB-dependent Th17 cell differentiation or Th17 cell–mediated EAE. (A) Analysis of RORγt, FOXP3, and effector cytokine expression in T lymphocytes from small intestine lamina propria of mice colonized with SFB. Unpaired t test analysis for one experiment with five control littermates and five Arkadia mutant mice. (B) EAE disease score in control littermates and Arkadia mutant mice. Data are from one experiment with 10 mice per group. EAE clinical scores were analyzed by two-way ANOVA. (C) Statistical analysis of indicated cell populations at peak of EAE disease. Data were collected from six control littermates and five Arkadia mutant mice (independent experiments from B). Differences between experimental groups were determined by unpaired t test. Error bars represent SD. Black circle, control littermates; red triangle, Arkadia mutant mice. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Arkadia degrades SKI/SnoN proteins to promote Treg cell differentiation
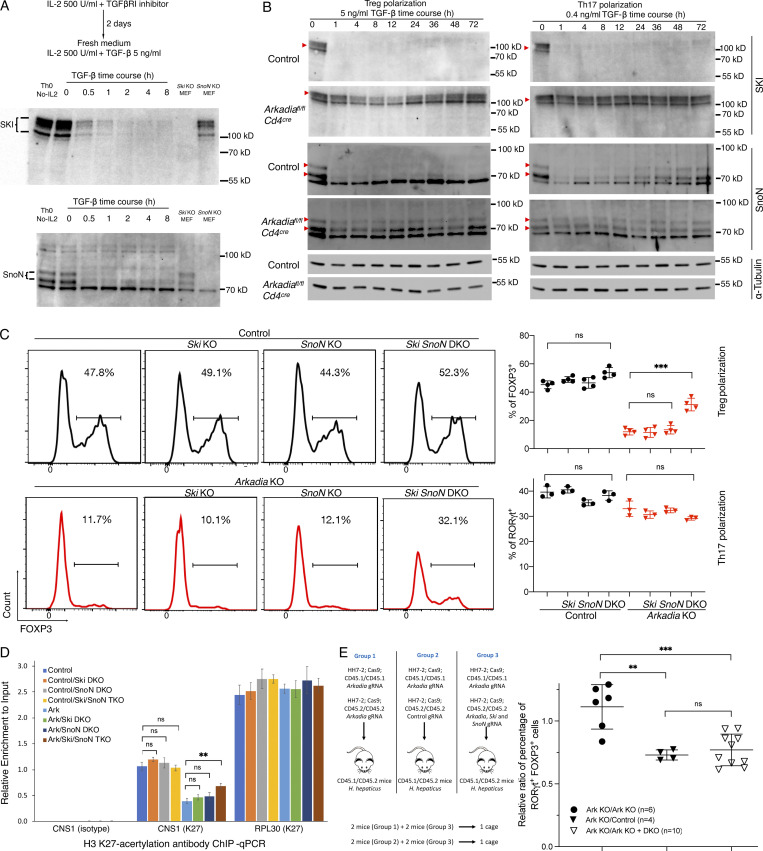
We next explored the mechanism by which Arkadia participates in iTreg cell differentiation and focused on Arkadia substrates. Only Ski and SnoN, among eight Ski family members, are expressed in Th17 and Treg cells (Table S1). We therefore assessed TGF-β–induced SKI and SnoN levels in Arkadia-deficient or sufficient differentiating Th17 and Treg cells and found rapid degradation of both in control, but not Arkadia-deficient, T cells (Fig. 5, A and B). Because SKI and SnoN are transcriptional repressors, we hypothesized that their inactivation may rescue Treg cell differentiation in Arkadia-null T cells. We therefore expressed Arkadia-targeting gRNA with gRNAs targeting Ski, SnoN, or both in naive T cells from Cas9-expressing mice, followed by Treg and Th17 differentiation. Targeting of both Ski and SnoN, but not either gene alone, rescued Treg differentiation in the absence of Arkadia, suggesting that SKI and SnoN have redundant roles in TGF-β–induced FOXP3 expression (Fig. 5 C). Consistent with their function of recruiting histone deacetylases (Nomura et al., 1999), loss of SKI and SnoN resulted in increased acetylation of histone H3K27 in the TGF-β–responsive CNS1 of Foxp3 (Fig. 5 D), indicative of transcriptional activation. By contrast, lack of SKI and SnoN had no influence on Th17 cell differentiation (Fig. 5 C).
Figure 5.
Arkadia degrades SKI and SnoN to regulate iTreg cell differentiation. (A) Validation of SKI and SnoN antibodies. T cells were activated by plate-bound anti-CD3ε and anti-CD28 and cytokine as indicated. Lysates from SKI- or SnoN-deficient MEFs were used as control. Multiple bands of SKI on immunoblot are due to phosphorylation (Marcelain and Hayman, 2005), and there are two isoforms of SnoN (76.4 kD and 71.1 kD). (B) Immunoblot of time course of degradation of SKI and SnoN in control and Arkadia mutant T cells differentiated under the indicated conditions. Red arrowheads indicate SKI and SnoN proteins detected by corresponding antibodies. α-Tubulin (∼50 kD) served as loading control. Data in A and B are representative of two independent experiments. (C) DKO of Ski and SnoN rescues Arkadia-deficient Treg cell differentiation in vitro. T cells electroporated with indicated gRNAs were cultured under Treg or Th17 differentiation conditions (Materials and methods). Left: Representative flow cytometry of Foxp3 expression in Treg conditions. Right: Technical replicates of one of three independent experiments with Treg and Th17 differentiation of CD4+ T cells electroporated with control (circles) or Arkadia-targeting (triangles) gRNAs plus control, Ski, SnoN, and Ski/SnoN (DKO) gRNAs. Statistical analyses were performed with unpaired t test. ***, P < 0.001. Error bars represent SD. (D) Arkadia regulates FOXP3 expression through histone modification at the Foxp3 locus. Differentiated Treg cells electroporated with indicated gRNAs were harvested for ChIP analysis using anti-acetyl-H3K27 or isotype control antibodies. Control, Ski, SnoN, and Ark represent KO of genes by sgRNAs. TKO, triple KO. CNS1, Foxp3 CNS1 region; RPL30, Rpl30 intron 2 region. Representatives of three independent experiments. Statistical analyses were performed with an unpaired t test. Error bars represent SD. **, P < 0.01. (E) DKO of Ski and SnoN rescues iTreg cell differentiation in Arkadia-deficient T cells in vivo. The experiment was performed as shown in the schematic and is described in Materials and methods. Relative ratios of iTreg frequency were calculated by dividing iTreg frequency of CD45.1/CD45.1 cells by that of CD45.2/CD45.2 cells in the recipient mice. To compare experimental groups, the relative ratio in each animal was calculated individually and combined for analysis with unpaired t test. Error bars represent SD. **, P < 0.01; ***, P < 0.001. Data shown are from one experiment with a total of 20 mice.
To investigate Ski and SnoN function in iTreg cells in vivo, we delivered Arkadia or Ski-SnoN-Arkadia triple-targeting gRNAs into isotype-marked naive T cells expressing Cas9 and the H. hepaticus–specific HH7-2 TCR (Fig. 5 E; Xu et al., 2018). The isotype-distinct cells were mixed at a 1:1 ratio and transferred into mice colonized with H. hepaticus, followed by analysis of differentiated donor-derived cells in the LILP. Consistent with the in vitro differentiation results, deficiency of both SKI and SnoN rescued antigen-specific iTreg cell differentiation in Arkadia-deficient T cells. Taken together, our data suggest that TGF-β–activated Arkadia degrades SKI and SnoN, allowing for unopposed acetylation of histone H3 in the Foxp3 locus and subsequent transcriptional activation.
The mammalian TGF-β superfamily is composed of more than 30 members, including TGF-βs, activins, NODAL, inhibins, and bone morphogenetic proteins, which play pleiotropic and indispensable roles in diverse biological processes (Massagué, 2012). In T lymphocyte differentiation, engagement of different signaling pathways explains how TGF-β contributes to distinct functional outcomes, but these pathways remain incompletely understood. Components of the canonical TGF-β pathway have essential roles in iTreg cell differentiation. These include TGF-β ligands, TGF-β receptors, and SMAD4, but SMAD2 and SMAD3 are functionally redundant in vivo. We found Arkadia indispensable for Treg cell differentiation in vitro, yet its loss in T cells in vivo resulted in a relatively mild phenotype compared with loss of TGF-β receptors, with no spontaneous inflammatory disease. The mutant mice were, however, prone to inflammation, which correlated with fewer iTreg cells. The incomplete ablation of iTreg cells could be due to SKI and SnoN degradation by other E3 ligases, such as anaphase-promoting complex (Stroschein et al., 2001; Wan et al., 2001) and Smurf2 (Bonni et al., 2001). Alternatively, SKI and SnoN repression of Foxp3 could be compensated by up-regulated transcriptional activators.
Although TGF-β and its receptors are vital for differentiation of both Th17 and iTreg cells, there was no defect in Th17 differentiation despite lack of SKI and SnoN degradation in Arkadia-deficient Th17 cells, which argues against SKI- and SnoN-mediated negative regulation of TGF-β signaling through binding to SMAD2/3 and SMAD4. One possibility may be attributed to unidentified SMADs-mediated suppressive signals that are not regulated by Arkadia.
Alternatively, this may reflect distinct TGF-β–induced SMAD-containing complexes (with or without SKI/SnoN proteins) having either transcriptional repression or activation activity, with active complexes bypassing repressive activity in Arkadia-deficient cells. Transcription factors SMAD2/3 and SMAD4 may be redundant activators in this scenario, since Th17 differentiation was intact in SMAD4 KO or SMAD2/3 DKO cells induced by TGF-β plus IL-6. In contrast, SMAD complexes would be predominantly repressive or lack transcriptional activity without TGF-β stimuli, explaining elevated RORγt expression with IL-6 alone in SMAD4-deficient T cells compared with WT cells. Further studies are needed to fully elucidate the mechanism of TGF-β–mediated Th17 cell differentiation.
To date, little is known about the regulation of Arkadia and its roles in different cell types that respond to activin, NODAL, and TGF-β stimuli. Furthermore, given that the upstream signals are conserved in many cell types, further dissection of the downstream TGF-β signaling components and their relative roles in different biological processes will be of utmost importance for future studies. Modulation of Arkadia may provide a platform to gain insight into multiple physiological processes mediated by TGF-β superfamily members.
Materials and methods
Mice
Mice were bred and maintained in the animal facility of the Skirball Institute (New York University School of Medicine) under SPF conditions. All animal procedures were performed in accordance with guidelines by the Institutional Animal Care and Usage Committee of New York University School of Medicine or National Institute of Allergy and Infectious Diseases. C57BL/6J, Cd4Cre (Tg(Cd4-cre)1Cwi/BfluJ), Cd45.1 (B6.SJL- Ptprca Pepcb/BoyJ), Rosa26-Cas9 knockin (Gt(ROSA)26Sortm1.1(CAG-cas9*,-EGFP)Fezh/J), Smad4fl/fl (Smad4tm2.1Cxd/J), and Tgfbr2 fl/fl (Tgfbr2tm1Karl/J) mice were obtained from The Jackson Laboratory. C57BL/6J mice were bred with Cd45.1 mice to produce Cd45.1/Cd45.2 WT control mice for bone marrow transfer experiments. Smad4fl/fl and Tgfbr2 fl/fl mice were bred with Cd4Cre mice to produce Smad4 and Tgfbr2 conditional DKO mice. HH7-2 TCR transgenic mice were previously described (Xu et al., 2018) and bred with Cd45.1 and Rosa26-Cas9 knockin mice for T cell transfer experiments. Arkadiafl/fl mice were generated by injection of Cas9 mRNA, single gRNA (sgRNA), and single-stranded oligo into C57BL/6J zygotes, resulting in the insertion of two LoxP sites flanking exon 13. Cas9 mRNA (L-7206) and single-stranded oligo (ultramer) were obtained from TriLink Biotechnologies and Integrated DNA Technologies, respectively. sgRNA was made in-house by adding T7 promoter (5′-TAATACGACTCACTATAGGG-3′) to the DNA fragment coding sgRNA, followed by in vitro transcription (Megashortscript; Invitrogen) and purification (Megaclear; Invitrogen). Arkadiafl/fl mice were backcrossed to C57BL/6J background for two generations and further bred with Cd4Cre mice for conditional KO of Arkadia in T cells. All experiments were performed with genetically modified mice and control littermates with matched sex and age. If not specified, mice used in all experiments were 6–12 wk old when treated with different stimuli.
Cell lines, antibodies for flow cytometry, and Western blotting
The following monoclonal antibodies (clone ID) were used for flow cytometry: CD4 (RM4-5), CD8 (53–6.7), CD25 (PC61.5), CD44 (IM7), CD45.1 (A20), CD45.2 (104), CD62L (MEL-14), TCRβ (H57-597), TCR Vβ6 (RR4-7), FOXP3 (FJK-16s), GATA3 (TWAJ), Helios (22F6), RORγt (B2D), T-bet (eBio4B10), IL-17A (eBio17B7), and IFN-γ (XMG1.2). DAPI or Live/Dead fixable blue (ThermoFisher) was used to discriminate live/dead cells. Anti-acetyl-H3K27 (#8173; Cell Signaling Technology) and IgG isotype control (#3900; Cell Signaling Technology) antibodies were employed in a chromatin immunoprecipitation (ChIP) assay. The following antibodies were used for Western blotting: SKI (sc-33693; Santa Cruz Biotechnology), SnoN (sc-9141; Santa Cruz Biotechnology), and α-tubulin (sc-69970; Santa Cruz Biotechnology). Protein concentration of cell lysate was determined by Bradford and Lowry methods (Bio-Rad), and 70 µg protein was loaded to 8% separating gel for Western blotting. SnoN−/− and Ski−/− MEFs are described elsewhere (Carthy et al., 2019 Preprint). MEFs were cultured in DMEM containing 10% FBS, 20 mM L-glutamine, and 1× penicillin/streptomycin.
In vitro T cell differentiation
Naive CD4+ T cells were defined as CD4+ CD8− CD25− CD44− CD62L+ cells and isolated on an Aria II (BD Biosciences) cell sorter. For polarization of T cells in vitro, goat-anti-hamster IgG (MP Biomedicals) was diluted (1:20) in PBS to coat 96-well plates overnight. T cells were activated by adding hamster anti-CD3ε (0.25 µg/ml) and anti-CD28 (1 µg/ml) monoclonal antibodies. Polarizations were performed in 300 µl RPMI medium supplemented with exogenous cytokines for different cell lineages: Th0 cells, 100 U/ml hIL-2 (Peprotech) and 10 µM TGF-β R1 inhibitor SB525334 (Selleckchem); Th1 cells, 100 U/ml hIL-2, 10 ng/ml IL-12 (Peprotech), and 2 µg/ml anti–IL-4 antibody (clone 11B11; BioXcell); Th17 cells, 0.2 ng/ml TGF-β (Peprotech), 20 ng/ml IL-6 (eBioscience), and 2 µg/ml anti–IL-4 and anti-IFNγ antibody (clone XMG 1.2; BioXcell), and 10 µM SB525334 was used to replace TGF-β to inhibit TGF-β signaling if required; and iTreg cells, 500 U/ml hIL-2 and 5 ng/ml TGF-β. For Th0 and Th1 cell polarization, 2 × 104 cells were plated in each well and transferred to uncoated 96-well plates after 2 d, followed by another 1 or 2 d of polarization. For Th17 and iTreg cell polarization, 104 cells were plated in each well and polarized for 3 d. For restimulation and cytokine analysis, cells were incubated at 37°C for 3 h in fresh medium with 50 ng/ml PMA (Sigma), 500 ng/ml ionomycin (Sigma), and 1 × GolgiStop (BD Biosciences).
For transcription factor staining, cells were stained for surface markers before nuclear factor staining following the manufacturer’s protocol (00–5523; eBioscience). For intracellular cytokine staining, Cytofix/Cytoperm buffer set (BD Biosciences) was used. Analysis of stained cells was performed on a LSR II (BD Biosciences) flow cytometer, and data were analyzed on FlowJo platform (BD Biosciences). FlowJo implements quadrant statistics through creating four nonoverlapping rectangular gates, named as "Q+number,” Q1 to Q4 for example. The same principle applies to other flow cytometry analyses when FlowJo is used.
Retrovirus packaging and infection of T cells
The retroviral plasmid pSIN-U6-sgRNA-EF1a-Thy1.1-P2A-Neo or MSCV-IRES-Thy1.1 was used for viral packaging and expression of gRNA or Arkadia, respectively. Plat-E cells (Cell Biolabs) and jetPRIME transfection reagent (Polyplus) were used to produce retrovirus, which was harvested 48 h after transfection. Before viral transduction in 96-well plates, 104 naive T cells in 100 µl medium were activated by anti-CD3ε and anti-CD28 cross-linking for 24 h. 100 µl retrovirus-containing medium was added to each well for viral transduction in the presence of 5 µg/ml polybrene. Cells were centrifuged at 2,200 rpm for 90 min at room temperature, followed by culture for 12 h in a cell incubator. Cells were then polarized under required conditions for 3 d. pSIN-U6-sgRNA-EF1a-Thy1.1-P2A-Neo was described in our previous study (Ng et al., 2019), and MSCV-IRES-Thy1.1 plasmid was a gift from Anjana Rao, La Jolla Institute, La Jolla, CA (catalog 17442; Addgene; Wu et al., 2006). The following gRNAs were used in this paper: Arkadia-gRNA1-forward: 5′-TGGTGGAAAAGGTGCATACA-3′; Arkadia-gRNA2-forward, 5′-GCATCTTTCAATTGTTCCCT-3′; Arkadia-gRNA3-forward, 5′-CATAGGATGCACCCAAACTA-3′; Arkadia-gRNA4-forward, 5′-ATCACGTAATCCACTTGAGG-3′; Arkadia-gRNA5-forward, 5′-GTGGGTGTGCTAATGCATGA-3′; Arkadia-gRNA6-forward, 5′-GGAAAAGGTGCATACATGGA-3′; Smad4-gRNA1-forward, 5′-AACTCTGTACAAAGACCGCG-3′; Smad4-gRNA2-forward, 5′-CAAAAGCGATCTCCTCCCGA-3′; Ski-gRNA1-forward, 5′-AGTCGCGCAGCACCGAGTTG-3′; Ski-gRNA2-forward, 5′-GGTACATGAGGCGGCAGTCG-3′; SnoN-gRNA1-forward, 5′-GGTCAACGTGATCAGCCCAC-3′; SnoN-gRNA2-forward, 5′-GAGATGGTGAAGACGACAAG-3′; Stat5-gRNA-forward, 5′-ACGGCTGGCTCTCGATCCAC-3′; Rorc-gRNA1-forward, 5′-GTTGCGGCTGGTTCGGTCAA-3′; Rorc-gRNA2-forward, 5′-GGTTGGTGCGCTGCCGTAGA-3′; Olfr2-gRNA-forward (control gRNA), 5′-AAATAGTAGTGCCCGCAGTG-3′.
Isolation of lymphocytes from small intestine lamina propria or LILP
Intestines were collected and carefully inspected to remove Peyer’s patches in small intestines and caecal patches in large intestines. After cleaning, intestines were sequentially treated with HBSS containing 1 mM DTT once at room temperature for 15 min, and HBSS containing 5 mM EDTA twice at 37°C for 30 min totally to remove epithelial cells. Tissues were minced in RPMI complete medium containing collagenase (1 mg/ml; Roche), DNase I (100 µg/ml; Sigma), dispase (0.05 U/ml; Worthington), followed by shaking for 50 min at 37°C. Leukocytes were isolated by 40%/80% Percoll gradient centrifugation (GE Healthcare). For restimulation ex vivo, isolated leukocytes were suspended in fresh medium containing 50 ng/ml PMA, 500 ng/ml ionomycin, and 1 × GolgiStop and incubated at 37°C for 3 h before staining. Thymus, LN, and spleen were mechanically disrupted directly on a 70-µm filter, followed by removal of blood cells with ammonium-chloride-potassium (ACK) lysis buffer.
H. hepaticus culture and oral infection with SFB and H. hepaticus
H. hepaticus was inoculated on blood agar plates (tryptic soy agar with 5% sheep blood; ThermoFisher) and cultured in a hypoxia chamber (Billups-Rothenberg) at 37°C for 4 to 5 d. The anaerobic atmosphere was achieved by aeration of gas mixture composed of 80% nitrogen, 10% hydrogen, and 10% carbon dioxide (Airgas). Before animal inoculation, H. hepaticus were suspended in Remel Brucella Broth (ThermoFisher). The concentration of bacteria was adjusted to an optical density of 1–1.5 measured at a wavelength of 600 nm. Each mouse was administered 0.2 ml/dose of bacterial suspension on days 0 and 4 by oral gavage. For SFB infection, one fecal pellet from Taconic mice was suspended in 200 µl PBS and passed through a 100-µm filter. The filtrate was given to one mouse by oral gavage. Each mouse received two doses totally with an interval of 3 d. T lymphocytes of intestinal lamina propria were recovered and analyzed 3–4 wk after the last gavage.
Bone marrow transfer
Recipient mice (CD45.1/CD45.1) were irradiated with 500 cGy + 500 cGy with an interval of 5 h 1 d before bone marrow transfer (day 0). On day 1, bone marrow was collected from congenic mice (WT CD45.1/CD45.2), control littermates, or Arkadia conditional KO mice (Arkadia cKO, CD45.2/CD45.2). Erythrocytes in bone marrow were removed by ACK lysis buffer, and T cells were depleted using anti-CD90.2 magnetic beads (Miltenyi), followed by determination of cell number. Equal numbers of bone marrow cells from control or Arkadia conditional KO mice (CD45.2/CD45.2) were mixed with cells from congenic mice (CD45.1/CD45.2), respectively, followed by transfer of 3 × 106 mixed cells into each lethally irradiated CD45.1/CD45.1 recipient mouse through retroorbital injection. Antibiotics (1 mg/ml sulfamethoxazole and 0.2 mg/ ml trimethoprim) were added to the drinking water and maintained for 2 wk. CD4+ TCRβ+ T lymphocytes from LILP were stained for T-bet, RORγt, and FOXP3 10 wk after transfer. Thymus, spleen, cecal-colonic draining LN (C1 LN), and inguinal LN were harvested for immune cells, followed by staining of nTreg cells (CD4+ CD8− Helios+ FOXP3+).
IL-10RA blockade and histology scoring on colon sections
Under SPF conditions, Arkadia conditional KO mice and control littermates were cohoused after birth and intraperitoneally administered one dose of 1 mg/ml anti–IL-10RA antibody at 10–12 wk old. Approximately 5 wk later, mice were sacrificed to isolate lamina propria lymphocytes or collect colonic samples for histology analysis. With H. hepaticus–colonized mice, one dose of 0.25 mg/ml anti–IL-10RA antibody was administered intraperitoneally. Mice were sacrificed to isolate LILP lymphocytes or collect colons for histology analysis 3 wk after the IL-10RA blockade. Colonic samples were sequentially fixed in 4% paraformaldehyde (Electron Microscopy Science), embedded in paraffin, and sectioned. Colon sections were stained with hematoxylin and eosin and scored for histopathology in a blinded fashion. Total score of colonic inflammation comprised individual scores from four categories: (1) number of goblet cells per high-power field (HPF; score of 1, 11–28 goblet cells per HPF; score of 2, 1–10 goblet cells per HPF; score of 3, <1 goblet cell per HPF); (2) submucosa edema (score of 0, no edema; score of 1, mild edema accounting for <50% of the diameter of the entire intestinal wall; score of 2, moderate edema involving 50–80% of the diameter of the entire intestinal wall; score of 3, profound edema involving >80% of the diameter of the entire intestinal wall); (3) depth of lymphocyte infiltration (score of 0, no infiltrate; score of 1, infiltration above muscularis mucosae; score of 2, infiltration extending to and including submucosa; score of 3, transmural Infiltration); and (4) epithelial changes (score of 0, no changes; score of 1, upper third or only surface epithelial missing; score of 2, moderate epithelial damage with intact base of crypts; score of 3, loss of crypts).
EAE
On day 0, the following reagents were prepared for injection: pertussis toxin (200 ng/mouse in 200 µl PBS), Mycobacteria tuberculosis H37RA suspended in CFA with a final concentration of 2 mg/ml (100 µl for each mouse) and myelin oligodendrocyte glycoprotein peptide (MOG 35-55 peptide, 200 µg/mouse in 100 µl PBS). An equal volume of M. tuberculosis H37RA-CFA was mixed with MOG/PBS in a 5 ml protein Lobind tube (Eppendorf), followed by emulsification in ice water through sonication. The mixture was centrifuged at 3,000 rpm for 2 min to confirm emulsion (no phase separation after centrifugation). 200 µl mixture was injected into each mouse subcutaneously with 27 G1/2 needle (BD Biosciences). On day 2, mice were administered a second dose of pertussis toxin (200 ng/mouse in 200 µl PBS). The disease score was measured daily 1 wk after the last pertussis toxin injection. A scoring system from 0 to 10 was applied to measure the severity of disease (0, no clinical signs; 1, limp tail; 2, paralyzed tail; 3, hindlimb paresis; 4, paralysis of one hindlimb; 5, paralysis of both hindlimb; 6, forelimb weakness; 7, paralysis of one forelimb; 8, paralysis of both forelimbs; 9, moribund; 10, death). The clinical scores were further analyzed by two-way ANOVA to determine difference between two mouse lines. For the analysis of T cell signatures at EAE disease peak, mice were sacrificed at day 16 following EAE induction. Total cells from spinal cords were isolated and stained for CD4, TCRβ, Helios, FOXP3, RORγt, and T-bet for analysis of T lymphocyte subpopulations.
KO of genes by electroporation
All gRNAs were expressed by transduction of retrovirus packaged with pSIN-U6-sgRNA-EF1a-Thy1.1-P2A-Neo plasmids. The gene-editing efficiencies of gRNAs were first confirmed by Western blotting or staining of targeted proteins in polarized T cells. For KO of genes by electroporation (4D-Nucleofector; Lonza), PCR products were amplified with primers (forward 5′-GAGGGCCTATTTCCCATGATTCC-3′ and reverse 5′-AAAAAAGCACCGACTCGGTGC-3′) flanking the hU6 promoter, sgRNA, and sgRNA scaffold in pSIN-U6-sgRNA-EF1a-Thy1.1-P2A-Neo plasmids. For each gRNA-expressing DNA, 400 µl PCR product was purified by Promega Wizard Gel/PCR purification columns, followed by NaAc-ethanol purification. Briefly, DNA products eluted from columns were mixed with 1/10 volume of 3 M NaAc pH5.2 and 2.5-fold volume of 100% ethanol. The mixture was placed at −20°C overnight and centrifuged for 30 min to precipitate DNA. The supernatant was removed and DNA was washed twice with 70% ethanol (precold at −20°C). 30 µl water was added to dissolve air-dried DNA. DNA concentration was determined by NanoDrop (the concentration was typically ∼0.5 µg/µl). Naive T cells expressing Cas9 were washed once with 1 ml prewarmed recovery medium (Mouse T Cell Nucleofector Medium; Lonza) before electroporation. 2 × 106 cells were suspended in 100 µl buffer (P3 Primary Cell 4D-Nucleofector X Kit; Lonza), mixed with 4 µg DNA, and transfected using the 4D-Nucleofector program DN-100. After electroporation, cells were suspended in 500 µl recovery medium and incubated in the incubator for 30 min. 2 × 104 cells were then seeded into each well of 96-well plates and incubated with plate-bound anti-CD3ε and anti-CD28 cross-linking for 24 h to ensure gene editing, followed by T cell differentiation for 3–4 d.
Rescue of Arkadia deficiency in vitro and ChIP
As described above, vectors encoding gRNAs targeting control, Arkadia, Ski, or SnoN were delivered into naive T cells expressing Cas9 through electroporation, followed by 24-h activation and polarization for 3 d in vitro. 4 µg DNA was used for each electroporation, in which 1.33 µg DNA was used for each gene when targeting three genes. Control gRNA was used to achieve a total of 4 µg DNA, when necessary, for example 2.67 µg control DNA plus 1.33 µg Arkadia-targeting DNA for Arkadia single KO. For ChIP analysis, anti-acetyl-H3K27 and isotype control antibodies were used to precipitate targeted DNA fragments (#9005; Cell Signaling Technology). Enrichment of genomic DNA fragments by ChIP was validated by real-time PCR (LightCycler 480; Roche) with primers targeting the Foxp3 CNS1 region. Primers (#7015; Cell Signaling Technology) targeting ribosomal protein L30 functioned as the control for the ChIP assay. The following primers were used for real-time PCR of Foxp3 CNS1 region: forward 5′-GTTTTGTGTTTTAAGTCTTTTGCACTTG-3′ and reverse 5′-CAGTAAATGGAAAAAATGAAGCCATA-3′.
Rescue of Arkadia deficiency in vivo
The protocol of electroporation to deliver DNA was the same as that for the ChIP assay. Briefly, vectors encoding Arkadia-targeting gRNA were delivered into naive T cells expressing Cas9, CD45.1/CD45.1, and HH7-2 TCR. Meanwhile, vectors encoding control, Arkadia, or Ski-SnoN-Arkadia triple-targeting gRNAs were delivered into naive T cells expressing Cas9, CD45.2/CD45.2, and HH7-2 TCR. Equal numbers of CD45.1/CD45.1 and CD45.2/CD45.2 naive cells (50,000 cells each) were mixed and designated as groups 1, 2, and 3 (Fig. 5 E). To exclude microbiome variation among cages, group 3 was used as internal control to groups 1 and 2. The mixed cells were resuspended in 100 µl medium containing 2% FBS and transferred by retroorbital injection into CD45.1/CD45.2 recipient mice colonized with H. helicobacter. Expression of RORγt and FOXP3 was examined in CD4+ TCRβ+ T lymphocytes from LILP 2 wk after cell transfer.
Statistical analysis
We used two-tailed P values calculated by unpaired t test (Welch’s t test) to determine the differences between experimental groups. In experiments with cotransferred T cells or bone marrow, the ratio in each mouse between the cotransferred cells was calculated individually and combined together for unpaired t test. For EAE disease comparison, the clinical scores were analyzed by two-way ANOVA. A P value <0.05 was reported as a significant difference (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). Details regarding the number of replicates and representative data can be found in the figure legends.
Online supplemental material
Fig. S1 shows Th17 and Treg cell differentiation with the deficiency of different components involved in TGF-β signaling. Fig. S2 shows the development of thymocytes and nTreg cells and expression of transcription factors and cytokines in T lymphocytes from Arkadia-deficient and sufficient mice under SPF conditions. Fig. S3 shows the comparison of colonic pathology and staining panels of T lymphocytes from LILP of Arkadia mutant mice and littermates. Table S1 lists the mRNA abundance of genes related to this research in various in vitro polarized T cells detected by bulk RNA sequencing.
Supplementary Material
lists gene expression detected by RNA sequencing in cells polarized in vitro (fragments per kilobase of exon per million mapped fragments).
Acknowledgments
We thank S.Y. Kim and the New York University Rodent Genetic Engineering Laboratory and the Experimental Pathology Research Laboratory for generating Arkadia mutant mice for histology processing and imaging. Both cores were partially supported by Cancer Center Support Grant P30CA016087.
This work was supported by the Irvington Institute fellowship program of the Cancer Research Institute (H. Xu); the Helen and Martin Kimmel Center for Biology and Medicine (D.R. Littman); the Howard Hughes Medical Institute (D.R. Littman); and National Institutes of Health grant 2R01AI080885 (D.R. Littman).
Author contributions: H. Xu and D.R. Littman designed experiments. H. Xu performed experiments and analyzed the data. L. Wu developed gRNA electroporation into T cells. H.H. Nguyen performed histology scoring. V. Raghavan contributed to EAE and colitis models. V. Episkopou provided advice and Arkadia reagents. H. Xu, K.R. Mesa, and D.R. Littman wrote the manuscript with input from co-authors. D.R. Littman supervised the research.
References
- Akiyoshi, S., Inoue H., Hanai J., Kusanagi K., Nemoto N., Miyazono K., and Kawabata M.. 1999. c-Ski acts as a transcriptional co-repressor in transforming growth factor-beta signaling through interaction with smads. J. Biol. Chem. 274:35269–35277. 10.1074/jbc.274.49.35269 [DOI] [PubMed] [Google Scholar]
- Bettelli, E., Carrier Y., Gao W., Korn T., Strom T.B., Oukka M., Weiner H.L., and Kuchroo V.K.. 2006. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 441:235–238. 10.1038/nature04753 [DOI] [PubMed] [Google Scholar]
- Bonni, S., Wang H.R., Causing C.G., Kavsak P., Stroschein S.L., Luo K., and Wrana J.L.. 2001. TGF-beta induces assembly of a Smad2-Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat. Cell Biol. 3:587–595. 10.1038/35078562 [DOI] [PubMed] [Google Scholar]
- Burchill, M.A., Yang J., Vogtenhuber C., Blazar B.R., and Farrar M.A.. 2007. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J. Immunol. 178:280–290. 10.4049/jimmunol.178.1.280 [DOI] [PubMed] [Google Scholar]
- Carthy, J.M., Ioannou M., and Episkopou V.. 2019. Arkadia via SNON enables NODAL-SMAD2/3 signaling effectors to transcribe different genes depending on their levels. bioRxiv. (Preprint posted November 7, 2019) 10.1101/487371 [DOI]
- Chen, W., Jin W., Hardegen N., Lei K.J., Li L., Marinos N., McGrady G., and Wahl S.M.. 2003. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 198:1875–1886. 10.1084/jem.20030152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani, M., Madar A., Galan C., Sellars M., Mace K., Pauli F., Agarwal A., Huang W., Parkhurst C.N., Muratet M., et al. 2012. A validated regulatory network for Th17 cell specification. Cell. 151:289–303. 10.1016/j.cell.2012.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, J., Ren G., Zhang L., Roberts A.I., Zhao X., Bothwell A.L., Van Kaer L., Shi Y., and Das G.. 2009. Transforming growth factor beta is dispensable for the molecular orchestration of Th17 cell differentiation. J. Exp. Med. 206:2407–2416. 10.1084/jem.20082286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Episkopou, V., Arkell R., Timmons P.M., Walsh J.J., Andrew R.L., and Swan D.. 2001. Induction of the mammalian node requires Arkadia function in the extraembryonic lineages. Nature. 410:825–830. 10.1038/35071095 [DOI] [PubMed] [Google Scholar]
- Feng, Y., Arvey A., Chinen T., van der Veeken J., Gasteiger G., and Rudensky A.Y.. 2014. Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell. 158:749–763. 10.1016/j.cell.2014.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoreschi, K., Laurence A., Yang X.P., Tato C.M., McGeachy M.J., Konkel J.E., Ramos H.L., Wei L., Davidson T.S., Bouladoux N., et al. 2010. Generation of pathogenic T(H)17 cells in the absence of TGF-β signalling. Nature. 467:967–971. 10.1038/nature09447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, T.J., Grosso J.F., Yen H.R., Xin H., Kortylewski M., Albesiano E., Hipkiss E.L., Getnet D., Goldberg M.V., Maris C.H., et al. 2007. Cutting edge: An in vivo requirement for STAT3 signaling in TH17 development and TH17-dependent autoimmunity. J. Immunol. 179:4313–4317. 10.4049/jimmunol.179.7.4313 [DOI] [PubMed] [Google Scholar]
- Ivanov, I.I., McKenzie B.S., Zhou L., Tadokoro C.E., Lepelley A., Lafaille J.J., Cua D.J., and Littman D.R.. 2006. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 126:1121–1133. 10.1016/j.cell.2006.07.035 [DOI] [PubMed] [Google Scholar]
- Ivanov, I.I., Atarashi K., Manel N., Brodie E.L., Shima T., Karaoz U., Wei D., Goldfarb K.C., Santee C.A., Lynch S.V., et al. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 139:485–498. 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz, S.Z., Lu L.F., and Rudensky A.Y.. 2012a. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 30:531–564. 10.1146/annurev.immunol.25.022106.141623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz, S.Z., Niec R.E., Kim H.Y., Treuting P., Chinen T., Zheng Y., Umetsu D.T., and Rudensky A.Y.. 2012b. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 482:395–399. 10.1038/nature10772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koinuma, D., Shinozaki M., Komuro A., Goto K., Saitoh M., Hanyu A., Ebina M., Nukiwa T., Miyazawa K., Imamura T., and Miyazono K.. 2003. Arkadia amplifies TGF-beta superfamily signalling through degradation of Smad7. EMBO J. 22:6458–6470. 10.1093/emboj/cdg632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn, T., Bettelli E., Oukka M., and Kuchroo V.K.. 2009. IL-17 and Th17 Cells. Annu. Rev. Immunol. 27:485–517. 10.1146/annurev.immunol.021908.132710 [DOI] [PubMed] [Google Scholar]
- Langrish, C.L., Chen Y., Blumenschein W.M., Mattson J., Basham B., Sedgwick J.D., McClanahan T., Kastelein R.A., and Cua D.J.. 2005. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J. Exp. Med. 201:233–240. 10.1084/jem.20041257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence, A., Tato C.M., Davidson T.S., Kanno Y., Chen Z., Yao Z., Blank R.B., Meylan F., Siegel R., Hennighausen L., et al. 2007. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 26:371–381. 10.1016/j.immuni.2007.02.009 [DOI] [PubMed] [Google Scholar]
- Levy, L., Howell M., Das D., Harkin S., Episkopou V., and Hill C.S.. 2007. Arkadia activates Smad3/Smad4-dependent transcription by triggering signal-induced SnoN degradation. Mol. Cell. Biol. 27:6068–6083. 10.1128/MCB.00664-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M.O., Sanjabi S., and Flavell R.A.. 2006. Transforming growth factor-beta controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity. 25:455–471. 10.1016/j.immuni.2006.07.011 [DOI] [PubMed] [Google Scholar]
- Liu, W., Rui H., Wang J., Lin S., He Y., Chen M., Li Q., Ye Z., Zhang S., Chan S.C., et al. 2006. Axin is a scaffold protein in TGF-beta signaling that promotes degradation of Smad7 by Arkadia. EMBO J. 25:1646–1658. 10.1038/sj.emboj.7601057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Zhang P., Li J., Kulkarni A.B., Perruche S., and Chen W.. 2008. A critical function for TGF-beta signaling in the development of natural CD4+CD25+Foxp3+ regulatory T cells. Nat. Immunol. 9:632–640. 10.1038/ni.1607 [DOI] [PubMed] [Google Scholar]
- Luo, K., Stroschein S.L., Wang W., Chen D., Martens E., Zhou S., and Zhou Q.. 1999. The Ski oncoprotein interacts with the Smad proteins to repress TGFbeta signaling. Genes Dev. 13:2196–2206. 10.1101/gad.13.17.2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan, P.R., Harrington L.E., O’Quinn D.B., Helms W.S., Bullard D.C., Elson C.O., Hatton R.D., Wahl S.M., Schoeb T.R., and Weaver C.T.. 2006. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 441:231–234. 10.1038/nature04754 [DOI] [PubMed] [Google Scholar]
- Marcelain, K., and Hayman M.J.. 2005. The Ski oncoprotein is upregulated and localized at the centrosomes and mitotic spindle during mitosis. Oncogene. 24:4321–4329. 10.1038/sj.onc.1208631 [DOI] [PubMed] [Google Scholar]
- Massagué, J. 2012. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 13:616–630. 10.1038/nrm3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrakis, K.J., Andrew R.L., Lee K.L., Petropoulou C., Dixon J.E., Navaratnam N., Norris D.P., and Episkopou V.. 2007. Arkadia enhances Nodal/TGF-beta signaling by coupling phospho-Smad2/3 activity and turnover. PLoS Biol. 5:e67. 10.1371/journal.pbio.0050067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeachy, M.J., Chen Y., Tato C.M., Laurence A., Joyce-Shaikh B., Blumenschein W.M., McClanahan T.K., O’Shea J.J., and Cua D.J.. 2009. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat. Immunol. 10:314–324. 10.1038/ni.1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano, Y., Mavrakis K.J., Lee K.L., Fujii T., Koinuma D., Sase H., Yuki K., Isogaya K., Saitoh M., Imamura T., et al. 2007. Arkadia induces degradation of SnoN and c-Ski to enhance transforming growth factor-beta signaling. J. Biol. Chem. 282:20492–20501. 10.1074/jbc.M701294200 [DOI] [PubMed] [Google Scholar]
- Ng, C., Aichinger M., Nguyen T., Au C., Najar T., Wu L., Mesa K.R., Liao W., Quivy J.P., Hubert B., et al. 2019. The histone chaperone CAF-1 cooperates with the DNA methyltransferases to maintain Cd4 silencing in cytotoxic T cells. Genes Dev. 33:669–683. 10.1101/gad.322024.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederländer, C., Walsh J.J., Episkopou V., and Jones C.M.. 2001. Arkadia enhances nodal-related signalling to induce mesendoderm. Nature. 410:830–834. 10.1038/35071103 [DOI] [PubMed] [Google Scholar]
- Nomura, T., Khan M.M., Kaul S.C., Dong H.D., Wadhwa R., Colmenares C., Kohno I., and Ishii S.. 1999. Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes Dev. 13:412–423. 10.1101/gad.13.4.412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, S.A., and Li M.O.. 2013. TGF-β: guardian of T cell function. J. Immunol. 191:3973–3979. 10.4049/jimmunol.1301843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi, S., Yamaguchi T., Nomura T., and Ono M.. 2008. Regulatory T cells and immune tolerance. Cell. 133:775–787. 10.1016/j.cell.2008.05.009 [DOI] [PubMed] [Google Scholar]
- Stroschein, S.L., Bonni S., Wrana J.L., and Luo K.. 2001. Smad3 recruits the anaphase-promoting complex for ubiquitination and degradation of SnoN. Genes Dev. 15:2822–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y., Liu X., Ng-Eaton E., Lodish H.F., and Weinberg R.A.. 1999. SnoN and Ski protooncoproteins are rapidly degraded in response to transforming growth factor beta signaling. Proc. Natl. Acad. Sci. USA. 96:12442–12447. 10.1073/pnas.96.22.12442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton, C., Brereton C., Keogh B., Mills K.H., and Lavelle E.C.. 2006. A crucial role for interleukin (IL)-1 in the induction of IL-17-producing T cells that mediate autoimmune encephalomyelitis. J. Exp. Med. 203:1685–1691. 10.1084/jem.20060285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto, T., Wakabayashi Y., Sekiya T., Inoue N., Morita R., Ichiyama K., Takahashi R., Asakawa M., Muto G., Mori T., et al. 2010. Smad2 and Smad3 are redundantly essential for the TGF-beta-mediated regulation of regulatory T plasticity and Th1 development. J. Immunol. 185:842–855. 10.4049/jimmunol.0904100 [DOI] [PubMed] [Google Scholar]
- Tone, Y., Furuuchi K., Kojima Y., Tykocinski M.L., Greene M.I., and Tone M.. 2008. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 9:194–202. 10.1038/ni1549 [DOI] [PubMed] [Google Scholar]
- Tsubakihara, Y., Hikita A., Yamamoto S., Matsushita S., Matsushita N., Oshima Y., Miyazawa K., and Imamura T.. 2015. Arkadia enhances BMP signalling through ubiquitylation and degradation of Smad6. J. Biochem. 158:61–71. 10.1093/jb/mvv024 [DOI] [PubMed] [Google Scholar]
- Veldhoen, M., Hocking R.J., Atkins C.J., Locksley R.M., and Stockinger B.. 2006a. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 24:179–189. 10.1016/j.immuni.2006.01.001 [DOI] [PubMed] [Google Scholar]
- Veldhoen, M., Hocking R.J., Flavell R.A., and Stockinger B.. 2006b. Signals mediated by transforming growth factor-beta initiate autoimmune encephalomyelitis, but chronic inflammation is needed to sustain disease. Nat. Immunol. 7:1151–1156. 10.1038/ni1391 [DOI] [PubMed] [Google Scholar]
- Wan, Y., Liu X., and Kirschner M.W.. 2001. The anaphase-promoting complex mediates TGF-beta signaling by targeting SnoN for destruction. Mol. Cell. 8:1027–1039. 10.1016/S1097-2765(01)00382-3 [DOI] [PubMed] [Google Scholar]
- Wu, Y., Borde M., Heissmeyer V., Feuerer M., Lapan A.D., Stroud J.C., Bates D.L., Guo L., Han A., Ziegler S.F., et al. 2006. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 126:375–387. 10.1016/j.cell.2006.05.042 [DOI] [PubMed] [Google Scholar]
- Xu, M., Pokrovskii M., Ding Y., Yi R., Au C., Harrison O.J., Galan C., Belkaid Y., Bonneau R., and Littman D.R.. 2018. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature. 554:373–377. 10.1038/nature25500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X.O., Panopoulos A.D., Nurieva R., Chang S.H., Wang D., Watowich S.S., and Dong C.. 2007. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J. Biol. Chem. 282:9358–9363. 10.1074/jbc.C600321200 [DOI] [PubMed] [Google Scholar]
- Yang, X.O., Nurieva R., Martinez G.J., Kang H.S., Chung Y., Pappu B.P., Shah B., Chang S.H., Schluns K.S., Watowich S.S., et al. 2008a. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 29:44–56. 10.1016/j.immuni.2008.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Xu J., Niu Y., Bromberg J.S., and Ding Y.. 2008b. T-bet and eomesodermin play critical roles in directing T cell differentiation to Th1 versus Th17. J. Immunol. 181:8700–8710. 10.4049/jimmunol.181.12.8700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, Z., Kanno Y., Kerenyi M., Stephens G., Durant L., Watford W.T., Laurence A., Robinson G.W., Shevach E.M., Moriggl R., et al. 2007. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 109:4368–4375. 10.1182/blood-2006-11-055756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Takaku M., Zou L., Gu A.D., Chou W.C., Zhang G., Wu B., Kong Q., Thomas S.Y., Serody J.S., et al. 2017. Reversing SKI-SMAD4-mediated suppression is essential for TH17 cell differentiation. Nature. 551:105–109. 10.1038/nature24283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, S.G., Wang J.H., Gray J.D., Soucier H., and Horwitz D.A.. 2004. Natural and induced CD4+CD25+ cells educate CD4+CD25- cells to develop suppressive activity: the role of IL-2, TGF-beta, and IL-10. J. Immunol. 172:5213–5221. 10.4049/jimmunol.172.9.5213 [DOI] [PubMed] [Google Scholar]
- Zheng, Y., Josefowicz S., Chaudhry A., Peng X.P., Forbush K., and Rudensky A.Y.. 2010. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 463:808–812. 10.1038/nature08750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, L., Lopes J.E., Chong M.M., Ivanov I.I., Min R., Victora G.D., Shen Y., Du J., Rubtsov Y.P., Rudensky A.Y., et al. 2008. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 453:236–240. 10.1038/nature06878 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists gene expression detected by RNA sequencing in cells polarized in vitro (fragments per kilobase of exon per million mapped fragments).