Abstract
Climate change is altering the frequency and severity of drought events. Recent evidence indicates that drought may produce legacy effects on soil microbial communities. However, it is unclear whether precedent drought events lead to ecological memory formation, i.e., the capacity of past events to influence current ecosystem response trajectories. Here, we utilize a long-term field experiment in a mountain grassland in central Austria with an experimental layout comparing 10 years of recurrent drought events to a single drought event and ambient conditions. We show that recurrent droughts increase the dissimilarity of microbial communities compared to control and single drought events, and enhance soil multifunctionality during drought (calculated via measurements of potential enzymatic activities, soil nutrients, microbial biomass stoichiometry and belowground net primary productivity). Our results indicate that soil microbial community composition changes in concert with its functioning, with consequences for soil processes. The formation of ecological memory in soil under recurrent drought may enhance the resilience of ecosystem functioning against future drought events.
Subject terms: Microbial ecology, Carbon cycle, Climate-change ecology
Legacies of past ecological disturbances are expected but challenging to demonstrate. Here the authors report a 10-year field experiment in a mountain grassland that shows ecological memory of soil microbial community and functioning in response to recurrent drought.
Introduction
The present functioning of ecosystems is determined by the temporal sequence of events that shapes interactions within communities. Yet, when trying to understand current ecosystem dynamics, history is an often-neglected aspect, even though highly relevant, especially in the light of rising climatic variability brought about by global change. In this context, emerging theoretical frameworks and recent evidence point at the important role of ecological memory1, defined as ‘the capacity of past states or experiences to influence present or future responses of a community’2. Indeed, the increase in frequency of a disturbance can create a memory of that disturbance, modifying the structure and organisation of ecological interactions within an ecosystem3. As global climatic change increases the frequency of extreme events, the temporal dynamic of ecosystem responses becomes a vital information to predict climate change consequences across temporal and spatial scales. For example, microorganisms and microbially mediated processes dominating biogeochemical cycles4 can be severely affected by drought events, which may lead to substantial alterations of soil carbon (C) and nutrient cycling5. Globally, the temporal variability of precipitation will likely become larger than ever6, which may lead to a higher frequency of drought events. The implications of increased drought frequency on soil greenhouse gas emissions and plant nutrient availability, and thus for ecosystem services as a whole7, can be severe. Understanding if soil community composition can shift in response to changing conditions8 and how this can affect ecosystem dynamics is, therefore, crucial to better predict the future alterations of biogeochemical cycles.
A prerequisite to create the memory of a disturbance such as drought is the presence of material or information legacies9. While material legacy refers to biotic or abiotic structures of a system (e.g. seed dispersal, soil pH, etc.), information legacy represents species life-history traits of a community. Field studies on legacy effects of drought on the soil microbial community are scarce, which has led many conclusions to be drawn from laboratory incubations. Furthermore, drought is a generic term referring to water shortage, which can either be a chronic or a short but intense precipitation reduction. Previous studies have shown that both types of drought can cause changes to the soil microbial community structure10–13 and that chronic precipitation reduction showed no legacy effects after rewetting of soil12,14. On the contrary, when drought was simulated as an intense stress event, it had significant long-lasting legacy effects on the microbial community, altering soil–plant feedback with changes in plant growth, nutrient acquisition, belowground C allocation and ecosystem functioning11,13,15–17. Such legacy effects were found at both during a consecutive drought event and after drought was terminated13,15,16, although it is not known yet how long these effects can last. Some studies have also investigated long-term effects of chronic drought in the field, but used laboratory incubation to disentangle short-term vs long-term effects12,18, potentially missing complex field dynamics. Results from field experiments comparing long-term recurrent drought events to single drought events are still lacking.
Here, we report the investigation of in situ drought effects of a system subjected to 10 years of recurrent drought events compared to a single drought event on the soil microbial community composition and the processes it mediates. The overarching goal of this work was to assess the formation of an ecological memory in soil microbial processes and the presence of legacies in the soil microbial community (in terms of composition, physiological parameters and stoichiometry). We hypothesized that: (i) recurrent drought events would cause larger shifts in the microbial community composition, compared to a single drought event, and (ii) recurrent drought events would lead to an ecological memory response of microbially mediated ecosystem processes.
This study is part of a long-term field drought experiment in a mountain grassland in Austria19. At this field site, drought is repeatedly implemented as an intense stress event during the growing season by completely excluding precipitation for about 2 consecutive months. In 2017, we compared grassland plots exposed to summer drought for 10 consecutive years with newly exposed ones and ambient controls. This allows us to disentangle drought responses, driven by microbial community shifts, to long-term recurrent drought from short-term physiological responses. Microbially driven carbon (C), nitrogen (N), phosphorous (P) and sulfur (S) cycling is assessed using potential enzymatic activity of 10 different enzymes. In addition, we analyse microbial biomass stoichiometry, microbial growth and carbon use efficiency (CUE), and community composition based on phospholipid fatty acids (PLFAs) and amplicon sequencing approaches. Additional soil data available at this site for plots subjected to 1, 2, 3, 4 and 5 years of recurrent summer drought events (data obtained in 2011 and 2012) is also analysed to understand the temporal aspects leading to an ecological memory development. To our knowledge, this is the first study that provides an integrative analysis of an ecological memory formation caused by recurrent drought events on the soil biogeochemical cycles mediated by the soil microbial community. We further describe insights into microbial community response to long-term drought, and we consider the implications for soil functionality in response to climate change. Specifically, our work shows two key findings: (i) recurrent drought strengthens shifts in the composition of microbial communities; and (ii) soil multifunctionality is buffered in response to drought by previous recurrent drought events. We further suggest that microbial community shifts and changes in the microbially mediated cycling of carbon and nutrients likely reflect community-dependent strategies to acclimate to drought, and likely controlling soil functions.
Results
Drought does not affect measured edaphic factors but modifies available C and N
Soil pH and total soil C, N and P contents (and their respective ratios) did not differ significantly between treatments (Supplementary Table 1). The experiment reduced soil water content throughout the experimental period (Supplementary Fig. 1, Supplementary Table 1 and Supplementary Fig. 2), with no differences between plots subjected to 10 years of recurrent drought events (10-year treatment) and plots exposed to a single drought event (1-year treatment). Similar results were obtained in the previous campaigns that were carried out in 2011 and 2012 (Supplementary Figs. 1 and 2). Independent of the number of drought events, extractable organic C (EOC) concentrations were higher compared to ambient controls. Among the measured available N pools, only NH4+ showed significant differences between treatments, where values followed the order (1 year > 10 years > control). Extractable inorganic and organic P (EIP and EOP) were not significantly affected by drought (Supplementary Table 1).
Ten years of recurrent drought events cause legacy effects on microbial biomass stoichiometry and community composition
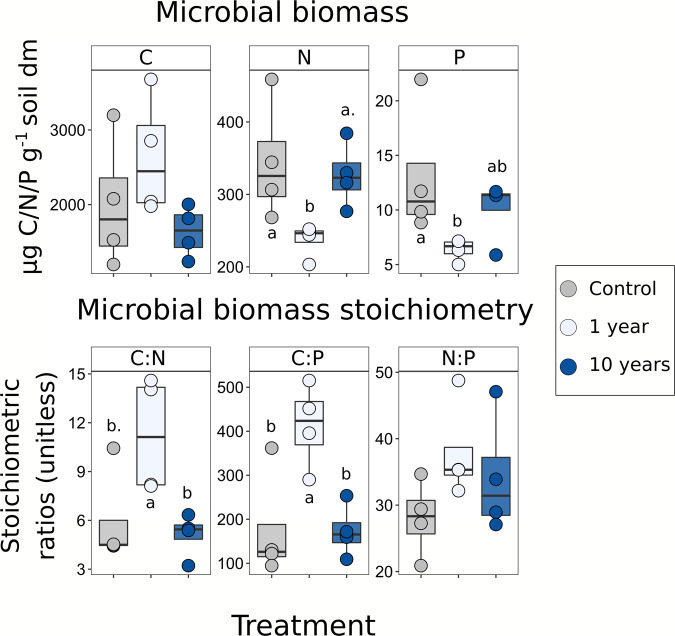
The magnitude and direction of responses of the microbial biomass and its stoichiometry differed significantly between the 1-year and 10-year treatments. A similar trend was found for all the measured variables where the 1-year treatment diverged from control values while the 10-year treatment had similar values to control. This trend was found significant for microbial biomass N (MBN, p = 0.025) and microbial biomass P (MBP, p = 0.046), whereas microbial biomass C (MBC) was not significant (Fig. 1, top panels). MBC and MBN were also analysed for the previous campaigns (2011 and 2012), with no clear drought effect up to 5 years of recurrent drought events (Supplementary Fig. 3). The ratios of C:N and C:P in microbial biomass significantly increased (p = 0.016 and p = 0.003, respectively) in response to the single drought event (1 year) but not after 10 years of recurrent drought, when compared to ambient plots (Fig. 1, bottom panels). The microbial N:P ratio followed a similar, although not significant, pattern.
Fig. 1. Drought effects on microbial biomass C, N and P.
Colour indicates soil treatment (control = grey, 1 year = light blue, 10 years = blue). The top graphs show microbial biomass carbon (MBC), nitrogen (MBN) and phosphorous (MBP). MBC was not significantly affected by drought (F-value: 2.02, p = 0.18), while MBN and MBP were (F-value: 5.7, p = 0.04, n = 4; F-value: 4.44, p = 0.06, n = 4). The same pattern was present in all the three values, where control and the 10-year treatment had similar values and the 1-year treatment was different. The bottom graphs show the stoichiometric ratios of microbial biomass. The ratio of C-to-N and C-to-P show similar results, where the 1-year treatment was significantly different from both control and 10 years (respectively, F-value: 5.72, p = 0.0249, n = 4; F-value: 8.06, p = 0.0098, n = 4). The ratio of N-to-P was not statistically affected by drought, and the 10-year treatment values ranged between the 1 year and control. Letters represent results from two-sided Tukey HSD post hoc test (a point next to letter represents a p-value on the 0.05 threshold). Box centre line represents median, box limits the upper and lower quartiles, whiskers the 1.5x interquartile range, while separated points represent outliers. The sample size ‘n’ represents biologically independent samples.
Although total microbial biomass (sum of all PLFA biomarkers), remained constant in all drought treatments compared to controls (Supplementary Table 2), drought had a clear impact on the microbial community composition (PERMANOVA: R2 = 0.38; p < 0.05; Supplementary Fig. 4), mostly driven by the 10-year treatment. Indeed, the 10-year drought clearly separated from the control, whereas the microbial community in the 1-year treatment overlapped with both control and 10-year treatment. Results from previous campaigns show that drought did not result in a significant difference between drought treatments and control in the 1-, 2- and 5-year treatments (Supplementary Fig. 5). The separation towards control plots was mostly driven by biomarkers belonging to arbuscular mycorrhizal fungi (AMF), Gram-positive and Gram-negative bacteria (e.g. a17:0, i17:0, 16:1ω5), while shifts in drought plots were related to fungal biomarkers (e.g. 18:2ω6,9). When individual biomarkers were used to estimate the biomass of main microbial groups, significant effects of drought were found only in Gram-negative and Gram-positive bacteria (p = 0.011 and p = 0.016; Supplementary Table 2) and in protozoa (p = 0.001; Supplementary Table 2), with a significant decrease. The NLFA (neutral lipid fatty acid) biomarker 16:1ω5 was assessed as an indicator of AMF biomass and no statistical difference was found between treatments and control (Supplementary Table 2).
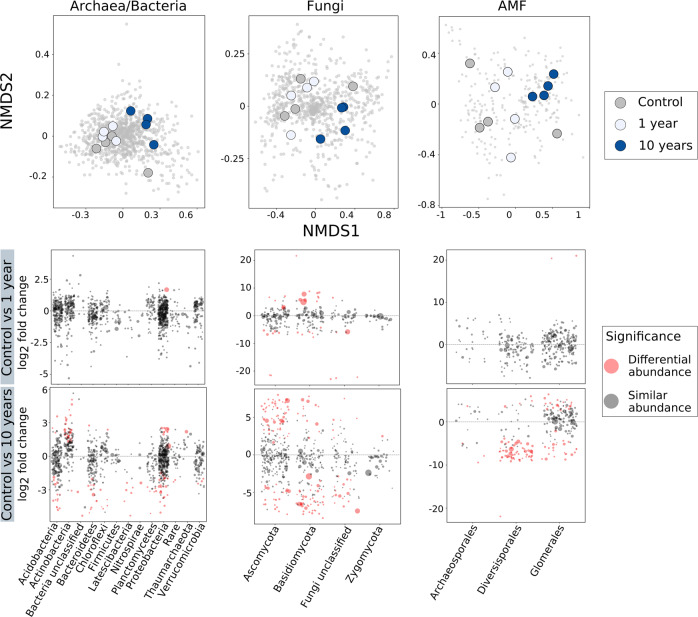
A more detailed picture of the microbial community response to drought, based on amplicon sequencing, revealed that 10 years of recurrent drought events had caused a significant microbial community shift, clearly separating it from single drought event (1 year) and control (Fig. 2, top panels), whereas there was no clear separation between control and 1 year. We analysed the V4 region of the SSU rRNA genes (for bacteria and archaea), the ITS1 region (for fungi in general) and AMF-ITS2 rDNA (for AMF). For all, bacteria, archaea, fungi and AMF, a significant effect of drought was found (PERMANOVA test: p = 0.014, p = 0.01, p = 0.015, respectively). To confirm the results from ITS1, the ITS2 region was also analysed showing very similar results (Supplementary Fig. 6). Differential abundance analysis further confirmed (Fig. 2, bottom panels) that the 10-year treatment led to a much higher number of differentially abundant taxa (compared to the control) than the 1-year treatment. The 10-year treatment reduced the relative abundances compared to controls of the groups: Acidobacteria, Bacteroidetes, Basidiomycota, fungi unclassified and Diversisporales, whereas Actinobacteria, Ascomycota and Glomerales showed higher relative abundance and Proteobacteria showed both higher and lower relative abundance (Supplementary Figs. 7, 8, 9, 10). No clear patterns were evident in the α-diversity indices (Richness, Chao1, ACE, Shannon and Inverse Simpson index) between treatments (Supplementary Fig. 11). We further analysed Legacy Response Groups (LRGs) and show the relative abundance of the main taxonomic groups that were either enriched or depleted in the 10-year vs the 1-year treatment (Supplementary Fig. 12). We specifically selected these two treatments to disentangle long-term vs short-term effects of drought. We found that most phyla (or orders) contributed at the same time to both types of LRGs (enriched and depleted groups). The main relative abundance taxa in the enriched LRGs belonged to the phyla Latescibacteria, Proteobacteria, Acidobacteria and Thaumarchaeota, within the bacterial and archeal community. Ascomycota followed by Basidiomycota represented the major enriched LRGs in the fungal community and the order Glomerales in the arbuscular mycorrhizae community.
Fig. 2. Drought effects on microbial community composition assessed by amplicon sequencing.
Colour indicates soil treatment (control = grey, 1 year = light blue, 10 years = blue). On top, graph NMDS representing archaea/bacteria, fungi and AMF (arbuscular mycorrhizal fungi). All the NMDS plots show the similar patterns, with an effect of drought (respectively, p = 0.014, p = 0.01, p = 0.015; PERMANOVA results, n = 4) and with a clear separation of the 10-year treatment from 1 year and control. The bottom graphs show results from the differential abundance analysis for control vs 1 year (top row) and control vs 10 years (bottom row). Circles in red represent taxa that had a significant differential abundance compared to the control, while in grey taxa that were not significant. Similarly to the NMDS plots, control vs 1 year results show little difference (only in the fungal community of Basidiomycota), while control vs 10 years shows strong differences at all levels (most affected groups being: Acidobacteria, Actinobacteria, Proteobacteria, Ascomycota, Basidiomycota, Diversisporales and Glomerales).
Ecological memory formation in the microbially mediated cycling of C and nutrients
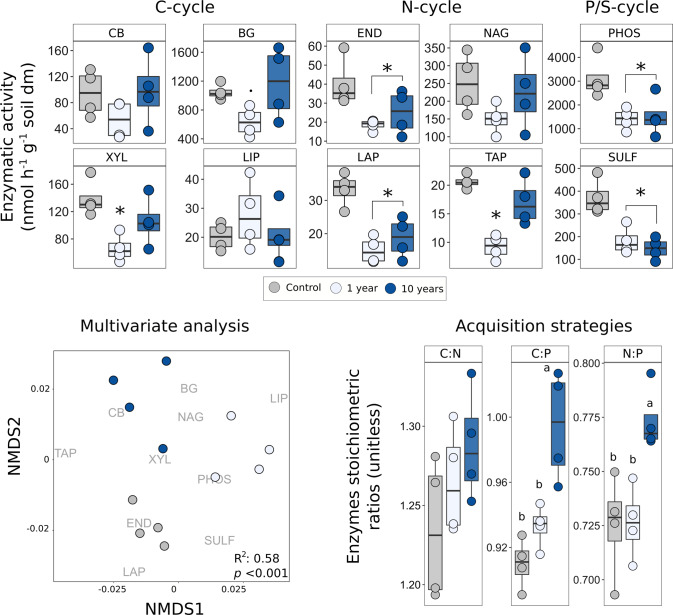
To analyse whether repeated droughts changed the potential of the microbial community functions and nutrient acquisition strategies in response to drought, we analysed a suite of extracellular enzymatic activities categorized as C-, N-, P-, S-cycle-related enzymes (see Supplementary Table 3). The enzymes were classified as C-related, when the enzymes were involved in the cycling of molecules that did not contain N, P or S, or related to one of the nutrients when involved in the depolymerization of molecules that contain N, P or S. It should be noted that, as all organic molecules contain C, it is impossible to completely separate the acquisition of C from the acquisition of nutrients contained in organic molecules (as in N-, P- and S-related enzymes, Supplementary Table 3). The drought treatments caused enzyme-specific responses (Fig. 3, top panels). C-cycle-related enzymes showed consistent patterns where the 10-year treatment had values always similar to the control, whereas the 1-year treatment was always different from both (although only β-xylosidase showed a significant effect of drought; p = 0.016). Similarly, in the N-cycle enzymes the 1-year drought treatment caused stronger reductions than the 10-year drought treatment when compared to the control (significant effects of drought were found for three enzymes: Endochitinase, Leucine aminopeptidase and Tyrosine-aminopeptidase, p = 0.03, p = 0.001 and p = 0.0006, respectively, Fig. 4). In contrast, S- and P-cycle enzymes showed a significant decrease in activity to both 1-year and 10-year treatments, compared to the control (Fig. 4). Enzymatic activity was also expressed per MBC, which led to very similar conclusions (Supplementary Fig. 13). Similar trends were also found in the previous sampling campaigns (2011 and 2012), for which we observed a statistically significant linear trend of Cellobiosidase towards control values with increasing years of drought (Supplementary Fig. 14), whereas Phosphatase, Endochitinase and Leucine aminopeptidase did not show a relationship with number of years of drought.
Fig. 3. Effects of drought on the potential enzymatic activity in soil.
Colour indicates soil treatment (control = grey, 1 year = light blue, 10 years = blue). Top graphs show the potential enzymatic activities of all measured enzymes, grouped for C-, N-, and P- and S-cycle-related enzymes (for full description of enzymes see Supplementary Table 1). C-cycle enzymes all show a similar trend, where 10 years have similar values to the control, and the 1 year is always different (only statistically significant in XYL: F-value = 6.68, p = 0.016, n = 4; and partially significant in BG: F-value = 3.2, p = 0.089, n = 4). N-cycle enzymes show different responses of the 10-year treatment, where, in two enzymes both drought treatments show a decrease (END: F-value = 4.9, p = 0.036, n = 4; LAP: F-value = 14.58, p = 0.0015, n = 4), and in the other two only the 1-year treatment shows decreased values, while 10 years show values similar to control (only significant in TAP: F-value = 18.79, p < 0.001, n = 4). P- and S-cycle enzymes show the same trend where both drought plots are significantly different from the control (PHOS: F-value = 6.53, p = 0.018, n = 4; SULF: F-value = 14.75, p = 0.0014, n = 4). Stars represent significant differences with the Tukey HSD post hoc test while a dot represents partially significant values (p < 0.1). Bottom left panel shows the NMDS of potential enzymatic activities for the year 2017, with a clear separation between all treatments (R2 = 0.58, p = 0.0003, n = 4, stress = 0.13). The bottom right panel represents acquisition strategies (stoichiometric ratios of the potential enzymatic activities), where a similar trend was found for all the ratios, although only statistically significant in C:P and N:P (respectively, F-value = 14.8, p = 0.001, n = 4; F-value = 8.6, p = 0.008, n = 4). Letters represent results from two-sided Tukey HSD post hoc test. Box centre line represents median, box limits the upper and lower quartiles, whiskers the 1.5x interquartile range, while separated points represent outliers. The sample size ‘n’ represents biologically independent samples.
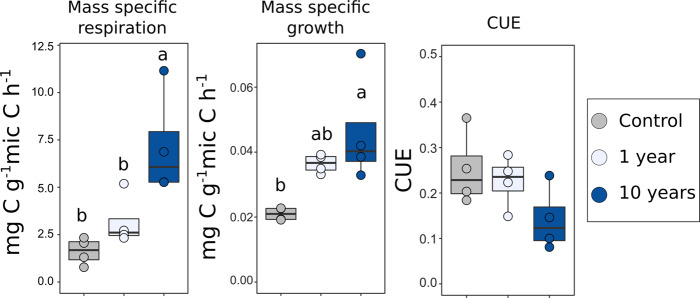
Fig. 4. Physiological parameters measured upon rewetting.
Colour indicates soil treatment (control = grey, 1 year = light blue, 10 years = blue). Drought had a significant effect on mass-specific respiration and growth rates (respectively, F-value = 9.67, p = 0.006, n = 4 and F-value = 14.55, p = 0.0015, n = 4), whereas it had no significant effect on CUE. The 10-year drought was significantly different from 1-year drought in mass-specific respiration rates. Letters represent results from two-sided Tukey HSD post hoc test. Box centre line represents median, box limits the upper and lower quartiles, whiskers the 1.5x interquartile range, while separated points represent outliers. The sample size ‘n’ represents biologically independent samples.
When enzymes were analysed in a multivariate analysis (non-metric multidimensional scaling, NMDS), drought had an overall significant effect across all potential enzymatic activities (PERMANOVA test p < 0.001; Fig. 3, bottom left panel), where responses to the 1 and 10 years of recurrent drought were clearly separated from each other as well as from the control. Treatment separation corresponded to specific enzymes, where control was related to the Endochitinase and Leucine aminopeptidase, the 10 years was related to Cellobiosidase, β-Glucosidase and Exochitinase, and the 1 year to Phosphatase and Lipase. In previous campaigns (2011 and 2012) we also observed a separation of treatments from control plots (although marginally significant), with no clear separation within the different drought treatments of 1, 2, 3, 4 and 5 years of recurrent droughts (Supplementary Fig. 14, bottom graphs).
We analysed the nutrient acquisition strategies of the overall community, calculated as stoichiometric ratio of enzymes. The C:N:P acquisition ratio is an integral feature to describe soil microbial community functions, linking environmental nutrient availability to the C:N:P stoichiometry of microbial biomass20–22. We found that 10 years of recurrent drought increased C:P and N:P acquisition ratios (p = 0.001 and p = 0.008, respectively) and a similar trend was also found for C:N (although not significant), whereas acquisition strategies were not affected by the 1-year treatment and remained similar to controls (Fig. 3, bottom right panel).
Shifts in microbial physiology and belowground multifunctionality in response to drought
We measured parameters related to microbial physiology (growth, respiration and CUE). Due to methodological reasons, it was necessary to add water to measure these parameters and therefore they represent the physiological rewetting response of the microbial communities subjected to drought. We found an increase in mass-specific respiration rates and growth rates (p = 0.006 and p = 0.017, respectively), but not in the CUE (Fig. 4). The 10-year drought led to average values of all parameters different from control and 1-year drought (Fig. 4), with lower CUE values and higher respiration and mass-specific growth rates, although only mass-specific growth rates were statistically significant.
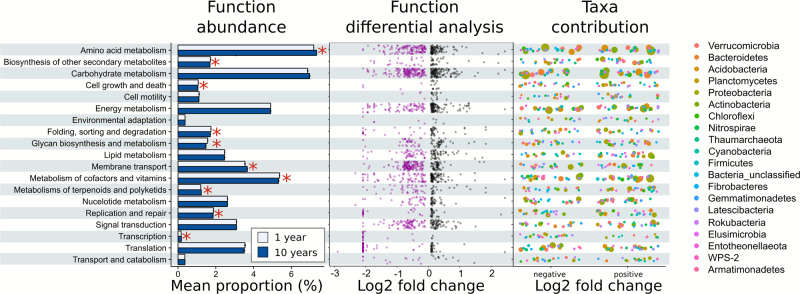
To obtain a substrate-independent measure of the effects of drought on microbial function and physiology, we predicted the functional potential of the soil bacterial community based on 16S rRNA gene sequencing using PICRUSt223. We specifically aimed at differentiating the response of the first (1 year) to the tenth (10 years) recurrent drought, in order to (i) disentangle shifts due to previous drought history from simple responses to drought, and also because (ii) there were very little differences between communities in control and 1-year treatment (see Fig. 2). We found that the abundance of major group of functions, such as amino acid metabolism, membrane transport, metabolism of cofactors and vitamins, and cell growth and death differed between the 1-year and 10-year treatments (Fig. 5, upper left graph). However, discussing broad classes of functions may be misleading as they represent multiple genes whose expression was concomitantly decreased and increased (Fig. 5, upper middle graph). For example, while we found no overall change in the total number of genes related to carbohydrates metabolism, hundreds of functions were both enriched and depleted (Fig. 5, middle panel). Therefore, we further explored individual gene responses, especially for processes known to be involved in the improvement of drought tolerance. We found that the number of genes related to the production of osmolytes (glycine betaine, glutamate, proline and choline), including enzymes such as betB, betI, gbsA and proA were enriched. A gene inducible via osmotic stress encoding a membrane transporter for K+ uptake (kdp) was found to be decreased in the 10-year drought; while the genes trkG and trlH, which are also involved in K+ uptake but are constitutive genes, were enriched. Other genes that are responsive to osmotic stress were found to be enriched, including ompC and ompF which express porins that facilitate the non-specific diffusion of small (≤500 Da) hydrophilic molecules24. We also found an enrichment of genes connected to the capsular or extracellular polysaccharides (EPS) metabolism (alg7, pslB and wbpW)25,26, although some were also reduced (e.g. pgaB). Moreover, we observed an enrichment in genes involved in peptidoglycan biosynthesis, like the murA and murB genes27. These constitute a shield for bacterial cells from abiotic stresses and help absorbing water28. Drought-tolerant bacteria also respond to low water activity by increasing the proportion of negatively charged phospholipids, including phosphatidylglycerol lipids, and we found that bacterial community in the 10-year treatment had higher relative abundance of gene encoding for the phosphatidylglycerophosphatase (pgpA gene). Lastly, ROS scavengers were observed to be enriched, like genes for enzymes belonging to the superoxide dismutase (gene SOD2) or catalase families. We also found evidence that bacterial communities may have increased their investment in resource acquisition traits to degrade more chemically complex and diverse substrates (such as the enrichment of genes expressing amylases, chitinases and enzymes related to lignin-degradation such as vdh), at the expense of pathways related to more simple compounds (glucose, fructose, mannose, etc.). We further used PICRUSt2 outputs to predict the contribution of amplicon sequence variants (ASVs) to individual functional categories. We observed that different microbial members of individual phyla contributed concomitantly in a positive or negative way to each predicted function.
Fig. 5. Potential functional capabilities and acquisition strategies of soil microbial communities.
The three graphs show results from PICRUSt2 comparing 1-year to 10-year drought. This revealed potential enrichment or depletion (respectively, positive and negative Log2 fold changes) of many functional traits of the microbial community related to important functions in response to drought history. The left graph shows the mean proportion of level 2 KEGG Orthologies within the bacterial community for the 1-year and 10-year treatments. Colour indicates soil treatment (1 year = light blue, 10 years = blue). Red asterisks indicate significant differential abundances of the overall group. The middle graphs represent the number of functions that were enriched (black circles) or depleted (purple circles) using DESeq2 (respectively, positive and negative Log2 fold changes). The right graph represents the microbial groups (bubble size represents bigger or smaller frequency of ASVs in each group of function within each phylum) contributing to enriched or depleted functions (respectively, positive or negative). Individual phylum colour are described in the figure.
Conclusions drawn from PICRUSt2 analyses should be interpreted cautiously as these represent a prediction of the functional potential of a community inferred from Illumina amplicon sequencing data. Nevertheless, our communities were well-characterized with regard to placement of ASVs in the per-default reference tree (containing 20,000 full-length 16S rRNA gene sequences), as indicated by the weighted nearest sequenced taxon index (NSTI). We found highly similar NSTI values over all the replicates and treatments ranging from 0.212 to 0.247, which was within the suggested range for well-characterized communities (0.1–0.523). This allowed us to carefully interpret the predicted metagenomes for potential functions and phyla that may react to repeated drought. We did not predict the functional potential of fungal communities due to the high variability of the ITS regions and previous findings indicating that predictions are just slightly better than random for fungal datasets23.
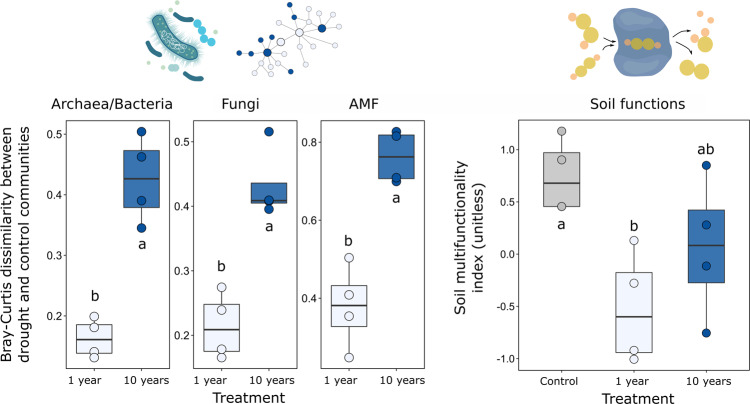
We further calculated two indices to summarize and quantify the changes in soil microbial community and soil functions. First, we calculated the Bray-Curtis dissimilarity index between drought treatments and control for each analysed microbial community group, as a quantitative indication of treatment differences between microbial communities (Fig. 6, left panels). The graph shows a clear increase in distance from the control with recurrent drought events. Second, we calculated a soil multifunctionality index to quantify the provision of multiple soil processes which are directly linked to ecosystem services29. This index is not the expression of a single parameter, but we utilized a wide set of soil parameters (enzymatic activities, soil C and nutrient stock, microbial biomass C, N and P, and root biomass productivity) to assess the response of multiple soil functions to recurrent drought events (Fig. 6, right panel). We show that a single drought event causes a strong decrease in soil functions, while with 10 years of recurrent drought events the index becomes more similar to ambient controls.
Fig. 6. Effects of drought on microbial community and soil functions.
Colour indicates soil treatment (control = grey, 1 year = light blue, 10 years = blue). Left panels show boxplots representing the Bray-Curtis dissimilarity index between drought treatments and control for archaea/bacteria (F = 44.969, p = 0.0005, n = 4), fungi (F = 32.86, p = 0.012, n = 4) and AMF (F = 23.002, p = 0.003, n = 4). The right panel shows boxplots representing the soil multifunctionality index (calculated by a wide set of soil parameters including enzymatic activities, soil C and nutrient stock, microbial biomass C, N and P and root biomass productivity) grouped by treatment (F = 5.514, p = 0.027, n = 4). Letters represent results from two-sided Tukey HSD post hoc test. Box centre line represents median, box limits the upper and lower quartiles, whiskers the 1.5x interquartile range, while separated points represents outliers.
Discussion
The concept of ecological memory is central to ecological theory. It determines an ecosystem’s response trajectory after recurrent stress events. Evidence of ecological memory references to a wide range of ecosystems and organisms subjected to climate change-related stress events30–32. With drought frequency and severity likely going to increase in many areas of the world6, knowing how ecosystems will respond to recurrent drought events is crucial. Most studies have focused on short-term effects of drought, while emerging evidence has highlighted the possible induction of legacy effects15,16,18,33, which are the prerequisite of ecological memory formation. Here we unravel ecological memory formation on microbially mediated soil functions, upon long-term recurrent drought in situ. Specifically, we show the development of biotic legacies, i.e., changes in the microbial community composition and effects on microbially mediated soil functions, with likely repercussions for the whole ecosystem.
We found a large shift in the community composition of bacteria, archaea and fungi, when exposed to 10 years of recurrent drought events, but not for a single drought event. This shift was confirmed by both PLFA analysis and amplicon sequencing of the 16S rDNA and ITS rDNA, at all investigated levels of the community (archaea, bacteria, fungi and AMF). This separation of the community from the control was developed only after multiple recurrent droughts, showing no clear effects up to 5 years of recurrent events (PLFA data only, Supplementary Fig. 5). The formation of an ecological memory relies on either information or material legacies8,17. Information legacies are difficult to evaluate in complex communities such as soil microorganisms with no a priori knowledge of the various organismic interactions, and were not analysed. Material legacies can be divided into abiotic and biotic. At our experimental site we did not find persistent changes in edaphic factors (soil pH, soil C and soil nutrient stocks), supporting the notion that it is a biotic legacy of drought that causes divergence from the expected responses to drought8. While we did find strong biotic legacies, in that we found strong shifts in the microbial community composition, we cannot completely exclude the influence of abiotic legacies, as there could be other relevant edaphic factors that we did not measure. It is also important to highlight that we did not assess whether the microbial community has reached a new stable state or whether community changes emerged only during the period of drought. Many examples of abrupt transitions to alternative states triggered by climate extremes exist34, although their implications for ecosystem functioning are not known.
It was recently shown that bacterial community’s changes in response to drought are consistent across many studies, indicating that the context of a particular location does not affect the phylogenetic pattern of response35. Drought resistance involves deeply conserved traits such as osmolyte production, cell wall features or spore formation36, which are present in similar groups within soil microbial communities. Indeed, similar to our study, Actinobacteria have previously been found to increase (or show no changes) in relative abundance with drought and Proteobacteria to decline35,37–40. Our results were also similar to previous reports on fungi, where Ascomycota were found to increase and Basidiomycota to decrease, indicating that they are likely more sensitive to drought33,41. These changes only became evident after 10 years of recurrent drought. We also found that AMF showed a trend in decreasing biomass in response to drought, similar to previous studies42, but without any change in composition in single drought42–44, whereas a clear shift was visible after 10 years of recurrent drought. This was especially evident in the fungi of order Diversisporales, which were strongly reduced. This result is particularly relevant given the important role of AMF in system productivity and C cycling45. We identified multiple LRGs (Supplementary Fig. 12), indicating phylogenetic groups that responded differently to a subsequent drought, when previously exposed to drought events. All the phyla (or orders) identified as LRGs showed both increased and decreased taxa. The increased taxa represented a large proportion of the total relative abundance, although this should be interpreted cautiously, as the decrease in relative abundance of some groups inevitably determines the increase of others.
Concomitantly with a biotic legacy of drought via altered soil microbial communities, we observed an ecological memory effect on the potential enzymatic activities, which only emerged after 10 years of recurrent drought events. Indeed, other treatments showed emerging trends, but no real ecological memory formation up to 5 years of recurrent drought events. Similar results were also found for microbial biomass stoichiometric ratios, showing an ecological memory effect. We explain these results via shifts of the microbial community composition in response to drought. After a single drought event we did not find indications of acclimatization of the microbial community structure and function (i.e., a similar composition and strongly altered functions compared to the control). Consequently, the community was unable to cope with the drought stress, which resulted in changes in biomass stoichiometry, as previously shown46, with no changes in the overall biomass. This is most likely because major pools of C- and N-based microbial metabolites are dynamic in their response to short-time drought, in which MBC was shown to increase and microbial amino acid content to decrease47. In contrast, when the microbial community had already repeatedly experienced drought events (i.e., in our 10 years recurrent drought treatment), the composition was significantly altered (representing a biotic legacy) and was better able to cope with drought, as evidenced by the functional capabilities which were more similar to controls than in the single-drought microbial community. We support this with a set of analyses related to community traits. First, we observed an effect of long-term recurrent drought on stoichiometric acquisition strategies. The C:N:P acquisition ratio represents an integral feature to describe soil microbial community demand, linking environmental nutrient availability to the C:N:P stoichiometry of microbial biomass20–22. We found that long-term recurrent drought events increased the acquisition strategy towards C- and N-compounds compared to P, while exposure to single drought maintained acquisition strategies similar to control levels. Community-level physiological parameters also revealed a memory effect, where the microbial community re-exposed to long-term drought showed faster growth and respiration rates and lower CUE values upon rewetting than those subjected to a single drought. This indicates that a shift in microbial community was selected by repeatedly occurring drought stress, allowing microbial communities to re-activate faster upon rewetting of soil.
We also analysed the functional potential of bacterial traits associated to drought resistance. We compared the effects of a single drought with 10 years of recurrent drought to disentangle short-term effects from long-term effects. The community exposed to 10 years of recurrent drought clearly revealed a shift in the bacterial community to being more tolerant to drought, indicated through a range of genes connected to amino acid, carbohydrate and lipid metabolism as well as membrane transporters and other protein families. In general, bacterial cells resist drought stress by decreasing energy consumption, preventing water loss and increasing water retention, and by protecting DNA and preventing protein damage through the accumulation and expression of osmoprotectants28,48. We found evidence of multiple genes connected to all these functions being enriched in the bacterial community subjected to 10 years of recurrent drought, which is indicative of a more drought-tolerant bacterial community. Indeed, previous studies have shown that microbial communities that acclimatize to drought usually enhance osmolyte production49,50, synthesis of capsules and exopolymeric substances to retain water28,51, dormancy and sporulation52. Recent evidence from a long-term drought experiment demonstrated that litter microbial community’s physiological response to drought shows a trade-off against growth, by redirecting investment from central metabolism to stress-coping mechanisms52. Within the bacterial community, we did not find a shift in the phyla contributing to such functions. Rather, we found that phyla could be connected to both enrichment and depletion at the same time (as previously shown35), indicating that the phylum does not represent a high enough level of resolution for identifying bacterial life strategies and responses to stressors53,54.
The results of this study illustrate several implications for ecosystem responses to drought. To summarize belowground responses to 10 years of recurrent drought as compared to a single drought event, we calculated a soil multifunctionality index which combines the expression of multiple microbial processes and functions11,55, including activities of 10 extracellular enzymes, soil and microbial biomass nutrients and root biomass productivity. We found that soil multifunctionality is buffered against drought effects when the system is subjected to a history of drought (Fig. 6). Given the pivotal importance of soil microbial communities for multiple ecosystem functions and services56, our results can have several implications. For example, it was shown that plant fitness is strongly linked to the rapid responses of soil microbial community structure to drought57, and that changes in the relative abundance of specific bacterial taxa are associated with increased drought tolerance of plants58. Moreover, the contribution of heterotrophic CO2 emissions from soil can become dominant during periods of drought followed by rewetting pulses59 and can account for up to 60% of net ecosystem CO2 release during a dry season60. As recently proposed, incorporating microbial community composition into ecosystem process models will help in predicting ecosystem responses to disturbances. This may be especially true for disturbances or other deviations from steady-state conditions, such as drought events61. Increasing climate variability6 will certainly add a further layer of complexity in predicting ecosystem response to disturbances and should be considered more explicitly in future studies.
The biotic legacy that we found at the microbial community level in our study, may be also connected to biotic legacies within the plant community. Indeed, a strong link exists at the root interface between plant and soil microbial community62. This is, for example, supported by the strong changes we observed in the arbuscular mycorrhizae community, which depends to a large extent on plants. At our experimental site, the plant community was indeed altered in response to recurrent droughts (Bahn et al. 2020, unpublished information). However, the influence between plant and microbes is bi-directional, where plants can shape the soil community and its drought response63 and in turn the soil microbes can have important consequences for the plant diversity, community composition and survival64,65. Therefore, it is not possible to conclude to which extent legacy effects in the plant community could determine legacies in soil microbes and vice versa. Drought can also modify plant C inputs into soil. For example, drought can alter the quantity and quality of root exudates66,67, and legacy effects have been shown to cause a reduction of belowground C inputs16. Plant litter chemistry might also be affected by direct drought effects and via changes in plant community caused by long-term drought68, which in turn might alter soil microbial communities69,70. A recent study showed that plant litter chemistry constrains microbial community gene expression and metabolite production associated with drought stress tolerance52. Although plant C input was not the focus of this experiment, plant community dynamics (and associated belowground C inputs) are thus likely an important player in the response of microbial communities to drought.
As drought events become more frequent, the response of an ecosystem to each new disturbance will likely be contingent on the history of precedent extreme events. Temporal phenomena are fundamental to ecology, as the enduring influences of past experiences and changing conditions often unfold over time71,72. In this context, ecological memory, i.e., ‘the capacity of past states or experiences to influence present or future responses of a community’2, is critical to predict a disturbance effect on ecosystems. The memory effect is transmitted as legacies of species and materials, which are prerequisites. We here examined the extent to which a soil microbial community subjected to long-term recurrent drought diverges from the response to a single drought event. Via a tailored experimental setting and repeated sampling campaigns, we were able to disentangle the effects of single drought vs recurrent drought events and assess whether ecological memory formation, induced by recurrent drought events, expresses during a subsequent drought. We show two key findings: (i) recurrent drought strengthens shifts in the composition of both bacterial and fungal communities; (ii) soil multifunctionality is buffered in response to drought by previous recurrent drought events. We further suggest that microbial community shifts and changes in the microbially mediated cycling of carbon and nutrients likely reflect community-dependent strategies in response to drought and likely controlling soil functions.
This study demonstrates how reinforcing interactions between consecutives stress events may affect the response trajectory of ecosystems to climate change. Such information is of utmost importance for understanding climate change feedbacks. Specifically, long-term exposure to recurrent drought events can select for a more tolerant soil microbial community and create a memory effect, modifying soil processes that mediate and enhance the resistance of soil functions to drought.
Methods
Site description
The study site is located in the Austrian Central Alps near Neustift, Stubai valley (47°07’45″N, 11°18’20″E; Supplementary Fig. 15) at 1850 m a.s.l. It is part of a mountain meadow that is annually cut for hay production at peak biomass (late July/early August) and fertilized every 3–4 years. The soil is a Dystric Cambisol (Food and Agriculture Organization-soil classification system) with a pH of 5.4 (in CaCl2). The vegetation is dominated by highly productive perennial grasses and herbs including Anthoxanthum odoratum, Festuca rubra, Alchemilla vulgaris, Leontodon hispidus and Trifolium repens and the meadow is generally characterized by a comparatively high primary productivity and high soil CO2 efflux rates73–75. The mean annual temperature and the mean annual precipitation are 3 °C and 1100 mm, respectively19,73–76.
Sampling campaigns and experimental set-up
Samples were collected from a unique drought experiment in a mountain grassland that was initiated in 2008. From 2008 to 2017 drought was simulated annually by equipping plots with rainout shelters (3 × 2 m, n = 4) for 8–10 consecutive weeks during the growing season and excluding approximately 34% of the annual precipitation76. In addition, in 2009, 2011 and 2017 new rainout shelters were installed to compare drought responses of plots subjected to a different number of drought events. Hence, in 2011 samples were collected from plots subjected to 1, 3 and 4 drought events, in 2012 from 2 and 5 drought events, and in 2017 for comparing 1 and 10 drought events (Supplementary Fig. 15). In 2017, drought plots were equipped with rainout shelters from May 17th until August 1st. During the drought period, the soil water content (SWC) in the 10-year and 1-year treatments ranged around 10% (volumetric soil water content at 10 cm depth), whereas it remained in the range of 25–50% in control plots (Supplementary Fig. 1). Drought was terminated by simulating a heavy rainfall event (20 mm of previously collected rainwater was applied to each of the plots) upon removal of the rainout shelters followed by subsequent exposure to natural precipitation. For each plot (n = 4 for each treatment) three replicate soil cores (2 cm diameter) were collected from a depth of 0–10 cm. Soil samples were collected at the end of July (after around 10 weeks of drought) for each campaign and transported to the laboratory (University of Vienna) where they were carefully sieved with a 2 mm sieve and roots were manually removed. Subsamples of soil were immediately frozen for further analysis of the soil microbial community through PLFA (in the 2011 campaign, only control and 1-year drought were analysed for PLFA). The remaining soil was immediately used to measure water content, potential enzymatic activity and for extractions of available nutrients, carbon and microbial biomass. In 2017, we expanded the set of analyses carried out. First, more enzymes were tested (Supplementary Table 3) and microbial communities were investigated by both PLFA and amplicon sequencing. Root biomass production during the 2017 campaign was also measured through in-growth cores. Cores were placed in the soil from May 31st to July 31st and root growth was measured over a period of 62 days. This data was only used to generate a soil multifunctionality index as it is part of another manuscript (Bahn et al., in preparation).
Analysis of soil microbial growth, respiration and carbon use efficiency
Soils were subjected to a laboratory incubation (24 h), where we applied 18O-labelled water to simulate a short time rewetting event and simultaneously study changes in microbial growth, respiration and CUE upon rewetting. We measured soil microbial activity (mass-specific gross growth rates, mass-specific respiration and CUE) by using 18O-H2O incorporation into microbial DNA77. Briefly, approximately 400 mg of fresh soil were incubated for 24 h at temperatures resembling the ambient field conditions during sampling (ca. 20 °C) after labelling them with 18O-H2O to reach an overall enrichment of 20 at% (97.0 at% water, Campro Scientific). Duplicates, which received the same volume of molecular-grade non-labelled water served as natural abundance controls. In order to calculate microbial respiration, gas samples were taken at the start and end of the incubation period and subsequently analysed for respective CO2 concentrations using a Trace GC Ultra (ThermoFischer, Waltham, USA). Following the collection of last gas sample, incubated soil samples were frozen in liquid nitrogen and stored at −80 °C. Consecutively, total microbial DNA was extracted via a DNA-extraction kit (FastDNA™ SPIN Kit for Soil, MP Biomedicals Santa Ana, USA) and quantified by PicoGreen assay (Quant-iT™ PicoGreen® dsDNA Assay Kit; ThermoFischer, Waltham, USA). The abundance of 18O in the extracted DNA was determined for labelled and control samples via a thermochemical elemental analyzer (TC/EA ThermoFisher) coupled via a Conflo III open split system (ThermoFisher) to an Isotope Ratio Mass Spectrometer (Delta V Advantage, ThermoFisher). The synthesis of new DNA was calculated as the difference in 18O abundance between the labelled and non-labelled samples (Oe as the 18O at% excess of the labelled sample), the 18O enrichment (Ol in at%) and the factor of 31.21 describing the proportional mass of O (%) in an average DNA molecule.
The microbial growth rate (the amount of produced microbial C) [ng C g−1 DW h−1] was calculated by using a conversion factor (fDNA = Cmic/DNAmic), which describes the relation between microbial DNA and microbial C, and the amount of newly produced DNA (for details see77).
We expressed the amount of carbon that was taken up by the microbial community [ng C g−1 DW h−1] as the sum of carbon allocated to respiration plus biomass production (growth).
The soil microbial CUE was calculated as the ratio of the C that was invested into growth to the total C that was taken up78.
We expressed mass-specific respiration rates [mg CO2-C g−1 Cmic h−1] as ratio between respiration-derived C and MBC over time, whereas the mass-specific growth rate [ng C ng Cmic−1 d−1] was calculated as the ratio of Cgrowth over Cmic.
Analysis of soil, microbial stoichiometry and available nutrients
All soil samples were analysed for gravimetric water content, microbial biomass C, N and P, extractable organic C (EOC), extractable organic nitrogen (EON), nitrate (NO3−), ammonium (NH4+), total free amino acids (TFAA), extractable organic P (EOP) and extractable inorganic P (EIP). Soil aliquots were dried at 60 °C for 72 h, weighed and finely ground for subsequent analysis of bulk total C and N contents by EA-IRMS (EA 1110; CE Instruments, Milan, Italy), coupled to a Finnigan MAT Delta Plus IRMS (ThermoFisher Scientific, Waltham, MA, USA). Soil P was measured photometrically based on phosphomolybdate blue reaction79 with prior ignition at 450 °C for 5 h in order to convert organic P to inorganic forms80. Microbial biomass C, N and P (MBC, MBN and MBP) were measured following the chloroform fumigation-extraction technique81, with some modifications. Briefly, 4 g of soil from each treatment was weighed in two different containers. To the first set of containers, 30 mL of 1 M KCl solution (for C and N) or 0.5 M NaHCO3 (for P) was added and shaken for 1 h, before filtering through ash-free cellulose filters and extracts kept frozen at −20 °C for later analyses. The other half of samples was fumigated in a desiccator with chloroform for 48 h and then extracted as the non-fumigated samples. Total C and N content of the KCl extracts was analysed by a TOC/TN analyzer (TOCVCPH E200V/TNM-122V; Shimadzu, Vienna, Austria). The difference between fumigated and non-fumigated samples was divided by 0.4581 and considered as the MBC, while a factor of 0.5482 was used to calculate MBN. Inorganic phosphate was measured photometrically based on phosphomolybdate blue reaction79 with or without prior alkaline persulfate oxidation for measuring total dissolved P and inorganic P, respectively83. MBP was calculated as the difference between the fumigated and non-fumigated samples. Ammonium and nitrate were determined with colorimetric methods, NH4+ with a modified indophenol reaction84 and NO3− with the VCl3/Griess assay85. EON was calculated by subtracting from N values of non-fumigated samples the sum of NH4+ and NO3−. Amino acid concentrations were quantified by a modified fluorometric OPAME procedure based on86, optimized for free amino acid measurement in protein hydrolysates87. Inorganic N, TFAA and P assays were run in a microtiter plate format (μQuant Qx200, Bio-Tek Instruments, Bad Friedrichshall, Germany).
Potential extracellular enzymatic activity
The potential extracellular enzymatic activity (EEA) of 10 different enzymes related to C-, N-, P- and S-cycle was measured (Supplementary Table 3) through a photometric method88, with some modifications. Briefly, 1 g of sieved soil was suspended in 100 mL of sodium acetate buffer (50 mM, pH 5) and ultrasonicated at low energy (total energy output of 0.35 kJ). Then, 200 μL of soil suspension and 50 μL of substrate were pipetted into black microtiter plates in six analytical replicates. Aminomethylcoumarin (AMC) was used for calibration of the proteases (Supplementary Table 3), while methylumbelliferyl (MUF) was used for calibration of all the remaining enzymes. Plates were incubated in the dark at 20 °C and fluorescence was measured every 30 min for six times at an excitation wavelength of 365 nm and an emission wavelength of 450 nm (Tecan Infinite M200 fluorimeter, Werfen, Austria). Activity was then calculated as the increase in fluorescence over time.
PLFA analysis
Soil microbial biomass and microbial community composition were estimated by extracting PLFAs from freeze-dried soil samples using the same procedure as described in89 with some modifications. Total lipids were extracted from soil using a chloroform/methanol/citric acid buffer and fractionated by solid-phase extraction. The NLFA fraction was collected by eluting samples with chloroform while the PLFA fraction was collected by eluting samples with methanol. After an internal standard (19:0) was added, NLFAs and PLFAs were converted to fatty acid methyl esters (FAMEs) by transesterification. Samples were analysed for identification and quantification using a GC (7890B GC System; Agilent, Santa Clara, CA, USA) connected to a TOF/MS (Pegasus HT; LECO Corporation, Saint-Joseph, MI, USA). Samples were injected in splitless mode (injector temperature 220 °C) and separated on a DB5 column (60 m × 0.25 mm × 0.25 μm; Agilent, Vienna, Austria) with 1.5 mL min−1 He as the carrier gas (GC program: 1 min at 80 °C, 30 °C min−1 until 150 °C, 1 min at 150 °C, 2 °C min−1 until 200 °C, 4 °C min−1 until 230 °C, 15 min at 230 °C, 30 °C min−1 until 290 °C and 5 min at 290 °C). FAMEs were identified using mixtures of bacterial and fungal FAMEs (Bacterial Acid Methyl Ester CP Mixture (Mtreya LLC, State College, PA, USA) and 37 Component FAME Mix (Supelco, Bellefonte, PA, USA)) and evaluated using NIST MS library for confirmative identification. FAMEs were quantified against the internal standard (19:0). We used the markers 18:1ω9, 18:2ω6,988 and 16:1ω5 for total fungi, although 16:1ω5 is a marker used for AMF it can also be present in bacteria90. Therefore, the NLFA 16:1ω5 was also measured, as this represents a more specific marker for AMF91. The sum of i15:0, a15:0, i16:0 a16:0, i17:0, a17:0 and a18:0 was used as general Gram‐positive bacterial marker and 16:1ω7, cy17:0 and cy19:0 for Gram‐negative bacteria92. The marker 10Me16:0 and 10Me17:0 were used for Actinobacteria92 and summed with general Gram‐positive, Gram‐negative and 14:0 and 15:0 to give total bacterial PLFAs93. For a number of minor peaks not present in the external standard, the double bond position could not be identified with confidence and therefore they were assigned to the general group altogether with the markers 16:0, 18:0, 17:1ω10 and 19:1w9.
Amplicon sequencing
DNA was extracted from freeze-dried samples using the FastDNA™ SPIN Kit for Soil (MP Biomedicals Santa Ana, USA). Extracted DNA was quantified (Quant-iT™ PicoGreen® dsDNA Assay Kit; ThermoFisher, Waltham, USA) and manually normalized to 5 ng µL−1 for each individual sample. Normalized DNA was used to generate amplicon libraries of archaeal, bacterial and fungal communities using a two-step barcoding approach94. In the first PCR step, target amplicons were generated with primers modified at the 5′ end to include a 16 bp head sequence [5′-GCTATGCGCGAGCTGC-3′]. In the second PCR step, products from the first reaction were amplified with barcoding primers that targeted the 16 bp head sequence and included a library-specific 8 bp barcode at the 5′ end. Archaeal and bacterial amplicons were generated using primers 515F95 and 806R96 that targeted the V4 region of the 16S rRNA genes. Fungal amplicons were generated for internal transcribed spacer region 1 (ITS1) using primers ITS1F and ITS297,98, additionally modified by99 and region 2 (ITS2) using primers gITS7ngs and ITS4ngs100. In-house mock communities for both SSU rRNA genes and ITS rDNA, and extraction blanks and water control were included for quality control of sequencing data. In addition, a nested PCR approach was used to target AMF. In the nested approach, a sequence of >2 kbp spanning the whole ITS rDNA region was amplified using a set of partially degenerate primers SSUmAf1-2 and LSUmAr1-4101. The resulting PCR product was then used as a template to amplify ITS2 using the 5′-modified primers 5.8S86 and ITS497,98. The complete overview of primers used in this study is given in Supplementary Table 4.
All oligonucleotides were obtained from Biomers.net (Ulm, Germany). First-step PCRs were performed in three analytical replicates of each sample (2 µL DNA, 5 ng µL−1) in a total reaction volume of 25 µL containing 1x Dream Taq Green Buffer including 2 mM MgCl2 (ThermoFisher Scientific), 0.2 mM dNTP (ThermoFisher Scientific), 0.1 mg mL−1 bovine serum albumin (ThermoFisher Scientific), 0.25 µM of each forward and reverse primer and 0.025 U Dream Taq DNA Polymerase (ThermoFisher Scientific). Amplification conditions for SSU rRNA-targeted PCRs (primers 515 F–806 R): 94 °C for 4 min, 25 cycles of 94 °C for 30 s, 52 °C for 30 s and 72 °C for 45 s, followed by final elongation at 72 °C for 10 min. Amplification conditions for ITS1-targeted PCRs (primers ITS1F–ITS2): 94 °C for 4 min, 25 cycles of 94 °C for 30 s, 52 °C for 30 s and 72 °C for 45 s, followed by final elongation at 72 °C for 10 min. Amplification conditions for ITS2-targeted PCRs (primers gITS7ngs–ITS4ngs): 94 °C for 4 min, 30 cycles of 94 °C for 30 s, 52 °C for 45 s, and 72 °C for 60 s, followed by final elongation at 72 °C for 10 min. Amplification conditions for SSU rRNA–ITS–LSU rRNA-targeted PCRs (primers SSUmAf1-2–LSUmAr1-4): 98 °C for 3 min, 30 cycles of 98 °C for 10 s, 58 °C for 30 s, and 72 °C for 3 min, followed by final elongation at 72 °C for 10 min. PCRs were performed in three analytical replicates of each sample (2 µL DNA, 5 ng µL−1) in a total reaction volume of 25 µL per sample containing 1x Phusion HF Buffer containing 1.5 mM MgCl2 (ThermoFisher Scientific), 0.2 mM dNTP (ThermoFisher Scientific), 1 µM of each forward and reverse primer, and 0.02 U Phusion DNA Polymerase (ThermoFisher Scientific).
Amplification conditions for AMF-ITS2-targeted PCRs (primers AM5.8S_ILfor–ITS4): 94 °C for 4 min, 25 cycles of 94 °C for 30 s, 52 °C for 45 s, and 72 °C for 60 s, followed by final elongation at 72 °C for 10 min. The first-step PCR was screened by agarose gel electrophoresis, pooled, purified using the SequalPrep™ Normalization Plate Kit (ThermoFisher Scientific) and used for second-step PCRs. Second-step PCRs were carried out in 50 µL reactions consisting of same concentration of reagents as the first-step PCRs while using 10 µL of template and 0.8 µM of barcoding primers (5′-BARCODE-HEAD-3′). PCR products were screened by agarose gel electrophoresis and purified and normalized using the SequalPrep™ Normalization Plate Kit resulting in an equimolar library. Samples were pooled and concentrated (innuPREP PCRpure PCR Cleanup Kit, analytikjena, Germany). The final pooled library was sent to Microsynth AG (Balgach, Switzerland) for sequencing on a MiSeq system (Illumina) using the TruSeq Nano DNA Library Prep Kit (Illumina, Cat FC-121-4001). The MiSeq was run in the 2 × 300 cycle configuration using the MiSeq Reagent kit v3 (Illumina, Cat MS-102-3003). Resulting datasets were deposited in the NCBI Sequence Read Archive under BioProject accession number PRJNA694357.
Paired reads were demultiplexed according to94 and were processed into merged ASVs using DADA2 v. 1.10.0102 with standard pooled processing (pool = TRUE) and with default parameters. ASVs predicted for fungi (ITS1, ITS2 and AMF-ITS2) were screened with103 ITSx v. 1.0.11. All ASVs were screened for chimeras using UCHIME104. ASVs were taxonomically classified using the RDPclassifier105 as implemented in Mothur106 v 1.39.5. 16S-V4 regions were classified using107 the SILVA SSU database 132 and ITS regions were classified using the Warcup training set V2108.
Calculations and statistical analysis
All analyses were performed in R V3.6.0109. Following amplicon sequencing, data handling and manipulation were undertaken using the phyloseq package110. Data was filtered by removing sequences matching ‘Mitochondria’, ‘Chloroplast’ and ‘Eukaryota’ from the archaeal and bacterial community, and by removing ‘Glomeromycota’ from the fungal community as the analysis of Glomeromycota was carried out with a specific primer. Also, in order to minimize the inflation of rare species in the community analysis, we removed ASVs that were not present more than two times in at least 20% of the samples. This filtered ASV matrix was used for all downstream analyses concerning amplicon sequencing data. For α-diversity analysis, all samples were subsampled (rarefied) to the minimum sample size using bootstrap subsampling at 1000 iterations to account for library size differences (5809 reads per sample for SSU V4, 9959 reads per sample for ITS1, 2270 reads per sample for ITS2, 2474 reads per sample for AMF-ITS2). α-Diversity indices were generated with the function ‘plot_richness’ from the package phyloseq. For β-diversity analysis, library size normalization was carried out using the geometric mean of pairwise ratios111. Detection of differentially abundant ASVs was carried out using the DESeq2 package112. Plots were generated using ggplot2 package113. We classified microbial ASVs into LRGs similarly to Meisner et al.33, but with some modifications. These are defined as groups of organisms with similar response to a change in their environment. Here, we defined groups of ASVs that differ between 10-year and 1-year treatments, which therefore identify groups of microorganisms that do not simply response to drought but that are specific to long-term drought effects. LRGs were extracted from the filtered ASV tables, as described in the ‘Methods’ section, in order to minimize the inflation of rare species in the community analysis. Detection of differentially abundant ASVs was carried out using the DESeq2 package112. LRGs were grouped for those ASVs showing either an enrichment in the 10-year treatment (positive) or a decrease (negative). The ASVs were then phylogenetically assigned to the corresponding phylum (or order for the AMF dataset) and expressed as relative abundance of the 10-year microbial community.
For all individual variables collected in 2017, we used a one-way ANOVA to test the main effects of treatments (control vs 1 year vs 10 years). We checked for homogeneity of variances and normality of residuals by inspecting the plot of standardized residuals vs predicted values, frequency histogram and QQ-plot114 (reported in Supplementary Statistical Report). Given the high number of tests presented, we corrected the p-values presented for false discovery rate (FDR), using the Benjamini-Hochberg (BH) procedure with the R function ‘p.adjust()’. We present the ANOVA results with and without correction in Supplementary Table 5. A post hoc test (Tukey HSD test) was used to compare differences between individual treatments. The package ‘effectsize’115 was used to calculate eta-squared effect size (η2) and 90% confidence intervals. Data integrating previous campaigns (2011 and 2012) was analysed by linear models using the function ‘lm’. Data from potential enzymatic activity, PLFA and amplicon sequencing was also analysed by means of permutational analysis of variance (9999 permutations) using the ‘adonis’ function in the package vegan116. Results from the permutational multivariate analyses were visualized by means of non-metric multidimensional scaling (NMDS). For this, we used the function ‘metaMDS’ from the package vegan.
To compare quantitative differences between control treatments and different years of drought subjection, we calculated the Bray-Curtis dissimilarity index with the function ‘vegdist’ of the package vegan. The index was calculated as an averaged pairwise comparison between plots subjected to drought and the control treatment. We calculated this index in the 2017 campaign from the amplicon sequencing data, for the 16S, ITS1 and AMF primer. We also calculated a response ratio, as the natural logarithm of drought treatments divided by the average of the control (Ln (RR)) for the sampling campaigns of 2011 and 2012. The response ratio is commonly used in ecological studies as a measure of experimental effect because it quantifies the proportionate change that results from an experimental manipulation117. Moreover, it allowed us to compare different variables (enzyme activities and acquisition strategies) in the 2011 and 2012 campaigns, and to relate changes within the control to previous years of drought. We calculated stoichiometric ratios of microbial acquisition strategies by dividing enzymes related to C-cycle (BG, CB, LIP and XYL) into either N-cycle-related enzymes (END, NAG, LAP, TAP) or P-cycle-related enzyme (PHOS) and also the ratio of N-cycle-related enzymes to P-cycle-related enzymes. For the 2011 and 2012 campaigns we used a reduced set as specified in Supplementary Table 3.
We also calculated a multifunctionality index, which represents the expression of multiple ecosystem processes and services together29. We calculated this index using only soil-related parameters including soil nutrients (soil C, N and P), microbial stoichiometry (biomass C, N and P), plant productivity (belowground net primary productivity) and microbially mediated organic matter decomposition (potential enzymatic activities). Thus, representing a soil multifunctionality index. In order to obtain a computable index for each treatment, we standardized each of the variables measured using the z-score transformation. These standardized soil-function values were then averaged to generate a multifunctionality index55.
We used the filtered ASV matrix to predict the soil bacterial metagenomes using PICRUSt223 with implemented tools HMMER (http://hmmer.org/), EPA-ng118, GAPPA119 and CASTOR120. The ASV file was converted into a.biom file using the R package biomformat121 and PICRUSt2 was executed to obtain the stratified output based on per-sequence contributions. We downloaded the KEGG Orthology (KO) from https://www.genome.jp/kegg-bin/get_htext and created a custom file that allowed to map KO numbers to broader categories as provided by the PICRUSt2 tool. The weighted nearest sequenced taxon index (NSTI) was found to be highly similar over all the replicates and treatments ranging from 0.212 to 0.247 and indicated a well-characterized community with regard to the placement of ASVs in the reference tree. This allowed us to carefully interpret the predicted metagenomes for potential functions and phyla that may react to repeated drought. We applied DESeq2 to identify the predicted functions that were differentially abundant among treatments including the phyla predicted to be contributing to these functions.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files.
Acknowledgements
We thank Margarete Watzka and Ludwig Seidl for assistance in the laboratory. We further thank Jan Jansa for guidance with AMF-targeted amplicon sequencing. The long-term drought treatment was maintained by projects funded by the Austrian Science Fund (FWF, project number P22214-B17 and I 1056) and the Austrian Academy of Sciences (ESS-programme, project ClimLUC). Open access funding provided by University of Vienna.
Author contributions
M.B. designed the experiment. M.B., A.R., L.F. and R.H. carried out the field campaigns, and A.C., H.S., L.F., D.Z., V.M., M.J. and P.G. the laboratory analyses. H.S. and C.W.H. processed amplicon sequencing data which was analysed by A.C. and H.S.; A.C. analysed the remaining data. A.C., A.R., H.S. and L.F. wrote the original draft, and all authors reviewed the manuscript.
Data availability
16S-V4 regions were classified using the SILVA SSU database 132 (https://www.arb-silva.de/). The data generated in this study (excluding data generated via the amplicon sequencing analysis) is provided in the Supplementary Information (Supplementary Data 1). The overall data generated in this study (including data generated via the amplicon sequencing analysis) has been deposited in the Zenodo repository database under accession code DOI: 10.5281/zenodo.5117955. The sequencing data generated in this study have been deposited in the NCBI Sequence Read Archive under BioProject accession number PRJNA694357.
Code availability
The R code supporting the findings presented here is available from the corresponding authors on request.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review informationNature Communications thanks Ji Chen, Adina Howe, and Ashish Malik for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Alberto Canarini, Email: alberto.canarini@hotmail.it.
Andreas Richter, Email: andreas.richter@univie.ac.at.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-021-25675-4.
References
- 1.Kannenberg SA, Schwalm CR, Anderegg WRL. Ghosts of the past: how drought legacy effects shape forest functioning and carbon cycling. Ecol. Lett. 2020;23:891–901. doi: 10.1111/ele.13485. [DOI] [PubMed] [Google Scholar]
- 2.Padisak J. Seasonal succession of phytoplankton in a large shallow lake (Balaton, Hungary)—a dynamic approach to ecological memory, its possible role and mechanisms. J. Ecol. 1992;80:217–230. doi: 10.2307/2261008. [DOI] [Google Scholar]
- 3.Power DA, et al. What can ecosystems learn? Expanding evolutionary ecology with learning theory. Biol. Direct. 2015;10:69. doi: 10.1186/s13062-015-0094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive Earth’s biogeochemical cycles. Science. 2008;320:1034–1039. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- 5.Reichstein M, et al. Climate extremes and the carbon cycle. Nature. 2013;500:287. doi: 10.1038/nature12350. [DOI] [PubMed] [Google Scholar]
- 6.Seneviratne, S. I. et al. Changes in climate extremes and their impacts on the natural physical environment. In Managing the Risks of Extreme Events and Disasters to Advance Climate Change Adaptation. A Special Report of Working Groups I and II of the Intergovernmental Panel on Climate Change (IPCC) (eds. Field, C.B. et al.) 109–230 (Cambridge University Press, 2017).
- 7.Pappas C, Mahecha MD, Frank DC, Babst F, Koutsoyiannis D. Ecosystem functioning is enveloped by hydrometeorological variability. Nat. Ecol. Evol. 2017;1:1263–1270. doi: 10.1038/s41559-017-0277-5. [DOI] [PubMed] [Google Scholar]
- 8.Hawkes CV, Keitt TH. Resilience vs. historical contingency in microbial responses to environmental change. Ecol. Lett. 2015;18:612–625. doi: 10.1111/ele.12451. [DOI] [PubMed] [Google Scholar]
- 9.Johnstone JF, et al. Changing disturbance regimes, ecological memory, and forest resilience. Front. Ecol. Environ. 2016;14:369–378. doi: 10.1002/fee.1311. [DOI] [Google Scholar]
- 10.Ochoa‐Hueso, R. et al. Drought consistently alters the composition of soil fungal and bacterial communities in grasslands from two continents. Glob. Chang. Biol. 24, 2818–2827 (2018). [DOI] [PubMed]
- 11.Bastida F, et al. Differential sensitivity of total and active soil microbial communities to drought and forest management. Glob. Chang. Biol. 2017;23:4185–4203. doi: 10.1111/gcb.13790. [DOI] [PubMed] [Google Scholar]
- 12.Evans SE, Wallenstein MD, Burke IC. Is bacterial moisture niche a good predictor of shifts in community composition under long-term drought? Ecology. 2014;95:110–122. doi: 10.1890/13-0500.1. [DOI] [PubMed] [Google Scholar]
- 13.de Vries FT, et al. Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 2018;9:3033. doi: 10.1038/s41467-018-05516-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rousk J, Smith AR, Jones DL. Investigating the long-term legacy of drought and warming on the soil microbial community across five European shrubland ecosystems. Glob. Chang. Biol. 2013;19:3872–3884. doi: 10.1111/gcb.12338. [DOI] [PubMed] [Google Scholar]
- 15.Kaisermann A, de Vries FT, Griffiths RI, Bardgett RD. Legacy effects of drought on plant–soil feedbacks and plant–plant interactions. New Phytol. 2017;215:1413–1424. doi: 10.1111/nph.14661. [DOI] [PubMed] [Google Scholar]
- 16.Fuchslueger L, et al. Drought history affects grassland plant and microbial carbon turnover during and after a subsequent drought event. J. Ecol. 2016;104:1453–1465. doi: 10.1111/1365-2745.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meisner A, De Deyn GB, de Boer W, van der Putten WH. Soil biotic legacy effects of extreme weather events influence plant invasiveness. Proc. Natl Acad. Sci. USA. 2013;110:9835–9838. doi: 10.1073/pnas.1300922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Nijs EA, Hicks LC, Leizeaga A, Tietema A, Rousk J. Soil microbial moisture dependences and responses to drying–rewetting: the legacy of 18 years drought. Glob. Chang. Biol. 2019;25:1005–1015. doi: 10.1111/gcb.14508. [DOI] [PubMed] [Google Scholar]
- 19.Fuchslueger L, Bahn M, Fritz K, Hasibeder R, Richter A. Experimental drought reduces the transfer of recently fixed plant carbon to soil microbes and alters the bacterial community composition in a mountain meadow. New Phytol. 2014;201:916–927. doi: 10.1111/nph.12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mooshammer M, Wanek W, Zechmeister-Boltenstern S, Richter A. Stoichiometric imbalances between terrestrial decomposer communities and their resources: mechanisms and implications of microbial adaptations to their resources. Front. Microbiol. 2014;5:22. doi: 10.3389/fmicb.2014.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waring BG, Weintraub SR, Sinsabaugh RL. Ecoenzymatic stoichiometry of microbial nutrient acquisition in tropical soils. Biogeochemistry. 2014;117:101–113. doi: 10.1007/s10533-013-9849-x. [DOI] [Google Scholar]
- 22.Sinsabaugh RL, et al. Stoichiometry of soil enzyme activity at global scale. Ecol. Lett. 2008;11:1252–1264. doi: 10.1111/j.1461-0248.2008.01245.x. [DOI] [PubMed] [Google Scholar]
- 23.Douglas GM, et al. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020;38:685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Csonka LN. Physiological and genetic responses of bacteria to osmotic stress. Microbiol. Mol. Biol. Rev. 1989;53:121–147. doi: 10.1128/mr.53.1.121-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitfield, G. B., Marmont, L. S. & Howell, P. L. Enzymatic modifications of exopolysaccharides enhance bacterial persistence. Front. Microbiol. 6, 471 (2015). [DOI] [PMC free article] [PubMed]
- 26.Byrd MS, et al. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol. 2009;73:622–638. doi: 10.1111/j.1365-2958.2009.06795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El Zoeiby A, Sanschagrin F, Levesque RC. Structure and function of the Mur enzymes: development of novel inhibitors. Mol. Microbiol. 2003;47:1–12. doi: 10.1046/j.1365-2958.2003.03289.x. [DOI] [PubMed] [Google Scholar]
- 28.Lebre PH, De Maayer P, Cowan DA. Xerotolerant bacteria: surviving through a dry spell. Nat. Rev. Microbiol. 2017;15:285–296. doi: 10.1038/nrmicro.2017.16. [DOI] [PubMed] [Google Scholar]
- 29.Delgado-Baquerizo M, et al. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 2016;7:10541. doi: 10.1038/ncomms10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes, T. P. et al. Ecological memory modifies the cumulative impact of recurrent climate extremes. Nat. Clim. Chang. 9, 40–43 (2019).
- 31.Seidl R, Donato DC, Raffa KF, Turner MG. Spatial variability in tree regeneration after wildfire delays and dampens future bark beetle outbreaks. Proc. Natl Acad. Sci. USA. 2016;113:13075–13080. doi: 10.1073/pnas.1615263113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hillebrand, H. & Kunze, C. Meta-analysis on pulse disturbances reveals differences in functional and compositional recovery across ecosystems. Ecol. Lett. 23, 575–585 (2020). [DOI] [PubMed]
- 33.Meisner, A., Jacquiod, S., Snoek, B. L., ten Hooven, F. C. & van der Putten, W. H. Drought legacy effects on the composition of soil fungal and prokaryote communities. Front. Microbiol. 9, 294 (2018). [DOI] [PMC free article] [PubMed]
- 34.Bardgett RD, Caruso T. Soil microbial community responses to climate extremes: resistance, resilience and transitions to alternative states. Philos. Trans. R. Soc. B Biol. Sci. 2020;375:20190112. doi: 10.1098/rstb.2019.0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Isobe K, Bouskill NJ, Brodie EL, Sudderth EA, Martiny JBH. Phylogenetic conservation of soil bacterial responses to simulated global changes. Philos. Trans. R. Soc. B Biol. Sci. 2020;375:20190242. doi: 10.1098/rstb.2019.0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barberán A, Caceres Velazquez H, Jones S, Fierer N. Hiding in plain sight: mining bacterial species records for phenotypic trait information. mSphere. 2017;2:e00237–17. doi: 10.1128/mSphere.00237-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouskill NJ, et al. Pre-exposure to drought increases the resistance of tropical forest soil bacterial communities to extended drought. ISME J. 2012;7:384. doi: 10.1038/ismej.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taketani RG, et al. Dry season constrains bacterial phylogenetic diversity in a semi-arid rhizosphere system. Microb. Ecol. 2017;73:153–161. doi: 10.1007/s00248-016-0835-4. [DOI] [PubMed] [Google Scholar]
- 39.Naylor D, DeGraaf S, Purdom E, Coleman-Derr D. Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J. 2017;11:2691. doi: 10.1038/ismej.2017.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu L, et al. Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria. Proc. Natl Acad. Sci. USA. 2018;115:E4284–E4293. doi: 10.1073/pnas.1717308115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taniguchi T, Kitajima K, Douhan GW, Yamanaka N, Allen MF. A pulse of summer precipitation after the dry season triggers changes in ectomycorrhizal formation, diversity, and community composition in a Mediterranean forest in California, USA. Mycorrhiza. 2018;28:665–677. doi: 10.1007/s00572-018-0859-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ren C, et al. Responses of soil total microbial biomass and community compositions to rainfall reductions. Soil Biol. Biochem. 2018;116:4–10. doi: 10.1016/j.soilbio.2017.09.028. [DOI] [Google Scholar]
- 43.Furze JR, et al. Resistance and resilience of root fungal communities to water limitation in a temperate agroecosystem. Ecol. Evol. 2017;7:3443–3454. doi: 10.1002/ece3.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deveautour C, Donn S, Power SA, Bennett AE, Powell JR. Experimentally altered rainfall regimes and host root traits affect grassland arbuscular mycorrhizal fungal communities. Mol. Ecol. 2018;27:2152–2163. doi: 10.1111/mec.14536. [DOI] [PubMed] [Google Scholar]
- 45.Averill C, Turner BL, Finzi AC. Mycorrhiza-mediated competition between plants and decomposers drives soil carbon storage. Nature. 2014;505:543–545. doi: 10.1038/nature12901. [DOI] [PubMed] [Google Scholar]
- 46.Dijkstra FA, He M, Johansen MP, Harrison JJ, Keitel C. Plant and microbial uptake of nitrogen and phosphorus affected by drought using 15N and 32P tracers. Soil Biol. Biochem. 2015;82:135–142. doi: 10.1016/j.soilbio.2014.12.021. [DOI] [Google Scholar]
- 47.Kakumanu ML, Ma L, Williams MA. Drought-induced soil microbial amino acid and polysaccharide change and their implications for C–N cycles in a climate change world. Sci. Rep. 2019;9:10968. doi: 10.1038/s41598-019-46984-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sleator RD, Hill C. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol. Rev. 2002;26:49–71. doi: 10.1111/j.1574-6976.2002.tb00598.x. [DOI] [PubMed] [Google Scholar]
- 49.Warren CR. Response of osmolytes in soil to drying and rewetting. Soil Biol. Biochem. 2014;70:22–32. doi: 10.1016/j.soilbio.2013.12.008. [DOI] [Google Scholar]
- 50.Bouskill NJ, et al. Belowground response to drought in a tropical forest soil. I. Changes in microbial functional potential and metabolism. Front. Microbiol. 2016;7:525. doi: 10.3389/fmicb.2016.00525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Flemming H-C, et al. Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 2016;14:563. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 52.Malik AA, et al. Drought and plant litter chemistry alter microbial gene expression and metabolite production. ISME J. 2020;14:2236–2247. doi: 10.1038/s41396-020-0683-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nunan, N., Raynaud, X. & Schmidt, H. The ecology of heterogeneity: soil bacterial communities and C dynamics. Philos. Trans. R. Soc. B Biol. Sci. 375, 20190249 (2020). [DOI] [PMC free article] [PubMed]
- 54.Li J, et al. Predictive genomic traits for bacterial growth in culture versus actual growth in soil. ISME J. 2019;13:2162–2172. doi: 10.1038/s41396-019-0422-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maestre FT, et al. Plant species richness and ecosystem multifunctionality in global drylands. Science. 2012;335:214–218. doi: 10.1126/science.1215442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grigulis K, et al. Relative contributions of plant traits and soil microbial properties to mountain grassland ecosystem services. J. Ecol. 2013;101:47–57. doi: 10.1111/1365-2745.12014. [DOI] [Google Scholar]
- 57.Lau JA, Lennon JT. Rapid responses of soil microorganisms improve plant fitness in novel environments. Proc. Natl Acad. Sci. USA. 2012;109:14058–14062. doi: 10.1073/pnas.1202319109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fitzpatrick CR, et al. Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl Acad. Sci. USA. 2018;115:E1157–E1165. doi: 10.1073/pnas.1717617115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang S, et al. Autotrophic and heterotrophic soil respiration responds asymmetrically to drought in a subtropical forest in the Southeast China. Soil Biol. Biochem. 2018;123:242–249. doi: 10.1016/j.soilbio.2018.04.029. [DOI] [Google Scholar]
- 60.López-Ballesteros A, et al. Enhancement of the net CO2 release of a semiarid grassland in SE Spain by rain pulses. J. Geophys. Res. Biogeosci. 2016;121:52–66. doi: 10.1002/2015JG003091. [DOI] [Google Scholar]
- 61.Schimel JP. Life in dry soils: effects of drought on soil microbial communities and processes. Annu. Rev. Ecol. Evol. Syst. 2018;49:409–432. doi: 10.1146/annurev-ecolsys-110617-062614. [DOI] [Google Scholar]
- 62.Canarini, A., Kaiser, C., Merchant, A., Richter, A. & Wanek, W. Root exudation of primary metabolites: mechanisms and their roles in plant responses to environmental stimuli. Front. Plant Sci. 10, 157 (2019). [DOI] [PMC free article] [PubMed]
- 63.de Vries FT, et al. Changes in root-exudate-induced respiration reveal a novel mechanism through which drought affects ecosystem carbon cycling. New Phytol. 2019;224:132–145. doi: 10.1111/nph.16001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Teste FP, et al. Plant-soil feedback and the maintenance of diversity in Mediterranean-climate shrublands. Science. 2017;355:173–176. doi: 10.1126/science.aai8291. [DOI] [PubMed] [Google Scholar]
- 65.Hu L, et al. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 2018;9:2738. doi: 10.1038/s41467-018-05122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Canarini, A., Merchant, A. & Dijkstra, F. A. Drought effects on Helianthus annuus and Glycine max metabolites: from phloem to root exudates. Rhizosphere2, 85–97 (2016).
- 67.Canarini, A. & Dijkstra, F. A. Dry-rewetting cycles regulate wheat carbon rhizodeposition, stabilization and nitrogen cycling. Soil Biol. Biochem. 81, 195–203 (2015).
- 68.Morecroft MD, et al. Changing precipitation patterns alter plant community dynamics and succession in an ex-arable grassland. Funct. Ecol. 2004;18:648–655. doi: 10.1111/j.0269-8463.2004.00896.x. [DOI] [Google Scholar]
- 69.Strickland MS, Osburn E, Lauber C, Fierer N, Bradford MA. Litter quality is in the eye of the beholder: initial decomposition rates as a function of inoculum characteristics. Funct. Ecol. 2009;23:627–636. doi: 10.1111/j.1365-2435.2008.01515.x. [DOI] [Google Scholar]
- 70.Allison SD, et al. Microbial abundance and composition influence litter decomposition response to environmental change. Ecology. 2013;94:714–725. doi: 10.1890/12-1243.1. [DOI] [PubMed] [Google Scholar]
- 71.Walker TWN, et al. A systemic overreaction to years versus decades of warming in a subarctic grassland ecosystem. Nat. Ecol. Evol. 2020;4:101–108. doi: 10.1038/s41559-019-1055-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ogle K, et al. Quantifying ecological memory in plant and ecosystem processes. Ecol. Lett. 2015;18:221–235. doi: 10.1111/ele.12399. [DOI] [PubMed] [Google Scholar]
- 73.Bahn M, Knapp M, Garajova Z, Pfahringer N, Cernusca A. Root respiration in temperate mountain grasslands differing in land use. Glob. Chang. Biol. 2006;12:995–1006. doi: 10.1111/j.1365-2486.2006.01144.x. [DOI] [Google Scholar]
- 74.Bahn M, et al. Soil respiration at mean annual temperature predicts annual total across vegetation types and biomes. Biogeosciences. 2010;7:2147. doi: 10.5194/bg-7-2147-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schmitt M, Bahn M, Wohlfahrt G, Tappeiner U, Cernusca A. Land use affects the net ecosystem CO2 exchange and its components in mountain grasslands. Biogeosciences. 2010;7:2297. doi: 10.5194/bg-7-2297-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Estiarte M, et al. Few multiyear precipitation–reduction experiments find a shift in the productivity–precipitation relationship. Glob. Chang. Biol. 2016;22:2570–2581. doi: 10.1111/gcb.13269. [DOI] [PubMed] [Google Scholar]
- 77.Spohn M, Klaus K, Wanek W, Richter A. Microbial carbon use efficiency and biomass turnover times depending on soil depth—Implications for carbon cycling. Soil Biol. Biochem. 2016;96:74–81. doi: 10.1016/j.soilbio.2016.01.016. [DOI] [Google Scholar]
- 78.Manzoni S, Taylor P, Richter A, Porporato A, Ågren GI. Environmental and stoichiometric controls on microbial carbon-use efficiency in soils. New Phytol. 2012;196:79–91. doi: 10.1111/j.1469-8137.2012.04225.x. [DOI] [PubMed] [Google Scholar]
- 79.Schinner F, Öhlinger R, Kandeler E, Margesin R, Kaiser P. Methods in soil biology. Bull. Inst. Pasteur. 1996;4:311–312. [Google Scholar]
- 80.Kuo, S. Phosphorus. In Methods of Soil Analysis, Part 3 (eds. Sparks, D. L. et al.) Ch. 32, 869–919 (SSSA, 1996).
- 81.Vance ED, Brookes PC, Jenkinson DS. An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 1987;19:703–707. doi: 10.1016/0038-0717(87)90052-6. [DOI] [Google Scholar]
- 82.Brookes PC, Landman A, Pruden G, Jenkinson DS. Chloroform fumigation and the release of soil nitrogen: a rapid direct extraction method to measure microbial biomass nitrogen in soil. Soil Biol. Biochem. 1985;17:837–842. doi: 10.1016/0038-0717(85)90144-0. [DOI] [Google Scholar]
- 83.Doyle A, Weintraub MN, Schimel JP. Persulfate digestion and simultaneous colorimetric analysis of carbon and nitrogen in soil extracts. Soil Sci. Soc. Am. J. 2004;68:669–676. doi: 10.2136/sssaj2004.6690. [DOI] [Google Scholar]
- 84.Kandeler E, Gerber H. Short-term assay of soil urease activity using colorimetric determination of ammonium. Biol. Fertil. Soils. 1988;6:68–72. doi: 10.1007/BF00257924. [DOI] [Google Scholar]
- 85.Hood-Nowotny R, Umana NH-N, Inselbacher E, Oswald- Lachouani P, Wanek W. Alternative methods for measuring inorganic, organic, and total dissolved nitrogen in soil. Soil Sci. Soc. Am. J. 2010;74:1018–1027. doi: 10.2136/sssaj2009.0389. [DOI] [Google Scholar]
- 86.Jones DL, Owen AG, Farrar JF. Simple method to enable the high resolution determination of total free amino acids in soil solutions and soil extracts. Soil Biol. Biochem. 2002;34:1893–1902. doi: 10.1016/S0038-0717(02)00203-1. [DOI] [Google Scholar]
- 87.Prommer J, et al. Biochar decelerates soil organic nitrogen cycling but stimulates soil nitrification in a temperate arable field trial. PLoS ONE. 2014;9:e86388. doi: 10.1371/journal.pone.0086388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kaiser C, Frank A, Wild B, Koranda M, Richter A. Negligible contribution from roots to soil-borne phospholipid fatty acid fungal biomarkers 18:2ω6,9 and 18:1ω9. Soil Biol. Biochem. 2010;42:1650–1652. doi: 10.1016/j.soilbio.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kaiser C, et al. Belowground carbon allocation by trees drives seasonal patterns of extracellular enzyme activities by altering microbial community composition in a beech forest soil. New Phytol. 2010;187:843–858. doi: 10.1111/j.1469-8137.2010.03321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Olsson PA. Signature fatty acids provide tools for determination of the distribution and interactions of mycorrhizal fungi in soil. FEMS Microbiol. Ecol. 1999;29:303–310. doi: 10.1111/j.1574-6941.1999.tb00621.x. [DOI] [Google Scholar]
- 91.Ngosong C, Gabriel E, Ruess L. Use of the signature fatty acid 16:1ω5 as a tool to determine the distribution of arbuscular mycorrhizal fungi in soil. J. Lipids. 2012;2012:236807. doi: 10.1155/2012/236807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Quideau, S. A. et al. Extraction and analysis of microbial phospholipid fatty acids in soils. J. Vis. Exp. 2016, 54360 (2016). [DOI] [PMC free article] [PubMed]
- 93.García-Orenes F, Morugán-Coronado A, Zornoza R, Cerdà A, Scow K. Changes in soil microbial community structure influenced by agricultural management practices in a mediterranean agro-ecosystem. PLoS ONE. 2013;8:e80522–e80522. doi: 10.1371/journal.pone.0080522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Herbold, C. et al. A flexible and economical barcoding approach for highly multiplexed amplicon sequencing of diverse target genes. Front. Microbiol. 6, 731 (2015). [DOI] [PMC free article] [PubMed]
- 95.Apprill A, McNally S, Parsons R, Weber L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015;75:129–137. doi: 10.3354/ame01753. [DOI] [Google Scholar]
- 96.Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016;18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 97.White, T. J., Bruns, T., Lee, S. & Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications 315–322 (Academic Press, 1990).
- 98.Paymaneh, Z., Sarcheshmehpour, M., Bukovská, P. & Jansa, J. Could indigenous arbuscular mycorrhizal communities be used to improve tolerance of pistachio to salinity and/or drought? Symbiosis79, 269–283 (2019).
- 99.Smith DP, Peay KG. Sequence depth, not PCR replication, improves ecological inference from next generation DNA sequencing. PLoS ONE. 2014;9:e90234. doi: 10.1371/journal.pone.0090234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tedersoo L, Lindahl B. Fungal identification biases in microbiome projects. Environ. Microbiol. Rep. 2016;8:774–779. doi: 10.1111/1758-2229.12438. [DOI] [PubMed] [Google Scholar]
- 101.Krüger M, Stockinger H, Krüger C, Schüßler A. DNA‐based species level detection of Glomeromycota: one PCR primer set for all arbuscular mycorrhizal fungi. New Phytol. 2009;183:212–223. doi: 10.1111/j.1469-8137.2009.02835.x. [DOI] [PubMed] [Google Scholar]
- 102.Callahan BJ, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bengtsson‐Palme J, et al. Improved software detection and extraction of ITS1 and ITS 2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 2013;4:914–919. [Google Scholar]
- 104.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Deshpande V, et al. Fungal identification using a Bayesian classifier and the Warcup training set of internal transcribed spacer sequences. Mycologia. 2016;108:1–5. doi: 10.3852/14-293. [DOI] [PubMed] [Google Scholar]
- 109.R Core Team. R: A Language and Environment for Statistical Computing (R Core Team, 2017).
- 110.McMurdie PJ, Holmes S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen L, et al. GMPR: a robust normalization method for zero-inflated count data with application to microbiome sequencing data. PeerJ. 2018;6:e4600–e4600. doi: 10.7717/peerj.4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2016).
- 114.Kozak M, Piepho H. What’s normal anyway? Residual plots are more telling than significance tests when checking ANOVA assumptions. J. Agron. Crop Sci. 2018;204:86–98. doi: 10.1111/jac.12220. [DOI] [Google Scholar]
- 115.Ben-Shachar MS, Lüdecke D, Makowski D. effectsize: Estimation of effect size indices and standardized parameters. J. Open Source Softw. 2020;5:2815. doi: 10.21105/joss.02815. [DOI] [Google Scholar]
- 116.Oksanen J, et al. Package ‘vegan’. Community Ecol. Packag. 2013;2:1–295. [Google Scholar]
- 117.Hedges LV, Gurevitch J, Curtis PS. The meta‐analysis of response ratios in experimental ecology. Ecology. 1999;80:1150–1156. doi: 10.1890/0012-9658(1999)080[1150:TMAORR]2.0.CO;2. [DOI] [Google Scholar]
- 118.Barbera P, et al. EPA-ng: massively parallel evolutionary placement of genetic sequences. Syst. Biol. 2019;68:365–369. doi: 10.1093/sysbio/syy054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Czech L, Stamatakis A. Scalable methods for analyzing and visualizing phylogenetic placement of metagenomic samples. PLoS ONE. 2019;14:e0217050–e0217050. doi: 10.1371/journal.pone.0217050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Louca S, Doebeli M. Efficient comparative phylogenetics on large trees. Bioinformatics. 2018;34:1053–1055. doi: 10.1093/bioinformatics/btx701. [DOI] [PubMed] [Google Scholar]
- 121.McMurdie, P. J. & Paulson, J. N. biomformat: An interface package for the BIOM file format. R/Bioconductor Package, version 1.0.0. (Bioconductor, 2015).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files.
Data Availability Statement
16S-V4 regions were classified using the SILVA SSU database 132 (https://www.arb-silva.de/). The data generated in this study (excluding data generated via the amplicon sequencing analysis) is provided in the Supplementary Information (Supplementary Data 1). The overall data generated in this study (including data generated via the amplicon sequencing analysis) has been deposited in the Zenodo repository database under accession code DOI: 10.5281/zenodo.5117955. The sequencing data generated in this study have been deposited in the NCBI Sequence Read Archive under BioProject accession number PRJNA694357.
The R code supporting the findings presented here is available from the corresponding authors on request.