Abstract
Background
Globally, one of the dominant causes of cancer-related mortality is liver cancer. Identification of potent biomarkers for initial stage diagnosis and prognosis is a key factor to ensure efficient therapy and reduce the mortality rate in liver cancer patients. REXO4 has been reported in neuropathic pain and familial isolated pituitary adenoma (FIPA), however, its relationship with liver cancer is still elusive.
Methods
In an attempt to scrutinize the expression of REXO4 in liver cancer, the Oncomine, and TCGA databases were analyzed. Real-time PCR was employed to identify the REXO4 mRNA levels in 45 patient tissue samples and western blot was used to detect the REXO4 protein levels in hepatocellular carcinoma (HCC) cells. Evaluation of the prognostic value of REXO4 in liver cancer was made using Univariate and multivariate Cox proportional hazards regression models and Kaplan–Meier plots. Tumor‐associated biological processes related to REXO4 were revealed by LinkedOmics. The correlation of REXO4 and immune infiltration was evaluated using the TIMER database.
Results
REXO4 is significantly up-regulated in liver cancer in comparison with the nontumor controls. Moreover, poor progression-free survival and overall survival is a frequent outcome related to high expression of REXO4, highlighting it as a risk factor in case of liver cancer. Additionally, the plausible role of REXO4 in tumor-immune interactions was also investigated and it was revealed that the immune infiltration and immune activation of liver cancer might have an association with REXO4.
Conclusions
REXO4 has a significant expression in liver cancer and could potentially become a predictor for the prognosis of liver cancers and a biomarker for targeted therapeutic regimens. Significant overexpression of REXO4 in HCC was revealed by the bioinformatics analysis, with REXO4 overexpression being related to a negative outcome in HCC patients, in addition, REXO4 might be associated with the immune infiltration in liver cancer. Such a vital understanding of the functioning of REXO4 may furnish a foundation for new targeted drug therapy as well as a new direction for additional investigation into the underlying mechanisms of REXO4 carcinogenesis in liver cancer.
Keywords: REXO4, prognostic value, hepatocellular carcinoma (HCC), biomarker
Introduction
There has been a rapid growth in the incidence of liver cancer worldwide, making it a predominant reason for morbidity and mortality around the globe (1). Because of the non-availability of highly specific and sensitive early diagnostic biomarkers and the high frequency of tumor metastasis, the improved diagnosis and treatment of hepatocellular carcinoma (HCC) are highly dependent on the identification of novel biomarkers and therapeutic targets which could further lead to an enhanced understanding of the pathology of HCC (2).
REXO4 is an unreported protein in the tumor, it was associated with inflammation in neuropathic pain and plays an important role in familial isolated pituitary adenoma (FIPA) (3,4). In an attempt to find the expression of REXO4 in multiple tumors, we employed public databases. It has been suggested by relevant research studies that in HCC, REXO4 acts as a potential biomarker for diagnosis as well as prognosis. Nevertheless, certainty associated with its role in HCC stays rather evasive. Therefore, the relation between REXO4 expression in HCC is necessary to be classified.
REXO4 expression and its importance in terms of prognosis of liver cancer patients have been comprehensively assessed in this study via the use of The Human Pathology Atlas, UALCAN, Gene Expression Profiling Interactive Analysis (GEPIA), Oncomine and tumor immune estimation resource (TIMER) public databases. Moreover, we compared ours results of detection of the expression of REXO4 in HCC tissues and cells, with those reported in the public database. The plausible biological functions and pathways of REXO4 in HCC were investigated by making use of LinkedOmics online website for analyzing REXO4 with respect to its functional network. Finally, we also employed TIMER for exploring the correlation among gene markers of immune infiltrating cells in HCC and REXO4 expression. The outcome of such analyses suggested that REXO4 may be a key biomarker for the prediction of prognosis in HCC. REXO4 can further potentially be utilized as a therapeutic target or diagnostic marker for HCC.
We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/jgo-21-98).
Methods
The Human Protein Atlas
Immunohistochemistry (IHC) data was obtained after a tissue microarray-based examination on various kinds of 44 normal tissue types is provided by the Human Pathology Atlas project (https://www.proteinatlas.org). It further provides the proteome analysis of 17 key types of cancer (5). Online data is conveniently available regarding the staining intensity, staining site, amount, and patients’ information in patients with the corresponding kinds of cancer. This study demonstrates the expression of REXO4 in normal tissues and typical protein expressions of IHC photographs of REXO4 were discovered in normal tissues and HCC in Human Protein Atlas.
TIMER
For a comprehensive analysis of tumor-infiltrating immune cells, TIMER is the most sought after web server (https://cistrome.shinyapps.io/timer/) (6,7). The difference of REXO4 between cancer and paracancerous tissues is evident from the database. Gene expression profiles can be employed to estimate the profusion of six immune infiltrates (B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells) using a statistical technique, which is confirmed by making use of pathological estimations. Additionally, the mode of expression of a gene pair in a certain specific type of cancer is illustrated by the “correlation” module which creates scatterplots and also generates estimated statistical significance and the Spearman correlation, that can be mediated by age or tumor purity (reflected by the fraction of cancerous cells in the admixture). We probed into the correlations among gene markers of immune infiltrating cells in HCC and REXO4 expression by employing the module.
Oncomine database
Oncomine database accessed through Oncomine online database (http://www.oncomine.com) was employed in this study to obtain the transcriptional expression profiles of REXO4 in HCC patients. Students’ t-test was used to compare the difference in transcriptional expression. Fold change and Cut-off value were as follows: Data type: mRNA; fold change =1.5; P-value =0.01, gene rank =10%.
GEPIA
GEPIA is an online database (http://gepia.cancerpku.cn/) (8) and a web server. A standard processing pipeline is adopted by GEPIA for analyzing the RNA-Seq expression data from TCGA and GTEx, which includes 9,736 tumor and 8,587 normal samples. Following sample procurement from TCGA, we used GEPIA to analyze the connections between disease-free survival rates and overall survival with REXO4 expression in HCC. Furthermore, for calculating the differential expression of REXO4, a boxplot employing the state of disease as a variable was graphed.
Kaplan–Meier Plotter database
The relation between the survival duration of patients with HCC and the expression of REXO4 was analyzed by performing K-M survival analysis (http://kmplot.com/analysis/) (9). For obtaining K-M survival plots, REXO4 was entered into the database. Calculations of the hazard ratio and log-rank p values were made. A P-value <0.05 was considered statistically significant.
UALCAN database analysis
Cancer OMICS data can be analyzed by using an interactive, user-friendly, and comprehensive online resource called UALCAN (10). It is characterized by its high-quality graphics and is built on PERL-CGI using javascript and CSS. Cancer OMICS data (TCGA, MET500, and CPTAC) available to the general public can be conveniently accessed using UALCAN.
LinkedOmics
Multi-omics data from all 32 TCGA Cancer types can be accessed by LinkedOmics which is an online portal made available for the public (11). Additionally, it comprises data based on mass spectrometry-based proteomics which has been generated by the Clinical Proteomics Tumor Analysis Consortium (CPTAC) for TCGA colorectal, breast, and ovarian tumors. Three analytical modules comprise the web application namely LinkFinder, LinkInterpreter, and LinkCompare. Of these, enrichment analysis is performed by the LinkInterpreter module, based on Gene Ontology, network modules, and biological pathways, along with related functional categories.
Real-time quantitative PCR and Western blot analyses
TRIzol® reagent (Invitrogen Life Technologies, USA) was used to extract Total RNA sequence from a total of 45 paired tumor and para-carcinoma normal samples. ddH2O with SYBR Green PCR Master Mix (Applied Biosystems, Japan) was used to dilute Primers. The fold change of REXO4 relative to β-actin determined the Transcriptional expression. PCR primers sequence for REXO4 are as follows: forward are 5-TCTCTTCCGGAGTCTTTTCCTG-3′ and reverse are 5′-CCTTCACTTGAGGCGAGGTC-3. The REXO4 mRNA expression was depicted as ΔCt=Ct (REXO4) −ΔCt (β-actin). Lysis of cells was brought about by a RIPA buffer (Sigma-Aldrich Chemie, Steinheim, Germany) comprising a protease inhibitor. The protein quantities were estimated using the BCA Protein Assay Kit (Pierce, Rockford, USA). Lysates upon separation by using SDS-PAGE were placed onto polyvinylidene difluoride membranes (Merck-Millipore, Darmstadt, Germany). Bovine serum albumin (BSA, 10%) was used to obstruct the membranes which were later incubated with primary antibodies against REXO4 (Abcam, #ab154941, Cambridge, USA), and β-actin (Proteintech, #60008) at 4 °C overnight. Incubation of the membranes with horseradish peroxidase (HRP)-conjugated secondary antibody then took place for 1 h at room temperature.
Patients and cell culture
Forty-five fresh HCC tissue samples along with matched normal tissue samples were obtained from the affiliated hospital of Nantong University from 2018–2019, they were not received therapy prior to surgery. Patients include 30 males and 15 females. Tissue samples were stored at ‒80 °C for RNA isolation. Informed consent was obtained from the patients, and the study was approved by the Ethics Committee of Affiliated Hospital of Nantong University (ethical code: 2018-L006). The study was conducted in accordance with the Declaration of Helsinki (as was revised in 2013). The normal human hepatic cell line QSG-7701 and four HCC cell lines Li-7, Hep3B, MHCC-97H, and HepG2 were purchased from the Cell Resource Center, Chinese Academy of Science Committee (Shanghai, China). Dulbecco’s modified Eagle’s medium supplemented with10% fetal bovine serum (FBS; GIBCO, Grand Island, USA), 100 µg/mL streptomycin, and 100 U/mL penicillin was used to culture these cells in a humidified incubator containing 5% (v/v) of CO2, at 37 °C.
Statistical analysis
Experimental data were written as the means ± SD of three independent experiments and analyzed in GraphPad Prism 7 software by Student’s t-test when drawing a comparison of only two groups or one-way analysis of variance when greater than two groups were compared. Overall survival of HCC patients was evaluated by the log-rank (Mantel-Cox) test; P<0.05 indicated a statistically significant difference.
Results
The level of expression of REXO4 expression in various types of human cancers and human tissues
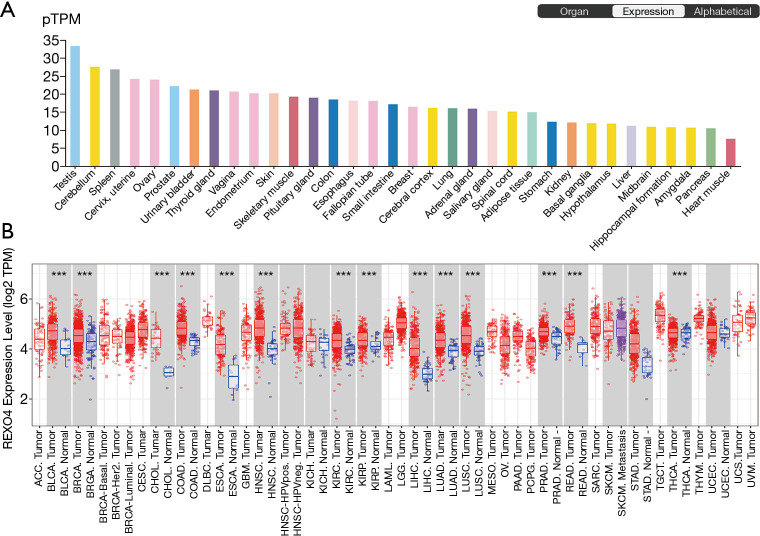
Determination of the REXO4 gene expression across tumor specimens as well as paired normal tissues was carried out using the Human Pathology Atlas and TIMER database. The expression of REXO4 was lower in normal liver tissue (Figure 1A). Figure 1B shows the differential expression level of REXO4, and analyses indicate that most cancers such as BLCA, LIHC, and LUAD manifest an elevated REXO4 expression, etc. Considering the low expression of REXO4 in normal liver tissue, but the high expression in HCC, we think that in HCC REXO4 expression might potentially function as a useful diagnostic indicator.
Figure 1.
Online databases showed REXO4 expression in various human tissues and cancers. (A) REXO4 expression levels in different normal tissues, data from Human Pathology Atlas database. (B) REXO4 expression was showing in human tumor samples and paired normal tissues. ***, P<0.001.
High expression of REXO4 in HCC patients and cells
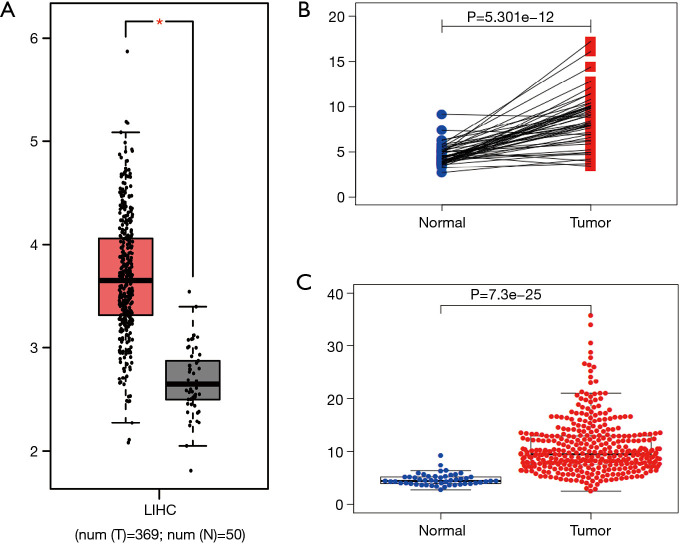
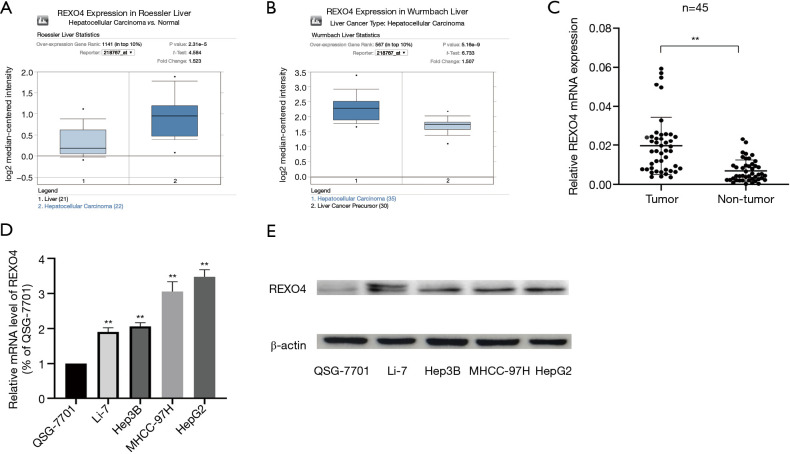
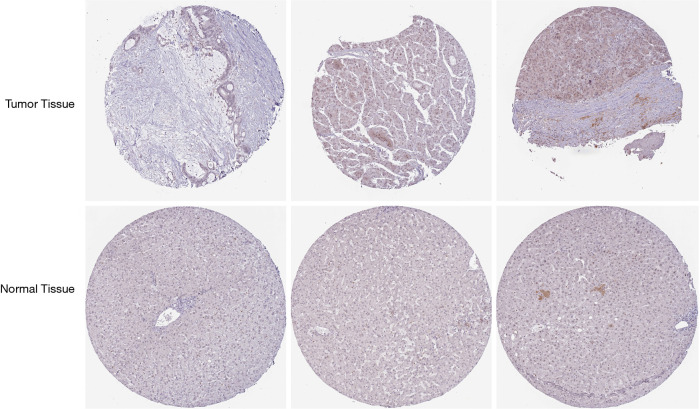
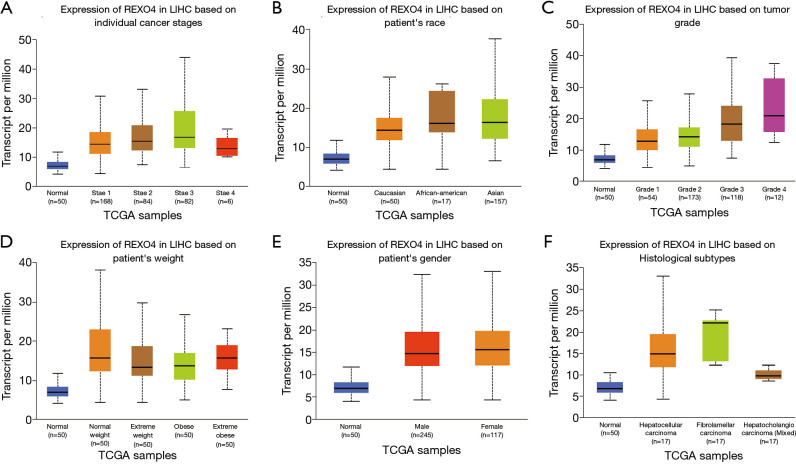
The published data of normal and cancer tissues obtained from TCGA databases was thoroughly assessed. According to the results, there is a significant upregulation of REXO4 in HCC (P<0.05, Figure 2A). The same results are shown by box plot and scatter plot (Figure 2B,C). The data obtained from the Oncomine database suggested that based on mRNA expression REXO4 ranked within the top 10% over-expression (Figure 3A,B). To further validate these results, an assessment of REXO4 mRNA expression was carried out using real-time PCR. As expected, a significant up-regulation of REXO4 was revealed in liver cancer tissues in comparison with matched adjacent nontumor tissues (Figure 3C). In addition, the expression of REXO4 of HCC cells was investigated by real-time PCR and western blot, which additionally demonstrated that in comparison to the peritumor tissues, REXO4 had a high expression in tumor tissues (Figure 3D,E). Besides that, immunohistochemical (IHC) staining of REXO4 protein in normal liver and HCC tissues was revealed by The Human Pathology Atlas online database (Figure 4). In an attempt to further examine the level of REXO4 expression for potential prognostic significance, we made use of the UALCAN database to carry out subgroup analyses of various clinical characteristics including those of stage, race, tumor grade, weight, gender, and histological subtypes. Subgroup analyses demonstrated that the expression levels of REXO4 were elevated in abnormal features than normal (Figure 5A-F). These findings suggest that, in the case of liver cancer, REXO4 expression levels can potentially act as a diagnostic biomarker.
Figure 2.
REXO4 is upregulated based on data from TCGA. (A) The expression of REXO4 in HCC tissues was significantly higher than that in normal tissues. Data from GEPIA. (B) Paired box plot showed that the expression of REXO4 was higher in cancer tissues. (C) Scatter plot showed that the expression of REXO4 was higher in HCC. *, P<0.05.
Figure 3.
The expression of REXO4 increased in HCC tissues and cells. (A,B) REXO4 different expressions in Roessler Liver, Wurmbach Liver respectively. (C) REXO4 expression was significantly increased in HCC compared with normal tissues. (D,E) REXO4 increased in HCC cells compared with normal liver cells. **, P<0.01. HCC, hepatocellular carcinoma.
Figure 4.
Representative images of immunohistochemistry staining of HCC patients in the Human Pathology Atlas database, ×200. (A) HCC female id2279. (B) HCC female id2766. (C) HCC male id2325. (D) liver female id3402. (E) liver female id3222. (F) liver male id1720.
Figure 5.
REXO4 expression levels in HCC based on clinical parameters depend on the UALCAN database. The relationship of stage (A), race (B), grade (C), weight (D), gender (E) and histological subtypes (F) with REXO4 expression was showing in liver cancer, respectively.
REXO4 expression is survival-associated
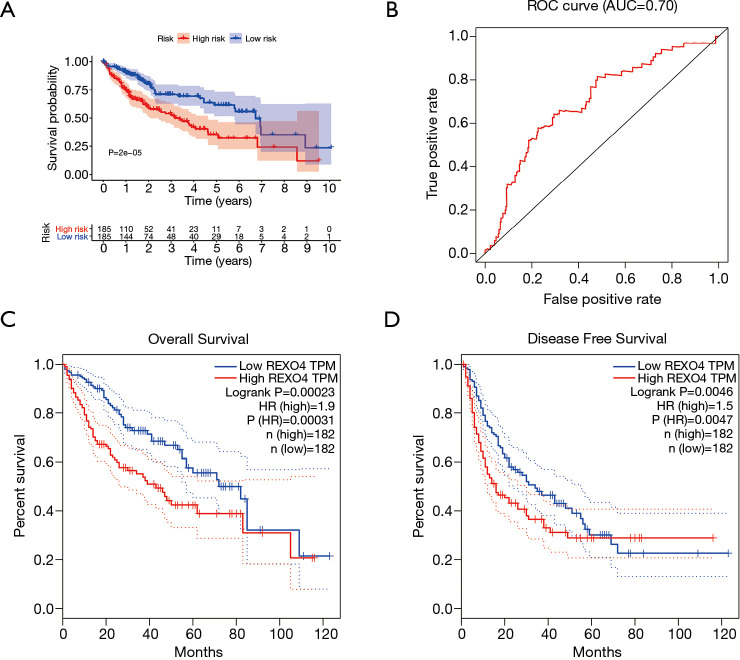
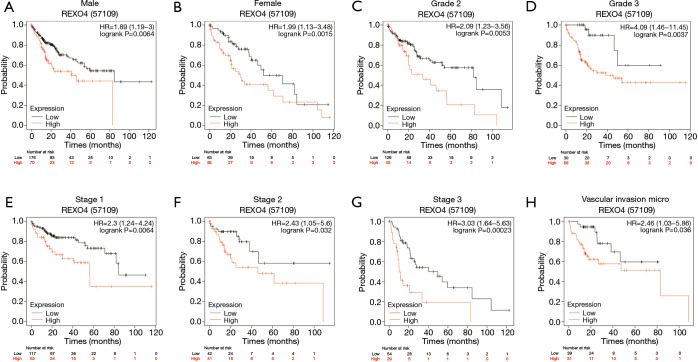
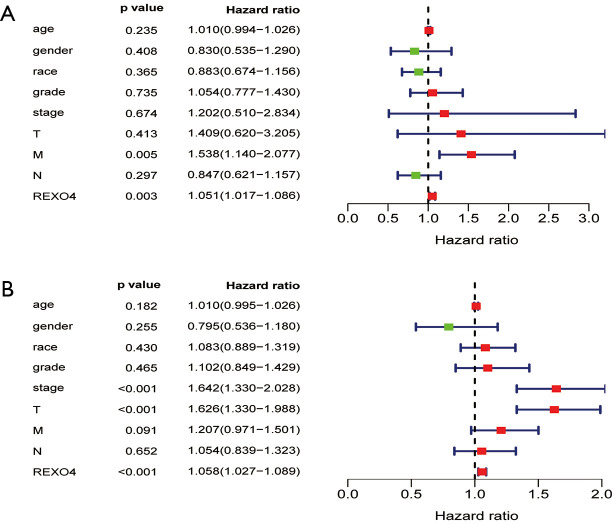
The association between the survival outcomes of HCC cohorts with available survival information and REXO4 expression was assessed using the Kaplan-Meier survival curves (Figure 6A). In the light of the median value of REXO4 expression level in individual cohorts, the patients were separated into two groups. In general, a significantly shorter overall survival (OS, P<0.05) was manifested by the high REXO4 expression group in comparison to the low expression group in LIHC. A promising prognostic power is demonstrated by REXO4 expression as the ROC curve revealed a value of 0.700 of AUC of REXO4 expression for predicting survival (Figure 6B). Next, the prognostic significance of REXO4 in cancers within the RNA sequencing data in TCGA was analyzed by making use of GEPIA. The outcome revealed that a poorer prognosis of OS and DFS was associated with REXO4 over-expression in HCC (Figure 6C,D). Different populations were taken into consideration for survival analyses. In male and female patients REXO4 up-regulation was associated with poorer survival as shown in Figure 7. Moreover, high REXO4 levels had a notable contribution towards worsening OS in HCC patients with grade II, grade III, stage I–III, and vascular invasion micro. The possibility of REXO4 was also verified as a prognostic biomarker using univariate and multivariate analysis. As illustrated in Figure 8, REXO4 had a prognostic significance in the TCGA cohorts.
Figure 6.
Diagnostic value of REXO4 in liver cancer. (A) Analysis of the relationship between REXO4 mRNA expression and overall survival in TCGA datasets. (B) ROC curves for HCC patients based on TCGA datasets. Data from TCGA database showed that high expression of REXO4 was associated with poor OS (C) and DFS (D). OS, overall survival; DFS, disease-free survival.
Figure 7.
The expression of REXO4 is related to the prognosis of HCC patients in different clinical manifestations. (A) male; (B) female; (C) grade 2; (D) grade 3; (E) stage 1; (F) stage 2; (G) stage 3; (H) vascular invasion micro. HCC, hepatocellular carcinoma.
Figure 8.
Univariate and multivariate Cox logistic regression analysis was performed to determine the association between clinical characters and OS in the TCGA data set. (A) multivariate analysis. (B) univariate analysis. OS, overall survival.
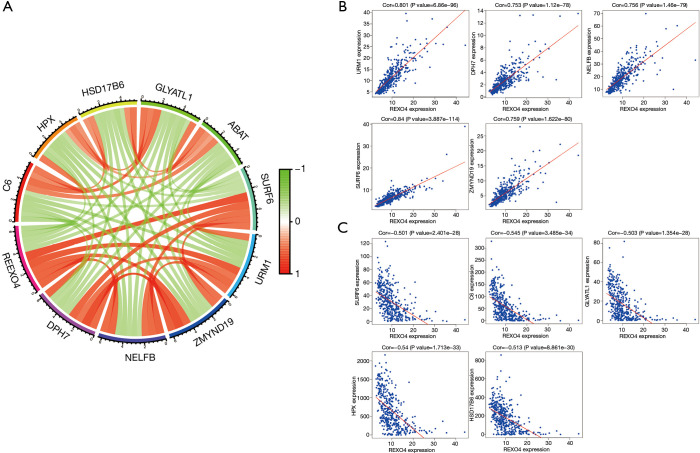
REXO4 co-expression genes in HCC
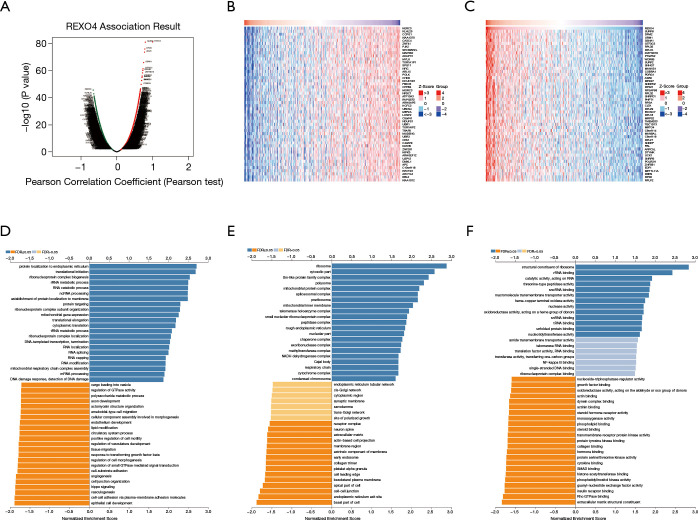
Ten genes related to REXO4 were shown in Figure 9, among them, HSD17B6, URM1, HPX, C6, ZMYND19 were playing a key part in the advancement of liver cancer (12-16). In an attempt to gain valuable insight regarding the biological importance of REXO4 in HCC, REXO4 co-expression mode in the LIHC cohort was examined using the function module of LinkedOmics. All target genes have been shown in Figure 10A, and the heat map reveals the top most significant genes having a negative and positive correlation with REXO4 (Figure 10B,C). Gene set enrichment analysis (GSEA) to annotate Significant Gene Ontology (GO) term showed that REXO4 co-expressed genes primarily participate in DNA damage response. RNA splicing, methyltransferase, snRNA binding, while the activities like lipid modification, cell-cell junction, SMAD binding, Rho GTPase binding were inhibited (Figure 10D-F). A rather widespread impact of REXO4 in HCC is thus suggested by these results.
Figure 9.
Significant related genes according to REXO4 in HCC. (A) Ten genes related to REXO4, five positive correlated genes and five negative correlated genes. (B) Five genes positively related to REXO4. (C) Five genes negatively associated with REX4. HCC, hepatocellular carcinoma.
Figure 10.
Most notable relevant genes and pathways according to REXO4 in HCC. (A) Volcano map shows 100 genes most related to REXO4. (B,C) Heat maps showing top 50 genes positively and negatively correlated with REXO4 in LIHC. Red dot on the right indicates positively correlated genes and green dot on the left indicates negatively correlated genes. (D-F) Identified significantly enriched GO and KEGG pathways according to REXO4 in HCC. HCC, hepatocellular carcinoma.
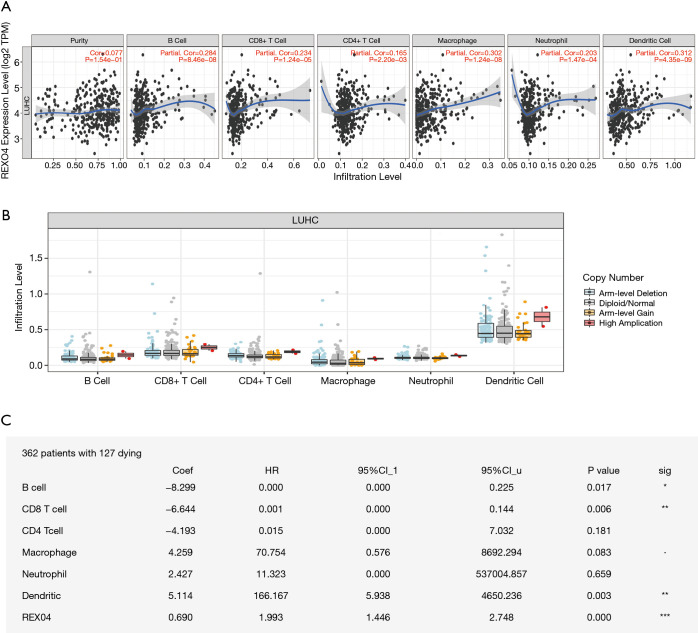
REXO4 expression is associated with immune infiltration in HCC
Immune infiltration levels may vary in cancer patients diagnosed with identical histology types, which could potentially lead to a variety of clinical outcomes (17,18). In primary tumor tissue, the elevated number of tumor-infiltrating lymphocytes associated with a good prognosis has been reported in several cancers, HCC being one of them (19,20). For the sake of confirming the relationship between immune infiltration of liver cancer and REXO4 expression, we employed TIMER to assess the correlations of tumor purity and infiltrating levels of immune cells with REXO4 expression. The outcome revealed that HCC tumor purity was uncorrelated with REXO4 expression. However, the REXO4 expression levels in HCC tissues has a correlation with tumor infiltration by B cells (r=0.284, P=8.46E-08), CD8+ T cells (r=0.234, P=1.24E-05), CD4+ T cells (r=0.165, P=2.20E-03), macrophages (r=0.302, P=1.24E-08), neutrophils (r=0.203, P=1.47E-04) and DCs (r=0.312, P=4.35E-09) (Figure 11A). Particularly, infiltrating levels of neutrophil cells have a notable correlation with REXO4 CNV (Figure 11B). In addition, we found that survival was related to immune infiltration (Figure 11C).
Figure 11.
REXO4 was related to immune infiltration level in HCC (A). REXO4 has significantly positively correlations with infiltrating levels of immune cells in HCC (B). REXO4 CNV affects the infiltrating levels of neutrophils cells in HCC (C). REXO4 is associated with tumor immunity. *, P<0.05; **, P<0.01; ***, P<0.001. HCC, hepatocellular carcinoma.
Discussion
Probably one of the most frequently occurring types of cancer is HCC and its prevalence has continued to rise in recent years (21). A series of steps are involved in the development of cancer cells which involve changes in growth, cell metabolism, proliferation, and adhesion (22,23). Several studies have provided information about the biomarker of liver cancer (24,25). The fact that REXO4 has been identified as a new biomarker for HCC, could aid in elucidating how alterations in cytokinesis and the immune environment stimulate the development of liver cancer. REXO4 has a specific correlation with the expression of numerous cell cycle regulatory genes and has the potential to be served as a plausible diagnostic marker in HCC. REXO4 expression demonstrated strong correlations with a variety of immune marker sets in HCC. The results suggest that REXO4 is correlated with immune infiltrating and prognosis in HCC.
REXO4 is a protein associated with inflammation in neuropathic pain and functions in FIPA (3,4), but it has not been reported in tumors. We made use of published data on liver cancer patients to estimate the expression and prognostic importance of REXO4. Besides that, the analysis of REXO4 shows the same result with published online databases. All the results show that REXO4 manifests an overexpression at mRNA levels in HCC tissues, and a raised REXO4 expression is notably related to a brief OS. Altogether, the potential of REXO4 as a prognostic marker and therapeutic target in HCC is highlighted by these data. In an attempt to comprehend how carcinogenesis is influenced by this alteration, we analyzed the genes that existed in co-occurrence with REXO4. Five negative and five positive genes have been shown in this study, among them, the expression of URM1 has been found to increase in liver cancer tissues, and knockdown of URM1 results in inhibiting the proliferation and migration of cancerous liver cells leading to accelerated apoptosis. In HCC patients, an elevated URM1 expression is found to be linked with poor prognosis (13). Besides that, HSD17B6, HPX, and C6 could be new biomarkers for the prognosis of HCC (12,14-16), and others need further experimental verification. We tested the REXO4 co-expression network by LinkedOmics to probe the signaling events in controlling the abnormal expression of REXO4. And the outcome points out that the functional consequence of REXO4 primarily includes DNA damage response, RNA splicing, methyltransferase, snRNA binding, while the activities like lipid modification, cell-cell junction, SMAD binding, and Rho GTPase binding were inhibited. The molecular pathways involved in HCC carcinogenesis are also consistent with these findings, but further studies are required to confirm this.
Tumor immunotherapy has been observed to undergo rapid development in recent years (17). There has been an increased understanding regarding the involvement of the immune processes in the development and progress of cancer (18). An active area of research includes the exploration of the tumor microenvironment as a therapy target as well as a prognostic and diagnostic biomarker. Numerous works have demonstrated that survival in liver cancer is influenced by immune cell infiltration (19-21). Other significant findings of the current study include the understanding of the role of REXO4 in tumor-immune interactions and that the expression of REXO4 has a notable correlation with the immune infiltration level in liver cancer. The outcome of our study demonstrated a most significant positive correlation between the level of B cells, CD8+ T cells, CD4+ T cells, macrophages, and DCs infiltration and CLEC3B expression in HCC. The infiltration of CD8+ T cells, B cells, and Dendritic cells generally influence the prognosis of patients.
Conclusions
In summary, multi-level evidence for the importance of hepatocarcinogenesis is demonstrated by this study. This work also provides evidence regarding the potential of REXO4 as a biomarker in HCC. Our results imply that upregulation of REXO4 in HCC may presumably have a far-reaching impact on DNA damage response, RNA splicing, methyltransferase, snRNA binding. Additionally, our findings indicate a novel potential immune regulatory role of REXO4 with respect to tumor immunity. These findings call for rather extensive research on HCC genomics and subsequently functional investigations.
Acknowledgments
Funding: This study was supported by grants from the National Natural Science Foundation of China (NO. 81871927).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as was revised in 2013). Informed consent was obtained from the patients, and this research was approved by the Ethics Committee of Affiliated Hospital of Nantong University (ethical code: 2018-L006).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Footnotes
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/jgo-21-98
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jgo-21-98
Peer Review File: Available at https://dx.doi.org/10.21037/jgo-21-98
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-21-98). The authors have no conflicts of interest to declare.
References
- 1.Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2021;7:6. 10.1038/s41572-020-00240-3 [DOI] [PubMed] [Google Scholar]
- 2.Kulik L, El-Serag HB. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterology 2019;156:477-491.e1. 10.1053/j.gastro.2018.08.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai G, Zhu Y, Zhao Y, et al. Network Analysis of miRNA and mRNA Changes in the Prelimbic Cortex of Rats With Chronic Neuropathic Pain: Pointing to Inflammation. Front Genet 2020;11:612. 10.3389/fgene.2020.00612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maddirevula S, Awartani K, Coskun S, et al. A genomics approach to females with infertility and recurrent pregnancy loss. Hum Genet 2020;139:605-13. 10.1007/s00439-020-02143-5 [DOI] [PubMed] [Google Scholar]
- 5.Uhlen M, Zhang C, Lee S, et al. A pathology atlas of the human cancer transcriptome. Science 2017;357:eaan2507. 10.1126/science.aan2507 [DOI] [PubMed] [Google Scholar]
- 6.Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. 10.1158/0008-5472.CAN-17-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li B, Severson E, Pignon JC, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol 2016;17:174. 10.1186/s13059-016-1028-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagy Á, Lánczky A, Menyhárt O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 2018;8:9227. 10.1038/s41598-018-27521-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46:D956-63. 10.1093/nar/gkx1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lv L, Zhao Y, Wei Q, et al. Downexpression of HSD17B6 correlates with clinical prognosis and tumor immune infiltrates in hepatocellular carcinoma. Cancer Cell Int 2020;20:210. 10.1186/s12935-020-01298-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng X, Zhang Y, Song F, et al. URM1 Promoted Tumor Growth and Suppressed Apoptosis via the JNK Signaling Pathway in Hepatocellular Carcinoma. Onco Targets Ther 2020;13:8011-25. 10.2147/OTT.S258843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barefoot ME, Varghese RS, Zhou Y, et al. Multi-omic Pathway and Network Analysis to Identify Biomarkers for Hepatocellular Carcinoma. Annu Int Conf IEEE Eng Med Biol Soc 2019;2019:1350-4. 10.1109/EMBC.2019.8856576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mu D, Qin F, Li B, et al. Identification of the Sixth Complement Component as Potential Key Genes in Hepatocellular Carcinoma via Bioinformatics Analysis. Biomed Res Int 2020;2020:7042124. 10.1155/2020/7042124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Q, Luo Z, Lu G, et al. LncRNA FABP5P3/miR-589-5p/ZMYND19 axis contributes to hepatocellular carcinoma cell proliferation, migration and invasion. Biochem Biophys Res Commun 2018;498:551-8. 10.1016/j.bbrc.2018.03.017 [DOI] [PubMed] [Google Scholar]
- 17.Antonangeli F, Natalini A, Garassino MC, et al. Regulation of PD-L1 Expression by NF-κB in Cancer. Front Immunol 2020;11:584626. 10.3389/fimmu.2020.584626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abousaway O, Rakhshandehroo T, Van den Abbeele AD, et al. Noninvasive Imaging of Cancer Immunotherapy. Nanotheranostics 2021;5:90-112. 10.7150/ntno.50860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Federico P, Petrillo A, Giordano P, et al. Immune Checkpoint Inhibitors in Hepatocellular Carcinoma: Current Status and Novel Perspectives. Cancers (Basel) 2020;12:3025. 10.3390/cancers12103025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishida N, Kudo M.Immune Phenotype and Immune Checkpoint Inhibitors for the Treatment of Human Hepatocellular Carcinoma. Cancers (Basel) 2020;12:1274. 10.3390/cancers12051274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kudo M.Recent Advances in Systemic Therapy for Hepatocellular Carcinoma in an Aging Society: 2020 Update. Liver Cancer 2020;9:640-62. 10.1159/000511001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ong JR, Bamodu OA, Khang NV, et al. SUMO-Activating Enzyme Subunit 1 (SAE1) Is a Promising Diagnostic Cancer Metabolism Biomarker of Hepatocellular Carcinoma. Cells 2021;10:178. 10.3390/cells10010178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S, Saviano A, Erstad DJ, et al. Risk Factors, Pathogenesis, and Strategies for Hepatocellular Carcinoma Prevention: Emphasis on Secondary Prevention and Its Translational Challenges. J Clin Med 2020;9:3817. 10.3390/jcm9123817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng JL, Wu JZ, Li GJ, et al. Identification of potential biomarkers of peripheral blood mononuclear cell in hepatocellular carcinoma using bioinformatic analysis: A protocol for systematic review and meta-analysis. Medicine (Baltimore) 2021;100:e24172. 10.1097/MD.0000000000024172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshio S, Kanto T. Macrophages as a source of fibrosis biomarkers for non-alcoholic fatty liver disease. Immunol Med 2021. [Epub ahead of print]. doi: . 10.1080/25785826.2020.1868664 [DOI] [PubMed] [Google Scholar]