Abstract
We evaluated the usefulness of PCR and antigen detection for the diagnosis of pulmonary aspergillosis. Forty-four serum samples from patients with pulmonary aspergillosis (33 with pulmonary aspergilloma, 4 with allergic bronchopulmonary aspergillosis, 4 with invasive pulmonary aspergillosis, and 3 with aspergillus pyothorax) were used in this study. PCR detection of Aspergillus DNA in serum samples was successful in 39 patients. Galactomannan antigen was detected by sandwich enzyme-linked immunosorbent assay in 25 patients and by latex agglutination test in 13 patients. Detection of Aspergillus DNA in serum samples by nested PCR had the highest sensitivity of the three methods tested for the diagnosis of pulmonary aspergillosis.
Substantial progress has been made during the past decade in the development of new approaches and methods for the serological diagnosis of mycoses. Clinically relevant antigens have been adapted for use in immunoassays, and the methods of detecting fungal antigens in body fluids have been progressively refined (5). Serodiagnosis relies on the detection of circulating antigens produced by Aspergillus fumigatus and other Aspergillus species. Galactomannan is one such antigen that can be measured in sera of patients with aspergillosis (4). Most serological tests use latex agglutination, but its sensitivity does not appear to be sufficient. To improve the sensitivity of galactomannan detection, a direct double sandwich enzyme-linked immunosorbent assay (ELISA) was designed (9).
PCR has been used successfully for the detection of specific Aspergillus DNA for the diagnosis of aspergillosis. For example, PCR has been used to detect DNA in bronchoalveolar lavage fluid of patients with invasive pulmonary aspergillosis (1, 10). However, bronchoalveolar lavage is not suitable for every patient and is even sometimes not recommended for certain patients, such as those with severe underlying diseases. The sampling of the bronchoalveolar fluid by using a bronchofiberscope is difficult in patients with neutropenia or thrombocytopenia after treatment with anticancer agents. The bronchofiberscopic examination is also hazardous in patients with hypoxia due to invasive pulmonary aspergillosis. Recently, a simpler diagnostic method has been established for the detection of specific DNA of Aspergillus species in serum samples by using the nested PCR assay in patients with invasive pulmonary aspergillosis (IPA) (11).
In the present study, we examined the usefulness of the PCR method for the detection of Aspergillus DNA in serum samples from patients with pulmonary aspergillosis. The results were compared with those obtained by immunodiagnostic methods, the sandwich ELISA or latex agglutination test for detection of galactomannan, and the clinical usefulness of these tests in the diagnosis of pulmonary aspergillosis was evaluated.
Participating patients were all those admitted to Nagasaki University Hospital between 1993 and 1996 and diagnosed with pulmonary aspergillosis (n = 44). The diagnosis of pulmonary aspergilloma was made based on the presence of a fungus ball detected on the chest X-ray, a positive result for aspergillus antibody in the serum, and isolation of Aspergillus spp. from the sputum or bronchial aspirate. The diagnosis of aspergillus pyothorax was made based on the isolation of Aspergillus spp. from a sample of pleural effusion. IPA was diagnosed by the histopathological findings for the lung or isolation of Aspergillus spp. from the sputum or sudden appearance of infiltration shadows in immunosuppressed patients, despite treatment with broad-spectrum antibiotics. The diagnosis of allergic bronchopulmonary aspergillosis (ABPA) was done according to the criteria reported by Rosenberg et al. (7). The diagnoses consisted of pulmonary aspergilloma (33 patients), ABPA (4 patients), aspergillus pyothorax (3 patients), and IPA (4 patients). Thirty-nine serum samples from patients with diagnoses other than pulmonary aspergillosis, consisting of lung cancer (six patients), pulmonary fibrosis (five patients), bacterial pneumonia (five patients), atypical mycobacteriosis (two patients), pulmonary cryptococcosis (six patients), and candidemia (two patients), and from patients with hemosputum (five patients) and healthy volunteers (eight volunteers) were used in this study for the evaluation for the specificity of PCR assay. Serum samples were stored at −80°C and thawed to 4°C within 12 h of testing.
Extraction of DNA from serum samples was performed according to the method described by Yamakami et al. (11). In the first step, 100 μl of the serum sample was combined with an equal volume of the lysis buffer containing 100 mM KCl, 20 mM Tris-HCl (pH 8.3), 5 mM MgCl2, 0.2 mg of gelatin per ml, and 0.9% polysorbate 20 solution. Proteinase K was added to a final concentration of 60 μg/ml. The mixture was incubated for 60 min at 55°C. Proteinase K was then inactivated by heating the mixture to 95°C for 10 min. The supernatant was used for PCR amplification following centrifugation at 12,000 × g for 10 min at 4°C. The oligonucleotide primer used in this study was based on the comparison of the sequences of 18S rRNA genes of Aspergillus species and other fungi deposited in the GenBank database. Nested PCR was performed with two sets of primers. The outer set consisted of M5c (5′-AGGGCACCACAAGGCGTGGA-3′) and M6b (5′-AAGAAGCCAGCGGCCCGCAA-3′), which together amplify a 209-bp sequence. The inner primer set consisted of M5cN (5′-GACTCAACACGGGGAAACTC-3′) and M6bN (5′-TAAGGGCC GAGGTCTCGTTC-3′), which together amplify a 136-bp sequence. The reaction mixture for PCR used in the present series of experiments consisted of 100 mM Tris-HCl (pH 8.3), 500 mM KCl, 25 mM MgCl2, 200 μM (each) deoxynucleotide triphosphates (dATP, dCTP, dGTP, and dTTP), 2.5 U of Taq DNA polymerase (AmpliTaq DNA polymerase; Perkin-Elmer), 30 pmol of each primer, and a DNA template solution. In a single PCR step, 30 pmol of each outer primer was combined with 10 μl of the prepared sample to yield a final volume of 50 μl. PCR was conducted in an automatic thermal cycler (GeneAmp PCR System 9600; Perkin-Elmer).
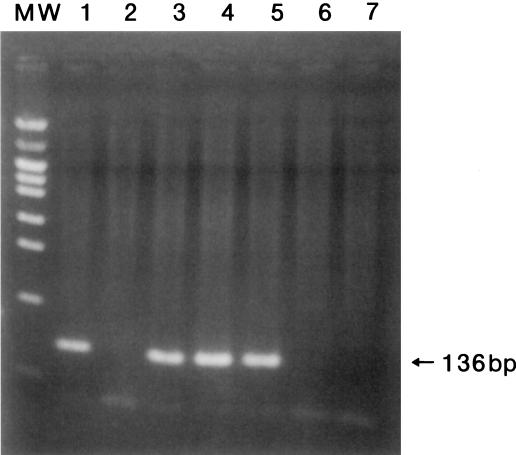
PCR was performed under the following conditions: denaturation at 98°C for 15 s, annealing at 57°C for 30 s, and extension at 72°C for 60 s for four cycles, and denaturation at 94°C for 15 s, annealing at 57°C for 30 s, and extension at 72°C for 60 s for 36 cycles. In the nested PCR step, 10 μl of the product obtained from the first amplification was added to a new reaction mixture with 30 pmol of each inner primer, and the process was performed under the following conditions: 30 cycles of 94°C for 15 s for denaturation, 60°C for 30 s for annealing, and 72°C for 60 s for extension. The total number of cycles in the first PCR was 40 with 30 additional cycles in the nested PCR. The total duration was 2.5 h in the first step plus 1.7 h in the nested step. To avoid possible contamination of the PCR mixture, all reactions were performed under stringent conditions as recommended by Kwok and Higuchi (3). The nested PCR products were electrophoresed on a 2% agarose gel containing ethidium bromide and photographed with a UV transillumination camera. A band of 136-bp size from the amplified PCR product was observed in agarose gel stained with ethidium bromide (Fig. 1).
FIG. 1.
A 2% agarose gel stained with ethidium bromide. Lane 1, positive control; lane 2, negative control; lanes 3 to 7, samples from patients diagnosed with invasive pulmonary aspergillosis, aspergilloma, aspergillus pyothorax, lung cancer, and pulmonary cryptococcosis, respectively. MW, molecular-weight marker.
Circulating Aspergillus galactomannan was detected by using two tests, a latex agglutination test (Pastorex Aspergillus; Sanofi Diagnostics Pasteur, Paris, France) and a sandwich immunocapture ELISA (Platelia Aspergillus; Sanofi Diagnostics). For both techniques, immune complexes were disrupted by adding 100 μl of treatment solution to 300 μl of undiluted serum in a tightly sealed Eppendorf tube which was heated to 100°C for 3 min and then centrifuged at 10,000 × g for 10 min. The supernatant was used for both tests. The latex agglutination test was performed by following the manufacturer’s protocol. The latex reagent was added and mixed with the supernatant. The results were recorded. A galactomannan control (75 ng/ml) included with the kit was tested with each batch of serum. For the ELISA technique, all reagents were provided by the manufacturer. The sensitized microtiter plates were filled with 50 μl of horseradish peroxidase-conjugated monoclonal antibody Eb-A2, a rat galactomannan, as the capture and detector antibody, in conjugate buffer followed by 50 μl of the supernatant of test solution. Plates were incubated at 37°C for 90 min. After thorough washings (five times), the reaction was carried out by incubation for 30 min in darkness with 100 μl of buffer containing o-phenylenediamine. The optical density (OD) was read at 450 and 620 nm. The result for each serum sample was calculated with the following ratio: cut off index (I) = OD of the sample/OD of serum with threshold concentration. The results were classified as follows: I ≦ 1.5, positive; 1 ≦ I ≦ 1.5, grey zone; and I < 1, negative.
The results are summarized in Table 1. The sensitivity of the PCR assay was the highest among the three tests conducted with the same serum samples. Aspergillus DNA was detected by PCR in 31 of the 33 patients with aspergilloma, all 4 of those with IPA, 3 of the 4 patients with aspergillus pyothorax, and 1 of the 4 patients with ABPA. All 39 samples from the patients with diagnoses other than pulmonary aspergillosis and the volunteers showed negative results both in the PCR assay and for the detection of galactomannan.
TABLE 1.
Numbers of patients diagnosed with aspergillosis with serum samples positive for Aspergillus DNA detected by PCR or for galactomannan antigen detected by ELISA or agglutination test
| Diagnosis | No. of patients | No. of patients with positive result for:
|
||
|---|---|---|---|---|
| PCR | ELISA (Platelia Aspergillus) | Latex aggluti-nation test (Pastorex Aspergillus) | ||
| Aspergilloma | 33 | 31 | 21 | 8 |
| Aspergillus pyothorax | 3 | 3 | 0 | 1 |
| IPA | 4 | 4 | 4 | 4 |
| ABPA | 4 | 1 | 0 | 0 |
| Total | 44 | 39 | 25 | 13 |
The sensitivity of the nested PCR was the highest for the detection of Aspergillus DNA in serum samples from patients with pulmonary aspergillosis. Early diagnosis of Aspergillus infections is still difficult, since useful serological tests have not been established. In the clinical laboratory, the latex agglutination test of galactomannan antigen is routinely the most commonly used test. The test requires a concentration of at least 15 ng of circulating galactomannan per ml for a positive result (8). The latex agglutination test for detection of galactomannan is highly specific, but the sensitivity does not appear to be sufficient (9). Another drawback of the latex agglutination test is that it yields a positive result only during advanced stages of infection in most patients with invasive pulmonary aspergillosis, and it thus does not aid in early diagnosis of the infection.
The new double sandwich immunosorbent assay can detect galactomannan at less than 1 ng per ml (8) and has been found to be clinically useful for the early initiation of antifungal therapy and monitoring of treatment in patients with clinically documented IPA (6). In the present study, the sensitivity of the ELISA was higher than that of the latex agglutination test. The detection of galactomannan antigen by ELISA seems to be a more sensitive method than latex agglutination detection. However, the latex agglutination test results were positive in all four patients with IPA. All patients with IPA in our study had advanced disease, and therefore studies of samples from more patients at the early stages of infection with IPA are probably necessary before the usefulness of the ELISA for the diagnosis of IPA can be fully assessed.
The diagnosis of pulmonary aspergilloma is usually based on radiological observation of fungus balls and detection of anti-aspergillus antibody in the serum. Most cases of aspergilloma are thought to arise from colonization and proliferation of the fungus in a preexisting pulmonary cavity. The histopathological characteristics of aspergilloma include an intracavitary mass of tangled mycelia (with both dead and live fungal elements) (2). Our investigation including PCR and detection of galactomannan antigen in patients with pulmonary aspergilloma indicated that the colonizing aspergilli might sometimes proliferate and invade the bloodstream. The colonizing aspergilli are thought to cause a progressive destruction of lung tissue during a long period of colonization in patients with pulmonary aspergilloma.
In conclusion, the PCR assay of Aspergillus DNA in serum samples was the most useful of the three laboratory tests for the detection of pulmonary aspergillosis.
REFERENCES
- 1.Bretagne S, Costa J M, Marmorat K A, Poron F, Cordonnier C, Vidaud M, Fleury F J. Detection of Aspergillus species DNA in bronchoalveolar lavage samples by competitive PCR. J Clin Microbiol. 1995;33:1164–1168. doi: 10.1128/jcm.33.5.1164-1168.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glimp R A, Bayer A S. Pulmonary aspergilloma. Diagnostic and therapeutic considerations. Arch Intern Med. 1983;146:303–308. doi: 10.1001/archinte.143.2.303. [DOI] [PubMed] [Google Scholar]
- 3.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature (London) 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 4.Latge J P, Kobayashi H, Debeaupuis J P, Diaquin M, Sarfati J, Wieruszeski J M, Parra E, Bouchara J P, Fournet B. Chemical and immunological characterization of the extracellular galactomannan of Aspergillus fumigatus. Infect Immun. 1994;62:5424–5433. doi: 10.1128/iai.62.12.5424-5433.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Repentigny L. Serological techniques for diagnosis of fungal infection. Eur J Clin Microbiol Infect Dis. 1989;8:362–375. doi: 10.1007/BF01963470. [DOI] [PubMed] [Google Scholar]
- 6.Rohrlich P, Sarfati J, Mariani P, Duval M, Carol A, Saint M C, Bingen E, Latge J P, Vilmer E. Prospective sandwich enzyme-linked immunosorbent assay for serum galactomannan: early predictive value and clinical use in invasive aspergillosis. Pediatr Infect Dis J. 1996;15:232–237. doi: 10.1097/00006454-199603000-00011. [DOI] [PubMed] [Google Scholar]
- 7.Rosenberg M, Patterson R, Mintzer R. Clinical and immunologic criteria for the diagnosis of allergic bronchopulmonary aspergillosis. Ann Intern Med. 1977;86:405. doi: 10.7326/0003-4819-86-4-405. [DOI] [PubMed] [Google Scholar]
- 8.Stynen D, Goris A, Sarfati J, Latge J P. A new sensitive sandwich enzyme-linked immunosorbent assay to detect galactofuran in patients with invasive aspergillosis. J Clin Microbiol. 1995;33:497–500. doi: 10.1128/jcm.33.2.497-500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sulahian A, Tabouret M, Ribaud P, Sarfati J, Gluckman E, Latge J P, Derouin F. Comparison of an enzyme immunoassay and latex agglutination test for detection of galactomannan in the diagnosis of invasive aspergillosis. Eur J Clin Microbiol Infect Dis. 1996;15:139–145. doi: 10.1007/BF01591487. [DOI] [PubMed] [Google Scholar]
- 10.Tang C M, Holden D W, Aufauvre B A, Cohen J. The detection of Aspergillus spp. by the polymerase chain reaction and its evaluation in bronchoalveolar lavage fluid. Am Rev Respir Dis. 1993;148:1313–1317. doi: 10.1164/ajrccm/148.5.1313. [DOI] [PubMed] [Google Scholar]
- 11.Yamakami Y, Hashimoto A, Tokimatsu I, Nasu M. PCR detection of DNA specific for Aspergillus species in serum of patients with invasive aspergillosis. J Clin Microbiol. 1996;34:2464–2468. doi: 10.1128/jcm.34.10.2464-2468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]