Abstract
V(D)J recombination is initiated by double-strand cleavage at recombination signal sequences (RSSs). DNA cleavage is mediated by the RAG1 and RAG2 proteins. Recent experiments describing RAG protein-RSS complexes, while defining the interaction of RAG1 with the nonamer, have not assigned contacts immediately adjacent to the site of DNA cleavage to either RAG polypeptide. Here we use UV cross-linking to define sequence- and site-specific interactions between RAG1 protein and both the heptamer element of the RSS and the coding flank DNA. Hence, RAG1-DNA contacts span the site of cleavage. We also detect cross-linking of RAG2 protein to some of the same nucleotides that cross-link to RAG1, indicating that, in the binding complex, both RAG proteins are in close proximity to the site of cleavage. These results suggest how the heptamer element, the recognition surface essential for DNA cleavage, is recognized by the RAG proteins and have implications for the stoichiometry and active site organization of the RAG1-RAG2-RSS complex.
The variable portions of antigen receptor genes are assembled from component gene segments during lymphocyte development by a process known as V(D)J recombination. The gene segments that undergo rearrangement are flanked by recombination signal sequences (RSSs), consisting of conserved heptamer (CACAGTG) and nonamer (ACAAAAACC) elements separated by a spacer of either 12 or 23 bp. Recombination brings two protein-coding regions together in an imprecise junction and joins the two RSSs with the heptamer elements fused head-to-head. Recombination occurs predominantly between gene segments flanked by RSSs with spacers of unequal lengths. This “12/23 rule” helps to restrict the reaction to developmentally useful combinations (19).
The products of the recombination activating genes, RAG1 and RAG2, are necessary for the initiation of V(D)J recombination (28, 41) and together catalyze the creation of a double-strand break at the border of an RSS (23, 50). They first introduce a nick 5′ to the first C nucleotide of the heptamer element. This exposes a 3′ hydroxyl which then attacks the opposite strand of the DNA, creating a 5′-phosphorylated blunt end on the signal side and a closed hairpin on the side that will form protein-coding sequence (see Fig. 1A).
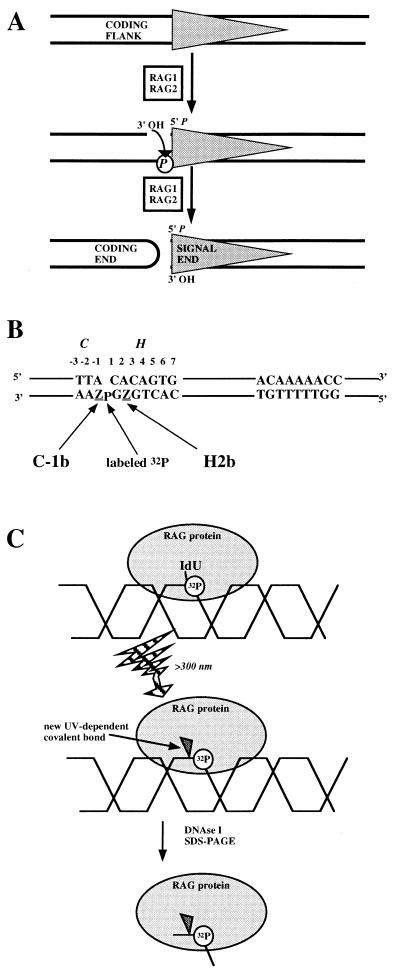
FIG. 1.
Experimental setup for detecting RAG protein-DNA interactions near the site of cleavage. (A) RSS cleavage by RAG proteins. The gray triangle represents CACAGTGN12/23ACAAAAACC. RAG1 and RAG2 catalyze first a nucleophilic attack by H2O on the top strand 5′ to the heptamer element, followed by use of the 3′ OH to attack the DNA phosphate (P) on the other strand. Coding flank refers to the DNA next to the RSS before cleavage. Signal end and coding end refer to the DNA species created by a double-strand break. (B) The coding flank (C) and heptamer (H) base pairs are numbered according to their distance from the site of DNA cleavage. IdU positions are named according to the element (C or H), the base pair, and the strand (top [t] or bottom [b]). In C-1b and H2b substrates the labeled phosphate is at the position indicated. (C) Schematic of cross-linking assay. The gray oval represents generic RAG protein binding to the DNA, represented as the interlocking “helix.” The lightning bolt arrow (UV irradiation) site-specifically cross-links RAG protein to the DNA through the iodo group. Surrounding DNA is removed by nucleases and TCA precipitation.
The sequence requirements for hairpin formation are more stringent than those for initial nicking. In cells and in crude extracts, hairpin formation requires the formation of a synaptic complex involving the two RSSs, while nicking can occur at an isolated signal (10, 11, 13, 44, 45). The RAG proteins intrinsically prefer to cleave a 12/23 pair of signals (52); however, the ubiquitous DNA-bending proteins HMG-1 and HMG-2 strengthen this preference by boosting cleavage at the RSS with a 23-bp spacer and by aiding in the formation of a synaptic complex (39, 49). In addition, in a single-signal context, some mutations of the heptamer element substantially inhibit hairpin formation without preventing initial nicking (7, 31).
Mutants of RAG1 have been identified as sensitive to the sequence of the coding flank DNA immediately adjacent to the heptamer (33, 36). The same coding flank sequences that prevent recombination with the mutant RAG1 (referred to as “bad flanks”) inhibit hairpin formation and not nicking when catalyzed by unmutated RAG1 and RAG2 in vitro. This preference, observed under conditions where the 12/23 rule is not being obeyed (in the presence of Mn2+), disappears when unpaired DNA is introduced into the coding flank or under conditions of coupled cleavage (7, 31, 52). This suggests that to catalyze hairpin formation, RAG1 and RAG2 must unwind the DNA at the heptamer-coding flank border and that synaptic complex formation promotes DNA unwinding.
The two-step reaction mechanism and stereochemistry of V(D)J signal cleavage have prompted comparisons with retroviral integration and transposition by elements such as Tn10 and Mu (51), and it was recently discovered that the Tn10 transposition mechanism includes the creation of hairpins at DNA termini (16). Transposases and retroviral integrases share not only a mechanism of phosphoryl transfer but also a common active-site architecture. In these systems, the active site is defined by three acidic amino acid residues that coordinate the divalent metal ions necessary for catalysis (3, 5, 38). For the RAG proteins, these amino acid residues have not been identified, leaving the issue of active-site architecture unresolved.
Individual roles for each of the RAG proteins in the catalysis of DNA cleavage have not been established. Neither protein has any obvious enzymatic activity on its own, but some progress has been made in identifying contacts between RAG1 protein and RSS DNA. Experiments using surface plasmon resonance and also an in vivo one-hybrid system provided evidence that RAG1 by itself has the ability to recognize and bind the RSS (8, 43). These studies suggested that the nonamer element is the more important DNA sequence feature required for binding and is recognized by the region of the RAG1 protein that has sequence similarity to the DNA-binding domain of Hin recombinase (hereafter called the nonamer-binding domain [NBD]). Later work that examined the formation of a stable complex containing RAG1, RAG2, and RSS DNA showed that RAG2 protein and both of the heptamer and nonamer elements were necessary for efficient and stable binding (2, 15).
The location of RAG2 in the RAG-RSS complex has been difficult to identify. Specific contacts between RAG1 and RSS DNA, as identified by dimethyl sulfate interference analysis, are found in the nonamer element and extend into the nonamer-proximal portion of the heptamer. One study has shown that the DNase I footprinting pattern obtained by using RAG1 alone is essentially the same as the pattern obtained with RAG1 and RAG2 (29). Another recent study found that when RAG1 and RAG2 were both bound to RSS DNA, the patterns of protection and binding interference extended further into the spacer and the nonamer-proximal portion of the heptamer than when only RAG1 was bound (46). Thus, it is possible that RAG2 makes weak contacts with the spacer and heptamer elements or that it does not contact the DNA at all, altering the DNA-binding tendencies of RAG1 indirectly.
In a single-RSS context, contacts between coding flank DNA and RAG proteins are required to stabilize a RAG-DNA complex (15). However, strong contacts between RAG1 or RAG2 and the nonamer-distal portion of the heptamer or coding flank DNA have not been identified. The presence of RAG2 in the RAG protein-RSS complex has been shown to encourage distortion of the DNA backbone at the heptamer-coding flank border (46). With the location of RAG2 unknown and RAG1’s location most firmly established at the nonamer element, it was unclear which of the RAG proteins contact the DNA near the site of DNA cleavage. To address this question, we have introduced the photolabile nucleotides 5-iododeoxyuridine (IdU) and 5-iododeoxycytosine (IdC) into RSS oligonucleotide substrates at specific positions near the heptamer-coding flank border (54). We reasoned that if the interactions between the RAG proteins and the DNA surrounding the site of cleavage are weak or transient, it still might be possible to capture the protein molecules interacting closely with the RSS DNA by UV cross-linking. We have detected sequence-specific interactions between RAG1 protein and both the nonamer-distal portion of the heptamer element and coding flank DNA. In addition, two of the iodinated nucleotide positions that cross-link to RAG1 protein also cross-link to RAG2 protein. Detection of both RAG1 and RAG2 proteins near the heptamer-coding flank border suggests that both proteins participate in direct recognition of the heptamer DNA and in construction of the enzymatic active site.
MATERIALS AND METHODS
Proteins.
The F2A1 cell line was generated by transfecting the B-cell lymphoma line M12 with heat shock-regulated RAG1 and RAG2 expression vectors as described previously (10, 18). The proteins expressed were murine RAG1 (amino acids 264 to 1008) and murine RAG2 (amino acids 1 to 387) with C-terminal extensions consisting of nine histidines and three copies of the c-myc epitope tag (24, 34, 35). A 30-liter portion of cultured cells was heat shocked for 6 min at 45°C, allowed to recover for 6 h, harvested, and frozen at −70°C. All subsequent steps were carried out on ice or at 4°C. Cell pellets were extracted with 100 ml of extraction buffer (325 mM NaCl; 3 mM MgCl2; 25 mM Tris-Cl, pH 7.5; 0.2 mM EDTA; 0.2 mM EGTA; 20% glycerol; 0.1% Nonidet P-40 [NP-40], 5 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride [PMSF]). After centrifugation at 30,000 × g for 30 min the supernatant was removed and diluted with 150 ml of buffer Q (20 mM Tris-Cl, pH 7.5; 20% glycerol, 5 mM DTT) plus 0.5 mM PMSF. This diluted extract was recentrifuged at 20,000 × g for 30 min and then loaded onto a 50-ml Q Sepharose Fast Flow column (Pharmacia) at 5 ml/min. The column was washed with 250 ml of buffer Q plus 140 mM NaCl and then eluted with 180 ml of buffer Q plus 350 mM NaCl. Proteins were precipitated by the addition of 72 g of (NH4)2SO4, recovered by centrifugation, resuspended in 25 ml of dialysis buffer, and dialyzed against 1 liter of buffer N7.5 (300 mM NaCl; 20 mM Na-HEPES, pH 7.5; 20 mM imidazole-Cl, pH 7.5; 20% glycerol; 0.1% NP-40; 7 mM β-mercaptoethanol) overnight. Proteins were loaded onto a 6-ml Ni2+-nitrilotriacetic acid (NTA) Superflow column (Qiagen) at 0.6 ml/min. The column was washed with 50 ml of buffer N7.5 and then with 25 ml of N6.2 (200 mM NaCl; 40 mM imidazole-Cl, pH 6.2; 20% glycerol; 7 mM β-mercaptoethanol). RAG proteins were eluted with a 100-ml gradient of imidazole (40 to 600 mM) in buffer N6.2. Fractions containing RAG proteins (determined by silver staining after sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE]) were pooled and dialyzed against buffer H (100 mM NaCl; 20 mM HEPES, pH 7.5; 3 mM MgCl2; 20% glycerol; 1 mM EDTA; 5 mM DTT) overnight. RAG proteins were further purified by loading them onto a 1-ml HiTrap heparin-Sepharose column (Pharmacia) at 0.6 ml/min, washing them with buffer H, and then eluting them in buffer H with a 10-ml gradient of NaCl (100 mM to 1.5 M), omitting MgCl2. The most active fractions (as determined by their ability to cleave the fr12x23 substrate [10]) were pooled, adjusted to 50% glycerol, and stored at −20°C. The preparation (30 μg each of RAG1 and RAG2 proteins per ml) is approximately 60% pure as determined by silver staining of SDS-PAGE gels.
Murine HMG2 protein (amino acids 1 to 185) lacking the C-terminal acidic domain and with an amino-terminal extension (MASHHHHHHSRTRRASVGPS) containing a polyhistidine region and protein kinase A phosphorylation site (55) was obtained by overexpression in Escherichia coli. Bacteria were lysed and sonicated briefly in buffer N7.5. After a thermal denaturation step at 72°C for 10 min, the bulk of the bacterial proteins was removed by centrifugation for 30 min at 30,000 × g and 0.45-μm (pore size) filtration. Further purification over a Ni2+-NTA column and elution with an imidazole gradient produced essentially pure protein, which was stored at −20°C in 50% glycerol.
Substrates.
The oligonucleotides used were synthesized by the Keck Foundation Biotechnology Resource Laboratory at Yale University on an Applied Biosystems 3948 synthesizer, with 5-iododeoxy-uridyl or -cytidyl phosphoramidites obtained from Glen Research. Unmodified oligonucleotides were purified by urea-PAGE. Sequences of the oligonucleotides used to make the unmutated C-1b and H2b substrates were as follows (z represents 5-iododeoxyuridine): QME27, 5′-TAAGACGTCGACGCGT-3′ (16 bases); QME28, 5′-zAAGACGTCGACGCGT-3′ (16 bases); QME29, 5′-GGATCCGGTTTTTGTTCAGGGCTGTATCACTGTG-3′ (34 bases); QME211, 5′-GGATCCGGTTTTTGTTCAGGGCTGTATCACTGzG-3′ (34 bases); and WTTOP, 5′-ACGCGTCGACGTCTTACACAGTGATACAGCCCTGAACAAAAACCGGATCC-3′ (50 bases).
All cross-linking substrates were prepared by 5′-end labeling 50 pmol of one oligonucleotide (in the C-1b and H2b substrates, QME28 and QME27, respectively) with an excess of [γ-32P]ATP and polynucleotide kinase. The labeled oligonucleotide was then annealed to a twofold excess of partner oligonucleotides (for C-1b and H2b, WTTOP and either QME29 or QME211 was used) and then ligated overnight at room temperature. Substrates were purified on native 10% polyacrylamide gels. Monitoring the efficiency of ligation on denaturing polyacrylamide gels revealed that >80% of the 32P-labeled DNA migrated at 50 nucleotides for C-1b and H2b substrates.
For C-1b and H2b substrates, mutation of the heptamer element replaced 5′-CACAGTG (top strand) with 5′-GAGCAGT. Mutation of the nonamer element replaced 5′-ACAAAAACC (top strand) with 5′-AGTCTCTGA. Mutation of both the heptamer and the nonamer replaced 5′-CACAGTG (top strand) with 5′-GAGCAGT and 5′-ACAAAAACC (top strand) with 5′-ACAAGGACC. 23-RSS substrates had identical coding flanks and had the following sequence for a spacer: 5′-ATACAGCCCTGATGTCTGGCTGT-3′.
For point mutants of the H2b and C-1b substrates, the sequence 5′-TTACACAGTG was changed to either 5′-TTAtACAGTG or 5′-ggACACAGTG (bad flank) (lowercase letters indicate altered residues). For C-1t substrates, the equivalent top-strand sequence was always 5′-TTCCACAGTG (underlined position was iodinated). The bottom-strand sequence was either complementary (5′-CACTGTGGAA) or not (5′-CACTGTGccA).
Substrates with iodomodifications at other positions were constructed analogously. For C-3t and C-2t substrates, the 32P label was placed 5′ to the iodinated nucleotide: top strand, 5′-pTTACACAGTG-3′ or 5′-TpTACACAGTG-3′, respectively. For H1t and H3t substrates, the 32P label was placed between the second and third positions of the heptamer: top strand, 5′-CApCAGTG. A mutant control for H3t was obtained by replacing CACAGTG with GCCCAGT (iodinated unmutated base-pair underlined). For H5b substrates, the 32P label was placed between the fifth and sixth positions of the heptamer: bottom strand, 3′-GTGTpCAC-5′. The mutant control for H5b replaced 5′-CACAGTG with 5′-GCGAGAC. For H6t substrates, the 32P label was placed between the fifth and sixth positions of the heptamer: top strand, 5′-CACAGpTG-3′. The mutant control for H6t replaced (top strand) 5′-CACAGTG with 5′-ACGCATT.
Cross-linking and cleavage reactions.
A standard cross-linking reaction mixture (75 mM Na-acetate; 2 mM Mg-diacetate; 20 mM Na-HEPES, pH 7.5; 10 μM ZnSO4; 2 mM DTT; 8% glycerol, 0.1 mg of bovine serum albumin per ml) contained 180 ng of each RAG protein and 2 × 106 cpm (1 pmol) of 32P-labeled DNA in 100 μl. Cross-linking reactions were set up at room temperature in polystyrene 96-well dishes. After the RAG proteins were added, reaction mixtures were incubated at 37°C for 10 min. Competitor DNA (5 μg of sheared salmon sperm DNA in most reactions), if used, was added after 5 min, and mixtures were incubated for 5 min more at 37°C. Cross-linking, shielded by the bottom of the polystyrene dish, took place for 12 min on a FotoPrep I 3-3500 UV illuminator with 312-nm bulbs. After the cross-linking, mixtures were transferred to Eppendorf tubes, heated to 68°C for 10 min, cooled, and adjusted to 10 mM MgCl2 and 1 mM CaCl2. The mixtures were digested by a mixture of 2 μg of DNase I and 0.2 μg of micrococcal nuclease for 1 h at 37°C. Mixtures were trichloroacetic acid (TCA) precipitated and subjected to SDS-PAGE with sample buffer containing β-mercaptoethanol. Separating gels were 7.5% (29:1) acrylamide-bisacrylamide, and the stacking gel contained 1% SDS. Gels were stained with Coomassie blue to identify protein markers before being dried for autoradiography. Cleavage reactions were performed in the same buffer as the standard cross-linking reaction.
Immunoprecipitations.
Cross-linking reaction mixtures to be immunoprecipitated were cross-linked as described above but were not heated to 68°C after UV irradiation. After digestion with nucleases, two cross-linking reaction mixtures containing twice the normal amount of RAG protein were pooled, adjusted to 0.1% SDS, and heated at 68°C for 10 min. Then, 500 μl of ice-cold immunoprecipitation (IP) wash buffer (1 M NaCl; 50 mM Tris, pH 7.5; 1% NP-40) was added, and the mixtures were precleared for 30 min at 4°C with 20 μl of protein G-agarose beads (Gibco-BRL). Next, 20 μg of anti-R1P7, anti-RAG2, or anti-R1P1 (a control, because the RAG1 protein used here does not have the R1P1 epitope) antibodies and an additional 20 μl of protein G-agarose beads were added. Mixtures were rotated overnight at 4°C and then centrifuged and washed five times with IP wash buffer. Precipitated proteins were eluted with 1% SDS at 55°C, TCA precipitated, and analyzed by SDS-PAGE as described above. The antibodies used here have been described elsewhere (1, 18, 25). The epitopes recognized included R1P7, RAG1 (amino acids 590 to 758); RAG2 (amino acids 70 to 516); and R1P1 (control), RAG1 (amino acids 56 to 123). The samples (see Fig. 8A) were transferred to a polyvinylidene difluoride membrane, probed with 9E10 (anti-myc tag) antibody, and developed by using alkaline phosphatase-conjugated secondary antibody and 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium (Jackson Immunoresearch).
FIG. 8.
Verification of the identities of 32P-labeled proteins with anti-RAG antibodies. (A) Mock binding reaction mixtures (no 32P-labeled DNA) were diluted with IP wash buffer, precleared, and immunoprecipitated overnight at 4°C. Immunoprecipitations were performed with antibodies to RAG1 (anti-R1) or RAG2 (anti-R2) or with a control antibody (see Materials and Methods). Lane 1 contains 10 ng of each RAG protein. Reaction mixtures in lanes 6 to 8 were adjusted to 0.1% SDS and incubated at 68°C for 10 minutes before dilution with cold IP wash buffer. Sizes of the protein markers are indicated in kilodaltons. It is unclear why RAG2 is not coprecipitated more efficiently in lane 2 (compare lanes 2 and 3). We note that the RAG1 antibody used recognizes epitopes within the region of RAG1 that interact with RAG2 (25) and is a different antibody than that used previously for coimmunoprecipitation studies of RAG1 and RAG2 (18). (B) Immunoprecipitations of cross-linking reactions, with wild-type 12-RSS substrates with IdU at the indicated positions, after nuclease treatment. The antibodies used for immunoprecipitation are indicated above each lane. (C) Immunoprecipitations of cross-linking reaction mixtures with anti-RAG2 antibodies. Lanes 1 to 4 use the same 23-RSS substrates and mutants as described in Fig. 5A, and lanes 5 to 6 use the 12-RSS C3t substrate and mutant used in Fig. 5B. Note that the immunoprecipitated samples shown here are enriched for RAG2 protein over the nonenriched samples in Fig. 5B. Because the efficiency of cross-linking to RAG1 is much higher than that to RAG2, small amounts of background RAG1 are visible in some lanes.
RESULTS
An assay for detecting cross-linking of RAG proteins to iodinated nucleotides in heptamer and coding-flank DNA.
We constructed 50-bp RSS substrates from three oligonucleotides, with a 32P label placed so that it is separated from the location of the iodinated nucleotide by only one or two phosphodiester bonds (Fig. 1B). The substrates are based upon a recombination signal with an optimized 12-bp spacer and 16 bp of coding flank DNA (30, 43). Incubation of RAG1 and RAG2 proteins with the iodomodified DNA followed by UV irradiation creates a novel covalent bond between the iodinated nucleotide and a polypeptide in close proximity to it (Fig. 1C). IdU and IdC are thought to cross-link preferentially to amino acids with aromatic and sulfur-containing side chains (26). It is important to recognize that failure to observe cross-linking to a particular nucleotide does not necessarily indicate the lack of protein contact altogether; instead, it may mean only that an appropriate amino acid is not close enough to the iodinated nucleotide. Subsequent nuclease treatment degrades un-cross-linked DNA and removes all but a few nucleotides from the modified protein (Fig. 1C). SDS-PAGE analysis and autoradiography allow visualization of the transfer of the 32P label to a polypeptide which, after cross-linking, will not differ substantially from its pre-cross-linking molecular weight (Fig. 1C). In this study we used a partially purified preparation of truncated RAG1 and RAG2 proteins, expressed in a mammalian cell line and modified with polyhistidine tags at the C terminus to facilitate their purification.
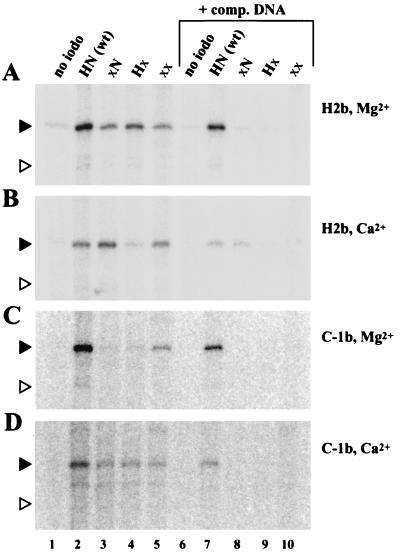
We initially used substrates with IdU introduced into the bottom strand, at the second position of the heptamer (H2b) and the first position of the coding flank (C-1b) immediately adjacent to the heptamer (Fig. 1B). Note that these two positions are on either side of the site of nucleophilic attack during hairpin formation; the 32P-labeled phosphate is the object of that attack. The major cross-linked protein migrates in an SDS-acrylamide gel (shown in Fig. 2) exactly as expected for the truncated RAG1 protein. No signal is observed with a noniodinated substrate (Fig. 2, lane 1), without UV irradiation (lanes 3 and 10), or with reactions lacking RAG protein (lanes 2 and 9). In agreement with previous results that showed that stable binding of RAG proteins to RSS DNA requires a divalent cation, the amount of cross-linked protein drops considerably when the 2 mM Mg2+ present in a standard binding reaction mixture is replaced with 0.5 mM EDTA (lanes 7 and 14). The cross-linking signal is also less when 2 mM Mg2+ is replaced by 2 mM Ca2+ (lanes 6 and 13). Although the overall efficiency of the cross-linking reaction is low (<5% of the cpm in the binding reaction mixture is transferred to nuclease-resistant material), binding seems to be stable, as little drop in the signal is seen when the preformed RAG-DNA complex is challenged before UV irradiation with a 100-fold excess of unlabeled nonspecific DNA (lanes 5 and 12). The immunoprecipitation experiments described below confirm that the 32P-labeled bands detected are RAG1 and RAG2.
FIG. 2.
RAG1 can be cross-linked to RSS DNA by using IdU and UV light. All three substrates used have consensus heptamer and nonamer sequences. See Fig. 1 for a graphic representation of the IdU and 32P positions. Components were added as indicated above the lanes. The divalent cations used were 2 mM MgCl2 (Mg), 2 mM CaCl2 (Ca), or 0.5 mM EDTA (−). Competitor (comp.) DNA was 5 μg of sheared salmon sperm DNA added after 5 min at 37°C. Binding reaction mixtures were incubated, cross-linked, treated with nuclease, and analyzed by SDS-PAGE as described in Materials and Methods. Positions of the protein markers are indicated on the left edge of the gel (sizes in kilodaltons). The black and white triangles indicate the expected positions of the truncated RAG1 and RAG2 proteins, respectively. The broad band at or above 200 kDa may be a highly cross-linked protein that does not completely enter the gel and has not been investigated further.
Sequence specificity of cross-linking.
To determine whether the protein-DNA interactions detected depended upon specific DNA sequences, we used cross-linking substrates that had severe mutations of either the heptamer element, the nonamer element, or both. In the absence of competitor DNA, mutations of the heptamer or nonamer elements do not eliminate cross-linking to the RAG1-sized protein. (Fig. 3, lanes 3 to 5). However, challenge with sheared salmon sperm DNA after incubating of the protein and labeled cross-linking substrate at 37°C for 5 min reveals that the interactions of RAG protein with the mutated substrates are labile (lanes 8 to 10), while challenge does not disrupt cross-linking to the wild-type substrate (lane 7). Stable interactions were sequence specific in the presence of Mg2+, requiring both the heptamer and the nonamer elements, when IdU was placed at either the H2b position (Fig. 3A) or the C-1b position (Fig. 3C). In addition to the cross-linking efficiency being generally poorer when Mg2+ is replaced by Ca2+ (Fig. 3B and D), mutating the heptamer element does not reduce the (already low) cross-linking signal detected in Ca2+ when the IdU is at the H2b position (Fig. 3B, lane 8). In this respect, our results resemble the observations made previously (29) of nonspecific DNA binding in the presence of Ca2+, although the interactions captured are still nonamer dependent.
FIG. 3.
Cross-linking of RAG1 to heptamer-coding flank DNA requires both heptamer and nonamer sequences. The 12-RSS substrates used are indicated above the lanes: HN (consensus heptamer and nonamer elements), xN (mutant heptamer), Hx (mutant nonamer), and xx (mutant heptamer and nonamer). The position of the IdU residue and the divalent cation used are indicated to the right of each panel. Competitor DNA (5 μg of sheared salmon sperm DNA) was added to the reaction mixtures for lanes 6 to 10. Reaction mixtures in lanes 1 and 6 contained a control substrate lacking IdU. The black and white triangles indicate the expected positions of the RAG1 and RAG2 proteins, respectively.
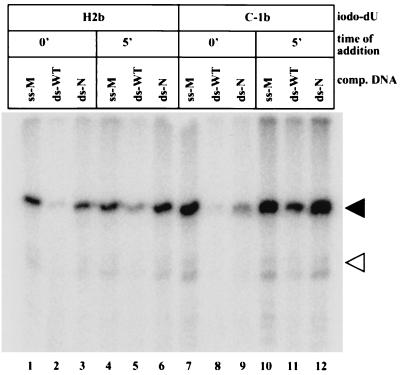
The specificity of DNA binding was examined further by using two other kinds of unlabeled competitor DNA: nonspecific single-stranded DNA and a consensus 12-RSS substrate. Competitor DNA was added to the 32P-labeled substrate DNA either before the RAG proteins (0 min) or 5 min after the RAG proteins were added (Fig. 4). RAG proteins could bind a 12-RSS substrate in the presence of an excess of genomic DNA (lanes 3 and 9). However, the cross-linking signal is reduced compared to when the competitor DNA is added after 5 min. We had previously determined that genomic DNA added after 5 min did not reduce the cross-linking signal (Fig. 2). With this comparison in mind, single-stranded oligonucleotides also did not diminish the cross-linking signal when added at t = 0 or 5 min (lanes 1, 4, 7, and 10). As expected from the results described by others, an intact 12-RSS substrate could effectively compete for DNA binding when present at t = 0 minutes (lanes 2 and 8), but it competes less effectively when added after complex formation (lanes 5 and 11) (15, 20). We conclude that nonspecific single-stranded DNA used as competitor by others (15, 29) does not interfere with cross-linking and that the RAG-DNA binding specificity, as measured indirectly here by the differing ability of 12-RSS and genomic DNA to compete for RAG proteins, is comparable to that observed by others (2, 15). Sheared genomic DNA added after 5 min was used in subsequent binding reactions because under these conditions the strongest cross-linking that still required both heptamer and nonamer sequences was observed.
FIG. 4.
Cross-linking to RAG1 can be blocked with specific competitor DNA. Binding reactions contained 2 mM Mg2+ and the indicated cross-linking substrate. Competitor DNA was added either before RAG protein (0′) or 5 min after RAG protein (5′) was added. The competitor DNAs used were 1 μM top-strand oligonucleotide used to make the substrate “xx” in Fig. 3 (ss-M), 1 μM unlabeled 50-bp double-stranded HN substrate (ds-WT), or 5 μg of sheared salmon sperm DNA, equivalent to ∼1.5 μM 50-bp DNA (ds-N). The black and white triangles indicate the expected positions of the RAG1 and RAG2 proteins, respectively.
Both 12-RSS and 23-RSS DNA cross-link like RAG proteins.
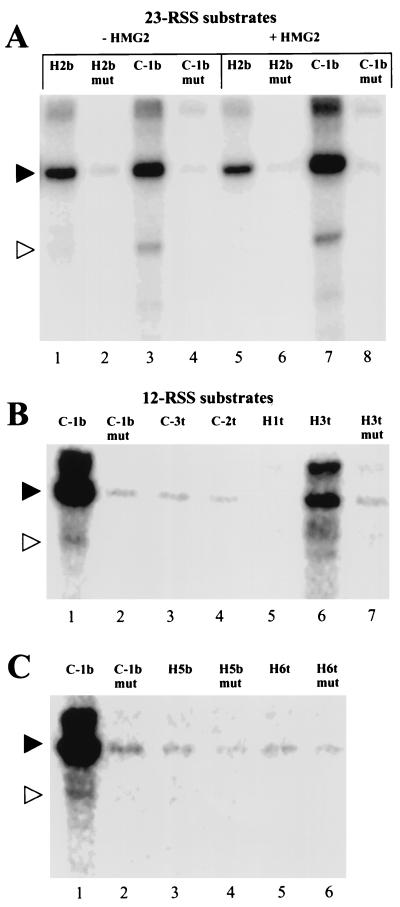
To determine whether RAG proteins would cross-link to DNA near the heptamer-coding flank border at 23-RSSs as well, we devised substrates that were 32P labeled and iodomodified at the C-1b and H2b positions as for the 12-RSS substrates described above (Fig. 5A). We observed weaker cross-linking to the RAG1-sized protein with the 23-RSS substrates compared to the 12-RSS substrates (data not shown), but the signal is still highly dependent upon heptamer and nonamer sequences (compare lanes 1 and 2 and lanes 3 and 4). HMG1 and HMG2 proteins, DNA-bending proteins that have an affinity for distorted and unpaired DNA sequences (4), have been reported to increase binding of the RAG proteins to a 23-RSS substrate. We observed that HMG2 did not uniformly enhance the interactions leading to cross-linking but instead resulted in decreased cross-linking to the heptamer H2b position and increased cross-linking to the coding-flank position C-1b (lanes 5 and 7). Interestingly, in reactions with substrates iodinated at the C-1b position, we observed a band, less prominent than the RAG1-sized band, that migrated at the expected size of RAG2 (white triangle, lanes 3 and 7). The identity of this band is addressed below.
FIG. 5.
Cross-linking reactions with 23-RSS substrates and 12-RSS substrates substituted at other heptamer-coding flank positions. All cross-linking reaction mixtures contain 2 mM Mg2+ and have sheared salmon sperm DNA added after 5 min. The black and white triangles indicate the expected positions of the RAG1 and RAG2 proteins, respectively. (A) 23-RSS substrates with IdU at the C-1b or H2b positions. “mut” indicates that both the heptamer and nonamer are mutated. In lanes 5 to 8, 50 ng of HMG2 protein was added before the RAG protein. (B and C) Cross-linking reactions with 12-RSS substrates with IdU or IdC at the indicated positions. Each mutant (“mut”) changes each base pair of the entire heptamer, except the modified nucleotide (see Materials and Methods), and leaves the nonamer intact. There are no mutants for C-3t, C-2t, and H1t because prior experimentation established that there was minimal cross-linking to those positions. Lanes 1 and 2 in each case are positive (C-1b) and negative (C-1b mut, “xx” from Fig. 3) control cross-linking reactions, respectively.
Evaluating other heptamer and coding-flank positions for RAG cross-linking.
We scanned through the heptamer and coding flank to find other pyrimidines that cross-link to RAG proteins. We used 12-RSS substrates with IdU or IdC incorporated at the C-3t, C-2t, H1t, H3t, H5b, and H6t positions (Fig. 5B and C; “t” and “b” indicate the top and bottom strands, respectively, as defined in Fig. 1B). With these substrates, the 32P label is placed so that a maximum of two phosphodiester bonds separates the labeled phosphate from the iodomodified base (see Materials and Methods). Cross-linking was detected when IdC was incorporated into the H3t position (Fig. 5B, lane 6), with the signal being somewhat weaker than the reference C-1b signal but still greatly reduced when the heptamer element is mutated (lane 7). Besides the specific signal at the H3t position, only very low-level cross-linking, which did not change when the RSS was mutated, was observed with the other substrates tested (Fig. 5B and C).
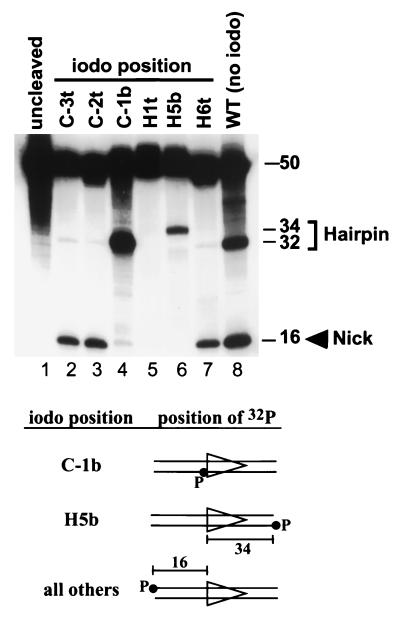
As noted above, failure to observe cross-linking at a particular DNA position is difficult to interpret. It might reflect the absence of a close protein-DNA interaction at that position, a close interaction involving amino acid residues inappropriate for cross-linking, or a perturbation of the binding interaction as a result of iodination of the RSS. To investigate the last possibility, RAG cleavage reactions were performed by using iodinated RSS substrates, and reaction products were visualized on denaturing polyacrylamide gels (Fig. 6). An iodine introduced at the C-1b and H5b positions (lanes 4 and 6) and H2b positions (data not shown) allowed hairpin formation (nicking could not be evaluated with these substrates because of the position of the 32P label; see bottom of Fig. 6). Iodination at positions H6t, C-2t, and C-3t permitted nicking but not hairpin formation (lanes 2, 3, and 7), while modification of the H1t position blocked all nicking and hairpin formation (lane 5). Because the iodo group lies in the major groove, these results suggest that to recognize the heptamer-coding flank region and to perform cleavage, the RAG proteins make close contacts with major groove recognition elements. These findings also suggest that our failure to observe specific cross-linking at a number of positions is due, at least in part, to interference with the RAG-RSS interaction by the iodine substitution.
FIG. 6.
Single RSS cleavage reactions with iodinated substrates. Cleavage was carried out in 2 mM Mg2+, and products were visualized by denaturing PAGE. Lane 1 is an uncleaved control, while lane 8 is a positive control (noniodinated wild-type RSS). Substrates have IdU or IdC introduced at the positions indicated above each lane and are labeled with 32P as indicated in the diagrams at the bottom of the figure. The C-1b and H5b substrates are labeled on the bottom strand, permitting only hairpin cleavage products (and not the nicked top strand) to be visualized on the gel. Hairpin formation with the H5b substrate generates a labeled 34-nucleotide signal end product, whereas for the other substrates, a labeled 32-nucleotide hairpin coding end product is generated.
Effects of heptamer mutations and suboptimal coding-flank sequence on cross-linking efficiency.
Cross-linking of RAG proteins to RSS DNA was more efficient in the presence of Mg2+, which supports catalysis, than in the presence of Ca2+, which does not. To examine further the link between catalysis and cross-linking, we made 12-RSS cross-linking substrates with a C-to-T mutation of the first position of the heptamer, a mutation which prevents hairpin formation but not nicking (10, 11, 31). With the iodo group at either the H2b or the C-1b position, this mutation reduced the amount of cross-linking (Fig. 7A, lanes 2, 3, 5, and 6). Therefore, the presence of the canonical C-G base pair at the first position of the heptamer is important for close interactions on both sides of the site of cleavage, as well as for hairpin formation.
FIG. 7.
Cross-linking to and cleavage of point mutant 12-RSS substrates. (A) Cross-linking reactions. IdU (C-1b and H2b)- or IdC (C-1t)-substituted substrates containing a consensus nonamer were used, as indicated above the lanes. Substrates contained either a consensus (w) or a mutant (m) heptamer (top strand, 5′-tACAGTG). The three nucleotides of the coding flank closest to the heptamer are shown above each lane (top-strand sequence as defined in Fig. 1A). 5′-TTA is a good flank, whereas 5′-ggA and 5′-TTc are bad flanks. The underlined nucleotides in lane 9 have no Watson-Crick base pairs on the bottom strand (the corresponding bottom-strand nucleotides are CC). The reaction in lane 1 contained a control substrate lacking IdU. The black and white triangles indicate the expected positions of the RAG1 and RAG2 proteins, respectively. (B) Single RSS cleavage reactions in 2 mM Mg2+ with noniodinated substrates. The structure of the substrate and the position of the 32P label are indicated at the top of the panel. Products were analyzed on a denaturing polyacrylamide gel. Nicking and hairpin formation result in 16- and 32-nucleotide products, respectively. Lane 1 is uncleaved substrate, and the sequences of the other substrates are indicated as in panel A. Lane 7 is an end-labeled 10-bp ladder (Gibco-BRL).
Hairpin formation in a single-RSS context is also sensitive to coding-flank sequence. “Bad” flanks support only nicking, while “good” flanks allow both nicking and hairpin formation (31, 36). The cross-linking substrates used in the experiments described above contained a good flank (5′-TTA-3′ immediately adjacent to the heptamer on the top strand [Fig. 1B]). Changing two residues to create the bad flank sequence 5′-GGA-3′ had different effects on cross-linking, depending on the site of the iodo group: at the C-1b position cross-linking was increased (Fig. 7A, lanes 2 and 4), while at the H2b position cross-linking was reduced (lanes 5 and 7). Therefore, a bad flank does not eliminate the interactions necessary for cross-linking. Instead, the results suggest that a bad flank selectively perturbs the DNA structure near the coding flank-heptamer border, impairing interactions between RAG1 and the second base pair of the heptamer.
We also examined cross-linking to the first position of the coding flank on the top strand (C-1t). Because an iodo group could be introduced only on a pyrimidine, which at this position invariably generates a bad flank, these experiments were performed only in the context of a bad flank. Interestingly, no cross-linking at this position was observed unless a noncomplementary sequence was introduced into the bottom strand, leading to unpairing of 2 bp of the coding flank (Fig. 7A, lanes 8 and 9; note that the bottom strand also has a bad-flank sequence). Unpairing has been found by others (7, 31) to restore hairpin formation on a bad-flank sequence, and analysis of cleavage reactions with the two bad-flank substrates confirms this (Fig. 7B, compare lanes 5 and 6). Therefore, cross-linking to the C-1t position correlates well with hairpin formation ability, perhaps because an unpaired structure at the heptamer-coding flank border allows the C-1t nucleotide to rotate into a new position that makes closer contacts with RAG1.
Finally, experiments were performed to confirm that the mutated single-RSS substrates used here yielded the expected cleavage products. For this purpose, 32P was introduced at the 5′ end of the top strand, and reaction products were visualized by denaturing PAGE (Fig. 7B; note that the substrates used in Fig. 7B are not iodinated). As predicted, mutation of the first position of the heptamer (lane 3), or bad-flank sequences (lanes 4 and 5), reduced hairpin formation but not nicking, and introduction of unpairing in the coding flank substantially increased hairpin formation with a bad-flank sequence (lane 6). These results were obtained in a Mg2+-containing buffer identical to that used in the cross-linking experiments, and similar results were obtained in Mn2+ (data not shown).
Immunoprecipitation analysis of 32P-labeled RAG proteins.
The major cross-linked protein in most reactions appeared to be RAG1, although in reactions with substrates with IdU at the C-1b position, a labeled species was visible at the expected position of RAG2 (Fig. 5A, lanes 3 and 7; Fig. 5B, lane 1; Fig. 5C, lane 1). To ascertain the identities of the cross-linked proteins, we used anti-RAG1 and anti-RAG2 antibodies to immunoprecipitate the proteins after cross-linking and nuclease digestion. These antibodies are specific for either RAG1 or RAG2 proteins and detect a single band in immunoblots of the RAG protein preparations used here (data not shown). To reduce coimmunoprecipitation of RAG1 with RAG2, which occurs in mock binding reactions (Fig. 8A, lane 3), we disrupted noncovalent interactions by adding 0.1% SDS and heating the binding reaction mixtures to 68°C before partially renaturing the proteins (enough to be recognized by the antibodies) in the presence of 1% NP-40. Under these conditions we observed efficient direct immunoprecipitation of RAG1 and RAG2 but not coimmunoprecipitation (lanes 6 and 7).
By using this direct immunoprecipitation assay and 12-RSS substrates, the 32P-labeled band which migrates at ∼100 kDa, the expected size of the truncated RAG1 protein, was brought down with anti-RAG1 antibodies (Fig. 8B, lanes 1 and 4) and at a background level with anti-RAG2 antibodies (compare lanes 2 and 5 to lane 3). This confirms that the upper cross-linked band is RAG1 protein. The 55-kDa RAG2-sized band precipitated with anti-RAG2 antibodies (lane 2, white triangle) and not RAG1 antibodies or control antibodies (lanes 1 and 3) in cross-linking reactions with C-1b modified substrate. This precipitated product was not visible with the H2b modified substrate (lane 5).
We then performed anti-RAG2 immunoprecipitations with 23-RSS substrates, their corresponding mutants, and with 12-RSS substrates with IdC at the H3t position (Fig. 8C). This confirmed RAG2 cross-linking to the C-1b but not to the H2b positions (lanes 1 and 3). The labeled RAG2 product was also visible when IdC was incorporated at the H3t position (lane 5), but only when the heptamer was intact (lane 6). There was a significant amount of 32P-labeled RAG1 protein that precipitated with anti-RAG2 antibodies in these experiments despite our denaturation-renaturation protocol (Fig. 8B, lane 2, and Fig. 8C, lanes 1 and 3; see legend to Fig. 8).
In summary, the 32P-labeled protein bands observed in cross-linking reactions with iodosubstituted RSSs have been identified by specific immunoprecipitation as the RAG1 and RAG2 proteins. RAG1 cross-links to the C-1b, H2b, and H3t positions, while RAG2 cross-links weakly to the C-1b and H3t positions.
DISCUSSION
Cross-linking of RAG proteins to RSS DNA near the site of cleavage.
Here we show, by UV-induced cross-linking, interactions between the RAG1 protein and both coding-flank and nearby heptamer element DNA. We also show that the RAG2 protein cross-links to a subset of the same positions. Several facts suggest that these interactions are significant for RAG-mediated RSS cleavage. First, they are sequence specific, requiring both heptamer and nonamer elements (Fig. 3). Second, they are stable to challenge by nonspecific competitor DNA (Fig. 4). Third, cross-linking appears to be site specific, occurring efficiently only when the iodonucleotide is placed at the C-1b, H2b, and H3t positions or at the C-1t position with an unpaired coding flank (although the absence of cross-linking at the other positions examined must be interpreted cautiously). Fourth, efficient binding and cross-linking depend upon the presence of a divalent cation (Fig. 2). Recent studies have suggested a direct role for divalent cations in guiding the DNA binding activities of RAG1 and RAG2 at the heptamer-coding flank border (37). Cross-linking can be detected in the presence of Ca2+, which does not support nicking or hairpin formation, suggesting that the observed interactions are not exclusively associated with catalysis. Ca2+ supports DNA binding, and Ca2+-containing RAG-DNA complexes can undergo cleavage when Mg2+ or Mn2+ is added (14, 15). Immunoprecipitation analysis revealed that both full-length and nicked top-strand DNAs are found associated with cross-linked bottom-strand DNA (data not shown). Thus, nicking is not required for cross-linking; neither is catalysis in general. However, conditions that favor catalysis, such as an unmutated heptamer, an unpaired coding flank, and the presence of Mg2+, promote interactions that result in cross-linking. Importantly, these results establish for the first time that both RAG1 and RAG2 proteins specifically interact with the DNA near the site of cleavage and reveal four nucleotides that the RAG proteins contact closely.
How is the heptamer element recognized?
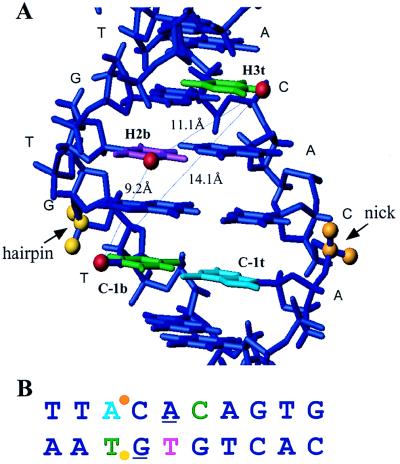
The three-dimensional placement of the IdU and IdC groups that cross-link to the RAG proteins suggests that RAG1 and RAG2 make contacts with the nonamer-distal portion of the heptamer DNA in the major groove, with individual sites of interaction marking a broad arc around the double helix (modeled in Fig. 9A). Our observation that some iodination positions, especially those near the site of cleavage, inhibit cleavage also points to major groove recognition. The palindromic sequence of the heptamer element (CAC//GTG) had raised the possibility that the heptamer might be recognized symmetrically, with a twofold axis through the fourth base pair. This possibility is unlikely given our failure to detect cross-linking at the H5b and H6t positions at intensities similar to those observed at the H3t and H2b positions. Asymmetric recognition of the two halves of the heptamer is also consistent with the higher conservation and greater functional importance of the first three nucleotides of the heptamer (13, 30, 31). How does the nonamer-proximal half of the heptamer contribute to recognition and cleavage of a RSS? The last 4 bp of the heptamer element have been shown to contribute to RAG1 binding, as shown by methylation interference and cleavage competition assays (29, 31, 46). In addition, the purine-pyrimidine alteration of the consensus heptamer might help to stabilize altered DNA structures that have been observed at CACA stretches (48). Indeed, a large number of biophysical and biochemical experiments have established that (CA)n sequences such as the heptamer element are more bendable and less thermally stable than other DNA sequences (6, 9, 12) and that they are capable of adopting a variety of conformations (40, 47, 48). This malleability may be an important part of heptamer recognition by RAG proteins (2, 46) because of the considerable distortion of the DNA backbone necessary for hairpin formation.
FIG. 9.
Map of cross-linking positions on heptamer-coding flank DNA. (A) Three-dimensional model developed by using MOLMOL (16a). Colors: green, pyrimidine base that when iodomodified cross-links to RAG1; purple, pyrimidine base that when iodomodified cross-links to RAG1 and RAG2; light blue, position that cross-links only when the base is an unpaired iodomodified pyrimidine; red, iodine; orange, target phosphate for nicking; yellow, target phosphate for hairpin formation; medium blue, other DNA, represented for simplicity as standard B-form DNA. (B) Linear model of heptamer-coding flank sequence, with a color scheme as in panel A. Purine bases necessary for hairpin formation (31) are underlined.
Unpairing of heptamer-coding flank DNA contributes to the efficiency of hairpin formation (7, 31), and our results show that such unpairing leads to a new cross-linking interaction between RAG1 and coding-flank DNA, immediately adjacent to the site of cleavage (position C-1t; Fig. 7A). In a fully paired DNA substrate, additional protein-DNA contacts would likely be necessary to drive the energetically unfavorable process of unpairing of coding-flank residues. We speculate that the interaction at C-1t may represent such an interaction.
Stoichiometry and interactions in higher-order RAG-DNA complexes.
The stronger and more widespread cross-linking to RAG1 protein, compared to RAG2 protein, indicates that RAG1 is the major player in recognizing the heptamer element. The detection of RAG1-DNA interactions at the nonamer-distal portion of the heptamer raises questions about the stoichiometry of the RAG protein-DNA complex. The distance between the position −1 of the coding flank and position 6 of the nonamer, modeled on undistorted B-form DNA, is more than 70 Å. Does the same RAG1 protein molecule bind both the heptamer and nonamer elements at the same time? While this is a possibility, it is tempting to think that heptamer and nonamer elements on the same RSS are bridged by a dimer of RAG1 molecules. Both the zinc-binding domain of RAG1 adjacent to the NBD (32) and the NBD itself (37) have been identified as capable of dimerization. Furthermore, a recently identified mutation of RAG1 that impairs NBD dimerization also reduces the efficiency of DNA cleavage at a single signal, implying that a multimer of RAG1 is the part of the actively cleaving protein-DNA complex (53). A number of laboratories have detected heterogeneous RAG-DNA complexes that differ in their electrophoretic mobilities (2, 37, 46, 53). The stoichiometries of these different RAG-DNA complexes have not been directly determined. The recent identification of one of these complexes as a two-signal synaptic complex (14) may enable the definition of protein-DNA contacts that are specific for a 12/23 pair of signals. One might expect, from comparisons with Mu transposase (17, 27), that in a two-signal RAG-DNA complex, the pattern of protein contacts would extend farther over the heptamer and coding flank than in a one-signal complex.
Contributions made by RAG2 protein: organization of the active site?
Cross-linking to RAG2 is always accompanied by cross-linking to RAG1. This suggests that RAG1 and RAG2 interact closely near heptamer-element DNA. Cross-linking to RAG2 can be detected at two positions which in undistorted B-form DNA are separated by 14 Å (Fig. 9A), raising the possibility that RAG1 and RAG2 form an extended interface in the vicinity of the heptamer. Alternatively, more than one RAG2 molecule may contact each RAG1 molecule, as has been suggested on the basis of immunoprecipitation data (18, 42). The more-restricted and less-efficient cross-linking to RAG2 that we observe may indicate a weak or limited set of DNA contacts and/or contacts via amino acids that are unsuitable for cross-linking. We note that we have not been able to detect efficient cross-linking to the C-1b or H2b positions with high concentrations of bacterially produced, catalytically active RAG1 protein in the absence of RAG2, despite the fact that this RAG1 protein exhibits specific RSS binding (32a). One reason for this might be that without the influence of RAG2, the part of RAG1 protein that makes contacts with the heptamer-coding flank border is disordered. We speculate that RAG2 alters the structure of this portion of RAG1 and that the bulk of RAG2 is located away from RSS DNA. The somewhat more extended pattern of binding interference and protection in the spacer region when RAG2 is present is consistent with this idea (46).
It is provocative that the strongest cross-linking of RAG2 occurs at the nucleotide position nearest to the target phosphate for hairpin formation, suggesting that both RAG1 and RAG2 play a role in forming the catalytic active site. Further investigations of the organization of the active site require identification of amino acid residues, such as aspartates or glutamates, that would coordinate divalent cations and whose mutation would disrupt catalysis but not formation of a RAG1-RAG2-RSS DNA complex. At this point the possibility that either RAG1 or RAG2 contributes catalytic amino acid residues remains open. It is also formally possible that separate active sites, analogous to the sites contained in the proteins TnsA and TnsB in the Tn7 system (22, 38), mediate nicking and hairpin formation. Sequencing of peptide fragments of RAG1 and RAG2 cross-linked to RSS DNA or other mapping techniques (21) might give hints leading to the identification of amino acid residues involved in catalysis.
ACKNOWLEDGMENTS
We thank Chia-Lun Tsai for assistance with RAG protein purification, Karla Rodgers for bacterial RAG1 protein and instruction on NAMOT and MOLMOL, the Schatz lab for patience with dimmed lights, Tad Koch for initial advice on cross-linking, and Eugenia Spanopoulou and Sankar Ghosh for helpful comments on the manuscript. We thank the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University for rapid oligonucleotide synthesis and the National Cell Culture Center (Minneapolis, Minn.) for large-scale F2A1 culture.
Q.M.E. was supported by a predoctoral fellowship from the National Science Foundation. I.J.V. was supported by an INSERM postdoctoral fellowship. This work was supported by Public Health Service grant AI-32524 from the National Institutes of Health. D.G.S. is an associate investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Agrawal A, Schatz D G. RAG1 and RAG2 form a stable post-cleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- 2.Akamatsu Y, Oettinger M. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Mol Cell Biol. 1998;18:4670–4678. doi: 10.1128/mcb.18.8.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker T A, Luo L. Identification of residues in the Mu transposase essential for catalysis. Proc Natl Acad Sci USA. 1994;91:6654–6658. doi: 10.1073/pnas.91.14.6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianchi M E, Beltrame M, Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989;243:1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- 5.Bolland S, Kleckner N. The three chemical steps of Tn10/IS10 transposition involve repeated utilization of a single active site. Cell. 1996;84:223–233. doi: 10.1016/s0092-8674(00)80977-0. [DOI] [PubMed] [Google Scholar]
- 6.Cheung S, Arndt K, Lu P. Correlation of the lac operator imino exchange kinetics with its function. Proc Natl Acad Sci USA. 1984;81:3665–3669. doi: 10.1073/pnas.81.12.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuomo C A, Mundy C L, Oettinger M A. DNA sequence and structure requirements for cleavage of V(D)J recombination signal sequences. Mol Cell Biol. 1996;16:5683–5690. doi: 10.1128/mcb.16.10.5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Difilippantonio M J, McMahan C J, Eastman Q M, Spanopoulou E, Schatz D G. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell. 1996;87:253–262. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- 9.Donlan M, Lu P. Transcriptional enhancer related DNA sequences: anomalous 1H NMR NOE crosspeaks. Nucleic Acids Res. 1992;20:525–532. doi: 10.1093/nar/20.3.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eastman Q M, Leu T M J, Schatz D G. Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature. 1996;380:85–88. doi: 10.1038/380085a0. [DOI] [PubMed] [Google Scholar]
- 11.Eastman Q M, Schatz D G. Nicking is asynchronous and stimulated by synapsis in 12/23-rule regulated V(D)J recombination. Nucleic Acids Res. 1997;25:4370–4378. doi: 10.1093/nar/25.21.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Folta-Stogniew E, Russu I. Sequence dependence of base-pair opening in a DNA dodecamer containing the CACA/GTGT sequence motif. Biochemistry. 1994;33:11016–11024. doi: 10.1021/bi00202a022. [DOI] [PubMed] [Google Scholar]
- 13.Hesse J E, Lieber M R, Mizuuchi K, Gellert M. V(D)J recombination: a functional definition of the joining signals. Genes Dev. 1989;3:1053–1061. doi: 10.1101/gad.3.7.1053. [DOI] [PubMed] [Google Scholar]
- 14.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 15.Hiom K, Gellert M. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 1997;88:65–72. doi: 10.1016/s0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- 16.Kennedy A K, Guhathakurta A, Kleckner N, Haniford D B. Tn10 excision via a DNA hairpin intermediate. Cell. 1998;95:125–134. doi: 10.1016/s0092-8674(00)81788-2. [DOI] [PubMed] [Google Scholar]
- 16a.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 17.Lavoie B D, Chan B S, Allison R G, Chaconas G. Structural aspects of a higher order nucleoprotein complex: induction of an altered DNA structure at the Mu-host junction of the Mu type 1 transpososome. EMBO J. 1991;10:3051–3059. doi: 10.1002/j.1460-2075.1991.tb07856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leu T M J, Schatz D G. rag-1 and rag-2 are components of a high-molecular-weight complex, and association of rag-2 with this complex is rag-1 dependent. Mol Cell Biol. 1995;15:5657–5670. doi: 10.1128/mcb.15.10.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis S M. The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 20.Li W, Swanson P, Desiderio S. RAG-1 and RAG-2-dependent assembly of functional complexes with V(D)J recombination substrates in solution. Mol Cell Biol. 1997;17:6932–6939. doi: 10.1128/mcb.17.12.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lykke-Andersen J, Garrett R, Kjems J. Mapping metal ions at the catalytic centres of two intron-encoded endonucleases. EMBO J. 1997;16:3272–3281. doi: 10.1093/emboj/16.11.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.May E W, Craig N L. Switching from cut-and-paste to replicative Tn7 transposition. Science. 1996;272:401–404. doi: 10.1126/science.272.5260.401. [DOI] [PubMed] [Google Scholar]
- 23.McBlane J F, van Gent D C, Ramsden D A, Romeo C, Cuomo C A, Gellert M, Oettinger M A. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 24.McMahan C J, Difilippantonio M J, Rao N, Spanopoulou E S, Schatz D G. A basic motif in the N-terminal region of RAG1 enhances recombination activity. Mol Cell Biol. 1997;17:4544–4552. doi: 10.1128/mcb.17.8.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMahan C J, Sadofsky M J, Schatz D G. Definition of a large region of RAG1 that is important for co-immunoprecipitation of RAG2. J Immunol. 1997;158:2202–2210. [PubMed] [Google Scholar]
- 26.Meisenheimer K M, Koch T H. Photocross-linking of nucleic acids to associated proteins. Crit Rev Biochem Mol Biol. 1997;32:101–140. doi: 10.3109/10409239709108550. [DOI] [PubMed] [Google Scholar]
- 27.Mizuuchi M, Baker T A, Mizuuchi K. DNase protection analysis of the stable synaptic complexes involved in Mu transposition. Proc Natl Acad Sci USA. 1991;88:9031–9035. doi: 10.1073/pnas.88.20.9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mombaerts P, Iacomini J, Johnson R S, Herrup K, Tonegawa S, Papaioannou V E. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 29.Nagawa F, Ishiguro K, Tsuboi A, Yoshida T, Ishikawa A, Takemori T, Otsuka A, Sakano H. Footprint analysis of the RAG protein recombination signal sequence complex for V(D)J type recombination. Mol Cell Biol. 1998;18:655–663. doi: 10.1128/mcb.18.1.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramsden D A, Baetz K, Wu G E. Conservation of sequence in recombination signal sequence spacers. Nucleic Acids Res. 1994;22:1785–1796. doi: 10.1093/nar/22.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramsden D A, McBlane J F, van Gent D C, Gellert M. Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J. 1996;15:3197–3206. [PMC free article] [PubMed] [Google Scholar]
- 32.Rodgers K K, Bu Z, Fleming K G, Schatz D G, Engelman D M, Coleman J E. A unique zinc-binding dimerization motif domain in RAG-1 includes the C3HC4 motif. J Mol Biol. 1996;260:70–84. doi: 10.1006/jmbi.1996.0382. [DOI] [PubMed] [Google Scholar]
- 32a.Rodgers, K. K., I. J. Villey, E. Corbett, D. G. Schatz, and J. E. Coleman. A dimer of RAG1 recognizes the recombination signal sequence and the complex stably incorporates HMG2. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 33.Roman C A J, Baltimore D. Genetic evidence that the RAG1 protein directly participates in V(D)J recombination through substrate recognition. Proc Natl Acad Sci USA. 1996;93:2333–2338. doi: 10.1073/pnas.93.6.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sadofsky M J, Hesse J E, Gellert M. Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res. 1994;22:1805–1809. doi: 10.1093/nar/22.10.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sadofsky M J, Hesse J E, McBlane J F, Gellert M. Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res. 1993;22:5644–5650. doi: 10.1093/nar/21.24.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sadofsky M J, Hesse J E, Vangent D C, Gellert M. RAG-1 mutations that affect the target specificity of V(D)J recombination—a possible direct role of RAG-1 in site recognition. Genes Dev. 1995;9:2193–2199. doi: 10.1101/gad.9.17.2193. [DOI] [PubMed] [Google Scholar]
- 37.Santagata S, Aidinis V, Spanopoulou E. The effect of Me++ cofactors at the initial stages of V(D)J recombination. J Biol Chem. 1998;273:16325–16331. doi: 10.1074/jbc.273.26.16325. [DOI] [PubMed] [Google Scholar]
- 38.Sarnovsky R J, May E W, Craig N L. The Tn7 transposase is a heteromeric complex in which DNA breakage and joining activities are distributed between different gene products. EMBO J. 1996;15:6348–6361. [PMC free article] [PubMed] [Google Scholar]
- 39.Sawchuk D, Weis-Garcia F, Malik S, Besmer E, Bustin M, Nussenzweig M, Cortes P. V(D)J recombination: modulation of RAG1 and RAG2 cleavage activity on 12/23 substrates by whole cell extract and DNA-bending proteins. J Exp Med. 1997;185:2025–2032. doi: 10.1084/jem.185.11.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schultz S, Shields G, Steitz T. Crystal structure of a CAP-DNA complex: the DNA is bent by 90°. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 41.Shinkai Y, Rathbun G, Kong-Peng L, Oltz E M, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall A M, Alt F W. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 42.Spanopoulou E, Cortes P, Shih C, Huang C M, Silver D P, Svec P, Baltimore D. Localization, interaction, and RNA binding properties of the V(D)J recombination-activating proteins RAG1 and RAG2. Immunity. 1995;3:715–726. doi: 10.1016/1074-7613(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 43.Spanopoulou E, Zaitseva F, Wang F-H, Santagata S, Baltimore D, Panayotou G. The homeodomain of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell. 1996;87:263–276. doi: 10.1016/s0092-8674(00)81344-6. [DOI] [PubMed] [Google Scholar]
- 44.Steen S B, Gomelsky L, Roth D B. The 12/23 rule is enforced at the cleavage step of V(D)J recombination in vivo. Genes Cells. 1996;1:543–553. doi: 10.1046/j.1365-2443.1996.d01-259.x. [DOI] [PubMed] [Google Scholar]
- 45.Steen S B, Gomelsky L, Speidel S L, Roth D B. Initiation of V(D)J recombination in vivo: role of recombination signal sequences in formation of single and paired double-strand breaks. EMBO J. 1997;16:2656–2664. doi: 10.1093/emboj/16.10.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swanson P, Desiderio S. V(D)J recombination signal recognition: distinct, overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity. 1998;9:115–125. doi: 10.1016/s1074-7613(00)80593-2. [DOI] [PubMed] [Google Scholar]
- 47.Tari L, Secco A. Base-pair opening and spermine binding B-DNA features displayed in crystal structure of a gal operon fragment: implications for protein-DNA recognition. Nucleic Acids Res. 1995;23:2065–2073. doi: 10.1093/nar/23.11.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Timsit Y, Vilbois E, Moras D. Base-pairing shift in the major groove of (CA)n tracts by B-DNA crystal structures. Nature. 1991;354:167–170. doi: 10.1038/354167a0. [DOI] [PubMed] [Google Scholar]
- 49.van Gent D C, Hiom K, Paull T T, Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Gent D C, McBlane J F, Ramsden D A, Sadofsky M J, Hesse J E, Gellert M. Initiation of V(D)J recombination in a cell-free system. Cell. 1995;81:925–934. doi: 10.1016/0092-8674(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 51.van Gent D C, Mizuuchi K, Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996;271:1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- 52.van Gent D C, Ramsden D A, Gellert M. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell. 1996;85:107–113. doi: 10.1016/s0092-8674(00)81086-7. [DOI] [PubMed] [Google Scholar]
- 53.Villa A, Santagata S, Bozzi F, Giliani S, Frattini A, Imberti L, Gatta L, Ochs H, Schwarz S, Notarangelo L, Vezzoni V, Spanopoulou E. Partial V(D)J recombination activity leads to Omenn’s syndrome. Cell. 1998;93:885–896. doi: 10.1016/s0092-8674(00)81448-8. [DOI] [PubMed] [Google Scholar]
- 54.Willis M C, Hicke B J, Uhlenbeck O C, Cech T R, Koch T H. Photocrosslinking of 5-iodouracil-substituted RNA and DNA to proteins. Science. 1993;262:1255–1257. doi: 10.1126/science.7694369. [DOI] [PubMed] [Google Scholar]
- 55.Zwilling S, Konig H, Wirth T. High mobility group protein 2 functionally interacts with the POU domains of octamer transcription factors. EMBO J. 1995;14:1198–1208. doi: 10.1002/j.1460-2075.1995.tb07103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]