Abstract
Background
The emergence of a new pathogen requires a rapid assessment of its transmissibility, to inform appropriate public health interventions.
Methods
The peer-reviewed literature published between 1 January and 30 April 2020 on COVID-19 in PubMed was searched. Estimates of the incubation period, serial interval and reproduction number for COVID-19 were obtained and compared.
Results
A total of 86 studies met the inclusion criteria. Of these, 33 estimated the mean incubation period (4–7 days) and 15 included estimates of the serial interval (mean 4–8 days; median length 4–5 days). Fifty-two studies estimated the reproduction number. Although reproduction number estimates ranged from 0.3 to 14.8, in 33 studies (63%), they fell between 2 and 3.
Discussion
Studies calculating the incubation period and effective reproduction number were published from the beginning of the pandemic until the end of the study period (30 April 2020); however, most of the studies calculating the serial interval were published in April 2020. The calculated incubation period was similar over the study period and in different settings, whereas estimates of the serial interval and effective reproduction number were setting-specific. Estimates of the serial interval were shorter at the end of the study period as increasing evidence of pre-symptomatic transmission was documented and as jurisdictions enacted outbreak control measures. Estimates of the effective reproduction number varied with the setting and the underlying model assumptions. Early analysis of epidemic parameters provides vital information to inform the outbreak response.
Coronavirus disease 2019 (COVID-19) presents an enormous challenge to public health. By 18 April 2020, 140 million cases had been reported across 222 countries and areas, with an estimate of 3 million people having died. (1) The overwhelming attention placed on COVID-19 and the volume of research published in the early months of this pandemic (over 4100 papers in PubMed to the end of April 2020) create challenges for public health responders attempting to understand the epidemiology of this disease. There is a need to distil and synthesize the findings that are most relevant to inform public health interventions.
Estimates of the transmission parameters of a pathogen are required as soon as practicable, to inform the public health response. With known pathogens, public health responders can use data and estimates from previous outbreaks to make evidence-based decisions. However, with an emerging pathogen, such as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), past outbreaks may provide limited utility; hence, epidemic parameters must be estimated from early cases and detected transmission events. A successful outbreak response is informed by rapid data collection and analysis, to understand the dynamics of disease spread and identify appropriate, informed interventions.
Understanding disease transmission of a new pathogen requires knowledge of the incubation period, serial interval and reproduction number. The basic reproduction number is the expected or average number of secondary cases that result from one infected person if no individuals in the population are immune to the pathogen and no measures are in place to reduce spread. In practice, pathogens rarely propagate freely through a population because individuals change their behaviour or governments enact public health interventions. The effective reproduction number is the expected or average number of secondary cases in a population where some individuals are immune or interventions to limit spread are in place.
The distribution of the incubation period is crucial for determining the length of quarantine for potentially exposed individuals and travellers. (2-4) Estimates of the serial interval provide public health responders with an idea of the time available to identify and isolate potential cases before they can spread the disease to others. (5, 6) The reproduction number of a disease provides a population-wide estimate of the scale of a potential outbreak and a baseline to test the effectiveness of different interventions in limiting disease transmission. (7-9) Although highly influential, early estimates of the incubation period, serial interval and reproduction number are generally based on small sample sizes that may not be representative of the wider population at risk. (7, 9, 10)
Although some literature reviews have reviewed the epidemiology of COVID-19, (11-14) they have not collated the estimates of epidemic parameters from the initial period of the COVID-19 pandemic. The aim of this study was to collate and compare the characteristics of the COVID-19 pandemic up to 30 April 2020.
Methods
Studies that describe or estimate the epidemic characteristics of the COVID-19 pandemic until 30 April 2020 were collected. Epidemiological parameters were limited to the incubation period, the serial interval and the reproduction number. The incubation period is the length of time experienced by an individual case from the point of infection to the start of symptom onset. The serial interval refers to the mean length of time between successive cases in a chain of transmission, measured as the length of time from symptom onset in a primary case to symptom onset in a secondary case. Both the incubation period and serial interval in this analysis are measured in days.
Over the course of the COVID-19 pandemic so far, governments have enacted public health interventions at different times and to different extents. Individual behaviours have changed at different rates as individuals have learned about COVID-19 and responded to media reports, government messaging and their understanding of risk. Several estimates of the reproduction number overlap periods when governments have enacted significant public health interventions. Although this study focuses on estimates from the early stages of the outbreak, when most of the population were susceptible and potentially not modifying their behaviour, this study refers to all estimates of the reproduction number as the effective reproduction number.
We searched peer-reviewed published research articles from PubMed using the terms “coronavirus” AND “novel” OR “new” OR “covid” OR “Wuhan” OR “ncp” OR “ncov” for articles published online until 30 April 2020. The literature search ran from 24 February 2020 to 12 May 2020. All articles were imported to Zotero 5.0.87 for review. Eligible articles were reviewed for date of online publication, study period, sample size, setting, method of calculating epidemic parameters, assumptions used to inform these calculations and output measures (including the approach to estimating uncertainty).
Studies were included in this review if they reported estimates of at least one of the relevant epidemic parameters and were written in English. Any articles published before 1 November 2019, pre-prints, grey literature and case reports were excluded.
Ethics and permissions
Ethical approval was not sought for this review of existing, publicly available peer-reviewed literature.
Results
The PubMed search returned 4426 articles published online up to 30 April 2020. Of these articles, 3581 were excluded at the screening assessment and a further 759 at the eligibility assessment, giving a total of 86 included studies. The results of the search and eligibility assessment are shown in Fig. 1.
Figure 1.
Preferred reporting items for systematic reviews and meta-analysis diagram of study selection
[insert Figure 1]
Of the 86 included studies, 15 calculated more than one epidemic parameter of interest. Sixty of the 86 studies used data from mainland China for part or all of their analysis, and 11 specifically analysed outbreak data from Hubei province or the city of Wuhan.
Incubation period
A total of 33 studies estimated the incubation period of COVID-19 (Table 1). Mean estimates were reported in 15 studies, ranging from 1.8 to 9.9 days; however, 44% of the mean estimates were 5–6 days. The shortest mean estimate (incubation period = 1.8 days) was calculated from returned travellers from Hubei province in China, using their last day of travel as their date of exposure. (29) One study’s mean estimate of 9.9 days was calculated from a series of 14 cases in Viet Nam. (33)
Table 1. Estimated incubation period of COVID-19 from included epidemiological parameters studies published between 1 January and 30 April 2020.
| Study authors | Online publication date |
Study period | Sample size | Setting | Estimate (days)* | Uncertainty estimate (days) |
Uncertainty measure |
|---|---|---|---|---|---|---|---|
| Chan et al. (15) | 24 January 2020 | 26 December 2019 –15 January 2020 |
5 | Mainland China | - | 3–6 | Range |
| Li et al. (16) | 29 January 2020 | Up to 22 January 2020 | 10 | Wuhan/Hubei | 5.2 | 4.1–7.0 | 95% CI |
| Backer, Klinkenberg and Wallinga (17) | 6 February 2020 | 20 January 2020 –28 January 2020 |
88 | International | 6.4 | 5.6–7.7 | 95% CrI |
| Ki and Task Force for 2019-nCoV (18) | 9 February 2020 | 20 January 2020 –8 February 2020 |
28 | Republic of Korea | 3.9; [3.0] | 0–15 | Range |
| Jiang, Rayner and Luo (19) | 13 February 2020 | Up to 8 February 2020 | 50 | Mainland China | 4.9 | 4.4–5.5 | 95% CI |
| Linton et al. (20) | 17 February 2020 | 17 December 2019 –31 January 2020 | 158 | International | 5.6; [4.6] | 4.4–7.4; 3.7–5.7 | 95% CrI |
| Xu et al. (21) | 19 February 2020 | 10 January 2020 –26 January 2020 |
56 | Mainland China | [4] | 3–5 | IQR |
| Tian et al. (22) | 27 February 2020 | 20 January 2020 –10 February 2020 |
203 | Mainland China | [6.7] | ± 5.2 | SD |
| Cai et al. (23) | 28 February 2020 | 19 January 2020 –3 February 2020 |
10 | Mainland China | 6.5 | 2–10 | Range |
| Guan et al. (24) | 28 February 2020 | Up to 23 January 2020 | 291 | Mainland China | [4] | 2–7 | IQR |
| Liu et al. (25) | 3 March 2020 | 1 January 2020 –5 February 2020 |
58 | Mainland China | 6.0; [5.0] | 3–8; 1–16 | IQR; Range |
| Lauer et al. (26) | 10 March 2020 | 4 January 2020 –24 February 2020 |
181 | International | [5.1] | 4.5–5.8 | 95% CI |
| Zhao et al. (27) | 12 March 2020 | 23 January 2020 –5 February 2020 |
19 | Mainland China | [8] | 6–11 | IQR |
| Pung et al. (28) | 16 March 2020 | 18 January 2020 –10 February 2020 |
17 | Singapore | [4] | 3–6; 1–11 | IQR; Range |
| Leung (29) | 18 March 2020 | 20 January 2020 –12 February 2020 |
105 | Mainland China (travelled to Hubei) |
1.8 | 1.0–2.7 | 95% CI |
| 70 | Mainland China (local transmission) | 7.2 | 6.1–8.4 | 95% CI | |||
| Chang et al. (30) | 23 March 2020 | 28 January 2020 –9 February 2020 |
15 | Mainland China | [5] | 1–6 | Range |
| Jin et al. (31) | 24 March 2020 | 17 January 2020 –8 February 2020 |
21 | Mainland China – GI symptoms | [4] | 3–7 | IQR |
| 195 | Mainland China – No GI symptoms | [5] | 3–8 | IQR | |||
| Zhang et al. (32) | 2 April 2020 | 19 January 2020 –17 February 2020 |
49 | Mainland China | 5.2 | 1.8–12.4 | 95% CI |
| Le et al. (33) | 2 April 2020 | 17 January 2020 –14 February 2020 |
12 | Viet Nam | 9.9 | ± 5.2 | SD |
| Zhu and Chen (34) | 2 April 2020 | 1 December 2019 –23 January 2020 |
Not specified | Mainland China, Hong Kong Special Administrative Region (SAR) China, Macau (SAR) China, Taiwan (China) | 5.67 | 1–14 | Range |
| Han et al.35 | 6 April 2020 | 31 January 2020 –16 February 2020 |
25 | Mainland China – adults | [5] | 3–12 | Range |
| 7 | Mainland China – children |
[4] | 2–12 | Range | |||
| Shen et al.36 | 7 April 2020 | 8 January 2020 –26 February 2020 |
6 | Mainland China | [7.5] | 1–16 | Range |
| Sanche et al.37 | 7 April 2020 | 15 January 2020 –30 January 2020 |
24 | Mainland China | 4.2 | 3.5–5.1 | 95% CI |
| Ghinai et al.38 | 8 April 2020 | February–March 2020 | 15 | United States of America | 4.3; [4] | 1–7 | Range |
| Huang et al.39 | 10 April 2020 | 23 January 2020 –20 February 2020 |
8 | Mainland China | [2] | 1–4 | Range |
| Zheng et al.40 | 10 April 2020 | 17 January 2020 –7 February 2020 |
161 | Mainland China | [6] | 3–8 | Range |
| Xia et al.41 | 12 April 2020 | 23 January 2020 –18 February 2020 |
10 | China incl. Hong Kong Special Administrative Region (SAR) China, Macau (SAR) China, Taiwan (China) | 7.0 | ± 2.59; 2–14 | SD; Range |
| Chen et al.42 | 14 April 2020 | 28 January 2020 –11 February 2020 |
12 | Mainland China | 8.0 | 1–13 | Range |
| Song et al.43 | 23 April 2020 | 16 January 2020 –29 January 2020 |
22 | Mainland China | - | 2–13 | Range |
| Jiang et al.44 | 23 April 2020 | 23 January 2020 –13 February 2020 |
4 | Mainland China | - | 9–13 | Range |
| Nie et al.45 | 27 April 2020 | 19 January 2020 –8 February 2020 |
2907 | Mainland China | [5] | 2–8 | IQR |
| Yu et al.46 | 29 April 2020 | Up to 19 February 2020 | 132 | Mainland China | [7.2] | 6.4–7.9 | 95% CI |
| Bi et al.47 | 30 April 2020 | 14 January 2020 –12 February 2020 |
138 | Mainland China | [4.8] | 4.2–5.4 | 95% CI |
*Mean estimates. Median estimates are shown in [square brackets]. Multiple estimates of incubation period for the same population within the same study are shown in the same row and separated by a semicolon. Estimates of the incubation period in the same study for different populations are shown in separate rows.
CI: confidence interval; CrI: credible interval; GI: gastrointestinal; IQR: interquartile range; SD: standard deviation.
Notes: Sample size reported in Table 1 is the sample size used to calculate the incubation period, not necessarily the whole study sample. All estimates are reported to one decimal place, except where stating findings from papers that did not provide that level of precision.
A further 22 estimates of the incubation period were summarized by their median. These studies were generally reporting on a specific cluster or outbreak investigation, and median estimates largely ranged from 4 to 7 days. Estimates outside of this range were calculated from case series; for example, a median range of 1–4 days was found among eight participants (35) and an estimated 8-day incubation period for a study involving 19 participants. (27) The distribution of the mean and median incubation estimates by sample size of the study is shown in Fig. 2.
Figure 2.
Incubation period estimates and sample size of study (n = 28 studies, 35 estimates) published between 1 January and 30 April 2020
[insert Figure 2]
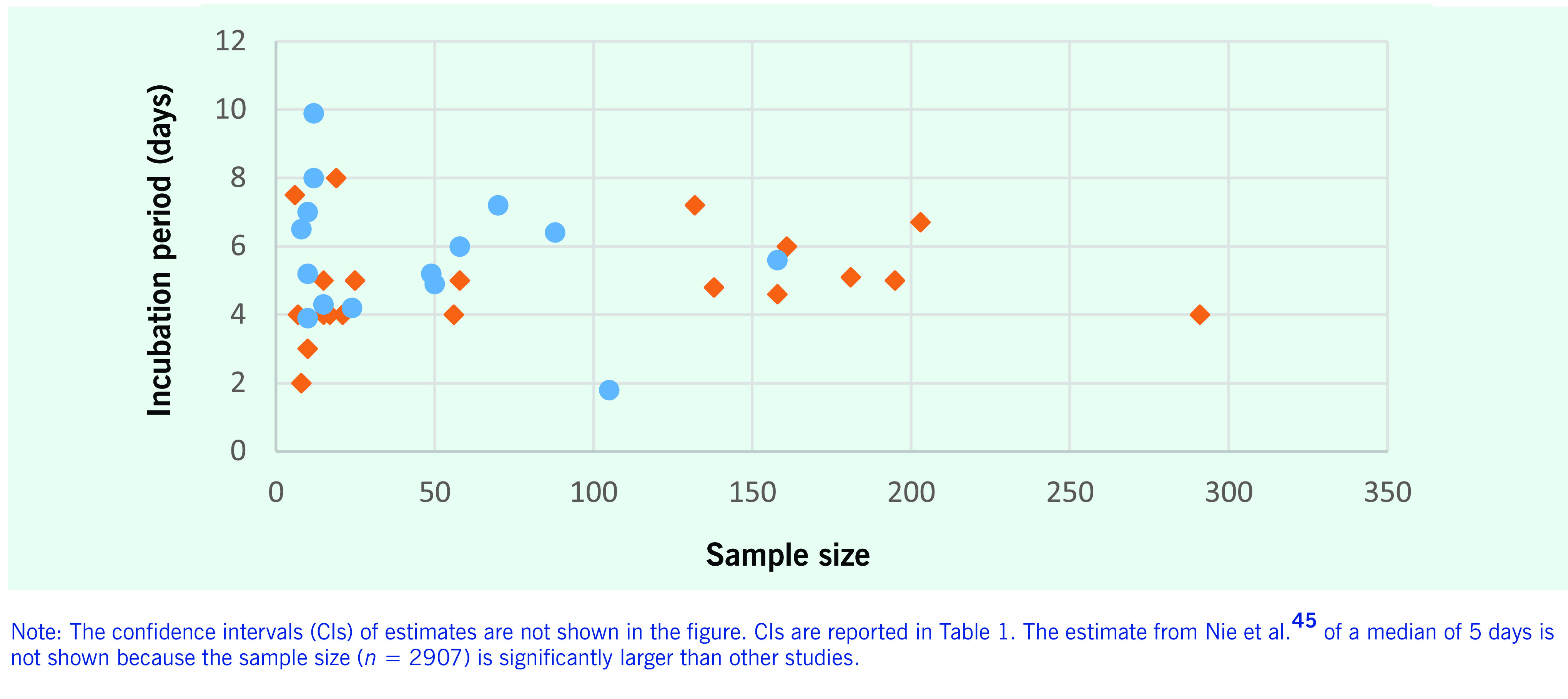
A further three studies only included a range of observed incubation periods. The longest incubation period from these studies was 16 days, recorded in an outbreak investigation in mainland China. (36) Additional estimates of the 95th percentile of the incubation period ranged from 10.3 days (95% confidence interval [CI]: 8.6–14.1) (17) to 14 days (95% CI: 12.2–15.9). (37)
Serial interval
Of the 15 studies that included a serial interval, eight were published in April 2020. Mean serial interval estimates were calculated in 14 studies and ranged from 3.1 to 7.5 days (Table 2).
Table 2. Estimated serial interval from included COVID-19 epidemiological parameters studies published between 1 January and 30 April 2020.
| Study authors |
Online publication date |
Study period | Sample size | Transmission pairs | Setting | Estimate (days)* | Uncertainty estimate (days) | Uncertainty measure |
|---|---|---|---|---|---|---|---|---|
| Li et al. (16) | 29 January 2020 | Up to 22 January 2020 | 10 | 6 | Wuhan/Hubei | 7.5 | 5.3–19.0 | 95% CI |
| Ki and Task Force for 2019-nCoV (18) | 9 February 2020 | 20 January 2020 –8 February 2020 |
28 | 12 | Republic of Korea | 6.6; [4.0] | 3–15 | Range |
| Liu et al. (25) | 3 March 2020 | 1 January 2020 –5 February 2020 |
15 single intracluster transmission cases | 12 clusters | Mainland China | 5.5 | - | - |
| 56 single co-exposure cases | 56 clusters | Mainland China | 3.1 | - | - | |||
| Nishiura et al. (38) | 4 March 2020 | Up to 12 February 2020 | Not specified |
28 – all pairs | International | [4.0] | 3.1–4.9 | 95% CrI |
| 18 – most certain pairs | International | [4.6] | 3.5–5.9 | 95% CrI | ||||
| Pung et al. (28) | 16 March 2020 | Up to 15 February 2020 | 4 | 3 | Singapore | 3–8 | Range | |
| Du et al. (39) | 19 March 2020 | 21 January 2020 –8 February 2020 | 752 | 468 | Mainland China | 4.0 | 3.5–4.4 | 95% CI |
| Wu et al. (40) | 19 March 2020 | 1 December 2019 –28 February 2020 | Not specified |
43 | International | 7 | 5.8–8.1 | 95% CI |
| Zhang et al. (32) | 2 April 2020 | 19 January 2020 –17 February 2020 |
63 | 35 | Mainland China | 5.1 | 3.1–11.6 | 95% CI |
| Ji et al. (41) | 7 April 2020 | 23 January 2020 –27 March 2020 | 51 | 32 | Wuhan/Hubei | 6.5 | 6.3 | SD |
| Huang et al. (35) | 10 April 2020 | 23 January 2020 –20 February 2020 | 9 | 8 | Mainland China | [1] | 0–4 | Range |
| Wang et al. (42) | 10 April 2020 | 11 January 2020 –16 February 2020 |
115 | 85 | Wuhan/Hubei | 5.5 | ± 2.7 | SD |
| He et al. (43) | 15 April 2020 | 7 January 2020 –4 March 2020 |
Not specified |
77 | International | 5.8; [5.2] | 4.8–6.8; 4.1–6.4 | 95% CI |
| Kwok et al. (44) | 23 April 2020 | 23 January 2020 –13 February 2020 |
38 | 26 | Hong Kong Special Administrative Region (SAR) China | 4.6 | 3.4–5.9 | 95% bCI |
| 26 – adjusted for right truncation | Hong Kong Special Administrative Region (SAR) China | 4.8 | 3.5–6.9 | 95% CrI | ||||
| Bi et al. (37) | 27 April 2020 | 14 January 2020 –12 February 2020 |
Not specified |
48 | Mainland China | 6.3; [5.4] | 5.2–7.6; 4.4–6.5 | 95% CI |
| Ganyani et al. (45) | 30 April 2020 | 14 January 2020 –27 February 2020 |
54 | 4 clusters | Singapore | 5.2 | –3.4–13.9 | 95% CrI |
| 114 | 16 clusters | Mainland China | 3.9 | –4.5–12.5 | 95% CrI |
*Mean estimates. Median estimates are shown in [square brackets]. Multiple estimates of serial interval for the same population within the same study are shown in the same row and separated by a semicolon. Estimates of the serial interval in the same study for different populations are shown in separate rows.
bCI: Bayesian confidence interval; CI: confidence interval; CrI: credible interval; SD: standard deviation.
Notes: Sample size reported is the sample size used to calculate the serial interval, not necessarily the whole study sample. All estimates are reported to one decimal place, except where stating findings from papers that did not provide that level of precision.
The estimated serial intervals were longer in studies published at the start than at the end of the study period, with a mean interval of 7.5 days in late January 2020 and a mean of 4–5 days in early March 2020. Estimates published from March 2020 onwards included transmission pairs with negative serial intervals, or intervals shorter than the incubation period, suggesting possible pre-symptomatic transmission. Mean estimates of the serial interval that included negative transmission pairs generally ranged from 3.9 to 5.8 days (Table 2).
The four median serial interval estimates ranged from 1.0 to 5.4 days. Excluding the estimate of 2 days from a case series of eight cases, (35) the median serial interval ranged from 4.0 to 5.4 days (Table 2).
Reproduction number
There were 90 estimates of the reproduction number from 52 studies across three World Health Organization (WHO) regions: Western Pacific Region, European Region and Region of the Americas. Reproduction number estimates ranged from 0.3 to 14.8. Of the 90 reported estimates, 33 estimates (37%) were between 2 and 3, and 20 estimates (22%) were between 3 and 4 (Table 3).
Table 3. Estimated reproduction number from included COVID-19 epidemiological parameters studies published between 1 January and 30 April 2020.
| Study authors | Online publication date |
Study period | Sample size | Method | Setting | Estimate | Uncertainty interval | Uncertainty measure |
|---|---|---|---|---|---|---|---|---|
| Wu et al. (46) | 23 January 2020 | 10 January 2020 –12 January 2020 | 41 | Zoonotic transmission – Cauchemez et al. 2013 (47) |
Wuhan/Hubei | 0.3 | 0.17–0.44 | 95% CI |
| Li et al. (16) | 29 January 2020 | Up to 22 January 2020 | 425 | Transmission model with renewal equations |
Wuhan/Hubei | 2.2 | 1.4–3.9 | 95% CI |
| Riou and Althaus (48) | 30 January 2020 | Up to 18 January 2020 | 50 | Stochastic transmission model |
Wuhan/Hubei | 2.2 | 1.4–3.8 | 90% HDI |
| Zhao et al. (49) | 30 January 2020 | 10 January 2020 –24 January 2020 |
2033 | Exponential growth model method | Mainland China | 2.24 –3.58 |
1.96–2.55 to 2.89–4.39 |
95% CI |
| Wu et al. (50) | 31 January 2020 | 1 December 2019 –28 January 2020 | 55 | Differential equation – SEIR compartment model |
International | 2.68 | 2.47–2.86 | 95% CrI |
| Zhao et al. (51) | 1 February 2020 | 1 December 2019 –24 January 2020 | 41 | Exponential growth model method | Mainland China | 2.56 | 2.49–2.63 | 95% CI |
| Tang et al. (52) | 7 February 2020 | 10 January 2020 –15 January 2020 | 41 | Differential equation – SEIR compartment model |
Mainland China | 6.47 | 5.71–7.23 | 95% CI |
| Ki and Task Force for 2019-nCoV (18) | 9 February 2020 | 20 January 2020 – 8 February 2020 |
26 | Estimated from transmission chains | Republic of Korea | 0.48 | 0.25–0.84 | 95% CI |
| Zhou et al. (53) | 12 February 2020 | Up to 25 January 2020 | 2820 | Differential equation – SEIR compartment model |
Mainland China | 2.83–3.28 | - | - |
| Jung et al. (54) | 14 February 2020 | 31 December 2019 –24 January 2020 | 92 | Exponential growth model method | Mainland China | 2.1; 3.2 | 2.0–2.2; 2.7–3.7 | 95% CI |
| Zhang et al. (55) | 22 February 2020 | Up to 16 February 2020 | 355 | Cori et al. methodology (56) | Cruise ship | 2.28 | 2.06–2.52 | 95% CI |
| Lai et al. (57) | 25 February 2020 | Up to 4 February 2020 | 52 | Coalescent-based exponential growth and a birth-death skyline method | Mainland China | 2.6 | 2.1–5.1 | 95% CI |
| Chen et al. (58) | 28 February 2020 | 7 December 2019 –1 January 2020 | Not specified | Bats-Hosts-Reservoir-People transmission network model | Wuhan/Hubei | 3.58 | - | - |
| Rocklov, Sjodin and Wilder-Smith (59) | 28 February 2020 | 21 January 2020 –19 February 2020 | 3700 | Differential equation – SEIR compartment model |
Cruise ship | 14.8 | - | - |
| Mizumoto and Chowell (60) | 29 February 2020 | 20 January 2020 –17 February 2020 | 3711 | Discrete time integral equation |
Cruise ship | 5.8 | 0.6–11.0 | 95% CrI |
| Fang, Nie and Penny (61) | 6 March 2020 | 20 January 2020 –29 February 2020 | 35 329 | Differential equation – SEIR compartment model |
Mainland China | 2.35–3.21 | - | - |
| Zhou et al.70 | 10 March 2020 | 10 January 2020 –31 January 2020 |
44 | Differential equation – SEIR compartment model |
Mainland China | 5.3167 | - | - |
| Kucharski et al.71 |
11 March 2020 | 1 December 2019 –11 February 2020 | Not specified |
Differential equation – SEIR compartment model |
Wuhan/Hubei | 2.35 | 1.15–4.77 | 95% CI |
| Yang and Wang72 | 11 March 2020 | 23 January 2020 –10 February 2020 | Not specified |
Differential equation – SEIR compartment model |
Wuhan/Hubei | 4.25 | - | - |
| Zhao and Chen73 | 11 March 2020 | 20 January 2020 –30 January 2020 |
Not specified |
Differential equation – SEIR compartment model |
Mainland China | 4.7092 | - | - |
| Choi and Ki74 | 12 March 2020 | 29 December 2019 –3 January 2020 |
Not specified |
Differential equation – SEIR compartment model |
Wuhan/Hubei | 4.028 | 4.010–4.046 | 95% CI |
| - | - | 20 January 2020 –17 February 2020 |
30 | - | Republic of Korea | 0.555 | 0.509–0.602 | 95% CI |
| Kuniya75 | 13 March 2020 | 15 January 2020 –29 February 2020 |
239 | Differential equation – SEIR compartment model |
Japan | 2.6 | 2.4–2.8 | 95% CI |
| Remuzzi and Remuzzi76 | 13 March 2020 | 19 February 2020 –8 March 2020 | Unclear | Exponential growth model method | Italy | 2.76–3.25 | - | - |
| Li et al.77 | 16 March 2020 | 10 January 2020 –23 January 2020 |
801 | Differential equation – SEIR compartment model |
Mainland China | 2.38 | 2.03–2.77 | 95% CrI |
| Shim et al.78 | 17 March 2020 | 20 January 2020 –26 February 2020 |
6284 | Generalized growth model | Republic of Korea | 1.5 | 1.4–1.6 | 95% CI |
| Du et al.49 | 19 March 2020 | 21 January 2020 –8 February 2020 |
752 | Not stated | Mainland China | 1.32 | 1.16–1.48 | 95% CI |
| Wu et al.50 | 19 March 2020 | 1 December 2019 –28 February 2020 |
45 771 | Differential equation – SEIR compartment model |
Wuhan/Hubei | 1.94 | 1.83–2.06 | 95% CrI |
| Yuan et al.79 | 28 March 2020 | 23 February 2020 –9 March 2020 |
Not specified |
Exponential growth model method; Wallinga time dependent method |
Italy | 3.27; 3.10 | 3.17–3.38; 2.21–4.11 | 95% CI |
| - | - | - | - | - | France | 6.32; 6.56 | 5.72–6.99; 2.04–12.26 | 95% CI |
| - | - | - | - | - | Spain | 5.08; 3.95 | 4.51–5.74; 0–10.19 | 95% CI |
| - | - | - | - | - | Germany | 6.07; 4.43 | 5.51–6.69; 1.83–7.92 | 95% CI |
| Anastassopoulou et al.80 | 31 March 2020 | 11 January 2020 –10 February 2020 |
Not specified |
Differential equation – SEIR compartment model |
Wuhan/Hubei | 4.6 | 3.56–5.65 | 90% CI |
| Ferretti et al.81 | 31 March 2020 | Up to end March 2020 | 40 transmission pairs |
Exponential growth model method | Mainland China | 2 | 1.7–2.5 | 90% CI |
| Huang et al.82 | 31 March 2020 | 13 January 2020 –9 March 2020 |
80 754 | Differential equation – SEIR compartment model |
Mainland China | 2.23–2.51 | - | - |
| Tian et al.83 | 31 March 2020 | 31 December 2019 –23 January 2020 | Not specified |
Differential equation – SEIR compartment model |
Mainland China | 3.15 | 3.04–3.26 | 95% BCI |
| Zhu and Chen34 | 2 April 2020 | 1 December 2019 –23 January 2020 |
Not specified |
Poisson Transmission Model |
Mainland China | 2.47 | 2.39–2.55 | 95% CI |
| Sanche et al.37 | 7 April 2020 | 15 January 2020 –30 January 2020 |
140 | Differential equation – SEIR compartment model |
Mainland China | 5.7 | 3.8–8.9 | 95% CI |
| Zhao et al.84 | 8 April 2020 | 1 December 2019 –8 January 2020 |
Not specified |
Differential equation – SEIR compartment model |
Wuhan/Hubei | 2.5 | 2.4–2.7 | 95% CI |
| Pan, Liu and Wang85 | 10 April 2020 | 5 December 2019 –8 March 2020 | 32 583 | Cori et al. methodology112 | Wuhan/Hubei | 3.82 | 3.72–3.93 | 95% CrI |
| Abbott et al.86 | 14 April 2020 | Up to 25 January 2020 | 1975 | Stochastic branching process model |
Mainland China | 2.8–3.8 | - | - |
| Puci et al. | 14 April 2020 | 22 March 2020 –29 March 2020 |
975 | Differential equation – SEIR compartment model |
Italy | 1.82 | 1.51–2.01 | 95% CI |
| Du et al.87 | 16 April 2020 | 1 December 2019 –22 January 2020 |
19 | Exponential growth method | Mainland China | 1.9 | 1.47–2.59 | 95% CrI |
| Torres-Roman et al.88 | 17 April 2020 | 6 March 2020 –15 March 2020 |
Not specified |
Cori et al. methodology112 |
Peru | 2.97 | - | - |
| Tsang et al.89 | 20 April 2020 | 15 January 2020 –3 March 2020 |
Not specified |
Exponential growth model | Mainland China | 2.8–3.5 | - | - |
| Muniz- Rodriguez et al.90 |
22 April 2020 | 19 February 2020 –19 March 2020 |
978 | Exponential growth model; renewal equations method | Islamic Republic of Iran | 4.4; 3.5 | 3.9–4.9; 1.3–8.1 | 95% CI |
| Zhuang et al.91 | 22 April 2020 | Up to 5 March 2020 | Not specified |
Stochastic model, maximum likelihood estimation approach |
Italy | 2.6; 3.3 | 2.3–2.9; 3.0–3.6 | 95% CI |
| - | - | - | - | - | Republic of Korea | 2.6; 3.2 | 2.3–2.9; 2.9–3.5 | 95% CI |
| Gatto et al.92 | 23 April 2020 | 24 February 2020 –23 March 2020 |
107 | Differential equation – SEIR compartment model |
Italy | 3.6 | 3.49–3.84 | 95% CI |
| Han et al.93 | 23 April 2020 | 21 January 2020 –15 February 2020 |
482 | Exponential growth model method | Mainland China | 2.9 | 1.8–4.5 | 95% CI |
| Caicedo-Ochoa et al.94 |
25 April 2020 | Up to 23 March 2020 (first 10 days after reaching 25 cases in each location) | Not specified |
Cori et al. methodology112 Two serial intervals used: 7.5 days; 4.7 days |
Spain | 6.48; 2.9 | 5.97–7.02; 2.67–3.14 | 95% CrI |
| - | - | - | - | - | Italy | 6.41; 2.83 | 6.11–6.71; 2.70–2.96 | 95% CrI |
| - | - | - | - | - | Ecuador | 12.86; 3.95 | 12.05–13.68; 3.70–4.21 | 95% CrI |
| - | - | - | - | - | Panama | 7.19; 3.67 | 6.37–8.08; 3.25–4.13 | 95% CrI |
| - | - | - | - | - | Brazil | 6.53; 2.91 | 5.85–7.25; 2.60–3.23 | 95% CrI |
| - | - | - | - | - | Chile | 5.79; 2.67 | 5.32–6.28; 2.45–2.89 | 95% CrI |
| - | - | - | - | - | Colombia | 5.65; 2.67 | 5.04–6.29; 2.38–2.98 | 95% CrI |
| - | - | - | - | - | Peru | 5.24; 2.36 | 4.68–5.83; 2.11–2.63 | 95% CrI |
| - | - | - | - | - | Mexico | 4.94; 2.42 | 4.37–5.56; 2.14–2.72 | 95% CrI |
| Bi et al.47 | 27 April 2020 | 14 January 2020 –12 February 2020 |
48 | Estimated from transmission chains | Mainland China | 0.4 | 0.3–0.5 | 95% CI |
| Distante et al.95 | 27 April 2020 | Up to 29 March 2020 | Not specified |
Exponential growth method | Italy | 3.6 | - | - |
| Ndairou et al.96 | 27 April 2020 | 4 January 2020 –9 March 2020 |
Not specified |
Differential equation – SEIR compartment model |
Wuhan/Hubei | 0.945 | - | - |
| Peirlinck et al.97 | 27 April 2020 | 21 January 2020 –4 April 2020 |
311 357 | Differential equation – SEIR compartment model |
United States of America | 5.3 | ± 0.95 | SD |
| Adegboye et al.98 |
28 April 2020 | 27 February 2020 –11 April 2020 |
318 | Cori et al. methodology112 |
Nigeria | 2.71 | - | - |
| Ganyani et al.55 | 30 April 2020 | 14 January 2020 –27 February 2020 |
91 | Exponential growth model method | Singapore | 1.25 | 1.17–1.34 | 95% CrI |
| - | - | - | 135 | Exponential growth model method | Mainland China | 1.41 | 1.26–1.58 | 95% CrI |
| Ivorra et al.99 | 30 April 2020 | 1 December 2019 –29 March 2020 |
Not specified |
Differential equation – SEIR compartment model |
Mainland China | 4.2732 | - | - |
Multiple estimates of the reproduction number for the same population within the same study are shown in the same row and separated by a semicolon. Estimates of the incubation period in the same study for different populations are shown in separate rows.
bCI: Bayesian confidence interval; CI: confidence interval; CrI: credible interval; HDI: high density interval; SD: standard deviation; SEIR: susceptible-exposed-infected-recovered.
Notes: Sample size reported is the sample size used to calculate the serial interval, not necessarily the whole study sample. All estimates are reported to the number of decimal places provided in each study.
The initial low estimate of 0.3 relied on the early assumption that the pathogen was primarily spread through zoonotic transmission. (46) Other estimates of the reproduction number under 1 were reported in jurisdictions with rapid public health interventions during the study period, including the Republic of Korea and Singapore. (18, 45, 62) The highest reproduction number estimate (14.8) was from analyses of transmission dynamics onboard the Diamond Princess cruise ship. (59)
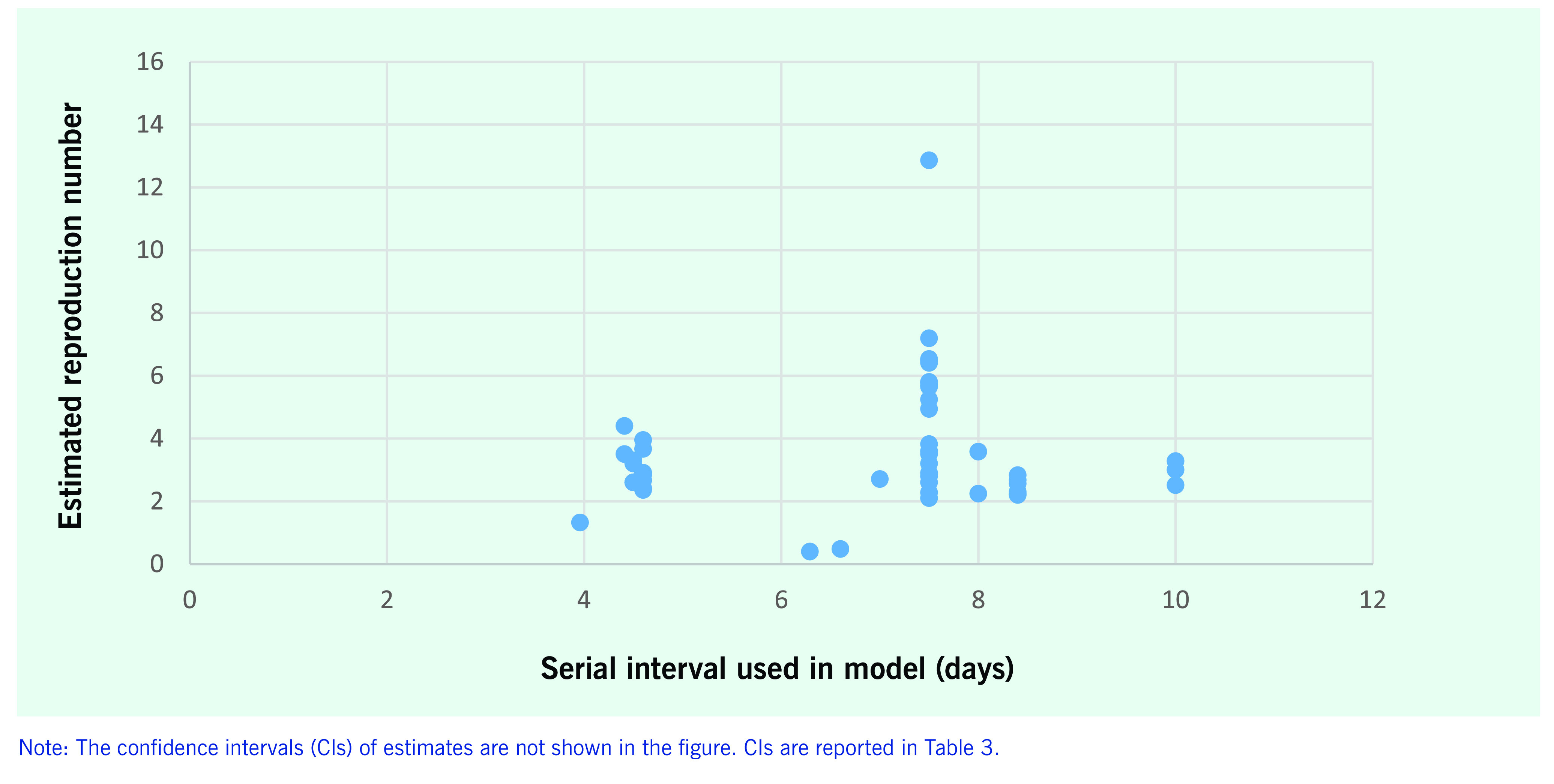
The distribution of reproduction number estimates by the assumed serial interval is shown in Fig. 3. Just over half (n = 50) of the 90 reproduction number results used an estimate of the serial interval to calculate the reproduction number. Serial interval estimates used to estimate the reproduction number ranged from 4 (39) to 10 days, with the latter taken from the estimated serial interval for severe acute respiratory syndrome (SARS) in early outbreaks. (63) Studies generally applied serial intervals from the earliest COVID-19 estimate of 7.5 days (16) and the accepted serial interval of SARS of 8.4 days. (63)
Figure 3.
Estimated reproduction number and serial interval of the model (n = 23 studies, 50 estimates) published between 1 January and 30 April 2020
[insert Figure 3]
Discussion
This study provides a review of estimated epidemic parameters of the COVID-19 outbreak up to 30 April 2020. Estimates of the incubation period were similar across the study period, with a mean estimated value of 5–6 days and a range of 2–14 days. Estimates of the serial interval shortened over the study period, from 7.5 days in late January 2020 to a mean of 4–5 days in early March 2020.
Estimates of the reproduction number varied in the studies collated up to 30 April 2020. Although some estimates of the reproduction number were as high as 14.8, over half were between 2 and 4. The higher estimates demonstrate the impact of the setting, individual behaviours and public health interventions – the highest estimates were associated with cruise ships, (55, 59, 60) whereas the lowest estimates were generally calculated in areas with a rapid response to an outbreak. (18, 45, 62, 64)
The incubation period reflects the growth of a virus in an individual, and thus is largely a biological function that would not be expected to vary with changes in human behaviour and wider public health interventions. Variations in the incubation period reported in this study may, in part, result from the study designs adopted. Several estimates of the incubation period were reported directly from cluster investigations, often with low sample sizes. Studies with more than 20 participants had less variation between estimates than studies with smaller sample sizes. The definition of exposure, including the potential for continuous exposure in a household, may also have influenced results by artificially lengthening or shortening the incubation period, depending on study design and differences in local epidemiological reporting protocols.
The serial interval and reproduction number are likely to be influenced by public health interventions, social behaviours and political decisions. Estimates of these two epidemic characteristics are therefore setting-specific, which may explain the variance across the results in this study. The serial interval estimates also changed as new information about the pathogen came to light, primarily the potential for pre-symptomatic and pauci-symptomatic transmission. (65-70) However, these revised estimates of the serial interval were rarely used to revise reproduction number estimates. A longer serial interval results in a higher estimate of the reproduction number. The earliest published estimate by Li et al.’s study (first published online on 29 January 2020) (16) of six transmission pairs in Wuhan was higher than most of the later estimates. That estimate was applied as an assumed serial interval in 10 studies published in March and April 2020, (54, 55, 57, 60, 71-76) despite not being used in Li et al.’s own calculation of the reproduction number. (16) These early studies have been used to inform national and regional responses to the COVID-19 pandemic, and they demonstrate the importance of and reliance on early estimates to inform future research and public health decision-making.
Variations in the estimated reproduction number may also occur due to other assumptions applied in calculations. The initial estimate of the reproduction number of 0.3 assumed zoonotic transmission as the primary mode of transmission, based on the information available at the time. (46) The method applied may also influence the final estimate of the reproduction number. This is evident in the studies estimating the reproduction number of the Wuhan outbreak from December 2019 to mid-February 2020, which increased in later publications that used the same data sources and time periods. The reproduction number was estimated to be 2.2 in studies published in January and February 2020, (16, 48) but increased to 4 in articles published in March and April 2020. (62, 77, 78)
The epidemiological parameters reviewed share some similarities to that of SARS and Middle East respiratory syndrome (MERS), two diseases caused by coronaviruses that have caused significant outbreaks in the early 21st century. The estimates of the range and mean of the incubation period of COVID-19 are similar to that of SARS (2–10 days, mean of 5–6 days) (2, 63, 79) and MERS (2–14 days, median of 5–6 days). (79, 80) However, the estimated serial interval for COVID-19 is shorter than the observed intervals for SARS (8.4 days) (63) and MERS (7.6–12.6 days). (80, 81) The later estimates of the COVID-19 serial interval published in April 2020 are shorter than the estimates for the incubation period, suggesting the potential for pre-symptomatic transmission, which has not been observed for SARS or MERS. (63, 80, 82) The estimated reproduction number of COVID-19 is similar to the estimates for the 2002–2003 SARS outbreak. (63)
This study has some important limitations. It provides a descriptive assessment and does not include meta-analysis or recalculations of results. The use of different methods and different outputs from each study limits the capacity for meta-analysis. This review may also be impaired by publication bias. Several included studies were based on small sample sizes, which led to imprecise results. The ongoing pandemic requires the active involvement of public health researchers to assess unfolding situations and advise on local responses. Fulfilling crucial roles as the pandemic unfolded may have limited the potential to publish findings, restricting our understanding of epidemic parameters in real time and reducing the representativeness of the results. This potential publication bias may also explain in part the overrepresentation of data from mainland China although COVID-19 has led to outbreaks worldwide. Nevertheless, the early published estimates included in this study have been used worldwide to inform public health responses, and they provide the best available evidence in the timeframe of this study.
Only studies written in English were included in this review. This excludes many early estimates written in Mandarin and Korean, which also limits the representativeness of this analysis. Furthermore, this analysis was limited to peer-reviewed published journal articles indexed in PubMed, which represents only a fraction of the literature published on the COVID-19 pandemic. The current pandemic has seen the proliferation of pre-print articles and increased attention on their results. Grey literature published by WHO, national governments and other organizations were also omitted. In times of emergency, pre-prints and grey literature may provide new information in a timely manner; however, this review focused only on estimations of epidemic parameters that have been subject to external peer review.
Pandemics are inherently uncertain times. The challenges of the ongoing COVID-19 pandemic are compounded by SARS-CoV-2 being a new pathogen, which public health and clinical professionals have had to rapidly assess, understand and respond to. Early estimates can provide useful interim guidance for public health decision-making. This is particularly true for transmission that is driven by biological characteristics, such as the incubation period. Epidemic characteristics that are influenced by human behaviours and public health interventions are less certain and require interpretation within the context of data collection and analysis of the study. Reliance on data from small sample sizes and specific settings is necessary in the context of an outbreak, but it also limits the generalizability of findings to other contexts.
Uncertainty in epidemic characteristics should not mean that we do not act. Although earlier estimates may rely on less-than-ideal sample sizes and sample structures, they are necessary to facilitate decision-making in a timely manner. However, reliance on the first estimates published may limit or bias our understanding of new data. The increasing availability of pre-print articles provides an outlet for urgent distribution of findings during an outbreak of a novel pathogen, provided preliminary findings are interpreted with caution before peer review. This study underscores the ongoing challenge and ever-present need for outbreak investigations and research to be both timely and frequently updated, to provide the best evidence to guide interventions. Further research is required to refine estimates of the serial interval and reproduction number, to improve our understanding of this pandemic in different contexts, and to provide reference values to enable a timely response to potential future outbreaks of COVID-19 and any future emerging coronaviruses and other potential pandemic diseases.
Conflicts of interest
None declared.
Funding
None.
References
- 1.Weekly epidemiological update on COVID-19 - 20 April 2021. Geneva: World Health Organization; 2021. Available from: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19 — 20-april-2021, accessed 23 April 2021.
- 2.Farewell VT, Herzberg AM, James KW, Ho LM, Leung GM. SARS incubation and quarantine times: when is an exposed individual known to be disease free? Stat Med. 2005November30;24(22):3431–45. 10.1002/sim.2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishiura H. Early efforts in modeling the incubation period of infectious diseases with an acute course of illness. Emerg Themes Epidemiol. 2007May11;4(1):2. 10.1186/1742-7622-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishiura H. Determination of the appropriate quarantine period following smallpox exposure: an objective approach using the incubation period distribution. Int J Hyg Environ Health. 2009January;212(1):97–104. 10.1016/j.ijheh.2007.10.003 [DOI] [PubMed] [Google Scholar]
- 5.Fine PEM. The interval between successive cases of an infectious disease. Am J Epidemiol. 2003December1;158(11):1039–47. 10.1093/aje/kwg251 [DOI] [PubMed] [Google Scholar]
- 6.Ma Y, Horsburgh CR Jr, White LF, Jenkins HE. Quantifying TB transmission: a systematic review of reproduction number and serial interval estimates for tuberculosis. Epidemiol Infect. 2018September;146(12):1478–94. 10.1017/S0950268818001760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker NG, Wang D, Clements M. Type and quantity of data needed for an early estimate of transmissibility when an infectious disease emerges. Euro Surveill. 2010July1;15(26):19603. 10.2807/ese.15.26.19603-en [DOI] [PubMed] [Google Scholar]
- 8.Caley P, Philp DJ, McCracken K. Quantifying social distancing arising from pandemic influenza. J R Soc Interface. 2008June6;5(23):631–9. 10.1098/rsif.2007.1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishiura H, Chowell G, Safan M, Castillo-Chavez C. Pros and cons of estimating the reproduction number from early epidemic growth rate of influenza A (H1N1) 2009. Theor Biol Med Model. 2010January7;7(1):1. 10.1186/1742-4682-7-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mercer GN, Glass K, Becker NG. Effective reproduction numbers are commonly overestimated early in a disease outbreak. Stat Med. 2011April30;30(9):984–94. 10.1002/sim.4174 [DOI] [PubMed] [Google Scholar]
- 11.Chang T-H, Wu J-L, Chang L-Y. Clinical characteristics and diagnostic challenges of pediatric COVID-19: A systematic review and meta-analysis. J Formos Med Assoc. 2020May;119(5):982–9. 10.1016/j.jfma.2020.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park M, Cook AR, Lim JT, Sun Y, Dickens BL. A systematic review of COVID-19 epidemiology based on current evidence. J Clin Med. 2020March31;9(4):967. 10.3390/jcm9040967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balla M, Merugu GP, Patel M, Koduri NM, Gayam V, Adapa S, et al. COVID-19, modern pandemic: A systematic review from front-line health care providers’ perspective. J Clin Med Res. 2020April;12(4):215–29. 10.14740/jocmr4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alimohamadi Y, Taghdir M, Sepandi M. Estimate of the basic reproduction number for COVID-19: A systematic review and meta-analysis. J Prev Med Public Health. 2020May;53(3):151–7. 10.3961/jpmph.20.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan JF-W, Yuan S, Kok K-H, To KK-W, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020February15;395(10223):514–23. 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020March26;382(13):1199–207. 10.1056/NEJMoa2001316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Backer JA, Klinkenberg D, Wallinga J. Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travellers from Wuhan, China, 20-28 January 2020. Euro Surveill. 2020February;25(5): 10.2807/1560-7917.ES.2020.25.5.2000062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ki M. Task Force for 2019-nCoV. Epidemiologic characteristics of early cases with 2019 novel coronavirus (2019-nCoV) disease in Republic of Korea. Epidemiol Health. 2020February9;42:e2020007. 10.4178/epih.e2020007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang X, Rayner S, Luo M-H. Does SARS-CoV-2 has a longer incubation period than SARS and MERS? J Med Virol. 2020May;92(5):476–8. 10.1002/jmv.25708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linton NM, Kobayashi T, Yang Y, Hayashi K, Akhmetzhanov AR, Jung S-M, et al. Incubation period and other epidemiological characteristics of 2019 novel coronavirus infections with right truncation: A statistical analysis of publicly available case data. J Clin Med. 2020February17;9(2):E538. 10.3390/jcm9020538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu X-W, Wu X-X, Jiang X-G, Xu K-J, Ying L-J, Ma C-L, et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: retrospective case series. BMJ. 2020February19;368:m606. 10.1136/bmj.m606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian S, Hu N, Lou J, Chen K, Kang X, Xiang Z, et al. Characteristics of COVID-19 infection in Beijing. J Infect. 2020April;80(4):401–6. 10.1016/j.jinf.2020.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiehao C, Jin X, Daojiong L, Zhi Y, Lei X, Zhenghai Q, et al. A case series of children with 2019 novel coronavirus infection: Clinical and epidemiological features. Clin Infect Dis. 2020September12;71(6):1547–51. 10.1093/cid/ciaa198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guan W-J, Ni Z-Y, Hu Y, Liang W-H, Ou C-Q, He J-X, et al. ; China Medical Treatment Expert Group for Covid-19. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020April30;382(18):1708–20. 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Liao X, Qian S, Yuan J, Wang F, Liu Y, et al. Community transmission of severe acute respiratory syndrome coronavirus 2, Shenzhen, China, 2020. Emerg Infect Dis. 2020June;26(6):1320–3. 10.3201/eid2606.200239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng Q, Meredith HR, et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: Estimation and application. Ann Intern Med. 2020May5;172(9):577–82. 10.7326/M20-0504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao D, Yao F, Wang L, Zheng L, Gao Y, Ye J, et al. A comparative study on the clinical features of COVID-19 pneumonia to other pneumonias. Clin Infect Dis. 2020July28;71(15):756–61. 10.1093/cid/ciaa247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pung R, Chiew CJ, Young BE, Chin S, Chen MI-C, Clapham HE, et al. ; Singapore 2019 Novel Coronavirus Outbreak Research Team. Investigation of three clusters of COVID-19 in Singapore: implications for surveillance and response measures. Lancet. 2020March28;395(10229):1039–46. 10.1016/S0140-6736(20)30528-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leung C. The difference in the incubation period of 2019 novel coronavirus (SARS-CoV-2) infection between travelers to Hubei and nontravelers: The need for a longer quarantine period. Infect Control Hosp Epidemiol. 2020May;41(5):594–6. 10.1017/ice.2020.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang D, Mo G, Yuan X, Tao Y, Peng X, Wang FS, et al. Time kinetics of viral clearance and resolution of symptoms in novel coronavirus infection. Am J Respir Crit Care Med. 2020May1;201(9):1150–2. 10.1164/rccm.202003-0524LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin X, Lian J-S, Hu J-H, Gao J, Zheng L, Zhang Y-M, et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. 2020June;69(6):1002–9. 10.1136/gutjnl-2020-320926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, Litvinova M, Wang W, Wang Y, Deng X, Chen X, et al. Evolving epidemiology and transmission dynamics of coronavirus disease 2019 outside Hubei province, China: a descriptive and modelling study. Lancet Infect Dis. 2020July;20(7):793–802. 10.1016/S1473-3099(20)30230-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le TQM, Takemura T, Moi ML, Nabeshima T, Nguyen LKH, Hoang VMP, et al. Severe acute respiratory syndrome coronavirus 2 shedding by travelers, Vietnam, 2020. Emerg Infect Dis. 2020July;26(7):1624–6. 10.3201/eid2607.200591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Y, Chen YQ. On a statistical transmission model in analysis of the early phase of COVID-19 outbreak. Stat Biosci. 2020April2;13(1):1–17. 10.1007/s12561-020-09277-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang L, Zhang X, Zhang X, Wei Z, Zhang L, Xu J, et al. Rapid asymptomatic transmission of COVID-19 during the incubation period demonstrating strong infectivity in a cluster of youngsters aged 16-23 years outside Wuhan and characteristics of young patients with COVID-19: A prospective contact-tracing study. J Infect. 2020June;80(6):e1–13. 10.1016/j.jinf.2020.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen Q, Guo W, Guo T, Li J, He W, Ni S, et al. Novel coronavirus infection in children outside of Wuhan, China. Pediatr Pulmonol. 2020June;55(6):1424–9. 10.1002/ppul.24762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bi Q, Wu Y, Mei S, Ye C, Zou X, Zhang Z, et al. Epidemiology and transmission of COVID-19 in 391 cases and 1286 of their close contacts in Shenzhen, China: a retrospective cohort study. Lancet Infect Dis. 2020August;20(8):911–9. 10.1016/S1473-3099(20)30287-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishiura H, Linton NM, Akhmetzhanov AR. Serial interval of novel coronavirus (COVID-19) infections. Int J Infect Dis. 2020April;93:284–6. 10.1016/j.ijid.2020.02.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du Z, Xu X, Wu Y, Wang L, Cowling BJ, Meyers LA. Serial interval of COVID-19 among publicly reported confirmed cases. Emerg Infect Dis. 2020June;26(6):1341–3. 10.3201/eid2606.200357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu JT, Leung K, Bushman M, Kishore N, Niehus R, de Salazar PM, et al. Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nat Med. 2020April;26(4):506–10. 10.1038/s41591-020-0822-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ji T, Chen H-L, Xu J, Wu L-N, Li J-J, Chen K, et al. Lockdown contained the spread of 2019 novel coronavirus disease in Huangshi city, China: Early epidemiological findings. Clin Infect Dis. 2020September12;71(6):1454–60. 10.1093/cid/ciaa390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Z, Ma W, Zheng X, Wu G, Zhang R. Household transmission of SARS-CoV-2. J Infect. 2020July;81(1):179–82. 10.1016/j.jinf.2020.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He X, Lau EHY, Wu P, Deng X, Wang J, Hao X, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020May;26(5):672–5. 10.1038/s41591-020-0869-5 [DOI] [PubMed] [Google Scholar]
- 44.Kwok KO, Wong VWY, Wei WI, Wong SYS, Tang JW-T. Epidemiological characteristics of the first 53 laboratory-confirmed cases of COVID-19 epidemic in Hong Kong, 13 February 2020. Euro Surveill. 2020April;25(16): 10.2807/1560-7917.ES.2020.25.16.2000155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ganyani T, Kremer C, Chen D, Torneri A, Faes C, Wallinga J, et al. Estimating the generation interval for coronavirus disease (COVID-19) based on symptom onset data, March 2020. Euro Surveill. 2020April;25(17):2000257. 10.2807/1560-7917.ES.2020.25.17.2000257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu P, Hao X, Lau EHY, Wong JY, Leung KSM, Wu JT, et al. Real-time tentative assessment of the epidemiological characteristics of novel coronavirus infections in Wuhan, China, as at 22 January 2020. Euro Surveill. 2020January;25(3): 10.2807/1560-7917.ES.2020.25.3.2000044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cauchemez S, Epperson S, Biggerstaff M, Swerdlow D, Finelli L, Ferguson NM. Using routine surveillance data to estimate the epidemic potential of emerging zoonoses: application to the emergence of US swine origin influenza A H3N2v virus. PLoS Med. 2013;10(3):e1001399. 10.1371/journal.pmed.1001399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riou J, Althaus CL. Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020. Euro Surveill. 2020January;25(4): 10.2807/1560-7917.ES.2020.25.4.2000058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao S, Lin Q, Ran J, Musa SS, Yang G, Wang W, et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: A data-driven analysis in the early phase of the outbreak. Int J Infect Dis. 2020March;92:214–7. 10.1016/j.ijid.2020.01.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020February29;395(10225):689–97. 10.1016/S0140-6736(20)30260-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao S, Musa SS, Lin Q, Ran J, Yang G, Wang W, et al. Estimating the unreported number of novel coronavirus (2019-nCoV) cases in China in the first half of January 2020: a data-driven modelling analysis of the early outbreak. J Clin Med. 2020February1;9(2):E388. 10.3390/jcm9020388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang B, Wang X, Li Q, Bragazzi NL, Tang S, Xiao Y, et al. Estimation of the transmission risk of the 2019-nCoV and its implication for public health interventions. J Clin Med. 2020February7;9(2):462. 10.3390/jcm9020462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou T, Liu Q, Yang Z, Liao J, Yang K, Bai W, et al. Preliminary prediction of the basic reproduction number of the Wuhan novel coronavirus 2019-nCoV. J Evid Based Med. 2020February;13(1):3–7. 10.1111/jebm.12376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jung S-M, Akhmetzhanov AR, Hayashi K, Linton NM, Yang Y, Yuan B, et al. Real-time estimation of the risk of death from novel coronavirus (COVID-19) infection: inference using exported cases. J Clin Med. 2020February14;9(2):E523. 10.3390/jcm9020523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang S, Diao M, Yu W, Pei L, Lin Z, Chen D. Estimation of the reproductive number of novel coronavirus (COVID-19) and the probable outbreak size on the Diamond Princess cruise ship: A data-driven analysis. Int J Infect Dis. 2020April;93:201–4. 10.1016/j.ijid.2020.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cori A, Ferguson NM, Fraser C, Cauchemez S. A new framework and software to estimate time-varying reproduction numbers during epidemics. Am J Epidemiol. 2013November1;178(9):1505–12. 10.1093/aje/kwt133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lai A, Bergna A, Acciarri C, Galli M, Zehender G. Early phylogenetic estimate of the effective reproduction number of SARS-CoV-2. J Med Virol. 2020June;92(6):675–9. 10.1002/jmv.25723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen T-M, Rui J, Wang Q-P, Zhao Z-Y, Cui J-A, Yin L. A mathematical model for simulating the phase-based transmissibility of a novel coronavirus. Infect Dis Poverty. 2020February28;9(1):24. 10.1186/s40249-020-00640-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rocklöv J, Sjödin H, Wilder-Smith A. COVID-19 outbreak on the Diamond Princess cruise ship: estimating the epidemic potential and effectiveness of public health countermeasures. J Travel Med. 2020 May 18;27(3):taaa030. doi: 10.1093/jtm/taaa030 pmid:32109273. 10.1093/jtm/taaa030 [DOI] [PMC free article] [PubMed]
- 60.Mizumoto K, Chowell G. Transmission potential of the novel coronavirus (COVID-19) onboard the diamond Princess Cruises Ship, 2020. Infect Dis Model. 2020February29;5:264–70. 10.1016/j.idm.2020.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fang Y, Nie Y, Penny M. Transmission dynamics of the COVID-19 outbreak and effectiveness of government interventions: A data-driven analysis. J Med Virol. 2020June;92(6):645–59. 10.1002/jmv.25750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choi SC, Ki M. Estimating the reproductive number and the outbreak size of novel coronavirus disease (COVID-19) using mathematical model in Republic of Korea. Epidemiol Health. 2020March12;42:e2020011. 10.4178/epih.e2020011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lipsitch M, Cohen T, Cooper B, Robins JM, Ma S, James L, et al. Transmission dynamics and control of severe acute respiratory syndrome. Science. 2003June20;300(5627):1966–70. 10.1126/science.1086616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shim E, Tariq A, Choi W, Lee Y, Chowell G. Transmission potential and severity of COVID-19 in South Korea. Int J Infect Dis. 2020April;93:339–44. 10.1016/j.ijid.2020.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arons MM, Hatfield KM, Reddy SC, Kimball A, James A, Jacobs JR, et al. ; Public Health–Seattle and King County and CDC COVID-19 Investigation Team. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N Engl J Med. 2020May28;382(22):2081–90. 10.1056/NEJMoa2008457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bae J-M. A Chinese case of COVID-19 did not show infectivity during the incubation period: Based on an epidemiological survey. J Prev Med Public Health. 2020March2;53(2):67–9. 10.3961/jpmph.20.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kimball A, Hatfield KM, Arons M, James A, Taylor J, Spicer K, et al. ; Public Health – Seattle & King County; CDC COVID-19 Investigation Team. Public Health – Seattle & King County; CDC COVID-19 Investigation Team. Asymptomatic and presymptomatic SARS-CoV-2 infections in residents of a long-term care skilled nursing facility - King County, Washington, March 2020. MMWR Morb Mortal Wkly Rep. 2020April3;69(13):377–81. 10.15585/mmwr.mm6913e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li P, Fu J-B, Li K-F, Liu JN, Wang H-L, Liu L-J, et al. Transmission of COVID-19 in the terminal stages of the incubation period: A familial cluster. Int J Infect Dis. 2020July;96:452–3. 10.1016/j.ijid.2020.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wei WE, Li Z, Chiew CJ, Yong SE, Toh MP, Lee VJ. Presymptomatic Transmission of SARS-CoV-2 - Singapore, January 23-March 16, 2020. MMWR Morb Mortal Wkly Rep. 2020April10;69(14):411–5. 10.15585/mmwr.mm6914e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tong Z-D, Tang A, Li K-F, Li P, Wang H-L, Yi J-P, et al. Potential presymptomatic transmission of SARS-CoV-2, Zhejiang Province, China, 2020. Emerg Infect Dis. 2020May;26(5):1052–4. 10.3201/eid2605.200198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sanche S, Lin YT, Xu C, Romero-Severson E, Hengartner N, Ke R. High contagiousness and rapid spread of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020July;26(7):1470–7. 10.3201/eid2607.200282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pan A, Liu L, Wang C, Guo H, Hao X, Wang Q, et al. Association of public health interventions with the epidemiology of the COVID-19 outbreak in Wuhan, China. JAMA. 2020May19;323(19):1915–23. 10.1001/jama.2020.6130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsang TK, Wu P, Lin Y, Lau EHY, Leung GM, Cowling BJ. Effect of changing case definitions for COVID-19 on the epidemic curve and transmission parameters in mainland China: a modelling study. Lancet Public Health. 2020May;5(5):e289–96. 10.1016/S2468-2667(20)30089-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Han Y, Liu Y, Zhou L, Chen E, Liu P, Pan X, et al. Epidemiological assessment of imported coronavirus disease 2019 (COVID-19) cases in the most affected city outside of Hubei Province, Wenzhou, China. JAMA Netw Open. 2020April1;3(4):e206785. 10.1001/jamanetworkopen.2020.6785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Caicedo-Ochoa Y, Rebellón-Sánchez DE, Peñaloza-Rallón M, Cortés-Motta HF, Méndez-Fandiño YR. Effective Reproductive Number estimation for initial stage of COVID-19 pandemic in Latin American Countries. Int J Infect Dis. 2020June;95:316–8. 10.1016/j.ijid.2020.04.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Distante C, Piscitelli P, Miani A. COVID-19 outbreak progression in Italian regions: approaching the peak by the end of March in northern Italy and first week of April in southern Italy. Int J Environ Res Public Health. 2020April27;17(9):E3025. 10.3390/ijerph17093025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang CY, Wang J. A mathematical model for the novel coronavirus epidemic in Wuhan, China. Math Biosci Eng. 2020March11;17(3):2708–24. 10.3934/mbe.2020148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anastassopoulou C, Russo L, Tsakris A, Siettos C. Data-based analysis, modelling and forecasting of the COVID-19 outbreak. PLoS One. 2020March31;15(3):e0230405. 10.1371/journal.pone.0230405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schneider E, Bermingham A, Pebody R, Watson J. SARS, MERS, and other coronavirus infections. In: Heymann D, editor. Control of Communicable Diseases Manual. 20th ed. Washington (DC): American Public Health Association Press; 2015., 10.2105/CCDM.2745.128 [DOI] [Google Scholar]
- 80.Cowling BJ, Park M, Fang VJ, Wu P, Leung GM, Wu JT. Preliminary epidemiological assessment of MERS-CoV outbreak in South Korea, May to June 2015. Euro Surveill. 2015June25;20(25):7–13. 10.2807/1560-7917.ES2015.20.25.21163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Assiri A, McGeer A, Perl TM, Price CS, Al Rabeeah AA, Cummings DAT, et al. ; KSA MERS-CoV Investigation Team. Hospital outbreak of Middle East respiratory syndrome coronavirus. N Engl J Med. 2013August1;369(5):407–16. 10.1056/NEJMoa1306742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zeng G, Xie S-Y, Li Q, Ou J-M. Infectivity of severe acute respiratory syndrome during its incubation period. Biomed Environ Sci. 2009December;22(6):502–10. 10.1016/S0895-3988(10)60008-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Han Y-N, Feng Z-W, Sun L-N, Ren X-X, Wang H, Xue Y-M, et al. A comparative-descriptive analysis of clinical characteristics in 2019-coronavirus-infected children and adults. J Med Virol. 2020September;92(9):1596–602. 10.1002/jmv.25835 [DOI] [PubMed] [Google Scholar]
- 84.Ghinai I, Woods S, Ritger KA, McPherson TD, Black SR, Sparrow L, et al. Community transmission of SARS-CoV-2 at two family gatherings - Chicago, Illinois, February-March 2020. MMWR Morb Mortal Wkly Rep. 2020April17;69(15):446–50. 10.15585/mmwr.mm6915e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zheng F, Tang W, Li H, Huang Y-X, Xie Y-L, Zhou Z-G. Clinical characteristics of 161 cases of corona virus disease 2019 (COVID-19) in Changsha. Eur Rev Med Pharmacol Sci. 2020March;24(6):3404–10. 10.26355/eurrev_202003_20711 [DOI] [PubMed] [Google Scholar]
- 86.Xia X-Y, Wu J, Liu H-L, Xia H, Jia B, Huang W-X. Epidemiological and initial clinical characteristics of patients with family aggregation of COVID-19. J Clin Virol. 2020June;127:104360. 10.1016/j.jcv.2020.104360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen J, Zhang Z-Z, Chen Y-K, Long Q-X, Tian W-G, Deng H-J, et al. The clinical and immunological features of pediatric COVID-19 patients in China. Genes Dis. 2020December;7(4):535–41. 10.1016/j.gendis.2020.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Song W, Li J, Zou N, Guan W, Pan J, Xu W. Clinical features of pediatric patients with coronavirus disease (COVID-19). J Clin Virol. 2020June;127:104377. 10.1016/j.jcv.2020.104377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jiang Y, Niu W, Wang Q, Zhao H, Meng L, Zhang C. Characteristics of a family cluster of severe acute respiratory syndrome coronavirus 2 in Henan, China. J Infect. 2020August;81(2):e46–8. 10.1016/j.jinf.2020.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nie X, Fan L, Mu G, Tan Q, Wang M, Xie Y, et al. Epidemiological characteristics and incubation period of 7,015 confirmed cases with coronavirus disease 2019 outside Hubei Province in China. J Infect Dis. 2020June16;222(1):26–33. 10.1093/infdis/jiaa211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu X, Sun X, Cui P, Pan H, Lin S, Han R, et al. Epidemiological and clinical characteristics of 333 confirmed cases with coronavirus disease 2019 in Shanghai, China. Transbound Emerg Dis. 2020July;67(4):1697–707. 10.1111/tbed.13604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhou WK, Wang AL, Xia F, Xiao YN, Tang SY. Effects of media reporting on mitigating spread of COVID-19 in the early phase of the outbreak. Math Biosci Eng. 2020March10;17(3):2693–707. 10.3934/mbe.2020147 [DOI] [PubMed] [Google Scholar]
- 93.Kucharski AJ, Russell TW, Diamond C, Liu Y, Edmunds J, Funk S, et al. ; Centre for Mathematical Modelling of Infectious Diseases COVID-19 working group. Early dynamics of transmission and control of COVID-19: a mathematical modelling study. Lancet Infect Dis. 2020May;20(5):553–8. 10.1016/S1473-3099(20)30144-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao S, Chen H. Modeling the epidemic dynamics and control of COVID-19 outbreak in China. Quant Biol. 2020March11;8(1):1–9. 10.1007/s40484-020-0199-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kuniya T. Prediction of the epidemic peak of coronavirus disease in Japan, 2020. J Clin Med. 2020March13;9(3):E789. 10.3390/jcm9030789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Remuzzi A, Remuzzi G. COVID-19 and Italy: what next? Lancet. 2020April11;395(10231):1225–8. 10.1016/S0140-6736(20)30627-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li R, Pei S, Chen B, Song Y, Zhang T, Yang W, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science. 2020May1;368(6490):489–93. 10.1126/science.abb3221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yuan J, Li M, Lv G, Lu ZK. Monitoring transmissibility and mortality of COVID-19 in Europe. Int J Infect Dis. 2020June;95:311–5. 10.1016/j.ijid.2020.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ferretti L, Wymant C, Kendall M, Zhao L, Nurtay A, Abeler-Dörner L, et al. Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science. 2020May8;368(6491):eabb6936. 10.1126/science.abb6936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huang R, Liu M, Ding Y. Spatial-temporal distribution of COVID-19 in China and its prediction: A data-driven modeling analysis. J Infect Dev Ctries. 2020March31;14(3):246–53. 10.3855/jidc.12585 [DOI] [PubMed] [Google Scholar]
- 101.Tian H, Liu Y, Li Y, Wu C-H, Chen B, Kraemer MUG, et al. An investigation of transmission control measures during the first 50 days of the COVID-19 epidemic in China. Science. 2020May8;368(6491):638–42. 10.1126/science.abb6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhao S, Stone L, Gao D, Musa SS, Chong MKC, He D, et al. Imitation dynamics in the mitigation of the novel coronavirus disease (COVID-19) outbreak in Wuhan, China from 2019 to 2020. Ann Transl Med. 2020April;8(7):448. 10.21037/atm.2020.03.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Abbott S, Hellewell J, Munday J, Funk S; CMMID nCoV working group. The transmissibility of novel Coronavirus in the early stages of the 2019-20 outbreak in Wuhan: Exploring initial point-source exposure sizes and durations using scenario analysis. Wellcome Open Res. 2020February3;5:17. 10.12688/wellcomeopenres.15718.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Du Z, Wang L, Cauchemez S, Xu X, Wang X, Cowling BJ, et al. Risk for transportation of 2019 novel coronavirus disease from Wuhan to other cities in China. Emerg Infect Dis. 2020May;26(5):1049–52. 10.3201/eid2605.200146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Torres-Roman JS, Kobiak IC, Valcarcel B, Diaz-Velez C, La Vecchia C. The reproductive number R0 of COVID-19 in Peru: An opportunity for effective changes. Travel Med Infect Dis. 2020Sep-Oct;37:101689. 10.1016/j.tmaid.2020.101689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Muniz-Rodriguez K, Fung IC-H, Ferdosi SR, Ofori SK, Lee Y, Tariq A, et al. Severe acute respiratory syndrome coronavirus 2 transmission potential, Iran, 2020. Emerg Infect Dis. 2020August;26(8):1915–7. 10.3201/eid2608.200536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhuang Z, Zhao S, Lin Q, Cao P, Lou Y, Yang L, et al. Preliminary estimates of the reproduction number of the coronavirus disease (COVID-19) outbreak in Republic of Korea and Italy by 5 March 2020. Int J Infect Dis. 2020June;95:308–10. 10.1016/j.ijid.2020.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gatto M, Bertuzzo E, Mari L, Miccoli S, Carraro L, Casagrandi R, et al. Spread and dynamics of the COVID-19 epidemic in Italy: Effects of emergency containment measures. Proc Natl Acad Sci USA. 2020May12;117(19):10484–91. 10.1073/pnas.2004978117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ndaïrou F, Area I, Nieto JJ, Torres DFM. Mathematical modeling of COVID-19 transmission dynamics with a case study of Wuhan. Chaos Solitons Fractals. 2020June;135:109846. 10.1016/j.chaos.2020.109846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Peirlinck M, Linka K, Sahli Costabal F, Kuhl E. Outbreak dynamics of COVID-19 in China and the United States. Biomech Model Mechanobiol. 2020December;19(6):2179–93. 10.1007/s10237-020-01332-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Adegboye OA, Adekunle AI, Gayawan E. Early transmission dynamics of novel coronavirus (COVID-19) in Nigeria. Int J Environ Res Public Health. 2020April28;17(9):E3054. 10.3390/ijerph17093054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ivorra B, Ferrández MR, Vela-Pérez M, Ramos AM. Mathematical modeling of the spread of the coronavirus disease 2019 (COVID-19) taking into account the undetected infections. The case of China. Commun Nonlinear Sci Numer Simul. 2020September;88:105303. 10.1016/j.cnsns.2020.105303 [DOI] [PMC free article] [PubMed] [Google Scholar]


