Abstract
Background
Circular RNAs (circRNAs) are a type of non-coding RNA which play an important role in the regulation of biological processes of various malignant tumors. However, the potential molecular mechanisms and roles of circRNAs in colorectal cancer (CRC) remain unelucidated.
Methods
In the current study, we analyzed the data of high-throughput microarray sequencing to screen differentially expressed circRNAs in CRC. Cell counting kit-8 (CCK-8), colony-formation, transwell, mouse xenograft models and immunohistochemistry assays were used to explore the function of circRNAs. TargetScan database, quantitative real-time polymerase chain reaction (qRT-PCR), double luciferase and RNA immunoprecipitation (RIP) were used to explore the molecular mechanism.
Results
We identified circMMP1 (hsa_circ_0024109) as a frequently upregulated circRNA in both CRC tissues and cells. Both in vitro and in vivo experiments demonstrated that circMMP1 knockdown significantly inhibited the proliferation and metastasis of CRC. The results demonstrated that circMMP1 promoted the growth and metastasis of CRC by sponging miR-1238 and upregulating the expression of matrix metalloproteinase 1 (MMP1), MMP2, and MMP9 expression.
Conclusions
In conclusion, our study identified the biological role of the circMMP1-miR-1238-MMP1/MMP2/MMP9 axis in the growth and metastasis of CRC, which is crucial for the monitoring and treatment of CRC. Further research on circMMP1 may provide a theoretical basis for diagnostic biomarkers of early CRC screening.
Keywords: circMMP1, circular RNAs (circRNAs), MMP, competing endogenous RNAs (ceRNAs), colorectal cancer (CRC)
Introduction
Colorectal cancer (CRC) is the second most common cause of cancer death in the United States. The incidence rate was 30 per 100,000 persons in Asia from 2012 to 2016. There incidence of CRC in young and middle-aged adults has been rising annually (1). Studies have shown that the 5-year survival rate of early CRC patients exceeds 90%, but the median survival time of those with advanced CRC is not more than 2 years (2). The diagnosis and treatment of CRC have become a pressing issue worldwide. If CRC patients can be diagnosed and treated early, their clinical outcomes will be significantly improved. For the early diagnosis of CRC, colonoscopy is an invasive examination method and its cost is relatively higher than other methods, so it is not suitable for population screening (3). Therefore, searching for a biodiagnostic marker with low invasiveness, high specificity, and sensitivity has a great significance for the clinical diagnosis and treatment of CRC.
Circular RNAs (circRNAs) are a kind of highly abundant non-coding covalently blocked RNA, and composed of reverse splicing sequences of precursor messenger RNAs (mRNA) of exons or introns without upstream heads or downstream tails (4). Compared to linear mRNA transcription, their circular structure makes them very stable and resistant to RNA exonuclease (5). As a new type of endogenous non-coding RNAs, they are widely expressed in tissues and cells. They can be used as miRNA sponges to regulate several key genes expression, alternatively splicing or acting as transcription factors encoding proteins (6).
With the development of circRNA-sequencing and bioinformatic technology, increasing circRNAs have been disclosed and studied (7). CircRNAs have been shown to participate in multiple biological processes of different diseases, such as heart failure, immune diseases, diabetes, or cancers (8). Among all the circRNAs that have been disclosed, ciRS-7/CDR1 is the most well-known. The CDR1AS has been demonstrated to promote the growth, migration, chemotherapy-resistance, and immunodeficiency by sponge miR-7 in many cancers (9-14). There have been few studies about the role of circRNAs in CRC. CircPP1R12A (hsa_circ_0000423) has been shown to enhance the malignant progression of colon cancer through the signal pathway of Hippo-YAP (15). Circ101555 may act as a competing endogenous RNA (ceRNA) for miR-597-5p, up-regulating CDK6 and RPA3 to promote CRC progression (16). CircDDX17 acts as a tumor suppressor in CRC (17). The expression of has_circ_0055625 is related to the pathological staging and metastasis of cancer, and promoting the growth and metastasis of colon cancer by acting as the sponge for miR-106b-5p (18). However, study about the role of circRNAs in CRC and its underlying molecular mechanism is still lacking.
In the current study, high-throughput microarray data from 5 pairs of CRC samples was analyzed to screen for the differentially upregulated circRNAs in CRC. The circMMP1 (hsa_circ_0024109) was identified as a frequently upregulated circRNA in both CRC tissues and cells. Validated by both in vitro and in vivo experiments, knockdown of circMMP1 remarkably inhibited the proliferation and metastasis ability of CRC. Dual luciferase and radioimmunoprecipitation assays (RIPA) were used to explore the underlying molecular mechanism. Overall, the discovery of the biological role of circKL-miR-1238- MMP1/MMP2/MMP9 axis in the growth and metastasis of CRC is important for the development of new therapeutic strategies. We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/atm-21-3930).
Methods
Tissue sample and ethics approval
Fresh colorectal tissues and adjacent tissues were obtained from the First Affiliated Hospital of Sun Yat-sen University. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the First Affiliated Hospital of Sun Yat-sen University (No.: 2019103-1) and informed consent was taken from all the patients. Experiments were performed under a project license (No.: 2019103) granted by the Ethics Committee of the First Affiliated Hospital of Sun Yat-sen University, in compliance with the guidelines of Institutional Animal Care and Use Committee of the First Affiliated Hospital of Sun Yat-sen University. A protocol was prepared before the study without registration.
Cell culture
The CRC cell SW480, SW620, HT-29, SW116 and colorectal epithelial cell NCM460 were all purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). All 5 cell lines were saved and cultured according to the instructions provided by ATCC. We occasionally used DNA fingerprinting to verify the authenticity of cells.
Quantitative reverse transcription polymerase chain reaction analysis
Total cellular RNA was extracted by TRIzol reagent (Sigma-Aldrich, St. Louis, MO, USA). We used SYBR Premix Ex Taq Kit (Takara, Kusatsu, Shiga, Japan) was used for quantitative reverse transcription polymerase chain reaction (RT-qPCR) assays. The primers for circMMP1 were F: 5'-CCTGGATAGGCAAGGGATAA-3', R: 5'-TTGGAAGGCTTTCTCAATGG-3'. The primers for matrix metalloproteinase 1 (MMP1) were F: 5'-GAGCAAACACATCTGAGGTACAGGA-3', R: 5'-TTGTCCCGATGATCTCCCCTGACA-3'. The primers for MMP2 were F: 5'-TACAGGATCATTGGCTACACACC-3', R: 5'-GGTCACATCGCTCCAGACT-3'. The primers for MMP9 were F: 5'-GCACGACGTCTTCCAGTACC-3', R: 5'-TCAACTCACTCCGGGAACTC-3'. The primers for MMP14 were F: 5'-GGCTACAGCAATATGGCTACC-3', R: 5'-GATGGCCGCTGAGAGTGAC-3'. The primers for MMP17 were F: 5'-GCACCAGCGACCACAAGAT-3', R: 5'-CGTTATTGTCCTTGAACACCCAGTA-3'. The primers for GAPDH were F: 5'-ACAACTTTGGTATCGTGGAAGG-3', R: 5'-GCCATCACGCCACAGTTTC-3'.
Western blot analysis
Cell protein was isolated using RIPA lysis and phenylmethylsulfonyl fluoride (PMSF) reagent. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel and polyvinylidene fluoride (PVDF) membranes were used to separate the target protein. The transfer condition was 300 mA for 2 h on the PVDF membranes. The membrane was treated with anti-MMP1, anti-MMP2, anti-MMP9, or anti-β-actin antibody (1:3,000) at 4 °C overnight, and incubated with the specific secondary antibody the next day at room temperature for about 2 h.
RNase R digestion assay
RNase R (8.5 U/µg) or control solution was used to treat the total RNA (5 µg) from CRC cell SW620 at 37 °C for 25 min, then the resulting RNA solution was purified and quantified by RT-qPCR analysis.
Actinomycin D assay
The linear mRNA transcription in CRC cell SW620 was degraded by exposing to 5 µg/mL actinomycin D for 0, 8, 16, or 24 h. The SW620 cell was harvested at a certain time. We then used RT-qPCR analysis to quantify the linear MMP1 mRNA and the circular circMMP1.
Cell Counting Kit-8 assay
The SW620 and SW480 CRC cells were digested and then resuspended, and si-circCTR transfected cancer cells and si-circMMP1 transfected cancer cells (5×103 cells/well) were seeded into the 96-well plate, and then incubated at 37 °C. Before measurement, Cell Counting Kit-8 (CCK-8) solution (10 µL) was added into each well and incubated at 37 °C for 1 h (19,20).
Transwell assay
The CRC cells (5×104 cells) were resuspended and then transferred into the upper chambers with serum-free medium, and the lower chambers were loaded with 20% fetal bovine serum (FBS) medium. Then, the cells were fixed by methanol, and 0.5% crystal violet was used to stain the cells. The cells were imaged and counted under microscope (21) (Olympus IX73, Japan).
Dual luciferase reporter assay
The SW620 and SW480 CRC cells were seeded into 96-well plates (5×103/well). The predicted sites of miR-1238 binding to circMMP1, and 3'-UTR of MMP1, MMP-2, MMP-9, MMp-14, and MMP-17 mRNAs were mutated. The constructed reporting vectors (circMMP1-wt/mut, MMP2 3'-UTR-wt/mut, MMP9 3'-UTR-wt/mut, MMP14 3'-UTR-wt/mut, or MMP17 3'-UTR-wt/mut), miRNA inhibitors, miRNA mimics, and their control were transfected into the CRC cells for 48 h.
RNA immunoprecipitation (RIP)
Anti-Ago2 antibody was used for RIP assays. The expression of circMMP1, MMP1, MMP-2, MMP-9 and miR-1238 was tested after RNA purification. The MS2bs-circMMP1 vector, MS2bs-circMMP1-mt vector and MS2bs-Rluc vector were transfected into SW620 cells and incubated for 48 h. Then, the RIP assay was performed. The abundance of miR-1238 was determined after the purification of RNA complexes.
Mouse xenograft assay
The SW620 CRC cells (2×107) stably inhibiting circMMP1 and its control were subcutaneously injected into nude mice. After 28 d, mice were euthanized and the tumors were weighed. The volume of tumors was estimated every 4 d by the formula 0.5 × width2 × length. The SW620 cells (3×105) were also injected into nude mice (tail veins). The lungs were excised after 8 weeks, then the metastatic sites were counted under microscope (22).
Statistical analysis
The software SPSS 23.0 (IBM Corp., Armonk, NY, USA) was used to analyze the data. All data were shown in the form of mean ± standard deviation (SD). Student’s t-test was used to analyze the data between groups. When the P value was less than 0.05, the difference was considered statistically significant.
Results
CircMMP1 is upregulated in CRC with circular features
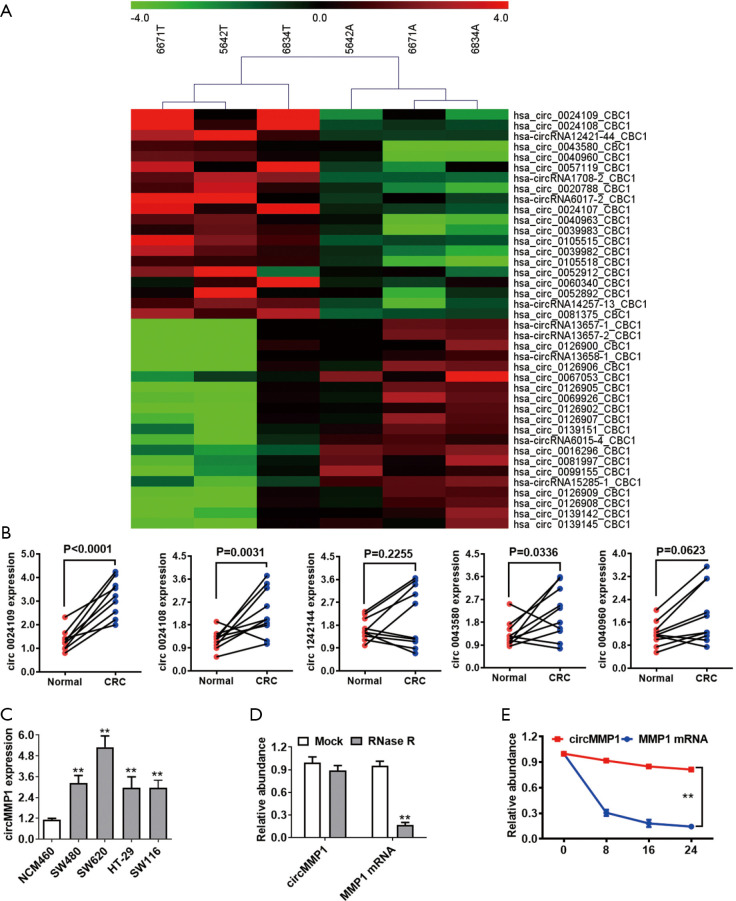
By analyzing the high-throughput microarray data from 5 pairs of CRC samples, we screened the 5 most differentially expressed circRNAs in CRC (Figure 1A). We used RT-qPCR to verify the expression of the top 5 upregulated circRNAs in 10 pairs of CRC tissue samples and their para-cancerous samples (Figure 1B). We found that circMMP1 was significantly upregulated in all CRC samples: circMMP1 was upregulated in CRC cell lines, especially in SW620 and SW480 cells, detected by qPCR analysis (Figure 1C). Then, actinomycin D and RNase R assays were used to examine the structure and stability of circMMP1. The results showed that circMMP1 was resistant to RNA exonuclease (RNase R) (Figure 1D). The circular form of circMMP1 had a longer half-life span, compared with the linear MMP1 mRNA (Figure 1E).
Figure 1.
CircMMP1 is upregulated in colorectal cancer with circular features. (A) High-throughput microarray sequencing from 5 pairs of CRC samples screened the significantly differentially expressed circRNAs. (B) Top 5 upregulated circRNA between CRC tissues and adjacent normal colorectal tissues was detected by qPCR analysis. (C) The relative expression level of circMMP1 in NCM460 and CRC cells. (D) The circular feature of circMMP1 was examined by RNase R assay in SW620. (E) Determined by actinomycin D treated assay, circular transcripts of circMMP1 were more stable than its linear MMP1 mRNA transcripts in SW620 CRC cell line. **P<0.01. CRC, colorectal cancer; qPCR, quantitative polymerase chain reaction.
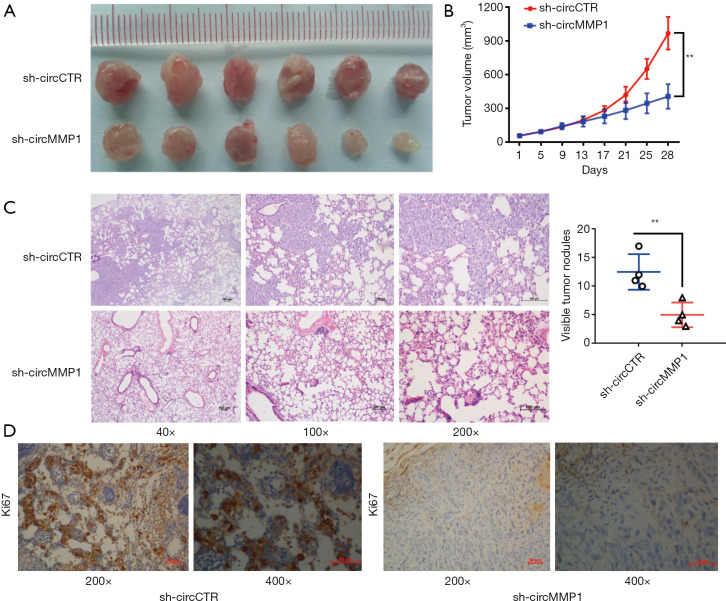
CircMMP1 knockdown inhibits the growth and metastasis of CRC in vivo
Tumor volume curves demonstrated that circMMP1 silencing could inhibit the growth of CRC (Figure 2A,2B). Moreover, we found that silencing of circMMP1 could also inhibit the metastatic ability of CRC cell SW620 in the lung metastasis experiment (Figure 2C). Additionally, immunohistochemistry (IHC) was used to analyze the protein level of proliferation-associated Ki67 in mouse xenografts tumors of 2 groups. The results demonstrated that Ki67 expression was significantly reduced in the sh-circMMP1 group (Figure 2D).
Figure 2.
CircMMP1 knockdown inhibits the growth and metastasis of CRC in vivo. (A) Mouse xenograft models of SW620 CRC cell line was established. Tumor volume was estimated every 4 d. (B) Tumor growth curve of circMMP1 knockout group and its control group. (C) The number of lung metastases was counted and recorded. The stained sections of lung metastases images are shown (HE staining). (D) The representative pictures of Ki-67 expression are shown (DAB staining). **P<0.01. CRC, colorectal cancer; HE, hematoxylin and eosin. HE, hematoxylin-eosin; DAB, 3,3’-diaminobenzidine.
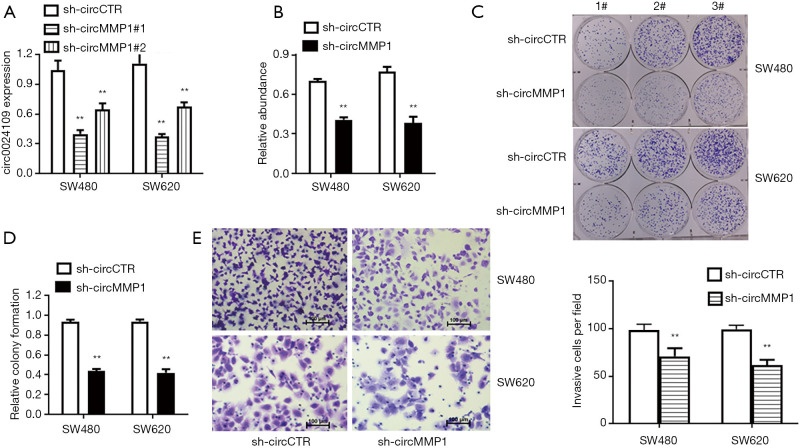
CircMMP1 silencing suppressed the proliferation and invasion of CRC cells in vitro
To analyze the function of circMMP1 in CRC development, circMMP1 expression was knocked down in SW480 and SW620 cells using the small interfering RNA (siRNA) targeting circMMP1 (si-circ_0024109). The knockdown efficacy of si-circMMP1 was verified in SW480 and SW620 CRC cell lines (Figure 3A). The result of CCK-8 assays showed that circMMP1 silencing significantly decreased proliferation of SW480 and SW620 cells (Figure 3B). A colony-formation assay was also conducted which yielded similar results as the proliferation assay (Figure 3C,3D). Invasion assays were then conducted to investigate whether circMMP1 silencing could suppress the invasion of CRC cells. Our results showed that circMMP1 silencing could decrease the invasion of both SW480 and SW620 cells (Figure 3E).
Figure 3.
CircMMP1 silencing suppressed the proliferation and invasion of CRC cells in vitro. (A) The efficacy of knockdown was verified in SW480 and SW620 CRC cell lines, assessing by qRT-PCR analysis. (B) The proliferation of cells in SW480 and SW620 was evaluated by CCK-8 assay. (C) Colony-formation assay was conducted with 3 different cell concentrations (0.5% crystal violet staining). (D) Statistical chart of Colony-formation assay. (E) The invasion ability of colorectal cancer cell SW620 was investigated by transwell assay (0.5% crystal violet staining). **P<0.01. CRC, colorectal cancer.
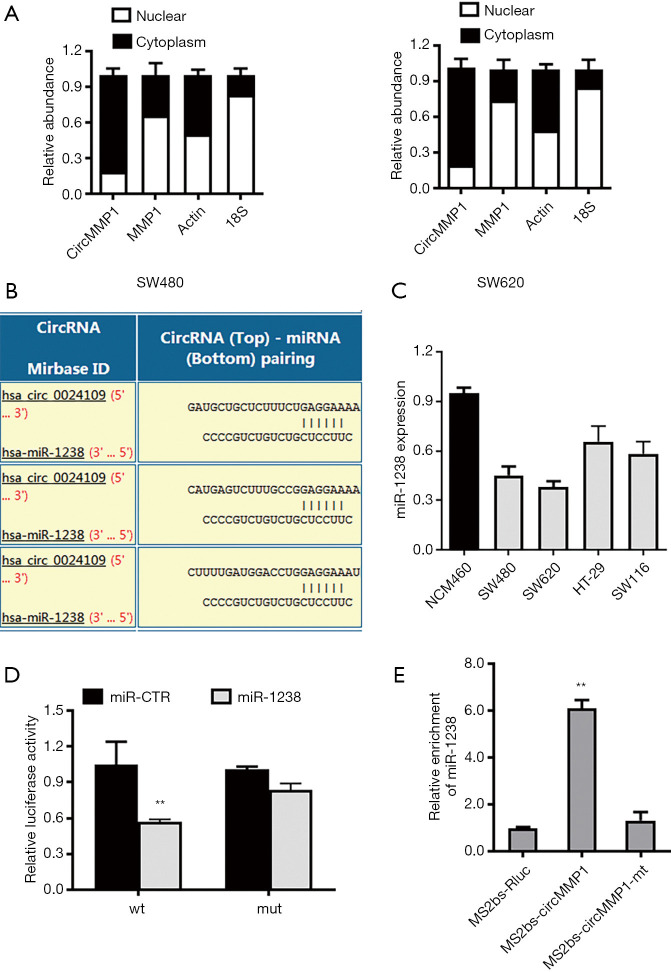
CircMMP1 is a sponge of miR-1238 in CRC
It was revealed that circMMP1 exists mainly in the cytoplasm of CRC cells by analysis of the different cellular proportions of SW480 and SW620 cells (Figure 4A). Then, the Circular RNA Interactome database was used to assess the potential interactions between circRNA and multiple miRNAs. Among all the miRNA candidates, miR-1238 was predicted to bind the circMMP1 sequence with 3 possible interacting sites (Figure 4B). Compared with normal colorectal cell NCM460, miR-1238 was all significantly downregulated in CRC cell lines SW480, SW620, HT29, and SW116, as detected by RT-qPCR analysis (Figure 4C). We then explored whether miR-1238 could bind to circMMP1. The result showed that the relative luciferase activity was significantly reduced when miR-1238 was co-transfected with WT 3'-circMMP1 plasmid, but not with MUT 3'-circMMP1 plasmid (Figure 4D). In order to verify the direct interaction between circMMP1 and miR-1238, we next performed RIP assay to pulldown circMMP1. The results revealed that miR-1238 was mainly concentrated in the MS2bs-circMMP1 overexpressed group in CRC cell SW620 (Figure 4E).
Figure 4.
CircMMP1 is a sponge of miR-1238 in CRC. (A) 18S, Actin, circMMP1, and MMP1 expression in nuclear and cytoplasmic fractions were analyzed in SW480 and SW620 cells by RT-qPCR. (B) Predicted binding sites of miR-1238 within the circMMP1 sequence. (C) The relative expression level of miR-1238 in CRC cells compared to normal colorectal cell NCM460. (D) Luciferase reporter assay of SW620 CRC cell co-transfected with miR-1238 mimics and the 3'-circMMP1 wild type or mutant luciferase reporter. (E) MS2-based RIP assay transfected with MS2bs-circMMP1 plasmid, MS2bs-circMMP1-mt plasmid or Rluc control plasmid. **P<0.01. CRC, colorectal cancer; RT-qPCR, quantitative reverse transcription polymerase chain reaction.
CircMMP1 promotes CRC progression through the circMMP1-miR-1238-MMP1/MMP2/MMP9 axis
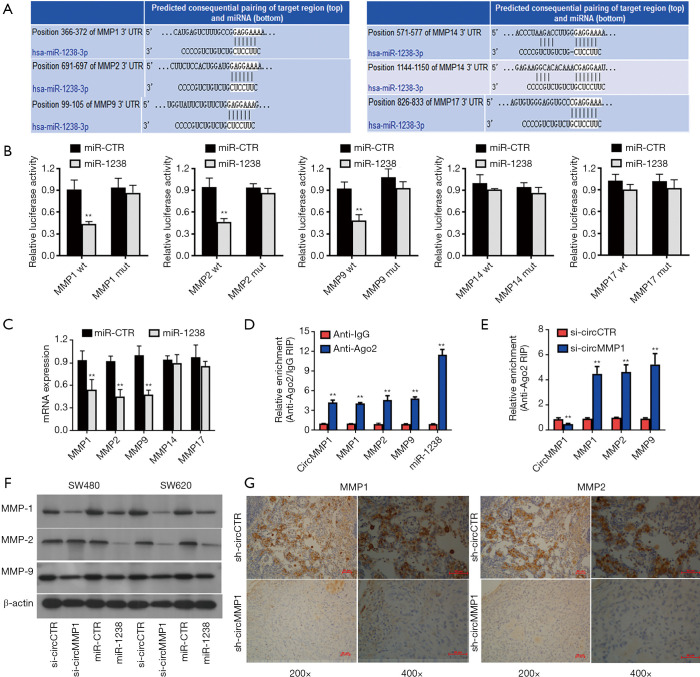
Next, TargetScan (http://www.targetscan.org) was used to further determine the potential targeting genes of miR-1238. Among the potential downstream genes, MMP1, MMP2, MMP9, MMP14, and MMP17 were identified as target genes of miR-1238 (Figure 5A). Then, we investigated whether miR-1238 could directly bind to the 3'-UTR of MMP1, MMP2, MMP9, MMP14, and MMP17 mRNA. The result indicated that the relative luciferase activity was significantly reduced when miR-1238 was co-transfected with WT 3'-UTR-MMP1, WT 3'-UTR-MMP2, or WT 3'-UTR-MMP9 plasmid, but not with WT 3'-UTR-MMP14 or WT 3'-UTR-MMP17 plasmid. Meanwhile, the result was not observed when a mutant vector was co-transfected (Figure 5B). Exogenous introduction of miR-1238 contributed to the reduction of MMP1, MMP2, and MMP9 mRNA expression levels, but not MMP14 and MMP17 (Figure 5C). The RIP assay showed that circMMP1, MMP1, MMP2, MMP9, and miR-1238 were all highly enriched in anti-AGO2 group in CRC cell SW620 (Figure 5D). Enrichment to RNA induced silencing complex (RISC) of MMP1, MMP2, or MMP9 was remarkably increased after circMMP1 silencing (Figure 5E). After transfecting with miR-1238 mimics in CRC cell lines SW480 and SW620, the protein level of MMP1, MMP2, and MMP9 were significantly decreased. Silencing of circMMP1 could also significantly inhibit the protein level of MMP1, MMP2, and MMP9 as revealed via western blot analysis (Figure 5F). The IHC was used to analyze MMP1 and MMP2 protein levels in mouse xenograft tumors of 2 groups. The results demonstrated that the MMP1 and MMP2 expression were remarkably decreased in the sh-circMMP1 group in tumor tissues (Figure 5G).
Figure 5.
CircMMP1 promotes CRC progression through the circMMP1-miR-1238-MMP1/MMP2/MMP9 axis. (A) MMP1, MMP2, MMP9, MMP14, and MMP17 were predicted as the downstream target of miR-1238, according to the TargetScan website. (B) Luciferase reporter assay of CRC cell SW620 co-transfected with miR-1238 and the 3'-UTR of MMP1/MMP2/MMP9/MMP14/MMP17 wild type/mutant luciferase reporter. (C) Overexpression of miR-1238 contributed to the reduction of MMP1, MMP2, and MMP9 expression in CRC cell SW620, verified by qPCR analysis. (D) Enrichment of circMMP1, MMP1, MMP2, MMP9, and miR-1238 on AGO2 as assessed by RIP assay. (E) Enrichment of MMP1, MMP2, and MMP9 to AGO2 was increased when circMMP1 was knocked down. (F) Knockdown of circMMP1 and overexpression of miR-1238 contributed to the reduction of MMP1, MMP2, and MMP9 expression in CRC cell lines SW480 and SW620, as detected by western blot analysis. (G) Knockdown of circMMP1 decreased the expression of MMP1 and MMP2 expression in xenograft tumors. The representative IHC images of MMP1 and MMP2 are showed (DAB staining). **P<0.01. CRC, colorectal cancer; IHC, immunohistochemistry.
Discussion
As a member of non-coding RNAs, circRNAs are widely expressed in mammalian tissues, and exhibit specificity in tissues or cells (23). Their covalently closed loop structure makes them indispensable components for a variety of biological processes, as opposed to useless products of mRNA pre-splicing (24). Nowadays, with the innovation of high-throughput technology, various circRNAs have been disclosed and deeply researched (25). It has been confirmed that circRNAs participate in a variety of cellular processes in cancer. Hundreds of circRNAs have been identified as predictive biomarkers and have the potential to be therapeutic targets for cancer therapy (26). For example, circAGO2 drives a variety of cancers progression through facilitating RISC HURs (27), and circ133 and circHIPK3 have been identified as important regulatory factors by the mechanism of ceRNA in CRC (28,29). Meanwhile, circFAM114A2 and circITCH have been identified as tumor suppressors in cancer (30,31), and circFAM120A has been confirmed to promote cell proliferation by preventing FAM120A from binding to IGF2BP2 (32). However, the potential molecular mechanism and biological role of circRNA in CRC have not been well investigated.
In this study, we analyzed the high-throughput circRNA microarray data from 5 pairs of CRC samples to screen for the differentially expressed circRNAs. The 5 most significantly upregulated circRNAs were hsa_circ_0024109, hsa_circ_0024108, hsa_circ_1242144, hsa_circ_0043580, and hsa_circ_0040960 were. We then verified their expression in 10 pairs of CRC patient samples and their adjacent normal colorectal samples by RT-qPCR, which revealed that circMMP1 (Hsa_circ_0024109) was the only circRNA all upregulated in 10 samples. Then, circMMP1 expression was knocked down to investigate the function in CRC. We found that circMMP1 knockdown significantly inhibited the proliferation and growth of CRC cells in vitro and in vivo. Therefore, we decided to explore its underlying molecular mechanism.
According to the ceRNA theory (33), circRNAs regulate and communicate with each other through competitive binding to shared miRNAs (34). In this research, circMMP1 was found to be a sponge of miR-1238 in CRC. This regulatory relationship was confirmed via dual luciferase reporter assay and RIP assay. Contrary to the expression of circMMP1, miR-1238 was all significantly downregulated in CRC, which further verified the combination of the two. It has been shown that miR-1238 inhibits a variety of processes in cancers, such as inhibition of cell proliferation by targeting LHX2 in non-small cell lung cancer (35), inhibition of the malignant progression of cutaneous squamous cell carcinoma (36), or suppression of the expression of LIM-Homeobox Gene 2 in prostate cancer cells (37). It has also been confirmed that miR-1238 is involved in the growth and metastasis of cholangiocarcinoma (38). In our study, we found that miR-1238 could bind to MMP1, MMP2, MMP9, MMP14, and MMP17 3'-UTRs, but only MMP1, MMP2, and MMP9 were verified as significantly downregulated by miR-1238. Research shows that extracellular matrix (ECM) and collagen can be degraded by MMPs that significantly contribute to tumor progression, especially the process of invasion, metastasis, and angiogenesis in cancer (39). In this study, we identified 3 important MMPs, including MMP1, MMP2, and MMP9, which are the direct target genes of miR-1238 in CRC cells. Silencing of circMMP1 and overexpression of miR-1238 could significantly inhibit the expression of MMP1, MMP2, and MMP9 in vitro, and circMMP1 silencing could remarkably decrease the expression of MMP1 and MMP2 in vivo. Collectively, circ_0024109 is capable of aggravating the malignant progression of CRC, which may be mediated by promoting of miR-1238-dependent upregulation of MMPs.
In conclusion, our study identified the biological role of circMMP1 in the growth and metastasis of CRC through the miR-1238-MMP1/MMP2/MMP9 axis. These findings are important for the development of new treatment strategies and potential prognostic implications for CRC.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was supported by the research grants from Project 5010 of The First Affiliated Hospital of Sun Yat-sen University (2018004).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the First Affiliated Hospital of Sun Yat-sen University (No.: 2019103-1) and informed consent was taken from all the patients. Experiments were performed under a project license (No.: 2019103) granted by the Ethics Committee of the First Affiliated Hospital of Sun Yat-sen University, in compliance with the guidelines of Institutional Animal Care and Use Committee of the First Affiliated Hospital of Sun Yat-sen University. A protocol was prepared before the study without registration.
Footnotes
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-3930
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-3930
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-3930). The authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin 2020;70:145-64. 10.3322/caac.21601 [DOI] [PubMed] [Google Scholar]
- 2.Fakih MG. Metastatic colorectal cancer: current state and future directions. J Clin Oncol 2015;33:1809-24. 10.1200/JCO.2014.59.7633 [DOI] [PubMed] [Google Scholar]
- 3.Buccafusca G, Proserpio I, Tralongo AC, et al. Early colorectal cancer: diagnosis, treatment and survivorship care. Crit Rev Oncol Hematol 2019;136:20-30. 10.1016/j.critrevonc.2019.01.023 [DOI] [PubMed] [Google Scholar]
- 4.Qu S, Yang X, Li X, et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett 2015;365:141-8. 10.1016/j.canlet.2015.06.003 [DOI] [PubMed] [Google Scholar]
- 5.Ebbesen KK, Hansen TB, Kjems J. Insights into circular RNA biology. RNA Biol 2017;14:1035-45. 10.1080/15476286.2016.1271524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer 2021;21:22-36. 10.1038/s41568-020-00306-0 [DOI] [PubMed] [Google Scholar]
- 7.Vo JN, Cieslik M, Zhang Y, et al. The Landscape of Circular RNA in Cancer. Cell 2019;176:869-881.e13. 10.1016/j.cell.2018.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacol Ther 2018;187:31-44. 10.1016/j.pharmthera.2018.01.010 [DOI] [PubMed] [Google Scholar]
- 9.Su Y, Lv X, Yin W, et al. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging (Albany NY) 2019;11:8183-203. 10.18632/aging.102312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang W, Yang X, Wang X, et al. Silencing CDR1as enhances the sensitivity of breast cancer cells to drug resistance by acting as a miR-7 sponge to down-regulate REGγ. J Cell Mol Med 2019;23:4921-32. 10.1111/jcmm.14305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zou Y, Zheng S, Deng X, et al. The Role of Circular RNA CDR1as/ciRS-7 in Regulating Tumor Microenvironment: A Pan-Cancer Analysis. Biomolecules 2019;9:429. 10.3390/biom9090429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 13.Zou Y, Zheng S, Deng X, et al. Diagnostic and prognostic value of circular RNA CDR1as/ciRS-7 for solid tumours: A systematic review and meta-analysis. J Cell Mol Med 2020;24:9507-17. 10.1111/jcmm.15619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang C, Zeng X, Shan R, et al. The Emerging Picture of the Roles of CircRNA-CDR1as in Cancer. Front Cell Dev Biol 2020;8:590478. 10.3389/fcell.2020.590478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng X, Chen L, Zhou Y, et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer 2019;18:47. 10.1186/s12943-019-1010-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z, Ren R, Wan D, et al. Hsa_circ_101555 functions as a competing endogenous RNA of miR-597-5p to promote colorectal cancer progression. Oncogene 2019;38:6017-34. 10.1038/s41388-019-0857-8 [DOI] [PubMed] [Google Scholar]
- 17.Li XN, Wang ZJ, Ye CX, et al. RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J Exp Clin Cancer Res 2018;37:325. 10.1186/s13046-018-1006-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Liu H, Zhao P, et al. Has_circ_0055625 from circRNA profile increases colon cancer cell growth by sponging miR-106b-5p. J Cell Biochem 2019;120:3027-37. 10.1002/jcb.27355 [DOI] [PubMed] [Google Scholar]
- 19.Peng F, Xiong L, Peng C. (-)-Sativan inhibits tumor development and regulates miR-200c/PD-L1 in triple negative breast cancer cells. Front Pharmacol 2020;11:251. 10.3389/fphar.2020.00251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, Peng F, Peng C, et al. Gut microbiota in tumor microenvironment: a critical regulator in cancer initiation and development as potential targets for chinese medicine. Am J Chin Med 2021;49:609-26. 10.1142/S0192415X21500270 [DOI] [PubMed] [Google Scholar]
- 21.Peng F, Xiong L, Xie X, et al. Isoliquiritigenin derivative regulates miR-374a/BAX axis to suppress triple-negative breast cancer tumorigenesis and development. Front Pharmacol 2020;11:378. 10.3389/fphar.2020.00378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng F, Tang H, Du J, et al. Isoliquiritigenin suppresses EMT-Induced metastasis in triple-negative breast cancer through miR-200c/C-JUN/ Formula: see text-Catenin. Am J Chin Med 2021;49:505-23. 10.1142/S0192415X21500233 [DOI] [PubMed] [Google Scholar]
- 23.Dong Y, He D, Peng Z, et al. Circular RNAs in cancer: an emerging key player. J Hematol Oncol 2017;10:2. 10.1186/s13045-016-0370-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cocquerelle C, Mascrez B, Hétuin D, et al. Mis-splicing yields circular RNA molecules. FASEB J 1993;7:155-60. 10.1096/fasebj.7.1.7678559 [DOI] [PubMed] [Google Scholar]
- 25.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453-61. 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li S, Han L. Circular RNAs as promising biomarkers in cancer: detection, function, and beyond. Genome Med 2019;11:15. 10.1186/s13073-019-0629-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, Yang F, Fang E, et al. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ 2019;26:1346-64. 10.1038/s41418-018-0220-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang H, Zhang H, Yang Y, et al. Hypoxia induced exosomal circRNA promotes metastasis of Colorectal Cancer via targeting GEF-H1/RhoA axis. Theranostics 2020;10:8211-26. 10.7150/thno.44419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis 2018;9:417. 10.1038/s41419-018-0454-8 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Liu T, Lu Q, Liu J, et al. Circular RNA FAM114A2 suppresses progression of bladder cancer via regulating ∆NP63 by sponging miR-762. Cell Death Dis 2020;11:47. 10.1038/s41419-020-2226-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Ge YZ, Xu L, et al. Circular RNA ITCH: A novel tumor suppressor in multiple cancers. Life Sci 2020;254:117176. 10.1016/j.lfs.2019.117176 [DOI] [PubMed] [Google Scholar]
- 32.Li S, Li X, Xue W, et al. Screening for functional circular RNAs using the CRISPR-Cas13 system. Nat Methods 2021;18:51-9. 10.1038/s41592-020-01011-4 [DOI] [PubMed] [Google Scholar]
- 33.Verduci L, Strano S, Yarden Y, et al. The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol Oncol 2019;13:669-80. 10.1002/1878-0261.12468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014;505:344-52. 10.1038/nature12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi X, Zhan L, Xiao C, et al. miR-1238 inhibits cell proliferation by targeting LHX2 in non-small cell lung cancer. Oncotarget 2015;6:19043-54. 10.18632/oncotarget.4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, Kong Y, Li H, et al. CircRNA circ_0067772 aggravates the malignant progression of cutaneous squamous cell carcinoma by regulating miR-1238-3p/FOXG1 axis. Genes Genomics 2021;43:491-501. 10.1007/s13258-021-01074-3 [DOI] [PubMed] [Google Scholar]
- 37.Shan G, Shao B, Liu Q, et al. circFMN2 Sponges miR-1238 to Promote the Expression of LIM-Homeobox Gene 2 in Prostate Cancer Cells. Mol Ther Nucleic Acids 2020;21:133-46. 10.1016/j.omtn.2020.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Xu Y, Yao Y, Liu Y, et al. Elevation of circular RNA circ_0005230 facilitates cell growth and metastasis via sponging miR-1238 and miR-1299 in cholangiocarcinoma. Aging (Albany NY) 2019;11:1907-17. 10.18632/aging.101872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao MQ, You AB, Zhu XD, et al. miR-182-5p promotes hepatocellular carcinoma progression by repressing FOXO3a. J Hematol Oncol 2018;11:12. 10.1186/s13045-018-0555-y [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as